Abstract
Studies have demonstrated that both mortality and severe illness rates exist significant difference in different gender COVID-19 patients, but the reasons are still very mysterious to date. Here, we firstly find that the survival outcome of female patients is better to male patients through analyzing the 3044 COVID-19 cases. Secondly, we identify many important master regulators [e.g. STAT1/STAT2 and zinc finger (ZNF) proteins], in particular female patients can express more ZNF proteins and stronger transcriptional activities than male patients in response to SARS-CoV-2 infection. Thirdly, we discover that ZNF protein activity is significantly negative correlation with the SARS-CoV-2 load of COVID-19 patients, and ZNF proteins as transcription factors can also activate their target genes to participate in anti-SARS-CoV-2 infection. Fourthly, we demonstrate that ZNF protein activity is positive correlation with the abundance of multiple immune cells of COVID-19 patients, implying that the highly ZNF protein activity might promote the abundance and the antiviral activity of multiple immune cells to effectively suppress SARS-CoV-2 infection. Taken together, our study proposes an underlying anti-SARS-COV-2 role of ZNF proteins, and differences in the amount and activity of ZNF proteins might be responsible for the distinct prognosis of different gender COVID-19 patients.
Keywords: SARS-CoV-2, COVID-19, master regulator, zinc finger protein, gender difference
Introduction
Currently, studies indicated that gender differences exist in SARS-CoV-2 infection and the survival rate of COVID-19 patients and the death and critical illness rate of males is higher than females [1–3]. However, the mechanism causing this different outcome is still unclear to date [1–3]. Therefore, a systematic elucidation of the underlying mechanisms is necessary for making the clinically gender-dependent treatment schedule.
The living habits, basic diseases and social activities were suggested to be the possible causes of the disparities in infection and death in different genders [3, 4], but few significant differences exist in the COVID-19 infection rate between different genders [5, 6], implying that those are not the main causes of resulting in the mortality difference. ACE2 and TMPRSS2 can function as receptors and transmembrane serine proteases to help SARS-CoV-2 to enter the host cell [7, 8], and their different expressions might result in the distinct survival outcome for different gender COVID-19 patients [9, 10]. Female patients can retain more immune cells than males, whereas males release more inflammatory factors than females after SARS-CoV-2 infection [11, 12], suggesting that the different reaction intensity of distinct signaling pathways between different gender patients might cause the different mortality. Our latest work identified 30 key master regulators (MRs) that can serve as anti-SARS-CoV-2 roles [13]. These above works suggest that differently expressed MRs might cause the difference outcome for different gender COVID-19 patients. To reveal this issue, we have embarked on the study.
Materials and methods
Clinical data source
The clinical data comes from 3044 COVID-19 patients who were hospitalized in Wuhan Huoshenshan Hospital from 4 February 2020 to 13 April 2020. All COVID-19 patients were diagnosed according to the ‘New Coronavirus Pneumonia Diagnosis and Treatment Plan’ (7th edition) issued by the National Health Commission of China. This study has been approved by the Research Ethics Committee of Wuhan Huoshenshan Hospital, China (HSSLL011). Written informed consent was obtained from each patient.
Public data source
The transcriptome data of 430 SARS-CoV-2 positive nasopharyngeal swabs and 54 negative controls are obtained from the GEO database (GSE152075) [14]. The PBMC and bronchoalveolar lavage fluid (BALF) single-cell transcriptome data comes from GSE149689 [15] and GSE145926 [16].
Differential expression analysis and MR inference
The edgeR R package is used to calculate differentially expressed genes [17, 18]. ARACNe and VIPER algorithms are combined to identify MRs [19, 20]. ARACNe algorithm is a method based on mutual information theory, which can infer the interaction between transcription factors (TFs) and target genes [19]. VIPER was used to infer the enrichment of each regulatory protein regulator in certain gene expression signature (GES) composed of full transcriptome data of healthy and COVID-19 patients [20]. These two algorithms have proved to be very effective for identifying the MR in the host’s response to viral infection [21, 22]. A method called Leading-edge Gene Set Enrichment Analysis implemented in the VIPER package was performed to identify the core target genes of MRs driving the enrichment on GES [20].
Gene function analysis and gene set activity calculation
The clusterProfiler package was used for gene ontology and KEGG pathway enrichment analysis [23]. The single-sample gene set enrichment analysis algorithm (ssGSEA) in the GSVA package was used to calculate the overall activity of ZNF proteins [24]. This algorithm can evaluate the overall activity of a predetermined gene set in a single sample based on the entire transcriptome expression. The 456 ZNF protein coding genes matched by the grep function were used as the predetermined antiviral gene set of ZNF proteins.
Single cell data analysis
The Seurat (version 3.0) was used for single-cell transcriptome data quality control, filtering, standardization and subsequent analysis [25]. All inclusion criterion and parameters for cell quality control and dimension reduction clustering were consistent with the data literature source [15, 16]. The canonical correlation analysis and anchors algorithm were used to remove batch effect [25] and high-dimensional cell data were visualized through uniform manifold approximation and projection (UMAP) [26]. We used cell scores to assess how well individual cells express a certain set of predetermined genes via AddModuleScore function in Seurat [25, 27].
Flow cytometry assay of lymphocyte subsets
A total of 548 individuals from 3044 COVID-19 patients were enrolled in this study, and measured with T cell, B cell and NK cell using flow cytometry. A total of 125 patients of them were measured with 30 types of lymphocyte subsets. The peripheral blood was collected from patients and all samples were tested within 6 h after being obtained. Briefly, 100 μL of fresh whole blood was incubated in 2 mL of VersaLyse (Beckman Coulter Life Science) between 20 and 30°C for 15 min to lyse erythrocytes, then being washed with 3 mL of 1 × PBS. Thereafter, the cell pellet was resuspended in 500 μL of 1 × PBS containing 0.8% IOTest 3 Fixative Solution. The above samples were ready for acquisition. To measure T cell, B cell, NK cell and lymphocyte subsets, stained all the 10 single color tubes from a single pouch of the Compensation Kit provided in the IM DuraClone IM cell subsets Tube, 25 tests (Table S1, see Supplementary Data available online at http://bib.oxfordjournals.org/), RUO with venous blood, measured by multiple-color flow cytometry according to the manufacturer’s instructions (Backman Coulter Life Science). The data were evaluated using the Kaluza Software of Beckman.
Results
The infection rate and prognosis of different genders
As shown in Figure 1, the infection rate of males is only 1.58 percentage points higher than females (Figure 1A), but males have a lower survival rate (Figure 1B) and a higher critical disease classification (Figure 1C) as well as a higher intensive care unit (ICU) admission ratio than females (Figure 1D). The death age for males is mainly at 60 ~ 90 years (Figure 1E), but 70 ~ 90 years for females (Figure 1F). The mortality rate of males in the same age group is about twice higher than females (Figure 1E and F), whereas only slight differences exist in the infection rate of different genders in the same age group except for the minimum or maximum age group with few subjects (Figure 1G and H).
Figure 1.
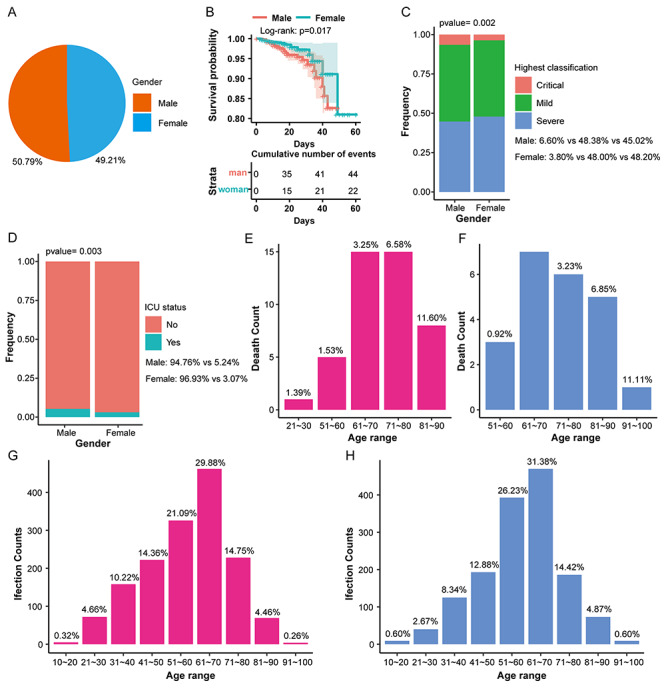
The distribution of infection and mortality of 3044 COVID-19 patients of different genders. (A) The pie chart shows the overall infection rate of patients of different genders. (B) The overall survival rate curve shows the survival rate of patients during the treatment of different genders. (C) Stacked histogram shows the highest disease classification ratio of patients of different genders. (D) Stacked histogram shows the proportion of ICU admissions of patients of different genders. (E) The histogram shows the mortality of males in different age groups. (F) The histogram shows that the mortality rate of females in different age groups. (G) The histogram shows the infection rate of males at different ages. (H) The histogram shows the infection rate of females in different age groups.
Identifying MRs of different gender COVID-19 patients
We applied the ARACNe algorithm to, respectively, obtain 113 676 and 103 621 transcriptional interactions for male and female patients and found that 839 TFs regulate 9427 target genes in males and 864 TFs regulate 9565 target genes in females. We next used the VIPER algorithm to identify the top 50 proteins as MRs for males and females, respectively (Figure 2A and B). We found that males and females share 16 common MRs, whereas 34 MRs are different (Figure S1, see Supplementary Data available online at http://bib.oxfordjournals.org/).
Figure 2.
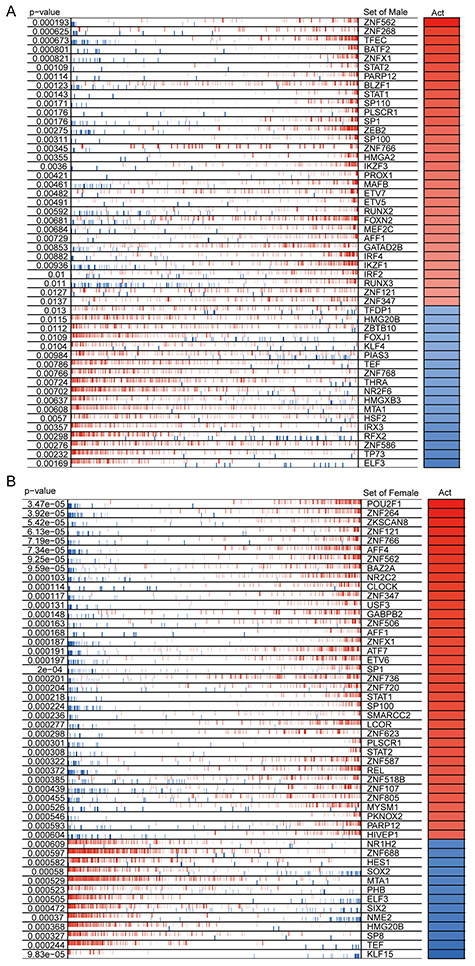
Similarities and differences of MRs in COVID-19 patients of different genders. (A) The heat map shows the enrichment of MRs in males in response to SARS-CoV-2 infection. (B) The heat map shows the MR enrichment of female in response to SARS-CoV-2 infection. The top 50 significantly enriched MRs are displayed. The P-value represents the significance of MR enrichment. Act represents the normalized enrichment score, which reflects the protein activity of the presumed MR.
GO annotation of MRs of different gender COVID-19 patients
As shown in Figure 3A, these MRs unique to male patients mainly participate in lymphocyte differentiation, regulation of leukocyte differentiation and T cell differentiation, but females are involved in steroid hormone-mediated signaling pathway and glucocorticoid receptor signaling pathway, whereas the 16 common MRs are enriched in defense response to virus, response to type I interferon, regulation of viral process, indicating that these common MRs involve in antiviral infection. Remarkably, the expression extent of 16 common MRs existed obvious differences between different gender patients (Figure 3B), and the fold change of STAT2, ZNFX1, ZNF121, ZNF347, ZNF562, AFF1 and SP1 of females is higher than males, respectively (Figure 3B), as well as the average protein activity of 16 common MRs in females is higher than males (Figure 3C).
Figure 3.
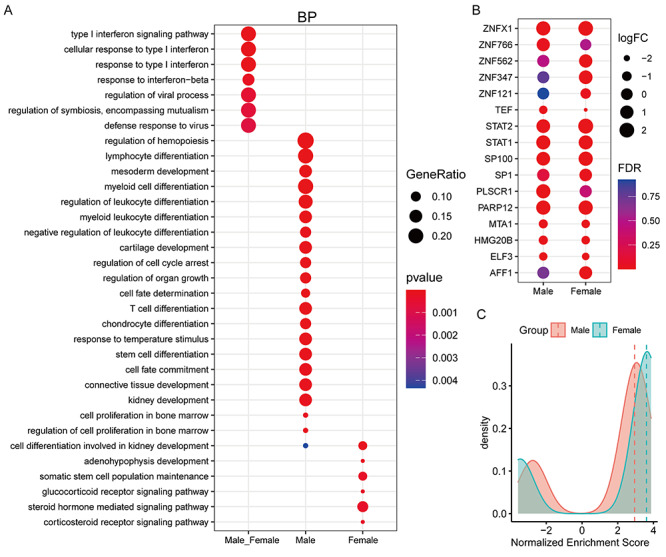
The similarities and differences in the function and activity of MRs in COVID-19 patients of different genders. (A) Functional enrichment analysis of common and specific MRs of different genders. BP is an acronym for biological process. (B) Changes in the mRNA levels of 16 shared MRs after different genders were infected with the coronavirus SARS-CoV-2. (C) Changes in the protein activity of 16 shared MRs after different sexes infected with the coronavirus SARS-CoV-2. The horizontal axis represents the normalized enrichment score used to estimate the protein activity of MRs. The larger the value, the stronger the protein activity. The vertical axis represents the number of MRs. The red and green lines represent the median value.
Female patients express more ZNF proteins and higher activities than males
Previous studies indicated that STAT1/STAT2 could activate the expressions of many ZNF proteins (Figure S3, see Supplementary Data available online at http://bib.oxfordjournals.org/) [28, 29]. Here, we demonstrated that mRNA expression levels and protein activities of STAT1 and STAT2 after SARS-CoV-2 infection are significantly upregulated than controls, in particular the upregulated level of SATA2 in female patients is higher than males (log2FC: 1.25 versus 2.14) (Figures 3B and S2, see Supplementary Data available online at http://bib.oxfordjournals.org/). We also found that female patients have 2 times more ZNF proteins than males among the top 50 MRs, and in addition to 5 shared ZNF proteins, females have 11 unique ZNF proteins, but only 3 in males (Figure 4A), as well as the overall protein activity of ZNF proteins is higher than males (Figure 4B). Moreover, 50 ZNF proteins were significantly up-regulated in females (log2FC > 1, FDR < 0.05), but only 12 in males, while these down-regulated ZNF proteins in females were equal to males (15 versus 15) (Figure 4C and D). And that the activity score value of ZNF proteins in females was higher than males (Figure 4E).
Figure 4.
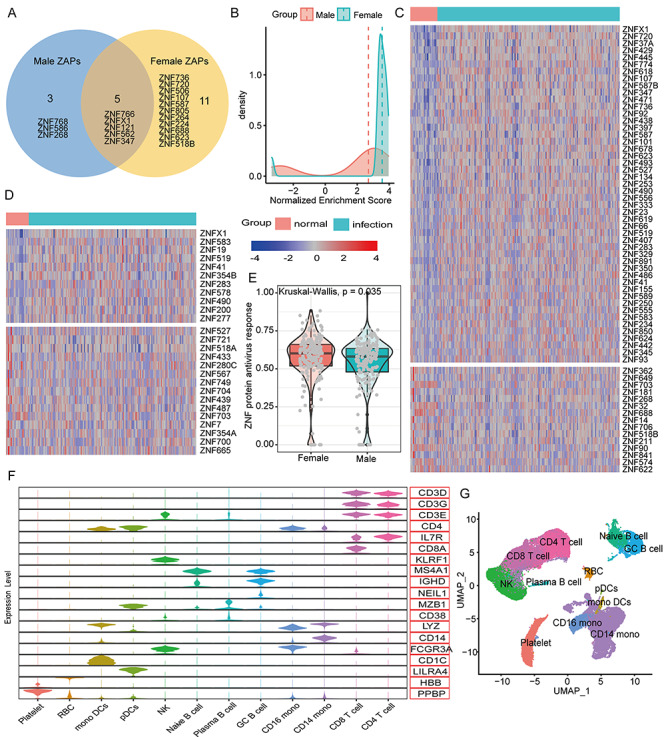
The similarities and differences in the functions and activities of ZNF proteins in COVID-19 patients of different genders. (A) The Venn diagram shows the common and specific ZNF proteins of different genders. (B) The difference in protein activity of the 5 shared ZNF proteins after different genders were infected with the coronavirus SARS-CoV-2. The horizontal axis represents the normalized enrichment score used to predict the protein activity of ZNF proteins. The larger the value, the stronger the protein activity. The vertical axis represents the number of MRs. The red and green lines represent the median value. (C) The heat map shows the expression changes of differentially expressed ZNF proteins in normal women and COVID-19 women. (D) The heat map shows the expression changes of differentially expressed ZNF proteins in normal women and COVID-19 men. (E) Differences in the overall ZNF protein activity among COVID-19 patients of different genders. The ssGSEA algorithm was used to calculate the overall expression activity of ZNF family genes. (F) The violin chart shows the expression of marker genes in different cell populations. For more information and marker combinations, please refer to Figure S4, see Supplementary Data available online at http://bib.oxfordjournals.org/, and its legend. (G) UMAP cluster diagram shows the cell population identification of single cell sequencing of all samples.
Total 43 102 single-cell data were further clustered into 27 clusters (Figure S4, see Supplementary Data available online at http://bib.oxfordjournals.org/). We used typical genes as immune cell markers to identify 12 main cell populations (Figures 4F and G and S5, see Supplementary Data available online at http://bib.oxfordjournals.org/). We found that male and female patients share the same immune cell types (Figure 5A), but females have much higher proportions of CD4 T cells, CD8 T cells and fewer monocytes than males (Figure 5B). Especially, the overall ZNF protein activity of females is higher than males whether in mild or severe patients (Figure 5C and D). Among 12 cell subgroups, the ZNF protein activity of mono DCs, GC B cells, CD14 monocytes and NK cells in mild female patients is, respectively, significantly higher than mild males (Figure 5E), in particular the ZNF protein activity differences of RBC, NK cells, GC B cells, CD8 T cells, CD4 T cells and CD14 monocytes are more obvious between severe female and male patients (Figure 5F). These above results indicated that female patients could express more ZNF proteins and stronger transcriptional activities than males do.
Figure 5.
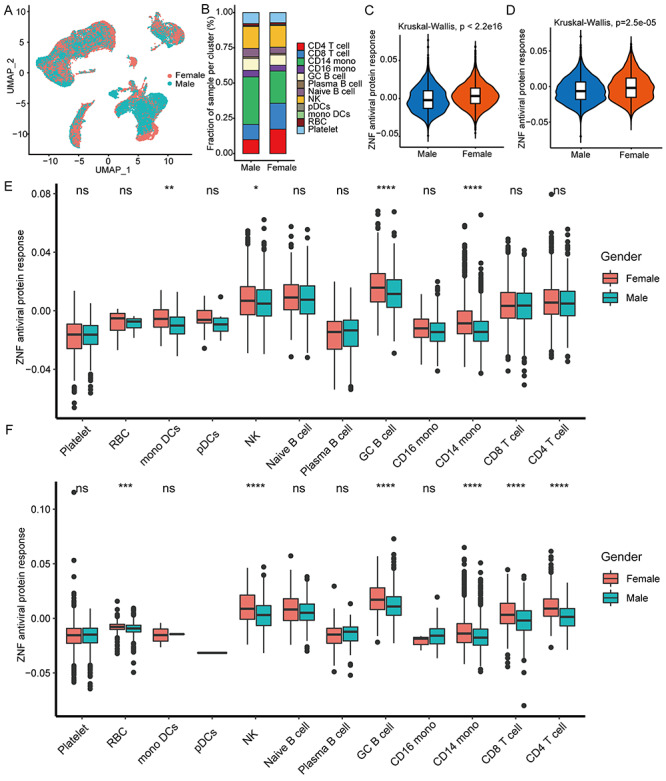
Female patients have higher antiviral activity of ZNF protein than male patients. (A) UMAP cluster diagram shows the single-cell distribution of COVID-19 patients of different genders. (B) Stacked histogram shows the cell ratio of COVID-19 patients of different genders. (C and D) Box diagram showing the antiviral activity of ZNF protein in mild and severe COVID-19 patients. (E) The box diagram shows the antiviral activity of ZNF protein in each cell cluster in mild COVID-19 patients. (F) The box diagram shows the antiviral activity of ZNF protein in each cell cluster in severe COVID-19 patients. Kruskal–Wallis test was used for statistical significance. Kruskal–Wallis test was used for statistical significance, ***P < 0.001, **P < 0.01, *P < 0.05, nsP > 0.05.
The activity of ZNF protein is related to anti-SARS-CoV-2 infection
Interestingly, the overall ZNF protein activity of COVID-19 patients was significantly higher than controls, and the ZNF protein activity of patients with high viral load was higher (Figure 6A). Remarkably, the increased ZNF protein activity was significantly negative correlation with the viral load of patients (Figure 6B), in particular the ZNFX1 is more significantly negative correlation with viral load (Figure 6C).
Figure 6.
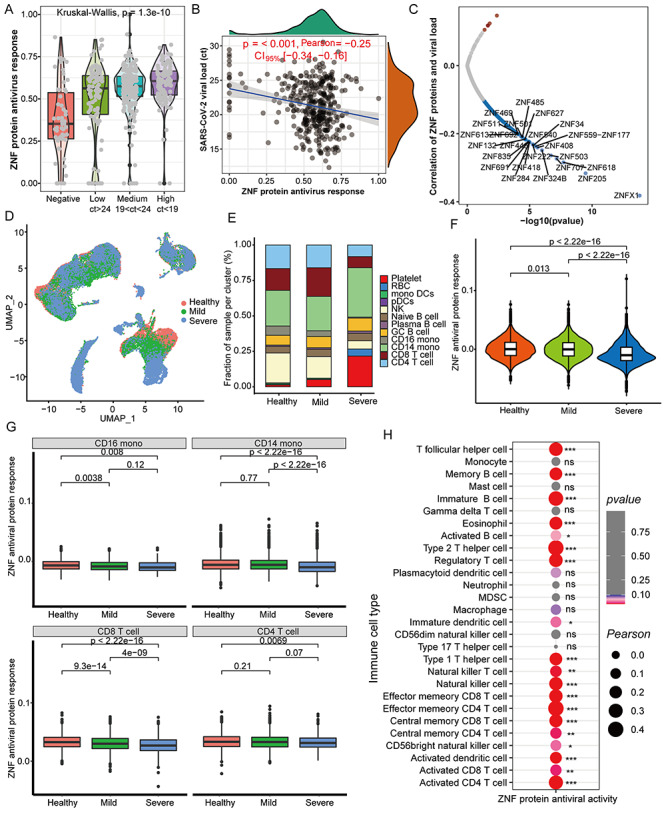
The ZNF protein activity is negatively correlated with virus loads but positively correlated with certain immune cells. (A) Differences in overall ZNF protein activity between COVID-19 patients and healthy controls with different viral loads in the pharynx. The Ct value represents the content of the SARS-CoV-2 pharynx, and the quartile value is used to determine the low, medium and high viral loads. (B) Correlation between pharynx viral loads and the overall ZNF protein activity in COVID-19 patients. (C) The correlation between the viral load of the pharynx and the mRNA level of a single ZNF family gene in COVID-19 patients. (D) UMAP cluster diagram shows the single-cell distribution of healthy control, mild and severe COVID-19 patients. (E) Stacked box chart shows the changes in cell proportions in healthy, mild and severe patients. (F) Box chart displays the overall ZNF protein antiviral activity of patients with different disease classes (healthy, mild and severe). (G) Box chart displays the overall ZNF protein antiviral activity in CD16 monocytes, CD14 monocytes, CD8 T cells and CD4 T cells of different disease classes (healthy, mild and severe). (H) Correlation between the abundance of throat immune cells and the overall ZNF protein activity in COVID-19 patients. Kruskal–Wallis test was used for statistical significance.
We further detected the overall ZNF protein activity in single cell level and found that healthy people and COVID-19 patients have the same cell subpopulation distribution (Figure 6D), but the proportion of CD4 and CD8 T cells in the serum of severe patients is, respectively, lower than healthy individuals and mild patients, and the monocytes are higher than healthy or mild ones (Figure 6E). The order of overall ZNF protein activity is healthy people > mild patient > severe patient (Figure 6F). We also found that the ZNF protein antiviral activity of CD4 T cells, CD8 T cells, CD14 and CD16 monocytes in severe patients are lower than healthy people and mild patients (Figure 6G), whereas the anti-SARS-CoV-2 activity of the other 8 cell subgroups has no obvious differences (Figure S6, see Supplementary Data available online at http://bib.oxfordjournals.org/). Remarkably, the ZNF protein activity is positively correlated with the abundance of 19 immune cells, especially T cells (Figure 6H), and it also is positively correlated with the abundance of T cells in scRNA-seq (Figure S7, see Supplementary Data available online at http://bib.oxfordjournals.org/).
ZNF proteins as TFs participate in anti-SARS-CoV-2 role
Interestingly, male and female patients shared five same ZNF TFs (Figure 4A), but their target genes were not identical, and females had more target genes than males (Figures S8 and S9, see Supplementary Data available online at http://bib.oxfordjournals.org/). Go enrichment analysis demonstrated that these target genes of male and female patients can significantly enrich in ‘interferon response’, ‘antiviral response’, ‘regulation of viral replication and cycle’ (Figure 7A) as well as a wide range of ‘RNA binding ability’ molecular function (Figure 7B), implying that the five ZNF TFs indeed involve in the anti-SARS-COV-2 infection. Importantly, we found that target genes of females could be enriched in some specific items, such as ‘myeloid cell differentiation’, ‘myeloid lymphocyte differentiation’, ‘interaction with host’ and ‘negative regulation of virus release’ (Figure 7A). In contrast, these target genes of males are mainly enriched in the ‘positive regulation cytokine production’, ‘cellular response to lipopolysaccharide’ and ‘cellular response to biotic stimulus’ (Figure 7B). Especially, some target genes of females can be enriched in some certain molecular function, such as helicase activities, ‘histone binding’ and ‘ubiquitin-like protein ligase binding’ (Figure 7B).
Figure 7.
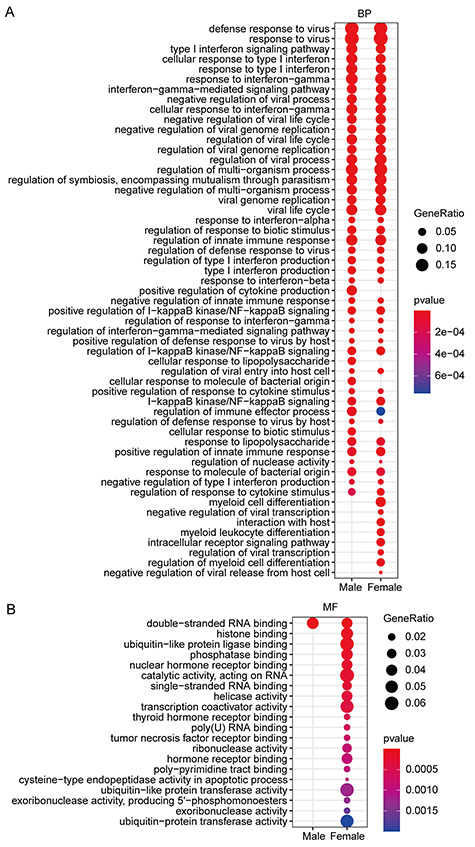
Functional enrichment analysis of target genes of ZNF TFs shared by different genders. (A) The biological process function enrichment analysis of the target genes of ZNF TFs shared by different genders. (B) Molecular function enrichment analysis of target genes of ZNF TFs shared by different genders.
BALF scRNA-seq reveals the anti-SARS-CoV-2 role of ZNF proteins
The 60 326 BALF single cells could be further clustered into 27 clusters (Figure S10, see Supplementary Data available online at http://bib.oxfordjournals.org/). According to the specific marker genes, 27 cell clusters were identified as 12 main cell types (Figures 8A and B and S11, see Supplementary Data available online at http://bib.oxfordjournals.org/). We found that the overall ZNF protein activity of different cell types undergone extensive changes after SARS-CoV-2 infection (Figure 8C). Compared with healthy people, the ZNF protein activity of the lung epithelial cells was significantly decreased in mild and severe patients (Figure 8C). Conversely, the ZNF activity of some immune cells, such as macrophages, CD8 T cells, CD4 T cells and NK cells, is, respectively, significantly increased in mild patients, but significantly reduced in severe patients (Figure 8C). Especially, the expression level and proportion of ZNFX1 in mild and severe patients were significantly increased, and similar expression trends also existed in other key immune genes (Figure 8D). The overall ZNF protein activity in the lung immune cells of mild and severe females was also higher than males, especially macrophages, T cells and NK cells (Figure 8E and F).
Figure 8.
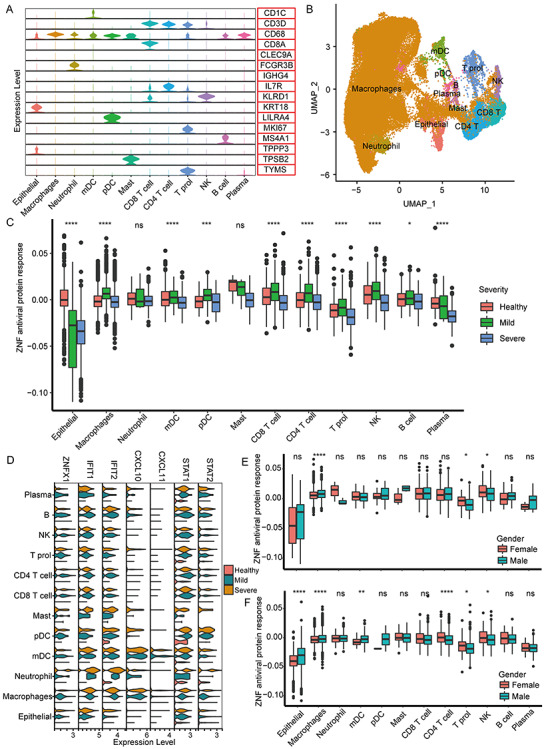
The antiviral activity of ZNF in different BALF cell populations. (A) The violin diagram shows the expression levels of marker genes of different cell types. (B) The UMAP cluster map shows the cell clustering of BALF single cells. (C) The box diagram shows the overall ZNF protein activity of different cell populations of mild and severe patients in healthy people. (D) The violin key antiviral gene ZNFX1, for example, figure displays IFIT1, IFIT2, CXCL10, CXCL11, STAT1 and STAT2 in healthy people, patients with mild and severe expression. (E) The box plot shows the overall ZNF protein activity of different cell populations of mild patients of different genders. (F) Box plot shows the overall ZNF protein activity of different cell populations in severe patients of different genders. Kruskal–Wallis test was used for statistical significance, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, nsP > 0.05.
Analysis of clinical indicators of male and female patients
Herein, we analyzed the TBNK cells data (T cells, B cells and NK cells) from 548 COVID-19 patients. Fascinatingly, female patients have higher immune abundance of CD 3+, CD4+ T cells, B cells and CD4/CD8 ratio than males (Figure 9A and E), but males have more NK cells than females (Figure 9F). Female patients have more naïve B cells and effectors CD8 T cells than males (Figure 9G and H). Although no significant differences existed in other 25 cell subsets (Figures S12–S14, see Supplementary Data available online at http://bib.oxfordjournals.org/), female patients do have higher proportion and stronger activity of lymphocytes than males.
Figure 9.
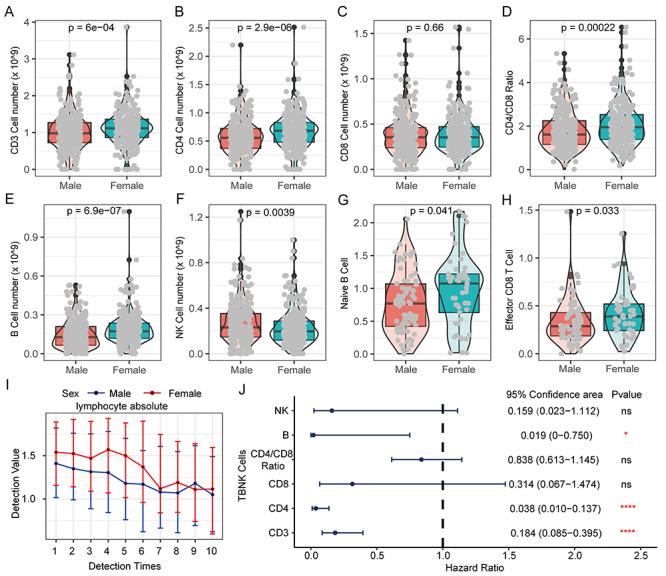
The difference of lymphocytes in COVID-19 patients of different genders and their prognostic effects. (A) The proportion of CD3 T cells in PBMC of COVID-19 patients of different genders. (B) The proportion of CD4 T cells in PBMCs of COVID-19 patients of different genders. (C) Proportion of CD8 T cells in PBMC of COVID-19 patients of different genders. (D) Differences in the ratio of CD4/CD8 in PBMC of COVID-19 patients of different genders. (E) Proportion of B cells in PBMC of COVID-19 patients of different genders. (F) The proportion of NK cells in PBMC of COVID-19 patients of different genders. (G) The proportion of Naive B cells in PBMCs of COVID-19 patients of different genders. (H) Proportion of Effector CD8 T cells in PBMC of COVID-19 patients of different genders. Kruskal–Wallis test was used for statistical significance. (I) The cumulative event rate shows that lymphocyte absolute value contributes to the prognosis of all COVID-19 patients. (J) The cumulative event rate shows the contribution of lymphocyte absolute value to the prognosis of male COVID-19 patients. (K) The cumulative event rate shows the contribution of lymphocyte absolute value to the prognosis of female COVID-19 patients. (L) The line chart shows the dynamic changes of lymphocyte absolute value of COVID-19 patients of different genders. With the progress of treatment and the passage of time, multiple test values during the patient’s hospitalization are used for comparison.
Herein, we further found that the incidence of critical illness and death patients is significantly increased with abnormal decrease of lymphocyte cell abundances (hazard ratio: 2.7330), indicating that lymphocytes could serve as a good protective factor for COVID-19 patients (Figure S15, see Supplementary Data available online at http://bib.oxfordjournals.org/). The lymphocyte abundance of male patients is always lower than females in earlier multiple tests, but the lymphocyte abundance gradually tends to be consistent between them with the treatment progresses (Figure 9I). Remarkably, lymphopenia is more detrimental to male COVID-19 patients than females with hazard ratio (male: 3.2977 versus female: 1.9776) (Figure S15, see Supplementary Data available online at http://bib.oxfordjournals.org/). Finally, we analyzed the effect of major lymphocyte subtypes on the prognosis of COVID-19 patients. Our results revealed that CD3, CD4, T cells and B cells as protective factors were significantly associated with the prognosis of COVID-19 patients, but CD8 T cells, NK cells and CD4/CD8 ratio were no significant effect on the prognosis of COVID-19 patients (Figure 9J).
Discussion
Elucidating the mechanism of the anti-SARS-CoV-2 infection is very important for the prevention and treatment of COVID-19 patients. Although many studies have revealed that certain genes or proteins play very key roles in anti-SARS-CoV-2 response [30–34], the mechanism of the anti-SARS-CoV-2 infection remains to be further studied. In this work, we demonstrated that female patients have better survival rates than males (Figure 1), which agrees with these reports [1–3]. The better prognosis of female patients to males was attributed to the lower expression of ACE2, TMPRSS2 and stronger autoimmunity [9–12]. However, the expression reduction of B cell and NK cell-specific transcripts as well as the suppression of NF-κB expression in male patients might be the reason of the poor prognosis [14], but this study only used gender as a batch correction without distinguishing the molecular differences between different genders. Therefore, the molecular mechanism that induces different prognostic outcomes between male and female patients is still a mystery up to now. In particular, it is unclear why women show stronger immunity and more immune cells than men after SARS-CoV-2 infection [35].
In our study, two key types of MRs (e.g. STAT1/STAT2 and ZNF proteins) are significantly upregulated after SARS-CoV-2 infection (Figures S2, see Supplementary Data available online at http://bib.oxfordjournals.org/, and 2–4), and the ZNF protein activity is strongly negative correlation with the SARS-CoV-2 load (Figure 6B), revealing that ZNF proteins might play critical anti-SARS-CoV-2 roles. Previous studies indicated that STAT1/STAT2 as the important members of the JAK–STAT signaling pathway could activate many ZNF protein expressions [28, 29, 36], and ZNF proteins could recognize and bind to the CpG site of SARS-CoV-2 to degrade or inhibit its replication and translation [37, 38]. In our work, the activity of ZNFX1 is significantly negative correlation with SARS-CoV-2 load (Figure 6C). Remarkably, this recent study has proven that SARS-CoV-2 can be restricted by zinc finger antiviral protein despite pre-adaptation to the low-CpG environment in humans, in particular type I, II and III IFNs all can strongly inhibit SARS-CoV-2 and further induce ZAP expressions, while these knockdown experiments also manifest that endogenous ZAPs can significantly restrict SARS-CoV-2 [37]. These above results seem to suggest that SARS-CoV-2 infection may activate STAT1/STAT2 expressions to promote multitudinous ZNF protein up-regulation to resist SARS-CoV-2 infection via binding to its CpG site [28, 29, 36, 37].
In addition, ZNF proteins can also serve as TFs to regulate target genes to involve in anti-SARS-CoV-2 infection (Figure 7A and B). Previous reports revealed that ZNF proteins as TFs can activate type I interferon signaling and multiple antiviral genes to suppress the virus [36, 39–41], which might support our results. Furthermore, studies indicated that ZNF TFs not only can promote the activation and maturation of immune cells [38, 42] but also directly or indirectly increase the content of antiviral antibodies [42, 43]. In our work, we find that the ZNF protein activity is significantly positive correlation with the abundances of 19 immune cells (Figure 6H), and the ZNF protein activity is more significantly positive correlation with the abundance of T cells of healthy and COVID-19 patients at single cell levels (r = 0.76, P = 0.0018) (Figure S7, see Supplementary Data available online at http://bib.oxfordjournals.org/). While our results also uncover that the ZNF protein antiviral activity of CD4 T cells, CD8 T cells, CD14 and CD16 monocytes in severe patients is lower than healthy people and mild patients (Figure 6G). Especially, the lymphocytes can act as a protective factor for the prognosis COVID-19 patients (Figure 8I and K). This cause may be that on the one hand, lymphocytes are the main source of interferon production to induce more ZNF protein expressions [42]. Taken together, these findings suggest that ZNF proteins and immune cells can promote each other to effectively suppress SARS-CoV-2 infection.
In conclusion, we have proposed a possible anti-SARS-CoV-2 mechanism of ZNF proteins (Figure 10), i.e. SARS-CoV-2 infection activates STAT1/STAT2 expressions to elevate multitudinous ZNF protein expressions (e.g. ISGs) to repress SARS-CoV-2 infection [28, 29, 44, 45]. On the one hand, ZNF proteins (e.g. ZNFX1) directly recognize the CpG site of SARS-CoV-2 and induce interferon production in the early stage of SARS-CoV-2 infection by activating RIG-I signaling pathway [37, 46, 47]. On the other hand, a large number of ZNF proteins can bind to the CpG site of SARS-CoV-2 to directly degrade SARS-CoV-2 or inhibit its replication and translation [37, 38, 47, 48]. While ZNF proteins can also function as TFs to regulate the expression of a large number of target genes including ISGs to enhance interferon signaling or directly promote the activation and mature of multiple types of immune cells against SARS-CoV-2 infection [36, 39–41, 43, 46, 49, 50]. Taken together, we here suggest that the highly expressed SATA2 in female patients might result in releasing more ZNF proteins and stronger ZNF protein activities to promote immune cell activities than males, and that these differentially expressed ZNF proteins and distinct activity might be responsible for the different survival outcome for different gender COVID-19 patients.
Figure 10.
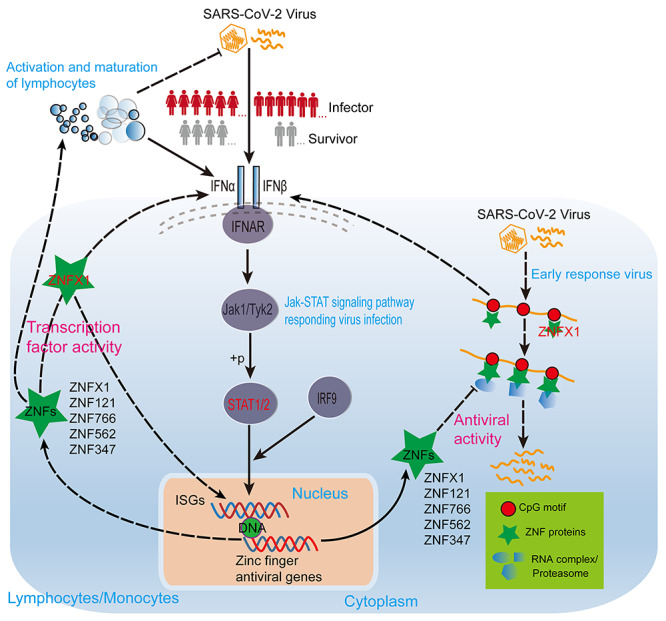
The model shows that ZNF proteins of COVID-19 patients of different genders resist SARS-CoV-2 infection through TF activity and antiviral activity. The solid line represents the well-known regulatory relationship, and the dashed line represents the regulatory relationship speculated based on our analysis results and existing literature. The sharp arrow indicates activation, and the flat arrow indicates inhibition.
Key Points
The systems biology algorithms ARACNe and VIPER are very effective for identifying major regulators (MRs) in response to viral infections.
We have identified MRs such as STAT1/STAT2, SP1, REL, etc., especially ZNF family proteins (also called zinc finger antiviral proteins), which respond to SARS-CoV-2 infection.
ZNF family proteins can inhibit SARS-CoV-2 and increase immune activity through transcription factor activity and as antiviral effector proteins.
Women have a better prognosis and higher ratio and activity of lymphocytes than men with COVID-19, which may be attributed to the higher ZNF activity in women.
Supplementary Material
Acknowledgements
We thank other researchers and experiments for sharing the data. We are also grateful to our peers for their suggestions and review of the manuscript. All the authors have agreed to the manuscript.
Shijie Qin is a PhD student in the Comparative Genomics and Bioinformatics Laboratory of the College of Life Sciences, Nanjing Normal University. His research interest is bioinformatics.
Weijun Xu is a clinical laboratory physician affiliated to the COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China. His research interest is molecular diagnosis of diseases.
Canbiao Wang is a graduate student in the Comparative Genomics and Bioinformatics Laboratory of the College of Life Sciences, Nanjing Normal University, China. His research interest is bioinformatics.
Sizhu Jiang is a student in the College of Arts and Sciences, Emory University, Atlanta, USA. His research interest is biostatistics and public health prevention.
Wei Dai is a postdoctoral researcher affiliated to the COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China. Her research interest is molecular diagnosis of diseases.
Yang Yang is a PhD student affiliated to the COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China. Her research interest is molecular diagnosis of diseases.
Jiawei Shen is a clinical laboratory physician affiliated to the COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China. Her research interest is molecular diagnosis of diseases.
Ping Jin is an associate professor in the Laboratory of Comparative Genomics and Bioinformatics, School of Life Sciences, Nanjing Normal University, China. Her research interests are innate immunity and the evolution of Branchiostoma.
Fei Ma is the team leader of the Laboratory of Comparative Genomics and Bioinformatics at the School of Life Sciences, Nanjing Normal University, China. His research interests include innate immunity, bioinformatics and the interaction between microRNA and transcription factors.
Xinyi Xia is the team leader of the COVID-19 Research Center, Institute of Laboratory Medicine, Jinling Hospital, Nanjing University School of Medicine; Nanjing Clinical College of Southern Medical University, Nanjing, China, and the member of Joint Expert Group for COVID-19, Department of Laboratory Medicine & Blood Transfusion, Wuhan Huoshenshan Hospital, China. His research interests are the pathological mechanism, prevention, diagnosis and treatment of COVID-19.
Contributor Information
Shijie Qin, Comparative Genomics and Bioinformatics Laboratory of the College of Life Sciences, Nanjing Normal University, China.
Weijun Xu, COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China.
Canbiao Wang, Comparative Genomics and Bioinformatics Laboratory of the College of Life Sciences, Nanjing Normal University, China.
Sizhu Jiang, College of Arts and Sciences, Emory University, Atlanta, USA.
Wei Dai, COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China.
Yang Yang, COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China.
Jiawei Shen, COVID-19 Research Center, Institute of laboratory medicine, Jinling Hospital, Nanjing University School of medicine, the first school of clinical medicine, Southern Medical University, China.
Ping Jin, Laboratory of Comparative Genomics and Bioinformatics, School of Life Sciences, Nanjing Normal University, China.
Fei Ma, Laboratory of Comparative Genomics and Bioinformatics at the School of Life Sciences, Nanjing Normal University, China.
Xinyi Xia, COVID-19 Research Center, Institute of Laboratory Medicine, Jinling Hospital, Nanjing University School of Medicine; Nanjing Clinical College of Southern Medical University, Nanjing, China.
Availability of Data and Materials
All the data supporting this article are available in the methods section and supplementary files of manuscript or can be obtained by contacting the author.
Authors’ contributions
S.Q., F.M. and X.X. conceived the study. S.Q. and C.W. collected omics data and conducted analysis; S.Q., C.W. and S.J. visualized diagrams. W.D. performed flow cytometry experiments and data collation. S.Q. and W.X. wrote the draft; F.M., S.Q. and P.J. revised the draft. X.X., W.X., Y.Y. and J.S. collected and provided COVID-19 clinical data; P.J. supervised the project progress. Y.Y. and J.S. participated in the commentary of the manuscript.
Funding
Key Foundation of Wuhan Huoshenshan Hospital (2020[18]); Key Research & Development Program of Jiangsu Province (BE2018713); Medical Innovation Project of Logistics Service (18JS005); Foundation of Jiangsu Population Association (JSPA2019017); Jiangsu Provincial Association for Maternal and Child Health Studies Commissioned Research Project Funding (JSFY202005); Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflicts of interest
All authors declare that there is no conflict of interest.
Reference
- 1. Yang P, Esper AM. Investigating the Sex Differences in COVID-19: Another Step Forward, But Many Unanswered Questions. Clin Infect Dis. 2020. Dec 3;71(9):2495–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alkhouli M, Nanjundappa A, Annie F, et al. Sex Differences in Case Fatality Rate of COVID-19: Insights From a Multinational Registry. Mayo Clin Proc. 2020. Aug;95(8):1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peckham H, de Gruijter NM, Raine C, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020. Dec 9;11(1):6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gasmi A, Noor S, Tippairote T, et al. Individual risk management strategy and potential therapeutic options for the COVID-19 pandemic. Clin Immunol. 2020. Jun;215:108409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J, Bai H, Liu J, et al. Distinct clinical characteristics and risk factors for mortality in female COVID-19 inpatients with Coronavirus Disease 2019 (COVID-19): a sex-stratified large-scale cohort study in Wuhan, China. Clin Infect Dis. 2020. Dec 15;71(12):3188–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pérez-López FR, Tajada M, Savirón-Cornudella R, et al. Coronavirus disease 2019 and gender-related mortality in European countries: a meta-analysis. Maturitas. 2020. Nov;141:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lukassen S, Chua RL, Trefzer T, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020. May 18;39(10):e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020. Apr;181(2):271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sama IE, Ravera A, Santema BT, et al. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin-angiotensin-aldosterone inhibitors. Eur Heart J. 2020. May 14;41(19):1810–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chanana N, Palmo T, Sharma K, et al. Sex-derived attributes contributing to SARS-CoV-2 mortality. Am J Physiol Endocrinol Metab 2020. Sep 1;319(3):E562–E567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahashi T, Ellingson MK, Wong P, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020 Dec;588(7837):315–320. [DOI] [PMC free article] [PubMed]
- 12. Qin L, Li X, Shi J, et al. Gendered effects on inflammation reaction and outcome of COVID-19 patients in Wuhan. J Med Virol. 2020. Nov;92(11):2684–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qin S, Xia X, Shi X, et al. Mechanistic insights into SARS-CoV-2 epidemic via revealing the features of SARS-CoV-2 coding proteins and host responses upon its infection. Bioinformatics. 2020 Aug 17:btaa725. [DOI] [PMC free article] [PubMed]
- 14. Lieberman NAP, Peddu V, Xie H, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol 2020. Sep 8;18(9):e3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee JS, Park S, Jeong HW, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020 Jul 10;5(49):eabd155.4. [DOI] [PMC free article] [PubMed]
- 16. Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020 Jun;26(6):842–844. [DOI] [PubMed] [Google Scholar]
- 17. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Jan 1;26(1):139–40. [DOI] [PMC free article] [PubMed]
- 18. McDermaid A, Monier B, Zhao J, et al. Interpretation of differential gene expression results of RNA-seq data: review and integration. Brief Bioinform. 2019 Nov 27;20(6):2044–2054. [DOI] [PMC free article] [PubMed]
- 19. Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006. Mar 20;7 Suppl 1(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alvarez MJ, Shen Y, Giorgi FM, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity Nat Genet. 2016 Aug;48(8):838–47. [DOI] [PMC free article] [PubMed]
- 21. Basso K, Saito M, Sumazin P, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010 Feb 4;115(5):975–84. [DOI] [PMC free article] [PubMed]
- 22. Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010 Jan 21;463(7279):318–25. [DOI] [PMC free article] [PubMed]
- 23. Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012. May;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013. Jan 16;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell 2019 Jun 13;177(7):1888-1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018 Dec 3. [DOI] [PubMed]
- 27. Zhang JY, Wang XM, Xing X, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol. 2020. Sep;21(9):1107–1118. [DOI] [PubMed] [Google Scholar]
- 28. Bick MJ, Carroll JW, Gao G, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003. Nov;77(21):11555–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 2002. Sep 6;297(5587):1703–6. [DOI] [PubMed] [Google Scholar]
- 30. Lukassen S, Chua RL, Trefzer T, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020. May 18;39(10):e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 April 16;181(2):271–280.e8. [DOI] [PMC free article] [PubMed]
- 32. Hoffmann M, Kleine-Weber H, Pöhlmann S. Multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020 May 21;78(4):779–784.e5. [DOI] [PMC free article] [PubMed]
- 33. Daly JL, Simonetti B, Klein K, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020 Nov 13;370(6518):861–865. [DOI] [PMC free article] [PubMed]
- 34. Cantuti-Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 Nov 13;370(6518):856–860. [DOI] [PMC free article] [PubMed]
- 35. Cattrini C, Bersanelli M, Latocca MM, et al. Sex hormones and hormone therapy during COVID-19 pandemic: implications for patients with cancer. Cancers (Basel). 2020 Aug 18;12(8):2325. [DOI] [PMC free article] [PubMed]
- 36. Wang G, Zheng C. Zinc finger proteins in the host-virus interplay: multifaceted functions based on their nucleic acid-binding property. FEMS Microbiol Rev. 2020 Nov 10:fuaa059. [DOI] [PubMed]
- 37. Nchioua R, Kmiec D, Müller JA, et al. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio. 2020 Oct 16;11(5):e01930–20. [DOI] [PMC free article] [PubMed]
- 38. Wang Y, Yuan S, Jia X, et al. Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat Cell Biol. 2019 Nov;21(11):1346–1356. [DOI] [PubMed]
- 39. Ozato K. PLZF outreach: a finger in interferon's pie. Immunity. 2009 Jun 19;30(6):757–8. [DOI] [PMC free article] [PubMed]
- 40. Najafabadi HS, Mnaimneh S, Schmitges FW, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol. 2015 May;33(5):555–62. [DOI] [PubMed]
- 41. Xu D, Holko M, Sadler AJ, et al. Promyelocytic leukemia zinc finger protein regulates interferon-mediated innate immunity. Immunity. 2009 Jun 19;30(6):802–16. [DOI] [PMC free article] [PubMed]
- 42. Zhu M, Zhou J, Liang Y, et al. CCCH-type zinc finger antiviral protein mediates antiviral immune response by activating T cells. J Leukoc Biol. 2020. Feb;107(2):299–307. [DOI] [PubMed] [Google Scholar]
- 43. Enders A, Short A, Miosge LA, et al. Zinc-finger protein ZFP318 is essential for expression of IgD, the alternatively spliced Igh product made by mature B lymphocytes. Proc Natl Acad Sci USA. 2014 Mar 25;111(12):4513–8. [DOI] [PMC free article] [PubMed]
- 44. Thiesen HJ. Multiple genes encoding zinc finger domains are expressed in human T cells. New Biol. 1990 Apr;2(4):363–74. [PubMed]
- 45. Huntley S, Baggott DM, Hamilton AT, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006 May;16(5):669–77. [DOI] [PMC free article] [PubMed]
- 46. Wang Y, Yuan S, Jia X, et al. Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat Cell Biol. 2019. Nov;21(11):1346–1356. [DOI] [PubMed] [Google Scholar]
- 47. Zhao Y, Song Z, Bai J, et al. ZAP, a CCCH-type zinc finger protein, inhibits porcine reproductive and respiratory syndrome virus replication and interacts with viral Nsp9. J Virol. 2019 May 1;93(10):e00001-19. [DOI] [PMC free article] [PubMed]
- 48. Kmiec D, Nchioua R, Sherrill-Mix S, et al. CpG frequency in the 5' third of the env gene determines sensitivity of primary HIV-1 strains to the Zinc-Finger antiviral protein. mBio. 2020 Jan 14;11(1):e02903–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu M, Zhou J, Liang Y, et al. CCCH-type zinc finger antiviral protein mediates antiviral immune response by activating T cells. J Leukoc Biol. 2020 Feb;107(2):299–307. [DOI] [PubMed]
- 50. Wang Y, Wang G, Wang H, et al. Chicken biliary exosomes enhance CD4(+)T proliferation and inhibit ALV-J replication in liver. Biochem Cell Biol. 2014 Apr;92(2):145–51. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting this article are available in the methods section and supplementary files of manuscript or can be obtained by contacting the author.


