Abstract
Background
Low initial severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody titers dropping to undetectable levels within months after infection have raised concerns about long-term immunity. Both the antibody levels and the avidity of the antibody–antigen interaction should be examined to understand the quality of the antibody response.
Methods
A testing-on-a-probe “plus” panel (TOP-Plus) was developed to include a newly developed avidity assay built into the previously described SARS-CoV-2 TOP assays that measured total antibody (TAb), surrogate neutralizing antibody (SNAb), IgM, and IgG on a versatile biosensor platform. TAb and SNAb levels were compared with avidity in previously infected individuals at 1.3 and 6.2 months after infection in paired samples from 80 patients with coronavirus disease 2019 (COVID-19). Sera from individuals vaccinated for SARS-CoV-2 were also evaluated for antibody avidity.
Results
The newly designed avidity assay in this TOP panel correlated well with a reference Bio-Layer Interferometry avidity assay (r = 0.88). The imprecision of the TOP avidity assay was <10%. Although TAb and neutralization activity (by SNAb) decreased between 1.3 and 6.2 months after infection, the antibody avidity increased significantly (P < 0.0001). Antibody avidity in 10 SARS-CoV-2 vaccinated individuals (median: 28 days after vaccination) was comparable to the measured antibody avidity in infected individuals (median: 26 days after infection).
Conclusions
This highly precise and versatile TOP-Plus panel with the ability to measure SARS-CoV-2 TAb, SNAb, IgG, and IgM antibody levels and avidity of individual sera on one sensor can become a valuable asset in monitoring not only patients infected with SARS-CoV-2 but also the status of individuals’ COVID-19 vaccination response.
Keywords: COVID-19, SARS-CoV-2, serological assay, antibody avidity, test-on-a-probe biosensors, vaccine
Introduction
Coronavirus disease 2019 (COVID-19), caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to crippling levels of morbidity and mortality around the world (1). Seroprevalence studies have begun to show a larger extent of SARS-CoV-2 infections than initially reported because of the high prevalence of infected individuals with mild or no symptoms (2, 3). However, lower SARS-CoV-2 IgG antibody levels have been reported in those with mild or no symptoms compared with those with severe COVID-19 (4–7). Furthermore, emerging evidence suggests that SARS-CoV-2 antibodies in some asymptomatic carriers may diminish over time to levels below detection (8–10). This decrease in antibody levels over time may include neutralizing SARS-CoV-2 antibodies, which play a vital role in viral clearance (11). These observations raise the question of whether acquired immunity may be short lived and herd immunity protection may be less durable than anticipated (12).
Although many studies focus on overall antibody titers, other factors are likely equally important in evaluating the humoral antibody response. Binding titers are determined by the antibody concentration and average affinity. Avidity can be defined as the strengthening of antibody binding through bi- or multivalency or as the functional affinity of the entire IgG, IgA, or IgM molecule, a net product of the intrinsic paratope–epitope affinity and valency (13). In this study, we use the term avidity in the latter sense. Low-avidity antibodies are typically produced early in the humoral immune response (14, 15). Over time, with affinity maturation, the intrinsic affinity of the antibody–antigen interaction strengthens and so does the functional affinity or avidity of bivalent IgG or classes of higher valency.
To evaluate whether these reported weak early antibody responses should be of clinical concern, various assays have emerged to help assess antibody avidity in the evaluation of the SARS-CoV-2 immune response (16–19). Antibody avidity may be measured in a variety of ways, including ELISAs, high-performance liquid chromatography, capillary electrophoresis, or single radial immunodiffusion. Although providing some insight into the functional affinity, these assays are often qualitative, labor intensive, and low-throughput and display low accuracy and precision. Therefore, biosensor technologies such as surface plasmon resonance and bio-layer interferometry (BLI) have become popular in monitoring the molecular binding between antigen and antibody in a real-time and cost-effective manner (20).
This study describes a similar but novel approach to evaluating the level and avidity of SARS-CoV-2 receptor-binding domain (RBD) antibodies using a testing-on-a-probe “plus” (TOP-Plus) panel that includes a newly developed avidity assay and the previously described SARS-CoV-2 TOP assays (total antibody [TAb], surrogated neutralizing antibody [SNAb]) on a single versatile biosensor platform. This fully automated assay panel was used in the current study to evaluate and describe the antibody response and antibody avidity approximately 1 month and 6 months after symptom onset in 80 individuals who were previously diagnosed with COVID-19 (21). The antibody avidity in 10 vaccinated individuals approximately 1 month after SARS-CoV-2 vaccination (first dose) was also evaluated as an early demonstration of its use for monitoring the response to vaccination.
Materials and Methods
Study Participants and Source of Specimens
The details of participant characteristics and associated COVID-19 symptoms have been described previously (21, 22). In summary, 80 adults aged 18–76 years who had been diagnosed with SARS-CoV-2 infection or who had a confirmed SARS-CoV-2 exposure had blood specimen collected at the Rockefeller University Hospital approximately 1.3 and 6.2 months after infection. Weill Cornell Medicine performed the antibody analyses as described below (in Evaluation of Clinical Utility and the online Supplemental Data).
Additional blood specimens were collected January 2–28, 2021, from a separate cohort of 10 individuals vaccinated for SARS-CoV-2 (Moderna, mRNA-1273 vaccine). Specimens were collected 25–28 days after administration of the first vaccine dose but before the second dose (median: 28 days after vaccination). Samples were analyzed on the TOP-Plus biosensor, as described below.
SARS-CoV-2 Antibody Avidity Assay Description
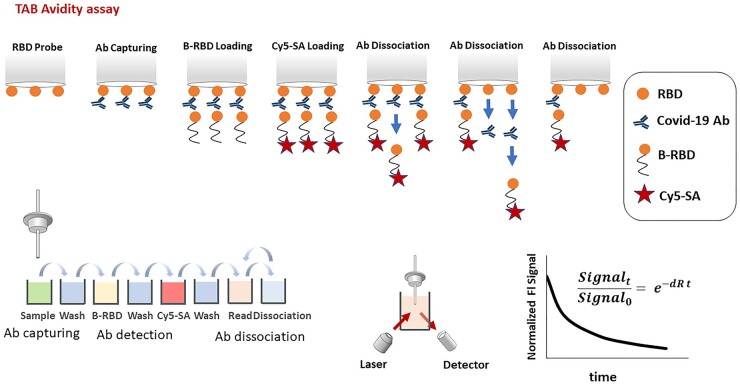
The principle of the SARS-CoV-2 antibody avidity assay is similar to a previously described technology (23) that measured SARS-COV-2 antibodies at the tip of an RBD-coated quartz probe and used a biotinylated RBD and a streptavidin-Cy5 conjugate as the signaling elements. However, the calculated relative dissociation rate (dR) allows for avidity testing in this new assay (Fig. 1). In short, an RBD-precoated probe is sequentially incubated in microwells containing the sample (to capture SARS-CoV-2–specific antibodies), biotinylated RBD and streptavidin-Cy5 conjugate along with washes between the incubation steps. After the initial fluorescent signal is measured (Signal_0), the probe with the immobilized immunocomplex enters into repetitive dissociation cycles with multiple incubations in PBS with Tween®-20 detergent (pH 7.4) as a dissociation buffer. After each incubation, the fluorescent signal is measured (Signal_t). Ultimately, a dissociation curve is constructed by plotting the normalized fluorescent signal (Signal_0/Signal_t) over time. The dR (1/s) is calculated from a function derived by fitting the dissociation curve, assuming first-order reaction kinetics.
Fig. 1.
SARS-CoV-2 antibody avidity assay principle. An RBD-precoated probe is sequentially incubated in microwells containing sample to capture SARS-CoV-2-specific antibodies, wash buffer, detection biotinylated RBD, wash buffer, and streptavidin-Cy5 conjugate. The fluorescent signal is then measured (Signal_0). After that, probe with the immobilized immunocomplex goes into repetitive dissociation cycles by multiple incubations in PBS with Tween®-20 detergent as dissociation buffer. After each incubation, the fluorescent signal is measured (Signal_t). At the end of measurement, the dissociation curve is constructed by plotting the normalized fluorescent signal (Signal_0/Signal_t) over time. The dR is then calculated by fitting first-order reaction kinetics to the dissociation curve.
The dissociation profile represents the rate of antibody dissociation from the RBD-coated probe. For an accurate measurement of relative antibody dR, a limited but adequate amount of antibody is loaded on the probe surface. Loading higher amounts of antibody, determined by getting a high initial fluorescent signal (Signal_0) over a certain threshold, causes the formation of a packed multilayer antibody construct on the probe surface. This leads to an inaccurate measurement because antibodies in a packed adsorbed layer cannot freely dissociate. Therefore, antibody packing density affects the dissociation measurement. In contrast, sensitive measurements require adequate antibody loading, as determined by the initial fluorescent signal above a certain level. Therefore, proper antibody loading must be within the proper range for accurate measurement. Samples with high antibody concentrations must be diluted for measurement. The appropriate initial fluorescent signal (which verifies optimal antibody loading) was practically determined through a titration study to be in the range of 20–615 relative fluorescence units (RFU), as discussed below under analytical validation. The dilution factor was determined by measuring the initial fluorescent signal to fall within this proper signal range.
Of note, a lower dR reflects both affinity maturation and multivalent binding development. Either a higher intrinsic binding strength of a paratope to RBD or addition of paratopes to the antibody structure results in a higher binding strength and a lower dR of a COVID-19 antibody–RBD pair.
Analytical Validation of SARS-CoV-2 Antibody Avidity
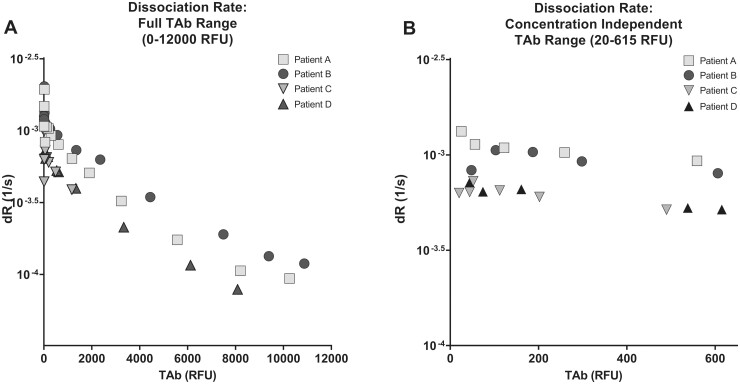
Titrations were performed to determine the proper range of antibody loading. Serum samples with different TAb levels (1171–10 872 RFU undiluted) were randomly selected from 4 patients with COVID-19 to perform titration studies. The pooled SARS-CoV-2 TAb-negative serum was used as diluent. Samples with initial fluorescent signal (Signal_0) in the range of 20–615 RFU showed consistent dR values, independent of the signal or concentration level (Fig. 2). We considered this fluorescent signal range as an indication for optimal antibody loading. Samples with a high fluorescent signal (Signal_0 > 615 RFU) were diluted accordingly for measurement. Samples with a low fluorescent signal (Signal_0 < 20 RFU) were identified as unmeasurable.
Fig. 2.
Determination of concentration independent range. (A), Dilution studies were performed using 4 randomly selected specimen with high TAb measurements (1171–10 872 RFU undiluted) and plotted against the disassociation rate. (B), The dissociation rate was independent of TAb concentration between 20 and 615 RFU of TAb.
The SARS-CoV-2 antibody avidity assay was evaluated with 12 different purified antibodies against SARS-CoV-2 that were purchased from various vendors (Supplemental Table 1). These are recombinant human, rabbit, or chimeric monoclonal and polyclonal antibodies of varying avidity levels to the RBD. A dissociation curve was generated utilizing pooled human sera from patients who were negative for SARS-CoV-2, with sera spiked with one of 5 antibodies and measured for avidity. The range of antibody concentrations used in the spike-in experiments are listed in Supplemental Table 2. The dRs were determined at varying levels of Signal_0 by spiking the negative pooled sera with one of 7 antibodies and measured for avidity.
Avidity Assay Precision and Interference
The imprecision of the avidity assay and interference studies, including cross-reactivity studies, are described in the online Supplemental Data.
BLI Comparison Study
BLI measurements by the Gator (Gator Bio) were used to compare the avidity of 12 different purified COVID-19 antibodies (Supplemental Table 1) with the TOP-Plus avidity assay. Gator koff (dR constant) measurement was performed at a fixed 10 μg/mL concentration level in a buffer containing 0.2% BSA and 0.02% Tween®-20. The TOP-Plus avidity assay measurements were performed using COVID-19–negative pooled serum spiked with one of these antibodies at a concentration level between 1 and 30 μg/mL (dependent on the appropriate fluorescence signal, range of 20–615 RFU, as described previously).
SARS-CoV-2 TAb and SNAb Assays
The SARS-CoV-2 TAb and SNAb assays were used to measure plasma TAb and SNAb antibodies against SARS-CoV-2. Plasma samples were assayed on the fully automated Pylon 3-D analyzer (ET HealthCare), as described previously (23, 24). Additional information may be found in the online Supplemental Data.
Statistical Analysis
Statistical analysis details are provided in the online Supplemental Data.
Results
Analytical Validation of SARS-CoV-2 Antibody Avidity
Determination of concentration-independent range in clinical specimens.
As described in the assay description, accurate antibody dR requires only sufficient amounts of antibody to be loaded onto the probe surface. To evaluate and minimize this potential source of artifact associated with these label-free methods, dilution studies were performed using 4 specimens with high TAb measurements (in the range 1171–10 872 RFU) and plotted against the dR (Fig. 2A). Based on these studies, it was determined that the concentration-independent range for this assay was between 20 and 615 RFU of TAb (Fig. 2B). Therefore, any specimen with a TAb >615 RFU was first diluted into the range of 20–615 RFU before determining the dR.
SARS-CoV-2 antibody avidity assay characterization with model COVID-19 purified antibodies.
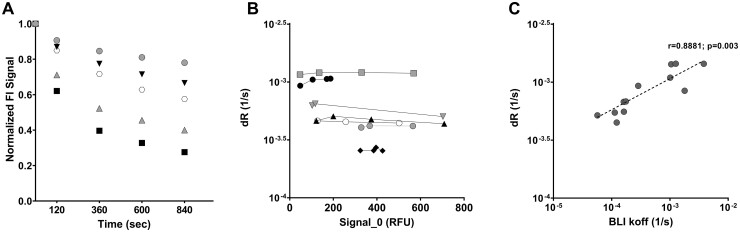
The dissociation profiles of 5 different antibodies over time are demonstrated in Fig. 3A. Antibodies of varying RBD binding strength (Supplemental Table 2) displayed different dRs and thus different dissociation profiles. The dRs were measured at proper antibody loading concentrations (0.06–30 μg/mL, varying for different antibody), and the avidity measurement was found to be independent of concentration (and, therefore, fluorescent signal) as long as the initial fluorescent signal (Signal_0) was in the proper range (Fig. 3B).
Fig. 3.
SARS-CoV-2 antibody avidity assay characterization with model COVID-19 purified antibodies. (A), Dissociation curve measurement of COVID-19–negative human serum spiked with 5 different COVID-19 antibodies. Signal_0/Signal_t is the normalized fluorescent signal on the y axis. (B), Relative dR measurement at varying levels of Signal_0. The COVID-19–negative human serum was spiked with 7 COVID-19 antibodies at different levels and measured. (C), Correlation between the SARS-CoV-2 avidity assay (dR measurement) and the BLI measurement (koff). Correlation between the 2 assays was assessed by the Spearman correlation coefficient.
Precision and interference.
The imprecision was determined by running the high and low levels of pooled patient samples (n = 5 to n = 10) 5 times per day on 5 different days. The imprecision of the TOP-Plus avidity assay was 7.5% and 9.8% at 2 dR levels of 7.23 ×10−4 1/s and 4.66 ×10−4 1/s, respectively. The stability of samples at 2–4 °C refrigerated conditions was at least 5 days (variation: <8%).
The TOP-Plus avidity assay was tested with common endogenous immunoassay interferences. Avidity of 2 SARS-CoV-2 model purified antibodies was measured in pooled SARS-CoV-2–negative serum, followed with spiking biotin, bilirubin, hemoglobin, or triglyceride and measuring antibody avidity in the presence of each potential interferent. The TOP-Plus avidity assay displayed no interference from the listed components up to the tested concentrations (Supplemental Table 3).
No cross-reactivity was displayed in sera from patients positive for HIV, Epstein–Barr virus, or rheumatoid factor. All 6 samples were negative (Supplemental Table 4) for TAb. Potential heterophilic antibody interference was further evaluated by performing spike-in experiments with a pooled human antimouse antibodies (HAMA) sample, which had 4 RFU (cutoff: 20 RFU), indicating no TAb assay interference by HAMA. There was no significant difference in values between the HAMA diluent and negative serum diluent, indicating that HAMA would not interfere with TAb and thus avidity measurements (Supplemental Table 5).
Correlation of TOP-Plus Avidity Assay to BLI
BLI is a well-established technique in avidity measurement (25, 26). To further validate the performance of the TOP-Plus avidity assay, Gator was used as a BLI reference method to measure the avidity of 12 purified COVID-19 antibodies. These values were compared with the avidity measured by the TOP-Plus avidity assay. It was found that the TOP-Plus avidity assay measurements correlated well (r = 0.88) with the Gator measurements (Fig. 3C).
Evaluation of Clinical Utility
TAb vs antibody avidity levels based on severity and persistence of symptoms.
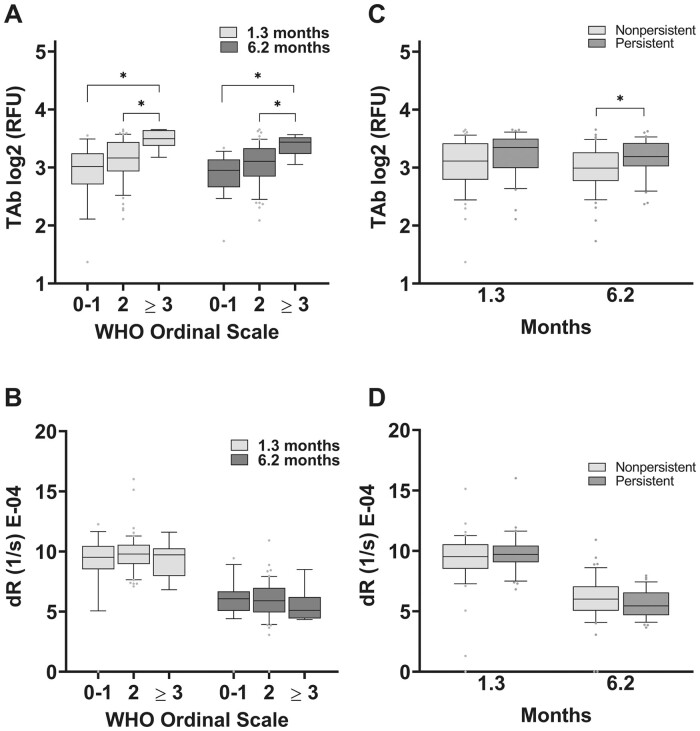
It had been previously shown that TAb levels were significantly lower in the outpatient population compared with the inpatient populations (23). For a better understanding of whether a similar relationship exists between TAb levels and severity of illness in this convalescent cohort, the individuals TAb levels were stratified by the severity of acute infection, as assessed by the WHO’s Ordinal Clinical Progression/Improvement Scale (27) (Supplemental Table 6). TAb levels were confirmed to be higher in individuals who had COVID-19 with limitations of their activities due to COVID-19 symptoms (WHO Ordinal Scale 2) and still higher in hospitalized patients (WHO Ordinal Scale ≥3). This was observed early in convalescence at 1.3 months and continued to 6.2 months after infection (Fig. 4A).
Fig. 4.
Antibody avidity is independent of prior COVID-19 severity (as determined by the WHO Ordinal Scale for Clinical Improvement) or persistence of symptoms. (A), TAb levels were higher in individuals with COVID-19 who had limitations in their activities due to COVID-19 symptoms (WHO Ordinal Scale 2) and higher still in hospitalized patients (WHO Ordinal Scale ≥ 3) at 1.3 and 6.2 months after infection. (B), At 1.3 and 6.2 months after infection, TAb levels were elevated in individuals who displayed persistence of symptoms beyond 6 weeks of symptom onset compared with those without persistence of symptoms. In contrast, antibody avidity remained unchanged across all WHO Ordinal Scales (C) or with persistence of COVID-19 symptoms (D). Persistent symptoms included fatigue, dyspnea, athletic deficit, or ≥3 solicited symptoms beyond 6 weeks of symptom onset.
Study participants had been asked about symptom persistence at their 6-month follow-up visit and were stratified retrospectively based on the responses (22). Persistent symptoms included fatigue, dyspnea, athletic deficit, or ≥3 solicited symptoms beyond 6 weeks of symptom onset. TAb levels were elevated in individuals who displayed persistence of symptoms beyond 6 weeks of symptom onset compared with those with no persistence of symptoms (Fig. 4B). Unlike TAb, antibody avidity remained unchanged across all WHO Ordinal Scales and in those with persistence of COVID-19 symptoms (Fig. 4, C and D).
SARS-CoV-2 antibody dynamics during early convalescence.
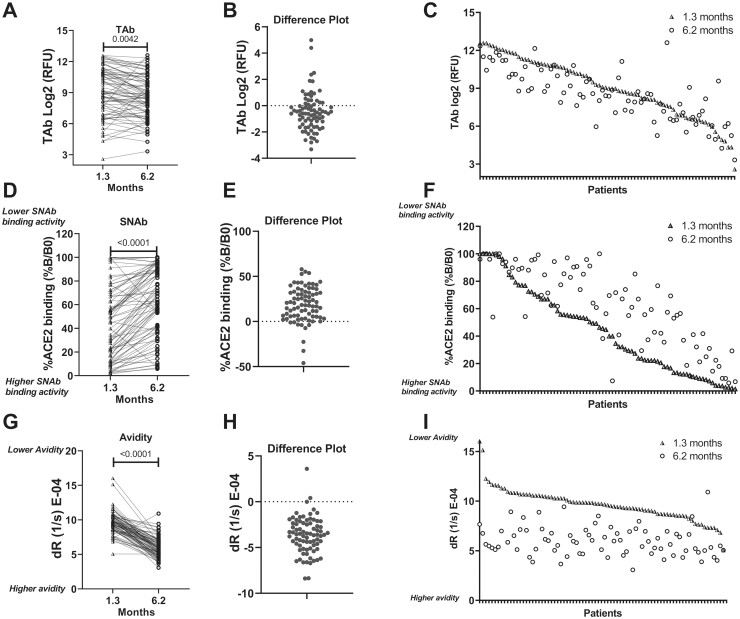
TAb was evaluated in 80 individuals with confirmed or suspected COVID-19 (21, 22) approximately 1.3 months and again approximately 6.2 months after the time that SARS-CoV-2 infection was first confirmed or suspected. TAb decreased over time in 58 of 80 individuals (Fig. 5C). The median TAb level at 1.3 months was 525 RFU (interquartile range [IQR]: 165.5–1943 RFU) compared with 380.5 RFU at 6.2 months (IQR: 136–1103; P = 0.0042) (Fig. 5, A and C). The decrease in TAb in this cohort mirrored the decreases in IgG and IgM levels that were measured using the same TOP biosensor (previously described in prior publications) (22, 24).
Fig. 5.
SARS-CoV-2 antibody dynamics during early convalescence. TAb, SNAb, and antibody avidity levels of 80 individuals who were positive for COVID-19 were plotted at 1.3 and 6.2 months (A, D, G). (B, E, H), The change in TAb, SNAb, and avidity over time (1.3 to 6.2 months). (C, F, I), Individual data pairs for TAb, SNAb, and avidity. TAb and SNAb activities (inversely associated with %B/B0) in the majority of individuals decreased over time. In contrast, the avidity (inversely associated with dR) increased during this time. The SNAb assay read-out is the percentage of RBD-ACE2 binding (%B/B0), which inversely correlates with the SNAb activity. The avidity assay measures the dR of SARS-CoV-2 antibodies from the RBD, which is inversely correlated with antibody avidity.
SNAb had been previously shown (23) to correlate well with both the plaque reduction neutralization test and the pseudovirus neutralization test, 2 well-established SARS-CoV-2 neutralization tests. In the SNAb assay, the percentage of RBD and angiotensin-converting enzyme 2 (ACE2) binding is defined as %B / B0 = (sample RFU / negative control RFU) × 100%. This current study found that the neutralization activity decreased over time in 69 of 80 individuals (Fig. 5F), as determined by the SNAb assay. The median percentage of ACE binding at 1.3 months was 42.32%B/B0 (IQR: 14.14–67.00) compared with 65.55%B/B0 (IQR: 38.85–89.71; P < 0.0001; Fig. 5, D and E). Together with the decrease in TAb, these results are indicative of not only overall SARS-CoV-S antibody levels diminishing over time but also a diminishment of the TAb neutralization activity.
In contrast, the antibody avidity increased in 76 of 80 individuals over this same time period (Fig. 5I), as indicated by a significant decrease in the median dR: 9.685 × 10−4/s at 1.3 months after infection to 5.830 × 10−4/s at 6.2 months after infection (P < 0.0001; Figs. 5G and 4H). This reflected a median increase of 3.85 × 10−4/s or 39.8%. (Fig. 5H).
Comparison of SARS-CoV-2 antibody avidity after vaccination to that of early convalescence.
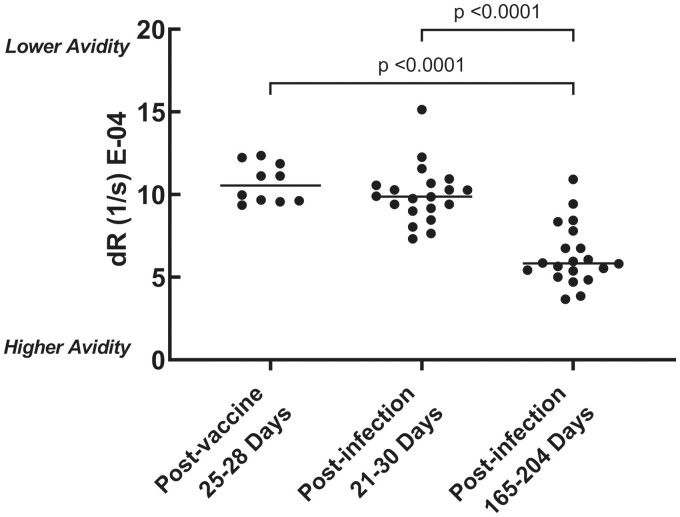
To demonstrate the avidity assay’s potential clinical utility in evaluating the antibody response to SARS-CoV-2 vaccination, antibody avidity was measured in individuals who were vaccinated with the mRNA-1273 vaccine approximately 1 month (25–28 days after vaccination; median: 28 days) after their first dose of the vaccine. These results were compared with those the 20 individuals in the previously described COVID-19–positive cohort that had specimen collected approximately 1 month after infection (21–30 days after symptom onset; median: 26 days). The antibody avidity levels of vaccinated individuals did not vary significantly from those of individuals with COVID-19 approximately 1 month after exposure (Fig. 6).
Fig. 6.
Comparison of antibody avidity in individuals previously infected compared with those who were vaccinated with the mRNA-1273 vaccine. Sera of 10 individuals, collected approximately 1 month (25–28 days after vaccination; median: 28 days) after their first dose of the mRNA-1273 vaccine, were analyzed for antibody avidity. These results were compared with those of the 20 individuals in the previously described COVID-19–positive cohort that had specimens collected approximately 1 month after infection (21–30 days after symptom onset; median: 26 days). For further comparison, the antibody avidity at approximately 6 months after infection in these 20 individuals who were COVID-19 positive were also displayed (165–204 days after vaccination; median: 183 days).
Discussion
Antibody avidity testing is not a new concept in the evaluation of an antibody response to infection or vaccination. Typically, antibodies generated early in a primary infection bind weakly to their respective antigen and exhibit low avidity or functional affinity (28). However, overall avidity toward an antigen increases as the response matures through somatic hypermutation, particularly of the variable loops of antigen-binding sites of B-cell receptors, and selective survival in the germinal center (29, 30). Because antibody avidity typically increases over time and is an indicator of a more mature antibody response, antibody avidity could be applied in assessing the efficacy of COVID-19 vaccination and immunity to SARS-CoV-2 and for screening donors for convalescent plasma antibody therapies.
Studies have tried to explain the SARS CoV-2 antibody response variability by focusing on antibody avidity (31). This study monitored avidity by measuring the relative dR of SARS-CoV-2–specific antibodies from RBD and compared it with TAb and SNAb, allowing for the assessment of the antibodies’ strength in binding to the virus. The dR inversely associates with the average antibody’s residence time at the epitope. Antibodies with lower dR values bind tightly to RBD and thus may be more efficient in clearing the virus and neutralizing infectivity (i.e., blocking entry into target cells) (32).
Antibody affinity reflects the rate constants of association and dissociation of an antibody with its target antigen [KD (M) = koff (1/s) / kon (1/Ms)]. In many serological applications, measurement of antibody–antigen interactions becomes a complicated process. Therefore, the most common approach is to disrupt the antibody–antigen binding by chaotropic agents (e.g., urea). The avidity is then assessed by measuring the change in the degree of release of antibody from the antigen by the chaotropic agent (17, 19). As a result, the assessed avidity of antibody depends on its resistance to the chaotropic agent and may not truly represent the avidity of antibody toward the antigen (13).
The TOP-Plus avidity assay presented measures the relative rate of dissociation of SARS-CoV-2 antibodies from the RBD antigen in plasma. However, this assay distinguishes itself from others in that it does not apply a chaotropic reagent. Therefore, the measured dR values better reflect the natural relative dR of antibodies from their target antigen than the conventional approaches in which chaotropes may alter the native structure of the antigen or antibody (13).
Previously no one assay could evaluate TAb levels, individual IgM and IgG levels, and avidity. The new TOP-Plus biosensor panel comprises 5 assays, allowing for TAb, SNAb, IgG, and IgM levels plus avidity testing on the same platform using the same biosensor principles with specific application applied for each assay. This probe was able to assess the overall decreasing trend in TAb and SNAb (Fig. 5) in addition to the previously reported decreases in IgG and IgM (22).
Our findings of the decay in total SARS-CoV-2 antibodies and neutralization antibody activities are consistent with previous studies (10, 16, 21, 33–37). However, SARS-CoV-2 antibody avidity did not show the same pattern of diminishment during the first 6 months of infection (Fig. 5). Our observation is congruent with the previous report (22) that memory B-cell responses continue to evolve and express antibodies with increased neutralizing potency and breadth. Therefore, the increased antibody avidity is indicative of continued evolution of the humoral response.
With the ongoing worldwide SARS-CoV-2 vaccination programs, such a panel could play a major role in monitoring the vaccination response in individuals and on a larger epidemiological scale. Because there are multiple dimensions in evaluating the humoral immune response to the SARS-CoV-2 vaccine, TOP-Plus has the potential to monitor adequate humoral immune response to the SARS-CoV-2 as a whole. Monitoring only for overall SARS-CoV-2 antibody levels or neutralization antibody levels could create a false impression of a diminishing immune response, whereas the TOP-Plus with its avidity assay may ensure appropriate immune response maturation. Indeed, our initial studies show that, at least in the early weeks after vaccination, vaccinated individuals display similar antibody avidity compared with those in a comparable period after infection, and it is hypothesized that the antibody avidity 6 months after vaccination will strengthen, as has been demonstrated after SARS-CoV-2 infection (Fig. 6). However, future studies would need to longitudinally follow vaccinated individuals to fully validate the assay for this purpose.
Different SARS-CoV-2 mutations raise concerns about the emergence of a more contagious or virulent variant. Therefore, there is a need for the development of assays that can properly characterize the humoral immune response to the SARS-CoV-2 vaccine and evaluate for immunity against these emerging virus variants. Although the current iteration of the TOP-Plus avidity assay measures the antibody avidity against the initially described SARS-CoV-2 RBD, the capability in measuring antibody avidity against other virus variants could be extended by replacing the RBD reagent of the assay (and probe) to the corresponding RBD of other virus variants.
The fact that only convalescent serum specimens were evaluated in this study is a limitation. The presented data cannot speak to the avidity maturation during the acute phase of infection and will require further studies to determine the full utility of the TOP-Plus avidity assay in patients who are acutely ill. Because the virus is newly evolved, it is expected that the antibody avidity for SARS-CoV-2 antigens during primary infection would be weak and that this avidity would increase over time. However, during the acute stages of infection, IgM could precede the IgG response, and it could be postulated that the overall avidity may display an initial spike during the acute stage of infection given the multimeric structure of the IgM antibody, masking the primary infection’s expected weaker avidity.
In conclusion, this TOP-Plus biosensor panel is a versatile sensing platform with high precision and an ability to measure SARS-CoV-2 TAb, SNAb, and individual IgG and IgM antibody levels along with the antibody’s long-term avidity. This combination of all-in-one testing will be a valuable asset in monitoring not only patients convalescing from COVID-19 but also the status of individuals’ COVID-19 vaccination response.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Acknowledgments
We thank all study participants who devoted time to our research; Drs. Barry Coller and Sarah Schlesinger, the Rockefeller University Hospital Clinical Research Support Office and nursing staff.
Nonstandard Abbreviations:
- COVID-19
Coronavirus disease 2019
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- BLI
bio-layer interferometry
- RBD
receptor-binding domain
- TOP-Plus
testing-on-a-probe “plus”
- TAb
total antibody
- SNAb
surrogate neutralizing antibody
- dR
dissociation rate
- RFU
relative fluorescence unit
- HAMA
human anti-mouse antibodies
- IQR
interquartile range
Author Declaration
A version of this paper was previously posted as a preprint on medrxiv.org (https://doi.org/10.1101/2021.02.03.21251089).
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
J. Yee and A. Sukhu performed the experiments. Y. Hao and S. Rand helped collect data. S.E. Racine-Brzostek wrote the manuscript, performed analysis, generated figures and aided in the review of the investigational findings. Z. Zhao oversaw the project, including conceptualization, interpretation of the data, statistical analysis and writing of the manuscript. H.S. Yang helped with editing the manuscript and interpretation of the data. A. Chadburn and M.M. Cushing helped review the manuscript. C. Gaebler collected specimens, analyze the patient data and review of the manuscript. R. Zuk oversaw the methodology for the TOP-Plus method development and conceptualization of the project. M. Karbaschi performed formal analysis for the TOP assay development, provided analytical data analysis and helped edit the manuscript. P.J. Klasse helped with editing the manuscript and performed a formal review. M. Caskey collected specimens, analyze the patient data and review of the manuscript. M.C. Nussenzweig collected specimens, analyze the patient data and review of the manuscript. Y. Shi helped with conceptualization of the project and design the experiments.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest: Employment or Leadership: M. Karbaschi, ET HealthCare Inc; R. Zuk, ET Healthcare Inc. Consultant or Advisory Role: M.M. Cushing, Octapharma Haemonetics. Stock Ownership: None declared. Honoraria: Z. Zhao, ET Healthcare Research Funding: Z. Zhao received seed instruments and research funding from ET Healthcare. Y. Hao was funded by China Scholarship Council. This work was supported by a COVID-19 research grant from Weill Cornell Medicine (M. Caskey), Weill Cornell Medicine Translational Research Program of the Department of Pathology and Laboratory Medicine at Weill Cornell Medicine (Z. Zhao). NIH grants P01 AI 110657 and R01 AI36082 (P.J. Klasse) and NIH grant P01-AI138398-S1 and 2U19AI111825 (M.C. Nussenzweig). C. Gaebler was supported by the Robert S. Wennett Post-Doctoral Fellowship, in part by the National Center for Advancing Translational Sciences (National Institutes of Health Clinical and Translational Science Award program, grant UL1 TR001866), and by the Shapiro-Silverberg Fund for the Advancement of Translational Research. M.C. Nussenzweig is a Howard Hughes Medical Institute Investigators. Expert Testimony: None declared. Patents: None declared. Role of Sponsor: The funding organizations played a direct role in the design of study, review and interpretation of data, preparation of manuscript, and final approval of manuscript. The funding organizations played no role in the choice of enrolled patients.
References
- 1.WHO. WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/?gclid=CjwKCAiAtK79BRAIEiwA4OskBtAbOw6T6wNgUS76dyazGbUP3AdVbHz7JZLTG4xKNRk_r3W9C9NQ-xoCRDoQAvD_BwE (Accessed April 9, 2021).
- 2.Sood N, Simon P, Ebner P, Eichner D, Reynolds J, Bendavid E, Bhattacharya J.. Seroprevalence of SARS-CoV-2-specific antibodies among adults in Los Angeles County, California, on April 10–11, 2020. JAMA 2020;323:2425–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havers FP, Reed C, Lim T, Montgomery JM, Klena JD, Hall AJ, et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23–May. JAMA Intern Med 2020;180:1576–86. [DOI] [PubMed] [Google Scholar]
- 4.Xie J, Ding C, Li J, Wang Y, Guo H, Lu Z, et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J Med Virol 2020;92:2004–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Zuiani A, Fischinger S, Mullur J, Atyeo C, Travers M, et al. Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production. Cell 2020;183:1496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K, Huang B, Wu M, Zhong A, Li L, Cai Y, et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun 2020;11:6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Y, Yan M, Wang L, Luan L, Liu J, Tian X, Wan N.. Analysis of the application value of serum antibody detection for staging of COVID-19 infection. J Med Virol 2021;93:899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel MM, Thornburg NJ, Stubblefield WB, Talbot HK, Coughlin MM, Feldstein LR, Self WH.. Change in antibodies to SARS-CoV-2 over 60 days among health care personnel in Nashville, Tennessee. JAMA 2020;324:1781–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020;26:1200–4. [DOI] [PubMed] [Google Scholar]
- 10.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020;370:1227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Long QX, Deng HJ, Hu J, Gao QZ, Zhang GJ, et al. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. [Epub ahead of print] Clin Infect Dis August 3, 2020. as doi:10.1093/cid/ciaa1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown TS, Walensky RP.. Serosurveillance and the COVID-19 epidemic in the US: undetected, uncertain, and out of control. JAMA 2020;324:749–51. [DOI] [PubMed] [Google Scholar]
- 13.Klasse PJ.How to assess the binding strength of antibodies elicited by vaccination against HIV and other viruses. Expert Rev Vaccines 2016;15:295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan PK, Lim PL, Liu EY, Cheung JL, Leung DT, Sung JJ.. Antibody avidity maturation during severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis 2005;192:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Victora GD, Nussenzweig MC.. Germinal centers. Annu Rev Immunol 2012;30:429–57. [DOI] [PubMed] [Google Scholar]
- 16.Luo YR, Chakraborty I, Yun C, Wu AHB, Lynch KL.. Kinetics of SARS-CoV-2 antibody avidity maturation and association with disease severity. [Epub ahead of print] Clin Infect Dis September 14, 2020. as doi:10.1093/cid/ciaa1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benner SE, Patel EU, Laeyendecker O, Pekosz A, Littlefield K, Eby Y, et al. SARS-CoV-2 antibody avidity responses in COVID-19 patients and convalescent plasma donors. J Infect Dis 2020;222:1974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valdivia A, Torres I, Huntley D, Alcaraz MJ, Albert E, Colomina J, et al. Qualitative assessment of SARS-CoV-2-specific antibody avidity by lateral flow immunochromatographic IgG/IgM antibody assay. J Med Virol 2021;93:1141–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T, Hsiung J, Zhao S, Kost J, Sreedhar D, Hanson CV, et al. Quantification of antibody avidities and accurate detection of SARS-CoV-2 antibodies in serum and saliva on plasmonic substrates. Nat Biomed Eng 2020;4:1188–96. [DOI] [PubMed] [Google Scholar]
- 20.Petersen RL.Strategies using bio-layer interferometry biosensor technology for vaccine research and development. Biosensors (Basel) 2017;7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020;584:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, et al. Evolution of antibody immunity to SARS-CoV-2. Nature 2021;591:639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang HS, Racine-Brzostek SE, Karbaschi M, Yee J, Dillard A, Steel PAD, et al. Testing-on-a-probe biosensors reveal association of early SARS-CoV-2 total antibodies and surrogate neutralizing antibodies with mortality in COVID-19 patients. Biosens Bioelectron 2021;178:113008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang HS, Racine-Brzostek SE, Lee WT, Hunt D, Yee J, Chen Z, et al. SARS-CoV-2 antibody characterization in emergency department, hospitalized and convalescent patients by two semi-quantitative immunoassays. Clin Chim Acta 2020;509:117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sultana A, Lee JE.. Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. Curr Protoc Protein Sci 2015;79:19.25.1–26. [DOI] [PubMed] [Google Scholar]
- 26.Kumaraswamy S, Tobias R.. Label-free kinetic analysis of an antibody-antigen interaction using biolayer interferometry. Methods Mol Biol 2015;1278:165–82. [DOI] [PubMed] [Google Scholar]
- 27.WHO. COVID-19 therapeutic trial synopsis. https://www.who.int/publications/i/item/covid-19-therapeutic-trial-synopsis (Accessed January 3, 2021).
- 28.Gutiérrez J, Maroto C.. Are IgG antibody avidity assays useful in the diagnosis of infectious diseases? A review. Microbios 1996;87:113–21. [PubMed] [Google Scholar]
- 29.Shlomchik MJ.Selection during antigen-driven B cell immune responses: the basis for high affinity antibody. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular biology of B cells. Burlington (MA: ): Academic Press; 2004. p. 339–48. [Google Scholar]
- 30.Merlo LMF, L Mandik-Nayak. Adaptive immunity. In: Prendergast GC, Jaffee EM, editors. Cancer immunotherapy. 2nd Ed. San Diego (CA: ): Academic Press; 2013. p. 25–40. [Google Scholar]
- 31.Bauer G.The variability of the serological response to SARS-corona virus-2: potential resolution of ambiguity through determination of avidity (functional affinity). J Med Virol 2021;93:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klasse PJ.Neutralization of virus infectivity by antibodies: old problems in new perspectives. Adv Biol 2014;2014:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muecksch F, Wise H, Batchelor B, Squires M, Semple E, Richardson C, et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis 2021;223:389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol 2020;5:1598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaudoin-Bussieres G, Laumaea A, Anand SP, Prevost J, Gasser R, Goyette G, et al. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio 2020;11:e02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crawford KHD, Dingens AS, Eguia R, Wolf CR, Wilcox N, Logue JK, et al. Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis 2021;223:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer AS, Jones FK, Nodoushani A, Kelly M, Becker M, Slater D, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol 2020;5:eabe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.