Abstract
Background
Classification and early detection of severe coronavirus disease 2019 (COVID-19) patients is required to establish an effective treatment. We tested the utility of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) to classify and predict the severity of COVID-19.
Methods
We used MALDI-TOF MS to analyze the serum peptidome from 72 patients with COVID-19 (training cohort), clinically classified as mild (28), severe (23), and critical (21), and 20 healthy controls. The resulting matrix of peak intensities was used for Machine Learning (ML) approaches to classify and predict COVID-19 severity of 22 independent patients (validation cohort). Finally, we analyzed all sera by liquid chromatography mass spectrometry (LC-MS/MS) to identify the most relevant proteins associated with disease severity.
Results
We found a clear variability of the serum peptidome profile depending on COVID-19 severity. Forty-two peaks exhibited a log fold change ≥1 and 17 were significantly different and at least 4-fold more intense in the set of critical patients than in the mild ones. The ML approach classified clinical stable patients according to their severity with 100% accuracy and correctly predicted the evolution of the nonstable patients in all cases. The LC-MS/MS identified 5 proteins that were significantly upregulated in the critical patients. They included the serum amyloid protein A2, which probably yielded the most intense peak detected by MALDI-TOF MS.
Conclusions
We demonstrate the potential of the MALDI-TOF MS as a bench to bedside technology to aid clinicians in their decision making regarding patients with COVID-19.
Keywords: COVID-19, machine learning, MALDI-TOF, serum peptidome
Our comparative analysis of serum from COVID-19 patients demonstrate the potential of the MALDI-TOF MS as a fast and clinically available technology to classify and predict COVID-19 severity in a clinical setting, helping clinicians to make decisions on COVID-19 patients.
Coronavirus infectious disease 2019 (COVID-19) was first reported in Wuhan, Hubei province, China as a new coronavirus disease caused by a positive-strand ribonucleic acid virus designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. This virus is responsible for a pandemic of unprecedented dimensions. Approximately 80% of COVID-19 cases are asymptomatic or present mild symptoms, such as fever, cough, fatigue, and dyspnea. In contrast, approximately 20% of patients with COVID-19 develop viral pneumonia, with an exaggerated host inflammatory response and hypoxia, requiring intubation and mechanical ventilation [2, 3]. These patients, classified as clinically severe or critical life-threatening infections, are mainly diagnosed empirically based on a set of clinical characteristics. However, patients with these symptoms have already evolved to a serious clinical condition that requires specialized intensive care. Therefore, it is essential to set up novel and rapid approaches to identify biomarkers for symptom onset and disease progression to facilitate triage of patients and establish appropriate treatments.
Peptidome-based studies using serum from patients and high-throughput spectrometric techniques promise to be valuable for the identification of COVID-19-associated biomarkers. Serum may contain proteins induced by the systemic effects or released to the lung as a result of the viral infection. Thus, patient serum can reflect the physiological or pathological state. Indeed, a proteomic and metabolomic analysis of serum from 46 patients with COVID-19 performed by Shen et al [4] demonstrated that by using serum proteins and metabolite biomarkers, it is possible not only to classify patients according to their grade of severity, but also to predict the progression to severe COVID-19. More recently, Messner et al [5] redesigned a high-throughput mass spectrometry platform that enabled the identification of up to 27 potential biomarkers that were differentially expressed depending on the severity grade of COVID-19. Although the technologies used in both studies are highly sensitive and provide robust results, they are time consuming, require specialized personnel, and, most importantly, they are not available in most of the hospitals, so their translation bench to bedside is limited.
In the present study, we used matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS), a simple and fast technology, available in most of the hospitals, to conduct a comparative analysis of serum from patients with COVID-19. Our results demonstrate the value and power of MALDI-TOF to classify and predict the progression of COVID-19 in a clinical setting.
METHODS
Patients Consent Statement
All human samples were taken after written consent of each participant. They were informed of the purposes of the study. The study was approved by the Institutional Review Board of Hospital Universitario Son Espases and the Regional Ethics Committee of Illes Balears.
Participants
This study included a total of 94 COVID-19 patients who attended Hospital Son Espases, the reference hospital of the Balearic Islands, between March 2020 and November 2020. The COVID-19 cases were confirmed based on the Chinese management guideline for COVID-19 [6]. Only patients who had a confirmed molecular diagnosis of SARS-CoV-2 ribonucleic acid polymerase chain reaction positive were enrolled.
The severity grade of COVID-19 was defined based on the above-mentioned guideline [6]. Accordingly, COVID-19 patients were classified into 3 subgroups: mild, severe, or critical. Mild included nonpneumonia and mild pneumonia cases. Severe was characterized by dyspnea, respiratory frequency ≥30/minute, blood oxygen saturation ≤93%, PaO2/FiO2 ratio <300, and/or lung infiltrates >50% within 24–48 hours. Critical cases were those that exhibited respiratory failure, septic shock, and/or multiple organ dysfunction/failure. Twenty healthy volunteers, including 13 who recovered from COVID-19, were also included in the study.
Sample Collection
Blood samples were collected into anticoagulant free-tubes. The tubes were centrifuged at 2500 rpm at 20ºC for 10 minutes within a 30-minute time frame. Serum from each patient sample was then collected, aliquoted, and stored at –80ºC. Each serum sample was heat inactivated at 56ºC for 90 minutes to inactivate the virus before analysis.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Serum Sample Preparation
The preparation of the serum and the analysis of the samples by MALDI-TOF MS were performed as previously described [7]. To obtain a better resolution and sensitivity in the MALDI-TOF analysis, serum samples were purified and concentrated using reversed-phase C18 tip, Pierce C18, following the manufacturer’s instructions. These are miniature reverse-phase columns packed into a 10-microliters pipet tips, with a micro-volume bed of reversed-phase medium fixed at its end, without dead volume. These tips are suitable for concentrating, desalting, and enriching protein/peptide samples before analysis. Samples are passed through activated reversed-phase C18 tips where the proteins/peptides are captured in the reverse-phase medium, salts are washed, and finally samples are eluted in a very small volume of solvent. In brief, 20 μL serum were mixed with 15 μL trifluoroacetic acid (TFA) 2%. Samples were passed through the Pierce C18 tip by pipetting up and down repeatedly (20 times), followed by the separation of the unbound protein solution. After washing the Pierce C18 tip with 10 μL of 0.1% TFA, the bound proteins/peptides were eluted with 4 μL of 0.1% TFA/CH3CN (1:1, v/v). The solution was passed through the Pierce C18 tip repetitively (8–10 times). The eluted protein/peptide solution was mixed with 4 μL a-cyano-4-hydroxycinnnamic acid (CHCA) matrix solution (10 mg of CHCA in 1 mL of 5% TFA/CH3CN, 1:1, v/v), and 1.5 μL of this mixture were spotted onto a MTP 384 target plate ground steel (Bruker Daltonics, Leipzig, Germany) and overlaid with 2 μL CHCA matrix and allowed to air dry. The analysis of each sample was conducted in triplicate.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight Analysis
Measurements were performed on an Autoflex III MALDI-TOF mass spectrometer (Bruker Daltonics) equipped with a 200 Hz smart beam laser using the MALDI Biotyper (MBT) Flex Control acquisition method (MBT_FC.pa), the same method used to identify microorganisms in many clinical microbiology laboratories. Spectra were generated by averaging 1000 single laser shots (100 shots at 10 different spot positions) at a laser frequency of 200 Hz and detected in linear positive mode. If the maximum intensity of the peaks was not greater than 5 × 103 (arbitrary units), more shots were recorded until reaching this minimum. The IS1 voltage was 20.1 kV, the IS2 voltage was maintained at 18.7 kV, the lens voltage was 8.4 kV, and the extraction delay time was 140 ns. Peaks between 2000 and 25 400 Da were selected for analysis. Mass accuracy was calibrated externally using the Bruker Bacterial Test Standard. Triplicates of each sample were obtained.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Data Processing
Raw mass spectra obtained by MALDI-TOF MS was analyzed using the MALDIquant R package [8]. Square root transformation, peak smoothing, baseline correction, and intensity normalization were performed on each mass spectrum. The average spectrum from the triplicates was obtained. Peaks were detected and binned across all average spectra with a signal-to-noise ratio of 5 and a tolerance of 0.002. Peaks presents in less than 15% of the spectra were rejected. All spectra from the groups under study were preprocessed, and peak detection was applied to obtain an intensity matrix. For every peak, the log fold-change intensity (logFC) between different groups was calculated using the median of the intensity for each group. Peaks with a logFC ≥1 were used for principal component analysis (PCA) [9] and Machine Learning (ML) analysis [10]. Five different ML algorithms were used to classify samples, namely, logistic regression, support vector machine with a linear kernel, naive Bayes, random forest, and decision tree. Results of testing the ML algorithms demonstrated that the models’ accuracies did not vary substantially among the ML methods (Supplementary Table 2); however, the Decision Tree algorithm was proven to be the best one. We decided to use the most frequent result obtained using all 5 ML algorithms to classify each sample.
Liquid Chromatography-Mass Sample Preparation
Three microliters of serum were diluted to 1 mL with 50 mM ammonium bicarbonate (0.2 µg/µL taking the average of plasma proteins as 80 mg/mL). One hundred microliters of the dilution were reduced with 11 µL of 50 mM dithiothreitol for 30 minutes at 56°C and were alkylated with 12.5 µL of iodoacetamide 20 mM for 20 minutes in the dark at 37°C. Total volume 123.5 µL, containing likely 24 µg of plasma protein, were digested with 10 µL of trypsin 100 ng/µL at 37°C overnight. Ten microliters of formic acid (FA) 5% were added to stop the digestion.
Liquid Chromatography-Mass Setup
The digested peptides were analyzed by liquid chromatography mass spectrometry with a nanoflow Agilent series 1200 LC system (Agilent Technologies, Waldbronn, Germany), with an autosampler equipped with an 8-µL capillary loop, coupled to a Q-Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) in data-dependent acquisition mode. For each acquisition, peptides were loaded onto a precolumn (ZORBAX 300 SB-C18, 5 µm, 5 mm *0.3 mm i.d.) at a flow rate of 15 µL/minute for 2 minutes and then analyzed using a 235-minute LC gradient (from 3% to 97% buffer B) at a flow rate of 250 nL/minute (analytical column, ZORBAX 300 SB-C18, 3.5 µm, 150 mm *0.075 mm i.d.). Buffer A was H2O containing FA 0.1%, and buffer B was acetonitrile with 0.1% FA. All reagents were MS grade. The m/z range of MS1 was 350–1650 with the resolution at 140 000 (at 200 m/z), automatic gain control (AGC) target of 3 × 106, and maximum ion injection time (max IT) of 250 ms. Top 10 precursors were selected for MS/MS experiment, with a resolution at 17 500 (at 200 m/z), AGC target of 5 × 104, and max IT of 200 ms. The isolation window of selected precursor was 4 m/z.
Liquid Chromatography-Mass Data Processing
The resultant mass spectrometric data were analyzed using Proteome Discoverer (version 2.2.0.388; Thermo Fisher Scientific) using a protein database composed of the Homo sapiens FASTA database downloaded from UniProtKB on 12 July 12, 2020, containing 20 304 reviewed protein sequences, and the SARS-CoV-2 virus FASTA downloaded from UniProtKB on May 20, 2020, containing 13 protein sequences. Enzyme was set to trypsin with 4 missed cleavage tolerance. Static modifications were set to carbamidomethylation (+57.02146) of cysteine, and variable modifications were set to oxidation (+15.99492) of methionine and dimethylation (+28.03075) of peptides N-termini. Precursor ion mass tolerance was set to 10 ppm, and product ion mass tolerance was set to 0.6 Da. The peptide-spectrum match allowed 1% target false-discovery rate (FDR) (strict) and 5% target FDR (relaxed). Normalization was performed against the total peptide amount. The other parameters followed the default setup.
RESULTS
We acquired MALDI mass spectra of 72 serum samples obtained from different COVID-19 patients: 28 samples were collected from mild COVID-19 patients, 23 were collected from severe patients, and 21 were collected from critical patients. To avoid the effect of the longitudinal immunological changes associated with this infection, we verified that patients did not change their clinical classification during at least 72 hours after the sample was taken. We also analyzed 20 serum samples obtained from healthy people, including 13 samples collected from individuals completely recovered, at least 1 month before, from COVID-19. The samples were randomized with respect to spotting and analysis. The relevant characteristics of each group are shown in the Supplementary Table 1.
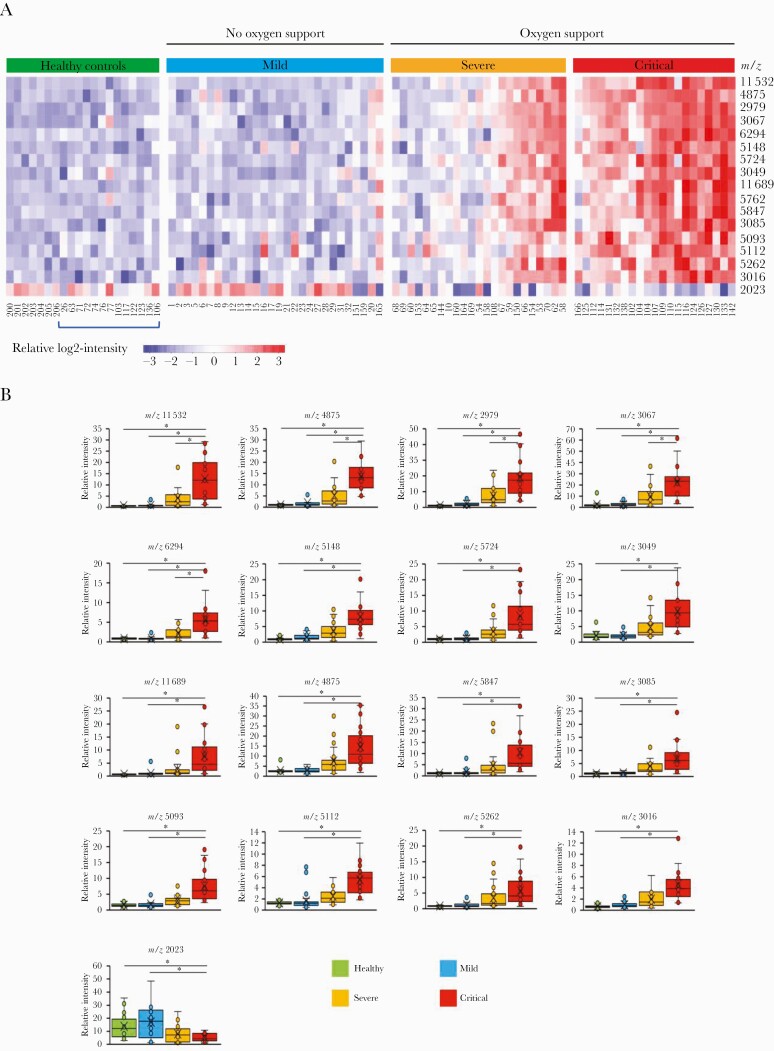
All spectra from the 4 groups under study were processed, and peak detection was applied to obtain an intensity matrix of 179 peaks in the mass range of 2000 to 25 000 daltons using MALDIquant. Representative mass spectra from each patient set are shown in Supplementary Figure S1. To select the most characteristic peaks distinguishing the groups classified according to COVID-19 severity, we calculated the median intensity of each peak for each group. Figure 1A illustrates the quantitative variability of those peaks that exhibited a log fold change ≥2 for COVID-19 severity on a heatmap. We found clear differences between critical and mild patients. Sixteen peaks were significantly different and at least 4-fold more intense in the set of critical patients than in the mild ones (Figure 1B). However, only 5 of those peaks (m/z; 11 532, 4875, 2979, 3067, and 6294) were significantly different between severe and critical patients.
Figure 1.
Matrix-assisted laser desorption ionization (MALDI) mass spectra of serum indicate clinical severity in coronavirus disease 2019 (COVID-19). (A) The heatmap illustrates peptidome profiles that inform on COVID-19 severity. The heatmap was generated using the ComplexHeatmap package [8] using those significantly different peaks based on unpaired 2 tailed t test (P < .05) and a log fold change ≥2 for COVID-19 severity. Groups were classified according to COVID-19 severity following the Chinese management guideline for COVID-19. Blue bracket below heatmap indicates healthy individuals who recovered from COVID-19. (B) Relative intensity of MALDI mass spectra peaks with major differences between groups. The boxes show the first and third quartiles as well as the median (middle), the mean (cross), and the outliers (circles outside the whiskers) of the relative intensity of the peaks that exhibited a log fold change ≥2 for COVID-19 severity. Asterisks indicate statistical significance based on unpaired 2-tailed t test (P < .05).
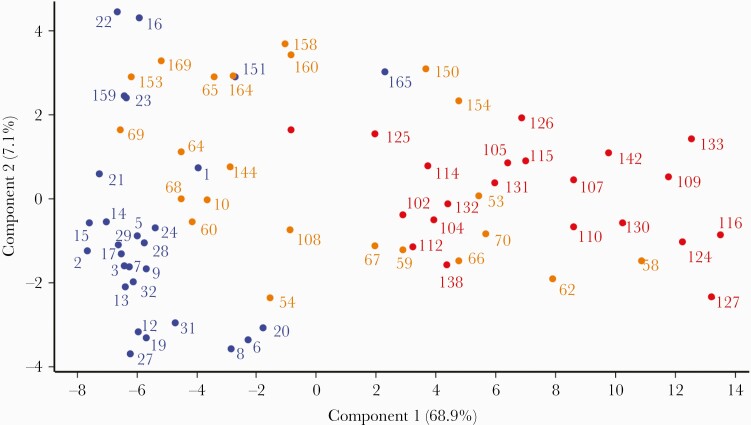
We then used PCA to compare samples from patients with different severity grade in a multidimensional space using the 42 peaks (m/z) that exhibited a log fold change ≥1 obtained from each sample by MALDI-TOF MS (Figure 2). All critical patients (Figure 2, red dots) were clearly separated from the mild patients (Figure 2, blue dots), except for the outlier sample 165. However, samples from severe patients were not separated from either mild or critical patients. In fact, samples from severe patients were distributed almost equally between the group of samples from mild and critical patients.
Figure 2.
Principal component analysis of the mass spectra of the serum samples from 72 coronavirus disease 2019 (COVID-19) patients analyzed by matrix-assisted laser desorption ionization time-of-flight. Groups were classified according to COVID-19 severity following the Chinese management guideline for COVID-19: mild (blue dots), severe (orange dots), and critical (red dots) patients. Only peaks with a log fold change ≥1 between groups were used for the analysis.
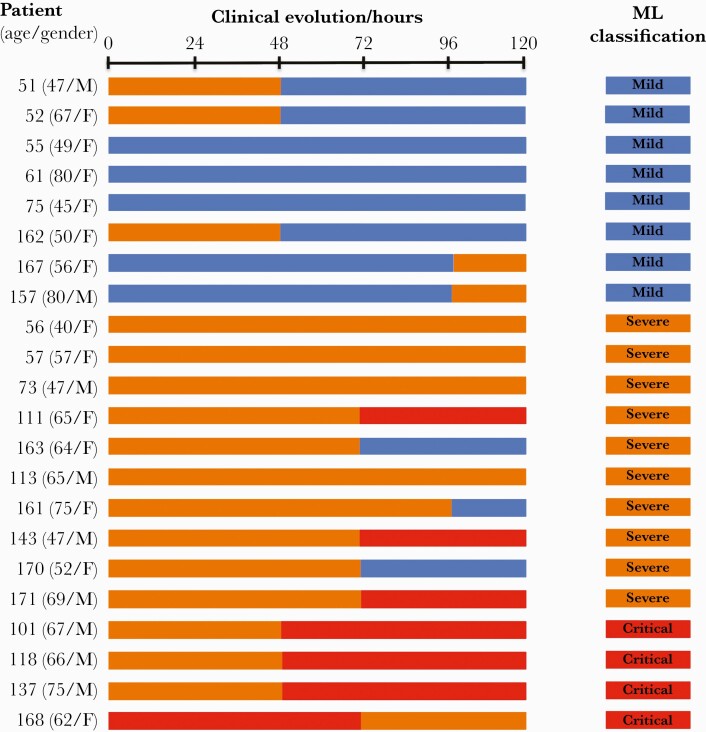
Next, we applied an ML approach to classify and predict COVID-19 severity. We built a support vector ML model using the same peaks (m/z) used in the PCA. Samples from the 72 patients were classified as mild, severe, and critical according to the Chinese guide and used as training cohort of the ML. The model was validated using a cohort of 22 independent patients (Figure 3).
Figure 3.
Coronavirus disease 2019 (COVID-19) patients and Machine Learning (ML) results. Clinical classification and ML results of COVID-19 patients used to validate the ML model setup with the training cohort described in Supplementary Table 1. Samples analyzed by matrix-assisted laser desorption ionization time-of-flight were collected at time 0. Groups were classified according to COVID-19 severity following the Chinese management guideline for COVID-19: mild (blue boxes), severe (orange boxes), and critical (red boxes) patients.
Case studies demonstrated the clinical utility of the peptidome profiles to classify or predict COVID-19 evolution (Figure 3). Thus, all patients who remained clinically stable during at least 72 hours (55, 56, 57, 61, 75, 73, 111, 113, 143, 157, 161, 163, 167, 168, 170, and 171) were correctly classified using ML. Three samples from patients classified as clinically severe (101, 118, and 137) were clustered in the group of critical patients using ML 48 hours before they clinically progressed from severe to critical. On the other hand, samples from patients 51, 52, and 162, classified as clinically severe (grade 2) when the samples were collected, which 48 hours later evolved to mild (grade 1), were clearly clustered in the group of mild patients using ML.
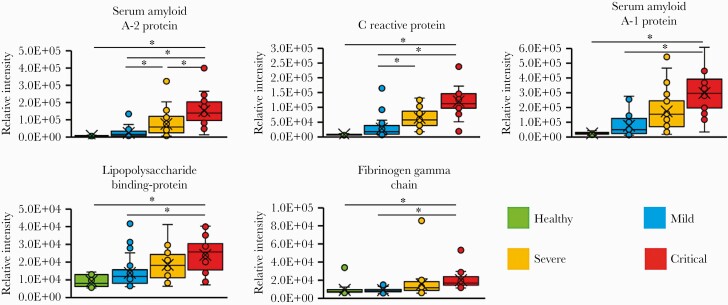
Most of the discriminating peaks identified by MALDI-TOF MS analysis had a low molecular weight (<5000 Da). They probably resulted from the fragmentation of proteins upregulated in severe and critical patients. It is interesting to note that the most substantial intensity difference was exhibited by the peak with m/z of 11 532, which might correspond to an unfragmented protein of the acute phase induced by the virus. To investigate this hypothesis, we performed a proteomic analysis of the samples by LC-MS/MS. We identified 5 proteins that were significantly upregulated according to the severity of the disease; the serum amyloid A2 protein (SAA2), the C-reactive protein (CRP), the serum amyloid protein A1 (SAA1), the lipopolysaccharide binding protein (LBP), and the gamma chain of the fibrinogen (FGG) (Figure 4 and Supplementary Figure 2). Only the serum level of SAA2 exhibited significant increments between mild and severe patients and between severe and critical patients. In addition, SAA1, CRP, LBP, and FGG were increased in the serum from the critical patients compared to mild patients, whereas only CRP was increased in the severe patients compare to mild patients. Given that the molecular weight of SAA2 is approximately 11.7 kDa, depending on the isoform [11], and that we found a good correlation between the level of both proteins and the intensity of the peak with m/z of 11 532, we suggest that this peak might correspond to the serum amyloid protein A2.
Figure 4.
Upregulated proteins according to coronavirus disease 2019 (COVID-19) severity. Groups were classified according to COVID-19 severity following the Chinese management guideline for COVID-19. Only proteins that were present in more than 70% of the samples identified by at least 5 peptides and significantly increased (P < .05) by a log-2-fold change >2 were included in the figure. The boxes show the first and third quartiles as well as the median (middle), the mean (cross), and the outliers (circles outside the whiskers).
DISCUSSION
In this study, we demonstrate that the molecular changes that occur in the sera of COVID-19 patients may be detected by MALDI-TOF MS analysis generating peptidome profiles that may be used as clinical classifiers. In addition, we show that it is possible to predict the progression of the disease using the peptidome signatures obtained with this technology. Finally, we provide strong evidence that serum amyloid A2 protein is one of the major biomarkers of severe COVID-19 disease.
To our knowledge, only 2 previous studies reported the use of mass spectrometry analysis of serum from COVID-19 patients to classify disease severity [4, 5]. However, both studies were performed using sophisticated technologies, which are not available in most of the hospitals. Our challenge was to test whether MALDI-TOF MS analysis, a simpler technology available in most of the clinical microbiology laboratories for identification of microbial species, was able to achieve similar results.
Our peptidome profile data identify the most important changes within the severe patients, upon which a patient is put on oxygen supply. This observation is consistent with the proteome analysis conducted by Messner et al [5], who found that at the molecular level the requirement of oxygen supply coincided with the progression to severe disease. In contrast, mild patients have a peptidome signature virtually identical to the healthy controls, which suggests that in nonsevere patients, changes are restricted to the site of infection, the respiratory tract, without significant molecular systemic alterations. As in previous studies [4, 5], major differences between critical and mild patients were due to the presence of upregulated proteins in the sera of the critical patients rather than the presence of downregulated proteins. Thus, among the peaks that exhibited major differences, only 1 peak (m/z 2023) was 4-fold more intense in the mild patients than in the critical patients.
The ML approach and analysis of clinical data demonstrated the clinical utility of the peptidome profiles to classify and predict COVID-19 evolution. Our study classified clinically stable patients with a high accuracy (100%), even higher to that obtained in a previous report (93%) [4]. In addition, the evolution of 100% of the patients was correctly predicted 48 hours before the clinical change. Overall, these results suggest that this technology is quite accurate to classify patients and to predict their prognostic. It would be interesting to conduct a longitudinal study with sequential daily samples from a cohort of patients at different grades of severity until their recovery to determine with more confidence the anticipation time of prediction.
One of the potential limitations of our study is that due to the rapid response required in the initial stages of the pandemic situation, we collected samples from the patients who were admitted to our hospital using as unique criteria that they were hospitalized due to a SARS-CoV-2 infection. Therefore, our study did not take into account some confounding factors, such as age. Nonetheless, the change of the intensity of the peaks between groups substantially exceeded the variability observed within each group with ages ranging from 33 to 89, suggesting that differences in the peptidomes profiles of different groups are poorly influenced by confounding factors.
The results of our comparative analysis of serum from COVID-19 patients with different grades of severity demonstrate the potential of the MALDI-TOF MS as a fast and clinically available technology to classify and predict the progression of this infectious disease in a clinical setting. Our workflow has a total hands-on time of less than 5 minute per sample from the inactivation of the serum samples, collected following standard procedures, to the acquisition of the mass spectra with the MALDI-TOF mass spectrometer. A single person can perform all of the procedures because the workflow is designed to reduce the pipetting to only 2 steps, mitigating variability. A key step concerns the cleanup and concentration of the samples using the reversed-phase C18 tips, which improves the resolution and sensitivity of the assay. One target plate with 96 positions can contain up to 30 serum samples per triplicate plus calibrating standards and can be ready for the mass spectrometric analysis in 2 hours. Next, a data acquisition scheme based on the MALDI Biotyper commonly used in the clinical microbiology laboratories for microbial identification is used. Therefore, this system could be implemented in most of the hospitals without major problems. However, if the available mass spectrometer does not have this acquisition system, the method should be optimized from a positive linear method available using the Bruker Bacterial Test Standard (BTS) calibrant. Finally, raw mass spectra obtained by MALDI-TOF MS can be analyzed using packages (MALDIquant R) [12] and ML algorithms that are free and available on the internet (https://scikit-learn.org/stable/ and https://pandas.pydata.org/). The inclusion of sample preparation controls on each plate enables cross-batch normalization, to correct batch effects in case these emerge at the sample preparation or the acquisition step of the workflow.
CONCLUSIONS
The laboratory can give clinicians the results of the mass spectra analysis in less than 3 hours. These results may support some clinical decisions including patient triage and early identification of patients at high risk of disease progression and severe illness to establish an effective treatment to avert progression to more serious illness, with the additional benefit of reducing the burden on healthcare systems. Furthermore, the data provided by the peptidome analysis could be useful to monitor the efficacy of the treatments and to manage the hospital resources, particularly, intensive care unit stays, which is particular important during the pandemic waves.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Sara Fernández for helpful discussions on statistical analysis.
Financial support. This study was funded by Instituto de Investigación Sanitaria de las Islas Baleares.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2019; 323:1061–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 2020; 182:59–72.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Messner CB, Demichev V, Wendisch D, et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst 2020; 11:11–24.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)—China, 2020. China CDC Wkly 2020; 2:113–22. [PMC free article] [PubMed] [Google Scholar]
- 7. Barceló F, Gomila R, de Paul I, et al. MALDI-TOF analysis of blood serum proteome can predict the presence of monoclonal gammopathy of undetermined significance. PLoS One 2018; 13:e0201793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. RStudio Team. RStudio: Integrated Development Environment for R [computer program]. Boston, MA: RStudio Team; 2015. [Google Scholar]
- 9. Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom Intell Lab Syst 1987; 2:37–52. [Google Scholar]
- 10. Larrañaga P, Calvo B, Santana R, et al. Machine learning in bioinformatics. Brief Bioinform 2006; 7:86–112. [DOI] [PubMed] [Google Scholar]
- 11. Li J, Xie Z, Shi L, et al. Purification, identification and profiling of serum amyloid A proteins from sera of advanced-stage cancer patients. J Chromatogr B Analyt Technol Biomed Life Sci 2012; 15:889–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibb S, Strimmer K. MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics 2012; 28:2270–1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.