Abstract
Background
Patients hospitalized with coronavirus disease 2019 (COVID-19) are at increased risk of health care–associated infections (HAIs), especially with prolonged hospital stays. We sought to identify incidence, antimicrobial susceptibilities, and outcomes associated with bacterial/fungal secondary infections in a large cohort of patients with COVID-19.
Methods
We evaluated adult patients diagnosed with COVID-19 between 2 March and 31 May 2020 and hospitalized >24 hours. Data extracted from medical records included diagnoses, vital signs, laboratory results, microbiological data, and antibiotic use. Microbiologically confirmed bacterial and fungal pathogens from clinical cultures were evaluated to characterize community- and health care–associated infections, including describing temporal changes in predominant organisms on presentation and throughout hospitalization. Univariable and multivariable logistic regression analyses were performed to investigate risk factors for HAIs.
Results
A total of 3028 patients were included and accounted for 899 positive clinical cultures. Overall, 516 (17%) patients with positive cultures met criteria for infection. Community-associated coinfections were identified in 183 (6%) patients, whereas HAIs occurred in 350 (12%) patients. Fifty-seven percent of HAIs were caused by gram-negative bacteria and 19% by fungi. Antibiotic resistance increased with longer hospital stays, with incremental increases in the proportion of vancomycin resistance among enterococci and ceftriaxone and carbapenem resistance among Enterobacterales. Intensive care unit stay, invasive mechanical ventilation, and steroids were associated with HAIs.
Conclusions
HAIs occur in a small proportion of patients hospitalized with COVID-19 and are most often caused by gram-negative and fungal pathogens. Antibiotic resistance is more prevalent with prolonged hospital stays. Antimicrobial stewardship is imperative in this population to minimize unnecessary broad-spectrum antibiotic use.
Keywords: bacterial infections, COVID-19, health care–associated infection
We comprehensively studied bacterial/fungal infections in 3028 COVID-19 patients. We identified salient temporal changes in predominant organisms and resistance over the course of patients’ hospitalizations. Independent predictors of health care–associated infection were intensive care unit stay, invasive mechanical ventilation, and steroids.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been accompanied by the widespread use of empiric broad-spectrum antimicrobial agents attributed to liberalization of antibiotic stewardship approaches in a time of clinical uncertainty and significant strain on health care systems [1, 2]. Development of bacterial or fungal superinfections are well-established risk factors for poor outcomes in patients with influenza pneumonia [3], such as intensive care unit (ICU) admission and mortality, and have also been reported in patients with respiratory syndromes due to other coronaviruses, including severe acute respiratory syndrome and Middle East respiratory syndrome [4–7]. However, the incidence and identity of bacteria and fungi causing secondary infections in patients with coronavirus disease 2019 (COVID-19) remain unclear.
In early reports from China, secondary infections were identified in up to 50% of patients who died from COVID-19, compared to only 15% of patients who did not [8]. However, the proportion of patients receiving empiric antibiotics has been shown to greatly exceed those with culture-proven infections. In a recent retrospective review of published studies (n = 806 patients), bacterial and fungal coinfections were reported in only 8% of patients, whereas >70% of patients received empiric antibacterial therapy [2]. Similarly, in a large meta-analysis of nearly 4000 hospitalized patients with COVID-19, predominantly from China, only 7% were found to have bacterial coinfections, which increased to 14% in critically ill patients [9].
Little is known about the characteristics of secondary infections in patients with COVID-19 [10]. Many published studies fail to adequately describe the timing of identified infections, making it difficult to distinguish between community-associated coinfection or health care–associated infection (HAI). In addition, few prior studies describe causative organisms and antibiotic susceptibilities, and follow-up time is often limited. The impact of immunomodulatory agents, such as corticosteroids and interleukin 6 (IL-6) receptor blockers, on the incidence of secondary infections is unknown, despite the widespread use of these agents to manage inflammatory complications of COVID-19 [11, 12].
Here we aimed to systematically characterize community- and health care–associated bacterial and fungal infections in hospitalized patients with COVID-19, including temporal changes in predominant organisms on presentation and throughout the hospital course. We further investigated clinical risk factors for HAIs.
MATERIALS AND METHODS
Setting and Study Design
This was a retrospective cohort study conducted at a quaternary medical center in New York City, which includes a large academic medical center and a small community hospital, both located in northern Manhattan. Patients were included if they were 18 years or older, presented to the emergency department between 2 March 2020 and 31 May 2020, tested positive for SARS-CoV-2 by reverse-transcription polymerase chain reaction from a nasopharyngeal swab sample, and were hospitalized >24 hours. We extracted clinical and demographic information from the patients’ electronic medical records. This included admission diagnoses, vital signs, laboratory results, microbiological data, antibiotic use data, and discharge dates. International Classification of Diseases (Ninth or Tenth Revision) codes were used to identify comorbidities for calculating the Charlson Comorbidity Index Score.
During the study period, patients with COVID-19 were managed according to internal guidelines, which underwent several updates during the study period to reflect the rapid evolution of available evidence and experience. Initially, this included the use of hydroxychloroquine with or without azithromycin in patients with severe disease. Additionally, patients with evidence of acute respiratory distress syndrome or other inflammatory complications were considered for receipt of low-dose methylprednisolone (0.5 mg/kg/dose every 12 hours) and/or the IL-6 receptor blocker tocilizumab. Patients meeting respective criteria were approached for enrollment in randomized clinical trials studying sarilumab (an alternative IL-6 receptor blocker), remdesivir, or convalescent plasma.
Patient Consent Statement
The Columbia University institutional review board approved this study and waived the need for informed consent.
Microbiological Data and Definitions of Infection
Culture and susceptibility results from all body sources were extracted from patients’ electronic medical records. Only bacterial and fungal organisms were considered. For each unique organism, only the first isolate collected at a given body site per patient was included in the analysis. Surveillance cultures from nares and rectal swabs and autopsy cultures were excluded. Community-associated coinfections were defined as those in which the positive culture was isolated in the first 72 hours of hospitalization or within 5 days prior to admission from an outpatient or emergency department visit. HAIs, in which the positive culture was obtained after hospital day 3, were further subdivided into infections detected on days 4–14, 15–28, or after hospital day 28.
Bacteremia was defined by a positive blood culture for any organism other than coagulase-negative staphylococci. Criteria for bacteremia due to coagulase-negative staphylococci included positive blood cultures for at least 2 consecutive days and receipt of an anti-staphylococcal antibiotic; if these criteria were not met, the blood culture was regarded as contaminated. All respiratory isolates except Candida and Enterococcus species were considered to indicate the presence of pneumonia, as protocols at our institution recommend culturing only in the setting of worsening symptoms or fever. Ventilator-associated pneumonia was defined by organism isolation from a respiratory source >2 calendar days after intubation. Organisms isolated from urinary cultures were challenging to evaluate for infection, given the difficulty of assessing symptoms in this critically ill patient population. All urinary isolates with an associated urinalysis with >10 white blood cells per high-power field were considered to constitute a urinary tract infection (UTI).
All drug susceptibilities were interpreted according to criteria outlined by the Clinical and Laboratory Standards Institute. To categorize drug susceptibilities and identify multidrug-resistant (MDR) organisms, we applied the following definitions: Staphylococcus aureus was considered methicillin resistant (MRSA) if the isolate was resistant to oxacillin; Enterococcus species resistant to vancomycin were categorized as vancomycin-resistant enterococci (VRE); MDR Enterobacterales were defined by resistance to ceftriaxone (Ceph-R) and carbapenem-resistant Enterobacterales (CRE) isolates were identified by a meropenem minimum inhibitory concentration (MIC) of ≥2 µg/mL; and carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii isolates were defined by a meropenem MIC of ≥4 µg/mL.
Statistical Analysis
Specific clinical parameters were described either as proportions or using the median and interquartile range (IQR), as appropriate. In univariable analyses, potential predictors of any infection vs no infection were compared between groups using the Fisher exact test or χ 2 for categorial variables, or unpaired t test or Mann-Whitney U test for continuous variables. Continuous variables were tested for normality utilizing the Kolmogorov-Smirnov test. We also assessed risk factors for development of HAI in adjusted and unadjusted logistic regression analyses. Variables that had a P value of < .1 in unadjusted analyses were included in multivariable logistic regression models assessing independent predictors of HAI. Candidate variables were removed from the full model if coefficient P values were nonsignificant and removal improved model fit, which was assessed by comparing pseudo R2 values. Data were analyzed using IBM SPSS Statistics for Windows, version 25.0 (IBM Corporation, Armonk, New York).
RESULTS
A total of 3028 adult patients confirmed to have COVID-19 were admitted for >24 hours during the study period. Among these, 899 positive clinical cultures were identified in 516 patients. Thus, 516 (17%) patients with positive cultures met criteria for infection. The characteristics of patients with COVID-19 with or without secondary infections are presented in Table 1.
Table 1.
Patient Characteristics With and Without Any Identified Infection in Patients Presenting to the Hospital With Coronavirus Disease 2019
| Characteristica | Total (N = 3028) | No Infection (n = 2512) | Infection (n = 516) | P Value |
|---|---|---|---|---|
| Age, y, median (IQR) | 64 (50–76) | 64 (48–76) | 67 (57–75) | <.001 |
| Male sex | 1623 (54) | 1333 (53) | 290 (56) | .210 |
| BMI, kg/m2, median (IQR) | 27.8 (24.2–32.6) | 28.0 (24.4–32.7) | 27.0 (23.4–32.3) | .027 |
| Comorbidities | ||||
| Hypertension | 1788 (59) | 1462 (58) | 326 (63) | .041 |
| Diabetes mellitus | 1165 (39) | 937 (37) | 228 (44) | .004 |
| Chronic kidney disease | 429 (14) | 349 (14) | 80 (16) | .375 |
| Liver disease | 166 (6) | 132 (5) | 34 (7) | .268 |
| Asthma | 400 (13) | 337 (13) | 63 (12) | .506 |
| COPD | 228 (8) | 169 (7) | 59 (11) | <.001 |
| HIV | 96 (3) | 83 (3) | 13 (3) | .430 |
| Solid organ transplant | 100 (3) | 80 (3) | 20 (4) | .506 |
| Days from symptom onset to hospital admission, median (IQR) | 6 (3–9) | 6 (3–10) | 5 (3–8) | .004 |
| Initial oxygen requirement | <.001 | |||
| None (room air) | 1927 (64) | 1681 (67) | 246 (48) | |
| Nasal cannula | 588 (19) | 110 (21) | 478 (19) | |
| Non-rebreather mask | 449 (15) | 321 (13) | 128 (25) | |
| Noninvasive ventilation | 8 (0.3) | 4 (0.2) | 4 (1) | |
| Invasive mechanical ventilation | 56 (2) | 28 (1) | 28 (5) | |
| Initial COVID-19 admission length of hospital stay, d, median (IQR) | 6 (3–11) | 6 (3–9) | 16 (7–41) | <.001 |
| Highest level of care | <.001 | |||
| Intensive care unit | 622 (21) | 318 (13) | 304 (59) | |
| General floor | 2279 (75) | 2070 (82) | 209 (41) | |
| Admitted and discharged from ED location | 127 (4) | 124 (5) | 3 (1) | |
| Invasive mechanical ventilation | 500 (17) | 221 (9) | 279 (54) | <.001 |
| Laboratory values on hospital presentation, median (IQR) | ||||
| C-reactive protein, mg/L | 114.0 (54.1–198.8) n = 2511 |
108.9 (49.4–187.8) n = 2024 |
146.9 (76.8–249.2) n = 487 |
<.001 |
| ESR, mm/h | 70 (48–97) n = 2358 |
69 (47–95) n = 1884 |
75 (51–101) n = 474 |
.002 |
| Ferritin, ng/mL | 672 (326–1256) n = 2480 |
653 (316–1209) n = 1998 |
781 (375–1549) n = 482 |
<.001 |
| Procalcitonin, ng/mL | 0.24 (0.11–0.62) n = 2503 |
0.21 (0.11–0.56) n = 2015 |
0.35 (0.16–1.07) n = 488 |
<.001 |
| D-dimer, μg/mL | 1.5 (0.8–3.3) n = 2265 |
1.4 (0.8–3.1) n = 1806 |
2.1 (1.0–4.7) n = 459 |
<.001 |
| Interleukin 6, pg/mL | 21.0 (7.3–49.9) n = 1946 |
17.4 (6.0–41.5) n = 1521 |
37.9 (14.2–92.5) n = 425 |
<.001 |
| Lactate dehydrogenase, U/L | 403 (294–564) n = 2480 |
391 (286–537) n = 1999 |
455 (341–679) n = 481 |
<.001 |
| Serum creatinine, mg/dL | 1.05 (0.79–1.63) n = 2852 |
1.04 (0.78–1.58) n = 2339 |
1.17 (0.83–1.90) n = 513 |
<.001 |
| WBC count, cells × 109/L | 7.5 (5.6–10.3) n = 2914 |
7.3 (5.5–9.8) n = 2401 |
8.6 (5.9–12.6) n = 513 |
<.001 |
| Neutrophils, % | 77 (68–84) n = 2660 |
76 (67–83) n = 2162 |
81 (73–86) n = 498 |
<.001 |
| Maximum laboratory values during hospitalization, median (IQR) | ||||
| C-reactive protein, mg/L | 161.4 (77.0–271.3) n = 2511 |
144.7 (66.8–237.5) n = 2024 |
274.7 (146.7–300.0) n = 487 |
<.001 |
| ESR, mm/h | 88 (60–117) n = 2357 |
82 (57–111) n = 1883 |
111 (82–130) n = 474 |
<.001 |
| Ferritin, ng/mL | 876 (392–1810) n = 2480 |
806 (371–1545) n = 1998 |
1403 (607–2909) n = 482 |
<.001 |
| Procalcitonin, ng/mL | 0.38 (0.14–1.72) n = 2503 |
0.30 (0.12–1.09) n = 2015 |
1.78 (0.49–8.93) n = 488 |
<.001 |
| D-dimer, μg/mL | 2.5 (1.0–9.5) n = 2265 |
1.8 (0.9–5.2) n = 1806 |
9.8 (3.1–20.0) n = 459 |
<.001 |
| Interleukin 6, pg/mL | 30.8 (10.0–94.2) n = 1946 |
23.5 (8.0–61.0) n = 1521 |
117.9 (37.4–157.5) n = 425 |
<.001 |
| Lactate dehydrogenase, U/L | 462 (331–675) n = 2480 |
435 (317–613) n = 1999 |
632 (421–925) n = 481 |
<.001 |
| Serum creatinine, mg/dL | 1.26 (0.89–2.51) n = 2852 |
1.16 (0.85–2.08) n = 2339 |
2.29 (1.24–5.35) n = 513 |
<.001 |
| WBC count, cells × 109/L | 10.8 (7.5–16.1) n = 2914 |
9.8 (7.1–13.8) n = 2401 |
19.2 (13.2–26.2) n = 513 |
<.001 |
| Neutrophils, % | 82 (73–88) n = 2660 |
80 (72–87) n = 2162 |
87 (81–91) n = 498 |
<.001 |
| COVID-19–specific therapy received | ||||
| Hydroxychloroquine | 1427 (47) | 1112 (44) | 315 (61) | <.001 |
| Remdesivir | 78 (3) | 53 (2) | 25 (5) | .001 |
| Tocilizumab | 166 (6) | 97 (4) | 69 (13) | <.001 |
| Steroids | 707 (23) | 477 (19) | 230 (47) | <.001 |
| Antibiotics received (any time) | ||||
| Any antibiotic | 2015 (67) | 1518 (60) | 497 (96) | <.001 |
| Anti-MRSA agentsb | 858 (28) | 490 (20) | 368 (71) | <.001 |
| Broad spectrum anti-Pseudomonas agentsc | 1141 (38) | 721 (29) | 420 (81) | <.001 |
| Hospital mortality | 647 (21) | 479 (19) | 168 (33) | <.001 |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: BMI, body mass index; COPD, chronic obstructive pulmonary disease; COVID-19, coronavirus disease 2019; ED, emergency department; ESR, erythrocyte sedimentation rate; HIV, human immunodeficiency virus; IQR, interquartile range; MRSA, methicillin-resistant Staphylococcus aureus; WBC, white blood cell.
aAll medians for continuous variables compared between groups using Mann-Whitney U test.
bAnti-MRSA agents: vancomycin (intravenous), linezolid, daptomycin, ceftaroline.
cBroad-spectrum anti-Pseudomonas agents: aminoglycosides, piperacillin/tazobactam, ceftazidime, cefepime, ceftolozane/tazobactam, ceftazidime/avibactam, imipenem/cilastatin, meropenem, meropenem/vaborbactam, aztreonam, levofloxacin, polymyxin B.
Community-Associated Coinfections
A total of 221 cultures from 183 patients (6%) were identified as coinfections within 72 hours of hospital admission. The majority were isolated from urine (112/221 [51%]), followed by blood (69/221 [31%]) and respiratory tract (26/221 [12%]). Fourteen isolates (6%) were identified from other sources, including 3 each from peritoneal fluid and wounds. The most common organisms identified were Escherichia coli (68/221 [31%]), S. aureus (25/221 [11%]), Proteus mirabilis (18/221 [8%]), and Klebsiella pneumoniae (17/221 [8%]). Escherichia coli accounted for 58 of 112 (52%) isolates from the urinary tract, whereas S. aureus was most frequently isolated from the respiratory tract (13/26 [50%]). Staphylococcus aureus and E. coli each accounted for 9 of 69 (13%) cases of community-associated bacteremias.
Health Care-Associated Infections
Health care–associated infections were identified in 350 (12%) patients, with the median onset of infection on hospital day 16 (IQR, 9–25). Among the 678 isolates identified as health care-associated, 385 (57%) were gram-negative bacteria, 167 (25%) gram-positive bacteria, and 126 (19%) fungi. The frequency of organisms and time of isolation are described in Table 2.
Table 2.
Description of Blood, Respiratory, and Urine Isolates by Organism Group, Hospital Day Onset After Admission, and Resistance Profile
| Source/Organism Group | Community-Acquired | Hospital Day 4–14 | Hospital Day 15–28 | Hospital Day >28 | Total |
|---|---|---|---|---|---|
| Blood isolates, No. (%) | |||||
| Total No. | 69 | 29 | 47 | 57 | 202 |
| Enterobacterales | 22 (32) | 4 (14) | 11 (23) | 19 (33) | 56 (28) |
| Ceftriaxone-resistant | 5/22 | 2/4 | 5/11 | 11/19 | 23/56 |
| Carbapenem-resistant | 0 | 0 | 0 | 4/19 | 4/56 |
| Staphylococcus species | 12 (17) | 10 (35) | 11 (23) | 7 (12) | 40 (20) |
| Methicillin-resistant | 1/12 | 1/10 | 2/11 | 2/7 | 6/40 |
| Candida species | 1 (1) | 6 (21) | 10 (21) | 12 (21) | 29 (14) |
| Fluconazole-resistant | 0 | 0 | 0 | 1/12 | 1/29 |
| Streptococcus species | 16 (23) | 5 (17) | 1 (2) | 2 (4) | 24 (12) |
| Enterococcus species | 4 (6) | 3 (10) | 5 (11) | 9 (16) | 21 (10) |
| Vancomycin-resistant | 1/4 | 2/3 | 2/5 | 2/9 | 7/21 |
| Miscellaneous gram-positives | 10 (15) | 0 (0) | 1 (2) | 1 (2) | 12 (6) |
| Pseudomonas species | 1 (1) | 0 (0) | 5 (11) | 4 (7) | 10 (5) |
| Acinetobacter species | 0 (0) | 0 (0) | 2 (4) | 1 (2) | 3 (2) |
| Gram-positive anaerobes | 1 (1) | 0 (0) | 1 (2) | 1 (2) | 3 (2) |
| Yeast, not otherwise specified | 0 (0) | 0 (0) | 0 (0) | 1 (2) | 1 (1) |
| Gram-negative anaerobes | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| Burkholderia species | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 1 (1) |
| Miscellaneous gram-negatives | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 1 (1) |
| Respiratory isolates, No. (%) | |||||
| Total No. | 26 | 99 | 114 | 85 | 324 |
| Enterobacterales | 3 (12) | 40 (40) | 59 (52) | 40 (47) | 142 (44) |
| Ceftriaxone-resistant | 1/3 | 18/40 | 31/59 | 22/40 | 72/142 |
| Carbapenem-resistant | 0 | 1/40 | 3/59 | 10/40 | 14/142 |
| Staphylococcus species | 13 (50) | 35 (35) | 22 (19) | 11 (13) | 81 (25) |
| Methicillin-resistant | 5/13 | 10/35 | 11/22 | 3/11 | 29/81 |
| Pseudomonas species | 6 (23) | 9 (9) | 23 (20) | 27 (32) | 65 (20) |
| Stenotrophomonas species | 0 (0) | 5 (5) | 4 (4) | 2 (2) | 11 (3) |
| Aspergillus species | 1 (4) | 2 (2) | 1 (1) | 1 (1) | 5 (2) |
| Streptococcus species | 1 (4) | 4 (4) | 0 (0) | 0 (0) | 5 (2) |
| Acinetobacter species | 1 (4) | 2 (2) | 1 (1) | 1 (1) | 5 (2) |
| Miscellaneous gram-positives | 0 (0) | 2 (2) | 1 (1) | 2 (2) | 5 (2) |
| Miscellaneous gram-negatives | 1 (4) | 0 (0) | 1 (1) | 1 (1) | 3 (1) |
| Scedosporium species | 0 (0) | 0 (0) | 1 (1) | 0 (0) | 1 (0.3) |
| Burkholderia species | 0 (0) | 0 (0) | 1 (1) | 0 (0) | 1 (0.3) |
| Urine isolates, No. (%) | |||||
| Total No. | 112 | 61 | 71 | 84 | 328 |
| Enterobacterales | 86 (77) | 18 (30) | 25 (35) | 48 (57) | 177 (54) |
| Ceftriaxone-resistant | 23/86 | 4/18 | 11/25 | 20/48 | 58/177 |
| Carbapenem-resistant | 1/86 | 0/18 | 1/25 | 5/48 | 7/177 |
| Candida species | 6 (5) | 33 (54) | 34 (48) | 17 (20) | 90 (27) |
| Fluconazole-resistant | NA | NA | NA | NA | NA |
| Enterococcus species | 9 (8) | 6 (10) | 2 (3) | 9 (11) | 26 (8) |
| Vancomycin-resistant | 2/9 | 1/6 | 1/2 | 5/9 | 9/26 |
| Pseudomonas species | 4 (4) | 2 (3) | 8 (11) | 7 (8) | 21 (6) |
| Staphylococcus species | 4 (4) | 1 (2) | 1 (1) | 0 (0) | 6 (2) |
| Methicillin-resistant | 0 | 0 | 0 | 0 | 0 |
| Yeast, not otherwise specified | 0 (0) | 0 (0) | 1 (1) | 3 (4) | 4 (1) |
| Streptococcus species | 1 (1) | 1 (2) | 0 (0) | 0 (0) | 2 (0.6) |
| Miscellaneous gram-positives | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 1 (0.3) |
| Burkholderia species | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 1 (0.3) |
Abbreviation: NA, not applicable.
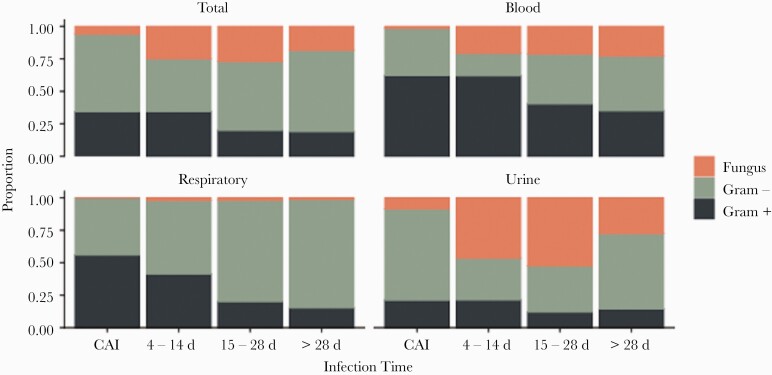
A total of 94 patients with COVID-19 accounted for 133 organisms isolated from blood cultures. The median time from hospital admission to first health care–associated bacteremia was 23 days (IQR, 15–37 days). Gram-positive organisms accounted for 42% of bacteremias, followed by gram-negative organisms (35%) and fungi (22%). All but 1 fungal pathogens isolated from blood cultures were Candida species. Overall, Enterobacterales was the most common organism group isolated from blood cultures (26%). However, bacteremias caused by staphylococci and streptococci were more frequent earlier in the hospital stay (Figure 1). They accounted for 15 of 29 (52%) bacteremias occurring within the first 14 days, compared to 12 of 47 (26%) occurring during hospital days 15–28 and 9 of 57 (16%) beyond hospital day 28. Enterobacterales were the most common cause of bacteremias occurring beyond hospital day 28 (19/57 [33%]).
Figure 1.
Proportion of organism group by source over the duration of hospital days. Abbreviation: CAI, community-associated infection.
There were 207 patients with 298 health care–associated respiratory isolates identified during the study period. Respiratory isolates were classified as hospital acquired in 17% and ventilator associated in 83%. Enterobacterales, staphylococci, and Pseudomonas species were the most common organism groups and accounted for 89% of respiratory pathogens (Table 2). Staphylococci were more commonly (35%) isolated early (within the first 14 days). Beyond day 14 of hospitalization, Enterobacterales and Pseudomonas species predominated, comprising 75% of respiratory isolates.
Urine isolates accounted for 216 health care–associated isolates, and consisted predominantly of Enterobacterales and enterococci (81%). An additional 31 isolates were identified from sites other than the blood, respiratory tract, or urine, including 17 (55%) from unspecified wound cultures. The majority of these isolates were collected beyond hospital day 28 (65%).
Antibiotic Use and Resistance
Among the entire cohort of 3028 patients, 2015 (67%) had exposure to at least 1 dose of an antibiotic. This includes 2512 patients without a microbiologically confirmed bacterial or fungal infection, of whom 1518 (60%) had any antibiotic exposure. Among those with HAIs, 340 (97%) had exposure to antibiotics. The most commonly used antibiotics prior to infection in patients who went on to develop an HAI included broad-spectrum penicillins (65%), cephalosporins (59%), anti-staphylococcal agents with activity against MRSA (56%), and tetracyclines (26%). Exposure to carbapenems (15%) and quinolones (5%) was less common.
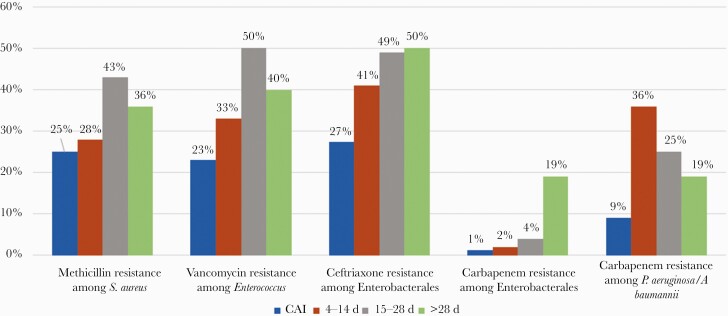
Drug susceptibilities differed by source of infection and duration of hospitalization, with the proportion of VRE, Ceph-R Enterobacterales, and CRE infections increasing with duration of hospitalization (Figure 2). MRSA was identified in 38 of 116 (33%) S. aureus isolates, all but 9 of which were isolated from the respiratory tract (76%). VRE accounted for 18 of 50 (36%) enterococci. Seven VRE bacteremias and 9 VRE urine isolates were identified, including 3 causing community-acquired infections. Cephalosporin-resistant Enterobacterales was the most common organism group with multidrug resistance, identified in 159 isolates. Ceph-R Enterobacterales accounted for a larger proportion of infections occurring on hospital days 15–28 (49%) and after hospital day 28 (50%), compared to community-acquired infections (27%) and infections on hospital days 4–14 (41%). Moreover, while the urine was the most common source of Ceph-R Enterobacterales community–associated infections (23/31 [74%]), the respiratory tract was the most common source of infection thereafter (71/128 [55%]). We identified 27 CRE isolates, most of which occurred after day 28 of hospitalization (78%) and were isolated from the respiratory tract (52%) [13]. Four bacteremias caused by CRE were identified and all 4 occurred after day 28 of hospitalization. Carbapenem-resistant Pseudomonas and Acinetobacter species were infrequent, and also consisted largely of respiratory tract infections occurring beyond day 14 of hospitalization.
Figure 2.
Frequency of resistant pathogens over hospital length of stay. Abbreviation: CAI, community-associated infection.
Risk Factors for Health Care-Associated Infection
Patients with HAI were significantly older and more likely to be male. These patients were also more likely to require ICU care, require invasive mechanical ventilation, receive COVID-19–specific therapies, and have more exposure to broad-spectrum antibiotics prior to infection (Table 3). In a multivariable model, requiring ICU care (odds ratio [OR], 4.140 [95% {CI}, 2.534–6.764]; P < .001), requiring invasive mechanical ventilation (OR, 6.044 [95% CI, 3.667–9.961]; P < .001), or receiving steroids (OR, 1.910 [95% CI, 1.419–2.571]; P < .001) was independently associated with development of an HAI. A post hoc sensitivity analysis logistic regression was performed focusing only on health care–associated bacteremia and respiratory infections as the dependent variable. The same variables (ICU care, invasive mechanical ventilation, and steroids) plus male sex and not receiving an anti-MRSA antibiotic were independently associated with HAI (Supplementary Table 1).
Table 3.
Unadjusted and Adjusted Analysis for Health Care–Associated Infection
| Characteristic | Unadjusted OR (95% CI) |
P Value | Adjusted OR (95% CI) |
P Value |
|---|---|---|---|---|
| Age, y | 1.006 (1.000–1.012) | .049 | 1.008 (.998–1.017) | .109 |
| Male sex | 1.436 (1.143–1.805) | .002 | 0.982 (.741–1.301) | .897 |
| BMI, kg/m2, each unit | 1.000 (.999–1.000) | .553 | … | |
| Comorbidities | ||||
| Hypertension | 1.237 (.982–1.558) | .071 | 1.204 (.888–1.634) | .232 |
| Diabetes mellitus | 1.174 (.936–1.473) | .165 | … | |
| Chronic kidney disease | 0.974 (.705–1.347) | .874 | … | |
| Liver disease | 1.093 (.682–1.752) | .713 | … | |
| Asthma | 0.791 (.557–1.124) | .191 | … | |
| COPD | 1.364 (.925–2.011) | .117 | … | |
| HIV | 0.774 (.386–1.553) | .471 | … | |
| Solid organ transplant | 1.145 (.632–2.076) | .654 | … | |
| Days from symptom onset to hospital admission, each day | 0.981 (.961–1.001) | .060 | … | |
| Initial oxygen requirement, each unit | 1.723 (1.547–1.918) | <.001 | … | |
| Nasal cannula (1 vs 0) | 1.016 (.767–1.345) | .914 | … | |
| Non-rebreather mask (2 vs 0) | 2.723 (2.096–3.537) | <.001 | 0.909 (.645–1.283) | .588 |
| Noninvasive ventilation (3 vs 0) | 7.529 (1.874–30.241) | .004 | 2.610 (.523–13.016) | .242 |
| Invasive mechanical ventilation (4 vs 0) | 4.881 (2.759–8.633) | <.001 | 0.690 (.365–1.303) | .252 |
| ICU level of care | 19.771 (15.147–25.806) | <.001 | 4.140 (2.534–6.764) | <.001 |
| Invasive mechanical ventilation prior to HAI | 17.521 (13.570–22.621) | <.001 | 6.044 (3.667–9.961) | <.001 |
| Laboratory values on hospital presentation, each unit | ||||
| C-reactive protein, mg/L | 1.004 (1.003–1.005) | <.001 | … | |
| ESR, mm/h | 1.005 (1.001–1.008) | <.001 | … | |
| Ferritin, ng/mL | 1.000 (1.000–1.000) | .038 | … | |
| Procalcitonin, ng/mL | 1.002 (.996–1.007) | .541 | … | |
| D-dimer, μg/mL | 1.001 (.997–1.006) | .622 | … | |
| Interleukin 6, pg/mL | 1.010 (1.008–1.012) | <.001 | … | |
| Lactate dehydrogenase, U/L | 1.001 (1.001–1.001) | <.001 | … | |
| Serum creatinine, mg/dL | 1.027 (.981–1.076) | .250 | … | |
| WBC count, cells × 109/L | 1.019 (1.004–1.035) | .015 | … | |
| Neutrophils, % | 1.035 (1.023–1.047) | <.001 | … | |
| COVID-19–specific therapy received | ||||
| Hydroxychloroquine | 0.330 (.259, .421) | <.001 | 1.142 (.840–1.553) | .398 |
| Remdesivir | 3.266 (1.981–5.386) | <.001 | 1.198 (.633–2.266) | .579 |
| Tocilizumab | 5.722 (4.092–8.003) | <.001 | 1.305 (.854–1.996) | .219 |
| Steroids | 5.014 (3.977–6.321) | <.001 | 1.910 (1.419–2.571) | <.001 |
| Antibiotics received prior to HAI | ||||
| Any antibiotic | 3.482 (2.566–4.723) | <.001 | … | |
| Anti-MRSA agentsa | 4.842 (3.843–6.102) | <.001 | 0.745 (.491–1.132) | .168 |
| Broad-spectrum anti-Pseudomonas agentsb | 5.271 (4.135–6.720) | <.001 | 1.162 (.754–1.793) | .496 |
Patients who died within 3 days of admission were excluded (n = 2959).
Abbreviations: BMI, body mass index; CI, confidence interval; COPD, chronic obstructive pulmonary disease; COVID-19, coronavirus disease 2019; ESR, erythrocyte sedimentation rate; HAI, health care–associated infection; HIV, human immunodeficiency virus; ICU, intensive care unit; MRSA, methicillin-resistant Staphylococcus aureus; OR, odds ratio; WBC, white blood cell.
aAnti-MRSA agents: vancomycin (intravenous), linezolid, daptomycin, ceftaroline.
bBroad-spectrum anti-Pseudomonas agents: aminoglycosides, piperacillin/tazobactam, ceftazidime, cefepime, ceftolozane/tazobactam, ceftazidime/avibactam, imipenem/cilastatin, meropenem, meropenem/vaborbactam, aztreonam, levofloxacin, polymyxin B.
DISCUSSION
Among this large cohort of COVID-19 patients, we detected notable temporal changes in the frequency of infection types and antimicrobial resistance patterns from the time of hospitalization to >28 days after admission. Only 6% presented with a community-associated bacterial or fungal coinfection, consisting largely of gram-negative UTIs. Microbiologically confirmed HAIs occurred in 12% of the patients over the course of the hospital stay. Gram-negative organisms accounted for 58% of all infections and increased in frequency with longer hospital stay, while staphylococci accounted for the majority of gram-positive organisms (57%). We also detected a relatively large proportion of fungal infections (15%), consisting primarily of hospital-associated Candida species. Importantly, the proportion of resistant isolates increased with duration of hospital stay, particularly with incremental increases in the proportion of vancomycin resistance among enterococci and ceftriaxone resistance and carbapenem resistance among Enterobacterales.
Previous studies have reported variable rates of both community- and hospital-associated secondary infections in adult patients of 0–17% and 0–43%, respectively [14–34]. Differences in the patient population, culture sources, and pathogens of interest are likely responsible for the wide variations reported. The 2 largest studies assessing for coinfection at the time of hospital admission reported incidence between 1% and 3% but included detection of viral respiratory pathogens and Mycoplasma [28, 32]. Among studies reporting the proportion of patients with hospital-associated infections, disease severity and follow-up time also varied. For example, 2 studies of ICU patients reported notably different secondary infections rates, with 1 study reporting that 37% of patients developed secondary bacteremia and the other study detecting hospital-associated bacterial or fungal infection in 14% of patients [17, 19]. Similarly, a review of patients over the age of 60 in which 54% were still hospitalized at day 28 reported 43% of patients developing secondary infection; however, the characteristics of these infections were not described [27].
While a relatively large proportion of patients in this study developed secondary infections (17%), we and others found that this was greatly exceeded by the proportion of patients receiving antibiotics, which reached 90% in some studies [33, 34]. In previous studies, antimicrobials were initiated upon initial presentation, despite the relatively low rates of bacterial coinfections reported. Our study documented antibiotic exposure at any point during the hospitalization in 60% of patients. High rates of antimicrobial use in conjunction with other risk factors, such as prolonged hospital and ICU stays, invasive devices, and the need for patient cohorting, may have contributed to the increasing incidence of MDR infections over the course of hospitalizations. MRSA was infrequent among S. aureus isolates within the first 14 days of hospitalization. Similarly, VRE was not common in the initial 14 days with both MRSA and VRE increasing to >40% beyond 14 days. This suggests that broad-spectrum gram-positive agents may not be necessary early on in the hospital course and therefore are potential targets for antimicrobial stewardship programs. Among gram-negative organisms, Ceph-R Enterobacterales were common urinary isolates early and likely reflect extended-spectrum β-lactamase–producing organisms present in the community. Of concern is the isolation of CRE among Enterobacterales at 4.2% during hospital days 15–28 and 19% beyond day 28. Population structure and mechanisms of resistance among carbapenemase-producing Enterobacterales in COVID-19 patients were previously reported from our institution [13]. The current study provides more clinical context on the true incidence of these pathogens over time. Two previous studies identified CRE [19, 29]. In the larger study, CRE was identified in 5% of respiratory and bloodstream isolates; however, the median length of stay was 13 days (IQR, 6–21 days) and only 28% of patients were still admitted at the time of analysis [29].
We found that Candida species accounted for 17% of all HAIs and consisted predominantly (72%) of UTIs. Increased use of antibiotics out of proportion to the number of microbiologically confirmed infections may have contributed to colonization and infection caused by Candida species in this population and deserves further exploration. However, while previous studies have raised concerns about fungal infections in patients with COVID-19 [18, 28, 29], the incidence remains low compared to other hospitalized patient populations [35]. Moreover, we likely overreported Candida species isolated from the urinary tract as we were unable to evaluate for symptomatic infections or the presence of urinary catheters. In this cohort, identification of Aspergillus species was highly uncommon, occurring in only 4 respiratory cultures. This number is much lower than recent reports identifying Aspergillus infections in up to 19% of COVID-19 ICU patients [36].
Predictors of HAIs identified were largely consistent with known risk factors for infections in hospitalized patients. We further identified a significant association between the use of steroids and development of an HAI. Although this association persisted after adjusting for other factors such as need for ICU admission and invasive mechanical ventilation, this finding may reflect use of immunomodulatory agents in patients with more severe COVID-19 or with prolonged hospitalizations. The scope of this study did not allow us to assess steroid dose or duration or the frequency of other opportunistic infections. Tocilizumab, an IL-6 receptor blocker, has been associated with increased risk of serious infections compared to other immunomodulatory agents in patients with rheumatoid arthritis [37]. In a study from Italy evaluating patients with COVID-19 treated with tocilizumab, 13% of patients in the treatment arm developed infections, compared to 4% who received standard of care without tocilizumab [38]. In the United States, superinfections were significantly higher in patients treated with tocilizumab (54%) than in controls (26%) [39]. We did not find tocilizumab to be independently associated with HAIs.
The strengths of this study include evaluation of both community-associated and health care–associated secondary infections, extended duration of follow-up, inclusion of all hospitalized patients, and evaluation for antimicrobial resistance. However, the study also has several limitations. Given the rapid influx of patients, ICU units were not clearly delineated and as such ICU admission was likely underreported. UTIs with pyuria were not evaluated in the context of urinary symptoms. Cultures were usually obtained in the setting of fever or presumed sepsis. While we may be overestimating pneumonias in ventilated patients, positive respiratory cultures were difficult to ignore in this setting and were clinically considered ventilator-associated pneumonias requiring treatment. While it is standard practice to obtain cultures prior to the initiation of antibiotics or before changing antibiotics, the appropriateness of this timing was not evaluated and could result in underreporting of infection rates. While more infections occurred in patients who received steroids, an extensive risk factor analysis in these patients was not completed as it was not the goal of this analysis. Antibiotic use was common, but we did not evaluate antibiotic duration or its relationship to the development of antibiotic resistance. We also could not determine the impact of efforts to limit health care worker patient exposure with the frequency of obtaining appropriate cultures. Finally, New York City was an early hot spot in the United States and the rapid influx of patients strained the health care system. As such, the generalizability of these results may be limited outside of a pandemic environment and each institution’s susceptibility patterns may differ, but they provide insight into the likelihood of bacterial and fungal infections and the need for broad-spectrum antibiotic use.
CONCLUSIONS
In our quaternary care hospital in New York City, 17% of patients with COVID-19 developed secondary bacterial or fungal infections during the peak of the pandemic. HAIs were not common, but the overwhelming majority were caused by gram-negative pathogens. Ceftriaxone-resistant and, more concerningly, carbapenem-resistant gram-negative organisms were increasingly isolated in patients with prolonged hospital stays. After adjusting for other factors, invasive mechanical ventilation, ICU care, and use of steroids were independent predictors of HAIs in patients hospitalized with COVID-19. Antimicrobial stewardship principles are of utmost importance early and throughout the hospitalization in these patients, and antimicrobials must be limited to suspected or confirmed bacterial infections. Additional studies are needed to evaluate the long-term consequences of steroid use, our antimicrobial decision-making, and ways to optimize antimicrobial use in patients hospitalized with COVID-19.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We would like to acknowledge the data in the COVID-CARE database based at New York-Presbyterian/Columbia University Irving Medical Center, Division of Infectious Diseases.
Author contributions. C. J. K., T. H. M., J. Z., B. N., A. G. S., and A. C. U. contributed to study conception, design, and analysis. J. Z. and M. M. contributed to data extraction. D. D. and T. H. M. performed necessary electronic medical record review. T. H. M. developed the initial draft of the manuscript. C. J. K., T. H. M., D. D., J. Z., B. N., M. M., E. I., L. B., J. S. S., M. S., A. G. S., and A. C. U. contributed to drafting, editing, and finalizing the manuscript. M. S., J. Z., and A. C. U. provided study supervision. All authors read and approved the final manuscript.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, NIH (grant numbers 5UM1AI069470-14 and supplement to M. E. S. and J. Z.; K23AI150378 and L30AI133789 to J. Z.; K08AI146284 to T. H. M.; and K23AI137316 to A. G. S.) and the Centers for Disease Control and Prevention (grant number U01CK000592-S1 to A. C. U. and J. Z.).
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Cummings MJ, Baldwin MR, Abrams D, et al. . Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet 2020; 395:1763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rawson TM, Moore LSP, Zhu N, et al. . Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis 2020; 71:2459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P, Liu Z, Chen Y, et al. . Bacterial and fungal infections in COVID-19 patients: a matter of concern. Infect Control Hosp Epidemiol 2020; 41: 1124–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arabi YM, Al-Omari A, Mandourah Y, et al. . Saudi Critical Care Trial Group . Critically ill patients with the Middle East respiratory syndrome: a multicenter retrospective cohort study. Crit Care Med 2017; 45:1683–95. [DOI] [PubMed] [Google Scholar]
- 5. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. . Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 2013; 13:752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zahariadis G, Gooley TA, Ryall P, et al. . Risk of ruling out severe acute respiratory syndrome by ruling in another diagnosis: variable incidence of atypical bacteria coinfection based on diagnostic assays. Can Respir J 2006; 13:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng Z, Chen R, Li Y, et al. . The clinical characteristics of secondary infection of lower respiratory in severe acute respiratory syndrome. Chin J Resp Crit Care Med 2003; 3:270–4. [Google Scholar]
- 8. Lu R, Zhao X, Li J, et al. . Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect 2020; 81:266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox MJ, Loman N, Bogaert D, O’Grady J. Co-infections: potentially lethal and unexplored in COVID-19 [manuscript published online ahead of print 24 April 2020]. Lancet Microbe 2020. doi:10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. RECOVERY Collaborative Group; Horby P, Lim WS, et al. . Dexamethasone in hospitalized patients with Covid-19. N Engl J Med 2021; 384:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guaraldi G, Meschiari M, Cozzi-Lepri A, et al. . Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol 2020; 2: e474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gomez-Simmonds A, Annavajhala MK, McConville TH, et al. . Carbapenemase-producing Enterobacterales causing secondary infections during the COVID-19 crisis at a New York City hospital. J Antimicrob Chemother 2021; 76:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hughes S, Troise O, Donaldson H, et al. . Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect 2020; 26:1395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen N, Zhou M, Dong X, et al. . Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arentz M, Yim E, Klaff L, et al. . Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA 2020; 323:1612–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zangrillo A, Beretta L, Scandroglio AM, et al. . COVID-BioB Study Group . Characteristics, treatment, outcomes and cause of death of invasively ventilated patients with COVID-19 ARDS in Milan, Italy. Crit Care Resusc 2020; 22:200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu X, Ge Y, Wu T, et al. . Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res 2020; 285:198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang X, Yu Y, Xu J, et al. . Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020; 8:475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou F, Yu T, Du R, et al. . Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395:1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goyal P, Choi JJ, Pinheiro LC, et al. . Clinical characteristics of Covid-19 in New York City. N Engl J Med 2020; 382:2372–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Z, Yang B, Li Q, et al. . Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis 2020; 71:769–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cai Q, Huang D, Ou P, et al. . COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy 2020; 75:1742–52. [DOI] [PubMed] [Google Scholar]
- 25. Du RH, Liang LR, Yang CQ, et al. . Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J 2020; 55:2000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng Y, Ling Y, Bai T, et al. . COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med 2020; 201:1380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L, He W, Yu X, et al. . Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J Infect 2020; 80:639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia-Vidal C, Sanjuan G, Moreno-García E, et al. . COVID-19 Researchers Group . Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect 2021; 27:83–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nori P, Cowman K, Chen V, et al. . Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol 2020; 42:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langford BJ, So M, Raybardhan S, et al. . Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect 2020; 26:1622–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect 2020; 81:266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen T, Dai Z, Mo P, et al. . Clinical characteristics and outcomes of older patients with coronavirus disease 2019 (COVID-19) in Wuhan, China (2019): a single-centered, retrospective study. J Gerontol A Biol Sci Med Sci 2020; 75:1788–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu C, Chen X, Cai Y, et al. . Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020; 180:934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu J, Li W, Shi X, et al. . Early antiviral treatment contributes to alleviate the severity and improve the prognosis of patients with novel coronavirus disease (COVID-19). J Intern Med 2020; 288:128–38. [DOI] [PubMed] [Google Scholar]
- 35. Magill SS, Edwards JR, Bamberg W, et al. . Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team . Multistate point-prevalence survey of health care–associated infections. N Engl J Med 2014; 370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Arkel ALE, Rijpstra TA, Belderbos HNA, et al. . COVID-19-associated pulmonary aspergillosis. Am J Respir Crit Care Med 2020; 202:132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pawar A, Desai RJ, Solomon DH, et al. . Risk of serious infections in tocilizumab versus other biologic drugs in patients with rheumatoid arthritis: a multidatabase cohort study. Ann Rheum Dis 2019; 78:456–64. [DOI] [PubMed] [Google Scholar]
- 38. Guaraldi G, Meschiari M, Cozzi-Lepri A, et al. . Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol 2020; 2:e474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Somers EC, Eschenauer GA, Troost JP, et al. . Tocilizumab for treatment of mechanically ventilated patients with COVID-19 [manuscript published online ahead of print 11 July 2020]. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.