Abstract
Aedes aegypti is the primary transmitter of the four viruses that have had the greatest impact on human health, the viruses causing yellow fever, dengue fever, chikungunya, and Zika fever. Because this mosquito is easy to rear in the laboratory and these viruses grow in laboratory tissue culture cells, many studies have been performed testing the relative competence of different populations of the mosquito to transmit many different strains of viruses. We review here this large literature including studies on the effect of the mosquito microbiota on competence. Because of the heterogeneity of both mosquito populations and virus strains used, as well as methods measuring potential to transmit, it is very difficult to perform detailed meta-analysis of the studies. However, a few conclusions can be drawn: (1) almost no population of Ae. aegypti is 100% naturally refractory to virus infection. Complete susceptibility to infection has been observed for Zika (ZIKV), dengue (DENV) and chikungunya (CHIKV), but not yellow fever viruses (YFV); (2) the dose of virus used is directly correlated to the rate of infection; (3) Brazilian populations of mosquito are particularly susceptible to DENV-2 infections; (4) the Asian lineage of ZIKV is less infective to Ae. aegypti populations from the American continent than is the African ZIKV lineage; (5) virus adaptation to different species of mosquitoes has been demonstrated with CHIKV; (6) co-infection with more than one virus sometimes causes displacement while in other cases has little effect; (7) the microbiota in the mosquito also has important effects on level of susceptibility to arboviral infection; (8) resistance to virus infection due to the microbiota may be direct (e.g., bacteria producing antiviral proteins) or indirect in activating the mosquito host innate immune system; (9) non-pathogenic insect specific viruses (ISVs) are also common in mosquitoes including genome insertions. These too have been shown to have an impact on the susceptibility of mosquitoes to pathogenic viruses.
One clear conclusion is that it would be a great advance in this type of research to implement standardized procedures in order to obtain comparable and reproducible results.
1. Background
There are hundreds of known arthropod-borne-viruses (arboviruses) of which about 30 are known to cause disease in humans (Cleton et al., 2012). Despite this diversity, only four arboviruses have caused by far the most human suffering, the viruses causing yellow fever, dengue, chikungunya and Zika. Not coincidently, one mosquito, Aedes aegypti, has historically been the primary vector in almost all major human epidemics of these four viruses. “Not coincidently” because these viruses are native to Africa, humans are a native African primate, and Ae. aegypti is a native African mosquito. It has been suggested that this long history together has allowed the viruses, mosquito, and primate host to coevolve in their native Africa before spreading around the world (Powell, 2018).
These four viruses are all single-stranded RNA viruses, known to have high mutation rates, which has likely aided their rapid evolution and adaptation to replicate in different hosts (Weaver, 2006; Rückert and Ebel, 2018). Three are flaviviruses, yellow fever virus (YFV), dengue viruses (DENVs), and Zika virus (ZIKV) and one an alphavirus, chikungunya virus (CHIKV). All cause similar symptoms in humans, high fever lasting 4–14 days and joint pain. Yet each has its unique pathology with high rates of mortality for YFV and sometimes DENVs, but rarely for CHIKV or ZIKV.
Fortuitously, Ae. aegypti is the easiest mosquito to rear and manipulate in the laboratory. The viruses can be grown in mosquito cell tissue cultures and either injected or added to blood used to feed females. This has led to a large number of laboratory studies of the relative competence (see definition below for vector competence) of mosquitoes from diverse geographic populations to transmit these viruses. The prevalence of diseases caused by these viruses is geographically heterogeneous likely, at least partly, due to variation in competence among local populations of Ae. aegypti.
Here we review studies of the ability of these four viruses to be transmitted by geographically diverse populations of Ae. aegypti We struggle with the issue of heterogeneity in laboratory procedures and virus strains used in an attempt to detect underlying patterns. How genetic diversity that affects phenotypes, such as vector competence, varies among populations remains an open question. However, the fact that populations of Ae. aegypti are genetically distinct (e.g., Gloria-Soria et al., 2016) makes it more likely that they vary in vector competence compared to genetically uniform species. We also consider the contribution of microbiota in vector competence. Microbiota is a normal part of the physiology of vectors and it is clear that these microbes can affect how mosquitoes react to infection with viruses. However, details of the interactions and how these interactions vary among genetically heterogeneous mosquito populations remain to be elucidated
1.1. Quantifying the epidemiological impact of Ae. aegypti
Aedes aegypti was first identified as vector for arbovirus in 1900 in Cuba by Walter Reed, Carlos Finlay and James Carroll (Reed and Carroll, 1901). A few years later (1906), Thomas Bancroft demonstrated that Ae. aegypti is able to also transmit DENVs and linked frequency of transmission to the diurnal biting habits of Ae. aegypti (Bancroft, 1906). The identification of the role of mosquitoes in the transmission cycle of human pathogens led scientists to the concept of vector control, that is, the control of pathogen transmission through the control of vectors. To formulate epidemiological predictions and assess the impact of vector control strategies, objective parameters have been proposed since the early 1900s that would mathematically link mosquito behaviors and their biological properties to pathogen transmission (Smith et al., 2012). The basic elements of the mathematical model of mosquito-borne disease were first conceptualized in the Ross-Mac-Donald “vectorial capacity” equation (Smith et al., 2012). Vectorial capacity defines the transmission potentials of a mosquito population and equals to VC = [ma2bpn]/−ln(p) where “m” is the density of vectors in relation to the host; “a” is the daily probability that the vector feeds on a host, this variable is raised to the second power because a mosquito needs to bite twice to perpetuate pathogen transmission; “b” is the intensity of transmission in relation to the initial infection rate, also called vector competence; “p” is the daily survival rate of a vector; “n” is the days it takes for a pathogen to move from the point of entry in the mosquito body (i.e. the mosquito midgut) to the point of exit (i.e. saliva), a parameter called “extrinsic incubation period” (EIP); and “1/ln(p)” is the probability of vector's surviving the EIP (Kauffman and Kramer, 2017; Rückert and Ebel, 2018).
Environmental and genetic factors of both the vector and the pathogen interact to influence the parameters of the VC equation. For instance, temperature influences EIP, the probability of mosquito survival, and may also indirectly affect adult density by impacting larval developmental time as amply discussed and reviewed elsewhere (Le Flohic et al., 2013; Gould and Higgs, 2009; Fish, 2008; Tabachnick, 2016; Kauffman and Kramer, 2017). Temperature also influences Ae. aegypti vector competence to DENVs (Carrington et al., 2013; Chepkorir et al., 2014; Gloria-Soria et al., 2017). Vector competence is defined as the capacity of a mosquito to acquire the pathogen and support its transmission; it is one of the most difficult parameters to compare among studies because no standardized procedures have been proposed and agreed upon by workers in the field to define viral transmission. An attempt to reduce the variability in vector competence estimates based on the genetic variability of the mosquito populations under test is to measure the heritability of viral titers in half-sibling experiments (i.e. Garcia-Luna et al., 2018; Vezzeille et al., 2016).
It has been challenging to identify a proxy for transmission given the difficulties in developing animal models for arboviral diseases that mimic pathogenesis and immunity in humans (Zompi and Harris, 2012). For instance, for DENVs, ZIKV and CHIKV various mouse models have been developed by genetically suppressing the mouse immune systems to allow viral replication and manifestation of disease symptoms (Na et al., 2017; Morrison and Diamond, 2017). However, these models are not applicable to all DENV serotypes (Na et al., 2017). YFV infects Indian crown and rhesus macaques that were used to develop early YFV vaccines (Beck and Barrett, 2015). In older literature, vector competence is often expressed in terms of infection and/or dissemination rate, that is the percentage of engorged females with virus detected in the head (as a proxy for the salivary glands, which are located at the base of the mosquito head) and/or in the whole body or legs. In more recent literature, the percentage of engorged females with viral particles in the saliva following the EIP (i.e. transmission rate) is often reported (Table 1). Viruses can be detected with various methods, primarily with RT-PCR using virus-specific primers and indirect immunofluorescent assays on head squashes. A few studies have tested transmission by inoculating tissue cultures (Aedes albopictus C6/36 and Ae. aegypti Aeg2 are the most used) with mosquito body extracts or saliva and doing plaque assays or testing for viral particles after an incubation period (Calvez et al., 2017; Agha et al., 2017); this confirms live virus particles are present in saliva, rather than simply viral RNA as detected by RT-PCR. Viral detection to test for transmission is mostly pursued between 7 and 14 days after viral infection (Table 1). Shorter incubation periods are used for CHIKV as this virus has a faster dissemination rate than DENVs (Dubrulle et al., 2009; Rückert and Ebel, 2018).
Table 1.
Summary of vector competence estimates across Ae. aegypti geographic populations to 1) DENVs, 2) ZIKV, 3) YFV; 4) CHIKV; 5) dual-infections and 6) infections with arboviruses other than DENVs, YFV, ZIKV and CHIKV.
| Vector Competence | ||||
|---|---|---|---|---|
| Reference | Mosquito origin | Virus genotype and strain | Infection Route, virus dose1 |
Results7 |
| 1) DENVs | ||||
| Calvez et al., 2018 | Noumea, NC | DENV-1 NC14-17022014-806 | BM2, 106 | IR in bodies 50 at 7 dpi, 10 at 14 dpi, 8 at 21 dpi; IR in the heads 60 at 7 dpi, 100 at 14 dpi, 100 at 21 dpi; TR 3 at 7dpi, 3 at 14 dpi, 8 at 21 dpi |
| Ouvea, NC | DENV-1 NC14-17022014-806 | BM, 106 | IR in bodies 53 at 7 dpi, 53 at 14 dpi, 33 at 21 dpi; IR in the heads 100 at 7 dpi, 87 at 14 dpi, 90 at 21 dpi; TR 3 at 7dpi, 13 at 14 dpi, 13 at 21 dpi | |
| Poindimie, NC | DENV-1 NC14-17022014-806 | BM, 106 | IR in bodies 33 at 7 dpi, 13 at 14 dpi, 17 at 21 dpi; IR in the heads 70 at 7 dpi, 100 at 14 dpi, 80 at 21 dpi; TR 0 at 7dpi, 3 at 14 dpi, 0 at 21 dpi | |
| Papeete, Thaiti Island | DENV-1 NC14-17022014-806 | BM, 106 | IR in bodies 47 at 21 dpi; IR in the heads 100 at 21 dpi; TR 3 at 7dpi, 35 at 21 dpi | |
| Serrato et al., 2017 | Valle Grande, Col | DENV-2 NG | BM, 108.1–107 | IR 68 at 15 dpi |
| Paso del Comercio, Col | DENV-2 NG | BM, 108.1–107 | IR 55 at 15 dpi | |
| Siloe, Col | DENV-2 NG | BM, 108.1–107 | IR 52 at 15 dpi | |
| Mariano Ramos | DENV-2 NG | BM, 108.1–107 | IR 52 at 15 dpi | |
| Hanoi, Viet8 | DENV-2 strain 6H, Hanoi Viet | BM, 2.8×107 | IR 4.2 at 25°C; 9.1 at 27°C; 80 at 32°C | |
| DENV-2 strain 434S, Long An Province, Viet | BM, 3.77×107 | IR 8.1 at 25°C; 13 at 27°C; 4.2 at 32°C | ||
| Ho Chi Minh City, Viet | DENV-2 strain 6H, Hanoi Viet | BM, 2.8×107 | IR 10.8 at 25°C; 2.8 at 27°C; 0 at 32°C | |
| DENV-2 strain 434S, Long An Province, Viet | BM, 3.77×107 | IR 24.6 at 25°C; 9.8 at 27°C; 7.7 at 32°C | ||
| Vazeille et al., 20169 | Center Cayenne, FG | DENV-1 isol. from a 2009 patient living in Cayenne | BM, 105–106 | IR 20 at 8 dpi, 〰35 at 10 dpi, 〰50 at 14 dpi; TR different from 0 only at 14 dpi, when it reached 〰10 |
| Center Cayenne, FG | DENV-4 isol. from a 2009 patient living in Cayenne | BM, 105–106 | IR 〰40 at 8 dpi, 〰60 at 10 dpi, 〰60 at 14 dpi; TR different from 0 only at 14 dpi, when it reached 〰8 | |
| Scattered housing area, Cayenne, FG | DENV-1 isol. from a 2009 patient living in Cayenne | BM, 105–106 | IR 〰20 at 8 dpi, 〰50 at 10 dpi, 〰78 at 14 dpi; TR was always 0 | |
| Scattered housing area, Cayenne, FG | DENV-4 isol. from a 2009 patient living in Cayenne | BM, 105–106 | IR 〰40 at 8 dpi, 〰35 at 10 dpi, 〰58 at 14 dpi; TR different from 0 only at 14dpi, when it reached 〰15 | |
| Guo et al., 2016 | Haikou strain, originally from Hainan province | DENV-2-FJ10 | BM, 1.75×105 | IR in midgut 0 up to 3 dpi; 5 from 5-7 dpi; 15 at 9 dpi, 25 at 15 dpi; IR in salivary glands 0 up to 5 dpi; 4 at 7 dpi, 15 at 9 dpi, 17 at 15 dpi |
| DENV-2-FJ11 | BM, 2×105 | IR in midgut 0 up to 3 dpi; 5 at 5 dpi, 10 at 7 dpi; 25 at 9 dpi, 35 at 15 dpi; IR in salivary glands 0 up to 5 dpi; 4 at 7 dpi, 10 at 9 dpi, 25 at 15 dpi | ||
| Fansiri et al., 2016 | Bangkok, Thai | 14 DENV-1 Thai isol. | BM, 1.5×105 −8.5 106 | IR 0 (B3 viral strain, experiment 2) - 100 (K15 and K4 viral strains experiment 1; B1, B76 and K25 viral strains experiment 2) |
| Kamphaeng Phet Province, Thai | 14 DENV-1 Thai isol. | BM, 1.5×105 −8.5 106 | IR 0 (K1 viral strain, experiment 2) - 100 (K25 viral strain experiment 1, B76 viral strain experiment 2) | |
| Fernandes da Moura et al., 2015 | Santiago Island, Capo Verde | DENV-1 42735/BR PE | BM, 5×104– 2×105 | IR 0 at 7 dpi, 74,9 at 14 dpi, 20 at 21 dpi in midguts; IR 24,3 at 7 dpi, 0 at 14 dpi, 67,5 at 21 dpi in whole body; TR 55 at 14 dpi |
| DENV-2 3808/BR-PE | BM, 1,4×105– 2×105 | IR 60 at 7 dpi, 80 at 14 dpi, 20 at 21 dpi in midguts; IR 0 at 7 dpi, 0 at 14 dpi, 92.5 at 21 dpi in whole body; TR 55 at 14 dpi | ||
| DENV-3 85469/BR-PE | BM, 106 | IR 12.5 at 7 dpi, 65 at 14 dpi, 75 at 21 dpi in midguts; IR 58,4 at 7dpi, 76,9 at 14 dpi, 93,8 at 21 dpi in whole body; TR 50 at 14 dpi | ||
| DENV-4 1385 (U1842) | BM, 106 | IR 0 at 7 dpi, 0 at 14 dpi, 9 at 21 dpi in midguts; IR 0 at 21 dpi in whole body; TR 0 at 14 dpi | ||
| Poole-Smith et al., 2015 | Patillas, PR | DENV-1 Hawaii | BM, 5–6 Log10 | IR 15, TR 3 |
| DENV-2 NG C | BM, 5–6 Log10 | IR 17, TR 5 | ||
| DENV-3 H87 | BM, 5–6 Log10 | IR 18, TR 2 | ||
| DENV-4 H241 | BM, 5–6 Log10 | IR 62, TR 42 | ||
| Dickson et al., 201410 | Fatick, S | DENV-2-75505 sylvatic genotype from S | BM, 1.5×106 | IR 61 |
| Bignona, S | DENV-2-75505 sylvatic genotype from S | BM, 1.5×106 | IR 29 | |
| Richard Toll, S | BM, 1.5×106 | IR 30 | ||
| DENV-2-75505 sylvatic genotype from S | ||||
| Goudiry, S | DENV-2-75505 sylvatic genotype from S | BM, 1.5×106 | IR 39 | |
| Aedes aegypti formosus Kedougou, S, sylvatic | DENV-2-75505 sylvatic genotype from S | BM, 1.5×106 | IR 60 | |
| Aedes aegypti formosus PK10, S, sylvatic | DENV-2-75505 sylvatic genotype from S | BM, 1.5×106 | IR 57 | |
| Mont Rolland, S | DENV-2-75505 sylvatic genotype from S | BM, 107 | IR 93 | |
| Rufisque, S | DENV-2-75505 sylvatic genotype from S | BM, 1.5×106 | IR 33 | |
| Gaye et al., 2014 | Sylvatic Aedes aegytpi formosus from Kedoungou, S | DENV-1 IbH28328 | BM3, 5×103.3 | IR 40 at 7 dpi, 30 at 15 dpi, 50 at 20 dpi |
| Sylvatic Ae.aegytpi formosus from Kedoungou, S | DENV3 H87 | BM3, 5×103.3 | IR 0 at 7 dpi, 8.3 at 15 dpi, | |
| Domestic Ae.aegypti from Dakar, S | DENV-1 IbH28328 | BM3, 5×103.3 | IR 0 at 7 dpi, 43.7 at 15 dpi, 30.8 at 20 dpi | |
| Domestic Ae. aegypti from Dakar, S | DENV3 H87 | BM3, 5×103.3 | IR 10 at 7 dpi, 15.2 at 15 dpi, 2.4 at 20 dpi | |
| Alto et al., 2014 | Key West, FL | DENV-1/US/BID-V852/2006 | BM, 6.8±0.5 log10 | IR 10 at 7 dpi and 6 at 14 dpi in midguts; 10 at 7 dpi and 88 at 14 dpi in whole body |
| DENV-2/US/BID-V1041/2006 | BM, 7.1±1.2 log10 | IR 28 at 7 dpi, at 14 dpi, 28 at 21 dpi in midguts; IR 12 at 7 dpi, 27 at 14 dpi in whole body | ||
| Gonçalves et al., 20149 | Belo Horizonte, BR | DENV-2 from a hs of a patient from Belo Horizonte in 1991 | BM, ntd | IR 60 and TR 58 in 2009; IR 78 and TR 55 in 2011 |
| Pongsiri et al., 2014 | Phet Province, Thai | six DENV-2 isol. from patients of the Phet Province in Thai | BM, 3.5–6 log10 | IR 20.9 at 7 dpi, 31.8 at 14 dpi |
| Ye et al., 20149 | Cairns, Aus | DENV-2 92-T strain isol. during a 1992 outbreak in Townsville | BM, 106 | IR 20-100 in midguts; 25-70 in heads |
| DENV-2 ET-300 strain isol. in Timor-Leste in 2000 | BM, 106 | IR 60-100 in midguts, 38-100 in heads | ||
| Rockhamton, Aus | DENV-2 92-T strain isol. during a 1992 outbreak in Townsville | BM, 106 | IR 85-100 in midguts; 35-100 in heads | |
| DENV-2 ET-300 strain isol. in Timor-Leste in 2000 | BM, 106 | IR 80-100 in midguts; 60-100 in heads | ||
| Chepkorir et al., 2014 | Nairobi, Kenya | DENV-2 from a hs (Sample N. 008/01/2012) | BM, 105.08 | mosquitoes kept at 26°C (Nairobi's average temperature) after infection, IR 12, disseminated infection 18 |
| DENV-2 from a hs (Sample N. 008/01/2012) | BM, 105.08 | mosquitoes kept at 30°C (Kilifi's average temperature) after infection, IR 20, disseminated infection 8 | ||
| Kifili, Kenya | DENV-2 from a hs (Sample N. 008/01/2012) | BM, 105.08 | mosquitoes kept at 26°C (Nairobi's average temperature) after infection IR 5, disseminated infection 35 | |
| DENV-2 from a hs (Sample N. 008/01/ 2012) | BM, 105.08 | mosquitoes kept at 30°C (Kilifi's average temperature) after infection IR 10, disseminated infection 42 | ||
| Guo et al., 2013 | Haiku strain, Chi | DENV-2 NG C | BM4, 7.7 log10 | IR in midguts at 1 dpi is 60; TR at 15 dpi 85.7 |
| DENV-2 43 | BM4, 7.2 log10 | IR in midguts at 1 dpi is 48.5; TR at 15 dpi 56.3 | ||
| Sim et al., 20139 | Rockefeller strain | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 100 |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 100 | ||
| Orlano strain | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 0 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 0 | ||
| Waco strain | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 15 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 10 | ||
| PR, field | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 30 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 25 | ||
| Saint Kitts, field | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 25 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 55 | ||
| Por Fin, field | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 28 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 10 | ||
| Puertp Triunfo, field | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 65 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 10 | ||
| Singapore, field | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 90 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 10 | ||
| Bangkok, field | DENV-2 NG C strain | BM, 106–7 | IR 7 dpi in midguts, 10 | |
| DENV4-WRAIR | BM, 106–7 | IR 7 dpi in midguts, 10 | ||
| Buckner et al., 2013 | Key West, FL | DENV-1 (strain BOLKW010) | BM, 6.3±0.2 Log10 | IR 93 in midguts, 80 in whole body |
| Carrington et al., 2013 | Kamphaeng Phet Province, Thai | DENV-1 | BM1, 3,09–4.16×105 | IR 28 |
| Lourenço-De-Oliveira et al., 2013 | Buenos Aires, Argentina | DENV-2 Thai 1974 | BM, 107 | IR in whole bodies 66.7 at 14 dpi and 78.1 at 21 dpi; TR 10.5 at 14 dpi and 6.7 at 21 dpi |
| Corrientes, Argentina | DENV-2 Thai 1974 | BM, 107 | IR in whole bodies 53.3 at 14 dpi and 76.7 at 21 dpi; TR 18.5 at 14 dpi and 36.4 at 21 dpi | |
| Salto, Uruguay | DENV-2 Thai 1974 | BM, 107 | IR in whole bodies 53.3 at 14 dpi and 76.7 at 21 dpi; TR 20 at 14 dpi and 17.9 at 21 dpi | |
| Richards et al., 2012 | Key West, FL | DENV-1 isol. BOL-KW010 | BM, 3.7 Log10 | IR 89 in the abdomen, 100 in legs; TR 0 when mosquitoes were kept at 28°C |
| Key West, FL | DENV-1 isol. BOL-KW010 | BM, 3.7 Log10 | IR 75 in the abdomen, 33 in legs; TR 0 when mosquitoes were kept at 30°C | |
| Stock Island, FL | DENV-1 isol. BOL-KW010 | BM, 3.7 Log10 | IR 75 in the abdomen, 100 in legs; TR 33 when mosquitoes were kept at 28°C | |
| Stock Island, FL | DENV-1 isol. BOL-KW010 | BM, 3.7 Log10 | IR 80 in the abdomen, 100 in legs; TR 0 when mosquitoes were kept at 30°C | |
| Carvalho-Leandro et al., 20129 | Petrolina, BR | DENV-2 3808/BR-PE | BM, 106–7 | IR 25 at 3 dpi, 70 at 7 dpi, 77 at 15 dpi, 50 at 21 dpi in midguts; IR 10 at 3 dpi, 20 at 7 dpi, 58 at 15 dpi and 100 at 21 dpi in fat; TR 40 at 7 dpi, 10 at 15 dpi, 40 at 21 dpi |
| Recife, BR | DENV-2 3808/BR-PE | BM, 106–7 | IR 5 at 3 dpi, 42,5 at 7 dpi, 20 at 15 dpi, 46.3 at 21 dpi in midguts; IR 0 at 3 dpi, 10 at 7 dpi, 70 at 15 dpi and 40 at 21 dpi in fat; TR 35 at 7 dpi, 60 at 15 dpi, 47.5 at 21 dpi | |
| Rec-L Recife Lab. strain | DENV-2 3808/BR-PE | BM, 106–7 | IR 5 at 3 dpi, 22 at 7 dpi, 20 at 15 dpi, 45 at 21 dpi in midguts; IR 0 at 3 dpi, 35 at 7 dpi, 35 at 15 dpi and 58 at 21 dpi in fat; TR 5 at 7 dpi, 20 at 15 dpi, 35 at 21 dpi | |
| Sylla et al., 2009 | D2MEB | DENV-2 JAM1409 | BM, 3.1×107–8 | IR 51.2 |
| D2S3 | DENV-2 JAM1409 | BM, 3.1×107–8 | IR 92.3 | |
| Schneider et al., 2007 | Bangkok, field | DENV-2 JaM1409 | BM, ntd | IR 32.22 +/− 8.56 |
| DS3 | DENV-2 JaM1409 | BM, ntd | IR 45.95 +/− 17.76 | |
| Form, Flavivirus refractory strC2:C83ain from Nigeria | DENV-2 JaM1409 | BM, ntd | IR 48.42 +/− 6.68 | |
| Ghana, field | DENV-2 JaM1409 | BM, ntd | IR 27.44 +/− 6.03 | |
| Ibo 11, Dengue refractory strain from Nigeria | DENV-2 JaM1409 | BM, ntd | IR 31.55 +/− 2.44 | |
| Mombasa, field | DENV-2 JaM1409 | BM, ntd | IR 30.23 +/− 3.14 | |
| MOYO-R | DENV-2 JaM1409 | BM, ntd | IR 19.54 +/− 9.73 | |
| MOYO-S, RED, mutant marker stock | DENV-2 JaM1409 | BM, ntd | IR 53.60 +/− 14.16 | |
| DENV-2 JaM1409 | BM, ntd | IR 38.79 +/− 14.17 | ||
| Trinidad, field | DENV-2 JaM1409 | BM, ntd | IR 34.92 +/− 29.27 | |
| Diallo et al., 200811 | Barkedji, S | sylvatic DENV-2 AdR 140875 | BM4, 1.6×107–106.5 | IR 7.4 |
| epidemic DENV-2 ArA 6894 | BM4, 1.6×107–106.5 | IR 1.74 | ||
| Dakar, S | sylvatic DENV-2 AdR 140875 | BM4, 1.6×107–106.5 | IR 7.8 | |
| epidemic DENV-2 ArA 6894 | BM4, 1.6×107–106.5 | IR 0 | ||
| Ngoye, S | sylvatic DENV-2 AdR 140875 | BM4, 1.6×107–106.5 | IR 17.2 | |
| epidemic DENV-2 ArA 6894 | BM4, 1.6×107–106.5 | IR 1.46 | ||
| Ndougoubene, S | sylvatic DENV-2 AdR 140875 | BM4, 1.6×107–106.5 | IR 9.3 | |
| epidemic DENV-2 ArA 6894 | BM4, 1.6×107–106.5 | IR 1.57 | ||
| Kedougou, S | sylvatic DENV-2 AdR 140875 | BM4, 1.6×107–106.5 | IR 1.35 | |
| epidemic DENV-2 ArA 6894 | BM4, 1.6×107–106.5 | IR 0 | ||
| Koung Koung, S | sylvatic DENV-2 AdR 140875 | BM4, 1.6×107–106.5 | IR 2.7 | |
| epidemic DENV-2 ArA 6894 | BM4, 1.6×107–106.5 | IR 1.85 | ||
| Knox et al., 2003 | Torres Strait, Aus | DENV-2 92T | BM5, 106.4 | IR 96 at 8 dpi, 100 at 12 and 16 dpi; TR 0 at 8 dpi; 8 at 12 dpi, 76 at 16 dpi |
| DENV-4 97B | BM5, 107 | IR 80 at 8 and 12 dpi, 84 at 16 dpi, 72 at 20 dpi; TR 0 at 8 and 12 dpi, 16 at 16 dpi, 16 at 20 dpi | ||
| Charters Towers, Aus | DENV-2 92T | BM5, 106.4 | IR 52 at 8 dpi, 60 at 8 dpi, 64 at 16 dpi; TR 8 at 8 dpi, 4 at 12 dpi, 24 at 16 dpi | |
| DENV-4 97B | BM5, 107 | IR 36 at 8 dpi, 16 at 12 dpi, 28 at 16 dpi, 32 at 20 dpi; TR 0 at 8,12 and 16 dpi, 8 at 20 dpi | ||
| Townsville, Aus | DENV-2 92T | BM5, 106.4 | IR 72 at 8 dpi, 90 at 8 dpi, 92 at 16 dpi; TR 0 at 8 dpi, 0 at 12 dpi, 28 at 16 dpi | |
| DENV-4 97B | BM5, 107 | IR 12 at 8 dpi, 28 at 12 dpi, 40 at 16 dpi, 32 at 20 dpi; TR 0 at 8, 12 and 16 dpi, 16 at 20 dpi | ||
| Cairns, Aus | DENV-2 92T | BM5, 106.4 | IR 80 at 8 dpi, 84 at 12 dpi, 80 at 16 dpi; 8 at 8 dpi, 4 at 12 dpi, 20 at 16 dpi | |
| DENV-4 97B | BM5, 107 | IR 16 at 8 dpi, 28 at 12 dpi, 36 at 16 and 20 dpi; TR 0 at 8 and 12 dpi, 4 at 16 and 20 dpi | ||
| Huber et al., 200312 | Ho Chi Minh City, (mosquitoes collected from 1975 to 1998) | DENV-2, strain not defined | BM, ntd | IR 94.8 +/− 3.61 |
| Ho Chi Minh City (mosquitoes collected from 1975 to 1998) | DENV-2, strain not defined | BM, ntd | IR 97.7 +/− 2.39 | |
| Paea strain, Thaiti | DENV-2, strain not defined | BM, ntd | IR 93.84 +/− 4.38 | |
| Lourenco-de-Oliveira et al., 2004 | Belém, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 96.3 |
| Ananindeua, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 94.23 | |
| Rio Branco, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 81.43 | |
| Porto Velho | DENV-2 Bangkok 1974 | BM, ntd | IR 83.19 | |
| Boa Vista, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 95,75 | |
| Salvador, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 81.48 | |
| Sao Luis, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 97,38 | |
| Feira de Santana, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 74,74 | |
| Milha, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 25,79 | |
| Pacuja, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 73,62 | |
| Quixeramobin, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 82,10 | |
| Represa dp Cigano, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 98,24 | |
| Tingua, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 84,85 | |
| Higienopolis, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 75,32 | |
| Moqueta, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 93,40 | |
| Rocinha, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 92,86 | |
| Comendador Soares, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 91,15 | |
| Cariacica, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 81,81 | |
| Potim, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 83,62 | |
| Leandro Ferreira, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 85,95 | |
| Foz de Iguacu, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 62,43 | |
| Maringa, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 73,6 | |
| Campo Grande, BR | DENV-2 Bangkok 1974 | BM, ntd | IR 72,73 | |
| Paea Lab. strain | DENV-2 Bangkok 1974 | BM, ntd | IR 93,34 +/− 4.63 | |
| Paupy et al., 200312 | Phon Penh City Center (Cambodia), mosquitoes collected in February | DENV-2 from a hs sample collected in Bangkok Thai in 1974 | BM3, 108.2 | IR 79,39 +/− 11,01 |
| Phon Penh City Center (Cambodia), mosquitoes collected in July | DENV-2 from a hs sample collected in Bangkok Thai in 1974 | BM3, 108.2 | IR 77,76 +/− 8,31 | |
| Phon Penh City suburbs north (Cambodia), mosquitoes collected in February | DENV-2 from a hs sample collected in Bangkok Thai in 1974 | BM3, 108.2 | IR 90,65 +/− 8,77 | |
| Phon Penh City suburbs west (Cambodia), mosquitoes collected in February | DENV-2 from a hs sample collected in Bangkok Thai in 1974 | BM3, 108.2 | IR 87 +/− 4,82 | |
| Phon Penh City suburbs south (Cambodia), mosquitoes collected in February | DENV-2 from a hs sample collected in Bangkok Thai in 1974 | BM3, 108.2 | IR 95,30 +/− 0.14 | |
| Paea strain, Thaiti | DENV-2 from a hs sample collected in Bangkok Thai in 1974 | BM3, 108.2 | IR 78.52 +/− 7.64 | |
| Thongrungkiat et al., 2003 | Chiang Rai, Thai | DENV-1 16007 | BM3, 108.1 | IR 19.4 |
| BM3, 1010 | IR 48.7 | |||
| DENV-2 16681 | BM3, 108.1 | IR 17.8 | ||
| BM3, 1010 | IR 25 | |||
| DENV-3 16562 | BM3, 108.1 | IR 3.8 | ||
| BM3, 1010 | IR 19.7 | |||
| DENV-4 1036 | BM3, 108.1 | IR 27.7 | ||
| BM3, 1010 | IR 54.8 | |||
| Nakhon Phanom, Thai | DENV-1 16007 | BM3, 108.1 | IR 16 | |
| BM3, 1010 | IR 48.2 | |||
| DENV-2 16681 | BM3, 108.1 | IR 15 | ||
| BM3, 1010 | IR 28 | |||
| DENV-3 16562 | BM3, 108.1 | IR 4.3 | ||
| BM3, 1010 | IR 18.5 | |||
| DENV-4 1036 | BM3, 108.1 | IR 15.6 | ||
| BM3, 1010 | IR 49.4 | |||
| Satun, Thai | DENV-1 16007 | BM3, 108.1 | IR 8.1 | |
| BM3, 1010 | IR 43.8 | |||
| DENV-2 16681 | BM3, 108.1 | IR 13.1 | ||
| BM3, 1010 | IR 27.6 | |||
| DENV-3 16562 | BM3, 108.1 | IR 0.9 | ||
| BM3, 1010 | IR 11.1 | |||
| DENV-4 1036 | BM3, 108.1 | IR 12.5 | ||
| BM3, 1010 | IR 54.5 | |||
| Bennett et al., 20029 | Hermosillo, Sonora, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | IR 45 |
| Guymas, Sonora, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 60 | |
| Culiacan, Sinaloa, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 80 | |
| Mazatlan, Sinaloa, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 65 | |
| Puerto Valarta, Jalisco, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 30 | |
| Manzanillo, Colima, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 55 | |
| Lazaro Cardenas, Michoacan, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 45, with a large standard deviation | |
| Ixtapa Zihuatanejo, Guerrero, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 42, with a large standard deviation | |
| Coyuca de Benitez, Guerrero, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 70 | |
| Puerto Excondido, Oaxaca, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 60 | |
| Tapachula, Chiapas, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 70 (two collections from Tapachula were tested giving one TR of 60, one of 80) | |
| Chetumal, Quintana Roo, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 80 | |
| Cancun, Quintana Roo, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 70 | |
| Merida, Yucatan, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 69 | |
| Campeche, Campeche, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 42 | |
| Ciudad del Carmen, Campeche, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 42 | |
| Villahermosa, Tabasco, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 58 | |
| Moloacan, Veracruz, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 58 | |
| Miguel Aleman, Tamaulipas, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 60 | |
| Nuevo Ladero, Tamaulipas, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 48 | |
| Monterey, Nuevo Leon, MX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 56 | |
| Huston, TX | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 40, with a great standard deviation | |
| Tucson, Arizona | DENV-2 JAM1409 | BM4, 107.5 to 108.5 | TR 68 | |
| Vazeille et al., 2001 | Mahaleja, Madagascar | DENV-2 Bangkok 1974 | BM3, 108.2 | IR 27.8 |
| Jeffreville, Madagascar | DENV-2 Bangkok 1974 | BM3, 108.2 | IR 32.5 | |
| Paea Lab. strain | DENV-2 Bangkok 1974 | BM3, 108.2 | IR 94 | |
| Tran et al., 1999 | Ho Chi Minh City | DENV-2 Bangkok 1974 | BM3, 108.2 | IR 96,16 +/− 3.35 |
| Watson & Kay, 199912 | Queensland, Aus Lab. strain | DENV-1 from hs of a patent in Townaville in 1990 | BM6, 0–6–3,6 Log10 | IR 31 +/− 23.34 |
| DENV-2 from hs of a patent in Townaville in 1992 | BM6, 1,2–4,2 Log10 | IR 35.5 +/− 25.67 | ||
| DENV-3 h87 | BM6, 0,9–3,9 Log10 | IR 42 +/− 27.72 | ||
| DENV-4 h241 | BM6, 0,6–3,6 Log10 | IR 36 +/− 22,02 | ||
| Jupp and Kemp, 199312 | Empangeni, SA | DENV-1 Cassim strain from Durban, SA | BM3, 7,2 Log10 | IR 100 at 8–10 dpi |
| Palm Beach, SA | DENV-1 Cassim strain from Durban, SA | BM, 6.1–7.1 Log10 | IR 15, TR 100 at 17–19 dpi; IR 28, TR 50 at 16–17 dpi | |
| DENV-2 BC 5007 strain from Taipei | BM3, 7.2–7.9 Log10 | IR 15.5 and TR 50 at 17–18 dpi; IR 25, TR 83 at 15 dpi | ||
| Durban, SA | DENV-1 Cassim strain from Durban, SA | BM3, 6.3–7.1 Log10 | IR 62.8, TR 92 at 17–19 dpi; IR 43, TR 73 at 13–15 dpi | |
| DENV-2 BC 5007 strain from Taipei | BM, 7–7.5 Log10 | IR 46, TR 75 at 14–15 dpi | ||
| Richards Bay, SA | DENV-1 Cassim strain from Durban, SA | BM3, 6.1–7.1 Log10 | IR 38, TR 69.5 at 17–19 dpi, | |
| DENV-2 BC 5007 strain from Taipei | BM3, 7.2–7.5 Log10 | IR 29.5; TR 69 at 14–20 dpi | ||
| Ndumu, SA | DENV-1 Cassim strain from Durban, SA | BM3, 6.3–7.1 Log10 | IR 36.5; TR 75 at 18–19 dpi | |
| DENV-2 BC 5007 strain from Taipei | BM, 7.1 Log10 | IR 41.67; TR 82 at 14–18 dpi | ||
| Skukuza, SA | DENV-1 Cassim strain from Durban, SA | BM3, 6.9–8.4 Log10 | IR 12.5; TR 100 at 14–20 dpi; | |
| DENV-2 BC 5007 strain from Taipei | BM3, 7–7.9 Log10 | IR 28; TR 66.5 at 16–19 dpi | ||
| Chen et al., 1993 | Kaohsiung, southern Taiwan | DENV-1 from a dengue patient during the dengue epideminc in Kaohsiung in 1987–1988 | IT | TR 50 at 14 dpi, 83.3 at 21 dpi |
| Bosio et al., 1998 | San Juan, PR | DENV-2PR-159, PR | BM, ntd | IR in midguts: 61 |
| Aedes aegytpi formosus from Ibo village, Nigeria | DENV-2PR-159, PR | BM, ntd | IR in midguts: 25 | |
| Mitchell et al., 1987 | Rexville strain from PR | DENV-1 1620, PR | BM3, 6.6–9.2 Log10 | IR 45 at 7 dpi, 605 at 14 dpi, TR 88 |
| DENV-2 1615, PR | BM3, 5.6–8.4 Log10 | IR 25 at 7 dpi, 28.67 at 13 dpi, 56.4 1t 14 dpi, TR 74 | ||
| DENV-3 1557, PR | BM3, 6.3–8.4 Log10 | IR 5 at 7 dpi, 58.2 at 14 dpi, TR 53 | ||
| DENV-4 1632, PR | BM3, 6.2–9.2 Log10 | IR 0 at 7 dpi, 19.67 at 13 dpi, 63 at 14 dpi, TR 42 | ||
| Boromisa et al., 1987 | Lab. strain from Huston, TX | DENV-1 YARU 40130, Fijii | BM3, 8.3 Log10 | IR 70 in midguts; 30 in whole body; TR 5 |
| Rosen et al., 1985 | Rockefeller strain | DENV-1 Hawaii 1944 | BM3, 107.8 | IR 16.7 |
| Niue strain from Niue Island | DENV-1 Hawaii 1944 | BM3, 107.8 | IR 0 | |
| DENV-1 Malay-1 (Malaysia 1965) | BM3, 107.8 | IR 0 | ||
| DENV-1 Malay-2 (Malaysia 1966) | BM3, 107.8 | IR 20 | ||
| DENV-1 Thai (Bangkok, 1971) | BM3, 107.8 | IR 25 | ||
| Rockefeller strain | DENV-2 NG 1944 | BM3, 107.8 | IR 50 | |
| DENV-2 Thaiti 1971 | BM3, 107.8 | IR 45 | ||
| Niue strain from Niue Island | DENV-2 Thaiti 1971 | BM3, 107.8 | IR 13.6 | |
| Tong strain from Tonga | DENV-2 Thaiti 1971 | BM3, 107.8 | IR 23.5 | |
| Rockefeller strain | DENV-3 H87 Manila, Phi 1956 | BM3, 107.8 | IR 26.7 | |
| DENV-3 Manila Manila Phi 1965 | BM3, 107.8 | IR 34.6 | ||
| DENV-3 Tahiti 1964 | BM3, 107.8 | IR 30.8 | ||
| DENV-3 Thai, Bangkok Thai 1971 | BM3, 107.8 | IR 36.8 | ||
| Trinidad strain from Trinidad | DENV-3 Manila Manila Phi 1965 | BM3, 107.8 | IR 20 | |
| DENV-3 Tahiti 1964 | BM3, 107.8 | IR 22.2 | ||
| DENV-3 Thai, Bangkok Thai 1971 | BM3, 107.8 | IR 71 | ||
| Rockefeller strain | DENV-4 H241 | BM3, 107.8 | IR 100–0 depending on viral dose inocula | |
| 2) ZIKV | ||||
| Calvez et al. (2018) | French Polynesia | NC-2014-5132, NC | BM, 107 TCID50/mL | IR: 53 at 6 dpi; 94 at 9 dpi; 97 at 14 dpi, 89 at 21 dpi; TR 0 between 6 and 9 dpi; 24 at 21 dpi |
| NC | IR: 88 at 6 dpi; 73 at 9 dpi; 77 at 14 dpi, 95 at 21 dpi; TR 0 at 6dpi, 3 at 9 dpi, 0 between 14 and 21 dpi | |||
| Samoa | IR: 33 at 6 dpi; 23 at 9 dpi; 50 at 14 dpi, 38 at 21 dpi; TR 0 between 6 and 9 dpi; 17 at 14 dpi and 30 at 21 dpi | |||
| Main et al. (2018) | Los Angeles, CA | PRVABC59, PR | BM, 5.4-6.4 log10 | IR: 85 at 14 dpi; 96 at 21 dpi; DR 78 at 7-14 dpi, TR 65 at 14 dpi, 74 at 21 dpi |
| MA66, P6-740, Maylasia | BM, 4.3-4.8 log10 | IR: 86 at 14 dpi; 96 at 21 dpi; DR 79 at 7 dpi, 91 at 14 dpi, TR 53 at 14 dpi, 87 at 21 dpi | ||
| BR15, SPH2015, BR | BM, 4.7 log10 | IR: 90; DR: 90; TR: 75 at 14 dpi | ||
| Garcia-Luna et al. (2018)12 | Apodaca, MX | PRVABC59, PR | BM, 1.5-1.8×106 | IR 79 at 7 dpi; 84 at 14 dpi; DR 71 at 7 dpi, 80 at 14 dpi; TR 15 at 7 dpi; 33 at 14 dpi |
| San Nicolas, MX | PRVABC59, PR | BM, 4×105-2×107 | IR 97 at 7 dpi; 93 at 14 dpi; DR 51 at 7 dpi, 88 at 14 dpi; TR 4 at 7 dpi; 27 at 14 dpi | |
| Monterey, MX | PRVABC59, PR | BM, 8×105-4×107 | IR 83 at 7 dpi; 63 at 14 dpi; DR 19 at 7 dpi, 45 at 14 dpi; TR 1 at 7 dpi; 14 at 14 dpi | |
| Cd. Madero, MX | PRVABC59, PR | BM, 6.2-8×105 | IR 53 at 7 dpi; 60 at 14 dpi; DR 28 at 7 dpi, 52 at 14 dpi; TR 7 at 7 dpi; 17 at 14 dpi | |
| Poza Rica, MX | PRVABC59, PR | BM, 1.4x105x1.8×107 | IR 100 at 7-14 dpi; DR 98 at 7 dpi, 100 at 14 dpi; TR 10 at 7 dpi; 52 at 14 dpi | |
| Minatitlan, MX | PRVABC59, PR | BM, 6.2×105-1.6×106 | IR 91 at 7dpi, 81 at 14 dpi; DR 72 at 7 dpi, 78 at 14 dpi; TR 10 at 7 dpi; 29 at 14 dpi | |
| Coatzacoalcos, MX | PRVABC59, PR | BM, 1.4×105-1.7×106 | IR 92 at 7dpi, 98 at 14 dpi; DR 73 at 7 dpi, 95 at 14 dpi; TR 24 at 7 dpi; 51 at 14 dpi | |
| Merida, MX | PRVABC59, PR | BM, 8×105-4.4×107 | IR 99 at 7dpi, 96 at 14 dpi; DR 74 at 7 dpi, 92 at 14 dpi; TR 10 at 7 dpi; 42 at 14 dpi | |
| Mazatan, MX | PRVABC59, PR | BM, 1.12-4.4×107 | IR 100 at 7-14dpi; DR 95 at 7 dpi, 100 at 14 dpi; TR 15 at 7 dpi; 23 at 14 dpi | |
| Guerrero, MX | PRVABC59, PR | BM, 2×106-1.8×107 | IR 98 at 7, 93 at 14dpi; DR 95 at 7 dpi, 93 at 14 dpi; TR 50 at 7 dpi; 42 at 14 dpi | |
| Dodson et al. (2018) | Rockefeller strain | PRVABC59, PR | BM, 2×108 | IR: 40.67 +/− 19; TR 2.67 +/− 4.62 |
| Roundy et al. (2017) | Salvador, BR | DAK AR 41525, S | BM/murine2, 104-6 | IR 100; TR100 |
| FSS 13025, Cambodia | BM/murine2, 104-6 | IR 75; TR 0 murine: IR 100; TR 40 | ||
| MEX1-7, MX | BM, 2×108 | IR 75; TR 0 | ||
| Dominican Republic | DAK AR 41525, S | BM, 2×108 | IR 100; TR100 | |
| FSS 13025, Cambodia | BM, 2×108 | IR 100; TR 18 | ||
| MEX1-7, MX | BM, 2×108 | IR 90; TR 20 | ||
| RioGrande Valley | DAK AR 41525, S | BM, 2×108 | IR 100; TR 30 | |
| FSS 13025, Cambodia | BM, 2×108 | IR 40; TR 0 | ||
| MEX1-7, MX | BM, 2×108 | IR 65; TR 0 | ||
| Kenney et al. (2017) | Poza Rica, MX, Lab. strain | PRV ABC59 | IT, 106 | IR 100; TR 67 |
| Heitmann et al., 2017 | Bayer company, Lab. strain | FB-GWUH-2016, Central America | BM, 107 | 18 °C: IR 55; TR 0 27 °C: IR 49; TR 22 |
| Fernandes et al. (2017) | Rio de Janeiro, BR | ZIKV strains from BR | BM, 106.36 | IR 68-100; |
| Guedes et al. (2017) | Fernando de Noronha, BR | BRPE 243/ 2015, BR | BM, 106 | IR 40 |
| Recife, Lab. strain | BRPE 243/ 2015, BR | BM, 106 | IR 44 | |
| Ciota et al. (2017) | Poza Rica, MX | CAM FSS130325, Cambodia | BM, 106.6-7.7 | IR 44; TR 33 |
| HND 2016-19,563, Honduras | BM, 106.6-7.7 | IR 47; TR 36 | ||
| Li et al. (2017)9 | HK strain from mosquitoes collected in Hainan province, Chi | SZ01/2016/Chi | BM, 3×105 | IR midguts: 80 at 2dpi, 80 at 4 dpi, 85 at 6 dpi, 90 at 8 dpi, 100 at 10 dpi, 90 at 12 dpi, 100 at 16,18 and 20 dpi |
| IR salivary glands: 58 at 2dpi, 78 at 4 dpi, 85 at 6 dpi, 90 at 8 dpi, 90 at 10 dpi, 100 at 12 dpi, 90 at 16,100 at 18 and 20 dpi | ||||
| RL strain from mosquitoes collected in Yunnan province, Chi | SZ01/2016/Chi | BM, 3×105 | IR midguts 100 at 2, 4, 6, 8, 10, 12, 16,18 and 20 dpi | |
| IR salivary glands: 60 at 2dpi, 80 at 4 dpi, 100 at 6 dpi, 90 at 8 dpi, 100 at 10, 12, 16, 18 and 20 dpi | ||||
| Ryckebusch et al. (2017) | Paea strain, Thaiti | PF-25013-18 | BM2, 2.5×107 | IR midguts 100 from 3 to 10 dpi, 85 at 13 dpi |
| IR in salivary glands 60 at 5, 6 and 8 dpi, 80 at 10 dpi and 7 at 14 dpi | ||||
| TR 11 at 8 dpi, 33 at 10 dpi, 16 at 14 dpi and 6.7 at 17 dpi | ||||
| Costa-da-Silva et al. (2017) | Rockefeller lab. Strain | ZIKVBR Isolated from a clinical case | BM; 2.2×106 | IR 95 in body and heads at 7 and 14 dpi; TR 10 at 7 dpi; 38 at 14 dpi |
| HWE Lab. strain | BM; 2.2×106 | IR 60 in body, 50 in heads at 7 dpi; 65 in body and head at 14 dpi; TR 0 at 7dpi, 35 at 14 dpi | ||
| RED lab. Strain | BM; 2.2×106 | IR 95 in body and 70 heads at 7 dpi; 95 in body and heads at 14 dpi; TR 0 at 7 dpi, 5 at 14 dpi | ||
| Weger-Lucarelli et al. (2016) | Poza Rica, MX | PRV ABC59, PR | BM, fresh 106.3 | IR 95, TR 70 |
| PRV ABC59, PR | BM, frozen 4 h 106.3 | IR 95, TR 65 | ||
| PRV ABC59, PR | BM, frozen 1 week 106.3 | IR 60, TR 22 | ||
| DAKAR 41525, S | BM, frozen 07.2 | IR 75, TR 55 | ||
| MR 766, Uganda | BM, frozen 107.2 | IR 58, TR 37 | ||
| Richard et al. (2016a) | Tahiti 2014 | PF13/2511013-18 Polynesia | BM4, 107 | BM: IR 85; TR 36 |
| Hall-Mendelin et al. (2016) | Queensland, Aus | MR 766, Uganda | BM4, 106.7 | BM: IR 57; TR 27 |
| Di Luca et al. (2016) | MX, Lab. strain | H/PF/2013 French Polynesia | BM, 106.4 | IR 40, TR 40 |
| Dutra et al. (2016) | Urca, Rio de Janeiro, BR | BRPE 243/2015 BR | BM, fresh 5×106 | IR 100, TR 100 |
| Alto et al. (2017) | Black eyed Liverpool, Lab. strain | PRV ABC59 | Murine 106.8 | IR 100; TR 24 |
| Boccolini et al. (2016) | Reynosa, MX, Lab. strain | H/PF/2013 French Polynesia | BM, 106.46 | IR 50; TR 38 |
| Chouin-Carneiro et al. (2016) | FG | NC-2014-5132, NC | BM4, 107 | 7 dpi: IR 100, TR 0 |
| Guadeloupe | NC-2014-5132, NC | BM4, 107 | 7 dpi: IR 87; TR 0 | |
| Martinique | NC-2014-5132, NC | BM4, 107 | 7 dpi I: IR 90; TR 0 | |
| Orlando, FL | NC-2014-5132, NC | BM4, 107 | 7 dpi: IR 93; TR nd | |
| Tubiacanga, BR | NC-2014-5132, NC | BM4, 107 | 7 dpi: IR 83; TR nd | |
| Li et al. (2012) | Singapore | MR 766, Uganda | BM4, 107 | BM: IR 100; TR 100 |
| Diagne et al. (2015)13 | Dakar, S, domestic | ArD 128,000 and 132,912, Kedougou | BM 6.4-7.6 log10 | IR+, DR+, TR 0 |
| Kedougou, S, sylvatic | ArD 128,000 and 132,912, Kedougou | BM 6.4-7.6 log10 | IR+, DR+, TR 0 | |
| Cornet and Robin (1979) | S-1971, Lab. strain | ArD 24,280, S | IT dose unknown 7-28 dpi | TR 91 |
| Boorman and Porterfield (1956) | Nigeria, Lab. strain | MR 766, Uganda | BM, 106.7 LD50 60 dpi | IR 100; TR 50 |
| 3)YFV | ||||
| Couto-Lima et al. (2017)12 | Goiania, BR | 74,018-1D from BR | BM, 106 | IR 0 at 3dpi, 〰 30 at 7dpi, 〰 80 at 14 dpi, 〰 70 at 14 dpi |
| 4408-1E from BR | BM, 106 | IR 0 at 3dpi, 〰 25 at 7dpi, 〰 78 at 14 dpi, 〰 10 at 14 dpi | ||
| S-79 from Senegal | BM, 106 | IR 0 at 3dpi, 〰 30 at 7dpi, 〰 80 at 14 dpi, 0 at 14 dpi | ||
| 74,018-1D from BR | BM, 106 | TR 0 at 3dpi, 0 at 7dpi, 〰 18 at 14 dpi, 0 at 14 dpi | ||
| 4408-1E from BR | BM, 106 | TR 0 at 3dpi, 0 at 7dpi, 〰 18 at 14 dpi, 58 at 14 dpi | ||
| S-79 from S | BM, 106 | TR 0 at 3dpi, 0 at 7dpi, 0 at 14 dpi, 0 at 14 dpi | ||
| Dickson et al. (2014) | Fatick | BA-55- West African Genyotype I, Nigeria | BM, 106 | IR 59 |
| Fatick | DAK -1279- West African Genyotype II, S | BM, 7.9×105 | IR 17 | |
| Bignona | BA-55- West African Genyotype I, Nigeria | BM, 106 | IR 13 | |
| Bignona | DAK -1279- West African Genyotype II, S | BM, 6.1×107 | IR 33 | |
| Richard Toll | BA-55- West African Genyotype I, Nigeria | BM, 2×106 BM, 7.9×105 | IR 10 | |
| Richard Toll | DAK -1279- West African Genyotype II, S | IR 57 | ||
| Goudiry | BA-55- West African Genyotype I, Nigeria | BM, 106 | IR 0 | |
| Goudiry | DAK -1279- West African Genyotype II, S | BM, 7.9×105 | IR 10 | |
| Ae aegypti formosus PK10, S, sylvatic | BA-55- West African Genyotype I, Nigeria | BM, 2×105 | IR 0 | |
| Ae aegypti formosus PK10, S, sylvatic | DAK -1279- West African Genyotype II, S | BM, 7.9×105 | IR 10 | |
| Ae aegypti formosus PK10, S, sylvatic | BA-55- West African Genyotype I, Nigeria | BM, 106 | IR 3 | |
| Ae aegypti formosus PK10, S, sylvatic | DAK -1279- West African Genyotype II, S | BM, 7.9×105 | IR 22 | |
| Mont Rolland | BA-55- West African Genyotype I, Nigeria | BM, 2×106 | IR 0 | |
| Mont Rolland | DAK -1279- West African Genyotype II, S | BM, 7.9×105 | IR 20 | |
| Rufisque | BA-55- West African Genyotype I, Nigeria | BM, 106 | IR 0 | |
| Rufisque | DAK -1279- West African Genyotype II, Senegal | BM, 7.9×105 | IR 11 | |
| Ellis et al. (2012) | Nairobi, Kenya | East African genotype (Sudan 2003) | BM, 6.7-7.5 log10 | IR 7 |
| Mariakani, Kenya | East African genotype (Sudan 2003) | BM, 6.7-7.5 log10 | IR 41 | |
| Kerio Valley, Kenya | East African genotype (Sudan 2003) | BM, 6.7-7.5 log10 | IR 11 | |
| Kakamega, Kenya | East African genotype (Sudan 2003) | BM, 6.7-7.5 log10 | IR 23 | |
| van den Hurk et al. (2011) | Cairns, Aus | African strain BA-55 (Nigeria 1955) | BM4, 107.2 | IR 80, TR 52 |
| South American strain, Cinetrop 28 (OBS 7549) Bolivia 1999 | BM4, 106.7 | IR 64, TR 64 | ||
| Asibi strain | BM4, 108 | IR 92, TR 80 | ||
| Townsville, Aus | African strain BA-55 (Nigeria 1955) | BM4, 107.2 | IR 72, TR 60 | |
| South American strain, Cinetrop 28 (OBS 7549) Bolivia 1999 | BM4, 106.7 | IR 36, TR 28 | ||
| Asibi strain | BM4, 108 | IR 96, TR 96 | ||
| RexD strain | African strain BA-55 (Nigeria 1955) | BM4, 107.2 | IR 82, TR 64 | |
| South American strain, Cinetrop 28 (OBS 7549) Bolivia 1999 | BM4, 106.7 | IR 40, TR 32 | ||
| Asibi strain | BM4, 108 | IR 76, TR 64 | ||
| Johnson et al. (2002) | Santos, Brazil | no. 71528 MG2001, from BR | BM, 7-7.8 log10 | IR 35, TR 25.5 |
| Lourenco-de-Oliveira et al. (2002) | Milhã, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 0 |
| Comendador Soares, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 0.9 | |
| Quixeramobim, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 1.7 | |
| Rocinha, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 3.3 | |
| Tinguá, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 4.9 | |
| Pacujá, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 5.6 | |
| Salvador, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 6.3 | |
| Higienópolis, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 6.7 | |
| Moquetá, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 7.6 | |
| Feira de Santana, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 10.6 | |
| Rio Branco, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 11.1 | |
| Leandro Ferreira, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 12.0 | |
| Cariacica, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 12.6 | |
| Boa Vista, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 12.9 | |
| Represa do Cigano, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 16.1 | |
| São Luis, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 19.6 | |
| Maringá, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 22.7 | |
| Porto Velho, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 24.4 | |
| Campo Grande, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 25 | |
| Potim, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 27.1 | |
| Belém, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 33.9 | |
| Ananindeua, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 46.4 | |
| Foz do Iguaçu, BR | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 48.6 | |
| Phnom Penh, Cambodia | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 64.4 | |
| Ho Chi Min | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 48.05 | |
| Maracay, Venezuela | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 13.6 | |
| West Palm Beach, FL | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 24.8 | |
| Ae. aegypti formosus Boulbinet Guinea | FIOCRUZ 74018/MG/01 | BM3, 108.7 | IR 3.3 | |
| Mitchell et al. (1987) | Rexville strain from PR | 788,379 | BM, 5.0-6.7 Log10 | IR 61 at 11 dpi, 80 at 14 dpi; TR 42 at 11 dpi, 38 at 14 dpi |
| Wallis et al. (1985) | Soufriere, Dominica | Asibi strain | BM, ntd | IR 17,17 +/− 13,50 |
| Tabachnick et al. (1985) | West Africa Sylvan, Dakar S, lab. Strain | Asibi strain | BM, ntd | IR 11 |
| West Africa Sylvan, N'Gove S, lab. Strain | Asibi strain | BM, ntd | IR 7 | |
| West Africa Sylvan, Gambia, lab. Strain | Asibi strain | BM, ntd | IR 27 | |
| East Africa Sylvan, Kampala Uganda, lab. Strain | Asibi strain | BM, ntd | IR 8 | |
| Asibi strain | BM, ntd | IR 34 | ||
| East Africa Sylvan, Kombeni, Kenya; lab. Strain | ||||
| East Africa Domestic, Kwa Dzivo Kenya; isofemale lines | Asibi strain | BM, ntd | IR 57 | |
| East Africa Domestic, Majengo Kenya; isofemale lines | Asibi strain | BM, ntd | IR 29 | |
| Asia-Pacific Domestic Bangalore India; lab. Strain | Asibi strain | BM, ntd | IR 23 | |
| Asia-Pacific Domestic Colombo Sri Lanka; lab. Strain | Asibi strain | BM, ntd | IR 21 | |
| Asia-Pacific Domestic Djakarta Java; lab. Strain | Asibi strain | BM, ntd | IR 32 | |
| Asia-Pacific Domestic Karachi Pakistan; lab. Strain | Asibi strain | BM, ntd | IR 30 | |
| Asia-Pacific Domestic Thai, Amphur strain | Asibi strain | BM, ntd | IR 28 | |
| Asia-Pacific Domestic Fiji; lab. Strain | Asibi strain | BM, ntd | IR 22 | |
| Domestic Austin, TX; isofemale lines | Asibi strain | BM, ntd | IR 29 | |
| Domestic Galveston, TX; lab. Strain | Asibi strain | BM, ntd | IR 16 | |
| Domestic Huston, TX; lab. Strain | Asibi strain | BM, ntd | IR 21 | |
| Domestic Welasco, Texas USA; lab. Strain | Asibi strain | BM, ntd | IR 15 | |
| Domestic Victoria, MX; isofemale lines | Asibi strain | BM, ntd | IR 20 | |
| Domestic Abbeville, Luisiana USA; lab. Strain | Asibi strain | BM, ntd | IR 12 | |
| Domestic Beamont, TX; lab. Strain | Asibi strain | BM, ntd | IR 26 | |
| Domestic Vero Beach, FL; field | Asibi strain | BM, ntd | IR 41 | |
| Domestic Esquintla, Guatemala; isofemale lines | Asibi strain | BM, ntd | IR 2 | |
| Domestic Malaga, Colombia; field | Asibi strain | BM, ntd | IR 46 | |
| Domestic Santa Cruz, Bolivia; isofemale lines | Asibi strain | BM, ntd | IR 31 | |
| Domestic Trinidad, West Indies; isofemale lines | Asibi strain | BM, ntd | IR 42 | |
| Domestic Arecibo, Puerto Rico; lab. Strain | Asibi strain | BM, ntd | IR 34 | |
| Domestic Limestone Bay, Anguilla; field | Asibi strain | BM, ntd | IR 39 | |
| Domestic Plymouth, Montserrat; field | Asibi strain | BM, ntd | IR 53 | |
| 4) CHIKV14 | ||||
| Agha et al. (2017) | Mombasa, Kenya | Lamu001 strain of and East/Central/South Africa lineage | BM, 105.6 | IR 0 at 5-7 dpi |
| BM, 105.9 | IR 6 at 5-7 dpi and 17 at 9 dpi | |||
| BM, 106.9 | IR 62 at 5-7 dpi | |||
| BM, 107.5 | IR 100 at 5-7 dpi and 75 at 14 dpi | |||
| Kisumu, Kenya | BM, 105.6 | IR 0 at 5-7 dpi and 0 at 14 dpi | ||
| BM, 105.9 | IR 20 at 5-7 dpi; 5 at 9 dpi and 6 at 14 dpi | |||
| BM, 106.9 | IR 40 at 5-7 dpi; 50 at 9 dpi and 63 at 14 dpi | |||
| Nairobi, Kenya | BM, 105.6 | IR 0 at 5-7 dpi and 17 at 14 dpi | ||
| BM, 105.9 | IR 7 at 5-7 dpi and 10 at 9 dpi | |||
| BM, 106.9 | IR 50 at 5-7 dpi and 57 at 9 dpi | |||
| BM, 107.5 | IR 71 at 5-7 dpi and 89 at 14 dpi | |||
| Alto et al. (2017) | Indian River/ St. Lucie County, FL | BM, 8 log10 | IR in legs 37 at 2dpi, 71 at 5 dpi, 28 at 12 dpi; TR 35 at 2 dpi, 66 at 5 dpi, 24 at 12 dpi | |
| Monroe County, FL | BM, 8 log10 | IR in legs 90 at 2dpi, 20 at 5 dpi, 54 at 12 dpi; TR 83 at 2 dpi, 18 at 5 dpi, 50 at 12 dpi | ||
| Manatee county, FL | BM, 8 log10 | IR in legs 71 at 2dpi, 68 at 5 dpi, 60 at 12 dpi; TR 58 at 2 dpi, 63 at 5 dpi, 51at 12 dpi | ||
| Dominican Repuublic | BM, 8 log10 | IR in legs 35 at 2dpi, 22 at 5 dpi, 18 at 12 dpi; TR 17 at 2 dpi, 19 at 5 dpi, 15 at 12 dpi | ||
| Ngoagouni et al. (2017) | Bangui, Central African Republic | ArB10262 | BM; 108 | IR 50 at 7 dpi, 27 at 14 dpi, TR 0 at 7 dpi, 28 at 14 dpi |
| Mbaika et al. (2016) | Coastal Kenya | South/Central Africa and Indian Ocean Genotype (Group III), subgroup IIIa and b | BM; 7.9×105 | IR tested in Midgut at 26 °C 26.41 7dpi; 33.96 10 dpi, 39.62 13 dpi; |
| IR tested in Midgut at 32 °C 26.41 7dpi; 33.96 10 dpi, 39.62 13 dpi; | ||||
| IR tested in legs at 26 °C 17.9 7dpi; 25.5 10 dpi, 17 13 dpi; | ||||
| IR tested in legs at 32 °C 6.8 7dpi; 20.4 10 dpi, 29.1 13 dpi; | ||||
| IR tested in heads at 26 °C 10.4 7dpi; 2.8 10 dpi, 2.8 13 dpi; | ||||
| IR tested in heads at 32 °C 2.9 7dpi; 16.5 10 dpi, 26.2 13 dpi; | ||||
| Western Kenya | South/Central Africa and Indian Ocean Genotype (Group III), subgroup IIIa and b | BM; 7.9×105 | IR tested in Midgut 26 °C 7.55 7dpi; 5,66 10 dpi, 18,88 13 dpi; | |
| IR tested in Midgut 32 °C 33,02 7dpi; 24,53 10 dpi, 24,53 13 dpi; | ||||
| IR tested in legs at 26 °C 26.5 7dpi; 11.8 10 dpi, 20.6 13 dpi; | ||||
| IR tested in legs at 32 °C 28.7 7dpi; 17.2 10 dpi, 26.4 13 dpi; | ||||
| IR tested in heads at 26 °C 26.5 7dpi; 17.6 10 dpi, 20.6 13 dpi; | ||||
| IR tested in heads at 32 °C 25.3 7dpi; 8 10 dpi, 23 13 dpi; | ||||
| Richard et al. (2016b) | districts of Toahotu, Thaiti Island | PF14/300914-109 | BM4, 7 log10 TCID50/mL | IR 78 at 6 dpi, 87 at 9 dpi, 90 at 14 dpi, 80 at 21 dpi |
| TR 5 at 2 dpi, 18 at 6 dpi, 34 at 9 dpi, 49 at 14 dpi abd 53 at 21 dpi | ||||
| Vega-Rua et al. (2014) | Vero Beach, FL | CHIKV 06.21 | BM 107.5 | IR 100 at 7 dpi, 100 at 10 dpi |
| CHIKV 05.115 | BM 107.5 | IR 100 | ||
| Chiapas, MX | CHIKV 06.21 | BM 107.5 | IR 96.7 at 7 dpi, 93.3 at 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 96.7 at 7 dpi, 100 at 10 dpi | ||
| Panama | CHIKV 06.21 | BM 107.5 | IR 96.7 at 7 dpi, 100 at 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 96.7 at 7 and 10 dpi | ||
| NC/2011-568 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Delta Amacuro, Venezuela | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Tumbes, Peru | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| Punchana, Peru | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Manaus, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| NC/2011-568 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Santarem, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| Parnamirin, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| Campos Belos,BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| Campos Grande, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Jurujuba, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Paqueta, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Vaz Lobo, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 dpi; 96,7 at 10 dpi | |
| Belford Roxo, BR | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| Santos, BR | CHIKV 06.21 | BM 107.5 | IR 93.3 at 7 dpi, 100 at 10 dpi | |
| Monteagudo, Bolivia | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Salto del Guaira, Paraguay | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| Asuncion, Paraguay | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 96.7 at 7 dpi, 93.3 at 10 dpi | ||
| Salto, Uruguay | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 and 10 dpi | ||
| Corrientes, Argentina | CHIKV 06.21 | BM 107.5 | IR 100 at 7 and 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 100 at 7 dpi, 96.7 at 10 dpi | ||
| Buenos Aires, Argentina | CHIKV 06.21 | BM 107.5 | IR 100 at 7 dpi, 96.7 at 10 dpi | |
| CHIKV 05.115 | BM 107.5 | IR 96.6 at 7 dpi, 100 at 10 dpi | ||
| NC/2011-568 | BM 107.5 | IR 96.9 at 7 dpi, 90 at 7 dpi | ||
| Dupont-Rouzeyrol et al. (2012) | Noumea, NC, mosquitoes had a 92% susceptibility to pyrethroids (pop 163/11) | NC/2011-568 | BM 107.5 | IR 53.3 at 3 dpi; 54.5 at 8 dpi; 66.7 at 14 dpi |
| Noumea, New Caledonia, mosquitoes had a 85% susceptibility to pyrethroids (pop 174/11) | BM 107.5 | IR 50 at 3 dpi; 64.3 at 8 dpi; 20 at 14 dpi | ||
| Noumea Laboratory strain, New Caledonia (pop 282/10) | BM 107.5 | IR 40 at 3 dpi; 58.8 at 8 dpi; 50 at 14 dpi | ||
| Noumea, NC, mosquitoes had a 92% susceptibility to pyrethroids (pop 163/11) | CHIKV-RE from Reunion Island (2005), also known as CHIKV 06.21 | BM 107.5 | IR 33.3 at 3 dpi; 57.1 at 8 dpi; 75 at 14 dpi | |
| Noumea, NC, mosquitoes had a 85% susceptibility to pyrethroids (pop 174/11) | BM 107.5 | IR 73.3 at 3 dpi; 46.2 at 8 dpi; 90 at 14 dpi | ||
| Noumea Lab.strain, NC(pop 282/10) | BM 107.5 | IR 40 at 3 dpi; 57.1 at 8 dpi; 66.7 at 14 dpi | ||
| Girod et al. (2011)15 | Pointe a Pitre, Carenage, Guadaloupe | CHIKV 06.21 | BM, 107.5 | IR 98 at 14 dpi in 2008; 96.6 at 7 dpi and 100 at 14 dpi in 2009 |
| Petit bourg, Prise d'eau, Guadalupe | CHIKV 06.21 | BM, 107.5 | IR 95.8 at 14 dpi in 2008; 97.9 at 14 dpi in 2009 | |
| Fort de France, Ermitage, Martinique | CHIKV 06.21 | BM, 107.5 | IR 98.9 at 14 dpi in 2008; 100 at 7 dpi and 96.8 at 14 dpi in 2009 | |
| Robert, Cafe, Martinique | CHIKV 06.21 | BM, 107.5 | IR 97.4 at 14 dpi in 2008; 88.9 at 7 dpi and 93.4 at 14 dpi in 2009 | |
| Cayenne, Centre Ville FG | CHIKV 06.21 | BM, 107.5 | IR 100 at 14 dpi in 2008; 97.5 at 7 dpi and 95.5 at 14 dpi in 2009 | |
| Cayenne, Madeleine, FG | CHIKV 06.21 | BM, 107.5 | IR 98.8 at 14 dpi in 2008; 94.7 at 7 dpi and 98.5 at 14 dpi in 2009 | |
| Pesko et al. (2009) | Palm Beach, FL | CHICK LR2006-OPY1, La Reunion Island | BM, 6.1 log10 | IR at 6 dpi 18.8 and 57.7 for mosquitoes feeding on pletdgets or water jackets membranes, respectively |
| BM, 5.2 log10 | IR at 6 dpi 4.5 and 23.8 for mosquitoes feeding on pletdgets or water jackets membranes, respectively | |||
| BM, 4.4 log10 | IR at 6 dpi 0 and 3.1 for mosquitoes feeding on pletdgets or water jackets membranes, respectively | |||
| BM, 3.6 og10 | IR at 6 dpi 0 and 0 for mosquitoes feeding on pletdgets or water jackets membranes, respectively results | |||
| 5) dual-infections | ||||
| Rückert et al. (2017)16 | Poza Rica, Mexico | CHIKV (strain 99,659) | BM 3.1×104-1.9×105 | IR 87; TR 20 at 3dpi, 30 at 7 dpi, 60 at 14 dpi |
| DENV-2 (strain Merida) | BM 3×103-7.4×105 | IR 87; TR 0 at 3 dpi, 15 at 7 dpi, 20 at 14 dpi | ||
| ZIKV (strain PRVABC59) | BM 1.7×104-5.4×105 | IR 48; TR 0 at 3 dpi, 8 at 7 dpi, 40 at 14 dpi | ||
| CHIKV (strain 99,659) + DENV-2 (strain Merida) | BM, as single | IR CHIKV 87; DENV-2 85; TR at 3 dpi CHIKV 10; DENV 0; at 7 dpi CHIKV 38; DENV 10; at 14dpi CHIKV 30, DENV 18 | ||
| CHIKV (strain 99,659) + ZIKV (strain PRVABC59) | BM, as single | IR CHIKV 90; ZIKV 45; TR at 3 dpi CHIKV 28; ZIKV 5; at 7 dpi CHIKV 45; ZIKV 8; at 14dpi CHIKV 40, ZIKV 38 | ||
| ZIKV (strain PRVABC59) + DENV-2 (strain Merida) | BM, as single | IR ZIKV 50; DENV-2 80; TR at 3 dpi DENV 028; ZIKV 0; at 7 dpi DENV 20; ZIKV 0; at 14dpi DENV 38, ZIKV 20 | ||
| Göertz et al. (2017) | Rockefeller strain | CHIKV strain 37,997 | BM 2×105 | IR 47.9, TR 10.4 |
| BM 2×106 | IR 66.7, TR 5.9 | |||
| BM 2×107 | IR 81.2, TR 21.2 | |||
| ZIK Suriname strain 011 V-01621 | BM 2×105 | IR 65.3, TR 34.7 | ||
| BM 2×106 | IR 92.2, TR 68.6 | |||
| BM 2×107 | IR 100, TR 68.3 | |||
| CHIKV (strain 37,997) + ZIKV Suriname strain | BM, as single | IR 84.4; TR 11.5 | ||
| 6) infections with arboviruses other than DENVs, YFV, ZIKV and CHIKV | ||||
| Wiggins et al. (2018)12 | Miami, FL | Mayaro virus, Tridinad strain TRVL 4675 | BM 7.5 log10 | IR 65 at 6 dpi; 80 at 6 dpi; 70 at 9-12 dpi; DR 44 at 3 dpi; 60 at 6 dpi; 80 at 9 dpi-12 dpi; TR < 10 at 3-9 dpi; 25 at 12 dpi |
| Wang et al. (2012) | Haikou strain, Chi | Western equine encephalomyelitis virus (WEEV), McMillian strain | BM, ntd | IR 25; TR 45 |
| Long et al. (2011) | Iquitos, Peru | Maroyo virus, strain IQT4235 | BM, 5.59-7.34 Log10 | IR 46.67±21.13; TR 83 +/− 23.44 |
| BM, 5.57-3.36 Log10 | IR 0.46 +/− 1.13; | |||
| Turell et al. (2007) | Kenya, collected as eggs in 1982 | Rift Valley Fever (RVFV) ZH501 from an Egyptian patient | BM, 〰107-7.8 | IR 100 at 3-10 dpi; 33 at 11-16 dpi |
| Rift Valley Fever ZH501 from an Egyptian patient | BM, 〰10 > 8 | IR 85 at 3-10 dpi; 75 at 11-16 dpi | ||
| Turell et al. (2001) | Rockefeller strain | West Nile virus Crow 397-99 | BM 107.2 | IR 16, TR < 16 |
| Kay et al. (1979) | Townsville colony, from northern Queesland in 1957 | Sindbis MRM39 | ||
| Getah N544 | BM, 4-6.5 Log ID50 | IR 64, TR 28.5, EIP 20 | ||
| Ross River T78 | BM, 4.9 Log ID50 | IR 100, TR 69, EIP 12 | ||
| Murray Valley Encephalitis MRM66 | BM, 5.1 Log ID50 | IR 96, TR 95, EIP 7-10 | ||
| Kunji MRM16 | BM, > 6.5 Log ID50 | IR 46, TR 38, EIP 20-27 | ||
| Kokobera MRM32 | BM, 4.2 Log ID50 | IR 100, TR 100, EIP 12 | ||
| Edge Hill C281 | BM, 2.7 Log ID50 | IR 89, TR 80, EIP 20 | ||
| Alfuy MRM3929 | BM, > 5.5 Log ID50 | IR 47, TR 21, EIP 10-15 | ||
| Corriparta MRM1 | BM, 2.1-2.9 Log ID50 | IR 100, TR 5, EIP 10-15 | ||
| Belmont Ch9824 | BM, ntd | IR 0, TR 0 | ||
| Ngaingan MRM14556 | BM, ntd | IR 10, TR 0 | ||
| CHIKV BKMS 459/64 | BM, 4.7 Log ID50 | IR 71, TR 57, EIP 15 | ||
| Kramer and Scherer (1976) | Laboratory strain | Venezuelan Encephalitis virus, epizootic strain subytoe I, variety B, 69TI597 | IT or BM | TR 60 at 14 dpi, 100 at 17 dpi, 50 at 21 and 27 dpi |
| Venezuelan Encephalitis virus, enzootic strain subytoe I, variety E, 63Z1 | IT or BM | TR 0 at all time points | ||
Abbreviations: BM, mosquitoes offered an infectious blood-meal; IT, mosquitoes were infected by intrathoracic inoculation; dpi, days post infection; IR, percentage of engorged females with viral particles in the head, legs and/or salivary glands; TR, transmission rate calculated as percentage of engorged females with viral particles in the saliva at 14 dpi, unless otherwise stated; PFU, plaque forming units, FFU, fluorescent focus unit, LD50, 50 infectious dose; TCID50, 50 tissue culture infectious dose; MID50, mosquito infectious dose for 50 of Ae. aegypti individuals; EIP, extrinsic incubation period; MX, Mexico; NC, New Caledonia; Col, Colombia; Viet, Vietnam; NG, New Guinea; FG, French Guiana; Thai, Thailand; S, S; PR, PR; BR, Brazil; Aus, Australia; Chi, China; Philippines, Phi; FL, Florida; South Africa, SA; Texas, TX; California, CA; isol., isolate; human serum, hs; lab. Strain, laboratory strain.
PFU/ml unless otherwise stated
FFU/ml
MID50/ml
TCID50/mL
CCID50/ml
PFU ingested per mosquito
expressed in unless otherwise stated
mosquitoes were tested for infections within the 9th generation after laboratory colonization
Infection and transmission rates reported here were extrapolated from a figure
wild-caught mosquitoes were adapted to the laboratory and tested at generation F10-15
Infection rates for DENV2 AdR 140,875 are mean over two infections experiments
results are mean over different experiments
mosquitoes were infected by all viruses strains and dissemination was studied for both strains
CHIKV 06.21 is the strain with the E1-226 V mutation and CHIKV 05.115 is the strain with the E1-226A mutation
experiments were carried out in two consecutive years (2008 and 2009); in 2009, two different concentrations of CHIKV were compared for infection rates at 7 dpi; only data for the highest concentration are shown here
mosquitoes of the F12_F14 after laboratory colonization were used in experimental infections.
1.2. Vector competence of Ae. aegypti populations for arboviruses
Despite the lack of uniformity in the procedures to test for vector competence and a focus on sampling mosquitoes in geographic areas with endemic arboviral infections or with significant epidemics (i.e. Thailand, Vietnam, New Caledonia, Mexico, Brazil, Florida, La Reunion island and Senegal), review of literature on infection, dissemination and transmission rates of arboviruses by Ae. aegypti mosquitoes support some general conclusions, data in Table 1. (1) Cases of complete refractoriness to arboviral infection are rare (Kay et al., 1979; Rosen et al., 1985; Diallo et al., 2008; Dickson et al., 2014; Agha et al., 2017). (2) Complete susceptibility to infection has been detected for Ae. aegypti populations from New Caledonia, Thailand, Australia, South Africa for DENVs; for Ae. aegypti populations from Dominican Republic, Brazil, China and Singapore for ZIKV; for populations from Mexico and Guadaloupe for CHIKV (Girod et al., 2011; Vega-Ruiz et al., 2014), but complete susceptibility was not observed for any population tested for YFV (Table 1); (3) Initial infection dose of virus positively correlates with infection rate. (4) Brazilian populations of Ae. aegypti are particularly susceptible to DENV-2 (Goncalves et al., 2014; Carvalho-Leandro et al., 2012; Lourenco-De-Oliveira et al., 2004). (5) The African lineage of ZIKV was shown to be more infective to Ae. aegytpi mosquitoes from the American continent than the ZIKV Asian lineage (Weger-Lucarelli et al., 2016; Roundy et al., 2017). (5) Virus adaptation to different mosquito species appears an important evolutionary force for CHIKV evolution, but its role in DENVs evolution is still controversial (Lambrechts et al., 2009; Tsetsarkin et al., 2011; Fansiri et al., 2016). The best-known example of vector-driven adaptation in an arbovirus is the emergence on La Reunion in 2005 of the A226V amino acid substitution in the E1 envelope glycoprotein of CHIKV that favors its replication in Aedes albopictus mosquitoes (Tsetsarkin et al., 2011). (6) Limited data are available on co-infections with different viruses or serotypes/genotypes of one viral species. Some co-infection experiments suggest competitive displacement of DENV-4 over DENV-1 (Vazeille et al., 2016) or superinfection interference (Muturi et al., 2017). Other studies indicate that Ae. aegypti infection with one arbovirus (i.e. CHIKV, DENV2 or ZIKV) only mildly affects infection with a subsequent infection with another (Rückert et al., 2017).
The most obvious and well accepted observation from reviewing literature on vector competence in Ae. aegypti is that there is great variability in susceptibility to arboviral infections across geographic populations and even for the same population with different viral species and strains; this variability includes comparisons between the domestic Ae. aegypti aegypti and the sylvatic Ae. aegypti formosus with respect to DENVs infections (Bosio et al., 1998; Gaye et al., 2014; Dickson et al., 2014). The great variation among geographic populations of mosquito is likely due to the fact that vector competence is a complex and evolving phenotype dependent on the tri-partite interaction among the host (i.e. mosquito), the pathogen, and host symbionts (Vasilakis and Tesh, 2015; Hedge et al., 2015). The high genetic structure among Ae. aegypti populations is also a likely contributing factor. This variation across populations suggests that the co-evolution between Ae. aegypti and arboviruses did not favor a single pathway/factor in the mosquito, likely because exposure to arboviral infection is the accidental consequence of hematophagy the primary purpose of which is to support egg development. Furthermore, it is unclear how great, or even if there is, any fitness cost to mosquitoes to transmit these viruses (see e.g., Padilha et al., 2018). Selection-driven variation is more likely to be on the virus.
Specific physiological and genetic factors in mosquitoes contributing to vector competence have been thoroughly reviewed elsewhere (Franz et al., 2015; Pando-Robles and Batista, 2017; Wang et al., 2017; Palmer et al., 2018).
1.3. Microbiota and vector competence
The gut of mosquitoes is colonized by a resident microbiota which influences key physiological processes related to pathogen transmission (Guégan et al., 2018; Pike et al., 2017). In Ae. aegypti, DENVs replication is significantly affected by gut bacterial flora (Xi et al., 2008; Ramirez et al., 2014), the depletion of which by antibiotics renders mosquitoes more susceptible (Xi et al., 2008). Oral reintroduction of specific bacterial species into the adult mosquito midgut results in decreased viral load in the vector (Ramirez et al., 2012, 2014). Mosquito gut bacteria are presumed to exert antiviral activity through either direct or indirect mechanisms (Dennison et al., 2014; Saraiva et al., 2016; Guégan et al., 2018). While these mechanisms are not completely understood, recent studies have demonstrated that indirect mechanisms rely mainly on the basal level activation of innate antiviral responses and antimicrobial peptides (AMPs) by the gut microbiota (Xi et al., 2008; Ramirez et al., 2012). On the other hand, antiviral activity may be directly mediated by bacterial antiviral compounds (Ramirez et al., 2014). Indeed, a Chromobacterium sp. isolated from the Ae. aegypti midgut in Panama (Csp_P) produces an aminopeptidase that can bind to envelope protein of DENVs and prevent viral attachment and further invasion/replication within the host cell (Saraiva et al., 2018). Interestingly, the same bacterium has been shown to be pathogenic to both Ae. aegypti and An. gambiae (Ramirez et al., 2014) via the production of hydrogen cyanide (Short et al., 2018). Besides, it is important to consider the massive increase of bacteria in the midgut of mosquito vectors after a blood meal, and the interference with physiological processes related to the control of midgut homeostasis, such as the production of Reactive Oxygen Species (ROS) and the peritrophic matrix (Kumar et al., 2010; Oliveira et al., 2011; Rodgers et al., 2017). These processes may potentially affect mosquito vector competence and should be further investigated.
The environment, especially the larval breeding water, is pivotal in determining the mosquito gut microbiota composition (Coon et al., 2014; Duguma et al., 2015; Gimonneau et al., 2014), which varies considerably among local habitats of geographically distinct populations (Coon et al., 2016). Most of the diversity found in the Ae. aegypti larvae gut is also present in the water where mosquitoes developed, with about half of it being transtadially transferred from larvae to adults (Coon et al., 2014). In addition to the environment, the mosquito genetic background also likely influences gut microbial diversity. While the mechanisms surrounding this interplay are largely unknown, concomitant decreases in both mosquito and bacterial genetic diversity have been observed in Ae. albopictus populations recently introduced in France (Minard et al., 2015).
It remains an open question of whether (and how) the gut microbial diversity influences mosquito competence to transmit human pathogenic arboviruses. Is the difference in vector competence among distinct mosquito populations due to their intrinsic microbiomes or genetic differences in the mosquitoes or, most likely, a combination/interaction of both factors? In this context, assessment of the gut bacteria repertoire of the genetically-selected DENV-resistant (MOYO-R) and -susceptible (MOYO-R) Ae. aegypti strains, identified some bacterial genera exclusively in either the resistant or in the susceptible strain (Charan et al., 2013). More recently, bacteria from the families Rhodobacteriaceae and Desulfuromonadaceae have been described as potential biomarkers of ZIKV infection in Ae. aegypti (Villegas et al., 2018). Exposure of germ-free Ae. aegypti larvae to different microbiota-derived bacterial species has been shown to result in variation in several mosquito life-history traits, including the load of DENVs disseminated to the insect head (Dickson et al., 2017). While these studies provide important insights on the interplay between mosquito microbiomes and vector competence, the relative contribution of mosquito genetics and its microbiome in the control of vector competence remains to be elucidated. This will almost certainly be key for understanding fundamental aspects of the variation in arbovirus transmission by different populations of Ae. aegypti.
1.4. Viriome and vector competence
The recent explosion of metagenomics studies led to the discovery of novel viral species, which are insect-specific and not able to replicate in vertebrate cells despite being phylogenetically-related to arboviruses (Vasilakis and Tesh, 2015; Bolling et al., 2015; Roundy et al., 2017). Insect-Specific Viruses (ISVs) identified so far in Ae. aegypti mosquitoes belong primarily to the Flaviviridae family, followed by the Negoviridae and Bunyaviridae families (Vasilakis and Tesh, 2015; Bolling et al., 2015; Hall et al., 2017). While the landscape of ISVs and their prevalence in natural mosquito populations vary greatly, the cell fusing agent virus (CFAV) appears to be the most common ISV in field-collected Ae. aegypti (Cook et al., 2006; Hall et al., 2017). Interestingly, CFAV transmits vertically and is absent in saliva and salivary glands of Ae. aegypti (Guegan et al., 2018). The impact of CFAV on Ae. aegypti vector competence has not been investigated yet, but heterologous interference was seen between Eilat virus and CHIKV in Ae. aegypti (Nasar et al., 2015). Eilat virus is an ISV of the Alphavirus genus, which was first isolated in Anopheles constani mosquitoes from Israel (Nasar et al., 2014). It readily infects Ae. aegypti (Nasar et al., 2014) and when used to infect mosquitoes prior to CHIKV infection, it delays CHIKV dissemination by 3 days (Nasar et al., 2015). Furthermore, it is possible that ISVs influence, to some extent, the mosquito's innate immune response, which could directly impact viral replication and the gut microbial diversity. These studies underscore the importance of expanding our knowledge of the viriome (the set of viruses in an organism) and highlight its possible application for the control of arboviral infections within mosquitoes (Hall et al., 2017).
Interaction between viruses and mosquitoes may include horizontal transfer of genetic material. The genome of Ae. aegypti is rich in sequences with similarities to ISVs of the Flavivirus and Rhabdovirus genera and Chuviruses (Chen et al., 2015; Palatini et al., 2017; Whitfiled et al., 2017). Sequences of viral origin are statistically enriched in piRNA clusters and encode for piRNAs, suggesting that they may function analogously to transposable element fragments within the piRNA pathway (Palatini et al., 2017, Whitefiled et al., 2017). In light of this, it has been proposed that viral integrations constitute a heritable immune signal and thus could be an additional factor shaping mosquito vector competence (Olson and Bonizzoni, 2017; Palatini et al., 2017; Whitfield et al., 2017).
2. Conclusions and perspective
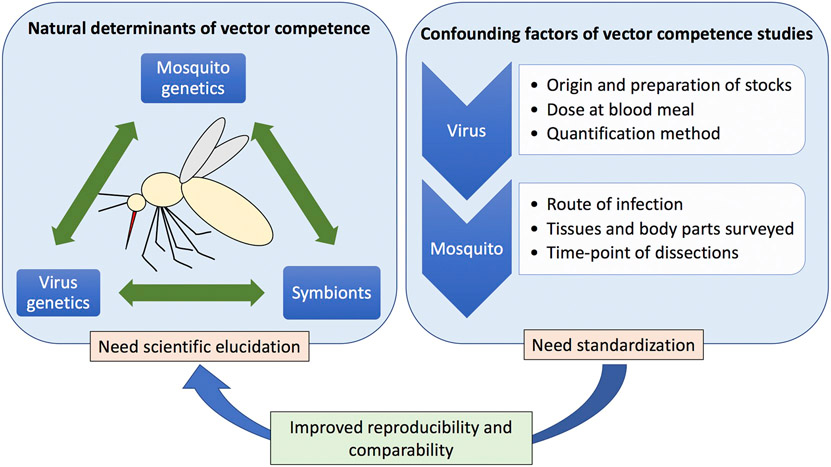
The recent emergence and spread of Zika, the current re-emergence of yellow fever in Brazil and Africa, the emergence of dengue in Europe, and the expansion of chikungunya to the New World brought vector-borne diseases to public attentions and fostered research. Despite great progress in the understanding of the interplay between arboviruses and vectors, the genetic and environmental elements that control vector competence in Ae. aegypti populations have yet to be fully elucidated. Here we reviewed historical and modern data on factors influencing vector competence in Ae. aegypti populations to four of the most prevalent arboviruses (i.e. DENVs, YFV, ZIKV and CHIKV). We identified no clear-cut distinctive natural factors associated with variation in vector competence among mosquito populations and/or viral species due primarily to the heterogeneity of materials (strains of mosquito and virus) and methods used in different studies. This highlights the need to standardize surveillance and laboratory procedures for assessing vector competence and to expand the range of mosquito populations and viral strains (and serotypes) tested (Fig. 1). While workers target populations and virus strains of interest to them, at the very least procedures to determine what are reported as infection rate, dissemination rate, and transmission rate should be standardized.
Fig. 1.
Natural and technical confounding factors related to arbovirus vector competence studies in Aedes aegypti. Despite progress in the understanding of the interplay between arboviruses and vectors, the genetic and environmental elements that control vector competence in Ae. aegypti populations have yet to be fully understood. Further elucidation is needed especially of co-evolutionary processes between arboviruses and vectors, as well as their symbionts. On the other hand, procedures used in vector competence studies should be standardized in order to improve reproducibility and comparability of scientific outputs. Together these will result in better understanding of genetic and microbial factors influencing arboviral transmission, which can lead to the development of new public health interventions.
While there is a clear influence of the microbiota on arboviral infection, the relative importance of mosquito genetics and microbial diversity, including the interplay between these factors, on vector competence remains largely unknown and deserves attention from the scientific community.
Acquisition of arboviruses by mosquitoes is a by-product of blood-feeding, which is a necessary physiological process for egg production. Even during active arboviral epidemics, the frequency of mosquitoes infected with the pathogenic virus is usually around 1%, but can vary from 0.05% to > 10% (Chow et al., 1998; Pham Thi et al., 2017; Perez-Castro et al., 2016; Medeiros et al., 2018). In addition to these human pathogenic viruses, blood-feeding exposes mosquitoes to a broad range of entities, including bacteria, fungi and other symbionts and parasites. Considering the essential role of blood-feeding, mosquitoes must be able to withstand these microbial challenges to survive. In this context, co-evolution between mosquitoes and viruses should be viewed as a by-product of diverse and possibly broad-range physiological processes. Some of these interactions may be deterministic and selection-driven while others may be stochastic (e.g., genetic drift) or indirect. In any case, it is clear that the genetic heterogeneity both within and among mosquito populations need to be considered in any attempts to identify genetic elements contributing to vector competence for arboviruses.
These studies have both basic science and applied importance. Unravelling the genetic components of vector competence means investigating the co-evolutionary processes between arboviruses and vectors, with the potential to identify factors that may be co-opted for genetic-based vector control strategies or identify steps in the transition from ISVs to arbovirus capable of infecting vertebraes. This should be possible in light of the fact that some ISVs are phylogeneticly ancestral to arboviruses in the same virus family (Marklewitz et al., 2015). Additionally, a better knowledge of the variability and interaction between mosquitoes and their microbiota could lead to novel vector control methods based on native and introduced mosquito symbionts (i.e. Asaia and Wolbachia spp.) (Ritchie et al., 2018).
Acknowledgements
We are grateful to Patrizia Chiari (University of Pavia) for providing assistance with the manuscript. This work was supported by the Human Frontier Science Program Research Grant RGP0007/2017 to M.B. and J.A.S.N.; by the Italian Ministry of Education, University and Research FARE project R1623HZAH5 to M.B.; by the São Paulo Research Foundation (FAPESP), Young Investigator Award 2013/11343-6 to J.A.S.N. J.R.P.'s research is supported by the US National Institutes of Health, NIAID.
References
- Agha SB, Chepkorir E, Mulwa F, Tigoi C, Arum S, Guarido MM, et al. , 2017. Vector competence of populations of Aedes aegypti from three distinct cities in Kenya for chikungunya virus. PLoS Negl. Trop. Dis 11, e0005860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto BW, Smartt CT, Shin D, Bettinardi D, Malicoate J, Anderson SL, et al. , 2014. Susceptibility of Florida Aedes aegypti and Aedes albopictus to dengue viruses from Puerto Rico. J. Vec. Ecol 39, 406–413. [DOI] [PubMed] [Google Scholar]
- Alto BW, Wiggins K, Eastmond B, Velez D, Lounibos LP, et al. , 2017. Transmission risk of two chikungunya lineages by invasive mosquito vectors from Florida and the Dominican Republic. PLoS Neg. Trop. Dis 11, e005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft TL, 1906. On the aetiology of dengue fever. Aust. Med. Gaz 25, 17–18. [Google Scholar]
- Beck AS, Barrett AD, 2015. Current status and future prospects of yellow fever vaccines. Expert Rev. Vaccines 14, 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett K, Olson K, Muno M, Fernandez-Salas I, Farfan J, et al. , 2002. Variation in vector competence for dengue-2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am. J. Trop. Med. Hyg 67, 84–92. [DOI] [PubMed] [Google Scholar]
- Boccolini D, Toma L, Luca M, Severini F, Romi R, Remoli ME, Sabbatucci M, Venturi G, Rezza G, Fortuna C, 2016. Euro. Surveill 21, 30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling BG, Weaver SC, Tesh RB, Vasilakis N, 2015. Insect-specific Virus Discovery: significance for the Arbovirus Community. Viruses 7, 4911–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorman JP, Porterfield JS, 1956. A simple technique for infection of mosquitoes with viruses; transmission of Zika virus. Trans. R. Soc. Trop. Med. Hyg 50, 238–242. [DOI] [PubMed] [Google Scholar]
- Boromisa RD, Rai KS, Grimstad PR, 1987. Variation in the vector competence of geographic strains of the Aedes albopictus for Dengue 1 virus. J. Am. Mos. Cont. Ass 3, 378–386. [PubMed] [Google Scholar]
- Bosio CF, Beaty BJ, Black WC, 1998. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am. J. Trop. Med. Hyg 59, 965–970. [DOI] [PubMed] [Google Scholar]
- Buckner EA, Alto BW, Lounibus PL, 2013. Vertical Transmission of Key West Dengue-1 Virus by Aedes aegypti and Aedes albopictus (Diptera: Culicidae) Mosquitoes from Florida. J. Med. Entomol 31, 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvez E, Guillaumot L, Girault D, Richard V, O'Connor O, Paoaafaite T, et al. , 2017. Dengue-1 virus and vector competence of Aedes aegypti (Diptera: Culicidae) populations from New Caledonia. Parasit. Vectors 10, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvez E, Mousson L, Vazeille M, O'Connor O, Cao-Lormeau VM, Mathieu-Daudé F, Pocquet N, Failloux AB, Dupont-Rouzeyrol M, 2018. Zika virus outbreak in the Pacific: Vector competence of regional vectors. PLoS Negl. Trop. Dis 12, e0006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington LB, Seifert SN, Armijos MV, Lambrechts L, Scott TW, 2013. Reduction of Aedes aegypti vector competence for dengue virus under large temperature fluctuations. Am J Trop Med Hyg 88, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Leandro D, Ayres CFJ, Guedes DRD, Suesdek L, Melo-Santos MAV, Oliveira CF, et al. , 2012. Immune transcript variations among Aedes aegypti populations with distinct susceptibility to dengue virus serotype 2. Acta Trop. 124, 113–119. [DOI] [PubMed] [Google Scholar]
- Charan SS, Pawar KD, Severson DW, Patole MS, Shouche YS, 2013. Comparative analysis of midgut bacterial communities of Aedes aegypti mosquito strains varying in vector competence to dengue virus. Parasitol. Res 112, 2627–2637. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Wei HL, Hsu EL, Chen ER, 1993. Vector competence of Aedes albopictus and Ae. aegypti (Diptera: Culicidae) to dengue 1 virus on Taiwan: development of the virus in orally and parenterally infected mosquitoes. J. Med. Entomol 30, 524–530. [DOI] [PubMed] [Google Scholar]
- Chen XG, Jiang X, Gu J, Xu M, Wu Y, Deng Y, Zhang C, Bonizzoni M, Dermauw W, Vontas J, Armbruster P, Huang X, Yang Y, Zhang H, He W, Peng H, Liu Y, Wu K, Chen J, Liraki M, Topalis P, Van Leeuwen T, Hall AB, Jiang X, Thorpe C, Mueller RL, Sun C, Waterhouse RM, Yan G, Tu ZJ, Fang X, James AA, 2015. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc. Natl. Acad. Sci. U. S. A 112, E5907–E5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepkorir E, Lutomiah J, Mutisya J, Mulwa F, Orindi B, et al. , 2014. The vector competence of Ae. aegypti mosquito populations from Kilifi and Nairobi for dengue-2 virus and the effect of temperature. Inter J Infect Dis 21, 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouin-Carneiro T, Vega-Rua A, Vazeille M, et al. , 2016. Differential susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika virus. PLoS Negl. Trop. Dis 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow VT, Chan YC, Yong R, Lee KM, Lim LK, Chung YK, Lam-Phua SG, Tan BT, 1998. Monitoring of dengue viruses in field-caught Aedes aegypti and Aedes albopictus mosquitoes by a type-specific polymerase chain reaction and cycle sequencing. Am J Trop Med Hyg 58, 578–586. [DOI] [PubMed] [Google Scholar]
- Ciota AT, Bialosuknia SM, Zink SD, et al. , 2017. Effects of Zika virus strain and Aedes mosquito species on vector competence. Emerg. Infect. Dis 23, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleton N, Koopmans M, Reimerink J, Godeke GJ, Reusken C, 2012. Come fly with me: review of clinically important arboviruses for global travelers. J. Clin. Virol 55, 191–203. [DOI] [PubMed] [Google Scholar]
- Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballeri X, 2006. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol 87, 735–748. [DOI] [PubMed] [Google Scholar]
- Coon KL, Vogel KJ, Brown MR, Strand MR, 2014. Mosquitoes rely on their gut microbiota for development. Mol. Ecol 23, 2727–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon KL, Brown MR, Strand MR, 2016. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol 25, 5806–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet M, Robin Y, 1979. Transmission experimentale comparee du virus Zika chez Aedes aegypti. Ent Med Parasitol 17, 47–53. [Google Scholar]
- Costa-Da-Silva AL, Ioshino RS, De Araújo HRC, Kojin BB, De Andrade Zanotto PM, et al. , 2017. Laboratory strains of Aedes aegypti are competent to Brazilian Zika virus. PLoS One 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto-Lima D, Madec Y, Bersot MI, Campos SS, Motta MDA, Dos Santos FB, et al. , 2017. Potential risk of re-emergence of urban transmission of Yellow fever virus in Brazil facilitated by competent Aedes populations. Sci. Rep 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Moura AJF, De Melo Santos MAV, Oliveira CMF, Guedes DRD, De Carvalho-Leandro D, et al. , 2015. Vector competence of the Aedes aegypti population from Santiago island, Cape Verde, to different serotypes of dengue virus. Parasit. Vectors 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison NJ, Jupatanakul N, Dimopoulos G, 2014. The mosquito microbiota influences vector competence for human pathogens. Curr Opin Insect Sci 3, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Luca M, Severini F, Toma L, et al. , 2016. Experimental studies of susceptibility of Italian Aedes albopictus to Zika virus. Euro Surveill. 21. [DOI] [PubMed] [Google Scholar]
- Diagne CT, Diallo D, Faye O, et al. , 2015. Potential of selected Senegalese Aedes spp. mosquitoes (Diptera: Culicidae) to transmit Zika virus. BMC Infect. Dis. 15, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo M, Ba Y, Faye O, Soumare ML, Dia I, Sall AA, 2008. Vector competence of Aedes aegypti populations from Senegal for sylvatic and epidemic dengue 2 virus isolated in West Africa. Trans of Royal Soc Trop Med Hyg 102, 493–498. [DOI] [PubMed] [Google Scholar]
- Dickson LB, Sanchez-Vargas I, Sylla M, Fleming K, Black WC, 2014. Vector Competence in West African Aedes aegypti is Flavivirus Species and Genotype Dependent. PLoS Negl. Trop. Dis 8, e3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson LB, Jiolle D, Minard G, Moltini-Conclois I, Volant S, Ghozlane A, Bouchier C, Ayala D, Paupy C, Valiente Moro C, Lambrechts L, 2017. Carryover effects of larval exposure to different environmental bacteria drive adult trait variation in a mosquito vector. Sci. Adv 3, e1700585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson BL, Pujhari S, Rasgon JL, 2018. Vector competence of selected north American Anopheles and Culex mosquitoes for Zika virus. PeerJ 6, e4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle M, Mousson L, Moutailler S, Vazeille M, Failloux AB, 2009. Chikungunya Virus and Aedes Mosquitoes: Saliva is Infectious as soon as two days after Oral Infection. PLoS One 4, e5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguma D, Hall MW, Rugman-Jones P, Stouthamer R, Terenius O, Neufeld JD, Walton WE, 2015. Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiol. 15, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont-Rouzeyrol M, Caro V, Guillaumot L, Vazeille M, D'Ortenzio E, et al. , 2012. Chikungunya Virus and the Mosquito Vector Aedes aegypti in New Caledonia (South Pacific Region). Vector-Borne Zoon Dis 12, 1036–1041. [DOI] [PubMed] [Google Scholar]
- Dutra HL, Rocha MN, Dias FB, Mansur SB, Caragata EP, et al. , 2016. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe 19, 771–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis BR, Sang RC, Horne KM, Higgs S, Wesso DM, 2012. Yellow fever virus susceptibility of two mosquito vectors from Kenya, East Africa. Trans Royal Soc Trop Med Hyg 106, 387–389. [DOI] [PubMed] [Google Scholar]
- Fansiri T, Pongsiri A, Klungthong C, Ponlawat A, Thaisomboonsuk B, Jarman RG, et al. , 2016. No evidence for local adaptation of dengue viruses to mosquito vector populations in Thailand. Evol. Appl 9, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes RS, Campos SS, Ribeiro PS, Raphael LM, Bonaldo MC, Lourenço-De-Oliveira R, 2017. Culex quinquefasciatus from areas with the highest incidence of microcephaly associated with Zika virus infections in the Northeast Region of Brazil are refractory to the virus. Mem. Inst. Oswaldo Cruz 112, 577–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish D, 2008. Why we do not understand the ecological connections between the environment and human health: the case for vector-borne disease. Vector Borne Dis 2008, 65–69. [Google Scholar]
- Franz AWE, Kantor AM, Passarelli AL, Clem RJ, 2015. Tissues barriers to arbovirus infection in mosquitoes. Viruses 7, 3741–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Luna SM, Weger-Lucarelli J, Rückert C, Murrieta RA, Young MC, Byas AD, Fauver JR, Perera R, Flores-Suarez AE, Ponce-Garcia G, Rodriguez AD, Ebel GD, Black WC 4th, 2018. Variation in competence for ZIKV transmission by Aedes aegypti and Aedes albopictus in Mexico. PLoS Negl. Trop. Dis 12, e0006599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaye A, Faye O, Diagne CT, Faye O, Diallo D, Weaver SC, et al. , 2014. Oral susceptibility of Aedes aegypti (Diptera: Culicidae) from Senegal for dengue serotypes 1 and 3 viruses. Tropical Med. Int. Health 19, 1355–1359. [DOI] [PubMed] [Google Scholar]
- Gimonneau G, Tchioffo MT, Abate L, Boissière A, Awono-Ambene PH, Nsango SE, Christen R, Morlais I, 2014. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect. Genet. Evol 28, 715–724. [DOI] [PubMed] [Google Scholar]
- Girod R, Gaborit P, Marrama L, Etienne M, Ramdini C, et al. , 2011. Viewpoint: High susceptibility to Chikungunya virus of Aedes aegypti from the French West Indies and French Guiana. Tropical Med. Int. Health 16, 134–139. [DOI] [PubMed] [Google Scholar]
- Gloria-Soria A, Ayala D, Bheecarry A, Calderon-Arguedas O, Chadee DD, Chiappero M, Coetzee M, Bin Elahee KB, Fernandez-Salas I, Kamal HA, Kamgang B, Khater EI, Kramer LD, Kramer V, Lopez-Solis A, Lutomiah J, Martins A Jr., Micieli MV, Paupy C, Ponlawat A, Rahola N, Rasheed SB, Richardson JB, Saleh AA, Sanchez-Casas RM, Seixas G, Sousa CA, Tabachnick WJ, Troyo A, Powell JR, 2016. Global genetic diversity of Aedes aegypti. Mol. Ecol 25:5377–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloria-Soria A, Armstrong PM, Powell JR, Turner PE, 2017. Infection rate of Aedes aegypti mosquitoes with dengue virus depends on the interaction between temperature and mosquito genotype. Proc. Biol. Sci 284 (1864). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göertz GP, Vogels CBF, Geertsema C, Koenraadt CJM, Pijlman GP, 2017. Mosquito co-infection with Zika and chikungunya virus allows simultaneous transmission without affecting vector competence of Aedes aegypti. PLoS Negl Trop Dis 11, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves CM, Melo FF, Bezerra JMT, Chaves BA, Silva BM, Silva LD, et al. , 2014. Distinct variation in vector competence among nine field populations of Aedes aegypti from a Brazilian dengue-endemic risk city. Parasit. Vectors 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EA, Higgs S, 2009. Impact of climate change and other factors on emerging arbovirus diseases. Trans. R. Soc. Trop. Med. Hyg 103, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes DR, Paiva MH, Donato MM, et al. , 2017. Zika virus replication in the mosquito Culex quinquefasciatus in Brazil. Emerg Microbes Infect (6), 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guégan M, Zouache K, Démichel C, Minard G, Potier P, Mavingui P, Moro CV, 2018. The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome 6, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XX, Zhu XJ, Li CX, Dong Y, De Zhang YM, Xing D, et al. , 2013. Vector competence of Aedes albopictus and Aedes aegypti (Diptera: Culicidae) for DEN2–43 and New Guinea C virus strains of dengue 2 virus. Acta Trop. 128, 566–570. [DOI] [PubMed] [Google Scholar]
- Hall RA, Bielefeldt-Ohmann H, McLean BJ, O'Brien CA, Colmant AM, Piyasena TB, Harrison JJ, Newton ND, Barnard RT, Prow NA, Deerain JM, Mah MG, Hobson-Peters J, 2017. Commensal Viruses of Mosquitoes: Host Restriction, Transmission, and Interaction with Arboviral Pathogens. Evol. Bioinformatics Online 12, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Mendelin S, Pyke AT, Moore PR, et al. , 2016. Assessment of local mosquito species incriminates Aedes aegypti as the potential vector of Zika virus in Australia. PLoS Negl. Trop. Dis 10, e004959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedge S, Rasgon JL, Huges GL, 2015. The microbiome modulates arbovirus transmission in mosquitoes. Curr Opin Virol 15, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmann A, Jansen S, Luhken R, et al. , 2017. Experimental transmission of Zika virus by mosquitoes from Central Europe. Euro Surveill. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber K, Le Loan L, Hoang TH, Tien TK, Rodhain F, Failloux AB, 2003. Aedes aegypti in South Vietnam: Ecology, genetic structure, vectorial competence and resistance to insecticides. Southeast Asian J Trop Med Public Health 34, 81–86. [PubMed] [Google Scholar]
- Johnson BW, Chambers TV, Crabtree MB, Filippis AMB, Vilarinhos PTR, et al. , 2002. Vector competence of Brazilian yellow fever virus isolate Aedes aegypti and Ae. abopictus for a Brazilian yellow fever virus isolate. Trans Royal Soc Trop Med Hyg 611–613. [DOI] [PubMed] [Google Scholar]
- Jupp PG, Kemp A, 1993. The potential for dengue in South Africa: Vector competence tests with dengue 1 and 2 viruses and 6 mosquito species. Trans Royal Soc Trop Med Hyg 87, 639–643. [DOI] [PubMed] [Google Scholar]
- Kauffman EB, Kramer LD, 2017. Zika Virus Mosquito Vectors: Competence, Biology, and Vector Control. J. Infect. Dis 216, 976–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BH, Carley JG, Fanning ID, Fillipic C, 1979. Quantitative studies of the vector competence of Aedes aegypti, Culex annulirostris and other mosquitoes (Diptera: Culicidae) with Murray Valley. J. Med. Entomol 16, 59–60. [DOI] [PubMed] [Google Scholar]
- Kenney JL, Romo H, Duggal NK, et al. , 2017. Transmission incompetence of Culex quinquefasciatus and Culex pipiens pipiens from North America for Zika virus. Am J Trop Med Hyg. 96, 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox TB, Kay BH, Hall R, Ryan PA, 2003. Enhanced vector competence of Aedes aegypti (Diptera: Culicidae) from the Torres Strait compared with mainland Australia for dengue 2 and 4 viruses. J. Med. Entomol 40, 950–956. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Scherer WF, 1976. Vector competence of mosquitoes as a marker to distinguish central American and Mexican epizootic from enzootic strains of Venezuelan enceph. Am J Trop Med Hyg. 25, 336–346. [DOI] [PubMed] [Google Scholar]
- Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C, 2010. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 327, 1644–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L, Chevillon C, Albright RG, Thaisomboonsuk B, Richardson JH, et al. , 2009. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol. Biol 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Flohic G, Porphyre V, Barbazan P, Gonzalez JP, 2013. Review of climate, landscape, and viral genetics as drivers of the Japanese encephalitis virus, ecology. PLoS Negl. Trop. Dis 7, e2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MI, Wong PS, Ng LC, Tan CH, 2012. Oral susceptibility of Singapore Aedes (Stegomyia) aegypti (Linnaeus) to Zika virus. PLoS Negl. Trop. Dis 6, e1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Guo XX, Deng YQ, Xing D, Sun AJ, Liu QM, Wu Q, Dong YD, Zhang YM, Zhang HD, Cao WC, Qin CF, Zhao TY, 2017. Vector competence and transovarial transmission of two Aedes aegypti strains to Zika virus. Emerg Microbes Infect 6, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KC, Ziegler SA, Thangamani S, Hausser NL, Kochel TJ, et al. , 2011. Experimental transmission of Mayaro virus by Aedes aegypti. Am J Trop Med Hyg. 85, 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco-De-Oliveira R, Vazeille M, Bispo De Filippis AM, Failloux AB, 2002. Oral susceptibility to yellow fever virus of Aedes aegypti from Brazil. Mem. Inst. Oswaldo Cruz 97, 437–439. [DOI] [PubMed] [Google Scholar]
- Lourenco-De-Oliveira R, Vazeille M, de Filippis AM, Failloux AB, 2004. Aedes aegypti in Brazil: genetically differentiated populations with highsusceptibility to dengue and yellow fever viruses. Trans Royal Soc Trop Med Hyg. 98, 43–44. [DOI] [PubMed] [Google Scholar]
- Lourenço-De-Oliveira R, Rua AV, Vezzani D, Willat G, Vazeille M, et al. , 2013. Aedes aegypti from temperate regions of South America are highly competent to transmit dengue virus. BMC Infect. Dis 13, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Nicholson J, Winokur OC, Steiner C, Riemersma KK, Stuart J, Takeshita R, Krasnec M, Barker CM, Coffey LL, 2018. Vector competence of Aedes aegypti, Culex tarsalis, and Culex quinquefasciatus from California for Zika virus. PLoS Negl. Trop. Dis 12, e0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklewitz M, Zirkel F, Kurth A, Drosten C, Junglen S, 2015. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl. Acad. Sci. U. S. A 112, 7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbaika S, Lutomiah J, Chepkorir E, Mulwa F, Khayeka-Wandabwa C, et al. , 2016. Vector competence of Aedes aegypti in transmitting Chikungunya virus: Effects and implications of extrinsic incubation temperature on dissemination and infection rates. Virol. J 13, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros AS, Costa DMP, Branco MSD, Sousa DMC, Monteiro JD, Galvao SPM, Azevedo PR, Fernandes JV, Araujo JMG, 2018. Dengue virus in Aedes aegypti and Aedes albopictus in urban areas in the state of Rio Granse do Norte, Brazil: Importance of virological and entomological surveillance. PLoS One 13, e0194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minard G, Tran FH, Van VT, Goubert C, Bellet C, Lambert G, Kim KLH, Thuy THT, Mavingui P, Valiente Moro C, 2015. French invasive Asian tiger mosquito populations harbor reduced bacterial microbiota and genetic diversity compared to Vietnamese autochthonous relatives. Front. Microbiol 6, e2836–e2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CJ, Miller BR, Gubler DJ, 1987. Vector competence of Aedes albopictus from Houston, Texas, for dengue serotypes 1to 4, yellow fever and Ross River viruses. J Am Mosquito Cont Ass 3, 460–465. [PubMed] [Google Scholar]
- Morrison TE, Diamond MS, 2017. Animal models of Zika virus infection pathogenesis and immunity. J Virol 91, e00009–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muturi EJ, Buckner E, Bara J, 2017. Superinfection interference between dengue-2 and dengue-4 viruses in Aedes aegypti mosquitoes. Tropical Med. Int. Health 22, 399–406. [DOI] [PubMed] [Google Scholar]
- Na W, Yeom M, Choi IK, Yook H, Song D, 2017. Animal models for dengue vaccine development and testing. Clin Exp Vaccine Res 6, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F, Haddow AD, Tesh RB, Weaver SC, 2014. Eilat virus displays a narrow mosquito vector range. Parasit. Vectors 7, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F, Erasmus JH, Haddow AD, Tesh RB, Weaver SC, 2015. Eilat virus induces both homologous and heterologous interference. Virology 484, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoagouni C, Kamgang B, Kazanji M, Paupy C, Nakouné E, 2017. Potential of Aedes aegypti and Aedes albopictus populations in the Central African Republic to transmit enzootic chikungunya virus strains. Parasit. Vectors 10, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira JHM, Gonçalves RLS, Lara FA, Dias FA, Gandara ACP, Menna-Barreto RFS, Edwards MC, Laurindo FRM, Silva-Neto MAC, Sorgine MHF, Oliveira PL, 2011. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 7, e1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KE, Bonizzoni M, 2017. Nonretroviral integrated RNA viruses in arthropod vectors: an occasional event or something more? Curr Opin Insect Sci. 22, 45–53. [DOI] [PubMed] [Google Scholar]
- Padilha KP, Resck MEB, Cunha OATD, Teles-de-Freitas R, Campos SS, Sorgine MHF, Lourenco-de-Oliveira R, Farnesi LC, Bruno RV, 2018. Zika infection decreases Aedes aegypti locomotor activity but does not influence egg production or viability. Mem. Inst. Oswaldo Cruz 113, e180290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatini U, Miesen P, Carballar-Lejarazu R, Ometto L, Rizzo E, Tu Z, van Rij RP, Bonizzoni M, 2017. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics 18, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer WH, Varghese F, van Rij R, 2018. Natural variation in resistance to virus infection in Dipteran insects. Viruses 10, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pando-Robles V, Batista CV, 2017. Aedes-borne virus-mosquito interactions: mass spectrometry strategies and findings. Vector-borne Zoon Dis 17, 361–375. [DOI] [PubMed] [Google Scholar]
- Paupy C, Chantha N, Vazeille M, Reynes JM, Rodhain F, et al. , 2003. Variation over space and time of Aedes aegypti in Phnom Penh (Cambodia): genetic structure and oral susceptibility to a dengue virus. Gen Res 82, 171–182. [DOI] [PubMed] [Google Scholar]
- Pérez-Castro R, Castellanos JE, Olano VA, Matiz MI, Jaramillo JF, Vargas SL, Sarmiento DM, Stenstrom TA, Overgaard HJ, 2016. Detection of all four dengue serotypes in Aedes aegypti female mosquitoes collected in a rural area in Colombia. Mem. Inst. Oswaldo Cruz 111, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesko K, Westbrook CJ, Mores CN, Lounibos LP, Reiskin MH, 2009. Effects of Infectious Virus Dose and Bloodmeal delivery Method on Susceptibility of Aedes aegypti and Aedes albopictus to Chikungunya Virus. J. Med. Entomol 46, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Thi KL, Briant L, Gavotte L, Labbe L, Perriat-Sanguinet M, et al. , 2017. Incidence of dengue and chikungunya viruses in mosquitoes and human patients in border provinces of Vientnam. Parasit. Vectors 10, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike A, Dong Y, Dizaji NB, Gacita A, Mongodin EF, Dimopoulos G, 2017. Changes in the microbiota cause genetically modified Anopheles to spread in a population. Science 357, 1396–1399. [DOI] [PubMed] [Google Scholar]
- Pongsiri A, Ponlawat A, Thaisomboonsuk B, Jarman RG, Scott TW, et al. , 2014. Differential susceptibility of two field Aedes aegypti populations to a low infectious dose of dengue virus. PLoS One 9, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole-Smith BK, Hemme RR, Delorey M, Felix G, Gonzalez AL, et al. , 2015. Comparison of Vector Competence of Aedes mediovittatus and Aedes aegypti for Dengue Virus: Implications for Dengue Control in the Caribbean. PLoS Negl. Trop. Dis 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, 2018. Mosquito-Borne Human Viral Diseases: why Aedes aegypti? Am J Trop Med Hyg. 98, 1563–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JL, Souza-Neto J, Torres Cosme R, Rovira J, Ortiz A, Pascale JM, Dimopoulos G, 2012. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis 6, e1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JL, Short SM, Bahia AC, Saraiva RG, Dong Y, Kang S, Tripathi A, Mlambo G, Dimopoulos G, 2014. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro antipathogen activities. PLoS Pathog. 10, e1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed W, Carroll J, 1901. The prevention of yellow fever. Public Health Pap Rep 27, 113–129. [PMC free article] [PubMed] [Google Scholar]
- Richard V, Paoaafaite T, Cao-Lormeau VM, 2016a. Vector Competence of French Polynesian Aedes aegypti and Aedes polynesiensis for Zika Virus. PLoS Negl. Trop. Dis 10, e0005024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard V, Paoaafaite T, Cao-Lormeau VM, 2016b. Vector Competence of Aedes aegypti and Aedes polynesiensis Populations from French Polynesia for Chikungunya Virus. PLoS Negl. Trop. Dis 10, e0004694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SL, Anderson SL, Alto BW, 2012. Vector competence of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) fordengue virus in the Florida Keys. J. Med. Entomol 49, 942–946. [DOI] [PubMed] [Google Scholar]
- Ritchie SA, van den Hurk AF, Smout MJ, Staunton KM, Hoffmann AA, 2018. Mission Accomplished? We need a Guide to the 'Post Release' World of Wolbachia for Aedes-borne Disease Control. Trends Parasitol. 34, 217–226. [DOI] [PubMed] [Google Scholar]
- Rodgers FH, Gendrin M, Wyer CAS, Christophides GK, 2017. Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes. PLoS Pathog. 13, e1006391–e1006392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen L, Roseboom LE, Gubler DJ, Lien JC, Chaniotis BN, 1985. Comparative susceptibility of mosquito species and strains to oral and parenteral infection with dengue and Japanese encephalitis viruses. Am. J. Trop. Med. Hyg 34, 603–615. [DOI] [PubMed] [Google Scholar]
- Roundy CM, Azar SR, Rossi SL, Huang JH, Leal G, Yun R, Fernandez-Salas I, Vitek CJ, Paploski IA, Kitron U, Ribeiro GS, Hanley KA, Weaver SC, Vasilakis N, 2017. Variation in Aedes aegypti Mosquito Competence for Zika Virus Transmission. Emerg. Infect. Dis 23, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert C, Ebel GD, 2018. How do Virus-Mosquito Interactions Lead to Viral Emergence? Trends Parasitol. 34, 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert C, Weger-Lucarelli J, Garcia-Luna SM, Young MC, Byas AD, et al. , 2017. Impact of simultaneous exposure to arboviruses on infection and transmission by Aedes aegypti mosquitoes. Nature Com 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckebusch F, Berthet M, Missé D, Choumet V, 2017. Infection of a French Population of Aedes albopictus and of Aedes aegypti (Paea Strain) with Zika Virus reveals Low Transmission rates to these Vectors' Saliva. J Mol Sci 18, 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva RG, Kang S, Simões ML, Angleró-Rodríguez YI, Dimopoulos G, 2016. Mosquito gut antiparasitic and antiviral immunity. Dev. Comp. Immunol 64, 53–64. [DOI] [PubMed] [Google Scholar]
- Saraiva RG, Fang J, Kang S, Angleró-Rodríguez YI, Dong Y, Dimopoulos G, 2018. Aminopeptidase secreted by Chromobacterium sp. Panama inhibits dengue virus infection by degrading the E protein. PLoS Negl. Trop. Dis 12, e0006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JR, Mori A, Romero-Severson J, Chadee DD, Severson DW, 2007. Investigations of dengue-2 susceptibility and body size among Aedes aegypti populations. Med. Vet. Entomol 21, 370–376. [DOI] [PubMed] [Google Scholar]
- Serrato IM, Caicedo PA, Orobio Y, Lowenberger C, Ocampo CB, 2017. Vector competence and innate immune responses to dengue virus infection in selected laboratory and field-collected Stegomyia aegypti (= Aedes aegypti). Med. Vet. Entomol 31, 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, van Tol S, MacLeod HJ, Dimopoulos G, 2018. Hydrogen cyanide produced by the soil bacterium Chromobacterium sp. Panama contributes to mortality in Anopheles gambiae mosquito larvae. Sci. Rep 8, 8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Jupatanakul N, Ramirez JL, Kang S, Romero-Vivas CM, et al. , 2013. Transcriptomic Profiling of Diverse Aedes aegypti Strains reveals increased Basal-level Immune Activation in Dengue Virus-refractory Populations and Identifies Novel Virus-vector Molecular Interactions. PLoS Negl. Trop. Dis 7, e2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Battle KE, Hay SI, Barker CM, Scott TW, McKenzie FE, 2012. Ross, Macdonald, and a theory for the dynamics and control of mosquito-transmitted pathogens. PLoS Pathog. 8, e1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylla M, Bosio C, Urdaneta-Marquez L, Ndiaye M, Black IVWC, 2009. Gene flow, subspecies composition, and dengue virus-2 susceptibility among Aedes aegypti collections in Senegal. PLoS Negl. Trop. Dis 3, e408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabachnick WJ, 2016. Ecological effects on arbovirus-mosquito cycles of transmission. Curr Opin Virol. 21, 124–131. [DOI] [PubMed] [Google Scholar]
- Tabachnick WJ, Wallis GP, Aitken TH, Miller BR, Amato GD, et al. , 1985. Oral infection of Aedes aegypti with yellow fever virus: geographic variation andgenetic considerations. Am J Trop Med Hyg 34, 1219–1224. [DOI] [PubMed] [Google Scholar]
- Thongrungkiat S, Jirakanjanaki N, Apiwathnasorn C, Prummongkol S, Samung Y, 2003. Comparative susceptibility to oral infection with dengue viruses among local strains of Aedes aegypti (Diptera : Culicidae) collected at different seasons of the year. J Vector Ecol 28, 166–170. [PubMed] [Google Scholar]
- Tran KT, Vazeille-Falcoz M, Mousson L, Tran HH, Rodhain F, et al. , 1999. Aedes aegypti in Ho Chi Minh City (Viet Nam): susceptibility to dengue 2 virus and genetic differentiation. Trans Royal Soc Trop Med Hyg. 93, 581–586. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin KA, Chen R, Sherman MB, Weaver SC, 2011. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol 1, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turell MJ, Guinn MLO, Dohm DJ, Jones JW, 2001. Vector Competence of north American Mosquitoes (Diptera: Culicidae) for West Nile Virus. J. Med. Entomol 38, 130–134. [DOI] [PubMed] [Google Scholar]
- Turell MJ, Lee JS, Richardson JH, Sang RC, Kioko EN, Agawo MO, Pecor J, O'Guinn ML, 2007. Vector competence of Kenyan Culex zombaensis and Culex quinquefasciatus mosquitoes for Rift Valley fever virus. J. Am. Mosq. Control Assoc 23, 378–382. [DOI] [PubMed] [Google Scholar]
- Van Den Hurk AF, McElroy K, Pyke AT, McGee CE, Hall-Mendelin S, et al. , 2011. Vector competence of Australian mosquitoes for yellow fever virus. Am J Trop Med Hyg 85, 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Tesh RB, 2015. Insect-specific viruses and their potential impact on arbovirus transmission. Curr Opin Virol 15, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazeille M, Mousson L, Rakatoarivony I, Villeret R, Rodhain F, et al. , 2001. Population genetic structure and competence as a vector for dengue type 2 virus of Aedes aegypti and Aedes albopictus from Madagascar. Am J Trop Med Hyg 65, 491–497. [DOI] [PubMed] [Google Scholar]
- Vazeille M, Gaborit P, Mousson L, Girod R, Failloux AB, 2016. Competitive advantage of a dengue 4 virus when co-infecting the mosquito Aedes aegypti with a dengue 1 virus. BMC Infect. Dis 16, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Rua A, Zouache K, Girod R, Failloux AB, Lourenco-De-Oliveira R, 2014. High Level of Vector Competence of Aedes aegypti and Aedes albopictus from ten American Countries as a crucial factor in the Spread of Chikungunya Virus. J. Virol 88, 6294–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas LEM, Campolina TB, Barnabe NR, Orfanó AS, Chaves BA, Norris DE, et al. , 2018. Zika virus infection modulates the bacterial diversity associated with Aedes aegypti as revealed by metagenomic analysis. PLoS One 13, e0190352–e0190366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis GP, Aitken TH, Beaty BJ, Lorenz L, Amato GD, et al. , 1985. Selection for susceptibility and refractoriness of Aedes aegypti to oralinfection with yellow fever vims. Am J Trop Med Hyg 34, 1225–1231. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang X, Zhang C, Xing Y, Wu DY, et al. , 2012. Vector Competence of five Common Mosquito Species in the People's Republic of China for Western Equine Encephalitis Virus. Vector-Borne Zoonotic Dis 12, 605–608. [DOI] [PubMed] [Google Scholar]
- Wang YH, Chang MM, Wang XL, Zheng AH, Zou Z, 2017. The immune strategies of mosquito Aedes aegypti against microbial infection. Dev. Comp. Immunol 1–10. [DOI] [PubMed] [Google Scholar]
- Watson TM, Kay BH, 1999. Vector competence of Aedes notoscriptus (Diptera: Culicidae) for Barmah Forest virus and of this species and Aedes aegypti (Diptera: Culicidae) for dengue 1–4 viruses in Queensland, Australia. J. Med. Entomol 36, 508–514. [DOI] [PubMed] [Google Scholar]
- Weaver SC, 2006. Evolutionary influences in arboviral disease. Curr Topics Microbiol Immunol 299, 285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger-Lucarelli J, Rückert C, Chotiwan N, Nguyen C, Garcia Luna SM, Fauver JR, Foy BD, Perera R, Black WC, Kading RC, Ebel GD, 2016. Vector Competence of American Mosquitoes for three Strains of Zika Virus. PLoS Negl. Trop. Dis 10, e0005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ZJ, Dolan PT, Kunitomi M, Tassetto M, Seetin MG, Oh S, Heiner C, Paxinos E, Andino R, 2017. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr Biol 27, 3511–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins K, Eastmond B, Alto BW, 2018. Transmission potential of Mayaro virus in Florida Aedes aegypti and Aedes albopictus mosquitoes. Med. Vet. Entomol 32, 436–442. [DOI] [PubMed] [Google Scholar]
- Xi Z, Ramirez JL, Dimopoulos G, 2008. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 4, e1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye YH, Ng TS, Frentiu FD, Walker T, Van Den Hurk AF, et al. , 2014. Comparative susceptibility of mosquito populations in North Queensland, Australia to oral infection with dengue virus. Am J Trop Med Hyg. 90, 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zompi S, Harris E, 2012. Animal models of dengue virus infection. Viruses 4, 62–82. [DOI] [PMC free article] [PubMed] [Google Scholar]