Abstract
Background
The determinants of coronavirus disease 2019 (COVID-19) disease severity and extrapulmonary complications (EPCs) are poorly understood. We characterized relationships between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNAemia and disease severity, clinical deterioration, and specific EPCs.
Methods
We used quantitative and digital polymerase chain reaction (qPCR and dPCR) to quantify SARS-CoV-2 RNA from plasma in 191 patients presenting to the emergency department with COVID-19. We recorded patient symptoms, laboratory markers, and clinical outcomes, with a focus on oxygen requirements over time. We collected longitudinal plasma samples from a subset of patients. We characterized the role of RNAemia in predicting clinical severity and EPCs using elastic net regression.
Results
Of SARS-CoV-2–positive patients, 23.0% (44 of 191) had viral RNA detected in plasma by dPCR, compared with 1.4% (2 of 147) by qPCR. Most patients with serial measurements had undetectable RNAemia within 10 days of symptom onset, reached maximum clinical severity within 16 days, and symptom resolution within 33 days. Initially RNAemic patients were more likely to manifest severe disease (odds ratio, 6.72 [95% confidence interval, 2.45–19.79]), worsening of disease severity (2.43 [1.07–5.38]), and EPCs (2.81 [1.26–6.36]). RNA loads were correlated with maximum severity (r = 0.47 [95% confidence interval, .20–.67]).
Conclusions
dPCR is more sensitive than qPCR for the detection of SARS-CoV-2 RNAemia, which is a robust predictor of eventual COVID-19 severity and oxygen requirements, as well as EPCs. Because many COVID-19 therapies are initiated on the basis of oxygen requirements, RNAemia on presentation might serve to direct early initiation of appropriate therapies for the patients most likely to deteriorate.
Keywords: SARS-CoV-2, RNAemia, digital PCR, severity prediction, extrapulmonary complications
Measurement of severe acute respiratory syndrome coronavirus 2 RNA from the plasma of patients with coronavirus disease 2019, using digital polymerase chain reaction, can predict disease severity, clinical deterioration, and extrapulmonary complications and may help guide patient triage and management.
As of April 2021, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 136 million infections and 2.9 million deaths [1]. Wide variation in clinical trajectories of coronavirus disease 2019 (COVID-19) poses challenges for clinicians seeking to identify patients at risk for deterioration. While COVID-19 often manifests as a viral pneumonia, extrapulmonary complications (EPCs) can produce more severe and recalcitrant disease [2, 3]. SARS-CoV-2, typically isolated from nasopharyngeal (NP) samples, has been detected in lower titers in whole blood [4–6], serum [6–10], plasma [11–14], and stool [6, 10, 15]. Histopathologic surveys have identified the virus in myocardial [16, 17], renal [16, 18], gastrointestinal [19], and neurologic tissues [16]. Is hematogenous spread of the virus or viral components associated with EPCs? The clinical and pathophysiologic significance of SARS-CoV-2 RNA in blood remains poorly understood.
SARS-CoV-2 RNAemia, detected with quantitative reverse-transcriptase real-time polymerase chain reaction (qPCR), has been correlated with severity of COVID-19 [12, 14, 20–24]. However, rates of RNAemia detected with qPCR have ranged from 0% to 41% [6–12, 20–22, 24, 25]. qPCR lacks the precision to measure low viral loads [26], and its sensitivity to protocol and threshold decisions hinders comparison across studies. Digital polymerase chain reaction (dPCR) has superior sensitivity, precision, and reproducibility, allowing absolute quantification of RNA without standard curves. Given its tolerance to inhibitors, dPCR facilitates detecting dilute targets in blood [27]. dPCR studies have accordingly reported higher rates of RNAemia (42.4% and 74.1%) and have linked RNAemia with disease severity [13, 14] and dysregulated host response [14].
In this prospective, longitudinal, observational study of patients with COVID-19 presenting to the emergency department (ED), we characterized relationships between SARS-CoV-2 RNAemia and clinical severity, future deterioration, and specific EPCs. We used array-based dPCR to maximize reliability and replicability and to simplify potential clinical adoption of RNAemia testing.
MATERIALS AND METHODS
Patients With COVID-19 and Specimen Collection
Beginning in April 2020, we collected blood and NP swab samples from patients enrolled in the Stanford University COVID-19 Biobank. Eligible patients were ≥18 years old, with a positive RT-PCR SARS-CoV-2 NP swab sample at initial ED presentation. We repeated blood sample collection on day 3, 7, and/or 30 in hospitalized patients, and asked discharged patients to return for repeated blood sampling. We collected blood in ethylenediaminetetraacetic acid–chelated vacutainers (Becton, Dickinson) and stored plasma at −80°C after 1200g centrifugation for 10 minutes at 25°C. We collected NP swabs in 1 mL of RNA Shield Stabilizing Solution (Zymo Research) and stored them at −80°C. We processed samples under biosafety level 2+ precautions (Stanford University APB-2551).
SARS-CoV-2 RNA Extraction
We extracted RNA from research NP swabs and plasma samples using the QIAamp Viral RNA Mini Kit and QIACube Connect (Qiagen). We extracted from 140 µL of sample solution, and eluted RNA in 50 µL of elution buffer after manual lysing for 10 minutes.
SARS-CoV-2 RNA Detection and Quantification
Multiplexed qPCR and dPCR reactions included extracted RNA, the |Q| Triplex Assay (Combinati), and the 4× RT-dPCR MM assay (Combinati). The |Q| Triplex Assay (Combinati) included primers and probes targeting N1 and N2 regions of the nucleoprotein gene, and the human ribonuclease P gene (RP). We divided the reactions as follows: 10 µL for qPCR using the QuantStudio 5 (Applied Biosystems by Thermo Fisher Scientific) and 9 µL for dPCR using the array-based |Q| assay (Combinati) (Supplementary Materials).
We considered qPCR specimens positive if cycle threshold (Ct) values for RP, N1, and N2 (all 3 values) were <40. For positive samples, we used the lesser of N1/N2 Ct for quantitative analysis. dPCR samples were positive if both N1 and N2 concentrations were ≥0.23 copy/µL, with the greater of the 2 used for quantitative analysis. We set negative qPCR results to 40, and negative dPCR results to 0. We repeated dPCR for inconclusive (N1 or N2, <0.23 copy/µL) and low results (N1 and N2, <1 copy/µL), and we used the results from the repeated tests. We calculated pairwise Pearson correlations between measures of qPCR Ct and log-transformed RNA concentrations from dPCR.
Patients with specimens collected after enrollment (day 0) typically had specimens collected on day 3 or 7, not both. Thus, we combined days 3 and 7 (henceforth “day 3/7”) for longitudinal analyses. For patients with samples from both days, we used the greater of the 2 values.
Clinical and Laboratory Measures
For each participant, we recorded disease severity at enrollment and at every blood sampling, and the maximum severity attributable to COVID-19 within 30 days of enrollment, using a modified World Health Organization (WHO) COVID-19 severity scale (Table 1 [28]). We documented dates for each severity score, and each participant’s diagnoses at discharge. We recorded demographic features, comorbid conditions, initial ED vital signs, presence of pneumonia at chest radiography or computed tomography, symptoms, and laboratory values (Supplementary Materials).
Table 1.
Modified World Health Organization COVID-19 Severity Score [28]
| WHO Score | Severity | Description |
|---|---|---|
| 1 | Mild | Asymptomatic infection not requiring admission |
| 2 | Mild | Symptomatic infection not requiring admission |
| 3 | Moderate | Admitted, not requiring supplemental oxygen |
| 4 | Moderate | Admitted, requiring oxygen by nasal cannula |
| 5 | Severe | Admitted, requiring oxygen by high-flow nasal cannula |
| 6 | Severe | Admitted, requiring mechanical ventilation |
| 7 | Severe | Admitted, requiring mechanical ventilation plus vasopressors or renal replacement therapy |
| 8 | Severe | Death from COVID-19 2019–related cause |
Abbreviations: COVID-19, coronavirus disease 2019; WHO, World Health Organization.
We dichotomized (low or high) vital signs as follows: mean arterial pressure <88 mm Hg (25th percentile for enrolled patients; no patients had a mean arterial pressure <65 mm Hg at enrollment) or >140 mm Hg (75th percentile); respiratory rate <8/min or >20/min; oxygen saturation by pulse oximetry <95% (25th percentile); and temperature <36°C or ≥38°C. We dichotomized laboratory values (Supplementary Material) based on our laboratory’s reference ranges. For predictive models, missing values were imputed nonparametrically for each participant with a random forests model, using 100 trees, and 10 iterations (R package missForest; version 1.4).
Defining EPCs
We created binary indicators for development of EPCs during each patient’s clinical course. Patients had neurologic involvement if they had a diagnosis of acute stroke, encephalitis, meningitis, neuroinflammatory disease, or delirium. Cardiovascular involvement included myocardial injury, acute coronary syndrome, cardiomyopathy, acute cor pulmonale, arrhythmia, and cardiogenic shock. Renal involvement was defined by acute kidney injury. Transaminitis or hyperbilirubinemia constituted hepatobiliary involvement. Hematologic involvement included new anemia, deep vein thrombosis, pulmonary embolus, myocardial infarction, acute stroke, acute limb ischemia, mesenteric ischemia, and catheter-related thrombosis. Finally, patients were considered to have immunologic involvement if they received a diagnosis of sepsis, septic shock, multiorgan failure, or secondary bacterial infection.
Characterizing Associations Between RNAemia and Clinical Severity
We calculated maximum severity scores for initially RNAemic and non-RNAemic patients, and compared mean scores with a 2-sample t-test. Among RNAemic patients, we calculated correlations between log-transformed RNA concentration and maximum severity (WHO scores, 1–8). We calculated proportions of RNAemic and non-RNAemic patients who manifested mild (WHO scores, 1–2), moderate (WHO scores, 3–4), or severe (WHO scores, 5–8) disease, who were hospitalized, who manifested EPCs, and whose condition worsened after presentation (maximum WHO score higher than score at enrollment), using χ 2 tests with continuity corrections. We calculated the odds ratio (OR) for deterioration, by RNAemia on enrollment (for consistency with logistic regression results), and we calculated exact 95% confidence intervals (CIs) [29]. We compared median length of hospitalization in days, and median degree of clinical worsening (difference between initial and maximum WHO scores), for RNAemic and non-RNAemic patients, using the Wilcoxon rank sum test with continuity correction.
Predictive Models for Severe Disease, EPCs, and RNAemia
We developed a predictive model for severe (WHO score, 5–8) disease, using as potential predictors all data available at ED presentation: demographic features, comorbid conditions, binary indicators of abnormal vital signs, pneumonia on chest imaging, patient-reported symptoms, and abnormal laboratory values. Because therapies such as remdesivir and dexamethasone were generally initiated based on oxygen requirements (the main component of the severity score), we excluded specific treatments from our models.
To prevent overfitting due to the large number of potential predictors, we selected variables via elastic net regularization (glmnet 4.0 in R software), using logistic models and 10-fold cross-validation, selecting the regularization parameter λ minimizing mean cross-validated error. We used the selected variables in a logistic model, and we estimated ORs and 95% CIs for prediction of severe disease. We calculated mean cross-validated area under the receiver operating characteristic curve (AUROC) of the resulting model.
We predicted development of EPCs in analogous fashion, excluding symptoms and laboratory markers potentially constitutive of EPCs. We again selected variables via cross-validated elastic net regularization, estimated ORs for the most robust predictors, and characterized overall predictive accuracy by AUROC.
We predicted the presence of RNAemia in analogous fashion, using as potential predictors demographic features, comorbid conditions, and symptoms, but excluding radiographic and laboratory findings, to approximate the predictability of RNAemia at initial presentation.
RESULTS
Patient Characteristics
We enrolled 191 COVID-19–positive ED patients and sampled their plasma on the day of enrollment (day 0). Some patients had additional NP or plasma samples collected at day 3, 7, and/or 30. Of these participants, 49.2% (94 of 191) were women. Their median age was 47 years (interquartile range, 34–61 years). Patients had a median (interquartile range) of 1 (0–3) comorbid condition and 4 (2–6) symptoms. Patient characteristics at enrollment are summarized in Table 2.
Table 2.
Patient Characteristics at Enrollment (N = 191)
| Characteristic | Value |
|---|---|
| Female sex, % (no./total) | 49.2 (94/191) |
| Age, median (IQR), y | 47 (34–61) |
| Medical history, % (no./total) | |
| Lung disease | 12.6 (24/191) |
| Cancer | 13.6 (26/191) |
| Diabetes | 26.7 (51/191) |
| Immunosuppression | 7.3 (14/191) |
| Heart disease | 11.0 (21/191) |
| Hypertension | 36.6 (70/191) |
| ACEI/ARB use | 18.3 (35/191) |
| Stroke | 4.2 (8/191) |
| Dementia | 4.7 (9/191) |
| DVT/PE | 5.8 (11/191) |
| Chronic kidney disease | 9.9 (19/191) |
| Smoking | 20.9 (40/191) |
| Symptoms at presentation, % (no./total) | |
| Fever | 64.4 (123/191) |
| Chills | 31.4 (60/191) |
| Cough | 67.5 (129/191) |
| Sore throat | 16.2 (31/191) |
| Congestion | 8.4 (16/191) |
| Shortness of breath | 63.4 (121/191) |
| Chest pain | 34.6 (66/191) |
| Myalgia | 34.6 (66/191) |
| Nausea, vomiting, or diarrhea | 40.8 (78/191) |
| Loss of taste | 38.7 (74/191) |
| Loss of smell | 27.2 (52/191) |
| Confusion | 2.6 (5/191) |
| Headache | 26.2 (50/191) |
Abbreviations: ACEI, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; DVT, deep vein thrombosis; IQR, interquartile range; PE, pulmonary embolus.
SARS-CoV-2 RNA Prevalence by Sample Type, Method, and Day of Collection
dPCR was more sensitive than qPCR, detecting RNAemia in 23.0% of patients (44 of 191) on day 0 (compared with 1.4% [2 of 147] for qPCR), 13.3% (6 of 45) on day 3 (0 for qPCR), and 6.8% (3 of 44) on day 7 (0 for qPCR). At day 30, neither platform detected RNAemia in 32 specimens. We describe the performance of the dPCR assay in the Supplementary Materials.
We observed a modest negative correlation (r = −0.30) between qPCR Ct values and dPCR RNA concentrations from the same plasma specimens (Supplementary Figure 1). Notably, NP and plasma dPCR values for 48 paired specimens were weakly correlated (r = 0.16). Plasma RNA by dPCR on day 0 was moderately correlated with concentrations on day 3/7 (r = 0.42).
Persistence of RNAemia
Of the 44 patients RNAemic by dPCR at enrollment, 27 had ≥1 follow-up sample. Of these, 92.6% (25 of 27) had highest RNA at enrollment, and 7.4% (2 of 27) on day 3/7; 22.2% (6 of 27) had detectable RNAemia at day 3/7. Of 50 COVID-19–positive patients not RNAemic at enrollment, only 1 was newly RNAemic on day 3/7.
RNAemia and Severity of Disease
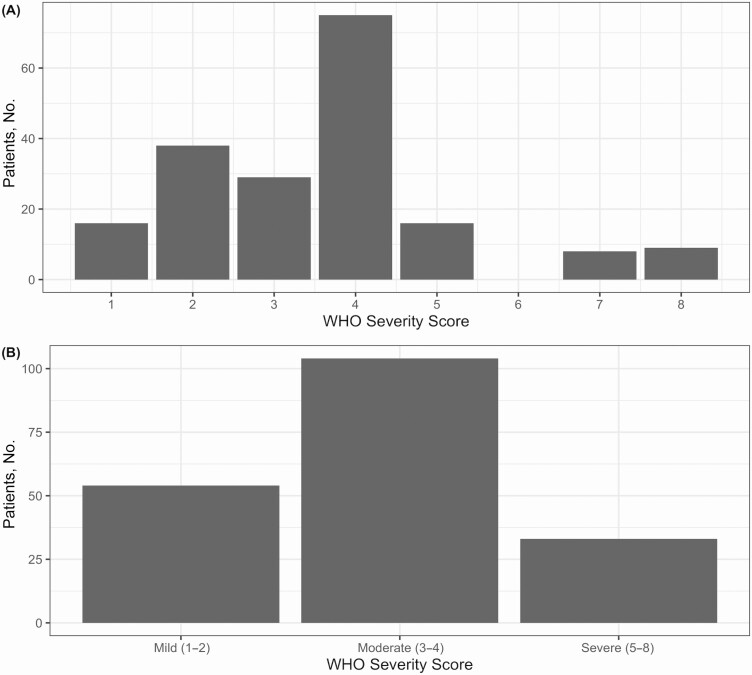
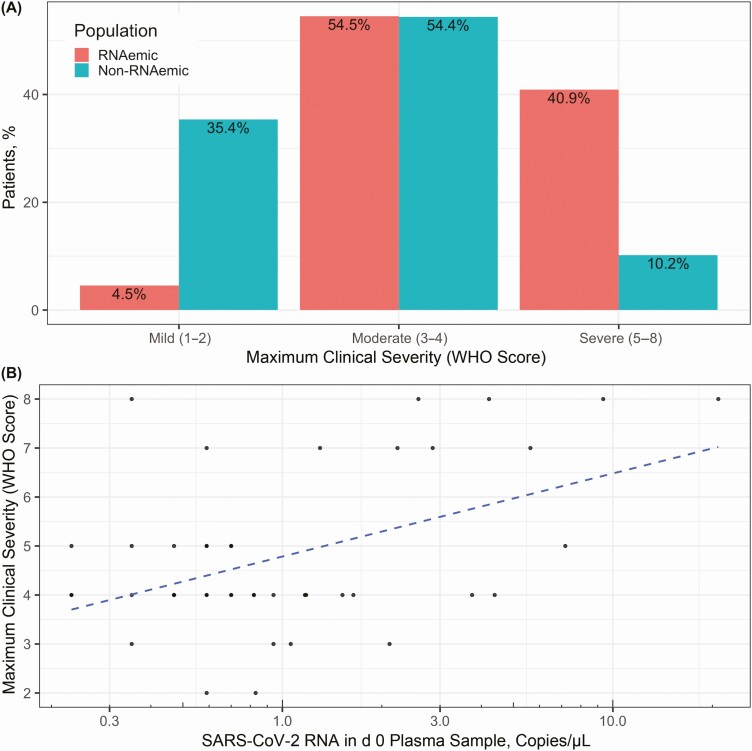
We classified patients’ disease as mild (n = 54), moderate (n = 104), or severe (n = 33) based on maximum WHO severity score (Figure 1). RNAemic patients had higher mean clinical severity (4.80) than non-RNAemic patients (3.24; difference, 1.56 [95% CI, 1.00–2.11]), and 40.9% of RNAemic patients developed severe disease, compared with 10.2% of non-RNAemic patients (difference, 30.7% [95% CI, 13.9%–47.5%]). Conversely, 4.5% of initially RNAemic patients had mild disease, compared with 35.4% of non-RNAemic patients (difference, 30.8% [95% CI, 19.5%–42.2%]) (Figure 2A). Among the 44 initially RNAemic patients, those with higher RNA concentrations manifested greater peak severity (r = 0.47 [95% CI, .20–0.67]) (Figure 2B). Severity trended higher in persistently RNAemic patients, compared with patients not RNAemic at day 3/7 (mean WHO score, 6.5 vs 5.0), but the difference was not significant (95% CI of difference, −.58 to 3.58).
Figure 1.
Distribution of discrete and binned World Health Organization (WHO) severity scores. We classified the maximum severity of 147 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) presentations using a modified WHO scoring system, with scores defined as follows: 1, asymptomatic infection; 2, symptomatic infection not requiring admission; 3, admitted without supplemental oxygen; 4, admitted, requiring oxygen by nasal cannula; 5, admitted, requiring oxygen by high-flow nasal cannula; 6, admitted, requiring mechanical ventilation; 7, admitted, requiring mechanical ventilation and vasopressors or renal replacement therapy; and 8, death from coronavirus disease 2019 (COVID-19)–related cause. A, Distribution of WHO scores. B, Distribution of binned (mild, moderate, and severe) scores.
Figure 2.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNAemia and clinical severity. A, RNAemic patients had higher mean maximum World Health Organization (WHO) scores (4.80) than non-RNAemic patients (3.24; difference, 1.56 [95% confidence interval [CI], 1.00–2.11]). Severe disease developed in 40.9% of RNAemic patients, compared with 10.2% of non-RNAemic patients (difference, 30.7% [95% CI, 13.9%–47.5%]). Of initially RNAemic patients, 4.5% had mild disease, compared with 35.4% of non-RNAemic patients (difference, 30.8% [95% CI, 19.5%–42.2%]). Equivalent proportions of RNAemic (54.5%) and non-RNAemic (54.4%) patients had disease of moderate severity. B, Among patients with detectable RNAemia at enrollment (n = 44), patients with higher plasma RNA concentrations manifested more severe disease (r = 0.47 [95% CI, .20–.67]). RNA concentrations in RNAemic patients were distributed approximately log normally, so were log scaled for depiction and calculation of correlation. Dashed blue line shows linear correlation between log-scaled plasma RNA concentration and maximum clinical severity.
In all, 90.9% of RNAemic patients (40 of 44) and 70.1% of non-RNAemic patients (103 of 147) required hospital admission (difference, 20.8% [95% CI, 8.1%–33.6%]). Among admitted patients, RNAemic patients had a longer median length of stay (7.6 vs 5.1 days; P < .01; Wilcoxon rank sum test).
In an elastic net–regularized, cross-validated logistic model of severe (WHO score, 5–8) disease, the significant predictors of severe disease were tobacco smoking, low oxygen saturation at presentation, and RNAemia (OR for severe disease, 6.72 [95% CI, 2.45–19.79]). The overall predictive performance of the model was good, with a mean cross-validated AUROC of 0.82 (Table 3).
Table 3.
Prediction of Severe Disease
| Predictora | OR (95% CI) |
|---|---|
| PMH: DM | 1.51 (.51–4.45) |
| Smoker status | 3.13 (1.08–9.38) |
| ED: MAP low | 2.59 (.92–7.47) |
| ED: SpO2 low | 5.36 (2.03–15.07) |
| ALC low | 3.12 (1.00–9.8) |
| Lactate high | 3.90 (.63–22.91) |
| Glucose high | 2.58 (.92–7.30) |
| RNAemia | 6.72 (2.45–19.79) |
Abbreviations: ALC, Absolute Lymphocyte Count; CI, confidence interval; DM, Diabetes Mellitus; ED, emergency department; MAP, mean arterial pressure; OR, odds ratio; PMH, Past Medical History; SpO2, oxygen saturation.
aPotential predictors of severe disease (World Health Organization score, 5–8) included demographic features (age ≥60 or ≥80 years and sex), past medical history features (lung disease, cancer, diabetes, immunosuppression, heart disease, hypertension, angiotensin-converting enzyme inhibitor or angiotensin receptor blocker use, stroke, dementia, deep venous thrombosis or pulmonary embolus, chronic kidney disease, tobacco smoking, and obesity), binary indicators of abnormal ED vital signs (low SpO2 and low or high MAP, heart rate, respiratory rate, and temperature), pneumonia on chest radiography or computed tomography, patient-reported symptoms (fever, chills, cough, sore throat, congestion, shortness of breath, chest pain, myalgias, nausea/vomiting/diarrhea, loss of taste, loss of smell, confusion, and headache), and binary indicators of abnormal laboratory values (high or low leukocyte or platelet count; low absolute lymphocyte count; low hemoglobin or fibrinogen levels; high levels of D-dimer, fibrinogen, C-reactive protein, lactate dehydrogenase, ferritin, troponin, lactate, serum urea nitrogen, creatinine, bilirubin, aspartate aminotransferase, alanine aminotransferase, and alkaline phosphatase; high prothrombin time or partial thromboplastin time; and high or low levels of sodium, potassium, chloride, bicarbonate, calcium, magnesium, and glucose).
To prevent overfitting, predictors were selected via elastic net regression of severe disease on these features with 10-fold cross-validation, selecting the regularization parameter λ minimizing mean cross-validated error, and yielding the features in the table above. In a logistic model regressing severe disease on these features, significant predictors of severe disease included tobacco smoking, SpO2, and RNAemia. RNAemia was associated with 6.7 times the odds of severe disease, adjusting for other features selected by elastic net–penalized regression, an association comparable in magnitude to the association of hypoxia at initial presentation with eventual severe disease. The mean cross-validated area under the receiver operating characteristic curve of the model in predicting severe disease was 0.82. The Akaike Information Criterion (AIC) was 134.74.
Dynamics of Infection Through Course of Illness
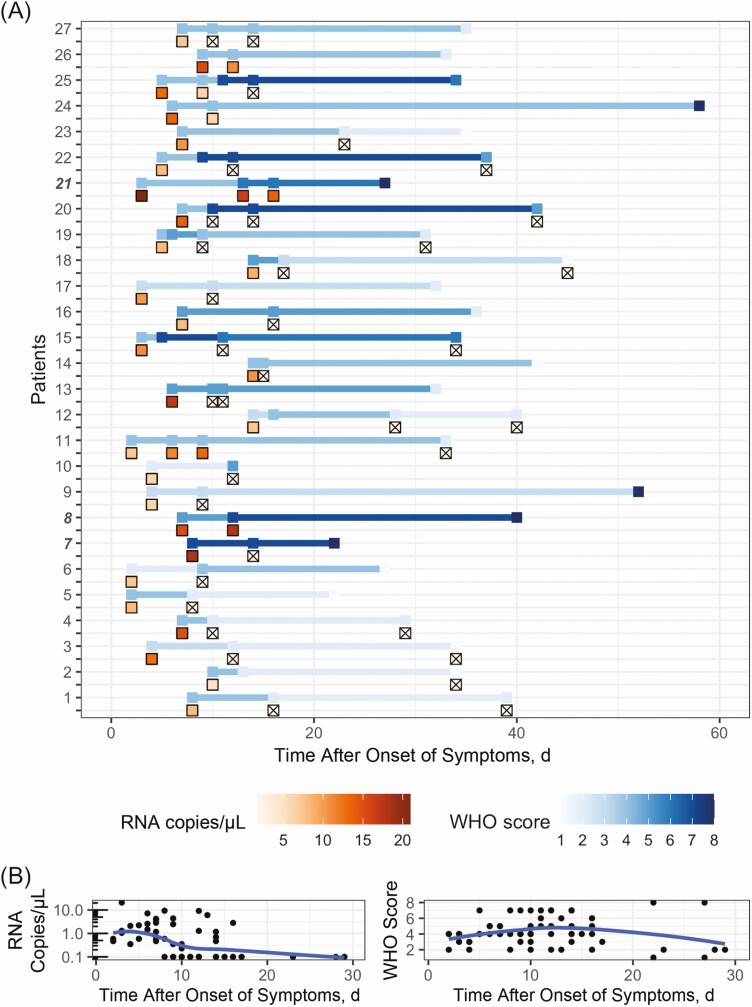
Twenty-seven initially RNAemic patients had subsequent plasma samples. Most (14 of 27) had undetectable plasma RNA within 10 days of symptom onset, reached maximum severity within 16 days, and had symptom resolution within 33 days (Figure 3). In the 27, disease was mild at enrollment in 2, moderate in 20, and severe in 5. Seventeen patients with initially moderate or severe disease recovered to mild severity, 6 remained in a severe condition, and 3 died in the hospital.
Figure 3.
Dynamics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNAemia and clinical severity, by modified World Health Organization (WHO) score. A, Serial plasma SARS-CoV-2 RNA concentrations and WHO scores for each of the 27 patients with longitudinal samples. Plasma RNA concentration (red gradient) and WHO scores (blue gradient) are shown with respect to the number of days since the reported onset of symptoms (not date of study enrollment) for each patient. Numbers of patients who died in the hospital are highlighted in boldface and italic. Specimens with undetectable RNAemia are represented with x’s. Fourteen of 27 patients had undetectable RNAemia by day 10, while the same proportion took 16 days to reach maximum severity, and 33 days for resolution of symptoms. B, Aggregate RNA and clinical dynamics in the 30 days after onset of symptoms. Loess regression curves represent trends in RNA and clinical dynamics. RNAemia peaked 3 days after symptom onset, while clinical severity peaked at 14 days.
Across all patients, 77.0% (147 of 191) manifested maximum severity at presentation, while 23.0% (44 of 191) continued to worsen (Supplementary Figure 2); 36.4% of initially RNAemic patients (16 of 44) and 19.0% of non-RNAemic patients (28 of 147) had worsening severity after initial presentation (difference, 17.4% [95% CI, .3%–34.4%]; OR, 2.43 [1.07–5.38]). Severity in RNAemic patients worsened by a median of 3 points on the modified WHO scale, compared with 1 point for non-RNAemic patients (P = .02; Wilcoxon rank sum test).
RNAemia Predicts EPCs
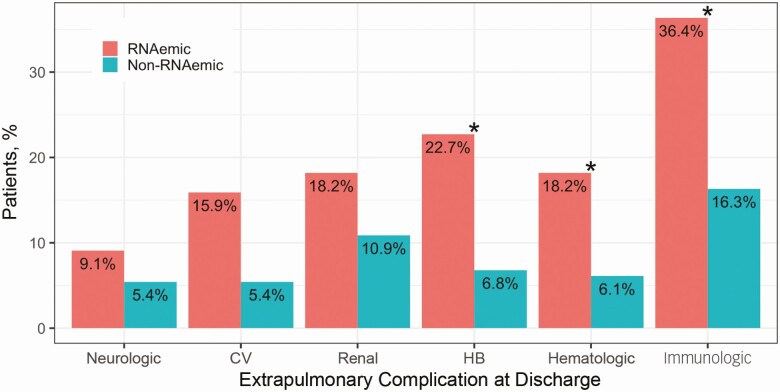
Of RNAemic patients, 56.8% (25 of 44) developed ≥1 EPC, compared with 30.6% (45 of 147) of non-RNAemic patients (difference in proportions, 26.2% [95% CI, 8.3%–44.1%]). RNAemic patients tended toward higher rates of all EPC categories (Figure 4), with significant differences in rates of hepatobiliary, hematologic, and immunologic complications (P < .05; χ 2 tests for equality of proportions with continuity correction). Supplementary Table 1 shows specific comorbid conditions, EPCs, SARS-CoV-2 RNA concentrations, and severity scores for all patients who were RNAemic at enrollment.
Figure 4.
Presence of extrapulmonary complications (EPCs), by RNAemia. Of patients RNAemic at enrollment, ≥1 EPC developed by hospital discharge in 56.8% (25 of 44), compared with 30.6% of non-RNAemic patients (45 of 147) (difference, 26.2% [95% confidence interval, 8.3%–44.1%]). RNAemic patients tended toward higher rates of EPCs across systems, though only differences in rates of hepatobiliary (HB), hematologic, and immunologic complications were individually significant; *P < .05 (χ 2 test for equality of proportions with continuity correction). Abbreviation: CV, cardiovascular.
In an elastic net–regularized, cross-validated logistic model, significant predictors for the development of ≥1 EPCs were chronic kidney disease, obesity, and RNAemia (OR, 2.81 [95% CI, 1.26–6.36]). The overall predictive performance of the model was fair, with a mean cross-validated AUROC of 0.73 (Table 4).
Table 4.
Prediction of Extrapulmonary Complications
| Predictora | OR (95% CI) |
|---|---|
| Age ≥80 y | 2.27 (.49–9.92) |
| PMH: heart disease | 2.13 (.63–7.41) |
| PMH: HTN | 1.74 (.81–3.69) |
| PMH: dementia | 3.60 (.58–25.33) |
| PMH: CKD | 4.56 (1.36–17.27) |
| Smoker | 1.88 (.80–4.42) |
| Obesity | 2.64 (1.23–5.84) |
| ED: RR high | 1.63 (.79–3.35) |
| ED: SpO2 low | 1.34 (.60–2.93) |
| RNAemia | 2.81 (1.26–6.36) |
Abbreviations: CI, confidence interval; CKD, chronic kidney disease; ED, emergency department; HTN, hypertension; OR, odds ratio; PMH, past medical history; RR, respiratory rate; SpO2, oxygen saturation.
aPotential predictors of extrapulmonary complications (EPCs) included demographic features (age ≥60 or ≥80 years; sex), past medical history features (lung disease, cancer, diabetes, immunosuppression, heart disease, HTN, angiotensin-converting enzyme inhibitor or angiotensin receptor blocker use, stroke, dementia, deep venous thrombosis or pulmonary embolus, CKD, tobacco smoking, and obesity), binary indicators of abnormal ED vital signs (low or high mean arterial pressure, heart rate, respiratory rate, and temperature; low SpO2), pneumonia on initial chest radiography or computed tomography, and patient-reported symptoms at enrollment, excluding those constitutive of extrapulmonary diagnosis (fever, chills, cough, sore throat, congestion, shortness of breath, chest pain, and myalgias). Laboratory values were not included, because many were constitutive of extrapulmonary diagnoses.
To prevent overfitting, predictors were selected via elastic net regression of EPCs (1 if a patient had ≥1 EPC; 0 if a patient had none) on these features with 10-fold cross-validation, selecting the regularization parameter λ minimizing mean cross-validated error, and yielding the features listed in Table 4. In a logistic model regressing EPCs on these features, significant predictors of EPC included CKD, obesity (body mass index >30 [calculated as weight in kilograms divided by height in meters squared]), and RNAemia. RNAemia was associated with 2.8 times the odds of EPC, comparable in magnitude to the association between obesity and development of EPCs. The mean cross-validated area under the receiver operating characteristic curve of the model in predicting EPC was 0.73. The Akaike Information Criterion (AIC) was 222.25.
Prediction of RNAemia on Presentation
We predicted RNAemia on presentation based on patient demographics, comorbid conditions, symptoms, and ED vital signs. In an analogous model (Supplementary Table 2), significant predictors of RNAemia were cough and hypoxia, though predictive performance was poor (AUROC, 0.66).
DISCUSSION
The pathogenesis of COVID-19, its temporal dynamics, and the determinants of disease severity and EPCs are incompletely understood. We explored the performance and clinical utility of dPCR in quantifying SARS-CoV-2 RNA in the nasopharynx and plasma, and characterized the relationships between RNAemia and disease severity, clinical deterioration, and EPCs. Array-based dPCR was much more sensitive than qPCR for the detection of SARS-CoV-2 in plasma, where the mean concentration of viral RNA was 3 orders of magnitude less than in the nasopharynx. RNAemia manifests early in the course of illness, while clinical manifestations peak later and are more prolonged. RNAemia at presentation predicts severe disease, ongoing clinical deterioration, and specific EPCs.
We found dPCR to be markedly more sensitive than qPCR, even with more stringent detection criteria (both N1 and N2 ≥0.23 copy/µL) than other studies (eg, N1 or N2 ≥0.1 copy/µL) [14]. dPCR was also more consistent in multiplex detection of both targets (N1 and N2), likely because dPCR’s partition format reduces preferential amplifications observed in bulk PCR [30]. Moreover, a microwell array dPCR platform enhances partition consistency, improves sensitivity by minimizing dead volume, and has a qPCR-like workflow well suited to clinical adoption. Early in an outbreak, or on emergence of novel variants, viral RNA standard curves are not widely available, making dPCR a natural choice for detecting novel pathogens.
RNAemia on presentation was a robust predictor of both severe disease and EPCs, after accounting for demographics, comorbid conditions, symptoms, vital signs, and a host of laboratory markers. Moreover, disease in RNAemic patients, compared with non-RNAemic patient, was more likely to worsen after presentation, and worsened by a greater degree. Previous studies have associated RNAemia with disease severity and mortality rates [12, 14, 23, 24], but reported associations with EPCs are more varied [2, 31, 32]. We included a more comprehensive scope of potential confounders than previous studies [14]. We also use cross-validation not only for model selection but also to assess the relative predictability of clinical severity (good; AUROC, 0.82), EPCs (fair; AUROC, 0.73), and RNAemia itself (poor; AUROC, 0.66). The poor predictability of RNAemia from patient features at presentation, and the weak correlations between NP and plasma RNA concentrations, suggest that RNAemia is not simply a consequence of sufficient viral load at the typical site of inoculation (which may also be subject to greater variation in sample quality) but may instead signal unique pathophysiologic and prognostic features [10, 33, 34].
The causes of SARS-CoV-2 RNAemia, and the mechanisms through which it affects disease severity and EPCs, require further investigation. RNAemia might arise from spillage from the respiratory tract, or from active viral replication in vascular endothelial [35] or perivascular cells [36]. Whether RNAemia represents genomic fragments, immunocomplexed or otherwise neutralized virus, or replication-competent intact virus cannot be determined from our data, and an attempt to culture SARS-CoV-2 from serum with low RNA levels was not successful [37]. SARS-CoV-1, however, has been found to replicate in circulating lymphocytes, monocytes, macrophages, and dendritic cells [38–40]. The RNAemia kinetics we observed follow a typical viral kinetic pattern, with high peak viral load early in the infection, followed by rapid decay (likely reflecting the innate immune response), before a slower clearance by acquired immunity [41]. Reduction of RNAemia has been correlated with appearance of antibodies [42], and worsening RNAemia with critical illness and death [43]. RNAemia before symptom onset has been anecdotally reported; more data is needed to better assess presymptomatic dynamics [37]. Because our findings suggest that RNAemia at presentation reflects the likelihood of subsequent disease progression, early testing for RNAemia could guide the initiation and monitoring of antiviral therapies [41].
We found a stronger association between RNAemia and EPCs (defined conservatively based on new diagnoses at discharge, rather than surrogate biomarkers) than previous studies [32]. Extrapulmonary injury could result from direct viral toxicity, endothelial cell damage and thromboinflammation, dysregulation of the immune response, or dysregulation of the renin–angiotensin–aldosterone system [2]. RNA load is associated with increased chemokines, interleukin 6, C-reactive protein, ferritin, coagulation activation, and tissue damage [14]. Transaminitis, frequently observed in RNAemic patients, might result from direct hepatocellular injury by SARS-CoV-2, from cytokine storm and hypoxia-associated metabolic derangement, or from antiviral drug-induced liver injury. The trend we observed toward higher incidence of acute kidney injury in RNAemic patients is consistent with prior evidence of renal tropism [16].
We found that SARS-CoV-2 RNAemia at initial ED presentation is a robust predictor of eventual clinical severity and EPCs. Despite the limited generalizability of a single-center study, the substantial predictive value of RNAemia in multiple aspects of the disease course suggests a role for plasma dPCR in triage and disposition. Because we use a measure of severity based primarily on oxygen requirements, and because many COVID-19 therapies are initiated on the basis of such requirements, RNAemia at presentation might serve to direct early initiation of appropriate therapies to the patients whose condition is most likely to deteriorate.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. Additional author members of the Stanford COVID-19 Biobank Study Group include Rosen Mann, Anita Visweswaran, Thanmayi Ranganath, Jonasel Roque, Monali Manohar, Hena Naz Din, Komal Kumar, Kathryn Jee, Brigit Noon, Jill Anderson, Bethany Fay, Donald Schreiber, Nancy Zhao, Rosemary Vergara, Julia McKechnie, Aaron Wilk, Lauren de la Parte, Kathleen Whittle Dantzler, Maureen Ty, Nimish Kathale, Arjun Rustagi, Giovanny Martinez-Colon, Geoff Ivison, Ruoxi Pi, Maddie Lee, Rachel Brewer, Taylor Hollis, Andrea Baird, Michele Ugur, Drina Bogusch, George R. Nahass, Kazim Haider, Kim Quyen Thi Tran, Laura Simpson, Michal Tal, Iris Chang, Evan Do, Andrea Fernandes, Alexandra S. Lee, Neera Ahuja, Theo Snow, and James Krempski.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants R01AI153133, R01AI137272, and 3U19AI057229–17W1 COVID SUPP 2), and by Eva Grove.
Potential conflicts of interest. A. J. R. reports being on the national pulmonary trials advisory committee for Merck and receiving honoraria for travel/time, during the last 36 months. C. A. B. reports grants from the National Institutes of Health and the Bill & Melinda Gates Foundation, outside the submitted work. K. C. N. reports support from National Institute of Allergy and Infectious Diseases, the National Heart, Lung, and Blood Institute, the National Institute of Environmental Health Sciences, and Food Allergy Research & Education, during the conduct of the study. K. C. N. also reports the following patents planned, issued, or pending: Inhibition of Allergic Reaction to Peanut Allergen Using an IL-33 Inhibitor, Special Oral Formula for Decreasing Food Allergy Risk and Treatment for Food Allergy, Basophil Activation Based Diagnostic Allergy Test, Granulocyte-Based Methods for Detecting and Monitoring Immune System Disorders, Methods and Assays for Detecting and Quantifying Pure Subpopulations of White Blood Cells in Immune System Disorders, Mixed Allergen Compositions and Methods for Using the Same, and Microfluidic Device and Diagnostic Methods for Allergy Testing Based on Detection of Basophil Activation (all outside the conduct of the study). K. C. N. is also a cofounder of Before Brands, Alladapt, Latitude, and IgGenix, outside the submitted work. E. A. reports consulting fees from Apple and Foresite Capital, reports participation on advisory boards for Personalis, DeepCell, Apple, and Silicon Valley Exercise Analytics, and reports being a nonexecutive director for AstraZeneca and holding stock for self with Personalis, all during the last 36 months. S. Y. is a scientific advisory board member for Combinati Inc. S. Y. reports being an existing stockholder as a scientific advisor for Combinati and also reports receipt of consumables and reagents and a 1-month loan of an instrument from Combinati to perform the current study, within the last 36 months. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Stanford COVID-19 Biobank Study Group:
Rosen Mann, Anita Visweswaran, Thanmayi Ranganath, Jonasel Roque, Monali Manohar, Hena Naz Din, Komal Kumar, Kathryn Jee, Brigit Noon, Jill Anderson, Bethany Fay, Donald Schreiber, Nancy Zhao, Rosemary Vergara, Julia McKechnie, Aaron Wilk, Lauren de la Parte, Kathleen Whittle Dantzler, Maureen Ty, Nimish Kathale, Arjun Rustagi, Giovanny Martinez-Colon, Geoff Ivison, Ruoxi Pi, Maddie Lee, Rachel Brewer, Taylor Hollis, Andrea Baird, Michele Ugur, Drina Bogusch, George R Nahass, Kazim Haider, Kim Quyen Thi Tran, Laura Simpson, Michal Tal, Iris Chang, Evan Do, Andrea Fernandes, Alexandra S Lee, Neera Ahuja, Theo Snow, and James Krempski
References
- 1. WHO coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int. Accessed 18 April 2021.
- 2. Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med 2020; 26:1017–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zheng KI, Feng G, Liu WY, Targher G, Byrne CD, Zheng MH. Extrapulmonary complications of COVID-19: a multisystem disease? J Med Virol 2020; 93:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peng L, Liu J, Xu W, et al. 2019 Novel coronavirus can be detected in urine, blood, anal swabs and oropharyngeal swabs samples. medRxiv [Preprint: not peer reviewed]. 25 February 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.02.21.20026179v1. [Google Scholar]
- 5. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 2020; 9:386–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020; 395:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lescure FX, Bouadma L, Nguyen D, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis 2020; 20:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ling Y, Xu SB, Lin YX, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 2020; 133:1039–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–69. [DOI] [PubMed] [Google Scholar]
- 11. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fajnzylber J, Regan J, Coxen K, et al. ; Massachusetts Consortium for Pathogen Readiness . SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 2020; 11:5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Veyer D, Kernéis S, Poulet G, et al. Highly sensitive quantification of plasma SARS-CoV-2 RNA sheds light on its potential clinical value. Clin Infect Dis 2021; 73:e2890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bermejo-Martin JF, González-Rivera M, Almansa R, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care 2020; 24:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 2020; 383:590–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tavazzi G, Pellegrini C, Maurelli M, et al. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur J Heart Fail 2020; 22:911–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Su H, Yang M, Wan C, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int 2020; 98:219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020; 158:1831–1833.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng S, Fan J, Yu F, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ 2020; 369:m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen X, Zhao B, Qu Y, et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis 2020; 71:1937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hogan CA, Stevens BA, Sahoo MK, et al. High frequency of SARS-CoV-2 RNAemia and association with severe disease. Clin Infect Dis 2020;ciaa1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eberhardt KA, Meyer-Schwickerath C, Heger E, et al. RNAemia corresponds to disease severity and antibody response in hospitalized COVID-19 patients. Viruses 2020; 12:1045. Available at: 10.3390/v12091045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawasuji H, Morinaga Y, Tani H, et al. SARS-CoV-2 RNAemia with higher nasopharyngeal viral load is strongly associated with severity and mortality in patients with COVID-19. medRxiv [Preprint: not peer reviewed]. 18 December 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.12.17.20248388v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. The COVID-19 Investigation Team. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 2020; 26:861–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dahdouh E, Lázaro-Perona F, Romero-Gómez MP, Mingorance J, García-Rodriguez J. Ct values from SARS-CoV-2 diagnostic PCR assays should not be used as direct estimates of viral load. J Infect 2020; 82:414–51; S0163445320306757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem 2015; 61:79–88. [DOI] [PubMed] [Google Scholar]
- 28. WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis 2020; 20:e192–e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cornfield J. A statistical problem arising from retrospective studies. Berkeley, CA: University of California Press, 1956:135–48. [Google Scholar]
- 30. Kuypers J, Jerome KR. Applications of digital PCR for clinical microbiology. J Clin Microbiol 2017; 55:1621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu D, Zhou F, Sun W, et al. Relationship between serum SARS-CoV-2 nucleic acid(RNAemia) and organ damage in COVID-19 patients: a cohort study. Clin Infect Dis 2020: ciaa1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Järhult JD, Hultström M, Bergqvist A, Frithiof R, Lipcsey M. The impact of viremia on organ failure, biomarkers and mortality in a Swedish cohort of critically ill COVID-19 patients. Sci Rep 2021; 11:7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020; 296:E32–E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prebensen C, Myhre PL, Jonassen C, et al. Severe acute respiratory syndrome coronavirus 2 RNA in plasma is associated with intensive care unit admission and mortality in patients hospitalized with coronavirus disease 2019. Clin Infect Dis 2020;ciaa1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 2004; 203:631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen L, Li X, Chen M, Feng Y, Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res 2020; 116:1097–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersson M, Arancibia-Cárcamo CV, Auckland K, et al. SARS-CoV-2 RNA detected in blood samples from patients with COVID-19 is not associated with infectious virus. medRxiv [Preprint: not peer reviewed]. 17 June 2020. Available from: https://www.medrxiv.org/content/10.1101/2020.05.21.20105486v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gu J, Gong E, Zhang B, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med 2005; 202:415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Law HK, Cheung CY, Ng HY, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 2005; 106:2366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li L, Wo J, Shao J, et al. SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J Clin Virol 2003; 28:239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goyal A, Cardozo-Ojeda EF, Schiffer JT. Potency and timing of antiviral therapy as determinants of duration of SARS-CoV-2 shedding and intensity of inflammatory response. Sci Adv 2020; 6:eabc7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Röltgen K, Powell AE, Wirz OF, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol 2020; 5:eabe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen L, Wang G, Long X, et al. Dynamics of blood viral load is strongly associated with clinical outcomes in coronavirus disease 2019 (COVID-19) patients. J Mol Diagn 2021; 23:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.