Abstract
Background
Serological testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) complements nucleic acid tests for patient diagnosis and enables monitoring of population susceptibility to inform the coronavirus disease 2019 (COVID-19) pandemic response. It is important to understand the reliability of assays with different antigen or antibody targets to detect humoral immunity after SARS-CoV-2 infection and to understand how antibody (Ab) binding assays compare to those detecting neutralizing antibody (nAb), particularly as we move into the era of vaccines.
Methods
We evaluated the performance of 6 commercially available enzyme-linked immunosorbent assays (ELISAs), including a surrogate virus neutralization test (sVNT), for detection of SARS-CoV-2 immunoglobulins (IgA, IgM, IgG), total or nAb. A result subset was compared with a cell culture–based microneutralization (MN) assay. We tested sera from patients with prior reverse transcription polymerase chain reaction–confirmed SARS-CoV-2 infection, prepandemic sera, and potential cross-reactive sera from patients with other non-COVID-19 acute infections.
Results
For sera collected >14 days post–symptom onset, the assay achieving the highest sensitivity was the Wantai total Ab at 100% (95% CI, 94.6%–100%), followed by 93.1% for Euroimmun NCP-IgG, 93.1% for GenScript sVNT, 90.3% for Euroimmun S1-IgG, 88.9% for Euroimmun S1-IgA, and 83.3% for Wantai IgM. Specificity for the best-performing assay was 99.5% for the Wantai total Ab, and for the lowest-performing assay it was 97.1% for sVNT (as per the Instructions for Use [IFU]). The Wantai Total Ab had the best agreement with MN at 98% followed by Euroimmun S1-IgA, Euro NCP-IgG, and sVNT (as per IFU) with 97%, 97% and 95%, respectively; Wantai IgM had the poorest agreement at 93%.
Conclusions
Performance characteristics of the SARS-CoV-2 serology assays detecting different antibody types are consistent with those found in previously published reports. Evaluation of the surrogate virus neutralization test in comparison to the Ab binding assays and a cell culture–based neutralization assay showed good result correlation between all assays. However, correlation between the cell-based neutralization test and some assays detecting Ab’s not specifically involved in neutralization was higher than with the sVNT. This study demonstrates the reliability of different assays to detect the humoral immune response following SARS-CoV-2 infection, which can be used to optimize serological test algorithms for assessing antibody responses post–SARS-CoV-2 infection or vaccination.
Keywords: ELISA, humoral immune response, neutralization test, SARS-CoV-2 antibodies, serology
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has infected >131 million people and caused >2.8 million deaths globally as of April 8, 2021 [1]. COVID-19 manifests as an acute respiratory illness, although asymptomatic infections occur [2].
Laboratory diagnosis of COVID-19 primarily relies on detection of viral RNA by reverse transcription polymerase chain reaction (RT-PCR) in respiratory tract samples or antigen tests, the sensitivity of which declines as infection resolves. In contrast, antibody (Ab) is detected in most individuals 10–15 days after onset of symptoms [3]. Serological testing can detect previous infection in people who have recovered without RT-PCR testing who are RT-PCR negative [4, 5] or whose RT-PCR results are difficult to interpret [6]. Serology may also inform our understanding of antibody longevity and quantification of neutralizing antibody (nAb) response in vaccine trials [6–8]. As the pandemic progresses, estimates of prior exposure and population prevalence from serological assays will play an increasingly important role for public health decision-making [9, 10]. However, there is substantial variation in assay performance depending on the antibody type detected and correlation to neutralizing antibodies [11].
In this study, we evaluate the sensitivity and specificity of 6 commercially available serological ELISA tests using either spike (S), nucleocapsid (NCP), or receptor-binding domain (RBD) antigens for detection of specific isotypes of SARS-CoV-2 (IgA, IgM, IgG), total Ab or nAb (using a surrogate virus neutralization test [sVNT]). We also compare a subset of ELISA results with microneutralization (MN), the current gold standard assay for SARS-CoV-2-specific antibody.
METHODS
Study Design
This was a retrospective study evaluating the sensitivity and specificity of commercially available ELISAs for detection of Abs to SARS-CoV-2 virus on well-pedigreed sera.
Evaluation Serum Samples
Sera from 3 patient groups were assembled to assess aspects of assay performance.
Euroimmun and sVNT assay sensitivity was assessed using a panel of 147 sera (panel A) from a group of patients with prior RT-PCR-confirmed SARS-CoV-2 infection.
A subset of 96 sera (panel B) were used to evaluate the Wantai assays. The Wantai total Ab assay requires 100 µL of sample; 51 panel A sera had insufficient volume for testing using the Wantai assays.
While only 6 (of the 51) sera were from convalescent patients collected >14 days post–symptom onset, the samples collected >14 days post–symptom onset represent 49% (72/147) of panel A and 69% (66/96) of panel B.
Specificity was calculated using 2 groups: a population group of sera representing the Victorian population collected prepandemic between 2011 and 2018 (n = 100 to n = 312, depending on the assay tested) and a cross-reactive group representing prepandemic sera to assess potential cross-reactivity from patients with seasonal coronavirus (HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1), SARS-CoV1, MERS-CoV, or other non-COVID-19 acute infections (n = 30 to n = 36, depending on the assay tested). See Tables 1 and 2 for the full list.
Table 1.
Description of Evaluation Samples for the Assays
| Group | Euro S1- IgA | Euro S1-IgG | Euro NCP-IgG | GenScript sVNT | Wantai IgM | Wantai Total Ab |
|---|---|---|---|---|---|---|
| Panel A | Panel B | |||||
| Sensitivity | ||||||
| Infected (RT-PCR positive) | ||||||
| <7 d | 41 | 41 | 40 | 41 | 13 | 13 |
| 7–14 d | 34 | 34 | 34 | 34 | 17 | 17 |
| >14 d | 72 | 72 | 72 | 72 | 66 | 66 |
| Total | 147 | 147 | 146 | 147 | 96 | 96 |
| Specificity | ||||||
| Population | 179 | 179 | 191 | 312 | 100 | 209 |
| Cross-reactive | 30 | 30 | 30 | 30 | 30 | 36 |
| Total | 209 | 209 | 221 | 342 | 130 | 245 |
| Overall total samples | 356 | 356 | 367 | 489 | 226 | 341 |
| Individuals | 330 | 330 | 342 | 463 | 224 | 339 |
Samples tested by Euro S1-IgA, Euro S1-IgG, Euro NCP-IgG, and GenScript sVNT referred to as panel A. Samples tested by Wantai IgM and Wantai Total Ab referred to as panel B.
Abbreviations: Ab, antibody; Euro, Euroimmun; Ig, immunoglobulin; NCP, nucleocapsid; RT-PCR, reverse transcription polymerase chain reaction; sVNT, surrogate virus neutralization test.
Table 2.
Cross-Reactivity Results for Non-COVID-19 Mixed Infection Prepandemic Group
| Antibodies Against | Euro S1-IgA | Euro S1-IgG | Euro NCP-IgG | GenScript sVNT | Wantai IgM | Wantai Total Ab |
|---|---|---|---|---|---|---|
| SARS-CoV-1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 |
| MERS-CoV | 0/9 | 0/9 | 1/9 | 0/9 | 0/9 | 0/9 |
| Seasonal hCoV | 0/5 | 0/5 | 0/5 | 0/4 | 0/2 | 0/2 |
| Mycoplasma | 1/4 | 1/4 | 2/4 | 0/4 | 0/2 | 0/2 |
| Parvovirus, parvo/HCV | 0/4 | 0/4 | 0/4 | 0/4 | 0/5 | 0/2 |
| CMV | 1/2 | 1/2 | 1/2 | 0/2 | 0/7 | 0/5 |
| Dengue | 1/5 | 0/5 | 0/5 | 0/5 | 0/6 | 0/7 |
| EBV | 0 | 0 | 0 | 0 | 0/4 | 0/4 |
| Flu A, adenovirus | 0 | 0 | 0 | 0 | 0/1 | 0/1 |
| Hep A, B, C, syphilis, HIV | 0 | 0 | 0 | 0 | 0/6 | 0/6 |
| Total | 4/30 | 3/30 | 5/30 | 1/30 | 1/30 | 1/36 |
| % positive | 13.3% | 10.0% | 16.7% | 3.3% | 3.3% | 2.8% |
Abbreviations: Ab, antibody; CMV, cytomegalovirus; EBV, Epstein-Barr virus; Euro, Euroimmun; Flu A, influenza A virus; HCV, hepatitis C virus; Hep, hepatitis; Ig, immunoglobulin; MERS-CoV, Middle East respiratory syndrome; NCP, nucleocapsid; Parvo, parvovirus; SARS-CoV-1, severe acute respiratory syndrome coronavirus 1.
Samples were obtained from the Victorian Infectious Diseases Reference Laboratory (VIDRL) at The Peter Doherty Institute for Infection and Immunity and Royal Melbourne Hospital (RMH), Victoria, Australia.
Testing Protocols
RT-PCR
SARS-CoV-2 RNA was detected in respiratory swabs by RT-PCR, and results were provided by both institutions. VIDRL used an in-house RT-PCR with previously published primers [12] targeting RdRP gene with positives then tested by a second assay targeting N gene and/or S gene. RMH utilized a commercial Coronavirus Typing assay (AusDiagnostics), a 2-step, heminested multiplex tandem PCR, with 7 coronavirus RNA targets plus a proprietary artificial sequence as an internal control [12] targeting OFR1a gene; positives were confirmed by GeneXpert, targeting N gene and E gene or tested by VIDRL’s in-house RT-PCR assay.
ELISA
ELISA testing was performed in the Serology Laboratory at VIDRL following the manufacturer’s Instructions for Use (IFU), with results reported semiquantitatively as either a signal/cutoff ratio (Euroimmun and Wantai) or percent inhibition (sVNT).
Assay performance characteristics were assessed using the panels described previously. Testing was performed manually, though high-throughput testing could be performed using robotic platforms.
Intra-assay variability was calculated by testing 10 in-house quality control sample replicates within the same microtiter plate and interassay variability by testing on different plates, on average, on 6 different days using the same kit lot number; results are reported as a coefficient of variation (CV).
Euroimmun Anti-SARS-CoV-2 S1-IgG, S1-IgA, and Anti-SARS-CoV-2 NCP-IgG (Euroimmun Medizinische Labordiagnostika, Lubeck, Germany)
These are indirect ELISAs for detection of immunoglobulin (Ig) class IgG or IgA against SARS-CoV-2 antigens. Wells are coated with either (i) S protein S1-domain or (ii) modified NCP. SARS-CoV-2 binding antibodies are detected using enzyme-labeled antihuman-IgG or antihuman-IgA conjugates and a colorimetric substrate and are read spectrophotometrically.
Wantai SARS-CoV-2 Total Ab (Beijing Wantai Biological Pharmacy Enterprise, Beijing, China)
This is a 2-step incubation antigen “sandwich” assay detecting total antibodies binding the SARS-CoV-2 RBD within the S1 subunit of S protein. Patient antibody to SARS-CoV-2 that binds antigen coated on the plate is bound by horseradish peroxidase (HRP) antigen conjugate, forming an antigen-antibody-antigen-HRP complex detectable by colorimetric substrate and read spectrophotometrically.
Wantai SARS-CoV-2 IgM
This is a capture ELISA for detection of IgM-class antibodies to SARS-CoV-2 virus. Anti-µ chain antibodies on the plate capture patient IgM antibodies; detection is by recombinant SARS-CoV-2 antigen-HRP-conjugate followed by a colorimetric substrate read spectrophotometrically.
GenScript SARS-CoV-2 Surrogate Virus Neutralization Test (GenScript USA, Inc., Piscataway, NJ, USA)
This is a species- and isotype-independent blocking ELISA that mimics virus neutralization, detecting circulating neutralizing SARS-CoV-2 antibodies that block the interaction between the viral spike RBD and host angiotensin-converting enzyme 2 (ACE2) cell surface receptor. HRP-conjugated recombinant SARS-CoV-2 RBD fragment binds any circulating nAb to RBD, preventing capture by the hACE2 protein on the well, which is subsequently removed in the following wash step. Substrate reaction incubation time is determined by temperature; ideal reaction temperature and time in the IFU are 25°C for 15 minutes. For temperatures lower than 25°C, the time can be extended. At 15 minutes, our control values did not meet the assay validity criteria, but at 20 minutes they fell within the acceptable ranges. Color intensity is inversely dependent on the titer of anti-SARS-CoV-2 nAbs. We report 3 types of alternative estimates: (i) 20% cutoff without retesting (as per the IFU), (ii) 20% cutoff with retesting for equivocal results (18%–22%), and (iii) 25% cutoff without retesting.
Microneutralization Assay
In a subset of sera, a comparison was performed using an in-house MN assay and the commercial ELISAs. To compare MN sensitivity and specificity with the Euroimmun and sVNT assays, 85 (panel A) samples from RT-PCR-positive patients were assessed. Seventy samples (panel B) were assessed for comparison of MN to the Wantai assays, which require a higher volume of sera than the other assays.
The MN assay was performed using an in-house assay as described previously [13]. Briefly, the ability of serial 2-fold dilutions of sera to neutralize the infectivity of 100 median tissue culture infectious doses of SARS-CoV-2 was assessed by inhibition of viral cytopathic effect in Vero cells.
The nAb titer was calculated using the Reed/Muench method [14, 15].
Statistical Analysis
All statistical analyses were conducted using R, version 4.0.2 [16]. Responses were assessed in each of the 3 patient groups (infected, population, cross-reactive). Sensitivity was estimated using the infected group separately for each of the 3 categories from time of symptom onset: <7 days, 7–14 days, >14 days. Each observation was treated as independent within each subgroup. Specificity was estimated separately for the population group and the cross-reactive group. The 95% CIs were generated using the exact binomial Clopper-Pearson method with the PropCIs R package [17]. Sensitivity estimates (point estimates and CI bounds) were averaged across the onset categories to give the “averaged” sensitivity estimate. Point estimates and interval bounds for the sensitivity and the population group specificity were used to calculate the corresponding estimates and bounds for positive predictive value (PPV) and negative predictive value (NPV) at different theoretical levels of population prevalence (0.1%, 0.5%, 1%, 5%, 10%, and 20%).
Patient Consent
Project ethical approval for RMH specimens was obtained from the Melbourne Health Human Research Ethics Committee (RMH HREC QA2020052); this included written patient consent. The in-house panel consisted of anonymized excess diagnostic specimens sent to VIDRL for COVID-19 testing and the “VIDRL Serum Reference Collection.”
RESULTS
Sensitivities and Specificities of 6 ELISA Assays
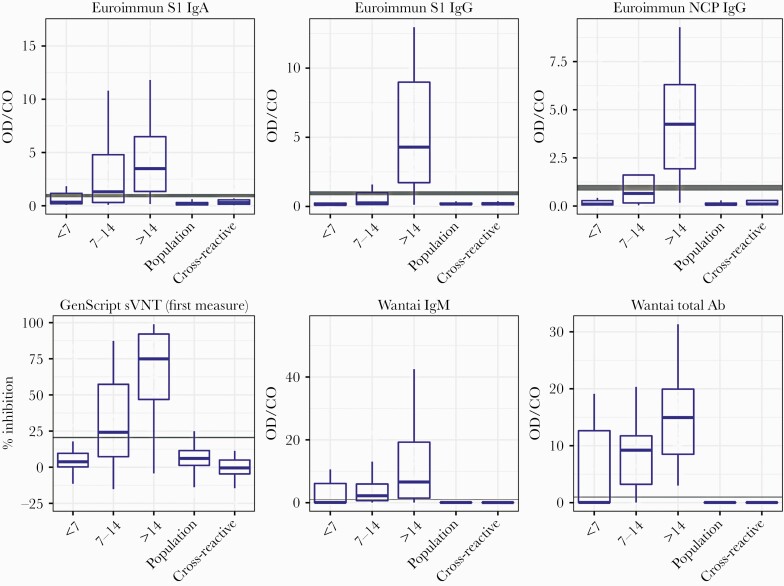
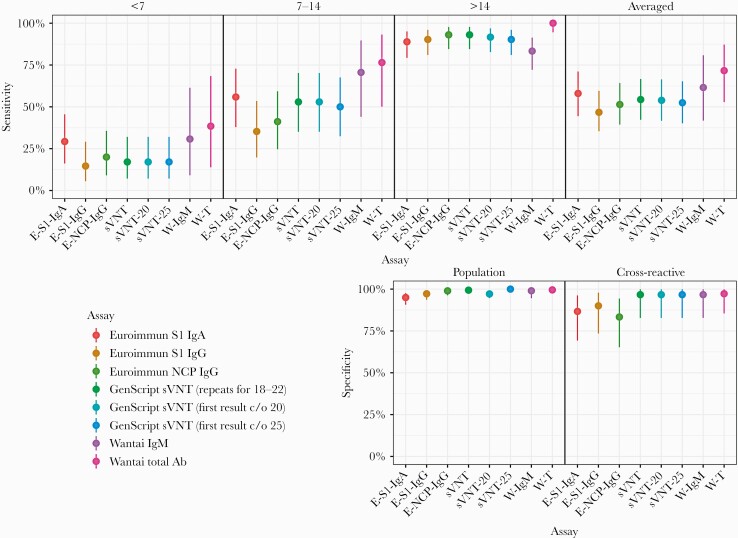
The performance characteristics of the 6 commercial assays are shown in Table 3. Increased sensitivity and increased antibody levels, reported as index values or percent inhibition, were observed across all time frames for all assays (Figure 1). For comparative assay sensitivity and specificity, see Figure 2.
Table 3.
Performance Characteristics of Assays With RT-PCR, All Time Points and Average
| Group | Euro | Euro | Euro | GenScript | Wantai | Wantai |
|---|---|---|---|---|---|---|
| S1 IgA | S1 IgG | NCP IgG | sVNT | IgM | Total Ab | |
| (20% c/o)a | ||||||
| (20% c/o/equ)b | ||||||
| (25% c/o)c | ||||||
| Sensitivity [95% CI], % | Panel A | Panel B | ||||
| Infected-RT-PCR pos | ||||||
| Averaged | 58.0 [44.4–71.1] | 46.7 [35.4–59.6] | 51.4 [39.4–64.2] | 54.4 [42.3–66.7]a,b,c | 61.6 [41.8–80.8] | 71.6 [52.8–87.2] |
| <7 d | 29.3 [16.1–45.5] | 14.6 [5.6–29.2] | 20.0 [9.1–35.6] | 17.1 [7.2–32.1]a,b,c | 30.8 [9.1–61.4] | 38.5 [13.9–68.4] |
| 7–14 d | 55.9 [37.9–72.8] | 35.3 [19.7–53.5] | 41.2 [24.6–59.3] | 52.9 [35.1–70.2]a,b,c | 70.6 [44–89.7] | 76.5 [50.1–93.2] |
| >14 d | 88.9 [79.3–95.1] | 90.3 [81.0–96.0] | 93.1 [84.5–97.7] | 93.1 [84.5–97.7]a,b,c | 83.3 [72.1–91.4] | 100 [94.6–100] |
| Specificity [95% CI], % | ||||||
| Population | 95 [90.7–97.7] | 97.2 [93.6–99.1] | 99 [96.3–99.9] | 97.1 [94.6–98.7]a | 99.0 [94.6–100] | 99.5 [97.4–100] |
| 99.4 [97.7–99.9]b | ||||||
| 100 [98.8–100]c | ||||||
| Cross-reactive assessment | 86.7 [69.3–96.2] | 90.0 [73.5–97.9] | 83.3 [65.3–94.4] | 96.7 [82.8–99.9]a,b,c | 96.7 [82.8–99.9] | 97.2 [85.5–99.9] |
| PPV % (best–worst) 0.5% prevalence >14 d | 8.2 [4.1–17.0] | 14.0 [6.0–34.6] | 30.9 [10.2–79.4] | 13.8 [7.1–26.8]a | 29.5 [6.2–94.8] | 51.2 [15.3–97.6] |
| 42.2 [15.6–86.3]b | ||||||
| 100 [25.7–100]c | ||||||
| NPV % (best–worst) 0.5% prevalence >14 d | 99.9 [99.8–100] | 99.9 [99.9–100] | 100 [99.9–100] | 100 [99.9–100]a,b,c | 99.9 [99.9–100] | 100 [100–100] |
| PPV (best–worst) 10% prevalence >14 d | 66.3 [48.6–82.0] | 78.2 [58.4–92.1] | 90.8 [71.6–98.8] | 77.9 [63.0–89.0]a | 90.3 [59.5–99.8] | 95.9 [79.9–99.1] |
| 94.2 [80.4–99.3]b | ||||||
| 100 [88.4–100]c | ||||||
| NPV (best–worst) 10% prevalence >14 d | 98.7 [97.5–99.4] | 98.9 [97.8–99.6] | 99.2 [98.2–99.7] | 97.8 [94.1–99.7]a,b,c | 98.2 [96.8–99.1] | 100 [99.4–100] |
For all calculations, equivocal results were treated as positive. Best–worst: best- and worst-case intervals—intervals obtained by setting the corresponding sensitivity and specificity estimates to their upper/lower interval bound); Averaged, average sensitivity across the symptom onset categories.
Abbreviations: Ab, antibody; c/o, cutoff; Euro, Euroimmun; Ig, immunoglobulin; NCP, nucleocapsid; NPV, negative predictive value; PPV, positive predictive value; sVNT, surrogate virus neutralization test.
a20% c/o; sVNT ≥20% cutoff, no repeats.
b20% c/o/equiv; sVNT ≥20% cutoff + repeats for equivocals (18%–22%).
c25% c/o; ≥25% cutoff, no repeats.
Figure 1.
Boxplots of data distribution as signal/cutoff value for each assay for the RT-PCR-positive and control sera by days post–symptom onset: <7 days, 7–14 days, and >14 days. Boxes represent median value and interquartile range, and whiskers represent largest and smallest values. Gray horizontal lines represent the cutoff value, and the shaded gray indicates the equivocal/borderline zone. Measure is index value (sample OD value/cutoff OD) for all assays except sVNT. sVNT measure is % inhibition, calculated as per IFU. Abbreviations: Ab, antibody; CO, cutoff; IFU, Instructions for Use; Ig, immunoglobulin; NCP, nucleocapsid; OD, optical density; RT-PCR, reverse transcription polymerase chain reaction; sVNT, GenScript surrogate virus neutralization test.
Figure 2.
Comparative analysis of assay sensitivity post–symptom onset <7 days, 7–14 days, ≥14 days, and averaged. Comparative analysis of assay specificity in cross-reactive assessment and population groups Abbreviations: Ab, antibody; E-S1-IgA, Euroimmun S1-IgA; E-S1-IgG, Euroimmun S1-IgG; E-NCP-IgG, Euroimmun NCP-IgG; Ig, immunoglobulin; NCP, nucleocapsid; sVNT, GenScript surrogate virus neutralization test (20% cutoff with repeat testing for equivocal results [18–22]; sVNT-20 with 20% cutoff with no repeat testing; sVNT-25 with 25% inhibition cutoff and no repeat testing); W-IgM, Wantai IgM, W-T, Wantai Total Ab.
Euroimmun S1 IgA, S1 IgG, and NCP IgG
Sensitivity was assessed using sera from subjects with RT-PCR-proven COVID-19 disease (Table 3).
When sera collected >14 days post–symptom onset were considered, the sensitivity was 88.9% (95% CI, 79.3%–95.1%) for S1-IgA, 90.3% (95% CI, 81.0%–96.0%) for S1-IgG, and 93.1% (95% CI, 84.5%–97.7%) for NCP-IgG. The averaged sensitivity across all time frames was 58.0% (95% CI, 44.4%–71.1%) for S1-IgA, 46.7% (95% CI, 35.4%–59.6%) for S1-IgG, and 51.4% (95% CI, 39.4%–64.2%) for NCP IgG.
Specificity was assessed using the population and cross-reactive groups (Table 3). Specificity when testing the prepandemic population group was lowest at 95.0% (95% CI, 90.7%–97.7%) using the S1-IgA, 97.2% (95% CI, 93.6%–99.1%) using S1-IgG, and highest at 99.0% (95% CI, 96.3%–99.9%) using NCP-IgG. In the cross-reactive assessment group, the lowest specificity was 83.3% (95% CI, 65.3%–94.4%) using NCP-IgG, 86.7% (95% CI, 69.3%–96.2%) using S1-IgA, and highest using S1-IgG at 90% (95% CI, 73.5%–97.9%). Initial testing of the S1-IgA kit gave poorer specificity than reported here; however, with the introduction of a new buffer by Euroimmun, retrospective testing of prepandemic samples showed an 8.5% specificity improvement.
Cross-reactivity was observed with sera containing anti-SARS-CoV-1 antibody (all tests), anti-MERS antibody (only NCP IgG, 1/9 samples), and sera positive for mycoplasma and CMV (all tests) and dengue (only S1-IgA, 1/5 samples) antibodies. No cross-reactivity was observed to sera positive for parvovirus-B19 or seasonal coronavirus antibodies (Table 2).
GenScript SARS-CoV-2 sVNT
For the COVID-19 RT-PCR-positive group, the sensitivity of the sVNT was 93.1% (95% CI, 84.5%–97.7%) when sera collected >14 days from symptom onset were considered and 54.4% (95% CI, 42.3%–66.7%) across all time periods (Table 3). The assay specificity was 97.1% (95% CI, 94.6%–98.7%) when the prepandemic population group was tested. All other assays required retesting of samples with results within 10%–20% of the assay cutoff (the “equivocal zone”), as described in each manufacturer’s IFU. Because the sVNT had similar intra-assay and interassay variability to the other assays, we wanted to see if specificity improved with retesting of samples within the 10% equivocal zone. When the prepandemic population group sera were tested with repeat testing of equivocal zones, the specificity increased to 99.4% (95% CI, 97.7%–99.9%), with 2 of the 312 samples being false positive (both MN negative). Both samples gave results close to the 20% inhibition cutoff (22.5%, 21%). Because the IFU suggests setting a population-specific cutoff, we considered a range of thresholds (Supplementary Figure 1a–c and Supplementary Table 1a and b). Using a cutoff value of 25% increased the specificity and PPV without changing the sensitivity and NPV (Table 3). In the cross-reactivity assessment group, cross-reactivity was observed with sera containing antibody to SARS-CoV-1 (Table 2).
Wantai Total Ab and IgM
The averaged sensitivity for the Wantai Total Ab was 71.6% (95% CI, 52.8%–87.2%), and for the IgM assay it was 61.6% (95% CI, 41.8%–80.8%), which improved to 100.0% (95% CI, 94.6%–100.0%) and 83.3% (95% CI, 72.1%–91.4%), respectively, when restricted to sera collected >14 days post–symptom onset (Table 3).
The specificity for the Wantai Total Ab was 99.5% (95% CI, 97.4%–100.0%), and for IgM it was 99.0% (95% CI, 94.6%–100.0%); in the population group and the cross-reactive group, these were 97.2% (95% CI, 85.8%–99.9%) and 96.7% (95% CI, 82.8%–99.9%), respectively. Cross-reactivity was observed with sera containing antibody to SARS-CoV-1 (Table 2).
Positive Predictive Value and Negative Predictive Value
Positive predictive value and negative predictive value were calculated across a range of population prevalence estimates: 0.1%, 0.5%, 1%, 10%, and 20% were calculated for each assay (Supplementary Table 2 and Supplementary Figure 2). In a low-prevalence setting like ours, assuming a population prevalence of 0.32% for Victoria and 0.11% for Australia [17], the PPV for the best-performing assay, Wantai Total Ab, was 40.2% and 18.7%, respectively, while the NPV was 100% for sera collected >14 days post–symptom onset.
Discordance Between Assays
In the RT-PCR-positive group, there were 41 discordant results, with discordance increasing with time since disease onset from 8/41 at <7 days postonset to 19/41 at >14 days postonset. In the population and cross-reactive assessment groups, there were 17 and 6 discordant results, respectively. In part, this appeared attributable to the type of antibody being detected, the antigen targeted in the assay, and the assay format. For samples collected >14 days postonset, 11 were Wantai IgM negative but Total Ab positive. All 11 samples were collected 21 to >30 days postonset, suggesting discordance attributable to IgM loss [18]. We retrospectively tested these samples for IgA (results not shown). Six of 11 (55%) were IgA positive, suggesting waning IgM antibody and to a lesser extent IgA (Supplementary Figure 3, heat map).
Intra- and Interassay Variability
Intra-assay variability was low for all assays, <10%, and ranged from CV = 3.1% for the Euroimmun S1-IgA to CV = 8.8% for the Euroimmun S1-IgG. Interassay variability ranged from Euroimmun NCP-IgG CV = 3.1% to Wantai IgM CV = 14.9% (Supplementary Table 3a and b).
Assay Comparison With Microneutralization
Eighty-five samples from RT-PCR-positive panel A were tested by MN (Table 4A). Seventy-three percent (62/85) were MN positive. Euroimmun S1-IgA had equal sensitivity, with 73% (62/85) samples positive, 67% (57/85) sVNT positive, 67% (56/84) Euroimmun NCP IgG positive, and 62% (53/85) Euroimmun S1 IgG positive. Hence MN and Euroimmun S1-IgA had the highest sensitivity. At >14 days post–symptom onset, Euroimmun S1 IgA and Euroimmun NCP-IgG had 100% (38/38) sensitivity, followed by MN and Euroimmun S1 IgG at 97% (37/38). sVNT was 95% (36/38) sensitive.
Table 4.
A, Microneutralization Comparison With ELISAsa; B, Microneutralization Comparison With ELISAs (% Agreement)b
| (a) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MN | sVNT | Euro S1-IgA | Euro S1-IgG | Euro NCP-IgG | MN | Wantai Total Ab | Wantai IgM | |||
| Group | Panel A | Panel B | ||||||||
| Sensitivity | No. | No | ||||||||
| Infected-RT-pos pos/total | 85 | 62/85 | 57/85 | 62/85 | 53/85 | 56/84b | 70 | 55/70 | 61/70 | 55/70 |
| (%) | (73) | (67) | (73) | (62) | (67) | (79) | (87) | (79) | ||
| <7 d pos/total | 21 | 9/21 | 5/21 | 8/21 | 4/21 | 5/20 | 9 | 3/9 | 4/9 | 3/9 |
| (%) | (43) | (24) | (38) | (19) | (25) | (33) | (44) | (33) | ||
| 7–14 d pos/total | 26 | 16/26 | 16/26 | 16/26 | 12/26 | 13/26 | 20 | 12/20 | 16/20 | 15/20 |
| (%) | (62) | (62) | (62) | (46) | (50) | (60) | (80) | (75) | ||
| >14 d pos/total | 38 | 37/38 | 36/38 | 38/38 | 37/38 | 38/38 | 41 | 40/41 | 41/41 | 36/41 |
| (%) | (97) | (95) | (100) | (97) | (100) | (98) | (100) | (88) | ||
| Specificity | No. | No | ||||||||
| Population | 20 | 0/20 | 0/20 | 11/20 | 2/20 | 0/20 | 14 | 0/14 | 0/14 | 0/14 |
| Cross-reactive assessment | 5 | 0/5 | 1/4 | 1/4 | 1/4 | 2/4 | 1 | 0/1 | 1/1 | 1/1 |
| Total pos/total | 25 | 0/25 | 1/24 | 10/24 | 3/24 | 2/24 | 17 | 0/15 | 1/15 | 1/15 |
| (%) | (0) | (4.2) | (41.7) | (12.5) | (8.3) | (0) | (7.3) | (7.3) |
| (b) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Group | Euro S1-IgA | Euro S1-IgG | Euro NCP-IgG | sVNT (20% c/o) | svnt-20 (20% c/o/equ) | svnt-25 (25% c/o) | Wantai IgM | Wantai Total Ab |
| Sensitivity | Overall % Agreement (% Best, % Worst) | |||||||
| Panel A | Panel B | |||||||
| Infected RT-PCR pos | 80% (62%, 91%) | 79% (61%, 92%) | 84% (66%, 95%) | 84% (65%, 95%) | 83% (64%, 94%) | 81% (62%, 94%) | 91% (67%, 99%) | 86% (62%, 97%) |
| Days from symptom onset | % Agreement (95% CI) [Pos/Total] | |||||||
| <7 days | 57% | 67% | 75% | 71% | 71% | 71% | 90% | 80% |
| (34%, 78%) | (43%, 85%) | (51%, 91%) | (48%, 89%) | (48%, 89%) | (48%, 89%) | (55%, 100%) | (44%, 97%) | |
| [12 / 21] | [14 / 21] | [15 / 20] | [15 / 21] | [15 / 21] | [15 / 21] | [9 / 10] | [8 / 10] | |
| 7–14 days | 85% | 77% | 81% | 85% | 85% | 81% | 89% | 79% |
| (65%, 96%) | (56%, 91%) | (61%, 93%) | (65%, 96%) | (65%, 96%) | (61%, 93%) | (67%, 99%) | (54%, 94%) | |
| [22 / 26] | [20 / 26] | [21 / 26] | [22 / 26] | [22 / 26] | [21 / 26] | [17 / 19] | [15 / 19] | |
| >14 days | 97% | 95% | 97% | 95% | 92% | 92% | 93% | 98% |
| (86%, 100%) | (82%, 99%) | (86%, 100%) | (82%, 99%) | (79%, 98%) | (79%, 98%) | (80%, 98%) | (87%, 100%) | |
| [37 / 38] | [36 / 38] | [37 / 38] | [36 / 38] | [35 / 38] | [35 / 38] | [38 / 41] | [40 / 41] | |
| Specificity | % Agreement (95% CI) [Pos/Total] | |||||||
| Population | 55% | 90% | 100% | 100% | 100% | 70% | 100% | 100% |
| (32%, 77%) | (68%, 99%) | (83%, 100%) | (83%, 100%) | (83%, 100%) | (46%, 88%) | (77%, 100%) | (77%, 100%) | |
| [11 / 20] | [18 / 20] | [20 / 20] | [20 / 20] | [20 / 20] | [14 / 20] | [14 / 14] | [14 / 14] | |
| Cross-reactive | 75% | 80% | 50% | 80% | 80% | 40% | 0% | 0% |
| (19%, 99%) | (28%, 99%) | (6.8%, 93%) | (28%, 99%) | (28%, 99%) | (5.3%, 85%) | (0%, 98%) | (0%, 98%) | |
| [3 / 4] | [4 / 5] | [2 / 4] | [4 / 5] | [4 / 5] | [2 / 5] | [0 / 1] | [0 / 1] |
Abbreviations: Ab, antibody; c/o, cutoff; equ, equivocal; Euro, Euroimmun; Ig, immunoglobulin; NCP, nucleocapsid; pos, positive; sVNT; GenScript surrogate virus neutralization test.
aGroup A—49% of sera (>14 days–post symptom onset); Group B—69% of sera (>14 days post–symptom onset).
bGroup A—49% of sera (>14 days post–symptom onset); Group B—69% of sera (>14 days post–symptom onset)
Seventy samples from RT-PCR-positive panel B were tested by MN. Seventy-nine percent (55/70) were MN positive. Wantai Total had the highest sensitivity, with 87% (61/70) positive. At >14 days post–symptom onset, Wantai Total Ab and MN had comparable sensitivities of 100% (41/41) and 98% (40/41), respectively, and Wantai IgM was 88% (36/41) sensitive (Table 4A).
The MN assay specificity for the population and cross-reactive assessment sera groups was 100%.
DISCUSSION
Reliable COVID-19 serosurveillance is important for guiding the pandemic response [9, 10]. At the individual level, serology can provide a tool for resolving the diagnosis for patients with infections not confirmed by RT-PCR. At the population level, it provides policy-makers with an assessment of the overall impact of the pandemic and vaccination efficacy.
Assay sensitivities ranged from 46.7% to 71.6%, and specificities from 95.0% to 99.5% for the prepandemic population group and from 83.3% to 97.2% for the cross-reactivity assessment group.
Sensitivity for sera collected >14 days post–symptom onset ranged from 83.3% to 100%. Consistent with the biology of an immune response, and as reported by others, when results from all time points were considered, sensitivity was low, but it increased substantially when only sera collected >14 days postonset were assessed [19], underlining the need to wait a sufficiently long period before confirming infection status by serology. The Euroimmun NCP-IgG and sVNT had good sensitivity; however, the highest sensitivity was achieved by Wantai Total Ab at 100%. The Euroimmun NCP-IgG and Wantai IgM had better prepandemic population group specificity at 99% than sVNT at 97.1%. Wantai Total Ab achieved the highest specificity in the cross-reactivity assessment group, at 97.2%.
We observed improved sVNT specificity when we varied from the IFU to use a +/-10% repeat equivocal zone; however, optimal specificity was achieved when we adjusted the cutoff to 25% without reducing the sensitivity and NPV (Table 3).
Cross-reactivity was observed in all assays with sera containing anti-SARS-CoV-1 antibody. Cross-reactivity was more commonly seen in the Euroimmun assays, and depending on the assay, in sera positive for MERS, mycoplasma, CMV, and dengue antibodies, consistent with other reports [19]. Assays challenged with seasonal coronavirus antibody–positive sera were not reactive, and no cross-reactivity to sera containing parvovirus B-19 antibodies was seen (Table 2).
The Wantai Total Ab and sVNT (using a modified population-specific cutoff of 25%) demonstrated the highest specificity. This may be because both assays detect Ab to the RBD within the S1-subunit, therefore increasing specificity by exclusion of cross-reacting epitopes outside this domain [19]. sVNT is the only immunoassay correlating with nAb [20, 21], although studies have shown that samples with Wantai Total Ab–positive results with an index ratio >10 had detectable levels of nAb by virus neutralization test [22]. Assay formats that utilize a single detector antibody, for example, Wantai Total Ab, may show greater specificity than those using 2 antibodies in an antigen-antibody-antibody format, such as the Euroimmun assays [23].
Except for the Wantai Total Ab, manufacturers for all assays reported higher sensitivities than we demonstrated. However, direct comparison of results is limited by differences in available sample cohorts and sampling time frames. Our results are consistent with prior reports of sensitivity for Wantai Total Ab of 100% for samples collected >14 days postonset [11]: Euroimmun S1-IgG sensitivity postonset: 43% (7–13 days), 67% (14–20 days), and 78% (≥21 days). The specificity was 96.0% [19]: Euroimmun NCP-IgG: 88.89% (>14 days) [21] and 95.2% (14–17 days). The specificity was 94.7% [24]. While other groups have reported slightly higher sVNT sensitivity (95%–100%) and specificity (100%) [21] than we observed, the sensitivity in our assessment was 93.1% (95% CI, 84.5%–97.7%) for sera collected >14 days post–symptom onset, with a specificity of 97.1% in the prepandemic population group. This improved to 100% with use of a modified 25% inhibition cutoff and 96.7% for the cross-reactive group.
We compared agreement between the ELISAs and an in-house MN assay, which is the current gold standard to assess protective immunity against SARS-CoV-2 [22]. The Wantai IgM assay had the highest agreement with MN at 91%, followed by Wantai Total Ab at 86%, and the lowest was with the Euroimmun S1-IgG at 79%. The sVNT performed according to the IFU gave the best concordance with MN at 84%, compared with 83% when an equivocal repeat zone was used and 81% when a 25% cutoff was used. At >14 days post–symptom onset, Wantai Total Ab had the best agreement with MN at 98%, followed by Euroimmun S1-IgA, Euro NCP-IgG, and sVNT (as per IFU) with 97%, 97%, and 95%, respectively, and Wantai IgM had the poorest agreement at 93%. MN and ELISA result concordance for sera from the prepandemic population group ranged from 55% to 100% and from 0% to 80% for the cross-reactive assessment group; MN was the only assay not to show reactivity to the SARS-CoV-1 antibody–positive sample. Interestingly, some of the Ab-binding assays showed better agreement with the MN than the sVNT.
The sVNT was not as sensitive as MN in our comparison. However, it is simpler to perform than a VNT and does not require a level 3 biocontainment laboratory or a highly skilled operator—nor does it have 5-day test turnaround time—and it has high specificity (if a population-adjusted cutoff is employed) and generally good correlation with VNT [21]. Further comparison is required to determine its suitability as an alternative to VNT in certain settings, particularly as we move into the postvaccination phase.
In our study, as reported by others, IgA and IgM were detected earlier than IgG, and IgA specificity was lower than IgM or IgG [18]. For clinical testing, the Wantai total antibody assay demonstrated the best overall performance with the highest sensitivity, specificity, PPV, and NPV. Detecting RBD-Ab, the strong linear correlation between S- and RBD-Ab and ACE-2 receptor binding suggests that nAb is being detected [21, 22]. It is also suitable for population screening where high specificity is required and high sensitivity is desirable. Other assays such as Euroimmun NCP-IgG have high sensitivity but lower specificity, with a low PPV. Further research on the utility of IgA and IgM detection is required, especially as indicators of recent infection. The high specificity of the sVNT assay was further improved to 100% with cutoff adjustment to 25% for our population. Although the sensitivity of the sVNT was lower than the Wantai Total Ab, its specificity makes it a reliable supplemental test to use with screening assays. The assays evaluated here detect antibodies to different viral targets—S, NCP, or RBD—and emerging humoral response dynamics studies suggest that these antibodies have different half-lives [25], an important consideration for serosurveillance. An additional important consideration for assay selection is understanding whether the assay detects non-nAb or nAb (particularly for testing postvaccination); however, further research is required.
The Wantai SARS-CoV-2 total Ab assay was alone among those tested in achieving sensitivity and specificity very close to that reported by the manufacturer (94.5% and 100%, respectively); 1 limitation of this assay is the requirement for 100 µL of serum vs 10 µL for the other assays. In low-prevalence populations and with low test PPVs, serological testing algorithms should use highly sensitive assays for screening such as Wantai Total Ab, followed by supplemental or confirmatory testing with highly specific assays such as sVNT or MN to ensure reliable results. An important component of this study is the comparison of the sensitivities of the ELISAs and MN, which should be considered in the context of discordant screening and supplemental results in testing algorithms.
The limitations of our study include the use of 2 different RT-PCR POS sample panels; however, there was sample overlap between the panels, with all of panel B included within panel A. Panel B also included more convalescent patient samples (69%) collected >14 days post–symptom onset compared with panel A, which contained 49%. A rise in index value/inhibition for all assays was observed over time (Figure 1); however, these measurements are semiquantitative, indicating antibody amount present for comparison between assays, not antibody titer, which can only be obtained by sample titration to “end point.”
In conclusion, our study provides a detailed comparison of 6 commercially available serology assays with performance consistent with what others have reported. The strength of our study is comparing these assays detecting different antibody types with a surrogate virus neutralization test and a gold standard virus neutralization assay. As we move into the postvaccination phase, consideration should be given to the use of GenScript sVNT in certain settings as an alternative to VNT. ELISA detection of anti-SARS-CoV-2-specific antibody provides evidence of prior SARS-CoV-2 infection or vaccination; however, further research is required to determine correlates of protection, antibody longevity, and the role played by the adaptive immune system.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors thank the Serology Laboratory at VIDRL, particularly Di Karamalakis, The Viral Identification Laboratory, and the Information Technology department, particularly Dallas Wilson. We thank Dr. Allen Cheng, Dr. Maryza Graham, and Megan Wieringa from Monash Medical Centre and Dr. Katherine Kedzierska from The Department of Microbiology and Immunology, The University of Melbourne at The Doherty Institute, for referring samples to VIDRL for COVID-19 investigation; Annette Fox from the WHO Collaborating Centre for Reference and Research on Influenza, VIDRL, Doherty Institute, for her contribution to the study; Prof. Lin Fa Wang for supplying us with the sVNT assay for evaluation. The Victorian Infectious Diseases Reference Laboratory is supported by the Victorian Department of Health and Human Services. The WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health.
Financial support. This work was supported by the Victorian Department of Health and Human Services and by the Australian Government Department of Health.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. European Centre for Disease Prevention and Control, Cohen A, Kessel B. False positives in reverse. COVID-19 situation update worldwide, as of 8 April 2020. Available at: https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases.
- 2. Gudbjartsson D, Norddahl G, Melsted K, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020; 383:1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seow J, Graham C, Merrick B, et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. Nat Microbiol 2020; 5:1598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 2020; 9:386–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urdea M. SARS-CoV-2 use cases white paper. White paper, Rockefeller Foundation, Arizona State University and Duke University. 2020. Available at: https://halteresassociates.com/wp-content/uploads/2020/08/Halteres-SARS-CoV-2-White-Paper-MUrdea-20200812-v08.pdf. Accessed 10 October 2020. [Google Scholar]
- 6. Yong SEF, Anderson DE, Wei WE, et al. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect Dis 2020; 20:809–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qiu H, Wu J, Hong L, et al. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis 2020; 20:689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Public Health England, Porton Down, Nuffield Department of Medicine, University of Oxford, Oxford University Hospitals NHS Foundation Trust. Evaluation of sensitivity and specificity of four commercially available SARS-CoV-2 antibody immunoassays. 2020. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/898437/Evaluation__of_sensitivity_and_specificity_of_4_commercially_available_SARS-CoV-2_antibody_immunoassays.pdf. Accessed date July 2020. [Google Scholar]
- 9. Halters Associates. Use cases for SARS-CoV-2 assays 2020. Available at: https://haltersassociates.com/halteres-sars-cov-2-use-case-tables/. Accessed 10 October 2020.
- 10. Lerner AM, Eisinger RW, Lowy DR, et al. The COVID-19 Serology Studies Workshop: recommendations and challenges. Immunity 2020; 53:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bond K, Nicholson S, Lim SM, et al. Evaluation of serological tests for SARS-CoV-2: implications for serology testing in a low-prevalence setting. J Infect Dis 2020; 222:1280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Houser KV, Gretebeck L, Ying T, et al. Prophylaxis with a Middle East respiratory syndrome coronavirus (MERS-CoV)-specific human monoclonal antibody protects rabbits from MERS-CoV infection. J Infect Dis 2016; 213:1 557–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subbarao K, McAuliffe J, Vogel L, et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol 2004; 78: 3572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 16. Ralph Scherer. PropCIs: various confidence interval methods for proportions. R package version 0.3-0. 2018. Available at: https://CRAN.R-project.org/package=PropCIs. Accessed 15 October 2020.
- 17.Australian Government Department of Health. Victorian COVID-19 data. Available at: https://www.health.gov.au/news/health-alerts/novel-coronavirus-2019-ncovhealth-alert/coronavirus-covid. Accessed 29 October 2020.
- 18. Isho B, Abe K, Zuo M, et al. Mucosal versus systemic antibody responses to SARS-CoV-2 antigens in COVID-19 patients. medRxiv 2020.08.01.20166553 [Preprint]. 29 August 2020. Available at: 10.1101/2020.08.01.20166553. Accessed 29 August 2020. [DOI] [Google Scholar]
- 19. Lassauniere R, Frische A, Harboe Z, et al. Evaluation of nine commercial SARS-C0V-2 immunoassays. medRxiv 2020.04.09.20056325 [Preprint]. 10 April 2020. Available at: 10.1101/2020.04.09.20056325. Accessed 11 October 2020. [DOI] [Google Scholar]
- 20. Wang LF, Ni Chi W, Wah Tan C, et al. Serological differentiation between COVID-19 and SARS infections. Emerg Microbes Infect 2020; 9:1497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan CW, Chia WN, Qin X, et al. A SARS-CoV-2 surrogate virus neutralization test (sVNT) based on antibody-mediated blockage of ACE2-spike (RBD) protein-protein interaction. Nature 2020; 38:1073–8. [DOI] [PubMed] [Google Scholar]
- 22. GeurtsvanKessel C, Okba N, Igloi Z, et al. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun 2020; 11:3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 2020; 26:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Elslande J, Decru B, Jonckheere S, et al. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin Microbiol Infect 2020; 26:1557.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grandjean L, Saso A, Ortiz A, et al. Humoral response dynamics following infection with SARS-CoV-2. medRxiv 2020.07.16.20155663 [Preprint]. 22 July 2020. Available at: 10.1101/2020.07.16.20155663. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.