Abstract
Objectives
To report our institutional experience in devising and implementing a pooling protocol and process for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reverse transcription polymerase chain reaction (RT-PCR) testing over a 3-month period in the fall of 2020.
Methods
The widespread testing implemented in the United States for detecting SARS-CoV-2 infection in response to the coronavirus disease 2019 pandemic has led to a significant shortage of testing supplies and therefore has become a major impediment to the public health response. To date, several institutions have implemented sample pooling, but publications documenting these experiences are sparse. Nasal and nasopharyngeal samples collected from low-positivity (<5%) areas were tested in pools of five on the Roche cobas 6800 analyzer system. Routine SARS-CoV-2 RT-PCR turnaround times between sample collection to result reporting were monitored and compared before and after sample pooling implementation.
Results
A total of 4,131 sample pools were tested over a 3-month period (during which 39,770 RT-PCR results were reported from the Roche system), allowing our laboratory to save 13,824 tests, equivalent to a conservation rate of 35%. A 48-hour or less turnaround time was generally maintained throughout the pooling period.
Conclusions
Sample pooling offers a viable means to mitigate shortfalls of PCR testing supplies in the ongoing pandemic without significantly compromising overall turnaround times.
Keywords: COVID-19, SARS-CoV-2, RT-PCR, PCR, Pooling, Pooled testing, Test reagents
Key Points.
Sample pooling is a viable solution to mitigate polymerase chain reaction (PCR) supply shortages due to unprecedented testing demands imposed by the ongoing COVID-19 pandemic.
Our institution has saved approximately 35% of tests needed to report 39,770 reverse transcription PCR results, while maintaining an excellent overall turnaround time, over a 3-month period in the fall of 2020.
We discuss both general considerations and specific issues related to all phases of testing when we implemented our pooling strategy.
The year 2020 has been overtaken by coronavirus disease 2019 (COVID-19) since emerging from Wuhan, China, infecting over 100 million individuals worldwide, with the United States leading in both infections (over 27 million) and deaths (over 460,000).1-3 The staff of the microbiology laboratories across the nation have been exceedingly challenged with detecting the virus in infected individuals, primarily by using real-time reverse transcription polymerase chain reaction (RT-PCR), which confirms the presence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA.4-6 Particularly in the United States, the staggering number of COVID-19 cases has created a commensurate demand for increased testing.7 Unfortunately, the demand has also highlighted numerous problems in the nation’s testing capabilities such as long turnaround times (TATs), the lack of testing kits, and shortages of testing supplies from reagents to pipetting tips.8,9
A potential solution to mitigate these limitations on laboratory testing, which emerged in the summer of 2020 but was anticipated and discussed as early as March, was sample pooling, a concept that is not novel but has shown practicality in past implementations with other infectious diseases such as syphilis and human immunodeficiency virus.10-14 However, while other countries have reported their experiences, very few US reports of implementing a COVID-19 pooling strategy on a large scale exist.15-20 As pointed out by the College of American Pathologists, there are both hurdles and potential disadvantages to sample pooling: (1) automated liquid handling and information systems customized for pooling that are neither well established nor readily available; (2) testing sensitivity that may be compromised by sample dilution; (3) the likely increase of TAT due to the additional manual labor needed for aliquoting, sample sorting, and result reporting; and (4) increased TAT for samples comprising a positive pool due to individual repeat testing of pooled specimens.21 Accordingly, the US Food and Drug Administration (FDA) has provided specific guidelines and recommendations for the implementation of pooled COVID-19 testing while recommending its use at laboratories processing large volumes of patient samples.22
In this study, we report our experience at the University of Chicago Medical Center (UCMC) Clinical Microbiology and Immunology Laboratory with validating and implementing a COVID-19 sample pooling strategy for the direct detection of SARS-CoV-2 RNA by RT-PCR on the Roche cobas 6800 system between July 31, 2020, and October 31, 2020. By comparing our testing strategy prior to pooling from March 15, 2020, to July 30, 2020, we hope our report can serve as a potential guide for laboratories contemplating the implementation of sample pooling, as well as demonstrating that pooling can conserve reagents and supplies while maintaining general testing capacity and TAT.
Materials and Methods
Validation of Pooling on the Roche cobas 6800 Analyzer System
Validation of pooling on the Roche cobas 6800 platform was conducted in accordance to FDA emergency use authorization (EUA) guidelines for COVID-19 specimen pooling.23 Archived nasopharyngeal patient samples with known COVID-19–positive or presumptive positive (ie, positive for only the pan-Sarbecovirus E-gene target) status exhibiting a range of cycle threshold (Ct) values that reflected that of our patient demographic were used. Twenty-four positive, 6 presumptive positive, and 30 negative samples were tested as part of the validation study. The positive agreement, or sensitivity, was 95.8% (23/24 positives) or 86.7% (with the inclusion of 3/6 presumptive positives for a total of 26/30 samples), which were both above the 85% positive agreement established by the FDA EUA guidelines. The negative agreement, or specificity, was 96.7% (29/30 negatives). For the 24 positive pools, the average Ct values increased by approximately 2.3 cycles. Patient sample testing combining five samples per pool was initiated on July 31, 2020. All patient samples were collected under a quality assurance protocol that qualifies for an institutional review board waiver.
Preanalytic Sample Processing
The Roche SARS-CoV-2 Assay uses the automated cobas 6800 analyzer system to test nasal or nasopharyngeal specimens collected with flocked swabs in liquid Amies Transport Medium (ATM or ESwab) or Universal Viral Transport Medium (UVTM), respectively. The standard volume of each ATM or UVTM collection tube is 1 mL or 3 mL, respectively, whereas an individual test performed on either sample type on the cobas 6800 requires 0.6 mL. In contrast, both the Cepheid Xpert Xpress SARS-CoV-2 Assay and BioFire FilmArray Respiratory Panel 2.1, which includes SARS-CoV-2 testing, require 0.3 mL of sample. All pooled sample handling was performed in a certified class II biological safety cabinet in a dedicated room housing the cobas 6800 instrument.
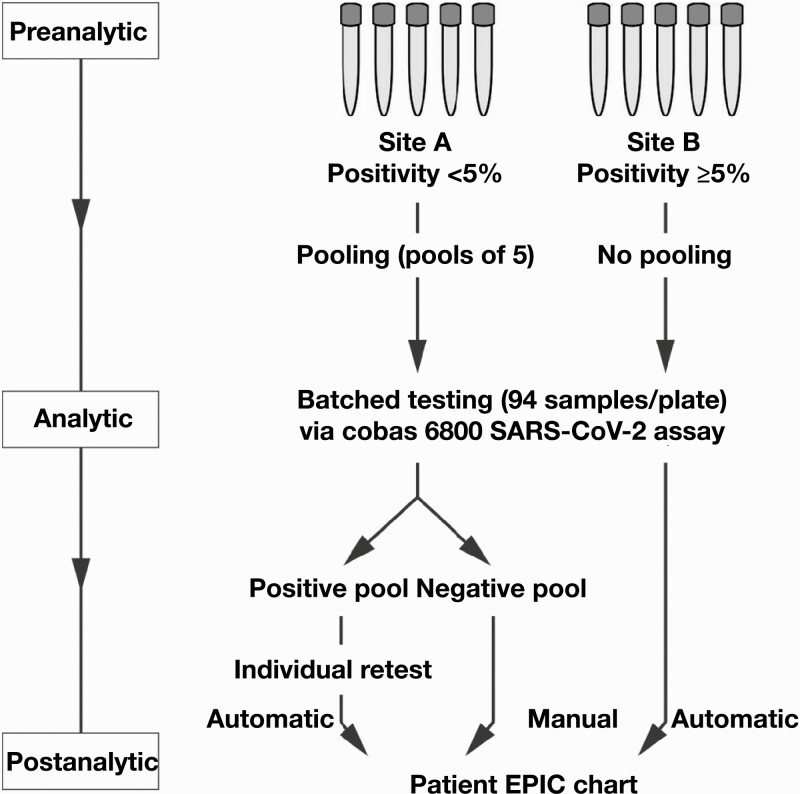
Following the implementation of specimen pooling for COVID-19 testing, all routine/nonstat specimens from both symptomatic and asymptomatic patients received by the UCMC Clinical Microbiology and Immunology Laboratory were stratified for individual or pooled testing based on the monitored COVID-19 positivity rate associated with the originating site (defined geographically across various regions in south Chicago and northwest Indiana) with a cutoff threshold of 5% Figure 1. Stat testing was generally performed for patients requiring emergent procedures, using either the Cepheid Xpert Xpress SARS-CoV-2 Assay or BioFire FilmArray Respiratory Panel 2.1, and therefore these specimens were not considered for pooling given the time-sensitive nature. Both the Cepheid and BioFire platforms served as backup systems for the cobas 6800 instrument.
Figure 1.
Sample pooling workflow at University of Chicago Medical Center for coronavirus disease 2019 testing on the Roche cobas 6800 analyzer system. Pooled testing was conducted on samples derived from sites with overall positivity rates of less than 5%. Positive pools necessitated repeat testing of the individual specimens making up the pool, and those results were automatically reported, whereas manual reporting of individual results was required for negative pools.
Pooling
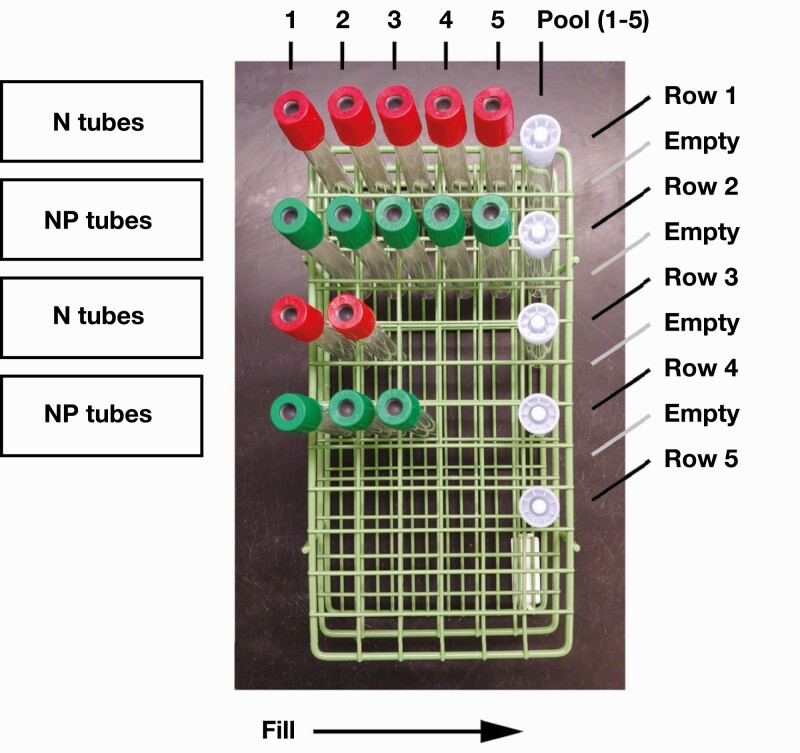
Specimens for pooling were systematically prearranged in rows of five in a 72-sample rack (6 × 12 slots). Nasal and nasopharyngeal samples were grouped separately. The sixth slot in each row was reserved for the pooled sample and was tracked by a barcode. Each pooled sample had a total volume of 1.00 to 1.25 mL, consisting of 200 to 250 µL of each of the five individual specimens in the corresponding row. If a row had fewer than five individual samples, then the final volume was evenly divided among the individual tubes to fill a total volume of approximately 1 mL (eg, 500 µL from two tubes or 350 µL from three tubes). All sample aliquoting was performed manually using disposable graduated transfer bulb pipettes. Samples appearing to be too viscous were vortexed briefly for several seconds before aliquoting. Multiple viscous samples were grouped with other samples to allow for less viscous pools, and these pools were also briefly vortexed. In practice, we took advantage of standard sample racks consisting of six slots per row as a simple way to organize and temporarily store the five specimens making up the pooled sample tube in position 6 for that row Figure 2. In addition to the physical rack position, the barcode identifier label on each tube was scanned and documented via an electronic spreadsheet to track the pooling status of each specimen, as well as the pooled sample tube they were aliquoted into.
Figure 2.
Mockup of rack organization for sample pooling. Empty rows were used to help organize and prevent mixing of sample tubes from separate pools. N and NP represent nasal and nasopharyngeal swab tubes, respectively. The sample pools consisted of either N or NP tubes but not a mix of both specimen types.
Several steps were carried out to reduce the risks of cross-contamination: (1) a single patient specimen tube was opened at any given time and reclosed before another specimen tube was opened, (2) a new bulb pipette was used for aliquoting from each specimen tube to a pooled tube and the bulb pipette was discarded after one use, (3) each pooled tube was immediately capped after aliquoting, and (4) each pooling row was separated by an empty row to avoid a mixup of nearby specimens.
Real-Time PCR and the Roche cobas 6800 Analyzer System
Real-time PCR detection of SARS-CoV-2 genetic material was conducted using the Roche cobas 6800 SARS-CoV-2 Assay performed on the fully automated cobas 6800 analyzer system. The assay integrates nucleic acid extraction and purification, reverse transcription, PCR amplification, and amplicon detection via a series of supply, transfer, processing, and analytic instrument modules within a single testing pipeline. While each patient sample is tested with the addition of an internal positive control RNA included in the PCR master mix, every batch of runs (up to 94 samples per plate) includes independent external positive and negative quality control samples. The runtime of the assay is approximately 3 hours. The data output is managed by the cobas 6800 software and is directly reported to EPIC for all individual, nonpooled specimens.
The cobas 6800 system uses a guanidine thiocyanate/detergent-based solution for viral lysis and magnetic glass particles for nucleic acid extraction, and the SARS-CoV-2 Assay primers are designed to detect a SARS-CoV-2–specific ORF1 a/b nonstructural region and a pan-Sarbecovirus conserved region in the protein envelope gene. The master mix consists of fluorescently labeled probes, deoxyribose nucleotide triphosphates (with deoxyuridine triphosphate, which is degraded by uracil-N-glycosylase, in place of deoxythimidine triphosphate), a DNA polymerase, and a variety of additives in a buffered solution. All reagents require refrigeration at 2°C to 8°C for storage.
Postanalytic Sample Processing and Reporting
A specific code was used in Sunquest Laboratory software to denote the specimens tested in a sample pool, and a Microsoft Excel electronic spreadsheet was used to identify and track all pooled specimens and samples. The results for all specimens from negative pools were reported manually to the patients’ electronic medical record (EMR) in EPIC via Sunquest, whereas each of the five specimens comprising a positive pool required individual testing. Individual testing was usually performed with the Cepheid and BioFire platforms; these require less sample volume compared with that required by the cobas 6800 system. All individually tested specimens automatically crossed over into the EMR. All data were backed up on a secure server to minimize the risk of data loss due to any unexpected instrument or power failure. Quality control reviews of positive pools were conducted postanalytically, after all individual repeat testing of a positive pool was completed. This retrospective process involved verifying that each positive pool indeed contained at least one positive specimen.
Personnel and Staffing
No extra personnel were recruited specifically to help implement COVID-19 pooling. Extra personnel were hired during pooling implementation but primarily to address preexisting and concurrent staffing issues. Adjustments were made with our current personnel to accommodate the extra work involved with pooling. In particular, the receiving staff were tasked with sorting incoming COVID-19 specimens by location and sample type (ie, nasal or nasopharyngeal) and organizing them in racks. The tech assigned to the COVID-19 work schedule would take those racks and aliquot the specimens into pools for testing using the Roche platform. Negative results were released by the COVID-19 tech, whereas the positive pools were identified for repeat individual specimen testing. Other staff members who were available would help in every step of the process, including sorting, pooling, and repeat testing.
TAT Monitoring
The TAT was defined from sample collection to result reporting and was tracked for all routine COVID-19 tests with a positive, presumptive positive, or negative result between March 15, 2020, and October 31, 2020. The TAT data were recorded for each individual sample but did not specify which platform was used. Since most routine/nonstat COVID-19 testing was performed on the cobas 6800 system, the TAT data are representative of the Roche. Data with less than 3.5 hours, or 210 minutes, between receiving the specimen and result reporting were rejected for analysis, since a typical run on the cobas 6800 instrument and preparation of samples would require a minimum 3.5 hours before the results were available.
Results
We performed a total of 123,727 COVID-19 tests across the Roche (75%), Cepheid (23%), and BioFire (2%) qualitative RT-PCR platforms between March 15, 2020, and October 31, 2020. While the overall positivity rate of all COVID-19 testing was 7.8%, the mean positivity rates pre- and postpooling were 8.6% and 6.7%, respectively. The Roche platform was the first to be implemented for clinical use and remained the primary platform for routine and, subsequently, pooled sample testing. The instrument TATs differentiated the three platforms: the Roche assay required approximately 3 hours of runtime, whereas the Cepheid and BioFire platforms required less than 1 hour.
Prior to implementing the pooling protocol, a total of 53,107 RT-PCR results (including all positive, presumptive positive, and negative results) were reported using the Roche platform between March 15, 2020, and July 30, 2020, which corresponded to a mean (SD) of 385 (208) tests per day (range, 10-1,009 tests). The Cepheid and BioFire platforms were implemented for clinical use on April 1, 2020, and May 27, 2020, respectively, which alleviated the testing volume demands on the Roche platform.
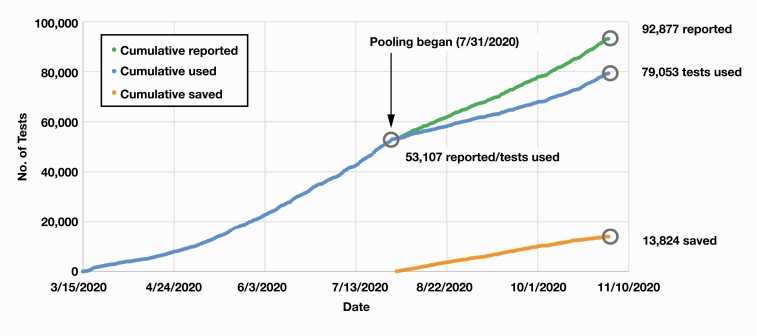
Pooled patient sample testing combining five samples per pool was initiated on July 31, 2020, beginning with 16 pools, which yielded zero positive samples and therefore amounted to 80 reported negative results from only 16 RT-PCR tests Figure 3. Only specimens originating from testing sites with COVID-19 positivity rates below 5% were pooled, whereas all other specimens were tested individually. The site-specific positivity rates were updated and monitored by the microbiology laboratory on a daily basis. Over the subsequent months from July 31, 2020, to October 31, 2020, a total of 39,770 RT-PCR results (including all positive, presumptive positive, and negative results) were reported from the Roche platform. Altogether, 4,131 sample pools were tested, corresponding to 20,655 individual test samples, of which 540 pools were positive for SARS-CoV-2, yielding 621 positive individual samples. Cumulatively, we were able to save 13,824 tests (ie, equivalent to a conservation rate of 35%) on the Roche platform over our pooling period, which represented a significant degree of conservation of reagents and consumables (Figure 3 and Figure 4).
Figure 3.
Cumulative coronavirus disease 2019 tests reported, used, and saved before and after sample pooling implementation. The cumulative number of tests reported, used, and saved (as a result of pooling) is shown in green, blue, and orange, respectively.
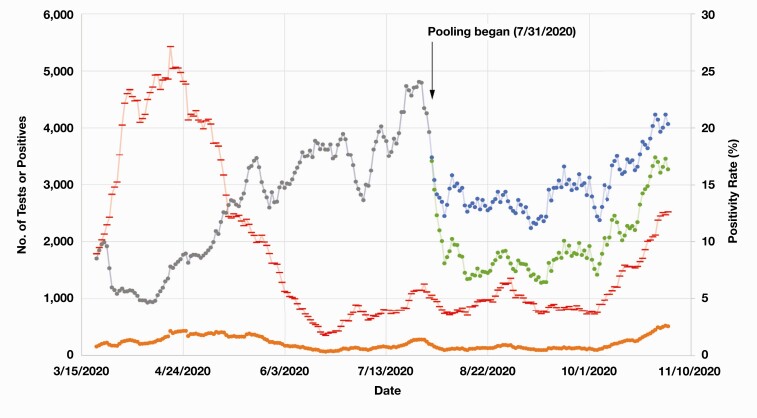
Figure 4.
Seven-day results, case positivity, and pooled tests of coronavirus disease 2019 (COVID-19) before and after implementation of sample pooling on the Roche cobas 6800 platform. The gray and blue lines represent the 7-day number of COVID-19 results reported before and after pooling, respectively, whereas the green line shows the 7-day number of COVID-19 tests used after pooling. Of note, the number of tests used and results reported were equivalent before pooling. The orange line represents the 7-day number of COVID-19–positive results, while the red line shows the 7-day COVID-19 positivity rate (or percent positive).
In our experience, the most significant challenges of implementing a pooling protocol occurred in the preanalytic and postanalytic phases of testing rather than in the analytic phase. The key preanalytic challenge was to determine and manage which samples were to be pooled and to create a robust system that would allow for efficient sorting and tracking of many samples among multiple medical technologists and sites on a daily basis. Additional challenges included (1) identifying the specimens comprising positive pools, (2) performing individual testing on these specimens, and (3) reporting these results in a timely manner. All of these processes were simultaneous with the continuous reception of new specimens, which had to be accessioned and sorted. The logistics and labor time required for retesting individual samples making up the positive pools and manually reporting the individual results of negative pools were key postanalytic challenges that needed to be addressed. Therefore, a notable challenge with sample pooling was the increased demand for these manual pre- and postanalytic processes, which also translated into an increased net TAT for testing.22,24
We were able to maintain an overall TAT of 48 hours or less for most of our test results at UCMC throughout the pooling period. Our mean (SD) TATs before and after pooling were 15.8 (12.6) hours, or 0.66 days, and 19.3 (11.1) hours, or 0.80 days, respectively. The percentage of results reported within 24 hours before and after pooling was 83.4% and 70.8%, respectively, whereas the percentage of results reported within 48 hours before and after pooling was 99.4% and 97.7%, respectively.
The mean (SD) TATs for positive results (includes presumptive positive results) before and after pooling were 16.1 (9.2) hours, or 0.67 days, and 20.4 (9.8) hours, or 0.85 days, respectively. The percentage of positive results reported within 24 hours before and after pooling was 82.7% and 63.3%, respectively. The percentage of positive results reported within 48 hours before and after pooling was 99.3% and 99.1%, respectively. Similarly, the mean (SD) TATs for negative results before and after pooling were 15.8 (12.9) hours, or 0.66 days, and 19.2 (11.2) hours, or 0.80 days, respectively. The percentage of negative results reported within 24 hours before and after pooling was 83.5% and 71.3%, respectively. The percentage of negative results reported within 48 hours before and after pooling was 99.4% and 97.6%, respectively.
Discussion
A significant challenge brought on by the COVID-19 pandemic was the upsurge of testing capabilities required to keep pace. Although many clinical tests for detecting SARS-CoV-2 have become available, RT-PCR remains the gold-standard test for detecting viral presence. However, the availability of materials required to perform RT-PCR, including laboratory consumables, test reagents, and kits, has remained a significant limitation to drastically expand testing capabilities. There is to date a growing body of literature comprising mathematical simulations and proof-of-concept experiments to support the implementation of pooled COVID-19 testing.25-27 On the other hand, despite the many EUA applications for pooled testing across a number of locations, there remains a paucity of documented experiences.22 Here we provide one of the first US reports documenting the challenges of developing and implementing a protocol for pooled COVID-19 testing. We present specific solutions devised as a result of our experiences, particularly to help our colleagues in this endeavor.
The UCMC Clinical Microbiology and Immunology Laboratory, like many other laboratories operating in a limited and confined space, also had been challenged by the Centers for Disease Control and Prevention’s COVID-19 mitigation guidelines emphasizing safe distancing and space management. Accordingly, we optimized our workflow not only to accommodate the cobas 6800 system and pooling protocol but also to keep our laboratory members safe while continuing to function with other duties. While not every process was automated (ie, separating samples and pooling aliquots), we were able to incorporate efficient manual solutions (ie, sorting specimens and bulb pipetting pools) into our workflow. The introduction of additional manual procedures into the COVID-19 workflow resulted in additional time, as reflected by an increase in our prepooling average TAT relative to our postpooling average TAT (15.8 hours vs 19.3 hours, respectively). Even so, we maintained a TAT goal of 48 hours or less and were able to exceed that goal with almost all specimens by reporting 99.4% of prepooling results and 97.7% of postpooling results within 48 hours. In comparison, the national average was 4 days (median, 3 days) in April 2020 and 2.7 days (median, 2 days) by September 2020.28 Our ability to effectively maintain a 48-hour or less TAT is a testament to our dedicated and hardworking staff and their collective efforts in striving to overcome logistics hurdles.
Our experiences demonstrate that despite the additional time that comes with pooling, laboratories can achieve a reliable and meaningful TAT. This also remained true for our positive pooled results requiring additional testing. Although the positive results indeed required additional time (approximately 4 more hours) compared with prepooled results, the average TAT was still below our set goal (99.3% prepooling and 99.1% postpooling results within 48 hours), further strengthening our previous point.
There is certainly the concern of diluting low positive/high Ct value samples during pooling, leading to a false-negative COVID-19 result. Our validation study had a single false-negative result (which we surmise to have been a low-positive sample given its high Ct value of 31.04) out of 24 positive pools, thereby yielding a sensitivity of 96% among the positive pooled samples. Several studies have suggested, however, the false-negative rate to be low for several different pool sizes. Yelin et al29 took previously tested nasal and throat COVID-19 swabs in transport media from Israeli health centers and mixed a positive sample into different-sized pools (1, 2, 4, 16, 32, 64) and found a 96% sensitivity for pools of 16. In pools of 10, Anderson et al30 used 494 previously tested nasopharyngeal swabs collected from a COVID-19 outbreak in Germany and ran the pools on a BioFire FilmArray, yielding a false-negative rate of 7%. Quest Diagnostics also conducted a study using a four-sample pooling approach using previously tested upper respiratory samples in the United States during May 2020 and July 2020 and stratified the specimens based on three positivity rates (1%-3%, 3%-6%, and 6%-10%) with each group containing two separate geographic locations, which per sensitivity analysis yielded a false-negative rate of 0.002%.31 Nevertheless, the possibility of missing these patients and thereby becoming a further issue during the pandemic is unsettling. However, the significance of missing a low positive patient is still debatable.32 A low positive detected by RT-PCR may correspond with the detection of SARS-CoV-2 RNA but not necessarily transmissible virus.33,34 This has been further supported by an in vitro study conducted by La Scola et al35 demonstrating specimens with viral loads of Ct more than 35 to be routinely nonculturable. Considering the current predicament of US COVID-19 testing, we decided the risk was tolerable and could be handled through other mitigation guidelines (ie, quarantine and retesting); the urgent priority was processing higher test volumes.
Overall, we view our pooling experience to be a resounding success. Our pooling strategy allowed us to save approximately 35 tests for every 100 results we reported, or a cost savings of 35% for this resource. The use of bulb pipettes allowed our laboratory to conserve precision pipette tips, and pooling required only one additional tube and bulb pipette for every five specimens. However, we do note that a downside to pooling is the notable increase in required efforts and resources on several fronts: sorting and aliquoting samples for pooling, deconvoluting negative pools for individual reporting, identifying individual specimens comprising positive pools for repeat testing, and tracking and maintaining records for each pool. Nevertheless, the primary motivation for pooling remained responsible stewardship of a scarce resource, allowing reagents to be allocated to those in need. At least as important, by expanding our testing volumes (the number of results reported per day postpooling compared with prepooling was 428 results/d vs 385 results/d, respectively) but still achieving a mean turnaround time of 19.3 hours, the Clinical Microbiology and Immunology Laboratory allowed more patients to know their COVID-19 status and isolate as necessary. The rapid TAT allowed physicians to render appropriate care and provided our institution and city a real-time picture of COVID-19 infections in the south side of Chicago. Among the future challenges we anticipated was the increasing positivity rate, which happened much more rapidly and sooner, forcing our laboratory to stop pooling. As mentioned, we set a cutoff prevalence rate of 5%, meaning approximately 77% (0.955) of our pools would be negative; however, starting in November 2020, our prevalence rate for many of our sites jumped to as high as 20%. While an argument could have been made to pool even with these high prevalence rates, had we continued at a prevalence rate of even 15%, our pools would have been negative 44% (0.855) of the time, begging the question of how much resources would be saved, if any, along with compromising our set TAT.
Another challenge we anticipated over the summer was the so-called twin-demic of both COVID-19 and influenza. While we validated the Roche COVID-19/Flu Combination Assay in anticipation of seasonal flu, we realized pooling would not be an option once influenza was widely circulating in the community. The microbiology staff are frontline workers, handling thousands of infectious specimens a day, in circumstances that are far from ideal. We empathize with other clinical microbiology personnel throughout the world; while pooling can seem a daunting endeavor, we hope our experience eases the way for others.
Acknowledgments
We thank the staff of the Clinical Microbiology and Immunology Laboratory at the University of Chicago Medical Center for their assistance and support.
References
- 1. Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu N, Zhang D, Wang W, et al. ; China Novel Coronavirus Investigating and Research Team . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu R, Zhao X, Li J, et al. . Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization. Laboratory testing strategy recommendations for COVID-19: interim guidance, 21 March 2020. https://apps.who.int/iris/handle/10665/331509. Accessed December 7, 2020. [Google Scholar]
- 6. World Health Organization. Diagnostic testing for SARS-CoV-2: interim guidance, 11 September 2020. https://apps.who.int/iris/handle/10665/334254. Accessed December 7, 2020. [Google Scholar]
- 7. Esbin MN, Whitney ON, Chong S, et al. . Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. American Society for Microbiology. Supply shortages impacting COVID-19 and non-COVID testing. https://asm.org/Articles/2020/September/Clinical-Microbiology-Supply-Shortage-Collecti-1. Accessed December 7, 2020. [Google Scholar]
- 9. Hagen A. Laboratory supply shortages are impacting COVID-19 and non-COVID diagnostic testing.https://asm.org/Articles/2020/September/Laboratory-Supply-Shortages-Are-Impacting-COVID-19. Accessed December 7, 2020.
- 10. Litvak E, Tu XM, Pagano M. Screening for the presence of a disease by pooling sera samples. J Am Stat Assoc. 1994;89:424-434. [Google Scholar]
- 11. Hogan CA, Sahoo MK, Pinsky BA. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323:1967-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emmanuel JC, Bassett MT, Smith HJ, et al. . Pooling of sera for human immunodeficiency virus (HIV) testing: an economical method for use in developing countries. J Clin Pathol. 1988;41:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewis JL, Lockary VM, Kobic S. Cost savings and increased efficiency using a stratified specimen pooling strategy for Chlamydia trachomatis and Neisseria gonorrhoeae. Sex Transm Dis. 2012;39:46-48. [DOI] [PubMed] [Google Scholar]
- 14. Salzer E, Nixon E, Drewes G, et al. . Screening pools of compounds against multiple endogenously expressed targets in a chemoproteomics binding assay. J Lab Autom. 2016;21:133-142. [DOI] [PubMed] [Google Scholar]
- 15. Lippi G, Henry BM, Sanchis-Gomar F, et al. . Updates on laboratory investigations in coronavirus disease 2019 (COVID-19). Acta Biomed. 2020;91:e2020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pilcher CD, Westreich D, Hudgens MG. Group testing for severe acute respiratory syndrome–coronavirus 2 to enable rapid scale-up of testing and real-time surveillance of incidence. J Infect Dis. 2020;222:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Salazar A, Aguilera A, Trastoy R, et al. . Sample pooling for SARS-CoV-2 RT-PCR screening. Clin Microbiol Infect. 2020;26:1687.e1-1687.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alcoba-Florez J, Gil-Campesino H, García-Martínez de Artola D, et al. . Increasing SARS-CoV-2 RT-qPCR testing capacity by sample pooling. Int J Infect Dis. 2021;103:19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garg J, Singh V, Pandey P, et al. . Evaluation of sample pooling for diagnosis of COVID-19 by real time-PCR: a resource-saving combat strategy [published online September 1, 2020]. J Med Virol. [DOI] [PubMed] [Google Scholar]
- 20. Praharaj I, Jain A, Singh M, et al. . Pooled testing for COVID-19 diagnosis by real-time RT-PCR: a multi-site comparative evaluation of 5- & 10-sample pooling. Indian J Med Res. 2020;152:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. College of American Pathologists. Pooled testing: guidance from the CAP’s Microbiology Committee. 2020; https://www.cap.org/covid-19/pooled-testing-guidance-from-cap-microbiology-committee. Accessed December 7, 2020.
- 22. US Food and Drug Administration. In vitro diagnostics EUAs. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas. Accessed December 11, 2020. [Google Scholar]
- 23. US Food and Drug Administration. Pooled sample testing and screening testing for COVID-19. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/pooled-sample-testing-and-screening-testing-covid-19. Accessed February 9, 2021. [Google Scholar]
- 24. Chong BSW, Tran T, Druce J, et al. . Sample pooling is a viable strategy for SARS-CoV-2 detection in low-prevalence settings. Pathology. 2020;52:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nalbantoglu OU. Group testing performance evaluation for SARS-CoV-2 massive scale screening and testing. BMC Med Res Methodol. 2020;20:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sinnott-Armstrong N, Klein D, Hickey B. Evaluation of group testing for SARS-CoV-2 RNA. medRxiv. 2020. [Google Scholar]
- 27. Cherif A, Grobe N, Wang X, et al. . Simulation of pool testing to identify patients with coronavirus disease 2019 under conditions of limited test availability. JAMA Netw Open. 2020;3:e2013075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. The State of the Nation. A 50-State COVID-19 Survey Report #17: COVID-19 test result times. https://covidstates.org/reports. Accessed December 7, 2020. [Google Scholar]
- 29. Yelin I, Aharony N, Tamar ES, et al. . Evaluation of COVID-19 RT-qPCR test in multi sample pools. Clin Infect Dis. 2020;71:2073-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson C, Castillo F, Koenig M, et al. . Pooling nasopharyngeal swab specimens to increase testing capacity for SARS-CoV-2. Med J (Ft Sam Houst Tex). 2021(PB 8-21-01/02/03):8-11. [PubMed] [Google Scholar]
- 31. Borillo GA, Kagan RM, Baumann RE, et al. . Pooling of upper respiratory specimens using a SARS-CoV-2 real-time RT-PCR assay authorized for emergency use in low-prevalence populations for high-throughput testing. Open Forum Infect Dis. 2020;7:ofaa466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perchetti GA, Sullivan KW, Pepper G, et al. . Pooling of SARS-CoV-2 samples to increase molecular testing throughput. J Clin Virol. 2020;131:104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wölfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465-469. [DOI] [PubMed] [Google Scholar]
- 34. Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249-2251. [DOI] [PubMed] [Google Scholar]
- 35. La Scola B, Le Bideau M, Andreani J, et al. . Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]