Abstract
Objective
To rapidly develop, validate, and implement a novel real-time mortality score for the COVID-19 pandemic that improves upon sequential organ failure assessment (SOFA) for decision support for a Crisis Standards of Care team.
Materials and Methods
We developed, verified, and deployed a stacked generalization model to predict mortality using data available in the electronic health record (EHR) by combining 5 previously validated scores and additional novel variables reported to be associated with COVID-19-specific mortality. We verified the model with prospectively collected data from 12 hospitals in Colorado between March 2020 and July 2020. We compared the area under the receiver operator curve (AUROC) for the new model to the SOFA score and the Charlson Comorbidity Index.
Results
The prospective cohort included 27 296 encounters, of which 1358 (5.0%) were positive for SARS-CoV-2, 4494 (16.5%) required intensive care unit care, 1480 (5.4%) required mechanical ventilation, and 717 (2.6%) ended in death. The Charlson Comorbidity Index and SOFA scores predicted mortality with an AUROC of 0.72 and 0.90, respectively. Our novel score predicted mortality with AUROC 0.94. In the subset of patients with COVID-19, the stacked model predicted mortality with AUROC 0.90, whereas SOFA had AUROC of 0.85.
Discussion
Stacked regression allows a flexible, updatable, live-implementable, ethically defensible predictive analytics tool for decision support that begins with validated models and includes only novel information that improves prediction.
Conclusion
We developed and validated an accurate in-hospital mortality prediction score in a live EHR for automatic and continuous calculation using a novel model that improved upon SOFA.
Keywords: crisis triage; mortality prediction; COVID-19; decision support systems, clinical, machine learning
INTRODUCTION
The SARS-CoV-2 virus has infected >70 million and killed >1.5 million people in the year since its origination (December 2019).1 The resulting pandemic has overwhelmed some regions’ health care systems and critical care resources, forcing the medical community to confront the possibility of rationing resources.2,3 In the United States, critical care triage guidance in the setting of resource scarcity is produced at the state-level through Crisis Standards of Care (CSC) protocols.4,5 These protocols attempt the difficult task of ethically allocating scarce resources to individuals most likely to benefit, with the aim of saving the most lives.6–8 To accomplish this, CSC protocols use organ dysfunction scores and chronic comorbidity scores to assess patient survivability. Ideally, scoring would avoid systematic bias and be generalizable, accurate, flexible to circumstance, and computable within electronic health record (EHR) systems with data collected in real time.9
At the foundation of most CSC protocols is the Sequential Organ Failure Assessment (SOFA) score.10,11 SOFA and other acuity scores (eg, Simplified Acute Physiology Score and APACHE) are well-validated but have significant limitations. They were developed over 20 years ago before widespread use of EHRs, are rigid regarding context, and were designed to measure severity of illness and predict mortality based on a few data points.12–17 Although SOFA predicts mortality from influenza pneumonia poorly, it was operationalized for use in patients with COVID-19.18,19 Optimizing the accuracy of mortality predictions is critical for medical triage because the decision to withhold or withdraw life-sustaining therapies is heavily influenced by a single score in many states’ CSC protocols.11
The COVID-19 pandemic created an emergent need for a novel, accurate, and location- and context-sensitive EHR-computable tool to predict mortality in hospitalized patients with and without COVID-19. Because developing a new score can take years, a predictive model must rely on well-validated scores. In contrast, COVID-19 is a novel disease for which existing scores may be of limited but unknown predictive value. As such, a predictive framework relying on multiple previously validated scores that can incorporate new information, but only keeps the new inputs that explicitly improve performance, is required. Stacked generalization provides a solution.20 A stacked model is built upon 1 or more baseline model(s) (eg, SOFA) and incorporates additional models only when they improve prediction.21
We rapidly developed, validated, and deployed a novel mortality score for triage of all hospitalized patients during the COVID-19 pandemic by stacking SOFA, qSOFA, a widely used pneumonia mortality score, an acute respiratory distress syndrome (ARDS) mortality model, and a comorbidity score.22–26 We then integrated recently reported predictors that may reflect COVID-19 pathophysiology. To test the novel model, we conducted a prospective cohort study of acutely ill adults with and without COVID-19 disease.
OBJECTIVE
To create a live, predictive analytics scoring system to support CSC (triage) decisions. The system should have the following characteristics: ethically defensible, continuously adaptable/updatable with new data and model information; temporally dependent; as personalized as possible; formed with both well-established/validated scoring models and novel models based on potentially preliminary data and information sources; quickly computable so that refreshed scores can be generated on the order of minutes; and computable with data available in a real-time EHR system.
MATERIALS AND METHODS
Because model development and training began before we had accumulated a large number of COVID-19 patients, we started by developing the novel mortality score using a multihospital retrospective cohort of 82 087 patient encounters (Figure 1B). As we accumulated COVID-19 patients, we conducted a prospective cohort study to validate the novel mortality score in patients with and without COVID-19. Our work was anchored by 4 goals. First, to use SOFA as a baseline and address its limitations through stacked generalization, adding other models with the potential to improve robustness and predictive performance. Second, to integrate and test potential COVID-19-specific predictors. Third, to rapidly deploy the new model in a live EHR across a 12-hospital system that serves more than 1.9 million patients. Fourth, to validate model performance prospectively. The Colorado Multiple Institutional Review Board approved this study.
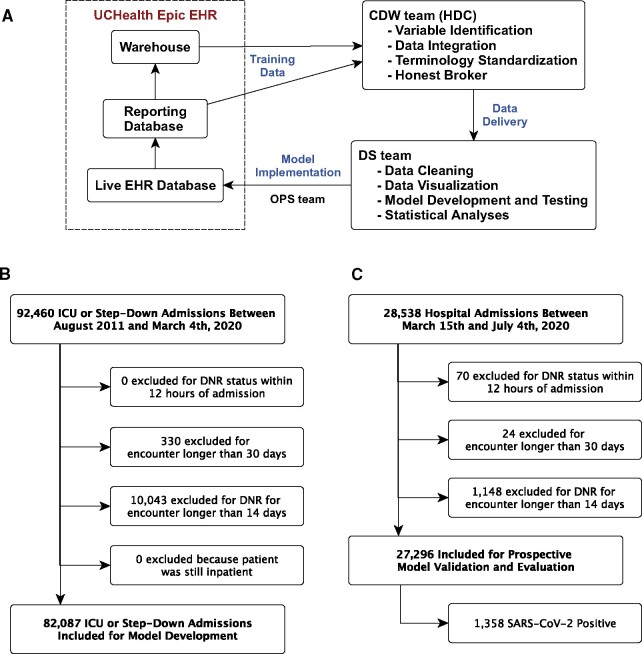
Figure 1.
Study data flow and cohort identification. A) Data flow through the EHR and research team, B) Retrospective Cohort selection for model development, C) Prospective Cohort selection for model evaluation and validation.
Study overview and model deployment
We originally developed, validated, and deployed the model using estimates from retrospective data, while simultaneously building technical capacity to transition to a model estimated on prospective data. The time from conception (March 2020) to deployment of the new model across the health system (April 2020) was 1 month. The model now generates a mortality risk estimate every 15 minutes for every inpatient across the health system. We then prospectively observed model performance through the end of July 2020. This study design is consistent with recent learning health system studies.27 Because of the rapidly evolving pandemic, we built a data pipeline for the stacked mortality model to update as new data were captured from the EHR.
Rapid development and implementation of a new score in a real-time EHR requires a full clinical and informatics pipeline including skilled data warehousing, data wrangling, machine learning, health system information technology (IT), and clinical and ethics personnel working in sync.28–30 All data flowed to the study team from UCHealth’s Epic instance through Health Data Compass, the enterprise data warehouse for the University of Colorado Anschutz Medical Campus (Figure 1A).31 HDC is a multiinstitutional data warehouse that links inpatient and outpatient electronic medical data, state-level all-payer claims data, and the Colorado Death Registry.
Retrospective cohort for initial model training
The retrospective cohort included all encounters of patients >14 years old hospitalized at any of UCHealth’s 12 acute care hospitals between August 2011 and March 4, 2020, whose hospital stay included admission to either an intensive care unit (ICU) or intermediate care unit. We restricted the retrospective data to encounters completed before March 5, 2020, the date of the first reported COVID-19 case in Colorado. We excluded encounters with a “do not attempt resuscitation” order placed within 12 hours of admission or a duration exceeding 14 days, as mortality after prolonged hospitalization likely represents different physiology than mortality from an acute event.
The retrospective cohort included 82 087 encounters by 63 290 unique patients. Of these encounters, 59 733 (72.8%) required ICU-level care, 14 847 (18.1%) required invasive mechanical ventilation, and 5726 (7.0%) ended in death. Patients had an average age of 58.1 ± 17.8 years and 35 826 (43.6%) were female. Demographics, clinical characteristics, and hospital course are shown in Supplementary eTable 1. Model inputs are shown in Supplementary eTables 2 and 3.
Prospective cohort
The prospective cohort included all encounters of patients >14 years old hospitalized at any of UCHealth’s 12 hospitals between March 15, 2020 (the date UCHealth halted elective procedures) through July 2020. Because CSC protocols apply to all hospitalized patients during a crisis, we included all inpatients regardless of level of care or COVID-19 status. We excluded encounters with a “do not attempt resuscitation” order placed within 12 hours of admission, patients who were still admitted, and encounters longer than 30 days.
The prospective cohort included a total of 28 538 encounters between March 15th, 2020 and July 2020 (Figure 1C). Of these, 1148 (4.0%) were excluded because the patient remained in hospital at the time of data censoring: in-hospital survival could not be assessed. Additionally, we excluded 70 and 24 encounters, respectively, due to active DNR and encounter length >30 days. Of the remaining 27 296 encounters, 1358 (5.0%) were positive for SARS-CoV-2, 4494 (16.5%) included intensive care unit (ICU)-level care, 1480 (5.4%) included invasive mechanical ventilation, and 717 (2.6%) died during the hospitalization. Of the 717 patients who received mechanical ventilation, 408 (27.6%) died. Additional demographics are shown in Table 1, Supplementary eTables 1 and 2.
Table 1.
Prospective cohort characteristics and hospital course
| All Encounters(N = 27 296) | COVID-19 Negative (N = 25 938) | COVID-19 Positive (N = 1358) | P value | |
|---|---|---|---|---|
| Age (SD) | 54.3 (20.4) | 54.2 (20.5) | 56.8 (18.4) | P < .001 |
| Female | 15 660 (57.4%) | 15 057 (58.0%) | 603 (44.4%) | P < .001 |
| Race | P < .001 | |||
| White or Caucasian | 20 430 (74.8%) | 19 848 (76.5%) | 582 (42.9%) | |
| Black or African American | 1964 (7.2%) | 1790 (6.9%) | 174 (12.8%) | |
| Other | 4481 (16.4%) | 3901 (15.0%) | 580 (42.7%) | |
| Unknown | 421 (1.5%) | 399 (1.5%) | 22 (1.6%) | |
| Ethnicity | P < .001 | |||
| Non-Hispanic | 22 496 (82.4%) | 21 755 (83.9%) | 741 (54.6%) | |
| Hispanic | 4398 (16.1%) | 3795 (14.6%) | 603 (44.4%) | |
| Unknown | 402 (1.5%) | 388 (1.5%) | 14 (1.0%) | |
| Supplemental O2 | 16 052 (58.8%) | 14 859 (57.3%) | 1193 (87.8%) | P < .001 |
| High Flow Nasal Cannula | 1398 (5.1%) | 1057 (4.1%) | 341 (25.1%) | P < .001 |
| Non-Invasive Ventilation | 1482 (5.4%) | 1382 (5.3%) | 100 (7.4%) | P < .001 |
| Median Hospital Days (IQR) | 3.0 (2.0, 5.2) | 3.0 (1.9, 5.0) | 5.5 (3.0, 9.6) | P < .001 |
| Overall Mortality | 717 (2.6%) | 551 (2.1%) | 166 (12.2%) | P < .001 |
| All Mechanical Ventilation | 1480 (5.4%) | 1241 (4.8%) | 239 (17.6%) | P < .001 |
| Median Hospital Days (IQR) | 8.4 (4.6, 15.1) | 7.7 (4.1, 13.3) | 15.2 (8.2, 21.0) | P < .001 |
| Median ICU Days (IQR) | 3.6 (1.6, 7.8) | 2.9 (1.4, 6.2) | 9.1 (5.3, 15.0) | P < .001 |
| Median Ventilator Days (IQR) | 1.8 (0.7, 5.7) | 1.4 (0.6, 3.9) | 7.5 (4.5, 12.6) | P < .001 |
| Mortality | 408 (27.6%) | 325 (26.2%) | 83 (34.7%) | P = .009 |
Note: Reported P values are to assess differences between COVID-19 negative and COVID-19 positive encounters.
Abbreviations: ICU, intensive care unit; IQR, interquartile range; SD, standard deviation.
Of the 1358 encounters positive for COVID-19, 407 (30.0%) received ICU-level care, 239 (17.6%) were intubated, and 166 (12.2%) patients died. Of the 239 patients requiring mechanical ventilation, 83 (34.7%) died.
Model methodology
We developed a model using stacked generalization to predict mortality.32–34 A stacked regression model takes other component models as covariates and estimates weights in accordance with their predictive power.34 We chose ridge regularized logistic regression as the top-level model to limit overfitting and to address correlation between the component models. Stacking allows for robust, accurate, and interpretable evaluation of the underlying models. In our case, because the second model level was a regularized logistic regression, we could observe the contribution of the first-level models explicitly. Importantly, the stacked model never performs worse than the most accurate component model by construction (see eMethods).20,34
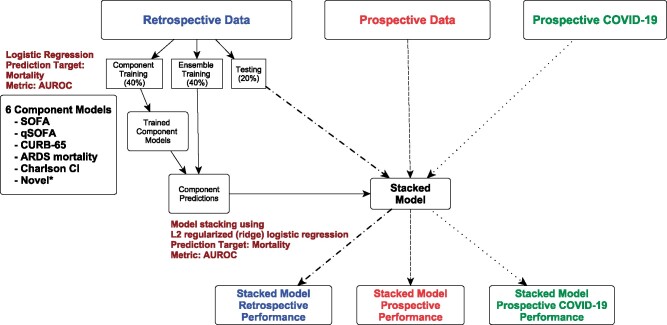
Our stacked regression construction takes 6 logistic regression mortality models as covariates (Figure 2). Four are validated organ dysfunction or pneumonia/ARDS mortality prediction tools, a fifth is a comorbidity score, and a sixth is novel and COVID-specific. These models include: (1) SOFA,12 (2) qSOFA,22 (3) the CURB-65 adult pneumonia mortality score,25 (4) a modified version of an ARDS mortality model,24 (5) a Charlson Comorbidity Index (CCI), and (6) a model made up of laboratory measures associated with COVID-19 disease severity or mortality (Supplementary eMethods, eTable 3).26,35 This model includes, for example, D-dimer, lactate dehydrogenase (LDH), absolute lymphocyte count, and creatinine kinase (CK) (Supplementary eMethods, eTable 3).36–38 The ARDS mortality model was attenuated to include the subset of predictors reliably available in structured form in live EHRs. We fit multiple forms of qSOFA, SOFA, and CURB-65 in an attempt to find the best balance of parsimony and knowledge gained (Figure 2). Variables such as gender, race, and disability status were not included in any models as per bioethics recommendations to avoid potential bias. Only the summary score from the CCI was included; no individual comorbidities were input into the models in order to avoid socioeconomic bias associated with some diagnoses (eg, diabetes).
Figure 2.
Stacked model development. Primary analysis/production model: we used retrospective data to train the component models (40%) and the ensemble/stacked model (40%) and to assess (blue) the ensemble/stacked model (20%). This ensemble/stacked model was used to predict mortality for the whole prospective (red) and prospective COVID-19 (green) datasets. Sensitivity Analysis 1 (not shown): same workflow as primary analysis but the prospective data were used to train and test the models (same 40/40/20 split). The final model was used to predict the entire prospective COVID-19 dataset. Sensitivity Analysis 2 (not shown): same workflow as primary analysis but the prospective COVID-19 data were used to train and test the models (same 40/40/20 split). We fit multiple qSOFA (4), SOFA (2), and CURB-65 (2) component models in health system-guided attempts for parsimony. The different forms of qSOFA, SOFA, and CURB-65 are shown in Supplementary eTable 3. All 11 component models were fed into the model stacking process. The novel COVID-19 model included laboratory results reported to be associated with COVID-19 mortality including D-dimer, LDH, absolute lymphocyte count, BUN, troponin, CK, ALT, and lactate(Supplementary eTable 2).
Training models to predict real-time mortality conservatively
Probability of mortality varies over the hospital course (Supplementary Appendix B) and can be estimated at any time during the hospitalization. In order to estimate and validate the model, we selected a single reference time point against which to make a prediction—when the SOFA score reached its maximum for the encounter. The retrospective data used to estimate the model included only patients with a definitive outcome—either discharge or death.
In order to train the models on retrospective data, we needed an effective “normalizing” point, a single point in time to predict eventual mortality, acknowledging that patients are nonstationary and enter the hospital in 1 state and continuously change until they leave in that state or another. If we estimated the models from retrospective data using every time point of every patient, we would impose a severe selection bias (patients with long stays would more heavily influence the model than those with short stays). Instead, we needed to select a single reference time point per patient to use to estimate the models. To be conservative and to avoid assuming knowledge about the future health trajectory of current patients the models were being applied to, we assumed that in production the current score (at each time step) of the current hospital patients was the worst SOFA score (most organ dysfunction) they would experience. To estimate the models to apply to that situation, we computed the SOFA for every patient in the retrospective training dataset along their entire stay. We then found the time point when the SOFA reached its peak for each patient. We then used that time point as the reference and trained the models using the covariates from that time point.
Operationally, this framework allows for real-time mortality prediction under the conservative assumption that the current measured state of the patient is the worst state the patient will experience. While this assumption will not be correct for all moments in time, it effectively underestimates the patient’s overall mortality, reducing the chance for premature limitation of critical care resources if used for triage decisions.
Model training, evaluation, and validation
We divided the retrospective data 40%-40%-20% for estimating the baseline logistic regression models, estimating the stacked model, and evaluating the stacked model, respectively (Figure 2). We estimated the stacked models with regularized (ridge) logistic regression and used 3-fold cross-validation to select a regularization parameter. The final stacked model was evaluated using empirical-bootstrap-estimated confidence intervals (CIs) and a primary metric of area under the receiver operator curve (AUROC). We validated the stacked model using the prospective cohort and the AUROC.
We chose AUROC as the accuracy metric because the primary goal of the mortality score was to generate a rank-ordered list of patients with associated survival probabilities to inform the allocation of scarce resources. The AUROC is an estimate of the probability of correctly ranking a case compared to a noncase. We also estimated other accuracy metrics including positive predictive value (PPV), sensitivity, specificity, accuracy, and F1 measure (Supplementary eTable 7), as well as area under the precision-recall curve (AUPRC, Table 3 and Supplementary eTable 5). We evaluated calibration using Brier’s score and Cox calibration regression (Supplementary eTable 8).
Table 3.
Model area under the receiver operator curve and precision recall curve for each of the component models and the final stacked model. Models were trained and validated on the initial retrospective cohort. The models were then validated on the prospective cohort and on the subset of patients with COVID-19. The AUROC and AUPRC for the retrospective cohort were based on a 20% holdout of the encounters for testing and evaluation. The prospective validation cohort reflects expected performance when running in a live EHR for both COVID-19 positive and negative patients. Bootstrapped 95% confidence intervals are shown for both AUROC and AUPRC.
|
Retrospective Validation Cohort (N = 16 418)
|
Prospective Validation Cohort (N = 27 296) |
COVID-19 Positive Validation Cohort (N = 1358) |
||||
|---|---|---|---|---|---|---|
| AUROC | AUPRC (baseline 0.07) | AUROC | AUPRC (baseline 0.03) | AUROC | AUPRC (baseline 0.12) | |
| SOFA | 0.90 (0.89, 0.90) | 0.55 (0.55, 0.57) | 0.90 (0.89, 0.91) | 0.42 (0.38, 0.46) | 0.85 (0.82, 0.88) | 0.56 (0.48, 0.63) |
| qSOFA | 0.83 (083, 0.84) | 0.35 (0.33, 0.36) | 0.84 (0.82, 0.86) | 0.26 (0.23, 0.29) | 0.79 (0.74, 0.83) | 0.43 (0.36, 0.51) |
| CURB-65 | 0.81 (0.81, 0.82) | 0.33 (0.31, 0.33) | 0.87 (0.86, 0.88) | 0.26 (0.23, 0.29) | 0.90 (0.87, 0.92) | 0.59 (0.52, 0.67) |
| ARDS Mortality | 0.85 (0.85, 0.86) | 0.51 (0.51, 0.54) | 0.88 (0.87, 0.90) | 0.40 (0.36, 0.44) | 0.86 (0.83, 0.89) | 0.60 (0.52, 0.67) |
| CCI | 0.63 (0.63, 0.66) | 0.11 (0.11, 0.12) | 0.72 (0.70, 0.73) | 0.05 (0.05, 0.06) | 0.75 (0.71, 0.78) | 0.26 (0.21, 0.33) |
| Novel Variables | 0.83 (0.83, 0.84) | 0.45 (0.44, 0.46) | 0.88 (0.87, 0.90) | 0.33 (0.29, 0.36) | 0.91 (0.89, 0.93) | 0.61 (0.54, 0.68) |
| Stacked Model | 0.93 (0.93, 0.94) | 0.65 (0.65, 0.67) | 0.94 (0.93, 0.95) | 0.54 (0.50, 0.57) | 0.90 (0.87, 0.92) | 0.65 (0.59, 0.71) |
Abbreviations: ARDS, acute respiratory distress syndrome; AUPRC, area under the precision-recall curve; AUROC, area under the receiver operator curve; CCI, Charlson Comorbidity Index; qSOFA, a widely used pneumonia mortality score; SOFA, sequential organ failure assessment.
To evaluate the impact of COVID-19 on mortality prediction, we retrainedthe model using the same training strategy but limited training data to the prospectively collected data (sensitivity analysis 1, Figure 2) and to the prospectively collected data from patients with COVID-19 (sensitivity analysis 2, Figure 2). We divided the cohort of patients with COVID-19 40%-40%-20% for estimating the baseline logistic regression models, estimating the stacked model, and evaluating the stacked model, respectively.
Data availability
The data underlying this article were provided by UCHealth by permission and cannot be shared. Analytic code will be made available on GitHub upon request to the corresponding author.
Ethical considerations
This novel score was developed with the purpose of optimizing mortality prediction for decision support for crisis triage. Consequently, the score parameters needed to fall with the ethical framework developed for crisis triage. In catastrophic circumstances, the goal of a resource allocation process should be to provide the most benefit to as many people as possible and to do so in ways that sustain social cohesion and trust in the healthcare system. To maintain trust, recommendations for rationing of resources must be made prospectively, transparently, and consistently across the institution and region and by decision-makers independent of the care team. For this reason, the target users at our health system were members of a triage team that would be activated if CSC became necessary. The triage team would be made up of a hospital administrator, a physician, a nurse, and an ethicist. Neither the physician nor the nurse would be part of any care teams at the time. The triage teams are shown the values of the various features but are not shown the model weights. The stacked model coefficients (Supplementary eTable 5) may be difficult for the average clinical or administrative user to understand in real time. Moreover, any decision to ration resources must embrace a commitment to fairness and a proscription against rationing based on nonclinical factors such as race, gender, sexual orientation, disability, religious beliefs, citizenship status, or “VIP,” socioeconomic, or insurance status.39–42 Consequently factors, such a race and potential proxies of race, were excluded from score development, even if they had the potential to improve accuracy. Ethical considerations for score development are more fully described in Supplementary Appendix C.
RESULTS
Prospective cohort characteristics and hospital course
Compared to patients without COVID-19, patients with COVID-19 were more likely to be male (55.6% vs 42.0%, P < .001); be Hispanic (44.4% vs 14.6%, P < .001); receive ICU-level care (30.0% vs 15.8%, P < .001); be intubated (17.6% vs 4.8%, P < .001); have a longer duration of mechanical ventilation (8.7 days vs 3.0 days, P < .001) and a longer hospital length of stay (7.6 days vs 4.3 days, P < .001); and not survive (12.2% vs 2.1%, P < .001). Patients with COVID-19 had higher SOFA and CURB-65 scores and LDH, ferritin, and D-dimer levels than patients without COVID-19 (all P < .05, Table 2). Mean troponin levels were lower in patients with COVID-19 compared to patients without COVID-19 (P = .002, Table 2). However, absolute lymphocyte count and CK levels were not dissimilar between groups (all P > .05, Table 2).
Table 2.
Mortality model inputs
| All encounters(N = 27 296) | COVID-19 negative (N = 25 938) | COVID-19 positive (N = 1358) | P value | |
|---|---|---|---|---|
| Scores | ||||
| Median qSOFA (IQR) | 0.0 (0.0, 1.0) | 0.0 (0.0, 1.0) | 0.1 (0.0, 1.0) | P < .001 |
| Median SOFA (IQR) | 2.0 (2.0, 4.0) | 2.0 (2.0, 3.0) | 3.0 (2.0, 5.0) | P < .001 |
| Median CURB-65 (IQR) | 1.0 (0.1, 2.0) | 1.0 (0.1, 2.0) | 1.0 (0.0, 2.0) | P = .44 |
| Charlson Comorbidity Index (IQR) | 1.0 (0.0, 3.0) | 1.0 (0.0, 3.0) | 1.0 (0.0, 2.0) | P = .38 |
| ARDS Mortality Model | ||||
| Transfusion FFP | 59 (0.2%) | 59 (0.2%) | 0 (0.0%) | P = .14 |
| Transfusion PRBC | 396 (1.5%) | 392 (1.5%) | 4 (0.3%) | P < .001 |
| GCS ≤ 8 | 264 (1.0%) | 246 (0.9%) | 18 (1.3%) | P = .21 |
| Lactate > 2 | 2676 (9.8%) | 2503 (9.6%) | 173 (12.7%) | P < .001 |
| Creatinine ≥ 2 | 2486 (9.1%) | 2323 (9.0%) | 163 (12.0%) | P < .001 |
| Mean Bilirubin (SD) | 0.7 ± 2.0 | 0.7 ± 2.0 | 0.6 ± 0.8 | P = .003 |
| Mean Arterial pH (SD) | 7.4 ± 0.0 | 7.4 ± 0.0 | 7.4 ± 0.1 | P = .001 |
| Mean PF (SD) | 335.7 ± 212.7 | 340.7 ± 215.8 | 239.6 ± 102.0 | P < .001 |
| Mean SpO2 (SD) | 94.7 ± 2.4 | 94.7 ± 2.4 | 93.4 ± 3.1 | P < .001 |
| Novel Model | ||||
| Mean D-Dimer (SD) | 405.0 ± 3,699.8 | 326.4 ± 2,440.3 | 1,906.2 ± 12,614.9 | P < .001 |
| Mean LDH (SD) | 229.1 ± 214.9 | 223.1 ± 207.4 | 343.5 ± 305.5 | P < .001 |
| Mean ALC (SD) | 1.4 ± 2.0 | 1.5 ± 2.0 | 1.3 ± 1.6 | P = .001 |
| Mean BUN (SD) | 19.4 ± 15.1 | 19.3 ± 14.9 | 21.2 ± 18.4 | P < .001 |
| Mean Troponin (SD) | 0.5 ± 9.0 | 0.6 ± 9.2 | 0.2 ± 3.9 | P = .002 |
| Mean CK (SD) | 173.7 ± 1,612.7 | 170.5 ± 1,567.2 | 235.4 ± 2,316.0 | P = .31 |
| Mean ALT (SD) | 21.1 ± 20.6 | 21.1 ± 21.0 | 20.9 ± 10.4 | P = .47 |
| Mean Lactate (SD) | 1.0 ± 1.1 | 1.0 ± 1.1 | 1.2 ± 1.6 | P < .001 |
In this table, the summary measures for the covariates of each component model in the stacked model are calculated at a single point in time—the time of maximum SOFA score for each encounter.
Abbreviations: ALC, absolute lymphocyte count; ALT, alanine aminotransferase; BUN, blood urea nitrogen; CK, creatinine kinase; FFP, fresh frozen plasma; GCS, Glasgow comas score; IQR, interquartile range; LDH, lactate dehydrogenase; PF, PaO2 to FiO2 ratio; PRBC, packed red blood cells, SD, standard deviation.
Compared to those in the retrospective cohort, patients in the prospective cohort were less likely to receive ICU-level care (16.5% vs 72.8%, P < .0001); less likely to be intubated (5.4% vs 18.1%, P < .0001); and less likely to die (2.6% vs 7.0% vs, P < .0001) (Supplementary eTable 1). This is likely because the prospective cohort included all admissions and not just ICU or intermediate care admissions.
Point-wise mortality estimates
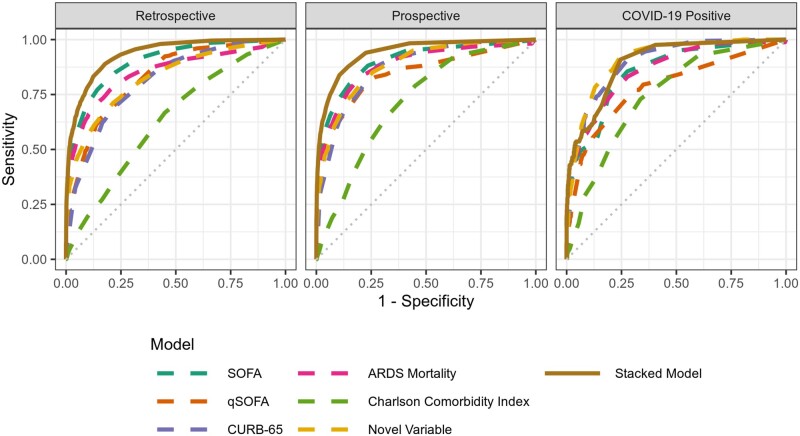
When validated using the prospective cohort, the individual component models predicted point-wise mortality (estimates of mortality risk ranging from 1%–99%) with AUROCs ranging from 0.72 (CCI) to 0.90 (SOFA) (Table 3). The stacked model predicted point-wise mortality better than any individual model: AUROC 0.94 (Figure 3). Most prospective encounters (95.7%) had predicted point-wise mortalities less than 10%. Within this group, observed mortality was only 1.0%, suggesting that the stacked model accurately identifies patients with low mortality (Supplementary eTable 4).
Figure 3.
Stacked model receiver operator characteristic curves. The retrospective cohort was used for training and validation (in a 40%-40%-20% split). The prospective and COVID-19 positive cohorts were used to validate the retrospectively trained model.
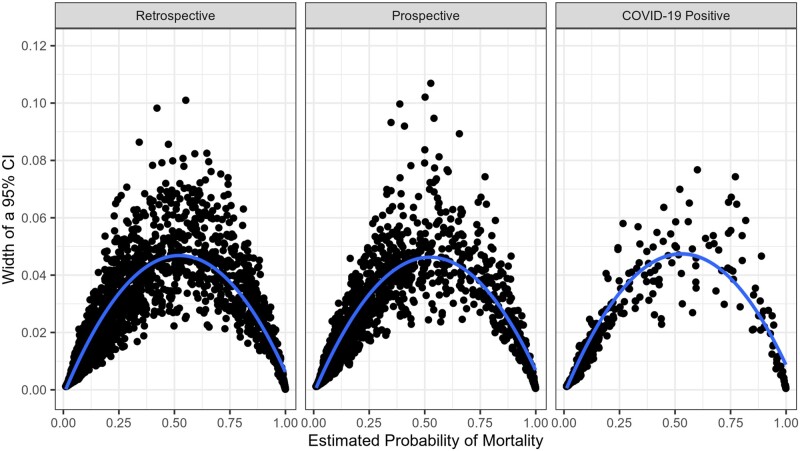
In patients with COVID-19, the AUROC for SOFA, CURB-65, the CCI, and novel variables was 0.85, 0.90, 0.75, and 0.91 respectively. In this subset of patients, the stacked model predicted mortality with an AUROC of 0.90. Additional performance metrics, including precision and recall, are shown in Figure 4. The stacked model predicted mortality with narrowest 95% CIs at the extremes of predicted mortality (Figure 5). Even at moderate predicted mortalities, 95% CIs were generally narrower than 10 percentage points.
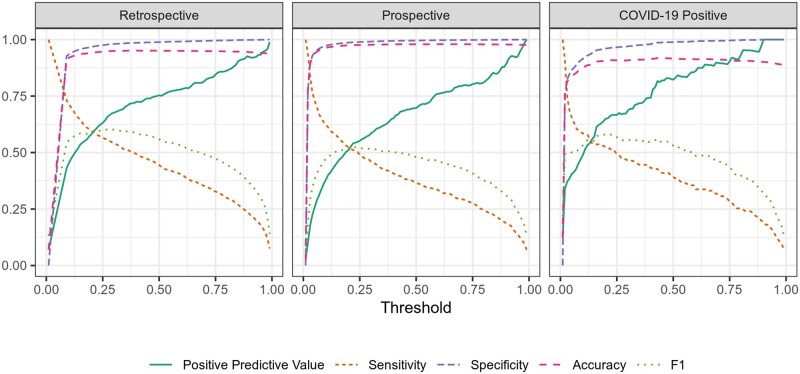
Figure 4.
Stacked model performance metrics across all potential probability thresholds. The purpose of the main stacked model was to create a ranked patient list by probability of mortality. If the model was to be used as part of a clinical decision support alert, then a threshold for the estimated probability would need to be used to define when an alert fires. Figure 4 shows common model performance metrics as a function of the threshold.
Figure 5.
Confidence intervals around point-wise predicted mortality. This figure shows the width of 95% confidence interval (y-axis) around the stacked model mortality probabilities estimates at each potential value for estimated probability. Confidence intervals were narrowest at the extremes of mortality probability (likely the most actionable predictions, thus the predictions with the highest stakes).
When trained with retrospective data and evaluated on patients with COVID-19, the novel model and the stacked model performed within their respective CIs (AUROCs of 0.91 and 0.90, respectively). However, retraining the stacked model only on patients with COVID-19 improved its COVID-19-specific AUROC to 0.95 (Supplementary Appendix B). The stacked model outperformed all other models for patients with COVID-19. This highlights the importance of flexible modeling constructs in highly fluid situations, such as at the onset of pandemics or when new diseases are encountered, and suggests that patients with COVID-19 have predictors of mortality that differ from average previously encountered patients.
Time-integrated mortality estimates
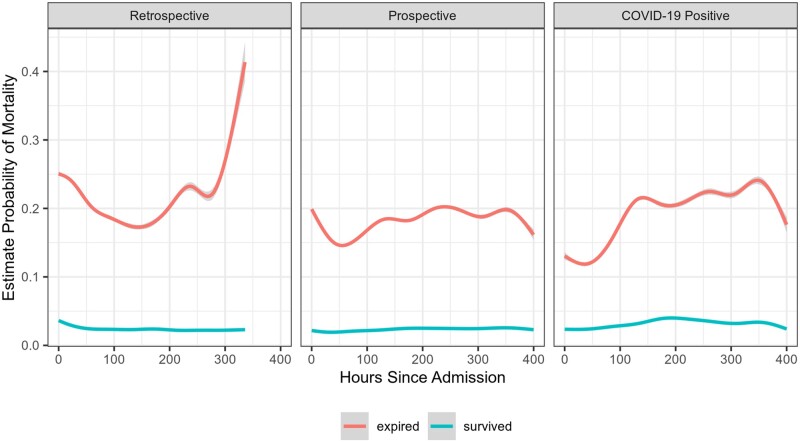
On average, patients who died had estimates of mortality probability that were high at admission and remained high (Figure 6). Patients who survived tended to have, on average, a much lower probability of mortality and a relatively smooth trajectory.
Figure 6.
Average predicted mortality over the course of the hospitalization, stratified by actual mortality. This figure shows smoothed average probability of mortality over the course of the hospitalization, stratified by actual mortality. On average, patients who died had mortality probability estimates much higher than those who did not die, even shortly after admission.
In sensitivity analyses, we generated models predicting mortality at 3 and 7 days after admission (instead of overall mortality). See Supplementary Appendix A/eMethods for the approach taken and Supplementary Appendix C, Ethical Considerations, for an explanation of the motivation behind these sensitivity analyses. These models performed relatively well, although worse than the point-wise stacked models: AUROC 0.84 (3 days) and 0.82 (7 days) in the retrospective cohort. In the prospective cohort and COVID-19 positive patients, accuracy was similar: AUROC 0.83 and 0.77 (3 days) and 0.84 and 0.80 at 7 days, respectively (Supplementary eTable 5).
DISCUSSION
We developed a new, accurate mortality prediction score that is adaptable to different diseases and settings. Improving upon SOFA and the CCI to predict mortality, our score allows more accurate and granular rank-ordering of patients likely to benefit from intensive care. We rapidly deployed the novel score in our EHR during the COVID-19 pandemic for potential real-time use in making triage decisions. We demonstrated that reliability was maintained in a prospective cohort of patients with and without COVID-19. Fortunately, we have not needed to use these scores for triage, but our development process forges a new path for leveraging EHRs, clinical expertise, and machine learning to provide real-time, situation-critical clinical decision support.
This article adds significantly to the literature regarding CSC and ethically allocating scarce medical resources. Like ours, most other scoring systems are based on the SOFA score, which was developed 20 years ago with simplicity, and not triage, in mind. SOFA does not always generalize well: for example, it predicted influenza H1N1 mortality poorly.18,19 While others have attempted to build novel scores that are simple and accurate ,6,7 our contribution is methodological. We show how to leverage many models—novel and well-worn—to create a robust, adaptable, model-averaged score. Our work builds on recent reports demonstrating in patients with COVID-19 that SOFA, CURB-65, Pneumonia Severity Index, Acute Physiology and Chronic Health Evaluation (APACHE II), and novel, COVID-specific COVID-GRAM scores predict mortality variably but reasonably well: AUROC 0.59–0.87, 0.84–0.85, 0.87, 0.96, and 0.78–0.88, respectively.43–47 Although APACHE II outperforms other scores, it includes data that are not easily extracted from an EHR in real-time. By stacking multiple models and using data extracted in real-time from the EHR, we demonstrate similar AUROC (0.94) to APACHE II in a large prospective cohort of patients for whom a CSC-based triage plan would operate: those with and without COVID-19. Finally, CSC protocols have collapsed SOFA scores to rank patients in just a few categories, reflecting the difficulty of knowing when SOFA scores are sufficiently different to make a meaningful difference for triage. Our approach generates 1%–99% risk of mortality and the ability to statistically differentiate between patients (or determine statistical ties) by calculating 95% CI for each score.
Our stacked model’s ability to predict mortality is tailored to our patient population in Colorado and could easily be tailored to smaller populations. This is important given the varied experiences with COVID-19. Our in-hospital (12% vs 21%) and ventilator mortality rates (35% vs 88%) were substantially lower than a New York cohort from the first wave of the pandemic.48 Our mortality rates approach those expected for moderate–severe ARDS.49,50 There are potentially many explanations for these differences, including younger age, difference in comorbidities, differences in therapeutic interventions, and learning from the experience of earlier effected areas. Moreover, the utilization of ICU level of care and mechanical ventilation varies widely across the world: in New York, 14.2% of patients were treated in an ICU and 12.2% of patient received mechanical ventilation. In contrast, in a cohort of patients in China, 50.6% of patients were admitted to an ICU and 42.2% received mechanical ventilation.37,38,43 Such differences may affect the predictive characteristics of a mortality score. Moreover, we found that patients with COVID-19 have unique characteristics and may benefit from specific mortality prediction models. Therefore, utilizing EHR data streams allows for flexibility to add additional components and retrain the stacked model as new knowledge and clinical experience accumulates. Importantly for generalization, the model can be tuned in real time to other local patient populations and disease characteristics.
Several aspects of the informatics infrastructure and workflow are important. First, such a rapid development process would have been impossible without a robust data warehouse staffed by experts with deep knowledge of EHR data and common clinical data models. The availability of high-quality data is known to be among the largest challenges in clinical applications of machine learning.51 Second, our data science team was in place and had substantial shared experience with data from the health system. It would be extremely challenging to either rapidly hire or outsource the necessary expertise during a pandemic. Third, our data science team already had access to highly capable cloud-based and on-premises HIPAA-compliant computational environments. Establishing the processes and controls for such an environment takes time and expert human resources; our campus had already made those investments. Fourth, our multidisciplinary team included leadership, a variety of potential end users, and experts from ethics, clinical informatics, machine learning, and clinical care.28 This diversity critically grounded the project in ethical principles and pragmatic clinical realties and allowed us to quickly iterate to a practical, implementable, and interpretable model. Because of urgent operational needs, we also had full institutional and regulatory support. Finally, we evaluated the model prospectively, an important gold-standard not often met by new machine learning-based informatics tools.28 Of note, there are many reports in the literature describing development of predictive models using EHR data, but very few reports of the implementation of those models in a live EHR for clinical use. In this case, the total elapsed time from including data extraction, model construction, and implementation to deployment within the EHR across the 12 UCHealth hospitals was 1 month, illustrating the potential capacity for novel predictive model development. Now that we have demonstrated a workflow to rapidly develop new informatics tools in our health system, we anticipate that many other tools will follow.
This manuscript has several limitations. First, all scores are calculated from EHR data. While this allows for real-time score calculation, it introduces the possibility of artifactual data skewing mortality prediction. This was partially addressed by placing acceptable ranges on physiologic variables (see Supplementary Appendix A). Second, missing data or data collected at different time intervals is inherent in the analysis of EHR data. To overcome this, we developed a system of imputation and last known value carry forward (see Supplementary Appendix A). Such assumptions may introduce systematic and unmeasured bias but are unavoidable operationally. Third, more sophisticated machine learning techniques (eg, Gaussian process regressions or attributable component analysis) may allow for more accurate mortality predictions.52–54 However, we chose methods that were robustly estimable and would allow for transparent interpretation of underlying model contributions to the overall score. Fourth, in-hospital mortality may not be the optimal metric to make triage decisions. One-year mortality or other related outcome measures may be a better metrics but, given the desire to validate a mortality predictor quickly, longer-term outcomes were not available. Fifth, our data and patient population are specific to Colorado, and results may differ geographically. Sixth, while a multidisciplinary group of experts designed this score to minimize potential bias from race, ethnicity, or socioeconomic status, this has not been rigorously validated and is the focus of ongoing research. Finally, some clinical indicators of illness severity were not included in the models (eg, prone positioning, continuous renal replacement therapy, and radiographic results). These data may improve mortality prediction but are difficult to routinely and reliably autoextract from the EHR.
CONCLUSION
We developed a novel and accurate in-hospital mortality score that was deployed in a live EHR and automatically and continuously calculated for real-time evaluation of patient mortality. The score can be tuned to a local population and updated to reflect emerging knowledge regarding COVID-19. Moreover, this score adheres to the ethical principles necessary for triaging.39–42 Further research to test multicenter score performance, refine mortality prediction over longer periods of time, and investigate the optimal methods to use such a score in a CSC protocol is needed.
FUNDING
PS is supported by NIH K23 HL 145001; DA and ML by NIH R01 LM012734; DK by NIH K08 HL125725; TB by NIH UL1 TR002535 and NIH UL1 TR002535 - 03S2.
AUTHOR CONTRIBUTIONS
TDB had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. PDS, DA, PED, SR, JS, and TDB contributed substantially to the study design, data acquisition, data analysis and interpretation, and the writing of the manuscript. DPK, BA, RM, and LL contributed substantially to data acquisition, verifying data integrity, and the writing of the manuscript. MEK contributed substantially to study design and the writing of the manuscript. DA, MEL, and PDS conceptualized and initially designed the statistical modeling framework. JSK, MKW, and JJG contributed substantially to the study design, data acquisition, and the writing of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge Sarah Davis, Michelle Edelmann, and Michael Kahn at Health Data Compass.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Dong E, Du H, Gardner L.. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20 (5): 533–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.IHME COVID-19 Forecasting Team. Modeling COVID-19 scenarios for the United States. Nat Med 2021; 27: 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Livingston E, Bucher K.. Coronavirus disease 2019 (COVID-19) in Italy. JAMA 2020; 323 (14): 1335. [DOI] [PubMed] [Google Scholar]
- 4.Colorado Department of Public Health and Environment. Colorado Crisis Standards of Care. https://cdphe.colorado.gov/colorado-crisis-standards-care Accessed February 22, 2021
- 5.Minnesota Department of Health. Minnesota Crisis Standards of Care. https://www.health.state.mn.us/communities/ep/surge/crisis/index.html Accessed February 22, 2021
- 6. Talmor D, Jones AE, Rubinson L, et al. Simple triage scoring system predicting death and the need for critical care resources for use during epidemics. Crit Care Med 2007; 35 (5): 1251–6. [DOI] [PubMed] [Google Scholar]
- 7. Adeniji KA, Cusack R.. The Simple Triage Scoring System (STSS) successfully predicts mortality and critical care resource utilization in H1N1 pandemic flu: a retrospective analysis. Crit Care 2011; 15 (1): R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grissom CK, Brown SM, Kuttler KG, et al. A modified sequential organ failure assessment score for critical care triage. Disaster Med Public Health Prep 2010; 4 (4): 277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wynia MK, Sottile PD.. Ethical triage demands a better triage survivability score. Am J Bioeth 2020; 20 (7): 75–7. [DOI] [PubMed] [Google Scholar]
- 10. Antommaria AHM, Gibb TS, McGuire AL, et al. Ventilator triage policies during the COVID-19 pandemic at US Hospitals associated with members of the Association of Bioethics Program Directors. Ann Intern Med 2020; 173 (3): 188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Piscitello GM, Kapania EM, Miller WD, et al. Variation in ventilator allocation guidelines by US state during the coronavirus disease 2019 pandemic: a systematic review. JAMA Netw Open 2020; 3 (6): e2012606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the European Society of Intensive Care Medicine. Intensive Care Med 1996; 22 (7): 707–10. [DOI] [PubMed] [Google Scholar]
- 13. Vincent JL, de Mendonça A, Cantraine F, et al. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: results of a multicenter, prospective study. Working group on “sepsis-related problems” of the European Society of Intensive Care Medicine. Crit Care Med 1998; 26 (11): 1793–800. [DOI] [PubMed] [Google Scholar]
- 14. Asai N, Watanabe H, Shiota A, et al. Efficacy and accuracy of qSOFA and SOFA scores as prognostic tools for community-acquired and healthcare-associated pneumonia. Int J Infect Dis 2019; 84: 89–96. [DOI] [PubMed] [Google Scholar]
- 15. Le Gall JR, Lemeshow S, Saulnier F.. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA 1993; 270 (24): 2957–63. [DOI] [PubMed] [Google Scholar]
- 16. Moreno R, Vincent JL, Matos R, et al. The use of maximum SOFA score to quantify organ dysfunction/failure in intensive care. Results of a prospective, multicentre study. Intensive Care Med 1999; 25 (7): 686–96. [DOI] [PubMed] [Google Scholar]
- 17. Ferreira FL. Serial evaluation of the SOFA score to predict outcome in critically ill patients. JAMA 2001; 286 (14): 1754. [DOI] [PubMed] [Google Scholar]
- 18. Guest T, Tantam G, Donlin N, et al. An observational cohort study of triage for critical care provision during pandemic influenza: “Clipboard physicians” or “evidenced based medicine”? Anaesthesia 2009; 64 (11): 1199–206. [DOI] [PubMed] [Google Scholar]
- 19. Khan Z, Hulme J, Sherwood N.. An assessment of the validity of SOFA score based triage in H1N1 critically ill patients during an influenza pandemic. Anaesthesia 2009; 64 (12): 1283–8. [DOI] [PubMed] [Google Scholar]
- 20. Wolpert DH. Stacked generalization. Neural Netw 1992; 5 (2): 241–59. [Google Scholar]
- 21. Hastie T, Tibshirani R, Friedman J.. The Elements of Statistical Learning. New York, NY: Springer; 2009. [Google Scholar]
- 22. Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis. JAMA 2016; 315 (8): 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song J-U, Sin CK, Park HK, et al. Performance of the quick Sequential (sepsis-related) Organ Failure Assessment score as a prognostic tool in infected patients outside the intensive care unit: a systematic review and meta-analysis. Crit Care 2018; 22 (1): 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cooke CR, Kahn JM, Caldwell E, et al. Predictors of hospital mortality in a population-based cohort of patients with acute lung injury. Crit Care Med 2008; 36: 1412–20. [DOI] [PubMed] [Google Scholar]
- 25. Pflug MA, Tiutan T, Wesemann T, et al. Short-term mortality of adult inpatients with community-acquired pneumonia: external validation of a modified CURB-65 score. Postgrad Med J 2015; 91 (1072): 77–82. [DOI] [PubMed] [Google Scholar]
- 26. Quan H, Li B, Couris CM, et al. Updating and validating the Charlson Comorbidity Index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am J Epidemiol 2011; 173 (6): 676–82. [DOI] [PubMed] [Google Scholar]
- 27. Semler MW, Self WH, Wanderer JP, et al. ; SMART Investigators and the Pragmatic Critical Care Research Group. Balanced crystalloids versus saline in critically ill adults. N Engl J Med 2018; 378 (9): 829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019; 25 (9): 1337–40. [DOI] [PubMed] [Google Scholar]
- 29. Rossetti SC, Knaplund C, Albers D, et al. Leveraging clinical expertise as a feature—not an outcome—of predictive models: evaluation of an early warning system use case. AMIA Annu Symp Proc 2019; 2019: 323–32. [PMC free article] [PubMed] [Google Scholar]
- 30. Collins SA, Cato K, Albers D, et al. Relationship between nursing documentation and patients’ mortality. Am J Crit Care 2013; 22 (4): 306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Health Data Compass. https://www.healthdatacompass.org/ Accessed May 17, 2020
- 32. Sill J, Takacs G, Mackey L, et al. Feature-weighted linear stacking. 2009. http://arxiv.org/abs/0911.0460 Accessed April 19, 2021.
- 33. Clark B. Comparing Bayes model averaging and stacking when model approximation error cannot be ignored. JMLR 2003; 4: 683–712. doi:10.1162/153244304773936090. [Google Scholar]
- 34. Breiman L. Stacked regressions. Mach Learn 1996; 24 (1): 49–64. [Google Scholar]
- 35. Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40 (5): 373–83. [DOI] [PubMed] [Google Scholar]
- 36. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020; 180 (7): 934–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395 (10229): 1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of Covid-19 in New York City. N Engl J Med 2020; 382 (24): 2372–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Persad G, Wertheimer A, Emanuel EJ.. Principles for allocation of scarce medical interventions. Lancet 2009; 373 (9661): 423–31. [DOI] [PubMed] [Google Scholar]
- 40. Truog RD, Mitchell C, Daley GQ.. The toughest triage—allocating ventilators in a pandemic. N Engl J Med 2020; 382 (21): 1973–5. [DOI] [PubMed] [Google Scholar]
- 41. Maves RC, Downar J, Dichter JR, et al. Triage of scarce critical care resources in COVID-19 an implementation guide for regional allocation: an expert panel report of the task force for mass critical care and the American College of Chest Physicians. Chest 2020; 158 (1): 212–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White DB, Katz MH, Luce JM, et al. Who should receive life support during a public health emergency? Using ethical principles to improve allocation decisions. Ann Intern Med 2009; 150 (2): 132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zou X, Li S, Fang M, et al. Acute Physiology and Chronic Health Evaluation II score as a predictor of hospital mortality in patients of coronavirus disease 2019. Crit Care Med 2020; 48 (8): e657–65–e665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi Y, Pandita A, Hardesty A, et al. Validation of pneumonia prognostic scores in a statewide cohort of hospitalised patients with COVID-19. Int J Clin Pract 2021; 75 (3): e13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. García Clemente MM, Herrero Huertas J, Fernández Fernández A, et al. Assessment of risk scores in Covid-19. Int J Clin Pract 2020; e13705. doi:10.1111/ijcp. 13705. [DOI] [PubMed] [Google Scholar]
- 46. Liang W, Liang H, Ou L, et al. ; China Medical Treatment Expert Group for COVID-19. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med 2020; 180 (8): 1081–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raschke RA, Agarwal S, Rangan P, et al. Discriminant accuracy of the SOFA score for determining the probable mortality of patients with COVID-19 pneumonia requiring mechanical ventilation. JAMA 2021; 325 (14): 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Richardson S, Hirsch JS, Narasimhan M, et al. ; and the Northwell COVID-19 Research Consortium. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA 2020; 323 (20): 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bellani G, Laffey JG, Pham T, et al. ; ESICM Trials Group. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 2016; 315 (8): 788–788. [DOI] [PubMed] [Google Scholar]
- 50. Ziehr DR, Alladina J, Petri CR, et al. Respiratory pathophysiology of mechanically ventilated patients with COVID-19: a cohort study. Am J Respir Crit Care Med 2020; 201 (12): 1560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rajkomar A, Dean J, Kohane I.. Machine learning in medicine. N Engl J Med 2019; 380 (14): 1347–58. [DOI] [PubMed] [Google Scholar]
- 52. Rasmussen CE, Williams CK.. Gaussian Processes in Machine Learning. Boston, MA: The MIT Press; 2006. [Google Scholar]
- 53. Tabak EG, Trigila G.. Conditional expectation estimation through attributable components. Inform Inference J IMA 2018; 7 (4): 727–54. [Google Scholar]
- 54. Mitchell EG, Tabak EG, Levine ME, et al. Enabling personalized decision support with patient-generated data and attributable components. J Biomed Inform 2021; 113: 103639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article were provided by UCHealth by permission and cannot be shared. Analytic code will be made available on GitHub upon request to the corresponding author.