Abstract
Brown adipose tissue can expend large amounts of energy, and therefore increasing its size or activity is a promising therapeutic approach to combat metabolic disease. In humans, major deposits of brown fat cells are found intimately associated with large blood vessels, corresponding to perivascular adipose tissue (PVAT). However, the cellular origins of PVAT are poorly understood. Here, we determine the identity of perivascular adipocyte progenitors in mice and humans. In mice, thoracic PVAT develops from a fibroblastic lineage, consisting of progenitor cells (Pdgfra+; Ly6a+; Pparg-) and preadipocytes (Pdgfra+; Ly6a+; Pparg+), which share transcriptional similarity with analogous cell types in white adipose tissue. Interestingly, the aortic adventitia of adult animals contains a population of adipogenic smooth muscle cells (SMCs) (Myh11+; Pdgfra-; Pparg+) that contribute to perivascular adipocyte formation. Similarly, human PVAT contains presumptive fibroblastic and SMC-like adipocyte progenitors, as revealed by single nucleus RNAseq. Taken together, these studies define distinct populations of progenitor cells for thermogenic PVAT, providing a foundation for developing strategies to augment brown fat activity.
Introduction
Brown adipose tissue (BAT) is specialized for heat production and has garnered much attention because of its capacity to counteract metabolic disease1–4. Brown adipocytes have large numbers of mitochondria that contain Uncoupling Protein 1 (UCP1). Upon activation, UCP1 uncouples the mitochondrial proton gradient from ATP synthesis, creating an electrochemical driving force to burn large amounts of fatty acids, glucose and other substrates for heat production5. Additional thermogenic mechanisms, including creatine- and calcium-driven futile cycles also operate in adipocytes6–8.
Multiple discrete thermogenic fat depots in humans can be activated by cold exposure or β-adrenergic activators9–12. Infants have a BAT depot in the interscapular region, analogous to the location of the largest BAT depot in many rodent species 2,13–16. However, the interscapular BAT pad regresses and becomes undetectable in adults 17. Adult humans retain BAT deposits in the supraclavicular region and in several depots surrounding large blood vessels, known as perivascular adipose tissue (PVAT) 1,18,19. For example, UCP1+ adipocytes are present within the carotid sheath and mediastinal cavity of humans18,20. PVAT in the thoracic peri-aortic region of human subjects increases glucose uptake in response to systemic administration of a β3-adrenergic receptor agonist, a key functional attribute of BAT1. Therefore, understanding the ontogeny of thermogenic PVAT may reveal therapeutic targets to expand brown fat mass for combating obesity and metabolic disease.
Here, we performed a detailed analysis of peri-aortic adipose tissue development and maintenance. We found that the aortic PVAT of mice forms perinatally and possesses characteristics of classical BAT. Single cell transcriptomic analysis of the aorta and associated adventitia in the perinatal period identified several distinct cell types, including preadipocytes and mesenchymal progenitor cells. Cell differentiation assays and genetic lineage tracing studies show that fibroblastic progenitor cells mediate aortic PVAT organogenesis. Interestingly, the aortic adventitia of adult mice lacks fibroblastic preadipocytes (Pdgfra+/Pparg+) but contains a population of adipogenic smooth muscle cells (SMCs) that contribute to adipocyte formation. Adult human PVAT similarly harbors presumptive fibroblastic and smooth muscle-like adipocyte progenitor cells. Together, these studies define multiple progenitor cell types for thermogenic adipocytes in developing and adult PVAT.
Results
Aortic PVAT expresses an EBF2-dependent classical brown fat program.
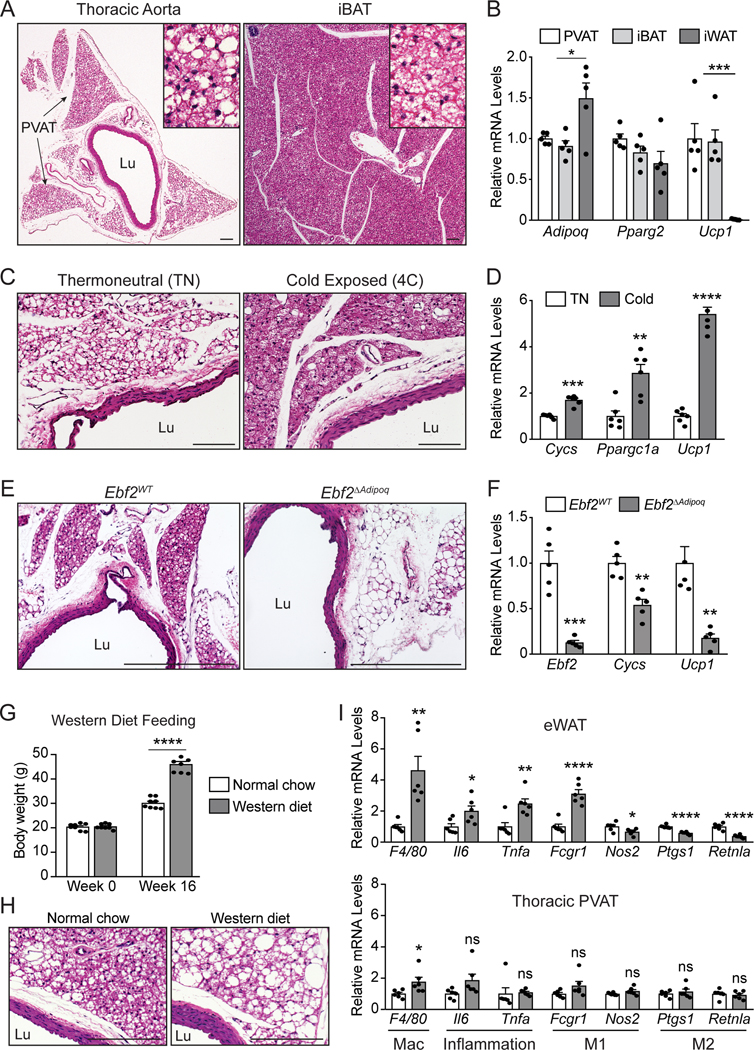
The aorta of adult mice is surrounded by multiple lobes of PVAT, all of which are enclosed by an anatomic compartment defining fascial layer (Fig. 1A). Histologically, perivascular adipocytes have densely eosin-stained cytoplasm and contain multiple lipid droplets, resembling brown adipocytes in interscapular BAT (iBAT) (Fig. 1A). PVAT from the thoracic aorta and iBAT expressed comparable levels of both common adipocyte genes (i.e. Adipoq and Pparg2) and the brown fat marker gene Ucp1 (Fig. 1B). The thermogenic character of aortic PVAT was further recruited by cold exposure. Aortic PVAT from cold-exposed (4°C for 1 week) mice had reduced lipid stores and expressed higher levels of thermogenesis-related genes (Cycs, Ppargc1a, and Ucp1), as compared to PVAT from mice housed at thermoneutrality (30°C) (Fig. 1C, D).
Fig.1. Aortic PVAT expresses an EBF2-regulated classical brown fat program.
(A) H&E staining of thoracic aorta and iBAT from C57/Bl6 mice (scale bar, 100 μm) Representative images of n=5 experiments.
(B) mRNA levels of indicated genes in thoracic PVAT, iBAT, and iWAT from C57/Bl6 mice. (n=5 mice per group; mean+/−SEM). One way ANOVA with Holm Sidak multiple corrections (Adipoq: p= 0.0228; Ucp1: p= 0.0005).
(C, D) H&E staining (C) and mRNA levels of indicated genes (D) in thoracic aorta from C57/Bl6 mice housed at thermoneutrality or 4°C for one week (n=6 mice per group; mean+/−SEM; scale bar, 100 μm). Two sample, two-sided t-test (Cycs: p= 0.0000188; Ppargc1a: p= 0.00171; Ucp1: p= 0.0000000747).
(E, F) H&E staining of thoracic aorta (E) and mRNA levels of indicated genes in aortic PVAT (F) from Ebf2WT and Ebf2ΔAdipoq mice (n=5 mice per group; mean+/−SEM; scale bar, 362.3 μm). Two sample, two-sided t-test (Ebf2: p=0.000194; Cycs: p=0.00144; Ucp1: p=0.00233).
(G) Body weights of C57/Bl6 mice fed normal chow or western diet for 16 weeks (n=8 mice per group; mean+/−SEM). Two way-repeated measures ANOVA with Holm Sidak multiple correction (p<0.0001).
(H, I) H&E staining of thoracic aorta (H) and mRNA levels of indicated genes in eWAT and thoracic PVAT (I) from above mice. (n=6 mice per group; mean+/− SEM). Two sample, two-sided t-test (PVAT [F4/80: p=0.0347]; eWAT [F4/80: p=0.00271; Il6: p=0.0246; Tnfa: p=0.00420; Fcgr1: p=0.000052; Nos2: p=0.0334; Ptgs1: p=0.000027; Retnla: p=0.000052). Statistical Testing: NS p>0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001. Lu: vessel lumen.
The transcription factor Early B Cell Factor-2 (EBF2) is a critical regulator of thermogenic adipocyte development 21–24. To determine if EBF2 controls PVAT fate, we analyzed thoracic aortas from mice lacking Ebf2 expression in adipocytes (Ebf2ΔAdipoq) and littermate control mice (Ebf2WT). Aortic PVAT from Ebf2ΔAdipoq mice displayed a whitened morphology, characterized by increased lipid deposition (Fig. 1E). Ebf2 mutant tissue also showed markedly reduced expression of brown fat-specific genes, including an ~80% reduction of Ucp1 and lower levels of Cycs (Fig. 1F). In contrast to WAT depots, iBAT is relatively resistant to inflammation triggered by a high fat or western diet25–28. To determine if PVAT is protected from diet-induced inflammation, we fed C57/Bl6 mice a western diet for 16 weeks (Fig. 1G). Aortic PVAT from western diet fed mice displayed enlarged lipid droplets (Fig. 1H)29. As expected, western diet feeding promoted inflammation in epididymal WAT, typified by the higher expression levels of macrophage and pro-inflammatory genes (“M1-like macrophage”) and decreased levels of anti-inflammatory (“M2-like macrophage”) genes (Fig. 1I). By contrast, western diet feeding did not affect the expression levels of various inflammatory genes in thoracic PVAT. The small rise of F4/80 levels is consistent with our prior study showing an increase in macrophages upon high fat diet feeding 29. Collectively, these findings demonstrate that aortic PVAT possesses many of the phenotypic properties of classical BAT.
Identification of fibroblast heterogeneity in developing aortic PVAT.
We next sought to identify the progenitor cells responsible for the genesis of PVAT. Histologic examination of thoracic aorta from embryonic day 18 (E18) embryos (just prior to birth) revealed an extensive network of adventitial fibroblasts and well-developed aortic SMCs but an absence of lipid-containing adipocytes (Fig. 2A). During the early postnatal period (P1-P3), prominent clusters of multilocular adipocytes formed around the aorta (Fig. 2A). Consistent with this observation, there was a dramatic rise in the expression levels of adipocyte- and brown fat-selective marker genes (Adipoq, Leptin (Lep), Pparg2, Ucp1) in thoracic aortas (which contain closely attached PVAT) between E18 and P3 (Fig. 2B).
Fig. 2. Identification of multiple fibroblast populations in developing aortic PVAT.
(A) H&E staining of retroperitoneum en bloc from E18 and P3 CD1 mice (scale bar, 517.6 μm). Representative of n=3 experiments.
(B) mRNA levels of indicated genes in thoracic aorta from E18 and P3 CD1 mice (n=3 biological replicates of pooled aortas; mean+/−SEM). Two sample, two-sided t-test (Adipoq: p= 0.000011; Lep: p= 0.000618; Pparg2: p= 0.000016; Ucp1: p= 0.0002).
(C) UMAP of gene expression in 17,957 cells from P3 thoracic aorta of P3 CD1 mice.
(D) UMAP showing expression of smooth muscle marker genes (left) and immunostaining of PDGFRa (red) and MYH11 (green) in sections of E18 thoracic aorta (right) (scale bar, 271.8 μm). Representative of n=2 experiments.
(E) UMAP showing expression of adipocyte marker genes (left) and immunostaining of PLIN1 (red) in P3 thoracic aorta. (scale bar, 543.5 μm). Representative of n=3 experiments.
(F) UMAP showing expression of preadipocyte marker genes and immunostaining of PPARg (red) and PDGFRa (green) in P3 thoracic aorta. Arrowheads show preadipocytes (scale bar, 135.9 μm). Representative of n=2 experiments.
(G) UMAP showing expression of intermediate cell genes and mRNA in-situ hybridization of Bace2 (red) in P3 thoracic aorta. Arrowheads indicate intermediate cells. (scale bar, 145 μm). Representative of n=2 experiments.
(H) UMAP showing expression of the progenitor marker gene Pi16 (left) and mRNA in-situ of hybridization of: (1) Pi16 (green) (middle); and (2) Clec11a (red) and Pi16 (green), (right) in P3 thoracic aorta. Arrowheads in [L] inset indicate progenitors. White arrowhead in [R] inset points to an intermediate cell. Yellow arrowhead shows progenitor cell (scale bar, 291 μm [L] and 145 μm [R]). Representative of n=2 experiments.
(I) Model for thoracic aorta tissue organization at P3.
Lu: vessel lumen; DAPI (blue) was used to stain nuclei. Statistical Testing: NS p>0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
To identify vascular adipocyte progenitor cells, we performed single cell RNA sequencing (scRNAseq) of all stromal-vascular cells isolated from digested thoracic aortas (including the associated adventia) at E18 and P3. We reasoned that these analyses would detect shifts in the cellular composition of aortic tissue as the adipose deposits form. However, integration of the E18 and P3 scRNAseq datasets revealed nearly identical cell population structures (Extended Data Fig. 1A-F) 30. The only notable difference was the emergence of greater numbers of Adipoq+ and Adipoq+/Ucp1+ cells at P3 relative to E18 (Extended Data Fig.1F).
We focused our subsequent analysis on the P3 dataset given its greater representation of Adipoq+/Ucp1+ adipocytes. Clustering analysis identified several expected cell populations, including endothelial cells, immune cells, mesothelium, and neuronal cells (Fig. 2C, Extended Data Fig. 1G). There was also a connected array of fibroblast cell clusters positioned between SMCs and adipocytes, including intermediate Cells, progenitor cells and preadipocytes (Fig. 2C). Clustering analysis identified three groups of SMCs, all marked by expression of the pan smooth muscle genes Myh11 and Acta2 (Fig 2D). MYH11+ SMCs were located around the aorta and did not express the fibroblast marker PDGFRa (Fig. 2D). Adipocytes were marked by the specific expression of Adipoq and Perilipin-1 (Plin1) (Fig. 2E). PLIN1+ adipocytes were exclusively located within discrete depots near the aorta (Fig 2E, right panel). Among the fibroblast cell groups, a putative population of preadipocytes clustered closest to adipocytes and expressed the adipogenic regulator Pparg and the fibroblast marker Pdgfra (Fig. 2F). Co-staining of PPARg and PDGFRa in thoracic aorta revealed that preadipocytes are mainly located at the periphery of adipose tissue lobes (Fig. 2F, right panel). Another population of fibroblastic cells, which we called intermediate cells, clustered close to SMCs and were marked by the selective expression of Bace2 and Clec11a (Fig. 2G). Intermediate cells also expressed Acta2, albeit at lower levels than SMCs (Fig. 2D). In situ mRNA hybridization analysis showed that Bace2-expressing cells are intimately associated with the outer wall of the aorta (Fig. 2G, right panel). The third fibroblast population, which we called progenitor cells, expressed enriched levels of Pi16 and Ly6a (Fig. 2H, Extended Data Fig. 1D). Pi16 mRNA was localized in adventitial cells surrounding the aorta. Co-staining of Pi16 mRNA with the intermediate cell marker, Clec11a, revealed that progenitor cells are located more distally to the aorta than intermediate cells (Fig. 2H). Altogether, these analyses identified multiple fibroblast cell subtypes residing in discrete anatomic compartments within the developing aorta and associated adventitia (Fig. 2I).
Isolation and characterization of adipogenic fibroblasts in perinatal aorta
We developed a fluorescence-activated cell sorting (FACS) strategy to isolate the three fibroblast populations (intermediate cells, progenitors, preadipocytes) and SMCs. Dissociated cells from aortas were stained with CD31, CD45, and Ter119 to separate away endothelial, immune, and erythroid cells, respectively (Extended Data Fig. 2A). Progenitors were then identified and sorted based on high expression of LY6A (Fig. 3A, Extended Data Fig. 2B). Preadipocytes were isolated from LY6A(−) cells based on their intermediate expression of CD142 and lack of CD200 expression (Fig. 3A, middle panel). SMCs were LY6A(−), CD200+ and CD142(−) (Fig. 3A (middle panel), Extended Data Fig. 2C). Intermediate cells were purified from the CD200+; CD142+ cells by gating against CD317+ mesothelial cells (Fig. 3A, right panel). We performed bulk RNA sequencing on the four sorted cell populations to validate the sorting strategy and to obtain deep expression profiles for pathway analyses. The cluster-defining genes identified from the single cell transcriptomes were mapped onto the sorted cell transcriptomes (see methods), revealing a high level of concordance (Fig. 3B, Extended Data Fig. 2D).
Fig. 3. Isolation of adipogenic fibroblasts from perinatal thoracic aorta.
(A) FACS isolation of fibroblast and SMC populations. Live (FVS510-); Lin- (CD45-,CD31-,Ter119-) cells were gated as shown to isolate the following cell populations: Progenitors [LY6A high], Preadipocytes (PreAd) [LY6A-; CD142 intermediate; CD200-], SMC [LY6A-; CD142-; CD200+], Intermediate Cells (Int) [LY6A-; CD142+; CD200+, CD317-]. Meso (Mesothelium) [LY6A-; CD142+; CD200+, CD317+]. (Representative plots from 10 separate experiments). Full strategy in Extended Data Fig. 2A.
(B) Expression heatmaps of Seurat-generated cluster-defining genes mapped on to sorted-cell RNA-Seq results (n=3 biological replicates per group).
(C) Pathway analysis of cluster-defining genes for each cell population. Cluster-defining genes are defined as significantly differentially expressed genes with log2FC>1.5 in every pairwise comparison from sorted-cell bulk RNA-seq (n=3 biological replicates). Graphs plot -log10 unadjusted p values. P values were calculated using EnrichR with WikiPathways2019 annotations and the hypergeometric test; Padjusted for multiple comparisons were calculated with Benjamini-Hochberg. Padjusted values in the order they appear on graphs: SMC (8.70E-06, 1.73E-04, 0.012645474, 0.078202676, 0.093946235, 0.093946235); Intermediate (0.03706, 0.093893, 0.129175 (all others)); Progenitor (5.81E-06, 8.70E-05, 7.50E-04, 0.019056, 0.025219, 0.060584); PreAd (1.97E-49, 6.31E-27, 2.74E-20, 9.95E-15, 5.46E-13, 1.37E-12)
(D) Bodipy (lipid; green) and Hoechst (DNA; blue) staining of cell cultures following differentiation with adipogenic cocktail (Representative image of 5 separate experiments). Scale Bars: Whole Well 500 μM, Zoom 100 μM.
(E) Quantification of adipocyte differentiation (Bodipy+ area above threshold/nuclei) (Data pooled from n=5 separate experiments, n=5 biologically independent replicates per group; mean+/−SEM). Two way-ANOVA followed by two-sided pairwise comparisons with Holm Sidak correction. Statistical Testing: ****p≤0.0001.
We compiled cell type-defining gene sets through differential gene expression analyses, consisting of genes that were significantly and highly enriched in a cell type relative to all others (Fig. 3C). Pathway analysis of the SMC gene set identified “Calcium regulation” and “Striated Muscle contraction”. Intermediate cells were enriched for the “Hedgehog signaling pathway”. Progenitor cells were enriched for genes involved in “Complement Activation” and “Endochondral ossification”. Preadipocytes were notable for their enriched expression of genes related to “electron transport chain”, “oxidative phosphorylation”, and “TCA cycle”, corresponding to gene programs that are critical for the activity of thermogenic adipocytes.
The adipogenic potential of sorted cell populations was investigated ex vivo. FACS-purified cell populations were plated and treated with a standard adipogenic differentiation cocktail. SMCs and intermediate cells displayed almost no capacity to undergo adipocyte differentiation, as assayed by Bodipy staining for lipid accumulation (Fig. 3D, E). By contrast, progenitor cells and preadipocytes efficiently formed adipocytes, with the preadipocytes displaying the highest levels of adipogenesis (Fig. 3D, E). Overall, these studies reveal a gradient of adipogenic competency from non-adipogenic SMCs to highly adipogenic preadipocytes.
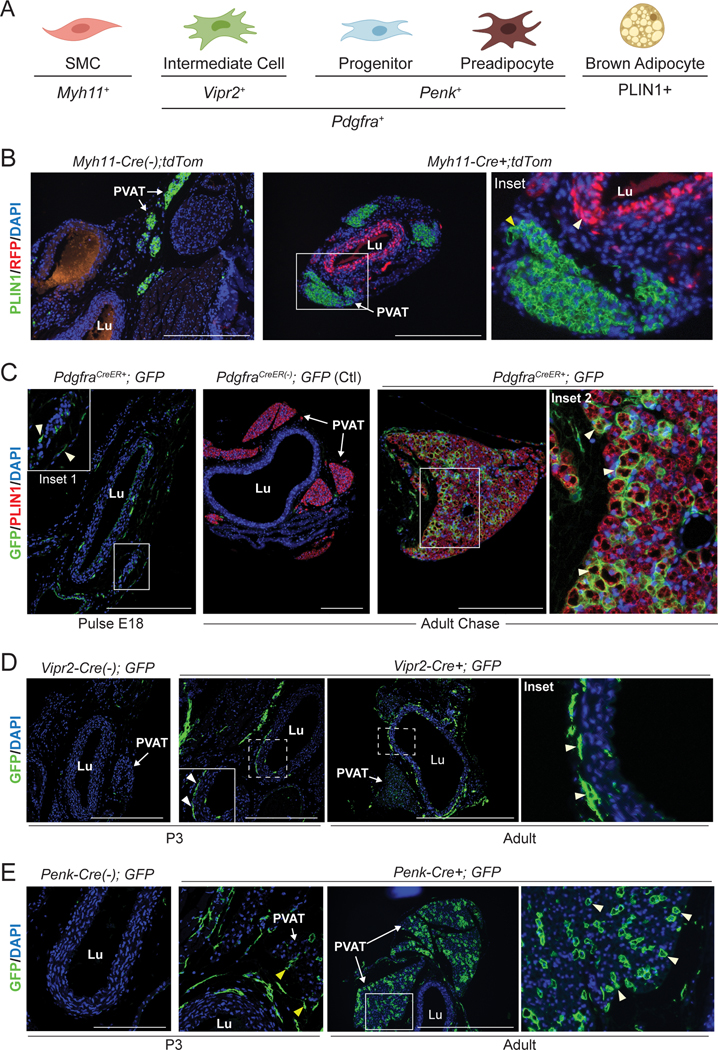
Adipogenic fibroblasts represent the major source of aortic PVAT in neonates.
The contribution of smooth muscle and fibroblastic cells to perivascular adipocyte development was examined in vivo using genetic lineage tracing models (Fig. 4A). We first used Myh11-Cre mice to test if SMCs give rise to peri-aortic adipocytes. Myh11 is a specific marker of SMCs31 and its expression was restricted to SMCs in our scRNAseq data set (Extended Data Fig. 3A). Analysis of Myh11-Cre; tdTomato reporter mice revealed uniform labeling of aortic smooth muscle cells (Fig. 4B). We did not detect any tdTomato+ fibroblasts or adipocytes around the aorta on postnatal day 3, implying that SMCs do not develop into adipocytes during the genesis of this depot (Fig. 4B).
Fig. 4. Adipogenic fibroblasts are the major source of aortic adipocytes in neonates.
(A) Schematic of cell types in perinatal aorta and corresponding marker genes. Created with BioRender.com.
(B) Staining of PLIN1 (green) and RFP (red) in sections of P3 thoracic aorta from Myh11-Cre(−); tdTom (control) and Myh11-Cre+; tdTom reporter mice. (Inset) White arrowhead indicates labeled SMCs. Yellow arrowhead shows unlabeled adipocytes. (n=2 Cre-; n=3 Cre+; scale bar, 271.8 μm).
(C) Staining of GFP (green) and PLIN1 (red) in sections of thoracic aorta from Pdgfra-CreER(−); mTmG (control) and Pdgfra-CreER+; mTmG mice harvested 24 hours (E18 Pulse) and 8 weeks (Adult Chase) after tamoxifen treatment. Arrowheads in inset 1 show labeled fibroblasts. Arrowheads in inset 2 show GFP+ adipocytes. (Pulse: n=1 Cre-; n=3 Cre+; Chase: n=1 Cre-; n=2 Cre+; scale bar, 271.8 μm).
(D) Staining of GFP (green) in sections of P3 and adult thoracic aorta from Vipr2-Cre(−); mTmG and Vipr2-Cre+; mTmG mice. Arrowheads indicate GFP expressing intermediate cells. (P3: n=3 mTmG-; n=4 mTmG+; Adult: n=1 mTmG-; n=4 mTmG+; scale bar, 290 μm (P3), 724.7 μm (adult)).
(E) Staining of GFP (green) in sections of P3 and adult thoracic aorta from Penk-Cre(−); mTmG and Penk-Cre+; mTmG+ mice. Yellow arrowheads show GFP+ fibroblasts. White arrowheads show GFP+ adipocytes. (P3: n=1 Cre-; n=3 Cre+; Adult: n=1 Cre-; n=3 Cre+; scale bar, 145 μm (P3), 724.7 μm (adult)).
PVAT: Perivascular Adipose Tissue; Lu: vessel lumen; DAPI (blue) was used to stain nuclei.
Pdgfra is expressed in all peri-aortic adventitial fibroblasts, including intermediate cells, progenitors and preadipocytes (Extended Data Fig. 3A). To evaluate the contribution of these cell populations to adipocytes, we utilized tamoxifen-inducible Pdgfra-CreER; GFP reporter mice (Pdgfra-CreER; mTmG). We administered one dose of tamoxifen to pregnant females on E18 to induce GFP expression in Pdgfra+ (fibroblast) cells. Analysis of embryos 24h later (pulse) revealed GFP expression in many fibroblastic cells surrounding the aorta (Fig. 4C). At later time points (chase), many mature adipocytes were marked by GFP expression, demonstrating that perivascular adipocytes initially develop from embryonic Pdgfra+ fibroblastic cells (Fig. 4C).
RNAseq analysis identified Vipr2 as a selective marker gene of intermediate cells relative to the other fibroblast subpopulations (Extended Data Fig. 3B). We used Vipr2-Cre; GFP (Vipr2-Cre; mTmG) reporter mice to determine if intermediate cells develop into adipocytes. At P3, GFP expression was confined to fibroblastic cells associated with the outer SMC layer of the aorta, consistent with an intermediate cell identity. In adult mice, we observed a similar pattern of GFP expression in fibroblast cells around the aorta, but an absence of GFP expression in adipocytes (Fig. 4D).
Penk expression was enriched in progenitors and preadipocytes, relative to other fibroblasts (Extended Data Fig. 3A). In P3 Penk-Cre:GFP (Penk-Cre; mTmG) reporter mice, GFP was expressed in peri-aortic fibroblastic cells, with GFP labeling of adipocytes becoming widespread in adulthood (Fig. 4E). Collectively, these experiments demonstrate that peri-aortic adipose tissue develops from fibroblastic progenitors and preadipocyte cells.
Comparative analysis of thermogenic and white adipogenic progenitors
We next sought to determine if perivascular progenitor cells were stably committed to the thermogenic lineage. The adipogenic cell populations from thoracic aorta (progenitor and preadipocyte) and preadipocytes from iWAT were plated and induced to undergo adipocyte differentiation. The PVAT and iWAT-derived cultures expressed similar levels of general adipocyte marker genes (Adipoq, Pparg2), indicating comparable adipogenic competency (Fig. 5A, Extended Data Fig. 4A). Notably, PVAT progenitors and preadipocytes but not iWAT preadipocytes adopted a thermogenic gene program (Dio2, Ucp1) following adipocyte differentiation (Fig. 5A, Extended Data Fig. 4A).
Fig. 5. Comparative gene expression profiling of white and brown adipogenic cells.
(A) mRNA levels of indicated genes in differentiated adipocytes derived from the following cell types of P3 CD1 mice: iWAT preadipocytes, thoracic perivascular progenitors and thoracic perivascular preadipocytes. (n=4 independent wells from pooled FACS populations per group; mean+/− SEM). One way-ANOVA followed by two-sided pairwise comparisons with Holm Sidak correction. P values in order: iWAT PreAd vs. Prog, iWAT PreAd vs. PreAd, Prog vs PreAd. Adipoq (0.4198, 0.3825, 0.7582); Pparg2 (0.0019, 0.3125, 0.0059); Dio2 (0.0002, 0.0021, 0.0705); Ucp1 (0.0583, 0.0003,0.0034).
(B) Principal component analysis (PCA) of the top 500 differentially expressed genes in sorted cell RNA Seq. (n=3 biological replicates per cell type). One PVAT PreAd datapoint includes two overlapping replicates.
(C) Venn diagram of genes enriched in progenitor populations relative to preadipocyte populations from iWAT and PVAT. Pathway analysis of genes commonly enriched in both depots (significantly differentially expressed and LFC>0). Graph plots -log10 unadjusted p values. P values were calculated using clusterProlifer with WikiPathways2019 annotations using the hypergeometric distribution. Padjusted for multiple comparisons was calculated with FDR. Padjusted in order: 0.002835, 0.01171, 0.01171, 0.01171, 0.028843, 0.070511, 0.077592.
(D) Venn diagram of genes enriched in preadipocyte populations relative to progenitor populations. Pathway analysis of genes commonly enriched in both depots (significantly differentially expressed and LFC>0) (left) and genes unique to PVAT (right). Graphs and p values calculated as in (C). P adjusted in order: Left (1.61E-16, 1.26E-13, 0.006022, 0.017277, 0.017277, 0.040287, 0.058059); Right (3.76E-43, 5.75E-36, 6.95E-26, 2.91E-21, 7.14E-13, 1.80E-09, 4.29E-07, 1.31E-06).
(E) Z-Score split heatmap of representative genes from GO analysis. Gene expression levels are calculated between cell types (i.e. PVAT Progenitors vs. PVAT PreAd) within a depot of origin (n=3 biological replicates per cell type).
(F) Model for conserved ontogeny of adipocyte development. Created with Biorender.com.
Statistical Testing: NS p>0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
Our previous work identified a lineage hierarchy in WAT, comprising multipotent progenitors and adipocyte-lineage committed preadipocytes32. We found that the developing PVAT contains transcriptionally similar adipogenic populations: progenitor cells expressing genes such as Pi16, Cd55 and Dpp4, and preadipocytes expressing commitment markers Pparg and Lpl 33.To search for conserved gene programs controlling these two cellular states, we performed bulk RNA sequencing on freshly sorted progenitors and preadipocytes from aortic PVAT and iWAT. Principal component analysis showed that tissue origin accounted for much of the variance in the expression of the 500 most variable genes (Fig. 5B). We defined a consensus gene signature of progenitors, consisting of genes selectively expressed in progenitors vs. preadipocytes from both PVAT and iWAT (Fig. 5C, Extended Data Fig. 4B). Pathway analysis of the progenitor consensus gene signature identified enrichment of Prostaglandin synthesis, TGFβ and WNT signaling (Fig. 5C). The consensus preadipocyte genes were enriched for “PPAR signaling”, “adipogenesis” (Pparg, Cd36, and Rxrg), as well as an unexpected signature of cell cycle activity (Mcm2, Cdc45, and Rpa1) (Fig. 5D, Extended Data Fig. 4C). PVAT preadipocytes specifically expressed genes related to electron transport chain, oxidative phosphorylation and the TCA cycle, including many genes important for the function (Cox5a, Cox7a2, and Ucp1) and transcriptional control (Ppargc1a and Esrra) of brown adipocytes (Fig. 5D,E, Extended Data Fig. 4D). These data support a model in which uncommitted progenitor cells (enriched for expression of anti-adipogenic WNT and TGFβ pathways) develop into preadipocytes, with specific induction of a thermogenic gene program in PVAT preadipocytes (Fig. 5F) 34–36.
Identification of adipogenic smooth muscle cells in adult PVAT
To determine if the cell types observed in developing PVAT were also present in adult mice, we performed scRNAseq analysis on the stromal-vascular fraction of thoracic aorta from 13-week-old mice. Many of the cell groups identified in adult aorta were analogous to perinatal cell types (Fig. 6A, Extended Data Fig. 5A). Clustering analysis revealed three populations of adult fibroblastic cells (all Pdgfra+), two of which were present in perinatal tissue: Pi16+/Ly6aHi progenitors and Bace2+/Clec11aHi intermediate cells (Fig. 6B). The third fibroblast population (“unknown fibroblasts”) was defined by selective expression of S100a4. In situ hybridization analysis of Ly6a, Pi16 and Bace2 showed that the spatial localization of intermediate and progenitor cells was conserved in adults, with intermediate cells located immediately adjacent to the aortic smooth muscle and progenitors located more distant radially (Extended Data Fig. 5B,C). Notably, we did not detect a fibroblastic preadipocyte (Pparg+/Pdgfra+) population in adult aorta. However, we identified a new cluster of SMCs (SMC 2) that expressed canonical smooth muscle markers Myh11, Acta2 and Tagln2, along with adipocyte markers like Pparg, Lpl and Fabp4 (Fig. 6C). Cd81, a recently described marker of smooth muscle-related beige fat progenitor cells in WAT,37 was widely expressed in multiple cell types, whereas the brown adipogenic regulator PRDM16 was selectively expressed in SMCs (Extended Data Fig. 5D).
Fig. 6. Identification of adipogenic smooth muscle cells (SMCs) in adult PVAT.
(A) UMAP of gene expression in 6,753 cells from adult thoracic aorta of pooled 13-week-old male CD1 mice.
(B) UMAPs showing expression of fibroblast marker genes.
(C) UMAPs showing expression of SMC marker genes.
(D) FACS isolation of fibroblast and SMC populations. Live Lin(−) cells were gated to isolate the following cell populations: Intermediate Cells [PDGFRa+,MCAM(−),CD200+], Progenitors [PDGFRa+,MCAM(−),CD200(−)], SMC-1 [PDGFRa(−),MCAM+,CD200+], SMC-2 [PDGFRa(−),MCAM+,CD200(−)]. (Representative image of 6 separate experiments). Full strategy in Fig Extended Data Fig. 5F.
(E) Expression heatmap of Seurat-generated cluster defining genes mapped on to sorted-cell RNA-Seq results (n=3 biological replicates per group).
(F) Pathway analysis of cluster-defining genes for each cell population. Cluster-defining genes were defined as significantly differentially expressed genes with log2FC>1.5 in every pairwise comparison (n=3 biological replicates). Graphs plot -log10 unadjusted p values. P values were calculated using clusterProlifer with WikiPathways2019 annotations and the hypergeometric distribution. Padjusted for multiple comparisons was calculated with FDR. Padjusted in order: Int cells (0.027783, 0.029414, 0.034167, 0.093761, 0.105357, 0.105357); Prog (0.026565, 0.026565, 0.029454, 0.078199, 0.078199, 0.181118); SMC1 (6.64E-05, 6.64E-05, 0.040965, 0.040965, 0.269701); SMC2 (0.000933, 0.001561, 0.004055, 0.008551, 0.047207, 0.047207).
We developed a FACS strategy to purify the following cell types from adult aortas for deep RNAseq and functional characterization: (1) intermediate cells (PDGFRa+;CD200+); (2) progenitors and unknown fibroblasts (PDGFRa+;CD200-); SMC 1 (MCAM+; CD200+); and SMC-2 (MCAM+;CD200-) (Fig. 6D, Extended Data Fig. 5E,F). The expression of cluster-defining genes from the single cell transcriptomes was mapped onto the sorted cell transcriptomes, demonstrating a high level of concordance (Fig. 6E, Extended Data Fig. 5G). Since genes differentially expressed between SMCs and fibroblasts dominate this comparison, we repeated this analysis using pairwise comparisons within fibroblasts (Int vs Prog) or within SMCs (SMC1 vs SMC2), and similarly found high concordance (Extended Data Fig. 5H,I). We determined cell type-defining gene sets by performing differential gene expression analysis. This analysis again demonstrated that intermediate cells were enriched for components of the “Hedgehog signaling” pathway while progenitor cells were enriched for the “WNT signaling” pathway (Fig. 6F). SMC-1 cells were enriched for “focal adhesion” and “integrin-mediated cell adhesion pathways”. Finally, SMC-2 cells were enriched for “Adipogenesis genes”, “Calcium regulation in the cardiac cell”, and “PPAR signaling (Fig. 6F).
Adipogenic activity of smooth muscle cells in adult PVAT
We investigated the adipogenic differentiation competency of FACS-purified SMC and fibroblastic populations isolated from peri-aortic adipose tissue of adult mice. CD200+ SMCs (SMC-1) did not undergo adipogenesis, whereas CD200(−) SMCs (SMC-2) efficiently differentiated into lipid droplet-containing adipocytes (Fig. 7A). The adult fibroblast populations displayed similar adipogenic activity to their neonatal counterparts, with progenitor cells undergoing robust adipocyte differentiation, and intermediate cells displaying no adipogenic potential (Fig. 7A).
Fig. 7. Adipogenic activity of smooth muscle cells (SMCs) in adult PVAT.
(A) Cell populations were FACS purified from adult aortas and induced to differentiate into adipocytes, followed by staining with Bodipy (lipid, green) and Hoechst (nuclei, blue) Quantification of adipocyte differentiation in cultures (right) (n=3 separate experiments; mean+/−SEM). Two way-ANOVA followed by all pairwise comparisons with Holm Sidak multiple correction. All comparisons p<0.0001, except SMC1 vs Int Cell p=0.44). Scale Bars 100 μm.
(B) Staining of RFP (red), PLIN1 (green), and DAPI (blue) in thoracic aorta from adult Myh11-Cre-; tdTom/+ and Myh11-Cre+; tdTom/+ mice treated with vehicle (DMSO) or rosiglitazone (Rosi). Arrowheads = mTomato-labeled adipocytes. (n=1 Cre-; n=2 DMSO, n=2 Rosi; scale bar, 271.8 μm).
(C) Flow cytometry analysis of cell populations in stripped aorta or PVAT alone from adult mice (n=2 biological replicates of pooled aortas; mean+/−SEM). Flow strategy in Extended Data Fig. 5F.
(D) UMAP of gene expression in 12,969 nuclei from adult human peri aortic PVAT (Subclustered to remove immune, endothelial, and mesothelial cells, n=3 humans, integrated analysis).
(E-G) UMAPs showing expression of adipocyte (E), Fibroblast (F) and SMC-related (G) marker genes in the dataset shown in (D).
(H) WikiPathways 2019 GO analysis of all genes differentially expressed (average logFC>0) in SMC-like cells vs. SMCs. Graph plots -log10 unadjusted p values. P values were calculated using clusterProlifer with WikiPathways2019 annotations and the hypergeometric distribution. Padjusted for multiple comparisons was calculated with FDR. Padjusted in order (0.000260333, 0.000781193, 0.001156153, 0.001880657, 0.002417556, 0.003143424, 0.004915838, 0.005278098, 0.005661329, 0.008085485, 0.013244988, 0.015799396).
(I) Model of thoracic aorta and associated tissue organization. Created with Biorender.com.
Statistical Testing: NS p>0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
To determine if SMCs can contribute to perivascular adipocyte development in adult animals, we analyzed the aortas of adult Myh11-Cre; tdTomato reporter animals. Under baseline conditions, tdTomato+ adipocytes were detected, albeit at low levels, in peri-aortic fat. To further assess the adipogenic activity of SMCs, we treated Myh11-Cre; TdTomato reporter mice with rosiglitazone to promote adipocyte turnover38. Rosiglitazone treatment greatly increased the number of tdTomato+; PLIN1+ adipocytes in thoracic PVAT (Fig. 7B). Importantly, rosiglitazone did not induce MYH11 expression in any adipocytes, strongly suggesting that SMCs underwent adipocyte differentiation (Extended Data Fig. 6A).
RNAseq analyses of freshly sorted cells identified Trpv1 as a gene expressed specifically in SMC2 cells compared to SMC1 cells and fibroblastic (Pdgfra+) cells (Extended Data Fig. 6B). We used Trpv1-Cre; GFP reporter mice to examine the adipogenic activity of SMC2 cells. In adult mice housed at thermoneutrality (30°C) or following cold exposure at 4°C for 1 week, GFP was detected in a subset of aortic PVAT adipocytes (Extended Data Fig. 6C). Altogether, these findings demonstrate that SMC2 cells contribute to perivascular adipocyte formation in adult animals.
We investigated the anatomic localization of the two SMC subtypes in thoracic aortic tissues. Immunostaining for the pan SMC marker TAGLN identified SMC cells within the adipose tissue proper as well as in the aortic media around the lumen (Extended Data Fig. 6D). Histologic analysis of the aorta in longitudinal section identified blood vessels coursing through the adipose tissue (Extended Data Fig.6E). To determine if the adipogenic Pparg+/CD200- SMCs (SMC-2) were selectively enriched in the aortic adventitia, we separated adventitia/adipose tissue and aortic media, and isolated cells for flow cytometry analysis (Fig. 7C). The adventitia was highly enriched for CD200(−) progenitor cells and the cleaned aorta was enriched for CD200+ intermediate cells, verifying the separation strategy. Notably, there was also striking enrichment of CD200- (adipogenic) SMCs (SMC-2) in the adventitia samples, whereas the cleaned aorta was enriched for CD200+ SMCs (SMC-1).
To test if fibroblastic cells also generate new adipocytes in adult PVAT, we tracked the fate of Pdgfra+ cells using Pdgfra-CreER; GFP reporter mice (Extended Data Fig. 6F). These mice were treated with tamoxifen to induce GFP-expression in fibroblasts, followed by treatment with rosiglitazone. Under these conditions, we observed many GFP-marked adipocytes in aortic PVAT, showing that fibroblastic cells can undergo de novo adipocyte formation (Extended Data Fig. 6F). Interestingly, cold exposure did not noticeably increase the development of new aortic PVAT adipocytes from either SMCs (Trpv1+) (Extended Data Fig. 6C) or fibroblastic cells (Pdgfra+) (Extended Data Fig. 6G), whereas Pdgfra+ cells generated many new adipocytes in iBAT (Extended Data Fig. 6G).
We identified the hedgehog-related gene Gli1 as a selective marker gene of intermediate fibroblast cells in adult aorta (Extended Data Fig. 6B). To determine if intermediate cells can generate adipocytes, we performed lineage analysis with Gli1-CreER;GFP mice (Extended Data Fig. 6H). Upon tamoxifen administration, GFP expression was induced in intermediate fibroblasts lining the aortic smooth muscle and was not expressed in any adipocytes (Extended Data Fig. 6H). Following rosiglitazone treatment, we did not detect GFP+ adipocytes, suggesting that intermediate cells are not a significant source of new adipocytes in adult animals.
Single nucleus transcriptomic analyses of human PVAT
To determine if human PVAT contains comparable cell types to those discovered in mice, we carried out single nucleus transcriptomic analysis on aortic PVAT samples. Integrated analysis of single nucleus transcriptomes from 3 subjects identified numerous cell clusters, many of which corresponded to readily identifiable cell types on the basis of marker gene expression (Extended Data Fig. 7A,B). For the subsequent analysis, we separated out the immune and endothelial cells, focusing our attention on adipocytes and any potential adipose progenitor groups. Mature adipocytes were marked by ADIPOQ expression, with a subset of these cells expressing the brown fat markers UCP1, EBF2, and PRDM16 (Fig. 7D,E, Extended Data Fig. 7D). We also identified 3 groups of PDGFRa-expressing fibroblasts (Fig. 7D,F). Among these, CD55+ fibroblasts clustered the farthest away from adipocytes and expressed enriched levels of several genes typical of mouse iWAT and PVAT progenitors, including CD55 and Pi16 (Fig. 7D,F, Extended Data Fig. 7F). The remaining 2 groups of fibroblasts expressed enriched levels of adipogenic genes including PPARg and clustered near adipocytes, likely corresponding to preadipocytes (i.e. PreAd-1 and −2). The 2 subgroups of preadipocytes expressed elevated levels of certain genes, exemplified by the selective expression of COL15A1 in PreAd-1 and COL4A4 in PreAd-2 (Extended Data Fig. 7D). ““Transitional cells”, expressing lower levels of PDGFRa and increasing levels of PPARg connected putative preadipocytes with adipocytes. These results suggest that human PVAT and mouse fat depots have remarkably similar fibroblastic -adipocyte precursor cell lineage hierarchies.
We also identified two closely connected clusters of SMC-related cells that we called “SMCs” and “PPARg-expressing SMC-like cells”. Both cell groups expressed many SMC-selective genes including NOTCH3, PDGFRb and TRPC633,39 (Fig. 7D,G, Extended Data Fig. 7C,D).Pathway analysis identified enrichment of smooth muscle-related terms in both cell groups, relative to all other groups (Extended Data Fig. 7E). The PPARg+ SMC-like cells were enriched for many adipogenesis pathways relative to SMCs (Fig. 7H). Interestingly, the SMC population specifically expressed the canonical smooth muscle gene MYH11 whereas the PPARg+ SMC-like cells expressed enriched levels of PPARg and other adipogenic genes (Fig. 7D,F,G, Extended Data Fig. 7C). Finally, we analyzed a publicly available single nucleus transcriptome dataset generated from deep neck BAT40, corresponding to PVAT near the common carotid artery. Like the aortic PVAT, deep neck BAT contained PPARg+ preadipocytes and presumptive progenitor cells marked by many common marker genes, including CD55 and PI16 (Extended Data Fig. 7G-J). We also detected a PPARg+ SMC-like population that expressed many SMC and adipogenic genes but lacked MYH11 expression (Extended Data Fig. 7G-J), similar to the aortic PVAT population. Taken together, these data suggest that PPARg+ SMC-like cells represent adipocyte progenitor cells that may be functionally analogous to the adipogenic SMC2 cells in mouse aorta.
Discussion
Aortic PVAT represents a unique type of thermogenic fat depot that is conserved in rodents and adult humans. This fat depot exhibits many of the defining properties of interscapular BAT in mice, including high levels of UCP1 expression, responsiveness to cold exposure, dependence on the brown fat transcriptional factor EBF2, and resistance to obesity-induced inflammation.
Aortic brown fat cells become organized into recognizable depots in the immediate postnatal period between E18 and P3. We posited that this period would involve large shifts in progenitor cell phenotypes, correlating with the burst of adipogenesis. Rather, we observed a striking similarity in the stromal cell composition of the depot immediately prior to birth (when no adipocytes were present) and after birth (when many adipocytes were present). This finding suggests that perivascular adipocyte progenitor cells are seeded earlier in development and remain poised to undergo adipocyte differentiation until receiving adipogenic cues. The period between E18 to P3 therefore presents an opportunity to identify the native signals that trigger adipogenesis.
Cell lineage tracking studies using the SMC-restricted Myh11-Cre driver indicated that differentiated SMCs do not contribute to aortic PVAT formation during the fetal and early postnatal period (Fig. 7I). This result contrasts with prior studies suggesting that perivascular adipocytes originate from SMCs based on lineage tracing studies using Tagln (Sm22)-Cre41,42. Tagln-Cre+ cells give rise to nearly all perivascular structures, including aortic smooth muscle, adventitial fibroblasts, and adipocytes. We assume that Tagln-Cre is activated during early embryonic development in a cell population that does not express Myh11 and is fated for a perivascular, but not an SMC restricted fate. Consistent with this notion, cells expressing Tagln at E8.5 develop into thermogenic adipocytes42.
Aortic adventitial tissue in perinatal mice contain two distinct groups of adipogenic fibroblasts, progenitors and preadipocytes. Progenitors were marked by expression of Cd55, Pi16, and Dpp4, whereas preadipocytes expressed adipocyte-related transcripts, including the adipogenic transcription factor Pparg. Analogous mesenchymal populations, expressing many of the same marker genes, are also present in subcutaneous WAT32. Pathway analysis identified enrichment of the anti-adipogenic WNT and TGFβ signaling pathways in both PVAT and WAT progenitors43–45. These pathways likely play crucial roles in maintaining progenitor cell identity, at least in part, via blocking adipocyte lineage progression.
We also identified a common gene signature of PVAT (brown) and WAT preadipocytes, including many genes associated with adipogenesis, such as Lpl and Cd36. The common preadipocyte signature was also significantly enriched for genes linked to cell cycle progression. In this regard, adipocyte differentiation of preadipocyte cell lines requires a round of post-confluent mitosis46,47. However, it remains unclear if the cell cycle gene profile identified in preadipocytes relates to cell cycle entry, exit or progression. Additionally, many genes associated with fatty acid oxidation and mitochondrial biogenesis were selectively expressed in PVAT (brown) vs. WAT preadipocytes, indicating that metabolic specialization is already established at the preadipocyte stage.
We identified another interesting fibroblastic cell type in aortas, which we termed intermediate cells due to their anatomic location in between the vascular smooth muscle cells and adventitial progenitors. Intermediate cells expressed enriched levels of Hedgehog signaling genes. Previous studies identified a discrete population of sonic hedgehog-responsive cells located underneath the aorta, likely corresponding to intermediate cells48,49. These cells did not undergo adipocyte differentiation in vitro or in vivo. Understanding the physiologic function(s) of intermediate cells remains a question of great interest. The intimate association of these cells with the outer layer of aortic smooth muscle suggests a potential role in modulating vascular biology. In this regard, a recent report suggests that fibroblastic cells surrounding the vasculature have the capacity to differentiate into SMCs upon severe vessel injury50. We did not detect these cells in human aortic PVAT, likely because of their close association with the vessel, which was not included in these samples.
Adult PVAT in mouse also possessed a distinctive subset of adipogenic SMCs that express Pparg and other adipocyte-related genes. This Pparg+ SMC population, marked by the expression of Trpv1 and Myh11, display robust potential to differentiate into adipocytes. Related to this finding, elegant work by the Graff group identified distinct lineages of embryonic and adult adipocyte progenitors in WAT51. Intriguingly, they showed that adult adipocyte progenitors in WAT arise via an Acta2-expressing mural or SMCs. It remains unclear if the Acta2+ cells in WAT or SMCs identified here in PVAT progress through a fibroblastic intermediate (i.e., preadipocyte) or differentiate directly into adipocytes. It is important to note that adult mouse PVAT also contains progenitor cells, a fibroblast population with the potential to produce adipocytes. Notably, cold exposure did not induce high levels of new adipocyte differentiation from either SMCs or progenitor cells in aortic PVAT, as compared to other fat depots52–54. This finding may suggest that PVAT expandability is somehow constrained. One possibility is that cold may act mainly on preadipocyte cells, which are depleted in adult mouse PVAT (Fig. 6A). Alternatively, these findings may suggest that the browning of the PVAT depot upon cold exposure is mainly driven by the activation of thermogenic programs in pre-existing mature fat cells. Taken together, our study suggests that both SMCs and progenitor cells contribute to the maintenance of adult PVAT in mice (Fig. 7I). Further studies are needed to determine if these populations are recruited by different stimuli.
Aortic PVAT in adult humans, unlike in mice, possesses two presumptive populations of fibroblastic “preadipocyte” cells, closely connected to mature adipocytes. A third type of fibroblast represents putative progenitor cells, clustering closely with preadipocytes and expressing many of the same marker genes as their counterparts in mice, including CD55, DPP4, and PI16. We also identified a presumptive population of adipogenic SMC-related cells in human PVAT. Interestingly however, these “PPARg+ SMC-like” cells in human PVAT, while clustering closely with SMCs and displaying enriched levels of many smooth muscle pathways, did not express the canonical smooth muscle marker Myh11. We speculate that SMCs may give rise to adipogenic “SMC-like” cells in human PVAT, coupled with the loss of some mature smooth muscle characteristics.
In summary, this work defines the progenitor cells responsible for the development and maintenance of PVAT, a distinct thermogenic depot conserved in rodents and humans. Based on pre-clinical studies in rodent models, BAT-targeted therapies for the treatment of human obesity and metabolic disease will depend on the ability to increase brown fat mass, either via transplantation of brown fat cells or by inducing brown fat cell formation in situ. The identification of the native progenitors and developmental ontogenies of thermogenic PVAT adipocytes, which are retained in adult humans, provides a critical foundation for therapeutic strategies aimed at augmenting brown fat.
Methods:
Mice:
All animal experiments were performed according to procedures approved by the University of Pennsylvania’s Institutional Animal Care and Use Committee (Approval #805649). Mice were housed under the care of University of Pennsylvania University Laboratory Animal Resources (ULAR). Animals were raised at room temperature on standard chow with a 12-hour light/dark cycle. All mouse housing and husbandry occurred at RT (22°C) unless specified otherwise. For thermoneutral acclimation, 4- to 5-week old mice were housed at 30°C for one month. For chronic cold exposure, 8- to 10-week-old mice were pair-housed in cages at 4°C for one week. Mice were fed a regular chow diet (LabDiet, 5010) or western diet composed of 40% fat and 0.15% cholesterol (Research Diets,D12079B). Tamoxifen (Sigma [T5648], stock 20 mg/mL in corn oil) was intraperitoneally injected at a dose of 100 mg/kg for one day or five consecutive days. Mice were treated with rosiglitazone (250 nMoles/d) (Cayman catalog #11884) for the experimental time course indicated in text. Experiments performed on embryonic and perinatal mice were conducted on male and female mice. Experiments at adult time points were performed in male and female mice between the ages of 8 to 12 weeks at the onset of the experiment.
C57/Bl6N (RRID:IMSR_TAC:B6) mice were obtained from Taconic. CD1 (RRID:IMSR_CRL:024) mice were obtained from Charles River. The following strains were obtained from the Jackson Laboratory: mTmG (strain name: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J, RRID:IMSR_JAX:007676), tdTom (strain name: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, RRID:IMSR_JAX:007914), Myh11-Cre (strain name: B6.Cg-Tg(Myh11-cre,-EGFP)2Mik/J, RRID:IMSR_JAX:007742), Pdgfra-CreER (strain name: B6.129S-Pdgfratm1.1(cre/ERT2)Blh/J, RRID:IMSR_JAX:032770), Gli1-CreER (strain name: Gli1tm3(cre/ERT2)Alj/J, RRID:IMSR_JAX:007913), Trpv1-Cre mice (B6.129-Trpv1tm1(cre)Bbm/J, RRID:IMSR_JAX:017769), Penk-Cre (strain name: B6;129S-Penktm2(cre)Hze/J, RRID:IMSR_JAX:025112), and Vipr2-Cre (strain name: B6.Cg-Vipr2em1.1(cre)Hze/J, RRID:IMSR_JAX:031332). The following stock was generated in the Seale lab: AdipoqCre; Ebf2 loxP/loxP (Angueira et al., 2020).
Histology and Immunofluorescence
Tissues were fixed in 4% PFA overnight, washed in PBS, dehydrated in ethanol, paraffin-embedded and sectioned. For en bloc retroperitoneal sections, perinatal mice were euthanized by decapitation, the thoracic viscera were removed, and the tissue was placed in 4% PFA overnight. Adult mice were perfused with 5 mL of 4% PFA and tissues were harvested as above. Slides were deparaffinized and heat-antigen retrieved in Bulls Eye Decloaking buffer (Biocare). Slides were incubated in indicated primary antibodies overnight, stained with secondary antibody, and developed with Tyramide Signal Amplification (TSA, Akoya Biosciences). For Trpv1-Cre tracing studies, sections were deparaffinized and rehydrated, followed by an antigen retrieval step in a modified citrate buffer (Dako Target Retrieval Solution, pH 6.1, Agilent). Sections were then incubated in Sudan Black (0.3% in 70% ethanol) to reduce autofluorescence signal. Blocking was performed in Millipore blocking reagent (EMD Millipore), followed by incubating with primary antibody overnight. Primary antibodies used for staining were: PLIN1: (1:200, CST: 3470, RRID:AB_2167268), PDGFRa: (1:50, Novus Biologicals: AF1062, RRID:AB_2236897), MYH11: (1:100, Abcam: ab53219, RRID:AB_2147146), PPARG: (1:500, Invitrogen: MA5–14889, RRID:AB_10985650), RFP: (1:250, VWR Scientific, 600–401-379, RRID:AB_2209751), GFP (1:500, Abcam: AB6673, RRID:AB_305643), CD200: (1:25, Novus Biologicals: AF2724, RRID:AB_416669), ACTA2: Sigma: (1:200, A2547, RRID:AB_476701), TAGLN: (1:100, Abcam: ab10135, RRID:AB_2255631), and DAPI (1:1000 Roche). RNA in situ hybridizations were performed using the RNAscope system (Advanced Cell Diagnostics; 323100, 323120) with the following probes: Bace2 (407151), Clec11a (583301), Pi16 (451311-C2), and Ly6a (427571-C2). Images were taken on a Leica TCS SP8 confocal microscope or Keyence inverted microscope.
RNA Extraction, qRT-PCR and RNA Sequencing analysis
Total RNA was extracted using TRIzol (Invitrogen) and Purelink RNA columns (Fisher). For processing of RNA from cultured primary adipocytes, RNA was extracted using Trizol (Invitrogen) combined with PicoPure RNA Isolation Kit (Fisher). mRNA was reverse transcribed into cDNA using the ABI High-Capacity cDNA Synthesis kit (ABI). Real-time PCR was performed on an ABI7900HT PCR machine using SYBR green fluorescent dye (Applied Biosystems). Fold changes were calculated in Microsoft Excel (2016) using the ΔΔCT method, with Tata Binding Protein (Tbp) mRNA serving as a normalization control and graphed in GraphPad Prism (Versions 7–9). RNA sequencing from sorted progenitors was performed at Genewiz with the following procedure. RNA was extracted using Trizol LS (Invitrogen) and quantified using Qubit (2.0, Life Technologies. Carlsbad, CA) and TapeStation. RNA sequencing libraries were made using the SMART-Seq v4 Ultra Low Input Kit. Samples were sequenced on an Illumina HiSeq 4000 with a 2×150 Paired End (PE) configuration.
FASTQ files were aligned to mm10 using STAR (2.5.2a) with quantMode = GeneCounts. The unstranded genecounts for each sample were analyzed for differential gene expression using the R (3.6.3) package DESeq2 (1.26.0). In general, for each experiment involving an RNA seq dataset, the DESeq2 model was built with the design (~CellType) where “CellType” refers to the different sorted populations, with all CellTypes included in the model. Pairwise differential expression was calculated using the DESeq2 function “results” with thresholds of alpha=0.1. Log2 Fold change thresholds were used as indicated. ENSEMBLID’s were converted to gene symbols using the R package org.Mm.eg.db (3.10.0). Pathway analysis was performed via WikiPathways with annotations for Mus Musculus using the package clusterProfiler (3.13.3) and rWikiPathways (1.6.1) (Figures 5,6 and 7, Extended Data Figs. 4, 7). Pathway analysis was performed using Enrichr (https://maayanlab.cloud/Enrichr/#), WikiPathways 2019 Mouse for Figure 3 55–58. Graphs plot -log10 unadjusted p values. P values were calculated using clusterProfiler or Enrichr with WikiPathways2019 annotations using a hypergeometric distribution and corrected for multiple comparisons using FDR (Figures 4,5,7, Extended Data Figs. 4, 7) or Benjamini-Hochberg (Fig. 3). Adjusted p values are reported in the figure captions.
To determine unique cell type/cluster defining genes from bulk RNA seq data, DESeq2 was used to generate pairwise comparisons between all cell types within an experiment (ex. Int vs Prog, Int vs SMC1, Int vs SMC2) and genes with a Log2FC>1.5 in every pairwise comparison were called “cluster defining” for a particular group.
Heatmaps represent either Log2 fold change versus row mean or row score as indicated as indicated. Underlying data for heatmaps came from the DESeq2 regularized log transform of the normalized count values using the DESeq2 function “rlog”. Figures showing side by side scRNA-seq and bulk RNA seq heatmaps contain the exact same genes in the same order on both heatmaps. For Figures 2B and 6E, “cluster defining genes” were generated using the Seuratv3 function FindAllMarkers; these genelists were thresholded at the indicated average logFC cutoff and then plotted using the function DimHeatmap. Analagous heatmaps were then generated to show DESeq2 analysis of bulk RNA-seq data of sorted cells. For Figures Extended Data Fig. 5G and H, a similar process was performed using pairwise comparisons between the indicated groups, using the Seurat function FindMarkers for scRNAseq data and a DESeq2 pairwise analysis for bulk RNA-seq data. Extended Data Figs. 2D and 5I correspond to the same data in Figures 2B and 6E respectively. They show the total number of genes plotted for each cluster in the heatmaps, as well as how many Seurat cluster defining genes were also upregulated in the bulk RNA-seq (Enriched/Concordant), and how many of the Seurat cluster defining genes were downregulated in the bulk RNA-seq data (De-enriched/ discordant).
scRNA Seq
Single cells were isolated from dissected aortas and flow sorted to isolate live (DAPI-) cells and remove debris. Cells were collected as either CD45+ (immune) and CD45- (non-immune) cells. For adult single cell RNA Sequencing, CD45+ cells were mixed with CD45- cells to achieve a proportion of ~20%. Single cell transcriptomes were collected using the 10X Genomics platform with v2 (perinatal) and v3 (adult) chemistry (10× Genomics, Pleasanton, CA) (Merrick et al., 2019). The libraries were sequenced using an Illumina HiSeq 4000. The reads were processed using the CellRanger pipeline and R Studio using Seuratv3 (Merrick et al., 2019, Stuart et al., 2019). Cells were filtered based on their expression of a minimum number of genes (E18/P3: 500 < nFeatureRNA < 3000; Adult: 250 < nFeature RNA < 4000) and mitochondrial genome (<10%) abundance for each dataset individually. Regression was performed using ScaleData function for the following variables: percentage of mitochondrial reads and cell cycle phase (G2M.Score/S.Score). Dimensionality reduction was performed in Seurat using UMAP and differential gene expression between clusters was performed using FindMarkers function. For all datasets, immune cells were collapsed into one cluster for simplicity. For the perinatal dataset, two clusters of mesothelial cells were collapsed to generate one mesothelial cluster. For the adult dataset, initial clustering revealed that SMC population 1 separated into two clusters, but the effect size of gene expression between the two groups was small, so they were collapsed to form SMC population 1. Feature-plots, dot-plots and heat-maps were generated using Seurat.
E18/P3 combined analysis
Dataset integration was performed using the Seurat functions FindIntegrationAnchors followed by IntegrateData. Scaling and regression were performed using the Seurat ScaleData function for the following variables: percentage of mitochondrial reads and cell cycle phase (G2M.Score/S.Score). Dimensionality reduction was performed in Seurat using UMAP and differential gene expression between clusters was performed using FindMarkers function. For all datasets, immune cells were collapsed into one cluster for simplicity. Single gene UMAP feature plots showing expression of select genes at E18 and P3 were generated for the RNA assay using a modified version of the Seurat FeaturePlot function. (This modified version plots both the E18 and P3 UMAPs on the same color/expression scale, while the default Seuratv3 function does not).
Human Aortic PVAT snRNA Seq: Individual Sample Analysis
The reads were processed using the CellRanger pipeline and R Studio using Seuratv3 (Merrick et al., 2019, Stuart et al., 2019). To exclude low quality nuclei and doublets, nuclei were filtered based on their expression of a minimum number of genes and mitochondrial abundance for each dataset individually PVAT1: (1000 < nFeatureRNA < 6000) & (0.025 < percent.mito < 1). PVAT2: (800 < nFeatureRNA < 5000) & (0.0.01 < percent.mito < 0.75). PVAT3: (900 < nFeatureRNA < 5000) & (0.0125 < percent.mito < 1). Regression was performed using ScaleData function for the following variables: percentage of mitochondrial reads, number of RNA features, and cell cycle phase (G2M.Score/S.Score). Dimensionality reduction was performed in Seurat using UMAP and differential gene expression between clusters was performed using FindMarkers function.
Human Aortic PVAT snRNA Seq: Combined Dataset Analysis
Dataset integration was performed using the Seurat functions FindIntegrationAnchors followed by IntegrateData. Scaling and regression were performed using the Seurat ScaleData function for the following variables: percentage of mitochondrial reads, number of RNA features, and cell cycle phase (G2M.Score/S.Score). Dimensionality reduction was performed in Seurat using UMAP and differential gene expression between clusters was performed using either the FindMarkers function (pairwise comparisons) or the FindAllMarkers (cluster defining genes) on the RNA assay for all comparisons.
On the integrated dataset, immune cells were collapsed into one immune cluster and endothelial cells were collapsed into one endothelial cluster for simplicity. Sub clustering analysis (Fig. 7) was performed by subsetting the Seurat object to remove endothelial and immune cells. Initial clustering revealed that adipocyte population separated into two clusters, but the effect size of gene expression differences between the two groups was small, so they were collapsed to form one adipocyte population. Dot plots, single gene UMAP feature plots, and single gene violin plots showing expression of select genes on the integrated dataset were generated for the RNA assay using Seurat DotPlot, FeaturePlot, and VlnPlot functions, respectively.
Deep Neck BAT Analysis
Previously published deep neck BAT snRNA-seq data were reanalyzed using the same protocol described here 40. Endothelial cells and immune cells that were identified by the Wolfrum group and by expression of PECAM1, VWF, CDH5 (Endothelial) and PTPRC, MRC1 (immune) were filtered out.
Differential gene expression between clusters was performed using FindAllMarkers function on the subsetted, reclustered Seurat object. This Seurat object was used to generate dot plots, single gene UMAP feature plots, and single gene violin plots using Seurat DotPlot, FeaturePlot, and VlnPlot functions respectively.
Generation of single cell suspensions from mouse aorta
Aorta were isolated from mice, minced and placed into digestion medium (DMEM, Collagenase D: 6.1mg/ml (Roche), Dispase II: 2.4 mg/ml (Roche) and placed at 37°C with constant agitation at 200 rpm. To enrich for cell populations with differential sensitivity to digestion, we utilized the following procedures. For embryonic/newborn mice, 20% of the digestion was quenched at 15 minutes, 20% at 20 minutes and the remaining 60% at 25 minutes (embryos/perinatal). For adult aortas 50% of the digestion was quenched at 30 minutes, 50% at 60 minutes. For stripped aorta flow cytometry analysis, perivascular adipose tissue was removed from the aorta and digested separately from the cleaned aorta. Tissue digestions were quenched with an equal volume of complete medium (DMEM/10% FBS). Dissociated cells were suspended using a P1000 pipette and filtered through a 100 μm filter. Cells were then pelleted at 400 g for 4 min and RBCs were lysed in 155mM NH4Cl, 12mM NaHCO3, and 0.1mM EDTA for 4 min. An equal volume of complete medium was added, and the cells were filtered through a 40um filter for downstream analyses.
Isolation of human aortic PVAT nuclei
Human peri-aortic fat tissue was obtained from patients undergoing coronary artery bypass grafting surgery (Male, Average age = 64 (Individual ages - 65,65,61), average BMI = 28.2 (individual BMI’s – 23.5, 25.9, 35.3). This work was performed at Maine Medical Center under the approval of Maine Medical Center IRB #1396116–4 and informed consent was obtained from all study subjects. The isolation and purification of nuclei as adapted from Hu et al 59. Briefly, 40–80 mg of frozen human PVAT was dounce homogenized using a glass homogenizer (Sigma-Aldrich, 7 mL) on ice in 2.5 mL homogenization buffer (0.32 M sucrose (Sigma, RNase & DNase free ultra-pure grade), 5 mM CaCl2 (Sigma), 3 mM MgAc2 (Sigma-Aldrich), 10 mM Tris-HCl pH 8.0 (FisherChem), 0.1% TritonX-100 (Amresco), 0.1 mM EDTA (Fischer), protease inhibitor cocktail (Roche)) and layered into ultracentrifuge tubes (Beckman Coulter) over 12 mL sucrose cushion (1.8 M sucrose, 10 mM Tris-HCl pH 8.0 (FisherChem), 3 mM MgAc2 (Sigma-Aldrich), protease inhibitor cocktail (Roche) and topped with 14.5mL homogenization buffer. Samples were ultracentrifuged (Beckman Coulter L-70K) using a swinging bucket rotor (Beckman Coulter SW28) at 25,000 rpm for 2 hrs at 4°C. The supernatant was aspirated to remove debris and the nuclear pellet was resuspended in 1 mL of DPBS (Corning) with RNase inhibitor (Lucigen). Following 20 mins incubation on ice, nuclei were filtered through a 100 μM cell strainer and pelleted at 5,000 rpm for 10 mins at 4°C. Nuclei were resuspended in 2% BSA (Gold biotechnology) in DPBS, spun again and filtered 2x through a 40 uM mini strainer (puriSelect). Nuclei were stained with Trypan blue, counted on a hemocytometer, and visually inspected for intact morphology before 10x loading at the Center for Applied Genomics Sequencing Core (Children’s Hospital of Philadelphia) using the following reagents: 10x genomics: v3.1 Next GEM kit, Sequencer: Illumina NovaSeq 6000 and Flow cell: SP 100 cycles v1.5.
Flow Cytometry and Sorting:
Isolated cells were suspended in FACS buffer (HBSS, 3% 0.45uM filtered FBS; Fischer). Cells were stained in FACS buffer with antibodies (see below) for 1 hr at 4°C in the dark. Cells were washed 2x with cold FACS buffer and sorted on a BD FACS Aria cell sorter (BD Biosciences) with a 100 μm nozzle as previously described 32. All compensation was performed at the time of acquisition in BD FACS DIVA software (8.0.1) using single stained cells or compensation beads (BioLegend catalog no. A10497). Downstream analysis and visualization of flow data was performed in FlowJo (10.6.2). The following antibodies were used for FACS protocols. P3 Aorta: LY6A-BV711 (1:100 Biolegend-108131 RRID:AB_2562241),CD317-PerCp-Cy5.5 (1:100 Biolegend-127022 RRID:AB_2566647), CD200-AF488 (1:100 Biorad-MCA1958A488), CD45-APC/Cy7 (1:1000, Biolegend-103115 RRID:AB_312980), CD31-APC/Fire750 (1:1000 Biolegend-102528 RRID:AB_2721491), Ter119-APC/Cy7 (1:1000 Biolegend 116223 RRID_AB:2137788), CD142-CF647 (1:1000 Sino Biological), FVS510 (1:1000 BD Biosciences 654406). P3 iWAT: CD45-APC/Cy7 (1:1000, Biolegend-103115 RRID:AB_312980), CD31-APC/Fire750 (1:1000 Biolegend-102528 RRID:AB_2721491), CD142-CF647 (1:1000 Sino Biological cat#50413-R001), CD26(DPP4)-FITC (Biolegend, San Diego, CA, cat# 302704 1:100); ICAM1-PE/Cy7 (Biolegend cat# 353116 1:100). Adult Aorta: CD200-AF488 (1:100 Biorad-MCA1958A488), CD45-APC/Cy7 (1:1000, Biolegend-103115 RRID:AB_312980), CD31-APC/Fire750 (1:1000 Biolegend-102528 RRID:AB_2721491), Ter119-APC/Cy7 (1:1000 Biolegend 116223 RRID_AB:2137788), MCAM-PerCP/Cy5.5 (1:100 Biolegend 134709 RRID:AB_11204083), PDGFRa-PE/Cy7 (1:200 Biolegend 135912 RRID:AB_2715974), and DAPI (1:10000 Roche 10236276001).
Cell Culture:
Sort-purified cells were plated on 384-well Cellbind plates (Sigma-Aldrich catalog no. CLS3770) and cultured in DMEM/F12 supplemented with 10% FBS and Primocin (50 μg/mL). Cells were plated at a density of 10,000–30,000 cells per well and induced to differentiate into adipocytes within 3 days of initial plating. Cell density and time until differentiation was kept consistent within each experiment. Adipogenic differentiation cocktail consisted of 1 μM dexamethasone (Sigma catalog no. D4902), 0.5 μM isobutylmethylxanthine (Sigma catalog no. I7018), 125 nM indomethacin, 20 nM insulin, and 1 nM T3 for 48 hours. Following two days of induction, media was changed to 20 nM insulin and 1 nM T3. Medium was changed every two days for six more days.
Adipogenesis Assays and Quantification:
Adipogenesis was assessed by staining with Bodipy 493/503 (Invitrogen catalog no. D3922) for lipid accumulation and Hoechst 33342 (Thermo Fisher catalog no. 62249) for nuclei32. Briefly, cells were differentiated in 384 well tissue culture plates (Sigma-Aldrich catalog no. CLS3770), fixed with 4% paraformaldehyde, stained, and imaged on a Keyence inverted microscope (BZX-710) using Keyence BZ-X Viewer Software (1.3.1.1) with the following filters: DAPI (ex, 360/40 nm; em, 460/50 nm; Keyence, OP-87762) and GFP (ex, 470/40 nm; em, 525/50 nm; Keyence, OP-87763) filters. Images were acquired at 20x in a 7×7 tiled grid and stitched to capture the entirety of each well. Tiling and stitching were performed with Keyence BZ-X Analyzer software (1.3.0.3). Image quantification was performed automatically in ImageJ (version 1.52E) using a macro which: 1) Split images into component channels, 2a) for the nuclei channel applied a 3-Sigma Gaussian blur, performed thresholding to identify signal above background, performed watershed to segmentation, and counted the number of nuclei, 2b) for the lipid channel applied a 2-Sigma Gaussian blurm performed thresholding to identify signal above background, and counted the area (#of pixels) with signal above threshold. The amount of adipogenesis was calculated as Lipid Area/#nuclei.
Statistical Analysis:
No power calculations were performed prior to initiation of the study. No mice were omitted from the study. All individual data points were plotted to assay normality. Experiments from a common pool of digested tissue or independent pools processed separately are indicated in the text. Statistical testing for qPCR and image analysis data were performed in GraphPad Prism (7–9). Two sample t tests were performed where comparisons between two groups were being assayed. One-way ANOVAs with pairwise comparisons corrected with the Holm Sidak method were used for comparisons between more than two groups. Two-way ANOVAs with multiple comparisons corrected with the Holm Sidak method were performed for adipogenesis experiments (within each experimental day compare between cell types). # indicates p value < 0.1; * indicates p value < 0.05, ** indicates p value < 0.01, *** indicates p value < 0.001, **** indicates p value < 0.0001.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Patrick Seale (sealep@pennmedicine.upenn.edu). Sequencing data and code are available on GEO (GSE164528). All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Extended Data
Extended Data Fig. 1. Single cell transcriptional profiling of aortas from E18 and P3 mice.
(A) UMAPs of gene expression in cells from E18 and P3 thoracic aorta of pooled CD1 mice.
(B-F) UMAPs showing expression of indicated genes for: smooth muscle cells (SMC) (B); Intermediate cells (C); Progenitors (D); Preadipocytes (E); and Adipocytes (F).
(G) Expression dotplot of indicated genes for cell clusters from P3 aorta.
Extended Data Fig. 2. Purification of smooth muscle and fibroblast cells from aorta (Related to Fig. 3).
(A) FACS isolation of fibroblastic and smooth muscle cell (SMC) populations. Dissociated cells were gated on: (1) SSC-A and FSC-A to exclude debris; (2) FSC-H vs. FSC-W then SSC-H vs. SSC-W to isolate single cells; and (3) live (FVS510-) Lin- (CD45-,CD31-,Ter119-) cells. Depicted sort gates were used to isolate the following populations: Progenitors [LY6A high], Preadipocytes (PreAd) [LY6A(−), CD142 mid; CD200(−)]; SMCs [LY6A(−); CD142(−); CD200+], Intermediate Cells (Int) [LY6A(−); CD142+; CD200+, CD317(−)], Meso (Mesothelium) [LY6A(−); CD142+; CD200+, CD317+] (Representative images from n=10 expts). Related to experiments shown in Figs. 3A and Fig 5. (B) Violin plots showing expression of genes used in the sorting strategy. (C) Staining of CD200 (red), MYH11 (green), and DAPI (blue) in P3 thoracic aorta. White arrowhead shows an Intermediate cell. Yellow arrowhead shows an SMC (scale bar, 65 μm; Lu: lumen). Representative of n=1 experiment. (D) Expression of Seurat cluster-defining genes in the sorted cell bulk RNAseq datasets. Gene levels in sorted cell populations were compared to the row mean and annotated as “enriched” or “de-enriched” (Log2 FC>0.25).
Extended Data Fig. 3. Expression profiles for Cre-driver mouse lines (Related to Fig. 4).
(A) UMAPs showing expression of genes used for Cre and CreER driver mouse strains.
(B) Vipr2 expression levels in indicated cell types from sorted cell bulk RNAseq analysis. (One way ANOVA followed by two sided pairwise comparisons with Holm Sidak correction. n=3 biologically independent samples per group; mean +/− SEM). Statistical Testing: ****p≤0.0001.
Extended Data Fig. 4. Gene profiling analyses of iWAT vs. PVAT progenitors and preadipocytes (Related to Fig. 5).
(A) mRNA levels of indicated genes in differentiated primary adipocytes from: iWAT preadipocytes, aortic PVAT progenitors, aortic PVAT preadipocyte cells (all from P3 CD1 mice). Second experimental replicate performed on different day (n=2 PVAT Pread; n=3 PVAT Progenitor; n=4 iWAT Pread independent wells per group from pooled FACS samples; mean+/− SEM). (B) Z-Score split heatmap of genes from overlaps depicted in Fig. 5C,D. Gene expression levels are calculated between cell types (i.e. PVAT Progenitors vs PVAT PreAd) within a tissue of origin (n=3 biological replicates). (C) Expression heatmap of all cell cycle genes (used by Seurat) in sorted progenitor and preadipocyte cells from iWAT and thoracic aortic PVAT. Data comes from DESeq2 normalized count data (bulk RNAseq). (D) Pathway analysis of genes enriched in preadipocytes from PVAT vs. iWAT (significantly differentially expressed and LFC>0). Graph plots -log10 unadjusted p values. P values were calculated using clusterProlifer with WikiPathways2019 annotations using the hypergeometric distribution. Padjusted for multiple comparisons was calculated with FDR. Padjusted in order (2.48E-34, 1.07E-20, 1.31E-15, 2.97E-13, 1.37E-12, 2.01E-10, 1.71E-07, 2.66E-07, 1.84E-05).
Extended Data Fig. 5. Purification and analysis of aorta-associated cells from adult animals (Related to Fig. 6).
(A) Expression dot plot of indicated genes in cell clusters from adult aorta. (B) mRNA in situ hybridization of Ly6a (green) in adult aorta. Arrowhead in inset shows progenitor cell. DAPI (blue) stains nuclei. (scale bar, 362.3 μm). Lu: lumen. Representative of n=2 experiments. (C) mRNA in situ hybridization of Pi16 (green) and Bace2 (red). White arrowhead shows progenitor cell. Yellow arrowhead shows intermediate cell (scale bar, 145 μm). Representative of n=2 experiments. (D) UMAPs showing Cd81 and Prdm16 expression. (E) Violin plots showing expression of indicated genes used for sorting. (F) FACS isolation of fibroblasts and smooth muscle cells (SMCs). To exclude debris and isolate single live cells, we gated on: (1) SSC-A and FSC-A; (2) FSC-H vs FSC-W; and (3) SSC-H vs SSC-W. We then gated on Live (DAPI-); Lin- (CD45-,CD31-,TER119-) cells. Further selection was used to isolate: Intermediate Cells [PDGFRa+, MCAM(−), CD200+], Progenitors [PDGFRa+, MCAM(−), CD200(−)], SMC1 [PDGFRa(−), MCAM+, CD200+], SMC2 [PDGFRa(−), MCAM+, CD200(−)]. (Representative image of 6 separate experiments). Related to experiments shown in Fig. 6D and Fig. 7A, C. (G) Expression of Seurat cluster-defining genes in the sorted cell bulk RNAseq datasets. Gene levels in sorted cell populations were compared to the row mean and annotated as “enriched” or “de-enriched” (Log2 FC>0.25). (H, I) Expression heatmap of Seurat cluster-defining genes mapped on to sorted-cell RNASeq results for fibroblast (H) and SMC populations (I) (n=3).
Extended Data Fig. 6. Analysis of aortic PVAT in adult mice (Related to Fig. 7).
(A) Staining for MYH11 (red) and PLIN (green) in aortas of Rosiglitazone (Rosi)-treated mice, scale bar 200 μm. Representative of n=1 experiment. Scale bar, 200 μm. (B) mRNA levels of Trpv1 and Gli1 in sorted cell populations. (C) Staining of GFP (green) and either PLIN1 or UCP1 (red) in aortas of male Trpv1-Cre; mTmG mice housed at thermoneutrality or following 1 week of 4C cold exposure. Scale bar, 100 μm. (D) Staining of TAGLN (red) in adult aorta. White arrowheads show adventitial SMCs. Yellow arrowhead indicates parenchymal SMCs of the aorta. Representative of n=2 experiments. Scale bar, 271.8 μm. (E) H&E staining of adult aorta. Arrowheads show adventitial blood vessel. (F) Staining of GFP (green) in aortas of Pdgfra-CreER+; mTmG+ mice following a 5-day pulse of Tamoxifen and a 2.5 week treatment with DMSO or Rosi. Arrowheads=GFP+ adipocytes (n=1 Cre-; n=4 DMSO; n=5 Rosi; scale bar, 543.5 μm). (G) Staining of GFP (green) in aortas of Pdgfra-CreER+; mTmG+ mice following a 5-day pulse of Tamoxifen and 2-week chase at room temperature or 4C (cold). Representative of n=1 experiment. Scale bar, 100 μm (H) Staining of GFP (green) in aortas of CreER- Ctl and Gli1-CreER+; mTmG+ mice following a 5-day pulse with Tamoxifen and 2.5-week treatment with DMSO or Rosi. (n=1 Cre-; n=3 Cre+; DMSO; n=3 Cre+; Rosi scale bar, 543.5 m). DAPI (blue) was used to stain nuclei.
Extended Data Fig. 7. Single nucleus transcriptomic analyses of human PVAT (Related to Fig. 7).
(A) UMAP of gene expression in 18,758 nuclei from adult human peri-aortic PVAT (Full dataset, n=3 humans, integrated analysis). (B) Expression dotplot of indicated genes in dataset from (A). (C, D) Violin plots (C) and UMAPs (D) showing expression of select genes, corresponding to the subclustered dataset in Fig. 7D. (E) Pathway analysis of cluster defining genes (average logFC>0.5) in SMC and PPARg+ SMC-like cells compared to all other clusters in the dataset from Fig. 7D. Graph plots -log10 unadjusted p values. P values were calculated using clusterProlifer with WikiPathways2019 annotations using the hypergeometric distribution. Padjusted for multiple comparisons was calculated with FDR. Padjusted in order SMC (1.59E-07, 2.50E-06, 2.75E-06, 0.000106107, 0.000968522, 0.000968522, 0.001899498); PPARg+ SMC Like Cells (5.75E-05, 0.000135397, 0.003396457, 0.00971142, 0.00971142, 0.010636022, 0.010636022). (F) Violin plots showing expression of select marker genes in the fibroblastic populations from Fig. 7D. (G) UMAP of gene expression in 7285 nuclei from adult human deep neck BAT (subclustered to remove immune and endothelial cells, n=16 humans, integrated analysis, re-analyzed data from: Sun et al 2020). (H-J) UMAPs (H), expression dotplot (I) and violin plots (J) showing expression of select genes in cells from the dataset above (G).
Acknowledgements
We thank Drs. Xin Bi, Daniel Rader, Mitchell Lazar, Mark Kahn, Jorge-Henao-Mejia, and members of the Seale lab for thoughtful discussion. We thank the University of Pennsylvania Diabetes Research Center for use of the Functional Genomics Core (P30-DK19525). We are grateful to Jacqueline Smiler, Renata Pellegrino Da Silva, Fernanda A. Mafra and James P. Garifallou in the Center for Applied Genomics Sequencing Core (Children’s Hospital of Philadelphia) for the single cell and single nucleus sequencing. This work was supported by NIH grants DK123356 and DK120982 to P.S; HL141149 to L.L; R01DK077097, R01DK102898, R01DK122808 (to Y.-H.T.), DK120062 to A.R.A., T32GM008216 to A.P.S, K01DK125608 (to F.S.), K01DK111714 (to M.D.L), P30-DK19525 (to Penn Diabetes Research Center) and P30DK036836 (to Joslin Diabetes Center’s Diabetes Research Center).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Cypess AM et al. Activation of human brown adipose tissue by a beta3-adrenergic receptor agonist. Cell Metab. 21, 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harms M. & Seale P. Brown and beige fat: development, function and therapeutic potential. Nat. Med 19, 1252–1263 (2013). [DOI] [PubMed] [Google Scholar]
- 3.O’Mara AE et al. Chronic mirabegron treatment increases human brown fat, HDL cholesterol, and insulin sensitivity. J. Clin. Invest 130, 2209–2219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoneshiro T. et al. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest 123, 3404–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon B. & Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev 84, 277–359 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Kazak L. et al. Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab. 26, 693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazak L. et al. A Creatine-Driven Substrate Cycle Enhances Energy Expenditure and Thermogenesis in Beige Fat. Cell (2015). doi: 10.1016/j.cell.2015.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda K. et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med 23, 1454–1465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sacks H. & Symonds ME Anatomical locations of human brown adipose tissue: functional relevance and implications in obesity and type 2 diabetes. Diabetes 62, 1783–1790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cypess AM et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med 360, 1509–1517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Marken Lichtenbelt WD et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med 360, 1500–1508 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Virtanen KA et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med 360, 1518–1525 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Aherne W. & Hull D. The site of heat production in the newborn infant. Proc. R. Soc. Med 57, 1172–1173 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heaton JM The distribution of brown adipose tissue in the human. J. Anat 112, 35–39 (1972). [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W. & Seale P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol 17, 691–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orava J. et al. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metab. 14, 272–279 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Lidell ME et al. Evidence for two types of brown adipose tissue in humans. Nat. Med 19, 631–634 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Cypess AM et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med 19, 635–639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huttunen P, Hirvonen J. & Kinnula V. The occurrence of brown adipose tissue in outdoor workers. Eur. J. Appl. Physiol. Occup. Physiol 46, 339–345 (1981). [DOI] [PubMed] [Google Scholar]
- 20.Cheung L. et al. Human mediastinal adipose tissue displays certain characteristics of brown fat. Nutr. Diabetes 3, e66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajakumari S. et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. (2013). doi: 10.1016/j.cmet.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angueira AR et al. Early B Cell Factor Activity Controls Developmental and Adaptive Thermogenic Gene Programming in Adipocytes. Cell Rep. 30, 2869–2878.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stine RR et al. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol. Metab 5, 57–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapira SN et al. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev. 31, 660–673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowal L. et al. Intrinsic Properties of Brown and White Adipocytes Have Differential Effects on Macrophage Inflammatory Responses. Mediators Inflamm. 2017, 9067049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lumeng CN, Bodzin JL & Saltiel AR Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest 117, 175–184 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian XY et al. Thermoneutral Housing Accelerates Metabolic Inflammation to Potentiate Atherosclerosis but Not Insulin Resistance. Cell Metab. 23, 165–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisberg SP et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J. Clin. Invest 116, 115–124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boucher JM et al. Pathological Conversion of Mouse Perivascular Adipose Tissue by Notch Activation. Arterioscler. Thromb. Vasc. Biol 40, 2227–2243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miano JM, Cserjesi P, Ligon KL, Periasamy M. & Olson EN Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ. Res 75, 803–812 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Merrick D. et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domenga V. et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 18, 2730–2735 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cristancho AG & Lazar MA Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol 12, 722–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross SE et al. Inhibition of adipogenesis by Wnt signaling. Science 289, 950–953 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Ignotz RA & Massagué J. Type beta transforming growth factor controls the adipogenic differentiation of 3T3 fibroblasts. Proc. Natl. Acad. Sci. U. S. A 82, 8530–8534 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oguri Y. et al. CD81 Controls Beige Fat Progenitor Cell Growth and Energy Balance via FAK Signaling. Cell 182, 563–577.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang W, Zeve D, Seo J, Jo A-Y & Graff JM Thiazolidinediones regulate adipose lineage dynamics. Cell Metab. 14, 116–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietrich A. et al. Increased vascular smooth muscle contractility in TRPC6−/− mice. Mol. Cell. Biol 25, 6980–6989 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun W. et al. snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Chang L. et al. Loss of perivascular adipose tissue on peroxisome proliferator-activated receptor-gamma deletion in smooth muscle cells impairs intravascular thermoregulation and enhances atherosclerosis. Circulation 126, 1067–1078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye M. et al. Developmental and functional characteristics of the thoracic aorta perivascular adipocyte. Cell. Mol. Life Sci 76, 777–789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longo KA et al. Wnt10b inhibits development of white and brown adipose tissues. J. Biol. Chem 279, 35503–35509 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Kennell JA & MacDougald OA Wnt signaling inhibits adipogenesis through beta-catenin-dependent and -independent mechanisms. J. Biol. Chem 280, 24004–24010 (2005). [DOI] [PubMed] [Google Scholar]
- 45.MacDougald OA & Mandrup S. Adipogenesis: forces that tip the scales. Trends Endocrinol. Metab 13, 5–11 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Tang Q-Q, Otto TC & Lane MD Mitotic clonal expansion: A synchronous process required for adipogenesis. Proc. Natl. Acad. Sci 100, 44 LP – 49 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang QQ & Lane MD Adipogenesis: From Stem Cell to Adipocyte. Annu. Rev. Biochem 81, 715–736 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Passman JN et al. A sonic hedgehog signaling domain in the arterial adventitia supports resident Sca1<sup>+</sup> smooth muscle progenitor cells. Proc. Natl. Acad. Sci 105, 9349 LP – 9354 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramann R. et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang J. et al. Arterial Sca1(+) Vascular Stem Cells Generate De Novo Smooth Muscle for Artery Repair and Regeneration. Cell Stem Cell 26, 81–96.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Jiang Y, Berry DC, Tang W. & Graff JM Independent stem cell lineages regulate adipose organogenesis and adipose homeostasis. Cell Rep. 9, 1007–1022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee Y-H, Petkova AP, Konkar AA & Granneman JG Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 29, 286–299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shao M. et al. Cellular Origins of Beige Fat Cells Revisited. Diabetes 68, 1874–1885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang QA, Tao C, Gupta RK & Scherer PE Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med 19, 1338–1344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu G, Wang L-G, Han Y. & He Q-Y clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martens M. et al. WikiPathways: connecting communities. Nucleic Acids Res. 49, D613–D621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen EY et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu P. et al. Dissecting Cell-Type Composition and Activity-Dependent Transcriptional State in Mammalian Brains by Massively Parallel Single-Nucleus RNA-Seq. Mol. Cell 68, 1006–1015.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]