Abstract
Birds display advanced behaviors, including vocal learning and problem-solving, yet lack a layered neocortex, a structure associated with complex behavior in mammals. To determine whether these behavioral similarities result from shared or distinct neural circuits, we used single-cell RNA sequencing to characterize the neuronal repertoire of the songbird song motor pathway. Glutamatergic vocal neurons had considerable transcriptional similarity to neocortical projection neurons; however, they displayed regulatory gene expression patterns more closely related to neurons in the ventral pallium. Moreover, while γ-aminobutyric acid–releasing neurons in this pathway appeared homologous to those in mammals and other amniotes, the most abundant avian class is largely absent in the neocortex. These data suggest that songbird vocal circuits and the mammalian neocortex have distinct developmental origins yet contain transcriptionally similar neurons.
Songbirds are one of the few animal groups that have specialized brain regions for the learning and production of vocalizations (1). Multiple components of the song-control system have well-established homologs in the mammalian brain (2). However, it has been challenging to identify homologs of song regions in the avian pallium, which has a markedly different structure compared to the mammalian pallium (3, 4). The two pallial regions of the song motor pathway, HVC (proper name) and RA (robust nucleus of the arcopallium), have been functionally compared to the mammalian neocortex (Fig. 1, A and B) (5), but it is not clear to what extent these similarities reflect conservation of shared elements from common ancestors (homologies) versus convergent evolution of non-homologous neural circuits.
Fig. 1. Cell classes and neuronal diversity in songbird pallial premotor regions.
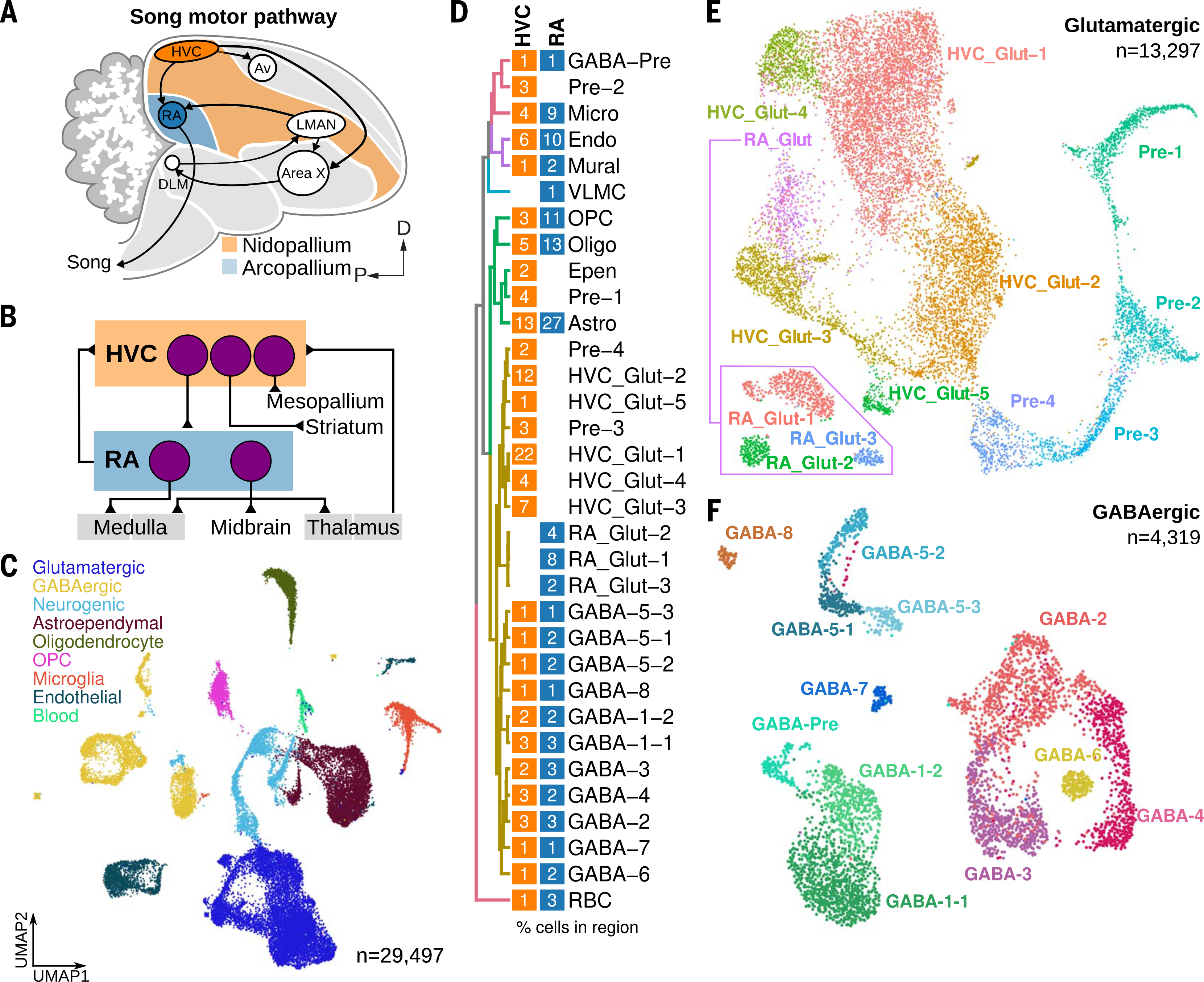
(A) Schematic highlighting the two forebrain nuclei of the song motor pathway: HVC (orange) and RA (blue). Each nucleus is a specialized substructure located in two large neuroanatomical domains: the nidopallium for HVC, and the arcopallium for RA. D, dorsal; P, posterior. (B) Schematic of the principal known projection classes in HVC and RA and their afferent and efferent connections. (C) UMAP (Uniform Manifold Approximation and Projection) plot of cells combined across species and regions colored by cell-type class. OPC, oligodendrocyte precursor cell. (D) Hierarchical clustering of average cluster expression profiles (see supplementary methods). Blocks correspond to HVC (orange) and RA (blue) and indicate the relative percentage of each cluster in each region. Values were rounded up to allow visualization of rare (<0.5%) but well-defined populations. GABA, GABAergic; Pre, neuronal precursor; Micro, microglia; Endo, endothelial; VLMC, vascular and leptomeningeal cells; Oligo, oligodendrocyte; Epen, ependymal; Astro, astrocyte; Glut, glutamatergic; RBC, red blood cell. (E and F) UMAP plots of (E) glutamatergic neurons (HVC_Glut-1 through HVC_Glut-5 and RA_Glut-1 through RA_Glut-3) and neurogenic lineage (Pre-1 through Pre-4) and (F) GABAergic neurons (GABA-1 through GABA-8) and GABAergic neurogenic lineage (GABA-Pre). Inset in (E) shows three distinct subclusters within RA.
Both HVC and RA reside in a large non-layered structure called the dorsal ventricular ridge (DVR), which is present in all sauropsids (birds and non-avian reptiles) but has no obvious corresponding region in mammals. Despite these anatomical differences, the DVR has functional, connectivity, and molecular properties similar to those of the neocortex (6, 7). One influential hypothesis proposes that nuclei within the DVR are homologous to layers of the neocortex (7–11). Alternatively, neurons in the DVR could exhibit similarities to neocortical neurons not because of homology between these two regions but through the evolution of similar functional cell types. For example, an alternative framework based on developmental gene expression patterns and topological organization argues that the DVR is an expansion of the ventral pallium, which in mammals includes olfactory areas, such as the piriform cortex, and the pallial amygdala (6, 12). Most comparative analyses to date have focused on gene expression in neuroanatomical regions (7, 11, 13–16), potentially masking insights that would arise from comparisons of individual cell types. To investigate these alternative hypotheses and directly compare the cell types of songbirds and other amniotes, we carried out large-scale single-cell RNA sequencing (RNA-seq) of neurons from the songbird song motor pathway and compared their gene expression profiles and regional distribution with those of neurons from mouse and turtle datasets.
Cell classes and neuronal diversity in song motor regions
We microdissected HVC and RA from adult males of two closely related species, zebra finches (Taeniopygia guttata) and Bengalese finches (Lonchura striata domestica) (Fig. 1A). Samples from each species were prepared independently, with single-nuclei sequencing performed on zebra finches and single-cell sequencing performed on Bengalese finches (fig. S1A and table S1). Each species dataset contained the same cellular classes with similar relative abundances (fig. S1, B to D, and table S2), and subsequent analyses were performed on combined data. Each region contained several major populations that could be classified by the expression of specific marker genes. These classes included neurons; neuroepithelial glia, including astrocytes (SLC15A2), ependymal cells (SPEF2), oligodendrocytes (PLP1), and oligodendrocyte precursor cells (PDGFRA); and non-neural cell types, including microglia (CSF1R), vascular endothelial cells (FLI1), mural cells (RGS5), vascular and leptomeningeal cells (LUM), and red blood cells (HBAD) (Fig. 1, C and D, and fig. S1E). Whereas most non-neuronal populations were highly similar between HVC and RA, glutamatergic neurons and neurogenic cell types were largely nonoverlapping, and γ-aminobutyric acid-releasing (GABAergic) neurons and astrocytes were similar between regions but displayed greater heterogeneity than non-neuronal types (Fig. 1D and fig. S1F). Among neuronal types, we identified eight populations of glutamatergic cells (SLC17A6), with five specific to HVC (HVC_Glut-1 through HVC_Glut-5) and three specific to RA (RA_Glut-1 through RA_Glut-3); and eight GABAergic populations (GAD1), each seen jointly in HVC and RA (GABA-1 through GABA-8) (Fig. 1, D to F, and fig. S1, E and I to L). Two of the GABAergic clusters could be further subclustered (GABA-1–1, GABA-1–2, GABA-5–1, GABA-5–2, and GABA-5–3) (Fig. 1F). A glutamatergic neurogenic lineage was present in the HVC dataset (Pre-1 through Pre-4) that progressed from adult neural stem cells (NR2E1) through intermediate neural precursors (SOX4) to migrating neuroblasts (DCX), consistent with well-characterized adult neurogenesis in this region (Fig. 1, D and E, and fig. S1, E, G, and H) (17–20). This lineage expressed several factors known to regulate neurogenesis in mammals, including NOTCH2, FABP7, TBR1, and NECTIN3 (fig. S1H) (21–23). Lastly, we identified a GABAergic neuroblast cluster (GABA-Pre) in both HVC and RA that likely represents a relatively uncharacterized inhibitory adult neurogenic population (Fig. 1, D and F, and fig. S1E) (24).
Glutamatergic neuron diversity in the song motor pathway
Prior work has extensively characterized the classes of glutamatergic neurons in the song motor pathway and their afferent and efferent connectivity patterns (Fig. 2A) (25). There are three known glutamatergic projection populations in HVC (25): one targeting RA (HVCra), one targeting the striatal song nucleus Area X (HVCX), and one targeting the mesopallial auditory nucleus Avalanche (HVCAv) (Fig. 2A). Our clustering of HVC glutamatergic neurons suggests that there are five distinct groups of neurons in the region, which could indicate that the three principal classes contain distinct transcriptional subclasses or that there are previously unreported glutamatergic subtypes in HVC. To first validate the HVC glutamatergic clusters (Fig. 2B), we identified genes with high cluster specificity (Fig. 2C, fig. S2A, and table S3; see supplementary methods) and carried out in situ hybridization assays using probes for the pan-glutamatergic marker SLC17A6 and for markers exhibiting high specificity for each of the five HVC clusters (Fig. 2, D and E, and fig. S2, B to D). These data confirm that each cluster is present in HVC, and that, in combination, these marker genes recover the five main single-cell glutamatergic clusters (Fig. 2, D and E).
Fig. 2. Diversity of songbird glutamatergic neurons.
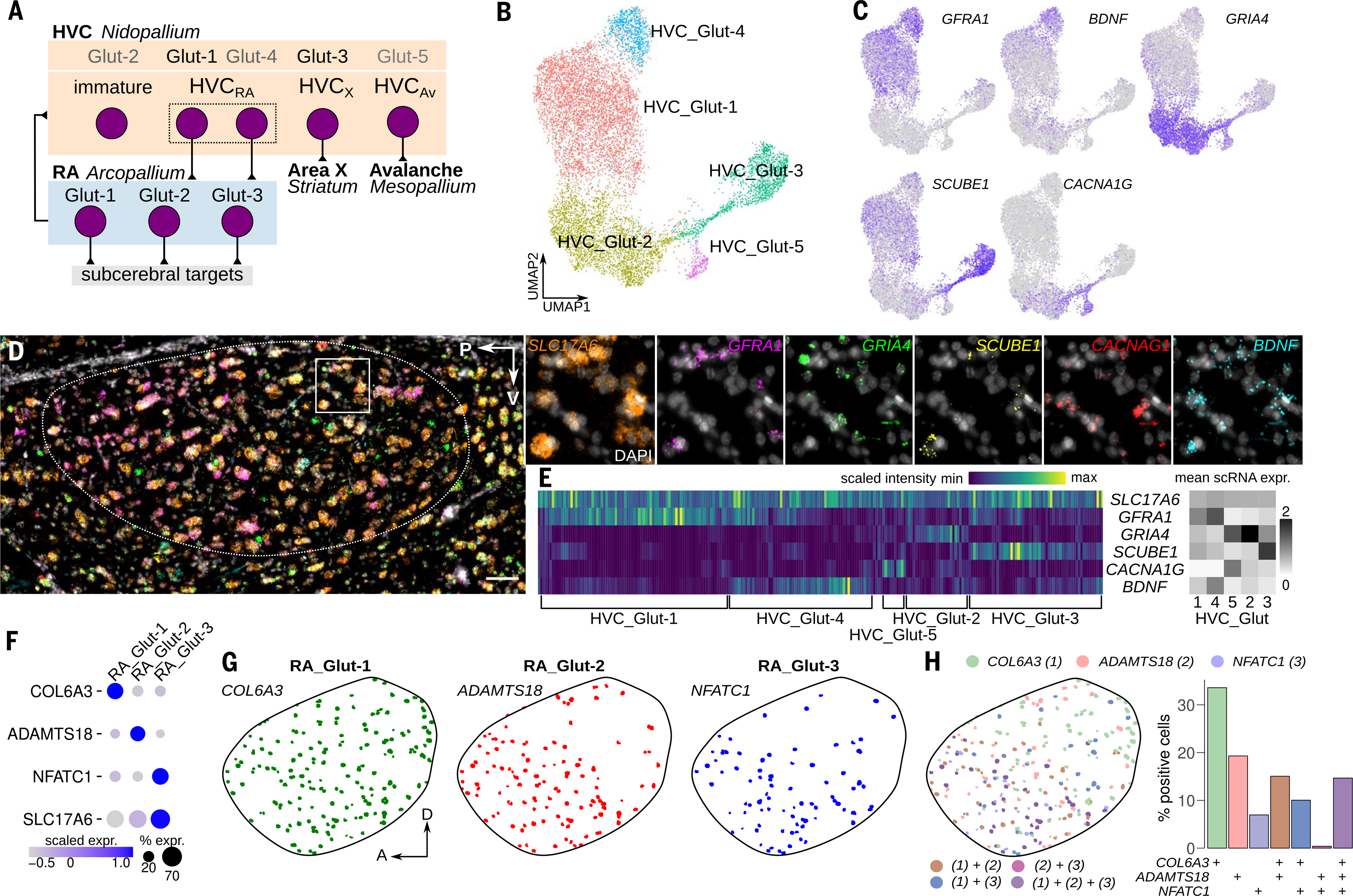
(A) Schematic of HVC and RA glutamatergic neuron classes with single-cell cluster assignments. These mappings were made using retrograde tracing combined with in situ hybridization (HVC_Glut-1 to HVCRA and HVC_Glut-3 to HVCX), transcriptional similarity to other classes (HVC_Glut-4 as a putative HVCRA subclass and HVC_Glut-2 as a putative immature neuron class), or relative abundance in the tissue (HVC_Glut-5 to HVCAv). (B) UMAP plot of HVC glutamatergic clusters and mappings to known glutamatergic projection classes. (C) UMAP plots of the expression of five marker genes GFRA1 (HVC_Glut-1/4), BDNF (HVC_Glut-4), GRIA4 (HVC_Glut-2), SCUBE1 (HVC_Glut-3), CACNA1G (HVC_Glut-5). (D) Example sagittal section of HVC (outlined) and signal from a six-channel in situ hybridization assay for HVC glutamatergic marker genes. Scale bar, 50 μm. Boxed region is enlarged at right and is split into single-channel images. (E) Quantification of in situ hybridization signal in SLC17A6-positive cells (n = 243) scaled by the maximum and minimum intensities for each gene. Heatmap columns are organized by hierarchical clustering. Labels below the heatmap indicate assignment of single-cell sequencing clusters to clusters derived from in situ hybridization data. For comparison, grayscale heatmap at right shows mean expression levels for each marker and cell type derived from sequencing. (F) Expression of SLC17A6 and marker genes in sequencing data for neurons from the three RA glutamatergic clusters. (G) Locations of RA glutamatergic clusters within RA, identified using in situ hybridizations against markers genes for each cluster: COL6A3 (RA_Glut-1), ADAMTS18 (RA_Glut-2), and NFATC1 (RA_Glut-3). A, anterior. (H) (Left) Overlaps of individual cell locations, colored by marker genes. (Right) Percentage of positive cells in each marker gene combination.
We additionally sought to determine which glutamatergic clusters correspond to neuron types that have previously been defined by their projection targets. We injected retrograde tracers into RA and Area X separately to label the cell bodies of the corresponding HVC projection populations (fig. S2E) and then used in situ hybridization probes to examine the co-localization of marker genes with labeled cells. The HVC_Glut-1 and HVC_Glut-4 marker gene GFRA1 was elevated in retrogradely identified HVCRA neurons (fig. S2, F and G), and these clusters express relatively high levels of the previously identified HVCRA marker UTS2B (26) (fig. S2, A and H). The expression of several activity-dependent genes (BDNF, HOMER1, and FOSL2) suggests that HVC_Glut-4 may represent an active population of HVC_Glut-1 neurons (Fig. 2C and fig. S2, A and I). However, electrophysiology and morphological characterizations of HVC projection neurons have described two groups of HVCRA neurons (27, 28). Consistent with the presence of two HVCra subclasses, HVC_Glut-1 and HVC_Glut-4 differentially express a moderate number of genes (n = 91) (fig. S2J and table S3), and the removal of previously defined singingand activity-related genes (29, 30) did not cause the HVC_Glut-4 cluster to collapse into HVC_Glut-1 (fig. S2K). The HVC_Glut-3 marker SCUBE1 was elevated in retrogradely labeled HVCX neurons (fig. S2, F and G), and HVC_Glut-3 was enriched for the known HVCX marker NTS (31) (fig. S2, A and H). Although marker genes for HVCAv have not yet been reported, the abundance of cells in HVC_Glut-5 (~2% of projection neurons) is comparable to that seen in a previous retrograde tracing analysis of HVCAv neurons (25) (fig. S1J). HVC_Glut-2 lies at the terminus of the neurogenic lineage (Fig. 1E) and is enriched for genes involved in dendritogenesis and synaptogenesis (fig. S1G) and therefore may constitute an immature population of HVCRA neurons, which are known to undergo replacement in adult songbirds (17–20). However, it is also possible that this cluster represents a previously undescribed mature glutamatergic population whose projection targets have not been defined or that does not project outside of HVC. In keeping with these interpretations, retrogradely labeled HVCRA and HVCX neurons do not show enriched expression of the HVC_Glut-2 marker gene GRIA4 relative to unlabeled neurons (fig. S2F).
Anatomical tract-tracing experiments have shown that glutamatergic neuron projections from RA are largely subcerebral, targeting motoneurons and premotor neurons in the medulla that control song production as well as the midbrain and thalamus (32–35) (Fig. 2A). Prior neurophysiological approaches have suggested the presence of one glutamatergic population (36–39), whereas morphology analysis has suggested the presence of three projection neuron classes in RA (40), consistent with the number of clusters in our single-cell analysis. As done for the HVC cluster validation, we identified genes with highly specific expression across the three RA glutamatergic clusters and performed in situ hybridization assays to validate their presence in RA (Fig. 2, F to H, and fig. S3, A to C). Expression of marker genes for RA_Glut-2 (ADAMTS18) and RA_Glut-3 (NFATC1) were ventrally biased, similar to populations that project to brainstem syringeal motoneurons, while cells expressing a marker gene for RA_Glut-1 (COL6A3) but not the RA_Glut-2 and RA_Glut-3 markers were dorsally biased, consistent with the distribution of neurons projecting to the mid-brain, respiratory premotor neurons in the medulla, and reciprocally back to HVC (32, 35, 41) (Fig. 2, F to H). Each glutamatergic cluster in both HVC and RA displayed different expression patterns of axon guidance-related genes, further supporting the mapping of each cluster onto distinct projection populations (fig. S3D). Together, these glutamatergic clusters represent three broad classes of connectivity patterns (Fig. 2A): two intratelencephalic groups [pallio-pallial (HVC_Glut-1/2/4/5) and pallio-striatal (HVC_Glut-3)] and one subcerebral (RA_Glut-1/2/3).
Pallial identities of vocal circuit neurons
The diversity of glutamatergic populations in HVC and RA offers an opportunity to examine current models of DVR homology at cellular resolution. The “nucleus-to-layer” model (Fig. 3A, “layer”) argues that neurons in the avian nidopallium, which contains HVC, are homologous to neocortical layer 2/3, and neurons in the arcopallium, which contains RA, are homologous to neurons in layer 5 (10, 15, 42). To assess cell-type similarities to the neocortex, we evaluated the transcriptional similarity of HVC and RA glutamatergic neurons to previously described mouse neocortical glutamatergic neurons (43) (Fig. 3B and fig. S4, A to C). All three RA glutamatergic neuron classes were similar to layer 5 subcerebral projection neurons (L5 pyramidal tract) and showed elevated levels of deep-layer markers (fig. S4D), in agreement with previous analyses of marker gene expression data that reported similarities between RA and layer 5 (11). However, HVC glutamatergic neurons did not map exclusively to layer 2/3 cortical neurons. Instead, they exhibited substantial diversity, with significant similarities between HVC neurons and intratelencephalic neurons from each of mouse cortical layers 2/3, 4, 5, and 6. Whereas HVC_Glut-1/4/2 exhibited greatest similarity to layer 2/3 cortical neurons, HVC_Glut-3 and HVC_Glut-5 had equal or greater similarity to deep-layer neurons, with the pallio-striatal cluster HVC_Glut3 (HVCX) showing no significant similarity to neurons in layer 2/3 (Fig. 3B and fig. S4A).
Fig. 3. Songbird glutamatergic neurons exhibit features of both ventral pallial and neocortical classes.
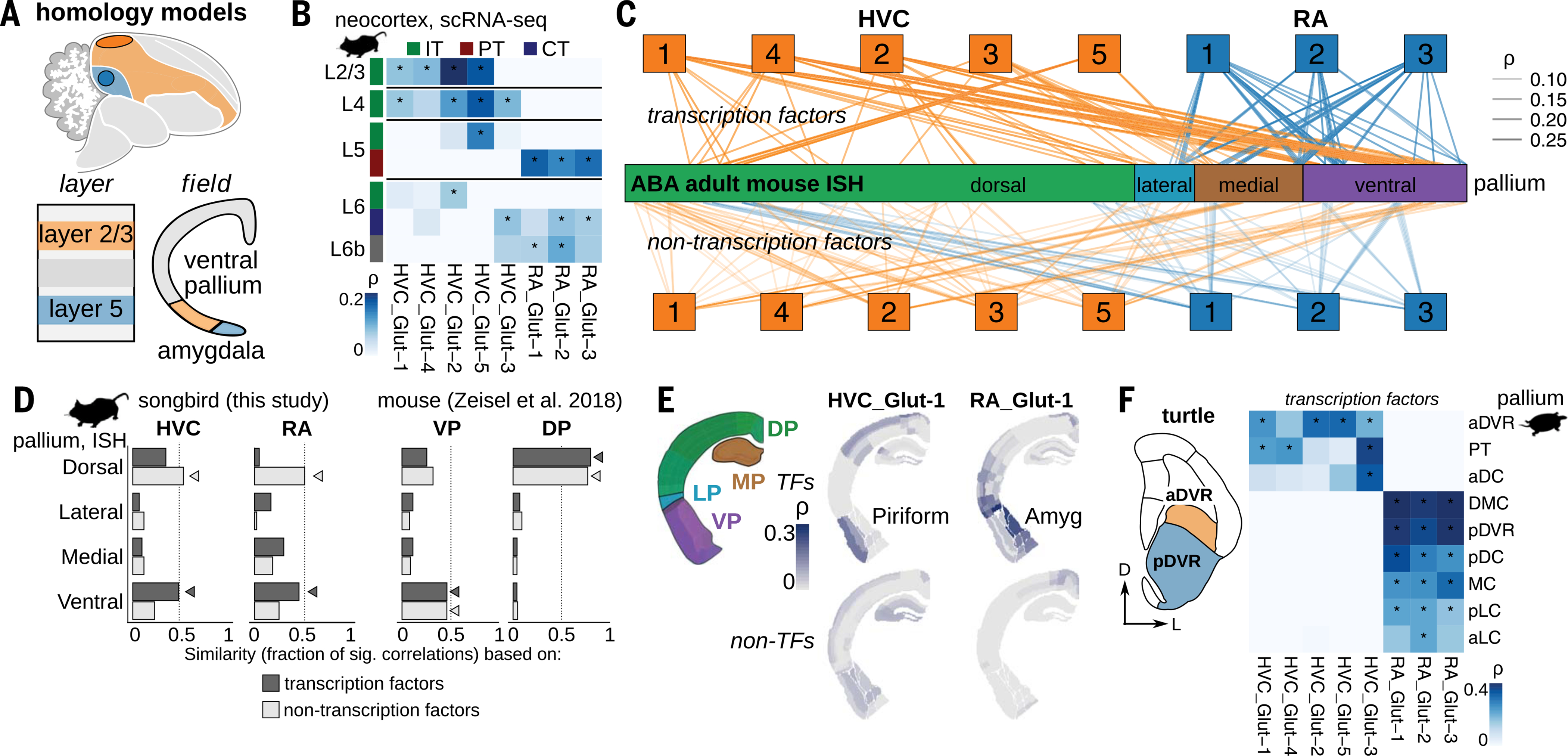
(A) Alternative models of homology between song motor pathway (SMP) regions in the nidopallium (orange) and arcopallium (blue) and the mammalian pallium. A “nucleus-to-layer” hypothesis (“layer”) argues that these regions are homologous to specific cell types or layers of the mammalian neocortex, while a “pallial field” hypothesis (“field”) proposes that these regions are homologous to derivatives of the mammalian ventral pallium. (B) Spearman correlations between scaled average expression profiles of SMP glutamatergic neurons and mouse neocortical glutamatergic neurons grouped by class (43). IT, intratelencephalic; CT, corticothalamic; PT, pyramidal tract. Negative correlations set to zero. Asterisk indicates shuffled P < 0.05. (C) Correlation analysis between transcription factor (top) and non-transcription factor (bottom) expression profiles in SMP glutamatergic neurons and regional in situ hybridization data across mouse pallial regions (ABA, Allen Brain Atlas). Lines represent Spearman correlations with shuffled P < 0.05. Mouse pallial organization as defined in (88). (D) Fraction of significant correlations (shuffled P < 0.05) between mouse pallial regions and (left) SMP glutamatergic neurons or (right) mouse forebrain glutamatergic neurons (44). Arrowheads indicate the pallial region with the highest similarity for each comparison. (E) Spearman correlations between SMP glutamatergic neurons (HVC_Glut-1 and RA_Glut-1) and adult mouse pallium mapped onto coronal sections from the Adult Mouse ABA. MP/DP/LP/VP, medial/dorsal/lateral/ventral pallium. (F) Hypothesized homologies between subregions of the DVR in turtles and songbirds (12). Schematic is a coronal section from the turtle forebrain [adapted from (49)]. Spearman correlations between scaled average transcription factor expression profiles of SMP glutamatergic neurons and glutamatergic neurons from the turtle pallium, grouped by brain region (49). a/pDC, anterior/posterior dorsal cortex; PT, pallial thickening; (D)MC, (dorsal) medial cortex; a/pLC, anterior/posterior lateral cortex; a/pDVR, anterior/posterior dorsal ventricular ridge. Negative correlations set to zero. Asterisk indicates shuffled P < 0.05.
To assess the robustness of these findings, we compared songbird glutamatergic clusters with neocortical glutamatergic neurons in two other mouse datasets (44, 45) (fig. S4, E and F). These analyses supported the broad conclusions from our original comparison. HVC_Glut-1/4/2 generally had greatest similarities to layer 2/3 neocortical neurons but also shared similarity with neurons in layers 4, 5, and 6. In contrast, HVC_Glut-5 and HVC_Glut-3 had the greatest similarities to neurons in deep layers, with the pallio-striatal cluster HVC_Glut-3 showing little or no similarity to layer 2/3. These comparisons argue against the nucleus-to-layer model in which HVC corresponds specifically to mammalian neocortical layer 2/3, and they instead indicate that HVC contains a diversity of glutamatergic neurons that have similarities to mammalian neurons across both upper and deep cortical layers. Furthermore, we examined whether song motor pathway glutamatergic neurons showed an association with particular neocortical regions, but we observed no differential similarities to primary motor cortex versus primary visual cortex (fig. S4C). Overall, these analyses show that neurons in the song motor pathway display stronger transcriptional similarity to neocortical neurons at the level of neuronal projection class—intratelencephalic for HVC and subcerebral for RA—than at the level of individual layers or specific cortical regions.
An alternative “pallial field homology” model (Fig. 3A, “field”) holds that the DVR is an expansion of the ventral pallium—a distinct developmental region of the pallium, which in mammals generates olfactory areas, the endopiriform nucleus, and the pallial amygdala (6, 12, 46). This sector is topologically distinct from the dorsal pallium, which gives rise to the neocortex in mammals. To assess this model, we broadened our analysis to include mammalian neurons from non-neocortical regions by comparing HVC and RA glutamatergic neuron gene expression to in situ hybridization data from anatomically defined regions of the adult mouse brain [Allen Adult Mouse Brain Atlas (47)] (Fig. 3, C to E, and fig. S5A) and two mouse single-cell RNA-seq datasets that contain non-neocortical pallial regions such as the piriform cortex, claustrum, and pallial amygdala in addition to the neocortex (44, 45) (figs. S4, E and F, and S5B). For both HVC and RA, these comparisons revealed different mappings, depending on which genes were used for analysis. We divided a list of orthologous genes between each species into transcription factors (TFs), whose expression may reflect regulatory networks that underlie conserved regional or cellular identity programs, and non-transcription factors (non-TFs), whose regulation may be more pliable over evolution and may more directly relate to neuronal function (48). Comparisons using non-TF expression profiles showed a bias toward dorsal pallial regions, including greatest similarities to different upper and deep-layer pyramidal neurons (Fig. 3, C and D, “non-transcription factors,” and fig. S5B), in agreement with the neocortex-only analysis described above. In contrast, comparisons using only TF expression showed a bias away from the dorsal pallium and toward the ventral pallium, including olfactory areas and the pallial amygdala (Fig. 3, C and D, “transcription factors”; fig. S5B; and table S4). In contrast to these findings indicating increased similarity of song system neurons to dorsal pallial neurons at the level of non-TF expression, similar comparisons between mouse forebrain glutamatergic neurons (44) and mouse pallial regions showed no dissociation between TF and non-TF expression patterns (Fig. 3D). We also found no association between song motor pathway neurons and the claustrum, which argues against early models that describe the nidopallium as homologous to this mammalian region in mammals (6) (fig. S4F). However, we did find similarities between several song system projection neurons and glutamatergic neurons in other non-neocortical regions, including the hippocampus (HVC_Glut-3 and RA_Glut-1/2/3) and cingulate deep-layer neurons (RA_Glut-1/2/3) (figs. S4, E and F, and S5, A and B). These songbird neurons show the strongest similarity to deep-layer neocortical neurons, which could indicate that deep-layer transcriptional signatures exist in neurons across multiple pallial regions.
To better understand how avian DVR neurons relate to what is known about the sauropsid pallium, we compared avian DVR data to previously published single-cell transcriptomic data from the DVR of the red-eared slider turtle (Trachemys scripta elegans), a non-avian reptile whose pallium has been extensively compared with the mammalian pallium (49). In general, we found that glutamatergic neurons of the song motor pathway were more similar to neurons of the reptilian DVR (Fig. 3F and fig. S5C) than to the anterior dorsal cortex, the pallial region in reptiles thought to be homologous to the neocortex (49).
The finding that glutamatergic neurons in the avian DVR show TF similarities to non-neocortical regions but effector gene expression similarities to neocortical neurons suggests that ventral pallial regulatory networks engage neocortical cell type-associated transcriptional programs (50). To examine this possibility in more detail, we used gene regulatory network inference to identify putative TF-target gene relationships in HVC and RA (fig. S6 and table S5). Correlations between TF-target gene associations revealed distinct modules of co-regulation that mapped onto the different song motor pathway projection neuron classes. Consistent with conclusions based on Allen Brain Atlas in situ expression patterns (Fig. 3, C to E, and fig. S5A), these modules were enriched for TFs with expression patterns biased toward the ventral pallium and away from the neocortex. However, several TFs associated with neocortical layer identity were embedded in each module, with upper-layer markers such as CUX1/2 and SATB1 (51) enriched in HVC_Glut-1/2/4/5 and deep-layer markers enriched in RA_Glut-1/2/3 and HVC_Glut-3. The thalamorecipient layer 4 TFs RORA and RORB were enriched in HVC_Glut-3/5, which could relate to inputs from the thalamic nucleus uvaeformis (Fig. 1B). Combined, these data show that glutamatergic neurons in the avian DVR engage regional-patterning TF networks characteristic of the ventral pallium as well as elements of dorsal pallial gene regulatory networks that likely contribute to neocortical-like effector gene expression.
Conserved classes of GABAergic neurons across amniotes
To validate the presence of the eight main GABAergic subtypes in the song motor pathway, we performed nine-channel in situ hybridizations for a pan-GABAergic marker (GAD1) and marker genes for each subtype that were selected from a list of genes with high cluster specificity (fig. S7A). In situ signal for each marker gene was present in HVC and RA and, in combination, labeled distinct GAD1-positive neurons (Fig. 4, A and B, and fig. S7, A to G). Unlike glutamatergic neuron classes, which differed transcriptionally between HVC and RA, GABAergic neuron classes were similar between regions (Fig. 1D and fig. S1, C, I, and J). The small number of genes that did show differential expression between regions (35 to 102 genes per cluster) were enriched for genes associated with synaptic transmission and neuron migration (fig. S8, A and B, and table S6). Hierarchical clustering of all subtypes together produced three main groupings, which, when mapped to the expression of mammalian GABAergic marker genes, aligned with the classification scheme used to organize mammalian GABAergic lineages by region of developmental origin: medial ganglionic eminence (MGE; GABA-2/3/4/6/7), caudal ganglionic eminence (CGE; GABA-5/8), and lateral ganglionic eminence (LGE; GABA-1/Pre) (52, 53) (Fig. 4C and fig. S7A). To more fully compare mammalian and songbird interneuron subtypes, we correlated the average expression levels of GABAergic clusters in songbirds with those in the mouse neocortex (43) (Fig. 4D and fig. S9A). We found strong similarities between bird and mouse GABAergic subtypes for both MGE class interneurons [songbird GABA-2/3/4/6 versus mouse somatostatin (Sst), parv-albumin (Pvalb), and lysosome-associated membrane family protein member 5 (Lamp5) class neurons] and CGE class interneurons [GABA-5 versus vasoactive intestinal peptide (Vip) and synuclein-γ (Sncg) class neurons], suggesting that the major interneuron classes are conserved and group by developmental origin across species. Despite these core similarities, two canonical interneuron marker genes were differentially expressed in songbird versus mouse neurons (fig. S8, C to F): SST was expressed not only in GABA-2 (which is comparable to mammalian Sst class) but also in GABA-3 and GABA-4 (which are otherwise comparable to Pvalb class), while VIP was not expressed in GABA-5 (otherwise comparable to mammalian Vip class) but instead in GABA-3 (otherwise comparable to mammalian Pvalb class).
Fig. 4. Diversity and organization of song motor pathway GABAergic neurons.
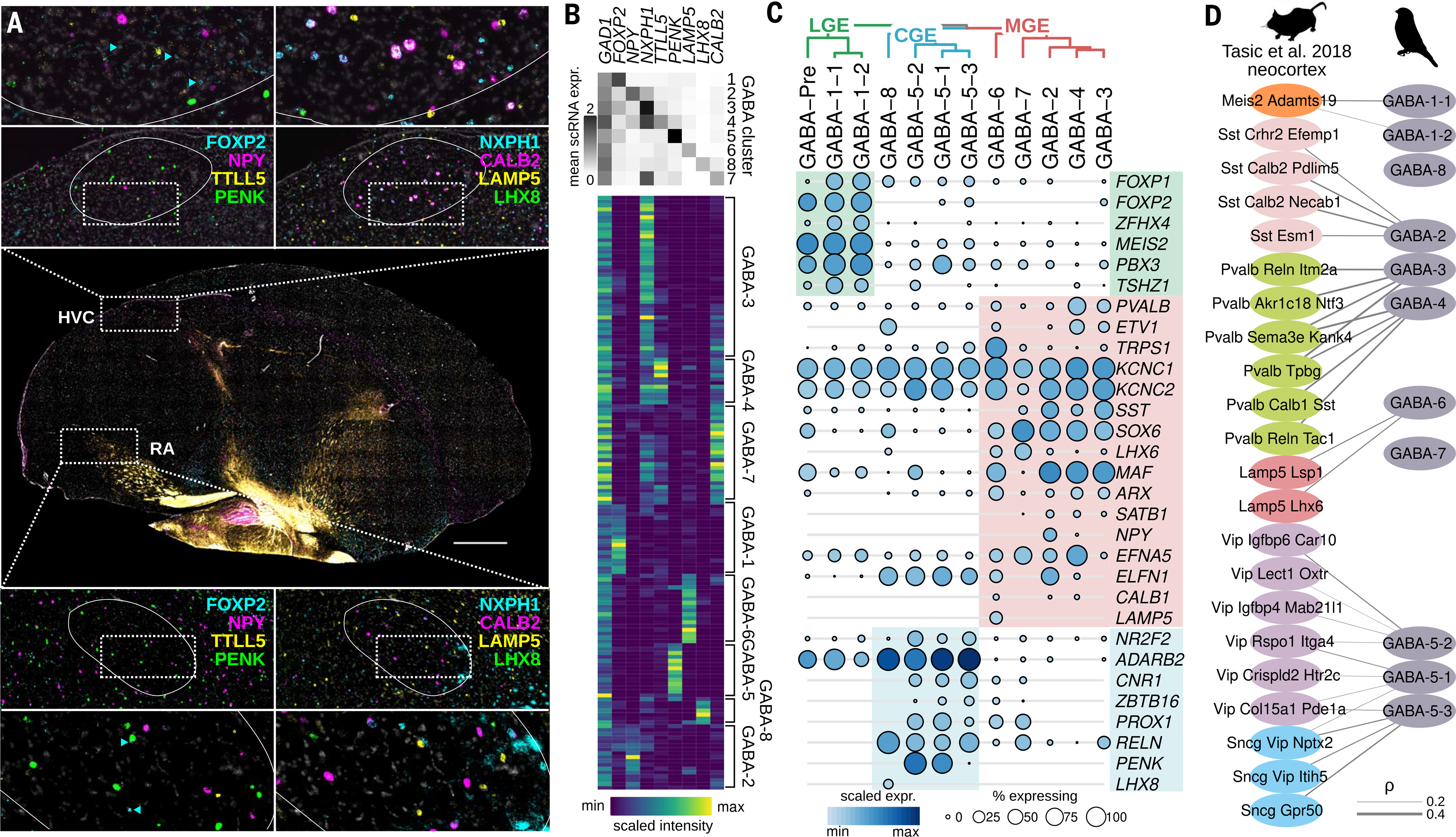
(A) In situ hybridization analysis of GAD1 and eight GABAergic subtype marker genes, FOXP2 (GABA-1), NPY (GABA-2), NXPH1 (GABA-3/4), TTLL5 (GABA-4), PENK (GABA-5), LAMP5 (GABA-6), CALB2 (GABA-7), and LHX8 (GABA-8). Shown is a sagittal Bengalese finch brain section with enlargements of HVC and RA. Each song motor pathway region is outlined in the enlarged images. Scale bar, 1 mm. Arrowheads in enlargements indicate examples of FOXP2- positive cells. (B) (Top) Average expression of GABAergic marker genes in single-cell clusters. (Bottom) Quantification of marker gene expression in GAD1-positive neurons from in situ hybridization in HVC and RA (n = 154), scaled by the maximum and minimum intensity values for each gene. Heatmap rows are organized by hierarchical clustering. Labels to the right of the heatmap indicate assignment of single-cell sequencing clusters to clusters derived from in situ hybridization data. (C) Hierarchical clustering and dotplot of songbird GABAergic neuron expression (sequencing data) shows organization by subpallial embryonic domain [markers from (49)]. Shade of dot represents mean expression within cluster, and size of dot represents percentage of cells within cluster expressing a given gene. LGE (lateral ganglionic eminence), green; MGE (medial ganglionic eminence), red; CGE (caudal ganglionic eminence), blue. (D) Correlation analysis between expression profiles of mouse neocortical (43) and song motor pathway GABAergic neurons. Lines represent Spearman correlations, with thickness scaled by correlation strength. Shown are the top four correlations for each comparison that are significant (shuffled P < 0.05) and are greater than 0.2. Labels correspond to the classification scheme used in the original publication.
Furthermore, several song motor pathway GABAergic classes—GABA-1, GABA-7, GABA-8, and GABA-Pre—did not directly map onto mouse neocortical interneuron classes. To determine whether these neurons show similarity to GABAergic neurons outside of the neocortex, we compared songbird neurons with two single-cell datasets that sampled a greater diversity of brain regions, one from the mouse forebrain (44) and one from the turtle pallium (49) (Fig. 5). As with the neocortical comparisons, songbird MGE and CGE clusters matched well with corresponding mouse and turtle GABAergic neuron types (Fig. 5, C and D, and fig. S9, B to D). However, GABA-1 showed the greatest similarity to non-neocortical LGE-class neurons, including striatal medium spiny neurons (MSNs), olfactory bulb granule cells (OBINH2), and turtle interneurons similar to amygdalar intercalated cells (i05 and i06) (49). Similarly, GABA-Pre showed strong similarity to olfactory bulb neuroblasts, which are precursors to adult-born granule cells (54). Likewise, GABA-7 and GABA-8 showed greatest similarity to two other non-neocortical interneuron types, olfactory bulb glomerular layer neurons and pallidal cholinergic neurons, respectively.
Fig. 5. Conserved GABAergic neuron identity across amniotes.

(A and B) Seurat anchor-based integration of mouse forebrain (44), turtle forebrain (49), and songbird motor pathway GABAergic neurons. UMAP plots of integrated datasets colored by (A) species and (B) cluster. For clarity, labels are shown only for mouse and finch datasets. OB, olfactory bulb; NGC, neurogliaform cells; MSNs, medium spiny neurons. Labels correspond to the classification schemes used in the original publications. (C and D) Predicted cross-species cluster similarities of songbird GABAergic neurons based on pairwise integration with (C) mouse and (D) turtle GABAergic neuron datasets. SST, somatostatin; Cck, cholecystokinin; PV, parvalbumin; HTR3A, serotonin receptor 3a; VIP, vasoactive intestinal peptide.
The strong molecular similarity of interneurons reported here to those in other species presents an opportunity to update models of song system function. For instance, a fast-spiking cortical interneuron that provides local inhibition between projection neurons has been characterized previously in songbirds (37, 39, 55) and likely corresponds to GABA-3 and/or GABA-4 owing to their transcriptional similarity to mammalian Pvalb-class interneurons (Fig. 4D and fig. S9A). More broadly, the known function of mammalian cell types suggests possible functions of each of their avian counterparts. In mammals, Sst-class interneurons (here corresponding to GABA-2) are implicated in circuit plasticity during reward-based tasks and motor learning (56). VIP-like neurons (here GABA-5) play a prominent disinhibitory role in regulating neocortical activity (57), raising the possibility that there is an analogous disinhibitory motif present in avian microcircuits. Cholinergic interneurons (here GABA-8) provide a local source of acetylcholine, a neuromodulator known to influence song properties (58). LGE-class interneurons (here GABA-1) establish sparsely active ensembles of odorant-driven neural activity in the olfactory bulb through feedforward and lateral inhibition (59, 60) and could potentially contribute similarly to sparsely patterned activity that is a hallmark of HVC projection neurons (61, 62). Additionally, LGE-class intercalated cells in the mammalian pallial amygdala form a disinhibitory circuit that is strongly modulated by dopamine (63), suggesting that this class could play a role in dopamine-dependent aspects of song learning (64).
Broad pallial distribution of LGE-class neurons in songbirds
Our comparative analysis indicates that the GABA-1 population is similar to LGE-class GABAergic neurons (Figs. 4C and 5, A to D, and fig. S9, B to D), which in mammals are mostly restricted to the subpallium, namely as MSNs of the striatum (65), and to ventral pallial structures such as the olfactory bulb (66) and pallial amygdala (67). To assess whether GABA-1 neurons are similar to other LGE-class neurons in birds, we combined the HVC and RA single-cell and single-nucleus data with a single-nucleus RNA-seq dataset from the striatal song system region Area X (68) (Fig. 6A and fig. S10, A and B). GABA-1 neurons clustered strongly with Area X LGE-class neurons (Fig. 6B and fig. S10, C and D), including MSNs and a neuron class that may be similar to arkypallidal neurons in mammals (69) (Fig. 6, B and C; fig. S10, C to E; and table S7). Consistent with LGE classification, GABA-1 neurons express several TFs associated with MSN identity, including FOXP1, FOXP2, and MEIS2 (70) (Fig. 6C and fig. S10E). In the mammalian neocortex, FOXP2 is expressed in deep-layer glutamatergic projection neurons (71, 72), but in the song motor pathway it is strongly enriched in GABA-1 neurons, such that the large majority of FOXP2-positive cells are GABAergic (98 to 100%) (fig. S11, A to D). Morphology analysis showed that GABA-1 neurons had similar spine densities and soma sizes to striatal LGE-class neurons (fig. S11, E and F).
Fig. 6. Broad distribution of LGE-derived interneurons in the songbird brain.
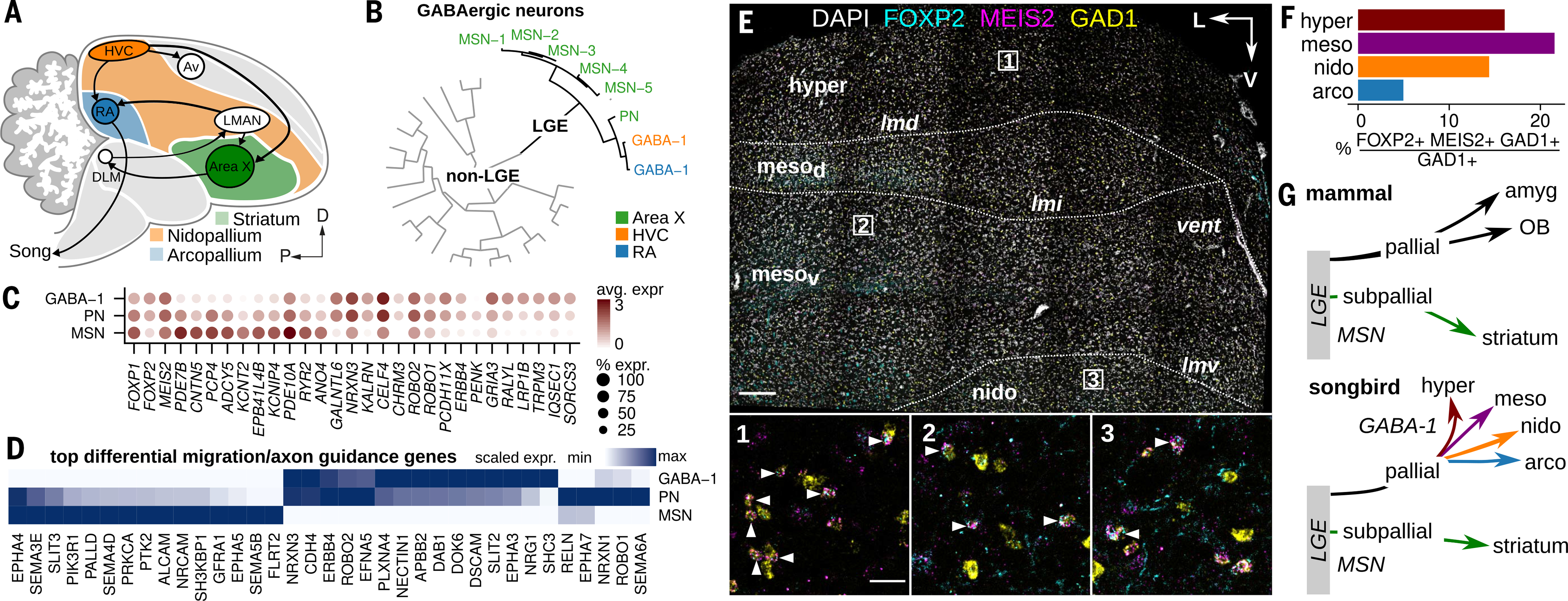
(A) Schematic illustrating the location of Area X within the avian striatum (green) in relation to the nidopallium (orange) and arcopallium (blue). (B) Hierarchical clustering of HVC, RA, and Area X GABAergic neuron expression profiles, divided by LGE-class and non-LGE-class neurons. PN, pallidal-like neuron. (C) Expression of top differentially expressed genes in LGE-derived GABAergic classes and three genes associated with striatal GABAergic neurons (FOXP1, FOXP2, and MEIS2). Shade of dot represents mean expression within cluster, and size of dot represents percentage of cells within the cluster expressing that gene. (D) Scaled expression of top differentially expressed axon guidance and neuron migration related genes across avian LGE-class neurons. (E) Three-color in situ hybridization of LGE-marker genes FOXP2 and MEIS2 and GABAergic marker gene GAD1. Shown is a coronal section from the anterior Bengalese finch brain. Major anatomical divisions: hyper, hyperpallium; mesod, dorsal mesopallium; mesov, ventral mesopallium; nido, nidopallium; Lamina: lmd, dorsal mesopallial lamina; lmi, intermediate mesopallial lamina; lmv, ventral mesopallial lamina. Nomenclature is as defined in (13). Scale bar, 200 μm. (Insets) Magnifications of three regions indicated in the main panel. Arrowheads indicate triple-positive cells. Scale bar, 50 μm. (F) Quantification of FOXP2/MEIS2/GAD1 in situ hybridization data, shown as percentage GAD1-positive cells in four pallial domains that coexpress FOXP2 and MEIS2. (G) Comparison of LGE-class GABAergic migration in mammals and songbirds. In mammals, the LGE contributes neurons in the pallial amygdala (intercalated cells), the olfactory bulb (granule and periglomerular interneurons), and subpallial structures such as the striatum (MSNs). Amyg, pallial amygdala.
The differential localization of songbird pallial GABA-1 neurons and subpallial MSNs suggests that each class engages distinct neuronal migration mechanisms. Indeed, ~50 axon guidance and neuronal migration genes are differentially expressed across songbird LGE-class neurons, including several with known roles in neocortical and olfactory bulb interneuron migration in mammals (e.g., ERBB4 and the ephrin, ROBO/SLIT, and semaphorin signaling systems) (73) (Fig. 6D). Finally, we analyzed the distribution of LGE-class neurons (FOXP2, MEIS2, and GAD1-positive) in several regions of the songbird pallium–including the arcopallium outside of RA, nidopallium ventral to HVC, mesopallium, and hyperpallium—and found that LGE-class neurons comprise a substantial fraction of the total number of GABAergic neurons in each region (Fig. 6, E and F).
Discussion
The evolutionary origin of the DVR, and the song motor pathway that is embedded within it, has long been unclear because of their structural differences with the mammalian neocortex. Our analysis supports the notion that the avian DVR is regionally homologous not to the neocortex but to non-neocortical structures of the ventral pallium (12). Yet we find that individual HVC and RA glutamatergic neurons show transcriptional similarities to neocortical projection neurons in multiple layers. In effect, the neural circuits across amniotes that enable complex behaviors engage overlapping yet distinct molecular and cellular elements, suggesting that diverse pallial architectures have evolved to support advanced behavioral repertoires.
Field-based models of pallial evolution describe a common set of gene regulatory mechanisms that specify a shared topological framework across amniotes (6, 12). Yet we demonstrate that field positioning across species is not the exclusive determinant of cellular identity. Different regions may generate similar neurons through evolutionary processes that integrate regional patterning programs with those specifying functional identity (48, 74). In both birds and mammals, pallial and neocortical circuits contain distinct classes of neurons characterized by thalamic input, intratelencephalic connectivity, and subcerebral output (50). We show that glutamatergic neurons in the song motor pathway exhibit greatest similarity to glutamatergic neurons in the mammalian neocortex at the level of projection class and by the division between superficial and deep-layer signatures, as has similarly been reported in turtles (49, 75). Alternatively, homologous regions, such as the anterior dorsal cortex of reptiles and the neocortex of mammals, may contain glutamatergic neurons that show divergent gene expression profiles between species (49). Thus, observed similarities in glutamatergic neurons could have arisen either through the retention of features from cell types present in the amniote pallium ancestral to birds and mammals (50, 76) or through convergent evolution (49, 75). An important step in clarifying these models will come through characterizing the extent to which similar functional cell types may have been distributed within ancestral regionalized pallia (77). The associations we report here between neurons in the avian DVR and neurons in multiple fields of the mammalian pallium suggest that neurons characterized by certain properties are not necessarily exclusive of a single pallial domain. A future challenge will be to identify the gene regulatory events at the interface between regional positioning and cellular identity that lead to the innovation of novel neural circuits and the behaviors that they support.
Beyond neuronal classes, evolution clearly has allowed for flexibility in pallial organization (76), in contrast to the subpallium, for which features have remained largely conserved in vertebrates for more than 500 million years (78). The correspondence of the sauropsid DVR with a homologous domain in mammals has eluded easy identification partly because its size and structure differ so markedly from anything present in the mammalian brain (3). However, it has recently been reported that sensory portions of the avian DVR contain an iterative layer-like arrangement of fibers that is reminiscent of cortical layer and column architectures (79). In contrast, the posterior DVR, in which HVC and RA are located, lacks such a clear structuring. These differences in anatomical structure across amniotes and within the avian brain, along with the presence of similar connectivity classes in the song motor pathway and the neocortex, suggest that neuronal connectivity rules can operate in diverse physical arrangements. The neocortex has a uniform, multilayered organization, whereas the DVR contains multiple distinct interconnected clusters of neurons. The DVR therefore represents an alternative system in which neural circuits have evolved to orchestrate behavior and integrate sensory information (80, 81).
We also report that such organizational flexibility extends to the migration and localization of GABAergic neurons in the pallium. The similarity of GABA-1 neurons to LGE-class neurons and their broad distribution in the songbird pallium suggest that the migration of LGE-class GABAergic neurons is more extensive in birds than in mammals (Fig. 6G). In both chickens and turtles, LGE-class neurons tangentially migrate into the DVR and dorsal pallium (82–85). However, transplantation of embryonic turtle LGE into mouse embryonic slices results in the migration of GABAergic cells into the piriform cortex and amygdala but not into the neocortex (84). Together, these results suggest that the avian DVR and mammalian ventral pallium express signaling factors that regulate the migration and retention of LGE-class neurons. Relatedly, avian MGE neurons implanted into the MGE of developing mouse embryos do not migrate into the cortical plate (which forms the neocortex) but instead remain in subcortical areas such as the piriform cortex and pallial amygdala (86). However, these neurons can integrate into the neocortex if implanted into the cortical plate directly, suggesting that mammalian-specific signals regulate their spatial distribution. Interneurons strongly influence information processing in neuronal microcircuits, and such differential control of GABAergic composition across phylogeny may confer species-specific computational properties.
Since its discovery in 1976 (87), the song motor pathway in songbirds has proven to be a powerful model for the neural control of complex, learned behaviors in vertebrates. This pathway often has been likened to components of the mammalian neocortex. Our cellular-level analysis provides further clarification of this relationship by showing that HVC and RA are not homologous as a field to layers or regions of neocortex, but rather that their identities are most closely aligned with the non-neocortical ventral pallium. Nevertheless, we find similarities at the level of individual cell types with neurons from each of the mammalian neocortical layers, and the song motor pathway also exhibits connectivity patterns that mirror features of neocortex, including thalamic inputs, subcortical and striatal projections, and interconnections with other telencephalic regions. Together, these cortical features of the song motor pathway suggest that this circuit, despite its regional similarity to the ventral pallium, may have evolved to perform computations in a manner that is functionally analogous to the neocortex. Integrating this cellular-resolution analysis of birdsong control circuitry with similar studies in other species will provide a framework for understanding the molecular and cellular innovations that underlie the development and evolution of behavioral diversity.
Materials and methods summary
Animal use
All Bengalese and zebra finches were from our breeding colonies at the University of California-San Francisco (UCSF) and the University of Texas Southwestern Medical Center (UTSW) or were purchased from approved vendors. Experiments were conducted in accordance with National Institutes of Health (NIH), UCSF, and UTSW policies governing animal use and welfare.
Single-cell and single-nucleus RNA sequencing library preparation and sequencing
HVC and RA were dissected from either adult male Bengalese finches or adult male zebra finches and prepared for single-cell or single-nucleus RNA sequencing, respectively, using library kits from 10X Genomics. Libraries were sequenced and then aligned to either the Bengalese or zebra finch genomes and associated gene annotations.
Data integration and clustering
Datasets from the two species were integrated using Seurat anchor-based mutual nearest neighbor integration and clustered using the Louvain algorithm with multilevel refinement. A low-resolution parameter (0.1 to 0.4) was first chosen to conservatively identify major clusters without oversplitting. The data was then subsetted for each major cell cluster, e.g., glutamatergic neurons, astrocytes, endothelial cells, etc., and the clustering procedure repeated to identify within-class subclusters, again selecting resolution parameters that favored well-separated clusters without oversplitting. Identities were assigned to each cluster on the basis of the expression of established marker genes.
Comparison with non-songbird datasets
Four single-cell datasets were downloaded for interspecies comparison: mouse neocortex (43), two datasets containing broader samplings of the mouse pallium (44, 45), and turtle forebrain (49). We used two approaches to compare the expression profiles of neurons between species: (i) correlations between mean expression profiles within clusters and (ii) integration of single-cell datasets (see supplementary methods for details).
Songbird single-cell cluster and mouse in situ hybridization comparison
In situ hybridization data for the Allen Adult Mouse Brain Atlas (47) was downloaded and filtered to retain only genes that were highly variable in the songbird dataset. Correlations were then performed as described above (see supplementary methods for details).
Fluorescent in situ hybridization (FISH)
FISH was performed using the RNAscope HiPlex system from ACDBio or the hairpin chain reaction system from Molecular Instruments (see supplementary methods for details).
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Veline and K. Li for technical assistance, and A. Alvarez-Buylla, B. Cooper, J. Johnson, M. Oldham, A. Pollen, J. Rubenstein, and M. Stryker for critical reading of the manuscript.
Funding:
Research reported in this publication was supported by the NINDS NIH under awards F32NS112557 to D.P.M.; F32NS098809 to B.M.C.; R01NS102488, R01NS108424, and R01DC014364 to T.F.R.; and UF1NS115821 and R21DC016340 to T.F.R. and G.K. Support was also provided by the Chan Zuckerberg Initiative, an advised fund of Silicon Valley Community Foundation (HCA-A-1704–01747) to G.K., NSF IOS-1457206 to T.F.R., the James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition (Scholar Award 220020467) to G.K., and HHMI Investigator award to M.S.B. G.K. is a Jon Heighten Scholar in Autism Research at UTSW.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Single-cell RNA-seq data are available in the Gene Expression Omnibus for Bengalese finches (GSE150486) and zebra finches (GSE153665). Processed data used for analyses are available at https://cloud.biohpc.swmed.edu/index.php/s/nLicEtkmjGGmRF8. Code used in key analyses and figure production is available at https://github.com/bradleycolquitt/songbird_cells (89).
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/371/6530/eabd9704/suppl/DC1
MDAR Reproducibility Checklist
View/request a protocol for this paper from Bio-protocol.
REFERENCES AND NOTES
- 1.Konopka G, Roberts TF, Insights into the neural and genetic basis of vocal communication. Cell 164, 1269–1276 (2016). doi: 10.1016/j.cell.2016.02.039; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brainard MS, Doupe AJ, Translating birdsong: Songbirds as a model for basic and applied medical research. Annu. Rev. Neurosci 36, 489–517 (2013). doi: 10.1146/annurev-neuro-060909-152826; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Striedter GF, Northcutt RG, Brains Through Time: A Natural History of Vertebrates (Oxford Univ. Press, 2020). [Google Scholar]
- 4.Gunturkun O, Bugnyar T, Cognition without cortex. Trends Cogn. Sci 20, 291–303 (2016). doi: 10.1016/j.tics.2016.02.001; [DOI] [PubMed] [Google Scholar]
- 5.Jarvis ED, Evolution of vocal learning and spoken language. Science 366, 50–54 (2019). doi: 10.1126/science.aax0287; [DOI] [PubMed] [Google Scholar]
- 6.Striedter GF, The telencephalon of tetrapods in evolution. Brain Behav. Evol 49, 179–213 (1997). doi: 10.1159/000112991; [DOI] [PubMed] [Google Scholar]
- 7.Reiner A, A new avian brain nomenclature: Why, how and what. Brain Res. Bull 66, 317–331 (2005). doi: 10.1016/j.brainresbull.2005.05.007; [DOI] [PubMed] [Google Scholar]
- 8.Karten HJ, The organization of the avian telencephalon and some speculations on the phylogeny of the amniote telencephalon. Ann. N. Y. Acad. Sci 167, 164–179 (1969). doi: 10.1111/j.1749-6632.1969.tb20442.x [DOI] [Google Scholar]
- 9.Karten HJ, Vertebrate brains and evolutionary connectomics: On the origins of the mammalian ‘neocortex’. Philos. Trans. R. Soc. London Ser. B 370, 20150060 (2015). doi: 10.1098/rstb.2015.0060; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis ED et al. , Avian brains and a new understanding of vertebrate brain evolution. Nat. Rev. Neurosci 6, 151–159 (2005). doi: 10.1038/nrn1606; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfenning AR et al. , Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346, 1256846 (2014). doi: 10.1126/science.1256846; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puelles L et al. , “The pallium in reptiles and birds in the light of the updated tetrapartite pallium model” in The Evolution of the Nervous Systems in Nonmammalian Vertebrates, Striedter G, Ed., vol. 1 of Evolution of Nervous Systems,Kaa JH, Ed. (Academic Press, ed. 2, 2017), pp. 519–555. [Google Scholar]
- 13.Jarvis ED et al. , Global view of the functional molecular organization of the avian cerebrum: Mirror images and functional columns. J. Comp. Neurol 521, 3614–3665 (2013). doi: 10.1002/cne.23404; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki IK, Kawasaki T, Gojobori T, Hirata T, The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Dev. Cell 22, 863–870 (2012). doi: 10.1016/j.devcel.2012.01.004; [DOI] [PubMed] [Google Scholar]
- 15.Dugas-Ford J, Rowell JJ, Ragsdale CW, Cell-type homologies and the origins of the neocortex. Proc. Natl. Acad. Sci. U.S.A 109, 16974–16979 (2012). doi: 10.1073/pnas.1204773109; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belgard TG et al. , Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc. Natl. Acad. Sci. U.S.A 110, 13150–13155 (2013). doi: 10.1073/pnas.1307444110; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez-Buylla A, Theelen M, Nottebohm F, Birth of projection neurons in the higher vocal center of the canary forebrain before, during, and after song learning. Proc. Natl. Acad. Sci. U.S.A 85, 8722–8726 (1988). doi: 10.1073/pnas.85.22.8722; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nordeen KW, Nordeen EJ, Projection neurons within a vocal motor pathway are born during song learning in zebra finches. Nature 334, 149–151 (1988). doi: 10.1038/334149a0; [DOI] [PubMed] [Google Scholar]
- 19.Scharff C, Kirn JR, Grossman M, Macklis JD, Nottebohm F, Targeted neuronal death affects neuronal replacement and vocal behavior in adult songbirds. Neuron 25, 481–492 (2000). doi: 10.1016/S0896-6273(00)80910-1; [DOI] [PubMed] [Google Scholar]
- 20.Alvarez-Buylla A, Kirn JR, Nottebohm F, Birth of projection neurons in adult avian brain may be related to perceptual or motor learning. Science 249, 1444–1446 (1990). doi: 10.1126/science.1698312; [DOI] [PubMed] [Google Scholar]
- 21.Obernier K, Alvarez-Buylla A, Neural stem cells: Origin, heterogeneity and regulation in the adult mammalian brain. Development 146, dev156059 (2019). doi: 10.1242/dev.156059; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK, The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol 18, 28–35 (2008). doi: 10.1016/j.conb.2008.05.006; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XX et al. , Nectin-3 modulates the structural plasticity of dentate granule cells and long-term memory. Transl. Psychiatry 7, e1228 (2017). doi: 10.1038/tp.2017.196; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott BB, Lois C, Developmental origin and identity of song system neurons born during vocal learning in songbirds. J. Comp. Neurol 502, 202–214 (2007). doi: 10.1002/cne.21296; [DOI] [PubMed] [Google Scholar]
- 25.Roberts TF et al. , Identification of a motor-to-auditory pathway important for vocal learning. Nat. Neurosci 20, 978–986 (2017). doi: 10.1038/nn.4563; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell ZW et al. , Urotensin-related gene transcripts mark developmental emergence of the male forebrain vocal control system in songbirds. Sci. Rep 9, 816 (2019). doi: 10.1038/s41598-018-37057-w; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egger R et al. , Local axonal conduction shapes the spatiotemporal properties of neural sequences. Cell 183, 537–548.e12 (2020). doi: 10.1016/j.cell.2020.09.019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shea SD, Koch H, Baleckaitis D, Ramirez J-M, Margoliash D, Neuron-specific cholinergic modulation of a forebrain song control nucleus. J. Neurophysiol 103, 733–745 (2010). doi: 10.1152/jn.00803.2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitney O et al. , Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science 346, 1256780 (2014). doi: 10.1126/science.1256780; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyssowski KM et al. , Different neuronal activity patterns induce different gene expression programs. Neuron 98, 530–546.e11 (2018). doi: 10.1016/j.neuron.2018.04.001; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez-Valpuesta M et al. , Corticobasal ganglia projecting neurons are required for juvenile vocal learning but not for adult vocal plasticity in songbirds. Proc. Natl. Acad. Sci. U.S.A 116, 22833–22843 (2019). doi: 10.1073/pnas.1913575116; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vicario DS, Organization of the zebra finch song control system: II. Functional organization of outputs from nucleus robustus archistriatalis. J. Comp. Neurol 309, 486–494 (1991). doi: 10.1002/cne.903090405; [DOI] [PubMed] [Google Scholar]
- 33.Wild JM, Descending projections of the songbird nucleus robustus archistriatalis. J. Comp. Neurol 338, 225–241 (1993). doi: 10.1002/cne.903380207; [DOI] [PubMed] [Google Scholar]
- 34.Goldberg JH, Fee MS, A cortical motor nucleus drives the basal ganglia-recipient thalamus in singing birds. Nat. Neurosci 15, 620–627 (2012). doi: 10.1038/nn.3047; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts TF, Klein ME, Kubke MF, Wild JM, Mooney R, Telencephalic neurons monosynaptically link brainstem and forebrain premotor networks necessary for song. J. Neurosci 28, 3479–3489 (2008). doi: 10.1523/JNEUROSCI.0177-08.2008; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonardo A, Fee MS, Ensemble coding of vocal control in birdsong. J. Neurosci 25, 652–661 (2005). doi: 10.1523/JNEUROSCI.3036-04.2005; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller MN, Cheung CYJ, Brainard MS, Vocal learning promotes patterned inhibitory connectivity. Nat. Commun 8, 2105 (2017). doi: 10.1038/s41467-017-01914-5; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sober SJ, Wohlgemuth MJ, Brainard MS, Central contributions to acoustic variation in birdsong. J. Neurosci 28, 10370–10379 (2008). doi: 10.1523/JNEUROSCI.2448-08.2008; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiro JE, Dalva MB, Mooney R, Long-range inhibition within the zebra finch song nucleus RA can coordinate the firing of multiple projection neurons. J. Neurophysiol 81, 3007–3020 (1999). doi: 10.1152/jn.1999.81.6.3007; [DOI] [PubMed] [Google Scholar]
- 40.DeVoogd TJ, Nottebohm F, Sex differences in dendritic morphology of a song control nucleus in the canary: A quantitative Golgi study. J. Comp. Neurol 196, 309–316 (1981). doi: 10.1002/cne.901960209; [DOI] [PubMed] [Google Scholar]
- 41.Vicario DS, A new brain stem pathway for vocal control in the zebra finch song system. Neuroreport 4, 983–986 (1993). doi: 10.1097/00001756-199307000-00037; [DOI] [PubMed] [Google Scholar]
- 42.Karten HJ, Homology and evolutionary origins of the ‘neocortex’. Brain Behav. Evol 38, 264–272 (1991). doi: 10.1159/000114393; [DOI] [PubMed] [Google Scholar]
- 43.Tasic B et al. , Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). doi: 10.1038/s41586-018-0654-5; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeisel A et al. , Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). doi: 10.1016/j.cell.2018.06.021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saunders A et al. , Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018). doi: 10.1016/j.cell.2018.07.028; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puelles L et al. , Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J. Comp. Neurol 424, 409–438 (2000). doi: ; [DOI] [PubMed] [Google Scholar]
- 47.Lein ES et al. , Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). doi: 10.1038/nature05453; [DOI] [PubMed] [Google Scholar]
- 48.Wagner GP, Homology, Genes, and Evolutionary Innovation (Princeton Univ. Press, 2014). [Google Scholar]
- 49.Tosches MA et al. , Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888 (2018). doi: 10.1126/science.aar4237; [DOI] [PubMed] [Google Scholar]
- 50.Briscoe SD, Ragsdale CW, Homology, neocortex, and the evolution of developmental mechanisms. Science 362, 190–193 (2018). doi: 10.1126/science.aau3711; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paolino A, Fenlon LR, Suarez R, Richards LJ, Transcriptional control of long-range cortical projections. Curr. Opin. Neurobiol 53, 57–65 (2018). doi: 10.1016/j.conb.2018.05.005; [DOI] [PubMed] [Google Scholar]
- 52.Hu JS, Vogt D, Sandberg M, Rubenstein JL, Cortical interneuron development: A tale of time and space. Development 144, 3867–3878 (2017). doi: 10.1242/dev.132852; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim L, Mi D, Llorca A, Marín O, Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018). doi: 10.1016/j.neuron.2018.10.009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim DA, Alvarez-Buylla A, The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harbor Perspect. Biol 8, a018820 (2016). doi: 10.1101/cshperspect.a018820; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mooney R, Prather JF, The HVC microcircuit: The synaptic basis for interactions between song motor and vocal plasticity pathways. J. Neurosci. 25, 1952–1964 (2005). doi: 10.1523/JNEUROSCI.3726-04.2005; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urban-Ciecko J, Barth AL, Somatostatin-expressing neurons in cortical networks. Nat. Rev. Neurosci 17, 401–409 (2016). doi: 10.1038/nrn.2016.53; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu Y, Kaneko M, Tang Y, Alvarez-Buylla A, Stryker MP, A cortical disinhibitory circuit for enhancing adult plasticity. eLife 4, e05558 (2015). doi: 10.7554/eLife.05558; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaffe PI, Brainard MS, Acetylcholine acts on songbird premotor circuitry to invigorate vocal output. eLife 9, e53288 (2020). doi: 10.7554/eLife.53288; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burton SD, Inhibitory circuits of the mammalian main olfactory bulb. J. Neurophysiol 118, 2034–2051 (2017). doi: 10.1152/jn.00109.2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gschwend O et al. , Neuronal pattern separation in the olfactory bulb improves odor discrimination learning. Nat. Neurosci 18, 1474–1482 (2015). doi: 10.1038/nn.4089; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hahnloser RHR, Kozhevnikov AA, Fee MS, An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature 419, 65–70 (2002). doi: 10.1038/nature00974; [DOI] [PubMed] [Google Scholar]
- 62.Kosche G, Vallentin D, Long MA, Interplay of inhibition and excitation shapes a premotor neural sequence. J. Neurosci 35, 1217–1227 (2015). doi: 10.1523/JNEUROSCI.4346-14.2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marowsky A, Yanagawa Y, Obata K, Vogt KE, A specialized subclass of interneurons mediates dopaminergic facilitation of amygdala function. Neuron 48, 1025–1037 (2005). doi: 10.1016/j.neuron.2005.10.029; [DOI] [PubMed] [Google Scholar]
- 64.Tanaka M, Sun F, Li Y, Mooney R, A mesocortical dopamine circuit enables the cultural transmission of vocal behaviour. Nature 563, 117–120 (2018). doi: 10.1038/s41586-018-0636-7; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waclaw RR, Wang B, Pei Z, Ehrman LA, Campbell K, Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron 63, 451–465 (2009). doi: 10.1016/j.neuron.2009.07.015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bandler RC, Mayer C, Fishell G, Cortical interneuron specification: The juncture of genes, time and geometry. Curr. Opin. Neurobiol 42, 17–24 (2017). doi: 10.1016/j.conb.2016.10.003; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waclaw RR, Ehrman LA, Pierani A, Campbell K, Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J. Neurosci 30, 6944–6953 (2010). doi: 10.1523/JNEUROSCI.5772-09.2010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao L et al. , Expression of FoxP2 in the basal ganglia regulates vocal motor sequences in the adult songbird. bioRxiv 2020.03.14.99104 [Preprint]. 16 October 2020. 10.1101/2020.03.14.991042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hegeman DJ, Hong ES, Hernández VM, Chan CS, The external globus pallidus: Progress and perspectives. Eur. J. Neurosci. 43, 1239–1265 (2016). doi: 10.1111/ejn.13196; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tucker ES et al. , Molecular specification and patterning of progenitor cells in the lateral and medial ganglionic eminences. J. Neurosci. 28, 9504–9518 (2008). doi: 10.1523/JNEUROSCI.2341-08.2008; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campbell P, Reep RL, Stoll ML, Ophir AG, Phelps SM, Conservation and diversity of Foxp2 expression in muroid rodents: Functional implications. J. Comp. Neurol 512, 84–100 (2009). doi: 10.1002/cne.21881; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Co M, Anderson AG, Konopka G, FOXP transcription factors in vertebrate brain development, function, and disorders. WIREs Dev. Biol 9, e375 (2020). doi: 10.1002/wdev.375; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faux C, Rakic S, Andrews W, Britto JM, Neurons on the move: Migration and lamination of cortical interneurons. Neurosignals 20, 168–189 (2012). doi: 10.1159/000334489; [DOI] [PubMed] [Google Scholar]
- 74.Tschopp P, Tabin CJ, Deep homology in the age of next-generation sequencing. Philos. Trans. R. Soc. London Ser. B 372, 20150475 (2017). doi: 10.1098/rstb.2015.0475; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tosches MA, Laurent G, Evolution of neuronal identity in the cerebral cortex. Curr. Opin. Neurobiol. 56, 199–208 (2019). doi: 10.1016/j.conb.2019.04.009; [DOI] [PubMed] [Google Scholar]
- 76.Briscoe SD, Ragsdale CW, Evolution of the chordate telencephalon. Curr. Biol 29, R647–R662 (2019). doi: 10.1016/j.cub.2019.05.026; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arendt D, The evolution of cell types in animals: Emerging principles from molecular studies. Nat. Rev. Genet 9, 868–882 (2008). doi: 10.1038/nrg2416; [DOI] [PubMed] [Google Scholar]
- 78.Grillner S, Robertson B, The basal ganglia over 500 million years. Curr. Biol 26, R1088–R1100 (2016). doi: 10.1016/j.cub.2016.06.041; [DOI] [PubMed] [Google Scholar]
- 79.Stacho M et al. , A cortex-like canonical circuit in the avian forebrain. Science 369, eabc5534 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Naumann RK, Laurent G, “Function and evolution of the reptilian cerebral cortex” in Evolutionary Neuroscience, Kaas JH, Ed. (Academic Press, ed. 2, 2020), pp. 213–245. [Google Scholar]
- 81.Laurent G, On the value of model diversity in neuroscience. Nat. Rev. Neurosci. 21, 395–396 (2020). doi: 10.1038/s41583-020-0323-1; [DOI] [PubMed] [Google Scholar]
- 82.Cobos I, Puelles L, Martínez S, The avian telencephalic subpallium originates inhibitory neurons that invade tangentially the pallium (dorsal ventricular ridge and cortical areas). Dev. Biol 239, 30–45 (2001). doi: 10.1006/dbio.2001.0422; [DOI] [PubMed] [Google Scholar]
- 83.Cobos I, Shimamura K, Rubenstein JL, Martínez S, Puelles L, Fate map of the avian anterior forebrain at the four-somite stage, based on the analysis of quail-chick chimeras. Dev. Biol 239, 46–67 (2001). doi: 10.1006/dbio.2001.0423; [DOI] [PubMed] [Google Scholar]
- 84.Métin C et al. , Conserved pattern of tangential neuronal migration during forebrain development. Development 134, 2815–2827 (2007). doi: 10.1242/dev.02869; [DOI] [PubMed] [Google Scholar]
- 85.Tuorto F, Alifragis P, Failla V, Parnavelas JG, Gulisano M, Tangential migration of cells from the basal to the dorsal telencephalic regions in the chick. Eur. J. Neurosci 18, 3388–3393 (2003). doi: 10.1111/j.1460-9568.2003.03059.x; [DOI] [PubMed] [Google Scholar]
- 86.Tanaka DH, Oiwa R, Sasaki E, Nakajima K, Changes in cortical interneuron migration contribute to the evolution of the neocortex. Proc. Natl. Acad. Sci. U.S.A 108, 8015–8020 (2011). doi: 10.1073/pnas.1102153108; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nottebohm F, Stokes TM, Leonard CM, Central control of song in the canary, Serinus canarius. J. Comp. Neurol 165, 457–486 (1976). doi: 10.1002/cne.901650405; [DOI] [PubMed] [Google Scholar]
- 88.Puelles L, Alonso A, García-Calero E, Martínez-de-la-Torre M, Concentric ring topology of mammalian cortical sectors and relevance for patterning studies. J. Comp. Neurol 527, 1731–1752 (2019). doi: 10.1002/cne.24650; [DOI] [PubMed] [Google Scholar]
- 89.Colquitt B, bradleycolquitt/songbird_cells: Revision, Version 1.0, Zenodo: (2020); 10.5281/zenodo.4289038. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


