Abstract
The cell cycle is a fundamental process that has been extensively studied in bacteria. However, many of its components and their interactions with machineries involved in other cellular processes are poorly understood. Furthermore, most knowledge relies on the study of a few models, but the real diversity of the cell division apparatus and its evolution are largely unknown. Here, we present a massive in-silico analysis of cell division and associated processes in around 1,000 genomes of the Firmicutes, a major bacterial phylum encompassing models (i.e. Bacillus subtilis, Streptococcus pneumoniae, and Staphylococcus aureus), as well as many important pathogens. We analyzed over 160 proteins by using an original approach combining phylogenetic reconciliation, phylogenetic profiles, and gene cluster survey. Our results reveal the presence of substantial differences among clades and pinpoints a number of evolutionary hotspots. In particular, the emergence of Bacilli coincides with an expansion of the gene repertoires involved in cell wall synthesis and remodeling. We also highlight major genomic rearrangements at the emergence of Streptococcaceae. We establish a functional network in Firmicutes that allows identifying new functional links inside one same process such as between FtsW (peptidoglycan polymerase) and a previously undescribed Penicilin-Binding Protein or between different processes, such as replication and cell wall synthesis. Finally, we identify new candidates involved in sporulation and cell wall synthesis. Our results provide a previously undescribed view on the diversity of the bacterial cell cycle, testable hypotheses for further experimental studies, and a methodological framework for the analysis of any other biological system.
Keywords: cell cycle, cell division, evolution, phylogenomics, Firmicutes, Bacteria, functional link, evolutionary scenario, phylogeny
Introduction
Understanding the functioning of cellular machines in both prokaryotic and eukaryotic is a major challenge in cell biology. For that, a diversity of experimental approaches is used, including genetics, cellular biology, interaction assays, structural biology, etc. Phylogenomics and omics data also provide a fertile ground to disclose new machines, components and functional links among system components, through the identification of co-transcribed genes, co-evolving genes, conserved synteny (i.e. gene neighborhood), gene fusion, gene co-occurrence, etc. (Galperin and Koonin 2000; Koonin et al. 2001; von Mering et al. 2003; Luciano et al. 2011; Chan et al. 2013; Agrebi et al. 2015; Poupel et al. 2016). Cell division is one of the most important and conserved cellular processes in Bacteria. Stricto sensu, it consists in the split of a cell, called the mother cell, into two or more cells, consequently named the daughter cells. This process relies on a complex cellular protein machinery called the divisome and requires numerous protein–protein interactions together with the coordination of a suite of biochemical reactions (Typas et al. 2012; Pinho et al. 2013). Cell division is tightly linked with others processes required for a successful cell cycle, which form together what we could define as the “Cell Division and Associated Processes” (CDAP). First, proteins of the divisome should be coordinated with proteins of the elongasome, the cell apparatus involved in elongation and the morphogenesis of the cell (Typas et al. 2012). Both divisome and elongasome are tightly linked to the synthesis, maturation, and recycling of the peptidoglycan, the major component of the cell wall (Typas et al. 2012; Pinho et al. 2013). Cell division is also directly interconnected to chromosome replication and segregation (Thanbichler 2010), because a strict temporal regulation between duplication and migration of chromosomes on both side of the septum is required before completion of cell constriction to avoid the so called “guillotine effect” over the nucleoid (Wu and Errington 2012). The replication process is itself linked to DNA repair to prevent chromosome defect during cell division (Thanbichler 2010). Other cellular processes, such as capsule synthesis and flagellar assembly, to cite a few, are also intimately coordinated with cell division (Aizawa and Kubori 1998; Nourikyan et al. 2015). Last but not least, sporulation, a developmental mode leading to the formation of a stress resistant structure called the spore that can form another viable cell when growth conditions improve is a particular type of cell division, sharing some processes such as cell wall synthesis and chromosomal segregation (Higgins and Dworkin 2012).
Delimitating and deciphering the respective function of each component of CDAP is a challenging task as these processes are tightly entangled and share many of their components. For example, FtsK participates to both cell division and chromosome segregation in Escherichia coli (Stouf et al. 2013). Similarly, RocS coordinates chromosome segregation and cell division in Streptococcus pneumoniae (Mercy et al. 2019). Moreover, despite apparent similarity and conservation of CDAP mechanisms, there are important differences among species. For instance, in rod-shape bacteria (e.g. Bacillus subtilis and E. coli), the new cell wall is inserted along the lateral side of the cell causing cell elongation and also at the cell center, where the division site locates, to build the cross-wall required for septation (division stricto sensu) (Typas et al. 2012). In contrast, both machineries co-localize and are coupled at the division site in cocci (Massidda et al. 2013; Pinho et al. 2013; Vollmer et al. 2019). Proteins involved in CDAP may also vary between species. For instance, the MIN system that contributes notably to the location of the division machinery at the cell center is found in B. subtilis. By contrast, this system is absent in Streptococcus pneumoniae which instead uses MapZ protein for this purpose (Garcia et al. 2016). Similarly, the nucleoid occlusion which contributes to the coordination between chromosome segregation and cell constriction is driven by the Noc in B. subtilis and SlmA in E. coli (Wu and Errington 2012).
As a consequence of the dynamic evolution of all these processes, our understanding of CDAP in bacteria remains partial. Furthermore, the precise role and activity of many important CDAP proteins remain unknown. For instance, the FtsQBL complex (believed to be the ortholog of the DivIBC-FtsL complex in B. subtilis) is essential for division in E. coli but its precise role is not fully understood (Choi et al. 2018). Finally, considering the numerous proteins of unknown function in proteomes, it is likely that the set of CDAP genes has not been fully disclosed. Illustrating this, it has been shown that Leuconostocaceae do not encode any described system involved in the placement of the Z-ring, suggesting that another, but yet unknown, system exists in these bacteria (Garcia et al. 2016).
Applying phylogenomic approaches to CDAP represents therefore an interesting and promising approach that provides a global picture of how CDAP evolved and may lead to the identification of new CDAP proteins and functional links. In this study, we focused on Firmicutes. This major bacterial phylum represents a perfect case study as it encompasses B. subtilis, S. pneumoniae, and Staphylococcus aureus, three important models regarding CDAP (Eswara and Ramamurthi 2017), and gathers a great diversity of cell shapes and membrane architecture that could reflect variations in CDAP. In this study, we analyzed more than 160 CDAP protein families. We found that most of them can be traced back to the ancestor of this phylum, defining the ancestral CDAP core of Firmicutes. Yet, beside this apparent conservation, we identified several evolutionary hot spots associated to important changes or rearrangements in the CDAP. We inferred a functional network that has revealed numbers of putative previously undescribed links between components and CDAP. Finally, the exhaustive survey of firmicutes proteomes allowed the identification of previously undescribed putative CDAP proteins and functional links.
Results
Distribution of CDAP Protein Families in Firmicutes
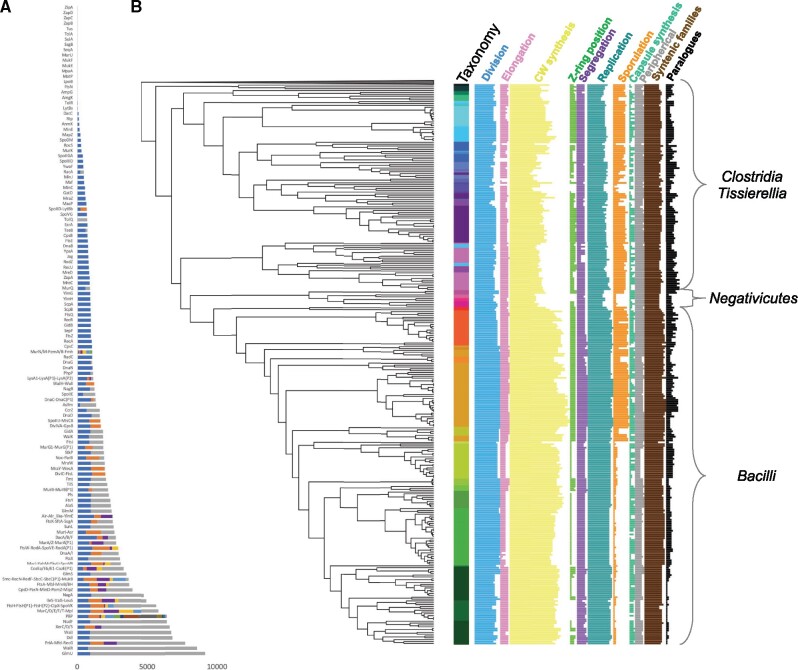
Based on a survey of the literature, we selected 162 proteins involved in CDAP such as cell elongation, peptidoglycan synthesis, chromosome replication and segregation, sporulation, and capsule synthesis in bacteria, as well as 7 proteins with possible role (e.g. GidA) or peripheric role (e.g. MraZ involved in regulation of transcription) in CDAP (supplementary table S1, Supplementary Material online). Among these 169 proteins, 124 correspond to proteins that have been characterized in Firmicutes members, while the others have been characterized in others bacterial phyla (e.g. Proteobacteria, Actinobacteria). This set of proteins contains Penicilin-Binding Proteins (PBPs) that are named according to a recent classification (Sauvage et al. 2008) (supplementary table S2, Supplementary Material online). To avoid biases linked to redundancy, proteins involved in several CDAP were classified in the category for which they have the most important role. A survey of 935 Firmicutes proteomes stricto sensu (De Vos et al. 2009) representing 304 species (supplementary data 1, Supplementary Material online), revealed homologues for 149 of the 169 proteins (fig. 1A). Without surprise, we found homologues of the 124 CDAP proteins characterized in Firmicutes. Regarding, the 45 proteins characterized in other bacterial phyla, 20 have no homologue in Firmicutes (e.g. ZipA), five have no orthologues in Firmicutes but may have distant homologues corresponding to other protein families (e.g. MipZ and PomZ orthologues are absent in Firmicutes proteomes, while their paralogues ParA-CpsD-MinD are present), and 20 have orthologues in Firmicutes (e.g. ZapA). Phylogenetic analyses of the 144 CDAP proteins with orthologues in Firmicutes showed that these proteins corresponded to 36 monogenic families (e.g. FtsZ, SepF) and 108 families from 58 multigenic families (e.g. CpsD, ParB, and MinD) (supplementary table S3, Supplementary Material online). Accurate analysis of the 58 multigenic families, disclosed 16 subfamilies tightly related to the studied CDAP proteins, referred as (Pi) for paralogue n°i (e.g. FtsH(P1)). Finally, a global survey of genetic contexts, highlighted 16 genes displaying a strongly conserved genomic neighborhood with the studied CDAP genes (e.g. ylmH). They correspond to four monogenic families and 12 families from five multigenic families. These 16 genes and the 16 paralogues have been added in the set of studied CDAP proteins. Altogether, this led to the study of 40 monogenic families and 136 families from 63 multigenic families.
Fig. 1.
Taxonomic distribution of CDAP protein families. (A) Size of protein families corresponding to CDAP proteins. Each line corresponds to either a monogenetic or a multigenetic family. More precisely, 40 CDAP protein families are monogenic, while 136 belong to 63 multigenic families. Families of multigenic families that do not correspond to CDAP are in grey, while other colours correspond to CDAP protein families. Name of the CDAP families are provided on the left. (B) Taxonomic distribution (presence/absence) of CDAP proteins in the 304 studied firmicutes species. The protein families are classified by cognate CDAP (see supplementary table S1, Supplementary Material online). Nodes on the phylogeny (and thus branch lengths) are ordered according to their relative dating proposed by Phylobayes. The coloured strip corresponds to family taxonomic rank (see supplementary fig.S1, Supplementary Material online).
The taxonomic distribution (presence/absence) of 176 CDAP proteins has been mapped on the phylogeny of the 304 firmicutes species (fig. 1B and supplementary fig.S1, Supplementary Material online). The number of CDAP proteins per species is highly variable, ranging from 146 in Bacillus weihenstephanensis to 71 in Mageeibacillus indolicus (mean: 111.8) (fig. 1B and supplementary fig.S2, Supplementary Material online). Moreover, while some CDAP proteins are universal in Firmicutes (e.g. FtsZ), others harbor patchy taxonomic distributions (e.g. sporulation genes).f
For each CDAP, we measured the component distribution variation across species using the coefficient of variation (i.e. the ratio of standard deviation on the mean of the number of components of the CDAP contained in firmicutes species). A low coefficient reflects small variations (i.e. homogeneity) in the number of components of the considered CDAP across species. Lowest coefficients of variation are observed for cell division, chromosome segregation and replication, and cell-wall synthesis (coefficients of variation = 0.104, 0.138, 0.179, and 0.106, respectively), contrasting with elongation (coefficient of variation = 0.324), and even more with Z-ring localization systems, sporulation, and capsule synthesis (coefficient of variation 0.591, 0.786, and 0.456, respectively). Without surprise, we observed a correlation between the number of CDAP proteins and the size of the proteomes (Pearson coefficient = 0.63, P-value < 0.05, supplementary fig.S2, Supplementary Material online), with Negativicutes and some Clostridia having the smallest set of CDAP proteins and the smallest proteomes. In contrast, Bacillales and especially the Bacillus cereus group and Paenibacillaceae have the largest proteomes and the highest number of CDAP proteins (supplementary fig.S2, Supplementary Material online). Altogether, these results indicate important fluctuations in the distribution of CDAP components, maybe linked to variations in proteome size, suggesting that important evolutionary events have occurred during the diversification of Firmicutes.
The Evolutionary History of CDAP in Firmicutes Revealed Several Hot Spots
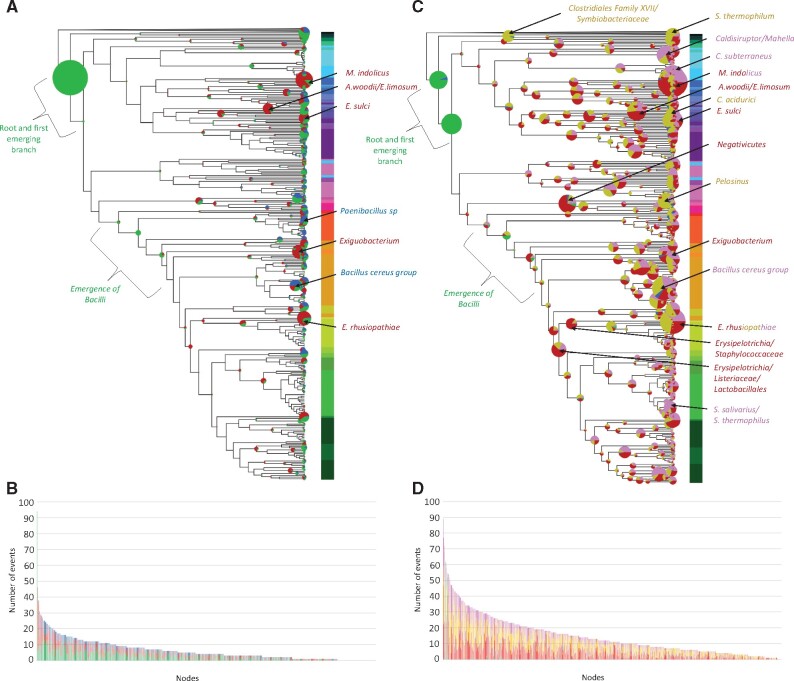
To go further, we studied the evolution of the CDAP proteins in Firmicutes using methods based on reconciliation (R) and phylogenetic profiles (PP). PP inferred globally less events (n = 3,865) compared to the reconciliation approach (n = 8,864) (Supplementary Table S4), but more gains and duplications (gains = 1,711 vs. 176 and duplications = 919 vs. 355, respectively). These differences reflect that reconciliation approach considers horizontal gene transfers (HGT), whereas phylogenetic profile-based approach considers secondary emergences as gains or duplications. Despite the significative difference in term of number of events per branch (Wilcoxon test P-value < 0.05), both methods are consistent (Pearson and Spearman coefficients = 0.65 and 0.61, P-value < 0.05, fig. 2 and supplementary fig.S3, Supplementary Material online). While most events were globally distributed across lineages, some branches could be identified as hot spots of events (i.e. concentrating a high number of events compared to the other branches) by both approaches (see below).
Fig. 2.
Evolutionary events that have impacted the 176 CDAP protein families during the diversification of Firmicutes.(A) Events inferred with the phylogenetic profile approach. (C) Events inferred with the reconciliation approach. Circles at branches correspond to inferred events. Their size is proportional to the number of events. Colors correspond to the type of evolutionary events: green = acquisitions, red = losses, blue = duplications, yellow = HGT, Pink = homologue replacements. Branches identified as hotspots are indicated by arrows. Colours associated to branch names correspond to the dominating type of events. (B) Distribution per branch of the number of events inferred with phylogenetic profiles. (D) Distribution per branch of the number of events inferred with reconciliation.
Portraying the Ancestor of Firmicutes
Most CDAP protein families are inferred at or right after the root of Firmicutes by both PP and reconciliation R (PP = 100 families, R = 114 families, intersection = 95 families), indicating they are ancient, likely ancestral, in this phylum (fig. 2, green circles and supplementary table S5, Supplementary Material online). The functions of these protein families may allow to sketch some features of the ancestor of Firmicutes. For instance, Mre proteins that are thought to be specifically present in rod-shape bacteria (particularly MreB) (Shi et al. 2018) are inferred in the ancestor of the Firmicutes. The inference of the proteins involved in MIN and NO indicates that these two systems could participate to the Z-ring positioning in the ancestor of Firmicutes, as in B. subtilis (Ortiz et al. 2016). Moreover, most of the analyzed sporulation genes can be traced back in the ancestor of Firmicutes, suggesting this cell may have been able to sporulate. Finally, we inferred as ancestral both chromosome segregation systems PAR and SMC, together with the FtsK protein, respectively involved in the segregation of the origin of replication, the bulk chromosome, and the terminal region. Altogether, these results suggest that the ancestor of Firmicutes could have been a rod-shape and sporulating bacterium, localizing its Z-ring notably by the MIN and NO system and segregating its chromosome using PAR and SMC systems, and FtsK.
The Emergence of Bacilli is Associated to an Extension of the CDAP Protein Family Set
Combining PP and R methods identified the five deepest branches of Bacilli class, one of the main lineages of Firmicutes, as a hot spot of CDAP protein acquisitions (fig. 2A and C). More precisely, both methods identified the acquisition of PbpA(P1), PpbA5, EzrA, SftA, RacA, MreBH, MurJ, MisCB, Spo0M, DnaI, DnaB, PbpB3, TseB, WalH, DacA, GpsB, CcrZ, RecU, PpbA3, YpsA, and Pfs (supplementary table S6, Supplementary Material online). Among them, an enrichment of the set of cell wall synthesis (PbpA3, PbpA5, PbpA(P1), PbpB3, DacA) and cell division components (EzrA, CcrZ and GpsB) was observed, despite the losses of PbpB(P1) and MurA(P2). We also inferred the acquisition of four replication proteins (DnaB, DnaI, RacA, RecU).
These results indicate that the emergence of Bacilli has been accompanied by several events impacting CDAP, especially cell-wall synthesis and cell division. The concomitant acquisition of specific DNA replication proteins and cell-wall synthesis deserve attention, raising the question of the role of such co-apparition from a functional perspective. Finally, acquisitions outnumber losses, suggesting that the emergence of Bacilli was accompanied by an enrichment or complexification of existing CDAP instead of the replacement of existing CDAP components.
Hot Spots of Gene Losses, Transfers, and Duplications
Beside Bacilli, we identified additional hot spots in Firmicutes. For instance, numerous losses of Min (Z-ring localisation), Mre (Elongation), Wal (Cell wall regulation), Spo (Sporulation), and Cps (Capsule synthesis) proteins, along with several PBPs and cell-wall recycling proteins were predicted by both PP and R approaches in specific lineages: Mageeibacillus indolicus (PP = 40, R = 47, intersection = 35), Exiguobacterium (PP = 27, R = 24, intersection = 17), Eubacterium sulci (PP = 19, R = 23, intersection = 11), Acetobacter woodii/Eubacterium limosum (PP = 25, R = 28, intersection = 14) and the facultative intracellular Erysipelotrichia rhusiopathiae (PP = 31, R = 22, intersection = 18) (fig. 2 and supplementary data 2, Supplementary Material online). Because some of these proteins are essential, such as the Z-ring anchoring protein, or Mre proteins, some losses may have been compensated by the recruitment of new, yet unidentified, components. For example, M. indolicus lacks both FtsA and SepF involved in the anchoring of FtsZ and supposed to be crucial for the assembly of the Z-ring (Ortiz et al. 2016). As a consequence, these bacteria, for which no experimental data are available, would represent interesting models to investigate cell division and associated processes. Reconciliation highlighted three additional hotspots of gene loss at the root of Negativicutes (37 events), Erysipelotrichia/Listeria/Lactobacillales (28 events), and Staphylococcaceae/Erysipelotrichia (29 events), that deserve further investigations.
Concerning HGT inferred by the reconciliation approach, two types of events have been distinguished: (i) de novo acquisitions and (ii) homologous replacements corresponding to the loss of a gene associated to the reacquisition of a homologous gene through HGT (supplementary table S4, Supplementary Material online). By making this distinction, we could identify 3,926 de novo acquisition and 2,388 homologous replacement events. A few lineages emerged as hot spots of de novo acquisitions: Symbiobacterium thermophilum (43 events), Erysipelotrichia rhusiopathiae (36 events), Pelosinus (39 events), Clostridium acidurici (35 events), and the stem of Clostridiales Family XVII/Symbiobacteriaceae (35 events) (fig. 2 and supplementary data 2, Supplementary Material online). In contrast, homologous replacements are mainly inferred in the stems of Caldanaerobacter subterranneus (41 events), Caldisiruptor and Mahella (33 events), M. indolicus (30 events), Erysipelotrichia rhusiopathiae (28 events), Streptococcus salivarius and Streptococcus thermophilus (28 events), and Bacillus toyonenis, Bacillus weinstephanensis, and Bacillus mycoides (Bacillus cereus group) (25 events) (fig. 2C and supplementary data 2, Supplementary Material online).
Finally, and contrasting with gene losses and HGT, gene duplication events are rarer. As expected, most of them are predicted by the PP approach. Branches harboring the highest number of duplication events correspond to the emergence of Bacillus cereus group (PP = 16, R = 4), and Paenibacillus sp. Y412MC10 (PP = 13, R = 4). Interestingly, the emergence of Bacillus cereus group corresponds mainly to the duplication of proteins involved in late stages of peptidoglycan synthesis: one duplication of PbpbA4, four of PbpB5, three of DacA, one of PbpA3, two of MurJ, and one of RodA, suggesting major changes of peptidoglycan synthesis in this group.
Evolution of CDAP Gene Syntenies in the Firmicutes
Functionally linked genes in prokaryotes are frequently associated in gene clusters, arrays of co-expressed and co-regulated genes (Wolf et al. 2001). As a consequence, conserved syntenies that are recurrently observed in genomes from distantly related prokaryotes can be used to predict functional interactions between genes and to infer function of uncharacterized genes (Moreno-Hagelsieb and Santoyo 2015; Koonin and Makarova 2019). Similarly, gene synteny disruptions or emergences can reflect changes in functional interactions. Accordingly, we analyzed the evolution of the syntenies of CDAP genes along the tree of Firmicutes.
Twenty Gene Clusters Are Inferred in the Ancestor of Firmicutes
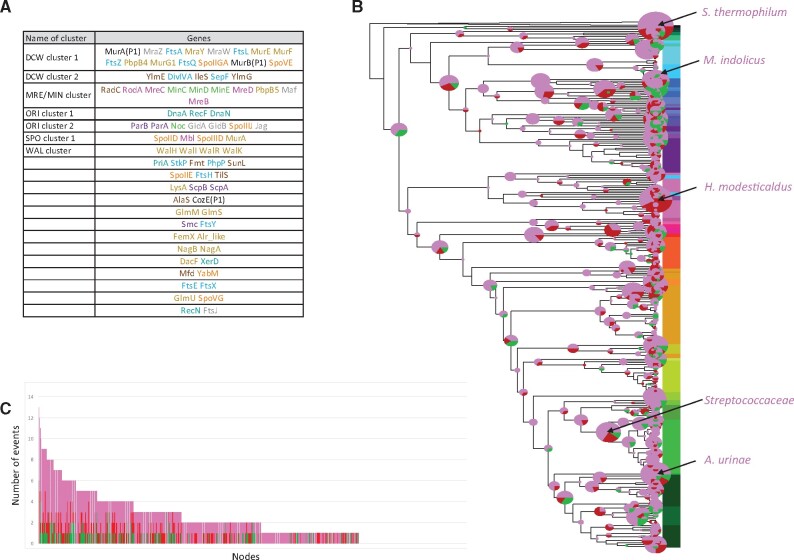
In genomes studied here, the number of gene clusters (at least 2 genes) ranges from 13 in Selenomonas sputigena ATCC 35185 to 29 in Bacillus megaterium WSH-002 (mean = 20.6). The evolutionary analyze of these clusters indicated that twenty gene clusters were likely present in the ancestor of Firmicutes (fig. 3A), some of them having been conserved throughout the diversification of this phylum. This is for instance the case of the MRE/MIN cluster that encompasses radC, rodA, mreC, minC, minD, minE, mreD, pbpB5, maf, and mreB (Levin et al. 1992) (fig. 3A and supplementary fig.S4, Supplementary Material online). This conserved neighborhood over large evolutionary scales likely indicate a strong functional link between Z-ring localization (Min proteins) and elongation (Mre proteins) (see below). Most members of this cluster have been lost independently at the emergence of a few clades, such as Streptococcaceae/Enterococcaceae and Staphylococcaceae/Erysipelotrichiae (supplementary fig.S4, Supplementary Material online).
Fig. 3.
Evolution of CDAP gene clusters during the diversification of Firmicutes. (A) gene clusters inferred at the root of Firmicutes. Color of family names corresponds to the CDAP (blue: cell division, pink: elongation, yellow: cell wall, green: Z-ring localization, violet: chromosomal segregation, cyan: chromosome replication, orange: sporulation, blue-green: capsule synthesis, grey: peripherical). (B) Evolutionary event affecting gene clusters. Circles at branches correspond to inferred events. Their size is proportional to the number of events. Colors correspond to the type of evolutionary events: green = creations, red = disruptions, and pink = modifications. Branches identified as hotspots are indicated by arrows. Colors associated to branch names correspond to the dominating type of events. (C) Distribution of the number of events per branch.
The DCW gene cluster, known to contain major cell division and cell-wall synthesis genes (Tamames et al. 2001), provided also interesting information. While a single DCW cluster is found in most Firmicutes, it is split into two distinct clusters DCW1 (murA(P1), mraZ, ftsA, mraY, mraW, ftsL, murE, murF, ftsZ, pbpB4, murG1, ftsQ, spoIIGA, murB(P1), and spoVE) and DCW2 (ylmE, sepF, ylmG, ylmH, divIVA, and ileS) in most Clostridia (supplementary fig.S5, Supplementary Material online). Without surprise, the split cluster was inferred as ancestral because it is mostly present in most deeply-branching firmicutes lineages. It should also be noted that two sporulation genes (spoIIGA and spoVE) are inferred to be present in the ancestral DCW1 cluster, consistently with the inference of sporulation in the Firmicutes ancestor, and confirming the strong links between sporulation and cell wall synthesis.
Finally, the gene cluster located at the origin of replication (hereafter referred as to ORI) and gathering dnaA, recF, dnaN, parA, parB, noc, gidA, gidB, spoIIIJ, and jag (EloR), is also inferred in the ancestor of Firmicutes (supplementary fig.S6, Supplementary Material online). This conserved cluster is dislocated in Streptococcaceae (see below).
Major Cluster Rearrangements
The accurate analysis of CDAP gene clusters in Firmicutes, disclosed 939 modification events corresponding to gene addition or removal from existing gene clusters and 154 events corresponding to de novo formation of gene clusters (fig. 3B–C). The number of events per branch ranges from zero to 13 (mean = 2.17). The stems of Streptococcaceae, Symbiobacterium thermophilum, Heliobacter modesticaldus, Aerococcus urinae, and Mageeibacillus indolicus are associated to 10 or more events affecting clusters otherwise largely conserved in Firmicutes (fig. 3 and supplementary figs.S5–S7). Among them, Streptococcaceae are interesting as they encompass the bacterial model Streptococcus pneumoniae. In Streptococcaceae, the DCW gene cluster, inferred to be ancestral in Firmicutes, is split into two distinct clusters, gathering mraW, ftsL, pbpB4, and mraY, on the one hand, and ylmE, ftsA, ylmG, ylmH, murD, ileS, sepF, murG1, ftsQ, divIVA, and ftsZ on the other hand. Furthermore, mraZ, notably involved in the regulation of transcription of the first eleven DCW cluster genes in E. coli (Eraso et al. 2014), has been lost (supplementary fig.S5, Supplementary Material online). The split of the DCW cluster in S. pneumoniae that has been reported elsewhere (Massidda et al. 1998) is in fact a common feature of all Streptococcaceae. Beside the DCW cluster, the strongly conserved ORI gene cluster has been dislocated into five pieces (gidB, jag/spoIIIJ1, dnaA/dnaN, parB, recF) in the stem of Streptococcaceae, while parA has been lost (supplementary fig.S6, Supplementary Material online). Additional events occurred, such as the break-up of the association of ftsY and smc, and the loss of xerD that was part of the scpA/spcB cluster (supplementary fig.S6). Finally, we observed the arrival of mapZ within the gene cluster gathering gpsB, pbpA3, recU, and ypsA, and the dissociation of dnaD from this cluster (supplementary fig.S7). All these rearrangements could reflect important changes in cell division and chromosomal replication and segregation in Streptococcaceae, as it is the case in S. pneumoniae. Indeed, S. pneumoniae, which is the most studied Streptococcaceae, presents noticeable differences with canonical bacterial models in term of cell division and chromosomal replication. For example, in S. pneumoniae elongation and division are coupled and located at the mid-cell (Massidda et al. 2013), MapZ and not the MIN system localizes the mid-cell (Fleurie, Lesterlin, et al. 2014), and RocS coordinates chromosome segregation and cell division (Mercy et al. 2019). Together with the fact that MapZ and RocS are mainly restricted to the Enterococcaceae and Streptococcaceae, these specificities could be potentially linked to the major observed rearrangements of gene clusters at the stem of Streptococcaceae, and reciprocally meaning that all Streptococcaceae could share these specificities.
Beside Streptococcaceae, gene cluster rearrangements are inferred in other Lactobacillales. For instance, the formation of new syntenies has been observed in Lactobacillaceae and Leuconostocaceae for which the DCW cluster is associated to several other clusters. First, the cluster encompassing macP (coding for a PBP regulator), nudF, and pfs became associated to the DCW gene cluster in the stem of Lactobacillaceae/Leuconostocaceae lineages (supplementary fig.S5, Supplementary Material online). Then, a synteny between DCW cluster and both MIN/MRE cluster and ezrA (localization of PBP) or ftsK (DNA translocase) occurred later in Lactobacillaceae lineages. Finally, during the diversification of the Leuconostocaceae, the synteny of macP, nudF, and pfs with the DCW cluster has been replaced by an association with the gene cluster encompassing stkP and phpP, two proteins involved in the control of cell elongation and septation (Fleurie, Manuse, et al. 2014). Again, these associations between different clusters could reflect changes in transcriptional regulation associated particularly to division and elongation processes.
Gene Co-evolution and Conserved Gene Neighborhood Highlight Putative Functional Links
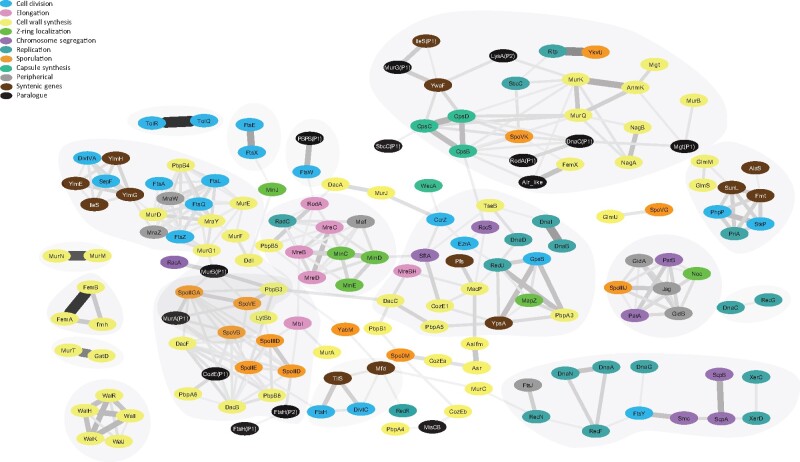
In order to unveil previously undescribed potential functional links among CDAP genes, we compared their evolutionary histories as well as their neighborhoods (synteny) using Jaccard coefficients. More precisely, for each pair of CDAP protein families, we computed a weighted and normalized Jaccard coefficient of common evolutionary events inferred either with phylogenetic profiles (Jwn-pp) or reconciliation-based approaches (Jwn-r). We also computed a Jaccard coefficient on shared syntenies (J-s). Without surprise, the correlation between Jwn-pp and Jwn-r was the highest (Pearson = 0.35 and Spearman = 0.51, all P-value < 0.05), because they both reflect gene co-evolution. In contrast, the correlation between J-s and either Jwn-pp or Jwn-r was rather weak (J-s/Jwn-pp: Pearson = 0.20 and Spearman = 0.16; J-s/Jwn-r: Pearson = 0.17 and Spearman = 0.22, all P-value < 0.05), probably because genes can co-evolve even if they are not syntenic, and reciprocally. Then, we calculated a score based on the three coefficients for each pair of CDAP protein families, which was used to build a correlation network. The network encompasses 160 nodes and 616 edges. As expected, the network tends to cluster CDAP proteins involved in the same cellular process (fig. 4 and supplementary table S1, Supplementary Material online). For instance, elongation proteins Mre are tightly linked together in the network. This is also the case of the Z-ring positioning proteins Min, sporulation proteins Spo, cell-wall synthesis Fem proteins, capsule synthesis Cps proteins, segregation Scp/Smc proteins, and cell-wall regulator Wal proteins. Also, the kinase StkP and its cognate phosphatase PhpP are closely connected in the network, as well as the PBPs involved in elongation (B5), division (B4), and sporulation (DacB, DacF, B3, A6) and their cognate processes (see supplementary table S3, Supplementary Material online). Altogether, this suggested that our approach is able to predict known functional links between CDAP protein families, yet it revealed also several associations between CDAP never reported previously.
Fig. 4.
Correlation network of CDAP proteins. Nodes represent CDAP protein families. They are colored according to their cognate CDAP. Edges represent the 2% highest Z-scores between pairs of CDAP protein families (see Material and methods). The color and the width of edges correspond to the strength of the Z-score. Main correlation clusters are indicated in grey. Families without any significative link with another family are not shown (Alr, ClpX, FtsK, LeuS, LysA, LysA(P1), LytB, MurI, MurZ, NudF, PbpA(P1), RecA, RodZ, ValS, XerS, ZapA).
MRE/MIN Cluster: connecting Elongation and Z-Ring Positioning
As expected, proteins of the MRE/MIN gene cluster (mreB, mreC, mreD, rodA, and pbpB5 (also known as pbp2a) involved in elongation on the one hand and minC, minD, and minE participating to the positioning of the Z-ring) inferred in the ancestor of Firmicutes (fig. 3A) are connected in the network (fig. 4). In the network, these proteins are tightly linked to radC involved in DNA damage repair (Saveson and Lovett 1999), and maf a nucleotide pyrrophosphatase involved in septum formation during division (Butler et al. 1993). Our analyses indicated that radC and maf were also present in the MRE/MIN cluster in the ancestor of Firmicutes (fig. 3A). While the synteny between mre, min, maf, and radC genes has been conserved in most Firmicutes, the synteny with rodA and pbpB5 has been lost at the emergence of Bacilli (supplementary fig.S4, Supplementary Material online). This event is concomitant with the hot spot of PBP turnover (i.e. the loss of PbpB(P1) and the acquisition of PbpA3, PbpA5, PbpA(P1), PbpB3, and DacA) mentioned above. RodA and PbpB5 being involved in the cell-wall synthesis during elongation, this strengthens the hypothesis that the emergence of Bacilli was accompanied by important changes in the synthesis of peptidoglycan.
Looking at the taxonomic distribution of the MRE/MIN cluster genes highlighted two groups of genes. More precisely, MreD, MreC, Pbp5, and RodA are present in most Firmicutes and lost together in a few lineages such as M. indolicus and Streptococcus pyogenes, while the taxonomic distribution of MinC, MinD, MinE, MreB and Maf is patchy (supplementary fig.S4, Supplementary Material online). MinE was lost in the stem of Bacilli, whereas the loss of MreB, MinC, and MinD occurred at the root of many cocci clades, such as Staphylococcaceae/Erysipelotrichia, Streptococcaceae/Enterococcaceae, and Pediococcus (supplementary fig.S4, Supplementary Material online). The lack of the Min components in cocci indicates that other Z-ring positioning systems should exist in these taxa. Strengthening this proposal, MapZ was reported to fulfil this function in Streptococcaceae/Enterococcaceae (Fleurie, Lesterlin, et al. 2014) but the systems in Staphylococcaceae/Erysipelotrichia and Pediococcus remain to be discovered. This is in line with the proposal of the existence of alternative mechanisms used to identify the mid-cell (Rodrigues and Harry 2012; Bailey et al. 2014). Altogether, these results indicate a strong functional link between elongation and the positioning of the Z-ring. The location of maf and radC within the MRE/MIN cluster is highly conserved across Firmicutes, suggesting a yet unreported functional link with elongation and/or Z-ring positioning in this phylum. Finally, our results also suggest that MreD, MreC, Pbp5, and RodA have an essential role in almost all Firmicutes, contrary to MinC, MinD, MinE, and MreB that seem to be specific to rod-shape bacteria. The taxonomic distribution of MreB appears to be more similar to Min proteins than with others Mre proteins, indicating that MreC and MreD could have a MreB-independent role in the cell.
A Coordination Between Segregation and Dimer Resolution
Our network highlighted also unexpected links between Smc, ScpA, and ScpB that form the chromosomal segregation SMC system (Nolivos and Sherratt 2014), XerC and XerD involved in the resolution of dimers of chromosomes (Thanbichler 2010), and FtsY, the translocase of the Signal Recognition Particle system involved in the co-translational protein addressing to the membrane (Seluanov and Bibi 1997) (fig. 4). These proteins are widely distributed in Firmicutes and inferred to be ancient in this phylum (supplementary fig.S6 and table S5, Supplementary Material online). ScpA and scpB are neighbour of xerD in most of Bacilli to the exception of Streptococcaceae (see below) and of xerC in a few other lineages such as Acidaminococcus (supplementary fig.S6, Supplementary Material online), while smc is systematically in the neighbourhood of ftsY. Strikingly, among the numerous events that occurred in the stem of Streptococcaceae mentioned above, the loss of xerC and xerD coincided with the disruption of the synteny between smc and ftsY (supplementary fig.S6, Supplementary Material online). According to the functional links between smc, scpA, and scpB and syntenies between scpA, scpB, xerC, and xerD on the one hand and smc and ftsY on the other hand, it is tempting to hypothesize that both events are linked. It is worthy to note that XerS that is also responsible for the resolution of dimer, was present in the ancestor of Streptococcaceae which could be a necessary condition for such changes.
A Link Between Cell Division, Cell-Wall Synthesis, and Recombination in Bacilli
The network shows links among GpsB, RecU, PbpA3, YpsA, and DnaD (supplementary fig.S7, Supplementary Material online). RecU is a Holliday junction resolvase involved in reparation of DNA during replication (Pereira et al. 2013), while DnaD has a role in the initiation of replication (Bonilla and Grossman 2012). In B. subtilis, GpsB localises PBP proteins, especially PbpA3 (PBP1) (Claessen et al. 2008), while YpsA seems to be involved in oxidative stress response and cell division regulation (Brzozowski, et al. 2019). These proteins are strongly conserved in Bacilli (supplementary fig.S7, Supplementary Material online). Our data indicate an acquisition of GpsB, RecU, PbpA3, and YpsA near the root of Bacilli. The situation is less clear for DnaD that is predicted to be acquired at root of Bacilli by PP but of more ancient origin by R. Regarding synteny, we inferred that ypsA and gpsB on one hand, and recU, pbpA3, and dnaD on the other hand formed first two distinct gene clusters in Firmicutes that were then associated into a single gene cluster suggesting the establishment of functional links between these five genes. This gene cluster has then been conserved during the diversification of Bacilli, to the exception of dnaD that moved out from the cluster at the emergence of Streptococcaceae, while mapZ joined it. At a larger scale, GpsB, RecU, PbpA3, YpsA, DnaD, and MapZ protein families are also linked to proteins associated to other CDAP (e.g. MacP, EzrA, Pfs, TseB, CcrZ, CozE1, PbpA5, RocS, SftA, MreBH, DacC, DnaI, and DnaB), which together form a larger cluster in the network (fig. 4). These links reflect similar taxonomic distribution and their co-acquisition at the root of Bacilli (see above) (supplementary fig.S8 and supplementary data 2–3, Supplementary Material online), highlighting again major changes and innovations in Bacilli regarding cell division, cell-wall synthesis, segregation and recombination.
New Insights on Sporulation
Ten on fifteen proteins involved in sporulation (six Spo proteins and four PBPs) are clustered together in the network (fig. 4 and supplementary fig.S8, Supplementary Material online) because they have similar histories. In fact, these proteins are inferred to be ancestral in Firmicutes and have been lost together and independently in different branches (supplementary fig.S9 and table S5, Supplementary Material online). These co-losses are all the more significant as these genes are not grouped in a single gene clusters (supplementary figs.S8 and S9, Supplementary Material online), and therefore observed co-losses cannot be explained by a single loss event. Interestingly, some CDAP protein families without known links with sporulation are connected to these ten sporulation proteins in the network. Among them, CozE(P1) and FtsH(P2) deserve attention. More precisely, the evolutionary history of CozE(P1) is especially linked with that of DacF (Jwn-pp = 0.83, Jwn-r = 0.82) and CozE(P1) displays a taxonomic distribution similar to several other sporulation protein families (i.e. SpoIID, SpoIIE, SpoIIGA, SpoIIID, SpoVB, SpoVE, PbpB6, DacB) (supplementary figs. S8 and S9, Supplementary Material online). CozE(P1) is a paralogue of CozE, a protein involved in PBPs regulation (Fenton et al. 2016), while DacF is a PBP associated to a carboxypeptidase activity involved in sporulation (Popham et al. 1999). Accordingly, it is tempting to speculate that CozE(P1) and DacF could be functionally linked, and that CozE(P1) is a regulator of DacF. Regarding FtsH(P2), its evolutionary history is similar to SpoIID, a major sporulation protein (Jwn-pp = 0.81, Jwn-r = 0.65). Because FtsH(P2) is a paralogue of FtsH, a protease involved notably in the degradation of the Z-ring, one could hypothesize that FtsH(P2) a protease specifically involved in sporulation.
Finally, Mbl is connected with, SpoIID, SpoIIID, and MurA (involved in sporulation and cell wall synthesis) in the network (supplementary figs.S8 and S9, Supplementary Material online), reflecting the synteny of their coding genes. The clustering of these genes is inferred as ancestral in Firmicutes (fig. 3). Mbl is a paralogue of MreB mainly involved in elongation during vegetative division, yet a role in sporulation has been reported in the actinomycete Streptomyces coelicolor (Heichlinger et al. 2011). Therefore, this might reflect a role of Mbl in sporulation also in the Firmicutes, as suggested in B. subtilis (Ojkic et al. 2016). If this link is confirmed in the future, this would represent a nice example of independent functional recruitment and bifunctional protein. Our results also suggest a functional link between MurA and sporulation proteins, and that among the multiple paralogues of the MurA/Z protein family, MurA could be the UDP-N-acetylglucosamine 1-carboxyvinyltransferase that intervenes in sporulation in Firmicutes, as previously suggested in B. subtilis (Vasudevan et al. 2007).
A Possible Link between FtsW and a Previously Undescribed PBP in Clostridia
In several Bacilli, FtsW, a peptidoglycan polymerase (Taguchi et al. 2019), interacts with the well-conserved PbpB4, such as PBP1 in Staphylococcus aureus, PBP2x in Streptococcus pneumoniae, and PBP2b in Bacillus subtilis (Gamba et al. 2016; Perez et al. 2019; Reichmann et al. 2019). Surprisingly, our network analysis strongly linked FtsW, not with PbpB4 but with a yet previously undescribed PBP homologue in Firmicutes, that we proposed to name PbpB(P1) (fig. 4). Indeed, when the gene coding for PbpB(P1) is present, it is systematically in synteny with ftsW, and even fused in Lachnoclostridium phytofermentans. In addition, when FtsW is absent, PbpB(P1) is also absent (Supplementary Figure S10). This strongly suggests a functional link between PbpB(P1) and FtsW. Furthermore, this functional link is likely ancient in Firmicutes because both genes were inferred near the root of this phylum (supplementary fig.S10 and supplementary table S5, Supplementary Material online). Interestingly, in the stem of Bacilli, PbpB(P1) has been lost while FtsW has been retained. Knowing that FtsW interacts with PbpB4 in Bacilli, it is possible that a change of functional partnership occurred in this lineage. Accordingly, it would be interesting to investigate experimentally the putative functional link between PbpB(P1) and FtsW.f
Is There a Link between CDAP and RNA-Related Processes?
It has been previously shown that the deletion of some proteins involved in RNA related processes (MraW, Maf, GidA, GidB, Jag, FtsJ) have an impact on cell division (von Meyenburg and Hansen 1980; Ogura et al. 1991; Tchigvintsev et al. 2013; Eraso et al. 2014; Gao, et al. 2016; Stamsas, et al. 2017). Still, some other studies highlighted the fact that the effect on cell division of some of them could be due to pleiotropic effects (i.e. GidA (Kinscherf and Willis 2002)) or polar effects (i.e. FtsJ (Bugl, et al. 2000)). Strikingly, our data showed highly conserved syntenies between the genes coding for these proteins and main CDAP gene clusters: mraW in the DCW cluster; maf in the MRE/MIN cluster, gidA, gidB, and jag in the ORI cluster; ftsJ with recN. Furthermore, five other genes involved in RNA related processes are also syntenic with CDAP genes: fmt and sunL are part of the stkP gene cluster, ileS of the DCW cluster, pfs of the macP cluster, and tilS of the ftsH cluster. These highly conserved associations suggest that a subtle and maybe indirect link exists between CDAP and RNA related processes. It would be tempting to hypothesize that some of these proteins could be involved in temporal regulation of CDAP, and therefore could represent interesting targets for exploratory studies.
Previously Undescribed Potential CDAP Proteins
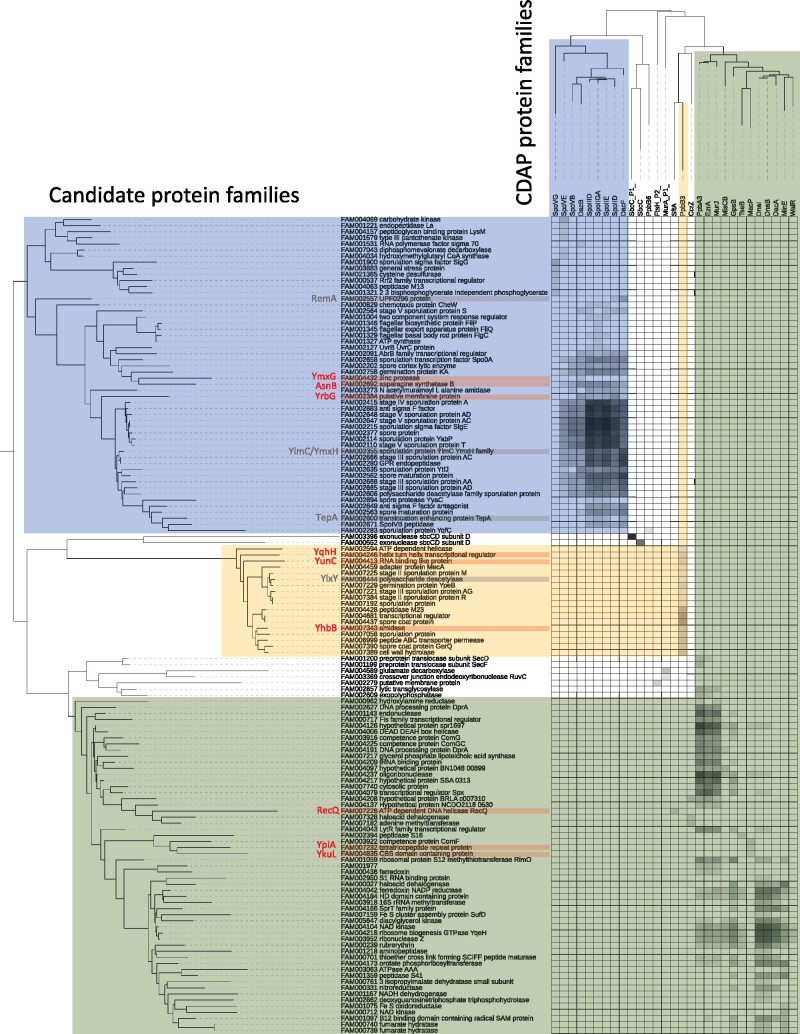
Finally, we sought to identify possible previously undescribed CDAP proteins using a comparative genomic approach. For this, we built a database of 187,348 protein families contained in the 304 firmicutes species and compared their phylogenetic profiles with those of the 176 studied CDAP protein families by using Mutual Information (MI). The 135 families showing a MI higher than 0.5 with at least one CDAP protein family were retained (supplementary data 12, Supplementary Material online). Three sets of candidate proteins can be clearly distinguished based on their taxonomic distribution (fig. 5). The first one includes proteins present in Clostridia and Bacillales, with phylogenetic profiles correlating with SpoVG, SpoVE, SpoVB, DacB, SpoIIID, SpoIIGA, SpoIIE, SpoIID, and DacF, all involved in sporulation. Without surprise, many of these candidate proteins were annotated as sporulation proteins. The second set encompassed less proteins, mainly present in Bacillales. These are also involved in sporulation and have a taxonomic distribution similar to that PbpB3, which is also involved in sporulation. These data highlight the existence of two different sets of sporulation protein families, one being ancient in Firmicutes, while the second is restricted to Bacillales. In comparison to a previous analysis aiming at identifying new sporulation genes by similar approaches, we recovered five of the eight candidates suggested (i.e. TepA, YlmC, YmxH, RemA, and YlxY) (Traag et al. 2013). However, we also identified six proteins never described before as involved in sporulation: YrbG (calcium-sodium antiporter, unknown function), YqhH (lipoprotein, unknown function), YunC (unknown function), YhbB (unknown function), YmxG (Zinc protease, unknown function), and AsnB (L-asparaginase) (fig. 5 and supplementary fig.S11, Supplementary Material online). These proteins could therefore represent potential new components of the sporulation machinery in the Firmicutes.
Fig. 5.
Mapping of correlation index of CDAP proteins and potential candidates. The heatmap represents the similarity between taxonomic distributions of studied CDAP protein families (top) and proteins families from firmicutes proteomes with the most similar taxonomic distribution (left). These proteins represent potential CDAP candidate families. The names of the leaves correspond to the major annotation of the family. The color of the squares corresponds to the strength of correlation (white = 0, black = 1). Three clusters are highlighted in color (blue and yellow correspond to sporulation clusters, green correspond to Bacilli cluster). New candidates are indicated in red, protein families highlighted elsewhere in grey (Traag et al. 2013).
The third set of proteins correlated with 12 CDAP families that originated in the stem of Bacilli (fig. 5). Comparison of the taxonomic distributions of these proteins indicate that some of them could have interesting links with specific CDAP protein families. For instance, SftA involved in chromosome segregation and RecQ involved in recombination display a similar taxonomic distribution, suggesting a functional link between the two proteins. Also, MacP, a protein involved in the regulation of PBP (Fenton et al. 2018), seems to have been co-acquired and co-lost with two proteins to unknown function corresponding to YkuL and YpiA. These could therefore represent potential partners of MacP.
Altogether, these results suggest the existence of previously undescribed putative CDAP families that will be worth investigating further.
Discussion
The cell division is one of the most important and most studied cellular processes. Yet, due to the extraordinary complexity of the underlying molecular mechanisms and its tightly connections with many other cell processes, the cell division remains partially understood. In bacteria, main steps of cell division and associated processes are globally conserved, underlying they derive from an ancestral common system. Yet, this ancestral system has been continuously adjusted and evolved through the addition or the loss of components, modifications in component functions, changes in regulation and interactions of components, etc. This led to a multitude of more or less pronounced variations in CDAP that cannot be caught and depicted by a few model organisms. In that context, phylogenomics is a valuable tool that allows to highlight parts of cellular processes or macromolecular systems that are shared or variable across lineages. Furthermore, by providing information on evolution of these systems, phylogenomics may help to disclose new functional links and new components.
Here, we presented an analysis conducted on the Firmicutes phylum based on more than 160 CDAP protein families. We showed that most of them were likely present in the ancestor of Firmicutes and have been conserved along the diversification of this major bacterial phylum. Many CDAP genes are organized in conserved gene clusters, most of them being likely present in the ancestor of Firmicutes and conserved in most firmicutes lineages, underlying the tightly and ancient links between these fundamental cellular processes. This set of genes represents the core of cell division and associated processes that was mostly conserved during the diversification of Firmicutes. On the opposite, other genes present a more complex evolutionary history, variations in their taxonomic distribution and gene neighborhood, illustrating the plasticity of cell the division machinery and its interactions with associated processes.
The reconstruction of CDAP protein evolutionary history highlights a continuous flow of gains, losses, duplications, HGTs, and homologous replacements of genes all along the diversification of the Firmicutes. Indeed, every branch is affected by such events, which indicates a continuous evolution of CDAP. Beyond this background noise, some branches emerge as hotspots as they concentrate more events. This is for instance the case of the stem of Bacilli or some species such as Mageeibacillus indolicus that could represent interesting case study. Also, some evolutionary convergences have been observed for similar phenotype emergence such as the cocci that have independently lost most of mre and min genes. Surprisingly and against all odds, we did not observe particular patterns between monoderms (Bacilli and most Clostridia) and diderms (i.e. Halanaerobiales and Negativicutes), to the exception of TolQ and TolR. These two proteins, involved in the invagination of the external membrane, are only present in diderms. This indicates that the independent emergence of monoderms lineages from a diderm ancestor (Antunes et al. 2016) has not been accompanied by major changes in the cell division apparatus, in agreement with a recent report (Taib et al. 2020).
Using an original approach, we inferred a network allowing visualizing functional links between CDAP components. As expected, known functional links are represented in the network confirming the interest of the approach. The network revealed also unreported links between CDAP components such as between a paralogue of CozE and sporulation proteins or between Smc and FtsY that represent interesting targets for experimental studies. Surprisingly, a few proteins involved in cell division do not present any evolutionary links and gene syntenies with other proteins. It is for example the case of FtsK that is extremely conserved in all Firmicutes and present no conserved neighborhood with any of divisome proteins, its biological partners. Worth to note, our study is focused on cell division. Accordingly, to our knowledge, all proteins known to be involved in this process but also in elongation and chromosomal segregation have been considered. In contrast, only a subset of the proteins involved in some other CDAP has been included (sporulation, chromosome replication, capsule synthesis). As a consequence, this network represents only a part of the functional interactions between CDAP and we expect that in the future, the enrichment of the network will reveal new information. Finally, using the evolutionary information contained in the studied CDAP components to screen firmicutes proteomes, we identified several proteins. Among them, around 10 may represent yet unidentified components of CDAP and thus interesting targets for future studies.
To conclude, this study brings never explored insights in cell division and associated processes evolution, filling a gap of fundamental knowledge about dynamics of such machineries and proposing several trails for more applied future works.
Materials and Methods
Dataset Assembly
A survey of the literature identified 29 proteins involved in cell division. This set of proteins was enriched by including 105 proteins involved in elongation, cell wall synthesis, and chromosomal segregation (supplementary table S1, Supplementary Material online). Most of them belong to Fts (Filamentous Temperature Sensitive), Min (MINicell), Mre (MuRein E), Mra (MuRein A), Mur (MURein), Rod (ROD-shape), Wal (cell WALl), Zap (Z-ring Associated Protein), Zip (Z-ring Interacting Protein), Dac (D-Alanine Carboxypeptidase), Div (DIVision), Glm (GLucosaMine), Nag (N-AcetylGlucosamine), PBP (Penicillin-Binding Protein), Fem (Factors Essential for Methicillin resistance), Tol (TOLerence), Muk (MUKaku), Par (PARtitioning), and Scp (Segregation and Condensation Protein) protein families. For PBPs, the classification by Sauvage and colleagues has been used (A1-6, B1-6, MGT, DacABCF) (Sauvage et al. 2008). Correspondences between common names in several reference strains are indicated in Table S2. In addition, a representative set of key proteins involved in cell division related processes such as replication (Dna (DNA), Xer (CER-specific recombination), Rec (RECombination)), sporulation (Spo (SPOrulation)), and capsule synthesis (Cps (Capsular Polysaccharide Synthesis)) but also seven proteins for which the precise function in CDAP is not fully understood (FtsJ, GidA, GidB, Jag, Maf, MraW, MraZ) have been added. Preliminary analyses identified sixteen conserved genes a priori not involved in CDAP but recurrently located in clusters of genes involved in cell division, and 16 very closely related paralogues of genes involved in cell division (annotated (Pi) for Paralog i) (see results). The corresponding proteins have been included in the analysis, leading to a set of 185 initial proteins.
Protein Family Assembly
About 937 complete proteomes (with completeness status “full”) of Firmicutes stricto sensu (De Vos et al. 2009) available at the NCBI FTP have been gathered in a local database (supplementary data 1). For each protein, this local database was queried with the BLASTP program from BLASTALL package 2.2.6 (Altschul et al. 1997) using a set of reference sequences as seed (Supplementary Table S3). Sequences corresponding to high scoring pairs with e-values lower than 10−6 were retrieved. The retrieved sequences have been aligned using MAFFT v7.123b (auto option) (Katoh and Standley 2013). The N-terminal and C-terminal variable regions have been removed using Gblocks version 0.91 b (Allowed Gap Positions = All, Minimum Number Of Sequences For A Flank Position = 0.6) (Castresana 2000). Kept positions have been used to build HMM profiles using HMMBUILD from the HMMER package v3.1b1 (Eddy 2011) and used to query the proteome database with HMMSEARCH (default options). Sequences with an E-value lower than 0.01 were retrieved and used to query the local database using BLASTP. Sequences were considered as homologues if the best hits corresponded to the set of proteins used to build the HMM profile. A new HMM profile including the newly identified homologues has been built and used to query the local database. This procedure has been repeated until no new homologues was identified. Finally, a survey of the genomic sequences corresponding to the 937 studied firmicutes was performed using TBLASTN to detect potential unannotated sequences (annotated as PSDG for pseudogene).
Each protein family was aligned using MAFFT (default option). The quality of the resulting multiple alignments has been manually verified using SEAVIEW (Gouy et al. 2010) and ALIVIEW (Larsson 2014). Doubtful sequences have been checked using reciprocal BLASTP and comparison of functional domain composition. Kept sequences have been realigned with MAFFT. Resulting multiple alignments have been trimmed with BMGE 1.1 (BLOSUM30 option) (Criscuolo and Gribaldo 2010) and used to infer phylogenetic trees with FASTTREE 2.1.8 (wag, cat = 4, gamma options) (Price et al. 2010).
The resulting trees show that among the 176 proteins analyzed, 40 correspond to monogenic protein families, while the 136 others corresponded to 63 multigenic superfamilies (supplementary data 4, Supplementary Material online). In that case, protein families have been delineated according to tree topology, taxonomic distribution, sequence length, functional protein domain composition, and genomic context. For initial proteins from non-firmicutes organisms, we checked the presence of orthologues in Firmicutes by performing an analysis on a database of 4,466 prokaryote proteomes.
Species Tree Construction
A species tree of Firmicutes has been inferred using ribosomal proteins (rprot). Sequences of the 53 bacterial rprot families have been identified in the 937 Firmicutes genomes using the engine of the RiboDB database (Jauffrit et al. 2016). Sequences from nine species (five Actinobacteria and four Cyanobacteria, supplementary data 1, Supplementary Material online) were used as outgroup. Then, to limit taxonomic redundancy and computation time, a subsample of 306 firmicutes strains has been selected by keeping randomly one strain per species (supplementary data 1 and 6, Supplementary Material online). Reach rprot family has been aligned using MAFFT version 7 (accurate option L-INS-i) (Katoh and Standley 2013). The resulting multiple alignments have been trimmed by BMGE 1.1 (BLOSUM30 option) (Criscuolo and Gribaldo 2010) and gathered to build a large supermatrix (315 sequences, 5,901 amino acid positions). A maximum-likelihood (ML) tree has been inferred using IQ-TREE 1.4.1 (Nguyen et al. 2015) (supplementary data 5, Supplementary Material online). We used a specific amino acid exchange matrix based on rprots from 1,634 bacterial proteomes from RiboDB using the ReplacementMatrix tool (Dang et al. 2011). To model the heterogeneity in the amino acid substitution process and evolutionary rate across sites, we used the PMSF (posterior mean site frequency) option with 20 mixture categories (Wang et al. 2018) and a gamma distribution with four categories of sites. The robustness of the branches of the resulting ML tree has been estimated using three methods: the Shimodaira–Hasegawa approximate likelihood ratio test (sh, 1,000 replicates), the abayes test (ab), and ultra-fast bootstrap (ub, 1,000 replicates) implemented in IQ-TREE (Nguyen et al. 2015). A global score of 3 was associated to a given branch when sh > 80%, ab > 0.8, and ub > 95%. A score of 2, 1, or 0 was attributed to the branch if one, two, or all the three supports were lower than those thresholds. The species phylogeny revealed that two strains, Coprothermobacter proteolyticus DSM 5265 and Thermodesulfobium narugense DSM 14796, do not belong to Firmicutes (branching in the external group). Accordingly, these strains were not considered for the analysis of the CDAP protein families, setting the number of considered Firmicutes species to 304. The tree agrees with the NCBI taxonomy (supplementary fig.S1, Supplementary Material online) and with the most recent ML tree of Firmicutes (Antunes et al. 2016).
The ML tree has been dated using Phylobayes 4.1c (Lartillot et al. 2009), with the same replacement matrix, a mixture model CAT with 20 categories, and a gamma distribution with four categories of sites (supplementary data 5, Supplementary Material online). A relaxed clock model with an uncorrelated gamma law has been used with a divergence of time prior following a birth death process. Two chains have been run for circa 17,000 iterations. Trees were sampled every 40 generations with a burn-in of 5,000 iterations. The convergence of chains has been checked using tracecomp and bpcomp (max divergence = 0 (< 0.3) and min effective size = 80 (> 50)). We also checked visually the convergence using Tracer (Rambaut et al. 2018). After convergence, the first chain has been used to generate an ultrametric chronogram using readdiv.
Inference of Evolutionary Events
Evolutionary events such as gains, losses, duplications of genes and horizontal gene transfers (altogether referred as gene events) have been inferred using two different methods. First, we used a ML phylogenetic profile-based approach to infer gene gains, losses, and duplications (supplementary data 2, 7 and 8, Supplementary Material online). For each protein family, we used PASTML (default parameters, Marginal Posterior Probabilities Approximation + F81) (Ishikawa et al. 2019) to infer the number of protein copy at each node of the species phylogeny. Gain, loss, and duplication events associated to a given branch have been inferred as follows: for each branch, if the corresponding node presents a lower number of copies than the ancestral node, the difference corresponds to the number of losses. When a higher number of copies is observed: if the ancestral node state is zero, it corresponds to gains, else, it corresponds to duplications.
The second approach relied on tree reconciliation to infer gene gains, losses, and duplications but also horizontal gene transfers (supplementary data 2, 9 and 10, Supplementary Material online). Briefly, for each protein family, a phylogenetic tree has been inferred with RAXML 0.5.0b (Stamatakis 2015) using the same sampling as the species tree (i.e. 304 Firmicutes). Sequences have been aligned using MAFFT (accurate option L-ins-I) and trimmed using BMGE 1.1 (BLOSUM30 option). MODELFINDER has been used to select the best-suited evolutionary model (Kalyaanamoorthy et al. 2017), applying the BIC criteria. The robustness of the resulting tree was measured using the rapid bootstrap procedure implemented in RAXML (100 replicates of the original dataset). Reconciliations between protein family phylogenies and the species tree have been performed using the amalgamation approach implemented ecceTERA 1.2.4 (Scornavacca et al. 2015). Bootstrap replicate trees generated by RAXML have been used to model the distribution of trees for the amalgamation. The costs of evolutionary events and the amalgamation weight have been estimated with the algorithm of cost estimation implemented in ecceTERA (one iteration) using default costs at the initiation step. Transfers from the dead have been authorized. Loss and HGT (in) at a same branch have been considered as homologue replacements.
Gene rearrangement events have been inferred using a parsimony-based approach (supplementary data 2 and 11, Supplementary Material online). Two genes were considered as neighbour if their distance on the chromosome was lower than 2 kb in a given genome. Syntenic gene clusters have been delineated by a single-linkage clustering approach. We built a two-character states matrix indicating for each pair of genes (columns) and for each genome (rows) whether these genes are part of the same syntenic gene cluster. Ancestral states have been inferred at each node of the species phylogeny by parsimony using a cost of 1 and 3 for the disruption or the appearance of a synteny between two genes, respectively. Ancestral syntenic gene clusters have been delineated at each node of the species phylogeny by applying a single-linkage clustering approach. Evolutionary events affecting ancestral syntenic gene clusters have been characterized in term of creation (emergence de novo of a cluster), modification (addition or withdrawal of gene from a cluster), and disruption (loss of synteny among all members of a cluster). To avoid overestimation of gene cluster events, we checked whether these events were the consequence of gene loss or gain (inferred by PP based approach) and considered only events that were not associated to such events.
Similarity between evolutionary histories and gene syntenies of protein families
The similarity between evolutionary histories and gene syntenies of protein families has been calculated using Jaccard coefficients (supplementary data 3, Supplementary Material online). More precisely for a given pair of proteins, we compared evolutionary events inferred with reconciliation, and phylogenetic profile approaches and the syntenic association of their genes along the species tree.
For evolutionary events (reconciliation and phylogenetic profiles):
Let A and B be protein families and E the set of events in every nodes of the species tree, the Jaccard coefficient between two families is
The Jaccard coefficient has been weighted by the number of common events as:
The weighted coefficient JwA, B has been normalized such as:
For the syntenies:
Let A and B be protein families, PX the set of species presenting a copy of X and NA, Bthe set of species presenting a neighbourhood between A and B (d 2,000 pb), the Jaccard coefficient between two families is:
Calculation of the final score:
Let A and B be protein families, Jwn-rA, B the normalized weighted Jaccard coefficient calculated by reconciliation events, Jwn-ppA, B the normalized weighted Jaccard coefficient calculated by phylogenetic profiles-based events and J-sA, B the Jaccard coefficient calculated by syntenies. A global score SA, B has been calculated as a mean of the three values. The Jwn-ppA, B and Jwn-rA, B relying on the same information (history of independent protein families), they have been weighted by a factor, compared to the J-sA, B. The score SA, B is then defined as:
A Z-score has then been calculated. Let and be the average and standard deviation of scores S between X and all other families:
The 2% of sequences presenting highest Z-score have been used to build the network.
Figures
Tree figures have been generated using iTOL (Letunic and Bork 2016) and genomic contexts have been generated by using GeneSpy 1.1 (Garcia et al. 2019). The network has been generated using Cytoscape v3.7.1 (Shannon et al. 2003).
Identification of Putative New CDAP Proteins
The proteomes of the 304 representative firmicutes strains were used to assemble a protein family database with SiLiX 1.2.9 (Miele et al. 2011) (identity threshold = 0.4 and sequence coverage = 0.8). The phylogenetic profile of each 187,348 assembled protein family has been compared to those of CDAP protein families using the Mutual information (supplementary data 12, Supplementary Material online).
Let A and B be protein families and Si the state (presence/absence) in the species i. The frequency of the state S of gene A is:
The joint frequency of states S(A) and S(B) is:
The mutual information between phylogenetic profiles of A and B is:
The values higher than 0.5 have been retained and used for the analysis. The families corresponding to the already studied CDAP proteins have been removed. The kept families have been annotated by the major annotation found in FAA database. The distance trees have been generated using fastme v2.1.6.1 (default options)(Lefort et al. 2015) and a matrix based on Mutual Information values.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work has been supported by the Centre National de la Recherche Scientifique (CNRS), The Université de Lyon, the Région Rhône-Alpes ARC1 (financial support for P.S.G.), the Agence National de la Recherche (15-CE32-0001-01 and 19-CE15-0011-01), the Bettencourt-Schueller Foundation, and the Investissement d’Avenir ‘Ancestrome’ (ANR-10-BINF-01-01). Calculations were performed using the computing facilities of the CC LBBE/PRABI.
Conflict of Interest statement
The authors declare no competing interests.
Data Availability
All data are incorporated into the article and its online supplementary material.
References
- Agrebi R, Wartel M, Brochier-Armanet C, Mignot T.. 2015. An evolutionary link between capsular biogenesis and surface motility in bacteria. Nat Rev Microbiol.13(5):318–326. [DOI] [PubMed] [Google Scholar]
- Aizawa SI, Kubori T.. 1998. Bacterial flagellation and cell division. Genes Cells 3(10):625–634. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res.25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LC, Poppleton D, Klingl A, Criscuolo A, Dupuy B, Brochier-Armanet C, Beloin C, Gribaldo S.. 2016. Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. Elife 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Mannik J.. 2014. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet.10(8):e1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla CY, Grossman AD.. 2012. The primosomal protein DnaD inhibits cooperative DNA binding by the replication initiator DnaA in Bacillus subtilis. J Bacteriol.194(18):5110–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski RS, Huber M, Burroughs AM, Graham G, Walker M, Alva SS, Aravind L, Eswara PJ.. 2019. Deciphering the role of a SLOG superfamily protein YpsA in Gram-positive bacteria. Front Microbiol.10:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U.. 2000. RNA methylation under heat shock control. Mol Cell.6(2):349–360. [DOI] [PubMed] [Google Scholar]
- Butler YX, Abhayawardhane Y, Stewart GC.. 1993. Amplification of the Bacillus subtilis maf gene results in arrested septum formation. J Bacteriol.175(10):3139–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol.17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Chan YB, Ranwez V, Scornavacca C.. 2013. Reconciliation-based detection of co-evolving gene families. BMC Bioinformatics.14(1):332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Kim J, Yoon HJ, Jin KS, Ryu S, Lee HH.. 2018. Structural insights into the FtsQ/FtsB/FtsL complex, a key component of the divisome. Sci Rep.8(1):18061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Emmins R, Hamoen LW, Daniel RA, Errington J, Edwards DH.. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol.68(4):1029–1046. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol.10(1):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CC, Lefort V, Le VS, Le QS, Gascuel O.. 2011. ReplacementMatrix: a web server for maximum-likelihood estimation of amino acid replacement rate matrices. Bioinformatics 27(19):2758–2760. [DOI] [PubMed] [Google Scholar]
- De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB.. 2009. The Firmicutes: Springer Nature.
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol.7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraso JM, Markillie LM, Mitchell HD, Taylor RC, Orr G, Margolin W.. 2014. The highly conserved MraZ protein is a transcriptional regulator in Escherichia coli. J Bacteriol.196(11):2053–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswara PJ, Ramamurthi KS.. 2017. Bacterial cell division: nonmodels poised to take the spotlight. Annu Rev Microbiol.71(1):393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AK, El Mortaji L, Lau DT, Rudner DZ, Bernhardt TG.. 2016. CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat Microbiol.2(3):16237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AK, Manuse S, Flores-Kim J, Garcia PS, Mercy C, Grangeasse C, Bernhardt TG, Rudner DZ.. 2018. Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proc Natl Acad Sci U S A.115(11):2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A, Lesterlin C, Manuse S, Zhao C, Cluzel C, Lavergne JP, Franz-Wachtel M, Macek B, Combet C, Kuru E, et al. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516(7530):259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A, Manuse S, Zhao C, Campo N, Cluzel C, Lavergne JP, Freton C, Combet C, Guiral S, Soufi B, et al. 2014. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet.10(4):e1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV.. 2000. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol.18(6):609–613. [DOI] [PubMed] [Google Scholar]
- Gamba P, Hamoen LW, Daniel RA.. 2016. Cooperative recruitment of FtsW to the division site of Bacillus subtilis. Front Microbiol.7:1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Tan M, Liu W, Zhang C, Zhang T, Zheng L, Zhu J, Li L, Zhou R.. 2016. GidA, a tRNA modification enzyme, contributes to the growth, and virulence of Streptococcus suis serotype 2. Front Cell Infect Microbiol.6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PS, Jauffrit F, Grangeasse C, Brochier-Armanet C.. 2019. GeneSpy, a user-friendly and flexible genomic context visualizer. Bioinformatics 35(2):329–331. [DOI] [PubMed] [Google Scholar]
- Garcia PS, Simorre JP, Brochier-Armanet C, Grangeasse C.. 2016. Cell division of Streptococcus pneumoniae: think positive!.Curr Opin Microbiol.34:18–23. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol.27(2):221–224. [DOI] [PubMed] [Google Scholar]
- Heichlinger A, Ammelburg M, Kleinschnitz EM, Latus A, Maldener I, Flardh K, Wohlleben W, Muth G.. 2011. The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. J Bacteriol.193(7):1533–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D, Dworkin J.. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev.36(1):131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa SA, Zhukova A, Iwasaki W, Gascuel O.. 2019. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol Biol Evol.36(9):2069–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauffrit F, Penel S, Delmotte S, Rey C, de Vienne DM, Gouy M, Charrier JP, Flandrois JP, Brochier-Armanet C.. 2016. RiboDB database: a comprehensive resource for prokaryotic systematics. Mol Biol Evol.33(8):2170–2172. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods.14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol.30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinscherf TG, Willis DK.. 2002. Global regulation by gidA in Pseudomonas syringae. JB 184(8):2281–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS.. 2019. Origins and evolution of CRISPR-Cas systems.Phil Trans R Soc B .374(1772):20180087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Aravind L.. 2001. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res.11(2):240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30(22):3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S.. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288. [DOI] [PubMed] [Google Scholar]
- Lefort V, Desper R, Gascuel O.. 2015. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol.32(10):2798–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res.44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Margolis PS, Setlow P, Losick R, Sun D.. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol.174(21):6717–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, Ducret A, Brochier-Armanet C, Mignot T.. 2011. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet.7(9):e1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda O, Anderluzzi D, Friedli L, Feger G.. 1998. Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology (Reading) 144(11):3069–3078. [DOI] [PubMed] [Google Scholar]
- Massidda O, Novakova L, Vollmer W.. 2013. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environ Microbiol.15(12):3133–3157. [DOI] [PubMed] [Google Scholar]
- Mercy C, Ducret A, Slager J, Lavergne JP, Freton C, Nagarajan SN, Garcia PS, Noirot-Gros MF, Dubarry N, Nourikyan J, et al. 2019. RocS drives chromosome segregation and nucleoid protection in Streptococcus pneumoniae. Nat Microbiol.4(10):1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele V, Penel S, Duret L.. 2011. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G, Santoyo G.. 2015. Predicting functional interactions among genes in prokaryotes by genomic context. Adv Exp Med Biol.883:97–106. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol.32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolivos S, Sherratt D.. 2014. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol Rev.38(3):380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourikyan J, Kjos M, Mercy C, Cluzel C, Morlot C, Noirot-Gros MF, Guiral S, Lavergne JP, Veening JW, Grangeasse C.. 2015. Autophosphorylation of the bacterial tyrosine-kinase CpsD connects capsule synthesis with the cell cycle in Streptococcus pneumoniae. PLoS Genet.11(9):e1005518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Tomoyasu T, Yuki T, Morimura S, Begg KJ, Donachie WD, Mori H, Niki H, Hiraga S.. 1991. Structure and function of the ftsH gene in Escherichia coli. Res Microbiol.142(2–3):279–282. [DOI] [PubMed] [Google Scholar]
- Ojkic N, Lopez-Garrido J, Pogliano K, Endres RG.. 2016. Cell-wall remodeling drives engulfment during Bacillus subtilis sporulation. Elife 5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C, Natale P, Cueto L, Vicente M.. 2016. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev.40(1):57–67. [DOI] [PubMed] [Google Scholar]
- Pereira AR, Reed P, Veiga H, Pinho MG.. 2013. The Holliday junction resolvase RecU is required for chromosome segregation and DNA damage repair in Staphylococcus aureus. BMC Microbiol.13(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AJ, Cesbron Y, Shaw SL, Bazan Villicana J, Tsui HT, Boersma MJ, Ye ZA, Tovpeko Y, Dekker C, Holden S, et al. 2019. Movement dynamics of divisome proteins and PBP2x:ftsW in cells of Streptococcus pneumoniae. Proc Natl Acad Sci U S A.116(8):3211–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho MG, Kjos M, Veening JW.. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol.11(9):601–614. [DOI] [PubMed] [Google Scholar]
- Popham DL, Gilmore ME, Setlow P.. 1999. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J Bacteriol.181(1):126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupel O, Moyat M, Groizeleau J, Antunes LC, Gribaldo S, Msadek T, Dubrac S.. 2016. Transcriptional analysis and subcellular protein localization reveal specific features of the essential WalKR system in Staphylococcus aureus. PLoS One 11(3):e0151449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarization in.Bayesian Phylogenetics Using Tracer 1.7. Syst Biol.67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann NT, Tavares AC, Saraiva BM, Jousselin A, Reed P, Pereira AR, Monteiro JM, Sobral RG, VanNieuwenhze MS, Fernandes F, et al. 2019. SEDS-bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat Microbiol.4(8):1368–1377. [DOI] [PubMed] [Google Scholar]
- Rodrigues CD, Harry EJ.. 2012. The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet.8(3):e1002561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P.. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev.32(2):234–258. [DOI] [PubMed] [Google Scholar]
- Saveson CJ, Lovett ST.. 1999. Tandem repeat recombination induced by replication fork defects in Escherichia coli requires a novel factor, RadC. Genetics 152(1):5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scornavacca C, Jacox E, Szöllősi GJ.. 2015. Joint amalgamation of most parsimonious reconciled gene trees. Bioinformatics 31(6):841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Bibi E.. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem.272(4):2053–2055. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Bratton BP, Gitai Z, Huang KC.. 2018. How to build a bacterial cell: mreB as the foreman of E. coli construction. Cell 172(6):1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2015. Using RAxML to infer phylogenies. Curr Protoc Bioinformatics 51:6– 14 11-16 14 14. [DOI] [PubMed] [Google Scholar]
- Stamsas GA, Straume D, Ruud Winther A, Kjos M, Frantzen CA, Havarstein LS.. 2017. Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae. Mol Microbiol.105(6):954–967. [DOI] [PubMed] [Google Scholar]
- Stouf M, Meile JC, Cornet F.. 2013. FtsK actively segregates sister chromosomes in Escherichia coli. Proc Natl Acad Sci U S A.110(27):11157–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, Kruse AC, Kahne D, Bernhardt TG, Walker S.. 2019. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat Microbiol.4(4):587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taib N, Megrian D, Witwinowski J, Adam P, Poppleton D, Borrel G, Beloin C, Gribaldo S.. 2020. Genome-wide analysis of the Firmicutes illuminates the diderm/monoderm transition. Nat Ecol Evol.4(12):1661–1672. [DOI] [PubMed] [Google Scholar]
- Tamames J, Gonzalez-Moreno M, Mingorance J, Valencia A, Vicente M.. 2001. Bringing gene order into bacterial shape. Trends Genet.17(3):124–126. [DOI] [PubMed] [Google Scholar]
- Tchigvintsev A, Tchigvintsev D, Flick R, Popovic A, Dong A, Xu X, Brown G, Lu W, Wu H, Cui H, et al. 2013. Biochemical and structural studies of conserved Maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides. Chem Biol.20(11):1386–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M. 2010. Synchronization of chromosome dynamics and cell division in bacteria. Cold Spring Harb Perspect Biol.2(1):a000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag BA, Pugliese A, Eisen JA, Losick R.. 2013. Gene conservation among endospore-forming bacteria reveals additional sporulation genes in Bacillus subtilis. J Bacteriol.195(2):253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W.. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol.10(2):123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, Popham DL.. 2007. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol.65(6):1582–1594. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Massidda O, Tomasz A.. 2019. The cell wall of Streptococcus pneumoniae. Microbiol Spectr. 7(3):GPP3-0018-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B.. 2003. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res.31(1):258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenburg K, Hansen FG.. 1980. The origin of replication, oriC, of the escherichia coli chromosome: genes near oriC and construction of oriC deletion mutations. In: Alberts B, editor. Mechanistic studies of DNA replication and genetic recombination.London: Academic Press. [Google Scholar]
- Wang HC, Minh BQ, Susko E, Roger AJ.. 2018. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst Biol.67(2):216–235. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV.. 2001. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res.11(3):356–372. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J.. 2012. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol.10(1):8–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are incorporated into the article and its online supplementary material.