Abstract
Endothermy is a typical convergent phenomenon which has evolved independently at least eight times in vertebrates, and is of significant advantage to organisms in extending their niches. However, how vertebrates other than mammals or birds, especially teleosts, achieve endothermy has not previously been fully understood. In this study, we sequenced the genomes of two billfishes (swordfish and sailfish), members of a representative lineage of endothermic teleosts. Convergent amino acid replacements were observed in proteins related to heat production and the visual system in two endothermic teleost lineages, billfishes and tunas. The billfish-specific genetic innovations were found to be associated with heat exchange, thermoregulation, and the specialized morphology, including elongated bill, enlarged dorsal fin in sailfish and loss of the pelvic fin in swordfish.
Keywords: endothermy, swordfish, sailfish, convergence, pkmb
Introduction
Endothermy, maintaining an elevated body temperature and controlling rate of heat production (Hulbert and Else 2000; IUPS Thermal Commission 2001; Koteja 2004; Clarke and Pörtner 2010; Lovegrove 2017), has been a revolutionary innovation for organisms, allowing them to overcome environmental constraints and adapt to new ecological niches (Clarke and Pörtner 2010; Hedrick and Hillman 2016). Endothermy is not only present in mammals and birds, it has also independently evolved at least five times in teleosts and chondrichthyans (Block and Finnerty 1994). In teleosts, it is represented by Istiophoriformes (billfishes), Scombriformes (tunas and butterfly kingfish), and Lampriformes (ophas) (Block and Finnerty 1994; Dickson and Graham 2004; Runcie et al. 2009) (fig. 1A). It is harder for organisms to keep warm in water than on land, because of the high heat capacity of water and the rapid rate of heat loss in aqueous environments (Block 1991; Tullis, Block and Sidell 1991). Several features have been found to be necessary for endothermy in teleosts and chondrichthyans, including large body size, a heat source (specific heat-generating organ or high basal metabolic rate), and heat exchangers (retia mirabilia) for conserving the heat (Block 1991).
Fig. 1.
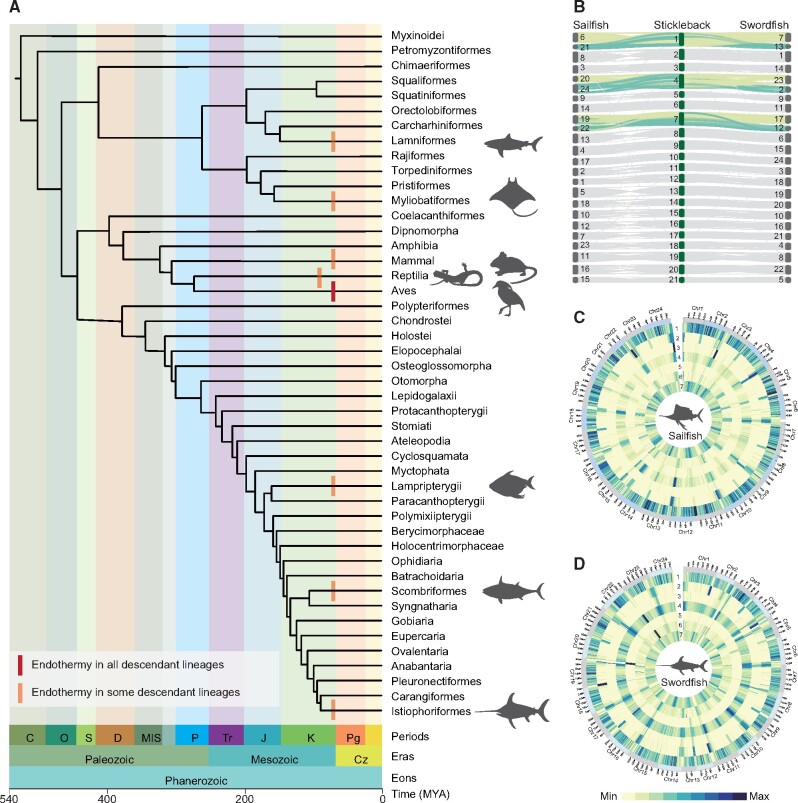
Origins of endothermy in vertebrates and genome assemblies for two billfishes. (A) Phylogenetic tree of vertebrate orders and major groups of endothermic animals (the backbone topology is from a previous study of bony fishes (Betancur-R et al. 2017), and the divergence time of each node were retrieved from the website TimeTree (http://www.timetree.org/; August24, 2020), which were estimated from a population of studies (Kumar et al. 2017)). Geological periods and ages (million years ago, MYA) are shown at the bottom. Cz: Cenozoic, Pg: Paleogene, K: Cretaceous, J: Jurassic, Tr: Triassic, P: Permian, MIS: Carboniferous, D: Devonian, S: Silurian, O: Ordovician, C: Cambrian. Endothermy is indicated with red vertical bars (see legend) at the branches. (B) Chromosomal syntenic relationship of two billfishes and stickleback; each line represents a syntenic block of 10 or more zones from the results of LAST with a similarity of 75% or more and length > 1,000 bp. The karyotype of billfishes shows that six of theirchromosomes correspond to three chromosomes in stickleback. (C and D) Circos plots showing the distributions of genomic components in sailfish and swordfish respectively, with 500,000 bp as a window. 1: Gene frequency, 2: Density of LINEs, 3: Density of LTRs, 4: Density of DNA, 5: Density of TRFs, 6: Density of SINEs, 7: Density of GC content.
The billfishes and tunas were among the first endothermic teleosts to be identified (Barrett and Hester 1964; Carey and Teal 1966; Carey 1982). Due to phenotypic similarities, including capability of heat production and conserving ability, rapid and sustained swimming, and large body (Dickson and Graham 2004), these two lineages were once thought to belong to the same suborder of Scombroidei (Johnson 1986). Subsequent studies based on molecular (Block et al. 1993) and physiological evidence have shown that their heating capacities are of independent origin, with billfish thought to be heated by heating organs (modified red muscle) and tuna by red muscle (Block 1991; Dickson and Graham 2004). Both ectothermic and endothermic vertebrates have red muscle, but only endotherms are capable of producing heat through futile cycles of calcium production in their myocytes (Block 1994; Bal and Periasamy 2020). More recent in-depth studies have shown the existence of specialized heating organs at least in the basal tuna species (Allothunnus fallai) (Sepulveda et al. 2007), and that the red muscles of both billfishes and tunas are capable of heating generation (Block et al. 2001; Stoehr et al. 2018). Although it has been shown that the physiological foundations of heat production in these two lineages evolved convergently, whether there is a common molecular mechanism has not been clarified to date.
Associated with their endothermic capacity, the billfishes have high metabolic rates, and a rapid turnover of nutrients to provide internal fuel supplies. The endothermic capacity of billfishes also ensured its well-developed vision system for scanning for prey (Fritsches, Brill and Warrant 2005). As well as possessing a long bill that can be used as a weapon, they are among the fastest teleosts at swimming short distances and are apex predators in the pelagic ocean (Dickson 1995; Watanabe et al. 2009; Lam et al. 2016).
In this study, we generated high-quality genomes for swordfish and sailfish, representing the two families of billfishes, the Xiphiidae and the Istiophoridae, respectively. We aim to: 1) validate the independent origin of endothermy in billfishes and tunas, and test whether convergent evolution occurred at the molecular level; 2) identify the most significant changes in the genomes of billfishes to investigate the mechanisms of endothermy in these species; 3) shed light on the molecular mechanisms underlying the specific phenotypes of billfishes, including their long bills, enhanced visual systems and changes in fin patterns.
Results
Chromosome-Level Assemblies and Genomic Characteristics of Billfishes
Using a combination of ONT long reads, BGI MGISEQ-2000 short reads, and Hi-C sequencing technologies (supplementary figs. S1 and S2, table S1, Supplementary Material online), we generated chromosome-level genome assemblies for sailfish (Istiophorus platypterus) and swordfish (Xiphias gladius). Both assemblies have 24 chromosomes and the size is 585.62 megabases (Mb) and 659.53 Mb for sailfish and swordfish respectively, with 98.62% and 96.64% of contigs anchored to the chromosomes (fig. 1B and supplementary fig. S3, Supplementary Material online). The assembled sizes of the two genomes are close to the genome sizes estimated by K-mer analysis (supplementary fig. S4, Supplementary Material online). The contig and chromosome N50 of the sailfish assembly are 11.30 Mb and 25.07 Mb, respectively, while they are 5.01 Mb and 27.87 Mb for the swordfish assembly, making them currently among the highest records for teleosts (supplementary tables S2 and S3, Supplementary Material online). Moreover, the genome assemblies of sailfish and swordfish have 95.70% and 97.30% complete BUSCO genes (supplementary table S3, Supplementary Material online).
A total of 26.70% and 36.35% bases were identified as repetitive sequences in the sailfish and swordfish genome respectively, consistent with the larger genome size of swordfish (fig. 1C and D, supplementary table S4, Supplementary Material online). Based on the high-quality genome assemblies, we identified totals of 22,465 and 21,632 protein-coding genes in sailfish and swordfish. The basic metrics for the genes of these two species, including gene number/length, exon number/length, and codon usage, are comparable to those of other teleost species (supplementary figs. S5–S7, supplementary table S5, Supplementary Material online).
Phylogenetic Relationship Between Billfishes and Tunas
A phylogenetic tree based on whole genome data is important to give a conclusion to whether billfishes and tunas are close or distant relatives. Here, we identified 4,150 one-to-one orthologous genes by the reciprocal best hit (RBH) method within billfishes and nine other teleost species. We applied two approaches to reconstruct the phylogenetic relationship. Firstly, the amino acids/codons/4D sites (Fourfold Degenerate Synonymous Site) of the 4,150 genes were concatenated, respectively, for generating maximum likelihood (ML) trees (supplementary figs. S8 and S9, Supplementary Material online). Then, we built the ML trees for each gene and applied the summary coalescent method to reconstruct the species tree for the 11 teleosts (supplementary fig. S9, Supplementary Material online). The reconstructed topologies from the above two methods are all consistent with the topology revealed by the mitochondrial genome in previous studies, that billfishes are closest to yellowtail amberjack (Seriola lalandi dorsalis) and then to barramundi perch (Lates calcarifer) (Little et al.2010; Betancur-R et al. 2017). To further demonstrate the reliability of this phylogenetic tree, we inspected the heterogeneity of the gene trees by DiscoVista (Sayyari et al. 2018). The major topology frequency (0.46) of 4,150 gene trees support the independent origin of billfishes and tunas (supplementary fig. S10, Supplementary Material online), and the other two alternative topologies were with a frequency of 0.31 and 0.23, respectively, which can be explained by incomplete lineage sorting. Given that the shared endothermy is not present in lineages other than Istiophoriformes and Scombriformes of the clade Percomorphaceae (Betancur-R et al. 2017), it is clear that endothermy is a result of convergent evolution in billfishes and tunas. Besides, we also estimated the divergence time of the 11 teleost species and found the two billfishes were diverged since 66 million years ago (MYA) (supplementary fig. S11, Supplementary Material online).
Signal of Convergent Evolution at Molecular Level
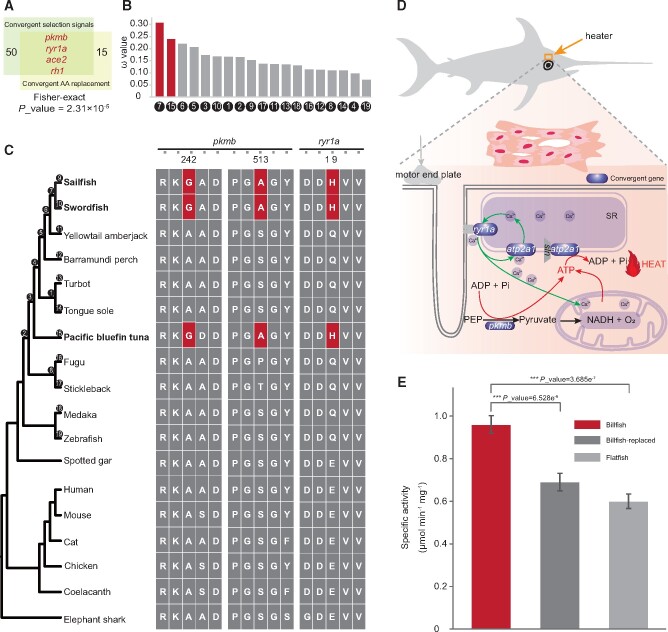
Based on the above results, the convergent evolution of endothermy between billfishes and tunas is confirmed at the level of phenotypes. To detect whether there is a convergent signal at molecular level, we have used two methods, including searching for convergent amino acid (AA) replacements as well as co-existing selection pressures. Following the convergence at conservative sites (CCS) method (Xu et al. 2017), 15 genes containing 16 convergent AA replacements were observed between billfishes and tunas (hereafter referred to asgenes with convergent AA replacement). These convergent sites were proved to be conserved in teleosts and even in all vertebrates investigated, except billfishes and tunas (supplementary figs. S12 and S13, Supplementary Material online). Simulations of AA replacements suggested that convergent events would happen at a very low level in random situations (supplementary fig. S14, Supplementary Material online). Subsequently, 50 genes were found to be under positive selection in the lineages of both billfishes and tunas (hereafter referred to asgenes with convergent positive selection signal) using the BUSTED model of HyPhy (Pond et al. 2005). In particular, four of these genes, ryr1a, pkmb, ace2 and rh1, possess both convergent AA replacements and convergent positive selection signal. Fisher’s exact test showed that the results obtained by these two methods (with convergent AA replacement or with convergent positive selection signal) were significantly correlated (P-value = 2.31 × 10−5), suggesting that the convergence phenomenon at the sequence level is not a result of random mutations (fig. 2A and B).
Fig. 2. Convergent evolution of heat production in billfishes and tunas. (A) Four genes have both positive convergent selection signals and convergent AA mutations. Fifty genes show positive convergent selection signals, and 15 genes have convergent AA mutations. Fisher’s exact test shows that the results obtained by these two methods are significantly correlated (P-value = 2.31 × 10−5). (B) The average ω (dN/dS) values for the 50 genes with positive convergent selection signals in 19 clades. (C) Pkmb has a G242A and an A513S convergent AA replacement in billfishes and tunas, which is conserved among vertebrates. Ryr1a has a Q19H AA convergent replacement, conserved among teleosts. (D) Schematic diagram of the mechanism of heat production by red muscle, where futile calcium cycles play a central role. Pyruvate kinase, encoded by pkmb, is the rate-limiting enzyme of glycolysis in muscle, supplying ATP and having a direct effect on mitochondrial metabolism. Ryr1a controls the release of calcium from the sarcoplasmic reticulum (SR), and atp2a1 is relevant to the pumping of calcium into the SR. (E) Specific activities of pkmb-billfish, pkmb-replaced and pkmb-flatfish orthologues. Data are means ± standard deviation (SD) of five independent experiments analyzed by a Student’s t-test.
Of the four genes with both a convergent AA replacement and a positive convergent selection signal, the gene pkmb, encoding pyruvate kinase, is an important enzyme for glycolysis in ATP synthesis (Mazurek et al. 1997; Dombrauckas et al. 2005). To test whether the two convergent AA replacement of pkmb in billfishes and tunas, S242A and A513G (fig. 2C and D) (located in the PK and PK_C domains, respectively), could affect its efficiency, we designed an enzyme activity experiment. Three gene sequences, pkmb-billfish (the original billfish sequence), pkmb-replaced (from the billfish sequence, but with the two convergent sites replaced by the AAs of flatfish), and pkmb-flatfish (the original flatfish sequence), were synthesized for testing their efficiency in vitro. The results of the enzyme activity experiment clearly showed that the efficiency of pkmb-billfish is significantly higher than that of pkmb-replaced and pkmb-flatfish (fig. 2E, supplementary fig. S15 Pond).
Genetic Innovation of Coding Genes
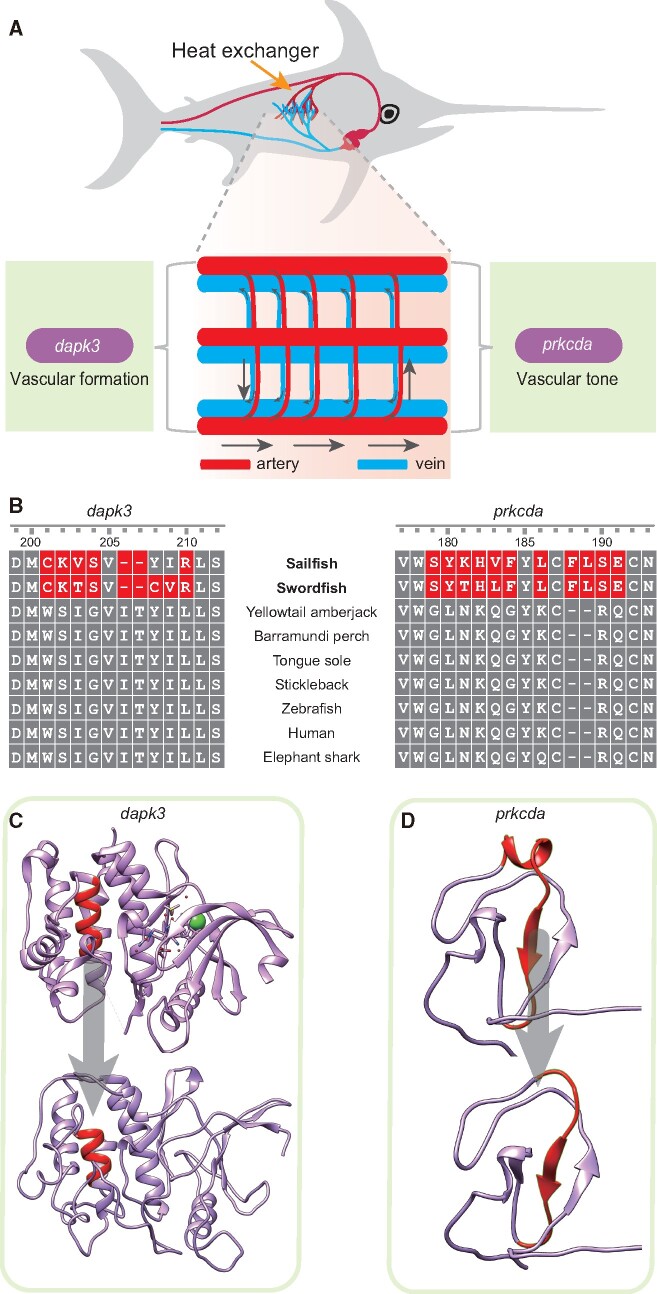
We further identified billfish lineage-specific innovations on the coding genes. Since these two billfishes have diverged for 66 million years, the shared genetic innovations that are highly conserved between them are likely to be under the pressure of purifying selection, and thus is more likely to be functionally important. Within the ortholog genes, nine genes were identified as positively selected genes (PSGs) at the billfish lineage (FDR adjusted P-value < 0.01, supplementary table S6 Pond) using the branch-site model of PAML (Yang 2007). Meanwhile, by synteny alignment, three motifs in coding genes were found to be highly diverged in billfishes, but are conserved in teleosts and even in vertebrates (supplementary figs. S16–S19 Pond). These three billfish-specific motifs are located within three genes, which are also the three PSGs with most significant P-values. The first of the three genes, dapk3, was found to be with a motif of 7 AAs (from the 201st to the 210th) in billfishes (fig. 3A and B, supplementary figs. S16 and S17, Supplementary Material online), and this is likely to affect the three-dimensional structure (fig. 3C). The second of the three significantly changed genes, prkcda, was observed with 11 (from the 179th to the 191st) billfish-specific AA replacements (fig. 3A and B, supplementary figs. S16 and S18, Supplementary Material online), and this also could affect the three-dimensional structure (fig. 3D). The last of the three genes, rfx4, had a motif of five continuous amino acids (from the 224th to the 228th) (supplementary figs. S16 and S19, Supplementary Material online). In addition to these three most special genes, the PSGs also include ggps1, with three positively selected sites (28th and 204th to 205th) (supplementary fig. S20, Supplementary Material online), and rp2, with two positively selected sites (279th and 310th) (supplementary fig. S21, Supplementary Material online). Besides, the gene families analysis revealed a total of 23/53 expanded/contracted gene families in the common ancestor (CA) of billfishes. The most significant expansion was in the fpr1 gene family (supplementary fig. S22, Supplementary Material online), which is important for innate immune system that operates in host defense and damage control (Dorward et al. 2015). We also identified 20 new genes that are of unknown function but are highly conserved in the two billfish genomes (supplementary table S7, Supplementary Material online).
Fig. 3. The genetic innovations associated with the heat exchanger. (A) Schematic diagram of the billfish heat exchanger. Red represents arteries. Blue represents veins. The directions of arrows indicate the directions of blood flow. (B) Sequence alignments of dapk3 and prkcda among vertebrates. (C) 3D structure of human dapk3 protein and structure simulated by a homologous approach for human dapk3 protein with the 7 AAs (from the 201st to the 210th) that are replaced in billfish, and these changes may be associated with the formation of vascular smooth muscle cells. (D) Three-dimensional structure of the C1 domain from human prkcd protein and structure simulated by a homologous approach of C1 domain for human prkcd protein with the 11 continuous (from the 179th to the 191st) billfish-specific AA replacements, which may promote the regulation of vascular tone and altered permeability for better heat exchange.
Divergent Conserved Non-Coding Elements (CNEs)
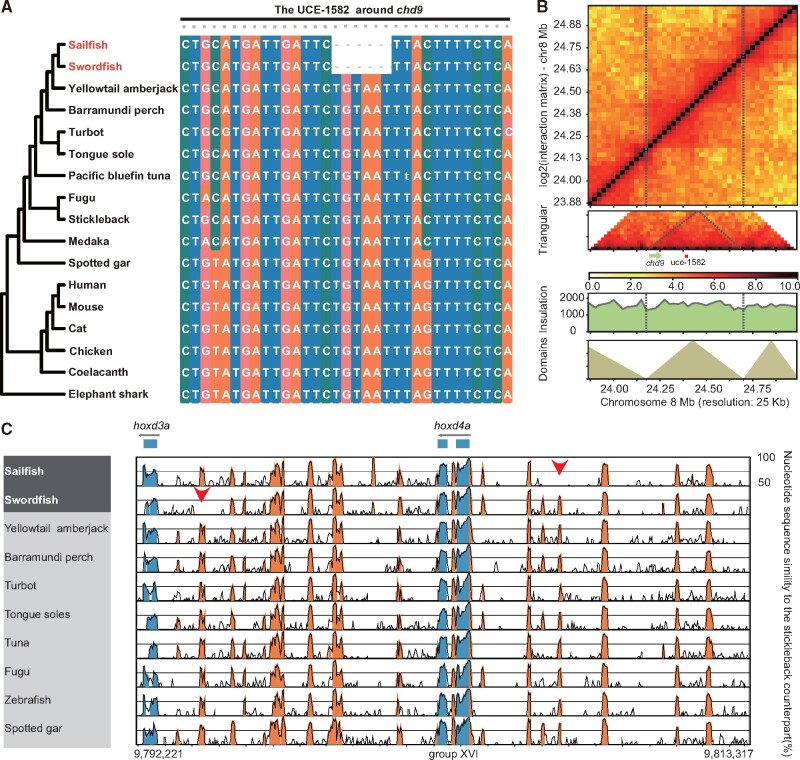
CNEs are usually cis-regulatory elements, including enhancers, repressors and insulators, in vertebrate genomes. A large number of CNEs were found to act as important roles in various functions (Navratilova et al. 2009; Attanasio et al. 2013; Lin et al. 2016), by regulating neighbor or distant genes (McLean et al. 2010). Through synteny alignment, we identified 13,451 conserved non-coding elements across nine teleosts (hereafter referred to as teleost-CNEs) (supplementary table S8, Supplementary Material online). Most of teleost-CNEs (13,159) could be well aligned against both billfish genomes with 95% coverage and 70% identity to the reference, only 32 and 36 of these CNEs were mis-assembled in the genomes of sailfish and swordfish, respectively, implying the high completeness of the billfishes’ genome assemblies. Within the teleost-CNEs, two of them were found to have billfish-specific deletions, 263 are specifically lost in swordfish and 29 are specifically lost in sailfish. For the two teleost-CNEs with billfish specific deletions, one (UCE-1582,) is located 172 kb upstream of chd9 with a 6-bp specific deletion in billfishes, and another is around bhlhb5. The UCE-1582 and its neighboring gene chd9 are also in the same topologically associating domain (TAD) predicted by Hi-C data (fig. 4A and B, supplementary fig. S23, Supplementary Material online). For the specifically lost CNEs, one CNE about 2 kb upstream of hoxd3a and one CNE about 3 kb upstream of hoxd4a were absent from swordfish and sailfish, respectively (fig. 4C), in comparison to the fact that the protein-coding genes and most CNEs within hox gene clusters are conserved in billfishes (supplementary fig. S24, Supplementary Material online). Besides, one CNE (∼190 bp) upstream of rpz was also found to be lost in the sailfish (supplementary fig. S25, Supplementary Material online). Moreover, six newly originated potential regulatory elements were identified in billfishes, and found to be highly conserved in the two billfish genomes but are absent in other vertebrates (supplementary table S9, Supplementary Material online). The above-mentioned genetic innovations are likely to be closely associated with the specific phenotypes of billfishes, as will be discussed in the sections below.
Fig. 4.
The long bill and fin specialization of billfishes. (A) A 6-bp billfish specific deletion islocated in UCE-1582, which is very highly conserved across vertebrates. It is a possible regulator of its nearest target gene chd9. (B) Hi-C maps for sailfish on chr 8 from 23.88 Mb to 25.00 Mb with 25,000 bp resolution; UCE-1582 and its nearest gene chd9 are in the same predicted TAD. (C) VISTA plots of the hoxd3a-hoxd4a cluster from 11 fishes; the stickleback sequence (ENSEMBL 98) was used as the reference to predict conserved regions with ≥ 70% identity and spanning ≥ 50 bp. Sequence alignment showed that one CNE has been lost at about 2,000 bp upstream of hoxd3a in swordfish, and another CNE has been lost at about 3,000 bp upstream of hoxd4a in sailfish, compared with the other 10 teleosts. Red arrows indicate missing CNEs.
Discussion
Convergent Evolution for Heat Production in Billfishes and Tunas
Previous studies show that the endothermy has been appeared at least three times in teleosts, including Istiophoriformes (billfishes), Scombriformes (tunas and butterfly kingfish) and Lampriformes (ophas) (Dickson and Graham 2004). Our phylogeny analysis based on whole genome data also validated the topology that billfishes and tunas are only distant relatives (Little et al. 2010; Betancur et al. 2017). Therefore, the endothermy in billfishes and tunas is indeed the result of convergent evolution at least at the level of morphology (Little et al. 2010; Betancur et al. 2017). Here, through genomic scanning, we further observed clear signals of convergent evolution at the molecular level. In particular, the convergent genes, that with convergent AAs replacement and/or positively selected signal, are highly related to heat production. Previous studies indicated that calcium futile cycles play a central role in heat production (Block 1991; Periasamy et al. 2017). The calcium futile cycles start with the nerve impulses, which initiate the release of calcium from the SR (the sarcoplasmic reticulum) to increase its concentration in the cytoplasm (Nowack et al. 2017; Periasamy et al. 2017). While one part of the ATPase (SERCA1) present in the SR transports Ca2+ from the cytosol to the SR, the other part of SERCA1 binds to sarcolipin, which prevents calcium recapture by the SR and allows the hydrolysis of ATP to generate heat through futile cycling (fig. 2D) (Block 1991; Rowland et al. 2015; Nowack et al. 2017; Periasamy et al. 2017). Meanwhile, the accumulation of Ca2+ stimulates ATP synthesis in mitochondria (Block 1991; Rowland et al. 2015; Nowack et al. 2017; Periasamy et al. 2017). Of the components of this process, pyruvate kinase, encoded by pkmb, is the rate-limiting enzyme for glycolysis in muscle, supplying it with ATP and having a direct effect on mitochondrial metabolism (Mazurek et al. 1997; Dombrauckas et al. 2005). The efficiency of pyruvate kinase has an important influence on the process of heat production, through enzyme activity experiments, we verified that the two convergent replacements of pkmb in billfishes and tunas could increase its efficiency (fig. 2E). Furthermore, the convergent replacement of the gene ryr1a, encoding calcium channel protein Ryr1a, may affect the efficiency of calcium pumping out of the SR. In addition, the atp2a1 and atp2a1l genes, which are relevant to pumping Ca2+ into the SR, were found to show a convergent positive selected signal in billfishes and tunas, and this may promote their efficiency. These convergently altered genes in heat production related pathways together contribute to the calcium futile cycle process for heat production (fig. 2D).
Genetic Innovations Associated with Heat Exchanger
Some of the billfish lineage-specific genetic innovations were found to be associated with other aspects of endothermy. The countercurrent heat exchangers, which consist of blood vessels, are crucial for endothermic teleosts for heat conserving. They can transport the vital heat being produced by heater organs and medial red muscle to the brain, eyes and swimming muscles, and prevent it from being dissipated at the gills (Carey and Teal 1966). The basic component of a countercurrent heat exchanger is two closely adjacent blood vessels, vein and artery, with different directions of blood flow; the heat is transferred from one vessel to its neighbor (fig. 3A) (Carey 1982; Brill et al. 1994). Interestingly, two PSGs of the three most special PSGs in billfishes are related to the genesis and regulation of the vasculature, respectively. The first PSG, dapk3, can mediate vascular structural remodeling via stimulation of the proliferation and migration of vascular smooth muscle cells (Usui et al. 2014). The change in dapk3 may be associated with the formation of vascular smooth muscle cells. The second PSG, prkcda, encodes Protein Kinase C delta, which is a regulator of vascular smooth muscle function and helps to maintain vascular tone (Malavez et al. 2011; Carracedo et al. 2014; Ringvold and Khalil 2017). In addition, for the six billfish-originated CNEs, one is located 13 kb downstream of fermt2, which encodes the protein kindlin-2. Previous work suggested that kindlin-2 interacts with endothelial adherens junctions to support vascular barrier integrity (Pluskota et al. 2017) and a knockdown of this gene increases neovascular permeability (Ying et al. 2018). The change in prkcda and the new CNE at the downstream of fermt2 could therefore be associated with the regulation of vascular tone and altered permeability for better heat exchange.
Potential Change in Thermoregulation
Thermoregulation is a key feature in warm-blooded animals. Without this capacity, they would not be able to cope with the external temperature changes brought about by the day-night cycle and environmental changes (Wheeler 1994; Westhoff et al. 2016). In mammals, the suprachiasmatic nucleus (SCN), a tiny region of the brain, is essential for controlling body temperature in response to the day-night cycle (Guzmán-Ruiz et al. 2015). The SCN can regulate body temperatures by releasing different hormones according to the level of daylight (Guzmán-Ruiz et al. 2015). However, in teleosts, it remains unclear how endothermic fishes regulate their body temperature. Coincidentally, the product of the last of the three special PSGs, rfx4, had been found is localized in the SCN of mouse (Araki et al. 2004). Although there is currently no research on this domain of rfx4, it is to be highly conserved across all the vertebrates investigated, including cartilaginous fishes and bony fishes. It is possible that the specific change in billfish rfx4 is related to the independent appearance of thermoregulatory capacity (supplementary fig. S19, Supplementary Material online). This result implies that SCN may be an important tissue for thermoregulation not only in mammals, but possibly also in teleosts, and the function of rfx4 in thermoregulation merits study in the future.
Characteristics of Aquatic Endothermic Vertebrates
The genetic innovations described above are consistent insights with previous studies, which have shown that the well-developed skeletal muscle system, completely closed circulatory system, and complex nervous system have laid the ground for the independent evolution of endothermy vertebrates (Reiber and McGaw 2009). One of the special features of aquatic endotherms is that the skeletal muscle of some lineages, such as billfishes and lamnid sharks, had evolved into highly efficient specialized heat organs (Ruben 1995; Bernal et al. 2001; Hedrick and Hillman 2016; Nowack et al. 2017). The other special feature is the special counter-current exchanger in many aquatic endotherms, such as endothermic teleosts, lamnid sharks, platypus, and leatherback sea turtle (Scholander 1957; Smith 1964; Grant and Dawso 1978; Bernal et al. 2001; Katz 2002; Davenport et al. 2015). In addition, compared to aquatic ectotherms, aquatic endotherms are usually characterized by a very active lifestyle. The endothermic teleosts and lamnid sharks are highly specialized pelagic predators with rapid and sustained swimming (Dickson and Graham 2004). Some studies on large extinct marine reptiles, such as mosasaurs, plesiosaurs and ichthyosaurs suggested that they also had similar characteristics (Bernard et al. 2010; Harrell et al. 2016; Fleischle et al. 2018). This may be an efficient and cost-saving adaptation strategy for aquatic endothermic vertebrates in response to the high heat capacity of the aqueous environment and the rapid heat loss.
Because endothermy is likely the evolutionary byproduct of many independent characters (Hayes and Garland 1995; Seebacher 2020), some adaptation strategies may differ between lineages. Taking body size as an example, while the endothermic capacity is enhanced by body size reduction in birds, endothermic teleosts, and chondrichthyans usually have large body sizes (Block 1991; Tullis et al. 1991; Dickson and Graham 2004). Recent studies on bird endotherms have revealed that a number of constraints might affect the endothermy strategy in birds, such as small body size, reduction in genome size and intron size, smaller cell size, and higher aerobic capacity (Waltari and Edwards 2002; Lee et al. 2014; Benson et al. 2018; Rezende et al. 2020). This implies that although we identified convergent evolution between two endothermic teleost lineages, this would not necessarily apply to other lineages. Further studies are still required to give a comprehensive understanding of aquatic ectotherms.
Enhanced Visual System in Billfishes and Pacific Bluefin Tuna
As the top predators in the pelagic ocean, billfishes, and pacific bluefin tuna have large eyes and excellent vision, both of which are vital for hunting (Fritsches et al. 2003; Matsumoto et al. 2012; Nakamura et al. 2013). In the dim light, rhodopsin (RH1) is the main receptor for acquiring visual information (Musilova et al. 2019), which is activated by phosphorylation of the 11-cis-retinal, and its sensitivity is affected by mutation (Berry et al. 2016, 2019). The convergent Y274W AA replacement in rh1 may promote their sensitivity under low light.
A previous study indicated that the high density and large dimensions of cone photoreceptors is one of the driving forces for the optical sensitivity of billfishes (Fritsches et al. 2003). The gene prdm1, which encodes Blimp1, controls the decision of cell fate in nascent photoreceptor cells. The bipolar cells are generated precociously in the absence of blimp1 (Brzezinski et al. 2010). We observed a gene duplication event for prdm1 in the common ancestor of billfishes (supplementary fig. S26, Supplementary Material online), and this may have helped billfishes to acquire a higher density of photoreceptor cells as compared to other more bipolar cells. Moreover, the PSG rp2, which can affect the development of photoreceptor cells (Li et al. 2015), was found to have two billfish-specific AA replacements which may be related to the large size of cone photoreceptors in these species.
Specialization of Bill and Fins in Billfishes
The most notable feature of billfishes is their spear-like snout or bill, which is a powerful weapon in predation. Combined with their distinctive swimming speed, the sharp bill can help them to pierce the bodies of agile prey (Kurvers et al. 2017). The bill of billfishes results from the specialization of the upper jaw bone. Previous studies have found that the gene runx2 was a major driver of craniofacial evolution in mammals, while chd9 was found to be a potential regulator for runx2 (Newton and Pask 2020). Interestingly, among the two CNEs with billfish-specific deletions, one (UCE-1582) is located 172 kb upstream of chd9 and they are also in the same TAD (fig. 4B and C, supplementary fig. S23, Supplementary Material online). It is possible that this deletion is related to the elongated bill in billfishes. Furthermore, the bill is vulnerable to damage during the capture of prey (supplementary fig. S27, Supplementary Material online), and billfishes have the ability to fully regenerate the bill (Atkins et al. 2014). Consistent with a previous study which reports that inhibition of ggps1 can accelerate the healing of bone fractures by activating the Bmp2-dependent Runx2 pathway (Dai et al. 2018), we also observed that ggps1 in billfishes was under positive selection. The three positively selected sites (28th and 204th to 205th) (supplementary fig. S21, Supplementary Material online), which are also conserved among vertebrates but billfishes, are on the “polyprenyl_synt” domain of ggps1, and this may be correlated with bill regeneration.
Although both the sailfish and swordfish are among the fastest teleosts at swimming short distances (Dickson 1995; Watanabe et al. 2009; Lam et al. 2016), each has a specific pattern of fin modifications; the sailfish has an unusually large dorsal fin, and the swordfish has lost its pelvic fin. The dorsal fin has been described as a stabilizer during steady swimming, as well as a pivot point when turning (Drucker and Lauder 2001). Pelvic fins are used as control surfaces during movement, combining with the anal and dorsal fin during turning or braking (Don et al. 2013). We have systematically scanned the hox gene clusters, which determine the anterior to posterior (A-P) polarity of early tetrapod limb buds or the fin buds of teleosts (Goodman 2002; Tarchini et al. 2006). While the genes and most CNEs of hox clusters in ray-finned fishes are conserved (supplementary fig. S24, Supplementary Material online), we found that a CNE about 2 kb upstream of hoxd3a and a CNE about 3 kb upstream of hoxd4a were absent from swordfish and sailfish respectively, deletions which may be correlated with the specific changes in the fin patterns (fig. 4D, Supplementary Material online). In addition to hox genes, the gene rpz has found to be vital for fin growth, and mutation of this gene results in zebrafish developing fin overgrowth (Goldsmith et al. 2003; Perathoner et al. 2014; Daane et al. 2018). We found that rpz is under positive selection in sailfish, with three species-specific AA mutations. And a CNE (∼190 bp) upstream of rpz was also found to be lost in the sailfish (supplementary fig. S25, Supplementary Material online). These genetic changes may be associated with the enlarged dorsal fin of sailfishes.
Conclusion
The endothermy in vertebrates is a typical case of convergent evolution and is crucial for species to overcome environmental constraints. Although there are plenty of studies related to terrestrial endothermy vertebrates, including mammals and birds, studies on aquatic endothermy vertebrates are still inadequate. In this study, the high-quality genome sequences of two billfishes validated the independent evolution of endothermy in billfishes and tunas, and also demonstrated evidence of convergent evolution at molecular level. Combining the convergent genes and lineage-specific innovations in billfishes, we found genetic changes associated with heat production, conservation and regulation might be essential for the endothermy of billfishes (supplementary fig. 28, Supplementary Material online). Meanwhile, we also revealed genetic changes underlying the specific bill, fins and eyes of billfishes. In future studies, the similarities and differences of different endothermy lineages and whether the convergent evolution of endothermy at molecular level is widespread among more distant lineages are worth to be explored.
Materials and Methods
Genome Sequencing and Assembly
Genomic DNA was extracted from the muscle and liver of a male swordfish and a male sailfish. A total of 52.51 gigabases of Nanopore reads, 36.50 gigabases of Illumina reads, and 62.90 gigabases of Hi-C reads for sailfish and 92.25 gigabases of Nanopore reads, 66.74 gigabases of Illumina reads, and 75.32 gigabases of Hi-C reads for swordfish were generated (supplementary table S1, Supplementary Material online). The Nanopore reads were obtained from the Oxford Nanopore GridIon platform. The Illumina reads (150 bp pair-end) were performed on a BGI MGISEQ-2000 with libraries produced using a TruSeqDNA PCR-Free kit. Hi-C reads were obtained on an Illumina HiSeq X Ten platform.
To predict the genome sizes of sailfish and swordfish, we firstly used SOAPec v2.0 (Luo et al. 2012) to count the kmer frequency with a kmer size of 17. The predicted genome size was around 598 Mb for sailfish and 704 Mb for swordfish (supplementary fig. S3, Supplementary Material online). The contigs were assembled by wtdbg2 v2.4.1 (Ruan and Li 2020) with standard parameters. The Illumina reads were aligned with BWA-MEM v.0.7.12-r1039 (Li 2013) with standard parameters and then polished with two rounds of NextPolish v1.0 (Hu et al. 2020), using the aligned files, which had been compressed, sorted and indexed by BWA. Finally, the assembled genomes were anchored by Hi-C reads via Juicer v1.5 (Durand, Shamim, et al. 2016) and 3 D-DNA (Dudchenko et al. 2017). Juicebox Assembly Tools (Durand, Robinson, et al. 2016) was then used to improve assembly quality. Finally, BUSCO v3.0.2 (Simão et al. 2015) was used with the library “actinopterygii_odb9”to analyze and evaluate the completeness of the gene set in our draft genome.
The mitochondrial genomes of sailfish and swordfish were assembled by NOVOPlasty v3.6 (Dierckxsens et al. 2017), based on the Illumina reads. The COI sequence of sailfish downloaded from NCBI (NC_012677) was used as the seed for the assembly.
Conservation and Syntenic Relationship with the Stickleback Genome
To evaluate the conversation and quality of the two assembled billfish genomes, we used the stickleback genome as a reference to study the synteny among these fishes. First, 24 chromosomes for both billfishes and 21 chromosomes for stickleback were aligned by LAST v942 (parameters: -E0.05) (Kielbasa et al. 2011). Second, the start and end positions of all the matching chromosomes were plotted by the python version MCscan (Tang et al. 2008).
Genome Annotation
We annotated the genomes of the two billfishes using a combination of ab initio gene predictions and homologous gene predictions (Wang et al. 2019). First, repeat sequences were annotated as follows: RepeatModeler v1.0.8 (Saha et al. 2008) was used to construct the de novo repeat library, RepeatMasker v3.3.0 (Tarailo-Graovac and Chen 2009) was used to construct the homology repeat library, and Tandem Repeats Finder v4.07 (Benson 1999) was used to match tandem repeat elements with the parameter “2 7 7 80 10 50 500 -d -h -ngs.”
After the repeat sequences had been annotated, we took the two genomes with repeat sequences soft-masked and used them to determine the coding gene structure of each. To begin with, Augustus v3.2.1 (Stanke et al. 2008), GeneID v1.4 (Alioto et al. 2018), and GlimmerHMM v3.0.4 (Majoros, Pertea and Salzberg 2004) were used to generate ab initio predictions with internal gene models. Next, the protein-coding sequences from seven species, turbot, tongue sole, yellowtail amberjack, barramundi perch, fugu,stickleback, and zebrafish (supplementary table S8, Supplementary Material online), were used to align genome sequences with tBLASTN (Altschul et al. 1990) to obtain the rough positions of homologous genes, and wise2.4.1 (Birney et al. 2004) was used for accurate positioning on the assembled genomes.Moreover, EVidenceModeler v1.1.1 (Haas et al. 2008) was run with default parameters to integrate the ab initio gene predictions and homologous gene prediction results. Finally, the gene set integrated by EVidenceModeler was translated into amino acid sequences, which were used to search for functional domains in the InterPro database with InterProScan v5.15 (Jones et al. 2014) to get Gene Ontology and PANTHER information. Then the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto 2000) was used for annotation of these gene sets.
Phylogeny Reconstruction
In addition to the two billfishes, another nine teleosts, turbot, tongue soles, yellowtail amberjack, barramundi perch, fugu,stickleback, zebrafish, medaka (all downloaded from ENSEMBL v98), and tuna (downloaded from http://nrifs.fra.affrc.go.jp; last accessed February 11, 2021) (supplementary table S5, Supplementary Material online), were used to reconstruct the phylogenetic relationships. First, OrthoFinder v2.3.4 (Emms and Kelly 2015) with default parameters and RBH method were used to cluster the homologous genes. In total, 4150 one-to-one orthologous genes were identified and aligned with MAFFT v7 (Katoh and Standley 2013), and the protein-coding sequences were also aligned by the program MAFFT to generate different sequence sets. Second, the assembled genome was divided into three datasets, the fourfold-degenerate sites (4D sites), the full amino acid sequences and the corresponding coding sequence alignments were used to construct the phylogeny trees by RAxML v8.2.4 (Stamatakis 2014). DiscoVista (Sayyari et al. 2018) was used to compute the frequency of the three topologies of ASTRAL species trees.
The mitochondrial genome sequences were divided into four groups, including the COI set, two rRNAs set, 13 amino acids set, the first base and second base of each codon set. Then all these datasets were used to construct the phylogenetic tree by RAXML v8.2.4 (Stamatakis 2014) with parameters “-m GTRGAMMA -f a -x 271828 -N 100 -p 12345.” Third, to calculate the time of divergence of both sailfish and swordfish from their relatives, MCMCTree was determined by PAML (Yang 2007) with the calibration time taken from www.timetree.org. Three softbound calibration time points were applied: tongue sole-turbot (49–81 Mya), medaka-stickleback (113–140 Mya), and medaka-zebrafish (205–255 Mya).
Demographic History
The demographic history of sailfish and swordfish was inferred with pairwise sequential Markovian coalescent (PSMC) analysis (Li and Durbin 2011),based on a substitution rate of 1.2e−9 per generation for swordfish and 2.2e−9 per generation for sailfish. We used the MEM algorithm of BWA v0.7.15 (Li 2013) to align the Illumina reads to the assembled genome with default parameters. Then the consensus diploid sequences were obtained by SAMtools and BCFtools (Li et al. 2009) with the parameters recommended by PSMC. Finally, 100 bootstraps were used to plot the effective population history.
Convergence with Tunas
To detect whether convergent signatures between billfishes and tunas could be found at the genome level, we used the dataset of aligned 4,150 one-to-one orthologues dataset to search the potential convergent amino acid replacements. The convergence at conservative sites (CCS) method (Xu et al. 2017) was used to detect the convergent sites at AA level by using the species tree of 11 teleosts. And then 10,000 simulations by PAML v4.9 (Yang 2007) were used for the ancestral sequence to eliminate the random convergence. We further applied the BUSTED model (Murrell et al. 2015) of HyPhy v2.5.15 (Pond et al. 2005) with the parameters “–code Universal –branches –tree –output” to detect the positively selected lineage for billfishes and tunas. Genes with convergent positive selection signals were selected when the adjusted P-value of the likelihood ratio tests (LRTs) for billfishes and tunas with less than 0.01. Then the ω value (non-synonymous/synonymous) of the selected gene sets was calculated by the ABSREL model with parameters “–code Universal –tree –output” based on the above tree.
Gene Family Expansion/Contraction
We used CAFE v3.1 (De Bie et al. 2006) with default parameters and the results from OrthoFinder to analyze the expansion and contraction of gene families in the two billfishes. Then the P-value of each gene family was calculated based on the results from CAFE. If the number of genes in one gene family was greater than 100, it was filtered. If the P-value was lower than 0.01, the gene family would be taken as having undergone significant expansion or contraction and others would be filtered. Pfam (Finn et al. 2016) with default parameters was used to annotate the protein-coding sequences for expanded domain analyzes. GO enrichment and KEGG enrichment were performed for the further functional analysis of these expanded and contracted gene families.
Positive Selection Analysis
To detect the evolutionary features of the billfish lineage, we used 11 teleosts (sailfish, swordfish, turbot, tongue sole, yellowtail amberjack, barramundi perch, fugu,stickleback, zebrafish, medaka, pacific bluefin tuna) to identify positive selection. All the orthologues from the results of the reciprocal best hit (RBH) method were used to test for positive selection for both sailfish and swordfish. The pipeline was as follows: one-to-one orthologues for the 11 species were aligned by PRANK v140603 (Löytynoja 2014) with parameters “-codon -f=fasta” and gaps were removed by Gblocks v0.91b (Talavera and Castresana 2007) with the parameter “-t = c.” Alignments with < 50 codons were discarded. Then sequences were converted to PAML format by a Perl script. Finally, PAML 4.9i (Yang 2007) was used for positive selection analysis with the maximum-likelihood method. The species tree generated by RAxML was used as the input tree for CodeML, with sailfish and swordfish selected as one clade. The branch-sites model in PAML was used to look for positive selection. The significance of differences between two models was evaluated using likelihood ratio tests (LRTs) by calculating twice the log-likelihood (2DL) of the difference following a chi-square distribution. Degrees of freedom were the differences in the number of free parameters between models. When the P-value of LRTs < 0.01 and at least one site had a posterior probability >0.8 according to Bayes Empirical Bayes (BEB) analysis of a gene, it would be selected as a candidate gene. Then we used the post hoc corrections for multiple testing for all the candidate genes by Benjamini and Hochberg's False Discovery Rate to decreases the false discovery rate.
Protein Structure Simulation
We first downloaded the protein structure files for the product of the human orthologue of each candidate gene from the PDB database (protein data bank, https://www.rcsb.org/; last accessed February 11, 2021). Then the relevant amino acid was modified to be that in our target gene without modification of other sites. After that, the modified sequences were simulated by Phyre2 (Kelley et al. 2015). Finally, the predicted result with the maximal score was selected as the best structure and visualized by UCSF Chimera (Pettersen et al. 2004).
Identification of Conserved Non-Coding Elements
Using the stickleback genome as a reference, whole genome alignments for 11 teleosts were generated. The genome data were downloaded from ENSEMBL (see phylogeny reconstruction for detail). First, the whole genome aligned result from LAST was submitted to the subprogram “maf-swap” of LAST (Kielbasa et al. 2011) to change the order of sequences in MAF-format alignments, then the subprogram “roast” of MULTIZ v3 (Blanchette et al. 2004) was used to extract the alignment of sequences at the same position on a chromosome in different species. Subprograms of PHAST (Siepel et al. 2005) were run for further analyzes. “msa_view” was used to identify aligned 4D sites of alignments with parameters “–in-format MAF –4d –features.”“phyloFit” (Zhou et al. 2011) was used to estimate a phylogenetic model for the 4D site datasets. “phyloBoot” (Zhou et al. 2011) was used to combine the separately estimated parameters by bootstrapping and support averaging of model parameters with parameters “–read-mods”. “phastCons” was used to estimate a model for conserved regions with parameters “–estimate-rho –no-post-probs –most-conserved –score”. After the conserved regions had been identified, ANNOVAR (Wang et al. 2010) was used to separate the coding regions and non-coding regions of each gene based on the GFF3 file of the stickleback genome. Then a series of python scripts were used to detect the divergent CNEs and divergent coding genes. The judgment criteria for the billfish specific CNEs is as follows. Firstly, CNEs with length less than 30 bp and similarity of all the 11 sequences less than 75% were filtered. For each site, if it was the same in sailfish and swordfish, and different from the corresponding site in the other nine teleosts which was the same in those species, it would be considered to be a divergent site. If the total value of the divergent site was more than 0.05%, it would be considered to be a potentially divergent CNE in sailfish and swordfish (billfis-specific CNE). Furthermore, six vertebrates (human, mouse, cat, chicken, coelacanth, and elephant shark) were used to check for conservation according to the above criteria; if the sequence was conserved in these species and all other teleosts but not sailfish and swordfish, then it would be considered to be a divergent CNE in the billfishes. Finally, we manually checked the candidate CNEs for accuracy; if the distance between the candidate CNE and the closest gene was less than 200,000 bp and the gene adjacent to the candidate CNE was the same gene in all the species, we considered the candidate CNE to be a potential cis-regulatory element for the adjacent gene.
We also used AVID v2.1 (Bray et al. 2003) to carry out alignments between each reference and query sequence with the parameters “-nm=both” to detect the lost CNEs. The input sequences were prepared by extending 50,000 bp upstream and downstream of the full-length gene. Then VISTA (Frazer et al. 2004) was used to plot the alignments. Finally, the interaction between CNEs and their nearby genes was plotted by HiCPlotter, with parameters “-pptd 1 -ptd 1,” to predict the TAD (Akdemir and Chin 2015).
Newly Original Elements
The gene sets for sailfish and swordfish were used as a query to align against 15 other vertebrate gene sets by BLASTP; if coverage > 50%, identity > 30%, or e value <e−3 was found in any gene set, the query gene was considered to have homologous ancestral sequences. After filtering, common genes for billfishes were checked against a transcriptome data set from a relative of swordfish (downloaded from http://www.swordfishomics.com/; last accessed February 11, 2021).
New CNEs were also detected by these methods. First, based on the results from LAST, these sequences conserved only for sailfish and swordfish, with > 95% identity and length > 50 bp, were used as a query to align with the other 15 vertebrate assembly genomes by BLASTN with the parameter “-word_size 6.” If coverage > 50%, identity > 30% or e value < e−3 was found in any genome, it was filtered. Then to exclude alignment errors, we extended by 100 bp at the start and end of the candidate UCE to align with more of the referencegenome using the BLASTN engine at NCBI. If the corresponding sequence could be found in this way, it was filtered.
Pkmb Protein Expression and Enzymatic Activity
Three pkmb protein sequences, pkmb-billfish, pkmb-flatfish and pkmb-replaced, were synthesized with codon optimization by FriendBio Technology Co., Ltd. (Wuhan, Hubei, China) (supplementary table S10, Supplementary Material online). The expression vectors, Frd-HIS-6736, Frd-HIS-6736M, and Frd-HIS-flatfish, which contain the coding regions of the above three sequences, respectively, were constructed using Nde I and EcoR I as described by Parkison et al. (1991) and Israelsen et al. (2013). Single colonies were picked into LB medium containing kanamycin, and 0.8 mM IPTG was used to induce expression when OD600 = 0.6 (37°C, 4 h), with uninduced Frd-HIS-6736 BL21 (DE3) and the induced Frd-HIS empty vectors as control. After centrifugation at 12,000 g for 1 min, the bacterial precipitate was collected, then an equal volume of 2× SDS-PAGE loading buffer was added and the sample was heated prior to analysis ofexpression by SDS-PAGE. Pyruvate kinase activities were measured using an NADH-LDH coupled assay with the Pyruvate Kinase Assay Kit from Beijing Solarbio Science & Technology Co., Ltd. with the addition of 10 mM fructose 1,6-bisphosphate (Ashizawa et al. 1991; Parkison et al. 1991). A spectrophotometer type 1510 from Thermo Fisher was used to measure the absorbance of a 200 μl reaction system, containing 6 μl 0.5 mg/ml pkmb protein, 20 μl fructose 1,6-bisphosphate and 174 μl buffer. The absorbance values A1 at 340 nm after 20 s and A2 after 2 min 20 s were recorded.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors are grateful for funding from the 1000 Talent Project of Shaanxi Province to Q.Q. and K.W.; the National Key Research and Development Program of China (2018YFC0309800); the National Natural Science Foundation of China (31972866 and 41876179); the Research Funds for Interdisciplinary Subject, NWPU (19SH030408) to Q.Q. and the Strategic Priority Research Program of CAS (XDB42000000) to S.H. We are also very grateful to Dr Yang Liu for his great help with our experiments, and the supports from Fish10K project.
Author Contributions
K.W. and S.H. designed and managed the project. H.J. collected the samples. M.H., W.X., Y.R., and Y.L. performed genome assembly and Hi-C chromosomal assembly. Y.Y. and Y.Z. performed the repeat annotation and gene annotation. B.W. and K.Y. performed the experiments. B.W., K.W., S.H., C.F., C.Z., W.X., W.W., and Q.Q. wrote and revised the manuscripts.
Conflict of Interest statement
The authors declare no competing interests.
Data and Code Availability Statement
The raw genome data for sailfish and swordfish have been deposited at NCBI under the project accession number PRJNA670841. The genome assembly files are under accession numbers JADGDB000000000 and JADGDC000000000.
References
- Akdemir KC, Chin L.. 2015. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 16(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioto T, Blanco E, Parra G, Guigó R.. 2018. Using geneid to identify genes. Curr Protoc Bioinformatics.64(1):e56. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Araki R, Takahashi H, Fukumura R, Sun F, Umeda N, Sujino M, Inouye ST, Saito T, Abe M.. 2004. Restricted expression and photic induction of a novel mouse regulatory factor X4 transcript in the suprachiasmatic nucleus. J Biol Chem. 279(11):10237–10242. [DOI] [PubMed] [Google Scholar]
- Ashizawa K, Willingham MC, Liang CM, Cheng SY.. 1991. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem. 266(25):16842–16846. [PubMed] [Google Scholar]
- Atkins A, Dean MN, Habegger ML, Motta PJ, Ofer L, Repp F, Shipov A, Weiner S, Currey JD, Shahar R.. 2014. Remodeling in bone without osteocytes: billfish challenge bone structure-function paradigms. Proc Natl Acad Sci U S A. 111(45):16047–16052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. 2013. Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342(6157):1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal NC, Periasamy M.. 2020. Uncoupling of sarcoendoplasmic reticulum calcium ATPase pump activity by sarcolipin as the basis for muscle non-shivering thermogenesis. Philos Trans R Soc Lond B Biol Sci. 375(1793):20190135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett I, Hester FJ.. 1964. Body temperature of yellowfin and skipjack tunas in relation to sea surface temperature. Nature 203(4940):96–97. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson RBJ, Hunt G, Carrano MT, Campione N, Mannion P.. 2018. Cope's rule and the adaptive landscape of dinosaur body size evolution. Palaeontology 61(1):13–48. [Google Scholar]
- Bernal D, Dickson KA, Shadwick RE, Graham JB.. 2001. Analysis of the evolutionary convergence for high performance swimming in lamnid sharks and tunas. Comp BiochemPhysiol A Mol Integr Physiol. 129(2–3):695–726. [DOI] [PubMed] [Google Scholar]
- Bernard A, Lécuyer C, Vincent P, Amiot R, Bardet N, Buffetaut E, Cuny G, Fourel F, Martineau F, Mazin JM, et al. 2010. Regulation of body temperature by some Mesozoic marine reptiles. Science 328(5984):1379–1382. [DOI] [PubMed] [Google Scholar]
- Berry J, Frederiksen R, Yao Y, Nymark S, Chen J, Cornwall C.. 2016. Effect of rhodopsin phosphorylation on dark adaptation in mouse rods. J Neurosci. 36(26):6973–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MH, Holt A, Salari A, Veit J, Visel M, Levitz J, Aghi K, Gaub BM, Sivyer B, Flannery JG, et al. 2019. Restoration of high-sensitivity and adapting vision with a cone opsin. Nat Commun. 10(1):1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R, Wiley EO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Ortí G.. 2017. Phylogenetic classification of bony fishes. BMC EvolBiol. 17(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14(4):708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block BA. 1991. Evolutionary novelties: how fish have built a heater out of muscle. Am Zool. 31(4):726–742. [Google Scholar]
- Block BA. 1994. Thermogenesis in muscle. Annu Rev Physiol. 56(1):535–577. [DOI] [PubMed] [Google Scholar]
- Block BA, Dewar H, Blackwell SB, Williams TD, Prince ED, Farwell CJ, Boustany A, Teo SLH, Seitz A, Walli A, et al. 2001. Migratory movements, depth preferences, and thermal biology of Atlantic bluefin tuna. Science 293(5533):1310–1314. [DOI] [PubMed] [Google Scholar]
- Block BA, Finnerty JR.. 1994. Endothermy in fishes: a phylogenetic analysis of constraints, predispositions, and selection pressures. Environ Biol Fish. 40(3):283–302. [Google Scholar]
- Block BA, Finnerty JR, Stewart AF, Kidd J.. 1993. Evolution of endothermy in fish: mapping physiological traits on a molecular phylogeny. Science 260(5105):210–214. [DOI] [PubMed] [Google Scholar]
- Bray N, Dubchak I, Pachter L.. 2003. AVID: a global alignment program. Genome Res. 13(1):97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill RW, Dewar H, Graham JB.. 1994. Basic concepts relevant to heat transfer in fishes, and their use in measuring the physiological thermoregulatory abilities of tunas. Environ Biol Fish. 40(2):109–124. [Google Scholar]
- Brzezinski JA, Lamba DA, Reh TA.. 2010. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137(4):619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey FG. 1982. A brain heater in the swordfish. Science 216(4552):1327–1329. [DOI] [PubMed] [Google Scholar]
- Carey FG, Teal JM.. 1966. Heat conservation in tuna fish muscle. Proc Natl Acad Sci U S A. 56(5):1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo S, Sacher F, Brandes G, Braun U, Leitges M.. 2014. Redundant role of protein kinase C delta and epsilon during mouse embryonic development. PLoS One 9(8):e103686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A, Pörtner HO.. 2010. Temperature, metabolic power and the evolution of endothermy. Biol Rev CambPhilosSoc. 85(4):703–727. [DOI] [PubMed] [Google Scholar]
- Daane JM, Lanni J, Rothenberg I, Seebohm G, Higdon CW, Johnson SL, Harris MP.. 2018. Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Sci Rep. 8(1):10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Li Q, Song X, Ge Y, Wu J, Zhang K, Wang C, Zhang Y, Teng H, Li C, et al. 2018. Knockdown of Ggps1 in chondrocyte expedites fracture healing by accelerating the progression of endochondral ossification in mice. J Bone Miner Metab. 36(2):133–147. [DOI] [PubMed] [Google Scholar]
- Davenport J, Jones TT, Work TM, Balazs GH.. 2015. Topsy-turvy: turning the counter-current heat exchange of leatherback turtles upside down. Biol Lett. 11(10):20150592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW.. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22(10):1269–1271. [DOI] [PubMed] [Google Scholar]
- Dickson KA. 1995. Unique adaptations of the metabolic biochemistry of tunas and billfishes for life in the pelagic environment. Environ Biol Fish. 42(1):65–97. [Google Scholar]
- Dickson KA, Graham JB. 2004. Evolution and consequences of endothermy in fishes. Physiol Biochem Zool. 77(6):998–1018. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD.. 2005. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 44(27):9417–9429. [DOI] [PubMed] [Google Scholar]
- Don EK, Currie PD, Cole NJ.. 2013. The evolutionary history of the development of the pelvic fin/hindlimb. J Anat. 222(1):114–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorward DA, Lucas CD, Chapman GB, Haslett C, Dhaliwal K, Rossi AG.. 2015. The role of formylated peptides and formyl peptide receptor 1 in governing neutrophil function during acute inflammation. Am J Pathol. 185(5):1172–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker EG, Lauder GV.. 2001. Locomotor function of the dorsal fin in teleost fishes: experimental analysis of wake forces in sunfish. J Exp Biol. 204(Pt 17):2943–2958. [DOI] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356(6333):92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL.. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3(1):99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL.. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3(1):95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischle CV, Wintrich T, Sander PM.. 2018. Quantitative histological models suggest endothermy in plesiosaurs. PeerJ.6:e4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I.. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server):W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsches KA, Brill RW, Warrant EJ.. 2005. Warm eyes provide superior vision in swordfishes. Curr Biol. 15(1):55–58. [DOI] [PubMed] [Google Scholar]
- Fritsches KA, Marshall NJ, Warrant EJ.. 2003. Retinal specializations in the blue marlin: eyes designed for sensitivity to low light levels. Mar Freshwater Res. 54(4):333–341. [Google Scholar]
- Goldsmith MI, Fisher S, Waterman R, Johnson SL.. 2003. Saltatory control of isometric growth in the zebrafish caudal fin is disrupted in long fin and rapunzel mutants. Dev Biol. 259(2):303–317. [DOI] [PubMed] [Google Scholar]
- Goodman FR. 2002. Limb malformations and the human HOX genes. Am J Med Genet. 112(3):256–265. [DOI] [PubMed] [Google Scholar]
- Grant TR, Dawso TJ.. 1978. Temperature regulation in the platypus, ornithorhynchus anatinus: production and loss of metabolic heat in air and water. Physiol Zool. 51(4):315–332. [Google Scholar]
- Guzmán-Ruiz MA, Ramirez-Corona A, Guerrero-Vargas NN, Sabath E, Ramirez-Plascencia OD, Fuentes-Romero R, León-Mercado LA, BasualdoSigales M, Escobar C, Buijs RM.. 2015. Role of the suprachiasmatic and arcuate nuclei in diurnal temperature regulation in the rat. J Neurosci. 35(46):15419–15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR.. 2008. Automated eukaryotic gene structure annotation using evidencemodeler and the program to assemble spliced alignments. Genome Biol. 9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell TL, Pérez-Huerta A, Suarez CA.. 2016. Endothermic mosasaurs? Possible thermoregulation of Late Cretaceous mosasaurs (Reptilia, Squamata) indicated by stable oxygen isotopes in fossil bioapatite in comparison with coeval marine fish and pelagic seabirds. Palaeontology 59(3):351–363. [Google Scholar]
- Hayes JP, Garland TJ.. 1995. The evolution of endothermy: testing the aerobic capacity model. Evolution 49(5):836–847. [DOI] [PubMed] [Google Scholar]
- Hedrick MS, Hillman SS.. 2016. What drove the evolution of endothermy? J Exp Biol. 219(3):300–301. [DOI] [PubMed] [Google Scholar]
- Hu J, Fan J, Sun Z, Liu S.. 2020. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics 36(7):2253–2255. [DOI] [PubMed] [Google Scholar]
- Hulbert AJ, Else PL.. 2000. Mechanisms underlying the cost of living in animals. Annu Rev Physiol. 62(1):207–235. [DOI] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. 2013. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155(2):397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUPS Thermal Commission. 2001. Glossary of terms for thermal physiology. Jpn J Physiol. 51(2):245–280. [Google Scholar]
- Johnson GD. 1986. Scombroid phylogeny: an alternative hypothesis. B Mar Sci. 39(1):1–41. [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.. 2000. KEGG: kyotoencyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz SL. 2002. Design of heterothermic muscle in fish. J Exp Biol. 205(Pt 15):2251–2266. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10(6):845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC.. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21(3):487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteja P. 2004. The evolution of concepts on the evolution of endothermy in birds and mammals. PhysiolBiochem Zool. 77(6):1043–1050. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Kurvers RHJM, Krause S, Viblanc PE, Herbert-Read JE, Zaslansky P, Domenici P, Marras S, Steffensen JF, Svendsen MBS, Wilson ADM, et al. 2017. The evolution of lateralization in group hunting sailfish. Curr Biol. 27(4):521–526. [DOI] [PubMed] [Google Scholar]
- Lam CH, Galuardi B, Mendillo A, Chandler E, Lutcavage ME.. 2016. Sailfish migrations connect productive coastal areas in the West Atlantic Ocean. Sci Rep. 6(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Cau A, Naish D, Dyke GJ.. 2014. Dinosaur evolution. Sustained miniaturization and anatomical innovation in the dinosaurian ancestors of birds. Science 345(6196):562–566. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at http://arxiv.org/abs/1303.3997.
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Rao KN, Zheng-Le Y, Hurd TW, Lillo C, Khanna H.. 2015. Loss of retinitis pigmentosa 2 (rp2) protein affects cone photoreceptor sensory cilium elongation in mice. Cytoskeleton 72(9):447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Fan S, Zhang Y, Xu M, Zhang H, Yang Y, Lee AP, Woltering JM, Ravi V, Gunter HM, et al. 2016. The seahorse genome and the evolution of its specialized morphology. Nature 540(7633):395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little AG, Lougheed SC, Moyes CD.. 2010. Evolutionary affinity of billfishes (Xiphiidae and Istiophoridae) and flatfishes (Plueronectiformes): independent and trans-subordinal origins of endothermy in teleost fishes. Mol Phylogenet Evol. 56(3):897–904. [DOI] [PubMed] [Google Scholar]
- Lovegrove BG. 2017. A phenology of the evolution of endothermy in birds and mammals. Biol Rev. 92(2):1213–1240. [DOI] [PubMed] [Google Scholar]
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 1079:155–170. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler.GigaScience 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL.. 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20(16):2878–2879. [DOI] [PubMed] [Google Scholar]
- Malavez Y, Gonzalez-Mejia ME, Doseff AI.. 2011. PRKCD (protein kinase C, delta). Atlas Genet Cytogenet Oncol Haematol. 13(1):28–42. [Google Scholar]
- Matsumoto T, Okada T, Sawada Y, Ishibashi Y.. 2012. Visual spectral sensitivity of photopic juvenile Pacific bluefin tuna (Thunnus orientalis). Fish PhysiolBiochem. 38(4):911–917. [DOI] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Eigenbrodt E.. 1997. The role of phosphometabolites in cell proliferation, energy metabolism, and tumor therapy. J BioenergBiomembr. 29(4):315–330. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G.. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 28(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, et al. 2015. Gene-wide identification of episodic selection. Mol Biol Evol. 32(5):1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova Z, Cortesi F, Matschiner M, Davies WIL, Patel JS, Stieb SM, de Busserolles F, Malmstrøm M, Tørresen OK, Brown CJ, et al. 2019. Vision using multiple distinct rod opsins in deep-sea fishes. Science 364(6440):588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Mori K, Saitoh K, Oshima K, Mekuchi M, Sugaya T, Shigenobu Y, Ojima N, Muta S, Fujiwara A, et al. 2013. Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna. Proc Natl Acad Sci U S A. 110(27):11061–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova P, Fredman D, Hawkins TA, Turner K, Lenhard B, Becker TS.. 2009. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev Biol. 327(2):526–540. [DOI] [PubMed] [Google Scholar]
- Newton AH, Pask AJ.. 2020. CHD9 upregulates RUNX2 and has a potential role in skeletal evolution. BMC Mol Cell Biol. 21(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack J, Giroud S, Arnold W, Ruf T.. 2017. Muscle non-shivering thermogenesis and its role in the evolution of endothermy. Front Physiol. 8:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkison C, Ashizawa K, McPhie P, Lin KH, Chen SY.. 1991. The monomer of pyruvate kinase, subtype M1, is both a kinase and a cytosolic thyroid hormone binding protein. BiochemBiophys Res Commun. 179(1):668–674. [DOI] [PubMed] [Google Scholar]
- Perathoner S, Daane JM, Henrion U, Seebohm G, Higdon CW, Johnson SL, Nusslein-Volhard C, Harris MP.. 2014. Bioelectric signaling regulates size in zebrafish fins. PLoSGenet. 10(1):e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy M, Herrera JL, Reis FCG.. 2017. Skeletal muscle thermogenesis and its role in whole body energy metabolism. Diabetes MetabJ. 41(5):327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J ComputChem. 25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Pluskota E, Bledzka KM, Bialkowska K, Szpak D, Soloviev DA, Jones SV, Verbovetskiy D, Plow EF.. 2017. Kindlin-2 interacts with endothelial adherens junctions to support vascular barrier integrity. J Physiol. 595(20):6443–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. [DOI] [PubMed] [Google Scholar]
- Reiber CL, McGaw IJ.. 2009. A review of the “open” and “closed” circulatory systems: new terminology for complex invertebrate circulatory systems in light of current findings. Int J Zool. 2009:1–8. [Google Scholar]
- Rezende EL, Bacigalupe LD, Nespolo RF, Bozinovic F.. 2020. Shrinking dinosaurs and the evolution of endothermy in birds. Sci Adv. 6(1):eaaw4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvold HC, Khalil RA.. 2017. Advances in Pharmacology Protein kinase C as regulator of vascular smooth muscle function and potential target in vascular disorders. New York: Academic Press. p.203–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland LA, Bal NC, Periasamy M.. 2015. The role of skeletal-muscle-based thermogenic mechanisms in vertebrate endothermy. Biol Rev. 90(4):1279–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H.. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17(2):155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben J. 1995. The evolution of endothermy in mammals and birds: from physiology to fossils. Annu Rev Physiol. 57(1):69–95. [DOI] [PubMed] [Google Scholar]
- Runcie RM, Dewar H, Hawn DR, Frank LR, Dickson KA.. 2009. Evidence for cranial endothermy in the opah (Lampris guttatus). J Exp Biol. 212(4):461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Bridges S, Magbanua ZV, Peterson DG.. 2008. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 36(7):2284–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayyari E, Whitfield JB, Mirarab S.. 2018. DiscoVista: interpretable visualizations of gene tree discordance. Mol Phylogenet Evol. 122:110–115. [DOI] [PubMed] [Google Scholar]
- Scholander PF. 1957. The wonderful net. Sci Am. 196(4):96–110. [Google Scholar]
- Seebacher F. 2020. Is endothermy an evolutionary by-product? Trends EcolEvol. 35(6):503–511. [DOI] [PubMed] [Google Scholar]
- Sepulveda CA, Dickson KA, Frank LR, Graham JB.. 2007. Cranial endothermy and a putative brain heater in the most basal tuna species, Allothunnus fallai. J Fish Biol. 70(6):1720–1733. [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith RE. 1964. Thermoregulatory and adaptive behavior of brown adipose tissue. Science 146(3652):1686–1689. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Stoehr A, Martin JS, Aalbers S, Sepulveda C, Bernal D.. 2018. Free-swimming swordfish, Xiphias gladius, alter the rate of whole body heat transfer: morphological and physiological specializations for thermoregulation. ICES J Mar Sci. 75(2):858–870. [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH.. 2008. Synteny and collinearity in plant genomes. Science 320(5875):486–488. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N.. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics.25(1):4–10. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Duboule D, Kmita M.. 2006. Regulatory constraints in the evolution of the tetrapod limb anterior-posterior polarity. Nature 443(7114):985–988. [DOI] [PubMed] [Google Scholar]
- Tullis A, Block BA, Sidell BD.. 1991. Activities of key metabolic enzymes in the heater organs of scombroid fish. J Exp Biol. 161(1):383–403. [DOI] [PubMed] [Google Scholar]
- Usui T, Sakatsume T, Nijima R, Otani K, Kazama K, Morita T, Kameshima S, Okada M, Yamawaki H.. 2014. Death-associated protein kinase 3 mediates vascular structural remodelling via stimulating smooth muscle cell proliferation and migration. Clin Sci (Lond). 127(8):539–548. [DOI] [PubMed] [Google Scholar]
- Waltari E, Edwards SV.. 2002. Evolutionary dynamics of intron size, genome size, and physiological correlates in archosaurs. Am Nat. 160(5):539–552. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Shen Y, Yang Y, Gan X, Liu G, Hu K, Li Y, Gao Z, Zhu L, Yan G, et al. 2019. Morphology and genome of a snailfish from the Mariana Trench provide insights into deep-sea adaptation. Nat Ecol Evol. 3(5):823–833. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Kubodera T, Yokawa K.. 2009. Feeding ecology of the swordfish Xiphias gladius in the subtropical region and transition zone of the western North Pacific. Mar Ecol Prog Ser. 396:111–122. [Google Scholar]
- Westhoff JT, Paukert C, Ettinger-Dietzel S, Dodd H, Siepker M.. 2016. Behavioural thermoregulation and bioenergetics of riverine smallmouth bass associated with ambient cold-period thermal refuge. Ecol Freshw Fish. 25(1):72–85. [Google Scholar]
- Wheeler PE. 1994. The thermoregulatory advantages of heat storage and shade-seeking behaviour to hominids foraging in equatorial savannah environments. J Hum Evol. 26(4):339–350. [Google Scholar]
- Xu S, He Z, Guo Z, Zhang Z, Wyckoff GJ, Greenberg A, Wu CI, Shi S.. 2017. Genome-wide convergence during evolution of mangroves from woody plants. Mol Biol Evol. 34(4):1008–1015. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Ying J, Luan W, Lu L, Zhang S, Qi F.. 2018. Knockdown of the KINDLIN-2 gene and reduced expression of kindlin-2 affects vascular permeability in angiogenesis in a mouse model of wound healing. Med Sci Monit. 24:5376–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS.. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39(suppl):W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw genome data for sailfish and swordfish have been deposited at NCBI under the project accession number PRJNA670841. The genome assembly files are under accession numbers JADGDB000000000 and JADGDC000000000.