Abstract
The field of evolutionary developmental biology can help address how morphological novelties evolve, a key question in evolutionary biology. In Arabidopsis thaliana, APETALA2 (AP2) plays a role in the development of key plant innovations including seeds, flowers, and fruits. AP2 belongs to the AP2/ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR family which has members in all viridiplantae, making it one of the oldest and most diverse gene lineages. One key subclade, present across vascular plants is the euAPETALA2 (euAP2) clade, whose founding member is AP2. We reconstructed the evolution of the euAP2 gene lineage in vascular plants to better understand its impact on the morphological evolution of plants, identifying seven major duplication events. We also performed spatiotemporal expression analyses of euAP2/TOE3 genes focusing on less explored vascular plant lineages, including ferns, gymnosperms, early diverging angiosperms and early diverging eudicots. Altogether, our data suggest that euAP2 genes originally contributed to spore and sporangium development, and were subsequently recruited to ovule, fruit and floral organ development. Finally, euAP2 protein sequences are highly conserved; therefore, changes in the role of euAP2 homologs during development are most likely due to changes in regulatory regions.
Keywords: AP2/TOE3, cooption, evo–devo, gene lineage evolution, morphological innovations, sporangium development
Introduction
One of the key questions in biology is how new structures evolve. Evolutionary developmental biology (evo–devo) which focusses on how changes in the genetic networks underlying development influence the evolution of taxa can help address this question. In particular, changes in cis- and trans- regulatory regions and variation in the protein sequences, or their interactors, have allowed the identification of preexisting genes or networks coopted for the development of novel traits (Jacob1977; Carroll 2000; Sanetra et al. 2005; Monteiro and Podlaha 2009). One powerful approach to assess how evolutionary novelties arise is the combined assessment of gene lineage evolution in parallel to the study of expression and function in a comparative manner across distantly related taxa. In plants, several morphological innovations, such as the seed, the flower, and the fruit, have evolved at different timepoints and correlate with the diversification and ecological dominance of specific plant lineages (Burleigh and Mathews 2004; Soltis and Soltis 2004; Zanne et al. 2014). Moreover, these structures are critical for plant reproduction as they provide protection and nourishment to the spores and sometimes to the zygote. To investigate how morphological innovations may evolve, we focus on the euAPETALA2 (euAP2) gene lineage known to be a key player in the development of seeds, flowers, and fruits in Arabidopsis thaliana (Arabidopsis) (Bowman et al. 1989; Jofuku et al. 1994; Yant et al. 2010; Ripoll et al. 2011).
The APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family is one of the most important gene lineages in plant development, with over 4,000 members present across Viridiplantae (Bowman et al. 1989; Jofuku et al. 1994; Ohme-Takagi and Shinshi 1995; Elliott et al. 1996; Moose and Sisco 1996; Kim et al. 2006; http://planttfdb.gao-lab.org/family.php?fam=AP2; last accessed May 14, 2020). According to the number of AP2 domains, the lineage has been divided into two classes; the AP2-like class encode proteins with two AP2 domains, namely AP2-R1 and AP2-R2, while the ERF-like class proteins only have the AP2-R1 domain (Jofuku et al. 1994; Elliott et al. 1996; Nole-Wilson et al. 2005; Yant et al. 2010; Li et al. 2016). These domains consist of three β sheets and one amphipathic α helix, which are important for DNA binding (Jofuku et al. 1994; Allen et al. 1998). Within seed plants, there are two sister clades: euAP2 and AINTEGUMENTA (ANT) (Kim et al. 2006). For the purpose of this paper, we will focus on the euAP2 lineage (sensuKim et al. 2006 followed by Zumajo-Cardona and Pabón-Mora 2016; called the AP2 group in Wang et al. 2016). This gene clade includes the canonical SCHLAFMUTZE (SMZ), SCHNARCHZAPFEN (SNZ), TARGET OF EAT1 (TOE1), TOE2, TOE3, and AP2. Gene evolution topologies from this clade recover two groups (Kim et al. 2006; Wang et al. 2016), one with TOE1/2, SMZ, and SNZ (TOE type sensuWang et al. 2016) and one with AP2 and TOE3 (AP2 type sensuWang et al. 2016). The latter subclade euAP2/TOE3 includes the two Arabidopsis paralogues AP2 and TOE3 as the result of a duplication event specific to Brassicaceae (Kim et al. 2006; Wang et al. 2016; Zumajo-Cardona and Pabón-Mora 2016). Two other major duplication events occurred in the euAP2/TOE3 lineage, one in monocots coinciding with the radiation of commelinids and another duplication previous to the diversification of basal eudicots (Zumajo-Cardona and Pabón-Mora 2016).
In Arabidopsis, AP2 is involved in the floral transition and contributes to sepal and petal identity (Koornneef et al. 1983; Jofuku et al. 1994; Yant et al. 2010). Mutant ap2 plants exhibit homeotic conversions of sepals and petals into leaf-like organs with stigmatic surfaces carrying ovules, and into stamens respectively (Pruitt et al. 1987; Komaki et al. 1988; Bowman et al. 1989; Kunst et al. 1989; Drews et al. 1991). Thereafter, AP2 was included in the ABC model for flower development, as an A-class gene (Bowman et al. 1991; Coen and Meyerowitz 1991). Originally A and C-class genes were thought to be mutually antagonistic in order to mark boundaries between the perianth and the stamens/carpels (Gustafson-Brown et al. 1994; Mandel and Yanofsky 1995). This was supported by the observations of AGAMOUS ectopic expression in the ap2 mutant (Bowman et al. 1993). In wild-type Arabidopsis, downregulation of AP2 in stamens and carpels primarily occurs via microRNA miR172 activity (Zhao et al. 2007; Wollmann et al. 2010). It is likely that a miR172-AP2 regulatory module is present throughout euphyllophytes, as miR172 has been identified in angiosperms, gymnosperms, and ferns (Axtell and Bartel 2005; Zhang et al. 2006). AP2 also functions in integument development and seed coat cell patterning as well as embryo development by controlling cell size and number (Jofuku et al. 1994; Ohto et al. 2005). In addition, AP2 also controls the reproductive transition via induction of AGAMOUS-LIKE 15 expression and by repression of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (Yant et al. 2010). Finally, AP2 is part of the Arabidopsis fruit development network, controlling replum growth and valve margin formation by a direct negative regulation of SHATTERPROOF, BREVIPEDICELLUS and REPLUMLESS (Ripoll et al. 2011).
Previous studies have identified homologues of euAP2/TOE3 across seed plants for which the protein sequences appear to be more similar to AP2 than to the TOE3 protein (Kim et al. 2006; Zumajo-Cardona and Pabón-Mora 2016; Najafi et al. 2018; Sharma et al. 2018; Kerstens et al. 2020), and have highlighted the pleiotropic roles of AP2 homologues in eudicots and monocots. In snapdragon (Antirrhinum majus, Plantaginaceae), the AP2 orthologues LIPLESS1 and 2 are expressed in bracts, sepals, the distal portions of the petals, and weakly in the carpels. The double lipless1/2 mutant shows homeotic conversion from sepals to bract-like organs and smaller petals (Keck et al. 2003). In petunia, PhAP2A (also known as REPRESSOR OF B-FUNCTION1, ROB1), is expressed in leaves and in all floral organ primordia, but phap2a mutants do not show any abnormal phenotype compared with wild-type plants (Maes et al. 2001; Morel et al. 2017). Additionally, PhAP2A expression is detected in the integuments during ovule development and the endosperm during seed development (Maes et al. 2001). Two additional paralogues have been identified in petunia, ROB2 and 3. Like PhAP2A, they too are broadly expressed in all floral whorls (Morel et al. 2017). Genetic analyses indicate redundancy among the three paralogues and only the triple mutant, rob1,2,3, shows defects in sepal, petal, and carpel development (Morel et al. 2017). In tomato, there are three paralogues SlAP2a,SlAP2b,and SlAP2c. RNAinterference-mediated repression targeting the three paralogs, show mutants with longer fused sepals and exhibit yellow-orange pigmentation linked to an increased β-carotene to lycopene ratio during fruit ripening. In addition, defects in the seed coat are also observed (Chung et al. 2010; Karlova et al. 2011). SlAP2a specific CRISPR-Cas9 knockouts confirm its function in fruit ripening and show that the sepal identity function is not dependent on AP2a alone (Wang et al. 2019). Finally, in kiwi, a natural mutant with a miR172 insensitive version of euAP2 results in aberrant flowers with multiple perianth whorls (Varkonyi-Gasic et al. 2012).
The AP2 homologues in monocots influence inflorescence architecture and the transition from the spikelet meristem to the floral meristem (Chuck et al. 2008; Lee and An 2012; Bommert and Whipple 2018). euAP2/TOE3 homologues include INDETERMINATE SPIKELET 1 (IDS1) and SISTER OF INDETERMINATE SPIKELET 1 (SID1) in maize (Chuck et al. 1998), SUPERNUMERARY BRACT (SNB), OsIDS1 (Lee et al., 2007; Lee and An, 2012) and RICE STARCH REGULATOR 1 (RSR1) in rice (Fu and Xue 2010), and Q (AP2L5) and AP2L2 in wheat (Faris et al. 2003; Debernardi et al., 2020). IDS1 in maize plays a role in inflorescence architecture by controlling the number of floral meristems produced (Chuck et al. 1998,2007,2008). On the other hand, single sid1 mutants do not have different phenotypes from the wild type plants, but both IDS1 and SID1 are needed for inflorescence meristem branching, spikelet determinacy, and floral meristem identity (Chuck et al. 2008). In rice, the snb mutant has a delayed floral transition and produces additional glumes (interpreted as bracts) before the formation of the floral meristem. The snb osids1 double mutant also has chimeric floral organs, with organ defects in paleas (sepal homologues) and lodicules (petal homologues) as well as homeotic conversion of lodicules into carpels (Lee et al. 2007; Lee and An 2012). Additionally, the rice RSR1 negatively regulates starch synthesis genes during the development of the endosperm (Fu and Xue 2010). The wheat Q locus controls many domestication-related traits that facilitate its harvest, such as the free-threshing character (Simons et al. 2006; Gil-Humanes et al. 2009), rachis fragility (Jantasuriyarat et al. 2004), spike length, plant height (Kato et al. 1998,2000) and flowering time (Kato et al. 1998). More recently, Q (AP2L5) and AP2L2 were shown to control the specification of axillary floral meristems, lemmas (bracts), paleas (sepals), and lodicules (petals), with conversion of lodicules into carpels (Debernardi et al. 2020).
Functional characterization of euAP2/TOE3 homologues is lacking in early diverging angiosperms, gymnosperms, and ferns. Nevertheless, real-time (RT)-PCR has been used to assess the expression of AP2 homologues in Amborella trichopoda, where it is found in all floral organs and leaves (Kim et al. 2006). In the gymnosperms,Larix marschlinsii and Picea thunbergii (with two paralogues each: LmAP1L1, LmAP2L2 and PtAP2L1, PtAP2L2, respectively), different paralogues are differentially expressed during somatic embryogenesis and the first stages of embryo development (Vahala et al. 2001; Shigyo and Ito 2004; Guillaumot et al. 2008). PtAP2L1 and PtAP2L2 expression has been also detected in the nucellus and the integument primordia (Shigyo and Ito 2004). Finally, in Ginkgo biloba, the expression of an AP2 homologue was detected in ovules and leaves (Zumajo-Cardona and Pabón-Mora 2016). In addition, ectopic expression of Picea abies homologues, PaAP2L1, PaAP2L2, and PaAP2L3, in an ap2 mutant background in Arabidopsis, was able to promote petal identity suggesting functional conservation at the molecular level across seed plants (Nilsson et al. 2007).
With the goal of assessing how euAP2/TOE3 homologues have acquired these functions over evolutionary time, we wanted to investigate their putative plesiomorphic roles outside core eudicots and monocots. Because functional characterization techniques are currently unavailable for many of the targeted taxa, we present detailed spatiotemporal expression analyses for AP2 homologues in the basal eudicot Bocconia frutescens (Papaveraceae), the early divergent angiosperm Aristolochia fimbriata (Aristolochiaceae), two gymnosperms, Ginkgo biloba (Ginkgoaceae) and Gnetum gnemon (Gnetaceae), and one fern, Ceratopteris richardii (Pteridaceae). These comprehensive expression analyses spanning major vascular plant lineages have enabled us to show that plesiomorphic roles for AP2-like homologues include sporangium and ovule development. Only more recently, in angiosperms, have these homologues been recruited into flowering transition, floral organ identity, and fruit development. Across vascular plants, there are few changes in the protein sequences of euAP2/TOE3 homologues, therefore the cooption of euAP2/TOE3 homologues in the development of morphological novelties are most likely due to independent evolution of cis- or trans-regulatory regions.
Results
euAP2/TOE3 Gene Evolution in Vascular Plants
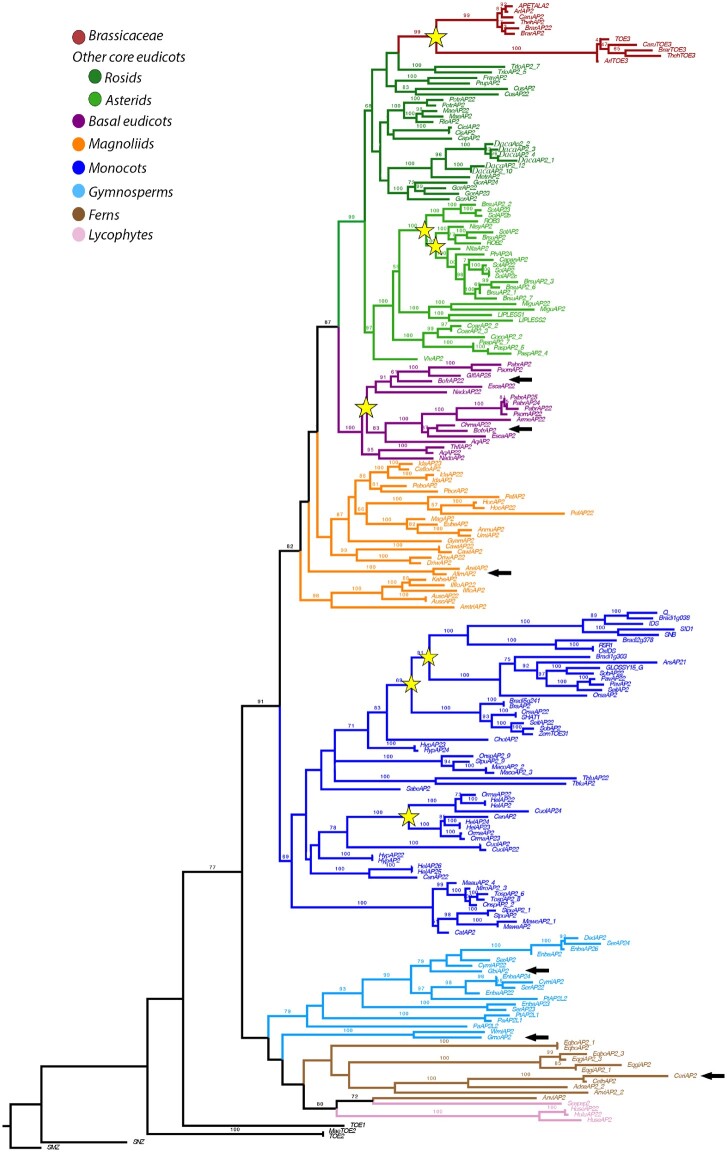
Here, we present an extended phylogenetic hypothesis of euAP2/TOE3 genes in tracheophytes. We isolated 84 genes from eudicots, 59 from monocots, 27 from early diverging angiosperms, 30 from gymnosperms, 25 from ferns, and 7 from lycophytes. The aligned matrix contained 750,384 characters. The gene tree obtained by ML analysis shows that euAP2/TOE3 homologues have remained as single copy in ferns, gymnosperms, early diverging angiosperms, and most core eudicots (fig. 1). Gene lineage duplications identified here include two major angiosperm duplication events previously described (Zumajo-Cardona and Pabón-Mora 2016), one in basal eudicots and another duplication event prior to the diversification of Brassicaceae (fig. 1). With extended sampling within Solanaceae, it was possible to identify two duplications specific to this family (fig. 1; supplementary fig. S3). Furthermore, here we present two additional duplication events within Poaceae (Bootstrap (BS) values= 89 and 79) and one corresponding to the diversification of Zingiberales (BS= 99). For the most part, the gene tree is consistent with known phylogenetic relationships among seed plants. However, the tree topology shows gymnosperm sequences forming a clade (AP2L clade sensuWang et al. 2016). In addition, we recover a clade with fern and lycophyte AP2 sequences as sister to gymnosperm homologues. Because the most recent hypothesis for vascular plants suggests the relations (lycophytes [ferns {gymnosperms + angiosperms}]), our topology may still require additional sampling to fully sort gene lineage evolution in vascular plants (fig. 1). Despite the sampling effort, no euAP2/TOE3 homologues were recovered from non-vascular plants.
Fig. 1.
ML tree of euAP2/TOE3 gene lineage across vascular plants. Yellow stars indicate large-scale duplication events. These duplications are (from top to bottom): 1) before the diversification of Brassicaceae; 2) two duplications specific to Solanaceae; 3) with the radiation of Papaveraceae; 4) two duplications before the radiation of Poales; 5) one within Zingiberales. BS values > 60% were placed at the nodes. Colors in the tree follow the top left conventions for each major group of seed plants. Black arrows point to the AP2 sequences where expression was analyzed by In Situ hybridization in each species.
The euAP2/TOE3 proteins are highly conserved across vascular plants, with the AP2-R1 and AP2-R2 binding domains present in all sequences analyzed (supplementary fig. S4; Kim et al. 2006). The miR172 binding site (AAASSFG[S/P]) was detected in ferns and seed plant homologues but not in lycophytes. When found, the miR172 motif is located at the C-terminus, except in two fern proteins, EqboAP2 and EqboAP2-1, from Equisetum bogotense, which have the miR172 motif in the N-terminus of the protein, before the AP2-R1 domain. In addition, the start motif (MW[D/N]LNxxP) is conserved and corresponds to an EAR repressor motif (DLNxxP; Ohta et al. 2001; Hiratsu et al. 2003; Kim et al. 2006; Guillaumot et al. 2008). Only the maize GLOSSY15 and a few gymnosperm, fern, and lycophyte proteins lack this EAR repressor motif. Toward the C-terminus of the AP2-R2 domain is another ERF-associated Amphiphilic Repression motif (EAR: LxLxLX; Ohta et al. 2001; Hiratsu et al. 2003) which is conserved across seed plants (Zumajo-Cardona and Pabón-Mora 2016) but is absent in GLOSSY15, most Equisetum giganteum homologues and in all lycophyte proteins (supplementary fig. S4). In general, all angiosperm protein sequences are shorter, with around 400–560 amino acids and have more conserved motifs, compared with gymnosperm, fern, and lycophyte homologues with around 550–770 amino acids and more variable regions.
In order to assess the possible roles of AP2 in plant development across tracheophytes, we carried out spatiotemporal expression analyses in selected species belonging to poorly explored plant lineages. Here, we present our results from six AP2 homologues across major vascular plant lineages namely, early diverging eudicots, early diverging angiosperms, gymnosperms, and ferns.
Expression of euAP2/TOE3 Homologues in the Basal Eudicots Eschscholzia californica and B occonia frutescens (Papaveraceae)
Two paralogues were identified in both Eschscholzia californica and Bocconia frutescens (Papaveraceae) as a result of the duplication event described above for basal eudicots (fig. 1). Eschscholzia californica flowers have two nonpersistent sepals, four petals, multiple stamens, and two carpels. In E. californica, the expression of the two paralogues named EscaAP2-1 and EscaAP2-2 was evaluated with RT-PCR and quantitative real-time (qRT)-PCR. The results show that EscaAP2-1 is more broadly expressed across different stages of flower development as well as in leaves; however, this paralogue is highly expressed in young floral buds, carpel in anthesis, and the mature fruit. Conversely, EscaAP2-2 has a more specific expression in the petals and the carpels during anthesis and the mature fruit (supplementary fig. S5).
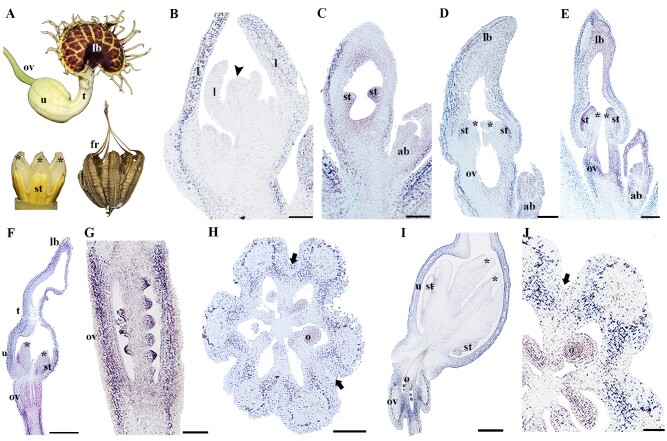
Bocconia frutescens flowers are also dimerous, with two nonpersistent sepals, apetalous, with two whorls of numerous stamens, and a bicarpellate gynoecium with a gynophore and a prominent bifurcated stigma (fig. 2A; Arango-Ocampo et al. 2016). The fruit has two valves, which at maturity fall apart leaving the single seed attached to a commissural ring-like structure (fig. 2A). Bocconia frutescens flower and fruit developmental stages have been previously described in detail (Zumajo-Cardona et al. 2017), the same stages are used here to describe the expression results obtained for the two AP2 paralogues isolated, named BofrAP2-1 and BofrAP2-2 (fig. 1). Our results show different expression patterns for the two copies. BofrAP2-1 is strongly expressed in the vegetative shoot apical meristem (SAM) and the young developing leaves (fig. 2B). BofrAP2-1 is slightly expressed in the floral primordia (fig. 2C). During early stages of flower development, BofrAP2-1 is not found to be expressed in sepals, stamens, or carpel primordia (fig. 2D and E). The expression of BofrAP2-1 is detected later in the sepals, stamens, carpel, and ovule as these start to elongate (fig. 2F) and it is maintained in the carpel tips as they overtop and close above the single ovule (fig. 2G). Later in flower development, as the style and stigma differentiate, the expression of BofrAP2-1 in the sepals is restricted to their growing tips and is maintained more homogenously in the other floral organs including stamens and carpels, as well as the single ovule (fig. 2H). In the anthetic flower, BofrAP2-1 expression becomes restricted to the epidermal layers of the carpel walls, to the growing tips of the stigma, the adaxial flanks of the massive stigmas, the transmitting tract, and the growing tips of the two integuments in the developing ovule (fig. 2Iand J). The expression of BofrAP2-1 persists in the young fruit, especifically in the medial inner region of the commissural ring, where the commissural ring meets the valve margins (fig. 2Kand L). BofrAP2-1 expression does not extend to the dehiscence zone through which the two valves split apart.
Fig. 2.
Expression analyses in Bocconia frutescens (A) of the two AP2 homologues: BofrAP2-1 and (B to L) and BofrAP2-2 (Mto U). (A:B) frutescens flower in anthesis and mature fruit.(B)Expression in the shoot apical meristem.(C to H)Floral stages3–9. (I) Stigmas of a preanthetic flower. (J) Ovary and stigma of a preanthetic flower. (K and L) Cross section of a young fruit. Note the expression is restricted to the commissural ring.(M)Leaf primordia. (N to R) Floral stages3–9. (S) Longitudinal section through the stigmas. (T and U) Cross section of a young fruit. Note the expression is restricted to the valves and commissural ring, no expression is detected in the dehiscence zone. Black arrowheads pointing to the dehiscence zone of the fruit; black arrow pointing to the shoot apical meristem; c, carpel; cr, commissural ring; fr, fruit; o, ovule; s, sepal; se, seed; st, stamen; si, stigma; sty, style; v, valve. Scales: 50 µm (C to E, N and O), 75 µm (P and Q), 100 µm (I,J, and S), 125 µm (Band M), 150 µm (F to H, R), 0.1 mm (K, T, and U).
Conversely, the paralogue, BofrAP2-2, is expressed in the SAM at lower levels compared with BofrAP2-1, and it cannot be detected in the young leaves (fig. 2M). In general, the expression of BofrAP2-2 during flower development is lower than the one identified for BofrAP2-1. BofrAP2-2 is not expressed in the floral primordia (fig. 2N), or when the stamen and carpel primordia start to develop (fig. 2O). However, BofrAP2-2 can be detected at the base and at the tip of the sepals, as well as at the growing tips of carpels as they overtop the single ovule (fig. 2P to R). The expression at the base of the sepals is maintained throughout flower development and corresponds to the sepal abscission zone active at anthesis (fig. 2P to R). When the style and stigma begin to differentiate, the expression of BofrAP2-2 marks the meeting point between the two stigmas on their adaxial flanks (fig. 2Q). This expression continues as the flower develops during stigmatic fusion (fig. 2R) and the formation of a massive style and during the differentiation of the transmitting tract (fig. 2S). Later in fruit development, BofrAP2-2 is broadly expressed in the valves, more strongly in the exocarp and the endocarp than in the mesocarp (fig. 2T). Finally, BofrAP2-2 is also found throughout the commissural ring, especially in the medial inner region, and like its paralogue it is lacking from the dehiscence zone (fig. 2T and U).
Expression of euAP2/TOE3 Homologues in the Early Divergent Angiosperm Aristolochia fimbriata (Aristolochiaceae)
The A. fimbriata flower grows solitary, axillary to each leaf. The flower consists of a perianth formed by three petaloid sepals fused to form a tubular structure divided in three regions: the proximal utricle, the medial tube, and the distal limb; petals are lacking (fig. 3A). Stamens and stigmas fuse forming a gynostemium crowning the inferior ovary (González and Stevenson 2000; Pabón-Mora et al. 2015). The fruit is an acropetally dehiscent septicidal capsule, leaving “baskets” full of seeds (fig. 3A). A single AP2 homologue was identified from the selected species A. fimbriata (fig. 1). We describe the expression results for AfimAP2, based on detailed descriptions of the developmental stages of flowers and fruits previously identified (Pabón-Mora et al. 2015). Our expression analyses show that AfimAP2 is not expressed in the SAM or in the floral bud prior to and during sepal initiation (fig. 3B). However, AfimAP2 is expressed in young leaves (fig 3B). AfimAP2 expression is detected in the sepals, in the stamen primordia, and in the ovary later in development (fig. 3C). Similar expression patterns are maintained throughout flower development, as the perianth curves and the thecae differentiate (fig. 3D and E), and later on when the utricle, tube and limb can be distinguished in the perianth (fig. 3E and F). When ovule differentiation begins, the expression of AfimAP2 can be seen in the ovary wall, in the nucellus, and in the two integuments (fig. 3G and H). In a preanthetic floral bud, AfimAP2 expression is no longer detected in the gynostemium, neither the stamens nor the stigmas, but it is maintained in the utricle and in the ovary (fig. 3I). Interestingly, AfimAP2 expression in the ovary is stronger around the main vascular trace and in the inner layers of the wall, and lacking from the septum (fig. 3H and J).
Fig. 3.
Expression of AfimAP2. (A) Aristolochia fimbriata flower in anthesis and mature fruit. (B) Inflorescence apex with floral buds in stage 1–2, note no expression floral and sepal primordia.(C) Flowers stage 3, expression in the adaxial side of the sepal, stamen primordia. (D and F) Floral buds at stages 4–7, note the expression in the ovary and ovules. (G) Ovary of a flower at stage 8.(H) Cross section through the ovary, AfimAP2 is expressed in the nucellus and integuments. (I) Utricle and ovary of a flower in preanthesis. (J) Cross section of the ovary of a flower in preanthesis. Note there is no expression in the septum. Asterisks indicate stigmas; black arrowhead pointing to the inflorescence meristem; black arrow pointing to the septum; ab, accessory bud; fr, fruit; l, leaf; lb, limb; o, ovule; ov, ovary; st, stamen; t, tube; u, utricle. Scales: 50 µm (H and I), 60 µm (J), 80 µm (G), 100 µm (B to F).
Expression of euAP2/TOE3 Homologues in the Gymnosperms: Ginkgo biloba andGnetum gnemon
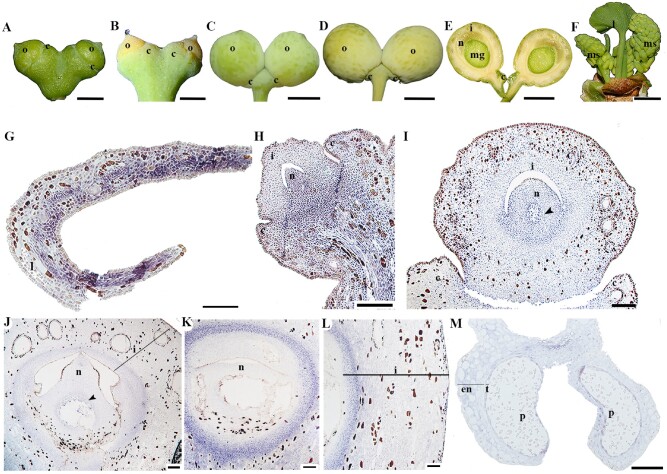
In Ginkgo biloba, the funiculus, ovule-bearing stalk, is initiated axillary to the leaf and it usually dichotomizes producing two ovules, which are characterized by having a single integument covering the nucellus. To perform the expression analyses, ovules of G.biloba at four different stages were collected. At the first stage, the ovules are immature with a thickened collar, a vegetative structure unique to Ginkgo, that develops from the hypodermal layer proximal to the ovule (Douglas et al. 2007; fig. 4A). At the second stage, when the ovules are ready to be fertilized, the integument forms a more conspicuous micropyle which releases the pollination drop (fig. 4B). The next stage is characterized by fleshy growth in the integument, largely linked to the formation of multiple mucilage canals; the collar remains as a small structure at the base of the ovules (fig. 4C). In the seed, the integument, now the fleshy testa turns yellow and ripens (fig. 4D and E). An immature pollen cone was included in the expression analysis as well (fig. 4F). One AP2 homologue was identified in G. biloba and was named GibiAP2 (fig. 1). The expression of GibiAP2 was evaluated in ovulate and pollen cones of G. biloba. One feature that is different in gymnosperms, compared with angiosperms, is that the tissue processing during the insitu hybridization experiment, results in brownish mucilage canal walls and tannin oxidation (fig. 4G to L; see also Lovisetto et al. 2013). Thus, brown color represents background and only the purple colorimetric reaction represents gene expression. GibiAP2 is expressed in the adaxial leaf surface (fig. 4G). Expression in the young developing ovule was detected in the vegetative tissues such as the stalk and collar, as well as the proximal portion of the ovule, the nucellus and slightly less in the integument (fig. 4H). As the ovule reaches the fertilization stage with a well-developed megagametophyte, two different integumentary layers are visible, the outer most layer is characterized by the presence of tannins and mucilage canals. At this point,GibiAP2 expression is detected in the integument, the nucellus, and the developing megagametophyte (fig. 4I). As the integument thickens, the expression of GibiAP2 is found in the integument and in the nucellus (fig. 4J). The same expression can be seen until the fertilized ovule transitions to a ripening seed (fig. 4K and L). In the pollen cone, weak expression of GibiAP2 is detected in the endothecium and the tapetum of the microsporangium and the microspores (fig. 4M).
Fig. 4.
Expression of GibiAP2 in Ginkgo biloba.(A to D). Ovules at different developmental stages. (E) Hand section of a mature ovule as shown in (D). (F) Young pollen cones in the axils of leaves. (G to M)GibiAP2 expression patterns. (G) Cross section of a leaf. (H to L)Ovule at different stages, note the expression in the integuments and nucellus is maintained throughout ovule development. (L) Close-up of theintegument of a fully mature ovule. (M) Pollen cone. Black arrowhead pointing to the megagametophyte; c, collar; en, endothecium; i, integument; l, leaf; mg, megagametophyte; ms, microsporangium; n, nucellus; o, ovule; p, pollen; st, sporogenous tissue; t, tapetum. Scales: 50 µm (J to L), 100 µm (G to I, M), 5 mm (A,B, F), 1 cm (C to E).
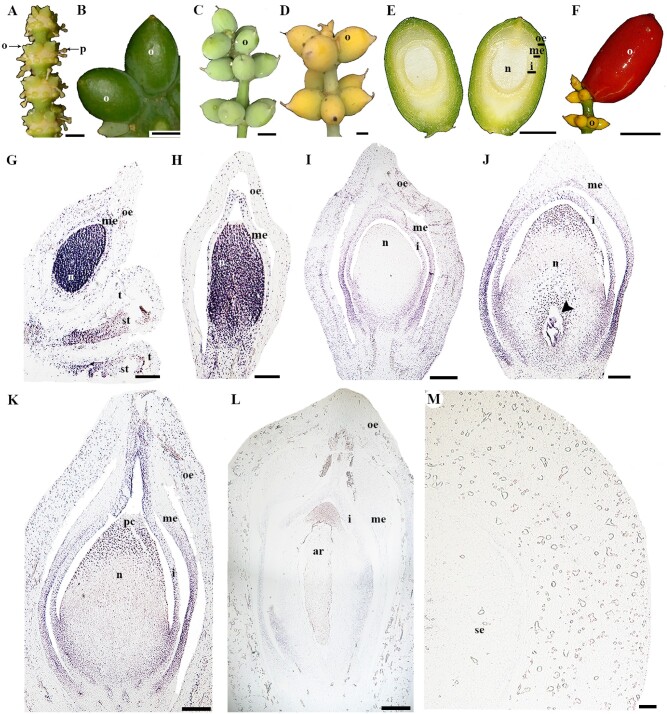
In the other gymnosperm species evaluated, Gnetum gnemon, the strobili develops axillary to the leaf and it has multiple nodes with rings of ovules (fig. 5A). The ovules are characterized by having an integument and two additional envelopes, for a total of three protective layers covering the megasporangium and later the megagametophyte. Thus, from the inside to the outside, the first layer corresponds to the integument, the second layer is the middle envelope, and the third layer is the outer envelope. The outer envelope becomes fleshy and colored, usually red (Endress1996; Rydin et al. 2006). Unlike, G. biloba, G. gnemon has bisexual cones, in each node of the strobili, an upper ring of ovules is formed and several rings of microsporangia develop basipetally, in the bisexual strobilus the ovules are usually abortive (fig. 5A;Pearson 2010). Ovules at seven different stages of development were collected, based on size and color. The first stage available was the ovule collected from a bisexual cone where the micropyle was not yet visible (fig. 5A). For the next two stages, the ovules have a green and coriaceous outer envelope (fig. 5B). The ovules at the next stage are still green but they have continued to grow, becoming fleshier with a clearly distinguishable micropyle (fig. 5C). At the following stage, the ovules turn yellow indicating they are competent for fertilization (fig. 5D). As the ovule grows, the micropyle closes (fig. 5E). Finally, the ovule turns into a red fleshy seed (fig. 5F). Like G. biloba, a single AP2 homologue was identified in G. gnemon and was named GneAP2 (fig. 1). In the earliest developmental stage of the bisexual cones sampled, the expression of GneAP2 was detected in the nucellus and the middle envelope of a young ovule, as well as in the sporogenous tissue of the microsporangia (fig. 5G). The expression of GneAP2 in the nucellus is maintained during early stages of ovule development (fig. 5H). As all three protective layers differentiate, the expression of GneAP2 is reduced in the nucellus but remains strong in the integument and in the proximal portion of the middle envelope (fig. 5I). GneAP2 is also expressed in the megagametophyte and in the pollen chamber, the region of the nucellus that receives the pollen (fig. 5J). These expression patterns are maintained as the ovule continues to develop, the expression is particularly strong in the integument including the region forming the micropyle (fig. 5K). Later on, GneAP2 expression is found in the proximal portion of the integument and in the archegonia of the megagametophyte (fig. 5L). The expression of GneAP2 in the mature seed is not detected (fig. 5M).
Fig. 5.
Expression analyses in Gnetum gnemon.(A) Bisexual cone with a ring of ovules and rings of pollen cones developing basipetally at each node. (B to F) Ovules at different stages of development. (E) Inside of the mature ovule showing three different layers protecting the nucellus. (F) Fully mature seed. (G) Pollen cone with ovule at the top. (H to L) Developing ovules, note the expression of GneAP2 is restricted to the nucellus, integument, the middle envelop and later in the archegonia and pollen chamber. (M) Cross section of a young seed. Black arrowhead pointing to the megagametophyte; ar, archegonia; i, integument; me, middle envelop; ms, microsporangium; n, nucellus; o, ovule; oe, outer envelope; p, pollen; pc, pollen chamber; se, seed; t, tapetum. Scales: 50 µm (J toM), 100 µm (G to I), 0.5 cm (A to D), 2 cm (E and F).
Expression of euAP2/TOE3 Homologues in the Fern Ceratopteris richardii
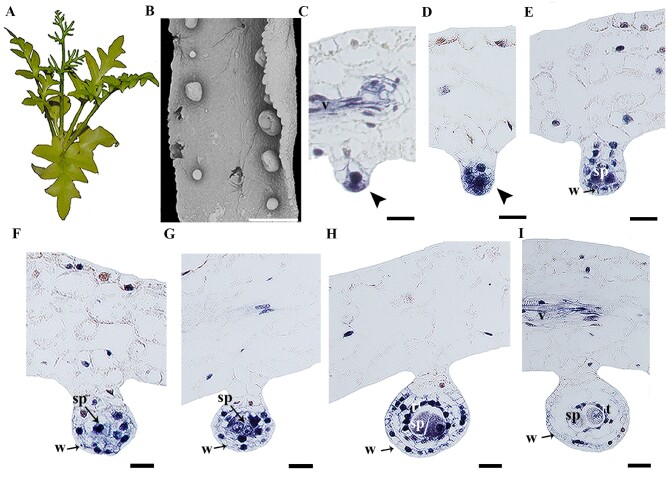
Ceratopteris richardii is a homosporous fern, meaning that there is no differentiation of female and male sporangia and spores, thus, all the spores are the same size and identity. The sporangia develop on the abaxial side of the leaf and as the plant matures, the leaf margins curve to cover the sporangia (Conway and Di Stilio 2020; fig. 6A and B). The development of the sporangium begins with the transverse division of a single superficial initial cell, with a series of subsequent cell divisions the lower cells will form the stalk and the apical cells will form the spore containing capsule or “head” of the sporangium (Foster and Gifford 1974). CerAP2 expression is detected at early stages of sporangium development (fig. 6C). As the inner cell develops, CerAP2 is expressed in the sporogenous tissue where the spores will develop and in the few cells forming the stalk (fig. 6D). Expression of CerAP2 in the sporangium wall and sporogenous tissue is maintained as the sporangia develops (fig. 6 D and E) but is no longer detected in the stalk (fig. 6F and G). Following the maturation of the sporangium, CerAP2 expression is maintained in the young spores, the inner wall of the sporangium (tapetum), and in the vasculature of the leaf (fig. 6H and I).
Fig. 6.
Expression analyses in Ceratopteris richardii.(A) Mature plant (note sterile leaves with wider laminae) (B) Scanning Electron Microscope Photograph of a fertile leaf (narrow lamina from A). (C to I) CerAP2 expression at different stages of sporangium development. Note the expression restricted to the sporangium, no expression is detected in the leaf. Black arrowhead pointing to the sporangium primordia; sp, sporogenous cells; t, tapetum; v, vasculature; w, sporangium wall. Scales: 500 µm (B); 10 µm (C to I).
Discussion
APETALA2 was first recognized as a perianth identity gene in Arabidopsis, and molecular genetic analyses placed it in the ABC model of floral development as an A-class gene (Bowman et al. 1989; Kunst et al. 1989; Bowman et al. 1991; Coen and Meyerowitz 1991; Bowman et al. 1993). AP2 homologues have been shown to play roles in fruit and seed development in Arabidopsis as well as in other core eudicots (Jofuku et al. 1994; Chung et al. 2010; Karlova et al. 2011; Ripoll et al. 2011), and have also been implicated in inflorescence patterning and yield in monocots (Chuck et al. 2008; Lee and An 2012; Bommert and Whipple 2018). Moreover, euAP2 genes are present in vascular plants but have remained little explored outside flowering plants. Importantly, the euAP2 genes are sister to the AINTEGUMENTA genes a lineage dating back to bryophytes as suggested by the presence of ANT homologues in Physcomitrella (Kim et al. 2006). This points to an ancient gene lineage with likely pleiotropic developmental roles in vascular plants. However, the data available so far does not explain why AP2 homologues perform so many different roles across angiosperms, nor does it clarify the ancestral roles of the gene lineage early in angiosperm evolution or in gymnosperms or ferns.
In order to infer the functional evolution of this gene lineage, we have: 1) updated the gene phylogeny with more inclusive gene sampling from vascular plants identifying seven major duplication events in the euAP2/TOE3 angiosperm subclade (Zumajo-Cardona and Pabón-Mora 2016), and homologues from gymnosperms, ferns, and lycophytes. These results differ from previous analyses in the finding of fern and lycophyte euAP2/TOE3 homologues (Kim et al. 2006; Dinh et al. 2012; Wang et al. 2016); 2) performed detailed spatiotemporal expression analyses in representative species from major vascular plant groups. Our results point to plesiomorphic roles of the gene lineage in the sporangia development in ferns and later recruitment of euAP2/TOE3 homologues in ovule and seed development in gymnosperms, with putative new roles in fruit development in angiosperms and their recruitment in perianth development in some angiosperms (fig. 7). Below, we discuss the implications of these findings in a gene lineage with pleiotropic developmental roles.
Fig. 7.
Summary of (A) duplication events of the euAP2/TOE3 gene lineage across vascular plantsand (B) Spatiotemporal expression of euAP2/TOE3 homologues during reproductive and vegetative phases, the purple color indicates the areas where expression has been reported for that clade. AP2 and TOE3 expression patterns in Arabidopsis as reported by previous studies (Würschum et al. 2006; Wollmann et al. 2010; Ripoll et al. 2011; Jung et al. 2014). Question mark denotes tissues for which expression information is not available . Black bars denote the absence of the tissue in that plant lineage; i, integument; ii, inner integument; n, nucellus; oi, outer integument; ov, ovule; sam, shoot apical meristem.
Early Recruitment of euAP2/TOE3 Homologues: From Sporangium Development in Ferns to Ovule Development in Seed Plants
Our expression analyses allow us to unequivocally place the ancestral role of euAP2/TOE3 homologues at the core of vascular plant reproduction. In the homosporous fern C.richardii, CerAP2 is expressed throughout sporangia development, from early sporangium development until the differentiation of the wall and the spores (fig. 6). Expression of euAP2/TOE3 homologues in the two gymnosperms G. biloba and G. gnemon indicate that they were retained over evolution as key factors in the development of the mega- and the microsporangia (figs. 4and 5). Perhaps more interesting is the fact that from early seed plant diversification, euAP2/TOE3 genes expand their expression domains to the protective integuments, forming around the megasporangia (figs. 4and 5), which are major innovations occurring in the seed plants (Brenner and Stevenson 2006). Another important observation is that in ferns and gymnosperms the expression of euAP2/TOE3 homologues in sporangia development is broader at first and at maturity becomes more restricted. Specifically expression of euAP2/TOE3 homologues remains closer to the tapetum and the spores in ferns, or in the nucellus and integument in gymnosperms. Because our data cannot be used to assess the function of the euAP2/TOE3 homologues, we can only hypothesize that they are associated with the nourishing and protective tissues of the meiotic products, the gametophytes, and perhaps even the young embryo(s). This is consistent with data in other gymnosperms like L. marschlinsii and P. abies, where euAP2/TOE3 homologues are expressed during the first stages of embryo development (Vahala et al. 2001; Shigyo and Ito 2004; Guillaumot et al. 2008).
The expression of AP2-like homologues in ovules and the two integuments was retained in angiosperms and it is clearly one of the plesiomorphic roles of the gene lineage present during flowering plant evolution. Evidence for this includes: 1) the expression of AfimAP2 in A.fimbriata (fig. 3F to L) and BofrAP2-1 in B. frutescens (fig. 2C to G) in the integuments and the nucellus; and 2) the recruitment of AP2 in Arabidopsis during integument development, as the ap2 mutant has in the first whorl carpel-like structures with placental tissue and ovules with defective or lacking integuments (Jofuku et al. 1994). Additionally, when studied with other ovule development genes, BELL1 and AG, the ovules developed in all whorls seem to be only enlarged cells that resemble a nucellus without actual integuments (Western and Haughn 1999). It is important to highlight that bryophytes (mosses, liverworts, and hornworts) also have sporangia but, no euAP2 homologues have been identified in this plant lineage so far (fig. 1; Kim et al. 2006). This could be because meristematic tissues vary between liverworts, mosses, hornworts, and vascular plants (Campbell1913; Ligrone et al. 2012). Liverworts have a sporangium or capsule, from which the spores develop, which grows by cell divisions in the absence of any localized area recognizable as a meristem (Gunning et al. 1978; Thomas1980). In mosses, an unifacial meristematic area is formed producing the sporangium primordium acropetally, this is a transient basal meristem (Renzaglia1978). On the other hand, hornworts also have a basal meristem but it remains active for the life of the sporophyte, a characteristic more similar to vascular plants (Campbell1913; Ligrone et al. 2012).
New Roles of euAP2/TOE3 Homologues in Flowering Plants Include Fruit Development
Our expression analyses of euAP2/TOE3 homologues in the early divergent angiosperm A. fimbriata and the basal eudicot B. frutescens point to their involvement in carpel and fruit development (figs. 2and 3). Carpels and fruits are major innovations in angiosperms which provided additional prezygotic barriers for egg cell fertilization in the ovules, as well as an efficient vessel for seed dispersal aiding in plant fitness. euAP2/TOE3 genes had already been identified as important players in fruit patterning of model core eudicot species (Alkio et al. 2012; Mühlhausen et al. 2013). In Arabidopsis, AP2 acts as an upstream regulator by directly repressing valve margin genes such as SHATTERPROOF (SHP) and replum identity genes such as BREVIPEDICELLUS and REPLUMLESS (Ripoll et al. 2011). In tomato, one of the AP2 homologues is involved in fruit ripening (Chung et al. 2010). It is remarkable that AP2-like copies in both A. fimbriata and B. frutescens are broadly expressed in the fruit walls (or the commissural ring), but always remain absent from the dehiscence zones, which are the regions through which the pericarp will break to release the seeds. Previous studies of RPL homologues in B. frutescens show mutually exclusive expression patterns with BofrAP2 copies, as BofrRPL is restricted to the dehiscence zone (Zumajo-Cardona et al. 2018). Taken together, the data suggest that the repression AP2–RPL may be a largely conserved module across angiosperms. Interestingly, SPATULA genes in B. frutescens share overlapping expression patterns with RPL homologues in the deshicence zone, suggesting that they too can be directly or indirectly repressed by AP2-like homologues. On the other hand, SHP is the result of a core-eudicot duplication, but, it has been suggested that its function in fruit development may be maintained by the paleoAGAMOUS, preduplication genes (Pabón-Mora et al. 2014). However, there is no evidence that paleoAG and AP2-like genes interact in any way so far. Thus, to confirm this hypothesis, more studies such as spatiotemporal expression analyses for paleoAGAMOUS are still required. As for early diverging angiosperms, it is less clear what are the putative genes controlling fruit histogenesis. However, in A. fimbriata, both AfimSPT (a SPATULA homologue) and AfimSTK (a SEEDSTICK homologue) have restricted expression to the septum and future dehiscence zones (Suárez-Baron et al. 2017; Peréz-Mesa et al. 2020). Altogether, the data does suggest that AP2-like homologues control aspects of fruit wall development and may repress other fruit development transcription factors to the deshisence zones in dry dehiscent fruits in noncore eudicots (figs 2and 3). Thus, AP2 function in fruit ripening seems to be present in dry dehiscent fruits and fleshy fruits, although further studies are required to determine if the function is conserved in dry indehiscent fruits (Ripoll et al. 2011; Wang et al. 2019).
Roles of euAP2/TOE3 Genes in Flower Development Are Broad, but Its Canonical A Function Appears Restricted to the Brassicaceae and Poaceae
In Arabidopsis, AP2 was classified as an A function gene based on its role in perianth development and the mutual antagonism of C function genes within the ABCE model of floral organ development (Bowman et al. 1991; Drews et al. 1991; Bowman et al. 1993; Tissier et al. 1999; Pelaz et al. 2000). Functional studies demonstrated that these dual roles of AP2 could be separated genetically (Bowman et al. 1991; Coen and Meyerowitz 1991). euAP2/TOE3 homologues have been unequivocally identified as A function genes by functional analyses in Arabidopsis and Brassica (Drews et al. 1991; Zhang et al. 2018). Canonical A function appears conserved in Poaceae, as molecular and genetic analyses in rice and wheat, show that AP2 orthologues are integral for perianth development and antagonism of C function; ap2 knockouts have homeotic conversions in the grass perianth (Lee et al, 2007; Lee and An 2012; Debernardi et al. 2020). euAP2/TOE3 gene expression has been detected in the floral meristem, sepals, petals, the ovary wall, and ovules in Arabidopsis, maize, petunia, rice, snapdragon, and wheat suggesting putative roles in all these organs (Bowman et al. 1989,1991,1993; Maes et al. 2001; Keck et al. 2003; Lee et al. 2007; Chuck et al. 2008; Lee and An 2012). Knockouts of euAP2/TOE3 in petunia and snapdragon affect perianth development, however, no homeotic conversion or ectopic expression of C class genes were detected in these mutants, indicating that the antagonism between A function, as specified by euAP2/TOE3, and C functions is not conserved in these species (Keck et al. 2003; Causier et al. 2010; Morel et al. 2017). The conservation of the ABC model of floral organ identity and, particularly, how the perianth whorl is specified across angiosperms is still unknown (Causier et al. 2010).
A conservation in the ABC model of floral organ development would suggest that the A and E function orthologues of APETALA1/FRUITFULL (AP1/FUL), euAP2/TOE3 (A class genes) and SEPALLATA (SEP, E class genes), would specify the perianth whorl (Pelaz et al. 2001; Causier et al. 2010). Surprisingly, we did not detect expression of euAP2/TOE3 orthologues in the perianth primordia of B. frutescens or A. fimbriata suggesting that, although they are expressed late in sepal development, these orthologues do not play a role in perianth specification or antagonism of C class genes early in floral development of these species (figs. 2 and 3). Bocconia frutescens has two caducous sepals and no petals. AP1/FUL and SEP orthologues are expressed in the sepals of B. frutescens which suggest these genes may specify this perianth whorl (Arango-Ocampo et al. 2016). It was hypothesized that the lack of petals in B. frutescens was due to a lack of a B function orthologue and expanded expression of the C function orthologue of AGAMOUS (AG; Arango-Ocampo et al. 2016). Aristolochia fimbriata has three petaloid sepals and also lacks petals. Expression analyses of AfimFUL, AfimSEP, and Afim AGAMOUS-Like 6 (AGL6) orthologues suggests that they are important for the specification of sepals in A. fimbriata (Pabón-Mora et al. 2015). Furthermore, the interaction of AfimSEP2 and AfimAGL6 proteins by yeast-2-hybrid indicate that these two protein orthologues are key players in perianth specification in A. fimbriata (Perez-Mesa et al. 2019) In turn, available data strongly suggest that AfimAP2 contribution may not be needed for perianth initiation. Knockouts of ABC floral organ identity orthologues are needed to better understand how the perianth is specified in these species.
Putative Functions of euAP2/TOE3 Homologs in Stem Cell Niche Maintenance in Flowering Plants
One of the key roles of euAP2/TOE3 genes is in reproductive transition and control of maintenance of the SAM as it has recently been included in the genetic pathway controlling meristem maintenance (Würschum et al. 2006; Balanzà et al. 2018). The mechanism of action of AP2 is the result of direct repression of FRUITFULL (FUL) thus, controlling global proliferative arrest (GPA), and in turn AP2 (and other euAP2 genes), directly or indirectly or controlling WUSCHEL maintaining the SAM (Würschum et al. 2006; Balanzà et al. 2018). Although this study does not include vegetative meristem versus reproductive meristem in all taxa, we detected expression of BofrAP2-1 in the SAM suggesting that it is conserved in basal eudicots and that its role in meristem development is putatively maintained (fig. 2B). In grasses, the spikelet is the basic unit of the inflorescence and AP2 homologues have been extensively reported to be involved in reproductive transition, from the spikelet meristem to the floral meristem (Chuck et al. 1998,2008; Lee and An 2012; Bommert and Whipple 2018; Debernardi et al. 2020). In maize IDS1 and SID1 are required to initiate the floral meristems and to control spikelet determinacy (Chuck et al. 1998; Chuck et al. 2008). A similar scenario is observed in wheat, where Q is involved in the initiation of the floral meristem (Debernardi et al. 2020). Altogether, it seems that the function of the AP2 homologues in SAM and reproductive transition is conserved across angiosperms, and it could be the ancestral role of the gene lineage like euAP2 genes (SMZ, SNZ, TOE1, 2, and 3) are also involved in flowering time (Schmid et al. 2003; Würschum et al. 2006; Huijser and Schmid 2011). Further studies in ferns and lycophytes are still required to predict if the transition to reproduction is conserved across vascular plants.
euAP2/TOE3 Gene Expression Domains Coincide with Major Seed Plant Innovations
Our results together with the data available on the euAP2/TOE3 genes allow us to provide different scenarios regarding functional evolution of the pleiotropic euAP2/TOE3 genes (fig. 7).
Our expression analyses suggest that the putative ancestral role of the euAP2/TOE3 gene lineage may be associated with sporangia development, as indicated by the expression of these genes in fern, gymnosperm, and angiosperm sporangia. Moreover, euAP2/TOE3 genes have been recruited in the formation of the integuments, a synapomorphy for seed plants, and in carpels and fruits, two major apomorphies for flowering plants (fig. 7). These results place the euAP2/TOE3 genes at the core of plant reproduction, linked to the occurrence of novel features across euphyllophytes. The underlying mechanisms controlling the shifting expression patterns of euAP2/TOE3 genes are still unknown, but we hypothesize that cis-regulatory elements and noncoding gene regions have likely diverged during the course of vascular plant evolution (Sharma et al. 2018).
Comparative sequence analyses show a conservation in the repression domains (RD, EAR domains) in AP2/TOE3 proteins and a conserved miR172 binding site in AP2/TOE3 genes across most euphyllophytes (Kim et al. 2006; Guillaumot et al. 2008). In turn, although the study did not include vegetative versus reproductive meristems for all taxa, we cannot rule out that the repression from vegetative to reproductive transition is part of the plesiomorphic roles of the euAP2/TOE3 gene lineage (fig. 7; Aukerman and Sakai 2003; Schmid et al. 2003; Chuck et al. 2007; Jung et al. 2007; Huijser and Schmid 2011; Balanzà et al. 2018; Debernardi et al. 2020). Even more, when TOE3 repression of AG is critical for floral patterning in Arabidopsis (Jung et al. 2014). It is well known that the RD functions through the recruitment of TOPLESS (TPL) and TPL-related corepressors, are also conserved across land plants (Ohta et al. 2001; Causier et al. 2012). Thus, it is possible that the repression of FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) or their homologues in euphyllophytes are conserved (Yant et al. 2010; Klintenäs et al. 2012).
The canonical A function of euAP2/TOE3 genes is characteristic of a large number of representatives in eudicots and Poaceae (fig. 7). The fact that most members of the euAP2/TOE3 gene clade have the miR172 binding sites suggest that this post-transcriptional regulation is conserved across euphyllophytes, and that the miR172 regulation is not the only mechanism associated with the floral organ identity function (supplementary fig. S4; Park et al. 2002; Griffiths‐Jones 2004; Axtell and Bartel 2005; Sunkar and Jagadeeswaran 2008; Luo et al. 2013; Zumajo-Cardona and Pabón-Mora 2016). In turn, the coupling of euAP2/TOE3 genes to floral organ identity roles may be linked to the acquisitions of specific and new protein–protein interactions in some angiosperms lineages.
Thus, it is clear that euAP2/TOE3 genes are part of the developmental toolkit that has allowed major innovations in seed plants: seeds, flowers, and fruits. We stress the importance of sequencing more genomes and the standardization of functional tools across land plants in their native context. Future studies need to focus on understanding shifting cis-regulatory elements that can help explain the expansion from sporangia expression into new domains in integuments, the carpels, the fruits and the perianth, as well as the changes in the genetic regulatory network integrating euAP2/TOE3 proteins across euphyllophytes.
Materials and Methods
Isolation of Homologues and Phylogenetic Analyses of the euAP2/TARGET of EAT 3 Gene Lineage
To reconstruct the evolution of the euAP2/TOE3 gene lineage across vascular plants we have included sequences from our own generated transcriptomes across flowering plants as follows: for several eudicots including Brugmansia suaveolens (Solanaceae,Hernández-Ciro and Pabón-Mora 2020), Coffea arabica, Condaminea corymbosa, Palicourea angustifolia (Rubiaceae, Salazar-Duque, Alzate, Urrea-Trujillo, Ferrandiz and Pabón-Mora In preparation) and Pilostylesboyacensis (Apodantaceae; González et al. 2020); for some monocots including Masdevallia coccinea, Masdevallia wendlandiana, Maxillaria aurea, Miltonia roezlii, Oncidium sp, Stelis pusilla, Tolumnia sp., (Orchidaceae, Madrigal et al. 2019); and for the ferns,Adiantum capillus-veneris (Pteridiaceae) Anemia villosa (Anemiaceae), E. bogotense, and Equisetum cf. giganteum (Equisetaceae, Rodríguez-Pelayo, Vasco, Ambrose and Pabón-Mora In preparation).Other fern and lycophyte homologues were retrieved from the OneKP transcriptome database (https://db.cngb.org/onekp/; last accessed January 9, 2021).
These sequences were recovered by a BLAST search using the euAP2/TOE3 sequences previously reported (Zumajo-Cardona and Pabón-Mora 2016), as well as the paralogues of Arabidopsis (AP2: AT4G36920 and TOE3: AT5G67180; supplementary table 1). The search was thoroughly performed in Physcomitrella patens (a moss), in the transcriptome available in cosmoss.org, as well as in other bryophytes and algae transcriptomes available in the OneKP database and in the genomes available through Phytozome. However, no euAP2/TOE3 homologues were retrieved outside vascular plants.
A total of 209 sequences were compiled and edited manually to keep only the open reading frame using AliView (Larsson2014) and then aligned using the online version of MAFFT (Katoh and Standley 2014) with a gap open penalty of 3.0, offset value of 0.8 and all the other default parameters. To find the nucleotide substitution model that best fits our data, according to Akaike Information Criterion (Akaike1974), we used the jModelTest 2 (Darriba et al. 2012), which identified the GTRGAMMA model as the best-fit model for our dataset. RAxML v.8.0.0 was used to estimate phylogenetic relationships under a maximum likelihood (ML) framework (Stamatakis et al. 2008; Stamatakis2014) using the complete nucleotide alignment of all homologues. The GTRGAMMA model was assigned and a full ML search was implemented, using the autoMRE bootstopping criterion to assess nodal support (-f a -# autoMRE option). Closely related genes from Arabidopsis: SMZ (AT3g54990), SNZ (AT2g39250), and TOE2 (AT5g60120), were used as the outgroup. The 44 newly isolated sequences were deposited in GenBank (GenBank numbers MW375853-MW375896).
To identify reported domains, new motifs, and obtain a comprehensive overview of protein sequence evolution in the lineage, a total of 103 sequences of euAP2/TOE3 were selected representing major vascular plant lineages (77 angiosperms; 22 gymnosperms; 11 ferns; 5 lycophytes). Complete sequences were permanently translated and uploaded as amino acids to the online Multiple Em for Motif Elicitation server (http://meme.nbcr.net; last accessed May 14, 2020) (Bailey et al. 2006), and run with all the default options.
Expression Analyses of Selected euAP2/TOE3 Homologues from Vascular Plants
To perform qRT-PCR, floral buds and flowers in anthesis from E. californica were collected from the grounds of the New York Botanical Garden (NYBG) and immediately frozen in liquid nitrogen. Total RNA extraction was carried out using TRIzol reagent (Invitrogen) from all dissected organs: floral bud, sepal, petal stamens, and carpels in preanthesis as well as in anthesis, immature fruit, mature fruit, and leaf. Genomic DNA contamination was removed using DNaseI (RNase-free, Austin, TX, USA) following the manufacturer’s instructions. First-strand cDNA was synthesized from 3ug of total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen, Grand Island, NY, USA) with oligodT20 primers, following manufacturer’s instructions. The resulting cDNA was diluted 1:4 and the target fragment was amplified using locus-specific primers for each paralogue with the help of online tools available at https://www.genscript.com/tools/real-time-pcr-taqman-primer-design-tool (last accessed May 14, 2020) (supplementary table 2).
For in situ hybridization analyses, inflorescences, floral buds, and fruits of B. frutescens were collected in the field (voucher: Colombia, Antioquia, Medellín, Km. 17 750, Vía El Escobero Las Palmas, sobre la via principal, January 2019, C. Zumajo-Cardona, F. Gonzalez and N. Pabón-Mora 14) and immediately fixed in formaldehyde–acetic acid–ethanol (FAA; 3.7% formaldehyde: 5% glacial acetic acid: 50% ethanol). Plants of A. fimbriata were grown in the laboratory with 15 h light. Developing SAMs in reproductive stages, floral buds and carpels were collected and fixed FAA. Ginkgo biloba young ovules and pollen cones were collected from the grounds of NYBG (Accession number: 1353/97) and immediately fixed in FAA. Gnetumgnemon young ovules and pollen cones were collected from the Nolen green houses at NYBG (Accession number: 2153/2002*C). The fern C.richardii strain RN3 was grown in a growth chamber under controlled conditions, 16-hour light at 26 °C (voucher: USA, New York, Pfizer lab, NYBG, November 2019, C. Zumajo-Cardona, T. Smalls and B. Ambrose 25 NY).
All samples were incubated in Formaldehide, Glacial Acetic Acid, and Ethanol (FAA) for 2–3 hours. Samples were then dehydrated in a standard ethanol series, embedded in paraffin (Paraplast-Xtra, Fisher Healthcare, Houston, TX, USA) and stored at 4 °C until use. The samples were sectioned at 10 μm with a Microm HM355 rotary microtome (Fisher Scientific, Pittsburgh, PA, USA). DNA templates for the synthesis of RNA probes were obtained by PCR amplification of 210–360 bp fragments with primers specific to each AP2 homologue (supplementary fig. S1; supplementary table 2). The fragments were cleaned using the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). Digoxigenin labeled RNA probes were prepared using T7 RNA polymerase (Roche, Switzerland), a murine RNAse inhibitor (New England Biolabs, Ipswich, MA, USA), and RNA labeling mix (Roche, Switzerland) according to each manufacturer’s protocol. In situ hybridization of RNA was carried out according to Ambrose et al. (2000) and Ferrandiz et al. (2000), and hybridized overnight at 55 °C. The probes were diluted 1:50 for all the experiments. Slides were permanently mounted in Permount (Fisher Scientific, Pittsburgh, PA, USA). Additionally, in situ hybridization experiments were performed with sense probes, to assess any nonspecific background signal (supplementary fig. S2). Sections were viewed on a Zeiss optical microscope and digitally photographed with a Nikon DXM1200C digital camera and ACT-1 software.
All flower and fruit developmental stages described here are based on previous descriptions for Bocconia frutescens (Zumajo-Cardona et al. 2017; Zumajo-Cardona et al. 2018) and A. fimbriata (Pabón-Mora et al. 2015).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgements
We thank Dr Dennis W. Stevenson for helpful discussion on gymnosperm morphology and anatomy. Natalí Hernández-Ciro (Universidad de Antioquia- Colombia) for help mining through databases for AP2 sequences in Solanaceae. The authors thank Marc Hachadourian Director of the NYBG’s Nolen Greenhouses for access to the Gnetum gnemon collections and plant care. NP-M was supported by the ExpoSeed (H2020 MSCA-RISE 2015-691109) EU grant, the Convocatoria Programáticas 2017-16302 and the Estrategia de Sostenibilidad 2018-2019, from the Universidad de Antioquia. The research was funded by the Eppley Foundation for Research, Inc. to BAA.
Data Availability
The data underlying this article are available in the GenBank Nucleotide Database with accession numbers provided in the methods and supplemental material. The alignment data underlying this article is available upon request to the corresponding author.
References
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automat Contr 19(6):716–723. [Google Scholar]
- Alkio M, Jonas U, Sprink T, van Nocker S, Knoche M.. 2012. Identification of putative candidate genes involved in cuticle formation in Prunus avium (sweet cherry) fruit. Ann Bot 110(1):101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M.. 1998. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. Embo J 17(18):5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ.. 2000. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification betweene udicots and monocots . Mol Cell 5(3):569–579. [DOI] [PubMed] [Google Scholar]
- Arango-Ocampo C, González F, Alzate JF, Pabón-Mora N.. 2016. The developmental and genetic bases of apetaly in Bocconia frutescens (Chelidonieae: papaveraceae). EvoDevo 7(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H.. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15(11):2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP.. 2005. Antiquity of MicroRNAs and their targets in land plants. Plant Cell 17(6):1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW.. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34(Web Server):W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanzà V, Martínez-Fernández I, Sato S, Yanofsky MF, Kaufmann K, Angenent GC, Bemer M, Ferrándiz C.. 2018. Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL-APETALA2 pathway. Nat Commun 9(1):565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Whipple C.. 2018. Grass inflorescence architecture and meristem determinacy. Semin Cell Dev Biol 79:37–47. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR.. 1993. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes Development 119: 721–743. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM.. 1989. Genes directing flower development in Arabidopsis. Plant Cell 1(1):37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM.. 1991. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112(1):1–20. [DOI] [PubMed] [Google Scholar]
- Brenner ED, Stevenson D.. 2006. Using genomics to study evolutionary origins of seeds. In: CG Williams, editor.Landscapes, genomics and transgenic conifers. Managing forest ecosystems. Dordrecht (Netherlands: ): Springer Netherlands. p. 85–106. [Google Scholar]
- Burleigh JG, Mathews S.. 2004. Phylogenetic signal in nucleotide data from seed plants: implications for resolving the seed plant tree of life. Am J Bot 91(10):1599–1613. [DOI] [PubMed] [Google Scholar]
- Campbell DH. 1913. The structure and development of mosses and ferns (Archegoniatae). New York: Macmillan [Google Scholar]
- Carroll SB. 2000. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101(6):577–580. [DOI] [PubMed] [Google Scholar]
- Causier B, Lloyd J, Stevens L, Davies B.. 2012. TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal Behav 7(3):325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Schwarz-Sommer Z, Davies B.. 2010. Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol 21(1):73–79. [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S.. 2008. Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135(18):3013–3019. [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S.. 2007. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet 39(12):1517–1521. [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S.. 1998. The control of maize spikelet meristem fate by theAPETALA2-like gene indeterminate spikelet1. Genes Dev 12(8):1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M-Y, Vrebalov J, Alba R, Lee J, McQuinn R, Chung J-D, Klein P, Giovannoni J.. 2010. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64(6):936–947. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM.. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353(6339):31–37. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Di Stilio VS.. 2020. An ontogenetic framework for functional studies in the model fern Ceratopteris richardii. Dev Biol 457(1):20–29. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Greenwood JR, Finnegan EJ, Jernstedt J, Dubcovsky J.. 2020. APETALA 2-like genes AP2L2 and Q specify lemma identity and axillary floral meristem development in wheat. Plant J 101(1):171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TT, Girke T, Liu X, Yant L, Schmid M, Chen X.. 2012. The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139(11):1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AW, Stevenson DW, Little DP.. 2007. Ovule development in Ginkgo biloba L., with emphasis on the collar and nucellus. Int J Plant Sci 168(9):1207–1236. [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM.. 1991. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65(6):991–1002. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR.. 1996. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8(2):155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 1996. Structure and function of female and bisexual organ complexes in gnetales. Int J Plant Sci 157(S6):S113–S125. [Google Scholar]
- Faris JD, Fellers JP, Brooks SA, Gill BS.. 2003. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 164(1):311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF.. 2000. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127(4):725–734. [DOI] [PubMed] [Google Scholar]
- Foster AS, , Gifford EM. 1974. Comparative Morphology of Vascular Plants. San Francisco: W. H. Freeman. [Google Scholar]
- Fu F-F, Xue H-W.. 2010. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154(2):927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Humanes J, Pistón F, Martín A, Barro F.. 2009. Comparative genomic analysis and expression of the APETALA2-like genes from barley, wheat, and barley-wheat amphiploids. BMC Plant Biol 9:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González AD, Pabón-Mora N, Alzate JF, González F.. 2020. Meristem genes in the highly reduced endoparasitic Pilostyles boyacensis (Apodanthaceae). Front Ecol Evol 8:209. [Google Scholar]
- González F, Stevenson DWm.. 2000. Perianth development and systematics of Aristolochia. Flora 195(4):370–391. [Google Scholar]
- Griffiths‐Jones S. 2004. The microRNA registry. Nucleic Acids Res 32:D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumot D, Lelu-Walter M-A, Germot A, Meytraud F, Gastinel L, Riou-Khamlichi C.. 2008. Expression patterns of LmAP2L1 and LmAP2L2 encoding two-APETALA2 domain proteins during somatic embryogenesis and germination of hybrid larch (Larix×marschlinsii). J Plant Physiol 165(9):1003–1010. [DOI] [PubMed] [Google Scholar]
- Gunning BES, Hughes JE, Hardham AR.. 1978. Formative and proliferative cell divisions, cell differentiation, and developmental changes in the meristem of Azolla roots. Planta 143(2):121–144. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge S, Yanofsky MF.. 1994. Regulation of the arabidopsis floral homeotic cene. Cell 76(1):131–143. [DOI] [PubMed] [Google Scholar]
- Hernández-Ciro N, Pabón-Mora N.. 2020. Anatomical and genetic bases underlying convergent evolution of fleshy and dry dehiscent fruits in Cestrum and Brugmansia (Solanaceae). Int J Dev Biol 52. Available from: www.ijdb.ehu.es/web/paper/200080np [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme‐Takagi M.. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, inArabidopsis. Plant J 34(5):733–739. [DOI] [PubMed] [Google Scholar]
- Huijser P, Schmid M.. 2011. The control of developmental phase transitions in plants. Development 138(19):4117–4129. [DOI] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 196(4295):1161–1166. [DOI] [PubMed] [Google Scholar]
- Jantasuriyarat C, Vales MI, Watson CJW, Riera-Lizarazu O.. 2004. Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat. Theor Appl Genet 108(2):261–273. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den BB, Montagu MV, Okamuro JK.. 1994. Control of arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6(9):1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M.. 2007. The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 225(3):575–588. [DOI] [PubMed] [Google Scholar]
- Jung J-H, Lee S, Yun J, Lee M, Park C-M.. 2014. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci 215–216:29–38. [DOI] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA.. 2011. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23(3):923–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Miura H, Akiyama M, Kuroshima M, Sawada S.. 1998. RFLP mapping of the three major genes, Vrn1, Q and B1, on the long arm of chromosome 5A of wheat. Euphytica 101(1):91–95. [Google Scholar]
- Kato K, Miura H, Sawada S.. 2000. Mapping QTLs controlling grain yield and its components on chromosome 5A of wheat. Theor Appl Genet 101:1114–1121. [Google Scholar]
- Katoh K, Standley DM.. 2014. MAFFT: iterative refinement and additional methods.In: Russell DJ, editor. Multiple sequence alignment methods. Methods in molecular biology. Totowa(NJ: ): Humana Press. p. 131–146. [DOI] [PubMed] [Google Scholar]
- Keck E, McSteen P, Carpenter R, Coen E.. 2003. Separation of genetic functions controlling organ identity in flowers. Embo J 22(5):1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstens MHL, Schranz ME, Bouwmeester K.. 2020. Phylogenomic analysis of the APETALA2 transcription factor subfamily across angiosperms reveals both deep conservation and lineage‐specific patterns. Plant J 103(4):1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Soltis PS, Wall K, Soltis DE.. 2006. Phylogeny and domain evolution in the APETALA2-like gene family. Mol Biol Evol 23(1):107–120. [DOI] [PubMed] [Google Scholar]
- Klintenäs M, Pin PA, Benlloch R, Ingvarsson PK, Nilsson O.. 2012. Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol 196(4):1260–1273. [DOI] [PubMed] [Google Scholar]
- Komaki MK, Okada K, Nishino E, Shimura Y.. 1988. Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development 104:195–203. [Google Scholar]
- Koornneef M, , van Eden J, , Hanhart CJ, , Stam P, , Braaksma FJ, , Feenstra WJ. 1983. Linkage map of Arabidopsis thaliana. J Hered 74(4):265–272. [Google Scholar]
- Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW.. 1989. AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1(12):1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30(22):3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-Y, An G.. 2012. Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69(3):445–461. [DOI] [PubMed] [Google Scholar]
- Lee D-Y, Lee J, Moon S, Park SY, An G.. 2007. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 49(1):64–78. [DOI] [PubMed] [Google Scholar]
- Li W, Wang T, Zhang Y, Li Y.. 2016. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. EXBOTJ 67(1):175–194. [DOI] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG, Renzaglia KS.. 2012. The origin of the sporophyte shoot in land plants: a bryological perspective. Ann Bot 110(5):935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovisetto A, Guzzo F, Busatto N, Casadoro G.. 2013. Gymnosperm B-sister genes may be involved in ovule/seed development and, in some species, in the growth of fleshy fruit-like structures. Ann Bot 112(3):535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Guo Z, Li L.. 2013. Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev Biol 380(2):133–144. [DOI] [PubMed] [Google Scholar]
- Madrigal Y, Alzate JF, González F, Pabón‐Mora N.. 2019. Evolution of RADIALIS and DIVARICATA gene lineages in flowering plants with an expanded sampling in non-core eudicots. Am J Bot 106(3):334–351. [DOI] [PubMed] [Google Scholar]
- Maes T, de Steene NV, Zethof J, Karimi M, D'Hauw M, Mares G, Montagu MV, Gerats T.. 2001. Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 13(2):229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF.. 1995. The arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7(11):1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A, Podlaha O.. 2009. Wings, horns, and butterfly eyespots: how do complex traits evolve? PLOS Biol 7(2):e1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Sisco PH.. 1996. Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10(23):3018–3027. [DOI] [PubMed] [Google Scholar]
- Morel P, Heijmans K, Rozier F, Zethof J, Chamot S, Bento SR, Vialette-Guiraud A, Chambrier P, Trehin C, Vandenbussche M.. 2017. Divergence of the floral A-function between an Asterid and a Rosid Species. Plant Cell 29(7):1605–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen A, Lenser T, Mummenhoff K, Theißen G.. 2013. Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. Plant J 73(5):824–835. [DOI] [PubMed] [Google Scholar]
- Najafi S, Sorkheh K, Nasernakhaei F.. 2018. Characterization of the APETALA2/ethylene-responsive factor (AP2/ERF) transcription factor family in sunflower. Sci Rep 8(1):11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Carlsbecker A, Sundås-Larsson A, Vahala T.. 2007. APETALA2 like genes from Picea abies show functional similarities to their Arabidopsis homologues. Planta 225(3):589–602. [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S, Tranby TL, Krizek BA.. 2005. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol 57(5):613–628. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H.. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7(2):173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M.. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13(8):1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M, Fischer RL, Goldberg RB, Nakamura K, Harada JJ.. 2005. Control of seed mass by APETALA2. Proc Natl Acad Sci 102(8):3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N, Suárez-Baron H, Ambrose BA, González F.. 2015. Flower development and perianth identity candidate genes in the basal angiosperm Aristolochia fimbriata (Piperales: aristolochiaceae). Front Plant Sci 6. Available from: http://journal.frontiersin.org/Article/10.3389/fpls.2015.01095/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N, Wong GK-S, Ambrose BA.. 2014. Evolution of fruit development genes in flowering plants. Front Plant Sci 5:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X.. 2002. CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12(17):1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson H. 2010 . Gnetales. Cambridge: Cambridge University Pres. [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF.. 2000. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405(6783):200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz S,, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF.. 2001. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26(4):385–394. [DOI] [PubMed] [Google Scholar]
- Peréz-Mesa P, Ortíz-Ramírez CI, González F, Ferrándiz C, Pabón-Mora N.. 2020. Expression of gynoecium patterning transcription factors in Aristolochia fimbriata (Aristolochiaceae) and their contribution to gynostemium development. EvoDevo 11(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt REChang CPang PPYMeyerowitz EM. 1987. . Molecular genetics and development of Arabidopsis. In: Loomis WR, editor. Genetic regulation of development. New York: A.R. Liss. p. 327–338. [Google Scholar]
- Perez-Mesa P, Suárez-Baron H, Ambrose BA, González F, Pabón-Mora N.. 2019. Floral MADS-box protein interactions in the early diverging angiosperm Aristolochia fimbriata Cham. (Aristolochiaceae: piperales. Evol Dev 21(2):96–110. [DOI] [PubMed] [Google Scholar]
- Renzaglia KS. 1978. Comparative morphology and developmental anatomy of the Anthocerotophyta. J. Hattori Bot. Lab. Available from: https://agris.fao.org/agris-search/search.do?recordID=US201301370475 [Google Scholar]
- Ripoll JJ, Roeder AHK, Ditta GS, Yanofsky MF.. 2011. A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138(23):5167–5176. [DOI] [PubMed] [Google Scholar]
- Rydin C, Pedersen KR, Crane PR, Friis EM.. 2006. Former diversity of Ephedra (Gnetales): evidence from early cretaceous seeds from Portugal and North America. Ann Bot 98(1):123–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanetra M, Begemann G, Becker M-B, Meyer A.. 2005. Conservation and co-option in developmental programmes: the importance of homology relationships. Front Zool 2(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU.. 2003. Dissection of floral induction pathways using global expression analysis. Development 130(24):6001–6012. [DOI] [PubMed] [Google Scholar]
- Sharma P, Watts A, Kumar V, Srinivasan R, Siwach P.. 2018. Cloning, characterization and expression analysis of APETALA2 genes of Brassica juncea (L.) Czern. Indian J Exp Biol 7. [Google Scholar]
- Shigyo M, Ito M.. 2004. Analysis of gymnosperm two-AP2-domain-containing genes. Dev Genes Evol 214(3):105–114. [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai Y-S, Gill BS, Faris JD.. 2006. Molecular characterization of the major wheat domestication Gene Q. Genetics 172(1):547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE.. 2004. The origin and diversification of angiosperms. Am J Bot 91(10):1614–1626. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57(5):758–771. [DOI] [PubMed] [Google Scholar]
- Suárez-Baron H, Pérez-Mesa P, Ambrose BA, González F, Pabón-Mora N.. 2017. Deep into the aristolochia flower: Expression of C, D, and E-class genes in Aristolochia fimbriata (Aristolochiaceae). J Exp Zool (Mol Dev Evol) 328(1–2):55–71. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G.. 2008. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RJ. 1980. Cell elongation in hepatics: the seta system. Bull Torrey Bot Club 107(3):339–345. [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JDG.. 1999. Multiple independent defective suppressor-mutator transposon insertions in arabidopsis: atool for functional genomics Plant Cell 11(10):1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahala T, Oxelman B, Arnold SV.. 2001. Two APETALA2‐like genes of Picea abies are differentially expressed during development. J Exp Bot 52(358):1111–1115. [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Lough RH, Moss SMA, Wu R, Hellens RP.. 2012. Kiwifruit floral gene APETALA2 is alternatively spliced and accumulates in aberrant indeterminate flowers in the absence of miR172. Plant Mol Biol 78(4–5):417–429. [DOI] [PubMed] [Google Scholar]
- Wang P, Cheng T, Lu M, Liu G, Li M, Shi J, Lu Y, Laux T, Chen J.. 2016. Expansion and functional divergence of AP2 group genes in spermatophytes determined by molecular evolution and Arabidopsis mutant analysis. Front Plant Sci 7:1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tavano ECDR, Lammers M, Martinelli AP, Angenent GC, de Maagd RA.. 2019. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci Rep 9(1):1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Haughn GW.. 1999. BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana: BEL1 and AG promote ovule identity. Plant J 18(3):329–336. [DOI] [PubMed] [Google Scholar]
- Wollmann H, Mica E, Todesco M, Long JA, Weigel D.. 2010. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137(21):3633–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Groß-Hardt R, Laux T.. 2006. APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18(2):295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M.. 2010. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22(7):2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, et al. 2014. Three keys to the radiation of angiosperms into freezing environments. Nature 506(7486):89–92. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA.. 2006. Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289(1):3–16. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang S, Wang X, Liu J, Guo X, Mu J, Tian J, Wang X.. 2018. Defective APETALA2 genes lead to sepal modification in brassica crops. Front Plant Sci 9:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X.. 2007. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J 51(5):840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumajo-Cardona C, Ambrose BA, Pabón-Mora N.. 2017. Evolution of the SPATULA/ALCATRAZ gene lineage and expression analyses in the basal eudicot, Bocconia frutescens L. (Papaveraceae). EvoDevo 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumajo-Cardona C, Pabón-Mora N.. 2016. Evolution of the APETALA2 gene lineage in seed plants. Mol Biol Evol 33(7):1818–1832. [DOI] [PubMed] [Google Scholar]
- Zumajo-Cardona C, Pabón-Mora N, Ambrose BA.. 2018. Duplication and diversification of REPLUMLESS – acase study in the Papaveraceae. Front Plant Sci 9:1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the GenBank Nucleotide Database with accession numbers provided in the methods and supplemental material. The alignment data underlying this article is available upon request to the corresponding author.