Abstract
Cultivated strawberry (Fragaria × ananassa) is one of our youngest domesticates, originating in early eighteenth-century Europe from spontaneous hybrids between wild allo-octoploid species (Fragaria chiloensis and Fragaria virginiana). The improvement of horticultural traits by 300 years of breeding has enabled the global expansion of strawberry production. Here, we describe the genomic history of strawberry domestication from the earliest hybrids to modern cultivars. We observed a significant increase in heterozygosity among interspecific hybrids and a decrease in heterozygosity among domesticated descendants of those hybrids. Selective sweeps were found across the genome in early and modern phases of domestication—59–76% of the selectively swept genes originated in the three less dominant ancestral subgenomes. Contrary to the tenet that genetic diversity is limited in cultivated strawberry, we found that the octoploid species harbor massive allelic diversity and that F. × ananassa harbors as much allelic diversity as either wild founder. We identified 41.8 M subgenome-specific DNA variants among resequenced wild and domesticated individuals. Strikingly, 98% of common alleles and 73% of total alleles were shared between wild and domesticated populations. Moreover, genome-wide estimates of nucleotide diversity were virtually identical in F. chiloensis,F. virginiana, and F. × ananassa (π = 0.0059–0.0060). We found, however, that nucleotide diversity and heterozygosity were significantly lower in modern F. × ananassa populations that have experienced significant genetic gains and have produced numerous agriculturally important cultivars.
Keywords: Fragaria, polyploid, genome evolution, selection, nucleotide diversity, linkage disequilibrium
Introduction
The popular garden plant commonly known as cultivated strawberry (Fragaria×ananassa) is a homoploid hybrid species with a unique domestication history spanning less than 300 years (Duchesne 1766; Darrow 1966). The first F.×ananassa individuals originated in western Europe in the early 1700 s as spontaneous hybrids between nonsympatric ecotypes of cross-compatible allo-octoploid (2n = 8× = 56) species Fragaria virginiana and Fragaria chiloensis native to North and South America, respectively. Wild-collected specimens of these species were introduced to Europe in the 1600s and 1700s and became established in common gardens where they freely hybridized (Darrow 1966). Offspring from these spontaneous hybrids were reported to be phenotypically unique and horticulturally superior to their parents. The interspecific origin of these hybrids, however, went undiscovered for nearly a half century (Duchesne 1766). The original hybrids and their descendants were disseminated and cultivated in Europe for nearly a century, far from the centers of diversity of F. chiloensis and F. virginiana (Barnet 1826; Millet 1898; Staudt 1962, 1989, 1999, 2003, 2009), before migrating to North America in the early nineteenth century and spreading worldwide (Fletcher 1917; Darrow 1937, 1966). Their horticultural superiority and phenotypic diversity drove the domestication and agricultural ascendancy of F.×ananassa over either parent species (Darrow 1966; Hancock et al.1999; Finn et al. 2013).
The process of domestication entails the selection and preferential propagation of specific phenotypes, which invariably leaves signatures of selection in the genomes of the improved individuals (Ross-Ibarra et al.2007; Purugganan and Fuller 2009; Meyer and Purugganan 2013; Gaut et al. 2018). Analyses of these genomic signatures can identify loci targeted by selection and shed light on the effects of selection, bottlenecks, migration, and admixture on genetic variation within and between populations (Tenaillon et al. 2001; Buckler et al.2006; Doebley et al.2006; Ross-Ibarra et al.2007; Purugganan and Fuller 2009, 2011; Gross and Olsen 2010; van Heerwaarden et al.2012; Hufford et al. 2012; Gaut et al. 2018). Selective sweeps arise in genomes when background selection decreases genetic variation among neutral loci that are swept or “hitchhike” in haploblocks harboring loci targeted by selection (Chen et al.2010; Schrider et al.2016; Gaut et al. 2018). The signatures of selection are typically strong across chronological and demographic boundaries in plants and animals, especially those with long domestication histories, for example, wine grape (Vitis vinifera; Zhou et al. 2017), wheat (Triticum aestivum; Akhunov et al. 2010; Balfourier et al. 2019; Pont et al. 2019), maize (Zea mays; Doebley 2004; Buckler et al.2006; Chia et al. 2012; Hufford et al. 2012; van Heerwaarden et al.2012), and dog (Canis lupus familiaris; Freedman et al. (2004).
The domestication of cultivated strawberry has been considerably shorter and faster than that of the aforementioned species and thus does not necessarily follow classic models of domestication (Doebley et al.2006; Purugganan and Fuller 2009; Gaut et al. 2018). Despite the recentness of domestication, we hypothesized that intense selection and population bottlenecks have profoundly altered genetic variation and left signatures of selection in the genomes of heirloom and modern cultivars. With less than 300 years since the beginning of F.×ananassa domestication, and a tradition of clonal preservation of heirloom and modern cultivars, extensive genetic resources exist to investigate how domestication and breeding have reshaped genetic diversity and population structure in F.×ananassa (Sjulin and Dale 1987; Horvath et al. 2011; Sánchez-Sevilla et al. 2015; Hardigan et al. 2018, 2020; Pincot et al. 2020). Over the last 50 years in particular, modern plant breeding has significantly and progressively improved several agriculturally important phenotypes and produced high yielding cultivars that have enabled the global expansion of strawberry production. While the phenotypic changes and genetic gains have been profound, the underlying genomic changes have largely remained a mystery, in part because subgenome-specific DNA variation could not be adequately analyzed on a genome-wide scale until an octoploid genome was assembled (Hardigan et al. 2018, 2020; Edger et al. 2019).
The development of a reference genome for F.×ananassa (Edger et al. 2019) was critical for addressing the questions tackled in the present study. Using the octoploid reference genome as a foundation, Hardigan et al. (2020) found that nucleotide diversity was sufficient to differentiate homoeologous DNA sequences and resolve subgenome-specific DNA variation across the genome. Most importantly from a technical perspective, they showed that 83.0% of short-read (paired-end 150 bp) DNA sequences could be unambiguously aligned to the octoploid reference genome. This technical hurdle, which limited earlier studies of genetic diversity in the octoploids, undoubtedly perpetuated the misconception that DNA variation was limited and could not be resolved in the octoploid (Gaston et al. 2020).
The narrative history of strawberry domestication has been chronicled by Darrow (1966) and others (Clausen 1915; Fletcher 1917; Sjulin and Dale 1987; Sjulin 2006); however, we only have a cursory understanding of the genomic history, and virtually no understanding of the magnitude and distribution of DNA variation in the wild progenitors and domesticated populations (Sjulin and Dale 1987; Sánchez-Sevilla et al. 2015; Hardigan et al. 2018). One of the curious dogmas to emerge from previous studies is that genetic variation is limited in F.×ananassa (Sjulin and Dale 1987; Hancock et al.1993; Horvath et al. 2011; Sánchez-Sevilla et al. 2015; Gaston et al. 2020). We suspect that this perception stems from the observation that a disproportionate fraction of the alleles found in certain domesticated populations have flowed through a small number of common ancestors (Sjulin and Dale 1987; Dale, Sjulin, and others 1990; Pincot et al. 2020). We examined this question in greater depth in the present study and in companion studies where we comparatively genetically mapped the genomes of F. chiloensis, F. virginiana, and F.×ananassa, identified DNA variants across the octoploid genome, developed high-density single nucleotide polymorphism (SNP) arrays using DNA sequences anchored to the octoploid reference genome, and reconstructed and analyzed the genealogy of cultivated strawberry from early hybrids to modern cultivars (Hardigan et al. 2020; Pincot et al. 2020). From the latter analysis, we identified 1,438 founders in the ancestry of F.×ananassa (Pincot et al. 2020). Although the contributions of common ancestors to genetic variation in domesticated populations were found to be unbalanced, the sheer number of founders suggested that domesticated populations might harbor significant genetic variation (Pincot et al. 2020), a hypothesis tested in the present study. The inferences in earlier studies were limited by the absence of genome-wide estimates of nucleotide diversity and heterozygosity, both of which were estimated in the present study from DNA variants identified by resequencing geographically and demographically diverse wild and domesticated individuals.
To elucidate the genetic structure of strawberry populations worldwide, several hundred wild and domesticated individuals were genotyped with 50 K or 850 K SNP arrays (Hardigan et al. 2020). The resequenced and genotyped individuals analyzed in the present study were predicted from earlier studies to effectively sample global genetic diversity (Horvath et al. 2011; Sánchez-Sevilla et al. 2015; Hardigan et al. 2018, 2020; Pincot et al. 2020), and included heirloom and modern cultivars developed in public breeding programs in North America, Europe, and Asia and wild individuals (ecotypes) collected across the geographic ranges of the octoploid founders. Here, we describe how domestication and breeding have reshaped genetic variation in F.×ananassa, present in-depth analyses of horticulturally important populations developed at the University of California, Davis (hereafter the “California” population) and the University of Florida (hereafter the “Florida” population), show that the octoploid species harbor massive allelic diversity, and further show that genetic variation has been broadly conserved in the global F.×ananassa population. Furthermore, we show that F. virginiana alleles have accumulated with greater frequency than F. chiloensis alleles in domesticated populations and that 59–76% of domestication-associated selective sweeps are found in the B, C, and D subgenomes (Edger et al. 2019). The importance of genetic variation in each of the four subgenomes is discussed in light of the transcriptional dominance of the A subgenome, the closest extant relative of the diploid progenitor being Fragaria vesca ssp. bracteata (Edger et al. 2019).
Results and Discussion
Chromosome Nomenclature and the Ancient Subgenomes of Octoploid Fragaria
Strawberry lacks a common language for homoeologous chromosomes and subgenomes analogous to those in allo-hexaploid wheat (Triticium aestivuum), allo-tetraploid peanut (Arachis hypogaea), and other allo-polyploid plants where universal chromosome and linkage group nomenclatures have long existed and empowered genetic and genomic studies (Pont et al. 2013; Salse 2016; Bertioli et al. 2019). Several independent linkage group nomenclatures with differing subgenome compositions have been applied in strawberry, challenging the integration of genetic and physical mapping information across studies (Hardigan et al. 2020). There is a broad consensus that F. vesca (A) and Fragariaiinumae (B) are two of the diploid ancestors of F. × ananassa; however, the subgenome assignment and chromosome nomenclature problem has persisted because of disagreements over the identities of the two remaining diploid ancestors, C and D (Tennessen et al. 2014; Edger et al. 2019, 2020; Liston et al. 2020). The ancestral ambiguity surrounding the nondominant C and D subgenomes stems from the multispecies origin of DNA comprising octoploid chromosomes (interspecific intra-chromosomal DNA variation), which have been evolutionarily reengineered through homoeologous exchanges (HEs), gene conversion events, and biased fractionation of ancestral gene contents (Edger et al. 2019). These phenomena are widespread and have been extensively documented in polyploid plants (Freeling et al. 2012; Renny-Byfield, Rodgers-Melnick, and Ross-Ibarra 2017; Bird et al. 2018; Edger et al. 2018a; Alger and Edger 2020). In the absence of these phenomena, assigning C and D subgenome origins would be straightforward, assuming the diploid ancestors or sufficiently close relatives existed (Edger et al. 2018a, 2019; Hardigan et al. 2020). Instead, these evolutionary processes created mosaics of ancestral DNA on the octoploid chromosomes, which we demonstrated through an analysis of the homology between transcripts from diploid speciesand genic sequences in the octoploid genome (Table 1; supplementary file S1, Supplementary Material online), and whole-genome alignment of F. vesca and F. × ananassa (fig. 1). We tackled the problem of assigning chromosomes to subgenomes in the present study using the transcriptomes of F. vesca subsp. bracteata, F. iinumae, Fragaria nipponica, and Fragaria viridis, which are the closest living diploid relatives of F. × ananassa identified by Edger et al. (2019). Our analyses shed light on the intrachromosomal composition of ancestral DNA, which we used in combination with genetic and physical mapping information to assign chromosomes to subgenomes.
Table 1.
Chromosome and subgenome assignments and nomenclature.
| Chromosome nomenclature |
Diploid transcript frequency (%)c |
|||||||
|---|---|---|---|---|---|---|---|---|
| Originala | Proposed | Sargent et al. (2016 ) | Tennessen et al. (2014 ) | Closest diploid relativeb | F. vesca | F. iinumae | F. nipponica | F. viridis |
| 1-4 | 1A | 1A | I-Av | F. vesca | 77.1 | 5.7 | 5.0 | 12.1 |
| 2-2 | 2A | 2A | II-Av | F. vesca | 81.4 | 7.1 | 3.8 | 7.7 |
| 3-4 | 3A | 3A | III-Av | F. vesca | 80.2 | 8.0 | 3.7 | 8.0 |
| 4-3 | 4A | 4A | IV-Av | F. vesca | 78.6 | 9.7 | 3.9 | 7.8 |
| 5-1 | 5A | 5A | V-Av | F. vesca | 80.8 | 7.6 | 3.5 | 8.1 |
| 6-1 | 6A | 6A | VI-Av | F. vesca | 77.2 | 7.8 | 5.3 | 9.7 |
| 7-2 | 7A | 7A | VII-Av | F. vesca | 74.2 | 11.2 | 4.5 | 10.1 |
| 1-2 | 1B | 1b | I-Bi | F. iinumae | 35.8 | 40.4 | 15.6 | 8.3 |
| 2-4 | 2B | 2b | II-Bi | F. iinumae | 35.3 | 39.7 | 11.5 | 13.5 |
| 3-2 | 3B | 3b | III-Bi | F. iinumae | 36.5 | 37.7 | 12.6 | 13.2 |
| 4-4 | 4B | 4b | IV-Bi | F. iinumae | 38.6 | 34.6 | 12.6 | 14.2 |
| 5-3 | 5B | 5b | V-Bi | F. iinumae | 27.6 | 44.9 | 14.1 | 13.5 |
| 6-3 | 6B | 6X2 | VI-Bi | F. iinumae | 33.7 | 40.6 | 13.7 | 12.0 |
| 7-3 | 7B | 7b | VII-Bi | F. iinumae | 37.5 | 37.5 | 10.0 | 15.0 |
| 1-3 | 1C | 1X2 | I-B2 | F. nipponica | 57.4 | 7.9 | 25.7 | 8.9 |
| 2-1 | 2C | 2X2 | II-B1 | F. nipponica | 64.4 | 2.9 | 14.4 | 18.3 |
| 3-3 | 3C | 3X2 | III-B1 | F. nipponica | 58.4 | 5.3 | 22.1 | 14.2 |
| 4-2 | 4C | 4X2 | IV-B2 | F. nipponica | 54.4 | 7.8 | 19.4 | 18.4 |
| 5-4 | 5C | 5X1 | V-B2 | F. nipponica | 56.2 | 3.8 | 21.9 | 18.1 |
| 6-2 | 6C | 6b | VI-B1 | F. nipponica | 58.2 | 2.0 | 22.2 | 17.6 |
| 7-1 | 7C | 7X2 | VII-B2 | F. nipponica | 44.1 | 5.1 | 32.2 | 18.6 |
| 1-1 | 1D | 1X1 | I-B1 | F. viridis | 56.9 | 5.6 | 20.8 | 16.7 |
| 2-3 | 2D | 2X1 | II-B2 | F. viridis | 55.5 | 10.9 | 13.6 | 20.0 |
| 3-1 | 3D | 3X1 | III-B2 | F. viridis | 53.2 | 3.2 | 17.7 | 25.8 |
| 4-1 | 4D | 4X1 | IV-B1 | F. viridis | 52.7 | 6.5 | 15.1 | 25.8 |
| 5-2 | 5D | 5X2 | V-B1 | F. viridis | 53.8 | 8.4 | 18.5 | 19.3 |
| 6-4 | 6D | 6X1 | VI-B2 | F. viridis | 54.7 | 5.4 | 19.6 | 20.3 |
| 7-4 | 7D | 7X1 | VII-B1 | F. viridis | 66.2 | 2.6 | 9.1 | 22.1 |
Original nomenclature proposed for chromosomes (pseudo-molecules) in the F.×ananassa“Camarosa” genome assembly (Edger et al. 2019).
Hypothesized closest living diploid relatives of ancient diploid donors of chromosomes found in F.×ananassa subgenomes.
Subgenome fractions were estimated from phylogenetic analysis of the transcriptomes of the hypothesized ancient diploid donors of genes found in the A, B, C, and D subgenomes of octoploid Fragaria.
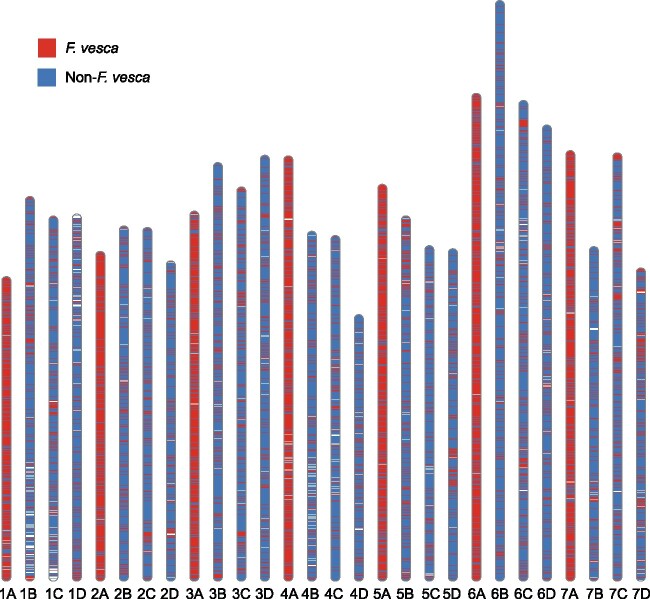
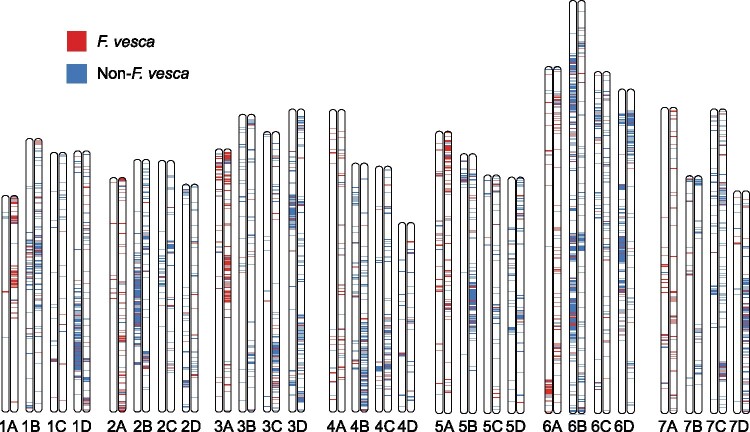
Fig. 1.
Distribution of F. vesca and non-F.vesca DNA sequences in the octoploid genome. The distribution of F. vesca DNA was ascertained by aligning the F. vesca“Hawaii” v4.0 genome assembly (Edger et al. 2018b) to the Fragaria × ananassa“Camarosa” v1.0 genome assembly (Edger et al. 2019). DNA sequence distributions in the octoploid genome were visualized using the R package RIdeogram (Hao et al. 2020).
Because homoeologous chromosomes are syntenic in octoploid strawberry (e.g., 1A, 1B, 1C, and 1D are syntenic), we oriented and numbered them according to their homology with F. vesca chromosomes (Shulaev et al. 2011; Edger et al. 2018b), similarly to most of the previous linkage group nomenclatures [reviewed by Hardigan et al. (2020)]. The assignment of homoeologous chromosomes to the A and B subgenomes was straightforward and unambiguous. F. vesca was the dominant source of genic DNA on 23 of 28 chromosomes, whereas F. iinumae was the dominant source of genic DNA on the other five chromosomes (Table 1; supplementary file S1, Supplementary Material online). F. vesca constituted 74.2–81.4% of the genic DNA on seven of the 28 chromosomes. Those chromosomes were assigned to the A subgenome (table 1; supplementary file S1, Supplementary Material online). F. iinumae constituted 34.6–44.9% of the genic DNA on seven of the remaining 21 chromosomes. The former were assigned to the B subgenome because F. iinumae genic DNA percentages ranged from 2.0 to 10.9 on the other 14 chromosomes. F. vesca DNA percentages ranged from 27.6 to 38.6 on the seven chromosomes we assigned to the B subgenome, in some cases being as prevalent as F. iinumae DNA on chromosomes of F. iinumaeorigin. This highlighted the mixed diploid ancestry of extant octoploid chromosomes, and transference of nonancestral DNA into the nondominant subgenomes.
With A and B subgenome assignments resolved, the remaining 14 chromosomes were assigned to the C and D subgenomes by applying the same rules and logic. After separating the F. vesca and F. iinumae DNA fractions, we were able to assign twelve of 14 chromosomes unequivocally to the C and D subgenomes based on whether the F. nipponica or F. viridis transcripts accounted for the largest remaining fraction of gene sequence (table 1; supplementary file S1, Supplementary Material online). Subgenome assignments for the other two chromosomes (specifically 2C and 1D) were equally straightforward but slightly more complicated. We assigned 2C to the C subgenome even though the estimated subgenome fraction was greater for F. viridis (18.3%) than F. nipponica (14.4%) because the estimated F. nipponica subgenome fraction for 2D (13.6%) was less than that for 2C (14.4%). Similarly, we assigned 1D to the D subgenome even though the estimated subgenome fraction was greater for F. nipponica (20.8%) than for F. viridis (16.7%) because the estimated F. viridis subgenome fraction for 1C (8.9%) was less than that for 1D (16.7%). The intrachromosomal patterns of mixed ancestral DNA variation that we observed highlight the complex evolutionary history of the octoploid genome (Edger et al. 2019, 2020).
Strikingly, our subgenome assignments for 24 of the 28 chromosomes (table 1; supplementary file S1, Supplementary Material online) were concordant with the subgenome assignments proposed by Sargent et al. (2016) from genetic mapping studies. The subgenome assignments proposed by Sargent et al. (2016) are shown in an updated and expanded version of the Rosetta Stone developed by Hardigan et al. (2020) to cross-reference linkage group nomenclatures (supplementary file S1, Supplementary Material online). Our A and B subgenome assignments are identical to those proposed by Tennessen et al. (2014). Other than chromosome 6B, the A and B subgenome assignments of Sargent et al. (2016) are identical to those proposed by Tennessen et al. (2014). The inconsistencies we identified between the Tennessen et al. (2014) and Sargent et al. (2016) linkage group nomenclatures were otherwise limited to the C and D subgenomes, which were consistent with the more complex and ambiguous ancestral origins of those subgenomes (table 1; supplementary file S1, Supplementary Material online). Our subgenome assignments are discordant with the linkage group assignments proposed by Tennessen et al. (2014) for four out of seven chromosomes in both the C and D subgenomes. Conversely, our C and D subgenome assignments were similar to those proposed by Sargent et al. (2016). The only discordance was for chromosomes 5C, 6C, and 5D, which were interchanged between the C and D subgenomes by Sargent et al. (2016); specifically, we discovered that chromosome 5C = 5X1, 6C = 6b, and 5D = 5X2, where B = b, C = X2, and D = X1 are the respective subgenome designations (supplementary file S1, Supplementary Material online). These comparisons highlight the quagmire of symbols, ciphers, and alphabets stymying the translation of results between genetic studies in strawberry. The proposed chromosome nomenclature provides a sound and logical framework for navigating the strawberry genome, which has been widely shown to be stably allo-octoploid (Rousseau-Gueutin et al. 2008; Tennessen et al. 2014; Sargent et al. 2016; Hardigan et al. 2020; Whitaker et al. 2020). We adopted the proposed chromosome orientations and nomenclature in the present study and for annotating newly developed phased chromosome-scale haploid assemblies of the F. × ananassa genome (unpublished data).
The identities of the C and D ancestors might never be resolved unequivocally. They could be extinct species, and their closest extant relatives may have become weak surrogates as the result of diploid species divergence and movement of nonancestral DNA into the C and D subgenomes (Edger et al. 2019, 2020). Liston et al. (2020) and Feng et al. (2021) challenged the validity of two of the closest available surrogates applied in our study (F. nipponica and F. viridis). Liston et al. (2020) argued that the C and D ancestors were two F. iinumae-like species that are either extinct, undiscovered, or never existed. Edger et al. (2020) sequenced and analyzed the F. iinumae genome and showed that only one of the octoploid strawberry subgenomes was F. iinumae-like, which cast serious doubt on the Liston et al. (2020) or Feng et al. (2021) hypotheses. Liston et al. (2020) and Feng et al. (2021) did not address the problem of intrachromosomal ancestral DNA variation in their phylogenetic approaches, nor did they identify other ancestors that could be used to assign chromosomes to subgenomes. They argued that the Edger et al. (2020) model was incorrect without providing an alternative model for the ancestors of subgenomes C and D. We acknowledge that those species could be extinct and thus unavailable for analysis. However, if their main challenge to the subgenome assignment problem is missing ancestral species, the best solution is to use information from the closest living C and D relatives.
Feng et al. (2021) used the octoploid short-read DNA sequences developed by Hardigan et al. (2020), the diploid F. iinumae genome developed by Edger et al. (2020), and an alignment-based approach to argue that F. viridis was not one of the diploid ancestors of F. × ananassa. Feng et al. (2021) stated that their “finding is in agreement with the results of Liston et al. (2020) but rejects the hypothesis of Edger et al. (2019, 2020),” and that their “results effectively resolve conflicting hypotheses regarding the putative diploid progenitors of the cultivated strawberry.”Feng et al. (2021) suggested that the lack of confirmation of F. viridis ancestry using their approach supported the Liston et al. (2020) hypothesis of three F. iinumae-like ancestors, without providing any evidence of F. iinumae ancestry for two additional subgenomes. Furthermore, they omitted any discussion of the counterarguments and genomic evidence presented by Edger et al. (2020), which ruled out additional F. iinumae-like ancestors. Edger et al. (2020), in a rebuttal to Liston et al. (2020), presented a chromosome-scale F. iinumae genome assembly and extensive phylogenetic and comparative genomic evidence to support four distinct diploid progenitor species, which was consistent with the findings of Yang and Davis (2017) and Edger et al. (2019). We assert that the Feng et al. (2021) analysis was deeply flawed because they failed to recognize: a) that the octoploid chromosomes are mosaics of DNA from four different diploid ancestors; b) that the ancestral diploid DNA fractions that have survived evolution are not equal; and c) that the chromosomes transmitted by the diploid ancestors are not intact in the octoploid. We provided evidence in the present study that DNA from other nonprogenitor diploid species can account for a majority of genes on the C and D chromosomes. Therefore, a chromosome-scale phylogenetic approach that assumes the closest living relatives of the C and D ancestors still comprise a majority of DNA on the extant C and D chromosomes is unlikely to identify their ancestral species. It has been approximately 1 million years since the formation of the octoploid, and an unknown period since the C and D ancestors merged (Edger et al. 2019). We recognize that F. nipponica and F. viridis might be distant relatives of the C and D diploids. The octoploid subgenomes are clearly not intact versions of those diploids. As the closest surrogates for the diploids comprising the remaining fraction of non-F.vesca and non-F. iinumae genes on the C and D subgenomes, they are currently the best available species for grouping the remaining chromosomes.
Whole-Genome Shotgun Sequencing Uncovered Millions of Subgenome Specific DNA Variants in Octoploid Strawberry
To develop insights into the effects of domestication on nucleotide diversity and population structure in cultivated strawberry, we whole-genome shotgun (WGS) sequenced the genomes of 99 F.×ananassa, 24 F. chiloensis, and 22 F. virginiana individuals (fig. 2; supplementary fig. S1, Supplementary Material online). These included 93 previously resequenced individuals (Hardigan et al. 2020) and 52 newly sequenced University of California, Davis (UCD) F.×ananassa individuals (supplementary file S2, Supplementary Material online). DNA sequences have been deposited in the NCBI Short Read Archive under BioProject PRJNA578384 (https://www.ncbi.nlm.nih.gov/sra/, last accessed December 1, 2020). The resequenced F.×ananassa individuals included historically important heirloom and modern cultivars and sampled allelic diversity across public germplasm collections and breeding programs worldwide. Their selection was informed by previous analyses of breeding history, genealogy, and population structure (Hardigan et al. 2018, 2020; Pincot et al. 2020). We split the global F.×ananassa population into “California” and “cosmopolitan” populations to study their unique demographic and breeding histories. The California population consisted solely of individuals developed in the UCD breeding program, which were historically important founders, advanced selections, and cultivars. The cosmopolitan population consisted of historically important cultivars and germplasm accessions developed in other public breeding programs worldwide (supplementary file S2, Supplementary Material online). The resequenced wild ecotypes sampled allelic diversity across the natural geographic ranges for seven of the eight subspecies of F. chiloensis and F. virginiana (Staudt 1962, 1989, 1999, 2009), all of which are known to have contributed allelic diversity to the F.×ananassa gene pool (Pincot et al. 2020). We found that 83.4% of short-read DNA sequences (150 bp paired-end or PE150 reads) uniquely aligned to the 0.81 Gbp “Camarosa” v1.0 genome, which was virtually identical to our previous estimate (Hardigan et al. 2020). Collectively, 41.8 M subgenome specific SNPs and INDELs were called and analyzed in the present study.
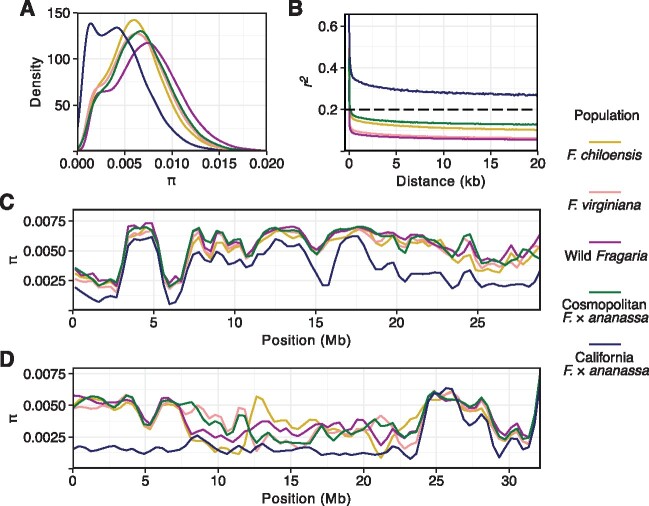
Fig. 2.
Subgenome nucleotide diversity and linkage disequilibrium.Genome-wide nucleotide diversity () and linkage disequilibrium (r2) statistics estimated from 41.8 M SNPs and INDELs. Statistics were estimated for the F. chiloensis, F. virginiana, California Fragaria × ananassa, and cosmopolitan Fragaria × ananassa populations, in addition to the combined F. chiloensis and F. virginiana populations (wild Fragaria). (A) Density plot of estimates for nonoverlapping 25 kb windows across the octoploid genome. (B) LD decay in the 0–20 kb range across the octoploid genome. The horizontal dashed line depicts the intercept used to report short-range LD decay (r20.2). (C) eEstimates for nonoverlapping 25 kb windows on chromosome 1B. (D) eEstimates for nonoverlapping 25 kb windows on chromosome 7C.
Wild Allelic Diversity Has Been Strongly Preserved in Domesticated Strawberry
Genetic diversity has not been significantly eroded by domestication in F.×ananassa (figs. 2 and 3; supplementary fig. S1, Supplementary Material online). We reached this conclusion by comparing shared and private allele frequencies, genome-wide estimates of nucleotide diversity, and heterozygosity in F. chiloensis, F. virginiana, and F.×ananassa populations. Genome-wide estimates of nucleotide diversity () were virtually identical for F. chiloensis (π = 0.0059), F. virginiana (π = 0.0060), and cosmopolitan F.×ananassa (π = 0.0059) populations. The patterns and physical distributions of nucleotide diversity were similar in these populations on nearly every chromosome (fig. 2; table 2; supplementary fig. S1, Supplementary Material online). Nucleotide diversity was significantly lower in the California population (π = 0.0040) than the other populations in our study. We attributed this to strong directional selection, breeding bottlenecks, and selective sweeps, which are explored in greater depth below (Chen, Patterson, and Reich 2010; Purugganan and Fuller 2011; Booker, Jackson, and Keightley 2017). The estimate distribution was left-skewed and bimodal in the California population, which starkly contrasted with the nearly overlapping distributions observed for the other populations (fig. 2). These differences among populations are illustrated for chromosomes 1B and 7C in fig. 2 and the other 27 chromosomes in supplementary fig. S1, Supplementary Material online. We attribute the secondary low-diversity peak in the distribution for the California population to the effects of strong selective sweeps, as shown on the entire upper arm of chromosome 7C (fig. 2D;supplementary fig. S1, Supplementary Material online). Haploblocks with decreased nucleotide diversity were found on at least 10 of the 28 chromosomes, many of which spanned entire or nearly entire chromosome arms (fig. 2; supplementary fig. S1, Supplementary Material online).
Table 2.
Nucleotide diversity () in the A, B, C, and D subgenomes of octoploid strawberry populations.
| Subgenome | Populationa | Chromosome |
Mean | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| A | California | 0.0043 | 0.0041 | 0.0042 | 0.0030 | 0.0032 | 0.0028 | 0.0015 | 0.0033 |
| Cosmopolitan | 0.0051 | 0.0054 | 0.0053 | 0.0046 | 0.0042 | 0.0044 | 0.0026 | 0.0045 | |
| F. chiloensis | 0.0049 | 0.0051 | 0.0052 | 0.0044 | 0.0043 | 0.0042 | 0.0027 | 0.0044 | |
| F. virginiana | 0.0048 | 0.0051 | 0.0055 | 0.0047 | 0.0045 | 0.0045 | 0.0029 | 0.0046 | |
| Mean | 0.0048 | 0.0049 | 0.0050 | 0.0042 | 0.0041 | 0.0040 | 0.0024 | 0.0042 | |
| B | California | 0.0033 | 0.0045 | 0.0055 | 0.0058 | 0.0058 | 0.0019 | 0.0040 | 0.0044 |
| Cosmopolitan | 0.0054 | 0.0071 | 0.0077 | 0.0072 | 0.0076 | 0.0043 | 0.0063 | 0.0065 | |
| F. chiloensis | 0.0049 | 0.0068 | 0.0068 | 0.0070 | 0.0065 | 0.0050 | 0.0055 | 0.0061 | |
| F. virginiana | 0.0059 | 0.0081 | 0.0080 | 0.0074 | 0.0075 | 0.0060 | 0.0066 | 0.0071 | |
| Mean | 0.0048 | 0.0066 | 0.0070 | 0.0069 | 0.0068 | 0.0043 | 0.0056 | 0.0060 | |
| C | California | 0.0032 | 0.0057 | 0.0049 | 0.0061 | 0.0056 | 0.0027 | 0.0018 | 0.0043 |
| Cosmopolitan | 0.0062 | 0.0078 | 0.0075 | 0.0074 | 0.0071 | 0.0057 | 0.0037 | 0.0065 | |
| F. chiloensis | 0.0053 | 0.0065 | 0.0059 | 0.0066 | 0.0061 | 0.0053 | 0.0036 | 0.0056 | |
| F. virginiana | 0.0062 | 0.0078 | 0.0073 | 0.0077 | 0.0069 | 0.0060 | 0.0043 | 0.0066 | |
| Mean | 0.0052 | 0.0069 | 0.0064 | 0.0070 | 0.0064 | 0.0049 | 0.0034 | 0.0058 | |
| D | California | 0.0038 | 0.0045 | 0.0046 | 0.0065 | 0.0042 | 0.0039 | 0.0038 | 0.0045 |
| Cosmopolitan | 0.0059 | 0.0070 | 0.0067 | 0.0076 | 0.0065 | 0.0065 | 0.0055 | 0.0065 | |
| F. chiloensis | 0.0049 | 0.0063 | 0.0061 | 0.0063 | 0.0055 | 0.0061 | 0.0048 | 0.0057 | |
| F. virginiana | 0.0061 | 0.0074 | 0.0071 | 0.0080 | 0.0069 | 0.0072 | 0.0053 | 0.0069 | |
| Mean | 0.0052 | 0.0063 | 0.0061 | 0.0071 | 0.0058 | 0.0059 | 0.0048 | 0.0059 | |
aThe germplasm accessions included in each population are identified in supplementary file S2, Supplementary Material online.
Nucleotide diversity was lowest in the A subgenome (π = 0.0042) and approximately 1.4-fold greater and virtually identical in the other three subgenomes (π = 0.0058 to 0.0060; table 2). This pattern was observed in both wild founder and domesticated populations, which suggests that the differences are unrelated to selection and other domestication forces. The less variable A subgenome was substantially derived from F. vesca (Edger et al. 2019), the transcriptionally dominant diploid ancestor (Edger et al. 2019). F. vesca is hypothesized to have fused with the genome of an unknown hexaploid ancestor and was the most recent diploid ancestor to merge with the other subgenomes in the octoploid nucleus (Edger et al. 2018a). This could explain the lower nucleotide diversity we found in the A subgenome (table 2). Another possibility is that purifying selection purged deleterious alleles from the A subgenome in the wild octoploid founders (Charlesworth, Morgan, and Charlesworth 1993; Cvijović, Good, and Desai 2018).
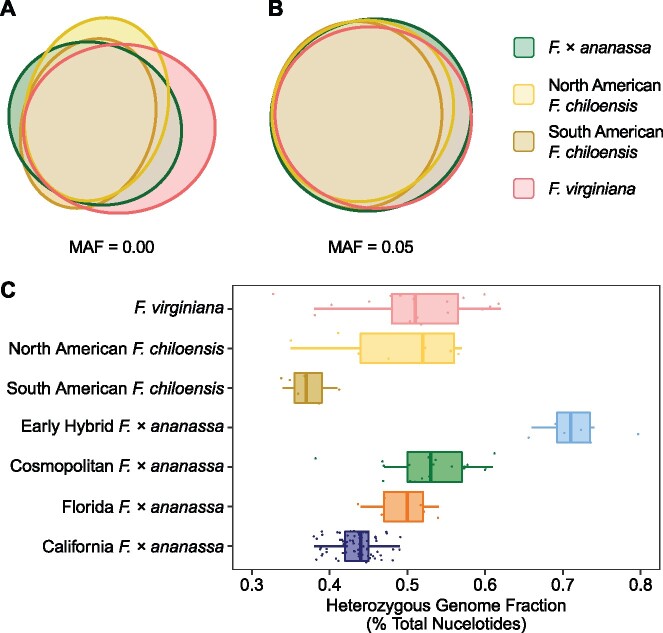
Strikingly, most of the ancestral alleles found in the phylogenetically and geographically diverse sample of F. chiloensis and F. virginiana ecotypes have persisted through the domestication of F.×ananassa (fig. 3). Using different minor allele frequency (MAF) thresholds, we discovered that private mutations were rare, for example, for MAF , we found that F.×ananassa harbored 98% of common alleles from the wild octoploids. For MAF , the percentage of total alleles (common and rare) retained in F.×ananassa was still 73% (fig. 3A, B). These shared-allele frequencies highlight the scarcity of private alleles and conservation of wild progenitor alleles in clonally preserved F.×ananassa individuals. We found that most F. chiloensis alleles exist in F. virginiana, a finding that is consistent with the hypothesized evolution of the octoploid species and subspecies (Dillenberger et al. 2018). Nucleotide diversity was only marginally greater in the combined wild population (π = 0.0068) relative to either species, supporting a high frequency of shared mutations among the wild progenitors.
Fig. 3.
Octoploid allele sharing and heterozygosity.(A, B) Euler diagrams depicting the distributions of shared and private alleles among South American F. chiloensis, North American F. chiloensis, F. virginiana, and Fragaria × ananassa individuals. Shared and private allele percentages and heterozygosities were estimated from 41.8 M SNPs and INDELs. (A) Euler diagram for a minor allele frequency (MAF) = 0.00, which depicts the overlap of both common and rare alleles. (B) Euler diagram for MAF = 0.05, which depicts the overlap of common alleles only for loci with MAF ≤ 0.05. (C) Box-and-whisker plot distribution of heterozygosity (H = v/n) estimates for individuals in F. chiloensis, F. virginiana, and Fragaria × ananassa populations, where v,the number of heterozygous SNPs and INDELs observed in an individual and n,the number of nongap nucleotides in the octoploid genome. The boxes span 1.0 standard deviation, whereas the whiskers span 2.0 standard deviations. The median heterozygosity for each group is depicted by the heavy vertical bar within the box.
The global population of cultivated F.×ananassa hybrids appears to have retained high levels of nucleotide diversity while harboring a majority of alleles, both common and rare, found in a geographically and phylogenetically diverse set of wild progenitor ecotypes. Thus, the broader F.×ananassa species complex does not appear to be strongly bottlenecked as a result of domestication.
Restructuring of Interspecific Heterozygosity from Early to Modern Hybrids
The earliest F.×ananassa hybrids originated from a single pair of F. chiloensis and F. virginiana individuals (founders). Although these first hybrids were important to early breeding, genetic diversity in the global F.×ananassa population has been repeatedly expanded and reshaped through introgression of alleles from numerous F. chiloensis and F. virginiana ecotypes (Darrow 1966; Hancock et al. 2001; Pincot et al. 2020). Pincot et al. (2020) identified 112 F. chiloensis, 65 F. virginiana, and 1,171 F.×ananassa founders in the genealogy of cultivated strawberry, and postulated that the number of wild founders might be higher because the wild founders of the 1,171 F.×ananassa founders were unknown. While the genetic contributions of these founders were unequal and individually small (Pincot et al. 2020), they collectively introduced significant allelic diversity into the primary gene pool of cultivated strawberry (figs. 2–5; supplementary fig. S1, Supplementary Material online). Our nucleotide diversity and heterozygosity estimates show that both wild founders (F. chiloensis and F. virginiana) and F.×ananassa harbor significant genetic diversity (figs. 2–5). We observed an average of one DNA variant every 140.9 bp among “early hybrids,” 196.1 bp among F. virginiana ecotypes, and 227.3 bp among F. chiloensis ecotypes.
Despite the interspecific origin of F.×ananassa, significant increases in heterozygosity were only observed among ’early’ hybrids (fig. 3C). Heterozygosities were greater for F. virginiana () than F. chiloensis (; p < 1e−5) and significantly greater for early hybrids () than F. virginiana (p < 1e−5) or F. chiloensis (p < 1e−6), where is the mean heterozygosity (H) of individuals in a population (fig. 3C). Heterozygosities were 1.4-fold or greater for early hybrids than for wild ecotypes. The most heterozygous individual in our study was the early hybrid ’White Carolina’ (). Heterozygosity in the cosmopolitan F.×ananassa population () was similar to that of F. virginiana (), while individuals in the commercially improved Florida () and California () populations were least heterozygous.
Concurrent with the reduction of interspecific heterozygosity, the genomic distribution of heterozygosity has been dramatically altered through breeding (Hardigan et al. 2020). Genetic mapping studies in several full-sib and S1 mapping populations have shown that DNA variation is limited in several chromosome regions in modern cultivars (Sargent et al. 2016; Pincot et al. 2018; Hardigan et al. 2020). Our analyses uncovered several haploblocks with reduced nucleotide diversity, some of which spanned entire chromosome arms (fig. 2; supplementary fig. S1, Supplementary Material online). Genetic and physical mapping of the F. chiloensis and F. virginiana genomes and genome-wide analyses of DNA variation have shown that the genomes of the wild octoploid founders of F. ×ananassa harbor massive diversity, for example, 1.9 M biallelic DNA variants were identified in the genome of a single F. chiloensis subsp. lucida individual (Del Norte; PI 551449; , compared to 1.6 M biallelic DNA variants in the genome of the F.×ananassa cultivar “Camarosa” (Hardigan et al. 2020). Heterozygous DNA variants were evenly distributed across the “Del Norte” genome, whereas heterozygous DNA variants were concentrated in 60% of the “Camarosa” genome. As we show below, these “genetic diversity deserts” are likely associated with selective sweeps (Chen et al.2010; Schrider et al.2016; Booker et al.2017; Gaut et al. 2018).
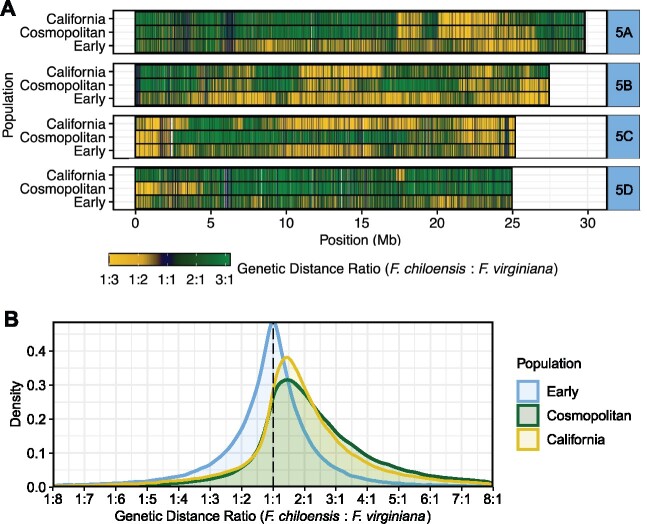
Species-preferential selection appears to have played an important role in the loss of interspecific heterozygosity from early to modern hybrids. We estimated the balance of wild founder allele dosage across the octoploid genome for early hybrid, cosmopolitan, and California F.×ananassa populations (fig. 4; supplementary fig. S2, Supplementary Material online). Because private F. chiloensis and F. virginiana alleles were scarce, ancestral allele dosages in F.×ananassa were estimated by the relative genetic distance of F.×ananassa to both wild progenitor species from DNA variants in nonoverlapping 10 kb windows. This revealed that large, contiguous chromosome segments in the F.×ananassa genome have undergone preferential selection for F. chiloensis or F. virginiana haplotypes (fig. 4A). As expected, the genomes of early hybrids harbored a relatively balanced distribution of F. chiloensis and F. virginiana diversity. By contrast, founder allele dosages were skewed towards F. virginiana in the genomes of individuals in cosmopolitan and California F.×ananassa populations (fig. 4B), which suggest that the genomic contribution from F. virginiana has been greater than that from F. chiloensis in domesticated hybrids. Several chromosomes (e.g., 2B and 4D) were almost entirely derived from F. virginiana in these populations (supplementary fig. S2, Supplementary Material online).
Fig. 4.
Genomic distribution of wild founder alleles in cultivated strawberry populations.(A) Heatmap displaying the relative contributions (dosages) of allelic diversity from wild octoploid progenitor species on chromosome 5 homoeologs in early hybrid, cosmopolitan, and California Fragaria × ananassa populations. Dosages were estimated from genetic distance ratios () in nonoverlapping 10 kb windows, where is the genetic distance between Fragaria × ananassa and F. chiloensis and is the genetic distance between Fragaria × ananassa and F. virginiana. (B) Kernel density plot displaying the distribution of ratios (allele dosages) estimated in nonoverlapping 10 kb windows across the octoploid genome in early hybrid, cosmopolitan, and California Fragaria × ananassa populations.
We uncovered several important gene functions that were over-represented in regions of species-preferential selection. We identified 4,289 genes in regions with five-fold F. chiloensis dosage bias and 5,425 genes in regions with five-fold F. virginiana dosage bias in both the cosmopolitan and California populations, and identified enriched Gene Ontology (GO) terms and protein (PFAM) domains relative to admixed genome regions (supplementary file S5, Supplementary Material online). F. chiloensis genes were enriched for functions related to photosynthetic light harvesting (GO: 0009765) and chlorophyll A/B binding (PF00504), as well as cell cycle functions including regulation of mitotic spindle organization (GO: 0060236), spindle assembly (GO: 0051225), microtubules (GO: 0005874), and cell cycle regulated microtubule-associated protein domains (PF12214). F. virginiana genes were enriched for self-pollen recognition (GO: 0048544) and S-locus glycoprotein domains (PF00954), critical functions for domestication due to the role of self-fertility in uniform fruit development. The total set of species-specific genes was enriched for transcription regulatory functions, including DNA-templated regulation of transcription (GO: 0006355), DNA-binding transcription factor activity (GO: 0003700), transcriptional repressor complex (GO: 0017053), methylation (GO: 0032259), and transcription coregulator activity (GO: 0003712). The reduced heterozygosity of modern hybrids, broad genomic regions of species-preferential selection (fig. 4; supplementary fig. S2, Supplementary Material online), and increased likelihood of species-preferential selection for several important gene functions, particularly transcription regulation, all support a hypothesis that F.×ananassa domestication relied on selection and fixation of beneficial alleles from an expanded pool of diversity, not by simply maximizing interspecific heterozygosity across the genome (Comai 2005).
Linkage Disequilibrium Rapidly Decayed in Octoploid Populations
Linkage disequilibrium (LD) rapidly decayed in the octoploid strawberry. Consistent with the evolutionary and domestication history of the populations under study, LD decayed more rapidly in the progenitors (F. virginiana and F. chiloensis) than F.×ananassa (fig. 2). Short-range LD () decayed at a mean distance of 20 bp in F. virginiana, 75 bp in F. chiloensis, and 120 bp in cosmopolitan F.×ananassa, similar to other outcrossing plant species (Tenaillon et al. 2001). Short-range LD decayed at 400 kb in California F.×ananassa, reflecting the prevalence of common haploblocks and identity-by-descent within the bottlenecked California population relative to the cosmopolitan population. We attributed these LD differences to breeding bottlenecks and directional selection as opposed to differences in recombination (Hartl et al.1997; Nordborg and Tavaré 2002; Gaut and Long 2003; Hardigan et al. 2020). LD in the genomes of wild octoploid taxa was correlated with phylogenetic divergence time and nucleotide diversity (fig. 2; Dillenberger et al. (2018). Dillenberger et al. (2018) estimated that monophyletic F. chiloensis evolved more recently (0.07–0.30 Ma) than paraphyletic F. virginiana (0.30–1.18 Ma)—the divergence time reported for F. virginiana was for the earliest branch in the phylogeny (Dillenberger et al. 2018). Genome-wide estimates of nucleotide diversity were similar for F. chiloensis ( = 0.0059) and F. virginiana ( = 0.0060) but lower for South American F. chiloensis ( = 0.0033) than North American F. chiloensis ( = 0.0056). These trends were consistent with theoretical expectations. South American F. chiloensis, which appears to have been introduced from North America, harbored fewer mutations than North American F. chiloensis (fig. 5).
Fig. 5.
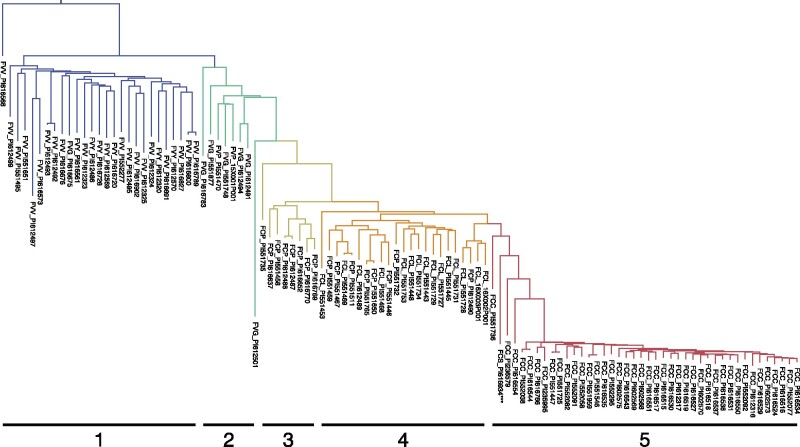
Cladogram for wild octoploid taxa.Genetic distances were estimated among 108 wild ecotypes from 1,905 array-genotyped SNP markers. Taxa are identified by three letter prefixes: F. virginiana subsp. glauca (FVG), F. virginiana subsp. grayana (FVY), F. virginiana subsp. platypetala (FVP), F. virginiana subsp. virginiana (FVV), F. chiloensis subsp. chiloensis (FCC), F. chiloensis subsp. lucida (FCL), F. chiloensis subsp. pacifica (FCP), and F. virginiana subsp. sandwichensis (FCS). Group 1 clades are comprised primarily of F. virginiana subspecific ecotypes originating east of the Continental Divide in North America (FVV, FVY). Group 2 clades are comprised of F. virginiana subspecific ecotypes originating in western North America (FVG, FVP). Group 3 clades are comprised of FCP ecotypes originating along the Pacific Coast of North America. Group 4 clades are comprised primarily of FCL ecotypes originating along the Pacific Coast of North America. Group 5 clades are comprised of South American FCC ecotypes, in addition to a Hawaiian FCS ecotype.
We observed that the closest relationships between octoploid species occurred between western North American F. virginiana subsp. glauca and platypetala (fig. 5; Clade 2) and North American F. chiloensis subsp. pacifica (fig. 5; Clade 3). Natural hybrids between the species have been documented in the Pacific Northwest of North America where their ranges overlap (Hancock and Bringhurst 1979; Staudt 1999). South American F. chiloensis subsp. chiloensis (fig. 5; Clade 5) was most closely related to North American clades containing a higher frequency of F. chiloensis subsp. lucida ecotypes (fig. 5; Clade 4). The sole F. chiloensis subsp. sandwichensis ecotype from Hawaii (PI616934) grouped roughly between the North and South American F. chiloensis clades. We used the software TreeMix (Pickrell and Pritchard 2012) to predict putative gene migrations between wild octoploid subspecies (supplementary fig. S3, Supplementary Material online) and found the strongest support for migration between western North American F. virginiana subspecies and North American F. chiloensis subspecies, which was consistent with the hypothesized evolutionary history of these taxa (Dillenberger et al. 2018).
Domestication of South American Beach Strawberry
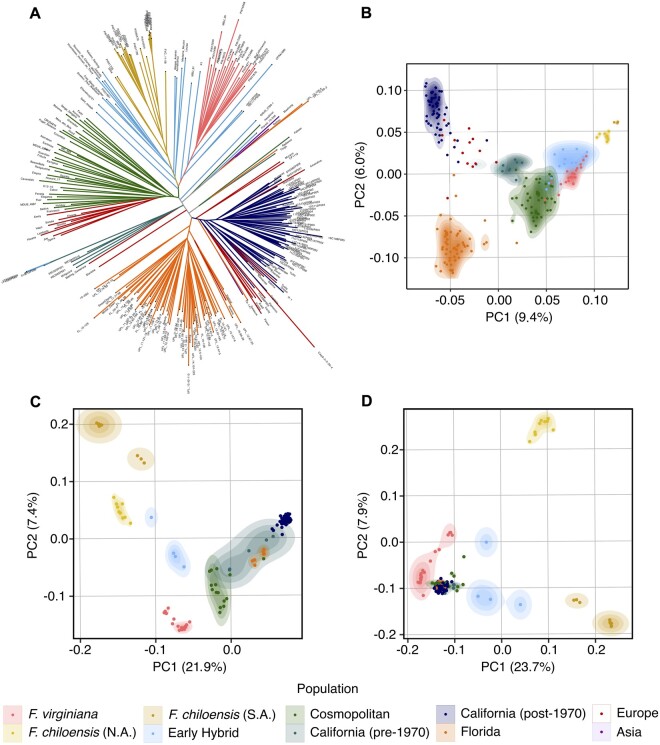
Hancock et al. (1999) and Finn et al. (2013) described the history of domestication of South American beach strawberry (F. chiloensis subsp. chiloensis). They reported that two native Chilean peoples, the Picunche and Mapuche, cultivated landraces as early as 1,000 years before present, and that landraces produced larger fruit than native wildtypes. We analyzed F. chiloensis subsp. chiloensis ecotypes across the entire geographic range in South America, which included individuals classified as landraces or cultivars in the USDA National Plant Germplasm System (https://www.ars-grin.gov/, last accessed December 1, 2020): PI551736 (Peruvian Ambato), PI616554 (Futalefu), and PI236579 (Darrow 72). Wild F. chiloensis subsp. chiloensis individuals formed a single clade (fig. 5) and WGS-based PCA cluster (fig. 6C, D), while “Ambato,”“Futalefu,” and “Darrow 72” were genetically distinct from the wild individuals. It was not clear that this resulted from population bottlenecks or directional selection; the cultivated individuals were located at the edge of the South American F. chiloensis clade, and their branch lengths (genetic distances) relative to other South American ecotypes supported past hybridization (fig. 5). PCA clusters generated from WGS variant calling placed cultivated beach strawberry individuals between wild F. chiloensis subsp. chiloensis and early hybrid F.×ananassa (fig. 6C, D). We suspect that “Peruvian Ambato,”“Futalefu,” and “Darrow 72” are not true F. chiloensis subsp. chiloensis cultigens, but descend from cryptic hybrids arising in South America between native F. chiloensis subsp. chiloensis and imported F.×ananassa.
Fig. 6.
Patterns of genetic diversity in wild and domesticated strawberry populations.(A) Maximum-likelihood phylogenetic tree of 259 octoploid individuals based on the 850 K SNP array G matrix. (B) Principal component analysis of 259 octoploid individuals based on the 850 K SNP array G matrix. (C) Principal component analysis of 145 octoploid individuals based on 41.8 M SNP and INDEL variants. (D) Principal component analysis of 46 wild octoploid individuals based on 41.8 M SNP and INDEL variants, with Fragaria × ananassa individuals projected onto wild-estimated PC axis.
Genetic Structure of Octoploid Strawberry Populations
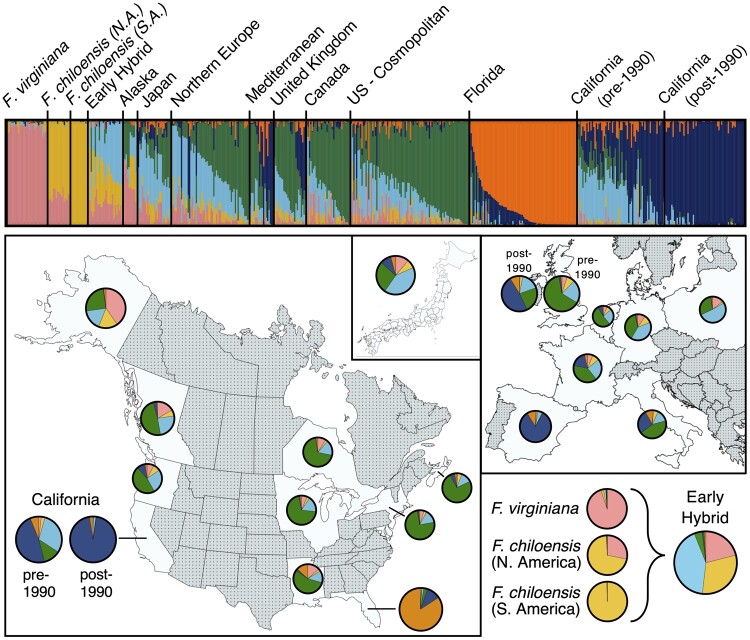
The complex pedigree networks underlying demographically and geographically unique populations of cultivated strawberry obscure their genetic structure, the result of a domestication history involving frequent and repeated admixture (Pincot et al. 2020). To resolve their hidden genetic structure, we applied cluster and principal component analyses to SNP genotype matrices () estimated from DNA variants called by WGS sequence alignment, or using 50 K (supplementary file S3, Supplementary Material online) and 850 K (supplementary file S4, Supplementary Material online) SNP arrays (Hardigan et al. 2020). Collectively, 1,569 individuals were genotyped with the 50 K SNP array, and 259 individuals were genotyped with the 850 K SNP array (Hardigan et al. 2020). These individuals included phylogenetically and demographically diverse F. chiloensis and F. virginiana ecotypes, early hybrids, and historically important heirloom and modern cultivars preserved by the USDA, UCD, and University of Florida (UF) germplasm collections (supplementary file S2, Supplementary Material online). We identified 41,932 polymorphic markers with the 50 K array and 446,644 polymorphic markers with the 850 K array. SNP genotypes for 41,932 codominant markers common to both arrays were combined and LD-pruned (r2 ≤ 0.70) for a global analysis of octoploid strawberry population structure (fig. 7).
Fig. 7.
Genetic structure of strawberry populations. Genetic structure shown was estimated for K = 6 populations among 1,637 wild and domesticated octoploid individuals genotyped with 50 K or 850 K SNP arrays (genotypes for SNP markers common to both arrays were analyzed). The upper panel displays admixture proportions for populations grouped by geographic origin with Fragaria ×ananassa individuals within each geographic group ordered by year of origin. The lower panels display K = 6 admixture proportions for populations originating in different states or countries.
Several insights emerged from these analyses. First, South American F. chiloensis, North American F. chiloensis, and F. virginiana populations formed distinct clusters correlated with demography and phylogeny (fig. 6). Second, early hybrids were roughly equidistant to North American F. chiloensis and F. virginiana clusters in the analysis of WGS-based DNA variants (fig. 6C, D), which lacked wild ascertainment bias. The early hybrids were furthest from the South American F. chiloensis cluster and closest to the F. virginiana cluster. Although the earliest F.×ananassa cultivars were interspecific hybrids between South American F. chiloensis and F. virginiana, North American alleles, many of which appear to be shared across native founder species (fig. 7), are more common than South American F. chiloensis alleles in the genetic background of F.×ananassa (fig. 6B–D).
Third, when the PCA coordinates for F.×ananassa individuals from across the globe were predicted using variable loadings from the wild octoploid taxa (projection of domesticated individuals onto wild PC axis), they formed a tight undifferentiated cluster equidistantly positioned between F. virginiana and early hybrids (fig. 6D). This supported our finding that F.×ananassa individuals harbor an excess of F. virginiana alleles, whereas early hybrids harbor an equal balance of F. chiloensis and F. virginiana alleles (fig. 4). The excess of F. virginiana alleles undoubtedly stems from a combination of migration and selection. The California and cosmopolitan populations both displayed a bias towards F. virginiana, which could plausibly be explained by an increase in F. virginiana allele dosage, in addition to directional selection for favorable F. virginiana alleles and selectively swept neutral loci (fig. 4; Chen, Patterson, and Reich 2010). Although the number of F. chiloensis founders () in the genealogy of cultivated strawberry was estimated to be two-fold greater than the number of F. virginiana founders (), the latter were estimated to have made larger genetic contributions (Pincot et al. 2020).
Fourth, the California and Florida populations formed distinct clusters highlighting their unique breeding histories and strong differentiation from the “cosmopolitan” population (fig. 6A, B). The California and Florida populations exhibited the greatest differentiation from wild ecotypes, which we attributed to directional selection and adaptation to their unique Mediterranean and subtropical environments (fig. 7; supplementary fig. S3, Supplementary Material online). Two-population estimates supported strong genetic restructuring within modern breeding populations. The California and Florida populations were both as divergent from cosmopolitan F.×ananassa as cosmopolitan F.×ananassa was from the wild founders (fig. 8). Population structure analysis with an admixture model supported this restructuring and predicted six octoploid subpopulations: one each for F. virginiana and F. chiloensis, and four for F.×ananassa, the latter roughly corresponding to the early hybrid, cosmopolitan, California, and Florida hybrids (fig. 7; supplementary fig. S3, Supplementary Material online). Outside of the California and Florida populations, we observed no clear population structure distinguishing F.×ananassa cultivars based on global geographic origin, that is, other North American, European, and Asian (mainly Japanese) cultivars were not sufficiently distinct to predict new continental subpopulations. We attributed this to global migrations and admixture that has characterized F.×ananassa breeding, such as the recent migration of California alleles into European hybrids (fig. 7; Pincot et al. 2020). Repeated introgression of wild ecotypes and migration of germplasm between breeding populations over the last three centuries were important determinants of F.×ananassa diversity and population structure, contributing to an admixed global population (Pincot et al. 2020). Accordingly, we observed no significant evidence of population structuring among North American, European, and Asian cultivars. Instead, population structure and genetic differentiation from wild ecotypes were strongest within the California and Florida populations, where intense directional selection under niche environments has produced important commercial cultivars, and where loss of diversity has been most significant.
Fig. 8.
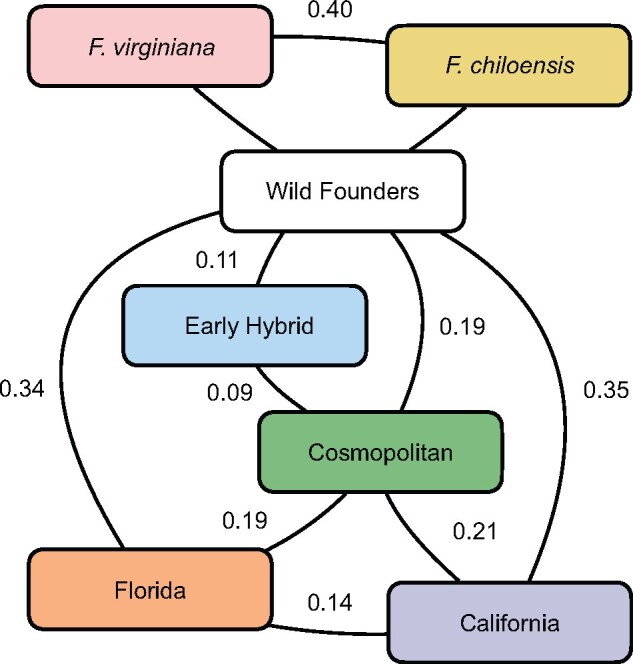
Octoploid strawberry population divergence.Two-population FST statistics estimating genetic divergence between wild, early hybrid Fragaria × ananassa, cosmopolitan Fragaria × ananassa, and the modern California and Florida Fragaria × ananassa populations. FSTstatistics were estimated using 259 octoploid individuals and 446,644 SNPs genotyped with the 850 K SNP array.
We validated the use of all available SNPs for analyzing population admixture by repeating the STRUCTURE analysis (K = 6) in a representative subset of individuals (n = 140) using total genomic SNPs subject to strict LD-pruning (r2 ≤ 0.5; n = 8,585), and neutral SNP sites subject to strict LD-pruning (r2 ≤ 0.5; n = 451). Despite relatively few remaining neutral sites, population admixture proportions predicted from total SNPs and neutral SNPs were nearly identical (supplementary fig. S4, Supplementary Material online). For each analysis, we generated pairwise matrices containing the admixture residual correlations between individuals using software evalAdmix (Garcia-Erill and Albrechtsen 2020) (supplementary fig. S4, Supplementary Material online). The uncorrected total SNP and neutral SNP matrices were highly similar (r2 = 0.96). The negligible bias when estimating population structure from total SNPs can be attributed to LD-pruning, and a low rate of coding sites on the octoploid SNP arrays; 72% of array SNPs were located in nonexonic regions. These results also demonstrated that kinship and admixture can be accurately predicted in octoploid strawberry with fewer than 500 disomic markers.
The maximum-likelihood tree generated by analysis of the 850 K SNP array matrix produced a clear picture of strawberry breeding history (fig. 6A). Early hybrids grouped closely to the wild octoploid founders. These “early” hybrids included several iconic heirloom cultivars in the genetic background of cultivated strawberry (Pincot et al. 2020), for example, “Vicomtesse Hericart de Thury,”“Jucunda,”“Ettersburg 121,” and “Madame Moutot” (light blue clades in fig. 6A). The cosmopolitan clade, closest to early hybrids, included several well-known and iconic cultivars, for example, “Senga Sengana,”“Howard 17,”“Earliglow,”“Mara des Bois,”“Hood,” and “Holiday.” Finally, the modern California population (post-1970) was found to have undergone the greatest differentiation and reduction in nucleotide diversity (figs. 2–6).
Selective Sweeps Associated with Strawberry Domestication
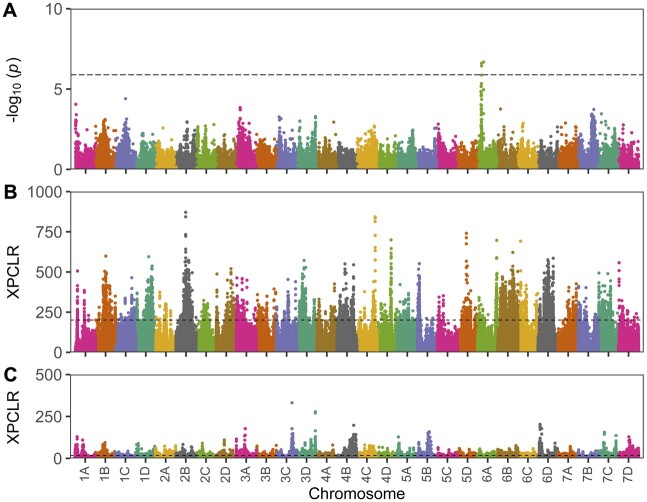
We observed a progressive decline in nucleotide diversity from the wild founders to early hybrids to heirloom cultivars to modern cultivars (figs. 2 and 3; supplementary fig. S3, Supplementary Material online). Hybrids between the wild octoploid founders were significantly more heterozygous than either parent; however, the initial increase in heterozygosity progressively decreased over the course of domestication. The estimated loss of genetic diversity was 11.3% in early and 37.0% in modern phases of domestication (fig. 9; table 3). We hypothesized that these decreases were primarily caused by directional selection and breeding bottlenecks. To explore this further, we scanned the genome for the presence of selective sweeps in early and modern phases of domestication using cross-population composite likelihood ratio (XP-CLR) analysis (Chen, Patterson, and Reich 2010; Hufford et al. 2012). We split individuals into wild founder, heirloom cultivar, and modern (post-1970) California cultivar groups and estimated XP-CLR statistics for the wild founder to heirloom cultivar (early phase) transition and heirloom to modern cultivar (modern phase) transition. We identified 4,064 selectively swept loci in the early phase of domestication (approximately 6.5% of the genome) and 5,248 selectively swept loci in the modern phase of domestication (approximately 6.0% of the genome) with negligible overlap between the two (table 3; fig. 10). The distribution of selectively swept loci mirrored population-specific differences in nucleotide diversity (figs. 2 and 10; supplementary fig. S1, Supplementary Material online; table 3).
Fig. 9.
Genome-wide association study and selective sweeps. (A) Genome-wide association study (GWAS) for fruit firmness (g/cm2) measured with a handheld penetrometer in a population of 466 wild and domesticated individuals. The horizontal dashed line delineates a Bonferroni-corrected P = 0.05 significance threshold. (B,C) Cross-population composite likelihood ratio (XP-CLR) statistics for DNA variants (loci) distributed across the octoploid strawberry genome. The dashed line identifies the upper 0.01 quantile of XP-CLR estimates. (B) XP-CLR statistics were estimated for the “early” phase of domestication by comparing DNA variants between wild ecotypes and both early hybrids and heirloom cultivars. (C) XP-CLR statistics were estimated for the “modern” phase of domestication by comparing DNA variants between modern cultivars and both early and heirloom cultivars.
Table 3.
Selective sweeps in the A, B, C, and D subgenomes of cultivated strawberry in early and modern phases of domestication.
| Subgenomea | Early-phase locib |
Modern-phase locic |
||
|---|---|---|---|---|
| Number | Percent | Number | Percent | |
| A | 1,034 | 23.6 | 1,998 | 35.9 |
| B | 1,673 | 38.2 | 1,280 | 23.0 |
| C | 361 | 8.3 | 900 | 16.2 |
| D | 1,175 | 26.9 | 1,112 | 20.0 |
| Unknown | 134 | 3.1 | 271 | 4.9 |
A, B, C, and D identify the subgenomes of F.×ananassa, which are admixed derivatives of the genomes of four diploid ancestors (see text and Edger et al. 2019).
Cross-population composite likelihood ratio (XP-CLR) statistics were estimated for loci in the early domestication phase by comparing DNA variants in wild octoploid founders (F. chiloensis and F. virginiana) to DNA variants in heirloom cultivars of F.×ananassa. The numbers and percentages of selected loci within each subgenome were estimated using an upper 1% cutoff in the XP-CLR distribution.
XP-CLR statistics were estimated for loci in the modern domestication phase by comparing heirloom to modern California cultivars.
Fig. 10.
Physical locations of selectively swept loci in the octoploid genome. The physical locations of selectively swept loci are shown in the Fragaria × ananassa“Camarosa” v1.0 genome (Edger et al. 2019) for the early phase of domestication (left-hand chromosome in each pair) and modern phase of domestication (right-hand chromosome in each pair). Loci homologous to F. vesca are shown in red. Loci homologous to other diploid ancestors are shown in blue. Chromosomes and locus positions were visualized using the R package RIdeogram (Hao et al. 2020).
The genome fractions that harbored selective sweeps in strawberry were comparable to those reported in maize (5%) and sunflower (Helianthus annuus) (7%) (Vigouroux et al. 2005; Chapman et al. 2008; Hufford et al. 2012). The selection coefficients () estimated for strawberry were, however, 10-fold smaller than those reported for maize and wheat, species with 10,000-year domestication histories (Purugganan and Fuller 2011; Hufford et al. 2012). The strength of selection was 2.5-fold greater in the early () than the modern () phase of domestication. Because of the incredibly short domestication history (<300 years), highly admixed nature of F.×ananassa lineages, and frequent infusion of allelic diversity from wild founders (migration), standing genetic variation should be a more important determinant of the strength of selection in strawberry than de novo mutations (Hermisson and Pennings 2005). Our results were consistent with this hypothesis.
Selective sweeps were observed on several chromosomes in each subgenome but were unequally distributed among the four subgenomes in both the early and modern phases of domestication (fig. 9; table 3, supplementary fig. S1, Supplementary Material online). The proportions of selectively swept loci within each subgenome, ordered from highest to lowest, were B > D > A > C in the early phase and A > B > D > C in the modern phase (fig. 9; table 3; supplementary fig. S1, Supplementary Material online). While the C subgenome harbored the smallest number of loci under selection, loci in each ancestral subgenome were targeted by selection (fig. 9; table 3). The A subgenome harbored roughly 23.6% of selectively swept loci in the early and 35.9% in the modern phase of domestication (table 3). Collectively, 73.4% and 59.2% of the selectively swept loci were found in the B, C, and D subgenomes in the early and modern phases of domestication, respectively. These results highlight the importance of allelic diversity tracing to both the dominant and nondominant diploid ancestors (Edger et al. 2019).
Unlike extensively investigated species with domestication histories spanning millenia, for example, tomato (Solanum lycopersicum), rice (Oryza sativa), maize, and wheat (Doebley 2004; Doebley, Gaut, and Smith 2006; Kovach, Sweeney, and McCouch 2007; Purugganan and Fuller 2009; Gross and Olsen 2010; Chia et al. 2012; Hufford et al. 2012), genes underlying strawberry domestication are largely unknown. To develop insights into the putative functions of genes targeted by selection, their functional categories were identified by GO term enrichment analysis (supplementary file S5, Supplementary Material online). We found that genes targeted by selection were more likely to affect fruit development, cell wall metabolism, the regulation of gene expression, and the regulation and coordination of hormone signaling pathways, including auxin, abscisic acid, and gibberellic acid pathways. The latter have been shown to regulate expansion and ripening in nonclimacteric fruit (Jia et al. 2011; Kang et al. 2013). Selection appears to have targeted genes encoding cell-wall-degrading enzymes known to affect fruit firmness and shelf life, for example, pectin lyases and polygalacturonases (Castillejo et al. 2004; Goulao and Oliveira 2008), in addition to genes encoding expansins and xyloglucan endotransglucosylases, which have been shown to affect fruit ripening and softening and other aspects of plant development (Marowa, Ding, and Kong 2016). Although many candidate genes within selective sweeps could have been targeted by selection, forward genetic studies have not yet uncovered genotype-to-phenotype associations for “domestication loci” (Purugganan and Fuller 2009).
We performed GWAS for two domestication traits, fruit firmness and fruit size, using a diverse set of octoploid individuals (). Narrow-sense genomic heritability estimates were 0.36 for fruit firmness and 0.55 for fruit size in the GWAS population. Statistically significant signals were not observed for fruit size (data not shown); however, we observed a significant signal for fruit firmness on chromosome 6A that overlapped with early-phase domestication sweeps (fig. 9). The most significant SNP associations (P ≤ 0.001) on 6A were observed from Mb 5.89–7.22. This chromosome segment was found to harbor a cluster of three polygalacturonase-encoding genes (FxaC_6-1g13880, FxaC_6-1g13900, and FxaC_6-1g13910) in a selective sweep spanning Mb 7.046–7.064. The enzymes encoded by this gene family are known to affect fruit firmness in apple (Malus domestica) and tomato (Kramer et al. 1992; Atkinson et al. 2012). Moreover, transgenic silencing of polygalacturonase genes in developing strawberry fruit has been shown to increase fruit firmness (Santiago-Doménech et al. 2008; Villarreal et al. 2008; Posé et al. 2015). The candidate polygalacturonase locus identified here could harbor causative mutations targeted by selection and thus represent an important “domestication” trait locus (fig. 9). Apart from this locus, which warrants further study, insights into other functionally important loci underlying the “domestication syndrome” of strawberry are limited. With the infrastructure in place to apply genome-informed approaches in octoploid populations, the opportunity exists to rapidly expand the catalog of loci and mutations associated with horticulturally important phenotypic diversity in strawberry (Doebley, Gaut, and Smith 2006; Purugganan and Fuller 2009).
Concluding Remarks
The present study was one of three companion studies we undertook to develop an in-depth understanding of DNA variation in the octoploid species, unravel the demographic, domestication, and breeding history of cultivated strawberry, and develop an understanding of the feasibility of bioinformatically resolving subgenome-specific DNA variation across the octoploid genome (Hardigan et al. 2020; Pincot et al. 2020). Among the most astonishing discoveries to emerge from these studies were the simplicity and completeness with which homoeologous DNA variants could be resolved and genetically and physically mapped in the octoploid genome using short-read DNA sequences, the presence of massive allelic diversity in wild founder and domesticated populations, and the preservation of significant genetic variation in domesticated populations, even those that have been strongly selected and bottlenecked (figs. 3–7). While diploid models have a logical place in biological studies (Gaston et al. 2020), our findings and many others highlight the feasibility and simplicity with which genetic and genomic approaches can be applied in allo-octoploid populations to discover genotype-to-phenotype associations, identify causal loci and mutations, understand and exploit natural genetic variation, and apply genome-informed breeding approaches (Liston et al.2014; Denoyes et al. 2017; Oh et al. 2019; Hardigan et al. 2020; Whitaker et al. 2020). We concluded from the small genome size (0.81 Gbp), high gene density (approximately 40%), and phenomenal nucleotide diversity that the navigation of the allo-octoploid strawberry genome might even be simpler than that of wheat, allo-tetraploid peanut, and many other paleopolyploid and allo-polyploid plants (Akhunov et al. 2010; Clevenger and Ozias-Akins 2015; Clevenger et al. 2017; Bertioli et al. 2019; Edger et al. 2019; Hardigan et al. 2020). To put octoploid strawberry nucleotide diversity into perspective, the four subgenomes appear to harbor similar diversity, with respect to SNP and INDEL variation, as paleopolyploid maize landraces (Tenaillon et al. 2001; Buckler, Gaut, and McMullen 2006; Gore et al. 2009). Most of this diversity appears to be preserved within domesticated individuals, assisted through clonal preservation of heirloom varieties. There are many unexplored aspects of this diversity, including a deeper exploration and characterization of the admixed genomic landscape and the effects of subgenome fractionation and other evolutionary forces (Freeling et al. 2012; Edger et al. 2018a; Alger and Edger 2020).
Our recent high-density comparative genetic mapping studies and others have substantiated allo-octoploid (disomic) meiotic pairing and segregation in F. chiloensis, F. virginiana, and F.×ananassa; hence, octoploid Fragaria follow the laws of diploid Mendelian genetics (Rousseau-Gueutin et al. 2008; Tennessen et al. 2014; Sargent et al. 2016; Hardigan et al. 2020; Whitaker et al. 2020). Although the four subgenomes independently recombine and segregate in present-day octoploids, they extensively recombined in ancient tetraploid, hexaploid, and octoploid ancestors and underwent subgenome fractionation and other changes as the ancestral polyploids formed and evolved (Freeling et al. 2012; Renny-Byfield et al.2017; Edger et al. 2018a, 2019, 2020). We proposed the A, B, C, and D subgenome nomenclature with the knowledge that none of the extant subgenomes are now purely derived from a single diploid ancestor (fig. 1; table 1; supplementary file S1, Supplementary Material online; Edger et al. (2019, 2020; Liston et al. 2020). These designations correspond to the four homoeologous chromosome complements found in present-day octoploids and provide a common language for identifying and labeling subgenomes, chromosomes, and linkage groups. The proposed nomenclature is agnostic to the origin of DNA on any one of the 28 chromosomes but consistent with observed subgenome fractionation, chromosome homology, and stable allo-octoploid (homoeolog-specific) recombination and segregation (Hardigan et al. 2020) (supplementary file S1, Supplementary Material online). With a logical and coherent genome-anchored nomenclature in place, we are advocating for the adoption of a single nomenclature to facilitate and expedite the exchange of information and future expansion of the catalog of functionally characterized loci, mutations, and genes underlying phenotypic diversity in the octoploid species (table 1; supplementary file S1, Supplementary Material online).
Materials and Methods
Plant Material
We analyzed 1,669 octoploid individuals (germplasm accessions) (supplementary file S2, Supplementary Material online), which included 37 F. chiloensis ecotypes, 40 F. virginiana ecotypes, and 1,592 F.×ananassa individuals. The data for these individuals were divided into subsets and populations as needed for specific analyses. These plant materials are preserved in clonal germplasm collections maintained at the University of California, Davis (UCD), University of Florida (UF), and US Department of Agriculture (USDA) National Plant Germplasm System (NPGS), National Clonal Germplasm Repository, Corvallis, Oregon, USA (https://www.ars.usda.gov/pacific-west-area/corvallis-or/national-clonal-germplasm-repository/, last accessed December 1, 2020). The F.×ananassa individuals included early interspecific hybrids, heirloom and modern cultivars, and unreleased individuals developed at UCD or UF. The UCD individuals analyzed spanned the entire history of the UCD breeding program (Hardigan et al. 2018; Pincot et al. 2020) (supplementary file S2, Supplementary Material online). Accession identification numbers, common names and aliases, sources, and other passport data are documented for every germplasm accession in supplementary file S2, Supplementary Material online.
Octoploid Strawberry Sequencing
We obtained Illumina WGS sequence data for 145 octoploid strawberry individuals (supplementary file S2, Supplementary Material online), including 24 F. chiloensis, 22 F. virginiana, and 99 F.×ananassa (supplementary file S2, Supplementary Material online). Newly emerging leaves were harvested from greenhouse or field grown plants (Davis or Winters, CA, USA). Genomic DNA was isolated from leaf tissue using the E-Z 96 Plant DNA kit (Omega Bio-Tek, Norcross, GA, USA) with Proteinase K added to the initial buffer and RNase treatment following lysate separation from the cellular debris. The manufacturers protocol was modified to include an additional spin step and incubation was carried out at 65 °C during elution. Paired-end sequencing libraries (PE150) were prepared using the KAPA Hyper Plus kit (Kapa Biosystems, Wilmington, MA, USA) and BIOO Nextflex adapters (BIOO Scientific, Austin, TX, USA). DNA was sheared using the Covaris E220 (Covaris Inc., Woburn, MA, USA) and size selected for an average insert size of 300-nt using magnetic beads (Mag-Bind® RxnPure Plus, Omega Bio-tek). Library QC was performed on a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Libraries were pooled and sequenced on a NovaSeq platform (Illumina, Inc., San Diego, CA, USA) at the UCSF Center for Advanced Technology, San Francisco, CA. Eight additional octoploid WGS datasets (PE100) were obtained from the NCBI SRA (SRR1513906, SRR1513893, SRR1513905, SRR1513903, SRR1513892, SRR1513904, SRR1513867, and SRR1513873). The resequenced F.×ananassa individuals included historically and commercially important heirloom and modern cultivars developed at UCD, UF, and other public institutions, in addition to historically important common ancestors of heirloom and modern cultivars identified by (Pincot et al. 2020) (supplementary file S2, Supplementary Material online).
WGS DNA Variant Calling
We called SNP and INDEL DNA variants using sequences that aligned uniquely to a single subgenome (A, B, C, or D) of the “Camarosa” v1.0 octoploid reference genome (Edger et al. 2019). Illumina reads were quality-trimmed using CutAdapt (v1.8) (Martin 2011) with default parameters and a minimum Phred score of 25. Trimmed reads were aligned to the “Camarosa” v1.0 genome using BWA-mem (v0.7.16) (Li 2013), processed to mark optical and PCR duplicates using Picard Tools (v2.18) (http://broadinstitute.github.io/picard, last accessed December 1, 2020), and INDEL-realigned using GATK (v3.8) (McKenna et al. 2010). Uniquely mapped reads (MapQ > 20) were used to predict variants with FreeBayes (v1.2) (Garrison and Marth 2012) and filtered with vcflib (https://github.com/vcflib/vcflib, last accessed December 1, 2020). A set of hard-filters was applied to remove variants with low site quality (vcflib: QUAL > 40), low contribution of allele observations to site quality (vcflib: QUAL/AO > 10), low read coverage (vcflib: DP > 500), strand bias (vcflib: SAF > 0 and SAR > 0), read-placement bias (RPR > 1 and RPL ≤ 1), unbalanced mapping quality of reference and alternate alleles (vcflib: 0.4 ≤ [MQM/MQMR] ≤ 2.5), unbalanced allele frequencies at heterozygous sites (vcflib: 0.2 ≤ AB ≤ 0.8), low end-placement probability score (EPP 3), and low strand-bias probability score (vcflib: SRP ≤ and SAP ≤ ). Individual sample genotypes were required to have individual read coverage , and at least two reads and a minimum of 0.20 read observations supporting each allele.
SNP Array Genotyping
We genotyped 1,387 individuals with 50 K SNP array only, 112 individuals with an 850 K SNP array only, and 144 individuals with both the 50 K and 850 K SNP arrays; hence, 1,643 individuals were genotyped with one or both SNP arrays (Affymetrix Inc., Santa Clara, CA, USA) (Hardigan et al. 2020). These included 32 F. chiloensis, 35 F. virginiana, and 1,576 F.×ananassaindividuals (supplementary file S2, Supplementary Material online). They were divided into subsets and populations as needed for specific analyses. Both SNP arrays were populated with probes that yield a high percentage of subgenome-specific codominant genotypic assays. Genomic DNA was isolated from samples using methods described for WGS sequencing libraries. CEL files containing sample fluorescence data were imported into the Affymetrix Axiom Analysis Suite (v1.1.1.66), and run in “polyploid” mode to predict marker genotype clusters.
Individual and Population Level Diversity Statistics
Population-level nucleotide diversity () and individual sample heterozygosity () estimates were calculated based on 41.8 M SNP and INDEL sequence variants using a custom perl script. Nucleotide diversity estimates were calculated genome-wide and in nonoverlapping 25 kb chromosome windows as the sum of pairwise diversity for all variant sites divided by total nongap (N) genomic nucleotides within a target region. Individual genome heterozygosity was calculated as the sum of heterozygous variant sites in a sample divided by total nongap (N) genomic nucleotides. Nei’s genetic distances were calculated using the “gendist” function in the PHYLIP (v3.69) software package ( https://evolution.genetics.washington.edu/phylip/, last accessed December 1, 2020; Nei and Li 1979; Nei 1987; Felsenstein 1989). For estimation of relative wild founder allele dosage, we reported the ratio of F.×ananassa genetic distance to F. chiloensis and F.×ananassa genetic distance to F. virginiana in nonoverlapping 10 kb chromosome windows. We generated two-population estimates using the R package SNPRelate (v1.6.4) (Zheng et al. 2012).
Population Structure Analysis
WGS, 850 K SNP array, and 50 K SNP array genotype matrices were LD-pruned (r2 ≤ 0.70) using the R package SNPRelate (v1.6.4) (Zheng et al. 2012). LD-pruned genotype matrices were used to evaluate population structure by principal component analysis (SNPRelate) and clustering with an admixture model using STRUCTURE (v.2.3.4) (https://web.stanford.edu/group/pritchardlab/structure.html, last accessed December 1, 2020; Pritchard, Stephens, and Donnelly 2000). The STRUCTURE analysis tested for K = 2–14 subpopulations (25,000 burn-in steps and 50,000 Markov-Chain Monte Carlo (MCMC) steps with 10 replicates per value. The optimal subpopulation () value was determined using the Evanno et al. (2005) method as applied by STRUCTURE HARVESTER (v0.6.94) (Earl and vonHoldt 2012) with sample orders calculated using CLUMPP (v1.1.2) (Jakobsson and Rosenberg 2007). We validated the use of all SNP sites for admixture prediction by repeating the STRUCTURE analysis in a representative subset of individuals (n = 140) using all SNPs subject to strict LD-pruning (r2 ≤ 0.5; n = 8,585), and neutral coding SNPs subject to strict LD-pruning (r2 ≤ 0.5; n = 451). The results were visualized using evalAdmix (Garcia-Erill and Albrechtsen 2020).
Phylogenetic Analysis
The wild octoploid subspecies cladogram was generated by merging the genotype matrix of wild individuals assayed in the present study with the 50 K SNP array and the genotype matrix of wild individuals assayed using the iStraw35 SNP array (Bassil et al. 2015; Verma et al. 2017) by Hardigan et al. (2018). We retained SNP markers tiled on both arrays and selected SNP markers that were polymorphic and contained no missing data. Using PHYLIP (v3.69) we generated 1,000 bootstrap datasets with the “seqboot” function, estimated genetic distance matrices using the “gendist” function, performed neighbor-joining analysis using the “neighbor” function, and generated the consensus tree using the “consense” function (Felsenstein 1989). A maximum-likelihood phylogenetic tree was constructed for 273 wild and domesticated individuals using the LD-pruned 850 K SNP array genotype matrix. The tree was generated in PHYLIP based on 1,000 bootstrap datasets using the “contml” function. Trees were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/, last accessed December 1, 2020).
Selective Sweep Analyses
Selective sweeps analyses were performed using the cross-population comparative likelihood ratio method implemented in XP-CLR (Chen et al.2010) with DNA variants (41.8 M SNPs and INDELs) called among WGS sequence alignments. We applied a fixed recombination rate of cM/bp as described by Tiley et al.(2015). We split individuals into three groups to scan the genome for selective sweeps in early and modern phases of domestication: wild ecotypes (), early hybrids and heirloom cultivars (), and modern cultivars () (supplementary file S2, Supplementary Material online). For the “early” domestication phase, we compared wild ecotypes to all pre-1970 cultivars (supplementary file S2, Supplementary Material online). For the “modern” domestication phase, we compared early hybrids and heirloom cultivars to modern (post-1970) UCD cultivars (supplementary file S2, Supplementary Material online). XP-CLR was run on overlapping 10 kb windows with a 1 kb step size. As recommended by (Chen, Patterson, and Reich 2010), we down-weighted variants in high LD (> 0.7). We selected the 1% of windows with highest selections coefficients as putative selective sweep windows. Windows without scores were removed and adjacent windows were merged to form single sweep regions. The coefficients of selection () were calculated as described by Chen et al.(2010).
Genome-Wide Association Study
Fruit size (g/berry) and fruit firmness (g/cm2) were measured on 466 wild and domesticated individuals in 2018 from unreplicated six-plant plots grown in Ventura, CA under commercial field conditions. The composition of the GWAS population is shown in supplementary file S2, Supplementary Material online. Fruit firmness was measured on six randomly selected berries/accession as maximum force with a QA Supplies FT2 handheld penetrometer equipped with a 3 mm probe (QA Supplies, Norfolk, VA, USA) (Abbott 1999). These individuals () were genotyped with a 50 K array SNP array (Hardigan et al. 2020). GWAS was performed in TASSEL (v5) (Bradbury et al. 2007) using a mixed linear model (MLM) analysis. Marker genotypes were imported in HapMap format and filtered using a MAF of 0.05. The kinship matrix was estimated using TASSEL. The sample Q-matrix was estimated using STRUCTURE and imported into TASSEL to account for population structure. Manhattan plots were produced by plotting -log10P-values for individual DNA marker loci by physical positions (Mb) in the “Camarosa” v1.0 reference genome, excluding loci with P.
Data Availability
WGS DNA sequences for resequenced individuals are available at the NCBI Short Read Archive under BioProject PRJNA578384. The octoploid (F.×ananassa ’Camarosa’ v1.0) reference genome (Edger et al. 2019) is available on DRYAD (https://doi.org/10.5061/dryad.b2c58pc, last accessed December 1, 2020) and the Genome Database for Rosaceae (https://www.rosaceae.org/species/fragaria_x_ananassa/genome_v1.0.a1, last accessed December 1, 2020). All supplemental files and custom scripts are available on DRYAD (https://doi.org/10.25338/B8RH0G, last accessed December 1, 2020).
Supplementary Material
Acknowledgments
This research was supported by grants to SJK from the United Stated Department of Agriculture (http://dx.doi.org/10.13039/100000199) National Institute of Food and Agriculture (NIFA) Specialty Crops Research Initiative (#2017-51181-26833), California Strawberry Commission (http://dx.doi.org/10.13039/100006760), and the University of California, Davis (http://dx.doi.org/10.13039/100007707), a USDA NIFA Postdoctoral Research Fellowship awarded to MAH (#2018-67012-27980), a USDA-ARS Small Fruit Crop Germplasm Committee grant awarded to NB (#2072-21000-049-06-S), and two USDA-NIFA (#1009804 and #2020-67013-30870) and one National Science Foundation (#2029959) grant awarded to PPE.
Supporting Information
Supplementary files S1–S5 are available online at DRYAD (https://doi.org/10.25338/B8RH0G, last accessed December 1, 2020). Supplementary figures S1–S3 are online at Molecular Biology and Evolution (http://www.mbe.oxfordjournals.org/, last accessed December 1, 2020). is the genetic distance between Fragaria ×ananassa and F is the genetic distance between Fragaria
References
- Abbott JA. 1999. Quality measurement of fruits and vegetables. Postharvest Biol. Tec. 15(3):207–225. [Google Scholar]
- Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Deal KR, et al. 2010. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics.11(1):702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alger EI, Edger PP.. 2020. One subgenome to rule them all: underlying mechanisms of subgenome dominance. Curr.Opin. Plant Biol. 54:108–113. [DOI] [PubMed] [Google Scholar]
- Atkinson RG, Sutherland PW, Johnston SL, Gunaseelan K, Hallett IC, Mitra D, Brummell DA, Schröder R, Johnston JW, Schaffer RJ.. 2012. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol.12:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfourier F, Bouchet S, Robert S, De Oliveira R, Rimbert H, Kitt J, Choulet F, Paux E, Consortium IWGS, Consortium B, International Wheat Genome Sequencing Consortium, et al. 2019. Worldwide phylogeography and history of wheat genetic diversity.Sci Adv. 5(5):eaav0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnet J. 1826. An account and description of the different varieties of strawberries which have been cultivated and examined in the Garden of the Horticultural Society of London.Trans. Hort. Soc. London.VI:145–224. [Google Scholar]
- Bassil NV, Davis TM, Zhang H, Ficklin S, Mittmann M, Webster T, Mahoney L, Wood D, Alperin ES, Rosyara UR, et al. 2015. Development and preliminary evaluation of a 90K Axiom®SNP array for the allo-octoploid cultivated strawberry Fragaria × ananassa. BMC Genomics.16(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli DJ, Jenkins J, Clevenger J, Dudchenko O, Gao D, Seijo G, Leal-Bertioli SCM, Ren L, Farmer AD, Pandey MK, et al. 2019. The genome sequence of segmental allotetraploid peanut Arachis hypogaea.Nat Genet. 51(5):877–884. [DOI] [PubMed] [Google Scholar]
- Bird KA, VanBuren R, Puzey JR, Edger PP.. 2018. The causes and consequences of subgenome dominance in hybrids and recent polyploids.New Phytol.220(1):87–93. [DOI] [PubMed] [Google Scholar]
- Booker TR, Jackson BC, Keightley PD.. 2017. Detecting positive selection in the genome.BMC Biol.15(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES.. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics.23(19):2633–2635. [DOI] [PubMed] [Google Scholar]
- Buckler ES, Gaut BS, McMullen MD.. 2006. Molecular and functional diversity of maize. Curr.Opin. Plant Biol. 9(2):172–176. [DOI] [PubMed] [Google Scholar]
- Byrne PF, Volk GM, Gardner C, Gore MA, Simon PW, Smith S.. 2018. Sustaining the future of plant breeding: the critical role of the USDA-ARS National Plant Germplasm System.Crop Sci.58(2):451–468. [Google Scholar]
- Castillejo C, de la Fuente JI, Iannetta P, Botella MÁ, Valpuesta V.. 2004. Pectin esterase gene family in strawberry fruit: study of FaPE1, a ripening-specific isoform. J. Exp. Bot. 55(398):909–918. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Pashley CH, Wenzler J, Hvala J, Tang S, Knapp SJ, Burke JM.. 2008. A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus). Plant Cell.20(11):2931–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D.. 1993. The effect of deleterious mutations on neutral molecular variation.Genetics.134(4):1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Patterson N, Reich D.. 2010. Population differentiation as a test for selective sweeps.Genome Res. 20(3):393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J-M, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, Elshire RJ, Gaut B, Geller L, Glaubitz JC, et al. 2012. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 44(7):803–807. [DOI] [PubMed] [Google Scholar]
- Clausen RE. 1915. Ettersburg strawberries. J. Hered. 6(7):324–331. [Google Scholar]
- Clevenger J, Chu Y, Chavarro C, Agarwal G, Bertioli DJ, Leal-Bertioli SCM, Pandey MK, Vaughn J, Abernathy B, Barkley NA, et al. 2017. Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Mol. Plant.10(2):309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger JP, Ozias-Akins P.. 2015. SWEEP: a tool for filtering high-quality SNPs in polyploid crops. G3.5:1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid.Nat Rev Genet.6(11):836–846. [DOI] [PubMed] [Google Scholar]
- Cvijović I, Good BH, Desai MM.. 2018. The effect of strong purifying selection on genetic diversity.Genetics.209(4):1235–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale A, Sjulin TM, others1990. Few cytoplasms contribute to North American strawberry cultivars.HortSci .25(11):1341–1342. [Google Scholar]
- Darrow GM. 1937. United States department of agriculture yearbook of agriculturestrawberry improvement. Washington(DC): United States Government Printing Office. p. 445–495. [Google Scholar]
- Darrow GM. 1966. The strawberry.History, breeding and physiology. New York: Holt, Rinehart & Winston. [Google Scholar]
- Denoyes B, Amaya I, Liston A, Tennessen J, Ashman T-L, Whitaker VM, Hytönen T, van de Weg E, Osorio S, Folta KM, et al. 2017. Genomics tools available for unravelling mechanisms underlying agronomical traits in strawberry with more to come.Acta Hortic. 1156(1156):13–24. [Google Scholar]
- Dillenberger MS, Wei N, Tennessen JA, Ashman T-L, Liston A.. 2018. Plastid genomes reveal recurrent formation of allopolyploid Fragaria. Am J Bot.105(5):862–874. [DOI] [PubMed] [Google Scholar]
- Doebley J. 2004. The genetics of maize evolution.Annu Rev Genet.38(1):37–59. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD.. 2006. The molecular genetics of crop domestication.Cell.127(7):1309–1321. [DOI] [PubMed] [Google Scholar]
- Duchesne A-N. 1766. Histoire naturelle des fraisiers. Paris, France: Didot le Jeune et C. J. Panckoucke. [Google Scholar]
- Earl DA, vonHoldt BM.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method.Conservation Genet Resour. 4(2):359–361. [Google Scholar]
- Edger PP, McKain MR, Bird KA, VanBuren R.. 2018a. Subgenome assignment in allopolyploids: challenges and future directions. Curr.Opin. Plant Biol. 42:76–80. [DOI] [PubMed] [Google Scholar]
- Edger PP, McKain MR, Yocca AE, Knapp SJ, Qiao Q, Zhang T.. 2020. Reply to: revisiting the origin of octoploid strawberry. Nat Genet. 52(1):5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Poorten TJ, VanBuren R, Hardigan MA, Colle M, McKain MR, Smith RD, Teresi SJ, Nelson ADL, Wai CM, et al. 2019. Origin and evolution of the octoploid strawberry genome.Nat Genet. 51(3):541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, VanBuren R, Colle M, Poorten TJ, Wai CM, Niederhuth CE, Alger EI, Ou S, Acharya CB, Wang J, et al. 2018b. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience.7(2):. gix124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study.Mol Ecol.14(8):2611–2620. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP - phylogeny inference package. Cladistics. 5:164–166. [Google Scholar]
- Feng C, Wang J, Harris AJ, Folta KM, Zhao M, Kang M.. 2021. Tracing the diploid ancestry of the cultivated octoploid strawberry.Mol. Biol. Evol. 38:478–785. doi: 10.1093/molbev/msaa238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn CE, Retamales JB, Lobos GA, Hancock JF.. 2013. The Chilean strawberry (Fragaria chiloensis): Over 1000 years of domestication.Horts .48(4):418–421. [Google Scholar]
- Fletcher SW. 1917. The Strawberry in North America: history, origin, botany, and breeding. New York: The Macmillan Company. [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, et al. 2004. Assessing the impact of population stratification on genetic association studies.Nat Genet. 36(4):388–393. [DOI] [PubMed] [Google Scholar]
- Freeling M, Woodhouse MR, Subramaniam S, Turco G, Lisch D, Schnable JC.. 2012. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr.Opin. Plant Biol. 15(2):131–139. [DOI] [PubMed] [Google Scholar]
- Garcia‐Erill G, Albrechtsen A.. 2020. Evaluation of model fit of inferred admixture proportions. Mol Ecol Resour. 20(4):936–949. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907.
- Gaston A, Osorio S, Denoyes B, Rothan C.. 2020. Applying the Solanaceae strategies to strawberry crop improvement.Trends Plant Sci. 25(2):130–140. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Long AD.. 2003. The lowdown on linkage disequilibrium. Plant Cell.15(7):1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Seymour DK, Liu Q, Zhou Y.. 2018. Demography and its effects on genomic variation in crop domestication.Nat. Plants. 4(8):512–520. [DOI] [PubMed] [Google Scholar]
- Gore MA, Chia J-M, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. 2009. A first-generation haplotype map of maize.Science.326(5956):1115–1117. [DOI] [PubMed] [Google Scholar]
- Goulao LF, Oliveira CM.. 2008. Cell wall modifications during fruit ripening: when a fruit is not the fruit. Trends Food Sci. Technol. 19(1):4–25. [Google Scholar]
- Gross BL, Olsen KM.. 2010. Genetic perspectives on crop domestication.Trends Plant Sci. 15(9):529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Luby J, Dale A.. 1993. Should we reconstitute the strawberry? Acta Hortic.348(348):86–93. [Google Scholar]
- Hancock JF, Callow PW, Dale A, Luby JJ, Finn CE, Hokanson SC, Hummer KE.. 2001. From the Andes to the Rockies: native strawberry collection and utilization.HortSci .36(2):221–224. [Google Scholar]
- Hancock JF, Lavín A, Retamales JB.. 1999. Our southern strawberry heritage: fragaria chiloensis of Chile.HortSci .34(5):814–816. [Google Scholar]
- Hancock JF, Bringhurst RS.. 1979. Ecological differentiation in perennial, octoploid species of Fragaria. Am. J. Bot. 66(4):367–375. [Google Scholar]
- Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G, Chen J.. 2020. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 6:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan MA, Feldmann MJ, Lorant A, Bird KA, Famula R, Acharya C, Cole G, Edger PP, Knapp SJ.. 2020. Genome synteny has been conserved among the octoploid progenitors of cultivated strawberry over millions of years of evolution. Front Plant Sci. 10:1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan MA, Poorten TJ, Acharya CB, Cole GS, Hummer KE, Bassil N, Edger PP, Knapp SJ.. 2018. Domestication of temperate and coastal hybrids with distinct ancestral gene selection in octoploid strawberry. Plant Genome.11(3):180049. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG, Clark AG.. 1997. Principles of population genetics. Sunderland(MA: ): Sinauer Associates. [Google Scholar]
- van Heerwaarden J, Hufford MB, Ross-Ibarra J.. 2012. Historical genomics of North American maize.Proc. Natl. Acad. Sci. U.S.A. 109(31):12420–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS.. 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics.169(4):2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Sánchez-Sevilla JF, Punelli F, Richard L, Sesmero-Carrasco R, Leone A, Höefer M, Chartier P, Balsemin E, Barreneche T, et al. 2011. Structured diversity in octoploid strawberry cultivars: importance of the old European germplasm. Ann. Appl. Biol. 159(3):358–371. [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia J-M, Cartwright RA, Elshire RJ, Glaubitz JC, Guill KE, Kaeppler SM, et al. 2012. Comparative population genomics of maize domestication and improvement.Nat Genet. 44(7):808–811., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA.. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics.23(14):1801–1806. [DOI] [PubMed] [Google Scholar]
- Jia H-F, Chai Y-M, Li C-L, Lu D, Luo J-J, Qin L, Shen Y-Y.. 2011. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol.157(1):188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z.. 2013. Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell.25(6):1960–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR.. 2007. New insights into the history of rice domestication.Trends Genet.23(11):578–587. [DOI] [PubMed] [Google Scholar]
- Kramer M, Sanders R, Bolkan H, Waters C, Sheeny RE, Hiatt WR.. 1992. Postharvest evaluation of transgenic tomatoes with reduced levels of polygalacturonase: processing, firmness and disease resistance. Postharvest Biol. Technol. 1(3):241–255. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997.
- Liston A, Cronn R, Ashman T-L.. 2014. Fragaria: a genus with deep historical roots and ripe for evolutionary and ecological insights. Amer. J. Bot. 101(10):1686–1699. [DOI] [PubMed] [Google Scholar]
- Liston A, Wei N, Tennessen JA, Li J, Dong M, Ashman T-L.. 2020. Revisiting the origin of octoploid strawberry. Nat Genet. 52(1):2–4. [DOI] [PubMed] [Google Scholar]
- Marowa P, Ding A, Kong Y.. 2016. Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 35(5):949–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads.Embnet J .17(1):10–12. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD.. 2013. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet.14(12):840–852. [DOI] [PubMed] [Google Scholar]
- Millet A. 1898. Les Fraisiers. Paris, France: Librarie Agricole de La Maison Rustique. [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Nei M, Li W-H.. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76(10):5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Tavaré S.. 2002. Linkage disequilibrium: what history has to tell us.Trends Genet.18(2):83–90. [DOI] [PubMed] [Google Scholar]
- Oh Y, Zurn JD, Bassil N, Edger PP, Knapp SJ, Whitaker VM, Lee S.. 2019. The strawberry DNA testing handbook.Horts .54(12):2267–2270. [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincot DDA, Ledda M, Feldmann MJ, Hardigan MA, Poorten TJ, Runcie DE, Heffelfinger C, Dellaporta S, Cole GS, Knapp SJ.. 2020. Social Network Analysis of the Genealogy of Strawberry: retracing the Wild Roots of Heirloom and Modern Cultivars. biorXiv. 10.1101/2020.09.30.320689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincot DDA, Poorten TJ, Hardigan MA, Harshman JM, Acharya CB, Cole GS, Gordon TR, Stueven M, Edger PP, Knapp SJ.. 2018. Genome-wide association mapping uncovers Fw1, a dominant gene conferring resistance to Fusarium wilt in strawberry. G3.8:1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont C, Murat F, Guizard S, Flores R, Foucrier S, Bidet Y, Quraishi UM, Alaux M, Doležel J, Fahima T, . et al. 2013. Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J. 76(6):1030–1044. [DOI] [PubMed] [Google Scholar]
- Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos-Korts D, Goué N, Balfourier F, et al. 2019. Tracing the ancestry of modern bread wheats. Nat Genet. 51(5):905–911. [DOI] [PubMed] [Google Scholar]
- Posé S, Kirby AR, Paniagua C, Waldron KW, Morris VJ, Quesada MA, Mercado JA.. 2015. The nanostructural characterization of strawberry pectins in pectate lyase or polygalacturonase silenced fruits elucidates their role in softening. Carbohydr.Polym.132:134–145. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics.155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Fuller DQ.. 2009. The nature of selection during plant domestication. Nature.457(7231):843–848. [DOI] [PubMed] [Google Scholar]
- Purugganan MD, Fuller DQ.. 2011. Archaeological data reveal slow rates of evolution during plant domestication. Evolution.65(1):171–183. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Rodgers-Melnick E, Ross-Ibarra J.. 2017. Gene fractionation and function in the ancient subgenomes of maize. Mol. Biol. Evol. 34(8):1825–1832. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra J, Morrell PL, Gaut BS.. 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. U.S.A. 104(Supplement 1):8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Lerceteau-Köhler E, Barrot L, Sargent DJ, Monfort A, Simpson D, Arus P, Guérin G, Denoyes-Rothan B.. 2008. Comparative genetic mapping between octoploid and diploid Fragaria species reveals a high level of colinearity between their genomes and the essentially disomic behavior of the cultivated octoploid strawberry. Genetics.179(4):2045–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J. 2016. Deciphering the evolutionary interplay between subgenomes following polyploidy: A paleogenomics approach in grasses. Am J Bot. 103(7):1167–1174. [DOI] [PubMed] [Google Scholar]
- Sánchez-Sevilla JF, Horvath A, Botella MA, Gaston A, Folta K, Kilian A, Denoyes B, Amaya I.. 2015. Diversity Arrays Technology (DArT) marker platforms for diversity analysis and linkage mapping in a complex crop, the octoploid cultivated strawberry (Fragaria × ananassa).PLoS One.10(12):e0144960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Doménech N, Jiménez-Bemúdez S, Matas AJ, Rose JKC, Muñoz-Blanco J, Mercado JA, Quesada MA.. 2008. Antisense inhibition of a pectate lyase gene supports a role for pectin depolymerization in strawberry fruit softening. J. Exp. Bot. 59(10):2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent DJ, Yang Y, Šurbanovski N, Bianco L, Buti M, Velasco R, Giongo L, Davis TM.. 2016. HaploSNP affinities and linkage map positions illuminate subgenome composition in the octoploid, cultivated strawberry (Fragaria × ananassa). Plant Sci. 242:140–150. [DOI] [PubMed] [Google Scholar]
- Schrider DR, Shanku AG, Kern AD.. 2016. Effects of linked selective sweeps on demographic inference and model selection. Genetics.204(3):1207–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, . et al. 2011. The genome of woodland strawberry (Fragaria vesca). Nat Genet. 43(2):109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjulin TM. 2006. Private strawberry breeders in California. HortSci .41(1):17–19. [Google Scholar]
- Sjulin TM, Dale A.. 1987. Genetic diversity of North American strawberry cultivars. J. Am. Soc. Hortic. Sci. 112: [Google Scholar]
- Staudt G. 1962. Taxonomic studies in the genus Fragaria typification of Fragaria species known at the time of Linnaeus. Can J Bot. 40(6):869–886. [Google Scholar]
- Staudt G. 1989. The species of Fragaria, their taxonomy and geographical distribution. Acta Hortic.265:23–34. [Google Scholar]
- Staudt G. 1999. Systematics and geographic distribution of the American strawberry species: taxonomic studies in the genus Fragaria (Rosaceae: potentilleae). Berkeley: University of California Press. [Google Scholar]
- Staudt G. 2003. Les dessins d’Antoine Nicolas Duchesne pour son Histoire naturelle des fraisiers. Paris, France: Publications scientifiques du Muséum. [Google Scholar]
- Staudt G. 2009. Strawberry biogeography, genetics and systematics. Acta Hortic.842(842):71–84. [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS.. 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. U.S.A. 98(16):9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Govindarajulu R, Ashman T-L, Liston A.. 2014. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol.6(12):3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiley GP, Burleigh JG, Burleigh G.. 2015. The relationship of recombination rate, genome structure, and patterns of molecular evolution across angiosperms. BMC Evol Biol.15:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Bassil NV, van de Weg E, Harrison RJ, Monfort A, Hidalgo JM, Amaya I, Denoyes B, Mahoney L, Davis TM, et al. 2017. Development and evaluation of the Axiom®IStraw35 384HT array for the allo-octoploid cultivated strawberry Fragaria × ananassa. Acta Hortic.1156(1156):75–82. [Google Scholar]
- Vigouroux Y, Mitchell S, Matsuoka Y, Hamblin M, Kresovich S, Smith JSC, Jaqueth J, Smith OS, Doebley J.. 2005. An analysis of genetic diversity across the maize genome using microsatellites. Genetics.169(3):1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal NM, Rosli HG, Martínez GA, Civello PM.. 2008. Polygalacturonase activity and expression of related genes during ripening of strawberry cultivars with contrasting fruit firmness. Postharvest Biol. Technol. 47(2):141–150. [Google Scholar]
- Whitaker VM, Knapp SJ, Hardigan MA, Edger PP, Slovin JP, Bassil NV, Hytönen T, Mackenzie KK, Lee S, Jung S.. 2020. A roadmap for research in octoploid strawberry.Hortic. Res. 7:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, , Davis TM. 2017. A New Perspective on Polyploid Fragaria (Strawberry) Genome Composition Based on Large-Scale, Multi-Locus Phylogenetic Analysis. Genome Biol Evol. 9(12):3433–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS.. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 28(24):3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Massonnet M, Sanjak JS, Cantu D, Gaut BS.. 2017. Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc. Natl. Acad. Sci. U S. A.114(44):11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS DNA sequences for resequenced individuals are available at the NCBI Short Read Archive under BioProject PRJNA578384. The octoploid (F.×ananassa ’Camarosa’ v1.0) reference genome (Edger et al. 2019) is available on DRYAD (https://doi.org/10.5061/dryad.b2c58pc, last accessed December 1, 2020) and the Genome Database for Rosaceae (https://www.rosaceae.org/species/fragaria_x_ananassa/genome_v1.0.a1, last accessed December 1, 2020). All supplemental files and custom scripts are available on DRYAD (https://doi.org/10.25338/B8RH0G, last accessed December 1, 2020).