Abstract
The genomes of inbred mice harbor around 50 endogenous murine leukemia virus (MLV) loci, although the specific complement varies greatly between strains. The Gv1 locus is known to control the transcription of endogenous MLVs and to be the dominant determinant of cell-surface presentation of MLV envelope, the GIX antigen. Here, we identify a single Krüppel-associated box zinc finger protein (ZFP) gene, Zfp998, as Gv1 and show it to be necessary and sufficient to determine the phenotype. By long-read sequencing of bacterial artificial chromosome clones from 129 mice, the prototypic strain, we reveal the source of sufficiency and deficiency as splice-acceptor variations and highlight the varying origins of the chromosomal region encompassing Gv1. Zfp998 becomes the second identified ZFP gene responsible for epigenetic suppression of endogenous MLVs in mice and further highlights the prominent role of this gene family in control of endogenous retroviruses.
Keywords: Gv1, Sgp3, KRAB ZFP, TRIM28, murine leukemia virus, endogenous retrovirus
Introduction
Exogenous murine leukemia viruses (MLVs) and endogenous MLV loci have long been studied in mice (Stoye2012). Although few laboratory mouse strains harbor MLVs capable of replication within young, immunocompetent mice, defective proviruses can still influence immune function (Young, Ploquin, et al. 2012) and cause disease (Paprotka et al. 2011; Young, Eksmond, et al. 2012). A variety of epigenetic mechanisms thus control the expression of endogenous retroviruses (ERVs) (Yang et al. 2015), including Krüppel-associated box zinc finger proteins (ZFPs), such as Zfp809 and Zfp708 (Wolf and Goff 2009; Seah et al. 2019), which recruit Trim28 and Setdb1 to ERVs to mediate their silencing (Jacobs et al. 2014; Ecco et al. 2017; Wolf et al. 2020).
Presentation of MLV surface (SU) glycoprotein, historically the “GIX antigen”, varies between strains of laboratory mice (Geering et al. 1966; Stockert et al. 1971; Obata et al. 1975; Tung, Fleissner, et al. 1975; Tung, Vitetta, et al. 1975). Crosses of (129) and (C57BL/6J, B6/J) strains revealed two unlinked loci controlling the phenotype, including Gv1, which influences tissue-specific (Levy et al. 1982) steady-state expression levels of multiple proviruses (Elder et al. 1977) and was proposed to encode a trans-acting regulatory factor with no linkage to known proviruses (Levy et al. 1985; Oliver1999). Backcrossing placed Gv1 alongside Sgp3 (otherwise known as Bxs6/Elspg2)(Oliver and Stoye 1999; Tucker et al. 2000; Haywood et al. 2001; Rankin et al. 2007) (fig. S1), which is analogously linked to serum gp70 levels and anti-gp70 immune responses, yet their relationship has not been conclusively addressed and the loci reportedly control different subsets of the ecotropic, modified-/polytropic, and xenotropic MLV subclasses (Oliver and Stoye 1999; Baudino et al. 2008; Yoshinobu et al. 2009; Treger et al. 2019).
Both 129- and B6- strains have previously been generated through serial backcrossing but have unfortunately been lost (Stockert et al. 1975). Extending recent work on Sgp3 (Treger et al. 2019), we have now sought to formally identify the gene corresponding to Gv1. We determine Zfp998 (2410141K09Rik) as necessary and sufficient to control the GIX phenotype and identify the source of deficiency at the locus in the prototypic strain, 129.
Results
Structural Variation around Gv1
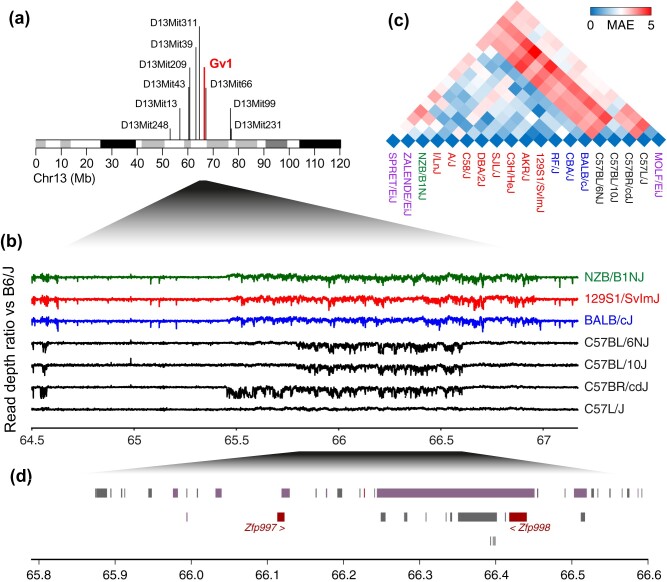
Given Gv1’smapping preceded publication of the mouse genome, we first reanalyzed previously-acquired large-scale backcross data (Oliver1999) to confirm its location. Assessment of nine markers across a 1,108-animal backcross (fig. S2) refined its location to the region between D13Mit311 and D13Mit66 (13:63.64–67.17 Mb) and identified peak linkage at 13:66.58 Mb (fig. 1a), within a large cluster of ZFPs (Kauzlaric et al. 2017). ZFP clusters form through local gene duplication events (Huntley et al. 2006) and extensive self-homology complicates their analysis (fig. S3); indeed, although many variations have been reported within this area (Henrichsen et al. 2009; Quinlan et al. 2010; Keane et al. 2011; Pezer et al. 2015), no two studies identified shared differences or grouped inbred strains consistently. Similarly, Mouse Genomes Project variant calls (Keane et al. 2011; Yalcin et al. 2011) revealed no consistent differences between and mouse strains or between Sgp3+ and Sgp3–strains within the candidate region.
Fig. 1.
The chromosomal context of Gv1.(a) Reanalysis of a large-scale backcross (fig. S2) highlighting the location of Gv1 on Chr 13 (GRCm38). The linkage peak was predicted by scaling predicted genetic distances to the flanking markers D13Mit311 and D13Mit66 to their intervening chromosomal distance. (b) Read depth ratios vs B6/J for exemplar strains (full data in fig. S4) showing CNVs within the area encompassing Gv1. C57-lineage strains are colored black and non-C57-lineage strains by phenotype: —blue, —red, and Sgp3+—green. (c)Heatmap of the median absolute error (MAE) between read depth ratio profiles for the strains in fig. S4. All non-C57-lineage strains group with ZALENDE/EiJ, whereas the C57-lineage group separately, and C57L/J with MOLF/EiJ. Coloring is according to (c), with wild-derived inbred strains in purple. (d) View of the genes within the area deleted within B6/N mice. Pseudogenes are colored gray, lincRNAs in purple, and protein coding genes in maroon. The protein coding ZFPs are annotated.
However, read depth analysis revealed surprising variation in comparison to B6/J. Most strains shared a ∼1.49 Mb region (13:65.47–66.96 Mb) of apparent copy-number variation (CNV) irrespective of their GIX or Sgp3 status (fig. 1b, fig. S4). Notably, C57-lineage strains (littermates of Lathrop’s F57 × M52 cross [Beck et al. 2000]) were distinct, although only C57L/J mice were equivalent to B6/J, as an ∼810 kb deletion was evident in C57BL/10J and C57BL/6N (B6/N) (13:65.79–66.60 Mb), which extended to ∼1.15 Mb in C57BR/cdJ (13:65.45–66.60 Mb). In comparison, areas of both increased and reduced depth within the non-C57-lineage strains was indicative of an alternate chromosomal conformation, potentially reflecting known differences in the contributions of different subspecies to the genomes of laboratory mice (Yang et al. 2007, 2011). Read depth profile clustering supported this theory, grouping C57-lineage mice with Mus musculus molossinus (MOLF/EiJ) and non-C57-lineage strains with M. m. domesticus (ZALENDE/EiJ) (fig. 1c, fig. S4). This was separately confirmed using the Mouse Phylogeny Viewer (Yang et al. 2011) (fig. S5). Importantly, non-C57-lineage strains also grouped with Mus spretus (SPRET/EiJ), which shares a common ancestor with the subspecies of M. musculus ∼2.3 million years ago and represents the ancestral state of the chromosome (Kumar et al. 2017).
Thus, the origin of the chromosomal region encompassing Gv1 differs between the prototypic strain, B6/J, and the majority of other laboratory strains. Additionally, several C57-lineage strains displayed large, unreported, deletions within the candidate region.
Zfp998 is Necessary and Sufficient to Determine GIX Status
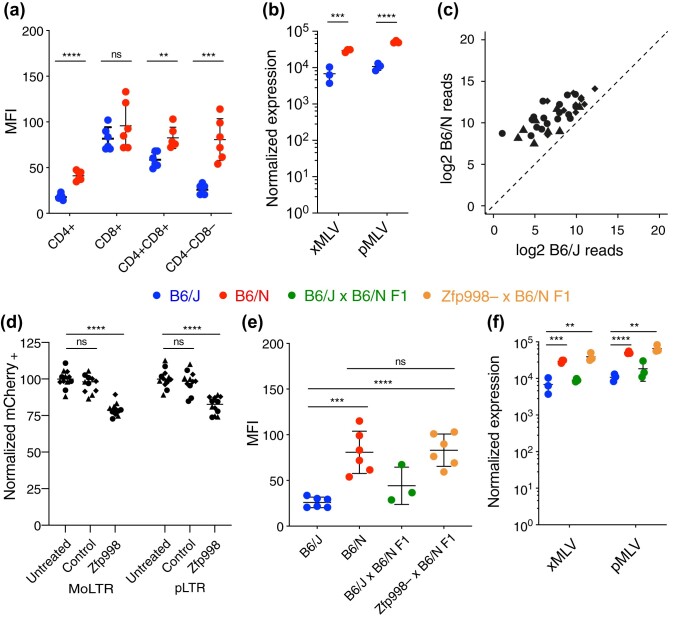
As the deletion within B6/N (fig 1b) fell within the area defined by the fine mapping of Gv1 (fig. 1a), we sought to determine the GIX status of this strain. Reflecting GIX’s historical usage as a T-cell marker (Obata et al. 1975), we stained for MLV SU across different populations of CD19–thymocytes (fig. S6). Significant differences between B6/N and B6/J were observed for three populations (fig. 2a) and most substantially for CD4–CD8– thymocytes. For a more holistic view, we confirmed this for whole-spleen samples by qRT-PCR (fig. 2b), where both pMLV and xMLV loci displayed significant upregulation, and by RNAseq, which revealed 14.5- and 5.3-fold average upregulation of the same groups, respectively (fig. 2c). No further significant stratifications, including on tRNA primer binding site (PBS) usage, were visible.
Fig. 2.
Zfp998 controls endogenous MLV expression in vitro and in vivo.(a) Median fluorescence intensity of 83A25 staining for MLV SU within the indicated subsets of CD19– thymocytes (six mice per group). (b) expression of x- and pMLV envelope measured by qRT-PCR and normalized relative to Hprt (three mice per group). Amplification of products for e- and mpMLV was negligible or absent after 40 cycles. (c)RNAseq expression analysis of endogenous MLVs in B6/N and B6/J (three mice per group). Significantly (q < 0.01, log2FC > 1) regulated MLV loci are indicated, with symbol shape denoting MLV subclass: triangle—mpMLV, circle—pMLV, and diamond—xMLV. (d) Numbers mCherry+ cells in otherwise untreated 293 T cells, the same cells transfected with a control plasmid, or with a Zfp998-containing expression plasmid. Symbol shape denotes the results of three independent experiments with 3/4 technical replicates. Significance values are from two-way ANOVA comparisons with no significant differences being determined between experiments or in experiment: treatment interactions. (e) Median fluorescence intensity of 83A25 staining for MLV SU within the indicated mouse genotypes (six, six, three, and six mice per group). (f)Expression of x- and pMLV envelope measured by qRT-PCR and normalized relative to Hprt (three mice per group). Amplification of products for e- and mpMLV was negligible or absent after 40 cycles. In (a), (b), (e), and (f), significance values are from Student’s t-tests with the indicated group sizes. ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
This placed Gv1 within the ∼810 kb deletion in B6/N, within which only two genes encoding KRAB-containing ZFPs are annotated: Zfp997 and Zfp998, with 19 and 14 zinc fingers, respectively (fig. 1d). These genes also formed the candidates for Sgp3 (Treger et al. 2019) and, although explicit identification was precluded by lack of separation in knockout animals, Zfp998 alone displayed specificity for the MLV LTR in electrophoretic mobility shift assays and was thus suggested as the most likely candidate. As Zfp998 was also the closest annotated protein-coding ZFP gene to the predicted point of peak linkage for Gv1 (fig. 1a), falling only 142 kb proximally, we chose to confirm its ability to control the expression of reporter-bearing MLV proviruses in a cell-based system, which was not previously assessed. Transient expression of Zfp998 significantly reduced numbers of mCherry+ cells in cultures bearing integrated viral genomes driving the reporter from either a Moloney-MLV or a consensus pMLV LTR (20.5% and 18.3% versus untreated controls, respectively) (fig 2d). This was equivalent to the difference in MLV SU staining observed for the CD8+ and CD4+CD8+ thymocyte populations (15.3 and 29.0%, respectively) (fig. 2a) and similarly representative of the 26–59% reductions seen upon Zfp708 treatment of RMER19B LTRs (Seah et al. 2019).
To next determine if individual disruption of Zfp998 was sufficient to recreate the phenotype in B6/J, we created knockout mice using CRISPR/Cas9 targeting. Two independent disruptions of the gene were confirmed by copy-number, split-read, and split-mate analyses of 10× PE150 whole genome sequencing (WGS) data, one truncating exon 4 (containing the zinc fingers) and one removing exons 1–3 (fig. S7). Importantly, no disruption of Zfp997 was noted in either case. Subsequent investigations of GIX phenotype were conducted using levels of MLV SU cell-surface staining and qRT-PCR. Within the control groups, as before, significantly higher levels of MLV SU were observed for B6/N than for B6/J mice (fig. 2e), which was reflected in elevated xMLV and pMLV expression (fig. 2f). Consistent with Gv1’s semi-dominant mode of action (Stockert et al. 1971), non-significant increases were also visible in B6/J × B6/N F1s. In comparison to this latter group, however, B6/J-Zfp998–× B6/N F1s fully recapitulated the phenotype of B6/N, both by MLV SU staining and qRT-PCR. Identical results were observed for both forms of the knockout, which are presented together.
Resolving the Source of Zfp998 Insufficiency
Together, these data confirmed that loss of Zfp998 was singularly sufficient to confer a phenotype within the B6 background and that its expression reduced expression of MLV proviruses in a biological system. Nevertheless, its chromosomal context differs substantially between C57- and non-C57-lineage laboratory mice and, amongst these, no groupings were visible that could define GIX status (fig 1c, fig. S4). Similarly, manual assessments of mouse genealogies (Beck et al. 2000) revealed no links in the origins of or strains.
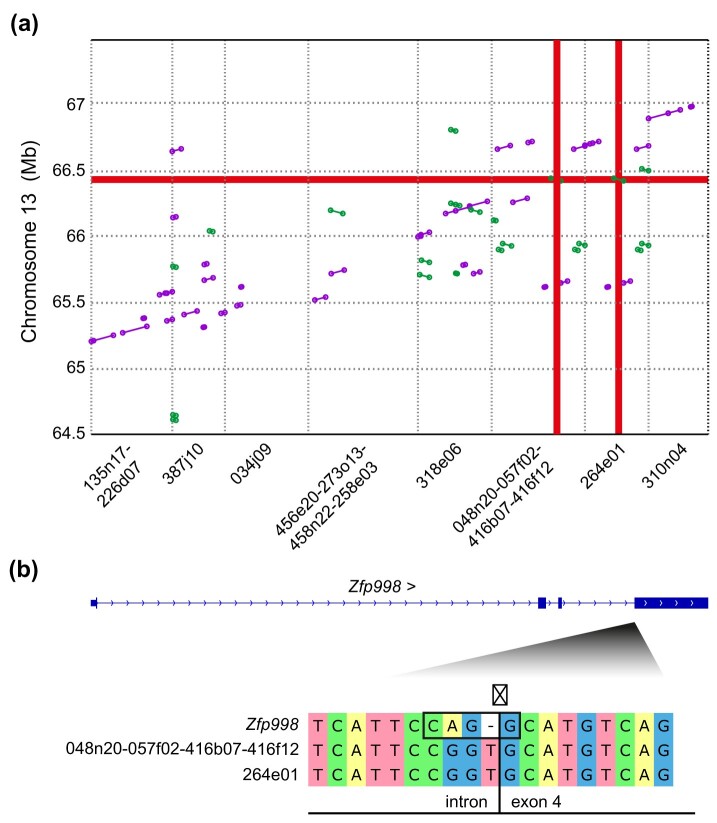
We therefore determined to clarify the status of the Gv1 locus within the prototypic strain, 129, which displays amongst the highest levels of cell-surface MLV SU (Stockert et al. 1971) and for which the AB2.2 ES cell line-derived bMQ bacterial artificial chromosome (BAC) library provides an excellent resource for comparative genetics (Adams et al. 2005). Sixteen BAC clones predicted to span the region were purchased, amplified in culture, and isolated using techniques to maintain their full lengths for subsequent Oxford Nanopore MinION long-read sequencing. This allowed for high coverages (median 2494×) to be achieved using only ≥1 kb reads and the full and contiguous assembly of all 16 BACs (fig. S8). Upon removal of pBACe3.6 vector sequences, 15 BAC sequences could be joined into eight scaffolds that aligned to the region (fig. 3a). Although a single scaffold could not be resolved, precluding orientation in relation to the B6/J-based GRCm38 reference and determination of the quantity of missing sequence between the areas assembled, large areas of sequence duplication (multiple X-Y paths occurring over the same Y axis ranges) were visible, as well as areas of unique sequence (X axis ranges with no corresponding X-Y path).
Fig. 3.
Nanopore sequencing allows assembly of the Gv1 gene region from 129 mice.(a) Alignment of the eight contigs resulting from assembly of the BACs against Chr 13. Diagonal lines represent only those areas ≥1 kb with ≥98% identity to the GRCm38 reference and are colored according to orientation (purple—forwards, green—reverse). Sequence names detail the scaffolding order and final sizes are shown above the alignment sections. Red highlighting shows the single position of Zfp998 in C57BL/6J (horizontal) intersecting with two independent copies within the BAC assemblies (vertical). (b) Comparison of the splice acceptor sequence (black box, with arrow marking point of splicing) for exon 4 of Zfp998 in comparison to the equivalent gene regions from the two assembled scaffolds. Area shown is the reverse complement of GRCm38 13:66432223-66432240.
Surprisingly, Zfp998 appeared not only present but apparently existed as two independent copies within the scaffolds (fig. 3a), both displaying 100% identity to the coding region of the Zfp998 reference. However, both exhibited defects at the splice acceptor for the terminal, zinc-finger-containing, exon, lacking the consensus mammalian splice acceptor sequence (N[C/T]AG↓G) (fig. 3b). Both variations in comparison to the Zfp998 reference sequence are classified as “high” impact splice acceptor variants by the Ensembl Variant Effect Predictor (McLaren et al. 2016) and would be predicted to occlude canonical splicing and production of functional protein. Full characterization of expression and splicing patterns for the identified loci would require complete, contiguous, assembly of the alternate chromosomal region derived from M. m. domesticus, as the level of variation observed (fig. 1b, fig. S4) otherwise precludes the non-spurious mapping of RNAseq data from non-C57-lineage strains to the B6/J-based mouse reference across this region.
Together, these data account for the prototypic GIXpositivity and negativity of 129 and B6/J, respectively. Likely through tandem duplication, the majority of laboratory mice contain two copies of the area in which Zfp998 exists within B6/J, although neither harbor functional copies of the gene in its reference state. Present in M. m. domesticus and M. spretus, this configuration represents the ancestral state within Mus. In contrast, the equivalent region from M. m. molossinus, present within B6/J, contains a single copy of the gene region that has been modified to allow splicing into the terminal exon, thereby creating a functional Zfp998 gene.
Discussion
The means by which hosts control the expression and, ultimately, the mobility and pathogenicity of endogenous repetitive elements is of great interest. Work detailing the complement and expression patterns of endogenous MLVs stems from the original creation of inbred lines by and for mouse fanciers, where the first detailed record-keeping allowed investigation of so-called “heritable cancers”(Russell1985). Following characterization of the GIX phenotype in the 1960s, the Gv1 locus was identified in 1971 (Stockert et al. 1971), although early mapping attempts were contradictory (Stockert et al. 1976) and the locus was only later localized to chromosome 13 (Oliver and Stoye 1999).
An analogous locus, Sgp3 was recently identified as either Zfp998 (dubbed Snerv1) or Zfp997 (Snerv2) (Treger et al. 2019) and we now positively identify Zfp998 as Gv1, making it highly probable that Gv1 and Sgp3 are differing phenotypic readouts of the same underlying gene, related to the specific mouse strains in which they have been studied. As such, it may be concluded that previously reported distinctions in the subclasses of MLVs regulated by the two loci likely reflect the differing complements of proviruses between strains—indeed, only ∼1/3 of MLV loci are commonly shared between any two backgrounds (Frankel et al. 1990), making this highly plausible—and we note that our data support a wider action for Gv1 than previously reported (Oliver and Stoye 1999).
Our work predicts that Zfp998 insufficiency is shared amongst non-C57-lineage mice and explains why the majority exhibit GIXpositivity, albeit to varying degrees (Stockert et al. 1971). Amongst the few non-C57-lineage strains historically genotyped as , it is further possible that negativity results from a lack of expression-competent proviruses and/or suitable transcriptional milieus, rather than from Zfp998 sufficiency. An exception must be seen in BALB/c mice; however, which also display linkage to the Gv1 locus in crosses to 129 (Oliver and Stoye 1999), suggesting that further work with the locus may yield additional insights. Additional research may also be required to clarify the mechanism of Zfp998-based control. In contrast to the previous identification of PBSGln1 as the binding site for Zfp998(Treger et al. 2019), our data suggest that the promoter activities of LTRs bearing either PBSGln1 or PBSPro are repressed to equivalent, physiological, extents.
This research highlights the difficulty of studying genes influencing multiple, insertionally-polymorphic, loci and underscores the necessity of working on defined backgrounds. High levels of homology within ZFP clusters hinder application of established wet- and dry-lab techniques and there is strong potential that further uncharacterized differences have significant bearings on inter-strain diversity. We note that even within the exceptionally well-characterized B6 substrains, we have revealed variations completely unexpected in magnitude that have eluded previous bioinformatic studies of the same WGS datasets (Simon et al. 2013). Overcoming these problems, we confirm another ZFP-based control of MLV expression alongside Zfp809(Wolf and Goff 2009), further highlighting the importance of epigenetic controls in the establishment of ERV repression.
Materials and Methods
Full methods are available as Supplementary Information.
Data Availability
All raw sequencing data generated in this study have been submitted to the European Nucleotide Archive under accessions PRJEB40145 and PRJEB40276.
Supplementary Material
Acknowledgments
We would like to acknowledge the Crick GEMS facility for CRISPR/Cas9 mouse work, along with the provisions of the BRF, FACS, HTS, and HPC Science Technology Platforms. We thank Dr Akiko Iwasaki for helpful discussions during the course of this study. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001162, FC001099), the UK Medical Research Council (FC001162, FC001099), and the Wellcome Trust (FC001162, FC001099). The funding bodies had no role in collection, analysis, or interpretation of the data presented.
References
- Adams DJ, Quail MA, Cox T, Van Der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, et al. 2005. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86(6):753–758. [DOI] [PubMed] [Google Scholar]
- Baudino L, Yoshinobu K, Morito N, Kikuchi S, Fossati-Jimack L, Morley BJ, Vyse TJ, Hirose S, Jørgensen TN, Tucker RM, et al. 2008. Dissection of genetic mechanisms governing the expression of serum retroviral gp70 implicated in murine lupus nephritis. J Immunol 181(4):2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MFW, Fisher EMC.. 2000. Genealogies of mouse inbred strains. Nat Genet 24(1):23–25. [DOI] [PubMed] [Google Scholar]
- Ecco G, Imbeault M, Trono D.. 2017. KRAB zinc finger proteins. Development 144(15):2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder JH, Jensen FC, Bryant ML, Lerner RA.. 1977. Polymorphism of the major envelope glycoprotein (gp70) of murine C-type viruses: virion associated and differentiation antigens encoded by a multi-gene family. Nature 267(5606):23–28. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Stoye JP, Taylor BA, Coffin JM.. 1990. A linkage map of endogenous murine leukemia proviruses. Genetics 124(2):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geering G, Old LJ, Boyse EA.. 1966. Antigens of leukemias induced by naturally occurring murine leukemia virus: their relation to the antigens of Gross virus and other murine leukemia viruses. J Exp Med 124(4):753–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood MEK, Vyse TJ, McDermott A, Thompson EM, Ida A, Walport MJ, Izui S, Morley BJ.. 2001. Autoantigen glycoprotein 70 expression is regulated by a single locus, which acts as a checkpoint for pathogenic anti-glycoprotein 70 autoantibody production and hence for the corresponding development of severe nephritis, in lupus-prone PXSB mice. J Immunol 167(3):1728–1733. [DOI] [PubMed] [Google Scholar]
- Henrichsen CN, Vinckenbosch N, Zöllner S, Chaignat E, Pradervand S, Schütz F, Ruedi M, Kaessmann H, Reymond A.. 2009. Segmental copy number variation shapes tissue transcriptomes. Nat Genet 41(4):424–429. [DOI] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L, Huntley S, et al. 2006. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 16(5):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D.. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516(7530):242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauzlaric A, Ecco G, Cassano M, Duc J, Imbeault M, Trono D.. 2017. The mouse genome displays highly dynamic populations of KRAB-zinc finger protein genes and related genetic units. PLoS One 12(3):e0173746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477(7364):289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: aresource for timelines, timetrees, and divergence times. Mol Biol Evol 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Levy DE, Lerner RA, Wilson MC.. 1982. A genetic locus regulates the expression of tissue-specific mRNAs from multiple transcription units. Proc Natl Acad Sci USA 79(19):5823–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Lerner RA, Wilson MC.. 1985. The Gv-1 locus coordinately regulates the expression of multiple endogenous murine retroviruses. Cell 41(1):289–299. [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F.. 2016. The Ensembl variant effect predictor. Genome Biol 17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y, Ikeda H, Stockert E, Boyse EA.. 1975. Relation of GIX antigen of thymocytes to envelope glycoprotein of murine leukemia virus. J Exp Med 141(1):188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL. 1999. Analysis of the control of endogenous murine retrovirus expression by Gv1[cited 2021Feb10]. Available from: https://discovery.ucl.ac.uk/id/eprint/10103009/
- Oliver PL, Stoye JP.. 1999. Genetic analysis of Gv1, a gene controlling transcription of endogenous murine polytropic proviruses. J Virol 73(10):8227–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung H-J, Tepper CG, Hu W-S, Fivash MJ, Coffin JM, Pathak VK.. 2011. Recombinant origin of the retrovirus XMRV. Science 333(6038):97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezer Ž, Harr B, Teschke M, Babiker H, Tautz D.. 2015. Divergence patterns of genic copy number variation in natural populations of the house mouse (Mus musculus domesticus) reveal three conserved genes with major population-specific expansions. Genome Res 25(8):1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, Hurles ME, Mell JC, Hall IM.. 2010. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res 20(5):623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin J, Boyle JJ, Rose SJ, Gabriel L, Lewis M, Thiruudaian V, Rogers NJ, Izui S, Morley BJ.. 2007. The Bxs6 locus of BXSB mice is sufficient for high-level expression of gp70 and the production of gp70 immune complexes. J Immunol 178(7):4395–4401. [DOI] [PubMed] [Google Scholar]
- Russell ER. 1985. A history of mouse genetics. Annu Rev Genet 19(1):1–28. [DOI] [PubMed] [Google Scholar]
- Seah MKY, Wang Y, Goy PA, Loh HM, Peh WJ, Low DHP, Han BY, Wong E, Leong EL, Wolf G, et al. 2019. The KRAB-zinc-finger protein ZFP708 mediates epigenetic repression at RMER19B retrotransposons. Dev 146:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MM, Greenaway S, White JK, Fuchs H, Gailus-Durner V, Wells S, Sorg T, Wong K, Bedu E, Cartwright EJ, et al. 2013. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol 14(7):R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockert E, Boyse EA, Obata Y, Ikeda H, Sarkar NH, Hoffman HA.. 1975. New mutant and congenic mouse stocks expressing the murine leukemia virus-associated thymocyte surface antigen GIX. J Exp Med 142(2):512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockert E, Boyse EA, Sato H, Itakura K.. 1976. Heredity of the GIX thymocyte antigen associated with murine leukemia virus: Segregation data simulating genetic linkage. Proc Natl Acad Sci USA 73(6):2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockert E, Old LJ, Boyse EA.. 1971. The G-IX system. A cell surface allo-antigen associated with murine leukemia virus; implications regarding chromosomal integration of the viral genome. J Exp Med 133(6):1334–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10(6):395–406. [DOI] [PubMed] [Google Scholar]
- Treger RS, Pope SD, Kong Y, Tokuyama M, Taura M, Iwasaki A.. 2019. The lupus susceptibility locus Sgp3 encodes the suppressor of endogenous retrovirus expression SNERV. Immunity 50(2):334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RM, Vyse TJ, Rozzo S, Roark CL, Izui S, Kotzin BL.. 2000. Genetic control of glycoprotein 70 autoantigen production and its influence on immune complex levels and nephritis in murine lupus. J Immunol 165(3):1665–1672. [DOI] [PubMed] [Google Scholar]
- Tung J-S, Fleissner E, Vitetta ES, Boyse EA.. 1975. Expression of murine leukemia virus envelope gycoprotein gp69/71 on mouse thymocytes. Evidence for two structural variants distinguished by presence vs absence of GIX antigen. J Exp Med 142(2):518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J-S, Vitetta ES, Fleissner E, Boyse EA.. 1975. Biochemical evidence linking the GIX thymocyte surface antigen to the gp69/71 envelope glycoprotein of murine leukemia virus. J Exp Med 141(1):198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP.. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458(7242):1201–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, de Iaco A, Sun MA, Bruno M, Tinkham M, Hoang D, Mitra A, Ralls S, Trono D, Macfarlan TS.. 2020. Krab-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. eLife 9:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellåker C, Goodstadt L, Nicod J, Bhomra A, et al. 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477(7364):326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YFS, Goh GYL, Neo SP, et al. 2015. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 163(1):230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel De Villena F.. 2007. On the subspecific origin of the laboratory mouse. Nat Genet 39(9):1100–1107. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH-T, Nachman MW, Pialek J, et al. 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43(7):648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinobu K, Baudino L, Santiago-Raber M-L, Morito N, Dunand-Sauthier I, Morley BJ, Evans LH, Izui S.. 2009. Selective up-regulation of intact, but not defective env RNAs of endogenous modified polytropic retrovirus by the Sgp3 locus of lupus-prone mice. J Immunol 182(12):8094–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G.. 2012. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491(7426):774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GR, Ploquin MJY, Eksmond U, Wadwa M, Stoye JP, Kassiotis G.. 2012. Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog 8(5):e1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data generated in this study have been submitted to the European Nucleotide Archive under accessions PRJEB40145 and PRJEB40276.