Abstract
After double fertilization, zygotic embryogenesis initiates a new life cycle, and stem cell homeostasis in the shoot apical meristem (SAM) and root apical meristem (RAM) allows plants to produce new tissues and organs continuously. Here, we report that mutations in DEAD-BOX RNA HELICASE 27 (RH27) affect zygote division and stem cell homeostasis in Arabidopsis (Arabidopsis thaliana). The strong mutant allele rh27-1 caused a zygote-lethal phenotype, while the weak mutant allele rh27-2 led to minor defects in embryogenesis and severely compromised stem cell homeostasis in the SAM and RAM. RH27 is expressed in embryos from the zygote stage, and in both the SAM and RAM, and RH27 is a nucleus-localized protein. The expression levels of genes related to stem cell homeostasis were elevated in rh27-2 plants, alongside down-regulation of their regulatory microRNAs (miRNAs). Further analyses of rh27-2 plants revealed reduced levels of a large subset of miRNAs and their pri-miRNAs in shoot apices and root tips. In addition, biochemical studies showed that RH27 associates with pri-miRNAs and interacts with miRNA-biogenesis components, including DAWDLE, HYPONASTIC LEAVES 1, and SERRATE. Therefore, we propose that RH27 is a component of the microprocessor complex and is critical for zygote division and stem cell homeostasis.
As a new component of the microprocessor complex in Arabidopsis, DEAD-BOX RNA HELICASE 27 regulates the initiation of zygotic embryogenesis and stem cell homeostasis in the shoot and root meristems.
Introduction
Zygote division is the initial step of embryogenesis in sexual organisms (Palovaara et al., 2016; Schulz and Harrison, 2019). In Arabidopsis (Arabidopsis thaliana), the zygote first elongates and then divides asymmetrically to produce a smaller apical cell and a larger basal cell. After two rounds of longitudinal divisions and one round of transverse division, the apical cell forms an eight-cell embryo. The embryo then undergoes periclinal divisions that separate the outer protoderm and the inner ground tissue cells at the dermatogen stage. Later, the shoot apical meristem (SAM), root apical meristem (RAM), and two cotyledons are initiated from the globular staged embryo (Palovaara et al., 2016). Genetic studies have shown that mutations of CULLIN1 (CUL1), EMBRYONIC FACTOR 1 (FAC1), CELL DIVISION CYCLE 5 (CDC5), ZEUS1 (ZEU1), DNA LIGASE I (LIG1), YAOZHE (YAO), ZAK IXIK (ZIX), GAMETE CELL DEFECTIVE 1 (GCD1), FAC19, ZYGOTE-ARREST 1 (ZYG1), or ZYG3 result in a zygote-lethal phenotype (Shen et al., 2002; Xu et al., 2005; Lin et al., 2007; Ronceret et al., 2008; Andreuzza et al., 2010; Li et al., 2010; Ngo et al., 2012; Wu et al., 2012; Yu et al., 2012; Guo et al., 2016; Yang et al., 2017), suggesting that these genes play a critical role in the initiation of zygote division.
Stem cells located in the SAM and RAM give rise to new tissues and organs continuously during post-embryonic development (Weigel and Jürgens, 2002). In Arabidopsis, the SAM is characterized by a convex structure that is divided into three zones: the central zone at the summit, the rib zone underneath, and the peripheral zone at the sides. Stem cells are located at the three outermost cell layers (L1–L3) in the central zone of the SAM, and the stem cell organizing center (SCOC) is located underneath and partially overlaps with the stem cell domain (Meyerowitz, 1997). A feedback regulatory loop, established via stem cell promotion by the homeodomain transcription factor WUSCHEL (WUS) and stem cell restriction by the CLAVATA3 (CLV3) peptide-CLV1/CLV2/CORYNE (CRN)/RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2) receptors, maintains the homeostasis of stem cells in the SAM (Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000; Müller et al., 2008;Kinoshita et al., 2010). HAIRY MERISTEM 1 (HAM1), a GRAS family transcription factor, acts as a co-factor of WUS to repress stem cell differentiation (Zhou et al., 2015). In the RAM, stem cells of different cell linages are in direct contact with four rarely dividing cells termed the quiescent center (QC), which is the SCOC in the RAM (Dolan et al., 1993). Similar to the scenario in the SAM, WUSCHEL-RELATED HOMEOBOX 5 (WOX5) and CLV3/EMBRYO SURROUNDING REGION-RELATED 40 (CLE40) peptide-ARABIDOPSIS CRINKLY4 (ACR4)/CLV1 receptor kinase complexes form a feedback regulatory loop that controls distal stem cell homeostasis in the RAM (Stahl et al., 2009, 2013).
MicroRNAs (miRNAs) play a critical role in maintaining stem cell homeostasis in the SAM and RAM by down-regulating the expression of their target genes (Li et al., 2017). MiR171 represses the expression of HAM1 (Llave et al., 2002), and ectopic expression of miR171-resistant HAM1 in the L1 layer of the SAM leads to reduced expression of CLV3, and consequently an enlarged SAM (Zhou et al., 2018). MiR164 down-regulates the expression of CUP-SHAPED COTYLEDON1 (CUC1) and CUC2, which are involved in defining the organ boundary of the SAM (Mallory et al., 2004a). In the stele, miR165/166 restricts the expression of PHABULOSA (PHB), which is critical for determination of the xylem cell fate in the RAM (Carlsbecker et al., 2010). In animals, miRNAs are generated through the cleavage and processing of primary miRNAs (pri-miRNAs) by Drosha, DGCG8, and Dicer (Grishok et al., 2001; Gregory et al., 2004). DEAD-box RNA helicases modulate the cleavage of pri-miRNAs by interacting with Drosha (Han et al., 2014; Yin et al., 2015; Zhao et al., 2016). In plants, miRNAs are generated by a microprocessor complex comprising DICER-LIKE 1 (DCL1), HYPONASTIC LEAVES 1 (HYL1), and SERRATE (SE; Wang et al., 2019b). In Arabidopsis, the DEAD-box RNA helicase family contains 58 members (Mingam et al., 2004), some of which are implicated in RNA processing events such as mRNA export, mRNA splicing, and rRNA processing (Gong et al., 2005; Liu et al., 2010; Guan et al., 2013; Paieri et al., 2018; Wang et al., 2019a). However, the role of DEAD-box RNA helicases in miRNA biogenesis has not been reported in plants.
In this study, we show that DEAD-BOX RNA HELICASE 27 (RH27) is critical for zygote division and stem cell homeostasis in Arabidopsis. The strong mutant allele of rh27-1 exhibited a zygote-lethal phenotype, while the weak allele of rh27-2 exhibited compromised stem cell homeostasis in both the SAM and RAM. In rh27-2 plants, the expression of several stem cell homeostasis-related genes, including HAM1, CUC1, CUC2, and PHB, was up-regulated, alongside reduced levels of miR171, miR164, and miR165/166. Small RNA sequencing (RNA-seq) showed reduced accumulation of a large subset of miRNAs in the rh27-2 line. In addition, biochemical studies revealed that RH27 is associated with pri-miRNAs and interacts with DAWDLE (DDL) and the microprocessor complex components HYL1 and SE, suggesting that RH27 regulates miRNA biogenesis and is required for zygote division and stem cell homeostasis.
Results
The zyg4-1 mutant with a point mutation in RH27 exhibits a zygote-lethal phenotype
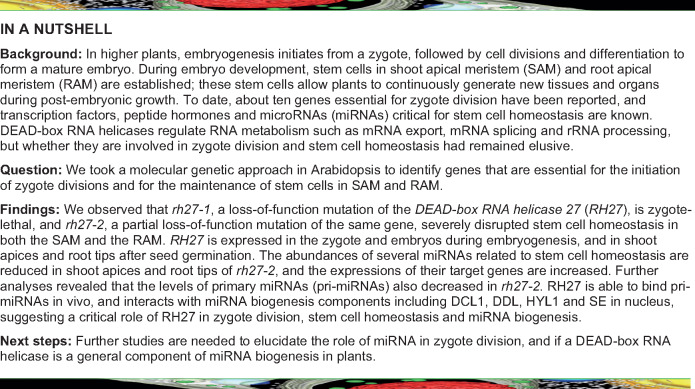
To identify genes that are essential for zygote division, a genetic screen was performed in an ethyl methanesulfonate-mutagenized M2 population of Arabidopsis, as described previously (Guo et al., 2016). This process enabled the identification of the zygote-lethal mutation zygote-arrest 4-1 (zyg4-1). A genetic analysis showed that 26.1% of ovules (n = 621) produced in the heterozygous zyg4-1/+ plants were aborted (Figure 1, A andTable 1), indicating that zyg4-1 is inherited as a single-gene recessive mutation. Reciprocal crosses between zyg4-1/+ and wild-type (Col-0) plants revealed that, when zyg4-1/+ was used as the female, 96 of the 174 BC1 plants genotyped were wild type and 78 were heterozygous. This finding is consistent with the ratio of 1:1 (χ2 test, P = 0.17), suggesting that the mutation of zyg4-1 did not affect female gametogenesis. The same result was observed when zyg4-1/+ was used as the pollen donor (n = 347, χ2 test, P = 0.63), indicating that the mutation of zyg4-1 did not affect male gametogenesis.
Figure 1.
Phenotypes of the zyg4 (rh27) Mutants. (A) Siliques of the following lines: wild-type (WT), zyg4-1 (rh27-1)/+ with early aborted ovules (white arrowheads), zyg4-1 (rh27-1)/+ carrying ProRH27:RH27-GUS, rh27-2/+ with late aborted ovules (yellow arrowheads), and zyg4-1 (rh27-1)/+ carrying ProRH27:ΔRH27-sYFP lines. Note the full complementation of the zyg4-1 (rh27-1) phenotype by ProRH27:RH27-GUS, while late aborted ovules (yellow arrowheads) were still observed in the siliques of zyg4-1 (rh27-1)/+ plants carrying ProRH27:ΔRH27-sYFP. ΔRH27 has a 310-bp deletion from the start codon of RH27, as found in rh27-2. Bar = 1 mm. (B) Nomarski images of whole-mount cleared ovules carrying wild-type and zyg4-1 (rh27-1) embryos at 2 DAP. Unlike the wild-type siblings at the octant stage, the zyg4-1 (rh27-1) embryos were arrested at the zygote, 1-cell, or 2-cell stage. The percentages of embryos arrested at different stages in aborted ovules are shown. Bar = 25 µm. (C) Schematic illustration of the RH27 gene. The open and black boxes indicate the untranslated and coding regions, respectively. The horizontal lines indicate introns. The G-to-A mutation in zyg4-1 (rh27-1) and the T-DNA insertion in rh27-2 are shown. The red arrows indicate positions of the primers used in (F) and (J). (D) Seedlings of the wild-type, rh27-2, zyg4-1 (rh27-1); ProRH27:RH27-GUS, and zyg4-1 (rh27-1); ProRH27:ΔRH27-sYFP lines at 6 DAG. Note that full complementation of zyg4-1 was achieved using ProRH27:RH27-GUS, but only a partial complementation (with a phenotype similar to that of rh27-2) was observed when ProRH27:ΔRH27-sYFP was used. Bar = 1 cm. (E) Nomarski images of whole-mount cleared ovules carrying wild-type or rh27-2 embryos at 14 DAP. Bar = 100 µm. (F) PCR-based genotyping of rh27-2 to show the successful insertion between the LP and RP primers. (G) Nomarski images showing abnormal cell division patterns in the rh27-2 line at 3 DAP, compared with their wild-type siblings in the same silique. Bar = 10 µm. (H) Primary root lengths of wild-type and rh27-2 plants in the first 10 DAG. For each genotype, data are shown as the mean ± SD of the primary root length (from at least 40 different roots). (I) Wild-type and rh27-2 plants at 40 DAG, showing that two shoots formed simultaneously in the rh27-2 plant. The numbers denote the frequencies of the phenotype shown. Bar = 10 cm. (J) RT-PCR analyses of RH27 transcripts in the rh27-2 line. Note that the full-length RH27 transcript was not detectable in rh27-2, but the transcripts upstream and downstream of the T-DNA insertion were detected. EIF4A was used as an internal control. The primers used are indicated in (C).
Table 1.
Genetic analyses of zyg4 (rh27) mutants
| Cross combinations |
Aborted ovules (%)* | Total ovules examined | P value** | |
|---|---|---|---|---|
| Female | Male | |||
| zyg4-1 (rh27-1)/+ | zyg4-1 (rh27-1)/+ | 26.1 | 621 | 0.53 |
| rh27-2/+ | rh27-2/+ | 24.4 | 1625 | 0.60 |
| zyg4-1 (rh27-1)/+ | rh27-2/+ | 25.9 | 984 | 0.51 |
| rh27-2/+ | zyg4-1 (rh27-1)/+ | 23.4 | 1068 | 0.23 |
| wild-type (Col-0) | wild-type (Col-0) | 2.9 | 137 | n.a. |
The percentages of total ovules that were aborted or developmentally delayed, as counted under a dissection microscope.
P-values were calculated from χ2 tests for the 3:1 segregation.
n.a.: not applicable.
Whole-mount clearing of early staged ovules from zyg4-1/+ plants, followed by Nomarski optical analyses, showed that 80.7% of the aborted embryos were arrested at the zygote stage, and the remainder were able to undergo one or two rounds of cell division (n = 186; Figure 1, B), indicating that ZYG4 is essential for zygote division.
Map-based cloning allowed us to localize the zyg4-1 mutation to chromosome 5, and a G-to-A substitution in the coding region of At5g65900, which led to conversion of the tryptophan (W) 417 residue to a stop codon, was identified (Figure 1, C). At5g65900 encodes the DEAD-box RNA helicase RH27 (Aubourg et al., 1999), which contains all 12 conserved domains present in DEAD-box RNA helicase proteins (Supplemental Figure S1).
To test whether the mutation in RH27 was responsible for the observed zygote-lethal phenotype of zyg4-1, we generated a construct that carried the full-length genomic DNA of RH27, including the 2,227-bp upstream region, in-frame fused to the coding sequence for the N-terminus of β-glucuronidase (GUS; ProRH27:RH27-GUS), and transformed it into wild-type Arabidopsis by Agrobacterium tumefaciens-mediated floral dip (Clough and Bent, 1998). Single-insertion homozygous ProRH27:RH27-GUS transgenic plants were then crossed with the zyg4-1/+ line. Among the F2 population examined, heterozygous zyg4-1/+ and homozygous zyg4-1 plants carrying ProRH27:RH27-GUS (Supplemental Figure S2) showed no defects in embryogenesis (Figure 1, A) or post-embryonic development (Figure 1, D), indicating that ProRH27:RH27-GUS was able to completely rescue the zygote-lethal phenotype of zyg4-1. We thus renamed zyg4-1 as rh27-1.
The weak mutant allele rh27-2 shows minor defects in embryogenesis and severely compromised stem cell homeostasis in the SAM and RAM
To decipher the function of RH27, we obtained the rh27-2 line, which carries a T-DNA insertion in the first exon of RH27 (SALK_068661, Figure 1, C), from the Arabidopsis Biological Resources Center. Heterozygous rh27-2/+ plants grew normally during the vegetative phase, but 24.4% of the seeds produced in these plants were shriveled or wrinkled (n = 1,625; Figure 1, A;Supplemental Figure S3; and Table 1), indicating that rh27-2 is a single-gene recessive mutant. Nomarski imaging at the time when the wild-type embryos were at the cotyledonary stage revealed that the embryos in the abnormal seeds from rh27-2/+ plants were at the globular to bent cotyledon stages, suggesting a late embryo-defective phenotype (Figure 1, E). A germination test showed that 77.5% of these abnormal seeds were able to germinate (n = 40) and were genotyped to be homozygous for rh27-2 (Figure 1, F). Whole-mount clearing of ovules at 3 days after pollination (DAP) revealed abnormal cell division patterns in rh27-2 embryos from the 16-cell stage onward (Figure 1, G).
Compared with the wild-type seedlings, the rh27-2 seedlings showed a short-root phenotype, with very slow growth of the primary roots (Figure 1, D and H). At the bolting stage, 58.9% of rh27-2 plants showed a multi-shoot phenotype, with two or occasionally three shoots formed simultaneously (n = 56), compared with one formed in the wild-type plants (n = 64, Figure 1, I). Reciprocal crosses between rh27-1/+ and rh27-2/+ plants showed that approximately one-quarter of the ovules produced in F1 siliques were aborted (Table 1), confirming that rh27-1 and rh27-2 are allelic.
Reverse transcription PCR (RT-PCR) analyses of different regions of RH27 transcripts in rh27-2 seedlings revealed that, although the full-length RH27 transcript was not detectable, transcripts upstream and downstream of the T-DNA insertion were detected (Figure 1, J). Since the insertion is located at the first exon of the gene, we speculated that it may have removed the non-conserved N-terminal region (Supplemental Figure S1), producing a truncated RH27 protein (ΔRH27) with partially compromised function. To test this hypothesis, a construct carrying the RH27 5′-regulatory sequence and ΔRH27 that was in-frame fused to the N-terminus of super Yellow Fluorescence Protein (sYFP; ProRH27:ΔRH27-sYFP) was generated and introduced to wild-type Arabidopsis through floral dip (Clough and Bent, 1998). A transgenic line with a homozygous single insertion was identified and crossed with the rh27-1/+ line. In siliques of rh27-1/+ plants carrying homozygous ProRH27:ΔRH27-sYFP, a late embryo-defective phenotype resembling that of the rh27-2/+ line was observed (Figure 1, A). In addition, homozygous rh27-1 seedlings carrying ProRH27:ΔRH27-sYFP (Supplemental Figure S2) were recovered and showed a rh27-2-like phenotype (Figure 1, D). Together, these data confirmed that ΔRH27 is a partially functional protein.
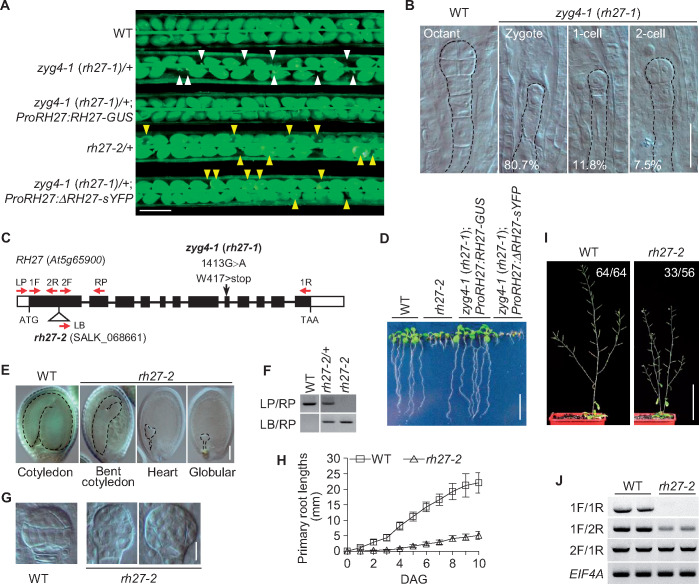
To elucidate the role of RH27 in post-embryonic growth and development, we performed detailed phenotypic analyses of rh27-2 seedlings. Whole-mount clearing of root tips at 5 days after germination (DAG) revealed that the length of the root meristematic zone in rh27-2 seedlings was much shorter than that in wild-type seedlings (Figure 2, A and B), and the meristem cell number, defined by the number of non-vacuolated cells in the cortex cell lineage (Casamitjana-Martı’nez et al., 2003), was also significantly lower in the mutant seedlings (Figure 2, C). By confocal examination of propidium iodide-stained roots, we observed an irregular organization of cell patterning in the meristematic zone of rh27-2 roots (Figure 2, D), as well as frequent cell death, indicated by intensive propidium iodide penetration (Truernit and Haseloff, 2008), in cells adjacent to and above the QC (Figure 2, D and E, indicated by open arrowheads).
Figure 2.
Abnormal root development and defective stem cell homeostasis in the rh27-2 line. (A) Nomarski images of root tips from wild-type (WT) and rh27-2 plants at 5 DAG. Brackets indicate the meristematic zone. Bar = 50 µm. (B) and (C) The average lengths of the meristematic zones (B, n = 20) and the average numbers of meristematic cells (C, defined by the number of non-vacuolated cells in the cortex cell linage, n = 40) in wild-type and rh27-2 roots at 5 DAG. Error bars indicate the SD and asterisks indicate significant differences between samples (two-tailed Student’s t test, **P < 0.01). (D) and (E) The cellular organization (D) and expression of ProWOX5:erGFP (green; E) in wild-type and rh27-2 root meristems at 5 DAG, examined under a confocal microscope. Propidium iodide (magenta) staining was used to show the cellular structure. Note the increased number of GFP-positive cells (yellow arrowheads), located above the QC (white arrowheads), in rh27-2 root meristems (E). Dead cells were observed frequently in rh27-2 root meristems (open arrowheads). Bar = 50 µm. (F) The numbers of cells expressing ProWOX5:erGFP in wild-type and rh27-2 root meristems. Data are shown as the mean ± SD of the number of ProWOX5:erGFP-positive cells in at least five roots. Asterisks indicate significant differences between the samples (two-tailed Student’s t test, **P < 0.01). (G) Examinations of QC25:GUS expression (blue) and starch grain accumulation (brownish blue) in wild-type and rh27-2 roots at 5 DAG. Starch grain accumulation was observed frequently in the expected CSC (eCSC) of rh27-2 roots but was not observed in CSC of the wild-type roots. The numbers denote the frequencies of CSC or eCSC with starch grains. CC, columella cell. Bar = 50 µm.
To analyze the cellular organization in rh27-2 roots further, we introduced the ProWOX5:erGFP marker, in which an endoplasmic reticulum-localized green fluorescent protein (erGFP) was expressed under the control of the WOX5 promoter (Sarkar et al., 2007), to the rh27-2 line by crossing. When examined under a confocal microscope at 1, 3, 5, and 7 DAG, instead of the well-defined ProWOX5:erGFP-positive QC cells observed in the wild-type roots, the pattern of ProWOX5:erGFP-positive cells in rh27-2 roots was altered greatly, with erGFP-positive cells located not only in QC cells, but also in those above the QC (Figure 2, E;Supplemental Figure S4). Next, we counted the number of erGFP-positive cells in serial confocal sections. In wild-type roots, an average of four erGFP-positive cells was observed consistently (Figure 2, F), while in rh27-2 roots, 6.6 ± 1.5 (n = 5), 11 ± 2.3 (n = 7), 9.7 ± 2.4 (n = 7), and 8.6 ± 3.0 (n = 8) erGFP-positive cells were observed at 1, 3, 5, and 7 DAG, respectively (Figure 2, F). We then used Lugol staining to examine the columella stem cells (CSCs) located underneath the QC, and observed that starch granules were accumulated in expected CSC regions in 85.4% of rh27-2 roots (n = 41; Figure 2, G), suggesting premature differentiation of the CSCs. By contrast, this phenomenon was not observed in wild-type roots (n = 29; Figure 2, G). These results suggest that stem cell homeostasis was disrupted in the RAM of the rh27-2 line.
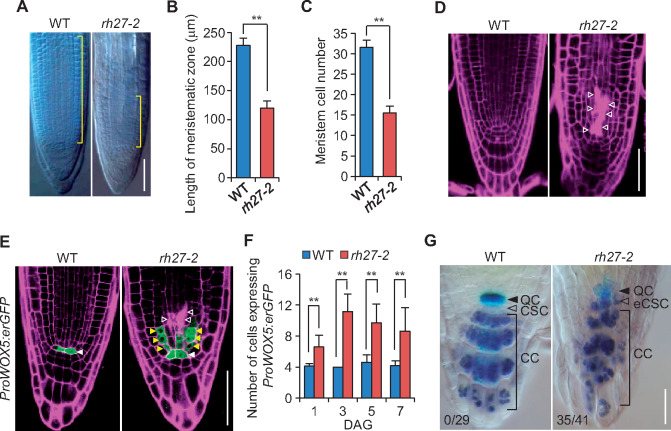
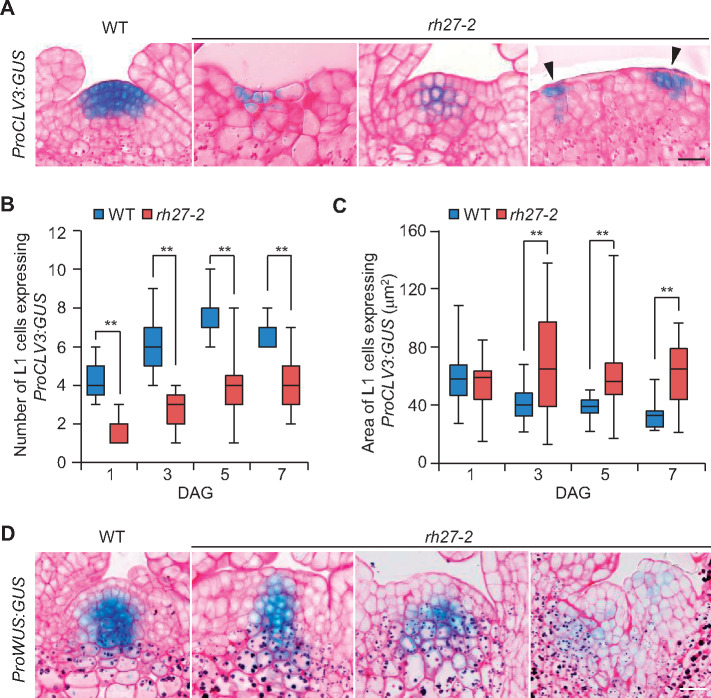
Next, we performed whole-mount clearing of shoot apices and observed that the SAMs in rh27-2 plants were small, flat, and occasionally displayed twin domes, unlike the single dome structures in the wild-type shoots (Supplemental Figure S5, A). Subsequently, we introduced ProCLV3:GUS, a marker for stem cells (Fletcher et al., 1999), and ProWUS:GUS, a marker for the SCOC (Mayer et al., 1998), to the rh27-2 line by crossing. A GUS assay revealed highly irregular GUS staining patterns in the SAMs of rh27-2 seedlings carrying ProCLV3:GUS, whereas wild-type seedlings displayed a well-defined stem cell domain (Supplemental Figure S5, B). Median sectioning across the SAM revealed that, compared with the wild-type plants, the rh27-2 plants carrying ProCLV3:GUS displayed a reduced number of ProCLV3:GUS-positive stem cells that were scattered in SAMs (Figure 3, A). Statistical analyses showed that, when examined at 1, 3, 5, and 7 DAG, the SAMs in rh27-2 plants had significantly fewer L1 cells with GUS signals than those in wild-type plants (Figure 3, B). However, ImageJ analyses (Schindelin et al., 2012) revealed that the GUS-positive cells in the L1 layer of rh27-2 plants were significantly larger than those in the wild-type plants (Figure 3, C). Similarly, GUS assays performed in wild-type and rh27-2 plants carrying ProWUS:GUS showed that, when examined at 3, 5, and 7 DAG, the intensity and pattern of the GUS signal varied greatly in the rh27-2 line (Figure 3, D andSupplemental Figure S6, A and B). A GUS signal was occasionally observed in the L1 layer of the SAMs in rh27-2 plants (Supplemental Figure S6, B, indicated by arrowheads), but was not observed in the corresponding region of wild-type plants. Taken together, these results suggest that stem cell homeostasis was disrupted in the RAM and SAM of rh27-2 plants.
Figure 3.
Stem cell homeostasis is defective in the SAM of rh27-2 plants. (A) Median sections of SAMs of wild-type (WT) and rh27-2 seedlings carrying ProCLV3:GUS, showing the presence of stem cells. Arrowheads indicate two GUS-positive domains in a SAM of a rh27-2 seedling. Sections from 24 shoot apices were examined for rh27-2. Bar = 20 µm. (B) and (C) Box plot diagrams showing the numbers (B) and areas (C) of GUS-positive L1 cells in median sections of SAMs from wild-type and rh27-2 plants carrying ProCLV3:GUS. Data are shown as the mean ± SD of the numbers of cells in at least 12 SAMs from each stage (B), and the areas of GUS-positive cells from at least 3 SAMs (C). Asterisks indicate significant differences between samples (two-tailed Student’s t test, **P < 0.01). (D) Median sections of wild-type and rh27-2 SAMs from 7 DAG seedlings carrying ProWUS:GUS, showing the SCOC in the SAM. Note the diffuse expression pattern of ProWUS:GUS in the rh27-2 SAMs. Sections from 12 shoot apices were observed for rh27-2. Bar = 20 µm. In (A) and (D), sections were stained with periodic acid-Schiff (pink) to show the cellular structure.
RH27 is expressed in embryos from the zygote stage onwards, and in the SAM and RAM
The introduction of a ProRH27:RH27-sYFP construct was able to complement the short-root phenotype of the rh27-2 mutant (Supplemental Figure S7, A). Immunoblot analyses using an anti-RH27 antibody showed that the full-length RH27-sYFP fusion protein was expressed in rh27-2; ProRH27:RH27-sYFP transgenic plants (Supplemental Figure S7, B), suggesting that the regulatory sequence used in ProRH27:RH27-sYFP carries all essential elements and that the RH27-sYFP fusion protein is functional. Confocal examination of ProRH27:RH27-sYFP transgenic plants showed that, during the reproductive stage, RH27-sYFP was expressed in mature ovules (Supplemental Figure S7, C), as well as unicellular and bicellular microspores, but was absent from mature pollen (Supplemental Figure S7, D).
During embryogenesis, RH27-sYFP expression was detected in the nuclei of the zygote staged embryos (Figure 4, A), and then in embryos at the 2-cell, 4-cell, globular, and torpedo-shaped stages (Figure 4, B–E). Examination of ovules excised from transgenic plants carrying the egg cell-specific marker pEC1:H2B-RFP (Ingouff et al., 2009) and pollinated by pollens from ProRH27:RH27-sYFP plants showed that expression of the paternal RH27-sYFP was activated at the zygote stage (Supplemental Figure S7, E).
Figure 4.
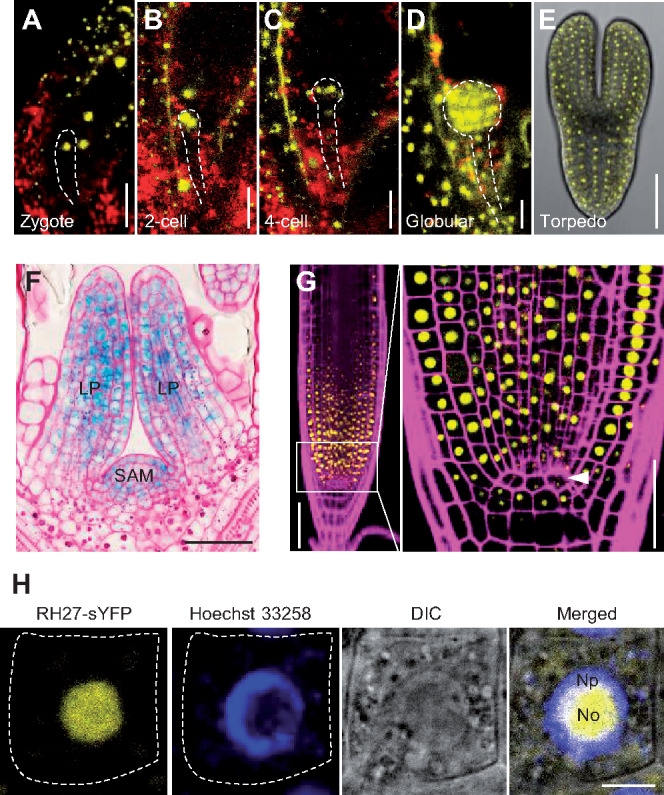
Expression and subcellular localization of RH27 in Arabidopsis. (A–E) Expression of ProRH27:RH27-sYFP (yellow) in embryos at the zygote (A), 2-cell (B), 4-cell (C), globular (D), and torpedo-shaped (E) stages. Note the nuclear localization of the RH27-sYFP signals. Red, auto-fluorescence. Dashed lines outline the embryos. Bars = 20 µm for (A) to (D), and 50 µm for (E). (F) Expression of ProRH27:RH27-GUS (blue) in the shoot apex, specifically the SAM and leaf primordia (LP). The section was stained with periodic acid-Schiff (pink) to show the cellular structure. Bar = 50 µm. (G) Expression of ProRH27:RH27-sYFP (yellow) in the RAM. Part of the meristematic zone (framed) is magnified (right) to show the presence of RH27-sYFP in meristematic cells, but not the QC (arrowhead). The root was stained with propidium iodide (magenta) to show the cellular structure. Bars = 50 µm. (H) Subcellular localization of the RH27-sYFP protein (yellow) in a root epidermal cell from a transgenic plant carrying ProRH27:RH27-sYFP. The nucleus was stained with Hoechst 33258 (blue). Note that RH27-sYFP was localized primarily in the nucleolus (No) and to a small degree in the nucleoplasmic region (Np). Dotted lines outline the cell boundary. Bar = 5 µm.
Next, the expression of RH27 during post-embryonic development was examined using ProRH27:RH27-GUS transgenic plants. RH27-GUS was actively expressed in shoot apices and root tips (Supplemental Figure S8, A). In the inflorescence, RH27-GUS signals were detected in floral primordia, young flowers, and pistils (Supplemental Figure S8, B). Semi-thin sectioning of shoot apices of ProRH27:RH27-GUS plants after GUS assays revealed that RH27-GUS was expressed in SAMs and young leaves (Figure 4, F). Confocal examination of roots from ProRH27:RH27-sYFP seedlings showed that RH27-sYFP signals were primarily located in the nuclei of RAM cells but were excluded from QC cells (Figure 4, G). Further examination of root epidermal cells in ProRH27:RH27-sYFP seedlings, counterstained with Hoechst 33258, showed that the RH27-sYFP protein was localized primarily in the nucleolus (Figure 4, H).
The rh27-2 mutant displays reduced levels of miRNAs and increased levels of their target genes in shoot apices and root tips
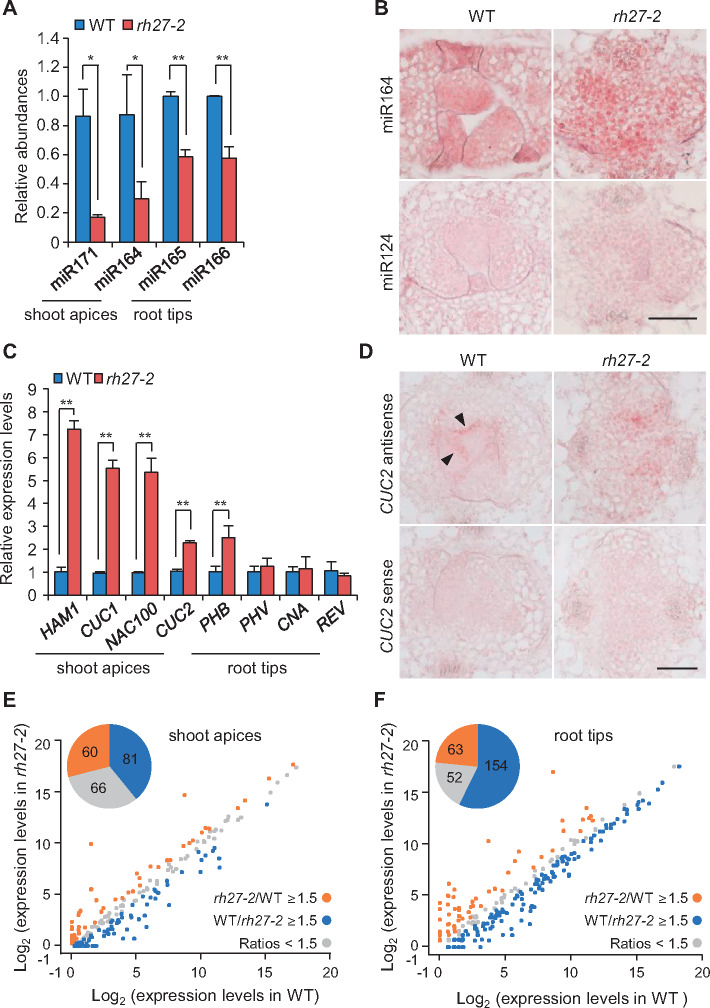
Since DEAD-box RNA helicases play an important role in miRNA biogenesis in animals (Han et al., 2014; Yin et al., 2015; Zhao et al., 2016), we used RT-quantitative PCR (RT-qPCR) analyses (Shi and Chiang, 2005) to examine the accumulations of miRNAs in rh27-2 plants. The abundances of the SAM-related miRNAs miR171 and miR164 in rh27-2 shoot apices were lower than those in wild-type shoot apices (Figure 5, A). Similarly, the abundances of the RAM-related miRNAs miR165 and miR166 in rh27-2 root tips were lower than those in wild-type root tips (Figure 5, A). Next, in situ hybridization was performed to examine the accumulation patterns of miRNAs in shoot apices using digoxigenin-labeled LNA oligo probes. In wild-type shoots, an antisense probe detected miR164 in cytoplasm-dense cells of the leaf primordia (Figure 5, B, left). By contrast, in rh27-2 shoots, miR164 was present in a dispersed pattern in vacuolated cells of the shoot apices (Figure 5, B, right).
Figure 5.
Accumulation and expression patterns of miRNAs and their target genes in rh27-2 seedlings. (A) Quantification of miRNAs in shoot apices and root tips of wild-type (WT) and rh27-2 seedlings by RT-qPCR. The expression levels were normalized to those of 5.8S rRNA (shoot apices) or U6 (root tips). (B) In situ hybridization analyses of miR164 in shoot apices from wild-type and rh27-2 plants at 5 DAG. Note the diffused expression of miR164 in vacuolated cells of rh27-2 shoots, compared with the specific expression pattern in cytoplasm-dense cells in the leaf primordia of wild-type shoots. A miR124 antisense probe from mouse was used as a negative control. Bar = 50 µm. (C) RT-qPCR analyses of the transcript levels of genes involved in stem cell homeostasis in the SAM and RAM. Note that the expression levels of HAM1 (a target of miR171), CUC1, CUC2, and NAC100 (targets of miR164), and PHB (a target of miR165/166) were up-regulated in rh27-2 plants. The expression levels were normalized to that of EIF4A. (D) In situ hybridization analyses using an antisense probe, showing the expression patterns of CUC2 in shoot apices from wild-type and rh27-2 seedlings. Note the diffused expression of CUC2 in rh27-2 shoots, compared with the boundary-specific expression (arrowheads) between the SAM and leaf primordia in wild-type shoots. A CUC2 sense probe was used as a negative control. Bar = 50 µm. (E) and (F) Comparison of the levels of miRNAs in wild-type and rh27-2 shoot apices (E) and root tips (F), as determined by small RNA-seq. Note that, compared with the wild-type shoot apices, 81 (blue) and 60 (orange) miRNAs were down-regulated and up-regulated (≥1.5-fold) in rh27-2 shoot apices, respectively (E). In addition, 154 (blue) and 63 (orange) miRNAs were down-regulated and up-regulated (≥1.5-fold) in rh27-2 root tips, respectively (F). In (A) and (C), the error bars indicate the SD from three independent samples. In each experiment, three independent repeats were conducted for each sample. Asterisks indicate significant differences between samples (two-tailed Student’s t test, *P < 0.05, **P < 0.01).
To determine whether the altered abundances of miRNAs in the rh27-2 line affect the expression of their target genes, RT-qPCR analyses were performed to examine the transcript levels of HAM1 and CUCs (CUC1, CUC2, and NAC100), which are targets of miR171 and miR164, respectively (Llave et al., 2002; Mallory et al., 2004a), in shoot apices from wild-type and rh27-2 seedlings at 5 DAG. In addition, the transcript levels of the HD-ZIP III family members PHB, PHV, CNA, and REV, which are targets of miR165/166 (Mallory et al., 2004b), were examined in root tips from both lines. Compared with those in shoot apices from wild-type plants, the levels of HAM1, CUC1, CUC2, and NAC100 were up-regulated in shoot apices from rh27-2 plants, and that of PHB was higher in root tips from rh27-2 plants than those from wild-type plants (Figure 5, C). In situ hybridization analyses of shoot apices using sense and antisense probes showed that CUC2 transcript was present at the boundaries between the SAM and leaf primordia (Figure 5, D, indicated by arrowheads), as reported previously (Ishida et al., 2000). However, in rh27-2 shoot apices, a diffused pattern and no evident boundary-specific expression were observed (Figure 5, D;Supplemental Figure S9).
To investigate the effect of the rh27-2 mutation on miRNA biogenesis in general, small RNA-seq was performed to analyze the levels of miRNAs in shoot apices and root tips excised from the wild-type and rh27-2 seedlings at 5 DAG. Data from three independent experiments for each set of samples were reproducible (Supplemental Figure S10). Among the 428 known miRNAs in Arabidopsis (http://www.mirbase.org/), 207 were detected in shoot apices and 81 were down-regulated in the rh27-2 line (Figure 5, E andSupplemental Data Set S1), including miR171 and miR394, which have well-known roles in stem cell homeostasis in the SAM (Knauer et al., 2013; Zhou et al., 2018). In root tips, 269 miRNAs were detected, of which 154 were down-regulated in the rh27-2 line (Figure 5, F andSupplemental Data Set S1). Notably, smaller numbers of miRNAs were up-regulated in the rh27-2 line: 60 and 63 in shoot apices and root tips, respectively. Furthermore, RNA-seq analyses of wild-type and rh27-2 seedlings at 5 DAG showed that 18 miRNA target genes were up-regulated in shoot apices and 31 were up-regulated in root tips of rh27-2 seedlings (Supplemental Figure S11, A and B).
RH27 is involved in pri-miRNA processing
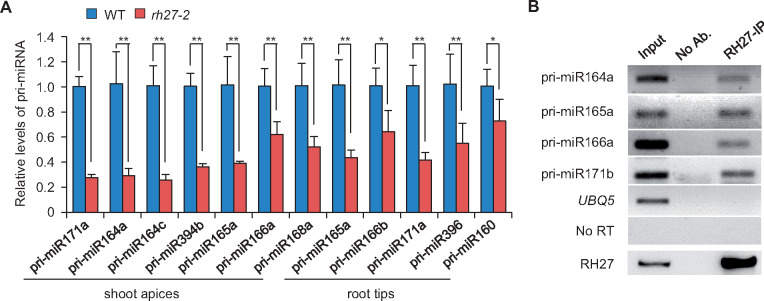
To explore the function of RH27 in miRNA biogenesis further, RT-qPCR analyses were performed to examine the levels of several pri-miRNAs in shoot apices and root tips of wild-type and rh27-2 seedlings at 5 DAG. All seven pri-miRNAs examined in the shoot apices were accumulated at lower levels in the rh27-2 than in the wild-type line (Figure 6, A). Similarly, five of the seven pri-miRNAs examined in root tips showed reduced levels of transcript in rh27-2 seedlings (Figure 6, A andSupplemental Figure S12, A). An RNA-seq analysis revealed that, among the 116 and 65 pri-miRNAs detected in shoot apices and root tips, 29 and 7 were down-regulated, respectively, in rh27-2 seedlings (Supplemental Figure S12, B and C).
Figure 6.
Involvement of RH27 in pri-miRNA processing. (A) RT-qPCR analyses of pri-miRNAs in shoot apices and root tips of wild-type (WT) and rh27-2 seedlings. Error bars indicate the SD of three independent samples. Asterisks indicate significant differences from the wild-type (two-tailed Student’s t test, *P < 0.05, **P < 0.01). (B) A RIP-RT-PCR assay using an anti-RH27 antibody to show the association of RH27 with pri-miR164a, pri-miR165a, pri-miR166a, and pri-miR171b in vivo. One-tenth of the RNA, as used in the RIP-RT-PCR analysis, was used as the input sample. No Ab.: No antibody control. In the bottom panel, the anti-RH27 antibody was used to show the presence of RH27 proteins in the input and RH27-IP samples. No RT: RIP-RT-PCR was performed with pri-miR166a primers without reverse transcription. UBQ5, negative control.
We then examined whether RH27 is associated with pri-miRNAs in vivo using RNA immunoprecipitation-coupled RT-PCR (RIP-RT-PCR). All four pri-miRNAs examined (pri-miR164a, pri-miR165a, pri-miR166a, and pri-miR171b) were co-precipitated by the anti-RH27 antibody (Figure 6, B), whereas no or very weak signals were obtained when no antibody was applied as a control. In addition, no precipitation signal was observed when UBIQUITIN 5 (UBQ5) primers were used as a negative control (Figure 6, B). Overall, these data suggest that RH27 is involved in pri-miRNA processing.
RH27 interacts with DDL, HYL1, and SE
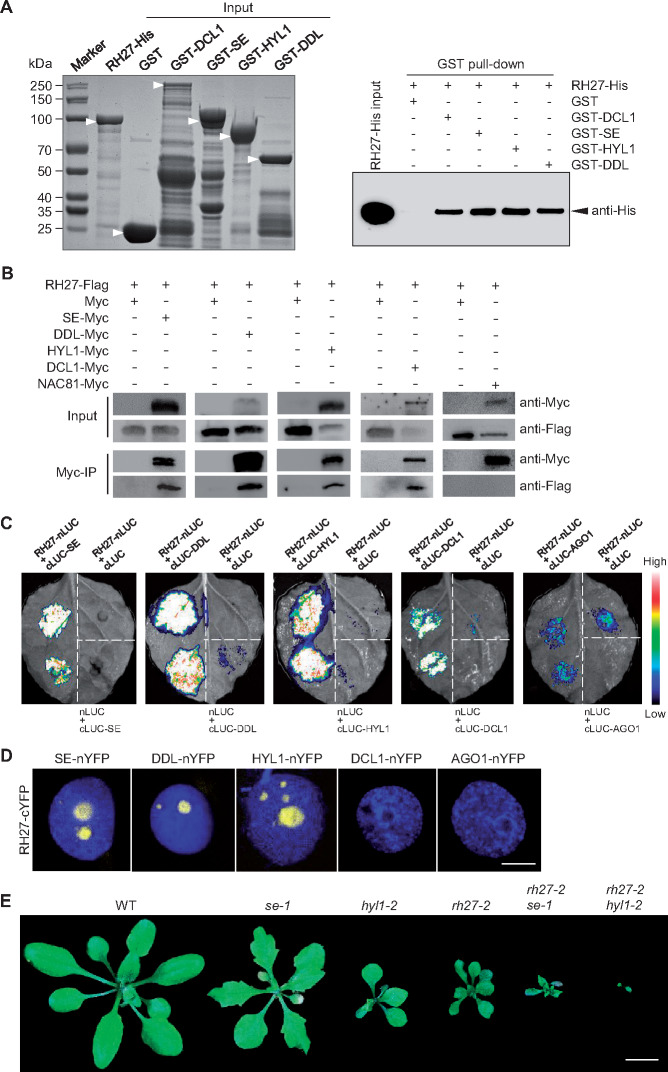
In animals, DEAD-box RNA helicases act as components of the microprocessor complex to regulate miRNA biogenesis (Gregory et al., 2004). In plants, the microprocessor complex consists of DCL1, SE, and HYL1. In addition, DDL interacts with DCL1 to facilitate the recruitment of substrates of the microprocessor complex (Yu et al., 2008). To determine whether RH27 interacts with these components, in vitro pull-down assays were performed using glutathione S-transferase (GST)-tagged DCL1, DDL, HYL1, or SE as the bait, and His-tagged RH27 as the prey. GST-DCL1, GST-DDL, GST-HYL, and GST-SE were able to pull-down RH27-His, whereas GST alone was not (Figure 7, A). Subsequently, a co-immunoprecipitation experiment was performed to examine the interactions of RH27 with DCL1, DDL, HYL1, and SE in vivo. In transient assays performed pair-wisely in Nicotiana benthamiana leaves, RH27-Flag was detected in precipitates of DCL1-Myc, DDL-Myc, HYL1-Myc, and SE-Myc. No interaction was detected when either the empty Myc vector or NAC81-Myc was used (Figure 7, B).
Figure 7.
RH27 interacts with DDL, HYL1, and SE. (A) In vitro pull-down assays using His-tagged RH27 (RH27-His) as prey, and GST-tagged DCL1, DDL, HYL1, or SE as bait, to examine the interaction of RH27 with miRNA biogenesis components. GST alone was used as a negative control. The left panel, a PAGE gel stained with Coomassie Bright Blue, shows heterologous production of input proteins with expected molecular weights (arrowheads) produced in Escherichia coli. The right panel shows that the RH27-His protein (arrowhead) was pulled-down by GST-DCL1, GST-DDL, GST-HYL1, and GST-SE, using glutathione MagBeads, as detected by the anti-His antibody. Three independent experiments were performed. (B) Co-immunoprecipitation experiments to examine the interaction of Flag-tagged RH27 with Myc-tagged DCL1, DDL, HYL1, or SE. The assays were performed by pair-wise co-infiltration of constructs in N. benthamiana leaves. Note that RH27-Flag was precipitated by DCL1-Myc, DDL-Myc, HYL1-Myc, and SE-Myc, but not by the controls: empty Myc vector (Myc) and NAC81 transcription factor fused with Myc (NAC81-Myc). Three independent experiments were performed. (C) LCI assays performed in N. benthamiana leaves to examine the interaction of RH27 with DCL1, DDL, HYL1, or SE. The indicated constructs were co-infiltrated. AGO1 was used as a negative control. Note the strong interactions between RH27-nLUC and cLUC-DDL, RH27-nLUC and cLUC-HYL1, and RH27-nLUC and cLUC-SE. A weak interaction was observed between RH27-nLUC and cLUC-DCL1. Three independent experiments were performed for each combination, and in each experiment, more than three leaves were infiltrated and examined. (D) A BiFC assay performed in N. benthamiana leaves, examined under a confocal microscope, to detect the interaction between RH27-cYFP and DCL1-nYFP, DDL-nYFP, HYL1-nYFP, or SE-nYFP. AGO1-nYFP was used as a negative control. Note that BiFC signals (yellow) were detected in the nuclei in combinations of RH27-cYFP and DDL-nYFP, RH27-cYFP and HYL1-nYFP, and RH27-cYFP and SE-nYFP. No interaction was detected for RH27-cYFP and DCL1-cYFP. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Three independent experiments were performed. Bar = 5µm. (E) Phenotypes of the se-1, hyl1-2, and rh27-2 single mutants, and the rh27-2 se-1 and rh27-2 hyl1-2 double mutants, at 25 DAG. Bar = 1 cm.
To validate the interactions in vivo, pair-wise firefly luciferase complementation imaging (LCI) assays (Chen et al., 2008) were performed in N. benthamiana using RH27-nLUC with one of the cLUC constructs (cLUC-DCL, cLUC-DDL, cLUC-HYL1, or cLUC-SE) or cLUC-AGO1 as a control. Strong interactions were observed for the combinations of RH27-nLUC and cLUC-DDL, RH27-nLUC and cLUC-HYL1, and RH27-nLUC and cLUC-SE (Figure 7, C). A weak interaction was also detected when RH27-nLUC and cLUC-DCL1 were co-infiltrated (Figure 7, C), while no interaction was detected when RH27-nLUC and cLUC-AGO1 were co-infiltrated (Figure 7, C). We then examined the interaction between RH27 and DCL1, DDL, HYL1, or SE using bimolecular fluorescence complementation (BiFC) assays (Walter et al., 2004). RH27 was able to interact with DDL, HYL1, and SE, but no interaction between RH27 and DCL1 was detected (Figure 7, D). It should be noted that the interaction signals were localized exclusively in discrete bodies, most likely nuclear dicing bodies (Fang and Spector, 2007; Song et al., 2007; Figure 7, D).
A further genetic analysis was performed by introducing the rh27-2 mutation into the se-1 or hyl1-2 mutant line by crossing. Phenotypic analysis of homozygous rh27-2 se-1 and rh27-2 hyl1-2 double mutants identified in the F2 progenies showed that the defects in growth and development were more severe in both double mutants than in any of the single mutants (Figure 7, E), suggesting an additive role of RH27, SE, and HYL1 in miRNA biogenesis.
Discussion
Studies performed over the past two decades have identified more than 10 genes that are essential for zygote division and are involved in cell cycle regulation, energy supply, and ribosomal RNA and tRNA processing (Shen et al., 2002; Xu et al., 2005; Lin et al., 2007; Ronceret et al., 2008; Andreuzza et al., 2010; Li et al., 2010; Ngo et al., 2012; Wu et al., 2012; Yu et al., 2012; Guo et al., 2016; Yang et al., 2017). In this study, we established that the zyg4-1 (rh27-1) mutant of Arabidopsis exhibits a zygote-lethal phenotype caused by a point mutation in the DEAD-box RNA helicase-coding gene RH27. Although this gene is not expressed in sperm cells of mature pollen, both the paternal and the maternal RH27 alleles are expressed immediately after fertilization. The contributions of maternal and paternal genomes to early zygotic embryogenesis have been disputed (Autran et al., 2011; Nodine and Bartel, 2012; Del Toro-De León et al., 2014; Zhao et al., 2019); however, the observed maternal and paternal RH27 expression in zygotes and the typical Mendelian inheritance of the zyg4-1 (rh27-1) mutation support the model of equal contributions of the two parental genomes.
As a family of highly conserved enzymes present in almost all living organisms, DEAD-box RNA helicases are involved in a range of processes, such as tumorigenesis, germ cell specification, and embryonic patterning, through the regulation of RNA metabolism (Schüpbach and Wieschaus, 1986; Botlagunta et al., 2008; Linder and Jankowsky, 2011; Cai et al., 2017). In plants, DEAD-box RNA helicases are implicated in drought tolerance, salinity sensitivity, and cold and heat responses (Gong et al., 2005; Khan et al., 2014; Zhu et al., 2015; Wang et al., 2016, 2019a). In this study, we observed that the weak allele of rh27-2, containing a T-DNA insertion in RH27 that causes truncation of the non-conserved N-terminal region of the RH27 protein, was defective in stem cell homeostasis in the SAM and RAM. Previous studies have demonstrated that stem cell-expressed CLV3 peptides, perceived by the CLV1/CLV2/RPK2 receptor kinases, establish a negative feedback regulation loop with the SCOC-expressed transcription factor WUS to maintain stem cell homeostasis in the SAM (Fletcher et al., 1999; Brand et al., 2000; Kinoshita et al., 2010). Here, homozygous rh27-2 seedlings showed short-root and multi-shoot phenotypes, in which the dome structure of SAM was lost and the expression patterns of the stem cell marker gene CLV3 and the SCOC marker gene WUS were irregular. Occasionally, multiple CLV3 and WUS expression domains were identified in rh27-2 plants, which is consistent with the formation of multiple shoots at the bolting stage. In the RAM, in contrast to the specific expression of WOX5 in four QC cells observed in wild-type plants, disorganized WOX5 expression, including increased WOX5-positive cell numbers and altered cell patterns, was observed in rh27-2 plants. Similarly, altered WOX5 expression has also been reported in plants expressing a mutant ACTIVE QUIESCENT CENTER1 (AQC1) gene, which encodes a tyrosylprotein sulfotransferase involved in peptide modifications during root stem cell maintenance (Zhou et al., 2010). In addition, CSCs, the stem cells for the columella cell linage, were prematurely differentiated in rh27-2 roots in this study. A similar phenotype has been observed following laser ablation of QC cells (van den Berg et al., 1997) or mutation of the WOX5 or AQC1 gene (Sarkar et al., 2007; Zhou et al., 2010). Overall, these results suggest that RH27 plays a critical role in stem cell homeostasis in both the SAM and RAM.
The rh27-2 mutant had reduced levels of miR164 and miR171, and increased levels of the CUC1, CUC2, NAC100, and HAM1 mRNAs in the shoot apices. CUC1 and CUC2, targets of miR164, are redundantly required to define organ boundaries in the SAM (Takada et al., 2001; Rhoades et al., 2002; Mallory et al., 2004a). In this study, in situ hybridization analyses revealed that CUC2 displayed a diffuse expression pattern in rh27-2 plants, unlike the boundary-specific expression seen in wild-type plants. Similarly, miR164 also displayed a dispersed expression pattern in the rh27-2 mutant, rather than the leaf primordium-specific expression seen in the wild-type line. It is possible that the altered expression pattern of CUC2 is a consequence of changes in the expression level and pattern of miR164, and may lead to defective functioning of the SAM in rh27-2 plants, as well as the multi-shoot phenotype. In addition, the up-regulation of HAM1, possibly caused by decreased expression of miR171, may also contribute to defective SAM function and stem cell homeostasis in the rh27-2 line. In agreement with this speculation, previous studies showed that mutations in miR164-coding genes, or expression of miR164-resistant CUC2 (resulting in expanded expression region of CUC2), lead to altered phyllotaxis in the inflorescence and organ boundaries in flowers (Laufs et al., 2004; Sieber et al., 2007). In roots, PHB is a gene essential for xylem cell fate specification, and a gain-of-function mutant of PHB containing a point mutation at the miR165/166 target site shows a short-root phenotype (Carlsbecker et al., 2010). Consistent with the short-root phenotype in the gain-of-function mutant of PHB, we observed that rh27-2 plants exhibited a similar short-root phenotype and an elevated expression level of PHB, alongside a reduced accumulation of miR165/166. It is reasonable to speculate that RH27-mediated miRNA biogenesis may control the expression of genes involved in stem cell homeostasis in the SAM and RAM (Figure 8).
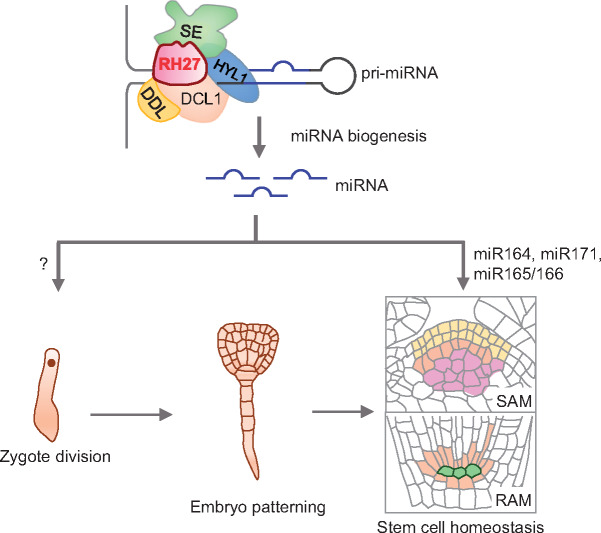
Figure 8.
A proposed model for the role of RH27 in zygote division and stem cell homeostasis. Based on the facts that mutations of RH27 caused a zygote-lethal phenotype (rh27-1) and defective stem cell homeostasis (rh27-2), and RH27 interacts with DDL, HYL1, and SE, associates with pri-miRNAs, and promotes the accumulations of miR164, miR171, and miR165/166, it is proposed that RH27 is a key player in zygote division, stem cell homeostasis and miRNA biogenesis in Arabidopsis. It should be noted that direct evidence for the involvement of miRNAs in zygote division is still lacking; therefore, a question mark is used.
In animals, DEAD-box RNA helicase is a component of the microprocessor complex (Gregory et al., 2004). In Arabidopsis, the microprocessor complex consisting of DCL1, HYL1, and SE (Wang et al., 2019b) recognizes and cleaves pri-miRNAs (Kurihara et al., 2006; Dong et al., 2008). The RNA-binding protein DDL is proposed to promote the access of DCL1 to its substrates (Yu et al., 2008). In this study, strong interactions of RH27 with DDL, HYL, and SE, and a weak interaction between RH27 and DCL1, were detected. It has been reported previously that DDL, HYL1, and SE bind to pri-miRNAs, and in this study, we established that RH27 binds to four different pri-miRNAs. These results led us to propose RH27 as a component of the microprocessor complex (Figure 8). Previous studies have shown that the dcl1-1 and se-4 mutant lines show embryo defects at the late globular stage (Schwartz et al., 1994; Lobbes et al., 2006), whereas the zyg4-1 (rh27-1) showed a zygote-lethal phenotype, suggesting that RH27 may also function in biological processes beyond miRNA biogenesis. This possibility is supported by the additive effects observed in genetic analysis of rh27-2, hyl1-2, and se-1 single and double mutants.
Mutation of DCL1, HYL1, or SE often results in reduced accumulation of miRNAs and increased levels of pri-miRNAs (Kurihara et al., 2006; Lobbes et al., 2006). However, in a genome-wide analysis of the se-2 mutant, although a large subset of miRNAs (71%) showed reduced accumulation, the levels of a small proportion (8%) were increased (Wang et al., 2018). Similar results were observed in this study: the levels of a large subset of miRNAs (39% in shoots, 57% in roots) were reduced in the shoot apices and root tips of the rh27-2 mutant, and the levels of a smaller proportion of miRNAs (29% in shoots, 23% in roots) were increased. Notably, we also observed reduced accumulations of pri-miRNAs in rh27-2 shoot apices and root tips, as reported previously for the ddl-1 mutant (Yu et al., 2008). We hypothesize that feedback regulation at the levels of miRNA transcription and processing may affect individual miRNAs and pri-miRNAs differently, as proposed before (Tang et al., 2012; Feng et al., 2020). Alternatively, since RH27 is a member of a family of 58 DEAD-box RNA helicases in Arabidopsis (Mingam et al., 2004), the observed changes in the levels of a subset of miRNAs in the rh27-2 mutant may represent only a fraction of the full complement of cellular miRNA biogenesis. Of course, with data obtained so far, we cannot exclude the possibility that RH27 may involve directly in miRNA transcription and stability.
Materials and methods
Plant materials and growth conditions
The Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was used as the wild-type, and unless otherwise described, all mutants were in the Col-0 background. The zyg4-1 (rh27-1) mutant was obtained by screening an ethyl methanesulfonate-mutagenized population, generated as described previously (Guo et al., 2016). The rh27-2 mutant (SALK_068661) was obtained from the Arabidopsis Biological Resources Center (https://abrc.osu.edu/). The mutant lines se-1 (Prigge and Wagner, 2001) and hyl1-2 (SALK_064863; Vazquez et al., 2004), and the reporter lines ProWOX5:erGFP (Blilou et al., 2005), QC25 (Sabatini et al., 2003), ProCLV3:GUS (Fletcher et al., 1999), ProWUS:GUS (Mayer et al., 1998), and pEC1:H2B-RFP (Ingouff et al., 2009) have been described previously. Plants were grown in commercially available humus soil at 21°C ± 2°C under a 16-h light (100 µmol photons m−2 s−1; supplied by white-light LED tubes) and 8-h dark cycle (Song et al., 2012). For seedling and RAM analyses, the surface-sterilized seeds were germinated on solid half-strength Murashige and Skoog (1/2 MS) salt medium containing 1% Suc and 1% agar and were vertically cultured. For SAM analyses, the seeds were germinated and cultured in liquid 1/2 MS medium on a rolling bank (Song et al., 2013).
Genetic analyses and molecular cloning
The rh27-1/+ and rh27-2 mutants were backcrossed to Col-0 for five and three generations, respectively, before further analyses. Map-based cloning of the mutation site in rh27-1 was performed in 2,700 F2 progenies from a cross between rh27-1/+ and Landsberg erecta (Ler). A G-to-A point mutation was detected in a rh27-1/+ plant after sequencing candidate genes. For complementation and expression analyses, the 2,227-bp 5′-upstream and the 2,914-bp genomic region (without the stop codon) of RH27 was fused to the GUS or sYFP reporter gene of the pPLV15 or pPLV18 vector (De Rybel et al., 2011) using a one-step cloning method (Gibson et al., 2009). The obtained ProRH27:RH27-GUS and ProRH27:RH27-sYFP constructs were transformed into wild-type plants using the Agrobacterium tumefaciens-floral dip method (Clough and Bent, 1998). The ProRH27:ΔRH27-sYFP construct was generated by inserting the 2,227-bp 5′-upstream and truncated 2,604-bp genomic region (ΔRH27, a 310-bp deletion from start codon onward) of RH27 into the pPLV18 vector, as described above. All primers used are listed in Supplemental Data Set S2.
Microscopic and histological analyses
Whole-mount clearing and observation of ovules, shoot apices, and root tips (Yu et al., 2012), GUS assays (Song et al., 2013), and Lugol staining (Zhou et al., 2010) were performed as described previously. For histological analyses after GUS assays, shoot apices were embedded in Technovit 7100 (Heraeus Kulzer), and 5 µm sections were cut on a Leica microtome (Leica EM UC7). The sections were stained with periodic acid-Schiff’s reagent (Baum, 2008) and photographed using a differential interference contrast-equipped microscope (Nikon 80i). The sizes (area) of the L1 cells in the SAM were measured and analyzed using ImageJ software (Schindelin et al., 2012). Transgenic plants carrying the fluorescent-reporter genes were examined under a Leica TCS SP5 or Zeiss LSM980 confocal laser scanning microscope. GFP was excited using a 488-nm laser line; sYFP was excited using a 514-nm laser line; and RFP was excited using a 561-nm laser line.
Reverse transcription quantitative PCR analyses
For RT-qPCR-based detection of pri-miRNAs and target genes of miRNAs, total RNAs were extracted from shoot apices and root tips (∼5 mm) of seedlings at 5 DAG using the RNAprep Pure Plant Kit (DP432, Tiangen). For shoot apices collection, cotyledons, the first pair of leaves, hypocotyls, and roots were cut off using a razor blade under a dissecting microscope. Reverse transcription was performed using the FastQuant RT Kit (KR106, Tiangen) and qPCR was performed with iQ SYBRGreen Supermix (1708886, BioRad) on the BioRad CFX96 system, using EIF4A as a normalization control. For quantification of mature miRNAs, small RNAs were extracted from shoot apices and root tips (∼5 mm) of seedlings at 5 DAG, collected as mentioned above, using the miRcute miRNA Isolation Kit (DP501, Tiangen), and reverse transcription was performed with the miRcute Plus miRNA First-Strand cDNA Kit (KR211, Tiangen). The qPCR analyses were performed with the miRcute Plus miRNA qPCR Kit (FP411, Tiangen) on the BioRad CFX96 system, using 5.8S rRNA or U6 snRNA as normalization controls. All data were analyzed using the method (Livak and Schmittgen, 2001). The primers used are listed in Supplemental Data Set S2.
RNA in situ hybridization
In situ hybridization analyses of miRNAs were performed according to the method described by Javelle and Timmermans (2012), with 3′-end digoxigenin-labeled LNA modified probes (Supplemental Data Set S2). Antisense miR164 and miR124 probes were used at a final concentration of 50 nM and hybridized at 42°C overnight. The mRNA in situ hybridization analyses were conducted as described previously (Xin et al., 2017). For RNA probes, a 568-bp CUC2 fragment was amplified from genomic DNA using gene-specific primers (Supplemental Data Set S2) and cloned into the pEASY Blunt3 vector. PCR products containing the T7 promoter sequence, amplified from recombinant plasmids, were used as templates for in vitro transcription. Slides were photographed under a bright field microscope (Nikon 80i).
RIP-RT-PCR
RIP-RT-PCR was performed as described previously (Ren et al., 2012). Briefly, wild-type seedlings at 10 DAG (approximately 2 g) were irradiated twice at 400 mJ/cm2 in ice-cold PBS buffer. The nuclei were then extracted and lysed, and the cross-linked RNA–protein complexes were immunoprecipitated using a rabbit polyclonal anti-RH27 antibody, which was generated against His-tagged full-length RH27 and then purified by protein G using affinity chromatography. Three percent immunoprecipitates were analyzed by immunoblotting and detected using a mouse anti-RH27 polyclonal serum (1:3,000) generated against the same recombinant RH27 protein as that used to generate the rabbit anti-RH27 antibody. RNAs were then purified from RH27–RNA complexes with acidic phenol:chloroform:isopentanol (25:24:1), precipitated with 100% ethanol, and then digested with DNase I (M0303, NEB). Reverse transcription was performed using the First Strand cDNA Synthesis Kit with random primers (FSK101, TOYOBO). RT-PCR analyses were then performed using the primers listed in Supplemental Data Set S2.
Small RNA-seq and data analysis
Three sets of shoot apices and root tips (∼3 mm) were harvested from wild-type and rh27-2 seedlings at 5 DAG, as described for the RT-qPCR analyses. Approximately 30 mg were collected for each sample and total RNAs were extracted from the samples using the miRNeasy Mini Kit (217004, Qiagen). To prepare libraries, small RNAs were enriched from 2 µg of total RNAs using PEG8000, following the standard protocol (Berry Genomics). The libraries were pooled and sequenced on the Illumina Nextseq CN500 platform. For each small RNA-seq library, the sequences of the 3ʹ-adaptors were removed and reads with sizes of 18–42 nt were selected using cutadapt (1.18; Martin, 2011). All clean reads were aligned to the Arabidopsis genome (TAIR10) using Bowtie2 (2.2.9; Langmead and Salzberg, 2012) with default parameters, and then alignments with up to one mismatch were selected. Reads were then aligned to the mature miRNA sequences from miRBase v22 using NCBI blast (2.2.26) with perfect matches. The aligned miRNAs were normalized against total mapped reads number of each library to calculate the reads per 30 million (RP30M) value. The Pearson correlation coefficient (r) of miRNA abundance was used to evaluate the consistency of replicate libraries. All replicate data sets were highly consistent (r > 0.95, P-value < 10−100). Subsequently, miRNAs with a fold change >1.5 were determined as differentially expressed.
RNA-seq and data analysis
Sample collections were performed as described for small RNA-seq. Total RNAs were extracted from the samples using the RNAprep Pure Plant Kit (DP432, Tiangen) and RNA libraries were prepared following the standard protocol (Berry Genomics). The libraries were pooled and sequenced on the Illumina Nextseq CN500 platform. RNA-seq reads containing a 3′-adaptor sequence were discarded using cutadapt (1.18; Martin, 2011). All clean reads were then aligned to the Arabidopsis genome (TAIR10) using HISAT2 (2.1.0; Kim et al., 2015) with default parameters. Reads with a unique genome location were used for subsequent analyses. The numbers of reads mapped to a single gene annotated in Araport11 were counted using htseq-count script from HTSeq (0.6.0; Anders et al., 2015). The expression level of each gene was normalized by FPKM (Fragment count Per Kilobase of exon per Million fragments mapped) and codes to analyze the data are available at GitHub (https://github.com/oneleaf425/rh27). Differentially expressed genes were detected using R package edgeR (Robinson et al., 2010). Genes with a false discovery rate value less than 0.01 and fold change greater than 2 were regarded as differentially expressed. To analyze pri-miRNA levels, 325 pri-miRNAs annotated in Araport11 were acquired and counted using htseq-count separately to avoid overlapping with protein coding genes (Wang et al., 2019c). A total of 131 previously validated miRNA-target pairs were used to analyze the expression of miRNA target genes (Ma et al., 2018).
BiFC and LCI assays
For BiFC assays, the coding sequence of RH27 was amplified from Arabidopsis and cloned in-frame to the coding sequence for the C-terminal region of YFP (cYFP) of the pSPYCE-35S vector. SE, DDL, HYL1, DCL1, or AGO1 was cloned in-frame to the coding sequence for the N-terminal region of YFP (nYFP) of the pSPYNE-35S vector (Walter et al., 2004). The cYFP- and nYFP-fusion constructs were transformed into A. tumefaciens and pair-wise combinations were co-infiltrated into Nicotiana benthamiana leaves. After incubation for 24 h in the dark, followed by an additional 16 h light/8 h dark photoperiod, YFP signals were detected as described in Microscopic and Histological Analyses. Three independent assays were performed. Primers used for the constructs are listed in Supplemental Data Set S2.
For LCI analyses, the coding sequence of RH27 was in-frame fused to the coding sequence for the N-terminus of nLUC in the pCAMBIA1300-nLUC vector, and the coding sequence of SE, DDL, HYL1, DCL1, or AGO1 was in-frame fused to the coding sequence for the C-terminus of cLUC in the pCAMBIA1300-cLUC vector. The nLUC- and cLUC-fused constructs were transformed into A. tumefaciens and co-infiltrated pair-wisely into the leaves of N. benthamiana. LUC signals were detected under a low-light cooled CCD camera (Tanon) (Chen et al., 2008). Three independent assays were performed, and at least three leaves were co-infiltrated for each combination per assay. The primers used for the constructs are listed in Supplemental Data Set S2.
Co-immunoprecipitation assay
The cDNA sequence of RH27 was cloned and fused in-frame to the coding sequence for the N-terminus of a Flag tag, and the cDNA sequences of SE, DDL, HYL1, DCL1, and NAC81 were cloned and fused to the coding sequence for the N-terminus of the Myc tag, by introducing into the pSuper1300 vector (Ni et al., 1995; Liu et al., 2017) to create the RH27-Flag, SE-Myc, DDL-Myc, HYL1-Myc, DCL1-Myc, and NAC81-Myc constructs. The primers used for the constructs are listed in Supplemental Data Set S2. A. tumefaciens strains carrying RH27-Flag were co-infiltrated pair-wisely with strains carrying the SE-Myc, DDL-Myc, HYL1-Myc, DCL1-Myc, or NAC81-Myc into 3-week-old N. benthamiana leaves. Leaf samples were collected after incubation for 24 h in the dark, followed by an additional 16 h light/8 h dark photoperiod, and were ground into a fine powder with liquid nitrogen. The leaf powder (0.5 g) was lysed with 1 mL NP-40 buffer (E124-01, GenStar) and incubated with anti-Myc beads (M047-11, MBL) according to the manufacturer’s instructions. The anti-Myc bead-bound proteins were washed three times with NP-40 and analyzed by immunoblotting using anti-Myc (C3956, Sigma, lot 016M4762V, 1:3,000) or anti-Flag (M185-7, MBL, lot 004, 1:3,000) antibodies.
In vitro pull-down assay
The coding sequence of DCL1, SE, HYL1, or DDL was in-frame fused with the coding sequence for the C-terminus of GST in the pGEX-6P-1 vector, and His-tagged RH27 was generated in the pET28SUMO vector. The GST-fusion constructs were then transformed into Escherichia coli strain BL21 (DE3), and expression of the recombinant proteins was induced by incubation with 0.2 mM isopropyl-β-D-thiogalactoside at 20°C overnight. GST-tagged proteins were purified by affinity chromatography with Glutathione Resin (L00206, GenScript), according to the manufacturer’s instructions. The RH27-His construct was transformed into the E. coli strain BL21 codon-plus RIL, and expression of the fusion protein was induced by incubation with 0.1 mM isopropyl-β-D-thiogalactoside at 16°C overnight. The induced cells were collected by centrifugation and lysed by sonication in binding buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, and 25 mM imidazole). Subsequently, the clarified lysate was incubated with the prepared Profinity™ IMAC Resins (156-0123, BioRad) at 4°C for 30 min. After washing the resin five times with binding buffer, the RH27-His protein was eluted with elution buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, and 500 mM imidazole). GST protein pull-down assays were based on Glutathione MagBeads instructions (L00327, GenScript). Briefly, a 30-μg sample of the GST, GST-DCL1, GST-SE, GST-HYL1, or GST-DDL fusion protein was incubated with Glutathione MagBeads. After washing out the non-bound GST-fused proteins, a 5-μg sample of RH27-His was incubated with the beads bound by the GST-fused protein in 0.5 mL pull-down buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, and 1% Triton X-100) for 1 h at 4°C with intermittent shaking. The beads were then washed three times with pull-down buffer and collected for detection by immunoblotting with an anti-His antibody (D291-7, MBL, lot 008, 1:3,000). The primers used for the constructs are listed in Supplemental Data Set S2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RH27 (At5g65900), WOX5 (At3g11260), CLV3 (At2g27250), WUS (At2g17950), HAM1 (At2g45160), CUC1 (At3g15170), CUC2 (At5g53950), NAC100 (At5g61430), PHB (At2g34710), PHV (At1g30490), CNA (At1g52150), REV (At5g60690), DCL1 (At1g01040), SE (At2g27100), HYL1 (At1g09700), DDL (At3g20550), AGO1 (At1g48410), NAC81 (AT5G08790), and EIF4A (At1g54270). The protein sequences of RH27 and its homologs in other species can be found in the National Center for Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov/) under the following accession numbers: NP_201391 (RH27, Arabidopsis), BAS86908 (OsRH27, Os03g0802700, Oryza sativa), NP_080136 (MmDDX18, Mus musculus), CAG33341 (HsDDX18, Homo sapiens), Q03532 (HAS1, Saccharomyces cerevisiae), and NP_415318 (RHLE, E. coli). The small RNA and RNA-seq data sets described in this study have been deposited in the GEO database under accession number GSE154473.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of RH27 and its homologs, showing conserved motifs (supports Figure 1, C).
Supplemental Figure S2. Genotypic analyses of zyg4-1 (rh27-1); ProRH27:RH27-GUS and zyg4-1 (rh27-1); ProRH27:ΔRH27-sYFP seedlings (supports Figure 1, D).
Supplemental Figure S3. Abnormal seeds produced in rh27-2/+ siliques (supports Figure 1, A).
Supplemental Figure S4. The expression of ProWOX5:erGFP in wild-type and rh27-2 RAMs at 1, 3, 5, and 7 DAG (supports Figure 2, E).
Supplemental Figure S5. Altered SAM morphology and ProCLV3:GUS expression in rh27-2 seedlings (supports Figure 3, A).
Supplemental Figure S6. The expression patterns of ProWUS:GUS in SAMs of wild type and rh27-2 (supports Figure 3, D).
Supplemental Figure S7. Complementation of the rh27-2 mutant by ProRH27:RH27-sYFP and expression analysis of the transgenic plants (supports Figure 4).
Supplemental Figure S8. The expression of ProRH27:RH27-GUS in seedlings and the inflorescence (supports Figure 4).
Supplemental Figure S9. In situ hybridization analyses of CUC2 in wild-type (WT) and rh27-2 shoot apices (supports Figure 5, D).
Supplemental Figure S10. Hierarchical clustering analysis showing the degree of reproducibility among different small RNA-seq libraries constructed for each genotype (supports Figure 5, E and F).
Supplemental Figure S11. Expression levels of miRNA target genes in shoot apices and root tips, as determined by RNA-seq analyses (supports Figure 5, E and F).
Supplemental Figure S12. Expression levels of pri-miRNAs in shoot apices and root tips (supports Figure 6, A).
Supplemental Data Set S1. Levels of miRNAs in shoot apices and root tips.
Supplemental Data Set S2. Sequences of the primers and probes used in this study.
Supplementary Material
Acknowledgments
The authors thank Dolf Weijers (Wageningen University) for sharing the pPLV vectors; Jian-Min Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the LCI vectors; Chongyi Xu (Institute of Botany, Chinese Academy of Sciences) for the NAC81-Myc construct; Shuxin Zhang (Shandong Agricultural University), Jin-Xin Liu (Institute of Botany, Chinese Academy of Sciences), Sheng-Yang Wu (Institute of Botany, Chinese Academy of Sciences), and Lei Guo (University of Maryland) for reading the manuscript and giving useful suggestions; Wei Xin (Institute of Botany, Chinese Academy of Sciences) for positive control probes and embedded blocks of RNA in situ hybridization; and Jingquan Li (Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences) for technical assistance with the confocal laser scanning microscope.
Funding
This work was supported by the National Transgenic Science and Technology Program (grant no. 2019ZX08010-003), the National Natural Science Foundation of China (grant nos. 31871455, 30625016 and 31788103), and the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Conflict of interest statement. None declared.
C.-M.L. designed and supervised the project. X.-L.H. performed most of the experiments. H.-Q.G. screened the mutant library and cloned the gene. W.-Q.C., Y.H., and Z.W. performed the biochemical experiments. J.S. analyzed the deep sequencing data. C.-M.L., X.-L.H., and X.-F.S. analyzed the data and wrote the manuscript. H.Z. and X.C. helped with revising the manuscript. All authors read and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: X.-F.S. (xfsong@ibcas.ac.cn) and C.-M.L. (cmliu@ibcas.ac.cn).
References
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreuzza S, Li J, Guitton AE, Faure JE, Casanova S, Park JS, Choi Y, Chen Z, Berger F (2010) DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development 137: 73–81 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Kreis M, Lecharny A (1999) The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res 27: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. (2011) Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 [DOI] [PubMed] [Google Scholar]
- Baum S (2008) The PAS reaction for staining cell walls. CSH Protoc 3: 1–3 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P, et al. (2008) Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 27: 3912–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Cai W, Xiong Chen Z, Rane G, Satendra Singh S, Choo Z, Wang C, Yuan Y, Zea Tan T, Arfuso F, Yap CT, et al. (2017) Wanted DEAD/H or alive: helicases winding up in cancers. J Natl Cancer Inst 109: 1–15 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamitjana-Martı’nez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Rybel B, van den Berg W, Lokerse A, Liao CY, van Mourik H, Möller B, Peris CL, Weijers D (2011) A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol 156: 1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Toro-De León G, García-Aguilar M, Gillmor CS (2014) Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514: 624–627 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dong Z, Han M-H, Fedoroff N (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL. (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yu Y, Zhou Y, Yang Y, Lei M, Lian J, He H, Zhang Y, Huang W, Chen Y (2020) A natural variant of miR397 mediates a feedback loop in circadian rhythm. Plant Physiol 182: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gong ZZ, Dong CH, Lee H, Zhu JH, Xiong LM, Gong DM, Stevenson B, Zhu JK (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, Zhu J (2013) A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 25: 342–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Jiang L, Zhang Y, Lu XL, Xie Q, Weijers D, Liu CM (2016) The anaphase-promoting complex initiates zygote division in Arabidopsis through degradation of cyclin B1. Plant J 86: 161–174 [DOI] [PubMed] [Google Scholar]
- Han C, Liu Y, Wan G, Choi HJ, Zhao L, Ivan C, He X, Sood AK, Zhang X, Lu X (2014) The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep 8: 1447–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Sakata T, Li J, Sprunck S, Dresselhaus T, Berger F (2009) The two male gametes share equal ability to fertilize the egg cell in Arabidopsis thaliana. Curr Biol 19: R19–R20 [DOI] [PubMed] [Google Scholar]
- Ishida T, Aida M, Takada S, Tasaka M (2000) Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol 41: 60–67 [DOI] [PubMed] [Google Scholar]
- Javelle M, Timmermans MCP (2012) In situ localization of small RNAs in plants by using LNA probes. Nat Protoc 7: 533–541 [DOI] [PubMed] [Google Scholar]
- Khan A, Garbelli A, Grossi S, Florentin A, Batelli G, Acuna T, Zolla G, Kaye Y, Paul LK, Zhu JK, et al. (2014) The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J 79: 28–43 [DOI] [PubMed] [Google Scholar]
- Knauer S, Holt Anna L, Rubio-Somoza I, Tucker Elise J, Hinze A, Pisch M, Javelle M, Timmermans Marja C, Tucker Matthew R, Laux T (2013) A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev Cell 24: 125–132 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131: 4311–4322 [DOI] [PubMed] [Google Scholar]
- Li HJ, Liu NY, Shi DQ, Liu J, Yang WC (2010) YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol 10: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Castillo-González C, Yu B, Zhang X (2017) The functions of plant small RNAs in development and in stress responses. Plant J 90: 654–670 [DOI] [PubMed] [Google Scholar]
- Lin Z, Yin K, Zhu D, Chen Z, Gu H, Qu LJ. (2007) AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res 17: 815–828 [DOI] [PubMed] [Google Scholar]
- Linder P, Jankowsky E (2011) From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12: 505–516 [DOI] [PubMed] [Google Scholar]
- Liu M, Shi DQ, Yuan L, Liu J, Yang WC (2010) SLOW WALKER3, encoding a putative DEAD-box RNA helicase, is essential for female gametogenesis in Arabidopsis. J Integr Plant Biol 52: 817–828 [DOI] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017) Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell 66: 117–128 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J (2006) SERRATE: a new player on the plant microRNA scene. EMBO Rep 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Liu C, Gu L, Mo B, Cao X, Chen X (2018) TarHunter, a tool for predicting conserved microRNA targets and target mimics in plants. Bioinformatics 34: 1574–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B (2004a) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang GL, Zamore PD, Barton MK, Bartel DP (2004b) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′-region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM (1997) Genetic control of cell division patterns in developing plants. Cell 88: 299–308 [DOI] [PubMed] [Google Scholar]
- Mingam A, Toffano-Nioche C, Brunaud V, Boudet N, Kreis M, Lecharny A (2004) DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J 2: 401–415 [DOI] [PubMed] [Google Scholar]
- Müller R,, Bleckmann A,, SimonR (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo QA, Baroux C, Guthörl D, Mozerov P, Collinge MA, Sundaresan V, Grossniklaus U (2012) The Armadillo repeat gene ZAK IXIK promotes Arabidopsis early embryo and endosperm development through a distinctive gametophytic maternal effect .Plant Cell 24: 4026–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB (1995) Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J 7: 661–676 [Google Scholar]
- Nodine MD, Bartel DP (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paieri F, Tadini L, Manavski N, Kleine T, Ferrari R, Morandini P, Pesaresi P, Meurer J, Leister D (2018) The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol 176: 634–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovaara J, de Zeeuw T, Weijers D (2016) Tissue and organ initiation in the plant embryo: a first time for everything. Annu Rev Cell Dev Biol 32: 47–75 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR (2001) The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13: 1263–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B (2012) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci U S A 109: 12817–12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A, Gadea-Vacas J, Guilleminot J, Lincker F, Delorme V, Lahmy S, Pelletier G, Chabouté ME, Devic M. (2008) The first zygotic division in Arabidopsis requires de novo transcription of thymidylate kinase. Plant J 53: 776–789 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Schulz KN, Harrison MM. (2019) Mechanisms regulating zygotic genome activation. Nat Rev Genet 20: 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E (1986) Maternal-effect mutations altering the anterior–posterior pattern of the Drosophila embryo. Roux Arch Dev Biol 195: 302–317 [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW (1994) Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235–3245 [DOI] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P (2002) Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell 13: 1916–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525 [DOI] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Song L, Han M-H, Lesicka J, Fedoroff N. (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A 104: 5437–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Guo P, Ren SC, Xu TT, Liu CM. (2013) Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiol 161: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Yu DL, Xu TT, Ren SC, Guo P, Liu CM (2012) Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol Plant 5: 515–523 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto Karine G, Kirschner Gwendolyn K, Schmid Julia B, Wink René H, Hülsewede A, et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371 [DOI] [PubMed] [Google Scholar]
- Tang R, Li L, Zhu D, Hou D, Cao T, Gu H, Zhang J, Chen J, Zhang C-Y, Zen K (2012) Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res 22: 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Hibara K-I, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Truernit E, Haseloff J (2008) A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang B, Chai H, Zhong Y, Shen Y, Yang W, Chen J, Xin Z, Shi H (2019a) The DEAD-box RNA helicase SHI2 functions in repression of salt-inducible genes and regulation of cold-inducible gene splicing. J Exp Bot 71: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Qin B, Li X, Tang D, Zhang Y, Cheng Z, Xue Y. (2016) Nucleolar DEAD-box RNA helicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice. PLoS Genet 12: e1005844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mei J, Ren G (2019b) Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci 10: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Quan L, Li S, You C, Zhang Y, Zhang Y, Gao L, Zeng L, Liu L, Qi Y, Mo B, Chen X (2019c) The PROTEIN PHOSPHATASE4 complex promotes transcription and processing of primary microRNAs in Arabidopsis. Plant Cell 31: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma Z, Castillogonzalez C, Sun D, Li Y, Yu B, Zhao B, Li P, Zhang X (2018) SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557: 516–521 [DOI] [PubMed] [Google Scholar]
- Weigel D, Jürgens G (2002) Stem cells that make stems. Nature 415: 751–754 [DOI] [PubMed] [Google Scholar]
- Wu JJ, Peng XB, Li WW, He R, Xin HP, Sun MX (2012) Mitochondrial GCD1 dysfunction reveals reciprocal cell-to-cell signaling during the maturation of Arabidopsis female gametes. Dev Cell 23: 1043–1058 [DOI] [PubMed] [Google Scholar]
- Xin W, Wang Z, Liang Y, Wang Y, Hu Y (2017) Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis. J Plant Physiol 214: 1–6 [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang HY, Xie CH, Xue HW, Dijkhuis P, Liu CM (2005) EMBRYONIC FACTOR 1 encodes an AMP deaminase and is essential for the zygote to embryo transition in Arabidopsis. Plant J 42: 743–756 [DOI] [PubMed] [Google Scholar]
- Yang KJ, Guo L, Hou XL, Gong HQ, Liu CM (2017) ZYGOTE-ARREST 3 that encodes the tRNA ligase is essential for zygote division in Arabidopsis. J Integr Plant Biol 59: 680–692 [DOI] [PubMed] [Google Scholar]
- Yin J, Park G, Lee JE, Choi EY, Park JY, Kim TH, Park N, Jin X, Jung JE, Shin D, et al. (2015) DEAD-box RNA helicase DDX23 modulates glioma malignancy via elevating miR-21 biogenesis. Brain 138: 2553–2570 [DOI] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, et al. (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Jiang L, Gong H, Liu CM (2012) EMBRYONIC FACTOR 19 encodes a pentatricopeptide repeat protein that is essential for the initiation of zygotic embryogenesis in Arabidopsis. J Integr Plant Biol 54: 55–64 [DOI] [PubMed] [Google Scholar]
- Zhao L, Mao Y, Zhao Y, He Y (2016) DDX3X promotes the biogenesis of a subset of miRNAs and the potential roles they played in cancer development. Sci Rep 6: 32739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Zhou X, Shen K, Liu Z, Cheng T, Liu D, Cheng Y, Peng X, Sun MX (2019) Two-step maternal-to-zygotic transition with two-phase parental genome contributions. Dev Cell 49: 882–893 [DOI] [PubMed] [Google Scholar]
- Zhou W, Wei L, Xu J, Zhai Q, Jiang H, Chen R, Chen Q, Sun J, Chu J, Zhu L, Liu CM, Li C (2010) Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yan A, Han H, Li T, Geng Y, Liu X, Meyerowitz EM (2018) HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 361: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen G, Dong T, Wang L, Zhang J, Zhao Z, Hu Z (2015) SlDEAD31, a putative DEAD-box RNA helicase gene, regulates salt and drought tolerance and stress-related genes in tomato. PLoS ONE 10: e0133849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.