Abstract
Cytochrome-P450-reductase transfers electrons to cytochrome-P450 through its flavin mononucleotide binding domain (FBD). Despite the importance of membrane-anchoring for FBD function, studies have focused on its soluble domain lacking the transmembrane-domain. Here we demonstrate that the reconstitution of FBD in nanodiscs enables high-resolution NMR measurements and renders a stable conformation.
Graphical Abstract
Cytochrome-P450 mediated electron transfer in liver microsomes is responsible for the oxidative metabolism of both endogenous (vitamins, fatty acids, prostaglandins, and steroids) and exogenous (therapeutic drugs and toxins) compounds.1–3 NADPH-cytochrome-P450-reductase (CPR) is an endoplasmic reticulum (ER) membrane‐bound protein that transfers electrons to different types of membrane-bound cytochrome-P450 enzymes to metabolize these compounds.4, 5 The ~78-kDa CPR consists of a ~5-kDa N-terminal region including a transmembrane domain and a ~72-kDa catalytic domain consisting of a flavin mononucleotide (FMN)-binding domain (FBD) and a flavin adenine dinucleotide (FAD)-binding domain.6, 7 The transmembrane and the catalytic domains are connected by a small ~1-kDa linker region. CPR is one of the two mammalian enzymes reported to possess both FAD and FMN prosthetic groups.8 It transfers two electrons one at a time from NADPH to cytochrome-P450 prosthetic heme iron via FAD and FMN prosthetic groups.8 CPR is also indispensable in donating electrons to other physiological electron acceptors such as microsomal heme oxygenase and cytochrome-b5. FBD of CPR is an immediate electron donor to cytochrome p450s and other oxygenases.8 Hence, many of the in vitro characterization studies especially the structural studies have been reported on FBD containing9 or lacking the transmembrane domain (denoted as truncated-FBD).10–12
CPR lacking the transmembrane domain is impaired in electron transfer to cytochrome-P450 signifying the importance of its transmembrane domain for the enzyme complex formation and function.3, 4, 13, 14 The interaction between the transmembrane domains of CPR and cytochrome-P450 in a lipid bilayer environment has been shown to be essential for cytochrome-P450’s enzymatic activities.3, 15–17 High-resolution studies on CPR and FBD have been challenging due to the hydrophobic nature and dynamics in the lipid interface. A solid-state NMR study on the lipid-anchored FBD (full-length FBD) revealed that the transmembrane domain in lipid bicelles adopts an α-helical conformation with a ~13° tilt of the helical axis from the lipid bilayer normal.7 However, the presence of detergent in bicelles has been shown to convert the functional cytochrome-P450 to an inactive P420.18, 19 Hence, detergent-based reconstitution approaches are not desirable for functional reconstitution of membrane-associated mammalian P450s.19 Although nanodiscs have been used to overcome this difficulty and to study the interaction between FBD and P450 in a lipid bilayer environment, lack of resonance assignment has limited such studies.20 In this study, we report a systematic assignment of resonances by performing a series of high-resolution 3D NMR experiments on the full-length FBD reconstituted in lipid nanodiscs, which is essential to investigate the interaction of FBD with P450 and lipid membrane in the presence and absence of substrates to better understand the enzymatic function of P450 at an atomic level.
The 13C- and 15N-labelled full-length FBD was successfully expressed in Escherichia coli C41 bacterial strain, and the protein was purified by DEAE anion-exchange and size-exclusion chromatography (SEC) as depicted in Figure 1A. The protein was reconstituted in 4F-DMPC nanodiscs and further purified by SEC (Fig. 1B). The nanodiscs with full-length FBD eluted earlier in SEC as compared to the protein-free 4F-DMPC nanodiscs suggesting the larger size of the nanodiscs containing FBD (Fig. 1B). This was confirmed by dynamic light-scattering (DLS) that showed a 0.5 nm increase in the hydrodynamic radius for the nanodiscs containing the full-length FBD (Figures 1D and S1).
Figure 1.
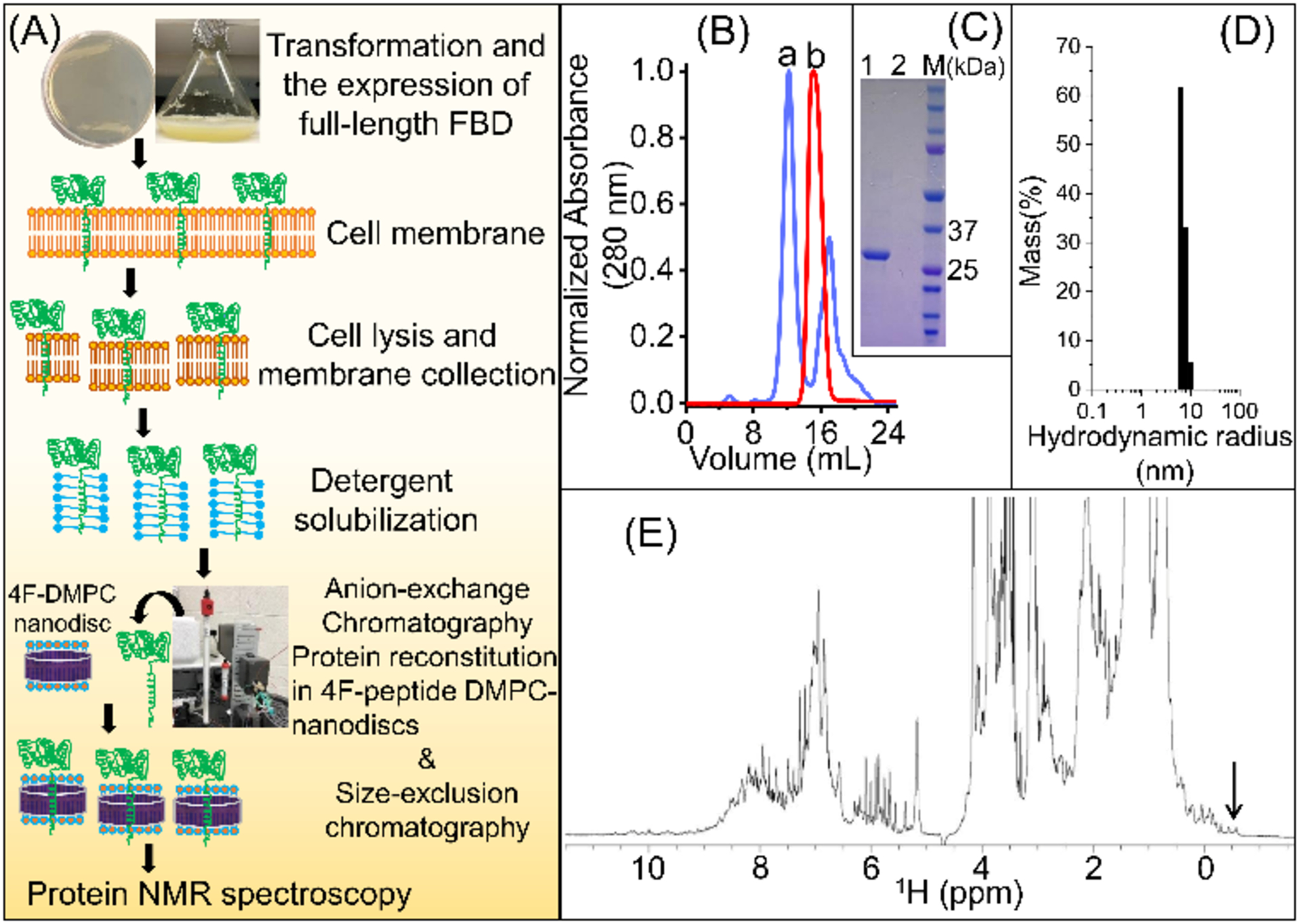
(A) Schematic of the expression and purification of full-length FBD. (B) A size-exclusion chromatogram designating the elution peaks from 4F-DMPC nanodiscs with (a) and without (b) the full-length FBD, respectively. (C) SDS-PAGE analysis of SEC-purified 4F-DMPC nanodiscs with (lane 1) and without (lane 2) full-length FBD, and M is the protein marker. (D) DLS size measurement of the SEC-purified full-length FBD reconstituted in 4F-DMPC nanodiscs. (E) 1H NMR spectrum of the full-length FBD reconstituted in 4F-DMPC nanodiscs (0.3 mM FBD in 40 mM potassium phosphate buffer (pH 7.4) containing 0.01% sodium azide) recorded at 25 °C on a Bruker 900 MHz NMR spectrometer. High-intensity peaks originated from DMPC lipids and 4F peptide. The methyl peaks between 0 to −0.6 ppm (indicated by an arrow) indicate a well-folded conformation for FBD in nanodiscs.
The suitability of sample conditions and stability for structural studies were examined using simple NMR experiments. A 1D 1H NMR spectrum showed reasonably well-dispersed peaks in the backbone amide region and the methyl peaks in the most upfield region between 0 to −0.6 ppm (Fig. 1E). High intensity peaks from the acyl chains of DMPC lipids and the 4F peptide in the nanodiscs also appeared in the 1H NMR spectrum (Fig. 1E). A 2D [15N-1H]-HSQC spectrum also showed well-dispersed backbone amide resonances in both 15N and 1H dimensions, indicating a well-folded conformation of the full-length FBD reconstituted in 4F-DMPC nanodiscs (Fig. 2).
Figure 2.
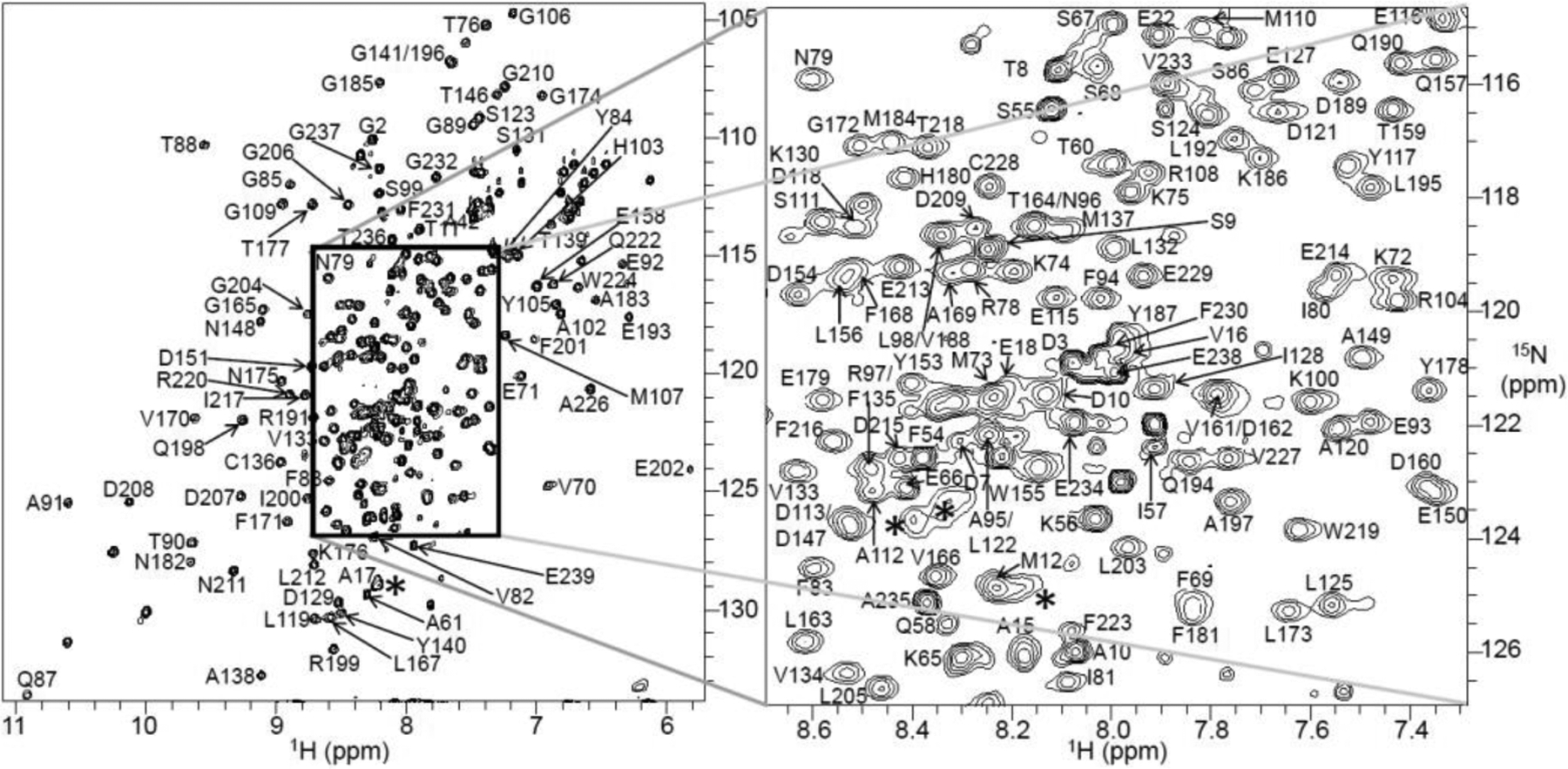
Two-dimensional [15N-1H]-HSQC spectrum of 0.3 mM full-length FBD reconstituted in 4F-DMPC nanodiscs recorded on a 600 MHz Bruker NMR spectrometer equipped with a cryogenically cooled triple-resonance probe operating at 25 °C. The spectrum was acquired with 16 scans and using 2048 and 256 data points for the 1H and 15N dimensions, respectively. The backbone NH assignments are annotated by the resonance peaks with one-letter amino acid codes followed by their position in the protein sequence. The region with resonance overlap is boxed and expanded on the right side. Line-broadening (*) and the appearance of low-intensity peaks from residues making close contacts with the lipid-bilayer and a minor conformation, respectively. The protein sample was prepared in 40 mM potassium phosphate buffer pH 7.4 containing 10 % 2H2O and 0.01% sodium azide.
The spectral quality and protein stability were further assessed using 2D [15N-1H]-HSQC and 2D [13C-1H]-HSQC NMR experiments before carrying out a series of 3D NMR experiments (Fig. S2). Except for a few peaks, uniform peak intensity was observed throughout the spectrum of the full-length FBD indicating the homogenous and monomeric form of the reconstituted protein (Fig. 2). The low-intensity and broad peaks could be arising from the residues in the N-terminal region and the regions associated with the lipid bilayer, respectively. New peaks were observed when comparing the spectra recorded on day-1 and day-6: in 7.6 to 8.6 ppm (1H) and 109 to 130 ppm (15N) regions in 2D [15N-1H]-HSQC (Fig. S2) and also in the different regions of 2D [13C-1H]-HSQC spectra (Fig. S2). The appearance of new peaks could be due to a partial denaturation of the protein with time. Despite these changes, the overall spectral dispersion was not affected, indicating good stability and suitability of the sample (i.e. full-length FBD in 4F-DMPC nanodiscs) for acquiring a series of 3D NMR spectra (Table S1).
Full-length FBD construct consisted of 239 amino acid residues, including 8 proline and 19 glycine residues, with a molecular weight of ~27-kDa. The sequence-specific resonance assignments were accomplished using a combination of conventional and TROSY-type triple-resonance HNCA, CBCA(CO)NH and HNCACB NMR spectra (Fig. S3). In these NMR experiments, ~95% of the expected cross-peaks were observed indicating the isotropic nature of the nanodiscs. A 2D [15N-1H]-HSQC spectrum with all the backbone amide assignments is shown in Figure 2. The Cα and Cβ chemical shifts of Cys residues are consistent with their reduced state (Table S2).21 An example of the assignment strip plots from 3D HNCACB and CBCA(CO)NH spectra are shown in Figure 3A and Figure S4. 98% amide (backbone, HN and NH) 100% Cα (backbone) and 99% Cβ (side-chain) assignments were determined for the 92% of the observed resonances (a total number of 202 residues including 18 Gly and 6 Pro residues) in the 2D [15N-1H]-HSQC spectrum. The assigned residues in the amino acid sequence are mapped onto the crystal structure of the soluble-domain of FBD lacking the transmembrane domain (Fig. 3B and 3C).13 The measured chemical shift values for the full-length FBD are listed in Table S2 and have been deposited in the BioMagResBank database (BMRB id: 50744). Except a few small differences, the secondary structures predicted from the NMR chemical shifts are in an excellent agreement with the crystal structure of the soluble FBD domain of CPR (Fig. S5).
Figure 3.
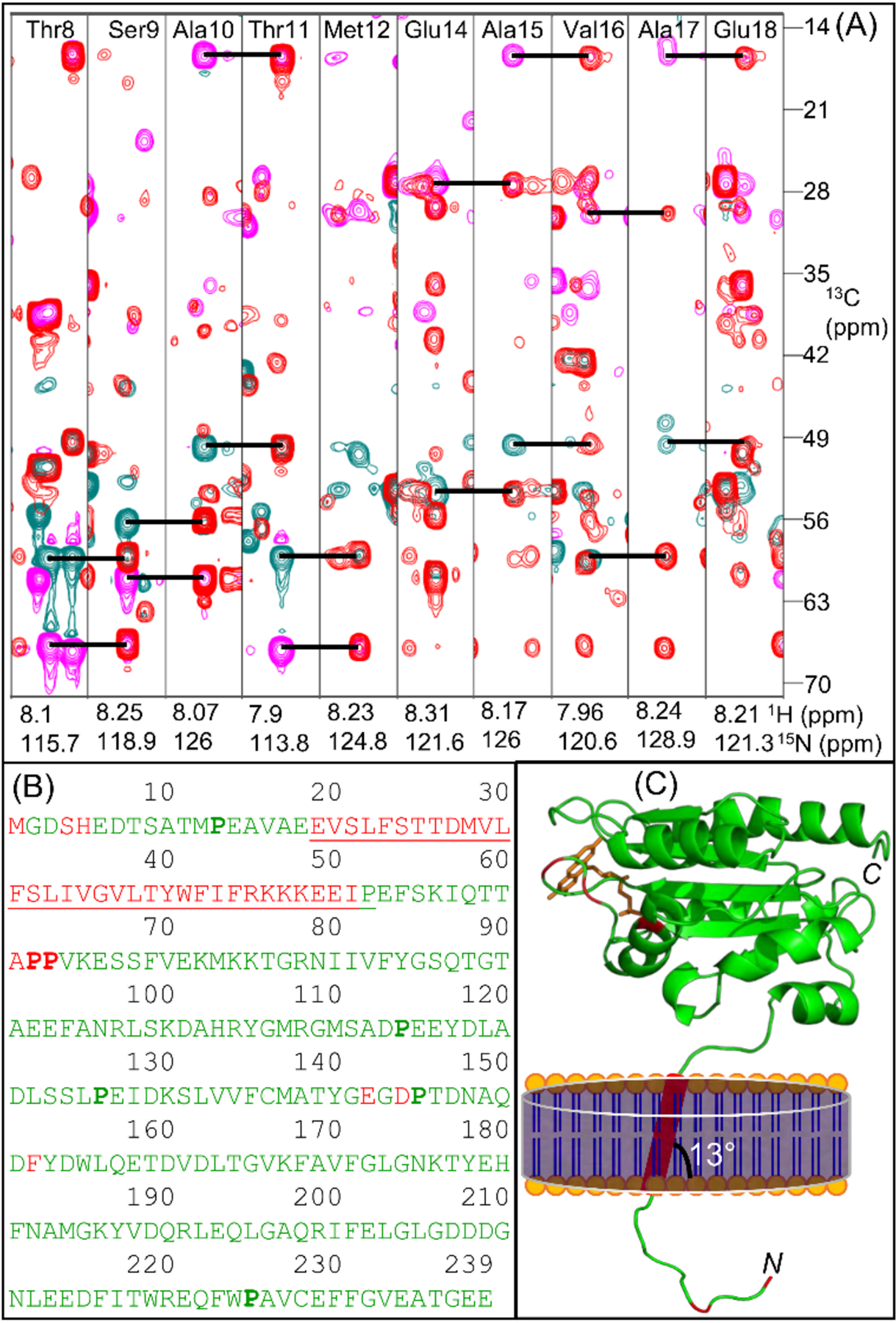
(A) Overlay of two-dimensional F1(13C)-F2(15N) strip plots obtained from three-dimensional HNCACB and CBCA(CO)NH spectra for the amino acid residues from Thr8 to Met12 and Glu14 to Glu18. Sequential intra-residue Cα (cyan) and Cβ (magenta) resonances in HNCACB are connected to corresponding inter-residue resonances in CBCA(CO)NH (red) using horizontal lines. NMR spectra were collected at 25 °C on a DMPC-nanodisc sample of 0.3 mM 13C-,15N-labelled full-length FBD in 40 mM potassium phosphate buffer (pH 7.4) containing 0.01% sodium azide. (B) Amino acid sequence of the full-length FBD highlighting the assigned residues colored in green, the unassigned residues colored in red and the transmembrane domain that is flanked by the short N-terminal region and the large C-terminal soluble domain is underlined. The proline residues are shown in bold. (C) Mapping the assigned residues on to the structure of FBD lacking the N-terminal region and the transmembrane domain (PDB id: 1AMO). The N-terminal region and the transmembrane domain are schematically shown. N- and C- indicate N- and C-termini, respectively. The prosthetic group ‘FMN’ is colored in orange.
The measured line-widths for the full-length FBD are slightly larger than those observed for the truncated-FBD (see Table S3 and Fig. S6), indicating a slower tumbling rate for full-length FBD reconstituted in nanodiscs. The 1H and 15N line-widths for the residues near the lipid bilayer of the nanodiscs were measured to be up to 41 and 34 Hz, respectively. These line-widths are nearly ~10 Hz larger than that observed for other residues in the soluble domain of the full-length FBD (Figures 4A and S6). The line-broadening observed for the residues Glu-14 to Glu-18 near the lipid bilayer surface (Table S3) is most likely due to their restricted motion and the enhanced spin-spin (T2) relaxation near the lipid bilayer. The poor chemical shift dispersion observed for the N-terminal residues 2–18 (<0.5 ppm) suggests that the N-terminal region is unstructured, which is correlated with the structure predicted based on chemical shifts (Fig. S5). The low-intensity peaks present in the 2D [15N-1H]-HSQC spectrum (Fig. S7) might be from a minor conformation of FBD as reported earlier.10 However, the number of low-intensity peaks observed for the full-length FBD is less than for the truncated-FBD spectrum (Fig. S7), indicating the improved homogeneity of the soluble domain conformation in nanodiscs. Due to peak overlap and low signal-to-noise ratio, the sequential NMR assignments for the low-intensity peaks could not be determined.
Figure 4.
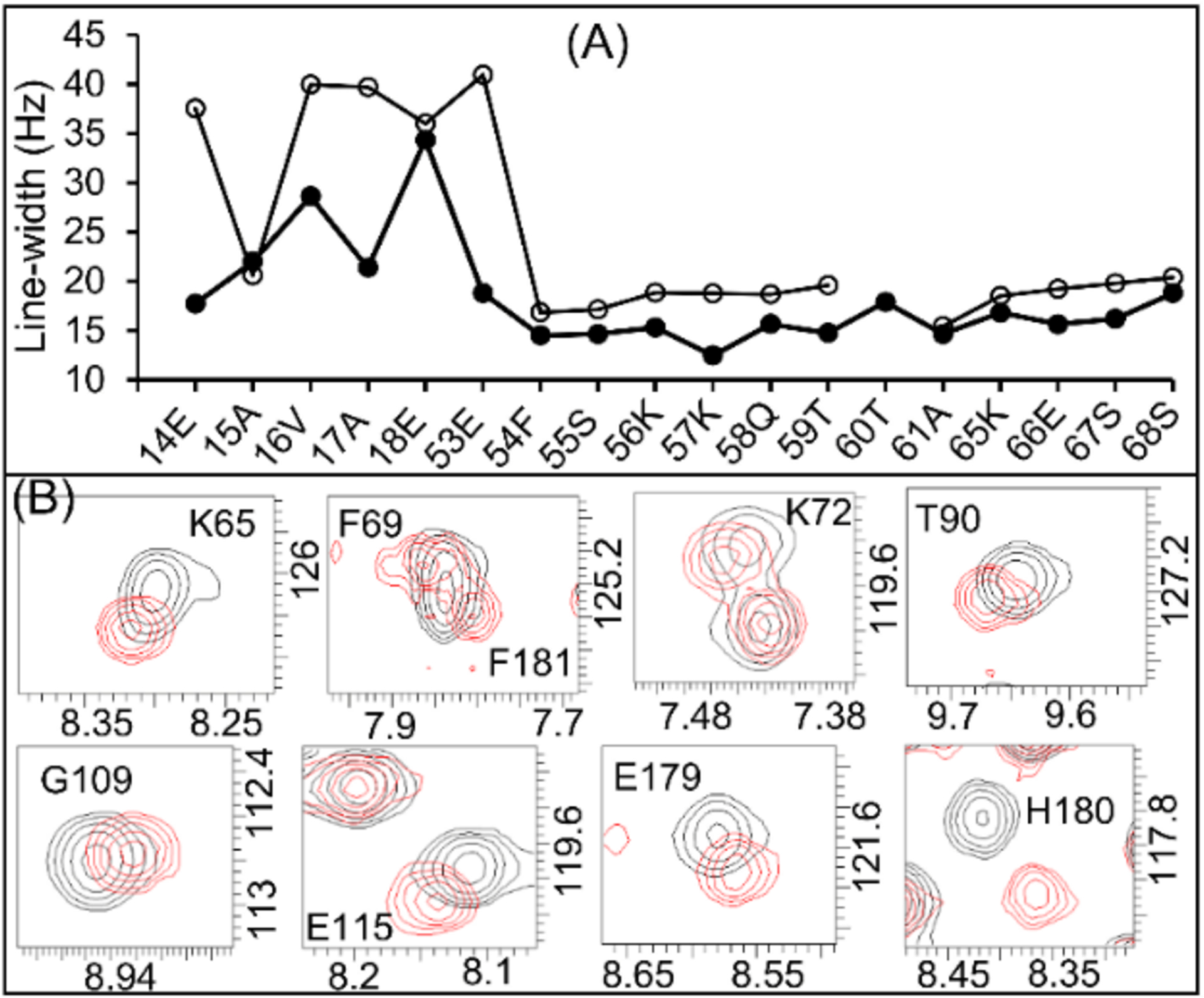
(A) 1H (open circles) and 15N (closed circles) line-widths of the amino acid residues near to the bilayered lipid-membrane (residues 14–18 and 53) compared to those in the soluble domain (residues 53–68) of the full-length FBD. (B) Expansion of 2D [15N-1H]-HSQC spectra of the full-length (black colored contours) and the truncated (red colored contours) FBD highlighting the perturbed resonances.
Despite few chemical shift perturbations (CSPs), the resonance assignments for the soluble domain of the full-length FBD are similar to those for the truncated-FBD (Fig. S7). The majority of the amino acid residues that showed CSPs (≥0.02 ppm) were found to be from the disordered N-terminus and those near the lipid bilayer and also from loop-3 (Figures S7 and S8). The observed perturbation to the resonances from loop-3 residues is likely due to the difference in the orientation of the FMN prosthetic group, which is non-covalently linked to the protein, when the protein is reconstituted in nanodiscs. Otherwise, the similar chemical shifts of full-length FBD and truncated-FBD indicate no structural changes due to reconstitution in lipid-nanodiscs. However, the CSPs suggest that the soluble domain of the full-length FBD would be making a few transient interactions with the lipid bilayer, which agrees with the previous solid-state NMR study.7 While the nanodiscs employed in this study used zwitterionic DMPC lipids, it would be useful to reconstitute the full-length FBD in nanodiscs containing the ER membrane lipids to obtain atomic-level insights on the interaction of the full-length FBD (or CPR) with the native lipid membrane in the absence and the presence of cytochrome-P450 and cytochrome-b5.17 Additionally, determining the ratio of different ER lipids specific to the cytochrome P450-FBD-b5 ternary complex using 31P NMR would be useful.17, 22 Since the NMR resonance assignments are now accomplished through this study, probing the roles of different lipid composition on the folding and mobility of the full-length-FBD and its interaction with P450 in the presence and absence of substrates can be measured by NMR.
Most of the structural and functional studies reported so far has been carried out with truncated-FBD that does not have a lipid membrane environment. However, the native protein has a transmembrane domain through which the large soluble-domain of CPR is anchored to the microsomal membrane (see the schematic in Fig. 3). Investigation of the interaction of the transmembrane domains of CPR and P450 in a lipid membrane is in progress using nanodiscs and solid-state NMR experiments. Our previous studies have reported the direct interaction of the transmembrane domain of cytochrome-P450 with its another redox partner, cytochrome-b5, using solid-state NMR.23
High-resolution NMR studies of membrane proteins require a stable, homogenous reconstitution in the near-native lipid membrane bilayer. Although the membrane-mimicking systems such as bicelles and liposomes have been extensively used for membrane protein studies, they all have some intrinsic limitations.24 Bicelles contains detergent molecules, which are detrimental to protein’s stability, and they are also sensitive to temperature and buffer conditions. On the other hand, liposomes are less stable and non-homogenous that would result in increased line-widths and thus poor quality NMR spectra. Hence, performing high-resolution NMR experiments that require long acquisition times using these reconstitution systems is not feasible. In contrast, as shown in this study, the reconstitution of the full-length FBD in 4F-DMPC nanodiscs provided a stable, homogenous conformation that rendered high-quality, high-resolution NMR data. Unlike the previously reported results for the soluble fragment of FBD in the absence of lipids, our results clearly demonstrate the feasibility of peptide-based lipid-nanodiscs to stabilize and study the native-like structural folding and dynamics of FBD. The NMR assignments for the full-length FBD reported here will be useful to study the structural interaction of FBD with cytochrome-P450 and the lipid membrane to better understand the structure, dynamics and enzymatic function of mammalian cytochrome-P450s by NMR, and will also be useful to probe the substrate-dependent competitive interaction of cytochrome-P450 with its redox partners cytochrome-b5 and CPR. Since nanodiscs are also used to investigate protein-ligand interactions25 and high-throughput screening in drug discovery26 in the presence of desired lipid composition, we expect this study to be useful to investigate the drug metabolism by cytochrome-P450.17, 27
Supplementary Material
Acknowledgements
This study was supported by NIH (R35GM139573 to A.R.). We thank Dr.Thirupathi Ravula and Dr.Mukesh Mahajan for help with sample preparations and for MM’s interest in the NMR data processing.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Guengerich FP, J. Biol. Chem, 2019, 294, 1671–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rendic S and Guengerich FP, Curr. Drug Metab, 2020, 21, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nath A, Grinkova YV, Sligar SG and Atkins WM, J. Biol. Chem, 2007, 282, 28309–28320. [DOI] [PubMed] [Google Scholar]
- 4.Xia C, Shen AL, Duangkaew P, Kotewong R, Rongnoparut P, Feix J and Kim J-JP, Biochemistry, 2019, 58, 2408–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backes WL and Kelley RW, Pharmacol. Ther 2003, 98, 221–233. [DOI] [PubMed] [Google Scholar]
- 6.Freeman SL, Martel A, Devos JM, Basran J, Raven EL and Roberts GCK, J. Biol. Chem, 2018, 293, 5210–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang R, Yamamoto K, Zhang M, Popovych N, Hung I, Im S-C, Gan Z, Waskell L and Ramamoorthy A, Biophys. J, 2014, 106, 2126–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermilion JL, Ballou DP, Massey V and Coon MJ, J. Biol. Chem, 1981, 256, 266–277. [PubMed] [Google Scholar]
- 9.Prade E, Mahajan M, Im S-C, Zhang M, Gentry KA, Anantharamaiah GM, Waskell L and Ramamoorthy A, Angew. Chem. Int. Ed, 2018, 57, 8458–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barsukov I, Modi S, Lian LY, Sze KH, Paine MJ, Wolf CR and Roberts GC, J. Biomol. NMR, 1997, 10, 63–75. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Q, Modi S, Smith G, Paine M, McDonagh PD, Wolf CR, Tew D, Lian LY, Roberts GC and Driessen HP, Protein Sci, 1999, 8, 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrada DF, Laurence JS and Scott EE, J. Biol. Chem, 2016, 291, 3990–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, Roberts DL, Paschke R, Shea TM, Masters BSS and Kim J-JP, Proc. Natl. Acad. Sci. USA, 1997, 94, 8411–8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black SD, French JS, Williams CH and Coon MJ, Biochem. Biophys. Res. Commun, 1979, 91, 1528–1535. [DOI] [PubMed] [Google Scholar]
- 15.Gideon DA, Kumari R, Lynn AM and Manoj KM, Cell Biochem. Biophys, 2012, 63, 35–45. [DOI] [PubMed] [Google Scholar]
- 16.Barnaba C and Ramamoorthy A, ChemPhysChem, 2018, 19, 2603–2613. [DOI] [PubMed] [Google Scholar]
- 17.Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im S-C, Anantharamaiah GM, Waskell L and Ramamoorthy A, Angew. Chem. Int. Ed, 2018, 57, 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravula T, Hardin NZ, Bai J, Im SC, Waskell L and Ramamoorthy A, ChemComm, 2018, 54, 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luthra A, Gregory M, Grinkova YV, Denisov IG and Sligar SG, Methods Mol. Biol, 2013, 987, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahajan M, Ravula T, Prade E, Anantharamaiah GM and Ramamoorthy A, ChemComm, 2019, 55, 5777–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma D and Rajarathnam K, J. Biomol. NMR, 2000, 18, 165–171. [DOI] [PubMed] [Google Scholar]
- 22.Krishnarjuna B, Ravula T and Ramamoorthy A, ChemComm, 2020, 56, 6511–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto K, Dürr UH, Xu J, Im SC, Waskell L and Ramamoorthy A, Sci. Rep, 2013, 3, 2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catoire LJ, Warnet XL and Warschawski DE, Membrane Proteins Production for Structural Analysis, 2014, 315–345. [Google Scholar]
- 25.Treuheit NA, Redhair M, Kwon H, McClary WD, Guttman M, Sumida JP and Atkins WM, Biochemistry, 2016, 55, 1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sligar SG and Denisov IG, Protein Sci, 2021, 30, 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denisov IG and Sligar SG, Nat. Struct. Mol. Biol, 2016, 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


