Abstract
A Gram-negative, strictly anaerobic mucin-degrading bacterium, which we designated strain E39T, was isolated from the rumen epithelium of Korean cattle. The cells were non-motile and had a coccus morphology. Growth of strain E39T was observed at 30–45°C (optimum, 39°C), pH 6.5–8.5 (optimum, pH 7.5), and in the presence of 0.0–1.0% (w/v) NaCl (optimum, 0.0–0.5%). Strain E39T contained C16:0, C18:0, C18:1 ω9c, iso-C15:0, and anteiso-C15:0 as the major fatty acids. The major polar lipids were phosphatidylethanolamine, unidentified aminophospholipid, and unidentified lipids. The major respiratory isoprenoid quinones were MK-8 and MK-9. The major fermented end-products of mucin were acetate and succinate. The G+C content of the genomic DNA was 46.4 mol%. Strain E39T was most closely related to Alloprevotella rava 81/4-12T with an 87.3% 16S rRNA gene sequence similarity. On the basis of phenotypic, chemotaxonomic, and molecular properties, strain E39T represents a novel genus of the family Prevotellaceae; as such, the name Pseudoprevotella muciniphila gen. nov., sp. nov. is proposed. A functional annotation of the whole genome sequences of P. muciniphila E39T revealed that this bacterium has a putative mucin-degrading pathway and biosynthetic pathways of extracellular polymeric substances and virulence factors which enable bacteria to adhere to the epithelial cells and avoid the host’s immune responses.
Introduction
Rumen, a forestomach of ruminants, is a complex symbiotic ecosystem. In the rumen, various types of microorganisms (archaea, bacteria, protozoa, fungi, and virus) exist and they are known as the rumen microbiota. The rumen microbiota provides nutrients for the host ruminants by digestion of feed particles, especially indigestive fibrous material, and biosynthesis of metabolites like amino acids, lipids, and vitamins [1]. The nutrients including volatile fatty acids, minerals, and other metabolites are absorbed and transported into the host’s blood vessels through the rumen epithelium [2]. Only small portion of rumen microbiota attaches to the rumen epithelium. Several rumen epithelial bacteria (epimural bacteria) and their functions in urea digestion, tissue recycling, and oxygen scavenging have been identified since 1960s [3, 4]. Recent studies showed the composition and expressed genes of the epimural community in response to subacute rumen acidosis challenges using meta-omics technologies [5, 6]. Although these culture-independent methods provide expansive information about the epimural community, culture-dependent studies about yet uncultured eprimural bacteria are essential to understand their functions in more depth.
Mucin is a host-derived glycoprotein composed of protein backbones and various oligosaccharides. Intestinal epithelial cells produce two types of mucins (secreted mucin and cell membrane-associated mucin). They have a role as barriers to prevent pathogens from penetrating the epithelial tissues [7]. Mucin-degrading bacteria can degrade mucin and utilize it as an energy source. Some of them penetrate into the host epithelial cells with their mucin-degrading ability and show pathogenicity, others play roles as commensal bacteria [8]. Studies on Akkermansia muciniphila, a commensal mucin-degrading bacterium, revealed that this bacterium improved the host metabolic disorders, such as obesity and insulin resistance, by controlling gut barrier permeability and inflammation [9, 10].
In the bovine rumen, there are salivary mucin secreted from salivary glands and cell-membrane associated mucins expressed on rumen epithelial cells [11]. In the 1960s, Fina et al. isolated five uncultured mucin-degrading rumen bacteria from rumen fluid and Mishra et al. determined mucin-degrading activity of several species of rumen bacteria (Butyrivibrio fibrisolvens, Selenomonas ruminantium, Streptococcus bovis, Peptostreptococcus elsdenii, and Bacteroides ruminicola) [12–14]. They focused on mucin-degrading bacteria floating in the rumen fluid and their roles in bloat syndrome. Subsequently, there has been few studies on mucin-degrading bacteria floating in the rumen fluid nor them attached to the rumen epithelium.
In this study, we used mucin as a novel carbon source to enrich previously uncultured mucin-degrading bacteria from the rumen epithelium of Korean cattle. We isolated a novel mucin-degrading bacterium, called strain E39T. Then, we performed phylogenetic analysis, phenotypic and chemotaxonomic characterization, and genome analysis. On the basis of phenotypic, chemotaxonomic, and molecular properties, strain E39T represents a novel genus of the family Prevotellaceae, for which we propose the designation Pseudoprevotella muciniphila gen. nov., sp. nov., strain E39T.
Materials and methods
Ethical statement
All experimental procedures were performed in accordance with the Animal Experimental Guidelines provided by the Seoul National University Institutional Animal Use and Care Committee, Republic of Korea. The experimental protocol was approved by the Seoul National University Institutional Animal Use and Care Committee (SNU-170626-1).
Enrichment and isolation of mucin-degrading bacteria
Strain E39T was isolated from the rumen epithelium of Korean cattle. Rumen epithelium tissue samples were excised from the ventral sacs of rumens immediately after slaughter at the abattoir in Bucheon (37°31’48.4"N 126°45’46.3"E), South Korea in 2017 and transported in a sterile container with rumen fluid. On arrival at the laboratory, the samples were moved into an anaerobic chamber and washed several times with anaerobic dilution solution (ADS; 3 g K2HPO4, 6 g NaCl, 3 g KH2PO4, 0.6 g CaCl2·2H2O, 0.6 g MgSO4·7H2O, 6 g (NH4)2SO4, 0.5 g cysteine-HCl, 0.5 g Na2S·9H2O, 0.625 g NaOH, and 1 mg resazurin per liter) to remove the rumen contents and non-adherent bacteria [15, 16]. Five grams of epithelial samples that were stripped from the muscle layer were homogenized in 30 ml of ADS and serially diluted (10-fold). Each 0.3 ml of dilution was inoculated into 30 ml of basal mucin medium in a butyl rubber stopped serum bottle and incubated at 39°C in an anaerobic atmosphere (95% CO2 5% H2) for 24 h for enrichment. The basal mucin medium was prepared by modifying medium 10 [17] and consisted of 2 g peptone, 0.5 g yeast extract, 2.5 g hog gastric mucin (Type Ⅲ; Sigma), 50 ml mineral solution 1 (6 g K2HPO4 per liter), 50 ml mineral solution 2 (12 g NaCl, 6 g KH2PO4, 1.2 g CaCl2·2H2O, 1.2 g MgSO4·7H2O, and 12 g (NH4)2SO4 per liter), 10 ml Pfenning’s solution (0.5 g EDTA, 0.1 g ZnSO4·7H2O, 0.03 g MnCl2·4H2O, 0.03 g H3BO3, 0.2 g CoCl2·6H2O, 0.01 g CuCl2·2H2O, 1.5 g FeCl2·4H2O, 0.02 g NiCl2·6H2O, 0.03 g Na2MoO4·2H2O, and 0.01 g Na2SeO3 per liter), 10 ml volatile fatty acid solution (700 ml 0.2 N NaOH, 17 ml acetic acid, 6 ml propionic acid, 4 ml butyric acid, 1 ml iso-butyric acid, 1 ml 2-metylbutyric acid, 1 ml valeric acid, and 1 ml isovaleric acid, pH 7.5 per liter), 1 ml hemin solution (0.5 g hemin and 10 ml 1N NaOH per liter), 1 ml resazurin (0.1%, w/v), 20 ml cysteine sulfide solution (6.25 g NaOH, 25 g cysteine-HCl, and 25 g Na2S·9H2O per liter), and 4 g NaHCO3 per liter. The enrichments were serially diluted with ADS, streaked onto basal mucin agar medium, and incubated at 39°C for 4 days under anaerobic conditions. Each colony was picked, inoculated into 5 ml of basal mucin medium in a Hungate tube, and incubated at 39°C for 24–48 h. Streaking, colony picking, and incubation were repeated until an isolate was pure. The isolates were preserved with 15% (v/v) glycerol stock solution and stored at –80°C.
Bacterial growth and genomic DNA extraction
Cells of strain E39T were grown in 250 ml basal mucin medium at 39°C for 24 h and harvested by centrifugation (10,000 rpm for 5 min). The genomic DNA of the cells was extracted, according to standard procedures, including phenol-chloroform extraction and ethanol precipitation [18].
16S rRNA gene based phylogeny
The 16S rRNA gene of strain E39T was amplified with PCR using the universal primers 10F (5′-AGT TTG ATC ATG GCT CAG ATT G-3′) and 1507R (5′-TAC CTT GTT ACG ACT TCA CCC CAG-3′) [19] and the resulting amplicons were sequenced using the universal primers 340F (5′-CCT ACG GGA GGC AGC AG-3′), 518R (5′-ATT ACC GCG GCT GCT GG-3′), and 805F (5′-GAT TAG ATA CCC TGG TAG TC-3′). The sequencing quality was checked, and the sequences were assembled using the Geneious program (ver. 11.0.4). The almost complete 16S rRNA gene sequence (1,476 nucleotides) of strain E39T was compared with that of all validated type strains using the Nucleotide Similarity Search program in the EzBioCloud server (http://www.ezbiocloud.net/identify/) [20]. Phylogenetic trees were constructed with MEGA (ver. 7.0.26) using the neighbor-joining (NJ), maximum-parsimony (MP), and maximum-likelihood (ML) methods [21]. The complete 16S rRNA gene sequence (1,540 nucleotides) from the genome sequence of strain E39T was used to construct the phylogenetic trees.
Phenotypic and chemotaxonomic characterization
The cell morphology of cells grown on basal mucin agar medium at 39°C for 3 days was investigated using phase-contrast microscopy and transmission electron microscopy (Talos L120C; FEI) at 120 kV. Gram staining was performed using a Sigma Gram staining kit following the manufacturer’s protocol. The growth of strain E39T, as determined from the optical density (OD) at a wavelength of 600 nm, was evaluated by culturing the cells in basal mucin medium, brain heart infusion (BHI) broth (BD), trypticase soy broth (TSB; BD), Columbia broth (Acumedia), and anaerobe basal broth (Oxoid) at 39°C for 48 h. The optimum temperature, pH, and NaCl concentration for growth were determined by culturing the cells on basal mucin medium for 48 h at different temperatures (5–45°C, at 5°C intervals), pH (5.0–9.0 at 0.5 pH unit intervals), and NaCl concentrations (0.0–2.0% at 0.5% intervals). To determine the optimum pH, different pH buffers were used in the appropriate pH range (Na2HPO4-NaH2PO4 buffer at pH 5.0–7.5; Tris-HCl buffer at pH 8.0–9.0) and the pH values were adjusted before and after autoclaving (121°C, 15 min) [22]. Oxygen tolerance was investigated by measuring growth (OD at 600 nm) in the absence of a reducing agent (cysteine sulfide solution) or in the aerobic condition on basal mucin medium. A. tannerae ATCC 51259T, A. rava 81/4-12T, Paraprevotella clara YIT 11840T and Prevotella melaninogenica ATCC 25845T, which is the type species of the genus Prevotella, were used as reference strains to compare enzyme profiles and cellular fatty acid composition. The enzyme profiles were determined using an API Rapid ID 32A identification kit (bioMérieux) following the manufacturer’s instructions. Analysis of cellular fatty acids was performed according to a standard MIDI protocol. All of the strains were cultivated in peptone-yeast extract-glucose (PYG) broth, except strain E39T, which was cultivated in basal mucin medium. Cells were harvested at the late exponential phase and cellular fatty acids were extracted from the cells following four steps (saponification, methylation, extraction, and base wash). Fatty acid methyl esters were analyzed by gas chromatography (Hewlett Packard 6890) and identified using the RTSBA6 database of the Microbial Identification System (Sherlock ver. 6.0B) [23]. The polar lipid profiles were analyzed by thin-layer chromatography following the Minnikin et al. method [24]. The following reagents were used to detect different types of polar lipids: 10% ethanolic molybdophosphoric acid (for total lipids), ninhydrin (for aminolipids), Dittmer-Lester reagent (for phospholipids), and α-naphthol (for glycolipids). The isoprenoid quinones of strain E39T, A. tannerae ATCC 51259T, A. rava 81/4-12T, P. clara YIT 11840T, and P. melaninogenica ATCC 25845T were extracted from their exponentially grown cells according to the procedure described by Jeon et al. [25] and analyzed at 40°C using an Agilent infinity 1290 UHPLC equipped with a photodiode array detector (PAD) and an Agilent 6550 ifunnel Q-TOF MS (Agilent Technologies, USA). Briefly, 2 μl of quinone samples were injected into an Agilent Eclipse Plus C-18 column (2.1 mm × 100 mm, 2.1 μm) and eluted at 40°C using water (A) and acetonitrile (B) containing 0.1% formic acid as a mobile phases with the following gradient: 0 min, 85% B; 30 min, 100% B; 40 min; and flow rate, 0.4 ml/min. Isoprenoid quinone peaks in the chromatograms were identified by their UV spectra generated by PAD and their molecular masses were assessed using Q-TOF MS. The mass spectrometry was performed under the following conditions: polarity, positive; gas temp, 250°C; nebulizer, 35 psi; capillary, (+) 4,000 V; MS range, 100–1,500 m/z.
Metabolite analysis using 1H NMR spectroscopy
Metabolic compounds including amino acids, monosaccharides, and organic acids in cultured broth of strain E39T were analyzed using 1H NMR spectroscopy, as described previously [26]. Briefly, the basal mucin medium (2.5 g hog gastric mucin per liter; no glucose), glucose medium (5 g glucose per liter; no hog gastric mucin), and mucin-glucose medium (2.5 g hog gastric mucin and 5 g glucose per liter) were prepared based on the basal mucin medium to investigate mucin and glucose utilization by strain E39T and their fermentation products. Strain E39T was cultured in 5 ml of each broth at 39°C for 0, 9, 18, 27, 36, and 54 h. The growth of the cells was monitored by measuring OD at 600 nm. The culture broths were centrifuged, filtered with a 0.45 μm syringe filter, and 0.3 ml of filtrate was mixed with 0.3 ml of 99.9% D2O (Sigma-Aldrich, USA) containing 5 mM sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS, 97%; Sigma-Aldrich). The mixtures were transferred into NMR tubes and their 1H NMR spectra were measured on Varian Inova 600-MHz NMR spectrometer (Varian, USA). Metabolic compounds were identified and quantified using the Chenomx NMR Suite program (ver. 6.1; Chenomx, Canada).
Genome sequencing and analysis
De novo genome sequencing was performed using a Pacific Biosciences (PacBio) RSII platform at Macrogen (Seoul, Korea; http://www.macrogen.com). A library was prepared using PacBio DNA Template Prep Kit 1.0. After sequencing, reads were trimmed to obtain high quality region and then assembled using RS hierarchical genome assembly process (HGAP ver. 3.0) [27]. The complete genome was annotated using a software tool Prokka with default parameter (ver. 1.12) [28].
For the bacterial core genes-based phylogenetic analysis, 92 up-to-date bacterial core genes were extracted from the genomes of strains in the family Prevotellaceae and multiple-aligned, and a phylogenetic tree was constructed using the up-to-date bacterial core gene (UBCG) tool ver. 3 (https://www.ezbiocloud.net/tools/ubcg) [29]. The average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values among the genomes of strain E39T and reference strains were calculated using a stand-alone software (http://www.ezbiocloud.net/sw/oat) [30] and the Genome-to-Genome Distance Calculator (GGDC) ver. 2.1 (http://ggdc.dsmz.de/distcalc2.php) [31], respectively. Functional annotation of predicted proteins was performed using BlastKOALA tool of Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.kegg.jp/blastkoala/) [32].
To predict putative mucin-degrading enzymes and carbohydrate active enzymes (CAZymes), the genome sequences of strain E39T and reference strains were submitted to a meta server for automated carbohydrate-active enzyme annotation (dbCAN2) (http://cys.bios.niu.edu/dbCAN2/) [33]. VRprofile (http://bioinfo-mml.sjtu.edu.cn/VRprofile) was used for the prediction of virulence and antibiotic resistant genes [34].
Nucleotide sequence accession number
The 16S rRNA gene and genome sequence of strain E39T were deposited in GenBank under MG763147 and CP033459, respectively. The genome data of reference strains: A. tannerae ATCC 51259T (GenBank acc. no., ACIJ00000000), A. rava 81/4-12T (GenBank acc. no., ACZK00000000), P. clara YIT 11840T (GenBank acc. no., AFFY00000000), P. melaninogenica ATCC 25845T (GenBank acc. no., CP002122-3), and Bacteroides thetaiotaomicron VPI 5482T (GenBank acc. No., AE015928) were obtained from GenBank for comparative analysis.
Results and discussion
16S rRNA gene and genome based phylogeny
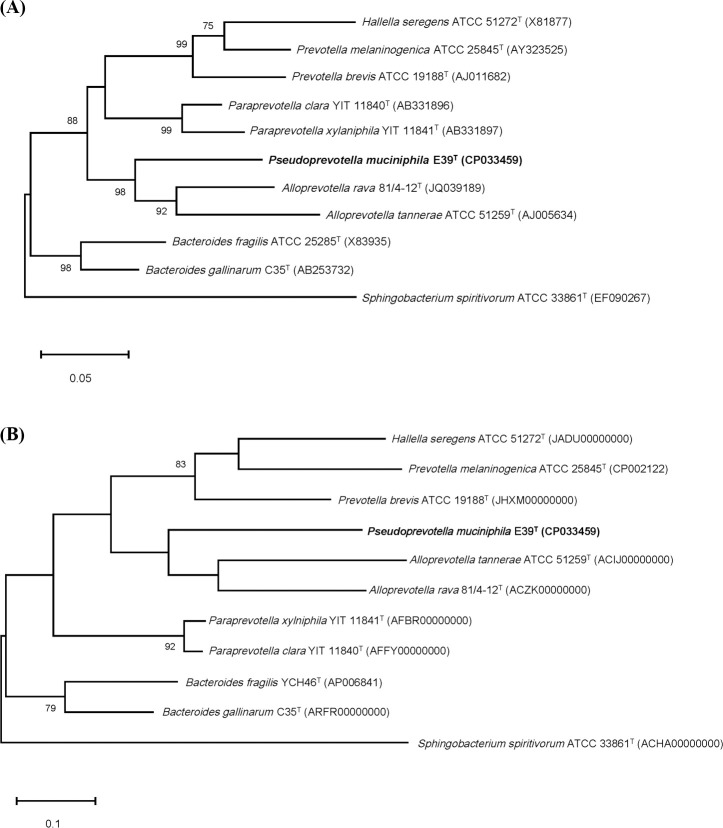
Comparative analysis of the 16S rRNA gene sequences revealed that strain E39T was closely related to the genera Alloprevotella, Paraprevotella, Prevotella, and Bacteroides. Alloprevotella rava 81/4-12T, Paraprevotella clara YIT 11840T, Paraprevotella xylaniphila YIT 11841T, and Bacteroides gallinarum JCM 13658T were most closely related to strain E39T with 87.3%, 86.6%, 86.3%, and 85.9% 16S rRNA gene sequence similarities, respectively. The phylogenetic trees based on the ML algorithm and 92 bacterial core genes showed that strain E39T was affiliated with the family Prevotellaceae and close to the genera Alloprevotella and Paraprevotella (Fig 1) [35–37]. Similarly, the phylogenetic trees based on the NJ and MP algorithms also showed that strain E39T was affiliated with the family Prevotellaceae (S1 and S2 Figs). However, all of the phylogenetic trees showed that strain E39T formed a distinct phylogenic lineage from the other genera. These molecular and phylogenetic analyses suggest that strain E39T represents a novel genus of the family Prevotellaceae.
Fig 1.
Phylogenetic relationship between strain E39T and closely related strains within the order Bacteroidales, based on 16S rRNA gene sequences (A) and 92 bacterial core genes of the genomes (B). (A) The 16S rRNA-based tree was constructed with the maximum-likelihood method. Bootstrap values are shown on nodes in percentages and are based on 2,000 replicates. Only the bootstrap values over 70% are shown. The scale bar equals 0.05 changes per nucleotide position. (B) Bacterial core genes-based tree was constructed with the UBCG (up-to-date bacterial core gene) tool. The scale bar equals 0.1 changes per nucleotide position. The 16S rRNA gene (GenBank accession no., EF090267) and genome (ACHA00000000) sequences of Sphingobacterium spiritivorum ATCC 33861T were used as an outgroup in the 16S rRNA gene- and bacterial core genes-based trees, respectively.
For additional conformation, ANI and dDDH were done with the genomes of strain E39T and 4 reference strains. As a result, strain E39T had lower ANI and dDDH values with A. tannerae (67.4%; 12.7%), A. rava (68.3%; 12.8%), P. clara (67.4%; 12.6%), and P. melaninogenica (66.7%; 12.7%) than single species thresholds (95–96% and 70%, respectively) [38].
Phenotypic and chemotaxonomic characterization
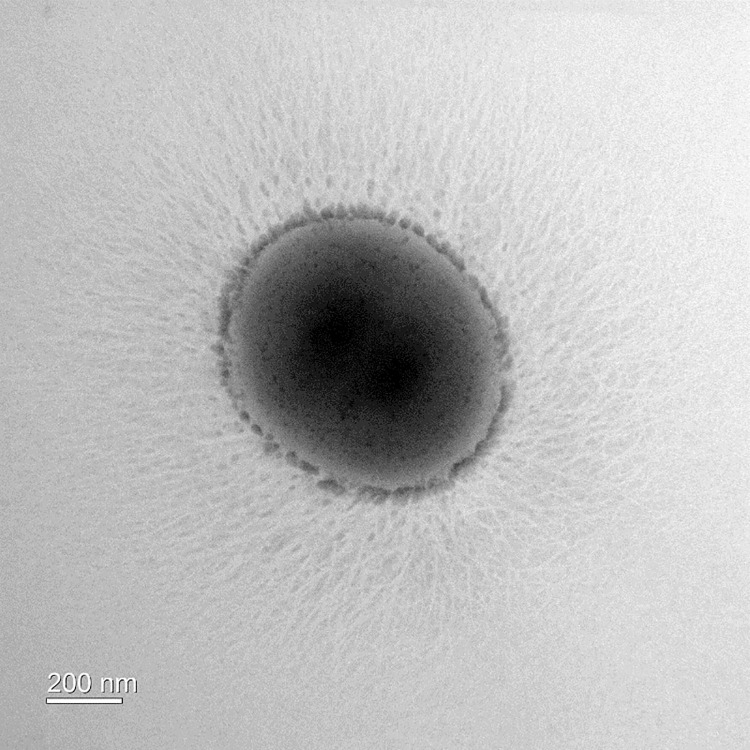
The transmission electron microscopic analyses showed that the cells of strain E39T were coccus in morphology (680–820 nm in diameter), and lacking in flagella (Fig 2). In addition, filamentous structures were observed from the cell surface. Among the various types of media, including BHI broth, TSB, Columbia broth, and anaerobe basal broth, strain E39T could only grow on basal mucin medium. Cells grew at temperatures between 30 and 45°C, pH between 6.5 and 8.5, and in NaCl concentration between 0.0 and 1.0%. When the headspaces were filled with anaerobic gas, growth was observed in the absence of a reducing agent, but at a lower rate than in its presence. Cells did not grow in an aerobic atmosphere regardless of whether a reducing agent was present or not. These results demonstrate that strain E39T prefers obligate anaerobic conditions but tolerates a trace amount of oxygen (S3 Fig).
Fig 2. A transmission electron micrograph showing the general morphology of negatively stained cells of strain E39T.
Cells were grown on basal mucin agar medium at 39°C for 3 days. Bar, 200 nm. Filamentous structures were observed from the cell surface.
In the API Rapid ID 32A panel, strain E39T had positive activities of mucin-degrading enzymes including β-galactosidase, N-acetyl-β-glucosaminidase, and α-fucosidase [8]. Lack of urease activity suggest that strain E39T may not attend to the digestion of urea, which is one of the roles of epimural bacteria. In particular, strain E39T was distinguished from the reference taxa by a positive activity of arginine dihydrolase (Table 1). The major cellular fatty acids (> 5% of the total fatty acids) of strain E39T were C16:0, C18:0, C18:1 ω9c, iso-C15:0, and anteiso-C15:0. Among strain E39T and the reference taxa of the family Prevotellaceae, iso-C15:0 and anteiso-C15:0 were commonly detected as major cellular fatty acids. Especially, strain E39T had higher portion of C16:0 and C18:0 compared with the reference taxa (Table 2). The major polar lipids of strain E39T were phosphatidylethanolamine (PE), unidentified aminophospholipid (APL), and three unidentified lipids (L) (S4 Fig). Strain E39T and the reference taxa of the family Prevotellaceae commonly contained PE and APL (except P. melaninogenica) (Table 1). Menaquinone (MK)-8 and MK-9 were identified from strain E39T as major respiratory quinones. However, MK-9 was identified from A. tanneare and A. rava, the most closely related strains, as the sole respiratory quinones, while MK-10 and 11 and MK-7 and 11 were identified from P. clara and P. melaninogenica, respectively (Table 1). It has been reported that members of the family Prevotellaceae contain MK of between 10 to 13 as isoprenoid numbers [37]. However, MK-9 was identified from strain E39T and Alloprevotella members and MK-8 was identified from only strain E39T (Table 1), which differentiated strain E39T from other genera of the family Prevotellaceae.
Table 1. Comparison of phenotype characteristics of strain E39T and closely related taxa.
| Characteristic | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Morphology | Coccus | Rod | Rod | Rod | Rod |
| Optimal temperature (°C) | 39 | ND | 35 | 37 | 37 |
| Optimal pH | 7.5 | ND | 7.0 | ND | ND |
| Enzyme activity (API Rapid ID 32a) * of | |||||
| Arginine arylamidase | – | – | – | – | + |
| β-glucuronidase | – | – | – | w | – |
| α-arabinosidase | – | – | – | + | – |
| α-glucosidase | – | + | – | – | + |
| Mannose fermentation | – | – | + | + | + |
| Raffinose fermentation | – | – | + | + | + |
| β-galactosidase 6 phosphate | w | – | w | – | + |
| Arginine dihydrolase | + | – | – | – | – |
| α-galactosidase | + | – | w | – | – |
| N-acetyl-β-glucosaminidase | + | – | + | – | + |
| α-fucosidases | + | + | – | – | + |
| Leucyl glycine arylamidase | + | + | – | – | + |
| Glutamyl glutamic acid arylamidase | + | + | – | – | + |
| Glutamic acid decarboxylase | + | + | – | w | w |
| Polar lipids†* | PE, APL, L | PE, APL | PE, APL, PL, L | PE, APL, L | PE, L |
| Major quinones* | MK-8, MK- 9 | MK-9 | MK-9 | MK-10, MK-11 | MK-7, MK-11 |
| DNA G+C content (mol%)‡ | 46.4 | 46.6 | 45.5 | 48.2 | 40.9 |
*Data were obtained from this study.
†PE, phosphatidylethanolamine; APL, unidentified aminophospholipid; PL, unidentified phospholipid; L, unidentified lipid.
‡The DNA G+C contents were calculated from the genome sequence.
Taxa: 1, strain E39T [this study]; 2, Alloprevotella tannerae ATCC 51259T [36]; 3, Alloprevotella rava 81/4-12T [35]; 4, Paraprevotella clara JCM 14859T [37]; 5, Prevotella melaninogenica ATCC 25845T [45]. All strains are positive for the following characteristics: activity* of β-galactosidase, alkaline phosphatase and alanine arylamidase. All strains are negative for the following characteristics: activity* of urease, β-glucosidase, proline arylamidase, phenylalanine arylamidase, leucine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, glycine arylamidase, histidine arylamidase and serine arylamidase, reduction of nitrates, and indole production. Symbols: +, positive;–, negative; w, weak; ND, not determined.
Table 2. Cellular fatty acid compositions (%) of strain E39T and closely related taxa.
| Fatty acid | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Saturated: | |||||
| C12:0 | 0.9 | tr | tr | tr | tr |
| C13:0 | 0.9 | – | – | – | – |
| C14:0 | 4.5 | 3.0 | 3.2 | 11.6 | 1.9 |
| C15:0 | – | – | – | – | – |
| C16:0 | 24.9 | 4.9 | 14.5 | 9.0 | 8.7 |
| C17:0 | 1.2 | tr | 1.7 | – | tr |
| C18:0 | 18.3 | 1.3 | 6.7 | 0.8 | 0.9 |
| C20:0 | 0.8 | tr | 0.6 | tr | tr |
| Unsaturated: | |||||
| C13:1 at 12–13 | 0.6 | tr | 2.2 | 1.5 | 2.2 |
| C18:1 ω9c | 10.3 | 0.9 | 6.7 | 23.2 | 23.1 |
| Hydroxy: | |||||
| C15:0 3-OH | 0.9 | tr | tr | tr | tr |
| C16:0 3-OH | 3.7 | 2.0 | 0.8 | 8.0 | 3.5 |
| C17:0 2-OH | tr | tr | 0.7 | tr | tr |
| iso-C17:0 3-OH | 2.1 | 9.4 | 13.3 | 0.6 | 3.6 |
| Branched: | |||||
| iso-C13:0 | 1.6 | 3.5 | 1.0 | tr | 0.8 |
| iso-C14:0 | – | 8.8 | – | 1.5 | 12.2 |
| iso-C15:0 | 9.0 | 26.6 | 8.4 | 11.0 | 5.6 |
| iso-C16:0 | 0.5 | 2.2 | 2.4 | tr | 1.0 |
| iso-C17:0 | 3.0 | 4.2 | 6.7 | tr | 1.4 |
| iso-C18:0 | – | tr | 1.0 | – | tr |
| iso-C19:0 | – | tr | 1.1 | – | 0.8 |
| anteiso-C13:0 | tr | 0.8 | tr | tr | tr |
| anteiso-C15:0 | 5.4 | 25.2 | 15.5 | 21.1 | 21.7 |
| anteiso-C17:0 | 0.9 | 1.9 | 2.4 | tr | 1.4 |
| Summed feature*: | |||||
| 3 | 0.9 | tr | 1.0 | 3.1 | 3.1 |
| 5 | 2.7 | tr | – | – | – |
| 8 | 3.5 | tr | 2.6 | 3.2 | 3.4 |
*Summed features represent groups of two fatty acids that cannot be separated by GLC with the MIDI system.
Summed feature 3, C16:1 ω7c and/or C16:1 ω6c; summed feature 5, C18:0 ante and/or C18:2 ω6,9c; summed feature 8, C18:1 ω7c and/or C18:1 ω6c.
Taxa: 1, strain E39T; 2, Alloprevotella tannerae ATCC 51259T; 3, Alloprevotella rava 81/4-12T; 4, Paraprevotella clara YIT 11840T; 5, Prevotella melaninogenica ATCC 25845T. All data were obtained from this study. Data are expressed as percentages of total fatty acids. In all strains, fatty acids that account for less than 0.5% are not shown. Major components (> 5.0%) are in bold. tr, trace amount (< 0.5%);–, not detected.
In conclusion, 16S rRNA gene and genome based analysis clearly supported identification of strain E39T as a novel genus of the family Prevotellaceae. On top of that, strain E39T was distinguished from the reference taxa by several traits, including coccus shape, growth only on media containing mucin, a positive activity for arginine dihydrolase, high portion of saturated fatty acids (C16:0 and C18:0),andtypical polar lipids (unidentified lipids), and MK-8 and MK-9 as major respiratory quinones (Table 3). Taken together, strain E39T is considered to represent a new genus within the family Prevotellaceae, for which the name Pseudoprevotella muciniphila gen. nov., sp. nov. is proposed.
Table 3. Comparison of characteristics between Pseudoprevotella gen. nov. and closely related genera within the family Prevotellaceae.
| Characteristic | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Habitat | Rumen epithelium | Human oral cavity | Gut intestinal tract of mammals | Rumen, gut intestinal tract, and vagina of mammals |
| Metabolic end products from glucose | Acetate, succinate*† | Acetate, succinate | Acetate, succinate | Acetate, succinate |
| Major cellular fatty acids | C16:0, C18:0* | anteiso-C15:0, iso-C15:0* | anteiso-C15:0, iso-C15:0, C18:1 ω9c | anteiso-C15:0 |
| Major quinones | MK-8, MK-9* | MK-9* | MK-10, MK-11 | MK-10~MK-13 |
| DNA G+C content (mol%) | 46.4 | 45–47 | 48–49 | 40–52 |
Metabolite changes during mucin and glucose fermentation
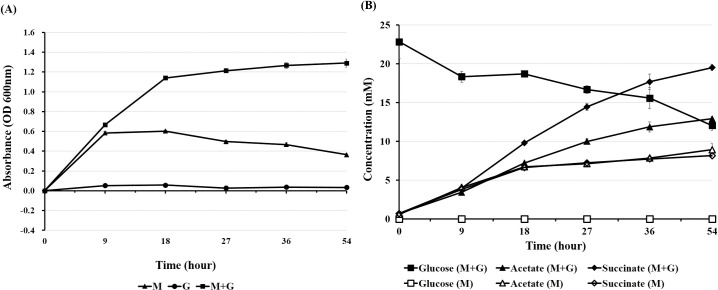
The growth of strain E39T was observed in the basal mucin medium and mucin-glucose medium, but not in glucose medium (Fig 3A). Strain E39T grew in the mucin-glucose medium better than in the basal mucin medium. Metabolites including free sugars, organic acids, and amino acids were analyzed by 1H NMR spectroscopy. The concentration of acetate and succinate increased continuously in the both media (Fig 3B). The concentration of mannose and N-acetylglucosamine which are sugar residues of mucin structure, increased in both media during the early fermentation (0–9 h), and then decreased after 9 h (S5 Fig). Alanine, glycine, and valine were detected as the major amino acids during the fermentation in the both media (S6 Fig). Strain E39T was not able to grow in media without mucin, and the growth of strain E39T was promoted by the addition of glucose. These results suggest that mucin is an essential growth nutrient for strain E39T and it can utilize glucose as an energy source only under the presence of mucin. Furthermore, it was shown that strain E39T might utilize mucin as an energy source during the early fermentation and produce acetate, succinate, and several amino acids as major fermented end-products.
Fig 3.
Growth of strain E39T in the basal mucin (M), glucose (G), and mucin-glucose (M+G) media (A) and profiles of glucose, acetate, and succinate in the basal (M) and mucin-glucose (M+G) media during fermentation (B). Data are presented as mean ± standard error from triplicates.
Genomic features of the complete genome of Pseudoprevotella muciniphila E39T
Annotation analysis revealed that the genome of strain E39T comprised 2,920,169 bases, 2,458 genes, 2,396 coding sequences (CDSs), 52 tRNA, 1 transfer-messenger RNA (tmRNA), and 9 rRNA (Table 4).
Table 4. Genomic features of Pseudoprevotella muciniphila E39T.
| Number of Contigs | 1 | ||
| Genome Size (bp) | 2,920,169 | ||
| G+C Content (mol%) | 46.4 | ||
| Number of Genes | 2,458 | Protein encoding genes | 2,396 |
| tRNA | 52 | ||
| tmRNA | 1 | ||
| rRNA | 9 | ||
| Proteins with Predicted Functions | 1,161 | ||
| Hypothetical or Uncharacterized Proteins | 1,235 | ||
| Proteins with KEGG Annotations | 971 | ||
| Carbohydrate-Active Enzymes with CAZyme Annotations | 128 | Glycoside Hydrolase | 64 |
| Carbohydrate-Binding Module | 5 | ||
| Glycosyl Transferase | 49 | ||
| Polysaccharide Lyase | 0 | ||
| Carbohydrate Esterase | 10 | ||
| Virulence Factors | 17 | ||
| Antibiotic Resistant Genes | 4 | ||
(1) Mucin degradation and utilization
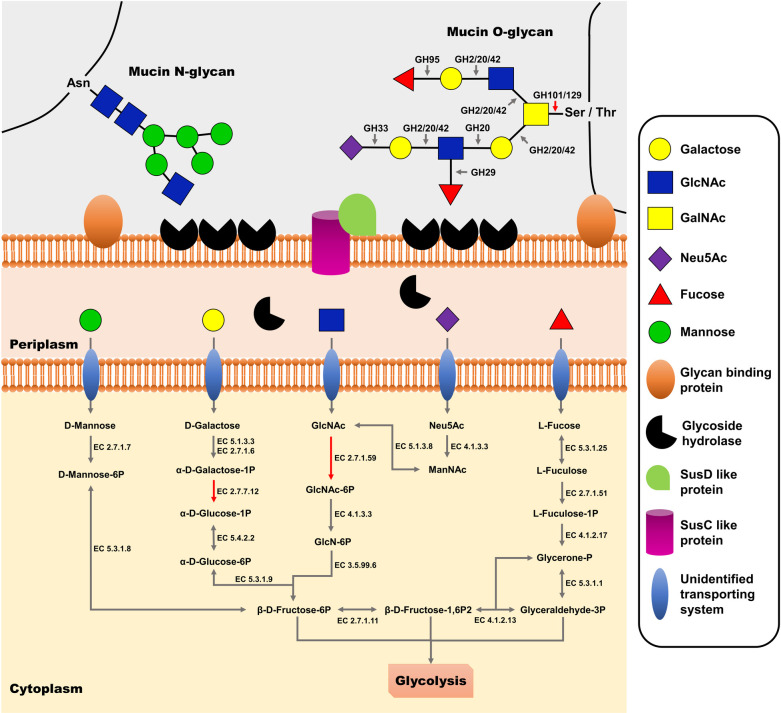
Mucin is host-derived glycoprotein composed of a protein backbone, lots of O-glycan, and a small number of N-glycan. N-acetlygalactosamine (GalNAc) is O-glycosylated to proline-threonine-serine (PTS) domain of a protein backbone and addition of galactose or N-acetlyglucosamine (GlcNAc) forms 8 types of O-glycan core structures. Addition of extended core (galactose and GlcNAc) and terminal residues (sialic acid and fucose) makes mucin structure more complex [39]. To utilize mucin as an energy source, bacteria need to have series of enzymes to degrade complex mucin structure. Several glycoside hydrolases (GHs) were known as enzymes involved in mucin degradation such as sialidases (GH33), α-fucosidases (GH29, GH95), exo- and endo-β-N-acetylglucosaminidases (GH84, GH85), β-galactosidases (GH2, GH20, GH42), α-N-acetylglucosaminidases (GH89), endo-β1,4-galactosidases (GH98), and α-N-acetylgalactosaminidases (GH101, GH129) [39, 40]. In this study, the result of CAZyme annotation using dbCAN2 tool showed that P. muciniphila E39T had 64 GHs, 5 carbohyrate-binding modules, 49 glycosyltransferases, 10 carbohydrate esterases, and no polysaccharide lyases (Table 4, S1 Dataset). Among 64 GHs, 28 GHs were putative enzymes involved in mucin degradation (Table 5). Compared to a Gram-negative mucin-degrading bacterium, B. thetaiotaomicron, P. muciniphila E39T had fewer but similar kinds of mucin-degrading GHs. Bacteroides thetaiotaomicron has Sus-like systems for utilization of mucin glycan [41]. Glycan binding proteins on the cell surface bind polysaccharide and GHs partially degrade glycan. And then, two outer membrane proteins, homologs of SusD and SusC, import oligosaccharide into periplasm. After transportation of glycan from extracellular place to periplasm, glycan is degraded into monosaccharides by GHs and transferred to cytoplasm through inner membrane transporters [40, 41]. We confirmed that P. muciniphila E39T also had homologs of SusC and SusD through a BlastP search against the genome of P. muciniphila E39T (S1 Dataset). Based on a KEGG pathway analysis and BlastP analysis, we constructed putative mucin degrading pathway of P. muciniphila E39T. There were genes associated with metabolism of carbon sources including galactose, sialic acid (Neu5Ac), fucose, GlcNAc, and mannose on the results of KEGG pathway analysis (Fig 4). However, the proposed metabolic pathway was incomplete because of the absence of two genes (galactose 1-phosphate uridyltransferase, EC 2.7.7.12; N-acetylglucosamine kinase, EC 2.7.1.59) involved in galactose and GlcNAc metabolisms, respectively. In addition, P. muciniphila E39T was negative in the mannose fermentation activity despite it harbored a mannose metabolic pathway. Further studies on mucin degrading pathway like monosaccharide transporting systems are needed to explain and understand mucin metabolic features of the P. muciniphila E39T.
Table 5. List of predicted mucin degrading enzymes in the complete genome of strain E39T.
| Mucin degrading enzymes | Glycoside hydrolases (GHs) | Locus IDa | KEGG ID | Prokka annotation |
|---|---|---|---|---|
| β-galactosidases | GH2 | L_00290 | K01190 (β-galactosidase) | β-galactosidase |
| L_01043 | K01190 (β-galactosidase) | β-galactosidase | ||
| L_01673 | K01190 (β-galactosidase) | Evolved β-galactosidase subunit α | ||
| GH20 | L_00323 | K12373 (hexosaminidase) | β-hexosaminidase | |
| L_00864 | – | β-N-acetylhexosaminidase | ||
| L_00865 | – | β-N-acetylhexosaminidase | ||
| L_01112 | K12373 (hexosaminidase) | β-hexosaminidase | ||
| L_01187 | K12373 (hexosaminidase) | β-hexosaminidase | ||
| L_01443 | K12373 (hexosaminidase) | β-hexosaminidase | ||
| L_01556 | K12373 (hexosaminidase) | β-hexosaminidase | ||
| L_01824 | K12373 (hexosaminidase) | β-hexosaminidase | ||
| GH42 | – | – | – | |
| Endo-β1,4-galactosidases | GH98 | – | – | – |
| α-N-acetylgalactosaminidases | GH101 | – | – | – |
| GH129 | – | – | – | |
| Exo- and endo-β-N-acetylglucosaminidases | GH84 | L_00360 | K01197 (hyaluronoglucosaminidase) | Hyaluronoglucosaminidase |
| L_01917 | K01197 (hyaluronoglucosaminidase) | O-GlcNAcase | ||
| GH85 | L_00833 | – | Hypothetical protein | |
| L_01606 | – | Hypothetical protein | ||
| α-N-acetylglucosaminidases | GH89 | L_01212 | K01205 (α-N-acetylglucosaminidase) | Hypothetical protein |
| L_01965 | K01205 (α-N-acetylglucosaminidase) | Hypothetical protein | ||
| Sialidases | GH33 | L_00875 | – | Hypothetical protein |
| L_01672 | K01186 (sialidase-1) | Sialidase | ||
| L_02283 | – | Sialidase | ||
| Fucosidases | GH29 | L_00284 | K01206 (α-L-fucosidase) | Hypothetical protein |
| L_00599 | K01206 (α-L-fucosidase) | Hypothetical protein | ||
| L_00887 | K01206 (α-L-fucosidase) | Hypothetical protein | ||
| L_00979 | K01206 (α-L-fucosidase) | Hypothetical protein | ||
| L_01701 | K01206 (α-L-fucosidase) | Hypothetical protein | ||
| GH95 | L_00982 | K15923 (α-L-fucosidase 2) | Hypothetical protein | |
| L_01250 | K15923 (α-L-fucosidase 2) | Hypothetical protein | ||
| L_02348 | K15923 (α-L-fucosidase 2) | Hypothetical protein |
aLocus IDs are the results of CAZyme annotation using dbCAN2 tool.
Fig 4. Proposed mucin degrading and utilizing pathways of the P. muciniphila E39T.
These putative pathways were constructed based on CAZyme annotation, KEGG pathway analysis and BlastP analysis. Mucin is initially degraded into oligo- or monosaccharides by mucin degrading glycoside hydrolases (GHs) followed by transportation into periplasm by Sus-like outer membrane proteins. Additional degradation is occurred by periplasmic glycoside hydrolases and mucin-derived monosaccharides are imported through unidentified transporters and utilized as carbon sources. Metabolic pathways that are present in the P. muciniphila E39T are depicted in gray, and metabolic pathways that are not present in the P. muciniphila E39T are depicted in red. GlcNAc: N-acetlyglucosamine, GalNAc: N-acetylgalactosamine, Neu5Ac: sialic acid, ManNAc: N-acetylmannosamine.
(2)Extracellular polymeric substances biosynthesis
Bacteria produce biofilms for various purposes. Biofilms are composed of extracellular polymeric substances including nucleic acids, lipids, proteins, and exopolysaccharides and this complex compounds have a wide range of roles in adhesion to other bacterial cells or host cells, protection from stresses such as antibiotic substances or harmful chemicals, and provision of structure for stratification against rapid environmental changes [42]. In the Wzx/Wzy-dependent pathway, one of exopolysaccharides biosynthesis pathways, glycosyltransferases assemble repeating units and a flippase (Wzx protein) translocates the units into the periplasmic place. The repeating units are elongated by a polymerase (Wzy protein) and transported across the outer membrane through a polysaccharide export protein [43]. P. muciniphila E39T also produced branch-shaped extracellular structures (Fig 2) and these structures are predicted to contribute to adhesion to mucin or host cells. We identified that P. muciniphila E39T had 1 putative extracellular polysaccharide biosynthesis locus (L_02166 –L_02195) through a BlastP search against the genome of P. muciniphila E39T (S1 Dataset). There were 8 putative glycosyltransferases (L_02166, L_02169, L_02175, L_02179, L_02180, L_02181, L_02183, L_02184), 1 polymerase (L_02174), 1 flippase (L_02189), 1 serine acetyltransferase (L_02182), 1 N-acetyltransferase (L_02182), 1 aminotransferase (L_02188), 1 polysaccharide pyruvyl transferase (L_02191), and 1 polysaccharide export protein (L_02193).
(3)Virulence factors
Bacterial virulence factors allow bacteria to survive in the host, participating in adhesion, colonization, invasion, evasion or inhibition of immune responses, etc [44]. P. muciniphila E39T had 17 putative virulence factors and they involved in adherence (KpsF, htpB, glf, hasB), antiphagocytosis (cps4J, cps4K, cps4L, hasB), immune evasion (tviC), stress reaction (clpC, clpP), O-antigen (galE, fcl), lipopolysaccharide (gmd), and metabolic adaptation (panD) (S1 Dataset). The virulence factors encoded in these genes may enable P. muciniphila E39T to survive on the rumen wall, a place which host’s defense mechanism is most active in the rumen, by attaching to epithelial cells and avoiding the host’s immune responses. The absence of genes encoding exotoxin and involved in invasion suggests that P. muciniphila E39T may have low pathogenicity. However, we cannot ignore the possibility that P. muciniphila E39T is a potential pathogen because of its endotoxin (lipopolysaccharide) and mucinolytic ability. Further researches at molecular level or in vivo studies are required to determine the pathogenicity of P. muciniphila E39T.
Conclusions
The genetic, physiological, and chemotaxonomic features support that strain E39T represents a novel genus of the family Prevotellaceae. As such, the name Pseudoprevotella muciniphila gen. nov., sp. nov. is proposed. Pseudoprevotella muciniphila E39T was isolated from the bovine rumen epithelium and this bacterium utilized mucin as a sole carbon source. The functional annotation of the complete genome of P. muciniphila E39T supported that P. muciniphila E39T possess a series of mucin degrading enzymes and putative mucin-degrading pathway. In addition, P. muciniphila E39T is predicted to have putative metabolisms to synthesize extracellular polymeric substances and virulence factors for adhering to rumen epithelial cells and evading the host’s immune responses. In short, this study contributes to discovery of a novel mucin-degrading bacterium which has a potential ability to significantly affect host’s physiology and its putative metabolic pathways which can assist to predict its function in epimural community.
Description of Pseudoprevotella muciniphila gen. nov., sp. nov.
Pseudoprevotella (Pseu.do.pre.vo.tel′la. Gr. adj. pseudês false; N.L. n. fem. n. Prevotella a bacterial generic name; N.L. fem. n. Pseudoprevotella false Prevotella).
The cells are strictly anaerobic, non-motile, Gram-negative and coccus in shape. The major cellular fatty acids are C16:0, C18:0, C18:1 ω9c, iso-C15:0, and anteiso-C15:0. The main polar lipids are phosphatidylethanolamine (PE), unidentified aminophospholipid (APL), and three unidentified lipids. The major respiratory quinones are MK-8 and MK-9. The major fermented end-products of mucin are acetate and succinate. The genus is a member of the family Prevotellaceae of the phylum Bacteroidetes. The type species is Pseudoprevotella muciniphila.
Description of Pseudoprevotella muciniphila sp. nov.
Pseudoprevotella muciniphila (mu.ci.ni′phi.la. N.L. neut. n. mucinum mucin; Gr. adj. philos loving; N.L. fem. adj. muciniphila mucin-loving).
In addition to the characteristics provided in the genus description above, this species grows at 30–45°C (optimum, 39°C), at pH 6.5–8.5 (optimum, pH 7.5), and in the presence of 0.0–1.0% (w/v) NaCl (optimum, 0.0–0.5%). This species had arginine dihydrolase, α-galactosidase, β-galactosidase, β-galactosidase-6-phosphate, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, α-fucosidase, alkaline phosphatase, leucyl glycine arylamidase, alanine arylamidase, and glutamyl glutamic acid arylamidase activity, but lacked urease, α-glucosidase, β-glucosidase, α-arabinosidase, β-glucuronidase, mannose fermentation, raffinose fermentation, reduction of nitrates, indole production, arginine arylamidase, proline arylamidase, phenylalanine arylamidase, leucine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, glycine arylamidase, histidine arylamidase, and serine arylamidase activity. This species has a DNA G+C content of 46.4 mol%. The type strain is E39T (KCTC 15717T = JCM 32621T), and it was isolated from the rumen epithelium of Korean cattle.
Supporting information
Bootstrap values over 70% are shown on the nodes as percentages of 2,000 replicates. Sphingobacterium spiritivorum ATCC 33861T (EF090267) was used as an outgroup. Bar indicates 0.02 changes per nucleotide position.
(DOCX)
Bootstrap values over 70% are shown on the nodes as percentages of 2,000 replicates. Sphingobacterium spiritivorum ATCC 33861T (EF090267) was used as an outgroup. Bar indicates 50 changes per nucleotide position.
(DOCX)
NROX: No reducing agent and aerobic headspace, NRAN: No reducing agent and anaerobic headspace, PROX: presence of reducing agent and aerobic headspace, PRAN: presence of reducing agent and anaerobic headspace. Data are presented as mean ± standard error from triplicates.
(DOCX)
Solvent systems: (I) chloroform-methanol-water (65:25:4, v/v/v); (II) chloroform-acetic acid-methanol-water (80:15:12:4, v/v/v/v). The TLC plates were sprayed with 10% ethanolic molybdatophosphoric acid. (A) strain E39T, (B) Alloprevotella tannerae, (C) Alloprevotella rava, (D) Paraprevotella clara, (E) Prevotella melaninogenica. PE, phosphatidylethanolamine; APL, unidentified aminophospholipids; PL, unidentified phospholipids; L, unidentified polar lipids.
(DOCX)
Profiles of galactose, N-acetylglucosamine, and mannose in the mucin-glucose (A) and basal mucin (B) media during fermentation. Data are presented as mean ± standard error from triplicates.
(DOCX)
Profile of major amino acids in the mucin-glucose (A) and basal mucin (B) media during fermentation. Data are presented as mean ± standard error from triplicates.
(DOCX)
(XLSX)
Data Availability
All 16S gene and genome sequence data of stain E39 are available from the GenBank database (accession numbers MG763147 and CP033459). Other reference strains' genome sequence data are available from the GenBank: A. tannerae ATCC 51259T (GenBank acc. no., ACIJ00000000), A. rava 81/4-12T (GenBank acc. no., ACZK00000000), P. clara YIT 11840T (GenBank acc. no., AFFY00000000), P. melaninogenica ATCC 25845T (GenBank acc. no., CP002122-3), and Bacteroides thetaiotaomicron VPI 5482T (GenBank acc. No., AE015928) were obtained from GenBank for comparative analysis. Other relevant data are within the manuscript and its supporting information files.
Funding Statement
This work was supported by a grant from the Next-Generation BioGreen 21 Program (PJ01114001), Rural Development Administration, Republic of Korea.
References
- 1.Puniya AK, Singh R, Kamra DN. Rumen microbiology: an overview. In: Choudhury PK, Salem AZM, Jena R, Kumar S, Singh R, Puniya AK, editors. Rumen Microbiology: From Evolution to Revolution. New Delhi: Springer; 2015. pp. 3–47. [Google Scholar]
- 2.Rémond D, Meschy F, Boivin R. Metabolites, water and mineral exchanges across the rumen wall: mechanisms and regulation. Ann Zootechn. 1996;45: 97–119. [Google Scholar]
- 3.Sadet S, Martin C, Meunier B, Morgavi DP. PCR-DGGE analysis reveals a distinct diversity in the bacterial population attached to the rumen epithelium. Animal. 2007;1: 939–944. 10.1017/S1751731107000304 [DOI] [PubMed] [Google Scholar]
- 4.Cheng KJ, McCowan RP, Costerton JW. Adherent epithelial bacteria in ruminants and their roles in digestive tract function. Am J Clin Nutr. 1979;32: 139–148. 10.1093/ajcn/32.1.139 [DOI] [PubMed] [Google Scholar]
- 5.Wetzels SU, Mann E, Pourazad P, Qumar M, Pinior B, Metzler-Zebeli BU, et al. Epimural bacterial community structure in the rumen of Holstein cows with different responses to a long-term subacute ruminal acidosis diet challenge. J Dairy Sci. 2017;100: 1829–1844. 10.3168/jds.2016-11620 [DOI] [PubMed] [Google Scholar]
- 6.Mann E, Wetzels SU, Wagner M, Zebeli Q, Schmitz-Esser S. Metatranscriptome sequencing reveals insights into the gene expression and functional potential of rumen wall bacteria. Front Microbiol. 2018;9: 43. 10.3389/fmicb.2018.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Micobiol. 2011;9: 265–278. 10.1038/nrmicro2538 [DOI] [PubMed] [Google Scholar]
- 8.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper R, de Vos W, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1: 254–268. 10.4161/gmic.1.4.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54: 1469–1476. 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- 10.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci. 2013;110: 9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoorens PR, Rinaldi M, Li RW, Goddeeris B, Claerebout E, Vercruysse J, et al. Genome wide analysis of the bovine mucin genes and their gastrointestinal transcription profile. BMC Genomics. 2011;12: 140. 10.1186/1471-2164-12-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fina LR, Hay CA, Bartley EE, Mishra B. Bloat in cattle. V. The role of rumen mucinolytic bacteria. J Anim Sci. 1961;20: 654–658. [DOI] [PubMed] [Google Scholar]
- 13.Mishra BD, Fina LR, Bartley EE, Claydon TJ. Bloat in cattle. XI. The role of rumen aerobic (facultative) mucinolytic bacteria. J Anim Sci. 1967;26: 606–612. 10.2527/jas1967.263606x [DOI] [PubMed] [Google Scholar]
- 14.Mishra BD, Bartley EE, Fina LR, Bryant MP. Bloat in cattle. XIV. Mucinolytic activity of several anaerobic rumen bacteria. J Anim Sci. 1968;27: 1651–1656. 10.2527/jas1968.2761651x [DOI] [PubMed] [Google Scholar]
- 15.Mead LJ, Jones GA. Isolation and presumptive identification of adherent epithelial bacteria (“Epimural” bacteria) from the ovine rumen wall. Appl Environ Microbiol. 1981;41: 1020–1028. 10.1128/AEM.41.4.1020-1028.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller RE, Iannotti EL, Asplund JM. Isolation and identification of adherent epimural bacteria during succession in young lambs. Appl Environ Microbiol. 1984;47: 724–730. 10.1128/AEM.47.4.724-730.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldwell DR, Bryant MP. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl Microbiol. 1966;14: 794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 19.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 20.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017;67: 1613–1617. 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomori G. Preparation of buffers for use in enzyme studies. Methods Enzymol. 1955;1: 138–146. [Google Scholar]
- 23.Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. Newark: MIDI Inc; 1990. [Google Scholar]
- 24.Minnikin DE, Patel PV, Alshamaony L, Goodfellow M. Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol. 1977;27:104–107. [Google Scholar]
- 25.Jeon CO, Lee DS, Park JM. Microbial communities in activated sludge performing enhanced biological phosphorus removal in a sequencing batch reactor. Water Res. 2003;37: 2195–2205. 10.1016/S0043-1354(02)00587-0 [DOI] [PubMed] [Google Scholar]
- 26.Han DM, Chun BH, Feng T, Kim HM, Jeon CO. Dynamics of microbial communities and metabolites in ganjang, a traditional Korean fermented soy sauce, during fermentation. Food Microbiol. 2020;92: 103591. 10.1016/j.fm.2020.103591 [DOI] [PubMed] [Google Scholar]
- 27.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10: 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 28.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30: 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 29.Na S, Kim YO, Yoon SH, Ha SM, Baek I, Chun J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol. 2018;56: 280–285. 10.1007/s12275-018-8014-6 [DOI] [PubMed] [Google Scholar]
- 30.Lee I, Kim YO, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66: 1100–1103. 10.1099/ijsem.0.000760 [DOI] [PubMed] [Google Scholar]
- 31.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 2013;14: 60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428: 726–731. 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46: W95–W101. 10.1093/nar/gky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Tai C, Deng Z, Zhong W, He Y, Ou HY. VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief Bioinform. 2018;19: 566–574. 10.1093/bib/bbw141 [DOI] [PubMed] [Google Scholar]
- 35.Downes J, Dewhirst FE, Tanner ACR, Wade WG. Description of Alloprevotella rava gen. nov., sp. nov., isolated from the human oral cavity, and reclassification of Prevotella tannerae gen. nov., comb. nov. Int J Syst Evol Microbiol. 2013;63: 1214–1218. 10.1099/ijs.0.041376-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore LV, Johnson JL, Moore WE. Descriptions of Prevotella tannerae sp. nov. and Prevotella enoeca sp. nov. from the human gingival crevice and emendation of the description of Prevotella zoogleoformans. Int J Syst Bacteriol. 1994;44: 599–602. 10.1099/00207713-44-4-599 [DOI] [PubMed] [Google Scholar]
- 37.Morotomi M, Nagai F, Sakon H, Tanaka R. Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family ’Prevotellaceae’ isolated from human faeces. Int J Syst Bacteriol. 2009;59: 1895–1900. 10.1099/ijs.0.008169-0 [DOI] [PubMed] [Google Scholar]
- 38.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106: 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6: 81–98. 10.3389/fgene.2015.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ndeh D, Gilbert HJ. Biochemistry of complex glycan depolymerisation by the human gut microbiota. FEMS Microbiol Rev. 2018;42: 146–164. 10.1093/femsre/fuy002 [DOI] [PubMed] [Google Scholar]
- 41.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4: 447–457. 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Limoli DH, Jones CJ, Wozniak DJ. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr. 2015;3. 10.1128/microbiolspec.MB-0011-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid J, Sieber V, Rehm B. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol. 2015;6: 496. 10.3389/fmicb.2015.00496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakour S, Sankar SA, Rathored J, Biagini P, Raoult D, Fournier PE. Identification of virulence factors and antibiotic resistance markers using bacterial genomics. Future Microbiol. 2016;11: 455–466. 10.2217/fmb.15.149 [DOI] [PubMed] [Google Scholar]
- 45.Shah HN, Collins DM. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Evol Microbiol. 1990;40: 205–208. 10.1099/00207713-40-2-205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bootstrap values over 70% are shown on the nodes as percentages of 2,000 replicates. Sphingobacterium spiritivorum ATCC 33861T (EF090267) was used as an outgroup. Bar indicates 0.02 changes per nucleotide position.
(DOCX)
Bootstrap values over 70% are shown on the nodes as percentages of 2,000 replicates. Sphingobacterium spiritivorum ATCC 33861T (EF090267) was used as an outgroup. Bar indicates 50 changes per nucleotide position.
(DOCX)
NROX: No reducing agent and aerobic headspace, NRAN: No reducing agent and anaerobic headspace, PROX: presence of reducing agent and aerobic headspace, PRAN: presence of reducing agent and anaerobic headspace. Data are presented as mean ± standard error from triplicates.
(DOCX)
Solvent systems: (I) chloroform-methanol-water (65:25:4, v/v/v); (II) chloroform-acetic acid-methanol-water (80:15:12:4, v/v/v/v). The TLC plates were sprayed with 10% ethanolic molybdatophosphoric acid. (A) strain E39T, (B) Alloprevotella tannerae, (C) Alloprevotella rava, (D) Paraprevotella clara, (E) Prevotella melaninogenica. PE, phosphatidylethanolamine; APL, unidentified aminophospholipids; PL, unidentified phospholipids; L, unidentified polar lipids.
(DOCX)
Profiles of galactose, N-acetylglucosamine, and mannose in the mucin-glucose (A) and basal mucin (B) media during fermentation. Data are presented as mean ± standard error from triplicates.
(DOCX)
Profile of major amino acids in the mucin-glucose (A) and basal mucin (B) media during fermentation. Data are presented as mean ± standard error from triplicates.
(DOCX)
(XLSX)
Data Availability Statement
All 16S gene and genome sequence data of stain E39 are available from the GenBank database (accession numbers MG763147 and CP033459). Other reference strains' genome sequence data are available from the GenBank: A. tannerae ATCC 51259T (GenBank acc. no., ACIJ00000000), A. rava 81/4-12T (GenBank acc. no., ACZK00000000), P. clara YIT 11840T (GenBank acc. no., AFFY00000000), P. melaninogenica ATCC 25845T (GenBank acc. no., CP002122-3), and Bacteroides thetaiotaomicron VPI 5482T (GenBank acc. No., AE015928) were obtained from GenBank for comparative analysis. Other relevant data are within the manuscript and its supporting information files.