Abstract
Known for its high genetic diversity and variation in genotypic presence in different regions of the world, hepatitis C virus (HCV) is estimated to infect about 71 million people globally. Selection of an appropriate therapeutic regimen largely depends on the identification of the genotype responsible for the infection. This systematic review and meta-analysis was conducted to provide a comprehensive view of HCV genotype and subtype distribution in Southeast Asia (SEA). The review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA). We searched five databases without year and language restrictions. Data from 90 eligible studies involving 15,089 genotypes and 9,646 subtypes representing 10 SEA countries were analyzed. The pooled estimates showed that genotype 1 (46.8%) [95% CI, 43.2–50.4; I2 = 92.77%; p < 0.001] was the most dominant HCV genotype in the region, followed by genotype 3 (23.1%) [95% CI, 19.4–27.2; I2 = 93.03%; p < 0.001], genotype 6 (16.5%) [95% CI, 13.8–19.6], genotype 2 (4.6%) [95% CI, 3.5–5.9], genotype 4 (1.1%) [95% CI, 0.7–1.5] and genotype 5 (0.8%) [95% CI, 0.4–1.3]. Philippines had the highest prevalence of genotypes 1 and 2. Genotype 6 became more prevalent after year 2000. Over 40 different subtypes were identified, with subtypes 1b (26.3%), 1a (21.3%), and 3a (14.3%) being the most prevalent of all the reported subtypes. Although on a global scale, genotype 6 is considered highly prevalent in SEA, evidence from this study reveals that it is the third most prevalent genotype within the region.
Introduction
Hepatitis C virus (HCV) infection constitutes a major health challenge globally. HCV is a principal cause of hepatocellular carcinoma (HCC), liver cirrhosis and liver failure, and it is thought to infect about 71 million people worldwide [1]. The virus, which belongs to the Flaviviridae family, consists of a positive-sense single-stranded RNA genome that spans approximately 9.6 kb [2, 3]. Flanked by 5’ and 3’ untranslated regions (UTRs), its long single open reading frame codes for structural (core [C] and envelope [E1 and E2]) and non-structural (P7, NS2, NS3, NS4A, NS4B, NS5A and NS5B) proteins [2, 3]. HCV exhibits a profound genetic diversity–an occurrence partly attributed to the absence of proofreading nuclease activity of its ribonucleic acid (RNA)-dependent RNA polymerase [4, 5]. Similar to human immunodeficiency virus (HIV), multiple quasispecies of HCV emerge quite frequently in vivo, posing significant threat to drug efficacy and vaccine development. Mutations that occur in the least conserved regions (i.e., hypervariable regions of the NS5A and envelope genes) of the HCV viral genome are thought to play a crucial role in immune evasion and the establishment of chronic infection [5].
Sequence and phylogenetic analysis of HCV genome has revealed at least six major genotypes (designated as genotype 1 to 6) and numerous subtypes (connoted with lowercase alphabets; e.g. 1a, 1b, 2a, etc.) [6]. Although HCV exhibits an extraordinary sequence diversity, all genotypes possess identical complement of co-linear genes of similar size within the open reading frame (ORF), and the known variants contain considerably similar sequence across the genome [7]. This makes it possible to classify them based on partial sequences from specific regions (e.g. core/E1 or NS5B) within the genome [7]. The distribution of HCV genotypes is largely dependent on geographical locations. Although genotypes 1, 2 and 3 seem to be distributed globally, ‘endemic’ strains of genotype 1 and 2 are mainly in West Africa while genotype 3 is dominant in South Asia [4]. Genotypes 4, 5, and 6 on the other hand, are predominant in the Middle East and Central Africa, Southern Africa, and Southeast Asia, respectively [4].
Distinct genotypes and subtypes of HCV vary in their degrees of resistance to some of the currently available antiviral drugs, making treatment decision genotype oriented [6, 8]. Determination of infecting genotype is thus fundamental to HCV therapy as it helps ascertain which antiviral drug to administer, its dosage, as well as treatment duration [9]. While there are several HCV genotyping methods, full genomic analysis or sequencing and phylogenetic analysis of informative and conserved regions (e.g. core/E1 or NS5B) of the genome remains the gold standard [5, 6, 10].
In the past, attempts have been made to provide global and regional distribution of HCV genotypes [4, 11–13]. However, there has been no detailed report on the actual distribution of the virus’ genotypes in Southeast Asia (SEA)–a multiethnic and socioculturally diverse Asian region spanning Brunei, Cambodia, East Timor, Indonesia, Laos, Malaysia, Myanmar, Philippines, Singapore, Thailand and Vietnam. This is further compounded by the fact that newer variants of the virus are continually being identified and characterized. In this article, we conducted a Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) compliant review of published articles reporting HCV genotypes in SEA since the virus was first identified in 1989 to provide an in-depth and up-to-date information on the distribution of HCV genotypes in the region.
Methods
Search strategy and selection criteria
We accessed five electronic databases (PubMed, Scopus, ScienceDirect, Google Scholar, and Web of Science) for studies reporting HCV genotypes in eleven SEA countries (Brunei, Cambodia, East Timor, Indonesia, Laos, Malaysia, Myanmar, Philippines, Singapore, Thailand, Vietnam). The databases were searched using a combination of terms (e.g., “hepatitis c”, “genotype”) related to the distribution of HCV genotypes in SEA without year or language restrictions. Details of the search strategy used is provided in S1 File. A final search was made on April 30, 2020.
Studies that conducted HCV genotyping and whose samples or analyzed data were collected in at least one of the SEA countries were included. We excluded studies (1) that are reviews, (2) with unknown sample origins (3) with only serological findings, (4) whose full text could not be obtained, or (5) contained duplicated data.
Quality assessment
The methodological quality of all the included studies was assessed using the Joanna Briggs Institute (JBI) critical appraisal checklist for prevalence data [14] (S2 File). A score of ‘1’ for ‘yes’ and ‘0’ for other parameters were assigned to attain a total quality score that ranged from 0 to 9. Studies with overall score of 7–9 were considered to be of sufficient quality. Two authors (A.A.I and Y.W.) independently assessed the studies. Studies were included if there was a consensus between the two reviewers. The quality assessment of the 90 included studies is provided in S1 Table.
Data extraction and analysis
From each of the included studies, three authors (A.A.I., N.A.M., and Y.W.) independently extracted information regarding country, year of publication, sample size, year of sample collection as well as reported HCV genotypes and subtypes. Data was also extracted for HCV genotypes reported without details of the subtypes. Disagreements were resolved by discussion and adjudication including a fourth author (R.H.S.). We converted HCV genotypes and variants published prior to 1994 to the consensus nomenclature proposed by Simmonds et al. [15] since a standardized genotype classification system was not available in the earlier years of HCV research. Where more than one article reported data from the same sample, record, or patient cohort, only one was counted and selected. Similarly, studies that genotyped and analyzed samples from more than one SEA country were categorized as “multi-country” rather than the individual countries included, and the data were extracted and analyzed in that form to avoid confusion. Data on ‘inconclusive’ or ‘untyped/undetermined’ genotypes and/or subtypes were neither extracted nor analyzed. Cases of more than one genotype and/or subtype from a single patient were tagged ‘mixed.’ Although identified, these mixed genotype data were excluded from the genotype and subtype analysis. In the case of immigrant workers, irrespective of their original countries, their data were pooled to reflect the countries they were working in when their samples were collected.
Data analysis was conducted using OpenMeta Analyst and meta (version 4.15–1) and metafor (version 2.4–0) packages of R (version 4.0.3) and RStudio (version 1.3.1093) [16]. The pooled prevalence of HCV genotypes was calculated, and subgroup analysis was done according to country and period of data collection. Random-effect model using the DerSimonian-Laird method of meta-analysis was employed to determine the pooled estimates of the reported HCV genotype and subtype proportions. Data were transformed using the logit transformation. A forest plot was subsequently generated to visually summarize details of the individual studies alongside the estimated common effect and degree of heterogeneity. Publication bias was examined using funnel plots (visual aid for detecting bias) and Egger’s regression test. The heterogeneities (i.e., variation in study outcomes between studies) of study-level estimates were evaluated by Cochran’s Q test and quantified using I2 statistics. I2 values of 25%, 50%, and 75% were considered low, moderate, and high heterogeneity, respectively [17]. Subgroup meta-analysis was used to analyze sources of heterogeneity. Sensitivity test was conducted using the leave-one-out analysis. P value of < 0.001 was considered to be statistically significant for all tests. A protocol was not lodged for this study.
Results
Search results and eligible studies
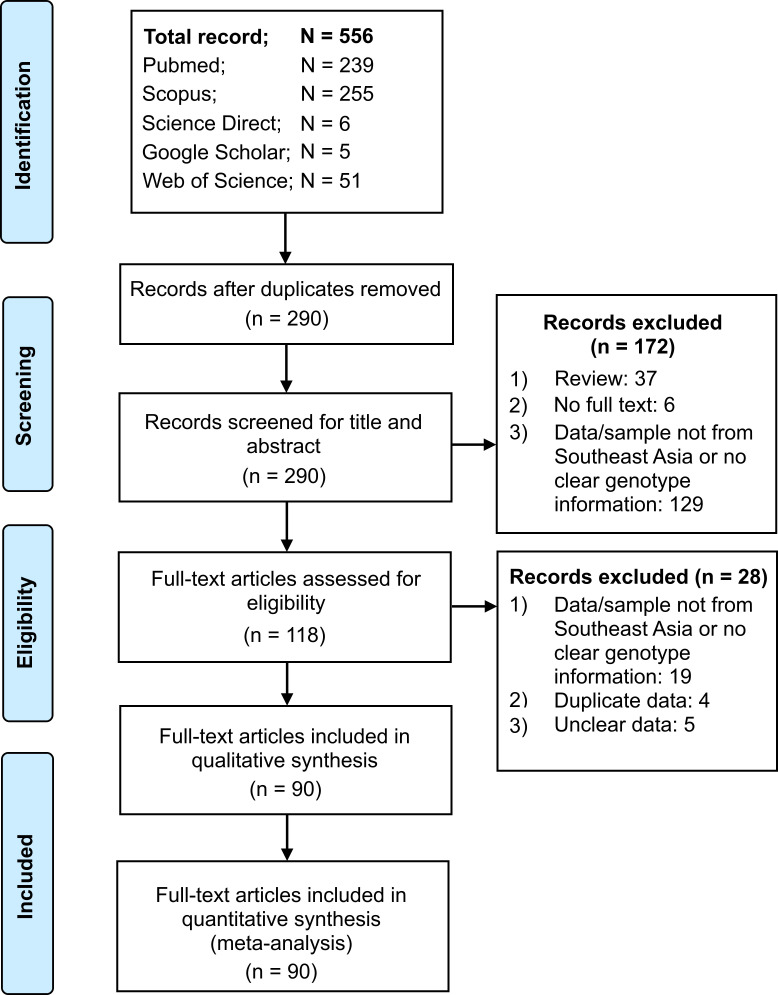
The article selection process for this study is shown in Fig 1. In brief, our search of five databases returned 556 records. After duplicate removal and exclusion of studies that did not meet the inclusion criteria, a total of 90 studies were eligible and thus included in quantitative synthesis (Fig 1).
Fig 1. Summary of article identification and selection process.
Characteristics of the eligible studies
All the eligible studies were of high methodological quality (S1 Table). Majority of the 90 studies included in this meta-analysis were reported from Thailand (n = 27). A total of 15,089 HCV genotypes were reported across the 90 studies and ranged from 6 (in Indonesia) to 3,020 (in Cambodia) (Table 1). Six HCV genotypes (genotype 1 to 6) were basically reported. The most targeted regions for genotyping analysis were the 5’UTR, core and NS5B regions, meanwhile, sequencing and phylogeny were the major genotyping methods utilized. While HCV genotype information was available for all included studies, only 69 studies provided data on HCV subtypes. No study met our search criteria for East Timor. As such, genotype data analyzed included studies from ten SEA countries. The reported genotypes and subtypes varied across countries.
Table 1. Characteristics of the selected studies reporting HCV genotypes in Southeast Asia.
| No. | Author | Country | Target region | Genotyping method | Total GT (n) | GT1 (n) | GT2 (n) | GT3 (n) | GT4 (n) | GT5 (n) | GT6 (n) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Chong et al., 2008 [18] | Brunei | – | sequencing | 7 | 3 | 1 | 3 | 0 | 0 | 0 |
| 2 | Budkowska et al., 2011 [19] | Cambodia | NS5B | phylogeny | 58 | 29 | 1 | 0 | 0 | 0 | 28 |
| 3 | De Weggheleire et al., 2017 [20] | Cambodia | – | LiPA | 87 | 46 | 4 | 1 | 0 | 0 | 36 |
| 4 | Lerolle et al., 2012 [21] * | Cambodia | NS5B | sequencing | 28 | 19 | 2 | 0 | 7 | 0 | 0 |
| 5 | Nouhin et al., 2019 [22] | Cambodia | NS5B | phylogeny | 3020 | 1444 | 134 | 0 | 0 | 0 | 1442 |
| 6 | Yamada et al., 2015 [23] | Cambodia | 5’NCR | – | 9 | 3 | 0 | 0 | 0 | 0 | 6 |
| 7 | Anggorowati et al., 2012 [24] | Indonesia | NS5B | phylogeny | 44 | 28 | 0 | 12 | 3 | 0 | 1 |
| 8 | Hadikusumo et al., 2016 [25] | Indonesia | NS5B | phylogeny | 6 | 2 | 0 | 4 | 0 | 0 | 0 |
| 9 | Hadiwandowo et al., 1994 [26] * | Indonesia | core | PCR | 81 | 39 | 24 | 17 | 0 | 1 | 0 |
| 10 | Handajani et al., 2019 [27] | Indonesia | 5’UTR, NS5B | phylogeny | 16 | 9 | 4 | 3 | 0 | 0 | 0 |
| 11 | Inoue et al., 2000 [28] # | Indonesia | 5’UTR | PCR, phylogeny | 57 | 38 | 12 | 7 | 0 | 0 | 0 |
| 12 | Juniastuti et al., 2014 [29] | Indonesia | 5’UTR, NS5B | sequencing | 99 | 57 | 2 | 39 | 1 | 0 | 0 |
| 13 | Kurniawan et al., 2018 [30] * | Indonesia | – | – | 274 | 199 | 30 | 32 | 12 | 0 | 1 |
| 14 | Lesmana et al., 1996 [31] | Indonesia | 5’NCR, core | PCR | 104 | 70 | 33 | 1 | 0 | 0 | 0 |
| 15 | Prasetyo et al., 2013 [32] | Indonesia | E1/E2, NS5B | phylogeny | 29 | 19 | 0 | 8 | 2 | 0 | 0 |
| 16 | Prasetyo et al., 2017 [33] | Indonesia | E1/E2 | phylogeny | 12 | 9 | 0 | 2 | 1 | 0 | 0 |
| 17 | Prasetyo et al., 2018 [34] | Indonesia | E1/E2 | phylogeny | 18 | 14 | 0 | 3 | 1 | 0 | 0 |
| 18 | Rinonce et al., 2013 [35] | Indonesia | NS5B | phylogeny | 100 | 98 | 0 | 2 | 0 | 0 | 0 |
| 19 | Sheng et al., 1994 [36] # | Indonesia | – | PCR | 66 | 51 | 13 | 0 | 2 | 0 | 0 |
| 20 | Soetjipto et al., 1996 [37] | Indonesia | 5’UTR, NS5B | sequencing | 80 | 60 | 18 | 1 | 1 | 0 | 0 |
| 21 | Tokita et al., 1996 [38] | Indonesia | core, NS5B | PCR, sequencing, phylogeny | 126 | 70 | 42 | 1 | 0 | 0 | 13 |
| 22 | Utama et al., 2008 [39] | Indonesia | 5’UTR, NS5B | phylogeny | 104 | 64 | 21 | 18 | 1 | 0 | 0 |
| 23 | Utama et al., 2010 [40] | Indonesia | 5’UTR, core, NS5B | phylogeny | 150 | 109 | 24 | 17 | 0 | 0 | 0 |
| 24 | Hübschen et al., 2011 [41] # | Laos | core/E1, NS5B | phylogeny | 45 | 2 | 0 | 0 | 0 | 0 | 43 |
| 25 | Hairul et al., 2012 [42] *,# | Malaysia | 5’UTR, NS5B | phylogeny | 28 | 7 | 0 | 19 | 1 | 0 | 1 |
| 26 | Ho et al., 2015 [43] * | Malaysia | – | linear array GT strip | 396 | 142 | 7 | 245 | 0 | 0 | 2 |
| 27 | Mohamed et al., 2013 [44] | Malaysia | NS5B | phylogeny | 37 | 10 | 0 | 27 | 0 | 0 | 0 |
| 28 | Ng et al., 2015 [45] | Malaysia | 5’UTR, NS5B | phylogeny | 126 | 52 | 0 | 72 | 0 | 0 | 2 |
| 29 | Tan et al., 2015 [46] * | Malaysia | – | LiPA | 45 | 12 | 0 | 33 | 0 | 0 | 0 |
| 30 | Zheng et al., 1996 [47] # | Malaysia | 5’NCR, core | PCR, sequencing | 7 | 4 | 0 | 3 | 0 | 0 | 0 |
| 31 | Bwa et al., 2019 [48] * | Myanmar | – | LiPA | 158 | 24 | 0 | 80 | 0 | 0 | 54 |
| 32 | Lwin et al., 2007 [49] | Myanmar | core, NS5B | phylogeny | 145 | 16 | 1 | 57 | 0 | 0 | 71 |
| 33 | Naing et al., 2015 [50] * | Myanmar | NS | PCR | 350 | 102 | 4 | 178 | 0 | 0 | 66 |
| 34 | Nakai et al., 2001 [51] | Myanmar | 5’UTR | PCR | 22 | 4 | 0 | 18 | 0 | 0 | 0 |
| 35 | Shinji et al., 2004 [52] | Myanmar | NS5B | phylogeny | 110 | 35 | 0 | 52 | 0 | 0 | 23 |
| 36 | Ye et al., 2019 [53] | Myanmar | core/E2, NS5B | phylogeny | 21 | 3 | 0 | 9 | 0 | 0 | 9 |
| 37 | Agdamag et al., 2005 [54] | Philippines | NS5B | phylogeny | 23 | 15 | 8 | 0 | 0 | 0 | 0 |
| 38 | Katayama et al., 1996 [55] | Philippines | 5’UTR, NS5B | sequencing | 30 | 27 | 3 | 0 | 0 | 0 | 0 |
| 39 | Durier et al., 2017 [56] # | Multi-country | 5’UTR, NS5B | PCR | 373 | 223 | 2 | 97 | 8 | 0 | 43 |
| 40 | Greene et al., 1995 [57] | Multi-country | E1, E2/NS1, NS4, NS5 | sequencing | 57 | 42 | 2 | 13 | 0 | 0 | 0 |
| 41 | Yusrina et al., 2018 [58] # | Multi-country | 5’UTR, core, NS5B | LiPA, PCR, phylogeny | 95 | 28 | 5 | 41 | 0 | 0 | 21 |
| 42 | Choy et al., 2019 [59] * | Singapore | – | LiPA | 116 | 77 | 0 | 39 | 0 | 0 | 0 |
| 43 | Lee et al., 1994 [60] * | Singapore | 5’NCR | sequencing | 40 | 35 | 2 | 3 | 0 | 0 | 0 |
| 44 | Soh et al., 2019 [61] | Singapore | – | LiPA | 63 | 24 | 1 | 27 | 4 | 0 | 7 |
| 45 | Akkarathamrongsin et al., 2013 [62] | Thailand | core, NS5B | phylogeny | 354 | 124 | 2 | 157 | 0 | 0 | 71 |
| 46 | Akkarathamrongsin et al., 2011 [63] | Thailand | core | phylogeny | 39 | 8 | 0 | 14 | 0 | 0 | 17 |
| 47 | Avihingsanon et al., 2014 [64] *,# | Thailand | 5’UTR, core, NS5B | sequencing | 370 | 128 | 2 | 176 | 0 | 1 | 63 |
| 48 | Barusrux et al., 2012 [65] | Thailand | 5’UTR, core | sequencing | 7 | 4 | 0 | 3 | 0 | 0 | 0 |
| 49 | Barusrux et al., 2014 [66] | Thailand | 5’UTR, core | phylogeny | 101 | 16 | 0 | 76 | 0 | 0 | 9 |
| 50 | Boonyarad et al., 2003 [67] | Thailand | core | LiPA, RFLP, phylogeny | 7 | 2 | 0 | 5 | 0 | 0 | 0 |
| 51 | Chuenjitkulthaworn et al., 2019 [68] | Thailand | – | – | 20 | 10 | 0 | 9 | 0 | 0 | 1 |
| 52 | Hansurabhanon et al., 2002 [69] # | Thailand | – | RFLP | 282 | 55 | 0 | 217 | 0 | 0 | 10 |
| 53 | Jutavijittum et al., 2009 [70] | Thailand | core/E1 | sequencing | 124 | 35 | 0 | 50 | 0 | 0 | 39 |
| 54 | Kanistanon et al., 1997 [71] | Thailand | 5’UTR, core | RHA | 216 | 76 | 0 | 99 | 0 | 0 | 41 |
| 55 | Kumthip et al., 2014 [72] | Thailand | core | LiPA, sequencing | 158 | 49 | 0 | 86 | 0 | 0 | 23 |
| 56 | Luengrojanakul et al., 1994 [73] *,# | Thailand | core | PCR | 83 | 14 | 21 | 3 | 0 | 43 | 2 |
| 57 | Martin et al., 2019 [74] | Thailand | HVR1 | phylogeny | 218 | 94 | 0 | 93 | 0 | 0 | 31 |
| 58 | Nakahira et al., 1995 [75] # | Thailand | 5’NCR | PCR | 42 | 42 | 0 | 0 | 0 | 0 | 0 |
| 59 | Netski et al., 2004 [76] | Thailand | core/E1 | phylogeny | 161 | 52 | 0 | 66 | 0 | 0 | 43 |
| 60 | Sirinawasatien et al., 2019 [77] * | Thailand | core | sequencing | 216 | 62 | 0 | 110 | 0 | 0 | 44 |
| 61 | Sirinawasatien et al., 2020 [78] * | Thailand | core | sequencing | 185 | 74 | 0 | 70 | 0 | 0 | 41 |
| 62 | Sistayanarain et al., 2011 [79] # | Thailand | core | PCR | 100 | 28 | 0 | 40 | 0 | 0 | 32 |
| 63 | Smolders et al., 2018 [80] *,# | Thailand | – | – | 98 | 38 | 0 | 46 | 0 | 0 | 14 |
| 64 | Songsivilai et al., 1996 [81] | Thailand | 5’NCR | RHA | 8 | 3 | 0 | 5 | 0 | 0 | 0 |
| 65 | Sugiyama et al., 1995 [82]* | Thailand | 5’UTR, core, NS5 | PCR sequencing | 13 | 7 | 2 | 4 | 0 | 0 | 0 |
| 66 | Sunanchaikarn et al., 2007 [83] | Thailand | 5’UTR, core | sequencing | 45 | 15 | 2 | 24 | 0 | 0 | 4 |
| 67 | Theamboonlers et al., 2000 [84] | Thailand | 5’NCR, core | RFLP | 124 | 61 | 0 | 54 | 0 | 0 | 9 |
| 68 | Tokita et al., 1995 [85] # | Thailand | core | PCR, sequencing, phylogeny | 79 | 33 | 3 | 43 | 0 | 0 | 0 |
| 69 | Wasitthankasem et al., 2015 [86] | Thailand | core, NS5B | phylogeny | 588 | 191 | 3 | 271 | 0 | 0 | 123 |
| 70 | Wasitthankasem et al., 2016 [87] | Thailand | core, NS5B | phylogeny | 23 | 4 | 0 | 11 | 0 | 0 | 8 |
| 71 | Wasitthankasem et al., 2017 [88] | Thailand | core | phylogeny | 234 | 74 | 0 | 73 | 0 | 0 | 87 |
| 72 | Do et al., 2015 [89] | Vietnam | – | – | 9 | 1 | 1 | 1 | 0 | 0 | 6 |
| 73 | Dunford et al., 2012 [90] | Vietnam | core/E1, NS5B | phylogeny | 282 | 169 | 1 | 5 | 0 | 0 | 107 |
| 74 | Duong et al., 2016 [91] | Vietnam | – | genotyping kit | 14 | 10 | 0 | 0 | 0 | 0 | 4 |
| 75 | Duong et al., 2019 [92] | Vietnam | – | genotyping kit | 18 | 13 | 0 | 0 | 0 | 0 | 5 |
| 76 | Kakumu et al., 1998 [93] # | Vietnam | core | PCR | 42 | 22 | 8 | 2 | 0 | 0 | 0 |
| 77 | Le Ngoc et al., 2019 [94] | Vietnam | 5’UTR, core, NS5B | phylogeny | 322 | 182 | 24 | 7 | 0 | 0 | 109 |
| 78 | Li et al., 2014 [95] | Vietnam | core/E1, NS5B | phylogeny | 236 | 77 | 34 | 0 | 0 | 0 | 125 |
| 79 | Lioznov et al., 2016 [96] * | Vietnam | – | – | 1604 | 806 | 308 | 0 | 0 | 0 | 490 |
| 80 | Minh 2015 [97] * | Vietnam | NS5B | phylogeny | 390 | 111 | 61 | 2 | 0 | 0 | 216 |
| 81 | Nadol et al., 2016 [98] * | Vietnam | 5’UTR | phylogeny | 41 | 30 | 1 | 0 | 0 | 0 | 10 |
| 82 | Nguyen et al., 2016 [99] | Vietnam | core, NS5B | sequencing | 93 | 64 | 0 | 5 | 0 | 0 | 24 |
| 83 | Nguyen et al., 2018 [100] * | Vietnam | – | – | 82 | 57 | 0 | 3 | 0 | 0 | 22 |
| 84 | Noppornpanth et al., 2006 [101] *,# | Vietnam | core, NS5B | phylogeny | 58 | 22 | 6 | 0 | 0 | 0 | 30 |
| 85 | Pham et al., 2009 [102] | Vietnam | core, NS5B | phylogeny | 70 | 33 | 0 | 4 | 0 | 0 | 33 |
| 86 | Pham et al., 2011 [103] | Vietnam | NS5B | sequencing | 842 | 256 | 128 | 0 | 0 | 0 | 458 |
| 87 | Song et al., 1994 [104] # | Vietnam | core | PCR | 47 | 43 | 4 | 0 | 0 | 0 | 0 |
| 88 | Tanimoto et al., 2010 [105] | Vietnam | 5’UTR/core, NS5B | phylogeny | 114 | 75 | 1 | 10 | 0 | 0 | 28 |
| 89 | Tokita et al., 1994 [106] # | Vietnam | – | PCR, phylogeny | 47 | 43 | 4 | 0 | 0 | 0 | 0 |
| 90 | Tran et al., 2003 [107] | Vietnam | 5’UTR | PCR | 21 | 9 | 8 | 0 | 1 | 0 | 3 |
GT, genotype; LiPA, line probe assay; RFLP, restriction-fragment length polymorphism; RHA, reverse hybridization assay; Multi-country, more than one Southeast Asian country.
*Study did not present data on HCV subtypes or the data could not be extracted.
#Study reports cases of mixed genotype and/or subtypes.
Pooled prevalence of HCV genotypes in Southeast Asia
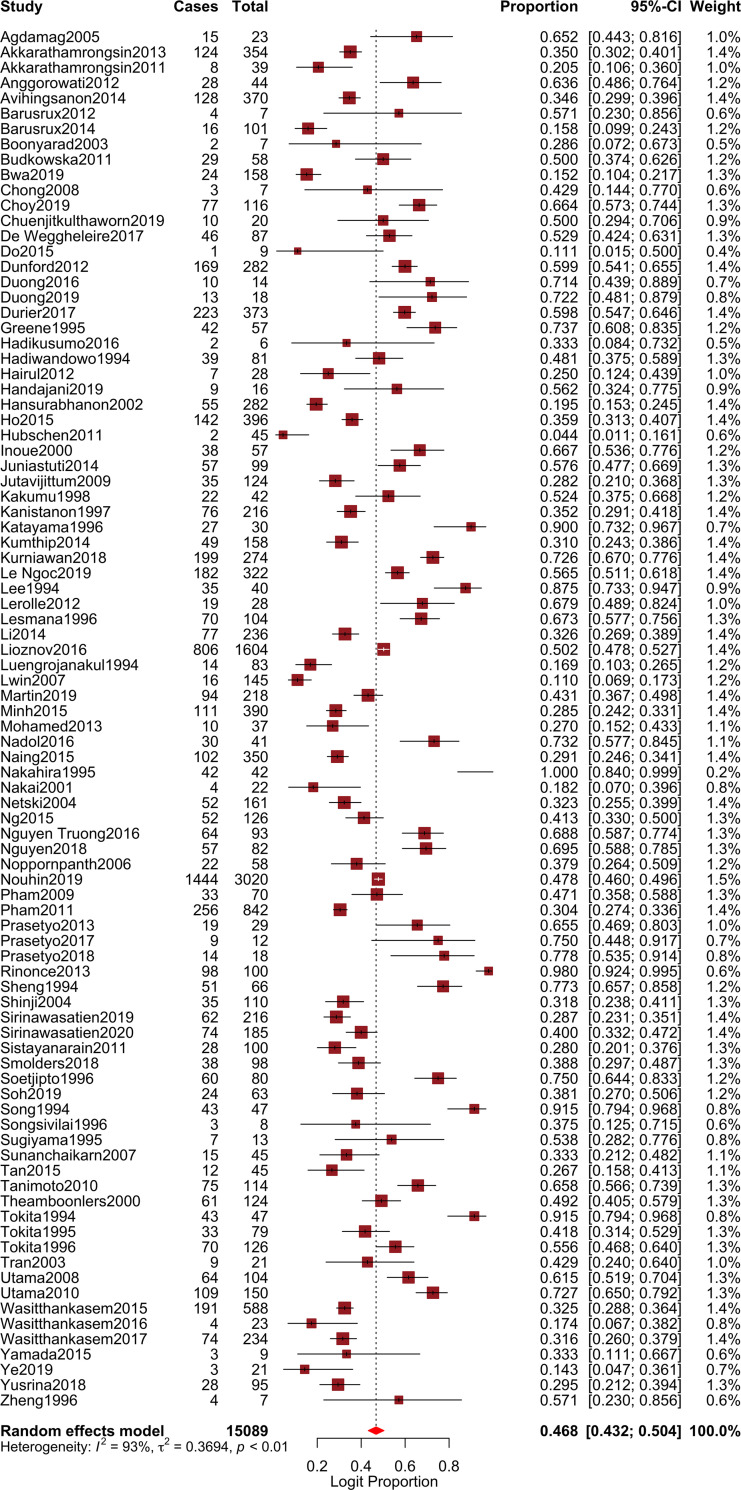
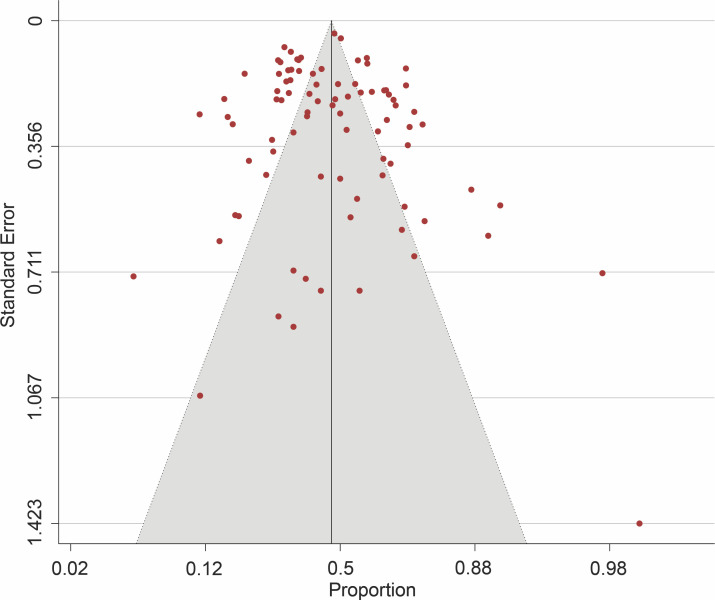
The pooled prevalence of genotype 1 (GT1) was estimated as 46.8% (95% CI, 43.2–50.4; I2 = 92.77%; p < 0.001) (Fig 2), genotype 2 (GT2) as 4.6% (95% CI, 3.5–5.9; I2 = 88.84%; p < 0.001), genotype 3 (GT3) as 23.1% (95% CI, 19.4–27.2; I2 = 93.03%; p < 0.001), genotype 4 (GT4) as 1.1% (95% CI, 0.7–1.5; I2 = 49.88%; p < 0.001), genotype 5 (GT5) as 0.8% (95% CI, 0.4–1.3; I2 = 79.25%; p < 0.001), and genotype 6 (GT6) as 16.5% (95% CI, 13.8–19.6; I2 = 93.98%; p < 0.001). Corresponding forest plots are shown in S1–S5 Figs. Except for genotype 4 with a moderate heterogeneity, the observed I2 statistics showed high heterogeneity (I2 > 75%) among studies reporting other HCV genotypes. Funnel plot for studies reporting the prevalence of HCV genotype 1 in Southeast Asia showed no publication bias (Fig 3) and Egger’s regression test revealed a non-significant p value (p = 0.495). However, there was evidence of publication bias in studies reporting other genotypes (Egger’s p < 0.0001) (S6–S10 Figs). Sensitivity assessment using the leave-one-out analysis did not reveal major changes to the estimate derived for all the HCV genotypes.
Fig 2. Forest plot showing pooled prevalence of HCV genotype 1 in Southeast Asia.
Fig 3. Funnel plot showing no publication bias for the studies reporting HCV genotype 1 in Southeast Asia.
Subgroup meta-analysis
Subgroup analysis was conducted to assess genotype distribution across the Southeast Asian countries and to identify possible source of heterogeneity among the studies. The result of subgroup meta-analysis by the distribution of HCV genotypes across countries revealed different degrees of variability in the studies (Table 2 and S11–S16 Figs). Overall, high genotype prevalence estimates were observed for genotypes 1, 3, and 6. For genotype 1, Philippines had the highest estimate (79.5%) although with fewer studies (n = 2) while Laos had the lowest (4.4%) with one study (Table 2). Studies from Malaysia and Cambodia showed low heterogeneity (I2 = 30.11% and 31.91%, respectively). For genotype 3, Malaysia had the highest estimate (63.1%) while Cambodia had the lowest (0.7%) (Table 2). Also, studies from Malaysia had low heterogeneity (I2 = 27.85%). For genotype 6, the highest estimate (95.6%) was from Laos, although with a single study, meanwhile, Indonesia and Malaysia had the lowest estimates (1.3% each). With the exception of Philippines and Malaysia, heterogeneity was of moderate to high in studies from other countries (Table 2).
Table 2. Subgroup analysis for comparison of genotype distribution across Southeast Asian countries.
| Country | Number of studies | Prevalence (%) | 95% CI | I2 (%) | Q | Heterogeneity test | |
|---|---|---|---|---|---|---|---|
| DF | p | ||||||
| Genotype 1 | |||||||
| Philippines | 2 | 79.5 | 45.6–94.7 | 77.16 | 4.378 | 1 | 0.036 |
| Thailand | 27 | 32.9 | 29.5–36.4 | 75.79 | 107.412 | 26 | < 0.001 |
| Indonesia | 17 | 62.7 | 61.4–72.6 | 73.77 | 61.005 | 16 | < 0.001 |
| Cambodia | 5 | 50.2 | 44.6–55.8 | 31.91 | 5.875 | 4 | 0.209 |
| Myanmar | 6 | 19.8 | 13.2–28.7 | 82.83 | 29.114 | 5 | < 0.001 |
| Brunei | 1 | 42.9 | 14.4–77.0 | – | – | – | – |
| Singapore | 3 | 65.7 | 38.3–85.5 | 91.71 | 24.122 | 2 | < 0.001 |
| Vietnam | 19 | 56.1 | 48.3–63.6 | 94.23 | 312.136 | 18 | < 0.001 |
| Multi-country* | 3 | 54.3 | 31.8–72.5 | 94.03 | 33.523 | 2 | < 0.001 |
| Malaysia | 6 | 34.9 | 29.5–40.7 | 30.11 | 7.154 | 5 | 0.209 |
| Laos | 1 | 4.4 | 1.1–16.1 | – | – | – | – |
| Overall | 90 | 46.8 | 43.2–50.4 | 92.77 | 1230.186 | 89 | < 0.001 |
| Genotype 2 | |||||||
| Philippines | 2 | 20.5 | 5.3–54.4 | 77.16 | 4.378 | 1 | 0.036 |
| Thailand | 27 | 1.1 | 0.5–2.6 | 82.46 | 148.233 | 26 | < 0.001 |
| Indonesia | 17 | 16.4 | 11.7–22.5 | 78.55 | 74.602 | 16 | < 0.001 |
| Cambodia | 5 | 4.4 | 3.8–5.2 | 0 | 1.417 | 4 | 0.841 |
| Myanmar | 6 | 1.0 | 0.5–2.1 | 0 | 1.851 | 5 | 0.869 |
| Brunei | 1 | 14.3 | 2.0–58.1 | – | – | – | – |
| Singapore | 3 | 2.2 | 0.6–7.8 | 28.18 | 2.785 | 2 | 0.248 |
| Vietnam | 19 | 10.6 | 7.9–14.0 | 80.40 | 91.830 | 18 | < 0.001 |
| Multi-country* | 3 | 2.3 | 0.6–8.6 | 74.12 | 7.729 | 2 | 0.021 |
| Malaysia | 6 | 1.7 | 0.9–3.1 | 0 | 2.081 | 5 | 0.838 |
| Laos | 1 | 1.1 | 0.1–15.1 | – | – | – | – |
| Overall | 90 | 4.6 | 3.5–5.9 | 88.84 | 797.228 | 89 | < 0.001 |
| Genotype 3 | |||||||
| Philippines | 2 | 1.8 | 0.3–11.9 | 0 | 0.017 | 1 | 0.897 |
| Thailand | 27 | 45.8 | 40.7–51.0 | 87.40 | 206.310 | 26 | < 0.001 |
| Indonesia | 17 | 13.0 | 8.4–19.6 | 83.56 | 97.306 | 16 | < 0.001 |
| Cambodia | 5 | 0.7 | 0.1–4.1 | 58.44 | 9.625 | 4 | 0.047 |
| Myanmar | 6 | 59.0 | 42.2–55.9 | 64.89 | 14.242 | 5 | 0.014 |
| Brunei | 1 | 42.9 | 14.4–77.0 | – | – | – | – |
| Singapore | 3 | 27.7 | 14.3–46.8 | 82.86 | 11.670 | 2 | 0.003 |
| Vietnam | 19 | 2.1 | 1.2–3.7 | 64.27 | 50.375 | 18 | < 0.001 |
| Multi-country* | 3 | 30.3 | 20.0–43.1 | 82.80 | 11.630 | 2 | 0.003 |
| Malaysia | 6 | 63.1 | 57.4–68.5 | 27.85 | 6.930 | 5 | 0.226 |
| Laos | 1 | 1.1 | 0.1–15.1 | – | – | – | – |
| Overall | 90 | 23.1 | 19.4–27.2 | 93.03 | 1277.648 | 89 | < 0.001 |
| Genotype 4 | |||||||
| Philippines | 2 | 1.8 | 0.3–11.9 | 0 | 0.017 | 1 | 0.897 |
| Thailand | 27 | 0.6 | 0.3–1.0 | 0 | 19.590 | 26 | 0.811 |
| Indonesia | 17 | 2.7 | 1.7–4.4 | 21.44 | 20.366 | 16 | 0.204 |
| Cambodia | 5 | 1.2 | 0.1–1.86 | 88.69 | 35.374 | 4 | < 0.001 |
| Myanmar | 6 | 0.6 | 0.2–1.7 | 0 | 3.158 | 5 | 0.676 |
| Brunei | 1 | 6.3 | 0.4–53.9 | – | – | – | – |
| Singapore | 3 | 2.2 | 0.4–11.8 | 52.26 | 4.189 | 2 | 0.123 |
| Vietnam | 19 | 0.6 | 0.3–1.2 | 11.77 | 20.402 | 18 | 0.311 |
| Multi-country* | 3 | 1.9 | 1.0–3.6 | 0 | 1.288 | 2 | 0.525 |
| Malaysia | 6 | 1.2 | 0.4–3.8 | 13.00 | 5.747 | 5 | 0.332 |
| Laos | 1 | 1.1 | 0.1–15.1 | – | – | – | – |
| Overall | 90 | 1.1 | 0.7–1.5 | 49.88 | 177.585 | 89 | < 0.001 |
| Genotype 5 | |||||||
| Philippines | 2 | 1.8 | 0.3–11.9 | 0 | 0.017 | 1 | 0.897 |
| Thailand | 27 | 0.8 | 0.2–2.8 | 89.12 | 238.885 | 26 | < 0.001 |
| Indonesia | 17 | 0.9 | 0.5–1.8 | 0 | 7.755 | 16 | 0.956 |
| Cambodia | 5 | 0.6 | 0.1–3.8 | 57.04 | 9.310 | 4 | 0.054 |
| Myanmar | 6 | 0.6 | 0.2–1.7 | 0 | 3.158 | 5 | 0.676 |
| Brunei | 1 | 6.3 | 0.4–53.9 | – | – | – | – |
| Singapore | 3 | 0.7 | 0.1–3.6 | 0 | 0.279 | 2 | 0.870 |
| Vietnam | 19 | 0.6 | 0.3–1.0 | 0 | 17.021 | 18 | 0.522 |
| Multi-country* | 3 | 0.4 | 0.1–1.9 | 0 | 0.933 | 2 | 0.627 |
| Malaysia | 6 | 0.9 | 0.3–2.9 | 0 | 4.451 | 5 | 0.486 |
| Laos | 1 | 1.1 | 0.1–15.1 | – | – | – | – |
| Overall | 90 | 0.8 | 0.4–1.3 | 79.25 | 428.906 | 89 | < 0.001 |
| Genotype 6 | |||||||
| Philippines | 2 | 1.8 | 0.3–11.9 | 0 | 0.017 | 1 | 0.897 |
| Thailand | 27 | 16.7 | 13.4–20.7 | 85.19 | 175.546 | 26 | < 0.001 |
| Indonesia | 17 | 1.3 | 0.6–3.1 | 57.39 | 37.553 | 16 | 0.002 |
| Cambodia | 5 | 45.7 | 36.7–54.9 | 61.09 | 10.280 | 4 | 0.036 |
| Myanmar | 6 | 29.0 | 18.1–43.0 | 90.90 | 54.930 | 5 | < 0.001 |
| Brunei | 1 | 6.3 | 0.4–59.3 | – | – | – | – |
| Singapore | 3 | 2.5 | 0.3–20.4 | 72.23 | 7.203 | 2 | 0.027 |
| Vietnam | 19 | 34.6 | 28.0–41.8 | 92.96 | 255.582 | 18 | < 0.001 |
| Multi-country* | 3 | 12.8 | 5.8–26.0 | 82.11 | 11.179 | 2 | 0.004 |
| Malaysia | 6 | 1.3 | 0.6–2.9 | 0 | 4.165 | 5 | 0.526 |
| Laos | 1 | 95.6 | 83.9–98.9 | – | – | – | – |
| Overall | 90 | 16.5 | 13.8–19.6 | 93.98 | 1479.448 | 89 | < 0.001 |
*Genotyped and analyzed samples from more than one Southeast Asian country.
Subgroup meta-analysis based on the period of data collection also revealed different degrees of variability in the studies (Table 3). Before year 2000, the prevalence estimates showed that genotype 1 (60.1%; 95% CI, 49.8–69.6; I2 = 91.50%) was the most prevalent HCV genotype followed by genotype 3 (15.5%; 95% CI, 9.8–23.6; I2 = 89.10%) (Table 3). Although with a reduced prevalence, the pooled estimates for year 2000–2009 also revealed genotype 1 (40.7%; 95% CI, 33.2–48.6; I2 = 93.05%) as the most dominant genotype. The estimate for genotype 3 (21.3%; 95% CI, 13.8–31.1; I2 = 94.52%) on the other hand was higher than the earlier decade. A high prevalence (20.5%; 95% CI, 13.9–29.2; I2 = 94.48%) was also observed for genotype 6 during the 2000–2009 study period. For the period of 2010–2020, the prevalence of genotypes 1 and 3 increased to 44.5% (95% CI, 40.1–49.0; I2 = 92.95) and 27.0% (95% CI, 21.9–32.9; I2 = 93.38) respectively, while genotype 6 decreased to 19.6% (95% CI, 15.8–24.1; I2 = 94.95). Except for genotype 4 and the periods between 2000–2020 of genotype 5, heterogeneity was generally high (I2 > 75%) in the study periods under consideration.
Table 3. Subgroup analysis for comparison of genotype distribution in Southeast Asia based on the period of data collection.
| Period of data collection | Number of studies | Prevalence (%) | 95% CI | I2 (%) | Q | Heterogeneity test | |
|---|---|---|---|---|---|---|---|
| DF | p | ||||||
| Genotype 1 | |||||||
| < 2000 | 22 | 60.1 | 49.8–69.6 | 91.50 | 246.973 | 21 | < 0.001 |
| 2000–2009 | 24 | 40.7 | 33.2–48.6 | 93.05 | 330.765 | 23 | < 0.001 |
| 2010–2020 | 44 | 44.5 | 40.1–49.0 | 92.95 | 609.605 | 43 | < 0.001 |
| Overall | 90 | 46.8 | 43.2–50.4 | 92.77 | 1230.186 | 89 | < 0.001 |
| Genotype 2 | |||||||
| < 2000 | 22 | 12.1 | 8.3–17.2 | 78.06 | 95.724 | 21 | < 0.001 |
| 2000–2009 | 24 | 4.2 | 2.4–7.0 | 82.94 | 134.849 | 23 | < 0.001 |
| 2010–2020 | 44 | 2.5 | 1.7–3.8 | 90.99 | 477.014 | 43 | < 0.001 |
| Overall | 90 | 4.6 | 3.5–5.9 | 88.84 | 797.228 | 89 | < 0.001 |
| Genotype 3 | |||||||
| < 2000 | 22 | 15.5 | 9.8–23.6 | 89.10 | 192.586 | 21 | < 0.001 |
| 2000–2009 | 24 | 21.2 | 13.8–31.1 | 94.52 | 419.506 | 23 | < 0.001 |
| 2010–2020 | 44 | 27.0 | 21.9–32.9 | 93.38 | 649.669 | 43 | < 0.001 |
| Overall | 90 | 23.1 | 19.4–27.2 | 93.03 | 1277.648 | 89 | < 0.001 |
| Genotype 4 | |||||||
| < 2000 | 22 | 1.2 | 0.7–2.0 | 0 | 9.890 | 21 | 0.980 |
| 2000–2009 | 24 | 1.0 | 0.4–2.4 | 67.89 | 71.628 | 23 | < 0.001 |
| 2010–2020 | 44 | 1.0 | 0.6–1.6 | 52.78 | 91.073 | 43 | < 0.00 |
| Overall | 90 | 1.1 | 0.7–1.5 | 49.88 | 177.585 | 89 | < 0.00 |
| Genotype 5 | |||||||
| < 2000 | 22 | 1.3 | 0.4–4.7 | 87.06 | 162.244 | 21 | < 0.001 |
| 2000–2009 | 24 | 0.7 | 0.4–1.2 | 0 | 14.872 | 23 | 0.899 |
| 2010–2020 | 44 | 0.6 | 0.4–0.9 | 0 | 43.161 | 43 | 0.464 |
| Overall | 90 | 0.8 | 0.4–1.3 | 79.25 | 428.906 | 89 | < 0.001 |
| Genotype 6 | |||||||
| < 2000 | 22 | 4.0 | 2.2–7.2 | 80.86 | 109.699 | 21 | < 0.001 |
| 2000–2009 | 24 | 20.5 | 13.9–29.2 | 94.48 | 416.986 | 23 | < 0.001 |
| 2010–2020 | 44 | 19.6 | 15.8–24.1 | 94.95 | 852.257 | 43 | < 0.001 |
| Overall | 90 | 16.5 | 13.8–19.6 | 93.98 | 1479.449 | 89 | < 0.001 |
Distribution of HCV subtypes in Southeast Asia
In this study, a total of 9,646 HCV subtypes were reported across the 69 studies and ranged from 3 (in Malaysia) to 2,950 (in Cambodia). Overall, data on HCV subtypes were available in studies from Brunei (n = 1), Cambodia (n = 4), Indonesia (n = 15), Laos (n = 1), Malaysia (n = 3), Myanmar (n = 4), Philippines (n = 2), Singapore (n = 1), Thailand (n = 21), and Vietnam (n = 14). Three other studies that contributed to the subtype data were studies categorized as “multi-country” (Table 1 and S3 File). The following subtypes were reported: subtype of genotype 1 (1a-1e), 2 (2a-2c, 2e, 2f, 2i-2k, and 2m), 3 (3a-3c, 3g, 3k), 4 (4a), and 6 (6a-6c, 6e, 6f, 6h-6v, 6xa-6xc, 6xf) (Table 4).
Table 4. Prevalence of HCV subtypes in Southeast Asia.
| HCV subtype | Pooled prevalence (%) | 95% CI |
|---|---|---|
| 1a | 21.3 | 17.0–26.4 |
| 1b | 26.3 | 22.1–30.9 |
| 1c | 2.0 | 1.3–3.1 |
| 1d | 0.9 | 0.6–1.3 |
| 1e | 0.9 | 0.7–1.2 |
| 2a | 3.4 | 2.4–4.8 |
| 2b | 1.1 | 0.7–1.8 |
| 2c | 1.1 | 0.7–1.5 |
| 2e | 1.2 | 0.9–1.7 |
| 2f | 1.0 | 0.7–1.3 |
| 2i | 1.1 | 0.8–1.5 |
| 2j | 0.9 | 0.6–1.2 |
| 2k | 0.9 | 0.6–1.2 |
| 2m | 1.1 | 0.8–1.5 |
| 3a | 14.3 | 11.0–18.5 |
| 3b | 3.6 | 2.5–5.1 |
| 3c | 0.9 | 0.7–1.3 |
| 3g | 0.9 | 0.7–1.3 |
| 3k | 1.4 | 0.9–2.3 |
| 4a | 1.2 | 0.8–1.6 |
| 6a | 2.8 | 1.9–4.1 |
| 6b | 1.1 | 0.8–1.6 |
| 6c | 0.9 | 0.7–1.2 |
| 6e | 2.6 | 1.8–3.6 |
| 6f | 1.8 | 1.2–2.7 |
| 6h | 1.1 | 0.8–1.6 |
| 6i | 1.5 | 1.1–2.0 |
| 6j | 1.1 | 0.8–1.5 |
| 6k | 0.9 | 0.7–1.3 |
| 6l | 1.3 | 0.9–1.8 |
| 6m | 1.1 | 0.7–1.6 |
| 6n | 1.6 | 1.0–2.5 |
| 6o | 1.0 | 0.7–1.2 |
| 6p | 1.7 | 1.4–2.1 |
| 6q | 1.1 | 0.8–1.6 |
| 6r | 1.1 | 0.7–1.8 |
| 6s | 1.6 | 1.3–1.9 |
| 6t | 0.9 | 0.6–1.2 |
| 6u | 0.8 | 0.6–1.1 |
| 6v | 0.9 | 0.6–1.2 |
| 6xa | 1.0 | 0.7–1.5 |
| 6xb | 0.9 | 0.6–1.2 |
| 6xc | 0.9 | 0.6–1.2 |
| 6xf | 1.6 | 1.3–2.0 |
The three most prevalent HCV subtypes in the Southeast Asian region were 1b (26.3%; 95% CI, 22.1–30.9), 1a (21.3%; 95% CI, 17.0–26.4), and 3a (14.3%; 95% CI, 11.0–18.5). The prevalence of other reported subtypes was less than 4% (Table 4).
Mixed genotypes and/or subtypes were identified in reports from some of the countries. They include Indonesia [(1b+2b+3b), (1a+1b), and (3a+3b)], Malaysia [(1+3), (3+4) and (1a+1b)], Thailand [(1+2), (1+3), (1+4), (1+5), (2+5), (3+6), (1+3+4), (1a+1b), (1a+3b), (1b+3a), (3a+3b) and (3a+6)], Vietnam [(1a+1b), (1a+2b), (1b+2a), (1a+1b+2b) and (1a+2b+3a)], and Multi-country (1a+3) (Table 1).
Discussion
The genotypic diversity of HCV has for several years been a major hurdle to drug and vaccine development. While there are effective antiviral drugs against HCV, there is yet to be a licensed vaccine. Thus, antiviral therapy remains the mainstay of managing HCV infection. Numerous studies have demonstrated that different antiviral compounds and drugs (including the more recent interferon-free direct-acting antiviral agents such as telaprevir, sofosbuvir, etc.) display variable antiviral activities against the infecting genotypes and subtypes of HCV [108–114], a concern largely attributed to resistance associated substitutions in HCV [115, 116]. A clear knowledge of the actual distribution of HCV genotype and subtypes in a region is pivotal not only to the treatment of the infection and the potential selection of candidate genotypic vaccine target for the region, but also to the development of appropriate government policies, interventions and programs.
In this study, we present a detailed and comprehensive view of HCV genotype distribution in SEA from available published regional data since the discovery of the virus in 1989. Apart from East Timor, genotype data was available for all the countries in this region. Most of the studies were from Thailand, Vietnam and Indonesia; possibly because of the burden of HCV infection [117–119] in relation to their high population. Similar to many other nations across the globe, we identified genotype 1 as the most dominant genotype in SEA. While this finding does not contradict the popular notion that genotype 6 is prevalent in SEA as supported by the extensive global study conducted by Messina et al. [4], it underscores the need for a cautious interpretation of results of studies conducted at global and regional levels. It is pertinent to clarify that genotype 6 is more prevalent in SEA than any other region in the world when discussed according to global prevalence. However, within SEA, genotype 1 is the most prevalent, followed by genotypes 3 and 6, respectively (Table 2). Combination of the pooled prevalence of these genotypes (1,3 and 6) accounted for more than 85% of the genotypes reported in the region, indicating that the three genotypes are the major HCV genotypes of concern. Notably, we found that the highest prevalence for genotype 1 (79.5%; 95% CI, 45.6–94.7), and genotype 2 (20.5; 95% CI, 5.3–54.4) occurred in Philippines and with an even distribution of the other genotypes. In addition, we observed that genotype 6 became more prevalent from the first decade of the twenty-first century. As suggested by previous reports [4, 118], our study showed that genotypes 4 and 5 were almost inexistent in SEA as only few isolated cases have been reported.
Although the prevalence of HCV subtypes appears to vary in this study, we identified more than forty different subtypes (Table 3), indicating the presence of a marked diversity of the virus’ subtypes in the region. Overall, subtypes 1b (26.3%; 95% CI, 22.1–30.9), 1a (21.3%; 95% CI, 17.0–26.4), and 3a (14.3%; 95% CI, 11.0–18.5) constitute the major subtypes in SEA identified by this study. The predominance of subtype 1b corroborates earlier reports on the subtype as the predominant HCV subtype across the globe [4, 11]. In addition to the cases of dual ‘mixed’ genotype infections identified in this study, several cases involving a mixture of more than two genotypes and/or subtypes in patients were reported in Indonesia, Vietnam, and Thailand [28, 80, 93]. Expectedly, such infected individuals would require more than the usual combinations of the available HCV drugs for optimal treatment outcomes. Given the high cost of treatment, the occurrence of ‘mixed’ genotypes and subtypes revealed in this study unveils an additional hurdle that should be considered in the current race for a vaccine, as agents capable of generating cross-reactive immunity would be highly invaluable.
This study however has its limitations. There was paucity of studies in some of the included SEA countries. For example, the estimates for Brunei and Laos were derived from one study each. This could amount to an underestimation of genotypes and subtypes. Another concern is the variations in the methods of genotyping, as well as the targeted region for genotyping. Furthermore, because we prioritized peer-reviewed publications in an attempt to limit the tendency of including studies with unreliable genotype reports, we could have missed some relevant studies. Nevertheless, a broad search of multiple databases was done to access as many studies as possible in order to curtail the impact of overlooked studies. Despite these limitations, we believe this study provides a comprehensive and up-to-date situation of HCV genotype distribution in SEA.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOC)
(PDF)
(PDF)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by Universiti Sains Malaysia in the form of grants awarded to RHS (304.PPSP.6316338, 304.PPSP.6316148). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. AAI and YW also acknowledge support from Universiti Sains Malaysia via the USM Fellowship Scheme.
References
- 1.Foung SKH, Baumert TF. Editorial: Current Progress and Challenges in the Development of a B Cell Based Hepatitis C Virus Vaccine. Front Immunol. 2018;9: 2577. 10.3389/fimmu.2018.02577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li D, Huang Z, Zhong J. Hepatitis C virus vaccine development: Old challenges and new opportunities. Natl Sci Rev. 2015;2: 285–295. 10.1093/nsr/nwv040 [DOI] [Google Scholar]
- 3.Lohmann V, Körner F, Koch J-O, Herian U, Theilmann L, Bartenschlager R. Replication of Subgenomic Hepatitis C Virus RNAs in a Hepatoma Cell Line. Science. 1999;285: 110–113. 10.1126/science.285.5424.110 [DOI] [PubMed] [Google Scholar]
- 4.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61: 77–87. 10.1002/hep.27259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weck K. Molecular methods of hepatitis C genotyping. Expert Rev Mol Diagn. 2005;5: 507–520. 10.1586/14737159.5.4.507 [DOI] [PubMed] [Google Scholar]
- 6.Cai Q, Zhao Z, Liu Y, Shao X, Gao Z. Comparison of three different HCV genotyping methods: Core, NS5B sequence analysis and line probe assay. Int J Mol Med. 2013;31: 347–352. 10.3892/ijmm.2012.1209 [DOI] [PubMed] [Google Scholar]
- 7.Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42: 962–973. 10.1002/hep.20819 [DOI] [PubMed] [Google Scholar]
- 8.Wyles DL, Luetkemeyer AF. Understanding hepatitis C virus drug resistance: clinical implications for current and future regimens. Top Antivir Med. 2017;25: 103–109. [PMC free article] [PubMed] [Google Scholar]
- 9.Hadziyannis SJ, Sette H, Morgan TR, Balan V, Diago M, Marcellin P, et al. Peginterferon-α2a and Ribavirin Combination Therapy in Chronic Hepatitis C. Ann Intern Med. 2004;140: 346–355. 10.7326/0003-4819-140-5-200403020-00010 [DOI] [PubMed] [Google Scholar]
- 10.Forns X, Maluenda MD, López-Labrador FX, Ampurdanès S, Olmedo E, Costa J, et al. Comparative study of three methods for genotyping hepatitis C virus strains in samples from Spanish patients. J Clin Microbiol. 1996;34: 2516–2521. 10.1128/JCM.34.10.2516-2521.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61: S45–57. 10.1016/j.jhep.2014.07.027 [DOI] [PubMed] [Google Scholar]
- 12.Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, et al. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011;31: 61–80. 10.1111/j.1478-3231.2011.02540.x [DOI] [PubMed] [Google Scholar]
- 13.Nguyen LH, Nguyen MH. Systematic review: Asian patients with chronic hepatitis C infection. Aliment Pharmacol Ther. 2013;37: 921–936. 10.1111/apt.12300 [DOI] [PubMed] [Google Scholar]
- 14.Munn Z, Moola S, Lisy K, Riitano D, Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int J Evid Based Healthc. 2015;13: 147–153. 10.1097/XEB.0000000000000054 [DOI] [PubMed] [Google Scholar]
- 15.Simmonds P, Alberti A, Alter HJ, Bonino F, Bradley DW, Brechot C, et al. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994;19: 1321–1324. [PubMed] [Google Scholar]
- 16.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36: 1–48. 10.18637/jss.v036.i03 [DOI] [Google Scholar]
- 17.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21: 1539–1558. 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 18.Chong VH, Zinna HS. Hepatitis C virus infection and haemodialysis: Experience of a district general hospital in Brunei Darussalam. Singapore Med J. 2008;49: 916–920. [PubMed] [Google Scholar]
- 19.Budkowska A, Kakkanas A, Nerrienet E, Kalinina O, Maillard P, Horm S V, et al. Synonymous mutations in the core gene are linked to unusual serological profile in hepatitis C virus infection. PLoS One. 2011;6. 10.1371/journal.pone.0015871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Weggheleire A, An S, De Baetselier I, Soeung P, Keath H, So V, et al. A cross-sectional study of hepatitis C among people living with HIV in Cambodia: Prevalence, risk factors, and potential for targeted screening. PLoS One. 2017;12(8): e0183530. 10.1371/journal.pone.0183530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerolle N, Limsreng S, Fournier-Nicolle I, Ly S, Nouhin J, Guillard B, et al. High frequency of advanced hepatic disease among HIV/HCV Co-infected patients in Cambodia: The HEPACAM study (ANRS 12267). J AIDS Clin Res. 2012;3:6. 10.4172/2155-6113.1000161 [DOI] [Google Scholar]
- 22.Nouhin J, Iwamoto M, Prak S, Dousset JP, Phon K, Heng S, et al. Molecular epidemiology of hepatitis C virus in Cambodia during 2016–2017. Sci Rep. 2019;9: 7314. 10.1038/s41598-019-43785-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada H, Fujimoto M, Svay S, Lim O, Hok S, Goto N, et al. Seroprevalence, genotypic distribution and potential risk factors of hepatitis B and C virus infections among adults in Siem Reap, Cambodia. Hepatol Res. 2015;45: 480–487. 10.1111/hepr.12367 [DOI] [PubMed] [Google Scholar]
- 24.Anggorowati N, Yano Y, Heriyanto DS, Rinonce HT, Utsumi T, Mulya DP, et al. Clinical and virological characteristics of hepatitis B or C virus co-infection with HIV in Indonesian patients. J Med Virol. 2012;84: 857–865. 10.1002/jmv.23293 [DOI] [PubMed] [Google Scholar]
- 25.Hadikusumo AA, Utsumi T, Amin M, Khairunisa SQ, Istimagfirah A, Wahyuni RM, et al. High rates of hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus infections and uncommon HBV genotype/subtype and HCV subtype distributions among transgender individuals in Surabaya, Indonesia. Japanese Journal of Infectious Diseases. 2016;69:493–499. 10.7883/yoken.JJID.2015.384 [DOI] [PubMed] [Google Scholar]
- 26.Hadiwandowo S, Tsuda F, Okamoto H, Tokita H, Wang Y, Tanaka T, et al. Hepatitis B virus subtypes and hepatitis C virus genotypes in patients with chronic liver disease or on maintenance hemodialysis in Indonesia. J Med Virol. 1994;43: 182–186. 10.1002/jmv.1890430216 [DOI] [PubMed] [Google Scholar]
- 27.Handajani R, Wungu CDK, Humairah I, Prabowo GI, Cholili U, Amin M, et al. Detection of Hepatitis C Virus (Hcv) Infection and Its Genotype in Patients at Hepatology Outpatient Clinic, Dr Soetomo General Hospital, Surabaya. IOP Conf Ser Earth Environ Sci. 2019;217. 10.1088/1755-1315/217/1/012051 [DOI] [Google Scholar]
- 28.Inoue Y, Sulaiman HA, Matsubayashi K, Julitasari, Iinuma K, Ansari A, et al. Genotypic analysis of hepatitis C virus in blood donors in Indonesia. Am J Trop Med Hyg. 2000;62: 92–98. 10.4269/ajtmh.2000.62.92 [DOI] [PubMed] [Google Scholar]
- 29.Juniastuti, Utsumi T, Nasronudin, Alimsardjono L, Amin M, Adianti M, et al. High rate of seronegative HCV infection in HIV-positive patients. Biomed reports. 2014;2: 79–84. 10.3892/br.2013.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurniawan J, Gani RA, Hasan I, Sulaiman AS, Lesmana CRA, Jasirwan COM, et al. Comparative efficacy of sofosbuvir-ribavirin versus sofosbuvir-daclatasvir for treatment of chronic hepatitis C in an area with limited NS5A inhibitor availability. Indian J Gastroenterol. 2018;37: 520–525. 10.1007/s12664-018-0921-2 [DOI] [PubMed] [Google Scholar]
- 31.Lesmana LA, Sulaiman HA, Noer HMS, Tsuda F, Okamoto H. Hepatitis C virus genotypes and co-infection with GB virus C in patients with anti-HCV-positive chronic liver disease in Jakarta, Indonesia. Int Hepatol Commun. 1996;6: 16–23. 10.1016/S0928-4346(96)00321-0 [DOI] [Google Scholar]
- 32.Prasetyo AA, Dirgahayu P, Sari Y, Hudiyono, Kageyama S. Molecular epidemiology of HIV, HBV, HCV, and HTLV-1/2 in drug abuser inmates in central Javan prisons, Indonesia. J Infect Dev Ctries. 2013;7: 453–467. 10.3855/jidc.2965 [DOI] [PubMed] [Google Scholar]
- 33.Prasetyo AA, Sari Y, Dharmawan R, Marwoto. Molecular status of human immunodeficiency virus, hepatitis B virus, and hepatitis C virus among transgender commercial sex workers in Surakarta, Indonesia. AIP Conf Proc. 2017;1813. 10.1063/1.4975949 [DOI] [Google Scholar]
- 34.Prasetyo AA, Marwoto, Adnan ZA, Hartono. Molecular status of Human Immunodeficiency Virus, Hepatitis B virus, and Hepatitis C virus among injecting drug male commercial sex workers in Surakarta, Indonesia. J Phys Conf Ser. 2018;1022. 10.1088/1742-6596/1022/1/012044 [DOI] [Google Scholar]
- 35.Rinonce HT, Yano Y, Utsumi T, Heriyanto DS, Anggorowati N, Widasari DI, et al. Hepatitis B and C virus infection among hemodialysis patients in yogyakarta, Indonesia: Prevalence and molecular evidence for nosocomial transmission. J Med Virol. 2013;85: 1348–1361. 10.1002/jmv.23581 [DOI] [PubMed] [Google Scholar]
- 36.Sheng L, Willems M, Widjaja S, Ali S, Simon S, Sulaiman A, et al. Geographical differences in HCV genotype distribution between Belgium and Indonesia. J Hepatol. 1994;21: S124. 10.1016/S0168-8278(05)81124-9 [DOI] [Google Scholar]
- 37.Soetjipto, Handajani R, Lusida MI, Darmadi S, Adi P, Soemarto, et al. Differential prevalence of hepatitis C virus subtypes in healthy blood donors, patients on maintenance hemodialysis, and patients with hepatocellular carcinoma in Surabaya, Indonesia. J Clin Microbiol. 1996;34: 2875–2880. 10.1128/JCM.34.12.2875-2880.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokita H, Okamoto H, Lizuka H, Kishimoto J, Tsuda F, Lesmana LA, et al. Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J Gen Virol. 1996;77: 293–301. 10.1099/0022-1317-77-2-293 [DOI] [PubMed] [Google Scholar]
- 39.Utama A, Budiarto BR, Monasari D, Octavia TI, Chandra IS, Gani RA, et al. Hepatitis C virus genotype in blood donors and associated liver disease in Indonesia. Intervirology. 2008;51: 410–416. 10.1159/000205515 [DOI] [PubMed] [Google Scholar]
- 40.Utama A, Tania NP, Dhenni R, Gani RA, Hasan I, Sanityoso A, et al. Genotype diversity of hepatitis C virus (HCV) in HCV-associated liver disease patients in Indonesia. Liver Int. 2010;30: 1152–1160. 10.1111/j.1478-3231.2010.02280.x [DOI] [PubMed] [Google Scholar]
- 41.Hübschen JM, Jutavijittum P, Thammavong T, Samountry B, Yousukh A, Toriyama K, et al. High genetic diversity including potential new subtypes of hepatitis C virus genotype 6 in Lao People’s Democratic Republic. Clin Microbiol Infect. 2011;17: E30–E34. 10.1111/j.1469-0691.2011.03665.x [DOI] [PubMed] [Google Scholar]
- 42.Hairul Aini H, Mustafa MIA, Seman MR, Nasuruddin BA. Mixed-genotypes infections with hepatitis C virus in hemodialysis subjects. Med J Malaysia. 2012;67: 199–203. [PubMed] [Google Scholar]
- 43.Ho SH, Ng KP, Kaur H, Goh KL. Genotype 3 is the predominant hepatitis C genotype in a multi-ethnic Asian population in Malaysia. Hepatobiliary Pancreat Dis Int. 2015;14: 281–286. 10.1016/s1499-3872(15)60363-0 [DOI] [PubMed] [Google Scholar]
- 44.Mohamed NA, Rashid ZZ, Wong KK, Abdullah SA, Rahman MM. Hepatitis C genotype and associated risks factors of patients at University Kebangsaan Malaysia Medical Centre. Pakistan J Med Sci. 2013;29: 1142–1146. 10.12669/pjms.295.3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng KT, Takebe Y, Chook JB, Chow WZ, Chan KG, Abed Al-Darraji HA, et al. Co-infections and transmission networks of HCV, HIV-1 and HPgV among people who inject drugs. Sci Rep. 2015;5: 15198. 10.1038/srep15198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan SS, Leong CL, Lee CKC. Treating hepatitis c in HIV/HCV co-infected patients in Malaysia- the outcomes and challenges. Med J Malaysia. 2015;70: 281–287. [PubMed] [Google Scholar]
- 47.Zheng W-Y, Hasebe F, Ali A, Sinniah M, Saraswathy TS, Ooi BG, et al. Genotype determination of hepatitis C virus strains in Malaysia. Trop Med. 1996;38: 51–60. [Google Scholar]
- 48.Bwa AH, Nangia G, Win STS, Maung ST, Han KAW, Htar SS, et al. Strategy and Efficacy of Generic and Pan-genotypic Sofosbuvir/Velpatasvir in Chronic Hepatitis C Virus: A Myanmar Experience. J Clin Exp Hepatol. 2019;9: 283–293. 10.1016/j.jceh.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lwin AA, Shinji T, Khin M, Win N, Obika M, Okada S, et al. Hepatitis C virus genotype distribution in Myanmar: Predominance of genotype 6 and existence of new genotype 6 subtype. Hepatol Res. 2007;37: 337–345. 10.1111/j.1872-034X.2007.00053.x [DOI] [PubMed] [Google Scholar]
- 50.Naing C, Sitt T, Aung AT, Aung K. Sustained virologic response to a dual peginterferon alfa-2a and ribavirin in treating chronic hepatitis c infection. Med. 2015;94: e1234. 10.1097/MD.0000000000001234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakai K, Win KM, Oo SS, Arakawa Y, Abe K. Molecular characteristic-based epidemiology of hepatitis B, C, and E viruses and GB virus C/hepatitis G virus in Myanmar. J Clin Microbiol. 2001;39: 1536–1539. 10.1128/JCM.39.4.1536-1539.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinji T, Yi YK, Gokan K, Tanaka Y, Ochi K, Kusano N, et al. Analysis of HCV genotypes from blood donors shows three new HCV type 6 subgroups exist in Myanmar. Acta Med Okayama. 2004;58: 135–142. 10.18926/AMO/32110 [DOI] [PubMed] [Google Scholar]
- 53.Ye M, Chen X, Wang Y, Duo L, Zhang C, Zheng Y-T. Identification of a new HCV subtype 6xg among injection drug users in kachin, myanmar. Front Microbiol. 2019;10: 814. 10.3389/fmicb.2019.00814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agdamag DM, Kageyama S, Alesna ET, Solante RM, Leaño PS, Heredia AML, et al. Rapid spread of hepatitis C virus among injecting-drug users in the Philippines: Implications for HIV epidemics. J Med Virol. 2005;77: 221–226. 10.1002/jmv.20439 [DOI] [PubMed] [Google Scholar]
- 55.Katayama Y, Barzaga NG, Allpio A, Soetjipto, Dol H, Ishido S, et al. Genotype analysis of hepatitis C virus among blood donors and inmates in Metro Manila, the Philippines. Microbiol Immunol. 1996;40: 525–529. 10.1111/j.1348-0421.1996.tb01104.x [DOI] [PubMed] [Google Scholar]
- 56.Durier N, Yunihastuti E, Ruxrungtham K, Kinh N V., Kamarulzaman A, Boettiger D, et al. Chronic hepatitis C infection and liver disease in HIV-coinfected patients in Asia. J Viral Hepat. 2017;24: 187–196. 10.1111/jvh.12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greene WK, Cheong MK, Ng V, Yap KW. Prevalence of hepatitis C virus sequence variants in south-east Asia. J Gen Virol. 1995;76: 211–215. 10.1099/0022-1317-76-1-211 [DOI] [PubMed] [Google Scholar]
- 58.Yusrina F, Chua CW, Lee CK, Chiu L, Png TS-Y, Khoo MJ, et al. Comparison of cobas HCV GT against Versant HCV Genotype 2.0 (LiPA) with confirmation by Sanger sequencing. J Virol Methods. 2018;255: 8–13. 10.1016/j.jviromet.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 59.Choy CY, Ang LW, Ng OT, Leo YS, Wong CS. Factors associated with hepatitis B and C co-infection among HIV-infected patients in Singapore, 2006–2017. Trop Med Infect Dis. 2019;4: 87. 10.3390/tropicalmed4020087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee ASG, Ng K-Y, Chee MK-L, Vathsala A, Choong H-L, Ng H-S, et al. Partial sequence analysis of the hepatitis C viral genome in Singapore patients. Biochem Biophys Res Commun. 1994;199: 37–40. 10.1006/bbrc.1994.1189 [DOI] [PubMed] [Google Scholar]
- 61.Soh BY-M, Kumar R, Ekstrom VS-M, Lin CY-H, Thangaraju SDO, Tan HH, et al. Prevalence of hepatitis C virus infection and the IL28B genotype polymorphism among blood donors and high-risk populations. Singapore Med J. 2019;60: 34–39. 10.11622/smedj.2018078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akkarathamrongsin S, Hacharoen P, Tangkijvanich P, Theamboonlers A, Tanaka Y, Mizokami M, et al. Molecular epidemiology and genetic history of hepatitis C virus subtype 3a infection in Thailand. Intervirology. 2013;56: 284–294. 10.1159/000351621 [DOI] [PubMed] [Google Scholar]
- 63.Akkarathamrongsin S, Praianantathavorn K, Hacharoen N, Theamboonlers A, Tangkijvanich P, Poovorawan Y. Seroprevalence and genotype of hepatitis C virus among immigrant workers from Cambodia and Myanmar in Thailand. Intervirology. 2011;54: 10–16. 10.1159/000318884 [DOI] [PubMed] [Google Scholar]
- 64.Avihingsanon A, Jitmitraparp S, Tangkijvanich P, Ramautarsing RA, Apornpong T, Jirajariyavej S, et al. Advanced liver fibrosis by transient elastography, Fibrosis 4, and alanine aminotransferase/platelet ratio index among Asian hepatitis C with and without human immunodeficiency virus infection: Role of vitamin D levels. J Gastroenterol Hepatol. 2014;29: 1706–1714. 10.1111/jgh.12613 [DOI] [PubMed] [Google Scholar]
- 65.Barusrux S, Nanok C, Puthisawas W, Pairojkul C, Poovorawan Y. Viral hepatitis B, C infection and genotype distribution among cholangiocarcinoma patients in northeast Thailand. Asian Pacific J Cancer Prev. 2012;13: 83–87. 10.7314/APJCP.2012.13.KKSuppl.83 [DOI] [PubMed] [Google Scholar]
- 66.Barusrux S, Sengthong C, Urwijitaroon Y. Epidemiology of hepatitis C virus genotypes in northeastern Thai blood samples. Asian Pacific J Cancer Prev. 2014;15: 8837–8842. 10.7314/apjcp.2014.15.20.8837 [DOI] [PubMed] [Google Scholar]
- 67.Boonyarad V, Chutaputti A, Choeichareon S, Bedi K, Theamboonlers A, Chinchai T, et al. Interspousal transmission of hepatitis C in Thailand. J Gastroenterol. 2003;38: 1053–1059. 10.1007/s00535-003-1205-9 [DOI] [PubMed] [Google Scholar]
- 68.Chuenjitkulthaworn T, Bandidniyamanon W, Ruchutrakool T, Chainuvati S. The natural history of hepatitis C viral infection and HCV genotypic distribution in Thai hemophilia patients at Siriraj hospital. J Med Assoc Thail. 2019;102(Suppl.10): 81–85. [Google Scholar]
- 69.Hansurabhanon T, Jiraphongsa C, Tunsakun P, Sukbunsung R, Bunyamanee B, Kuirat P, et al. Infection with hepatitis C virus among intravenous-drug users: Prevalence, genotypes and risk-factor-associated behaviour patterns in Thailand. Ann Trop Med Parasitol. 2002;96: 615–625. 10.1179/000349802125001465 [DOI] [PubMed] [Google Scholar]
- 70.Jutavijittum P, Jiviriyawat Y, Yousukh A, Pantip C, Maneekarn N, Toriyama K. Genotypic distribution of hepatitis c virus in voluntary blood donors of northern Thailand. Southeast Asian J Trop Med Public Health. 2009;40: 471–479. [PubMed] [Google Scholar]
- 71.Kanistanon D, Neelamek M, Dharakul T, Songsivilai S. Genotypic distribution of hepatitis C virus in different regions of Thailand. J Clin Microbiol. 1997;35: 1772–1776. 10.1128/JCM.35.7.1772-1776.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumthip K, Chusri P, Pantip C, Thongsawat S, O’Brien A, Nelson KE, et al. Hepatitis C virus genotypes circulating in patients with chronic hepatitis C in Thailand and their responses to combined PEG-IFN and RBV therapy. J Med Virol. 2014;86: 1360–1365. 10.1002/jmv.23962 [DOI] [PubMed] [Google Scholar]
- 73.Luengrojanakul P, Vareesangthip K, Chainuvati T, Murata K, Tsuda F, Tokita H, et al. Hepatitis C virus infection in patients with chronic liver disease or chronic renal failure and blood donors in Thailand. J Med Virol. 1994;44: 287–292. 10.1002/jmv.1890440313 [DOI] [PubMed] [Google Scholar]
- 74.Martin M, Vanichseni S, Leelawiwat W, Anekvorapong R, Raengsakulrach B, Cherdtrakulkiat T, et al. Hepatitis C virus infection among people who inject drugs in Bangkok, Thailand, 2005–2010. WHO South-East Asia J public Heal. 2019;8: 50–55. 10.4103/2224-3151.255350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakahira S, Tanprasert S, Nuchprayoon C, Mazda T, Nakata K, O’Charoen R, et al. Characterization of hepatitis C virus antibody positive blood donors in Japan and Thailand: a comparative study. Int Hepatol Commun. 1995;4: 216–222. 10.1016/0928-4346(95)00249-9 [DOI] [Google Scholar]
- 76.Netski DM, Wang X-H, Mehta SH, Nelson K, Celentano D, Thongsawat S, et al. Hepatitis C Virus (HCV) Core Antigen Assay to Detect Ongoing HCV Infection in Thai Injection Drug Users. J Clin Microbiol. 2004;42: 1631–1636. 10.1128/jcm.42.4.1631-1636.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sirinawasatien A, Techasirioangkun T. Prevalence and predictive factors of liver cirrhosis identified by vibration-controlled transient elastography (Fibroscan®) in chronic viral hepatitis c patients at rajavithi hospital. J Med Assoc Thail. 2019;102: 33–39. [Google Scholar]
- 78.Sirinawasatien A, Techasirioangkun T. Sofosbuvir-based regimens in the treatment of patients with chronic hepatitis C virus infection: Real-world efficacy in Thailand. PLoS One. 2020;15: e0229517. 10.1371/journal.pone.0229517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sistayanarain A, Kunthalert D, Vipsoongnern Y. A shift in the Hepatitis C virus genotype dominance in blood donor samples from Thailand. Mol Biol Rep. 2011;38: 4287–4290. 10.1007/s11033-010-0552-x [DOI] [PubMed] [Google Scholar]
- 80.Smolders EJ, Thammajaruk N, de Kanter CTMM, Colbers A, Chaiyahong P, Cuprasitrut T, et al. Peg-interferon and ribavirin treatment in HIV/HCV co-infected patients in Thailand: efficacy, safety and pharmacokinetics. Trop Med Int Heal. 2018;23: 295–305. 10.1111/tmi.13027 [DOI] [PubMed] [Google Scholar]
- 81.Songsivilai S, Dharakul T, Kanistanon D. Hepatitis C virus genotypes in patients with hepatocellular carcinoma and cholangiocarcinoma in Thailand. Trans R Soc Trop Med Hyg. 1996;90: 505–507. 10.1016/s0035-9203(96)90296-5 [DOI] [PubMed] [Google Scholar]
- 82.Sugiyama K, Kato N, Nakazawa T, Yonemura Y, Phornphutkul K, Kunakorn M, et al. Novel genotypes of hepatitis C virus in Thailand. J Gen Virol. 1995;76: 2323–2327. 10.1099/0022-1317-76-9-2323 [DOI] [PubMed] [Google Scholar]
- 83.Sunanchaikarn S, Theamboonlers A, Chongsrisawat V, Yoocharoen P, Tharmaphornpilas P, Warinsathien P, et al. Seroepidemiology and genotypes of hepatitis C virus in Thailand. Asian Pacific J Allergy Immunol. 2007;25: 175–182. [PubMed] [Google Scholar]
- 84.Theamboonlers A, Kaew-In N, Hirsch P, Poovorawan Y. Determination of the genotypes of hepatitis C virus in Thailand, from restriction-fragment length polymorphisms. Ann Trop Med Parasitol. 2000;94: 525–527. 10.1080/00034983.2000.11813573 [DOI] [PubMed] [Google Scholar]
- 85.Tokita H, Okamoto H, Luengrojanakul P, Vareesangthip K, Chainuvati T, Iizuka H, et al. Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d) and ninth (9b, 9c) major genetic groups. J Gen Virol. 1995;76: 2329–2335. 10.1099/0022-1317-76-9-2329 [DOI] [PubMed] [Google Scholar]
- 86.Wasitthankasem R, Vongpunsawad S, Siripon N, Suya C, Chulothok P, Chaiear K, et al. Genotypic distribution of hepatitis C virus in Thailand and Southeast Asia. PLoS One. 2015;10: e0126764. 10.1371/journal.pone.0126764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wasitthankasem R, Posuwan N, Vichaiwattana P, Theamboonlers A, Klinfueng S, Vuthitanachot V, et al. Decreasing hepatitis C virus infection in Thailand in the past decade: Evidence from the 2014 national survey. PLoS One. 2016;11: e0149362. 10.1371/journal.pone.0149362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wasitthankasem R, Vichaiwattana P, Siripon N, Posuwan N, Auphimai C, Klinfueng S, et al. Assessment of hepatitis C virus infection in two adjacent Thai provinces with drastically different seroprevalence. PLoS One. 2017;12: e0177022. 10.1371/journal.pone.0177022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Do SH, Yamada H, Fujimoto M, Ohisa M, Matsuo J, Akita T, et al. High prevalences of hepatitis B and C virus infections among adults living in Binh Thuan province, Vietnam. Hepatol Res. 2015;45: 259–268. 10.1111/hepr.12350 [DOI] [PubMed] [Google Scholar]
- 90.Dunford L, Carr MJ, Dean J, Waters A, Nguyen LT, Ta Thi TH, et al. Hepatitis C virus in vietnam: High prevalence of infection in dialysis and multi-transfused patients involving diverse and novel virus variants. PLoS One. 2012;7: e41266. 10.1371/journal.pone.0041266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Duong CM, McLaws ML. An investigation of an outbreak of hepatitis C virus infections in a low-resourced hemodialysis unit in Vietnam. Am J Infect Control. 2016;44: 560–566. 10.1016/j.ajic.2016.01.014 [DOI] [PubMed] [Google Scholar]
- 92.Duong MC, McLaws M-L. Screening haemodialysis patients for hepatitis C in Vietnam: The inconsistency between common hepatitis C virus serological and virological tests. J Viral Hepat. 2019;26: 25–29. 10.1111/jvh.12994 [DOI] [PubMed] [Google Scholar]
- 93.Kakumu S, Sato K, Morishita T, Anh TK, Binh NH, Dien B V, et al. Prevalence of hepatitis B, hepatitis C, and GB virus C/hepatitis G virus infections in liver disease patients and inhabitants in Ho Chi Minh, Vietnam. J Med Virol. 1998;54: 243–248. [PubMed] [Google Scholar]
- 94.Le Ngoc C, Thanh TTT, Lan PTT, Mai TN, Hoa TN, My NN, et al. Differential prevalence and geographic distribution of hepatitis C virus genotypes in acute and chronic hepatitis C patients in Vietnam. PLoS One. 2019;14: e0212734. 10.1371/journal.pone.0212734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li C, Yuan M, Lu L, Lu T, Xia W, Pham VH, et al. The genetic diversity and evolutionary history of hepatitis C virus in Vietnam. Virology. 2014;468–470: 197–206. 10.1016/j.virol.2014.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lioznov DA, Chung NH, Nikolaenko SL, Trung TB, Lan FT, Phong NZ. Clinical and laboratory characteristics of chronic hepatitis C in Vietnam on the example of Ho Chi Minh City Hepatology Clinic. J Infektologii. 2016;8: 72–78. 10.22625/2072-6732-2016-8-4-72-78 [DOI] [Google Scholar]
- 97.Minh NH. The Distribution Pattern of Hepatitis C Virus Genotypes in Vietnam Based on the Non-structural 5B Region. Clin Gastroenterol Hepatol. 2015;13: 1385. 10.1016/j.cgh.2015.04.041 [DOI] [Google Scholar]
- 98.Nadol P, O’Connor S, Duong H, Mixson-Hayden T, Tram TH, Xia G-L, et al. High hepatitis C virus (HCV) prevalence among men who have sex with men (MSM) in Vietnam and associated risk factors: 2010 Vietnam Integrated Behavioural and Biologic Cross-Sectional Survey. Sex Transm Infect. 2016;92: 542–549. 10.1136/sextrans-2015-052518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nguyen Truong T, Laureillard D, Lacombe K, Duong Thi H, Pham Thi Hanh P, Truong Thi Xuan L, et al. High Proportion of HIV-HCV Coinfected Patients with Advanced Liver Fibrosis Requiring Hepatitis C Treatment in Haiphong, Northern Vietnam (ANRS 12262). PLoS One. 2016;11: e0153744. 10.1371/journal.pone.0153744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguyen TT, Lemee V, Bollore K, Vu H V, Lacombe K, Thi XLT, et al. Confirmation of HCV viremia using HCV RNA and core antigen testing on dried blood spot in HIV infected peoples who inject drugs in Vietnam. BMC Infect Dis. 2018;18: 622. 10.1186/s12879-018-3529-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Noppornpanth S, Lien TX, Poovorawan Y, Smits SL, Osterhaus ADME, Haagmans BL. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol. 2006;80: 7569–7577. 10.1128/JVI.00312-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pham DA, Leuangwutiwong P, Jittmittraphap A, Luplertlop N, Hoa KB, Akkarathamrongsin S, et al. High prevalence of hepatitis C virus genotype 6 in Vietnam. Asian Pacific J Allergy Immunol. 2009;27: 153–160. [PubMed] [Google Scholar]
- 103.Pham VH, Nguyen HDP, Ho PT, Banh D V, Pham HLT, Pham PH, et al. Very high prevalence of hepatitis C virus genotype 6 variants in southern vietnam: Large-scale survey based on sequence determination. Jpn J Infect Dis. 2011;64: 537–539. [PMC free article] [PubMed] [Google Scholar]
- 104.Song P, Duc DD, Hien B, Nakata S, Chosa T, Watanabe J, et al. Markers of hepatitis C and B virus infections among blood donors in Ho Chi Minh City and Hanoi, Vietnam. Clin Diagn Lab Immunol. 1994;1: 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tanimoto T, Cuong NH, Ishizaki A, Chung PTT, Huyen HTT, Trung N V, et al. Multiple routes of hepatitis C virus transmission among injection drug users in Hai Phong, Northern Vietnam. J Med Virol. 2010;82: 1355–1363. 10.1002/jmv.21787 [DOI] [PubMed] [Google Scholar]
- 106.Tokita H, Okamoto H, Tsuda F, Song P, Nakata S, Chosa T, et al. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc Natl Acad Sci U S A. 1994;91: 11022–11026. 10.1073/pnas.91.23.11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tran HT-T, Ushijima H, Quang VX, Phuong N, Li T-C, Hayashi S, et al. Prevalence of hepatitis virus types B through E and genotypic distribution of HBV and HCV in Ho Chi Minh City, Vietnam. Hepatol Res. 2003;26: 275–280. 10.1016/s1386-6346(03)00166-9 [DOI] [PubMed] [Google Scholar]
- 108.Reesink HW, Zeuzem S, Weegink CJ, Forestier N, van Vliet A, van de Wetering de Rooij J, et al. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology. 2006;131: 997–1002. 10.1053/j.gastro.2006.07.013 [DOI] [PubMed] [Google Scholar]
- 109.Foster GR, Hézode C, Bronowicki J-P, Carosi G, Weiland O, Verlinden L, et al. Telaprevir alone or with peginterferon and ribavirin reduces HCV RNA in patients with chronic genotype 2 but not genotype 3 infections. Gastroenterology. 2011;141: 881–889.e1. 10.1053/j.gastro.2011.05.046 [DOI] [PubMed] [Google Scholar]
- 110.Cooper C, Lawitz EJ, Ghali P, Rodriguez-Torres M, Anderson FH, Lee SS, et al. Evaluation of VCH-759 monotherapy in hepatitis C infection. J Hepatol. 2009;51: 39–46. 10.1016/j.jhep.2009.03.015 [DOI] [PubMed] [Google Scholar]
- 111.Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Müh U, Welker M, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132: 1767–1777. 10.1053/j.gastro.2007.02.037 [DOI] [PubMed] [Google Scholar]
- 112.McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360: 1827–1838. 10.1056/NEJMoa0806104 [DOI] [PubMed] [Google Scholar]
- 113.Ramirez S, Mikkelsen LS, Gottwein JM, Bukh J. Robust HCV Genotype 3a Infectious Cell Culture System Permits Identification of Escape Variants With Resistance to Sofosbuvir. Gastroenterology. 2016;151: 973–985.e2. 10.1053/j.gastro.2016.07.013 [DOI] [PubMed] [Google Scholar]
- 114.Larrey D, Lohse AW, Trepo C, Bronowicki J-P, Arastéh K, Bourlière M, et al. Antiviral Effect, Safety, and Pharmacokinetics of Five-Day Oral Administration of Deleobuvir (BI 207127), an Investigational Hepatitis C Virus RNA Polymerase Inhibitor, in Patients with Chronic Hepatitis C. Antimicrob Agents Chemother. 2013;57: 4727–4735. 10.1128/AAC.00565-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pawlotsky J-M. Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology. 2016;151: 70–86. 10.1053/j.gastro.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 116.Sarrazin C. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol. 2016;64: 486–504. 10.1016/j.jhep.2015.09.011 [DOI] [PubMed] [Google Scholar]
- 117.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2011;17: 107–115. 10.1111/j.1469-0691.2010.03432.x [DOI] [PubMed] [Google Scholar]
- 118.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22: 7824–7840. 10.3748/wjg.v22.i34.7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57: 1333–1342. 10.1002/hep.26141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOC)
(PDF)
(PDF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.