Abstract
Since its initial characterization, Escherichia coli RNase I has been described as a single-strand specific RNA endonuclease that cleaves its substrate in a largely sequence independent manner. Here, we describe a strong calcium (Ca2+)-dependent activity of RNase I on double-stranded RNA (dsRNA), and a Ca2+-dependent novel hybridase activity, digesting the RNA strand in a DNA:RNA hybrid. Surprisingly, Ca2+ does not affect the activity of RNase I on single stranded RNA (ssRNA), suggesting a specific role for Ca2+ in the modulation of RNase I activity. Mutation of a previously overlooked Ca2+ binding site on RNase I resulted in a gain-of-function enzyme that is highly active on dsRNA and could no longer be stimulated by the metal. In summary, our data imply that native RNase I contains a bound Ca2+, allowing it to target both single- and double-stranded RNAs, thus having a broader substrate specificity than originally proposed for this traditional enzyme. In addition, the finding that the dsRNase activity, and not the ssRNase activity, is associated with the Ca2+-dependency of RNase I may be useful as a tool in applied molecular biology.
INTRODUCTION
Escherichia coli RNase I (EC 3.1.27.6) is a 27 kDa periplasmic endoribonuclease belonging to the RNase T2 family of enzymes (reviewed in Deshpande and Shankar (1)). Among the members of this family are the prototypical fungal RNase T2 enzymes and the plant self-incompatibility RNases. These enzymes are characterized by the presence of two conserved active-site segments (CAS1 and CAS2), which contain two histidine residues required for cleavage (2). Unlike the smaller and more well-studied RNase A and RNase T1 enzymes, which preferentially cleave at the 3′ of pyrimidines and guanosines, respectively, RNase I is one of the few ribonucleases that can cleave ssRNA between any two nucleotides [reviewed in (3) and (4)]. RNase I was first described and isolated in 1959 and 1961 from E. coli ribosomes, which released the enzyme after treatment with lysozyme and EDTA (5,6). Subsequent work has determined that the ribosomal association was an artifact of cell disruption and that the enzyme is normally localized to the periplasm (7,8), even when over-expressed (9). Consistent with this localization, the pre-processed form of RNase I has a 23-aa leader sequence which conforms to signal peptide consensus rules (10) and is absent from the periplasmic enzyme (11). Interestingly, a cytoplasmic variant of RNase I, stemming from the same gene origin, was found to exhibit differences in thermal denaturation and pH stability compared to the periplasmic RNase I variant (10,12).
Hydrolysis of RNA by E. coli RNase I yields nucleoside 3′-phosphates via 2′,3′-cyclic phosphate intermediates (5). The enzyme is active in the presence of EDTA and does not require any metal cofactors to degrade single stranded RNA (ssRNA) (7). It contains eight cysteines, all of which have been shown to be paired and form four disulfide bonds (13). Historically, RNase I has been reported to exhibit a significant preference for ssRNA over double stranded RNA (dsRNA) (5) and to bind, but not cleave DNA. It is not uncommon for nucleases to preferentially cleave one substrate but demonstrate reduced activity against secondary substrates, particularly at high enzyme:substrate (E:S) ratios. As the concentration of RNase I is increased, the apparent specificity of the enzyme has been reported to decrease, with all cellular RNA becoming susceptible to hydrolysis at high enzyme concentrations in vitro (12,14).
Here, we describe a Ca2+-dependent strong dsRNase activity for RNase I, and a novel function as an RNA:DNA hybridase, exclusively cleaving the RNA strand of a double-stranded RNA:DNA hybrid substrate. Purifying, storing, or reacting RNase I in the presence of EDTA leads to a significant loss of its dsRNase activity if Ca2+ is omitted from the reaction, while its activity against ssRNA is largely unaffected by Ca2+. The crystal structure of E. coli RNase I revealed a Ca2+ ion bound to the enzyme, even though no metal was added during crystallization. By mutating an amino acid residue that is important for the metal ion coordination, we were able to simulate the Ca2+-induced activation of the enzyme in the absence of the metal, further supporting the presence of a previously uncharacterized and functional metal-binding site in E. coli RNase I. Our findings shine light on native functions of RNase I that previously have been obscured by common purification methods and have significant implications on and expand the potential use of RNase I in current and future methods in molecular biology.
MATERIALS AND METHODS
Substrates
Synthetic RNA and DNA oligos (Integrated DNA Technologies) were reconstituted in RNase-free Milli-Q water as 100 μM stock solutions. Double-stranded substrates were generated by annealing complementary strands in Buffer A [10 mM Tris–HCl, 50 mM NaCl, 1 mM DTT, pH 8.0] by denaturing the strands for 2 min at 94°C, followed by cooling to 25°C via a temperature gradient at the rate of 0.1°C/s with subsequent storage at –20°C. Double-stranded 33-mer substrates consisted of 31-paired bases and two single-stranded base overhangs on each 3′ end. All other substrates were designed to form blunt ends when annealed. The unannealed top (t) strands of oligos were used as single-stranded substrates. Sequences of the synthetic oligos and the chemical nature of their 5′ and 3′ termini are provided in Supplementary Material (Supplementary Table S1).
Protein expression and purification
Recombinant wild-type E. coli RNase I enzyme was periplasmically expressed with its native signal peptide as an N-terminal MBP fusion protein (75.1 kDa) and stored in a glycerol-containing buffer [10 mM Tris–HCl, 100 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 50% glycerol, pH 8.0]. RNase I mutants were generated using the Q5 Site-Directed Mutagenesis Kit (NEB E0554). Expression of wild-type and mutant RNase I variants was induced with 0.1 mM IPTG, and the enzymes were expressed from pMal-p2X-RNase I-containing plasmids in T7 Express lysY Competent E. coli [MiniF lysY (CamR)/fhuA2 lacZ::T7 gene1 [lon] ompT gal sulA11 R(mcr-73::miniTn10–TetS)2 [dcm] R(zgb-210::Tn10–TetS) endA1 Δ(mcrC-mrr)114::IS10] (NEB C3010) for 16 h at 16°C. After breaking the cells by sonication in the presence of protease inhibitors (1 mM PMSF, 0.5 nM Leupeptin, 2.75 mM benzamidine, 2 nM pepstatin) and removal of cell debris by centrifugation at 21 000 × g for 45 min, the recombinant proteins were purified from the crude extract using 10 ml BioRad Econo-Pac Disposable Chromatography Columns packed with 3 ml Amylose Resin (NEB E8021), followed by dialysis and storage in RNase I storage buffer (10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 0.5 mM EDTA and 50% glycerol) at –20°C. For the purification of the RNase Iwt (–DTT/–EDTA) variant, EDTA and DTT were omitted from all buffers. The amino acid annotation is based on a mature, untagged RNase Iwt enzyme and in accordance with the previously published crystal structure of E. coli RNase I (15).
Enzymatic assays
In a typical RNase I digestion assay, 1.5 pmol of single- or double-stranded substrate was cleaved with varying amounts of enzyme in Buffer A supplemented with 0.5 mM EDTA [10 mM Tris–HCl, 50 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH8.0] in a total volume of 15 μl. Reactions with the RNase Iwt (–DTT/–EDTA) variant did not contain EDTA or DTT, unless otherwise indicated. Enzymes were added as the last component of the reaction mixture, and all RNase I stocks and dilutions were made with RNase storage buffer (with or without DTT/EDTA as indicated). After assembling the reactions on ice, they were incubated for 15 min at 37°C and stopped by adding 0.8 U Proteinase K (NEB P8107) on ice, followed by 15 min incubation at 37°C. After addition of 3 μl 6× Gel Loading Dye Purple, no SDS (NEB B7025), samples were immediately loaded onto a non-denaturing (10% polyacrylamide TBE) gels and run in 1× TBE buffer at 180 V for 30 min. Gels were visualized using the Cy2 channel of an Amersham Typhoon Laser Scanner after staining with SYBR Gold (1/20 000 dilution, Thermo Fisher Scientific S11494). Gel bands of three independent replicas per experiment were quantified and analyzed using ImageQuant TL software, unless otherwise indicated. Time course assays were performed as described in the Supplementary Data.
RNA mass spectrometry
For intact mass analysis, 0.5 μM of dsRNA substrate was incubated with the indicated RNase I variant and concentration for 15 min at 37°C in a 20 μl reaction. Reactions were stopped by the addition of 20 μl formamide and analyzed without further purification. Intact mass analysis was performed by tandem liquid chromatography–mass spectrometry (LC–MS/MS) on an Vanquish Horizon UHPLC System equipped with a diode array detector and a Thermo Q-Exactive Plus mass spectrometer operating under negative electrospray ionization mode (–ESI). UHPLC was performed using a Thermo DNAPac™ RP Column (2.1 × 50 mm, 4 μm) at 70°C and 0.3 ml/min flow rate, with a gradient mobile phase from 18% to 55% solvent B in 6 min (Solvent A: 1% HFIP, 0.1% DIEA, 1 mM EDTA in H2O; Solvent B: 0.075% HFIP, 0.0375% DIEA, 1 mM EDTA in 80% MeOH in H2O). UV signal was recorded at 260 nm. MS data acquisition was performed in the scan mode (600–2500 m/z) at 70 000 mass resolution with the following parameters: capillary temperature of 320°C, spray voltage of 2.7 kV and sheath, and auxiliary and sweep gases at 35, 8, 0 arbitrary units, respectively. ESI-MS raw data was deconvoluted using ProMass HR (Novatia, LCC).
RESULTS
Ca2+ stimulates the degradation of synthetic dsRNA oligos by RNase I
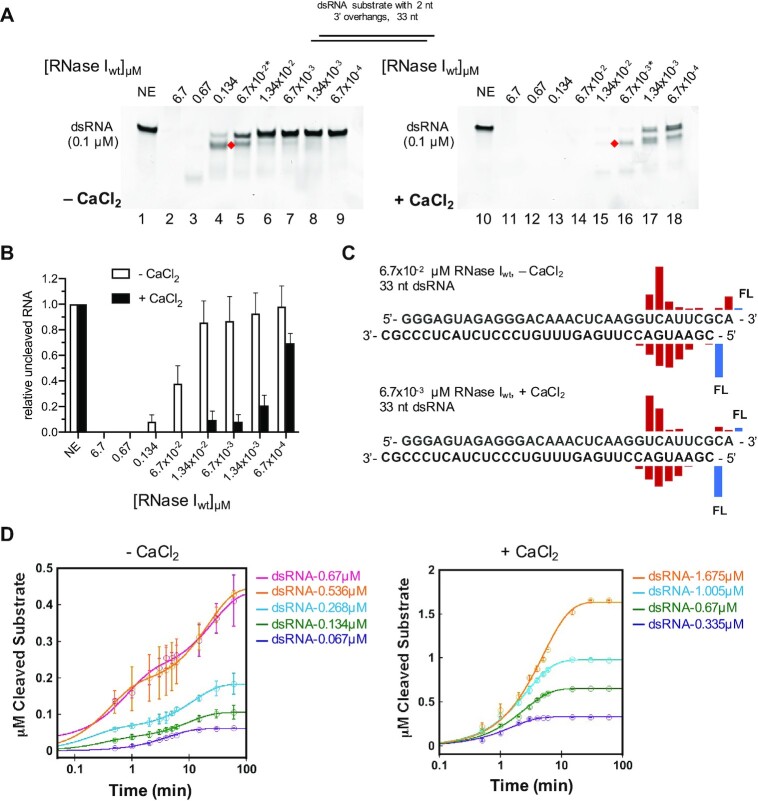
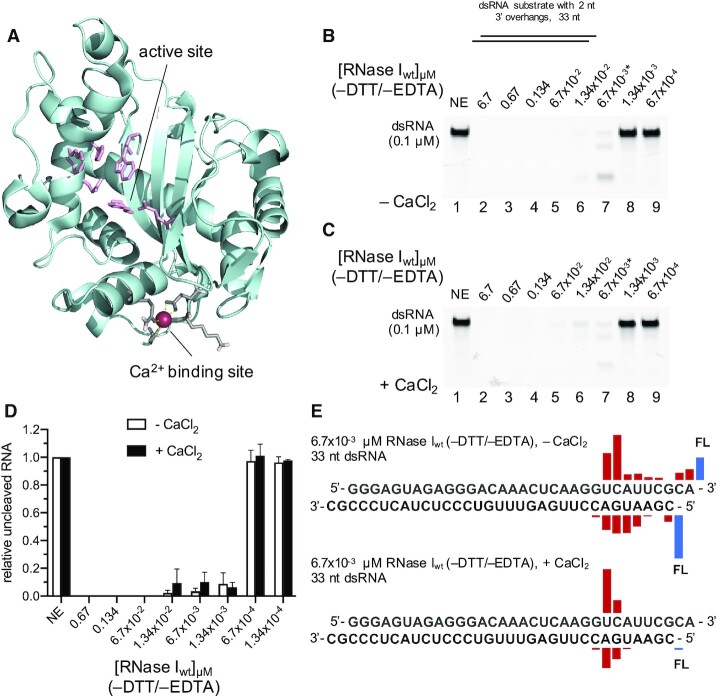
RNase I has traditionally been described as a ssRNA specific endonuclease that does not require any cofactors for its activity. Non-specific cleavage (i.e. cleavage of something other than a ssRNA substrate) has been previously reported for RNase I as a function of enzyme concentration, with high E:S ratios leading to the degradation of all cellular RNA (12). In this study, we aimed to characterize the effect of metal ions on RNase I activity, and initially purified RNase I from E. coli in the presence of 1 mM EDTA (see Materials and Methods). The enzymatic reactions contained a final concentration of 0.5 mM EDTA to chelate metal ions that may have been co-purified with the enzyme. In agreement with previous studies, we found that RNase I was able to degrade a double-stranded synthetic RNA (dsRNA) substrate possessing 3′ overhangs at the highest E:S ratios tested (Figure 1A, lanes 2–3), whereas at E:S ratios of 1.34:1 and lower, RNase I failed to fully degrade the dsRNA substrate (lanes 4–9). The addition of 4 mM CaCl2 to the reaction significantly increased RNase I activity against dsRNA, now enabling RNase I to cleave the majority of the dsRNA substrate at an E:S ratio of 0.134:1 (Figure 1A, lane 15 and B).
Figure 1.
Calcium significantly stimulates RNase I activity on dsRNA. (A) 33mer dsRNA (0.1 μM) substrate with 2 nt 3′ overhangs was incubated at 37°C for 15 min without (NE) or with various concentrations of RNase I as indicated above the lanes (from 6.7 μM to 6.7 × 10−4 μM) in either the absence (left panel) or presence (right panel) of 4 mM CaCl2. Asterisks indicate the RNase I concentrations used for the subsequent MS analysis. Red diamonds mark primary cleavage products. (B) Quantification of the relative amount of uncleaved RNA substrate ± CaCl2. (C) MS results showing the location of the cleavage (location of the bar above the sequence) and the relative abundance of the resulting fragment (height of the bar) for reactions containing 67 nM RNase I without Ca2+ (top) and 6.7 nM RNase I with Ca2+ (bottom). FL, full-length oligonucleotide (uncleaved substrate). (D) 6.7 × 10−3 μM (left panel) or 1.34 × 10−3 μM (right panel) of RNase I were incubated in the absence or presence of Ca2+, respectively, and various concentrations of dsRNA as indicated. Reactions were quenched at the indicated timepoints and the amount of digested full-length dsRNA over time (circles) was plotted. Solid traces are exponential curve fits to the data.
In order to characterize the effect of Ca2+ on the catalytic efficiency of RNase I in more detail, we performed time course analysis in the absence and presence of CaCl2 (Figure 1D). To replicate the conditions tested in the endpoint experiments shown in Figure 1, we selected the same dsRNA substrate with 3′ overhangs as substrate. Since the RNase activity did not allow for the detection of a singular cleavage event, we quantified the reduction of the band intensity of the full-length dsRNA substrate over time to measure activity. In the absence of Ca2+, 6.7 × 10−3 μM RNase I was incubated with dsRNA at E:S ratios of 1:10 to 1:100 (Supplementary Figure S2A), while 1.34 × 10−3 μM RNase I was tested with dsRNA at E:S ratios of 1:250 to 1:1250 (Supplementary Figure S2B). Reactions were quenched by the addition of Proteinase K at timepoints between 0 and 60 min as indicated in Supplementary Figure S2. The amount of digested full-length dsRNA over time was plotted and exponential fits were used to describe the course of the reaction (Figure 1D). The turnover number (kcat) of RNase I in the presence of CaCl2 is ∼ 90-fold higher than in the absence of the metal, confirming its role in activation of the enzyme toward dsRNA (Supplementary Figure S2).
We observed a primary cleavage product on RNA gels (Figure 1A, red diamonds), represented by a band running slightly faster than the substrate, suggesting a preferred cleavage position for RNase I on the dsRNA substrate. This was rather surprising, as RNase I had previously been described as a sequence independent ribonuclease (reviewed in (1)). To identify the primary cleavage products, we subjected cleaved RNA samples to intact mass spectrometry (MS) analysis. RNase I reactions were carried out in the absence or presence of 4 mM CaCl2 with 67 or 6.7 nM RNase I, respectively, to adjust for the different cleavage efficiencies to test comparable amounts of the primary cleavage products.
We observed similar cleavage patterns for both conditions, with a main cleavage site 8 to 9 nt upstream of the 3′ end on the top strand, leaving a 2′,3′-cyclic phosphate end. On the bottom strand, the main cleavage site was located 5–6 nt downstream of the 5′ end (Figure 1 and Supplementary Figure S12, and Supplementary Table S2). FAM-labeling of the top strand 3′ end and bottom strand 5′ end allowed us to detect the corresponding short cleavage products in the range of 5–12 nt in length (Supplementary Figures S3A-C and S13). These data indicate that RNase I function as an endoribonuclease rather than an exoribonuclease, generating a cut in a region close to the end of one RNA strand, and then the respective region of the other strand, before fully digesting it. Interestingly, we observed that one strand was always cleaved at higher efficiency than the opposing strand, suggesting a staggered cleavage mechanism, where one strand is cleaved first, followed by the other strand. Furthermore, the data show that Ca2+, or the lack thereof, does not affect the substrate preference nor the cleavage mechanism of RNase I, as we observed similar cleavage patterns in the presence and absence of Ca2+.
To validate our findings, we tested other short, synthetic substrates consisting of either fully base-paired (blunt-ended) 31mer RNA or a double-stranded 41mer DNA-RNA-DNA chimera, consisting of a 27 nt dsRNA flanked on each end by 7 nt dsDNA. Supporting our initial data, we found that the addition of Ca2+ is required for RNase I to fully degrade both substrates (Supplementary Figure S4), showing that the stimulatory effect of Ca2+ on dsRNA is not dependent on freely available ssRNA ends. Despite a general decrease in cleavage efficiency when RNase I was presented with the double-stranded DNA-RNA-DNA chimera, RNase I activity was significantly stimulated by the addition of Ca2+ to the reaction (Supplementary Figure S4B and D). Moreover, the Ca2+-dependency on the degradation of an RNA substrate with DNA ends by RNase I corroborates the MS data (Figure 1C) against exoribonucleolytic activity. To exclude the possibility of DNase contamination (causing removal of the DNA ends), we showed that RNase I was able to bind and not cleave single- and double-stranded DNA substrates, exhibiting a higher affinity for ssDNA (Supplementary Figure S5A, B). Furthermore, analysis of fluorescently labeled ssDNA and dsDNA substrates by capillary electrophoresis (16) after incubation with either RNase I or its dilution buffer (buffer A), both in the absence and presence of 4 mM CaCl2, showed no evidence for DNase activity (Supplementary Figure S5C).
To test if the primary cleavage pattern was a sole feature of the specific dsRNA template used in the experiment, we analyzed by intact MS the cleavage products of two independent blunt-ended dsRNA substrates in the absence and presence of Ca2+ (Supplementary Figures S6, S14 and S15). We found that the main cleavage site in the short dsRNA (26mer) was 2–3 nt upstream of the 3′ end on the top strand and 2–4 nt downstream of the 5′ end on the bottom strand (Supplementary Figure S6A). As for the long dsRNA (40mer), the main cleavage site was 8 to 9 nt upstream of the 3′ end on the top strand and 7–9 nt downstream of the 5′ end on the bottom strand (Supplementary Figure S6B). No particular sequence cleavage preferences were observed. Interestingly, secondary cleavage products were detected at the opposite ends of each strand, leaving either a 2′,3′-cyclic phosphate or a 3′-phosphate end.
To exclude the possibility that the MBP-tag had an impact on RNase I substrate specificity, and thus was responsible for the observed stimulatory effect of Ca2+, we tested two commercially available tagless RNase I enzymes and observed similar results (Supplementary Figure S7). Both tagless RNase I variants exhibited a significant increase in activity on dsRNA when supplemented with Ca2+ (5.6-fold increase for the 1:25 dilution of RNase IA and 25-fold increase for a 1:125 dilution of RNase IB), showing that the MBP-tag did not affect RNase I function nor its dependency on Ca2+ for dsRNase activity.
Ca2+does not affect RNase I activity on single-stranded RNA
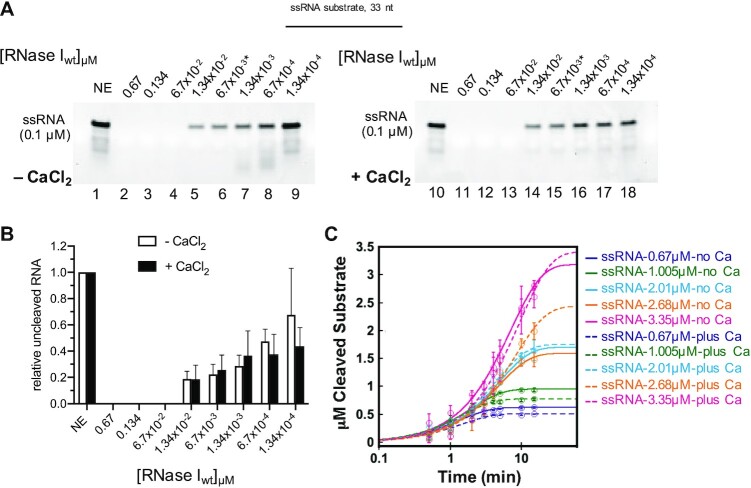
We wondered whether the addition of Ca2+ generally increased the activity of RNase I or if the observed stimulation was specific to dsRNA substrates. Thus, we monitored the RNase I-mediated degradation of a 33mer ssRNA (corresponding to the top strand of the 33mer dsRNA substrate tested above) in the presence and absence of CaCl2 (Figure 2). To our surprise, we found no significant difference in the cleavage activity between the two conditions, suggesting that the single-strand endonuclease activity of RNase I is unaffected by Ca2+ (Figure 2A, B). This finding was confirmed when we accessed the amount of full length ssRNA digested by RNase I over time (Supplementary Figure S8). 6.7 × 10−4 μM RNase I was incubated with ssRNA at E:S ratios of 1:1000 to 1:5000 in the absence and presence of Ca2+. The resulting traces were almost superimposable, suggesting that Ca2+ has no effect on the enzyme's targeting of ssRNA substrate (Figure 2C). This was reflected in the comparable kcat values for the enzyme under both conditions (Supplementary Figure S8C, Supplementary Table S4). While we saw a potential primary cleavage product of ssRNA at various timepoints and E:S ratios (Supplementary Figure S8), we were unable to detect any major degradation products by MS. Taken together, these data support our finding that RNase I generates primary cleavage products with dsRNA substrates, whereas it degrades ssRNA largely in a non-specific manner and independently of Ca2+. Comparison of the kcat values suggest that ssRNA is ∼ 2.5-fold more favored than dsRNA, whether or not Ca2+ is present in the ssRNA reactions, while dsRNA in the absence of Ca2+ is the least favored substrate (Supplementary Table S4).
Figure 2.
Calcium is not required for RNase I activity on ssRNA. Reactions in (A) contained a 33mer ssRNA (0.1 μM) substrate with either no enzyme (NE) or various concentrations of RNase I (from 0.67 to 1.34 × 10−4 μM) for 15 minutes at 37°C. The relative amount of uncleaved RNA is shown in (B). (C) 6.7 × 10−4 μM of RNase I were incubated with various concentrations of ssRNA from 0.67 to 3.35 μM. Reactions were quenched at the indicated timepoints and the amount of digested full length ssRNA over time (circles) was plotted. Solid (no Ca2+) and dashed (plus Ca2+) traces are exponential curve fits to the data.
RNase I degrades the RNA strand of an RNA:DNA hybrid in a Ca2+-dependent manner
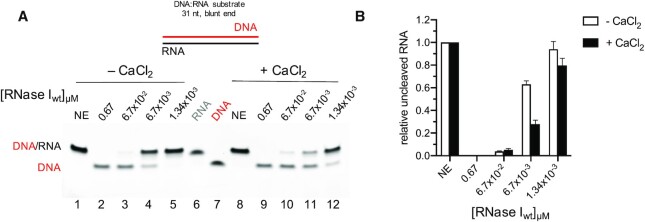
RNase I has been reported to bind, but not cleave DNA, and both of those findings have been reproduced in this study (Supplementary Figure S5). Given the ability of RNase I to cleave dsRNA substrates in the presence of Ca2+, we asked whether the enzyme would degrade the RNA strand in a blunt-ended 31mer RNA:DNA hybrid. RNase I exhibited a strong, Ca2+-inducible activity on the RNA:DNA hybrid that was comparable or even slightly higher than on its dsRNA counterpart (Figure 3). This hybridase activity of RNase I has not been reported before and expands the potential applications of this enzyme as a tool in molecular biology.
Figure 3.
RNase I exhibits hybridase activity. Calcium induced activation of RNase I against 0.1 μM of a double stranded 31mer hybrid substrate containing a DNA top and an RNA bottom strand as shown above the gel. (A) 0.67 μM to 1.34 × 10−3 μM of RNase I were incubated with the substrate in the absence (lanes 1–5) or presence of 4 mM CaCl2 (lanes 8–12). Lanes 6 and 7 show the separate RNA and DNA strands, respectively. (B) Quantification of the uncleaved RNA.
dsRNA substrates are not generally more susceptible to digestion by ssRNases in the presence of Ca2+
Since Ca2+ enhanced the activity of RNase I for dsRNA, but not for ssRNA, we sought to test if our observations were a result of Ca2+ affecting the properties of the dsRNA substrates. In particular, the double-stranded substrates may be more prone to breathing in the presence of Ca2+, resulting in accessible ssRNA ends and thus an increased susceptibility to RNase I as a ssRNA endonuclease. To address this possibility, we investigated if pancreatic RNase A, a ssRNA specific endonuclease, displayed increased activity against a dsRNA substrate in the presence of Ca2+. RNase A is a metal-independent endoribonuclease with a strong substrate preference for ssRNA over dsRNA (17). Accordingly, RNase A was not able to efficiently cleave a 33mer dsRNA substrate, neither in the absence, nor in the presence of Ca2+ (Supplementary Figure S9A, upper panel). To exclude a potential inhibitory effect of CaCl2, we carried out the reaction with a 33mer ssRNA substrate and found that Ca2+ does not interfere with RNase A activity (Supplementary Figure S9A, lower panel). Differential scanning fluorimetry (DSF) was utilized to further verify the effect of Ca2+ on the dsRNA stability. We found comparable Tm values for the dissociation of the 33mer dsRNA substrate in the absence or presence of 1 mM CaCl2 (Supplementary Figure S9B), a Ca2+ concentration sufficient to fully stimulate RNase I activity (Figure 4). A similar Tm was also found for incubation of the dsRNA with 1 mM MgCl2. Thus, our data suggest that there is no significant structural change of the dsRNA substrate in the presence of these metal ions. Taken together, these observations argue against Ca2+ affecting the dsRNA substrate and rather imply that the stimulation of dsRNA degradation is due to an interaction of Ca2+ with RNase I.
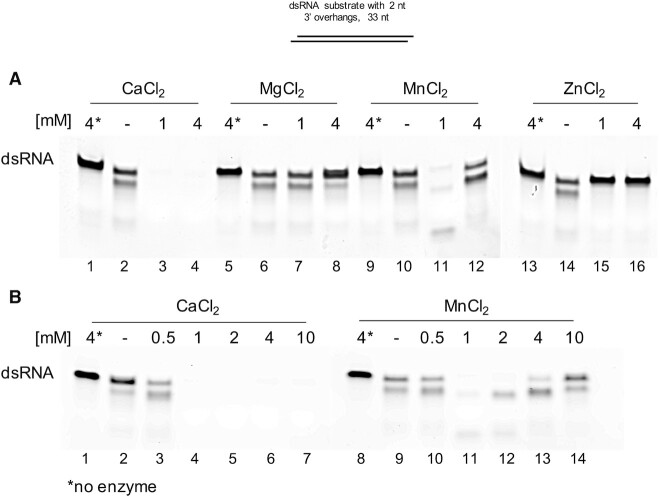
Figure 4.
Effect of metal ions on RNase I activity against dsRNA. (A) To test whether other divalent cations exhibit a similar stimulatory effect on dsRNA cleavage by RNase I, 0.1 μM dsRNA substrate was incubated with 0.067 μM RNase I in the presence of 0.5 mM EDTA and either 1 mM or 4 mM CaCl2, MgCl2, MnCl2 or ZnCl2. (B) Broader range titration of CaCl2 and MnCl2 from 0.5 to 10 mM. Asterisks indicate no-enzyme control reactions containing 4 mM of the respective metal ion.
Ca2+ cannot be substituted by other metals to stimulate dsRNA degradation by RNase I
Next, we investigated whether the dsRNA endonucleolytic activity of RNase I would be retained in the presence of other divalent metal ions. It is not uncommon for metalloenzymes to display a preference for one metal cofactor but also bind other metals of similar properties. However, binding of alternate metals may result in the formation of an inactive complex or in an enzyme with altered or reduced activities. We compared the effect on dsRNA degradation by RNase I in the presence of 1 mM or 4 mM Ca2+, Mg2+, Mn2+ and Zn2+. As expected, Ca2+ significantly stimulated degradation of dsRNA at any concentration tested (Figure 4A, lanes 2–4). The addition of Zn2+ even at 1 mM completely inhibited RNase activity (Figure 4A, lanes 14–16). While addition of 1 mM MgCl2 did not have any effect on RNase I activity (Figure 4A, lanes 6–7), the presence of 4 mM Mg2+ dramatically inhibited dsRNA degradation (Figure 4A, lane 8). This was a rather surprising finding, as Ca2+ has been shown to functionally substitute for Mg2+ in other divalent cation binding proteins (18,19). Interestingly, we found that 1 mM Mn2+ also enhanced the ability of RNase I to degrade dsRNA (Figure 4A, lane 11). However, titrating MnCl2 from 0.5 mM to 10 mM revealed that while 1 to 2 mM Mn2+ stimulated RNase I activity, concentrations of 4 mM or above had no stimulatory effect on dsRNA degradation (Figure 4B, lanes 9–14). In contrast, Ca2+ concentrations of 1 mM or higher all activated the degradation of dsRNA (Figure 4B, lanes 2–7). Thus, we conclude that the stimulatory effect on RNase I-mediated dsRNA degradation is specific to Ca2+, even though Mn2+ also stimulated dsRNase activity at low concentrations.
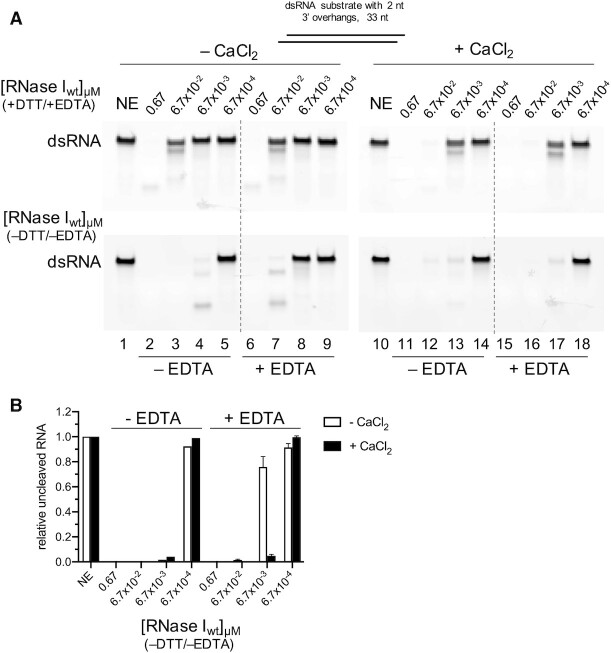
RNase I purified without EDTA does not require Ca2+ for its dsRNase activity
In order to find an explanation for our findings, we examined the previously published crystal structure of E. coli RNase I and discovered a calcium ion in the structure whose presence or potential function was not discussed before [2PQX](15) (Figure 5A). We hypothesized that the Ca2+-bound state of RNase I, and thus its ability to cleave both, single and double-stranded RNA substrates with high affinity, may be its inherent state. As previous in vitro studies on RNase I enzymes had been done with enzymes that were purified in buffers containing EDTA to inactivate potentially co-purifying metal-dependent nucleases and metalloproteases, we wondered if purification and storage in EDTA containing buffers could be sufficient to remove the Ca2+ from RNase I and thereby obscure its intrinsic dsRNase activity. We thus purified RNase I with buffers lacking DTT and EDTA, resulting in a highly pure enzyme (Supplementary Figure S1) that, in stark contrast to the previous preparation, exhibited high activity on a 33mer dsRNA substrate both in the presence and absence of Ca2+ (Figure 5B–D). 6.7 nM RNase Iwt (–DTT/–EDTA) showed comparable levels of activity as 6.7 nM Ca2+-activated RNase I purified in the presence of EDTA and DTT, regardless if Ca2+ was added to or omitted from the reaction (compare Figure 1B lane 6 and 1C with Figures 5B–C, lane 7 and 5D). However, RNase Iwt (–DTT/–EDTA) at lower concentrations failed to cleave the substrate, while the RNase I from the original preparation (+DTT/+EDTA), when supplied with CaCl2, was still able to partially digest dsRNA. This could be explained by the lack of stabilizing DTT in the reaction rendering the RNase I four methionine residues susceptible to oxidation and ultimately leading to inactivation of the enzyme (20,21). Collectively, our data supports the hypothesis that endogenous RNase I contains a bound Ca2+, which in turn allows it to efficiently cleave double- and single-stranded RNA.
Figure 5.
RNase I purified in the absence of EDTA exhibits an inherent strong dsRNase activity. (A) The crystal structure of RNase I (2PQX; Rodriguez et al., 2008) contained a previously overlooked Ca2+ binding site (colored in grey, coordinating Ca2+ represented as magenta sphere) that is located approximately 18 Å away from the enzyme's active site (colored in light pink). 0.1 μM dsRNA was incubated with various concentrations of RNase I as indicated above the lanes (from 6.7 to 6.7 × 10−4 μM) in the absence (B) or presence (C) of 4 mM CaCl2. No DTT or EDTA was present in the reactions shown in (B) and (C). The asterisks indicate the RNase I dilutions selected for MS analysis. The uncleaved RNA was quantified and plotted in (D). (E) MS results showing the location of the cleavage and the relative abundance of the resulting fragment for reactions containing 6.7 nM RNase I without (top) and with Ca2+ (bottom). FL, full-length oligonucleotide (uncleaved substrate).
Cleavage products of both 6.7 nM RNase Iwt (–DTT/–EDTA) ± CaCl2 reactions were further subjected to MS analysis. We observed similar cleavage patterns for RNase Iwt (–DTT/–EDTA) compared to RNase I (+DTT/+EDTA), with a main cleavage site at 8 nt (– CaCl2) or 9 nt (+ CaCl2) upstream of the top strand 3′ end. The opposite main cleavage sites were located 5–6 nt (– CaCl2) or 7 nt (+ CaCl2) downstream of the bottom strand 5′ end (Figure 5E, Supplementary Figure S16, and Supplementary Table S3A, B). These preferred cleavage sites were shared by both RNase I preparations and confirm the stimulatory effect of Ca2+.
If endogenous RNase I does contain Ca2+, we wondered if the metal could be removed post-purification by challenging it with various concentrations of EDTA. We incubated 0.1 μM dsRNA with various concentrations of RNase Iwt (–DTT/–EDTA) either in the absence or presence of 0.5 mM EDTA, both in the plus and minus 4 mM CaCl2 conditions. While the RNase Iwt (–DTT/–EDTA) was highly active in the absence of EDTA ± CaCl2 (Figure 6A, lower panel, lanes 2–4, 11–13), adding 0.5 mM EDTA strongly reduced the activity in the absence of Ca2+ (Figure 6A, lower panel, lanes 6–9, and Figure 6B). However, if Ca2+ was supplemented to a EDTA:Ca2+ ratio of 1:8, RNase I activity was fully recovered (Figure 6A, lower panel, lanes 15–18, and Figure 6B). The effect of EDTA was concentration dependent, with 4 mM EDTA having a stronger inhibitory effect in the absence of Ca2+ than with 0.5 mM EDTA (Supplementary Figure S10A, left panel, and Supplementary Figure S10B). However, at a 1:1 EDTA:Ca2+ ratio, EDTA did not affect RNase Iwt (–DTT/–EDTA) activity (Supplementary Figure S10A, right panel), suggesting Ca2+ is still bound to the enzyme. In agreement with our previous observations, neither Ca2+, nor challenging the reaction with up to 4 mM EDTA had a significant impact on the activity of RNase Iwt (–DTT/–EDTA) toward ssRNA (Supplementary Figure S10C). Surprisingly, we found that 4 mM EDTA even seemed to slightly, but reproducibly, stimulate ssRNase function at 13.4 nM RNase I in absence of CaCl2 (Supplementary Figure S10D). This stimulatory effect was lost when Ca2+ was added to the reaction, again strongly supporting the that the ssRNase activity is independent of Ca2+. Taken together these data suggest that endogenous RNase I contains a bound Ca2+ and that while this bound Ca2+ is required for its efficient dsRNase activity, RNase I ssRNase activity is completely metal independent.
Figure 6.
EDTA removes RNase I bound Ca2+ leading to the inhibition of its dsRNase activity. (A) 0.1 μM dsRNA and various concentrations of RNase I purified in the presence (top panel) or absence (bottom panel) of DTT and EDTA (–DTT/–EDTA) were incubated without or with 0.5 mM EDTA in the reaction, as indicated below the gels. Reactions in lanes 10–18 contained 4 mM CaCl2. The quantification of the uncleaved RNA substrate is shown in (B). NE, no enzyme control.
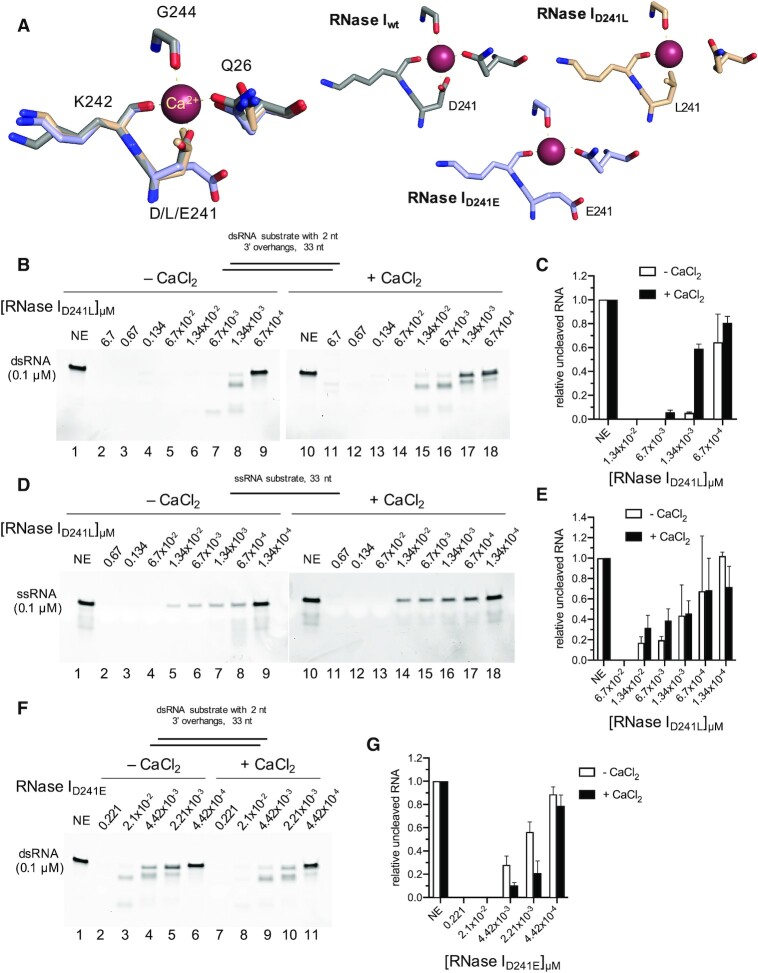
An aspartate to leucine substitution in the Ca2+ coordinating site on RNase I mimics the effect of Ca2+ binding
Ca2+ is coordinated by four amino acids in its binding site on RNase I: two via side chains (glutamine Q26 and aspartate D241), two via backbone carbonyl groups (lysine K242 and glycine G244), and two additional coordination sites are provided by water molecules [2PQX] (15) (Figure 5A, Supplementary Figure S11A). We reasoned that if Ca2+ binding to the enzyme was important for its dsRNase activity, manipulation of the metal binding site should have an impact on RNase I response to Ca2+. We designed a series of mutant RNase I variants containing substitutions at position D241, as we hypothesized that the aspartate with its two contact sites might be essential for Ca2+ coordination. We chose amino acids whose side chains would either likely disrupt Ca2+ binding (D241 to leucine, L; RNase ID241L) or would be more similar to the native amino acid being substituted (D241 to glutamate, E; RNase ID241E). Molecular modelling using MODELLER to generate homology models of both mutants suggested that in the D241L mutant, another coordinating residue, Q26, is positioned farther away from the Ca2+ binding site compared to its location in the wild type enzyme (Figure 7A). This in addition to the loss of two interaction sites due to the D to L substitution suggested impaired Ca2+ binding by the D241L mutant. Substituting D241 with glutamate did not affect Q26 positioning. However, the two oxygens in E241 of the RNase ID241E variant were facing away from the Ca2+ coordinating site. These two oxygens may still be able to support the binding of Ca2+, albeit their location suggests a weakened interaction and thus potentially altered response to Ca2+. Both MBP-tagged mutant proteins were purified and stored in buffers containing DTT (1 mM) and EDTA (0.5 mM) (Supplementary Figure S1). Enzymatic assays were conducted for both mutated enzymes as described for the wild-type RNase I.
Figure 7.
Activity of RNase I D241L and D241E mutants. RNase I cleavage of synthetic dsRNA or ssRNA using dilution series of RNase I mutant enzymes. (A) Structure (wt) and homology models (D241L/E) of the Ca2+ coordinating site on RNase I. Either a dsRNA substrate (B, F) or a ssRNA substrate (D) was cleaved by the indicated amount of RNase ID241L (B, D) or RNase ID241E (F) in the absence or presence of Ca2+. The concentration for each enzyme is indicated at the top of each lane. (C, E, G) show the respective quantifications of the uncleaved RNA substrates in (B, D, F). NE, no enzyme control.
Strikingly, RNase ID241L exhibited a dramatic increase in activity on dsRNA substrates in the absence of Ca2+ over RNase Iwt. About 97% of the full-length 33mer dsRNA substrate (featuring 2 nt 3′ overhangs) was degraded by 20 fmol of RNase ID241L versus only 8% by RNase Iwt (Figure 7B, lane 6 and Figure 7C versus Figure 1A, lane 8 and Figure 1C). Substitution of the aspartate for a leucine resulted in a mutant whose activity resembled that of RNase Iwt (–DTT/–EDTA) and that of RNase Iwt (+DTT/+EDTA) in presence of Ca2+ (Figure 7B, lane 7 and Figure 7C versus Figure 1B, lane 6 and Figure 1C versus Figure 5B, lane 7 and Figure 5D). Surprisingly, addition of Ca2+ to the reaction resulted in a slight reduction of dsRNA degradation (Figure 7B, lanes 10–18 and Figure 7C), suggesting that the D241L mutation may block the enzyme from responding to calcium in the same fashion as RNase Iwt. Similar results were obtained for treatment of the blunted-ended 31mer dsRNA substrate with RNase ID241L (Supplementary Figure S11B–D). Despite the dramatic increase in dsRNase activity in the absence of Ca2+, the ssRNase activity of RNase ID241L remained similar to that of RNase Iwt (Figure 7D–E versus Figure 2). Supplementing Ca2+ did not change RNase ID241L activity on ssRNA, suggesting that the D241L mutation is a gain-of-function mutation that exclusively affects dsRNA degradation.
Similar to what was observed for D241L, substituting D241 with glutamate (D241E), a more similar amino acid to aspartate, resulted in an enzyme with increased activity against dsRNA relative to RNase Iwt (Figure 7F, lanes 2–6 and Figure 7G versus Figure 1A,C). However, unlike the D241L mutant, cleavage of dsRNA by RNase ID241E could be activated by CaCl2, although to a significantly lesser extent than by RNase Iwt. For instance, Ca2+ addition led to a decrease of uncleaved RNA from ∼55% to ∼20% at an E:S ratio of 0.022:1 for the D241E mutant (Figure 7F, lanes 5, 10 and Figure 7G). For the wild-type enzyme, the amount of uncleaved RNA was reduced from ∼93% to ∼19% at an E:S ratio of 0.0134:1 (Figure 1A, lane 8, Figure 1B, lane 8, and Figure 1C). We speculate that, in contrast to D241L, the D241E mutation does not fully disrupt the Ca2+ interaction with the enzyme. In summary, mutations in the Ca2+ binding site strongly affected RNase I activity on dsRNA and its response to supplemented Ca2+, again supporting the finding that a bound Ca2+ is required for inherent dsRNase function.
DISCUSSION
RNase I has previously been described as a metal-independent and non-sequence specific endoribonuclease and has been used as a tool in molecular biology for removing ssRNA from reactions for decades. Inspired by early work on Enterobacter ribonuclease, which showed that the addition of Ca2+ resulted in a substantial increase in degradation of yeast tRNA and suggested that Ca2+ may have a role in regulating dsRNA degradation by RNases (13), we investigated the effect of Ca2+ and other divalent cations on the activity of E. coli RNase I. Our findings suggest that endogenous E. coli RNase I contains a bound Ca2+ that is not essential for its ssRNase activity, but critical for the previously overlooked strong inherent dsRNase and hybridase activities of RNase I.
RNase Iwt purified in EDTA-containing buffers required supplementary Ca2+ in order to efficiently cleave dsRNA or the RNA strand of a double-stranded DNA:RNA hybrid. In contrast, RNase Iwt purified in the absence of EDTA and DTT retained its inherent dsRNase activity independently of supplementary Ca2+, unless challenged with EDTA. Our experimental data indicate that RNase I is Ca2+-bound in its native state, allowing the enzyme to target double- and single-stranded RNA substrates with high efficiency. In agreement, we identified the presence of a bound Ca2+ in a previously published crystal structure of E. coli RNase I. However, most buffers used for protein purification contain the metal-chelating EDTA in order to inhibit metal-dependent nucleases and proteases (22), which has likely precluded a more in depth understanding of the multiple function of RNase I. Our data demonstrates that by capturing Ca2+, EDTA dramatically reduces RNase I dsRNase activity.
Of particular interest was a mutation in the Ca2+ coordinating site on RNase I. Altering the Ca2+ binding site by removing two Ca2+ coordinating oxygens and slight structural rearrangement of a third Ca2+-contacting residue resulted in a gain-of-function variant of RNase I that displayed a dramatic increase in dsRNase activity relative to that of the wild-type enzyme purified with DTT and EDTA-containing buffers. We speculate that the structural change induced by the D241L mutation mimics a Ca2+ bound-like conformation that also is not able to actually bind Ca2+ anymore. This is supported by the finding that supplementary Ca2+ did not result in an increased activity towards either double-stranded or single-stranded RNA substrates. Interestingly, the D241E mutation, wherein glutamate is replaced with an amino acid of similar properties, yielded an RNase I variant with higher activity on dsRNA than RNase Iwt but was still inducible by the addition of Ca2+, although not to the same extent as the wild type.
However, it is still unclear how a Ca2+-binding site located ∼18 Å away from the catalytic site (Supplementary Figure S11E), in a fairly nonconserved region of the enzyme, can have such a dramatic influence on substrate specificity. One possibility is that Ca2+ binding stabilizes the enzyme in a way that it can accept the binding or retain double-stranded substrates in its active site. The center of RNase I is formed by a main β-sheet that harbors four of the seven active site residues (15). Of the six β-strands, β1 is directly connected to the α1 helix containing the Ca2+-coordinating Q26. In addition, β6 is positioned directly upstream of the Ca2+ binding amino acid positions D241 and L242. Binding of Ca2+ (or a D241L mutation) may result in a subtle conformational rearrangement of the β-sheet, enough to alter the specificity of the enzyme. A similar mechanism was reported by Pan and Lazarus (23) in human DNase I, a calcium-sensitive nuclease, where the binding of Ca2+ to a portion of the enzyme resulted in the stabilization of an adjacent secondary structure, thus allowing an extended activity of the enzyme over a wider range of substrates. Doucet et al. (24) reported an external loop stabilization in RNase A that caused an increased substrate release, thus significantly influencing the activity of the enzyme. While not directly supporting catalysis, a bound Ca2+ ion was found to reduce the local negative charge density and thereby promoting stable binding of dsRNA binding to E. coli RNase III (25). We speculate that only Ca2+-bound RNase I can accommodate and coordinate a stretch of dsRNA in its active site, while the loss of Ca2+ leads to structural and/or surface charge changes in the active site that either impair substrate binding, retention, or the subsequent catalytic processes.
Our MS data showed that RNase I displayed a strong preference for one end over the other in all dsRNA substrates tested. dsRNA degradation starts with an initial cleavage event close to the preferred end, resulting in a primary cleavage product, before cleavage will eventually occur at the other end. The presence of a primary cleavage product was very surprising to us, as RNase I has historically been described as a non-sequence specific endoribonuclease. Even more striking were our MS data, showing that no matter what dsRNA template we tested, the initial cleavage event occurred close to the end in one strand, and was always followed by cleavage of the opposite nucleotide in the complimentary strand. There was no apparent sequence or structural similarity between the dsRNA substrates tested, initially suggesting that the primary cleavage is not caused by a preference of the enzyme for a sequence or three-dimensional fold. We speculate that the primary cleavage product is a result of a specific binding orientation of the dsRNA to the enzyme, which is facilitated by the Ca2+-bound state of RNase I or its D241L mutant. However, it is unclear a) why the enzyme displays such a strong preference for one particular end of all dsRNA substrates tested, b) why the third and fourth cuts occurred at the opposite dsRNA end, and c) if the dsRNA substrate is released and rebound between these two events. Adding FAM-labels to the preferably cleaved end of the dsRNA did alter the preferred cleavage site by shifting it 2–3 nucleotides closer to the end. Surprisingly, the addition of a bulky fluorophore did not change the affinity of RNase I to cleave at that particular end (compare Figure 1C with Supplementary Figure S3), suggesting that a yet to be determined sequence feature dictates the cleavage preference for one end over the other. This feature may provide an RNA fold that is preferably accommodated by the active site of Ca2+-bound RNase I. We hypothesize that such a feature could either be only partially (substrates in Supplementary Figure S6) or not (substrate in Figure 1C) present in the opposing end of the substrates, explaining why we do or do not observe a secondary cleavage event at that respective end. Further investigation is needed to answer these questions and to understand the detailed mechanism of dsRNA cleavage by RNase I.
Interestingly, we find that low concentrations of Mn2+ also have a stimulatory effect on RNase I, initially suggesting that Mn2+ and Ca2+ may be interchangeable in inducing its dsRNase activity. It is common for enzymes that bind Mg2+ and Zn2+ to also bind Ca2+; however, far fewer enzymes can tolerate metal exchange and still retain full activity. For example, many metal-dependent ribonucleases, including RNase III and RNase H, require Mg2+ but are inactive when Mg2+ is substituted with Ca2+. This is also true for many DNA endonucleases, where binding and structural studies rely on the ability of Ca2+ to form a site-specific, non-catalytic enzyme complex that would be otherwise inaccessible for analysis (26–29). Ca2+ concentrations of 1 mM and higher consistently stimulate RNase I dsRNase activity. In contrast, and rather surprisingly, Mn2+concentrations of 2 mM or higher gradually lose their activating effect. One possible explanation is that the Mn2+-induced stimulatory effect on dsRNA cleavage is due to Mn2+ substituting Ca2+ in the metal binding site of RNase I, while the reversal of activation at high Mn2+ concentrations is caused by a secondary, yet unknown, effect.
A fascinating question to ask is if RNase I strong dependency on Ca2+ for its dsRNase activity has biological significance in a sense that it may be used to regulate RNase I function in vivo. Our preliminary kinetic measurements of the enzymatic efficiency of RNase I suggest that ssRNA is the preferred substrate for RNase I, closely followed by dsRNA when Ca2+ is present. The function of Ca2+ in bacteria has been difficult to determine and only recently have studies begun to shed light on the regulatory role of the metal in prokaryotes as well as in eukaryotes (reviewed by Dominguez (30)). High concentrations of free Ca2+ have been reported in the E. coli periplasm (31), indicating that availability of Ca2+ may not be a limiting factor. While there still is some uncertainty on the cellular role of RNase I (reviewed by Deutscher (32)), new studies showed that bacterial RNase I is involved in the regulation of a multitude of processes from motility, metabolism, and resistance (33,34). These regulatory functions have been linked to the generation of 2′,3′-cyclic nucleotide monophosphates (2′,3′-cNMPs) by RNase I through RNA scavenging (35,36), where the ability to efficiently degrade single-stranded as well as double-stranded or highly structured RNA is highly desirable. Indeed, we found that RNase Iwt (–DTT/–EDTA) exhibited higher activity on a pre-folded Leu-tRNA than the RNase Iwt variant that had been purified in the presence of EDTA (data not shown). Our finding that Ca2+-bound RNase I can process ssRNA and dsRNA at comparable rates supports the role of RNase I as an RNA scavenging enzyme and the hypothesis that functional RNase I in the cell is Ca2+-bound.
The finding that the addition of CaCl2 to an in vitro reaction significantly expands the substrate specificity of RNase I paves the way for new applications of this enzyme in molecular biology. RNase I is predominantly used to remove RNA from DNA solutions or protein preparations, as well as in Ribonuclease protection assays. Much more efficient removal of highly structured dsRNA substrates, which are typically much harder to degrade, may now be enabled by supplementing Ca2+ to a reaction containing commercially available RNase I preparations. Our findings also suggest that RNase protection assays, or all assays in which RNase I is used to enrich for RNA-containing double-stranded substrates, should be performed strictly in the absence of Ca2+ or Mn2+ to avoid degradation of the dsRNA.
Here, we present the previously undescribed Ca2+-dependent dsRNA degradation and hybridase activities of E. coli RNase I, 61 years after its discovery. While not affecting the endoribonuclease activity on ssRNA, Ca2+ binding to RNase I is required to efficiently degrade dsRNA. Our data suggest that the Ca2+-bound state is the endogenous form of RNase I, allowing the enzyme to target a broad selection of single- or double-stranded RNA substrates.
Supplementary Material
ACKNOWLEDGEMENTS
The authors want to thank Dr. Chudi Guan for valuable advice and discussion during the initial phase of this project.
Contributor Information
Sebastian Grünberg, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Baptiste Coxam, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Tien-Hao Chen, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Nan Dai, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Lana Saleh, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Ivan R Corrêa, Jr., New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA
Nicole M Nichols, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
Erbay Yigit, New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs, Inc. Funding for open access charge: New England Biolabs, Inc.
Conflict of interest statement. S.G., T-H.C., N.D., L.S., I.R.C., N.M.N. and E.Y. are employees of New England Biolabs, Inc. New England Biolabs is a manufacturer and vendor of molecular biology reagents, including several enzymes and buffers used in this study. This affiliation does not affect the authors impartiality, adherence to journal standards and policies, or availability of data.
REFERENCES
- 1. Deshpande R.A., Shankar V.. Ribonucleases from T2 family. Crit. Rev. Microbiol. 2002; 28:79–122. [DOI] [PubMed] [Google Scholar]
- 2. Irie M. Structure-function relationships of acid ribonucleases: lysosomal, vacuolar, and periplasmic enzymes. Pharmacol. Ther. 1999; 81:77–89. [DOI] [PubMed] [Google Scholar]
- 3. Aravind L., Koonin E.V.. A natural classification of ribonucleases. Methods Enzymol. 2001; 341:3–28. [DOI] [PubMed] [Google Scholar]
- 4. Kennell D. Processing endoribonucleases and mRNA degradation in bacteria. J. Bacteriol. 2002; 184:4645–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spahr P.F., Hollingworth B.R.. Purification and mechanism of action of ribonuclease from Escherichia coli ribosomes. J. Biol. Chem. 1961; 236:823–831. [Google Scholar]
- 6. Elson D. Latent enzymic activity of a ribonucleoprotein isolated from Escherichia coli. Biochim. Biophys. Acta. 1959; 36:372–386. [DOI] [PubMed] [Google Scholar]
- 7. Neu H.C., Heppel L.A.. The release of ribonuclease into the medium when Escherichia coli cells are converted to speroplasts. J. Biol. Chem. 1964; 239:3893–3900. [PubMed] [Google Scholar]
- 8. Neu H.C., Heppel L.A.. Some observations on the ‘Latent’ ribonuclease of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1964; 51:1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu L., Deutscher M.P.. The Escherichia coli rna gene encoding RNase I: sequence and unusual promoter structure. Gene. 1992; 119:101–106. [DOI] [PubMed] [Google Scholar]
- 10. Meador J. 3rd, Kennell D.. Cloning and sequencing the gene encoding Escherichia coli ribonuclease I: exact physical mapping using the genome library. Gene. 1990; 95:1–7. [DOI] [PubMed] [Google Scholar]
- 11. Padmanabhan S., Zhou K., Chu C.Y., Lim R.W., Lim L.W.. Overexpression, biophysical characterization, and crystallization of ribonuclease I from Escherichia coli, a broad-specificity enzyme in the RNase T2 family. Arch. Biochem. Biophys. 2001; 390:42–50. [DOI] [PubMed] [Google Scholar]
- 12. Cannistraro V.J., Kennell D.. Ribonuclease YI*, RNA structure studies, and variable single-strand specificities of RNases. Methods Enzymol. 2001; 341:175–185. [DOI] [PubMed] [Google Scholar]
- 13. Frank J.J., Hawk I.A., Levy C.C.. Peptides isolated from Enterobacter nuclease as potential polyamine binding sites. Biochim. Biophys. Acta. 1976; 432:369–380. [DOI] [PubMed] [Google Scholar]
- 14. Cannistraro V.J., Kennell D.. RNase YI* and RNA structure studies. Nucleic Acids Res. 1997; 25:1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodriguez S.M., Panjikar S., Van Belle K., Wyns L., Messens J., Loris R.. Nonspecific base recognition mediated by water bridges and hydrophobic stacking in ribonuclease I from Escherichia coli. Protein Sci. 2008; 17:681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Greenough L., Schermerhorn K.M., Mazzola L., Bybee J., Rivizzigno D., Cantin E., Slatko B.E., Gardner A.F.. Adapting capillary gel electrophoresis as a sensitive, high-throughput method to accelerate characterization of nucleic acid metabolic enzymes. Nucleic Acids Res. 2016; 44:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Libonati M., Sorrentino S.. Revisiting the action of bovine ribonuclease A and pancreatic-type ribonucleases on double-stranded RNA. Mol. Cell. Biochem. 1992; 117:139–151. [DOI] [PubMed] [Google Scholar]
- 18. Ralec C., Henry E., Lemor M., Killelea T., Henneke G.. Calcium-driven DNA synthesis by a high-fidelity DNA polymerase. Nucleic Acids Res. 2017; 45:12425–12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swan M.K., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009; 16:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liang X., Kaya A., Zhang Y., Le D.T., Hua D., Gladyshev V.N.. Characterization of methionine oxidation and methionine sulfoxide reduction using methionine-rich cysteine-free proteins. BMC Biochem. 2012; 13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim G., Weiss S.J., Levine R.L.. Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta. 2014; 1840:901–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cox J.A., Wnuk W., Stein E.A.. Isolation and properties of a sarcoplasmic calcium-binding protein from crayfish. Biochemistry. 1976; 15:2613–2618. [DOI] [PubMed] [Google Scholar]
- 23. Pan C.Q., Lazarus R.A.. Ca2+-dependent activity of human DNase I and its hyperactive variants. Protein Sci. 1999; 8:1780–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doucet N., Watt E.D., Loria J.P.. The flexibility of a distant loop modulates active site motion and product release in ribonuclease A. Biochemistry. 2009; 48:7160–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Nicholson A.W.. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical footprinting using catalytically inactive RNase III mutants. EMBO J. 1996; 15:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 26. Cao W. Binding kinetics and footprinting of TaqI endonuclease: effects of metal cofactors on sequence-specific interactions. Biochemistry. 1999; 38:8080–8087. [DOI] [PubMed] [Google Scholar]
- 27. Daniels L.E., Wood K.M., Scott D.J., Halford S.E.. Subunit assembly for DNA cleavage by restriction endonuclease SgrAI. J. Mol. Biol. 2003; 327:579–591. [DOI] [PubMed] [Google Scholar]
- 28. Embleton M.L., Williams S.A., Watson M.A., Halford S.E.. Specificity from the synapsis of DNA elements by the Sfi I endonuclease. J. Mol. Biol. 1999; 289:785–797. [DOI] [PubMed] [Google Scholar]
- 29. Vipond I.B., Halford S.E.. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995; 34:1113–1119. [DOI] [PubMed] [Google Scholar]
- 30. Dominguez D.C. Calcium signalling in bacteria. Mol. Microbiol. 2004; 54:291–297. [DOI] [PubMed] [Google Scholar]
- 31. Jones H.E., Holland I.B., Campbell A.K.. Direct measurement of free Ca(2+) shows different regulation of Ca(2+) between the periplasm and the cytosol of Escherichia coli. Cell Calc. 2002; 32:183–192. [DOI] [PubMed] [Google Scholar]
- 32. Deutscher M.P. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006; 34:659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duggal Y., Fontaine B.M., Dailey D.M., Ning G., Weinert E.E.. RNase I modulates Escherichia coli motility, metabolism, and resistance. ACS Chem. Biol. 2020; 15:1996–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fontaine B.M., Martin K.S., Garcia-Rodriguez J.M., Jung C., Briggs L., Southwell J.E., Jia X., Weinert E.E.. RNase I regulates Escherichia coli 2′,3′-cyclic nucleotide monophosphate levels and biofilm formation. Biochem. J. 2018; 475:1491–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nicholson A.W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999; 23:371–390. [DOI] [PubMed] [Google Scholar]
- 36. Condon C., Putzer H.. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002; 30:5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.