Abstract
G-quadruplexes (G4s), higher-order DNA and RNA secondary structures featuring guanine-rich nucleic acid sequences with various conformations, are widely distributed in the human genome. These structural motifs are known to participate in basic cellular processes, including transcription, splicing, and translation, and their functions related to health and disease are becoming increasingly recognized. In this review, we summarize the landscape of G4s involved in major neurodegenerative disorders, describing the genes that contain G4-forming sequences and proteins that have high affinity for G4-containing elements. The functions of G4s are diverse, with potentially protective or deleterious effects in the pathogenic cascades of various neurological diseases. While the studies of the functions of G4s in vivo, including those involved in pathophysiology, are still in their early stages, we will nevertheless discuss the evidence pointing to their biological relevance. A better understanding of this unique structural element in the biological context is important for unveiling its potential roles in the pathogenesis of diseases such as neurodegeneration and for designing new diagnostic and therapeutic strategies.
INTRODUCTION
Nucleic acids, including DNA and RNA, represent the basic molecular code of life. The functions of nucleic acids are determined not only by their primary sequences but also their secondary or higher-order structures. The most abundant and thus the most studied secondary nucleic acid structures are formed by base-pairing through conventional Watson–Crick hydrogen bonding. However, alternative stable structures can also arise, such as G-quadruplexes (G4s), in which guanine bases are connected with Hoogsteen hydrogen bonds (1,2). Originally identified in in vitro experimental settings, G4s are four-stranded secondary structures formed in guanine-rich DNA or RNA sequences. The resulting structures exhibit high thermodynamic stability under near-physiological conditions as well as resistance to nuclease activity (3,4). G4s are increasingly recognized for playing many different roles in cellular environments associated with both normal physiology and pathology. There have been several reviews on the disease relevance of G4s (5–7), with oncological studies in particular drawing attention to these structural elements as candidates for therapeutic intervention (8,9). Interestingly, G4s have increasingly also been associated with a diverse set of genes and pathways implicated in neurological disorders. While the involvement of G4s in the regulation of selected neurological diseases and non-coding RNAs has been described previously (10), what has been lacking is a comprehensive discussion of the role of G4s within many neurological disorders, with an emphasis on the predicted outcomes and established mechanisms underlying the G4-induced pathological cascade. Here, we summarize and discuss the roles of G4s associated with various genetic players in neurodegeneration, such as Alzheimer's disease (AD), fragile X syndrome (FXS), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Parkinson's disease (PD), and prion disease (Table 1).
Table 1.
Summary of the roles and predicted effects of G-quadruplexes in neurodegenerative diseases
| Disease | Gene/protein | Role of G4s |
|---|---|---|
| Alzheimer's disease (AD) | APP | Overproduction is suppressed by a 3′ UTR G4 motif (101). |
| ADAM10 | Translation is inhibited by a parallel 5′ UTR G4 (80). | |
| BACE1 | Full-length (501) transcript production is activated by a G4 motif in 3rd exon during alternative splicing (106). | |
| Fragile X Syndrome (FXS) | FMR1/FMRP | Inhibits translational machinery of SMNDC1 (97), Shank1 (95), PSD-95(98) via interactions with G4 motifs to prevent pathogenic upregulation. Alternative splicing is negatively regulated by G4s found in the coding region of the FMR1 mRNA (105). |
| Amyotrophic Lateral Sclerosis/Frontal-temporal Dementia (ALS/FTD) | C9orf72 | Transcription is negatively regulated by DNA G4s formed at C9orf72 HRE (37). C9orf72 RNA G4s are implicated in regulation of many cellular processes. |
| TDP-43 | Transport of mRNA to neurites for translation is dependent on the presence of 3′UTR G4s in mRNAs (124). | |
| ANG | Cleavage product-induced G4 formation is critical for stress granule formation and translational inhibition (131). | |
| Parkinson's Disease (PD) | SNCA | 5′ UTR G4 motifs suppress cap-dependent translation (79) and influence cap-independent translation. |
| Prion Disease | PrP | Deleterious conversion to PrPSc is triggered by PrP binding to G4 motifs (136). |
| Progressive Myoclonus Epilepsy Type I (PME1) | CSTB | Dodecamer repeat forms parallel G4s at physiological pH (138). |
Formation of G-quadruplex structures
The unique structural formation of G4s governs their role in the cell. Four guanine bases bonded together, often referred to as a G-quartet or G-tetrad, form a square planar arrangement in which each of the four bases acts as a donor and acceptor of two hydrogen bonds (Figure 1A) (11). G4s are the result of two or more G-quartets stacked sequentially. Depending on the topology and sources of the folding strands, G4s can adopt parallel, antiparallel, or hybrid complexes intra- or inter-molecularly (Figure 1B). G4s can be formed by DNA, RNA or a hybrid of both. Several distinctive features and structures contribute to the stability of G4s. First, G4s are stabilized by Hoogsteen hydrogen bonds between the G-rich strands of the nucleotides (2) and by π-orbital interactions among the stacked quartets (12). One of the most important factors involved in stabilizing G4s is the coordination of a monovalent metal ion, usually K+ (13). Metal ion coordination occurs when metal ions enter an interior channel, formed by Hoogsteen binding patterns and stacking interactions, and interact with the oxygen at position 6 (O6) of each guanine atom throughout the length of the structure, counteracting the negative electrostatic effects of carbonyl groups (14). Several factors dictate which physiologically relevant cation stabilizes the G4 structures, including the ionic radius, hydration energy, and binding strength toward O6. K+ is typically the favored cation because of its smaller effective ionic radius and lesser dehydration free energy. Accordingly, physiologically relevant concentrations of K+ have been shown to stabilize G4s in vitro (15).
Figure 1.
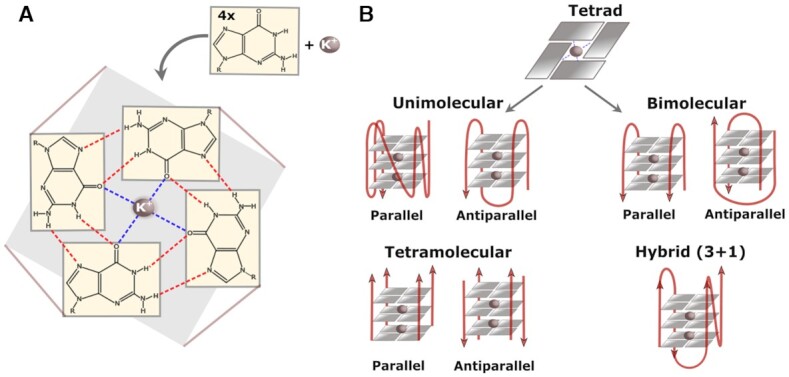
Experimentally observed G-quadruplex structures, orientations, and conformations. (A) G4s are nucleic acid secondary structures consisting of four guanine bases organized in a square planar arrangement and connected through Hoogsteen hydrogen bonds. Within the central channel, dipole interactions between guanine O6 and a cation, usually K+, lend additional stability to the G4 structure. (B) There is great structural heterogeneity in the G4s, with these motifs capable of adopting parallel, antiparallel, or hybrid conformations in a unimolecular, bimolecular or tetramolecular manner.
Multifaceted cellular functions of G-quadruplexes
There are hundreds of thousands of sites in the human genome containing G-rich sequences that are predicted by computational methods to form G4s (16,17). The chemical properties of G4s have been extensively characterized in vitro, although the nature and prevalence of these structures in vivo remain a subject of debate (18). However, with the recent development of G4 structure-specific antibodies, small molecule ligands, and chemical probing, DNA and RNA G4s have been increasingly detected in cells under physiological conditions (19–22). Studies of the biological implications of G4s can be classified into three categories of research that provide different levels of evidence: (i) bioinformatic and computational predictions of G4 motifs, (ii) in vitro studies of G4s and (iii) in vivo studies of G4s. Although the study of G4 function in a cellular context is ideally elucidated through in vivo evidence, the purpose of our review is to highlight and offer timely discussion concerning the interesting roles and functions of G4s that are revealed through all three types of research, in order to promote further study of the implications of G4s in neurological disorders. To provide context for our later discussion, we will first present an overview of the impact of G4s on transcription, translation, splicing and other cellular functions.
G4s are thought to play an important role in regulating transcription (Figure 2A). G4 motifs are enriched in promoter regions of the human genome, with their regulation of oncogenic promoters especially well characterized (17,23). G4-specific antibodies have been utilized in chromatin immunoprecipitation (ChIP) and immunohistochemical analyses to detect the in vivo formation of G4s in the cell. For example, a G4-specific antibody hf2 or BG4 has been used to demonstrate the influence of G4s on transcription (24,25). By analyzing gene expression patterns associated with DNA G4s using ChIP analysis, G4s have been shown to be capable of enhancing or inhibiting transcription (25,26). The extent of studies elucidating mechanisms by which G4s affect transcription is still limited, but we will discuss the possible mechanistic scenarios in the following sections on DNA G4s and transcription.
Figure 2.
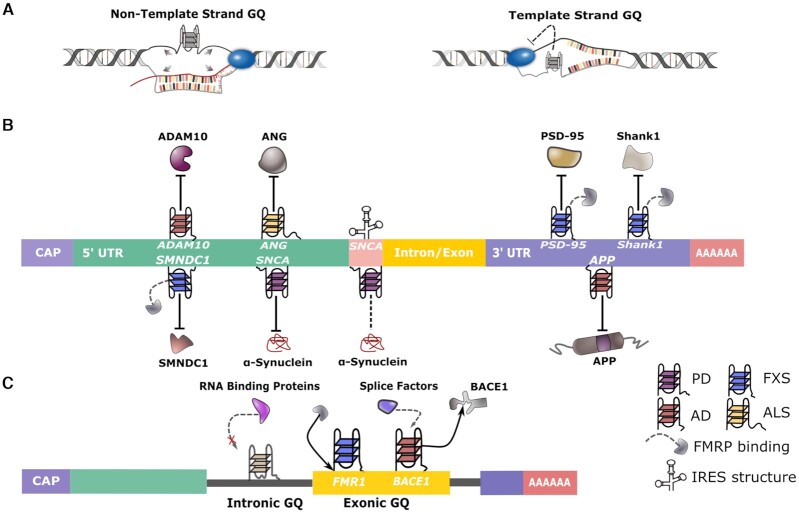
Proposed G-quadruplex regulation of biological processes. G4s regulate transcription, translation, and splicing. (A) Non-template strand G4s can regulate transcription by keeping the template strand single-stranded or by promoting formation of R-loops. Template strand G4s could regulate transcription by blocking RNA polymerase progression. (B) G4s found in mRNAs, particularly in regulatory regions such as the 5′-UTR, IRES and 3′-UTR, can potentially regulate the translation of proteins associated with AD, FXS, ALS and PD. G4s may influence both cap-dependent and cap-independent translation, and G4-interacting proteins such as FMRP are also involved. Arrows indicate inhibition of translation. (C) G4 motifs are found upstream and downstream of splice junctions. G4 structures may prevent access to RNA protein binding sites, mediate the binding of regulatory proteins such as FMRP, or recruit splice factors to influence alternative splicing, such as the selective generation of BACE1 isoforms.
G4s can also influence translation (Figure 2B). G4s that are enriched in the regulatory regions of mRNAs, such as the 5′ untranslated region (5′-UTR) and the 3′-UTR, can exert control over the translation process (27,28). Studies focused on the most regulated stage of translation, translation initiation, have identified G4s as possible inhibitors of initiation that could act to prevent cap-dependent translation from occurring. In various mRNAs, G4s have also been identified in the internal ribosome entry site (IRES), an RNA element allowing for the initiation of cap-independent translation. Regulation of IRES-mediated, cap-independent translation by G4 motifs is well documented although, as we discuss later, the exact effect of the G4 motifs remains controversial. The well-established binding between fragile X mental retardation 1 protein (FMRP) and G4s has also hinted at a potential regulatory loop in which G4s found in FMRP-binding mRNAs help suppress the translation of these mRNAs, which as discussed below has been linked to the pathogenesis of FXS or other neurological diseases.
RNA G4s are also involved in the regulation of splicing (Figure 2C). These secondary structures have been implicated as regulators of pre-mRNA processing, including adenylation and alternative splicing (29). Although studies elucidating the exact mechanisms through which G4-involved splicing occur are limited, as we discuss in our later section on G4’s regulation of splicing in both cis- and trans-acting mechanisms, G4 binding to splicing factors is a common theme shared by the examples we provide.
G4s have also been identified as regulatory players in other cellular processes. Given the multifaceted influences of G4s and their high prevalence within the cell, they are likely involved in a myriad of cellular functions. As selected examples, we will discuss the influence of G4s in RNA-protein interactions, including those involving membraneless RNP granules, nucleocytoplasmic transport, the tRNA stress response, and more, when we take a deeper look at the mechanisms of G4-dependent regulations relevant to several neurological diseases in the sections below.
DNA G-QUADRUPLEXES AND TRANSCRIPTION
Several studies have pointed to a role for DNA G4s in the regulation of transcription. For the transcription-linked disease genes that we discuss in this section, the most thoroughly studied role of G4s in transcription belongs to their effect on C9orf72, which has been implicated in ALS and FTD (Figure 3). ALS is a neurological disorder marked by progressive degeneration of motor neurons within the brain and spinal cord. Closely related to ALS is FTD, a disease characterized by continuous neuronal loss in the frontal and temporal cortices. Given their shared genetic causes and other neuropathological similarities, ALS and FTD are thought to exist within the same spectrum of disease (30,31). The most common cause of ALS-FTD is a hexanucleotide repeat expansion (HRE), (GGGGCC)n, in a non-coding region of the chromosome 9 open reading frame 72 (C9orf72) gene (32,33). ALS-FTD patients typically harbor thousands of HRE repeats, in contrast to healthy controls that normally possess fewer than 25 repeats (32). The proposed mechanisms of pathogenesis in ALS-FTD include loss of C9orf72 function, toxicity of the HRE RNA, and generation of aberrant poly-dipeptides through repeat-associated non-ATG-dependent translation (34). It has been established that the C9orf72 HRE adopts stable G4 structures, potentially implicating these motifs in the pathology of ALS and FTD (35–37).
Figure 3.
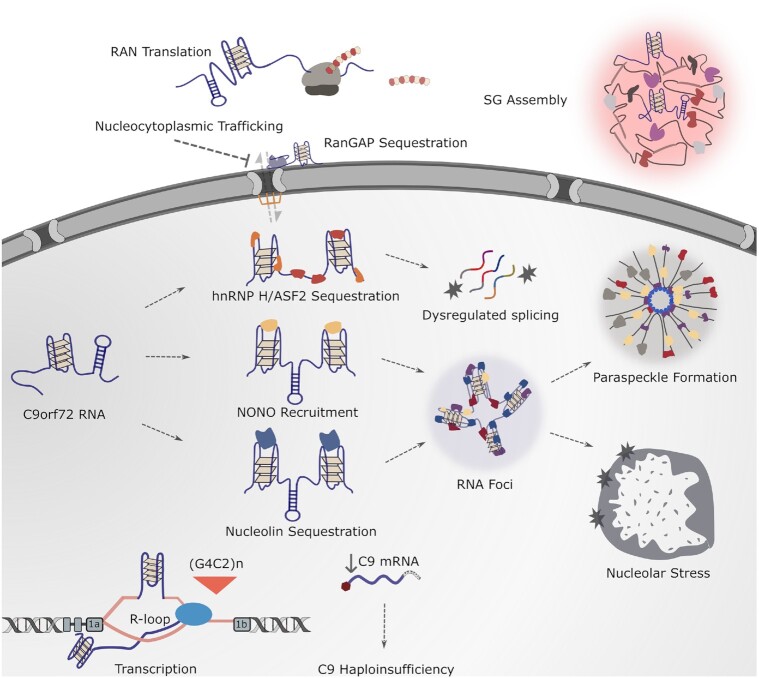
The role of G-quadruplexes in pathogenic cascades associated with the hexanucleotide repeat expansion in C9orf72. The C9orf72 DNA and RNA HRE are involved in a range of molecular cascades underlying ALS-FTD pathology. DNA G4s stabilize R-loop formation, which together results in the suppression of transcription. RNA G4s recruit several protein factors, including the splicing factors hnRNP H and ASF2, the paraspeckle protein NONO, and the nucleolar component nucleolin. As a result, a variety of functional consequences arise, including splicing dysregulation, RNA foci formation, paraspeckle dysregulation, and nucleolar stress. C9orf72 RNA G4s are also involved in nucleocytoplasmic trafficking, stress granule assembly, and possibly non-canonical repeat-associated translation.
DNA G-quadruplex-mediated negative regulation of transcription
It has been demonstrated through in vitro transcriptional assays that DNA G4s formed at the C9orf72 HRE block transcription by impairing RNA polymerase processivity, leading to decreased expression of the C9orf72 gene (Figure 3) (37). In addition to C9orf72, DNA G4s have also been shown to inhibit transcription in BRCA1, a gene encoding a critical DNA repair factor that has also been linked to AD because of its reduced levels in AD mouse models and patient tissues (38,39), and in the prion protein gene (PRNP). In the case of BRCA1, Pyridostatin, a G4-binding small molecule, stabilizes G4s in the BRCA1 promoter and represses BRCA1 transcription in rat cortical neurons, compromising double-stranded break repair activity and inducing neurotoxicity resulting from cumulative insults to genomic integrity (40). A recent study investigating interactions between G4s and the prion protein has reported the presence of two G4 motifs in the promoter region of PRNP that can form hybrid G4 structures (41). One of the G4 motifs has been shown to inhibit transcription, and prion protein (PrP) apparently has the capacity to interact with and induce the unfolding of that same G4, suggesting that PrP can bind to G4 motifs in its own promoter to auto-regulate transcription (41). Given the enrichment of G4 motifs at the promoters in the human genome (17,24), it is likely that many genes whose transcription is regulated by these secondary structures are relevant to the neurological diseases discussed here.
The functions of G4s as transcriptional switches likely depend on their conformation and stability, which can be influenced by their repeat and flanking sequences, the lengths of the repeats, and environmental factors. For example, the repeat length of the C9orf72 HRE DNA, d(G4C2)n, influences whether the C9orf72 HRE adopts a parallel or antiparallel topology (37,42). In vitro, the dominant and most stable conformation formed from the C9orf72 HRE DNA, an antiparallel G4, arises from d(G4C2)4 and leads to decreased transcription (37). The antiparallel G4 formed from the d(G4C2)4 repeat has a monomeric chair-type conformation with a characteristic antiparallel G-tetra core and three edgewise loops, but with distinct 4-layer stacking (43). Other repeat lengths, such as d(G4C2)2, d(G4C2)3 and d(G4C2)5, have been shown to form heterogeneous G4 mixtures with parallel and antiparallel topologies (37,43,44). A recent study using homogenous d(G4C2)2 samples isolated through anion exchange chromatography has demonstrated that the d(G4C2)2 repeat can form intermolecular G4s with a parallel conformation that fold as symmetric tetramers or with an antiparallel conformation that fold as asymmetric dimers (44). While the topologies of G4 structures formed in the PRNP gene have not been elucidated to the same extent as those in C9orf72, there is evidence to indicate that the G4 motif found at the PRNP promoter region likely adopts a hybrid (3+1) topology. Circular dichroism (CD) spectroscopy has revealed a CD profile with positive peaks around 260 and 290 nm and a negative peak around 240 nm (41), indicating a hybrid structure with three parallel strands and one antiparallel strand (45). To date, most studies on G4 and related structures are limited to in vitro analyses; probing these structures under native conditions in the cells awaits new technologies and has yet to be achieved.
While the mechanisms explaining the regulation of transcription by G4s have not been completely resolved yet, there are several possible scenarios by which G4s may regulate transcription that are suggested by past studies. After transcription is initiated, the transcription bubble generates positive or negative supercoiling regions that can propagate and induce stress, eventually forming single-stranded segments that can fold into G4s (46,47). It is possible that G4s that form upstream of the transcription start site (TSS) can impede transcription by acting as an obstacle to block RNA polymerase in the transcribed region (48–51), by recruiting G4-binding proteins that inhibit transcription (47,52), or by failing to maintain the open DNA conformation that facilitates transcription re-initiation (46,52–54). G4s that form downstream of the TSS can cause transcription reinitiation if located on the coding or sense strand by keeping the template strand single-stranded, or they can block RNA polymerase progression if located on the template or antisense strand by serving as a physical obstacle (46,47).
The G-quadruplex and R-loop feedback loop
In the context of double-stranded DNAs, one of the consequences of G4s formed on one DNA strand, such as the non-template strand in the case of the C9orf72 HRE, is the formation of an R-loop, a three-stranded structure composed of the displaced single-stranded DNA together with a DNA:RNA hybrid formed through Watson-Crick base pairing that involves the other DNA strand (55). The C9orf72 HRE has been shown to readily form R-loops in vitro (Figure 3) (37,56). The G4C2 repeats can adopt stable secondary DNA structures, including G4s and hairpins, which can stabilize the displaced strand and therefore the R-loops. Recent studies of non-C9orf72 model G4 motifs have pointed to the interplay between G4s and R-loop formation as a potential positive feedback loop. For example, results from single-molecule fluorescence assays that have modeled the G4s and R-loops in vitro suggest that R-loop formation can occur prior to G4 formation and subsequently stabilize the R-loops, via positive feedback, during further rounds of transcription (57,58).
R-loops have been extensively linked to the regulation of transcription, with these secondary structures thought to regulate transcription activation, elongation, and termination (59). Studies have shown that the formation of R-loops leads to transcriptional stalling, and therefore decreased gene expression in vitro (60,61). R-loops formed from the C9orf72 HRE have also been shown to stall RNA polymerase II and increase abortive transcription in vitro (37). The influence of G4 and R-loop formation on C9orf72 gene expression can presumably initiate a number of different pathological cascades, including those related to bidirectional transcription and epigenetic modification, in which R-loops have been reported to participate (61,62).
DNA methylation affects G-quadruplex stability
The stability of G4s can be affected by DNA methylation. For example, in the case of the C9orf72 HRE, methylation of its DNA is present in most patients harboring more than 90 repeats (63). Both 5-methylcytosine (5mC) and its oxidized form, 5-hydroxymethylcytosine (5hmC), have been suggested as the forms in which C9orf72 HRE DNA methylation occurs. It has been reported that both the G-rich strand and the C-rich antisense strand of the C9orf72 (G4C2)n DNA repeat can form four-stranded quadruplex structures, and that the 5mC and 5hmC modifications have differential effects on the stability and protein binding of the DNA structures (64). In another case, the 5mC modification of a DNA G4 found in the promoter region of bcl-2 has been shown to increase the stability of the G4 (65). Furthermore, DNA methylation of G4 motifs in VEGF has been reported to decrease the initial elongation efficiency of PCR, which could indicate G4 stabilization (66). Another study investigating the effect of CpG methylation on the binding affinity of G4s to associated proteins has found that the influence of CpG methylation on the binding affinity also simultaneously affects G4 structure and topology (67). In addition, R-loop structures, which have been found at CpG islands, have been proposed to suppress DNA methylation. R-loop formation has been suggested to maintain the unmethylated state of CpG islands, and R-loop formation also provides protection from DNMT3B1-mediated DNA methylation (68).
DNA secondary structures, including G4s as well as their companion R-loop structures, may have a profound effect on transcription activity and epigenetic modifications at the HRE or gene locus, consistent with multiple reports showing that DNA methylation plays a role in reduced C9orf72 gene expression (69–71). Consequently, the dysregulation of gene expression that occurs as a result of modifications at the transcriptional level may contribute to the development of neurodegenerative diseases that involve the loss of C9orf72 expression and production of aberrant repeat-containing RNAs and polypeptides. However, analyses of the correlation between CpG hypermethylation of the C9orf72 promoter and disease progression have not yielded consistent results (72,73). Given the complicated roles that DNA methylation is likely to play in disease development, further molecular studies and analysis of larger clinical datasets are clearly needed in the future.
Other functions of DNA G-quadruplexes
G4s are also linked to the disruption of DNA replication as well as the induction of DNA damage and genome instability (74,75). These higher-order structures may contribute to repeat instability, a common feature of most repeat expansion diseases, in which expansion or contraction of the repeat can occur. The human C9orf72 allele contains 2–25 units of the G4C2 repeat, whereas ALS- and FTD-linked alleles have been found to contain up to thousands of the repeats. Varying lengths of the C9orf72 repeats have been observed across generations or in cells from different tissues of individual patients; thus, the repeat instability could arise during meiosis, somatic cell division, or post-mitotic stages. The G4C2 repeat has been shown to affect DNA replication in a length- and orientation-dependent manner in human cells (76). The formation of R-loops at the C9orf72 G4C2 repeat has also been linked to repeat instability. In addition, the C9orf72 G4C2 repeat has been shown to be capable of forming bidirectionally transcribed double R-loops in vitro, with these double R-loops more prone to repeat instability than are single R-loops (56). Understanding the roles of G4s and R-loops may help elucidate the mechanisms of repeat instability in the neurodegenerative diseases.
RNA G-QUADRUPLEXES AND TRANSLATION
RNA structures play critical roles in biological processes that include translation. The bioinformatic observation that G4s are enriched in the non-coding regions but rarely found in coding sequences suggests that exons may have evolved to avoid the stable higher-order structures that likely impede ribosome scanning (77). At the same time, RNA G4s located in the 5′-UTR and 3′-UTR regions have been shown to play important regulatory roles in enhancing or inhibiting translation of various proteins associated with a range of neurological diseases, including AD, PD and FXS (Figure 2B).
RNA G-quadruplexes inhibit translation initiation
Within the 5′-UTR of mRNAs, G4s have been identified as structural elements responsible for regulating the initiation stage of the translation process. The presence of Lewy bodies, cytoplasmic inclusions consisting of the protein α-synuclein (encoded by SNCA), within dopaminergic neurons is a hallmark of PD, the second most common neurodegenerative disorder after AD. Because the overaccumulation and aggregation of the protein α-synuclein is thought to initiate a cascade that imparts neurotoxicity to neurons and other cells, understanding the regulation of α-synuclein translation could shed light on the pathogenic process in PD (78).
Bioinformatic analysis has revealed the presence of three non-overlapping G4 motifs at the proximal 5′-UTR of SNCA that operate together to repress SNCA translation (79). These G4 motifs are only effective at suppressing translation when the inhibitory effect is measured cumulatively. Analysis of the mutated G4 motifs found within the 5′-UTR SNCA has revealed that two of the mutated G4 motifs exhibit enhanced translation, along with higher mRNA levels, when compared to the wild-type control (79). Hence, mutating these two G4 motifs should promote SNCA translation at least in part by either enhancing transcription or stabilizing the SNCA 5′-UTR reporter mRNA. However, because the third mutated G4 motif did not exhibit altered mRNA levels when translation was enhanced, it is likely that translation initiation is directly affected.
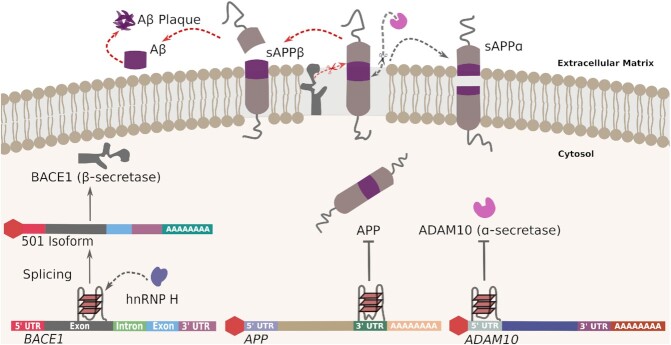
The involvement of G4s in the regulation of translation initiation may not be limited to SNCA, since it has been proposed that G4s found in the 5′-UTR of the metalloprotease ADAM10 can potentially affect translation initiation (Figure 4). Effective elimination of neurotoxic Aβ plaques, which are characteristic of AD, relies on amyloid precursor protein (APP) processing by ADAM10, an α-secretase that cleaves APP within its Aβ domain and releases the neuroprotective N-terminal portion of APP, sAPP-α (80,81). A unimolecular, parallel G4 formed in the 5′-UTR of ADAM10 mRNA has been shown to inhibit ADAM10 translation in vitro, and mutations of the 5′-UTR G4 exhibited enhanced ADAM10 translation, secretion of sAPP-α, and anti-amyloidogenic processing of APP (80). A subsequent study found that a methylquinolinium derivative known as compound 24 binds to the G4-forming sequence of ADAM10 via a high-affinity interaction, with compound 24 exposure promoting ADAM10 translation and decreasing Aβ production in cells (81). These studies demonstrate a role for the 5′-UTR G4 in the translation of disease-relevant enzymes such as ADAM10 and suggest that their RNA secondary structure could be a potential therapeutic target.
Figure 4.
Proposed involvement of G-quadruplexes in Alzheimer's disease. Through the differential cleavage of APP, G4s participate in the regulation of Aβ biogenesis, either by affecting either the expression of APP itself or that of APP-processing enzymes. A G4 motif in the 3′-UTR of APP mRNA has been found to negatively regulate APP translation. A G4 motif within the coding region of BACE1 mRNA may act as a recruitment site for splice regulator hnRNP H, resulting in favored production of the full-length BACE1 501 transcript isoform and leading to enhanced APP proteolysis and Aβ production. Also, a G4 motif within the 5′-UTR of ADAM10 mRNA negatively regulates the production of ADAM10 α-secretase activity, thereby suppressing cleavage of APP and contributing to Aβ production.
The roles that G4s play in the regulation of translation initiation are particularly important to elucidate because it is believed that most regulation of translation occurs at the initiation stage in eukaryotes (82). Although the evidence thus far points to an inhibitory role for G4s in translation, the relevant mechanisms are not limited to the impediment of ribosome scanning at 5′-UTRs of mRNAs, as exemplified by the G4-containing tRNA fragments discussed in the section below regarding stress granules. Future studies that further elucidate the mechanisms by which G4s affect translation initiation may clarify the role of G4s in neurodegenerative diseases with pathological features associated with altered cap-dependent translation.
G-quadruplexes in non-canonical translation
Unlike most eukaryotic translation initiation, which requires an m7G cap at the 5′ end of the mRNA to initiate ribosome scanning in order to locate the start codon, there are alternative non-canonical mechanisms that initiate translation without these standard features. IRES is an RNA element that allows for internal ribosome entry to initiate translation in a non-cap-dependent manner. G4 motifs have been identified within IRES sequences and have been clearly shown to exert an influence, albeit with varied effects, on IRES activity. Several studies have shown that intramolecular G4 motifs found within the IRES promote IRES-mediated translation (83,84) and that removal of the G4 motif prevents IRES initiation (85), whereas another study has shown that G4 motif stabilization inhibits IRES-mediated translation (86). Of relevance to neurodegeneration is the fact that an IRES element located in the 5′-UTR of SNCA has been shown to enhance SNCA translation and synthesis in response to cellular stress (79). However, mutations in the three G4 motifs that we mentioned in our discussion of G4s and translation initiation did not affect the IRES activity and thus their role in the translation remain unclear (79). Thus, although the role of G4 elements in negatively regulating cap-dependent translation is established (87,88), the precise interplay of these elements in stress-induced cap-independent translation remains in need of future elucidation.
C9orf72 repeat-associated non-ATG (RAN) translation refers to the observation that the C9orf72 HRE repeats in a non-coding region can still enable translation in the absence of an ATG start codon when the repeat length exceeds certain limits (89,90). C9orf72 HRE-containing sense and antisense transcripts undergo RAN translation and produce aggregation-prone dipeptide repeat proteins. The dipeptide repeat proteins are toxic and induce neurodegeneration in experimental models. G4s are formed on the C9orf72 HRE RNAs, but a direct role for the quadruplex structures in the regulation of the RAN translation remains to be established. Nonetheless, the potential involvement of G4s in pathologically relevant RAN translation may provide a target for therapeutic intervention.
G-quadruplex-dependent translational regulation in FXS
Fragile X syndrome (FXS) is the most common inherited intellectual disability and is induced by the silencing of the RNA binding protein known as fragile X mental retardation 1 protein (FMRP) (91). The expansion and hypermethylation of trinucleotide (CGG)n repeats within the 5′-UTR of FMR1 mRNA, along with aberrant CpG island hypermethylation preceding the open reading frame of FMR1, underlie FMRP repression in FXS. FMRP is an mRNA-binding protein that is deemed essential for normal neurological function because of its suggested role in dendritic mRNA transport and postsynaptic translation (92). Phosphorylated FMRP generally operates as a cis-acting translational repressor of a subset of dendritically localized RNAs, inhibiting their premature translation during neuronal trafficking (93). In FXS pathology, the absence of FMRP-mediated translational blockade can result in translational overactivation, leading to aberrant and constitutive production of synaptic proteins, internalization of receptors, and impaired synaptic plasticity (94).
The binding affinity of FMRP for its mRNA targets depends on the presence of specific secondary structures, including G4s. It has been shown that the RGG box domain of FMRP binds with high affinity to G4s of its target mRNAs and regulates their expression (95,96). FMRP binds to a subset of its mRNA targets through G4 motifs located in the various regions of the transcripts.
FMRP has also been shown to bind to G4s located in the 5′-UTR and 3′-UTR of its other target mRNAs. For example, the FMRP RGG box has been observed to bind to an intramolecular, parallel G4 formed in the 5′-UTR of survival motor neuron domain containing 1 (SMNDC1) in vitro (97). G4 motifs formed in the 5′-UTR of mRNAs typically operate as inhibitory translational elements, and FMRP binding may repress translation by impeding the accessibility of the translation initiation sites. At 3′-UTRs, FMRP binding to G4s can also promote translational repression through less clear mechanisms that may be mediated by microRNAs in some cases. It has been shown that FMRP binds to two sequential parallel 3′-UTR G4 structures in Shank1 mRNA and presumably contributes to its translational repression (95). FMRP has been implicated in G4-microRNA interplay through an observation that FMRP can negatively regulate the translation of postsynaptic density protein 95 (PSD-95) via a microRNA-dependent mechanism involving the 3′ UTR G4 motifs found in PSD-95. The guanine-rich region of the 3′UTR PSD-95 mRNA is capable of folding into two alternate parallel G4 conformations that exist in equilibrium with each other, with the dominant G4 conformation exposing the complementary nucleotide seed sequence of miR-125a. The two G4 structures, connected by a linker region, bind to miR-125a and form a stable complex within the 3′UTR PSD-95 mRNA (98). Subsequently, phosphorylated FMRP and miR-125a operate in conjunction to repress PSD-95 translation (99).
Interestingly, fused in sarcoma (FUS), a protein linked to ALS/FTD that plays a role in synaptic function regulation and local translation, has been shown to bind with high specificity to parallel G4s formed in the mRNA of PSD-95 or Shank1 (100). The nanomolar dissociation constants for complexes containing FUS and PSD-95 or Shank1 G4s are comparable to those of complexes containing FMRP, suggesting that FUS can potentially compete with FMRP binding to the G4s in these mRNAs and thereby influence their translation.
G4s have been shown to regulate the translation of APP, the key protein in AD; a G4 motif in the 3′-UTR of APP mRNA has been found to negatively regulate APP translation (Figure 4) (101). FMRP has also been linked to the regulation of APP translation, with FMRP shown to repress APP translation by recruiting APP mRNA to processing bodies, where non-translating mRNAs are stored or degraded (102). FMRP was also observed to bind to the coding region of APP mRNA at a guanine, G-quartet-like sequence and regulate the translation of APP in a manner dependent on the activation of a subtype of glutamate receptor (103). Heterogeneous nuclear ribonucleoprotein C (hnRNP C) has been found to compete with FMRP for binding at the coding region, with hnRNP C binding promoting APP translation by displacing FMRP and alleviating its translational blockade (102). Therefore, FMRP and hnRNP C appear to work in concert to regulate APP translation through a mechanism that may suggest a role for G4s in the process.
RNA G-QUADRUPLEXES AND SPLICING
RNA G-quadruplexes regulates alternative splicing as cis elements
G4s found in the exonic regions of FMR1 and BACE1 mRNAs (which are implicated in FXS and AD, respectively) have been shown to regulate alternative splicing by acting as a molecular switch, controlling the distribution of the various isoforms produced.
Interestingly, it has been demonstrated that a selective mRNA target of FMRP is FMR1 itself. FMRP has been shown to bind to a specific and high-affinity site located in the nucleotide sequence of the 3′ terminal coding region of FMR1 mRNA that is responsible for encoding the RGG domain of FMRP. It was initially thought that through binding to the coding region site, FMRP could repress its own translation through a negative-feedback loop (104). However, a follow-up study has shown that the coding region site within FMR1 mRNA, which contains two alternative G4 structures, is a potent exonic splicing enhancer (105). The exonic splicing enhancer activity of the coding region site was nullified when G4 formation was inhibited, indicating that the splicing activity of the coding region site is dependent on G4 formation. FMRP binding to the coding region site has been shown to control the relative amounts of short and long FMRP isoforms produced by alternative splicing of exon 15. Importantly, G4 structures are thought to mediate the binding between FMRP and the coding region site, since removal of both G4s reduces the FMRP binding to nonspecific levels (105). It has been suggested that increased expression of the full-length FMRP isoform alters FMR1 splicing events around the coding region site to favor short-isoform production. Hence, the presence of G4s at the coding region site can potentially allow for an additional layer of FMRP self-regulation through splicing, mediating its expression through a negative autoregulatory loop.
BACE1 encodes a transmembrane protease that is responsible for the production of the first cleavage product in the conversion of APP to Aβ. Alternative splicing of BACE1 yields six shorter isoforms, but only the full-length transcript, known as isoform 501, is involved in APP proteolysis. It has been shown that BACE1 mRNA harbors a G-rich sequence on its third exon that has G4-forming potential (106). This G-rich sequence has been reported to recruit splicing regulator heterogeneous nuclear ribonucleoprotein H (hnRNP H) to BACE1 mRNA, thereby facilitating the alternative splicing of BACE1 mRNA isoforms. The G-rich sequence-dependent hnRNP H recruitment selectively activates the production of isoform 501, while simultaneously inhibiting generation of the shorter isoforms (Figure 4). Accordingly, deletion of the G-rich sequence has been found to silence the production of isoform 501, whereas knockdown of hnRNP H results in reduced isoform 501 translation, repressed APP proteolysis, and a consequent decrease in Aβ (106).
RNA G-quadruplexes sequester splicing factors
C9orf72 HRE G4s have been shown to sequester proteins involved in splicing regulation, leading to dysregulated splicing and RNA toxicity via a trans-acting mechanism (Figure 3). In vitro cellular studies have revealed that C9orf72 RNA G4s bind to the splicing regulator ASF/SF2, inducing RNA toxicity through protein-binding interactions (36). Similarly, G4 formation is responsible for high-affinity associations between C9orf72 HRE RNA and the essential splicing factor hnRNP H in vitro (107). Dysregulated splicing of several hnRNP H target transcripts, as a result of the sequestration of the protein by G4s formed from the C9orf72 HRE, contributes to neurodegeneration in C9orf72 HRE-carrying patients’ brains (107). Other splicing factors, such as hnRNP A3, have been found to associate with RNAs containing hexanucleotide G4C2 repeats (108), potentially contributing to the splicing dysregulations in the patients.
G-QUADRUPLEX-DEPENDENT RNA-PROTEIN INTERACTIONS
As mentioned in the discussion of splicing dysregulation above, RNA G4s have been shown to recruit and sequester cellular proteins from their normal functions, leading to pathological consequences that can include neurodegeneration. In fact, RNA G4s formed from the C9orf72 HRE repeat have been shown to interact with a wide range of different types of proteins and influence multiple cellular processes, including nucleolar stress, paraspeckle formation, phase separation, and stress granule formation. Here we highlight several cellular events that are pathologically relevant, with the understanding that the pathways mediated by G4 RNA-protein interactions in neurological disease are likely much more diverse than what we discuss here.
RNA G-quadruplexes mediate the formation of membraneless RNP structures
The RNA- and protein-containing membraneless organelles referred to as RNP granules are maintained through protein-protein, protein-RNA, and RNA-RNA interactions (109). These membraneless organelles, which include both nuclear and cytoplasmic bodies such as stress granules, nucleoli, Cajal bodies, P-bodies, and paraspeckles (110), are formed by phase separation of components, allowing for the condensation of proteins into subcellular membraneless compartments (111). Phase separation, which has been reviewed extensively (112,113), has been used to conceptualize the formation of membraneless organelles that behave as liquid droplets (111).
The C9orf72 HRE RNA forms foci in patients’ brains that are potentially pathogenic (32). The neurotoxic consequences of the sequestration of proteins in HRE RNA foci are seen in the mislocalization of nucleolin, an essential nuclear protein that is critical for the function of the nucleolus. Nucleolin is known to preferentially bind to C9orf72 HRE RNA in a G4-dependent manner (37). The C9orf72 HRE RNA colocalizes with nucleolin in the nucleoli of patients’ brains to impair nucleolar function and induce nucleolar stress, linking abnormal C9orf72 HRE nucleic acid structures to ALS pathology induced by nucleolar stress (Figure 3) (37). C9orf72 RNA G4s may have promise as a therapeutic target given that small molecules stabilizing C9orf72 RNA G4s have been shown to reduce levels of RNA foci and dipeptide repeat proteins in Drosophila and neuronal models (114).
Paraspeckles, which are nuclear ribonuclear bodies assembled on the long non-coding RNA (lncRNA) NEAT1, have been linked to ALS when paraspeckle formation is observed in the early stages of ALS pathology, and the ALS-linked proteins TDP-43 and FUS, among others, are found to be enriched in paraspeckles (115,116). A potential role for paraspeckles in C9orf72 pathology was recognized when RNA foci from the C9orf72 HRE were shown to possess paraspeckle-like characteristics. Like paraspeckles, C9orf72 RNA foci co-localize with the paraspeckle proteins SFPQ, NONO, RBM14, hnRNP H and FUS (117). Interestingly, it has been observed that C9orf72 RNA foci can also form paraspeckle-like bodies in a NEAT1-independent manner, suggesting that the C9orf72 RNA HRE can act as a scaffold for paraspeckle-like structures (117). Notably, the assembly of endogenous paraspeckles may be driven by the G4 structures on its scaffold, lncRNA NEAT1. An abundance of G4 motifs has been observed on NEAT1, and paraspeckle proteins such as NONO have been found to bind to NEAT1 in vitro and in vivo through these G4 motifs (118). Furthermore, the enrichment of G4 motifs is conserved among NEAT1 homologs despite their low sequence homology, highlighting the potential role for G4s as a structural element that recruits NONO and contributes to the seeding of paraspeckle assembly. C9orf72 HRE RNAs compete with NEAT1 for the binding of paraspeckle proteins such as NONO in a G4-dependent manner (118), suggesting that the HRE RNAs can potentially cause disruption of paraspeckle function as part of the pathogenic cascades in the disease (Figure 3).
Stress granules are cytoplasmic foci composed of proteins and RNA, including translationally stalled mRNAs, that are formed under stressful conditions and have been proposed to lead to pathological protein aggregates in neurodegenerative diseases that include AD, ALS, and FTD (119). The C9orf72 HRE RNA has been shown to affect stress granule dynamics. A study determining the effect of the HRE RNA on phase separation, a process considered important for RNA granule formation because it enables the compartmentalization of proteins, has found that the C9orf72 HRE RNA can enhance the formation of both stress granules and nuclear foci, as well as promote the condensation of RNA granule proteins and phase separations in vitro (120). C9orf72 RNA HRE-mediated condensation is reported to respond to ionic conditions, which can directly contribute to G4 stabilization. Indeed, G4s formed from the C9orf72 HRE RNA have been shown to promote phase transitions in vitro and in cells. It has therefore been proposed that C9orf72 G4s can phase-separate in vitro through the condensation of RNA granule components, leading to stress granule formation (Figure 3) (120).
RNA G-quadruplexes and nucleocytoplasmic transport
The trafficking of proteins and RNAs across the nuclear envelope through the nuclear pores is a major transport system in the cell, and pathologies of nucleocytoplasmic transport have been increasingly observed in neurodegenerative diseases. C9orf72 HRE RNA G4s have been linked to disrupted nucleocytoplasmic transport through the observation that RanGAP, a regulator of nucleocytoplasmic transport, influences HRE-mediated neurodegeneration in experimental models (Figure 3) (121). RanGAP exhibits a higher binding affinity for HRE sense-strand G4s than for hairpin structures in vitro, suggesting that RanGAP may operate by preferentially binding the sense RNA G4 formed from the C9orf72 HRE. It has been reported that RanGAP function is impaired in C9orf72 ALS iPS neurons and, correspondingly, that C9orf72 HREs decrease nuclear import (121). Treatment of TMPyP4, a compound that destabilizes RNA G4s, decreases the affinity of RanGAP for the G4s and rescues the nuclear import defects, suggesting that unfolding the G4s can suppress nuclear transport deficits caused by the C9orf72 HRE.
Recognition of G-quadruplexes by TDP-43
TAR DNA-binding protein 43 (TDP-43) is a DNA/RNA-binding protein that is responsible for long-distance mRNA transport and the regulation of local protein synthesis related to conserving neural cell polarity and synaptic plasticity (122). The TDP-43 proteinopathy, the deposition of TDP-43 protein in ubiquitinated and hyperphosphorylated aggregates, is present in the majority of ALS cases and nearly 50% of all FTD cases (123). Both the loss of TDP-43’s RNA processing functions and the gain-of-toxicity from the TDP-43 proteinopathy have been proposed to underlie the pathogenesis of the relevant diseases.
When RNA motifs for TDP-43 binding have been explored through in vitro systematic evolution of ligands by exponential enrichment (SELEX) screening, all the top RNA targets of TDP-43 have been found to harbor G4 motifs (124). TDP-43 has been confirmed to bind to natural DNAs and RNAs in a parallel G4-specific manner, and its targets include PSD-95 and CaMKIIα mRNAs, which are transported on the basis of the recognition of their 3′UTR G4 motifs (96,124), suggesting that TDP-43 binds to and transports these G4-containing mRNAs into neurites for local translation. TDP-43 has also been found to bind to the C9orf72 HRE RNA in vitro, and pre-incubation of TDP-43 with G4C2 repeat RNAs decreases TDP-43’s binding affinity for G4-containing PSD-95 and CamKIIa mRNAs, suggesting that sequestration of TDP-43 by the C9orf72 HRE RNAs may impair TDP-43’s RNA transport function (124).
Interestingly, mutant ALS-associated TDP-43M337V exhibits a lower binding affinity for G4-containing mRNAs, indicating that Met337 within the C‐terminal Gly‐rich domain of TDP‐43 is critical for G4 binding (124). A follow-up study from the same group has expanded upon these findings by showing that the Gly-rich region of TDP-43 is the region that is responsible for recognition and binding of specific G4-containing mRNAs (125). The implications of these observations for ALS-FTD pathology are potentially significant, given that as much as 30% of neuronal mRNAs harbor 3′-UTR G4 motifs (126).
G-quadruplexes are integral to the tRNA-mediated stress response
G4 structures on tRNAs may also play a regulatory role in stress granule formation and translation. A subset of ALS in patients is linked to missense mutations in angiogenin, a ribonuclease with the ability to cleave both rRNAs and tRNAs. In response to stress stimuli, angiogenin cleaves mature tRNAs into 5′ and 3′ tRNA-derived stress-induced RNAs (tiRNAs) in order to suppress unwanted translation (127,128). ALS-linked mutant angiogenin has been found to exhibit limited catalytic activity and to fail to induce tRNA cleavage (129). Under conditions of stress, certain tiRNA fragments, i.e. 5′-tiRNAAla and 5′-tiRNACys, inhibit protein synthesis by displacing cap-binding eIF4F complexes from capped mRNA, leading to the formation of stress granules and inhibition of translation (127). The ability of these tiRNA fragments to perform these functions depends on a 5′ terminal oligoguanine (5′TOG) motif, a stretch of guanine residues located at the 5′ end, and a secondary structure characterized by the presence of a stem–loop sandwiched between the 5′ and 3′ regions (127,130). The 5′TOG motif found in 5′-tiRNAAla has been shown to be capable of forming a highly symmetric, parallel tetramolecular RNA G4 with five tetrad layers (131). The formation of G4s within the 5′TOG motif is correlated with the association of 5′-tiRNAAla and 5′-tiRNACys with the translational repressor Y-box binding protein 1 (128); furthermore, the removal of G4 from the 5′TOG motif prevents stress granule formation (131), with both findings indicating a requisite role for G4s in the tRNA-mediated stress response that results in stress granule formation and translational inhibition.
Concerning the mechanism governing the tiRNA G4-triggered stress granule formation, it has been shown that eIF2α phosphorylation, the canonical trigger of stress granule formation, is not involved in the tiRNA-induced stress response. Rather, G4s formed from the 5′TOG motif of 5′-tiRNAAla directly interact with eIF4G, inhibiting the assembly of the eIF4F translation initiation complex and in turn stimulating stress granule formation (132). A larger pool of tiRNAs of varying fragment sizes with the propensity to form G4s has been shown to contain the 5′TOG motif, indicating that the number of potential tiRNAs capable of inhibiting translation is larger than initially suspected (133).
A potential role for G-quadruplexes in oxidative stress
G4s formed by G4C2 DNAs and RNAs from the C9orf72 repeat region have been reported to bind to heme, an iron-containing compound of the porphyrin class, and form tight complexes under physiologically relevant conditions (134). This heme-quadruplex complex containing G4C2 repeat nucleotides exhibits both peroxidase and oxidase activity in a quadruplex-dependent manner (37,134). These in vitro observations have raised the possibility that the C9orf72 HRE-induced heme-quadruplex complexes may catalyze aberrant oxidative reactions or interfere with iron homeostasis or mitochondrial function, in which heme plays an important role, leading to cell damages responsible for neurodegeneration. Recently, physiological evidence for the binding of G4 and heme has been gleaned outside of a C9orf72-specific context, in experiments that indirectly detected the release of heme from G4s by analyzing how treatment with a G4 ligand (predicted to displace sequestered heme) affected the expression of genes involved in heme catabolism and iron homeostasis. These experiments showed that PhenDC3, a G4 ligand, displaces G4-bound heme in vitro and causes heme oxidase 1 induction in human cells, supporting the concept that G4s sequester heme in the cell (135). Such heme-quadruplex complexes may have physiological functions in living cells that need to be further explored.
G-QUADRUPLEXES IN OTHER NEUROLOGICAL DISEASES
Prion diseases
Prion proteins are thought to be responsible for transmissible spongiform encephalopathies (TSEs), or prion diseases, which are characterized by the deleterious transition from the normal, soluble, alpha-helix-rich cellular form of prion protein (PrPC) to the insoluble, beta-sheet-rich prion proteins susceptible to aggregation (PrPSc). In addition to the abovementioned G4 motifs at the promoter of the PrP gene, which may mediate autoregulation at the transcriptional level (41), there are putative G4 motifs in the PrP mRNA as well. In the human PrP protein, the octa-repeat domain, which is found in the N-terminal domain of PrP and is crucial for aggregation, consists of five repeats of an octapeptide sequence. The underlying mRNA sequence harbors five putative G4 motifs. The secondary structure of this RNA segment is dynamic and assumes various conformations, including hairpins and G4s. It was initially determined, when short sequences (24 nt) of the PrP mRNA segment were tested, that G4s could be formed, and that these structures may interfere with translation (136). PrPc binding to the G4s in PrP mRNA has been suggested to regulate the co-translational folding of the PrP protein and thus potentially lead to spontaneous conversion of PrPSc (136). However, a later study has demonstrated that the full sequence containing the five G4 motifs in the PrP RNA segment exhibits preferential double-stranded A-helical hairpin structures instead of G4s, suggesting that the sequence context may influence the confirmation or stability of the secondary structures (137). Therefore, future studies are needed to ascertain the pathological consequences of the structures and functions of PrP mRNA in vivo.
Progressive myoclonus epilepsy
The most common cause of progressive myoclonus epilepsy type I, also known as Unverricht-Lundborg disease, is a d(C4GC4GCG)-d(CGCG4CG4) dodecamer repeat expansion at the cystatin B (CSTB) promoter on chromosome 21q22.3 (138). The dodecamer repeat mutation leads to significantly lower CSTB mRNA levels in patients, which in turn leads to a loss-of-function of CSTB as a cysteine protease inhibitor. Emerging evidence points to a possible role for G4 secondary structures in relation to the functions of the dodecamer repeat. The G-rich bottom strand of the promoter region has been shown to form stable secondary structures featuring parallel G4s under physiologically relevant conditions in vitro (138). Although a clear link has been established between secondary structures and the CSTB promoter, further study is needed to elucidate the direct effects and regulatory mechanism of G4s on the functions of the dodecamer repeat in the disease.
CONCLUSION
Research on G4s as they relate to biology and neurodegeneration remains one of the most fascinating fields as well as one of the most complex, given the concurrent prevalence of G4 involvement in neurological diseases and the existing uncertainties regarding in vivo functions and biological mechanisms. G4s can act as a regulatory structural element in neurological diseases, initiating neuroprotective and neurotoxic cascades by influencing a plethora of cellular processes. Given their unique structural properties, G4s can serve as effective targets for therapeutic interventions employing aptamers, antisense oligonucleotides, or small molecules. For these approaches to be therapeutically viable, however, they must achieve a great degree of specificity in recognizing unique genomic G4s that possess high variability in their sequence, orientation, and stability. Future studies are needed to uncover the dynamics of G4s and the genetic players involved in neurodegeneration. These approaches may reveal previously unknown culprits in the intricate etiology of neurodegenerative disorders, while supporting the development of novel therapeutic strategies.
ACKNOWLEDGEMENTS
We thank members of Wang lab for discussion. We apologize for not being able to cite all relevant publications due to space constraints.
Author contribution: E.W., R.T., Y.S., R.L. and J.W. wrote the paper. E.W. and R.T. designed the figures with suggestions from J.W.
Contributor Information
Ernest Wang, Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore MD, 21205, USA; Department of Neuroscience, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Ravi Thombre, Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore MD, 21205, USA; Department of Neuroscience, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Yajas Shah, Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore MD, 21205, USA; Department of Neuroscience, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Rachel Latanich, Department of Medicine, Division of Gastroenterology and Hepatology, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
Jiou Wang, Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore MD, 21205, USA; Department of Neuroscience, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA.
FUNDING
NIH [NS074324, NS089616, NS110098]. Funding for open access charge: Federal or private funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gellert M., Lipsett M.N., Davies D.R.. Helix formation by guanylic acid. PNAS. 1962; 48:2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364. [DOI] [PubMed] [Google Scholar]
- 3. Cao Z., Huang C.-C., Tan W.. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal. Chem. 2006; 78:1478–1484. [DOI] [PubMed] [Google Scholar]
- 4. Lane A.N., Chaires J.B., Gray R.D., Trent J.O.. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008; 36:5482–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Y., Brosh R.M.. G-quadruplex nucleic acids and human disease. FEBS J. 2010; 277:3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Métifiot M., Amrane S., Litvak S., Andreola M.-L.. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014; 42:12352–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cammas A., Millevoi S.. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017; 45:1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Islam M.K., Jackson P.J., Rahman K.M., Thurston D.E.. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016; 8:1259–1290. [DOI] [PubMed] [Google Scholar]
- 9. Patel D.J., Phan A.T.n., Kuryavyi V.. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simone R., Fratta P., Neidle S., Parkinson G.N., Isaacs A.M.. G-quadruplexes: emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015; 589:1653–1668. [DOI] [PubMed] [Google Scholar]
- 11. Otero R., Schöck M., Molina L.M., Lægsgaard E., Stensgaard I., Hammer B., Besenbacher F.. Guanine quartet networks stabilized by cooperative hydrogen bonds. Angew. Chem. Int. Ed. 2005; 44:2270–2275. [DOI] [PubMed] [Google Scholar]
- 12. Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chemistry. 2007; 13:9738–9745. [DOI] [PubMed] [Google Scholar]
- 13. Bhattacharyya D., Mirihana Arachchilage G., Basu S.. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016; 4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ida R., Wu G.. Direct NMR detection of alkali metal ions bound to G-quadruplex DNA. J. Am. Chem. Soc. 2008; 130:3590–3602. [DOI] [PubMed] [Google Scholar]
- 15. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877. [DOI] [PubMed] [Google Scholar]
- 17. Huppert J.L., Balasubramanian S.. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007; 35:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353:aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biffi G., Di Antonio M., Tannahill D., Balasubramanian S.. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014; 6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M., Sen D., Lansdorp P.M.. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2013; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang S.Y., Lejault P., Chevrier S., Boidot R., Robertson A.G., Wong J.M., Monchaud D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018; 9:4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discovery. 2011; 10:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lam E.Y.N., Beraldi D., Tannahill D., Balasubramanian S.. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013; 4:1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M.et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 26. González V., Guo K., Hurley L., Sun D.. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009; 284:23622–23635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bugaut A., Balasubramanian S.. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012; 40:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beaudoin J.-D., Perreault J.-P.. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013; 41:5898–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soemedi R., Cygan K.J., Rhine C.L., Glidden D.T., Taggart A.J., Lin C.-L., Fredericks A.M., Fairbrother W.G.. The effects of structure on pre-mRNA processing and stability. Methods. 2017; 125:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lomen-Hoerth C., Anderson T., Miller B.. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002; 59:1077–1079. [DOI] [PubMed] [Google Scholar]
- 31. Umoh M.E., Dammer E.B., Dai J., Duong D.M., Lah J.J., Levey A.I., Gearing M., Glass J.D., Seyfried N.T.. A proteomic network approach across the ALS-FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Mol. Med. 2018; 10:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J.et al.. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011; 72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L.et al.. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011; 72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gitler A.D., Tsuiji H.. There has been an awakening: emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res. 2016; 1647:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M., Parkinson G., Isaacs A.M.. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2012; 2:1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reddy K., Zamiri B., Stanley S.Y., Macgregor R.B., Pearson C.E.. The disease-associated r (GGGGCC) n repeat from the C9orf72 gene forms tract length-dependent uni-and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013; 288:9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R.et al.. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mano T., Nagata K., Nonaka T., Tarutani A., Imamura T., Hashimoto T., Bannai T., Koshi-Mano K., Tsuchida T., Ohtomo R.et al.. Neuron-specific methylome analysis reveals epigenetic regulation and tau-related dysfunction of BRCA1 in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E9645–E9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suberbielle E., Djukic B., Evans M., Kim D.H., Taneja P., Wang X., Finucane M., Knox J., Ho K., Devidze N.et al.. DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nat. Commun. 2015; 6:8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moruno-Manchon J.F., Koellhoffer E.C., Gopakumar J., Hambarde S., Kim N., McCullough L.D., Tsvetkov A.S.. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging (Albany NY). 2017; 9:1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pradhan P., Srivastava A., Singh J., Biswas B., Saini A., Siddique I., Kumari P., Khan M.A., Mishra A., Yadav P.K.et al.. Prion protein transcription is auto-regulated through dynamic interactions with G-quadruplex motifs in its own promoter. Biochim. Biophys. Acta Gene Regul. Mech. 2020; 1863:194479. [DOI] [PubMed] [Google Scholar]
- 42. Liu C., Geng Y., Miao H., Shi X., You Y., Xu N., Zhou B., Zhu G.. G-quadruplex structures formed by human telomeric DNA and C9orf72 hexanucleotide repeats. Biophys. Rev. 2019; 11:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou B., Liu C., Geng Y., Zhu G.. Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci. Rep. 2015; 5:16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou B., Geng Y., Liu C., Miao H., Ren Y., Xu N., Shi X., You Y., Lee T., Zhu G.. Characterizations of distinct parallel and antiparallel G-quadruplexes formed by two-repeat ALS and FTD related GGGGCC sequence. Sci. Rep. 2018; 8:2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Biswas B., Kandpal M., Vivekanandan P.. A G-quadruplex motif in an envelope gene promoter regulates transcription and virion secretion in HBV genotype B. Nucleic Acids Res. 2017; 45:11268–11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Du Z., Zhao Y., Li N.. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008; 18:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Armas P., David A., Calcaterra N.B.. Transcriptional control by G-quadruplexes: In vivo roles and perspectives for specific intervention. Transcription. 2017; 8:21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Belotserkovskii B.P., Liu R., Tornaletti S., Krasilnikova M.M., Mirkin S.M., Hanawalt P.C.. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:12816–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Broxson C., Beckett J., Tornaletti S.. Transcription arrest by a G quadruplex forming-trinucleotide repeat sequence from the human c-myb gene. Biochemistry. 2011; 50:4162–4172. [DOI] [PubMed] [Google Scholar]
- 50. Belotserkovskii B.P., Soo Shin J.H., Hanawalt P.C.. Strong transcription blockage mediated by R-loop formation within a G-rich homopurine–homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res. 2017; 45:6589–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim N. The interplay between G-quadruplex and transcription. Curr. Med. Chem. 2019; 26:2898–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. David A.P., Margarit E., Domizi P., Banchio C., Armas P., Calcaterra N.B.. G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 2016; 44:4163–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smestad J.A., Maher L.J.. Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med. Genet. 2015; 16:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang J., Haeusler A.R., Simko E.A.. Emerging role of RNA• DNA hybrids in C9orf72-linked neurodegeneration. Cell Cycle. 2015; 14:526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reddy K., Schmidt M.H., Geist J.M., Thakkar N.P., Panigrahi G.B., Wang Y.-H., Pearson C.E.. Processing of double-R-loops in (CAG)·(CTG) and C9orf72 (GGGGCC)·(GGCCCC) repeats causes instability. Nucleic Acids Res. 2014; 42:10473–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lim G., Hohng S.. Single-molecule fluorescence studies on cotranscriptional G-quadruplex formation coupled with R-loop formation. Nucleic Acids Res. 2020; 48:9195–9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee C.-Y., McNerney C., Ma K., Zhao W., Wang A., Myong S.. R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat. Commun. 2020; 11:3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 60. Groh M., Lufino M.M., Wade-Martins R., Gromak N.. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 2014; 10:e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Skourti-Stathaki K., Kamieniarz-Gdula K., Proudfoot N.J.. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014; 516:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chan Y.A., Aristizabal M.J., Lu P.Y., Luo Z., Hamza A., Kobor M.S., Stirling P.C., Hieter P.. Genome-wide profiling of yeast DNA: RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014; 10:e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xi Z., Zhang M., Bruni A.C., Maletta R.G., Colao R., Fratta P., Polke J.M., Sweeney M.G., Mudanohwo E., Nacmias B.et al.. The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol. 2015; 129:715–727. [DOI] [PubMed] [Google Scholar]
- 64. Zamiri B., Mirceta M., Bomsztyk K., Macgregor R.B. Jr, Pearson C.E. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC) 8•(GGCCCC) 8 repeat: effect of CpG methylation. Nucleic Acids Res. 2015; 43:10055–10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin J., Hou J.-q., Xiang H.-d., Yan Y.-y., Gu Y.-c., Tan J.-h., Li D., Gu L.-q., Ou T.-m., Huang Z.-s.. Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: implications for epigenetic regulation. Biochem. Biophys. Res. Commun. 2013; 433:368–373. [DOI] [PubMed] [Google Scholar]
- 66. Yoshida W., Yoshioka H., Bay D.H., Iida K., Ikebukuro K., Nagasawa K., Karube I.. Detection of DNA methylation of G-quadruplex and i-motif-forming sequences by measuring the initial elongation efficiency of polymerase chain reaction. Anal. Chem. 2016; 88:7101–7107. [DOI] [PubMed] [Google Scholar]
- 67. Tsukakoshi K., Saito S., Yoshida W., Goto S., Ikebukuro K.. CpG methylation changes G-quadruplex structures derived from gene promoters and interaction with VEGF and SP1. Molecules. 2018; 23:944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chédin F.. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu E.Y., Russ J., Wu K., Neal D., Suh E., McNally A.G., Irwin D.J., Van Deerlin V.M., Lee E.B.. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. (Berl). 2014; 128:525–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Belzil V.V., Bauer P.O., Prudencio M., Gendron T.F., Stetler C.T., Yan I.K., Pregent L., Daughrity L., Baker M.C., Rademakers R.et al.. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013; 126:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jackson J.L., Finch N.A., Baker M.C., Kachergus J.M., DeJesus-Hernandez M., Pereira K., Christopher E., Prudencio M., Heckman M.G., Thompson E.A.et al.. Elevated methylation levels, reduced expression levels, and frequent contractions in a clinical cohort of C9orf72 expansion carriers. Mol Neurodegener. 2020; 15:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xi Z., Zinman L., Moreno D., Schymick J., Liang Y., Sato C., Zheng Y., Ghani M., Dib S., Keith J.et al.. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am. J. Hum. Genet. 2013; 92:981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Russ J., Liu E.Y., Wu K., Neal D., Suh E., Irwin D.J., McMillan C.T., Harms M.B., Cairns N.J., Wood E.M.et al.. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol. 2015; 129:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lerner L.K., Sale J.E.. Replication of G quadruplex DNA. Genes. 2019; 10:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. De Magis A., Manzo S.G., Russo M., Marinello J., Morigi R., Sordet O., Capranico G.. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Thys R.G., Wang Y.-H.. DNA replication dynamics of the GGGGCC repeat of the C9orf72 gene. J. Biol. Chem. 2015; 290:28953–28962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lu B., Gehrke S., Wu Z.. RNA metabolism in the pathogenesis of Parkinson's disease. Brain Res. 2014; 1584:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koukouraki P., Doxakis E.. Constitutive translation of human α-synuclein is mediated by the 5′-untranslated region. Open Biology. 2016; 6:160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lammich S., Kamp F., Wagner J., Nuscher B., Zilow S., Ludwig A.-K., Willem M., Haass C.. Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5′-untranslated region. J. Biol. Chem. 2011; 286:45063–45072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dai J., Liu Z.Q., Wang X.Q., Lin J., Yao P.F., Huang S.L., Ou T.M., Tan J.H., Li D., Gu L.Q.et al.. Discovery of small molecules for uprRegulating the translation of antiamyloidogenic secretase, a disintegrin and metalloproteinase 10 (ADAM10), by binding to the G-quadruplex-forming sequence in the 5′ untranslated region (UTR) of its mRNA. J. Med. Chem. 2015; 58:3875–3891. [DOI] [PubMed] [Google Scholar]
- 82. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bonnal S., Schaeffer C., Créancier L., Clamens S., Moine H., Prats A.-C., Vagner S.. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003; 278:39330–39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Baird S.D., Turcotte M., Korneluk R.G., Holcik M.. Searching for IRES. RNA. 2006; 12:1755–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S.. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010; 132:17831–17839. [DOI] [PubMed] [Google Scholar]
- 86. Cammas A., Dubrac A., Morel B., Lamaa A., Touriol C., Teulade-Fichou M.-P., Prats H., Millevoi S.. Stabilization of the G-quadruplex at the VEGF IRES represses cap-independent translation. RNA Biology. 2015; 12:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Arora A., Dutkiewicz M., Scaria V., Hariharan M., Maiti S., Kurreck J.. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008; 14:1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Oyaghire S.N., Cherubim C.J., Telmer C.A., Martinez J.A., Bruchez M.P., Armitage B.A.. RNA G-quadruplex invasion and translation inhibition by antisense γ-peptide nucleic acid oligomers. Biochemistry. 2016; 55:1977–1988. [DOI] [PubMed] [Google Scholar]
- 89. Mori K., Weng S.M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C.et al.. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013; 339:1335–1338. [DOI] [PubMed] [Google Scholar]
- 90. Ash P.E., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W. 3rd, Rademakers R.et al.. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013; 77:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Darnell J.C., Klann E.. The translation of translational control by FMRP: therapeutic targets for FXS. Nat. Neurosci. 2013; 16:1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Siomi H., Siomi M.C., Nussbaum R.L., Dreyfuss G.. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993; 74:291–298. [DOI] [PubMed] [Google Scholar]
- 93. Zalfa F., Giorgi M., Primerano B., Moro A., Di Penta A., Reis S., Oostra B., Bagni C.. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003; 112:317–327. [DOI] [PubMed] [Google Scholar]
- 94. Antar L., Bassell G.. Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron. 2003; 37:555–558. [DOI] [PubMed] [Google Scholar]
- 95. Zhang Y., Gaetano C.M., Williams K.R., Bassell G.J., Mihailescu M.R.. FMRP interacts with G-quadruplex structures in the 3′-UTR of its dendritic target Shank1 mRNA. RNA Biology. 2014; 11:1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B.. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001; 107:489–499. [DOI] [PubMed] [Google Scholar]
- 97. McAninch D.S., Heinaman A.M., Lang C.N., Moss K.R., Bassell G.J., Mihailescu M.R., Evans T.L.. Fragile X mental retardation protein recognizes a G quadruplex structure within the survival motor neuron domain containing 1 mRNA 5′-UTR. Mol. Biosyst. 2017; 13:1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Stefanovic S., Bassell G.J., Mihailescu M.R.. G quadruplex RNA structures in PSD-95 mRNA: potential regulators of miR-125a seed binding site accessibility. RNA. 2015; 21:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. DeMarco B., Stefanovic S., Williams A., Moss K.R., Anderson B.R., Bassell G.J., Mihailescu M.R.. FMRP-G-quadruplex mRNA-miR-125a interactions: Implications for miR-125a mediated translation regulation of PSD-95 mRNA. PLoS One. 2019; 14:e0217275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Imperatore J.A., McAninch D.S., Valdez-Sinon A.N., Bassell G.J., Mihailescu M.R.. FUS Recognizes G quadruplex structures within neuronal mRNAs. Front. Mol. Biosci. 2020; 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Crenshaw E., Leung B.P., Kwok C.K., Sharoni M., Olson K., Sebastian N.P., Ansaloni S., Schweitzer-Stenner R., Akins M.R., Bevilacqua P.C.et al.. Amyloid precursor protein translation is regulated by a 3′UTR guanine quadruplex. PLoS One. 2015; 10:e0143160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lee E.K., Kim H.H., Kuwano Y., Abdelmohsen K., Srikantan S., Subaran S.S., Gleichmann M., Mughal M.R., Martindale J.L., Yang X.et al.. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 2010; 17:732–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Westmark C.J., Malter J.S.. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007; 5:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H.. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001; 20:4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Didiot M.-C., Tian Z., Schaeffer C., Subramanian M., Mandel J.-L., Moine H.. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008; 36:4902–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Fisette J.F., Montagna D.R., Mihailescu M.R., Wolfe M.S.. A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J. Neurochem. 2012; 121:763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L.. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016; 5:e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mori K., Lammich S., Mackenzie I.R., Forne I., Zilow S., Kretzschmar H., Edbauer D., Janssens J., Kleinberger G., Cruts M.et al.. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013; 125:413–423. [DOI] [PubMed] [Google Scholar]
- 109. An H., Shelkovnikova T.A. Stress granules regulate paraspeckles: RNP granule continuum at work. Cell Stress. 2019; 3:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gomes E., Shorter J.. The molecular language of membraneless organelles. J. Biol. Chem. 2019; 294:7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. van Leeuwen W., Rabouille C.. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic. 2019; 20:623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hyman A.A., Weber C.A., Jülicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 113. Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L.et al.. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018; 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Simone R., Balendra R., Moens T.G., Preza E., Wilson K.M., Heslegrave A., Woodling N.S., Niccoli T., Gilbert-Jaramillo J., Abdelkarim S.et al.. G‐quadruplex‐binding small molecules ameliorate C9orf72FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 2018; 10:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shelkovnikova T.A., Kukharsky M.S., An H., Dimasi P., Alexeeva S., Shabir O., Heath P.R., Buchman V.L.. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018; 13:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nishimoto Y., Nakagawa S., Hirose T., Okano H.J., Takao M., Shibata S., Suyama S., Kuwako K., Imai T., Murayama S.et al.. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 2013; 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bajc Cesnik A., Darovic S., Prpar Mihevc S., Stalekar M., Malnar M., Motaln H., Lee Y.B., Mazej J., Pohleven J., Grosch M.et al.. Nuclear RNA foci from C9ORF72 expansion mutation form paraspeckle-like bodies. J. Cell Sci. 2019; 132:jcs224303. [DOI] [PubMed] [Google Scholar]
- 118. Simko E.A., Liu H., Zhang T., Velasquez A., Teli S., Haeusler A.R., Wang J.. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 2020; 48:7421–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wolozin B., Ivanov P.. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019; 20:649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Fay M.M., Anderson P.J., Ivanov P.. ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 2017; 21:3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S.et al.. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015; 525:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mackenzie I.R., Rademakers R., Neumann M.. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010; 9:995–1007. [DOI] [PubMed] [Google Scholar]
- 123. Majumder V., Gregory J.M., Barria M.A., Green A., Pal S.. TDP-43 as a potential biomarker for amyotrophic lateral sclerosis: a systematic review and meta-analysis. BMC Neurol. 2018; 18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ishiguro A., Kimura N., Watanabe Y., Watanabe S., Ishihama A.. TDP-43 binds and transports G-quadruplex-containing m RNA s into neurites for local translation. Genes Cells. 2016; 21:466–481. [DOI] [PubMed] [Google Scholar]
- 125. Ishiguro A., Kimura N., Noma T., Shimo-Kon R., Ishihama A., Kon T.. Molecular dissection of ALS-linked TDP-43: involvement of the Gly-rich domain in interaction with G-quadruplex mRNA. FEBS Lett. 2020; 594:2254–2265. [DOI] [PubMed] [Google Scholar]
- 126. Subramanian M., Rage F., Tabet R., Flatter E., Mandel J.L., Moine H.. G–quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011; 12:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ivanov P., O’Day E., Emara M.M., Wagner G., Lieberman J., Anderson P.. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Crabtree B., Thiyagarajan N., Prior S.H., Wilson P., Iyer S., Ferns T., Shapiro R., Brew K., Subramanian V., Acharya K.R.. Characterization of human angiogenin variants implicated in amyotrophic lateral sclerosis. Biochemistry. 2007; 46:11810–11818. [DOI] [PubMed] [Google Scholar]
- 130. Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G.-F., Anderson P.. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010; 285:10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lyons S.M., Gudanis D., Coyne S.M., Gdaniec Z., Ivanov P.. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 2017; 8:1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lyons S.M., Kharel P., Akiyama Y., Ojha S., Dave D., Tsvetkov V., Merrick W., Ivanov P., Anderson P.. eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic Acids Res. 2020; 48:6223–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jackowiak P., Hojka-Osinska A., Gasiorek K., Stelmaszczuk M., Gudanis D., Gdaniec Z., Figlerowicz M.. Effects of G-quadruplex topology on translational inhibition by tRNA fragments in mammalian and plant systems in vitro. Int. J. Biochem. Cell Biol. 2017; 92:148–154. [DOI] [PubMed] [Google Scholar]
- 134. Grigg J.C., Shumayrikh N., Sen D.. G-quadruplex structures formed by expanded hexanucleotide repeat RNA and DNA from the neurodegenerative disease-linked C9orf72 gene efficiently sequester and activate heme. PLoS One. 2014; 9:e106449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Gray L.T., Lombardi E.P., Verga D., Nicolas A., Teulade-Fichou M.-P., Londoño-Vallejo A., Maizels N.. G-quadruplexes sequester free heme in living cells. Cell Chem. Biol. 2019; 26:1681–1691. [DOI] [PubMed] [Google Scholar]
- 136. Olsthoorn R.C. G-quadruplexes within prion mRNA: the missing link in prion disease?. Nucleic Acids Res. 2014; 42:9327–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Czech A., Konarev P.V., Goebel I., Svergun D.I., Wills P.R., Ignatova Z.. Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes. Sci. Rep. 2019; 9:2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Saha T., Usdin K.. Tetraplex formation by the progressive myoclonus epilepsy type-1 repeat: implications for instability in the repeat expansion diseases. FEBS Lett. 2001; 491:184–187. [DOI] [PubMed] [Google Scholar]