Abstract
The transition from meiotic spermatocytes to postmeiotic haploid germ cells constitutes an essential step in spermatogenesis. The epigenomic regulatory mechanisms underlying this transition remain unclear. Here, we find a prominent transcriptomic switch from the late spermatocytes to the early round spermatids during the meiotic-to-postmeiotic transition, which is associated with robust histone acetylation changes across the genome. Among histone deacetylases (HDACs) and acetyltransferases, we find that HDAC3 is selectively expressed in the late meiotic and early haploid stages. Three independent mouse lines with the testis-specific knockout of HDAC3 show infertility and defects in meiotic exit with an arrest at the late stage of meiosis or early stage of round spermatids. Stage-specific RNA-seq and histone acetylation ChIP-seq analyses reveal that HDAC3 represses meiotic/spermatogonial genes and activates postmeiotic haploid gene programs during meiotic exit, with associated histone acetylation alterations. Unexpectedly, abolishing HDAC3 catalytic activity by missense mutations in the nuclear receptor corepressor (NCOR or SMRT) does not cause infertility, despite causing histone hyperacetylation as HDAC3 knockout, demonstrating that HDAC3 enzyme activity is not required for spermatogenesis. Motif analysis of the HDAC3 cistrome in the testes identified SOX30, which has a similar spatiotemporal expression pattern as HDAC3 during spermatogenesis. Depletion of SOX30 in the testes abolishes the genomic recruitment of the HDAC3 to the binding sites. Collectively, these results establish the SOX30/HDAC3 signaling as a key regulator of the transcriptional program in a deacetylase-independent manner during the meiotic-to-postmeiotic transition in spermatogenesis.
INTRODUCTION
Mammalian spermatogenesis is precisely regulated at multiple stages, including spermatogonial self-renewal and differentiation, meiosis, and a postmeiotic round spermatid development process known as spermiogenesis. This whole developmental process starts with the proliferation of undifferentiated spermatogonia (Type A spermatogonia), followed by mitotic divisions into Type B spermatogonia, and generation of pre-leptotene spermatocytes prior to meiosis. Meiosis starts with prophase I that, in turn, is divided into substages of leptotene, zygotene, pachytene, diplotene, and diakinesis. At the early stages of meiotic prophase I, genes encoding components of the synaptonemal complex are highly expressed in preparation for meiotic chromosome events, including chromosomal synapsis and recombination (1–3).
There are several major transitions during mammalian spermatogenesis. In the mitosis-to-meiosis transition or meiotic entry decision, it is known that retinoic acid (RA) signaling plays an important role in remodeling of diploid mitotic cells into primary spermatocytes through inducing expression of the key meiotic regulator STRA8 (stimulated by retinoic acid 8) (4–6). Specifically, STRA8 coordinates with the transcription factor MEIOSIN to drive meiotic gene activation for establishing meiosis-specific chromosome events (7), whereas the Doublesex and mab-3 related transcription factor DMRT1 negatively regulates the entry into meiosis through repressing RA responsiveness and Stra8 transcription (8,9). Recent studies demonstrated that a genome-wide reorganization of super-enhancers occurs at the mitosis-to-meiosis transition (10,11).
The meiotic-to-postmeiotic transition, or meiotic exit, is another important step during spermatogenesis, but the underlying epigenomic regulatory mechanism is not clear (12). After prophase, spermatocytes progress through two reductive meiotic divisions into haploid round spermatids (RS), which further experience profound chromatin condensation and nucleus reorganization (13), and eventually generate mature spermatozoa. During the transition from meiosis to postmeiotic round spermatids, germ cells undergo extensive epigenomic reprogramming to establish male germline stage-specific cell identity (12,14,15). Such epigenomic changes are associated with gene expression changes, resulting in the RS-specific transcriptome that is distinct from the mitotic or meiotic phase of germ cells (16,17). It is unclear how the chromatin-associated coregulator complexes drive transcriptional regulation during this transition. Histone acetylation is a general marker for epigenomic remodeling. Through unbiased profiling, we found histone acetylation changes during the meiotic spermatocytes-to-postmeiotic RS transition.
Histone acetylation is controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs) (18). Through gene expression analysis, we found that HDAC3 is selectively expressed in later stages of spermatocytes and early RS. HDAC3 is a Class I HDAC and functions in nuclear repressor complexes that contain either NCOR (nuclear receptor corepressor) or its homolog SMRT (silencing mediator of retinoic and thyroid receptors) (19,20). The deacetylase activity for HDAC3 requires association with the deacetylase activation domain (DAD) of NCOR and SMRT (21). The binding of DAD with HDAC3 causes a conformational change of HDAC3 protein, which allows the substrate to access the catalytic site (22). Knock-in mice bearing missense mutations in the DAD of both NCOR and SMRT (Y478A in NCOR, Y470A in SMRT) are referred as NS-DAD mutant (NS-DADm) and have no detectable or drastically abolished HDAC3 enzymatic activity in multiple tissues (23–26). In this study, we use conditional Hdac3 knockout mice with Vasa-cre, Neurog3-cre or Stra8-cre as well as the whole-body NS-DADm knock-in mice to investigate HDAC3 function in male germline development. We discovered an unexpected regulatory mechanism involving HDAC3 in meiosis exit.
MATERIALS AND METHODS
Animals
C57BL/6 Hdac3fl/fl mice were generously provided by Mitchell A Lazar at the University of Pennsylvania (27). This floxed Hdac3 allele (Hdac3fl) was generated using homologous recombination in embryonic stem cells. In the targeted allele, exons 4–7 were flanked by loxP sites. Stra8-cre mice (Stock number: 008208), Neurog3-cre mice (Stock number: 006333), and Vasa-cre mice (Stock number: J006954) were from the Jackson Laboratory. Sox30 knockout mice were generated through CRISPR/Cas9 with two sgRNAs targeting the exon 2, which contains the DNA binding HMG-box domain. All animals were maintained and used according to the guidelines of the Institutional Animal Care and Use Committee of Nanjing Medical University. NS-DADm mice were previously described (23). NS-DADm mice harbored mutations in the DAD of both alleles of Ncor1 (Y478A in NCOR) and Ncor2 (Y470A in SMRT), and were generated by crossing the Ncor1 DAD mutant mice (N-DADm) with the Ncor2 DAD mutant mice (S-DADm) (23). N-DADm (the Ncor1 Y478A knockin allele) was generated through gene targeting in ES cells (28). The targeting vector contains the Y478A mutation within the DAD of NCOR and a loxP flanked neomycin selection cassette was inserted downstream from exon 13 for Cre-mediated removal. A similar strategy was used for the generation of S-DADm (the Ncor2 Y470A mutation) (23). NS-DADm mice were maintained according to the guidelines of the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Histological analysis
Testes and epididymides were freshly fixed in Bouin's solution (Sigma, SLBJ3855V) for overnight, dehydrated in graded ethanol (70%, 80%, 90%,100%) and embedded in paraffin. 5 μm cross-sections were prepared and stained with hematoxylin and eosin.
Immunofluorescence and chromosome spread
Immunofluorescence assay was previously described (29). Tissues were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. Tissues were dehydrated with ethanol (70%, 80%, 90%, 100%), cleared in xylene, embedded in paraffin, and cut into 5 μm sections. Sections were washed in xylene twice and rehydrated in ethanol (100%, 95%, 70%, 50%, 0%). Slides were boiled in 10 mM sodium citrate for 15 min and washed in PBS twice. Slides were blocked with blocking buffer (10% FBS, 1% Trixon X-100 in PBS) for 1 h at room temperature and incubated with primary antibodies at 4°C overnight. The primary antibodies were as follows: anti-HDAC3 (1:200, Abcam, ab7030), anti-SYCP3 (1:200, Abcam, ab97672), anti-PLZF (1:200, R&D, AF2944), SOX30 (1:200, ABclonal, A11759). For immunofluorescence analysis for acrosome, cryosections were incubated for 1 h at 37°C with FITC-conjugated peanut agglutinin (RL-1072, Vector Labs). Nuclear DNA was stained with DAPI (F6057; Sigma). Chromosome spreads were prepared as previously described (30) and were further incubated with anti-SYCP1 (1:200, Abcam, ab15090) and anti-SYCP3 (1:200, Abcam, ab97672). All the staining of testis sections and spread confocal images were visualized on a confocal microscope (Carl Zeiss, LSM700).
Spermatogenic cell isolation
Pachytene spermatocytes and round spermatids were isolated from adult mouse testes (Cells from six control mice or 10 KO mice were pooled for one biological replicate. RNA-seq libraries were prepared in biological triplicates). The average purity of pachytene spermatocytes isolated from adult Hdac3fl/+ and Stra8-cre/Hdac3fl/– mice was 86% and 82%, respectively. The average purity of round spermatids from each genotype was 92% and 84%, respectively. For the assays for HDAC3 protein level in isolated spermatogenic populations, spermatogonia were isolated from young mice at postnatal days 6–8 when testes consist of spermatogonia and Sertoli cells. Different populations of germ cells were sorted by the STA-PUT method as described (31). Briefly, tissues were digested with collagenase IV (1 mg/ml) at 37°C for 10–20 min, centrifuged, and washed with DMEM. Cell suspension was subsequently digested with 0.25% Trypsin containing DNase I (1 mg/ml) 37°C for 5 min to prepare single-cell suspensions. Single-cell suspensions were further loaded into a cell separation apparatus (ProScience Inc. Canada) and followed by 2–4% bovine serum albumin (BSA) gradient (2% BSA and 4%BSA in DMEM were loaded into the separation apparatus chamber). We harvested the cell fractions after 1.5–3 h sedimentation and identified their distinct cell types based on their morphological characteristics, cell diameters and DAPI staining pattern under a light microscope.
Immunoprecipitation, immunoblot and HDAC assay
Tissues were rinsed with PBS and lysed in cold RIPA buffer supplemented with protease inhibitor cocktail tablets (Roche, 4693124001). Homogenized lysates were rotated at 4°C for 1 h, and centrifuged at 12 800 rpm for 40 min at 4°C. Protein concentration of collected supernatant was determined by bicinchoninic acid (BCA) assay. 20 μg proteins for each lane were separated by sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE). For immunoprecipitation, tissue mass (80 mg) were dounced 30 strokes or 1 × 107 293T cells were lysed in 500 μl lysis buffer (20 mM Tris–Cl pH = 7.4, 150 mM Nacl, 0.5% Triton X-100, 0.5% sodium dexycholate, 1 mM DTT, 1× protease inhibitor) and left on ice for 10 mins. Lysates were further diluted with 500 μl of dilution buffer (20 mM Tris pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 1 mM DTT, protease inhibitor). Lysates was centrifuged and the supernatant was collected. The supernatant was pre-cleared with washed protein A beads (Milipore, 16–156) for 2 h, then incubated with 10 μg indicated antibodies (anti-HDAC3, Abcam, ab7030), anti-SOX30 (ABclonal, A11759) overnight at 4°C with rotation. 25 μl washed protein A beads were loaded into the mixture and incubated for 3 h at 4°C with rotation. Bead complexes were pelleted and washed three times with wash buffer [20 mM Tris (pH 7.4), 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA]). The protein complex was eluted off the beads into 50 μl 2× loading buffer (4% SDS, 20% glycerol, 120 mM Tris–HCl pH 6.8, 10% β-mercaptoethanol, 0.02% bromophenol blue). HDAC assay was conducted using fluorescence kit (Active Motif) following manufacturer's instruction.
Vector construction and cell transfections
Truncated Sox30 expression constructs with N-terminus (aa1–364), C-terminus (aa436–782), HMG domain absence (aa1–346, 436–782) and C-terminus absence (aa1–435) were generated by PCR amplification using mouse testis cDNA. These fragments were inserted into PRK5 with FLAG or HA tag to synthesize recombinant protein. Each required tagged protein expression construct was transfected in HEK293T cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were collected 48 hrs after transfection, followed by co-immunoprecipitation with anti-FLAG and anti-HA antibodies.
Dual luciferase assays
The Nkapl fragment containing binding sites was PCR-amplified from mouse testes cDNA and cloned into PGL4.17-Luciferase vector. HEK 293T cells were transfected with PGL4.17-Luciferase vector (500 ng) and Renilla luciferase vector (2.0 ng) plus expression constructs (500 ng): PRK5/HDAC3, PRK5/SOX30, PRK5/SOX30/HDAC3 or the control vector PRK5/FLAG. Cell extracts were harvested 48 hrs after transfection, and luciferase assays were performed according to the manufacturer's protocol (Promega).
Quantitative RT-PCR Assay and RNA-seq
Total RNA was extracted from the samples using TRIzol reagent. For isolated spermatogenic cells, total RNA was isolated from pachytene spermatocytes and round spermatids from Hdac3fl/+ and Stra8-cre/Hdac3fl/– mice (Cells from six control mice or 10 Stra8-cre/Hdac3fl/– mice were pooled for one biological replicate. RNA-seq libraries were prepared in biological triplicates). The concentration and purity of RNA were determined by absorbance at 260/280 nm. 1 μg of total RNA was reverse transcribed using a PrimeScript™RT Master Mix (TaKaRa, RR036A). The cDNA was diluted by 5–6 times and 1 μl cDNA were used for each reaction using SYBR Green Premix Ex Taq II (RR820A, TaKaRa, Japan). A standard reaction contained 200 nmol/l of forward and reverse primer, 1 μl cDNA and the final reaction volume was 20 μl. The reaction was initiated by preheating at 50°C for 2 min, followed by 95°C for 10 min. Subsequently, 40 amplification cycles were carried out with 15 s denaturation at 95°C and 30 s annealing and extension at 60°C. This assay was performed in a thermocycler StepOnePlus™ Real-Time PCR system (Applied Biosystems) and all reactions were performed in triplicate. Gene expression was normalized to Arbp. All the primers were listed in Supplementary Table S1. For RNA-seq, strand-specific libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation kit (Illumina) according to the manufacturer's instructions before submitting to the Illumina Hiseq X ten system. RNA-seq library preparation and sequencing were performed at Omics Core of Bio-Med Big Data Center, Chinese Academy of Sciences (Shanghai, China). Clean data were obtained by trimming the adaptor sequence and removing sequences with low quality. Clean reads were mapped to the mouse genome (mm10) with HISAT2 (v.2.1.0; Johns Hopkins University, Baltimore, MD, USA) with a GTF file download from the UCSC database (University of California, Santa Cruz). The aligned reads of genes were counted using HTSeq followed by DESeq2 normalization to evaluate gene expression as normalized counts per million. Significant differentially expressed genes were identified as those with a P-value or false discovery rate (FDR) value above the threshold (pachytene spermatocytes and round spermatids: P < 0.01) and fold- change >1.5 using DESeq2 software.
ChIP-seq and data analysis
Fresh testes were collected from wild-type and Sox30 KO mice at postnatal day 20, ground into powder in liquid N2 and cross-linked by 1% formaldehyde, followed by quenching with glycine solution. Chromatin was fragmented by sonication in ChIP SDS lysis buffer (50 mM Tris–Cl, pH 8; 10 mM EDTA, pH 8; 1% SDS) using the Covaris-S220 sonicator. Cross-linked chromatin was incubated with indicated antibodies (anti-HDAC3, Abcam, ab7030; anti-SOX30, ABclonal, A11759; anti-H3K9ac, Milipore, 07–352; anti-H3K27ac, Abcam ab4729) in ChIP dilution buffer (50 mM HEPES, pH7.5, 155 mM NaCl, 1.1% Triton X-100, 0.11% Na-deoxycholate) with protease inhibitors overnight. Cross-linking was reversed overnight at 65°C, and DNA was extracted by using phenol/chloroform/isoamyl alcohol. Precipitated DNA was amplified for deep-sequencing or analyzed by quantitative PCR with SYBR Green Premix Ex Taq II (RR820A, TaKaRa). For ChIP-seq, DNA was amplified according to the ChIP Sequencing Sample Preparation Guide provided by Illumina using adaptors and primers. Deep sequencing was performed by Computational Biology Omics Core of the CAS-MPG Partner Institute using Illumina Hiseq X-10 (2 × 150). ChIP-seq reads were aligned to the mouse genome (mm10) using BOWTIE software (bowtie2 version 2.3.4.3). The aligned reads were further filtered to remove those with low mapping quality and duplicated arising as an artifact of sample amplification or sequencing. Peak calling was carried out by using MACS2 (version 2.1.2, with options'-mfold 5, 50-p 0.0001'), on ChIP file against the input file. Genome-wide normalized signal coverage tracks were created by bamCoverage in deepTools (version 3.3.0) and visualized in the Integrative Genomics Viewer (IGV version 2.5.0). Peaks were annotated to the genomic region and the nearest genes within 2 kb of TSS using Bioconductor package ChIPSeeker (version 1.16.1). Peaks overlapping by at least 1 nt with unique gene model promoters (±3 kb of each unique gene model Transcription Starting Site) were considered as promoter located. De novo motif searches of ChIP-seq peaks were performed using Homer (version v4.11.1) with default parameters (32). ChIP-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GSE153065) database.
Statistical analysis
All data are reported as mean ± SD unless otherwise noted in the figure legends. Significance was tested by using the two-tailed unpaired Student's t-test (*P < 0.05; ** P < 0.01; *** P < 0.001) using Prism 7.0 (GraphPad Software, La Jolla, CA, USA).
RESULTS
Histone acetylation correlates with gene expression changes from meiotic to postmeiotic stages
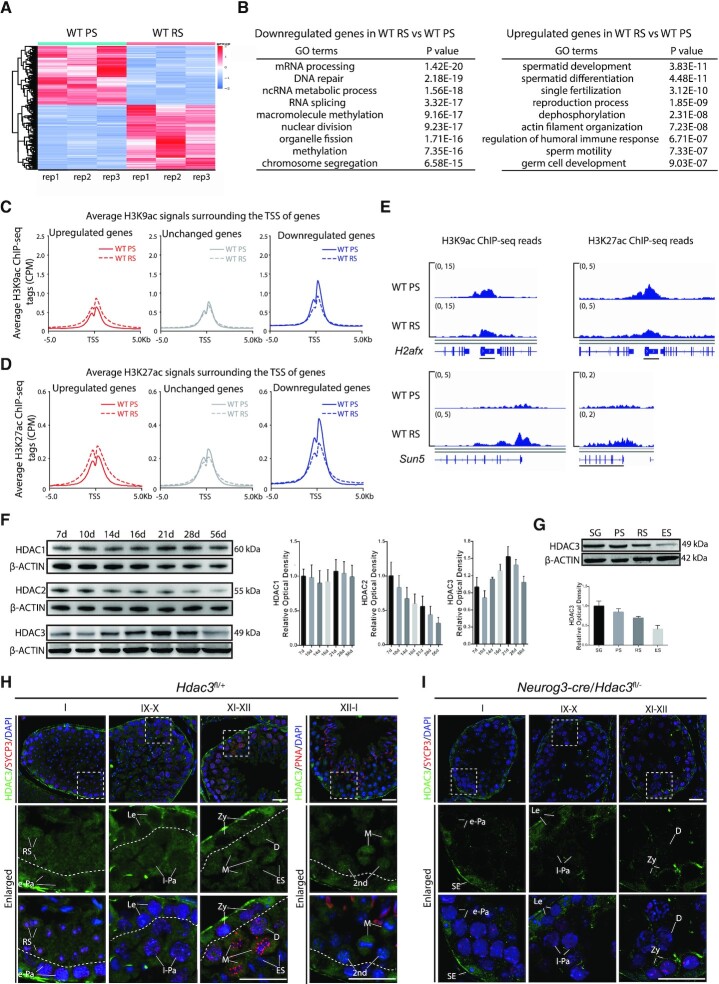
The meiotic spermatocytes at the pachytene stage and haploid round spermatids are two major cell populations at meiotic and postmeiotic stages with unique features in the male germline. Here, we isolated these two populations from the adult mice at 7–8 weeks old. We found that the transcriptomes of pachytene spermatocytes and postmeiotic round spermatids were remarkably distinct (Figure 1A). Many genes involved in spermatid development and differentiation are expressed at higher levels in postmeiotic round spermatids, whereas genes in DNA repair, RNA splicing, nuclear division, chromosome segregation, and organelle fission are expressed at higher levels in pachytene spermatocytes (Figure 1B). These gene ontology terms were similarly enriched either in upregulated or downregulated genes in two recent studies with single-cell RNA transcriptomes of mouse male germ cells (17,33). A key mechanism by which the germ cells establish stage-specific transcriptome is through epigenomic modifications of the chromatin, and histone acetylation is a major epigenomic marker during the process (12,17,34,35). We performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) with antibodies specific for acetylated histone H3 lysine (H3K9ac) and acetylated histone H3 lysine 27 (H3K27ac) using stage-specific pachytene spermatocytes (PS) and round spermatids (RS) from the adult mice at 7–8 week-old. Concomitant with transcriptional alterations, we observed enhanced accumulation of H3K9ac at genes that were upregulated at RS stage in comparison to the PS stage (Figure 1C and E), especially at the regions near the transcriptional start site (TSS). Higher H3K27ac was similarly observed for those upregulated genes (Figure 1D and E). The binding of H3K9ac and H3K27ac remained constant or was weaker at genes that were unchanged or downregulated at RS stage, compared with the PS stage (Figure 1C–E). These results show that H3K9ac and H3K27ac levels correlate with the upregulated transcription of genes in pachytene spermatocytes or round spermatids.
Figure 1.
Transcriptomic changes during the meiotic spermatocytes-to-postmeiotic RS transition are correlated with the genome-wide histone acetylation levels. (A) A heatmap depicting gene expression pattern at round spermatid stage versus pachytene spermatocyte stage in wild-type. Fold change >1.5 up (red) or down (green) with P < 0.01. PS, pachytene spermatocytes; RS, round spermatids. (B) Top nine enriched biological processes for the genes expressed at higher levels or lower levels in RS compared to PS in wild-type. (C) Average H3K9ac profiles at upregulated (red), unchanged (gray), and downregulated (blue) genes in RS versus PS in adult wild-type. Average H3K9ac signals, represented by counts per million, from –5 to + 5 kb surrounding the TSS of genes were shown. (D) Average H3K27ac profiles at upregulated (red), unchanged (gray), and downregulated (blue) genes in RS versus PS in adult wild-type. Average H3K27ac signals, represented by counts per million, from –5 to + 5 kb surrounding the TSS of genes were shown. (E) Genome browser tracks depicting reads accumulation of H3K9ac and H3K27ac on representative meiotic gene H2afx and postmeiotic haploid gene Sun5. (F) Western blot analysis of HDAC1, HDAC2, and HDAC3 during spermatogenesis. β-Actin serves as a loading control. The image is a representation of two independent experiments with similar results. The corresponding optical density readings for biological duplicates are shown. (G) The protein amount of HDAC3 in different fractions of spermatogenic cells. SG: spermatogonia; ES: elongating spermatids. PS, RS, and ES were from mice at age 7–8 weeks (n = 8), and spermatogonia (SG) were isolated from mice at postnatal day 6–8 using the STA-PUT method (n = 20). Quantification of HDAC3 protein with the corresponding optical density readings are shown. The representative image of biological duplicates is shown. (H) Immunofluorescent staining of HDAC3 (green), SYCP3 (lateral elements of the synaptonemal complex, red), and Hoechst (DNA, blue) on frozen sections from adult wild-type. Areas within the rectangles were enlarged in the following panel, and different stages of seminiferous tubules were shown. Abbreviations: SE: somatic Sertoli cells; SG: spermatogonia; RS: round spermatids. Spermatocytes were categorized into the following groups: Le: leptotene; Zy: zygotene; e-Pa: early pachytene; m-Pa: mid pachytene; l-Pa: late pachytene; D: diplotene; M: Metaphase; 2nd: secondary spermatocytes. Scale bars, 25 μm. (I) Immunofluorescent staining of HDAC3 (green), SYCP3 (red) and Hoechst (DNA, blue) on frozen sections from adult Neurog3-cre/Hdac3fl/– testes. Scale bars, 25 μm.
Elongation of round spermatids during late spermatogenesis coincides with a genome-wide histone hyperacetylation, which triggers transcription-independent histone-to-protamine replacement (35–40). This suggests a possible mechanism that suppresses histone acetylation in late meiosis and early RS stages. Histone deacetylation is driven by HDACs. There are 18 mammalian HDACs that belong to four classes. To address which particular HDAC plays a major role in spermatogenesis, we compared the expression levels of HDACs in meiotic spermatocytes based on the previously reported transcriptomes of isolated spermatogenic cell types (14), and found that the mRNA transcripts of Hdac1, Hdac2, Hdac3, Sirt1, Sirt2, Sirt3 and Sirt4 were more abundant than other Hdac genes (Supplementary Figure S1A). Sirtuins are Class III NAD+-dependent deacetylases. SIRT1 specifically regulates acrosome biogenesis during spermiogenesis, suggesting that it functions in the window later than the early RS stage (41–44). Both Sirt3 KO or Sirt4 KO mice are fertile (45), and no obvious phenotypes were observed in Sirt2 depleted testes (46). Among other groups of HDACs with high expression levels, only HDAC3 protein exhibited a dynamic pattern of expression in germ cell development (Figure 1F). HDAC3 protein increased significantly in testis with the highest abundance amount at postnatal day 16 to 21 (P16–21), and its expression decreased thereafter throughout spermatogenesis (Figure 1F), albeit consistently expressed Hdac3 mRNA levels throughout spermatogenesis (14). Western blot analysis using protein extracts of sorted germ cell populations by STA-PUT Velocity Sedimentation revealed that HDAC3 protein was highly enriched in spermatogonia (SG), PS and RS, but dramatically reduced in elongating spermatids (Figure 1G), when histone hyperacetylation occurs. The purities of distinct germ cell populations were assessed according to their distinct morphological features (Supplementary Figure S1B) and further validated by immunostaining with stage-specific molecular markers γH2AX and PNA, which distinguishes the XY body in pachytene spermatocytes and the acrosome in round spermatids (Supplementary Figure S1C), respectively. Subcellular fractionation showed that HDAC3 protein was abundant in the chromatin fraction but also found in the cytoplasm fraction in testes (Supplementary Figure S1D). Immunofluorescence staining of HDAC3 and SYCP3 (a component of the synaptonemal complex) in adult testes revealed a dynamic stage-specific expression pattern (Figure 1H). HDAC3 was readily detected in the somatic Sertoli cells and spermatogonia, but was not detectable in leptotene and zygotene spermatocytes at stage IX–X and XI–XII or in early pachytene spermatocytes at stage I (Figure 1H). Detailed analysis indicated that HDAC3 protein expressed in the nuclei of late pachytene spermatocytes (stage IX–X), and reached its highest level in the nuclei of diplotene (stage XII), metaphase cells (stage XII), and secondary spermatocytes (stage XII–I) (Figure 1H).
Postmeiotic spermatid development encompasses 16 steps (steps 1–16), as defined by morphological changes in acrosome and nuclear structure. Immunostaining with HDAC3 and PNA (an acrosome marker) indicated that HDAC3 was expressed in the nuclei of round spermatids at step 1 (stage I tubules) (Supplementary Figure S1E). When round spermatids advanced to spermiogenic steps 2–3, perinuclear foci-like structures with HDAC3 signals were observed (step 2–3, stage II–III), although the nuclear expression of HDAC3 was still present (Supplementary Figure S1E). During later stages of round spermatid development at step 7–8, stage VII–VIII, these cytoplasmic HDAC3 foci were enlarged, concomitant with a decrease in its nuclear signal (Supplementary Figure S1E). The nuclear signal of HDAC3 was dramatically decreased at spermiogenic step 9 and became completely absent at the following step 10 when spermatids start to elongate (Supplementary Figure S1E). The HDAC3 signals in the cytoplasm persisted until spermiogenic step 12 and then decreased in elongated spermatids after step 12 and in mature spermatozoa (Supplementary Figure S1E). To provide negative controls for the HDAC3 immunofluorescence staining, we sought to obtain HDAC3 knockout tissues. Because the global deletion of Hdac3 is embryonic lethal, we bred mice with a floxed HDAC3 allele (27) with mice harboring germline-specific Neurog3-cre. The HDAC3 immunofluorescence signals were absent in spermatocytes during later stages and round spermatids (Figure 1I and Supplementary Figure S1F), but remained at similar levels in the somatic Sertoli cells in adult Neurog3-cre/Hdac3fl/– littermates (Figure 1I). These results demonstrate that this antibody is specific for HDAC3.
HDAC3 is required for meiotic exit and transition to round spermatid
To systemically address the function of HDAC3 in spermatogenesis, we bred mice with a floxed HDAC3 allele (27) with mice harboring Vasa-cre, Neurog3-cre, and Stra8-cre to generate testis-specific knockouts of Hdac3 (Supplementary Figure S2A). The Vasa-cre begins to express in germ cells at embryonic day 15 (47). The Neurog3-cre starts to express in the testis at postnatal day 7 (48), resulting in almost complete recombination at postnatal day 9 (leptotene/zygotene spermatocytes) (49). Stra8-cre starts to express in differentiating spermatogonia with the peak expression in leptotene spermatocytes (50). The conditional HDAC3 knockout mice (Vasa-cre/Hdac3fl/–, Neurog3-cre/Hdac3fl/– and Stra8-cre/Hdac3fl/–, respectively) are all viable. We use cre/Hdac3fl/+ male mice and Hdac3fl/fl female mice to generate experimental cohorts. The littermate Hdac3fl/+ serves as the control. Western blot analysis showed that HDAC3 protein was reduced markedly in testes of Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– mice at postnatal day 18 (Supplementary Figure S2B). The residual expression is probably due to somatic Sertoli cells that also express HDAC3.
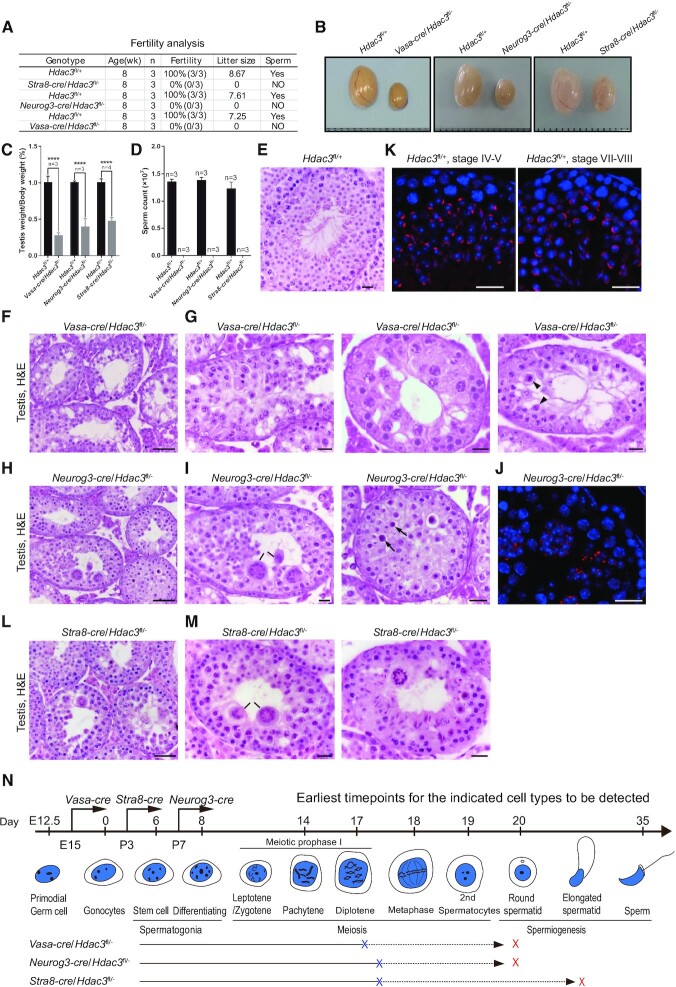
Strikingly, all the Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– males were infertile (Figure 2A). In adulthood, the testes from Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– mice were apparently smaller than the control (Figure 2B) and weighed at <40% of the control (Figure 2C). Mature sperm were absent in the cauda epididymis of all three HDAC3 knockouts (Figure 2D). While control Hdac3fl/– littermates produced an average of 7.8 offsprings per litter, no pups were obtained from adult Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– males when mating with normal fertile females (Figure 2A). These results demonstrate that HDAC3 is required for male fertility.
Figure 2.
Hdac3-deficiency causes male sterility and germ cell arrest at the late stages of prophase I and early RS stage. (A) Fertility data collected from adult Vasa-cre, Neurog3-cre, and Stra8-cre/Hdac3fl/– mice. n = 3 for each genotype. (B) Representative images of testes from adult Vasa-cre, Neurog3-cre, and Stra8-cre/Hdac3fl/– mice and their age-matched wild-types. (C) Testes weights were dramatically reduced in Vasa-cre, Neurog3-cre, and Stra8-cre/Hdac3fl/– mice at 8-week-old. Data presented is the ratio of testes weight and body weight (n = 3, *** P < 0.001, Student's t test). (D) Comparison of epididymal sperm counts between adult Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– mice and their age-matched wild types. n = 3 for each genotype. *** P < 0.001, Student's t test. (E–I, L–M) Hematoxylin and eosin staining of testes from adult wild type (E), Vasa-cre/Hdac3fl/– (F, G), Neurog3-cre/Hdac3fl/– (H, I) and Stra8-cre/Hdac3fl/– (L, M). Arrowheads in (G) point to metaphase cells with apoptosis in stage XII tubules of Vasa-cre/Hdac3fl/–. Black lines in Figure 2I and M point to multinucleated cell clusters of round spermatids. Arrows in Figure 2I indicate apoptotic cells during the transition from metaphase cells into secondary spermatocytes in stage XII tubules of Neurog3-cre/Hdac3fl/–. (J, K) Immunofluorescent staining of PNA on testis sections from wild type (K) and Neurog3-cre/Hdac3fl/– (J). PNA is the acrosome marker, and acrosome formation at developmental step 4 and step 8 was showed in wild-type (K), but acrosome beyond step 2–3 was completely absent in Neurog3-cre/Hdac3fl/– males (J). Scale bars, 20 μm. (N) A diagram representing arrested stages of germ cell development in Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– mice. Blue and red crosses on lines indicate the earliest and ultimate time point of spermatogenic arrest, respectively.
We performed histological analyses to assess the germ-cell development defects due to HDAC3 deficiency. Seminiferous tubules of control testes were full of spermatogenic cells representative of different stages, including mitotic spermatogonia, meiotic spermatocytes, postmeiotic round spermatids, elongating spermatids, and spermatozoa with highly compacted chromatin (Figure 2E). In contrast, elongating/condensing spermatids and normal spermatozoa were completely absent in seminiferous tubules of both the Vasa-cre/Hdac3fl/– mice (Figure 2F) and Neurog3-cre/Hdac3fl/– mice (Figure 2H). In Vasa-cre/Hdac3fl/– testes, many tubules were arrested at later stages of meiotic prophase I (diplotene stage) or early round spermatid stages (Figure 2G). Some Vasa-cre-mediated KO spermatocytes progressed to metaphase cells and were arrested at the M phase with apoptosis in stage XII tubules (Figure 2G, right panel, arrowheads).
In Neurog3-cre/Hdac3fl/– testes, over 70% of tubules were arrested at the round spermatid stage (Figure 2I, left panel). We found that these round spermatids in Neurog3-cre/Hdac3fl/– failed to elongate or condense, resulting in their formation into multinucleated cell clusters (Figure 2I, left panel, black lines). Immunostaining with the acrosome marker PNA (peanut agglutinin) indicated that the round spermatids in Neurog3-cre/Hdac3fl/– were arrested at early stages, as confirmed by the absence of the acrosomal structure in round spermatids beyond step 2–3 (Figure 2J), in contrast to differentiated round spermatids at step 4–5 and step 7–8 observed in wild-type testes tubules (Figure 2K). Additionally, a subset of Neurog3-cre/Hdac3fl/– tubules exhibited defects in the transition from metaphase cells into secondary spermatocytes in stage XII tubules (Figure 2I, right panel, arrows).
Similar to that of Neurog3-cre/Hdac3fl/–, over 60% seminiferous tubules in Stra8-cre /Hdac3fl/– testes were arrested at the round spermatid stage (Figure 2L and M). The remaining tubules were arrested either at later stages of meiotic prophase I or elongating spermatid stage (Figure 2M, right panel). Immunostaining indicates that tubules of Vasa-cre, Neurog3-cre, and Stra8-cre/Hdac3fl/– mice contained similar numbers of PLZF-positive undifferentiated spermatogonia compared to wild-types (Supplementary Figure S3A and S3B and data not shown). Consistent with the above defects, the epididymal tubule from all the Vasa-cre, Neurog3-cre, and Stra8-cre/Hdac3fl/– mice were depleted of mature spermatozoa (Supplementary Figure S3C). Although some germ cells in Stra8-cre HDAC3 KO mice differentiated into elongating spermatids, all the conditional HDAC3 knockouts using Vasa-cre, Neurog3-cre and Stra8-cre shared phenotypical similarities, with the earliest defects detected in the late stages of spermatocytes and eventually mainly arrested at early RS (Figure 2N).
We further examined the first meiotic prophase I by immunostaining the spread nuclei with SYCP3 and SYCP1, the lateral and central elements of the synaptonemal complex (51–54). The spermatocytes were staged based on the distribution of SYCP3 and SYCP1 proteins, and many stages of meiotic prophase I (leptotene, zygotene, pachytene, and diplotene) prior to the metaphase stage were observed in Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– (Supplementary Figure S3D). Collectively, these experiments demonstrate that HDAC3 functions in the time window when spermatocytes exit meiosis to become round spermatids. In the absence of HDAC3, germ cells exhibited defects during the latter stages of prophase I and the transition to the round spermatid.
HDAC3 represses early meiotic genes but activates spermatid genes
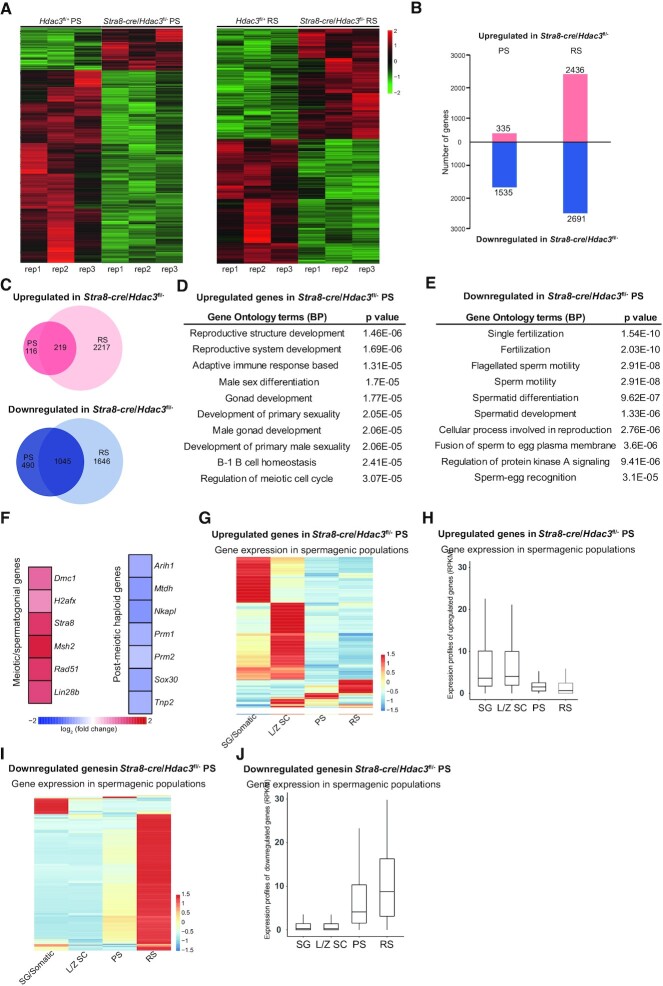
To profile stage-specific transcriptomic alterations upon HDAC3 deletion, we performed RNA-sequencing (RNA-seq) on purified pachytene spermatocytes (PS) and round spermatids (RS) from control and Stra8-cre/Hdac3fl/– mice. The purity of the PS and RS isolations was validated with γH2AX and PNA immunostaining, respectively (Supplementary Figure S4A and B). In PS, we found 335 and 1535 genes significantly up- and downregulated (P < 0.01, fold change > 1.5), respectively, in Stra8-cre/Hdac3fl/– compared to control (Figure 3A and B). In RS, 2409 and 2716 genes were significantly up- and downregulated, respectively (Figure 3A and B). Both upregulated and downregulated genes exhibited considerable overlap, with the downregulated genes showing more overlap (Figure 3C). These results suggest that, despite being generally known as a transcriptional repressor, HDAC3 has both positive and negative effects on gene expression in testes at the meiotic and postmeiotic stages.
Figure 3.
HDAC3 regulates transcriptomic programming during the meiotic-to-postmeiotic transition in the male germline. (A) Heatmap of RNA-seq data showing differentially expressed genes (DEGs) in PS (left) and RS (right) from wild-type and Stra8-cre/Hdac3fl/– testes. PS and RS were isolated from adult Hdac3fl/+ (n = 6) and Stra8-cre/Hdac3fl/– (n = 10) for each set. Experiments were performed in biological triplicates. (B) The number of DEGs (P < 0.01, fold change > 1.5). (C) Venn diagram showing the overlap among the upregulated transcripts (up) and down-regulated transcripts (bottom) in Stra8-cre/Hdac3fl/– cells compared to WT at both PS and RS stages. (D, E) The top 10 most enriched GO biological processes based on their P-values for upregulated genes and downregulated genes in Stra8-cre/Hdac3fl/– PS were shown. (F) Heat map depicting upregulated genes and downregulated genes identified in Gene Ontology analysis. (G, I) Upregulated genes or downregulated genes in Stra8-cre/Hdac3fl/– PS were used to produce a heatmap depicting their expression profile in stage-specific spermatogenic cells. SG/Somatic: spermatogonial and somatic cells, LZ-SC: leptotene/zygotene spermatocytes. (H, J) Average expression levels of upregulated genes or downregulated genes in Stra8-cre/Hdac3fl/– PS across stage-specific spermatogenic cells.
The upregulated genes in Stra8-cre/Hdac3fl/– showed a strong enrichment for early male development, including reproductive system development, male gonad development, and regulation of meiosis (Figure 3D). These include genes involved in meiotic initiation, meiotic recombination, and spermatogonial self-renewal, such as Rad51, H2afx, Dmc1, Stra8, Msh2 and Lin28b (Figure 3F). The upregulation of these target genes was further validated by RT-quantitative PCR (RT-qPCR) in Stra8-cre/Hdac3fl/– PS and RS (Supplementary Figure S4C). The expression of these genes normally declines as meiocytes progress from leptonema/zygonema to pachytene spermatocytes or spermatids (2). The downregulated genes in Stra8-cre/Hdac3fl/– PS or RS were enriched in spermatid differentiation, including flagellated sperm motility and spermatid development (Figure 3E). Many genes are essential for haploid development and acrosomal formation, including Mtdh1, Nkapl, Sox30, Arih1, Tnp2, Prm1 and Prm2 (Figure 3F). The downregulation of these genes was further confirmed by RT-qPCR (Supplementary Figure S4D).
Genes upregulated in Stra8-cre/Hdac3fl/– PS and RS were highly enriched with spermatocytes- or spermatogonial cell-specific genes, when compared to the previously reported transcriptomes of isolated stage-specific germ cells, such as spermatogonial/somatic cells, leptotene/zygotene spermatocytes, PS and RS (2) (Figure 3G). These genes were normally suppressed in wild-type mice after the cells enter the PS or RS stage (Figure 3H). For example, Rad51, H2afx, Dmc1, Stra8, Msh2 and Lin28b were downregulated during meiotic progression from leptonema/zygonema to pachynema and spermatids. By contrast, the majority of downregulated genes in Stra8-cre/Hdac3fl/– PS and RS were preferentially expressed at the postmeiotic spermatid stage (Figure 3I and J). Thus, HDAC3 establishes the male germline gene transcriptional program at meiosis-to-postmeiosis transition by suppressing early meiotic genes and activating haploid spermatid genes.
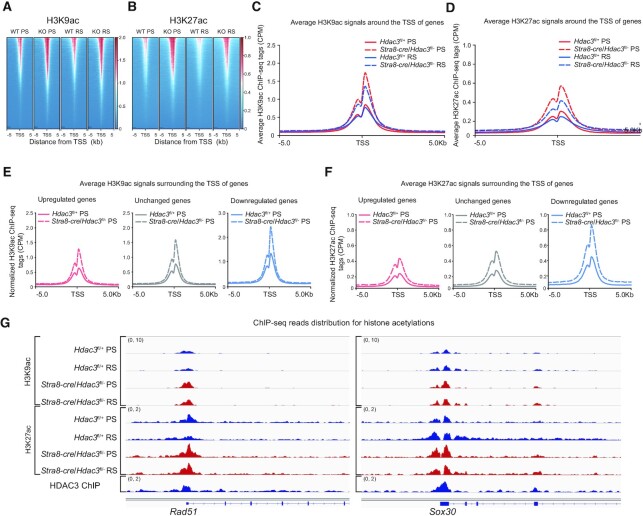
To elucidate the molecular basis through which HDAC3 suppresses the meiotic transcriptional program and drives the expression of haploid genes, we next assessed the effect of HDAC3 depletion on acetylation dynamics during germ cell development. Ablation of HDAC3 led to increased recruitment of H3K9ac at many genes near their promoter regions in PS and RS (Figure 4A and C). This enhanced recruitment of H3K9ac was also observed at HDAC3 binding sites near promoter regions (Supplementary Figure S5A). Similarly, genome-wide H3K27ac levels were high in HDAC3-depleted PS and RS (Figure 4B and D and Supplementary Figure S5B). Surprisingly, the increases of H3K9ac occupancy at HDAC3 binding sites were also pronounced near genes that were downregulated or unchanged upon HDAC3 depletion in both PS (Figure 4E) and RS (Supplementary Figure S5C). ChIP-seq analysis further confirmed that loss of HDAC3 dramatically enhanced the H3K27ac level at many HDAC3 binding sites even at genes that were repressed or unchanged upon HDAC3 depletion (Figure 4F and Supplementary Figure S5D). Consistent with the above nonbiased acetylation profiles, the H3K9ac and H3K27ac levels in HDAC3-depleted PS was increased substantially compared to WT cells not only at upregulated genes in KO versus WT (Rad51, H2afx, Sycp1, Dmc1 and Sycp3) (Figure 4G and Supplementary Figure S5E), but also at many previously determined repression genes (Tnp2, Prm3, Oaz1, Nkapl and Sox30) (Figure 4G and Supplementary Figure S5E). The fact that HDAC3 depletion in testis suppressed the expression of postmeiotic spermatid genes despite histone hyperacetylation at these sites suggests that histone acetylation does not necessarily cause gene activation and that HDAC3 might mediate transcriptional regulation beyond its deacetylase activity.
Figure 4.
Histone acetylation and gene expression uncoupled at PS and RS stages. (A) Heat map of H3K9ac binding signal in wild type and Stra8-cre/Hdac3fl/- from –5 to +5 kb surrounding the TSS of genes at the pachytene spermatocyte and round spermatid stage. ChIP-seq with antibody against H3K9ac was performed in pachytene spermatocytes (PS) and round spermatids (RS) isolated from adult wild type and Stra8-cre/Hdac3fl/–. (B) Heat map of H3K27ac binding signal in wild type and Stra8-cre/Hdac3fl/- from –5 to +5 kb surrounding the TSS of genes. ChIP-seq with antibody against H3K27ac was performed in pachytene spermatocytes (PS) and round spermatids (RS) isolated from adult wild type and Stra8-cre/Hdac3fl/–. (C, D) Average H3K9ac (C) or H3K27ac (D) ChIP signals of Stra8-cre/Hdac3fl/– PS and RS versus wild-type PS and RS within –5 kb/+5 kb of TSS. CPM, counts per million. (E, F) Average H3K9ac or H3K27ac profiles at upregulated (red), unchanged (gray) and downregulated (blue) genes in Stra8-cre/Hdac3fl/– versus their wild-types at PS stage. CPM, counts per million. (G) Genome browser tracks depicting reads accumulation of H3K9ac and H3K27ac on representative meiotic gene Rad51 and postmeiotic haploid gene Sox30 in WT and Stra8-cre/Hdac3fl/– at PS and RS stages. Y axis scales in brackets represent reads per million.
Abolishing HDAC3 enzyme activity does not affect male fertility
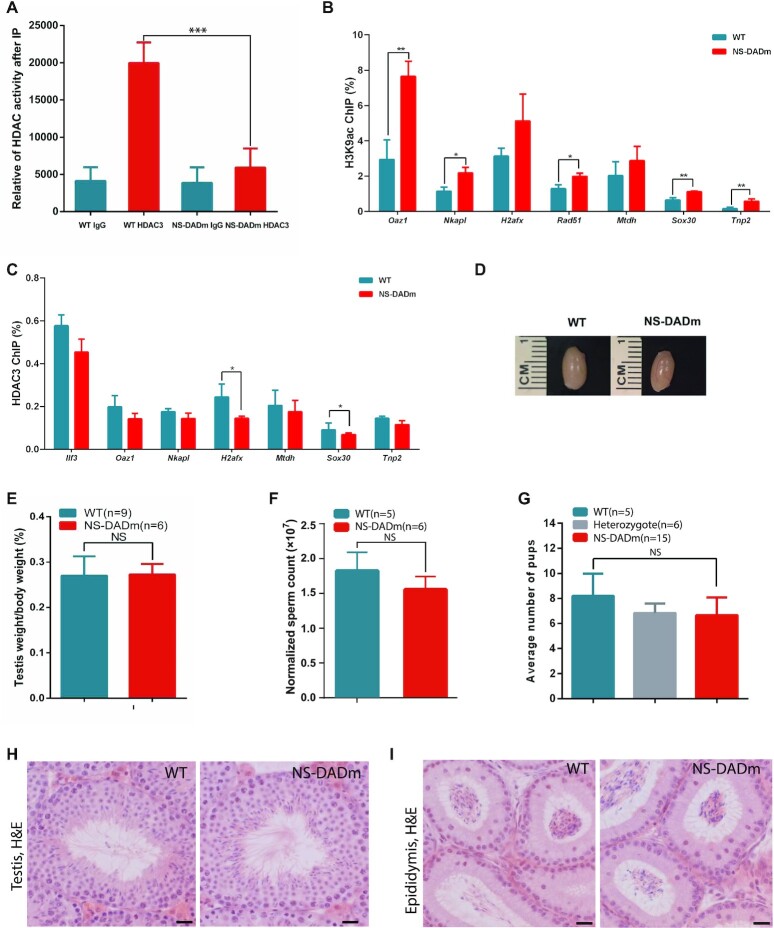
We next investigated the role of HDAC3 catalytic activity in mouse testis and determined whether the spermatogenic arrest at the meiotic exit in HDAC3 knockouts results from disruption of its catalytic activity. To address this possibility, we aim to abolish the enzyme activity of HDAC3 without affecting its protein level. Purified HDAC3 protein does not show detectable deacetylase enzyme activity. Instead, the enzyme activity of HDAC3 becomes detectable only after it forms a complex with the conserved DAD domain of NCOR and SMRT (21,55). Disruption of this interaction with missense whole-body knock-in mutations in the DAD of both NCOR and SMRT (NS-DADm) abolished the catalytic activity in HDAC3 in all the tissue or organs examined so far, including the liver, muscle, and brain (23,25,56). We immunoprecipitated HDAC3 protein from testes lysates extracted from wild-type and NS-DAD mutants and performed the HDAC assay. HDAC3 catalytic activity was readily detectable in WT testes but was undetectable in testes from NS-DADm mice (Figure 5A). We then measured histone acetylation levels at HDAC3-binding sites by performing H3K9ac ChIP-qPCR in NS-DADm and WT testes. Consistent with the undetectable deacetylase activity of HDAC3, the enrichment of H3K9ac at the previously-determined sites was significantly enhanced in NS-DADm testes compared to WT (Figure 5B). HDAC3 ChIP-qPCR analysis at these sites revealed that HDAC3 still bound to these target sites in NS-DADm testes (Figure 5C). Previous studies showed that a second DAD-independent domain of NCOR/SMRT also binds HDAC3, although it does not activate the HDAC3 enzyme activity (21,23,57,58). Our results suggest that the interaction with NCOR/SMRT through the second domain is largely sufficient for recruiting HDAC3 to the genome in mouse testes.
Figure 5.
HDAC3 regulates spermatogenesis through deacetylase-independent mechanisms. (A) Testes tissue lysates were immunoprecipitated with either HDAC3 or IgG antibodies, and subjected to HDAC assay. n = 3. (B) The mouse testes from wild type and NS-DADm at postnatal day 20 were subjected to ChIP with H3K9ac antibodies, followed by qPCR analysis at the indicated sites. n = 4 for each genotype. * P < 0.05, ** P < 0.01, Student's t test. (C) ChIP-qPCR analysis of HDAC3 recruitment at the previously-determined sites in wild type and NS-DADm mice at postnatal day 20. n = 4 for each genotype. * P < 0.05, Student's t test. (D) Testis size from 8-week-old wild-type (WT) and NS-DADm. (E) The ratio of testis weight to body weight in WT and NS-DADm. NS, not significant. Wild type, n = 9; NS-DADm, n = 6. (F) The sperm counts obtained from epididymides of 8-week-old wild-type and NS-DADm. Wild type, n = 5; NS-DADm, n = 6. (G) Fertility analysis for adult wild-type, heterozygote, and NS-DAD mutant males. n = 5 for WT; n = 6 for heterozygous; n = 15 for NS-DAD mutants. (H) Hematoxylin and eosin-stained testis sections from 8-week-old wild-type and NS-DADm showed a full spectrum of spermatogenic cells. Scale bar, 20 μm. (I) H&E staining of cauda epididymis from adult wild-type and NS-DADm. Scale bar, 20 μm.
In contrast to Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/- males, NS-DADm male mice were fertile. Both homozygous knock-in (NS-DADm) and heterozygous knock-in male mice were able to father litters and displayed normal mating behavior (Figure 5G). The average litter size from NS-DADm homozygous males was comparable to that of heterozygous mutants and wild-type males (Figure 5G). NS-DADm testes also displayed a similar size (Figure 5D) and weight proportional to their body weight (Figure 5E). The sperm count from mutant males was still abundant in the epididymal tubule (Figure 5F). In contrast to Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– males with spermatogenesis predominantly being arrested at late stages of meiosis or early round spermatids, histology of NS-DAD mutant testes appeared to be normal, with a full spectrum of spermatogenic cells at various developmental stages (Figure 5H) and the presence of abundant mature sperm in the epididymis (Figure 5I). These results demonstrate that the deacetylase enzymatic activity is largely not required for the in vivo function of HDAC3 in spermatogenesis.
HDAC3 associates with SOX30 and in testes co-localizes with SOX30 on genomic sites
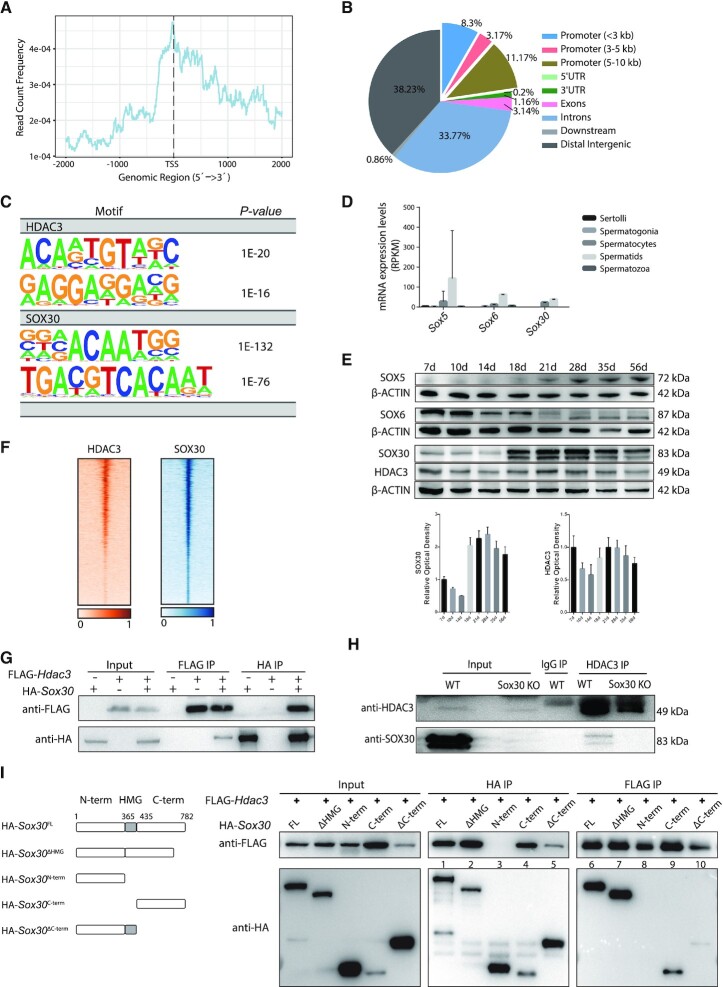
Since HDAC3 lacks an intrinsic DNA-binding ability, we seek to identify transcription factors that recruit HDAC3 to the genome in testes. Previous studies showed that HDAC3 forms a complex with NCOR/SMRT and is recruited to chromatin by tissue-specific transcription factors along with NCOR (59–61). We prepared biologically duplicate ChIP-seq libraries of HDAC3 from testes at P20 when HDAC3 is abundantly expressed and when the late-stage spermatocytes and early RS are highly represented in testes (62). We identified 3976 HDAC3 binding sites. Genomic annotation of ChIP reads revealed that 23% of HDAC3 sites resided in promoter regions of annotated genes within 10 kb of their TSS. The other HDAC3 sites were intergenic regions (38%), introns (34%) and exons (3%) (Figure 6A and B). De novo motif analysis identified the top enriched motif at HDAC3 sites contained a major consensus sequence ‘ACAAT’, which is the common binding motif for all Sox proteins (Figure 6C). Sox family transcriptional factors contain a conserved DNA-binding HMG box (63), and they are well-established regulators of cell fate decisions during differentiation initiation and development (64). There are 20 mammalian Sox proteins that belong to ten groups termed A to H. Among them, mRNA transcripts of Sox5, Sox6 and Sox30 increased in meiotic spermatocytes and expressed highly in RS (Figure 6D) (14). Western blot analysis further revealed that only SOX30 shared a similar expression pattern with HDAC3 during spermatogenesis (Figure 6E). The expression of both SOX30 and HDAC3 in testes increased at P18 when pachytene spermatocytes appear, reached the highest level at P21 when later stages of spermatocytes and early RS are enriched, and started to decline by P28 (Figure 6E). SOX30 is a testis-specific transcription regulator for activating postmeiotic haploid gene program starting at late stages of spermatocytes and through round spermatids development (17,65–67).
Figure 6.
HDAC3 and SOX30 co-localize on the genome in testes. (A) Normalized HDAC3 ChIP-seq reads on UCSC mm10 RefSeq gene bodies. (B) Annotations of HDAC3 ChIP-seq peaks in wild-type testes at P20. (C) Top enriched motifs in the binding sites for HDAC3 and SOX30 using Homer. Sequences within ± 200 bp from the centers of all the binding sites were used for de novo motif analysis. (D) mRNA transcripts levels of Sox members across stage-specific spermatogenic cells were assessed by RNA-seq (14). (E) Western blot analysis for the protein levels of SOX5, SOX6, SOX30, and HDAC3 during spermatogenesis. The image is a representation of two independent experiments with similar results. The corresponding optical density readings for biological duplicates are shown. (F) Heat map depicting HDAC3 and SOX30 co-localization at many binding sites. (G) FLAG-Hdac3 and/or HA-Sox30 expression constructs were transfected in HEK 293T cells and subjected to immunoprecipitation analysis. (H) Testes protein lysates were immunoprecipitated either with HDAC3 or normal IgG antibodies followed by immunoblot analysis. Protein lysates were prepared from wild type and Sox30 KO at postnatal day 20. (I) HA-tagged SOX30 mutants with deleted fragments were coexpressed with FLAG-HDAC3 in HEK 293T cells. Lysates were immunoprecipitated with anti-FLAG antibodies, and the immunoprecipitated protein complex was examined for HA (SOX30).
We postulate that SOX30 plays a major role in recruiting HDAC3 in the late meiotic stage. We performed ChIP-seq for SOX30 and HDAC3 in mouse testes at P20 when both HDAC3 and SOX30 expressed at their highest levels. HDAC3 and SOX30 binding strongly co-localized on a genome-wide scale (Figure 6F). As expected, the top enriched motifs discovered at SOX30 binding sites contained a consensus sequence ‘ACAAT’ (Figure 6C).
Immunoprecipitation assay in HEK293T cells co-transfected with FLAG-tagged HDAC3 and HA-tagged SOX30 showed the association between exogenous expressed HDAC3 and SOX30 (Figure 6G). Co-immunoprecipitation analysis also demonstrated that HDAC3 pulled down endogenous SOX30 in wild-type testes at P20 (Figure 6H). SOX30 was not observed in HDAC3 immunoprecipitates in Sox30 KO testes, and SOX30 protein was also absent in immunoprecipitates with the normal IgG antibody (Figure 6H). The interaction between HDAC3 and SOX30 was abolished using the lysis buffer containing SDS (data not shown), suggesting that the interaction is dynamic and of low affinity. We further mapped the sites of interaction between HDAC3 and SOX30 by a series of co-immunoprecipitations using tagged truncation constructs. SOX30 protein was truncated into three regions: SOX30 N terminus, the DNA-binding HMG domain, and SOX30 C terminus. The SOX30-HDAC3 interaction was disrupted upon the deletion of the SOX30 C terminus (Figure 6I). Conversely, the SOX30 C terminus alone was sufficient to bind HDAC3 (Figure 6I). These results suggest that the C terminus of SOX30 mainly mediated its association with HDAC3.
HDAC3 coordinates with SOX30 in transcriptional activation of postmeiotic genes during reprogramming
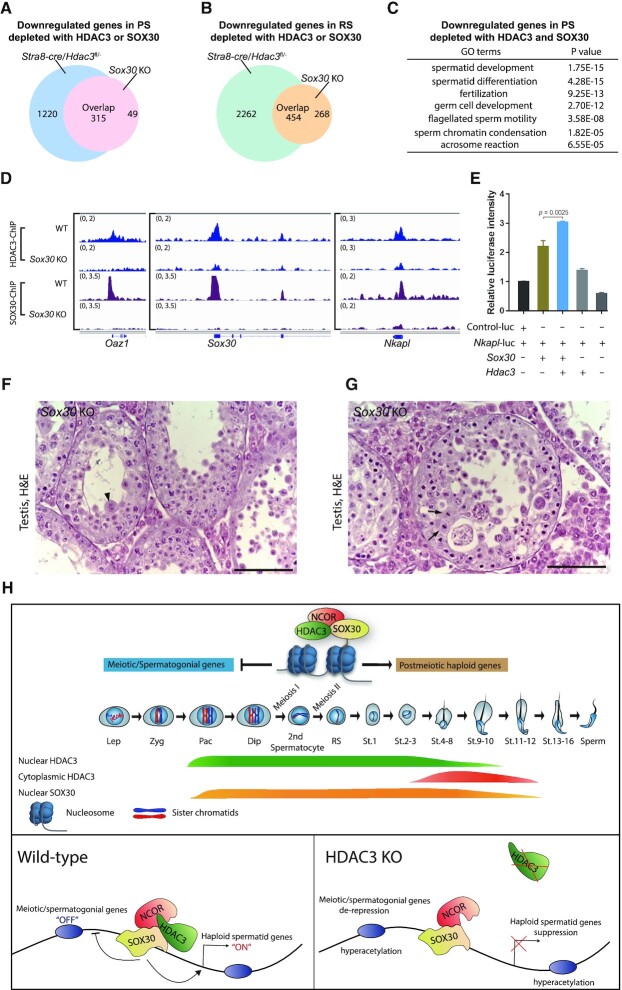
To investigate the co-regulation of gene expression by HDAC3 and SOX30, we analyzed RNA-seq results from Stra8-cre/Hdac3fl/– mice and Sox30 null mice in isolated pachytene spermatocytes and round spermatids. Transcriptome profiling revealed that among the 364 genes that were downregulated by SOX30 KO, 315 genes (86.5%) showed reduced expression with Stra8-cre/Hdac3fl/– in pachytene spermatocytes (Figure 7A). Transcriptomic changes for the downregulated genes by either SOX30 KO or Stra8-cre/Hdac3fl/– were also similar in round spermatids (Figure 7B). Moreover, genes downregulated in both SOX30 KO and Stra8-cre/Hdac3fl/– at the PS stage exhibited a considerable overlap (74.9%) with genes downregulated in both knockouts at the RS stage. These results suggest that HDAC3 serves as a co-regulator for SOX30-mediated transcriptional activation.
Figure 7.
HDAC3 functions as a SOX30 coactivator in testes. (A) 315 genes out of 364 downregulated genes upon Sox30 KO were also downregulated in Stra8-cre/Hdac3fl/– at the pachytene spermatocyte stage. (B) Among 722 downregulated genes in Sox30 KO at RS stage, 454 genes were also downregulated in Stra8-cre/Hdac3fl/–. (C) Gene ontology analysis of the co-target genes downregulated in both Sox30 KO and Stra8-cre/Hdac3fl/- at PS stage. (D) Genome browser tracks showing HDAC3 ChIP-seq and SOX30 ChIP-seq in wild-type and Sox30 KO testes at P20-P21. (E) Luciferase reporter assay showing HDAC3 co-activates the SOX30 transcriptional activity on the Nkapl promoter. (F, G) H&E staining of testes from Sox30 KO mice at 8-week-old. Arrowheads indicate multinucleated cells, and arrows point to aberrant metaphase cells. (H) Summary model for the regulatory roles of SOX30/HDAC3 signaling during meiotic exit.
Analysis of genes downregulated in pachytene spermatocytes depletion with either HDAC3 or SOX30 revealed a strong enrichment for spermatid differentiation-associated events, including chromatin condensation, the formation of an acrosome, and assembly of the flagellum (Figure 7C). For example, the common target sites of both HDAC3 and SOX30 in P20 testes contained the promoter regions of genes involved in haploid round spermatid differentiation, such as Oaz1, Nkapl, and the transcription factor Sox30 itself (Figure 7D).
To evaluate whether SOX30 mediates HDAC3 recruitment to the genome, we performed ChIP-seq of HDAC3 in WT and Sox30 KO testes at P20 (Figure 7D). HDAC3 binding to these co-targets was significantly lower in testes from mice lacking Sox30 than from WT mice. The diminished enrichment of HDAC3 on common target genes, including Nkapl, Oaz1 and Sox30, was shown in Sox30 KO testes (Figure 7D). Further, HDAC3 substantially increased the transcriptional activity of SOX30 at the proximal promoters of Nkapl, which contained predicted binding sites of co-localized HDAC3 and SOX30 (Figure 7E).
To explore the functional interplay between HDAC3 and SOX30 during germ cell development, we compared the phenotype of the HDAC3 knockout and SOX30 null mice. Genetic ablation of Sox30 rendered spermatogenesis arrested at round spermatid stage at step 2–3 (Figure 7F), a phenotype similar to Vasa-cre, Neurog3-cre, and Stra8-cre/Hdac3fl/– males (Figure 2F, H and L). This is in line with recent findings (17,65–67). A closer examination of Sox30 KO tubules also reveals the presence of aberrant metaphase cells or secondary spermatocytes at stage XI-XII (Figure 7G), resembling the transition defects observed in testes from Vasa-cre, Neurog3-cre and Stra8-cre/Hdac3fl/– (Figure 2G, I and M). Collectively, these data suggest that HDAC3 coordinates with SOX30 for driving haploid gene expression programs at late spermatocytes-to-postmeiotic transition.
DISCUSSION
Our study delineates an intricate regulatory network that governs the transcriptional transition from meiosis into postmeiotic haploid development in the male germline. During meiosis initiation, RA signals induce the transcriptional activation of many meiotic genes at the early spermatocyte stage, which prepares the formation of the synaptonemal complex and the subsequent chromosomal events. This wave of gene expression is normally repressed at late meiosis and postmeiotic haploid differentiation, concomitant with activation of the haploid spermatid transcriptomic program. The absence of HDAC3 results in de-repression of these meiotic spermatocyte genes and downregulation of postmeiotic spermatid genes, suggesting the failure to properly exist meiosis and to enter postmeiotic spermatid stages. Histology studies confirmed that conditional HDAC3 knockouts using Vasa-cre, Neurog3-cre, or Stra8-cre exhibited spermatogenesis arrest at late stages of meiosis or early round spermatids. Thus, we demonstrated an essential role of HDAC3 in the transition from late stages of spermatocytes into haploid round spermatids (Figure 7H).
Our data suggest a functional interdependence of SOX30 and HDAC3 during meiotic exit in spermatogenesis. Both HDAC3 and SOX30 appear in the nuclei of middle pachytene spermatocytes, abundantly expressed from later stages of spermatocytes to early round spermatids at step 1 (Supplementary Figure S6). Genetic knockout of Sox30 leads to the downregulation of postmeiotic haploid genes and impaired meiotic exit, resembling the transcriptomic changes and the phenotype in testis-specific HDAC3 knockouts. Consistent with this, the SOX30 motif was the top hit in HDAC3 binding sites. SOX30 protein contains an HMG domain, one of the highly conserved nucleic acid binding domains with the ability to recognize and bind DNA (63). The binding of HDAC3 at targeted genomic loci was reduced in Sox30 null testes. These results suggest that SOX30 guides HDAC3 in targeting specific genomic loci to regulate gene expression through consensus sequences (Figure 7H). Of note, the binding of HDAC3 at some genomic loci was not completely abolished in the absence of SOX30, suggesting that other transcription factors may also be involved. Intriguingly, the expression patterns of HDAC3 and SOX30 start to differ after the timing of meiosis exit. HDAC3 protein shuttles to perinuclear foci-like structures when round spermatids advanced to spermiogenic steps 2–3, whereas the nuclear signal of SOX30 persisted in round spermatids afterward with its high level at spermiogenic step 8–9 (Supplementary Figure S6). Thus, this pattern of HDAC3 synchronizes its unique role in priming the transcription factor SOX30 to ensure transcriptional reprogramming during meiosis exit, but SOX30 protein also has an HDAC3-independent function in late stages of round spermatids during spermatogenesis.
The role of HDACs in mammalian spermatogenesis is not well understood in vivo, although SIRT1, SIRT6 and HDAC6 that functions during the late stages of spermiogenesis (41,42,44,68–70). A recent study reported that conditional knockout of the histone acetyltransferase GCN5 resulted in defects in nucleosome eviction in spermiogenesis (40). Conditional knockout of HDAC3 in testes elevated meiotic/spermatogonial genes and suppressed haploid genes at meiotic-to-postmeiotic transition stages. These data suggest that HDAC3 can be both a coactivator and corepressor. This is further supported by the evidence that HDAC3 was recruited to haploid genes and increased the transcriptional activity of SOX30 at targeted genes. Consistent with our findings, HDAC3 functions as a coactivator of estrogen-related receptor α (ERRα) to drive UCP1 and the thermogenic transcriptional program in brown adipose tissue (60). Such a non-canonical function of HDAC3 is found in the small intestine, where it coactivates ERRα to induce microbiota-dependent rhythmic transcription of the lipid transporter genes (71). A genome-wide mapping study in eukaryotic organisms revealed the recruitment of both HATs and HDACs on active and silent genes (72,73), suggesting the HAT/HDAC binding is transient and can be involved in both gene activation and silencing. Of course, it is also possible that the downregulation of gene expression in testis-specific HDAC3 KO is indirect and is mediated by upregulating a secondary suppressor.
All the conditional HDAC3 knockouts using Vasa-cre, Neurog3-cre and Stra8-cre exhibit defects in meiotic exit, with an arrest at the late stage of meiosis or early stage of round spermatids. However, HDAC3 catalytic mutants (NS-DADm) are fertile and do not recapitulate the phenotypes of testis-specific HDAC3 knockouts. The enzyme-independent function of HDAC3 in spermatogenesis is surprising because the histone acetylation profiles correlated with gene expression at meiotic and postmeiotic stages in the wild-type. However, it is consistent with several recent studies. Re-expression of deacetylase-dead HDAC3 mutants is sufficient to rescue the hepatosteatosis phenotype and repress lipogenic genes in HDAC3-depleted liver to a similar degree as wild-type HDAC3 (58), demonstrating that the enzyme-independent action of HDAC3 in hepatic lipid metabolism in vivo. Deletion of Rpd3, an HDAC3 ortholog in yeast, has minimal effects on the transcription of many genes despite an increase in their acetylation profiles at binding sites (74). HDAC3 was shown to be expressed highly in mouse granulosa cells (GCs) and reported as an essential component of the inhibitory mechanism in GCs to maintain oocyte meiosis arrest before the luteinizing hormone (LH) surge (75). With the LH induction, decreased HDAC3 in GCs releases its retraining effects on gene expression to facilitate oocyte maturation. Nevertheless, gene expression changes elicited by the HDAC3 inhibitor HDACi 4b does not resemble those caused by the genetic depletion of HDAC3 in GCs in this study. Our study demonstrated knockout of HDAC3 in the male germline increased the H3K9ac and H3K27ac levels at many genes whose expression is downregulated or unchanged in PS and RS upon HDAC3 depletion (Figure 4E and F). This hyperacetylation profile is similarly observed in NS-DADm, despite the fact that NS-DADm mice are fertile with a full spectrum of spermatogenic cells. Thus, these results demonstrate that histone hyperacetylation is insufficient to activate transcription, and suggest a deacetylase-independent role of HDAC3 in mouse testis. What mediates the deacetylase-independent role of HDAC3? It is possible that a large and dynamic protein-protein complex is assembled at the timing of meiotic exit. The functions of transcription factors, including SOX30, require HDAC3 as a cofactor to maintain complex integrity and regulate gene transcription. Collectively, these results suggest that histone acetylation may be a bystander outcome of the open chromatin that is associated with active transcription rather than the primary cause of the transcription activation. Nevertheless, the overall correlation of histone acetylation and gene expression suggests that histone acetylation can still serve as a marker for open chromatin or active enhancers. In summary, our results elucidate a deacetylase-independent role of HDAC3 in coordinating with SOX30 to achieve transcriptional reprogramming during the meiotic-to-postmeiotic transition.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mitchell A. Lazar at the University of Pennsylvania for providing Hdac3fl/fl mice.
Contributor Information
Huiqi Yin, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Zhenlong Kang, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Yingwen Zhang, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Yingyun Gong, Department of Medicine, Baylor College of Medicine, Houston, TX 77030, USA; Department of Endocrinology and Metabolism, The First Affiliated Hospital of Nanjing Medical University, Nanjing 210029, People's Republic of China.
Mengrou Liu, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Yanfeng Xue, Department of Medicine, Baylor College of Medicine, Houston, TX 77030, USA.
Wenxiu He, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Yanfeng Wang, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Shuya Zhang, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Qiushi Xu, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Kaiqiang Fu, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Bangjin Zheng, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Jie Xie, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Jinwen Zhang, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Yuanyuan Wang, Department of Neurobiology, School of Basic Medical Science, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Mingyan Lin, Department of Neurobiology, School of Basic Medical Science, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Yihan Zhang, CAS Key Laboratory of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences.
Hua Feng, CAS Key Laboratory of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences.
Changpeng Xin, Center for Reproductive Medicine, the Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Peoples' Republic of China.
Yichun Guan, Center for Reproductive Medicine, the Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Peoples' Republic of China.
Chaoyang Huang, Department of Cardiology, the First Affiliated Hospital, Zhejiang University School of Medicine, People's Republic of China.
Xuejiang Guo, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
P Jeremy Wang, Department of Biomedical Sciences, University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA 19104, USA.
Joseph A Baur, Institute for Diabetes, Obesity, and Metabolism and Department of Physiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Ke Zheng, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
Zheng Sun, Department of Medicine, Baylor College of Medicine, Houston, TX 77030, USA.
Lan Ye, State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing 211166, People's Republic of China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFC1003500, 2016YFA0500902]; National Natural Science Foundation of China [31771653, 31970791 to K.Z.]; National Natural Science Foundation of China [31871503, 32070843to L.Y.]; NIH/National Institute of General Medical Sciences [R35GM118052 to P.J.W]; NIH [ES027544, DK111436, HL153320, AG069966 to Z.S.]; John S. Dunn Foundation; Mrs. Clifford Elder White Graham Endowed Research Fund. Funding for open access charge: National Natural Science Foundation of China Grant [31871503, 31771653, 31970791].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wang P.J., McCarrey J.R., Yang F., Page D.C.. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001; 27:422–426. [DOI] [PubMed] [Google Scholar]
- 2. da Cruz I., Rodriguez-Casuriaga R., Santinaque F.F., Farias J., Curti G., Capoano C.A., Folle G.A., Benavente R., Sotelo-Silveira J.R., Geisinger A.. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics. 2016; 17:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davis M.P., Carrieri C., Saini H.K., van Dongen S., Leonardi T., Bussotti G., Monahan J.M., Auchynnikava T., Bitetti A., Rappsilber J.et al.. Transposon-driven transcription is a conserved feature of vertebrate spermatogenesis and transcript evolution. EMBO Rep. 2017; 18:1231–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M.J., Rossant J.et al.. Retinoid signaling determines germ cell fate in mice. Science. 2006; 312:596–600. [DOI] [PubMed] [Google Scholar]
- 5. Koubova J., Menke D.B., Zhou Q., Capel B., Griswold M.D., Page D.C.. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. PNAS. 2006; 103:2474–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson E.L., Baltus A.E., Roepers-Gajadien H.L., Hassold T.J., de Rooij D.G., van Pelt A.M., Page D.C.. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. PNAS. 2008; 105:14976–14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishiguro K.I., Matsuura K., Tani N., Takeda N., Usuki S., Yamane M., Sugimoto M., Fujimura S., Hosokawa M., Chuma S.et al.. MEIOSIN Directs the Switch from Mitosis to Meiosis in Mammalian Germ Cells. Dev. Cell. 2020; 52:429–445. [DOI] [PubMed] [Google Scholar]
- 8. Matson C.K., Murphy M.W., Griswold M.D., Yoshida S., Bardwell V.J., Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev. Cell. 2010; 19:612–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murphy M.W., Sarver A.L., Rice D., Hatzi K., Ye K., Melnick A., Heckert L.L., Zarkower D., Bardwell V.J.. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. PNAS. 2010; 107:13360–13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maezawa S., Sakashita A., Yukawa M., Chen X., Takahashi K., Alavattam K.G., Nakata I., Weirauch M.T., Barski A., Namekawa S.H.. Super-enhancer switching drives a burst in gene expression at the mitosis-to-meiosis transition. Nat. Struct. Mol. Biol. 2020; 27:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maezawa S., Yukawa M., Alavattam K.G., Barski A., Namekawa S.H.. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2018; 46:593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammoud S.S., Low D.H., Yi C., Carrell D.T., Guccione E., Cairns B.R.. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014; 15:239–253. [DOI] [PubMed] [Google Scholar]
- 13. Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002; 296:2176–2178. [DOI] [PubMed] [Google Scholar]
- 14. Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., Gu H., Barthes P., Kokkinaki M., Nef S., Gnirke A.et al.. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013; 3:2179–2190. [DOI] [PubMed] [Google Scholar]
- 15. Gan H., Wen L., Liao S., Lin X., Ma T., Liu J., Song C.X., Wang M., He C., Han C.et al.. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 2013; 4:1995. [DOI] [PubMed] [Google Scholar]
- 16. Pattabiraman S., Baumann C., Guisado D., Eppig J.J., Schimenti J.C., De La Fuente R.. Mouse BRWD1 is critical for spermatid postmeiotic transcription and female meiotic chromosome stability. J. Cell Biol. 2015; 208:53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Y., Zheng Y., Gao Y., Lin Z., Yang S., Wang T., Wang Q., Xie N., Hua R., Liu M.et al.. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018; 28:879–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strahl B.D., Allis C.D.. The language of covalent histone modifications. Nature. 2000; 403:41–45. [DOI] [PubMed] [Google Scholar]
- 19. Goodson M., Jonas B.A., Privalsky M.A.. Corepressors: custom tailoring and alterations while you wait. Nuclear Receptor Signal. 2005; 3:e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perissi V., Jepsen K., Glass C.K., Rosenfeld M.G.. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 2010; 11:109–123. [DOI] [PubMed] [Google Scholar]
- 21. Guenther M.G., Barak O., Lazar M.A.. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 2001; 21:6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watson P.J., Fairall L., Santos G.M., Schwabe J.W.. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012; 481:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. You S.H., Lim H.W., Sun Z., Broache M., Won K.J., Lazar M.A.. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 2013; 20:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang L., He X., Liu L., Jiang M., Zhao C., Wang H., He D., Zheng T., Zhou X., Hassan A.et al.. Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev. Cell. 2016; 36:316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song S., Wen Y., Tong H., Loro E., Gong Y., Liu J., Hong S., Li L., Khurana T.S., Chu M.et al.. The HDAC3 enzymatic activity regulates skeletal muscle fuel metabolism. J. Mol. Cell Biol. 2019; 11:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koerner M.V., FitzPatrick L., Selfridge J., Guy J., De Sousa D., Tillotson R., Kerr A., Sun Z., Lazar M.A., Lyst M.J.et al.. Toxicity of overexpressed MeCP2 is independent of HDAC3 activity. Genes Dev. 2018; 32:1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mullican S.E., Gaddis C.A., Alenghat T., Nair M.G., Giacomin P.R., Everett L.J., Feng D., Steger D.J., Schug J., Artis D.et al.. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011; 25:2480–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alenghat T., Meyers K., Mullican S.E., Leitner K., Adeniji-Adele A., Avila J., Bucan M., Ahima R.S., Kaestner K.H., Lazar M.A.. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008; 456:997–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng K., Xiol J., Reuter M., Eckardt S., Leu N.A., McLaughlin K.J., Stark A., Sachidanandam R., Pillai R.S., Wang P.J.. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl. Acad. Sci. U. S. A. 2010; 107:11841–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kolas N.K., Marcon E., Crackower M.A., Hoog C., Penninger J.M., Spyropoulos B., Moens P.B.. Mutant meiotic chromosome core components in mice can cause apparent sexual dimorphic endpoints at prophase or X-Y defective male-specific sterility. Chromosoma. 2005; 114:92–102. [DOI] [PubMed] [Google Scholar]
- 31. Bellve A.R. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993; 225:84–113. [DOI] [PubMed] [Google Scholar]
- 32. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang W., Tan H., Sun M., Han Y., Chen W., Qiu S., Zheng K., Wei G., Ni T.. Independent component analysis based gene co-expression network inference (ICAnet) to decipher functional modules for better single-cell clustering and batch integration. Nucleic Acids Res. 2021; gkab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y., Wang H., Zhang Y., Du Z., Si W., Fan S., Qin D., Wang M., Duan Y., Li L.et al.. Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol. Cell. 2019; 73:547–561. [DOI] [PubMed] [Google Scholar]
- 35. Gaucher J., Boussouar F., Montellier E., Curtet S., Buchou T., Bertrand S., Hery P., Jounier S., Depaux A., Vitte A.L.et al.. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012; 31:3809–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaucher J., Reynoird N., Montellier E., Boussouar F., Rousseaux S., Khochbin S.. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 2010; 277:599–604. [DOI] [PubMed] [Google Scholar]
- 37. Boussouar F., Goudarzi A., Buchou T., Shiota H., Barral S., Debernardi A., Guardiola P., Brindle P., Martinez G., Arnoult C.et al.. A specific CBP/p300-dependent gene expression programme drives the metabolic remodelling in late stages of spermatogenesis. Andrology. 2014; 2:351–359. [DOI] [PubMed] [Google Scholar]
- 38. Shiota H., Barral S., Buchou T., Tan M., Coute Y., Charbonnier G., Reynoird N., Boussouar F., Gerard M., Zhu M.et al.. Nut directs p300-dependent, genome-wide H4 hyperacetylation in male germ cells. Cell Rep. 2018; 24:3477–3487. [DOI] [PubMed] [Google Scholar]
- 39. Qian M.X., Pang Y., Liu C.H., Haratake K., Du B.Y., Ji D.Y., Wang G.F., Zhu Q.Q., Song W., Yu Y.et al.. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013; 153:1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luense L.J., Donahue G., Lin-Shiao E., Rangel R., Weller A.H., Bartolomei M.S., Berger S.L.. Gcn5-mediated histone acetylation governs nucleosome dynamics in spermiogenesis. Dev. Cell. 2019; 51:745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu C., Song Z., Wang L., Yu H., Liu W., Shang Y., Xu Z., Zhao H., Gao F., Wen J.et al.. Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development. 2017; 144:441–451. [DOI] [PubMed] [Google Scholar]
- 42. Coussens M., Maresh J.G., Yanagimachi R., Maeda G., Allsopp R.. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One. 2008; 3:e1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McBurney M.W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J.R., Lansdorp P.M., Lemieux M.. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 2003; 23:38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bell E.L., Nagamori I., Williams E.O., Del Rosario A.M., Bryson B.D., Watson N., White F.M., Sassone-Corsi P., Guarente L. SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development. 2014; 141:3495–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tatone C., Di Emidio G., Barbonetti A., Carta G., Luciano A.M., Falone S., Amicarelli F.. Sirtuins in gamete biology and reproductive physiology: emerging roles and therapeutic potential in female and male infertility. Hum. Reprod. Update. 2018; 24:267–289. [DOI] [PubMed] [Google Scholar]
- 46. Tang X., Chen X.F., Wang N.Y., Wang X.M., Liang S.T., Zheng W., Lu Y.B., Zhao X., Hao D.L., Zhang Z.Q.et al.. SIRT2 acts as a cardioprotective deacetylase in pathological cardiac hypertrophy. Circulation. 2017; 136:2051–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gallardo T., Shirley L., John G.B., Castrillon D.H.. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007; 45:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Korhonen H.M., Meikar O., Yadav R.P., Papaioannou M.D., Romero Y., Da Ros M., Herrera P.L., Toppari J., Nef S., Kotaja N.. Dicer is required for haploid male germ cell differentiation in mice. PLoS One. 2011; 6:e24821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng K., Wang P.J.. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLos Genet. 2012; 8:e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sadate-Ngatchou P.I., Payne C.J., Dearth A.T., Braun R.E.. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008; 46:738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schalk J.A., Dietrich A.J., Vink A.C., Offenberg H.H., van Aalderen M., Heyting C.. Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma. 1998; 107:540–548. [DOI] [PubMed] [Google Scholar]
- 52. de Vries F.A., de Boer E., van den Bosch M., Baarends W.M., Ooms M., Yuan L., Liu J.G., van Zeeland A.A., Heyting C., Pastink A.. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005; 19:1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yuan L., Liu J.G., Zhao J., Brundell E., Daneholt B., Hoog C.. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000; 5:73–83. [DOI] [PubMed] [Google Scholar]
- 54. Yang F., De La Fuente R., Leu N.A., Baumann C., McLaughlin K.J., Wang P.J.. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 2006; 173:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ishizuka T., Lazar M.A.. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol. Endocrinol. 2005; 19:1443–1451. [DOI] [PubMed] [Google Scholar]
- 56. Zhou W., He Y., Rehman A.U., Kong Y., Hong S., Ding G., Yalamanchili H.K., Wan Y.W., Paul B., Wang C.et al.. Loss of function of NCOR1 and NCOR2 impairs memory through a novel GABAergic hypothalamus-CA3 projection. Nat. Neurosci. 2019; 22:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Privalsky M.L. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu. Rev. Physiol. 2004; 66:315–360. [DOI] [PubMed] [Google Scholar]
- 58. Sun Z., Feng D., Fang B., Mullican S.E., You S.H., Lim H.W., Everett L.J., Nabel C.S., Li Y., Selvakumaran V.et al.. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell. 2013; 52:769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sun Z., Miller R.A., Patel R.T., Chen J., Dhir R., Wang H., Zhang D., Graham M.J., Unterman T.G., Shulman G.I.et al.. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 2012; 18:934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Emmett M.J., Lim H.W., Jager J., Richter H.J., Adlanmerini M., Peed L.C., Briggs E.R., Steger D.J., Ma T., Sims C.A.et al.. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017; 546:544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feng D., Liu T., Sun Z., Bugge A., Mullican S.E., Alenghat T., Liu X.S., Lazar M.A.. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011; 331:1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ernst C., Eling N., Martinez-Jimenez C.P., Marioni J.C., Odom D.T.. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 2019; 10:1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Osaki E., Nishina Y., Inazawa J., Copeland N.G., Gilbert D.J., Jenkins N.A., Ohsugi M., Tezuka T., Yoshida M., Semba K.. Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic Acids Res. 1999; 27:2503–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sarkar A., Hochedlinger K.. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013; 12:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang D., Xie D., Lin X., Ma L., Chen J., Wang Y., Duo S., Feng Y., Zheng C., Jiang B.et al.. The transcription factor SOX30 is a key regulator of mouse spermiogenesis. Development. 2018; 145:164723–164733. [DOI] [PubMed] [Google Scholar]
- 66. Feng C.A., Spiller C., Merriner D.J., O’Bryan M.K., Bowles J., Koopman P.. SOX30 is required for male fertility in mice. Sci. Rep. 2017; 7:17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bai S., Fu K., Yin H., Cui Y., Yue Q., Li W., Cheng L., Tan H., Liu X., Guo Y.et al.. Sox30 initiates transcription of haploid genes during late meiosis and spermiogenesis in mouse testes. Development. 2018; 145:164855–164869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wei H., Khawar M.B., Tang W., Wang L., Liu C., Jiang H., Li W.. Sirt6 is required for spermatogenesis in mice. Aging (Albany NY). 2020; 12:17099–17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y., Kwon S., Yamaguchi T., Cubizolles F., Rousseaux S., Kneissel M., Cao C., Li N., Cheng H.L., Chua K.et al.. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 2008; 28:1688–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Parab S., Shetty O., Gaonkar R., Balasinor N., Khole V., Parte P.. HDAC6 deacetylates alpha tubulin in sperm and modulates sperm motility in Holtzman rat. Cell Tissue Res. 2015; 359:665–678. [DOI] [PubMed] [Google Scholar]
- 71. Kuang Z., Wang Y., Li Y., Ye C., Ruhn K.A., Behrendt C.L., Olson E.N., Hooper L.V.. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. 2019; 365:1428–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K.. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009; 138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hughes M.E., DiTacchio L., Hayes K.R., Vollmers C., Pulivarthy S., Baggs J.E., Panda S., Hogenesch J.B.. Harmonics of circadian gene transcription in mammals. PLos Genet. 2009; 5:e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kurdistani S.K., Robyr D., Tavazoie S., Grunstein M.. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 2002; 31:248–254. [DOI] [PubMed] [Google Scholar]
- 75. Wang H., Cai H., Wang X., Zhang M., Liu B., Chen Z., Yang T., Fang J., Zhang Y., Liu W.et al.. HDAC3 maintains oocyte meiosis arrest by repressing amphiregulin expression before the LH surge. Nat. Commun. 2019; 10:5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.