Abstract
The bipolar mitotic spindle is a highly conserved structure among eukaryotes that mediates chromosome alignment and segregation. Spindle assembly and size control are facilitated by force-generating microtubule-dependent motor proteins known as kinesins. In animals, kinesin-12 cooperates with kinesin-5 to produce outward-directed forces necessary for spindle assembly. In plants, the relevant molecular mechanisms for spindle formation are poorly defined. While an Arabidopsis thaliana kinesin-5 ortholog has been identified, the kinesin-12 ortholog in plants remains elusive. In this study, we provide experimental evidence for the function of Arabidopsis KINESIN-12E in spindle assembly. In kinesin-12e mutants, a delay in spindle assembly is accompanied by the reduction of spindle size, demonstrating that KINESIN-12E contributes to mitotic spindle architecture. Kinesin-12E localization is mitosis-stage specific, beginning with its perinuclear accumulation during prophase. Upon nuclear envelope breakdown, KINESIN-12E decorates subpopulations of microtubules in the spindle and becomes progressively enriched in the spindle midzone. Furthermore, during cytokinesis, KINESIN-12E shares its localization at the phragmoplast midzone with several functionally diversified Arabidopsis KINESIN-12 members. Changes in the kinetochore and in prophase and metaphase spindle dynamics occur in the absence of KINESIN-12E, suggest it might play an evolutionarily conserved role during spindle formation similar to its spindle-localized animal kinesin-12 orthologs.
KINESIN-12E aids in the rapid alignment of chromosomes during cell division in Arabidopsis.
Introduction
Kinesins are molecular nanomotors that mediate microtubule-based cellular processes, such as the unidirectional transport of vesicles/organelles, microtubule organization, signal transduction, and cell division (Klinman and Holzbaur, 2018; Nebenführ and Dixit, 2018). One of the critical functions of kinesin is the assembly of the mitotic bipolar spindle. Spindle bipolarity is achieved by the orchestrated antiparallel arrangement of higher-ordered microtubule bundles at a central position (Zhang and Dawe, 2011), although different mechanisms evolved between animals and land plants. In the absence of canonical centrosomes, most land plants form acentrosomal bipolar spindles using well-conserved spindle assembly factors (Zhang and Dawe, 2011; Yamada and Goshima, 2017). During late prophase, before the disassembly of the preprophase band, microtubule reorganization and nucleation at the surface of the nuclear envelope initiate spindle assembly (Masoud et al., 2013). Dynamic microtubules that generate the spindle are progressively sorted, converging into two poles at an axis perpendicular to the preprophase band and encapsulating the nucleus (Ambrose and Cyr, 2008). After nuclear envelope breakdown in both plant and animal cells, numerous microtubule bundles emanate from the spindle poles and connect to kinetochores, forming the kinetochore–microtubule bundles (kinetochore-fibers; Smirnova and Bajer, 1998). During prometaphase and metaphase, the progressive shrinking and lateral expansion of kinetochore-fibers lead to chromosome alignment in the metaphase plate. Within the spindle midzone, interpolar and antiparallel microtubules interdigitate, with the majority of microtubule plus-ends facing, and minus-ends pointing away from the chromosomes (Euteneuer et al., 1982). The pole-ward movement of the chromosomes during anaphase is promoted by the lengthening of interpolar microtubules and the concomitant shrinking of opposing kinetochore-fibers from either their plus- or minus-ends, depending on the organism/cell type (Hyman and Karsenti, 1996; Zhang and Dawe, 2011; Masoud et al., 2013; Yamada and Goshima, 2017).
In animal cells, the most important kinesins that facilitate the bipolar spindle arrangement belong to the microtubule plus-end directed kinesin-5 and kinesin-12 classes (Ferenz et al., 2010; Hancock, 2014). Evidence suggests that kinesin-5 motors are functionally conserved in vascular plants. The loss of Arabidopsis thaliana KINESIN-RELATED PROTEIN125C (AtKRP125C), one of four kinesins-5 class motor proteins in Arabidopsis, severely compromised spindle organization, resulting in the generation of monopolar and aberrant spindles similar to those observed in animals in the absence of the animal kinesin-5 Eg5 (Bannigan et al., 2007). In cultured human cells, eg5 spindle defects were alleviated by overexpressing human HsKif15 (Tanenbaum et al., 2009; Vanneste et al., 2009). HsKif15 binds to kinetochore-fibers, likely via a nonmotor microtubule-binding site, and its knockout results in reduced spindle length, suggesting that the dimeric motor controls spindle size by sliding parallel microtubules (Sturgill and Ohi, 2013; Sturgill et al., 2014). Under certain conditions, HsKif15 forms tetramers in vitro and in vivo, switches tracks at microtubule intersections, and binds to antiparallel microtubules, potentially allowing for the reorganization of spindle microtubules (Drechsler et al. 2014; Drechsler and McAinsh, 2016; Mann et al., 2017).
In plants, although kinesin-12 orthologs in the model moss Physcomitrium patens reportedly localize to the spindle, the function of kinesin-12 activity in spindle assembly is unclear (Miki et al., 2014). Among the six kinesin-12 family members in A. thaliana, four have been functionally characterized. The redundantly acting PHRAGMOPLAST-ASSOCIATED KINESIN (PAKRP) 1/KINESIN-12A and PAKRP1-LIKE (PAKRP1L)/KINESIN-12B decorate the phragmoplast midzone, and their simultaneous loss abolishes the first postmeiotic cytokinesis in male gametophytes due to disorganized microtubules and the consequent failure to form the cell plate (Lee and Liu, 2000; Pan et al., 2004; Lee et al., 2007). The PHRAGMOPLAST ORIENTING KINESINS (POK)1/KINESIN-12C and POK2/KINESIN-12D pair also perform redundant functions in maintaining the division site throughout mitosis (Müller et al., 2006). In particular, they colocalize with the preprophase band during prophase and persist at the division site until the cessation of cytokinesis (Lipka et al., 2014; Herrmann et al., 2018; Müller and Livanos, 2019). In addition, POK2 emerges at the phragmoplast midzone in a motility-dependent, microtubule plus-end directed manner where it interacts with MICROTUBULE-ASSOCIATED PROTEIN 65 (MAP65) proteins and promotes phragmoplast expansion (Herrmann et al., 2018). Phylogenetic analysis has suggested these two kinesin-12 pairs belong to two distinct classes, but the roles of these family members in spindle assembly are unclear (Shen et al., 2012; Müller and Livanos, 2019).
Here, we report the functional characterization of KINESIN-12E, the closest relative of POK1/POK2 KINESIN-12 (Müller and Livanos, 2019). Although KINESIN-12E shares presence at the phragmoplast with KINESIN-12A/B and POK2 during cytokinesis, during mitosis, its localization pattern differs markedly from other plant kinesin-12 proteins, as it exhibits perinuclear accumulation during prophase and associates with microtubule subpopulations during prometaphase and specifically with the spindle midzone during metaphase. Consistent with this distribution pattern, kin12e mutants exhibit a delay in spindle organization and form shorter and less dynamic spindles than the wild type. We propose that KINESIN-12E contributes to spindle assembly, sharing functional similarities with the animal kinesin-12 orthologs.
Results
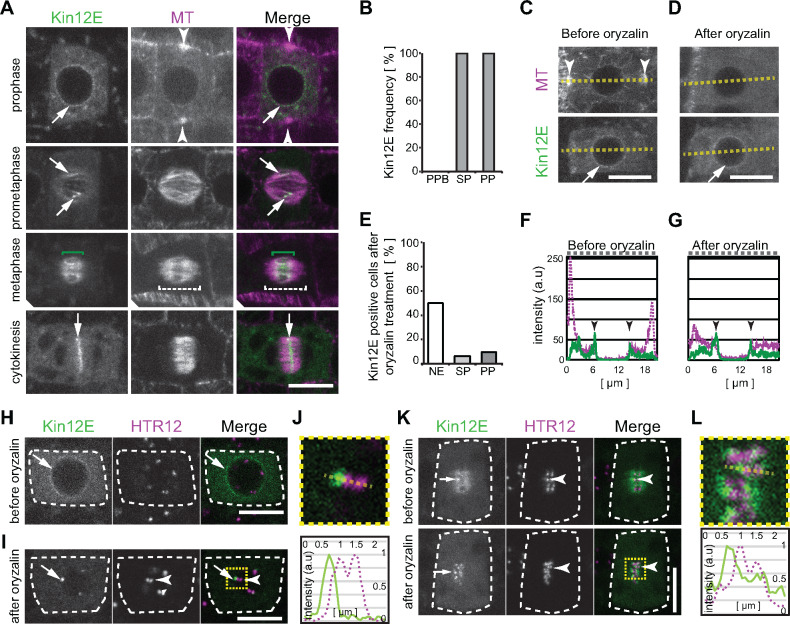
Gene expression analysis using native promoter-driven reporter assays revealed KINESIN-12E expression in meristematic tissues (Figure 1A, Supplemental Figure 1A–C). To investigate the spatio-temporal localization of Arabidopsis KINESIN-12E during different cell cycle stages, we analyzed root meristems of 4- to 6-day-old seedlings in several plant lines expressing fusions between green fluorescent protein (GFP) and genomic KINESIN-12E as well as the microtubule reporter, pUB10:RFP-MBD (Lipka et al., 2014). During prophase, KINESIN-12E accumulated in the cytosol and displayed a perinuclear distribution but did not colocalize with preprophase band microtubules (Figure 1A and B, Supplemental Figure 1D and G). Following nuclear envelope breakdown, KINESIN-12E decorated prophase spindle microtubules and was pronouncedly enriched in regions of microtubule overlap (Figure 1A, Supplemental Figure 1G, Supplemental Movie 1). During metaphase and anaphase, KINESIN-12E was restricted to the spindle midzone (Figure 1A and B, Supplemental Figure 1G, Supplemental Movies 1 and 2), roughly complementing the localization of Arabidopsis TPX2, a key component of spindle bipolarity (Supplemental Figure 1E and 1F; Petrovská et al., 2013). The confinement of KINESIN-12E to the spindle midzone suggests either the specific regulation of its microtubule affinity or reflects tip-tracking behavior and/or indicates the affinity for interaction partners, i.e., midzone-localized proteins. Alternatively, KINESIN-12E might preferentially bind to antiparallel microtubules, like MAP65-3 (Ho et al., 2011).
Figure 1.
GFP-KINESIN-12E shows cell cycle-specific subcellular localization and differential microtubule dependencies in root meristems. (A) Dividing plant cells coexpressing pkin12E:GFP-kinesin12E (Kin12E) and the microtubule (MT) reporter pUBN:RFP-MBD. Cell cycle stages are indicated. During prophase, GFP-Kin12E localizes to the nuclear envelope (arrow). The arrowheads mark the preprophase band. After nuclear envelope breakdown during prometaphase, GFP-Kin12E associates with microtubules of the central region of the forming spindle (arrows). During metaphase, GFP-Kin12E accumulates in the midzone (green brackets) of the spindle (dashed brackets). During cytokinesis, GFP-Kin12E localizes exclusively to the phragmoplast midzone (arrows). (B) Frequency of GFP-Kin12E colocalization with mitotic microtubule arrays. Preprophase band (PPB; 0%, n = 26 cells), spindle (SP; 100%, n = 48 cells), phragmoplast (PP; 100%, n = 34 cells). (C–G) GFP-Kin12E maintenance at the nuclear envelope is independent of microtubules. Prophase cell (C) before and (D) after treatment with 10 µM oryzalin for 5 min. Note that the preprophase band microtubules (arrowheads) depolymerized upon treatment, while GFP-Kin12E at the nuclear envelope persisted. Dashed line corresponds to plot profiles in (F and G). a.u., arbitrary units. E, Percentage of cells displaying GFP-Kin12E signal 10 min after treatment with 10 µM oryzalin (pooled data from two drug treatments). Nuclear envelope (NE, 50%, n = 20 cells), spindles (SP, 6%, n = 16 cells), phragmoplasts (PP, 10%, n = 21 cells). All cells displayed GFP-Kin12E signal before treatment. (F and G) Fluorescence intensity profile plot along the dashed line in (C) and (D), depicting RFP-MBD (dashed, magenta) and GFP-Kin12E (continuous, green) distribution before and after oryzalin. Note the persistence of GFP-Kin12E peaks (arrowheads) in (G). Drug treatment was performed twice. H–L, Kin12E accumulates close to kinetochores. Dividing plant cells coexpressing pkin12E:GFP-Kin12E and pUBN:RFP-HTR12 (HTR12). (H) Untreated prophase cell (dashed outline) displaying GFP-Kin12E at the nuclear envelope (arrow) and HTR12 puncta in the nucleus. (I) Cell in prometaphase after nuclear envelope breakdown in the presence of 10 µM oryzalin. GFP-Kin12E forms puncta (arrow) in close proximity to HRT12 ones (arrowhead). (J) The upper panel shows four-fold magnification of boxed area in (I), and the lower panel shows the respective intensity plot profile (arbitrary units, a.u.) of GFP-Kin12E (continuous line) and HTR12 (dashed line) along the dashed line in the panel above. Intensity values are plotted along the y-axis and distance is plotted along the x-axis. K, Metaphase cell before (upper panels) and 5 min after the application of 10 µM oryzalin (lower panels). GFP-Kin12E shows diffuse accumulation at the spindle and occasionally forms puncta (arrow) close to paired HTR12 puncta (arrowheads). Upon oryzalin treatment, GFP-Kin12E puncta become pronounced (treatment was performed twice). L, Upper panel shows four-fold magnification of boxed area in (K), and the lower panel shows the intensity plot profile along the dashed line depicted in the panel above. Continuous line indicates GFP-Kin12E intensity (arbitrary units, a.u.) and dashed line indicates HTR12 intensities. Intensity values are plotted along the y-axis and distance is plotted along x-axis. Maximum intensity z-projections of confocal images are shown in (A, C, D), and the scale bar indicates 10 µm. Single plane confocal images are shown in (H–L), and scale bar indicates 5 µm.
During cytokinesis, KINESIN-12E decorated the phragmoplast midzone akin to its paralog POK2 and the two other close relatives, Arabidopsis KINESIN-12A/B (Figure 1A and 2A, Supplemental Figure 1D, Supplemental Movies 1 and 2; Lee and Liu, 2000; Herrmann et al., 2018). The localization at the phragmoplast midzone could potentially reflect a general capacity of KINESIN-12 to bind selectively to antiparallel microtubules. However, exclusive binding to kinetochore-fibers akin to HsKif15 (Mann et al., 2017) was not observed for KINESIN-12E.
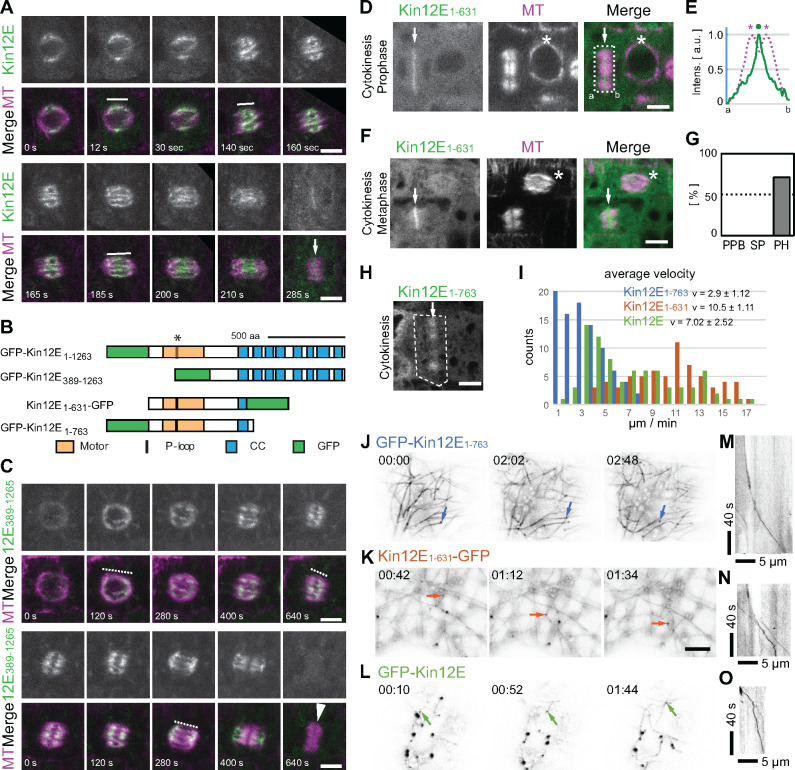
Figure 2.
Mitotic localization and microtubule association of KINESIN-12E and KINESIN-12E deletion mutants. (A and C) Time series of dividing cells coexpressing the microtubule reporter pUBN:RFP-MBD (MT) and either (A) pKin12E:GFP-kinesin-12E (Kin12E) or (C) pUBN:GFP-Kin12E389-1263 (Kin12E389-1263). (A) Note that Kin12E decorates the central region of the prophase spindle and remains at the spindle midzone during metaphase and anaphase (representatively indicated by white lines). During cytokinesis, Kin12E localizes to the phragmoplast midzone (white arrow). (B) Protein domain organization of full-length GFP-Kin12E (GFP-Kin12E1-1263), and of deletion mutants GFP-Kin12E389-1263, GFP-Kin12E1-763, and Kin12E1-631–GFP; coiled coil (CC), Motor domain (Motor), P-loop motif/ATP-binding site. The asterisk indicates the position of the T181N mutation within the P-loop motif. (C) Kin12E389-1263 is uniformly distributed (representatively indicated by dashed lines) along microtubules in all spindle stages but disappears after anaphase. The white arrowhead points to the phragmoplast. (D and F) Mitotic cells coexpressing p35S:Kin12E1-631–GFP (Kin12E1-631) and pUBN:RFP-MBD. (D) Cells during prophase (asterisk) and cytokinesis, displaying Kin12E1-631 localization at the phragmoplast midzone (arrow). (E) Normalized average intensity (arbitrary units, a.u.) plot profile of boxed area in (D). Dot marks Kin12E1-631 intensity peak (line), asterisks mark RFP-MBD (MT) intensity peaks (dashed line). Intensity peaks are clearly separated. (F) Cells showing Kin12E1-631 localization during cytokinesis (arrow) and prometaphase (asterisk). (G) Frequency (%) of Kin12E1-631 colocalization with preprophase bands (0%, n = 8 cells), spindles (0%, n = 12 cells) and phragmoplasts (70%, n = 27 cells). Note that Kin12E1-631 accumulates at the phragmoplast midzone during cytokinesis, but remains cytosolic in all other cell cycle stages. (H) Cell (dashed box) during cytokinesis showing Kin12E1-763 localization along phragmoplast microtubules (arrow). (I) Graph shows velocity frequency distribution and average velocity (v) of p35S:GFP-Kin12E (n = 4 seedlings, 16 cells, 66 counts, green), p35S:Kin12E1-631-GFP (n = 5 seedlings, 15 cells, 61 counts, orange), and p35S:GFP-Kin12E1-763 (n = 4 seedlings, 17 cells, 89 counts, blue). (J–L) Individual time frames (as indicated) from time-lapse movies showing (J) p35S:GFP-Kin12E1-763, (K) p35S:Kin12E1-631-GFP, and (L) p35S:GFP-Kin12E. Arrows indicate successive positions of individual motile, microtubule track-switching clusters. (M–O) Kymographs of clusters shown in (J–L). Letters in (D, E) indicate distance from a to b. Single plane images are shown in (A, C), and maximum intensity z-projections (z = three images) are shown in (D, F). Scale bars indicate 5 µm.
To determine whether the subcellular localization of KINESIN-12E is microtubule-dependent, we treated seedlings expressing GFP-KINESIN-12E with the microtubule polymerization inhibitor oryzalin and imaged root cells before and after treatment (Figure 1C–G). The treatment did not interfere with KINESIN-12E distribution at the perinuclear region during prophase (Figure 1C, D, F, and G). In approximately half of the treated cells (50%, n = 20 cells), GFP-KINESIN-12E remained at the nuclear periphery, pointing to an association with the nuclear envelope and not with perinuclear microtubules (Figure 1E). The other 50% of cells transitioned into prometaphase before the drug exerted its full effect. By contrast, GFP-KINESIN-12E located at the spindle midzone (6%, n = 16 cells) and phragmoplast (10%, n = 21 cells) was sensitive to the drug (Figure 1E). Notably, in untreated cells, we detected conspicuous puncta of high signal intensity reminiscent of kinetochores (Figure 1K), accompanying the otherwise filamentous and diffuse GFP-KINESIN-12E signal (Figure 1, Supplemental Movie 2).
To examine the possibility that KINESIN-12E associates with kinetochore-fibers, we used the kinetochore reporter CENTROMERIC HISTONE H3 (CENH3/HTR12; hereafter referred to as HTR12; Lermontova et al., 2011). During prophase, RFP-HTR12 dwelled in characteristic punctate structures inside the nucleus that aligned pairwise during metaphase (Figure 1H, K, and L). Intriguingly, when we applied oryzalin, in prophase cells undergoing nuclear envelope breakdown, KINESIN-12E initially remained closely associated with the HTR12-tagged kinetochores (44%, n = 59 kinetochores in 7 spindles in 7 seedlings) and in metaphase cells (91%, n = 78 kinetochores in 9 spindles in 8 seedlings; Figure 1I and J). These GFP-KINESIN-12E puncta vanished after prolonged treatment, but their disappearance was delayed relative to GFP-KINESIN-12E along spindle microtubules. This difference in sensitivity to oryzalin, together with the close association to kinetochores, indicates microtubule plus-end directed motility of KINESIN-12E.
Next, we assessed the contribution of motor- and nonmotor protein domains on the KINESIN-12E localization pattern in root meristems. We created constructs encoding the nonmotor domain mutant GFP-KINESIN-12E389-1263 and expressed these constructs, pUB10:GFP-kinesin-12E389-1263 (Figure 2C) and p35S:GFP-kinesin-12E389-1263 (Supplemental Figure 1H), in Arabidopsis seedlings. Similar to full-length, native promoter-driven GFP-KINESIN-12E (Figure 2A–B, Supplemental Movie 1), GFP-KINESIN-12E389-1263 localized to the perinuclear region just prior to nuclear envelope breakdown and then continuously decorated the spindle, except for the pole regions during prometaphase to anaphase (Figure 2B and C, Supplemental Figure 1H). After chromosome segregation, GFP-KINESIN-12E389-1263 translocated toward the poles and its signals became faint during phragmoplast formation (Figure 2C, Supplemental Figure 1H). Intriguingly, this nonmotor domain is associated with microtubules despite the lack of canonical microtubule-binding sites (Figure 2B). However, the C-terminal association with microtubules in Arabidopsis might be mediated by a specific interaction partner.
Furthermore, we expressed two motor-domain mutant constructs p35S:kinesin-12E1-631-GFP and p35S:GFP-kinesin-12E1-763, in Arabidopsis (Figure 2B, Supplemental Figure 1I and J). In stably transformed Arabidopsis seedlings, both KINESIN-12E1-631-GFP and GFP-KINESIN-12E1-763 appeared in the cytosol in all cells of the root meristem, likely due to their 35S promoter-driven ubiquitous expression (Figure 2D–H). Nevertheless, KINESIN-12E1-631-GFP associated predominantly with the phragmoplast midzone (Figure 2D–G), similar to the full-length protein localization during cytokinesis (Figure 1, Supplemental Figure 1). GFP-KINESIN-12E1-763 assumed a broader, more dynamic distribution at the phragmoplast (Figure 2H, Supplemental Movie 2).
To examine whether KINESIN-12E modulates microtubule behavior, we transiently expressed p35S:GFP-kinesin-12E, p35S:kinesin-12E1-631-GFP, and p35S:GFP-kinesin-12E1-763 in Nicotiana benthamiana seedlings and investigated their localization in cotyledon epidermal cells. The three fusion-proteins displayed similar dynamic clusters and more or less faint filaments resembling microtubules (Figure 2J–L, Supplemental Movie 3). When coexpressed with microtubule-reporter proteins (mCherry-TUA6, RFP-MBD), we regularly observed only the KINESIN-12E proteins along microtubules or the reporter, impeding investigation of changes in microtubule dynamicity by GFP-KINESIN-12E variants. On rare occasions, coexpression of the motor-domain mutants and microtubule reporters was detected, confirming the association of KINESIN-12E with microtubules, which in these cases were static and continuously labeled by the motors (Supplemental Figure 1I and J). Nevertheless, we successfully extracted velocities from time-lapse recordings of motile clusters of these single expressed GFP-KINESIN-12E variants in N. benthamiana seedlings using the Kymograph plugin in ImageJ (Figure 2I, M–O; Livanos et al., 2017). GFP-KINESIN-12E1-763 moved more slowly, with an average velocity of 2.9 ± 1.12 µm/min, compared to KINESIN-12E1-631-GFP (10.5 ± 1.11 µm/min) and full-length GFP-KINESIN-12E (7.02 ± 2.52 µm/min), which showed an intermediate velocity frequency distribution (Figure 2I). The average velocity of GFP-KINESIN-12E was comparable to that of HsKif15 (128.7 nm/s equals 7.7 µm/min, Mann et al., 2017) and faster than that of the POK2 motor (4.4 µm/min, Chugh et al., 2018). Notably, the velocities of the KINESIN-12E variants varied widely but did not correlate with cluster size (Supplemental Figure 1I–K, Supplemental Movie 3). Remarkably, for all GFP-KINESIN-12E variants, we observed clusters switching tracks and dragging along faintly labeled microtubules similar to the track switching HsKif15 (Figure 2J–L, Supplemental Movie 3; Mann et al., 2017).
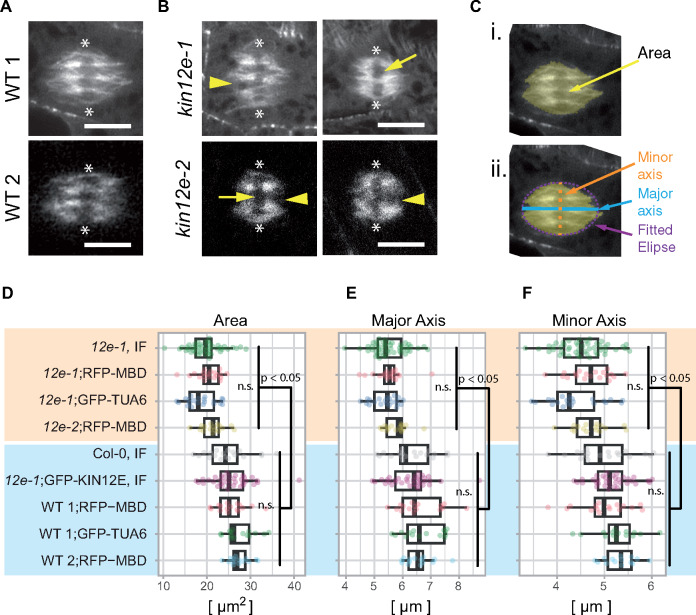
To examine the function of KINESIN-12E, we investigated two T-DNA knock-out mutant lines, kinesin-12e (kin12e)-1 and kin12e-2 (Supplemental Figure 2A and B). Seedlings and adult plant phenotypes were indistinguishable from the wild type (Supplemental Figure 2C and D), but we detected size differences in metaphase spindles (Figure 3, Supplemental Figure 3). We imaged kin12e-1 and kin12e-2 and wild-type root cells expressing either the microtubule reporter RFP-MBD or GFP-TUA6 (Figure 3A and B). To rule out any contribution of the microtubule reporters to the observed phenotypes, we also collected images from tubulin immunofluorescence detection experiments (Figure 3D, Supplemental Figure 3A and B). Using ImageJ, we segmented metaphase spindles in confocal images and measured the areas (Figure 3C (i.)). To approximate spindle shapes, an ellipse was fitted to the selected spindles, and the ellipse’s major and minor axis lengths were determined (Figure 3C (ii.)). Compared to wild type, the mutant kin12e microtubule spindles occupied smaller areas consistent with the reduced lengths of both the major and minor axes of the fitted ellipses (Figure 3D–F, Supplemental Table 1). Spindles of the kin12e mutants frequently showed large microtubule-free gaps in the equatorial plane (Figure 3B). Spindle bipolarity was not affected in the mutants, although on occasion, the spindle poles appeared more lacerated (Figure 3A and B, Supplemental Figure 3B). The expression of GFP-KINESIN-12E in kin12e-1 reduced its spindle defects, emphasizing the role played by KINESIN-12E in maintaining metaphase spindle architecture (Figure 3D–F).
Figure 3.
Loss of KINESIN-12E causes defects in metaphase spindle organization. A and B, Metaphase spindles of (A) wild type (WT) 1 (Col-0) and WT 2 (obtained from a population segregating kin12e-2) and (B) of mutants kin12e-1 and kin12e-2. Note gaps/lacerated regions at spindle poles (arrowheads) and large gaps (arrows) in the regions of metaphase plates/aligned chromosomes (axis between asterisks) indicative of fewer microtubule bundles. Scale bars indicate 5 µm. (C) The parameters quantified in (D–F) from segmented metaphase spindles (I). For segmentation, the last image before anaphase was selected (live cell imaging, single median plane) or maximum intensity z-projections (3–4 images, z = 1 µm for immunodetection) were used. The area was measured using segmentation of microtubule signal. An ellipse was fitted to the segmented spindle, and the major and minor axes (ii.) were measured in ImageJ. (D–F) Data summaries are shown as boxplots, with the box indicating the interquartile range (IQR), the whiskers showing the range of values that are within 1.5 × IQR, and the centerline indicating the median. The color-coded data are shown as jittered dots. Sample numbers are as follows (kin12e is abbreviated to 12e for simplicity): 12e-1 IF (n = 48 cells), 12e-1; RFP-MBD (n = 19 cells), 12e-1; GFP-TUA6 (n = 21 cells), 12e-2; RFP-MBD (n = 16 cells), Col-0 IF, n = 19 cells), 12e-1; GFP-KIN12E IF (n = 35 cells), WT 1; RFP-MBD (n = 16 cells), WT 1; GFP-TUA6 (n = 15 cells), and WT 2; RFP-MBD (n = 16 cells). IF indicates data acquired from immunofluorescence detection. A clear difference (P < 0.05, ANOVA post hoc, Bonferroni simultaneous pairwise comparison) is visible between mutants (highlighted in yellow) and wild type (highlighted in blue) for (D) area, (E) major axis, and (F) minor axis values (means ± SDs are given in Supplemental Table 1). Images in (A–C) were recorded with Airyscan detector with the exception of kin12e-2.
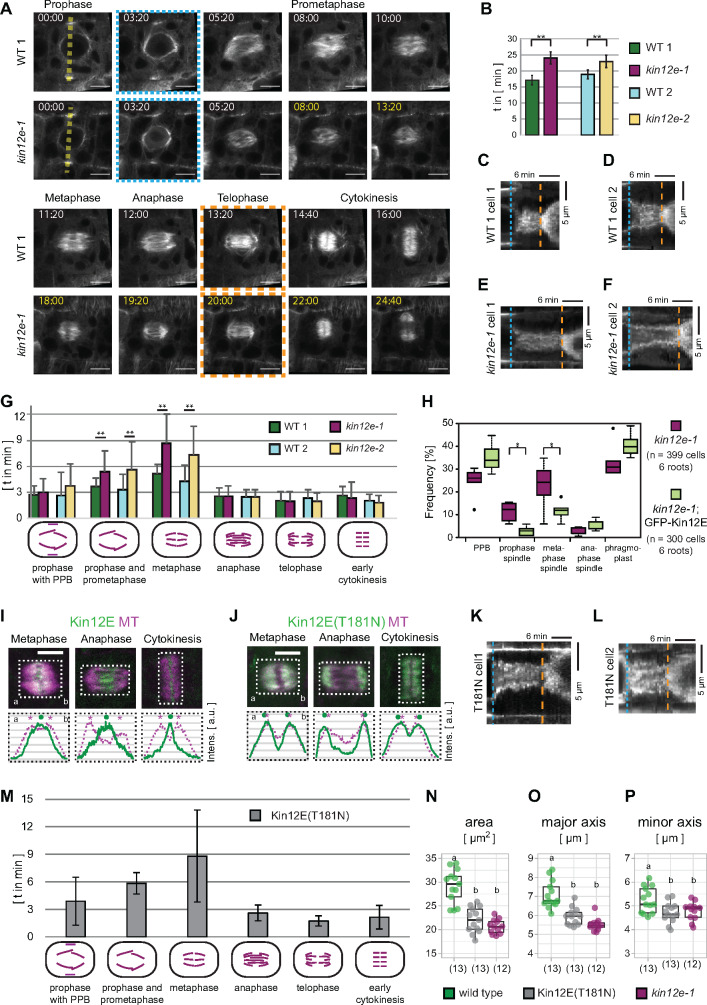
To determine the impact of KINESIN12-E loss during cell division, we monitored mitotic progression in wild type and mutant seedlings expressing microtubule reporters. The time-lapse recordings revealed an extension of mitosis in both mutant alleles compared to wild type (Figure 4A and B). In the kin12e mutant alleles, we detected (on average) a 6-min delay between nuclear envelope breakdown (blue dashed line in Figure 4A) and telophase (orange dashed line in Figure 4A and B). In particular, the duration of prometaphase and metaphase was prolonged in both kin12e alleles, while the progression from anaphase to early cytokinesis (anaphase/telophase spindle, disk phragmoplast) was unaffected compared to wild type (Figure 4G, Supplemental Figure 3C, Supplemental Table 2). Consistent with the delay in prometaphase and metaphase, the frequency of prophase and metaphase spindles increased in mutants compared to the complementation line (Figure 4H). Time-lapse imaging of mitotic cells also revealed that the sorting and bundling of microtubules during the assembly of the prophase and metaphase spindle were compromised, potentially causing the delayed mitotic progression in the mutant (Supplemental Movie 4).
Figure 4.
Loss of KINESIN-12E prolongs the transition from prophase to prometaphase and metaphase. (A) Comparison of mitotic duration in kinesin12e-1 (kin12e-1) and its segregating wild type (WT) 1 in representative time-lapse recordings. Plants expressed the microtubule reporter pUBN:RFP-MBD. Median plane images were recorded at 40 s time intervals. The time-lapse images were aligned (blue punctate box, 3:20 min) to show equivalent cell cycle stages. At 5:20 min, both genotypes displayed prophase spindles. However, prometaphase and metaphase lasted longer in the mutant (determined as time between blue and orange boxed stages). At 12:00 min, the WT 1 formed an anaphase spindle, while anaphase spindle emergence was delayed until 19:20 min in the kin12e-1 mutant. Progression through anaphase and subsequent stages were unaffected. A disk phragmoplast formed after 14:40 min and 22:00 min in WT 1 and the mutant, respectively, before phragmoplast expansion commenced (orange dashed box). (B) The duration of cell division from prophase until early cytokinesis was prolonged in kin12e mutant alleles (kin12e-1: 29.01 min ± 11.36 min, *P < 0.01, n = 16 cells; kin12e-2: 27.97 min ± 11.65 min, *P < 0.01, n = 17 cells) compared to their respective WT controls (WT 1: 23.90 min ± 8.49 min, n = 16 cells; WT 2: 22.13 min ± 8.90 min, n = 26 cells). Data were compiled from short time-lapse recordings, and only complete stages were scored. (C–F) Kymographs of RFP-MBD signal distribution. Blue punctate line indicates nuclear envelope breakdown (NEB), orange dashed line indicates the beginning of phragmoplast expansion. (C) Kymograph along yellow dashed line shown in (A) WT 1 prophase. (D) Additional kymograph, representative of WT 1, independent of (A). (E) Kymograph along yellow dashed line, shown in (A) kin12e-1 prophase. (F) Additional kymograph, representative of kin12e-1 and independent of (A). Note the differences in cell cycle progression, indicated by yellow time stamps in (A), and by the differences between NEB (blue punctate line) and phragmoplast expansion (orange dashed line) along the time axis in C–F. (G) Duration of cell cycle stages classified according to the respective mitotic microtubule arrays (shown in the x-axis in a graphical representation). Note the prolonged duration of prophase and metaphase in kin12e-1 and kin12e-2 compared to WT. Mean values and SDs are shown in Supplemental Table 2. (H) Boxplot shows data summary of meristematic root cells in different cell cycle stages in kin12e-1 mutants and kin12e-1 expressing pkin12E:GFP-Kinesin-12E (Kin12E). The boxes indicate the interquartile range (IQR), the whiskers show the range of values that are within 1.5 × IQR, and the center line indicates the median. Significant differences were determined by one-way ANOVA post hoc Tukey HSD. Note the increased frequencies of prophase spindles and metaphase spindles in kin12e-1 mutants. Data were collected from whole-mount seedling immunofluorescence detection of tubulin. Single plane images are shown in (A, C) and maximum intensity z-projections (z = three images) are shown in (D, E, I, J). Scale bars indicate 5 µm. I and J, Comparison of (I) Kin12E localization and motor-dead mutant (J) Kin12E(T181N), both expressed from the p35S promoter in different cell cycle stages as indicated. Lower panels depict normalized average intensity plot profiles of boxed areas in the upper panel images. Dots mark Kin12E1-631 intensity peaks (continuous line), asterisks mark RFP-MBD (MT) intensity peaks (dashed line). Compare the well-separated intensity peaks in (I) with the overlapping intensity peaks in (J). Letters a–b in (D, F, I, J) indicate reference points for the plot profiles. (K and L) Representative kymographs of mitotic cells expressing motor-dead p35S:GFP-Kin12E(T181N). M, Duration of individual mitotic stages in kin12e-1 root cells expressing motor-dead p35S:GFP-Kin12E(T181N; G). Note prolonged duration of prophase and metaphase in wild type (G). Mean values and SDs are shown in Supplemental Table 1. (N–P) Bar graphs showing mean (N) spindle area, (O) major, and (P) minor spindle axis of wild type compared to motor dead kinesin12E(T181N);kin12e-1 and to kin12e-1. Different lowercase letters indicate significant differences between groups (b is P ≤ 0.001 in (N) and (O), and b is P ≤ 0.5 in (P) by one-way ANOVA post hoc Tukey HSD). Mean values and SD are given in Supplemental Table 2. Time-lapse imaging at 40 s and 2 min time intervals from prophase to cytokinesis was performed on multiple days using different seedlings in (B, G, H, M).
To test whether the localization pattern of KINESIN-12E is motility-dependent, we generated an ATP hydrolysis-deficient mutant expressing p35S:GFP-kinesin-12E(T181N) (GFP-KINESIN-12E(T181N), Figure 4J–P). This motor-dead kinesin associated with all microtubule arrays in interphase and prophase cells. In cells undergoing metaphase and anaphase, the mutant decorated the entire spindles, as well as the phragmoplasts undergoing cytokinesis (Figure 4I and J, Supplemental Figure 1L and N, Supplemental Movie 5). To exclude the possibility that expression driven by the p35S promoter caused artificial spindle decoration with the motor-dead mutant, we analyzed plant lines expressing p35S:GFP-kinesin-12E for direct comparison. These lines showed localization patterns similar to pkinesin12E:GFP-kinesin-12E expressing lines (Supplemental Figure 1D–F, L, M, and O). Notably, expressing p35S:GFP-kinesin-12E(T181N) in the kin12e-1 mutant background neither complemented its delay in mitosis nor its spindle phenotype (Figure 4M–P). Collectively, these results reveal the role of KINESIN-12E in prophase and metaphase spindle assembly and consequently in the timely progression of mitosis.
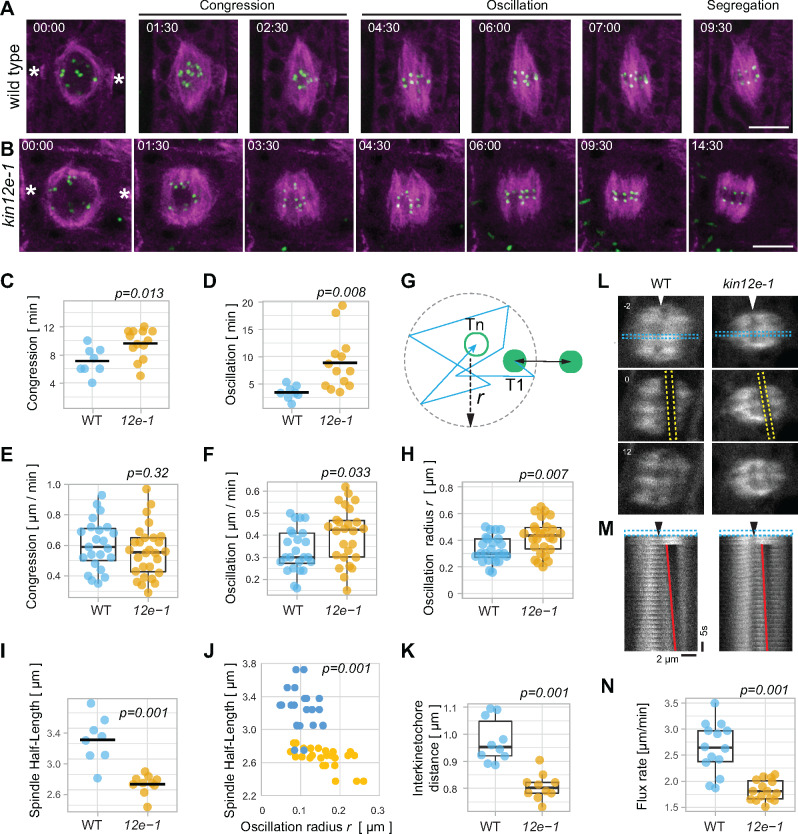
Since KINESIN-12E appears to stabilize microtubules during spindle assembly, we investigated whether its absence affected kinetochore-fiber dynamics by tracking RFP-HTR12-labeled kinetochores (Figure 5). Time-lapse imaging uncovered a delay in chromatid/kinetochore congression in kin12e, although the average velocities of kinetochore movements were comparable to those of the wild type (Figure 5A–C and E). However, during the oscillation of chromosomes in the equatorial plane in metaphase, kinetochore movements were somewhat accelerated in kin12e (Figure 5F).
Figure 5.
Spindle and kinetochore dynamics are altered in kinesin-12e (kin-12e) mutants. Individual frames of (A) wild type and (B) kin12-e taken from time-lapse series, depicting stages of chromatid congression and oscillation (visualized by the HTR12 kinetochore marker, green) during prometaphase and metaphase and segregation during anaphase. The microtubule marker TUA6 is shown in magenta. Scale bars indicate 5 µm. Note the delay in segregation of kinetochores of sister chromatids. The asterisks indicate the preprophase band. (C and D) (C) Congression and (D) oscillation of kinetochores are markedly prolonged in kin12e (congression 9.6 ± 2.1 min; oscillation 8.85 ± 5.06 min; n = 13 cells) compared with WT (congression 7.08 ± 1.94 min; oscillation 3.44 ± 1.25 min; n = 8 cells). Bars in the Jitter plots depict means. (E) Velocities of congression are similar in WT (0.6 ± 0.16 µm/min, n = 23 kinetochores from 8 cells in 8 plants) and kin-12e (0.56 ± 0.17 µm/min, n = 28 kinetochores from 10 cells in 10 plants). (F) Oscillation velocities in WT (0.33 ± 0.1 µm/min, n = 24 kinetochores from eight cells in eight plants) and kin-12e (0.4 ± 0.12 µm/min, n = 26 kinetochores from 10 cells in 10 plants) differ significantly. (G) Schematic representation of kinetochore oscillation radius definition and kinetochore distance. The track (zigzag arrow) of one kinetochore per pair (green disks) between time points T1 (green disc) and Tn (magenta circle) was fitted with a circle (dashed circle) and its radius r in ImageJ (relates to (H)). Double arrow depicts distance between paired kinetochores. (H) Jitter plots of oscillation radius r as defined in (G), relates to (J). (I) Jitter plots of spindle half-lengths (half-length of pole-to-pole axis) of spindles that were used to determine the oscillation radius r in (H) relates to (J). (J) Scatter plot showing the oscillation radius r of kinetochores (normalized by their spindle half-length) plotted against the spindle half-length (absolute values as in (I) of WT (dark blue dots, n = 24 kinetochores in eight spindles of eight plants) and kin-12e (bright yellow dots, n = 26 kinetochores from 10 spindles in 10 plants). Note the shift of kin-12e kinetochores toward increasing oscillation radius. (K) Box plot showing distances between kinetochore pairs were decreased during oscillation in kin-12e (0.8 ± 0.04 µm. Each data point is the average distance of n = 10 cells in 10 plants, from 170 individual distances measurements) compared to WT (0.98 ± 0.08 µm, each data point is the average distance of n = 10 cells in 10 plants, from 102 distances measurements). Raw distances are shown in Supplemental Figure 3D. Bar shows the median value. L, FRAP of GFP-TUA6 in metaphase spindles. Frames before FRAP (−2 s), immediately after FRAP (0 s) and after FRAP (12 s) are shown. Yellow dashed line outlines FRAP region in frame 0 s. Blue dashed line (in −2 s) delineates the region used to assemble the montage in (M). (M) Montage of FRAP time series (time interval 40 s) from Frame 1 to Frame 50 with time increment 2. Red line indicates FRAP region. Arrowheads in (L) and (M) point to the metaphase plate. (N) Box plot of flux rate of GFP-TUA6 calculated from FRAP experiments, decreased in kin-12e (1.83 µm/min ± 0.2, n = 15 spindles, six plants) compared to WT (2.64 µm/min ± 0.49, n = 14 spindles, five plants). Velocities are means ± STDV. P-values indicated in the graphs were calculated by one-way ANOVA with post hoc Tukey HSD.
To further analyze kinetochore dynamics during oscillation, we determined the radius r of a circle fitted onto the kinetochore tracks (Figure 5G). In the kin12e mutants, the radius increased despite their shorter spindle axis, indicating that kinetochore-fibers were less stable in the absence of kinesin12E (Figure 5H–J). Together, these findings indicate that kinetochore-fiber stability is reduced in the mutants. This finding is consistent with the loss of the dynamic instability of microtubules upon transient overexpression of the GFP-KINESIN-12E motor domains (Supplemental Figure 1I and J). In such a scenario, compromised kinetochore-fiber dynamics would affect the distance between paired kinetochores. Thus, we measured the interkinetochore distances during congression and oscillation. Indeed, the average distance between paired kinetochores decreased in kin12e, suggesting that kinetochores in the mutant were either under less tension or chromosome attachment to kinetochore fibers was reduced compared to the wild type (Figure 5K, Supplemental Figure 3D). Furthermore, we measured the poleward flux, i.e., the poleward movement of microtubules, by monitoring the fluorescence recovery after photobleaching (FRAP) in a rectangular region parallel to the metaphase plate (Figure 5L and M; Buster et al., 2007; Leong et al., 2020). We observed a decrease in the flux rate in kin12e (Figure 5N), suggesting that KINESIN-12E modulates microtubule dynamics during metaphase. As reduced flux entails altered microtubule dynamics, we quantified the turnover of spindle microtubules from the FRAP experiments. For the wild type, the average half-time of microtubule turnover (t1/2 = 31 s ± 15, n = 15) was comparable to previously reported turnover times in plant spindles (Hush et al., 1994). In the mutant, the half-time appeared to be reduced (kin12e, t1/2 = 22 s ± 11, n = 15), but the difference was not significant (Supplemental Figure 3E). Nonetheless, the observed defects in microtubule and kinetochore dynamics did not compromise chromatid segregation, as ploidy levels were comparable in wild type and mutant alleles (Supplemental Figure 3F and G). The latter observation implies that the loss of KINESIN-12E function is counterbalanced by the slowing of mitosis and redundant or compensatory activities that are likely accomplished by KINESIN-5.
Discussion
Phylogenetic analysis of selected plant kinesin-12 family members revealed two clusters of plant kinesin-12 paralogs (Shen et al., 2012; Müller and Livanos, 2019). Of the six Arabidopsis homologs, the four KINESIN-12 paralogs characterized to date, i.e., class I POK1 and POK2, and Class II KINESIN-12A and KINESIN-12B, perform strictly plant-specific functions in division plane maintenance, phragmoplast guidance, and stability (Müller et al., 2006; Lee et al., 2007; Lipka et al., 2014; Müller and Livanos, 2019). In this study, we provide insight into the function of Class I KINESIN-12E. In brief, during prophase, KINESIN-12E gathers in the perinuclear region, and during cytokinesis, it accumulates at the phragmoplast midzone, a pattern similar to that of several of the abovementioned plant KINESIN-12 family members. Therefore, KINESIN-12E might act redundantly with other plant KINESIN-12 family members at the phragmoplast, serving plant-specific functions that need to be further characterized. Nevertheless, despite grouping with division site residents POK1 and POK2 in the same phylogenetic class (Shen et al., 2012; Müller and Livanos, 2019), KINESIN-12E localizes to the spindle midzone and promotes spindle assembly, most likely facilitating microtubule reorganization and the stabilization of kinetochore-fibers during prometaphase and metaphase. Spindle-related functions for kinesin-12 have also been shown in metazoans (Kapoor, 2017).
How does KINESIN-12E contribute to spindle assembly? Our mutant analysis revealed that KINESIN-12E functions in controlling metaphase spindle size, while it is nonessential for bipolarity (Figures 3 and 4). A similar short spindle phenotype was reported for a knock-out mutant of the human kinesin-12 ortholog HsKif15 (Supplemental Figure 4), which was attributed to compromised HsKif15’s sliding activity, counteracting inward-directed forces within the spindle (Sturgill and Ohi, 2013; Mann et al., 2017). In this study, the loss of KINESIN-12E compromised efficient microtubule sorting and lateral bundling, which are relevant for spindle shape and rapid chromosome alignment (Figures 3 and 5, Supplemental Movie 4; Musacchio and Salmon, 2007; Kajtez et al., 2016). In particular, we found prolonged chromosome congression and oscillation and at the same time reduced interkinetochore distances, suggesting that unstable kinetochore-fibers in the kin12e mutant exert less tension on kinetochores or that the attachment of chromosomes is reduced (Figure 5; Musacchio and Salmon, 2007). Thus, KINESIN-12E might be a good candidate protein that generates part of the outward-directed forces in the plant spindle via microtubule sliding.
Consistently, the transiently expressed KINESIN-12E displayed motility and track-switching and occasionally dragging along microtubules, pointing to the potential role of KINESIN-12E in reorganizing microtubule arrays in planta (Figure 2, Supplemental Movie 3). Track-switching has also been observed for HsKif15, which forms tetramers in vitro (Drechsler et al., 2014) and in mammalian cell extracts (Mann et al., 2017). Other studies revealed the presence of a secondary microtubule-binding site in the carboxy-terminal tail of the HsKif15 domain that facilitates microtubule sliding in vitro and in vivo (Sturgill and Ohi, 2013). The inhibitory activity of that tail allows for the autonomous targeting and preferential binding of kinetochore-fibers (Sturgill et al., 2014). Our work shows that the carboxy-terminal stalk domain of KINESIN-12E alone still associates with the entire spindle in planta (Figure 2), making the existence of a cryptic microtubule-binding site in KINESIN-12E plausible. In addition, the different mean velocities between motor-domain and full-length proteins hint at the regulation of KINESIN-12E motility via the nonmotor region (Figure 2, Supplemental Movie 3), as has been described recently for Girardia kinesin-14 (Tseng et al., 2020). Besides, direct binding to specific microtubule populations (e.g., to post-translationally modified tubulin) or to specific interaction partners such as the MAP65 microtubule cross-linker might be responsible for the distinct localization of KINESIN-12E at the spindle. Interestingly, the initial perinuclear localization of KINESIN-12E during prophase is independent of microtubules, and its functional significance is still unclear (Figure 1), implying that the association of KINESIN-12E with specific subcellular regions, i.e., the microtubule-overlap in mitotic and cytokinetic bipolar arrays, is mediated by motor-dependent and motor-independent mechanisms.
Does KINESIN-12E associate with kinetochore-fibers? KINESIN-12E motility is likely microtubule plus-end directed, similar to its plant homolog POK2, which localizes to the microtubule-plus ends-enriched phragmoplast midzone (Smertenko et al., 2011; Chugh et al., 2018; Herrmann et al., 2018). The absence of midzone labeling by the motor-dead KINESIN-12E(T181N) mutant corroborates microtubule plus-end directionality. Furthermore, its inability to rescue the knock-out mutant underscores the importance of motility for functionality, i.e., microtubule sliding activity (Figure 2). Oryzalin experiments suggested that KINESIN-12E might be tightly associated with kinetochore-fiber plus-ends (Figure 1). Indeed, in HeLa cells, HsKIF15 protects kinetochore-fibers from cold-induced depolymerization (Vanneste et al., 2009; Sturgill and Ohi, 2013), and high concentrations of HsKif15 molecules reduce catastrophe at the microtubule plus ends in vitro (Drechsler and McAinsh, 2016). Similarly, transient expression of KINESIN-12E motor-domain mutants at high levels abolished dynamic microtubule instability (Supplemental Figure 1), suggesting that KINESIN-12E shield kinetochore-fibers plus-ends from depolymerization and helps sustain stable connections with the kinetochores during chromosome alignment.
Indeed, interkinetochore distances were reduced in the kin12e mutants, also indicating reduced tension. The reduction in interkinetochore distance could be a consequence of the diminished poleward flux rate and/or kinetochore-fiber stability (Figure 5). Slower flux could be due to slower polymerization and/or a slower depolymerization rate (Buster et al., 2007). The lower flux in the smaller spindle of the mutant might explain the efficient chromosome separation and the absence of ploidy defects (Supplemental Figure 3). Moreover, overlapping antiparallel microtubules exhibit flux and often pass close to kinetochores, serving as platforms for sliding microtubule plus-end-directed kinesins (Maiato et al., 2017). In metazoans, the kinesin Eg5 is the major driver of poleward flux, although additional plus-end directed kinesins, i.e., kinesin-12, also contribute to this process (Mitchison, 2005; Buster et al., 2007). However, in mammals, the functional significance of kinesin-12 for spindle integrity becomes obvious only in the absence of Eg5 (Tanenbaum et al., 2009; Hancock, 2014; Kapoor, 2017). Consistently, the absence of a growth, or more severe cellular kinesin-12 mutant phenotype in plants may be due to redundancy with intact kinesin-5 activity in the spindle, similar to the overlapping functions of kinesin-12 and kinesin-5 observed in mammalian cells (Tanenbaum et al., 2009; Hancock, 2014). The spindle defects in kin12e might also reflect the notion that excessive inward-directed forces of microtubule minus-end directed kinesin-14, i.e., ATK5, are no longer counterbalanced effectively (Ambrose and Cyr, 2007, Yoshida et al., 2019).
In summary, based on our findings, KINESIN-12E contributes to spindle assembly reminiscent of its animal kinesin-12 orthologs (Sturgill and Ohi, 2013; Drechsler et al., 2014; Mann et al., 2017). However, cross-species experiments are required to examine the extent of the possible functional conservation of these proteins. How exactly KINESIN-12E contributes to microtubule bundling and sliding requires further investigation. Nevertheless, the data collectively support three possible scenarios explaining the mode of KINESIN-12E activity during metaphase spindle assembly: KINESIN-12E might regulate microtubule dynamics through direct binding to the microtubule lattice, indirectly, e.g., by regulating the attachment of microtubules to kinetochores, or both.
Methods
Plant material
Arabidopsis thaliana ecotype Columbia (Col-0, wild type) plants were used throughout the study. The kinesin-12e (kin12e, At3g44050) mutant lines are T-DNA insertion lines obtained from the Wisconsin T-DNA library and GABI-KAT. The T-DNA insertion in WiscDsLox4G07 (kin12e-1; Woody et al., 2007) maps to exon 13 and that in GABI-KAT_429E11 (kin12e-2; Kleinboelting et al., 2011) maps to exon 1; both were sequence verified. Genotyping primers are listed in Supplemental Table 3. All the other mutant lines were obtained from The European Arabidopsis Stock Centre (NASC). The different transgenic plant lines were generated via Agrobacterium tumefaciens–mediated transformation as previously described (Müller et al., 2006). For transgenic plants, multiple independent T1 plants were screened for each construct (5 ≤ n ≥ 7). To visualize the microtubule cytoskeleton, the microtubule reporter lines pUBN:RFP-MBD (Lipka et al., 2014) and pUBN:RFP-EB1b (At5g62500) (Chugh et al., 2018) were used. The p35S:GFP-TUA6 (At4g14960) line (N6551) was obtained from NASC. To visualize kinetochores, we generated pUBN:RFP-HTR12 harboring the kinetochore reporter CENH3/HTR12 (At1g01370) and transformed it into Col-0.
Plant cultivation and growth conditions
Seeds were surface sterilized using 6% sodium hypochlorite for 5 min, followed by three washes with ddH2O. Sterilized seeds were placed on plates containing half-strength Murashige and Skoog basal medium (Murashige and Skoog, 1962), 0.1% 2-(N-morpholino) ethanesulfonic acid (MES), and 1% agar. After 2–4 days of stratification at 4°C in the dark, the plates were transferred to growth chambers under long-day conditions (cycling 16 h light/8 h dark; Osram L18W/840 cool-white lamps; 200 µmol m − 2 s − 1) at 22°C and 40% humidity. For reproduction and crosses, 2- to 3-week-old seedlings were transferred onto soil.
Generation of cDNA, amplification of PCR products, and ligation
The cDNA was generated as described in Müller et al. (2006), using Superscript Reverse Transcriptase II (Invitrogen, 18064-22). All enzymes used for restriction digests were purchased from Fermentas. PCR fragments used for cloning purposes were amplified with Phusion DNA Polymerase (New England Biolabs, M0530L) and the oligonucleotides listed in Supplemental Table 3. All amplified or digested PCR fragments were purified using a PCR purification kit (Bioline, BIO-52060) or they were first extracted from agarose gels and purified afterward. The generated constructs were sequenced (GATC/Eurofins).
Generation of entry clones
Restriction sites marked in bold are insert specific. Primer combinations used for insert amplification are listed in Supplemental Table 3. All inserts were sequence verified.
pENTR:kin12E398-1263
The cDNA extracted from inflorescences was used to amplify the respective coding sequence (Subclone 2 and Subclone 3) and Subclone 2 was cloned into pGEM T-easy vector (Promega, A1360). Then, Subclone 3 was cloned with ClaI/NotI into the pGEM_kinesin-12EC_Subclone2 backbone and afterward, the insert was cloned into the Gateway (Invitrogen, Thermo Fisher Scientific) compatible pENTR2B vector via EcoRI/NotI sites.
pENTR:pkin12E:genGFP-kin12E1-1263
The full-length KINESIN-12E GFP fusion construct driven by its own promoter was cloned in multiple steps, using pENTR:kin12E398-1263 as a backbone. In pENTR:kin12E398-1263, the cDNA fragment was replaced by a genomic DNA fragment amplified from Col-0 using the same primer combination and restriction enzymes, creating pENTR:genkin12E398-1263. A genomic fragment (Kin12E_1-3313 bp) was amplified and cloned into pENTR:genkin12E398-1263 via Acc65I/EcoRI to create pENTR:genkin12E1-1263, which contained the full genomic DNA sequence of KINESIN12E.
Next, extension PCR of three overlapping fragments was performed as described in Atanassov et al. (2009). The three fragments (Fragment I: pkin12E; Fragment II: GFP-linker; Fragment III: linker-kin12E-992bp) were fused stepwise. Fragments I and II were fused in a volume of 30 μL containing 14 μL mixture of both PCR fragments (7 μL each; ∼200–800 ng; molar ratio 1:1), 6 μL of 5× Phusion HF Buffer, 3 μL of 2 mM dNTP mix, 0.3 μL Phusion DNA Polymerase (2 U/μL) and 6.7 μL H2O). The fragments were fused in a thermocycler at 98°C for 10 min, 60°C for 1 min, and 72°C for 10 min. This cycle was repeated five times, followed by a final extension step at 72°C for 10 min. Half of the reaction mix was used as a template for an enrichment PCR using primers flanking Fragments I and II. After confirming successful amplification of Fragment I+II, Fragment III was added by fusion PCR following the procedure described above. The final PCR product pkin12E:GFP-kin12E-992 was cloned into pGEM. Lastly, the insert of pGEM:pkin12E:GFP-kin12E-992 was ligated into pENTR:genkin12E1-1263 via Acc65I/MlsI restriction sites to create pENTR:pkin12E:GFP-kin12E1-1263.
pENTR:kin12E(Hybrid)
A genomic fragment of 3.2 kb (Subclone 1) starting at the first ATG codon was amplified and cloned into pENTR:kin12E398-1263 via Acc65I/EcoRI. The resulting clone contains genomic DNA and cDNA.
pENTR:HTR12
The coding sequence of HTR12 was amplified using seedling cDNA and cloned into pENTR3C (Invitrogen) via EcoRI/EcoRI digestion and subsequent ligation.
pENTR:TPX2
The full-length genomic sequence of TPX2 was amplified from genomic DNA extracted from leaves and cloned into the pGEM T-easy vector. Afterward, it was cloned into pENTR3C using EcoRI.
pENTR3C kin12E1-763
A 1,828 bp genomic fragment downstream of the KINESIN-12E motor domain sequence was amplified using oligonucleotides Kat301F and POK-likeMD(1-763aa)_Stop_Not_R and introduced into the pENTR:kin12E(Hybrid) backbone following digestion with EcoRI and NotI and ligation.
Generation of expression vectors
Recombination of pENTR clones with destination vectors was performed using LR Clonase (Invitrogen, 11791-020). The vector/insert borders were sequenced to confirm the correct reading frames.
pkin12E:GFP-kin12E
The pENTR:pkin12E:GFP-genkin12E1-1263 full-length construct was introduced into the binary vector pMDC99.
pUBN:GFP-kin12E
pENTR:kin12E(Hybrid) was introduced into the binary vector pUBN:GFP-Dest.
GFP-kin12E398-1263
pENTR:kin12E398-1263 was introduced into the binary vector pUBN:GFP-Dest and into pFK241p35S:GFP-Dest.
p35S:kin12E-GFP1-631
A 3,222 bp genomic fragment containing the KINESIN-12E motor domain sequence was amplified with Not/AscI restrictions sites added and cloned into vector pOCC12:GFP-6xHIS (Chugh et al., 2018). The pOCC12:kin12E1-631-GFP-6HIS was digested with DraI and the binary vector pFK241 (Lipka et al., 2014; Herrmann et al., 2018) was digested with BsrGI; subsequently, both were blunt-ended and digested with XbaI. Due to an additional XbaI restriction site in kin12E-GFP1-631, a 700 bp fragment had to be reintegrated into the XbaI site by a subsequent cloning step to generate pFK241p35S:kin12E1-631-GFP. The insert was sequence-verified.
p35S:GFP-kin12E1-763
In a Gateway LR reaction the pENTR:kin12E1-763 construct was introduced into pFK241p35S:GFP-Dest.
RFP-HTR12
In a Gateway LR reaction, the pENTR:HTR12 construct was introduced into pUBN:RFP-Dest.
RFP-TPX2
In a Gateway LR reaction, pENTR:TPX2 was cloned into pUBN:RFP-Dest.
β-glucuronidase-staining
For gene expression analysis, a 954 bp fragment upstream of the translation initiation site of KINESIN-12E (promoter of KINESIN-12E) was cloned in-frame (HindIII/BamHI) with glucuronidase in the pWD137 vector. The experimental setup and analysis were performed as previously described in (Müller et al., 2006). The 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc, Peqlab) was used as a substrate. Images were taken with a Zeiss Axiophot epifluorescence microscope equipped with a digital camera.
Confocal imaging and image processing
Four to seven days after germination, seedlings were mounted on slides or in LabTek chambers according to Peterson and Torii (2012). Imaging was performed on Leica TCS SP8 with Resonant Scanner and Zeiss LSM880 Airyscan inverted confocal laser-scanning microscopes. Both microscopes are equipped with Argon/Krypton mixed gas laser and 561 nm solid-state laser. Water immersion objectives (63×, numerical aperture 1.20) were used for image acquisition. GFP was excited with the 488 nm laser line from the argon/krypton laser, and Leica Hybrid detectors recorded emission between 492 and 530 nm. Red fluorescent protein was excited with a 561 nm laser and fluorescent signal detected with a standard photomultiplier tube with a detection window between 570 and 630 nm. Imaging on Zeiss LSM880 was performed using dichroic filter set 488/561. Room temperature was constant at 22°C.
Raw images were initially processed with ImageJ/Fiji (http://rsb.info.nih.gov/ij/). Sample drift was corrected using the StackReg plugin. Kymographs were generated using the Multiple kymograph plugin. The color merges were performed with ImageJ and Adobe Photoshop CS5 v12.0.4 (Adobe Systems), and figures were assembled with Adobe Illustrator CS5 v15.0.2. Time-lapse for analysis of kinetochore dynamics was performed using lines expressing GFP-TUA6 and RFP-HTR12. Images stacks of 5–6 images with 2 µm z-interval were taken at 30 s and 40 s time intervals using an LSM 880 Airyscan. Plants expressing GFP-TUA6 were used for FRAP experiments where images were acquired every 0.5 s with a Leica TCS SP8 confocal microscope.
Oryzalin treatment
Seedlings coexpressing GFP-KINESIN-12E and RFP-MBD were imaged and subsequently treated by replacing mounting medium with 10 µM oryzalin (Supelco, PS-410). Cells in different cell cycle stages were analyzed during oryzalin treatment at 2 min intervals for 10–50 min.
Immunolocalization
Immunofluorescence staining of 4- to 5-day-old seedlings was performed as previously described (Stöckle et al., 2016). Imaging was performed with a Leica TCS SP8 using a 63× objective, 16× line averaging, and 4× zoom.
Transient expression of p35S driven and GFP-tagged KINESIN-12E variants in N. benthamiana seedlings
Nicotiana benthamiana transformation was performed as described by Li et al. (2009). In brief, N. benthamiana seeds were sterilized with 6% (v/v) sodium hypochlorite and plated on MS ½ + 0.1% MES (pH 5.7) containing agar plates. Agrobacterium tumefaciens (AGL-1 strain) liquid overnight cultures were diluted to OD600 0.1 with fresh LB medium and grow until OD600 0.8–1. The Agrobacterium cells were harvested and the pellets resuspended in infiltration medium (0.005% Silwet L-77, 0.5% sucrose, 100 μM acetosyringone) to a final OD600 0.02. Six-day-old N. benthamiana seedlings were immersed in well plates containing infiltration medium. The penetration of Agrobacteria into the tissues was facilitated by vacuum infiltration. Following vacuum infiltration, the excess medium was removed from the seedlings with paper towels, and the seedlings were placed on Paul’s medium plates (full-strength MS, 0.1% sucrose, pH 5.8 plus 0.5% [w/v] Phytagel; Buschmann, 2016). The seedlings were grown under standard conditions for 72 h after infiltration and subsequently imaged under a Leica TCS SP8 with Resonant Scanner at 2-s intervals.
Nuclei fixation for flow cytometry
Seeds were plated on 1% agar (Serva) in ½ Murashige and Skoog basal salt mixture (Duchefa), stratified for 4 days, and transferred to the growth chamber (16 h day/8 h night cycle). Roots of 5-day-old seedlings were collected into six-well microtiter plates and nuclei were isolated according to (Wang et al., 2015). Tissues were fixed with 1% formaldehyde in MC buffer (10 mM potassium phosphate [Roth 3904.2], pH 7.0; 50 mM NaCl [Roth3957.1], 0.1 M sucrose [Roth 9286.2], 0.01% Triton X-100 [Roth 3051.1]). Constant vacuum was applied for 20 min at 100 mbar using a BÜCHI B-171 Vacobox. The fixative was removed and exchanged with 0.15 mM glycine (Roth 3908.1) in MC buffer for 10 min under a vacuum. The samples were rinsed twice with MC buffer and once with H2O, 5 min each. Excess water was gently drained with paper towels. The samples were immediately mixed with 1 mL nuclei isolation buffer (general-purpose buffer [GBP]: 0.5 mM spermine 4HCl [Sigma-Aldrich S2576], 30 mM sodium citrate [Roth HN13.1], 20 mM MOPS [Sigma-Aldrich M1254], 80 mM KCl [Roth 6781.1], 20 mM NaCl, pH 7, 0.5% Triton X-100; Loureiro et al., 2007). Using a razor blade, the tissues were chopped to a slurry and filtered through a 70 µm cell strainer (Greiner 542070). Nuclei flow-through was stained with 1 μg/mL 4',6-diamidino-2-phenylindole (DAPI).
Flow cytometry
Flow cytometry was performed using a Beckman Coulter MoFlo XDP cell sorter according to Zhu et al. (2017) with the following modifications: FL10 (529/28) and total nuclei were scored from the final endo-specific gates. The mean endoreduplication and mean C-value was calculated according to Barrow and Jovtech (2007).
Sequence alignment
Sequences were aligned using Multiple alignment program MAFFT version 7 online (Kuraku et al., 2013; Katoh et al., 2017) with default settings. The alignment was exported to Jalview for visualization (Waterhouse et al., 2009).
Data analysis
Spindle size
To determine spindle area and major and minor axis, spindle images derived from live-cell imaging and immune-localization experiments were processed. For live-cell images, plants expressing GFP-TUA6 and RFP-MBD were used. Single median plane images or z-projections of small stacks were segmented with ImageJ (adjust > threshold). Only minor adjustments were made to segment the entire spindle. The Analyze particle plugin was used to segment the spindle. In Set measurements > Fit ellipse was selected. The areas of the segmented region as well as the major and minor axes of the fitted ellipse were then measured.
Analysis of congression and oscillation velocities
Plants expressing GFP-TUA6 and HTR12 were used for imaging. Only one kinetochore (HTR12 dot) of each pair was manually tracked in time-lapse series. Congression was defined as the time between the frame immediately before kinetochores started moving toward the cell center and the frame showing apparent alignment of kinetochores in the metaphase plate. Oscillation started with the subsequent frame and ended with the frame immediately before kinetochore segregation.
Oscillation dynamics
As a measure of kinetochore dynamicity, the oscillation radius of one kinetochore per pair was determined. For this, a circle was manually fitted onto the entire track of the kinetochore during oscillation, circumscribing the mobility area. The radius of this area was normalized by the respective spindle’s half-length (pole-to-pole distance) and then plotted against the absolute spindle half-length.
Interkinetochore distance
Using Fiji, the distances between paired kinetochores (distance between the center of mass) within the same z-frame were measured at consecutive time points during the oscillation phase. For each cell, the average distance between paired kinetochores was determined, and this cell average constitutes one data point. The average distance was then calculated from ten cells in 10 plants per genotype. Raw data were plotted and are shown in Supplemental Figure 3.
Spindle microtubule flux rate
GFP-TUA6-expressing wild type and kinesin-12e lines were used for the analysis. Prometaphase and metaphase spindles were imaged every 0.5 s on the Leica TCS SP8, before and after bleaching of a rectangular region of interest (ROI) roughly parallel to the cell equator. For efficient bleaching, 10 frames were bleached with the 405 nm laser at 100%. For the illustration of the bleached region in Figure 5, from every third time frame, 10-pixel wide slices along the pole-to-pole axis were arranged using the montage tool in Fiji. Kymographs of the equivalent regions were generated to calculate the microtubule flux rate using the kymograph tool in Fiji. The method is adapted from Buster et al. (2007) and Leong et al. (2020).
FRAP analysis
FRAP analysis was performed using Stowers Plugins collection (https://research.stowers.org/imagejplugins/). In brief, background-subtracted (rolling ball 100 pixel) FRAP time series were opened in Fiji and the bleached region (ROI) was selected using the rectangular selection tool. Applying create spectrum jru v1 Plugin, signal intensity in ROI was measured and plotted automatically for individual time points in each FRAP experiment. Plots of individual FRAP experiments were combined (combine trajectories jru v1) and normalized (normalize trajectories jru v1, option min_max). The microtubule turnover (half time of signal recovery, tau, τ) was computed for each experiment using batch FRAP jru v1, and average half-time (t1/2) and SDs for each condition were calculated in Excel. Plots of average normalized signal recovery were assembled in Excel. Significance of values was determined with one-way ANOVA.
Boxplots and jitter plots were assembled in Plots of Data (https://huygens.science.uva.nl/PlotsOfData/). One-way ANOVA was performed using (https://astatsa.com/OneWay_Anova_with_TukeyHSD/), and for statistics of ploidy experiments, One-way ANOVA with Fisher’s LSD (least-squares difference) was performed using JMP 14 Statistics Analysis Software. Tests and parameters are given in Supplemental File 1.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: KINESIN-12E (AT3G44050), EB1b (AT5G62500) TUA6 (AT4G14960), CENH3/HTR12 (AT1G01370), TPX2 (AT1G03780).
Supplemental data
Supplemental Figure 1. The localization of KINESIN-12E is mitosis-specific.
Supplemental Figure 2. Characterization of kinesin-12e (kin-12e) mutants.
Supplemental Figure 3. Characterization of spindle phenotypes and ploidy levels.
Supplemental Figure 4. Multiple sequence alignment of selected KINESIN-12 orthologs.
Supplemental Table 1. Quantification of spindle phenotypes.
Supplemental Table 2. Average duration of individual mitotic stages.
Supplemental Table 3. Oligonucleotides used in the study.
Supplemental Movie 1. Cell cycle-dependent localization of GFP-KINESIN-12E.
Supplemental Movie 2. KINESIN-12E localizes to the nuclear envelope, spindle, and phragmoplast midzone.
Supplemental Movie 3. Motile KINESIN-12E clusters switch microtubule tracks in N. benthamiana transient expression experiments.
Supplemental Movie 4. Spindle assembly in wild type and kinesin12e-1.
Supplemental Movie 5. KINESIN-12E (T181N) irreversibly binds to microtubules.
Supplemental File 1. ANOVA tables.
Supplementary Material
Acknowledgments
We appreciate the help of Molly A.G. Perchlik for critical reading and editing. We acknowledge Theresa Lauster for photography of seedlings and Nan Wang and Chang Liu for providing the protocol and assisting with flow cytometry. We would like to thank Berenike Walter for help with cloning of p35S:GFP-kin12E389-1263.
Funding
We are grateful for funding from the Deutsche Forschungsgemeinschaft to S.M. (grant no. SFB 1101-A04, MU3133/6-1, MU3133/7-1) and generous support from ZMBP at the University of Tübingen.
Conflict of interest. The authors declare that they have no conflict of interest.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dr. Sabine Müller (sabine.mueller@zmbp.uni-tuebingen.de).
A.H. and S.M. conceptualized and performed experiments and analyzed data, and wrote the initial manuscript. E.L., S.Z., P.L., K.B., and L.R. performed experiments. S.M. and P.L. edited the final manuscript with input from all coauthors.
References
- Ambrose JC, Cyr R (2007) The kinesin ATK5 functions in early spindle assembly in Arabidopsis. Plant Cell 19: 226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose JC, Cyr R (2008) Mitotic spindle organization by the preprophase band. Mol Plant 1: 950–960 [DOI] [PubMed] [Google Scholar]
- Atanassov II, Atanassov II, Etchells J, Turner SR (2009) A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan A, Scheible W-R, Lukowitz W, Fagerstrom C, Wadsworth P, Somerville C, Baskin TI (2007) A conserved role for kinesin-5 in plant mitosis. J Cell Sci 120: 2819–2827 [DOI] [PubMed] [Google Scholar]
- Barow M, Jovtchev G (2007) Endopolyploidy in plants and its analysis by flow cytometry. In J Dolezel, J Greilhuber, J Suda, ed, Flow Cytometry with Plant Cells, WILEY-VCH Verlag, Weinheim, pp. 349–372
- Buschmann H (2016) Plant cell division analyzed by transient agrobacterium-mediated transformation of tobacco BY-2 cells. Methods Mol Biol 1370: 17–25 [DOI] [PubMed] [Google Scholar]
- Buster DW, Zhang D, Sharp DJ (2007) Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell 18: 3094–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh M, Reißner M, Bugiel M, Lipka E, Herrmann A, Roy B, Müller S, Schäffer E (2018) Phragmoplast orienting kinesin 2 is a weak motor switching between processive and diffusive modes. Biophys J 115: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McAinsh AD (2016) Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci USA 113: E1635–E1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD (2014) The Kinesin-12 Kif15 is a processive track-switching tetramer. eLife 3: e01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, Jackson WT, McIntosh JR (1982) Polarity of spindle microtubules in Haemanthus endosperm. J Cell Biol 94: 644–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Gable A, Wadsworth P (2010) Mitotic functions of kinesin-5. Semin Cell Dev Biol 21: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock WO (2014) Mitotic kinesins: a reason to delve into kinesin-12. Curr Biol 24: R968–R970 [DOI] [PubMed] [Google Scholar]
- Herrmann A, Livanos P, Lipka E, Gadeyne A, Hauser MT, Damme DV, Müller S (2018) Dual localized kinesin‐12 POK2 plays multiple roles during cell division and interacts with MAP65‐3. EMBO Reports 19: e46085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C-MK, Hotta T, Guo F, Roberson RW, Lee Y-RJ, Liu B (2011) Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell 23: 2909–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Karsenti E (1996) Morphogenetic properties of microtubules and mitotic spindle assembly. Cell 84: 401–410 [DOI] [PubMed] [Google Scholar]
- Hush JM, Wadsworth P, Callaham DA, Hepler PK (1994) Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J Cell Sci 107: 775–784 [DOI] [PubMed] [Google Scholar]
- Kajtez J, Solomatina A, Novak M, Polak B, Vukušić K, Rüdiger J, Cojoc G, Milas A, Šumanovac Šestak I, et al. (2016) Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat Commun 7: 10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T (2017) Metaphase spindle assembly. Biology 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinformatics 20: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2011) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman E, Holzbaur ELF (2018) Walking forward with kinesin. Trends Neurosci 41: 555–556 [DOI] [PubMed] [Google Scholar]
- Kuraku S, Zmasek CM, Nishimura O, Katoh K (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res 41: W22–W28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Li Y, Liu B (2007) Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell 19: 2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Liu B (2000) Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr Biol 10: 797–800 [DOI] [PubMed] [Google Scholar]
- Leong SY, Edzuka T, Goshima G, Yamada M (2020) Kinesin-13 and Kinesin-8 function during cell growth and division in the moss Physcomitrella patens. Plant Cell 32: 683–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Rutten T, Schubert I (2011) Deposition, turnover, and release of CENH3 at Arabidopsis centromeres. Chromosoma 120: 633–640 [DOI] [PubMed] [Google Scholar]
- Li JF, Park E, von Arnim AG, Nebenführ A (2009) The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka E, Gadeyne A, Stöckle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Muller S (2014) The phragmoplast-orienting Kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26: 2617–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livanos P, Chugh M, Müller S (2017) Analysis of phragmoplast kinetics during plant cytokinesis. In Jiang L, ed, Plant Protein Secretion: Methods and Protocols. Springer New York, New York, NY, pp. 137–150 [DOI] [PubMed] [Google Scholar]
- Loureiro J, Rodriguez E, Dolezel J, Santos C (2007) Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot 100: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Gomes AM, Sousa F, Barisic M (2017) Mechanisms of chromosome congression during mitosis. Biology 2017, 6, 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BJ, Balchand SK, Wadsworth P, Zheng Y (2017) Regulation of Kif15 localization and motility by the C-terminus of TPX2 and microtubule dynamics. Mol Biol Cell 28: 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoud K, Herzog E, Chabouté M-E, Schmit A-C (2013) Microtubule nucleation and establishment of the mitotic spindle in vascular plant cells. Plant J 75: 245–257 [DOI] [PubMed] [Google Scholar]
- Miki T, Naito H, Nishina M, Goshima G (2014) Endogenous localizome identifies 43 mitotic kinesins in a plant cell. Proc Natl Acad Sci USA 111: E1053–E1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ (2005) Mechanism and function of poleward flux in Xenopus extract meiotic spindles. Philos Trans R Soc Lond B Biol Sci 360: 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Livanos P (2019) Plant Kinesin-12: localization heterogeneity and functional implications. Int J Mol Sci 20: 4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Han S, Smith LG (2006) Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr Biol 16: 888–894 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Dixit R (2018) Kinesins and myosins: molecular motors that coordinate cellular functions in plants. Annu Rev Plant Biol 69: 329–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Lee YR, Liu B (2004) Localization of two homologous Arabidopsis kinesin-related proteins in the phragmoplast. Planta 220: 156–164 [DOI] [PubMed] [Google Scholar]
- Peterson KM, Torii KU (2012) Long-term, high-resolution confocal time lapse imaging of arabidopsis cotyledon epidermis during germination. J. Vis. Exp. (70), e4426, doi:10.3791/4426 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovská B, Jeřábková H, Kohoutová L, Cenklová V, Pochylová Ž, Gelová Z, Kočárová G, Váchová L, Kurejová M, Tomaštíková E, et al. (2013) Overexpressed TPX2 causes ectopic formation of microtubular arrays in the nuclei of acentrosomal plant cells. J Exp Bot 64: 4575–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Collatos AR, Bibeau JP, Furt F, Vidali L (2012) Phylogenetic analysis of the Kinesin superfamily from physcomitrella. Front Plant Sci 3: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Piette B, Hussey PJ (2011) The origin of phragmoplast asymmetry. Curr Biol 21: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Smirnova EA, Bajer AS (1998) Early stages of spindle formation and independence of chromosome and microtubule cycles in Haemanthus endosperm. Cell Motility 40: 22–37 [DOI] [PubMed] [Google Scholar]
- Stöckle D, Herrmann A, Lipka E, Lauster T, Gavidia R, Zimmermann S, Muller S (2016) Putative RopGAPs impact division plane selection and interact with kinesin-12 POK1. Nat Plants 2: 16120. [DOI] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R (2013) Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol 23: 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Das D K, Takizawa Y, Shin Y, Collier S E, Ohi M D, Hwang W, Lang MJ, Ohi R (2014) Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism. Curr Biol 24: 2307–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macůrek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema RH (2009) Kif15 cooperates with Eg5 to promote bipolar spindle assembly. Curr Biol 19: 1703–1711 [DOI] [PubMed] [Google Scholar]
- Tseng KF, Mickolajczyk KJ, Feng G, Feng Q, Kwok ES, Howe J, Barbar EJ, Dawson SC, Hancock WO, Qiu W (2020) The tail of Kinesin-14a in giardia is a dual regulator of motility. Curr Biol 30: 3664–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I (2009) The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol 19: 1712–1717 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D (2015) Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res 25: 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ (2007) The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Yamada M, Goshima G (2017) Mitotic spindle assembly in land plants: molecules and mechanisms. Biology 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida MW, Yamada M, Goshima G (2019) Moss Kinesin-14 KCBP accelerates chromatid motility in anaphase. Cell Struct Funct 44: 95–104 [DOI] [PubMed] [Google Scholar]
- Zhang H, Dawe RK (2011) Mechanisms of plant spindle formation. Chromosome Res 19: 335–344 [DOI] [PubMed] [Google Scholar]
- Zhu W, Hu B, Becker C, Doğan ES, Berendzen KW, Weigel D, Liu C (2017) Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol 18: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.