Abstract
Both inter- and intra-specific diversity has been described for trichome patterning in fruits, which is presumably involved in plant adaptation. However, the mechanisms underlying this developmental trait have been hardly addressed. Here we examined natural populations of Arabidopsis (Arabidopsis thaliana) that develop trichomes in fruits and pedicels, phenotypes previously not reported in the Arabidopsis genus. Genetic analyses identified five loci, MALAMBRUNO 1–5 (MAU1–5), with MAU2, MAU3, and MAU5 showing strong epistatic interactions that are necessary and sufficient to display these traits. Functional characterization of these three loci revealed cis-regulatory mutations in TRICHOMELESS1 and TRIPTYCHON, as well as a structural mutation in GLABRA1. Therefore, the multiple mechanisms controlled by three MYB transcription factors of the core regulatory network for trichome patterning have jointly been modulated to trigger trichome development in fruits. Furthermore, analyses of worldwide accessions showed that these traits and mutations only occur in a highly differentiated relict lineage from the Iberian Peninsula. In addition, these traits and alleles were associated with low spring precipitation, which suggests that trichome development in fruits and pedicels might be involved in climatic adaptation. Thus, we show that the combination of synergistic mutations in a gene regulatory circuit has driven evolutionary innovations in fruit trichome patterning in Arabidopsis.
Arabidopsis has evolved trichome formation in fruits by combining mutations in three transcription factors of the regulatory network underlying trichome patterning.
Introduction
Trichomes, or plant hairs, are epidermal cells highly differentiated as outgrowths involved in adaptation to multiple abiotic and biotic environmental factors (Hauser, 2014). They directly protect from sunlight, heat, and UV radiation, while indirectly influence transpiration, water use efficiency, and photosynthesis (Bickford, 2016). In addition, trichomes provide a defensive barrier against a wide range of herbivores (Dalin et al., 2008; Fürstenberg-Hägg et al., 2013). In the past two decades, the development of trichomes has extensively been studied to disentangle the mechanisms controlling cellular morphogenesis and differentiation, including cell cycle and fate (Ishida et al., 2008; Balkunde et al., 2010; Grebe, 2012).
Trichomes are developed in leaves of most plants, but they also can be found in other aerial organs, such as stems, pedicels, sepals, petals, or fruits. The distribution of trichomes in different organs has classically been used as a morphological character to describe inter- and intra-specific patterns of variation in Angiosperms (Judd et al., 1999). In particular, the presence of trichomes in fruits provides a qualitative trait for plant taxonomy that reflects a differential regulation in leaves and fruits, with a developmental boundary between reproductive and vegetative organs (Tutin et al., 1993; Hülskamp and Schnittger, 1998; Serna and Martin, 2006; Ó'Maoiléidigh et al., 2013). The qualitative robustness of trichome production in different organs is compatible with the quantitative phenotypic plasticity observed for the density of trichomes in response to environmental factors, such as herbivores or water stress (Guimil and Dunand, 2006; Züst et al., 2012; Bloomer et al., 2014; Bickford, 2016). Accordingly, genetic and molecular studies in the model plant Arabidopsis have revealed a complex regulatory network that integrates endogenous signals from hormones such as jasmonic acid and gibberellins (Pattanaik et al., 2014). Currently, more than 40 proteins have been identified as positive or negative regulators of trichome patterning (Balkunde et al., 2010; Doroshkov et al., 2019). In particular, MYB, bHLH, and WDR transcription factors play an essential role in trichome initiation because they form a central trimeric complex encoded by GLABRA1 (GL1), GLABRA3 (GL3)/ENHACER of GL3 (EGL3), and TRANSPARENT TESTA GLABRA1 (TTG1). This complex induces trichome initiation by activating the expression of the homeodomein protein GLABRA2 (GL2; Grebe, 2012; Pattanaik et al., 2014). In addition, seven single repeat R3 MYB transcription factors have been reported to repress trichome formation by disrupting the function of the trimeric complex (Wang and Chen, 2014).
Despite the tight regulation of trichome patterning, substantial intraspecific variation has also been described for leaf trichome density, which is presumably involved in adaptation to different environments (Hauser, 2014). In particular, GL1 accounts for the glabrous phenotype occurring occasionally in multiple Brassicaceae plants (Hauser et al., 2001; Bloomer et al., 2012; Li et al., 2013). In addition, ENHANCER of TRIPTYCHON and CAPRICE 2 (ETC2) and ATMYC1 contribute to the quantitative natural variation for leaf trichome density in Arabidopsis (Hilscher et al., 2009; Symonds et al., 2011). However, the molecular mechanisms underlying the intra- and interspecific diversity for trichome patterning in other organs remain mostly unknown.
Like many other wild and crop plants, all species of the Arabidopsis genus have taxonomically been characterized by the absence of trichomes in fruits (Al-Shehbaz and O’Kane, 2002). However, artificial gene perturbations have led to fruit trichome formation, indicating that the underlying regulatory pathway is not completely absent in Arabidopsis (Schnittger et al., 1998; Ó‘Maoiléidigh et al., 2013). In agreement with the differential trichome patterns of fruits and leaves, these studies have also shown that the key regulator of reproductive organ formation, AGAMOUS (AG), suppresses the leaf and trichome developmental programs during flower development (Ó’Maoiléidigh et al., 2013). Therefore, Arabidopsis trichome formation is precisely restricted to sepals, while it is silenced in other flower organs. Refuting this archetype, here we identify Arabidopsis natural populations that develop trichomes in fruits and pedicels. As new qualitative traits likely involved in adaptation, such phenotypes appear as evolutionary innovations for this wild plant (Wagner, 2011).
In this study, we address the genetic, molecular, and evolutionary mechanisms underlying Arabidopsis diversity for fruit and pedicel trichome patterning. Functional dissection of these traits identified regulatory and structural mutations in three genes encoding MYB transcription factors, TCL1, TRY, and GL1, which showed strong synergistic interactions. In addition, we found that these traits and mutations are restricted to an ancient relict lineage of Arabidopsis, and they correlate with climate precipitation. Together, our results reveal that such novel fruit trichome patterns have evolved from standing and new variation at the core regulatory network.
Results
Fruit trichome development has evolved in the Arabidopsis relict lineage
Analysis of a worldwide collection of 420 Arabidopsis accessions identified 24 wild strains that develop trichomes in fruits, all coming from local populations in the Iberian Peninsula (Figure 1). Branched and simple trichomes were initiated during flower development, since they appear fully developed in carpels at anthesis (Figure 1, A–C). All but three of these accessions also displayed trichomes in pedicels, whereas 15 additional genotypes from the same world region had trichomes in pedicels but not in fruits (Figure 1, D and F). Genome-wide analysis of the genetic structure of 235 Iberian accessions included in this study showed that 88% of the populations with trichomes in fruits belong to the highly differentiated genetic group previously described as relict in this region (Figure 1F; 1001 Genomes Consortium, 2016; Tabas-Madrid et al., 2018). Relict accessions define an ancient lineage that originated in Africa and was isolated from the nonrelict Eurasian populations about 90 kya (Durvasula et al., 2017). However, all relict accessions analyzed from Africa, as well as 40% of the relict Iberian accessions, did not bear trichomes in fruits or pedicels. Therefore, fruit trichome development appears as a new trait evolved within the Iberian relict lineage.
Figure 1.

Genetic bases and geographic distribution of trichome pattern variation in fruits and pedicels of Arabidopsis. A–D, Photographs of carpels (A, B), fruits (C), and pedicels (D) of Don-0 and Ler accessions taken under stereomicroscope (A, C, D) or by scanning electron microscopy (B). In (D), Moj-0 accession is also included to illustrate the development of trichomes in pedicels but not in fruits. E, QTL mapping of fruit trichome pattern in the Don-0/Ler RIL population. Genetic maps of Arabidopsis linkage groups are shown in the abscissa and LOD scores in the ordinate. The LOD threshold used for QTL detection is shown as a hatched horizontal line, and the 2-LOD support intervals of the detected QTL are depicted as gray horizontal lines on the genetic maps. For each QTL, its name and the percentage of explained phenotypic variance is included. F, Geographic distribution of Iberian populations classified according to their genetic group (relicts and nonrelicts) and the development of trichomes in fruits and pedicels. The number of accessions in each class is indicated in the legend. G, Trichome number in the first fruit (FTN; upper panel), and pedicel trichome pattern measured as the number of hairy pedicels in the first 30 fruits (PTP; lower panel), of ILs differing in MAU2, MAU3, and MAU5 Don-0/Ler alleles. Dots and bars correspond to means ± 0.95 confidence intervals of three lines per genotype (10–24 plants per line). Graphical genotypes of ILs are depicted in the lower part of the panel. Differences among genotypes were tested by mixed linear models, and the same or different letters indicate nonsignificant and significant differences as tested by Tukey’s test (P <0.05).
Development of trichomes in fruits requires multiple MAU loci interacting synergistically
To determine the genetic bases of Arabidopsis fruit trichome patterning, we carried out quantitative trait locus (QTL) mapping in a population of recombinant inbred lines (RILs) derived from an accession showing trichomes in fruits, Don-0, and the reference strain Landsberg erecta (Ler; Figure 1E). To this end, we qualitatively phenotyped 375 RILs for the absence/presence of trichomes in fruits and compared these data with the 118 markers of the Ler/Don-0 genetic map (Mendez-Vigo et al., 2016). A total of five loci were identified and named as MALAMBRUNO (MAU) 1–5 (after Cervantes’ sorcerer who gave beards to the countess and her maidservants in Don Quixote). Don-0 alleles in all MAU loci increased fruit trichome density, but MAU2, MAU3, and MAU5 showed the largest effects. To characterize these major loci, we developed introgression lines (ILs) carrying Don-0 alleles at MAU2, MAU3, and/or MAU5 in a Ler genetic background. Phenotypic analyses showed that fruit trichome development requires Don-0 alleles in all three MAU loci (Figure 1G), although lines with Don-0 alleles in MAU5 and MAU2 or MAU3 occasionally displayed a few trichomes (<5) in the first fruits. These results indicated that the three loci show strong synergistic epistasis (P <0.001; Supplemental Data Set 1) to control fruit trichome formation. In contrast, the development of trichomes in pedicels was mainly determined by the large additive effect of MAU2 and its genetic interaction with MAU3 (Figure 1G;Supplemental Data Set 1). Thus, the same loci account for the distinct genetic architectures of fruit and pedicel trichome traits.
Regulatory variation causing hypofunction of TCL1 underlies MAU2
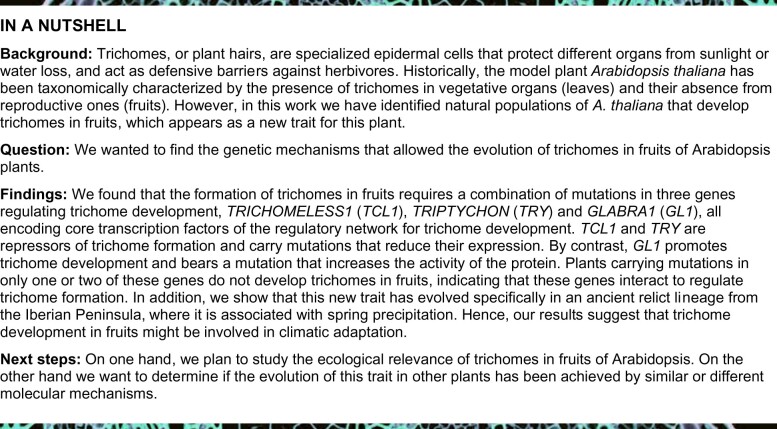
To find candidate genes for the MAU loci, we carried out genome-wide association (GWA) analyses for fruit and pedicel trichome traits using 235 Iberian accessions (Figure 2;Supplemental Figure S1). Comparison of the qualitative fruit trichome pattern and the 2.2 million single nucleotide polymorphisms (SNPs) segregating in these accessions detected several significant genomic regions around the locations of MAU2 and MAU5. However, as expected for the complex epistatic architecture of this trait, the most prominent statistical significances did not overlap with MAU mapping regions (Supplemental Figure S1). On the contrary, in agreement with a simpler genetic basis, the most significant genomic region detected for pedicel trichome pattern overlapped with MAU2, which suggests the presence of a frequent allele with large additive effect (Figure 2). The strongest associations were located around a cluster of three genes, TRICHOMELESS1 (TCL1), TCL2, and ETC2 (Figure 2B), all encoding R3 MYB transcription factors that negatively regulate trichome formation (Wang and Chen, 2014). A previous study identified an ETC2 missense mutation contributing to the natural variation for leaf trichome density (Hilscher et al., 2009). Although this polymorphism was segregating in the Iberian Peninsula, Don-0 and Ler accessions carried the same allele, and it was not significantly associated with fruit or pedicel trichome traits. In contrast, the most significant SNPs in this cluster were located around TCL1 (At2g30432), whose artificial loss-of-function mutant specifically develops trichomes in pedicels (Wang et al., 2007; Supplemental Figure S2). Furthermore, gene expression analyses showed that only TCL1 differed significantly between the parental accessions in reproductive organs (Figure 3;Supplemental Figure S3).
Figure 2.
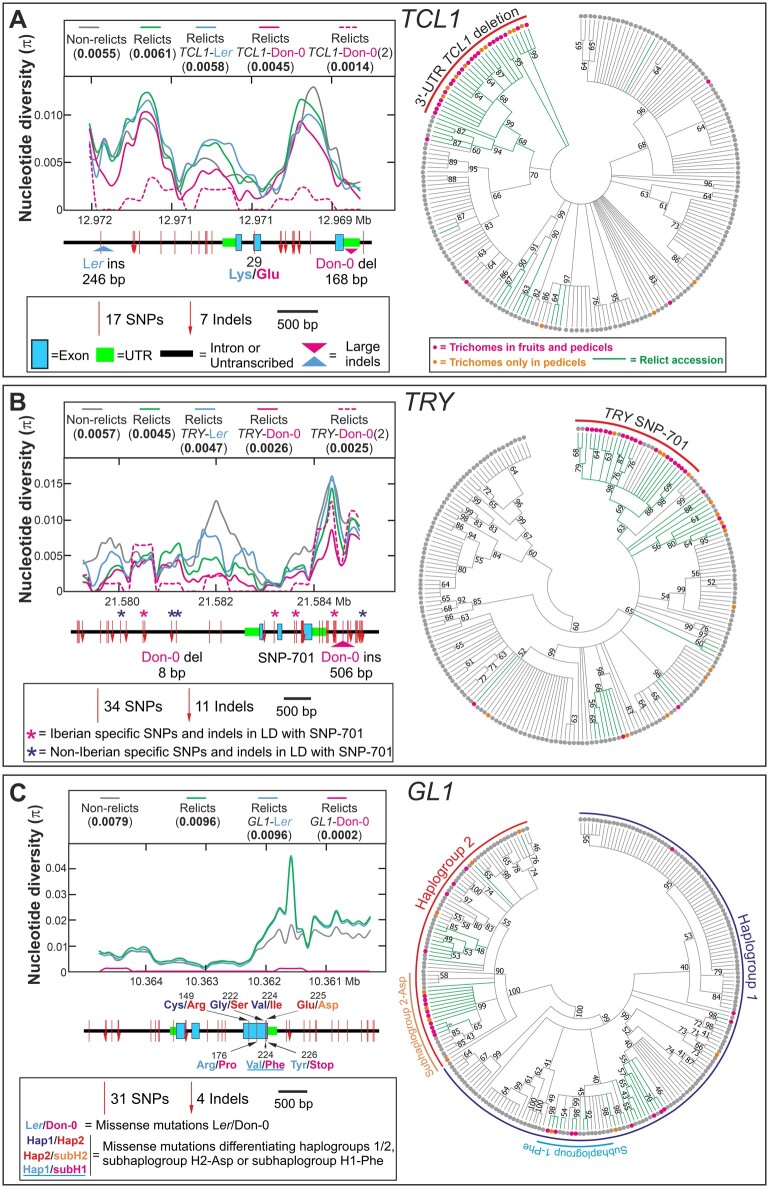
GWA analyses of pedicel trichome pattern. A, Manhattan plots from a GWA study carried out with 235 Iberian accessions phenotyped qualitatively for the absence/presence of trichomes in pedicels. The heritability of the trait, as well as the number of associated SNPs and genes, are indicated in the upper part of the panel. Mapping intervals of MAU2, MAU3, and MAU5 are shown as red boxes. B and C, Zooms of Manhattan plots in MAU2 genomic region (B) and along TCL1 gene (C). In (B), genes across the ∼50-kb MAU2 region displaying the most significant associations are depicted in the lower part of the panel, including the gene cluster of ETC2, TCL2, and TCL1. In the lower part of (C), TCL1 structure in introns, exons, and UTRs is presented, depicting the SNPs, indels, and missense mutations found between Don-0 and Ler. The effect of the 3′-UTR deletion of TCL1 (Don-0 del 168 bp) is included in (C) but not in (A) and (B), which were based on segregating SNPs. The red dotted line of each panel indicates the significance threshold of –log(P)=7.64 (corresponding to 5% with Bonferroni correction for multiple testing); SNPs above this threshold are red colored in (B) and (C).
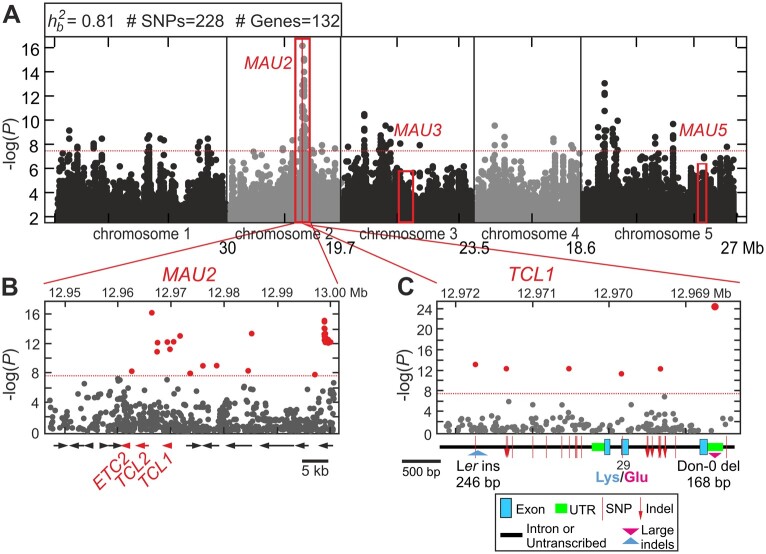
Figure 3.
Expression and fruit trichome pattern of candidate genes in parental and introgression lines. A, TCL1, TRY, and GL1 expression in vegetative and reproductive organs of parental accessions. B, TCL1, TRY, and GL1 expression in reproductive organs of ILs bearing Don-0 alleles in MAU2, MAU3, and/or MAU5. In (A) and (B), each bar depicts the mean ± se of three biological replicates, and values of all lines are relative to Ler vegetative expression. C, Effect of try mutant allele on fruit trichome number (number of trichomes in the first fruit) when combined with Don-0 alleles in MAU2 and/or MAU3. Dots and bars represent means ± 0.95 confidence intervals of two to three lines per genotype (7–10 plants per line) homozygous for Don-0 or Ler alleles in MAU2 and MAU3, as well as for TRY wild-type or try mutant alleles. A fruit of a homozygous try plant carrying Don-0 alleles at MAU2 and MAU3 is shown in the right side of the panel to illustrate the highly branched and aggregated trichomes of this genotype. In (B) and (C), graphical genotypes of ILs are depicted in the lower part of panels. Gene expression or phenotypic differences among genotypes were statistically tested by mixed linear models, the same or different letters indicating nonsignificant or significant differences, as tested by Tukey’s test (P < 0.05).
To test the functionality of TCL1 Don-0 and Ler alleles, we developed genomic constructs including the promoter, coding and 3′-untranslated region (3′-UTR) regions from each accession, and we used them to generate transgenic lines in tcl1 mutant genetic background (Figure 4). Phenotypic characterization of 12–14 T3 homozygous independent transgenic lines for each construct showed that both alleles reduced the number of pedicels developing trichomes. However, the two types of transgenic lines differed significantly in their pedicel trichome pattern (P <0.001; Supplemental Data Set 1). These results indicate that both TCL1 alleles, Don-0 and Ler, are functional, but Don-0 carries a partial loss-of-function (or hypomorphic) allele compared with Ler and Col backgrounds. To further determine if TCL1 is MAU2, we also transformed the IL-235. We selected this genetic background because Don-0 alleles in MAU2, MAU3, and MAU5 are required to develop high fruit trichome density. Since TCL1 is a negative regulator of trichome development, it can be expected that TCL1-Ler and TCL1-Don-0 will differentially reduce fruit trichome number (FTN). Analysis of 6–7 homozygous transgenic lines showed significant differences in pedicel trichome pattern (PTP), similar to those observed in the tcl1 background (Supplemental Figure S4B). In addition, both classes of transgenic lines differed quantitatively in FTN, TCL1-Ler showing, on average, a five-fold larger reduction than TCL1-Don-0 (Figure 4C). Hence, we concluded that TCL1 underlies MAU2 effects on pedicel and fruit trichome patterning.
Figure 4.
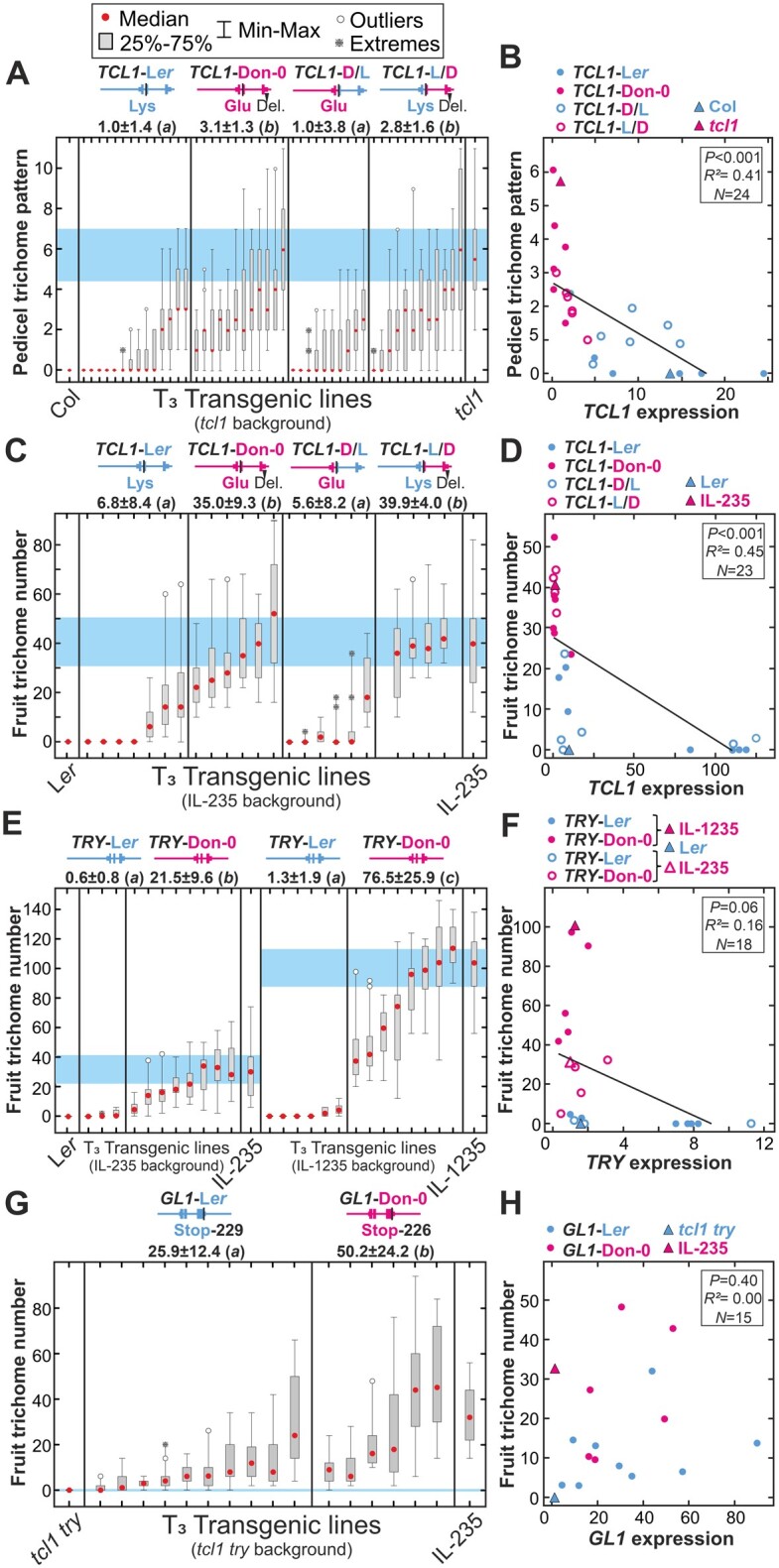
Trichome pattern and gene expression of transgenic lines for TCL1, TRY, and GL1. A, C, Pedicel (A) and fruit (C) trichome phenotypes of independent homozygous transgenic lines carrying parental (left side) or chimeric (right side) genomic constructs of TCL1, in tcl1 mutant (A) or IL-235 (C) genetic backgrounds. B, D, Linear regressions between pedicel or fruit trichome patterns and TCL1 expression in flower buds, for TCL1 transgenic lines in tcl1 (B) or IL-235 (D) backgrounds. E, G, Fruit trichome number of independent homozygous transgenic lines carrying Ler or Don-0 parental genomic constructs of TRY (E) or GL1 (G) in IL-235 (left side of E), IL-1235 (right side of E), or tcl1 try (G) genetic backgrounds. F, H, Relationship between fruit trichome number and TRY (F) or GL1 (H) expression in flower buds, for TRY (F) or GL1 (H) transgenic lines. Gene expressions of transgenic lines are relative to the expression of untransformed controls. Pedicel trichome pattern was measured as the number of hairy pedicels in the first 30 fruits (A, B), whereas fruit trichome number (FTN) is the number of trichomes in the first fruit (C–H). In (A, C, E, and G), transgenic lines are arranged from low to high mean FTN, and 95% confidence intervals for untransformed controls are shown as blue-shaded areas. Phenotypic differences among genotypes were statistically tested by mixed linear models; the same or different letters on top of each panel indicate nonsignificant or significant differences, as tested by Tukey’s test (P < 0.05).
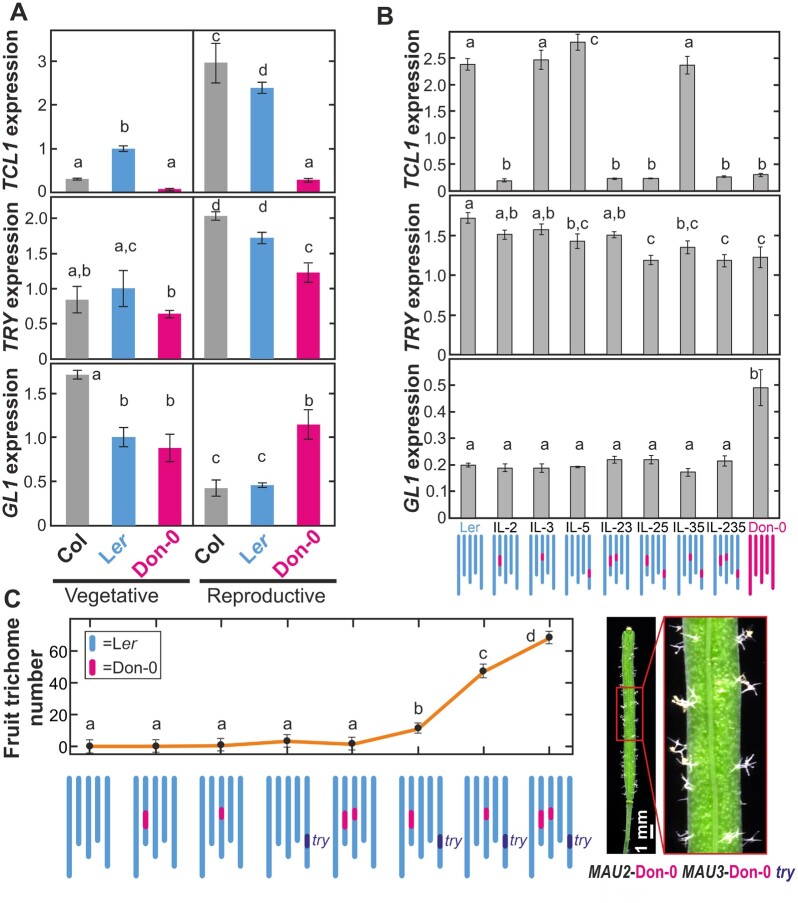
To reveal if the partial loss of function of TCL1-Don-0 is caused by structural or regulatory polymorphisms, we analyzed TCL1 nucleotide diversity. Don-0 and Ler parents differed only in one amino acid substitution (Lys29 to Glu29), but also in numerous polymorphisms across noncoding regions (Figure 5). In particular, Don-0 carried a 168-bp deletion spanning almost the complete 3′-UTR, which was in strong linkage disequilibrium (LD) with the missense mutation, a small indel, and three other noncoding SNPs (Supplemental Table S1). Clustering analysis of TCL1 sequences showed that these six polymorphisms define a haplogroup that includes most accessions developing trichomes in fruits or pedicels (Figure 5A). However, several additional analyses supported TCL1-Don-0 deletion as a cis-regulatory causal polymorphism. First, association tests including Don-0/Ler indels in the 235 Iberian accessions showed that TCL1-Don-0 deletion was the most significant polymorphism linked to pedicel trichome pattern (−log(P)=24.3; Figure 2C). In fact, all accessions carrying TCL1-Don-0 deletion developed trichomes in pedicles, and 67% of them also in fruits. Second, Don-0 and Ler parents strongly differed in gene expression, especially in the reproductive organs where TCL1 had two- to three-fold higher expression than in leaves (Figure 3A). This expression variation was largely determined by MAU2/TCL1 as shown by the analysis of ILs (Figure 3B;Supplemental Data Set 1). To distinguish the effects of the missense mutation and the 3′-UTR deletion, we developed two chimeric genomic constructs between Don-0 and Ler TCL1 alleles, which split both polymorphisms (Figure 4, A–D;Supplemental Figure S4, B and C). Phenotypic analyses of transgenic lines for the chimeric alleles, in tcl1 and IL-235 backgrounds, showed that transgenes carrying only the TCL1 3′-UTR deletion behave similar to lines carrying the complete TCL1-Don-0 allele. Quantitative differences among transgenic lines correlated with TCL1 expression (Figure 4B, D;Supplemental Figure S4C), TCL1-Don-0 deletion accounting for 41%–68% of the variation for fruit and pedicel trichome pattern, as well as for TCL1 expression (Supplemental Data Set 1). Therefore, the partial loss of function of the TCL1-Don-0 allele is caused by a 3′-UTR regulatory deletion, which reduces gene expression and the repression of trichome development in pedicels and fruits. In agreement with the geographic distribution of accessions developing trichomes in fruits, this TCL1 deletion is present only in Iberian relicts; it was not found in any other Arabidopsis genome from Eurasia or Africa (Supplemental Table S1).
Figure 5.
Genetic diversity of TCL1, TRY, and GL1. A–C, Nucleotide diversity in Iberian accessions (left panels), and neighbor-joining (NJ) trees displaying the genetic relationships among accessions (right panels), for TCL1 (A), TRY (B), and GL1 (C). Left panels show sliding window plots of the nucleotide diversity in nonrelict accessions, and relicts classified according to their Don-0/Ler alleles. The average nucleotide diversity (π) of each group of accessions is shown in round brackets on top of each graph. To enable comparisons of relict accessions with Don-0 alleles among the three genes, Don-0(2) shows the diversity in Don-0 and Bon-61 accessions, the only genotypes with such alleles in GL1. The genomic structure of each gene, as well as the location and the total number of Ler/Don-0 SNPs and indels, are shown below the abscissa axes of left panels. In (B), TRY polymorphisms found in strong LD with the SNP in position 701 (SNP-701) are marked with asterisks, colors indicating their regional Iberian specificity or not. In (C), Ler/Don-0 missense and nonsense mutations are shown below GL1 genomic structure, whereas missense polymorphisms differentiating the major GL1 haplogroups 1 and 2, or subhaplogroups found within major haplogroups, are displayed above the gene. Note that the underlined Val/Phe-224 amino acid substitution differentiating subhaplogroup 1-Phe is polymorphic between Ler and Don-0 accessions. In NJ trees, branches corresponding to partitions reproduced in <50% (A, B) or <40% (C) bootstrap replicates are collapsed, and branches corresponding to relict accessions are colored in green. A lower cut-off value was used for GL1 tree condensation due to its larger nucleotide diversity. Accessions developing trichomes in fruits, pedicels, or none of these organs, are depicted as magenta, orange, or gray colored dots, respectively. Clusters of accessions differentiated by TCL1 or TRY mutations associated with fruit trichome patterning (A, B), or major GL1 haplogroups differentiated by missense mutations (C), are highlighted with colored lines.
Regulatory variation causing TRY hypofunction underlies MAU5
As a first step to find the gene underlying MAU5, we fine mapped this locus by genotyping a segregating family of 1,350 plants with 18 genetic markers around MAU5 and subsequently phenotyping FTN in the offspring of 83 recombinant plants selected from this population (Supplemental Figure S5, A–C). MAU5 was thus located in a small genomic region of 3.5 kb where the genome sequence of Arabidopsis Col-0 reference strain predicts that there is only an open reading frame, which corresponds to TRY (At5g53200; Supplemental Figure S5, A–C). Like TCL1, TRY encodes for another R3 MYB transcription factor that negatively regulates trichome development (Schellmann et al., 2002). However, an induced try null mutation affects mainly trichome development in leaves, leading to increased trichome branching and density with an aggregated pattern (Supplemental Figure S2). To test if a TRY loss-of-function allele may underlie MAU5, we introgressed the try mutation into a Ler line carrying Don-0 alleles of MAU2 and MAU3 (Supplemental Figure S6). Phenotypic analyses of homozygous ILs showed that try causes fruit trichome formation when combined with natural Don-0 alleles in MAU2 and/or MAU3 (Figure 3C). Similar to MAU5-Don-0 alleles, try interacted significantly with MAU2 and MAU3 (Supplemental Data Set 1). However, try mutation had a stronger effect than the natural MAU5-Don-0 allele, because try lines displayed not only fruit trichomes with an aggregated pattern, but also higher fruit trichome density.
To validate that TRY from Don-0 and Ler differ functionally, we developed two genomic constructs corresponding to Don-0 and Ler alleles, and we used them to generate transgenic lines in try mutant background (Supplemental Figure S4D). The two transgenes complemented the aggregated trichome pattern observed in try leaves, indicating that both natural TRY alleles are functional. In addition, we tested the differential function of TRY Don-0 and Ler alleles in the reproductive phase by generating transgenic lines in IL-235 and IL-1235 backgrounds with high FTN. Phenotypic analyses of 3–8 homozygous independent lines for each transgene and background showed significant differences in FTN between both TRY alleles (P < 0.008; Figure 4E and Supplemental Data Set 1). Overall, TRY-Don-0 transgenes showed a weaker reduction of fruit and pedicel trichome number than TRY-Ler lines (Supplemental Figure S4E). Hence, we concluded that TRY underlies MAU5, for which Don-0 carries a partial loss-of-function allele.
Analyses of TRY sequences showed that Don-0 and Ler do not differ in any missense mutation, indicating that their functional variation is not caused by structural polymorphisms (Figure 5B). In agreement, Don-0 displayed significantly lower TRY expression than Ler mainly in reproductive organs, with a two-fold higher TRY expression than in leaves (Figure 3A). Analysis of gene expression in ILs differing in MAU2, MAU3, or MAU5 alleles further showed that the lower TRY-Don-0 expression is mostly determined by MAU5/TRY (Figure 3B). Likewise, transgenic lines differed in TRY expression (P = 0.01), which correlated marginally with FTN (Figure 4F and Supplemental Data Set 1). In addition, TRY expression was also regulated by TCL1/MAU2 because MAU2-Don-0 ILs had significantly lower expression than MAU2-Ler (P < 0.001; Figure 3B and Supplemental Data Set 1). Col and the tcl1 mutant, however, did not differ in TRY expression, suggesting that TRY trans-regulation depends on the TCL1 allele and/or the genetic background (Supplemental Figure S2D). Therefore, Don-0 and Ler not only vary in TRY cis-regulatory polymorphisms, but also in TRY trans-regulation mediated by TCL1.
To identify potential cis-regulatory TRY polymorphisms, we carried out an association analysis for fruit trichome pattern including all Don-0/Ler SNPs and indels segregating in the Iberian Peninsula. An intronic SNP located 701-bp downstream from the start codon (SNP-701) displayed the strongest association (−log(P)=11.0; Supplemental Figure S1E). This SNP showed strong LD with seven other polymorphisms, which together defined a TRY haplogroup present in 75% of the accessions developing trichomes in fruits (Figure 5B). Don-0 alleles in all these SNPs show high frequency in Iberian relict accessions, whereas they are not present in African relict populations and are nearly absent in the rest of Eurasia (Supplemental Table S1). Thus, Don-0 allele at SNP-701, and/or other Iberian relict polymorphisms in LD, likely reduce TRY expression in the reproductive organs, leading to trichome development in fruits.
Structural variation causing GL1 hyperfunction underlies MAU3
Fine mapping using a segregating population of 362 plants located MAU3 within a genomic region of 38 kb, which included the positive regulator of trichome development GL1 (Supplemental Figure S5, D–F). GL1 (At3g27920) encodes a R2R3 MYB transcription factor whose multiple natural loss-of-function mutations account for the glabrous phenotype observed at very low frequency across Arabidopsis geographic range (Hauser et al., 2001; Bloomer et al., 2012). Sequence analysis showed that GL1 from Don-0 carries a nonsense mutation (Tyr226 to Stop226) producing a truncated protein that lacks the last three amino acids. In addition, Don-0 and Ler also differed in two missense mutations located in GL1 region encoding the C-terminal domain, as well as in other polymorphisms in noncoding regions (Figure 5C).
To functionally characterize GL1 from Don-0 and Ler we developed genomic constructs including the promoter, coding and 3′-regions of each accession. Transgenic lines generated with these constructs in the genetic background of the null mutant gl1 showed that Don-0 and Ler GL1 alleles are active, because both transgenes complemented the gl1 glabrous phenotype (Supplemental Figures S2C and S4G). We then aimed to test by genetic reconstruction of Don-0 fruit trichome patterning, if GL1-Don-0 might be a gain-of-function allele. Since this trait requires loss-of-function alleles of TCL1 and TRY, we first obtained the double mutant tcl1 try, which shows the additive effects of both mutations, but absence of trichomes in fruits (Supplemental Figure S4H). We used this genetic background to generate 6–10 homozygous independent transgenic lines for each GL1 genomic construct, and we quantified them for FTN (Figure 4G). The two classes of transgenic lines developed at least a few trichomes per fruit, in agreement with previous gene perturbation studies (Schnittger et al., 1998; Ó'Maoiléidigh et al., 2013). Hence, GL1 overexpression produced by doubling the number of natural GL1 alleles is sufficient to trigger the development of trichomes in fruits, in the absence of the repression mediated by TCL1 and TRY. Furthermore, quantitative analyses showed that GL1 alleles from Don-0 and Ler are functionally different because GL1-Don-0 induced significantly more trichomes than GL1-Ler (P = 0.016; Figure 4G;Supplemental Figure S4H and Supplemental Data Set 1). We thus concluded that GL1 is MAU3, for which Don-0 carries a gain-of-function (or hypermorphic) allele.
To explore whether the Don-0 allele is caused by structural or regulatory polymorphisms, we also analyzed GL1 expression in parental accessions, as well as in introgression and transgenic lines. In contrast to the reference strains Ler and Col showing about half the expression in reproductive than vegetative organs, Don-0 accession had similar expression in leaves and fruits (Figure 3A). Accordingly, Don-0 displayed twice the expression of Ler in reproductive organs. However, all introgression and transgenic lines showed comparable GL1 levels (Figures 3B and 4H), indicating that the differential GL1 expression between Don-0 and Ler is not due to GL1 cis-regulatory polymorphisms or trans-regulation mediated by TCL1/MAU2 or TRY/MAU5, but by another unknown locus. Therefore, mutations altering the structure of GL1 protein most likely account for GL1-Don-0 gain of function.
Comparative analysis of GL1 protein in 13 Brassicacea species showed a high conservation of the last three amino acids (Supplemental Figure S7), thus supporting GL1-Don-0 nonsense mutation as the functional polymorphism. This mutation was found only in another Iberian accession (Bon-61) that also develops trichomes in fruits and that was collected 8-km distant from Don-0 (Figure 1F). In addition, GL1 showed higher nucleotide diversity than TCL1 or TRY, with most nonsynonymous and silent variation occurring in the 3′-region (Figure 5C). However, the two accessions carrying the nonsense mutation had substantially lower GL1 diversity than the rest of relict accessions, indicating that Don-0 and Bon-61 alleles are very similar and diverged rather recently (Figure 5C). Clustering analysis of Iberian GL1 diversity identified two main haplogroups, 1 and 2, previously described (Bloomer et al., 2012), as well as two subhaplogrous, all differentiated by missense mutations (Figure 5C;Supplemental Figure S7). The two haplogroups differed in GL1 amino acid substitutions in positions 149, 222, and 224, and they are distributed at high frequency (0.19–0.43) throughout Africa and Eurasia (Supplemental Table S1). Subhaplogroups were differentiated by substitutions Val224 to Phe224 (subhaplogroup 1-Phe) and Glu225 to Asp225 (subhaplogroup 2-Asp), but these appeared to be segregating only in Iberia. In fact, 37% of the accessions developing trichomes in fruits belong to subhaplogroup 2-Asp, suggesting that the structural mutations differentiating this allele might also produce GL1 gain of function. However, contrary to TCL1 and TRY, no GL1 polymorphism was associated with fruit trichome pattern (−log(P) <3.1; Supplemental Figure S1A). Low GL1 allele frequency and/or allelic heterogeneity might, nevertheless, strongly reduce the GWA statistical power to detect large-effect alleles in Arabidopsis (Atwell et al., 2010; Barboza et al., 2013). Hence, several GL1 gain-of-function alleles, likely caused by independent structural mutations, might have occurred in the Iberian relict lineage.
Fruit trichome development is associated with low precipitation climates
To identify environmental factors that might drive Arabidopsis evolution of fruit and pedicel trichome patterning, we analyzed the relationships between both traits and climate parameters from the population locations of the 235 Iberian accessions (Figure 6). Autologistic regressions showed that fruit trichome pattern correlates negatively with all climate variables measuring winter and spring precipitation, whereas it correlates less and positively with spring and summer temperatures (P < 0.01; Figure 6A). Furthermore, pedicel trichome pattern also showed maximum correlations with spring precipitations. These associations were not determined by the overall geographic distribution of relict populations in Iberia because similar results were found when analyzing only these populations (Supplemental Figure S8 and Supplemental Data Set 2). In agreement with these results, both trichome traits were also significantly associated with spring precipitation when including the genetic structure as covariate in general linear models (GLMs; P < 10−4; Supplemental Data Set 2). Moreover, genotype–environment associations showed that Don-0 alleles at TCL1 3′-UTR and TRY SNP-701 were distributed mainly in locations with low spring precipitation (Figure 6C). Don-0 alleles for GL1 nonsense and Asp225 missense polymorphisms were also linked to low precipitation, although their low frequency hampered statistical tests (Supplemental Data Set 2). Finally, we tested gene–environment associations by applying the latent factor mixed model (LFMM) method aimed to remove the confounding effects of genetic structure in these correlations. Genome-wide analysis of the 2.2 million SNPs segregating in the Iberian accessions detected 119 genes significantly associated with spring precipitation, including TCL1 (−log(P)=5.04) but not GL1 or TRY. Thus, trichome development in fruits and pedicels, as well as the TCL1 gene, appear to be associated with low precipitation in spring season, which is the reproductive period for these relict populations (Exposito-Alonso et al., 2018).
Figure 6.
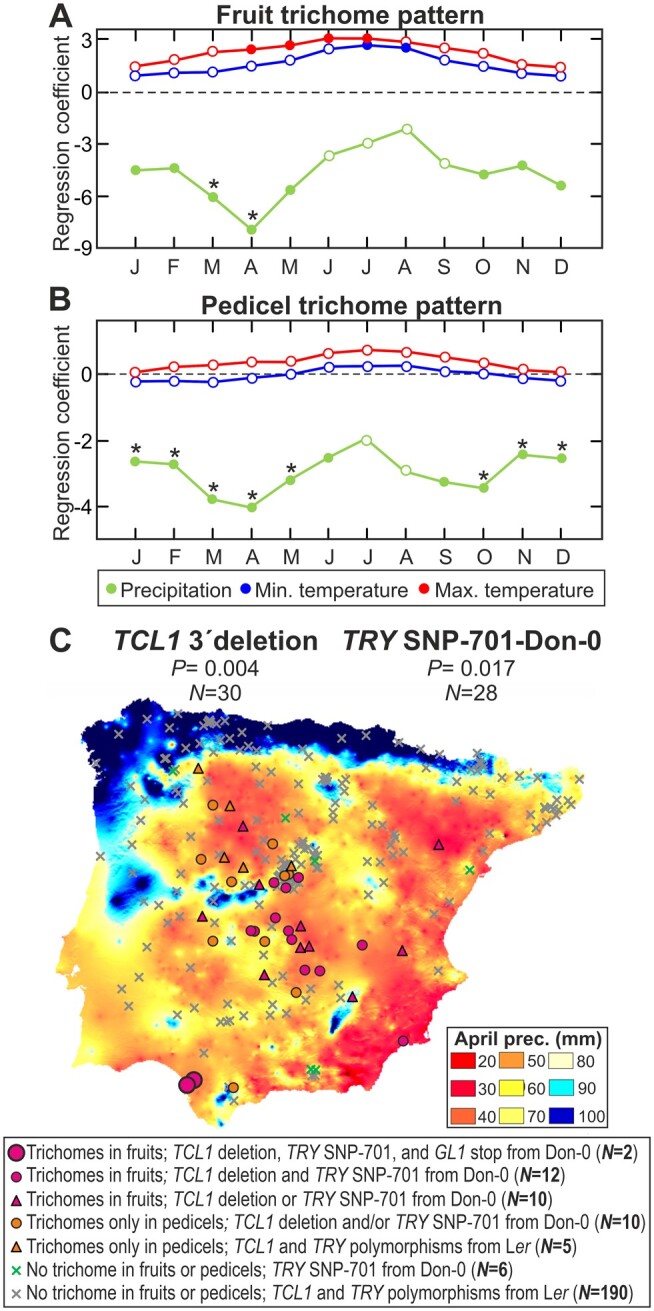
Relationship between fruit or pedicel trichome pattern and climate in Arabidopsis. A, B, Logistic regression coefficients between fruit (A) or pedicel (B) trichome pattern, and monthly precipitation (green line), minimum temperature (blue line), or maximum temperature (red line), along the year. Months in the abscissa are indicated with the first letter of the month. Filled circles and asterisks depict significant regressions (P < 0.05 or 0.01, respectively), while white circles are nonsignificant coefficients. C, Geographic and climatic distribution of TCL1 and TRY polymorphisms associated with fruit trichome pattern and April precipitation. Map shows the distribution of populations classified according to trichome development in fruits and/or pedicels, and their alleles in TCL1 3′-UTR deletion and TRY SNP-701 (see legend below the map). Number of accessions with Don-0 allele, and statistical significances of the logistic regressions between polymorphisms and spring precipitation, are shown over the map.
Discussion
Evolution of fruit trichome patterning by synergistic mutations in the core regulatory network
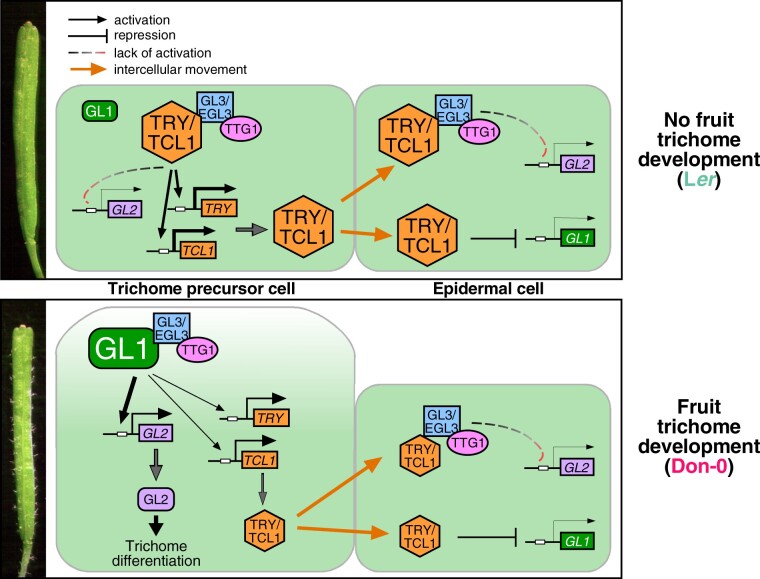
In this study, we demonstrate that Arabidopsis has evolved trichome development in fruits and pedicels, which has not previously been found within the Arabidopsis genus. Genetic analyses showed that the combination of synergistic alleles in three epistatic loci, MAU2/TCL1, MAU3/GL1, and MAU5/TRY, is necessary and sufficient for the evolution of these traits in Don-0 population. These alleles release the leaf trichome program in reproductive organs, as indicated by the branched morphology of trichomes of carpels, fruits and pedicels, in contrast to the simple trichomes of stems and sepals (Hülskamp and Schnittger, 1998; Ó‘Maoiléidigh et al., 2013). The isolation of these MAU loci reveals mutations in the core transcription factors of the regulatory network underlying trichome patterning (Figure 7). In particular, the development of trichomes in fruits involves the concurrence of cis-regulatory mutations causing partial loss of function in two negative regulators, TCL1 and TRY, together with a structural gain-of-function mutation in the positive regulator GL1. The R2R3 MYB transcription factor encoded by GL1 is part of the trimeric complex that activates the expression of GL2, a downstream homeodomain gene inducing trichome differentiation (Ishida et al., 2008; Pattanaik et al., 2014). The C-terminal region of GL1 contains the transcription-activating domain (Wang and Chen, 2008), supporting the conclusion that the GL1-Don-0 nonsense mutation may alter the transcriptional activation properties. In addition, this carboxy region appears to be a target for GL1 structural evolution, as shown by the large intra- and inter-specific diversity. In contrast to GL1, the function of the small single R3 MYB transcription factors encoded by TCL1 and TRY has been modified through regulatory mutations that reduce their expression in reproductive organs. Both proteins are thought to move intercellularly and negatively regulate trichome differentiation in neighbor epidermal cells by competing with GL1 for GL3/EGL3 binding (Figure 7; Pattanaik et al., 2014; Pesch et al., 2014; Wang and Chen, 2014). However, regulatory mutations in these repressor genes affect different developmental components of trichome repression. This is illustrated by the large additive effect displayed specifically by TCL1-Don-0 allele on pedicel trichome patterning. In addition, TCL1 represses GL1 expression (Wang et al., 2007), whereas we have also found that TCL1 may regulate TRY (Figure 3B) and TRY might affect GL1 expression (Supplemental Figure S2). Regulatory feedbacks among these transcription factors further depend on the plant organ, allele, and/or genetic background, in agreement with the complex regulation of R3 MYB genes (Wang et al., 2008; Pesch et al., 2014; Wang and Chen, 2014). Interestingly, it has been shown that the master regulator of flower organ identity AG suppresses trichome initiation during carpel development by controlling the expression of GL1 and TCL1, but not that of TRY, which represses carpel trichome formation in parallel with AG (Ó’Maoiléidigh et al., 2013). Thus, the multiple mechanisms regulating trichome initiation through the MYB transcription factors GL1, TCL1, and TRY need to be jointly modulated to trigger trichome development in fruits of Arabidopsis. On the contrary, a reduction of only TCL1 expression is sufficient for trichome formation in pedicels, indicating that a weaker repression is acting in this structure.
Figure 7.
Molecular model for the natural variation in the regulation of fruit trichome patterning in Arabidopsis. Panels depict the main regulators of trichome differentiation in epidermal precursor and neighboring cells (reviewed in Pattanaik et al., 2014; Wang and Chen, 2014) of wild accessions without (upper panel), or with (lower panel) trichomes in fruits. The molecular mechanisms underlying natural alleles of TCL1, TRY, and GL1 are represented by the size of protein symbols and transcriptional arrows, larger sizes depicting increased amount (TCL1 and TRY) or protein activity (GL1).
Adaptation of the Arabidopsis relict lineage by evolution of fruit trichome patterning
As shown by the geographic distribution and genomic patterns of populations, the development of trichomes in fruits has specifically evolved in the Iberian relict lineage after its split from the North Africa relicts about 70–45 kya (Durvasula et al., 2017). In contrast to nonrelict accessions often found in agricultural and urban landscapes, relict populations live in less disturbed environments, such as holm oak and pine tree Mediterranean forests (Marcer et al., 2016; 1001 Genomes Consortium, 2016). Supporting the ancient demographic history of Iberian relicts in this region, relict habitat suitability has been correlated with stable vegetation dynamics since the last glacial maximum and during Holocene (Toledo et al., 2020). Hence, the evolution of these new fruit trichome patterns in relict populations further supports an old adaptive history of this lineage, while it provides mechanisms for its adaptation. In addition, comparisons of traits, genes, and climate variables showed significant associations suggesting that trichomes in fruits and pedicels might be involved in local adaptation to low precipitation during Arabidopsis reproductive season. Accordingly, trichomes might be maintained in these populations as fruit protection from water stress or defense against herbivores showing a climate-dependent distribution. We cannot, nevertheless, discard that these traits are neutral in their populations, but most likely they are selected against in other locations due to trade-offs between trichome formation and fitness related traits (Mauricio, 1998; Züst and Agrawal, 2017). Demonstration of the ecological role of trichome development in fruits and pedicels awaits further studies addressing their effects on fitness under natural conditions differing in potential selective forces, such as water availability, temperature stress, or herbivory.
Several results indicate that local adaptation mediated by fruit trichome patterning likely occurred from standing genetic variation in TCL1 and TRY, together with new mutations in GL1. First, Don-0 alleles at TCL1 and TRY segregate among relict accessions without trichomes in fruits (Figure 6C), which is in agreement with the trigenic epistasis underlying fruit trichome development. Although the causal mutations at each gene might be neutral when they are separated, the TCL1-Don-0 allele is probably maintained by natural selection acting on the presence of trichomes in pedicels. Second, in contrast to TCL1 and TRY, the premature stop codon of GL1-Don-0 is restricted to two nearby local populations. As expected for local adaptation from new mutations (Barrett and Schluter, 2008), accessions bearing GL1-Don-0 alleles showed a drastic reduction in GL1 nucleotide diversity, which is 10-fold larger than the reduction observed in accessions with Don-0 alleles in TCL1 or TRY (Figure 5).
Furthermore, despite the fact that genetic variation in TCL1, TRY, and GL1 largely accounts for the evolution of fruit trichome patterning in Arabidopsis, our results indicate that other loci also fine tune this trait. In particular, MAU1 and MAU4 will likely explain some relict populations showing trichomes in fruits but carrying Don-0 mutations only in TCL1 or TRY. Moreover, additional alleles of GL1, TCL1, or TRY might affect gene function and contribute to the development of trichomes in fruits and pedicels. This is suggested by the segregation of several GL1 structural polymorphisms specifically among Iberian relict accessions, and by the occurrence of a few accessions showing trichomes in pedicels but lacking TCL1-Don-0 allele. Finally, the development of trichomes in fruits is not an exclusive trait of Arabidopsis, but it has also evolved in plants of multiple dicotyledonous families, including other Brassicaceae (e.g. hoary stock, black mustard), Rosaceae (e.g. peach), or Cucurbitaceae (e.g. cucumber), where MYB transcription factors have been involved in fruit trichome formation (Vendramin et al., 2014; Yang et al., 2018). Hence, future studies are needed to assess the convergence of mechanisms underlying fruit trichome development in different wild and crop species (Serna and Martin, 2006) and to uncover the precise evolutionary processes maintaining this trait in nature.
Materials and methods
Plant material
We analyzed an Arabidopsis collection of 235 genetically distinct wild accessions from the Iberian Peninsula, including 170 previously described genotypes (Manzano-Piedras et al., 2014; Vidigal et al., 2016; Tabas-Madrid et al., 2018) and 65 new samples (Supplemental Data Set 3). Each accession comes from a different georeferenced local population, except a pair that comes from the same location (<1-km distance; Supplemental Data Set 3). Together they span a region of 800 km × 700 km and an altitudinal range between 1 and 2,662-m above sea level. Populations showed an average pairwise distance of 353 ± 196 km, with a minimum and maximum of 1.5 and 1,042 km, respectively. These accessions are publicly available through Nottingham Arabidopsis Stock Centre (NASC; http://arabidopsis.info). In addition, we phenotyped a collection of 185 Arabidopsis accessions from the rest of world, which included most native areas of Eurasia and Africa (Supplemental Data Set 4).
For QTL mapping, we used a population of 375 RILs derived from a cross between Don-0 accession, with trichomes in fruits, and the reference laboratory strain Ler (Mendez-Vigo et al., 2016; Supplemental Data Set 5).
The gl1-1 spontaneous mutant (N1688) and the T-DNA insertion mutants tcl1 (N675198) and try (N6518), all in the Col background, were obtained from NASC. The double mutant tcl1 try was selected from a F2 population derived from a cross between both single mutants, which was genotyped using mutant allele-specific markers (Supplemental Table S2).
Growth conditions and phenotypic analyses
Plants were grown in pots with soil and vermiculite at 3:1 proportion using growth chambers at 21°C and a long-day photoperiod (16 h of cool-white fluorescent light, photon flux of 100 µmol/m2 s).
For phenotypic analyses, all RILs, ILs, accessions, or transgenic lines carrying transgenes of the same gene were grown simultaneously in the same experiment. To this end, we used two (RILs and accessions) or three (ILs and transgenic lines) complete block designs with randomization, each block containing one pot with six plants per line.
Overall, first developed fruits displayed the highest trichome density in valves and pedicels, which decreased in an acropetal fashion. For GWA analysis and QTL mapping, trichome patterning in fruits and pedicels was scored in a qualitative manner. Fruits were classified in three categories, corresponding to absence of trichomes in fruit valves, low trichome density (1–30 trichomes per fruit), and high trichome density (>30 trichomes per fruit), respectively. Similarly, plants were classified according to the absence or the presence of trichomes in the pedicels of all first five fruits or more. In introgression and transgenic lines, trichome patterning was analyzed quantitatively. FTN was scored as the number of trichomes in the two valves of the first fully fertile fruit (positions 1–3 of the main inflorescence), which was counted under a stereomicroscope (×30 magnification). PTP was quantified as the number of pedicels developing trichomes in the first 30 flowers of the main inflorescence.
Cryo-scanning electron microscopy
Carpels and fruits of Ler and Don-0 were analyzed in a cryo-scanning electron microscope (cryo-SEM) Zeiss 960, using the protocol described elsewhere (Ascaso et al., 2003). Briefly, fresh flowers were collected and dissected one day after anthesis and carpels were individually placed on the sample holder of the CT1500 Cryotrans instrument (Oxford). Samples were plunge-frozen immediately in supercooled nitrogen and transferred to the pre-chamber of the microscope precooled at −180°C. Subsequently, samples were sputter-coated with gold and transferred to the cryo-SEM chamber.
QTL mapping and development of ILs
QTL mapping was carried out using qualitative estimates of fruit trichome pattern derived from 5 to 12 plants per RIL (Supplemental Data Set 5). The multiple-QTL-model method was applied as implemented in MapQTL v.4.0 software (van Ooijen, 2000) on the 118 genetic markers of the available genetic map of Ler/Don-0 RIL population (Mendez-Vigo et al., 2016). QTLs were detected with a logarithm of the odss (LOD) threshold of 2.5, which corresponds to a genome-wide significance α = 0.05, as estimated with MapQTL permutation test. The additive allele effects, the percentage of variance explained by each QTL, and the total variance explained by the additive effects of all detected QTL were obtained from the final multiple QTL model.
Fine mapping of MAU5 and MAU3 was carried out using self-progenies of 1,350 and 362 plants derived from RIL-380 and IL-253het, respectively (Supplemental Methods and Supplemental Figure S5).
ILs carrying Don-0 alleles of MAU2, MAU3, or MAU5, in a Ler genetic background, were developed by phenotypic and genotypic selection during recurrent backcrossing of four Ler/Don-0 RILs, to Ler (Supplemental Methods and Supplemental Figure S9). We obtained a total of 21 ILs, which correspond to three independent lines for each of the seven different combinations of one to three MAU genomic regions from Don-0. ILs were named as IL followed by the numbers of the MAU loci carrying Don-0 alleles (IL-2, IL-3, IL-5, IL-23, IL-25, IL-35, and IL-235). An additional IL carrying Don-0 alleles in MAU1, MAU2, MAU3, and MAU5 was obtained and named as IL-1235.
The try mutation from Col accession was introgressed into Ler background by three successive backcross generations using Ler as female parent. In each generation, a heterozygous try plant was selected by genotyping with an allele-specific marker (Supplemental Table S2). A single BC3 plant was then crossed to IL-23 carrying Don-0 alleles at MAU2 and MAU3 in Ler genetic background, to derive a F2BC4(IL-23 × try) population of 344 plants (Supplemental Figure S6). ILs carrying different alleles in MAU2, MAU3, and try were obtained from this population by genotypic selection (Supplemental Table S3).
Genome sequencing, genetic structure, and GWA analyses
DNA for genome sequencing was isolated from mature leaves as previously described (Mendez-Vigo et al., 2016). Genome sequences of 65 wild accessions from the Iberian Peninsula were obtained from paired-end libraries using a HiSeq 3000 sequencer (Illumina, San Diego) through the genomic facility of the Centre for Gene Regulation (https://www.crg.eu/) and are available at NCBI SRA under the BioProject accession number PRJNA646494. These sequences were analyzed by the Service of Bioinformatics for Genomics and Proteomics (CNB-CSIC) together with similar sequences previously generated for the remaining 170 Iberian accessions (1001 Genomes Consortium, 2016; Supplemental Data Set 3). Quality control, SNP calling and genotyping, as well as functional annotation of each genome were carried out following the pipelines described in Tabas-Madrid et al. (2018; Supplemental Methods). Thus, we generated a variant call format (VCF) file with 2186721 SNPs for GWA analyses using 235 Iberian accessions.
The population structure of the 235 Iberian accessions was estimated by the model-based clustering algorithm implemented in ADMIXTURE (Alexander et al., 2009) using the 761133 nonsingleton SNPs with no missing data (Supplemental Methods). In agreement with previous structure studies of Iberian genomes (1001 Genomes Consortium, 2016; Tabas-Madrid et al., 2018), this analysis identified 48 accessions belonging to the highly differentiated genetic group named as relict and 187 genomes fitting into three nonrelict Iberian genetic groups (Supplemental Data Set 3).
GWA analyses were carried out using the standard mixed linear model implemented in Tassel v.5 (Bradbury et al., 2007). The genetic kinship matrix included as covariate to control for population structure was estimated from the proportion of shared alleles (Atwell et al., 2010). A high significance threshold of −log(P)=7.64, corresponding to 5% with Bonferroni correction for multiple tests, was applied to detect associations. Given the low frequency of accessions with trichomes in fruits (10%) or pedicels (15%), an excess of SNPs were found as showing significant associations. Therefore, these analyses were mainly applied to detect candidate genes within QTL mapping intervals. Additional GWA analyses were carried out for TCL1, TRY, and GL1 including the deletions and insertions segregating between Don-0 and Ler as detected by standard Sanger sequencing of parental accessions. Structural variants in these genes were genotyped in the 235 Iberian accessions by reanalyzing the alignments in the nucleotide positions of Don-0/Ler polymorphisms. Allele frequencies of TCL1, TRY, and GL1 polymorphisms were also estimated in the 1,135 worldwide genomes (1001 Genomes Consortium, 2016) and the 80 African genomes (Durvasula et al., 2017) previously sequenced.
The number of genes detected by GWA analyses was derived from positions of significant SNPs, including the two flanking genes when SNPs were located in intergenic regions. Broad sense heritabilities () of trichome traits, explained by the kinship matrix, were estimated by genomic best linear unbiased prediction as implemented in Tassel (Bradbury et al., 2007).
Genomic constructs and transgenic lines
Genomic fragments of 3.8, 6.7, and 5.1 kb from TCL1, TRY, and GL1, respectively, were sequenced from Don-0 and Ler accessions, including the following regions: 2.0, 3.9, and 1.7 kb of promoter and 5′-UTR; 1.2, 1.0, and 1.5 of coding sequences; and 0.6, 1.8, and 1.9 kb of the 3′-regions of each gene, respectively. For sequencing, 6–13 overlapping fragments of 0.8–1.2 kb were PCR amplified (Supplemental Table S2) and products were sequenced using an ABI PRISM 3730xl DNA analyser. DNA sequences were aligned using DNASTAR v.15.0 (Lasergene) and alignments were inspected and edited by hand with GENEDOC (Nicholas et al., 1997).
The six TCL1, TRY, and GL1 genomic fragments sequenced from Don-0 and Ler were cloned in the pCAMBIA 1300 binary vector (CAMBIA, Canberra, Australia) by standard molecular biology techniques. These fragments were PCR amplified using Phusion high fidelity DNA polymerase (New England Biolabs, Beverly, USA), cloned in KpnI/BamHI, SacI/SalI or BamHI/SalI cloning sites, and checked by sequencing (Supplemental Table S2). Two additional TCL1 chimeric constructs were developed by reciprocally changing a 3′-fragment of 1.3 kb released with BglI/XbaI restriction enzymes, between Don-0 and Ler TCL1 constructs.
The eight genomic constructs were transferred by electroporation to AGL0 Agrobacterium tumefaciens strain (Lazo et al., 1991) and plants of the following Arabidopsis genotypes were transformed by the floral dip method: mutant tcl1 and line IL-235 were transformed with the four TCL1 genomic constructs; try mutant, as well as lines IL-235 and IL-1235, were used for the two TRY genomic constructs; mutant lines gl1 and tcl1 try, for the two GL1 genomic constructs. T1 transformants were screened by hygromycin resistance and lines carrying single insertions were selected based on resistance segregation in T2 families. Three to 14 independent homozygous T3 lines were selected for each construct and genetic background, their transgene and endogenous TCL1, TRY, or GL1 alleles being verified by PCR before phenotypic analyses (Supplemental Tables S2 and S3).
Phylogenetic analyses
The intraspecific relationships among TCL1, TRY, or GL1 genomic sequences derived from the 235 Iberian genomes analyzed in this study (Supplemental Data Set 3) were determined by constructing neighbor-joining trees using MEGA v.7 (Tamura et al., 2011) and applying 10,000 bootstrap permutations for statistical significances.
GL1 sequences from Brassicaceae plants were obtained from Phytozome v.12 (https://phytozome.jgi.doe.gov; A. thaliana, A. lyrata, A. halleri, Boechera stricta, Capsella rubella, Capsella grandiflora and Brassica rapa); GenBank (Camelina sativa, Arabis alpina, Brassica napus, and Brassica villosa); or from Cardamine hirsuta genome project (http://chi.mpipz.mpg.de/). The alignment of proteins was obtained with CLUSTAL (https://www.ebi.ac.uk/Tools/msa/clustalo/) and edited by hand with GENEDOC (Nicholas et al., 1997; Supplemental Data Set 6).
Gene expression
To measure the expression of TCL1, TRY, and GL1 genes, plants were grown as described for phenotypic analyses, but pots contained ∼50 or 9 plants for vegetative or reproductive samples, respectively. After sowing, pots were placed at 4°C and short-day photoperiod (8 h light:16 h darkness) for seed stratification. Thereafter, pots were transferred to a growth chamber with long-day photoperiod and 21°C. Genotypes to be compared (parental accessions, ILs, or transgenic lines with transgenes of the same gene) were grown simultaneously in a single experiment, including three pots per genotype (or nine pots for controls), organized in three randomized blocks. For vegetative samples, 14- to 18-day-old rosettes were harvested, whereas for reproductive samples the flower buds of the main inflorescences were collected 3–5 days after flowering initiation (30- to 40-day-old plants depending on the genetic background). Tissue from the three blocks of each genotype was mixed before RNA isolation using TRIzol reagent according to manufacturer’s protocol (Invitrogen). Potential DNA contamination was removed by DNAse digestion and subsequent RNA purification was carried out with high pure RNA isolation kit (Roche). cDNA was synthesized from 3 µg of total RNA using AMV reverse transcriptase (Invitrogen) and dT15 oligonucleotides. TCL1, TRY, and GL1 expressions were analyzed by RT-quantitative PCR (Supplemental Table S2). To avoid amplification differences caused by DNA polymorphisms, primers were designed in gene regions carrying no polymorphism among Col, Ler, and Don-0. The genes encoding ubiquitin-conjugating enzyme 21 (UBC; At5g25760) or protein phosphatase 2A (PP2A; At1g13320), described as highly steady among organs and environments (Czechowski et al., 2005), were used as endogenous control for RNA sample standardization (Supplemental Table S2). All genes were amplified with Power SYBR green mix in a 7300 real-time PCR system (Applied Biosystem) and quantified using the standard curve method. Mean and standard errors were derived from three biological replicates (RNA isolated from plants grown in different pots) for parental accessions and ILs, or from three technical replicates (RT-quantitative PCR wells from the same cDNA sample) for transgenic lines.
Environmental and statistical analyses
A total of 87 environmental variables were obtained from a geographic information system previously developed for the Iberian collection of Arabidopsis accessions (Manzano-Piedras et al., 2014), which contains climate, habitat, and soil pH variables from population locations. The relationship between qualitative (binary) trichome traits and the environmental variables was tested by autologistic regression models (Dormann, 2007) using SAM software v.3.1 (Rangel et al., 2010). These models follow the principles of classical logistic regression but include an additional explanatory variable, the autocovariate, to correct for the effect of spatial autocorrelation of the response variable. Autocovariate values depend on the response variable and the geographical distances among observations (Dormann, 2007). Thus, standardized regression coefficients were estimated for the environmental variable and the spatial autocorrelation in the same model. The overall fit of the autologistic model is given as a pseudo R2, which corresponds to the estimated McFadden’s Rho-squared value (×100). Regression models were applied to the complete Iberian collection and to the set of relict populations (Supplemental Data Set 2). The association between binary nucleotide polymorphisms and environmental variables was tested similarly (Supplemental Data Set 2).
The associations between the most significant environmental variables (spring precipitation as dependent variable) and trichome traits (explanatory variable) were also tested by GLMs that include the ancestry membership to genetic groups as covariates to reduce the confounding effect of genetic structure. To ensure independence, only three out of the four genetic groups detected with Admixture were used as covariates in these models.
Gene–environment association analyses were also carried out by applying the LFMM method (Caye et al., 2019) on spring precipitation variables and the genome-wide data set of 2186721 SNPs described above. In the LFMM method, the allele frequency at a locus is the dependent variable explained by a fixed environmental factor and by the random effects of hidden (latent) factors representing residual levels of population structure (François et al., 2016). LFMMs were applied using the LFMM 2 method as implemented in the R package lffm, with K = 4 latent factors because this is the number of genetic groups estimated in the Iberian collection. Adjusted P-values were obtained for each SNP using the genome inflation factor method, and Benjamini–Hochberg algorithm was applied at a false discovery rate (FDR) = 0.1 to correct for multiple testing (François et al., 2016). This resulted in a significance threshold of −log(P)=4.86 for detection of potential environmental associations. The number of genes affected by associated SNPs was estimated as described for GWA analyses.
Phenotypic and gene expression differences between accessions and organs were tested by mixed GLMs including genotypes and organs as fixed-effect factors and replicates as random effect factor. Differences between transgenic lines were also tested by mixed GLMs including transgenes as fixed factor, and lines (nested within transgenes) as random factor (Supplemental Data Set 1). In ILs, the additive and interaction effects of MAU2, MAU3, and MAU5 loci were tested by GLMs including the three loci as fixed effect factors. MAU2, MAU3, and try effects were tested similarly in the F2BC4(IL-23 × try) and derived ILs. These analyses were carried out with the statistical packages SPSS v.24 or Statistica v.8.
Accession numbers
Raw sequences of the 65 Arabidopsis genomes generated in this article are available at NCBI SRA repository under the BioProject accession number PRJNA646494. Gene sequence data from this article can be found in the GenBank/EMBL libraries under the accession numbers MT439837–MT439842.
Supplemental Data
Supplemental Figure S1. Genome-wide association analysis of fruit trichome pattern.
Supplemental Figure S2. tcl1, try, and gl1-1 mutants used in this study.
Supplemental Figure S3. Expression of TCL1, TCL2, and ETC2 in parental accessions.
Supplemental Figure S4. Phenotypes and gene expressions in transgenic lines for TCL1, TRY, and GL1.
Supplemental Figure S5. Fine mapping of MAU5 and MAU3.
Supplemental Figure S6. Frequency distribution of fruit trichome number in a F2BC4(IL-23 x try) population.
Supplemental Figure S7. Sequence alignment of GL1 proteins from Brassicaceae.
Supplemental Figure S8. Relationship between trichome pattern and climate in relict accessions.
Supplemental Figure S9. Development of MAU introgression lines.
Supplemental Table S1. Phenotypic and geographic distribution of TCL1, TRY, and GL1 polymorphisms.
Supplemental Table S2. Oligonucleotides used for TCL1, TRY, and GL1 sequencing, cloning, genotyping, and expression analyses.
Supplemental Table S3. Molecular genetic markers used for the development of introgression lines, fine mapping, and validation of transgenic lines.
Supplemental Methods. Fine mapping, development of ILs, and genome sequence analyses.
Supplemental Data Set 1. Statistical analyses in parental, introgression, and transgenic lines.
Supplemental Data Set 2. Association analyses between environmental variables and trichome traits or nucleotide polymorphisms.
Supplemental Data Set 3. Information on the Iberian Arabidopsis accessions.
Supplemental Data Set 4. Information on worldwide Arabidopsis accessions.
Supplemental Data Set 5. Fruit trichome pattern of the Ler/Don-0 RIL population.
Supplemental Data Set 6. Alignment of GL1 proteins from Brassicaceae.
Supplementary Material
Acknowledgements
Authors thank Mercedes Ramiro for technical assistance. A.F.-P. was recipient of a PhD fellowship (BES-2017-080063) from the Ministerio de Economía, Industria y Competitividad (MINECO) of Spain.
Funding
This work has been funded by grants PID2019-104249GB-I00 and BIO2016-75754-P from the Agencia Estatal de Investigación of Spain (AEI)/10.13039/501100011033 and FEDER (UE) to C.A.-B.
Conflict of interest statement. None declared.
C.A.-B. conceived the project and designed the research. N.A., M.S., and B.M.-V. carried out most of the experiments. R.T.-P. and J.C.O. analyzed the Iberian genome sequences. A.F.-P. participated in the genome-wide association assays. F.X.P. carried out the environmental analyses. C.A.-B. wrote the article with inputs from all authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) is: Carlos Alonso-Blanco (calonso@cnb.csic.es).
References
- Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz I, O’Kane SL (2002) Taxonomy and phylogeny of Arabidopsis (Brassicaceae). Arabidopsis Book, doi: 10.1199/tab.0001. Published September 30, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascaso C, Souza-Egipsy V, Sancho LG (2003) Locating water in the dehydrated thallus of lichens from extreme microhabitats (Antarctica). Lichenologica 86: 213–223 [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed]
- Balkunde R, Pesch M, Hulskamp M (2010). Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr Top Dev Biol 91: 299–321 [DOI] [PubMed] [Google Scholar]
- Barboza L, Effgen S, Alonso-Blanco C, Kooke R, Keurentjes JJ, Koornneef M, Alcazar R (2013). Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proc Natl Acad Sci USA 110: 15818–15823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Schluter D (2008) Adaptation from standing genetic variation. Trends Ecol Evol 23: 38–44 [DOI] [PubMed] [Google Scholar]
- Bickford CP (2016) Ecophysiology of leaf trichomes. Funct Plant Biol dx.doi.org/10.1071/FP16095. Published June 3, 2016 [DOI] [PubMed] [Google Scholar]
- Bloomer RH, Juenger TE, Symonds VV (2012) Natural variation in GL1 and its effects on trichome density in Arabidopsis thaliana. Mol Ecol 21: 3501–3515 [DOI] [PubMed] [Google Scholar]
- Bloomer RH, Lloyd AM, Symonds VV (2014) The genetic architecture of constitutive and induced trichome density in two new recombinant inbred line populations of Arabidopsis thaliana: phenotypic plasticity, epistasis, and bidirectional leaf damage response. BMC Plant Biol 14: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Caye K, Jumentier B, Lepeule J, François O (2019) LFMM 2: fast and accurate inference of gene-environment associations in genome-wide studies. Mol Biol Evol 36: 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalin P, Agren J, Bjorkman C, Huttunen P, Karkkainen K (2008) Leaf trichome formation and plant resistance to herbivory. In, Schaller A, ed, Induced Plant Resistance. Springer. Sttugart, Germany. [Google Scholar]
- Dormann C (2007) Assessing the validity of autologistic regression. Ecol Model 207: 234–242 [Google Scholar]
- Doroshkov AV, Konstantinov DK, Afonnikov DA, Gunbin KV (2019) The evolution of gene regulatory networks controlling Arabidopsis thaliana L. trichome development. BMC Plant Biol 19: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, Tsuchimatsu T, Burbano HA, Pico FX, Alonso-Blanco C, Hancock AM (2017) African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: 5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M, Brennan AC, Alonso-Blanco C, Pico FX (2018) Spatio-temporal variation in fitness responses to contrasting environments in Arabidopsis thaliana. Evolution 72: 1570–1586 [DOI] [PubMed] [Google Scholar]
- François O, Martins H, Caye K, Schoville SD (2016) Controlling false discoveries in genome scans for selection. Mol Ecol 25: 454–469 [DOI] [PubMed] [Google Scholar]
- Fürstenberg-Hägg J, Zagrobelny M, Bak S (2013) Plant defense against insect herbivores. Int J Mol Sci 14: 10242–10297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M (2012) The patterning of epidermal hairs in Arabidopsis—updated. Curr Opin Plant Biol 15: 31–37 [DOI] [PubMed] [Google Scholar]
- Guimil S, Dunand C (2006) Patterning of Arabidopsis epidermal cells: epigenetic factors regulate the complex epidermal cell fate pathway. Trends Plant Sci 11: 601–609 [DOI] [PubMed] [Google Scholar]
- Hauser MT (2014) Molecular basis of natural variation and environmental control of trichome patterning. Front Plant Sci 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Harr B, Schlotterer C (2001) Trichome distribution in Arabidopsis thaliana and its close relative Arabidopsis lyrata: molecular analysis of the candidate gene GLABROUS1. Mol Biol Evol 18: 1754–1763 [DOI] [PubMed] [Google Scholar]
- Hilscher J, Schlotterer C, Hauser MT (2009) A single amino acid replacement in ETC2 shapes trichome patterning in natural Arabidopsis populations. Curr Biol 19: 1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Schnittger A (1998) Spatial regulation of trichome formation in Arabidopsis thaliana. Semin Cell Dev Biol 9: 213–220 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Judd WS, Campbell CS, Kellogg EA, Stevens PF (1999) Plant Sytematics, A Phylogenetic Approach, Sinauer Assocates, Massachusetts, USA [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Li F, Zou Z, Yong HY, Kitashiba H, Nishio T (2013) Nucleotide sequence variation of GLABRA1 contributing to phenotypic variation of leaf hairiness in Brassicaceae vegetables. Theor Appl Genet 126: 1227–1236 [DOI] [PubMed] [Google Scholar]
- Manzano-Piedras E, Marcer A, Alonso-Blanco C, Pico FX (2014) Deciphering the adjustment between environment and life history in annuals: lessons from a geographically-explicit approach in Arabidopsis thaliana. PloS One 9: e87836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcer A, Méndez-Vigo B, Alonso-Blanco C, Picó FX (2016) Tackling intraspecific genetic structure in distribution models better reflects species geographical range. Ecol Evol 26: 2084–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio R (1998) Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am Nat 151: 20–28 [DOI] [PubMed] [Google Scholar]
- Mendez-Vigo B, Savic M, Ausin I, Ramiro M, Martin B, Pico FX, Alonso-Blanco C (2016) Environmental and genetic interactions reveal FLOWERING LOCUS C as a modulator of the natural variation for the plasticity of flowering in Arabidopsis. Plant Cell Environ 39: 282–294 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HBJ, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4: 1–4 [Google Scholar]
- Ó'Maoiléidigh DS, et al. (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen JW (2000) MapQTL Version 4.0: user friendly power in QTL mapping: addendum to the manual of version 3.0. Plant Research International. Wageningen, The Netherlands [Google Scholar]
- Pesch M, Dartan B, Birkenbihl R, Somssich IE, Hulskamp M (2014) Arabidopsis TTG2 regulates TRY expression through enhancement of activator complex-triggered activation. Plant Cell 26: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanaik S, Patra B, Singh SK, Yuan L (2014) An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front Plant Sci 5: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel TF, Diniz-Filho JAF, Bini LM (2010) SAM: a comprehensive application for Spatial Analysis in Macroecology. Ecography 33: 46–50 [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Jurgens G, Hulskamp M (1998) Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125: 2283–2289 [DOI] [PubMed] [Google Scholar]
- Serna L, Martin C (2006) Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci 11: 274–280 [DOI] [PubMed] [Google Scholar]
- Symonds VV, Hatlestad G, Lloyd AM (2011) Natural allelic variation defines a role for ATMYC1: trichome cell fate determination. PLoS Genet 7: e1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas-Madrid D, Mendez-Vigo B, Arteaga N, Marcer A, Pascual-Montano A, Weigel D, Xavier Pico F, Alonso-Blanco C (2018) Genome-wide signatures of flowering adaptation to climate temperature: Regional analyses in a highly diverse native range of Arabidopsis thaliana. Plant Cell Environ 41: 1806–1820 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo B, Marcer A, Méndez-Vigo B, Alonso-Blanco C, Picó FX (2020) An ecological history of the relict genetic lineage of Arabidopsis thaliana. Env Exp Bot 170: 103800 [Google Scholar]
- Tutin TG, Burges NA, Chater AO, Edmondson JR, Heywood VH, Moore DM, Valentine DH, Walters SM, Webb DA, Akeroyd JR, et al. (1993) Flora Europaea, Cambridge University Press [Google Scholar]
- Vendramin E, Pea G, Dondini L, Pacheco I, Dettori MT, Gazza L, Scalabrin S, Strozzi F, Tartarini S, Bassi D, et al. (2014) A unique mutation in a MYB gene cosegregates with the nectarine phenotype in peach. PloS One 9: e90574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal DS, Marques AC, Willems LA, Buijs G, Mendez-Vigo B, Hilhorst HW, Bentsink L, Pico FX, Alonso-Blanco C (2016) Altitudinal and climatic associations of seed dormancy and flowering traits evidence adaptation of annual life cycle timing in Arabidopsis thaliana. Plant Cell Environ 39: 1737–1748 [DOI] [PubMed] [Google Scholar]
- Wagner A (2011) The molecular origins of evolutionary innovations. Trends Genet 27: 397–410 [DOI] [PubMed] [Google Scholar]
- Wang S, Chen JG (2008) Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol 49: 1792–1804 [DOI] [PubMed] [Google Scholar]
- Wang S, Chen JG (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci 5: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG (2007) TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Yang S, Cai Y, Liu X, Dong M, Zhang Y, Chen S, Zhang W, Li Y, Tang M, Zhai X, et al. (2018) A CsMYB6-CsTRY module regulates fruit trichome initiation in cucumber. J Exp Bot 69: 1887–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA (2012) Natural enemies drive geographic variation in plant defenses. Science 338: 116–119 [DOI] [PubMed] [Google Scholar]
- Züst T, Agrawal AA (2017) Trade-offs between plant growth and defense against insect herbivory: an emerging mechanistic synthesis. Ann Rev Plant Biol 68: 513–534 [DOI] [PubMed] [Google Scholar]
- 1001 Genomes Consortium (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.