Abstract
Ethylene is an important phytohormone with pleotropic roles in plant growth, development, and stress responses. ETHYLENE INSENSITIVE2 (EIN2) mediates the transduction of the ethylene signal from the endoplasmic reticulum membrane to the nucleus, where its C-terminus (EIN2-C) regulates histone acetylation to mediate transcriptional regulation by EIN3. However, no direct interaction between EIN2-C and EIN3 has been detected. To determine how EIN2-C and EIN3 act together, we followed a synthetic approach and engineered a chimeric EIN2-C with EIN3 DNA-binding activity but lacking its transactivation activity (EIN2C–EIN3DB). The overexpression of EIN2C–EIN3DB in either wild-type or in the ethylene-insensitive mutant ein3-1 eil1-1 led to a partial constitutive ethylene response. Chromatin immunoprecipitation sequencing showed that EIN2C–EIN3DB has DNA-binding activity, indicating that EIN3DB is functional in EIN2C–EIN3DB. Furthermore, native EIN3 protein levels determine EIN2C–EIN3DB binding activity and binding targets in a positive feedback loop by interacting with EIN2C–EIN3DB to form a heterodimer. Additionally, although EIN3 does not direct affect histone acetylation levels in the absence of EIN2, it is required for the ethylene-induced elevation of H3K14Ac and H3K23Ac in the presence of EIN2. Together, we reveal efficient and specific DNA-binding by dimerized EIN3 in the presence of ethylene to mediate positive feedback regulation, which is required for EIN2-directed elevation of histone acetylation to integrate into an EIN3-dependent transcriptional activation.
The DNA-binding activity and specificity are enhanced by dimerized EIN3, which promotes the ethylene-induced elevation of H3K14Ac and H3K23Ac in the presence of EIN2, through a positive feedback regulation.
Introduction
Ethylene, a gaseous plant hormone, is important for a myriad of physiological and developmental processes including seed germination, plant growth, fruit ripening, organ abscission, and senescence. It is also involved in responses to stresses such as drought, cold, flooding, and infections (Alonso et al., 1999; Pierik et al., 2006; Ju and Chang, 2015). The common aquatic ancestor of plants, which existed about 450 million years ago, possessed an ethylene signaling pathway that was similar to that of modern Arabidopsis (Arabidopsis thaliana; Ju et al., 2015). The receptors for ethylene are located at the endoplasmic reticulum (ER) membrane (Chen et al., 2002). The factors involved in transmission of the signal from the ER membrane to the nucleus have been previously identified based on decades of genetic and molecular biology research (Chang et al., 1993; Kieber et al., 1993; Chao et al., 1997; Hua and Meyerowitz, 1998; Hua et al., 1998; Alonso et al., 1999; Chen et al., 2002; Huang et al., 2003; Grefen et al., 2008; Ju et al., 2012; Qiao et al., 2012; Wang and Qiao, 2019). ETHYLENE INSENSITIVE2 (EIN2) has been shown to be the key mediator in this process (Ju et al., 2012; Qiao et al., 2012). The Arabidopsis EIN2 gene encodes a 1,294 amino acid protein with a predicted 12-fold hydrophobic transmembrane domain at its N terminus that shows a high sequence similarity to the conserved natural resistance-associated macrophage protein family of metal ion transporters (Alonso et al., 1999), although no metal transport activity has been shown for EIN2 to date (Alonso et al., 1999). In the presence of ethylene, the hydrophilic C terminus of EIN2 (EIN2-C) is dephosphorylated and cleaved from the rest of the protein, and cleaved EIN2-C translocates to the P body to suppress the translation of the two F-box proteins EIN3-BINDING F BOX PROTEIN1 (EBF1) and EBF2 (Li et al., 2015; Merchante et al., 2015), and to the nucleus to activate key transcription factors EIN3/EIN3-LIKE1 (EIL1)-dependent transcription (Qiao et al., 2012).
Recent studies revealed that the histone acetylation marks H3K14Ac and H3K23Ac are positively associated with gene expression in the response to ethylene, and that the ethylene-induced change in histone acetylation depends on EIN2 levels (Zhang et al., 2016b; Wang and Qiao, 2019; Wang et al., 2019). By ectopically expressing EIN2-C in an ein2-5 mutant using clustered regularly interspaced short palindromic repeats (CRISPRs) combined with a dead version of the CRISPR-associated nuclease (dCas9), we demonstrated that EIN2-C directly regulates histone acetylation (Zhang et al., 2017). Detailed mechanistic studies showed that in the presence of ethylene, nucleus-localized EIN2-C interacts with EIN2 NUCLEAR-ASSOCIATED PROTEIN1 (ENAP1), a histone-binding protein, to associate with the chromatin of ethylene-responsive genes to regulate acetylation at H3K14 and H3K23, leading to EIN3-dependent transcriptional regulation (Zhang et al., 2016b, 2017).
It is now well established that many of the effects exerted by transcription factors in eukaryotes are through their interaction with co-regulators that modify the chromatin state, which results in a more open (in the case of activation) or a more closed (in the case of repression) conformation. In ethylene signaling, EIN3 and EIL1 are the key transcription factors that are necessary and sufficient for the ethylene response (Chao et al., 1997; Guo and Ecker, 2003; Chang et al., 2013). EIN3 has been shown to form a homodimer in the absence of DNA in vitro, and the EIN3 binding element was identified after analysis of genes such as ETHYLENE RESPONSE FACTOR1 (ERF1) and ETHYLENE RESPONSE DNA-BINDING FACTOR2 (EDF2), whose expression is highly upregulated by ethylene (Ohme-Takagi and Shinshi, 1990; Eyal et al., 1993; Meller et al., 1993; Sessa et al., 1995; Shinshi et al., 1995; Sato et al., 1996; Solano et al., 1998). Our previous study demonstrated that EIN3 is partially required for the EIN2-directed elevation of histone acetylation in response to ethylene. Yet, how EIN3 is involved in EIN2-C-mediated histone acetylation, and how the regulation of histone acetylation integrates into EIN3-dependent transcriptional regulation in response to ethylene is still largely unknown. We have found that mutations in EIN3 lead to a decrease in both histone acetylation levels and the transcriptional activities (Zhang et al., 2016a). Both EIN2-C and EIN3 interact with ENAP1 (Zhang et al., 2016a); however, no direct interaction between EIN2-C and EIN3 has been detected by multiple approaches. Given that both transcriptional regulation and histone acetylation are impaired simultaneously in ein3 mutants, it is not possible to uncouple the function of EIN3 in the regulation of histone acetylation and transcriptional regulation by using ein3 mutants. In this study, to determine how EIN3 functions in EIN2-directed histone acetylation and transcriptional regulation in response to ethylene, we engineered a chimeric protein that fuses EIN2-C to the EIN3 DNA-binding domain (EIN2C–EIN3DB). The overexpression of EIN2C–EIN3DB in wild-type Arabidopsis (Col-0 accession) and ein3-1 eil1-1 seedlings leads to a partial constitutive ethylene response. Chromatin immunoprecipitation sequencing (ChIP-seq) showed that EIN2C–EIN3DB binds DNA in vivo and that native EIN3 protein levels are crucial for EIN2C–EIN3DB binding activity and specificity. Pull-down and co-immunoprecipitation assays also demonstrated that EIN2C–EIN3DB interacts with native EIN3 both in vitro and in vivo, and that the DNA-binding activity of EIN2C–EIN3DB depends on the presence of native EIN3. By examining the levels of H3K14Ac and H3K23Ac in ein3-1 eil1-1 EIN2S645A and ein2-5 EIN3ox seedlings, we found that EIN3 does not directly affect the levels of H3K14Ac and H3K23Ac in the absence of EIN2. However, EIN3 is required for the ethylene-induced elevation of H3K14Ac and H3K23Ac mediated by EIN2-C. In summary, in the presence of ethylene, efficient and specific DNA binding by a potentially dimerized EIN3 mediates a positive feedback regulation, which is required for EIN2-directed elevation of histone acetylation at H3K14Ac and H3K23Ac to integrate into an EIN3-dependent transcriptional regulation.
Results
A fusion of EIN2-C and the EIN3 DNA-binding domain can regulate the ethylene response
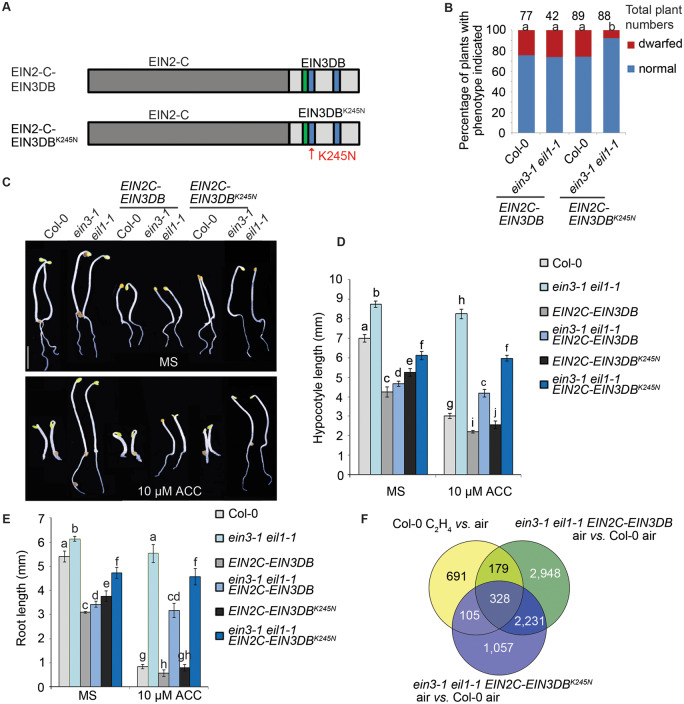
It has been proposed that transcription factors guide chromatin remodeling complexes to certain regions of the genome through their sequence-specific DNA-binding domains. However, how exactly EIN3 functions in the EIN2-dependent regulation of histone acetylation in response to ethylene is unknown. Our previous study has shown that both EIN2-C and EIN3 interact with ENAP1 (Zhang et al., 2016a), yet no direct interaction between EIN2-C and EIN3 has been detected by different approaches. To investigate whether and how the EIN3 DNA-binding domain plays such a role, we fused the sequence encoding the EIN3 DNA-binding domain (EIN3DB) with the sequence encoding EIN2-C in both orientations (EIN2C–EIN3DB and EIN3DB–EIN2C; Figure 1A;Supplemental Figure 1A). Additionally, EIN2-C was fused with a EIN3DB point mutant carrying a K to N mutation at amino acid 245 (K245N) that completely abolishes EIN3 DNA-binding activity (Chao et al., 1997; Solano et al., 1998) in both orientations (EIN2C–EIN3DBK245N and EIN3DBK245N–EIN2C; Figure 1A;Supplemental Figure 1A). We then introduced all constructs into Col-0 and into the ethylene insensitive mutant ein3-1 eil1-1, which lacks both EIN3 and EIL1 (Alonso et al., 2003). About 20% of T1 plants that carried wild-type EIN3DB (EIN2C–EIN3DB or EIN3DB–EIN2C) in both backgrounds showed a constitutive ethylene-responsive adult plant phenotype, as did about 20% of T1 plants that carried EIN3DBK245N (EIN2C–EIN3DBK245N or EIN3DBK245N–EIN2C) in Col-0 (Figure 1B;Supplemental Figure 1B). However, only 10% of T1 plants with the EIN3DBK245N transgenes (EIN2C–EIN3DBK245N and EIN3DBK245N–EIN2C) showed a constitutive ethylene-responsive phenotype in the ein3-1 eil1-1 background (Figure 1B;Supplemental Figure 1B). Since EIN3DB–EIN2C and EIN2C–EIN3DB resulted in similar phenotypes with similar percentages of dwarf T1 plants (Figure 1B;Supplemental Figure 1B), we performed subsequent analyzes using EIN2C–EIN3DB and EIN2C–EIN3DBK245N.
Figure 1.
Overexpression of engineered EIN2C–EIN3DB leads to a partial constitutive ethylene-responsive phenotype. (A) Diagrams showing the fusion of EIN2C with EIN3DB. Green indicates the proline-rich region; blue indicates the second and third basic motifs in EIN3DB. (B) The ratio of adult EIN2C–EIN3DB and ein3-1 eil1-1 EIN2C–EIN3DB plants that displayed a dwarf phenotype. Eight-week-old plants were grown under 16 h light and 8 h dark before investigation. Different letters indicate significant differences, determined by hypergeometric probability (P <0.05). (C) The phenotypes of 3-day-old Col-0 and ein3-1 eil1-1 seedlings that express EIN2C–EIN3DB or EIN2C–EIN3DBK245N. Seedlings were grown on MS medium with or without 10-μM ACC. Bar indicates 2.5 mm. (D and E) Lengths of (D) hypocotyls and (E) roots of 3-day-old etiolated seedlings of the indicated phenotypes. Data are means ± SD of at least 30 seedlings. Different letters indicate significant differences, determined using one-way ANOVA (Analysis of Variation) Scheffe method (P < 0.05). (F) Venn diagrams showing ethylene-regulated genes that are differentially expressed in ein3-1 eil1-1 EIN2C–EIN3DB or ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings compared to Col-0. Total RNA was prepared from 3-day-old etiolated seedlings treated with air. Differentially expressed genes were identified by reads per kilobase per million reads (RPKM) ≥1 with twofold change and q ≤ 0.05.
We then obtained homozygous transgenic lines and examined the phenotypes of 3-day-old etiolated seedlings. Consistent with our observations in T1 transgenic plants, overexpression of EIN2C–EIN3DB or EIN2C–EIN3DBK245N led to a partial ethylene-responsive phenotype in Col-0 and ein3-1 eil1-1 seedlings (Figure 1, C–E;Supplemental Figure S1, C–E). Interestingly, although EIN2C–EIN3DB and EIN2C–EIN3DBK245N protein levels were comparable in both Col-0 and ein3-1 eil1-1 (Supplemental Figure 1F), the partial constitutive ethylene-responsive phenotype observed in EIN2C–EIN3DBK245N overexpression lines was weaker than that resulting from the overexpression of EIN2C–EIN3DB in both Col-0 and ein3-1 eil1-1 (Figure 1, C–E;Supplemental Figure 1, C–E). Furthermore, the phenotype caused by the overexpression of EIN2C–EIN3DB or EIN2C–EIN3DBK245N in ein3-1 eil1-1 was weaker than in Col-0 (Figure 1, C–E;Supplemental Figure 1, C–E). To further confirm if the phenotype of ein3-1 eil1-1 EIN2C–EIN3DBK245N and ein3-1 eil1-1 EIN2C–EIN3DB is ethylene related, we conducted a transcriptome deep sequencing analysis. Compared to the levels in Col-0 seedlings, more genes were differentially expressed in ein3-1 eil1-1 EIN2C–EIN3DB seedlings (5,686 genes) than in ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings (3,721 genes). About 40% of all ethylene-regulated genes evaluated had an altered expression level in ein3-1 eil1-1 EIN2C–EIN3DB seedlings (Figure 1F), as compared to 33% in ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings (Figure 1F). When further compared to ein3-1 eil-1, we found 22% of ethylene-regulated genes with an altered expression in ein3-1 eil1-1 EIN2C–EIN3DB seedlings (Supplemental Figure 1, G–J), against only 12% in ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings (Supplemental Figure 1, G–J). Most of the shared genes in ein3-1 eil1-1 EIN2C–EIN3DB vs. ein3-1 eil1-1 were regulated in the same direction as those responding to ethylene in Col-0 (Supplemental Figure 1, H–J). These analyses confirmed the ethylene-responsive phenotype of ein3-1 eil1-1 EIN2C–EIN3DB and ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings at a molecular level. The fact that fewer genes were differentially expressed upon expression of EIN2C–EIN3DBK245N than upon expression of EIN2C–EIN3DB may possibly be due to the interruption of EIN3 DNA-binding activity, which could potentially lead to a weaker ethylene-responsive phenotype associated with the overexpression of EIN2C-EIN3DBK245N (Figure 1F;Supplemental Figure 1, G–J).
EIN2C–EIN3DB retains DNA-binding activity and is enhanced by native EIN3
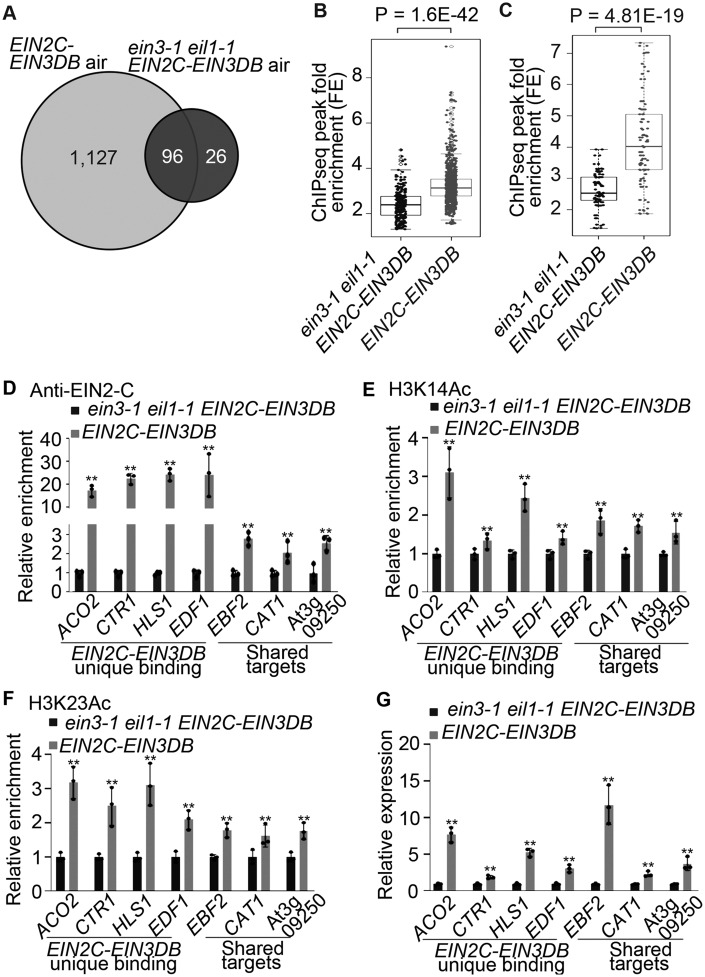
To evaluate EIN3 DNA-binding ability in the context of the engineered EIN2C–EIN3DB protein, we conducted chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) using an anti-EIN2-C antibody in EIN2C–EIN3DB (in Col-0), ein3-1 eil1-1 EIN2C–EIN3DB, EIN2C–EIN3DBK245N (in Col-0), and ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings treated with air (Figure 2A). We detected more than 1,000 genes whose promoters were bound by EIN2C–EIN3DB in EIN2C–EIN3DB seedlings (Figure 2A). In contrast, only about 100 genes had their promoters bound by EIN2C–EIN3DB in ein3-1 eil1-1 EIN2C–EIN3DB seedlings (Figure 2A). The EIN2C–EIN3DB ChIP-seq signals from EIN2C–EIN3DB seedlings were significantly higher than those from ein3-1 eil1-1 EIN2C–EIN3DB seedlings (Figure 2B). We obtained a similar result when comparing the ChIP-seq signals in shared peaks between the two genotypes (Figure 2C). No ChIP signals were detected in EIN2C–EIN3DBK245N or ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings. These results suggest that the DNA-binding domain of EIN2C–EIN3DB has DNA-binding activity and that K245 is necessary for this activity, which is consistent with previous studies (Chao et al., 1997; Solano et al., 1998). Our data also suggest that endogenous EIN3 potentially regulates EIN2C–EIN3DB binding activity.
Figure 2.
Endogenous EIN3 protein regulates EIN2C–EIN3DB binding activity, histone acetylation, and gene expression. (A) Venn diagram showing EIN2C–EIN3DB-bound targets in the Col-0 and ein3-1 eil1-1 backgrounds. ChIP-seq was conducted on 3-day-old etiolated EIN2C–EIN3DB and ein3-1 eil1-1 EIN2C–EIN3DB seedlings treated with air. (B and C) Boxplots and dotplots of peak signals in EIN2C–EIN3DB and ein3-1 eil1-1 EIN2C–EIN3DB seedlings determined by analyses of (B) signals from the peaks identified in EIN2C–EIN3DB and ein3-1 eil1-1 EIN2C–EIN3DB or (C) signals from shared peaks. P-value was calculated by one-way ANOVA and indicated in the figure. (D–F) ChIP-qPCR assays of (D) the binding of EIN2C–EIN3DB, (E) H3K14Ac levels, and (F) H3K23Ac in selected loci from 3-day-old etiolated seedlings of the indicated backgrounds. Data represent relative fold changes. G, RT-qPCR quantification of gene expression in 3-day-old etiolated seedlings. Shared targets indicate binding targets that are shared between ein3-1 eil1-1 EIN2C–EIN3DB and EIN2C–EIN3DB. Each ChIP-qPCR or RT-qPCR was repeated at least three times. Values are means ± SD of three biological replicates. **P< 0.05 when compare to the control in Col-0 background by one-tailed unpaired t test.
These results, however, also raised a question: how can overexpression of EIN2C–EIN3DBK245N, which has no DNA-binding activity, cause a weak ethylene responsive phenotype even in the absence of EIN3 and EIL1? We speculated that both EIN2C–EIN3DB and EIN2C–EIN3DBK245N associate with ENAP1, allowing EIN2-C to regulate histone acetylation above a basal level, therefore leading to an alteration in the expression of a subset of ethylene-responsive genes. To test this hypothesis, we examined the interaction between ENAP1 with EIN2C–EIN3DB and ENAP1 with EIN2C–EIN3DBK245N. As expected, we detected an interaction between ENAP1 and EIN2C–EIN3DB and with EIN2C–EIN3DBK245N by yeast two hybrid, with the interaction between EIN2-C and ENAP1 serving as a positive control (Supplemental Figure 2A). We also compared the binding targets of EIN2C–EIN3DB and ENAP1: about 41% of EIN2C–EIN3DB binding targets overlapped with those of ENAP1 (Supplemental Figure 2B), which further validated our hypothesis.
As more genes were bound by EIN2C–EIN3DB in Col-0 than in ein3-1 eil1-1 seedlings, we speculated that the native EIN3 protein enhances the binding activity of EIN2C–EIN3DB. To test this idea, we compared EIN2C–EIN3DB ChIP-seq signals in Col-0 with those of ein3-1 eil1-1 seedlings. There were significantly more binding targets identified in Col-0 seedlings (1,223) than in ein3-1 eil1-1 seedlings (122; Figure 2A). A chromatin immunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR) assay further confirmed that there was increased enrichment of EIN2C–EIN3DB binding detected in the Col-0 background relative to ein3-1 eil1-1 (Figure 2D). This observation strongly indicates that native EIN3 protein enhances EIN2C–EIN3DB binding.
Given the fact that EIN2-C is the key factor that regulates histone acetylation during the ethylene response (Zhang et al., 2017) and that EIN2C–EIN3DB confers an ethylene-responsive phenotype, we then conducted ChIP-qPCR of H3K14Ac and H3K23Ac to examine the association between EIN2C–EIN3DB binding and histone acetylation levels. The levels of H3K14Ac and H3K23Ac were higher in EIN2C–EIN3DB seedlings than in ein3-1 eil1-1 EIN2C–EIN3DB seedlings (Figure 2, E and F). Furthermore, reverse transcription (RT-qPCR) showed that gene expression is positively correlated with the levels of H3K14Ac and H3K23Ac (Figure 2G). These data suggest that native EIN3 protein regulates EIN2C–EIN3DB binding and that the levels of H3K14Ac and H3K23Ac are positively associated with EIN2C–EIN3DB binding activity.
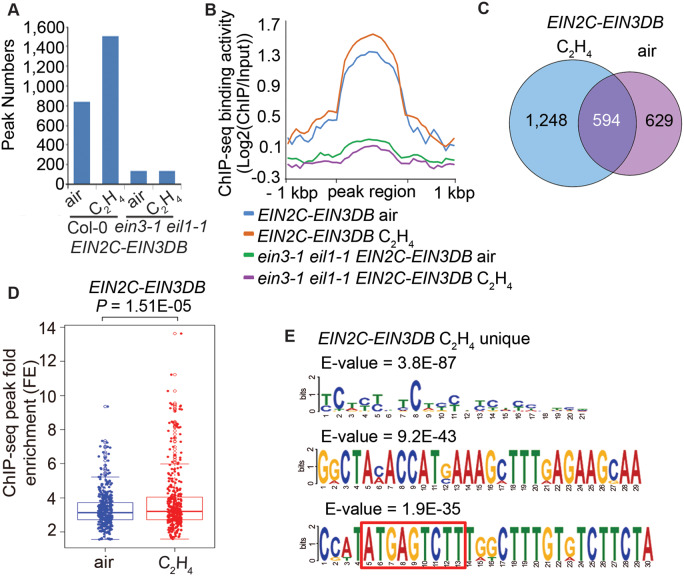
To further confirm the roles of native EIN3 protein in the regulation of EIN2C–EIN3DB binding activity, we compared the binding of EIN2C–EIN3DB under three conditions: in ein3-1 eil1-1 EIN2C–EIN3DB seedlings without native EIN3 protein; in EIN2C–EIN3DB seedlings without ethylene treatment, corresponding to EIN3 basal levels; in EIN2C–EIN3DB after 4 h of ethylene treatment, when EIN3 has accumulated (Guo and Ecker, 2003; Potuschak et al., 2003). We detected about 1,000 peaks in EIN2C–EIN3DB seedlings without ethylene treatment and about 1,500 binding peaks after ethylene treatment (Figure 3, A and B). We obtained a similar number of binding peaks (∼130) in ein3-1 eil1-1 EIN2C–EIN3DB seedlings with or without ethylene treatment, and differences in peak numbers or in ChIP-seq signals were not significant (Figure 3, A and B). The peak numbers and ChIP-seq signals were thus significantly higher in EIN2C–EIN3DB seedlings than in ein3-1 eil1-1 EIN2C–EIN3DB seedlings (Figure 3, A and B). We then compared the EIN2C–EIN3DB binding targets in Col-0 seedlings with and without ethylene treatment (Figure 3C). Almost 50% of genes bound by EIN2C–EIN3DB without ethylene treatment were also bound by EIN2C–EIN3DB with ethylene treatment (Figure 3C), and the enrichment of EIN2C–EIN3DB ChIP-seq signals were significantly higher with ethylene treatment than in the absence of ethylene treatment (Figure 3D), which indicates that EIN2C–EIN3DB binding strength is positively correlated with native EIN3 protein levels.
Figure 3.
EIN2C–EIN3DB binding activity is positively associated with endogenous EIN3 protein levels. (A) Number of EIN2C–EIN3DB ChIP peaks identified from 3-day-old EIN2C–EIN3DB and ein3-1 eil1-1 EIN2C–EIN3DB etiolated seedlings treated with or without ethylene. (B) ChIP-seq signals of 1-kbp upstream and downstream of peak regions from EIN2C–EIN3DB and ein3-1 eil1-1 EIN2C–EIN3DB seedlings treated with or without ethylene. (C) Venn diagrams showing the number of EIN2C–EIN3DB-bound genes in EIN2C–EIN3DB seedlings with and without ethylene treatment. (D) Boxplots and dotplots of ChIP-seq signal in EIN2C–EIN3DB seedlings with and without ethylene treatment. P-value was calculated by one-way ANOVA and indicated in the figure. (E) DNA-binding motifs identified from EIN2C–EIN3DB-bound regions uniquely targeted under ethylene treatment. E-values are indicated. Motifs were identified using MEME-ChIP (http://meme-suite.org/tools/meme-chip) by the analysis of 300-bp centered on each EIN2C–EIN3DB binding peak. Red box highlights the motif with similarity to the known EIN3 binding motif. The canonical EIN3 binding motif sequence is: (A/T)(T/C)G(A/C/T)A(T/C/G)(C/G)T(T/G).
We then performed several motif searches: using the peak sequences bound by EIN2C–EIN3DB in Col-0 seedlings without ethylene treatment; with the peak sequences from the shared targets with ethylene treatment; with the peak sequences from the shared targets without ethylene treatment; and with the peak sequences specifically after ethylene treatment. No classical EIN3 binding element was identified from the peaks without ethylene treatment or from the peaks shared between with and without ethylene treatments (Supplemental Figure 3, A–C). However, we did identify the previously established EIN3 binding element from the peaks sequences that specifically appeared upon ethylene treatment (Figure 3E; Chang et al., 2013). Taken together, these data strongly suggest that native EIN3 protein levels regulate EIN2C–EIN3DB binding specificity, and that the positive correlation between EIN3 protein levels and EIN2C–EIN3DB binding activity indicates the existence of a positive feedback regulation in response to ethylene.
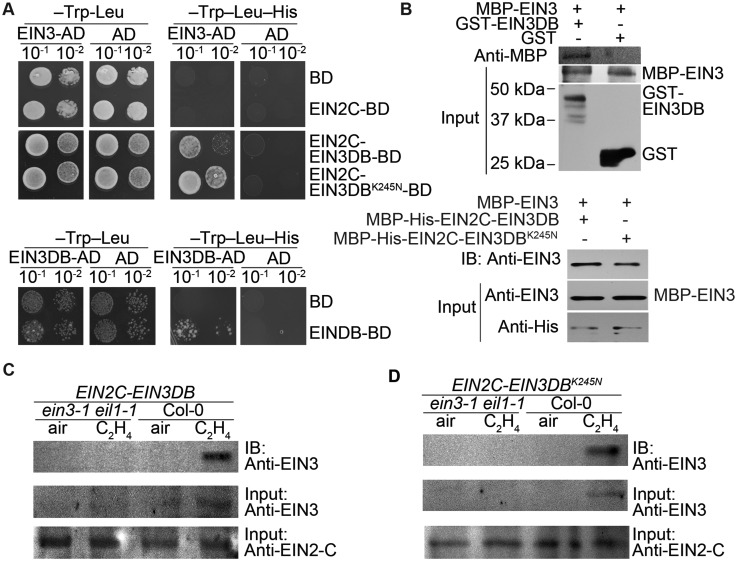
EIN2C–EIN3DB interacts with EIN3 both in vivo and in vitro to form dimers
The positive correlation between EIN2C–EIN3DB binding and native EIN3 protein levels led us to speculate that native EIN3 interacts with EIN2C–EIN3DB. To test this hypothesis, we conducted yeast two-hybrid assays and in vitro pull-down assays. Both EIN2C–EIN3DB and EIN2C–EIN3DBK245N interacted with EIN3 (Figure 4A, upper panel), demonstrating that the point mutation in the EIN3 DNA-binding domain has no effect on the interaction between EIN2C–EIN3DB and EIN3. Interestingly, we found that EIN3DB can form a homodimer (Figure 4A, lower panel), which indicates that the EIN3DB moiety may potentially mediate the interaction between EIN3 and EIN3DB, although no such interaction was detected by yeast two-hybrid assay (Supplemental Figure 4A). To further test the interaction between EIN3 and EIN3DB, we conducted a pull-down assay using recombinant maltose-binding protein (MBP)-EIN3 and glutathione S-transferase (GST)-EIN3DB. The result showed that recombinant EIN3 interacts with EIN3DB (Figure 4B, upper panel), suggesting that EIN3DB is sufficient to mediate the interaction between EIN3 and EIN3DB. A previous study has shown that EIN3 can form dimers in yeast two-hybrid assays (Solano et al., 1998). However, it is normally impossible to dissect the contribution of EIN3 dimerization with EIN3 function in vivo because the EIN3 DNA-binding domain overlaps with the dimerization domain. Indeed, when dimerization is abolished, the DNA-binding activity of EIN3 will be largely reduced. However, EIN2C–EIN3DB provides a perfect tool to explore the role of EIN3 dimerization in vivo. We first examined the interaction between EIN3 and EIN2C–EIN3DB or EIN2C–EIN3DBK245N by pull-down assay. Consistent with the yeast two-hybrid result, EIN3 interacted with both EIN2C–EIN3DB and EIN2C–EIN3DBK245N (Figure 4B, a lower panel). We next evaluated these interactions by in vivo immunoprecipitation with an anti-EIN2 antibody in EIN2C–EIN3DB, ein3-1 eil1-1 EIN2C–EIN3DB, EIN2C–EIN3DBK245N, and ein3-1 eil1-1 EIN2C–EIN3DBK245N seedlings both with and without ethylene treatment. We detected an interaction between EIN3 and EIN2C–EIN3DB in EIN2C–EIN3DB seedlings as well as between EIN3 and EIN2C–EIN3DBK245N in EIN2C–EIN3DBK245N seedlings with the presence of ethylene (Figure 4, C and D). These interactions were confirmed by in vivo immunoprecipitation assays in Arabidopsis protoplasts derived from ein2-5 EIN3ox and ein2-5 etiolated seedlings and transfected with 35Spro:EIN2C–EIN3DB or 35Spro:EIN2C–EIN3DBK245N (Supplemental Figure 4, B and C).
Figure 4.
EIN3 interacts with EIN2C–EIN3DB to regulate its binding activity. (A) Yeast two-hybrid assays to examine the interaction between EIN3 and EIN2C–EIN3DB (upper panel), and the dimerization of EIN3DB (lower panel). Yeast cells co-expressing the indicated combinations of constructs were grown on nonselective (left) or selective (right) medium. Different dilutions of cultures indicated in the figure were inoculated on each spot. (B) Pull-down assays to examine the interaction between EIN3 and EIN3DB, EIN2C–EIN3DB, or EIN2C–EIN3DBK245N. Reactions were performed using recombinant MBP-EIN3, GST-EIN3DB, and MBP-His fusion proteins (MBP-His-EIN2C-EIN3DB and MBP-His-EIN2C-EIN3DBK245N). Proteins were separated by SDS–PAGE and detected with anti-EIN3 or anti-MBP antibody. (C and D) In vivo immunoprecipitation assay to examine the interaction between EIN3 and (C) EIN2C–EIN3DB or (D) EIN2C–EIN3DBK245N in total protein extracts from 3-day-old etiolated seedlings of the indicated genotypes treated with or without ethylene.
EIN2 regulates histone acetylation in the absence of EIN3 and EIL1
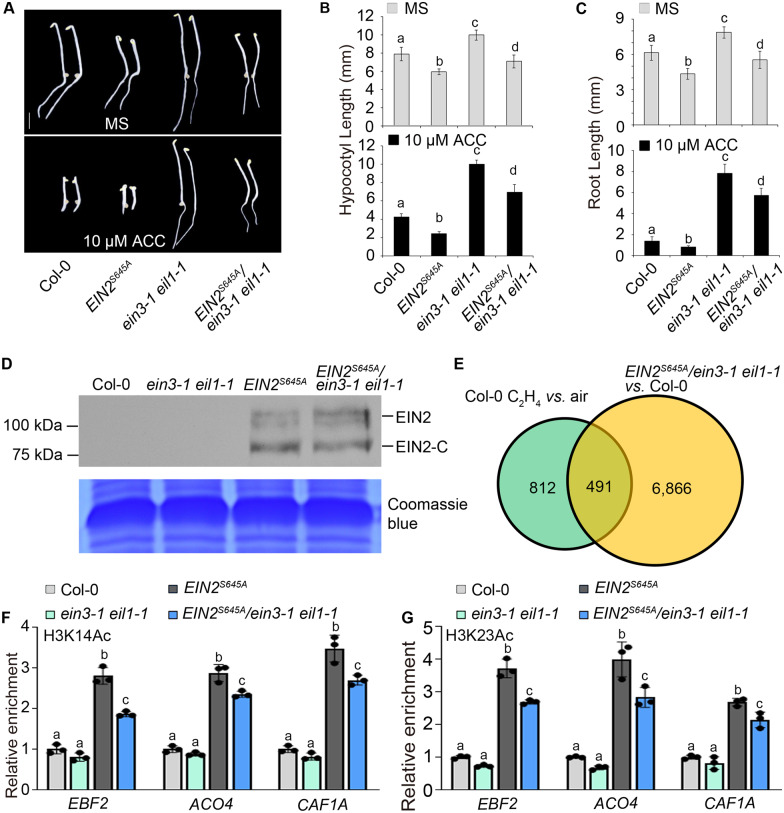
To further investigate the function of EIN3 in EIN2-C-mediated regulation of histone acetylation during ethylene response, we introduced the previously generated EIN2S645Aox transgene, in which the C terminus of EIN2 is constitutively localized to the nucleus and causes a constitutive ethylene-response phenotype (Qiao et al., 2012), into ein3-1 eil1-1 mutants by crossing to generate ein3-1 eil1-1 EIN2S645A. Etiolated seedlings for ein3-1 eil1-1 EIN2S645A displayed a partial ethylene-responsive phenotype in shoots and roots when compared to that of ein3-1 eil1-1 and Col-0 in the absence of ethylene treatment, and the phenotype of ein3-1 eil1-1 EIN2S645A was not altered by ethylene exposure (Figure 5, A–C). Further testing showed that EIN2S645A protein levels were similar in EIN2S645A and in ein3-1 eil1-1 EIN2S645A (Figure 5D). To study the association between the ein3-1 eil1-1 EIN2S645A phenotype and ethylene responsiveness at a molecular level, we analyzed gene expression levels in ein3-1 eil1-1 EIN2S645A and Col-0 seedlings by RNA-seq. About 7,000 genes were differentially expressed in ein3-1 eil1-1 EIN2S645A seedlings when compared to Col-0 (Figure 5E), and about 38% (491/1,303) of ethylene-regulated genes were altered in ein3-1 eil1-1 EIN2S645A seedlings compared to Col-0 seedlings (Figure 5E). When compared to ein3-1 eil1-1, we found that about 5,000 genes were differentially expressed in ein3-1 eil1-1 EIN2S645A seedlings and that about 29% of ethylene-regulated genes were altered in ein3-1 eil1-1 EIN2S645A seedlings (Supplemental Figure 5, A and B). We validated these results by RT-qPCR confirmed for selected genes (Supplemental Figure 5C), showing that sufficient levels of EIN2-C can alter the expression of a subset of ethylene-responsive genes in the absence of EIN3 and EIL1.
Figure 5.
EIN2-mediated elevation of histone modifications is independent of EIN3. (A–C) The seedling phenotypes of Col-0, ein3-1 eil1-1, EIN2S645A, and ein3-1 eil1-1 EIN2S645A etiolated seedlings treated with or without 10-μM ACC. Different letters indicate significant differences determined using one-way ANOVA Scheffe method (P <0.05). Bar indicates 2.5 mm.
We next evaluated H3K14Ac and H3K23Ac levels for the subset of genes that was elevated in ein3-1 eil1-1 EIN2S645A seedlings relative to ein3-1 eil1-1 seedlings. H3K23Ac and H3K14Ac levels were significantly higher in ein3-1 eil1-1 EIN2S645A seedlings than in ein3-1 eil1-1, but lower than in EIN2S645A seedlings (Figure 5, F and G). This result suggests that in the absence of EIN3, EIN2-C acts to elevate H3K14Ac and H3K23Ac levels, while EIN3 is important for ethylene-induced elevation in H3K14Ac and H3K23Ac.
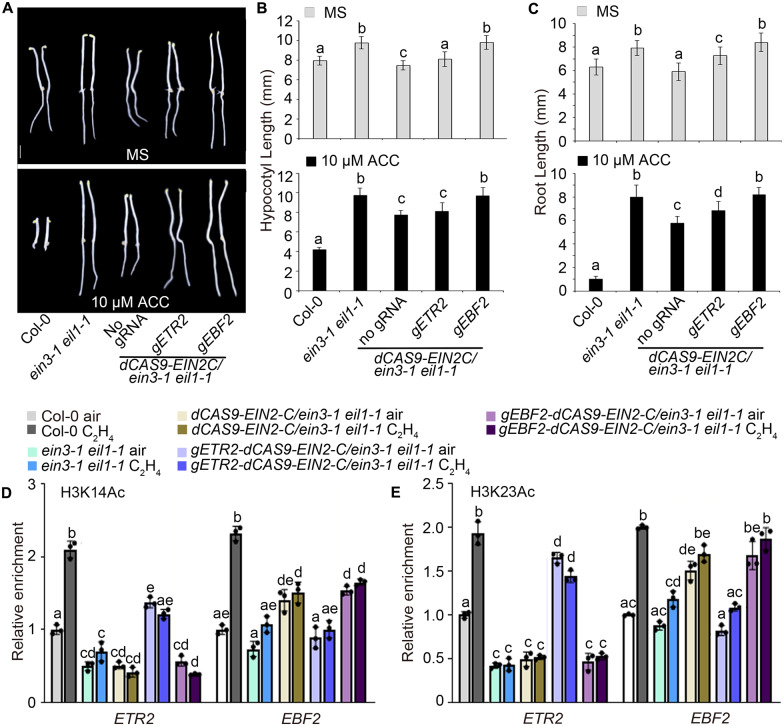
We previously demonstrated that EIN2-C directly regulates histone acetylation by using the gEBF2-dCAS9-EIN2C transgene, in which EIN2-C is targeted to the promotor region of EBF2, a typical ethylene elevated gene, by the gRNA/dCAS9 system (Zhang et al., 2017). To further investigate the roles of EIN3 in the regulation of histone acetylation in the promoter regions of its target genes, we introduced gEBF2-dCAS9-EIN2C and gETR2-dCAS9-EIN2C, derived from another well-known EIN3 target (Chang et al., 2013), into ein3-1 eil1-1 mutants by crossing. We found that in the absence of EIN3 and EIL1, seedlings displayed partial ethylene-responsive phenotypes (Figure 6, A–C; Supplemental Figure 6A), similar to those of gEBF2-dCAS9-EIN2C and gETR2-dCAS9-EIN2C in Col-0 or ein2-5 mutant backgrounds (Zhang et al., 2017). H3K14Ac and H3K23Ac levels were elevated in both ein3-1 eil1-1 gEBF2-dCAS9-EIN2C and ein3-1 eil1-1 gETR2-dCAS9-EIN2C when compared to ein3-1 eil1-1 seedlings (Figure 6, D and E); the expression of genes targeted by the guide RNA was also elevated (Supplemental Figure 6B). There was no significant ethylene-induced elevation in either H3K14Ac or H3K23Ac levels in ein3-1 eil1-1 gEBF2-dCAS9-EIN2C or ein3-1 eil1-1 gETR2-dCAS9-EIN2C seedlings (Figure 6, D and E). This result further confirmed that EIN2-C regulates histone acetylation in the absence of EIN3, while EIN3 is required for EIN2C-mediated elevation of H3K14Ac and H3K23Ac in response to ethylene.
Figure 6.
Histone acetylation induced by EIN2 does not depend on EIN3. (A) The phenotypes of 3-day-old dCas9-EIN2-C/ein3-1 eil1-1, gETR2-dCas9-EIN2-C ein3-1 eil1-1, and gEBF2-dCas9-EIN2-C ein3-1 eil1-1 transgenic etiolated seedlings grown on medium with or without 10-μM ACC. Bar indicates 2.5 mm. (B and C) Lengths of hypocotyls (B) and roots (C) of 3-day-old etiolated seedlings grown with or without 10-μM ACC. Values are means ± sd of at least 30 seedlings. Different letters indicate significant differences determined using one-way ANOVA Scheffe method (P < 0.05). (D–E) Levels of (D) H3K14Ac and (E) H3K23Ac in the guide RNA target loci in 3-day-old etiolated seedlings. Data are fold-changes relative to levels in Col-0 etiolated seedlings grown in air. Each ChIP-qPCR or RT-qPCR was repeated at least three times. Values are means ± sd of three biological replicates. Different letters indicate significant differences determined using one-way ANOVA Scheffe method (P <0.05).
EIN3 does not directly regulate histone acetylation in the absence of EIN2
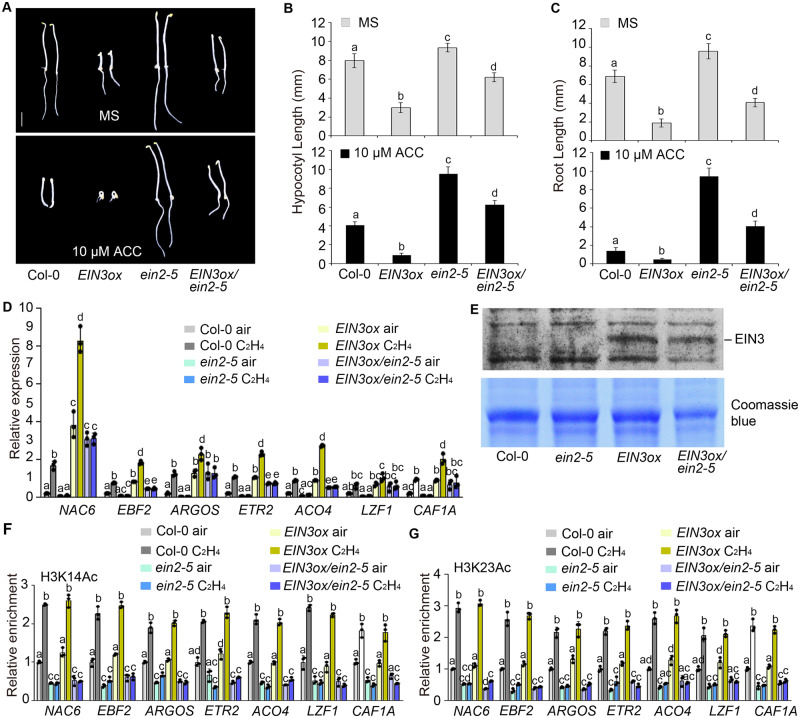
To further study EIN3-regulated histone acetylation in the absence of EIN2, we introduced the EIN3 overexpression transgene in the ein2-5 mutant to generate ein2-5 EIN3ox by crossing. The constitutive ethylene-responsive phenotype of EIN3ox seedlings was partially suppressed by ein2-5. However, these seedlings still displayed a partial ethylene-responsive phenotype when compared to the ein2-5 mutant, whether with or in the absence of ethylene treatment (Figure 7, A–C;Supplemental Figure 7A). The expression of ethylene-responsive genes was significantly reduced in ein2-5 EIN3ox seedlings compared to the levels in EIN3ox seedlings, again with and without ethylene treatment (Figure 7D). In contrast, EIN3 protein levels were comparable in EIN3ox and in ein2-5 EIN3ox seedlings (Figure 7E). These results suggest that EIN2 plays a key role in EIN3-dependent transcriptional regulation.
Figure 7.
EIN3 does not directly regulate histone acetylation in the absence of EIN2. (A) The phenotypes of Col-0, ein2-5, EIN3ox, and ein2-5 EIN3ox etiolated seedlings treated with or without 10-μM ACC. Bar indicates 2.5 mm. (B and C) The lengths of (B) hypocotyls and (C) roots in 3-day-old etiolated seedlings for Col-0, ein2-5, EIN3ox, and ein2-5 EIN3ox grown with or without 10-μM ACC. Values are means ± sd of at least 30 seedlings. Different letters indicate significant differences determined using one-way ANOVA Scheffe method (P <0.05). (D) RT-qPCR analysis of the indicated genes. Total RNAs were extracted from Col-0, ein2-5, EIN3ox, and ein2-5 EIN3ox 3-day-old etiolated seedlings treated with or without ethylene. Different letters indicate significant differences determined using one-way ANOVA Scheffe method (P < 0.05). (E) Immunoblot assay for EIN3 protein levels in Col-0, ein2-5, EIN3ox, and ein2-5 EIN3ox seedlings using anti-EIN3 antibody. Total proteins were extracted from 3-day-old etiolated seedlings. (F and G) ChIP-qPCR assays to examine (F) H3K14Ac and (G) H3K23Ac levels at the indicated loci in 3-day-old etiolated seedlings of Col-0, ein2-5, EIN3ox, and ein2-5 EIN3ox treated with or without ethylene. Data are fold-changes relative to Col-0 grown in air. Values are means ± sd of three biological replicates. Each ChIP-qPCR or RT-qPCR was repeated at least three times. Different letters indicate significant differences determined using one-way ANOVA Scheffe method (P < 0.05).
We then examined H3K14Ac and H3K23Ac levels in the promoters of ethylene-responsive genes. The H3K14Ac and H3K23Ac levels in ein2-5 EIN3ox seedlings were reduced when compared to those in EIN3ox seedlings, while the levels were comparable to that in ein2-5 (Figure 7, F and G), demonstrating that histone acetylation levels are not directly correlated with EIN3 protein levels in the absence of EIN2. Taken together, these results indicate that EIN3 is not sufficient to regulate histone acetylation in the absence of EIN2. However, EIN3 is required for the EIN2-mediated elevation in histone acetylation in response to ethylene.
Discussion
We determined that EIN2 interacts with the histone-binding protein ENAP1 (Zhang et al., 2016a) to regulate histone acetylation, and that EIN3 is potentially involved in the regulation of histone acetylation through its interaction with ENAP1. However, the role of EIN3 in histone modification is still largely unknown. We speculated that the DNA-binding activity of EIN3 is important for EIN2-directed histone acetylation. Although both EIN2-C and EIN3 interact with ENAP1, no interaction between EIN2-C and EIN3 has been detected, thus it is almost impossible to directly study how EIN3 regulates EIN2-C-mediated histone acetylation in response to ethylene. In this research, we propose a new strategy to study how EIN2-C and EIN3 function together by fusing EIN2-C with the DNA-binding domain from EIN3 to create an EIN2-C variant with EIN3 DNA-binding activity. The synthetic protein EIN2C–EIN3DB provides a perfect system to achieve our goal because the protein preserves both EIN3 DNA-binding activity and EIN2-C function. In addition, EIN2C–EIN3DB can interact with EIN3 through the EIN3 DNA-binding domain and largely fulfilled their typical functions. Thus, this approach provides a great opportunity to dissect the function of EIN3 in EIN2-mediated histone acetylation and gene expression in vivo. We believe this approach can also be applied more broadly to other proteins.
Here, we provide multiple lines of evidence showing that native EIN3 is important to determine the target for histone modification, and that the positive feedback loop mediated by dimerized EIN3 is required for the EIN2-mediated elevation of histone acetylation in response to ethylene. First, by fusing EIN2-C to EIN3DB, we found that a mutation in EIN3DB that disrupts DNA binding (EIN3DBK245N) also leads to a weaker ethylene-responsive phenotype than with wild-type EIN3DB. Second, we found that EIN2C–EIN3DB DNA-binding activity is positively correlated with EIN3 protein levels and that the EIN2C–EIN3DB binding signal is correlated with H3K14Ac and H3K23Ac levels and with expression of ethylene-responsive genes. Third, we found that EIN3 and EIN2C–EIN3DB can interact and that the binding of EIN2C–EIN3DB to target genes is enhanced by native EIN3 and that the binding specificity also increased by native EIN3 in vivo. Fourth, in the absence of EIN3, overexpression of EIN2-C can elevate histone acetylation compared to without EIN2-C; however, EIN3 is required for the ethylene-induced elevation of H3K14Ac and H3K23Ac levels. Finally, we provide evidence that EIN3 alone is not sufficient to regulate H3K14Ac or H3K23Ac levels in response to ethylene. Based on these results, we propose that in the presence of ethylene, the EIN2-C-induced elevation of H3K14Ac and H3K23Ac levels requires a positive feedback regulation mediated by EIN3, potentially through its dimerization (Supplemental Figure 7B). This positive feedback regulation integrates EIN2-C-mediated histone acetylation regulation into EIN3-dependent transcriptional regulation in response to ethylene.
In the absence of EIN3, overexpression of EIN2C–EIN3DB resulted in a partial constitutive ethylene-responsive phenotype. More importantly, expression of approximately 30% of ethylene-regulated genes was altered in ein3-1 eil1-1 EIN2C–EIN3DB seedlings. Meanwhile, transcriptional activation is a stepwise process that requires the creation and maintenance of an open chromatin structure, assembly of the preinitiation complex, and finally the transition to productive elongation. Most likely, in the absence of EIN3, EIN2-C interacts with ENAP1, which shares target genes with EIN3, to regulate histone acetylation, by creating an open chromatin structure that leads to an elevation above basal levels for a subset of genes. EIN3 potentially determines where the preinitiation complex forms for transcriptional regulation; therefore, in the absence of EIN3, EIN2C–EIN3DB binding efficiency and specificity are poor, and ethylene-induced transcription regulation is impaired.
Dimerization of transcription factors enhances DNA binding and increases binding specificity. For example, most auxin-responsive factors consist of an N-terminal B3-type DNA-binding domain, a variable middle region that can function as either an activation or repressor domain, and a C-terminal dimerization domain that is involved in protein-protein interaction. Dimerization mediated by the C-terminal domain determines binding site selection (Kim et al., 1997; Guilfoyle and Hagen, 2007, 2012; Boer et al., 2014; Hewezi et al., 2014). Consistent with this model, abscisic acid-responsive element binding factors determine abscisic acid-mediated transcriptional regulation of stress response genes, and the specificity of their transcriptional regulation is determined by dimerization among these factors (Choi et al., 2000; Uno et al., 2000; Jakoby et al., 2002). Interaction between EIN3 and other transcription factors such as JASMONATE ZIM DOMAIN, MYC2, OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF 59, and PHYTOCHROME INTERACTING FACTOR 3 participates in crosstalk with jasmonate and light signaling pathways (Zhu et al., 2011; He et al., 2017; Liu et al., 2017). EIN3 homodimerization was previously demonstrated in vitro, but it appears that binding DNA is not necessary for this dimerization (Solano et al., 1998). It should be noted that EIN3 dimerization has yet to be replicated in vivo. Given that the DNA-binding domain and the dimerization domain of EIN3 overlap, it is normally impossible to uncouple the dimerization and DNA-binding activity of EIN3 by mutating EIN3 to prevent its dimerization (Solano et al., 1998). The EIN2C–EIN3DB fusion protein provides here a unique opportunity to address the question of in vivo relevance of this dimerization, because the protein preserves both EIN3 DNA-binding activity and EIN2-C function. The EIN2C–EIN3DB fusion protein can also interact with EIN3 through the EIN3 DNA-binding domain. Although we observed no DNA binding for EIN2-C using an anti-EIN2-C antibody under a standard ChIP-seq method, we succeeded in detecting DNA binding by EIN2C–EIN3DB using an anti-EIN2-C antibody. Therefore, we identified EIN2C–EIN3DB binding targets without interference from native EIN2 and EIN3 proteins by performing our analyses in the ein3-1 eil1-1 mutant background. Importantly, we observed an interaction between EIN2C–EIN3DB and EIN3 in vivo and examined how native EIN3 protein works to regulate EIN2C–EIN3DB binding in various backgrounds. Our results clearly demonstrated that the binding specificity of EIN2C–EIN3DB is enhanced by EIN3, and that the interaction of EIN2C–EIN3DB with EIN3 plays an important role in histone acetylation at H3K14Ac and H3K23Ac of target genes.
Due to the tight link between histone modification and transcriptional regulation, it is a great challenge to dissect how transcription factors and their transcriptional activity effect histone modifications in vivo. The synthetic approach of using chimeric proteins to reconstruct a signaling pathway provides a powerful tool for functional dissection and has been widely applied to studies in cell signaling transduction, plant growth and development. For example, in order to test whether the recruitment of the chromatin-remodeling complex BRAHMA (BRM)/SPLAYED (SYD) leads to the induction of MONOPTEROS (MP, also called AUXIN RESPONSE FACTOR 5) target genes, the authors tethered the chromatin remodeling complex to MP target genes by fusing the DNA-binding domain to BUSHY, a shared component of the BRM and the SYD chromatin remodeling complex. The chimeric protein can rescue the flower primordium initiation phenotype in the hypomorph mp-S319 mutant and thus carries out the function(s) of its wild-type counterpart (Wu et al., 2015). LONG HYPOCOTYL 5 (HY5) is an important transcription factor that plays pleotropic roles in light signaling. To determine its function, Burko et al. applied the synthetic approach to analyze chimeric proteins between HY5 and either a constitutive transcriptional activator (VP16) or repressor (SRDX) in Arabidopsis and tomato (Solanum lycopersicum; Burko et al., 2020). They demonstrated that the primary activity of HY5 is to promote transcription (Burko et al., 2020). The CRISPR-dCas9 system has been used as a synthetic transcriptional regulator to direct the expression of a target gene or multiple genes by fusing dCas9 with a transcription repression domain such as the KRAB domain, or via fusion with transcription activation domains such as VP16 (Konermann et al., 2015; Park et al., 2017; Parsi et al., 2017; Moradpour and Abdulah, 2020). Yet, the chimeric proteins generated by this synthetic approach potentially will have some problems: the 3D protein structure of the chimera may potentially be altered relative to the native protein and modify its function. Therefore, the evaluation on the proteins by a genetic approach is also necessary and critical to its success.
Materials and methods
Plant growth conditions
Arabidopsis thaliana seeds were surface sterilized in 50% bleach with 0.01% Triton X-100 for 15 min and washed five times with sterile, double-distilled H2O before plating on Murashige and Skoog (MS) medium (4.3-g MS salts, 10-g sucrose, pH 5.7, 8 g phyto agar per liter) with or without the addition of 10-µM 1-aminocyclopropane-1-carboxylic acid (Sigma), the biosynthetic precursor for ethylene. After 3–4 days of stratification at 4°C in the dark, the plates were wrapped in foil and kept at 24°C in an incubator before the seedling phenotypes were analyzed. For propagation, seedlings were transferred from plates to soil (Pro-mix-HP) and grown to maturity at 22°C under 16 h light (F32T8/SPP50/ECO 5000K, SKU: GE-66350, General Electric)/8-h dark cycles. For all gene expression assays, protein level assays, RNA-seq, and ChIP-seq assays, ethylene treatments of Arabidopsis seedlings were performed by growing seedlings on MS plates in air-tight containers in the dark for 4 h, supplied with either a flow of hydrocarbon-free air (Zero grade air, AirGas) or hydrocarbon-free air with 10 p.p.m. ethylene, as previously described (Kieber et al., 1993; Zhang et al., 2018). For hypocotyl and root length measurements, 3-day-old seedlings were scanned using an Epson Perfection V700 Photo scanner, and hypocotyl and root lengths were measured using Image J (https://imagej.nih.gov/ij/).
Cloning of EIN2C–EIN3DB, EIN3DB–EIN2C, EIN2C–EIN3DBK245N, and EIN3DBK245N–EIN2C
All primers are listed in Supplemental Table 1.
For EIN2C–EIN3DB, the EIN2-C sequence was PCR-amplified by Phusion High-Fidelity DNA Polymerase (M0530S, NEB) using the primers EIN2-kpn1 and EIN2CEIN3DB-R, the EIN3DB sequence was amplified using the primers EIN2CEIN3DB-F and EIN3DB-sal1, using the first strand Arabidopsis cDNA from Col-0 as template. The PCR products were further purified, mixed together and used as template for PCR using the primers EIN2-kpn1 and EIN3DB-sal1 to generate EIN2C–EIN3DB. For EIN3DB–EIN2C, the EIN3DB sequence was amplified using the primers EIN3DB-kpn1 and EIN3DB–EIN2C-R, the EIN2-C sequence was amplified using the primers EIN3DBEIN2C-F and EIN2C-sal1, using the first strand Arabidopsis cDNA from Col-0 as template. The PCR products were further purified, mixed together, and used as template for PCR using the primers EIN3DB-kpn1 and EIN2C-sal1 to generate EIN3DB–EIN2C sequence. For EIN2C–EIN3DBK245N, the DNA fragments were amplified using the primers EIN2-kpn1 and EIN3K245N-R, EIN3K245N-F, and EIN3DB-sal1, using the EIN2C–EIN3DB fragment as template, and the PCR products were further purified, mixed together and used as template for PCR using EIN2-kpn1 and EIN3DB-sal1 to generate EIN2C–EIN3DBK245N. For EIN3DBK245N–EIN2C, the DNA fragments were amplified using the primers EIN3DB-kpn1 and EIN3K245N-R, EIN3K245N-F, and EIN2C-sal1, using the EIN3DB–EIN2C fragments as template, and the PCR products were further purified, mixed together and served as template for PCR using EIN3DB-kpn1 and EIN2C-sal1 to generate EIN3DBK245N–EIN2C. The EIN2C–EIN3DB, EIN3DB–EIN2C, EIN2C–EIN3DBK245N, EIN3DBK245N–EIN2C PCR fragments were further digested by Kpn1 and Sal1 and cloned into the pCHF3 vector.
RNA extraction and real-time PCR
Total RNA was extracted using the RNeasy Plant Kit (Qiagen) from 3-day-old etiolated seedlings treated for 4 h with air or ethylene gas, as described above. First strand cDNA was synthesized using Superscript III First-Strand cDNA Synthesis Kit (Invitrogen). Real-time quantitative PCR was performed with the LightCycler 480 SYBR Green I Master (Roche) following the manufacturer’s instructions. PCR reactions were performed in triplicate on a Roche 96 LightCycler. Expression levels were normalized to ACT2. Primers used for qPCR are listed in Supplemental Table 1.
RNA-seq processing and analysis
Three-day-old etiolated seedlings treated with air or ethylene gas for 4 h were harvested, and total RNA was extracted using Plant RNA Purification Reagent (Invitrogen) as described previously (Qiao et al., 2012). Total RNA (4 µg) was used to prepare RNA-seq libraries using TruSeq RNA Library Prep Kit (Illumina; Zhang et al., 2016a, 2016b; Wang et al., 2017). Multiplexed libraries were sequenced on an Illumina NextSeq. Raw RNA-seq reads were aligned to the TAIR10 genome release using TopHat version 2.0.9 (Kim et al., 2013; Wang et al., 2018) with default parameters. Differentially expressed genes were identified using Cufflinks version 2.1.1 following the workflow with default parameters described previously (Trapnell et al., 2012). The genes showing |log2(fold-change)| > 1, reads per kilobase per million reads values > 1 and q < 0.05 were considered to be differentially expressed.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed using the ProQuest™ Two-Hybrid System (Invitrogen) following a previously published method (Zhang et al., 2016b). Briefly, the coding sequences of EIN2-C, EIN3DB, EIN2C–EIN3DB, and EIN2C–EIN3DBK245N were introduced into the pDBLeu vector, and the coding sequences of EIN3 and ENAP1 were introduced into pEXP-AD502. The resulting pDBLeu and pEXP-AD502 constructs were co-transformed into yeast (Saccharomyces cerevisiae) strain AH109. The transformants were grown on SD–Trp–Leu medium or SD–Trp–Leu –His medium. Growth on SD–Trp–Leu–His medium indicates interaction between the corresponding proteins.
Immunoblots
Total proteins were extracted from 3-day-old etiolated seedlings with extraction buffer (50-mM Tris–HCl, pH 8.0, 150-mM NaCl, 1-mM EDTA, 0.1% Triton X-100, and protease inhibitor cocktail). After incubation on ice for 30 min, plant extracts were centrifuged at 15,000g for 15 min at 4°C. Proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and electroblotted onto a nitrocellulose membrane. The membrane was blocked using 1 × TBST (20-mM Tris, 150-mM NaCl, 0.1% Tween-20) with 5% w/v dry milk and was probed with the indicated primary antibodies (diluted in 1 × TBST with 5% w/v dry milk). After washing three times with 1 × TBST buffer (15 min each), the membrane was then probed with secondary goat anti-rabbit (Bio-Rad 170-6515) or goat anti-mouse (Bio-Rad 170-6516) antibodies conjugated with horseradish peroxidase. The signals were detected by a chemiluminescence reaction using the SuperSignal kit (Pierce). Polyclonal anti-EIN2-C (Qiao et al., 2009, 2012) and anti-EIN3 antibodies developed in house according to Guo and Ecker, 2003 were used at dilutions of 1:2,000.
Pull-down assays
The coding sequences of EIN2C–EIN3DB and EIN2C–EIN3DBK245N were introduced into the pVP13 vector (to generate MBP-His-EIN2C-EIN3DB and MBP-His-EIN2C-EIN3DBK245N). These MBP-His fusion proteins were purified using amylose resin (New England Biolabs) and re-purified using Ni-NTA Agarose (Qiagen) before being washed using pull-down buffer (50-mM Tris–HCl, pH 8.0, 150-mM NaCl, 0.5-mM EDTA, 0.5% Tween-20, and protease inhibitor cocktail). GST-EIN3DB was purified using Glutathione Sepharose (GE Healthcare). MBP-tagged EIN3 (MBP-EIN3, purified using amylose resin) was added and incubated for 1 h at 4°C. After washing five times with pull-down buffer, Sepharose beads were collected by brief centrifugation (2,000 g, 2 min) and then resuspended in protein extraction buffer (50-mM Tris–HCl, pH 8.0, 150-mM NaCl, 1-mM EDTA, 0.1% Triton X-100, and protease inhibitor cocktail). Proteins were separated by SDS–PAGE and detected with the anti-EIN3 at a 1:2,000 dilution (Guo and Ecker, 2003) or anti-MBP (NEB; catalog number E8032S, lot number 0101501) antibody at a 1:25,000 dilution.
Immunoprecipitation assays
Total proteins extracted from EIN2C–EIN3DB, ein3-1 eil1-1 EIN2C–EIN3DB, EIN2C–EIN3DBK245N, and ein3-1 eil1-1 EIN2C–EIN3DBK245N plants were immunoprecipitated using anti-EIN2-C antibody (Qiao et al., 2012). Samples were incubated overnight at 4°C and washed three times before analysis by immunoblot as above.
ChIP-seq and ChIP-qPCR
Briefly, 3-day-old etiolated seedlings treated with air or ethylene for EIN2C–EIN3DB (one replicate representing the mixed tissues from two independent collections of pooled seedlings) and ein3-1 eil1-1 EIN2C–EIN3DB (two biological replicates representing two independent collections from pooled seedlings) were harvested and cross-linked in 1% formaldehyde, and the chromatin was isolated and sonicated using Bioruptor (setting H, 0.5 min on and 2 min off, 25 cycles at 4°C; put tubes on ice for 1 min every five cycles). The indicated antibodies were added to the sonicated chromatin, followed by incubation overnight to precipitate bound DNA fragments (Zhang et al., 2016b). Two-microgram antibodies were used for each sample, including anti-EIN2-C antibody (Qiao et al., 2012), anti-H3K14Ac (Millipore; catalog number 07-353; lot number 3259085), and anti-H3K23Ac (Millipore; catalog number 07-355; lot number 3249938). DNA was eluted and amplified by PCR with primers corresponding to genes of interest. Primers used for ChIP-qPCR are listed in Supplemental Table 1.
Chromatin-immunoprecipitated DNA was sequenced using an Illumina NextSeq platform according to standard operating procedures. Initial quality-control analysis was performed using FastQC (Andrews, 2013). The genome-wide average depth of coverage obtained for each library is provided in Supplemental Table 2. Single-end reads were first mapped to the Arabidopsis genome (TAIR10; Lamesch et al., 2012) using bowtie software (version 0.7.1; Langmead et al., 2009) with default parameters. Peaks significantly enriched in ChIP-seq tags were identified using MACS2 (version 2.1.0.20150603; Zhang et al., 2008). Peaks within 2 kbp of a gene body were assigned to the gene. Genes present in both biological replicates were retained as binding targets. Peaks were merged when the maximum gap between two peaks was <200 bp, and shared peaks were used for subsequent analysis. R (version 3.2.2) scripts were used to generate of Venn diagrams.
Accession numbers
The Arabidopsis Genome Initiative numbers for the genes mentioned in this article are as follows: EIN2 (At5g03280); ENAP1 (At3g11100); EIN3 (At3g20770); EIL1 (At2g27050); ERF1, (At3g23240); EDF2 (At1g68840); ACT2 (At3g18780); ETR2, (At3g23150); EBF2 (At5g25350); NAC6 (At5g39610); ARGOS (At3g59900); ACO4 (At1g05010); LZF1 (At1g78600); CAF1A (At3g44260); ACO2 (At1g62380); CTR1 (At5g03730); HLS1 (At4g37580); EDF1 (At1g25560); CAT1 (At1g20630); NTF2 family member (At3g09250). RNA-seq and ChIP-seq data reported in this study have been deposited in the Gene Expression Omnibus database under Accession Number GSE138154.
Supplemental data
Supplemental Figure 1. Engineered EIN3DB–EIN2C leads to partial ethylene-responsive phenotype.
Supplemental Figure 2. Interaction test between ENAP1 and EIN2C–EIN3DB.
Supplemental Figure 3. Motifs identified from EIN2C–EIN3DB binding targets in different conditions.
Supplemental Figure 4. Yeast two hybrid assay to examine interaction between EIN3 and EIN3DB.
Supplemental Figure 5. The roles of EIN2S645A in the absence of EIN3 and EIL1.
Supplemental Figure 6. Immunoblots to detect the expression of dCas9-EIN2-C protein levels.
Supplemental Figure 7. Model of EIN3 mediated positive feedback regulation in EIN2-directed histone acetylation and transcriptional regulation
Supplemental Table 1. Primers used in this paper.
Supplemental Table 2. Aligned reads from chromatin immunoprecipitation sequencing libraries.
Supplemental Data Set 1. ANOVA tables.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing seeds. We thank Dr Enamul Huq and Karen Browning for comments on the manuscript. We thank the UT Austin sequencing co-facility for all the sequencing work.
Funding
This work was supported by grants from the National Institute of Health (NIH-R01GM115879).
L.W, Z.Z, F.Z., and H.Q designed the project; L.W, Z.Z, F.Z, Z.S, B.Z, A.H., and F.V.Z performed the experiments; L.W. analyzed the high throughput data; H.Q wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hong Qiao (hqiao@austin.utexas.edu).
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2013) FastQC: a quality control tool for high throughput sequence data. www.bioinformatics.babraham.ac.uk/projects/fastqc
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, Lopez-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589 [DOI] [PubMed] [Google Scholar]
- Burko Y, Seluzicki A, Zander M, Pedmale UV, Ecker JR, Chory J (2020) Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant cell 32: 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao QM, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Eyal Y, Meller Y, Lev-Yadun S, Fluhr R (1993) A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. Plant J 4: 225–234 [DOI] [PubMed] [Google Scholar]
- Grefen C, Stadele K, Ruzicka K, Obrdlik P, Harter K, Horak J (2008) Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol Plant 1: 308–320 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2012) Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci 190: 82–88 [DOI] [PubMed] [Google Scholar]
- Guo HW, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- He X, Jiang J, Wang CQ, Dehesh K (2017) ORA59 and EIN3 interaction couples jasmonate-ethylene synergistic action to antagonistic salicylic acid regulation of PDF expression. J Integr Plant Biol 59: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Piya S, Richard G, Rice JH (2014) Spatial and temporal expression patterns of auxin response transcription factors in the syncytium induced by the beet cyst nematode Heterodera schachtii in Arabidopsis. Mol Plant Pathol 15: 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Ju C, Chang C (2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol 169: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Van de Poel B, Cooper ED, Thierer JH, Gibbons TR, Delwiche CF, Chang C (2015) Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat Plants 1: 14004. [DOI] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H (2015) EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683 [DOI] [PubMed] [Google Scholar]
- Liu X, Liu R, Li Y, Shen X, Zhong S, Shi H (2017) EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell 29: 3051–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller Y, Sessa G, Eyal Y, Fluhr R (1993) DNA-protein interactions on a cis-DNA element essential for ethylene regulation. Plant Mol Biol 23: 453–463 [DOI] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enriquez P, Binder BM, Heber S, Stepanova AN, Alonso JM (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697 [DOI] [PubMed] [Google Scholar]
- Moradpour M, Abdulah SNA (2020) CRISPR/dCas9 platforms in plants: strategies and applications beyond genome editing. Plant Biotechnol J 18: 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1990) Structure and expression of a tobacco beta-1,3-glucanase gene. Plant Mol Biol 15: 941–946 [DOI] [PubMed] [Google Scholar]
- Park J-J, Dempewolf E, Zhang W, Wang Z-Y (2017) RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PloS One 12: e0179410–e0179410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsi KM, Hennessy E, Kearns N, Maehr R (2017) Using an inducible CRISPR-dCas9-KRAB effector system to dissect transcriptional regulation in human embryonic stem cells. Methods Molec Biol 1507: 221–233 [DOI] [PubMed] [Google Scholar]
- Pierik R, Tholen D, Poorter H, Visser EJ, Voesenek LA (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 11: 176–183 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Kitajima S, Koyama T, Yamada Y (1996) Ethylene-induced gene expression of osmotin-like protein, a neutral isoform of tobacco PR-5, is mediated by the AGCCGCC cis-sequence. Plant Cell Physiol 37: 249–255 [DOI] [PubMed] [Google Scholar]
- Sessa G, Meller Y, Fluhr R (1995) A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol Biol 28: 145–153 [DOI] [PubMed] [Google Scholar]
- Shinshi H, Usami S, Ohme-Takagi M (1995) Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol Biol 27: 923–932 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Qiao H (2019) New insights in transcriptional regulation of the ethylene response in Arabidopsis. Front Plant Sci 10: 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xi Y, Sung S, Qiao H (2018) RNA-seq assistant: machine learning based methods to identify more transcriptional regulated genes. BMC Genomics 19: 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang F, Rode S, Chin KK, Ko EE, Kim J, Iyer VR, Qiao H (2017) Ethylene induces combinatorial effects of histone H3 acetylation in gene expression in Arabidopsis. BMC Genom 18: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Zhang F, Qiao H (2019) Chromatin regulation in the response of ethylene: nuclear events in ethylene signaling. Small Methods, doi: 10.1002/smtd.201900288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M-F, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D (2015) Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4: e09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang L, Ko EE, Shao K, Qiao H (2018) Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang L, Lim JY, Kim T, Pyo Y, Sung S, Shin C, Qiao H (2016a) Phosphorylation of CBP20 links microRNA to root growth in the ethylene response. PLoS Genet 12: e1006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Qi B, Wang L, Zhao B, Rode S, Riggan ND, Ecker JR, Qiao H (2016b) EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling. Nat Commun 7: 13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang L, Qi B, Zhao B, Ko EE, Riggan ND, Chin K, Qiao H (2017) EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc Natl Acad Sci USA 114: 10274–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, Mu A, Jiang Z, Kim JM, To TK, Li W, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.