The parallel ability of the Arabidopsis glucosyltransferase UGT76B1 to conjugate and inactivate three signaling compounds mutually balances their activity and limits the immune response.
Abstract
Glucosylation modulates the biological activity of small molecules and frequently leads to their inactivation. The Arabidopsis thaliana glucosyltransferase UGT76B1 is involved in conjugating the stress hormone salicylic acid (SA) as well as isoleucic acid (ILA). Here, we show that UGT76B1 also glucosylates N-hydroxypipecolic acid (NHP), which is synthesized by FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1) and activates systemic acquired resistance (SAR). Upon pathogen attack, Arabidopsis leaves generate two distinct NHP hexose conjugates, NHP-O-β-glucoside and NHP glucose ester, whereupon only NHP-O-β-glucoside formation requires a functional SA pathway. The ugt76b1 mutants specifically fail to generate the NHP-O-β-glucoside, and recombinant UGT76B1 synthesizes NHP-O-β-glucoside in vitro in competition with SA and ILA. The loss of UGT76B1 elevates the endogenous levels of NHP, SA, and ILA and establishes a constitutive SAR-like immune status. Introgression of the fmo1 mutant lacking NHP biosynthesis into the ugt76b1 background abolishes this SAR-like resistance. Moreover, overexpression of UGT76B1 in Arabidopsis shifts the NHP and SA pools toward O-β-glucoside formation and abrogates pathogen-induced SAR. Our results further indicate that NHP-triggered immunity is SA-dependent and relies on UGT76B1 as a common metabolic hub. Thereby, UGT76B1-mediated glucosylation controls the levels of active NHP, SA, and ILA in concert to balance the plant immune status.
Introduction
Defense against pathogens by plants is induced at the site of inoculation to combat the infection in situ. In addition, systemic acquired resistance (SAR) can be activated by systemic tissues protecting against subsequent infections (Vlot et al., 2009; Fu and Dong, 2013; Shah and Zeier, 2013; Klessig et al., 2018; Vlot et al., 2020). A variety of small molecules are employed by plants to initiate and regulate these defense responses upon pathogen attack. Salicylic acid (SA) is a key signaling molecule required for both local and systemic defense. Upon pathogen infection, SA is synthesized primarily via the precursor isochorismate by ISOCHORISMATE SYNTHASE 1/SALICYLIC ACID INDUCTION DEFICIENT 2 (ICS1/SID2) in chloroplasts and exported by ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5) to the cytosol, where it initiates defense responses. A major route involves the establishment of a lower redox potential leading to NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1)-dependent changes in gene expression (Mou et al., 2003; Ding et al., 2018).
Different SA conjugates accumulate in response to pathogen inoculation, with glucoside formation being the dominant SA modification. SA glucosylation is considered crucial for controlling the level of unconjugated SA and thereby attenuating the defense responses (Vlot et al., 2009; Dempsey et al., 2011; Dempsey and Klessig, 2017). Three small-molecule glucosyltransferases of Arabidopsis thaliana, UGT74F1, UGT74F2, and UGT76B1, have been identified as SA glucosyltransferases (Dean and Delany, 2008; Song et al., 2008; von Saint Paul et al., 2011; Noutoshi et al., 2012; Li et al., 2015).
Interestingly, UGT76B1 also glucosylates another small molecule, isoleucic acid (ILA; 2-hydroxy-3-methyl pentanoic acid) (von Saint Paul et al., 2011). ILA competitively inhibits SA glucosylation by UGT76B1. Therefore, the enhanced pathogen resistance synergistically induced by exogenous application of ILA and SA suggested the suppression of the attenuating SA glucosylation by ILA (Noutoshi et al., 2012; Bauer et al., 2020). The biosynthesis of ILA has not been resolved in plants; however, it is likely linked to the biosynthesis or catabolism of the branched-chain amino acid isoleucine via reduction of its deaminated derivative 2-keto-3-methyl pentanoic acid (Maksym et al., 2018).
The lysine-derived amino acid, pipecolic acid (Pip) and its oxidized derivative N-hydroxypipecolic acid (NHP) are two other small molecules important to pathogen defense, in particular for the establishment of SAR (Návarová et al., 2012; Bernsdorff et al., 2016; Chen et al., 2018; Hartmann et al., 2018). Two major enzymes, the l-Lys-α-aminotransferase AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1) and the reductase SAR-DEFICIENT4 (SARD4) are involved in Pip biosynthesis from lysine (Ding et al., 2016; Hartmann et al., 2017; Hartmann and Zeier, 2018). Characterization of Arabidopsis ald1 mutants defective in the endogenous accumulation of Pip and exogenous feeding of Pip in various scenarios indicated its importance in plant defense responses and showed the Pip-dependency of SAR (Návarová et al., 2012; Vogel-Adghough et al., 2013).
However, mutants affecting FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1) were completely compromised in SAR and failed to acquire resistance in response to exogenous Pip feeding, suggesting the requirement of another molecule downstream of FMO1 for SAR (Mishina and Zeier, 2006; Návarová et al., 2012). FMO1 was biochemically characterized in vitro and as a pipecolate N-hydroxylase that catalyzes the conversion of Pip to NHP (Chen et al., 2018; Hartmann et al., 2018). Upon pathogen attack, NHP accumulates systemically in the Arabidopsis foliage. In addition, exogenous feeding with NHP potently activated SAR to bacterial and oomycete infection and abrogated the SAR-defect of the NHP-deficient fmo1 mutant, indicating that NHP is the actual SAR-inducing compound of this resistance pathway (Chen et al., 2018; Hartmann et al., 2018).
Similar to SA and ILA, a glycosylated form of NHP was detected in Arabidopsis leaves after pathogen inoculation or exogenous application of NHP. However, the uridine diphosphate (UDP)-dependent glycosyltransferase(s) (UGT(s)) involved in NHP hexoside (NHP-H) formation and the functional implication of this conjugation remained elusive. It had been speculated that NHP-H could be an inactivated storage form and/or a mobile entity in SAR (Chen et al., 2018; Hartmann and Zeier, 2018).
Transcriptome analyses of Pip-treated Arabidopsis plants or systemic SAR tissue revealed a strong induction of genes involved in SA, Pip, and NHP biosynthesis, such as ICS1, EDS5, ALD1, SARD4, and FMO1. The strong upregulation of these genes indicates that the molecular implementation of the Pip-induced defense responses, including a positive feedback amplification of Pip, relies on links between these signaling molecules. Defense gene induction by Pip was dependent on functional FMO1, suggesting the requirement of NHP formation in SAR-related gene expression (Hartmann et al., 2018). The same Pip-, FMO1-, and SAR-dependent expression pattern was also found for UGT76B1, the gene encoding the SA- and ILA-glucosyltransferase (Hartmann et al., 2018).
Therefore, we aimed at examining the role of UGT76B1 in the Pip-induced defense and its regulation via NHP. We addressed the effect of Pip application on ugt76b1 loss-of-function mutants, which show enhanced resistance toward biotrophic pathogens, and on plants constitutively overexpressing (OE) UGT76B1, which are more susceptible than the wild-type (Wt; von Saint Paul et al., 2011). A comparative metabolic analysis revealed that the wild type (Wt) and UGT76B1-OE plants accumulated an NHP-O-glucoside termed NHP-H2, which was completely absent from ugt76b1. This suggested a direct additional activity of UGT76B1 as an NHP glucosyltransferase, which could be confirmed in vitro and in vivo. Importantly, NHP-H2 was distinct from a previously reported NHP-H (Hartmann and Zeier, 2018), here termed as NHP-H1, which is formed in a UGT76B1-independent manner. Mass spectrometry and digestion experiments suggest that this conjugate represents an NHP glucose ester. We show that UGT76B1 orchestrates the glucosylation of the three defense activators SA, ILA, and NHP and thereby controls basal defense and SAR. The loss of UGT76B1 leads to a SAR-like, NHP-dependent enhanced immune status, whereas overexpression of UGT76B1 depletes the levels of free NHP and SA and compromises basal immunity and SAR.
Results
The formation of an NHP-H is abolished in the ugt76b1 mutants
Previous studies have shown that exogenous application of Pip strongly enhances the transcripts of the Arabidopsis SA glucosyltransferase encoding UGT76B1 along with several SAR-related genes including genes involved in SA and NHP biosynthesis. This transcriptional response to Pip was lost by fmo1 plants indicating a positive impact of FMO1 or its product NHP on UGT76B1 expression (Hartmann et al., 2018; Hartmann and Zeier, 2019). To conversely investigate a potential impact of UGT76B1 on the metabolic response to exogenous Pip, we performed comparative liquid chromatography - mass spectrometry (LC–MS)-based metabolite analyses of leaf extracts of an ugt76b1 loss-of-function mutant, a UGT76B1-OE line, and the Wt. Exogenous Pip application was associated with a considerable uptake and relocation of Pip to rosette leaves by all three genotypes (Figure 1A). In addition, Wt and UGT76B1-OE plants accumulated low amounts of the Pip derivative NHP in response to Pip application (Figure 1B). Notably, the leaves of ugt76b1 plants showed a by far higher NHP level upon Pip treatment than the other lines. Moreover, untreated ugt76b1 exhibited a strong endogenous accumulation of both Pip and, in particular, of NHP (Figure 1, A and B). This indicated a role of UGT76B1 in suppressing the levels of both Pip and NHP. Furthermore, a metabolite with m/z 308.1346 was enhanced upon Pip application by the Wt and UGT76B1-OE plants (Figure 1C). Intriguingly, this metabolic feature was undetectable in leaf extracts of the ugt76b1 mutant, strongly suggesting an indispensable role of UGT76B1 in its formation (Figure 1C).
Figure 1.
Accumulation of an NHP-hexoside following treatment with resistance enhancers is dependent on UGT76B1. A–C, Pip, NHP, and m/z 308.1346 content in rosette leaves of 5-week-old Wt, ugt76b1, and a UGT76B1-OE line 48 h after watering with control solution or 1 mM dl-Pip. Rosette leaves were harvested, extracted, and analyzed by LC–MS. Bars are means ± sd, n = 4; significant differences are indicated by letters according to one-way ANOVA (P.adj. < 0.05). D–F, Detection of Pip, NHP, and m/z 308.1346 in leaf extracts of 3-week-old plants sprayed with a control solution or 1 mM BTH. Leaves were harvested 48 h after the treatment and analyzed by LC–MS. Bars are means ± sd, n = 4; significant differences are indicated by letters according to one-way ANOVA (P.adj. < 0.05). G, MS/MS fragmentation of m/z 308.1346 ± 0.005 identified in plant extracts. H, Detection of m/z 308.1346 in extracts of Wt Col and of ugt76b1, fmo1, and fmo1 ugt76b1 mutant lines using LC–MS.
To further investigate the role of UGT76B1 in defense-related metabolism, we comparatively analyzed extracts of Wt, ugt76b1, and UGT76B1-OE plants treated with the priming-inducer benzothiadiazole (BTH) by LC–MS. The levels of Pip and NHP were induced ten-fold in Wt leaves 48 h after BTH treatment. The UGT76B1-OE line showed strongly reduced accumulation of Pip and NHP following BTH application, whereas the ugt76b1 mutant over-accumulated both metabolites (Figure 1, D and E). As observed above for the Pip treatments, the ugt76b1 mutant plants were incapable of basal and BTH-induced formation of the m/z 308.1346 feature (Figure 1F). In contrast, the Wt and UGT76B1-OE plants substantially accumulated m/z 308.1346 after BTH application (Figure 1F; Supplemental Figure S1A).
The LC–MS [M + H]+ ion at m/z 308.1346 (±0.005) fragmented a main feature at m/z 146.0814, which is characteristic for an [NHP+H]+ ion, and the loss of a hexose unit (m/z difference of 162). This suggested an NHP-H structure for this substance (Figure 1G; Chen et al., 2018). To genetically substantiate that m/z 308.1346 is an NHP-derived hexoside, we further analyzed its occurrence in extracts of the fmo1 mutant and an fmo1 ugt76b1 double mutant. The fmo1 background renders these lines incapable of NHP biosynthesis because of its defect in NHP-synthesizing FMO1 (Hartmann et al., 2018). Consistent with the proposed NHP-H structure, the m/z 308.1346 peak was undetectable in extracts of the fmo1 and fmo1 ugt76b1 lines (Figure 1H). Thus, our comparative LC–MS metabolite analyses indicate that UGT76B1 mediates the formation of an NHP-H by Arabidopsis and thereby prevents over-accumulation of unconjugated NHP.
Arabidopsis accumulates two NHP-H peaks in an FMO1-dependent manner
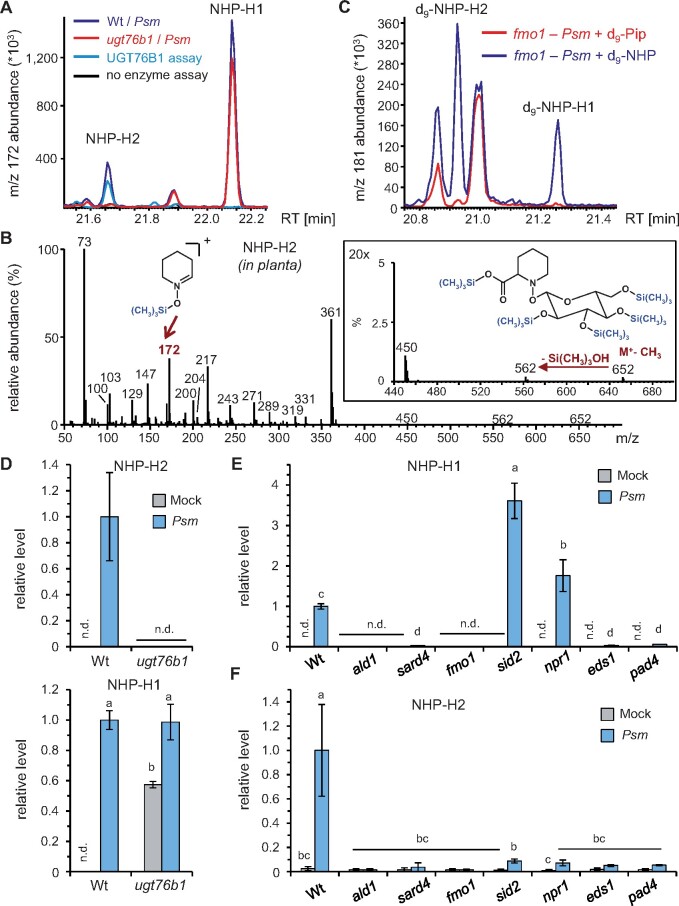
Previous gas chromatography-mass spectrometry (GC–MS)-based studies indicate that inoculation with a bacterial pathogen such as Pseudomonas syringae induces the formation of NHP and an NHP hexose conjugate (Hartmann et al., 2018; Hartmann and Zeier, 2018). In addition, an NHP-H with features similar to m/z 308.1346 (Figure 1G) was reported to accumulate in Arabidopsis in response to P. syringae attack (Chen et al., 2018). However, it was unresolved whether the NHP-H detected via LC–MS would correspond to the NHP hexose conjugate described by Hartmann and Zeier (2018) via GC–MS analyses, or whether two distinct NHP derivatives were identified in these studies.
To clarify this issue, we performed comparative metabolite analyses of leaf extracts from mock- and P. syringae pv. maculicola (Psm)-inoculated Wt and ugt76b1 plants using GC–MS analysis after trimethylsilylation of the extracted analytes according to previous work (Hartmann et al., 2018). Notably, we identified two putative NHP-hexoside peaks that both showed the m/z 172 mass spectral trimethylsilyl-N-hydroxypiperidine fragment characteristic for NHP. In addition, the mass spectra of both compounds displayed an m/z 652 in the higher spectrum range, which is consistent with an M+-CH3 ion of a penta-trimethylsilyated NHP-hexose conjugate, a thereof derived m/z 562 fragment, and further fragment ions characteristic for per-trimethlysilylated hexose conjugates (Figure 2, A and B; Ehmann, 1974; Hartmann and Zeier, 2018).
Figure 2.
Inoculation with Psm induces accumulation of two distinct NHP-H derivatives dependent on a functional NHP biosynthetic pathway. A, GC–MS analysis reveals the accumulation of two distinct NHP-hexose conjugates (NHP-H1 and NHP-H2) in leaves of Psm-inoculated Col-0 plants. Overlaid ion chromatograms (m/z 172) are shown (blue: Wt, red: ugt76b1). NHP-H1 accumulates independently of UGT76B1 and represents the NHP-hexose-conjugate previously described by Hartmann and Zeier (2018). The accumulation of NHP-H2 is absent in ugt76b1 mutants. Moreover, NHP-H2 is synthesized in vitro by recombinant UGT76B1 (green: full enzyme assay with recombinant UGT76B1, black control assay without enzyme). Sample derivatization by trimethylsilylation was performed prior to GC–MS analyses. B, Mass spectrum of the penta-trimethylsilylated NHP-hexoside NHP-H2 (molecular weight: 667 g mol−1) from plant extracts. The M+-CH3 ion at m/z 652, which produces an m/z 562 ion by loss of Si(CH3)3OH, is clearly discernible. The structure of the trimethylsilyl-N-hydroxypiperidine fragment at m/z 172 is indicated. Fragment ions characteristic for per-trimethlysilylated hexose conjugates are m/z 450, m/z 361, m/z 271, m/z 217, and m/z 204 (Ehmann, 1974). Note that the mass spectrum of penta-trimethylsilylated NHP-H1 exhibits similar fragmentation patterns, including a more prominent m/z 172 (Hartmann and Zeier, 2018). C, Both NHP-H2 and NHP-H1 are biosynthetically derived from NHP. Feeding of deuterated D9-NHP to Psm-inoculated fmo1 plants results, in contrast to feeding with D9-Pip, in the formation of D9-labeled NHP-H2 and D9-labeled NHP-H1. Ion chromatograms of m/z 181 are depicted. The m/z 181 ion corresponds to a D9-trimethlysilyl-hydroxypiperidine fragment. GC–MS analyses as described in (A) and (B). D, Relative levels of NHP-H2 and NHP-H1 in Psm-inoculated or Mock-treated (MgCl2-infiltrated) leaves of Wt and ugt76b1 plants at 48 hpi. Data represent the mean ± sd of three biological replicates. Mean levels in Psm-inoculated Col-0 leaves are set to 1. Different letters denote significant differences (P < 0.05, ANOVA and post hoc Tukey HSD test). E and F, Relative levels of NHP-H1 (E) and NHP-H2 (F) in Psm-inoculated or Mock-treated leaves of Wt plants, NHP biosynthesis-defective mutants (ald1, sard4, and fmo1), SA pathway mutants (sid2 and npr1), as well as eds1 and pad4 mutants at 24 hpi. Other details as described in (D).
One of the two peaks present in the extracts from Psm-inoculated leaves was accumulating independently of UGT76B1. In fact, this peak represented the Psm-inducible NHP-hexoside previously described by Hartmann and Zeier (2018). Therefore, this substance was termed as NHP-H1 (Figure 2A). In contrast, the second GC–MS-detected NHP-hexose conjugate, which we named NHP-H2, was only detected in extracts from infected Wt leaves, yet absent from ugt76b1 mutant plants (Figure 2, A and B). The UGT76B1-dependent accumulation of NHP-H2 detected by the GC–MS analyses was reminiscent of the accumulation pattern of the above-described m/z 308.1346 LC–MS peak, suggesting that NHP-H2 corresponds to the NHP-H identified by our LC–MS analyses (Figure 1). In contrast to the GC–MS analyses, a second UGT76B1-independent peak such as NHP-H1 was not unequivocally detected by our LC–MS settings.
To substantiate the formation and identity of the two NHP-Hs by GC–MS, deuterium-labeled D9-Pip or D9-NHP was fed to Psm-inoculated fmo1 plants. Both hexosides were absent from inoculated, D9-Pip-fed fmo1 plants, indicating that their formation requires the FMO1-dependent Pip-to-NHP conversion (Figure 2C). Notably, deuterated NHP-H1 and NHP-H2 were detected in fmo1 plants after D9-NHP feeding and subsequent Psm inoculation, corroborating their direct derivation from NHP (Figure 2C). Moreover, the Wt accumulated both natural NHP-Hs in parallel with unbound NHP upon Psm infection, whereas they were virtually absent from noninoculated control plants. However, NHP-H2 was UGT76B1-dependent and not detected (n.d.) in extracts of Psm-inoculated ugt76b1 plants, whereas the level of NHP-H1 was similar in leaf extracts of Psm-inoculated Wt and ugt76b1 mutant plants (Figure 2D). Similar to the basal levels of free NHP, the basal levels of NHP-H1 were also strongly enhanced by naïve ugt76b1 plants (Figure 2D). Together, our findings demonstrate the accumulation of two distinct NHP hexose derivatives upon pathogen inoculation, the UGT76B1-independent NHP-H1, and the UGT76B1-dependent NHP-H2. A time course analysis of accumulation of NHP, NHP-H1, and NHP-H2 in Psm-inoculated Wt leaves suggests that NHP glycosylation relatively promptly follows the initial induction of NHP biosynthesis between 10 and 16-h post-inoculation (hpi; Supplemental Figure S2).
We next examined the regulatory principles of the pathogen-inducible accumulation of these distinct NHP-Hs. Consistent with their biochemical derivation from NHP (Hartmann et al., 2018), the Psm-triggered accumulation of both NHP-H1 and NHP-H2 was absent from ald1 and fmo1, and strongly diminished by sard4 (Figure 2, E and F). The accumulation of free NHP is also tightly regulated by the two immune-regulatory genes EDS1 and PHYTOALEXIN-DEFICIENT4 (PAD4) (Hartmann et al., 2018). A similar EDS1- and PAD4-dependency exists for the pathogen-induced biosynthesis of the two NHP-Hs, because the accumulation of both NHP-H1 and NHP-H2 was greatly suppressed by eds1 and pad4 mutant plants (Figure 2, E and F). Previous analyses also revealed an over-accumulation of NHP by the Arabidopsis SA biosynthetic mutant sid2 and by the SA-perception-defective mutant npr1 during the course of Psm infection, indicating a modulation of NHP accumulation by an intact SA pathway (Hartmann et al., 2018). Interestingly, the accumulation of NHP-H2 was strongly dependent on intact SID2 and NPR1 genes, whereas NHP-H1 showed similar accumulation characteristics as free NHP and over-accumulated by sid2 and npr1 mutants (Figure 2, E and F). These analyses show that the inducible biosynthesis of the two identified NHP-Hs proceeds via an intact NHP biosynthetic pathway and, just like NHP biosynthesis, is boosted by the EDS1/PAD4-immune regulatory node. Notably, the pathogen-inducible generation of the two hexosides is oppositely regulated by SA signaling: whereas the accumulation of NHP-H1 and free NHP is negatively affected by a functional SA pathway, NHP-H2 accumulates in strong dependence of intact SA biosynthesis and signaling.
UGT76B1 glucosylates NHP to NHP-H2 in vitro
The complete loss of NHP-H2 in the ugt76b1 mutants suggested that UGT76B1, known to glucosylate SA and ILA, may glycosylate NHP as an additional substrate. A homology model of UGT76B1 was created based on the crystal structure of the Arabidopsis SA glucosyltransferase UGT74F2 using the phyre2 software (Kelley et al., 2015; George Thompson et al., 2017). Interestingly, the predicted aglycon-binding pocket of UGT76B1 is larger than that of UGT74F2 and, therefore, it is able to accommodate ligands that are more spacious. The in silico analysis predicts that NHP can alternatively bind to the active site of UGT76B1 in addition to the known aglyca SA and ILA (Supplemental Figure S3).
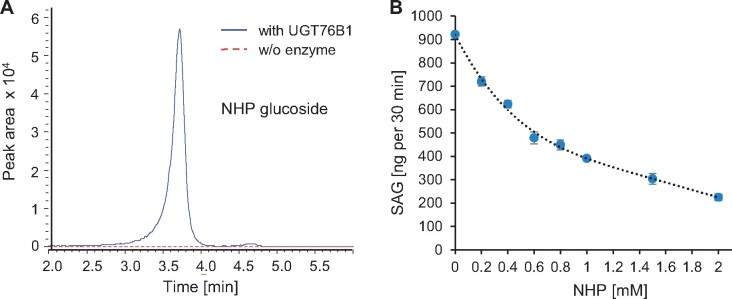
The recombinantly produced UGT76B1 enzyme was incubated with NHP in the presence of UDP glucose to test whether it is able to catalyze the glucosylation of NHP in vitro. Indeed, LC–MS analysis of the enzymatic reactions revealed a peak with m/z 308.1346 co-eluting with the respective peak of plant extracts, whereas it was absent from the control incubation without the enzyme (Figure 3A;Supplemental Figure S1B). Fragmentation of the in vitro synthesized m/z 308.1346 showed diagnostic fragments of an NHP-H, as observed for the metabolite from plant extracts (Figure 1G;Supplemental Figure S1C), supporting the formation of NHP glucoside by UGT76B1 in vitro. The NHP glucoside produced in vitro also co-eluted with the detected NHP-H2 when analyzed by GC–MS, and showed the same mass spectral fragmentation pattern (Figure 2, A and B; Supplemental Figure S4). Thus, the LC–MS feature m/z 308.1346 and the GC–MS peak NHP-H2 were coinciding in both analytical settings with the NHP glucoside produced in vitro by UGT76B1.
Figure 3.
UGT76B1 glucosylates NHP in vitro. A, LC–MS separation of the in vitro reaction of UGT76B1 (blue curve; control in stippled red) with NHP and UDP glucose producing an NHP glucoside with m/z 308.1346, which shows the correct MS/MS fragmentation pattern (Supplemental Figure S1C). B, NHP inhibits the UGT76B1-dependent SA glucosylation. Means ± sd, n = 3. Additional mutual inhibitions of the UGT76B1 substrates SA, NHP, and ILA are shown in Supplemental Figure S5.
The capability of UGT76B1 to glucosylate NHP in addition to SA and ILA suggested a mutual interference of the conjugation of these substrates. Maksym et al. (2018) had previously shown that ILA competitively inhibited the glucosylation of SA by UGT76B1. In vitro competition assay using different combinations of substrates were employed to further investigate the mutual interference. Importantly, the newly identified substrate NHP inhibits the glucosylation of SA and ILA (Figure 3B;Supplemental Figure S5A). Conversely, SA and ILA inhibit UGT76B1-mediated NHP glucosylation (Supplemental Figure S5B).
Together, these analyses show that the UGT76B1 glucosyltransferase catalyzes the glucosylation of NHP in vitro. The in vitro reaction product is identical to the detected, UGT76B1-dependent NHP-H2 hexoside. The usage of UDP-glucose as a substrate in our in vitro assays further reveals that NHP-H2 is an NHP glucose conjugate. The three UGT76B1 substrates SA, ILA, and NHP mutually compete with each other when analyzed with the glucosyltransferase in vitro, suggesting a regulatory interplay of these three immune-active metabolites via UGT76B1-dependent glucosylation.
Enzyme digestion assays and mass spectrometric properties suggest that NHP-H1 is NHP glucose ester and NHP-H2 is NHP-O-β-glucoside
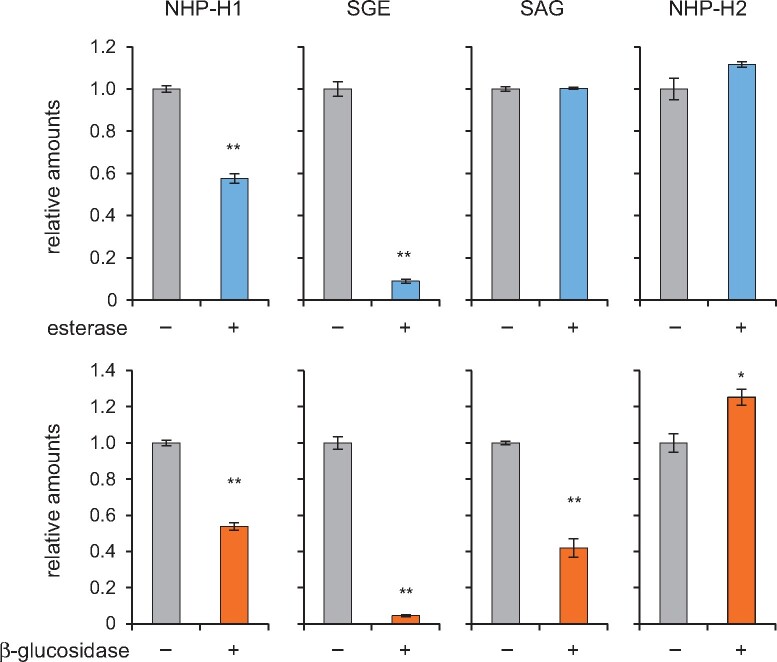
NHP possesses two reactive groups, a carboxylic acid and an N-OH hydroxyl group, which are alternatively amenable to sugar conjugation resulting in an ester bond and an O-β-glucoside, respectively. To get further insight into the nature of conjugation of NHP-H1 and NHP-H2, we employed hydrolytic digestions of leaf extracts with an esterase and a β-glucosidase. Besides NHP-H1 and NHP-H2, the extracts from P. syringae-inoculated Wt leaves contained substantial levels of the SA glucose derivatives SA glucose ester (SGE) and SA-O-β-glucoside (SAG), which provided intrinsic ester- and O-β-glucoside-bound reference metabolites in this assay.
All four hexose conjugates were stably detectable by GC–MS when incubated overnight with a sodium phosphate buffer of pH 6 (Figure 4). As expected, esterase strongly diminished the level of the reference metabolite SGE, whereas SAG remained unchanged. Consistent with previous digestion assays (Edwards, 1994), β-glucosidase treatment resulted in the partial cleavage of both the glucosidic bond of SAG and the ester linkage of SGE (Figure 4). Notably, NHP-H1 showed a similar sensitivity to the esterase- and β-glucosidase treatments as SGE, supporting a glucose ester structure for this conjugate (Figure 4). Consistent with the partial hydrolysis of NHP-H1, the levels of unconjugated NHP in the extracts increased upon incubation with esterase or β-glucosidase (Supplemental Figure S6).
Figure 4.
Effects of esterase and β-glucosidase treatments on NHP-H1, NHP-H2, SGE, and SAG. An extract of Arabidopsis leaves inoculated with Psm that accumulated NHP- and SA-conjugates was buffered to pH 6.0 with 0.1 mM sodium phosphate, and aliquots thereof incubated for 15 h with 10 U mL−1 esterase (+), 10 U mL−1 β-glucosidase (+), or with buffer only (−). Samples were analyzed by GC–MS (Figure 2), and amounts of analytes were related to ribitol as internal standard. All four hexose conjugates were stably detectable by GC–MS when incubated overnight at pH 6.0. Values are expressed relative to the means of the buffer only condition (n = 3). Asterisks indicate significant differences between buffer only (−) and enzyme-treatments (**P < 0.001 and *P < 0.01 (two-tailed Student’s t test).
In contrast, NHP-H2 showed resistance to esterase treatments (Figure 4;Supplemental Figure S7), which was in line with the presumed identity of NHP-H2 as NHP-O-β-glucoside. However, β-glucosidase treatments did not diminish the NHP-H2 level either (Figure 4; Supplemental Figure S7). This may be explained by a resistance of the special N-O-glucosidic linkage present in NHP-O-β-glucoside to the employed almond β-glucosidase, which chemically differs from the common C-O-glucosidic bond of SAG and other characterized β-glucosidase substrates. Nevertheless, the biochemistry of UGT76B1 catalyzing O-β-glucoside formation of ILA and SA clearly supports an NHP glucoside structure of NHP-H2 (Supplemental Figure S7).
Furthermore, the GC–MS-derived mass spectra of the silylated NHP conjugates showed a more prominent m/z 172 trimethylsilyl-N-hydroxypiperidine fragment in the NHP-H1 than in the NHP-H2 spectrum (Figure 2B; Hartmann and Zeier, 2018). This further supports an NHP glucose ester structure for NHP-H1 and an NHP-O-β-glucoside structure for NHP-H2, since the glucose ester can be directly cleaved into m/z 172, whereas the glucoside can only produce this fragment by an additional rearrangement. Together, the digestion assays and mass spectrometric results support an NHP glucose ester structure for NHP-H1 and an NHP-O-β-glucoside structure for the UGT76B1-dependent NHP-H2.
Exogenous ILA promotes NHP accumulation
Exogenous ILA had previously been shown to induce defense responses and synergistically enhance the SA-dependent defense pathway (von Saint Paul et al., 2011; Bauer et al., 2020). After the identification of NHP as the third substrate of UGT76B1, we were interested to know how ILA would functionally interact with NHP and SA in vivo. Therefore, we treated Wt plants with 500 µM ILA and determined the change in the levels of SA- and Pip-related metabolites. Interestingly, exogenously applied ILA significantly induced the amount of Pip and NHP 48 h after the treatment (Figure 5, A and B). In addition, NHP-H2 tended to be enhanced by the ILA treatment (Supplemental Figure S8A).
Figure 5.
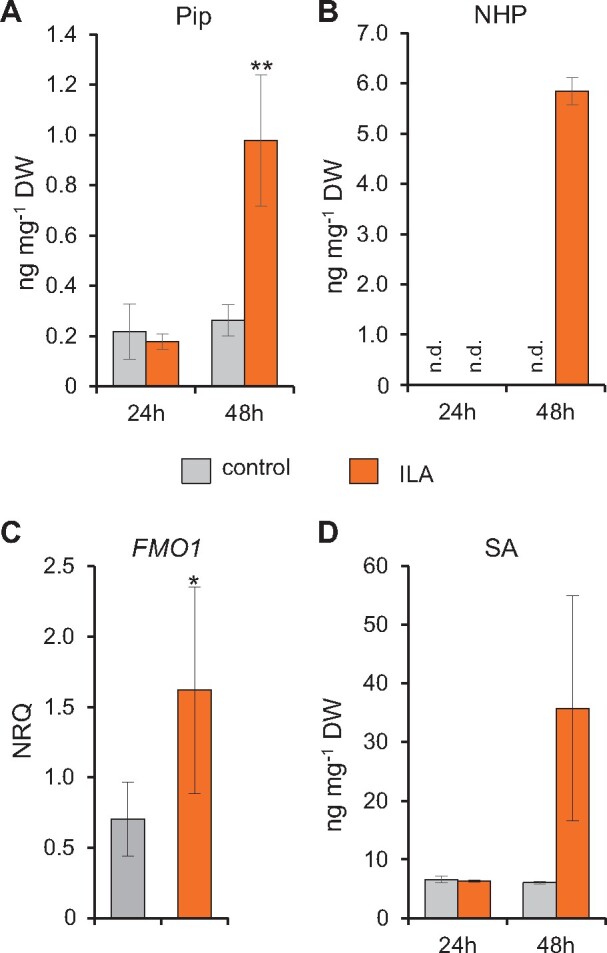
Exogenous ILA enhances the accumulation of SA, Pip, and NHP, and induces expression of FMO1. A and B, Pip and NHP levels of leaves of 12-day-old Wt seedlings 24 and 48 h after incubation in half MS medium without (gray bar) and with 500 µM ILA (black bars). Bars represent means ± sd; n = 3–4. Differences between treated or untreated plants were analyzed by Welch two-sample t test; *P < 0.05. C, Transcript abundance of FMO1 of leaves of 14-day-old plantlets grown in liquid culture was measured by RT-qPCR 48 h after the application of 500 µM ILA. Gene expression was normalized to S16 and UBQ5; bars are means ± sd; n = 4. Differences between treated or untreated plants were analyzed by Welch two-sample t test; *P < 0.05. D, SA levels of leaves of 12-day-old Wt seedlings as described for (A and B).
To determine whether the NHP accumulation also involves the activation of its biosynthetic pathway, we analyzed the expression of the NHP biosynthetic gene FMO1. Indeed, the transcript level of FMO1 was induced 48 h after ILA application (Figure 5C). The transcript level of UGT76B1 was slightly enhanced (Supplemental Figure S8D).
Similarly, we investigated the change in the abundance of SA-related metabolites after exogenous ILA application. SA, SAG, and SGE were enhanced by the treatment after 48 h (Figure 5D;Supplemental Figure S8, B and C). Thus, ILA positively affects both the NHP and SA defense pathways, suggesting a functional interaction between the UGT76B1 substrates ILA, SA, and NHP in plant immunity.
Enhanced pathogen resistance of ugt76b1 requires intact FMO1
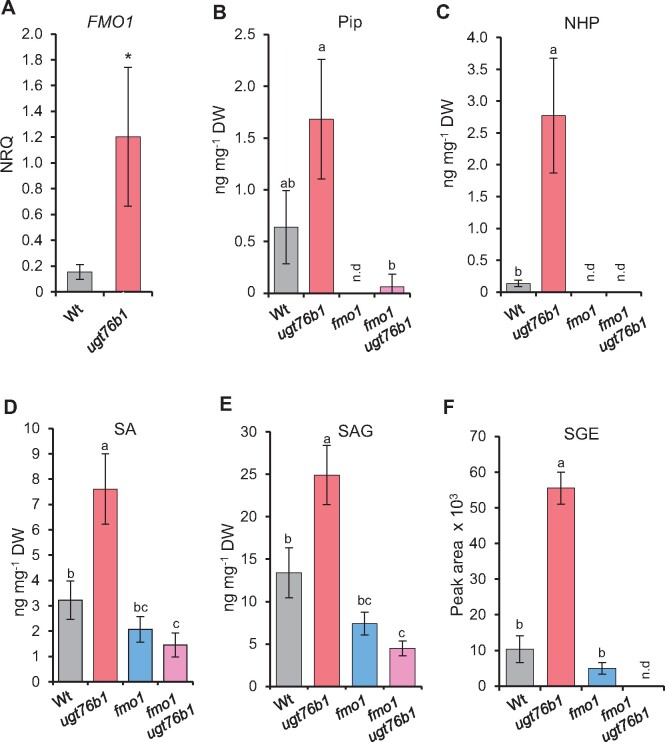
The ugt76b1 mutant plants are more resistant to biotrophic pathogen attack than the Wt (von Saint Paul et al., 2011). This phenotype had been related to the constitutively elevated level of SA and the upregulation of SA-dependent marker genes observed in this mutant, which, in turn, was attributed to the loss of UGT76B1 as an SA glucosyltransferase (von Saint Paul et al., 2011). However, the loss of UGT76B1 also enhanced the levels of the defense-related metabolites Pip and NHP (Figure 1). Indeed, the NHP biosynthetic gene FMO1 and the SA-biosynthetic gene SID2 were constitutively upregulated by ugt76b1 plants (Figure 6A;Supplemental Figure S9A).
Figure 6.
Enhanced accumulation of defense-related transcripts and metabolites by the ugt76b1 mutant is dependent on FMO1. A, RT-qPCR analysis of FMO1 transcript abundance of leaves of 4-week-old Wt and ugt76b1 plants grown under short-day conditions; the normalized relative quantity was determined based on the UBQ5 and S16 internal standards; bars show means ± sd, n = 4; differences between genotypes were analyzed by Welch two-sample t test; *P < 0.05. B–F, Pip- and SA-related metabolites were determined using leaf extracts of 4-week-old Wt, ugt76b1, fmo1, and fmo1 ugt76b1 plants grown under short-day conditions using LC–MS. Bars show means ± sd, n = 4; significant differences between genotypes were analyzed by Welch two-sample t test; *P < 0.05 (C) and by one-way ANOVA with post hoc Lincon test as indicated by letters (P.adj. < 0.05) (B and D–F).
To examine the role of NHP in the enhanced resistance of ugt76b1, we employed the fmo1 ugt76b1 double mutant line lacking NHP biosynthesis. Notably, the transcript abundances of SID2 and of the SA-responsive genes PR1 and NPR1 were reduced to Wt levels by the fmo1 ugt76b1 double mutant (Supplemental Figure S9, B and C). Furthermore, the enhanced levels of SA, SAG, SGE, and Pip found in ugt76b1 leaves were reduced to Wt levels by the introgression of fmo1, whereas the accumulation of NHP observed for ugt76b1 was completely abolished by the fmo1 ugt76b1 mutant background (Figure 6, C–F). NHP-H2 was absent from all these mutants in accordance with the role of UGT76B1 and FMO1 in its biosynthesis (Figure 1H).
To directly examine the significance of accumulating NHP for the enhanced resistance phenotype of ugt76b1, we comparatively assayed basal resistance of Wt, fmo1, ugt76b1, and fmo1 ugt76b1 lines toward P. syringae pv. tomato DC3000 (Pst). The loss of FMO1 (and NHP) abolished the enhanced resistance phenotype of ugt76b1, since the fmo1 ugt76b1 double mutant displayed a basal susceptibility similar to the fmo1 single mutant or the Wt (Figure 7A). Together, this indicates that the enhanced defense status of ugt76b1, which includes enhanced accumulation of the NHP precursor Pip, activation of the SA defense pathway, and a SAR-like resistance phenotype, is triggered by the constitutive elevation of unconjugated NHP by ugt76b1 plants.
Figure 7.
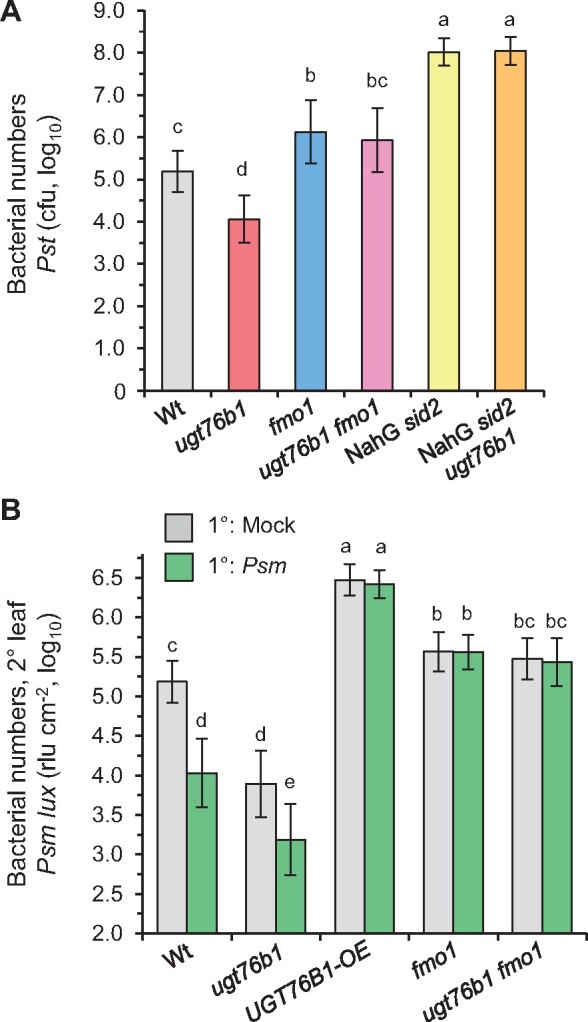
Local and systemic immunity is enhanced by ugt76b1 loss-of-function, whereas overexpression of UGT76B1 compromises SAR. A, Susceptibility of ugt76b1 introgressed into fmo1 and NahG sid2 toward Pst DC3000. Four-week-old Wt, ugt76b1, fmo1, fmo1 ugt76b1, NahG sid2, and NahG sid2 ugt76b1 plants were infiltrated with 5 × 104 cfu (OD600 = 0.0001) of Pst DC3000. Bacterial growth was monitored after 72 h. Bars are means ± sd of 15 replicates from three independent experiments, each experiment consisting of five biological replicates. The presented values are log10-transformed. Different letters denote significant differences (P < 0.01, ANOVA and post hoc Tukey HSD test). The smaller growth of ugt76b1 in comparison to Wt is ameliorated by the introgression of fmo1 or NahG sid2 (Supplemental Figure S10). B, The ugt76b1 mutant is able to induce SAR, whereas the UGT76B1-OE line is deficient in SAR. To assess SAR, three 1° leaves of a plant were mock-treated or Psm-inoculated (OD600 = 0.005). Two days later, three 2° leaves were challenge-infected with a bioluminescent Psm strain (Psm lux; OD600 = 0.001), and growth of Psm lux was assessed after 2.5 days by luminescence measurements. Bacterial numbers were determined as rlus per leaf area (rlu cm−2). The presented values are log10-transformed. Bars are the mean ± sd of 12 or more replicate leaf samples from 6 to 7 different plants. Different letters denote significant differences (P < 0.01, ANOVA and post hoc Tukey HSD test). Independent experiments yielded similar results with some variability of the SAR phenotype of ugt76b1 (Supplemental Figure S11).
In addition, the SA-depleted NahG sid2 and NahG sid2 ugt76b1 lines were employed to address the role of SA in the ugt76b1-associated resistance. The depletion of SA strongly raised the pathogen susceptibility of NahG sid2 (Figure 7A). Notably, the introgression of the ugt76b1 background into the NahG sid2 line did not alter the resistance to bacterial infection, since NahG sid2 ugt76b1 plants showed similar susceptibly to Pst as the SA-deficient NahG sid2 line (Figure 7A). Together, these resistance assays indicate that the elevated NHP level of ugt76b1 is the trigger for the primed defense status and enhanced basal resistance phenotype to bacterial infection. Elevated NHP activates SA biosynthesis in ugt76b1, yet SA is finally required for the NHP-initiated resistance response.
Overexpression of UGT76B1 in Arabidopsis depletes the levels of free NHP and SA, promotes formation of the respective O-β-glucosides, and compromises SAR
NHP was previously identified as a main metabolic regulator of SAR in Arabidopsis and other plant species (Chen et al., 2018; Hartmann et al., 2018; Schnake et al., 2020). To examine the impact of UGT76B1-mediated NHP glucosylation on SAR, we compared the SAR response of Wt, UGT76B1-OE, ugt76b1, ugt76b1 fmo1, and fmo1 plants. Lower (1°) rosette leaves of a subset of plants were therefore pre-inoculated with Psm to induce SAR, whereas the 1° leaves of other plants were mock-treated for the examination of an appropriate non-induced control. Two days later, all the plants were challenge-infected with a bioluminescent Psm strain (Psm lux, Fan et al., 2008) and resistance to the bacterial infection was assessed by bioluminescence measurements 2.5 days later (Hartmann et al., 2017; Gruner et al., 2018). Inducing pre-inoculations with Psm limited bacterial growth in challenged 2° leaves of the Wt compared to those of mock-control plants by 15- to 20-fold, indicating a strong SAR establishment (Figure 7B;Supplemental Figure S11). Strikingly, both mock- and Psm-pre-treated UGT76B1-OE plants exhibited a high susceptibility to the challenge infection, indicating a complete incompetence of SAR induction and a lowered basal immunity of this line. Conversely, control plants of the ugt76b1 mutant showed a marked constitutive SAR that was further enhanced by a Psm-pre-inoculation in two out of three experiments (Figure 7B;Supplemental Figure S11). In contrast, the ugt76b1 fmo1 line lacked both the pathogen-inducible SAR and the constitutive resistance phenotype observed for ugt76b1, and thus behaved similar as the fmo1 single mutant (Figure 7B;Supplemental Figure S11A).
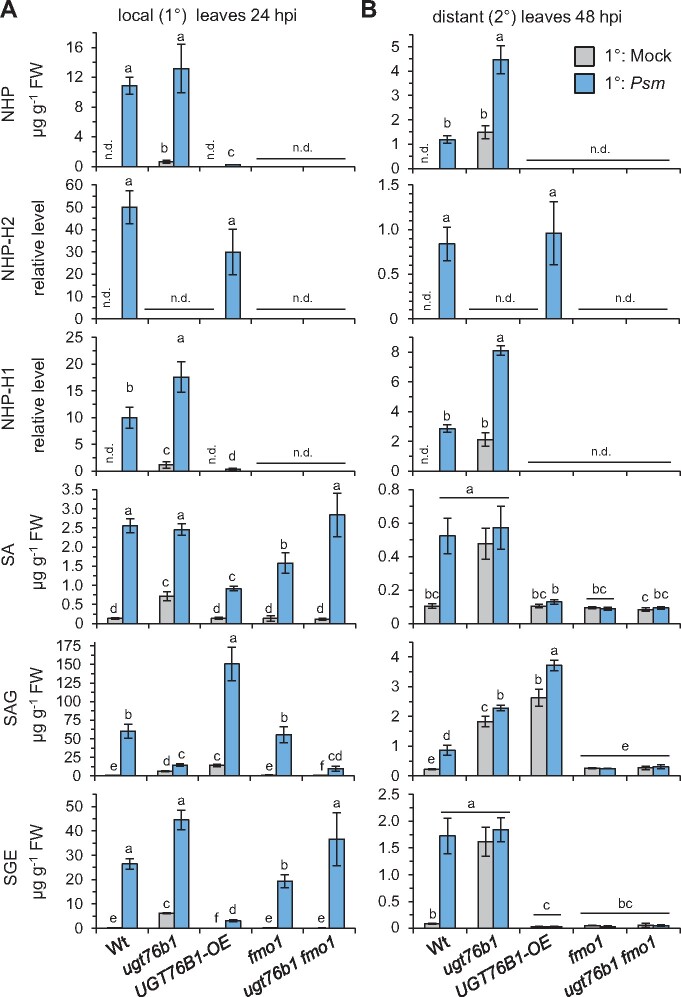
A hallmark of SAR is the accumulation of the SAR-related metabolites NHP and SA in the 1°-inoculated leaves and in the distal, noninoculated leaves following pathogen attack (Hartmann et al., 2018; Vlot et al., 2021). Interestingly, both the O-β-glucoside NHP-H2 and the glucose ester NHP-H1 accumulated in the locally inoculated and the distal leaves of Wt plants alongside with NHP upon Psm inoculation (Figure 8). Similarly, the levels of SA, the O-β-glucoside SAG, and the glucose ester SGE increased systemically in the Psm-inoculated Wt. Consistent with the function of UGT76B1 as an NHP and SA glucosyltransferase, the UGT76B1-OE line systemically accumulated substantial amounts of the O-β-glucosides NHP-H2 and SAG upon Psm attack, whereas accumulation of NHP and SA as well as of their respective glucose esters NHP-H1 and SGE were low in the 1°-inoculated and fully absent from the 2°-distant leaves (Figure 8). The depletion of the levels of free NHP and SA by the enhanced UGT76B1 O-β-glucosylation activity of the UGT76B1-OE plants, therefore, resulted in a complete lack of accumulation of the unbound, presumably signal-active metabolites in the 2° leaf tissue (Figure 8), which is consistent with the SAR-defect of UGT76B1-OE plants (Figure 7B).
Figure 8.
The Psm-induced local and systemic accumulation of NHP, SA, and the respective glucose conjugates is deranged in ugt76b1 knockout and UGT76B1 OE lines. A, Levels of NHP, NHP-H2, NHP-H1, SA, SAG, and SGE in Psm-inoculated or Mock-treated leaves of the indicated Arabidopsis lines at 24 hpi. Bars represent means ± sd of four biological replicates Different letters denote significant differences (P < 0.05, Kruskal–Wallis H test). B, Levels of defense metabolites in 2°-distant leaves upon Psm- or Mock-inoculation of 1° leaves at 48 hpi. Details as described in (A). The accumulation of Pip in these scenarios is depicted in Supplemental Figure S12.
The ugt76b1 mutant contained elevated basal levels of NHP, SA, and their glucosylated derivatives with the exception of the solely UGT76B1-dependent product NHP-H2 (Figure 8). In contrast, the glucose esters NHP-H1 and SGE over-accumulated by the ugt76b1 mutant, whereas the Psm-induced accumulation of the O-β-glucosides NHP-H2 and SAG was fully absent or strongly compromised, respectively. Moreover, ugt76b1 plants were able to further increase the levels of NHP and NHP-H1 after Psm inoculation in the distal, systemic tissue (Figure 8B). These metabolite data are consistent with the observed constitutive SAR phenotype and a further pathogen-inducible increase of the SAR response of ugt76b1 (Figure 7B). In addition, it again corroborates the in vivo O-β-glycosylation activity of UGT76B1 toward NHP and SA. Consistent with the previously characterized crucial function of FMO1 for the biosynthesis of NHP (Hartmann et al., 2018), the ugt76 fmo1 and the fmo1 mutants failed to locally and systemically elevate NHP and both NHP glucose conjugates upon Psm attack (Figure 8). Moreover, both mutants were incapable of accumulating SA and its derivatives specifically in the distant tissue and were compromised to activate SAR (Figures 7, B and 8, B).
In conclusion, our resistance assays and metabolite analyses indicate that the O-β-glycosylation activity of UGT76B1 balances the levels of free NHP and SA both in the absence of pathogen infestation and in the course of SAR induction following pathogen attack. Whereas a loss of UGT76B1 function leads to a hyper-activated SAR-like status, the unregulated, constitutive UGT76B1 expression abrogates the systemic accumulation of free NHP and SA and prevents SAR. Together, our results are in accordance with the notion that the pathogen-induced accumulation of unconjugated NHP functions as an initial, decisive trigger for the induction of SAR.
Discussion
UGT76B1 glucosylates NHP, SA, and ILA to generate the respective O-β-glucosides
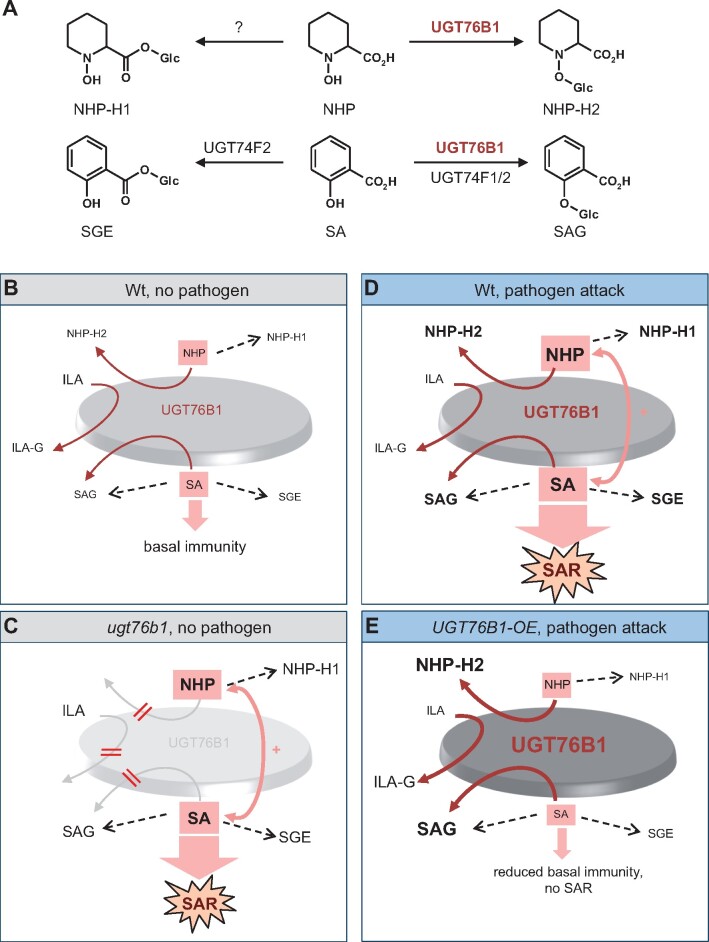
Previous in vitro and in vivo studies have shown that UGT76B1 catalyzes the glucosylation of SA and ILA (von Saint Paul et al., 2011; Noutoshi et al., 2012; Maksym et al., 2018; Bauer et al., 2020). Here, we identify NHP as a third substrate of the Arabidopsis UGT76B1 protein and provide evidence by in vitro and analyses that UGT76B1 catalyzes the pathogen-inducible biosynthesis of NHP-O-β-glucoside, which we named as NHP-H2 (Figures 2, 3, and 9A). The enzyme activity of UGT76B1 toward three different substrates suggests a relatively low substrate specificity. SA, ILA, and NHP share a carboxylic acid and a neighboring hydroxyl group as common features for the conjugation with the activated sugar molecule, although the planar structure of the aromatic SA differs from the aliphatic ILA and the heterocyclic NHP moieties.
Figure 9.
Biochemistry and modulatory action of UGT76B1 in plant basal immunity and SAR. A, In addition to ILA, UGT76B1 glycosylates the two SAR-regulatory metabolites NHP and SA in parallel to form the respective O-β-glucosides NHP-H2 and SAG. The glucosyltransferases UGT74F1 and UGT74F2 also contribute to SA glucosylation. The enzyme catalyzing esterification of NHP to NHP-H1 is not yet characterized. B–E, Model for the function of UGT76B1 as a central hub in the regulation of plant basal immunity and SAR. The model illustrates the four major defense scenarios addressed in this study. Relative metabolite levels are depicted by the size of the letters of aglyca and glucosides, which also subsumes the enhanced or repressed biosynthetic activities. Increasing darkness of the central disc, sizes of letters, and widths of the dark red arrows symbolize the activity of UGT76B1 for each scenario. The strength of immune/SAR signaling is indicated by the sizes of purple arrows. B, Naïve Wt is in a state of contained basal defense. UGT76B1 provides a metabolic hub controlling the levels of the unconjugated, immune-stimulating NHP and SA. Both substrates can be alternatively glucosylated by UGT76B1 to the putatively inactive SAG and NHP-H2. Thereby, the mutual amplification loops of NHP and SA (+) and, consequently, basal defense are contained. ILA is an additional, competing substrate that inhibits SAG and NHP-H2 formation. C, The loss of UGT76B1 glucosylation releases this control. NHP and SA mutually enforce each other and promote a SAR-like, NHP- and SA-dependent enhanced basal immune status. The loss of UGT76B1 abolishes the formation of ILA glucoside and NHP-H2, whereas SAG and SGE are still produced due to the presence of other Arabidopsis SA glucosyltransferases. In addition, the elevated NHP level is accompanied by the enhanced formation of the UGT76B1-independent NHP-H1. D, The pathogen-induced activation of FMO1-mediated NHP biosynthesis and ICS1-regulated SA biosynthesis triggers SAR. In the Wt, UGT76B1 is transcriptionally induced as well. This inducible UGT76B1 expression modulates the levels of free NHP and SA and thereby dampens immune signaling once SAR is activated. E, In UGT76B1-OE plants, a constitutively de-regulated UGT76B1 expression shifts the basal und pathogen-inducible NHP and SA metabolic pools toward O-β-glucoside (NHP-H2, SAG) formation. This compromises NHP and SA accumulation, reduces basal immunity, and abrogates SAR.
The structural modeling of the UGT76B1 protein shows the presence of a wide substrate-binding pocket due to the small amino acid side chains that face the catalytic site, which may promiscuously allow the binding of a variety of substrates (Figure 3A;Supplemental Figure S3). Specifically, the modeling supports our experimental findings that SA, ILA, and NHP are alternative substrates of UGT76B1. Consequently, these substrates compete with each other in vitro. The reaction of one substrate mutually inhibits the glucosylation of the respective other aglyca, such as the inhibition of SA and NHP glucosylation by ILA and the mutual inhibition of SA and NHP glucosylation by NHP and SA, respectively (Figures 3, B and 9, A; Supplemental Figures S3 and S5) (Maksym et al., 2018). Thereby, SA-, NHP-, and ILA-O-glucosides, but not the respective glucose esters are formed (Figure 4; Supplemental Figure S7; see below; Li et al., 2015). Both ILA glucoside and NHP-H2 formation are strictly dependent on UGT76B1. In contrast, SAG can be also formed by other glucosyltransferases in vitro and in vivo, namely by AtUGT74F1 and AtUGT74F2 (Dean and Delaney, 2008; von Saint Paul et al., 2011; Noutoshi et al., 2012; George Thompson et al., 2017) (Figure 9A).
Arabidopsis accumulates the UGT76B1-generated β-glucoside NHP-H2 and the UGT76B1-independent glucose ester NHP-H1 upon pathogen attack
Arabidopsis leaves accumulate NHP-H2 in strict dependence of a functional UGT76B1 gene upon inoculation with the bacterial pathogen P. syringae, in response to treatment with the priming-inducing chemical BTH, and by exogenous feeding with Pip. In contrast, the previously identified NHP-H NHP-H1 accumulates independently of functional UGT76B1 in response to P. syringae inoculation (Figures 2, D and 8; Hartmann and Zeier, 2018). conversion of D9-labeled NHP into D9-labeled NHP-H1 and D9-labeled NHP-H2 and the strict dependency of the accumulation of both NHP-Hs on the NHP biosynthetic genes ALD1, SARD4, and FMO1 demonstrate their direct derivation from NHP (Figure 2). Digestion assays with esterase and glucosidase, the mass spectra of the silylated NHP-Hs, and the known O-β-glucoside-catalyzing activity of UGT76B1 suggest that NHP-H1 and NHP-H2 are NHP glucose ester and NHP-O-β-glucoside, respectively (Figure 9A). Based on the LC–MS-derived characteristics (Figure 1; Supplemental Figure S1), NHP-H2 may be identical to the previously LC–MS-detected NHP-H described by Chen et al. (2018). Recent methylation-based analysis from the same research group corroborates that this hexoside corresponds to the NHP-O-β-glucoside (Holmes et al., 2021). The conjugation modes of NHP are therefore analogous to those of SA, for which the O-glucoside SAG and the SGE exist (Figure 9A; Dean and Delaney, 2008; George Thompson et al., 2017).
Transcripts of the biosynthetic genes responsible for l-Lys to NHP conversion (ALD1, SARD4, and FMO1) and NHP-O-β-glucoside formation (UGT76B1) accumulate following microbial attack, indicating that this l-Lys catabolic pathway is regulated at the level of transcription (Hartmann and Zeier, 2019). Moreover, the immune regulators EDS1 and PAD4 positively influence Pip, NHP, and NHP glucoside formation (Figure 2, E and F; Návarová et al., 2012; Hartmann et al., 2018). Interestingly, the two identified NHP hexose derivatives differ with respect to the regulation of their biosynthesis by SA: the accumulation of NHP-H2 requires intact SA biosynthesis (ICS1/SID2) and perception (NPR1), whereas NHP-H1 generation is negatively influenced by an intact SA pathway. Furthermore, NHP-H1 also hyper-accumulates in conjunction with the ugt76b1-enhanced basal defense and shows an accumulation pattern similar to free NHP (Figures 2, E and F and 8; Hartmann et al., 2018).
The reduced accumulation of NHP-H2 by SA pathway mutants could also be the result of a reduced expression of UGT76B1. Indeed, the transcript abundance of UGT76B1 was repressed by the sid2 mutant following mock and Psm treatments and enhanced by exogenous SA treatment (Gruner et al., 2013; Bernsdorff et al., 2016). Thus, the over-accumulation of NHP and NHP-H1 by SA pathway-defective mutants may result from an attenuated NHP-H2 production. However, the functional implications of these different regulatory patterns are not yet resolved.
UGT76B1 contains the levels of free NHP to modulate basal immunity
Unstressed plants keep NHP at very low basal levels and exhibit only modest basal immunity against infection by adapted pathogens (Figures 1, 7, 8, and 9B; Hartmann et al., 2018; Schnake et al., 2020). Upon pathogen contact, however, Wt plants can induce SAR to subsequent infection and thereby achieve a primed state of enhanced systemic immunity that allows a boosted activation of defense responses (Figure 9D;Fu and Dong, 2013; Shah and Zeier, 2013). The pathogen-induced activation of the pipecolate pathway, which entails accumulation of Pip and NHP in locally inoculated and distant leaf tissue, is a key and indispensable event to trigger SAR and the associated primed state (Návarová et al., 2012; Bernsdorff et al., 2016; Chen et al., 2018; Hartmann et al., 2018; Schnake et al., 2020).
Our results show that the loss of UGT76B1 leads to a markedly elevated endogenous level of NHP already in the absence of pathogen contact. This goes hand in hand with enhanced basal resistance to P. syringae and increased basal levels of SA, SA derivatives, and defense-related gene expression of this mutant (Figures 1 and 6–8; Supplemental Figure S9). Therefore, the ugt76b1 mutant exhibits a constitutively primed, SAR-like state (Figure 9C). However, ugt76b1 plants also show a slightly reduced rosette size under normal growth conditions as a negative impact (von Saint Paul et al., 2011; Supplemental Figure S10). Notably, basal resistance, SA accumulation, and defense-related gene expression of the ugt76b1 fmo1 double mutant, which is entirely blocked in NHP biosynthesis, is similar to or even below Wt levels (Figures 1, 6, and 7; Supplemental Figures S9 and S11). This indicates that elevated NHP triggers immune priming of ugt76b1. Thus, UGT76B1 as an SA, ILA, and in addition, NHP glucosyltransferase suppresses such an NHP-activated and SA-dependent defense status of Wt plants (Figure 9, B and C; see below).
Our results on the SAR-like phenotype of ugt76b1 and the NHP-O- β-glucosylating activity of UGT76B1 are consistent with the results of another parallel study (Mohnike et al., 2021). In addition to impaired biochemical inactivation of NHP, as observed by ugt76b1, de-regulated overproduction of NHP in Arabidopsis defective in the transcription factor CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3 (CAMTA3) or plants OE the calcium-dependent protein kinase CPK5 can cause resistance-enhancing phenotypes and a disturbed balance of growth and defense (Guerra et al., 2020; Kim et al., 2020; Sun et al., 2020).
UGT76B1 regulates the levels of SAR-active metabolites and coordinates SAR
The pathogen-induced SAR establishment of Arabidopsis is closely associated with the accumulation of the immune activators NHP and SA in the locally inoculated and in distantly located leaves (Hartmann et al., 2018). In addition, the glucose conjugates NHP-H2, NHP-H1, SAG, and SGE systemically accumulate in response to a local P. syringae attack (Figures 8 and 9D). The incapability of NHP-H2 formation by ugt76b1 not only increased the basal levels of free NHP and SA, but also prompted plants to systemically accumulate enhanced levels of NHP after pathogen inoculation (Figure 8B). In addition, the constitutive SAR of ugt76b1 can be further enhanced following pathogen attack. However, the degree of a further pathogen-inducible SAR effect of ugt76b1 plants may thereby depend on the strength of the constitutive SAR that is already activated in the course of development (Figure 7B; Supplemental Figure S11). The constitutive and the pathogen-inducible SAR effects observed for ugt76b1 were absent from the ugt76b1 fmo1 double mutant, indicating that they depended on (constitutive and inducible) NHP accumulation.
Importantly, the overexpression of UGT76B1 caused a metabolic imbalance that abolished SAR. The pool of all three NHP derivatives that accumulate after pathogen attack of the Wt was entirely shifted toward the O-β-glucoside NHP-H2 in local and in systemic leaves by the constitutive overexpression of UGT76B1. Similarly, SAG accumulation was enhanced and SA and SGE formation were reduced in the UGT76B1-OE line (Figure 8). On the one hand, this result is consistent with the biochemical function of UGT76B1 as an NHP- and SA-O-β-glucosyltransferase. On the other hand, the concomitant loss of SAR in the UGT76B1-OE line suggests that the O-β-glucosides NHP-H2 and SAG are SAR-inactive compounds (Figure 7B). SAG is considered as an inactive form of SA that is potentially stored in the vacuole and might be remobilized to free SA (Klessig et al., 2018). It is unclear, whether a similar function of NHP-H2 as a hydrolyzable reservoir for NHP exists.
The loss of pathogen-induced SAR by the UGT76B1-OE plants can be explained by their compromised local and systemic accumulation of SAR-active, free NHP, and SA, which is caused by excessive O-β-glucosylation (Figure 9E). Consistent with our results on Arabidopsis, a parallel study by Holmes et al. (2021) reports similar tendencies on genetically induced SAR in tomato (Solanum lycopersicum). In their study, SAR was induced by transient expression of the Arabidopsis NHP biosynthetic genes ALD1 and FMO1 in tomato; however, co-expression of the Arabidopsis UGT76B1 triggered NHP to NHP-O-β-glucoside conversion and abolished SAR. In the Arabidopsis Wt situation, UGT76B1 transcripts systemically accumulate after pathogen attack, and NHP biosynthesis and NHP glucosylation are timely coordinated events (Supplemental Figure S2). In sum, this suggest that the UGT76B1-mediated NHP- and SA-O-β-glucosylation coordinates, together with a pathogen-induced activation of the NHP and SA biosynthetic pathways, the levels and homeostasis of SAR-inducing metabolites, and the timing of SAR. The role of the glucose ester NHP-H1 in SAR and the glycosyltransferase responsible for its biosynthesis remains to be determined (Figure 9A).
UGT76B1 functions as a regulatory hub integrating the immune-active small molecules NHP, ILA, and SA
UGT76B1-dependent SA glucosylation is an important means to negatively control plant immune responses, in conjunction with at least two other SA glucosyltransferases of Arabidopsis, AtUGT74F1 and AtUGT74F2 (Dean and Delaney, 2008; von Saint Paul et al., 2011; Noutoshi et al., 2012; George Thompson et al., 2017). Plants unable to accumulate SA display a strongly impaired local resistance to diverse virulent and avirulent pathogens and are compromised in SAR (Gaffney et al., 1993; Volko et al., 1998; Nawrath and Métraux, 1999; Bernsdorff et al., 2016). Moreover, NHP will only be able to induce a strong SAR, if plants possess an intact SA biosynthesis pathway, indicating a close cooperation and mutual activation of NHP and SA in SAR induction (Bernsdorff et al., 2016; Hartmann et al., 2018; Figure 9, B–E).
Consistent with this, the ugt76b1 mutation that elevates free NHP levels no longer causes induction of acquired resistance when introgressed to the SA-depleted NahG sid2 background. In fact, NahG sid2 and NahG sid2 ugt76b1 plants display a similar and strongly attenuated resistance to P. syringae, which is more severely compromised than the resistance of fmo1 and ugt76b1 fmo1 lines (Figure 7A). This illustrates the key function of SA for both basal immunity and acquired resistance and indicates that SA is necessary for the NHP-triggered resistance response observed by ugt76b1 (Figure 9). However, the SA pathway can be also activated independently of NHP at pathogen inoculation sites, as shown for Pseudomonas-inoculated ald1 mutants devoid of Pip and NHP (Bernsdorff et al., 2016). This scenario implies that there are additional interactions regulating NHP and SA levels.
ILA is a further player in the UGT76B1-associated immune hub. Exogenous application of ILA activates defense in a synergistic manner with SA (Bauer et al., 2020) and it increases NHP and SA levels (Figure 5). In vitro, ILA inhibits the UGT76B1-mediated glucosylation of NHP and SA, which may contribute to the increased levels of both immune modulators in vivo (Figures 3 and 9; Supplemental Figure S5; Maksym et al., 2018). The deficiency of ILA glucosylation of the ugt76b1 mutant is in line with its increased endogenous ILA levels (von Saint Paul et al., 2011; Maksym et al., 2018), which, in turn, may also contribute to the primed state of ugt76b1. However, ILA may not enhance defense apart from this UGT76B1-mediated action, because its exogenous application to ugt76b1 mutants did no more amplify SA-induced expression of defense marker genes (Bauer et al., 2020). On the other hand, ILA may play a role in mitigating defense responses. At the sites of Pst inoculation, the endogenous ILA concentration is lowered in response to infection (Maksym et al., 2018). In this scenario, the ILA-mediated inhibition of NHP and SA glucosylation by UGT76B1 would be reduced as suggested by the in vitro studies. Consequently, an enhanced glucosylation of NHP and SA would attenuate the defense signaling toward a balanced immune status in the course of infection. Currently, these roles of ILA cannot be ultimately resolved in vivo, since the biosynthetic pathway and corresponding loss-of-function mutants have not yet been identified.
In conclusion, UGT76B1 provides a common regulatory hub orchestrating basal defense and SAR by the glucosylation of the immune-active small molecules NHP, ILA, and SA. NHP and SA foster defense and mutually amplify each other, while their action may be further promoted by ILA (Figure 9; Bernsdorff et al., 2016; Chen et al., 2018; Wang et al., 2018; Bauer et al., 2020). The conjugation by UGT76B1 contains the levels of active NHP and SA in an interactive manner and, as our in vitro studies indicate, is associated with the mutual competition of the aglycon substrates, while, in addition, NHP and SA positively affect each other (Figure 9). The loss of functional UGT76B1 releases this control and leads to an activated state of basal defense, which is triggered by NHP, further enhanced by ILA, and eventually dependent on SA as the central downstream signal. Thus, ugt76b1 establishes a SAR-like status in the absence of a pathogen infestation (Figure 9C). In contrast, overexpression of UGT76B1 eliminates SAR (Figure 9E). In Wt plants, the promiscuous glucosylating activity of the pathogen-inducible, functional UGT76B1 maintains a nonactivated, steady state of basal defense of naïve plants and enables the dynamic, interactive control of the immune-active compounds during SAR activation in response to pathogens (Figure 9D).
Materials and methods
Plant materials and growth conditions
Wt A. thaliana accession Columbia (WT), ugt76b1-1 knockout mutant (SAIL_1171A11), a line constitutively overexpressing UGT76B1 (von Saint Paul et al., 2011), ald1 (SALK_007673, Návarová et al., 2012), sard4-5 (GK_428E01; Rosso et al., 2003; Hartmann et al., 2017), fmo1-1 (SALK_026163; Bartsch et al., 2006; Mishina and Zeier, 2006), npr1-2 (NASC ID: N3801; Cao et al., 1997), pad4-1 (NASC ID: N3806 Glazebrook et al., 1996), eds1-2 (Bartsch et al., 2006), the SA-depleted double mutant NahG sid2 (Veeragoni et al., 2020), and NahG sid2 ugt76b1 triple mutants were used. fmo1 ugt76b1 was generated from the above single mutants and provided by Wei Zhang. Multiple mutants were obtained by genetic crossing.
Mutants were obtained from the Arabidopsis seed stock center (Scholl et al., 2000) except for NahG and sid2-1 (Gaffney et al., 1993; Nawrath and Métraux, 1999). Plants were grown in a controlled growth chamber (light/dark regime 10/14 h at 20/16°C, 80/65% relative humidity, light at 100 µmol m−2 s−1, type 840 lamps (Osram, Garching, Germany) were used for illumination) on a peat moss-based substrate (Floragard Multiplication substrate, Oldenburg, Germany): quartz sand (8: 1) mixture or using similar conditions (21°C/18°C) for Psm inoculations and SAR experiments as described by Gruner et al. (2018). For liquid cultures, half MS medium with 1% sucrose was used and the seedling was germinated and grown while shaking at 100 rpm in the same conditions described above.
Application of chemicals and sample collection
BTH (Syngenta, Basel, Switzerland) was applied by spraying a 1 mM aqueous solution onto 3-week-old plants; 20 rosettes were harvested after 24 or 48 h. For Pip treatment 5-week-old plants were either watered with 10 mL water or 1 mM d/l-Pip (Sigma-Aldrich, St Louis, MO, USA) solution as described before (Návarová et al., 2012); leaves of five rosettes were harvested 48 h after the onset of the treatment for one biological sample. For ILA (Interchim, Montluçon, France) treatment, ∼20 Wt seedlings were germinated and grown for 12 days in six-well plates under conditions described above. Then, the media of one half of the samples was replaced with fresh half MS and the remaining half with 500 µM ILA containing half MS medium. The treatment was applied for 24 and 48 h. Finally, the shoots were harvested after washing with distilled water and blotting on tissue paper. In all cases, the harvested samples were immediately frozen in liquid nitrogen.
labeling experiments with isotope-labeled metabolites
Treatments with isotope-labeled metabolites were essentially performed as described in Hartmann et al. (2018). Three to four mature leaves of 5-week-old soil-grown Arabidopsis Wt or fmo1 plants were infiltrated in the morning either with Psm (OD600 = 0.005) or 10 MgCl2 (Mock) as described above. Four to 6 h after the inoculation event, the same leaves were infiltrated with a 2 mM solution of d9-labeled NHP prepared freshly in HPLC-grade water. Water infiltrations served as control treatments. DL-2-piperidine-d9 carboxylic acid (D9-Pip; Sigma-Aldrich 688444) was co-infiltrated at a final concentration of 2 mM as part of the final bacterial suspension or Mock-solution. Infiltrated leaves were harvested at 48 hpi (in reference to the first infiltration event with Psm or mock solutions) and GC–MS analysis of trimethylsilyl-derivatized metabolites was carried out according to the described analytical protocols (section: GC–MS analyses). Authentic d9-labeled 1-hydroxypiperidine-2-carboxylic acid (d9-NHP) was synthesized according to a protocol of Murahashi and Shiota (1987), as described in Hartmann et al. (2018) using piperidine-d11 (CAS Number: 143317-90-2) as reactant.
UGT enzyme assay
The coding sequence of UGT76B1 was amplified from a cDNA clone using the oligonucleotides UGT76B1_Popin_F and UGT76B1_Popin_R (Supplemental Table S1), cloned into pOPINS3C vector (Berrow et al., 2007), and transformed into Escherichia coli pRareGold. The recombinant protein was purified from a 4-l culture in auto induction medium (Studier, 2005) by affinity chromatography using a His-Trap column (GE Healthcare, Chicago, IL, USA). Sample was eluted with an imidazole-buffer gradient from 15 to 300 mM. Pooled UGT76B1-containing fractions were incubated with precision protease (1:100 ratio) to cleave off the His6-SUMO tag. The cleaved sample was diluted (to lower salt concentration to 50 mM NaCl) and applied on a Q column (GE Healthcare). The protein was eluted from the Q column with a gradient of 50–1,000 mM NaCl in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) 7.5, 2% (v/v) glycerol, and 2 mM DTT. The fractions containing pure UGT76B1 were pooled, concentrated to 5 mL, and applied onto a size-exclusion Superdex200 column (GE Healthcare) equilibrated with 20 mM HEPES 7.5, 200 mM NaCl, 2 mM DTT. The tag-free UGT76B1-containing fractions were pooled and concentrated.
Enzyme activity assays were performed as described (Meßner et al., 2003) using 0.5-µg recombinant UGT protein, the aglyca SA, NHP, and ILA at the indicated concentrations and combinations, and 4-mM UDP glucose (2 mM in case of 14C-UDP glucose (Hartmann Analytics, Braunschweig, Germany) labeling at 30°C for 30 min unless otherwise noted. The product formation was analyzed by HPLC to measure SAG through absorption at 302 nm or radiolabeled products, and by LC–MS for all other cases. Preparative reactions to produce NHP-H2 using recombinant UGT76B1 and SGE-containing SA glucosides using UGT74F2 (AT2G43820) were incubated at least 2 h at 30°C.
Esterase and β-glucosidase digestion
Leaf extracts from 5-week-old Psm ES4326-inoculated Arabidopsis Col-0 plants (48 hpi) accumulating the NHP- and SA-conjugates of interest were used for esterase and β-glucosidase digestion experiments. Shock-frozen leaf tissue was freshly-ground to a fine powder and extracted twice with ice-cold MeOH/H2O (80:20, v/v) in a ratio of 1 mL per 100 mg leaf tissue. The resulting extract was then evaporated to dryness and re-suspended in water. Aliquots of this solution were incubated in sodium phosphate buffer (50 mM final, pH 6.0) for 15 h at 30°C, either in the presence of 50 U mL−1 porcine liver esterase (E3019; Sigma-Aldrich) or 0.5 U mL−1 β-glucosidase from almonds (7512.1; Carl Roth, Karlsruhe, Germany). Aliquots incubated in buffer without the respective enzymes served as control treatments. Reactions were stopped by addition of an excess amount of MeOH/H2O solution (80:20, v/v) that was supplemented with an internal standard mix. Samples were then immediately analyzed according to the GC–MS-based analytical procedure described below. The amounts of NHP-H1, NHP-H2, SAG, and SGE were quantified in relation to ribitol (CAS N°: 488-81-3, A5502; Sigma-Aldrich) as internal standard.
The hydrolytic experiments were independently repeated using the in vitro produced NHP-H2 and an NHP-H2 containing leaf extracts (prepared from 4-week-old Wt plants spray-treated with 1 mM BTH for 48 h) using LC–MS analytics. Aliquots of the in vitro reaction or freeze-dried extracts dissolved in water were treated with 15 U mL−1 porcine liver esterase (pH 8.0) or 0.9 U mL−1 almond β-glucosidase (pH 5.0) (E2884, G0395; Sigma-Aldrich) for 5 h incubation at 30°C. SAG and SGE contained in the extracts were used as internal controls of the hydrolysis by glucosidase and esterase; in fact, SGE was even susceptible to the alkaline condition (pH 8) used for esterase incubation.
LC–MS analyses
For determination of SA- and Pip-related metabolites, frozen-dried materials were crushed to powder and lyophilized. The extraction and measurement of the compounds were carried out as described before with minor modifications (Wenig et al., 2019). Briefly, ∼20 mg of freeze-dried material was resuspended in 1.5 mL 70% methanol, vigorously shaken at 4°C for 1 h, and subsequently centrifuged at 18,000g for 10 min. Supernatants were then transferred to new 2-mL tubes, concentrated in vacuo (RVC 2-25 CDplus, Christ) for 2 h, and shortly frozen at −80°C. The samples were freeze-dried overnight, re-dissolved in 100-µL acetonitrile: H2O (1:1, v/v), and then centrifuged for 5 min at 18,000g at 4°C. Next, 90 µL of supernatants were transferred to microwell plates fitted with 0.2-µm-type polyvinylidene difluoride (PVDF) filters and centrifuged. Five microliters of the extract was injected twice as technical replicate for the LC–MS analyses. Pip, NHP, and NHP-H2 were detected by positive ionization mode, whereas SA, SAG, and SGE were measured by negative ionization mode. Mass spectra were acquired in a mass range of 50–1300 m/z. SA (Sigma-Aldrich), SAG (Santa Cruz Biotechnology Dallas, TX, USA), SGE, Pip (Sigma-Aldrich), and NHP were identified using authentic standards. SGE was produced using recombinant Arabidopsis UGT74F2 expressed as a GST fusion protein (Lim et al., 2002; Meßner et al., 2003). NHP was synthesized according to Hartmann et al. (2018) and confirmed by GC–MS analyses or by LC mass spectrometry. LC–MS/MS fragmentation patterns of NHP and NHP-H2 were in accordance with Chen et al. (2018).
SA and SAG were quantified against an internal standard curve with ten calibration points and two internal standards (p-nitrophenol, Fluka; camphorsulfonic acid, Sigma-Aldrich) at 1 mg L−1. Pip and NHP were quantified against external standard curves of (d,l-piperidine-2-carboxylic acid, Sigma-Aldrich) and NHP with six calibration points. Normalized peak areas were used as a semi-quantitative estimate of NHP-H2 and SGE, since the reference compounds were obtained as unpurified reaction products of the recombinant glucosyltransferases UGT76B1 and UGT74F2, respectively. SGE peaks were normalized with the internal standard camphorsulfonic acid. Normalization of Pip, NHP, and NHP-H2 was performed using the total ion chromatogram during the gradient elution. Retention times and m/z values: SAG 8.0–8.5 min, 299.0750; SGE 9.2–9.5 min, 299.0750; SA 10.0–10.2 min, 137.0250; internal standards p-nitrophenol 10.0–10.1 min, 138.0195; camphorsulfonic acid 9.0 min, 231.0695; Pip 2.1–2.4 min, 130.0860; NHP 1.4–1.7 min, 146.0817; NHP-H2: 3.3–3.7 min, 308.1346.
GC–MS analyses
GC–MS-based analyses of plant metabolites (NHP-H1, NHP-H2, SAG, and SGE) was performed as detailed in Hartmann et al. (2018) and Stahl et al. (2019) with minor modifications. Briefly, 50 mg of pulverized, frozen leaf samples were extracted twice with 1 mL of MeOH/H2O (80:20, v/v). For internal standardization, 1 µg of salicin and ribitol were added. Six hundred microliters of the extract were evaporated to dryness, re-dissolved in 100 µL of 100-mM sodium phosphate (pH 6.0), and the solvent removed again. Free hydroxyl groups of the analytes were converted into their trimethylsilyl derivatives by adding 20 µL of pyridine, 20 µL of N-methyl-N-trimethylsilyltrifluoroacetamide containing 1% trimethylchlorosilane (v/v), and 60 µL of hexane. The mixture was heated to 70°C for 30 min. After cooling, samples were diluted with hexane, and 2 µL of the solution was separated on a gas chromatograph (GC 7890A; Agilent Technologies) equipped with a Phenomenex ZB-35 (30 m × 0.25 mm × 0.25 µm) capillary column. The following GC temperature settings were used: 70°C for 2 min, with 10°C/min to 250°C, followed by 2°C/min to 270°C and 10–320°C, finished by a final hold step at 320°C for 5 min. Mass spectra were recorded in the electron ionization mode between m/z 50 and m/z 750 with a 5975C mass spectrometric detector (Agilent Technologies). Metabolites were analyzed using the Agilent MSD ChemStation software. For quantification, substance peaks of selected ion chromatograms (NHP-H1: m/z 172; NHP-H2: m/z 172; SAG: m/z 267; SGE: m/z 193) were integrated, and peak areas of plant metabolites were related to those of the internal standards salicin (m/z 268) or ribitol (m/z 307).
RT-qPCR
For determination of the expression of defense-related genes, leaf samples were collected from soil-grown 4-week-old Wt, ugt76b1, fmo1, and fmo1 ugt76b1 lines grown under conditions described above. Rosettes of three plants were pooled to represent one biological sample and each genotype was analyzed in four replicates. For determination of FMO1 expression of ILA-treated Wt seedlings, treatments and sample collection was carried out as described above. RNA extraction, cDNA synthesis, and expression analysis of the genes was performed according to Bauer et al. (2020).
Modeling
A homology model of UGT76B1 was created on the template of UGT74F2 with SA and UDP glucose (PDB ID: 5U6M, George Thompson et al., 2017) using the phyre2 software (http://www.sbg.bio.ic.ac.uk/phyre2; Kelley et al., 2015) with manual adjustments. The position of UDP-glucose in the structure was deduced by superposition of UGT76B1 model with flavonoid 3-O glucosyltransferase (PDB ID: 2C1Z; Offen et al., 2006). The 3D models of N-OH Pip, SA, and ILA acid were placed into the protein using the docking program PatchDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/index.html) and analyzed using COOT (Emsley et al., 2010). The localization near UDP-glucose was used as an additional criterion for positioning the aglycon substrates.
Bacterial inoculation and infection assays
Full-grown leaves of 4–5-week-old Wt, ugt76b1, fmo1, fmo1 ugt76b1, NahG sid2, and NahG sid2 ugt76b1 plants were gently pressure infiltrated either with 10 mM MgCl2 as control, or with suspensions of Pst DC3000 or Psm strain ES4326 (Psm;Hartmann et al., 2017) in 10 mM MgCl2. For basal resistance assays, plants inoculated with Pst (OD600nm = 0.0001) were grown for 3 days (72 h) at growth conditions described above. Three leaf discs from three independent plants, representing one biological sample, were pooled 2 and 72 h after the inoculation and immersed in 500 µL of 10 mM MgCl2 solution containing 0.01% Silwet L70 (Lehle Seeds). The analysis of the bacterial growth was carried out as described before (Katagari et al., 2002). Each treatment was replicated 5 times and the complete experiment was independently repeated 2 times. Combined data of all experiments are displayed. For the determination of metabolite accumulation in response to Psm, a bacterial suspension of OD600nm = 0.005 was used for leaf inoculation. Mock- and Psm-inoculated leaves were harvested at different times after the treatment. Six leaves from two different plants were routinely pooled for one biological replicate, and three to four biological replicates per treatment analyzed by GC–MS.
SAR assays
To induce SAR, three 1° rosette leaves of 5-week-old, short-day-grown Arabidopsis plants were inoculated with Psm by gently syringe-infiltrating bacterial suspensions in 10 mM MgCl2 (OD600 = 0.005) into the abaxial sides of the leaves (Gruner et al., 2018). Analogous infiltration of a 10 mM MgCl2 solution served as a mock-control treatment. Two days later, three upper (2°) rosette leaves of the plants were challenge-inoculated by infiltration with a suspension (OD600 = 0.001) of Psm carrying the Photorhabdus luminescens luxCDABE operon (Psm lux; Fan et al., 2008). Bacterial growth was assessed 2.5 days later by punching leaf discs from inoculated leaf areas and determining the bioluminescence of Psm lux with a Sirius FB12 luminometer (Berthold Detection Systems). Bacterial numbers were expressed as relative light units (rlus) per cm2 leaf area, which show strong positive correlations with colony-forming units (cfu cm−2) assessed by the more traditional, plating-based assays (Gruner et al., 2018).
Statistics
Statistical analyses of Figures 1, 5, and 6 and of Supplemental Figures S8, S9, and S10 were performed in R version 3.5.1 for Windows (https://www.r-project.org/). The WRS2 package based on Wilcox’ WRS function was used. One-way multiple group comparisons were tested in R using the robust one-way analysis of variance (ANOVA) function t1way with Lincon post hoc test. P-values were Holm-corrected and adjusted P-values were used for analysis (Mair and Wilcox, 2018). Two groups were compared using Welch two-sample t tests (Figures 5, A and 6, C; Supplemental Figure S8) or t test in case of samples passing normality and equal variance tests (Supplemental Figure S8D).
To test for statistical differences in datasets of bacterial growth experiments (Figure 7; Supplemental Figure S11) and GC–MS-based metabolite measurements (Figures 2 and 8; Supplemental Figure S12), log10-transformed measuring values were subject to ANOVA and a post hoc Tukey’s HSD test or a Kruskal–Wallis H test using the IBM™ SPSS™ statistics software version 26 (Hartmann et al., 2018). For pairwise comparison of metabolite data, a two-tailed Student’s t test was applied (Figure 4; Supplemental Figures S2 and S6).
Accession numbers
ALD1 (AT2G13810), EDS1 (AT3G48090), FMO1 (AT1G19250), ICS1/SID2 (AT1G74710), NPR1 (AT1G64280), PAD4 (AT3G52430), SARD4 (AT5G52810), and UGT76B1 (AT3G11340).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. UGT76B1 loss-of-function abolished the occurrence of m/z 308.1246 that co-elutes with the in vitro produced NHP glucoside (supports Figures 1 and 3).
Supplemental Figure S2. Time course of accumulation of NHP and NHP glucose conjugates in Psm-inoculated leaves of Arabidopsis Col plants (supports Figure 2).
Supplemental Figure S3. Structural modeling of aglyca at the substrate-binding pocket of UGT76B1 (supports Figure 3).
Supplemental Figure S4. The GC/MS-mass spectra of NHP-H2 detected in vitro and are identical (supports Figures 2 and 3).
Supplemental Figure S5. SA and ILA inhibit NHP-H2 formation, whereas NHP inhibits the formation of ILA-O-β-glucoside by UGT76B1 in vitro (supports Figure 3).
Supplemental Figure S6. Effects of esterase and glucosidase treatments on the levels of free NHP (supports Figure 4).
Supplemental Figure S7. Esterase and ß-glucosidase treatments of plant extracted NHP, ILA, and SA glycosides (supports Figure 4).
Supplemental Figure S8. Exogenous ILA enhances the accumulation of NHP-H2, SAG, and SGE. UGT76B1 is slightly induced by exogenous ILA application (supports Figure 5).
Supplemental Figure S9. Introgression of fmo1 mutation into ugt76b1 reverses the expression of SID2 and defense marker genes to Wt level (supports Figure 6).
Supplemental Figure S10. Rosette size of Wt, ugt76b1, fmo1, fmo1 ugt76b1, NahG sid2, and NahG sid2 ugt76b1 plants (supports Figure 7).
Supplemental Figure S11. Additional SAR assays for Wt plants, ugt76b1 knockout, and UGT76B1-OE lines (supports Figure 7B).
Supplemental Figure S12. Local and systemic accumulation of Pip by ugt76b1 knockout and UGT76B1-OE lines (supports Figure 8).
Supplemental Table S1. Oligonucleotides used for RT-qPCR analyses and cloning.
Supplementary Material
Acknowledgments
We are grateful to Wei Zhang for discussion, critical reading of the manuscript, and generation of the fmo1 ugt76b1 double mutant. Elisabeth Georgii supported us by discussion and statistical analyses. Corina Vlot-Schuster and Jörg Durner were instrumental throughout the project. We thank Daniel Moser for assisting I.Y. in inoculation assays. The work was supported by the Helmholtz Zentrum München (A.R.S) and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) via a DFG grant to J.Z. (ZE467/6-2) and Germany’s Excellence Strategy (EXC 2048/1 – Project ID: 390686111).
D.W.M., S.B., M.H., and I.Y. performed experiments. M.H. and J.Z. analyzed metabolites by GC–MS, B.G. contributed expression analyses, and B.L. performed LC–MS experiments. R.J. modeled the binding of substrates at the active site. D.W.M., S.B., M.H., J.Z., and A.R.S. conceived the project and designed experiments; D.W.M., S.B., J.Z., and A.R.S. wrote the paper. All authors read and approved the final version of the article.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) are Anton R. Schäffner (schaeffner@helmholtz-muenchen.de) and Jürgen Zeier (juergen.zeier@uni-duesseldorf.de).
References
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid–independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Mekonnen DW, Geist B, Lange B, Ghirardo A, Zhang W, Schäffner AR (2020) The isoleucic acid triad: distinct impacts on plant defense, root growth, and formation of reactive oxygen species. J Exp Bot 71: 4258–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsdorff F, Döring AC, Gruner K, Schuck S, Bräutigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28: 102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res 35: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chen YC, Holmes EC, Rajniak J, Kim JG, Tang S, Fischer CR, Mudgett MB, Sattely ES (2018) N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc Natl Acad Sci USA 115: E4920–E4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JV, Delaney SP (2008) Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol Plant 132: 417–425 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF (2017) How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol 15: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P, Rekhter D, Ding Y, Feussner K, Busta L, Haroth S, Xu S, Li X, Jetter R, Feussner I. et al. (2016) Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 28: 2603–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467 [DOI] [PubMed] [Google Scholar]
- Edwards R (1994) Conjugation and metabolism of salicylic acid in tobacco. J Plant Physiol 143: 609–614 [Google Scholar]
- Ehmann A (1974) Identification of 2-O-(indole-3-acetyl)-D-glucopyranose, 4-O-(indole-3-acetyl)-d-glucopyranose and 6-O-(indole-3-acetyl)-d-glucopyranose from kernels of Zea mays. Carbohydr Res 34: 99–114 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystall Section D 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KW (2008) Novel genetically encoded biosensors using firefly luciferase. ACS Chem Biol 3: 346–351 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Ann Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- George Thompson AM, Iancu CV, Neet KE, Dean JV, Choe JY (2017) Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci Rep 7: 46629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Návarová H, Attaran E, Zeier J (2013) Reprogramming of plants during systemic acquired resistance. Front Plant Sci 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Zeier T, Aretz C, Zeier J (2018) A critical role for Arabidopsis MILDEW RESISTANCE LOCUS O2 in systemic acquired resistance. Plant J 94: 1064–1082 [DOI] [PubMed] [Google Scholar]
- Guerra T, Schilling S, Hake K, Gorzolka K, Sylvester FP, Conrads B, Westermann B, Romeis T (2020) Calcium-dependent protein kinase 5 links calcium signaling with N-hydroxy-l-pipecolic acid- and SARD1-dependent immune memory in systemic acquired resistance. New Phytol 225: 310–325 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier J (2018) l-lysine metabolism to N-hydroxypipecolic acid: an integral immune-activating pathway in plants. Plant J 96: 5–21 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier J (2019) N-hydroxypipecolic acid and salicylic acid: a metabolic duo for systemic acquired resistance. Curr Opin Plant Biol 50: 44–57 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Kim D, Bernsdorff F, Ajami-Rashidi Z, Scholten N, Schreiber S, Zeier T, Schuck S, Reichel-Deland V, Zeier J (2017) Biochemical principles and functional aspects of pipecolic acid biosynthesis in plant immunity. Plant Physiol 174: 124–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Zeier T, Bernsdorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Bräutigam A, Hölzel T,. et al. (2018) Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plants immunity. Cell 173: 456–469.e16 [DOI] [PubMed] [Google Scholar]
- Holmes EC, Chen YC, Mudgett MB, Sattely ES (2021) Arabidopsis UGT76B1 glycosylates N-hydroxy-pipecolic acid and inactivates systemic acquired resistance in tomato. Plant Cell 33: 750--765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagari F, Thlimony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book 1: e0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The phyre2 web portal for protein modeling, prediction and analysis. Nat Prot 10: 845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gilmour SJ, Chao L, Park S, Thomashow MF (2020) Arabidopsis CAMTA transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes. Mol Plant 13: 157–168 [DOI] [PubMed] [Google Scholar]
- Klessig DF, Choi HW, Dempsey DA (2018) Systemic acquired resistance and salicylic acid: past, present, and future. Mol Plant Microbe Interact 31: 871–888 [DOI] [PubMed] [Google Scholar]
- Li W, Zhang F, Chang Y, Zhao T, Schranz ME, Wang G (2015) Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 27: 1907–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles DJ (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem 277: 586–592 [DOI] [PubMed] [Google Scholar]
- Mair P, Wilcox R (2018) Robust Statistical Methods Using WRS2. The WRS2 Package. https://cran.r-project.org/web/packages/WRS2/ accessed from December 2019 until November 2020 [DOI] [PubMed]
- Maksym RP, Ghirardo A, Zhang W, von Saint Paul V, Lange B, Geist B, Hajirezaei MR, Schnitzler JP, Schäffner AR (2018) The defense-related isoleucic acid differentially accumulates in Arabidopsis among branched-chain amino acid-related 2-hydroxy carboxylic acids. Front Plant Sci 9: 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meßner B, Thulke O, Schäffner AR (2003) Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 217: 138–146 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnike L, Rekhter D, Huang W, Feussner K, Tian H, Herrfurth C, Zhang Y, Feussner I (2021) The glycosyltransferase UGT76B1 is critical for plant immunity as it governs the homeostasis of N-hydroxy-pipecolic acid. Plant Cell 33: 735--749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Murahashi SI, Shiota T (1987) Short-step synthesis of amino acids and N-hydroxyamino acids from amines. Tetrahedron Lett 28: 6469–6472 [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutoshi Y, Okazaki M, Kida T, Nishina Y, Morishita Y, Ogawa T, Suzuki H, Shibata D, Jikumaru Y, Hanada A,. et al. (2012) Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24: 3795–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25: 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schnake A, Hartmann M, Schreiber S, Malik J, Brahmann L, Yildiz I, von Dahlen J, Rose LE, Schaffrath U, Zeier J (2020) Inducible biosynthesis and immune function of the systemic acquired resistance inducer N-hydroxypipecolic acid in monocotyledonous and dicotyledonous plants. J Exp Bot 71: 6444–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Zeier J (2013) Long-distance communication and signal amplification in systemic acquired resistance. Front Plant Sci 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Koo YJ, Seo HS, Kim MC, Choi YD, Kim JH (2008) Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae. Phytochemistry 69: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Stahl E, Hartmann M, Scholten N, Zeier J (2019) A role for tocopherol biosynthesis in Arabidopsis thaliana basal immunity to bacterial infection. Plant Physiol 181: 1008–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Exp Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Sun T, Huang J, Xu Y, Verma V, Jing B, Sun Y, Orduna AR, Tian H, Huang X, Xia S,. et al. (2020) Redundant CAMTA transcription factors negatively regulate the biosynthesis of salicylic acid and N-hydroxypipecolic acid by modulating the expression of SARD1 and CBP60g. Mol Plant 13: 144–156 [DOI] [PubMed] [Google Scholar]
- Veeragoni SR, Lange B, Serrano M, Nawrath C, Bauer S, Schäffner AR, Thordal-Christensen H, Durner J, Gaupels F (2020) Mutant muddle: some Arabidopsis eds5 mutant lines have a previously unnoticed second-site mutation in FAH1. Plant Physiol 182: 460–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Ann Rev Phytopath 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Sales JH, Lenk M, Bauer K, Brambilla A, Sommer A, Chen Y, Wenig M, Nayem S (2021) Systemic propagation of immunity in plants. New Phytol 229: 1234–1250 10.1111/nph.16953 [DOI] [PubMed] [Google Scholar]
- Vogel-Adghough D, Stahl E, Návarová H, Zeier J (2013) Pipecolic acid enhances resistance to bacterial infection and primes salicylic acid and nicotine accumulation in tobacco. Plant Sign Behav 8: e26366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volko SM, Boller T, Ausubel FM (1998) Isolation of new Arabidopsis mutants with enhanced disease susceptibility to Pseudomonas syringae by direct screening. Genetics 149: 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Saint Paul V, Zhang W, Kanawati B, Geist B, Faus-Keßler T, Schmitt-Kopplin P, Schäffner AR (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23: 4124–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, , Schuck S, , Wu J, , Yang P, , Döring A-C, , Zeier J, , Tsuda K ( 2018) A MPK3/6-WRKY33-ALD1-pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 30: 2480–2494 10.1105/tpc.18.00547 30228125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenig M, Ghirardo A, Sales JH, Pabst ES, Breitenbach HH, Antritter F, Weber B, Lange B, Lenk M, Cameron RK,. et al. (2019) Systemic acquired resistance networks amplify airborne defense cues. Nat Commun 10: 3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.