Abstract
Systemic acquired resistance (SAR) is a mechanism that plants utilize to connect a local pathogen infection to global defense responses. N-hydroxy-pipecolic acid (NHP) and a glycosylated derivative are produced during SAR, yet their individual roles in this process are currently unclear. Here, we report that Arabidopsis thaliana UGT76B1 generated glycosylated NHP (NHP-Glc) in vitro and when transiently expressed alongside Arabidopsis NHP biosynthetic genes in two Solanaceous plants. During infection, Arabidopsis ugt76b1 mutants did not accumulate NHP-Glc and accumulated less glycosylated salicylic acid (SA-Glc) than wild-type plants. The metabolic changes in ugt76b1 plants were accompanied by enhanced defense to the bacterial pathogen Pseudomonas syringae, suggesting that glycosylation of the SAR molecules NHP and salicylic acid by UGT76B1 plays an important role in modulating defense responses. Transient expression of Arabidopsis UGT76B1 with the Arabidopsis NHP biosynthesis genes ALD1 and FMO1 in tomato (Solanum lycopersicum) increased NHP-Glc production and reduced NHP accumulation in local tissue and abolished the systemic resistance seen when expressing NHP-biosynthetic genes alone. These findings reveal that the glycosylation of NHP by UGT76B1 alters defense priming in systemic tissue and provide further evidence for the role of the NHP aglycone as the active metabolite in SAR signaling.
The Arabidopsis UDP-glycosyltransferase UGT76B1 glycosylates the systemic acquired resistance-signaling metabolite NHP and can inactivate systemic defense responses when expressed in tomato.
Introduction
Systemic acquired immunity in plants is a coordinated defense response that leads to heightened disease protection throughout the plant body following an initial, localized pathogen attack. Several small molecules help orchestrate this process, including the ubiquitous hormone salicylic acid (SA; Klessig et al., 2018) and the recently discovered signaling metabolite N-hydroxy-pipecolic acid (N-hydroxy-pipecolic acid [NHP]; Chen et al., 2018; Hartmann et al., 2018), which is thought to play a lead role in systemic acquired resistance (SAR). The enzyme flavin monooxygenase 1 (FMO1) catalyzes the N-hydroxylation of the nonproteinogenic amino acid pipecolic acid (Pip) during the biosynthesis of NHP and is required for the initiation and amplification of SAR signaling (Chen et al., 2018; Hartmann et al., 2018).
Both SA and NHP can be isolated from plants with several metabolic modifications, most notably as glycosylated derivatives. In prior work using Arabidopsis thaliana (Arabidopsis) plants, we and others observed that both NHP and its hexose-conjugated derivative (NHP-Glc) accumulate after bacterial infection in seedlings and leaves (Chen et al., 2018; Hartmann and Zeier, 2018). Both NHP-Glc and the aglycone are absent from unelicited plants and pathway mutants deficient in FMO1, prompting questions about the role of NHP glycosylation in SAR. The glycosyltransferase required for the generation of NHP-Glc, however, has remained elusive.
Structural modifications of small plant signaling molecules appear to have evolved as a dynamic mechanism to modulate the activity of these chemical signals. Common enzymatic modifications to base hormone scaffolds include hydroxylation, carboxylation, sulfation, acetylation, methylation, amino acid conjugation, and glycosylation (Westfall et al., 2013). Some hormones, such as the defense hormone jasmonic acid (JA), undergo multiple enzymatic modifications (Wasternack and Hause, 2013) to create bioactive (Staswick and Tiryaki, 2004), inactive (Smirnova et al., 2017), and differentially active (Nakamura et al., 2011) compounds. Often, loss-of-function mutations of these modifying enzymes can have severe impacts on plant physiology, leading to developmental phenotypes in the case of auxins (Nakazawa et al., 2001; Takase et al., 2004; Staswick et al., 2005), brassinosteroids (Choi et al., 2013), and gibberellins (Wang et al., 2012) and to altered responses to environmental stresses in the case of abscisic acid (Liu et al., 2015), JA (Caarls et al., 2017; Smirnova et al., 2017), and SA (Liu et al., 2009; Boachon et al., 2014). In some instances, hormone conjugation appears to serve as a reservoir of a molecule for fast deployment, whereas in other cases, it seems to be a metabolic mechanism for attenuating the activity and depleting the active form of the hormone (Piotrowska and Bajguz, 2011).
Several lines of evidence suggest that the NHP aglycone is sufficient to initiate SAR signaling but have not yet revealed a functional role for glycosylation. For example, the treatment of Arabidopsis (Chen et al., 2018; Hartmann et al., 2018), sweet pepper (Capsicum annuum; Holmes et al., 2019), or tomato (Solanum lycopersicum; Holmes et al., 2019) leaves with synthetic NHP induces resistance against bacterial infection in distal tissues not treated with NHP. Furthermore, transient overexpression of the Arabidopsis NHP biosynthetic enzymes AGD2-LIKE DEFENSE PROTEIN1 (ALD1; Navarova et al., 2012) and FMO1 (Chen et al., 2018; Hartmann et al., 2018) leads to the production of NHP in tomato leaves and results in enhanced resistance to bacterial infection in distal tissues (Holmes et al., 2019). Notably, NHP-Glc was not detected in the NHP-treated tomato leaves, suggesting that NHP-Glc biosynthesis and/or accumulation may not occur in tomato. These data, coupled with the observation that NHP-Glc does not accumulate in the absence of infection in Arabidopsis, suggest that NHP-Glc is not simply a storage form of NHP.
Despite the clear role of NHP biosynthesis in the initiation of systemic resistance, several open questions remain regarding (i) the active form of NHP metabolites; (ii) the potential role of NHP glycosylation in modulating SAR signaling; and (iii) more broadly, mechanisms of signal initiation, transport, and attenuation in plant systemic resistance. In an effort to better understand the potential role of NHP-Glc in the SAR response, we sought to establish the genetic and biochemical basis for NHP glycosylation in Arabidopsis and test the influence of the putative glycosylating enzyme(s) in SAR-mediated disease resistance. Here, we report that the Arabidopsis UDP-glycosyltransferase UGT76B1 can generate glycosylated NHP (NHP-Glc) in vitro and when transiently expressed alongside Arabidopsis NHP biosynthetic genes in two Solanaceous plants. Our results provide important insights into how plants use specific metabolic transformations to alter the behavior of the key signaling molecule NHP during systemic defense responses.
Results
Heterologously expressed Arabidopsis UGT76B1 glycosylates NHP in planta
Previous studies have indicated that NHP-Glc accumulates in Arabidopsis after pathogen infection (Chen et al., 2018; Hartmann and Zeier, 2018). We hypothesized that a dedicated NHP-glycosyltransferase may be highly expressed under pathogen stress conditions (Figure 1, A). We analyzed a set of publicly available microarray datasets for the mRNA expression patterns of 103 Arabidopsis UDP-dependent glycosyltransferase genes (UGTs) under various biotic stress conditions (Supplemental Figure S1). We prioritized testing of candidate UGTs based upon their high levels of mRNA abundance across all biotic stress conditions and selected a few others based upon their high levels of mRNA abundance under a specific pathogen stress. For the initial screen, we selected 14 UGTs from this microarray analysis (Supplemental Figure S1) as well as four additional UGTs (UGT73B2, UGT73B3, UGT73C3, and UGT73C5) based on expression profiles in RNA sequencing experiments (Bernsdorff et al., 2016; Hartmann et al., 2018). Our goal was to first identify Arabidopsis UGTs that could generate NHP-Glc during heterologous expression and to subsequently determine the roles of any candidates in Arabidopsis.
Figure 1.
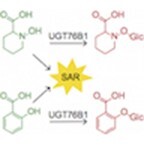
Screen of 18 Arabidopsis UGTs for their ability to glycosylate NHP. A, Biosynthetic pathway for the production of NHP-Glc from l-Lys in Arabidopsis. The biosynthetic activity of UGT76B1 was characterized in this work. B, Abundance of NHP-Glc measured with LC–MS after transient expression of GFP or respective Arabidopsis UGTs alongside Arabidopsis ALD1 + FMO1 in N. benthamiana leaves. In the initial screen, Agrobacterium strains harboring distinct UGTs were combined in equal proportions and co-infiltrated with Agrobacterium strains harboring ALD1 and FMO1. In the second screen, Agrobacterium strains harboring UGT76B1, UGT76F2, or UGT85A1 were separately co-infiltrated with Agrobacterium strains harboring ALD1 and FMO1. Total inoculum (OD600) was kept constant in both experiments by including Agrobacteria harboring GFP as a control. Bars represent the mean ± sd (n = 3 independent biological replicates). Values reported as zero indicate no detection of metabolites. Asterisks indicate significant differences in NHP-Glc levels (one-tailed t test; *P < 0.05, **P < 0.01). The experiment was repeated two times with similar results. C, Representative LC–MS chromatograms of NHP-Glc (m/z = 308.134) in extracts from N. benthamiana (black) and Arabidopsis adult leaves (blue) transiently expressing ALD1 + FMO1 + UGT76B1 after infiltration with 1 mM NHP synthetic standard. D, Comparative MS/MS spectra of NHP-Glc in extracts from N. benthamiana (black) and Arabidopsis adult leaves (blue) transiently expressing ALD1 + FMO1 + UGT76B1 after infiltration with 1 mM NHP synthetic standard at collision energies of 10 and 40 V.
In previous studies, we used Agrobacterium-mediated transient expression in Nicotiana benthamiana (Kapila et al., 1997) as a heterologous expression platform to produce NHP in planta (Chen et al., 2018; Holmes et al., 2019). Under these transient expression conditions, NHP-Glc was not detected in extracts from N. benthamiana leaves (Chen et al., 2018; Holmes et al., 2019). We hypothesized that this heterologous expression system could be used to screen Arabidopsis UGT candidates with minimal background signal from native enzymes. We cloned 18 candidate UGT cDNAs and transiently expressed them with Arabidopsis ALD1 and FMO1 (the minimal set of genes required for NHP biosynthesis; Holmes et al., 2019) in N. benthamiana leaves. To expedite testing and metabolite analysis, we expressed our candidate enzymes in groups of three by combining Agrobacterium strains harboring separate UGT candidates and GFP in equal proportions and co-infiltrated them with Agrobacteria harboring ALD1 and FMO1.
Liquid chromatography–mass spectrometry (LC–MS) analysis of methanolic extracts from these leaves revealed that one set of genes (UGT76B1, UGT76F2, and UGT85A1) led to significant accumulation of NHP-Glc when coexpressed with ALD1 and FMO1 compared with coexpression of ALD1 and FMO1 with GFP (Figure 1, B). We then transiently expressed each of these respective UGTs with ALD1 and FMO1 and found that leaves expressing UGT76B1 (At3g11340) were the only ones that accumulated a significant amount of NHP-Glc (Figure 1, B). Nicotiana benthamiana leaves transiently expressing ALD1, FMO1, and UGT76B1 accumulated significantly less free NHP (as measured using LC–MS) than did leaves expressing ALD1 and FMO1 alone, indicating a high conversion rate from NHP to NHP-Glc by UGT76B1 (Supplemental Figure S2). The compound produced in N. benthamiana had the same LC–MS retention time (Figure 1, C) and MS/MS fragmentation pattern (Figure 1, D) as NHP-Glc produced in adult Arabidopsis leaves, suggesting that Arabidopsis UGT76B1 produced the same glycosylated NHP derivative as the compound that natively accumulates in Arabidopsis.
In vitro biochemistry of UGT76B1 expressed in N. benthamiana and E. coli
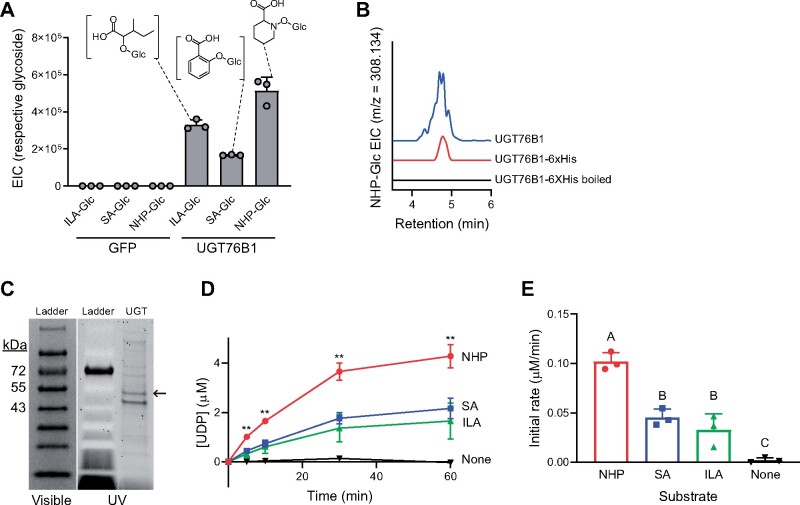
UGT76B1 glycosylates the plant hormone SA and the isoleucine catabolite 2-hydroxy-3-methyl-pentanoic acid (ILA) in vitro and contributes to the accumulation of their respective glycosides in planta (von Saint Paul et al., 2011; Noutoshi et al., 2012; Maksym et al., 2018; Bauer et al., 2020a). Our results in N. benthamiana suggested that UGT76B1 could glycosylate a third defense-related metabolite, NHP. To confirm these previous findings and determine that the NHP-Glc we detected in N. benthamiana was a direct result of UGT76B1 activity, we spiked crude protein extracts from N. benthamiana leaves expressing GFP or UGT76B1 with UDP-glucose and the aglycone substrates ILA, SA, or NHP. Protein extracts from leaves expressing GFP did not produce any of the respective glycosides, whereas extracts from leaves expressing UGT76B1 produced all three compounds (Figure 2, A). Furthermore, we transiently expressed His-tagged UGT76B1 in N. benthamiana leaves and enriched for UGT76B1 using Ni-NTA affinity purification. Partially purified UGT76B1-6xHis from N. benthamiana catalyzed the synthesis of NHP-Glc in vitro, while denatured protein did not (Figure 2, B).
Figure 2.
In vitro characterization of Arabidopsis UGT76B1. A, GFP or Arabidopsis UGT76B1 was transiently expressed in N. benthamiana leaves and crude protein extracts were incubated with 5 mM UDP glucose and 1 mM aglycone substrates (2-hydroxy-3-methylvaleric acid [ILA], SA, or NHP). Levels indicate abundances of glycosides measured with LC–MS after 3 h incubation. Bars represent the mean ± sd (n = 3 independent biological replicates). m/z used for quantification are: ILA-Glc ([M-H]− = 293.124), SA-Glc ([M-H] − = 299.077), and NHP-Glc ([M+H]+ = 308.134). The experiment was repeated two times with similar results. B, Representative LC–MS chromatograms of NHP-Glc (m/z = 308.134) from crude extract from N. benthamiana leaves transiently expressing Arabidopsis UGT76B1 (blue) and enriched (red) or denatured (black) UGT76B1-6xHis purified from N. benthamiana leaves. C, SDS–PAGE gel of enriched UGT76B1-6xHis purified from E. coli. The same gel imaged under visible light and UV light is included to better visualize the ladder bands. Expected mass of UGT76B1-6xHis is ∼51 kDa. D, Enriched UGT76B1-6xHis from E. coli was incubated with NHP (red), SA (blue), ILA (green), or no substrate (black) in vitro. Aliquots were quenched at increasing time points, and free UDP liberated from the reaction of UGT76B1 with its respective substrates was measured using an enzyme-linked assay. Asterisks indicate a significant difference (two-tailed t test; **P < 0.01). Points represent the mean ± sd (n = 3 independent biological replicates). The experiment was repeated two times with similar results. E, Initial rate of reaction from (D) was measured as the slope from t = 0 to t = 5 min. Different letters indicate significantly different groups using two-tailed t tests (P < 0.01). Bars represent the mean ± sd (n = 3 independent biological replicates).
Given the promiscuity of some plant UGTs on structurally similar substrates (Lim et al., 2002), it is not surprising that UGT76B1 can glycosylate ILA, SA, and NHP. To better understand the ability of this enzyme to glycosylate these substrates, we expressed and enriched UGT76B1-6xHis from E. coli (Figure 2, C) and tested its activity using an enzyme-coupled assay (Zegzouti et al., 2013). During each UGT catalytic reaction, UDP is released and the concentration of free UDP in a given reaction can be directly measured using this assay. Reactions with NHP generated significantly more UDP over the course of 1 h than was generated with SA or ILA as substrates (Figure 2, D). The initial rate of reaction with NHP was also approximately 2× faster than that with either SA or ILA (Figure 2, E). These results confirm the notion that UGT76B1 acts on ILA, SA, and NHP and indicate that it is more active on NHP as a substrate under these conditions.
To determine where glucose conjugation is occurring on NHP, we derivatized synthetic NHP and an extract from N. benthamiana leaves transiently expressing ALD, FMO1, and UGT76B1 with trimethylsilyldiazomethane (TMSD), a reagent commonly used to selectively methylate carboxylic acids (Kühnel et al., 2007; Topolewska et al., 2015). Derivatization of synthetic NHP generated a major, singly methylated product and a minor doubly methylated product (Supplemental Figure S3, A). We hypothesize that the singly methylated product is NHP methyl ester based upon MS/MS fragmentation and the reported activity of TMSD (Supplemental Figure S3, A). Derivatization of the N. benthamiana extract revealed a methylated NHP-Glc product with an MS/MS fragmentation pattern that matches that of NHP methyl ester (Supplemental Figure S3, B), suggesting that UGT76B1 is generating NHP-β-D glucoside. UGT76B1 is also known to generate the β-d-glucoside of SA (SA-Glc; von Saint Paul et al., 2011; Noutoshi et al., 2012). A synthetic standard of NHP-Glc (which is currently unavailable) is required to definitively elucidate the structure of the glycosylated NHP produced by UGT76B1.
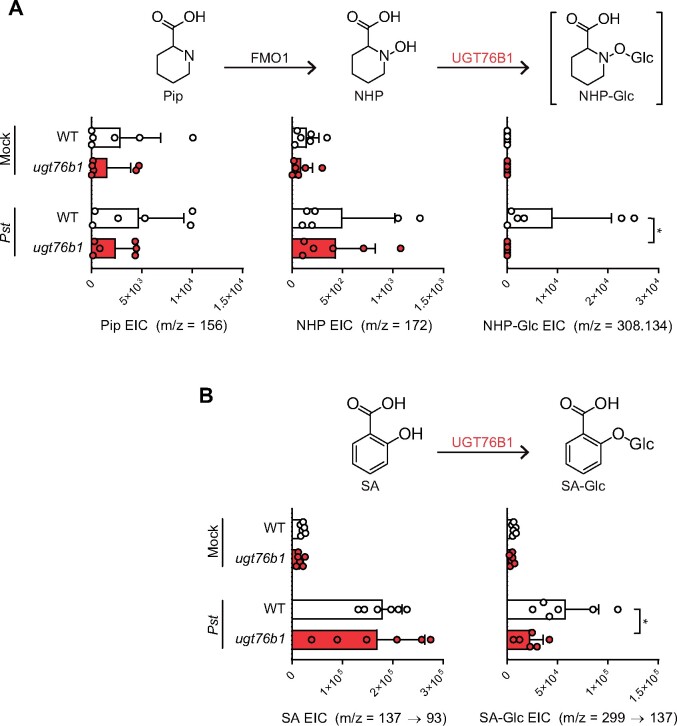
Arabidopsis ugt76b1 mutants are impaired in NHP-Glc and SA-Glc production
Given that UGT76B1 is capable of glycosylating NHP when expressed heterologously in N. benthamiana, we next sought to determine its native function in Arabidopsis. We obtained the Syngenta Arabidopsis Insertion Library (SAIL; Sessions et al., 2002) T-DNA insertional line SAIL_1171_A11 (ugt76b1-1; hereafter ugt76b1) from the Arabidopsis Biological Resource Center (ABRC). This mutant line was previously used to study the function of UGT76B1 (von Saint Paul et al., 2011). To quantify NHP and SA derivatives, we grew wild-type Arabidopsis Col-0 (hereafter WT) and ugt76b1 plants axenically in hydroponic medium for 2 weeks, treated the seedlings with 10 mM MgCl2 (mock), Pseudomonas syringae pathovar tomato DC3000 (Pst), 1 mM NHP, or 100 µM SA, and measured metabolite levels using GC–MS and LC–MS (Figure 3 and Supplemental Figure S4). WT plants had significantly higher abundance of NHP-Glc and SA-Glc than ugt76b1 in Pst-treated plants (Figure 3), highlighting the important contribution from UGT76B1 in the glycosylation of NHP and SA during infection. While NHP-treated ugt76b1 plants did contain detectable NHP-Glc, the abundance was reduced over 99% compared with WT plants, suggesting that UGT76B1 is the primary NHP glycosyltransferase in Arabidopsis (Supplemental Figure S4). There may be other minor enzymes that contribute to NHP glycosylation, but NHP-Glc was only detectable in ugt76b1 plants when they were supplemented with a high concentration of NHP (Supplemental Figure S4) and not when the plants were treated with Pst (Figure 3). The abundance of SA-Glc was reduced approximately 60% in ugt76b1 plants compared with WT when supplemented with SA (Supplemental Figure S4), suggesting that the activity of UGT76B1 may also contribute significantly to the glycosylation of SA in Arabidopsis. Our findings also corroborate recent work reported in this issue showing that UGT76B1 is required for the production of NHP-Glc in Arabidopsis (Bauer et al., 2020b; Mohnike et al., 2020). In particular, Mohnike et al. (2020) demonstrate that other mutant alleles (ugt76b1-3 and ugt76b1-4) are deficient in NHP-Glc accumulation, providing evidence that this effect is not due to an unknown second site mutation in ugt76b1-1 (Mohnike et al., 2020).
Figure 3.
Abundance of NHP- and SA-related metabolites in WT and ugt76b1 mutant seedlings. Arabidopsis WT (white bars) and ugt76b1 (red bars) seedlings were grown axenically in hydroponic medium for 2 weeks and treated with 10 mM MgCl2 (mock) or a suspension of Pst at OD600 of 0.01. After 24 h, the seedlings were harvested and analyzed for NHP-related metabolites (A) and SA-related metabolites (B). Pip and NHP were measured as trimethylsilyl (TMS) and 2-TMS derivatives, respectively, using GC–MS. NHP-Glc, SA, and SA-Glc were measured using LC–MS. Bars represent the means ± sd (n = 6 independent biological replicates). Values reported as zero indicate no detection of metabolites. Asterisks indicate a significant difference in metabolite levels (one-tailed t test; *P < 0.05). The experiment was repeated two times with similar results.
Arabidopsis ugt76b1 mutants are more resistant to bacterial infection than the wild type
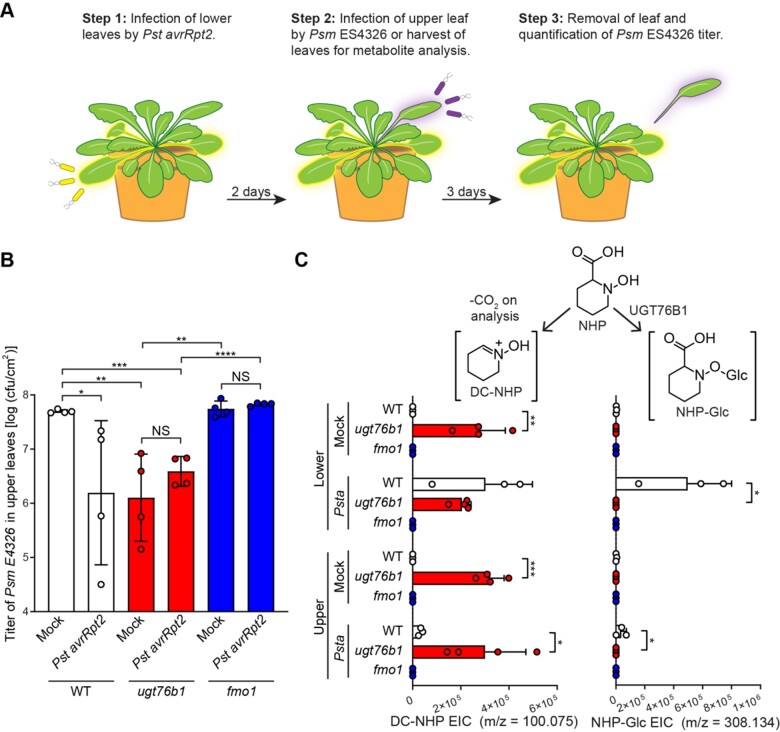
A previous study showed that Arabidopsis ugt76b1 mutants are more resistant to the biotrophic pathogen Pst and more susceptible to the necrotrophic pathogen Alternaria brassicicola than the wild type (von Saint Paul et al., 2011), indicating that UGT76B1 plays a critical role in regulating disease resistance signaling. Given that UGT76B1 can glycosylate NHP in Arabidopsis (Figure 3 and Supplemental Figure S4), we hypothesized that UGT76B1 might regulate the abundance of NHP that is available to initiate and sustain defense priming during SAR. To test this hypothesis and explore the functions of NHP-Glc and UGT76B1, we performed SAR experiments as previously described (Chen et al., 2018; Hartmann et al., 2018). Briefly, three lower leaves (leaf number 5–7) of 4-week-old WT, ugt76b1, and fmo1 (a NHP- and SAR-deficient mutant) plants were infiltrated with 10 mM MgCl2 (mock) or a 5 × 106 cfu/mL suspension of Pst avrRpt2, an avirulent strain that induces a strong defense response in WT plants. Two days later, an upper leaf of each plant was challenged with a 1 × 105 cfu/mL suspension of Psm ES4326, a virulent strain (Figure 4, A and Supplemental Figure S5, A). Disease symptoms and titers of Psm ES4326 in the infected upper leaves were then photographed and quantified at 3 days post infiltration (dpi), respectively (Figure 4, B and Supplemental Figure S5, B).
Figure 4.
SAR assays in Arabidopsis WT, ugt76b1, and fmo1 plants. A, Design of SAR assays in Arabidopsis. Three lower leaves (leaf numbers 5–7) of each plant were infiltrated with a 5 × 106 cfu/mL suspension of Pst avrRpt2 (Psta; local infection) or 10 mM MgCl2 as a mock control. For the bacterial growth assays in (B): 2 days after local infection, one upper leaf (leaf number 10) of each plant was challenged with 1 × 105 cfu/mL suspension of Psm ES4326 (distal infection). Three days later, the disease symptoms of upper leaves were photographed and the titer of Psm ES4326 was determined. For metabolite analysis in (C): 2 days after local infection with Pst avrRpt2, the three lower infected leaves and three upper uninfected leaves (leaf numbers 8–10) were harvested and separately pooled for metabolite analysis. B, Titer of Psm ES4326 in upper, challenged leaves of WT (white bars), ugt76b1 (red bars), and fmo1 (blue bars) plants. Bars represent the mean ± sd (n = 4 independent biological replicates). Asterisks indicate a significant difference in bacterial titer (one-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS—not significant). The experiment was repeated three times with similar results. Repetition 2 of this experiment is reported in Supplemental Figure S6. C, Extracted ion abundances of DC-NHP (a degradation product of NHP) and NHP-Glc in methanolic tissue extracts from lower and upper leaves of WT (white bars), ugt76b1 (red bars), and fmo1 (blue bars) plants. Bars represent the means ± sd (n = 3 or 4 independent biological replicates). DC-NHP and NHP-Glc were measured using LC–MS. Values reported as zero indicate no detection of metabolites. Asterisks indicate a significant difference in metabolite levels (one-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001). The experiment was repeated two times with similar results.
Upper leaves from WT plants initially treated with Pst avrRpt2 harbored significantly less growth of Psm ES4326 than WT plants under mock treatment (Figure 4, B and Supplemental Figure S6, A). These plants also developed fewer disease symptoms (e.g. bacterial speck and chlorosis; Supplemental Figure S5, B), indicating the establishment of SAR. By contrast, SAR protection was abolished in fmo1 plants (Figure 4, B and Supplemental Figure S5, B). Notably, the titers of Psm ES4326 in the upper leaves of mock-treated ugt76b1 plants were significantly lower than those of mock-treated WT plants. In addition, the titers of Psm ES4326 in the upper leaves of ugt76b1 plants under mock and Pst avrRpt2 treatment were similar (Figure 4, B), indicating that an initial pathogen infection was not required for disease resistance in the ugt76b1 leaves. Finally, all lower leaves (mock or Pst avrRpt2) of ugt76b1 plants showed early senescence on the leaf margin, and leaves of uninfected ugt76b1 plants were consistently smaller than those of WT plants (Supplemental Figure S5, B), which is consistent with a previous report (von Saint Paul et al., 2011). Taken together, these findings indicate that the mutation of UGT76B1 leads to enhanced resistance regardless of an initial pathogen infection.
Based on our observations that ugt76b1 seedlings have altered abundances of NHP, SA, and their glycosylated forms (Figure 3 and Supplemental Figure S4), we measured determined the abundance of these metabolites using a modified SAR assay. We used the same experimental setup; however, we did not challenge the plants with Psm ES4326. Instead, we harvested lower and upper leaves 2 days after mock or Pst avrRpt2 treatment for metabolite analysis. We detected high background levels of Pip, SA, and SA-Glc in mock-treated ugt76b1 plants, indicating that these plants are already primed with both NHP- and SA-related metabolites (Supplemental Figure S5, C and D). Neither fmo1 nor ugt76b1 plants accumulated any NHP-Glc, while WT plants showed significant increases NHP-Glc levels in both the lower and upper leaves (Figure 4, C and Supplemental Figure S6, B), confirming the requirement for these two enzymes in the NHP-Glc biosynthetic pathway. As previously reported (von Saint Paul et al., 2011), ugt76b1 plants contained significantly more SA-Glc than WT plants under mock conditions, suggesting that other UGTs are still able to generate SA-Glc at appreciable levels in this context (Supplemental Figure S5, D). We did not directly detect any free NHP by GC–MS in this experiment, perhaps due to the instability of the molecule (Chen et al., 2018). The abundance of decarboxylated NHP (DC-NHP; which has been reported as a degradation product of NHP; Chen et al., 2018) was significantly elevated in mock- and Pst avrRpt2-treated ugt76b1 plants versus the WT (Figure 4, C and Supplemental Figure S6, B), pointing to the constitutive accumulation of NHP and its subsequent degradation (either in planta or during the metabolite extraction process).
To provide further structural evidence for DC-NHP, we synthesized 2,3,4,5-tetrahydropyridine N-oxide (DC-NHP) using two separate chemical methods (Stappers et al., 2002; Gella et al., 2009; Chen et al., 2018; Supplemental Figure S7, A). Previous evidence suggests that this molecule spontaneously dimerizes and exists primarily in its dimer form (Alford et al., 1966; Alsbaiee and Ali, 2008). This observation is consistent with the co-eluting m/z features at 199.144 (DC-NHP dimer) and 100.075 (DC-NHP) that we observed in both ugt76b1 plants and the product of our chemical synthesis (Supplemental Figure S7, B and C). This finding is also consistent with the results of MS/MS of the dimer mass, which fragments exclusively into the monomer mass at a collision-induced voltage of 10 V (Supplemental Figure S7, D). The MS/MS fragmentation patterns of the plant-derived molecule and synthesized DC-NHP are consistent with one another (Supplemental Figure S7, E), providing further evidence for this structural characterization. Taken together, these results indicate that enhanced resistance in ugt76b1 is associated with elevated abundance of NHP- and SA-related metabolites under uninduced conditions.
Expression of UGT76B1 abolishes NHP-induced protection in tomato
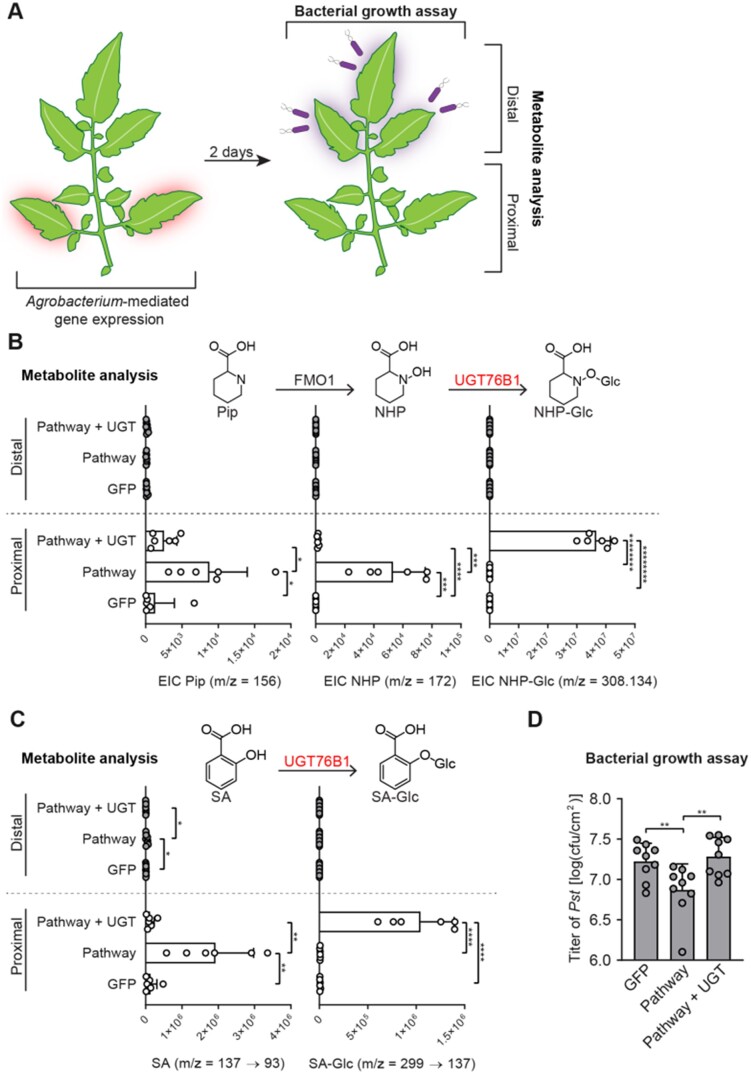
The enhanced resistance exhibited by Arabidopsis ugt76b1 mutants with significantly reduced levels of NHP-Glc suggested that the glycosylation of NHP reduces its bioactivity as a SAR signaling molecule. We next turned to tomato as a model system to further investigate the role of NHP glycosylation using Agrobacterium-mediated transient expression of UGT76B1 in the context of a pathogen infection. We previously used this approach to establish that NHP can induce a SAR response in a tomato. Here, we reasoned that phenotypic analysis of tomato leaves expressing UGT76B1 in addition to the NHP pathway would allow us to better isolate the role of NHP glycosylation, independently of its role in SA glycosylation, and avoid the complex metabolic dynamics observed in adult Arabidopsis ugt76b1 mutants grown in soil (including the constitutive accumulation of NHP-derived metabolites). We previously determined that transient expression of Arabidopsis ALD1 and FMO1 in tomato leaflets proximal to the main stem is sufficient to induce the production of NHP and inhibit the growth of Pst in infected distal leaflets (Holmes et al., 2019). Notably, altering NHP levels alone in tomato did not lead to the production of NHP-Glc (Holmes et al., 2019), suggesting that background activity of native tomato UGTs would not interfere with our interpretation of the results of the assay.
We hypothesized that overexpressing Arabidopsis UGT76B1 with ALD1 and FMO1 would increase the ratio of NHP-Glc to NHP in proximal tomato leaflets and decrease the SAR response in distal leaflets infected with Pst. To test this hypothesis, we infiltrated the two proximal leaflets of a fully expanded tomato leaf with Agrobacterium strains harboring GFP or GFP + ALD1 + FMO1 (Pathway), or Pathway + UGT76B1 (Figure 5, A). Two days post-infiltration, we harvested both proximal and distal leaflets for metabolite analysis (Figure 5, A–C). Proximal leaflets accumulated significantly less NHP and SA when expressing UGT76B1 alongside the NHP biosynthetic genes than did leaflets expressing NHP biosynthetic genes alone (Figure 5, B and C). Conversely, these leaflets accumulated significantly more NHP-Glc and SA-Glc, than the control, suggesting that the aglycones had been directly converted (Figure 5, B and C). The only significant metabolic change that we measured in distal leaflets was the accumulation of free SA in leaves expressing only the NHP metabolic pathway enzymes (Supplemental Figure S8).
Figure 5.
Transient expression of Arabidopsis UGT76B1 with ALD1 and FMO1 in tomato leaves. A, Design of the transient SAR assays in tomato. Two leaflets of a tomato leaf proximal to the main stem (highlighted in red) were inoculated with Agrobacteria harboring GFP (GFP) or a combination of strains harboring GFP + Arabidopsis ALD1 + Arabidopsis FMO1 (Pathway) without and with Arabidopsis UGT76B1 (Pathway + UGT). For the bacterial growth assay in (B): 2 dpi with Agrobacteria, distal leaflets (highlighted in purple) were inoculated with a 1 × 105 cfu/mL suspension of Pst. Four dpi, distal leaves were harvested for quantification of Pst titers. For metabolite analysis in (C) and (D): 2 dpi with Agrobacteria, both proximal leaflets infiltrated with Agrobacteria and distal, untreated leaflets were harvested independently for analysis. B, Abundances of Pip, NHP, and NHP-Glc in tomato leaflets expressing GFP, Pathway, and Pathway + UGT (white bars) and leaflets distal to those infiltrated with Agrobacteria (gray bars). Bars for proximal leaflets represent means ± sd (two leaflets each from n = 3 independent plants). Bars for distal leaflets represent means ± sd (three leaflets each from n = 3 independent plants). Pip and NHP were measured as TMS and 2-TMS derivatives, respectively, using GC–MS. NHP-Glc was measured using LC–MS. Values reported as zero indicate no detection of metabolites. Asterisks indicate a significant difference in metabolite levels (one-tailed t test; *P < 0.05, ***P < 0.001, ****P < 0.0001, *******P < 1 × 10−6, ********P < 1 × 10−7). The experiment was repeated two times with similar results. C, Abundances of SA and SA-Glc in tomato leaflets expressing GFP, Pathway, and Pathway + UGT (white bars) and leaflets distal to those infiltrated with Agrobacteria (gray bars). Bars for proximal leaflets represent means ± sd (two leaflets each from n = 3 independent plants). Bars for distal leaflets represent means ± sd (three leaflets each from n = 3 independent plants). SA and SA-Glc were measured using LC–MS. Values reported as zero indicate no detection of metabolites. Asterisks indicate a significant metabolite difference in metabolite levels (one-tailed t test; **P < 0.01, ****P < 0.0001). The experiment was repeated two times with similar results. D, Titer of Pst in distal leaflets 4 dpi. Bars represent mean log cfu/cm2 ± sd (three leaflets each from n = 3 independent plants). Asterisks indicate a significant difference (one-tailed t test; **P < 0.01).
Using the same experimental design that we used for metabolite profiling, we inoculated two proximal tomato leaflets with Agrobacterium strains harboring GFP or GFP + ALD1 + FMO1 (Pathway), or Pathway + UGT76B1 (Figure 5, A). At 48-h post inoculation, we challenged three distal leaflets with a 1 × 105 cfu/mL suspension of Pst. Consistent with a previous report (Holmes et al., 2019), transient expression of the NHP pathway in proximal leaflets resulted in significant protection against Pst in challenged distal leaflets compared with leaflets transiently expressing GFP alone (Figure 5, D). Notably, this systemic resistance was compromised when UGT76B1 was overexpressed alongside the NHP pathway in proximal leaflets. Expressing UGT76B1 alone did not alter protection compared with expressing GFP alone (Supplemental Figure S9). Together, these results demonstrate that overexpressing UGT76B1 is sufficient to convert NHP to NHP-Glc. Moreover, these data indicate that increasing the abundance of NHP-Glc is not sufficient to induce defense priming, suggesting that NHP, and not NHP-Glc, is the bioactive signal for SAR.
Discussion
UGTs are a highly expanded class of biosynthetic enzymes in plants, with 120 UGT genes in the Arabidopsis genome (Paquette et al., 2003). Characterized UGTs from Arabidopsis play diverse roles, including the detoxification of xenobiotic substrates and the regulation of active hormone levels. Substrate promiscuity is a feature of some plant UGTs, allowing them to conjugate diverse xenobiotic substrates (Osmani et al., 2009) while others have evolved to be far more specific, including the glycosyltransferases UGT74F1 and UGT74F2, which glycosylate SA in a regiospecific manner (George Thompson et al., 2017). The role of hormone-specific UGTs is often to generate inactive yet stable storage forms of a hormone that, in some cases, may be hydrolyzed back into the active molecule (Westfall et al., 2013). Our results indicate that NHP-Glc is an inactive or less active derivative of NHP in Arabidopsis and tomato. It is unknown whether NHP-Glc can be enzymatically hydrolyzed back into NHP to reactivate immune signaling; however, little to no NHP-Glc accumulates in Arabidopsis plants in the absence of infection (Chen et al., 2018), which suggests that NHP biosynthesis is the primary mechanism to initiate NHP-dependent SAR signaling.
Given the proposed role of UGT76B1 in NHP glycosylation, one might expect that the loss of a glycosylation step in ugt76b1 plants would cause a reciprocal increase in the levels of aglycone substrates of UGT76B1. While we could not measure this trend directly for NHP due to its instability in planta and in extracts (Chen et al., 2018), we did observe a reciprocal increase in the level of DC-NHP, a downstream product of NHP instability (Supplemental Figure S7), in the upper leaves of adult ugt76b1 plants treated locally with Pst avrRpt2 (Figure 4, B). This suggests that, under these conditions, NHP does accumulate, but only transiently due to its instability. We did not observe this reciprocal increase under all conditions, however (e.g. in seedlings [Figure 3, A] or local leaves of adult plants [Figure 4, B]). One possible reason that seedlings do not accumulate aglycone products is due to their physiological and developmental state. The accumulation of NHP and SA in ugt76b1 plants likely occurs over time and may cause positive feedback for their biosynthesis (Chen et al., 2018; Hartmann and Zeier, 2019), such that their accumulation does not occur linearly over time. Adult ugt76b1 plants treated locally with Pst avrRpt2 also did not accumulate more aglycone than WT plants (Figure 4, B and Supplemental Figure S5, D), pointing to the existence of complex regulatory mechanisms that maintain an optimal abundance of active signaling molecules in plants under pathogen stress. These metabolite measurements are also a snapshot over the course of the infection process and therefore do not provide a complete picture of the dynamics of NHP accumulation, interconversion, and degradation. Time course studies and methods for measuring NHP-specific responses could provide more information on the complex dynamics of NHP accumulation in ugt76b1 plants.
By mining publicly available Arabidopsis mRNA expression data, we found that the core NHP biosynthetic genes ALD1 and FMO1 appear to be tightly co-regulated. Even though UGT76B1 is induced under many pathogen stress conditions, its expression is not as highly correlated with the expression of core pathway genes across these same conditions (Supplemental Figure S10). We observed a similar phenomenon with known SA UGTs (Supplemental Figure S10). This suggests that the differential expression of hormone-modifying UGTs with different pathogen stressors may help coordinate dynamic immune responses. This highlights the importance of further investigation into how transcriptional and/or posttranscriptional regulation of UGT76B1 expression affects the abundance of bioactive metabolites during SAR. UGT76B1 is constitutively expressed in roots (von Saint Paul et al., 2011), suggesting that it may also play a role in the tissue-specific regulation of metabolite levels.
While NHP-Glc is the primary form of NHP detected in Arabidopsis and in the closely related plant rapeseed (Brassica rapa; Chen et al., 2018; Holmes et al., 2019), we did not observe NHP-Glc accumulation in N. benthamiana or tomato without ectopic expression of Arabidopsis UGT76B1. These data differ from the patterns of accumulation observed for SA-Glc, which has been detected in diverse plant families, including the Solanaceae (Lee et al., 1995). We showed that the expression of UGT76B1 can inactivate NHP-related pathogen defense responses in tomato (Figure 5), which raises the question of how, or if, tomato can natively modulate the abundance of NHP, the active SAR signal. Other compounds downstream of NHP have been detected during the transient expression of NHP biosynthetic enzymes in N. benthamiana (Chen et al., 2018; Holmes et al., 2019), perhaps representing distinct mechanisms that have evolved to modulate the abundance of active hormones during defense in other plant species.
ugt76b1 plants have increased resistance to Pst and altered SA-dependent gene expression in local tissues (von Saint Paul et al., 2011). Our results reveal that ugt76b1 plants have a basal level of resistance to Psm ES4326 infection equivalent to an SAR response induced in WT plants (Figure 4). We hypothesize that the increased availability of free NHP may be a driver of this phenotype in ugt76b1, as these plants have little to no ability to glycosylate NHP (Figure 3 and Supplemental Figure S4), and NHP is known to be a potent modulator of defense responses (Chen et al., 2018; Hartmann et al., 2018).
Many aspects of plant defense are intimately intertwined. Complex regulatory mechanisms underlie responses to different pathogens (Glazebrook, 2005) and the coordination of SAR (Shah et al., 2014). Vital components of the Arabidopsis signaling network include NHP and SA, which are both required to establish functional SAR (Klessig et al., 2018; Hartmann and Zeier, 2019). RNA sequencing has uncovered a large overlap between NHP- and SA-dependent gene regulation, as well as the presence of SA-independent regulation of SAR (Bernsdorff et al., 2016; Hartmann et al., 2018; Hartmann and Zeier, 2019). Our biochemical studies demonstrated that the enzyme previously known to glycosylate SA and ILA (von Saint Paul et al., 2011; Maksym et al., 2018) can also metabolize NHP (Supplemental Figures S4, S5), further connecting these signaling molecules. It is possible that the true biological function of UGT76B1 is to glycosylate a set of small molecules and that all of these molecules play distinct roles in defense. One potential mechanism for how UGT76B1 achieves specificity in the glycosylation of signaling metabolites is through spatial or temporal expression patterns. It is possible that the changes in local concentrations of small molecule substrates over time and physical proximity with metabolic enzymes enhance the underlying specificity we observe in vitro. Experiments that track the movement of these metabolites could help elucidate the role of spatial patterning across plant tissue in signaling in the future.
Notably, our experiments in tomato provided additional evidence that NHP is a bioactive signaling molecule in SAR and revealed that glycosylation can be used to modulate this systemic response. Simply by expressing UGT76B1 alongside the NHP biosynthetic enzymes in tomato, the beneficial effect of producing NHP was abolished (Figure 5, B). This finding raises the question of whether NHP metabolism is used in other plants to regulate signaling. Such information will also be valuable for engineering approaches that seek to tune enhanced resistance in tomato. Improving resistance using synthetic chemicals has been challenging due to an inherent imbalance of plant defense and growth in the presence of inducers (Heil et al., 2000; Huot et al., 2014). In order to engineer immunity using synthetic approaches, defense-yield tradeoffs that naturally occur in plants to balance limited resources will need to be addressed (Mauch et al., 2001; Ning et al., 2017). While NHP may be protective in the context of infection, constitutive expression of NHP would likely cause unintended growth defects, and any stable system would require inducible control of pathway enzymes and a mechanism to attenuate the signal in the absence of infection. We have shown that UGT76B1 can eliminate the NHP-dependent SAR signal in tomato (Figure 5); this activity could be leveraged to engineer dynamic control over crop defense.
In closing, our results reveal that metabolism by the UDP-glycosyltransferase UGT76B1 plays critical role in modulating immunity in Arabidopsis by glycosylating NHP, the key chemical initiator of SAR. We anticipate that uncovering the association of UGT76B1 with NHP signaling will more broadly contribute to our understanding of how plants use metabolic transformations of small plant signals to tune the dynamics, tissue specificity, and spatial regulation of defense responses.
Materials and methods
Gene expression and correlation analysis
Arabidopsis microarray datasets were obtained from the NASCArrays database (Craigon et al., 2004; indexed experiments can be found at http://arabidopsis.info/affy/link_to_iplant.html). Log-scaled gene expression ratios were calculated from experiments 120, 122, 123, 167, 169, 330, 415, and 447 as previously described (Rajniak et al., 2015). Pearson’s r correlation coefficients between genes were calculated from log2 normalized expression data from these microarray datasets.
Plant materials and growth conditions
For experiments with hydroponically grown seedlings, A. thaliana ecotype Col-0 (WT), homozygous SAIL (Sessions et al., 2002), or T-DNA insertional line (SAIL_1171_A11; ugt76b1-1; Col-0 background) seeds were surface sterilized with 50% ethanol for 1 min, followed by 50% bleach for 10 min, washed three times in sterile water, and resuspended in 1× Murashige–Skoog (MS) medium with vitamins (PhytoTechnology Laboratories; pH 5.7). The seeds were placed into 3 mL of MS medium + 5 g/L sucrose in wells of six-well culture plates (five seeds/well). The plates were sealed with micropore tape (3 M), vernalized at 4°C for 48 h, and transferred to a growth chamber at 50% humidity, 22°C, and 100 µmol/m2/s photon flux (Philips F17T8/TL841 17 W bulbs) under a 16-h/8-h day/night cycle. After 1 week, spent medium was removed and replaced with 3 mL of fresh MS medium + 5 g/L sucrose. Plants were elicited after an additional week of growth. For experiments with adult Arabidopsis plants, Col-0, fmo1-1 (SALK_026163; Col-0 background), and ugt76b1 plants were grown in soil in a growth chamber at 80% humidity, 22°C, and 100 µmol/m2/s photon flux (Philips F32T8/TL941 32W bulbs) under a 10-h/14-h day/night cycle. For experiments with tomato (S. lycopersicum cultivar VF36), plants were grown in a greenhouse (14-h/10-h day/night cycle, 25°C–28°C, natural light) for 4–5 weeks. Nicotiana benthamiana plants were grown in soil on a growth shelf at room temperature and humidity, and 140 µmol/m2/s photon flux (Fluence Bioengineering RAZR3 LED array) under a 16-h/8-h day/night cycle for 4 weeks prior to transient transformation.
Cloning of Arabidopsis UGT candidate genes
Agrobacterium tumefaciens GV3101 and C58C1 pCH32 strains harboring Arabidopsis ALD1 and FMO1 genes in the pEAQ-HT vector (Peyret and Lomonossoff, 2013) were constructed previously (Holmes et al., 2019). Arabidopsis UGT candidates were PCR-amplified from Arabidopsis WT complementary DNA (cDNA) using gene-specific primers (Supplemental Table S1), cloned into pEAQ-HT between AgeI and SmaI cut sites using Gibson assembly, and transformed into E. coli 10-β. Sequence-confirmed plasmids were then transformed into A. tumefaciens GV3101 using heat shock. To create the His-tagged construct, Arabidopsis UGT76B1 was PCR-amplified from WT cDNA using gene-specific primers (Supplemental Table S1), cloned into the pET24b vector under the control of the T7 promoter, and transformed into E. coli BL21.
Transient expression in N. benthamiana
Agrobacterium strains were grown on LB agar plates with the appropriate antibiotics for 24 h. Cells were scraped from the plates with an inoculation loop, washed three times with Agrobacterium induction medium (10 mM MES buffer, 10 mM MgCl2, and 150 µM acetosyringone [pH 5.7]), resuspended in Agrobacterium induction medium, and incubated at room temperature for 2 h with agitation. To screen candidate UGTs, Agrobacteria harboring ALD1, FMO1, and the respective UGT genes were combined in equal proportions with each culture at an OD600 of 0.1. In all cases, Agrobacteria harboring GFP were added to ensure an equal final OD600 of 0.6. These solutions were infiltrated into the leaves of 4-week-old N. benthamiana plants using needleless syringes. Plants were incubated for 72 h on growth shelves under a 16-h light/8-h dark cycle prior to sample harvest.
Sample harvest and derivatizations
For all metabolomics experiments, plant tissue was harvested, lyophilized to dryness, and homogenized using a ball mill (Retsch MM 400) at 25 Hz for 2 min. Single-well Arabidopsis hydroponics samples were resuspended in 500 µL of 80:20 MeOH:H2O and incubated at 4°C for 10 min. Nicotiana benthamiana and tomato samples were resuspended in 20 µL of 80:20 MeOH:H2O per mg dry tissue and incubated at 4°C for 10 min. The liquid fraction of each sample was split for LC–MS and GC–MS analysis, respectively. Samples for GC–MS analysis were further derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA; Holmes et al., 2019). Samples were derivatized with TMSD using previously established methods (Topolewska et al., 2015). Briefly, 200 µL dried methanolic extracts were resuspended in 125 µL, methanol, 50 µL toluene, and 50 µL 2M TMSD in hexane, incubated for 1 h at room temperature, dried under N2, and resuspended in 200 µL AcN + 0.1% formic acid (FA) for LC–MS analysis.
LC–MS analysis
NHP, NHP-Glc, and TMSD-derivatized NHP and NHP-Glc were measured using previously published methods on an Agilent 1260 HPLC coupled to an Agilent 6520 quadrupole time-of-flight electrospray ionization (Q-TOF ESI) mass spectrometer (Chen et al., 2018). DC-NHP was previously identified as an NHP-degradation product using untargeted metabolomics and was measured using previously published methods on an Agilent 1260 HPLC coupled to an Agilent 6520 Q-TOF ESI mass spectrometer (Chen et al., 2018). For in vitro metabolomics experiments, SA-Glc and ILA-Glc were measured using the same parameters except in negative ionization mode. NHP-Glc and TMSD-derivatized NHP compounds were fragmented using a collision-induced dissociation (CID) energy of 10 V. TMSD-derivatized NHP-Glc was fragmented using a CID of 40 V. Extracted ion chromatogram (EIC) values were determined by extracting chromatograms with a 20-ppm error and integrating peak areas using MassHunter software (Agilent).
SA and SA-Glc were measured using an Agilent 1290 Infinity II UHPLC coupled to an Agilent 6470 triple quadrupole (QQQ) mass spectrometer. A 1.8-µm, 2.1 × 50 mm Zorbax RRHD Eclipse Plus C18 column was used for reverse phase chromatography with mobile phases of A [water with 0.1% FA] and B [acetonitrile (AcN) with 0.1% FA]. The following gradient was used for separation with a flow rate of 0.6 mL/min (percentages indicate percent buffer B): 0–0.2 min (5%), 0.2–4.2 min (5%–95%), and 4.2–5.2 min (95%–100%). The MS was run in negative mode with the following parameters: gas temperature, 250°C; gas flow rate, 12 L/min; nebulizer, 25 psig. SA was measured using monitored transitions with the following parameters: Precursor ion, 137.0239; product ions, 93 and 65.1; dwell, 150 ms; fragmentor voltage, 158 V; collision energy, 20 and 32 V, respectively, cell accelerator voltage, 4 V. SA-Glc was measured using monitored transitions with the following parameters: Precursor ion, 299.0767; product ions, 137 and 93; dwell, 150 ms; fragmentor voltage, 158 V; collision energy, 5 and 20 V, respectively, cell accelerator voltage, 4 V.
GC–MS analysis
TMS-derivatized samples were measured for Pip and NHP using published methods on an Agilent 7820A gas chromatograph coupled to an Agilent 5977B mass spectrometer (Holmes et al., 2019).
Bacterial strains and growth conditions
Escherichia coli strain 10-β, P. syringae strains pv. tomato DC3000 (Pst), pv. maculicola ES4326 (Psm ES4326), and pv. tomato harboring the avirulence gene avrRpt2 (Pst avrRpt2), and A. tumefaciens strains GV3101 and C58C1 pCH32 were used in this study. Escherichia coli strains were grown in lysogeny broth (LB) agar containing the appropriate antibiotics at 37°C. Pseudomonas strains were grown at 28°C on nutrient yeast glycerol agar (NYGA) medium containing rifampicin (100 μg/mL). Agrobacterium strains were grown at 28°C on LB agar containing rifampicin (100 μg/mL), tetracycline (5 μg/mL), and kanamycin (50 μg/mL) for C58C1 pCH32 and gentamycin (100 μg/mL) and kanamycin (50 μg/mL) for GV3101.
Elicitation methods
For hydroponics experiments, Pst was grown on LB agar plates at 30°C. A single colony was grown in liquid LB medium to an OD600 of ∼0.5, washed three times, and resuspended to an OD600 of 0.1 in MS medium + 5 g/L sucrose. Thirty microliters of 1 M MgCl2 (mock), 100 mM NHP, 10 mM SA, or the Pst solution were used for elicitations.
SAR assays in Arabidopsis
SAR bacterial growth assays were performed as described (Chen et al., 2018). A 30–32-day-old Col-0, fmo1, and ugt76b1 plants were used in this assay. Briefly, three lower leaves (leaf numbers 5–7) of each plant were infiltrated with 10 mM MgCl2 or a 5 × 106 cfu/mL suspension of Pst avrRpt2 in 10 mM MgCl2. Two days later, one upper leaf (leaf number 10) of each plant was inoculated with a 1 × 105 cfu/mL suspension of Psm ES4326, and the plants were kept under a dome to maintain humidity. The disease symptoms of Psm ES4326-infected upper leaves were photographed at 3 dpi, and the titer of Psm ES4326 in these leaves was quantified by homogenizing leaves discs in 1 mL of 10 mM MgCl2, plating appropriate dilutions on NYGA medium with rifampicin (100 μg/mL). Plates were incubated at 28°C for 2 days prior to counting bacterial colonies.
Metabolic profiling of defense priming in Arabidopsis
Three lower leaves (leaf numbers 5–7) of 30–32-day-old Col-0, fmo1, and ugt76b1 Arabidopsis plants were infiltrated with 10 mM MgCl2 and a 5 × 106 cfu/mL suspension of Pst avrRpt2 in 10 mM MgCl2. Forty-eight hour later, the three treated lower leaves and three untreated upper leaves (leaf numbers 8–10) were harvested, pooled, and frozen in liquid nitrogen for metabolic profiling by GC–MS, LC–MS, and triple quadrupole (QQQ)-MS analysis.
Transient expression and SAR assays in tomato
Transient expression and SAR assays were performed as previously (Holmes et al., 2019). Briefly, Agrobacterium C58C1 pCH32 strains harboring combinations of GFP, FMO1, ALD1, and UGT76B1 were infiltrated into two proximal (bottom) leaflets of the third and fourth compound leaves of 4–5-week-old tomato plants for 48 h. For metabolic profiling, two proximal and three distal leaflets of the third compound leaf were harvested. For the bacterial growth assays, the three distal leaflets of the fourth compound leaves were inoculated with a 1 × 105 cfu/mL suspension of Pst. Plants were incubated for four additional days, and the titer of Pst was then determined by plating serial dilutions (Holmes et al., 2019).
In vitro assays
Crude protein was extracted from 80 mg of fresh tissue from N. benthamiana leaves transiently expressing GFP, UGT76B1, or UGT76B1-6xHis using a P-PER plant protein extraction kit (Pierce). Crude extracts of GFP and UGT76B1 were used directly in in vitro metabolomics assays. Protein concentrations were determined using a bicinchoninic acid assay kit (Pierce). All in vitro metabolomics assays with protein from N. benthamiana were performed in 200 µL reaction volumes at room temperature with the following concentrations of reagents: 0.1 M Tris–HCl pH 7.5, 5 mM UDP-Glucose, 1 µg total protein, and 0.5 mM aglycone (NHP, SA, or ILA). The reactions were quenched by adding 50 µL reaction to 150 µL AcN.
Escherichia coli BL21 strains harboring His-tagged UGT76B1 were grown overnight at 37°C in LB. Two milliliters of overnight culture was inoculated into 25 mL LB and cultures were grown at 37°C to an OD600 of 0.6. The cultures were then induced with 0.5 mM IPTG and grown for an additional 5 h at 28°C. The cells were harvested and disrupted using an EmulsiFlex B15 (Avestin). The soluble fractions were enriched using gravity flow through Ni-NTA agarose resin and eluted with increasing concentrations of imidazole. Proteins were concentrated using 30 kDa centrifugal filters, buffer-exchanged into 50 mM Tris–HCl, pH 8 with 10% glycerol, and kept at −80°C for long-term storage.
In vitro time course experiments were performed with 1 µM enriched E. coli UGT76B1-6xHis protein fraction in 200 µL reactions containing 0.1 M Tris–HCl pH 7.5, 0.5 mM UDP-Glc, and aglycone substrates at a concentration of 0.5 mM. The reactions were monitored as a time course at 5, 10, 30, and 60 min. Free UDP was measured as a proxy for reaction progress using a UDP-Glo enzyme assay kit (Promega; Zegzouti et al., 2013).
Synthesis of DC-NHP
2,3,4,5-Tetrahydropyridine N-oxide (DC-NHP) was synthesized from pyridine using previously established methods (Gella et al., 2009). Briefly, 2 mmol piperidine was added to 3.5 mL of a 4:1 AcN:tetrahydrofuran (THF) mixture on ice. 2.8 mL of 0.1 M aqueous Na2EDTA was added and stirred, followed by 0.84 g NaHCO3. The solution was kept on ice with constant stirring and 1.29 g Oxone (potassium peroxymonosulfate) was added over the course of 2 h. After the addition of Oxone, the solution was stirred on ice for one additional hour prior to purification. The final solution was extracted twice with 10 mL ethyl acetate and the organic fraction was dried and used for downstream analyses. DC-NHP was also synthesized from N-hydroxypiperidine as previously described (Chen et al., 2018). Briefly, two equivalents of meta-Chloroperoxybenzoic acid (mCPBA) were added dropwise to a solution of N-hydroxypiperidine in dichloromethane on ice, with constant stirring. The solution was stirred on ice for 5 min, followed by 30 min at 20°C. The structure of DC-NHP is known to exist in its dimeric form, which is consistent with our observations in planta and with synthesized DC-NHP (Alford et al., 1966; Alsbaiee and Ali, 2008). The large in-source fragment of m/z = 100.075 was used throughout to quantify the abundance of DC-NHP in plant extracts.
Statistical analyses
One-tailed or two-tailed Student’s t tests were performed to determine statistically significant differences between experimental and control groups. One versus two-tailed t tests were determined based upon the null hypothesis and are indicated in the figure legends. P-values <0.05 were considered statistically significant, and all significance levels are indicated in the figure legends (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001). Statistical testing results are shown in Supplemental Data Set S1).
Accession numbers
The sequence data for this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: UGT71B4 (AT4G15260), UGT73B2 (AT4G34135), UGT73B3 (AT4G34131), UGT73C5 (AT2G36800), UGT73D1 (AT3G53150), UGT74F2 (AT2G43820), UGT76B1 (AT3G11340), UGT76F2 (AT3g55700), UGT85A1 (AT1G22400), UGT85A7 (AT1G22340), UGT86A2 (AT2G28080), UGT89A2 (AT5G03490), UGT73C3 (AT2G36780), UGT92A1 (AT5G12890), UGT73B4 (AT2G15490), UGT87A2 (AT2G30140), UGT76E12 (AT3G46660), ALD1 (AT2G13810), and FMO1 (AT1G19250). Germplasm used in this study includes fmo1-1 (SALK_026163) and ugt76b1-1 (SAIL_1171_A11). LC–MS and GC–MS data integral to the conclusions in the manuscript have been deposited in Mass Spectrometry Interactive Virtual Environment (MassIVE; https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under accession numbers MSV000086046, MSV000086048, and MSV000086056.
Supplemental data
Supplemental Figure S1. mRNA expression profiles of candidate Arabidopsis UGT genes.
Supplemental Figure S2. Arabidopsis UGT76B1 glycosylates NHP in N. benthamiana.
Supplemental Figure S3. TMSD derivatization of NHP and NHP-Glc.
Supplemental Figure S4. Metabolic profiling of Arabidopsis WT and ugt76b1 mutant seedlings.
Supplemental Figure S5. Metabolic profiling of Arabidopsis WT, fmo1, and ugt76b1 plants in SAR assays.
Supplemental Figure S6. Trial 2 bacterial growth and log-scaled abundances of DC-NHP and NHP-Glc in Arabidopsis WT, ugt76b1, and fmo1 plants.
Supplemental Figure S7. Synthesis and characterization of DC-NHP.
Supplemental Figure S8. Abundance of SA in distal leaflets of tomato during transient expression of NHP-Glc pathway genes.
Supplemental Figure S9. Effect of transient expression of Arabidopsis UGT76B1 in tomato leaves used for transient SAR analysis.
Supplemental Figure S10. mRNA expression and coexpression analysis of SA-Glc and NHP-Glc biosynthetic genes in Arabidopsis obtained from publicly available microarray data.
Supplemental Table S1. Primers used in this study
Supplemental Data Set S1. Results of statistical analyses.
Supplementary Material
Acknowledgments
The authors thank George Lomonossoff (John Innes Centre) for providing the pEAQ plasmid. They also thank K. Smith and J. Foret (Stanford) for helpful discussions.
Funding
This work was supported by a Howard Hughes Medical Institute-Simons Faculty Scholar award (to E.S.S.), the National Science Foundation Graduate Research Fellowship DGE-1656518 (to E.C.H.), National Science Foundation IOS-1555957 and Binational Science Foundation Grant 2011069 (to M.B.M.), and Ministry of Science and Technology of Taiwan-105-2917-I-564-093 (to Y.-C.C.).
Conflict of interest statement. None declared.
E.C.H., Y.-C.C., E.S.S., and M.B.M designed the research. E.C.H. and Y.-C.C. performed the research. E.C.H., Y.-C.C., E.S.S., and M.B.M. analyzed the data. E.C.H., Y.-C.C., E.S.S., and M.B.M. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) are: Elizabeth S. Sattely (sattely@stanford.edu) and Mary Beth Mudgett (mudgett@stanford.edu).
References
- Alford EJ, Hall JA, Rogers MAT (1966) Aliphatic hydroxylamines. Part IV: N-hydroxyhexamethyleneimine. J Chem Soc C Org 1103–1107 [Google Scholar]
- Alsbaiee A, Ali SA (2008) The face selectivity of 1,3-dipolar cycloaddition reactions of 4-butyloxycarbonyl-3,4,5,6-tetrahydropyridine 1-oxide. Tetrahedron 64:6635–6644 [Google Scholar]
- Bauer S, Mekonnen DW, Geist B, Lange B, Ghirardo A, Zhang W, Schäffner AR (2020a) The isoleucic acid triad: distinct impacts on plant defense, root growth, and formation of reactive oxygen species. J Exp Bot 71:4258–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Mekonnen DW, Hartmann M, Janowski R, Lange B, Geist B, Zeier J, Schäffner AR (2020b) UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid and controls basal pathogen defense. Plant Cell ( 10.1101/2020.07.12.199356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsdorff F, Doring AC, Gruner K, Schuck S, Brautigam A, Zeier J (2016) Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28:102–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boachon B, Gamir J, Pastor V, Erb M, Dean JV, Flors V, Mauch-Mani B (2014) Role of two UDP-glycosyltransferases from the L group of Arabidopsis in resistance against Pseudomonas syringae. Eur J Plant Pathol 139:707–720 [Google Scholar]
- Caarls L, Elberse J, Awwanah M, Ludwig NR, de Vries M, Zeilmaker T, Van Wees SCM, Schuurink RC, Van den Ackerveken G (2017) Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc Natl Acad Sci U S A 114:6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Holmes EC, Rajniak J, Kim J-G, Tang S, Fischer CR, Mudgett MB, Sattely ES (2018) N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc Natl Acad Sci U S A 115: E4920–E4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Cho Y-H, Kim K, Matsui M, Son S-H, Kim S-K, Fujioka S, Hwang I (2013) BAT1, a putative acyltransferase, modulates brassinosteroid levels in Arabidopsis. Plant J 73:380–391 [DOI] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res 32:D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gella C, Ferrer È, Alibés R, Busqué F, de March P, Figueredo M, Font J (2009) A metal-free general procedure for oxidation of secondary amines to nitrones. J Org Chem 74:6365–6367 [DOI] [PubMed] [Google Scholar]
- George Thompson AM, Iancu CV, Neet KE, Dean JV, Choe J-Y (2017) Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci Rep 7:46629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier J (2018) L-lysine metabolism to N-hydroxypipecolic acid: an integral immune-activating pathway in plants. Plant J 96:5–21 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier J (2019) N-hydroxypipecolic acid and salicylic acid: a metabolic duo for systemic acquired resistance. Curr Opin Plant Biol 50:44–57 [DOI] [PubMed] [Google Scholar]
- Hartmann M, Zeier T, Berndorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Brautigam A, Holzel T, et al. (2018) Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173:1–14 [DOI] [PubMed] [Google Scholar]
- Heil M, Hilpert A, Kaiser W, Linsenmair KE (2000) Reduced growth and seed set following chemical induction of pathogen defence: does systemic acquired resistance (SAR) incur allocation costs? J Ecol 88:645–654 [Google Scholar]
- Holmes EC, Chen Y-C, Sattely ES, Mudgett MB (2019) An engineered pathway for N-hydroxy-pipecolic acid synthesis enhances systemic acquired resistance in tomato. Sci Signal 12:eaay3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY (2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7:1267–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila J, De Rycke R, Van Montagu M, Angenon G (1997) An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci 122:101–108 [Google Scholar]
- Klessig DF, Choi HW, Dempsey DMA (2018) Systemic acquired resistance and salicylic acid: past, present, and future. Mol Plant Microbe Interact 31:871–888 [DOI] [PubMed] [Google Scholar]
- Kühnel E, Laffan DDP, Lloyd-Jones GC, Martínez del Campo T, Shepperson IR, Slaughter JL (2007) Mechanism of methyl esterification of carboxylic acids by trimethylsilyldiazomethane. Angew Chem 46:7075–7078 [DOI] [PubMed] [Google Scholar]
- Lee HI, León J, Raskin I (1995) Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci U S A 92:4076–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E-K, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles DJ (2002) The activity of arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem 277:586–592 [DOI] [PubMed] [Google Scholar]
- Liu P-P, Yang Y, Pichersky E, Klessig DF (2009) Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in Arabidopsis. Mol Plant Microbe Interact 23:82–90 [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan J-P, Li D-K, Luo Q, Yan Q, Liu Z-B, Ye L-M, Wang J-M, Li X-F, Yang Y (2015). UDP-glucosyltransferase71C5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol 167:1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksym RP, Ghirardo A, Zhang W, von Saint Paul V, Lange B, Geist B, Hajirezaei M-R, Schnitzler J-P, Schäffner AR (2018) The defense-related isoleucic acid differentially accumulates in Arabidopsis among branched-chain amino acid-related 2-hydroxy carboxylic acids. Front Plant Sci 9:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C (2001) Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J 25:67–77 [DOI] [PubMed] [Google Scholar]
- Mohnike L, Rekhter D, Huang W, Feussner K, Tian H, Herrfurth C, Zhang Y, Feussner I (2021) The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 33: 735--749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Mithöfer A, Kombrink E, Boland W, Hamamoto S, Uozumi N, Tohma K, Ueda M (2011) 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol 155:1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25:213–221 [DOI] [PubMed] [Google Scholar]
- Navarova H, Bernsdorff F, Doring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24:5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Liu W, Wang G-L (2017) Balancing immunity and yield in crop plants. Trends Plant Sci 22:1069–1079 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Okazaki M, Kida T, Nishina Y, Morishita Y, Ogawa T, Suzuki H, Shibata D, Jikumaru Y, Hanada A, et al. (2012) Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24:3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Møller BL (2009) Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70:325–347 [DOI] [PubMed] [Google Scholar]
- Paquette S, Møller BL, Bak S (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62:399–413 [DOI] [PubMed] [Google Scholar]
- Peyret H, Lomonossoff GP (2013) The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol Biol 83:51–58 [DOI] [PubMed] [Google Scholar]
- Piotrowska A, Bajguz A (2011) Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 72:2097–2112 [DOI] [PubMed] [Google Scholar]
- Rajniak J, Barco B, Clay NK, Sattely ES (2015) A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 525:376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14:2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Chaturvedi R, Chowdhury Z, Venables B, Petros RA (2014) Signaling by small metabolites in systemic acquired resistance. Plant J 79:645–658 [DOI] [PubMed] [Google Scholar]
- Smirnova E, Marquis V, Poirier L, Aubert Y, Zumsteg J, Ménard R, Miesch L, Heitz T (2017) Jasmonic acid oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea infection. Mol Plant 10:1159–1173 [DOI] [PubMed] [Google Scholar]
- Stappers F, Broeckx R, Leurs S, Van Den Bergh L, Agten J, Lambrechts A, Van den Heuvel D, De Smaele D (2002) Development of a safe and scalable amine-to-nitrone oxidation: a key step in the synthesis of R107500. Org Process Res Dev 6:911–914 [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16:2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J 37:471–483 [DOI] [PubMed] [Google Scholar]
- Topolewska A, Czarnowska K, Haliński ŁP, Stepnowski P (2015) Evaluation of four derivatization methods for the analysis of fatty acids from green leafy vegetables by gas chromatography. J Chromatogr B 990:150–157 [DOI] [PubMed] [Google Scholar]
- von Saint Paul V, Zhang W, Kanawati B, Geist B, Faus-Keßler T, Schmitt-Kopplin P, Schäffner AR (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23:4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Jin S-H, Hu H-Q, Sun Y-G, Wang Y-W, Han P, Hou B-K (2012) UGT87A2, an Arabidopsis glycosyltransferase, regulates flowering time via FLOWERING LOCUS C. New Phytol 194:666–675 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall CS, Muehler AM, Jez JM (2013) Enzyme action in the regulation of plant hormone responses. J Biol Chem 288:19304–19311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Engel L, Vidugiris G, Goueli S (2013) Detection of glycosyltransferase activities with homogenous bioluminescent UDP detection assay. Glycobiology 23:1340–1341 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.