Abstract
Lignans/neolignans are generally synthesized from coniferyl alcohol (CA) in the cinnamate/monolignol pathway by oxidation to generate the corresponding radicals with subsequent stereoselective dimerization aided by dirigent proteins (DIRs). Genes encoding oxidases and DIRs for neolignan biosynthesis have not been identified previously. In Arabidopsis thaliana, the DIR AtDP1/AtDIR12 plays an essential role in the 8-O-4′ coupling in neolignan biosynthesis by unequivocal structural determination of the compound missing in the atdp1 mutant as a sinapoylcholine (SC)-conjugated neolignan, erythro-3-{4-[2-hydroxy-2-(4-hydroxy-3-methoxyphenyl)-1-hydroxymethylethoxy]-3,5-dimethoxyphenyl}acryloylcholine. Phylogenetic analyses showed that AtDP1/AtDIR12 belongs to the DIR-a subfamily composed of DIRs for 8-8′ coupling of monolignol radicals. AtDP1/AtDIR12 is specifically expressed in outer integument 1 cells in developing seeds. As a putative oxidase for neolignan biosynthesis, we focused on AtLAC5, a laccase gene coexpressed with AtDP1/AtDIR12. In lac5 mutants, the abundance of feruloylcholine (FC)-conjugated neolignans decreased to a level comparable to those in the atdp1 mutant. In addition, SC/FC-conjugated neolignans were missing in the seeds of mutants defective in SCT/SCPL19, an enzyme that synthesizes SC. These results strongly suggest that AtDP1/AtDIR12 and AtLAC5 are involved in neolignan biosynthesis via SC/FC. A tetrazolium penetration assay showed that seed coat permeability increased in atdp1 mutants, suggesting a protective role of neolignans in A. thaliana seeds.
The dirigent protein AtDP1/AtDIR12 and the laccase AtLAC5 play essential roles in the biosynthesis of seed-coat protective neolignans via sinapoylcholine/feruloylcholine in Arabidopsis.
Introduction
Phenolic compounds including lignins, lignans, neolignans, and flavonoids are major plant secondary metabolites (also known as specialized metabolites; Petersen et al., 2010; Ralph et al., 2019). These metabolites are derived from the phenylpropanoid metabolic pathway and are widely distributed in the plant kingdom (Weng and Chapple, 2010). Phenolic compounds play an important role in the structural enhancement of the plant body, biological defense, environmental stress tolerance, and interactions between plants and other organisms, and also contribute to human health as pharmaceuticals, dietary supplements, flavors, and pigments (Vogt, 2010).
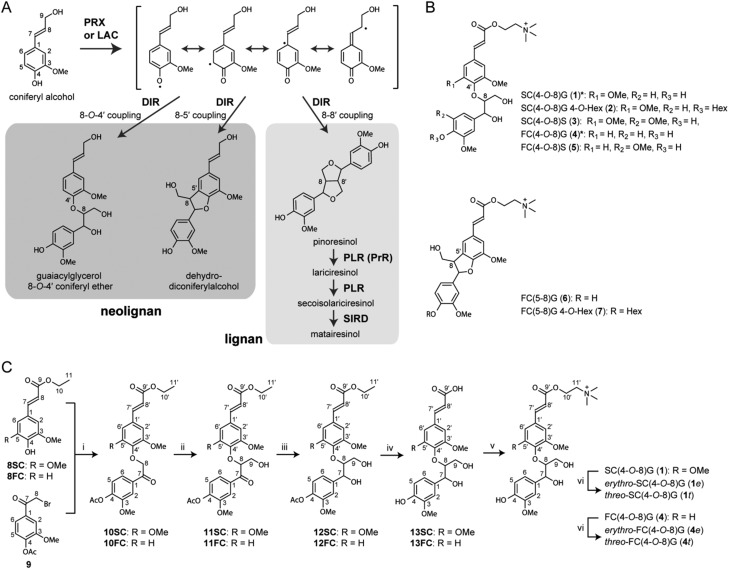
Lignans are a class of dimeric phenylpropanoid metabolites [(C6C3)2] with a C8-C8′ linkage; compounds with all other types of linkages are called neolignans (Moss, 2000; Umezawa, 2003a, 2003b). The biosynthesis of lignans such as pinoresinol, matairesinol, lariciresinol, and secoisolariciresinol is initiated with coniferyl alcohol (CA) in the cinnamate/monolignol pathway and has been well-studied. CA is oxidized by oxidases such as peroxidases and laccases (LACs) to give rise to the corresponding radical, which is then dimerized stereoselectively at the C8 and C8′ positions to a lignan, pinoresinol, with the aid of dirigent proteins (DIRs; Davin and Lewis, 2003; Umezawa, 2003a; Suzuki and Umezawa, 2007; Figure 1A). Pinoresinol is further converted to matairesinol via lariciresinol and secoisolariciresinol by pinoresinol/lariciresinol reductase and secoisolariciresinol dehydrogenase (Davin and Lewis, 2003; Umezawa, 2003a; Suzuki and Umezawa, 2007; Figure 1A). In Arabidopsis (Arabidopsis thaliana), pinoresinol is converted to lariciresinol by pinoresinol reductase (Nakatsubo et al., 2008). The biosynthetic pathway from CA to matairesinol is common to a number of plant species, and a wide variety of lignans are derived from pinoresinol, lariciresinol, secoisolariciresinol, and matairesinol by aromatic substituent modification (Umezawa, 2003a).
Figure 1.
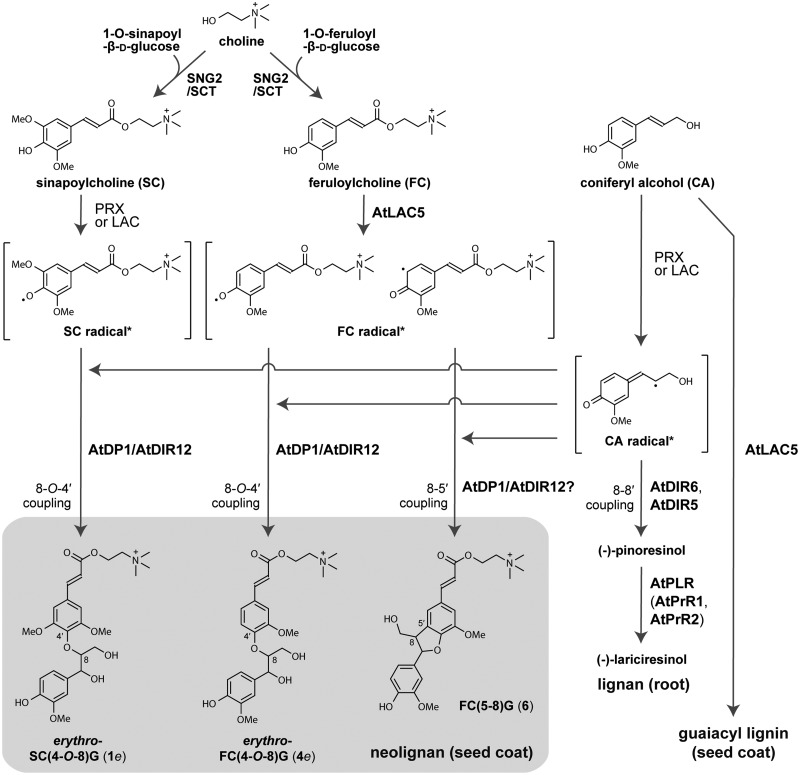
Lignans and neolignans. (A) The biosynthetic pathways of lignans and neolignans. DIR, dirigent protein; LAC, laccase; PLR, pinoresinol/lariciresinol reductase; PrR, pinoresinol reductase; PRX, peroxidase; SIRD, secoisolariciresinol dehydrogenase. (B) Structures of neolignans in Arabidopsis seeds (see also Supplemental Table 1). G, Guaiacyl moiety; S, Syringyl moiety; SC, sinapoylcholine; FC, feruloylcholine; Hex, hexose. Asterisks indicate the compounds authenticated with chemically synthesized standards. (C) Synthetic scheme for neolignans SC(4-O-8)G (1) and FC(4-O-8)G (4). Reaction conditions: (i) acetone/K2CO3/reflux; (ii) dioxane/K2CO3/formaldehyde/r.t.; (iii) ethanol/NaBH4/r.t.; (iv) NaOH aq./r.t.; (v) DMF/bromocholine bromide/NaOH/90°C; (vi) preparative HPLC. r.t., room temperature.
In contrast, the biosynthetic pathway for neolignans remains largely unknown. Neolignans are also derived from monolignols generated in the cinnamate/monolignol pathway (Umezawa et al., 2011). The biosynthetic pathways for producing lignan and neolignans diverge at the step for oxidative coupling of monolignols (Figure 1A). Several studies dealing with crude enzyme preparations mediating the formation of neolignans from phenylpropanoid monomers have been reported (Orr and Lynn, 1992; Katayama and Kado, 1998; Sartorelli et al., 2001; Lourith et al., 2005; Umezawa et al., 2011); however, there are no published reports of genes encoding oxidases and DIRs involved in neolignan biosynthesis.
DIR was originally identified as a protein for guiding the stereoselective coupling of CA radicals in the production of (+)-pinoresinol, a lignan, in Forsythia sp. (Davin et al., 1997). Subsequently, genes encoding (+)- or (−)-pinoresinol-forming DIRs were found in a variety of seed plants including Arabidopsis (Davin et al., 1997; Kim et al., 2002, 2012, 2015; Ralph et al., 2007; Pickel et al., 2010; Dalisay et al., 2015; Seneviratne et al., 2015; Gasper et al., 2016). DIR-mediated enantioselective bimolecular coupling is not restricted to the CA radical. Atroposelective formation of (+)-gossypol from hemigossypol, a sesquiterpene phenol, is also mediated by DIRs in Gossypium spp. (Effenberger et al., 2015, 2017). Moreover, pterocarpan synthases with dirigent-like domains have also been reported to occur in several plants (Uchida et al., 2017; Meng et al., 2020).
DIRs belong to a multi-gene family that is classified into six subfamilies (DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-f, and DIR-g) based on phylogenetic analyses (Ralph et al., 2007). The DIRs involved in lignan biosynthesis (guiding 8-8′ coupling) belong to the DIR-a subfamily and are functionally conserved among plant species.
In Arabidopsis, lignans and neolignans are localized in leaves and roots (Böttcher et al., 2008; Nakatsubo et al., 2008; Okazawa et al., 2011; Dima et al., 2015) and neolignans are also present in seeds (Böttcher et al., 2008). Arabidopsis contains 25 DIR genes (AtDIR1–AtDIR25) that are distributed in the DIR-a, DIR-b/d, and DIR-e subfamilies in addition to an atypical type, OVULE ABORTION 2 (At5g49030.3; Paniagua et al., 2017). AtDIR6 in the DIR-a subfamily was identified as a (−)-pinoresinol-forming DIR based on overexpression experiments, RNAi approaches, and recombinant protein assays (Pickel et al., 2010; Kim et al., 2012). Recombinant AtDIR5 protein (DIR-a subfamily) also has a (−)-pinoresinol-forming activity in vitro (Kim et al., 2012). ENHANCED SUBERIN 1 (ESB1, AtDIR10) in the DIR-e subfamily is essential for building the extracellular lignin-based Casparian strip (Hosmani et al., 2013; Barbosa et al., 2019); however, the physiological functions of the other Arabidopsis DIRs are still unknown.
Based on the correlation analyses of gene expression and metabolite accumulation, we found an orphan DIR gene, AtDP1/AtDIR12 (At4g11180, DIR-a subfamily), that showed no significant correlation with putative lignans and neolignans that accumulate in roots and other DIR genes (Matsuda et al., 2010). The preliminary data suggested that the T-DNA inserted atdp1 mutant lacked a putative neolignan compound with a peak at m/z 668 (Matsuda et al., 2010); however, the chemical structure of the compound missing in the mutant and AtDP1/AtDIR12 function remained to be verified.
Here, we report the identification of a sinapoylcholine (SC)-conjugated neolignan, erythro-3-{4-[2-hydroxy-2-(4-hydroxy-3-methoxyphenyl)-1-hydroxymethylethoxy]-3,5-dimethoxyphenyl}acryloylcholine, SC(4-O-8)G (1e; Figure 1) as the compound missing in the atdp1 mutant. The chemically synthesized compound was used for confirmation of this result. A complementation test of the atdp1 mutant with AtDP1/AtDIR12 genomic clones demonstrated that AtDP1/AtDIR12 is essential for the biosynthesis of 8-O-4′-type neolignans in seeds. Transgenic plants expressing AtDP1/AtDIR12 promoter–reporter constructs show that AtDP1/AtDIR12 is localized in the outer integument 1 (oi1) cells of seeds. In addition, we found that mutants deficient in AtLAC5, a LAC co-expressed with AtDP1/AtDIR12 showed significant metabolic changes among feruloylcholine (FC)-conjugated neolignans, including erythro-3-{4-[2-hydroxy-2-(4-hydroxy-3-methoxyphenyl)-1-hydroxymethylethoxy]-3-methoxyphenyl}acryloylcholine, FC(4-O-8)G (4e in Figure 1). Neolignan analysis of the mutants defective in the SC biosynthesis ability showed that SC- and FC-conjugated neolignans are synthesized from SC/FC and CA. A tetrazolium salt assay showed that seed coat permeability was increased in the atdp1 mutants. These data indicate that AtDP1/AtDIR12 and AtLAC5 are involved in the neolignan biosynthetic pathway and suggest that neolignans play an important role in protection from external factors.
Results
The atdp1 mutants lack putative neolignans
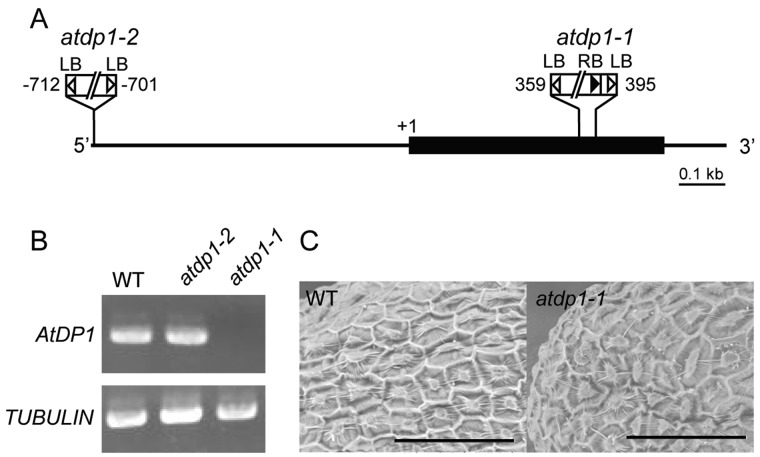
We previously reported that the T-DNA insertion mutant designated dp1 (SAIL_60_D04) lacked a compound having a peak at m/z 668 in seeds (Matsuda et al., 2010). An additional homozygous T-DNA insertion mutant, SALK_062238, was isolated and designated as atdp1-2, and dp1 (SAIL_60_D04) was renamed atdp1-1 (Figure 2A). The T-DNA was inserted between −711 and −702 base pairs of the AtDP1/AtDIR12 promoter region in atdp1-2. Reverse transcription-polymerase chain reaction (RT-PCR) analysis detected AtDP1/AtDIR12 transcripts in atdp1-2, whereas the transcript was not detected in atdp1-1 (Figure 2B), indicating that atdp1-1 is a null allele of the AtDP1/AtDIR12 gene. No obvious phenotypic differences were observed in the appearances of wild-type and atdp1-1 mutant seeds by scanning electron microscopy (Figure 2C).
Figure 2.
T-DNA insertion mutants of AtDP1/AtDIR12. (A) Schematic representation of AtDP1/AtDIR12 with two T-DNA insertion mutants used in this study. The thick line indicates the coding region and the thinner line indicates the 5′- and 3′-untranslated regions. AtDP1/AtDIR12 has no introns. White and black triangles show the left and right borders, respectively. Numbers indicate the position of the T-DNA insertion. LB, left border; RB, right border. (B) RT-PCR analysis of transcripts in wild-type (WT, Col-0) and two independent homozygous mutant lines (atdp1-1 and atdp1-2). C, Scanning electron micrograph of epidermal cells from a dry seed of WT (Col-0) and the homozygous atdp1-1 mutant. Scale bars = 100 µm.
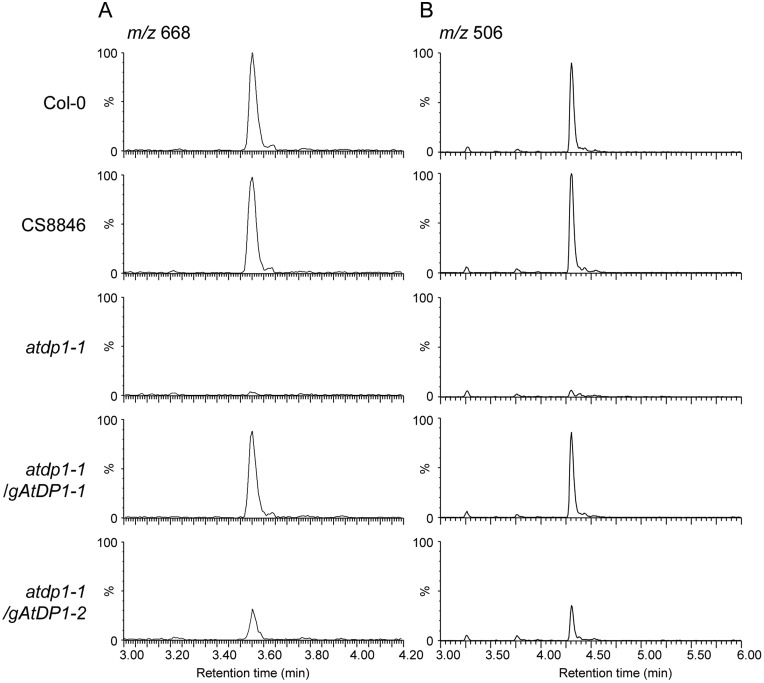
Metabolome analyses showed that a compound with a peak at m/z 506 corresponding to a new putative neolignan compound and the previously reported putative neolignan compound (m/z = 668; Matsuda et al., 2010) were both missing in seeds of the atdp1-1 mutant (Figure 3). To assess whether the AtDP1/AtDIR12 mutation was responsible for the absence of the two compounds observed in the mutant, we performed a complementation test of the atdp1-1 mutant. The mutant was transformed with two types of AtDP1/AtDIR12 genomic clones (gAtDP1-1 and gAtDP1-2, 3.0 kb/1.9 kb AtDP1/AtDIR12 genomic fragments containing 1.8-kb/0.72-kb promoter regions, respectively). Independent transgenic lines carrying gAtDP1-1 and gAtDP1-2 had essentially the same metabolic profile as wild-type plants, although the levels of the target compound were lower in transgenic lines carrying gAtDP1-2 (Figure 3). These data demonstrated that AtDP1/AtDIR12 is crucial for the biosynthesis of these two putative neolignan compounds in seeds, and that a 1.9-kb AtDP1/AtDIR12 genomic fragment containing 0.72 kb of the promoter region contains at least the minimum region necessary for functional complementation of the metabolic defect in atdp1-1.
Figure 3.
UPLC-MS analyses of the atdp1 mutant lines. Extracted ion chromatograms at m/z 668 (A) and 506 (B) of wild-type (Col-0), a parental line of atdp1-1 (CS8846), the atdp1-deficient mutant (atdp1-1), and the atdp1-deficient mutant complemented with 3.0-kb and 1.9-kb DP1 genomic fragments (atdp1-1/gAtDP1-1 and atdp1-1/gAtDP1-2, respectively).
Structural identification of a seed-specific neolignan missing in the atdp1 mutant
The structure of the putative neolignan compound (m/z = 668) missing in the atdp1-1 mutant was proposed to be 3-{4-[2-hydroxy-2-(4-hexosyloxy-3-methoxyphenyl)-1-hydroxymethylethoxy]-3,5-dimethoxyphenyl}acryloylcholine, SC(4-O-8)G 4-O-Hex (2; Figure 1B), based on mass spectral (MS/MS) data of a chemical class of compounds and/or spectral similarity to known compounds of a chemical class (Matsuda et al., 2010). Similarly, the other missing compound (m/z = 506) was proposed to be 3-{4-[2-hydroxy-2-(4-hydroxy-3-methoxyphenyl)-1-hydroxymethylethoxy]-3,5-dimethoxyphenyl}acryloylcholine, SC(4-O-8)G (1; Figure 1B; Matsuda et al., 2010).
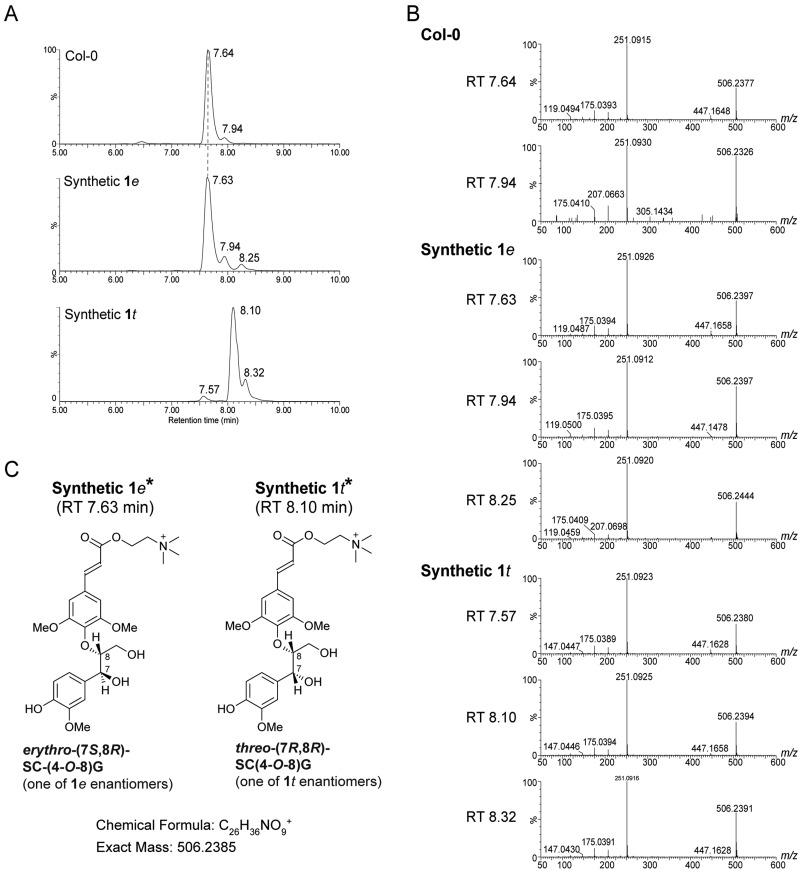
To determine the structures of the putatively annotated neolignan compounds missing in the atdp1 mutant, we chemically synthesized authentic standards (racemates) for the two stereoisomers of neolignan 1, namely erythro-SC(4-O-8)G (1e) and threo-SC(4-O-8)G (1t), as shown in Figure 1C and further described in Materials and methods section. The structural identities of the synthesized compounds were validated by nuclear magnetic resonance (NMR) spectroscopy with 1D 1H and 13C and 2D correlation spectroscopy (COSY), heteronuclear single quantum correlation (HSQC), and heteronuclear multiple bond coherence (HMBC) pulse schemes (see Materials and methods section and Supplemental Data Set 1). The stereochemistry of 1e and 1t were based on close comparisons of their NMR chemical shift and coupling constant data with those reported in the literature (Ralph and Helm, 1991; Helm and Ralph, 1992; Wolfram et al., 2010). The retention time on ultra performance liquid chromatography (UPLC; 7.64 min), mass and MS/MS spectra of the missing compound (m/z = 506) in the atdp1-1 mutant were identical to those of the authentic standard 1e (Figure 4). In addition, we confirmed that MS/MS spectra of the products (Figure 4) were identical with those of SC(4-O-8)G (1) previously isolated from transgenic Brassica napus seeds (Wolfram et al., 2010). UPLC–MS/MS data suggested that there is no detectable amount of the threo isomer 1t in the Col-0 control seeds. A minor peak (retention time 7.94 min; Figure 4) with MS/MS spectra identical with that of 1e may correspond to the cis-form of 1e because trans-cinnamic acid derivatives are known to isomerize to the cis-form after exposure to sunlight or UV light (Turner et al., 1993; Yang et al., 1999; Wong et al., 2005), and the ratio of the minor peak increased during the purification process in the light.
Figure 4.
Identification of neolignan SC(4-O-8)G (1) in Arabidopsis seeds. UPLC chromatograms (A) and MS/MS spectra (B) of synthetic standards of erythro-SC(4-O-8)G (1e) and threo-SC(4-O-8)G (1t) and the corresponding product from Col-0 are shown. The structural interpretation of fragment ions was shown in Böttcher et al. (2008). (C) Structures of synthetic 1e and 1t. *Synthetic 1e (RT 7.63 min) and 1t (RT 8.10 min) are each racemic mixtures. Only the (7S, 8R) enantiomer of 1e and the (7R, 8R) enantiomer of 1t are depicted. RT, retention time.
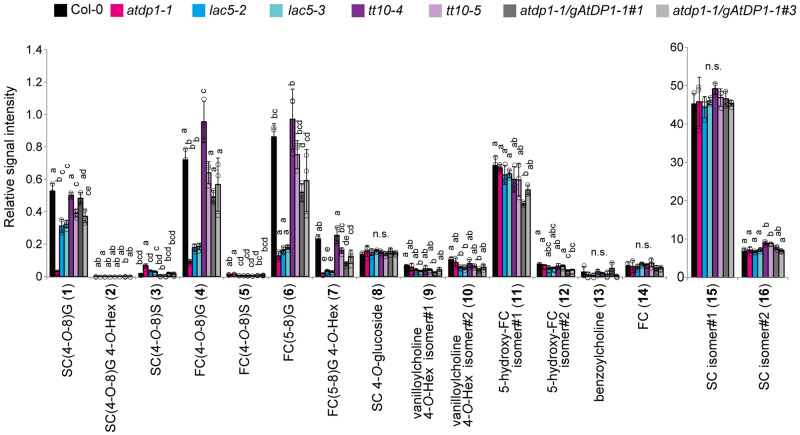
Further comprehensive analyses of neolignans, and SC and FC derivatives suggested that, along with the SC-conjugated neolignan 1e, the level of several FC-conjugated neolignans [FC(4-O-8)G (4), FC(5-8)G (6), and FC(5-8)G 4-O-Hex (7; Figure 1B)] in the atdp1-1 mutant were significantly less than those in the wild type (Figure 5; Supplemental Table 1, Supplemental Data Set 2). Accordingly, we chemically synthesized the authentic erythro- and threo-isomers of neolignan 4, i.e. erythro-FC(4-O-8)G (4e) and threo-FC(4-O-8)G (4t), respectively, in a manner similar to that described above for the SC(4-O-8)G isomers 1e and 1t (Figure 1C, and also see the Materials and methods section). As a consequence, we established that FC(4-O-8)G depleted in the atdp1-1 mutant was the erythro isomer 4e (Böttcher et al., 2008; Supplemental Figure 1). As we observed for SC(4-O-8)G, the Col-0 seeds exclusively contain the erythro isomer 4e with no detectable threo isomer 4t (Supplemental Figure 1). A putative cis-form of 4e (retention time 6.91 min and 6.90 min, Supplemental Figure 1, A, B, and F) was also detected. Taken together, our results indicate that AtDP1/AtDIR12 specifically mediates the formation of SC- and FC-conjugated 8-O-4′-type neolignans 1e and 4e in Arabidopsis. In addition, the depletion of FC(5-8)G (6) and FC(5-8)G 4-O-Hex (7) in the atdp1-1 mutant suggested that AtDP1/AtDIR12 may also be involved in the formation of FC-conjugated 8-5′-type neolignans. Interestingly, the level of SC(4-O-8)S (3) is higher in the atdp1-1 mutant (Figure 5; Supplemental Table 1, Supplemental Data Set 2). This finding suggests that, in the atdp1-1 mutant, random coupling of SC radicals with CA/sinapyl alcohol radicals may occur and/or excess amounts of substrates such as SC radicals may be utilized by other putative DIR(s) involved in SC(4-O-8)S biosynthesis.
Figure 5.
UPLC–QTOF–MS analyses of neolignans and choline derivatives in the mutants. The structures of compounds 1–7 are shown in Figure 1. Data represent the means ± sd (three biological replicates per sample). The means were compared by a one-way analysis of variance (ANOVA). Statistically significant differences (P < 0.05) were identified by Tukey’s test and are indicated by lowercase letters to represent differences between groups. G, guaiacyl moiety; S, syringyl moiety; SC, sinapoylcholine; FC, feruloylcholine; Hex, hexose; n.s., not significant.
Phylogenetic analysis of AtDP1/AtDIR12
DIRs from Forsythia intermedia, western red cedar (Thuja plicata), Arabidopsis, Schizandra chinensis, pea (Pisum sativa), and flax (Linum usitatissimum) were previously shown to be involved in the biosynthesis of lignans, (+)- or (−)-pinoresinol, via 8-8′ phenoxy radical coupling and belong to the DIR-a subfamily (Davin et al., 1997; Gang et al., 1999; Kim et al., 2002, 2012, 2015; Ralph et al., 2007; Pickel et al., 2010; Dalisay et al., 2015). The phylogenetic tree of the functionally identified DIRs indicated that these DIRs can be classified into two clusters based on their stereoselectivity, i.e. (+)- and (−)-pinoresinol-forming DIRs; AtDP1/AtDIR12 belongs to the cluster of (−)-pinoresinol-forming DIRs (Figure 6; Supplemental Figure 2). The sequence identities between (+)- or (−)-pinoresinol-forming DIRs are 57%–67% or 55%–63% at the amino acid level, respectively, except in case of genes from the same plant species. AtDP1/AtDIR12 had a lower sequence identity with (+)- and (−)-pinoresinol-forming DIRs (40%–48% and 47%–50%, respectively, at the amino acid level), but had higher sequence identities with functionally unidentified DIR-a genes (63% and 68% for AtDIR13 and AtDIR14, respectively) and (−)-pinoresinol-forming DIRs (62% and 56% for AtDIR6 and AtDIR5, respectively) from Arabidopsis.
Figure 6.
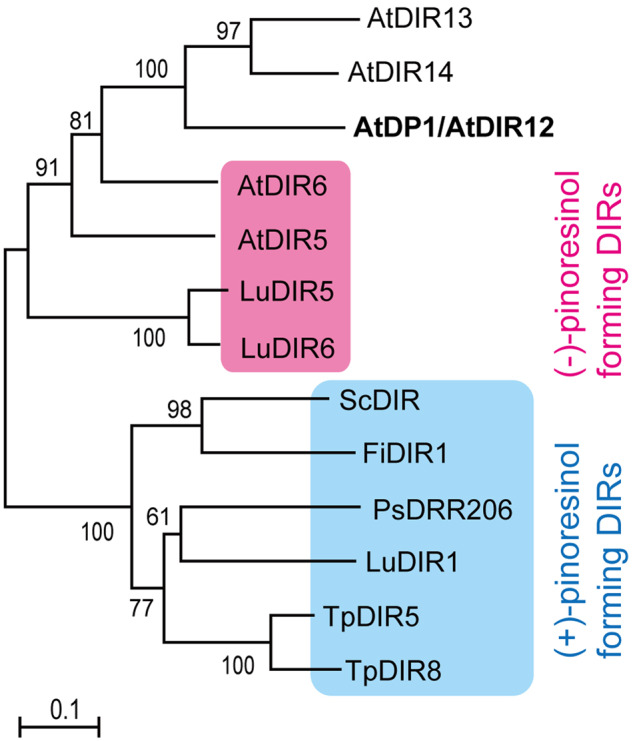
Nonrooted molecular phylogenetic tree of functionally identified DIRs. (+)- and (−)-pinoresinol-forming DIRs are shaded by blue and magenta, respectively. The tree was constructed as described in the Materials and methods section. The alignment used for this analysis is shown in Supplemental Figure 2. Bar = 0.1 amino acid substitutions per site. The GenBank accession numbers for the sequences are shown in parentheses: AtDP1/AtDIR12 (At4g11180, BT004016); AtDIR5 (At1g64160, BT010486); AtDIR6 (At4g23690, BT002439); AtDIR13 (At4g11190, BT015628); AtDIR14 (At4g11210, BT015626); FiDIR1 (AF210061); LuDIR1 (KM433751); LuDIR5 (KM433753); LuDIR6 (KM433752); PsDRR206 (U11716); ScDIR (HQ428029); TpDIR5 (AF210067); TpDIR8 (AF210070). At, Arabidopsis thaliana; Fi, Forsythia intermedia; Lu, Linum usitatissimum; Ps, Pisum sativum; Sc, Schizandra chinensis; Tp, Thuja plicata.
Structural comparison of AtDP1/AtDIR12 and DIRs for lignan biosynthesis
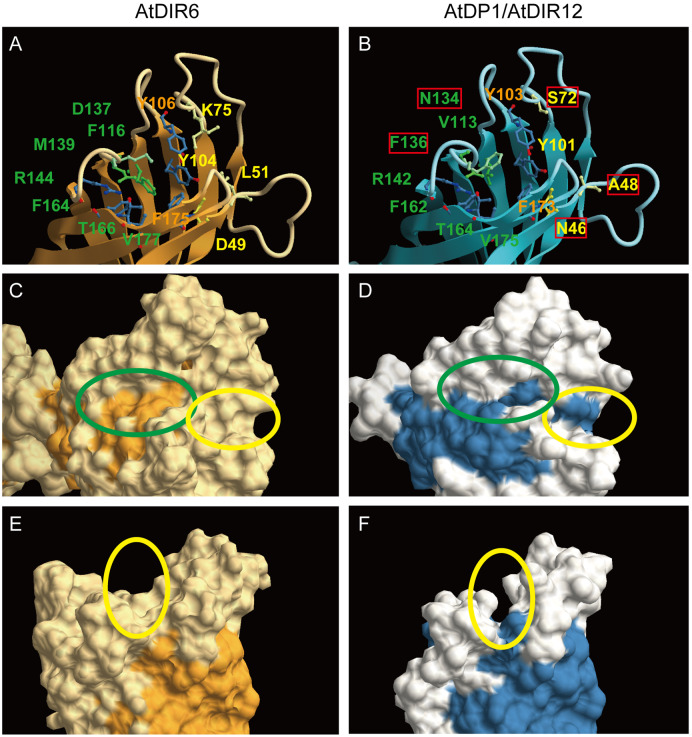
Multiple alignments of DIRs suggest the amino acids in AtDP1/AtDIR12 that may partly contribute to catalysis for neolignan biosynthesis. Several amino acid residues unique to AtDP1/AtDIR12 but differently conserved in (+)- and/or (−)-pinoresinol-forming DIRs (Asn46, Ile79, Asn80, Asn104, Val113, Trp115, Asp132, Asn134, Leu154, Lys165, and Cys174) were found (Supplemental Figure 2). Of the five polar residues (corresponding to His39, Asp40, Thr84, Ser91, and Arg141 in PsDRR206) that are highly conserved in both (+)- and (−)-pinoresinol-forming DIRs and lined the edge of the putative substrate binding sites (Kim et al., 2015), four residues (His45, Thr84, Ser91, and Arg142 in AtDP1/AtDIR12) are also conserved, but Asp40 (Asn46 in AtDP1/AtDIR12) is unique to AtDP1/AtDIR12 (Supplemental Figure 2). Crystal structures of a (−)-pinoresinol-forming AtDIR6 showed that active sites are formed by two lobes (pockets A and B; Gasper et al., 2016). Seven of the 13 amino acid residues forming pocket A (Phe116, Asp137, Met139, Arg144, Phe164, Thr166, and Val177), pocket B (Asp49, Leu51, Lys75, and Tyr104), and a region between the pockets (Tyr106 and Phe175) are conserved in AtDP1/AtDIR12 (Gasper et al., 2016; Paniagua et al., 2017; Figure 7). However, two of the six residues proposed to be essential for the activity through hydrogen bond formation or acid catalysis in AtDIR6 (Asp137 and Asp49 of Asp137, Arg144, Thr166, Asp49, Tyr104, and Tyr106) are altered to Asn134 and Asn46 in AtDP1/AtDIR12, respectively. Homology modeling of AtDP1/AtDIR12 based on the AtDIR6 structure showed that substitution of the residues in pocket A (Phe116, Asp137, and Met139 in AtDIR6 to Val113, Asn134, and Phe136 in AtDP1) and pocket B (Asp49, Leu51, and Lys75 in AtDIR6 to Asn46, Ala48, and Ser72 in AtDP1/AtDIR12) conferred a larger and deeper cavity for catalysis (Figure 7; Gasper et al., 2016; Paniagua et al., 2017). Substitutions to Ser72 and Ala48 in AtDP1/AtDIR12 also contribute to slit formation between pockets A and B (Figure 7, E and F). These results may reflect the ability of AtDP1/AtDIR12 to accept bulkier substrates than AtDIR5 and AtDIR6.
Figure 7.
Homology modeling of AtDP1/AtDIR12 based on the crystal structure of AtDIR6. Active site of AtDIR6 (A) and AtDP1/AtDIR12 (B). Amino acids residues for pocket A (green), pocket B (yellow), and the region between the pockets (orange) are shown. Residues unique to AtDP1/AtDIR12 are shown in red boxes. Molecular surface structures of AtDIR6 (C, E) and AtDP1/AtDIR12 (D, F) are shown. Pockets A and B are shown within green and yellow ovals, respectively. Homology modeling was conducted using the SWISS-MODEL Web site (https://swissmodel.expasy.org/) with AtDIR6 (PDB: 5LAL) as a template. CueMol2 was used for the macromolecular structure visualization (http://www.cuemol.org/en/).
AtDP1/AtDIR12 is expressed in the outer integument cells of seeds
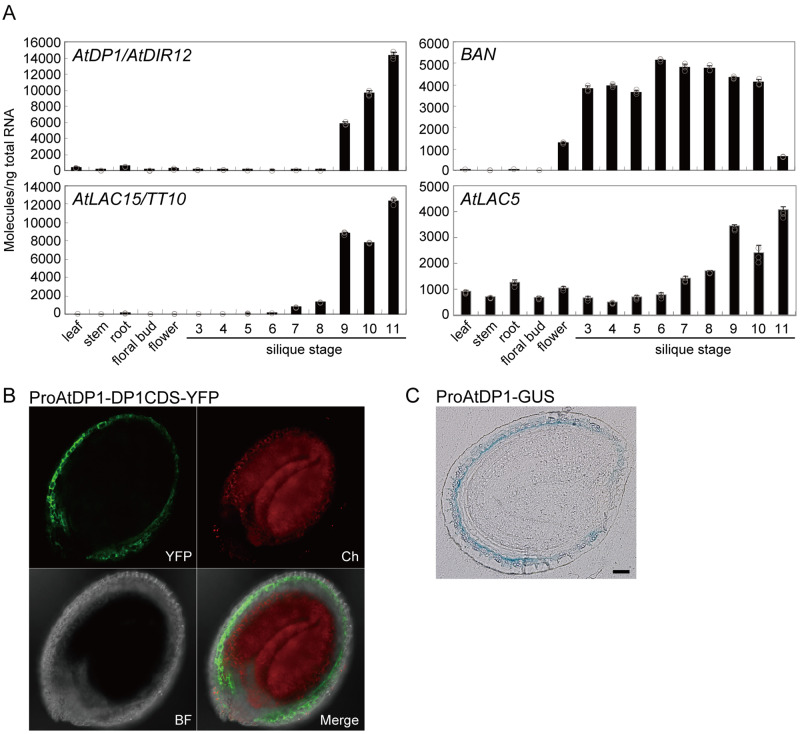
The accumulation pattern of AtDP1/AtDIR12 transcripts in various organs including seeds at nine developmental stages was investigated by reverse transcription quantitative PCR (RT-qPCR). AtDP1/AtDIR12 transcripts predominantly accumulated in seeds at developmental stages 9 to 11, but not in other organs (Figure 8A). These data are consistent with the seed-specific accumulation of erythro-SC(4-O-8)G (1e) and the expression profile of AtDP1/AtDIR12 archived on the Arabidopsis eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). The expression profile of anthocyanidin reductase encoded by BANYULS (BAN), a key enzyme in the proanthocyanidin (PA) biosynthetic pathway, was clearly distinct from that of AtDP1/AtDIR12 during seed development. Compared to AtDP1/AtDIR12, BAN transcripts accumulated in seeds across a broader range of developmental stages (stages 3–10; Figure 8A), suggesting that the two phenylpropanoid-derivative biosynthetic pathways for neolignan and PA are distinctively regulated at the transcriptional level in Arabidopsis seeds.
Figure 8.
Expression pattern of AtDP1/AtDIR12. (A) Transcript abundance of AtDP1/AtDIR12, BAN, AtLAC15/TT10, and AtLAC5 in organs of Arabidopsis wild type (Col-0). Error bars represent sd of three technical replicates per sample. BAN, anthocyanin reductase. (B) Confocal microscopy images depicting the localization of YFP in transgenic plants transformed with a 720-bp promoter of AtDP1 fused to AtDP1 cDNA with YFP (ProAtDP1-AtDP1CDS-YFP). YFP fluorescence (YFP); chlorophyll autofluorescence (Ch); a bright-field image (BF) and a merged image of the three channels (merge) are shown. Seed stage, 7 days after flowering (corresponding to silique stage 11). (C) Sections (5 µm) of GUS-stained seeds expressing the GUS reporter gene driven by the AtDP1 promoter. Seed stage, 7 days after flowering (corresponding to silique stage 11). Scale bars = 50 µm.
To assess the cellular localization patterns of AtDP1/AtDIR12 in developing seeds, transgenic plants harboring a 720-bp promoter region of AtDP1/AtDIR12 fused to AtDP1/AtDIR12 cDNA with yellow fluorescent protein (YFP; ProAtDP1-AtDP1CDS-YFP) were generated. The YFP fluorescence was specifically observed in the outer integument 1 (oi1) of seeds expressing ProAtDP1-AtDP1CDS-YFP (Figure 8B). Additionally, we generated transgenic plants harboring the 720-bp promoter of AtDP1 fused to GUS (ProAtDP1-GUS). The blue coloration from GUS staining was observed in seed oi1 cells of plants expressing ProAtDP1-GUS (Figure 8C). These data indicate that AtDP1/AtDIR12 localized in oi1 cells of developing seeds in which flavonols also accumulated (Pourcel et al., 2005). TargetP-2.0 (http://www.cbs.dtu.dk/services/TargetP/) predicts the presence of an N-terminal signal peptide in AtDP1/AtDIR12 (likelihood 0.9957), suggesting that AtDP1/AtDIR12 may sort to compartments in the secretory pathway.
Oxidases for neolignan biosynthesis
Next, we focused on identifying oxidases required for the formation of the monolignol radicals. LACs and/or peroxidases have been suggested to generate the monolignol radicals for lignan/neolignan; however, no specific genes for neolignans were known. Davin et al. reported that a LAC-like oxidase mediated the formation of enantioselective (+)-pinoresinol in the presence of Forsythia DIR (Davin et al., 1997; Gang et al., 1999). Therefore, we gave priority to LACs. We hypothesized that the target LAC(s) would need to be simultaneously expressed with AtDP1/AtDIR12 to perform the function. Among 17 Arabidopsis LACs, positive correlations (r > 0.3) of coordinated expression were noted between AtDP1/AtDIR12 and two LACs, i.e. AtLAC15/TRANSPARENT TESTA 10 (TT10; At5g48100, r = 0.708) and AtLAC5 (At2g40370, r = 0.399). AtLAC15/TT10 and AtLAC5 are major LACs expressed in seeds (Schmid et al., 2005). RT-qPCR showed that transcripts of AtLAC15/TT10 and AtLAC5 are abundant in seeds at developmental stages 9 to 11 as with AtDP1/AtDIR12. The abundance of AtLAC5 transcripts was relatively low compared to those of AtDP1/AtDIR12 and AtLAC15/TT10 (Figure 8A).
In Arabidopsis, AtLAC15/TT10 is involved in the oxidative polymerization of flavonoids/PAs and lignins (Pourcel et al., 2005; Liang et al., 2006). To date, several plant LACs have been functionally identified as LACs that polymerize lignin (AtLAC4, AtLAC11, and AtLAC17; Berthet et al., 2011; Zhao et al., 2013; Tobimatsu and Schuetz, 2019), PAs and/or phenolic compounds (BnTT10, GaLAC1, PtLAC3, SofLAC, and ZmLAC3; Ranocha et al., 2002; Caparros-Ruiz et al., 2006; Wang et al., 2008; Cesarino et al., 2013; Zhang et al., 2013). Phylogenetic comparison of the 17 AtLACs with functionally identified LACs from other plants showed that AtLAC15/TT10 belongs to a cluster that includes BnTT10, ZmLAC3, and GaLAC1. AtLAC5 belongs to another cluster with AtLACs of unknown function that includes AtLAC3, AtLAC12, and AtLAC13 (Figure 9; Supplemental Figure 3).
Figure 9.
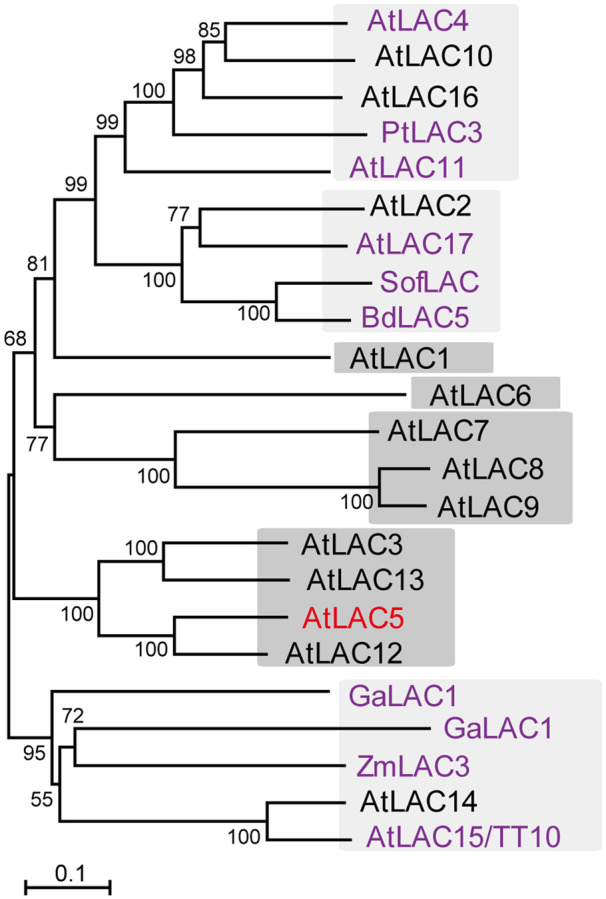
Nonrooted molecular phylogenetic tree of Arabidopsis LACs and functionally identified LACs. Functionally identified LACs are shown in purple. The clusters with unknown LACs only are shown with a dark grey background. The tree was constructed as described in Materials and methods section. Bar = 0.1 amino acid substitutions per site. The GenBank accession numbers for the sequences are shown in parentheses: AtLAC1 (At1g18140, NP_173252); AtLAC2 (At2g29130, NP_180477); AtLAC3 (At2g30210, NP_180580); AtLAC4 (At2g38380, NP_565881); AtLAC5 (At2g40370, NP_181568); AtLAC6 (At46570, NP_182180); AtLAC7 (At3g09220, NP_187533); AtLAC8 (At5g01040, NP_195724); AtLAC9 (At5g01050, NP_195725); AtLAC10 (At5g01190, NP_195739); AtLAC11 (At5g03260, NP_195946); AtLAC12 (At5g05390, NP_196158); AtLAC13 (At5g07130, NP_196330); AtLAC14 (At5g09360, NP_196498); AtLAC15 (At5g48100, NP_199621); AtLAC16 (At5g58910, NP_200699); AtLAC17 (At5g60020, NP_200810); BdLAC5 (Bradi1g66720, XP_003558240); BnTT10 (NP_001302959); GaLAC1 (NP_001316972); PtLAC3 (CAC14719); SofLAC (SCVPRZ3027A08.g and SCUTST3084C11.g); ZmLAC3 (CAJ30499). At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Bn, Brassica napus; Ga, Gossypium arboreum; Pt, Populus trichocarpa; Sof, Saccharum officinarum; Zm, Zea mays. The alignment used for this analysis is shown in Supplemental Figure 3.
To elucidate the function of AtLAC15/TT10 and AtLAC5, T-DNA inserted knockout lines for AtLAC15/TT10, SALK_002972 (tt10-4), and SALK_128292 (tt10-5; Pang et al., 2013) and those for AtLAC5, SALK_092440 (lac5-2), and SALK_093534 (lac5-3), were isolated. No transcripts of AtLAC5 in the lac5-2 and lac5-3 lines were found by RT-PCR analysis (Supplemental Figure 4).
The analyses of neolignans, and SC and FC derivatives showed that the abundance of FC-conjugated neolignans [erythro-FC(4-O-8)G (4e), FC(5-8)G (6), and FC(5-8)G 4-O-Hex (7; Figure 1B; Supplemental Figure 1)] in the lac5 mutants was as low as those in the atdp1-1 mutant (Figure 5; Supplemental Table 1, Supplemental Data Set 2). Interestingly, the abundance of erythro-SC(4-O-8)G (1e) in the lac5 mutants may be slightly lower than that in wild type, but the accumulation level was much higher than that in the atdp1 mutant. Neolignans in tt10 mutants showed no significant changes in abundance. The content of choline derivatives (8–16, Figure 5) was similar in wild type, and the lac5 and tt10 mutants. These data suggested that AtLAC5 is predominantly involved in the formation of FC-conjugated neolignans, despite the linkage types between monolignol and choline derivatives, but is not required for SC-conjugated neolignan biosynthesis.
Localization of AtLAC5 in seeds
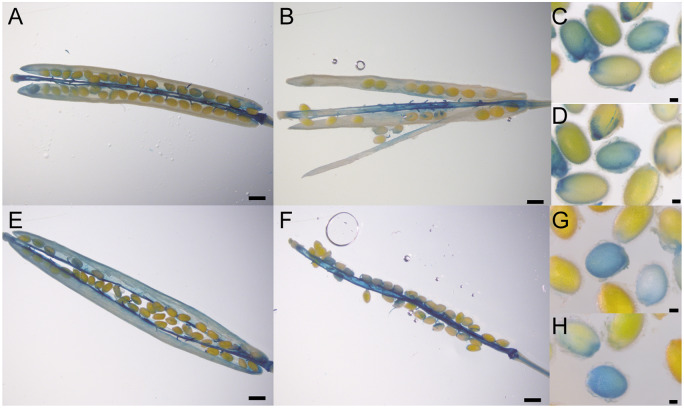
Experiments using the AtLAC5 promoter (2,358 bp)-GUS fusion construct suggested that AtLAC5 is localized in the replum and abscission zone in siliques and seed embryos (Turlapati et al., 2011). As the AtLAC5 promoter region was estimated to be 4,140 bp (Turlapati et al., 2011), we transformed plants with constructs containing either the 4,140-bp or a 2,000-bp promoter region of AtLAC5 fused to GUS (ProAtLAC5-4140-GUS, ProAtLAC5-2000-GUS). The blue coloration by GUS staining was predominantly observed in the replum, septum, and funiculus and to a lesser extent in the valves of transgenic plants, using either promoter (Figure 10, A, B, E, and F). Staining of the seeds was also observed but it was not clear (Figure 10, C, D, G, and H).
Figure 10.
Expression pattern of AtLAC5. GUS staining of transgenic plants harboring the 4,140-bp (A–D) or 2,000-bp (E–H) promoter region of AtLAC5 fused to GUS. Siliques at 7 (B), 8 (F), 9 (A and E) days after flowering and immature seeds at 10 days after flowering (C, D, G, and H) were used for the analyses. Scale bars=0.64 mm (A, B, E, and F) or 0.1 mm (C, D, G, and H).
TargetP-2.0 (http://www.cbs.dtu.dk/services/TargetP/) predicts the presence of an N-terminal signal peptide in AtLAC5 (likelihood 0.9998), suggesting that AtLAC5 may sort to compartments in the secretory pathway as is also the case for AtDP1/AtDIR12. Transcriptome data obtained from the Arabidopsis eFP browser showed that AtLAC5 transcripts accumulate exclusively in seeds and particularly in seed coats of mature green seeds (Supplemental Figure 5). The Arabidopsis Seed Coat eFP Browser showed that the accumulation level of AtLAC5 transcripts in ap2-7 mutants is lower compared to wild-type (Supplemental Figure 5). AP2 is a transcription factor required for differentiation of the epidermal and subepidermal palisade layers of the outer integument corresponding to the oi2 and oi1 cell layers, respectively (Western et al., 2001; Dean et al., 2011). These data suggest that AtLAC5 is expressed in the outer integument of seeds. The accumulation level of AtDP1 transcripts in the ap2-7 mutants is also lower than that of wild-type (Supplemental Figure 5). The Arabidopsis Seed Coat eFP browser also suggests that AtDP1/AtDIR12 is expressed earlier than AtLAC5, an observation that is somewhat contradictory to our RT-qPCR data showing that the expression of AtLAC5 is similar to that of AtDP1/AtDIR12 (Figure 8). AtDP1/AtDIR12 may be ready for precise radical coupling prior to the appearance of AtLAC5 or tissue dissection may trigger additional AtDP1/AtDIR12 expression.
The biosynthetic pathway for SC/FC-conjugated neolignans
To date, there has been almost no information about the biosynthetic pathway for SC- and FC-conjugated neolignans. As a possible route to synthesize SC- and FC-conjugated neolignans, it is conceivable that choline derivatives (SC and FC) may directly couple with CA via 8-O-4′ or 8-5′ couplings (Figure 11). SC is synthesized by sinapoylglucose: choline sinapoyltransferase (SCT), SCT/SCPL19 (At5g09640), an enzyme that transfers a sinapoyl moiety from 1-O-sinapoyl-β-d-glucose to choline (Shirley et al., 2001). SCT/SCPL19 also has a feruloylglucose: choline feruloyltransferase activity (Shirley and Chapple, 2003). A mutant defective in the SCT/SCPL19 locus was designated as sinapoylglucose accumulator 2 (sng2). SC levels are decreased and levels of 1-O-sinapoyl-β-d-glucose and choline are increased in ethyl methanesulfonate-induced sng2 mutant seeds (Shirley et al., 2001). We isolated two homozygous sng2 T-DNA insertion mutants, sng2-3 (SALK_053495C) and sng2-4 (SALK_018120C) to compare with a known sng2-2 mutant (SALK_002255C; Lee et al., 2012; Figure 12; Supplemental Data Set 3). The T-DNA in sng2-3 and sng2-4 was inserted in the exon of SNG2 but between positions +787 to +808 and +1884 to +1894 base pairs, respectively, from the start codon of SCT/SCPL19. The analyses of neolignans and choline derivatives showed that SC- and FC-conjugated neolignans were clearly missing in the three sng2 mutant seeds as with SC and FC (Figure 12; Supplemental Data Set 3). On the other hand, the accumulation level of benzoylcholine in the three sng2 mutants was similar to that in Col-0. This finding is consistent with the previously published data using the sng2-2 mutant (Lee et al., 2012). Collectively, these data support our notions that SCT/SCPL19 also functions as a feruloylglucose: choline feruloyltransferase in planta, and SC/FC are coupled to CA to yield the SC- and FC-conjugated neolignans (Figure 11).
Figure 11.
The proposed biosynthetic pathways of neolignans, lignans, and lignins in Arabidopsis. Asterisks indicate the reader’s convenience, each phenolic radical is depicted with its resonance form chosen to allow the radical coupling, 8-O-4′ and 8-5′, to be rationalized; the two FC● structures shown, for example, are simply two resonance forms of the same phenolic radical. DIR, dirigent protein; laccase, LAC; PRX, peroxidase; PLR, pinoresinol/lariciresinol reductase; PrR, pinoresinol reductase; G, guaiacyl moiety; SC, sinapoylcholine; FC, feruloylcholine; CA, coniferyl alcohol.
Figure 12.
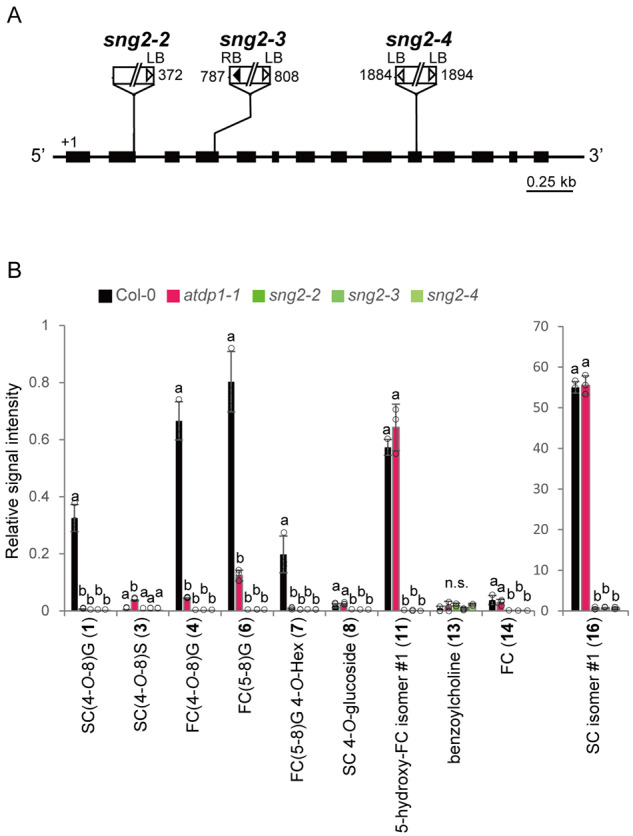
T-DNA insertion mutants of SCT, sng2. (A) Schematic representation of SCT showing the three T-DNA insertion mutants used in this study. The thick line indicates the coding region and the thinner line indicates the 5′- and 3′-untranslated regions. White and black triangles show the left and right borders, respectively. Numbers indicate the position of the T-DNA insertion. LB, left border; RB, right border. (B) UPLC–QTOF–MS analyses of neolignans and choline derivatives in the mutants. The structures of compounds (1, 3, 4, 6–8, 11, 13, 14, 16) are shown in Figure 1. Data represent the means ± sd (three biological replicates per sample). The means were compared by a one-way ANOVA. Statistically significant differences (P < 0.05) were identified by Tukey’s test and are indicated by lowercase letters to represent differences between groups. G, Guaiacyl moiety; S, Syringyl moiety; SC, sinapoylcholine; FC, feruloylcholine; Hex, hexose; n.s., not significant.
Neolignans affect seed coat permeability
Phenolic compounds such as flavonoids are known to be antioxidants that can prevent the generation of free radicals and subsequent oxidative damage to cells (Sano et al., 2016). Arabidopsis transparent testa mutants that are unable to synthesize flavonoids, including PAs, showed a significant increase in seed coat permeability and reduced germination capacity during long-term seed storage (Debeaujon et al., 2000).
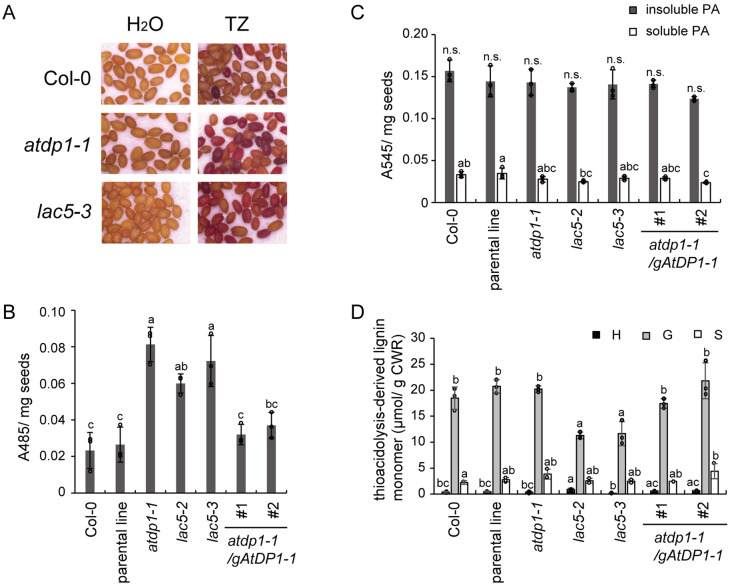
To investigate the effect of decreasing the neolignan content of seeds, we tested the permeability of seed coats by the tetrazolium penetration assay (Berridge et al., 1996; Vishwanath et al., 2013, 2014). Tetrazolium salts are colorless compounds, but they are converted to red products called formazans by endogenous NADH-dependent reductases after they penetrate the seed coat (Berridge et al., 1996). The intensity of red coloration is directly proportional to the seed coat permeability (Vishwanath et al., 2014). Quantitative analyses by extraction of the formazans in 95% ethanol showed that accumulation of formazans in atdp1-1 was the highest and that accumulation in lac5-2, lac5-3, wild-type, and the parental line was 73.7% ± 6.4%, 88.8% ± 17.2%, 28.7% ± 12.2%, 32.5% ± 11.8% of atdp1-1, respectively (Figure 13B; Supplemental Data Set 4). The permeability of the atdp1-1 mutant complemented with genomic AtDP1/AtDIR12 was similar to that of the wild-type.
Figure 13.
Seed coat permeability of atdp1 and lac5 mutants. Seed coat permeability was tested by measuring the conversion of tetrazolium salts to red products called formazans. (A) Seeds of wild type (Col-0) and the mutants (atdp1-1 and lac5-3) stained in a tetrazolium salt (TZ) solution or H2O after 48 h incubation. (B) The absorbance of ethanol extracts containing formazan from the seeds stained with tetrazolium salts at 485 nm. The intensity of A485 is directly proportional to the permeability of the seed coat. (C) Soluble and insoluble proanthocyanidins were analyzed by acid hydrolysis as described in Materials and methods section. No significant difference (P < 0.05) was detected for the insoluble proanthocyanidins. (D) The yield of thioacidolysis-derived p-hydroxyphenyl (H)-, guaiacyl (G)-, and syringyl (S)-type trithioethylpropane monomers released from H-, G-, and S-type lignin polymer units. Data represent the means ± sd (three biological replicates per sample). The means were compared by a one-way ANOVA. Statistically significant differences (P < 0.05) were identified by Tukey’s test and are indicated by lowercase letters to represent differences between groups. n.s., not significant; CWR, cell wall residue.
We also analyzed the PAs and lignin content to test the possibility that loss of function of AtDP1/AtDIR12 or AtLAC5 might also affect the levels of these compounds, thereby resulting in higher seed coat permeability in the mutants. The PA content as determined by the butanol-HCl assay (Tohge et al., 2005; Kitamura et al., 2010) showed no significant difference between Col-0 and the atdp1 and lac5 mutants in which the neolignan content was substantially lower than wild type (Figure 13C; Supplemental Data Set 4). The relative lignin content of Col-0 and the mutants was evaluated by the yield of lignin monomers released by analytical thioacidolysis, a reaction that cleaves the quantifiable lignin monomers by chemical cleavage of the major β-O-4 linkages in lignin polymers (Lapierre et al., 1986). Consequently, we detected no lignin reduction in atdp1-1, supporting our notion that the decrease in the neolignan content primarily led to the high permeability of the seed coat in this mutant (Figure 13D; Supplemental Data Set 4). Additionally, the two lac5 mutants (lac5-2 and lac5-3), also had reduced yields of guaiacyl-type lignin monomers, whereas yields of syringyl-type lignin monomers in the lac5 mutants were similar to those of wild type (Figure 13D; Supplemental Data Set 4). This result suggests that AtLAC5 may be involved not only in FC-conjugated neolignan biosynthesis but also in guaiacyl-type lignin biosynthesis in Arabidopsis seeds. This result supports the hypothesis that the high permeability of the lac5 seed coats may be attributed to a reduced content of both neolignan and lignin.
Discussion
AtDP1/AtDIR12 is essential for neolignan biosynthesis
Our results demonstrated that AtDP1/AtDIR12 is essential for the biosynthesis of SC- and FC-conjugated 8-O-4′-type neolignans in Arabidopsis (Figures 3, 5). Although the absolute configurations of the AtDP1/AtDIR12-derived neolignans 1e and 4e (Figure 1C), i.e. whether they have (8S, 7R) and/or (7S, 8R) configurations, are yet to be determined, it is plausible that AtDP1/AtDIR12 guides the regio- and stereo-selective 8-O-4′ coupling of CA with SC and FC to form optically active products. Intriguingly, the wild-type seeds appeared to accumulate the erythro isomers 1e and 4e exclusively with no detectable amounts of the corresponding threo isomers 1t and 4t (Figure 4; Supplemental Figure 1). This observation strongly suggests that AtDP1/AtDIR12 may guide not only the radical coupling step but also that in the subsequent rearomatization step in which the quinone methide intermediates are attacked by water during the formation of neolignans 1e and 4e. This intriguing stereochemical mechanism, including which enantiomer of erythro compounds is accumulated and how these reactions are mediated by AtDP1/AtDIR12, should be further addressed together with a detailed three-dimensional protein structure.
To date, several DIRs involved in the stereoselective (enantioselective) 8-8′ coupling of CA to produce lignan, (+) or (−)-pinoresinol, have been identified (Davin et al., 1997; Kim et al., 2002, 2012, 2015; Ralph et al., 2007; Pickel et al., 2010; Dalisay et al., 2015), but there have been no reports of DIRs involved in the 8-O-4′ couplings of monolignols or their analogs for neolignan biosynthesis. Of the six DIR subfamilies, AtDP1/AtDIR12 belongs to the DIR-a family with other DIRs involved in lignan biosynthesis. Phylogenetic analysis of functionally identified DIR-a genes revealed that DIR-a can be classified into two clusters based on their stereoselectivity, (+)- and (−)-pinoresinol-forming DIRs. AtDP1/AtDIR12, AtDIR13, and AtDIR14 belong to a cluster of (−)-pinoresinol-forming DIRs that include AtDIR6 and AtDIR5 (Figure 6). Evolutionary relationships of the Arabidopsis DIR-a genes proposed by Wang et al. (2013) suggest that AtDIR5 was an ancestral gene and AtDP1/AtDIR12 and AtDIR6 were derived from AtDIR5 by the whole-genome duplication event β or AtDP1/AtDIR12 was derived from AtDIR6 by the whole-genome duplication event α (Supplemental Figure 6; Wang et al., 2013). These data suggest that AtDP1/AtDIR12 may have diverged from (−)-pinoresinol-forming DIRs.
The neolignan biosynthetic pathway in Arabidopsis
Our results using the sng2 mutants that are defective in the ability to synthesize SC revealed that SC- and FC-conjugated neolignans are synthesized from SC/FC and CA, and AtDP1/AtDIR12 is essential for producing the 8-O-4′-type neolignans, erythro-SC(4-O-8)G (1e) and erythro-FC(4-O-8)G (4e), in Arabidopsis seeds. Notably, in seeds of the atdp1-1 mutant, the abundance of other neolignans putatively annotated as FC-monolignol conjugate FC(5-8)G (6) and its glycoside FC(5-8)G 4-O-Hex (7; Figure 1B) were also coordinately reduced (Figure 5), suggesting that AtDP1/AtDIR12 may be also able to guide the 8-5′-coupling of monolignol radicals with FC radicals (Figure 11).
Arabidopsis contains derivatives of lignans, neolignans, and oligolignols in leaves/shoots and lignan derivatives in roots (Nakatsubo et al., 2008; Okazawa et al., 2011; Dima et al., 2015). In leaves, 8-O-4′- and 8-5′-type neolignans composed of ferulic acid/feruloyl malate/sinapoyl malate and monolignols have been identified (Dima et al., 2015). In the inflorescence stem of Arabidopsis, a pathway to produce feruloylmalate(4-O-8)G from ferulic acid and CA via ferulic acid(4-O-8)G and feruloylhexose(4-O-8)G has been suggested (Vanholme et al., 2012). Given that AtDP1/AtDIR12 is specifically expressed in seeds, other DIR(s) may guide 8-O-4′ and/or 8-5′ radical coupling of monolignols and sinapoyl/feruloyl derivatives in organs other than seeds. The enzyme responsible for the final step in the synthesis of sinapoylmalate, sinapoylglucose:malate SMT, was identified in Arabidopsis previously (Lehfeldt et al., 2000). Future reciprocal studies focusing on chemical characterization and genetic identification of other AtDIRs and the neolignan analyses of the smt mutants will further elucidate the neolignan biosynthetic pathway in Arabidopsis.
In addition, a variety of (neo)lignans, including aglycones and glycosides, are reportedly stored in Arabidopsis leaf vacuoles (Dima et al., 2015). On the other hand, in flaxseed (Linum usitatissimum), lignans are mainly localized in the secondary walls of sclerite cells in the seed outer integument (Attoumbre et al., 2010). AtDP1/AtDIR12, AtLAC5, and SCT/SCPL19 that are all involved in seed neolignan biosynthesis were predicted to have signal peptides, suggesting that these proteins may be secreted and sorted into compartments such as vacuoles, tonoplasts, plasma membranes, and cell walls by the secretory pathway; however, the intracellular localization patterns of AtDP1/AtDIR12, AtLAC5, and SCT/SCPL19 are unknown. All three flaxseed DIRs involved in lignan biosynthesis, LuDIR1, LuDIR5, and LuDIR6, also have signal peptides (Corbin et al., 2018). In Forsythia intermedia stems, DIR(s) are localized to the cell wall (Burlat et al., 2001). Arabidopsis LACs for lignin biosynthesis, AtLAC4 and AtLAC17, are reportedly localized to secondary cell walls (Schuetz et al., 2014), and AtLAC15/TT10 for proanthocyanidin biosynthesis is localized to vacuoles (Pang et al., 2013). Serine carboxypeptidase-like (SCPL) proteins are known to localize in vacuoles (Hause et al., 2002; Mugford et al., 2013). Further investigation is required to elucidate the intracellular localization of AtDP1/AtDIR12, AtLAC5, and SCT/SCPL19 and the sites for neolignan biosynthesis and storage.
AtLAC5 is required for the biosynthesis of FC-conjugated neolignans
Peroxidases and/or LACs have been reported to be probably involved in monolignol oxidation in lignin and lignan biosynthesis, but no oxidases for neolignan biosynthesis have been identified. Here, we revealed that AtLAC5 is also essential for neolignan biosynthesis. Interestingly, AtLAC5 is involved in the synthesis of the FC-conjugated 8-O-4′-type as well as the 8-5′-type neolignans but is not involved in the synthesis of SC-conjugated neolignans (Figure 5). This finding suggests that AtLAC5 is responsible for the oxidation of FC but not for SC; other oxidases may oxidize SC to form SC-conjugated neolignans with CA. LACs are frequently able to catalyze the oxidation of a wide range of substrates in vitro (Sterjiades et al., 1992; Bao et al., 1993); however, the substrate specificity may be more strict in vivo. A seed coat-specific LAC from Cleome hassleriana, ChLAC8, is capable of oxidizing caffeoyl alcohol and sinapyl alcohol but not CA (Wang et al., 2020). This finding is consistent with our hypothesis.
In addition, our lignin analysis data based on thioacidolysis suggested that AtLAC5 is also involved in the oxidation of CA to form guaiacyl-type lignins but is not involved in the oxidation of sinapyl alcohol for syringyl-type lignins in Arabidopsis seed coats (Figure 13D). These data suggest that AtLAC5 preferentially oxidizes guaiacyl-type substrates, e.g. FC and CA, over syringyl-type substrates, e.g. SC and sinapyl alcohol, although further biochemical studies are needed to support this hypothesis.
The in vitro assays using developing seeds showed that tt10 mutant seeds had a reduced polymerization activity to produce possible 8-O-4′, 8-8′, and/or 8-5′ dimers of CA compared to wild-type seeds (Pourcel et al., 2005; Liang et al., 2006). TT10 is localized in oi1 cells (Pourcel et al., 2005); however, no significant changes in the neolignan content were observed in the tt10 (atlac15) mutants. Our results indicated that TT10/AtLAC15 is not involved in neolignan biosynthesis and suggest that the substrate specificity of TT10/AtLAC15 may be also strict in vivo, or TT10/AtLAC15 may be separated spatially from monolignols in the cells.
Physiological roles of neolignans in Arabidopsis seeds
AtDP1/AtDIR12 is specifically expressed in seeds. Analyses using transgenic plants harboring an AtDP1/AtDIR12 promoter fused to AtDP1/AtDIR12 cDNA with YFP or GUS showed that AtDP1/AtDIR12 is localized in the oi1 cells of developing seeds (Figure 8). These results suggest that AtDP1/AtDIR12-guided neolignans are synthesized in oi1 cells that surround the embryo. The tetrazolium salt assay showed that higher seed coat permeability was observed in the atdp1-1 mutant that is deficient in neolignans (Figure 5) but has comparable accumulation levels of PAs and lignins as the wild type (Figure 13). This result suggests that neolignans play a role in protecting seeds against environmental factors. Furthermore, considering the remarkable biological activities of neolignans and lignans as toxins and deterrents for insects and microorganisms (Nitao et al., 1992; Schroeder et al., 2006; Kulik et al., 2014), neolignans in Arabidopsis seeds may also contribute to chemical defense against herbivores and pathogenic microorganisms by their toxicity.
How neolignans affect seed coat permeability is unknown. Arabidopsis mutants unable to synthesize PAs also had a significant increase in seed coat permeability to tetrazolium salts (Debeaujon et al., 2000). PAs accumulate in the innermost layer of the inner integument (endothelium or inner integument 1 layer; Debeaujon et al., 2003). Seeds are firmly protected by phenolic compounds such as neolignans and PAs in at least two cell layers. These compounds, along with lignin in testa cell walls, may form the barriers to penetration.
Our findings herein shed light on new aspects of DIRs and LACs in neolignan biosynthesis. By regulating the stereoselective radical coupling of phenolic compounds, DIRs and LACs are able to produce structurally diverse, specialized phenolic metabolites for chemical defense.
Materials and methods
Plant materials
Arabidopsis thaliana (accession Columbia-0, Lehle seeds) was used as the wild type in this study. The T-DNA inserted mutants, SAIL_60_D04 (atdp1-1) and SALK_062238 (atdp1-2) for AtDP1/AtDIR12, SALK_002972 (tt10-4) and SALK_128292 (tt10-5) for AtLAC15/TT10, SALK_092440 (lac5-2) and SALK_093534 (lac5-3) for AtLAC5, SALK_002255C (sng2-2), SALK_053495C (sng2-3), and SALK_018120C (sng2-4) for SNG2 were obtained from the Arabidopsis Biological Resource Center (ABRC, https://abrc.osu.edu/). As the parental line of SAIL_60_D04 (atdp1-1), CS8846 was used for comparison. T-DNA insertion lines, SALK_062238 (atdp1-2), SALK_092440 (lac5-2), SALK_093534 (lac5-3), SALK_053495C (sng2-3), and SALK_018120C (sng2-4) were screened by PCR using specific primers for AtDP1/AtDIR12 (At4g11180-6818953f and At4g11180-6819803r), AtLAC5 (At2g40370/886f and At2g40370/1826r), SNG2 (At5g09640_SCPL19_-8F and At5g09640_SCPL19_2519R), and T-DNA (LBa1 and RBa1; Supplemental Table 2). PCR products were sequenced to determine the exact insertion points. RT-qPCR was performed as described previously (Yonekura-Sakakibara et al., 2007) with primers At4g11180-14f and At4g11180-607r for the atdp1 mutants, and At2g40370/875f and At2g40370/1826r for the lac5 mutants (Supplemental Table 2). Homozygous knockout lines of SAIL_60_D04 (atdp1-1), SALK_002972 (tt10-4), SALK_128292 (tt10-5), and SALK_002255C (sng2-2), were isolated as described previously (Matsuda et al., 2010; Lee et al., 2012; Pang et al., 2013). Plants were grown in soil at 22°C under 16/8 h light (40–75 μmol m−2 s−1 of fluorescent lamp)/dark cycle.
Evaluation of T-DNA insertion mutants
For complementation tests, ca. 3.0-kb and 1.9-kb genomic fragments covering 1.8 kb and 0.72 kb of the promoter region, the entire AtDP1/AtDIR12 coding region and 0.6 kb of the 3′-noncoding region were amplified by PCR using primers, gDP1-Rv2 and CACC-gDP1-Fw1 or CACC-gDP1-Fw2 (Supplemental Table 2). Amplified fragments were cloned into the pENTR/D-TOPO vector (Invitrogen) as the entry vector and sequenced to confirm the absence of PCR errors. pGWB1 was used as the destination vector, and the LR reactions for the binary vectors gDP1-1 and gDP1-2 were catalyzed using the Gateway LR ClonaseTMII enzyme mix (Invitrogen). gDP1-1 (pGWB1/3.0-kb AtDP1/AtDIR12 genomic fragment, pSPB3566) and gDP1-2 (pGWB1/1.9 kb AtDP1/AtDIR12 genomic fragment, pSPB3567) were transformed into Agrobacterium tumefaciens strain EHA105, and Arabidopsis plants were transformed by the floral-dip method (Clough and Bent, 1998). T2 plants were selected on half-strength Murashige and Skoog medium containing 50 µM glufosinate ammonium (Sigma cat#45520), 50 mg L−1 kanamycin sulfate, and 50 mg L−1 carbenicillin, sodium salt.
Neolignan analysis using LC–ESI–Q-Tof–MS
The seeds (3.0 mg) were homogenized in 50 volumes of the extraction solvent, 50% (v/v) MeOH with 0.5-mg L−1 lidocaine, and 10-camphor sulfonic acid, using a mixer mill (MM300, Retsch) with zirconia beads for 10 min at 20 Hz. After centrifugation at 15,000g and filtration (HLB µElution plate, Waters), the extracts (3 µL) were applied to a liquid chromatography (LC)–mass spectrometry (MS) system with an electrospray ionization (ESI) interface (LC, Waters Aquity UPLC system; MS, Waters Q-Tof Premier). For analyses of the putative neolignan compounds (m/z = 668 or 506), LC system conditions (positive analysis) were as described previously (Matsuda et al., 2009).
For analyses of neolignans and choline derivatives, the dried samples were extracted with 50 µL of 80% (v/v) MeOH containing 2.5 mM lidocaine and 10-camphor sulfonic acid per mg dry weight using a mixer mill with zirconia beads for 7 min at 18 Hz and 4°C. After centrifugation for 10 min, the supernatant fraction was filtered using an HLB µElution plate (Waters). The extracts (1 µL) were analyzed using an LC–Q-Tof–MS (LC, Waters Acquity UPLC system; MS, Waters Xevo G2 Q-Tof). Analytical conditions were as follows: LC column, Acquity bridged ethyl hybrid (BEH) C18 (1.7 µm, 2.1 mm × 100 mm, Waters); solvent system, solvent A [water including 0.1% (v/v) formic acid] and solvent B [acetonitrile including 0.1% (v/v) formic acid]; gradient program, 99.5%A/0.5%B at 0 min, 99.5%A/0.5%B at 0.1 min, 75%A/25%B at 10 min, 0.5%A/99.5%B at 10.1 min, 0.5%A/99.5%B at 12.0 min, 99.5%A/0.5%B at 12.1 min, and 99.5%A/0.5%B at 15.0 min; flow rate, 0.3 mL/min at 0 min, 0.3 mL/min at 10 min, 0.4 mL/min at 10.1 min, 0.4 mL/min at 14.4 min, and 0.3 mL/min at 14.5 min; column temperature, 40°C; MS detection: capillary voltage, +3.0 keV, cone voltage, 25.0 V, source temperature, 120°C, desolvation temperature, 450°C, cone gas flow, 50 L/h; desolvation gas flow, 800 L/h; collision energy, 6 V; mass range, m/z 50‒1,500; scan duration, 0.5 s; interscan delay, 0.014 s; data acquisition, centroid mode; polarity, positive; Lockspray (Leucine enkephalin): scan duration, 1.0 s; interscan delay, 0.1 s. MS/MS data were acquired in the ramp mode with the following analytical conditions: (1) MS: mass range, m/z 50–1,500; scan duration, 0.5 s; inter-scan delay, 0.014 s; data acquisition, centroid mode; polarity, positive and (2) MS/MS: mass range, m/z 50–1,500; scan duration, 0.1 s; inter-scan delay, 0.014 s; data acquisition, centroid mode; polarity, positive; collision energy, ramped from 10 to 50 V. In this mode, MS/MS spectra of the top 10 ions (>1,000 counts) in an MS scan were automatically obtained. If the ion intensity was less than 1,000, MS/MS data were not collected and the instrument moved to the next top 10 ions. Original peak intensity values were divided with those of the internal standard (lidocaine: m/z = 235.181, retention time, 6.497 min) to normalize the peak intensity values and were shown as relative peak intensities.
Chemical synthesis of authentic standards for SC(4-O-8)G and FC(4-O-8)G
The authentic, racemic standards for neolignans SC(4-O-8)G (1) and FC(4-O-8)G (4) were synthesized as shown in Figure 1C. The starting compounds, i.e. ethyl sinapate 8SC and ethyl ferulate 8FC were synthesized according to methods described in the literature (Sakakibara et al., 2007), and 1-(4-acetoxy-3-methoxyphenyl)-1-ethanone was prepared from acetovanillone using standard acetylation procedures with pyridine and acetic anhydride. Other chemicals were purchased from Wako Pure Chemical (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan) and were used as received. Flash chromatography was performed with Redi Sep Rf silica cartridges on a Combi Flash Rf system (Teledyne ISCO, Lincoln, NE, USA) using an ethyl acetate (EtOAc)/hexane gradient as the eluent and a UV detector (at 254 nm). Preparative HPLC was performed with an XBridge BEH C18 OBD Prep column (130 Å, 5 mm, 10 mm × 150 mm; Waters Co., Milford, USA) on a Shimadzu 20A HPLC system (Shimadzu Co. Ltd., Kyoto, Japan) using an acetonitrile/water gradient as the eluent and a UV detector (at 254 nm). NMR spectra were recorded on a JEOL JNM-LA400MT FT-NMR system (400 MHz, JEOL, Tokyo, Japan) or a Bruker Biospin AVANCE III 800US system (800 MHz, Bruker Biospin, Billerica, MA, USA). The central solvent peaks were used as internal references (δC/δH, acetone-d6, 29.84/2.04; chloroform-d, 77.00/7.26; methanol-d4, 49.00/3.31). Standard 1D and 2D (COSY, HSQC, and HMBC) NMR experiments were used for structural assignments. Determination of erythro and threo configurations was based on the comparison of NMR chemical shifts and coupling constant data reported for identical or analogous erythro and threo 8-O-4-type dimer models as reported by Ralph and Helm (1991) and Helm and Ralph (1992).
Compound 9
To a stirred solution of 1-(4-acetoxy-3-methoxyphenyl)-1-ethanone (9.6 mmol) in EtOAc (10 mL), pyridinium bromide perbromide (9.6 mmol) was added. After 2 h of stirring at room temperature, the reaction mixture was neutralized with 1 M NaHCO3 on ice and extracted three times with EtOAc over distilled water. The combined organic layer was washed with brine (a saturated aqueous solution of NaCl), dried over Na2SO4, and evaporated in vacuo. The crude product was then purified by flash chromatography to produce a white solid of compound 9 (48.9% yield). 1H-NMR (400 MHz, acetone-d6): δ = 7.74 (1H, d, J 8.8 Hz, H-6), 7.73 (1H, s, H-2), 7.27 (1H, d, J 8.8 Hz, H-5), 4.82 (2H, s, H-8), 3.95 (3H, s, OMe), 2.31 (3H, s, OAc).
Compound 10SC
To a suspension of compound 8SC (3.48 mmol) and K2CO3 (3.48 mmol) in 10 mL acetone, compound 9 (3.87 mmol) in 20 mL acetone was added dropwise at room temperature and then refluxed for 16 h. After cooling to room temperature, the reaction mixture was filtered and added to EtOAc/hexane (1:2, v/v). The organic phase was washed with distilled water and brine, dried over Na2SO4, and evaporated in vacuo to a residue that was purified by flash chromatography to produce a colorless solid of compound 10SC (57.4% yield). 1H-NMR (400 MHz, chloroform-d): δ = 7.70 (1H, s, H-2), 7.63 (1H, d, J 7.2 Hz, H-6) 7.60 (1H, d, J 15.6 Hz, H-7′), 7.13 (1H, d, J 8.0 Hz, H-5), 6.75 (2H, s, H-2′/6′), 6.35 (1H, d, J 15.8 Hz, H-8′), 5.24 (2H, s, H-8), 4.27 (2H, d, J 7.1 Hz, H-10′), 3.90 (3H, s, C-3OMe), 3.84 (6H, s, C-3′/5′OMe), 2.33 (3H, s, OAc), 1.34 (3H, t, J 7.1 Hz, H-11′).
Compound 10FC
Compound 10FC was synthesized from compounds 8FC and 9 using the above described procedure for compound 10SC and was isolated as a colorless solid (73.3% yield). 1H-NMR (400 MHz, chloroform-d): δ = 7.66 (1H, s, H-2), 7.64-7.56 (2H, overlapped, H-6, H-7′), 7.16 (1H, d, J 7.8 Hz, H-5), 7.08 (1H, s, H-2′), 7.04 (1H, d, J 8.8 Hz, H-6′), 6.78 (1H, d, J 8.8 Hz, H-5′), 6.32 (1H, d, J 16.6 Hz, H-8′), 5.37 (1H, s, H-8), 4.26 (2H, q, J 7.1 Hz, H-10′), 3.93 (3H, s, C-3OMe), 3.90 (3H, s, C-3′OMe), 2.35 (3H, s, OAc), 1.34 (3H, t, J 7.1 Hz, H-11′).
Compound 11SC
To a stirred suspension of compound 10SC (1.96 mmol) in 30 mL of dioxane, K2CO3 (14 mmol) and 37% (v/v) formaldehyde (3.92 mmol) were added. After 5 h of stirring at room temperature, the reaction mixture was filtered and concentrated in vacuo to a residue that was purified by flash chromatography to produce a colorless solid of compound 11SC (76.2% yield). 1H-NMR (400 MHz, chloroform-d): δ = 7.72 (1H, s, H-2), 7.67 (1H, d, J 8.8 Hz, H-6), 7.58 (1H, d, J 15.6 Hz, H-7′), 7.12 (1H, d, J 8.0 Hz, H-5), 6.75 (2H, s, H-2′/6′), 6.35 (1H, d, J 15.6 Hz, H-8′), 5.15–5.16 (1H, m, H-8), 4.26 (2H, q, J 7.4 Hz, H-10′), 3.98–4.04 (2H, br m, H-9), 3.89 (3H, s, C-3OMe), 3.76 (6H, s, 3.76, C-3′/5′OMe), 2.34 (3H, s, OAc), 1.33 (3H, t, J 7.4 Hz, H-11′).
Compound 11FC
Compound 11FC was synthesized from compound 10FC using the above described procedure for compound 11SC and was isolated as colorless solid (86.3% yield). 1H-NMR (400 MHz, chloroform-d): δ = 7.70 (1H, d, J 7.8 Hz, H-6), 7.68 (1H, s, H-2), 7.58 (1H, d, J 16.6 Hz, H-7′), 7.14 (1H, d, J 7.8 Hz, H-5), 7.06 (1H, s, H-2′), 7.00 (1H, d, J 8.8Hz, H-6′), 6.75 (1H, d, J 7.8 Hz, H-5′), 6.30 (1H, d, J 16.6 Hz, H-8′), 5.52 (1H, d, J 5.9Hz, H-8), 4.25 (2H, q, J 7.1 Hz, H-10′), 4.08–4.16 (2H, m, H-9), 3.87 (3H, s, C-3OMe), 3.86 (3H, s, C-3′OMe), 2.34 (3H, s, OAc), 1.33 (3H, t, J 7.1 Hz, H-11′).
Compound 12SC
To a solution of compound 12SC (1.45 mmol) in 25 mL ethanol, NaBH4 (10.5 mmol) was added at room temperature. The reaction mixture was stirred under nitrogen gas at room temperature for 5 min, and then the excessive NaBH4 was quenched by adding acetic acid (1 mL). The reaction mixture was concentrated in vacuo and extracted three times with EtOAc (10 mL) over water (10 mL). The combined organic layer was washed with brine, dried over Na2SO4, and evaporated in vacuo to a crude oil that was purified by flash chromatography to produce a colorless solid of compound 12SC as a mixture of erythro and threo isomers (95.8% yield, erythro/threo isomer ratio = 7:3 as determined by 1H-NMR). 12SC (erythro isomer): 1H-NMR (400 MHz, acetone-d6): δ = 7.65 (1H, d, J 15.6 Hz, H-7′), 7.25 (1H, s, H-2), 7.13 (2H, s, H-2′/6′), 7.08–7.11 (1H, br d, H-6), 7.01–7.04 (1H, br d, H-5), 6.53 (1H, d, J 15.6 Hz, H-8′), 5.12 (1H, br m, H-7), 4.29–4.34 (1H, m, H-8), 4.25 (2H, q, J 6.8 Hz, H-10′), 3.96 (6H, s, C-3′/5′OMe), 3.87 (3H, s, C-3OMe), 3.76–3.82 (1H, m, H-9A), 3.49–3.58 (1H, m, H-9B), 2.27 (3H, s, OAc), 1.33 (3H, t, J 6.8 Hz, H-11′). 12SC (threo isomer): 1H-NMR δH (acetone-d6) δ = 7.65 (1H, d, J 15.6 Hz, H-7′), 7.27 (1H, s, H-2), 7.13 (2H, s, H-2′/6′), 7.08–7.11 (1H, br, H-6), 7.01–7.04 (1H, br, H-5), 6.58 (1H, d, J 15.6 Hz, H-8′), 5.16 (1H, br m, H-7), 4.25 (2H, q, J 6.8 Hz, H-10′), 4.16–4.20 (1H, m, H-8), 3.99 (6H, s, C-3′/5′OMe), 3.84 (3H, s, C-3OMe), 3.61–3.70 (1H, m, H-9A), 3.40–3.48 (1H, m, H-9B), 2.27 (3H, s, OAc), 1.33 (3H, t, J 6.8 Hz, H-11′).
Compound 12FC
Compound 12FC was synthesized from 11FC using the above described procedure for compound 12SC and was isolated as a mixture of erythro and threo isomers in the form of a colorless solid (86.3% yield, erythro/threo isomer ratio = 6:4 as determined by 1H-NMR). 12FC (erythro isomer): 1H-NMR (400 MHz, acetone-d6): δ = 7.65 (1H, d, J 15.6 Hz, H-7′), 7.37 (1H, s, H-2′), 7.33 (1H, s, H-2), 7.07–7.25 (overlapped, 4H, H-5, H-6, H-5′, and H-6′), 6.47 (1H, d, J 16.4 Hz, H-8′), 5.07 (1H, m, H-7), 4.49–4.53 (2H, br, H-8), 4.21–4.29 (2H, q, J 6.8 Hz, H-10′), 3.98 (3H, s, C-3′OMe), 3.94 (3H, s, C-3OMe), 3.82–3.90 (1H, m, H-9A), 3.61–3.65 (1H, m, H-9B), 2.27 (3H, s, OAc), 1.33 (3H, t, J 6.8 Hz, H-11′). 12FC (threo isomer): 1H-NMR (400 MHz, acetone-d6): δ = 7.63 (1H, d, J 15.6 Hz, H-7′), 7.41 (1H, s, H-2′), 7.33 (1H, s, H-2), 7.07–7.25 (overlapped, 4H, H-5, H-6, H-5′, and H-6′), 6.49 (1H, d, J 16.4 Hz, H-8′), 5.07 (1H, m, H-7), 4.54–4.58 (2H, m, H-8), 4.21–4.29 (2H, q, J 6.4 Hz, H-10′), 3.98 (3H, s, C-3′OMe), 3.94 (3H, s, C-3OMe), 3.82–3.90 (1H, m, H-9A), 3.61–3.65 (1H, m, H-9B), 2.27 (3H, s, OAc), 1.33 (3H, t, J 6.4 Hz, H-11′).
Compound 13SC
Compound 12SC (1.12 mmol) was dissolved in 20 mL of 2 M NaOH. After stirring for 10 min at room temperature, the reaction mixture was acidified with 3 M HCl, and extracted three times with CH2Cl2-EtOAc (1:1, v/v). The combined organic layer was washed with brine, dried over Na2SO4, and evaporated in vacuo. The crude product was purified by flash chromatography to give a colorless oil of compound 13SC as a mixture of erythro and threo isomers (48.2% yield, erythro/threo isomer ratio = 7:3 as determined by 1H-NMR). 13SC (erythro isomer): 1H-NMR (400 MHz, methanol-d4): δ = 7.63 (1H, d, J 15.6, H-7′), 7.02 (1H, s, H-2), 6.96 (2H, s, H-2′/6′), 6.83 (1H, d, J 8.7 Hz, H-6), 6.78 (1H, d, J 8.7 Hz, H-5), 6.47 (1H, d, J 15.6 Hz, H-8′), 5.04 (1H, br d, H-7), 4.36–4.39 (1H, m, H-8), 3.95–3.98 (1H, m, H-9A), 3.90 (3H, s, C-3OMe), 3.86 (6H, s, C-3′/5′OMe), 3.63–3.67 (1H, m, H-9B). 13SC (threo isomer): 1H-NMR (400 MHz, methanol-d4): δ = 7.64 (1H, d, J 15.6 Hz, H-7′), 7.06 (1H, s, H-2), 7.02 (2H, s, H-2′/6′), 6.91 (1H, d, J 8.8 Hz, H-6), 6.79 (1H, d, J 8.8 Hz, H-5), 6.49 (1H, d, J 15.6 Hz, H-8′), 5.04 (1H, br d, H-7), 4.18–4.22 (1H, m, H-8), 3.82–3.85 (1H, m, H-9A), 3.93 (3H, s, C-3OMe), 3.87 (6H, s, C-3′/5′OMe), 3.40–3.42 (1H, m, H-9B).
Compound 13FC
Compound 13FC was synthesized from 12FC using the above described procedure for compound 7SC and was isolated as a mixture of erythro and threo isomers in the form of a colorless oil (97.0% yield, erythro/threo isomer ratio = 6:4 as determined by 1H-NMR). 13FC (erythro isomer): 1H-NMR (400 MHz, methanol-d4): δ = 7.62 (1H, d, J 15.6 Hz, H-7′), 7.36 (1H, s, H-2′), 7.21 (1H, d, J 8.8 Hz, H-6′), 7.12–7.14 (overlapped, 2H, H-2, and H-5′), 6.92 (1H, d, J 8.8 Hz, H-6), 6.78 (1H, d, J 8.8 Hz, H-5), 6.44 (1H, d, J 15.6 Hz, H-8′), 4.91–4.92 (1H, m, H-7), 4.37–4.41 (1H, m, H-8), 3.95 (3H, s, C-3′OMe), 3.83 (3H, s, C-3OMe), 3.72–3.77 (1H, m, H-9A), 3.53–3.58 (1H, m, H-9B). 13FC (threo isomer): 1H-NMR (400 MHz, methanol-d4): δ = 7.58 (1H, d, J 15.6 Hz, H-7′), 7.31 (1H, s, H-2′), 7.19 (1H, d, J 8.8 Hz, H-6′), 7.12–7.14 (1H, br s, H-2), 7.05 (1H, d, J 8.8 Hz, H-5’), 6.92 (1H, d, J 8.8 Hz, H-6), 6.76 (1H, br d, H-5), 6.42 (1H, d, J 15.6 Hz, H-8′), 4.91–4.92 (1H, m, H-7), 4.46–4.49 (1H, m, H-8), 3.95 (3H, s, C-3′OMe), 3.83 (3H, s, C-3OMe), 3.72–3.77 (1H, m, H-9A), 3.53–3.58 (1H, m, H-9B).
3-{4-[2-Hydroxy-2-(4-hydroxy-3-methoxyphenyl)-1-hydroxymethylethoxy]-3′,5′-dimethoxyphenyl}acryloylcholine [SC(4-O-8)G (1)]
Choline esterification of compound 13SC was carried out according to the method of Böttcher et al. (2008). Briefly, to a solution of compound 13SC (0.116 mmol) in DMF (220 µL), 1 M NaOH (110 µL), and 1 M bromocholine bromide in water (220 µL) were successively added at room temperature. The reaction mixture was then heated to 90°C for 24 h. After cooling to room temperature, the reaction mixture was diluted with water (2 mL), eluted on a Strata-X-CW solid-phase extraction cartridge (Phenomenex, Torrance, USA), and concentrated in vacuo, to give a crude mixture of erythro and threo isomers 1e and 1t as a colorless oil (yield 65.6%). A portion of this crude mixture was dissolved in H2O-MeOH (9:1, v/v), and subjected to preparative HPLC to isolate pure compounds 1e and 1t. erythro-SC(4-O-8)G (1e): 1H-NMR (800 MHz, methanol-d4): δ = 7.70 (1H, d, J 15.9 Hz, H-7′), 6.99 (1H, d, J 1.9 Hz H-2), 6.95 (2H, s, H-2′/6′), 6.79 (1H, dd, J 8.2, 1.9 Hz, H-6), 6.73 (1H, d, J 8.2 Hz, H-5), 6.55 (1H, d, J 15.9 Hz, H-8′), 4.90 (1H, d, J 5.2 Hz, H-7), 4.69-4.65 (2H, m, H-10′), 4.37–4.34 (1H, m, H-8), 3.91 (1H, dd, J 12.0, 5.4 Hz, H-9A), 3.85 (6H, s, C-3′/5′OMe), 3.83 (3H, s, C-3OMe), 3.79–3.77 (2H, m, H-11′), 3.62 (1H, br dd, H-9B), 3.26 (9H, s, H-12′NMe); 13C-NMR (200 MHz, methanol-d4): δ = 167.47 (C-9), 154.80 (C-3′/5′), 148.68 (C-3), 147.53 (C-7′), 146.96 (C-4), 139.57 (C-4′), 133.85 (C-1), 131.27 (C-1′), 120.80 (C-6), 117.11 (C-8′), 115.68 (C-5), 111.58 (C-2), 107.01 (C-2′/6′), 87. 52 (C-8), 74.18 (C-7), 66.25 (C-11′), 61.80 (C-9), 58.89 (C-10′), 56.80 (C-3′/5′OMe), 56.37 (C-3OMe), 54.48 (C-12′NMe). threo-SC(4-O-8)G (1t): 1H-NMR (800 MHz, methanol-d4): δ = 7.71 (1H, d, J 15.9 Hz, H-7′), 7.01 (1H, d, J 1.9 Hz, H-2), 6.98 (2H, s, H-2′/6′), 6.87 (1H, dd, J 8.4, 1.9 Hz, H-6), 6.75 (1H, d, J 8.2 Hz, H-5), 6.57 (1H, d, J 15.9 Hz, H-8′), 4.99 (1H, d, J 6.6 Hz, H-7), 4.69–4.65 (2H, m, H-10′), 4.22–4.19 (1H, m, H-8), 3.99 (6H, s, C-3′/5′OMe), 3.83 (3H, s, C-3OMe), 3.80–3.76 (3H, overlapped, H-9A and H-11′), 3.37 (1H, dd, J 11.9, 3.5 Hz, H-9B), 3.26 (9H, s, H-12′NMe); 13C-NMR (200 MHz, methanol-d4): δ = 167.49 (C-9), 154.60 (C-3′/5′), 148.75 (C-3), 147.21 (C-7′), 147.21 (C-4), 139.96 (C-4′), 133.54 (C-1), 131.43 (C-1′), 120.83 (C-6), 117.30 (C-8′), 115.84 (C-5), 111.77 (C-2), 106.99 (C-2′/6′), 88. 88 (C-8), 74.41 (C-7), 66.24 (C-11′), 61.92 (C-9), 58.92 (C-10′), 56.83 (C-3′/5′OMe), 56.38 (C-3OMe), 54.51 (C-12′NMe).
3-{4-[2-Hydroxy-2-(4-hydroxy-3-methoxyphenyl)-1-hydroxymethylethoxy]-3′-methoxyphenyl}acryloylcholine [FC(4-O-8)G (4)]
Compound 4 as a crude mixture of erythro and threo isomers 4e and 4t (colorless oil, 46.3% yield) was prepared from 13FC using the above described procedure for compound 1. Pure isomers 4e and 4t were then isolated by preparative HPLC. erythro-FC(4-O-8)G (4e): 1H-NMR (800 MHz, methanol-d4): δ = 7.66 (1H, d, J 15.9 Hz, H-7′), 7.18 (1H, d, J 2.0 Hz, H-2′), 7.10 (1H, dd, J 8.4, 2.0 Hz, H-6′), 7.03 (1H, d, J 2.0 Hz, H-2), 6.97 (1H, dd, J 8.2, 2.0 Hz, H-5′), 6.84 (1H, dd, J 8.2, 2.0 Hz, H-6), 6.70 (1H, d, J 8.2 Hz, H-5), 6.45 (1H, d, J 15.9, H-8′), 4.81 (1H, d, J 6.0 Hz, H-7), 4.67–4.64 (2H, m, H-10′), 4.51 (1H, m, H-8), 3.84 (1H, br, H-9A), 3.83 (1H, br, H-9B), 3.82 (3H, s, C-3′OMe), 3.80 (3H, s, C-3OMe), 3.79–3.76 (2H, m, H-11′), 3.25 (9H, s, H-12′NMe); 13C-NMR (200 MHz, methanol-d4): δ = 167.70 (C9′), 152.24 (C-4′), 151.82 (C-3′), 148.67 (C-3), 147.58 (C-7′), 147.10 (C-4), 133.99 (C-1), 129.14 (C-1′), 123.81 (C-6′), 121.17 (C-6), 117.40 (C-5′), 115.61 (C-5), 115.46 (C-8′), 112.54 (C-2′), 112.06 (C-2), 85.47 (C-8), 74.11 (C-7), 66.27 (C-11′), 62.48 (C-9), 58.78 (C-10′), 56.63 (C-3′OMe), 56.34 (C-3OMe), 54.47 (C-12′NMe). threo-FC(4-O-8)G (4t): 1H-NMR (800 MHz, methanol-d4): δ = 7.69 (1H, d, J 15.9 Hz, H-7′), 7.25 (1H, d, J 2.0 Hz, H-2′), 7.14 (1H, dd, J 8.4, 2.0 Hz, H-6′), 7.06 (1H, d, J 8.4 Hz, H-5′), 7.03 (1H, d, J 2.0 Hz, H-2), 6.85 (1H, dd, J 8.1, 2.0 Hz, H-6), 6.75 (1H, d, J 8.1 Hz, H-5), 6.47 (1H, d, J 15.9 Hz, H-8′), 4.89 (1H, d, J 5.4 Hz, H-7), 4.68–4.63 (2H, m, H-10′), 4.48–4.45 (1H, m, H-8), 3.90 (3H, s, C-3′OMe), 3.82 (3H, s, C-3OMe), 3.79–3.75 (3H, overlapped, H-9A and H-11′), 3.53 (1H, dd, J 11.9, 5.8 Hz, H-9B), 3.25 (9H, s, H-12′NMe); 13C-NMR (200 MHz, methanol-d4): δ = 167.69 (C9′), 152.49 (C-4′), 151.74 (C-3′), 148.85 (C-3), 147.53 (C-7′), 147.22 (C-4), 133.81 (C-1), 129.27 (C-1′), 123.96 (C-6′), 120.66 (C-6), 117.40 (C-5′), 115.85 (C-5), 115.60 (C-8′), 112.51 (C-2′), 111.74 (C-2), 86. 21 (C-8), 73.91 (C-7), 66.27 (C-11′), 62.09 (C-9), 58.80 (C-10′), 56.68 (C-3′OMe), 56.35 (C-3OMe), 54.48 (C-12′NMe).
Phylogenetic analysis
Deduced amino acid sequences of DIRs or LACs were aligned using the CLUSTAL W program in MEGA X (version 10.1, https://www.megasoftware.net/; Kumar et al., 2018). A phylogenetic tree was constructed using the Neighbor-Joining method (Saitou and Nei, 1987) in MEGA X with the following parameters: bootstrap test (500 replicates), Poisson model, uniform rates, and complete deletion. Machine-readable files of the phylogenetic analyses used to for Figures 6 and 9 are provided as Supplemental Files 1 and 2, respectively.
Reverse transcription quantitative PCR
RNA extraction, cDNA synthesis, and reverse transcription quantitative PCR (RT-qPCR) were performed as described previously (Yonekura-Sakakibara et al., 2014). The developmental stages of leaves, stems, roots, floral buds, and flowers used for analyses were as described previously (Yonekura-Sakakibara et al., 2007). The developmental stages of siliques were as follows: stage 3, 24–36 h after flowering (HAF); stage 4, 36–48 HAF; stage 5, 48–72 HAF; stage 6, 72–96 HAF; stage 7, 96–108 HAF; stage 8, 108–120 HAF; stage 9, 120–132 HAF; stage 10, 132–144 HAF; stage 11, 144–192 HAF. Primers used are described in Supplemental Table 2 (At4g11180cDNA-RT101f and At4g11180cDNA-RT186r for AtDP1/AtDIR12, At1g61720pda14333_RT107f andAt1g61720pda14333_RT179r for BANYULS, At5g48100pda12625_RT478f and At5g48100pda12625_RT552r for AtLAC15/TT10, and At2g40370qPCRNo2_1090F and At2g40370qPCRNo2_1107R for AtLAC5) and were checked for specific product formation using a dissociation program. Plasmid DNAs containing the corresponding genes were used as templates for calibration. RT-qPCR was performed in triplicate.
Generation and analysis of YFP/GUS reporter lines
A 720-bp fragment of the AtDP1/AtDIR12 promoter region or the promoter region including the AtDP1/AtDIR12 coding region was amplified by PCR using primers, At4g11180_CCAC+promoter F and At4g11180_promoter R or At4g11180_R (Supplemental Table 2). Amplified fragments were cloned into the pENTR/D-TOPO vector (Invitrogen) as the entry vector. The resulting plasmid was sequenced to confirm the absence of PCR errors. pHGY, pH35GY derivatives from which the CaMV 35S promoter sequence is deleted (Endo et al., 2009) and pBGGUS (Funakoshi, Japan) were used as the destination vectors. The LR reactions for the binary vectors pKYS405 (pHGY/ProAtDP1-AtDP1CDS-YFP) and pKYS415 (pBGGUS/ProAtDP1-GUS) were catalyzed using the Gateway LR ClonaseTMII enzyme mix (Invitrogen).
The 4,140-bp or 2,000-bp fragments of the AtLAC5 promoter region were amplified by PCR using primers, At2g40370pro_CCAC + 4140F or At2g40370pro_CCAC + 2000F and At2g40370ATG_R (Supplemental Table 2). Amplified fragments were cloned into the pENTR/D-TOPO vector (Invitrogen) as the entry vector. The binary vectors, pKYS540 (pBGGUS/ProAtLAC5-4140-GUS) and pKYS541 (pBGGUS/ProAtLAC5-2000-GUS), were constructed using pBGGUS (Funakoshi, Japan) as the destination vector as described above.pKYS405 (pHGY/ProAtDP1-AtDP1CDS-YFP), pKYS415 (pBGGUS/ProAtDP1-GUS), pKYS540 (pBGGUS/ProAtLAC5-4140-GUS), and pKYS541 (pBGGUS/ProAtLAC5-2000-GUS) were transformed into Agrobacterium and subsequently into Arabidopsis plants as described above. Transgenic T2 plants for pKYS405 were selected on half-strength Murashige and Skoog medium containing 25 mg L−1 hygromycin B and 50 mg L−1 carbenicillin disodium salt and those for pKYS415, pKYS540, and pKYS541 were selected on Murashige and Skoog medium containing 50 μM glufosinate ammonium and 50 mg L−1 carbenicillin disodium salt.
Confocal laser scanning microscopy, light microscopy, and electron microscopy
Fluorescent samples were observed using a confocal laser scanning microscope system (Model LSM510 META, Axioplan2 imaging, a Plan-Apochromat lens (63X/1.4 oil DIC; optical slice, 1 μm); Carl Zeiss) with 488-nm excitation and a 505–530 nm band-pass filter for YFP, 488-nm excitation and a 650-nm long-pass filter for chlorophyll autofluorescence and a 25-mV argon laser. Composite figures were prepared using the Zeiss LSM Image Browser software.
For histochemical GUS assays, tissues were treated with ice-cold 90% (v/v) acetone for 20 min on ice and stained in 100 mM sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% (v/v) Triton X-100, 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl glucuronide, 2 mM potassium ferricyanide, and 2 mM potassium ferrocyanide. After 60 min vacuum infiltration, samples were incubated overnight at 37°C and subsequently de-stained by a series of washes in 70% (v/v) ethanol, ethanol/acetic acid (6:1 v/v ratio), 70% (v/v) ethanol. For sectioning, tissue samples were fixed with 2% (v/v) glutaraldehyde and 1% (v/v) formaldehyde in 0.1 M phosphate buffer, pH 7.0, for 24 h at 4°C, washed in phosphate buffer, dehydrated using a series of graded ethanol solutions [30%, 40%, 60%, 70%, 85%, 95%, 100% (v/v)] and embedded in Technovit 7100 resin (Heraeus Kulzer, Wehrheim, Germany) according to the manufacturer’s instructions. Tissue sections were sliced into 5-µm transverse sections using a rotary microtome (Leica RM2125) and tungsten carbide disposable blades (Leica TC-65). Sections were observed using an Olympus BX53 microscope.
Surfaces of dry seeds were observed using a scanning electron microscope (Hitachi TM-1000).
Coexpression analyses
Coexpression analyses were conducted as described previously (Yonekura-Sakakibara et al., 2007, 2008; Saito et al., 2008). The genes encoding LACs (r > 0.3) were extracted from the list of genes co-expressed with AtDP1/AtDIR12.
Seed coat permeability test
Seed coat permeability was tested by a tetrazolium penetration assay (Vishwanath et al., 2013, 2014). Dried seeds (10 mg) were incubated in 250 μL of an aqueous solution with or without 1% (w/v) tetrazolium red (2,3,5-triphenyltetrazolium chloride, Wako, Japan) at 30°C for 48 h in the dark. After incubation, the samples were washed with water, resuspended in 1 mL 95% (v/v) ethanol, and finely ground with a mortar and pestle to extract the formazans. After centrifugation at 15,000g for 3 min, the supernatant fraction was recovered and the absorbance at 485 nm was measured. Three biological replicates were used for the analyses.
Proanthocyanidin analysis
PA extraction and acid hydrolysis were performed in triplicate as described previously (Tohge et al., 2005; Kitamura et al., 2010). Mature seeds (10 mg) were homogenized in 0.75 mL of 70% acetone containing 5.26 mM Na2S2O5 in a mixer mill (Qiagen Retsch MM300 TissueLyser) for 1 min at 20 Hz, followed by sonication for 20 min. After centrifugation at 15,000g for 5 min, the supernatant fraction was evaporated and resuspended in 1 mL of HCl: butanol: 70% acetone (2:10:3). The absorbance of the solutions before/after hydrolysis at 95°C for 60 min was measured at 545 nm and the difference was treated as the soluble PA fraction. The pellet after extraction with 70% acetone was also evaporated, suspended in the HCl: butanol: 70% acetone solution and hydrolyzed as the insoluble PA fraction.
Lignin analysis
Mature seeds (40 mg) were pulverized, extracted successively with chloroform–methanol (2:1, v/v), 100% methanol, and water, followed by freeze-drying to obtain the seed cell-wall fractions (Tobimatsu et al., 2013). The seed cell-wall fractions were then subjected to analytical thioacidolysis (Lapierre et al., 1986) using the modified method described by Yamamura et al., 2012. The released lignin monomers were derivatized with N,O-bis(trimethylsilyl)acetamide and quantified by gas chromatography/mass spectrometry using 4,4′-ethylenebisphenol as an internal standard (Yue et al., 2012).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL libraries under the following accession numbers: AtDP1/AtDIR12 (At4g11180, BT004016), AtDIR5 (At1g64160, BT010486), AtDIR6 (At4g23690, BT002439), AtDIR13 (At4g11190, BT015628), AtDIR14 (At4g11210, BT015626), FiDIR1 (AF210061), LuDIR1 (KM433751), LuDIR5 (KM433753), LuDIR6 (KM433752), PsDRR206 (U11716), ScDIR (HQ428029), TpDIR5 (AF210067), TpDIR6 (AF210070), AtLAC1 (At1g18140, NP_173252); AtLAC2 (At2g29130, NP_180477); AtLAC3 (At2g30210, NP_180580); AtLAC4 (At2g38380, NP_565881); AtLAC5 (At2g40370, NP_181568); AtLAC6 (At46570, NP_182180); AtLAC7 (At3g09220, NP_187533); AtLAC8 (At5g01040, NP_195724); AtLAC9 (At5g01050, NP_195725); AtLAC10 (At5g01190, NP_195739); AtLAC11 (At5g03260, NP_195946); AtLAC12 (At5g05390, NP_196158); AtLAC13 (At5g07130, NP_196330); AtLAC14 (At5g09360, NP_196498); AtLAC15 (At5g48100, NP_199621); AtLAC16 (At5g58910, NP_200699); AtLAC17 (At5g60020, NP_200810); BdLAC5 (Bradi1g66720, XP_003558240); BnTT10 (NP_001302959); GaLAC1 (NP_001316972); PtLAC3 (CAC14719); SofLAC (SCVPRZ3027A08.g and SCUTST3084C11.g); ZmLAC3 (CAJ30499).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure 1 . Identification of neolignan FC(4-O-8)G (4) in Arabidopsis seeds.
Supplemental Figure 2. Multiple alignment of DIRs in the DIR-a family.
Supplemental Figure 3 . Multiple alignment of Arabidopsis LACs and functionally identified LACs.
Supplemental Figure 4 . T-DNA insertion mutants of AtLAC5.
Supplemental Figure 5 . Expression pattern of AtLAC5 and AtDP1/AtDIR12 using the Arabidopsis eFP browser (Winter et al., 2007) and Arabidopsis Seed Coat eFP browser (Dean et al., 2011).
Supplemental Figure 6 . A proposed model for gene duplication of the Arabidopsis DIR-a gene family.
Supplemental Table 1. Metabolites detected in seeds of wild-type and the tested mutants.
Supplemental Table 2. Primers used in this study.
Supplemental Data Set 1. 1H and 13C NMR spectra of synthetic neolignans SC(4-O-8)G and FC(4-O-8)G.
Supplemental Data Set 2. One-way analysis of variance for Figure 5.
Supplemental Data Set 3. One-way analysis of variance for Figure 12.
Supplemental Data Set 4. One-way analysis of variance for Figure 13.
Supplemental File 1. Alignment corresponding to the phylogenetic analysis in Figure 6.
Supplemental File 2. Alignment corresponding to the phylogenetic analysis in Figure 9.
Supplementary Material
Acknowledgments
We thank Dr T. Nakagawa (Shimane University) for kindly providing the pGWB1 vector, Dr K. Toyooka (RIKEN) for technical support on YFP analysis, and Mr M. Suzuki for his technical support for LC analyses. We also thank Dr H. Kaji and Ms K. Yamada for their assistance in NMR analysis and Dr S. Suzuki for assistance in preparing the standard compounds. A part of this study was conducted using System for Development and Assessment of Sustainable Humanosphere (Research Institute for Sustainable Humanosphere and Center of Ecological Research, Kyoto University) and the NMR spectrometer at JURC (Institute for Chemical Research, Kyoto University).
Conflict of interest statement. None declared.
These authors contributed equally to this work (K.Y.-S., M.Y.).
K.Y.-S., T.U., and K.S. designed research. K.Y.-S., M.Y., F.M., E.O., R.N., S.S., T.M., and Y.T. performed research. K.Y.-S., M.Y., F.M., R.N., T.M., and Y.T. analyzed the data. K.Y.-S., M.Y., Y.T., T.U., and K.S. wrote the article.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) are: Kazuki Saito (kazuki.saito@riken.jp), and Toshiaki Umezawa (tumezawa@rish.kyoto-u.ac.jp).
References
- Attoumbre J, Bienaime C, Dubois F, Fliniaux MA, Chabbert B, Baltora-Rosset S (2010) Development of antibodies against secoisolariciresinol–application to the immunolocalization of lignans in Linum usitatissimum seeds. Phytochemistry 71: 1979–1987 [DOI] [PubMed] [Google Scholar]
- Bao W, O'Malley DM, Whetten R, Sederoff RR (1993) A laccase associated with lignification in loblolly pine xylem. Science 260: 672–674 [DOI] [PubMed] [Google Scholar]
- Barbosa ICR, Rojas-Murcia N, Geldner N (2019) The Casparian strip-one ring to bring cell biology to lignification? Curr Opin Biotechnol 56: 121–129 [DOI] [PubMed] [Google Scholar]
- Berridge MV, Tan AS, McCoy KD, Wang R (1996) The biochemical and cellular basis of cell proliferation assays that use tetrazolium salts. Biochemica 4: 15–20 [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cezard L, Le Bris P, Borrega N, Herve J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C, von Roepenack-Lahaye E, Schmidt J, Schmotz C, Neumann S, Scheel D, Clemens S (2008) Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol 147: 2107–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlat V, Kwon M, Davin LB, Lewis NG (2001) Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57: 883–897 [DOI] [PubMed] [Google Scholar]
- Caparros-Ruiz D, Fornale S, Civardi L, Puigdomenech P, Rigau J (2006) Isolation and characterisation of a family of laccases in maize. Plant Sci 171: 217–225 [Google Scholar]
- Cesarino I, Araujo P, Sampaio Mayer JL, Vicentini R, Berthet S, Demedts B, Vanholme B, Boerjan W, Mazzafera P (2013) Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J Exp Bot 64: 1769–1781 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbin C, Drouet S, Markulin L, Auguin D, Laine E, Davin LB, Cort JR, Lewis NG, Hano C (2018) A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: from gene identification and evolution to differential regulation. Plant Mol Biol 97: 73–101 [DOI] [PubMed] [Google Scholar]
- Dalisay DS, Kim KW, Lee C, Yang H, Rubel O, Bowen BP, Davin LB, Lewis NG (2015) Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: integrated omics and MALDI mass spectrometry imaging. J Nat Prod 78: 1231–1242 [DOI] [PubMed] [Google Scholar]
- Davin LB, Lewis NG (2003) An historical perspective on lignan biosynthesis: monolignol, allylphenol and hydroxycinnamic acid coupling and downstream metabolism. Phytochem Rev 2: 257–288 [Google Scholar]
- Davin LB, Wang HB, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275: 362–366 [DOI] [PubMed] [Google Scholar]
- Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4: 1074–1091 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima O, Morreel K, Vanholme B, Kim H, Ralph J, Boerjan W (2015) Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27: 695–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effenberger I, Harport M, Pfannstiel J, Klaiber I, Schaller A (2017) Expression in Pichia pastoris and characterization of two novel dirigent proteins for atropselective formation of gossypol. Appl Microbiol Biotechnol 101: 2021–2032 [DOI] [PubMed] [Google Scholar]
- Effenberger I, Zhang B, Li L, Wang Q, Liu Y, Klaiber I, Pfannstiel J, Wang Q, Schaller A (2015) Dirigent proteins from cotton (Gossypium sp.) for the atropselective synthesis of gossypol. Angew Chem Int Ed Engl 54: 14660–14663 [DOI] [PubMed] [Google Scholar]
- Endo S, Pesquet E, Yamaguchi M, Tashiro G, Sato M, Toyooka K, Nishikubo N, Udagawa-Motose M, Kubo M, Fukuda H, et al. (2009) Identifying new components participating in the secondary cell wall formation of vessel elements in zinnia and Arabidopsis. Plant Cell 21: 1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Costa MA, Fujita M, Dinkova-Kostova AT, Wang HB, Burlat V, Martin W, Sarkanen S, Davin LB, Lewis NG (1999) Regiochemical control of monolignol radical coupling: a new paradigm for lignin and lignan biosynthesis. Chem Biol 6: 143–151 [DOI] [PubMed] [Google Scholar]
- Gasper R, Effenberger I, Kolesinski P, Terlecka B, Hofmann E, Schaller A (2016) Dirigent protein mode of action revealed by the crystal structure of AtDIR6. Plant Physiol 172: 2165–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Meyer K, Viitanen PV, Chapple C, Strack D (2002) Immunolocalization of 1-O-sinapoylglucose:malate sinapoyltransferase in Arabidopsis thaliana. Planta 215: 26–32 [DOI] [PubMed] [Google Scholar]
- Helm RF, Ralph J (1992) Lignin-hydroxycinnamyl model compounds related to forage cell wall structure. 1. Ether-linked structures. J Agric Food Chem 40: 2167–2175 [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE (2013) Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci USA 110: 14498–14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Kado Y (1998) Formation of optically active neolignans from achiral coniferyl alcohol by cell-free extracts of Eucommia ulmoides. J Wood Sci 44: 244–246 [Google Scholar]
- Kim KW, Moinuddin SG, Atwell KM, Costa MA, Davin LB, Lewis NG (2012) Opposite stereoselectivities of dirigent proteins in Arabidopsis and schizandra species. J Biol Chem 287: 33957–33972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Smith CA, Daily MD, Cort JR, Davin LB, Lewis NG (2015). Trimeric structure of (+)-pinoresinol-forming dirigent protein at 1.95: a resolution with three isolated active sites. J Biol Chem 290: 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Jeon JH, Fujita M, Davin LB, Lewis NG (2002) The western red cedar (Thuja plicata) 8-8' DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol Biol 49: 199–214 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Matsuda F, Tohge T, Yonekura-Sakakibara K, Yamazaki M, Saito K, Narumi I (2010) Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J 62: 549–559 [DOI] [PubMed] [Google Scholar]
- Kulik T, Busko M, Pszczolkowska A, Perkowski J, Okorski A (2014) Plant lignans inhibit growth and trichothecene biosynthesis in Fusarium graminearum. Lett Appl Microbiol 59: 99–107 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C (1986) Preparative thioacidolysis of spruce lignin: isolation and identification of main monomeric products. Holzforschung 40: 47–50 [Google Scholar]
- Lee S, Kaminaga Y, Cooper B, Pichersky E, Dudareva N, Chapple C (2012) Benzoylation and sinapoylation of glucosinolate R-groups in Arabidopsis. Plant J 72: 411–422 [DOI] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Davis E, Gardner D, Cai X, Wu Y (2006) Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Lourith N, Katayama T, Ishikawa K, Suzuki T (2005) Biosynthesis of a syringyl 8-O-4 ‘ neolignan in Eucommia ulmoides: formation of syringylglycerol-8-O-4 ’-(sinapyl alcohol) ether from sinapyl alcohol. J Wood Sci 51: 379–386 [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K (2009) MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J 57: 555–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Hirai MY, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart NJ, Sakurai T, Shimada Y, Saito K (2010) AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol 152: 566–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Moinuddin SGA, Kim SJ, Bedgar DL, Costa MA, Thomas DG, Young RP, Smith C, Cort JR, Davin LB, et al. (2020) Pterocarpan synthase (PTS) structures suggest a common quinone methide-stabilizing function in dirigent proteins and proteins with dirigent-like domains. J Biol Chem 295: 11584–11601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss GP (2000) Nomenclature of lignans and neolignans (IUPAC Recommendations 2000). Pure Appl Chem 72: 1493–1523 [Google Scholar]
- Mugford ST, Louveau T, Melton R, Qi X, Bakht S, Hill L, Tsurushima T, Honkanen S, Rosser SJ, Lomonossoff GP, et al. (2013) Modularity of plant metabolic gene clusters: a trio of linked genes that are collectively required for acylation of triterpenes in oat. Plant Cell 25: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsubo T, Mizutani M, Suzuki S, Hattori T, Umezawa T (2008) Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J Biol Chem 283: 15550–15557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitao JK, Johnson KS, Scriber JM, Nair MG (1992) Magnolia virginiana Neolignan compounds as chemical barriers to swallowtail butterfly host use. J Chem Ecol 18: 1661–1671 [DOI] [PubMed] [Google Scholar]
- Okazawa A, Hori K, Okumura R, Izumi Y, Hata N, Bamba T, Fukusaki E, Ono E, Satake H, Kobayashi A (2011) Simultaneous quantification of lignans in Arabidopsis thaliana by highly sensitive capillary liquid chromatography-electrospray ionization-ion trap mass spectrometry. Plant Biotechnol 28: 287–293 [Google Scholar]
- Orr JD, Lynn DG (1992). Biosynthesis of dehydrodiconiferyl alcohol glucosides: implications for the control of tobacco cell growth. Plant Physiol 98: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Cheng X, Huhman DV, Ma J, Peel GJ, Yonekura-Sakakibara K, Saito K, Shen G, Sumner LW, Tang Y, et al. (2013). Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta 238: 139–154 [DOI] [PubMed] [Google Scholar]
- Paniagua C, Bilkova A, Jackson P, Dabravolski S, Riber W, Didi V, Houser J, Gigli-Bisceglia N, Wimmerova M, Budinska E, et al. (2017) Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J Exp Bot 68: 3287–3301 [DOI] [PubMed] [Google Scholar]
- Petersen M, Hans J, Matern U (2010). Biosynthesis of phenylpropanoids and related compounds. In Wink M, ed, Annual Plant Reviews. Wiley-Blackwell, Chichester, West Sussex; Ames, IA, pp 182–257 [Google Scholar]
- Pickel B, Constantin MA, Pfannstiel J, Conrad J, Beifuss U, Schaller A (2010) An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew Chem Int Ed Engl 49: 202–204 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Helm RF (1991) Rapid proton NMR method for determination of threo:erythro ratios in lignin model compounds and examination of reduction stereochemistry. J Agric Food Chem 39: 705–709 [Google Scholar]
- Ralph J, Lapierre C, Boerjan W (2019) Lignin structure and its engineering. Curr Opin Biotechnol 56: 240–249. [DOI] [PubMed] [Google Scholar]
- Ralph SG, Jancsik S, Bohlmann J (2007) Dirigent proteins in conifer defense II: extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 68: 1975–1991 [DOI] [PubMed] [Google Scholar]
- Ranocha P, Chabannes M, Chamayou S, Danoun S, Jauneau A, Boudet AM, Goffner D (2002) Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol 129: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K (2008) Decoding genes with coexpression networks and metabolomics – ‘majority report by precogs’. Trends Plant Sci 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakakibara N, Nakatsubo T, Suzuki S, Shibata D, Shimada M, Umezawa T (2007) Metabolic analysis of the cinnamate/monolignol pathway in Carthamus tinctorius seeds by a stable-isotope-dilution method. Org Biomol Chem 5: 802–815 [DOI] [PubMed] [Google Scholar]
- Sano N, Rajjou L, North HM, Debeaujon I, Marion-Poll A, Seo M (2016). Staying alive: molecular aspects of seed longevity. Plant Cell Physiol 57: 660–674 [DOI] [PubMed] [Google Scholar]
- Sartorelli P, Benevides PJC, Ellensohn RM, Rocha MVAF, Moreno PRH, Kato MJ (2001) Enantioselective conversion of p-hydroxypropenylbenzene to (+)-conocarpan in Piper regnellii. Plant Sci 161: 1083–1088 [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schroeder FC, del Campo ML, Grant JB, Weibel DB, Smedley SR, Bolton KL, Meinwald J, Eisner T (2006). Pinoresinol: a lignol of plant origin serving for defense in a caterpillar. Proc Natl Acad Sci USA 103: 15497–15501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, Demura T, Ellis B, Samuels AL (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol 166: 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne HK, Dalisay DS, Kim KW, Moinuddin SG, Yang H, Hartshorn CM, Davin LB, Lewis NG (2015) Non-host disease resistance response in pea (Pisum sativum) pods: biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 113: 140–148 [DOI] [PubMed] [Google Scholar]
- Shirley AM, Chapple C (2003) Biochemical characterization of sinapoylglucose:choline sinapoyltransferase, a serine carboxypeptidase-like protein that functions as an acyltransferase in plant secondary metabolism. J Biol Chem 278: 19870–19877 [DOI] [PubMed] [Google Scholar]
- Shirley AM, McMichael CM, Chapple C (2001) The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J 28: 83–94 [DOI] [PubMed] [Google Scholar]
- Sterjiades R, Dean JF, Eriksson KE (1992). Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol 99: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Umezawa T (2007). Biosynthesis of lignans and norlignans. J Wood Sci 53: 273–284 [Google Scholar]
- Tobimatsu Y, Schuetz M (2019) Lignin polymerization: how do plants manage the chemistry so well? Curr Opin Biotechnol 56: 75–81 [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Chen F, Nakashima J, Escamilla-Trevino LL, Jackson L, Dixon RA, Ralph J (2013) Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Matsui K, Ohme-Takagi M, Yamazaki M, Saito K (2005) Enhanced radical scavenging activity of genetically modified Arabidopsis seeds. Biotechnol Lett 27: 297–303 [DOI] [PubMed] [Google Scholar]
- Turlapati PV, Kim KW, Davin LB, Lewis NG (2011) The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233: 439–470 [DOI] [PubMed] [Google Scholar]
- Turner LB, Mueller-Harvey I, McAllan AB (1993) Light-induced isomerization and dimerization of cinnamic acid derivatives in cell walls. Phytochemisty 33: 791–796 [Google Scholar]
- Uchida K, Akashi T, Aoki T (2017) The missing link in leguminous pterocarpan biosynthesis is a dirigent domain-containing protein with isoflavanol dehydratase activity. Plant Cell Physiol 58: 398–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T (2003a) Diversity in lignan biosynthesis. Phytochem Rev 2: 371–390 [Google Scholar]
- Umezawa T (2003b) Phylogenetic distribution of lignan producing plants. Wood Res 90: 27–110 [Google Scholar]
- Umezawa T, Yamamura M, Nakatsubo T, Suzuki S, Hattori T (2011). Stereoselectivity of the biosynthesis of norlignans and related compounds. In Gang D ed, The Biological Activity of Phytochemicals. Springer, New York, pp 179–197 [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanath SJ, Domergue F, Rowland D (2014) Seed coat permeability test: tetrazolium penetration assay. In Bio-protocol. pp e1173. [Google Scholar]
- Vishwanath SJ, Kosma DK, Pulsifer IP, Scandola S, Pascal S, Joubes J, Dittrich-Domergue F, Lessire R, Rowland O, Domergue F (2013). Suberin-associated fatty alcohols in Arabidopsis: distributions in roots and contributions to seed coat barrier properties. Plant Physiol 163: 1118–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T (2010). Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang CL, Zhu ML, Yu Y, Zhang YB, Wei ZM (2008) Generation and characterization of transgenic poplar plants overexpressing a cotton laccase gene. Plant Cell Tissue Organ Cult 93: 303–310 [Google Scholar]
- Wang X, Zhuo C, Xiao X, Wang X, Docampo-Palacios M, Chen F, Dixon RA (2020) Substrate specificity of LACCASE8 facilitates polymerization of caffeyl alcohol for C-lignin biosynthesis in the seed coat of Cleome hassleriana. Plant Cell 32: 3825–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Tan X, Paterson AH (2013) Different patterns of gene structure divergence following gene duplication in Arabidopsis. BMC Genomics 14: 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Chapple C (2010). The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLOS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram K, Schmidt J, Wray V, Milkowski C, Schliemann W, Strack D (2010) Profiling of phenylpropanoids in transgenic low-sinapine oilseed rape (Brassica napus). Phytochemistry 71: 1076–1084 [DOI] [PubMed] [Google Scholar]
- Wong WS, Guo D, Wang XL, Yin ZQ, Xia B, Li N (2005). Study of cis-cinnamic acid in Arabidopsis thaliana. Plant Physiol Biochem 43: 929–937 [DOI] [PubMed] [Google Scholar]
- Yamamura M, Hattori T, Suzuki S, Shibata D, Umezawa T (2012) Microscale thioacidolysis method for the rapid analysis of β-O-4 substructures in lignin. Plant Biotechnol 29: 419–423 [Google Scholar]
- Yang XX, Choi HW, Yang SF, Li N (1999) A UV-light activated cinnamic acid isomer regulates plant growth and gravitropism via an ethylene receptor-independent pathway. Aust J Plant Physiol 26: 325–335 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K (2007) Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J Biol Chem 282: 14932–14941 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Nakabayashi R, Sugawara S, Tohge T, Ito T, Koyanagi M, Kitajima M, Takayama H, Saito K (2014) A flavonoid 3-O-glucoside:2"-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana. Plant J 79: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Lu F, Sun R-C, Ralph J (2012) Syntheses of lignin-derived thioacidolysis monomers and their uses as quantitation standards. J Agric Food Chem 60: 922–928 [DOI] [PubMed] [Google Scholar]
- Zhang K, Lu K, Qu C, Liang Y, Wang R, Chai Y, Li J (2013) Gene silencing of BnTT10 family genes causes retarded pigmentation and lignin reduction in the seed coat of Brassica napus. PLoS One 8: e61247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.