Abstract
The leaf vasculature plays a key role in solute translocation. Veins consist of at least seven distinct cell types, with specific roles in transport, metabolism, and signaling. Little is known about leaf vascular cells, in particular the phloem parenchyma (PP). PP effluxes sucrose into the apoplasm as a basis for phloem loading, yet PP has been characterized only microscopically. Here, we enriched vascular cells from Arabidopsis leaves to generate a single-cell transcriptome atlas of leaf vasculature. We identified at least 19 cell clusters, encompassing epidermis, guard cells, hydathodes, mesophyll, and all vascular cell types, and used metabolic pathway analysis to define their roles. Clusters comprising PP cells were enriched for transporters, including SWEET11 and SWEET12 sucrose and UmamiT amino acid efflux carriers. We provide evidence that PP development occurs independently from ALTERED PHLOEM DEVELOPMENT, a transcription factor required for phloem differentiation. PP cells have a unique pattern of amino acid metabolism activity distinct from companion cells (CCs), explaining differential distribution/metabolism of amino acids in veins. The kinship relation of the vascular clusters is strikingly similar to the vein morphology, except for a clear separation of CC from the other vascular cells including PP. In summary, our single-cell RNA-sequencing analysis provides a wide range of information into the leaf vasculature and the role and relationship of the leaf cell types.
A leaf scRNA-seq analysis provides insight into leaf vascular cell types and critical resources to build hypotheses and strategies related to the transport of ions, metabolites, and signals.
Introduction
A key feature of multicellularity is the division of labor. During evolution, when plants moved from aquatic to terrestrial environments, new mechanisms were required for the exchange of nutrients—both photoassimilates from aerial organs to soil-anchored sections, and water and essential nutrients from the soil to the aerial, photosynthetic organs. Terrestrial plants developed complex vascular systems to provide, for example, roots with photoassimilates and to provide a photosynthetic organism with essential nutrients. Cells had to differentiate to acquire unique identities by establishing differential transcriptional networks. The vasculature serves both transport and communication between organs. Arabidopsis leaf veins are conjoint, collateral open and closed bundles, with xylem on the adaxial and phloem on the abaxial side (Haritatos et al., 2000). The veins consist of at least seven different cell types with unique features identifiable by light and electron microscopy. In Arabidopsis, the abaxial phloem is composed of enucleate sieve elements (SEs), as the actual conduits, which are coupled to companion cells (CCs), and a third, poorly understood cell type, the phloem parenchyma (PP). When mature, the adaxial xylem consists of dead tracheary elements that are accompanied by xylem parenchyma (XP). The vascular parenchyma (VP) is often located at the interface between phloem and xylem. At earlier stages of development, xylem and phloem are separated by meristematic cells (procambium; open-type vasculature), that differentiate toward the tip of the leaf where procambium is absent (closed-type; Kang and Dengler, 2004). Phloem and xylem exchange water and solutes in complex ways, and thus the cells must be equipped with specific sets of transporters. Moreover, it is likely that the different cell types have specialized metabolic activities. In addition, the vasculature plays important roles in communication by translocating hormones, small RNAs, and even proteins; and also appear to be involved in electrical signaling (Nguyen et al., 2018).
PP is one of the poorly defined and least characterized vascular cell types. In Arabidopsis, PP has so-called cell wall ingrowths with a transfer cell appearance which are thought to play a role in amplifying the surface area to allow for higher transport rates (Edwards et al., 2010; Arun Chinnappa et al., 2013). Since in many plant species the interface between CC and SE (SE/CC) contains only few plasmodesmata (PD), photoassimilate translocation requires an apoplasmic transport route (Haritatos et al., 2000). SE/CC loading is mediated by the H+-sucrose symporter SUT1 (originally identified in potato; named SUC2 in Arabidopsis), which imports sucrose from the cell wall space (Riesmeier et al., 1994). We recently identified sucrose uniporters of the SWEET family in a specific subset of cells in the phloem that likely represent PP (Chen et al., 2012). SWEET expression in PP was confirmed using translational GFP fusions, using a new confocal microscopy method that enabled unambiguous PP identification (Cayla et al., 2019). A key goal of this work was to characterize the role of PP in more detail by identifying its mRNA outfit. This could serve, for example, as a basis for identifying transporters involved in phloem loading of other nutrients, as well as metabolic pathways active in these cells.
Understanding the complexity of plant cells at a single cell level has long been an active area of interest in plant biology. Comprehension at this level is crucial to determine subtle distinctions between, for example, the multitude of complex vascular cell types. Recently, high-throughput single-cell RNA-sequencing (scRNA-seq) has been used to provide new insights into root cell biology and development (Denyer et al., 2019; Ryu et al., 2019; Shulse et al., 2019; Zhang et al., 2019; Wendrich et al., 2020). These latest technologies have allowed us to build on the resolution of prior atlases derived from the isolation of specific cell types, using fluorescent-activated cell sorting of fluorescently labeled protoplasts, and/or laser-capture microdissection (Birnbaum et al., 2003). To gain insights into the specific transcript profiles of the leaf vasculature, we optimized protoplast isolation protocols to enrich for leaf vascular cells. Using scRNA-seq, we identified unique and characteristic mRNA signatures, which revealed fundamental differences in amino acid metabolism and transport pathways in PP and CC important for phloem translocation as well as differences in hormone and glucosinolate metabolism.
Results
Enrichment of vascular protoplasts
Standard protoplast isolation protocols are efficient for protoplasting mesophyll cells for downstream applications such as transient expression. However, these procedures do not efficiently release cells from the vasculature. Here, we developed a methodology for isolating vascular-enriched protoplast populations from mature Arabidopsis leaves (Figure 1; Supplemental Figure S1). Initially, a fraction of the nonvascular cell types were removed using the “tape-sandwich” method which effectively eliminated trichomes, guard cells, and epidermis from the abaxial leaf surface (Supplemental Figure S1B; Wu et al., 2009). Cuts along the midvein further facilitated access of cell wall digesting enzymes to the vasculature (Figure 1A;Supplemental Figure S1B). Elevated concentrations of mannitol lead to an increased release of protoplasts (Figure 1B;Supplemental Figure 1, C and D). We monitored the release by light microscopy, reverse transcription- quantitative polymerase chain reaction (RT-qPCR), and fluorescence microscopy (Figure 1, D and E; Supplemental Figure 1, E and F). The mRNA levels of marker genes were used to evaluate the enrichment of vascular protoplasts during optimization of the protocols (Figure 1, A–C). In addition, the release of intact vascular protoplasts was verified microscopically by monitoring fluorescently labeled cells using stably transformed Arabidopsis lines expressing pAtSWEET11:AtSWEET11-GFP, a tentative marker for PP (Chen et al., 2012), and Q0990, a procambial (PC) marker (Radoeva et al., 2016; Figure 1, D and E; Supplemental Figure S1E). The bulk leaf (not subjected to protoplasting) transcriptome showed high correlation with bulk protoplast transcriptomes (Pearson’s correlation coefficient of 0.9; Supplemental Figure 2A and Supplemental Data Set 1). The high correlation indicates that the protoplasting protocol used here did not impact the relative abundance of cells in the leaf and supported the notion that the scRNA-seq data derived from the protoplasts should cover essentially all cell types present in a mature leaf (the term enriched here thus refers to enrichment over standard protocols).
Figure 1.
Strategies to enrich vascular protoplasts. (A–C) Box plot representation of the RT-qPCR analysis results showing the relative transcript level of the marker for the PP, SWEET11, and the marker for XP, GLR3;6. UBQ10 was used as an internal control for normalization. The abaxial epidermis was removed from leaves harvested from 6-weeks-old plants grown under short day conditions and was treated as indicated. The data shown are from a single experiment with three technical replicates (mean ± se, n = 3). (A) Leaves from which the abaxial epidermis had been stripped were either cut on each side of the main vein before enzymatic digestion of the cell wall (wounded, W) or directly placed in the digesting enzyme solution (nonwounded, NW). (B) Leaves from which the abaxial epidermis had been stripped were cut on each side of the main vein and incubated in the enzyme solution containing 0.4 M or 0.6 M mannitol. (C) Protoplasts isolated from leaf sample prepared as in (B) were incubated in an enzyme solution composed of cell wall degrading enzymes; cellulase Onozuka R-10, macerozyme R-10 obtained from Yakult (Tokyo), and Duchefa (Haarlem). (D) GFP (Green fluorescent protein) fluorescence (cyan) marking PP cells in leaf protoplasts. Protoplasts were isolated from 6-week-old pAtSWEET11:AtSWEET11-GFP plants expressing a PP cell-specific marker. Magenta, chlorophyll autofluorescence. Scale bar: 10 µm. (E) GFP fluorescence (cyan) marking procambium cells in leaf protoplasts. Protoplasts were isolated from 6-week-old Q0990 plants expressing a procambium cell-specific marker. Magenta, chlorophyll autofluorescence. Scale bar: 20 µm.
scRNA-seq of vascular-enriched leaf protoplast population
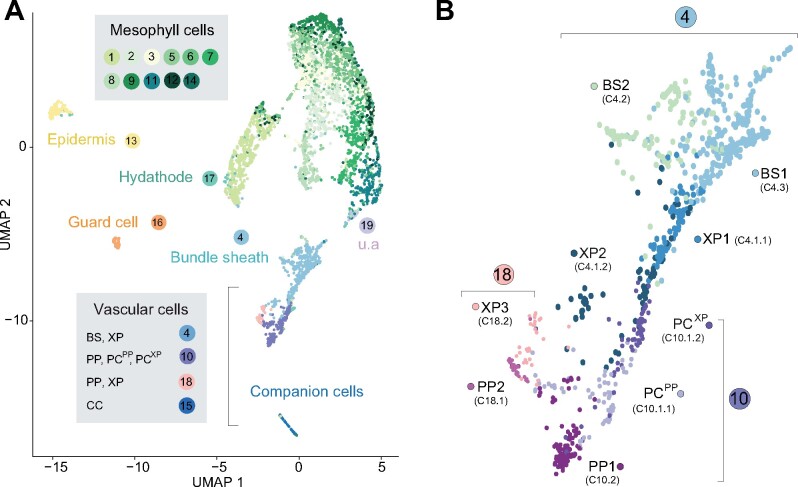
scRNA-seq libraries were produced from vasculature-enriched leaf protoplast populations, with 5,230 single-cell transcriptomes obtained from two biological replicates. Sequencing to a depth of approximately 96,000 reads per cell was undertaken, and identified a median number of 3,342 genes, and 27,159 unique molecular identifiers (representing unique transcripts), per cell (Supplemental Figure 2B). Unsupervised clustering using Seurat (Satija et al., 2015) identified 19 distinct cell clusters (Figure 2; Supplemental Movie 1 and Supplemental Data Set 2). Plotting the transcriptomes from the two replicates in two dimensions using Uniform Manifold Approximation and Projection (UMAP; McInnes et al., 2018) revealed an overlapping distributions of cells, and a similar proportion of cell identities (Supplemental Figure 2C). mRNA profiles and relative cell numbers in the clusters were highly correlated in the replicates (Supplemental Data Set 1). To assign cell identity to the clusters, we examined the specificity of transcripts of known marker genes (Figure 3 and Supplemental Figure 3, Supplemental Data Sets 3 and 4).
Figure 2.
Assignment of cellular identity to clusters. (A) UMAP dimensional reduction projection of 5,230 Arabidopsis leaf cells. Cells were grouped into 19 distinct clusters using Seurat (Butler et al., 2018). The cluster number is shown and colored based on the colors assigned to each cell type (u.a.—unassigned, i.e. cluster could not be assigned to a known cell type). Each dot indicates individual cells colored according to the cell type assigned. (B) Magnification of C4, C18, and C10 subclusters. Different colors indicate distinct cell identities. BS, bundle sheath; PC, procambium; PP, phloem parenchyma; PCXP, procambium; XP, xylem parenchyma cells with features relating to xylem differentiation. PCPP, procambium cells with features relating to phloem differentiation.
Figure 3.
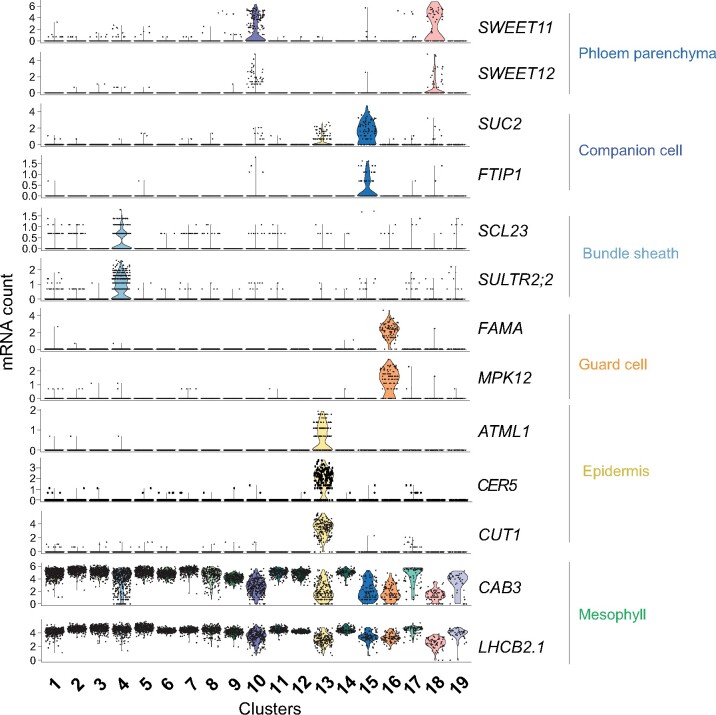
mRNA levels of marker genes in clusters used to assign cell types. Violin plots showing transcript enrichment of known cell type-specific marker genes across clusters. Clusters are indicated on the x-axis. The name of the cell type assigned to each cluster is indicated on the right side of the violin plots.
The 19 clusters cover all major cell types of the leaf. The population was dominated by mesophyll cells (∼75% of all cells), as indicated by the enrichment of known mesophyll markers (CAB3, LHCB2.1, and CA1; Sawchuk et al., 2008; Endo et al., 2014; Figure 3; Supplemental Figure S3A). The mesophyll group comprised 12 clusters (Clusters, or C1, 2, 3, 5, 6-9, 11, 12, 14; Figures 2A and 3; Supplemental Data Set 4) and formed three major branches on the UMAP plot. Naively, two separate mesophyll clusters can be expected, one for palisade and one for spongy parenchyma. FIL (YAB1) transcripts, markers for spongy parenchyma were enriched in C2 and C7, possibly indicating that these clusters may represent spongy parenchyma (Supplemental Figure S3A; Uemoto et al., 2018). Among the mesophyll clusters, C6 and C9 were enriched for rRNAs (Supplemental Data Set 4). We did not remove rRNA before sample preparation and did not filter cells that showed higher rRNA transcript levels since rRNA levels could vary between cells. Whether C6 and C9 represent unique biologically relevant cell populations in the leaf or are due to artifacts was not further evaluated. While the mesophyll clusters likely contain interesting information, we did not characterize them further, as we decided to focus on the vasculature.
Beyond the mesophyll, epidermal cells were identified in C13, based on enrichment of the epidermis-specific transcription factor AtML1 (Takada et al., 2013) and wax-related genes (Figure 3). Epidermal guard cells were found in C16, indicated by the specific enrichment of FAMA and guard cell-specific MAP kinases (Figure 3; Supplemental Figure 3A). Enrichment of transcripts of the purine transporter PUP1 and the homolog of carrot chitinase, EP3, indicated that C17 comprises hydathodes cells (Supplemental Figure 3A; Bürkle et al., 2003).
Several clusters showed a vasculature identity, based on the enrichment of known marker genes, indicating that the enrichment for these cell types was successful. The vasculature is composed of phloem and xylem, often separated by a meristematic layer, the procambium, which initiates and maintains vascular organization. We assigned clusters to the seven known cell types present in leaves (Figure 2). Additionally, we identified cell types with identities that had properties of two cell types, e.g. xylem and procambium.
Cluster 4 was found to contain bundle sheath (BS) and xylem cells and was subsequently subclustered into three cell populations, C4.1, C4.2, and C4.3 (Figure 2B;Supplemental Data Set 5). Subclusters C4.2 and C4.3 were enriched for the BS marker SCL23, and the sulfate transporter, SULTR2;2 (Figure 3; Supplemental Figure 3, B and C). C4.2 was enriched for genes involved in photosynthetic processes, possibly indicating a differentiation of BS cells (Supplemental Data Set 5). The existence of two BS clusters is consistent with morphological descriptions (Haritatos et al., 2000). C4.1, C10, C15, and C18, with a total of 478 cells, were assigned as vascular cell types (Figures 2 and 3; Supplemental Figure 3A, Supplemental Data Sets 2 and 3).
Degree of kinship relations among BS, XP, PC and PP
Clusters composed of BS, XP, and PC, and PP cells show a J-shaped relatedness in our UMAP plot (Figure 2). BS is related to two types of xylem cells (XP1 and XP2), that abut the PC. PC subclusters into two subsets closely related to XP1 (PCXP) and PP1 (PCPP), respectively. The PCPP is adjacent to the two PP clusters PP1 and PP2. XP3 and PP2 form a separate group, which together with XP2, form the hook of the J shape in our UMAP plot (Figure 2B).
Xylem-related clusters
The enrichment of GLR3;6 (Nguyen et al., 2018) and ACL5 markers identified C4.1 as parenchymatic xylem cells (Supplemental Figures 3, A and D–I). Further clustering of C4.1 resulted in two subgroups, XP1 and XP2, distinguished by the relative enrichment of photosynthetic genes (Supplemental Data Set 5). An additional subcluster with xylem identity, XP3, was observed as a subgroup of cells from C18. XP3 was enriched with xylem markers as well as those for VP (Endo et al., 2008; Supplemental Figure 3, D–I; Supplemental Figure 4). Transport proteins were enriched in the xylem clusters (XP1–XP3), as one may expect from their role in supplying tracheal elements with ions and nutrients for xylem translocation. These included amino acid transporters, such as the H+/amino acid symporter AAP6 and the UmamiT amino acid exporter family member UmamiT22 (Okumoto et al., 2002; Pilot et al., 2004; Dinkeloo et al., 2018). The transcript of amino acid export protein GDU4 was also enriched in the xylem clusters. Multiple SULTR low-affinity sulfate transporters, which play pivotal roles in sulfate transport in XP cells, were detected in the xylem cells as well as the boron transporter NIP6.1, the glucosinolate importer GTR2/NPF2.11 (Nour-Eldin et al., 2012; Madsen et al., 2014), the heavy metal transporter HMA2, and the plastidic bile acid transporter BAT5/BASS5 (Supplemental Data Set 6).
Phloem- and xylem-related subpopulations of the PC subclusters
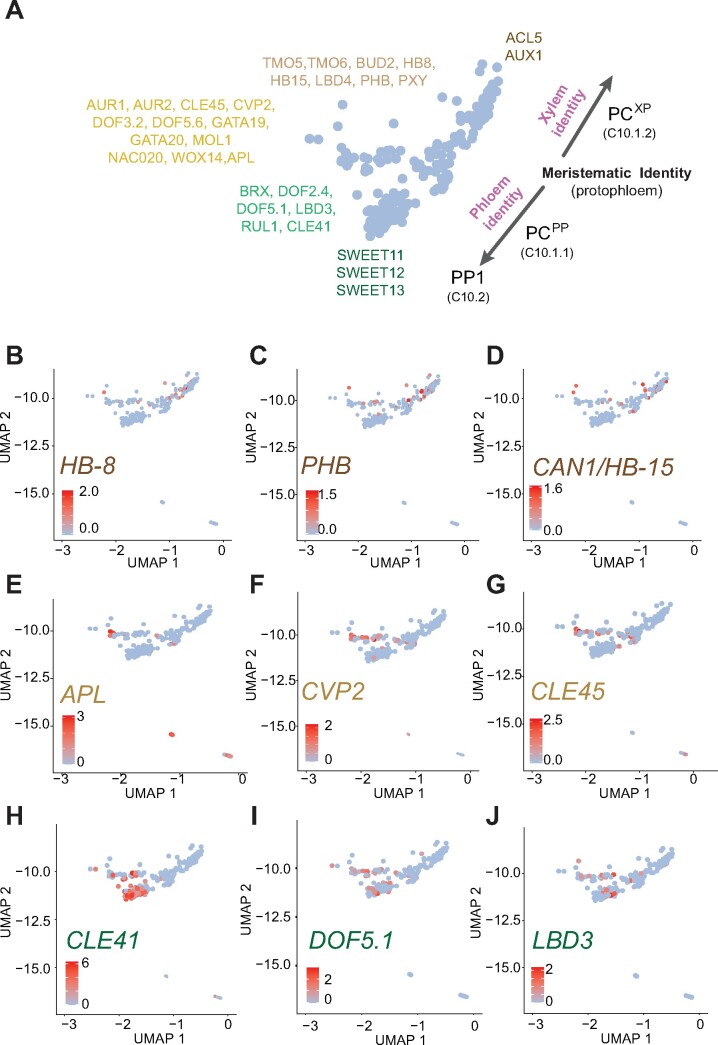
Arabidopsis procambium constitutes a bifacial stem cell population producing xylem on the adaxial pole and phloem on the abaxial pole (Elo et al., 2009; Sanchez et al., 2012). On our UMAP plot, the XP clusters are in proximity to Cluster 10. Cluster 10 could be further subdivided into three subclusters: the PP1 (C10.2) and two distinct PC populations, C10.1.1 and C10.1.2 (Figures 2 and 4A; Supplemental Data Sets 7 and 8). While cells in PCPP correspond to PC cells involved in the maintenance of meristematic identity and the differentiation of phloem cells, cells of PCXP appear to be more closely related to the formation of xylem cells, and is located closer to the XP clusters (Figure 4; Supplemental Data Sets 7 and 8).
Figure 4.
Identification of the procambium cell cluster with distinct procambium cell identities. (A) Schematics representing subpopulations of Cluster 10. Genes enriched in the subpopulations are indicated. (B–D) UMAP showing enrichment of transcripts of genes related to xylem differentiation. (E–G) UMAP showing enrichment of transcripts of genes related to maintenance of protophloem pluripotency and differentiation. (H–J) UMAP showing the distribution transcripts related to phloem differentiation.
Cells in the PCXP cluster were enriched with transcripts for homeodomain leucine-zipper (HD-Zip) transcription factor, HOMEOBOX GENE 8 (HB-8; Baima et al., 2001) transcripts, known to trigger xylem differentiation (Figure 4B). Adaxial HD-ZIP transcription factors, such as REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV/IFL1), PHABULOSA (PHB), and CORONA (CNA/AtHB15), were also enriched in this cluster (Figure 4, C and D; Supplemental Figure S5). Conversely, the PCPP cluster closest to the PCXP was enriched with factors involved in the maintenance of protophloem identity and pluripotency in roots and stem, such as COTYLEDON VASCULAR PATTERN 2 (CVP2), CVP2 LIKE 1 (CVL1; Rodriguez-Villalon et al., 2015), BARELY ANY MERISTEM 3 (BAM3), and CLAVATA3/EMBRYO SURROUNDING REGION-RELATED 45 (CLE45;Rodriguez-Villalon et al., 2014; Gujas et al., 2020; Figure 4, E–G; Supplemental Figure S5). BRX and OCTOPUS (Truernit et al., 2012), involved in protophloem differentiation at different ends of the developing protophloem cells, were also enriched in the nonoverlapping cells in PCPP. ALTERED PHLOEM DEVELOPMENT (APL), a major phloem marker, which inhibits xylem differentiation, was also detected in this region (Bonke et al., 2003; Figure 4E). Transcripts related to phloem differentiation, including CLE41, DOF5.1, and LBD3 were enriched in the lower region of PCPP (Figure 4, H–J; Supplemental Figure S5). Strikingly, the arrangement in the UMAP plot, of the PCPP cluster facing the PP cluster (C10.2) and the PCXP facing the XP clusters, is analogous to the morphological positioning. However, as the markers used for inferring the clusters are based not only on leaf markers, but also root and stems due to the lack of leaf procambium cell markers, further verification will be required to provide a comprehensive understanding of procambium cells in the mature leaf. Feature plots of additional procambium marker genes in the subpopulations of C10 are shown in Supplemental Figure S5.
PP clusters
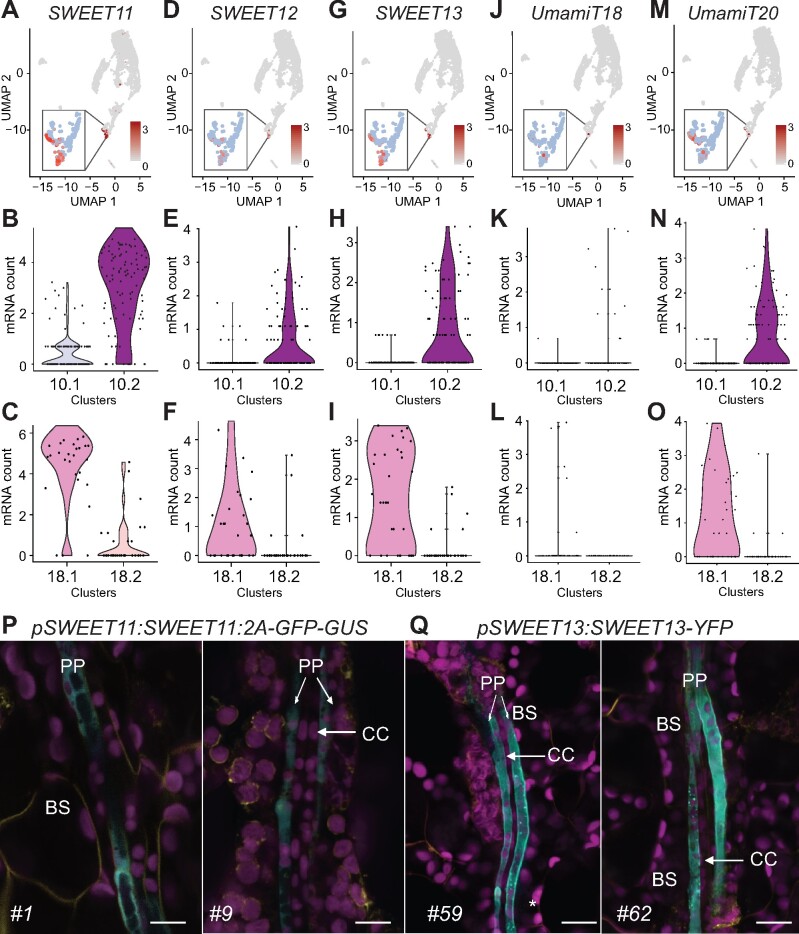
SWEET11 and SWEET12 efflux transporters had previously been reported to be expressed in the PP (Chen et al., 2012; Cayla et al., 2019). Sucrose released by the SWEETs into the apoplasm is taken up into the neighboring SE/CC via the sucrose/H+ symporter SUC2/SUT1 (Riesmeier et al., 1994; Gottwald et al., 2000). C10.2 (PP1) and C18.1 (PP2) were specifically enriched for SWEET11 and SWEET12 (Figures 3 and 5, A–F). Although the top marker genes were not mutually exclusive in either of the clusters (Supplemental Figure 6, A and B), genes related to callose deposition and cell wall thickening were over-represented in PP1, reflecting transfer cell identities (Supplemental Figure 6, C and D; Maeda et al., 2014). Among them, NAC056, a transcription factor known to play a role in cell wall ingrowth deposition in the PP, was enriched in PP1 in comparison to PP2 (Supplemental Figure 6C; Wu et al., 2018). On the other hand, genes involving photosynthesis were enriched in C18.1 indicating that C18.1 and C10.2 could be cells undergoing differential photosynthetic activity (Supplemental Figure 6E).
Both C10.2 and C18.1 are enriched for transcripts related to transport processes, reflecting the roles of PP as a critical cell type required for loading diverse substrates into the phloem (Supplemental Data Sets 7, 9–12). We identified an additional member of the Clade III SWEET family, SWEET13, in the cells that expressed SWEET11 and SWEET12 (Figure 5, G–I). SWEET13 transcripts show compensatory accumulation in sweet11;sweet12 double knockout mutants, indicating redundant roles with SWEET11 and SWEET12 (Chen et al., 2012). As it is challenging to unambiguously distinguish phloem cell types by cross-sections, we used a modified protocol from a recently developed confocal imaging method (Cayla et al., 2019), which allows us to distinguish PP cells from the CC, SE, and BS, based on the position of chloroplasts, presence of cell wall ingrowths, and cell size. Using this method, we were able to validate the presence of SWEET11 and SWEET13 in the PP cells in pSWEET11:SWEET11-2A-GFP (Figure 5P) and pSWEET13:SWEET13-YFP (Figures 5Q) expressing lines.
Figure 5.
Three SWEET sucrose transporters and UmamiT amino acid transporters mark the PP cluster. (A–O) UMAP and violin plots of C10 and C18 subclusters showing enrichment of SWEET11 (A–C), SWEET12 (D–F), SWEET13 (G–I), UmamiT18/SIAR1 (J–L), and UmamiT20 (M–O) transcripts in PP clusters. Subcluster 10.1 corresponds to PC, 18.2 to XP3, and 10.2 and 18.1 to PP. Inset show magnification of C10 and C18. (P and Q) Confocal microscopy images of pSWEET11:SWEET11-2A-GFP-GUS (P) and pSWEET13:SWEET13-YFP (Q) leaf showing GFP (P) or YFP (Q) fluorescence specific to PP. Magenta, chlorophyll autofluorescence. Yellow, FM4-64FX. Cyan GFP fluorescence (P) or YFP fluorescence (Q). Scale bars: 10 µm. BS, bundle sheath; CC, companion cell; PP, phloem parenchyma cells are marked. Numbers on the bottom left indicate independent transgenic lines.
UmamiT family members have been described as cellular exporters for amino acids (Ladwig et al., 2012). We thus speculated that UmamiTs might play analogous roles for the efflux of amino acids from PP as the SWEETs do for sucrose. Consistent with our hypothesis, we found UmamiT18/SIAR1 mRNA in PP (Figure 5, J–L). Vascular expression in leaves is supported by UmamiT18/SIAR1 transcriptional fusion lines (Ladwig et al., 2012). In addition, transcripts for six other members of the UmamiT family (UmamiT12, 17, 20, 21, 28, 30) were enriched in the two PP clusters (Figure 5, M–O; Supplemental Figure 7, A–F; Supplemental Data Sets 7 and 12). Notably, many of the PP-specific UmamiTs were coexpressed with each other as well as SWEET11 and 12 (Supplemental Figure 7G and Supplemental Data Set 13).
As many transcripts related to transport processes were specifically enriched in the PP clusters, and many of them were coregulated (Supplemental Data Set 13), this may indicate that they are subject to control by the same transcriptional networks. We therefore searched for transcription factors that might be responsible for coregulation of the functionally related transporters. We identified multiple leucine zipper genes (bZIP) either specific to the PP clusters (bZIP6, bZIP7, bZIP9), or present in PP and other clusters (TGA7, bZIP11; Supplemental Figure 8). We were able to confirm that bZIP9 promoter activity was specific for the PP cells in the leaf vasculature, using a transcriptional fusion, pbZIP9:GFP-GUS (Figure 6, A–D). Strong GUS activity was also detected in cell types known to be involved in apoplasmic transport steps, such as the unloading zones of anthers and seeds, and the veins of petals, receptacles, ovules, transmitting tract, and funiculi (Supplemental Figure S9). The cell specificity of bZIP9 is consistent with a possible role in the activation of target genes involved in transport process across different tissues. However, as previously reported, bzip9 single mutants did not display phenotypic differences compared with wild type (WT) plants (Supplemental Figure S10, A and B; Silveira et al., 2007; Veerabagu et al., 2014) and transcript level of PP markers were not significantly changed in the bzip9 mutant (Supplemental Figure S10C). As bZIP family members act as dimers or heteromers, hence are functionally connected, it may be necessary to characterize mutants in multiple PP-expressed bZIP family members to elucidate their role.
Figure 6.
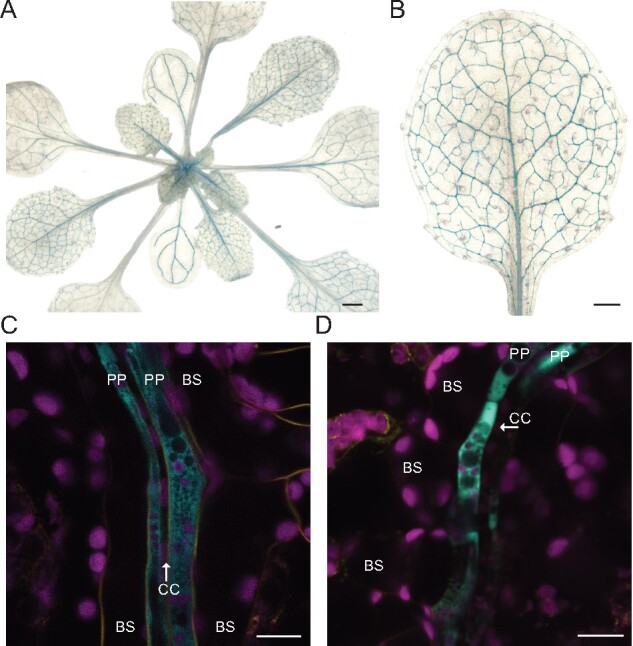
Reporter gene analysis of pbZIP9:GFP-GUS plants. (A and B) GUS-stained transgenic plants expressing the transcriptional pbZIP9:GFP-GUS reporter construct show GUS activity in the leaf vasculature. Four-week-old plants grown in LD conditions were used for GUS histochemistry. A magnified image of the seventh leaf is shown in b. Scale bars: 1 mm (A) and 0.5 mm (B). (C and D) Confocal microscopy images of two independent pbZIP9:GFP-GUS reporter lines showing GFP fluorescence specific for PP. Magenta, chlorophyll autofluorescence. Yellow, FM4-64FX, Cyan, GFP fluorescence. BS, bundle sheath; CC, companion cell; PP, phloem parenchyma cells are marked. Scale bar: 10 µm.
The CC cluster
CCs, which acquire carbon and nitrogen from the adjacent PP, but also maintain the functionality of the enucleate SEs, cluster separately from all other cell types. C15 was designated as CC based on the enrichment of transcripts for the sucrose/H+ symporter SUC2/SUT1, the H+-ATPase AHA3, and genes such as FTIP and APL (Figure 3;Supplemental Figure S3A). CCs were enriched for transcripts of various transporters, including the amino acid/H+ symporter AAP2 (Zhang et al., 2010), the potassium channel KAT1 (Schachtman et al., 1992), the hexose uniporters SWEET1 and SWEET4 (Chen et al., 2010), and the tonoplast peptide transporter PTR4/NPF8.4 (Weichert et al., 2012, p. 4; Supplemental Data Set 12). All these markers are also enriched in the CC translatome (Mustroph et al., 2009). A subset of cells in CC is specialized for synthesizing FT, a small mobile protein that acts as florigen (Chen et al., 2018). Although we could neither detect FT mRNA nor the presence of specialized CC cells in our data set, many genes involved in the regulation of FT and control of flowering, for instance, FTIP, CDF5, EFM, VOZ1, were enriched in CC (Liu et al., 2012; Yan et al., 2014; Henriques et al., 2017; Kumar et al., 2018; Supplemental Data Set 3).
PP and CC cells undergo differentiation through distinct pathways
The clear separation of PP and CC in the UMAP was striking and indicates the presence of distinct differentiation pathways. The MYB coiled-coil-type transcription factor APL plays a key role for the definition of phloem identity (Bonke et al., 2003). In our data set, APL was preferentially expressed in CC, but also detected in the PCPP cluster (Figure 4E;Supplemental Figure 3A). As APL function is necessary for the development of the SE/CC complex, one may propose two hypotheses: (i) APL serves as a master regulator of all phloem cells or (ii) PP develops independently of the SE/CC differentiation. While apl mutants are characterized by drastic reduction of CC marker SUC2, apl retains expression of multiple PP markers (Figure 7, A and B); PP marker transcripts were even > four-fold higher in the apl mutant (Figure 7B; Bonke et al., 2003). Whether the increase of PP-marker transcripts in apl is due to a compensatory mechanism in which APL represses PP differentiation, as suggested for xylem differentiation (Bonke et al., 2003), remains to be elucidated.
Figure 7.
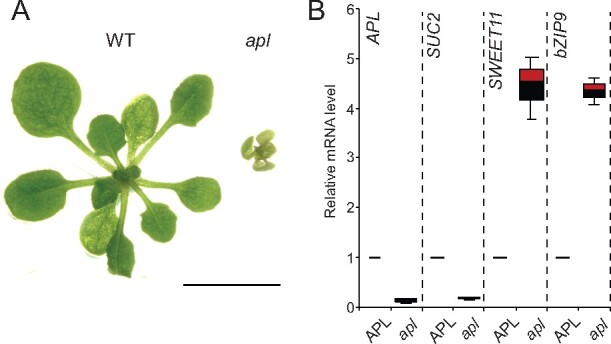
Distinct PP and CC marker gene expression in the apl mutant. (A) Morphology of apl mutant (right) grown on LD conditions for 2 weeks. WT plant grown under the same condition is shown on the left side. Scale bar: 1 cm. (B) RT-qPCR analysis of CC-marker gene (SUC2) and PP- marker genes (SWEET11 and bZIP9). Segregating seeds from heterozygous parents were plated on MS for 2 weeks. The first and second leaves from plants homozygous for the APL mutation (apl) and heterozygous for the mutation or WT (APL) were collected for RNA extraction and RT-qPCR. Three independent replicates showed similar results and a representative experiment with three technical replicates are shown (mean ± se, n = 3).
The existence or function of PP has not been described for Arabidopsis roots. One may hypothesize that the root phloem-pole pericycle plays a role in the unloading of small molecules and thus ontogenetically or functionally be related to leaf PP (Ross-Elliott et al., 2017). Cell clusters in the roots with transcript profiles related to leaf PP1 do not appear to correspond to phloem pole pericycle identity. The root single-cell transcriptome atlas from Wendrich et al. (2020), which includes the highest proportion of vascular cells, had identified the phloem and pericycle as distinct clusters. A comparison shows that 19% of the phloem-pole pericycle markers overlap with pericycle markers in root single-cell sequencing data set, indicating that PP and pericycle are distinct (Brady et al., 2007; Wendrich et al., 2020). Furthermore, none of the recent single-cell sequencing analyses of roots provide a detailed analysis of different phloem cells (Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shulse et al., 2019; Zhang et al., 2019; Wendrich et al., 2020). Therefore, we compared the markers from our leaf phloem clusters with the phloem markers identified by single-cell sequencing from roots (Wendrich et al., 2020). Surprisingly, root phloem markers had the highest overlap with C10.2 (Supplemental Figure 11A). Markers enriched in leaf C10.2 and the root phloem cluster include genes related to callose deposition, as found for leaf PP1 (Supplemental Figure 11B). This finding may indicate the existence of a yet unidentified cell type in roots with a leaf PP identity. Whether PP cells are present in the root and whether they play analogous roles as in the leaf remains to be elucidated.
Unique and complementary metabolic landscapes of PP and CC
Amino acid transporters of the AAP and UmamiT families are relatively nonselective and transport many of the proteogenic and other amino acids (Fischer et al., 1995; Dinkeloo et al., 2018). One may therefore assume that the translocation of most amino acids is similar and that relative amino acid levels are mainly determined by the relative rates of biosynthesis, that the amino acids all enter the translocation stream in similar ways and that the relative levels do not change substantially during translocation. However, labeling studies have shown that amino acids behave very differently, with some being effectively metabolized along the path of source-to-sink partitioning, while other stay largely unmetabolized (Atkins, 2000). Surprisingly, the amino acid distribution in stems is also varying for different amino acids (Metzler et al., 1995). These phenomena might be explained by differential distribution of metabolic activities in different vascular cell types. We therefore carried out a pathway activity analysis to define a pathway activity score (PAS) that quantifies the average expression of the pathway’s genes in one cell type relative to the other cell types (Xiao et al., 2019). The PAS values of the CC and PP were strikingly different for both amino acid biosynthetic and degradation pathways (Figure 8). While the activity of biosynthetic amino acid pathways was low in CC, PP-containing clusters showed elevated metabolic activity. A comprehensive analysis of pathway activities across all clusters revealed many other interesting features unique to specific cell types (Supplemental Figure 13). For instance, the epidermis cluster (C13) was enriched with biosynthetic activities for wax esters, cuticular wax, the suberin monomer, and very long chain fatty acids. These substances function as coatings in the epidermis, serving as a moisture barrier and protecting plants from pathogens. The PP-including cluster, C18, showed high activity of hormone pathways such as abscicic acid (ABA), ethylene, jasmonic acid (JA), and giberellic acid (GA), and the BS and XP clusters (C4) and the two PP-including clusters (C10 and C18) were enriched for glucosinolate biosynthesis activity (Supplemental Figures 12–15).
Figure 8.
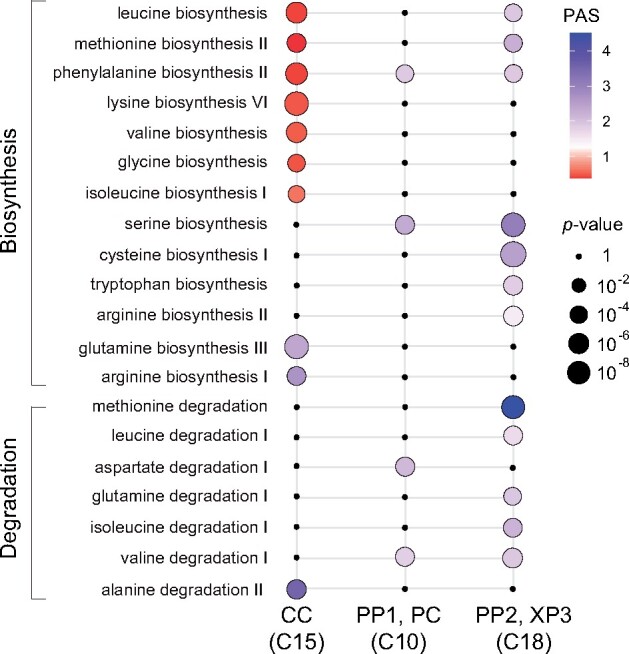
Amino acid biosynthesis and degradation pathways are differentially represented in the CC, PP, PC, and XP3 cells. Metabolic pathway activities of amino acid biosynthesis and degradation pathways in Clusters 15, 10, and 18. Statistical significance is represented as differences in dot size. Statistically insignificant values are shown as black dots (random permutation test, P >0.05). Colors represent the PAS; a score 1 (violet) indicates a higher activity. Activities were compared between all clusters in the scRNA-seq data set.
Cluster 19 as a candidate for putative S-cell cluster
Glucosinolate-rich cell types (S-cells) are found in floral stems of Arabidopsis but also described as sulfur-rich cells in the PP of the leaf (Koroleva et al., 2010). S-cells contain more than 10-fold higher sulfur levels , high levels of glucosinolate and act as functional shield of the plant vascular system from herbivores and pathogens. As S-cells were suggested to be present in the abaxial area of PP in leaves, we questioned whether S-cells are present in the PP clusters. Markers for S-cells are presently unavailable, therefore, we searched for markers that are known to be absent in S-cells. Proteomic analysis revealed that glucosinolates are not produced in S-cells, as biosynthetic enzymes could not be detected in isolated S-cell extracts. The promoter activities of key enzymes and markers for glucosinolate biosynthesis such as CYTOCHROME P450 83A1(CYP83A1) and BRANCHED-CHAIN AMINOTRANSFERASE 4 (BCAT4) were not detected in S-cells (Li et al., 2010; Nintemann et al., 2018; Hunziker et al., 2019). We detected transcripts of CYP83A1, BCAT4, as well as other transcripts encoding proteins involved in the synthesis of aliphatic and indolic glucosinolates in the BS, xylem (C4) and PP/PC/XP (C10, C18) clusters (Supplemental Figure 14). The enrichment of HIG1 transcription factor which activate glucosinolate biosynthetic genes (Gigolashvili et al., 2007; Supplemental Data sets 4 and 7) also exclude the cells in the PP clusters (C10.2, C18.1) from containing S-cells and suggest the PP, procambium, xylem, and BS cells as sites for glucosinolate biosynthesis.
A unique characteristic of the S-cells is that these cells undergo programmed cell death at the early stages of differentiation. We searched for cells that could be enriched with the transcripts related to programmed cell death and identified Cluster 19, a cluster distinct from, but closely spaced to the BS cells. This cluster was enriched with transcripts related to programmed cell death, hypersensitive response, defense, and immune response (Supplemental Data set 14). This cluster also showed high activity scores in insect chewing-induced glucosinolate breakdown pathway (Supplemental Figure 12B). This result is in line with the primary role of S-cells in releasing toxic compounds through glucosinolate breakdown upon chewing induced-mechanical disruption. Although we cannot rule out that these subset cells were clustered based on the stress response from the protoplasting process, they could serve as candidates as putative S-cells.
Cell-type-specific expression of plasmodesmatal proteins
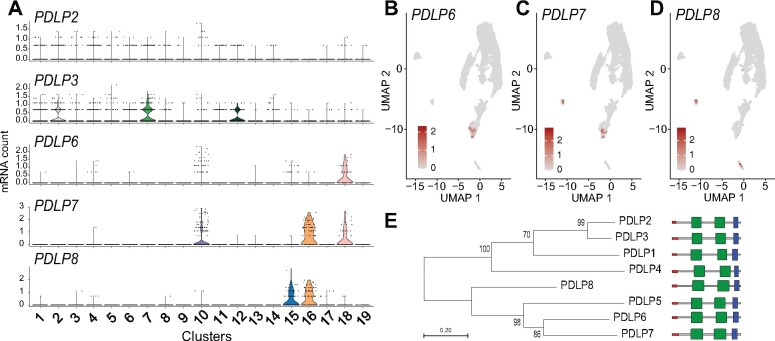
PDs are highly complex channels that interconnect plant cells, likely transporting solutes, metabolites, small RNAs, and proteins. Different cell types have unique types of PD, including that which connects CC to enucleate SEs to supply all components necessary for function. The PDs typically are branched on the CC side and have a single pore on the SE side. We here found that transcripts of PD genes such as the PLAMODESMATA-LOCATED PROTEIN (PDLP)s and MULTIPLE C2 DOMAINS AND TRNAMEMBRANE REGION PROTEIN (MCTP)s showed distinct expression patterns in different cell types (Figure 9, A–D; Supplemental Figure 16, A and B; Lee et al., 2011; Brault et al., 2019). For example, PDLP6, 7, and 8 transcripts were found in PP and CC, while PDLP2 and 3 transcripts were present in mesophyll cells (Figure 9, A–D;Supplemental Figure 16A). The cell-type specificity of PDLPs was correlated with their phylogenetic relationships (Figure 9E). Our results are consistent with data from GUS reporter fusions for the PDLP7 and 8 promoters (Tanvir, 2016). MCTP1 (FTIP1) and 3 transcripts were mainly enriched in the vasculature (Figure 3;Supplemental Figure 16B). In particular, MCTP1 (FTIP1) transcript was specifically enriched in CC, consistent with a role in trafficking the florigen protein FLOWERING LOCUS T (FT), from CC to SEs (Liu et al., 2012). In comparison, MCTP4, MCTP6, and MCTP15 are ubiquitously expressed in vascular and mesophyll cells (Supplemental Figure 16B). Overall, our data support the concept of PD-type-specific assembly of PDs.
Figure 9.
Transcript enrichment of PDLP genes in vascular cell types. (A) Violin plot showing transcript enrichment of PDLPs. (B–D) UMAP plot showing the enrichment of PDLP6 (B), PDLP7 (C), and PDLP8 (D) transcripts in the PP clusters (B and C), guard cell cluster (C and D), and CC cluster (D). (E) Phylogenetic analysis of PDLPs in Arabidopsis. The phylogenetic tree was generated with the maximum likelihood method implemented in PhyML (Lemoine et al., 2019). Percent support values from 1,000 bootstrap samples are shown. Protein motifs predictions are based on the SMART database (http://smart.embl-heidelberg.de). DUF26 domains, transmembrane region, and signal peptide are shown in green, blue, and red, respectively.
Discussion
Despite the advances in scRNA-seq technologies, application to plant cells still faces challenges since the cell walls of plant cells must be removed for the release of individual cells. Different cell types are likely under different osmotic pressure in the intact plant, which might result in breakage if the osmotic conditions are not adequate. Recent studies using Arabidopsis (Liu et al., 2020), rice (Wang et al., 2020), as well as maize (Bezrutczyk et al., 2021) aerial tissues clearly demonstrate difficulty in capturing vascular cell types reflecting the need for careful strategy development. A key cell type in the vasculature, the PP, was not identified in any of the recently published scRNA-seq data from roots (Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shulse et al., 2019; Zhang et al., 2019; Wendrich et al., 2020).
Here, we systematically optimized protoplast isolation protocols to enrich vascular cell types and produced a single-cell transcriptome and metabolic activity score atlas that covers essentially all known cell types in the Arabidopsis leaf. The scRNA-seq confirmed that except for trichomes, abaxial epidermis, and guard cells, which were intentionally removed, all known nucleus-containing cells of the leaf were represented in the dataset. In particular, all cells from the leaf vasculature were identified.
The vascular cells had identities clearly distinct from those of the epidermis, guard cells, and mesophyll. The BS formed a supercluster together with all vascular cells including the vascular meristem, except the CCs. A major finding within this atlas was that the kinship relation of vascular cells was highly similar to the actual morphology of the vasculature, with the exception of the CC, which formed a unique island separate from BS, xylem, procambium, and the PP. Surprisingly, although we used 6-weeks-old plants for the analysis, in which the developmental processes were expected to be completed, the arrangement of the XP–PCXP–PCPP–PP clusters in the UMAP plot reflected a potential developmental trajectory. Future pseudotime trajectory analyses from the dataset, or comparison to data from younger leaves, will allow us to better understand the transition from meristematic cells to differentiated phloem or xylem cell types and trans-differentiation processes from PP into transfer cells. Some clusters showed gradients, indicating a transition, in which transcriptional modules (e.g. for xylem identity) decreased in favor of phloem modules. One example is the PC, in which cells closer to the xylem shared some xylem properties, while those closer to PP shared PP properties. A similar behavior was seen for the VP. It is conceivable that the well-studied dorsoventral cues, or cues from neighboring cells, are responsible for these gradients.
From their positioning inside the phloem, one may naively have expected the PP and CC to cluster together. CCs are unique since they have to fulfill their own tasks, e.g. import sucrose from PP, and at the same time maintain the function of the adjacent enucleate SEs, living cells that act as conduits for assimilate translocation (Bel and Knoblauch, 2000). Based on this dual role, it is possibly not surprising that they form such a distinct island. Whether PP and CC derive from a common ancestor remain unclear since the ontogeny of PP is unknown. The ontogenesis of CC is much better understood (Bonke et al., 2003). SE/CC mother cells divide asymmetrically to produce CC and SE. The APL transcription factor is a master regulator for SE/CC development, and a repressor of xylem identity (Bonke et al., 2003). Based on our data, it seems unlikely that APL is responsible for driving PP ontogeny but may implicate alternative pathways for PP differentiation.
A major discovery was the assignment of Clusters 10.2 and 18.1 as PP, providing insights into the PP transcriptome. Although C10.2 and 18.1 appear to represent different states, C10.2 having transfer cell identity and C18.1 being enriched in photosynthetic activity, it will be interesting to determine if these clusters represent spatially different cells types or developmental trajectories. The PP has essential roles in sucrose transfer to the SE/CC, but also likely many other functions in transport, metabolism and signaling. SWEET13 is a gene that may act in a compensatory role, as evidenced by higher mRNA levels in sweet11;12 mutants. Here, we detected SWEET13 in the same cells as SWEET11 and 12 (Chen et al., 2012). Notably, in a parallel study, we found that while in main veins and rank-1 intermediate veins of maize, the orthologs in maize, named ZmSWEET13a, b and c, are likely also expressed in PP, in the C4-specific rank-2 intermediate veins that are mainly responsible for phloem loading, the three ZmSWEET13s are not in PP, but in two abaxial BS cells (Bezrutczyk et al., 2021). These data indicated to us that maize uses a different path for phloem loading of sucrose as compared to, for example, Arabidopsis or potato (Kühn et al., 1997).
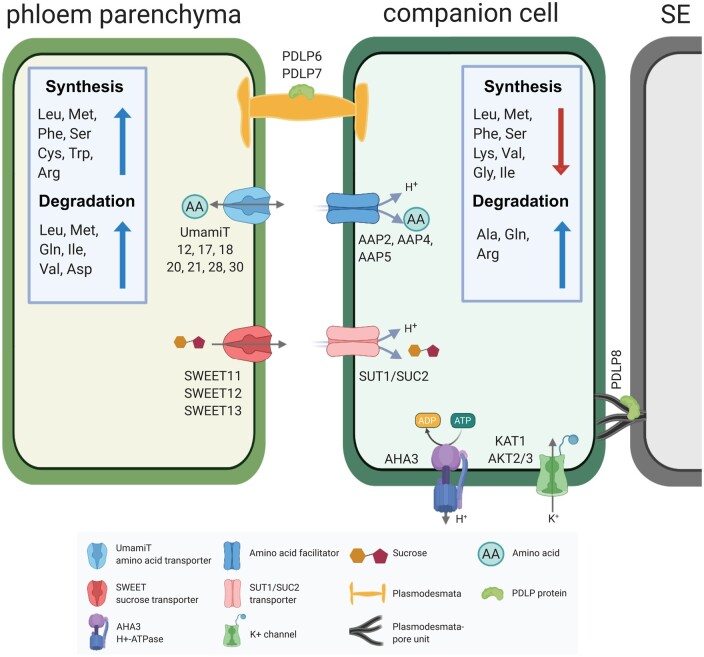
In addition to the clade III SWEETs, scRNA-seq provided extensive insights into other genes enriched in PP, in particular, identifying transcripts of seven members of the UmamiT amino acid transporter family. UmamiTs are rather nonselective transporters involved at sites where cellular amino acid efflux is required. For example, UmamiT18/SIAR1 transcripts, similar to SWEET11 and 12, were detected in the chalazal region of developing seed (Ladwig et al., 2012; Chen et al., 2015). Knockout mutants showed reduced accumulation of amino acids in seeds, likely due to both a reduction in efflux from leaves and reduced efflux from the chalaza (Ladwig et al., 2012). Of note, the SWEETs and UmamiTs showed high coexpression indices. One may thus hypothesize that the genes share transcriptional control, and thus it will be interesting to analyze whether common transcription factors binding sites are present in the PP-specific genes. SWEETs and SUTs seem to act as a pair, one responsible for sucrose efflux from PP, the other for active import into the SE/CC. UmamiTs also seem to act in pairs, with UmamiTs in PP and AAP amino acid H+/symporters including AAP2, 4 and 5 in the CC (Fischer et al., 1998; Figure 10). These complementary functions are consistent with the clear separation of the clusters in the UMAP plots. A striking result from the comparison of PP and CC was that their metabolism was also highly differentiated. In particular, we found major differences regarding amino acid metabolic pathways. Both isotope labeling studies as well as amino acid localization by 2D-NMR had indicated that amino acids behave very differently regarding entry into metabolism along with the translocation pathway and entry into the phloem (Metzler et al., 1995; Atkins, 2000). This difference could thus likely be explained by the metabolic activities along the path, rather than their nonselective transporters.
Figure 10.
Hypothetical phloem loading process in an Arabidopsis leaf. Sucrose produced during photosynthesis in the mesophyll cells is transported across the bundle sheath to the PP. Biosynthesis and catabolism of multiple amino acids is highly active in the PP. Transporters present in the PP secrete sucrose (SWEET11, 12, 13) or amino acids (UmamiT11, 12, 17, 18, 20, 21, 28, 30) into the apoplasm. H+/sucrose cotransporters import sucrose (SUT1/SUC2) and amino acids (AAP2, 4, 5) into the SE/CC. The H+ gradient required for the active import of sucrose and amino acids into the SE/CC is provided by plasma membrane H+-ATPases. The membrane potential is maintained by the potassium channels (KAT1 and AKT2/3). Symplasmic transport is mediated by PDLP6 and PDLP7 in the PD in PP cells and PDLP8 enriched in the PD-pore unit of the CC. Note that the schematic is based on transcript levels and the distribution could differ at the protein level. Schematics was made in © BioRender—https://biorender.com.
Cell type-specific metabolic pathway analysis revealed many other interesting aspects. For example, hormone metabolism varied between cell types. Cell types responsible for hormones biosynthesis are largely unknown. The metabolic analysis provides a map that may guide identification of hormone biosynthesis and transport pathways in leaves. For instance, high activities of ABA, ethylene, JA, and GA biosynthesis pathways were detected in C18 (which includes the PP), consistent with the high number of ABA, JA, and GA transporters in the phloem (Kuromori et al., 2014; Kanno et al., 2016, p. 13; Ye et al., 2016; Nguyen et al., 2017). As most transporters show functional redundancy and display diverse substrate specificity, this data set serves as a source to identify cell type-specific redundant family members for the generation of multiple mutants, to eliminate or verify interacting partners, and to identify yet-unknown transporters.
Besides the insights into both metabolism and into apoplasmic transport pathways, the study also showed that cell types with unique types of PD express particular paralogs of PD-specific proteins, such as PDLPs or MCTPs. The combined analysis of sym- and apoplasmic fluxes of ions, metabolites, and signaling molecules at the cellular level will likely enable a much better understanding of the physiology of the leaf.
In summary, scRNA-seq enabled us to identify unique and distinct features of the different vascular cell types present the leaf. Through transcriptomic and metabolic pathway analyses at the single-cell level, we identified potential roles of PP not only in the transport of sugars, but also in amino acid transport. Importantly, we also identified unexpected roles of PP in hormone biosynthesis and defense-related responses. The information in this study provides key resources to develop strategies influencing the flux of ions, metabolites, and signals.
Materials and methods
Enrichment of vascular protoplasts
Protoplasts were isolated from mature leaves of 6-weeks-old Arabidopsis Col-0 plants grown under short-day (8-h light/16-h dark) conditions at a PAR of 60 µmol m−2 s−1. For protoplast isolation, the tape-sandwich method (Wu et al., 2009) was modified and applied for removal of the abaxial epidermis, guard cells, and trichomes. The adaxial side of the fully developed leaves was stabilized by placing on the time tape, and the abaxial side was adhered to the 3M Magic tape. The abaxial epidermis was removed by pulling off the 3M Magic tape. Two cuts were made on each side of the major vein of the peeled leaves using a razor blade (Swann-Morton, Sheffield). Seven to nine abaxial epidermis-peeled and cut leaves attached to time tape were immediately immersed in the petri dish containing 15 mL freshly prepared protoplast isolation solution (1% Cellulase Onuzuka R-10 (Duchefa, Haarlem), 0.3% macerozyme R-10, (Duchefa, Haarlem), 0.6 M Mannitol, 20 mM MES (2-(N-Morpholino)ethanesulfonic acid hydrate) pH 5.7, 20 mM KCL, 1 mM dithiothreitol (DTT), 10 mM CaCl2, and 0.1% bovine serum albumin, BSA). DTT and BSA were added to the enzyme solution to protect the protoplasts. Leaves were shaken at 30 rpm (IKA Rocker 3D orbital shaker). on a platform shaker for 2 h. The release of the protoplasts was monitored every 30 min by checking released cells under the microscope or by monitoring what was left on the time tape. After confirming the release of the protoplasts into the solution, 10 mL of wash buffer (0.6 M Mannitol, 20 mM MES pH 5.7, 20 mM KCl, 1 mM DTT, 10 mM CaCl2, and 0.1% BSA) was slowly added to the petri dish containing the protoplasts. The solution was filtered into a round bottom 20 mL tube using a 70-µm pore size filter (Corning, NY, USA). The protoplast solution was centrifuged at 100 x g for 3 min in a swinging rotor. The protoplasts were washed four times with 20 mL washing buffer. After the final wash step, protoplasts were slowly resuspended in 1 mL wash buffer and gently filtered twice using a 40-µm Flowmi cell strainer (Bel-Art SP Scienceware, NJ, USA). The number of protoplasts was counted with the C-Chip Neubauer improved hemocytometer (NanoEnTek, Seoul) under a light microscope (Leica VT1000, Wetzlar). The viability of protoplasts was determined using trypan blue solution (Gibco; Thermo Fisher Scientific, MA, USA).
RT-qPCR
Total RNA was extracted using the RNeasy Kit (Qiagen, Hilden). cDNA synthesis and gDNA removal steps were performed using QuantiTect Reverse Transcription Kit (Qiagen, Hilden), qPCR was performed on Stratagene Mx3000P (Agilent Technologies, CA, USA) using the Lightcycler 480 SYBR Green I Master Mix (Roche, Penzberg). Transcript levels were quantified using the relative standard curve method. Values were normalized to the level of the internal control, UBQ10. Primers used for RT-qPCR are listed in Supplemental Table 1. For RT-qPCR of apl mutants, segregating seeds from heterozygous parents were sown on MS media. RNA was extracted using 2-weeks-old plants grown under LD conditions.
Single cell RNA-seq library preparation and sequencing
Two biological replicates were performed with the aim of capturing approximately 7,000 leaf protoplasts for each replicate. Two batches of freshly isolated protoplasts were obtained from eight leaves from different plants (per batch) adjusted to 700–900 cells/μL and loaded into the 10× Genomics Chromium single cell microfluidics device according to the Single Cell 3′ Reagent Kit v2 protocol (10× Genomics, CA, USA). Eleven cycles were used for cDNA amplification and 12 cycles were used for final PCR amplification of the adapter-ligated libraries. The quality and size of the final library was verified on a DNA High Sensitivity Bioanalyzer Chip (Agilent Technologies, CA, USA), and libraries were quantified using the NEBNext Library Quantification Kit for Illumina (New England Biolabs, MA, USA). scRNA-seq library sequencing was performed on a NextSeq platform (Illumina Inc, CA USA), using the sequencing parameters 26,8,0,98 (c.ATG, Tübingen).
Generation of single-cell expression matrices
Reads were aligned to the Arabidopsis thaliana reference genome (Araport 11) using Cell Ranger 3.0.2 (10× Genomics, CA, USA) with default parameters. The output files for the two replicates were aggregated into one gene–cell expression matrix using Cell Ranger aggregate with the mapped read depth normalization option.
Dimensionality reduction, UMAP visualization, cell clustering analysis, and correlation analysis
The Seurat R package (version 3.1.0; Satija et al., 2015; Butler et al., 2018) was used for dimensionality reduction analysis. The SCTransform option was used for normalization, scaling the data and finding variable genes using default parameters (Hafemeister and Satija, 2019). During normalization, potential variation due to mitochondrial mapping percentage was removed. We did not remove rRNA before sample preparation and did not filter cells with higher rRNA transcript levels, since rRNA levels can vary among cells. Fifty principal components were selected as input for a graph-based approach to cluster cells by cell type using a resolution value of 0.8 in all clustering analyses. UMAP dimensional reduction (McInnes et al., 2018) was used for two-dimensional visualization using 10 principal components, 30 neighboring points and a minimum distance of 0.1. Subclustering was performed using the same parameters. For the correlation analysis between single-cell replicates across individual clusters, the average expression of cells within a cluster was calculated and the Pearson correlation coefficient was determined.
Identification of differentially expressed genes and cluster-specific marker genes
Genes differentially expressed across clusters or subclusters were identified by comparing average transcript levels in cells of a given cluster to that of cells in all other clusters using the Seurat package likelihood ratio test (Bimod). The following cutoffs were applied: average expression difference ≥0.25 natural Log and q < 0.01. Cluster-specific marker genes were selected from among the differentially expressed genes based on the criteria that marker genes must be expressed in >10% of cells within the cluster (PCT1, percentage of cells where the feature is detected in the first group), and <10% of cells across all other clusters (PCT2, percentage of cells where the feature is detected in the second group).
Bulk RNA-seq library preparation and sequencing
Total RNA was extracted from leaves (not protoplasted) and leaf protoplasts isolated using the same method for the single-cell sequencing using the RNeasy Kit (Qiagen, Hilden). On-column DNase treatment was performed to remove residual gDNA using the RNase-free DNase kit (Qiagen, Hilden) following manufacturer recommendations. Two biological replicates were made for leaf and leaf protoplast samples. The integrity of the RNA was confirmed using Agilent RNA 6000 Nano Chip (Agilent Technologies, CA, USA) and LabChip GX (PerkinElmer, MA, USA). RNA concentration was measured using the Qubit Fluorometer using the RNA broad range quantification kit (Thermo Fisher, MA, USA). For mRNA poly-A enrichment, 5 µg of total RNA was purified using the Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, MA, USA). Libraries were constructed using the ULTRA II directional library kit (New England Biolab, MA, USA), and size selection was done using SPRI beads (New England Biolab, MA, USA) following the manufacturer manual with the following exceptions: 7 min of fragmentation time and 26.5 and 10 µL SPRI beads pre-PCR were used to enrich ∼400 bp inserts. Library amplification included 10 PCR cycles, and 0.7 volumes of (35 µL) of SPRI beads were used for post-PCR purification. QC-tested libraries (Agilent Technologies, CA, USA) were sequenced on an Illumina HiSeq 2500 lane with 150-bp paired-end (Novogene, Beijing).
Bulk RNA-seq analysis
Paired-end reads (150 bp) were aligned to the A. thaliana reference genome (Araport11) using STAR (Dobin et al., 2012; maximum intron length of 2 kbp). Differential expression analysis was carried out in R (v3.6.1) using Bioconductor (v3.9) and DESeq2 (v1.24; absolute Log2FC ≥1 and q-value <0.05). For correlation analysis of gene expression between protoplasted and nonprotoplasted bulk tissues, the Log2 (mean FPKM + 1) expression values were calculated for each gene. Pearson correlation coefficient was determined in R.
Pathway activities in different cell types
We followed the formulation introduced in Xiao et al. (2019) to calculate a PAS, which depends on the mRNA level of its constituent genes. The mRNA count of gene i in cell k was denoted as gi,k. We first normalized the mRNA count of a gene in a cell by the average for all genes in the cell, and denoted this normalized level as g'i,k. The mean transcript level Ei,j of gene i in cell type j is then defined as the mean g'i,k over all cells of that type, , where nj is the number of cells classified as cell type j, and k is the index for individual cells. Ei,j is normalized by the average transcript level of gene i across all cell types to become the relative transcript level ri,j of gene i in cell type j: , where N is the total number of cell types. The PAS of pathway t in cell type j, denoted as pt,j, is then a weighted average of the relative transcript levels across the pathway genes: ; here, mt is the number of genes in pathway t, and the weight wi of gene i is defined as the reciprocal of the number of pathways that include gene i, to ensure a stronger influence of pathway-specific genes. As the relative mRNA levels ri,j are centered around 1, the same is true for the pathway activity pt,j, with pt,j <1 corresponding to underrepresentation of pathway t in cell type j relative to the pathway’s activity across all cell types; conversely, pt,j >1 indicates a higher than average activity in cell type j. To assess the statistical significance of a pt,j value, we performed a permutation test, shuffling the cell type labels of the genes a thousand times to simulate the null distribution of pt,j under the assumption of no systematic cell type-specific pathway activity; we defined an empirical P-value by comparing pt,j to this null distribution. AraCyc pathway lists (Mueller et al., 2003) used for the analysis can be found in Supplemental Data Set 15.
Transgenic plants and T-DNA insertion lines
For the generation of pbZIP9:GFP-GUS lines, a 2,289-bp fragment of the bZIP9 gene promoter was amplified using bZIP9pro attB1 and bZIP9pro attB2 primers using Col-0 genomic DNA as template. The corresponding PCR fragment was purified and used for GATEWAY BP reaction into pDONR221 and cloned into the destination vector pBGWFS7,0 through LR reaction. For generating pSWEET11:SWEET11-2A-GFP-GUS lines, a 4,784-bp fragment consisting of promoter and SWEET11 genomic region including all exons and introns was amplified using SWEET11-2A-attB1 and SWEET11-2A-attB2 primers. The SWEET11-2A-attB2 reverse primer contained the 2A cleavage sequence. The genomic SWEET11 with the 2A cleavage site was cloned into pDONR221 and subcloned into the pKGWFS7,0 vector through LR reaction. To generate pSWEET13:SWEET13-YFP, a 3,941-bp fragment including SWEET13 promoter and its genomic region was amplified using SWEET13attB1 and SWEET13attB2 primers and cloned into the donor vector pDONR221-f1 by BP reaction. Subsequently, the fragment was sub-cloned into a gateway-compatible vector, pEG-TW1 (Kim et al., 2019) to generate pSWEET13:SWEET13-YPF construct by LR reaction. Transformation of plants was performed using the floral dip method (Zhang et al., 2006). Primers used for amplification are listed in Supplemental Table 1. The bzip9 mutant (salk_093416c) was obtained from the Nottingham Arabidopsis Stock Centre. Homozygous mutants were identified by PCR as described (http://signal.salk.edu/tdnaprimers.2.html) using the primer pairs listed in Supplemental Table 1.
GUS histochemistry
GUS staining was performed as described with minor modifications (Martin et al., 1992). Tissues were fixed with 90% ice-cold acetone. After applying vacuum for 10 min, acetone was removed and replaced by prestaining solution [1 mM EDTA, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 100 mM sodium phosphate (pH 7.0), 1% Triton-X-100]. After 10 min vacuum, the prestaining solution was replaced with staining solution containing 1 mM EDTA, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 100 mM sodium phosphate (pH 7.0), 1% Triton-X-100, and 2 mM X-Gluc and further vacuum infiltrated for 15 min in the dark. The tissues in staining solutions containing X-Gluc were incubated at 37°C in the dark for 4 h. Ethanol series were performed from 25% to 70% ethanol in 30 min steps.
Sample preparation for imaging leaf vasculature
Rosette leaves were excised by cutting the leaf petiole from 5-weeks-old plants grown under SD conditions. Double-sided tape was used to attach the petiole and the upper lamina of the leaf on the slide glass (abaxial side facing upward, as in the tape-sandwich method). The abaxial epidermis was peeled using the 3M Magic tape and immediately covered with water. The region of interest was excised with a razor blade and moved to a new slide glass. Samples were applied with FM4-64FX (Sigma-Aldrich, Missouri; 5 µg/mL) for 5 min for staining the cell membrane and imaged immediately. PP cells were identified based on several criteria only clearly detectable in longitudinal sections, i.e. plasma membrane invaginations, and cell diameter (about 8–10 μm). CCs were distinguished from the PP cells based on the regularly aligned chloroplast strings. BS cells were identified by their large size and chloroplast alignment on one side of the cell (Cayla et al., 2019).
Confocal imaging
Fluorescence images were captured using a Leica TCS SP8 confocal microscope with a 20× or 40× objective with water immersion. GFP, YFP, FM4-64FX, and chlorophyll autofluorescence signals were acquired using the following settings: GFP, excitation 488 nm (white-light laser) and emission 492–552 nm; YFP, excitation 514 nm and emission 510–565 nm; FM4-64FX, excitation 561 nm and emission 599–680 nm; chlorophyll autofluorescence, excitation 638 nm, and emission 645–738 nm.
Phylogenetic analysis
Reported amino acid sequences from A. thaliana PDLPs were aligned using the tool MAFFT with 0.123 and 1.53 gap extend penalty and gap opening penalty values, respectively. Maximum likelihood tree was constructed with full-length amino acid sequences with the software NGPhylogeny.fr. The sequence alignment file and the tree file are provided in Supplemental Files 1 and 2. Bootstrap percentages at the branch points were estimated from 1,000 bootstrap replications. The evolutionary distances were computed using the amino acid replacement LG matrix and the units are the number of amino acid substitutions per site.
Data availability
All sequencing data, both raw and processed, have been deposited in Gene Expression Omnibus GEO (www.ncbi.nlm.nih.gov/geo/) under the accession number GSE161482. The Arabidopsis leaf scRNA-seq data set will be deposited in the PscB (https://www.zmbp-resources.uni-tuebingen.de/timmermans/plant-single-cell-browser/; Ma et al., 2020) for visualization.
Supplemental data
Supplemental Figure 1. Optimization of vascular protoplast enrichment.
Supplemental Figure 2. Reliability of Arabidopsis leaf scRNA-seq data set.
Supplemental Figure 3. Enrichment of known marker genes for assigning cell types to clusters.
Supplemental Figure 4. Vascular parenchyma-specific marker genes enriched in Cluster 18 (XP3).
Supplemental Figure 5. Distinct procambium cell identities in C10 subclusters.
Supplemental Figure 6. Distinct identities of PP1 and PP2.
Supplemental Figure 7. Transcripts of UmamiT amino acid transport family members are enriched in the PP.
Supplemental Figure 8. Transcript enrichment of multiple bZIP transcription factors in the PP clusters.
Supplemental Figure 9. GUS staining of pbZIP9:GFP-GUS plants showed promoter activity in multiple tissues and organs.
Supplemental Figure 10. Phenotypic analysis of bzip9.
Supplemental Figure 11. Comparison of root and leaf phloem markers.
Supplemental Figure 12. Hormone and glucosinolate pathway activity across clusters.
Supplemental Figure 13. Metabolic pathway activity across all cell types.
Supplemental Figure 14. Enrichment of transcripts related to glucosinolate biosynthesis and transport.
Supplemental Figure 15. Enrichment of transcripts related to glucosinolate transport.
Supplemental Figure 16. Cell type-specific transcript enrichment of plasmodesmatal proteins.
Supplemental Table 1. Primers used in this study.
Supplemental Movie 1. Three-dimensional UMAP plot of Arabidopsis leaf single cells.
Supplemental File 1. Arabidopsis thaliana PDLPs amino acid sequence alignment file (fasta).
Supplemental File 2. Arabidopsis thaliana PDLPs tree file (nhx).
Supplemental Data Set 1. Overview of the RNA-seq data generated in this study and list of differentially expressed genes (DEGs) in bulk RNA-seq of protoplasted and nonprotoplasted leaves.
Supplemental Data Set 2. Correlation of two scRNA-seq biological replicates and the number of cells in the clusters and subclusters.
Supplemental Data Set 3. Known marker genes used for cluster annotaton.
Supplemental Data Set 4. Cluster specific marker genes and DEGs across all clusters.
Supplemental Data Set 5. Cluster 4 subcluster markers.
Supplemental Data Set 6. List of genes enriched in the xylem clusters (XP1, XP2, XP3).
Supplemental Data Set 7. Cluster 10 subcluster markers.
Supplemental Data Set 8. Cluster 10.1 subcluster markers.
Supplemental Data Set 9. Cluster 18 subcluster markers.
Supplemental Data Set 10. GO enrichment analysis of C18.1 vs. C10.2 markers.
Supplemental Data Set 11. GO enrichment analysis of Cluster 10 marker genes.
Supplemental Data Set 12. List of genes related with transport processes enriched in the CCs (C15) and phloem parenchyma (C10.2, C18.1).
Supplemental Data Set 13. List of genes coexpressed with UmamiT20 and SWEET11.
Supplemental Data Set 14. GO enrichment analysis of Cluster 19 marker genes.
Supplemental Data Set 15. Gene-to pathway table.
Supplementary Material
Acknowledgments
We thank Colin P. S. Kruse (Los Alamos National Laboratory, USA) for constructive comments on the metabolic pathway activity analysis, Sebastian Hänsch for assistance with confocal microscope imaging (Center for Advanced Imaging, HHU, Germany), Ykä Helariutta and Pawel Roszak for sharing APL/apl seeds (University of Cambridge, UK). We also thank Xiaoqing Qu (Bio-Protocol, China) for helping with the generation of pSWEET13:SWEET13-YFP construct and Sylvie Dinant (INRA, France) for sharing information on live-cell imaging of phloem cells.
Funding
This research was supported by the National Science Foundation (SECRETome Project: Systematic Evaluation of CellulaR ExporT from plant cells, IOS-1546879), Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC-2048/1—project ID 390686111 and SFB 1208—Project-ID 267205415, as well as the Alexander von Humboldt Professorship to W.B.F.
Conflict of interest statement. None declared.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) are: Wolf B. Frommer (frommew@hhu.de) and Ji-Yun Kim (jiyun.kim@hhu.de).
References
- Arun Chinnappa KS, Nguyen TTS, Hou J, Wu Y, McCurdy DW (2013) Phloem parenchyma transfer cells in Arabidopsis - an experimental system to identify transcriptional regulators of wall ingrowth formation. Front Plant Sci 4:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CA (2000) Biochemical aspects of assimilate transfers along the phloem path: N-solutes in lupins. Austr J Plant Physiol 27: 531–537 [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJE, Knoblauch M (2000) Sieve element and companion cell: the story of the comatose patient and the hyperactive nurse. Funct Plant Biol 27: 477–487 [Google Scholar]
- Bezrutczyk M, Zöllner NR, Kruse CPS, Hartwig T, Lautwein T, Köhrer K, Frommer WB, Kim J-Y (2021) Evidence for phloem loading via the abaxial bundle sheath cells in maize leaves. The Plant Cell 33: 531--547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mähönen AP, Hauser M-T, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee J-Y, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brault ML,, Petit JD,, Immel F,, Nicolas WJ,, Glavier M,, Brocard L,, Gaston A,, Fouché M,, Hawkins TJ, Crowet J‐M, et al. (2019) Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep 20: e47182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle L, Cedzich A, Döpke C, Stransky H, Okumoto S, Gillissen B, Kühn K, Frommer WB (2003) Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J 34: 13–26 [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotech 36: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayla T, Le Hir R, Dinant S (2019) Live-cell imaging of fluorescently tagged phloem proteins with confocal microscopy. Methods Mol Biol 2014: 95–108 [DOI] [PubMed] [Google Scholar]
- Chen LQ,, Hou B-H, Lalonde S, Takanaga H, Hartung ML, Qu X-Q, Guo W-J, Kim J-G, Underwood W, Chaudhuri B. et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Lin IW, Qu X-Q, Sosso D, McFarlane HE, Londoño A, Samuels AL, Frommer WB (2015) A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27: 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu X-Q, Hou B-H, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207. [DOI] [PubMed] [Google Scholar]
- Chen Q, Payyavula RS, Chen L, Zhang J, Zhang C, Turgeon R (2018) FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proc Natl Acad Sci USA 115: 2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K, Timmermans MCP (2019) Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev Cell 48: 840–852.e5 [DOI] [PubMed] [Google Scholar]
- Dinkeloo K, Boyd S, Pilot G (2018) Update on amino acid transporter functions and on possible amino acid sensing mechanisms in plants. Sem Cell Dev Biol 74: 105–113 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2012) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Martin AP, Andriunas F, Offler CE, Patrick JW, McCurdy DW (2010) GIGANTEA is a component of a regulatory pathway determining wall ingrowth deposition in phloem parenchyma transfer cells of Arabidopsis thaliana. Plant J 63: 651–661 [DOI] [PubMed] [Google Scholar]
- Elo A, Immanen J, Nieminen K, Helariutta Y (2009) Stem cell function during plant vascular development. Sem Cell Dev Biol 20: 1097–1106 [DOI] [PubMed] [Google Scholar]
- Endo A, Sawada Y, Takahashi H, Okamoto M, Ikegami K, Koiwai H, Seo M, Toyomasu T, Mitsuhashi W,, Shinozaki K,. et al. (2008) Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol 147: 1984–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA (2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W-N, André B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, Frommer WB (1998) Amino acid transport in plants. Trends Plant Sci 3: 188–195 [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB (1995) Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem 270: 16315–16320 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock H-P, Müller C, Weisshaar B, Flügge U-I (2007) The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 50: 886–901 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujas B, Kastanaki E, Sturchler A, Cruz TMD, Ruiz-Sola MA, Dreos R, Eicke S, Truernit E, Rodriguez-Villalon A (2020) A reservoir of pluripotent phloem cells safeguards the linear developmental trajectory of protophloem sieve elements. Curr Biol 30: 755–766.e4 [DOI] [PubMed] [Google Scholar]
- Hafemeister C,, Satija R (2019) Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritatos E, Medville R, Turgeon R (2000) Minor vein structure and sugar transport in Arabidopsis thaliana. Planta 211: 105–111 [DOI] [PubMed] [Google Scholar]
- Henriques R, Wang H, Liu J, Boix M, Huang L-F, Chua N-H (2017) The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytologist 216: 854–867 [DOI] [PubMed] [Google Scholar]
- Hunziker P, Halkier BA, Schulz A (2019) Arabidopsis glucosinolate storage cells transform into phloem fibres at late stages of development. J Exp Bot 70: 4305–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, Dorrity MW, Saunders L, Bubb KL, Trapnell C, Fields S, Queitsch C, Cuperus JT (2019) Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell 31: 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J,, Dengler N (2004) Vein pattern development in adult leaves of Arabidopsis thaliana. Int J Plant Sci 165: 231–242 [Google Scholar]
- Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, Koshiba T, Kamiya Y, Ueda M, Seo M (2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun 7: 13245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E-J, Lee S-H, Park C-H, Kim S-H, Hsu C-C, Xu S, Wang Z-Y, Kim S-K, Kim T-W (2019) Plant U-Box40 mediates degradation of the brassinosteroid-responsive transcription factor BZR1 in Arabidopsis Roots. Plant Cell 31: 791–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Gibson TM, Cramer R, Stain C (2010) Glucosinolate-accumulating S-cells in Arabidopsis leaves and flower stalks undergo programmed cell death at early stages of differentiation. Plant J 64: 456–469 [DOI] [PubMed] [Google Scholar]
- Kühn C, Franceschi VR, Schulz A, Lemoine R, Frommer WB (1997) Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275: 1298–1300 [DOI] [PubMed] [Google Scholar]
- Kumar S, Choudhary P, Gupta M, Nath U (2018) VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1) and VOZ2 interact with CONSTANS and promote photoperiodic flowering transition. Plant Physiol 176: 2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Sugimoto E, Shinozaki K (2014) Intertissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiol 164: 1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwig F, Stahl M, Ludewig U, Hirner AA, Hammes UZ, Stadler R, Harter K, Koch W (2012) Siliques are Red1 from Arabidopsis acts as a bidirectional amino acid transporter that is crucial for the amino acid homeostasis of siliques. Plant Physiol 158: 1643–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F, , Correia D, , Lefort V, , Doppelt-Azeroual O, , Mareuil F, , Cohen-Boulakia S, , Gascuel O ( 2019) NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Research 47: W260–W265. 10.1093/nar/gkz303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kristiansen KA, Hansen BG, Halkier BA (2010) Cellular and subcellular localization of flavin-monooxygenases involved in glucosinolate biosynthesis. J Exp Bot 62: 1337–1346 [DOI] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H (2012) FTIP1 Is an essential regulator required for florigen transport. PLoS Biol 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou Y, Guo J, Li J, Tian Z, Zhu Z, Wang J, Wu R, Zhang B,, Hu Y,. et al. (2020) Global dynamic molecular profiling of stomatal lineage cell development by single-cell RNA sequencing. Mol Plant 13: 1178–1193 [DOI] [PubMed] [Google Scholar]
- Ma X, , Denyer T, , Timmermans MCP (2020) PscB: A Browser to Explore Plant Single Cell RNA-Sequencing Data Sets. Plant Physiol 183: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen SR, Olsen CE, Nour-Eldin HH, Halkier BA (2014) Elucidating the role of transport processes in leaf glucosinolate distribution. Plant Physiol 166: 1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Song W, Sage T, DellaPenna D (2014) Role of callose synthases in transfer cell wall development in tocopherol deficient Arabidopsis mutants. Front Plant Sci 5: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Wöhner RV, Hummel S, Willmitzer L, Frommer WB (1992) The GUS reporter system as a tool to study plant gene expression. In Gallagher SR, ed, GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. Academic Press, San Diego, pp 23–43 [Google Scholar]
- McInnes L, Healy J, Melville J (2018) UMAP: uniform manifold approximation and projection for dimension reduction. arXiv eprint arXiv:1802.03426
- Metzler A, Izquierdo M, Ziegler A, Köckenberger W, Komor E, von Kienlin M, Haase A, Decorps M (1995) Plant histochemistry by correlation peak imaging. Proc Natl Acad Sci USA 92: 11912–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CT, Kurenda A, Stolz S, Chételat A, Farmer EE (2018) Identification of cell populations necessary for leaf-to-leaf electrical signaling in a wounded plant. Proc Natl Acad Sci USA 115: 10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CT, Martinoia E, Farmer EE (2017) Emerging jasmonate transporters. Mol Plant 10: 659–661 [DOI] [PubMed] [Google Scholar]
- Nintemann SJ, Hunziker P, Andersen TG, Schulz A, Burow M, Halkier BA (2018) Localization of the glucosinolate biosynthetic enzymes reveals distinct spatial patterns for the biosynthesis of indole and aliphatic glucosinolates. Physiol Plant 163: 138–154 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jørgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488: 531–534 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W (2002) High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J Biol Chem 277: 45338–45346 [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB (2004) Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoeva T, ten Hove CA, Saiga S, Weijers D (2016) Molecular characterization of Arabidopsis GAL4/UAS enhancer trap lines identifies novel cell-type-specific promoters. Plant Physiol 171: 1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB (1994) Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J 13: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Villalon A, Gujas B, Kang YH, Breda AS, Cattaneo P, Depuydt S, Hardtke CS (2014) Molecular genetic framework for protophloem formation. Proc Natl Acad Sci USA 111: 11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Villalon A, Gujas B, van Wijk R, Munnik T, Hardtke CS (2015) Primary root protophloem differentiation requires balanced phosphatidylinositol-4,5-biphosphate levels and systemically affects root branching. Development 142: 1437. [DOI] [PubMed] [Google Scholar]
- Ross-Elliott TJ, Jensen KH, Haaning KS, Wager BM, Knoblauch J, Howell AH, Mullendore DL, Monteith AG, Paultre D,, Yan D,. et al. (2017) Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. eLife 6: e24125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KH, Huang L, Kang HM, Schiefelbein J (2019) Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol 179: 1444–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, Nehlin L, Greb T (2012) From thin to thick: major transitions during stem development. Trends Plant Sci 17: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A (2015) Spatial reconstruction of single-cell gene expression data. Nat Biotech 33: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchuk MG, Donner TJ, Head P, Scarpella E (2008) Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol 148: 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI, Lucas WJ, Anderson JA, Gaber RF (1992) Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258: 1654–1658 [DOI] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Ciobanu D, Lin J, Yoshinaga Y, Gouran M, Turco GM, Zhu Y, O’Malley RC, Brady SM, et al . (2019) High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep 27: 2241–2247.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira AB, Gauer L, Tomaz JP, Cardoso PR, Carmello-Guerreiro S, Vincentz M (2007) The Arabidopsis AtbZIP9 protein fused to the VP16 transcriptional activation domain alters leaf and vascular development. Plant Sci 172: 1148–1156 [Google Scholar]
- Takada S, Takada N, Yoshida A (2013) ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140: 1919. [DOI] [PubMed] [Google Scholar]
- Tanvir Z (2016) Expression domain analysis of four members of the plasmodesmata-localized protein family in Arabidopsis. University of Delaware, Delaware [Google Scholar]
- Truernit E, Bauby H, Belcram K, Barthélémy J, Palauqui J-C (2012) OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 139: 1306. [DOI] [PubMed] [Google Scholar]
- Uemoto K, Araki T, Endo M (2018) Isolation of Arabidopsis palisade and spongy mesophyll cells. Methods Mol Biol 1830: 141–148 [DOI] [PubMed] [Google Scholar]
- Veerabagu M, Kirchler T, Elgass K, Stadelhofer B, Stahl M, Harter K, Mira-Rodado V, Chaban C (2014) The interaction of the Arabidopsis Response Regulator ARR18 with bZIP63 mediates the regulation of PROLINE DEHYDROGENASE expression. Mol Plant 7: 1560–1577 [DOI] [PubMed] [Google Scholar]
- Wang Y, Huan Q, Chu X, Li K, Qian W (2020) Single-cell transcriptome analyses recapitulate the cellular and developmental responses to abiotic stresses in rice. bioRxiv: 2020.01.30.926329
- Weichert A, Brinkmann C, Komarova NY, Dietrich D, Thor K, Meier S, Suter Grotemeyer M, Rentsch D (2012) AtPTR4 and AtPTR6 are differentially expressed, tonoplast-localized members of the peptide transporter/nitrate transporter 1 (PTR/NRT1) family. Planta 235: 311–323 [DOI] [PubMed] [Google Scholar]
- Wendrich JR, Yang BJ, Vandamme N, Verstaen K, Smet W, Van de Velde C, Minne M, Wybouw B, Mor E,, Arents HE,. et al. (2020) Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370: eaay4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F-H, Shen S-C, Lee L-Y, Lee S-H, Chan M-T, Lin C-S (2009) Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hou J, Yu F, Nguyen STT, McCurdy DW (2018) Transcript profiling identifies NAC-domain genes involved in regulating wall ingrowth deposition in phloem parenchyma transfer cells of Arabidopsis thaliana. Front Plant Sci 9: 341–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Dai Z, Locasale JW (2019) Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun 10: 3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Shen L, Chen Y, Bao S, Thong Z, Yu H (2014) A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev Cell 30: 437–448 [DOI] [PubMed] [Google Scholar]
- Ye Z-W, Lung S-C, Hu T-H, Chen Q-F, Suen Y-L, Wang M, Hoffmann-Benning S, Yeung E, Chye M-L (2016) Arabidopsis acyl-CoA-binding protein ACBP6 localizes in the phloem and affects jasmonate composition. Plant Mol Biol 92: 717–730 [DOI] [PubMed] [Google Scholar]
- Zhang L, Tan Q, Lee R, Trethewy A, Lee Y-H, Tegeder M (2010) Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22: 3603–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T-Q, Xu Z-G, Shang G-D, Wang J-W (2019) A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol Plant 12: 648–660 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protocols 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data, both raw and processed, have been deposited in Gene Expression Omnibus GEO (www.ncbi.nlm.nih.gov/geo/) under the accession number GSE161482. The Arabidopsis leaf scRNA-seq data set will be deposited in the PscB (https://www.zmbp-resources.uni-tuebingen.de/timmermans/plant-single-cell-browser/; Ma et al., 2020) for visualization.