Abstract
The external application of nitrogen (N) fertilizers is an important practice for increasing crop production. However, the excessive use of fertilizers significantly increases production costs and causes environmental problems, making the improvement of crop N-use efficiency (NUE) crucial for sustainable agriculture in the future. Here we show that the rice (Oryza sativa) NUE quantitative trait locus DULL NITROGEN RESPONSE1 (qDNR1), which is involved in auxin homeostasis, reflects the differences in nitrate () uptake, N assimilation, and yield enhancement between indica and japonica rice varieties. Rice plants carrying the DNR1indica allele exhibit reduced N-responsive transcription and protein abundance of DNR1. This, in turn, promotes auxin biosynthesis, thereby inducing AUXIN RESPONSE FACTOR-mediated activation of transporter and N-metabolism genes, resulting in improved NUE and grain yield. We also show that a loss-of-function mutation at the DNR1 locus is associated with increased N uptake and assimilation, resulting in improved rice yield under moderate levels of N fertilizer input. Therefore, modulating the DNR1-mediated auxin response represents a promising strategy for achieving environmentally sustainable improvements in rice yield.
DULL NITROGEN RESPONSE1-mediated auxin homeostasis improves N metabolism and yield in rice via auxin response factors, making this pathway a promising target for sustainable agriculture in the future.
Introduction
Nitrogen (N) is one of the most important nutrients for plant growth and grain yield. Accordingly, the application of N fertilizers has led to a dramatic boost in global crop yields over the last half-century (Godfray et al., 2010; Liu et al., 2013; Chen et al., 2014; Li et al., 2017). Excess N input, however, causes significant environmental damage and reduces biodiversity (Guo et al., 2010; Kaiser, 2018). Therefore, developing new crop varieties that combine high yields with improved N-use efficiency (NUE) is an urgent goal for achieving more sustainable agriculture with a minimum demand for N (Luo et al., 2020). Although ammonium () is considered to be a favorable source of N for plants growing under anaerobic conditions, a significant amount of nitrate (; accounting for as much as 40% of total available N) is absorbed and utilized by paddy field-grown rice (Oryza sativa) due to the occurrence of nitrification in the rhizosphere (Li et al., 2008; Xu et al., 2012; Chen and Ma, 2015; Yang et al., 2015; Gao et al., 2019).
To date, major components involved in acquisition, transport, assimilation, and signal transduction in rice have been well-characterized. uptake is mainly mediated by the dual-affinity transporter OsNRT1.1B (Chen and Ma, 2015; Duan and Zhang, 2015; Hu et al., 2015a; Fan et al., 2016), the low-affinity transporter OsNPF2.4 (Xia et al., 2015), and the high-affinity transporters OsNRT2.1, OsNRT2.2, and OsNRT2.3a, which require interactions with the partner protein OsNAR2.1 (Yan et al., 2011). OsNRT1.1B and OsNPF2.4 also function in long-distance transport, whereas redistribution is mediated by OsNPF6.1 (Tang et al., 2019). Following uptake, is reduced to nitrite () by the OsNR2-encoded NADH/NADPH-dependent reductase (NR) for assimilation (Gao et al., 2019).
Accumulating evidence indicates that crosstalk between plant N metabolism and several phytohormone signaling pathways, including gibberellin, auxin, cytokinin, abscisic acid, and strigolactone, affects plant growth and development (Sakakibara et al., 2006; Kiba et al., 2011; Luo et al., 2018). Therefore, adjusting the concentrations of particular phytohormones and/or modifying their signaling mediators have emerged as potential strategies for enhancing plant NUE. For example, two gibberellin-regulated transcription factors in rice, GROWTH-REGULATING FACTOR 4 (GRF4) and NITROGEN-MEDIATED TILLER GROWTH RESPONSE5 (NGR5), globally activate plant N metabolic processes and enhance N-mediated tillering, respectively, serving as targets for improving crop NUE and yield (Li et al., 2018; Wu et al., 2020). Similarly, the crosstalk between auxin and N metabolism has been investigated since the 1930s. Early studies suggested that N deficiency reduces auxin content in the above-ground tissues of plants (Avery et al., 1937; Avery and Pottorf, 1945), whereas more recently, the opposite pattern was observed in roots. Specifically, low levels lead to auxin accumulation in roots in Arabidopsis thaliana, soybean (Glycine max), wheat (Triticum aestivum), and maize (Zea mays), suggesting that auxin plays an important role in relaying external N availability to root growth responses (Caba et al., 2000; Walch-Liu et al., 2006; Tian et al., 2008; Ma et al., 2014). Indeed, high N levels reduce root auxin contents by modifying the transcript levels of genes involved in auxin influx (e.g. AUX1, LAX2, and LAX3) and efflux (e.g. PIN1, PIN2, PIN4, and PIN7). In contrast, exogenous treatment with the auxins 1-naphthaleneacetic acid (NAA) and indole-3-acetic acid (IAA) restored primary root growth in maize, even under high conditions that are inhibitory to root elongation (Tian et al., 2008).
In Arabidopsis, the dual-affinity transceptor (transporter/receptor) NRT1.1 can also transport auxin and is essential for initiating lateral root proliferation in soil patches with high levels (Krouk et al., 2010). On the other hand, lateral root emergence and growth triggered by low N availability is dependent on the functional auxin biosynthetic enzyme TRYPTOPHAN (Trp) AMINOTRANSFERASE-RELATED 2 (Ma et al., 2014). Auxin signaling components have also been identified as N-responsive regulators of root architecture. downregulates the expression of the microRNA miR167 while upregulating the expression of miR393, which targets the Auxin Response Factor (ARF) gene ARF8 and the auxin receptor gene AFB3, respectively, in Arabidopsis. Consequently, mutants lacking either ARF8 or AFB3 exhibit a compromised root developmental response to external N (Wu et al., 2006; Gifford et al., 2008; Vidal et al., 2010). Nevertheless, although the genes involved in auxin biosynthetic and signaling pathways have been extensively studied, our knowledge of how auxin-related genes respond to the fluctuations in N availability is still very limited. Moreover, whether auxin, in turn, regulates plant N metabolism and could therefore be utilized to improve crop NUE remains to be determined.
Here, we carried out quantitative trait locus (QTL) analysis of uptake with a set of 71 single-segment substitution lines (SSSLs) obtained by crossing rice varieties Hua–Jing–Xian (HJX74) and IRAP9. This analysis revealed DULL NITROGEN RESPONSE1 (DNR1), a candidate gene that might be involved in modulating auxin homeostasis. DNR1 diverges between indica and japonica rice varieties, which might contribute to the difference in uptake activity between the two subspecies. The indica DNR1 variant (DNR1indica) confers reduced DNR1 protein abundance, increased auxin accumulation, and enhanced uptake and assimilation. Further analysis revealed that the auxin-responsive OsARFs promote NUE and grain yield by trans-activating the expression of genes related to metabolism . Therefore, the DNR1–auxin–OsARFs module represents an excellent target for simultaneously improving NUE and yield potential in rice.
Results
Identification of qDNR1 as a major QTL associated with uptake in rice
We previously examined the 15 uptake rates of 36 semi-dwarf1 (sd1)-containing indica rice varieties and identified GRF4 as a regulator of NUE (Li et al., 2018). In this study, we expanded our survey to include both indica and japonica sd1-containing varieties and found significant variations in their 15 uptake rates. For example, indica rice variety HJX74 exhibited much higher uptake activity than japonica variety IRAP9 (Figure 1A). Therefore, we constructed a set of 71 SSSLs by crossing IRAP9 (the donor parent) with HJX74 (the recurrent parent). Each SSSL contained a single chromosome segment from IRAP9 substituted into the HJX74 genetic background (Xi et al., 2006). Subsequent QTL analysis of these 71 SSSLs revealed two loci responsible for the reduced uptake rate in IRAP9. One of these loci, qDNR1, was identified on the short arm of chromosome 1 defined by the markers R15 and STS-1-20.2 (Figure 1, B and D; Supplemental Data Set 1). The other locus, qDNR10, is located on chromosome 10 and coincides (in map position) with OsNRT1.1B. The variation in OsNRT1.1B in indica and japonica rice subspecies has been reported to contribute to the difference in their uptake capacities (Hu et al., 2015a).
Figure 1.
DNR1, a major QTL for uptake in rice, represses auxin accumulation and is N-responsive. (A) 15 uptake rates of the parental plants indica HJX74 and japonica IRAP9. P-values were generated from two-sided Student’s t tests. Data are mean ± sem (n = 6). (B) QTL analysis of a set of 71 SSSLs, each containing a single IRAP9 chromosomal segment in the HJX74 background. The positions of the two major QTLs are shown. (C) Genotyping of progeny homozygous for qDNR1 delimited the locus to ∼132-kb stretch defined by the markers R15 and STS-1-20.2. Chromosomal segments homozygous for IRAP9 and HJX74 alleles are represented by filled and open bars, respectively. (D) 15 uptake rates for the 16 SSSLs. 15 uptake rate of HJX74 served as a control. Different letters denote significant differences (P < 0.05) from Duncan’s multiple range test. Data are mean ± sem (n = 6). (E) Morphology of mature NIL plants. Scale bar, 15 cm. (F) DNR1 transcript abundance in roots. Transcript abundance was measured relative to NIL-DNR1HJX74 (set to 1). P-values were generated from two-sided Student’s t tests. Data are mean ± sem (n = 3). (G) DNR1 protein abundance in roots. The two DNR1 bands represent proteins encoded by two different DNR1 transcripts, one encoding a protein containing 394 amino acids and the other encoding a C-terminally truncated protein with 208 amino acids. Data are representative of three independent experiments, with similar results. (H) Auxin content in roots. P-values were generated from two-sided Student’s t tests. Data are mean ± sem (n = 3). (I) Trp content in roots. P-values were generated from two-sided Student’s t tests. Data are mean ± sem (n = 3). (J) 15 uptake rate in roots. P-values were generated from two-sided Student’s t tests. Data are mean ± sem (n = 6). (K) DNR1 mRNA levels in plants supplied with increasing levels of N (0.15 N, 0.1875 mM NH4NO3; 0.3 N, 0.375 mM NH4NO3; 0.6 N, 0.75 mM NH4NO3; 1 N, 1.25 mM NH4NO3). Transcript abundance was measured relative to 1N (set to 1). Different letters denote significant differences (P < 0.05) from Duncan’s multiple range test. Data are mean ± sem (n = 3). (L) DNR1 protein abundance in response to increasing N supply. The two DNR1 bands represent proteins encoded by two different DNR1 transcripts, one encoding a protein containing 394 amino acids and the other encoding a C-terminally truncated protein with 208 amino acids. Data are representative of three independent experiments, with similar results.
We used 144 BC1F2 and 947 BC2F2 plants developed from a backcross between 27-055 (donor parent) and HJX74 (recurrent parent) for fine-scale mapping of qDNR1. The candidate region was narrowed down to ∼11-kb segment flanked by the markers DN11 and DN12, which harbors the gene promoter region of the LOC_Os01g08270 locus (Supplemental Figure S1A). Sequence comparison of the candidate gene DNR1 revealed multiple single nucleotide polymorphisms (SNPs) and small indels (insertions/deletions) of a few base pairs (bp) between HJX74 and IRAP9, as well as a 520-bp deletion (−1,728 to −1,209 in the promoter region) in HJX74 (Supplemental Figure S1, A and B). This 520-bp segment was present in all japonica varieties but was absent in all indica varieties from our collection, which coincides with the generally lower DNR1 transcript levels and higher uptake rates in indica compared to japonica (Supplemental Figure S2). Therefore, the downregulation of DNR1, likely conferred by the truncated indica DNR1 promoter, appears to promote uptake in rice.
Phylogenetic analysis suggested that DNR1 is closely related to Arabidopsis VAS1, which encodes an aminotransferase that catalyzes the conversion of indole-3-pyruvate to L-Trp, thus antagonizing auxin biosynthesis (Supplemental Figure S3A; Zheng et al., 2013; Pieck et al., 2015). The homologous relationship between these two genes was also confirmed by a similar phylogenetic analysis reported recently (Matsuo et al., 2020). Moreover, amino acid sequence alignment indicated that DNR1 and VAS1 share high sequence similarity (Supplemental Figure S3B). Interestingly, although a previous study showed that Arabidopsis VAS1-green fluorescent protein (GFP) fusion protein localizes to the cytoplasm (Zheng et al., 2013), our analysis of both rice protoplasts transfected with DNR1 fused with GFP (DNR1–GFP) and transgenic rice plant carrying the p35S:DNR1-GFP construct showed that DNR1 is present in both the nucleus and cytoplasm in rice (Supplemental Figure S4, A and B). Isolation of nuclear and cellular protein extracts confirmed the presence of the DNR1–GFP fusion protein in both compartments, with the nucleus-localized transcription factor GRF4 serving as a control (Supplemental Figure S4C; Che et al., 2015; Duan et al., 2015; Hu et al., 2015b).
To investigate how the two DNR1 variant alleles confer the variation in uptake observed in the two rice subspecies, we developed a near-isogenic line (NIL) in the HJX74 background that carries the japonica DNR1 allele from IRAP9 (NIL-DNR1IRAP9; Figure 1E). NIL-DNR1IRAP9 exhibited semi-dwarfism, increased tillering and reduced panicle branching, grain number, and grain yield due to the increased abundance of DNR1 mRNA and DNR1 protein compared to NIL-DNR1HJX74 (Figure 1, F and G; Supplemental Figure S5, A–G). In addition, NIL-DNR1IRAP9 plants had a relatively low auxin content but high Trp content, which is consistent with the predicted function of DNR1 (Figure 1, H and I). NIL-DNR1IRAP9 plants also exhibited reduced uptake and reduced NR activity (Figure 1J;Supplemental Figure S5H). These results indicate that DNR1indica is a reduced function allele conferring agronomically desirable traits. We examined the expression pattern of DNR1 by quantitative polymerase chain reaction with reverse transcription (RT-qPCR) and found that it is mainly expressed in root, leaf, and node tissues (Supplemental Figure S4D). This expression pattern is consistent with the abovementioned phenotypic changes in NIL-DNR1IRAP9 with regard to both root uptake and shoot architecture. Moreover, in addition to regulating uptake and reduction, we discovered that DNR1 is regulated by N: the transcript level of DNR1 increased with increasing N supply, which led to increased DNR1 protein abundance (Figure 1, K and L).
DNR1-mediated auxin homeostasis regulates N metabolism
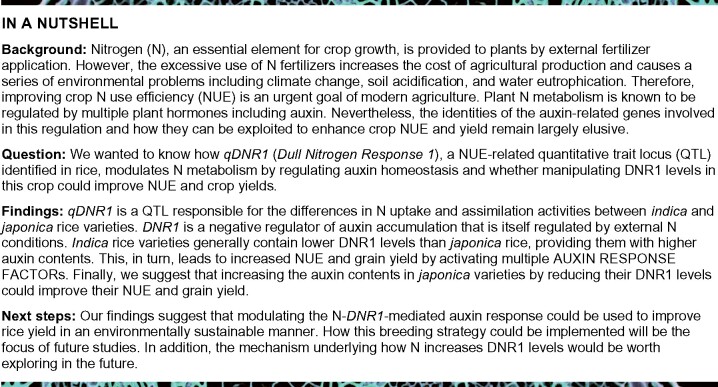
To further explore the phenotypic consequences of varying DNR1 expression and therefore auxin concentration in rice, we constructed pAct:DNR1-Flag overexpression lines and dnr1 mutants in the Zhonghua 11 (ZH11; a japonica rice variety) background (Figure 2A;Supplemental Figure S6). The dnr1 mutation was generated by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), which resulted in a 2-bp deletion in the second exon of DNR1 and therefore likely disrupts its normal gene function (Supplemental Figure S6, C and D). Notably, dnr1 plants exhibited a tall stature, reduced tiller number, and increased IAA but reduced Trp content, whereas the opposite phenotypes were observed in pAct:DNR1-Flag overexpression lines (Figure 2, B–E).
Figure 2.
DNR1 regulates uptake and assimilation by modulating plant auxin level. (A) Morphology of mature ZH11, ZH11 pAct:DNR1-Flag-1, ZH11 pAct:DNR1-Flag-2, and dnr1 plants. Scale bar, 20 cm. (B, C) Plant height (B) and number of tillers per plant (C) of ZH11, ZH11 pAct:DNR1-Flag-1, ZH11 pAct:DNR1-Flag-2, and dnr1 plants. Data are mean ± sem (n = 16). (D, E) Auxin content (D), and Trp content (E) in the roots of ZH11, ZH11 pAct:DNR1-Flag-1, ZH11 pAct:DNR1-Flag-2, and dnr1 plants. Data are mean ± sem (n = 3). (F, G) 15 uptake rate (F) and NR activities (G) of ZH11, ZH11 pAct:DNR1-Flag-1, ZH11 pAct:DNR1-Flag-2, and dnr1 plants under mock and external IAA treatments (1 μM). Data are mean ± sem (n = 3). H, I, mRNA abundances of OsNRT1.1B (H) and OsNRT2.3a (I) in roots relative to ZH11 (set to 1). Data are mean ± sem (n = 3). (J, K) mRNA abundances of OsNPF2.4 (J) and OsNIA2 (K) in shoots relative to ZH11 (set to 1). Data are mean ± sem (n = 3). (B–K) Different letters denote significant differences (P < 0.05) from Duncan’s multiple range test.
We also examined whether altered DNR1 levels lead to changes in root architecture. Compared to ZH11 and dnr1, the overexpression of DNR1 repressed lateral root initiation and proliferation (Supplemental Figure S7). These results suggest that the upregulation of DNR1 reduces root auxin contents, leading to a reduction in the size and production of lateral roots (Figure 2D;Supplemental Figure S7). Importantly, the lack of DNR1 promoted both uptake and NR activity compared to ZH11, whereas plants overexpressing DNR1 exhibited the lowest metabolic activities (Figure 2, F and G). The repressive effects of DNR1 overexpression on metabolism was reversed by exogenous IAA treatment, which increased both the uptake rate and downstream NR activity to a level similar to those in ZH11, pAct:DNR1-Flag overexpression lines, and dnr1 mutants (Figure 2, F and G). Taken together, these results confirm the notion that DNR1-mediated changes in auxin content affect plant growth and N metabolism.
To unravel the underlying mechanism of how DNR1 and auxin modulate metabolism, we compared the transcript abundance of mRNAs encoding uptake transporters (e.g. OsNRT1.1B, OsNRT2.3a, and OsNPF2.4) and downstream assimilation enzymes (e.g. OsNIA2) by RT-qPCR. The expression levels of OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 were higher in the dnr1 mutants and lower in the pAct:DNR1-Flag overexpression lines compared to wild-type ZH11 (Figure 2, H–K). Therefore, DNR1 reduces auxin contents in rice, thereby repressing NUE by downregulating genes involved in metabolism.
OsARFs mediate the promotion of N metabolism by auxin
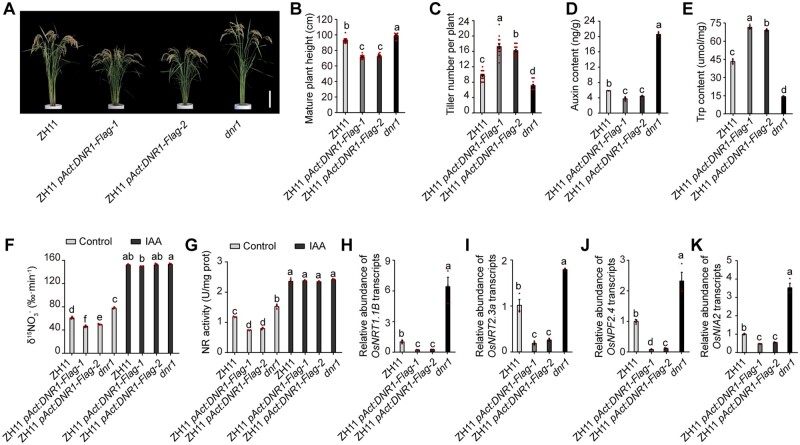
ARFs are a family of transcription factors that regulate the expression of auxin-responsive genes in a partially distinct manner (Li et al., 2016). In this study, RT-qPCR analyses identified seven OsARF members (OsARF1, OsARF5, OsARF6, OsARF17, OsARF19, OsARF24, and OsARF25) that were upregulated in dnr1 but downregulated in pAct:DNR1-Flag overexpression plants compared to ZH11, as well as being repressed by high N supply (Supplemental Figure S8). We thus speculated that DNR1 modulates plant N metabolism via these key auxin signaling components. To test this hypothesis, we generated single knock-out mutants for these OsARFs using CRISPR/Cas9 in the ZH11 background (Supplemental Data Set 2) and measured their 15 uptake rates. Compared to the wild-type ZH11 control, the majority of the osarf mutations compromised 15 uptake capacity to various extents, suggesting that these OsARFs are involved in regulating uptake in rice (Figure 3A).
Figure 3.
OsARFs regulate plant growth and promote the expression of multiple N-metabolism genes. (A) 15 uptake rate in roots of osarf mutants. Data are mean ± sem (n = 3). (B) Morphology of mature ZH11, osarf6, and osarf17 plants. Scale bar, 15 cm. (C, D) Plant height (C), and the number of tillers per plant (D) in ZH11, osarf6, and osarf17 plants. Data are mean ± sem (n = 12). (E, F) mRNA abundances of OsNRT1.1B (E) and OsNRT2.3a (F) in roots relative to ZH11 (set to 1). Data are mean ± sem (n = 3). (G, H) mRNA abundances of OsNPF2.4 (G) and OsNIA2 (H) in shoots relative to ZH11 (set to 1). Data are mean ± sem (n = 3). I, J, OsARF6-Flag and OsARF17-Flag-mediated ChIP-qPCR enrichment (relative to Input) of TGTCTC-containing promoter fragments (marked with an asterisk) from OsNRT1.1B. Data are mean ± sem (n = 3). (K–N) OsARF6 and OsARF17 activate OsNRT1.1B (K), OsNRT2.3a (L), OsNPF2.4 (M), and OsNIA2 (N) promoter-LUC fusion constructs in transient transactivation assays. The LUC/REN activity obtained from a co-transfection with an empty effector construct and indicated reporter constructs was set to 1. Data are mean ± sem (n = 3). (A, C–N) Different letters denote significant differences (P < 0.05) from Duncan’s multiple range test.
We selected osarf6 and osarf17, whose 15 uptake rates were most affected, for further analysis. Both osarf6 and osarf17 mutants displayed a dwarf stature and enhanced tillering compared to ZH11 (Figure 3, B–D). These phenotypes are consistent with the phenotypes observed in the DNR1 overexpression lines and in partially auxin-deficient plants. To confirm the regulatory effects of DNR1 and OsARFs on rice tillering, we compared the transcript levels of several known N-responsive regulators of tillering (OsSPL14, D3, OsTB1, and D14 as repressors and NGR5 as a promoter of tillering) in wild-type and the corresponding mutant plants (Takeda et al., 2003; Ishikawa et al., 2005; Arite et al., 2009; Jiao et al., 2010; Miura et al., 2010; Jiang et al., 2013; Zhou et al., 2013; Wang et al., 2017). Compared to ZH11, the expression levels of OsSPL14, D3, and OsTB1 were reduced by DNR1 overexpression and by the osarf6 or osarf17 loss-of-function mutations but increased in the dnr1 mutant. No significant changes in D14 or NGR5 transcript abundance were detected in these plants (Supplemental Figure S9). Therefore, the enhanced tillering observed in DNR1 overexpression, osarf6, and osarf17 plants is likely caused by the inhibition of tillering repressors OsSPL14, D3, and OsTB1.
RT-qPCR assays demonstrated that OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 mRNA levels were reduced in both osarf6 and osarf17 mutants, presumably due to a defective auxin signaling pathway (Figure 3, E–H). ARF transcription factors recognize and bind to TGTCTC/GAGACA sites through their highly conserved B3-type DNA-binding domain (Kim et al., 1997; Tiwari et al., 2003; Piya et al., 2014; Chandler, 2016). Accordingly, chromatin-immunoprecipitation qPCR (ChIP-qPCR) and electrophoretic mobility shift assays (EMSAs) confirmed that both OsARF6 and OsARF17 bind directly to the TGTCTC/GAGACA-containing segments within the promoter regions of genes involved in uptake and assimilation (Figure 3, I and J; Supplemental Figures S10 and S11). Consequently, OsARF6 and OsARF17 activated transcription from the OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 promoters in transient transactivation assays performed in rice protoplasts (Figure 3, K–N).
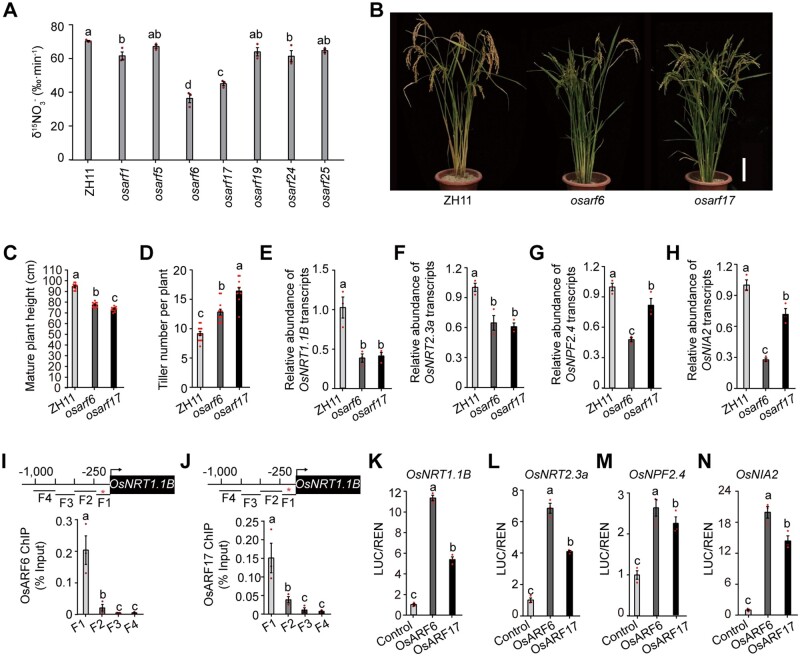
Next, we investigated whether exogenous IAA application would enhance uptake through OsARFs (Figure 4A). OsARF6, OsARF17, and metabolism-related genes (OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2) were upregulated by IAA treatment (Figure 4, B–D). Specifically, IAA promoted ChIP-qPCR enrichment of TGTCTC/GAGACA motif-containing fragments from the OsNRT1.1B promoter, resulting in its elevated transcriptional activation compared to mock treatment (Figure 4, E and F; Supplemental Figure S12). Accordingly, the inhibited uptake in osarf6 and osarf17 was partially reverted by exogenous IAA treatment (Figure 4G). Furthermore, overexpression of OsARF6 or OsARF17 significantly restored the deficiency in absorption caused by DNR1 overexpression, although not to the same level as ZH11 (Figure 4H). Taken together, these observations suggest that although OsARFs share a certain degree of functional redundancy, neither OsARF6 nor OsARF17 is dispensable for promoting uptake in rice.
Figure 4.
Auxin promotes the expression of N metabolism-related genes, and uptake is mediated by OsARFs. (A) 15 uptake rate in the roots of two-week-old plants under mock and external IAA treatment (1 μM). P-values were generated from two-sided Student’s t tests. Data are mean ± sem (n = 3). (B, C) mRNA abundance of OsARF6 and OsARF17 in roots under IAA treatment relative to the control (set to 1). P-values were generated from two-sided Student’s t tests. Data are mean ± sem (n = 3). (D) mRNA abundance in roots and shoots under IAA treatment relative to the control (set to 1). ** indicates the least significant difference at 0.01 probability level, generated from Student’s t tests. Data are mean ± sem (n = 3). (E, F) Extent of OsARF6 and OsARF17-mediated ChIP-qPCR enrichment (relative to Input) of TGTCTC-containing promoter fragments from OsNRT1.1B (fragment 1; shown in Figure 3, I and J). P-values were generated from two-sided Student’s t-tests. Data are mean ± sem (n = 3). (G) 15 uptake rate in the roots of ZH11, osarf6, and osarf17 plants under mock and external IAA treatment (1 μM). Different letters denote significant differences (P < 0.05) from Duncan’s multiple range test. Data are mean ± sem (n = 3). (H) 15 uptake rate in the roots of OsARF6 and OsARF17 overexpression lines in the ZH11 pAct:DNR1-Flag-1 background. Different letters denote significant differences (P < 0.05) from Duncan’s multiple range test. Data are mean ± sem (n = 6).
Increased auxin content promotes NUE and grain yield
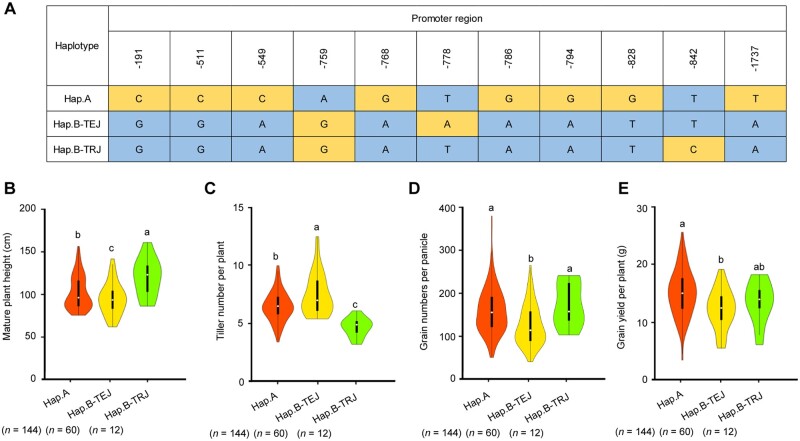
As mentioned above, among our collection of 14 indica and 12 japonica varieties, their DNR1 alleles are characterized by the absence and presence of a 520-bp promoter segment, respectively, which contribute to their differences in DNR1 transcript levels and uptake activity (Supplemental Figure S2). On closer inspection, we discovered that this indel is linked to 11 SNPs in the promoter region of this gene (Supplemental Data Set 3), which also differentiate the indica and japonica DNR1 alleles. To investigate if this applies more broadly to other varieties of these two subspecies, we conducted a haplotype analysis of the DNR1 promoter in 216 rice accessions (Yu et al., 2017) and detected three distinct haplotypes (Haplotype A: 144 indica varieties; Haplotype B-TEJ: 60 temperate japonica varieties; Haplotype B-TRJ: 12 tropical japonica varieties). It is worth noting that haplotype A of the DNR1 promoter contains the aforementioned 11 SNPs specific to indica. Furthermore, although haplotype A- and haplotype B-TEJ-containing varieties are commonly grown under similar planting conditions, Haplotype A is associated with increased plant height, reduced tillering, increased grain number, and higher grain yield potential, which is consistent with phenotypes caused by repressed DNR1 expression and auxin accumulation (Figure 5).
Figure 5.
Natural allelic variation of DNR1 is associated with variation in plant and grain morphology and grain yield performance. (A) DNA polymorphisms in the promoter region of DNR1. (B–E) Violin plots for plant height (B), number of tillers per plant (C), number of grains per panicle (D), and grain yield (E) of rice varieties carrying different DNR1 promoter haplotypes (Haplotype A, Haplotype B-TEJ, or Haplotype B-TRJ). All data are from plants grown under normal paddy-field fertilization conditions. Data are mean ± sem (Haplotype A, n = 144; Haplotype B-TEJ, n = 60; Haplotype B-TRJ, n = 12). Different letters indicate statistically significant differences between groups (P < 0.05) from Tukey’s honestly significant difference test.
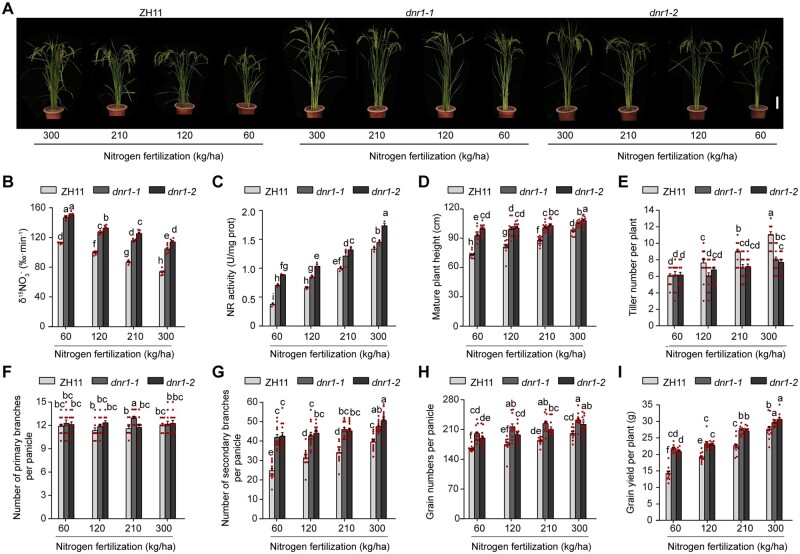
Since we demonstrated that the relatively low DNR1 transcript levels and the resulting high auxin levels in indica rice varieties promote N metabolism, we aimed to comprehensively examine the agronomic consequences of increasing auxin content in the japonica ZH11 background by knocking out DNR1. Compared to ZH11 control plants, dnr1 rice had enhanced metabolism, a taller stature, and lower tiller number under both high and low levels of N supply (Figure 6, A–E). Notably, although panicle primary branching was not detectably affected, grain yield per plant was promoted in the dnr1 mutants, especially under lower levels of N supply, due to increases in the number of secondary branches and grains per panicle (Figure 6, F–I). This is likely due to the greater uptake and assimilation capacities exhibited by dnr1, which is evident from the higher total above-ground N content in the dnr1 mutants relative to ZH11 (Supplemental Figure S13A). Nevertheless, the distribution ratio of N in the four above-ground tissues examined (Brown rice; Husk, Rachis, and Peduncle; Leaves; Culm) was not significantly altered by the lack of DNR1 (Supplemental Figure S13B).
Figure 6.
dnr1 mutants have increased grain yield and NUE. (A) Morphology of mature ZH11, dnr1-1, and dnr1-2 plants supplied with different levels of N. Scale bar, 15 cm. (B) 15 uptake rate in roots. Data are mean ± sem (n = 6). (C) NR activity. Data are mean ± sem (n = 3). (D) Plant height. Data are mean ± sem (n = 16). (E–I) Number of tillers per plant (E), number of primary branches per plant (F), number of secondary branches per plant (G), number of grains per panicle (H), and grain yield per plant (I) of ZH11, dnr1-1, and dnr1-2 plants. Data are mean ± sem (n = 12). (B–I) Different letters denote significant differences (P < 0.05) from Duncan’s multiple range test.
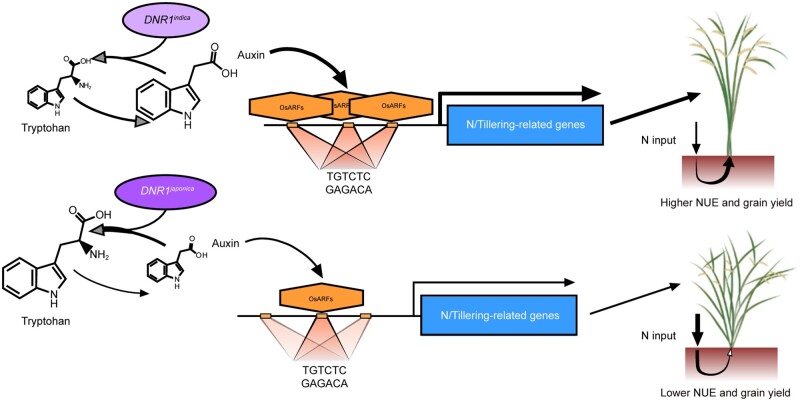
In conclusion, our results collectively suggest that the difference in auxin content in indica and japonica rice varieties controlled by their respective DNR1 variant alleles contributes to their differences in NUE and yield (Figure 7). Thus, manipulating auxin content via DNR1 represents a promising strategy for sustainably increasing grain yield while reducing environmentally degrading levels of agricultural N input in rice.
Figure 7.
Model of how rice NUE and yield are regulated by the DNR1–auxin–OsARFs module in indica and japonica rice varieties. The DNR1indica variant allele leads to reduced transcript and protein abundances of DNR1, and therefore the accumulation of auxin. This consequently induces OsARF-mediated activation of genes involved in both N metabolism and the inhibition of tillering. As a result, rice plants carrying the DNR1indica allele exhibit high yield with reduced N requirement. On the contrary, the DNR1japonica allele confers increased tillering and N demand due to a reduced auxin level.
Discussion
Current global crop productivity is governed by the excessive use of nitrogenous fertilizers, which have detrimental effects on both the environment and human health (Chen et al., 2014). Therefore, developing new crop varieties with high yields under low N conditions would be highly desirable in the near future. To achieve this goal, we need to understand the complex regulatory network controlling plant N metabolism. Increasing evidence suggests that external N status affects the biosynthetic and signal transduction pathways of auxin, although little is known about the identities of the genes that are N-responsive and whether they, in turn, regulate N metabolism. In this study, we discovered and characterized DNR1, a regulator of NUE in rice, expanding the current knowledge of the crosstalk between auxin signaling and N metabolism.
We identified qDNR1 as an important QTL for uptake in rice. This gene encodes an enzyme responsible for maintaining auxin homeostasis. The DNR1 locus diverges in DNA sequence between indica and japonica rice subspecies. The lack of a 520-bp promoter segment of DNR1indica results in lower DNR1 protein abundance, increased auxin content, and higher uptake in indica compared to japonica varieties (Figure 1; Supplemental Figures S1 and S2). Furthermore, a loss-of-function dnr1 mutation confers auxin overaccumulation and enhances metabolism by upregulating genes encoding transporters and NR (Figure 2). Notably, DNR1 transcript abundance is induced by N, therefore forming a negative feedback regulatory loop (Figure 1, K and L). Taken together, we demonstrated that DNR1 functions as a mediator of plant N metabolism by antagonizing auxin accumulation and is itself regulated by N availability.
ARF family transcription factors play key roles in relaying auxin signals to alter plant growth and developmental processes. ARF8 is essential for mediating normal root developmental responses to external N in Arabidopsis (Wu et al., 2006). Interestingly, we showed that in rice, auxin-mediated promotion of uptake is also controlled by members of the OsARF family, since some osarf mutations, especially osarf6 and osarf17, lead to significantly reduced uptake rates. We confirmed that OsARF6 and OsARF17, which belong to the same subclass as Arabidopsis ARF8 (Wang et al., 2007), act as transactivators of N metabolism-related genes by directly binding to the TGTCTC/GAGACA-containing segments within their promoter regions (Figure 3; Supplemental Figures S10 and S11). Accordingly, the overexpression of OsARF6 or OsARF17 counteracted the repressive activity of DNR1 on uptake (Figure 4H). However, this restoration was incomplete in both cases, and exogenous IAA treatment only partially recovered the uptake rates of osarf6 and osarf17 mutants (Figure 4G). These results suggest that OsARF6 and OsARF17 synergistically promote metabolism and that the uptake rate in a rice mutant lacking both OsARF6 and OsARF17 might have a strongly reduced response to auxin.
Members of the ARF family can be divided into transcriptional activators and repressors, depending on the amino acid composition of the nonconserved middle region subdomain; both OsARF6 and OsARF17 are categorized as transcriptional activators (Shen et al., 2010). However, our mutant analysis indicated that a number of OsARFs, including both transcriptional activators (OsARF5, OsARF6, OsARF17, OsARF19, and OsARF25) and repressors (OsARF1 and OsARF24), are positive regulators of uptake in roots (Figure 3A). For example, OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 transcript levels were slightly reduced in the rice osarf1 and osarf24 mutants compared to the wild-type (Supplemental Figure S14). Further investigations are therefore needed to resolve whether some OsARFs, especially those belonging to the transcriptional repressor category, activate uptake through a distinct molecular mechanism.
Finally, we constructed dnr1 mutant lines in the japonica ZH11 background, which mimic the generally lower DNR1 transcript levels caused by the DNR1indica allele in indica rice varieties, and investigated their agronomic traits. Briefly, the above-ground architecture was moderately altered by the dnr1 mutation due to increased auxin accumulation, but crucially, both NUE and grain yield were significantly enhanced in the mutant lines compared to ZH11, without detectably affecting N distribution (Figure 6; Supplemental Figure S13). Therefore, we propose that the DNR1-Auxin-OsARFs cascade represents an important target that could be exploited to enhance crop NUE, thereby facilitating sustainable farming in the future (Figure 7).
Materials and methods
Plant materials and growth conditions in the field
Detailed information about the rice (O. sativa) germplasm used in this study for positional cloning and haplotype analysis was reported previously (Sun et al., 2014; Wang et al., 2017; Yu et al., 2017; Li et al., 2018). A set of 71 SSSLs were derived from a cross between HJX74 (an elite indica variety from South China) as the recipient parent and IRAP9 (a japonica variety from Brazil) as the donor parent. Each SSSL contains a single substituted segment from IRAP9 in the HJX74 genetic background, which was used for QTL analysis and map-based cloning. NIL-DNR1IRAP9 plants were generated by crossing the HJX74 × 27-055 F1 hybrid with HJX74 (the recurrent parent) at least 6 times. Field-grown rice plants were raised under standard paddy conditions at experimental stations in Lingshui (Hainan Province) and Hefei (Anhui Province), China, as previously described (Sun et al., 2014; Li et al., 2018;). Phenotypic parameters of rice, such as plant height, tiller number per plant, and grain yield per plant, were measured as previously described (Hu et al., 2015a, 2015b).
Hydroponic culture of plants
Seeds were sterilized with 20% NaClO solution for 30 min, and uniformly growing individuals after germination were selected for further analyses as described previously (Li et al., 2018). Seven-day-old seedlings were transferred to 40 L of nutrient solution (1.25 mM NH4NO3, 0.3 mM NaH2PO4·2H2O, 0.35 mM K2SO4, 1 mM CaCl2, 1 mM MgSO4·7H2O, 20 µM EDTA (ethylenediaminetetraacetic acid)–Fe, 0.5 mM Na2SiO3, 9 µM MnCl2, 20 µM H3BO3, 0.77 µM ZnSO4, 0.32 µM CuSO4, and 0.39 µM (NH4)6Mo7O24, pH 5.5) in PVC (polyvinyl chloride) pots and grown for 4 weeks. When required, N concentrations were varied by replacing 1.25 mM NH4NO3 (1 N) with 0.75 mM NH4NO3 (0.6 N), 0.375 mM NH4NO3 (0.3 N), or 0.1875 mM NH4NO3 (0.15 N). All nutrient solutions were replaced twice per week, and the pH was adjusted to 5.5 daily. The temperature was maintained at 16 h light (30°C) and 8 h dark (22°C) with an ∼400 μmol/s/m2 photon density and a relative humidity of 60%.
QTL analysis and fine mapping of DNR1
Seventy-one SSSLs were used for QTL analysis, from which two main QTLs for uptake were identified. One hundred and forty-four BC1F2 and 947 BC2F2, derived from 27-055 and HJX74, were used for fine mapping. The primer sequences used for map-based cloning and genotyping assays are listed in Supplemental Data Set 1.
Phylogenetic analysis
The amino acid sequences of DNR1 and its homologs in other plants (15 amino acid sequences) were downloaded from PLAZA (https://bioinformatics.psb.ugent.be/plaza/) and are provided in Supplemental File 1. Phylogenetic analyses were carried out in MEGA7 (Kumar et al., 2016), and the phylogenetic tree was constructed using the Neighbor-Joining method (500-bp replications; Felsenstein, 1985). The evolutionary distances were calculated using the Poisson correction method and represent the number of amino acid substitutions per site. After discarding all ambiguous positions for each sequence pair, 548 positions were included in the final dataset.
Transgene constructs
The full-length DNR1, OsARF6, and OsARF17 cDNAs were amplified from ZH11 before being inserted into pAct:Flag-nos using Gateway recombination cloning technology according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA; 11789100 and 11791100) , to generate the pAct:DNR1-Flag, pAct:OsARF6-Flag, and pAct:OsARF17-Flag constructs. Similarly, full-length DNR1 was inserted into p35S:GFP-nos to generate the p35S:DNR1-GFP construct (Wang et al., 2012). gRNA constructs used to produce the CRISPR/Cas9-generated DNR1, OsARF1, OsARF5, OsARF6, OsARF17, OsARF19, OsARF24, and OsARF25 loss-of-function alleles in the ZH11 genetic background were generated as described elsewhere (He et al., 2018). Transgenic rice plants were generated by Agrobacterium-mediated transformation as described elsewhere (Huang et al., 2009). Target information for CRISPR/Cas9-generated mutants is listed in Supplemental Data Set 2. Relevant primer sequences are listed in Supplemental Data Set 4.
RT-qPCR analysis
Total RNAs were extracted from different plant tissues using TRIzol reagent (Ambion, Austin, TX, USA), and full-length cDNAs were reverse transcribed using a cDNA synthesis kit (Vazyme, Nanjing, China; R323-01). qPCR was performed on a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Each reaction contained 5 μL diluted cDNA, 0.5-μL primers, and 10 μL SYBR Green Master Mix in a total volume of 20 μL, according to the manufacturer’s instructions (Vazyme; Q111-02). The program included a melting step at 95°C for 30 s and an amplification step consisting of 40 cycles of 95°C for 5 s followed by 60°C for 20 s. All RT-qPCR experiments were performed with three biological replicates (obtained from independent extraction processes using separate plant materials). The values of transcript abundance represent the average of three technical replicates (separate qPCRs using the same cDNA). The rice ACTIN1 gene was used as an internal control. Relevant RT-qPCR primer sequences are listed in Supplemental Data Set 5.
Immunoblot analysis
Extraction buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40 detergent, 10% glycerol, 1 mM DTT (dithiothreitol) and protease inhibitor cocktail (Roche LifeScience, Penzberg, Germany) was used to extract total protein. Protein samples were separated by 10% SDS–PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) and transferred to a nitrocellulose membrane under wet transfer conditions. DNR1 protein was detected by probing the membrane with anti-DNR1 (ABclonal, Woburn, MA, USA) and anti-DDDDK-tag antibodies (MBL, Woburn, MA, USA, M185-11, and Lot.007). The results of immunoblotting were visualized on the Tanon-5200 Chemiluminescent Imaging System (Tanon Science and Technology).
Preparation of nuclear and cellular protein extracts
Extraction buffer containing Ficoll (25 mg/mL), Dextran T 40 (50 mg/mL), sucrose (137 mg/mL), 25 mM Tris–HCl (pH 7.4), 10 mM MgCl2, 0.5 mM DTT, and protein inhibitor cocktail (Roche LifeScience) was used to separate cytoplasmic and nuclear proteins. The DNR1–GFP, GRF4-GFP, and free GFP proteins were detected by probing the membrane with anti-GFP antibody (MBL, 598-7, and Lot.005). The results of immunoblotting were visualized on the Tanon-5200 Chemiluminescent Imaging System (Tanon Science and Technology).
ChIP-qPCR assays
Details of ChIP-qPCR analysis have been described elsewhere (O’Geen et al., 2010). Briefly, ∼2 g samples of 2-week-old wild-type and transgenic rice seedlings (pAct:OsARF6-Flag and pAct:OsARF17-Flag) were fixed with 1% formaldehyde, subjected to cross-linking under a vacuum for 15 min to cross-link protein–DNA complexes, and ground into a fine powder in liquid N. Following isolation and lysis of nuclei, chromatin was isolated and fragmented by ultrasonication into ∼500-bp fragments. The sonicated chromatin was incubated with 7 μg of antibody-Flag (Sigma, St Louis, MO, USA; F1804) overnight at 4°C for immunoprecipitations. The next day, washing, elution, reverse cross-linking, and DNA purification steps were performed. Enrichment of DNA fragments was determined by RT-qPCR analysis performed on three biological replicates (obtained from independent extraction processes using separate plant materials). Relevant PCR primer sequences are listed in Supplemental Data Set 6.
EMSA
Full-length OsAFR6 and OsARF17 cDNAs were amplified and cloned into the pCold-TF vector (Takara, Kyoto, Japan). His-OsAFR6 and His-OsAFR17 recombinant proteins were purified using Ni-NTA agarose (Qiagen, Hilden, Germany; 30210) following the manufacturer’s instructions. DNA probes were artificially amplified and labeled using a biotin label kit (Sangon Biotech, Shanghai, China). DNA gel shift assays were performed using a LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, Waltham, MA, USA; 20148). Relevant primer sequences are listed in Supplemental Data Set 7.
In vitro transient transactivation assays
Approximately 2-kb DNA promoter fragments from OsNRT1.1B, OsNRT2.3a, OsNPF2.4, and OsNIA2 were amplified from ZH11 and inserted into the EcoRV and XbalI sites of pUC19 containing the firefly luciferase (LUC) reporter gene driven by the 35S minimal TATA box and 5×GAL4 binding elements. Full-length cDNAs of OsARF6 and OsARF17 were amplified and fused to the sequence encoding GAL4BD by inserting them into the BamHI and EcoRI sites of pRTBD to generate the effector plasmids pRTBD-OsARF6 and pRTBD-OsARF17, respectively. Transient transactivation assays were performed using rice protoplasts as described elsewhere (Wang et al., 2015). The Dual-LUC Reporter Assay System (Promega, Madison, WI, USA; E1960) and the GloMax™ 20-20 Luminometer were used for the LUC activity assays, with the Renilla (REN) LUC gene used as an internal control. Relevant PCR primer sequences are listed in Supplemental Data Set 4.
Determination of IAA content
Samples of ∼50-mg root tip tissue were ground into a powder in liquid N and extracted with methanol/water/formic acid (15:4:1, V/V/V). The combined extracts were evaporated to dryness under an N-gas stream, reconstituted in 80% methanol (V/V), filtrated (PTFE, 0.22 μm, Anpel), and analyzed using an LC–ESI–MS/MS (liquid chromatography-electrospray ionization-tandem mass spectrometry) system and ESI-triple quadrupole-linear ion trap–MS system (Flokova et al., 2014; Cui et al., 2015; Simura et al., 2018; Xiao et al., 2018).
Determination of Trp content
Samples of ∼25-mg plant tissue were treated with methanol/acetonitrile/water (2:2:1, V/V/V), ground into a powder, and ultrasonically cracked in an ice bath for 10 min (Power: 80 Hz). Following centrifugation for 15 min (25,000 g, 4°C), 500 μL of supernatant was freeze-dried. The drained supernatant was re-dissolved in 500 μL 10% methanol solution and analyzed using the LC–MS/MS Sciex5500-Transcend II system.
15N uptake analysis
After growth in hydroponic culture for 4 weeks, 15 influx into roots was measured as described elsewhere (Ho et al., 2009; Li et al., 2018). Roots and shoots were separated, dried at 80°C for 72 h, and the 15N content measured using the Isoprime 100 elemental analyzer (Elementar, Langenselbold, Germany).
Measurement of NR activity
Fresh plant material (∼1 g) from individual rice plant supplied with four different N levels was used to measure NR activity, following the instruction manual of the NR Kit (Solarbio LIFE SCIENCES, Beijing, China; BC0080).
Determination of plant N concentration
Samples from different plant organs were dried in an oven at 80°C for 72 h. Following tissue homogenization, N content was measured using an elemental analyzer (IsoPrime100; Elementar).
Accession numbers
Sequence data of the rice genes studied in this article can be found in The Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/) under the following accession numbers: DNR1 (LOC_Os01g08270), OsNRT1.1B (LOC_Os10g40600), OsNRT2.3a (LOC_Os01g50820), OsNPF2.4 (LOC_Os01g36720), OsNIA2 (LOC_Os08g36500), OsARF1 (LOC_Os01g13520), OsARF5 (LOC_Os02g04810), OsARF6 (LOC_Os02g06910), OsARF17 (LOC_Os06g46410), OsARF19 (LOC_Os06g48950), OsARF24 (LOC_Os12g29520), and OsARF25 (LOC_Os12g41950).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of DNR1 by fine-scale mapping, and sequence divergence in this gene between rice varieties HJX74 and IRAP9.
Supplemental Figure S2. The 520-bp INDEL in the DNR1 promoter underlies the differences in DNR1 transcript level and uptake rate between indica and japonica varieties.
Supplemental Figure S3. Phylogenetic tree of DNR1 and its homologous genes, and amino acid sequence alignment of DNR1 and Arabidopsis VAS1.
Supplemental Figure S4. Subcellular localization and expression pattern of DNR1.
Supplemental Figure S5. Shoot architecture and NR activity of mature NIL plants.
Supplemental Figure S6. DNR1 transcript abundance and DNR1 protein accumulation in ZH11 pAct:DNR1-Flag and dnr1 plants compared to ZH11.
Supplemental Figure S7. Reducing auxin contents by increasing DNR1 abundance disturbs lateral root development.
Supplemental Figure S8. The seven OsARFs whose transcript abundances are significantly increased in dnr1 plants.
Supplemental Figure S9. Transcript abundances of key regulators of tillering in ZH11, ZH11 pAct:DNR1-Flag, dnr1, osarf6, and osarf17 plants.
Supplemental Figure S10. OsARF-mediated ChIP-qPCR enrichment of TGTCTC-containing promoter fragments from genes involved in metabolism.
Supplemental Figure S11. His-OsARFs bind directly to the promoters of genes related to metabolism.
Supplemental Figure S12. Auxin promotes the transactivation of OsARF6 and OsARF17 toward OsNRT1.1B.
Supplemental Figure S13. N distributions in different organs of above-ground parts of ZH11 and dnr1 rice plants.
Supplemental Figure S14. Transcript abundances of N-related genes in ZH11, osarf1, and osarf24 plants.
Supplemental Data Set 1. Primer sequences used for map-based cloning and genotyping assays.
Supplemental Data Set 2. Information about osarf mutants.
Supplemental Data Set 3. SNPs in the OsDNR1 promoter region of indica and japonica rice varieties.
Supplemental Data Set 4. Primer sequences used for transgene construction.
Supplemental Data Set 5. Primer sequences used for qPCR assays.
Supplemental Data Set 6. Primer sequences used for ChIP assays.
Supplemental Data Set 7. Primer sequences used for EMSAs.
Supplemental File 1. Amino acid sequences of DNR1 and its homologs in other plants used for phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Caifu Jiang (China Agricultural University) for critical suggestions and Xinyu Jiang (Nanjing Agricultural University) for help with making the violin plots.
Funding
This work was supported by grants from the Young Elite Scientists Sponsorship Program by CAST (grant no. 2019QNRC001), Nanjing Agricultural University Start-up Funding, Fundamental Research Funds for the Central Universities (grant no. JCQY201903), and Jiangsu Collaborative Innovation Center for Modern Crop Production. Work in NPH’s laboratory was supported by the BBSRC-Newton “Rice” Initiative (grant no. BB/N013611/1), and also by BBSRC Response Modes grant no. BB/S013741/1.
Conflict of interest statement. The authors declare no competing interests.
S.Z., L.Z., C.S., Z. J., and S.L. designed the research. S.Z., L.Z., and C.S. performed most of the experiments. S.Z., L.Z., C.S., and Z.J. conducted QTL analysis and map-based cloning. S.Z., L.Z., C.S., H.Z., Y.T., and K.W. performed field experiments. Y.L. conducted ChIP-qPCR analysis and phylogenetic analysis. S.Z. performed nuclear and cellular protein separation. T.Z., N.Y., and Y.H. constructed transgenic rice plants. J.Y. performed haplotype analysis. S.W. provided SSSL lines. Z.J. and S.L. wrote the manuscript. J.W., N.P.H., Y.Z., and X.F. revised the manuscript. All authors discussed the results and contributed to the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) are Shan Li (shanli@njau.edu.cn) and Shaokui Wang (shaokuiwang@scau.edu.cn).
References
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50:1416–1424 [DOI] [PubMed] [Google Scholar]
- Avery GS, Burkholder PR, Creighton HB (1937) Nutrient deficiencies and growth hormone concentration in Helianthus and Nicotiana. Am J Bot 24:553–557 [Google Scholar]
- Avery GS, Pottorf L (1945) Auxin and nitrogen relationships in green plants. Am J Bot 32:666–669 [Google Scholar]
- Caba JM, Centeno ML, Fernández B, Gresshoff PM, Ligero F (2000) Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta 211:98–104 [DOI] [PubMed] [Google Scholar]
- Chandler JW (2016) Auxin response factors. Plant Cell Environ 39:1014–1028 [DOI] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu S, Liu D, Xiao Y, Hu B, Liu L. et al. (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2:15195. [DOI] [PubMed] [Google Scholar]
- Chen X, Cui Z, Fan M, Vitousek P, Zhao M, Ma W, Wang Z, Zhang W, Yan X, Yang J. et al. (2014) Producing more grain with lower environmental costs. Nature 514:486–489 [DOI] [PubMed] [Google Scholar]
- Chen ZC, Ma JF (2015) Improving nitrogen use efficiency in rice through enhancing root nitrate uptake mediated by a nitrate transporter, NRT1.1B. J Genet Genomics 42:463–465 [DOI] [PubMed] [Google Scholar]
- Cui K, Lin Y, Zhou X, Li S, Liu H, Zeng F, Zhu F, Ouyang G, Zeng Z (2015) Comparison of sample pretreatment methods for the determination of multiple phytohormones in plant samples by liquid chromatography–electrospray ionization-tandem mass spectrometry. Microchem J 121:25–31 [Google Scholar]
- Duan D, Zhang H (2015) A single SNP in NRT1.1B has a major impact on nitrogen use efficiency in rice. Sci China Life Sci 58:827–828 [DOI] [PubMed] [Google Scholar]
- Duan P, Shen N, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y (2015) Regulation of OsGRF4 by osmiR396 controls grain size and yield in rice. Nat Plants 2:15203. [DOI] [PubMed] [Google Scholar]
- Fan X, Feng H, Tan Y, Xu Y, Miao Q, Xu G (2016) A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J Integr Plant Biol 58:590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- Flokova K, Tarkowska D, Miersch O, Strnad M, Wasternack C, Novak O (2014) UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 105: 147–157 [DOI] [PubMed] [Google Scholar]
- Gao Z, Wang Y, Chen G, Zhang A, Yang S, Shang L, Wang D, Ruan B, Liu C, Jiang H. et al. (2019) The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat Commun 10:5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327:812–818 [DOI] [PubMed] [Google Scholar]
- Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, Christie P, Goulding KW, Vitousek PM, Zhang FS (2010) Significant acidification in major Chinese croplands. Science 327:1008–1010 [DOI] [PubMed] [Google Scholar]
- He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y (2018) Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant 11:1210–1213 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138:1184–1194 [DOI] [PubMed] [Google Scholar]
- Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y. et al. (2015a) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47:834–838 [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D. et al. (2015b) A rare allele of GS2 enhances grain size and grain yeld in rice. Mol Plant 8:1455–1465 [DOI] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41:494–497 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46:79–86 [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y. et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504:401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X. et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544 [DOI] [PubMed] [Google Scholar]
- Kaiser J (2018) Too much of a good thing? Science 359:1346–1347 [DOI] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H (2011) Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J Exp Bot 62: 1399–1409 [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K. et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hu B, Chu C (2017) Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J Exp Bot 68: 2477–2488 [DOI] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y. et al. (2018) Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SB, Xie ZZ, Hu CG, Zhang JZ (2016) A review of auxin response factors (ARFs) in plants. Front Plant Sci 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YL, Fan XR, Shen QR (2008) The relationship between rhizosphere nitrification and nitrogen-use efficiency in rice plants. Plant Cell Environ 31: 73–85 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Y, Han W, Tang A, Shen J, Cui Z, Vitousek P, Erisman JW, Goulding K, Christie P. et al. (2013) Enhanced nitrogen deposition over China. Nature 494: 459–462 [DOI] [PubMed] [Google Scholar]
- Luo L, Wang H, Liu X, Hu J, Zhu X, Pan S, Qin R, Wang Y, Zhao P, Fan X. et al. (2018) Strigolactones affect the translocation of nitrogen in rice. Plant Sci 270: 190–197 [DOI] [PubMed] [Google Scholar]
- Luo L, Zhang Y, Xu G (2020) How does nitrogen shape plant architecture? J Exp Bot 71: 4415–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Li J, Qu B, He X, Zhao X, Li B, Fu X, Tong Y (2014) Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J 78: 70–79 [DOI] [PubMed] [Google Scholar]
- Matsuo S, Miyatake K, Endo M, Urshimo S, Kawanishi T, Negoro S, Shimakoshi S, Fukuoka H (2020) Loss of function of the Pad-1 aminotransferase gene, which is involved in auxin homeostasis, induces parthenocarpy in Solanaceae plants. Proc Natl Acad Sci USA 117: 12784–12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42: 545–549 [DOI] [PubMed] [Google Scholar]
- O’Geen H, Frietze S, Farnham PJ (2010) Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol Biol 649: 437–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieck M, Yuan Y, Godfrey J, Fisher C, Zolj S, Vaughan D, Thomas N, Wu C, Ramos J, Lee N. et al. (2015) Auxin and tryptophan homeostasis are facilitated by the ISS1/VAS1 aromatic aminotransferase in Arabidopsis. Genetics 201: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piya S, Shrestha SK, Binder B, Stewart CN, Hewezi T (2014) Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5: 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Shen C, Wang S, Bai Y, Wu Y, Zhang S, Chen M, Guilfoyle TJ, Wu P, Qi Y (2010) Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L). J Exp Bot 61: 3971–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simura J, Antoniadi I, Siroka J, Tarkowska D, Strnad M, Ljung K, Novak O (2018) Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiol 177: 476–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Qian Q, Wu K, Luo J, Wang S, Zhang C, Ma Y, Liu Q, Huang X, Yuan Q. et al. (2014) Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet 46: 652–656 [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33: 513–520 [DOI] [PubMed] [Google Scholar]
- Tang W, Ye J, Yao X, Zhao P, Xuan W, Tian Y, Zhang Y, Xu S, An H, Chen G. et al. (2019) Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat Commun 10: 5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Chen F, Liu J, Zhang F, Mi G (2008) Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J Plant Physiol 165: 942–951 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutierrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG (2006) Nitrogen regulation of root branching. Ann Bot 97: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24 [DOI] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C. et al. (2015) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47: 949–954 [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Qian Q, Liu Q, Li Q, Pan Y, Ye Y, Liu X, Wang J, Zhang J. et al. (2017) Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res 27: 1142–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q. et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44: 950–954 [DOI] [PubMed] [Google Scholar]
- Wu K, Wang S, Song W, Zhang J, Wang Y, Liu Q, Yu J, Ye Y, Li S, Chen J. et al. (2020) Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367: eaaz2046. [DOI] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133:4211–4218 [DOI] [PubMed] [Google Scholar]
- Xi ZY, He FH, Zeng RZ, Zhang ZM, Ding XH, Li WT, Zhang GQ (2006) Development of a wide population of chromosome single-segment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.). Genome 49:476–484 [DOI] [PubMed] [Google Scholar]
- Xia X, Fan X, Wei J, Feng H, Qu H, Xie D, Miller AJ, Xu G (2015) Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J Exp Bot 66:317–331 [DOI] [PubMed] [Google Scholar]
- Xiao HM, Cai WJ, Ye TT, Ding J, Feng YQ (2018) Spatio-temporal profiling of abscisic acid, indoleacetic acid and jasmonic acid in single rice seed during seed germination. Anal Chim Acta 1031:119–127 [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182 [DOI] [PubMed] [Google Scholar]
- Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ 34:1360–1372 [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang J, Cai Z (2015) Nitrification activities and N mineralization in paddy soils are insensitive to oxygen concentration. Acta Agric Scand 66:272–281 [Google Scholar]
- Yu J, Xiong H, Zhu X, Zhang H, Li H, Miao J, Wang W, Tang Z, Zhang Z, Yao G. et al. (2017) OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol 15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novak O, Dai X, Zhao Y, Ljung K, Noel JP, Chory J (2013) Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9:244–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L. et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504:406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.