Abstract
In wheat (Triticum aestivum L.), breeding efforts have focused intensively on improving grain yield and quality. For quality, the content and composition of seed storage proteins (SSPs) determine the elasticity of wheat dough and flour processing quality. Moreover, starch levels in seeds are associated with yield. However, little is known about the mechanisms that coordinate SSP and starch accumulation in wheat. In this study, we explored the role of the endosperm-specific NAC transcription factor TaNAC019 in coordinating SSP and starch accumulation. TaNAC019 binds to the promoters of TaGlu-1 loci, encoding high molecular weight glutenin (HMW-GS), and of starch metabolism genes. Triple knock-out mutants of all three TaNAC019 homoeologs exhibited reduced transcript levels for all SSP types and genes involved in starch metabolism, leading to lower gluten and starch contents, and in flour processing quality parameters. TaNAC019 directly activated the expression of HMW-GS genes by binding to a specific motif in their promoters and interacting with the TaGlu-1 regulator TaGAMyb. TaNAC019 also indirectly regulated the expression of TaSPA, an ortholog of maize Opaque2 that activates SSP accumulation. Therefore, TaNAC019 regulation of starch- and SSP-related genes has key roles in wheat grain quality. Finally, we identified an elite allele (TaNAC019-BI) associated with flour processing quality, providing a candidate gene for breeding wheat with improved quality.
The wheat seed-specific transcription factor TaNAC019 regulates storage protein and starch accumulation by cooperating with TaGAMyb, and the TaNAC019-BI allele is candidate for higher processing quality in breeding.
Introduction
Bread wheat (Triticum aestivum L.) is one of the most widely cultivated and consumed crops worldwide. Wheat flour is used to produce a wide range of products, such as leavened and unleavened breads, steamed buns, noodles, pancakes, cakes, and cookies (Veraverbeke and Delcour, 2002). The end-use value of wheat flour mainly depends on the properties of seed storage proteins (SSPs), especially gluten proteins, which give wheat dough products their unique viscoelastic properties (Payne et al., 1987; Biesiekierski, 2017).
Gluten is a complex mixture of two types of proteins: polymeric glutenins and monomeric gliadins. Glutenins are subdivided into high molecular weight (HMW-GSs) and low molecular weight (LMW-GSs) glutenin subunits, both of which confer elasticity to dough. Although HMW-GSs account for only about 10% of gluten proteins, the variation in their composition explains up to 70% of the genetic variation in dough processing quality (Payne et al., 1987; Eagles et al., 2002; Liu et al., 2005). Compared with HMW-GSs, higher levels of LMW-GSs are present in the grain of bread wheat; the LMW-GS content is associated with dough resistance and extensibility (Dong et al., 2010). Gliadin content is also associated with dough extensibility (Shewry et al., 2002). The quantitative effects of individual HMW-GSs (Ragupathy et al., 2008; Li et al., 2015; Cho et al., 2017), as well as the ratios of HMW-GSs/LMW-GSs and HMW-GSs/gliadins, are relevant for different uses of dough (Veraverbeke and Delcour, 2002; Rasheed et al., 2014).
Wheat HMW-GSs are encoded by TaGlu-1 loci on the long arms of homoeologous chromosomes 1A, 1B, and 1D. Each locus contains two tightly linked genes encoding X- and Y-type subunits, i.e. TaGlu-1Ax, TaGlu-1Ay, TaGlu-1Bx, TaGlu-1By, TaGlu-1Dx, and TaGlu-1Dy (Galili and Feldman, 1985). LMW-GSs are encoded by Glu-3 loci (i.e. Glu-A3, Glu-B3, and Glu-D3) located on the short arm of homoeologous group 1 chromosomes, and each locus contains multiple gene copies (Singh and Shepherd, 1988; Zhang et al., 2011; Lee et al., 2016). Gliadins are encoded by genes in two genomic regions: the Gli-1 loci, located on the short arms of group 1 chromosomes, encoding γ-, δ-, ω-gliadins, and the Gli-2 loci, located on the short arms of homoeologous group 6 chromosomes, encoding α-gliadins (Singh and Shepherd, 1988; Huo et al., 2018).
Conserved cis-regulatory motifs have been identified in the promoters of genes encoding SSPs (Makai et al., 2015), and a number of transcription factors (TFs) involved in SSP regulation have been characterized. Wheat storage protein activator (TaSPA) binds to a general control nondepressible 4 (GCN4)-like binding motif in the promoters of genes encoding glutenins and gliadins and activates their transcription (Albani et al., 1997; Ravel et al., 2014), whereas SPA heterodimerizing protein (SHP) suppresses glutenin gene expression (Boudet et al., 2019). TaFUSCA3, a B3-Superfamily TF that also interacts with TaSPA, activates TaGlu-1Bx7 by binding to the RY-box motif in its promoter (Sun et al., 2017). The DNA-binding with one finger (Dof)-type prolamin-box binding factor (PBF) binds to the prolamin-box (P-box) in the promoter regions of wheat α-gliadin and LMW-GS genes (Dong et al., 2007) and binds to the TaGlu-1By8 and -1Dx2 loci, leading to promoter hypomethylation (Zhu et al., 2018). Finally, the GA-dependent MYB TF TaGAMyb activates HMW-GS gene expression by directly binding to the TaGlu promoter and by recruiting the GCN5-like histone acetyltransferase (HAT; Guo et al., 2015). Despite the identification of these TFs, little is known about their functional interactions or the intricate mechanisms of the SSP regulatory pathway.
High baking quality is associated with the protein composition of wheat grain, but negatively correlated with high yield, which is itself associated with starch content (Rharrabti et al., 2001; Zorb et al., 2018). Although TFs that regulate the expression of genes encoding wheat starch biosynthetic enzymes have been reported (Kang et al., 2013; Kumar et al., 2018), how TFs regulate starch biosynthesis remains unknown. In addition, little is known, in any plant species, about how TFs simultaneously regulate SSP and starch accumulation, thus coordinating the balance between quality and yield.
A recent report indicated that ZmNAC128 and ZmNAC130, which belong to the NAC (NAM [No Apical Meristem]–ATAF [Arabidopsis Transcription Activation Factor]–CUC [Cup-shaped Cotyledons]) superfamily, act redundantly to modulate starch and protein accumulation during maize (Zea mays) endosperm development (Zhang et al., 2019). The NAC domain superfamily is one of the largest groups of plant-specific TFs. NAC TFs are involved in various developmental and growth processes, although they are mainly known for their roles in plant responses to biotic and abiotic stress (Olsen et al., 2005; Mathew and Agarwal, 2018). Rice (Oryza sativa) OsNAP (NAC-LIKE, ACTIVATED BY AP3/PI) and wheat NAM-B1 play indirect roles in regulating protein content in grains, by accelerating leaf senescence and nitrogen accumulation in aerial parts (Uauy et al., 2006; Liang et al., 2014). In addition, NAC genes that are specifically expressed in barley (Hordeum vulgare) and rice seeds may directly regulate seed size, but little is known about the underlying mechanism (Christiansen et al., 2011; Mathew et al., 2016).
In this study, we provide evidence that TaNAC019, a seed-specific NAC TF, regulates both SSP and starch accumulation in wheat grain. We explored the mechanism underlying TaNAC019-mediated transcriptional regulation of gene expression, in which TaNAC019 directly binds to and activates the expression of its target genes, including HMW-GS genes, the starch synthase gene TaSSIIa, and the sucrose synthase gene TaSuSy1. TaNAC019 also affects TaSPA expression, and physically interacts and cooperates with TaGAMyb to activate TaGlu expression. Finally, we identified a specific natural genetic variant of TaNAC019 (TaNAC019-BI) that is associated with higher wheat processing quality, thus providing a promising candidate gene for wheat quality improvement in breeding programs.
Results
Identification of the endosperm-specific nuclear protein TaNAC019, which binds to the promoters of glutenin genes
With the aim of identifying TFs that regulate the expression of glutenin genes, we performed a yeast one-hybrid assay using the TaGlu-1Dy promoter as bait against a wheat cDNA expression library. Among the 347 positive clones, three matched TraesCS3A02G077900, as revealed by Basic Local Alignment Search Tool for DNA (BLASTN) analysis; this gene encodes a member of the wheat NAC TF family. Phylogenetic analyses of wheat NAC TF family members indicated that TraesCS3A02G077900 is an ortholog of the seed-specific barley gene HvNAC019 (Christiansen et al., 2011; Borrill et al., 2017; Guerin et al., 2019), whose function is still unknown. However, reciprocal BLAST analysis (Borrill et al., 2017; Guerin et al., 2019) indicated that TraesCS3A02G077900 is not an ortholog of the endosperm-specific rice NAC TF genes, Os01g01470 or Os01g29840, which are associated with seed size (Mathew et al., 2016), or their maize orthologs, ZmNAC128 and ZmNAC130, which directly regulate protein and starch accumulation (Zhang et al., 2019). Accordingly, we named TraesCS3A02G077900 as TaNAC019.
Bread wheat is an allohexaploid species whose genes usually have homoeologs in all three subgenomes (Glover et al., 2016). Among transcripts sharing high sequence similarity with TraesCS3A02G077900, the two loci TraesCS3B02G092800 and TraesCS3D02G078500 mapped to chromosome 3 of subgenomes B and D, respectively. The NAC proteins encoded by these three genes grouped within the same phylogenetic cluster, along with most of the wheat seed-specific NACs identified to date (Guerin et al., 2019; Supplemental Figure S1). We confirmed the genomic locations of the three genes using a set of Chinese Spring (CS) nullisomic–tetrasomic lines and homoeolog-specific primers (Supplemental Figure S2). The three homoeologous sequences were named TaNAC019-A, TaNAC019-B, and TaNAC019-D.
Although most NAC TFs are soluble proteins found in the nucleus, several NACs are membrane-bound TFs or localize to the cytoplasm (Mathew and Agarwal, 2018). Analysis of protein fusion constructs between TaNAC019 homoeologs and the green fluorescent protein (GFP), transiently expressed in Nicotiana benthamiana leaves, indicated that all three homoeologs are localized to the nucleus and cytoplasm (Supplemental Figure S3).
Reverse transcription-quantitative PCR (RT-qPCR) analysis showed that the TaNAC019 homoeologs are expressed almost exclusively in the endosperm and that TaNAC019-B is expressed at higher levels than TaNAC019-A or TaNAC019-D (Figure 1, A). During kernel development, TaNAC019-A/B/D transcripts were undetectable until 12 days after pollination (DAP), and the highest transcript levels were detected at ∼25 DAP (Figure 1, B). The glutenin gene transcripts started to accumulate before TaNAC019 transcripts and reached their highest peak at 25 DAP, as measured using a primer combination that detects the total mRNA levels of the four TaGlu genes expressed in the wheat cultivar Nongda3338 (i.e. TaGlu-1Bx/1By and TaGlu-1Dx/1Dy; TaGlu-1Ax/1Ay loci are not expressed in this cultivar; Supplemental Figure S4 and Figure 1, B).
Figure 1.
TaNAC019 expression in wheat seeds. A, Relative transcript levels of the three TaNAC019 homoeologs in the indicated tissues, as determined by RT-qPCR. Data were normalized to wheat TaACTIN, shown as mean ± sd (n = 3). B, Relative transcript levels of the three TaNAC019 homoeologs (y-axis on the left) and TaGlu-All (y-axis on the right), estimated using total RNA extracted from whole seeds from 4 to 7 DAP and endosperm (from 12 to 45 DAP). TaGlu-All transcript levels were measured using a primer combination allowing mRNA amplification of four TaGlu genes (i.e. TaGlu-1Bx, TaGlu-1By, TaGlu-1Dx, and TaGlu-1Dy). C, As described in B; primer combinations specific for other wheat seed specific NACs (Supplemental Figure S1) were employed. For clarity, only data obtained for the homoeolog located on the A subgenome for each locus are shown, as mean ± sd (n = 3).
Immunoblot analysis with an antibody that recognizes the three TaNAC019 proteins validated the expression patterns observed by RT-qPCR (Supplemental Figure S5, A and C). These results suggest that TaNAC019 is positively associated with the expression of TaGlu genes. A comparison of TaNAC019 to other wheat seed-specific NAC transcripts (Supplemental Figure S1) indicated that they have different expression patterns during seed development, with most other transcripts accumulating to higher levels during two distinct developmental stages, while TaNAC019 reached its highest expression level from 20 to 25 DAP and TraesCS7A02G0569100 was expressed early and reached its highest expression level from 25 to 30 DAP (Figure 1, C). These results suggest that different seed-specific TaNACs might have distinct, or not fully overlapping, functions.
TaNAC019 regulates glutenin accumulation, gluten formation, and wheat processing quality
To investigate the role of TaNAC019, we generated knock-out mutants in the wheat cultivar Fielder background via CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9) gene editing. To simultaneously knock out all three TaNAC019 homoeologs, we designed a single guide RNA (sgRNA) that targets a conserved region within the first exon of these three genes (Supplemental Figure S6). We obtained two independent lines (nac019-1 and nac019-2) with frameshift mutations leading to premature termination in all three TaNAC019 homoeologs. TaNAC019 protein was undetectable in lines nac019-1 and nac019-2, as revealed by immunoblot analysis (Supplemental Figure S5, B).
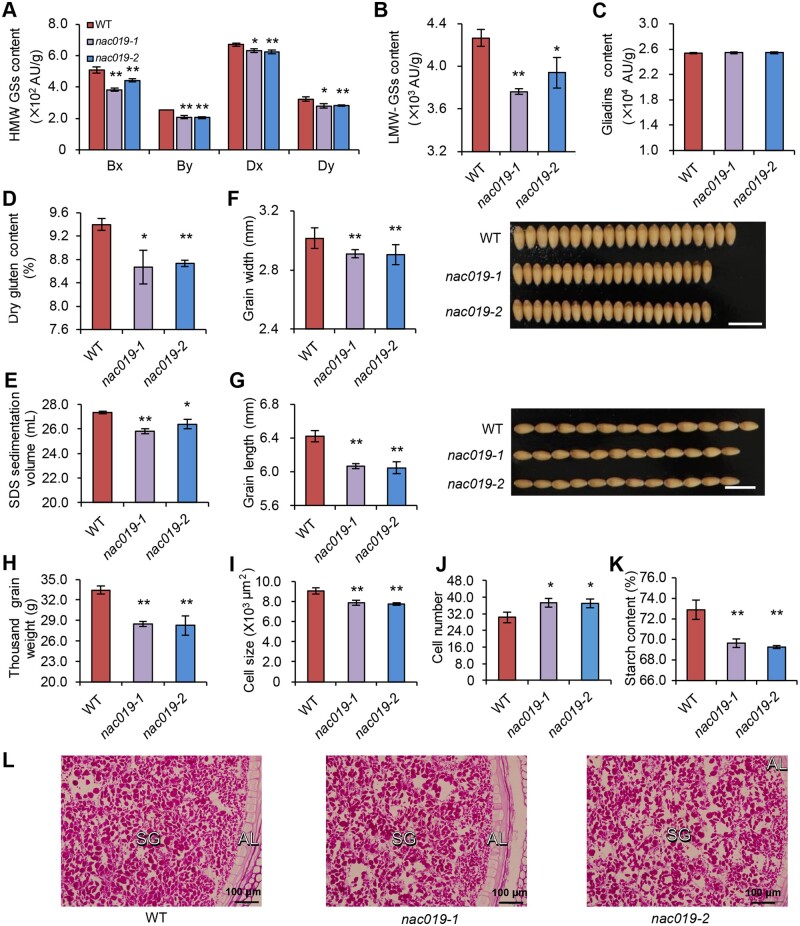
Since TaNAC019 is a putative regulator of TaGlu-1Dy, we performed reverse-phase high-performance liquid chromatography (RP-HPLC) to measure HMW-GS contents in mature seeds. The levels of all subunits including TaGlu-1Bx, TaGlu-1By, TaGlu-1Dx, and TaGlu-1Dy were reduced in the nac019 mutants compared with the wild type (Figure 2, A). This difference was confirmed by protein gel analysis, followed by silver staining (Supplemental Figure S7). The LMW-GS levels were also reduced in the nac019 mutants (Figure 2, B), whereas gliadin levels in the mutants were similar to those in the wild type (Figure 2, C). In addition, the loss of TaNAC019 function was associated with a decrease in dry gluten contents (Figure 2, D) and SDS sedimentation volume (Figure 2, E), an indicator of processing and end-product quality. Starch levels and accumulation of starch granules were also reduced in the nac019 mutants (Figure 2, K and L). These results indicate that TaNAC019 regulates both glutenin and starch accumulation.
Figure 2.
Seed phenotypes of wheat nac019 knock-out lines. A–D, Results from RP-HPLC analysis of HMW-GS (A), LMW-GS (B), and gliadin (C) contents and from measuring dry gluten content (D) and SDS sedimentation volume (E). F–K, Seed size-related phenotypes of nac019 mutants: grain width (F), grain length (G), TGW (H), cell size (I), cell number (J), and starch content (K). Three repeats were carried out for each sample to measure the total amounts of HMW-GSs, LMW-GSs, gliadins, dry gluten content, SDS sedimentation values, and starch content. For kernel weight, grain width, and grain length, nine replicates were used, each with approximately 25 g seeds. The single and double asterisks represent significant differences compared with WT and determined by Student’s t test at P < 0.05 and P < 0.01, respectively. L, Starch granules analysis of nac019 mutants and wild type. SG, starch granules; AL, aleurone. Three seeds were used for histological analysis for starch granules staining.
The nac019 mutants had smaller kernels than the wild type. Indeed, grain width and length (Figure 2, F and G), as well as thousand-grain weight (TGW; Figure 2, H) were lower in the mutants. We measured the cell size and cell number of developing endosperm from nac019 mutants and wild type, and the results indicated that cell size decreased while cell number increased in nac019 mutants when compared with the wild type (Figure 2, I and J and Supplemental Figure S8). Conversely, and in agreement with the seed-specific expression of TaNAC019, no visible phenotypic differences were detected in nac019 mutant mature plants relative to wild type, including plant height, tiller number, spike number, and flowering time (Supplemental Figure S8). These results indicate that TaNAC019 regulates glutenin and starch accumulation, gluten formation, and seed size and weight, thus influencing the processing quality of wheat flour.
TaNAC019 regulates the expression of various genes, including genes involved in SSP and starch accumulation
We identified the downstream targets of TaNAC019 by transcriptome deep sequencing (RNA-seq), using RNA extracted from wild-type Fielder and nac019-2 endosperms at 25 DAP. Among the 36,456 genes detected by RNA-seq, a total of 2,260 genes were downregulated and 3,580 were upregulated in nac019-2 compared with wild-type Fielder (Supplemental Data Set S1). Among the downregulated genes, we identified the three TaNAC019 homoeologs, indicating that the CRISPR/Cas9 gene editing affected not only their protein biosynthesis, but also their mRNA production or stability. Importantly, TaSPA was also downregulated in the nac019-2 mutant (Figure 3, A and Supplemental Data Set S1), indicating that although TaNAC019 binds directly to TaGlu-1Dy in the yeast one-hybrid system, it might also regulate SSP accumulation indirectly by modulating TaSPA expression. Indeed, all four TaGlu-1 loci expressed in Fielder (TaGlu-1Bx, TaGlu-1By, TaGlu-1Dx, and TaGlu-1Dy), as well as all 17 genes encoding LMW-GS and 66 of 78 genes encoding gliadins, were downregulated in nac019-2 (Supplemental Data Set S2).
Figure 3.
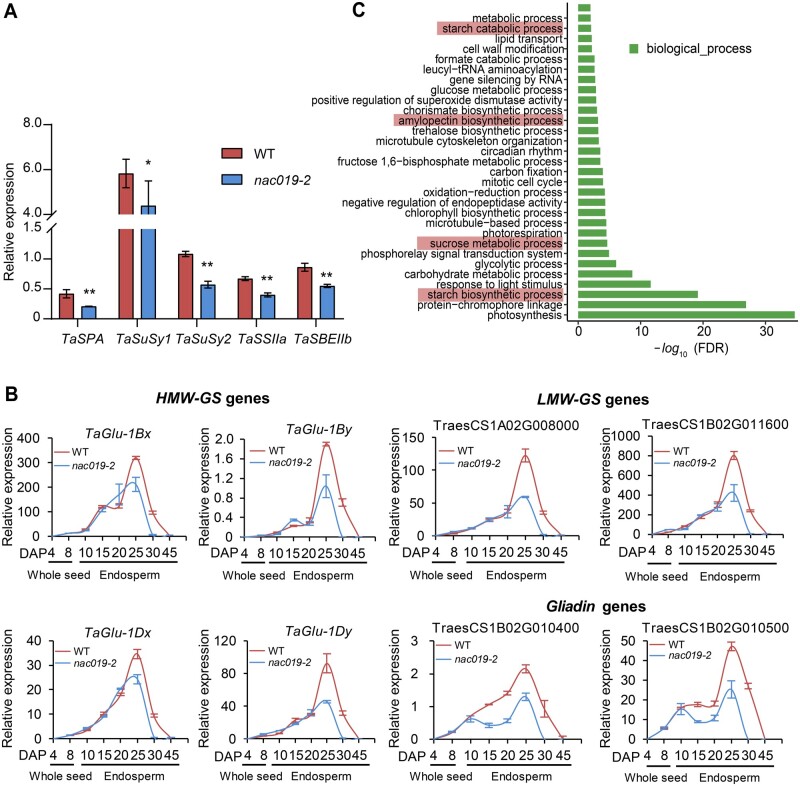
Genes and processes regulated by TaNAC019. A, RT-qPCR analysis of TaSPA, TaSBEIIb (starch branching enzyme IIb), TaSuSy1 (sucrose synthase), TaSuSy2 (sucrose synthase), and TaSSIIa (starch synthase 2a) expression in nac019-2 and wild-type endosperm at 25 DAP. RT-qPCR data were normalized to wheat TaACTIN. The data from three replicates are shown as ±sd. The single and double asterisks represent significant differences determined by Student’s t test at P < 0.05 and P < 0.01, respectively. B, Expression of the indicated HMW-GS, LMW-GS, and gliadin genes during seed development, analyzed in nac019-2 and wild type. RT-qPCR data were normalized to wheat TaACTIN, and each bar in the graph corresponds to the mean value of three data points from one representative biological replicate. C, GO analysis of downregulated genes in the nac019-2 transcriptome compared with wild type. P-values were adjusted by the Benjamini–Hochberg correction, and only significant categories (FDR < 0.05) are displayed. The red rectangles indicate the pathways related to starch metabolism.
Since RP-HPLC indicated that the levels of HMWs and LMW proteins were reduced in mature nac019 seeds, whereas the levels of gliadins were not (Figure 2, B–D), we investigated the effects of the nac019-2 mutation on SSP gene expression during seed development. Specifically, we used RT-qPCR to compare the transcript levels of the four TaGlu-1 genes, two LMW-GS genes, and two gliadin genes in nac019-2 versus the wild type. In agreement with the RNA-seq data, all of these genes were downregulated in 25 DAP endosperm from nac019-2 compared with the wild type (Figure 3, B). However, while expression of the genes encoding HMW-GS and LMW-GS started to decrease after 20 DAP, gliadin gene expression started to decrease earlier during seed development. Overall, these results indicate that TaNAC019 activates expression of glutenin- and gliadin-encoding genes during specific stages of endosperm development. However, in mature seeds of the nac019 knock-out mutants, the amounts of some SSPs, especially gliadins, were restored to wild-type levels, possibly due to the previously documented phenomenon of compensation between glutenin and gliadin levels that occurs when the level of one of these types of SSPs is altered (Scossa et al., 2008; Chen et al., 2019).
Gene ontology (GO) analysis revealed that the genes downregulated in nac019 are highly enriched for starch and amylopectin biosynthetic processes, as well as sucrose metabolic and starch catabolic processes (Figure 3, C). In agreement with the reduced starch levels in nac019, the downregulated genes included 45 genes related to starch metabolism (Supplemental Figure S9 and Supplemental Data Set S3). The downregulation of four genes that are key factors related to sucrose and starch metabolism, including TaSBEIIb (encoding starch branching enzyme IIb), TaSuSy1 (encoding sucrose synthase), TaSuSy2 (also encoding sucrose synthase), and TaSSIIa (encoding starch synthase IIa) was confirmed by RT-qPCR (Figure 3, A). Genes upregulated in nac019 were enriched in GO terms associated with oxidation–reduction processes, chitin catabolic process, and regulation of transcription (Supplemental Figure S10). Together, these results indicate that TaNAC019 concomitantly regulates both SSP and starch accumulation.
TaNAC019 binds to a specific motif in its target genes and activates their expression
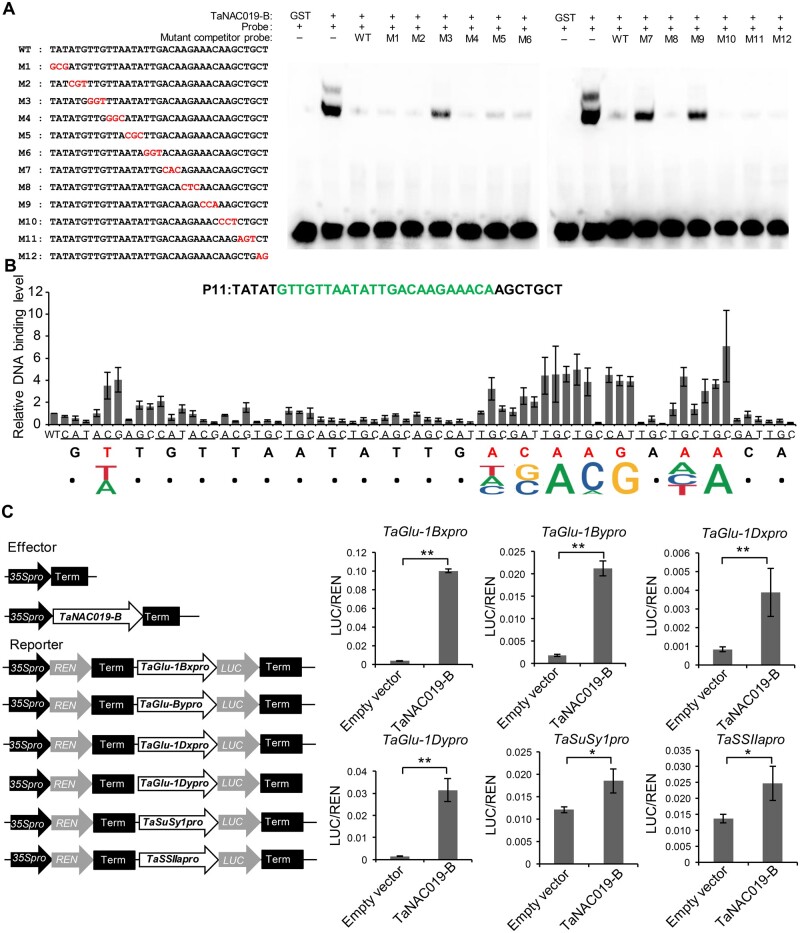
To explore the mechanism of TaNAC019-mediated regulation, we investigated the TaNAC019 binding motif in its target genes. We used 60 fragments covering a 1.6-kbp region upstream of the transcriptional start site (TSS) of TaGlu-1Dy to detect the DNA-binding ability of TaNAC019 by electrophoretic mobility shift assays (EMSAs). Four fragments were bound by a bacterially expressed glutathione S-transferase (GST)-TaNAC019-B recombinant protein, with regions P11 and P43 showing the strongest binding (Supplemental Figure S11). We validated the binding of TaNAC019-B to the P11 region by yeast one-hybrid assay (Supplemental Figure S12). To identify the precise TaNAC019-binding site, we used a series of oligonucleotides spanning the P11 region harboring mutations as competitor probes in EMSA. The probes M3, M7, and M9 failed to prevent TaNAC019 binding, indicating that the nucleotides mutated in these probes are required for the binding of TaNAC019 to its targets (Figure 4, A). We then performed EMSAs using competitor probes with single-nucleotide mutations covering the region between M3 and M9. Comparison to the wild-type probe revealed that the binding sequence was [AT]NNNNNNNNNNN[ATC][CG]A[CA]GN[ACT]A (Figure 4, B). To verify the specificity of this motif, we generated a P11 fragment harboring mutations in the predicted TaNAC019-binding site (MP11). TaNAC019-B recombinant protein failed to bind to this fragment in both yeast one-hybrid assays (Supplemental Figure S12) and EMSAs (Supplemental Figure S13).
Figure 4.
TaNAC019 binds to a specific motif in the TaGlu promoters. A, A three-nucleotide mutation series spanning the P11 oligonucleotide sequence was generated (highlighted in red) and used as competitors of the P11 probe in EMSAs with TaNAC019-B-GST recombinant protein. “+” and “−” indicate the presence and absence, respectively, of the corresponding probes and proteins. B, Mononucleotide mutations were introduced into the P11 core region (highlighted in green) and used as competitor probes in EMSAs. Each nucleotide of the core region was converted into the three other nucleotides. The average values of TaNAC019-B-binding levels from three independent replicates are shown on the y-axis. Nucleotides required for TaNAC019-B binding are indicated in red and the deduced binding motif is shown along the x-axis. C, Dual luciferase transcriptional activity assay to assess the ability of TaNAC019-B to transactivate target gene expression. Schematic diagrams of the effector and reporter plasmids. On the y-axis, LUC/REN indicates the ratio of the signal detected for firefly luciferase (LUC) versus Renilla reniformis luciferase (REN) activity. Error bars represent standard deviation for three replicates. The single and double asterisks indicate statistically significant differences estimated by Student’s t test at P < 0.05 and P < 0.01, respectively.
We searched for the TaNAC019-binding sequence in the promoter regions (2.5 kbp upstream of the TSS) of the genes that were differentially expressed in nac019-2 versus wild type. The TaNAC019-binding motif was found in 37.1% and 39.1% of down- and upregulated genes, respectively (Supplemental Data Set S1). Importantly, the TaSPA promoter lacked this motif. The TaNAC019-binding site was detected in all four HMW-GS genes expressed in Fielder, whereas only 3 out of 17 LMW-GS and 18 out of 69 gliadin genes contained this sequence (Supplemental Data Set S2). In addition, among the 48 downregulated genes involved in starch metabolism, 27 contained the TaNAC019-binding motif in their promoter regions (Supplemental Data Set S3). The binding of TaNAC019 to the TaGlu-1Bx, TaGlu-1By, TaGlu-1Dx, TaSuSy1, and TaSSIIa promoters was confirmed by EMSA (Supplemental Figure S13). We then assessed the transactivation of four TaGlu genes, TaSuSy1, and TaSSIIa by TaNAC019 in a dual luciferase transcriptional activity assay, which established that TaNAC019 activates the expression of these genes in vivo (Figure 4, C).
Overall, these findings suggest that TaNAC019 binds to and directly activates the expression of a subset of its targets, whereas other targets might be regulated via indirect mechanisms. Alternatively, TaNAC019 might bind to a subset of its targets at a different motif from that found in TaGlu-1Dy, or perhaps it binds to the same motif but located more than 2.5 kbp away from the TSS.
TaNAC019 and TaGAMyb physically interact and additively cooperate to activate TaGlu expression
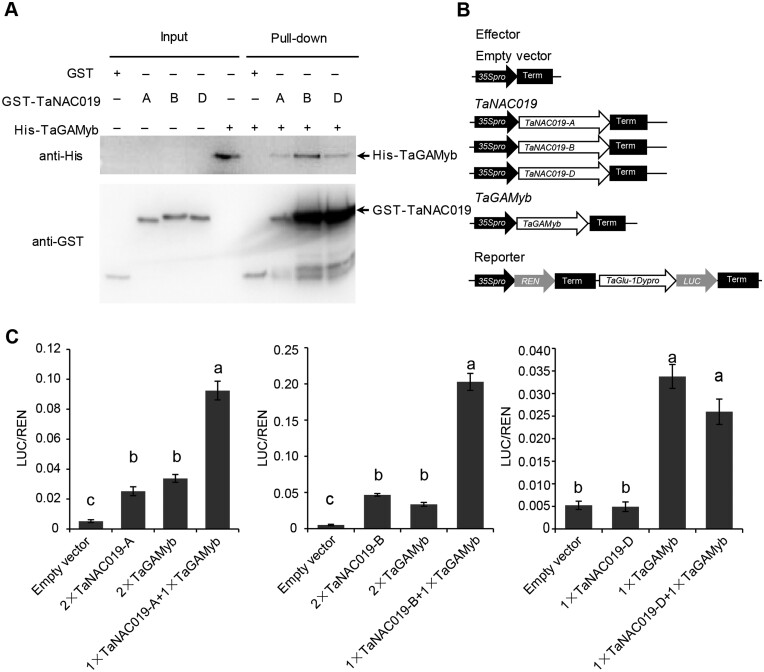
NAC TF activity sometimes requires interactions with paralogous proteins (Mendes et al., 2013; Mathew et al., 2016) or other TFs (Xu et al., 2013). We therefore assessed whether TaNAC019 homoeologs can interact with one another using the yeast two-hybrid system. However, none of the proteins interacted with each other in yeast two-hybrid assays (Supplemental Figure S14). Subsequently, we tested the interaction between each TaNAC019 homoeolog and several TFs involved in SSP regulation, such as TaSPA, TaGAMyb, and the TaGCN5-like protein HAT. TaNAC019 interacted with TaGAMyb, but not with TaGCN5 or TaSPA in this assay (Supplemental Figure S14). The physical interaction between TaNAC019-A/B/D and TaGAMyb was confirmed in a pull-down assay using recombinant proteins fused with an epitope tag (Figure 5, A).
Figure 5.
TaNAC019 and TaGAMyb physically interact and additively cooperate to drive TaGlu-1Dy promoter expression. A, Pull-down experiments to verify the interactions between the three TaNAC019 homoeologs and TaGAMyb. B and C, Dual luciferase transcriptional activity assays to assess the ability of TaNAC019 homoeologs and TaGAMyb to transactivate TaGlu-1Dy promoter expression. B, Schematic diagrams of the effector and reporter plasmids. C, On the y-axis, LUC/REN indicates the ratio of the signals detected for firefly luciferase (LUC) and R. reniformis luciferase (REN) activity. Error bars represent standard deviation for six replicates. The same letters above each bar indicate that the means did not differ significantly by one-way ANOVA and Tukey’s multiple comparison test at P < 0.05. “2×” and “1×” indicate that the final concentration of Agrobacteria containing the corresponding construct was OD600 0.2 and OD600 0.1, respectively.
To investigate the biological significance of the interaction between TaNAC019 and TaGAMyb, we assessed their ability to transactivate the TaGlu-1Dy promoter in a dual luciferase transcriptional activity assay (Figure 5, B). TaNAC019-A and TaNAC019-B, but not TaNAC019-D, activated the TaGlu-1Dy promoter (Figure 5, C). TaGAMyb also activated the promoter when expressed alone (Figure 5, C). Interestingly, co-expression of TaNAC019-A or TaNAC019-B with TaGAMyb led to a significant increase in TaGlu-1Dy promoter activation compared with the expression of the single effector constructs. Conversely, co-expressing TaNAC019-D and TaGAMyb activated the TaGlu-1Dy promoter to a level similar to that observed when TaGAMyb was expressed alone (Figure 5, C). Collectively, these results indicate that the three TaNAC019 homoeologs physically interact with TaGAMyb and that two of these TFs, TaNAC019-A and TaNAC019-B, cooperate and have additive effects with TaGAMyb in driving TaGlu-1Dy expression.
The three TaNAC019 homoeologs exhibit functional divergence
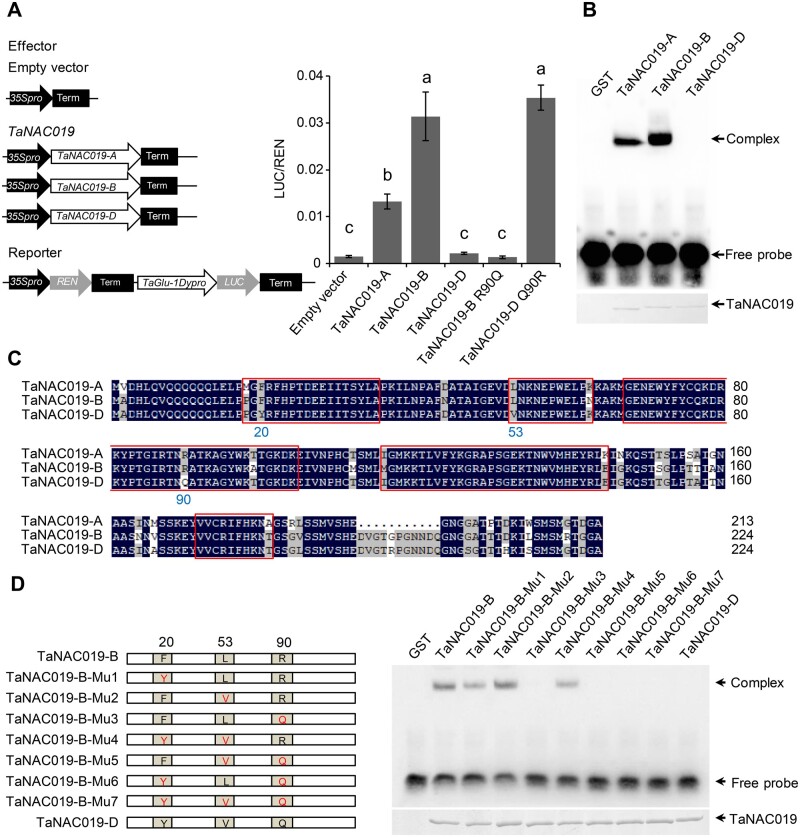
The combination and interactions of the three subgenomes of allohexaploid wheat have given rise to phenotypic variation, which is sometimes associated with the generation of functional divergence among homoeologous genes (Feldman et al., 2012). To test for possible functional divergence among TaNAC019-A, TaNAC019-B, and TaNAC019-D, we performed a dual luciferase transcriptional activity assay to compare their ability to activate TaGlu-1Dy expression. LUC activity was 9-fold and 21-fold higher in the presence of TaNAC019-A and TaNAC019-B, respectively, compared with the empty vector control, whereas no significant changes were detected in the presence of TaNAC019-D (Figure 6, A), which was consistent with results of Figure 5, C. Similar results were obtained when we analyzed the activation of the TaGlu-1By and TaGlu-1Dx promoters (Supplemental Figure S15). We also compared the ability of the three TaNAC019 homoeologs to bind to the TaGlu-1Dy promoter. Consistent with the results obtained from our transactivation analysis, TaNAC019-B showed a stronger binding affinity to the TaGlu-1Dy promoter than TaNAC019-A, whereas TaNAC019-D did not bind to this promoter (Figure 6, B).
Figure 6.
The three TaNAC019 homoeologs show functional divergence. A, Dual luciferase transcriptional activity assays to compare the ability of the three TaNAC019 homoeologs to transactivate TaGlu-1Dy promoter expression. R90Q and Q90R indicate the R to Q mutation at amino acid 90 of TaNAC019-B and the Q to R mutation at amino acid 90 of TaNAC019-D, respectively. On the y-axis, LUC/REN indicates the ratio of the signals detected for firefly luciferase (LUC) and R. reniformis luciferase (REN) activity. Error bars represent standard deviation for six replicates. The same letters above each bar indicate that the means did not differ significantly by one-way ANOVA and Tukey’s multiple comparison test at P < 0.05. B, EMSAs to compare TaGlu-1Dy promoter binding between the three TaNAC019 homoeologs. C, Amino acid sequence alignment of the three TaNAC019 homoeologs. The core NAC DNA-binding domains are highlighted with red rectangles. The amino acids where TaNAC019-D differs from TaNAC019-A and TaNAC019-B at positions 20, 53, and 90 are labeled. D, The TaNAC019-B sequence was mutated to generate proteins carrying different combinations of the three amino acid residues characterizing TaNAC019-D, and each GST-fused recombinant protein was used in EMSAs to assess the binding of these proteins to the TaGlu-1Dy promoter.
A comparison of the amino acid sequences of TaNAC019 homoeologs revealed a high level of similarity (92.9%). However, the NAC domain of TaNAC019-D differs from TaNAC019-A and TaNAC019-B in three amino acid residues located at positions 20, 53, and 90 (Figure 6, C). We introduced these three mutations into TaNAC019-B and used the resulting GST-fused recombinant proteins in EMSAs. The R90RQ mutation was sufficient to impair the ability of the protein to bind to the TaGlu-1Dy promoter (Figure 6, D). The R90Q mutation also affected the transactivation ability of TaNAC019-B in a dual luciferase assay, whereas the Q90R mutation in TaNAC019-D restored the transactivation activity of this protein (Figure 6, A). Overall, these results indicated that the abilities of the three homoeologs to bind to and activate the TaGlu-1Dy promoter differ, with TaNAC019-D showing no activity.
The elite allele TaNAC019-BI improves wheat processing quality
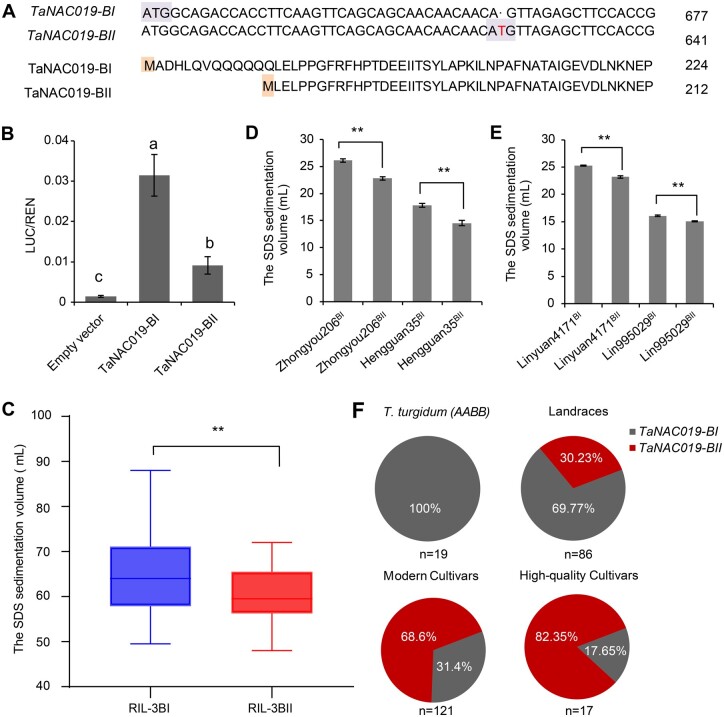
Finally, we used single-nucleotide polymorphism (SNP) analysis to investigate the genetic variation of the three homoeologs in a natural population of 226 wheat cultivars from China and foreign countries, including 19 tetraploid wheat (AABB) accessions, 86 landraces corresponding to the original pool of diversity in China, 121 elite modern cultivars registered after the Green Revolution, and 17 cultivars with high bread-baking quality. This analysis uncovered two TaNAC019-B haplotypes in this population (Supplemental Data Set S4). The first haplotype, named TaNAC019-BI, encoded a 224 amino acid protein (Figure 7, A). The second haplotype, named TaNAC019-BII, carried a single T insertion 39 nucleotides downstream of the translational start site. This insertion led to a premature translation termination, but also generated a new translational start site, thus producing a shorter protein lacking 12 N-terminal amino acids (a glutamine-rich activation domain; Escher et al., 2000). Dual luciferase transcriptional activity assays showed that TaNAC019-BI activates the TaGlu-1Dy promoter more strongly than TaNAC019-BII does (Figure 7, B).
Figure 7.
Natural variation of TaNAC019-B. A, Nucleotide and amino acid sequence alignments of TaNAC019-BI and TaNAC019-BII haplotypes indicating the detected polymorphism. The T insertion located 39 nucleotides downstream of the translation start site of TaNAC019-BII is shown in red. B, Dual luciferase transcriptional activity assay (performed as described in Figures 5, C and 6, A) to compare the efficiency of TaNAC019-BI and TaNAC019-BII in activating TaGlu-1Dy promoter. On the y-axis, LUC/REN indicates the ratio of the signals detected for firefly luciferase (LUC) and R. reniformis luciferase (REN) activity. Error bars represent standard deviation for six replicates. The different letters above each bar indicate that means differ significantly by one-way ANOVA and Tukey’s multiple comparison test at P < 0.05. C, Comparison of SDS sedimentation volume between RILs containing TaNAC019-BI and TaNAC019-BII, generated from Jimai44 (TaNAC019-BI) and Jimai229 (TaNAC019-BII). Lines in the box plots indicate the median. The 10th/90th percentiles of outliers are shown. The double asterisks represent significant differences, as determined by Student’s t test at P < 0.01. D, SDS sedimentation volume, measured in backcrossed lines in the Zhongyou206 and Hengguan35 genetic backgrounds containing TaNAC019-BII or TaNAC019-BI. Double asterisks indicate statistically significant differences estimated by Student’s t test at P < 0.01. E, As described in D, except that the SDS sedimentation volume was measured in backcrossed lines in the Linyuan4171 and Lin995029 genetic backgrounds containing TaNAC019-BII or TaNAC019-BI. F, Distribution of TaNAC019-BII and TaNAC019-BI haplotypes in tetraploid wheat, landraces, elite modern cultivars, and cultivars with high bread-baking quality. n indicates the sample size.
To compare the contributions of TaNAC019-BI and TaNAC019-BII to wheat processing quality, we examined recombinant inbred lines (RILs) generated from the parents Jimai44 (carrying TaNAC019-BI) and Jimai229 (carrying TaNAC019-BII). RILs containing TaNAC019-BI had better processing quality than RILs containing TaNAC019-BII, as revealed by higher SDS sedimentation volumes (Figure 7, C). To investigate the value of TaNAC019-BI for quality improvement during wheat breeding, we generated backcrossed lines containing TaNAC019-BII in the genetic background of two cultivars (Zhongyou206 and Hengguan35) originally harboring the TaNAC019-BI variant (Supplemental Figure S16). The SDS sedimentation volume was significantly lower in backcrossed lines (BC3F3) Zhongyou206BII and Hengguan35BII compared with Zhongyou206BI and Hengguan35BI (Figure 7, D). Conversely, the production of backcrossed lines (BC3F3) with TaNAC019-BI in the genetic background of two cultivars (Linyuan4171 and Lin995029) originally containing TaNAC019-BII led to an increase in the SDS sedimentation volume (Figure 7, E). Moreover, kernel weight increased in association with TaNAC019-BII, but only in the Hengguan35 genetic background (Supplemental Figure S17). These results suggest that the TaNAC019-BI allelic variant is associated with higher wheat quality compared with TaNAC019-BII.
All tetraploid ancestors tested here contained the TaNAC019-BI haplotype (Figure 7, F), indicating that this haplotype originated from tetraploid wheat. Curiously, 69.8% of the 86 landraces contained TaNAC019-BI, but only 31.4% of elite modern cultivars and 17.65% of wheat cultivars with high bread-making quality had this haplotype (Figure 7, F). These observations suggest that TaNAC019-BII was selected during the modern wheat breeding process. Collectively, these findings identify TaNAC019-BI as a novel marker for wheat processing quality that could be used in breeding programs.
Discussion
Mechanism of TaNAC019-mediated regulation of SSP and starch accumulation in wheat seeds
In this study, we showed that nac019 knock-out mutants of all three TaNAC019 homoeologs exhibited the following characteristics: (i) reduced transcript levels of almost all genes encoding SSPs (HMW-GS, LMW-GS, and gliadins) and TaSPA, a well-known SSP activator (Albani et al., 1997; Ravel et al., 2014), in 25 DAP endosperm, resulting in decreased levels of HMW-GS and LMW-GS but not gliadin in mature seeds; (ii) reduced transcript levels of many genes involved in starch metabolism in 25 DAP endosperm, resulting in lower starch contents in mature seeds; and (iii) reduced mature seed size and weight, as well as reduced parameters associated with processing quality. In addition, TaNAC019 bound to a specific motif in the TaGlu-Dy promoters, and activated their expression; it also physically interacted with and additively cooperated with TaGAMyb for TaGlu-Dy transactivation. Overall, these findings indicate that TaNAC019 plays an important role in the concomitant regulation of SSP and starch contents.
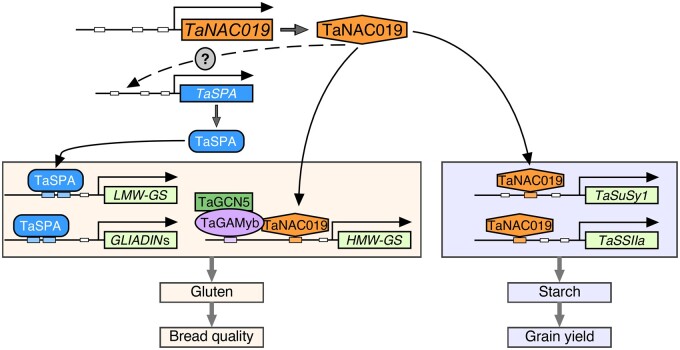
We propose a model describing how TaNAC019 regulates SSP and starch levels in wheat grain (Figure 8). According to this model, TaNAC019 regulates the expression of SSP genes in two ways. First, it directly binds to and activates SSP promoters. Second, it activates TaSPA expression, which might occur indirectly, as no TaNAC019-binding site was detected in the 2.5-kbp sequence upstream of the TSS of TaSPA. TaNAC019 binds to the promoters and activates the expression of HMW-GS genes via additive cooperation with TaGAMyb. Since TaGAMyb recruits the TaGCN5 HAT (Guo et al., 2015), it is likely that chromatin-related regulatory mechanisms are also involved in this process. In addition, SSP genes are expressed earlier than TaNAC019 genes, suggesting that other regulatory factors regulate SSP genes during early seed developmental stage, but TaNAC019 is required during later stage. TaNAC019-mediated activation of genes encoding LMW-GS and gliadin might primarily occur indirectly through the activation of TaSPA and/or other still unknown regulatory factors. Indeed, the TaNAC019-binding site identified in the TaGlu-1 promoters was only found in a few genes encoding LMW-GS and gliadin. Notably, in mature seeds, the LMW-GS and HMW-GS protein contents were lower in nac019 compared with wild type, whereas similar levels of gliadin were detected in these seeds. Perhaps this was due to compensation, which occurs when the protein level of one of the two SSP types is altered (Galili et al., 1986; Dumur et al., 2004; Gil-Humanes et al., 2012; Chen et al., 2019). Since gliadin transcript levels were lower in nac019 versus wild type even during late endosperm development, i.e. after the peak of gliadin gene expression, the restoration of gliadin proteins to wild-type levels in the nac019 mutants appears to be related to a post-transcriptional mechanism. A similar phenomenon was observed in the maize reas1-ref mutant, where the expression of zein genes was downregulated, but their translation level was increased significantly (Qi et al., 2016). TaNAC019-mediated regulation of starch accumulation likely occurs via a similar mechanism (Figure 8). TaNAC019 was able to directly bind the TaSuSy1 and TaSSIIa promoters and activate their expression in vivo (Figure 4, C and Supplemental Figure S13). Since the TFs and mechanisms involved in regulating wheat starch metabolism have not yet been clarified (Kang et al., 2013; Kumar et al., 2018), it is unclear whether, in addition to binding to and activating various starch metabolism genes, TaNAC019 also modulates the expression of specific regulatory factors. In addition, transcriptome analysis indicated that TaNAC019 is not only involved in SSP and starch gene regulation; it may also regulate genes involved in cell division or other processes controlling cell number.
Figure 8.
Model of the role of TaNAC019 in regulating SSP and starch accumulation in wheat seeds. See the text for details. Dotted arrows indicate indirect activation, while solid arrows represent direct activation due to the presence of a TaNAC019-binding site within the promoter. The question mark indicates the possible TaNAC019-mediated regulation of still unknown regulatory factors. TaNAC019 might regulate other genes involved in cell division or other processes controlling cell number, which were not shown in this model.
A recent study reported that overexpression of TaNAC019-A reduces seed size and starch content (Liu et al., 2020), which is similar to the phenotype of the nac019 mutant. TaNAC019 is specifically expressed in the endosperm; therefore, the reduced seed size and starch content might be a secondary effect of TaNAC019 overexpression outside of the seed. Moreover, overexpression of a TF perturbs the stoichiometry of TF-containing complexes, which can affect the regulation of their target genes (Levine and Tjian, 2003). By contrast, the precise elimination of a TF through gene editing represents a more specific strategy relative to overexpression. In addition, Liu et al. (2020) reported that TaNAC019-A repressed the expression of ADP-Glucose Pyrophosphorylase Small subunit 1 (TaAGPS1), which was downregulated in the overexpression line. We found that TaAGPS1 was also downregulated in the nac019 mutant (Supplemental Figure S9). TaNAC019 may act by both activating and repressing the expression of different genes and intra- and intermolecular interactions are relevant for NAC-mediated gene activation or repression (Xu et al., 2013). These observations might also indicate functional divergence among the three homoeologs, considering that TaNAC019-B showed stronger binding affinity for the TaGlu-1Dy promoter than TaNAC019-A (Figure 6, A and B), and TaNAC019-A might play an important role in TaAGPS1 regulation.
Finally, there are six additional NAC TF genes in wheat with seed-specific expression, whose functions are currently unknown. Since SSP and starch production are two major processes that occur during seed formation, it is reasonable to assume that other seed-specific NACs are involved in their regulation. We found that the mRNA expression patterns of these NACs differ, with specific transcripts accumulating at distinct developmental stages, suggesting that they have distinct and/or stage-specific activity. A comprehensive functional characterization of each seed-specific NAC will provide definitive information about their biological roles. However, these NACs probably form an elaborate network that functions during different stages of seed development and might interact at the genetic or physical levels to optimize SSP and starch levels.
Peculiarities of TaNAC019-mediated regulation
A recent study (Zhang et al., 2019) provided evidence that the NAC paralogs ZmNAC128 and ZmNAC130, which are specifically expressed in maize seed, regulate both SSP and starch accumulation. These maize NACs are the orthologs of two rice NACs, Os01g01470 and Os01g29840. In previous phylogenetic analyses (Borrill et al., 2017; Guerin et al., 2019) and in our data (Supplemental Figure S1), a reciprocal BLAST strategy was used to identify wheat NAC orthologs across rice and barley. In this strategy, if the reciprocal BLAST analysis did not identify the same pair of genes in both directions, the genes in question were ruled out as orthologs. Although some rice orthologs of wheat NACs were found using this strategy, no TaNAC019 orthologs were identified in rice. Hence, although ZmNAC128/ZmNAC130 and TaNAC019 play similar roles in regulating SSP and starch accumulation, they are not orthologs. However, it is worth noting that ZmNAC128/ZmNAC130 and Os01g01470/Os01g29840 do not have any other orthologs among seed-specific NACs in wheat. This finding suggests that seed-specific rice/maize NACs differentiated from wheat NACs, likely reflecting the evolutionary distance between these species. In this context, it is relevant that wheat contains specific SSP classes, such as HMWs, which may require specific regulatory factors not present in maize or rice.
Aside from possible evolutionarily related differentiation, ZmNAC128/ZmNAC130 and TaNAC019 also differ in the number of their SSP and starch-related targets. ZmNAC128/ZmNAC130 regulate the expression of some but not all SSP genes (Zhang et al., 2019), whereas TaNAC019 modulates the expression of genes for all SSP types. In addition, ZmNAC128/ZmNAC130 activate the expression of seven genes involved in starch metabolism (Zhang et al., 2019). Six of these genes have wheat orthologs that are also downregulated in nac019-2, but 10 additional starch-related genes were downregulated in this mutant (Supplemental Figure S9). In conclusion, it appears that TaNAC019 has a broader effect than ZmNAC128/ZmNAC130 on the expression of genes associated with SSP and starch production.
Another TaNAC019 feature is the presence of three homoeologs exhibiting functional divergence, which represents an intrinsic peculiarity of allohexaploid bread wheat, where different subgenome combinations and interactions have given rise to phenotypic variation (Feldman et al., 2012). We found that one of the three TaNAC019 homoeologs, TaNAC019-D, can interact with TaGAMyb but does not bind to or activate TaGlu promoters. The very low transcript levels of TaNAC019-D suggest that it might be a pseudogene without any residual relevant function. However, this notion contrasts with the finding that the R90Q mutation, which impairs the binding capacity of TaNAC019-D, is fully conserved in all hexaploid wheat accessions (Supplemental Data Set S4). The TaNAC019-D sequence in the seven diploid goat grass (Aegilops tauschii) accessions, which are among the D genome donors for hexaploid wheat, also contains the Q residue, suggesting that this mutation originated in A. tauschii (Supplemental Data Set S4). Similar to other NACs (Mendes et al., 2013; Mathew et al., 2016), TaNAC019 homoeologs, including TaNAC019-D, interact with other regulatory factors such as TaGAMyb. The interaction between TaNAC019-D and TaGAMyb did not affect the TaGAMyb-related activation of TaGlu in dual luciferase transactivation assays. However, TaNAC019-D might act as a modulator of gene expression with other TFs.
In this study, we focused on the roles of TaNAC019 proteins as activators of SSPs and starch accumulation. However, transcriptome analysis revealed a similar number of down- and upregulated genes in nac019-2 versus the wild type, and a similar percentage of TaNAC019-binding sites was found in the two groups of genes. Indeed, a NAC can act both as an activator and repressor, and the conformational interactions between the N-terminal NAC domain, harboring a NAC repression domain (NARD), and the C-terminal transcriptional activation region (TAR) of NAC proteins are crucial for their activating or repressive activities (Xu et al., 2013). Accordingly, the capacity of TaNAC019 to function as a transcriptional repressor or activator might depend on its conformational assembly at the binding site. Different assemblies could occur depending on interactions with other proteins, as well as the promoter context, which could provide different stabilization contacts for the DNA–protein interaction.
TaNAC019 is a new candidate gene for improving processing quality in wheat
Two TFs were shown to be involved in the concomitant regulation of SSP and starch accumulation in rice and maize seeds (Wang et al., 2013; Zhang et al., 2019). TaNAC019 is the first wheat TF with this function identified to date in wheat. Starch content is related to yield, while SSP content and composition are associated with quality (Rharrabti et al., 2001; Zorb et al., 2018). Therefore, the finding that TaNAC019 regulates both starch and SSP contents and affects the HMW/gliadin and LMW/gliadin ratios in mature seeds makes TaNAC019 an excellent candidate for developing wheat cultivars with both high yields and good quality. Overexpression of TaNAC019-A led to reduced starch content, kernel weight, and kernel width (Liu et al., 2020), indicating that specific levels of TaNAC019 might be required for yield and quality improvement. Biotechnological approaches may be used to precisely control TaNAC019 expression in seeds. In addition, our analysis of the genetic variability of TaNAC019 in natural populations provided evidence that a specific haplotype, TaNAC019-BI, is positively associated with wheat processing quality. Hence, TaNAC019-BI represents a genetic marker that may be used in breeding programs.
Importantly, only 30% of modern cultivars carry a TaNAC019-BI allele, whereas this allele was detected in 70% of the landraces examined, suggesting that this allele has not yet been well exploited by breeding programs. During the Green Revolution, bread wheat underwent a dramatic transformation, resulting in a significant reduction in genetic diversity, while landrace germplasms remained largely unexploited (Balfourier et al., 2019). As demonstrated by the Reduced height (Rht) dwarfing and Fusarium head blight 1 (Fhb1) disease resistance genes (Hedden, 2003; Su et al., 2019), these landrace accessions are a valuable reservoir of novel alleles. Perhaps the TaNAC019-BI haplotype from landraces could be exploited, providing another example of how this germplasm could contribute to breeding programs.
Materials and methods
Plant materials
The names and geographical origins of all wheat (T. aestivum) cultivars, landraces and Triticum turgidum (AABB) accessions used in this study are listed in Supplemental Data Set S4. All wheat plants were grown in the experimental field of China Agricultural University in Beijing (39°57ʹN, 116°17ʹE) and in a greenhouse at a relative humidity of 75% and 26/20°C day/night temperatures, with a light intensity of 3,000 lux (Master GreenPower CG T 400W E40; Philips). For gene expression analysis, root, leaf, and stem tissues were harvested from greenhouse-grown plants 10 days after seed germination, and young spikes (2 cm), embryos at 15 DAP, whole seeds at the watery ripe stage (4 and 7 DAP), and endosperm at the milky stage (12, 15, and 18 DAP), soft dough stage (20 and 25 DAP), and hard dough stage (30 and 45 DAP) were harvested from plants grown in the field. The endosperm and embryos were manually obtained by dissecting grains. For each sample, developing seeds from three different plants within one planting plot were mixed to represent one biological replicate and at least three biological replicates from different plot or different year (2017–2019) were used. All samples were immediately frozen in liquid nitrogen and stored at −80°C.
Backcrossed lines were obtained as shown in Supplemental Figure S15. The restriction endonuclease PciI (NEB, R0655L) was used to distinguish between TaNAC019-BI and TaNAC019-BII by the Cleaved Amplified Polymorphic Sequence (CAPS) method, which was also used for genotyping during backcrossed lines generation. The RIL population was generated from two parents (Jimai44 and Jimai229) with selfing for five generations.
Yeast one-hybrid screening and assay
For yeast one-hybrid screening, a cDNA expression library derived from wheat seeds mixed from various pollination days in the pGADT7 vector was used as prey. The promoter fragment for TaGlu-1Dy was amplified and cloned into the pAbAi vector. This construct was linearized by the restriction enzyme BstBI and then integrated into the genome of the Y1HGold yeast strain to obtain a bait-specific reporter strain. Yeast one-hybrid screening was performed following the manufacturer’s instructions (Takara, 630491).
For the detection of TaNAC019 binding to a specific TaGlu-1Dy promoter region, the 35 bp P11 sequence and the corresponding MP11-mutated sequence as three repeats were synthesized and inserted into the pAbAi vector, respectively (BGI, China). The construct was linearized by BstBI digestion and transformed into Y1HGold cells to generate the Y1H bait strain. Full-length TaNAC019-BI cDNA was inserted into the pGADT7 vector and transformed into the Y1H bait strain. The transformants were selected on synthetic dropout −Ura and −Leu medium containing 125 mM AbA.
Phylogenetic analysis
We obtained protein sequences with a NAM domain (PF02365) in barley (122), wheat (454), and rice (94) from Ensembl Plants with the BioMart tool from Phytozome (Supplemental File S2). Maize sequences were reported by Zhang et al. (2019; Supplemental File S2). We aligned the amino acid sequences corresponding to the NAM domain with ClustalW and constructed a phylogenetic tree using the neighbor-joining method in MEGA 7 software with default parameters (Kumar et al., 2016). The evolutionary distances were calculated using the Poisson model. The phylogeny test was computed using bootstrap method with 10,000 replications (Supplemental File S3).
Subcellular localization
The full-length open-reading frames (ORFs) of TaNAC019-A/B/D were amplified using specific primers and Tks Gflex DNA polymerase (TaKaRa, R060A) and cloned upstream of the 5′-end of the GFP gene (GFP) in the pCAMBIA1300 vector containing the mannopine synthase promoter (maspro). The resulting construct maspro:TaNAC019-GFP and the maspro:GFP control were separately introduced into young leaves of 4-week-old N. benthamiana plants by Agrobacterium (Agrobacterium tumefaciens)-mediated transient infiltration (Liu et al., 2010). The infiltrated plants were cultured at 22°C for 48 h, and GFP signals observed on a confocal microscope (Eclipse TE2000; Nikon).
Production of nac019 knock-out mutants
For genome editing via CRISPR/Cas9, a sgRNA was designed based on the first exon sequence of TaNAC019 using the E-CRISP Design website (http://www.e-crisp.org/E-CRISP/designcrispr.html). Two reverse complementary sgRNA sequences with BsaI cohesive ends were synthesized. Double-stranded oligonucleotides were annealed by slowly cooling from 100°C to room temperature and inserted into the expression cassette of the pBUE411 vector (Xing et al., 2014). The resulting plasmid was transformed into wheat cultivar Fielder via Agrobacterium-mediated (strain EHA105) transformation (Ishida et al., 2015). Homozygous nac019-1 and nac019-2 mutants were obtained by selfing of T0 plants for four generations. At each generation, sequences of all three homoeologs from representative transgenic seedlings were analyzed. The lines with homozygous editing at TaNAC019-A/B/D were selected for selfing. For example, lines homozygous for edited alleles at TaNAC019-A and TaNAC019-B were obtained in the T2 generation of nac019-1 mutant, and homozygous edited alleles at TaNAC019-D were obtained in the T3 generation.
Phenotypic analysis
Wild-type and nac019 seeds were planted in a block containing 10 rows (1.5 m long) at a spacing of 20 cm. Mature seeds from the plot were harvested and seeds from every 10 plants were mixed for phenotypic assessment. After adjusting the moisture content of seeds to 14%, the seeds were milled into flour using a Brabender Quadrumat Junior laboratory mill (Brabender, Duisburg, Germany). The contents of HMW-GSs, LMW-GSs, and gliadins were detected by RP-HPLC as previously described with some modifications (Zhang et al., 2014): the Agilent Technologies 1260 Infinity II RP-HPLC system and an Agilent ZORBAX 300SB-C18 column (150 mm × 4.6 mm, 5 μm) were used. The elution solvents were water containing 0.6 mL/L trifluoroacetic acid (solvent A) and acetonitrile containing 0.6 mL/L trifluoroacetic acid (solvent B). The elution gradient for glutenins was as follows: 0 − 2 min, 21% B; 2 − 52 min, from 21% to 53.5% B; 52 − 54 min, from 53.5% to 21% B; 54 − 60 min, 21% B. The elution gradient for gliadins was as follows: 0 − 2 min, 25% B; 2 − 27 min, from 25% to 50% B; 27 − 35 min, from 50% to 25% B; 35 − 50 min, 25% B. The column temperature was 60°C and the flow rate was 1 mL/min. The injection volume was 8 μL and the eluent was monitored at 210 nm. The total amounts of HMW-GSs, LMW-GSs, and gliadins were estimated by integrating the relevant RP-HPLC peaks present in the chromatograms. Three replications were performed for each sample. Dry gluten content was evaluated using the voluntary standard method of the National Standard of China 5506.4-2008 (GB/T5506.4-2008). SDS sedimentation values were determined using 2 g of flour to detect sedimentation volume for 5 min as described by Chen et al. (2019). Three repeats were carried out for each sample.
A Wanshen SC-G seed detector (Hangzhou Wanshen Detection Technology Co., Ltd.) was used to measure kernel weight, grain width, and grain length. Nine replicates were used, each with approximately 25 g seeds. Starch content was determined using Megazyme Total Starch Assay Kit (Megazyme; Catalog no.: KTSTA-50A) based on the use of thermostable α-amylase and amyloglucosidase. Three repeats were carried out for each sample.
Histological analysis
Immature wild-type and nac019 mutant seeds at 6, 15, and 20 DAP were sampled from spikes. Seeds were fixed in FAA solution (63% ethanol, 5% acetic acid, 2% formaldehyde) for 24 h at room temperature. After dehydration in a graded ethanol series, sections were slowly infiltrated with Steedman’s wax, embedded in paraffin, sectioned to 4 μm thickness with a microtome, and dewaxed in absolute ethanol. Sections (4 μm) were stained with Toluidine Blue. Images were collected using a OLYMPUS BX53 Microscope. Endosperm cell number was manually calculated for three seeds and the differences between WT and the nac019 mutants were determined by one-way ANOVA. Starch granules from three seeds of each line were stained by Periodic Acid–Schiff.
RT-qPCR
Total RNA was extracted from samples using TransZol Plant (TransGen Biotech, ET121-01), and first-strand cDNAs (20 μL) were synthesized from 1 μg starting total RNA using a reverse transcription kit (Vazyme Biotech, R223-01) according to the manufacturer’s instructions. The first-strand cDNAs was diluted with H2O to 80 μL. For RT-qPCR, the reaction mixture was composed of the 1 μL cDNA template, 0.2 mΜ primers, and 5 μL SYBR Green Mix (Vazyme Biotech, Q121-02/03) in a final volume of 10 μL. Amplification was performed using a CFX96 real-time system (BioRad). Differences in relative transcript levels were calculated using the 2−ΔCT method (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_040980.pdf) relative to wheat TaACTIN (TraesCS5B02G124100). RT-qPCR was performed as technical triplicates per sample. Three biological replicates were performed, with similar results; the results from one replicate are shown in the figures.
Electrophoretic mobility shift assay
TaNAC019 was expressed in Escherichia coli (BL21 DE3) by cloning the TaNAC019 coding sequences into the pGEX6P-1 vector (Solarbio, P0300) to produce a GST fusion protein. Recombinant protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in Luria Bertani (LB) buffer overnight at 16°C. The cells were harvested, sonicated in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) containing 1 mM phenylmethanesulfonyl fluoride (PMSF) and 1/2 of a tablet of protease inhibitor cocktail (Roche) for 1 h, and centrifuged at 13,000 × g for 45 min. The supernatant was mixed with GST MAG Agarose Beads (Novagen, http://www.novagen.com) and rocked overnight at 4°C. The fusion protein was eluted from the beads with 50 mM Tris–HCl (pH 8.0) containing 10 mM reduced glutathione.
The double-stranded probes (labeled with biotin at their 5′-end) were annealed from complementary oligonucleotides by cooling from 100°C to room temperature in annealing buffer. The sequences of the probes are listed in Supplemental Data Set S5. The DNA-binding reactions were performed in a solution containing 20% binding buffer (100 mM Tris, 500 mM KCl, 10 mM DTT; pH 7.5), 2.5% glycerol, 0.2 mM EDTA, and 50 ng/μL poly(dI–dC). Following incubation at room temperature for 20 min, the samples were loaded onto a 6% native polyacrylamide gel. Transfer to a nylon membrane and detection of biotin-labeled DNA were performed according to the manufacturer’s instructions (Thermo Scientific 20148 LightShift Chemiluminescent EMSA Kit). Three replicates were performed, with similar results; the results from one replicate are shown in the figures.
Yeast two-hybrid assay
The coding sequences of TaSPA, TaNAC019-A, TaNAC019-B, and TaNAC019-D were cloned into the pGADT7 vector, while TaGAMyb, TaGCN5, TaNAC019-A, and TaNAC019-B were cloned into the pGBKT7 vector. For the yeast two-hybrid assays, the plasmids were transformed into yeast strain AH109 using the lithium acetate method, and the cells were grown on minimal medium (−Leu −Trp) according to the manufacturer’s instructions (Clontech). The positive colonies with T-antigen and P53 were plated onto minimal medium (−Leu −Trp −His −Ade) to test for possible interactions.
Dual luciferase transcriptional activity assay
Dual luciferase transcriptional activity assays were performed as previously described (Guan et al., 2014). Fragments corresponding to 1,600 bp upstream of the TSS of TaGlu-1Bx, TaGlu-1By, TaGlu-1Dx, TaGlu-1Dy, TaSuSy1, and TaSSIIa were PCR-amplified from CS genomic DNA and cloned into the pGreenII 0800-LUC vector as reporter plasmids (Hellens et al., 2005). The ORFs of TaNAC019 and TaGAMyb were cloned into the pHB vector as effectors. Young leaves of 4-week-old N. benthamiana plants were co-infiltrated with Agrobacterium (strain GV3101) harboring different combinations of these plasmids. Firefly luciferase activity derived from the TaGlu-1Bx (TaGlu-1Bxpro:LUC), TaGlu-1Dy (TaGlu-1Dypro:LUC), TaGlu-1By (TaGlu-1Bypro:LUC), TaGlu-1Dx (TaGlu-1Dxpro:LUC), TaSuSy1 (TaSuSy1pro:LUC), or TaSSIIa (TaSSIIapro:LUC) promoters and Renilla luciferase under the control of the 35S promoter (35Spro:REN) were quantified using the Dual-Luciferase Reporter Assay system (Promega) with a Synergy 2 Multi-Detection Microplate Reader (BioTek Instruments). Normalized data are presented as the ratio of luminescent signal intensity for reporter versus internal control reporter (35Spro:REN) from three independent biological samples.
TaNAC019 antibody production
The full-length TaNAC019-B sequence was used to produce a specific rabbit polyclonal antibody by Wuxi Pharma Tech Company (China).
Immunoblot analysis
Harvested seeds and endosperm were ground in liquid nitrogen, and proteins were extracted in 4× loading buffer (200 mM Tris–HCl pH 6.8, 40% [v/v] glycerol, 8% [w/v] SDS, and 20% [v/v] β-mercaptoethanol) at 100°C for 10 min. The solution was placed on ice for 2 min and centrifuged for 10 min at 12,000 rpm at room temperature. Protein extracts were separated by 10% (v/v) SDS-PAGE and transferred to a polyvinylidene fluoride membrane (cat. no. 162-0177; Bio-Rad) for 60 min at 300 mA under temperature-controlled conditions. The membrane was blocked using 5% (w/v) fat-free milk for 1 h and incubated overnight at 4°C in 1× Tris-buffered saline containing Tween 20 (TBST; 1× TBS with 0.1% [v/v] Tween 20) and anti-TaNAC019-B antibody (diluted 1:1,000). After washing three times with 1× TBST for 10 min each time, the membrane was incubated in 5% (w/v) fat-free milk in 1× TBST solution containing the secondary antibody (diluted 1:3,000) for 1 h at room temperature, followed by three washes (10 min each time) with 1× TBST. Two-component Reagent-clarity Western ECL Substrate (cat. no. 170-5060; Bio-Rad) was used for detection. The signals were detected using a Chemiluminescence Imaging system (ChemiScope 6000). Three biological replicates were performed, with similar results; the results from one replicate are shown in the figures.
Pull-down assay
To generate His-tagged TaGAMyb protein, the TaGAMyb ORF was amplified and cloned into the EcoRI/SalI sites of the pET-32a vector (Novagen). To generate GST-tagged TaNAC019-A, TaNAC019-B, and TaNAC019-D, the respective ORFs were amplified and cloned separately into the BamHI/EcoRI sites of the pGEX-6P-1 vector (Solarbio, P0300). Recombinant protein production was induced following the manufacturer’s manual. For the GST pull-down assay, approximately 5 mL E. coli lysates containing GST or GST-TaNAC019 fusion protein were mixed with 400 μL immobilized glutathione agarose (Thermo Scientific) for 2 h at 4°C. The beads were washed five times with 1 mL PBS and incubated with His-tagged TaGAMyb fusion protein lysates for more than 2 h at 4°C. The protein complex–immobilized glutathione agarose conjugates were eluted in 100 μL 4× loading buffer (200 mM of Tris–HCl at pH 6.8, 40% [v/v] glycerol, 8% [w/v] SDS, and 20% [v/v] β-mercaptoethanol) at 100°C for 10 min. The bound proteins were analyzed by 10% PAGE and immunoblot analysis was performed with anti-His or anti-GST antibodies (diluted 1:1,000; Abcam). Three replicates were performed, with similar results; the results from one replicate are shown in the figures.
Transcriptome deep sequencing (RNA-seq)
Three biological replicates were performed for each sample. RNA-seq libraries were prepared with a TruSeq RNA Sample Preparation Kit v2 (Illumina) according to the manufacturer’s instructions and sequenced to generate 150-bp paired-end reads on the HiSeq platform. Approximately 72–102 million reads were generated per sample. Raw reads obtained from Illumina sequencing were processed and filtered using Trimmomatic (Bolger et al., 2014) with “SLIDINGWINDOW:4:20 MINLEN:40” to generate high-quality reads. The RNA-seq reads were aligned to the CS reference genome (International Wheat Genome Sequencing Consortium RefSeq v1.1) downloaded from Ensembl Plants using STAR with default parameters (Dobin et al., 2013). FeatureCounts was used to count the reads mapped to each gene (Liao et al., 2014). The read counts were normalized to FPKM values to show the relative gene expression levels. After using a gene expression criterion of FPKM value ≥ 1 in at least one sample, DESeq2 was used for differential gene expression analysis (Love et al., 2014). The genes showing an absolute value of log2 (fold-change) > 0.8 and adjusted P-value (false discovery rate) of <0.05 were considered to be differentially expressed genes.
Significantly enriched GO categories of up- and downregulated genes (adjusted P-value [false discovery rate] <0.05) were identified using ClusterProfiler (Yu et al., 2012). FIMO with default parameters was used to identify TaNAC019-binding motifs within 2.5 kbp upstream of the TSS of each gene (Grant et al., 2011).
Statistical analysis
Quantitative data, including three biological and at least three technical replicates, are presented in the form of mean ± sd. Means of two samples were compared using Student’s two-tailed t tests. Analysis of variance (one-way ANOVA) was conducted using Graphpad Prism 8.0.2 software with default parameters. Significant differences were determined by one-way ANOVA or the Student’s t test (Supplemental File S1): *P < 0.05 and **P < 0.01. Tukey’s multiple comparison test was used to determine exactly which group means were different.
Data availability
Accession numbers
The cDNA sequences of TaNAC019-A, TaNAC019-BI, TaNAC019-BII, and TaNAC019-D have been deposited in GenBank under accession numbers MN685185, MN685186, MN685187, and MN685188, respectively. RNA sequencing data are available at the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA656089.
Supplemental data
Supplemental Figure S1. Phylogenetic analysis of wheat, maize, rice, and barley NACs.
Supplemental Figure S2. Genomic locations of TaNAC019 homoeologs.
Supplemental Figure S3. Subcellular localization of TaNAC019 homoeologs.
Supplemental Figure S4. TaGlu expression during wheat seed development.
Supplemental Figure S5. Characterization of anti-TaNAC019 antibody and analysis of TaNAC019 levels.
Supplemental Figure S6. Strategy used to produce the nac019 knockout lines.
Supplemental Figure S7. HMW-GS from nac019-1, nac019-2, and WT in a silver-stained protein gel.
Supplemental Figure S8. Histological analysis of developing seeds of nac019 and wild type and phenotypes of mature nac019 and wild-type plants.
Supplemental Figure S9. Starch metabolism genes that are downregulated in nac019-2 compared with the wild type.
Supplemental Figure S10. GO analysis of genes up-regulated in the nac019-2 transcriptome compared with the wild type.
Supplemental Figure S11. Analysis of the binding of TaNAC019-B to the TaGlu-1Dy promoter via EMSA.
Supplemental Figure S12. Analysis of the binding of TaNAC019-B to the TaGlu-1Dy promoter via yeast one-hybrid analysis.
Supplemental Figure S13. Validation of the TaNAC019-binding motif identified using EMSA assay.
Supplemental Figure S14. Yeast two-hybrid analysis of TaNAC019 protein interactions.
Supplemental Figure S15. Dual luciferase transcriptional activity assays to detect TaNAC019-induced activation of TaGlu-1Dx and TaGlu-1Bx promoters.
Supplemental Figure S16. Schematic representation of the method used to generate backcrossed lines.
Supplemental Figure S17. TGWs of the backcrossed lines.
Supplemental Data Set S1. List of differentially expressed transcripts in the nac019 mutant and wild type.
Supplemental Data Set S2. Expression of SSP genes in the nac019 and wild type.
Supplemental Data Set S3. Expression of genes related to starch biosynthesis in nac019 and the wild type.
Supplemental Data Set S4. Names and geographical origins of all wheats used in this research, and haplotype analysis of TaNAC019-B and TaNAC019-D.
Supplemental Data Set S5. Probes sequences and location on the promoter of TaGlu-1Dy used for EMSA.
Supplemental Data Set S6. Primers used for genes expression and vectors construction.
Supplemental File S1. Summary of statistical analyses.
Supplemental File S2. Protein sequences for the phylogenetic tree shown in Supplemental Figure S1.
Supplemental File S3. Protein sequence alignment for the phylogenetic tree shown in Supplemental Figure S1.
Supplemental File S4. Newick format of the phylogenetic tree of Supplemental Figure S4.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Rentao Song and Jinshen Lai (China Agricultural University) for helpful discussions and comments on the text. They thank Dr. Xianchun Xia and Shuanghe Cao (Chinese Academy of Agricultural Sciences) for assistance with RP-HPLC analysis, Dr. Rentao Song (China Agricultural University) for providing the transient expression vectors (pUC18-35S and pGreenII 0800-LUC), Dr. Tingjie Cao (Henan Academy of Agricultural Sciences) for providing the wheat cultivars from Henan province, China, and Dr. Jianru Zuo (Chinese Academy of Sciences) for suggestions about EMSA optimization.
Funding
This work was supported by the National Natural Science Foundation of China [Grant No. 91935304], the National Key Research and Development Program of China [2020YFE0202300], and the Beijing Outstanding University Discipline Program, TaiShan industrial Experts Programme [No. tscy20190106].
Conflict of interest statement. The authors declare that they have no competing interests.
Y.G. and K.A. performed most of the experiments. W.G. contributed to the yeast two-hybrid assays. Y.C. and F.J. analyzed the data. R.Z., X.Z., and S.C. contributed to the phenotypic analysis. V.R. helped revise the manuscript. X.C., M.X., H.P., Z.H., W.G., and J.D. performed some of the experiments. Y.Y., V.R., Z.N., and Q.S. designed the research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) is: Yingyin Yao (yingyin@cau.edu.cn).
References
- Albani D, Hammond-Kosack MC, Smith C, Conlan S, Colot V, Holdsworth M, Bevan MW (1997) The wheat transcriptional activator SPA: a seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 9: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfourier F, Bouchet S, Robert S, De Oliveira R, Rimbert H, Kitt J, Choulet F, International Wheat Genome Sequencing C., BreedWheat C, Paux E (2019) Worldwide phylogeography and history of wheat genetic diversity. Sci Adv 5: eaav0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiekierski JR (2017) What is gluten? J Gastroenterol Hepatol 32: 78–81 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrill P, Harrington SA, Uauy C (2017) Genome-wide sequence and expression analysis of the NAC transcription factor family in polyploid wheat. G3 7: 3019–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet J, Merlino M, Plessis A, Gaudin JC, Dardevet M, Perrochon S, Alvarez D, Risacher T, Martre P, Ravel C (2019) The bZIP transcription factor SPA heterodimerizing protein represses glutenin synthesis in Triticum aestivum. Plant J 97: 858–871 [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhang W, Gao Y, Yang C, Gao X, Peng H, Hu Z, Xin M, Ni Z, Zhang P, et al. (2019) High molecular weight glutenin subunits 1Bx7 and 1By9 encoded by Glu-B1 locus affect wheat dough properties and sponge cake quality. J Agric Food Chem 67: 11796–11804 [DOI] [PubMed] [Google Scholar]
- Cho S-W, Roy SK, Chun J-B, Cho K, Park CS (2017) Overexpression of the Bx7 high molecular weight glutenin subunit on the Glu-B1 locus in a Korean wheat landrace. Plant Biotechnol Rep 11: 97–105 [Google Scholar]
- Christiansen MW, Holm PB, Gregersen PL (2011) Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res Notes 4: 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Ni Z, Yao Y, Nie X, Sun Q (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol Biol 63: 73–84 [DOI] [PubMed] [Google Scholar]
- Dong L, Zhang X, Liu D, Fan H, Sun J, Zhang Z, Qin H, Li B, Hao S, Li Z, et al. (2010) New insights into the organization, recombination, expression and functional mechanism of low molecular weight glutenin subunit genes in bread wheat. PLoS ONE 5: e13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumur J, Jahier J, Bancel E, Lauriere M, Bernard M, Branlard G (2004) Proteomic analysis of aneuploid lines in the homeologous group 1 of the hexaploid wheat cultivar Courtot. Proteomics 4: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Eagles HA, Hollamby GJ, Eastwood RF (2002) Genetic and environmental variation for grain quality traits routinely evaluated in southern Australian wheat breeding programs. Aust J Agric Res 53: 1047–1057 [Google Scholar]
- Escher D, Bodmer-Glavas M, Barberis A, Schaffner W (2000) Conservation of glutamine-rich transactivation function between yeast and humans. Mol Cell Biol 20: 2774–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Levy AA, Fahima T, Korol A (2012) Genomic asymmetry in allopolyploid plants: wheat as a model. J Exp Bot 63: 5045–5059 [DOI] [PubMed] [Google Scholar]
- Galili G, Feldman M (1985) Genetic control of endosperm proteins in wheat: 3. Allocation to chromosomes and differential expression of high molecular weight glutenin and gliadin genes in intervarietal substitution lines of common wheat. TAG. Theoretical and applied genetics. Theor Angew Genet 69: 583–589 [DOI] [PubMed] [Google Scholar]
- Galili G, Levy AA, Feldman M (1986) Gene-dosage compensation of endosperm proteins in hexaploid wheat Triticum aestivum. Proc Natl Acad Sci U S A 83: 6524–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Humanes J, Piston F, Gimenez MJ, Martin A, Barro F (2012) The introgression of RNAi silencing of gamma-gliadins into commercial lines of bread wheat changes the mixing and technological properties of the dough. PLoS ONE 7: e45937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover NM, Redestig H, Dessimoz C (2016) Homoeologs: what are they and how do we infer them? Trends Plant Sci 21: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Yue X, Zeng H, Zhu J (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26: 438–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin C, Roche J, Allard V, Ravel C, Mouzeyar S, Bouzidi MF (2019) Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. aestivum L.). PLoS ONE 14: e0213390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Yang H, Liu Y, Gao Y, Ni Z, Peng H, Xin M, Hu Z, Sun Q, Yao Y (2015) The wheat transcription factor TaGAMyb recruits histone acetyltransferase and activates the expression of a high-molecular-weight glutenin subunit gene. Plant J 84: 347–359 [DOI] [PubMed] [Google Scholar]
- Hedden P (2003) The genes of the Green Revolution. Trends Genet 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo N, Zhu T, Altenbach S, Dong L, Wang Y, Mohr T, Liu Z, Dvorak J, Luo MC, Gu YQ (2018) Dynamic evolution of alpha-gliadin prolamin gene family in homeologous genomes of hexaploid wheat. Sci Rep 8: 5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Tsunashima M, Hiei Y, Komari T (2015) Wheat (Triticum aestivum L.) transformation using immature embryos. In: Wang K, ed., Agrobacterium Protocols, Vol 1, Springer New York, New York, NY, pp. 189–198 [DOI] [PubMed] [Google Scholar]
- Kang GZ, Xu W, Liu GQ, Peng XQ, Guo TC (2013) Comprehensive analysis of the transcription of starch synthesis genes and the transcription factor RSR1 in wheat (Triticum aestivum) endosperm. Genome 56: 115–122 [DOI] [PubMed] [Google Scholar]
- Kumar P, Mishra A, Sharma H, Sharma D, Rahim MS, Sharma M, Parveen A, Jain P, Verma SK, Rishi V, et al. (2018) Pivotal role of bZIPs in amylose biosynthesis by genome survey and transcriptome analysis in wheat ( Triticum aestivum L.) mutants. Sci Rep 8: 17240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Beom HR, Altenbach SB, Lim SH, Kim YT, Kang CS, Yoon UH, Gupta R, Kim ST, Ahn SN, et al. (2016) Comprehensive identification of LMW-GS genes and their protein products in a common wheat variety. Funct Integr Genomics 16: 269–279 [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R (2003) Transcription regulation and animal diversity. Nature 424: 147–151 [DOI] [PubMed] [Google Scholar]
- Li Y, An X, Yang R, Guo X, Yue G, Fan R, Li B, Li Z, Zhang K, Dong Z, et al. (2015) Dissecting and enhancing the contributions of high-molecular-weight glutenin subunits to dough functionality and bread quality. Mol Plant 8: 332–334 [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci U S A 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liu L, He Z, Yan J, Zhang Y, Xia X, Peña RJ (2005) Allelic variation at the Glu-1 and Glu-3 loci, presence of the 1B.1R translocation, and their effects on mixographic properties in Chinese bread wheats. Euphytica 142: 197–204 [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Liu Y, Hou J, Wang X, Li T, Majeed U, Hao C, Zhang X (2020) The NAC transcription factor NAC019-A1 is a negative regulator of starch synthesis in wheat developing endosperm. J Exp Bot 19: 5794–5807 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makai S, Eva C, Tamas L, Juhasz A (2015) Multiple elements controlling the expression of wheat high molecular weight glutenin paralogs. Funct Integr Genomics 15: 661–672 [DOI] [PubMed] [Google Scholar]
- Mathew IE, Agarwal P (2018) May the fittest protein evolve: favoring the plant-specific origin and expansion of NAC transcription factors. BioEssays 40: e1800018. [DOI] [PubMed] [Google Scholar]
- Mathew IE, Das S, Mahto A, Agarwal P (2016) Three rice NAC transcription factors heteromerize and are associated with seed size. Front Plant Sci 7: 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragao FJ, Fontes EP (2013) GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci U S A 110: 19627–19632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Payne PI, Nightingale MA, Krattiger AF, Holt LM (1987) The relationship between HMW glutenin subunit composition and the bread-making quality of British-grown wheat varieties. J Sci Food Agric 40: 51–65 [Google Scholar]
- Qi W, Zhu J, Wu Q, Wang Q, Li X, Yao D, Jin Y, Wang G, Wang G, Song R (2016) Maize reas1 mutant stimulates ribosome use efficiency and triggers distinct transcriptional and translational responses. Plant Physiol 170: 971–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragupathy R, Naeem HA, Reimer E, Lukow OM, Sapirstein HD, Cloutier S (2008) Evolutionary origin of the segmental duplication encompassing the wheat GLU-B1 locus encoding the overexpressed Bx7 (Bx7OE) high molecular weight glutenin subunit. TAG. Theoretical and applied genetics. Theor Angew Genet 116: 283–296 [DOI] [PubMed] [Google Scholar]
- Rasheed A, Xia X, Yan Y, Appels R, Mahmood T, He Z (2014) Wheat seed storage proteins: advances in molecular genetics, diversity and breeding applications. J Cereal Sci 60: 11–24 [Google Scholar]
- Ravel C, Fiquet S, Boudet J, Dardevet M, Vincent J, Merlino M, Michard R, Martre P (2014) Conserved cis-regulatory modules in promoters of genes encoding wheat high-molecular-weight glutenin subunits. Front Plant Sci 5: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rharrabti Y, Elhani S, Martos-Nunez V, Garcia Del Moral LF (2001) Protein and lysine content, grain yield, and other technological traits in durum wheat under Mediterranean conditions. J Agric Food Chem 49: 3802–3807 [DOI] [PubMed] [Google Scholar]
- Scossa F, Laudencia-Chingcuanco D, Anderson OD, Vensel WH, Lafiandra D, D’Ovidio R, Masci S (2008) Comparative proteomic and transcriptional profiling of a bread wheat cultivar and its derived transgenic line overexpressing a low molecular weight glutenin subunit gene in the endosperm. Proteomics 8: 2948–2966 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG, Belton PS, Tatham AS (2002) The structure and properties of gluten: an elastic protein from wheat grain. Phil Trans R Soc Lond B Biol Sci 357: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Shepherd KW (1988) Linkage mapping of genes controlling endosperm storage proteins in wheat. Theor Appl Genet 75: 628–641 [Google Scholar]
- Su Z, Bernardo A, Tian B, Chen H, Wang S, Ma H, Cai S, Liu D, Zhang D, Li T, et al. (2019) A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat Genet 51: 1099–1105 [DOI] [PubMed] [Google Scholar]
- Sun F, Liu X, Wei Q, Liu J, Yang T, Jia L, Wang Y, Yang G, He G (2017) Functional characterization of TaFUSCA3, a B3-superfamily transcription factor gene in the wheat. Front Plant Sci 8: 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraverbeke WS, Delcour JA (2002) Wheat protein composition and properties of wheat glutenin in relation to breadmaking functionality. Crit Rev Food Sci Nutr 42: 179–208 [DOI] [PubMed] [Google Scholar]
- Wang JC, Xu H, Zhu Y, Liu QQ, Cai XL (2013) OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot 64: 3453–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon do Y, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, et al. (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25: 4708–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Jondiko TO, Tilley M, Awika JM (2014) Effect of high molecular weight glutenin subunit composition in common wheat on dough properties and steamed bread quality. J Sci Food Agric 94: 2801–2806 [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu D, Jiang W, Guo X, Yang W, Sun J, Ling H, Zhang A (2011) PCR-based isolation and identification of full-length low-molecular-weight glutenin subunit genes in bread wheat (Triticum aestivum L.). Theor Appl Genet 123: 1293–1305 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Dong J, Ji C, Wu Y, Messing J (2019. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci U S A 116: 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fang L, Yu J, Zhao Y, Chen F, Xia G (2018) 5-Azacytidine treatment and TaPBF-D over-expression increases glutenin accumulation within the wheat grain by hypomethylating the Glu-1 promoters. Theor Appl Genet 131: 735–746 [DOI] [PubMed] [Google Scholar]
- Zorb C, Ludewig U, Hawkesford MJ (2018) Perspective on wheat yield and quality with reduced nitrogen supply. Trends Plant Sci 23: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers
The cDNA sequences of TaNAC019-A, TaNAC019-BI, TaNAC019-BII, and TaNAC019-D have been deposited in GenBank under accession numbers MN685185, MN685186, MN685187, and MN685188, respectively. RNA sequencing data are available at the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA656089.