Abstract
Grain filling in maize (Zea mays) is regulated by a group of spatiotemporally synchronized transcription factors (TFs), but the factors that coordinate their expression remain unknown. We used the promoter of the grain filling-specific TF gene Opaque2 (O2) to screen upstream regulatory factors and identified a B3 domain TF, ZmABI19, that directly binds to the O2 promoter for transactivation. zmabi19 mutants displayed developmental defects in the endosperm and embryo, and mature kernels were opaque and reduced in size. The accumulation of zeins, starch and lipids dramatically decreased in zmabi19 mutants. RNA sequencing revealed an alteration of the nutrient reservoir activity and starch and sucrose metabolism in zmabi19 endosperms, and plant phytohormone signal transduction and lipid metabolism in zmabi19 embryos. Chromatin immunoprecipitation followed by sequencing coupled with differential expression analysis identified 106 high-confidence direct ZmABI19 targets. ZmABI19 directly regulates multiple key grain filling TFs including O2, Prolamine-box binding factor 1, ZmbZIP22, NAC130, and Opaque11 in the endosperm and Viviparous1 in the embryo. A number of phytohormone-related genes were also bound and regulated by ZmABI19. Our results demonstrate that ZmABI19 functions as a grain filling initiation regulator. ZmABI19 roles in coupling early endosperm and embryo development are also discussed.
ZmABI19 directly regulates the key factors that coordinate endosperm and embryo development and grain filling in maize.
Introduction
Transcriptional regulation of seed development, particularly the embryo, appears to be conserved in dicots and monocots (Sreenivasulu and Wobus, 2013). Embryogenesis and seed maturation in Arabidopsis (Arabidopsis thaliana) are well characterized and are regulated by LAFL family transcription factors (TFs), named after the three representative B3 domain TFs LEAFY COTYLEDON 2 (LEC2), ABSCISIC ACID INSENSITIVE 3 (ABI3), and FUSCA3 (FUS3) and the Heme Activator Protein 3 (HAP3) family CCAAT-box binding factor, LEC1 (Santos-Mendoza et al., 2008; Sreenivasulu and Wobus, 2013; Lepiniec et al., 2018). B3 domain TFs are plant-specific proteins. The first identified and cloned B3 TF was the maize (Zea mays) TF Viviparous1 (Vp1), a homolog of Arabidopsis ABI3 (Mccarty et al., 1991; Giraudat et al., 1992). LAFL TFs have redundant but apparently divergent functions in the regulation of seed development (Roscoe et al., 2015). LEC1 is mainly expressed from the globular stage to the bent-cotyledon stage, whereas the highest LEC2 mRNA levels are detected between the preglobular and bent-cotyledon stages (Santos-Mendoza et al., 2008; Fatihi et al., 2016). By contrast, FUS3 and ABI3 transcripts accumulate at the late stage of embryogenesis. LAFL family TFs also regulate the expression of downstream factors by affecting plant hormone biosynthesis (Gazzarrini et al., 2004; Suzuki and McCarty, 2008; Carbonero et al., 2016). Furthermore, B3 domain TFs bind to the RY motif (whose core sequence is 5′-CATG-3′) and transactivate genes involved in seed storage proteins and oil accumulation, and TFs such as MYB DOMAIN PROTEIN 118 (MYB118), WRINKLED1 (WRI1), and AGAMOUS-LIKE 15 (AGL15) (Kroj et al., 2003; Braybrook et al., 2006; Wang and Perry, 2013; Baud et al., 2016). Interaction and activation networks have been established between LAFL TFs (Santos-Mendoza et al., 2008; Baud et al., 2016; Fatihi et al., 2016; Boulard et al., 2017). For instance, LEC2 directly transactivates the expression of ABI3 and FUS3, whose encoded proteins have autoregulation capacity (Braybrook et al., 2006; Wang and Perry, 2013).
The main storage proteins in the Arabidopsis embryo are 2S albumins and 12S cruciferins. Their encoding genes are transcriptionally regulated by basic Leucine Zipper TFs (bZIP10 and bZIP25) and B3 TFs (LEC2, ABI3, and FUS3) (Giraudat et al., 1992; Bäumlein et al., 1994; Parcy et al., 1994; Stone et al., 2001; Lara et al., 2003). In the maize embryo, VP1 transactivates the expression of globulin1 and globulin2, which encode primary embryo storage proteins. Mutation in Vp1 causes a dramatic reduction in the biosynthesis of globulin proteins (Kriz, 1989; Zheng et al., 2019). This observation indicates that ABI3/VP1 homologs have conserved functions in the transactivation of storage protein-encoding genes in dicots and monocots. LAFL transcripts are also detected in the endosperm: one such LAFL gene, FUS3, is expressed in the aleurone layer. Aleurone is the sole residual endosperm tissue in Arabidopsis seeds and accumulates storage proteins and lipids during seed maturation (Tsuchiya et al., 2004). Although the involvement of the LAFL network in embryo development has been extensively studied in Arabidopsis, its role in endosperm development and filling is poorly understood.
Maize seeds exhibit the largest caryopsis of the grass family, as the endosperm accounts for 90% of the total dry seed weight. The major storage reserves in maize endosperm are starch and proteins, accounting for 70% and 10% of the endosperm dry mass, respectively. The main storage proteins are prolamins, called zeins, which account for more than 60% of all endosperm proteins. Based on their structure and sequence similarity, zein proteins are classified into four families: α (19 and 22 kDa), γ (50, 27, and 16 kDa), β (15 kDa), and δ (18 and 10 kDa) (Esen, 1987; Coleman and Larkins, 1999). Starch and protein biosynthesis in the endosperm is regulated by a complex network involving coordinated expression of genes in multiple processes (Li and Song, 2020).
Opaque2 (O2) is a classic bZIP-type TF that was first identified as a transcriptional regulator for 22-kD α-zein and 15-kD β-zein genes, and was later shown to transactivate the expression of all zein genes except for 16-kD γ-zein (Schmidt et al., 1987; Cord Neto et al., 1995; Li et al., 2015). O2 also functions as a regulator of starch biosynthesis. Indeed, O2 regulates the expression of pyruvate orthophosphate 1 (PPDK1), PPDK2, and starch synthase III (SS3), whose encoded proteins function as critical components in the starch biosynthetic enzyme complex (Zhang et al., 2016). Recently, O2 was shown to transactivate three sucrose synthase-encoding genes (Shrunken1 [Sh1], Sus1, and Sus2). Sucrose synthase converts sucrose and uridine diphosphate (UDP) to fructose and UDP-glucose, providing the substrates for starch and protein biosynthesis (Deng et al., 2020). In this respect, O2 appears to act as a central regulator in endosperm filling by transcriptionally regulating genes involved in sucrose cleavage, starch biosynthesis, and protein storage. Prolamin-box factor 1 (PBF1) is another endosperm-specific TF belonging to the Dof (DNA-binding with one finger) family (Vicente-Carbajosa et al., 1997). In contrast to O2, PBF1 alone has much milder transactivation activity upon zein, PPDK, and SS3 expression, but it can interact with O2, resulting in synergistic transactivation of their downstream genes (Zhang et al., 2015, 2016; Yang et al., 2016). The NAM, ATAF, and CUC (NAC) TFs ZmNAC128 and ZmNAC130 accumulate specifically in the endosperm and regulate the transcription of Brittle2 (Bt2, encoding the small subunit of adenosine diphosphate glucose pyrophosphorylase) and 16-kD γ-zein (Zhang et al., 2019b). Silencing of ZmNAC128 and ZmNAC130 transcripts by RNA interference (RNAi) resulted in a shrunken kernel phenotype with a significant reduction in starch and protein content. ZmbZIP22 is a newly identified bZIP-type TF that regulates the expression of 27-kD γ-zein together with O2 heterodimerizing proteins (OHPs), O2 and PBF1 (Zhang et al., 2015; Li et al., 2018). Unlike O2 and Pbf1 but similar to MADS47, Ohp1, and Ohp2 are generally expressed in maize tissues (Zhang et al., 2015; Qiao et al., 2016). The MADS47 protein interacts with O2 to regulate the expression of α-zeins and 50-kD γ-zein. Naked endosperm1 (NKD1) and NKD2 are duplicate INDETERMINATE DOMAIN TFs and appear to function as central regulators of gene expression in developing maize endosperm. They directly regulate O2 and Vp1 expression (Gontarek et al., 2016). Opaque11 (O11) is an endosperm-specific basic Helix–Loop–Helix (bHLH) TF. The O11 gene is activated as early as 3 days after pollination (DAP) and regulates a variety of biological processes in maize endosperm development (Feng et al., 2018). O11 regulates endosperm development by transactivating the expression of Nkd2 and ZmDof3 as well as nutrient metabolism by transactivating Pbf1 and repressing O2. O11 also regulates the expression of PPDK genes and genes encoding multiple carbohydrate metabolic enzymes. Taken together, these results indicate that endosperm filling is regulated by a group of TFs, of which O2, PBF1, ZmNAC128, ZmNAC130, and O11 are specifically expressed in the endosperm, indicating a synchronized regulation of starch and protein biosynthesis. These results also suggest that these TFs are coordinately regulated.
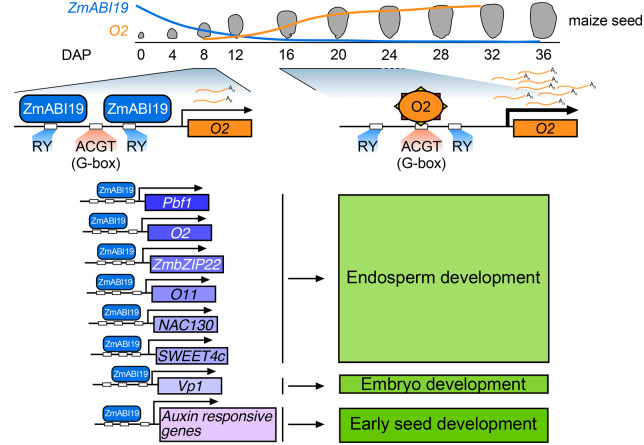
To understand the mechanisms responsible for the synchronized expression patterns of grain filling TFs, we identified their upstream regulators using O2 as a test case. Accordingly, we identified the B3 domain TF ZmABI19 based on its ability to bind to the two RY motifs in the O2 promoter and significantly transactivate gene expression. ZmABI19 also directly recognizes and transactivates Pbf1, ZmbZIP22, ZmNAC130, and O11 in the endosperm and Vp1 in the embryo. In addition, ZmABI19 is a transactivator of the sugar transport SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTER 4c (SWEET4c). zmabi19 mutants showed an opaque and small kernel phenotype with altered endosperm filling and embryo lethality. A combination of chromatin immunoprecipitation (ChIP) followed by deep-sequencing (ChIP-seq) and deep sequencing of the transcriptome (RNA-seq) analyses determined the ZmABI19 regulatory network in the coordination of endosperm and embryo development, nutrient reservoir activity, starch and sucrose metabolism, and plant hormone signal transduction in maize seeds.
Results
Identification of key transcriptional cis-elements in the O2 promoter
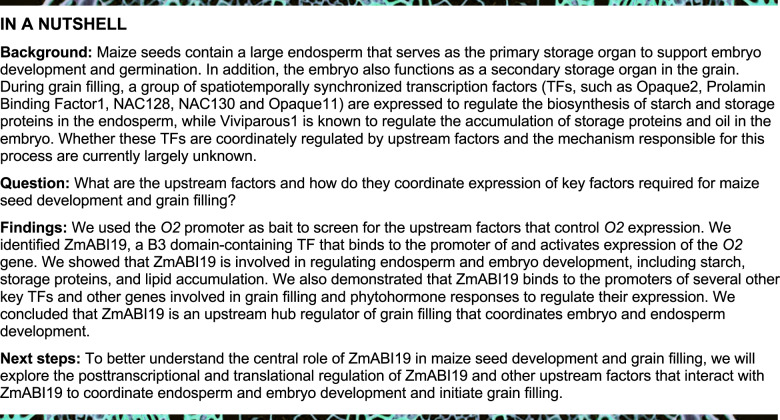
To identify key transcriptional cis-elements in the O2 promoter, we investigated the transcriptional activity of the promoter through a promoter deletion series driving expression of a reporter gene. To this end, we amplified four fragments of the O2 promoter with different lengths (300-, 400-, 500-, and 1,000-bp upstream of the start codon). A dual-luciferase (LUC) reporter (DLR) assay was used to test the transactivation activity of these fragments. In this system, the Renilla LUC (REN) reporter gene was driven by the cauliflower mosaic virus (CaMV) 35S promoter and used as an internal control, while the O2 promoter fragments were placed upstream of firefly LUC. These constructs were designated O2pro300:LUC, O2pro400:LUC, O2pro500:LUC, and O2pro100:LUC (Figure 1, A). The reporter plasmids were introduced into endosperm cells by particle bombardment. LUC activities from O2pro300:LUC and O2pro400:LUC were similar, but LUC activity derived from O2pro500:LUC and O2pro1000:LUC increased by approximately 38.5% and 36.6% compared with O2pro400:LUC, respectively (Figure 1, A). Thus, the sequence between −400 and −500 bp upstream of the start codon likely contains a key cis-element required for transactivation of the O2 promoter.
Figure 1.
Identification of cis-elements and the associated TF for O2 promoter transactivation. (A) Transcriptional activities of different O2 promoter fragments tested via particle bombardment. (Top) The schematic diagrams of the reporter constructs and (bottom) the dual luciferase reporter (DLR) assay. **P < 0.01; ns, no significant difference. (B) Yeast one-hybrid assay showing that ZmABI19 binds to the two RY motifs in the O2 promoter. (Top) The bait sequences with normal (in magenta) or mutated (in green) RY motifs and (bottom) the yeast one-hybrid assay. The Aureobasidin A (AbA) concentration was 250 ng/mL. (C) Transactivation of the O2 promoter by ZmABI19. The relative ratio of LUC/REN tested in Arabidopsis protoplasts via co-transforming the reporter plasmids with the effector construct. **P < 0.01; ns, no significant difference. (D) EMSA of the specific binding of ZmABI19-His to the two RY motifs in the O2 promoter. The normal and mutant probes shown in (B) were labeled with biotin. Unlabeled intact probes were used for competition. Comp, competing probes unlabeled with biotin.
We then searched this region for known cis-acting regulatory elements through the Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). This search resulted in the identification of nine separate motifs: an as-1 motif, a CGTCA motif, a TGACG motif, two RY motifs, a MYB motif, a MYC motif, an ABA-responsive element (ABRE), a G-box or O2 box (overlapping), and a CAAT-box (Supplemental Table S1).
Identification of candidate TFs that bind the O2 promoter
The RY motif is a cis-acting regulatory element involved in seed-specific regulation and is recognized by plant-specific TFs containing a B3 domain. We used a candidate approach to identify possible B3 TFs that recognize the O2 promoter. The maize genome is predicted to encode 51 proteins with B3 domains, collectively named VP1 and ZmABI2 to ZmABI51 (https://www.grassius.org/family.php?family=ABI3-VP1&species=Maize). Of these encoding genes, 37 were expressed in seeds, based on public RNA-seq datasets (Supplemental Data Set S1) (Chen et al., 2014). To explore which B3 TF might recognize the RY motifs in the O2 promoter, we conducted yeast one-hybrid assays. We cloned the full-length coding sequence (CDS) of the 37 B3 domain protein-encoding genes into the pGADT7-Rec plasmid and expressed them as fusion proteins with the yeast GAL4 AD (activation domain) in the Y1HGold host yeast (Saccharomyces cerevisiae) strain. A bait fragment (−400 to −500 bp upstream of the start codon, containing two RY motifs, named A and B) was cloned into the pAbAi vector and integrated into the yeast genome (Figure 1, B). Only ZmABI4 (Zm00001d033313) and ZmABI19 (Zm00001d011712) recognized the bait sequence (Supplemental Figure S1).
To validate the observed protein–DNA interaction, we used modified fragments with mutations in either the first (aB), the second (Ab), or both (ab) RY motifs in yeast one-hybrid assays (Figure 1, B). ZmABI4 recognized the aB and ab constructs, but not Ab (Supplemental Figure S2, A), indicating that the interaction between ZmABI4 and the O2 promoter fragment is likely nonspecific. We also performed a DLR assay to test the transactivation activity of ZmABI4 onto the O2 promoter in the endosperm via particle bombardment. We generated an effector construct by placing the full-length CDS of ZmABI4 under the control of the CaMV 35S promoter (35Spro:ZmABI4) (Supplemental Figure S2, B) and tested with the O2pro1000:LUC reporter construct (abbreviated O2pro) created above (Figure 1, A). Co-expression of 35Spro:ZmABI4 failed to increase the transactivation of the O2 promoter over levels from the negative control consisting of the empty pRI101 vector (Supplemental Figure S2, B).
We then turned to ZmABI19 and the O2 promoter. In yeast one-hybrid assays, ZmABI19 recognized the aB and Ab fragments, but failed to recognize the ab fragment, indicating that both RYs motif are functional for ZmABI19-mediated binding (Figure 1, B). In DLR assays, co-expression of 35Spro:ZmABI19 with O2pro in the endosperm resulted in significantly increased LUC activity compared with the negative control (P-value = 0.0003, Supplemental Figure S3). Both RY motifs were required to produce maximum transactivation by ZmABI19, as mutation in either motif led to lower transactivation than the wild-type promoter. When both RY motifs were mutated, transactivation by ZmABI19 was abolished, resulting in LUC activity similar to that of the negative control (Supplemental Figure S3). To validate this result, we also conducted the DLR assay in Arabidopsis protoplasts, and obtained results consistent with the particle bombardment data (Figure 1, C).
Next, we performed an electrophoretic mobility shift assay (EMSA) to test ZmABI19 binding to the two RY motifs in vitro. Oligonucleotides (from −400 to −500 bp upstream of the start codon) containing the two RY motifs labeled with biotin were used as probes. As shown in Figure 1, D, binding of the ZmABI19-His recombinant protein to the normal probes was evident as a retarded band in the gel. Because of the strong affinity of ZmABI19 for the RY motifs, we applied high concentrations of unlabeled normal probes (200 and 500×) for the competition and observed a gradual decrease in the signal intensity of the retarded bands. By contrast, the retarded bands were not apparently affected by the addition of unlabeled mutant probes. When either RY motif was mutated, ZmABI19-His still bound the probes. However, when both RY motifs were mutated, ZmABI19-His did not recognize the probes, in agreement with the yeast one-hybrid assay results (Figure 1, B). We observed that the Ab or aB cold probes (200×) failed to compete with labeled AB probes for binding, suggesting that ZmABI19 has an overwhelmingly higher affinity for the probe with two functional RY motifs. Taken together, these results demonstrate that the B3 DNA-binding protein ZmABI19 specifically recognizes the two RY motifs in the O2 promoter and causes transcriptional activation.
DNase I footprinting and transient assays previously showed that O2 can bind to and activate its own promoter (Lohmer et al., 1991). However, other research also suggested that O2 had no effect on its own transcription (Ciceri et al., 1999). To resolve this discrepancy, we performed EMSA on the O2 box positioned at −462 bp upstream of the start codon. The results demonstrated that O2 specifically bound to the O2 box (Supplemental Figure S4, A). DLR assays further indicated that LUC activity from the O2pro:LUC reporter increased by approximately 9.6-fold in the presence of 35Spro:O2 compared with the control, supporting the first finding that O2 exerts auto-transactivation by recognizing the O2 box in its own promoter (Supplemental Figure S4, B).
O11 was recently proposed to repress O2 activity by competing for the same binding site in the O2 promoter (Feng et al., 2018). Since the RY motifs and O2 box are closely located in the O2 promoter, we performed a LUC complementation imaging (LCI) assay to examine whether ZmABI19 and O11 physically interact. We fused ZmABI19 and O11 to the N- and C-terminal domains of LUC (NLUC and CLUC), respectively. The results showed that co-infiltration of ZmABI19-NLUC with O11-CLUC in Nicotiana benthamiana leaves produced strong LUC activity, whereas infiltration of either vector alone with the corresponding empty constructs did not yield a visible signal (Supplemental Figure S5, A). Bimolecular fluorescence complementation (BiFC) assay was used to further confirm the association between ZmABI19 and O11 in Arabidopsis protoplasts. The YFP fluorescence signal was effectively reconstituted when protoplasts were transfected with nYFP-ZmABI19 and cYFP-O11, while no signal was detected between nYFP-ZmABI19 and the negative control (an unrelated protein, cell wall invertase MINIATURE 1) or empty vector, confirming the protein–protein interaction between ZmABI19 and O11 in vivo (Supplemental Figure S5, B). The DLR assay showed that O11 alone had no apparent activation or repression effect on the O2 promoter in Arabidopsis protoplasts. Compared with co-transfection of ZmABI19 with an empty-vector control or an unrelated TF (ZmIAA10), co-expression of ZmABI19 with O11 significantly lessened the transactivation activity by ZmABI19 (Supplemental Figure S5, C). These results demonstrated that O11 physically interacts with ZmABI19 and represses ZmABI19-mediated transactivation, consistent with the dual activator/repressor activity profiles of O11 function (Feng et al., 2018).
Phylogenetic analysis of ZmABI19 protein homologs
We constructed a phylogenetic tree based on the full-length protein sequences of ZmABI19 and its homologous proteins in purple false brome (Brachypodium distachyon), barley (Hordeum vulgare), rice (Oryza sativa), sorghum (Sorghum bicolor), foxtail millet (Setaria italica), wheat (Triticum aestivum), A. thaliana, camelina (Camelina sativa), desert poplar (Populus euphratica), wild strawberry (Fragaria vesca), wild cabbage (Brassica oleracea), rapeseed (Brassica napus), and the moss Physcomitrium patens. The phylogenetic tree revealed that ZmABI19 was highly conserved in monocots and evolutionarily related to Arabidopsis FUS3 (Figure 2, A and Supplemental Files S1 and S2). However, protein sequence alignment analysis demonstrated that ZmABI19 shares only 24.6% similarity with FUS3, mainly in the B3 domain region (Supplemental Figure S6).
Figure 2.
Phylogenetic analysis of ZmABI19 and expression pattern of ZmABI19. (A) Phylogenetic tree of ZmABI19 and its homologs. Evolutionary distances were estimated with a neighbor-joining algorithm. The numbers above the branches indicate the percentage from 1,000 bootstrap. The scale bar indicates the average number of amino acid substitutions per site. The homologous protein of P. patens was used as an outgroup. (B) and (C) RT-qPCR analysis of ZmABI19 expression in different tissues (B) and during seed development (C). En, endosperm; Em, embryo. All expression levels were normalized to that of Actin. Error bars represent SD of three biological replicates. (D) Immunoblot analysis of ZmABI19 in developing seeds. ACTIN was used as control. (E) and (F) RNA in situ hybridization of ZmABI19. Longitudinal sections of (E) 8- and (F) 16-DAP seeds were hybridized with an antisense ZmABI19 probe. (G) The endosperm at higher resolution from (E). (H) The embryo at higher resolution from (F). (I) The BETL at higher resolution from (F). (J) No visible signals were detected when the 16-DAP seed was hybridized with a sense probe. En, endosperm; Em, embryo. Bars = 1 mm in (E), (F), and (J); Bars = 250 μm in (G)–(I).
The plant-specific B3 superfamily comprises the well-characterized auxin response factor (ARF) and LAV (LEC2, ABI3, VP1/ABI3-like [VAL]) families, and the less-understood RELATED TO ABI3/VP1 (RAV) and REPRODUCTIVE MERISTEM (REM) families (Swaminathan et al., 2008). In Arabidopsis, the LAV family is composed of three AFL (LEC2, FUS3, and ABI3) and three VAL (VAL1, VAL2, and VAL3) TFs. To further investigate the potential conservation of B3 domain-containing TFs in maize and Arabidopsis, we constructed a phylogenetic tree using 50 B3 TFs from maize and six LAV proteins from Arabidopsis. As the protein sequence of ZmABI6 is too short with only 38 amino acids, it was excluded from the phylogenetic analysis. VP1 and ABI3 clustered in the same clade (Supplemental Figure S7 and Supplemental Files S3 and S4), consistent with the observation that VP1 could complement the abi3 mutants in Arabidopsis (Zhang et al., 2019a). VAL proteins play a role in the repression of embryonic development, and they clustered with ZmABI22, ZmABI32, and ZmABI39 (Supplemental Figure S7 and Supplemental Files S3 and S4). By contrast, ZmABI19 did not cluster with Arabidopsis FUS3 or LEC2, indicating rapid sequence divergence since the split between monocots and dicots.
Temporal and spatial expression patterns of ZmABI19 during maize seed development
To explore the tissue-specific expression pattern of ZmABI19, mRNAs extracted from different tissues were used for reverse transcription quantitative PCR (RT-qPCR). Although ZmABI19 was detected in all examined tissues, it was predominantly expressed in developing seeds (Figure 2, B). We further investigated the expression profile of ZmABI19 during seed development. Since it is difficult to separate the endosperm and embryo at early developmental stages, we used whole seeds from 2 to 8 DAP for RNA extraction. At later stages, the endosperm and embryo were dissected for examination. RT-qPCR revealed that ZmABI19 was highly expressed during early seed development, peaking at 2 DAP and gradually decreasing thereafter (Figure 2, C). From 10 to 24 DAP, when the endosperm and embryo in the seed were examined separately, ZmABI19 transcript levels in the embryo continued to decrease, but were always much higher than in the endosperm at each time point. We also examined ZmABI19 protein levels in whole seeds from 1 to 24 DAP. Immunoblot analysis revealed that ZmABI19 accumulated at a much higher levels in the early (1–8 DAP) stages of seed development relative to the filling (10–24 DAP) stage (Figure 2, D). The protein accumulation pattern did not strictly follow the gradual reduction seen for ZmABI19 transcripts at each stage, suggesting that ZmABI19 might be regulated at both transcriptional and translational levels.
To examine the ZmABI19 spatial expression pattern in maize seeds, we performed RNA in situ hybridization with a ZmABI19 antisense probe at 8 and 16 DAP. The results revealed that ZmABI19 was expressed throughout the kernels, including the maternal tissues (pericarp and nucellus), endosperm and embryo (Figure 2, E and F). The hybridization signal was strongly detected in the entire endosperm at 8 DAP (Figure 2, E and G), but appeared to decrease in the endosperm at the filling stage (Figure 2, F). At 16 DAP, ZmABI19 was abundantly expressed in the basal section of the scutellum in the embryo, and strong signals were also detected in the basal endosperm transfer layer (BETL) (Figure 2, H and I). By then, only residual signals were observed in starchy endosperm cells (Figure 2, F). In control experiments, the ZmABI19 sense probe did not produce a visible signal (Figure 2, J). These results suggest that ZmABI19 has regulatory functions in both endosperm and embryo development.
To determine its subcellular localization, we fused full-length ZmABI19 to the N-terminus of the enhanced green fluorescent protein (eGFP). Free eGFP was used as a control. Both gene cassettes were driven by the constitutive 35S promoter. The resulting constructs were transiently expressed in Arabidopsis leaf mesophyll protoplasts. Similar to the free eGFP construct, we detected signal from ZmABI19-eGFP in both nuclei and the cytoplasm (Figure 3, A). Similar nuclear and cytosolic localization of ZmABI19-eGFP was also observed in N. benthamiana leaves, which were infiltrated with Agrobacterium harboring the ZmABI19-eGFP construct (Figure 3, B). To further confirm the location of ZmABI19, we generated stable transgenic Arabidopsis plants in the Columbia-0 background. Observation of root tip cells from T2 plants showed that ZmABI19 also localized to nuclei and the cytoplasm (Supplemental Figure S8). Finally, we isolated cytoplasmic and nuclear proteins from 4-DAP maize seeds: immunoblot analysis revealed that ZmABI19 was present in both fractions (Figure 3, C).
Figure 3.
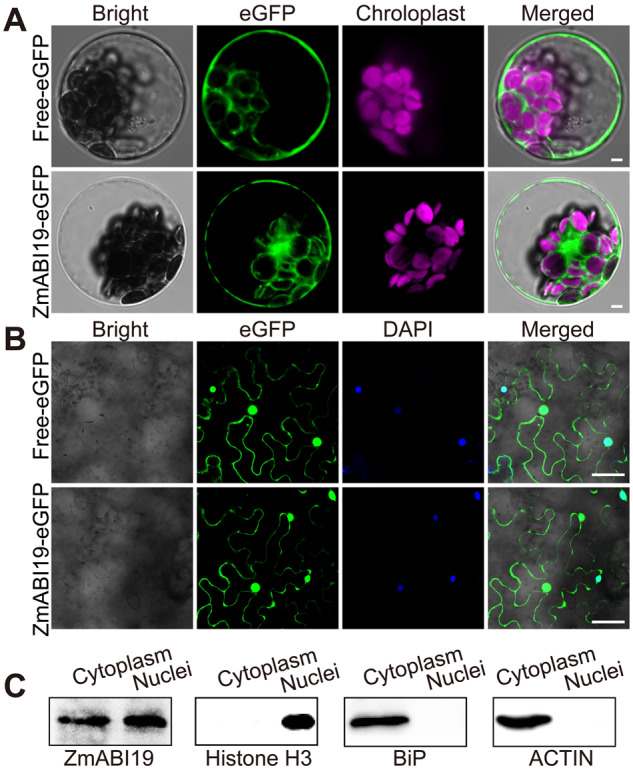
Subcellular localization of ZmABI19. (A) Subcellular localization of ZmABI19-eGFP in Arabidopsis protoplasts. (Top) Free-eGFP used as a control and (bottom) ZmABI19-eGFP. Bars = 25 μm. (B) Subcellular localization of ZmABI19-eGFP in N. benthamiana leaves. (Top) Free-eGFP used as control and (bottom) ZmABI19-eGFP. Bars = 20 μm. (C) Immunoblot analysis of cytoplasmic and nuclear fractions obtained from 4-DAP B104 maize seeds. Antibodies against ZmABI19, histone H3 (nuclear marker), BiP and ACTIN (cytoplasmic markers) were used.
Genetic confirmation of ZmABI19 function
To validate the regulatory function of ZmABI19 genetically, null mutants were required to demonstrate the effect on O2 gene expression. We identified a mutant stock (UFMu-09010, see “Materials and methods” section) with a Mu7 Cy transposable element in the last exon downstream of the encoded B3 domain region, but this stock did not disrupt gene expression, protein accumulation, or function (Supplemental Figure S9). Based on sequence analysis, the predicted variant protein (designated ZmABI19-Mu) and normal ZmABI19 had a similar molecular mass (Supplemental Figure S10) and the activation domain was located before the altered C-terminal region (caused by the Mu insertion) in ZmABI19-Mu (Supplemental Figure S11, A), thereby resulting in a minimal effect, if any, on transcriptional transactivation capacity. Consistent with this hypothesis, this allele did not cause any visible phenotype in zmabi19-Mu homozygous seeds.
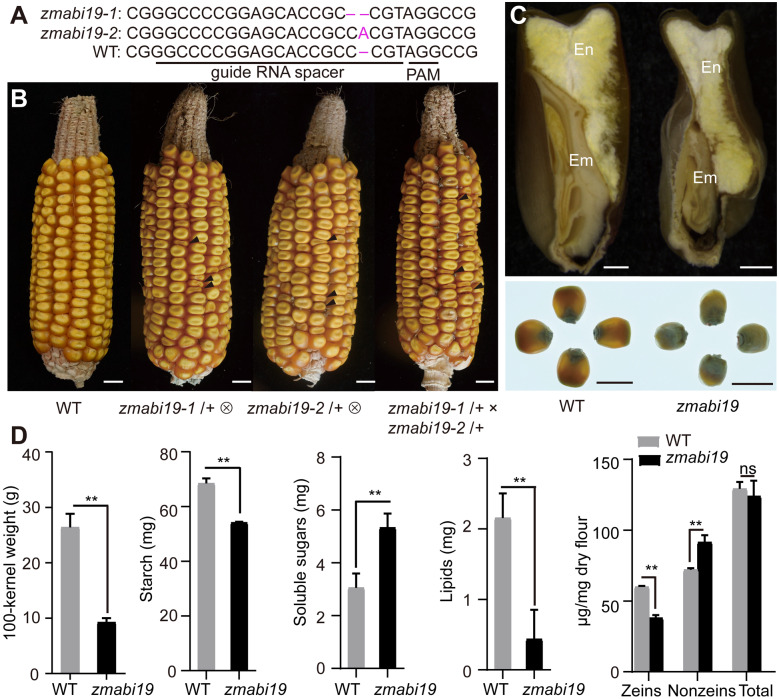
We then employed a genome editing approach with clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) to knock out this gene in the B104 inbred line. The guide RNA was designed to target the first exon of ZmABI19. We recovered 15 CRISPR/Cas9 edited plants, from which we identified two types of DNA editing events. The zmabi19-1 mutants had a single bp deletion of a cytosine at position +122 bp relative to the start codon, while the zmabi19-2 mutants had a single bp insertion of an adenine at position +123 bp. Both alleles resulted in a premature stop codon 25 and 2 bp downstream of the deletion and insertion, respectively (Figure 4, A). The positive plants were crossed with the wild type to segregate out the CRISPR/Cas9 construct and then the construct-free plants were self-pollinated. Representative ears for each self-pollinated zmabi19-1/+ and zmabi19-2/+ mutants are shown in Figure 4B, where one quarter of the seeds exhibited a small-size and flattened shape (Supplemental Table S2). We determined the genotypes of 24 flattened seeds from each of the self-pollinated zmabi19-1/+ and zmabi19-2/+ ears by PCR amplification and sequencing, which revealed that they were all homozygous for the “C” deletion and “A” insertion, respectively, indicating that the small seed phenotype was caused by the gene-edited mutations in ZmABI19. Immunoblot analysis showed that ZmABI19 was absent in zmabi19-1 and zmabi19-2 homozygous seeds, indicating these are null alleles (Supplemental Figure S12). Because the mutant seeds did not germinate (Supplemental Figure S13), we performed an allelic test between the heterozygous plants. The cross of zmabi19-1/+ and zmabi19-2/+ failed to complement the mutant phenotype in the resulting ears, as they segregated normal and mutant seeds in a 3:1 ratio, confirming that zmabi19-1 and zmabi19-2 are allelic (Figure 4, B). Because the phenotypes of the two alleles were nearly identical, we used the zmabi19-1 allele as a representative to study the function of ZmABI19 in subsequent experiments and abbreviated it zmabi19.zmabi19 seeds were much smaller than wild-type seeds and exhibited a starchy endosperm phenotype. They appeared non-vitreous when observed on a light box (Figure 4, C). The 100-kernel weight of the mutant was 9.3 g, only ∼35.1% of wild-type values (Figure 4, D). The content of total starch was decreased (P-value = 0.0002, Figure 4, D), whereas the level of soluble sugars was significantly increased in zmabi19 seeds (P-value = 0.007, Figure 4, D). Total protein levels were not overtly altered, while the lipid content was dramatically reduced compared with that of the wild type (P-value = 0.0054, Figure 4, D). These results indicate that loss of function in ZmABI19 has broadly adverse effects on seed development and filling.
Figure 4.
Phenotypes and storage-reserve contents of wild-type and zmabi19 seeds. (A) The CRISPR/Cas9-edited sequences in zmabi19 alleles. The zmabi19-1 and zmabi19-2 mutants have a “C” deletion and “A” insertion, respectively. The guide RNA spacer and protospacer adjacent motif (PAM) site are indicated. (B) Ear phenotypes of the wild type, self-pollinated zmabi19-1/+ and zmabi19-2/+, and the cross between zmabi19-1/+ and zmabi19-2/+. The black arrowheads indicate the zmabi19 seeds. Bar = 1 cm. (C) Seed phenotypes of wild type and zmabi19-1. (Top) longitudinal sections of wild-type and mutant seeds and (bottom) wild-type and mutant seeds observed on a light box. Em, embryo; En, endosperm. Bar = 1 cm. (D) Measurement of 100-kernel weight and contents of starch, soluble sugars, lipids, and proteins in wild-type and zmabi19-1 seeds. **P < 0.01; ns, no significant difference.
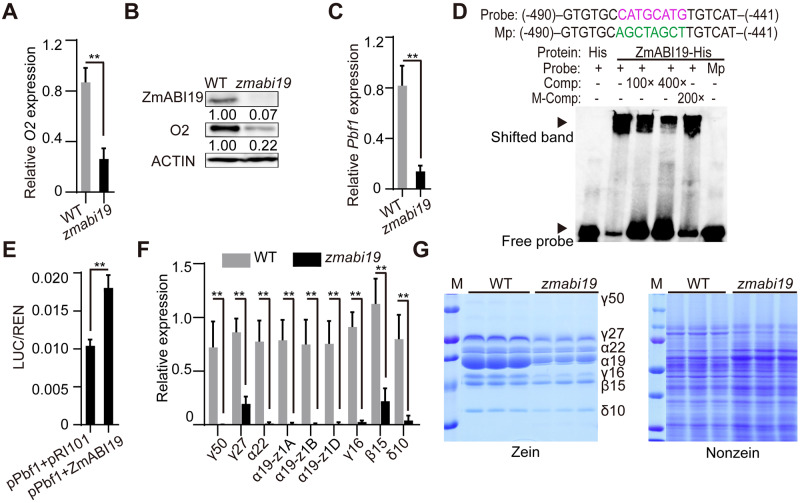
We examined whether zmabi19 affected the mRNA and protein levels of O2. RT-qPCR analysis revealed that O2 transcript levels in zmabi19 were reduced to about 30.2% of wild-type levels at 16 DAP (Figure 5, A), at which time storage reserves are actively synthesized. Consistent with transcript levels, O2 protein abundance was markedly reduced in zmabi19 endosperm (Figure 5, B).
Figure 5.
Mutation in ZmABI19 Affects the expression of O2, Pbf1, and zein genes. (A), (C), and (F) RT-qPCR analysis of O2, Pbf1, and zein expression in 16-DAP wild-type and zmabi19-1 endosperms. All expression levels were normalized to that of Actin. Error bars represent SD of three biological replicates. **P < 0.01. (B) Immunoblot analysis of ZmABI19 and O2 proteins in wild-type and zmabi19-1 seeds. The number below each lane indicates the protein levels relative to the values in wild type set as 1, which were all normalized against ACTIN. (D) EMSA of the specific binding of ZmABI19-His to the RY motif in the Pbf1 promoter. The normal and mutated RY motifs are indicated in magenta and green, respectively. Mp, mutated probes; M-Comp, mutated competitor. (E) DLR assay of the transactivation of the Pbf1 promoter by ZmABI19. Shown in the relative ratio of LUC/REN tested in Arabidopsis protoplasts via co-transfecting the reporter plasmid with effector constructs. **P < 0.01. (G) SDS-PAGE analysis of (left) zein and (right) non-zein proteins in wild-type and zmabi19-1 seeds. The size of each zein protein band is indicated beside it. γ50, 50-kD γ-zein; γ27, 27-kD γ-zein; α22, 22-kD α-zein; α19, 19-kD α-zein; γ16, 16-kD γ-zein; β15, 15-kD β-zein; and δ10, 10-kD δ-zein.
Since the temporal and spatial expression patterns of O2 and Pbf1 are nearly identical, they may be coordinately regulated by common factors. As with O2, the expression of Pbf1 was significantly reduced in zmabi19 endosperm (P-value = 0.0021, Figure 5, C). We searched the Pbf1 promoter for putative DNA cis-elements and found an RY motif at position −458 bp upstream of the start codon (Figure 5, D). EMSA and DLR assays revealed that ZmABI19 recognized and transactivated the Pbf1 promoter (Figure 5, D and E), confirming that Pbf1 is directly regulated by ZmABI19.
We then examined zein gene expression and found that transcripts and protein levels of all zeins, including 16-kD γ-zein, which is not directly regulated by O2 or PBF1, were dramatically reduced compared with wild-type levels (Figure 5, F and G), indicating that other factors involved in the regulation of zein genes are directly or indirectly affected by zmabi19. Due to proteome rebalancing, the levels of nonzein proteins increased in zmabi19 to compensate for the zein reduction, thereby resulting in wild-type total protein levels in zmabi19 (Figure 4, D and G).
Developmental and filling defects in zmabi19 seeds
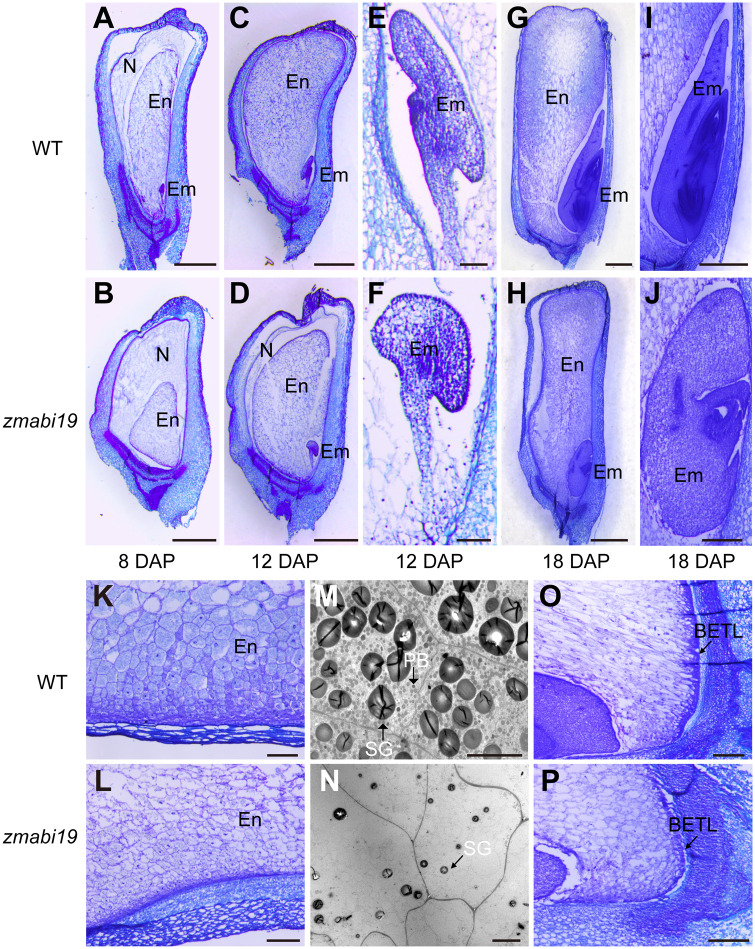
zmabi19 seeds, which appear smaller and paler during seed development, were clearly distinguished from wild-type seeds on the cob as early as 8 DAP (Supplemental Figure S14). To characterize their developmental defects, we examined paraffin sections of wild-type and mutant seeds at 8, 12, and 18 DAP, representing the time points for before initiation, after initiation, and the height of endosperm filling, respectively, by light microscopy. At 8 DAP, endosperm cells underwent rapid proliferation and differentiation. The seed and endosperm in zmabi19 were smaller than those of the wild type (Figure 6, A and B). zmabi19 endosperm was surrounded by a larger nucellus. Wild-type embryos developed into the transition stage, whereas mutant embryos were too small to be observed. At 12 DAP, the nucellus in wild-type seeds was completely degraded (Figure 6, C), but a layer of residual nucleus was still evident in zmabi19 (Figure 6, D). As a result, wild-type seeds contained a larger endosperm than zmabi19 mutant seeds. The embryo proper and scutellum began to differentiate in the wild type, forming obvious organ boundaries (Figure 6, E). By contrast, the zmabi19 embryo proceeded from the proembryo to the transition stage and developed abnormally (Figure 6, F). By 18 DAP, when storage reserves were actively synthesized in wild-type seeds, both the endosperm and embryo in zmabi19 showed developmental defects (Figure 6, G and H). In contrast to the starchy endosperm cells in the wild type, which contained a denser cytoplasm, zmabi19 cells were severely impaired and had obvious lesions (Figure 6, K and L). Transmission electron microscopy revealed that the size and number of starch granules and protein bodies in zmabi19 endosperm were drastically reduced compared with those of the wild type (Figure 6, M and N). Abnormalities in BETL were also observed in zmabi19, which formed less cell wall ingrowth relative to the wild type (Figure 6, O and P). For embryo development, typical embryonic structures, including the shoot apical meristem (SAM), root apical meristem, coleoptile, coleorhiza, leaf primordia, and scutellum, were established in the wild type (Figure 6, I). By contrast, embryogenesis in zmabi19 was arrested at the L1 stage. The scutellum was abnormal and there was no root meristem formed in zmabi19 embryos (Figure 6, J). Taken together, the results demonstrated that mutation of ZmABI19 causes profound defects in seed development and endosperm filling.
Figure 6.
Light and transmission electron microscopy observations of wild-type and zmabi19 seeds during seed development. (A)–(L) and (O) and (P) Light microscopy observations of wild-type and zmabi19-1 seeds. (A), (C), (E), (G), (I), (K) and (O) Wild-type seeds; (B), (D), (F), (H), (J), (L) and (P) zmabi19-1 seeds. WT, wild type; N, nucellus; En, endosperm; Em, embryo;. Bars = 1 mm in (A)–(D), (G) and (H); Bars = 100 μm in (E) and (F); Bars = 250 μm in (I) to (L) and (O) and (P). (M) and (N) Transmission electron microscopy observations of starchy endosperm cells of wild-type and zmabi19-1 seeds. PB, protein body; SG, starch granule. Bar = 10 μm.
Transcriptomic alterations in zmabi19 seeds at the filling stage
To globally explore the regulatory roles of ZmABI19 in seed development and storage reserve biosynthesis, we performed transcriptome deep sequencing (RNA-seq) of wild-type and zmabi19 endosperms and embryos at 16 DAP. After read counting and normalization, we detected 20,791 and 21,445 genes expressed in 16-DAP endosperms and embryos, respectively (Figure 7, A).
Figure 7.
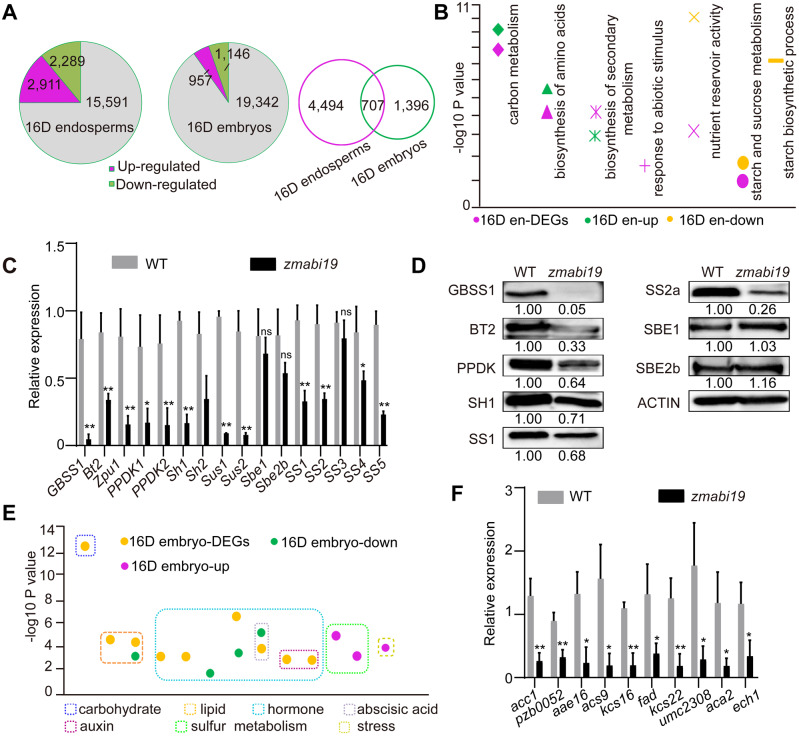
Comparison of transcriptomes between wild-type and zmabi19-1 endosperms and embryos at 16 DAP. (A) Pie chart of DEGs between (left) wild-type and zmabi19-1 endosperms and (middle) embryos and Venn diagram showing the overlap between DEGs in (right) endosperms and embryos. (B) Enrichment analysis of DEGs in 16-DAP endosperms. 16D en-DEGs, all DEGs; 16D en-up, upregulated DEGs in zmabi19; 16D en-down, downregulated DEGs in zmabi19-1; D, DAP; en, endosperm. (C) RT-qPCR analysis of starch biosynthetic genes in wild-type and zmabi19-1 seeds. All expression levels were normalized to that of Actin. Error bars represent SD of three biological replicates. *P < 0.05; **P < 0.01; ns, no significant difference. (D) Immunoblot analysis of starch biosynthetic proteins in wild-type and zmabi19-1 seeds at 16 DAP. The number below each lane indicates the protein levels relative to the values in wild type set as 1, which were all normalized against ACTIN. (E) Enrichment analysis of DEGs in 16-DAP embryos. 16D embryo-DEGs, all DEGs; 16D embryo-down, downregulated DEGs in zmabi19-1; 16D embryo-up, upregulated DEGs in zmabi19-1; D, DAP. (F) RT-qPCR analysis of genes involved in lipid biosynthesis and metabolism in wild-type and zmabi19-1 seeds at 16 DAP. All expression levels were normalized to that of Actin. Error bars represent SD of three biological replicates. *P < 0.05; **P < 0.01.
Differentially expressed genes (DEGs) were identified by comparing gene expression between the wild type and zmabi19 (P-value < 0.05, absolute fold change > 2.0). We detected 5,200 DEGs (2,289 downregulated and 2,911 upregulated) between wild-type and zmabi19 endosperms (Figure 7, A and Supplemental Data Set S2). To determine the biological processes affected by mutation of ZmABI19 in the endosperm, we subjected DEGs to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. The modules for carbon metabolism and biosynthesis of amino acids were highly enriched (Figure 7, B). As expected, the GO term associated with nutrient reservoir activity was apparently enriched, with zein genes among the involved DEGs (Figure 7, B and Supplemental Data Set S3). Consistent with the observation in Figure 5, F, transcripts of almost all the zein-coding genes were dramatically decreased in 16-DAP endosperms (Supplemental Data Set S3). The zmabi19 mutation also affected the process of starch and sucrose metabolism (Figure 7, B and Supplemental Data Set S3). We performed RT-qPCR to analyze the expression of all known starch biosynthetic genes, PPDK1 and PPDK2. The transcript levels of granule-bound starch synthase 1 (GBSS1), Bt2, Z. mays pullulanase-type debranching enzyme (Zpu1), PPDK1, PPDK2, Sh1, Sh2, Sus1, Sus2, sucrose synthase 1 (SS1), SS2, SS4, and SS5 were significantly reduced, of which GBSS1, Sus1, and Sus2 appeared to be the most affected (Figure 7, C). By contrast, transcript levels of starch branching enzyme 1 (Sbe1), Sbe2b, and SS3 were not reduced. SS3 is transcriptionally regulated by O2 and PBF1 (Zhang et al., 2016). The expression of SS3 was not affected by the downregulation of O2 or Pbf1 in zmabi19 endosperm, suggesting the existence of a complementary mechanism regulated by other changed factors in this mutant. Consistent with the mRNA levels, immunoblot analysis barely detected the GBSS1 protein, and the protein levels of BT2, PPDK, SH1, SS1, and SS2a were dramatically reduced in zmabi19 (Figure 7, D). The protein accumulation of SBE1 and SBE2b showed no apparent differences between wild-type and zmabi19 endosperms (Figure 7, D). Taken together, these results demonstrated that protein and starch biosynthesis was dramatically affected in zmabi19 endosperms.
We next characterized the 2,103 DEGs in zmabi19 embryos, of which 1,146 were downregulated and 957 were upregulated relative to the wild type (Figure 7, A and Supplemental Data Set S4). When the two groups of DEGs in 16-DAP endosperms and embryos were compared, 707 DEGs were identified as common DEGs, suggesting that they have conserved functions in the two filial structures (Figure 7, A). Combined GO and KEGG analyses revealed drastic alterations in carbohydrate metabolism and phytohormone-mediated signaling pathways in 16-DAP embryos (Figure 7, E and Supplemental Data Set S5). Furthermore, abscisic acid (ABA) and auxin-activated signaling pathways were notably affected. The pathways associated with lipid storage and metabolism were also enriched. Consistent with the RNA-seq data, RT-qPCR confirmed that the transcript levels of lipid biosynthesis-related genes were significantly reduced in zmabi19 embryos (Figure 7, F).
Identification of ZmABI19 target genes using ChIP-seq coupled with differential expression analysis
ZmABI19 appears to play crucial roles in both early seed development and late grain filling. Eight DAP is defined as the time point when early endosperm development is completed and endosperm cells are undergoing rapid mitotic division and differentiation for later storage reserve biosynthesis. To maximize the identification of the genes that are directly recognized and transactivated by ZmABI19, we used 8-DAP whole grains to perform RNA-seq and ChIP followed by deep sequencing (ChIP-seq) analyses. RNA-seq was performed as described above. We obtained 2,621 DEGs (1,309 downregulated and 1,312 upregulated) when comparing the wild-type and zmabi19 transcriptomes (Figure 8, A and Supplemental Data Set S6). When the three groups of DEGs (16-DAP endosperms and embryos and 8-DAP grains) were compared, we identified 700 common DEGs between 8-DAP grains and 16-DAP endosperms, and 351 common DEGs between 8-DAP grains and 16-DAP embryos, indicating that a significant number of genes are conserved at these two developmental stages (Figure 8, B).
Figure 8.
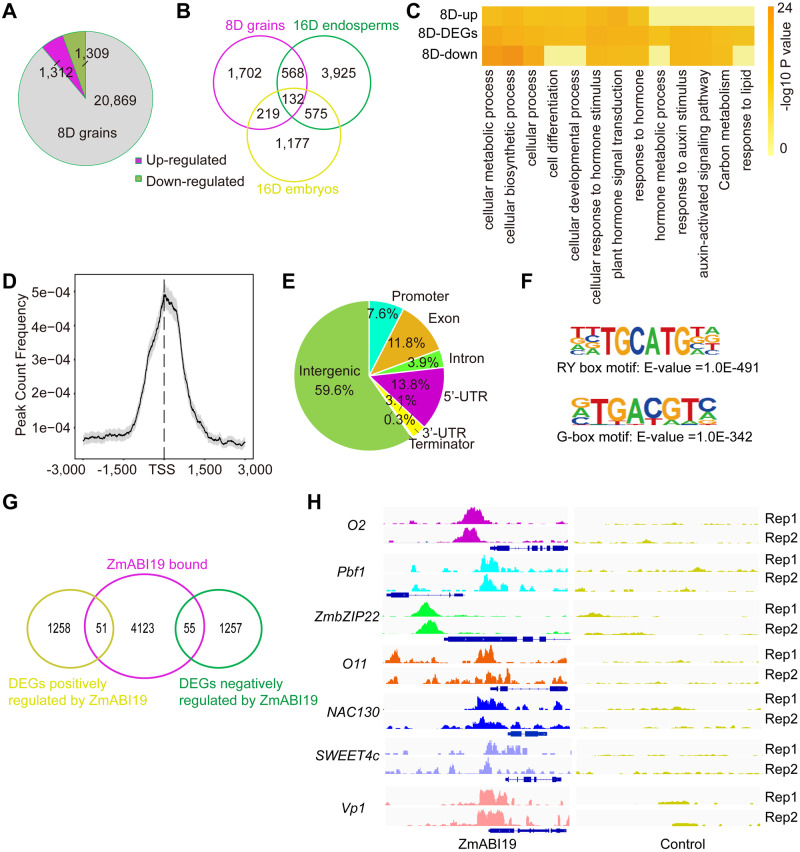
Summary of the RNA-seq and ChIP-seq analyses at 8 DAP. (A) Pie chart of DEGs in wild-type and zmabi19-1 grains. (B) Venn diagram showing common DEGs in 8-DAP grains, 16-DAP endosperms, and 16-DAP embryos. (C) Enrichment analysis of DEGs in 8-DAP whole grains. 8D-up, upregulated DEGs in zmabi19-1; 8D-DEGs, all DEGs; 8D-down, downregulated DEGs in zmabi19-1; and D, DAP. (D) Distribution of ZmABI19 binding sites corresponding to the −3,000 to +3,000 bp region flanking the TSS. (E) Distribution of ZmABI19-binding regions in the maize genome. Promoter, −3 kbp to +100 bp of the TSS and Terminator, –100 bp to +1 kbp of the transcription terminator site (TTS). (F) ZmABI19-binding motifs identified in the 1-kbp flanking sequences around the genic peaks. The enrichment P-value was calculated using Fisher’s exact test. E indicates 10 raised to a power in scientific notation. (G) Venn diagram showing the overlap of the 4,229 genes bound by ZmABI19 with the DEGs identified from the RNA-seq data at 8 DAP. (H) Peak distribution of ZmABI19-binding sites for O2, Pbf1, ZmbZIP22, O11, NAC130, SWEET4c, and Vp1. The loci are shown using the Integrated Genome Browser (IGV). Rep1 and Rep2 indicate the peaks in two replicates of ChIP-seq data.
To determine the biological processes affected by zmabi19 in 8-DAP grains, we subjected the 2,621 DEGs to GO and KEGG analyses. Cellular metabolic and biosynthetic processes were enriched and most related genes were downregulated in zmabi19 embryos (Figure 8, C and Supplemental Data Set S7). In addition, plant hormone signal transduction and cellular response to phytohormone stimulus were remarkably affected in zmabi19. In 8-DAP zmabi19 grains, 63 and 102 DEGs were identified in the categories of plant hormone signal transduction and cellular response to hormone stimulus, respectively. Among them, GO analysis showed that highly enriched functions were in response to auxin stimulus and auxin-activated signaling pathway (Figure 8, C and Supplemental Data Set S7). Additionally, significantly altered expression patterns were also observed for carbon metabolism and responses to lipid (Figure 8, C).
ChIP-seq assays were performed using antibodies raised against ZmABI19. The specificity of ZmABI19 antibodies was verified by immunoblot analysis (Supplemental Figure S12). We extracted chromatin from 8-DAP whole grains; we used the IgG antibody as a negative control. Binding sites were identified from consistent peaks generated from two independent biological replicates. Using model-based analysis of ChIP-Seq (MACS2) with a cutoff q-value < 0.05, we detected 22,676 peaks corresponding to putative ZmABI19-bound sites in the maize genome (Supplemental Data Set S8). Distribution of the binding peaks in the genome revealed that all the 10 chromosomes were mapped (Supplemental Figure S15). Among all detected peaks, highly concentrated binding peaks were located around 1 kbp from the transcription start site (TSS) regions (Figure 8, D). We determined the distribution pattern of the bound peaks in genic regions, containing the gene bodies and the regions from 3-kbp upstream of the TSS to 1-kbp downstream of the transcription termination site (TTS). A total of 40.4% of bound peaks were located in genic regions. Among them, 7.6% were located in the promoter regions (−3 kbp to +100 bp of the TSS), and 13.8% and 3.1% were located in the 5′-untranslated region (5′-UTR) and 3′-UTR sequences, respectively (Figure 8, E). The remaining bound peaks located to exons (11.8%), introns (3.9%), and terminator regions (0.3%, –100 bp to +1 kbp of the TTS). Annotation of bound peaks in the genic regions showed that 4,229 genes were identified as putative ZmABI19-bound targets (Supplemental Data Set S9).
The 1 kbp flanking sequences around all genic peaks were analyzed to identify novel ZmABI19 binding motifs. The RY motif with the core sequence TGCATG was highly overrepresented (E-value = 1.0E−491; Figure 8, F). In addition, G-box elements with the core sequence ACGT were also identified (E-value = 1.0E−342; Figure 8, F). To further obtain high-confidence target genes for ZmABI19, we overlaid ZmABI19-bound genes in the promoter regions with the DEGs in 8-DAP grains from the wild type versus zmabi19 comparison. This analysis resulted in the identification of 106 putatively direct ZmABI19 targets, including 51 downregulated and 55 upregulated DEGs (Figure 8, G and Supplemental Data Set S10). KEGG enrichment analysis demonstrated that the overlapping targets were significantly enriched in plant hormone signal transduction and starch and sucrose metabolism (Supplemental Figure S16 and Supplemental Data Set S11). As examples of the accuracy of our identification of ZmABI19 target genes, the O2 and Pbf1 promoter regions were significantly enriched in the identified ZmABI19 binding sites (Figure 8, H). ChIP-qPCR assays in the O2 and Pbf1 promoter regions were carried out using the eluted DNA from the ChIP assay. Chromatin segments around the two RY motifs in the O2 promoter and the RY motif in the Pbf1 promoter were significantly enriched in the DNA pool bound by ZmABI19 antibodies (P-value = 0.0002 for RY-A and 0.0005 for RY-B in the O2 promoter; P-value = 0.0032 for the RY in the Pbf1 promoter; Supplemental Figure S17, A and B). We also found that a few known grain filling regulatory factors, such as O11, ZmbZIP22, NAC130, and Vp1, and the BETL-specific transporter gene SWEET4c were bound and regulated by ZmABI19 (Figure 8, H). In addition, a number of genes involved in plant hormone signal transduction were identified as ZmABI19 targets (Supplemental Data Sets S10 and S12).
ZmABI19 directly regulates key grain filling factors in the endosperm and embryo
To validate that these genes were truly recognized and transactivated by ZmABI19, we performed a series of biochemical experiments to test them individually.
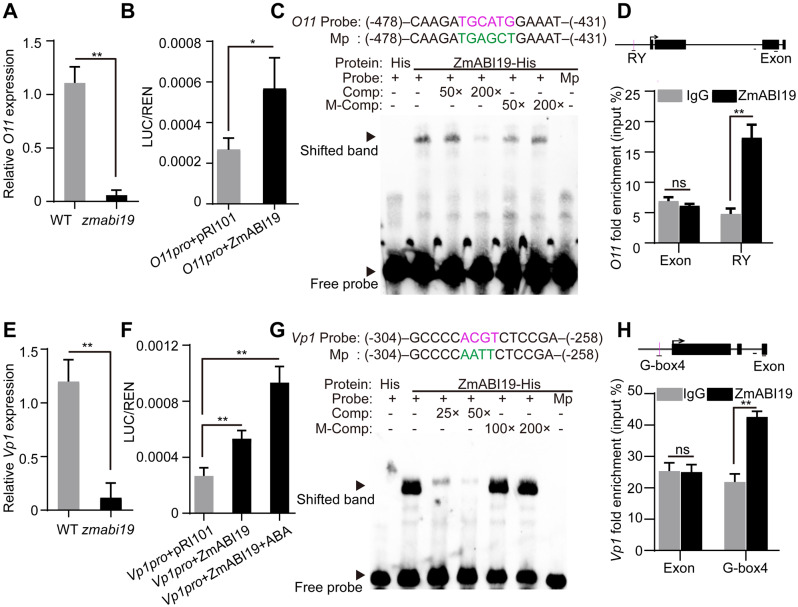
ChIP-seq revealed that the O11 promoter was directly bound by ZmABI19 and significantly enriched in ZmABI19-binding sites (Figure 8, H). Consistent with the RNA-seq data at 8 DAP, RT-qPCR demonstrated that the expression of O11 was apparently downregulated in zmabi19 (Figure 9, A and Supplemental Data Sets S6 and S9). DLR assay showed a significant transactivation of O11pro:LUC when co-transfected with 35Spro:ZmABI19 (P-value = 0.033, Figure 9, B). Searching the O11 promoter revealed an RY motif located −448 bp upstream of the start codon. EMSA confirmed direct binding of ZmABI19 to this RY motif in vitro (Figure 9, C), consistent with the ChIP-qPCR result that showed enrichment of the O11 promoter around the RY motif (Figure 9, D).
Figure 9.
ZmABI19 binds to the promoters of O11 and Vp1 for transactivation. (A) and (E) RT-qPCR analysis of (A) O11 and (E) Vp1 in wild-type and zmabi19-1 grain at 8 DAP. Error bar represented ±SD (n = 3). P-values were determined by Student’s t test. **P < 0.01. (B) and (F) DLR assay of the transactivation of the (B) O11 and (F) Vp1 promoters by ZmABI19. Shown is the LUC/REN ratio tested in Arabidopsis protoplasts via co-transfecting the reporter plasmids and the effector construct. *P < 0.05; **P < 0.01. (C) and (G) EMSA of the binding of ZmABI19-His to the RY motif in the (C) O11 promoter and (G) the G-box in the Vp1 promoter. The normal motifs are indicated in magenta and mutated ones in green. Mp, mutated probes; Comp, competing probes unlabeled with biotin; M-Comp, mutated competitor. (D) and (H) ChIP-qPCR assay of the binding of ZmABI19 to the RY motif in the (D) O11 promoter and (H) the G-box in Vp1 promoter. The examined regions are indicated under the schematic diagrams of the O11 and Vp1 loci by the horizontal lines. Arrows indicate the TSS. Black boxes indicate the exons. Vertical magenta lines indicate the RY motif and G-box. The bars below the intron regions represent 100 bp. **P < 0.01; ns, no significant difference.
Analysis of the bound targets revealed that the Vp1 promoter was enriched among ZmABI19-binding sites (Figure 8, H). RT-qPCR showed that Vp1 transcript levels were remarkably reduced in zmabi19 (Figure 9, E). DLR assay further demonstrated that the Vp1 promoter was transactivated by ZmABI19, and that transactivation activity was significantly enhanced when 10-μM ABA was added to this assay (P-value = 0.0009, Figure 9F). Analysis of the Vp1 promoter revealed that an RY motif and five G-boxes (G-box1 to G-box5) were located in the −1.5 kbp region upstream of the start codon (Supplemental Figure S18). EMSA showed a band shift for the G-box4 probe in the gel (Supplemental Figure S18). Furthermore, the binding specificity was verified by competition experiments, where gradually reduced signals were observed with the addition of unlabeled intact probes and a mutated probe failed to be recognized by ZmABI19 (Figure 9, G). ChIP-qPCR also revealed the enrichment of the sequence around G-box4 in the Vp1 promoter (Figure 9, H).
We also tested other ZmABI19 target genes by DLR assay and found that the promoters of ZmbZIP22, NAC130, NAC86, NAC134, and SWEET4c were significantly transactivated by ZmABI19 (Supplemental Figures S19, A–E).
ZmABI19 directly regulates genes related to plant hormone signal transduction
ARFs are effectors of the auxin response and transfer the auxin signal to regulate a defined set of genes (Roosjen et al., 2018). At low auxin levels, the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) TFs prevent ARFs from controlling auxin-responsive genes. We found that ZmABI19 bound the promoter region or 5′-UTR of nine ARF genes that were also differentially expressed in the 8-DAP RNA-seq data (Supplemental Data Set S12). In addition, 11 Aux/IAA TFs were also identified among the bound targets. The transcript levels of IAA5, IAA8, IAA9, IAA19, IAA24, and IAA38 decreased, while those for IAA12, IAA36, IAA43, and IAA44 increased in the 8-DAP RNA-seq data (Supplemental Data Set S12). The ARF4 and ARF26 promoters were selected to test their transactivation activity using the DLR assay. LUC activity was significantly increased when driven by the two ARF promoters when the reporter constructs were co-transfected with 35Spro:ZmABI19 (P-value = 0.018 for ARF4 and 0.005 for ARF26, Supplemental Figures S19, F and 19, G), indicating that they are direct targets of ZmABI19.
Discussion
Conserved functions of FUS3 homologs in seed development
Seed development is elaborately directed by a regulatory network of TFs controlling downstream target genes. In dicotyledonous species, such as Arabidopsis, the endosperm is degraded to a single cell layer (i.e. the aleurone layer) at an early stage, and the nutrients are mainly stored in the embryo (Costa et al., 2004; Sabelli and Larkins, 2009b), while monocotyledonous species, such as maize, contain a large and permanent endosperm with a cavity where a smaller embryo develops (Costa et al., 2004). The endosperm accumulates abundant starch and storage proteins, and the embryo stores proteins and lipids (Kroj et al., 2003; Sabelli and Larkins, 2009b; Larkins et al., 2017). The RY motif has been identified as the target for central regulators of seed development, such as LEC2, ABI3, and FUS3. Mutations in any of these factors result in a dramatic reduction in the accumulation of storage materials (proteins and lipids) in the embryo. The RY motif appears to be functionally conserved in the promoters of many spore- and seed-specific genes across a wide range of species, from ferns through gymnosperms to angiosperms (Schallau et al., 2007). The fern spore may be regarded as a storage tissue, reminiscent of dicotyledonous embryos, in terms of the expression of a storage globulin-like protein (Shutov et al., 1998).
Recent research has demonstrated that FUS3 is also expressed in the chalaza and funiculus of mature ovules and plays a critical role in reproductive development before fertilization. After fertilization, both FUS3 transcript and protein accumulate in the embryo during the early stages of embryogenesis, whereas FUS3 expression is repressed by class I BASIC PENTACYSTEINE (BPC) proteins in most of the endosperm. Repression of FUS3 by BPC proteins in the endosperm is proposed to be required to reduce the rate of endosperm nucleus proliferation and promote endosperm differentiation and embryo growth (Wu et al., 2020). ZmABI19 and FUS3 diverge greatly in their protein sequence (sharing only 24.6% similarity, Supplemental Figure S6), even though they are the most evolutionarily related of the B3 TFs between Arabidopsis and maize (Supplemental Figure S7). FUS3 is highly expressed in the embryo and also detected in the sole residual endosperm tissue, aleurone (Keith et al., 1994; Tiedemann et al., 2008), while ZmABI19 is highly expressed in whole seeds at the early stage (Figure 2, E). At the filling stage, ZmABI19 expression was weakly detected in starchy endosperm cells, but strongly detected in the BETL of endosperm and the basal section of the scutellum in the embryo (Figures 2, F–I), indicating that ZmABI19 plays an important role in maize endosperm development. Investigation of the zmabi19 and fus3 mutants revealed some phenotypic similarities in the embryo. fus3 mutants display seed abortion with a shrunken crown and distorted coat (Keith et al., 1994; Tiedemann et al., 2008; Chan et al., 2017). Gene-edited homozygous zmabi19 mutant was seed-lethal with a defective endosperm and embryo (Figure 6). Similar to fus3, zmabi19 seeds accumulated lower levels of lipids and storage proteins than the wild type (Figure 4, D). Orthologous genes to FUS3 and ZmABI19 were characterized in other monocotyledonous species, such as HvFUS3 in barley and TaFUSCA3 in wheat (Moreno-Risueno et al., 2007; Sun et al., 2017). Expression of HvFUS3 and TaFUSCA3 in the fus3 mutant background rescued the mutant phenotypes, indicating that FUS3 orthologs in monocots and dicots are functionally conserved in embryo development.
Since Arabidopsis contains only a transient endosperm, the function of FUS3 homologs in later stages of endosperm development in monocots remains unknown. De-repression or ectopic expression of FUS3 in the Arabidopsis endosperm increases cell proliferation, resulting in enlarged endosperm and larger seeds at the expense of embryo development (Wu et al., 2020). This suggests that FUS3 promotes both embryo and endosperm development in the seed. Previous research showed that HvFus3 transcripts also accumulate in barley endosperm and transcriptionally activate the endosperm-specific genes Hor2 and Itr1, which encode a B-hordein storage protein and the trypsin inhibitor BTI-CMe, respectively, indicating the essential roles of FUS3 homologs in cereal endosperm development and illustrating a conserved regulatory apparatus for seed development (Moreno-Risueno et al., 2007).
ZmABI19 functions as a grain filling initiator
The endosperm-specific and development-dependent activation of a number of grain filling regulators indicates the presence of common cis-regulatory motifs and TFs that interact to coordinate their expression. Since O2 acts as a master regulator for endosperm filling (Li et al., 2015; Zhang et al., 2016; Deng et al., 2020), we employed the O2 promoter to determine the cis-motif(s) and TF(s) responsible for the coordinated expression of grain filling TFs. We identified a key transcriptional fragment or element in the O2 promoter and demonstrated that ZmABI19 binds the RY motifs and transactivate the O2 promoter (Figure 1).
When the expression patterns of ZmABI19 and O2 were compared, they showed different tendencies throughout seed development. ZmABI19 was expressed at remarkably higher levels in the endosperm at the early phase (1–8 DAP) than at mid phase (10–24 DAP) (Figure 2, C). By contrast, O2 transcripts were first detected at 8 DAP, and then O2 expression levels increased rapidly, reaching a peak at 25 DAP (Zhang et al., 2015). Interestingly, the expression discrepancy was also observed for Vp1 and its regulated genes (Zheng et al., 2019). Two possibilities may be considered to explain this discrepancy. First, ZmABI19 is regulated at both the transcriptional and posttranscriptional levels. Indeed, although ZmABI19 transcript levels exhibited a diminishing expression pattern during grain filling, the protein appeared to constantly accumulate during the early (1–8 DAP) and mid developmental phases (10–24 DAP, Figure 2, D). Such protein accumulation may sustain steady transactivation of O2 expression, when the silent chromatin and DNA modifications in the O2 promoter are removed during the early- to mid-phase transition. In leaves, O2 targets, such as 22-kD α-zein genes, show high levels of cytosine methylation, which is accompanied by transcriptionally silent chromatin that is enriched in histone H3 dimethylated at Lys-9 (H3K9me2) and Lys-27 (H3K27me2). The switch between transcriptional silencing and activation in the endosperm is a two-step process involving an early potentiated state and a later activated state, where the epigenetic marks are erased and RNA Polymerase II (RNPII), O2, and the transcriptional ADAptor2 (Ada2)/General Control Nonderepressible 5 (Gcn5) coactivator bind to the target sites (Locatelli et al., 2009). It is likely that transcriptional activation of the O2 promoter also occurs through such a process at approximately 8 DAP. Second, ZmABI19 primarily functions to initiate the expression of O2 at 8 DAP, and later the O2 protein joins to transactivate its own promoter. Indeed, we demonstrated that O2 binds to the O2 box in its promoter and exerts auto-activation (Supplemental Figure S4), which is consistent with previous observations (Lohmer et al., 1991). Previous ChIP-seq data also revealed that O2 binds to its own promoter (Li et al., 2015; Zhan et al., 2018).
Exploring the regulatory network of zein genes is important for the nutritional improvement of maize grains. Several factors have been identified that directly regulate zein gene expression, including O2, Pbf1, Ohp1, Ohp2, ZmbZIP22, ZmMADS47, NAC128, and NAC130 (Schmidt et al., 1992; Pysh and Schmidt, 1996; Vicente-Carbajosa et al., 1997; Wu and Messing, 2012; Zhang et al., 2015, 2016, 2019b; Qiao et al., 2016; Yang et al., 2016; Li et al., 2018). In general, these genes additively and cooperatively regulate the expression of zein genes. O2 interacting with PBF1 plays a major role in the transactivation of 19- and 22-kD α-zein genes. OHPs interacting with PBF1 primarily transactivate 27-kD γ-zein (Zhang et al., 2015; Yang et al., 2016). ZmbZIP22 co-regulates the transcription of 27-kD γ-zein with O2, PBF1, and OHPs (Li et al., 2018). NAC128 and NAC130 transactivate 16-kD γ-zein and Bt2 (Zhang et al., 2019b). In this study, we demonstrated that the Pbf1, ZmbZIP22, NAC130, and O11 promoters were directly bound and transactivated by ZmABI19 as well (Figures 5, D and E, 8, H, and 9, A–D, and Supplemental Figures 17, B and 19, A and B). Co-regulation of these grain filling regulators is essential for synchronized initiation of zein protein and starch biosynthesis in starchy endosperm cells.
Transportation of soluble sugars into the developing seeds directly influences the development process and determines the seed size and weight. The structure of cell wall ingrowths of BETL is critical for nutrient allocation and is viewed as an important interface regulating sugar entrance into the developing seed (Chourey and Hueros, 2017). SWEET4c, encoding a hexose transporter, is responsible for transferring hexoses across the BETL as an important step for endosperm filling (Koch, 2004; Sosso et al., 2015). SWEET4c is predominately expressed in the BETL and considered as a master regulator for BETL cell fate (Doll et al., 2017). The ChIP-seq data and subsequent experiments demonstrated that SWEET4c was a direct target bound and transactivated by ZmABI19 (Figure 8,H and Supplemental Figure S19, E), indicating that ZmABI19 is essential for BETL development and sink strength because of its transcriptional regulation of SWEET4c.
VP1 plays a pivotal role in scutellum development and protein reallocation from endosperm to embryo (Zheng et al., 2019). Transcriptional regulation by Vp1 is involved in ABA signaling (McCarty et al., 1989; Mccarty et al., 1991; McCarty, 1995). ABA signaling is generally coupled with LAFL TFs controlling embryo development and maturation (Zheng et al., 2019). RNA-seq of 16-DAP embryos revealed that genes related to ABA signal transduction were significantly affected by the loss of ZmABI19 function (Figure 7, E). ChIP-seq and DLR analyses revealed that ZmABI19 directly bound to the Vp1 promoter and that the transactivation activity was increased when ABA was added to the reaction (Figures 8, H and 9, F). Previous reports showed that B3 domain TFs could directly bind the RY motif and G-box (Vasil et al., 1995; Monke et al., 2012). Consistent with this observation, we found that ZmABI19 also recognized the two kinds of DNA elements (Figure 8, F). While ZmABI19 binds the RY motif in most cases, such as for O2, Pbf1, and O11, it recognizes G-box4 in the Vp1 promoter (Figure 9, G and H, and Supplemental Figure S18). The expression of Vp1 begins at 3–5 DAP and reaches a peak at 10 DAP (Yi et al., 2019; Zheng et al., 2019). The expression of ZmABI19 was also highly enriched in early seed development (1–8 DAP). Similar to the Vp1 mRNA signal, the ZmABI19 signal was also observed in the scutellum (Figure 2, H). The overlapping temporal and spatial expression trends support the hypothesis that ZmABI19 directly regulates Vp1 expression to promote embryogenesis and facilitate nutrition reallocation from the endosperm to the embryo at the filling stage.
ZmABI19 is a key regulator during early seed development
After double fertilization, the primary endosperm cell undergoes several rounds of nuclear divisions, resulting in a coenocyte (0–2 DAP) (Sabelli and Larkins, 2009b). The coenocyte is cellularized through the formation of radial microtubule systems around the nuclei (2–4 DAP) (Sabelli and Larkins, 2009a; SabelLi et al., 2018). Then, these cells undergo rapid proliferation and differentiate into distinguishable cell types (4–8 DAP). These early mitotic divisions of the endosperm are a critical step to determine seed size and weight because most cells in the mature endosperm are created during this stage (Sabelli and Larkins, 2009a; SabelLi et al., 2018). ZmABI19 was highly expressed during early seed development and expressed at a low level in the endosperm at the filling stage (Figure 2, C). Observations of paraffin sections at 8 DAP revealed that endosperm development in zmabi19 was severely delayed (Figure 6, A and B). At seed maturity, the 100-kernel weight was greatly reduced to approximately 35.1% of the wild type in zmabi19 (Figure 4, D).
Eight DAP is a key point to illustrate the connected processes of early seed development and graining filling. RNA-seq analysis at 8 DAP revealed that plant hormone signal transduction was remarkably affected in zmabi19 (Figure 8, C). Auxin plays pivotal roles in growth and development throughout the entire plant life cycle. Auxin is a main contributor in establishing and patterning the embryo development process and regulating seed coat and endosperm development (Lau et al., 2012; Figueiredo et al., 2015; Figueiredo and Köhler, 2018; Robert et al., 2018). In maize, auxin increases at the beginning of endoreduplication and remains at high levels throughout kernel development (Lur and Setter, 1993; Doll et al., 2017). The auxin response is mediated by ARF TFs, which recognize specific sequences in the promoters of auxin-responsive genes to activate or repress transcription (Ulmasov et al., 1999). Aux/IAA TFs act as repressors by binding and sequestering ARF TFs and are degraded upon perception of auxin (Kim et al., 1997; Guilfoyle and Hagen, 2007). ChIP-seq analysis revealed that ZmABI19 probably directly regulates a number of ARF and Aux/IAA TFs, indicating that ZmABI19 responds to auxin signals to regulate different auxin-related genes to balance the developmental process in maize seed (Supplemental Data Set S12).
In summary, ZmABI19 regulates seed development and grain filling by directly binding to the promoters of and transactivating the expression of key factors in the endosperm and embryo. During early seed development, ZmABI19 transcripts highly accumulate in the whole grain. ZmABI19 directly regulates auxin-responsive genes to promote seed growth and development (Figure 10). BETL cell fate is established at approximately 5 DAP. SWEET4c transcripts are highly enriched at early stage and considered as a critical factor for BETL cell fate. SWEET4c is transactivated by ZmABI19, promoting nutrient transport to developing seeds. At 8 DAP, O2 and Pbf1 begin to be expressed in the endosperm. ZmABI19 directly binds to their promoters to initiate transcription. Due to the low expression of ZmABI19 at the filling stage, O2 is able to transactivate its own promoter, which rapidly increases its expression. Moreover, ZmABI19 directly recognizes the RY motif in the O11 promoter and transactivates gene expression. ZmABI19 and O11 interact and antagonistically regulate the expression of O2. The promoters of Pbf1, ZmbZIP22, and NAC130 are also bound and transactivated by ZmABI19. These grain filling factors are coordinated to regulate protein and starch synthesis, leading to a strict spatiotemporal expression pattern in the endosperm. In the embryo, ZmABI19 directly regulates the expression of Vp1 to promote embryogenesis and strengthen nutrient allocation from endosperm to embryo. When ZmABI19 is inactivated, both endosperm and embryo development are severely affected. We provide strong evidence to support the idea that ZmABI19 plays crucial roles in coordinating embryo and endosperm development and functions as an initiator for grain filling.
Figure 10.
A proposed model for the regulatory network of ZmABI19 in seed development. O2 begins to be transcribed at 8 DAP. At this time, ZmABI19 binds to the two RY motifs of the O2 promoter to initiate O2 expression. When entering the endosperm filling stage, the expression of ZmABI19 decreases in the endosperm. Instead, O2 binds to the O2 box to promote auto-activation. ZmABI19 directly regulates other key factors in the endosperm (Pbf1, ZmbZIP22, O11, NAC130, and SWEET4c) and in the embryo (Vp1) for coordinated regulation of graining filling. ZmABI19 also regulates auxin responsive genes in early seed development.
Materials and methods
Plant materials and growth conditions
Maize plants were cultivated in the greenhouse and the Song Jiang experimental field in Shanghai, China, or the field in Sanya, China. Wild-type N. benthamiana and Arabidopsis (A. thaliana) of the Columbia-0 (Col-0) accession were grown in the greenhouse at 22°C and 70% relative humidity under 16-h light and 8-h dark photoperiod (90 μmol/m2/s).
We collected tissues [root, stem, leaf, SAM, tassel, ear, and 3-DAP seeds] from the maize inbred B104 to determine the specific expression pattern of ZmABI19. Immature kernels were harvested at 1, 2, 3, 4, 6, 8, 10, 12, 14, 16, 18, 20, and 24 DAP, immediately frozen in liquid nitrogen and stored at −80°C until use for RNA and protein extraction. Samples were collected from three individual plants for each developmental stage.
The Mu-insertion mutant (UFMu-09010) was obtained from the UniformMu Stock Center (https://www.maizegdb.org/uniformmu).
The CRISPR/Cas9 transgenic lines (in the B104 background) were created through Agrobacterium (Agrobacterium tumefaciens)-mediated transformation as previously described (Scott, 2009). The 20-bp guide RNA target editing sequence was selected in the first exon of ZmABI19. Resistant calli were selected on selection medium containing bialaphos for three rounds. In total, 15 independent knockout plants were obtained and crossed with wild type to obtain construct-free materials.
Genotyping
To identify the Mu and transgenic materials, leaves were harvested and placed into a 2-mL tube containing a stainless bead. The genomic DNA was extracted using the cetyl trimethylammonium bromide (CTAB) method. The Mu materials were genotyped using three primers: one pair flanking the Mu insertion and the last primer specific to the Mu terminal inverted repeats. The partial sequences for Bar and ZmABI19 were amplified from genomic DNA using specific primers to genotype the transgenic materials. Construct-free samples were further analyzed by PCR amplification. The genome-edited sites were genotyped by PCR amplification and sequencing.
Protein accumulation analysis
Mature kernels of each sample were ground into a fine powder in a steel pot. A total of 50 mg for each sample was weighed for zein extraction in a 0.5-mL zein extraction buffer (3.75 mM sodium borate, 2% 2-mercaptoethanol [v/v], 0.3% SDS [w/v], and 70% ethanol [v/v]; pH 10). Non-zein proteins were extracted in a 0.5-mL non-zein buffer (12.5 mM sodium borate, 2% 2-mercaptoethanol [v/v], and 5% SDS [w/v]; pH 10). Three biological replicates from three plants for each sample were analyzed. The zein and non-zein accumulation patterns were analyzed by electrophoresis on 15% and 10% SDS-PAGE gels, respectively. Quantification of protein levels was performed using a BCA Protein Assay Kit (Thermo Scientific™) following the manufacturers’ protocols.
Immunoblotting
The specific antibodies of ZmABI19 and O2 were produced using unique sequences in maize. The antibodies were produced in rabbits by Abclonal, China. Starch biosynthesis-related antibodies were generated previously (Zhang et al., 2016). The secondary antibody to detect the synthesized antibodies was goat anti-rabbit IgG-horseradish peroxidase (HRP) (Abmart Cat# M21002L). The ACTIN antibody (Abmart Cat# M20009L) and the related goat anti-mouse IgG-HRP (Abmart Cat# M21001L) were purchased. Total proteins were extracted in a 0.4-mL lysis buffer (50 mM HEPES (4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid) pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 [v/v], 0.1% deoxycholate [w/v], and 1× proteinase inhibitor cocktail), separated on a 10% SDS-PAGE gel and transferred to a PVDF (polyvinylidene fluoride) membrane. Samples were incubated with primary antibodies (1:1,000 dilution) for 1 h at room temperature and washed with Tris-buffered saline (TBS)-0.1% TWEEN 20 buffer three times. Secondary antibodies (1:5,000 dilution) were then incubated for 1 h. The membrane was washed three times in TBST buffer. The signal was detected using the NovexECL Chemiluminescent Substrate Reagent Kit (Invitrogen) and visualized using a Tanon-5200 Chemiluminescent Imaging system (Tanon Science and Technology).
RNA extraction, RT-qPCR, and 3′-RACE
Immature seeds, separated endosperms and embryos were placed in a 2.0-mL tube and suspended in 0.3-mL RNA extraction buffer (150 mM LiCl, 50 mM Tris pH 8.0, 5 mM EDTA pH 8.0, and 1% SDS [w/v]). The homogenized samples were further extracted twice using 0.3 mL phenol–chloroform (pH 4.2) and once with 0.3 mL chloroform. The aqueous supernatant was mixed with 1 mL TRIzol regent (Invitrogen™). The extracted total RNA was further purified using RNeasy Mini Kit with DNaseI digestion (Qiagen). RNA purity and concentration were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific™). We used 1 μg of total RNA for reverse transcription with the ImProm-II™ Reverse Transcription System (Promega). Real-time PCR was performed using TB Green™ Premix Ex Taq™ II (Takara) on a Bio-Rad CFX-96 PCR thermocycler. The relative expression levels were calculated using the 2–ΔΔCt method, in which the maize Actin gene was used as the internal control (Livak and Schmittgen, 2001). All primers used in this experiment are listed in Supplemental Data Set S13.
The Mu insertion type was identified by the 3′-RACE System (Invitrogen™). The nested PCR technology was applied to obtain the full-length transcribed Mu sequence in zmabi19-Mu; the primers used are listed in Supplemental Data Set S13. The resulting PCR products were purified, cloned into pMD18-T vector (Takara), and sequenced.
RNA in situ hybridization
Developing B104 kernels at 8 and 16 DAP were used for RNA in situ hybridization. A ZmABI19 fragment was amplified by PCR and inserted into the pSPT18 vector. Primers are listed in Supplemental Data Set S13. The sense and antisense RNA probes were produced in vitro using T7 and SP6 RNA polymerase with DIG RNA Labeling Mixture (Roche), respectively. Tissue processing and detailed procedures using 10 μm sections have been described previously (Zhang et al., 2015).
Cytological analysis
For light microscopy analysis, wild-type and zmabi19 seeds at 8, 12, and 18 DAP were collected from the same plant for paraffin preparation. The seeds were fixed in FAA buffer (formaldehyde:acetic acid:ethanol:water = 10:5:50:35, v/v/v/v) and vacuumed for 30 min. After dehydration with an ethanol series, seeds were embedded in paraffin. Sections (10 μm) were rehydrated in ethanol gradient series of 100%, 95%, 85%, 70%, 50%, and 30% ethanol, followed by water. The sections were then stained using toluidine blue solution (0.1%) and photographed under a bright field using a Leica microscope.
For transmission electron microscopy, endosperms at 18 DAP were sliced and fixed in 2.5% (w/v) glutaraldehyde in 0.2 M phosphate buffer at pH 7.2. The samples were then fixed in 2.5% (w/v) OsO4, dehydrated, and embedded in epoxy resin. Thin sections were obtained using a Leica microtome, and stained with uranyl acetate and lead citrate. Observations were performed using a transmission electron microscope.
Yeast one-hybrid assay
Yeast one-hybrid assays were performed according to the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech). Briefly, the bait sequences were synthesized, cloned into pAbAi vector, and integrated into the genome of the Y1HGold yeast strain. The prey sequences were amplified, cloned into the pGADT7-Rec vector, and fused with the bait yeast strain. The primers used are listed in Supplemental Data Set S13. To determine protein and DNA interaction, the bait yeast strains were selected on medium lacking uracil with different concentrations of the Aureobasidin A (AbA) antibiotic.
Transcriptional activation assay
The full-length coding sequence (CDS) and truncated sequences of ZmABI19 were cloned in the plasmid pGBKT7. The resulting plasmids were separately transformed into yeast Y2HGold strain according to the procedure of Matchmaker Gold Yeast Two-Hybrid System (Clontech). The yeast strains were spotted on selection medium lacking tryptophan (Trp) or lacking Trp, histidine (His), and adenine (Ade).
DLR assay
The reporter plasmids were generated by inserting promoter sequences into the cloning site upstream of the LUC CDS in the vector pGreen0080. Mutations of the promoter sequence were created using PCR amplification and validated by sequencing. The primers used are listed in Supplemental Data Set S13. The effector plasmids were prepared by inserting the CDS into the pRI101 vector containing the 35S promoter. Arabidopsis mesophyll protoplasts were isolated and transformed using polyethylene glycol (PEG)-CaCl2 methods. After an incubation of 12 h, total protein was extracted and analyzed on a luminometer (Promega 20/20). A commercial LUC analysis kit was used according to the manufacturer’s protocols (Promega). Three biological replicates were performed for each experiment.
Particle bombardment
We harvested, separated, and placed 8-DAP B73 endosperms on half-strength Murashige and Skoog (MS) medium. Gold powder (Alfa Aesar) was cleaned twice with ethanol and kept at a final concentration of 60 mg mL−1. A total of 0.1 mL cleaned gold powder for each sample was coated with DNA from the reporter and effector plasmids. The gold powder mixed with vectors was transiently transformed into the endosperm using a Bio-Rad PDS-1000/He™ biolistic particle delivery system (Bio-Rad, Hercules, CA, USA). After incubation for 16 h, total protein was extracted and analyzed on a luminometer (Promega 20/20). Three biological replicates were performed for each experiment.
LCI assay
The full-length CDS of ZmABI19 and O11 were cloned into JW771 (N-terminal half of luciferase, NLUC) and JW772 (C-terminal half of luciferase, CLUC) to produce ZmABI19-NLUC and O11-CLUC, respectively. The resulting plasmids were introduced separately into Agrobacterium strain GV3101. Individual colonies were grown in LB medium overnight with the appropriate antibiotics, collected by centrifugation and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 150 mM acetosyringone) at a final OD600 = 1.0. The cell suspensions were mixed in equal volume and injected into N. benthamiana leaves. After 2 days, leaves were injected with 0.8 mM luciferin and LUC signals were observed using a Tanon 5200 chemiluminescent Imaging system.
BiFC assay
The full-length CDS of ZmABI19 and O11 were cloned into pSAT4-nEYFP and pSAT4-cEYFP plasmids, respectively. Arabidopsis mesophyll protoplasts were isolated and transformed as described in the procedure for the DLR assay. Co-transfected protoplasts were incubated for 18 h. Fluorescence was monitored using an excitation wavelength of 488 nm on a LSM880 confocal microscope.
Phylogenetic analysis
Homologous sequences were identified in the National Center for Biotechnology Information (NCBI) nr (nonredundant protein sequences) database by performing a Basic Local Alignment Search Tool for protein (BLASTp) search with the ZmABI19 protein sequence. All B3 domain-containing TFs in maize were obtained from the Grassius database (https://www.grassius.org/family.php?family=ABI3-VP1&species=Maize). The protein sequences of the LAV family members in Arabidopsis were downloaded from the Arabidopssi Information Rresource (TAIR) database (https://www.arabidopsis.org/). Protein sequences were aligned with MUSCLE in the MEGA7.0 software. Evolutionary distances were evaluated using the neighbor-joining algorithm. The phylogeny testing applied the bootstrap method with 1,000 replicates.
Subcellular localization
The ZmABI19 CDS was cloned into the pCAMBIA1300-35S-eGFP vector. The primers used are listed in Supplemental Data Set S13. The resulting vectors were introduced into Agrobacterium strain GV3101 and the bacterial suspensions infiltrated into about 3-week-old N. benthamiana leaves. After 2 days, the infiltrated leaves were collected and imaged on a LSM880 confocal microscope (Zeiss, Jena, Germany). To observe the nucleus, 1 μg mL−1 of 4′,6-diamidino-2-phenylindole (DAPI) solutions (Roche) was injected. In addition, the same plasmids were transfected into Arabidopsis mesophyll protoplasts. After a 16-h incubation, the transfected protoplasts were imaged on a LSM880 confocal microscope. The constructs were also transformed into Arabidopsis plants (Col-0) using Agrobacterium-mediated transformation; T2 plants were used to observe the GFP signal. The cytoplasmic and nuclear protein fractions were isolated from 4-DAP seeds as previously described (Feng et al., 2018).
Recombinant protein purification and EMSA
The full-length CDSs of ZmABI19 and O2 were inserted into the cloning sites of the pColdTF expression vector (Takara). Recombinant proteins were produced in Escherichia coli DE3 (BL21) cells. Protein production was induced for 24 h by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 0.5 mM when OD600 of the cells reached 0.6. The cells were then sonicated and the supernatants containing the fusion proteins were purified with Ni-HA beads (QIAGEN). Oligonucleotide probes were synthesized and labeled with biotin at the 5′ or 3′-end. The probes and recombinant proteins were mixed in the DNA binding reactions (1× binding buffer, 2.5% glycerol, 5 mM MgCl2, 50 ng/mL poly [dI•dC], and 0.05% NP-40). The mixture was loaded onto a 6% native PAGE in 0.5× Tris/borate/EDTA buffer. After electrophoresis, the binding reactions were transferred to a nylon membrane, and then the transferred DNA was crosslinked to the membrane by UV light. Subsequent detection was performed using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) on a Tanon 5200 chemiluminescent Imaging System (Tanon Science and Technology). All oligonucleotides and primers are listed in Supplemental Data Set S13.
Measurement of starch and soluble sugar
Starch content was determined with the Megazyme Total Starch Assay Kit (#KTSTA-50A) according to the manufacturer’s instructions. Briefly, 100 mg of mature kernel flour was weighed and added to 0.2 mL aqueous ethanol (80% v/v) to aid dispersion. The pellets were resuspended in 2 mL of 2 M KOH and stirred in an ice/water bath for 20 min. After adding 8 mL of 1.2 M sodium acetate buffer pH 3.8, we added 0.1 mL thermostable α-amylase and 0.1 mL amyloglucosidase and mixed intermittently in a water bath at 50°C for 30 min. Then 1.0 mL of the above reaction was diluted with 9 mL water and 3.0 mL GOPOD (glucose oxidase/ peroxidase) reagent was added to 0.1 mL of the resulting diluted solution. After incubating the tubes at 50°C for 20 min, the absorbances for each sample and the d-glucose control were read at 510 nm against the blank reagent. All measurements were performed at least three times.
A total of 100 mg mature kernel flour was weighed for each sample and used for measurement of soluble sugar content, which was determined using the anthrone method as previously described (Wang et al., 2013).
Measurement of lipids
The mature seeds for wild type and zmabi19 were dried at 45°C for 60 h, then ground to a fine powder using a tissue grinder. A total of 200 mg seed powder for each sample was mixed with 4 mL of a methanol and acetyl chloride mixture (10:1, v/v). Then 5 mL of C19:0 internal standard was added and incubated in a water bath at 80°C for 2 h. After cooling, 5 mL of 7% K2CO3 was added to the mixture, and the supernatant was injected into a gas chromatograph with standard manipulation procedures.
Transcriptome deep-sequencing (RNA-seq) analysis
Total RNA was extracted from 8-DAP whole grains, 16-DAP endosperm, and 16-DAP embryo as described above. Three independent biological replicates from three ears were collected. Sequencing libraries were conducted using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) and sequenced on the Illumina NovaSeq 6000 platform as paired-end reads. Quality control of the raw data was checked using the FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The adapters and unpaired sequences were removed using the Trimmomatic software (Bolger et al., 2014). The cleaned reads were mapped to the maize reference genome B73v4 using Hisat2 (v2.0.5) (Pertea et al., 2016). Differentially expression genes were identified with an adjusted P-value < 0.05 and absolute fold change greater than 2.0 using the edgeR (v3.30.0) software package in R (Robinson et al., 2010). The GO and KEGG analyses were performed using the database g:Profile (http://biit.cs.ut.ee/gprofiler/). The KEGG enrichment profile was also performed using KEGG Mapper (https://www.kegg.jp/kegg/tool/map_pathway2.html).
ChIP-seq and ChIP-qPCR
ChIP-seq with ZmABI19-specific antibodies was conducted using a previously described method with minor modifications (Li et al., 2015; Feng et al., 2018). Two independent biological replicates were performed. In brief, 8-DAP grains were dissected and immediately crosslinked with 1% formaldehyde (Sigma) in phosphate buffer (10 mM sodium phosphate pH 7.0, 50 mM NaCl, and 0.1 M sucrose). The samples were kept on ice under vacuum infiltration twice for 15 min. Fixation was stopped using 0.125 M glycine, and the fixed materials were washed three times with phosphate buffer, dried with blotting paper, frozen in liquid nitrogen, and stored at −80°C. About 2.5 g fixed samples were ground into a fine powder and homogenized in 20 mL M1 buffer (10 mM sodium phosphate pH 7.0, 0.1 M NaCl, 1 M 2-methyl 2,4-pentanediol, 10 mM β-mercaptoethanol), and filtered through two layers of Miracloth prior to nuclei extraction. The homogenate was further washed with M2 buffer (10 mM sodium phosphate, pH 7.0, 0.1 M NaCl, 1 M 2-methyl 2,4-pentanediol, 10 mM β-mercaptoethanol, 10 mM MgCl2, and 0.5% Triton X-100) and M3 buffer (10 mM sodium phosphate, pH 7.0, 0.1 M NaCl, and 10 mM β-mercaptoethanol). Nuclei-enriched extracts were resuspended in 1 mL lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 [v/v], 0.1% deoxycholate [w/v], and proteinase inhibitor cocktail) and then sonicated for 15 cycles of 30 s at high amplitude, and cooled in ice water for 30 s between pulses using a Bioruptor UCD-200 (Diagenode). The size of chromatin fragments was estimated using agarose gel electrophoresis from 200 to 500 bp fragment size. Antibodies against ZmABI19 and Rabbit IgG control (Abclonal, China) were used for immunoprecipitation. The chromatin solutions were pre-cleared using 40 μL Pierce™ Protein A Magnetic Beads (ThermoFisher) for 1 h at 4°C. The mixture was then incubated overnight with 10 μL ZmABI19 antibody and IgG control, respectively, followed by incubation with 60 μL Pierce™ Protein A Magnetic Beads for 1 h on a rotating tube shaker. The beads were washed three times using wash buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10 μM ZnSO4, 1% Triton X-100, and 0.05% SDS) and eluted by elution buffer (200 mM NaHCO3 and 2% SDS). The prepared DNA was recovered using a QIAquick PCR purification kit (Qiagen), and quality was checked using an Agilent 2100 Bioanalyzer. The DNA libraries were constructed using the NEXTflex Rapid DNA Sequencing Kit (Bioo Scientific Corp.) and sequenced on the Illumina NovaSeq 6000 platform.
For ChIP-qPCR analysis, the obtained DNA was diluted and analyzed using specific DNA primers (Supplemental Data Set S13). The amplification was performed using TB Green™ Premix Ex Taq™ II (Takara) on the Bio-Rad CFX-96 PCR thermocycler. The ChIP values were normalized to their relative input DNA values, and the fold-changes were calculated based on the relative enrichment between ZmABI19 and IgG immunoprecipitates.
ChIP-seq analysis
Raw data of ChIP-seq was quality-controlled using FastQC software and cleaned through the Trimmomatic software. The clean reads were aligned to the maize reference genome B73v4 using Bowtie2 (2.2.3) with default parameters (Langmead and Salzberg, 2012). Mapped reads were used for ChIP-seq peak calling by MACS2 (v2.1.0) with the cutoff q-value < 0.05. Peak-related genes were annotated with ChIPseeker in R (https://bioconductor.org/packages/release/bioc/html/ChIPseeker.html). The enrichment of sequence motifs was detected by HOMER (http://homer.ucsd.edu/homer/motif/). The KEGG enrichment analysis was implemented by the g:Profile (http://biit.cs.ut.ee/gprofiler/) and KEGG Mapper (https://www.kegg.jp/kegg/tool/map_pathway2.html).
Statistical analysis
Microsoft Excel (2016) was used to calculate P-value by paired two-tailed Student t test methods. Detailed statistical analysis is shown in Supplemental Data Set S14.
Accession numbers
O2 (Zm00001d018971); Pbf1 (Zm00001d005100); ZmABI19 (Zm00001d011712); ZmABI4 (Zm00001d033313); ZmbZIP22 (Zm00001d021191); SWEET4c (Zm00001d015912); NAC130 (Zm00001d008403); Vp1 (Zm00001d042396; O11 (Zm00001d003677); Actin (Zm00001d010159); acc1 (Zm00001d024998); pzb0052 (Zm00001d021046); aae16 (Zm00001d034832); acs9 (Zm00001d053009); kcs16 (Zm00001d039053); fad (Zm00001d051636); kcs22 (Zm00001d045660); umc2308 (Zm00001d018487); aca2 (Zm00001d008432); ech1 (Zm00001d009182). Sequences used for phylogenetic analysis can be found in Supplemental Files S1 and S3, Supplemental Figure S2, A, and Supplemental Data Set S1. The RNA-seq and ChIP-seq data in this article are available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series entries PRJNA629420.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Yeast one-hybrid assay of the binding of B3 domain TFs to the O2 promoter.
Supplemental Figure S2. Transactivation of the O2 promoter by ZmABI4.
Supplemental Figure S3. DLR assay of the transactivation of the O2 promoter by ZmABI19.
Supplemental Figure S4. Auto-transactivation of the O2 promoter by O2 through recognizing the O2 box.
Supplemental Figure S5. Antagonistic roles of ZmABI19 and O11 in transactivation of the O2 promoter.
Supplemental Figure S6. Amino acid alignment of FUS3 and ZmABI19-B73.
Supplemental Figure S7. Phylogenetic analysis of B3 transcription factors in maize and Arabidopsis.
Supplemental Figure S8. Subcellular localization of ZmABI19-eGFP in Arabidopsis transgenic plants.
Supplemental Figure S9. The Mu insertion does not disrupt ZmABI19 function.
Supplemental Figure S10. Amino acid alignment of ZmABI19-W22 and ZmABI19-Mu.
Supplemental Figure S11. Identification of the transcriptional activation domain of ZmABI19.
Supplemental Figure S12. Immunoblot analysis of the specificity of the ZmABI19 antibodies and protein levels of ZmABI19 in wild-type and zmabi19 seeds.
Supplemental Figure S13. Germination of wild-type and zmabi19 seeds.
Supplemental Figure S14. Self-pollinated zmabi19/+ ears at different developmental stages.
Supplemental Figure S15. Distribution of the binding peaks in the genome from the ZmABI19 ChIP-seq data.
Supplemental Figure S16. KEGG enrichment analysis of the common targets by overlapping the ZmABI19-bound genes from the ChIP-seq data with the DEGs from the RNA-seq data of 8-DAP grains of the wild type versus zmabi19.
Supplemental Figure S17. ChIP-qPCR assay of the binding of ZmABI19 to the RY motifs in the O2 and Pbf1 promoters.
Supplemental Figure S18. EMSA to explore the binding region for ZmABI19 in the Vp1 promoter.
Supplemental Figure S19. DLR assay of the transactivation of the potential target promoters by ZmABI19.
Supplemental Table S1. Predicted cis-elements in the O2 promoter between −400 and –500 bp upstream of the Start Codon.
Supplemental Table S2. Chi-squared (χ2) test showing the single gene recessive inheritance of zmabi19 in segregating ears.
Supplemental Data Set S1. B3 domain transcription factor genes expressed in maize seeds based on the public RNA-seq data.
Supplemental Data Set S2. DEGs identified from the RNA-Seq data of 16-DAP endosperms.
Supplemental Data Set S3. Enrichment analysis of the DEGs of 16-DAP endosperms.
Supplemental Data Set S4. DEGs identified from the RNA-Seq data of 16-DAP embryos.
Supplemental Data Set S5. Enrichment analysis of the DEGs of 16-DAP embryos.
Supplementary Data Set S6. DEGs identified from the RNA-Seq data of 8-DAP grains.
Supplemental Data Set S7. Enrichment analysis of DEGs from the RNA-Seq data of 8-DAP grains.
Supplemental Data Set S8. Peak distribution obtained from the ZmABI19 ChIP-seq data.
Supplemental Data Set S9. Target genes obtained from the ZmABI19 ChIP-seq data.
Supplemental Data Set S10. High-confidence potential ZmABI19 targets identified by overlapping the genes bound by ZmABI19 in the promoter region with the DEGs from the RNA-Seq data of 8-DAP grains.
Supplemental Data Set S11. Enrichment analysis of high-confidence potential ZmABI19 targets identified by overlapping the genes bound by ZmABI19 in the promoter region with the DEGs from the RNA-Seq data of 8-DAP grains.
Supplemental Data Set S12. Target genes from the ZmABI19 ChIP-seq data for the genes related to plant hormone signal transduction pathway.
Supplemental Data Set S13. Primers used in this study.
Supplemental Data Set S14. Detailed statistical analysis in this study.
Supplemental File S1. Files of the alignment used to generate the phylogenetic tree in Figure 2, A.
Supplemental File S2. Newick tree format of the phylogenetic tree data from Figure 2, A.
Supplemental File S3. Files of the alignment used to generate the phylogenetic tree in Supplemental Figure S7.
Supplemental File S4. Newick tree format of the phylogenetic tree data from Supplemental Figure S7.
Supplementary Material
Acknowledgments
The authors thank Zan Wu from the Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University, for bioinformatic analysis of RNA-seq and ChIP-seq data; Prof. Zeng Tao from the College of Agriculture and Biotechnology, Zhejiang University for Arabidopsis-related experiment; Jiqin Li from the CAS Center for Excellence in Molecular Plant Sciences for the technique support of electron microscopy.
Funding
This work was supported by the Chinese Academy of Sciences [XDB27010201 to Y.W.] and the National Natural Science Foundation of China [91935305, 31830063 and 31925030 to Y.W.].
Conflict of interest statement. None declared.
References
- Baud S, Kelemen Z, Thévenin J, Boulard C, Blanchet S, To A, Payre M, Berger N, Effroy-Cuzzi D, Franco-Zorrilla JM, et al. (2016) Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by L-AFL proteins in Arabidopsis seed. Plant Physiol 171: 1099–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumlein H, Miséra S, Luerßen H, Kölle K, Horstmann C, Wobus U, Müller AJ (1994) The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J 6: 379–387 [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard C, Fatihi A, Lepiniec L, Dubreucq B (2017) Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits. Biochim Biophys Acta 1860: 1069–1078 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci U S A 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonero P, Iglesias-Fernández R, Vicente-Carbajosa J (2016) The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds. J Exp Bot 68: 871–880 [DOI] [PubMed] [Google Scholar]
- Chan A, Carianopol C, Tsai AY, Varatharajah K, Chiu RS, Gazzarrini S (2017) SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J Exp Bot 68: 4219–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J (2014) Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol 166: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Hueros G (2017). The basal endosperm transfer layer (BETL): Gateway to the maize kernel. Maize Kernel Development, (Larkins BA): 56–67 [Google Scholar]
- Ciceri P, Locatelli F, Genga A, Viotti A, Schmidt RJ (1999) The activity of the maize Opaque2 transcriptional activator is regulated diurnally. Plant Physiol 121: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C, Larkins B (1999) The prolamins of maize. In Shewry P, Casey R, eds, Seed Proteins. Kluwer Academic Publisher, Dordrecht, The Netherlands, pp 109–139 [Google Scholar]
- Cord Neto G, Yunes JA, da Silva MJ, Vettore AL, Arruda P, Leite A (1995) The involvement of Opaque 2 on beta-prolamin gene regulation in maize and Coix suggests a more general role for this transcriptional activator. Plant Mol Biol 27: 1015–1029 [DOI] [PubMed] [Google Scholar]
- Costa LM, Gutièrrez-Marcos JF, Dickinson HG (2004) More than a yolk: the short life and complex times of the plant endosperm. Trends Plant Sci 9: 507–514 [DOI] [PubMed] [Google Scholar]
- Deng Y, Wang J, Zhang Z, Wu Y (2020) Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol J 18: 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll NM, Depège-Fargeix N, Rogowsky PM, Widiez T (2017) Signaling in early maize kernel development. Mol Plant 10: 375–388 [DOI] [PubMed] [Google Scholar]
- Esen A (1987) A proposed nomenclature for the alcohol-soluble proteins (zeins) of maize (Zea mays L.). J Cereal Sci 5: 117–128 [Google Scholar]
- Fatihi A, Boulard C, Bouyer D, Baud S, Dubreucq B, Lepiniec L (2016) Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Sci 250: 198–204 [DOI] [PubMed] [Google Scholar]
- Feng F, Qi W, Lv Y, Yan S, Xu L, Yang W, Yuan Y, Chen Y, Zhao H, Song R. (2018) OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell 30: 375–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Köhler C (2018) Auxin: a molecular trigger of seed development. Genes Dev 32: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C (2015) Auxin production couples endosperm development to fertilization. Nat Plants 1: 1–6 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontarek BC, Neelakandan AK, Wu H, Becraft PW (2016) NKD transcription factors are central regulators of maize endosperm development. Plant Cell 28: 2916–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci U S A 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Kriz AL (1989) Characterization of embryo globulins encoded by the maize Glb genes. Biochem Genet 27: 239–251 [DOI] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P, Onatesanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, Vicentecarbajosa JJJoBC (2003). Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Larkins BA, Wu Y, Song R, Messing J (2017) Maize seed storage proteins. Maize Kernel Dev. 175–189 [Google Scholar]
- Lau S, Slane D, Herud O, Kong J, Jurgens G (2012) Early embryogenesis in flowering plants: setting up the basic body pattern. Annu Rev Plant Biol 63: 483–506 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Devic M, Roscoe TJ, Bouyer D, Zhou DX, Boulard C, Baud S, Dubreucq B (2018) Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod 31: 291–307 [DOI] [PubMed] [Google Scholar]
- Li C, Song R (2020) The regulation of zein biosynthesis in maize endosperm. Theor Appl Genet 133: 1443–1453 [DOI] [PubMed] [Google Scholar]
- Li C, Yue Y, Chen H, Qi W, Song R (2018) The ZmbZIP22 transcription factor regulates 27-kD gamma-zein gene transcription during maize endosperm development. Plant Cell 30: 2402–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H, et al. (2015) Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize. Plant Cell 27: 532–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TDJM (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Locatelli S, Piatti P, Motto M, Rossi V (2009) Chromatin and DNA modifications in the Opaque2-mediated regulation of gene transcription during maize endosperm development. Plant Cell 21: 1410–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmer S, Maddaloni M, Motto M, Di Fonzo N, Hartings H, Salamini F, Thompson RD (1991) The maize regulatory locus Opaque‐2 encodes a DNA‐binding protein which activates the transcription of the b‐32 gene. EMBO J 10: 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lur H, Setter TL (1993) Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol 103: 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR (1995) Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Biol 46: 71–93 [Google Scholar]
- McCarty DR, Carson CB, Stinard PS, Robertson DS (1989) Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell 1: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Monke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hahnel U, Junker A, Weisshaar B, Conrad U, Baumlein H, et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MÁ, González N, Díaz I, Parcy F, Carbonero P, Vicente-Carbajosa J (2007) FUSCA3 from barley unveils a common transcriptional regulation of seed-specific genes between cereals and Arabidopsis. Plant J 53: 882–894 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Schmidt RJ (1996) Characterization of the maize OHP1 gene: evidence of gene copy variability among inbreds. Gene 177: 203–208 [DOI] [PubMed] [Google Scholar]
- Qiao Z, Qi W, Wang Q, Feng Y, Yang Q, Zhang N, Wang S, Tang Y, Song R (2016). ZmMADS47 regulates zein gene transcription through interaction with opaque2. PLoS Genet 12: e1005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutierrez CL, Wojcikowska B, Pěncik A, Novak O, Chen J, Grunewald W, Dresselhaus T, Friml J (2018) Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants 4: 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Mccarthy DJ, Smyth GKJB (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosjen M, Paque S, Weijers D (2018) Auxin response factors: output control in auxin biology. J Exp Bot 69: 179–188 [DOI] [PubMed] [Google Scholar]
- Roscoe TT, Guilleminot J, Bessoule J-J, Berger F, Devic M (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol 56: 1215–1228 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2009a) The contribution of cell cycle regulation to endosperm development. Sex Plant Reprod 22: 207–219 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2009b) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Nguyen H, Larkins BA (2018) Cell cycle and endosperm development. Annu Plant Rev 22: 294–310 [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schallau A, Kakhovskaya I, Tewes A, Czihal A, Tiedemann J, Mohr M, Grosse I, Manteuffel R, Baumlein H (2007) Phylogenetic footprints in fern spore- and seed-specific gene promoters. Plant J 53: 414–424 [DOI] [PubMed] [Google Scholar]
- Schmidt R, Burr F, Burr B (1987) Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science 238: 960–963 [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G (1992) Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4: 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP (2009) Transgenic maize: methods and protocols. Ann Bot 109: 7 [Google Scholar]
- Shutov AD, Braun H, Chesnokov YV, Baumlein H (1998) A gene encoding a vicilin-like protein is specifically expressed in fern spores. Evolutionary pathway of seed storage globulins. Eur J Biochem 252: 79–89 [DOI] [PubMed] [Google Scholar]
- Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, et al. (2015) Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 47: 1489–1493 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Wobus U. (2013) Seed-development programs: a systems biology-based comparison between dicots and monocots. Annu Rev Plant Biol 64: 189–217 [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci U S A 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Liu X, Wei Q, Liu J, Yang T, Jia L, Wang Y, Yang G, He G (2017) Functional characterization of TaFUSCA3, a B3-superfamily transcription factor gene in the wheat. Front Plant Sci 8: 1133–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR (2008) Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Peterson KJ, Jack TJTiPS (2008) The plant B3 superfamily. Trends Plant Sci 13: 647–655 [DOI] [PubMed] [Google Scholar]
- Tiedemann J, Rutten T, Mönke G, Vorwieger A, Rolletschek H, Meissner D, Milkowski C, Petereck S, Mock H-P, Zank T, et al. (2008) Dissection of a complex seed phenotype: novel insights of FUSCA3 regulated developmental processes. Dev Biol 317: 1–12 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nambara E, Naito S, McCourt P (2004) The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J 37: 73–81 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci U S A 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V, Marcotte WR, Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, McCarty DR (1995) Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 7: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci U S A 94: 7685–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-C, Xu H, Zhu Y, Liu Q-Q, Cai X-L (2013). OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot 64: 3453–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Mohamed D, Dowhanik S, Petrella R, Gregis V, Li J, Wu L, Gazzarrini S (2020) Spatiotemporal restriction of FUSCA3 expression by class I BPCs promotes ovule development and coordinates embryo and endosperm growth. Plant Cell 32: 1886–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Messing J (2012) Rapid divergence of prolamin gene promoters of maize after gene amplification and dispersal. Genetics 192: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ji C, Wu YR (2016) Divergent transactivation of maize storage protein zein genes by the transcription factors opaque2 and OHPs. Genetics 204: 581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Gu W, Chen J, Song N, Gao X, Zhang X, Zhou Y, Ma X, Song W, Zhao H, et al. (2019). High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31: 974–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Li G, Ryu C-H, Ma C, Zhang S, Lloyd A, Hunter BG, Larkins BA, Drews GN, Wang X, et al. (2018) Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell 30: 2425–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun Q, Zhang C, Hao G, Wang C, Dirk LMA, Downie AB, Zhao T (2019a) Maize VIVIPAROUS1 interacts with ABA INSENSITIVE5 to regulate GALACTINOL SYNTHASE2 expression controlling seed raffinose accumulation. J Agric Food Chem 67: 4214–4223 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yang J, Wu Y (2015) Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zheng X, Yang J, Messing J, Wu Y (2016) Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc Natl Acad Sci U S A 113: 10842–10847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dong J, Ji C, Wu Y, Messing J (2019b) NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci U S A 116: 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Li Q, Li C, An D, Xiao Q, Wang W, Wu Y (2019) Intra-kernel reallocation of proteins in maize depends on VP1-mediated scutellum development and nutrient assimilation. Plant Cell 3: 2613–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.