Abstract
A reduction in pod shattering is one of the main components of grain legume domestication. Despite this, many domesticated legumes suffer serious yield losses due to shattering, particularly under arid conditions. Mutations related to pod shattering modify the twisting force of pod walls or the structural strength of the dehiscence zone in pod sutures. At a molecular level, a growing body of evidence indicates that these changes are controlled by a relatively small number of key genes that have been selected in parallel across grain legume species, supporting partial molecular convergence. Legume homologs of Arabidopsis thaliana silique shattering genes play only minor roles in legume pod shattering. Most domesticated grain legume species contain multiple shattering-resistance genes, with mutants of each gene typically showing only partial shattering resistance. Hence, crosses between varieties with different genes lead to transgressive segregation of shattering alleles, producing plants with either enhanced shattering resistance or atavistic susceptibility to the trait. The frequency of these resistance pod-shattering alleles is often positively correlated with environmental aridity. The continued development of pod-shattering-related functional information will be vital for breeding crops that are suited to the increasingly arid conditions expected in the coming decades.
Recent genetic, genomic, and phenotypic studies of pod shattering in grain legumes lay the foundation for breeding crops suited for increasingly arid conditions.
Introduction
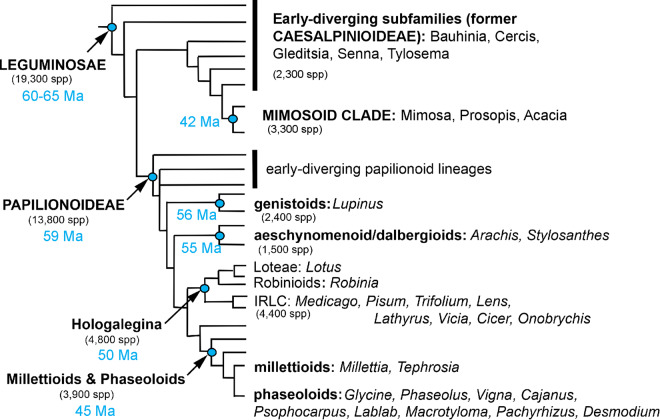
Flowering plants evolved numerous novel mechanisms for seed dispersal during their immense radiation, which Darwin famously called an abominable mystery. Seed dispersal in flowering plants has been modified frequently throughout history due to changing selection pressure and symbioses. A primary unifying feature of the legume family, or Fabaceae, is a unicarpellate fruit (legumen in Latin or pods), which produces seeds along a single ventral suture (Esau, 1977). Seeds are typically dispersed by the explosive dehiscence of the pod at fruit maturity, a process known as pod shattering (Simpson, 2019). This form of dispersal has been highly successful for wild species. The legume family is the third largest family of flowering plants in terms of species number, with at least 19,300 species (LPWG, 2017; Figure 1). Pod dehiscence in legumes can lead to devastating yield losses in agricultural environments. Humans have, therefore, selected strongly against pod shattering in domesticated legumes (Ogutcen et al., 2018; Di Vittori et al., 2019). A reduction in seed dispersal and the loss of seed dormancy are two fundamental domestication traits among seed-propagated crops, including grain legumes (Koinange et al., 1996).
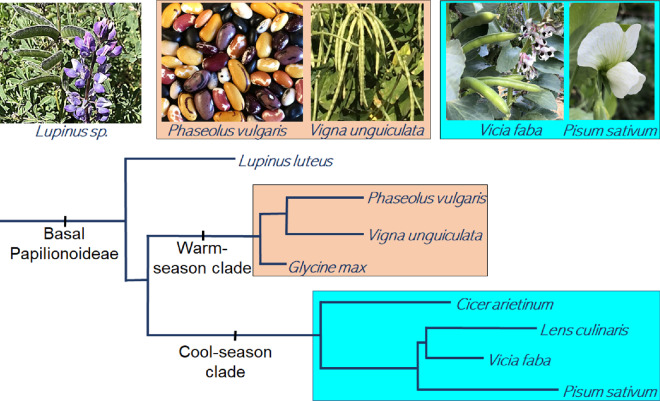
Figure 1.
A schematic representation of relationships between major legume taxa. The Fabaceae family is the third largest in terms of species among plants, and is widespread from the sub-arctic to the tropics. Approximately 40–41 species have been clearly domesticated in the Fabaceae, the most of any plant family (Harlan, 1992; Hammer and Khoshbakht, 2015). The dehiscent legume fruit type is a key unifying feature of the Fabaceae family and occurs in nearly all wild members of the family with above-ground fruit. Selection against pod dehiscence has occurred in parallel in dozens of domesticated legume species. This presents a set of replicated natural experiments on the molecular basis of parallel evolution, and also has major implications for crop improvement. Major clades and a selection of representative genera are displayed. Figure from Gepts et al. (2005) with minor nomenclature changes based on LPWG (2017). All nodes other than those separating the genistoids, the aeschynomenoid/dalbergioid clade, and the core papilionoids received Bayesian posterior probabilities of 1.0 according to Wojciechowski et al. (2004) and LPWG (2017)
The Fabaceae are the second most economically important family of flowering plants after the Poaceae (LPWG, 2017). The pervasive utility of the Fabaceae in agriculture is partly due to their associations with nitrogen-fixing bacteria, which make them well-suited to complement nitrogen-hungry crops such as cereals and for cultivation in some nutritionally depleted soils. The fixed nitrogen is used to produce protein-rich seeds. In addition, these seeds have amino acid profiles that make up for the essential amino acid deficiencies of other crops (Gepts and Bliss, 1984). Members of the Fabaceae grown for these dry seeds are known as grain legumes or pulses to distinguish them from other members grown as vegetables, forage, or for other uses. Approximately 40–41 species in the Fabaceae have been domesticated as human food crops, which is more than any other plant family (Harlan, 1992; Hammer and Khoshbakht, 2015). The value of domestication for studying evolutionary biology was central to the arguments of Darwin (1859), and the strong parallel selection that has occurred during legume domestication is a highly replicated natural experiment in biological evolution (Bitocchi et al., 2017). Selection for the indehiscent trait has occurred in parallel in the Fabaceae family several dozen times, offering a unique opportunity to study the molecular basis of parallel evolution.
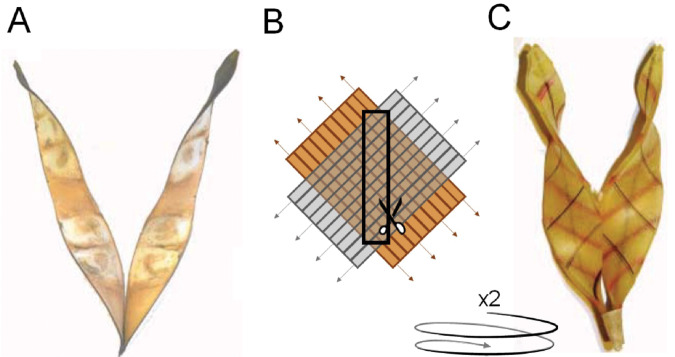
At maturity, wild legumes disperse their seeds through pod shattering, which is the result of interacting forces between several structures (Figure 2). As pods dry, differences in moisture content lead to differential contraction between pod wall layers (Buckovic, 1952; Armon et al., 2011). The wall fiber layers, which are oriented at an oblique angle through the pod, create tension that pulls at the sutures from both sides in a plane perpendicular to that of the fiber axis. In the legume Bauhinia variegata, pod walls contain two layers that contract in perpendicular directions during the drying process. This contraction leads to the helical coiling of each wall. The torsional force associated with this coiling increases as the pods become drier, applying strain where the two halves of the pod meet at the sutures. Mechanical experiments have shown that the coiling pattern of legume pod valves can be closely replicated mechanically using contractile materials that mimic legume pod structure (Armon et al., 2011; Figure 3). The coiling of pod walls also has a strong positive relationship with the thickness of the wall fiber layer in domesticated legumes (Takahashi et al., 2020). Therefore, increased deposition of wall fiber leads to yield losses in two ways: by promoting pod shattering and by competing with seeds for photosynthate (Assefa et al., 2013).
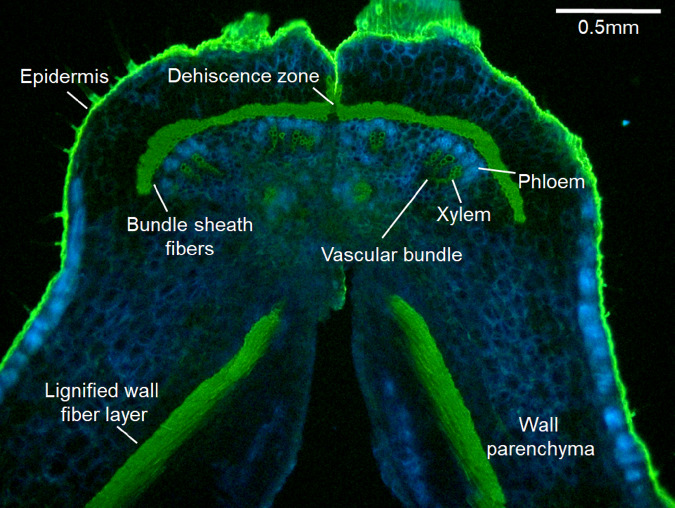
Figure 2.
A detailed view of the anatomy of pod sutures, transverse section. Pod sutures include vascular bundles with xylem and phloem; on the outer side of these is a layer of fibers called the bundle sheath. Pod dehiscence occurs along the plane of symmetry, through the dehiscence zone. This occurs when tension caused by the contraction of wall fibers overcomes the load limit of the dehiscence zone. Pod of Phaseolus acutifolius (tepary bean) stained with 0.01% auramine and 0.007% calcofluor. Figure credit: T. Parker and S. Lo.
Figure 3.
Mechanical representation of pod wall patterns and forces associated with legume shattering. (A) Dehisced pods of Bauhinia variegata. Pod dehiscence in legumes has long been hypothesized to result from differential contraction between perpendicular and obliquely oriented pod wall layers. (B) To replicate this, two sheets are created that can expand in only one axis. These sheets are glued together so that their expansion axes are perpendicular. A central section at an oblique angle is removed and the process is repeated a second time. (C) When allowed to contract, the sheets display the same twisting pattern as legume pods. Figure adapted with permission from Armon et al. (2011).
The medial portions of legume pod sutures contain several forms of weak cells, including a nonlignified abscission layer that extends into the vascular bundle sheath and a dehiscence zone with cells that lack secondary cell wall thickening (Figures 2 and 4). In wild legumes, the sutures are not strong enough to withstand the tension produced by the pod walls, and violent dehiscence occurs. This explosive breaking is known as pod shattering. Ballistic seed dispersal in wild legumes routinely spreads seeds over a 1–5 m radius (Lee, 1984; Malo, 2004; Ambrose and Ellis, 2008).
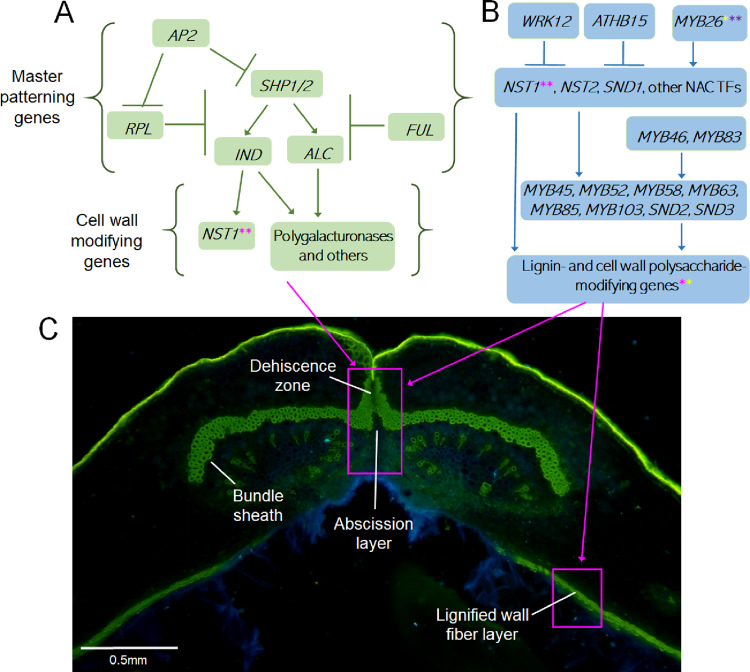
Figure 4.
Genetic models of fruit patterning and cell wall development. (A) In Arabidopsis, pod development in the area around the sutures is controlled by several master patterning genes and their downstream cell-wall-modifying genes. Homologs of the Arabidopsis fruit patterning regulators have frequently been used as candidate genes in dehiscence studies, but so far little experimental evidence exists for their roles in grain legume domestication. Gene abbreviations: see text. (B) A detailed view of the cell-wall-modifying pathway in Arabidopsis, which includes homologs of several candidates for legume shattering. Asterisks indicate genes with relatively well-characterized roles in pod shattering in Glycine max (red asterisks; Dong et al., 2014; Funatsuki et al., 2014; Zhang and Singh, 2020), P. vulgaris (yellow asterisks; Rau et al., 2019; Parker et al., 2020a, 2020b), and Vigna spp. (purple asterisks; Lo et al., 2018; Takahashi et al., 2020). (C) Dehiscence-related structures in V. unguiculata ventral sutures. In wild-types, the lignified pod wall fiber layers pull at the suture from both sides. If the dehiscence zone lacks the structural integrity to withstand the tension, dehiscence occurs along the abscission layer and the central part of the bundle sheath. Pods stained with 0.01% auramine and 0.007% calcofluor. Lignified cell walls and the hydrophobic cuticle are stained green, nonlignified cell walls are blue. (C) image credit: T Parker and S. Lo.
Domestication-related reductions in pod shattering have occurred by modifying the tension imposed by wall fibers and the strength of the sutures. These transitions have followed strongly parallel trajectories in terms of both microscopic and macroscopic pod structure (Table 1). This is an example of a Vavilovian homologous series (Vavilov, 1922), in which a highly parallel range of phenotypes has been selected in a group of related but independently selected organisms. Unraveling the genetic and biochemical nature of these mutations is a rapidly evolving field. Rau et al. (2019) proposed that nonorthologous mechanisms were responsible for the loss of pod shattering in legumes, accurately reflecting the state of research at the time. Since then, an increasing body of evidence suggests that homologous genes can often govern variation in this trait between species (Di Vittori et al., 2020; Parker et al., 2020a, 2020b; Takahashi et al., 2020; Zhang and Singh 2020), although several genes and mechanisms are responsible for this trait (Lenser and Theißen, 2013).
Table 1.
Pod traits as a Vavilovian homologous series in domesticated legumes
| Anatomical state | Shattering phenotype | Example species | References |
|---|---|---|---|
| Wild-type pods | Full shattering | All with above-ground pods and extant wild relatives | Ogutcen et al. (2018), Di Vittori et al. (2019) |
| Reduced twisting force of pod wall (disrupted fiber orientation, biochemistry, etc.) | Reduced shattering | C. arietinum, G. max, L. angustifolius, L. digitatus, L. sativus, P. vulgaris, P. sativum, V. sativa, V. angularis, V. radiata, V. stipulacea, V. umbellata | Gladstones (1967), Koinange et al. (1996), Isemura et al. (2010, 2012), Talukdar (2011), Funatsuki et al. (2014), Murgia et al. (2017), Dong et al. (2017a, 2017b), Takahashi et al. (2019, 2020), Aguilar-Benitez (2020) |
| Strengthening of the dehiscence zone | Reduced shattering | G. max, L. angustifolius, L. luteus, L. digitatus, V. sativa, V. stipulacea | Gladstones (1967), Dong et al. (2014, 2017a), Takahashi et al. (2019) |
| The absence of pod wall fiber | Loss of shattering, pod nontwisting, dehiscence not leading to seed release | P. vulgaris, P. sativum, V. villosa, V. Unguiculata | Emerson (1904), White (1917), Blixt (1978), Koinange et al. (1996), Myers et al. (2001), Murgia et al. (2017), Kissing Kucek et al. (2020) |
| Major reduction in suture fiber | Loss of shattering, string formation often temperature-dependent | P. vulgaris, P. sativum | Emerson (1904), Wellensiek (1971), McGee and Baggett (1992), Koinange et al. (1996), Hagerty et al. (2016) |
Different domesticated legume populations show strong variation in shattering. In addition, this shattering is greatly exacerbated by environmental dryness (Zhang et al., 2018). In soybean (Glycine max), typical yield losses to pod shattering range from 53 to 319 kg·ha-1, with average losses of over 100 kg·ha-1 (Philbrook and Oplinger, 1989; Tukamuhabwa et al., 2002). In arid climates, yield losses in soybean can be as high as 50–100% (Bhor et al., 2014). Caviness (1965) evaluated the critical moisture content that leads to pod shattering across one wild soybean and three domesticated types. The wild soybean shattered rapidly at 60% relative humidity. One domesticated type (cv. “Rokusun”) also shattered at this humidity, although more slowly than the wild-type. A 30% relative humidity was sufficient to cause shattering in cv. “Ogden”, but cv. “Lee” required prolonged exposure to 15% relative humidity. This interaction between genotype and environment is paralleled throughout many legume crops. For example, Anderson (1955) determined that birdsfoot trefoil (Lotus corniculatus) pods shattered readily at a relative humidity under 40%, leading to yield losses ranging from 5% to 71%. Understanding the genetic and molecular basis of shattering resistance and its environmental dependency will be important for minimizing these yield losses. Climate change is predicted to increase average global aridity (Sherwood and Fu, 2014), thereby exacerbating shattering in susceptible varieties. Unless this can be mitigated by introgressing dehiscence-resistance alleles into these varieties, yield losses to pod shattering will increase.
To date, the genetic basis of pod shattering resistance has been only partially resolved, and plant biologists are rapidly gaining the tools needed to study and improve this trait in many species. A thorough understanding of the genetic and environmental control of pod shattering will be essential for attaining maximum yields in this important crop family. In this review, we summarize the available information on the genetics of pod shattering in model systems such as Arabidopsis thaliana (Arabidopsis), as well as major crop grain legumes.
Arabidopsis
Pod shattering has been studied in several legume and nonlegume species. The first and best studied of these is Arabidopsis, a member of the Brassicaceae. While members of the Fabaceae have a single carpel per fruit, members of the Brassicaceae produce siliques with two longitudinally fused carpels. These siliques show certain characteristics similar to those of legumes, including a lignified endocarp layer in the pod wall, a lignified layer at the suture, and a separation layer immediately adjacent to the lignification layer along which dehiscence occurs. The separation layer is analogous to the dehiscence zone of legumes. Like legumes, the separation of Arabidopsis fruit occurs due to torsion applied by the endocarp fibers on the suture. The identity of each of these structures is regulated by several genes, including APETALA2 (Jofuku et al., 1994), SHATTERPROOF1/2 (Liljegren et al., 2000), INDEHISCENT (IND) (Liljegren et al., 2004), ALCATRAZ (ALC) (Rajani and Sundaresan, 2001), FRUITFULL (Gu et al., 1998), and REPLUMLESS (Roeder et al., 2003), which interact to produce each component (Figure 4A). Several genes, including IND and ALC, are the products of gene duplications in the Brassicaceae, implying that their roles in legumes may be considerably different (Ballester and Ferrándiz, 2017). These master regulators or patterning genes specify the existence of major pod structures and tissues, with major changes in fruit patterning observed in mutants of these genes. Downstream of these genes, several transcription factors control cell wall biosynthesis genes, ultimately regulating the quantity and biochemical qualities of the cell walls. These downstream cell-wall-modifying genes include NAC SECONDARY THICKENING1 (NST1), NST2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 (Mitsuda et al., 2005; Ogawa et al., 2009). In Arabidopsis, MYB26, NST1/2, and several other transcription factors interact to activate secondary cell wall biosynthesis and lignification, leading to the dehiscence of anthers (Yang and Wang, 2016; Yang et al., 2017). Many elements of this pathway, including NST1/2, are widely expressed in lignifying tissues in Arabidopsis (Figure 4B), including siliques. These structures are related to those relevant for legume pod shattering (Dong and Wang 2015; Figure 4C).
Basal grain legumes
Domesticates in the early-diverging lineages of the legume subfamily Papilionoideae include peanut (Arachis hypogea) and lupins (Lupinus sp.; LPWG, 2017; Figure 5). The fruit of peanut is geocarpous, and a reduction in pod shattering was not involved in the domestication of this species. In contrast, reduced pod shattering has been very important for lupin domestication.
Figure 5.
A schematic of relationships between selected Papilionoid species that underwent a reduction in shattering during domestication. Most domesticated grain legumes fall into either the warm-season clade (Millettioids and Phaseoloids) or the cool-season clade (Hologalegina), with a few basal domesticates including Lupinus spp. and Arachis hypogea (not shown). Most cool-season grain legumes are of Near Eastern origin, while the majority of warm-season clade domesticates are from the tropics of Africa, the Americas, and Asia. Figure developed from a 95% majority-rule Bayesian consensus tree based on 81 plastid-encoded peptides, adapted from LPWG 2017 (CC-BY 4.0), with posterior probabilities of all nodes except the Lens/Vicia/Pisum divergences supported with Bayesian posterior probabilities of 1.0 by Wojciechowski et al. (2004) and LPWG (2017). Source of plant photos: T. Parker and S. Lo.
Lupinus spp
The lupin genus (Lupinus) is relatively large, with approximately 280 species (Wolko et al., 2011). Four lupin species have been domesticated: the two ancient domesticates white lupin (Lupinus albus) of the Mediterranean, tarwi (Lupinus mutabilis Sweet) of the Andes, as two European species domesticated since the 19th century, annual yellow lupin (Lupinus luteus) and narrow-leaf lupin (Lupinus angustifolius). A reduction in pod dehiscence has occurred in all of these species, which has often been highly parallel (Wolko et al., 2011; Atchison et al., 2016; Gresta et al., 2017).
In L. luteus, von Sengbusch (1938) identified the recessive mutation invulnerabilis, which prevents pod shattering. Varieties with this mutation lack a dehiscence zone in the pod suture and, instead, have thickly lignified cells through the vascular sheath (von Sengbusch, 1938). This mutation was subsequently used widely, as many varieties of L. luteus have included this allele (Wolko et al., 2011). The resistance to shattering provided by invulnerabilis is sufficient for the relatively humid climates of northern Europe but does not provide strong resistance in the comparatively arid climates of the Mediterranean or western Australia (Wolko et al., 2011). Recent studies suggest that a second pod shattering-related allele may exist in the yellow lupin population (Iqbal et al., 2020), although this allele has not yet been mapped or described in detail. Due to the incomplete resistance to pod shattering provided by invulnerabilis, the pyramiding of multiple shattering resistance alleles is required in L. luteus.
Despite its relatively recent origin, L. angustifolius is currently the most widely cultivated lupin species (Gresta et al., 2017). Through mutant screens, Gladstones (1967) identified two distinct mutations that prevent shattering in this species. The first of these, lentus, disrupts the parallel orientation of the wall fibers of the pod endocarp, reducing the torsion forces on the dehiscence zone. Under semi-arid conditions, pods with the lentus mutation alone still dehisce but do so more slowly than the wild-type. This dehiscence is often not forceful enough to detach seeds from the pod. The lentus mutation is also associated with a change in pod color to a subtle purple. The gene model Lup018336 has been proposed as a candidate for lentus based on eQTL mapping (Plewiński et al., 2019). Lup018336 encodes a protein carrying a DUF1218 domain that is closely related to proteins that organize foliar symmetry and modify pod fiber deposition in Arabidopsis. This gene is downregulated in nonshattering types, while the neighboring gene model Lup018348 is upregulated in these plants. Lup018348 is a member of the MATE family; some MATE family proteins regulate anthocyanin and proanthocyanidin transport and flavonoid metabolism. The tight linkage of these genes, along with their expression patterns, may explain the strong relationship between purple pod color and the loss of shattering due to lentus (Plewiński et al., 2019).
The second mutation identified in L. angustifolius by Gladstones, named tardus, leads to the strengthening of the normal dehiscence zone at the sutures in a manner that closely parallels invulnerabilis of L. luteus. Individuals with this mutation display a fusion of the two halves of the vascular sheath fiber bundles at the sutures. If dehiscence does occur in these individuals, it often occurs outside of the lignified medial portions of the pod sutures. Occasionally, this dehiscence occurs along the obliquely oriented endocarp fibers, which still produce considerable torsion in tardus mutants that lack the lentus mutation. The gene model Lup002448 has been suggested as a candidate for tardus based on eQTL mapping. Lup002448 encodes a G family ATP-binding ABC transporter; some members of this family regulate Arabidopsis fiber development through the differential transport of monolignols (Plewiński et al., 2019). Maximum resistance to pod shattering can be achieved in L. angustifolius by combining the lentus and tardus mutations. Double-mutant individuals produce pods with very weak pod torsion and highly strengthened sutures, leading to a nearly complete loss of pod shattering (Gladstones, 1967).
Gladstones (1967) also investigated the pod shattering of a less widely cultivated species of lupin, L. digitatus. In a dramatic example of parallel evolution, two mutants were found with striking similarity to those of L. angustifolius. The conjunctus mutation leads to strengthening of the dehiscence zone like tardus and invulnerabilis, and single mutants often show transverse cracking in the pod walls. The macer mutation leads to a major reduction in wall fiber deposition and pod torsion, like lentus. Resistance to pod shattering is widespread in the ancient domesticates L. albus (Kazimierski, 1964) and L. mutabilis (Wolko et al., 2011). This resistance is due to changes in pod structure that closely match those of macer mutants in L. digitatus (Gladstones, 1967). The evolution of similar mechanisms to resist pod shattering in Lupinus is a striking example of phenotypic parallelism and reflects trends seen in the legume family in general.
Warm season legumes
Soybean
Soybean is the most widely produced grain legume globally [Food and Agriculture Organization of the United Nations {FAOSTAT}, 2020]. Pod dehiscence has been better studied in this species than in any other legume, and major genes with well-characterized effects on the process have been identified. Bailey et al. (1997) identified a major pod dehiscence QTL (quantitative trait locus), which was fine-mapped to chromosome Gm16 of the soybean genome (Funatsuki et al., 2006; Kang et al., 2009; Yamada et al., 2009). The underlying gene, known as POD DEHISCENCE 1 (PDH1), includes a premature stop codon in shattering-resistant types of soybean (Funatsuki et al., 2014). Genetic complementation has shown that the wild-type variant is sufficient to cause shattering in a fully shattering-resistant genetic background (Funatsuki et al., 2014). Allele frequencies at this gene are strongly related to the ecogeographic origins of landraces (Funatsuki et al., 2014; Bandillo et al., 2017; Zhang and Singh, 2020). Despite this, the gene is not associated with any known anatomical variation in pods, indicating that it may influence pod biochemistry or composition (Suzuki et al., 2009). PDH1 encodes a dirigent-type protein that guides the radical coupling process during the polymerization of monolignols. The gene is highly expressed in the pod endocarp layer; plants with the mutant allele display highly reduced pod twisting (Funatsuki et al., 2014). The effect of PDH1 on pod dehiscence is generally larger than that of any other gene (Bandillo et al., 2017; Hu et al., 2019; Zhang and Singh, 2020). The strong effect of PDH1 may be responsible for its extremely low allele frequency in wild populations, in contrast to other soybean shattering resistance alleles (Zhang and Singh, 2020). Molecular markers have recently been developed to introgress the useful allele into shattering varieties (Miranda et al., 2019).
A gain-of-function mutation of the No Apical Meristem (NAM), Arabidopsis thaliana Activating Factor (ATAF)1/2, and Cup-shaped Cotyledon (CUC)2 (NAC) family transcription factor gene SHATTERING1-5 (SHAT1-5) is also related to a reduction in soybean pod dehiscence (Dong et al., 2014). Mutant varieties have a 20-bp deletion in a repressor element upstream of the gene. This leads to 15-fold increases in the expression of this gene in the dehiscence zone, strengthening cell walls in the area. The mutant allele is associated with a 116-kb selective sweep around the gene (Dong et al., 2014). Despite this, Zhang and Singh (2020) determined that the overall effect of the mutant allele may be small compared to that of other loci. The wild-type allele still predominates in Asian soybean landraces and cultivars in the United States.
Zhang and Singh (2020) recently used genome-wide association studies to identify another gene, NST1A (Glyma.07G050600), with a potential role in soybean pod shattering. NST1A is a NAC family transcription factor and a paralog of SHAT1-5. The authors identified an indel in its coding sequence leading to a premature stop codon. Paradoxically, this may lead to a gain-of-function for the gene, like the shattering-resistant allele of the paralogous SHAT1-5 (Zhang and Singh, 2020). Premature stop codons have been documented to cause gain-of-function effects in related NAC-family transcription factors in Arabidopsis due to changes in protein localization (Li et al., 2011). Epistatic analyses showed that NST1A works with PDH1 to provide durable resistance to pod shattering. The combination of the paralogs NST1A and SHAT1-5 with PDH1 to increase suture strength and reduce wall tension parallels patterns found in many other legumes.
Another shattering-associated candidate gene, Glyma09g06290, was recently described by Hu et al. (2019). Glyma09g06290 encodes a basic helix-loop-helix protein and is a relative of the Arabidopsis genes IND and ALC. This gene is highly expressed in developing pods, and molecular markers have been developed to assist in its introgression (Hu et al., 2019).
The soybean lineage underwent a whole genome duplication event approximately 13 million years ago, since its divergence from all other domesticated pulses. This duplication was followed by the divergence of chromosomes into an allotetraploid with disomic inheritance, in which paralogous gene pairs were retained (Schmutz et al., 2010). The disomic inheritance has allowed some recessive, loss-of-function alleles to be selected, as illustrated by pdh1 for pod indehiscence (Funatsuki et al., 2014) and dt1 for determinacy (Tian et al., 2010). Nevertheless, the allotetraploidization may have affected the genetic inheritance of pod shattering, as genes with redundant paralogs may complement loss-of-function alleles. The genes related to reduced pod shattering in soybean may therefore not be fully representative of those found in other species.
Common bean
Common bean (Phaseolus vulgaris) is the primary grain legume used for direct human consumption (Gepts et al., 2008; Singh , 2013). This species has a variety of uses and was independently domesticated in Middle America and the Andes (Gepts et al., 1986; Kwak and Gepts, 2009; Bitocchi et al., 2013; Ariani et al., 2018). Each domesticated gene pool is divided into several ecogeographic races, each of which is adapted to particular environments (Singh et al., 1991; Kwak and Gepts, 2009). Numerous genes have major effects on pod dehiscence in common bean.
Domesticated common beans are divided into two major commercial types: dry beans and snap beans. Dry bean varieties must be sufficiently resistant to pod shattering to maintain high yields while maintaining the ability to open and release seeds at harvest. Snap beans are grown for their edible green pods that are used as vegetables. These types of beans have been bred to produce very little fiber in the pod sutures (also known as pod strings) and pod walls; as a consequence, they possess extreme resistance to pod shattering. Snap beans are descended from dry beans and therefore harbor the shattering resistance loci of dry beans as well as unique snap bean loci. The diverse uses, repeated domestications, and wide climatic adaptations of common beans have led to a series of pod traits that are among the most diverse in the legume family.
Murgia et al. (2017) evaluated phenotypic variation in pod shattering across a spectrum of common bean pod phenotypes. Using a population of backcrossed lines ultimately derived from a cross between Midas (stringless snap bean, Andean origin) and G12873 (Middle American wild bean from Morelos state in Mexico) (Koinange et al., 1996), the authors developed a systematic framework to analyze each component of the variation associated with pod shattering in the species. They found that stringless and fully indehiscent individuals had significantly lower levels of fiber, lignin, cellulose, and hemicellulose than those with dehiscence. In their sample, the degree of pod twisting was only weakly related to pod shattering.
In dry beans of the Middle American gene pool, Parker et al. (2020a) identified a major locus (PvPdh1) associated with strong shattering resistance. QTL mapping determined that PvPdh1 (Phvul.003G252100), the common bean ortholog of GmPDH1, showed complete co-segregation with the pod shattering phenotype in a Middle American recombinant inbred population (n = 226; Berny Mier y Teran et al., 2019). A genome-wide association study of 278 diverse varieties of Middle American beans (Moghaddam et al., 2016) further validated this result, as the most significant single-nucleotide polymorphism (SNP) from this analysis was located 5.7 kb from PvPdh1. Sanger sequencing identified a point mutation in a highly conserved active site of PvPdh1. The wild-type SNP encodes an amino acid that has been strictly conserved in the protein family for 400 million years, highlighting its functional importance. The resistance allele is found in members of the race Durango, which are native to highland, semi-arid regions with relatively low precipitation in northern Mexico. Race Mesoamerica types, which are native to lowland, higher-humidity regions, typically have the wild-type susceptible allele. Market classes in which the shattering-resistant PvPdh1 allele predominates have the lowest levels of shattering found in dry beans (Parker et al., 2020b). This allele is associated with a selection sweep and a reduction in pod twisting, and markers have been developed to facilitate its introgression (Parker et al., 2020b).
In dry beans of the Andean gene pool, several major loci control resistance to pod shattering. Using a diverse assemblage of 208 Andean dry bean varieties (Cichy et al., 2015), loci on chromosomes Pv03, Pv05, Pv08, and Pv09 were all found to be strongly associated with pod shattering (Parker et al., 2020a). The Pv05 locus colocalizes to a region identified in parallel in a snap bean × wild bean population (Rau et al., 2019, explained in the next paragraph). Since snap beans are derived from domesticated dry beans, the studies likely identified the same gene on Pv05. The loci on Pv03, Pv08, and Pv09 may be related to cell-wall-modifying transcription factors and enzymes, such as MYB transcription factors, cellulose synthases, or polygalacturonases (Parker et al., 2020a). Close homologs of the master regulator genes of Arabidopsis carpels are absent in these regions. The Pv09 locus includes Phvul.009G238800, the common bean ortholog of Glyma09g06290, which has been implicated in soybean shattering (Hu et al., 2019).
Using a population derived from the Andean snap bean cultivar “Midas” and the wild bean accession G12873 (Koinange et al., 1996), Rau et al. (2019) identified a large-effect QTL on chromosome Pv05 in the immediate vicinity of PvMYB26. Smaller-effect QTLs were found on chromosomes Pv04 and Pv09. PvMYB26 is more highly expressed in shattering individuals than in nonshattering types at 5–7 days after pod set (Di Vittori et al., 2020). At 9–11 days after pod set, this expression pattern is reversed. In Arabidopsis, MYB26 is a direct regulator of NST1, homologs of which control pod shattering in soybean as SHAT1-5 and NST1A. A mounting body of evidence suggests that closely related MYB26 orthologs regulate pod shattering in the genus Vigna (Suanum et al., 2016; Lo et al., 2018; Takahashi et al., 2020).
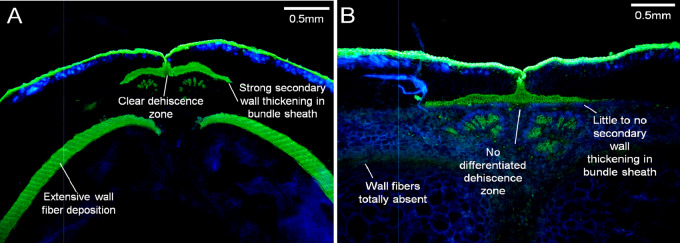
The study of pod strings and wall fibers, which are greatly reduced in snap beans, predates the study of the other pod traits of common bean. Most modern snap bean varieties have no wall fiber deposition and dramatically reduced secondary cell wall thickening in the vascular sheath at the pod sutures. In dry beans, this fiber bundle can be removed from the pod and is referred to as the pod string. These snap bean pod character states represent some of the widest divergences from wild-types that occur among domesticated legumes (Figure 6).
Figure 6.
Extremes of pod fiber development in common bean. The range in pod fiber between (A) wild-type and (B) stringless accessions of P. vulgaris is among the greatest variation found in any species of the legume family. Wild-type accessions of P. vulgaris and most other species have strong wall fiber deposition, strong secondary wall thickening in the bundle sheath, and a clear dehiscence zone without secondary thickening is present. In stringless snap bean varieties, wall fiber may be totally absent, little to no secondary cell wall thickening occurs in the bundle sheath, and the dehiscence zone is not well differentiated. Pods stained with 0.01% auramine and 0.007% calcofluor. Figure credit: T. Parker and S. Lo.
In the 1880s, Calvin Keeney identified the first known stringless beans through mutant screens of the variety “Refugee wax” (Barnes, 1910). Emerson (1904) applied the recently re-discovered principles of Gregor Mendel to evaluate this trait genetically. He noted that the stringless character was dominant in crosses between certain parents, while in others it was incompletely dominant or recessive. Stringless F1 plants produced F2 populations with a typical 3:1 ratio of stringless: stringy types. This large-effect and largely qualitative gene was later named Stringless (St) by Prakken (1934), a name that has subsequently been used extensively. By contrast, the segregation pattern of F2 populations derived from stringy F1 plants could not be explained by any Mendelian pattern. Wellensiek (1922) confirmed the dominance of the stringless character and its single-gene inheritance. Joosten (1927) continued working with Wellensiek’s populations and was the first to analyze pod strings quantitatively. He recognized that some amount of string deposition is found in all types, including those described as stringless, and that this trait could be influenced by environmental factors. Currence (1930) proposed a two-gene model for the inheritance of pod strings and named the genes T and S. In his model, dominant S prevents the production of pod strings, while T can epistatically overcome this to create pod strings. This two-gene model is like that of Drijfhout (1970, 1978). Drijfhout’s model proposes St as a dominant allele leading to the stringless character and Ts for temperature-sensitive partial suture strings. This model only differs from the model of Currence (1930) in terms of the temperature-sensitive effect of Ts and its incomplete string formation. Some of the confusing patterns related to pod string formation could be explained by Drijfhout’s digenic and environmentally influenced model.
Prakken (1934) thoroughly evaluated the quantitative genetic variation in pod fiber development. Like Joosten, he used a 1–10 scale to categorize pod string strength and correlated these to cell types in the bundle sheaths (see Figure 6 for extremes of this range). All pods in his evaluations developed lignified vascular bundle sheaths of roughly the same size, but the ratios between cell types within the bundle sheaths varied. The cellular composition of vascular bundle sheaths was correlated with string strength. In varieties with weak pod strings, vascular sheaths contained few strong fiber cells, which included lignified primary and secondary cell walls. Instead, the vascular sheaths consisted primarily of relatively weak cells with only primary cell walls, which Prakken referred to as wood cells. In types with a strong string, fiber cells predominated through nearly the entire bundle sheath, and the weaker wood cells were found only in a narrow strip at the dehiscence zone. A wide range of partial-string patterns existed between these extremes. From the perspective of modern molecular biology, the weak string appears to result of overspecification of the dehiscence zone outside of its typically restrained area in wild-type individuals.
Koinange et al. (1996) were the first to genetically map pod fiber traits in common bean. In their population, which was derived from the stringless snap bean cv. “Midas” and wild accession G12873, the locus for the existence of pod strings (St) mapped to linkage group Pv02 (Freyre et al., 1998). Pv02 includes the common bean homolog of INDEHISCENT (PvIND), but recombination is occurring between PvIND and the stringless trait (Gioia et al., 2013). Furthermore, no causal polymorphism could be identified in the gene or its promoter sequence (Gioia et al., 2013). The 7.8 cM of recombination space between St and PvIND identified by Gioia et al. indicates that the polymorphism should be several megabases from the gene, based on known recombination rates in the area (Schmutz et al., 2014). Despite this, Hagerty et al. (2016) reaffirmed that the causal polymorphism for St must be near PvIND, as the 500-kb region between their flanking markers includes the candidate gene. The approximate colocalization of PvIND and St is not compelling evidence that they are the same locus (i.e. that PvIND is the molecular basis of the phenotypic St locus). Further work is required to understand the inheritance and molecular basis of pod strings.
Using a population descended from cvs. “Minuette” and “OSU 5630,” Davis et al. (2006) mapped pod string formation to chromosome Pv06, even though both parents are considered stringless. This surprising result is consistent with the multigene models suggested by Currence (1930) and Drijfhout (1970, 1978). Pv06 was subsequently shown to include the common bean homologs of SHATTERPROOF1/2 (Nanni et al., 2011). These genes specify suture identity in Arabidopsis, although no candidate polymorphisms could be identified in their common bean equivalents. The genes related to pod strings on both Pv02 and Pv06 remain elusive.
Several studies have investigated the inheritance of pod wall fibers in common bean, with dramatically different results. These studies have typically proposed one major factor (Emerson, 1904; Koinange et al., 1996), along with secondary modifiers (Tschermak, 1901, 1902; Lamprecht, 1932; Prakken, 1934), that alter the thickness and extent of the fibers. Prakken (1934) used To to describe the gene controlling wall fibers. Whereas Prakken (1934) and Hagerty et al. (2016) determined that wall fiber and pod strings were unlinked, Koinange et al. (1996) found complete co-segregation between the two traits. Koinange et al. (1996) mapped both To and St to chromosome Pv02 in a Midas/G12873 population. Hagerty et al. (2016) identified a locus that pleiotropically modifies pod wall fiber and pod dimensions on chromosome Pv04. Both parents of their population produce limited wall fibers, suggesting that the QTL on chromosome Pv04 might represent one of the wall-fiber-modifying genes.
The reversion of pod fiber traits, including wall fibers and suture strings, is common in snap beans (Emerson, 1904) and occurs at a rate of approximately 0.5% to 2.25% (Hagerty et al., 2016). These revertants include chimeric plants with individual revertant branches (Prakken, 1934). The production of snap bean seeds requires manual verification that each plant has not reverted to the stringy character, which is labor-intensive and costly. The unusual properties of snap bean pod traits, including their frequent reversions, unusual segregation ratios, recalcitrance to mapping, and inconsistent interactions between alleles suggest that they could be governed by non-Mendelian genetic mechanisms. Whether transposons, epigenetic modifications, gametophytic selection, or other factors are involved in pod development in snap bean remains to be resolved.
Lima bean
Lima bean (Phaseolus lunatus), like common bean, was domesticated twice: once in Middle America and once in the Andes (Gutiérrez-Salgado et al., 1995; Motta-Aldana et al., 2010). Chacón-Sánchez and Martínez-Castillo (2017) identified strong divergence between wild and domesticated Middle American lima beans on chromosome 9 of the species. The lima bean homolog of AT5G60910 (FRUITFULL, or FUL) is found in this region. In Arabidopsis, FUL is associated with the specification of silique wall identity and therefore functions upstream of genes regulating wall fiber development and lignification. This chromosome is also syntenic to common bean chromosome 9, which is known to regulate shattering in that species (Rau et al., 2019; Parker et al., 2020a). The relationship between this lima bean locus and pod shattering is not yet well resolved.
More recently, Garcia et al. (2020) identified a difference in the expression level of the lima bean ortholog of PvPdh1 (PlPdh1) between wild and domesticated lima beans. In the Middle American wild-type accession G25230 (from Colima, Mexico), the expression level of this gene in full-length pods immediately before seed-fill was double that of the Middle American domesticated type G27455 (from Sucre, Colombia). This could be an example of parallel evolution among soybean, common bean, and lima bean, with three different types of mutations (premature stop codon, amino acid substitution, and change in gene transcription) responsible for the reduced function of this gene. The authors also found evidence that the lima bean homolog of ALC is differentially expressed between the wild and domesticated types. Despite this, the lima bean homolog of NST1, which functions downstream of ALC in the Arabidopsis pod development pathway, was not differentially expressed between the groups. If the lima bean homologs of ALC and FUL are responsible for regulating pod dehiscence in lima bean, they would represent relatively rare examples of master cell-fate specifying genes controlling pod shattering during legume domestication.
Cowpea and yardlong bean
Pod shattering in cowpea (Vigna unguiculata) is related to the strength and thickness of the pod fiber layer (Lush and Evans, 1981). The pod walls of dehiscent cowpea varieties have two adjacent but distinct fiber layers, including one on the inner surface of the pod walls and another immediately contiguous toward the outer surface of the pod.
Genetic studies have led to the identification of QTLs for pod shattering in cowpea using bi-parental and backcrossed mapping populations (Andargie et al., 2011; Suanum et al., 2016; Lo et al., 2018; Takahashi et al., 2020). In brief, Andargie et al. (2011) measured pod shattering based on pod fiber layer thickness and reported four QTLs (qps1, qps6.1, qps6.2, and qps10) controlling this trait. A recent study from Lo et al. (2018) identified two QTLs controlling the presence or absence of pod shattering (CPshat3 and CPshat5). None of these QTLs coincide with the ones reported by Andargie et al. (2011), while CPshat5 overlaps with the main pod shattering and pod fiber contents region reported in yardlong bean (Suanum et al., 2016) and common bean (Rau et al., 2019; Di Vittori et al., 2020). A cowpea reference genome was recently developed (Lonardi et al., 2019), which allowed Lo et al. (2018) to investigate the candidate genes underlying CPshat3 and CPshat5. Among these, a few stand out in the context of pod shattering and were reported as interesting candidates. Vigun03g306000 (VuNAC007) encodes a NAC domain transcription factor involved in secondary cell wall biosynthesis (Figure 4; Wang et al., 2011). Comparative sequence analyses revealed high protein sequence similarity between VuNAC007 and SHAT1-5, which affects pod shattering in soybean (see above). Vigun05g273500 (VuMYB26) encodes a MYB domain protein homologous to Arabidopsis MYB26. AtMYB26 acts upstream of the lignin biosynthesis pathway and is required for anther dehiscence (Figure 4; Yang and Wang, 2016; Yang et al., 2017).
In V. unguiculata subsp. sesquipedalis (yardlong bean), QTLs for pod shattering based on pod twists (qPdt1.1 and qPdt7.1) were identified by Suanum et al. (2016). Moreover, Suanum et al. reported co-localization of QTLs for pod fiber contents (cellulose, hemicellulose, and lignin) and pod shattering. Based on comparative genome analysis with adzuki bean, Suanum et al. suggested that the QTL region for pod fiber content and pod shattering in yardlong bean contains genes encoding the MYB transcription factor MYB83, which regulates cellulose and lignin biosynthesis. MYB83 functions immediately downstream of NST1 (Figure 4B). Pod fiber content is related to the existence of a pod wall fiber layer, which is absent in yardlong bean. Fine mapping together with whole-genome sequencing of wild cowpea revealed Vigun05g273500 (VuMYB26) as the key factor for the formation of this wall fiber (Takahashi et al., 2020). Furthermore, Takahashi et al. (2020) found that in individuals with a reduction or loss of wall fiber, VuMYB26 encodes a truncated protein due to a premature stop codon. Watcharatpong et al. (2020) recently narrowed down the same QTL region reported by Suanum et al. (2016) to identify candidate genes for pod fiber and pod shattering. Using the cowpea reference genome (Lonardi et al., 2019), Watcharatpong et al. (2020) reported VuMYB26, in parallel with Takahashi et al. (2020), and also identified Vigun05g266600 (VuBGLU12) as a potential candidate for pod fiber contents and shattering. VuBGLU12 encodes a beta-glucosidase predicted to modify the properties of cell wall material. Hence, VuNAC007, VuBGLU12, and VuMYB26 are strong candidates for further molecular investigation of pod shattering in cowpea.
Adzuki bean
In adzuki bean (Vigna angularis (Willd.) Ohwi & H. Ohashi), the loss of pod shattering has been associated with a reduction or loss of pod sclerenchyma (Takahashi et al., 2020). Early studies have reported QTLs controlling pod shattering based on the number of twists on the pod after opening (Isemura et al., 2007; Kaga et al., 2008). Both studies suggested that a single recessive gene controls the loss of pod shattering in domesticated adzuki bean, and both independently identified the same QTL region on chromosome 7. Takahashi et al. (2020) recently performed fine-mapping with backcross populations and narrowed down the QTL region to 4 kbp. A single gene encoding a MYB domain protein (VaMYB26) exists in this region. VaMYB26 is an ortholog of candidate genes for pod shattering in common bean (PvMYB26) and cowpea/yardlong bean (VuMYB26) (Lo et al., 2018; Rau et al., 2019; Di Vittori et al., 2020; Parker et al., 2020a; Watcharatpong et al., 2020; see above). Sequence comparison of VaMYB26 between wild and domesticated adzuki bean revealed a thymine insertion in the coding sequence of domesticated types, leading to a frameshift and premature stop codon. This stop codon causes the loss of 125 amino acids found in the wild-type protein. VaMYB26 regulates pod wall tension by causing a major reduction in wall fiber in mutant types. The repeated selection of MYB26 mutants in the Vigna–Phaseolus complex suggests that MYB26 orthologs are good targets for further characterization in other species.
Other Vigna spp
Like cowpea and adzuki bean, QTLs controlling pod shattering have been reported in mung bean (Vigna radiata R. Wilczek). Isemura et al. (2012) examined pod shattering based on the number of twists along the length of shattered pods and the percentage of shattered pods at several time points after harvesting. The authors reported two QTLs for number of twists and three co-located QTLs for shattering pods from each time point. Furthermore, Isemura et al. (2012) found that the QTL positions for shattering pods co-localized with one of the QTLs for number of twists. Further studies will be needed to fine-map these QTLs and to isolate the underlying genes.
Pod shattering has also been examined in rice bean (Vigna umbellata (Thunb.) Ohwi and H. Ohash). Isemura et al. (2010) reported one QTL controlling the number of twists in shattered pods. Yundaeng et al. (2019) measured pod shattering in moth bean (Vigna aconitifolia (Jacq.) Maréchal) based on the number of twists and the percentage of dehiscent pods (Yundaeng et al., 2019). The authors reported three QTLs for this trait, including co-localized QTLs for number of twists and percentage of dehiscent pods on chromosomes 1 and 7 of the species. Furthermore, Yundaeng et al. (2019) found that one of the QTLs appeared to be similar to those reported in mungbean and yardlong bean, whereas the main QTL is considered common to the ones reported in adzuki bean, mungbean, rice bean, and yardlong bean (discussed above). The identification of a single shattering-related QTL across all five species of domesticated Vigna studied (whether they originated in Africa (subgenus Vigna; Vaillancourt et al., 1993)) or in Asia (subgenus Ceratotropis; Tomooka et al., 2002; Javadi et al., 2011) strongly indicates that a common genetic basis for the trait exists in this genus. The independent evolution of similar characteristics has been achieved through selection at orthologous loci.
Takahashi et al. (2019) recently evaluated pod shattering in the biotic stress-resistant legume Vigna stipulacea using mutant populations. The authors identified mutant lines with reduced shattering and selected a mutant (rps1) with a total loss of pod shattering. The complete loss of the shattering habit in this mutant was related to the suppressed formation of the dehiscence zone between valves. The pods also lacked the coiling habit of wild-type individuals. Takahashi et al. suggested that the mutation in rps1 might be a gene involved in the SHAT1-5 pathway reported in soybean (Dong et al., 2014). At an anatomical level, this suture strengthening closely parallels the invulnerabilis, tardus, and conjunctus mutations selected in mutant screens of Lupinus in the past century (Gladstones, 1967).
Cool season legumes
Pea
Pea (Pisum sativum) is among the earliest domesticated plants and was a component of the original founder crop complex from the Near East (Harlan, 1992). Several genes are known to regulate pod shattering in pea. A major factor known as Dpo or Dpo1 (Marx, 1971; Blixt, 1972) has been mapped to linkage group III of the pea genome (Weeden, 2002). Mutations in Dpo1 are thought to have been independently selected in each of the two domesticated subspecies of pea, ssp. sativum and ssp. abyssinicum (Weeden, 2018). Dpo1 is a large-effect gene believed to be involved in the earliest reduction in dehiscence in pea, as the allele is polymorphic between wild-types and primitive landraces, but not between landraces and modern cultivars (Weeden, 2007). A cell-wall-modifying extensin gene co-located with Dpo1 is differentially expressed between shattering-susceptible and shattering-resistant pea varieties (Prokesova et al., 2016; Hradilova et al., 2017). Dpo1 is related to a reduction in pod wall twisting and tension strength (Waines, 1975). Dpo2 is located on LG VII and is responsible for a further post-domestication reduction in pod shattering. Green pod (Gp) was one of the original pea genes described by Gregor Mendel (Mendel, 1866; Reid and Ross, 2011). Gp or a gene co-located with it also has a large effect on pod dehiscence in pea and is even epistatic over the effect of dpo1 (Weeden, 2007). Whether this gene pleiotropically affects the structure of the pod wall has been disputed (e.g. Price, 1988; Weeden, 2007). Gp specifically affects plastid development in the pod mesocarp (Price, 1988), but the role of this process in pod dehiscence is currently unclear. Another mutation related to pod dehiscence in pea is Np. Np leads to the development of neoplasm (excessive and aberrant parenchyma in pods) in plants grown in the greenhouse. The role of this mutation in field settings is relatively minor.
The P, V, and N genes of pea control the existence of pod wall fibers and eliminate pod shattering (White, 1917; Blixt, 1978; Myers et al., 2001). P and V each individually reduce endocarp fiber deposition, and together they eliminate all wall fiber deposition (Lamprecht, 1948). This also eliminates shattering, although minor dehiscence along the suture may occur. N serves to reduce the thickness of the pod wall (Wellensiek, 1925).
Pisum sativum also includes stringless varieties. A mutant with this characteristic was first identified by Lamprecht (1938) as Sin but was subsequently lost (McGee and Baggett, 1992). A second mutant was identified by Wellensiek (1971) and called Sin-2. The trait shows some similarities to common bean stringlessness, including temperature sensitivity, unusual segregation ratios, and recalcitrance to mapping. Several of these traits are explained by the mutation’s pleiotropic reduction in pollen tube growth rate, which distorts segregation in progeny (McGee and Baggett, 1992). Despite this, the recessiveness and lack of reversion found in the stringless pea allele contrast strongly with that of common bean.
Lentil
The Pod indehiscence (Pi) gene is a primary regulator of pod shattering that was selected during the domestication of lentil (Lens culinaris Medikus). The recessive mutation that provides shattering resistance has been mapped to lentil linkage group IV (Ladizinsky, 1979a; Tahir et al., 1993; Tahir and Muehlbauer, 1994; Fratini et al., 2007). This large-effect gene is found in a syntenic region to that of Dpo1 of pea (Weeden et al., 1992, 2002), suggesting that homologous genes may be responsible for this trait. Many domesticated lentil varieties bearing this mutation nonetheless display pod shattering in the semi-arid conditions of the Middle East (Ladizinsky, 1979a; Erskine , 1985). More recently, at least two additional QTLs associated with pod dehiscence have been identified in lentil (Fratini et al., 2007), as was originally suggested by Ladizinsky (1979a). These QTLs could be important for minimizing yield losses in hot, dry environments where pod shattering continues to be problematic (Laghetti, 2008).
Chickpea
Unlike most pulses, chickpea (Cicer arietinum) only underwent a small reduction in pod shattering during domestication. The wild relative C. reticulatum has pods that are only weakly dehiscent (Ladizinsky, 1979b). In the wild, these pods sometimes remain intact until the entire pod abscises from the stem, after which the pod shatters to disperse seeds (Ladizinsky, 2014). These indehiscent pods represent a major pre-adaptation for domestication (Ladizinsky, 1979b; Abbo et al., 2003). Kazan et al. (1993) identified a single large-effect locus called Pin, which contributed to the comparatively small reduction in pod dehiscence during the domestication of chickpea. Ladizinsky (1979b) suggested that pod dehiscence is under oligogenic control in chickpea. This mode of control was supported by a recent study by Aguilar-Benitez et al. (2020). The authors identified several loci that contribute to pod dehiscence, including the chickpea ortholog of PDH1, making it the third PDH1 ortholog associated with pod shattering in the legume family, along with its orthologs in soybean and common bean (Funatsuki et al., 2014; Parker et al., 2020a, 2020b). The genetic basis of several other loci has not yet been resolved. Domesticated chickpeas continues to experience yield reductions due to pod shattering when left in the field for a prolonged period of time (Abbo et al., 2014), making these alleles useful for future crop improvement.
Grass pea
Talukdar (2011) identified and mapped two major mutations regulating pod dehiscence in grass pea (Lathyrus sativus). One gene includes two allelic mutations, pod1 and pod2, each of which leads to a major reduction in pod dehiscence and the complete elimination of pod twisting. This locus was mapped to linkage group LTr VII in a region syntenic to Dpo2 of P. sativum. The second locus, which includes the mutant allele pod3, is located on LTr IV and is not syntenic to any other dehiscence-related genes. The pod3 mutation reduces pod twisting, although to a lesser degree than pod1/pod2. Mutations at either gene lead to reduced amounts of total pod wall material, which improves the harvest index, as seen in other species (Talukdar, 2011; Assefa et al., 2013; Murgia et al., 2017; Takahashi et al., 2020).
Vicia spp
Faba bean (syn: fava bean, broad bean, or horse bean, Vicia faba) is one of the earliest domesticated legumes, but its origins remain obscure (Ladizinsky, 1975; Maxted , 1991; Caracuta, 2016). Wild forms of faba bean have not been identified in living populations. Archaeological evidence indicates that humans harvested wild faba beans 3,000 years before their domestication, but these populations are thought to be extinct today (Caracuta et al., 2015; Caracuta et al., 2016). Vicia faba has a different number of chromosomes (2n = 12) from its closest counterparts in the Narbonensis complex of the genus (2n = 14), preventing successful hybridization between V. faba and its wild relatives (Maalouf et al., 2019). This trait will make the identification of domestication-related loci in faba bean challenging. Nevertheless, domesticated faba beans show different levels of susceptibility to shattering, and breeding programs continue to seek improvement in this trait (Bishnoi et al., 2018).
Common vetch (Vicia sativa) is grown as a cover crop and for fodder globally. Shattering is a major issue for seed production in this crop, and shattering-susceptible varieties have the greatest potential to become problematic weeds for subsequent plantings (Abd El-Moneim, 1993). Paradoxically, Abd El-Moneim (1993) identified shattering-resistance plants in three wild-type populations of common vetch and transferred this trait to shattering-susceptible cultivated forms through backcrossing. Shattering resistance in crown vetch was determined to be controlled by the recessive allele ns.
Dong et al. (2017a) determined that shattering-resistant varieties of V. sativa lacked an abscission zone in the bundle sheath, leading to the formation of strong sutures. Gene expression analysis revealed that most differentially expressed genes between shattering phenotypic categories were related to cell wall modifications (Dong et al., 2017b). Shattering-resistant varieties also had fewer than half the number of twists as shattering-susceptible varieties. This difference resulted from changes in the cell walls of the lignified wall layer (e.g. Funatsuki et al., 2014; Takahashi, 2020), pointing to the possibility that multiple genes may be responsible for resistance to pod shattering in V. sativa. Dong et al. (2017a) also determined that the thickness of the external valve margin layer is also correlated with shattering, although the identity of this structure is not yet thoroughly understood.
The loss of pod valve twisting and the formation of shriveled corrugated pod walls around seeds at maturity have also been documented in the closely related plant hairy vetch (Vicia villosa Roth; Kissing Kucek et al., 2020). This finding provides evidence for the loss of wall fiber in this species in a manner that parallels the patterns found in P. vulgaris, P. sativum, and V. unguiculata.
Parallel evolution and molecular convergence
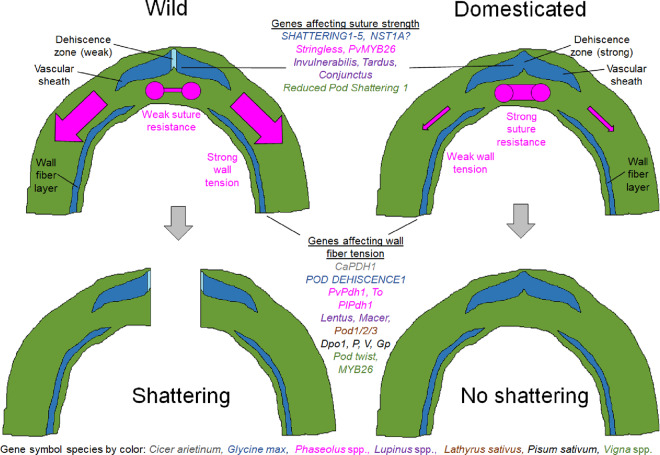
Pod shattering in legumes offers a highly replicated natural experiment to explore the molecular basis of parallel selection. Across the legume family, pod shattering is controlled by a mixture of orthologous and nonorthologous loci (Figure 7). Closely related taxa are typically more likely to share highly similar molecular means to the same phenotypic ends. Some of these patterns are particularly strong within genera. QTL mapping of the pod-shattering trait in five species of Vigna identified a single locus with an effect across all five species (Yundaeng et al., 2019). Fine mapping in V. angularis and V. unguiculata indicated that orthologs of MYB26 are likely responsible for this trait, which is related to shattering in the Andean gene pool of P. vulgaris. Different genes of the same pathway were targeted in Glycine max, including SHAT1-5 and NST1A, which are close paralogs. The whole-genome duplication of the soybean lineage since its divergence from Phaseolus and Vigna could have led to reduced orthology in the genetic control of shattering due to new genetic redundancy, subfunctionalization, or other changes. The soybean-shattering resistance gene PDH1 is orthologous to PvPdh1 in common bean. PlPDH1 has also been downregulated during lima bean evolution. In the cool season legumes, Dpo1 of P. sativum and Pi of L. culinaris are found in syntenic regions. Similarly, the pod1 and pod2 mutations of L. sativus are found in a locus syntenic with that of Dpo2 of P. sativum. Together, these findings indicate that several genes regulate variation in pod shattering in legumes and that these genes have often been selected repeatedly across species.
Figure 7.
Schematic diagrams of shattering-related pod mechanics in legumes. In wild types, shattering occurs because the tension of the pod walls (red arrows) exceeds the tensile capacity of the suture (horizontal red line). Mutations that reduce wall tension or increase suture strength lead to reduced shattering. Several well-known genes affecting pod shattering are indicated and color-coded by species.
Conversely, however, nonorthologous genes are also involved in pod shattering. The clearest example of this situation is the inheritance differences in pod shattering in the Andean versus Middle American gene pools of common bean. Although the two gene pools belong to the same species, different, nonhomologous genes for pod indehiscence were selected in the two gene pools (Parker et al., 2020a). A similar observation was made for the bush growth habit in common bean. Bush growth habits have been selected in both domesticated gene pools. In the Andean domesticated gene pool, the bush growth habit is based primarily on a determinate growth habit controlled by several loss-of-function mutations in the PvTFL1y gene (Kwak et al., 2012); other traits were selected to achieve a bush growth habit in the Middle American domesticated gene pool (Moghaddam et al., 2016). More generally, both Schmutz et al. (2014) and Rendón-Anaya et al. (2017) identified contrasting gene expression patterns in the Andean and Middle American gene pools, respectively.
The genetic bases controlling pod shattering and a bush growth habit in Andean versus Middle American common bean are quite distinct. This contradicts the idea that molecular convergence underlies phenotypic convergence in phylogenetically related taxa. Lenser and Theißen (2013) proposed five factors that promote molecular convergence. These include (1) the nodal positioning of transcription factors in gene networks such as flowering pathways; (2) simple metabolic pathways such as seed starch pathways; (3) minimal pleiotropic effects such as those of MYB-transcription factors; (4) selection on standing variation; and (5) phylogenetic similarity. More information is needed to ascertain to what extent each of these factors plays a role, if any, across crops and domestication traits.
Surprisingly, homologs of the major Arabidopsis fruit patterning genes appear to be of little significance for legume domestication (Figure 4A). A possible exception to this general trend is PvIND, the common bean homolog of INDEHISCENT, which maps near the St or Stringless locus, although it may not be causally responsible for variation in this trait. The master regulators of fruit patterning in Arabidopsis cause gross changes in phenotype, which may be more dramatic than what is ideal for grain legume production. For example, the stringless trait in common bean is a gross mutation with major changes in cell fate identity across several pod structures. This mutation, however, leads to morphological changes so dramatic that seeds are difficult to separate from pods. The stringless trait is therefore absent in all common bean varieties bred for dry bean production and is found only in snap beans grown as a vegetable. In general, selections for nonshattering in grain legumes have focused on downstream cell-wall-modifying genes (Figure 4B) rather than the major patterning regulators like those of Arabidopsis. The extent of conservation of pod patterning pathways between the Brassicaceae and the Fabaceae is also unclear.
Gene function and environmental effects
The exact roles of several major candidate shattering genes are still unresolved, in part due to experimental limitations in the Fabaceae. While in vitro culture is possible for some groups, especially those in the cool-season clade, legumes tend to be recalcitrant to tissue culture and regeneration, which limits the range of experimental approaches that can be used to evaluate them (Pratap et al., 2018). Genetic studies are therefore typically based on combinations of fine-mapping, sequencing of large populations, expression data, anatomical, and biochemical analyses, and in some cases transformation of species such as Arabidopsis with candidate legume genes (e.g. Kwak et al., 2012; Repinski et al., 2012). Genes identified based on low-resolution mapping or differential expression alone, for example, should therefore be seen as candidates in need of further validation. In some cases, the identification of candidate genes by multiple approaches has led to very strong support for specific genes or gene families, but their exact roles remain unclear.
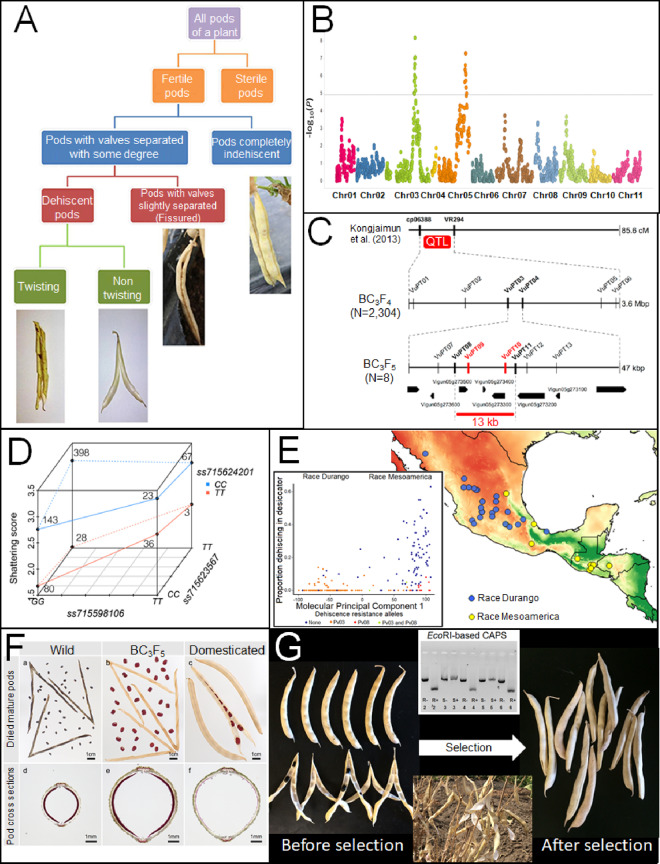
Members of the MYB26 gene clade appear to have been targeted by selection at least six times during the domestication of Vigna and Phaseolus, but the roles of these genes in pod development are not yet clear. Further work will be required to identify where these genes are expressed in pods and to relate them to mechanical models of pod dehiscence. Similarly, the relationship between PDH1 and pod dehiscence has not yet been fully explained. Dirigent genes such as PDH1 are known to affect molecular chirality during lignin synthesis, but the chirality of legume pod coiling can be fully explained by the differential contraction of wall layers. Furthermore, the two valves of a legume pod have opposite chirality, while the protein product of PDH1 guides the synthesis of only a single chiral isomer. The role of PDH1 in directly creating both chiralities would be very difficult to explain. Instead, it is likely that PDH1 and its orthologs play an indirect role in pod shattering, exerting their influence by changing the extent of cell contraction rather than its directionality. Linking the biochemical role of PDH1 with the mechanics of fruit shattering represents an important area for future study. Resolving these gaps in the current knowledge will require the use of a combination of approaches. These include improved study of the biochemistry and biophysics of pod fibers, precise methods for phenotyping that allow the roles of distinct genes to be separated, high-resolution mapping, interspecific and intergeneric comparisons, analysis of epistasis, evaluation of the relationships between alleles and the environment, and development of high-throughput methods for selection (Figure 8). These methods will be complemented by continued advances in genomics, which will continue to revolutionize a range of biological disciplines. These studies will have significant value for the applied sciences and will shed further light on the genetic basis of parallel evolution.
Figure 8.
Future directions for studying pod shattering in legumes. (A) Characterize the range of diversity within the target species. This allows for the precise separation of relevant factors during phenotyping. (B) Map and (C) Fine-map loci associated with each trait to find candidate genes and mutations. This will be facilitated by rapidly improving genomic tools across legumes and a better understanding of molecular pathways relevant to pod development. (D) Determine the epistatic relationship between shattering resistance alleles. (E) Evaluate patterns between shattering resistance, ecogeography, and alleles in diverse germplasm. (F) Correlate anatomical and mechanical patterns caused by each allele. Together, (A–F) can be used to build a global model for pod shattering within a species. (G) Use all available data to develop genotyping and phenotyping tools that will optimize selection for the nonshattering trait in the target environment. Images reproduced under CC-BY 4.0 license from Murgia et al. (2017) (A), Lo et al. (2018) (B), Takahashi et al. (2020) (C and G), Zhang and Singh (2020) (D), and with permission of the publisher from Parker et al. (2020a) (F) and Parker et al. (2020b) (G).
Recent studies have called into question the idea of single large-effect domestication genes that clearly delineate wild and domesticated populations (e.g. Zhang and Singh, 2020). Instead, there may be a range of loci and alleles that affect suitability for a particular environment, including the human-mediated environment. Crosses between crop varieties with different loci can lead to transgressive segregation, including improved adaptability to the cultivated environment or atavistic reversion to wild characters (Parker et al., 2020a). For many genes, the “wild” and “domesticated” alleles may be relatively common in both domesticated and wild populations but at different frequencies. In some cases, large-effect shattering resistance alleles do not even reach 50% frequency among domesticated landraces (Zhang and Singh, 2020). This could call into question the utility of genome-wide screening for domestication genes based solely on bioinformatic data (e.g. Schmutz et al., 2014). It could also fundamentally affect the way domestication genes are viewed in terms of crop evolution. For example, the distinction between domestication genes and improvement genes is becoming increasingly tenuous as our understanding of selection and adaptation to a broad range of cultivated environments increases.
Pod shattering has a strong relationship with the environment of origin. Several studies have demonstrated the strong relationship between the PDH1 allele in soybean and aridity (Funatsuki et al., 2014; Bandillo et al., 2017; Zhang and Singh, 2020). Because shattering is the result of pod dryness, selection against shattering alleles may have been strongest in arid regions. In common bean, the mutant allele of the orthologous PvPdh1 predominates in members of the ecogeographic race Durango, which is adapted to highland, semi-arid environments in northern Mexico (Singh et al., 1991). The wild-type allele of this gene is found in members of the lowland, humid-adapted race Mesoamerica. In cowpea, susceptibility to pod shattering is strongly correlated with the humidity in the area of origin of landraces (Lush et al., 1980). Alleles that predominate in arid environments could be useful for breeding legumes suited for areas that are predicted to become drier or more variable in future climate models.
Developing crops suited to rapidly evolving, adverse environmental conditions will be a major hurdle in the coming years. Many legume crops display extreme variation in pod shattering, with the greatest losses occurring when crops are grown under arid conditions. In many species, these losses can be overcome by pyramiding multiple genes that interact to provide resistance to pod shattering. The ranges of phenotypes resulting from each mutation form a strongly homologous series. Typically, individual shattering-resistance mutations either strengthen the sutures or reduce the torsion of pod walls (Figure 7). These isolated mutations may be suitable for humid climates, especially if the crop can be reliably harvested immediately upon maturity. However, these individual alleles are often insufficient in dry climates or when the timing of harvest cannot be assured. In these cases, multiple shattering resistance alleles are required for improved resistance. Combining these alleles will require a thorough understanding of the genetic control of pod shattering, which has only recently begun to be clarified.
After more than 10,000 years of selection, pod shattering remains problematic in many legume taxa. The strong effect of environmental aridity on this trait indicates that it will be a growing issue in the coming decades. Recent genetic advances have shed light on the control of pod dehiscence and have shown that key genes in a relatively small number of genetic pathways are responsible for its control. This genetic knowledge will contribute to the development of new crop varieties that are resilient to pod shattering across environments.
Acknowledgments
The authors would like to thank Judy Jernstedt for providing sectioning and microscopy equipment used to generate parts of Figures 2, 4, and 6. M. I. Chacón-Sánchez and J. Duitama also generously provided preliminary results on lima bean prior to publication of those results. T.A.P. was supported by the Clif Bar Family Foundation/Seed Matters and Lundberg Family Farms, and Sassoum Lo by the University of California President’s Postdoctoral Fellowship Program. Funding for studies on pod shattering in the PG group was provided by Lundberg Family Farm, Richvale, CA.
Funding
T.A.P was supported by the Clif Bar Family Foundation/Seed Matters and Lundberg Family Farms, and Sassoum Lo by the University of California President’s Postdoctoral Fellowship Program. Funding for studies on pod shattering in the PG group was provided by Lundberg Family Farm, Richvale, CA, the Department of Plant Sciences, and the Plant Sciences Field Facility.
Conflict of interest statement. The authors have no relevant interests to disclose.
All the authors contributed to the review of available literature, drafting and editing the manuscript, and developing figures. All authors have read and agreed to the published version of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Paul Gepts (plgepts@ucdavis.edu).
References
- Abbo S, Shtienberg D, Lichtenzveig J, Lev-Yadun S, Gopher A (2003) The chickpea, summer cropping, and a new model for pulse domestication in the ancient Near East. Quart Rev Biol 78: 435–448 [DOI] [PubMed] [Google Scholar]
- Abbo S, van-Oss RP, Gopher A, Saranga Y, Ofner I, Peleg Z (2014) Plant domestication versus crop evolution: a conceptual framework for cereals and grain legumes. Trends Plant Sci 19: 351–360 [DOI] [PubMed] [Google Scholar]
- Abd El-Moneim AM (1993) Selection for non-shattering common vetch, Vicia sativa L. Plant Breed 110: 168–171 [Google Scholar]
- Aguilar-Benitez D, Rubio J, Millán T, Gil J, Die JV, Castro P (2020) Genetic analysis reveals PDH1 as a candidate gene for control of pod dehiscence in chickpea. Molec Breed 40: 1–12 [Google Scholar]
- Ambrose M, Ellis T (2008) Ballistic seed dispersal and associated seed shadow in wild Pisum germplasm. Pisum Genet 40: 5–10 [Google Scholar]
- Andargie M, Pasquet RS, Gowda BS, Muluvi GM, Timko MP (2011) Construction of a SSR-based genetic map and identification of QTL for domestication traits using recombinant inbred lines from a cross between wild and cultivated cowpea (V. unguiculata (L.) Walp.). Molec Breed 28: 413–420 [Google Scholar]
- Anderson S (1955) Development of pods and seeds of birdsfoot trefoil, Lotus corniculatus L., as related to maturity and to seed yields. Agron J 47: 483–487 [Google Scholar]
- Ariani A, Berny Mier y Teran JC, Gepts P (2018) Spatial and temporal scales of range expansion in wild Phaseolus vulgaris. Mol Biol Evol 35: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armon S, Efrati E, Kupferman R, Sharon E (2011) Geometry and mechanics in the opening of chiral seed pods. Science 333: 1726–1730 [DOI] [PubMed] [Google Scholar]
- Assefa T, Beebe SE, Rao IM, Cuasquer JB, Duque MC, Rivera M, Battisti A, Lucchin M (2013) Pod harvest index as a selection criterion to improve drought resistance in white pea bean. Field Crops Res 148: 24–33 [Google Scholar]
- Atchison GW, Nevado B, Eastwood RJ, Contreras‐Ortiz N, Reynel C, Madriñán S, Filatov DA, Hughes CE (2016) Lost crops of the Incas: origins of domestication of the Andean pulse crop tarwi, Lupinus mutabilis. Am J Bot 103: 1592–1606 [DOI] [PubMed] [Google Scholar]
- Bailey M, Mian M, Carter T, Ashley D, Boerma H (1997) Pod dehiscence of soybean: identification of quantitative trait loci. J Hered 88: 152–154 [Google Scholar]
- Ballester P, Ferrándiz C (2017) Shattering fruits: variations on a dehiscent theme. Curr Opin Plant Biol 35: 68–75 [DOI] [PubMed] [Google Scholar]
- Bandillo NB, Anderson JE, Kantar MB, Stupar RM, Specht JE, Graef GL, Lorenz AJ (2017) Dissecting the genetic basis of local adaptation in soybean. Scient Rep 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PT (1910) The string-bean man and his work. Plant World 13: 299–301 [Google Scholar]
- Berny Mier y Teran JC, Konzen ER, Palkovic A, Tsai SM, Rao IM, Beebe S, Gepts P (2019) Effect of drought stress on the genetic architecture of photosynthate allocation and remobilization in pods of common bean (Phaseolus vulgaris L.), a key species for food security. BMC Plant Biol 19: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhor T, Chimote V, Deshmukh M (2014) Inheritance of pod shattering in soybean [Glycine max (L.) Merrill]. Electron J Plant Breed 5: 671–676 [Google Scholar]
- Bishnoi S, Hooda J, Sharma P (2018) Analysis of gene effects for yield and yield component traits in faba bean (Vicia faba L.) genotypes. J Animal Plant Sci 28: 187–196 [Google Scholar]
- Bitocchi E, , Bellucci E, , Giardini A, , Rau D, , Rodriguez M, , Biagetti E, , Santilocchi R, , Spagnoletti Zeuli P, , Gioia T, et al. (2013) Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytologist 197 (1): 300–313. 10.1111/j.1469-8137.2012.04377.x [DOI] [PubMed] [Google Scholar]
- Bitocchi E, Rau D, Bellucci E, Rodriguez M, Murgia ML, Gioia T, Santo D, Nanni L, Attene G, Papa R (2017) Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front Plant Sci 8: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt S (1972) Mutation genetics in Pisum. Agri Hort Genet 30: 1–293 [Google Scholar]
- Blixt S (1978) Some genes of importance for the evolution of the pea in cultivation (and a short presentation of the Weibullshom-PGA pea collection). In Conference Proceedings on Broadening the Genetic Base of Crop Production, Wageningen, pp 195–202
- Buckovic RG (1952) Some of the morphologic and agronomic factors associated with pod dehiscence in Lotus corniculatus. MS thesis, Oregon State College, Corvallis, OR, USA
- Caracuta V, Weinstein-Evron M, Kaufman D, Yeshurun R, Silvent J, Boaretto E (2016) 14,000-year-old seeds indicate the levantine origin of the lost progenitor of faba bean. Scient Rep 6: 37399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuta V, Barzilai O, Khalaily H, Milevski I, Paz Y, Vardi J, Regev L, Boaretto E (2015) The onset of faba bean farming in the southern levant. Scient Rep 5: 14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness C (1965) Effects of relative humidity on pod dehiscence in soybeans. Crop Sci 5: 511–513 [Google Scholar]
- Chacón-Sánchez MI, Martínez-Castillo J (2017) Testing domestication scenarios of lima bean (Phaseolus lunatus L.) in Mesoamerica: insights from genome-wide genetic markers. Front Plant Sci 8: 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichy KA, Porch TG, Beaver JS, Cregan P, Fourie D, Glahn RP, Grusak MA, Kamfwa K, Katuuramu DN, McClean P, et al. (2015) A Phaseolus vulgaris diversity panel for Andean bean improvement. Crop Sci 55: 2149–2160 [Google Scholar]
- Currence TM (1930) Inheritance studies in Phaseolus vulgaris. Tech Bull Minnesota Agric Exp Stn 68: 3–28 [Google Scholar]
- Darwin C (1859) The Origin of Species. John Murray, London, UK [Google Scholar]
- Davis J, Kean D, Yorgey B, Fourie D, Miklas P, Myers J (2006) A molecular marker linkage map of snap bean (Phaseolus vulgaris). Annu Rep Bean Improv Coop 49: 73 [Google Scholar]
- Di Vittori V, Gioia T, Rodriguez M, Bellucci E, Bitocchi E, Nanni L, Attene G, Rau D, Papa R (2019) Convergent evolution of the seed shattering trait. Genes 10: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Vittori V, Bitocchi E, Rodriguez M, Alseekh S, Bellucci E, Nanni L, Gioia T, Marzario S, Logozzo G, Rossato M, et al. (2020) Pod indehiscence in common bean is associated to the fine regulation of PvMYB26 and a non-functional abscission layer. bioRxiv: 2020.2004.2002.021972 [DOI] [PMC free article] [PubMed]
- Dong D, Yan L, Dong R, Liu W, Wang Y, Liu Z (2017a) Evaluation and analysis of pod dehiscence factors in shatter-susceptible and shatter-resistant common vetch. Crop Sci 57: 2770–2776 [Google Scholar]
- Dong R, Dong D, Luo D, Zhou Q, Chai X, Zhang J, Xie W, Liu W, Dong Y, Wang Y (2017b) Transcriptome analyses reveal candidate pod shattering-associated genes involved in the pod ventral sutures of common vetch (Vicia sativa L.). Front Plant Sci 8: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Wang Y-Z (2015) Seed shattering: from models to crops. Front Plant Sci 6: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang B-H, Liu B-L, Wang Y-Z (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5: 3352. [DOI] [PubMed] [Google Scholar]
- Drijfhout E (1970) Influence of temperature on string formation of beans (Phaseolus vulgaris). Euphytica 19: 145–151 [Google Scholar]
- Drijfhout E (1978) Inheritance of temperature-dependent string formation in common bean. Annu Rep Bean Improv Coop 21: 33–34 [Google Scholar]
- Emerson R (1904) Heredity in bean hybrids. Ann Rep Nebr Agric Exp St 17: 33–78 [Google Scholar]
- Erskine W (1985) Selection for pod retention and pod indehiscence in lentils. Euphytica 34: 105–112 [Google Scholar]
- Esau K (1977) Anatomy of Seed Plants, Ed 2.Wiley, New York [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAOSTAT) (2020) Food and agriculture data. www.fao.org/faostat/en/#search/soybean (March 27, 2020)
- Fratini R, Vinagre MYD, García PG, de la Vega MP (2007) Identification of quantitative trait loci (QTL) for plant structure, growth habit and yield in lentil. Spanish J Agric Res 3: 348–356 [Google Scholar]
- Freyre R, Skroch PW, Geffroy V, Adam-Blondon A-F, Shirmohamadali A, Johnson WC, Llaca V, Nodari R, Pereira P, Tsai S-M, et al. (1998) Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theoret Appl Genet 97: 847–856 [Google Scholar]
- Funatsuki H, Ishimoto M, Tsuji H, Kawaguchi K, Hajika M, Fujino K (2006) Simple sequence repeat markers linked to a major QTL controlling pod shattering in soybean. Plant Breed 125: 195–197 [Google Scholar]
- Funatsuki H, Suzuki M, Hirose A, Inaba H, Yamada T, Hajika M, Komatsu K, Katayama T, Sayama T, Ishimoto M (2014) Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci 111: 17797–17802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia T, Duitama J, Smolenski Zullo S, Gil J, Ariani A, Dohle S, Palkovic A, Skeen P, Bermudez C, Debouck DG, et al. (2020) Comprehensive genomic resources related to domestication and crop improvement traits in lima bean, https://www.researchsquare.com/article/rs-95762/v1. NatureResearch, ResearchSquare, pre-print server [DOI] [PMC free article] [PubMed]
- Gepts P, Bliss F (1984) Enhanced available methionine concentration associated with higher phaseolin levels in common bean seeds. Theoret Appl Genet 69: 47–53 [DOI] [PubMed] [Google Scholar]
- Gepts P, Osborn T, Rashka K, Bliss F (1986) Phaseolin-protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris): evidence for multiple centers of domestication. Econ Bot 40: 451–468 [Google Scholar]
- Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND (2005) Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol 137: 1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P, Aragão FJL, Barros E, Blair MW, Brondani R, Broughton W, Galasso I, Hernández G, Kami J, Lariguet P, et al. (2008) Genomics of Phaseolus beans, a major source of dietary protein and micronutrients in the tropics. InMoore PH, Ming R, eds, Genomics of Tropical Crop Plants. Springer New York, New York, NY, pp 113–143 [Google Scholar]
- Gioia T, Logozzo G, Kami J, Spagnoletti Zeuli P, Gepts P (2013) Identification and characterization of a homologue to the Arabidopsis INDEHISCENT gene in common bean. J Hered 104: 273–286 [DOI] [PubMed] [Google Scholar]
- Gladstones J (1967) Selection for economic characters in Lupinus angustifolius and L. digitatus. Aust J Exp Agric 7: 360–366 [Google Scholar]
- Gresta F, Wink M, Prins U, Abberton M, Capraro J, Scarafoni A, Hill G (2017) Lupins in European cropping systems. In Murphy-Bokern D, Stoddard F, Watson C, eds, Legumes in Cropping Systems. CABI Publishing, Wallingford, UK, pp 88–108 [Google Scholar]
- Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Salgado A, Gepts P, Debouck DG (1995) Evidence for two gene pools of the Lima bean, Phaseolus lunatus L., in the Americas. Genet Resour Crop Evol 42: 15–28 [Google Scholar]
- Hagerty CH, Cuesta-Marcos A, Cregan P, Song Q, McClean P, Myers JR (2016) Mapping snap bean pod and color traits, in a dry bean × snap bean recombinant inbred population. J Am Soc Horticult Sci 141: 131–138 [Google Scholar]
- Hammer K, Khoshbakht K (2015) A domestication assessment of the big five plant families. Genet Resour Crop Evol 62: 665–689 [Google Scholar]
- Harlan JR (1992) Crops and Man. American Society of Agronomy, Madison, WI [Google Scholar]
- Hradilova I, Trněný O, Valkova M, Cechova M, Janska A, Prokešová L, Aamir K, Krezdorn N, Rotter B, Winter P (2017) A combined comparative transcriptomic, metabolomic, and anatomical analyses of two key domestication traits: pod dehiscence and seed dormancy in pea (Pisum sp.). Front Plant Sci 8: 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Kan G, Hu W, Li Y, Hao D, Li X, Yang H, Yang Z, He X, Huang F, et al. (2019) Identification of loci and candidate genes responsible for pod dehiscence in soybean via genome-wide association analysis across multiple environments. Front Plant Sci 10: 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal MM, Erskine W, Berger JD, Nelson MN (2020) Phenotypic characterisation and linkage mapping of domestication syndrome traits in yellow lupin (Lupinus luteus L.). Theoret Appl Genet 133: 2975–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemura T, Kaga A, Tomooka N, Shimizu T, Vaughan DA (2010) The genetics of domestication of rice bean, Vigna umbellata. Ann Bot 106: 927–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemura T, Kaga A, Konishi S, Ando T, Tomooka N, Han OK, Vaughan DA (2007) Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm-season legumes. Ann Bot 100: 1053–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T, Jo U, Vaughan DA, Tomooka N (2012) Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PloS One 7: e41304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javadi F, Tun YT, Kawase M, Guan K, Yamaguchi H (2011) Molecular phylogeny of the subgenus Ceratotropis (genus Vigna, Leguminosae) reveals three eco-geographical groups and late pliocene–pleistocene diversification: evidence from four plastid DNA region sequences. Ann Bot 108: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, Den Boer B, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten J (1927) Een onderzoek naar het kenmerk der “draadloosheid” bij verschillende boonenrassen [Investigations concerning the “stringlessness” of varieties of beans]. Mededelingen van de Landbouwhogeschool Wageningen 31: 1–49 [Google Scholar]
- Kaga A, Isemura T, Tomooka N, Vaughan DA (2008) The genetics of domestication of the azuki bean (Vigna angularis). Genetics 178: 1013–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S-T, Kwak M, Kim H-K, Choung M-G, Han W-Y, Baek I-Y, Kim MY, Van K, Lee S-H (2009) Population-specific QTLs and their different epistatic interactions for pod dehiscence in soybean [Glycine max (L.) Merr.]. Euphytica 166: 15 [Google Scholar]
- Kazan K, Muehlbauer F, Weeden N, Ladizinsky G (1993) Inheritance and linkage relationships of morphological and isozyme loci in chickpea (Cicer arietinum L.). Theoret Appl Genet 86: 417–426 [DOI] [PubMed] [Google Scholar]
- Kazimierski T (1964) Inheritance of certain characters in the Lupinus albus L. x L. graecus Boiss hybrid. Genet Polon 5: 309 [Google Scholar]
- Kissing Kucek L, Riday H, Rufener BP, Burke AN, Eagen SS, Ehlke N, Krogman S, Mirsky SB, Reberg-Horton C, Ryan MR (2020) Pod dehiscence in hairy vetch (Vicia villosa Roth). Front Plant Sci 11: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinange EM, Singh SP, Gepts P (1996) Genetic control of the domestication syndrome in common bean. Crop Sci 36: 1037–1045 [Google Scholar]
- Kwak M, Gepts P (2009) Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theoret Appl Genet 118: 979–992 [DOI] [PubMed] [Google Scholar]
- Kwak M, Toro O, Debouck DG, Gepts P (2012) Multiple origins of the determinate growth habit in domesticated common bean (Phaseolus vulgaris). Ann Bot 110: 1573–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladizinsky G (1975) On the origin of the broad bean, Vicia faba L. Israel J Bot 24: 80–88 [Google Scholar]
- Ladizinsky G (1979a) The genetics of several morphological traits in the lentil. J Hered 70: 135–137 [Google Scholar]
- Ladizinsky G (1979b) Seed dispersal in relation to the domestication of Middle East legumes. Econ Bot 33: 284–289 [Google Scholar]
- Ladizinsky G (1989) Origin and domestication of the Southwest Asian grain legumes. InHarris DR, Hillman GC, eds, Foraging and Farming: The Evolution of Plant Exploitation. Unwin Hyman, London, pp. 374–389 [Google Scholar]
- Laghetti G, Piergiovanni AR, Sonnante G, Lioi L, Pignone D (2008). The Italian lentil genetic resources: a worthy basic tool for breeders. Eur J Plant Sci Biotechnol 2: 48–59 [Google Scholar]
- Lamprecht H (1932) Beiträge zur Genetik von Phaseolus vulgaris. II. Über Vererbung von Hülsenfarbe und Hülsenform [Contribution to the genetics of Phaseolus vulgaris. II. On the inheritance of pod color and pod shape]. Hereditas 16: 295–340 [Google Scholar]
- Lamprecht H (1938) Über Hülseneigenschaften bei Pisum, ihre Vererbung und ihr züchterischer Wert [On the pod properties of Pisum, their inheritance and breeding value]. Der Züchter 10: 150–157 [Google Scholar]
- Lee TD (1984) Effects of seed number per fruit on seed dispersal in Cassia fasciculata (Caesalpiniaceae). Bot Gazette 145: 136–139 [Google Scholar]
- Lenser T, Theißen G (2013) Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci 18: 704–714 [DOI] [PubMed] [Google Scholar]
- Li P, Wind JJ, Shi X, Zhang H, Hanson J, Smeekens SC, Teng S (2011) Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc Natl Acad Sci 108: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF (2004) Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Lo S, Muñoz-Amatriaín M, Boukar O, Herniter I, Cisse N, Guo Y-N, Roberts PA, Xu S, Fatokun C, Close TJ (2018) Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Scient Rep 8: 6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardi S, Muñoz-Amatriaín M, Liang Q, Shu S, Wanamaker SI, Lo S, Tanskanen J, Schulman AH, Zhu T, Luo M-C, et al. (2019) The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J 98: 767–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legume Phylogeny Working Group (LPWG) (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66: 44–77 [Google Scholar]
- Lush W, Evans L (1981) The domestication and improvement of cowpeas (Vigna unguiculata (L.) Walp.). Euphytica 30: 579–587 [Google Scholar]
- Lush W, Evans L, Wien H (1980) Environmental adaptation of wild and domesticated cowpeas (Vigna unguiculata (L.) Walp.). Field Crops Res 3: 173–187 [Google Scholar]
- Maalouf F, Hu J, O'Sullivan DM, Zong X, Hamwieh A, Kumar S, Baum M (2019) Breeding and genomics status in faba bean (Vicia faba). Plant Breed 138: 465–473 [Google Scholar]
- Malo JE (2004) Potential ballistic dispersal of Cytisus scoparius (Fabaceae) seeds. Aust J Bot 52: 653–658 [Google Scholar]
- Marx G (1971) New linkage relations for chromosome III of Pisum. Pisum Newslett 3: 18–19 [Google Scholar]
- Maxted N (1991) The newly discovered relatives of Vicia faba L. do little to resolve the enigma of its origin. Bot Chron 10: 435–465 [Google Scholar]
- McGee RJ, Baggett JR (1992). Unequal growth rate of pollen tubes from normal and stringless pea genotypes. HortScience 27: 833–834 [Google Scholar]
- Mendel G (1866) Versuche über Pflanzen-Hybriden [Experiments on plant hybrids]. Verhandlungen des naturforschenden Vereins in Brünn IV, pp 3–47 [Google Scholar]
- Miranda C, , Culp C,, Škrabišová M, , Joshi T, , Belzile F, , Grant DM,, Bilyeu K (2019) Molecular tools for detecting Pdh1 can improve soybean breeding efficiency by reducing yield losses due to pod shatter. Mol Breeding 39: 27. 10.1007/s11032-019-0935-1 [DOI] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam SM, Mamidi S, Osorno JM, Lee R, Brick M, Kelly J, Miklas P, Urrea C, Song Q, Cregan P (2016) Genome-wide association study identifies candidate loci underlying agronomic traits in a Middle American diversity panel of common bean. Plant Genome 9: 3. [DOI] [PubMed] [Google Scholar]
- Motta-Aldana JR, Serrano-Serrano ML, Hernández-Torres J, Castillo-Villamizar G, Debouck DG, Chacóns MI (2010) Multiple origins of lima bean landraces in the Americas: evidence from chloroplast and nuclear DNA polymorphisms Crop Sci 50: 1773–1787 [Google Scholar]
- Murgia ML, Attene G, Rodriguez M, Bitocchi E, Bellucci E, Fois D, Nanni L, Gioia T, Albani DM, Papa R (2017) A comprehensive phenotypic investigation of the “pod-shattering syndrome” in common bean. Front Plant Sci 8: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J, Baggett J, Lamborn C (2001) Origin, history, and genetic improvement of the snap pea (Pisum sativum L.). Plant Breed Rev 21: 93 [Google Scholar]
- Nanni L, Bitocchi E, Bellucci E, Rossi M, Rau D, Attene G, Gepts P, Papa R (2011) Nucleotide diversity of a genomic sequence similar to SHATTERPROOF (PvSHP1) in domesticated and wild common bean (Phaseolus vulgaris L.). Theoret Appl Genet 123: 1341–1357 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogutcen E, Pandey A, Khan MK, Marques E, Penmetsa RV, Kahraman A, von Wettberg EJ (2018) Pod shattering: a homologous series of variation underlying domestication and an avenue for crop improvement. Agronomy 8: 137 [Google Scholar]
- Parker TA, Berny Mier y Teran JC, Palkovic A, Jernstedt J, Gepts P (2020a) Pod indehiscence is a domestication and aridity resilience trait in common bean. New Phytol 225: 558–570 [DOI] [PubMed] [Google Scholar]
- Parker T, Sousa LL, Floriani TO, Palkovic A, Gepts P (2020b) Towards the introgression of PvPdh1 for increased resistance to pod shattering in common bean. Theoret Appl Genet, doi: 10.1007/s00122-020-03698-7 (01 November 2020 accessed) [DOI] [PubMed] [Google Scholar]
- Philbrook B, Oplinger E (1989) Soybean field losses as influenced by harvest delays. Agron J 81: 251–258 [Google Scholar]
- Plewiński P, Książkiewicz M, Rychel-Bielska S, Rudy E, Wolko B (2019) Candidate domestication-related genes revealed by expression quantitative trait loci mapping of narrow-leafed lupin (Lupinus angustifolius L.). Int J Mol Sci 20: 5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakken R (1934) Inheritance of colours and pod characters in Phaseolus vulgaris L. Genetica 16: 177–296 [Google Scholar]
- Pratap A, Prajapati U, Singh CM, Gupta S, Rathore M, Malviya N, Tomar R, Gupta AK, Tripathi S, Singh NP (2018) Potential, constraints and applications of in vitro methods in improving grain legumes. Plant Breed 137: 235–249 [Google Scholar]
- Price D, Smith C, Hedley C (1988) The effect of the gp gene on fruit development in Pisum sativum L. I. Structural and physical aspects. New Phytol 110: 261–269 [Google Scholar]
- Prokesova L, Dedicova L, Janova A, Cevelova L, Lonova K, Hanacek P (2016) Study of extensin gene expression as a candidate responsible for pea pod dehiscence. In MendelNet 2016 Conference Proceedings, pp 752–756
- Rajani S, Sundaresan V (2001) The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol 11: 1914–1922 [DOI] [PubMed] [Google Scholar]
- Rau D, Murgia ML, Rodriguez M, Bitocchi E, Bellucci E, Fois D, Albani D, Nanni L, Gioia T, Santo D, et al. (2019) Genomic dissection of pod shattering in common bean: mutations at nonorthologous loci at the basis of convergent phenotypic evolution under domestication of leguminous species. Plant J 97: 693–714 [DOI] [PubMed] [Google Scholar]
- Reid JB, Ross JJ (2011) Mendel’s genes: toward a full molecular characterization. Genetics 189: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendón-Anaya M, Montero-Vargas JM, Saburido-Alvarez S, Vlasova A, Capella-Gutiérrez S, Ordaz-Ortiz JJ, Aguilar OM, Vianello-Brondani RP, Santalla M, Delaye L, et al. (2017) Genomic history of the origin and domestications of common bean in the Americas unveils its closest sister species. Genome Biol 18: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repinski SL, Kwak M, Gepts P (2012) The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theoret Appl Genet 124: 1539–1547 [DOI] [PubMed] [Google Scholar]
- Roeder AH, Ferrándiz C, Yanofsky MF (2003) The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol 13: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean P, Mamidi S, Wu G, Cannon S, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, et al. (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood S, Fu Q (2014) A drier future? Science 343: 737–739 [DOI] [PubMed] [Google Scholar]
- Simpson MG (2019) Plant Systematics. Academic Press, San Diego, CA [Google Scholar]
- Singh SP (2013) Common Bean Improvement in the Twenty-first Century. Springer Science & Business Media, Dordrecht, The Netherlands [Google Scholar]
- Singh SP, Gepts P, Debouck DG (1991). Races of common bean (Phaseolus vulgaris, Fabaceae). Econ Bot 45: 379–396 [Google Scholar]
- Suanum W, Somta P, Kongjaimun A, Yimram T, Kaga A, Tomooka N, Takahashi Y, Srinives P (2016) Co-localization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol Breed 36: 1–11 [Google Scholar]
- Suzuki M, Fujino K, Funatsuki H (2009) A major soybean QTL, qPDH1, controls pod dehiscence without marked morphological change. Plant Prod Sci 12: 217–223 [Google Scholar]
- Tahir M, Muehlbauer F (1994) Gene mapping in lentil with recombinant inbred lines. J Hered 85: 306–310 [Google Scholar]
- Tahir M, Simon C, Muehlbauer F (1993) Gene map of lentil: a review. Lens Newslett 20: 3–10 [Google Scholar]
- Takahashi Y, Sakai H, Yoshitsu Y, Muto C, Anai T, Pandiyan M, Senthil N, Tomooka N, Naito K (2019) Domesticating Vigna stipulacea: a potential legume crop with broad resistance to biotic stresses. Front Plant Sci 10: 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Kongjaimun A, Muto C, Kobayashi Y, Kumagai M, Sakai H, Satou K, Teruya K, Shiroma A, Shimoji M, et al. (2020) Same locus for non-shattering seed pod in two independently domesticated legumes, Vigna angularis and Vigna unguiculata. Front Genet 11: 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar D (2011) Genetics of pod indehiscence in grass pea (Lathyrus sativus L.). J Crop Improve 25: 161–175 [Google Scholar]
- Tian Z,, Wang X,, Lee R,, Li Y,, Specht JE,, Nelson RL,, McClean PE,, Qiu L,, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci 107: 8563–8568. 10.1073/pnas.1000088107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomooka N, Vaughan DA, Moss H, Maxted N (2002) The Asian Vigna: Genus Vigna Subgenus Ceratotropis Genetic Resources. Springer Science+Business, Dordrecht, The Netherlands [Google Scholar]
- Tschermak E (1901) Weitere Beiträge über Verschiedenwertigkeit der Merkmale bei Kreuzung von Erbsen und Bohnen [Further contributions on character-state differences of traits in crosses of peas and beans]. Bericht Deutsch Botan Gesell XIX: 35–51 [Google Scholar]
- Tschermak E (1902) Über die gesetzmässige Gestaltungsweise der Mischlinge [About the pattern-following segregation of hybrids]. Zeitsch landwirtsch Versuch Oester 5: 781–860 [Google Scholar]
- Tukamuhabwa P, Dashiell KE, Rubaihayo P, Nabasirye M (2002) Determination of field yield loss and effect of environment on pod shattering in soybean. Afr Crop Sci J 10: 203–209 [Google Scholar]
- Vaillancourt RE, Weeden NF, Bruneau A, Doyle JJ (1993) Chloroplast DNA phylogeny of Old World Vigna (Leguminosae). Syst Bot 18: 642–651 [Google Scholar]
- Vavilov NI (1922) The law of homologous series in variation. J Genet 12: 47–89 [Google Scholar]
- von Sengbusch R (1938) The breeding of sweet lupins. Herbage Rev 6: 1–9 [Google Scholar]
- Waines JG (1975) The biosystematics and domestication of peas (Pisum L.). Bull Torrey Bot Club 102: 385–395 [Google Scholar]
- Wang H, Zhao Q, Chen F, Wang M, Dixon RA (2011) NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J 68: 1104–1114 [DOI] [PubMed] [Google Scholar]
- Watcharatpong P, Kaga A, Chen X, Somta P (2020) Narrowing down a major QTL region conferring pod fiber contents in yardlong bean (Vigna unguiculata), a vegetable cowpea. Genes 11: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden N, Muehlbauer F, Ladizinsky G (1992) Extensive conservation of linkage relationships between pea and lentil genetic maps. J Hered 83: 123–129 [Google Scholar]
- Weeden NF (2007) Genetic changes accompanying the domestication of Pisum sativum: is there a common genetic basis to the ‘domestication syndrome’ for legumes? Ann Bot 100: 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden NF (2018) Domestication of pea (Pisum sativum L.): the case of the Abyssinian pea. Front Plant Sci 9: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden NF, Brauner S, Przyborowski JA (2002) Genetic analysis of pod dehiscence in pea (Pisum sativum L.). Cell Mol Biol Lett 7: 657–664 [PubMed] [Google Scholar]
- Wellensiek S (1925) Pisum-crosses. I. Genetica 7: 1–64 [DOI] [PubMed] [Google Scholar]
- Wellensiek S (1971) Lamprecht’s gene sin for stringless. Pisum Newslett 3: 48 [Google Scholar]
- White OE (1917) Studies of inheritance in Pisum. II. The present state of knowledge of heredity and variation in peas. Proc Am Philos Soc 56: 487–588 [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well‐supported subclades within the family. Am J Bot 91: 1846–1862 [DOI] [PubMed] [Google Scholar]
- Wolko B, Clements JC, Naganowska B, Nelson MN (2011) Lupinus. InWild Crop Relatives: Genomic and Breeding Resources. Springer, New York, pp 153–206 [Google Scholar]
- Yamada T, Funatsuki H, Hagihara S, Fujita S, Tanaka Y, Tsuji H, Ishimoto M, Fujino K, Hajika M (2009) A major QTL, qPDH1, is commonly involved in shattering resistance of soybean cultivars. Breed Sci 59: 435–440 [Google Scholar]
- Yang C, Song J, Ferguson AC, Klisch D, Simpson K, Mo R, Taylor B, Mitsuda N, Wilson ZA (2017) Transcription factor MYB26 is key to spatial specificity in anther secondary thickening formation. Plant Physiol 175: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Wang H (2016) Molecular mechanisms for vascular development and secondary cell wall formation. Front Plant Sci 7: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yundaeng C, Somta P, Amkul K, Kongjaimun A, Kaga A, Tomooka N (2019) Construction of genetic linkage map and genome dissection of domestication-related traits of moth bean (Vigna aconitifolia), a legume crop of arid areas. Mol Genet Genom 294: 621–635 [DOI] [PubMed] [Google Scholar]
- Zhang J, Singh AK (2020) Genetic control and geo-climate adaptation of pod dehiscence provide novel insights into soybean domestication. G3: Genes Genom Genet 10: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Tu B, Liu C, Liu X (2018) Pod anatomy, morphology and dehiscing forces in pod dehiscence of soybean (Glycine max (L.) Merrill). Flora 248: 48–53 [Google Scholar]