Abstract
Plants can contain biosynthetic gene clusters (BGCs) that nominally resemble those found in microbes. However, while horizontal gene transmission is often observed in microbes, plants are limited to vertical gene transmission, implying that their BGCs may exhibit distinct inheritance patterns. Rice (Oryza sativa) contains two unlinked BGCs involved in diterpenoid phytoalexin metabolism, with one clearly required for momilactone biosynthesis, while the other is associated with production of phytocassanes. Here, in the process of elucidating momilactone biosynthesis, genetic evidence was found demonstrating a role for a cytochrome P450 (CYP) from the other “phytocassane” BGC. This CYP76M8 acts after the CYP99A2/3 from the “momilactone” BGC, producing a hemiacetal intermediate that is oxidized to the eponymous lactone by a short-chain alcohol dehydrogenase also from this BGC. Thus, the “momilactone” BGC is not only incomplete, but also fractured by the need for CYP76M8 to act in between steps catalyzed by enzymes from this BGC. Moreover, as supported by similar activity observed with orthologs from the momilactone-producing wild-rice species Oryza punctata, the presence of CYP76M8 in the other “phytocassane” BGC indicates interdependent evolution of these two BGCs, highlighting the distinct nature of BGC assembly in plants.
Investigation of momilactone production in rice demonstrates roles for two unlinked biosynthetic clusters, requiring interdependent evolution and highlighting the distinct nature of their assembly.
Introduction
All organisms produce specialized metabolites that largely reflect their current (and evolutionarily recent past) ecological interactions. The selective pressure for production of these compounds sometimes leads to genomic clustering of the genes encoding the enzymes for particular biosynthetic pathways/networks. This has long been recognized in prokaryotes, where such organization is widespread, driven at least in part by the prevalence of horizontal gene transfer (Lawrence, 1999). Such biosynthetic gene clusters (BGCs) can also be found in fungi, although these are much less prevalent in this kingdom, and the role of horizontal gene transfer in their evolution is less clear (Rokas et al., 2018). Intriguingly, nominally similar BGCs have recently been found in plant genomes as well (Nutzmann et al., 2016). These plant BGCs did not originate via horizontal gene transfer from microbes, but rather by a poorly understood process of gene “recruitment” (Nutzmann et al., 2018), which must function within the limitations of purely vertical gene transmission.
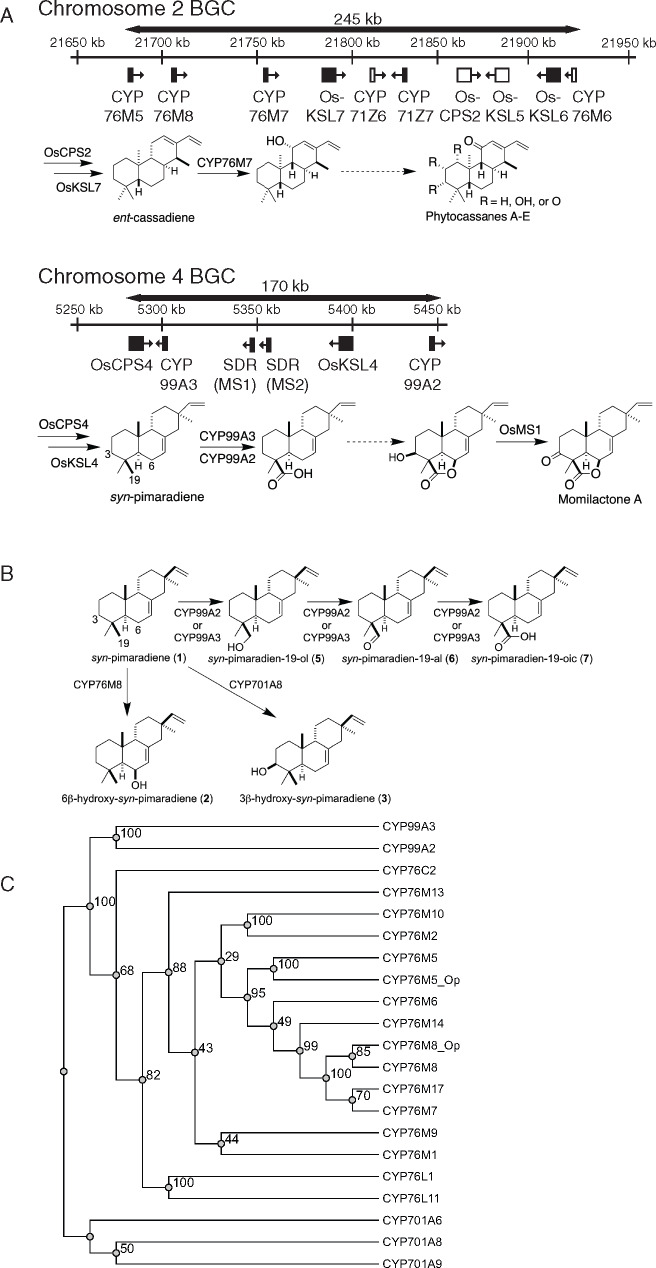
Rice (Oryza sativa) contains at least two BGCs involved in the production of phytoalexins (Figure 1, A). Each of these BGCs has been nominally assigned to production of a specific family of phytoalexins, with that on chromosome 4 (c4BGC) assigned to momilactone production and that on chromosome 2 (c2BGC) nominally associated with phytocassane production (Miyamoto et al., 2016). As is true for the majority of rice phytoalexins (Peters, 2006), the momilactones and phytocassanes are labdane-related diterpenoids characterized by the bicyclization reaction catalyzed by class II diterpene cyclases, which is typically followed by further cyclization catalyzed by class I diterpene synthases (Peters, 2010). Both rice BGCs encode not only consecutively acting diterpene cyclases and synthases (Prisic et al., 2004; Wilderman et al., 2004; Kanno et al., 2006), but also subsequently acting cytochrome P450 (CYP) mono-oxygenases (Shimura et al., 2007; Swaminathan et al., 2009; Wang et al., 2011, 2012b; Wu et al., 2011, 2013; Kitaoka et al., 2015b), with c4BGC also containing short-chain alcohol dehydrogenases/reductases (SDRs).
Figure 1.
Biosynthetic gene clusters (BGCs) and labdane-related diterpenoid metabolism in rice. (A) Schematic of rice BGCs and associated metabolic pathways. Filled black boxes represent genes induced by chitin elicitation, while empty white boxes represent those not induced (Okada et al., 2007), with arrowheads indicating the direction of transcription. The reactions catalyzed in the characteristic biosynthetic pathways by enzymes encoded within each BGC are indicated as described in the text, while dotted arrows represent multiple unknown steps. (B) Known activities of rice CYPs with syn-pimaradiene (1). (C) Phylogenetic tree for the rice CYP76M sub-family, along with other CYPs associated with momilactone biosynthesis in rice (numbers indicate bootstrap values).
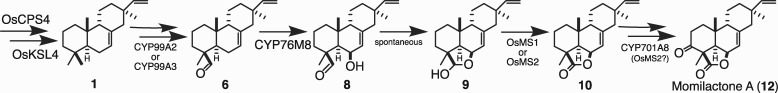
Genetic evidence demonstrates that the diterpene cyclase and synthase from c4BGC (Xu et al., 2012), as well as both CYPs (CYP99A2 and CYP99A3) from this gene cluster (Shimura et al., 2007) are involved in momilactone biosynthesis. The two SDRs can catalyze the oxidation of 3β-hydroxy-syn-pimaradien-19,6β-olide to form the characteristic carbon-3 (C3) keto group and, hence, have been termed momilactone A synthases (OsMS1 and OsMS2; Shimura et al., 2007; Kitaoka et al., 2016). Accordingly, c4BGC seems to be dedicated to momilactone production (i.e. is a “momilactone” BGC), with the diterpene cyclase (OsCPS4) and synthase (OsKSL4) together producing the olefinic precursor syn-pimaradiene (Otomo et al., 2004a, 2004b; Wilderman et al., 2004; Xu et al., 2004), which can be further transformed into syn-pimaradien-19-oic acid by CYP99A2 and/or CYP99A3 (Wang et al., 2011; Kitaoka et al., 2015b; Figure 1, A). However, it is clear that additional enzymes are required to complete this biosynthetic pathway. In particular, formation of the eponymous lactone ring clearly requires oxidation at the C6β position, while hydroxylation at C3β is also required.
Indeed, other CYPs can react with syn-pimaradiene at these latter two positions (Figure 1, B), with CYP701A8 catalyzing hydroxylation at C3β (Kitaoka et al., 2015b) and CYP76M8 catalyzing hydroxylation at C6β (Wang et al., 2012b). Intriguingly, CYP76M8 is found in the c2BGC, which also contains the ent-cassadiene synthase gene OsKLS7 that initiates phytocassane biosynthesis (Cho et al., 2004), as well as the upstream acting diterpene cyclase gene OsCPS2 (Otomo et al., 2004a; Prisic et al., 2004). The c2BGC also contains the closely related CYP76M7, encoding an enzyme that catalyzes the hydroxylation at C11α of ent-cassadiene required for phytocassane production (Swaminathan et al., 2009). Thus, the c2BGC has been termed the “phytocassane” BGC (Miyamoto et al., 2016), although it clearly contains genes required for the production of other labdane-related diterpenoids, such as OsKSL6, which is involved in biosynthesis of oryzalides (Toyomasu, 2008).
Notably, CYP76M8 falls within a phylogenetic clade of the CYP76M sub-family that, in addition to CYP76M5, CYP76M6, and CYP76M7, which also are found in the c2BGC (Figure 1, A), has members located elsewhere in the rice genome. Specifically, these members include CYP76M14, located on chromosome 1, and CYP76M17, located on chromosome 6, both of which are more closely related to CYP76M8 and CYP76M7 than are the co-clustered CYP76M5 or CYP76M6 (Figure 1, C). Although RNAi knock-down of both CYP76M7 and CYP76M8 reduced momilactone as well as phytocassane production it had minimal effects on expression of the more distantly related genes CYP76M5 and CYP76M6. However, because CYP76M14 and CYP76M17 were not examined (Ye et al., 2018), it remains possible that these paralogs might be involved in momilactone and/or phytocassane biosynthesis.
Here, further investigation of momilactone biosynthesis is reported, including genetic analysis. While several of the rice CYP76M sub-family members exhibit analogous biochemical activity, the genetic studies indicated that CYP76M8 is primarily responsible for momilactone biosynthesis, while CYP76M7 is primarily responsible for phytocassane production. CYP76M8 acts after CYP99A2 and/or CYP99A3 from the “momilactone” BGC, with the resulting hemiacetal further oxidized to the eponymous lactone by the OsMS1 and/or OsMS2 also from this BGC. Thus, the “momilactone” c4BGC is both incomplete and fractured by the need for CYP76M8 to act in between CYP99A2/3 and OsMS1/2. In addition, the analogous activity observed with an orthologous CYP76M sub-family member in the momilactone-producing wild-rice species Oryza punctata supports interdependent evolution of the “momilactone” c4BGC with the more complex c2BGC. The implications of this finding for assembly and evolution of the rice BGCs, as well as those in plants more generally, are then discussed.
Results
Co-expression analysis of genes for momilactone biosynthesis
Co-expression is the best indicator of shared roles of enzymes in plant specialized metabolism pathways and is sufficient to recover those that are known to be encoded by BGCs, including the two BCGs from rice, which were further found to be largely co-regulated (Wisecaver et al., 2017). Indeed, chitin induces the “momilactone” c4BGC and most of the genes in the c2BGC in a similar fashion, particularly including CYP76M5, CYP76M7, and CYP76M8, along with CYP76M14, as well as CYP701A8 and the closely related paralog CYP701A9, from outside the BGCs (Okada et al., 2007). Queries of the RiceXPro and RiceFREND databases further support co-expression of these genes (Sato et al., 2013a, 2013b), as well as CYP76M17, including in response to jasmonic acid (Supplemental Figure S1, A), which has been shown to induce momilactone production (Nojiri et al., 1996). Heavy metals have also been shown to elicit momilactone biosynthesis (Kodama et al., 1988). Investigation of the fast-growing cultivar (cv.) Kitaake used here, via a previously developed liquid chromatography–tandem mass spectrometry (LC–MS/MS) method focused on analysis of the known rice labdane-related diterpenoids (Lu et al., 2018), found that treatment with copper chloride (CuCl2) yielded greater amounts and numbers of these natural products than induction with methyl jasmonate. In addition, qRT-PCR analysis revealed that this CuCl2 treatment induced similar increases in the mRNA levels of CYP76M7 and CYP76M8 as seen with the genes from the “momilactone” c4BGC, as well as CYP701A8 and CYP701A9 (Supplemental Figure S1, B). Accordingly, co-expression analysis supports a role in momilactone biosynthesis for CYP701A8 and/or its closely related paralog CYP701A9, along with CYP76M7 and/or CYP76M8 from the c2BGC, as well as their closely related paralogs CYP76M14 and/or CYP76M17.
Biochemical evidence for a role of CYP76M8-related clade members in momilactone biosynthesis
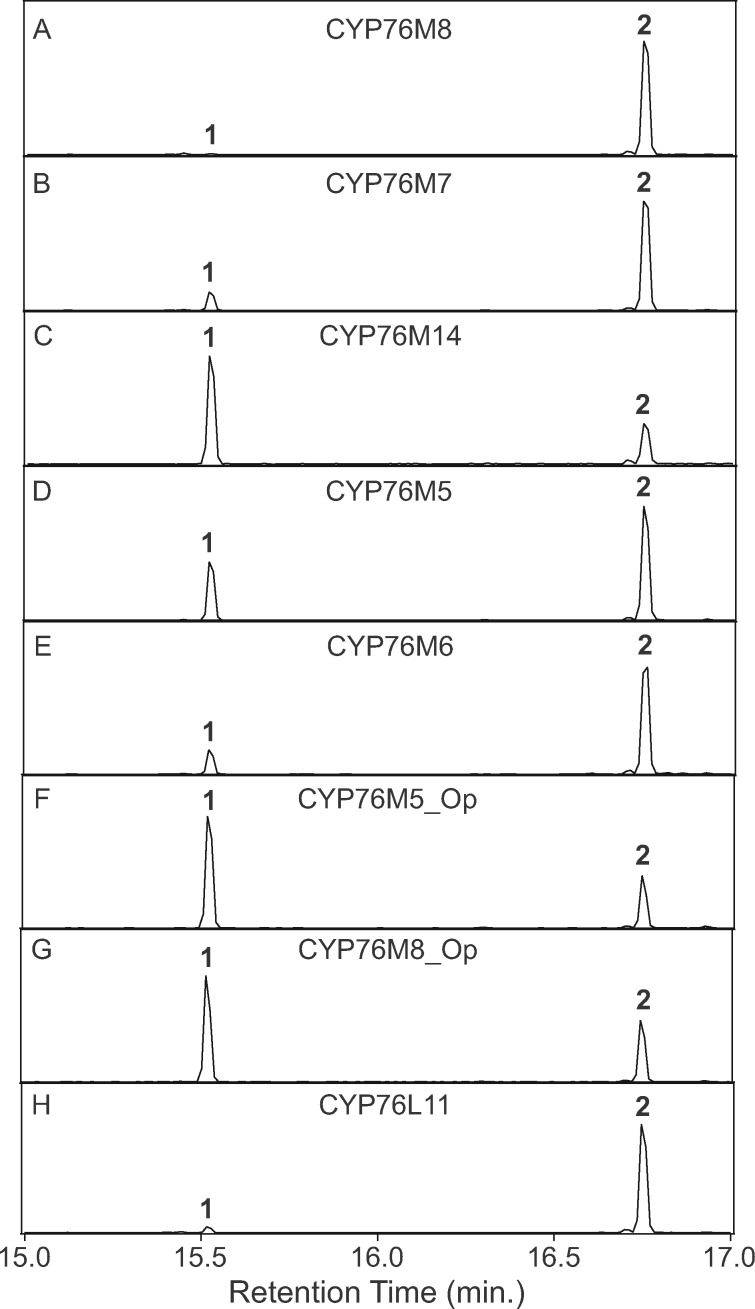
While a previous report suggested that CYP76M7 only reacts with ent-cassadiene and, hence, is dedicated to phytocassane biosynthesis, that work utilized the native gene for recombinant expression in Escherichia coli (Swaminathan et al., 2009). It has since been demonstrated that complete recoding to optimize codon usage for such bacterial expression can significantly increase the observed activity (Wang et al., 2011). Thus, it seemed possible that CYP76M7 might be able to catalyze the same 6β-hydroxylation of syn-pimaradiene (1) as CYP76M8, which would indicate potential redundancy between these enzymes for momilactone biosynthesis. This possibility was examined here using an optimized version of CYP76M7 in a synthetic biology approach (Kitaoka et al., 2015a) that relies on a previously developed modular metabolic engineering system for E. coli (Cyr et al., 2007). Specifically, the optimized CYP76M7 was co-expressed with a CYP reductase (CPR) in E. coli also engineered to produce 1 and the resulting product compared with that from CYP76M8 (Figure 2, A and B). Notably, this experiment revealed that CYP76M7 efficiently converts 1 to the same 6β-hydroxy-syn-pimaradiene (2) produced by CYP76M8.
Figure 2.
Analogous 6β-hydroxylase activity of the CYP76M clade, as well as CYP76L11, with syn-pimaradiene (1) to produce 6β-hydroxy-syn-pimaradiene (2). GC–MS extracted ion (m/z = 257, 273) chromatograms of extracts from E. coli engineered to produce 1 and co-expressing the indicated CYP76 family member, along with the requisite CPR redox partner. (A) CYP7M8. (B) CYP76M7. (C) CYP76M14. (D) CYP76M5. (E) CYP76M6. (F) CYP76M5_Op. (G) CYP76M8_Op. (H) CYP76L11.
Given the potential redundancy between CYP76M7 and CYP76M8 for momilactone biosynthesis, it seemed prudent to further investigate if the closely related CYP76M14 and/or CYP76M17 might also exhibit such activity. Accordingly, their ability to act on 1 was investigated via the same approach. Indeed, both enzymes also readily react with 1, albeit only CYP76M14 produces 2 (Figure 2, C), while CYP76M17 instead primarily produces an oxo-hydroxy derivative, along with smaller amounts of an oxo derivative (Supplemental Figure S2). Based on further optimization of recombinant expression of rice CYPs in bacteria (Kitaoka et al., 2015b), re-investigation of CYP76M5 and CYP76M6 revealed that these enzymes can also react with 1 to produce 2, albeit the more distantly related CYP76M5 does so rather inefficiently (Figure 2, D and E).
In addition, the wild-rice species O. punctata produces momilactones, but not phytocassanes, and, while the equivalent region to the rice c2BGC has lost most of constituent genes, it still contains orthologs of CYP76M5 and CYP76M8 (Miyamoto et al., 2016). To investigate the possibility that these genes (CYP76M5_Op and CYP76M8_Op) might be involved in momilactone biosynthesis in O. punctata, the ability of CYP76M5_Op and CYP76M8_Op to react with 1 also was assessed here. Indeed, both produced 2 (Figure 2, F and G). This finding further supports a potential role for CYP76M sub-family members from the rice c2BGC in momilactone biosynthesis.
Beyond the Oryza genus, barnyard grass (Echinochloa crus-galli), also a member of the Poaceae family but in the relatively distant Panicoideae sub-family, contains a BGC orthologous to the rice “momilactone” c4BGC, yet it also contains an additional CYP (Guo et al., 2017). Although this was originally suggested to be a member of the CYP76M sub-family, it actually has been assigned as CYP76L11 and, hence, falls within a separate CYP sub-family (Figure 1, C). Nevertheless, it has been hypothesized that this also might act as a 6β-hydroxylase in momilactone biosynthesis (Peters, 2020). This hypothesis was assessed here by investigating the ability of CYP76L11 to react with 1, demonstrating that it also efficiently produces 2 as well (Figure 2, H).
Distinct roles for CYP76M7 and CYP76M8
Although the previous RNAi investigation ruled out significant roles for CYP76M5 and CYP76M6 in momilactone biosynthesis (Ye et al., 2018), this still leaves the potential for genetic redundancy in this pathway. Particularly intriguing is the possibility that CYP76M7 and/or CYP76M8 from the c2BGC might play a role in momilactone production, which is otherwise largely encoded by the c4BGC. To determine the roles of CYP76M7 and CYP76M8 in rice labdane-related diterpenoid biosynthesis, these genes were targeted for inactivating mutagenesis by CRISPR-Cas9 using a previously described approach (Bi and Yang, 2017). Both genes were targeted in cv. Kitaake rice, and three homozygous mutant lines selected in the T1 generation for both genes, with each line containing mutations that lead to premature termination (Supplemental Figure S3). All experiments described hereafter were carried out using seeds derived from these lines (i.e. T2 or later generation plants). All of these cyp76m7 and cyp76m8 lines exhibited normal growth and development relative to parental/wild-type (WT) plants.
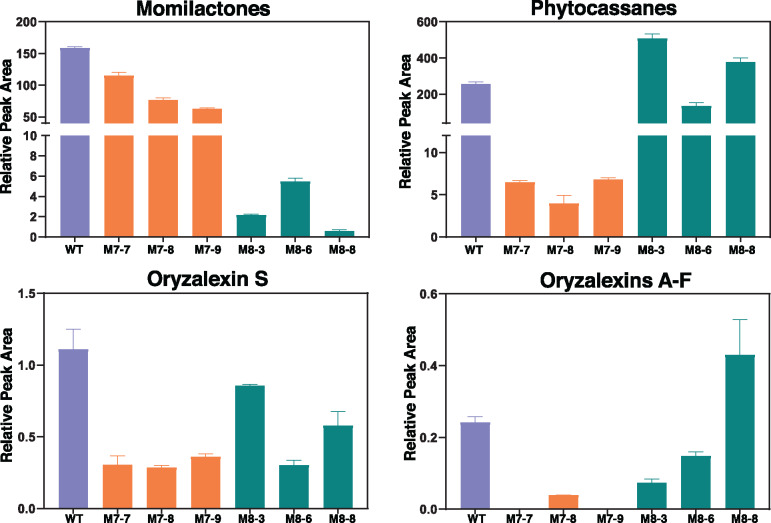
To investigate the effects of knocking-out these genes on rice labdane-related diterpenoid metabolism, the mutant lines were analyzed by LC–MS/MS. Strikingly, this targeted chemotypic analysis revealed distinct effects for each mutant, particularly on phytocassane and momilactone biosynthesis (Figure 3). Specifically, while phytocassane production was almost completely abolished in the cyp76m7 lines, these plants exhibited only moderate reductions in their accumulation of momilactones. By contrast, in the cyp76m8 lines, momilactone production was much more significantly reduced, but these plants had no uniform change in their accumulation of phytocassanes. In addition, knocking out these genes had effects on the production of other rice labdane-related diterpenoids. For example, the cyp76m7 lines also had severely reduced levels of oryzalexins (although only oryzalexins D and E were observed in cv. Kitaake), as well as moderate reductions in production of the structurally distinct oryzalexin S. The cyp76m8 lines exhibited similar moderate reductions in the accumulation of oryzalexin S but did not exhibit a uniform effect on the production of oryzalexins D and E.
Figure 3.
Effect of knocking-out CYP76M7 (M7) or CYP76M8 (M8) on labdane-related diterpenoid metabolism in rice relative to wild-type (WT) plants (average from three plants with error bars indicating standard deviation).
To verify that knocking-out CYP76M7 or CYP76M8 did not affect expression of the other genes involved in momilactone biosynthesis, their transcripts levels were analyzed by qRT-PCR in a selected line for each gene. In particular, expression of all the genes from the “momilactone” c4BGC and several from the c2BGC, including all those from the CYP76M sub-family, as well as CYP701A8 and the closely related CYP701A9, were examined. This analysis indicated that all the relevant genes were still induced by CuCl2 (Supplemental Figure S4). Therefore, the observed chemotypic effects can be attributed to just to the loss of the targeted gene and the biochemical role of the encoded cytochrome P450.
Characterization of CYP76M8 activity in momilactone biosynthesis
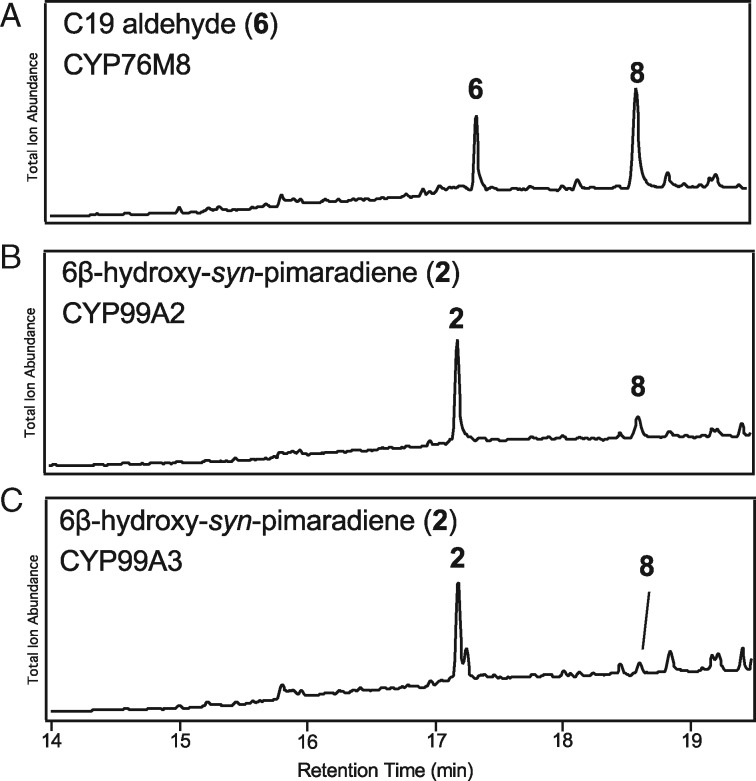
Given that CYP76M8 is particularly important for momilactone biosynthesis, its role was further explored using an extension of the synthetic biology approach described above. In particular, CYP76M8 was co-expressed with either CYP99A2, CYP99A3, or CYP701A8 (along with a CPR) in E. coli also engineered to produce 1. When co-expressed with CYP701A8, essentially only the CYP76M8 product 2 was observed, along with just trace amounts of the CYP701A8 product 3β-hydroxy-syn-pimaradiene (3) and small amounts of a novel product, presumably a diol based on the observed molecular ion of m/z = 304 (Supplemental Figure S5). The diol was verified to be 3β,6β-dihydroxy-syn-pimaradiene (4) by feeding 3 to CYP76M8 (with CPR) in cell-free assays to provide sufficient amounts to isolate for structural analysis by NMR spectroscopy (Supplemental Table S1 and Supplemental Figures S6–S12). However, feeding 4 to CYP99A2 or CYP99A3 in similar cell-free assays did not lead to any further transformation. Similarly, co-expression of CYP701A8 with either CYP99A2 or CYP99A3 in E. coli also engineered to produce syn-pimaradiene did not lead to any novel products.
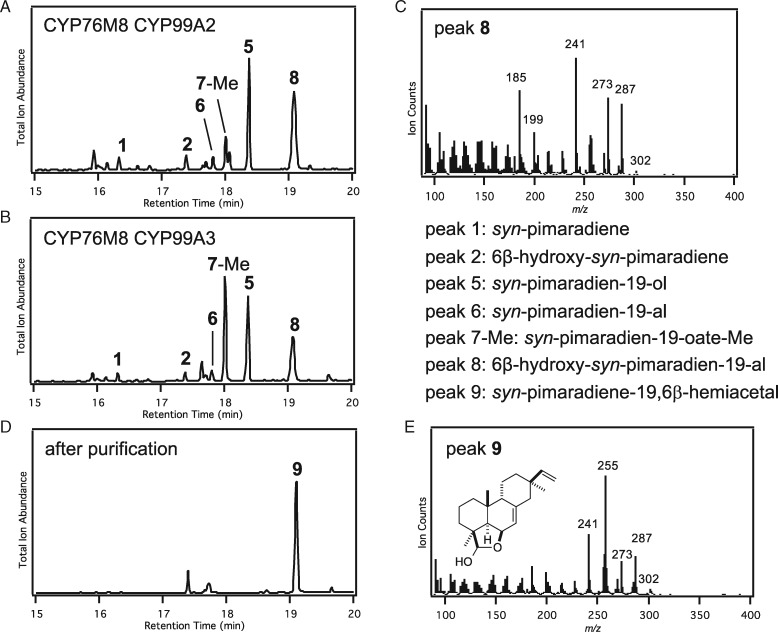
By contrast, co-expression of CYP76M8 with CYP99A2 or CYP99A3 led to observation of significant amounts of a novel product (i.e. in addition to 2 and the sequentially produced C19 alcohol (5), aldehyde (6), and acid (7) derivatives of 1 formed by CYP99A2/3; Figure 1). This novel product appeared to be an oxo-hydroxy derivative of 1 based on the apparent molecular ion of m/z = 302 (Figure 4, A–C). While verified to be the expected 6β-hydroxy-syn-pimaradien-19-al (8), this was only indirectly observed. In particular, during purification, 8 underwent spontaneous conversion to a different compound (9) that retained an apparent molecular ion of m/z = 302 (Figure 4, D and E). This derivative was determined by NMR structural analysis (Supplemental Table S2 and Supplemental Figures S13–S19) to contain a 19,6β-hemiacetal, stemming from intramolecular addition of the 6β-hydroxyl to the 19-aldehyde in 8.
Figure 4.
Coupled activity of CYP76M8 with CYP99A2 or CYP99A3. (A) and (B) GC–MS chromatogram of extract from E. coli engineered to produce syn-pimaradiene and co-expressing CYP76M8 and (A) CYP99A2 or (B) CYP99A3, along with the requisite CPR redox partner. (C) GC–MS chromatogram of novel product after purification. Peaks numbered as described in the text and shown in the figure (note that -Me indicates methylated carboxylate). (D) Mass spectrum of 8. (E) Mass spectrum of 9, along with the corresponding chemical structure, as determined by NMR.
To investigate the order in which these CYPs act, cell-free assays were carried out feeding either the CYP76M8 product 2 to CYP99A2 or CYP99A3, or the CYP99A2/3 aldehyde product 6 to CYP76M8 (Figure 5). While both of these potential intermediates could be converted to the further elaborated 8, CYP99A2 and CYP99A3 reacted relatively poorly with 2, as only small amounts of 8 were observed. By contrast, CYP76M8 reacted much more efficiently with 6, suggesting preferential biosynthesis via the production of 6 from 1 by CYP99A2/3, with subsequent transformation by CYP76M8 to 8, followed by heterocyclization to 9, which seems to occur spontaneously.
Figure 5.
Ordering CYP activity. GC–MS chromatograms from cell-free assays of (A) CYP76M8 with syn-pimaradien-19-al (6); (B) CYP99A2 or (C) CYP99A3 with 6β-hydroxy-syn-pimaradiene (2); with 6β-hydroxy-syn-pimaradien-19-al (8) product, as indicated.
Lactonization of hemiacetal intermediate by OsMS1/2
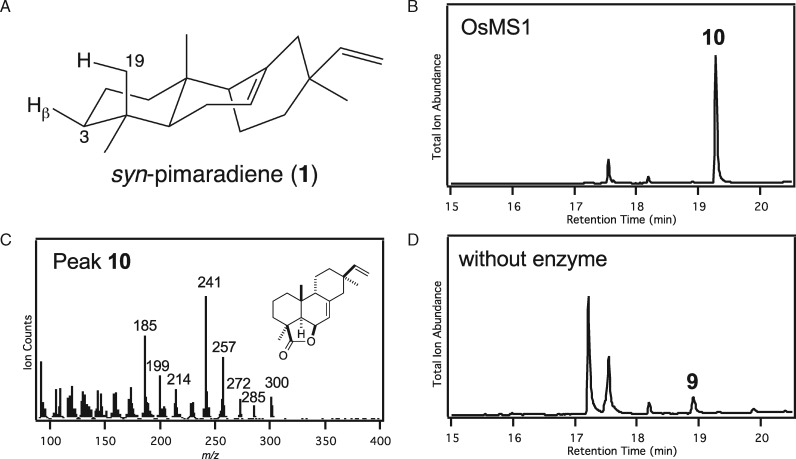
In order for 9 to serve as an intermediate in momilactone biosynthesis, its 19,6β-hemiacetal must be further oxidized to the eponymous lactone. Such oxidation is often catalyzed by SDRs. Not only OsMS1 and OsMS2, but also other members of the SDR110C sub-family in rice, exhibit inducible transcription (i.e. OsMS3, OsMI2, and OsMI3), with OsMS1–3 and OsMI3 exhibiting activity relevant to rice labdane-related diterpenoid phytoalexin biosynthesis (Kitaoka et al., 2016). Of particular relevance here, these latter four SDRs were all shown to oxidize the 3β-hydroxy of 3 to the corresponding 3-keto group, which is proximal to C19 (Figure 6, A), suggesting they might be able to oxidize the C19-hydroxy in the hemiacetal of 9. Accordingly, the ability of all five of these inducible rice SDR100C sub-family members to react with 9 was investigated here via in vitro reactions. Notably, OsMS1, OsMS2, and OsMS3 all reacted with 9, forming the same product in each case, which exhibited a molecular ion of m/z = 300, indicating oxidation (Figure 6, B–D). These assays could be readily scaled-up, enabling sufficient amounts of this product to be isolated for structural analysis by NMR (Supplemental Table S3 and Supplemental Figures S20–S26), which verified that it was the expected syn-pimaradien-19,6β-olide (10).
Figure 6.
SDR oxidation from hemiacetal to lactone. (A) Three-dimensional rendering of syn-pimaradiene (1) indicating the proximity of known 3β position targeted by SDRs and C19 requiring oxidation in syn-pimaradien-19,6β-hemiacetal (9). (B)–(D) GC–MS analysis of in vitro assays with 9. (B) Chromatogram of OsMS1 catalyzed oxidation to syn-pimaradien-19,6β-olide (10). (C) Mass spectrum of 10, along with the corresponding chemical structure, as determined by NMR. (D) Chromatogram of the negative control reaction (* indicates degradation products of 9 that are particularly prevalent following incubation in aqueous solution—e.g. these are present in much smaller amounts in the originally purified 9—see Figure 5, C).
Consistent with a role for 10 in rice momilactone metabolism, this lactone derivative can be found in rice hulls (Supplemental Figure S27), the tissue from which the momilactones were originally isolated (Kato et al., 1973). To further investigate the relevance of the observed activity, kinetic analysis was carried out (Table 1). Intriguingly, OsMS1 and OsMS2 exhibited considerably higher kinetic efficiency with the hemiacetal 9 than with their originally reported substrate 3β-hydroxy-syn-pimaradien-19,6β-olide (11). Nevertheless, OsMS1 exhibited substantially higher catalytic efficiency with 9 than 11, while OsMS2 reacted with 11 much more efficiently than did OsMS1. These results suggest emerging sub-functionalization of these two closely related SDRs, which seem to have arisen via recent tandem gene duplication from a presumably promiscuous precursor. Regardless, the high catalytic efficiency of OsMS1 and OsMS2 with 9, as well as the presence of the resulting 10 in rice, supports use of this hemiacetal-containing intermediate in momilactone biosynthesis.
Table 1.
OsSDR110C kinetic parameters with biosynthetic intermediates
| Substrate | SDR110C- | K M (µM) | k cat (s−1) | k cat/KM (s−1M−1) |
|---|---|---|---|---|
| syn-pimaradiene-19,6β-hemiacetal (9) | MS1 | 44 ± 17 | (1.9 ± 0.2) × 10−1 | 4 × 103 |
| MS2 | 12 ± 4 | (1.0 ± 0.1) × 10−1 | 8 × 103 | |
| MS3 | 420 ± 220 | (1.9 ± 0.1) × 10−1 | 5 × 102 | |
| 3β-hydroxy-syn-pimaradien-19,6β-olidea | MS1 | 900 ± 400 | (8 ± 3) × 10−2 | 9 × 101 |
| MS2 | 200 ± 100 | (4 ± 1) × 10−1 | 2 × 103 |
Data from previous report (Kitaoka et al., 2016).
CYP701A8 can convert syn-pimaradien-19,6 β-olide (10) to momilactone A
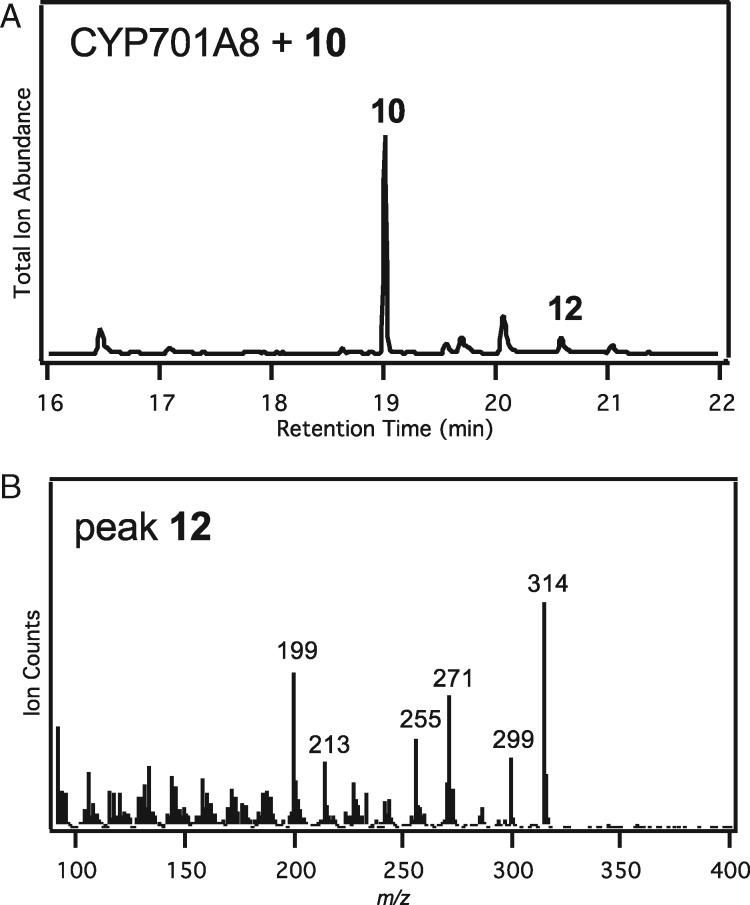
Given the already known ability of CYP701A8 to produce the C3β-hydroxy derivative of 1 (Kitaoka et al., 2015b), this was an obvious candidate to carry out equivalent hydroxylation of 10. Moreover, the proximity of C3β to C19 noted above (Figure 6, A) was used to first suggest the C3β-hydroxylation activity found with CYP701A8. In turn, this proximity was used here to suggest that, conversely, CYP99A2/3 might be able to carry out such hydroxylation with elaborated derivatives (i.e. in addition to acting on C19 with 1). Accordingly, the ability of all three of these CYPs to react with 10 was investigated via cell-free assays. Notably, while CYP99A2 and CYP99A3 were unable to further react with 10, CYP701A8 was able to produce small amounts of momilactone A (Figure 7), suggesting that it can further react beyond initial production of the C3β-hydroxy derivative to produce the characteristic C3-keto group of momilactone A (12).
Figure 7.
Production of momilactone A by CYP701A8. Verified by comparing retention time and mass spectra to an authentic standard. (A) GC–MS chromatogram from a cell-free assay of CYP701A8 with syn-pimaradien-19,6β-olide (10), with the production of momilactone A (12). (B) Mass spectrum of 12.
Discussion
The biochemical results reported here indicate that the eponymous lactone ring of the momilactones is produced by a somewhat extended biosynthetic scheme (Figure 8). In particular, rather than being more simply formed via dehydration of adjacent hydroxyl and carboxylic acid moieties, as has been indicated for other lactone-ring containing terpenoids (Ikezawa et al., 2011; Gou et al., 2018), the results here indicate initial formation of a hemiacetal, which then requires further oxidation. Indeed, both OsMS1 and OsMS2 encoded in the “momilactone” c4BGC react more efficiently with the hemiacetal containing intermediate 9 than the penultimate momilactone A precursor 11.
Figure 8.
Proposed momilactone A biosynthetic pathway. This pathway is initiated by the cyclization reactions catalyzed by OsCPS4 and OsKSL4 to form 1, with subsequent formation of the 19-aldehyde derivative 6 catalyzed by CYP99A2 or CYP99A3, then further 6β-hydroxylation catalyzed by CYP7M8 forming 8, followed by spontaneous formation of the 19,6β-hemiacetal 9, then oxidization to the 19,6β-lactone 10 catalyzed by OsMS1 or OsMS2, with the final 3β-hydroxylation catalyzed by CYP701A8, which may also catalyze further oxidation to the 3-keto containing momilactone A (12), a step that can also be catalyzed by OsMS2.
Perhaps more interestingly, the genetic evidence provided here demonstrates that CYP76M8 plays an important role in momilactone biosynthesis, suggesting interdependent evolution of the two BGCs in rice. This hypothesis is supported by the finding that the wild-rice species O. punctata, which produces momilactones but not phytocassanes, has an orthologous “momilactone” BGC and, while the equivalent region to the O. sativa c2BGC only has orthologs of CYP76M5 and CYP76M8 (Miyamoto et al., 2016), as shown here, both CYP76M5_Op and CYP76M8_Op catalyze the same C6β-hydroxylation of 1 as CYP76M8, providing equivalent functionality for momilactone biosynthesis.
Strikingly, despite falling within a separate plant sub-family, barnyard grass (E. crus-galli) contains a very similar “momilactone” BGC, with orthologs of OsCPS4, OsKSL4, and OsMS, along with a CYP99A sub-family member, but also CYP76L11 as well (Guo et al., 2017). The biochemical equivalence shown here between CYP76L11 and the rice CYP76M clade members suggests that this was “recruited” to serve the same function in momilactone biosynthesis as shown here for CYP76M8 in O. sativa. This further highlights the incomplete nature of the rice “momilactone” BGC, which clearly does not contain all the genes needed for even biosynthesis of the less elaborated momilactone A. Indeed, the contrast between the E. crus-galli and rice “momilactone” BGCs in recruitment of the relevant C6β-hydroxylase further supports interdependent evolution of the c2BGC with the “momilactone” c4BGC in rice.
Notably, the rice “momilactone” c4BGC is not only incomplete but also fractured, needing CYP76M8 to operate on the syn-pimaradien-19-al (6) produced by the OsCPS4, OsKSL4, and CYP99A2/3 all encoded within the “momilactone” c4BGC before OsMS1/2 (also encoded within this c4BGC) can act on the resulting hemiacetal 9. Moreover, to the extent that OsMS2 is important for the final oxidation reaction that forms the characteristic keto moiety, the rice “momilactone” c4BGC is further fractured by the need for CYP701A8 to first carry out the preceding hydroxylation at C3β. This contrasts with other plant BGCs shown to be incomplete, such as various triterpenoid BGCs in Arabidopsis thaliana, where peripheral genes encoding promiscuous enzymes that function in multiple biosynthetic pathways and act after those encoded by the genes in the relevant BGC are also required (Huang et al., 2019).
The incomplete and fractured nature of the rice “momilactone” c4BGC provides an informative contrast to the generally self-contained nature of microbial BGCs. This presumably reflects the vertical nature of genetic transmission in plants versus the more frequent horizontal gene transfer that occurs in microbes. Hence, in plants, the inheritance of more specialized metabolic pathways, such that for the momilactones, can be assured by genetic linkage (e.g., physical proximity) not only of the relevant genes to each other but, alternatively, to other essential genes. The latter presumably explains the absence of CYP701A8 in the rice “momilactone” BGC, as this is a paralog of CYP701A6, encoding the ent-kaurene oxidase required for gibberellin biosynthesis, and these are found in a tightly linked tandem array composed of all five paralogs in rice (Wang et al., 2012a). Such CYP701A gene duplication and diversion of a paralog to more specialized metabolism seems to be widespread in the Poaceae (Ding et al., 2019), which presumably explains the lack of recruitment of these genes to the “momilactone” BGCs in this plant family. Indeed, it is possible that the CYP701A sub-family member that acts as the requisite ent-kaurene oxidase for gibberellin biosynthesis might serve this function, as CYP701A3 from Arabidopsis can also can catalyze C3β-hydroxylation of 1 (Mafu et al., 2016).
On the other hand, the results reported here support interdependent evolution of the two unlinked BGCs in rice. Based on extensive phylogenomic analysis, it has been suggested that the “phytocassane” c2BGC was assembled first, with subsequent assembly of the “momilactone” c4BGC (Miyamoto et al., 2016). This order of appearance may underlie the lack of recruitment of CYP76M8 from the “phytocassane” c2BGC to the “momilactone” c4BGC. In particular, CYP76M8 seems to have arisen via tandem gene duplication, specifically along with the neighboring CYP76M7, which is essential for phytocassane biosynthesis. This suggests that the ancestral CYP76M sub-family member was recruited to the “phytocassane” c2BGC for its role in the biosynthesis of these labdane-related diterpenoid phytoalexins and then served a promiscuous “moon-lighting” role in momilactone biosynthesis during the evolution of this pathway. However, inclusion of the subsequent duplicate that became more dedicated to momilactone metabolism in this original “phytocassane” c2BGC seems to have provided sufficient selective pressure for its retention to alleviate any need to recruit this gene to the emerging “momilactone” c4BGC. Loss of the remainder of the “phytocassane” c2BGC in the O. punctata lineage may have occurred recently enough that there simply may not have been sufficient time to recruit the CYP76M ortholog to the “momilactone” BGC. By contrast, the orthologous “momilactone” BGC in E. crus-galli, which has been suggested to have been acquired via hybridization and introgression (Peters, 2020), evolved in the absence of a “phytocassane” BGC. This is then consistent with its recruitment of CYP76L11 to serve a similar role as CYP76M8 in momilactone biosynthesis. However, it must be noted that the recently uncovered “momilactone” BGC in the early diverging bryophyte Calohypnum plumiforme independently evolved and also does not seem to contain a 6β-hydroxylase (Mao et al., 2020). This has led to speculation that phytotoxicity of certain intermediates provided negative selection pressure that drove the assembly of these “momilactone” BGCs (Zhang and Peters, 2020). Regardless, the use of CYP76M8 for momilactone biosynthesis in both O. sativa and O. punctata demonstrates interdependent evolution of the “momilactone” c4BGC with the pre-existing “phytocassane” c2BGC in Oryza.
In conclusion, the formal elucidation of momilactone A biosynthesis reported here not only clarifies formation of the eponymous lactone ring, but also the incomplete and fractured nature of the “momilactone” c4BGC, which provides some insight into the evolutionary pressures that shape plant BGCs. While frequent horizontal gene transfer has led to the generally self-contained nature of microbial BGCs, plants are essentially limited to vertical genetic transmission, alleviating the strict need for such completeness per se. Indeed, there appear to be two additional genetic loci beyond the “momilactone” c4BGC that are required for biosynthesis of these labdane-related diterpenoids in rice. As discussed above, one is the tandem array of CYP701A sub-family members, with co-inheritance of the divergent CYP701A8 functioning in such more specialized metabolism ensured by its linkage to the essential ent-kaurene oxidase gene CYP701A6 required for gibberellin biosynthesis. The other is the earlier assembled, more complex “phytocassane” c2BGC, demonstration of which reveals interdependent evolution of plant BGCs. Accordingly, this study highlights the distinct evolutionary constraints on plant BGC assembly, which should be useful in future studies on the intriguing effect more specialized metabolism can have on genome organization.
Materials and methods
General
Due to previous usage based on physical proximity within the c2BGC, i.e. to CYP76M5, CYP76M6, and CYP76M8 (Swaminathan et al., 2009), here CYP76M7 corresponds to Os02g056990 and CYP76M17 to Os06g0599200, which is the reverse of their official assignments. Unless otherwise noted, chemicals were purchased from Fisher Scientific and molecular biology reagents from Invitrogen. The rice (O. sativa) plants used here are all from the sub-species japonica, specifically cv. Kitaake, grown as previously described (Zhang et al., 2020).
GC analyses were performed with a Varian 3900 GC with Saturn 2100 ion trap MS in electron ionization (70 eV) mode for GC–MS analyses, or with an Agilent 6890N GC with flame-ionization detection (FID) for GC–FID analyses. Samples (5 μL) were injected in splitless mode at 50°C and, after holding for 3 min at 50°C, the oven temperature was increased at a rate of 14°C/min to 300°C, where it was held for an additional 3 min. MS data from 90 to 600 m/z were collected starting at 12 min after injection until the end of the run.
HPLC was carried out with an Agilent 1100 system equipped with an auto-sampler, fraction collector, and diode-array detector, run in reversed phase at 0.5 mL/min with a ZORBAX Eclipse XDB-C8 column (150 × 4.6 mm, 5 µm), using deionized water and acetonitrile. The column was preequilibrated with 50% acetonitrile/water (0–2 min) and eluted with 50%–100% acetonitrile gradient (2–30 min), followed by a 100% acetonitrile wash (30–45 min).
NMR spectra were recorded at 25°C in deuterated chloroform (CDCl3) or deuterated benzene (C6D6). NMR spectra were collected using a Bruker Avance 700 spectrometer equipped with a 5-mm HCN cryogenic probe. Structural analysis was carried out using 1D 1H, double-quantum-filtered correlation spectroscopy (DQF-COSY), heteronuclear single-quantum coherence (HSQC), heteronuclear multiple bond correlation (HMBC), and nuclear overhauser effect correlated spectroscopy (NOESY) spectra acquired at 700 MHz and 1D 13C spectra acquired at 174 MHz using standard experiments from Bruker TopSpin v1.3 software. Chemical shifts were referenced using known deuterated chloroform (CDCl3; 13C 77.23 ppm, 1H 7.24 ppm) or deuterated benzene (C6D6; 13C 128.0 ppm, 1H 7.15 ppm) signals offset from tetramethylsilane (TMS).
Phylogenetic analysis
The phylogenetic analysis was based on the amino acid sequences of the rice CYP76 family members, as found in GenBank (see Accession Numbers). The family members from O. punctata and E. crus-galli were translated from the coding sequences found in the RiceRelativesGD database (Mao et al., 2019); specifically CYP76M5_Op (Opunc02g19190), CYP76M8_Op (Opunc02g19200), and CYP76L11 (scaffold290.14). In addition to CYP76L11, CYP76L1 and CYP76C2 were included as outgroup sequences for the CYP76M sub-family. Also included were the other rice CYP that might play a role in momilactone biosynthesis as described in the text. These protein sequences were aligned with CLC Main Workbench 20.0.4 (Qiagen) using a gap open cost = 10.0, gap extension cost = 1.0, and end gap cost = as any other, in the very accurate alignment mode. The presented phylogenetic tree (Figure 1, C) was generated from this alignment using the Maximum-Likelihood algorithm via the PhyML 3.0 webserver (www.atgc-montpellier.fr/phyml/) with automatic model selection by SMS, no starting tree, NNI tree improvement, and 100 replicate bootstrapping (Guindon et al., 2010; Lefort et al., 2017). The tree was rooted above the separation of the CYP76 family from the other CYP families presented here and ordered to present the CYP76M clade at the top.
Recombinant constructs
For recombinant expression in E. coli, the CYP76M5, CYP76M8, CYP76M14, CYP76M17, CYP99A2, CYP99A3, and CYP701A8 constructs used in this study were the synthetic fully codon-optimized and N-terminally modified constructs described previously (Wang et al., 2011, 2012a, 2012b). Analogous constructs were synthesized for CYP76M6, CYP76M7, CYP76M17, CYP76M5_Op, CYP76M8_Op, and CYP76L11 (Supplemental Figures S28–S33). These genes were closed into the second multiple cloning site (MCS), using the NdeI and XhoI restriction sites, of a pETDuet-1 (Novagen) vector into which a Gateway (Invitrogen) DEST cassette had been inserted into the first MCS, as previously described (Cyr et al., 2007), and a CPR from Arabidopsis thaliana (AtCPR1) installed (Kitaoka et al., 2015b). To enable dual CYP expression, the base pETDuet-1/DEST was used. CYP99A2 or CYP99A3 were inserted into the second MCS of pETDuet-1/DEST using the NdeI and XhoI restriction sites, and CYP76M8 was inserted via directional recombination into the DEST cassette, creating pETDuet-1/DEST::CYP76M8/(CYP99A2 or CYP99A3). For dual expression of CYP701A8 and CYP76M8, the pETDuet-1/CYP701A8/CYP76M8 construct was created by sub-cloning CYP701A8 into the first MCS using the NcoI and BamHI restriction sites, while CYP76M8 was sub-cloned into the second MCS using the NdeI and XhoI restriction sites. For in vitro cell-free assays, the C41 strain was transformed with pETDuet-1/DEST:AtCPR1/(CYP76M8, CYP99A2, CYP99A3, or CYP701A8), or pCDFDuet-1/DEST::AtCPR1 (as a negative control), all of which were previously described (Kitaoka et al., 2015b). The SDR expression constructs also were those previously described (Kitaoka et al., 2016,).
Pathway reconstruction via metabolic engineering
To screen for CYP diterpenoid production in our metabolic engineering system, genes for the relevant pathway were transformed into the C41 OverExpress strain of E. coli (Lucigen). Thus, the relevant CYPs were co-expressed using one of the pETDuet-1/DEST::CYP/CYP vectors and pETDuet-1/CYP/CYP vectors described above, with a GGPP synthase and OsCPS4 carried on the compatible pGGsC (Cyr et al., 2007), as well as OsKSL4 and AtCPR1 carried on the further compatible pCDFDuet1/DEST::OsKSL4/AtCPR1 (Kitaoka et al., 2015b). The resulting recombinant bacteria were cultured in 5-mL Terrific Broth (TB) medium (pH 7.0) containing the appropriate antibiotics at 37°C overnight. These cultures (2 mL) were used to inoculate TB medium (pH 7.5, 50 mL in 250-mL Fernbach flasks), and also shaken at 37°C until their OD600 reached 0.6–0.8. The cultures were then shifted to 16°C, supplemented with 5 mg/L riboflavin and 75 mg/L 5-aminolevulinic acid, and induced with 1-mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) after 1 h. After 72 h, the resulting diterpenoids were extracted with an equal volume of 10% ethyl acetate/n-hexanes. The organic extract was separated, dried under a gentle stream of N2 gas, and dissolved in 0.1-mL n-hexanes for GC–MS analysis (1-μL injection volumes). For the samples where CYP99A2 or CYP99A3 were co-expressed, the extract was methylated with diazomethane before GC–MS analysis.
Cell-free lysate preparation expressing CYPs and in vitro enzyme assay
CYP preparations were carried out essentially as previously described (Kitaoka et al., 2015b). Briefly, the CYPs were co-expressed with AtCPR1 using one of the various CYP expression vectors described above, which were generally co-transformed with the compatible pCDFDuet-1/AtCPR1 vector into the C43 OverExpress strain of E. coli (Lucigen). The resulting recombinant bacteria were cultured in 5-mL TB medium (pH 7.5) containing the appropriate antibiotics at 37°C overnight. These cultures (4 mL) were used to inoculate TB medium (pH 7.0, 4 × 100 mL in 250-mL Fernbach flasks), and also shaken at 37°C until their OD600 reached 0.6–0.8. The cultures were then shifted to 16°C, supplemented with 5 mg/L riboflavin and 75 mg/L 5-aminolevulinic acid, and induced with 1-mM IPTG after 1 h. After 72 h, the cells were harvested via centrifugation (10 min 5,000 × g), resuspended in 30 mL buffer (0.1 M Tris/HCl, pH 7.5, 0.5 mM EDTA, 20% glycerol), and passed twice through a French press homogenizer (Emulsiflex-C5: Avestin) at 1,500 psi. The resulting lysates were clarified via centrifugation (20 min 14,000 × g), and the concentration of CYP in the resulting supernatant (cell-free lysate) was quantified by carbon monoxide (CO)-binding difference spectra using the standard extinction coefficient of 91 mM−1 cm−1 (Omura and Sato, 1964).
CYP reactions were carried out in 1 mL buffer (0.1 M Tris/HCl, pH 7.5, 0.5 mM EDTA, 20% glycerol) containing 0.2 mM NADPH, 10 μM substrate, and cell-free lysate from recombinant cultures containing 10 nM CYP. The assays were initiated by adding substrate and incubated at 32°C for 30 min. The reactions were halted by incubation at 90°C for 5 min. Reaction product was extracted with 10% ethyl acetate/n-hexanes (3 × 2 mL). The organic extracts were separated, combined, dried under a gentle stream of N2 gas, and the residue dissolved in 100-μL n-hexanes for GC–MS analysis.
Isolation and structural analysis of 3β,6β-dihydroxy-syn-pimaradiene (4)
To obtain the novel derivative of syn-pimaradiene (1) produced by CYP76M8 and CYP701A8, in vitro assays were carried out with 3β-hydroxy-syn-pimaradiene (3), produced via metabolic engineering with CYP701A8 as previously described (Kitaoka et al., 2015b), to 30 mL of cell-free lysate containing 60-nM CYP76M8 (and AtCPR1) in the presence of 67 µM NADPH. After incubation at 32°C overnight, the reaction product was extracted twice with equal volumes of 10% ethyl acetate/n-hexanes. These organic extracts were pooled, dried by rotary evaporation, and the resulting residue dissolved in water. The resulting solution was applied to a Bond Elut C18 cartridge column (1 g, Agilent), and 3β,6β-dihydroxy-syn-pimaradiene (4) was eluted with 80% acetonitrile. After evaporation, 4 was purified via HPLC. Fractions containing 4 were identified by GC–MS analysis, pooled, and dried under N2 gas. The purified compound was dissolved in 0.5-mL C6D6 and placed in an NMR microtube for NMR analysis (Supplemental Table S1 and Supplemental Figures S5–S11). Notably, the COSY correlations of H-5 (δH 0.99) and H-7 (δH 5.27) with H-6 (δH 4.31) suggested the presence of a hydroxyl group at C-6. NOESY spectra provided a Nuclear Overhauser Effect (NOE) cross-peak signal between H-6 (δH 4.31) and H-5 (δH 0.99) to assign the stereochemistry at C-6.
Isolation and structural analysis of syn-pimaradiene-19,6β-hemiacetal (9)
To obtain the novel derivative of 1 produced by CYP76M8 and CYP99A3, the volumes of the corresponding recombinant cultures were increased (8 × 500 mL in 2.8-L Fernbach flasks). After fermentation as described above, these cultures were extracted twice with equal volumes of 10% ethyl acetate/n-hexanes and the organic extracts separated, combined, and dried by rotary evaporation. The resulting residue was dissolved in 5 mL of n-hexanes and fractioned via flash chromatography over a 4-g silica column using a Reveleris system with UV detection and automated fraction collector (Grace), with an n-hexanes to acetone step gradient (100% n-hexanes, 90% n-hexanes/acetone, and 80% n-hexanes/acetone) as the mobile phase. syn-Pimaradiene-19,6β-hemiacetal (9) eluted in the 80% n-hexanes/acetone fraction. Further purification was carried out using HPLC. Fractions containing 9 were identified by GC–MS analysis, pooled, and dried under N2 gas. The purified compound was dissolved in 0.5-mL CDCl3 and placed in an NMR microtube (Shigemi) for NMR analysis. Connections between protonated carbons were obtained from DQF-COSY, and correlations from the HMBC spectra were used to complete the partial structure (Supplemental Table S2 and Supplemental Figure S12–S18). Notably, the presence of a hemiacetal ring was indicated by the HMBC correlation of H-19 (δH 5.34) with C-6 (δC 73.9). The stereochemistry at C-6 was assigned based on the NOE cross-peak signal between H-6 (δH 5.34) and H-18 (δH 5.34).
Enzymatic assays for SDRs
The recombinant SDRs (OsMS1, OsMS2, OsMS3, OsMI2, and OsMI3) were prepared as previous reported (Kitaoka et al., 2016). Initial assays were conducted with 0.5 µM SDR, 50 µM substrate, and 1 mM NAD+ in 0.5 mL Tris–HCl buffer (100-mM Tris–HCl, pH 8.0). After incubating at 30°C for 1 h, the reaction mixture was extracted three times with 0.5 mL 10% ethyl acetate/n-hexanes. The organic extracts were combined, dried under N2 gas, and resuspended in 100 µL n-hexanes for analysis by GC–MS.
Kinetic analysis was carried out using 10–40 nM recombinant SDR in 0.5 mL assays run for 5–10 min at 30°C, using the conditions determined by preliminary analyses to fall within the linear response range. The oxygenated diterpene substrates were added in varying concentrations (2–300 µM). To stop the reaction, assay vials were placed on ice and the enzymatic products immediately extracted with 10% ethyl acetate/n-hexanes as described above, and turnover quantified by analysis with GC-FID.
Isolation and structural analysis of syn-pimaradien-19,6β-olide (10)
The metabolic engineering system was employed to obtain 0.7 mg of syn-pimaradiene-19,6β-hemiacetal (9), which was then oxidized by OsMS1 in vitro. This reaction was conducted with 0.5 µM OsMS1 and 1 mM NAD+ in 15-mL Tris–HCl buffer (100 mM Tris–HCl, pH 8.0). After incubating at 30°C for 3 h, the reaction mixture was extracted three times with an equal volume of n-hexanes. The organic extracts were separated, combined, dried under N2 gas, and the residue dissolved in 50% acetonitrile/H2O for purification by HPLC. Fractions containing syn-pimaradien-19,6β-olide (10) were identified by GC–MS analysis, pooled, and dried under N2 gas. Purified 10 was dissolved in 0.5-mL CDCl3 and placed in an NMR microtube (Shigemi) for NMR analysis. Connections between protonated carbons were obtained from DQF-COSY, and correlations from the HMBC spectra were used to complete the partial structure (Supplemental Table S3 and Supplemental Figures S19–S25). Notably, the presence of the lactone ring was indicated by the HMBC correlation of H-6 (δH 4.85) with C-19 (δC 183.0).
Identification of syn-pimaradien-19,6β-olide (10) in planta
Rice hulls (ca. 100 g) were extracted with methanol (1 L) by stirring overnight at room temperature. After filtration, the crude methanol extract was evaporated under reduced pressure. The residue was purified by silica gel column chromatography with step gradient (10% ethyl acetate/n-hexanes 100 mL, 20% ethyl acetate/n-hexanes 100 mL, and 40% ethyl acetate/n-hexanes 100 mL) as the mobile phase. The 40% ethyl actate/n-hexanes fraction was evaporated under reduced pressure, resuspended in 1 mL n-hexanes, and analyzed by GC–MS.
Construction of CRISPR/Cas9 vectors
The CRISPR/Cas9 vectors were constructed basically as previously described (Zhou et al., 2014). Briefly, guide RNA (gRNA) was assembled from oligonucleotides (Supplemental Table S4), with two selected for each target and incorporated into the BtgZI and BsaI cloning sites in the intermediate vector pENTR-gRNA. The resulting vectors were confirmed by sequencing of the gRNA inserts, which were then transferred to pBY2-Cas9 using Gateway LR Clonase. Escherichia coli strain XL1-Blue was used for molecular cloning and Agrobacterium tumefaciens strain EHA105 for transformation of the japonica rice cv. Kitaake, which was carried out by the plant transformation facility at Iowa State University, as previously described (Toki, 1997).
Genotyping
Genomic DNA was extracted from leaves of each individual T0 transgenic rice seedling using Plant DNAzol Reagent (Invitrogen). The relevant regions were amplified by PCR with specific primers flanking the target sites (Supplemental Table S4), and the reactions treated with ExoSAP-IT (Affymetrix, Santa Clara, CA, USA) prior to sequencing. The sequencing chromatograms were carefully examined for patterns indicating heterozygosity, as well as to identify homozygous mutations. For heterozygous/diallelic mutations, T1 generation plants were also genotyped. All homozygous mutant lines were also screened by PCR to select for loss of the CRISPR/Cas9-2gRNA construct, with T3 plants from homozygous lines used in the following experiments (genotyping data can be found in Supplemental Figure S3). Of the plants analyzed here, all the cyp76m8 lines, although only cyp76m7-7 of the cyp76m7 lines, were CRISPR/Cas9-2gRNA free.
Expression analysis
Total RNA was isolated from the samples using an RNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA). To remove gDNA, a Turbo DNA-free kit (Thermo Scientific) was used. Reverse transcription (RT) was performed using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific). For qRT-PCR analysis, a Step OnePlus thermocycler was employed, Power UpTM SYBR Green Master Mix (Thermo Scientific) was used. The Primer Express program 3.0 (Applied Biosystems, Foster City, CA, USA) was used to design the primers for the genes chosen (Supplemental Table S5), and the ACTIN gene (LOC_Os03g61970) was chosen as an internal control to normalize the data. The thermal cycling conditions were 30 s at 95°C, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s, with melting curves then analyzed over 60–95°C. Each sample was analyzed in triplicate, with the relative quantification method used to evaluate quantitative variation between these technical replicates.
Chemotyping
Leaves from 3-week-old rice plants were cut into 5-cm pieces, weighed, and trimmed to 0.1 g. These sections were induced by floating on water with 0.5 mM CuCl2. After 72 h, the leaf tissues were frozen in liquid N2 and ground into a fine power, which was extracted by shaking in 3 mL methanol for 72 h in the cold room. Sclareol (2.31 µg) was added to each sample as an internal standard. The methanol extract was filtered (0.2-µM nylon filter; Thermo) using a glass syringe, dried under N2, and resuspended in 1 mL methanol.
These clarified extracts were subjected to LC–MS/MS analysis, with 15 µL injections run over a Supelco (Sigma–Aldrich, St. Louis, MO, USA) Ascentis C18 column (10 cm × 2.1 mm; 3 μm) using an Agilent Technologies 1100 Series HPLC system coupled to both a UV–Vis diode array detector and an Agilent Technologies Mass Selective Trap SL detector located in the Iowa State University W.M. Keck Metabolomics Research Laboratory. A binary gradient was used, consisting of water with 0.1% (v/v) acetic acid (buffer A) and acetonitrile with 0.1% (v/v) acetic acid (buffer B). The solvent gradient elution was programmed as follows: initial condition, 40% buffer B; 0–13 min, a linear gradient from 40% buffer B to 85% buffer B; 13–14 min, a linear gradient from 85% buffer B to 100% buffer B; 14–14.5 min, a linear gradient from 100% buffer B to 40% buffer B. The rice labdane-related diterpenoids were identified based on comparison to authentic standards, matching both retention times and the calculated monoisotopic mass of the molecular ion for each [M + H]+ with an isolation selection window of ±0.5 m/z. The isolated masses were reionized and the resulting mass spectra recorded, with quantification based on selected secondary ions as previously described (Lu et al., 2018), using the quantAnalysis function of the 6300 Series Ion Trap LC–MS software package (version 1.8, Bruker).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: CYP76M1 (XP_015650125), CYP76M2 (XP_015648069), CYP76M5 (XP_015624125), CYP76M6 (XP_015625955), CYP76M7 (XP_015623593), CYP76M8 (XP_015623630), CYP76M9 (XP_015641878), CYP76M10 (XP_015648813), CYP76M13 (XP_015619510), CYP76M14 (XP_015628496), CYP76M17 (XP_015641474), CYP76L1 (XP_015610661), CYP76L1 (XP_015610661), CYP76C2 (NP_182081), CYP99A2 (XP_015633968), CYP99A3 (XP_015634021), CYP701A6 (XP_015643248), CYP701A8 (AAT46567), and CYP701A9 (XP_015641629).
Supplemental data
Supplemental Figure S1. Co-expression analysis of genes from the rice BGCs and other CYP genes.
Supplemental Figure S2. Divergent activity of CYP76M17 with syn-pimaradiene (1).
Supplemental Figure S3. Genotyping alignments.
Supplemental Figure S4. CuCl2 induction of genes potentially involved in momilactone biosynthesis in cyp76m7 or cyp76m8 mutant plants.
Supplemental Figure S5. Dihydroxylation catalyzed by CYP76M8 and CYP701A8.
Supplemental Figure S6. Schematic summary of NMR structural analysis of 3β,6β-dihydroxy-syn-pimaradiene (4).
Supplemental Figure S7. 1H spectrum of 3β,6β-dihydroxy-syn-pimaradiene (4).
Supplemental Figure S8. 13C spectrum of 3β,6β-dihydroxy-syn-pimaradiene (4).
Supplemental Figure S9. COSY spectrum of 3β,6β-dihydroxy-syn-pimaradiene (4).
Supplemental Figure S10. HSQC spectrum of 3β,6β-dihydroxy-syn-pimaradiene (4).
Supplemental Figure S11. HMBC spectrum of 3β,6β-dihydroxy-syn-pimaradiene (4).
Supplemental Figure S12. NOESY spectrum of 3β,6β-dihydroxy-syn-pimaradiene (4).
Supplemental Figure S13. Schematic summary of NMR structural analysis of syn-pimaradiene-19,6β-hemiacetal (9).
Supplemental Figure S14. 1H spectrum of syn-pimaradiene-19,6β-hemiacetal (9).
Supplemental Figure S15. 13C spectrum of syn-pimaradiene-19,6β-hemiacetal (9).
Supplemental Figure S16. COSY spectrum of syn-pimaradiene-19,6β-hemiacetal (9).
Supplemental Figure S17. HSQC spectrum of syn-pimaradiene-19,6β-hemiacetal (9).
Supplemental Figure S18. HMBC spectrum of syn-pimaradiene-19,6β-hemiacetal (9).
Supplemental Figure S19. NOESY spectrum of syn-pimaradiene-19,6β-hemiacetal (9).
Supplemental Figure S20. Schematic summary of NMR structural analysis of syn-pimaradien-19,6β-olide (10).
Supplemental Figure S21. 1H spectrum of syn-pimaradien-19,6β-olide (10).
Supplemental Figure S22. 13C spectrum of syn-pimaradien-19,6β-olide (10).
Supplemental Figure S23. COSY spectrum of syn-pimaradien-19,6β-olide (10).
Supplemental Figure S24. HSQC spectrum of syn-pimaradien-19,6β-olide (10).
Supplemental Figure S25. HMBC spectrum of syn-pimaradien-19,6β-olide (10).
Supplemental Figure S26. NOESY spectrum of syn-pimaradien-19,6β-olide (10).
Supplemental Figure S27. Presence of syn-pimaradien-19,6β-olide (10) in planta.
Supplemental Figure S28. Sequence of synthetic CYP76M6.
Supplemental Figure S29. Sequence of synthetic CYP76M7.
Supplemental Figure S30. Sequence of synthetic CYP76M17.
Supplemental Figure S31. Sequence of synthetic CYP76M5_Op.
Supplemental Figure S32. Sequence of synthetic CYP76M8_Op.
Supplemental Figure S33. Sequence of synthetic CYP76L11.
Supplemental Table S1. NMR data for 3β,6β-dihydroxy-syn-pimaradiene (4) (C6D6)
Supplemental Table S2. NMR data for syn-pimaradiene-19,6β-hemiacetal (9) (CDCl3)
Supplemental Table S3. NMR data for syn-pimaradien-19,6β-olide (10) (CDCl3)
Supplemental Table S4. Cloning and genotyping primers
Supplemental Table S5. qRT-PCR primers
Supplementary Material
Acknowledgments
The authors thank Prof. David Nelson (Univ. Tenn.) for the assignment of CYP76L11.
Funding
This work was support by grants from the United States Department of Agriculture - National Institute of Food and Agriculture (grant no. 2020-67013-32557 to R.J.P. and B.Y.) and the National Institutes of Health (grant no. GM131885 to R.J.P.), as well as a fellowship from the International Postdoctoral Exchange Fellowship Program (grant no. 20170057 to J.Z.).
Conflict of interest statement. R.J.P. is a member of the scientific advisory board of Manus Bio, Inc.
N.K., J.Z., R.K.O., B.B., and Y.W. were involved in designing and carrying out experiments. B.Y., Z.L., and R.J.P. were involved in conceptual design and obtaining financial support. N.K., J.Z., and R.J.P. were primarily responsible for writing the manuscript and was responsible for distribution of materials.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Zhaohu Li (lizhaohu@cau.edu.cn) and Reuben J. Peters (rjpeters@iastate.edu).
References
- Bi H, Yang B (2017) Gene editing with TALEN and CRISPR/Cas in rice. Prog Mol Biol Transl Sci 149: 81–98 [DOI] [PubMed] [Google Scholar]
- Cho E-M, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, et al. (2004) Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J 37: 1–8 [DOI] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc 129: 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Murphy KM, Poretsky E, Mafu S, Yang B, Char SN, Christensen SA, Saldivar E, Wu M, Wang Q, et al. (2019) Multiple genes recruited from hormone pathways partition maize diterpenoid defences. Nat Plants 5: 1043–1056 [DOI] [PubMed] [Google Scholar]
- Gou J, Hao F, Huang C, Kwon M, Chen F, Li C, Liu C, Ro DK, Tang H, Zhang Y (2018) Discovery of a non-stereoselective cytochrome P450 catalyzing either 8alpha- or 8beta-hydroxylation of germacrene A acid from the Chinese medicinal plant, Inula hupehensis. Plant J 93: 92–106 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Guo L, Qiu J, Ye C, Jin G, Mao L, Zhang H, Yang X, Peng Q, Wang Y, Jia L, et al. (2017) Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat Commun 8: 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Jiang T, Liu YX, Bai YC, Reed J, Qu B, Goossens A, Nutzmann HW, Bai Y, Osbourn A (2019) A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364: eaau6389. [DOI] [PubMed] [Google Scholar]
- Ikezawa N, Gopfert JC, Nguyen DT, Kim SU, O’Maille PE, Spring O, Ro DK (2011) Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio- and stereoselective hydroxylations in sesquiterpene lactone metabolism. J Biol Chem 286: 21601–21611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T (2006) Characterization of a rice gene family encoding type-A diterpene cyclases. Biosci Biotechnol Biochem 70: 1702–1710 [DOI] [PubMed] [Google Scholar]
- Kato T, Kabuto C, Sasaki N, Tsunagawa M, Aizawa H, Fujita K, Kato Y, Kitahara Y, Takahashi N (1973) Momilactones, growth inhibitors from rice, Oryza sativa L. Tetrahedron Lett 14: 3861–3864 [Google Scholar]
- Kitaoka N, Lu X, Yang B, Peters RJ (2015a) The application of synthetic biology to elucidation of plant mono-, sesqui-, and diterpenoid metabolism. Mol Plant 8: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka N, Wu Y, Xu M, Peters RJ (2015b) Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl Microbiol Biotechnol 99: 7549–7558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka N, Wu Y, Zi J, Peters RJ (2016) Investigating inducible short-chain alcohol dehydrogenases/reductases clarifies rice oryzalexin biosynthesis. Plant J 88: 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama O, Yamada A, Yamamoto A, Takemotot T, Akatsuka T (1988) Induction of phytoalexins with heavy metal ions in rice leaves. J Pesticide Sci 13: 615–617 [Google Scholar]
- Lawrence J (1999) Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr Opin Genet Dev 9: 642–648 [DOI] [PubMed] [Google Scholar]
- Lefort V, Longueville JE, Gascuel O (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34: 2422–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhang J, Brown B, Li R, Rodriguez-Romero J, Berasategui A, Liu B, Xu M, Luo D, Pan Z, et al. (2018) Inferring roles in defense from metabolic allocation of rice diterpenoids. Plant Cell 30: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafu S, Jia M, Zi J, Xu M, Morrone D, Wu Y, Peters RJ (2016) Probing the promiscuity of ent-kaurene oxidase via combinatorial biosynthesis. Proc Natl Acad Sci U S A 113: 2526–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Chen M, Chu Q, Jia L, Sultana MH, Wu D, Kong X, Qiu J, Ye CY, Zhu QH, et al. (2019) RiceRelativesGD: a genomic database of rice relatives for rice research. Database (Oxford) 2019: baz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Kawaide H, Higuchi T, Chen M, Miyamoto K, Hirata Y, Kimura H, Miyazaki S, Teruya M, Fujiwara K. et al. (2020) Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants. Proc Natl Acad Sci U S A 117: 12472–12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Fujita M, Shenton MR, Akashi S, Sugawara C, Sakai A, Horie K, Hasegawa M, Kawaide H, Mitsuhashi W, et al. (2016) Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. Plant J 87: 293–304 [DOI] [PubMed] [Google Scholar]
- Nojiri H, Sugimora M, Yamane H, Nishimura Y, Yamada A, Shibuya N, Kodama O, Murofushi N, Omori T (1996) Involvement of jasmonic acid in elicitor-induced phytoalexin production in suspension-cultured rice cells. Plant Physiol 110: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutzmann HW, Huang A, Osbourn A (2016) Plant metabolic clusters—from genetics to genomics. New Phytol 211: 771–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutzmann HW, Scazzocchio C, Osbourn A (2018) Metabolic gene clusters in eukaryotes. Annu Rev Genet 52: 159–183 [DOI] [PubMed] [Google Scholar]
- Okada A, Shimizu T, Okada K, Kuzuyama T, Koga J, Shibuya N, Nojiri H, Yamane H (2007) Elicitor induced activation of the methylerythritol phosphate pathway towards phytoalexin biosynthesis in rice. Plant Mol Biol 65: 177–187 [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R (1964) The carbon monoxide-binding pigment of liver microsome II. Solubilization, purification, and properties. J Biol Chem 239: 2379–2385 [PubMed] [Google Scholar]
- Otomo K, Kenmoku H, Oikawa H, Konig WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T (2004a) Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J 39: 886–893 [DOI] [PubMed] [Google Scholar]
- Otomo K, Kanno Y, Motegi A, Kenmoku H, Yamane H, Mitsuhashi W, Oikawa H, Toshima H, Itoh H, Matsuoka M. et al. (2004b) Diterpene cyclases responsible for the biosynthesis of phytoalexins, momilactones A, B, and oryzalexins A-F in rice. Biosci Biotechnol Biochem 68: 2001–2006 [DOI] [PubMed] [Google Scholar]
- Peters RJ (2006) Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317 [DOI] [PubMed] [Google Scholar]
- Peters RJ (2010) Two rings in them all: The labdane-related diterpenoids. Nat Prod Rep 27: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ (2020) Doing the gene shuffle to close synteny: dynamic assembly of biosynthetic gene clusters. New Phytol 227: 992–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136: 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Wisecaver JH, Lind AL (2018) The birth, evolution and death of metabolic gene clusters in fungi. Nat Rev Microbiol 16: 731–744 [DOI] [PubMed] [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y (2013a) RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res 41: D1206–D1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Namiki N, Takehisa H, Kamatsuki K, Minami H, Ikawa H, Ohyanagi H, Sugimoto K, Itoh J, Antonio BA, et al. (2013b) RiceFREND: a platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res 41: D1214–D1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko K-W, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N. et al. (2007) Identification of a biosynthetic gene cluster in rice for momilactones. J Biol Chem 282: 34013–34018 [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15: 16–21 [Google Scholar]
- Toyomasu T (2008) Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci Biotechnol Biochem 72: 1168–1175 [DOI] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Peters RJ (2011) CYP99A3: functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J 65: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Wu Y, Peters RJ (2012a) CYP701A8: A rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol 158: 1418–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Okada K, Yamazaki K, Wu Y, Swaminathan S, Yamane H, Peters RJ (2012b) Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J Biol Chem 287: 6159–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ (2004) Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Borowsky AT, Tzin V, Jander G, Kliebenstein DJ, Rokas A (2017) A global coexpression network approach for connecting genes to specialized metabolic pathways in plants. Plant Cell 29: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Hillwig ML, Wang Q, Peters RJ (2011) Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization of CYP71Z6 & 7. FEBS Lett 585: 3446–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang Q, Hillwig ML, Peters RJ (2013) Picking sides: distinct roles for CYP76M6 and -8 in rice oryzalexin biosynthesis. Biochem J 454: 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39: 309–318 [DOI] [PubMed] [Google Scholar]
- Xu M, Galhano R, Wiemann P, Bueno E, Tiernan M, Wu W, Chung IM, Gershenzon J, Tudzynski B, Sesma A, et al. (2012) Genetic evidence for natural product-mediated plant–plant allelopathy in rice (Oryza sativa). New Phytol 193: 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Yamazaki K, Minoda H, Miyamoto K, Miyazaki S, Kawaide H, Yajima A, Nojiri H, Yamane H, Okada K, et al. (2018) In planta functions of cytochrome P450 monooxygenase genes in the phytocassane biosynthetic gene cluster on rice chromosome 2. Biosci Biotechnol Biochem 82: 1021–1030 [DOI] [PubMed] [Google Scholar]
- Zhang J, Peters RJ (2020) Why are momilactones always associated with biosynthetic gene clusters in plants? Proc Natl Acad Sci U S A 117: 13867–13869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, Xing J, Yu H, Zhang R, Tian X, Duan L, Zhang M, Peters RJ, Li Z (2020) Introducing selective agrochemical manipulation of gibberellin metabolism into a cereal crop. Nat Plants 6: 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42: 10903–10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.