Abstract
Background
The berberine (Ber) is an isoquinoline alkaloid compound extracted from Rhizoma coptidis and has the effect that reduces adipose. MicroRNA‐192 (miR‐192) is related to fat metabolism. However, the relevant mechanism of berberine on lipid metabolism during in vitro maturation (IVM) of porcine oocytes remains unclear.
Objectives
In this study, we investigated the molecular mechanism by which berberine promotes the IVM and lipid metabolism of porcine oocytes via miR‐192.
Methods
Ber was added to IVM medium of porcine oocytes. MiR‐192 agomir, miR‐192 antagomir and negative control fragment were microinjected into the cytoplasm of oocytes without Ber. Rates of oocyte IVM and embryonic development in each group were observed. The content of lipid droplets in IVM oocytes in each group was analyzed by Nile red staining. Expression levels of miR‐192 and FABP3, SREBF1 and PPARG, were detected by qPCR and western blotting. The target genes of miR‐192 were determined by luciferase reporter assays.
Results and Conclusions
We found that Ber significantly increased the rate of oocytes IVM and blastocyst development, and decreased the area and numbers of lipid droplets in IVM oocytes. Ber significantly increased the expression of miR‐192 in IVM oocytes, and significantly decreased the expression of SREBF1 and PPARG, which were target genes of miR‐192. This study indicates that Ber promotes lipid metabolism in porcine oocytes by activating the expression of miR‐192 and down‐regulating SREBF1 and PPARG, thus, improving IVM of porcine oocytes.
Keywords: berberine, in vitro maturation, lipid metabolism, miR‐192, porcine oocyte
This study investigated the molecular mechanism of Berberine (Ber) promoting the in vitro maturation (IVM) and lipid metabolism of porcine oocytes via miR‐192. The results suggest that Ber promotes lipid metabolism in IVM porcine oocytes and improves oocyte IVM by activating miR‐192 and downregulating SREBF1 and PPARG.

1. INTRODUCTION
The procedures of embryo production in vitro can greatly improve the utilization efficiency of valuable domestic animals, accelerate the selection and expansion of beneficial livestock breeds and genetics, and provide the foundation for human assisted reproductive technology. Compared with other species, porcine oocytes have a large amount of lipids in the cytoplasm, which has a great impact on in vitro maturation (IVM), fertilization, embryo development and cryopreservation of porcine oocytes (Hoyos‐Marulanda et al., 2019; Sun et al., 2001). It is reported that resveratrol decreased the lipid content in porcine oocytes and significantly increased blastocyst development after parthenogenetic activation by enhancing the expression of SIRT1 (sirtuin 1) in porcine oocytes (Itami et al., 2015). The melatonin regulated the gene of lipogenesis (acetyl‐coa carboxylase 1, fatty acid synthase, peroxisome proliferator‐activated receptor gamma, sterol regulatory element binding transcription factor 1) and lipolysis (adipose triglyceride lipase, comparative gene identification‐58, hormone‐sensitive lipase, perilipin 2) abundantly expressed, which can increase the content of fatty acids (FA) and ATP during IVM of porcine oocytes to provide energy basis for the further development of oocytes, and further increase the rate of blastocyst development after in vitro fertilization (IVF) of oocytes (Jin et al., 2017). Therefore, it is important to investigate molecular mechanism of lipid metabolism in porcine oocytes for efficient IVM and embryonic development.
MicroRNA (miRNA) is a non‐coding RNA with a length of about 20–24 nucleotides (Jochen et al., 2018). During IVM of oocytes, the frequency of miRNA associated with all other small RNA was found to decrease markedly from the GV (21.9%) to the MII (5.6%) stage, among which miR‐574 decreased significantly and miR‐21 increased significantly, indicating that they play a major role in the IVM of porcine oocytes (Yang et al., 2012). MiR‐93, miR‐191 and miR‐26a were stably expressed in porcine oocytes during IVM. These results suggest that these miRNAs may play roles in regulating cell proliferation and differentiation during the development of oocytes and early embryos (Mahdi et al., 2015). Stowe et al. found that individual in vivo (IVO) and IVF produced embryos for the transcription level of miR‐24 differed for culture conditions which in lower level detected in the IVO embryos, making this miRNA expression level reflect embryo quality (Stowe et al., 2012). Dionysios et al. produced obese mice using a high‐fat diet for a long duration and checked the expression profile of miRNA in adipose tissue. The abundance of miR‐192 was observably decreased in the adipose tissue of obese mice, so miR‐192 may inhibit fat formation and accumulation (Dionysios et al., 2012). Pan et al. determined miRNAs in fat tissues from two breeds of sheep by high‐throughput sequencing, and found that miR‐192 was positively correlated with fat metabolism in sheep (Pan et al., 2018). However, the mechanism of miRNA actions on IVM of porcine oocytes and lipid metabolism remained unclear.
Berberine (Ber) is a isoquinoline alkaloid compound extracted from Rhizoma coptidis (Kim et al., 2018; Wang et al., 2018). Our prophase study showed that the content of lipid droplets in porcine oocytes matured in MⅡ phase was significantly lower than that in GV stage porcine oocytes. Therefore, during IVM of porcine oocytes, lipid droplets in porcine oocytes were gradually decomposed, providing an energy and material basis for oocyte maturation and embryo development after fertilization. Using a miRNA array, we found that Ber upregulated the expression of miR‐192 in porcine oocytes during IVM after adding Ber to the porcine oocyte IVM medium. We predicted the target genes of miR‐192, such as FABP3 (fatty acid binding protein 3), SREBF1 (steroid regulatory element binding transcription factor 1) and PPARG (peroxisome proliferator‐activated receptor gamma), and used GO analysis to predict potential target gene‐related pathways including cell metabolism and substance metabolism (Huang et al., 2017). Whether or not Ber plays a role in porcine oocyte in vitro development by regulating miR‐192 and the mechanism of miR‐192 actions remained unknown and required further study.
This study was conducted to explore the molecular mechanism of Ber promoting the IVM efficiency and lipid metabolism of porcine oocytes via miR‐192. We determined the relationship between miR‐192 and lipid droplets in porcine oocytes and the molecular pathway of Ber affecting porcine oocytes during IVM. This study will provide new research ideas for further solving the problem of cryopreservation of porcine oocytes and embryos, promote the preservation of pig germplasm resources and the development of pig industry, and provide experimental basis for human biomedical research.
2. MATERIALS AND METHODS
2.1. Experimental materials
Fresh healthy porcine ovaries (Sows are 6–7 months old and prepubescent) were collected from the Beijing Fifth Meat Union Factory, kept in normal saline with streptomycin and penicillin (Hebei Yuanzheng Pharmaceutical Co., Ltd.) and sent to the laboratory within 1 hr at 35°C. Fresh semen from Landrace boar was collected from SPF boar station in northern China and sent to the laboratory within 1 hr at a controlled temperature of 17°C. The same Landrace boar semen was used throughout the experiment.
Unless otherwise stated, the chemicals used in this study were purchased from Sigma‐Aldrich USA.
2.2. Porcine oocyte collection and IVM
Cumulus–oocyte complexes (COCs) were aspirated from 3 to 6 mm ovarian follicles using a 12‐gauge needle attached to a 10‐ml syringe. Then, the COCs were washed three times with TL‐HEPES‐PVA solution, and COCs with more than two layers of cumulus cells and a dense and homogeneous cytoplasm were selected under stereomicroscopy. The COCs were transferred to IVM medium for the control group, and to IVM medium with Ber (0.1 μg/ml, purity ≥ 99.97%; National Institute for the Control of Pharmaceutical and Biological Products) for the Ber group (Huang et al., 2017), and cultured in an incubator at 39°C with 5% CO2, 95% air and 100% relative humidity for 44 hr. IVM medium (TCM199 supplemented with 26‐mmol/L sodium bicarbonate, 3.05 mmol/L glucose, 0.91 mmol/L sodium pyruvate, 0.57 mmol/L L‐Cysteine, 10 ng/ml epidermal growth factor, 0.1% polyvinyl alcohol, 10% porcine follicular fluid, 75 μg/ml peniclin and 50 μg/ml streptomycin) with 1 μg/ml luteinizing hormone (LH) and 1 μg/ml follicle stimulating hormone (FSH) was used for 0–22 hr, then without LH and FSH at 22–44 hr.
After IVM for 44 hr, the cumulus cells were removed from COCs using 0.1% hyaluronidase. Oocytes with an obvious perivitelline space, intact membrane and extruding first polar body were selected for following experiment, and the first polation rate was calculated. The first polar body rate (MII oocyte rate) = the number of oocytes extruding the first polar body/ total number of oocytes cultured in vitro.
2.3. Microinjection of miR‐192 agomir and antagomir
In order to study the effect of miR‐192 on porcine oocytes, oocytes without Ber at 22 hr IVM were randomly selected to remove cumulus cells with hyaluronidase and placed in 50‐μl culture medium droplets covered with mineral oil. Microinjections were performed using an Olympus IX71 (Olympus) inverted microscope equipped with Narishige MMO‐202ND oil hydraulic 3D micromanipulators (Narishige, Japan). The 100 nmol/L miR‐192 agomir (miR‐192 overexpression group), 100 nmol/L miR‐192 antagomir (miR‐192 low expression group) and 100 nmol/L negative control fragment (negative control group) were injected into the cytoplasm of Ber‐free oocytes respectively. Optimal concentrations of the miR‐192 overexpression group and low expression group were screened in a pre‐experiment. The negative control fragment is the vector gene for constructing miR‐192 agonist. The injection volume in each group was 10–11 L. Microinjection was not performed in the control group and Ber group. MiR‐192 antagomir and miR‐192 agomir sequence are shown in Table 1.
TABLE 1.
miR‐192 agomir and miR‐192 antagomir sequence
| Gene Name | Sequence | |
|---|---|---|
| Sense (5′ to 3′) | Antisense (5′ to 3′) | |
| Negative control fragment | CAGUACUUUUGUGUAGUACAA | |
| MiR‐192 agomir | CUGACCUAUGAAUUGACAGCC | CUGUCAAUUCAUAGGUCAGUU |
| MiR‐192 antagomir | GGCUGUCAAUUCAUAGGUCAG | |
2.4. IVF of porcine oocytes
In order to study the effects of Ber and miR‐192 on embryo development of IVF, fresh semen from the boar was heated for 30 min using a water bath at 37°C, then centrifuged (1,000 r/min, 5 min). Concentrated semen was transferred to capacitated fluid (Wang et al., 2019) and incubated for 30 min at 39°C under 5% CO2. Oocytes with the first polar body were transferred to 50‐μl mTBM fertiliser droplets for 30 min. Then, the capacitated sperm (1.5 × 106/ml) was added to the mature oocytes in mTBM fertilization droplets and incubated for 6 hr.
2.5. In vitro development of porcine zygotes
In order to study the influences of Ber and miR‐192 on development of IVF embryos, the porcine zygotes from IVF were washed three times with the NCSU‐23 embryo culture medium (0.4% BSA) with or without Ber (0.1 μg/ml), then transferred into 50 μl droplets of NCSU‐23 embryo culture medium pre‐incubated in a CO2 incubator for 4 hr (about 15–20 zygotes per droplet were cultured in vitro). Using Olympus stereomicroscope (SZX16, Japan), morphological changes in embryos in each period and the rate of embryo development was observed and counted every 24 or 48 hr. The 2‐cell, 4‐cell, 8‐cell, morula and blastocyst stages were observed at 48, 72, 96, 120 and 168 hr, respectively, and culture media were replaced at 48 hr intervals.
2.6. Lipid staining of porcine oocytes
In order to detect the influences of Ber and miR‐192 on lipid droplets of porcine oocytes, the area and quantity of lipid droplets in IVM porcine oocytes were measured. Using the Nile red staining kit (GMS12196, GENMED, Shanghai, China), 2‐μl Nile red staining solution was mixed with 2‐ml PBS and stored in an ice tank. The mature oocytes in each group were washed with PBS and then treated with diluted Nile red staining solution for 10 min while avoiding light. Individual oocytes on a glass slide were examined using a laser confocal microscope (488 nm wavelength, FV‐1000, Olympus), by scanning and with continuous photography to determine the fluorescence distribution of the lipid droplets. Then, we collected the image and determined the number of lipid droplets and the area of fluorescence on the image.
2.7. Quantitative real‐time PCR (qPCR)
For detecting the influences of Ber and miR‐192 on lipid droplets in porcine oocytes, the animal total RNA extraction kit (SN0313D, SENO Biotechnologies Co.) was used to extract the total RNA from the mature oocytes of each group (200 oocytes in each group) and NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) was used for qualitative analysis. All primer sequences, accession numbers, product length and primer positions for qPCR were showed in Table 2. Primer sequences were synthesized by Shanghai Bioengineering Co., Ltd. Quantitative amplification of cDNA was performed in 96‐well optical reaction plates using cDNA first‐chain rapid synthesis kit (DF8004, SENO Biotechnologies Co.). The qPCR was performed using an Biored Chrome 4 Real‐Time qPCR System (Biored). The qPCR mix (20 μl) included 10 μl of SYBR green premix, 1 μl of each forward and reverse primer (10 μmol/L), 1 μl of cDNA and 7 μl of dH2O. The qPCR conditions were as follows: 30 s denaturation at 95°C, 40 cycles of PCR for the quantitative analysis (95°C for 5 s and 60°C for 30 s), one cycle for the melting curve analysis (95°C for 5 s, 60°C for 1 min, 95°C for 1 s) and cooling at 4°C. The relative expression level for each gene was calculated using the 2−ΔΔCT method. The qPCR analysis was performed three times for each group sample. Additionally, we defined the gene expression cut‐off as a mean Ct value of 35. GAPDH was used as the reference gene for FABP3, SREBF1 and PPARG. U6 was used as the reference gene for miR‐192.
TABLE 2.
List of primers and polymerase chain reaction (PCR) parameters used in the present study
| Primer | Primer pair sequences (5’ to 3’) | Primer positions | Accession numbers | Product length |
|---|---|---|---|---|
| GAPDH | F: TGAAGGTCGGAGTGAACGGATT | 105–126 | NM_001206359.1 | 120 |
| R: CCATGTAGTGGAGGTCAATGAAGG | 224–201 | |||
| PPARG | F: CATTCGCATCTTTCAGGG | 931–948 | NM_214379 | 134 |
| R: GGACGCCATACTTTAGGA | 1064–1047 | |||
| FABP3 | F: CAAGCCTACCACAATCATCG | 154–173 | NM_001099931.1 | 279 |
| R: TCGTAAGTGCGAGTGCAAAC | 224–243 | |||
| SREBF1 | F: ACGCCGCCTCCTTCCACCAT | 338–357 | NM_214157.1 | 134 |
| R: AAGGCAGGCACCGACGGGTA | 756–775 | |||
| miR‐192 | F:CTGTGACCTATGGAATTGGC | 7–56 | NC_010444.4 | 69 |
| R:GGCTCTGACCTATGAATTGAC | ||||
| U6 | F: GTGCTCACTTAGGCAGCACA | 1–20 | XR_003955558.1 | 107 |
| R:ATTTGCATGTCATTCTTGCAC |
F, forward; R, reverse
2.8. Western blotting
To further determine the effects of Ber and miR‐192 on lipid droplets of porcine oocytes, we examined the translation levels of FABP3, SREBF1 and PPARG in porcine oocytes matured in vitro. Matured oocytes (400–500 oocytes/group) were collected and mixed with radioimmunoprecipitation buffer (RIPA) solution containing protease and phosphatase inhibitors. The mixture was incubated at 4°C for 30 min and mixed every 1 min. The extracted proteins were diluted and boiled 5 min for SDS‐PAGE on the condition of 5% stacking gel at 80 V for 20 min and 12% separating gel at 120 V for 2 hr. Then, the proteins were transferred onto a nitrocellulose membrane using a electrical transfer system at 300 mA for 30 min. The membranes were then blocked using 1% BSA (BD Biosciences, China) for 2 hr at room temperature. After that, membranes were incubated with primary antibody at 4°C overnight. The primary antibodies included β‐actin (ZSGB‐BIO, Beijing), and PPARG, FABP3 and SREBF1 (all 1:1,000 dilution, abcam). The next day, membranes were washed and then incubated with secondary antibody (1:5,000) at room temperature for 50 min. After washing with TBST, the membranes were incubated with ECL chromogenic solution for 1 min. Signals were detected by enhanced chemiluminescence. Quantity One v.4.6.2 software was used to quantify the gray values of the protein bands. The experiment was repeated three times.
2.9. Luciferase reporter assay
In order to check the target gene of miR‐192, the wild‐type (WT) and mutant porcine FABP3 3′UTR and SREBF1 3′UTR, and the WT porcine PPARG 3′UTR containing putative binding sites for miR‐192 were cloned into the pmirGlo vector (LP0041, OXOID). The mutant sites were synthesized and shown in Figure 4. We did not predict binding sites between miR‐192 and porcine PPARG 3′UTR, so we did not synthesize a mutant PPARG 3′UTR. Five ng of each pmirGlo construct was co‐transfected with ssc‐miR‐192 mimic or a negative control into HEK293 cells in a 24‐well plate using Lipofectamine 2000 (11668‐019, Invitrogen). The transfection was incubated in a 37°C, 5% CO2 incubator for 5 hr. The transfection solution was desorbed and complete culture medium containing 10% FBS was added, the cells were incubated at 37°C with 5% CO2 for 24 hr. The cell extracts were harvested, and the firefly and Renilla luciferase activity were measured using the Dual‐Luciferase reporter system (E1910, Promega) based on the manufacturer's instructions.
FIGURE 4.
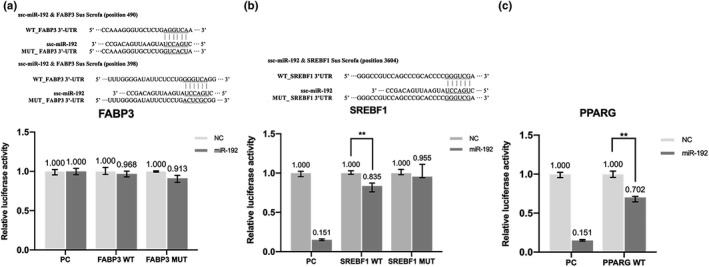
The result of dual‐luciferase reporter gene assay of transfected vector plasmid. (a) The result of dual‐luciferase reporter gene assay of transfected FABP3 vector plasmid. (b) The result of dual‐luciferase reporter gene assay of transfected PPARG vector plasmid. (c) The result of dual‐luciferase reporter gene assay of transfected SREBF1 vector plasmid. PC, positive control; WT, wild‐type; MUT, mutant type; NC, negative control. **Indicates significant difference (p < 0.01), *Indicates significant difference (p < 0.05), no sign indicates no significant difference
2.10. Statistical analysis
All data analyzed by SPSS 20.0 software were shown as mean ± SD. The statistical significance was determined by One‐way ANOVA and SNK multiple comparison methods. Image J Image Analysis Software was used to calculate the number of lipid droplets and fluorescence area. A p‐value less than 0.05 was considered significant, whereas a p‐value less than 0.01 was considered extremely significant.
3. RESULTS
3.1. Effects of Ber and miR‐192 on IVM and lipid droplet content of porcine oocytes
3.1.1. Effects of Ber and miR‐192 on IVM of porcine oocytes and in vitro development of IVF embryos
As seen in Table 3, the rate of first polar body in Ber group was significantly higher than those in the control group (p < 0.01). There was no significant difference between the miR‐192 overexpression group and the control group (p > 0.05). There was significant difference among miR‐192 overexpression group, negative control group and low expression group (p < 0.01). There was significant difference between miR‐192 overexpression group and miR‐192 low expression group (p < 0.01). The low expression group was significantly lower than the control group (p < 0.01). The negative control group was significantly lower than the control group (p < 0.01).
TABLE 3.
Effect of Ber and miR‐192 on porcine oocyte in vitro maturation
| Group | No. of replicates | No. of oocytes | Pb. I rate/% |
|---|---|---|---|
| Control | 9 | 804 | 59.50 ± 1.06A |
| Negative control | 9 | 1,279 | 50.70 ± 1.20B |
| MiR‐192 overexpression | 9 | 1,154 | 56.47 ± 1.64A |
| MiR‐192 low expression | 9 | 1,340 | 35.46 ± 5.27C |
| Ber | 9 | 816 | 74.28 ± 1.26B |
The values denoted by different capital letters within the same column represent significant difference at 0.01 level (p < 0.01), the same letters indicate no significant difference (p > 0.05).
As seen in Table 4, compared with other groups, the addition of Ber significantly increased blastocyst rate (p < 0.01) of IVF embryos. There was no significant difference between the miR‐192 overexpression group and the control group (p > 0.05), whereas there was significant difference among the miR‐192 overexpression group, the negative control group and the miR‐192 low expression group (p < 0.01). The miR‐192 low expression group was significantly lower than the control group (p < 0.01). The negative control group was significantly lower than the control group (p < 0.01). It is concluded that overexpression of miR‐192 significantly improved the IVM and embryo development rate of porcine oocytes, and Ber further promoted the in vitro development of porcine oocytes and IVF embryos.
TABLE 4.
Effect of Ber and miR‐192 on porcine embryo development in vitro
| Group | No. of replicates | No. of MII oocytes | 48‐hr Cleavage rate/% | 72‐hr 4‐cell rate/% | 96‐hr 8‐cell rate/% | 120‐hr Morula rate/% | 168‐hr Blastocyst rate/% |
|---|---|---|---|---|---|---|---|
| Control | 6 | 240 | 58.41 ± 2.47Aa | 48.51 ± 2.26A | 40.18 ± 1.52Aa | 31.56 ± 3.56A | 19.79 ± 0.82A |
| Negative control | 6 | 237 | 48.18 ± 2.53B | 40.73 ± 3.87B | 32.21 ± 3.27Bc | 24.43 ± 2.50B | 14.80 ± 2.17B |
| MiR‐192 overexpression | 6 | 241 | 55.47 ± 2.98Ab | 45.61 ± 3.97A | 35.59 ± 4.51Ab | 28.06 ± 4.16A | 19.26 ± 1.94A |
| MiR‐192 low expression | 6 | 247 | 40.30 ± 3.21C | 33.08 ± 3.38C | 27.27 ± 2.73C | 21.05 ± 2.73C | 10.85 ± 0.37C |
| Ber | 6 | 236 | 78.47 ± 1.35D | 68.10 ± 1.17D | 58.01 ± 2.50D | 46.86 ± 1.13D | 29.22 ± 1.46D |
The values denoted by different lowercase and capital letters within the same column represent significant differences at the p < 0.05 and p < 0.01 levels respectively. And the same letters indicate no significant difference (p > 0.05). The embryo development rate in each period is based on the number of mature oocytes.
3.1.2. Effects of Ber and miR‐192 on the content and area of lipid droplets in porcine oocytes matured in vitro
As seen in Table 5 and Figure 1, after microinjection, the number and area of oocyte lipid droplets in the miR‐192 overexpression group were not significantly different from those in the Ber group, but were significantly lower than those in control group, negative control group and miR‐192 low expression group (p < 0.01). There was no significant difference between the control group and the negative control group (p > 0.05). The number of lipid droplets in the miR‐192 low expression group was significantly higher than that in the control group and negative control group (p < 0.05), but the area of lipid droplets in the miR‐192 low expression group was not significantly different from that of control group and negative control group (p > 0.05). These results show that lipid droplets decreased gradually during IVM of porcine oocytes, and the content of lipid droplets in oocytes decreased significantly after the addition of Ber and the miR‐192 overexpression agent. Therefore, Ber and miR‐192 are likely to be involved in lipid metabolism in porcine oocytes.
TABLE 5.
Changes in the number of lipid droplets in different groups of oocytes after IVM
| Group | No. of MII oocytes | No. of lipid droplets |
|---|---|---|
| Control | 32 | 271.56 ± 30.03Aa |
| Negative control | 30 | 221.36 ± 38.26Aa |
| MiR‐192 overexpression | 31 | 196.61 ± 46.97B |
| MiR‐192 low expression | 24 | 304.83 ± 38.2Ab |
| Ber | 28 | 190.07 ± 24.68B |
The values denoted by different lowercase and capital letters within the same column represent significant differences at the p < 0.05 and p < 0.01 levels respectively. And the same letters indicate no significant difference (p < 0.05).
FIGURE 1.
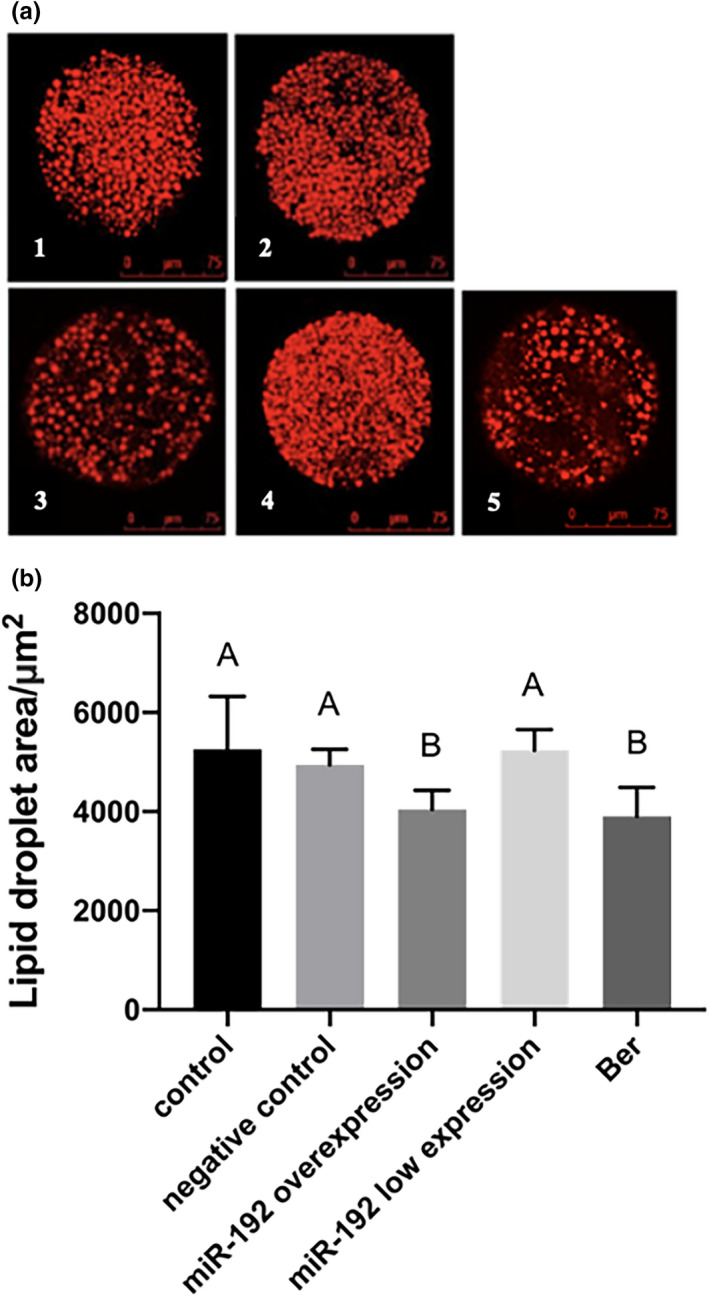
The results of each group by Nile Red dye. (a) The picture of each group oocyte by Nile Red dye. Scale bar indicates 75 μm. 1. Control; 2. negative control; 3. miR‐192 overexpression; 4. miR‐192 low expression and 5. Ber. (b) The effect of Ber and miR‐192 of lipid droplets area of porcine oocytes. The values denoted by different capital letters represent significant difference at p < 0.01 level, the same letters indicate no significant difference (p > 0.05)
3.2. Regulation of lipid droplet metabolism‐related genes by miR‐192 in porcine oocytes
3.2.1. Detection of miR‐192 expression
There was no significant difference in miR‐192 expression between the Ber group and miR‐192 overexpression group (p > 0.05, Figure 2). The miR‐192 overexpression group was significantly higher than the control group, negative control group and miR‐192 low expression group (p < 0.01). There was no significant difference between the control and negative control groups (p > 0.05), but the expression of miR‐192 in the low expression group was significantly lower than that in the control and negative control group (p < 0.01). These results show that the miR‐192 overexpression/low expression significantly increased and decreased, respectively, the expression of miR‐192 in porcine oocytes, and Ber also increased oocyte expression of miR‐192.
FIGURE 2.

The relative expression levels of miR‐192 in porcine IVM oocytes from different groups. The values denoted by different capital letters represent significant difference at p < 0.01 level, the same letters indicate no significant difference (p > 0.05)
3.2.2. Effect of Ber and miR‐192 overexpression/low expression on FABP3, SREBF1 and PPARG expression levels
The expression levels of FABP3, SREBF1 and PPARG transcripts in the miR‐192 overexpression group and Ber group were significantly lower than those in the control group and negative control group (p < 0.01, Figure 3a). The expression of miR‐192 in the low expression group was significantly higher than that in other groups (p < 0.01). There was no significant difference between the control group and the negative control group (p > 0.05). The expression levels of FABP3 and PPARG transcripts in the miR‐192 overexpression group were not significantly different from those in the Ber group (p > 0.05). The level of SREBF1 transcript expression in the Ber group was significantly lower than that in the miR‐192 overexpression group (p < 0.05). Therefore, the expression of FABP3, SREBF1 and PPARG appeared to be correlated with the expression of miR‐192, and the effect of Ber on lipid droplet metabolism in porcine oocytes may be related to the expression of miR‐192.
FIGURE 3.

Effect of Ber and miR‐192 on gene expression level related to lipid metabolism. (a) Effect of Ber and miR‐192 on the transcription level of genes related to lipid metabolism. (b) Effect of Ber and miR‐192 on the translation level of proteins related to lipid metabolism. The values denoted by different lowercase and capital letters represent significant difference at p < 0.05 level and p < 0.01 level respectively. And the same letters indicate no significant difference (p > 0.05)
The expression levels of FABP3, SREBF1 and PPARG proteins in the miR‐192 overexpression group and Ber group were significantly lower than those in the control and negative control group (p < 0.01, Figure 3b). The expression levels of these proteins in the miR‐192 low expression group were significantly higher than those in the other groups (p < 0.01). There was no significantly difference between the control and negative control group (p > 0.05). There was no significant difference in the SREBF1 and PPARG protein expression levels between the miR‐192 overexpression group and Ber group (p > 0.05). The expression of FABP3 in the Ber group was significantly lower than that in miR‐192 overexpression group (p < .05). These findings indicate that Ber may affect lipid droplet metabolism of porcine oocytes through the FABP3, SREBF1 and PPARG genes.
3.2.3. Detection of miR‐192 target genes by luciferase reporter gene analysis
There are two putative binding sequences for miR‐192 in the porcine FABP3 3′UTR, with wild‐type (WT) and mutant (MUT) miR‐192 sites shown in Figure 4a. Compared with the negative control (NC) group, cells transfected with miR‐192 mimics and the FABP3 WT 3′UTR vector showed no significant difference in reporter gene expression (p > 0.05, Figure 4a). The putative WT and MUT miR‐192 binding sequences in the porcine SREBF1 3′UTR were shown in Figure 4b. Cells transfected with miR‐192 mimics and SREBF1 3′UTR vector had significantly decreased (22%, p < 0.01) luciferase activity compared with the NC group (Figure 4b). To confirm the specificity of binding, we performed the same assay with the SREBF1 3′UTR MUT (or SREBF1 3′UTR MUT vector with miR‐192 inhibitor) and found that the luciferase activity was partially restored. Cells transfected with miR‐192 mimics and PPARG 3′UTR vector had significantly decreased (29%, p < 0.01) luciferase activity compared with the NC group (Figure 4c). It was concluded that SREBF1 and PPARG are the target genes of miR‐192.
4. DISCUSSION
In the last 10 years, the influence of Ber on lipid lowering has been explored in considerable detail. Ber has been proved to regulate lipid metabolism disorders (Zhang et al., 2018) and reduce the level of plasma triglyceride, total cholesterol and low density lipoprotein (LDL) in mice fed a high‐fat diet (Esfandiar et al., 2014; Feng et al., 2018). In HEK293T and HepG2 cell lines, Ber upregulated miR‐373 through RAC‐alpha serine/threonine‐protein kinase (AKT1), which increased LDL receptor expression to reduce the LDL level (Li, Tang et al., 2018). Our study showed that Ber decreased the lipid content of porcine oocytes and increased the maturation rate of oocytes. Therefore, we speculated that Ber may be involved in the lipid metabolism in porcine oocytes and promote the maturation of porcine oocytes. This study provides an important experimental basis for optimizing the in vitro culture system of porcine oocytes and embryos, and promoting the development of cryopreservation technology.
At present, Ber has been shown to regulate the expression of several miRNAs (Li, Tang et al., 2018). Studies indicated that Ber upregulated miR‐22‐3p expression and decreased Bcl‐2 levels (Chen et al., 2016), but reduced the expression level of miR‐122 and decreased the level of glucose and lipid metabolism, inhibiting the proliferation of hepatoma cells (Wei et al., 2016). In this study, it was found that both miR‐192 and Ber could significantly reduce the lipid content of porcine oocytes and significantly increase the maturation rate of oocytes. Ber upregulated the expression of miR‐192, providing strong evidence that Ber regulated lipid metabolism of porcine oocytes matured in vitro through miR‐192. Studies had shown that after miR‐192 was upregulated, the transcription level of FABP3 and other genes associated with lipid metabolism in Caco‐2 cells were inhibited (Gil‐Zamorano et al., 2014). Past work showed that miR‐192 upregulated the expression of SREBF1 in mouse hepatocytes to decrease the fatty degeneration and lipid content of hepatocytes (Lin et al., 2017). Our experiments showed that Ber reduced the expression of FABP3, SREBF1 and PPARG genes related to lipid metabolism in porcine oocytes matured in vitro. Therefore, we propose that Ber may decrease the lipid content in oocytes by affecting miR‐192 and its downstream genes related to lipid metabolism to increase porcine oocyte–embryonic development.
The FABP3, PPARG and SREBF1 proteins play the vital role in the regulating of the lipid metabolism. FABP3 is a member of the fatty acid binding family. Previous results showed that FABP3 gene polymorphisms were significantly related to intramuscular fat in pigs (Sweeney et al., 2015; Xu et al., 2018). Maite et al. found that the increasing lipid content in bovine oocytes during IVM was due to the transport of FABP3 from granulosa cells into oocytes (Del et al., 2017). PPARG is one subtype of the peroxisome proliferator‐activated receptor which plays a major role in adipocyte differentiation (Yang, Yang et al., 2018) and lipid metabolism (Choi et al., 2016; Gao et al., 2018; Li, Mihalcioiu et al., 2018). Gorga et al. found that murine Sertoli cells incubated with 10‐μM rosiglitazone (a PPARG activator) exhibited increased triacylglyceride levels and lipid droplet content and elevated proteins expression of genes for involved in fatty acid storage, such as the fatty acid transporter Cd36, glycerol‐3‐phosphate‐acyltransferases 1 and 3, diacylglycerol acyltransferase 1 and perilipins 1, 2 and 3 (proteins that participate in lipid droplet formation and stabilization) (Gorga et al., 2017). Other work showed that ginsenoside Rc inhibited the expression of PPARG, reduced the number of adipocytes and inhibited the accumulation of lipid (Yang & Kim, 2015). SREBF1 is the main regulator of fat synthesis (Shimano & Sato, 2017). Low expression of SREBF1 inhibits the accumulation of lipids in many cancer cells, thus, slowing down the metastasis and progression of cancers, such as lung cancer (Luo et al., 2017), endometrial carcinoma (Shafiee et al., 2017) and renal cell carcinoma (Yang, Zhang et al., 2018). Previous work showed that miR‐25 reduced the lipid content in goat mammary epithelial cells by inhibiting the expression of SREBF1‐ and PPARG‐related lipid metabolism genes (Ma et al., 2018).
Hence, the mRNA and protein expression levels of FABP3, PPARG and SREBF1 were detected, and our results showed that miR‐192 decreased the expression levels of FABP3, PPARG and SREBF1. This finding indicated that miR‐192 could reduce the number of lipid droplets in porcine oocytes and inhibit the fat formation through regulating FABP3, PPARG and SREBF1. It was proved that PPARG and SREBF1 were target genes of miR‐192 by luciferase reporter gene technology, and we speculated that PPARG and SREBF1 were major targets of miR‐192 actions regulated by Ber, FABP3 may be an indirect gene. Therefore, our results suggested that miR‐192 actions mediated by Ber can reduce the lipid content of porcine oocytes during IVM mainly through PPARG and SREBF1, thus, promoting oocyte maturation and embryo development. The precise mechanism of PPARG and SREBF1 actions on the IVM and lipid metabolism of porcine oocytes needs further study.
To sum up, Ber significantly increased the IVM rate of porcine oocytes and the in vitro development rate of blastocysts, and decreased the number and area of lipid droplets in oocytes. Ber significantly increased the expression of miR‐192 and decreased the expression of FABP3, SREBF1 and PPARG in mature oocytes. The target genes of miR‐192 were shown to be SREBF1 and PPARG. These results indicate that Ber promotes lipid metabolism in porcine oocytes and improves the IVM of oocytes by activating expression of miR‐192 that mainly acts on SREBF1 and PPARG.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
JiaGe Dai: Data curation; Formal analysis; Investigation; Software; Validation; Visualization; Writing‐original draft; Writing‐review & editing. XiaoMeng Huang: Formal analysis; Investigation; Validation; Writing‐original draft. Chao Zhang: Conceptualization; Investigation; Methodology. XiaoFei Luo: Formal analysis; Investigation; Validation. SuYing Cao: Investigation; Validation. JunLi Wang: Investigation; Validation. Bing Liu: Investigation; Validation. JianMing Gao: Conceptualization; Data curation; Funding acquisition; Methodology; Project administration; Resources; Supervision; Validation; Writing‐original draft; Writing‐review & editing.
ETHICAL STATEMENT
Animal welfare and experimental procedures were implemented in accordance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006), and ratified by the animal ethics committee of the Beijing University of Agriculture.
ACKNOWLEDGEMENTS
This work was supported by grants from National Natural Science Foundation of China (No.31572402) and the Beijing Scientific Research Program (No. KZ20160020017). We thank Beijing Key Laboratory of Traditional Chinese Veterinary Medicine Beijing University of Agriculture and Key Laboratory of Urban Agriculture (North China), Ministry of Agriculture, P. R. China, for technical support.
Dai J, Huang X, Zhang C, et al. Berberine regulates lipid metabolism via miR‐192 in porcine oocytes matured in vitro. Vet Med Sci.2021;7:950–959. 10.1002/vms3.393
JiaGe Dai and XiaoMeng Huang are contributed equally to this work.
Funding information
National Natural Science Foundation of China, Grant/Award Number: 31572402; Beijing Scientific Research Program, Grant/Award Number: KZ20160020017.
DATA AVAILABILITY STATEMENT
Data available on request from the authors. The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Chen, J. , Wu, F. X. , Luo, H. L. , Liu, J. J. , Luo, T. , Bai, T. , & Fan, X. H. (2016). Berberine upregulates miR‐22‐3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. American Journal of Translational Research, 8, 4932–4941. [PMC free article] [PubMed] [Google Scholar]
- Choi, S. S. , Kim, E. S. , Jung, J. E. , Marciano, D. P. , Jo, A. , Koo, J. Y. , & Choi, J. H. (2016). PPARG antagonist Gleevec improves insulin sensitivity and promotes the browning of white adipose tissue. Diabetes, 65, 829–839. 10.2337/db15-1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del, C. M. , Silveira, J. C. , Sangalli, J. R. , Andrade, G. M. , Sousa, L. R. D. S. , Silva, L. A. , & Perecin, F. (2017). Fatty acid binding protein 3 and transzonal projections are involved in lipid accumulation during in vitro maturation of bovine oocytes. Scientific Reports, 7, 2645. 10.1038/s41598-017-02467-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionysios, V. C. , Apostolos, Z. , Panos, G. Z. , Ralitsa, P. I. , Agathoklis, I. P. , Venetsana, E. K. , & Ioannis, G. H. (2012). Differential expression of MicroRNAs in adipose tissue after long‐term high‐fat diet‐induced obesity in mice. PLoS One, 7, e34872. 10.1371/journal.pone.0034872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiar, H. , Mahmoud, R. K. , Abolfazle, K. , & Morteza, B. (2014). Metabolic effects of berberine on liver phosphatidate phosphohydrolase in rats fed on high lipogenic diet: An additional mechanism for the hypolipidemic effects of berberine. Asian Pacific Journal of Tropical Medicine, Z1, 429–435. 10.12980/APJTB.4.2014C474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W. W. , Kuang, S. Y. , Tu, C. , Ma, Z. J. , Pang, J. Y. , Wang, Y. H. , & Xiao, X. H. (2018). Natural products berberine and curcumin exhibited better ameliorative effects on rats with non‐alcohol fatty liver disease than lovastatin. Biomedicine & Pharmacotherapy, 99, 325–333. 10.1016/j.biopha.2018.01.071 [DOI] [PubMed] [Google Scholar]
- Gao, R. , Chen, W. , Yan, H. , Xie, X. , Liu, D. , Wu, C. , & Wang, L. (2018). PPARG agonist rosiglitazone switches fuel preference to lipids in promoting thermogenesis under cold exposure in C57BL/6 mice. Journal of Proteomics, 176, 24–36. 10.1016/j.jprot.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Gil‐Zamorano, J. , Martin, R. , Daimiel, L. , Richardson, K. , Giordano, E. , Nicod, N. , García‐Carrasco, B. , Soares, S. M. A. , Iglesias‐Gutiérrez, E. , Lasunción, M. A. , Sala‐Vila, A. , Ros, E. , Ordovás, J. M. , Visioli, F. , & Dávalos, A. (2014). Docosahexaenoic acid modulates the enterocyte caco‐2 cell expression of microRNAs involved in lipid metabolism. Journal of Nutrition, 144, 575–585. 10.3945/jn.113.189050 [DOI] [PubMed] [Google Scholar]
- Gorga, A. , Rindone, G. M. , Regueira, M. , Pellizzari, E. H. , Camberos, M. C. , Cigorraga, S. B. , & Meroni, S. B. (2017). PPARG activation regulates lipid droplet formation and lactate production in rat Sertoli cells. Cell and Tissue Research, 369, 611–624. 10.1007/s00441-017-2615-y [DOI] [PubMed] [Google Scholar]
- Hoyos‐Marulanda, V. , Alves, B. S. , Rosa, P. R. A. , Vieira, A. D. , Gasperin, B. G. , Mondadori, R. G. , & Lucia, T. Jr (2019). Effects of polyunsaturated fatty acids on the development of pig oocytes in vitro following parthenogenetic activation and on the lipid content of oocytes and embryos. Animal Reproduction Science, 205, 150–155. 10.1016/j.anireprosci.2019.05.003 [DOI] [PubMed] [Google Scholar]
- Huang, X. M. , Zhang, C. , Wu, L. , Liu, B. , Wang, J. L. , Cao, S. Y. , & Gao, J. M. (2017). Effects of berberine on miRNA and lipid metabolism related genes in vitro maturation of porcine oocytes. Chinese Journal of Animal Science, 10, 47–51. 10.19556/j.0258-7033.2017-10-047 [DOI] [Google Scholar]
- Itami, N. , Shirasuna, K. , Kuwayama, T. , & Iwata, H. (2015). Resveratrol improves the quality of pig oocytes derived from early antral follicles through sirtuin1 activation. Theriogenology, 83, 1360–1367. 10.1016/j.theriogenology.2015.01.029 [DOI] [PubMed] [Google Scholar]
- Jin, J. X. , Lee, S. , Taweechaipaisankul, A. , Kim, G. A. , & Lee, B. C. (2017). Melatonin regulates lipid metabolism in porcine oocytes. Journal of Pineal Research, 62, e12388. 10.1111/jpi.12388 [DOI] [PubMed] [Google Scholar]
- Jochen, T. B. , Veronika, L. F. , Mark, D. R. , Stefan, B. , & Susanne, E. U. (2018). Small RNA‐seq analysis of single porcine blastocysts revealed that maternal estradiol‐17beta exposure does not affect miRNAisoform (isomiR) expression. BMC Genomics, 19, 1–16. 10.1186/s12864-018-4954-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Lee, J. , You, D. , Jeong, Y. , Jeon, M. , Yu, J. , Kim, S. W. , Nam, S. J. , & Lee, J. E. (2018). Berberine suppresses cell motility through downregulation of TGF‐β1 in triple negative breast cancer cells. Cellular Physiology and Biochemistry, 45, 795–807. 10.1159/000487171 [DOI] [PubMed] [Google Scholar]
- Li, C. H. , Tang, S. C. , Wong, C. H. , Wang, Y. , Jiang, J. D. , & Chen, Y. (2018). Berberine induces miR‐373 expression in hepatocytes to inactivate hepatic steatosis associated AKT‐S6 kinase pathway. European Journal of Pharmacology, 825, 107–118. 10.1016/j.ejphar.2018.02.035 [DOI] [PubMed] [Google Scholar]
- Li, J. , Mihalcioiu, M. , Li, L. , Zakikhani, M. , Camirand, A. , & Kremer, R. (2018). Vitamin D prevents lipid accumulation in murine muscle through regulation of PPARG and perilipin‐2 expression. The Journal of Steroid Biochemistry and Molecular Biology, 177, 116–124. 10.1016/j.jsbmb.2017.10.010 [DOI] [PubMed] [Google Scholar]
- Lin, Y. , Ding, D. , Huang, Q. , Liu, Q. , Lu, H. , Lu, Y. , Dong, S. (2017). Downregulation of miR‐192 causes hepatic steatosis and lipid accumulation by inducing SREBF1: Novel mechanism for bisphenol A‐triggered non‐alcoholic fatty liver disease. Biochimica Et Biophysica Acta (BBA) ‐ Molecular and Cell Biology of Lipids, 1862, 869–882. 10.1016/j.bbalip.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Luo, D. , Xiao, H. W. , Dong, J. L. , Li, Y. , Feng, G. X. , Cui, M. , & Fan, S. J. (2017). B7–H3 regulates lipid metabolism of lung cancer through SREBF1‐mediated expression of FASN. Biochemical and Biophysical Research Communications, 482, 1246–1251. 10.1016/j.bbrc.2016.12.021 [DOI] [PubMed] [Google Scholar]
- Ma, L. , Qiu, H. L. , Chen, Z. , Li, L. , Zeng, Y. , Luo, J. , & Gou, D. M. (2018). MiR‐25 modulates triacylglycerol and lipid accumulation in goat mammary epithelial cells by repressing PGC‐1beta. Journal of Animal Science and Biotechnology, 9, 868–877. 10.1186/s40104-018-0262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi, M. , Helena, T. A. T. , Tom, A. E. S. , & Bernard, A. J. R. (2015). Validating reference microRNAs for normalizing qRT‐PCR data in bovine oocytes and preimplantation embryos. BMC Developmental Biology, 15, 25–34. 10.1186/s12861-015-0075-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y. Y. , Jing, J. J. , Zhao, J. X. , Jia, X. L. , Qiao, L. Y. , An, L. X. , & Liu, W. Z. (2018). MicroRNA expression patterns in tail fat of different breeds of sheep. Livestock Science, 207, 7–14. 10.1038/srep18470 [DOI] [Google Scholar]
- Shafiee, M. N. , Mongan, N. , Seedhouse, C. , Chapman, C. , Deen, S. , Abu, J. , & Atiomo, W. (2017). Sterol regulatory element binding protein‐1 (SREBF1) gene expression is similarly increased in polycystic ovary syndrome and endometrial cancer. Acta Obstetricia Et Gynecologica Scandinavica, 96, 556–562. 10.1111/aogs.13106 [DOI] [PubMed] [Google Scholar]
- Shimano, H. , & Sato, R. (2017). SREBP‐regulated lipid metabolism: Convergent physiology‐divergent pathophysiology. Nature Reviews Endocrinology, 13, 710–730. 10.1038/nrendo.2017.91 [DOI] [PubMed] [Google Scholar]
- Stowe, H. M. , Curry, E. , Calcatera, S. M. , Krisher, R. L. , Paczkowski, M. , & Pratt, S. L. (2012). Cloning and expression of porcine Dicer and the impact of developmental stage and culture conditions on MicroRNA expression in porcine embryos. Gene, 501(2), 198–205. 10.1016/j.gene.2012.03.058 [DOI] [PubMed] [Google Scholar]
- Sun, Q. Y. , Wu, G. M. , Lai, L. , Park, K. W. , Cabot, R. , Cheong, H. T. , Day, B. N. , Prather, R. S. , & Schatten, H. (2001). Translocation of active mitochondria during pig oocyte maturation, fertilization and early embryo development in vitro. Reproduction, 122, 155–163. 10.1530/rep.0.1220155 [DOI] [PubMed] [Google Scholar]
- Sweeney, T. , O'Halloran, A. M. , Hamill, R. M. , Davey, G. C. , Gil, M. , Southwood, O. I. , & Ryan, M. T. (2015). Novel variation in the FABP3 promoter and its association with fatness traits in pigs. Meat Science, 100, 32–40. 10.1016/j.meatsci.2014.09.014 [DOI] [PubMed] [Google Scholar]
- Wang, J. L. , Zhang, C. , Liu, B. , Huang, X. M. , Dai, J. G. , Tian, J. H. , & Gao, J. M. (2019). Function of berberine on porcine in vitro fertilization embryo development and differential expression analysis of microRNAs. Evidence‐based Reproduction in Domestic Animals, 54, 520–530. 10.1111/rda.13397 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Feng, S. , Ding, N. , He, Y. , Li, C. , Li, M. , Ding, X. , Ding, H. , Li, J. , Wu, J. , & Li, Y. U. (2018). Anti‐inflammatory effects of berberine hydrochloride in an LPS‐induced murine model of mastitis. Evidence‐based Complementary and Alternative Medicine, 2018, 1–9. 10.1155/2018/5164314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S. , Zhang, M. , Yu, Y. , Lan, X. , Yao, F. , Yan, X. , & Hatch, G. M. (2016). Berberine attenuates development of the hepatic gluconeogenesis and lipid metabolism disorder in Type 2 diabetic mice and in palmitate‐incubated HepG2 cells through suppression of the HNF‐4alpha miR122 pathway. PLoS One, 11, e0152097. 10.1371/journal.pone.0152097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. G. , Wang, C. , Jin, E. , Gu, Y. , Li, S. , & Li, Q. (2018). Identification of differentially expressed genes in longissimus dorsi muscle between Wei and Yorkshire pigs using RNA sequencing. Genes & Genomics, 40, 413–421. 10.1007/s13258-017-0643-3 [DOI] [PubMed] [Google Scholar]
- Yang, C. X. , Du, Z. Q. , Elane, C. W. , Max, F. R. , Randall, S. P. , & Jason, W. R. (2012). Small RNA profile of the cumulus‐oocyte complex and early embryos in the pig. Biology of Reproduction, 87, 1–11. 10.1095/biolreprod.111.096669 [DOI] [PubMed] [Google Scholar]
- Yang, H. , Zhang, X. , Liu, F. , Fan, J. , Wang, B. L. , & Dong, C. J. (2018). SREBF1‐driven lipid desaturation supports clear cell renal cell carcinoma growth through regulation of NF‐kB signaling. Biochemical and Biophysical Research Communications, 495, 1383–1388. 10.1016/j.bbrc.2017.11.163 [DOI] [PubMed] [Google Scholar]
- Yang, J. W. , & Kim, S. S. (2015). Ginsenoside Rc Promotes anti‐adipogenic activity on 3T3‐L1 adipocytes by down‐regulating C/EBPα and PPARG. Molecules, 20, 1293–1303. 10.3390/molecules20011293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , Yang, C. , Luo, J. , Wei, Y. , Wang, W. , & Zhong, Y. (2018). Adiponectin promotes preadipocyte differentiation via the PPARG pathway. Molecular Medicine Reports, 17, 428–435. 10.3892/mmr.2017.7881 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Kishi, H. , & Kobayashi, S. K. (2018). Add‐on therapy with traditional Chinese medicine: An efficacious approach T for lipid metabolism disorders. Pharmacological Research, 134, 200–211. 10.1016/j.phrs.2018.06.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available on request from the authors. The data that support the findings of this study are available from the corresponding author upon reasonable request.


