Abstract
Background
Respiratory syncytial virus (RSV) is a leading cause of infant respiratory disease. Infant airway microbiota has been associated with respiratory disease risk and severity. The extent to which interactions between RSV and microbiota occur in the airway, and their impact on respiratory disease susceptibility and severity, are unknown.
Methods
We carried out 16S rRNA microbiota profiling of infants in the first year of life from (1) a cross-sectional cohort of 89 RSV-infected infants sampled during illness and 102 matched healthy controls, and (2) a matched longitudinal cohort of 12 infants who developed RSV infection and 12 who did not, sampled before, during, and after infection.
Results
We identified 12 taxa significantly associated with RSV infection. All 12 taxa were differentially abundant during infection, with 8 associated with disease severity. Nasal microbiota composition was more discriminative of healthy vs infected than of disease severity.
Conclusions
Our findings elucidate the chronology of nasal microbiota dysbiosis and suggest an altered developmental trajectory associated with RSV infection. Microbial temporal dynamics reveal indicators of disease risk, correlates of illness and severity, and impact of RSV infection on microbiota composition.
Keywords: microbiota, RSV, infant respiratory disease
The findings of our study provide novel insights into the developmental dynamics of the nasal microbiome in the first year of life as they relate to susceptibility, acute illness, severity, and convalescence associated with first-time RSV infection.
((See the Major Article by Chu et al, on pages 1639–49.)
The composition and function of host-associated microbial communities are associated with many aspects of health and disease [1]. These relationships between the microbiome and host biology exhibit spatial and temporal dependencies, with relevant interactions manifest by the microbiota of specific body sites during critical periods of host development, environmental exposure, pathogenesis, illness, or convalescence [2–6]. Specifically, there is a growing body of evidence that the microbiome influences immune maturation and function [7–9], mucosal surface physiology [10, 11], and the risk and severity of acute and chronic respiratory diseases [12–16].
Respiratory syncytial virus (RSV) is the most significant respiratory tract infection affecting infants. It is the most frequent cause of acute lower respiratory infections in children younger than 5 years of age, and a common cause of hospitalization in children younger than 2 years of age [17–19]. Approximately one-half of infants are infected with RSV during their first year of life, and nearly all have been infected by 2 years of age. Severe disease requiring hospitalization occurs in 1%–3% of those infected, and in most cases is not accompanied by any of the known risk factors such as age at infection, preterm birth, underlying cardiopulmonary disease, or immunosuppression [20–22]. Additionally, RSV infection in early life has been linked to subsequent development of asthma and chronic obstructive lung disease [19, 23].
Recent studies have identified associations between nasopharyngeal microbiota and RSV clinical manifestations, including severity [24, 25]. Nasopharyngeal microbiota composition has been shown to be altered during periods of acute RSV infection and the abundance of certain bacterial taxa have been associated with immune response and disease severity [13, 24–26]. While these findings suggest that respiratory microbiota may play an important role in RSV infection, the spatial and temporal scope of a relationship remains unclear. Specifically, whether associations between RSV infection and microbiota composition are limited to the nasopharynx, and in what sequence and duration they manifest, are not well understood [27, 28].
Here, we analyze the nasal microbiota of 2 cohorts of infants to elucidate the relationship between airway microbial communities and RSV infection. We used a large cross-sectional cohort of infants composed of an RSV-infected case group sampled during acute illness and a matching healthy control group to characterize the nasal microbiota of acute RSV infection and identify associations with disease severity. To assess associations with the nasal microbiota that may exist before or after RSV infection, we used a smaller longitudinal cohort composed of a group of infants that developed RSV infection during their first year of life and another group that did not, with each group sampled at matching time points corresponding to before, during, and after acute illness.
METHODS
Clinical Methods
All study procedures were approved by the University of Rochester Medical Center (URMC) Research Subjects Internal Review Board (protocol number RPRC00045470) and all subjects’ caregivers provided informed consent. The infants included in the study were from the University of Rochester Respiratory Pathogens Research Center AsPIRES (Assessing and Predicting Infant RSV Effects and Severity) [29] and Prematurity, Respiratory Outcomes, Immune System, and Microbiome (PRISM) studies and cared for prior to discharge in the URMC Golisano Children’s Hospital and Rochester General Hospital newborn nurseries and birthing centers. For the cross-sectional cohort (Table 1), we analyzed 191 nasal samples from 89 subjects with acute RSV infection and 102 healthy subjects. For the longitudinal cohort (Table 2), we collected 72 nasal samples from 12 RSV positive subjects and 12 healthy subjects. Healthy controls were asymptomatic and had not been exposed to antibiotics within 30 days of enrollment or at the time of sampling. Additional selection criteria and seasonal time frames for subjects in both cohorts are detailed in Supplementary Methods and Supplementary Material. Patient metadata for the cross-sectional and longitudinal cohorts is included in Supplementary Table 1.
Table 1.
Summary Characteristics of Cross-Sectional Cohort
| Variables | Aggregate (n = 191) | Control (n = 102) | Case (n = 89) | P Value |
|---|---|---|---|---|
| Sex, No. male/female | 105/86 | 61/41 | 44/45 | .19 |
| Mode of delivery, No. vaginal/cesarean | 128/63 | 68/34 | 60/29 | 1.00 |
| Gestational age at birth, wk, mean ± SD | 39.23 ± 1.17 | 39.27 ± 1.13 | 39.18 ± 1.23 | .58 |
| Age at sampling, mo, mean ± SD | 3.09 ± 2.21 | 3.01 ± 2.19 | 3.18 ± 2.24 | .59 |
| Any antibiotics to date, No. | 0 | 0 | 21 | <.01 |
| Severity score, mean ± SD | 2.24 ± 3.01 | 0 ± 0 | 4.81± 2.65 | NA |
| Severity group, mild/severe | NA | NA | 30/59 | NA |
Abbreviation: NA, not applicable.
Table 2.
Summary Characteristics of Longitudinal Cohort
| Variables (mean ± SD or n) | Aggregate (n = 24) | Control (n = 12) | Case (n = 12) | P Value |
|---|---|---|---|---|
| Sex, male/female | 8/16 | 4/8 | 4/8 | 1.00 |
| Mode of delivery, vaginal/cesarean | 14/10 | 6/6 | 8/4 | 0.68 |
| Gestational age at birth, wk | 39.17 ± 1.05 | 39.58 ± 0.90 | 38.75 ± 1.06 | 0.05 |
| Age at initial sampling, mo | 1.18 ± 0.29 | 1.26 ± 0.36 | 1.10 ± 0.19 | 0.19 |
| Age at second sampling, mo | 2.70 ± 0.79 | 2.65 ± 0.76 | 2.74 ± 0.86 | 0.78 |
| Age at final sampling, mo | 3.67 ± 0.80 | 3.83 ± 0.70 | 3.51 ± 0.89 | 0.35 |
| Severity score | 0.89 ± 1.30 | 0 ± 0 | 1.77± 1.34 | NA |
| Severity group, mild/severe | NA | NA | 10/2 | NA |
Abbreviation: NA, not applicable.
16s rRNA Amplicon Sequencing
Genomic DNA was extracted and the V1–V3 16S rRNA hypervariable region was sequenced as described previously [4]. Bioinformatics processing was performed with QIIME 2 [30], using DADA2 [31] for denoising and the GreenGenes reference database [32, 33] as the basis of taxonomic classification. Additional methodological details are in Supplementary Methods.
Associations of Taxon Abundance With RSV Infection and Disease Severity
Because differential abundance testing of high-throughput sequencing-based microbial community profiling data is relatively immature, with no consensus methodology [34, 35], we applied 4 prominent univariate and multivariate algorithms that were selected to be complementary in terms of their strengths and technical limitations. We required that significant results be corroborated across multiple methods to be accepted. Details of diversity and machine learning classification/regression analyses are in Supplementary Methods.
RESULTS
Overview of Infant Cohort
The cross-sectional case-control cohort yielded 191 nasal samples with 16S rRNA sequencing data from 89 subjects with acute RSV infection and 102 matched healthy subjects (Table 1). The average number of reads per sample was 64 320 with 180 samples having at least 5000 reads. All subjects were full term and younger than 10 months, and the ill and healthy groups matched at the population level in terms of sex, gestational age at birth, mode of delivery, and age at the time of sampling. Infected subjects were divided into mild and severe based on a threshold global respiratory severity score (GRSS) of 3.5, yielding groups of 30 and 59, respectively [36]. Severity scores and additional patient metadata for the cross-sectional and longitudinal cohorts are in Supplementary Table 1.
The longitudinal cohort yielded 72 nasal samples with 16S rRNA sequencing data corresponding to 12 healthy controls and 12 RSV-positive subjects sampled at 3 time points: 1 month of age, during acute illness (and corresponding age for healthy controls), and 1 month after illness (Table 2). The average number of reads per sample was 47 745, with 67 samples having at least 5000 reads. Healthy controls closely matched RSV-positive subjects in terms of sex, mode of delivery, gestational age at birth, and age at the time of sampling. All subjects were full term and younger than 1 year. Healthy controls did not develop symptomatic respiratory infection between birth and at least 10 days after their last sample was taken. Notably, only 2 of the RSV cases in this cohort exhibited severe disease (GRSS > 3.5).
Microbiota Diversity and Associations With RSV Infection and Severity
In the cross-sectional cohort, α diversity as measured by Faith index was elevated in RSV positive subjects at the time of infection relative to age matched healthy controls (P = .039), with a greater difference observed in the group of subjects with severe disease (mean Faiths index of healthy = 1.933, mild illness = 2.176, and severe illness = 2.250). The difference between subjects with mild and severe infection was not significant and the severity score was not significantly correlated with Faith index (r = 0.134; P = .065). There were no significant differences in α diversity as measured by the Shannon index, suggesting that the observed differences reflect increased phylogenetic heterogeneity in the subjects with infection, as opposed to a greater number of total species or more even distributions of species’ relative abundances.
In the longitudinal cohort, weighted and unweighted UniFrac distances were used to assess β diversity at each visit, and to assess the magnitude of change within individuals from visit to visit (Figure 1A). At all 3 time points, significant differences were found between the group that developed RSV infection and the group that did not, based on the weighted UniFrac metric (initial visit P = .032, illness and age matched healthy visit P = .009, and follow-up visit P = .012). By unweighted UniFrac, these differences were significant at the initial visit (P = .035) and the illness visit (P = .011), and approached significance at the postillness visit (P = .078). By both metrics, the largest, most significant difference was observed at the illness visit (and the corresponding time point for the healthy controls). Further examination of β diversity during illness using the cross-sectional cohort (Figure 1B) revealed more significant differences between healthy subjects and severely ill subjects (unweighted UniFrac P = .003, weighted UniFrac P = .003) than between healthy subjects and subjects with mild disease (unweighted UniFrac P = .036, weighted UniFrac P = .005), as well as apparently greater differences between healthy and RSV-infected infants when the infection occurred at younger ages: among subject 0–3 months old, UniFrac permutational multivariate analysis of variance (PERMANOVA) healthy vs mildly ill P = .538 (unweighted) and P = .084 (weighted), healthy vs severely ill P = .001 (unweighted) and P = .001 (weighted); among subjects > 6 months old, healthy vs mildly ill P = .931 (unweighted) and P = .191 (weighted), healthy vs severely ill P = .309 (unweighted) and P = .389 (weighted).
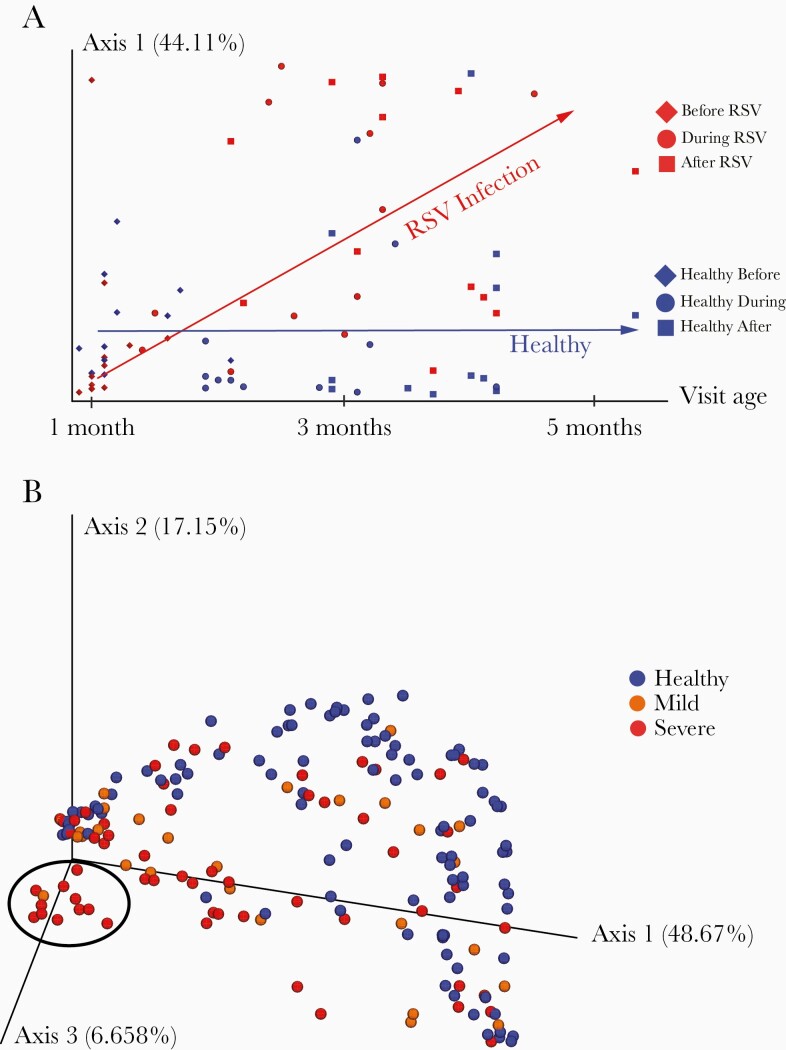
Figure 1.
Principal coordinate analysis (PCoA) of weighted UniFrac distances was used to visualize relationships between the nasal microbiota of infants with respect to respiratory syncytial virus (RSV) infection, illness severity, and time. Weighted UniFrac distances quantify the compositional dissimilarity between microbial communities, incorporating information about the phylogenetic relatedness between bacteria observed across samples. PCoA provides a summary representation of overall similarity/dissimilarity relationships among a set of samples, capturing as much information as possible using the fewest number of dimensions/principal coordinates. The proportion of overall variation represented along a single axis is indicated as a percentage in the axis label. A, From the longitudinal cohort only, samples are plotted with principal coordinate 1 on the y-axis and infant age at the time of sampling on the x-axis. Samples are colored red or blue based on whether or not an infant developed RSV infection (red) at any point during the period of observation, and their shape indicates the time-point at which the sample was taken: initial healthy/preillness visit (diamond), illness visit/age-matched healthy visit (circle), or postillness/age-matched final healthy visit (square). The red and blue arrows indicate observed longitudinal trends within the group of subjects that developed RSV infections and the group that stayed healthy, respectively. B, From the cross-sectional cohort only, samples are plotted in 3 dimensions using the first 3 principal coordinates. Samples are colored according to RSV infection status and severity: healthy (blue), mild RSV infection (orange), or severe RSV infection (red). A cluster of subjects in the foreground on the left, notable for dominant abundance of Haemophilus influenzae, is circled in black. While no clear segregation is observed between mild and severe illness, healthy samples occupy a notable crescent shaped structure around the illness samples, with the H. influenzae dominated cluster furthest away from this crescent.
Notably, as there were 70 healthy and 54 ill infants 0–3 months of age, and only 10 healthy and 10 ill infants > 6 months old, unequal sample sizes may be driving these apparent differences.
Assessing the magnitude of longitudinal changes by the unweighted UniFrac metric, the within-subject change from the initial visit to the illness visit, and the corresponding time point in healthy subjects, was larger among the subjects that developed infection than those that remained healthy (P = .061). All computed α and β diversity values are in Supplementary Table 2.
Longitudinal Abundance Patterns of RSV-Associated Taxa
The relative abundance of 12 distinct taxa exhibited significant associations with the occurrence of RSV infection according to multiple corroborative statistical assessments. While all 12 taxa were differentially abundant between RSV-infected and healthy infants during illness, and the corresponding time point in healthy subjects, they exhibited distinguishable patterns of temporal dynamics, pre- and postillness occurrence, and associations with illness severity (Table 3). Notably, associations between nasal microbiota and RSV infection were not confined to the period of acute infection: all but 1 (Haemophilus) of the 12 taxa associated with RSV infection were significantly differentially abundant between groups either before or after illness, or both. Most of the taxa (7/12) that were differentially abundant between RSV-infected and healthy infants during illness were differentially abundant at the initial visit at 1 month of age, prior to illness. Similarly, most of the taxa (8/12) that were differentially abundant during illness were differentially abundant after illness. However, persistent differential abundance between groups across all 3 time points was observed only in a minority (4/12) of taxa. Furthermore, the microbiota differences between health and RSV infection were not simply categorical but varied in magnitude with illness, as most of the taxa (8/12) that were differentially abundant during illness were associated with illness severity. Additionally, most of these severity-associated taxa (6/8) exhibited persistent differences beyond the period of acute illness and were differentially abundant during and after illness, while half (4/8) were differentially abundant prior to illness. Finally, the abundances of most RSV-associated taxa were positively associated with the disease, and only a minority of taxa (5/12) that differed in abundance between groups were elevated in healthy infants.
Table 3.
Taxa Associated with Respiratory Syncytial Virus (RSV) Infection and Illness Severity and the Time Points at Which They Differ Significantly Between Healthy Infants and Those Who Become Ill With RSV
| Taxon | Positive Association | Before Illness | During Illness | After Illness | Illness Severity |
|---|---|---|---|---|---|
| Alphaproteobacteria | Disease | + | + | + | + |
| Gammaproteobacteria | Disease | + | + | + | + |
| Pseudomonadales (Pseudomonas) | Disease | + | + | + | (+) |
| Moraxella | Disease | + | + | + | − |
| Corynebacterium | Health | + | + | − | − |
| Gluconacetobacter | Disease | + | + | − | + |
| Anaerococcus | Health | + | + | − | − |
| Staphylococcus | Health | − | + | + | + |
| Betaproteobacteria (Burkholderiales) | Disease | − | + | + | (+) |
| Bacilli | Health | − | + | + | + |
| Clostridia | Health | − | + | + | − |
| Haemophilus (influenzae) | Disease | − | + | − | (+) |
Member clades in parentheses are the primary drivers of the association indicated in parentheses.
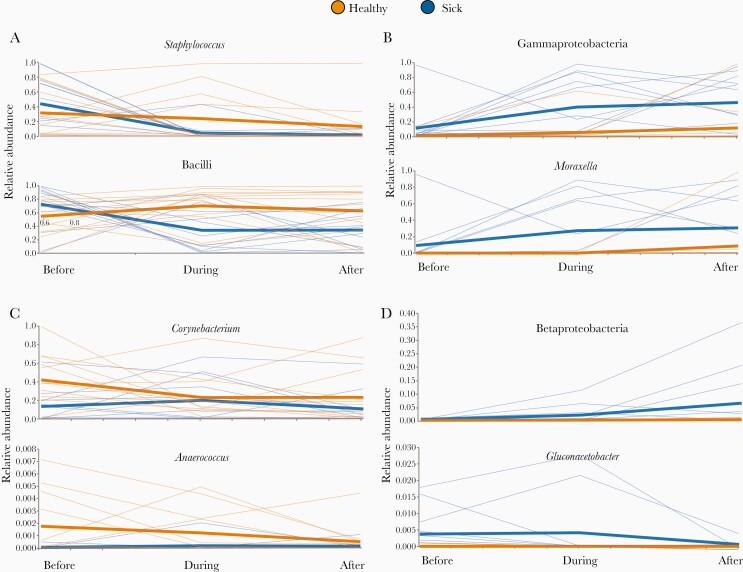
Staphylococcus, Clostridia (not shown), and Bacilli each exhibited a similar temporal pattern; ubiquitous and comparable in abundance between groups prior to illness, significantly diminished during illness (P ≤ .001, .013, and .005, respectively) and remained so 1 month later (Figure 2A; P = .011, .033, and .039, respectively). The classes Alphaproteobacteria (not shown) and Gammaproteobacteria, and Gammaproteobacteria member clades Pseudomonadales (not shown) and Moraxella, also exhibited a common pattern in that all 4 were significantly elevated in the infants that developed RSV infection before (P ≤ .001, .033, .003, and .001, respectively), during (P ≤ .036, .002, .001, and .001, respectively), and after illness (Figure 2B; P = .044, .009, .038, and .003, respectively). By contrast, Corynebacterium and Anaerococcus were elevated in infants that did not develop RSV infections at the preillness time point (P = .008 and P = .020, respectively) and the illness time point (P < .001 and P = .008, respectively) but did not differ between groups at the postillness time point (Figure 2C). The difference in Corynebacterium abundance between health and illness was more pronounced in the cross-sectional cohort (Supplementary Figure 1). Finally, 3 taxa exhibited unique temporal trends with respect to illness: Betaproteobacteria increased in abundance over time in the RSV group only (Figure 2D), being significantly elevated during (P = .006) and after (P = .039) illness, while Haemophilus was significantly more abundant in the infected group during illness (P < .001) and minimally abundant in both groups before and after (Supplementary Figure 2). Gluconacetobacter exhibited a distinct temporal pattern in that it was elevated in the RSV group before (P = .003) and during (P < .001) illness, but no difference was observed between groups after illness (Figure 2D). The composition of all cross-sectional and longitudinal samples summarized at all taxonomic levels is in Supplementary Tables 3 and 4.
Figure 2.
Relative abundances (y-axes) of select taxa at all 3 time points (x-axes) in the longitudinal cohort. Each thin line corresponds to the abundance of a given taxon in a particular individual, while the thick lines show the mean abundance of each group at each time point. Members of the healthy group are orange and members of the group that developed infection are blue. Significant taxa were grouped based on different temporal patterns of abundance with respect to illness, and each panel contains examples from a different group: (A) similar abundance between infection and healthy groups prior to illness, but decreased during and after illness in subjects that become infected; (B) consistently elevated in the illness group; (C) elevated in the healthy group before and during illness, but not after; and (D) idiosyncratic temporal dynamics observed in each taxon. Of the members of the fourth group shown here, Betaproteobacteria is nearly absent from all subjects at the preillness time point, and then becomes increasingly abundant during and after illness in the infection group while remaining nearly absent from the healthy group. Gluconacetobacter is elevated in the infection group prior to and during illness, and substantially diminishes in abundance with convalescence.
Abundance of Taxa Associated With Severity in Acute Illness
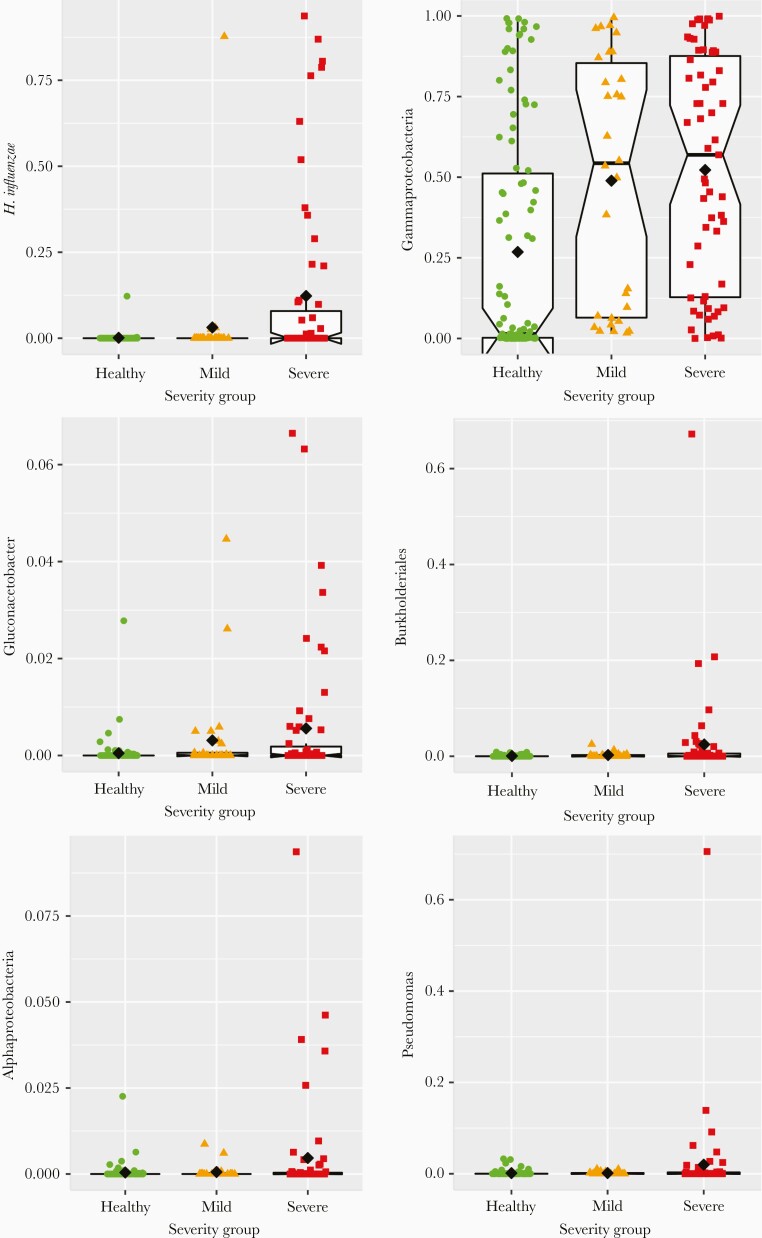
The abundance of 6 of the taxa associated with RSV infection were positively associated with severity at the time of acute illness: Alphaproteobacteria (P = .026), Gammaproteobacteria (P < .001), Pseudomonas (P < .001), Gluconacetobacter (P < .001), Burkholderiales (P = .015), and Haemophilus (P < .001), with exceptionally high levels of Haemophilus influenzae very strongly associated (P < .001) with severe disease (Figure 3). Pseudomonas and Burkholderiales were the primary drivers of associations between severity and their corresponding clades, Pseudomonadales and Betaproteobacteria. The abundance of Bacilli (P < .001) and Staphylococcus (P < .001), conversely, were negatively associated with disease severity at the time of illness. As described above, Moraxella, Corynebacterium, Anaerococcus, and Clostridia were associated with the occurrence of RSV infection (or lack thereof), but they were not associated with severity of disease.
Figure 3.
Relative abundances (y-axes) of select taxa significantly associated with more severe disease in the cross-sectional cohort, with samples grouped by dichotomizing illness based on severity into mild and severe groups (x-axes), using a severity score threshold of 3.5. Each colored point represents the relative abundance of a given taxon in a single individual, with columns (left to right), shapes (circle, triangle, square), and colors (green, orange, red) distinguishing between healthy, mild illness, and severe illness groups, respectively. The black diamonds indicate the group mean for each group. Box plots are overlaid on each group, centered on the group median, with notches indicating an approximately 95% confidence interval, boxes indicating boundaries of the first and third quartiles, and whiskers extending to the largest and smallest values no further than 1.5 × interquartile range from the boxes. Points beyond the whiskers are commonly considered outliers, which in this case would suggest that many of the observed associations between taxon relative abundance and illness severity are driven primarily by outliers, or that taxon abundance in severely ill infants comprises more than 1 underlying distribution.
Predicting RSV Infection Status and Illness Severity From Microbiota Composition
To further assess the relationship between nasal microbiota and RSV infection using the cross-sectional cohort, gradient tree boosting machine learning models were trained and applied to predict the RSV infection status of a subject using the composition of their nasal microbiota, where status was defined in 3 ways: RSV infected vs healthy; healthy vs mild RSV infection vs severe RSV infection; and severity score (with all healthy subjects having a score of 0). Five-fold cross-validation was employed, with 20% of samples being held out during training and then used to test the accuracy of the trained model. This approach can indicate how much information about a subject’s status is reflected in the composition of their nasal microbiota. Performance was best when distinguishing infected from healthy, which could be done with 90% accuracy using compositional profiles at the level of exact sequence variants. Distinguishing healthy, mild disease, and severe disease was less effective, with an accuracy of 77% being achieved using compositional profiles summarized based on taxonomic assignment at the level of species. Prediction of the continuous valued severity score exhibited the worst performance, with 38% of the variance in severity score being explained by the model, also using species level compositional profiles. Sequence variants classified as Moraxella, Staphylococcus, Corynebacterium, or Streptococcus comprised the top 3 most informative features across all 3 models. Model result summaries are in Supplementary Tables 5–7.
DISCUSSION
In this study, we characterized signatures of dysbiosis associated with RSV infection and illness severity in infant nasal microbiota. We identified differences in measures of microbial diversity and in the abundance of specific bacterial taxa between infants who develop RSV infections and those who did not, and showed that these differences manifested longitudinally before, during, and after illness in a number of distinct patterns. While these associations are consistent with observations made previously of nasopharyngeal microbiota during acute illness [13, 26], the findings reported here elucidate the temporal sequence and persistence of these phenomena beyond the period of acute illness, and demonstrate their occurrence in the nasal cavity.
Based on the observed temporal patterns of differential abundance before, during, and after illness, the relationships between most of the taxa identified as significant and RSV infection were assigned to 1 of 3 general categories. The first category includes taxa that change during and after illness relative to healthy controls of the same age. The dynamics of these taxa are consistent with RSV infection influencing the abundance of normal flora that diminish in a persistent way as a result of infection (Clostridia, Bacilli, and Staphylococcus). The second category consists of taxa increased in abundance in infants who develop RSV infection at all time points: before, during, and after illness. The patterns of occurrence of these taxa imply that they either are indicative of underlying factors making the host more susceptible to infection or directly contribute to infection susceptibility (Alphaproteobacteria, Gammaproteobacteria, Pseudomonadales, and Moraxella). The third category is composed of taxa significantly elevated in the healthy control subjects at the initial visit and the time point corresponding to illness. Such taxa (Corynebacterium and Anaerococcus) likely either reflect underlying protective qualities of the host, promote such qualities, or are directly protective themselves. The remaining 3 taxa identified as significant each exhibit unique patterns of differential abundance between groups and do not fit well into any of the 3 categories. The interpretations of their associations with RSV infection are less clear, but the occurrence patterns of Betaproteobacteria and Haemophilus could be explained by an opportunistic or synergistic relationship, wherein RSV infection produces circumstances conducive to their increasing in abundance, while their increased abundance may contribute to infection severity. The occurrence patterns of Gluconacetobacter suggest that it may reflect or promote infection susceptibility, but convalescence corresponds to an unfavorable host environment and it diminishes in abundance.
Most RSV-associated taxa are associated both with the presence or absence of infection, and also with disease severity. This implies that the biological underpinnings of these associations exist to varying degrees as opposed to being categorically distinct, and that this variation is reflected in illness severity during infection. However, the fact that our infected vs healthy classifier outperforms our severe illness vs mild illness vs healthy classifier and the severity score regressor, and furthermore all significant taxa are associated with illness while only 8 of them are associated with severity, suggest that the composition of nasal microbiota is more strongly associated with the difference between RSV infected and uninfected than it is with continuous variation along the gradient from health to severe illness.
The taxa that are differentially abundant at 1 month of age—prior to infection and illness—present intriguing possibilities. At a basic level, it may be possible to predict an infant’s risk of RSV infection in the first year of life based on the presence and abundance of these taxa at 1 month. Furthermore, understanding the mechanism by which these bacteria are associated with infection risk could provide valuable insights into immunological development or mucosal function. More speculatively, the possibility exists that the association is causal, which would suggest that these taxa may be suitable targets for prebiotic, probiotic, or antimicrobial interventions. Similar reasoning could be applied to H. influenzae and Betaproteobacteria Burkholderiales, which are not differentially abundant prior to illness but are associated with illness severity, and which could be targeted or assayed during infection to mitigate or predict severity. Whether these microbes merely reflect underlying factors that influence infection susceptibility, severity, and resistance, or contribute to them directly, the clinical significance of RSV infection in the short term, and respiratory infection-associated asthma and atopy in the long term, make these bacteria and their relationship to respiratory health important targets for translationally oriented study.
We recognize a number of limitations of this study. Notably, our longitudinal cohort was substantially smaller than our cross-sectional cohort and incidentally it only contained 2 severely ill subjects. This prevented us from assessing associations with severity at the pre- and postillness time points, which would be desirable. Similarly, analyses of the pre- and postillness time points were not as powerful as those of the acute illness/cross-sectional time point, and the different numbers of significant results identified at each time point may be reflective of this disparity in power. We were also limited to short read amplicon sequencing to profile the bacterial communities that were sampled, which inherently limits our ability to resolve species and strains of bacteria. Furthermore, marker gene assays contain no functional information about the microbial communities and no immunological information about the host. More comprehensive assays such as shotgun metagenomic sequencing and flow cytometry would greatly enrich our understanding of the systems of interest. Finally, all of our subjects were younger than 1 year, had not been previously infected with RSV, and no subject was sampled more than approximately 1 month after illness, which prevented us from examining microbiota-RSV associations among infants who became infected in their second year of life or who had recurrent infections, and made it impossible to determine how long the associations we observed persisted after illness. Similarly, our earliest samples were at approximately 1 month of age and already showed differences between subjects who went on to acquire RSV infections and those who did not, so we were unable to determine how early those differences manifested. Nevertheless, our findings provide novel insight into the developmental dynamics of the nasal microbiome in the first year of life as they relate to susceptibility, acute illness, severity, and convalescence associated with first-time RSV infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. Microbiome sequencing in this study was completed by the University of Rochester Genomics Research Center. We thank Amy Murphy R.N., Mary Criddle R.N., and Doreen Francis R.N. for assistance in recruiting and following study subjects.
Author Contributions. S. R. G., E. E. W., and M. T. C. conceptualized the study. S. R. G., E. E. W., T. J. M., M. T. C., and A. G. designed the experiments. E. E. W., M. T. C., and A. R. F. developed the cohort, and collected the specimens and clinical data. J. H.-W., S. B., J. J., and A. C. facilitated data organization, management, and analysis. T. J. M., M. T. C., E. E. W., A. L. G., H. A. K., S. R. G., J. H. W., J. J., S. B., A. C., and A. G. generated, analyzed, and interpreted the data. S. R. G. and A. G. wrote and/or revised the manuscript. All authors read and approved the final manuscript.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant number HHSN272201200005C).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data availability. All phenotypic data, 16S rRNA sequence reads, and generated datasets are publicly available through the database of Genotypes and Phenotypes (dbGaP) accession phs001201.v2.p1. Overlapping datasets from the AsPIRES study utilized for our study were also used for 2 related studies (Chu et al [37] and Wang et al [38]).
References
- 1. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148:1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biol 2011; 12:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Faust K, Sathirapongsasuti JF, Izard J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 2012; 8:e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grier A, McDavid A, Wang B, et al. Neonatal gut and respiratory microbiota: coordinated development through time and space. Microbiome 2018; 6:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hofstra JJ, Matamoros S, van de Pol MA, et al. Changes in microbiota during experimental human Rhinovirus infection. BMC Infect Dis 2015; 15:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu Q, Wischmeyer J, Gonzalez E, Pichichero ME. Nasopharyngeal polymicrobial colonization during health, viral upper respiratory infection and upper respiratory bacterial infection. J Infect 2017; 75:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol 2017; 18:851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gollwitzer ES, Saglani S, Trompette A, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med 2014; 20:642–7. [DOI] [PubMed] [Google Scholar]
- 9. Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017; 15:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am J Respir Crit Care Med 2017; 196:1582–90. [DOI] [PubMed] [Google Scholar]
- 13. de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Steenhuijsen Piters WA, Sanders EA, Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc Lond B Biol Sci 2015; 370:20140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hilty M, Qi W, Brugger SD, et al. Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis 2012; 205:1048–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 2012; 78:6262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hall CB, Weinberg GA, Blumkin AK, et al. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics 2013; 132:e341–8. [DOI] [PubMed] [Google Scholar]
- 18. Stockman LJ, Curns AT, Anderson LJ, Fischer-Langley G. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997–2006. Pediatr Infect Dis J 2012; 31:5–9. [DOI] [PubMed] [Google Scholar]
- 19. Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev 2010; 23:74–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boyce TG, Mellen BG, Mitchel EF Jr, Wright PF, Griffin MR. Rates of hospitalization for respiratory syncytial virus infection among children in medicaid. J Pediatr 2000; 137:865–70. [DOI] [PubMed] [Google Scholar]
- 21. García CG, Bhore R, Soriano-Fallas A, et al. Risk factors in children hospitalized with RSV bronchiolitis versus non-RSV bronchiolitis. Pediatrics 2010; 126:e1453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 1986; 140:543–6. [DOI] [PubMed] [Google Scholar]
- 23. Proud D, Chow CW. Role of viral infections in asthma and chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 2006; 35:513–8. [DOI] [PubMed] [Google Scholar]
- 24. Fonseca W, Lukacs NW, Ptaschinski C. Factors affecting the immunity to respiratory syncytial virus: from epigenetics to microbiome. Front Immunol 2018; 9:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosas-Salazar C, Shilts MH, Tovchigrechko A, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis 2016; 214:1924–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biesbroek G, Tsivtsivadze E, Sanders EA, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014; 190:1283–92. [DOI] [PubMed] [Google Scholar]
- 28. Lynch JP, Sikder MA, Curren BF, et al. The influence of the microbiome on early-life severe viral lower respiratory infections and asthma-food for thought? Front Immunol 2017; 8:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walsh EE, Mariani TJ, Chu C, et al. Aims, study design, and enrollment results from the assessing predictors of infant respiratory syncytial virus effects and severity study. JMIR Res Protoc 2019; 8:e12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019; 37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hawinkel S, Mattiello F, Bijnens L, Thas O. A broken promise: microbiome differential abundance methods do not control the false discovery rate. Brief Bioinform 2019; 20:210–21. [DOI] [PubMed] [Google Scholar]
- 35. Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017; 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caserta MT, Qiu X, Tesini B, et al. Development of a global respiratory severity score for respiratory syncytial virus infection in infants. J Infect Dis 2017; 215:750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chu CY, Qiu X, McCall MN, et al. Airway gene expression correlates of RSV severity and microbiome composition in infants. 14 September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang L, Chu CY, McCall MN, et al. Airway gene-expression classifiers for respiratory syncytial virus (RSV) disease severity in infants. bioRxiv 628701 [Preprint]. 9. May 2020. [cited 16 September 2020]. Available from: 10.1101/628701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.