Abstract
Background
Respiratory syncytial virus (RSV) is the leading cause of severe respiratory disease in infants. The causes and correlates of severe illness in the majority of infants are poorly defined.
Methods
We recruited a cohort of RSV-infected infants and simultaneously assayed the molecular status of their airways and the presence of airway microbiota. We used rigorous statistical approaches to identify gene expression patterns associated with disease severity and microbiota composition, separately and in combination.
Results
We measured comprehensive airway gene expression patterns in 106 infants with primary RSV infection. We identified an airway gene expression signature of severe illness dominated by excessive chemokine expression. We also found an association between Haemophilus influenzae, disease severity, and airway lymphocyte accumulation. Exploring the time of onset of clinical symptoms revealed acute activation of interferon signaling following RSV infection in infants with mild or moderate illness, which was absent in subjects with severe illness.
Conclusions
Our data reveal that airway gene expression patterns distinguish mild/moderate from severe illness. Furthermore, our data identify biomarkers that may be therapeutic targets or useful for measuring efficacy of intervention responses.
Keywords: RSV, airway transcriptome, host response, nasal transcriptome, severity
Our data suggest that acute, interferon-mediated intracellular signaling in airway cells is critical for normal responses to RSV infection in infants.
(See the Major Article by Grier et al, on pages 1650–8.)
Respiratory Syncytial Virus (RSV), a negative-strand RNA virus in the Pneumoviridae family, is the most important cause of respiratory tract infection during infancy, causing annual winter outbreaks [1–6]. In the United States, approximately half of the 4 million newborns are infected with RSV during their first winter, with 1%–3% hospitalized and an additional 4%–7% and 10%–16% seen in emergency departments or physician offices, respectively [7]. Mortality is uncommon in the United States (approximately 50 deaths annually); however, in developing countries it is estimated that annually RSV causes 118 thousand deaths, 6 million cases of severe acute lower respiratory illness, and 3 million hospitalizations in children younger than 5 years [8, 9]. Currently, there is no available vaccine for RSV, although several candidate vaccines are in clinical trials.
Long-established major risk factors for severe RSV illness include prematurity, chronic lung disease of prematurity, congenital heart disease, neuromuscular disease, and immune compromise [4, 10]. However, approximately 70% of hospitalized infants in the United States have no overt risk factors for severe illness, although young age at infection, environmental influences such as tobacco smoke exposure, viral load and strain, low levels of maternally derived RSV-neutralizing antibody, as well as a multitude of genetic host factors have been associated with severe disease in some but not all studies [4, 11–20]. Recently, the presence of Haemophilus influenzae and Streptococcus pneumoniae in the nasal microbiota during RSV infection has been associated with greater severity [21–23].
Finally, and importantly, the infant’s immune response to RSV is thought to be a major driver of disease pathogenesis, especially during primary infection [24, 25]. Several studies in infants suggest that T helper 2 (Th2) biased responses and Th17 responses during primary infection may contribute to a more inflammatory and severe outcome [26–28]. Innate immune responses by immune cells, such as neutrophils, and respiratory epithelial cells are also likely to play pivotal roles in both eliminating virus replication as well as enhancing or moderating the inflammatory response [29, 30].
The AsPIRES (Assessing and Predicting Infant RSV Effects and Severity) study is a comprehensive study designed to identify factors associated with disease severity in full-term healthy infants younger than 10 months undergoing primary RSV infection. In this report we analyze gene expression of nasal respiratory epithelial cells, in addition to the influence of respiratory microbiota, in relation to illness severity during primary RSV infection.
METHODS
Please see Supplementary Methods for details regarding the study cohort, procedures, and methods used for data analysis. The transcriptional and microbiota data described in this manuscript are available in the database of Genotypes and Phenotypes (dbGaP) accession phs001201.v2.p1. An analysis of the predictive potential of nasal gene expression, using an overlapping and related transcriptional data set, is described in Wang et al [31]. A study of the chronology of airway microbiota dysbiosis associated with RSV infection, using an microbiota data set, is described in Grier et al [32].
Study Subjects
RSV infected infants were identified and enrolled into the AsPIRES cohort as previously described [33, 34]. Briefly, RSV was identified in a prospectively enrolled birth cohort, a second group of infants with respiratory illness seen in pediatric offices and emergency rooms, and a hospital cohort diagnosed with RSV on admission. Ill subjects were previously healthy full-term infants (>36 weeks’ gestation at birth) younger than 10 months of age at the time of primary RSV infection. Parental informed consent was obtained and the study was approved by the institutional review boards of the University of Rochester and Rochester General Hospital.
Study Protocol and Procedures
Initial RSV infection was confirmed by quantitative reverse transcription polymerase chain reaction (RT-PCR) as described [11]. For the prospectively enrolled birth cohort a diagnostic nasal swab was performed in the research laboratory. For the hospital cohort, RSV was identified by standard-of-care RT-PCR in the clinical laboratory. Infants enrolled in office or emergency rooms were diagnosed by either standard-of-care or a research assay. Once identified as RSV infected, all enrolled infants had a nasal swab for analysis of respiratory microbiota from one nostril and a nasal wash followed by collection of nasal epithelial cells for nasal transcriptome studies from the contralateral nostril performed within 24 hours of initial viral diagnosis, as previously described [35]. Nasal samples were collected with a flocked swab by gentle rubbing at the level of the inferior turbinate.
Defining Illness Severity
Illness severity was defined on a continuous scale (from 0 to 10) using a global respiratory severity score (GRSS) [33]. For some secondary analyses, severe illness was dichotomously defined as GRSS > 3.5.
Library Preparation and Sequencing
We have previously described methods for sequencing infant nasal RNA samples [35]. Sequences were aligned to reference genome GRCh38, normalized to FPKM (fragments per kilobase of exon per million reads), and filtered for nominal expression. Twenty-five samples were removed from the analytical data set for poor quality. We used ComBat [36] to remove batch effects. To avoid spurious findings due to outliers, we also winsorized the data at 1% and 99% levels.
Gene Significance Analyses
Univariate and multivariate significance analyses were conducted using the R package limma [37] with a robust M-estimator [38]. Benjamini-Hochberg multiple testing correction was applied to control the false discovery rate (FDR) at .05 level [39].
Functional Classification
Ingenuity Pathway Analysis [40], ToppGene Suite [41] and CTen [42] were used for ontological analyses.
Gene Expression Validation
Quantitative real-time polymerase chain reaction (qPCR) was performed as we have previously described [35].
Cytokine Determination Multiplex Assay
Nasal wash samples were tested for cytokines and chemokines using R&D Systems Human Luminex assay following the manufacturer’s protocol.
Detection of Pathogenic Virus and Bacteria
TaqMan Array Card (TAC) technology was used to detect common respiratory pathogens as well as Hemophilus influenzae and Streptococcus pneumoniae, as previously described [43–45].
Microbiome Analysis
Airway microbiota analysis from infants was performed essentially as previously described [35]. Reads were analyzed using phylogenetic and operational taxonomic unit (OTU) methods in the Quantitative Insights into Microbial Ecology software [46], and normalized using the cumulative sum stabilization method from the metagenomicSeq R package [47].
Transcriptome and Microbiome Integration
We limited analyses to those 83 subjects for whom both transcriptome and microbiome samples were available, and who were not on antibiotics. A multivariate linear regression was conducted on GRSS as the outcome variable including visit age, a single OTU marginally associated with GRSS (H. Influenzae), and the expression profiles of each gene marginally associated with GRSS (1185 genes in total).
Statistical Analysis of Clinical Data
Descriptive characteristics of the study cohort were reported in Table 1. For binary variables, percentages and frequencies were reported; for continuous variables, means and standard deviations were reported. Appropriate statistical tests were performed to test the association between each clinical variable and disease severity. Specifically, for dichotomous severity (mild vs severe), we used 2-sample Welch t test and Fisher exact test for continuous and binary variables, respectively; for continuous severity (GRSS), 2-sample Welch t test and Pearson correlation test were used for the binary and continuous clinical variables, respectively. A P value < .05 was considered statistically significant. All analyses were performed with SAS (version 9.3; SAS Institute) and the R programming language (version 3.5; R Foundation for Statistical Computing).
Table 1.
Subject Demographics
| Characteristic | Total (n = 106) | Mild (n = 42) | Severe (n = 64) | Mild vs Severe | Correlation With GRSS | ||
|---|---|---|---|---|---|---|---|
| t Statistics | P Value | Pearson Correlation Coefficient | P Value | ||||
| Continuous variable, mean ± SD |
|||||||
| Age, mo | 3.35 ± 2.22 | 3.52 ± 1.99 | 3.24 ± 2.37 | 0.6579 | .5122 | −0.2081 | .0323 |
| Gestational age, wk | 38.90 ± 1.37 | 39.05 ± 1.25 | 38.8 ± 1.44 | 0.9515 | .3437 | −0.1003 | .3064 |
| Birth weight, kg | 3.34 ± 0.61 | 3.32 ± 0.68 | 3.36 ± 0.57 | −0.3240 | .7468 | 0.0721 | .4625 |
| Family size | 4.16 ± 2.25 | 4.43 ± 2.86 | 3.98 ± 1.73 | 0.9027 | .3703 | −0.0494 | .6150 |
| Viral load, log10 NS titer | 2.43 ± 1.16 | 2.48 ± 1.11 | 2.40 ± 1.20 | 0.3191 | .7504 | 0.0209 | .8346 |
| Mild vs Severe | Comparison With GRSS | ||||||
| Fisher Odds Ratio | P Value | t Statistics | P Value | ||||
| Categorical variable, No. (%) | |||||||
| Sex, female | 54 (51) | 19 (45) | 35 (55) | 0.687 | .4275 | 0.5493 | .584 |
| White | 62 (58) | 23 (55) | 39 (61) | 1.5276 | .3115 | −0.5511 | .5832 |
| Tobacco smoke exposure, true | 36 (34) | 14 (33) | 22 (34) | 1.0472 | 1 | −0.7605 | .4492 |
| RSV strain, A | 59 (56) | 23 (55) | 36 (56) | 0.9939 | 1 | 0.207 | .8365 |
| Bacterial colonization, true | 79 (75) | 30 (71) | 49 (77) | 1.3033 | .6498 | −1.2747 | .2092 |
| Viral coinfection, truea | 18 (17) | 9 (21) | 9 (14) | 0.603 | .4285 | 0.6068 | .5495 |
Data represent samples collected at the time of acute infection (1–10 d after onset of clinical symptoms) from 106 subjects. Data are mean ± SE for continuous variables and the frequency in both groups for categorical variables. P values were computed by statistical tests that are appropriate to the nature of the variables (see section “Statistical Analysis of Clinical Data”).
Abbreviations: GRSS, global respiratory severity score; NS, neutralization serum; RSV, respiratory syncytial virus.
aCoinfecting viruses were adenovirus (3), bocavirus (1), coronaviruses (11), human metapneumovirus (1), parechovirus (1), and rhinovirus (6).
RESULTS
Subject Demographics
We sought to understand target organ resident cell responses during primary RSV infection in infants displaying the full spectrum of illness severity. We used a nasal cell sampling procedure [35, 48] to measure infant airway transcriptional responses in 106 subjects from the AsPIRES study [33, 34]. Demographic data for these 106 infants are described in Table 1. Subjects were assigned a GRSS [33]. There was no association between severity and RSV strain, family-reported environmental tobacco smoke exposure, or the presence of other viral pathogens or bacterial colonization based on a positive RT-PCR. The severity was only associated with subject age at the time of infection, and only when the severity was defined as a continuous variable.
Airway Transcriptional Correlates of Disease Severity
We assessed the expression of 13 688 genes by RNAseq following standard quality control-based sample and gene filtering [26, 35]. Gene expression was enriched in canonical (eg, CDH1, EPCAM) and upper airway-specific (eg, BPIFs, MUCs) marker genes, with lower levels of expression for leukocyte markers (Supplementary Figure 2). Univariate analysis indicated that many variables were not associated with significant differences in gene expression after appropriate adjustments for multiple testing (Table 2). As previously reported [35], the presence of known bacterial pathogens had a large impact on airway gene expression (n = 470 genes), appearing to be driven more by the presence of S. pneumoniae (n = 691 genes) than Moraxella or H. influenzae when identified by TAC. Interestingly, the presence of any additional pathogen during acute infection, defined by TAC technology, was not significantly associated with clinical severity (P = .319, data not shown). Notably, the time elapsed from the onset of clinical symptoms (time) also had a major impact on gene expression (n = 216 genes). No gene was marginally identified as significantly associated with severity when defined dichotomously (mild vs severe). However, 142 genes were significantly associated with GRSS when used as a continuous score. We completed a multivariate analysis including sex, race, bacterial pathogen colonization, and time since onset of clinical symptoms. This analysis identified 252 genes significantly associated with GRSS, 728 genes associated with time since illness onset, and 135 genes associated with the presence of any bacterial pathogen (Table 2 and Supplementary Figure 3).
Table 2.
Number of Genes With Significant Changes Associated With Individual Clinical and Demographic Variables
| Clinical Variable | Value (n = 106) |
No. of Associated Genes | |
|---|---|---|---|
| Univariate, BH P < .05 | Multivariate, BH P < .05 | ||
| Continuous variables, mean ± SD | |||
| Clinical severity, GRSS | 4.35 ± 2.66 | 142 | 252 |
| Days since onset of illness | 4.60 ± 0.20 | 216 | 728 |
| Gestational age, wk | 38.90 ± 1.37 | 0 | 0 |
| Age, months | 3.35 ± 2.22 | 5 | 1 |
| Categorical variables, No. (%) | |||
| GRSS, mild | 42 (40) | 0 | … |
| Sex, female | 54 (51) | 18 | 60 |
| White, true | 62 (58) | 13 | 17 |
| Tobacco smoke exposure | 36 (34) | 0 | 0 |
| Viral coinfection | 18 (17) | 7 | 3 |
| Bacterial colonization | 79 (75) | 470 | 135 |
| Streptococcus pneumoniae | 39 (37) | 691 | … |
| Haemophilus influenzae | 29 (27) | 22 | … |
| Moraxella | 67 (63) | 0 | … |
We performed univariate and multivariate regression model to identify gene expression changes associated with intrinsic (age, GRSS, days since onset of illness) and extrinsic (tobacco smoke exposure and bacterial colonization or viral coinfection) factors. Shown are the number of genes identified as significant for each variable in the model.
Abbreviations: BH, Benjamini-Hochberg; GRSS, global respiratory severity score.
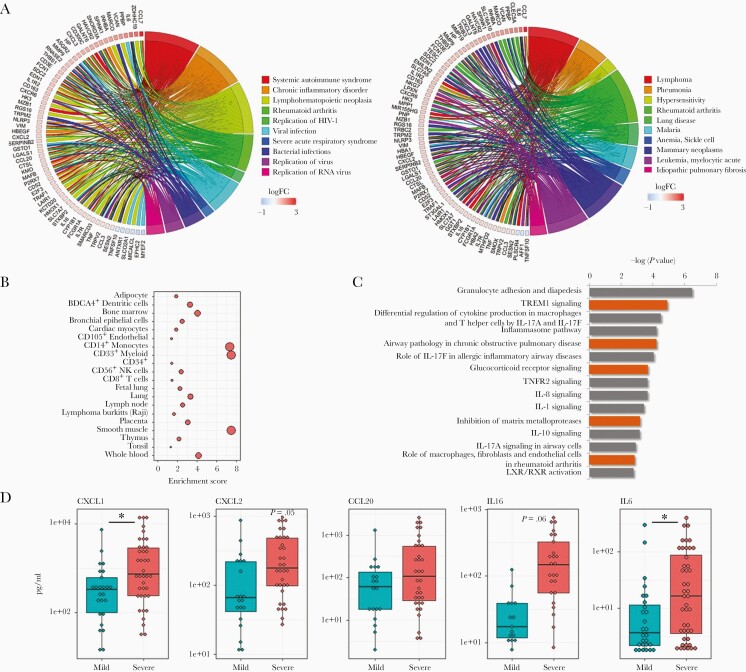
Genes significantly associated with severity were rich in chemokines, cytokines, and interleukin-related molecules (Supplementary Figure 4). Ontological analysis of this gene set identified multiple biological functions related to viral infection (Figure 1A), and suggested biomarkers of severity are associated with pneumonia (Figure 1A). This observation is consistent with the lower respiratory track pathogenesis of severe RSV infections. We next assessed cell signatures in these severity biomarkers. Unlike the epithelial predominant signature of the healthy asymptomatic infant [35], severe responses to RSV infection were associated with signatures of multiple immune cells (Figure 1B). The most predominant cell signatures were related to CD14+ and CD33+ myeloid lineages. Pathway analysis confirmed increases in expression of myeloid cell-related genes in severe cases, and strongly implicated interleukin-17 (IL-17)-related responses (Figure 1C).
Figure 1.
Airway gene expression patterns associated with clinical severity in RSV-infected infants. Gene expression from nasal/airway samples, collected from 106 infants infected with RSV, was assessed by high-throughput RNA sequencing. A, Circos plots displaying biological function (left) and disease (right) ontologies associated with severity. Shown are a subset of genes with expression patterns significantly associated (FDR < .05, fold change > 1.5) with clinical severity, following adjustment for other variables in a multivariate model, and key ontologies significantly associated with severity (n = 629 genes, FDR < .05). Biological functions are enriched in viral infection-related ontologies, and disease ontologies include pneumonia, a lower airway pathology. B, Cell type enrichment plots indicate severity genes are associated with multiple hematopoietic lineages, predominantly CD14+ monocytes, CD33+ myeloid cells, and smooth muscle cells. C, Canonical pathway analysis of severity-associated genes suggests alterations in immune signaling in infants with severe clinical symptoms, including IL-17. D, Multiplex ELISA analysis of nasal washes from RSV-infected infants confirms that the production of select immune-related proteins are increased in infants with severe clinical symptoms. Asterisk, P value < .05; Boxes, upper and lower quartiles of data; Whiskers, the highest and lowest data point. Abbreviations: ELISA, enzyme-linked immunosorbent assay; FDR, false discovery rate; IL-17, interleukin-17; RSV, respiratory syncytial virus.
We validated severity-associated gene expression changes by qPCR (Supplementary Table 1). We also performed multiplex enzyme-linked immunosorbent assay (ELISA) analysis using nasal washes. As shown in Figure 1D, we validated significant differences in the level of multiple inflammatory biomarkers associated with severity (eg, CXCL1, CXCL2, IL-6), and nonsignificant differences in others (eg, IL-16).
Effects of Airway Microbiota on Severity Biomarkers
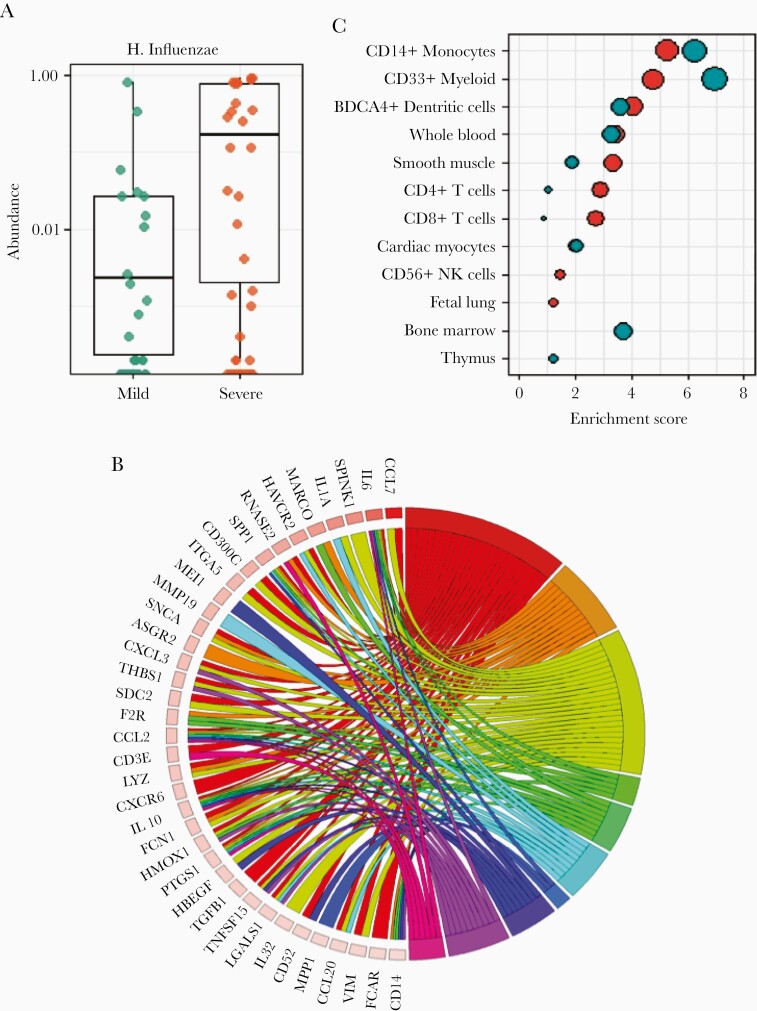
Recent work by others has demonstrated the specific presence of H. influenzae in the airway as a correlate to severe clinical responses in infants with RSV. We completed unbiased microbiota analysis of the airway of our subjects. H. influenzae (Supplementary Figure 5), was the only OTU significantly associated with severity following adjustment for multiple testing in this cohort (Figure 2A and Supplementary Table 2). This presumably represents nontypable H. influenzae because the incidence of H. influenzae type B is uncommon. Interestingly, 9 OTUs (not including H. influenzae) were significantly associated with age, suggesting the nasal microbial communities evolve extensively during infancy in this cohort [49].
Figure 2.
Microbiota effects on airway gene expression in RSV-infected infants. 16s rRNA sequencing was used to interrogate the airway microbiome of 106 infants infected with RSV. A, Only Haemophilus influenzae showed a significant association between relative abundance and clinical severity. B, Airway gene expression in RSV-infected infants was reassessed in a multivariate model including H. influenzae. A circos plot displays disease ontologies associated with severity. Shown are a subset of genes with expression patterns significantly associated (FDR < .05, fold change > 1.5) with clinical severity when H. influenzae is used as a variable in a multivariate model, and key disease ontologies significantly associated with severity (n = 643 genes, FDR < .05). C, Cell type enrichment analysis was used to examine genes significantly associated with severity including H. influenzae as compared to the same model without H. influenzae. This analysis showed adding H. influenzae to the model suggested increases in CD4+ and CD8+ T-cell signatures, and decreases in myeloid cells and monocyte signatures. Asterisk, P value < .05; Boxes, upper and lower quartiles of data; Whiskers, the highest and lowest data point. Abbreviations: FDR, false discovery rate; Hflu, Haemophilus influenzae; RSV, respiratory syncytial virus.
Given the impact of bacterial pathogens on airway gene expression [35] (Table 2), and the singular association of H. influenzae with severity, we sought to identify severity-associated gene expression independent of the impact of H. influenzae. We modified our multivariate analysis by including the relative abundance of H. influenzae in the model. We identified 643 genes significantly associated with GRSS independent of the presence of H. influenzae (and visit age). Many of these genes, pathways (Supplementary Figures 6–8), and cell types were identified in our analysis not including H. influenzae. However, ontological analysis of this gene set identified a greater association with bacterial infection (Figure 2B). Additionally, when including H. influenzae, we note that cell type signatures were attenuated for myeloid lineages and inflated for both CD4+ and CD8+ lymphocyte lineages (Figure 2C). These data suggest that H. influenzae influences the clinical severity of RSV infection in infants by modifying the nature of the inflammatory response.
Effects of Timing on Severity Biomarkers
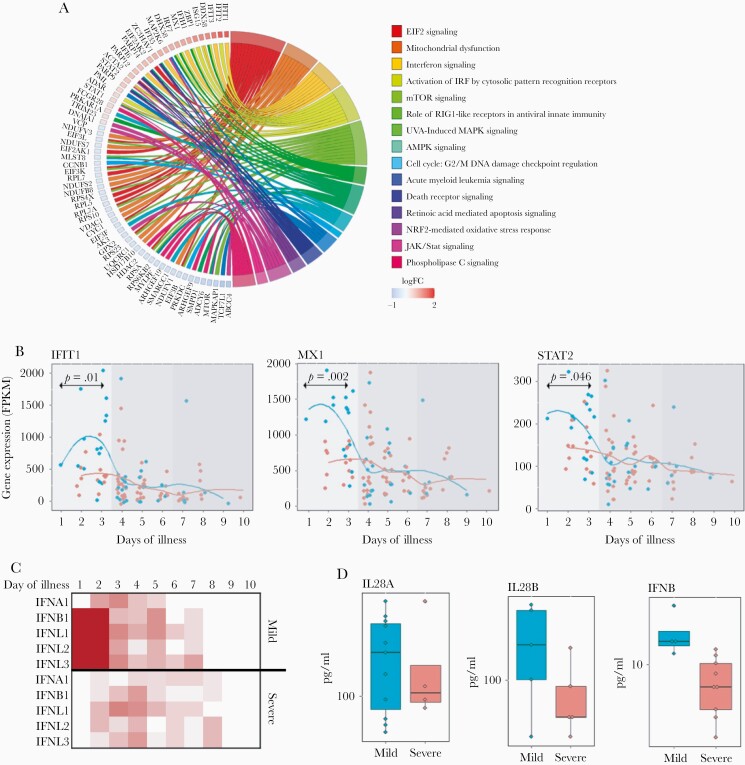
As indicated above, the samples were collected across the acute phase of infection, following the onset of clinical symptoms, and the timing of sample collection had a significant impact on gene expression (Table 2 and Supplementary Figures 9). We interrogated timing biomarkers to better understand the airway response to infection. Pathway analysis (Figure 3A and Supplementary Figures 10 and 11) identified increasing EIF2 and mTOR signaling, and alterations in mitochondrial and oxidative phosphorylation activity. There was also diminishing innate interferon signaling over time. The genes contributing to this signature included intracellular interferon (IFN) signaling molecules (MX1, IFIT1, STAT2), but not the ligands or receptors themselves. We performed exploratory analysis of these IFN pathway-related genes, which revealed a surprising dichotomy between severe and nonsevere subjects (Figure 3B). Significant increases in IFN pathway gene expression in nonsevere subjects occurred only during the first 3–4 days following the onset of clinical symptoms, whereas little to no evidence for interferon signaling was noted in any subjects at later time points. qPCR confirmed a complex relationship between IFN pathway gene expression, clinical severity, and the ontogeny of illness (Supplementary Table 3).
Figure 3.
Alterations in interferon pathway activity define clinical symptoms. Airway gene expression in 106 RSV-infected infants was assessed for significant association with time since onset of clinical symptoms. A, A circos plot displays signaling pathway ontologies associated with severity. Shown are a subset of genes with expression patterns significantly associated (FDR < .05, fold change > 1.5) with time since onset of clinical symptoms in a multivariate model, and key signaling pathway ontologies significantly associated with severity (n = 692 genes, FDR < .05). B, Pseudotime series plots display gene expression patterns for key interferon signaling pathway genes significantly associated with the time since onset of clinical symptoms. IFIT1, MX1, and STAT2 display distinct patterns of temporal expression in RSV-infected infants, with significantly higher expression in infants with mild as compared to severe symptoms. C, Type 1/3 interferon ligand gene expression over time (day 1–10) was assessed separately in mild and severely affected infants. Shown is the estimated expression level (more red equals higher expression) for individual ligand genes at each day following the onset of clinical symptoms. These data are consistent with an early activation in type 1/3 interferon pathway activity in mild subjects only. D, Multiplex ELISA analysis of nasal washes from RSV infected infants confirms that the production of type 1/3 interferon ligands is increased in infants with mild clinical symptoms. Asterisk, P value < .05; Boxes, upper and lower quartiles of data; Whiskers, the highest and lowest data point. Abbreviations: ELISA, enzyme-linked immunosorbent assay; FDR, false discovery rate; FPKM, fragments per kilobase of exon per million reads; RSV, respiratory syncytial virus.
Analysis of regulators for timing biomarkers implicated both type 3 (IFNL1; P < 10−35) and type 1 (IFNA2; P < 10−30) ligands, as well as the canonical interferon-associated transcription factors IRF-3, -7, and -5 (P < 10−15) (Supplementary Figure 12). We performed a post hoc analysis and found evidence for induced expression for both type 1 and 3 IFN ligands (particularly IFNB1, IFNL1, IFNL2, IFNL3) in nonsevere subjects, but not in severe subjects (Figure 3C). Multiplex ELISA of nasal washings confirmed induced levels of IFNB and IFNL in nonsevere subjects (Figure 3D). These data suggest that interferon signaling responses and/or interferon production, within the first few days of infection, contribute to establishing clinical severity in RSV-infected infants.
DISCUSSION
We applied our novel approach to comprehensive molecular analysis of the airways to understand correlates of clinical severity for infant RSV illness. Our data support the utility of this biospecimen as a reasonable surrogate for pathophysiological responses occurring in the lower airway. In addition, the data provide novel insight into disease responses related to the severity of clinical symptoms, the presence of co-occurring microbes and the timeline of clinical symptoms.
We were not surprised to identify many chemokines that are reflective of a more robust inflammatory response in subjects with severe illness. Somewhat more insightful is the ontological analysis that implicates airway changes in IL-1, -8, -10, and -17 signaling in severe patients. Our data also suggest the inflammatory response in the airway differs in infants with severe RSV-associated illness, including an enrichment in CD14+ and CD133+ myeloid cells.
We were interested in understanding the role of the airway microbiota in clinical responses to RSV in infants. We failed to find a strong association between the presence/absence of microbial pathogens and severity. However, we confirmed a report published during our studies indicating a higher burden of H. influenzae in infants with severe clinical symptoms requiring hospitalization [23]. Although other taxa were associated with airway gene expression, we were somewhat surprised to find no other OTUs were significantly associated with severity. It is worth noting microbiome analysis was unable to identify S. pneumoniae at the species level. Given our prior studies revealing a large impact of the microbiota upon gene expression in the airway, we studied the interrelationships between the airway microbiome, airway gene expression, and clinical severity. In general, these analyses confirmed that the presence of known pathogens, particularly bacteria, are strongly associated with gene expression in asymptomatic [35, 48] and symptomatic [48] infants. We considered the specific effect of the microbiota H. influenzae on the relationship between gene expression and RSV severity. The results are consistent with H. influenzae altering the inflammatory response, from myeloid-predominant to CD4/8 T-lymphocyte predominant. These data provide a currently untested hypothesis for a potential mechanism whereby H. influenzae contributes to severe responses at the molecular and cellular level, which is a focus of current investigation.
Our identification of distinct, time-dependent changes in IFN signaling only in nonseverely affected infants is both novel and consistent with the fundamental biology of viral responses. Perhaps this was not readily apparent in prior studies focusing on severely affected infants as they show limited evidence for activation of this pathway. We also draw attention to the narrow time window in which differences in IFN signaling between these groups is evident. Even in our own data, differences in expression do not reach the level of statistical significance without restricting our analyses to the first few days following the onset of clinical symptoms. We should emphasize that our primary observation is not that severity is associated with IFN ligand production, but with estimated levels of signaling activation in the airway. However, we also show differences in IFN ligand expression at both the RNA and protein levels.
The above results are consistent with a pathogenesis model of RSV in which early airway neutrophil infiltration mediated by IL-8 and the emergence of T-cell responses including Th17 CD4 cells playing a role at the peak of disease severity. This is not surprising given the association of Th17 responses and wheezing, a hallmark of severe RSV infection. Given the relationship between H. influenzae abundance and disease severity, and the influence of bacterial colonization on airway epithelia gene expression, one might speculate about the effect on RSV severity by preventing early infancy colonization by this organism. Finally, the finding that early induction of genes in the innate IFN pathway is associated with less severe disease is notable because the RSV NS1 and NS2 proteins are potent and early inhibitors of antiviral type I IFN [50]. Whether this is related to viral strain differences or intrinsic host differences should be explored.
Our evaluation of human biospecimen-derived RNAseq data has utilized multiple analytical and statistical approaches, in an effort to thoroughly interrogate the in vivo responses they reflect. Critically, we have applied appropriately conservative corrections for multiple testing in each case. We believe that use of these analytical approaches is justified, as they each provide distinct yet important insight. Univariate analyses are used to estimate the direct associations between one covariate and individual genes. These analyses are easy to perform and interpret, and do not suffer from potential collinearity issues among covariates. As such, they are useful to initially explore the overall pattern of associations between clinical covariates and transcriptome profiles. On the other hand, a multivariate model considers the influence of several covariates on gene expression simultaneously; therefore, the estimated association between the response variable and a given covariate is less likely to be affected by interdependences. Arguably, a multivariate model is more natural than multiple univariate models because it more closely approximates the complex biological processes that collectively influence the transcriptome. That being said, more caution must be exercised when using complex multivariate models. When uninformative covariates are included in a multivariate model, they may mask true associations and reduce the statistical power to detect informative associations. A more serious issue is the potential collinearity among the covariates, which may prevent modeling certain combinations of covariates and produce unstable estimates. Therefore, we believe it is a practical strategy to first use univariate analyses to identify a subset of important covariates, and then use them to build a lean and robust multivariate model.
We acknowledge this study is not without limitations. We have focused our efforts in identifying gene expression responses that distinguish infants severely affected by RSV infection as compared to infants who are not severely ill and have not included noninfected controls. Therefore, we are unable to address the specificity of the responses we observe and report. Other studies by our group [48] and unpublished data suggest that responses to viral infection can be robustly identified. Another limitation is the associative nature of the in vivo data presented. One must resist presuming that the correlative observations we report are mechanistic, rather than hypothesis generating.
In conclusion, analysis of gene expression by RNAseq in nasal epithelial cells coupled with measurement of the upper airway microbiota at the time of infection offers a minimally invasive method to understand the complex pathogenesis of a common pediatric respiratory pathogen. Results can be used to propose potential interventions that may moderate disease severity.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The University of Rochester Genomics Research Center supported generation of the sequencing data. The authors would like to thank the AsPIRES team for critical assistance with subject recruitment and sample collection. We are indebted to the patients and families who agreed to participate in these studies.
Author contributions. T. J. M., E. E. W., S. R. G., and M. T. C. conceptualized the study. T. J. M., E. E. W., M. T. C., S. R. G., and C. C. designed the experiments. E. E. W., M. T. C., A. R. F., and D. J. T. developed the cohort, and collected the specimens and clinical data. J. H. W. and A. C. facilitated data organization, management, and analysis. T. J. M., M. T. C., E. E. W., X. Q., M. N. M., C. C., L. W., J. H.-W., A. C., A. G., C. S., and D. J. T. generated, analyzed and interpreted the data. T. J. M., E. E. W., M. T. C., X. Q., M. N. M., C. C., L. W., S. R. G., A. G., G. S. P., and J. H.-W. wrote and/or revised the manuscript.
Financial support. This work was supported by National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant number HHSN272201200005C); University of Rochester School of Medicine and Dentistry Scientific Advisory Committee Incubator Grant; and University of Rochester Center for Clinical and Translational Science Institute (grant number UL1 TR002001).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data availability. Complete molecular and microbiota data for these studies are available in the database of Genotypes and Phenotypes (dbGaP) accession phs001201.v2.p1.
References
- 1. Bloom-Feshbach K, Alonso WJ, Charu V, et al. Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): a global comparative review. PLoS One 2013; 8:e54445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hall CB, Weinberg GA, Iwane MK, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med 2009; 360:588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haynes AK, Manangan AP, Iwane MK, et al. Respiratory syncytial virus circulation in seven countries with global disease detection regional centers. J Infect Dis 2013; 208(suppl 3):S246–54. [DOI] [PubMed] [Google Scholar]
- 4. Meissner HC. Viral bronchiolitis in children. N Engl J Med 2016; 374:62–72. [DOI] [PubMed] [Google Scholar]
- 5. Midgley CM, Haynes AK, Baumgardner JL, et al. Determining the seasonality of respiratory syncytial virus in the United States: the impact of increased molecular testing. J Infect Dis 2017; 216:345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mufson MA, Levine HD, Wasil RE, Mocega-Gonzalez HE, Krause HE. Epidemiology of respiratory syncytial virus infection among infants and children in Chicago. Am J Epidemiol 1973; 98:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall CB, Weinberg GA, Blumkin AK, et al. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics 2013; 132:e341–8. [DOI] [PubMed] [Google Scholar]
- 8. Shi T, McAllister DA, O’Brien KL, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 2017; 390:946–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Byington CL, Wilkes J, Korgenski K, Sheng X. Respiratory syncytial virus-associated mortality in hospitalized infants and young children. Pediatrics 2015; 135:e24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hall CB, Powell KR, MacDonald NE, et al. Respiratory syncytial viral infection in children with compromised immune function. N Engl J Med 1986; 315:77–81. [DOI] [PubMed] [Google Scholar]
- 11. Walsh EE, Wang L, Falsey AR, et al. Virus-specific antibody, viral load, and disease severity in respiratory syncytial virus infection. J Infect Dis 2018; 218:208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capella C, Chaiwatpongsakorn S, Gorrell E, et al. Prefusion F, postfusion F, G antibodies, and disease severity in infants and young children with acute respiratory syncytial virus infection. J Infect Dis 2017; 216:1398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stensballe LG, Ravn H, Kristensen K, et al. Respiratory syncytial virus neutralizing antibodies in cord blood, respiratory syncytial virus hospitalization, and recurrent wheeze. J Allergy Clin Immunol 2009; 123:398–403. [DOI] [PubMed] [Google Scholar]
- 14. Glezen WP, Paredes A, Allison JE, Taber LH, Frank AL. Risk of respiratory syncytial virus infection for infants from low-income families in relationship to age, sex, ethnic group, and maternal antibody level. J Pediatr 1981; 98:708–15. [DOI] [PubMed] [Google Scholar]
- 15. McNally JD, Sampson M, Matheson LA, Hutton B, Little J. Vitamin D receptor (VDR) polymorphisms and severe RSV bronchiolitis: a systematic review and meta-analysis. Pediatr Pulmonol 2014; 49:790–9. [DOI] [PubMed] [Google Scholar]
- 16. Alvarez AE, Marson FAL, Bertuzzo CS, et al. Association between single nucleotide polymorphisms in TLR4, TLR2, TLR9, VDR, NOS2 and CCL5 genes with acute viral bronchiolitis. Gene 2018; 645:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyairi I, DeVincenzo JP. Human genetic factors and respiratory syncytial virus disease severity. Clin Microbiol Rev 2008; 21:686–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodriguez-Fernandez R, Tapia LI, Yang CF, et al. Respiratory syncytial virus genotypes, host immune profiles, and disease severity in young children hospitalized with bronchiolitis. J Infect Dis 2017; 217:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis 2011; 204:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vandini S, Biagi C, Lanari M. Respiratory syncytial virus: the influence of serotype and genotype variability on clinical course of infection. Int J Mol Sci 2017; 18:1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hasegawa K, Linnemann RW, Mansbach JM, et al. Nasal airway microbiota profile and severe bronchiolitis in infants: a case-control study. Pediatr Infect Dis J 2017; 36:1044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 2013; 372:3–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Collins PL, Graham BS. Viral and host factors in human respiratory syncytial virus pathogenesis. J Virol 2008; 82:2040–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mariani TJ, Qiu X, Chu C, et al. Association of dynamic changes in the CD4 T-cell transcriptome with disease severity during primary respiratory syncytial virus infection in young infants. J Infect Dis 2017; 216:1027–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russell CD, Unger SA, Walton M, Schwarze J. The human immune response to respiratory syncytial virus infection. Clin Microbiol Rev 2017; 30:481–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Legg JP, Hussain IR, Warner JA, Johnston SL, Warner JO. Type 1 and type 2 cytokine imbalance in acute respiratory syncytial virus bronchiolitis. Am J Respir Crit Care Med 2003; 168:633–9. [DOI] [PubMed] [Google Scholar]
- 29. Deng Y, Herbert JA, Smith CM, Smyth RL. An in vitro transepithelial migration assay to evaluate the role of neutrophils in respiratory syncytial virus (RSV) induced epithelial damage. Sci Rep 2018; 8:6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Do LAH, Pellet J, van Doorn HR, et al. Host transcription profile in nasal epithelium and whole blood of hospitalized children under 2 years of age with respiratory syncytial virus infection. J Infect Dis 2017; 217:134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Chu CY, McCall MN, et al. Airway gene-expression classifiers for respiratory syncytial virus (RSV) disease severity in infants. bioRxiv 628701 [Preprint]. 9. May 2020. [cited 16 September 2020]. Available from: 10.1101/628701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grier A, Gill AL, Kessler HA, et al. Temporal dysbiosis of infant nasal microbiota relative to respiratory syncytial virus infection [published online ahead of print 14 September 2020]. J Infect Dis 2020. 2021; 223:1650–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caserta MT, Qiu X, Tesini B, et al. Development of a global respiratory severity score for respiratory syncytial virus infection in infants. J Infect Dis 2017; 215:750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walsh EE, Mariani TJ, Chu C, et al. Aims, study design and enrollment results from the assessing predictors of infant respiratory syncytial virus effects and severity study. JMIR 2019; 8:e12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chu CY, Qiu X, Wang L, et al. The healthy infant nasal transcriptome: a benchmark study. Sci Rep 2016; 6:33994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8:118–27. [DOI] [PubMed] [Google Scholar]
- 37. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 2016; 10:946–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995; 57:289–300. [Google Scholar]
- 40. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014; 30:523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009; 37:W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shoemaker JE, Lopes TJ, Ghosh S, Matsuoka Y, Kawaoka Y, Kitano H. CTen: a web-based platform for identifying enriched cell types from heterogeneous microarray data. BMC Genomics 2012; 13:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Caserta MT, Yang H, Gill SR, Holden-Wiltse J, Pryhuber G. Viral respiratory infections in preterm infants during and after hospitalization. J Pediatr 2017; 182:53–8.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harvey JJ, Chester S, Burke SA, et al. Comparative analytical evaluation of the respiratory TaqMan Array Card with real-time PCR and commercial multi-pathogen assays. J Virol Methods 2016; 228:151–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kodani M, Yang G, Conklin LM, et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol 2011; 49:2175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 2013; 10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu J, Peterson DR, Baran AM, et al. Host gene expression in nose and blood for the diagnosis of viral respiratory infection. J Infect Dis 2019; 219:1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grier A, McDavid A, Wang B, et al. Neonatal gut and respiratory microbiota: coordinated development through time and space. Microbiome 2018; 6:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thwaites RS, Coates M, Ito K, et al. Reduced nasal viral load and IFN responses in infants with respiratory syncytial virus bronchiolitis and respiratory failure. Am J Respir Crit Care Med 2018; 198:1074–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.