Abstract
Background:
Osteosarcoma represents the most common malignant bone tumor with high metastatic potential and inferior prognosis. RNA methylation (N6-methyladenosine [m6A]) is a prevalent RNA modification that epigenetically influences numerous biological processes including tumorigenesis. This study aims to determine that m6A regulators are significant biomarkers for osteosarcoma, and establish a prognostic model to predict the survival of patients.
Methods:
In this study, we comprehensively analyzed the underlying associations between m6A regulators’ mRNA expressions and metastasis as well as prognosis of osteosarcoma patients in the Cancer Genome Atlas. Multivariate Cox-regression analysis was used to screen regulators that were significantly associated with overall survival of osteosarcoma patients. Least absolute shrinkage and selection operator (LASSO) Cox-regression analysis was used for constructing m6A regulator-based osteosarcoma prognostic signature.
Results:
Some of the regulators exhibited aberrant mRNA levels between osteosarcoma samples with and without metastasis. Multivariate Cox-regression analysis identified several regulators with potential prognostic significance. A risk score formula consisted of methyltransferase-like 3, YTH domains of Homo sapiens, and fat mass and obesity-associated protein was obtained through which patients could be prognostically stratified independently of potential confounding factors. The signature was also significantly associated with the metastatic potential of osteosarcoma. All the analyses could be well reproduced in another independent osteosarcoma cohort from the Gene Expression Omnibus.
Conclusions:
In conclusion, this study first revealed potential roles of m6A regulators in osteosarcoma metastasis and prognosis, which should be helpful for its clinical decision-making.
Keywords: Gene Expression Omnibus, m6A regulators, metastasis, osteosarcoma, prognosis, the Cancer Genome Atlas
1. Introduction
Osteosarcoma is the most common primary malignancy of bone.[1] The 5-year event free survival for patients with localized osteosarcoma is approximately 70% to 80%, however, for patients with metastases and relapses, the survival rate is less than 20%.[2] Current treatment approach of osteosarcoma is surgical treatment for the gross tumors while controlling the metastases with systemic chemotherapy.[3] The development of new treatment strategies has always been at the forefront of osteosarcoma research. Osteosarcoma is extremely heterogeneous in both its origins and manifestations, which is associated with various genomic alterations.[4] Conventional osteosarcoma shows extreme genetic instability, and around 80% of the cases show loss of heterozygosity and genomic instability signatures.[4] Since 2008, new genetic indicators have been identified every year.[5] Unfortunately, despite the progress in this field, the survival rate has not been quite improved, which highlights the need for further exploration.[6]
Genome integrity can be affected by various endogenous or exogenous events, and prevention and/or repair of genomic DNA damage can respond to these stressors to maintain cellular survival.[7] The defective component in DNA damage and repair machinery is one of the core factors that influence treatment outcome in osteosarcoma.[8] Previous reports have indicated that RNA-mediated DNA repair system plays a direct and active role in genome modification and remodeling, which may have important implications in gene targeting and gene therapy.[9,10] N6-methyladenosine (m6A) is an RNA modification that can alter RNA structure and regulate RNA stability as well as RNA metabolism.[11–13] Moreover, m6A has been associated with the tumorigenesis and development of several tumor types, such as leukemia, glioblastoma, lung, and liver cancers,[14,15] however, the role of m6A in osteosarcoma remains poorly understood. Recently, m6A RNA methylation is proved to regulate the UV-induced DNA damage response through triggering the recruitment of DNA repair proteins.[16] Thus, we propose that it is significant to clarify the correlation between m6A regulators and the metastasis and prognosis of osteosarcoma.
In this study, the information of 88 osteosarcoma tumor samples was obtained from the Cancer Genome Atlas database. We analyzed the underlying association between the expression of m6A regulators and metastasis as well as prognosis, and identified the key regulators with potential prognostic value, and constructed a prognosis signature based on these regulators. This signature was also tested using the osteosarcoma data from the Gene Expression Omnibus database.
2. Materials and methods
2.1. Study population
All the subjects in this study were obtained from the publicly accessible sources. Training set was acquired from the Cancer Genome Atlas which consisted of 88 osteosarcoma tumor samples. Three samples were excluded from the training set for lack of complete survival information. Testing set was obtained from the Gene Expression Omnibus with the accession number of GSE21257 which included 53 osteosarcoma tumor samples. All samples of this study were from primary tumors, and the clinicopathological characteristics of osteosarcoma patients in the training and testing sets were shown in Table S1, Supplemental Digital Content. As all the subjects here were obtained from the publicly accessible sources, and no human or animal subjects were included, ethical approval was not necessary for our study.
2.2. Expression data
Expression levels of mRNA from osteosarcoma tumor tissues in the training and testing sets were quantified through high-throughput RNA-sequencing and Illumina human-6 v2.0 expression beadchip, respectively. A total of 12 and 9 m6A regulators were detected in the training and testing set, respectively, with 8 overlaps. The mRNA levels of those regulators between osteosarcoma samples with different clinicopathological features were analyzed.
2.3. Construction of m6A regulator-based prognostic signature for osteosarcoma
Multivariate Cox-regression analysis was applied to assess the associations between m6A regulators and overall survival (OS) probability of osteosarcoma patients in the training set. Regulators exhibiting significant prognostic relevance were retained and used in LASSO Cox-regression analysis to construct the osteosarcoma prognostic signature through which each sample could be assigned a risk score.
2.4. Survival analysis
We used Kaplan–Meier method to estimate osteosarcoma patients’ OS probability. The difference in OS probability between osteosarcoma subgroups was determined by log-rank test. P-value < .05 was used as the significant threshold.
2.5. Statistical analysis
Comparison of mRNA levels between subgroups of osteosarcoma patients was performed by Wilcoxon rank sum test. Multivariate Cox-regression analysis was used to adjust the effects of confounding factors on OS probability. All the statistical analyses in this study were performed in R version 3.4.1.
3. Results
3.1. m6A regulators are associated with osteosarcoma metastatic status
The number of m6A regulators identified in both training and testing datasets was shown in Figure S1, Supplemental Digital Content. The mRNA levels of m6A regulators were compared between osteosarcoma samples with and without metastasis in both training (Fig. 1A) and testing (Fig. 1C) sets. As a result, methyltransferase-like 14 (METTL14) exhibited significantly decreased mRNA level in osteosarcoma samples with metastasis compared with those without metastasis in the training set as shown in Figure 1B. Another 2 regulators including fat mass and obesity-associated protein (FTO), and zinc finger CCCH domain-containing protein 13 (ZC3H13) could achieve marginal significance in the training set. Four out of the 9 regulators including protein virilizer homolog (KIAA1429), methyltransferase-like 3 (METTL3), Wilms’ tumor 1-associated protein (WTAP), and YTH domains of Homo sapiens (YTHDC1) were significantly differentially expressed in osteosarcoma samples with metastasis compared with those without metastasis in the testing set as shown in Figure 1D.
Figure 1.
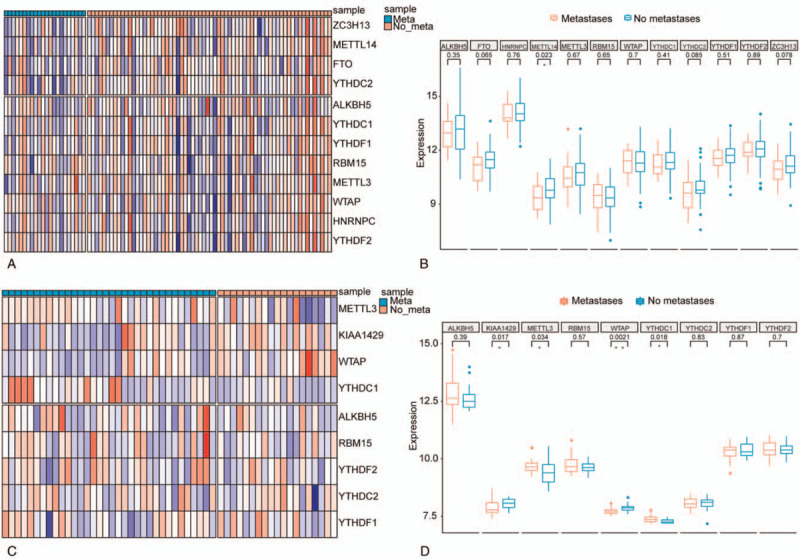
Expression landscape of the 12 m6A regulators in osteosarcoma samples. (A) Heatmap illustrating mRNA expression values of the 12 m6A regulators across the 88 osteosarcoma samples from TCGA (training set). (B) Boxplots illustrating comparison of each m6A regulator's mRNA expression values between osteosarcoma samples with and without metastasis from TCGA (training set). Wilcoxon P values were provided above the boxplots. (C) Heatmap illustrating mRNA expression values of the 9 m6A regulators that were profiled by Illumina expression microarray across the 53 osteosarcoma samples from GSE21257 dataset (testing set). (D) Boxplots illustrating the comparison of each m6A regulator's mRNA expression values between osteosarcoma samples with and without metastasis from GSE21257 dataset (testing set). Wilcoxon P values were provided above the boxplots. “Meta” represents osteosarcoma samples with metastasis, while “no meta” represents osteosarcoma samples without metastasis; ∗ and ∗∗ represents P-value < .05 and .01, respectively. TCGA = the Cancer Genome Atlas.
3.2. m6A regulators have prognostic significance
Osteosarcoma samples in training set were clustered into 2 subgroups via K-means method based on their Euclidean distance that was calculated through the 13 m6A regulators’ mRNA expression values (Fig. 2A). Besides, principle component analysis could definitely separate the samples within the 2 subgroups as shown in Figure 2B, which should indicate the vital roles of the 13 regulators in defining samples. What's more, the 2 groups exhibited significantly different OS probabilities (Fig. 2C), which implied the prognostic significance of these 13 m6A regulators in osteosarcoma.
Figure 2.
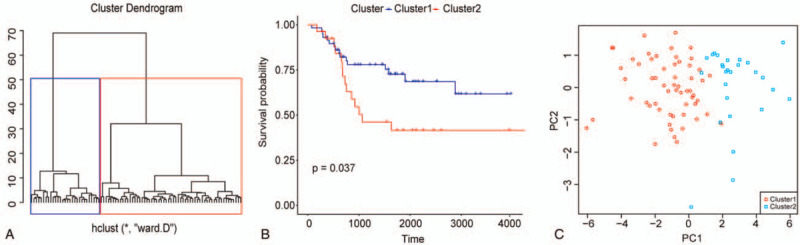
m6A regulators could stratify osteosarcoma patients that were prognostically different. (A) Clustering of osteosarcoma samples from TCGA based on their mRNA expression values of the 12 m6A regulators via K-means method. (B) Kaplan–Meier survival curves of osteosarcoma patients from TCGA stratified by cluster. (C) PCA scatter plot of osteosarcoma samples from TCGA along first 2 principal components. Scatter shapes and colors are used to distinguish samples within different K-means clusters. PCA = principle component analysis, TCGA = the Cancer Genome Atlas.
3.3. Prognostic signature
Multivariate Cox-regression analysis identified METTL3, YTHDC1, and FTO as significant osteosarcoma OS probability-associated markers in training set (Fig. 3A). Prognostic signature was built through LASSO Cox-regression analysis based on the mRNA expressions of these 3 significant regulators through which each sample could be assigned a risk score. Figure 3B illustrated distribution of risk score of osteosarcoma samples in training set. Besides, higher risk score was associated with inferior OS probability of osteosarcoma in both training and testing sets as shown in Figure 3C and D.
Figure 3.
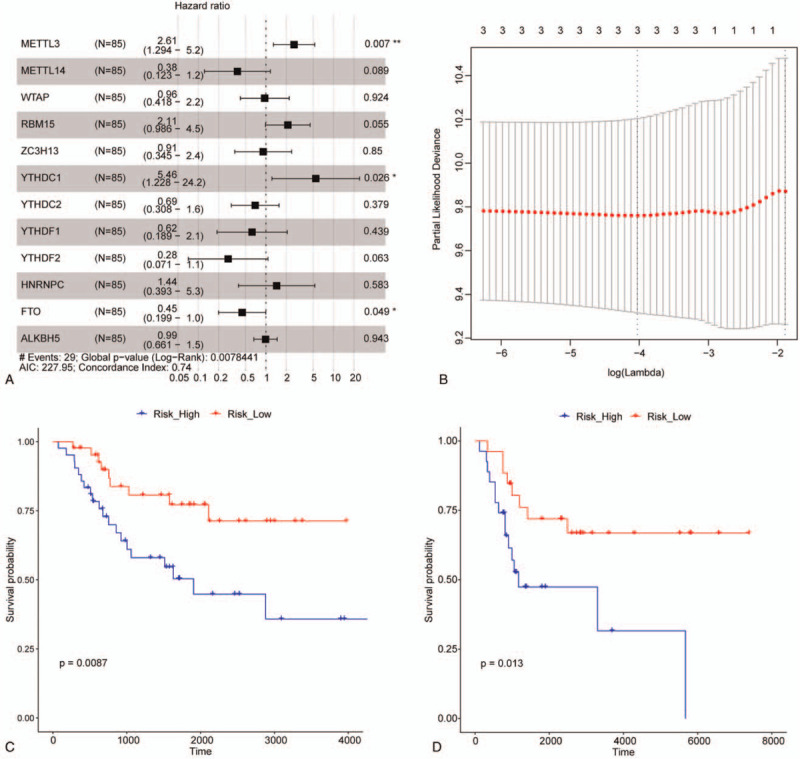
Construction of m6A regulator-based prognostic signature for osteosarcoma. (A) Forest plot of multivariate Cox-regression analysis illustrates associations between each m6A regulator and overall survival of osteosarcoma patients from TCGA. Square data indicates hazard ratio and error bar is 95% confidence interval. ∗ and ∗∗ represents P-value < .05 and .01, respectively. (B) Risk score distribution of osteosarcoma samples in training set. (C) and (D) are the Kaplan–Meier survival curves of osteosarcoma patients from TCGA (training set) and GSE21257 (testing set), respectively. Samples are stratified by the median risk score. TCGA = the Cancer Genome Atlas.
3.4. Clinical relevance of prognostic signature
Here we proposed to explore the potential associations between prognostic signature and common clinicopathological features of osteosarcoma patients including age, gender, and metastatic status. As a result, there was no significant difference in risk score between female and male osteosarcoma samples in both training and testing set (Fig. 4B and F). Strikingly, significant difference in the risk score was observed between osteosarcoma samples that stratified by age of 15 in testing set (Fig. 4E), but not in training set (Fig. 4A). Risk score in osteosarcoma samples with metastasis was significantly higher than that in samples without metastasis in both training and testing sets as shown in Figure 4C and G. What's more, risk score was still an unfavorable factor for the OS probability of osteosarcoma after adjusting for the effects of age, gender, and metastatic status in both training and testing sets (Fig. 4D and H).
Figure 4.
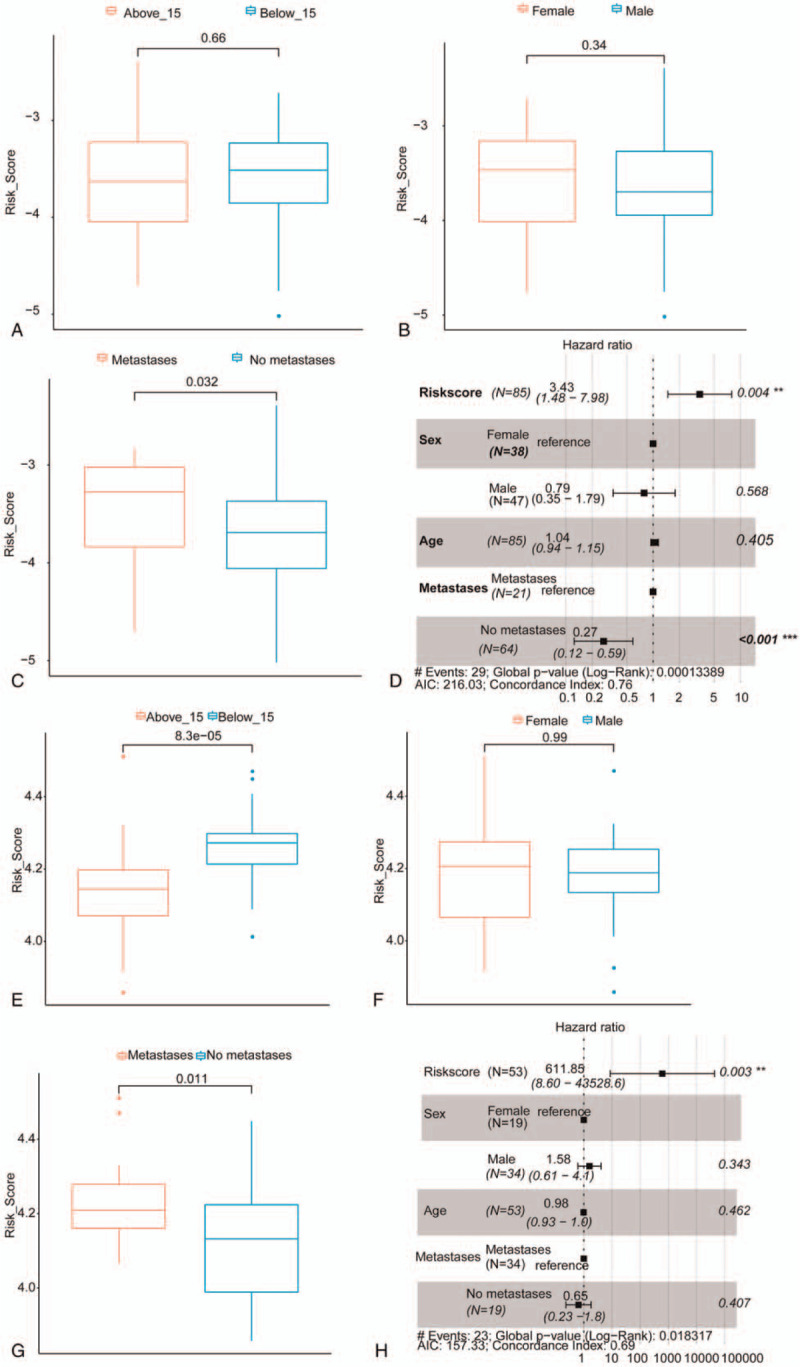
Clinical relevance of risk score. (A–C) Boxplots illustrating comparison of risk score for osteosarcoma samples from TCGA (training set) stratified by their age, gender, and metastatic status. Wilcoxon P values are provided above the boxplots. (D) Forest plot of multivariate Cox-regression analysis indicates risk score as an independent prognostic signature for osteosarcoma patients from TCGA (training set) after adjusting for confounding factors including age, gender, and metastatic status. (E–G) Boxplots illustrating comparison of risk score for osteosarcoma samples from GSE21257 dataset (testing set) stratified by their age, gender, and metastatic status. Wilcoxon P values are provided above the boxplots. (H) Forest plot of multivariate Cox-regression analysis indicates risk score as an independent prognostic signature for osteosarcoma patients from GSE21257 dataset (testing set) after adjusting for confounding factors including age, gender, and metastatic status. TCGA = the Cancer Genome Atlas.
4. Discussion
In this study, we identified several m6A regulators whose expressions were significantly different between osteosarcoma samples with and without metastasis, including METTL14, FTO, ZC3H13, KIAA1429, METTL3, WTAP, and YTHDC1 (Fig. 1), suggesting that m6A RNA modifications should be involved in the metastasis of osteosarcoma. The m6A regulators can be divided into writers, erasers, and readers by their functions of adding, removing, or recognizing m6A-modification, respectively.[17] The prominent m6A regulators include METTL3, WTAP, RNA binding motif protein 15 (RBM15), ZC3H13, YTHDC1, YTHDC2, YTHDF1, YTHDF2, heterogeneous nuclear ribonucleoprotein C (HNRNPC), FTO, α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5), METTL14, and KIAA1429, whose expressions have been studied in gliomas.[18] METTL3, METTL14, and WTAP are the crucial components of the m6A methyltransferase complex.[19] METTL3 methylates target sequence in single strand RNA,[20,21] with a potential function in the regulation of mRNA metabolism.[22] The main function of METTL14 is providing a scaffold for RNA binding, thereby promoting the catalytic activity of METTL3.[23–25] Additionally, METTL14 also plays a key role in various biological processes including embryonic development, gametogenesis, and neurogenesis.[26–28] Another member of m6A methyltransferase complex, WTAP, regulates gene transcription via the interaction with METTL3 and METTL14[29] and involves in the regulation of cell apoptosis.[22] KIAA1429 and ZC3H13 are also required in the complex for m6A methylation.[30,31] KIAA1429 exhibits an essential function in the regulation of methyltransferase complex by recruiting core components of catalyzing,[30] while ZC3H13 participates in the regulation of embryonic stem cell renewal via promoting m6A methylation, which plays an adaptive role between RBM15/Nito and m6A machinery.[31] RBM15 functions in methylation by binding the complex of WTAP-METTL3 and shows a selective activity towards X-inactive specific transcript (XIST).[32] YTHDC1 is one of the m6A readers and regulates splicing as well as nuclear export of mRNA modified by m6A.[32,33] YTHDC2 is larger than other members of YTH family, thus it is endowed diverse functions, such as regulation of RNA binding and structure, and interaction with other complexes.[34,35] An interaction between YTHDF1 and initiation of translating is observed, which improves the translation efficiency.[36] While YTHDF2 could enhance degrading effects of the transcripts which are modified by m6A through recruitment of deadenylase complex.[37] HNRNPC mediates RNA transcripts processing, in which RNA structure is altered by m6A to make RNA transcripts accessible for HNRNPC binding.[38] FTO is a demethylase that demethylates m6A in RNA and DNA,[39,40] and its function is adaptive in various substrates. ALKBH5, localizing to nucleus, is the second identified demethylase. It works in a specific condition in which m6A is the preferential sequence.[41] We performed this study in an attempt to investigate the expression and potential role of these 13 regulators in osteosarcoma. We found that in osteosarcoma samples with metastasis, METTL14 exhibited significantly decreased expression, and WTAP were significantly differentially expressed. The regulation of METTL14 expression on the metastatic behavior of tumor cells varies in different tumors. METTL14 down-regulation would promote the metastatic potential of tumor cells through modulating the primary microRNA 126 process in hepatocellular carcinoma,[42] while it was reported as an oncogenic signal in acute myeloid leukemia.[43] In osteosarcoma, the expression of METTL14 was downregulated in osteosarcoma multidrug-resistant cells as well as tumor-initiating cell,[44] suggesting that it may play a vital role in the emergence and maintaining of osteosarcoma cells.
Through analyzing the prognosis information of osteosarcoma patients, METTL3, YTHDC1, and FTO were identified as significant markers associated with OS probability. METTL3 is a m6A writer and involved in multiple cancers. In hepatoblastoma, METTL3 is associated with cancer development through Wnt/β-catenin signalling pathway and considered as a prognosis predictor. Miao W's group have reported that METTL3 were able to promote osteosarcoma progression by regulating the m6A level of lymphoid enhancer-binding factor 1 (LEF1), and silencing of METTL3 contributed to decrease in m6A methylation and mRNA level of LEF1, thereby suppressing Wnt/β-catenin signaling pathway.[45] Thus it is speculated that METTL3 may associate with osteosarcoma prognosis via Wnt/β-catenin signaling pathway. FTO is a m6A eraser and has been demonstrated to promote acute myeloid leukemia, melanoma, and cervical cancer cells.[46–48] It was reported that, the expression of FTO was significantly decreased in the process of honokiol-induced osteosarcoma cell apoptosis,[28] however, its expression appeared to be independent of the drug-resistance induced by doxorubicin of osteosarcoma cells.[44] In papillary thyroid carcinoma, a prognostic signature including FTO showed good performance in prognosis prediction,[49] but its prognostic value and underlying mechanism in osteosarcoma has not been investigated yet. YTHDC1, a m6A reader, is a member of the YT521-B homology (YTH) domain family, which identifies the m6A group to exercise different downstream effects,[50,51] and was demonstrated to be associated with the progression of prostate cancer.[52] YTHDC1 was proved to be a prognostic factor in ovarian cancer with TP53 mutation,[53] but its prognostic role in osteosarcoma is still unknown. To our knowledge, we are the first to report that osteosarcoma patients could be prognostically stratified using METTL3, YTHDC1, and FTO, which is independent of potential confounding factors including age, gender, and metastatic status. However, the underlying mechanism is still not confirmed and warrants further study.
Due to the complexity of tumor epigenetic regulation, the transfer of m6A biomarkers to the daily clinical practice still faces many challenges, and there are also some limitations in this study. First, we just analyzed the relationship between expression of m6A regulators and OS of osteosarcoma patients due to the limited data. Second, we clustered the osteosarcoma samples into 2 subgroups based on Euclidean distance that was calculated through the expressions of m6A regulators, however, we did not perform in-depth analysis of differences in their biological processes. Based on the results of this study, we would collect clinical samples to further investigate the relationship of m6A regulators with the effect of osteosarcoma chemotherapy, and explore the regulatory mechanisms of the key m6A regulators at the cellular level.
5. Conclusion
In this study, we comprehensively analyzed associations between mRNA expressions of m6A regulators with the metastasis and prognosis of osteosarcoma patients, and found that several m6A regulators exhibited aberrant mRNA levels with metastatic and prognostic significance. METTL3, YTHDC1, and FTO were identified as key regulators, and the osteosarcoma prognostic signature based on these m6A regulator could prognostically stratify the patients independently of potential confounding factors. This study highlighted the important role of RNA modification in screening of new therapeutic targets and prognosis prediction of osteosarcoma.
Author contributions
Conceptualization: Quanbin Zhang.
Data curation: Wenpeng Zhang, Quanbin Zhang.
Formal analysis: Quanbin Zhang.
Methodology: Wenpeng Zhang, Quanbin Zhang.
Project administration: Quanbin Zhang.
Software: Wenpeng Zhang, Lina Wang, Ping Zhang, Quanbin Zhang.
Supervision: Wenpeng Zhang, Lina Wang, Ping Zhang, Quanbin Zhang.
Validation: Wenpeng Zhang, Lina Wang, Ping Zhang, Quanbin Zhang.
Visualization: Wenpeng Zhang, Lina Wang, Ping Zhang, Quanbin Zhang.
Writing – original draft: Wenpeng Zhang, Quanbin Zhang.
Writing – review & editing: Wenpeng Zhang, Quanbin Zhang.
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: METTL14 = methyltransferase-like 14, METTL3 = methyltransferase-like 3, OS = overall survival, WTAP = Wilms’ tumor 1-associated protein.
How to cite this article: Zhang W, Wang L, Zhang P, Zhang Q. m6A regulators are associated with osteosarcoma metastasis and have prognostic significance: a study based on public databases. Medicine. 2021;100:20(e25952).
All the co-authors agree to the publication of the article.
The authors have no conflicts of interest to disclose.
The datasets analyzed for this study can be found in the TCGA (https://tcga-data.nci.nih.gov/tcga), GEO (https://www.ncbi.nlm.nih.gov/geo/).
The datasets generated during and/or analyzed during the current study are publicly available.
Supplemental digital content is available for this article.
References
- [1].Durfee RA, Mohammed M, Luu HH. Review of osteosarcoma and current management. Rheumatol Ther 2016;3:221–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am 2016;47:283–92. [DOI] [PubMed] [Google Scholar]
- [3].Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother 2015;16:2727–36. [DOI] [PubMed] [Google Scholar]
- [4].Suehara Y, Alex D, Bowman A, et al. Clinical genomic sequencing of pediatric and adult osteosarcoma reveals distinct molecular subsets with potentially targetable alterations. Clin Cancer Res 2019;25:6346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Evola FR, Costarella L, Pavone V, et al. Biomarkers of osteosarcoma, chondrosarcoma, and ewing sarcoma. Front Pharmacol 2017;8:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther 2018;18:39–50. [DOI] [PubMed] [Google Scholar]
- [7].Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017;168:644–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Goricar K, Kovac V, Jazbec J, Zakotnik B, Lamovec J, Dolzan V. Genetic variability of DNA repair mechanisms and glutathione-S-transferase genes influences treatment outcome in osteosarcoma. Cancer Epidemiol 2015;39:182–8. [DOI] [PubMed] [Google Scholar]
- [9].Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 2008;451:153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Storici F. RNA-mediated DNA modifications and RNA-templated DNA repair. Curr Opin Mol Ther 2008;10:224–30. [PubMed] [Google Scholar]
- [11].Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 2017;45:6051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis 2018;9:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res 2018;28:507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254–70. [DOI] [PubMed] [Google Scholar]
- [16].Xiang Y, Laurent B, Hsu CH, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017;543:573–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Erson-Bensan AE, Begik O. m6A modification and implications for microRNAs. Microrna 2017;6:97–101. [DOI] [PubMed] [Google Scholar]
- [18].Chai RC, Wu F, Wang QX, et al. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY) 2019;11:1204–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Scholler E, Weichmann F, Treiber T, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 2018;24:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kan L, Grozhik AV, Vedanayagam J, et al. The m(6)A pathway facilitates sex determination in Drosophila. Nat Commun 2017;8:15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rottman FM, Bokar JA, Narayan P, Shambaugh ME, Ludwiczak R. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie 1994;76:1109–14. [DOI] [PubMed] [Google Scholar]
- [22].Ping XL, Sun BF, Wang L, et al. is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014;24:177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 2016;5:e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang X, Feng J, Xue Y, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016;534:575–8. [DOI] [PubMed] [Google Scholar]
- [25].Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell 2016;63:306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014;16:191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lin Z, Hsu PJ, Xing X, et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res 2017;27:1216–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yoon KJ, Ringeling FR, Vissers C, et al. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 2017;171:877–89.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Horiuchi K, Kawamura T, Iwanari H, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 2013;288:33292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep 2014;8:284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Knuckles P, Lence T, Haussmann IU, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev 2018;32:415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016;537:369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 2017;6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hsu PJ, Zhu Y, Ma H, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 2017;27:1115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tanabe A, Tanikawa K, Tsunetomi M, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1alpha mRNA is translated. Cancer Lett 2016;376:34–42. [DOI] [PubMed] [Google Scholar]
- [36].Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 2015;161:1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Du H, Zhao Y, He J, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 2016;7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015;518:560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007;318:1469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jia G, Yang CG, Yang S, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett 2008;582:3313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017;65:529–43. [DOI] [PubMed] [Google Scholar]
- [43].Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 2018;22:191–205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang Y, Zeng L, Liang C, et al. Integrated analysis of transcriptome-wide m(6)A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics 2019;11:1693–715. [DOI] [PubMed] [Google Scholar]
- [45].Miao W, Chen J, Jia L, Ma J, Song D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun 2019;516:719–25. [DOI] [PubMed] [Google Scholar]
- [46].Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 2017;31:127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yang S, Wei J, Cui YH, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun 2019;10:2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zou D, Dong L, Li C, Yin Z, Rao S, Zhou Q. The m(6)A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int 2019;19:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hou J, Shan H, Zhang Y, Fan Y, Wu B. m(6)A RNA methylation regulators have prognostic value in papillary thyroid carcinoma. Am J Otolaryngol 2020;41:102547. [DOI] [PubMed] [Google Scholar]
- [50].Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol 2017;33:319–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhuang M, Li X, Zhu J, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res 2019;47:4765–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Luxton HJ, Simpson BS, Mills IG, et al. The oncogene metadherin interacts with the known splicing proteins YTHDC1, Sam68 and T-STAR and plays a novel role in alternative mRNA splicing. Cancers (Basel) 2019;11:1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang Q, Zhang Q, Li Q, Zhang J, Zhang J. Clinicopathological and immunological characterization of RNA m(6) A methylation regulators in ovarian cancer. Mol Genet Genomic Med 2021;9:e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


