Abstract
Student's t test is valid for statistical inference under the normality assumption or asymptotically. By contrast, although the bootstrap t test was proposed in 1993, it is seldom adopted in medical research. We aim to demonstrate that the bootstrap t test outperforms Student's t test under normality in data. Using random data samples from normal distributions, we evaluated the testing performance, in terms of true-positive rate (TPR) and false-positive rate and diagnostic abilities, in terms of the area under the curve (AUC), of the bootstrap t test and Student's t test. We explore the AUC of both tests with varying sample size and coefficient of variation. We compare the testing outcomes using the COVID-19 serial interval (SI) data in Shenzhen and Hong Kong, China, for demonstration. With fixed TPR, the bootstrap t test maintained the equivalent accuracy in TPR, but significantly improved the true-negative rate from the Student's t test. With varying TPR, the diagnostic ability of bootstrap t test outperformed or equivalently performed as Student's t test in terms of the AUC. The equivalent performances are possible but rarely occur in practice. We find that the bootstrap t test outperforms by successfully detecting the difference in COVID-19 SI, which is defined as the time interval between consecutive transmission generations, due to sex and non-pharmaceutical interventions against the Student's t test. We demonstrated that the bootstrap t test outperforms Student's t test, and it is recommended to replace Student's t test in medical data analysis regardless of sample size.
Key words: Bootstrap t test, clinical epidemiology, COVID-19, serial interval, statistical hypothesis testing
Introduction
Statistical hypotheses testing is an essential approach adopted for medical and healthcare data analysis [1]. Student's t test is one of the crucial tests that is widely used to conduct statistical inference for normally (or approximately normally) distributed dataset or those with sufficiently large sample size when the central limit theorem (CLT) is applicable [2, 3]. Student's t test may yield unsatisfactory testing outcome when samples are skewed [4], mostly likely with small sample size. Bootstrap methods have been proposed in 1970s, and have been used to analyse such as not normally distributed data [5, 6]. It is (asymptotically) more accurate than the standard estimates using sample variance and based on the assumptions of normality [7, 8]. Although a bootstrap t test was proposed by Efron and Tibshirani in 1993 [9], it was considered the percentile of bootstrapped test statistic samples at the significant level. To avoid repetition, we omit the algorithm of bootstrap t test in this study since the detailed algorithm was already introduced in [9]. This improved version of t test is seldom adopted in medical research.
Objectives
As mentioned, it is commonly accepted to use Student's t test when normality of the data suffices, whereas the bootstrap approach could be adopted to resolve the situation without normality. In this study, we demonstrated that for data from normal population, the bootstrap t test outperforms Student's t test in terms of different measures of the testing accuracy. We explored the general features of the data sample with which bootstrap t tests are likely to have more plausible testing outcome.
Methods
The details of the testing procedures of bootstrap t test could be found in [9]. The pairwise two-sample t tests are conducted based on the null hypothesis, H0, that assumes the means of the two populations equal. Data samples are randomly generated from normally distributed populations, which will be used to compare the testing outcome based on data samples and the facts of populations. We evaluated the testing performance in two scenarios including
-
•
scenario (i): the H0 is true; and
-
•
scenario (ii): the H0 is false.
Then, the possibility that H0 was not rejected in scenario (i) is the true-positive rate (TPR), i.e. sensitivity. The possibility that H0 was rejected in scenario (ii) is the true-negative rate (TNR), i.e. specificity. Theoretically, the TPR is (1 − α), where α is known as the rate of the type I error, i.e. false-alarm rate, and similarly, TNR is (1 − β), where β is the rate of the type II error, i.e. miss rate. It is a common practice to set α at 5%, and the test is formulated with the aim to minimise β [1, 10].
Fixed TPR
With TPR = (1 − α) = 95%, i.e. α = 5%, we evaluated
-
(i)
the consistency in TPR,
-
(ii)
the levels of TNR and
-
(iii)
informedness (i.e. Youden's J statistic)
of two types of t tests with varying sample size and coefficient of variation (CV) = s.d./difference in mean, in the samples [11]. Here, the informedness = TPR + TNR − 1, ranging from 0 to 1 (inclusive), is a single statistic that estimates the probability of an informed decision [12], which evaluates the performance of diagnostic tests. The informedness is 0 when a diagnostic test gives the same proportion of positive results for both true and false groups, which implies the testing outcome is totally uninformed. The informedness 1 indicates an ideal situation that TPR = TNR = 1, which implies that the testing outcome is perfectly informed. Since the test statistic of t test is mainly determined by CV and sample size, these two factors are thus included in the testing performance evaluation.
Varying TPR
With varying TPR, i.e. (1 − α), we could measure the diagnostic performance of both tests by using TPR and TNR in pairs. With all pairs of TPR and TNR, we could construct the receiver operating characteristic (ROC) curve to illustrate the diagnostic abilities of the two t tests in terms of the area under the curve (AUC).
Testing performance evaluation
For each set of sample size, CV and α, we tested 10 000 pairs of random-generated data samples to estimate the TPR and TNR, and then to calculate the informedness and AUC. We ran 1000 bootstrap samples to conduct the bootstrap t test. We also ran 1000 bootstrap samples in the testing outcomes of the two t test to generate the 95% confidence intervals (CIs) of the estimated metrics.
For demonstration, we compare the testing outcomes by using the COVID-19 serial interval (SI), which is defined as the time interval between consecutive transmission generations, data in Shenzhen and Hong Kong, China. This demonstrative example is considered as a part of results (instead of methodology), and thus elaborated in the next section.
Results and discussion
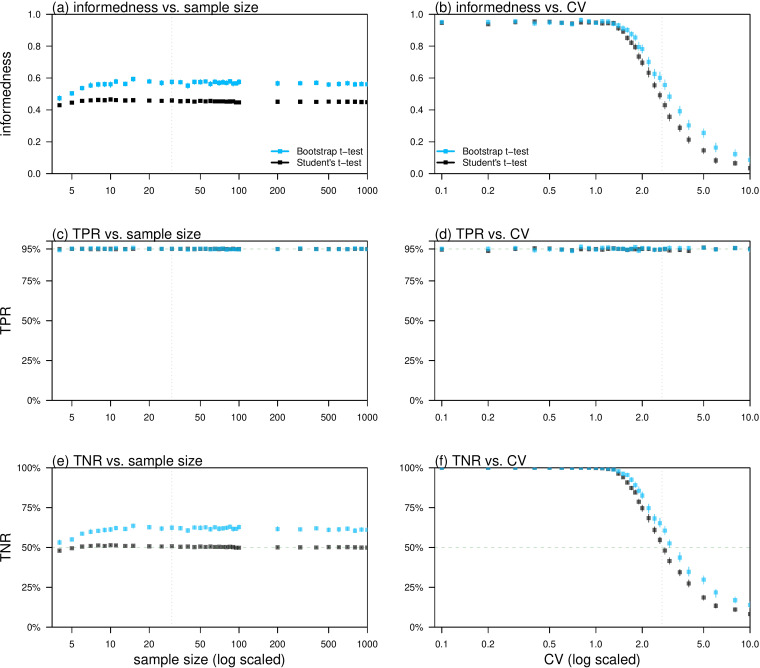
We found that the informedness of bootstrap t test outperformed Student's t test for both a wide range of varying sample sizes and CVs, see Figure 1(a) and (b). Since the TPRs were consistently stabilised at 95%, see Figure 1(c) and (d), the difference in the informedness was due to the differences in the TNRs, see Figure 1(e) and (f). With fixed α, the bootstrap t test maintained the equivalent accuracy in TPR, but significantly improved the TNR compared to the Student's t test, see Figure 1(c)–(f). This can be interpreted as the bootstrap t test is more likely to exclude the unrealistic hypothesis, when H0 is false, compared to the Student's t test and meanwhile maintained its judgement to the true statement, when H0 is true. Since the null hypothesis is known a priori to be false [13], H0 is commonly expected to be rejected based on sufficient (statistical) evidence [1, 4, 10]. Thus, the improvement in TNR was remarkably desirable.
Fig. 1.
Relations between the testing accuracies of the bootstrap t test (blue) and Student's t test (black), including informedness, TPR and TNR, and the features of the data samples including sample size and CV. Panels (a) and (b) show the relations between informedness ( = TPR + TNR − 1) and sample size and CV respectively. Panels (c) and (d) show the relations between TPR and sample size and CV respectively. Panels (e) and (f) show the relations between TNR and sample size and CV respectively. The CVs (of the data samples) were determined by 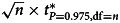 , where n denotes the sample size, t* is the quantile of the t distribution and ‘df’ is the degree of freedom, in panels (a), (c) and (e). The sample size was fixed to be 30 in panels (b), (d) and (f). The level of α was fixed to be 5% in all panels. The vertical bars in each panel represent the 95% CIs.
, where n denotes the sample size, t* is the quantile of the t distribution and ‘df’ is the degree of freedom, in panels (a), (c) and (e). The sample size was fixed to be 30 in panels (b), (d) and (f). The level of α was fixed to be 5% in all panels. The vertical bars in each panel represent the 95% CIs.
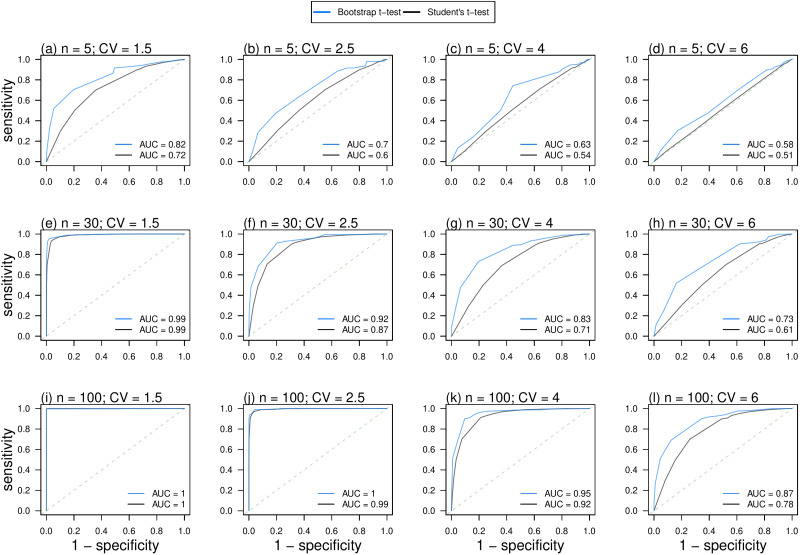
In Figure 2, the diagnostic ability of bootstrap t test outperformed or equivalently performed as Student's t test in terms of the AUC. The diagnostic ability of bootstrap t test outperformed Student's t test not only when the sample size is small, e.g. see Figure 2(b) and (c), but also when the sample size becomes large, e.g. see Figure 2(i) and (k). Although Student's t test can be conducted with sufficiently large sample size when the CLT is applicable [2, 3], we found that bootstrap t test outperformed or equivalently performed as Student's t test regardless of the sample size.
Fig. 2.
ROC curves and AUCs of the bootstrap t test (blue) and Student's t test (black) with varying sample sizes, n, and CVs of the data samples. The diagonal dashed lines show the testing performance of a random classifier.
On one hand, the AUC of Student's t test approached that of bootstrap t test, i.e. equivalent performance, when the sample size became larger and CV became smaller, e.g. see Figure 2(e) and (j). Under these circumstances, the distributions of samples to be tested are distinguishably separated, and thus straightforwardly, the two tests could yield ‘to reject H0’ outcomes equivalently. This finding indicated that given sufficiently large sample size, Student's t test was capable of achieving equivalent diagnostic ability as bootstrap t test when the two datasets were discriminative in the central tendency and had low dispersion. It is also interesting to note that the equivalent performance only appears when the values of the AUC of two tests equal to 0.5, i.e. random classifier, or 1, i.e. perfect classifier. Either AUC = 0.5 or AUC = 1 would rarely occur due to the unusual features of the testing datasets, e.g. extremely large sample size and small CV or extremely small sample size and large CV.
On the other hand, when the sample size is small and CV is large, e.g. see Figure 2(b), (c), (d), (g) and (h), the distributions of samples to be tested are difficult to differentiate. In these situations, the diagnostic ability of bootstrap t test outperformed Student's t test in terms of the AUC.
In summary, for data samples from normally distributed populations, both testing performance and diagnostic abilities of the bootstrap t test outperformed Student's t test regardless of varying sample size and CV. We have summarised our findings and the situation when normality fails in Table 1. Specially, for small samples, when data fail to meet normality assumption, other non-parametric tests and their bootstrap versions are also recommended to fit the study purpose.
Table 1.
Summary of the situations to be tested and the recommendation of Student's or Bootstrap t tests
| Normality | Sample size | Dispersion | Student's or Bootstrap t test | Remark | Reference |
|---|---|---|---|---|---|
| No | Small | Small | Bootstrap t test | Non-parametric tests are also preferred | 7, 14, 15 |
| Large | |||||
| Large | Small | None | |||
| Large | None | ||||
| Yes | Small | Small | Bootstrap t test | None | This study |
| Large | Bootstrap t test | None | |||
| Large | Small | Both | Equivalent AUC | ||
| Large | Bootstrap t test | None |
Note: The ‘dispersion’ in this study is measured by the CV.
Demonstrative example of COVID-19
We demonstrate the performance of bootstrap t test against the Student's t test by using the COVID-19 SI dataset from the early outbreaks in Shenzhen and Hong Kong, two neighbour cities on the southeast coast of China. In infectious disease transmission, the SI is defined as the difference between the onset date of a secondary case and that of its associated primary case in a consecutive transmission chain [16]. With the pathogen's transmissibility fixed, a shorter SI implies that the disease may transmit more rapidly in terms of the epidemiological outcomes at the population scale, e.g. number of cases. The SI is one of the key epidemiological parameters to characterise the disease transmission process, and it is of importance in determining the changing patterns of the epidemic curve [17–20]. The SI can be inferred from the contact tracing surveillance data and reconstruction of the transmission chains, which is well studied in previous studies [21–33], and widely adopted in modelling analysis [34–47].
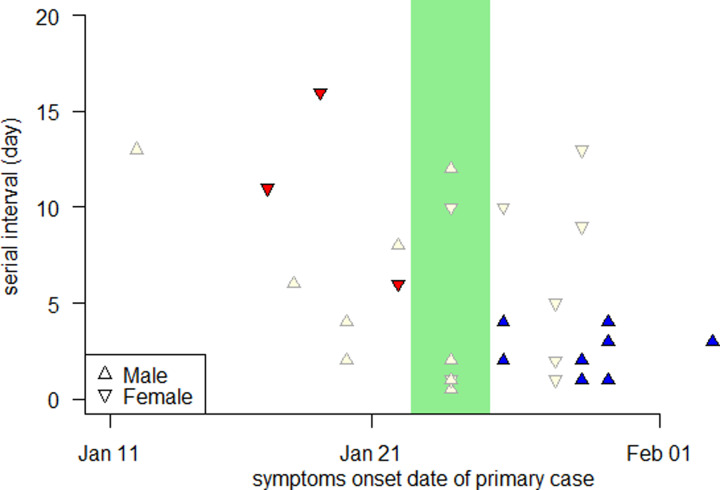
The SI data were collected via the public domains until 22 February 2020 for Shenzhen, and until 15 February 2020 for Hong Kong. The study periods cover the major epidemic wave in Shenzhen and the first-epidemic wave in Hong Kong. This dataset was published previously in [48, 49] as well as studied in [50]. We extract transmission pairs, i.e. one secondary case is epidemiologically linked to one and only one primary case, with no missing information of the primary case's sex. We obtained a total of 34 transmission pairs including 22 (14 male and 8 female primary cases) from Shenzhen, and 12 (6 male and 6 female primary cases) from Hong Kong. There were 33 (out of a total of 34) transmission pairs with primary cases' symptoms onset date in January 2020, see Figure 3.
Fig. 3.
SIs of the COVID-19 transmission pairs in Shenzhen and Hong Kong, China, during the early outbreaks. The SI with male or female primary case is represented by upward or downward triangle, respectively. The hollow or filled (red for female and blue for male) triangle represents the SI data excluded or included in the t tests, respectively. The green shading area highlights the CLNY.
We evaluate the two t tests by examining whether they are able to identify the difference in COVID-19 SI due to sex and non-pharmaceutical interventions (NPIs). Thus, we conduct the t test on two groups of SI samples separated from the original dataset based on two epidemiological evidences. They include
-
•
evidence (i): according to the previous studies [28, 50], a female COVID-19 primary case is likely having longer SI than male; and
-
•
evidence (ii): due to non-NPIs, e.g. social distancing, city lockdown, travel suspension, wearing face mask, regular sterilisation, the SI was shortened, i.e. became smaller, over time [31, 50].
Hence, we divide the COVID-19 SI samples based on the sex of primary case, and Chinese Lunar New Year (CLNY) from 23 to 26 January 2020 [51], after which most of the NPIs (including city lockdown) were implemented and enhanced. Two groups of SI samples are selected for the t tests. They are
-
•
samples from population (i): SI samples with female primary case whose symptoms onset was before CLNY (sample size is 3, see red dots in Fig. 3), and
-
•
samples from population (ii): SI samples with male primary cases whose symptoms onset was after CLNY (sample size is 10, see blue dots in Fig. 3).
Straightforwardly, the mean SI of population (i) is expected higher than the mean SI of population (ii), which is also supported by the evidence found in previous studies [28, 31, 50].
As for the outcomes from the t tests, we report the one-side bootstrap t test yields a P value = 0.04 of statistical significance, whereas the one-side Student's t test yields a P value = 0.05. Therefore, we demonstrate that the bootstrap t test outperforms the Student's t test by successfully detecting the difference in COVID-19 SI due to sex and NPIs.
Limitations
This comparison analysis study has limitations. As one of the classic drawbacks mentioned in [52], for the bootstrap on samples from a population without a finite variance, the bootstrap will be unlikely to converge. However, medical data samples are (commonly) from the real-world samples and thus the variance are expected to be finite. Although we have demonstrated the testing performances by using large sets of randomly generated data samples, the study would benefit from real-world examples that have different conclusions from the bootstrap t test and Student's t test, respectively.
Conclusions
We demonstrated that the bootstrap t test outperforms Student's t test, and it is recommended to replace Student's t test in medical data analysis regardless of sample size.
Acknowledgement
The authors thank G. Yang and Y. Han, both from the Chinese University of Hong Kong, for their helpful discussion at the very early stage of this study.
Author contributions
SZ conceptualised the study, conducted the analysis, drafted the manuscript and critically revised the contents. SZ, ZY and DH discussed the results. All authors had full access to the data, contributed to the study, approved the final version for publication and take responsibility for its accuracy and integrity.
Financial support
DH was supported by General Research Fund (Grant Number 15205119) of the Research Grants Council (RGC) of Hong Kong, China, and an Alibaba (China) Co., Ltd. Collaborative Research grant.
Ethical standards
There was no experiment conducted. This research was based on the computer simulation, and publicly available dataset. Hence, the ethics approval was not applicable.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268821001047.
click here to view supplementary material
Conflict of interest
DH received support from an Alibaba (China) Co., Ltd. Collaborative Research grant. MHW is a shareholder of Beth Bioinformatics Co., Ltd. Other authors have no conflict of interest.
Data availability statement
The COVID-19 data used in this study are available via the Supplementary materials.
References
- 1.Guyatt G et al. (1995) Basic statistics for clinicians: 1. Hypothesis testing. Canadian Medical Association Journal 152, 27. [PMC free article] [PubMed] [Google Scholar]
- 2.Bland M (2015) An Introduction to Medical Statistics. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Kirkwood BR and Sterne JA (2010) Essential Medical Statistics. Malden, Massachusetts, USA: John Wiley & Sons. [Google Scholar]
- 4.Wilcox R (2017) Modern Statistics for the Social and Behavioral Sciences: A Practical introduction. Boca Raton, Florida, USA: Chapman and Hall/CRC. [Google Scholar]
- 5.Efron B (1992) Bootstrap methods: another look at the jackknife. In Kotz S and Johnson N (eds), Breakthroughs in Statistics. New York City, New York, USA: Springer, pp. 569–593. [Google Scholar]
- 6.Dwivedi AK, Mallawaarachchi I and Alvarado LA (2017) Analysis of small sample size studies using nonparametric bootstrap test with pooled resampling method. Statistics in Medicine 36, 2187–2205. [DOI] [PubMed] [Google Scholar]
- 7.DiCiccio TJ and Efron B (1996) Bootstrap confidence intervals. Statistical Science 11, 189–212. [Google Scholar]
- 8.Carpenter J and Bithell J (2000) Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Statistics in Medicine 19, 1141–1164. [DOI] [PubMed] [Google Scholar]
- 9.Efron B and Tibshirani RJ (1994) An Introduction to the Bootstrap. Boca Raton, Florida, USA: Chapman and Hall/CRC Press. [Google Scholar]
- 10.Anderson DR, Burnham KP and Thompson WL (2000) Null hypothesis testing: problems, prevalence, and an alternative. The Journal of Wildlife Management 64, 912–923. [Google Scholar]
- 11.Schisterman EF et al. (2005) Optimal cut-point and its corresponding Youden index to discriminate individuals using pooled blood samples. Epidemiology (Cambridge, Mass.) 16, 73–81. [DOI] [PubMed] [Google Scholar]
- 12.Youden WJ (1950) Index for rating diagnostic tests. Cancer 3, 32–35. [DOI] [PubMed] [Google Scholar]
- 13.Dushoff J, Kain MP and Bolker BM (2018) I can see clearly now: reinterpreting statistical significance. Methods in Ecology Evolution 10(6), 756–759. [Google Scholar]
- 14.Adèr HJ (2008) Advising on Research Methods: A Consultant's Companion. BV, Rosmalen, The Nederland: Johannes van Kessel Publishing. [Google Scholar]
- 15.Goodhue DL, Lewis W and Thompson R (2012) Does PLS have advantages for small sample size or non-normal data? MIS Quarterly 36, 981–1001. [Google Scholar]
- 16.Fine PEM (2003) The interval between successive cases of an infectious disease. American Journal of Epidemiology 158, 1039–1047. [DOI] [PubMed] [Google Scholar]
- 17.Wallinga J and Lipsitch M (2007) How generation intervals shape the relationship between growth rates and reproductive numbers. Proceedings: Biological Sciences 274, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao S et al. (2020) Serial interval in determining the estimation of reproduction number of the novel coronavirus disease (COVID-19) during the early outbreak. Journal of Travel Medicine 27, taaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grassly NC and Fraser C (2008) Mathematical models of infectious disease transmission. Nature Reviews Microbiology 6, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiura H et al. (2010) Pros and cons of estimating the reproduction number from early epidemic growth rate of influenza A (H1N1) 2009. Theoretical Biology and Medical Modelling 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu XK et al. (2020) Reconstruction of transmission pairs for novel coronavirus disease 2019 (COVID-19) in mainland China: estimation of superspreading events, serial interval, and hazard of infection. Clinical Infectious Diseases 71, 3163–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tindale LC et al. (2020) Evidence for transmission of COVID-19 prior to symptom onset. eLife 9, e57149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Z et al. (2020) Serial interval of COVID-19 among publicly reported confirmed cases. Emerging Infectious Diseases 26, 1341–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganyani T et al. (2020) Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, march 2020. EuroSurveillance 25, 2000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S et al. (2020) Estimating the serial interval of the novel coronavirus disease (COVID-19): a statistical analysis using the public data in Hong Kong from January 16 to February 15, 2020. Frontiers in Physics 8, 347. [Google Scholar]
- 26.You C et al. (2020) Estimation of the time-varying reproduction number of COVID-19 outbreak in China. International Journal of Hygiene and Environmental Health 228, 113555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishiura H, Linton NM and Akhmetzhanov AR (2020) Serial interval of novel coronavirus (COVID-19) infections. International Journal of Infectious Diseases 93, 284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma S et al. (2020) Epidemiological parameters of COVID-19: case series study. Journal of Medical Internet Research 22, e19994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q et al. (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. New England Journal of Medicine 382, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adam DC et al. (2020) Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nature Medicine 26, 1714–1719. [DOI] [PubMed] [Google Scholar]
- 31.Ali ST et al. (2020) Serial interval of SARS-CoV-2 was shortened over time by nonpharmaceutical interventions. Science (New York, N.Y.) 369, 1106–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferretti L et al. (2020) Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science (New York, N.Y.) 368, eabb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowling BJ et al. (2009) Estimation of the serial interval of influenza. Epidemiology (Cambridge, Mass.) 20, 344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chinazzi M et al. (2020) The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science (New York, N.Y.) 368, 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chong KC et al. (2020) Monitoring disease transmissibility of 2019 novel coronavirus disease in Zhejiang, China. International Journal of Infectious Diseases 96, 128–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kucharski AJ et al. (2020) Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infectious Diseases 20, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung K et al. (2020) First-wave COVID-19 transmissibility and severity in China outside Hubei after control measures, and second-wave scenario planning: a modelling impact assessment. Lancet (London, England) 395, 1382–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizumoto K and Chowell G (2020) Transmission potential of the novel coronavirus (COVID-19) onboard the diamond Princess Cruises Ship, 2020. Infectious Disease Modelling 5, 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musa SS et al. (2020) Estimation of exponential growth rate and basic reproduction number of the coronavirus disease 2019 (COVID-19) in Africa. Infectious Diseases of Poverty 9, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiura H et al. (2020) The rate of underascertainment of novel coronavirus (2019-nCoV) infection: estimation using Japanese passengers data on evacuation flights. Journal of Clinical Medicine 9, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran J et al. (2020) A re-analysis in exploring the association between temperature and COVID-19 transmissibility: an ecological study with 154 Chinese cities. European Respiratory Journal 56, 2001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riou J and Althaus CL (2020) Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. EuroSurveillance 25, 2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tuite AR and Fisman DN (2020) Reporting, epidemic growth, and reproduction numbers for the 2019 novel coronavirus (2019-nCoV) epidemic. Annals of Internal Medicine 172, 567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volz E et al. (2021) Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184, 64–75, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu JT et al. (2020) Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nature Medicine 26, 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao S et al. (2020) Estimating the unreported number of novel coronavirus (2019-nCoV) cases in China in the first half of January 2020: a data-driven modelling analysis of the early outbreak. Journal of Clinical Medicine 9, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao S et al. (2020) Imitation dynamics in the mitigation of the novel coronavirus disease (COVID-19) outbreak in Wuhan, China from 2019 to 2020. Annals of Translational Medicine 8, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang K et al. (2020) Estimating the serial interval of the novel coronavirus disease (COVID-19) based on the public surveillance data in Shenzhen, China from January 19 to February 22, 2020. Transboundary and Emerging Diseases 67(6), 2818–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao S et al. (2020) Estimating the serial interval of the novel coronavirus disease (COVID-19): a statistical analysis using the public data in Hong Kong from January 16 to February 15, 2020. Frontiers in Physics 8, 347. [Google Scholar]
- 50.Zhao S et al. (2020) COVID-19 and gender-specific difference: analysis of public surveillance data in Hong Kong and Shenzhen, China, from January 10 to February 15, 2020. Infection Control & Hospital Epidemiology 41, 750–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leung GM, Cowling BJ and Wu JT (2020) From a sprint to a marathon in Hong Kong. New England Journal of Medicine 382, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Athreya K (1987) Bootstrap of the mean in the infinite variance case. The Annals of Statistics 15, 724–731. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268821001047.
click here to view supplementary material
Data Availability Statement
The COVID-19 data used in this study are available via the Supplementary materials.