Abstract
Cardiomyopathy occurs at significantly higher rates in survivors of childhood cancer than the general population, but few studies have evaluated racial or ethnic disparities, and none have assessed potential genetic factors contributing to this outcome. In this study, childhood cancer survivors of African ancestry exposed to cardiotoxic therapies (anthracyclines and/or heart radiation) (n=246) were compared to cardiotoxic-exposed survivors of European ancestry (n=1645) in the St. Jude Lifetime Cohort. Genetic variants were examined using whole-genome sequencing data among survivors of African ancestry, first based on ejection fraction (EF) as a continuous outcome, followed by clinical history of cardiomyopathy. Survivors of African ancestry showed 1.53- and 2.47-fold risks of CTCAE grade 2–4 and grade 3–4 cardiomyopathy than survivors of European ancestry. A novel locus at 1p13.2 showed significant association with EF [rs6689879*C: EF reduction=4.2%; P=2.8×10−8] in 246 survivors of African ancestry, which was successfully replicated in 1645 survivors of European ancestry but with attenuated magnitude (EF reduction=0.4%; P=0.042). In survivors of African ancestry, rs6689879*C showed a 5.43-fold risk of cardiomyopathy and 1.31-fold risk in those of European ancestry. Among survivors of African ancestry with rs6689879*C and CTCAE grade 2–4 cardiomyopathy, the PHTF1 promoter region was hypomethylated. Similar results were observed in survivors of European ancestry, albeit with reduced magnitudes of hypomethylation among those with rs6689879*C and CTCAE grade 2–4 cardiomyopathy. PHTF1 was upregulated in human-induced pluripotent stem cell-derived cardiomyocytes from patients with doxorubicin-induced cardiomyopathy. These findings have potential implications for long-term cardiac surveillance and up-front cancer care for patients of African ancestry.
Keywords: Cardiomyopathy, Childhood cancer survivors, African American, Anthracyclines, Whole-genome sequencing
Introduction
Cardiotoxicity, a leading cause of morbidity and mortality among adult survivors of childhood cancer, is strongly associated with exposure to anthracycline chemotherapies and radiation therapy involving the heart. Survivors in the Childhood Cancer Survivor Study (CCSS) were nearly six times more likely to report being diagnosed with congestive heart failure compared to their siblings(1). Among survivors clinically-assessed in the St. Jude Lifetime Cohort (SJLIFE)(2), those exposed to a cumulative anthracycline dose ≥250 mg/m2 [odds ratio (OR)=2.7; 95% confidence interval (CI)=1.1–6.9] or cardiac radiation ≥15Gy (OR=1.9; 95% CI=1.1–3.7) were more likely to have evidence of cardiomyopathy (ejection fraction (EF)<50%) than unexposed survivors.
These risk estimates have largely been based upon studies conducted among survivors of European ancestry and therefore obscure differences in cardiotoxicity risk across racial groups. In the general population, the prevalence of non-ischemic cardiomyopathy is higher among individuals of African ancestry(3,4) and has been associated with genetic variants(5). Studies of racial/ethnic differences among childhood cancer survivors are limited but have reported a higher prevalence of cardiovascular risk factors and an elevated overall risk for cardiovascular disease among survivors of African ancestry. Adjustment for socioeconomic and cardiovascular risk factors while modifying the risk in some studies, has not consistently abrogated the risks(6), suggesting potential genetic factors. While there are a few studies evaluating genetic risk factors for cardiomyopathy in childhood cancer survivors, these have predominantly been restricted to survivors of European ancestry(7–11). The genetic basis of therapy-related cardiotoxicity in survivors of African ancestry has not been studied.
We aimed to identify genetic risk factors associated with EF and clinically-diagnosed cardiomyopathy among childhood cancer survivors of African ancestry exposed to cardiotoxic therapies (anthracycline chemotherapy and/or heart radiotherapy) and to understand the underlying mechanisms using survivor-specific DNA methylation data. Additionally, we also explored published RNA-sequencing data on human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) from six female breast patients(12), with respect to the identified genetic association(s).
Materials and Methods
Study cohort
The SJLIFE(13) study is a retrospectively-constructed cohort with prospective clinical follow-up and ongoing enrollment of ≥5 year survivors of childhood cancer treated at St. Jude Children’s Research Hospital (SJCRH). This study was approved by the institutional review board of the SJCRH and all participants provided written informed consent.
Ejection fraction and cardiomyopathy
Two-dimensional Doppler echocardiography was performed using a VIVID-7 machine (General Electric Medical Systems, Milwaukee, WI) with 3-dimensional imaging of left ventricular volumes, according to American Society of Echocardiography guidelines(14). The presence of cardiomyopathy was clinically assessed and graded using a modified version of the National Cancer Institute’s Common Terminology Criteria for Adverse Events (CTCAE) version 4.03(15), and classified as mild (grade 1), moderate (grade 2), severe or disabling (grade 3), life-threatening (grade 4), or fatal (grade 5).
Cardiomyopathy risk by ancestry
We used a multivariable logistic regression to examine potential risk difference of cardiomyopathy by ancestry, adjusting for sex, age at cancer diagnosis, age at last follow-up, cumulative anthracycline dose (expressed in doxorubicin equivalents(16)) and average heart radiation dose. Logistic regression was chosen over Cox regression and other time-to-event analytic approaches because the exact time of onset of cardiomyopathy is not known in this study. Cardiomyopathy in most survivors is detected by echocardiography performed during the SJLIFE campus visit. Before the SJLIFE visit, these survivors are generally asymptomatic and not receiving screening by their local healthcare providers. Thus, the onset time of cardiomyopathy would be sometime before the SJLIFE visit, which we do not know. To assess potential confounding by known cardiovascular risk factors(17,18), the analysis was further adjusted for hypertension, abnormal glucose metabolism, hypercholesteremia, hypertriglyceridemia, sedentary behavior, risky alcohol drinking and smoking. Definitions of cardiovascular risk factors are described in Appendix.
Whole-genome sequencing (WGS)
WGS data (36.8X) were obtained from a larger effort to sequence whole genomes of 3006 SJLIFE survivors(19). Details of DNA sample extraction, WGS, quality control (QC), mapping, variant identification and annotation are described previously(20,21) as well as in Appendix. Of the 2986 WGS samples that passed QC, data for EF, cardiomyopathy and childhood cancer treatment exposures were available for 2332 survivors. Of these, ancestry was classified based on genotype data as African and European in 246 and 1645 survivors exposed to cardiotoxic therapies (anthracycline chemotherapy and/or heart radiotherapy), respectively (Supplementary Figure 1 and Appendix). All these survivors had complete data on EF, cardiomyopathy and adjustment covariates such as demographic and treatment variables, and thus were included in the downstream analyses.
Discovery analysis for EF
Considering the relatively small sample size of 246 survivors of African ancestry, our discovery analysis used EF as the phenotype because it is a quantitative trait and an intermediate phenotype that is used to diagnose and monitor cardiomyopathy. Association analyses were performed as described previously(20,21). Briefly, 9.3 million common [minor allele frequency (MAF)≥0.05] variants were examined using linear regression assuming an additive genetic model, adjusting for sex, age at cancer diagnosis, age at last follow-up, cumulative anthracycline dose, average heart radiation dose and the top 10 principal components. Optimized Sequence Kernel Association Test (SKAT-O) implemented in EPACTS(22) was used to assess 10.2 million rare/low-frequency (MAF<0.05 and minor allele count≥3) variants aggregated by their functional class/types with respect to the RefSeq gene model and agnostic 4 kb sliding window across the genome (Appendix). Common variants with P<5×10−8 were considered significant at the genome-wide level. Significance threshold for rare/low-frequency variants was set as P<7.3×10−8 accounting for 686,236 tests (Appendix).
Associations with cardiomyopathy
Next, we evaluated genome-wide significant associations of the EF analysis above with cardiomyopathy. Considering the relatively small number of survivors with cardiomyopathy and the number of covariates in the model, we used the method of propensity score analysis to assess the genetic associations with cardiomyopathy. Specifically, we first calculated propensity scores by regressing each SNP (dichotomized based on presence or absence of the variant) on all the adjustment covariates. We then assessed genetic associations with both moderate-to-life-threatening (CTCAE grade 2–4) and severe-to-life-threatening (CTCAE grade 3–4) cardiomyopathy using conditional logistic regression, where we matched on the propensity score deciles, eliminating the need to estimate their effects.
Replication
Our replication cohort included 1645 SJLIFE survivors of European ancestry exposed to cardiotoxic therapies. Replication analyses for EF were performed using the same statistical framework as in the discovery analysis. Genetic associations for cardiomyopathy were carried out using logistic regression models with the same adjustment covariates as the EF analysis.
Stratified analyses by anthracyclines only or heart radiotherapy only
We explored effects of genome-wide significant loci among survivors treated with either anthracyclines alone or heart radiation alone. Of the 246 survivors of African ancestry, 93 were treated with anthracyclines only including 12 and 6 with CTCAE grade 2–4 and grade 3–4 cardiomyopathy, respectively. Furthermore, 62 were treated with treated with heart radiotherapy only, including 10 (grade 2–4) and 5 (grade 3–4), respectively. Due to a limited number of survivors with cardiomyopathy, we performed stratified analyses with EF only (EF is a continuous outcome and thus its analysis has higher power than that of cardiomyopathy, a binary outcome) using the same linear regression and adjustment covariates considered in analyses of all survivors.
Specificity of genetic effects
To examine whether the identified genetic variants are specific to childhood cancer survivors exposed to cardiotoxic therapies, we evaluated them in independent SJLIFE survivors of African (n=55) and European (n=225) ancestries who were not treated with either anthracycline chemotherapy or heart radiotherapy. Considering relatively small number of unexposed survivors, we assessed the genetic associations only with EF. Furthermore, we evaluated the genetic variants in individuals of African ancestry with sickle-cell disease enrolled in the lifetime cohort study Sickle Cell Clinical Research and Intervention Program (SCCRIP)(23) and in individuals of European ancestry from the UK Biobank(24) (Appendix). The SCCRIP participants are mostly children with normal EF and more closely resemble the general African-American population.
Extended bioinformatic and functional analyses
For the genome-wide significant locus for EF with successful replication, we first performed conditional analysis to determine presence of one or multiple independent signals (Appendix). We then performed various bioinformatic and functional analyses to gain insights into the underlying molecular mechanisms.
GTEx expression and splice quantitative trait loci
We searched the GTEx portal(25) (accessed on October 7, 2020) to see if the index SNP represents expression qualitative trait locus (eQTL) and/or splice qualitative trait locus (sQTL) in multiple human tissues.
DNA methylation in childhood cancer survivors
Samples for DNA methylation among SJLIFE survivors were collected during their SJLIFE campus visits. Details of DNA methylation experiments, data processing and QC steps are described in Appendix. Whole blood DNA methylation (DNAm) data were available on 265 SJLIFE survivors of African ancestry, including 39 (91%) with CTCAE grade 2–4 cardiomyopathy. Among the 265 survivors of African ancestry with a median age at last follow-up of 34.5 years (range, 13.7 to 65.9 years), the median time to collection of samples for DNA methylation from cancer diagnosis was 25.1 years (range, 0.1 to 50.0 years). We extracted M-values (log2-tranformed ratio of methylated to unmethylated probe intensities) of DNAm probes +/− 250 kb of the index SNP and examined its association with DNA methylation levels, using linear regression adjusting for age at DNA collection, sex and DNA extraction methods. Multiple testing correction was performed controlling for the false discovery rate (FDR) at 0.10 by Benjamini-Hochberg’s procedure(26). For the DNAm probes significantly associated with the index SNP (FDR<0.10), we then examined their associations with cardiomyopathy, using linear regression adjusting for age at DNA collection, sex and DNA extraction methods.
DNAm probes showing significant associations with the index SNP and cardiomyopathy in survivors of African ancestry were also examined in 1460 survivors of European ancestry for whom DNA methylation data were available. Specifically, we examined association of M-values of the DNAm probe(s) with the index SNP genotype and CTCAE grade 2–4 cardiomyopathy, using the linear regression and the same adjustment covariates as above. Among the 1460 survivors of European ancestry with a median age at follow-up of 37.0 years (range, 9.2 to 70.2 years), the median time to collection of samples for DNA methylation from cancer diagnosis was 22.5 years (range, 2.0 to 50.1 years).
Transcriptomic response to doxorubicin-induced cardiotoxicity (DIC) in human induced pluripotent stem cell-derived cardiomyocytes
We obtained previously published RNA-sequencing data(12) on hiPSC-CMs from six female breast cancer patients exposed (at least one year prior) to 240 mg/m2 doxorubicin or equivalent, of whom three developed DIC (post-treatment EF=10–45%) and the remaining three did not (post-treatment EF>55%). The RNA-sequencing was performed after treating the lines with 0 and 1 μM doxorubicin. Counts per million reads (cpm) were calculated using ‘cpm’ function in the edgeR R package(27). Samples with <107 exonic reads and genes with median log2(cpm)<0 were excluded. Following quantile normalization, data for gene(s) prioritized by molecular QTLs were extracted and their mean expressions with respect to DIC were compared using the Student’s t-tests.
Associations of previously reported loci for cardiomyopathy in the general population among survivors of African ancestry
We searched the GWAS catalog(28) (on October 7, 2020) for studies on cardiomyopathy in the general population and found four studies(29–32) that reported a total of 10 loci. Of these, 9 were identified in European populations(29–31) and 1 locus was identified in African Americans(32). We examined associations of these 10 loci with cardiomyopathy in survivors of African ancestry using logistic regression adjusting for the same covariates used in the discovery GWA analysis.
Data and Code Availability
Aligned binary alignment map files for all the survivors included in this study and their joint genotype calls are accessible through the St. Jude Cloud (https://stjude.cloud). All data generated or analyzed during the study are included in the published article. Summary statistics from the discovery sample are available from the corresponding author upon request.
Results
Demographic and treatment characteristics of SJLIFE childhood cancer survivors of African and European ancestry in discovery and replication cohorts, respectively, are provided in Supplementary Table 1. EF in both cohorts were comparable with median of 56% (range: 16% to 68%) in the discovery cohort and 56% (range: 18% to 77%) in the replication cohort. Multivariable analyses estimated that survivors of African ancestry have a 1.38-fold (P=0.10) and 2.30-fold (P=3.9×10−3) increased risk of grade 2–4 and grade 3–4 cardiomyopathy, respectively (Table 1). These risks were greater (OR=1.53; P=0.049 and OR=2.47; P=4.3×10−3) after adjusting for known cardiac risk factors. Similar results showing increased risks of cardiomyopathy were observed in Black survivors identified by self-reporting of race compared to Caucasian survivors (Supplementary Table 2). We also observed greater proportions (unadjusted) of both CTACE grade 2–4 and CTCAE grade 3–4 cardiomyopathy among survivors of Hodgkin lymphoma, non-Hodgkin lymphoma, and bone sarcomas and after 31 to 40 years since primary cancer diagnosis whereas survivors of leukemia had lower proportions (Supplementary Tables 3 and 4).
Table 1 |.
Multivariable analyses assessing association between genetic ancestry and risk of cardiomyopathy in childhood cancer survivors exposed to cardiotoxic therapies from the St. Jude Lifetime Cohort (SJLIFE)
| Cardiomyopathy | Genetic ancestry | Survivor cases (N) | Survivor controls (N) | Adjusted for demographic and treatment exposures* | Further adjusted for known cardiac risk factors** | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | ||||
| CTCAE grade 2–4 | European | 211 | 1434 | Reference | |||
| African | 40 | 206 | 1.38 (0.94–2.03) | 0.10 | 1.53 (1.00–2.34) | 0.049 | |
| Others | 13 | 132 | 0.86 (0.47–1.58) | 0.63 | 0.89 (0.48–1.65) | 0.71 | |
| CTCAE grade 3–4 | European | 65 | 1434 | Reference | |||
| African | 19 | 206 | 2.30 (1.30–4.05) | 0.0039 | 2.47 (1.33–4.59) | 0.0043 | |
| Others | 2 | 132 | 0.44 (0.10–1.85) | 0.26 | 0.47 (0.11–2.04) | 0.31 | |
Cases, survivors affected by cardiomyopathy; controls, survivors free from cardiomyopathy
analyses adjusted for age at cancer diagnosis, age at last follow-up, sex, cumulative anthracycline dose and average heart radiation
analyses further adjusted for known cardiac risk factors including body mass index, hypertension, diabetes, hypercholesterolemia, hypertriglyceridemia, sedentary behavior, risk alcohol drinking and smoking
Genetic associations for EF
Common variant analysis in survivors of African ancestry identified two loci on 1p13.2 and 15q25.3 for EF with genome-wide significance (Table 2, Supplementary Figure 2 and Supplementary Table 5). The quantile-quantile plot (Supplementary Figure 3) and a genomic inflation factor of 0.994 indicate no evidence of population stratification. The 1p13.2 locus was marked by nine SNPs located within MAGI3 (membrane associated guanylate kinase, WW and PDZ domain containing protein 3) with their minor alleles genome-wide significantly associated with approximately 4% reduction [standard error (se)~0.007] in EF per allele (Table 2, Supplementary Table 3 and Figure 1a). The 15q25.3 locus was marked by rs9788776 near AGBL1 (ATP/GTP binding protein-like 1) with its minor allele G conferring 5.9% reduction in EF (se=0.01; P=3.5×10−8) (Table 2 and Figure 1b). A total of 31 analytical sets of rare/low-frequency variants showed significant associations (P<7.3×10−8) with EF in survivors of African ancestry (Supplementary Table 6).
Table 2 |.
Novel risk loci for EF and cardiomyopathy identified in childhood cancer survivors exposed to cardiotoxic therapies
| Chr | SNP | Position | EA/OA | Survivor cohorts | EF |
CTCAE grade 2–4 cardiomyopathy |
CTCAE grade 3–4 cardiomyopathy |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | BETA | SE | P | EAFsurvivor cases/EAFsurvivor controls | OR (95% CI) | P | EAFsurvivor cases/EAFsurvivor controls | OR (95% CI) | P | |||||
| 1 | rs6689879 | 113559097 | C/T | African discovery | 0.17 | −0.042 | 0.007 | 2.8×10−8 | 0.26/0.15 | 3.71 (1.87–7.37) | 1.8×10−4 | 0.35/0.15 | 5.43 (2.20–13.43) | 2.5×10−4 |
| European Replication | 0.30 | −0.004 | 0.002 | 0.042 | 0.34/0.30 | 1.24 (0.98–1.56) | 0.071 | 0.35/0.30 | 1.31 (0.88–1.94) | 0.19 | ||||
| 15 | rs9788776 | 87090779 | G/A | African discovery | 0.08 | −0.059 | 0.01 | 3.5×10−8 | 0.21/0.06 | 5.24 (2.30–11.92) | 7.9×10−5 | 0.18/0.06 | 4.51 (1.52–13.37) | 6.6×10−3 |
| European Replication | Monomorphic | |||||||||||||
Chr, chromosome; Genomic position is based on GRCh38 genome build; EA, effect allele; OA, other allele; EAF, effect allele frequency; BETA, effect size; SE, standard error; OR, odds ratio; CI, confidence interval; Cardiotoxic therapies (either anthracycline chemotherapy and/or heart radiotherapy); Cases are survivors affected with cardiomyopathy; Controls are free from cardiomyopathy; Discovery cohort included 246 survivors of African ancestry exposed to cardiotoxic therapies, and the replication cohort included 1645 survivors of European ancestry exposed to cardiotoxic therapies
Figure 1 |.
LocusZoom plots of the novel risk loci for left ventricular ejection fraction (EF) and cardiomyopathy in survivors of African ancestry of childhood cancer showing associations with EF expressed as –log10(P value) obtained from analysis of common variants in the discovery sample. Results for the SNPs within +/− 500kb of the index SNPs (a) rs6689879 within MAGI3 and (b) rs9788776 near AGBL1 are shown. SNPs are shown as circles, with the index SNP represented by purple diamond. All other SNPs are color coded according to the strength of LD with the top SNP (as measured by r2 in the African 1000 Genomes project data).
Of the nine genome-wide significant SNPs at the 1p13.2 locus, the association of rs6689879 with EF replicated (0.4% reduction; se=0.002; P=0.042) (Table 2 and Supplementary Table 5) in 1645 survivors of European ancestry. The remaining eight SNPs did not show significant associations (P>0.42). The associations of additional seven SNPs in high LD (r2>0.81) with rs6689879 also replicated with similar effect sizes and statistical significance (beta~0.4%; se=0.002) to that of rs6689879 (Supplementary Table 5). SNP rs9788776 at the 15q25.3 locus was monomorphic in survivors of European ancestry and hence its association with EF could not be independently evaluated. Of the 31 significant analytical sets including rare/low frequency variants, none showed significant associations (P>0.058) (Supplementary Table 6) although not all the variants initially examined for each analytical set in the survivors of African ancestry were present in the replication cohort including survivors of European ancestry.
Associations with cardiomyopathy
We examined rs6689879 at the 1p13.2 locus for its associations with cardiomyopathy in the 246 survivors of African ancestry in the discovery cohort. Our results showed significant associations of rs6689879 with 3.71-fold (P=1.8×104) and 5.43-fold (P=2.5×104) risk of grade 2–4 (n=40) and grade 3–4 (n=19) cardiomyopathy, respectively (Table 2). Similarly, rs9788776 was associated with 5.24-fold (P=7.9×10−5) and 4.51-fold (P=6.6×10−3) increased risk of grade 2–4 and grade 3–4 cardiomyopathy, respectively (Table 2). In 1645 survivors of European ancestry in the replication cohort, associations of rs6689879 with grade 2–4 (n=211; OR=1.24; P=0.071) and 3–4 (n=65; OR=1.31; P=0.19) cardiomyopathy were also observed, but without statistical significance (Table 2). Association of rs9788776 at the 15q25.3 locus with cardiomyopathy could not be assessed because it was monomorphic in survivors of European ancestry.
We did not observe significant associations between the two novel loci and EF in survivors of both African [rs6689879 (beta=−0.027; se=0.018; P=0.15) and rs9788776 (beta=−0.04; se=0.036; P=0.25)] and European ancestry [rs6689879 (beta=0.007; se=0.007; P=0.28)] not exposed to cardiotoxic therapies. No associations with EF were also observed at both 1p13.2 (MAF=0.13; beta=1.71; se=1.44; P=0.24) and 15q25.3 (MAF=0.08; beta=0.51; se=1.67; P=0.76) loci in the individuals with sickle-cell disease. In the UKBB individuals of European ancestry, the 1p13.2 locus did not show a significant association with cardiomyopathy (MAF=0.31; OR=0.99; P=0.30).
Genetic effects with respect to exposure to anthracyclines alone or heart radiation alone
Among survivors of African ancestry exposed to anthracyclines only, both rs6689879 and rs9788776 showed similar effects on EF [3.0% reduction (P=0.044) and 4.4% reduction (P=0.022), respectively] to those obtained among all survivors (Table 2). Among survivors exposed to radiation only, rs6689879 conferred slightly reduced effect (2.4% reduction; P=0.14) whereas rs9788776 showed a stronger effect (8.1% reduction; P=5.4×10−4).
Proportion of racial disparities explained by the two risk loci
We evaluated the joint contribution of rs6689879 and rs9788776 to the observed cardiomyopathy risk difference between survivors of African and European ancestries, using the same multivariable logistic regression adjusting for demographic, treatment and cardiovascular risk factors. Adding the two SNPs, risks of CTCAE grade 2–4 and CTCAE grade 3–4 cardiomyopathy in survivors of African compared to those of European ancestry reduced the ORs to 1.26 (P=0.34) and 2.15 (P=0.028) from 1.53 (P=0.049) and 2.47 (P=4.3×10−3), respectively. Thus, the two loci together may explain 17.6% (grade 2–4) and 13.0% (grade 3–4) of the racial disparity in cardiomyopathy risk between survivors of African and European ancestries.
QTLs at the 1p13.2 locus
Conditional analysis did not identify any secondary association signal at the 1p13.2 locus. Based on the GTEx data, the rs6689879 is an eQTL for MAGI3, PHTF1 (putative homeodomain transcription factor 1), and two non-coding RNAs (RP5–1073O3.2 and RP4–730K3.3) and a sQTL for PHTF1 and DCLRE1B (DNA cross-link repair 1B) in multiple human tissues (Supplementary Figures 4 and 5).
The 1p13.2 locus regulates DNA methylation among survivors of childhood cancer
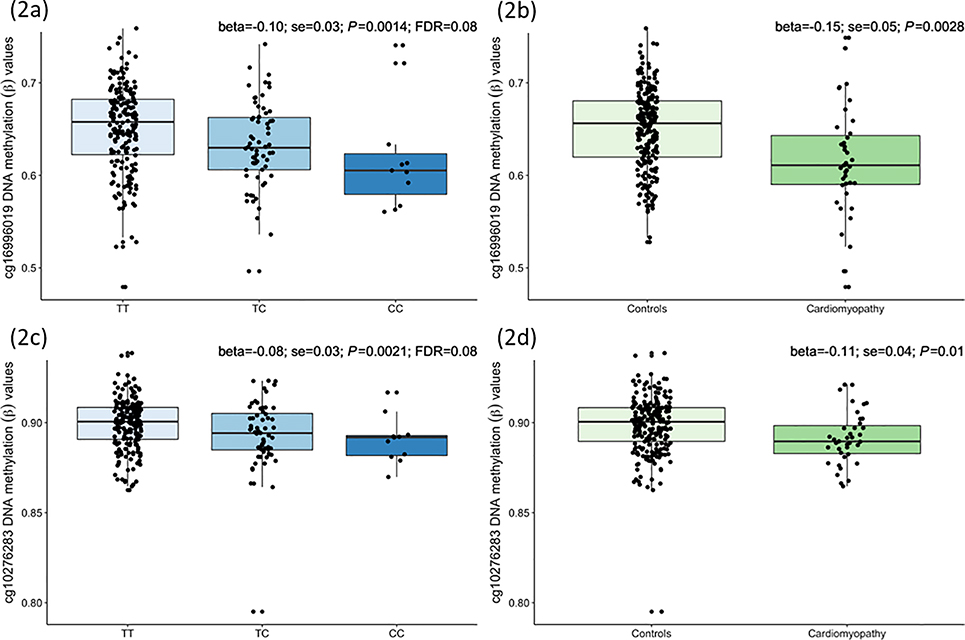
In 265 survivors of African-descent, we found the risk allele C of rs6689879 was significantly correlated with decreased methylation levels of cg16996019 (beta=−0.10; P=0.0014; FDR=0.08) and cg10276283 (beta=−0.08; P=0.0021; FDR=0.08) (Figures 2a, 2c and Supplementary Table 7). The cg16996019 is located 634 bp upstream of transcription start site of PHTF1 within the promoter region, and cg10276283 is located 937 bp away from an enhancer within intron 4 of RSBN1 (round spermatid basic protein 1) that interacts(33) with its protomer and another enhancer within intron 2 of RSBN1 also interacts with the promoter of PHTF1 (Supplementary Figure 6). Notably, both cg16996019 (beta=−0.15; P=0.0028) and cg10276283 (beta=−0.11; P=0.01) were hypomethylated in survivors of African ancestry with grade 2–4 cardiomyopathy (Figures 2b and 2d).
Figure 2 |.
Childhood cancer survivors of African ancestry-specific DNA methylation levels of cg16996019 [located 634 bp upstream of transcription start site of PHTF1 (putative homeodomain transcription factor 1) within the promoter region] and cg10276283 [located 937 bp away from an enhancer within intron 4 of RSBN1 (round spermatid basic protein 1)] with respect to three genotypes of rs6689879 near MAGI3 and between survivors with (n=39) and without (n=226) moderate-to-life-threatening cardiomyopathy in the discovery cohort. Boxplots show beta values (ratio of methylated to unmethylated probe intensities) of cg16996019 with respect to (a) rs6689879’s genotypes and (b) cardiomyopathy status, and of cg10276283 with respect to (c) rs6689879’s genotypes and (d) cardiomyopathy status. Results from linear regression models examining associations of cg16996019 and cg10276283 with rs6689879 (additively coded) and cardiomyopathy status, adjusted for age at DNA collection, sex and DNA source are shown on top right corners of each boxplot.
In 1460 survivors of European ancestry, we found the risk allele C of rs6689879 was also significantly associated with decreased methylation levels of cg16996019 (beta=−0.028; P=0.014). The cg16996019 located within the promoter region of PHTF1 was also hypomethylated, albeit not statistically significantly, in survivors of European ancestry with grade 2–4 cardiomyopathy (beta=−0.033; P=0.13). The cg10276283 did not show statistically significant associations with rs6689879 genotype or cardiomyopathy risk.
Transcriptomic response to DIC
In untreated hiPSC-CMs samples derived from breast cancer patients who received doxorubicin and experienced DIC, PHTF1 was significantly overexpressed (beta=0.45; P=0.028) compared to those who received doxorubicin but did not develop DIC (Figure 3).
Figure 3 |.
Dot plots showing PHTF1 normalized expression after log2-transformation of counts per million reads (cpm) measured in human induced pluripotent stem cell-derived cardiomyocytes generated from six breast cancer patients who were exposed to 240 mg/m2 doxorubicin at least one year prior to their transcriptome measurement of whom three developed doxorubicin-induced cardiotoxicity (DIC) and the other three did not (no_DIC). RNA-sequencing was performed on these six patients after 1 μM doxorubicin treatment as well as without exposure (0 μM).
Cardiomyopathy loci in the general population and their associations in survivors
In survivors of African ancestry, associations of 7 of the 10 loci were directionally consistent with those observed for cardiomyopathy in the general population, with 1 variant on CACNB4 that was previously identified in African Americans reaching nominal significance for both grade 2–4 (OR=0.15; P=1.6×10−3) and grade 3–4 (OR=0.10; P=8.9×10−3) cardiomyopathy (Supplementary Table 8). Both of these ORs of CACNB4 variant were appreciably larger than that observed for cardiomyopathy in the general population (OR=0.50)(32). Another variant on ALPK3 also showed stronger associations, especially for grade 2–4 cardiomyopathy, in survivors of African ancestry with borderline statistical significance (OR=6.73; P=0.075).
Discussion
Using the systematic clinically assessed SJLIFE cohort of adult survivors of childhood cancer, we identified an increased risk for cardiomyopathy among survivors of African ancestry compared to those of European ancestry and, importantly, identified genetic variants significantly associated with this disparity. The risk for moderate-to-life threatening (grade 2–4) and severe-to-life threatening (grade 3–4) cardiomyopathy was 1.53 and 2.47-fold higher, respectively, among survivors of African compared to European ancestry, despite adjustment for cardiovascular risk factors. Two genetic loci on chromosomes 1p13.2 and 15q25.3 conferred significantly increased risk for survivors of African ancestry but the 1p13.2 showed an attenuated effect and variant at the 15q25.3 was absent in survivors of European ancestry. Together, the two loci explained 17.6% and 13.0% of the racial disparity in risk of grade 2–4 and grade 3–4 cardiomyopathy, respectively, between survivors of African and European ancestries. These novel findings may elucidate disparities in the pathophysiology of adverse treatment-related cardiac outcomes among childhood cancer survivors of African ancestry, leading to improved risk stratification in frontline therapy. Our results suggest an opportunity for survivorship-care providers to consider a more rigorous cardiac surveillance strategy in African American survivors.
While adult survivors of childhood cancer are known to be at an increased risk for late cardiac morbidity and mortality, disparities across racial/ethnic groups have been understudied. Analyzing survivors of childhood, adolescent, and young adult cancers (diagnosed 0–35 years) in the Surveillance, Epidemiology, and End Results registry, Berkman and colleagues identified an elevated risk for all-cause [hazard ratio (HR)=1.75; 95% CI=1.70–1.79] and cardiovascular (HR=2.13; 95% CI=1.85–2.46) mortality among Black survivors compared to White survivors(34). When restricted to survivors diagnosed 0–14 years, this racial disparity persisted for all-cause death (HR=1.26; 95% CI=1.18–1.35) but abated for cardiovascular death (HR=1.08; 95% CI=0.62–1.89). Studying survivors of adolescent and young adult cancers diagnosed between the ages of 15–39 years, Keegan et al.(6) found an increased risk for cardiovascular disease among African-American survivors compared to European-American (HR=1.55; 95% CI=1.33–1.81), despite adjustment for demographics, socioeconomic factors, and cancer type. In a report from the CCSS, non-Hispanic Black survivors were more likely to report grade 3–5 congestive heart failure [relative rate (RR)=1.8; 95% CI=1.1–2.9] with attenuation after adjustments for socioeconomic status (RR=1.6; 95% CI=1.0–2.4) and cardiovascular risk factors (RR=1.4; 95% CI=0.9–2.3)(35). To our knowledge, our study is the first to investigate genetic variants associated with cancer therapy-related cardiomyopathy specifically in survivors of African ancestry. Risk loci on chromosomes 1 and 15 were significantly associated with reduced EF and increasing severity of cardiomyopathy among survivors of African ancestry. The chromosome 1 locus conferred over 5-fold risk in survivors of African ancestry but only 1.3-fold in survivors of European ancestry. The chromosome 15 locus was present with MAF of 8% in survivors of African ancestry, but monomorphic in survivors of European ancestry, which is consistent with its frequency of less than 0.05% in individuals of European ancestry in the 1000 Genomes project. Results in childhood cancer survivors not exposed to cardiotoxic therapies and in non-childhood cancer survivors, including individuals of African ancestry, suggesting that both risk loci may be specific to pediatric cancer survivors exposed to either anthracyclines and/or heart radiation, although additional studies are required to further confirm these observations.
We did not observe greater or reduced effects of either locus on EF when analyses were stratified to include survivors exposed to anthracyclines only or heart radiotherapy only, except that rs9788776 on chromosome 15 showed a stronger effect on EF (8.1% reduction; P=5.4×10−4) among survivors exposed to radiation only. Considering relatively small sample sizes in our analyses, additional studies are required to further investigate these exploratory findings. We observed limited replication of loci for cardiomyopathy identified in the general population among survivors of African ancestry in this study. This is likely due to: (i) all survivors of African ancestry in this study were previously exposed to cytotoxic cancer therapies and therefore, different mechanisms may underlie cardiomyopathy in survivors than in the general population(36); and (ii) because of the widespread differences in allele frequencies and linkage disequilibrium patterns in the genomes of European and African populations, it is possible that genetic variants identified in European individuals are not associated in individuals of African ancestry. This hypothesis was supported by strong replication of only one locus that was also identified in the general population of African Americans.
We found the risk allele C of rs6689879 modulates expression and alternative splicing of PHTF1 in multiple human tissues, suggesting its strong regulatory activity. PHTF1 encodes a putative homeodomain transcript factor which is mainly involved in DNA-dependent transcription and regulation of biological processes. Results from DNA methylation data among survivors of both African and European ancestries further corroborated the functional role of rs6689879 showing its association with hypomethylation of the promoter of PHTF1. Furthermore, in survivors of both African and European ancestries with cardiomyopathy, the PHTF1 promoter was also hypomethylated. Notably, the magnitudes of hypomethylation of PHTF1 promoter with respect to the risk allele C of rs6689879 and cardiomyopathy in survivors of European ancestry were 3- and 5-fold smaller, respectively, relative to those observed in survivors of African ancestry. These DNA methylation results are consistent with the attenuated effect of rs6689879 on cardiomyopathy risk in survivors of European than those of African ancestry, and provide epigenetic evidence supporting dysregulation of PHTF1 as the possible explanation underlying the rs6689879’s association with cardiomyopathy. We speculate that larger effects of the risk allele of rs6689879 on cardiomyopathy risk in survivors of African than those of European ancestry despite the allele frequency being higher in survivors of European ancestry (MAF 30%) than in survivors of African ancestry (MAF 17%) may be due to purifying selection(37,38). Based on this hypothesis, risk alleles that increase an individual’s susceptibility to diseases are kept at lower frequencies in populations. Thus, it is possible that the C allele of rs6689879 has a stronger effect on cardiomyopathy in survivors of African ancestry and is less frequent in this population, in comparison to survivors of European ancestry. Potential role of PHTF1 in cardiomyopathy was further supported by its upregulation with respect to doxorubicin-induced cardiotoxicity, specifically in the untreated hiPSC-CMs samples derived from breast cancer patients who were previously exposed to doxorubicin and experienced cardiotoxicity. All breast cancer patients were at least one year off treatment at the time of their RNA-sequencing(12) and therefore the overexpression of PHTF1 in patients with cardiotoxicity may represent long-term molecular response to doxorubicin, mimicking those in survivors of childhood cancer. Additional functional studies using relevant tissues such as hiPSC-CMs generated from survivors of African ancestry are warranted to confirm the involvement of PHTF1 in cardiomyopathy, to identify the target genes and pathways underlying the chromosome 15 locus following independent replication, and to further probe into molecular mechanisms. Moreover, research efforts should also focus on conducting larger studies including survivors of African ancestry to identify additional genetic loci that may further explain the biological basis of racial disparities in cardiomyopathy risk.
Our study is limited by lack of replication of findings in an independent cohort of survivors of African ancestry of childhood cancer; however, such a cohort with both clinically-defined cardiomyopathy and genome-wide data does not currently exist. Notably, SNP rs6689879 was replicated in survivors of European ancestry in the same direction, albeit with a smaller magnitude of effect. Studies evaluating racial disparities in childhood cancer survivors have been challenged by the limited number of minority survivors represented in cohort studies and the availability of biological specimens to support genetic investigations(39). Consequently, sample sizes are limited precluding discovery of genetic variants with modest effect sizes and/or lower allele frequencies.
In conclusion, we identified childhood cancer survivors of African ancestry have a risk of cardiomyopathy that exceeds that of survivors of European ancestry, despite adjustment for cardiovascular risk factors. A novel locus on chromosome 1 was significantly associated with cardiomyopathy risk specifically in survivors of African ancestry and our molecular data suggested dysregulation of PHTF1 may be an explanation underlying this genetic association. Future investigations will help further elucidate these findings, clarify risk factors, and identify targets for potential therapeutic interventions.
Supplementary Material
Statement of Significance:
Childhood cancer survivors of African ancestry are at higher risk of cardiomyopathy than those of European ancestry, and a novel locus at 1p13.2 is associated with therapy-related cardiomyopathy specifically in African-American survivors.
Acknowledgements/Funding:
The St. Jude Lifetime Cohort (SJLIFE) study is supported by the National Cancer Institute at the National Institutes of Health (U01 CA195547: MMH and LLR, Principal Investigators; Cancer Center Support CORE grant CA21765: C. Roberts, Principal Investigator). This work is also supported by R01 CA216354 (YY and JZ, Principal Investigators) from the National Cancer Institute at the National Institutes of Health and the American Lebanese Syrian Associated Charities, Memphis, Tennessee.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Mulrooney DA, Yeazel MW, Kawashima T, Mertens AC, Mitby P, Stovall M, et al. Cardiac outcomes in a cohort of adult survivors of childhood and adolescent cancer: retrospective analysis of the Childhood Cancer Survivor Study cohort. BMJ 2009;339:b4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulrooney DA, Armstrong GT, Huang S, Ness KK, Ehrhardt MJ, Joshi VM, et al. Cardiac Outcomes in Adult Survivors of Childhood Cancer Exposed to Cardiotoxic Therapy: A Cross-sectional Study. Ann Intern Med 2016;164:93–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loehr LR, Rosamond WD, Chang PP, Folsom AR, Chambless LE. Heart failure incidence and survival (from the atherosclerosis risk in communities study). Am J Cardiol 2008;101:1016–22 [DOI] [PubMed] [Google Scholar]
- 4.Yancy CW. Heart failure in African Americans: A cardiovascular enigma. J Card Fail 2000;6:183–6 [DOI] [PubMed] [Google Scholar]
- 5.Myers VD, Gerhard GS, McNamara DM, Tomar D, Madesh M, Kaniper S, et al. Association of Variants in BAG3 With Cardiomyopathy Outcomes in African American Individuals. JAMA Cardiol 2018;3:929–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keegan THM, Kushi LH, Li Q, Brunson A, Chawla X, Chew HK, et al. Cardiovascular disease incidence in adolescent and young adult cancer survivors: a retrospective cohort study. J Cancer Surviv 2018;12:388–97 [DOI] [PubMed] [Google Scholar]
- 7.Aminkeng F, Bhavsar AP, Visscher H, Rassekh SR, Li Y, Lee JW, et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat Genet 2015;47:1079–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco JG, Sun CL, Landier W, Chen L, Esparza-Duran D, Leisenring W, et al. Anthracycline-related cardiomyopathy after childhood cancer: role of polymorphisms in carbonyl reductase genes--a report from the Children’s Oncology Group. J Clin Oncol 2012;30:1415–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Liu W, Sun CL, Armenian SH, Hakonarson H, Hageman L, et al. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the children’s oncology group. J Clin Oncol 2014;32:647–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Sun CL, Quinones-Lombrana A, Singh P, Landier W, Hageman L, et al. CELF4 Variant and Anthracycline-Related Cardiomyopathy: A Children’s Oncology Group Genome-Wide Association Study. J Clin Oncol 2016;34:863–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Pavia P, Kim Y, Restrepo-Cordoba MA, Lunde IG, Wakimoto H, Smith AM, et al. Genetic Variants Associated With Cancer Therapy-Induced Cardiomyopathy. Circulation 2019;140:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burridge PW, Li YF, Matsa E, Wu HD, Ong SG, Sharma A, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med 2016;22:547–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson MM, Ness KK, Nolan VG, Armstrong GT, Green DM, Morris EB, et al. Prospective medical assessment of adults surviving childhood cancer: study design, cohort characteristics, and feasibility of the St. Jude Lifetime Cohort study. Pediatr Blood Cancer 2011;56:825–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang RM, Bierig M, Devereux RB, Flachskampf FA, Foster E, Pellikka PA, et al. Recommendations for chamber quantification: a report from the American Society of Echocardiography’s Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J Am Soc Echocardiogr 2005;18:1440–63 [DOI] [PubMed] [Google Scholar]
- 15.Hudson MM, Ehrhardt MJ, Bhakta N, Baassiri M, Eissa H, Chemaitilly W, et al. Approach for Classification and Severity Grading of Long-term and Late-Onset Health Events among Childhood Cancer Survivors in the St. Jude Lifetime Cohort. Cancer Epidem Biomar 2017;26:666–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feijen EA, Leisenring WM, Stratton KL, Ness KK, van der Pal HJ, Caron HN, et al. Equivalence Ratio for Daunorubicin to Doxorubicin in Relation to Late Heart Failure in Survivors of Childhood Cancer. J Clin Oncol 2015;33:3774–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanna A, Pequeno P, Gupta S, Thavendiranathan P, Lee DS, Abdel-Qadir H, et al. Increased Risk of All Cardiovascular Disease Subtypes Among Childhood Cancer Survivors Population-Based Matched Cohort Study. Circulation 2019;140:1041–3 [DOI] [PubMed] [Google Scholar]
- 18.Armstrong GT, Oeffinger KC, Chen Y, Kawashima T, Yasui Y, Leisenring W, et al. Modifiable Risk Factors and Major Cardiac Events Among Adult Survivors of Childhood Cancer. Journal of Clinical Oncology 2013;31:3673–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZM, Wilson CL, Easton J, Thrasher A, Mulder H, Liu Q, et al. Genetic Risk for Subsequent Neoplasms Among Long-Term Survivors of Childhood Cancer. Journal of Clinical Oncology 2018;36:2078–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sapkota Y, Cheung YT, Moon W, Shelton K, Wilson CL, Wang Z, et al. Whole-Genome Sequencing of Childhood Cancer Survivors Treated with Cranial Radiation Therapy Identifies 5p15.33 Locus for Stroke: A Report from the St. Jude Lifetime Cohort Study. Clin Cancer Res 2019;25:6700–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapkota Y, Wilson CL, Zaidi AK, Moon W, Fon Tacer K, Lu L, et al. A novel locus predicts spermatogenic recovery among childhood cancer survivors exposed to alkylating agents. Cancer Res 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.EPACTS: Efficient and Parallelizable Association Container Toolbox.
- 23.Hankins JS, Estepp JH, Hodges JR, Villavicencio MA, Robison LL, Weiss MJ, et al. Sickle Cell Clinical Research and Intervention Program (SCCRIP): A lifespan cohort study for sickle cell disease progression from the pediatric stage into adulthood. Pediatr Blood Cancer 2018;65. [DOI] [PubMed] [Google Scholar]
- 24.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. Plos Med 2015;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, et al. Genetic effects on gene expression across human tissues. Nature 2017;550:204–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met 1995;57:289–300 [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47:D1005–D12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esslinger U, Garnier S, Korniat A, Proust C, Kararigas G, Muller-Nurasyid M, et al. Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy (vol 12, e0172995, 2017). Plos One 2020;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meder B, Ruhle F, Weis T, Homuth G, Keller A, Franke J, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J 2014;35:1069−+ [DOI] [PubMed] [Google Scholar]
- 31.Villard E, Perret C, Gary F, Proust C, Dilanian G, Isnard R, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J 2011;32:26– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu HC, Dorn GW, Shetty A, Parihar A, Dave T, Robinson SW, et al. A Genome-Wide Association Study of Idiopathic Dilated Cardiomyopathy in African Americans. J Pers Med 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Stein TI, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database-Oxford 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berkman AM, Brewster AM, Jones LW, Yu J, Lee JJ, Peng SA, et al. Racial Differences in 20-Year Cardiovascular Mortality Risk Among Childhood and Young Adult Cancer Survivors. J Adolesc Young Adult Oncol 2017;6:414–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Leisenring WM, Ness KK, Robison LL, Armstrong GT, Yasui Y, et al. Racial/Ethnic Differences in Adverse Outcomes Among Childhood Cancer Survivors: The Childhood Cancer Survivor Study. Journal of Clinical Oncology 2016;34:1634–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Im C, Qin N, Wang Z, Qiu W, Howell CR, Sapkota Y, et al. Generalizability of “GWAS Hits” in Clinical Populations: Lessons from Childhood Cancer Survivors. Am J Hum Genet 2020;107:636–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, et al. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet 2013;14:460–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson MR, Wegmann D, Ehm MG, Kessner D, Jean PS, Verzilli C, et al. An Abundance of Rare Functional Variants in 202 Drug Target Genes Sequenced in 14,002 People. Science 2012;337:100–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatia S, Gibson TM, Ness KK, Liu Q, Oeffinger KC, Krull KR, et al. Childhood cancer survivorship research in minority populations: A position paper from the Childhood Cancer Survivor Study. Cancer-Am Cancer Soc 2016;122:2426–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Aligned binary alignment map files for all the survivors included in this study and their joint genotype calls are accessible through the St. Jude Cloud (https://stjude.cloud). All data generated or analyzed during the study are included in the published article. Summary statistics from the discovery sample are available from the corresponding author upon request.