Abstract
The failure of once promising target-specific therapeutic strategies often arises from redundancies in gene expression pathways. Even with new melanoma treatments, many patients are not responsive or develop resistance, leading to disease progression in terms of growth and metastasis. We previously discovered that the transcription factors ETS1 and PAX3 drive melanoma growth and metastasis by promoting the expression of the MET receptor. Here, we find that there are multiple ETS family members expressed in melanoma and that these factors have redundant functions. The small molecule YK-4–279, initially developed to target the ETS gene-containing translocation product EWS-FLI1, significantly inhibited cellular growth, invasion, and ETS factor function in melanoma cell lines and a clinically relevant transgenic mouse model, BrafCA;Tyr-CreERT2;Ptenf/f. One of the anti-tumor effects of YK-4–279 in melanoma is achieved via interference of multiple ETS family members with PAX3 and the expression of the PAX3-ETS downstream gene MET. Expression of exogenous MET provided partial rescue of the effects of YK-4–279, further supporting that MET loss is a significant contributor to the anti-tumor effects of the drug. This is the first study identifying multiple overlapping functions of the ETS family promoting melanoma. Additionally, targeting all factors, rather than individual members, demonstrated impactful deleterious consequences in melanoma progression. Given that multiple ETS factors are known to have oncogenic functions in other malignancies, these findings have a high therapeutic impact.
Introduction
Melanoma is an aggressive cancer type, with a high propensity for invasion and metastasis early in the disease process. There are several factors that actively drive melanoma progression including MET, a tyrosine kinase receptor overexpressed in melanoma and implicated in tumor growth, invasion, and drug resistance (1). One driver for overexpression of the MET gene is through transcriptional upregulation by PAX3 and ETS1. Both PAX3 and ETS1 transcription factors are essential in melanocyte development, and form transcriptional complexes that regulate MET expression through upstream genomic enhancers (2). ETS1, while having undetectable expression in normal skin melanocytes and common nevi, is overexpressed in primary and metastatic melanoma (3, 4). In melanoma, ETS1 levels are elevated due to protection from the deubiquinase Usp9x and subsequent protein degradation (5). Additionally, ETS1 levels and/or activity also increased in response to Vemerafenib drug treatment, UV response, and ER stress (5–7). One mechanism where elevated levels of ETS1, and other ETS proteins, promotes melanoma progression is through their transcription factor function, due to mutations creating de novo ETS binding sites within enhancer regions of cancer promoting genes (such as the TERT loci) and driving the expression of these genes (8, 9). Given the action of ETS1 on several pro-tumor genes, ETS1 would be an advantageous target for melanoma therapy.
Great advances in melanoma therapeutics have been made in recent years, including immunotherapeutics such as anti-PD-1 antibodies (nivolumab, pembrolizumab), and small molecule therapeutics including BRAF(V600E)-targeting drugs (vemurafenib, dabrafenib), either as a monotherapy or in combination with MEK inhibitors (trametinib, cobimetinib) (10). However, these treatments have limitations in response rates and durability and additional therapeutic options are needed. The small molecule YK-4–279, though initially discovered to inhibit the translocation product EWS-FLI1 in Ewing sarcoma cells, can also affect other ETS-family factors besides FLI1, including ERG and ETV1 (11–13). Several mechanisms of action for YK-4–279 have been described, including disrupting the interaction of ETS-domain interactions between RNA helicase A and p68 DDX5 (11, 14). This work provides solid clinical promise for YK-4–279, and is currently being tested in a Phase I clinical trial in relapsed or refractory Ewing Sarcoma (NCT02657005) using the chemical analog TK216.
We have identified, herein, that multiple ETS factors are commonly expressed in both melanoma cell lines and patient tissues, have at least partial functional redundancy, and are active drivers of cancer progression. Although YK-4–279 was designed to target just one specific translocation product, we discovered that this compound affects multiple ETS-containing proteins. YK-4–279 inhibits ETS factors in terms of transcriptional abilities, interaction with PAX3, and the promotion of potent downstream effector genes such as the tyrosine kinase receptor MET. The use of YK-4–279 has a highly significant impact the inhibition of melanoma growth and invasion in culture and in the clinically relevant BrafCA;Tyr-CreERT2;Ptenf/f melanoma mouse model. This novel use of a transcription factor-targeting small molecule in melanoma may provide a viable new option for therapy.
Materials and Methods
Cell lines and culture conditions.
The studies were performed with at least two out of three representative melanoma cell lines: MEL624 (RRID:CVCL_8054), A375 (ATCCCat#CRL-7904, RRID:CVCL_0132), and SKMEL28 (ATCCCat#HTB-72, RRID:CVCL_0526). These cell lines were chosen as they are commonly used in the literature, have typical genetic mutations found in melanoma, and demonstrate growth levels and morphology typical for cultured melanoma cell lines. MEL-ST cells are immortalized non-transformed human melanocytes (generously provided by Neil Ganem (Boston University) and Robert Weinberg (Whitehead Institute) (15)), and hMEL cells are primary human melanocytes (generously provided by Andrey Sharov (Boston University)). Other human melanoma cell lines for the multi-cell screen (Figure 1, Supplementary Figure S1) were purchased from either ATCC or the NCI Tumor Repository. MCF10a and MCF7 cells were generously provided by Drs. Margaret Gardel and Tong-Chuan He (University of Chicago). All cells were maintained at 37°C with 5% CO2 and grown in Dulbecco’s Modified Eagle Media and supplemented with 10% Fetal Bovine Serum (Sigma) except for MEL-ST cells that were enriched with 5% FBS and 1% penicillin/ streptomycin. Morphology and melanoma marker testing were utilized to verify that the cells are from a melanocytic origin, and validity that cells were negative for the presence of Mycoplasma followed standard laboratory protocols. For drug treatments, 10 mM stock solutions of YK-4–279 or SCH772984 (Apexbio Technology #A3946 and #A3805 respectively) were prepared in DMSO and diluted in media as indicated. Drug levels used in studies followed prior protocols and demonstrated similar toxicity levels as previously reported ((16) and Supplementary Figure S2A,B)).
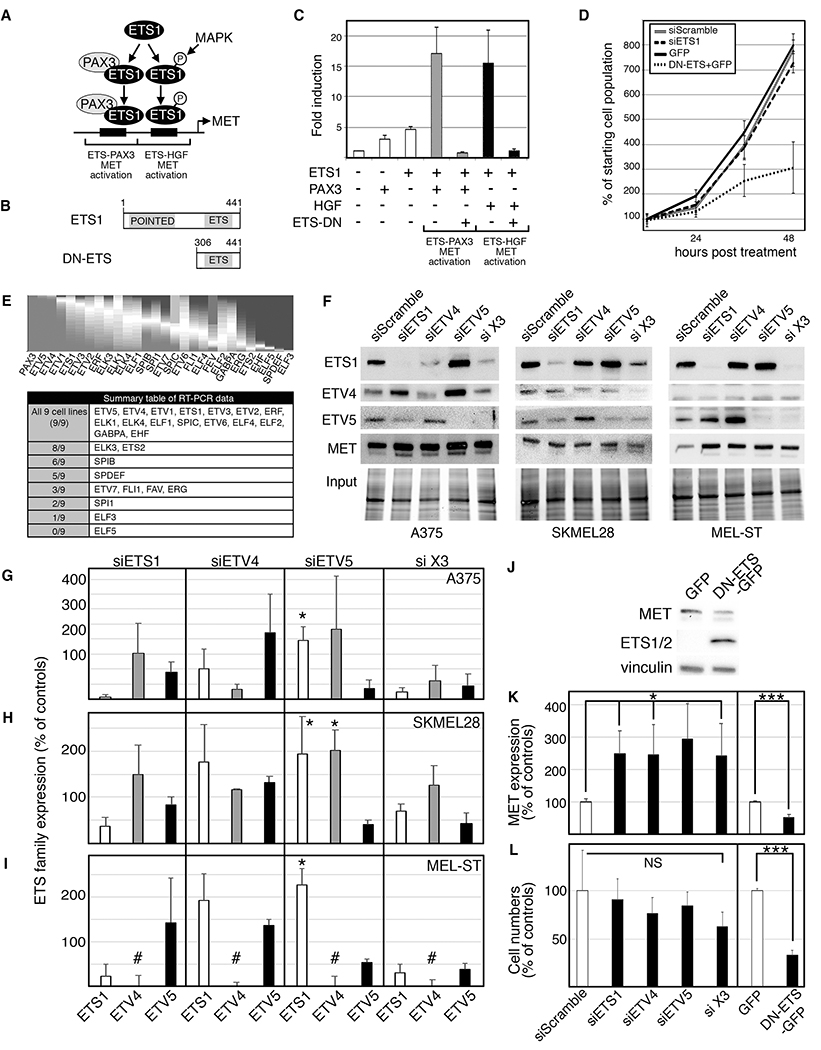
Figure 1.
A dominant-negative ETS protein inhibits growth and expression of MET in melanoma cells. A. A schematic of dual ETS1 regulation of the MET promoter. ETS1 (with PAX3) drives MET gene expression through an ETS-PAX binding site, and a phosphorylated ETS1 protein promotes MET expression through a PAX-independent mechanism and a separate “ETS-HGF” binding site. B. A diagram of a full-length ETS1 protein with both Pointed and ETS domains, and a dominant-negative ETS1 (DN-ETS) containing only the ETS domain. C. DN-ETS inhibits ETS1 activation of the MET promoter through both PAX3- and HGF-dependent synergistic mechanisms. Luciferase assays of 293T cells transfected with a MET promoter luciferase reporter construct (METpm) in the absence (−) or presence (+) of ETS1 expression with or without PAX3, DN-ETS and/or HGF. Data are presented as fold induction over light units of METpm alone. D. Expression of DN-ETS, but not siETS1, significantly attenuates melanoma growth. A375 cells are transfected with ETS1 siRNA, siScramble control, or DN-ETS or GFP expression constructs, and counted over a time-course. E. Multiple ETS family members are commonly and variably expressed in human melanoma samples and melanoma cell lines. A gene expression profile heatmap of PAX3 and ETS family members using RNASeq data from 473 melanoma patients was created utilizing the UCSC Cancer Genomics Browser (top panel). The data are depicted as a proportions plot with higher expression (grey shading from bottom) and lower expression (dark grey shading from top) in melanoma compared to other cancer subtypes. In addition, multiple ETS family members are expressed in the panel of nine melanoma cell lines (summary table of RT-PCR screen shown in table). F-I. Inhibition of ETS factors influences the expression of other family members in A375 and SKMEL28 melanoma cells, and MEL-ST melanocyte cells. The data represent inhibition with siETS1, siETV4, siETV5, or all three (si X3) as indicated on column top, and bars (in G-I) represent western blot densitometry for ETS1 (white bars), ETV4 (grey), or ETV5 (black) as indicated on column bottom. The Y-axis is densitometry readings normalized to siScramble controls (set at 100%). Data shown are western analysis from three independent experiments. (∗=p≤0.05, #=no detectable expression of ETV4 in MEL-ST cells). J. Expression of a dominant negative ETS protein (DN-ETS-GFP) decreased MET expression in A375 cells, when compared to mock transfected cells (GFP). K. MET expression is increased when ETS family members are inhibited and decreased with expression of DN-ETS-GFP in A375 cells. Densitometry of three independent western analyses show a significant change in MET expression (representative western blots shown in F,J), ∗=p≤0.05, ∗∗∗=p≤0.0005. L. Inhibition of ETS family members with DN-ETS-GFP significantly attenuated cell numbers but ETS-specific siRNA did not. Cell counts of three independent experiments at 48 hours post treatment were normalized to control groups (siScramble or GFP), which were set at 100% (∗∗∗=p≤0.0005).
Small interfering RNA inhibition.
Melanoma cells were transfected following prior methods (2) with siRNA (Ambion/Thermo-Fisher) targeting ETS1 (cat. #4392421, ID 54847), ETV4 (ID 54860), and ETV5 (ID 34864) or scramble control (100 pmol each siRNA) with Lipofectamine-2000 (Invitrogen). Transfection efficiency for each siRNA was determined to be at levels greater than 90% loss of target protein expression as determined by western analyses.
In vivo melanoma model and treatment.
BrafCA;Tyr-CreERT2;Ptenf/f mice were previously described ((17), Jackson Laboratory stock number 013590, C57BL/6J background). Mouse colonies were maintained under specific pathogen-free conditions and experimental procedures were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC). Six- to 7-week-old BrafCA;Tyr-CreERT2;Ptenf/f mice were surgically implanted with osmotic minipump (Alzet model 1004) with a release rate of drug of 0.11 uL/hour. Pumps were filled with either 50% of DMSO (control) or with 1.12 mM YK-4–279 total/final concentration in the pump. For mice of approximately 25g body weight, it was estimated that the pumps release 2.6 to 3 uM YK-4–279 drug/day. Following pump transplantation (day 0), the back of the mice was shaved and 4mM tamoxifen (Sigma Aldrich) was applied topically during three consecutive days (day 1,2,3). If necessary, a second minipump containing drug was implanted when needed at day 28 or 29 after the first pump implantation, since pumps are rated to last for a 4-week duration. Mice are observed daily, and melanoma initiation and progression days were recorded as described (18). The determination of tumor initiation day was when pigment cell aggregates were clearly visible from the skin surface (Figure 3A,B). The determination of progression day was that when tumor lesions form a palpable lesion by touch and form visible nodular lesions (Figure 3E,F). Mice were removed from study for tissue collection at day 35 (females), day 52 (males), or when tumor burden exceeded 1cm2. Tumor were analyzed and measured for thickness by a dermatopathologist blinded to the sample group.
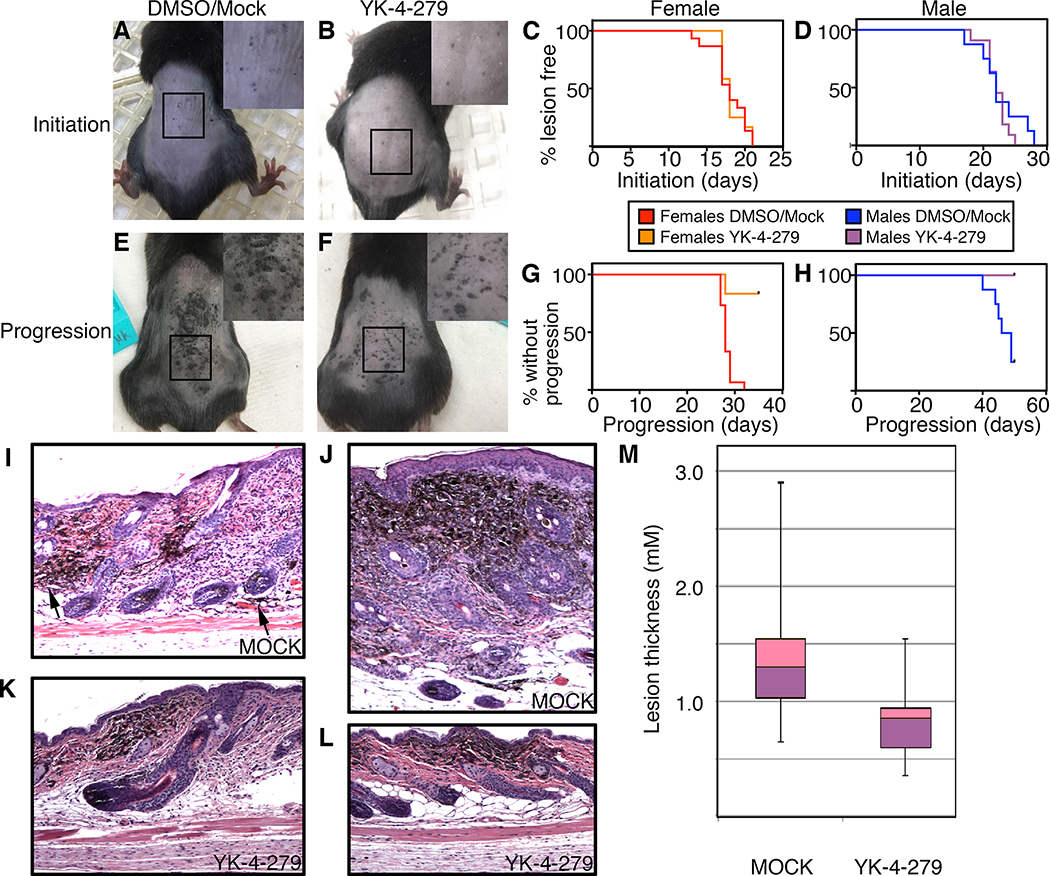
Figure 3.
YK-4–279 attenuates melanoma progression in a mouse model of melanoma. The melanoma model, BrafCA;Tyr-CreERT2;Ptenf/f, was transplanted with osmotic pumps containing DMSO alone or 1.12 mM YK-4–279 total/final concentration that released a constant level of 1.6 mg/Kg of drug. A,B. Photographs of control (A) and drug (B) group mice with initiation lesions, with magnification (2X) in insets. C,D. Kaplan-Meier survival graphs of initiation in female (C) and male (D) experimental and control matched groups. There were no significant differences between curves (p=0.45). E,F. Photographs of control (E) and drug (F) group mice with progressed tumors, with magnification (2X) in insets. G,H. YK-4–279 treatment attenuates tumor progression. There were significant differences between Kaplan-Meir curves (p<0.0001 females, p=0.004 males). I-L. Cutaneous pigmented lesions of mock treated mice (I,J) were full thickness tumors, while lesions in YK-4–279 treated mice (K,L) were superficial. In mock treated skin, even relatively flat lesions have cells invading into the subdermis (I, arrowheads). The more typical mock treated lesion is thick (>1mm, J). Sections shown are either parallel to the anterior-posterior axis (I,K,L) or transverse section (J). M. Mock treated mice have thicker pigmented lesions than YK-4–279 treated mice. In the measurement of the thickest lesion per slide, lesions were 1.37±0.55 mm and 0.82±0.32 mm for mock and drug treated mice, respectively (p=0.00098, n=16 mice/group). Each box represents the upper and lower quartile with a line at the median, whisker lines extend from maximum to minimum thicknesses, unpaired 2-tailed t test.
Western analysis.
Cells were lysed in M-PER (Mammalian Protein Extraction Buffer) buffer. Proteins (50 μg) were separated on a 4–15% Bio-Rad stain free gels (10 well 4–15% Biorad-Stain-free-gel cat no 4568084), transferred to a nitrocellulose membrane, and probed with antibodies against MET (Cell Signaling Technology Cat#8198, RRID:AB_10858224), ETS1 (Santa Cruz Cat#sc-55581, RRID:AB_831289), ETS1/2 (for detecting DN-ETS, (Santa Cruz Cat#sc-374509, RRID:AB_10987669), ETV4 (Proteintech Cat#10684–1-AP, RRID:AB_2100984), ETV5 Sigma-Aldrich Cat#WH0002119M2, RRID:AB_1841526). GAPDH (Cell Signaling Cat#5174, RRID:AB_10622025) or total protein input (Stain Free System, Bio-Rad) was used as a loading control. ImageLab (BioRad) was utilized to quantify band intensity that was normalized to loading controls. All western analyses were repeated minimally in triplicate.
Histology.
Skin tissue was collected at the end time point of experiment, formalin-fixed, paraffin-embedded, sectioned into 5 micron thick sections, and stained with hematoxylin and eosin (H&E). Samples were anonymized and assessed for lesion thickness by a dermatopathologist. For measuring tumor thickness, H&E stained samples were digitally scanned and pigmented lesions were measured from the top of the granular cell layer in the epidermis to the deepest point of invasion. At least 5 tumors from each mouse specimen were measured and the largest number was used as the representative thickness of the specimen.
Immunofluorescence staining.
For tissue sections, samples were treated with heat-induced antigen retrieval with citrate buffer (pH 6.0, Abcam). Fixed cells or tissue were permeablized with a 0.25% TritonX-100 solution, blocked with Duolink Blocking solution, and incubated with primary antibodies: PAX3 (1:100) (Abcam), ETS1(1:50) (Santa Cruz), ETV5 (1:200)(R&D Systems) and/or MET (1:200), (Ab-1003, Sigma Aldrich). The samples were incubated with F594 labeled anti-rabbit IgG and F488 labeled anti-mouse IgG (Cell Signaling) secondary antibody, and cover slipped with duolink in situ mounting medium with DAPI or Hardset Antifade Mounting Medium with DAPI (Vector Laboratories). For analysis of MET expression, sections were examined and scored as negative, low MET expression (<10% MET positive cells of lesion) or abundant MET expression (>10% MET in tumor) under a fluorescence microscope in comparison to both positive control and negative control. At least 5 anonymized tumors from two or more sections of each mouse specimen were examined, and a final score was given after examination.
Statistical analysis.
Statistical tests were performed using GraphPad Prism6 (RRID:SCR_002798). For luciferase assays, the means of the fold-change induction for each group was compared to the reporter alone using one-way ANOVA with a Dunnett’s multiple comparison test. Kaplan-Meier survival plots were generated on Prism6 and analyzed using Log-rank (Mantel-Cox) tests with one degree of freedom. Significance of correlation in MET expression between drug versus mock treated groups was determined with 2-tailed Fisher exact tests, with nominal data (abundant versus low/absent) analyzed with X2 analysis (CI=95%). Sample sizes were calculated using power analysis, with power >85% for mouse experiments and >90% for immunofluorescence scoring methods. All scoring of staining was performed by a “blinded” observer to the identity of sample group. All error bars are calculated as standard deviation (SD) unless noted. For analysis of significance where multiple sets are included for each group, error bars are calculated by standard error of the mean (SEM) as indicated. Findings are presented as significant if the p values were less than or equal to 0.05. All experiments were performed minimally in triplicate.
Study approval.
Animal experiments were performed following the NIH guidelines and were approved by the IACUC of Boston University.
Additional methods (Vectors, Luciferase assays, Data mining, Reverse-transcriptase PCR, MTT/MTS assays, Invasion assays, Zymograms, Proximity Ligation Assays (PLA), Co-immunoprecipitation analysis, MET or ETS1 rescue experiments, and phosphorylated receptor tyrosine kinase (RTK) assays) are included in Supplemental methods.
Results
A dominant negative ETS protein, but not an ETS1 siRNA, inhibits ETS1 and PAX3 function and melanoma cell growth.
In melanoma, ETS1 drives MET gene expression directly through enhancer elements within the MET 5’ proximal promoter through two pathways (2). The first mechanism is through recruitment of ETS1 by PAX3 to a specific “ETS-PAX” binding site, and the second is through a PAX-independent enhancer “ETS-HGF” binding site that is more active when ETS1 is phosphorylated (Figure 1A). Previously, our group utilized a dominant-negative ETS1 (DN-ETS) protein, commonly used in developmental studies, which contains only the C-terminal DNA-binding domain and acts as a dominant-negative protein (2). In parallel to our prior findings, DN-ETS significantly attenuated the ability of ETS1 to activate a MET enhancer reporter construct synergistically with either PAX3 or HGF/MAPK-mediated phosphorylation (Figure 1B,C, p<0.0005). DN-ETS also significantly inhibited melanoma growth in vivo (2) as well as in culture (Figure 1D, dotted line versus solid line mock control group, p=0.026 at 48 hr). However, direct inhibition of ETS1 transcript through siRNA targeting did not result in any significant difference between controls in A375 melanoma cells (Figure 1D, dashed line versus double line mock control group). This suggests that the DN-ETS-mediated inhibition on ETS1 function and melanoma growth is not purely through blocking ETS1.
The presence and functional redundancy of other ETS family members in melanoma provides one explanation for why a dominant negative ETS1, but not an ETS1-targeted siRNA, blocked transcriptional activity and cell growth. ETS1 is part of 28-member ETS protein family that all share an ETS DNA binding domain. Since all ETS factors bind to very similar cis regulatory sites, the DN-ETS most likely inhibits these other ETS proteins as well. Although ETS factors in melanoma have not been extensively studied, there is some evidence that ETV1, ETV4, and ETV5 have oncogenic function (19–21). To determine which ETS factors are commonly expressed in melanoma, in silico and reverse transcription polymerase chain reaction (RT-PCR) analysis was performed. A gene expression profile of ETS family members using RNASeq data from 473 melanoma patients was created utilizing the UCSC Cancer Genomics Browser (Figure 1E). It supports that, in patient-derived tumor tissue, multiple ETS family members are expressed. In addition, expression of ETS family members was screened in melanoma cell lines. To determine if the factors had measurable transcript levels from ETS genes, a panel of nine lines was screened with family-specific primers by RT-PCR. Expression of ETS factors was common, with 16 expressed in all nine of the cell lines and only one factor (ELF5) not expressed in any of the melanoma lines analyzed (Figure 1E, Supplementary Figure S1). These findings support that multiple ETS factors are expressed in melanoma cells.
To determine if inhibition of another common ETS factor in melanoma would alter MET expression, expression of ETS1 and two of the most overexpressed factors in patient samples, ETV4 and ETV5, was inhibited. ETS1, ETV4, and ETV5 were targeted singularly and in combination, and ETS factor and MET levels were measured by western analysis in two melanoma lines (A375 and SKMEL28) as well as the melanocyte line MEL-ST (Figure 1F). These experiments revealed two surprising findings: 1) inhibition of one ETS factor effected the expression of other family members, and 2) blocking the expression of ETS family proteins increased MET levels, while DN-ETS decreased levels. ETS1, ETV4, and ETV5 were expressed in both melanoma cell lines, while the MEL-ST cells expressed ETS1 and ETV5 but had no detectable levels of ETV4. Overall, the trend after inhibition of a single ETS factor was an increase in the other two family members, with a significant multifold increase in ETS1 and ETV4 after inhibition of ETV5 (Figure 1G–I). In terms of MET expression, while DN-ETS protein significantly decreased MET protein levels in A375 cells (Figure 1J,K), MET levels increased when the expression of single ETS family members (or all three in tandem) was inhibited (Figure 1F,K). Further, while the DN-ETS protein inhibited cell number expansion, there was no effect on any of the cells with ETS family targeted siRNA (Figure 1D,L). These findings support that targeting ETS1, ETV4, and ETV5 singularly or in combination fails to phenotype the impact of DN-ETS on melanoma cell growth and MET expression.
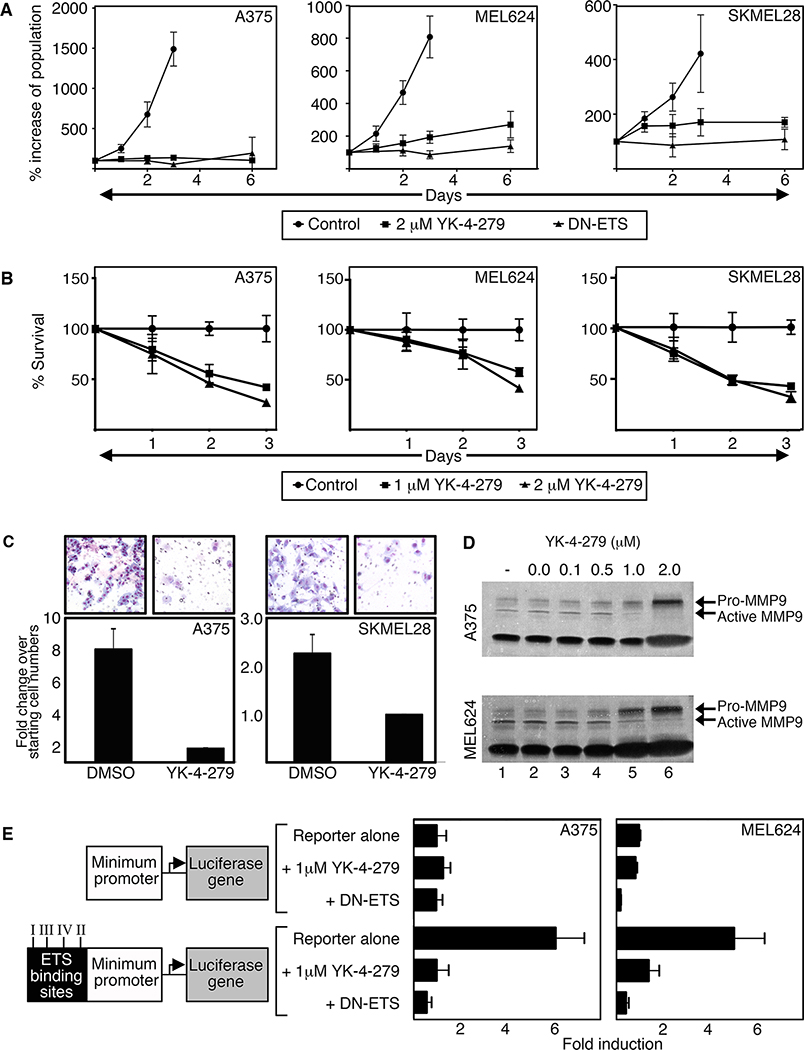
The small molecule drug YK-4–279 mimics the effects of DN-ETS for cell proliferation, survival, invasion, and ETS function.
Specific targeted therapy against ETS factors may hold promise as a melanoma treatment. Recently, the small molecule inhibitor YK-4–279 has been utilized to efficiently target the translocation product EWS-FLI1 in Ewing sarcoma (11, 22). This compound also efficiently inhibits other EWS translocation products with ETS family members (ERG, ETV1) specifically through the ETS domain (12). The addition of YK-4–279 attenuated the growth and viability of three different melanoma cell lines and mimicked the growth effects of DN-ETS (Figure 2A,B). YK-4–279 also significantly reduced melanoma cell invasion through matrigel-coated transwells (Figure 2C). One mechanism for melanoma invasion is through activation of the gelatinase MMP9 (23). Treatment of YK-4–279 resulted in decreased activation of MMP9 protein in a dose dependent manner (Figure 2D). To determine if YK-4–279 blocks ETS transcription factor activity, we utilized a luciferase reporter assay. Two reporters, one containing a minimal promoter upstream of a luciferase gene and another with four distinct but highly related ETS binding sites (24) ligated upstream of the minimum promoter, were transfected into A375 and MEL624 cells. While endogenous activation of the reporter construct with the ETS sites was detectable and significant in comparison to controls (6.10±1.24 (A375) and 5.12±1.34 (MEL624) fold over reporter without ETS binding site), introduction of either DN-ETS or 1uM YK-4–279 significantly attenuated ETS transcription factor activity (Figure 2E). Taken together, YK-4–279 inhibits melanoma cell growth, survival, and invasion in cellulo through inhibition of ETS activity.
Figure 2.
YK-4–279 inhibits melanoma cell growth, invasion, and ETS activity. A. YK-4–279 and DN-ETS comparably reduce cell growth. Melanoma cells were transfected with DN-ETS-GFP or treated with 2μM YK-4–279 and cell numbers quantified on days 0,1,2,3, and 6. B. YK-4–279 inhibits melanoma cell proliferation and survival. Cells were treated with 1μM or 2μM YK-4–279. MTT assays were used to assess cell survival compared to respective untreated controls. C. YK-4–279 inhibits melanoma cell invasion. Cells were seeded onto collagen-coated transwells and treated with either DMSO or YK-2–479. Cell invasion was examined 16h post drug treatment. DMSO cells migrated at rates 7.84±1.43 (A375) or 2.27±0.38 (SKMEL28) fold more than cells treated with YK-4–279. D. YK-4–279 inhibits MMP9 activation in a dose dependent manner. Media from cells treated with (0.1–2.0μM, lanes 3–6) or without (untreated or DMSO alone, lanes 1,2) YK-4–279 for 7 days were collected, concentrated, and run on a gelatin gel to examine MMP2/9 activity. E. YK-4–279 inhibits ETS transcriptional activity. Cells were transfected with a reporter construct containing a minimal promoter without (top three bars) or with (bottom three bars) the addition of a 60 base-pair sequence containing four different ETS binding sites. With minimum promoter without ETS sites serving as control (first top bar, 1 fold), the reporter with ETS sites produced 6.10±1.24 (A375, p<0.001) or 5.12±1.34 (MEL624, p<0.01) fold more light units than control levels. Luciferase activity was reduced to base levels with DN-ETS or drug.
YK-4–279 attenuates melanoma progression in a mouse model of melanoma.
To test YK-4–279 potential in vivo, the melanoma mouse model BrafCA;Tyr-CreERT2;Ptenf/f was utilized since it demonstrates melanoma tumor progression that parallels human disease in terms of kinetics, genetics, and histology (17, 25). To determine if YK-4–279 had any significant effect in this model, three time-points were documented: induction, initiation, and progression. Induction was activation of the model with tamoxifen/4-hydroxy-tamoxifen (4-OHT) and recorded as defined by day 1, initiation was the first sighting of a pigmented lesion (Figure 3A,B), and progression occurred when the lesion progresses to a palpable lesion (Figure 3E,F). Previous reports found initiation of pigmented lesions at approximately two weeks that progressed to nodular melanomas in the following two to four weeks (17). In our experiments, we found a significant sex bias in this melanoma model (18). In control experiments, initiation occurred at 17.9±2.3 days in female mice, and 22.7±3.6 days in males, and progression at 28.3±1.3 days and 46.6±3.5 days for females and males respectively. For this reason, each drug group was paired with sex-matched controls. At day 0, mice were transplanted with osmotic pumps, either with YK-4–279 (releasing drug at 1.6mg/Kg) or DMSO alone. There was no significant difference in initiation of pigmented lesions between experimental or control groups for either females or males (Figure 3C,D), with initiation days in 17.9±2.3 days and 18.3±1.3 days in females, and 22.7 days±3.6 days and 22.1 days±1.9 days in males in control and experimental groups, respectively. Additionally, there was a lack of significance between survival curves (X2= 0.03, p=0.86 females, X2=0.58, p=0.45 log rank tests).
However, significant differences were noted between control and YK-4–279 groups in terms of progression of pigmented lesions to nodular tumors. All control group females progressed before or by day 32, but progression only occurred in two (out of 12) of the drug group (Figure 3G) (X2= 18.8, p<0.0001, log rank tests, 1df). Males followed a similar pattern, with 75% progression in controls but with 0% in the YK-4–279 treated group (Figure 3H) (X2= 12.4, p=0.004, log rank tests, 1df). This model is prone to develop secondary non-pigmented tumors (25) so any surviving mice were collected at day 52 due to secondary tumor burden in control mice. In the mock treated mice, there was evidence of invasion into the subdermal layer even in relatively thin lesions (Figure 3I), with the majority of lesions presented as nodular melanomas with evident pigmented cells throughout the dermis and epidermis (Figure 3J), the YK-4–279 treated mice had pigmented growths that were superficial both grossly and histologically in superficial dermis (Figure 3K,L). Measurements of the thickest section of lesions on day 52 post induction demonstrated a significant difference between groups, 1.37±0.55mm for mock and 0.82±0.32mm for YK-4–279 treated groups (p=0.00098, n=16 samples, Figure 3M). The majority of mock lesions presented as full thickness with tumor cells in the epidermis, dermis, and subdermal layer, while the drug treated lesions lacked any signs of invasion. While there was no difference in initiation of pigmented lesions detected in the transgenic model, there was a significant block of progression in terms of palpable lesions, tumor thickness, and notable invasion into the subdermis.
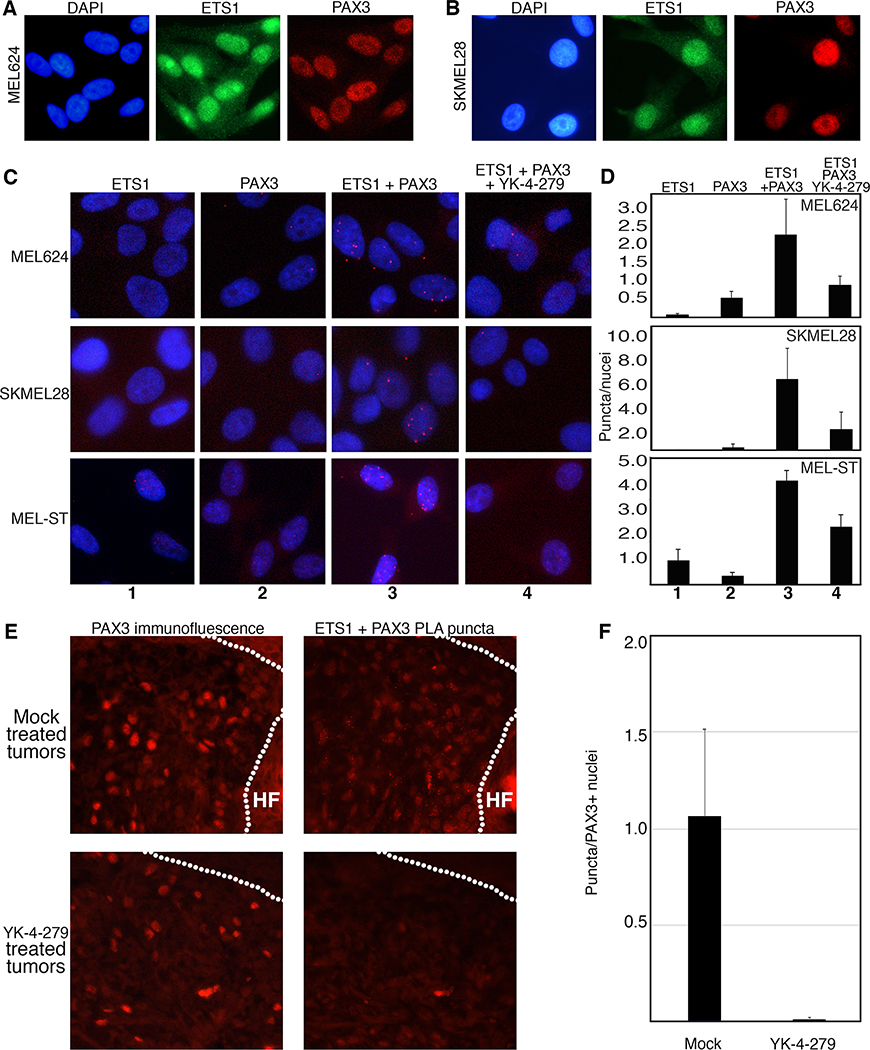
YK-4–279 inhibits ETS1 and PAX3 interaction in melanoma cells.
One possible mechanism of YK-4–279 action on ETS factor function on in vivo and in cellulo growth, migration, and invasion is through interaction with PAX3. Our group and others discovered PAX3 to be overexpressed and actively driving melanoma growth and migration ((26–28) and Figure 1E). Loss of PAX3 expression in melanoma leads to a drastic decrease in cell viability, while conversely increasing PAX3 promotes growth (28). In addition, PAX3 is a known partner of ETS1 in melanoma (2). PAX3 and ETS1 are expressed in melanoma cells and directly interact biochemically (2) and in situ through proximity ligation assays (PLA)(Figure 4A–D). The addition of YK-4–279 disrupted this interaction, reducing the number of puncta/cell in melanoma cells from 2.30±1.00 to 0.90±0.25 (MEL624, p=0.0050) and 6.26±2.82 to 2.26±1.30 (SKMEL28, p=0.018), and in melanocyte cells from 4.33±0.41 to 1.33±0.59 (MEL-ST, p=0.015) (Figure 4D). These findings in cell culture were paralleled in the in vivo melanoma mouse model BrafCA;Tyr-CreERT2;Ptenf/f, where mice treated chronically with YK-4–279 have significantly less interaction between ETS1 and PAX3 as measured by puncta (Figure 4E). The number of puncta per tumor sample was normalized by PAX3 expressing cells, and while the ETS1-PAX3 was abundant in mock treated mice (1.06±0.45 puncta/PAX3 expressing nuclei), the treatment with YK-4–279 significantly decreased the number of puncta (0.010±0.008 puncta/PAX3 expressing nuclei, p=0.0008)(Figure 4F). These findings support that YK-4–279 inhibits the interaction of ETS1 with PAX3.
Figure 4.
YK-4–279 inhibits ETS1 and PAX3 interaction in melanoma cells. A,B. ETS1 and PAX3 are expressed in MEL624 (A) and SKMEL28 (B) cells by immunofluorescence. C,D. ETS1 and PAX3 interact in situ in melanoma cells but this complex is inhibited by YK-4–279. ETS1 and PAX3 antibody alone did not result in significant number of puncta generated through proximity ligase assay (PLA) (columns (C) or bars (D) 1 and 2). Significant PLA puncta were produced with both antibodies (columns 3), 2.3±1.00 (MEL624), 6.26±2.82 (SKMEL28) and 4.33±0.41 (MEL-ST) puncta/nuclei. YK-4–279 significantly reduced the number to 0.9±0.25 (MEL624), 2.26±1.3 (SKMEL28) and 1.33±0.59 (MEL-ST) puncta/nuclei, (columns 4, all p<0.05). E,F. ETS1 and PAX3 interact in mouse melanoma tumors and this complex was inhibited after YK-4–279 treatment. For each sample on sequential sections, tumor cells were identified by PAX3 immunofluorescence, and PAX3-ETS1 interaction was detected by PLA puncta (E). ETS1-PAX3 PLA puncta were reduced in melanoma tissue from 1.06±0.45 puncta/PAX3 expressing nuclei in tumors of mock treated mice to 0.010±0.008 in YK-4–279 mice (error expressed as SEM) with the number of puncta normalized to PAX3 positive expressing nuclei in the sample (p=0.0008 2-tailed t-test, graphed data in F).
ETS1 and PAX3 interaction was not significantly disrupted in melanoma cells with MEK inhibition.
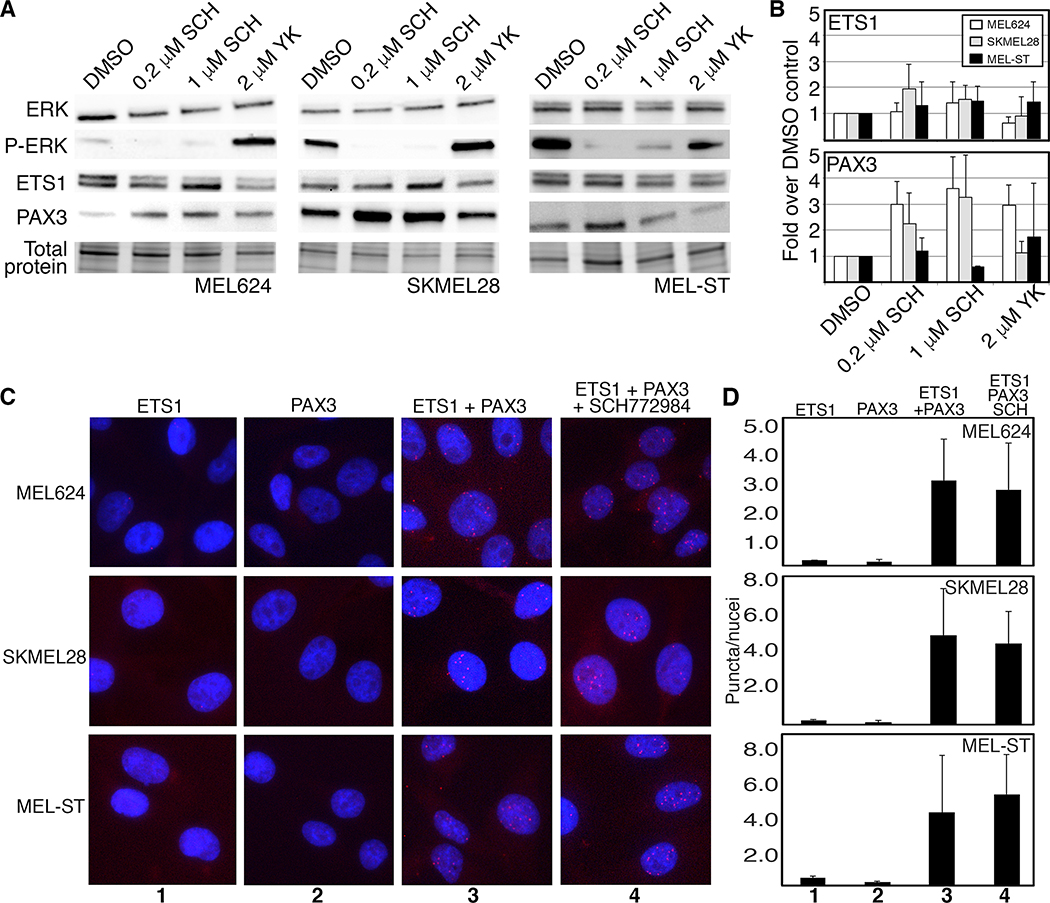
The MAPK pathway is frequently hyperactive in melanoma cells, and is a frequent target for therapeutics. Both ETS1 and PAX3 are downstream targets of this pathway, and PAX3 levels increase in response to pathway inhibitors vemerafenib and selumetinib (7, 29). To test ETS1 and PAX3 response to MAPK pathway inhibition, cells were treated with the ERK1/2 small molecule inhibitor SCH772984. In melanoma cells MEL624 and SKMEL28, and in the melanocyte cell line MEL-ST, there was no difference in ETS1 protein levels with treatment of either YK-4–279 or SCH772984 (Figure 5A,B). PAX3 levels increased significantly with ERK1/2 inhibition in both melanoma cell lines (3.61±1.19 fold over control levels (MEL624), and 3.27±1.61 fold (SKMEL28), both p=0.01 for 1μM SCH772984) in parallel to previous reports (29). MEL-ST cells had no significant changes in PAX3 levels in the drug concentrations tested. Further, there was also an increase in PAX3 levels in MEL624 cells with YK-4–279 treatment (2.96±0.78 fold over control levels, p=0.006). To determine if ERK1/2 inhibition impacted PAX3-ETS1 interactions in situ, PLA was performed on SCH772984 treated and control cells (Figure 5C,D). For the three cell lines tested, there was no significant difference in the number of puncta counted between the PLA DMSO and drug treated groups. Attenuation of the MAPK pathway through SCH772984 ERK1/2 small molecule inhibition did not have a significant impact on PAX3-ETS1 interaction in our experiments.
Figure 5.
ERK inhibition, via SCH772984 treatment, had little to no effect on PAX3 and ETS1 interaction in melanoma cells. A,B. ERK inhibition had minimal effect on ETS1 protein levels, but increased PAX3 in melanoma cells. PAX3 levels rose 3.61±1.19 fold over control levels (MEL624), and 3.27±1.61 fold (SKMEL28), both p=0.01 for 1μM SCH772984. C,D. Treatment with 2μM SCH772984 did not reduce PAX3-ETS1 interactions in melanoma cells. Number of PLA puncta per nuclei between DMSO and SCH772984 treated cells were not significantly different between groups for all cell lines tested.
YK-4–279 inhibits the interaction of PAX3 with ETS family members.
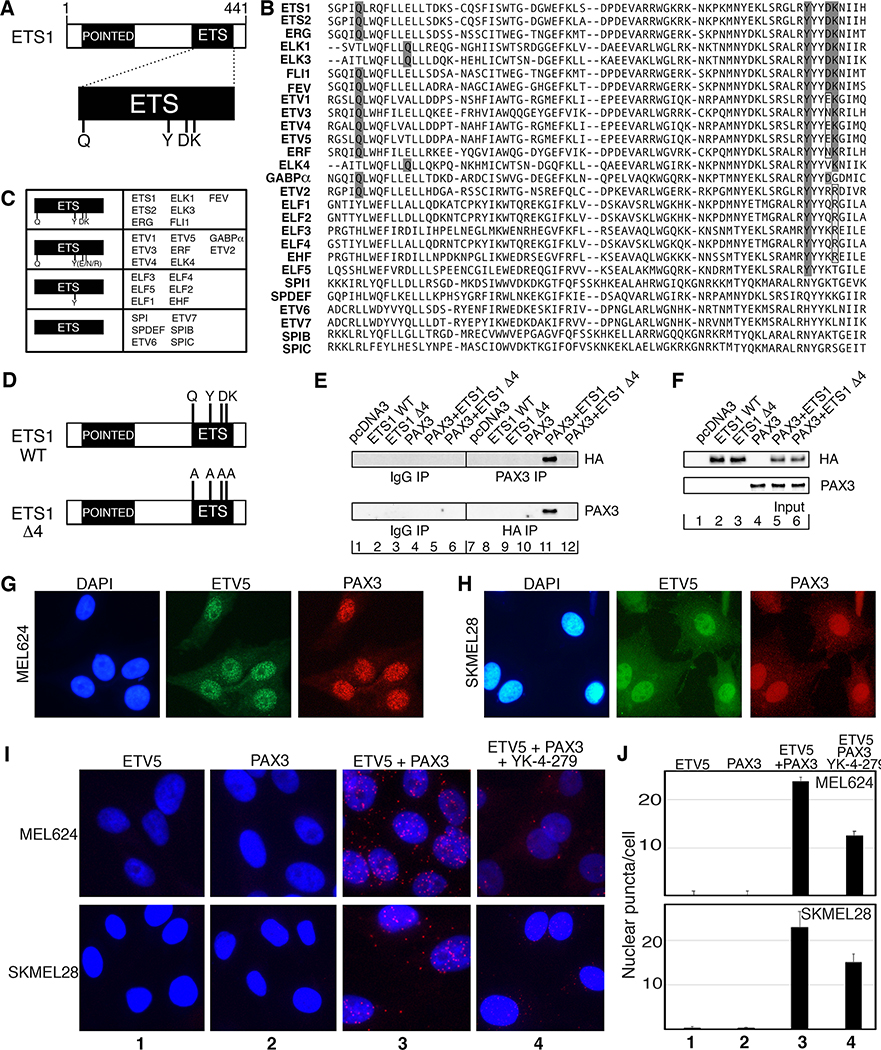
Our data suggest that there are functional redundancies among ETS family members and there are many other ETS factors expressed in the melanoma cells in addition to ETS1 (Figure 1). In addition, it is not known if PAX3 interacts with any other ETS factors besides ETS1. There is support that PAX3 binds to other ETS factors from studies focused on PAX5-ETS1 interactions (2, 30), where it was determined that ETS1 binds to PAX5 though four amino acid epitopes within the ETS DNA binding domain, Q, Y, D, K, which corresponds to amino acids 336, 395, 398, and 399 of ETS1 ((31) and Figure 6A). Fifteen ETS family members have identical or similar epitopes: 5 with identical epitopes (ETS1, ETS2, ERG, FLI1, FEV), 3 with atypical Q epitopes but retaining surrounding leucine/isoleucine (ELK1, ELK3, ELK4), and 8 with different but similar amino acids replacing D and/or K epitopes (Figure 6B,C). It is predicted that these ETS family members should bind to PAX3. The 12 remaining ETS family members only share the Y epitope or none of the epitopes. To determine if ETS1 interacts with PAX3 through the corresponding amino acid epitopes, mutant ETS1 was generated by replacing Q336, Y395, D398 and K399 with alanines (A) (Figure 6D, mutant construct ETS1-Δ4). ETS1-Δ4 lost the ability to interact with PAX3 after both proteins were co-expressed exogenously in 293T cells (Figure 6E,F). This suggests that Q336, Y395, D398 and/or K399 are necessary amino acid epitopes for ETS1-PAX3 interaction. These findings provide evidence that other ETS factors that possess these epitopes have the potential to interact with PAX3.
Figure 6.
PAX3 binds to ETS1 through four epitopes that are similar in some but not all ETS factors including ETV5. A. ETS1 contains four previously identified epitopes within the ETS DNA binding domain (31) that interact with PAX proteins, Q336, Y395, D398 and K399. B. Amino acid sequences of ETS family ETS DNA domains, with PAX-interacting epitopes highlighted. C. Summary chart of sequences from (B), with ETS family members with all four epitopes the same (first group) or similar (second group), with only Y395 (third group), or without any shared epitopes (fourth group). D. Schematic of wild type (ETS1 WT) and mutant ETS1 with four PAX3 interacting epitopes changed to alanines (ETS1 Δ4). E. 293T cells were transfected with PAX3 and/or ETS1-WT or ETS1-Δ4 expression constructs or empty vector, and cell lysates were immunoprecipitated with anti-HA or anti-PAX3 antibodies (lanes 7–12) or control antibodies (lanes 1–6). Immunoprecipitants were probed by western analysis for ETS1 (anti-HA, top) or PAX3 (anti-PAX3, bottom). F. Input control samples for experiments shown in (E). Input proteins were probed for ETS1 (anti-HA, top) or PAX3 (anti-PAX3, bottom). G,H. ETV5 and PAX3 are expressed in MEL624 (G) and SKMEL28 (H) cells by immunofluorescence. I,J. ETV5 and PAX3 interact in melanoma cells but this complex was inhibited with YK-4–279. ETV5 and PAX3 antibody alone do not result in significant number of puncta generation (columns (I) or bars (J) 1 and 2). Significant PLA puncta were produced with both antibodies (columns 3), 23.69±4.71 (MEL624) and 22.93±3.62 (SKMEL28) puncta/nuclei. YK-4–279 significantly reduced the number of puncta (columns 4), 12.5±2.78 (MEL624, p=0.0031) and 15.01±1.93 (SKMEL28, p=0.050) puncta/nuclei.
To determine if PAX3 binds to other ETS family members, and if so, whether YK-4–279 disrupts this interaction, the in cellulo experiments from Figure 4 were repeated with another candidate ETS factor. For these experiments, ETV5 was selected since 1) it has similar but not identical PAX3-interacting epitopes to ETS1 (in group 2, Figure 6C), 2) ETV5 and other PEA3-ETS factors were identified to be expressed and active in melanoma (32, 33), and 3) ETV5 was the most common ETS family member in the UCSD melanoma dataset (Figure 1E). We discovered that ETV5, like ETS1, was expressed in melanoma cells (Figure 1E–G, Figure 6G,H) and binds to PAX3 (Figure 6I column 3). The addition of YK-4–279 reduced the number of puncta per cell from 23.69±4.71 to 12.5±2.78 (MEL624, p=0.0031) and 22.93±3.62 to 15.01±1.93 (SKMEL28, p=0.0050)(Figure 6I column 4, Figure 6J). Accordingly, we found that multiple ETS factors, or at least ETS1 and ETV5, bind to PAX3 and that YK-4–279 inhibits this interaction.
YK-4–279 reduces MET expression in cellulo and in vivo.
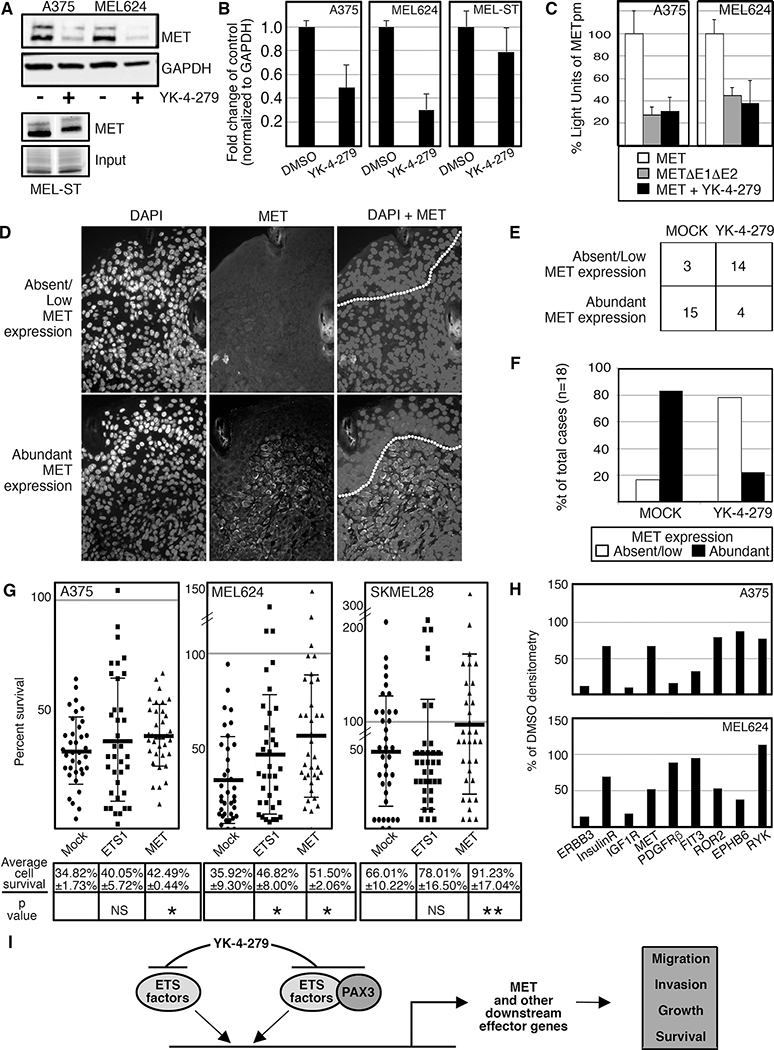
YK-4–279 inhibited ETS factor function (Figure 2E), ETS1-PAX3 interaction (Figure 4), and demonstrated a reduction in melanoma growth in cellulo and in vivo (Figures 2,3). The receptor tyrosine kinase MET is a downstream target of ETS1 and PAX3 (Figure 1A and (2)) with active roles in melanoma cell proliferation and survival (1). In melanoma cells, MET protein levels decreased significantly after treatment with YK-4–279 (0.49±0.19 fold p=0.044 (A375), and 0.30±0.13 fold p=0.011 (MEL624), Figure 7A,B). In the melanocyte line MEL-ST there was a downward trend in MET expression but not significantly (0.79±0.20 fold p=0.13). YK-4–279 also significantly decreased luciferase expression driven by a MET promoter containing two ETS binding sites (30.40±13.2% p=0012 (A375), 37.80±22.5% p=0.0043 (MEL624), Figure 7C). This decrease in luciferase expression was similar to the outcome of mutating both ETS binding sites in the promoter. To determine if there was a difference in MET expression between mock and drug treated groups in mouse tumors, MET expression in pigmented lesions was detected by immunofluorescence and scored as “Absent/Low MET expression” (0–10% MET expressing cells in lesion) or “Abundant MET expression” (>10% cells expressing MET)(Figure 7D) and scored with the group anonymized. With 18 specimens per group, most mock treated lesions expressed abundant MET (15/18 samples, 83.3%) while YK-4–279 lesions were predominantly MET low or absent with only 4/18 with abundant MET expression (22.2%, Figure 7E,F). The correlation of MET expression between drug versus mock treated groups was determined to be significant (p=0.00018, >85% power) with 2-tailed Fisher exact tests, with nominal data (abundant versus low/absent) analyzed with X2 analysis (CI=95%). Melanoma cells treated with YK-4–279 express significantly less MET receptor levels both in culture and in vivo.
Figure 7.
YK-4–279 reduces MET expression in cellulo and in vivo. A,B. YK-4–279 reduces MET expression in melanoma cells by western analysis (A) with densitometry readings of three independent western blots (B). There was significant MET reduction with YK-4–279 treatment, 0.49±0.17 fold (A375, p=0.044) and 0.30±0.13 fold (MEL624, p=0.011). C. Both enhancer mutation and YK-4–279 reduce MET promoter activity in melanoma cells. A MET reporter vector containing two ETS binding sites (shown schematically in Figure 1A) expression was reduced if the ETS sites are mutated (27.62 ±8.82%, A375, 44.76±6.13% MEL624) or in the presence of YK-4–279 (30.40±13.20, A375, 37.80±22.5%, MEL624). All reductions of luciferase activity were significant, p<0.005. D. MET expression in BrafCA;Tyr-CreERT2;Ptenf/f mouse tumor lesions. Immunofluorescence of nuclei (DAPI, first column), MET (middle column), and combined DAPI/MET (last column, dotted line indicating epithelial/dermal junction). Top row is an example of a lesion with low or absent MET expression, bottom row a lesion with abundant (>10% cells positive) MET. E. Summary of anonymized scoring of MET expression in BrafCA;Tyr-CreERT2;Ptenf/f mouse tumors. Tumors from DMSO (MOCK, first column) or drug (YK-4–279, second column) treated mice are scored for absent/low expression of MET (top row) or abundant (>10%, bottom row), n=18 for each group. F. Graph of data from (E). Mock treated lesions expressed abundant MET (15/18 samples, 83.3%) while YK-4–279 lesions were predominantly MET low or absent with only 4/18 with abundant MET expression (22.2%). The correlation of MET expression between drug versus mock treated groups was significant (p=0.00018, >85% power). G. Transfection of ETS1 and MET partially rescues YK-4–279 cell loss (ETS1 in MEL624, MET in all three lines) in melanoma cell lines. Percent survival after 48h of YK-4–279 treatment for each group (mock, ETS1, MET, x-axis) is shown from representative experiments from cell lines A375, MEL624, and SKMEL28, with the average of three independent experiments shown below each column with significant differences between mock and transfected groups determined by ANOVA (p values as shown, NS=not significant, *=p<0.05, **=p<0.005). Differences in cell numbers between YK-4–279 and DMSO cells are compared within transfected groups (mock, ETS1, MET) to determine percent survival (y-axis). The grey line indicates 100% of DMSO treated cell numbers, or levels for a complete rescue from YK-4–279 treatment. For each experiment, at least 400 cells were counted/group. H. Densitometry from Phospho-RTK (receptor tyrosine kinase) array from mock and 2μM YK-4–279 treated A375 and MEL624 melanoma cells. Representative arrays are shown in Supplementary Figure S3A–C. RTKs with differential densitometry between groups for at least one cell line is graphed where the genes are listed on the X-axis and percent of DMSO controls is indicated on the Y-axis, with DMSO levels set at 100%. I. Simplified schematic of the molecular pathways affected by YK-4–279 treatment on melanoma cells. The drug inhibits ETS1 and other ETS factors and blocks ETS domain function and binding to protein partners such as PAX3. This leads to an attenuation of ETS-dependent activation of downstream effector genes such as MET, and a decrease in pro-tumor function including migration and invasion.
To determine if YK-4–279 loss of cells is dependent (fully or partially) or independent of ETS1 or MET, three melanoma cell lines (A375, MEL624, SKMEL28) were transfected with empty vector (mock) or constructs expressing either ETS1 or MET (Figure 7G). Each transfection set was treated with either YK-4–279 or DMSO, and cell numbers after 48 hours in the DMSO group was set at 100%. It was discovered that in two lines (A375, SKMEL28), ETS1 transfection did not significantly rescue YK-4–279 cell loss. However, ETS1 did provide partial protection for MEL624 cells (p=0.023). Addition of MET resulted in a significant partial rescue in all three cell lines, with a near total rescue in SKMEL28 cells. These findings support that at least part of the growth inhibitory effect of YK-4–279 treatment is due to a loss of MET expression. However, this also suggests that there are, most likely, other factors involved with YK-4–279 cell loss. Due to what is known about YK-4–279, it is likely that this compound has pleiotropic effects. Since receptor tyrosine kinases (RTKs), including MET, are major factors involved with cellular signaling, a non-biased screen using a phospho-RTK assay array was utilized to measure overall changes in RTK phosphorylation. After YK-4–279 treatment, the overall trend was that there was no change in phosphorylation for the majority of the represented RTKs except for nine that decreased (≤75% of control levels). In the nine factors (listed in Figure 7H, with array locations shown in Supplementary Figure S3A,B), five decreased in both cell lines (ERBB3, Insulin receptor, IGF1R, MET, and ROR2), three in A375 (PDGF, FIT3, RYK), and one in MEL624 (EPHB6). Of note, MET was reduced to approximately half the levels of the controls with YK-4–279 treatment in both cell lines, in a similar pattern to the reduction to protein levels (Figure 7A,B). YK-4–279 was found to reduce the phosphorylation of a subset of RTK factors within a representative phosphorylation array, including MET.
We discovered a molecular pathway of ETS family members functioning as transcription factors, synergistically activating genes with PAX3, and actively promoting melanoma growth and invasion through the regulation of downstream effector genes such as MET (Figure 7I). The ETS-inhibitor drug YK-4–279 inhibits ETS factor function, interaction with PAX3, and melanoma tumor progression. This work supports YK-4–279 as a potential therapeutic option in melanoma.
Discussion
In our presented work, we find multiple ETS family members expressed in melanoma (Figure 2). ETS factors are often deregulated in cancers, through overexpression, chromosomal mutations, translocations, and post-transcriptional modification. In melanoma, there are prior reports of ETS family overexpression, including ETS1, ETV1, ETV4, ETV5, ETV6, and GABPA (3, 4, 19, 21, 34–36). In addition, ETS genes are amplified due to large chromosomal rearrangements, including ETV1 gene amplification in melanomas and ETV6 translocation with the NTRK3 gene in Spitz Nevus (19, 20, 37). ETS factors are regulated by phosphorylation, sumoylation, and ubiquination. Some, but not all, of the ETS factors (21 members) are phosphorylated by ERK2, and this pathway is commonly overactive in melanoma (38, 39). Some of the ETS factors are also targets for COP1, part of an E3 ubiquitin complex, and this factor may be deleted in melanoma (40). In addition to the loss of COP1, the deubiquinase Usp9x also protects ETS factors from proteasomal degradation in melanoma (5). ETS1 and other ETS factors contain the Usp9x recognition site (MNYEK*LSR, located at the C terminal end of the ETS domain, (Figure 6)). Our data support some redundancy of function of the ETS factors (Figure 1,3) such as interaction with PAX3 (Figure 6). Our data and others support that there are regulatory pathways and functions that are shared and divergent among ETS family members.
Due to the function of ETS factors, we hypothesized that YK-4–279 has the potential to bind and inhibit the ETS domain and have a significant effect against melanoma. YK-4–279 treatment of the BrafCA;Tyr-CreERT2;Ptenf/f mouse model led to a block of tumor progression (Figure 3). This may be due to an inhibition of growth, increased cell death, decreased migratory ability, and/or a block in invasion, and this is supported by the functional data in Figure 3. ETS factors have been implicated in tumor growth (19). However, in our model system there was not a complete block of growth, since tumor initiation was not affected (Figure 4A–D) and there was an evident proliferation of pigmented cells histologically (Figure 4J–L). ETS factors are linked to migration in normal neurogenesis (41, 42). Additionally, in epithelial tumors, ETS/AP1 binding sites were located in RAS-inducible genes that are linked to cell migration (38). ETS factors also activate genes that correlate with metastatic spread, or are demonstrated to actively drive metastasis. This includes CEACAM1 and cathepsin (43, 44), MMPs (21), and MET (2). Therefore, the effectiveness of YK-4–279 in inhibiting melanoma progression in our studies is most likely due to a block of ETS-dependent promotion of pro-tumor downstream effector genes.
We discovered that YK-4–279 has anti-cancer effects in melanoma cell lines and mouse models (Figure 2,3). Other studies support these findings, as YK-4–279 appears capable of targeting a wide variety of cancers (16). Our data indicate that YK-4–279 suppresses melanoma invasion in cellulo and in vivo (Figure 2C,D, Figure 3I–M). These findings are in parallel with a recent prostate cancer mouse xenograft model study, where YK-4–279 treatment demonstrated decreased expression of ETV1 metastasis-related target genes MMP7, GLYATL2 and FKBP10 (13). In these studies, it is not clear if the impact of YK-4–279 was purely dependent on ETV1 or if the drug also blocked other factors independent of ETV1. YK-4–279 inhibited growth in cell culture (Figure 2A), and reduced melanocytic proliferation in mouse models (Figure 3). Further, YK-4–279 decreased MET levels as well as the phosphorylation of a number of RTKs including MET (Figure 7). In our studies, we revealed that MET is likely a significant part of the YK-4–279 anti-tumor effect, since reintroduction of exogenous MET at least partially protected melanoma cells from the negative impact of the drug on cell numbers (Figure 7G). In Ewing sarcoma, YK-4–279 induces G2-M cell cycle arrest and increases the level of CyclinB1 by reducing EWS-FLI1-mediated expression of target gene ubiquitin ligase UBE2C that promotes the degradation of CyclinB1 (45). YK-4–279 promotes cell death by inhibiting the alternative splicing function of EWS-FLI1, which increases the proapoptotic isoforms of MCL1 and BCL2 (45). In neuroblastoma cells, without affecting CyclinB1 levels, YK-4–279 induces mitotic arrest at G2/M phase by disrupting spindle assembly, and increases caspase3-dependent apoptosis (16). We do not exclude such mechanisms that may also be involved in YK-4–279 melanoma inhibition.
In our studies, we used a transgenic rather than a xenograft model, as the BrafCA;Tyr-CreERT2;Ptenf/f mouse more closely mimics melanoma found in patients in terms of progression, intact microenvironments and immune systems, and is a better predictor of translational therapeutic outcomes (46). Additionally, we utilized osmotic pumps, which allowed 1) a constant dose of the drug that circumvented problems with the quick clearance of the drug, 2) a reduction in stress on the mice by avoiding daily handling and injections, and 3) an ability to use a relatively low dose of drug compared to other studies. Of note, pharmacokinetic studies find that YK-4–279 has a short half-life in blood plasma and continuous infusion of the compound was required for tumor regression in the rat model of Ewing sarcoma (47). This short plasma half-life is remedied with the use of osmotic pumps, which have been shown as a promising systems for controlled drug delivery (48). The big advantage of using osmotic pumps is that the drug release is independent of the pH and hydrodynamics of the dissolution medium, which assures a steady drug concentration and activity level into the blood plasma (48). Another advantage is that the osmotic pump is able to maintain drug concentration for 28 days. Therefore, the treatment process only requires two implantations. The drug dose in our study (1.6 mg/kg) is far less than the levels that have been used by injection method (75–150mg/kg) (11, 13) and still shows promise in inhibiting tumor progression, suggesting that a higher therapeutic potential of YK-4–279 may be achieved with increased dose in future investigation using osmotic pumps as drug delivery system.
BRAF-V600E mutant tumors targeted with small molecule inhibitors such as Vemerafenib or Debrafenib, as a single agent or in combination with MEK inhibitors, demonstrated good response rates but low durability to treatment (10). In our findings and as reported by others, PAX3 and ETS1 levels and/or activity were altered in response to MAPK pathway inhibition (Figure 5 and (7, 29)). The use of YK-4–279 has promise as a combination therapy, where mutant BRAF inhibitor drugs attenuate BRAF hyperactivation and YK-4–279 counteracts the upregulation and function of PAX3. In summary, our work, utilizing a mouse model mimicking human disease using physiologically clinically relevant levels of drug, supports that YK-4–279 has potential as a melanoma therapeutic through the targeting of ETS protein family function as an activator of pro-tumorigenic genes.
Supplementary Material
Significance.
These findings identify YK-4–279 as a promising therapeutic agent against melanoma by targeting multiple ETS family members and blocking their ability to act as transcription factors.
Acknowledgements
We thank Jennifer D. Kubic and Rhoda Alani for scientific input and discussion. We thank Margaret Gardel and Tong-Chuan He (University of Chicago), Neil Ganem and Andrey Sharov (Boston University), and Robert Weinberg (Whitehead Institute) for cell lines and reagents. This work was supported by grants from the National Institutes of Health, grant numbers R01CA184001, R01AR062547, T32GM007183, and 1UL1TR001430, the American Cancer Society, grant number RSG-CSM-121505, the Wendy Will Case Foundation, and the Friends of Dermatology-Chicago, the American Skin Association Daneen and Charles Stiefel Investigative Scientist Award, the Harry J. Lloyd Charitable Trust, the Falanga Scholar endowment (Boston University), Boston University Dermatology Scholars award, and the Department of Dermatology at Boston University.
Footnotes
Conflict of interest statement: The authors have declared that no conflict of interest exists.
References
- 1.Czyz M HGF/c-MET Signaling in Melanocytes and Melanoma. Int J Mol Sci. 2018;19(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubic JD, Little EC, Lui JW, Iizuka T, and Lang D. PAX3 and ETS1 synergistically activate MET expression in melanoma cells. Oncogene. 2015;34(38):4964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keehn CA, Smoller BR, and Morgan MB. Expression of the ets-1 proto-oncogene in melanocytic lesions. Mod Pathol. 2003;16(8):772–7. [DOI] [PubMed] [Google Scholar]
- 4.Rothhammer T, Hahne JC, Florin A, Poser I, Soncin F, Wernert N, et al. The Ets-1 transcription factor is involved in the development and invasion of malignant melanoma. Cell Mol Life Sci. 2004;61(1):118–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potu H, Peterson LF, Kandarpa M, Pal A, Sun H, Durham A, et al. Usp9x regulates Ets-1 ubiquitination and stability to control NRAS expression and tumorigenicity in melanoma. Nature communications. 2017;8:14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong L, Jiang CC, Thorne RF, Croft A, Yang F, Liu H, et al. Ets-1 mediates upregulation of Mcl-1 downstream of XBP-1 in human melanoma cells upon ER stress. Oncogene. 2011;30(34):3716–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foulds CE, Nelson ML, Blaszczak AG, and Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24(24):10954–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61. [DOI] [PubMed] [Google Scholar]
- 9.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, and Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luther C, Swami U, Zhang J, Milhem M, and Zakharia Y. Advanced stage melanoma therapies: Detailing the present and exploring the future. Crit Rev Oncol Hematol. 2019;133:99–111. [DOI] [PubMed] [Google Scholar]
- 11.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat Med. 2009;15(7):750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahim S, Beauchamp EM, Kong Y, Brown ML, Toretsky JA, and Uren A. YK-4–279 inhibits ERG and ETV1 mediated prostate cancer cell invasion. PLoS ONE. 2011;6(4):e19343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahim S, Minas T, Hong SH, Justvig S, Celik H, Kont YS, et al. A small molecule inhibitor of ETV1, YK-4–279, prevents prostate cancer growth and metastasis in a mouse xenograft model. PLoS One. 2014;9(12):e114260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selvanathan SP, Graham GT, Erkizan HV, Dirksen U, Natarajan TG, Dakic A, et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc Natl Acad Sci U S A. 2015;112(11):E1307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37(10):1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kollareddy M, Sherrard A, Park JH, Szemes M, Gallacher K, Melegh Z, et al. The small molecule inhibitor YK-4–279 disrupts mitotic progression of neuroblastoma cells, overcomes drug resistance and synergizes with inhibitors of mitosis. Cancer Lett. 2017;403:74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr., et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai Y, Haresi AJ, Huang L, and Lang D. Differences in tumor initiation and progression of melanoma in the Braf(CA); Tyr-CreERT2;Pten(f/f) model between male and female mice. Pigment Cell Melanoma Res. 2020;33(1):119–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jane-Valbuena J, Widlund HR, Perner S, Johnson LA, Dibner AC, Lin WM, et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70(5):2075–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehra R, Dhanasekaran SM, Palanisamy N, Vats P, Cao X, Kim JH, et al. Comprehensive Analysis of ETS Family Members in Melanoma by Fluorescence In Situ Hybridization Reveals Recurrent ETV1 Amplification. Translational oncology. 2013;6(4):405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hata H, Kitamura T, Higashino F, Hida K, Yoshida K, Ohiro Y, et al. Expression of E1AF, an ets-oncogene transcription factor, highly correlates with malignant phenotype of malignant melanoma through up-regulation of the membrane-type-1 matrix metalloproteinase gene. Oncology reports. 2008;19(5):1093–8. [PubMed] [Google Scholar]
- 22.Barber-Rotenberg JS, Selvanathan SP, Kong Y, Erkizan HV, Snyder TM, Hong SP, et al. Single enantiomer of YK-4–279 demonstrates specificity in targeting the oncogene EWS-FLI1. Oncotarget. 2012;3(2):172–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDougall JR, Bani MR, Lin Y, Rak J, and Kerbel RS. The 92-kDa gelatinase B is expressed by advanced stage melanoma cells: suppression by somatic cell hybridization with early stage melanoma cells. Cancer Res. 1995;55(18):4174–81. [PubMed] [Google Scholar]
- 24.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. Embo J. 2010;29(13):2147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooijkaas AI, Gadiot J, van der Valk M, Mooi WJ, and Blank CU. Targeting BRAFV600E in an inducible murine model of melanoma. Am J Pathol. 2012;181(3):785–94. [DOI] [PubMed] [Google Scholar]
- 26.Scholl FA, Kamarashev J, Murmann OV, Geertsen R, Dummer R, and Schafer BW. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001;61(3):823–6. [PubMed] [Google Scholar]
- 27.Vachtenheim J, and Novotna H. Expression of genes for microphthalmia isoforms, Pax3 and MSG1, in human melanomas. Cell Mol Biol (Noisy-le-grand). 1999;45(7):1075–82. [PubMed] [Google Scholar]
- 28.Kubic JD, Mascarenhas JB, Iizuka T, Wolfgeher D, and Lang D. GSK-3 promotes cell survival, growth, and PAX3 levels in human melanoma cells. Mol Cancer Res. 2012;10(8):1065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith MP, Brunton H, Rowling EJ, Ferguson J, Arozarena I, Miskolczi Z, et al. Inhibiting Drivers of Non-mutational Drug Tolerance Is a Salvage Strategy for Targeted Melanoma Therapy. Cancer Cell. 2016;29(3):270–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitzsimmons D, Hodsdon W, Wheat W, Maira S, Wasylyk B, and Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary comlexes on a B-cell-specific promoter. Genes & Develop. 1996;10:2198–211. [DOI] [PubMed] [Google Scholar]
- 31.Garvie CW, Hagman J, and Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell. 2001;8(6):1267–76. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y, Zheng C, Zhang J, Lu D, Zhuang J, Xing S, et al. Recognition of CD146 as an ERM-binding protein offers novel mechanisms for melanoma cell migration. Oncogene. 2012;31(3):306–21. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y, Cao Z, Wong EW, Guan Y, Ma W, Zhang JQ, et al. COP1/DET1/ETS axis regulates ERK transcriptome and sensitivity to MAPK inhibitors. J Clin Invest. 2018;128(4):1442–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dissanayake K, Toth R, Blakey J, Olsson O, Campbell DG, Prescott AR, et al. ERK/p90(RSK)/14–3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicua. Biochem J. 2011;433(3):515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schick N, Oakeley EJ, Hynes NE, and Badache A. TEL/ETV6 is a signal transducer and activator of transcription 3 (Stat3)-induced repressor of Stat3 activity. J Biol Chem. 2004;279(37):38787–96. [DOI] [PubMed] [Google Scholar]
- 36.Zhang T, Xu M, Makowski MM, Lee C, Kovacs M, Fang J, et al. SDHD Promoter Mutations Ablate GABP Transcription Factor Binding in Melanoma. Cancer Res. 2017;77(7):1649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh I, Tee MK, Botton T, Shain AH, Sparatta AJ, Gagnon A, et al. NTRK3 kinase fusions in Spitz tumours. J Pathol. 2016;240(3):282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plotnik JP, Budka JA, Ferris MW, and Hollenhorst PC. ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res. 2014;42(19):11928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selvaraj N, Kedage V, and Hollenhorst PC. Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun Signal. 2015;13:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu G, Zhang Q, Huang Y, Song J, Tomaino R, Ehrenberger T, et al. Phosphorylation of ETS1 by Src family kinases prevents its recognition by the COP1 tumor suppressor. Cancer Cell. 2014;26(2):222–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arber S, Ladle DR, Lin JH, Frank E, and Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101(5):485–98. [DOI] [PubMed] [Google Scholar]
- 42.Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, et al. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35(5):877–92. [DOI] [PubMed] [Google Scholar]
- 43.Kfir-Elirachman K, Ortenberg R, Vizel B, Besser MJ, Barshack I, Schachter J, et al. Regulation of CEACAM1 Protein Expression by the Transcription Factor ETS-1 in BRAF-Mutant Human Metastatic Melanoma Cells. Neoplasia. 2018;20(4):401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tripathi R, Fiore LS, Richards DL, Yang Y, Liu J, Wang C, et al. Abl and Arg mediate cysteine cathepsin secretion to facilitate melanoma invasion and metastasis. Sci Signal. 2018;11(518). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zöllner SK, Selvanathan SP, Graham GT, Commins RMT, Hong SH, Moseley E, et al. Inhibition of the oncogenic fusion protein EWS-FLI1 causes G2-M cell cycle arrest and enhanced vincristine sensitivity in Ewing’s sarcoma. Sci Signal. 2017;10(499). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merlino G, Flaherty K, Acquavella N, Day CP, Aplin A, Holmen S, et al. Meeting report: The future of preclinical mouse models in melanoma treatment is now. Pigment Cell Melanoma Res. 2013;26(4):E8–E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamhamedi-Cherradi SE, Menegaz BA, Ramamoorthy V, Aiyer RA, Maywald RL, Buford AS, et al. An Oral Formulation of YK-4–279: Preclinical Efficacy and Acquired Resistance Patterns in Ewing Sarcoma. Mol Cancer Ther. 2015;14(7):1591–604. [DOI] [PubMed] [Google Scholar]
- 48.Keraliya RA, Patel C, Patel P, Keraliya V, Soni TG, Patel RC, et al. Osmotic drug delivery system as a part of modified release dosage form. ISRN Pharm. 2012;2012:528079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.