Abstract
GRP78 (Glucose-regulated protein, 78 kDa) is a key regulator of ER (endoplasmic reticulum) stress signaling. Cancer cells are highly proliferative and have high demand for protein synthesis and folding, which results in significant stress on the ER. To respond to ER stress and maintain cellular homeostasis, cells activate the unfolded protein response (UPR) that promotes either survival or apoptotic death. Cancer cells utilize the UPR to promote survival and growth. In this study, we describe the discovery of a series of novel hydroxyquinoline GRP78 inhibitors. A representative analog, YUM70, inhibited pancreatic cancer cell growth in vitro and showed in vivo efficacy in a pancreatic cancer xenograft model with no toxicity to normal tissues. YUM70 directly bound GRP78 and inactivated its function, resulting in ER stress-mediated apoptosis. A YUM70 analog conjugated with BODIPY show co-localization of the compound with GRP78 in the ER. Moreover, a YUM70-PROTAC (PROteolysis TArgeting Chimera) was synthesized to force degradation of GRP78 in pancreatic cancer cells. YUM70 showed a strong synergistic cytotoxicity with topotecan and vorinostat. Together, our study demonstrates that YUM70 is a novel inducer of ER stress with preclinical efficacy as a monotherapy or in combination with topoisomerase and HDAC inhibitors in pancreatic cancer.
Keywords: GRP78, ER stress, apoptosis, pancreatic cancer, PROTAC
INTRODUCTION
Pancreatic cancer remains one of the deadliest human diseases and options for effective systemic therapy are limited (1,2). Thus, there is an urgent need to develop new and more impactful therapies for this disease. Currently, gemcitabine-based regimens are considered the standard of care treatment for pancreatic cancer patients. Two front line regimens, gemcitabine/nab-paclitaxel, and folfirinox have shown a survival benefit but at the expense of significant side effects (3). Moreover, lack of response and development of resistance to treatment limit the use of the front line regimens. Thus, novel treatment options are needed to overcome drug resistance when used as a single agent or in combination with standard of care.
The endoplasmic reticulum (ER) is a multifunctional cellular organelle responsible for the proper folding of newly synthesized proteins, degradation of misfolded proteins, and maintenance of cellular homeostasis. Cancer cells are subject to intrinsic stress as they are highly proliferative and have a higher demand for protein synthesis and folding. Additionally, cancer cells are subject to extrinsic stress in the cancer microenvironment including hypoxia, low pH, and nutrient deprivation (4). Such conditions contribute to ER stress and impaired ER functions. As a result, cells activate the unfolded protein response (UPR) to mitigate the consequences of ER stress and to maintain cellular homeostasis. The UPR has dual functions; it can either mitigate the deleterious effect of ER stress or activate apoptosis (5,6). Cancer cells are known to direct the UPR to promote survival and growth. Thus, redirecting the UPR response to apoptosis in cancer cells is a promising approach for cancer therapy.
Glucose-regulated protein, 78 kDa (GRP78), also referred to as HSPA5/BiP, is a key molecular chaperone in the ER and also a master regulator of ER stress signaling (7). Under normal conditions, GRP78 associates with ER transmembrane receptors, protein kinase RNA-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme-1 (IRE1), and activating transcription factor 6 (ATF6) and maintains these sensors in an inactive state (8). Under stress, unfolded proteins accumulate in the ER resulting in GRP78 dissociation from the transmembrane receptors and causing activation of PERK, IRE1, and ATF6 (9). Activated PERK leads to phosphorylation of the α-subunit of eukaryotic initiation factor 2 (eIF2α) that in turn shuts off global mRNA translation reducing the protein load on the ER. This event protects cells from ER stress-related damage. However, prolonged ER stress leads to the activation of transcription factor 4 (ATF4) by the phosphorylated eIF2α resulting in the subsequent transcriptional upregulation of C/EBP homologous protein (CHOP) and growth arrest and DNA damage-inducible 34 (GADD34) (8). CHOP translocates to the nucleus and facilitates programmed cell death by upregulating its proapoptotic target genes. Activated IRE1 promotes the splicing of a retained intron from the mRNA encoding the transcription factor X box-binding protein 1 (XBP1) in the cytoplasm (10). The generated splicing variants, XBP1s, move to the nucleus and induce the transcription of genes coding for ER chaperones which protect the cells from the deleterious effects of ER stress (11). In response to ER stress, ATF6 dissociates from the ER membrane and moves to the Golgi apparatus, where its cytoplasmic domain undergoes proteolytic cleavage to form an active transcription factor. This active version of ATF6 translocates to the nucleus and promotes the transcription of several UPR genes encoding GRP78, GRP94, protein disulfide isomerase (PDI), and XBP1 allowing the cells to re-establish initial homeostasis (12). Thus, GRP78 regulates UPR by activating above mentioned ER transmembrane sensors and play important roles in regulating various cellular process required for tumorigenesis. Several murine cancer models confirm GRP78 requirement for tumorigenesis (13). Moreover, GRP78 interacts with and suppresses the activation of caspase-7 to prevent apoptosis (14), promoting cytoprotection and modulating chemosensitivity (15). Conversely, inhibition of GRP78 triggers UPR and causes caspase-4 mediated apoptosis (16). In cancer cells, ER stress inducers (such as thapsigargin and tunicamycin) cause UPR-mediated apoptosis (17). In mutant KRAS-driven pancreatic cancer in mice, GRP78 haploinsufficiency suppresses acinar-to-ductal metaplasia and oncogenic signaling (18). Thus, inhibition of GRP78 is an effective approach to disrupt ER homeostasis and suppress its anti-apoptotic properties. Furthermore, GRP78 induction in tumor, stromal, and dormant cancer cells, as an adaptive response to ER stress, promotes therapeutic resistance in cancer (19); therefore, inhibition of GRP78 overcomes resistance to multiple anti-cancer treatments (13). Moreover, increased GRP78 expression levels in patient tumor tissues correlate with poor survival in several cancers (20,21). In conclusion, GRP78 inhibitors could be efficacious in suppressing tumor growth, including pancreatic cancer, and overcome resistance.
In the present study, we designed and synthesized a series of novel hydroxyquinolines targeting GRP78. The analog, YUM70, showed significant efficacy in a pancreatic cancer xenograft model with no detectable toxicity to normal tissues. YUM70 treatment upregulates ER stress-related genes, induces apoptosis, and demonstrates synergy with the FDA approved drugs topotecan and vorinostat in killing pancreatic cancer cells.
MATERIALS and METHODS
Reagents and Drugs
The following reagents and drugs were used: DMSO (Sigma, D2438), DFO (Sigma, D9533), Tosedostat (Cayman, 23395), Topotecan (Selleckchem, S1231), Vorinostat (LC Laboratories, V-8477), MG132 (Cayman, 10012628), Trypsin (Promega, V5280), VER (Tocris, 3803/10), Actinomycin D (Cayman, 11421), Cycloheximide (Sigma, C6255), CellTracker ™ Green CMFDA dye (Thermo Fisher Scientific, C2925), Tunicamycin (Cayman Chemicals, 11445), ER-Tracker™ Red (Invitrogen, E34250), Gemcitabine (Tocris, 3259), 5-Fluorouracil (LKT Laboratories, F4480), Paclitaxel (LKT Laboratories, P0092).
Cell lines and culture conditions
Pancreatic cancer cells (MIA PaCa-2, PANC-1, and BxPC-3) and colon cancer cells (HCT116 p53+/+, HT29) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Human Pancreatic Nestin-expressing (HPNE) and UM59 (22) cells were kindly provided by Dr. Diane Simeone (University of Michigan, Ann Arbor, MI, USA). Other cell lines, HCT116 p53−/− and H1299 were obtained from John Hopkins, A549, MCF-7, OVCAR-8, OVCAR-3, and Skov-3 were obtained from NCI, SHEP-1 and SHSY-5Y were obtained from Erika Newman, U of M, WM115 was obtained from Fallahi-Sichani Lab, U of M. Cell lines were maintained in the appropriate growth media containing 10% heat-inactivated FBS (Gibco) at 37 °C in a humidified atmosphere of 5% CO2. All cell lines used were maintained in culture under 35 (10 for HPNE) passages and tested regularly for mycoplasma contamination using Plasmo Test™ kit (InvivoGen, rep-pt1). Individual cell line authentication was regularly performed twice a year using short tandem repeat (STR) analysis at the institutional core facility.
Western blot
MIA PaCa-2, PANC-1, and BxPC-3 (4×105/well) cells were cultured in 6-well tissue culture plates. Cells were treated with compounds at desired concentrations for specified times. After treatment, cells were lysed with M-PER (Thermo Fisher Scientific, 78503) at 4°C for 20 min and centrifuged [12000 rpm (Jouan, rotor radius 84 mm), 10 min, 4 °C]. Protein concentrations of supernatants were measured with the BCA protein detection kit (Thermo Fisher Scientific, Rockford, IL). Protein samples (30 μg) were subjected to SDS-PAGE analysis. 4–20% polyacrylamide gels with Tris/glycine/SDS running buffer (Bio-Rad, Hercules, CA) were used for the separation of proteins. Proteins were electro-transferred to methanol-activated Immobilon-FL PVDF membranes (EMD Millipore, Billerica, MA). Membranes were blocked with blocking buffer (Thermo Fisher Scientific, Rockford, IL) for 1 hr at room temperature and incubated with primary antibodies (Cell Signaling Technology, Danvers, MA) in 5% BSA overnight at 4 °C. The following primary antibodies were used for immunoblotting. Cell Signaling Technology antibody: actin (4970S), CHOP (2895S), cleaved PARP (5625), p-eIF2α (9721), cleaved caspase 3 (9664), caspase 3 (14220), PARP (9542), PERK (5683P), IRE1α (3294); GRP78 (Santa Cruz/Proteintech, SC13968/PA519503), CHAC-1 (GenTex, GTX120775), DDIT4 (Thermo Fisher Scientific, PA5–13253) and FAM129A (Sigma, SAB4300570), UPP1 (Sigma, SAB1402388) ATF4 (Proteintech, 10835–1-AP). Membranes were then washed with TBST three times for 10 min each and incubated with Dylight 800-conjugated secondary antibodies (Thermo Fisher Scientific) at 1:7500 dilution in 5% milk for 1 h at room temperature. Membranes were washed with TBST (3 times for 5 min) and TBS (5 min). The fluorescent signal was detected by Odyssey Imaging Systems (LI-COR Biosciences, Lincoln, NE).
Immunofluorescence
Cells were treated with tunicamycin 2 hr before YUM488 (15 μM) treatment for 16 hr. After treatment cells were washed with PBS then stained with ER tracker™ according to the manufacturer’s protocol. Cells were then fixed with 4% paraformaldehyde followed by permeabilization with 0.2% Triton X100. Cells were then blocked with blocked in 5% BSA and probed with the GRP78 antibody overnight at 4 °C. Following washes and incubation with secondary antibodies, cells were then stained with Hoechst (Invitrogen, 33342) and slides were mounted using ProLong™Gold (Thermo Fisher Scientific). Imaged using a Zeiss microscope.
3D spheroid assay
Cell were resuspended in media containing 10% FBS and collagen (0.015mg/ml, Stemcell Technologies). 3000–4000 tumor cells/well were seeded in U-bottomed 96-well plates (Corning), allowed to attach overnight, and then received indicated treatments. On day 6, Images of the spheroids were taken with an Olympus IX83 inverted microscope at 10X magnification. The cell viability of 3D spheroids was determined with CellTiter-Glo® 3D cell viability assay (Promega, G9683), following the manufacturer’s recommendations. Luminescence was measured in Multi-Mode Microplate Reader (Synergy H1, BioTek).
Bru-seq
Bru-seq analysis was performed as previously reported (23). Briefly, 4× 106 MIA PaCa-2 cells seeded placed in 10 cm dishes on Day 1. On Day 2, cells were treated with DMSO or YUM70 at 5 μM for 24 hr. Bromouridine, at a final concentration of 2 mM, was added to the media to label newly synthesized nascent RNA in the last 30 min of treatment. Cells were then collected and lysed in TRIZOL and total RNA was isolated. Bromouridine-containing RNA was isolated using anti-BrdU antibodies conjugated to magnetic beads, converted into cDNA libraries, and deep sequenced. Sequencing reads were mapped to the HG38 reference genome.
ATPase assay
ATP turnover and ADP generation was measured using the ADP-Glo™ Kinase Assay kit (Promega, V6930). Reaction mixtures were prepared in 384-well white OptiPlate (Perkin Elmer) and contained 0.1 μg His-tagged recombinant protein (full length or ATPase domain) and increasing concentrations of respective compounds in standard ATPase assay buffer. Reactions were pre-incubated with compounds for 30 min at 37 °C, followed by the addition of 2 μM ATP and further 2 hr incubation. Luminescence was read on a plate reader (Synergy H1, BioTek).
Thermal shift assay
The fluorescence-based thermal shift assay was carried out using the Thermofluor instrument from Johnson & Johnson (New Brunswick, NJ). The thermal shift of purified GRP78 (0.5 mg/ml in 50 mM Tris-HCl pH 8.0 buffer) in the presence or absence of YUM70 was determined as described (24). Briefly, 5 μl protein-dye (1,8 ANS, 0.3 mM) solutions were dispensed in each well of 384-well microplate (Thermo Scientific, AB1384K) and an equal volume of the test compound solutions was dispensed to each well, then, 3 μl of silicone oil (Sigma) was added to each well to avoid evaporation. 1% DMSO (no test compound) in buffer was used as a control. The plate was heated at a temperature range from 25 to 80 °C with 0.5 °C/min. Fluorescence was measured by fiber optics and fluorescence emission was detected by measuring light intensity using CCD camera. Compounds were tested in triplicate.
Cellular thermal shift assay (CETSA)
CETSA was carried out in PANC-1 cell lysate as previously described (24). Briefly, cells were harvested, washed with PBS, and diluted with cell lysis buffer (25 mM Tris-HCl pH 7.5 and 10 mM MgCl2) supplemented with the complete protease inhibitor cocktail. The cell suspension was freeze-thawed three times in liquid nitrogen. The soluble fraction was separated from debris by spinning down at 20000 × g for 20 min. The cell lysate was diluted with lysate buffer and treated with YUM70 (100 μM) and DMSO separately. After 30 min incubation at room temperature, the respective lysates were divided into smaller aliquots (50 μl) and heated individually at different temperatures for 3 minutes (Veriti thermal cycler, Applied Biosystems) followed by cooling for 3 minutes at room temperature. The heated lysates were centrifuged at 20000 × g for 20 minutes at 4 °C to separate the soluble fractions from precipitates. Supernatants were transferred to a new microfuge tube, quantified, and analyzed by SDS-PAGE followed by Western blot.
Caspase activity assay
Cells were plated in 384 well plates at 4000 cells/well. The next day cells were treated with YUM70 or tunicamycin at indicated doses for the indicated times. At the end of the treatment Caspase-3/7-GLO reagent (25 μl, Promega, G8090) was added to the well and incubated for 30 min at room temperature. Luciferase activity was measured using luminometer (Synergy H1, BioTek).
Annexin V–FITC apoptosis assay
MIA PaCa-2, PANC-1, and BxPC-3 cells (1–2×105)/well were seeded in 6 well plates, allowed to attach overnight, and then received indicated treatments for 48 hrs. Cells were washed with cold PBS, then resuspended and stained with Propidium iodide (PI) and Annexin V-FITC using Annexin V–FITC Apoptosis Detection Kit (BD Pharmigen, 556547) according to the manufacturer’s protocol. The resulting fluorescence was measured by a flow cytometer (Bio-Rad ZE5 Analyzer).
Xenograft study
MIA PaCa-2 cells (2.0 × 106, 100 μl) in PBS, were injected subcutaneously into the dorsal flank of 8-week old female NCr nude mice (Taconic Bioscience, Bar Harbor, Maine). All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee. Tumor size was monitored twice a week through caliper measurement and tumor volumes were calculated using the formula: 0.5 × D × d2, where D and d were the longest and shortest perpendicular diameters, respectively. Mice were randomly grouped (n = 5 in the control group and n = 5 in the treatment group) when the average tumor size reached 50 mm3. Control mice (n = 5) received vehicle (10% DMSO, 60% propylene glycol and 30% saline v/v, 100 μL) alone. YUM70 (30 mg/kg in 10% DMSO, 60% propylene glycol and 30% saline v/v, 100 μL) was administrated by intraperitoneal injection 5 days a week. Tumor volumes and body weights were measured twice a week to monitor tumor burden and weight loss during treatment. The study was concluded when the tumor size in the control group reached 1000 mm3. At the end of the experiment, animals were euthanized and tumor, heart, pancreas, liver, kidney, lung, and spleen were collected, fixed, and paraffin-embedded for histology. Tumor volumes were compared using the unpaired Student’s t-test.
QUANTIFICATION AND STATISTICAL ANALYSIS
Enzyme and cell data (IC50 values) were determined from at least three independent experiments and are presented as mean ± SD. All lysate, in vitro cell CETSA data were expressed as means ± SD. Tumor burden and body weight change data are represented as mean ± SEM. GraphPad Prism 8 was used for visual analysis. Statistical tests were performed using the unpaired two-tailed Student’s t-test.
Detailed methods for other experimental procedures are provided in the Supplementary Material section.
RESULTS
YUM70 is cytotoxic to pancreatic cancer cells
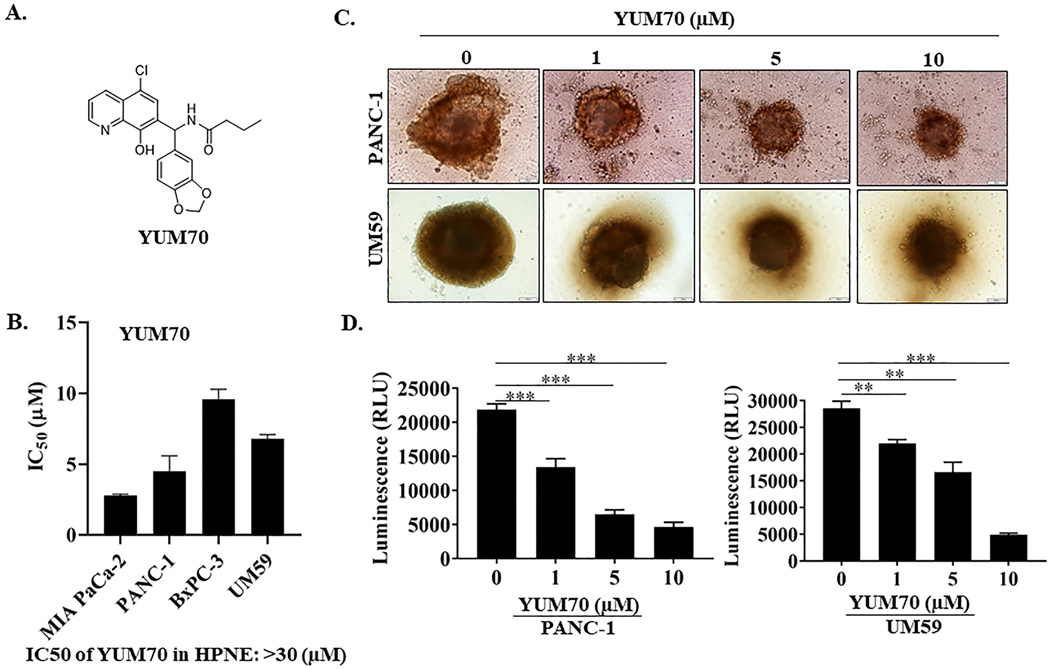
YUM70, a derivative of 8-hydroxyquinoline (Figure 1A), was identified in a phenotypic screen for cytotoxicity from 40,000 in-house drug-like compounds (25). YUM70 showed selective cytotoxicity for pancreatic cancer cell lines and pancreatic cancer 3D spheroids generated from PANC-1 and UM59 cell lines over normal pancreatic tissue-derived HPNE cells (Figures 1B- 1D). UM59 is a patient-derived cell line with a KRAS mutation (26). Both PANC-1 and UM59 cells formed compact spheroids (661.73 ± 31.98 and 537.83 ± 16.35 μm in diameter, Figure 1C; Supplementary Fig. S1A). The spheroid size was reduced by 1.8 and 1.7 fold for PANC-1 and UM59 respectively (p-value < 0.001, n = 6) upon YUM70 treatment. Importantly, when we measured ATP by luminescence using CellTiter-Glo® 3D cell viability assay, we observed that viability was reduced 4.7 and 5.8 times (p-value <0.0001, n = 6) for PANC-1 and UM59, respectively. These results demonstrate that YUM70 is not only cytotoxic to pancreatic cancer cells grown as a monolayer but also cytotoxic to tumorspheres. Additionally, YUM70 was cytotoxic in a larger panel of cell lines in 2D culture (Supplementary Fig. S1B) and in a 3D culture model generated from ovarian cancer cells (Supplementary Fig. S1C). Altogether, cytotoxicity results demonstrate that YUM70 is effective against various tumor cell types grown either in 2D or 3D. Interestingly, we observed IC50 of YUM70 varied among pancreatic cancer cell lines. IC50 of YUM70 in BxPC-3 is approximately three times higher than MIA PaCa-2 cells. PANC-1 and UM59 cells are also more sensitive to YUM70 than BxPC-3 cells. MIA PaCa-2, PANC-1, and UM59 cells have mutated KRAS while BxPC-3 cells have wild-type KRAS (26). Since > 90% of pancreatic cancer patients have mutated KRAS, we primarily focused on pancreatic cancer.
Figure 1. YUM70 is cytotoxic to pancreatic cancer cell lines.
A. Chemical structure of YUM70. B. Cytotoxicity of YUM70 in a panel of pancreatic cancer cell lines and normal pancreatic tissue-derived cells (HPNE) measured by the MTT assay. IC50 presented as mean ± SD of three independent experiments performed in duplicate. C. YUM70 dose-dependently decreased PANC-1 and UM59 cell proliferation in 3D-culture systems. D. Quantification of cell viability of 3D spheroids was performed with CellTiter-Glo® 3D cell viability assay. Data are presented as mean ± SD of three or more spheroids from three independent experiments. *p < 0.01, **p < 0.001, ***p < 0.0001
We synthesized a series of 8-hydroxyquinoline analogs to determine the importance of various substitutions at positions R1, R2, and R3 for cytotoxicity. The detailed chemistry is described in the Supplementary Material section. All synthesized compounds were tested for cytotoxicity by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay in a panel of pancreatic cancer cells (Supplementary Table S1). Structure-activity relationship studies suggested that the hydroxyl group (OH group) at position X is important for activity; replacement of OH with OCH3 decreased potency (YUM117, YUM245). Among all the tested compounds, we selected YUM70 for mechanistic studies because of its superior solubility, microsomal stability, and PK properties over some of the more cytotoxic compounds. YUM70 showed promising t1/2 in an in vitro human liver microsomal study (t1/2 > 60 min) and a PK study (t1/2 > 84 min) in mice (Supplementary Fig. S2A–S2C).
YUM70 induces ER stress-mediated apoptosis
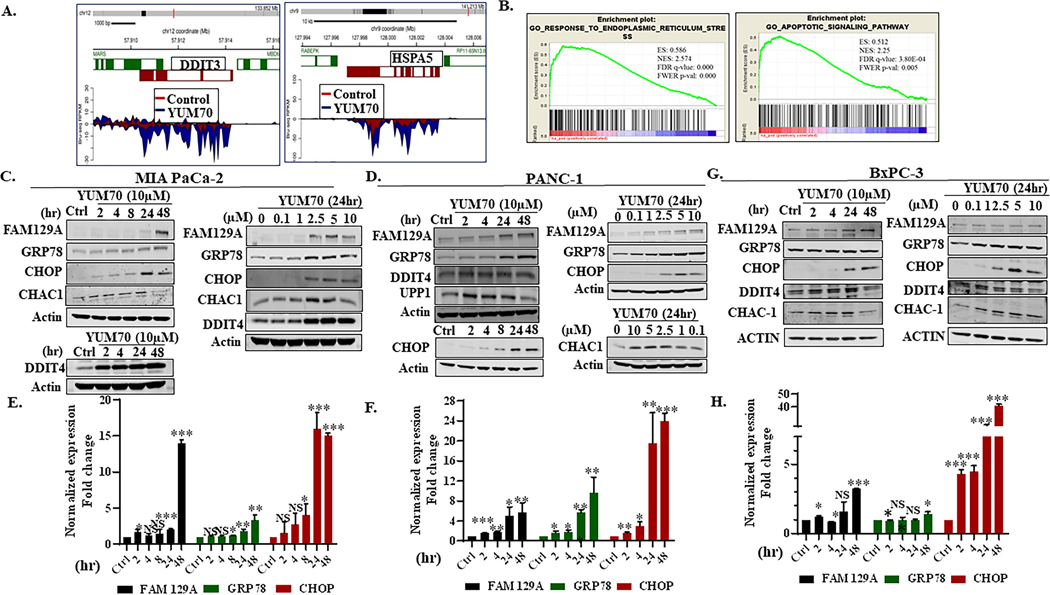
To understand the potential mechanism of action of YUM70, we performed nascent RNA sequencing (Bru-seq) (23) to study the changes in transcription in MIA PaCa-2 cells treated with 5 μM YUM70. We observed that after YUM70 treatment, many of the upregulated genes were involved in the UPR. The top 25 upregulated and downregulated genes are listed in Supplementary Tables S2 and S3, respectively. FAM129A and GDF15 are the top upregulated genes induced by YUM70 that are known to respond to ER stress (27,28). YUM70 treatment induces the transcription of both TRIB3 and SLC1A4 by 13.9 and 13.7 fold respectively (Supplementary Table S2). CHAC1 and ATF3 are also induced by YUM70 and ER stress (29,30). Another upregulated gene is UNC5B, a tumor suppressor gene and a direct transcriptional target of p53 that can be activated by various insults. UNC5B induces apoptosis when unbound by netrin-1 (31). Other upregulated genes, for example, DDIT4, SESN2, JDP2, BBC3, DDIT3, are all implicated in ER stress response (32,33). The upregulation of SESN2 in response to ER stress is dependent on both PERK and IRE1/XBP1 arms of the UPR (34). Genes like PHGDH, AKNA, UPP1 are not well studied as ER stress response mediators. Altogether, these results suggest that YUM70 treatment induces ER stress.
DAVID functional annotation analysis revealed that the transcriptionally upregulated genes are involved in ER stress, cell death, and apoptosis; and downregulated genes are associated with the process of DNA binding and chromatin assembly (Supplementary Fig. S3A–S3B). Transcription of GRP78 (HSPA5) and CHOP (DDIT3, a key ER stress marker) was enhanced by YUM70 treatment (Figure 2A) and correlated with protein and mRNA expression (Figure 2; Supplementary Fig. S4A–S4D). We also performed gene set enrichment analysis (GSEA) on all genes ranked by log2fold change. GSEA revealed enrichment of genes involved in UPR, hypoxia, apoptosis, and the p53 pathway in the Hallmark gene set (Supplementary Table S4). Additionally, we observed enriched gene sets related to ER stress, cellular response to topologically incorrect protein, intrinsic apoptotic signaling pathway in response to ER stress in the Gene Ontology (GO) biological process database (Figure 2B and Supplementary Table S5). GSEA of transcription factors revealed that the major upregulated transcription factors are ATF3, CHOP, and CREB (Supplementary Table S6) that are well known to be activated upon ER stress. Of note, we performed Bru-seq in two biological replicates and we observed a similar set of genes up and downregulated and similar pathways involvement in GSEA analysis (Supplementary Fig. S5A–S5C). Important microRNAs and lncRNAs are listed in Supplementary Tables S7–S10. STRING analysis (https://string-db.org/) on upregulated genes also revealed the biological process: cellular response to unfolded protein, ER unfolded protein response, regulation of ER stress-induced intrinsic apoptotic signaling pathway, IRE1-mediated UPR, PERK-mediated UPR, and ATF6-mediated UPR as major readouts (Supplementary Tables S11–S14). Taken together, findings from DAVID, GSEA, and STRING analysis are consistent with the hypothesis that YUM70 induces a strong ER stress response.
Figure 2. YUM70 upregulates UPR target proteins and induces ER stress and apoptosis.
A. Expressions of DDIT3 (CHOP) and HSPA5 (GRP78) are upregulated by YUM70 treatment (Bru-seq data). B. GSEA analysis of YUM70 treatment reveals enrichment of ER stress and apoptosis pathway. C–D. Lysates of MIA PaCa-2 and PANC-1 cells treated with YUM70 at indicated doses and times were immunoblotted with indicated antibodies. A representative data set from three independent experiments is shown. E-F. Relative expression (fold change) of FAM129A, GRP78, and CHOP normalized to GAPDH/Actin in MIA PaCa-2 and PANC-1 cells were calculated from the band intensity of three independent experiments and are presented as mean fold change at indicated time (hr). G-H. Representative immunoblot and relative expression (mean fold change) of the above stated proteins in BxPC-3 cells are presented. DMSO control indicated as Ctrl. Error bars represent standard deviation. The band intensity was quantified in Image Studio Ver 3.1 software. *p < 0.05, **p < 0.01, ***p < 0.001.
To further validate the hypothesis that YUM70 induces ER stress, we performed a connectivity map (CMap) analysis on YUM70 Bru-seq data. The top bioactive compounds are listed in Supplementary Table S15. Known ER-stress inducers such as tunicamycin and thapsigargin showed a high similarity score (99.9 and 99.8, respectively) (Supplementary Table S15). Additionally, proteasome inhibitors such as MG-132 (99.3), SCH-79797 (99.3), MLN-2238 (98.4), z-leu3-VS (98.3); HDAC inhibitors such as JNJ-26854165 (98.2), THM-I-94 (97.6), panobinostat (97.5), and vorinostat (92.23); and topoisomerase inhibitors such as irinotecan (99.4) and topotecan (93.3), also showed similar nascent RNA transcriptome signatures to YUM70 with >90 similarity scores (Supplementary Table S15). Moreover, CMap showed a strong similarity between the transcriptional signature of HSPA5 (GRP78) knockdown (Supplementary Table S15) and YUM70 treatment. Altogether, these results suggest that YUM70 treatment in pancreatic cancer cells results in ER stress and has a signature that is similar to GRP78 knockdown.
While upregulated genes display a strong ER stress signature, downregulated genes showed significant enrichment of pathways related to E2F targets, G2M checkpoint, MYC targets, and pathways related to the mitotic spindle, and chromosome segregation (Supplementary Tables S16–S18). To assess the potential effects of YUM70 on cell cycle progression, we performed cell cycle analysis by flow cytometry. No significant effects on cell cycle distribution were observed following a 24 hr treatment using YUM70 concentrations of up to 3 times the IC50. However, treatments with a high dose (15 μM) of YUM70 for 24 and 48 hr increased the S-phase population, with a decrease in the G0/G1 population (Supplementary Fig. S6A–S6C). The result suggests that at the high dose the accumulation of cells in the S phase may be due to an intra-S phase checkpoints activation which is not allowing cells to enter the next phase of the cell cycle. GSEA of transcription factors also revealed that E2F is the predominant transcription factor downregulated upon YUM70 treatment (Supplementary Table S18). E2F plays a major role during the G1/S transition in the cell cycle. Moreover, downregulated expression of E2F family members has been shown to induce both inappropriate S phase entry and apoptosis (35). Protein-protein interaction (PPI) network analysis using the STRING online database also revealed similar pathway enrichment for upregulated and downregulated genes (Supplementary Tables S19–S22).
YUM70 induces ER stress-mediated apoptosis via phosphorylation of eIF2α
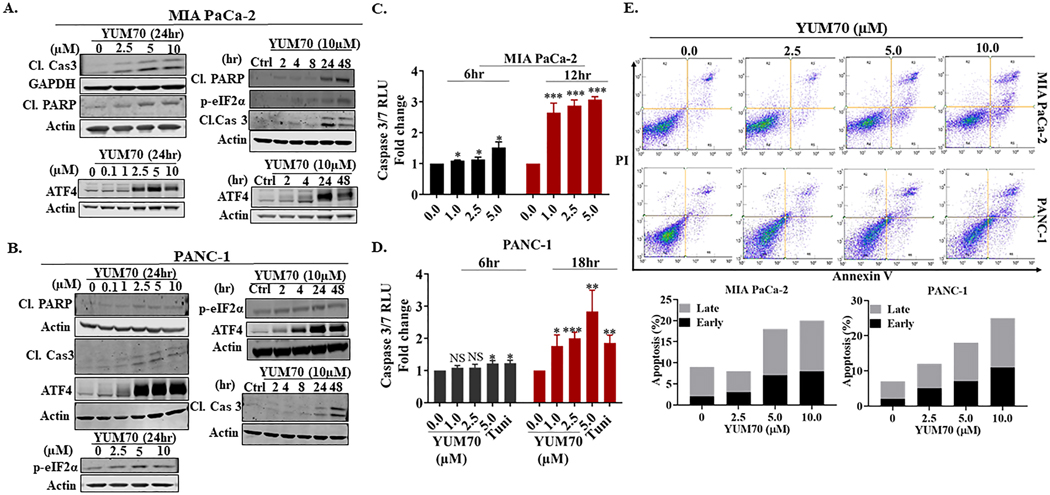
To characterize ER stress-mediated apoptosis, we performed Western blot on lysates from MIA PaCa-2, PANC-1, and BxPC-3 cells treated with YUM70 and assessed selected UPR markers. YUM70 treatment increased the protein levels of FAM129A, DDIT3, CHAC-1, DDIT4, UPP1, and GRP78 in a dose- and time-dependent manner (Figures 2C–2D). Upregulation of pro-apoptotic gene FAM129A (Supplementary Table S2) and an increase in its protein levels (Figures 2E–2F) suggest that YUM70 may induce apoptosis via ER stress. We also observed an increase in mRNA levels for GRP78 and CHOP (Supplementary Fig. S4). In BxPC-3 cells, we didn’t observe a significant increase in GRP78 or FAM129A (Figures 2G–2H). However, we observed a significant increase in the protein levels of CHOP. Significant upregulation of TRIB3 and SLC1A4 (mRNA) suggested that YUM70 may induce apoptosis by eIF2α phosphorylation, followed by activation of ATF4 (30,36). Indeed, we observed induction of ATF4 (Figures 3A–3B) and upregulation of downstream targets of ATF4 and CHOP (Supplementary Table S2, Figures 2C–2H). We also observed increased protein expression of CHAC1 and DDIT4. During ER stress PERK-mediated phosphorylation of eIF2 is known to inhibit global translation while it allows translation of ATF4 mRNA. We observed the induction of phosphorylation of eIF2α in a dose and time-dependent manner (Figures 3A–3B, Supplementary Fig. S7A) upon YUM70 treatment. As the PERK/ATF4/CHOP signaling pathway plays a crucial role in the induction of cell apoptosis, we assessed apoptosis markers like cleaved caspase 3 and cleaved PARP upon YUM70 treatment. We observed increased levels of cleaved caspase 3 and cleaved PARP at the indicated doses and times (Figures 3A–3B, Supplementary Fig. S7A–S7D). In BxPC-3 cells, we did not observe increased levels of cleaved PARP with varying doses of YUM70. However, we observed increased levels of cleaved caspase 3, ATF4, and p-eIF2α. Therefore, the effect of YUM70 is different in BxPC-3 cells than in PANC-1 and MIA PaCa-2 cells. We confirmed that YUM70 induces apoptosis by increasing caspase 3/7 activity (Figures 3C–3D). We used tunicamycin (Tu) as a positive control for the caspase 3/7 activity assay since it is a known ER stress inducer that promotes apoptosis. Mechanistically, tunicamycin disrupts protein maturation by blocking N-linked glycosylation in the endoplasmic reticulum (ER) of eukaryotic cells (37). Induction of apoptosis by YUM70 in a dose-dependent manner was confirmed in Annexin V-FITC assay (Figure 3E; Supplementary Fig. S7). However, a fewer number of apoptotic cells were found at a higher dose of YUM70 in BxPC-3 cells than in the other two cell lines.
Figure 3. YUM70 upregulates proteins involved in the apoptotic pathway.
A-B. Lysates of MIA PaCa-2 and PANC-1 cells were immunoblotted with indicated antibodies. A representative experiment out of three is shown. The actin blot used in Figure 3A is the same as in Figure 2C. C-D. Caspase 3/7 activity levels in MIAPaCa-2 and PANC-1 cells, following treatment with YUM70. Data are presented as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. E. YUM70 dose-dependently causes apoptosis in MIA PaCa-2 and PANC-1 cells. Top, cells in the bottom left quadrant of each panel (Annexin V–negative, PI-negative) are viable, whereas cells in the bottom right quadrant (Annexin V–positive, PI-negative) are in an early stage of apoptosis, and cells in the top right quadrant (Annexin V–positive, PI-positive) are in a late stage of apoptosis/necrosis. Bottom, the percentage of apoptotic cells is shown in a histogram. A representative image of three independent experiments is shown.
Together, treatment with YUM70 resulted in phosphorylation of eIF2α and increases in ATF4 and CHOP leading to apoptosis induction as indicated by PARP and caspase 3 cleavage, and increased caspase 3/7 activity. YUM70 is an ER stress inducer and kills cells by activating the apoptosis pathway.
YUM70 targets GRP78
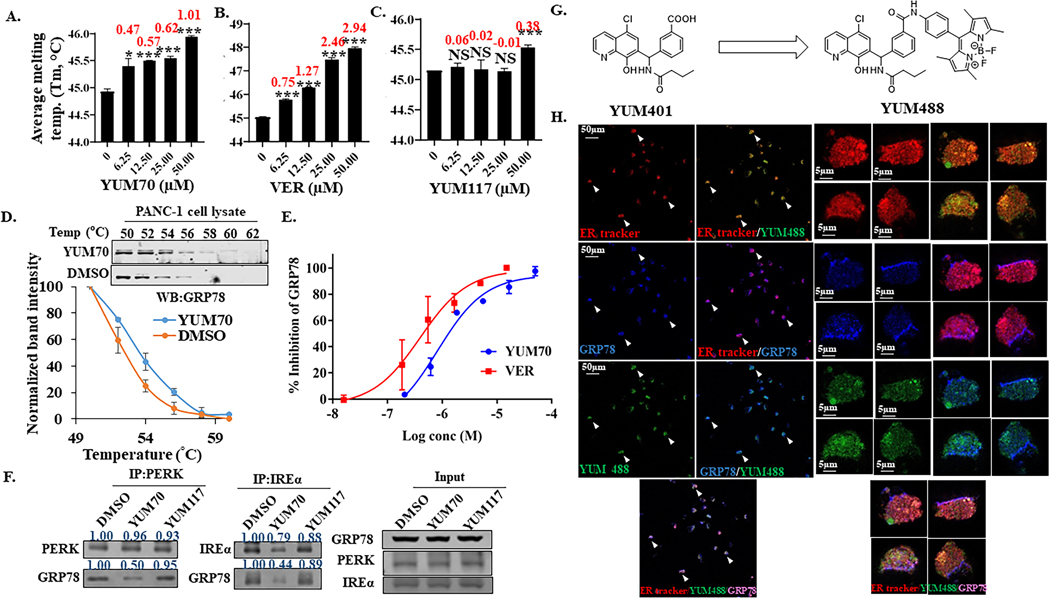
Since GRP78 dissociation from ER sensors is a crucial event to initiate the UPR and YUM70 treatment produces a gene signature similar to GRP78 knock-down, we hypothesized that GRP78 may be the target of YUM70. Treatment with YUM70 increased the protein levels of CHOP (DDIT3) and GRP78 (HSPA5) (Figure 2A) perhaps due to the induction of UPR. To determine whether YUM70 binds to GRP78, we performed a thermal shift assay using purified full-length GRP78. YUM70 binds to full-length GRP78 and causes a positive shift in the melting temperature (Tm) of GRP78 in a dose-dependent manner (Figure 4A; Supplementary Fig. S8A). The positive control VER155008 (VER), an HSP70/GRP78 inhibitor, stabilized GRP78 as expected (Figure 4B; Supplementary Fig. S8B). In contrast, YUM117, an inactive analog of YUM70, did not produce any Tm shift (Figure 4C; Supplementary Fig. S8C).
Figure 4. YUM70 targets GRP78.
Apparent melting temperature (Tm) derived from thermal shift assay and the corresponding thermal shift were determined at various concentrations of A. YUM70 B. VER and C. YUM117. 1% DMSO was used as control. Data are presented as mean ± SD. *p < 0.01, **p < 0.001, ***p < 0.0001. Numbers in red are thermal shift at respective concentrations. D. CETSA melt curves for GRP78 in PANC-1 cell lysates treated with YUM70 (100 μM). 1% DMSO was used as control. Data are presented as mean ± SD of three independent experiments. E. Inhibition of GRP78 enzymatic activity in the presence or absence of YUM70 and VER. The results are presented as mean % inhibition of GRP78 of three independent experiments. Error bars represent standard deviation. F. PERK and IRE1α were immunoprecipitated overnight from PANC-1 cell lysates in the presence of DMSO, YUM70 or YUM117. Western blotting was performed with indicated antibodies. A representative experiment out of three is shown. G. Chemical structure of YUM401 and YUM488 (BODIPY-conjugated YUM401). H. YUM488 co-localizes with GRP78 in the ER. Immunofluorescence image of MIA PaCa-2 treated with tunicamycin for 2 hr followed by 15 μM YUM488 treatment for 16 hr. ER was labeled with ER-Tracker (detection wavelength 566–626 nm), GRP78 was labeled with antibody (detection wavelength 656–700 nm), and YUM488 was detected at wavelength 497–564 nm.
To further validate these findings, we performed a cellular thermal shift assay (CETSA) to assess target engagement in cells. PANC-1 cell lysates were either treated with DMSO alone or with YUM70 or VER, followed by heating at different temperatures (Figure 4D; Supplementary Fig. S8D). YUM70 stabilized cellular GRP78 (Figure 4D). Disruption of GRP78 ATPase activity inhibits the release of bound peptides, resulting in the accumulation of unfolded or misfolded proteins in the ER (38). YUM70 inhibits GRP78 ATPase activity of the full-length protein (IC50 of YUM70 1.5±0.3 μM, VER 0.7±0.3 μM, Figure 4E) as measured by ATP turnover and ADP generation. Taken together, the results confirmed that YUM70 binds to GRP78 and inhibits its enzymatic activity. Thus, we suggest that upon YUM70 treatment, GRP78 dissociates from its ER partners, PERK, IREα, and ATF6, leading to the induction of the UPR. Co-immunoprecipitation showed that incubation of PANC-1 cell lysates with YUM70 decreased the interactions between GRP78 and both PERK and IREα (Figure 4F). While YUM70 was able to hinder these interactions, inactive compound YUM117 was unable to do so (Figure 4F), suggesting a direct effect of YUM70 on the complexes. Moreover, CMap showed a strong similarity between the transcriptional signatures of HSPA5 (GRP78) knockdown (Supplementary Table S15) and YUM70 treatment. We knocked down GRP78 using two individual siRNAs in MIAPaCa-2 cells (Supplementary Fig. S8E). In human PDAC cell lines, knockdown of GRP78 resulted in decreased proliferation (39), therefore we performed partial knockdown of GRP78 that resulted in increased YUM70 cytotoxicity by 2 fold (Supplementary Fig. S8F). To test the selectivity of the compound to GRP78, we performed a binding study for other ER proteins PDI, GSTO1, and HSP70 (Supplementary Fig. S8G–S8I). We performed a competitive binding assay in the presence of specific fluorescence probes for GSTO1 (24) and PDI (40). No binding was observed at 1 and 10 μM YUM70, but there was a reduction in band intensity at 100 μM. YUM70 did not inhibit PDI reductase activity at 40 μM (Supplementary Fig. S8G). However, a non-selective analog, YUM79, which has similar cytotoxicity, showed 94% inhibition of PDI activity. YUM79 also binds to PDI and GSTO1 as shown in Supplementary Fig. S8G–S8H. VER nonselectively inhibits both HSP70 and GRP78 due to the structural similarity between these two proteins. Our data show that YUM70 did not bind to HSP70 (Supplementary Fig. S8I). These observations suggest that YUM70 selectively binds to GRP78 over PDI, GSTO1, and HSP70.
Next, we used immunofluorescent microscopy to investigate whether our inhibitors and GRP78 co-localize in the cell. We synthesized a fluorescent BODIPY conjugate of a YUM70 analog (Figure 4G) to study its subcellular distribution. Due to the lack of a substitution site on YUM70, we used the close analog YUM401 that showed a similar cytotoxicity/inhibitory profile. As shown in Figure 4H, BODIPY-conjugated YUM401 (YUM488) showed a strong co-localization with the GRP78 antibody and ER-tracker. This result supports that YUM70 analog, YUM401 targets GRP78 in the ER.
Proteolysis targeting chimera (PROTAC) is a powerful technology for targeted protein degradation (41). To selectively degrade GRP78, we synthesized a PROTAC by incorporating YUM70, a linker, and an E3-ligase recruiting ligand. The detailed chemistry is described in the Supplementary Material section. To evaluate the extent of GRP78 degradation, we treated MIA PaCa-2 cells with YUM70-PROTAC (DX2–145, Supplementary Fig. S9A) for 24 hr at various doses and the level of GRP78 protein was assessed by Western blot. DX2–145 elicited a concentration-dependent degradation of GRP78 (Supplementary Fig. S9B–S9C) and increased CHOP expression (Supplementary Fig. S9B). We observed maximum degradation at 10 μM and a decrease in degradation at 20 μM due to the hook effect, a typical property of PROTACs. In a separate experiment, we show that MG132, a proteasome inhibitor, blocked DX2–145-mediated degradation of GRP78 (Supplementary Fig. S9D). This data demonstrates that DX2–145 degrades GRP78 in a proteasome-dependent manner. We observed a similar pattern of degradation with the PROTAC YUM513 (YUM401-PROTAC) (Supplementary Fig. S9E–S9G). In a separate experiment, we treated MIA PaCa-2 cells with YUM513 for 4 hr or 24 hr and observed a decrease in intensity of a band of approximately 70 kD by SDS-PAGE followed by Coomassie blue staining (Supplementary Fig. S9H). Analysis of this band using LC-MS/MS confirmed the degradation of GRP78 (Supplementary Fig. S9I). DX2–145 is more effective in degrading GRP78 than YUM513 and was selected for further studies. When cells were pre-treated with the parent compounds for 2 hr followed by DX2–145 (10 μM), we observed an increase in GRP78 levels in YUM70 pre-treated cells as expected. Pretreatment with pomalidomide at doses up to 10 μM did not show complete blockade of GRP78 degradation (Supplementary Fig. S10A–S10C). We observed similar patterns with YUM401 (Supplementary Fig. S10D–S10F). In contrast, pre-treatment with YUM117, an inactive analog, was unable to increase GRP78 levels in a dose-dependent manner. YUM117 was also unable to significantly change GRP78 protein levels in the presence or absence of DX2–145 (Supplementary Fig. S10D–S10E). Altogether, these results directly show that YUM70 and its analogs target GRP78 and clearly demonstrate that GRP78 can be degraded through cereblon-based PROTACs.
The data above suggest that YUM70 binds to GRP78 and inhibits its enzymatic activity. Moreover, YUM70 can up-regulate GRP78 transcription and protein levels as shown by Bru-seq, RT-PCR, and Western blot analysis. To further validate these observations, we blocked the transcription or translation machinery by actinomycin D (ACD: 10 nM for MIA PaCa-2 and 20 nM for PANC-1) and cycloheximide (CHX: 2.5 μM) pre-treatment for 2 hr followed by 24 hr YUM70 (5 and 10 μM) treatment. We observed a significant decrease in GRP78 protein expression in the presence of CHX (Supplementary Fig. S11A–S11B). We did not observe a significant decrease in GRP78 protein levels in the presence of ACD as compared to YUM70 alone. This observation suggests that YUM70 up-regulates GRP78 primarily at the translational level rather than at the transcriptional level, which is in agreement with the classical concept that UPR mainly affects translation.
The result indirectly shows that although YUM70 inhibits GRP78 enzymatic activity; it increases the expression of GRP78 by increasing the chaperone translation mechanism.
Molecular docking predicts that YUM70 may bind to the C-terminal substrate-binding domain of GRP78
Although YUM70 binds to the full-length GRP78 and inhibits its enzymatic activity, we did not observe a similar effect using a truncated protein containing the ATPase domain (GRP78ATPase) (Supplementary Fig. S12A–S12G). Thus, we hypothesized that YUM70 may bind a site other than the active site of GRP78. Molecular docking of YUM70 was performed on the substrate-binding domain (SBD) of GRP78 (PDB: 5E85) to identify a putative binding site. There are two possible binding sites on the substrate-binding domain: the peptide binding site and the site corresponding to the novolactone-binding domain (NBD) in HSP70 (PDB: 4WV7) (42). YUM70 and its inactive analog YUM117 were docked into both sites of GRP78. In the peptide-binding site, YUM70 interacts with several important substrate-binding residues, including Ser452, Gln458, Val461, Thr462, Ile463, and Lys464 (Supplementary Fig. S13A–S13F). Ser452 and Gln458 formed H-bonds with the hydroxyl group of YUM70. The amide carbonyl forms two H-bonds with Thr462. The phenyl ring formed a pi-cation interaction with Lys464 and the quinoline ring is packed into a hydrophobic pocket formed by Ile426, Phe451, and Ile463. When we docked the inactive analog YUM117, it failed to form an H-bond with Ser452 or Gln458. The hydroxyl group of YUM70 is replaced by a methoxy group in YUM117, which likely explains this observation. Docking scores (GOLD fitness score) of YUM70 and YUM117 on the peptide binding site are 61.17 and 54.57, respectively. While protein-ligand interactions at the substrate-binding site may explain the different ATPase inhibitory activities and cytotoxicities exerted by YUM70 and YUM117 in pancreatic cancer cells, interactions at the novolactone-binding site fail to explain the activity difference. Finally, the docking score of YUM70 is higher at the substrate-binding site than at the novolactone-binding site (61.17 vs 54.83). Altogether, docking suggests that YUM70 may bind to the substrate-binding site of GRP78.
Next, we used partial proteolysis to investigate the proteolytic pattern of GRP78 in the presence or absence of YUM70, ATP, and ADP. Partial proteolysis using trypsin and visualization through Coomassie blue staining revealed a unique proteolytic degradation pattern of GRP78 (Supplementary Fig. S13G). In the presence of YUM70, we observed clear protection of GRP78 from proteolytic degradation by trypsin. GRP78 has two main domains (NBD and SBD) and functionally active GRP78 undergoes two major conformational changes (43), known as open (ATP-bound) and closed (ADP-bound) conformations. It is reported that during closed conformation when peptides tightly bind to the pocket of SBD, ADP is simultaneously bound to NBD (44). In trypsin digestion experiments, we also observed a weak stabilization after adding ADP. We observed protection of GRP78 in the presence of YUM70 alone as well as with ATP and ADP. However, more protection of GRP78 was observed by YUM70 in the presence of ADP than ATP. These results suggest that YUM70 may cause a conformation change in GPR78.
FAM129A is a potential biomarker for ER stress induced by YUM70 in pancreatic cancer
Knockdown of GRP78 (GRP78 siRNA-1), similar to YUM70 treatment, increased the expression of FAM129A, CHOP, and DDIT4 in MIA PaCa-2 and PANC-1 cells (Supplementary Fig. S14A). However, GRP78 knockdown did not alter CHAC1 levels. Interestingly, GRP78 siRNA-1 was unable to decrease GRP78 levels in BxPC-3 cells, thus we used another siRNA (GRP78 siRNA-2) targeting a different sequence of GRP78. siRNA-2 decreased GRP78 levels > 80 % in BXPC3 cell and > 95% in MIAPaCa-2 and PANC-1 cells (Supplementary Fig. S14B). Both siRNAs decreased GRP78 levels >95% in HT29 colon cancer cells with wild type KRAS. We observed a similar pattern with siRNA-1 and siRNA-2 in MIAPaCa-2 and PANC-1 cells. BxPC3 showed different patterns of CHAC-1, DDIT4, and CHOP protein levels than MIAPaCa-2 and PANC1 cells upon GRP78 knockdown. Similar to BxPC3, in HT29 cells, we did not observe a significant change in levels of DDIT4 and CHAC-1 upon GRP78 knockdown; however, we observed a significant increase in CHOP levels upon GRP78 knockdown in HT29 cells.
The expression of FAM129A was significantly elevated by YUM70 and knockdown of GRP78. FAM129A is a proapoptotic gene and one of the ATF4 target genes that regulate PERK and eIF2α as part of a feedback loop (45). We showed that YUM70 treatment increased the levels of ATF4 as well as phosphorylation of eIF2α. The result suggests that FAM129A positively regulates the PERK-eIF2α axis in pancreatic cancer and causes apoptosis mediated cell death. Thus, FAM129A transcript and protein levels can be used to evaluate the response to GRP78 inhibitors in pancreatic cancer.
Interestingly, the protein level of FAM129A was increased by YUM70 treatment in MIA PaCa-2 and PANC-1 cell lines in a time- and dose-dependent manner. However, in BxPC-3 cells, we only observed an increased expression of FAM129A in a time-dependent manner. We also did not observe significant changes in GRP78 expression in BxPC-3 cells. YUM70 was more potent in KRAS mutant cells than wild-type cells (Figure 1B). MIA PaCa-2 cells were the most sensitive, while BxPC-3 cells were the most resistant of the three pancreatic cancer cell lines to YUM70. In an Annexin V-FITC assay, we observed more apoptosis by YUM70 in MIA PaCa-2 and PANC-1 than BxPC-3 cells (Figure 3E; Supplementary Fig. S7). Although, the result suggests mutated KRAS cells are more sensitive to YUM70 than wild-type cells, such observations need to be expanded to a larger panel of cell lines and more in-depth mechanistic studies to better define the relationship between KRAS and GRP78.
YUM70 reduces tumor growth in a MIA PaCa-2 xenograft model
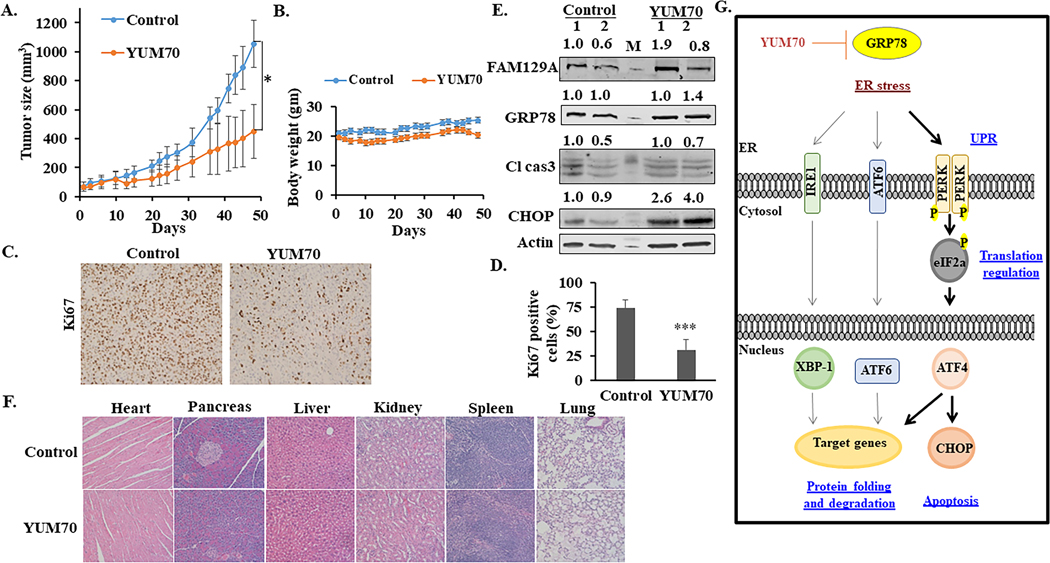
To evaluate the in vivo efficacy of YUM70, we performed xenograft studies in NCr nude female mice. Subcutaneous human pancreatic cancer xenografts were established using MIA PaCa-2 cells on the dorsal flank of immune-deficient mice. The mice were injected intraperitoneally with either YUM70 (30 mg/kg) or vehicle (10% DMSO, 70% PG, 20% saline) 5 days a week for 7 weeks. A significant tumor growth delay (p < 0.05) was observed (Figure 5A) with no significant change in body weight during the course of treatment (Figure 5B). Consistent with our in vitro data, YUM70 treatment suppressed tumor cell proliferation as confirmed by the significant decrease in Ki67 staining (Figures 5C–5D) and the increased expression levels of CHOP and FAM129A in the treatment group (Figure 5E). Immunohistochemical (IHC) staining revealed significantly increased levels of cleaved caspase 3 in YUM70 treated tumor tissues (Supplementary Fig. S15A). Additionally, we observed strong GRP78 staining in control tumors as compared to the YUM70 treated tumors (Supplementary Fig. S15B). However, no significant changes in GRP78 mRNA levels were observed (Supplementary Fig. S15C) in control and treated tumor tissues. No gross toxicity was observed in the heart, pancreas, liver, kidney, lung, and spleen (Figure 5F). These results demonstrate that YUM70 treatment induced ER stress-mediated apoptosis in the tumor cells implanted in mice without major toxicity to normal tissues. A plausible model for the anticancer mechanism of YUM70 in pancreatic cancer is presented in Figure 5G.
Figure 5. YUM70 inhibits pancreatic tumor growth in vivo.
A. Tumor growth curves of MIA PaCa-2 tumor-bearing nude mice treated with vehicle (n = 5) or YUM70 (n = 5). Data are shown as mean tumor volumes (error bars, SEM). A significant reduction in tumor volumes was observed upon YUM70 treatment (*p < 0.05). B. Evaluation of mouse weights during the xenograft experiment. Error bars indicate mean ± SEM. C. Ki67 immunohistochemistry staining in tumor sections. D. Percent of Ki67 positive cells were calculated as the fraction of Ki67 positive cells compared to the total number of cells in the field × 100. (n = 10; five fields of view from two tumors per group). Graphical data is presented as Mean ± SD, ***p < 0.0001. E. Lysate from two tumors per group was blotted for FAM129A, GRP78, CHOP, cleaved caspase 3. ‘M’ is the molecular weight marker. Normalized relative densities computed using ImageJ (NIH) are shown above. F. YUM70 did not show systemic toxicity in vivo. Representative micrographs of hematoxylin and eosin (H&E)-stained organ tissue sections. Images were taken with an Olympus IX83 inverted microscope at 20X magnification. G. Mechanism of YUM70 mediated cell death. YUM70 is cytotoxic to pancreatic cancer cells via a mechanism involving the allosteric inhibition of GRP78 activity, leading to an increase in ER stress and induction of protein levels of UPR target genes. As a result, YUM70 treatment causes prolonged ER stress-mediated apoptosis.
YUM70 induces synergistic cell death with topotecan and vorinostat
YUM70 showed moderate potency in vitro and in vivo as a single agent. YUM70 is more potent than 5FU but less potent than gemcitabine and paclitaxel (Supplementary Fig. S16A). To find novel and effective therapies for pancreatic cancer, we performed clonogenic survival assays using combination of YUM70 with several clinically approved drugs, drugs in clinical trials, and preclinical agents. YUM70 showed a synergistic effect when combined with MG132 (carbobenzoxy-Leu-Leu-leucinal) (Supplementary Fig. S16B–S16C), topotecan or vorinostat (Figures 6A–6E), and an additive effect with 5-FU (fluorouracil), tosedostat, and DFO (deferoxamine mesylate) (Supplementary Fig. S16D–S16I). We did not observe a significant additive effect or synergy by combining YUM70 with gemcitabine.
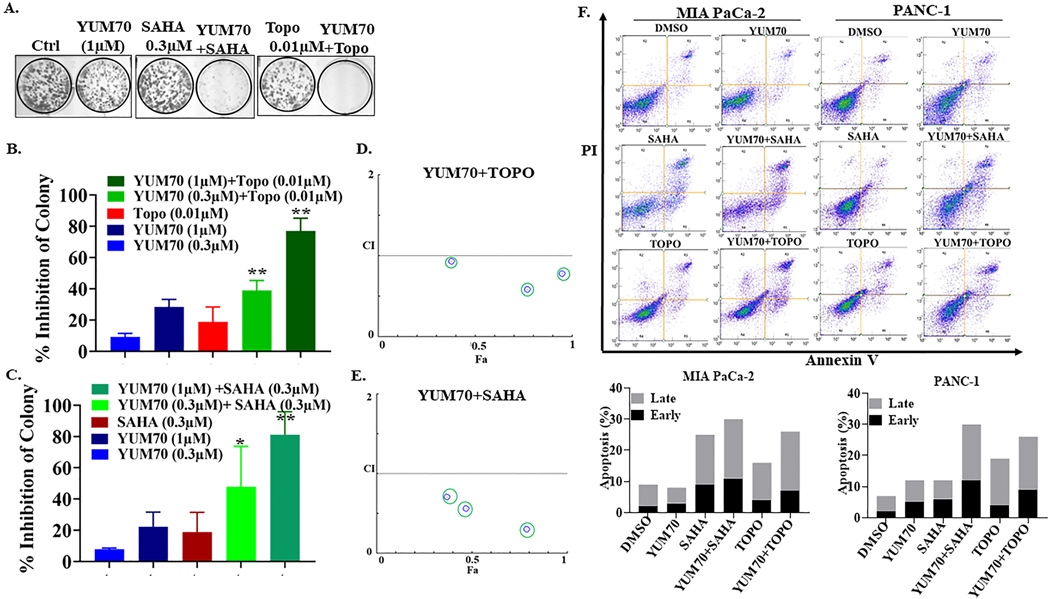
Figure 6. Synergistic effect of YUM70 in combination with topotecan and vorinostat.
MIA PaCa-2 cells were treated with YUM70 with or without topotecan (Topo) and vorinostat (SAHA), at stated concentrations and kept in culture until colonies were observed in DMSO treated control. A. A representative image is shown (one concentration). B and C. The number of colonies was quantified using Image Studio ver3.1 software from three independent experiments (more than one concentration). Graphical data is presented as mean ± SD, *p < 0.05, **p < 0.01. The p-value of the combination was calculated and compared to YUM70 alone. D and E. The combined effect was calculated using CompuSyn software. CI <1 is defined as synergism. F. Combination regimen causes apoptosis in MIA PaCa-2 and PANC-1 cells. Top, cells in the bottom left quadrant of each panel (Annexin V–negative, PI-negative) are viable, whereas cells in the bottom right quadrant (Annexin V–positive, PI-negative) are in the early stage of apoptosis, and cells in the top right quadrant (Annexin V–positive, PI-positive) are in the late stage of apoptosis/necrosis. Bottom, the percentage of apoptotic cells is shown in a histogram. A representative image of three independent experiments is shown.
YUM70-topotecan combination showed strong synergy with a combination index (CI) of 0.59 at 1μM YUM70 and 0.01 μM topotecan (Figure 6D and Supplementary Table S23). YUM70-SAHA combination was also able to significantly enhance cytotoxicity with CI 0.29 at a dose of 1 μM YUM70 and 0.3 μM vorinostat (Figure 6E and Supplementary Table S23). YUM70-MG132 combination also showed synergy in the clonogenic assay, at non-cytotoxic concentrations (Supplementary Fig. S16B–S16C). The synergistic effect of YUM70 and topotecan, vorinostat, and MG132 can be attributed to apoptosis enhancement. To test this hypothesis, we performed annexin V apoptosis assays (Figure 6F) and found an increase in early and late-stage apoptosis for each drug combination. Previous reports have shown that MG132 induces apoptosis by ER stress and subsequent activation of a mitochondria-dependent caspase cascade (46). Collectively, the reported study and our data suggest that the synergistic effect of these combinations was due to apoptosis enhancement. Such combinations can be further optimized for future clinical applications.
DISCUSSION
Despite recent advancements in the molecular, pathological, and biological understanding of pancreatic cancer, it remains a devastating disease with limited options for effective treatments. Thus, there is an urgent need to develop new therapies for this disease. In this study, we identified YUM70 as a lead compound showing in vitro cytotoxicity in both 2D and 3D pancreatic cancer culture systems and significant synergy with topotecan and vorinostat. Importantly, YUM70 showed significant anticancer activity in an in vivo pancreatic cancer xenograft model with no observed toxicity to normal tissues. However, the anticancer effect of YUM70 is modest as a single agent and normal tissue toxicity was measured for the treatment period only.
Mechanistically, we demonstrate that YUM70 suppresses proliferation and induces ER stress and apoptosis in pancreatic cancer cells. Acute ER stress leads to transient activation of the UPR signaling network to restore ER homeostasis. However, prolonged UPR activation promotes cell death by activating apoptosis. CHOP, a transcription factor known to be involved in ER stress-induced apoptosis, is distinctly overexpressed in response to ER stress through IRE1-, PERK- and ATF6-dependent transcriptional induction (47). These factors are activated by GRP78 when it dissociates from these ER sensors/receptors that in turn increase expression of GRP78. The induced expression of ATF4, CHOP, and GRP78, and phosphorylation of eIF2α following YUM70 treatment (Figures 2 and 3), strongly suggests induction of PERK/ATF4/CHOP mediated cell death. Bioinformatics analysis also supports these findings. Interestingly, YUM70 treatment increases the levels of the ER stress regulatory proteins DDIT4 and UPP1 as early as 2 hrs, suggesting a role for these proteins in ER stress initiation. YUM70 also increased the transcription of SLC1A4 and CHAC1 (glutathione-specific gamma-glutamylcyclotransferase 1). CHAC1 is a pro-apoptotic component of ER stress that has glutamylcyclotransferase activity specific for glutathione (GSH). GSH synthesis requires glycine, cysteine and glutamate. It has been reported that cystine-glutamate exchange leads to the activation of an ER stress response and upregulation of CHAC1 (29). SLC1A4 is a neutral amino acid transporter, selective for alanine, serine, and cysteine. Other reports also showed increased expression of CHAC1 and SLC1A4 upon ER stress (48). Together, these data suggest that YUM70 exacerbates ER stress leading to cell death (Figure 5G). These observations also suggest SLC1A4 may have a role in GSH transport. However, further studies are needed to better elucidate the role of CHAC1 and SLC1A4 under ER stress.
Previously, several small molecules were identified that are non-selective inhibitors of GRP78 (49,50). YUM70 is selective for GRP78 over other ER proteins including GSTO1, PDI (Supplementary Fig. S8G and S8H) and HSP70 (Supplementary Fig. S8I). We demonstrated in multiple complementary assays that YUM70 binds to recombinant and cellular GRP78. Importantly, using YUM70 as a warhead, we synthesized the first PROTAC to degrade GRP78 through a cereblon-mediated E3 ligase mechanism. Using LC-MS/MS-based proteomics, we confirmed the degradation of GRP78 (Supplementary Fig. S9H, and S9I) by our PROTAC. DX2–145 degrades GRP78 in a dose-dependent manner, and MG132 completely blocked its degradation. Pretreatment with parent compounds and pomalidomide decreased GRP78 degradation by DX2–145, but was unable to completely block degradation of GRP78 at the tested concentrations (Supplementary Fig. S10C and S10F). We did not observe an increased cytotoxicity of DX2–145 over YUM70, suggesting that our first-generation PROTAC needs further optimization. The potency of PROTAC can be improved by optimizing the ligand of E3 ubiquitin ligase, or the linker, or using a more potent warhead. Since YUM70 treatment increases GRP78 levels, a degradation strategy via a PROTAC will be a more efficient approach to target GRP78 in tumors.
Oncogenic mutations of KRAS, a key driver of tumor growth, are found in most pancreatic cancer patients. We observed that mutant KRAS pancreatic cancer cells are more susceptible to apoptosis induced by YUM70 than cells carrying wild-type KRAS. Similarly, mutant KRAS-driven colon cancer cells were sensitive to the inhibition of heat shock protein HSP90 and prone to ER stress (51). Previously, it was shown that KRAS-transfected cells have higher basal levels of ER-stress related gene expression than the wild-type cells (52). A possible reason may be that the activation of KRAS in pancreatic cancer increases protein synthesis resulting in constitutive ER stress, similar to BRAF oncogenic activation in melanoma (53). Importantly, Lee et al showed that 50% reduced expression of GRP78 is sufficient to inhibit tumor growth in KRAS-driven pancreatic cancer mouse models (18). These results are in agreement with prior reports that ER stress inducers sensitize KRAS mutant cells to apoptosis (51,52). Therefore, KRAS mutation can serve as a predictive biomarker to select patients who may benefit from treatment with GRP78 inhibitors or degraders. Our study suggest that inhibition or degradation of GRP78 can be a promising approach for the treatment of KRAS mutated pancreatic cancer.
YUM70 treatment significantly increased FAM129A protein levels in vitro and in vivo. Additionally, knockdown of GRP78 increased the expression of FAM129A. Although FAM129A is upregulated in prostate cancer and knockdown of FAM129A decreases cell growth in vitro, a recent study showed that FAM129A is a differentially expressed gene in pancreatic cancer (54). Previous reports and our study suggest that FAM129A positively regulates the ATF4- PERK-eIF2α axis in pancreatic cancer leading to apoptosis-mediated cell death. Thus, FAM129A could be used as a pharmacodynamic (PD) marker to evaluate the response to GRP78 inhibitors in pancreatic cancer.
Current evidence suggests that GRP78 haploinsufficiency has no major deleterious effect on organ homeostasis in young as well as aged mice (55) consistent with the notion that normal organ function requires only a low basal level of GRP78 for maintenance, while cancer cells require high levels of active GRP78 for growth, survival, invasion and therapeutic resistance. This is in agreement with our observation that YUM70 preferentially blocks the growth of cancer but not normal pancreatic cells. To our knowledge, YUM70 is among the first small molecule inhibitors that directly bind to GRP78, suppresses its ATPase activity, and causes its dissociation from the ER stress sensors, leading to activation of the UPR and apoptosis. Another compound, HA15 that targets GRP78 among other proteins, triggers ER stress and autophagy and overcomes BRAF inhibitor resistance in melanoma and other cancer cells (56). A ruthenium compound, KP1339 (IT-139) that inhibits GRP78 and disrupt ER homeostasis, is currently under Phase I clinical investigation (57). Thus, ER stress inducers and GRP78 inhibitors hold promise for the treatment of pancreatic cancer (58).
In this study, we observed a synergistic cell killing effect of YUM70 with topotecan, a topoisomerase I inhibitor, and vorinostat, an HDAC inhibitor. Topotecan induces apoptosis by p53 activation (59). It is currently used to treat small cell lung cancer and ovarian cancer (60) and is the first topoisomerase I inhibitor approved for oral use (Hycamtin Capsules, GlaxoSmithKline). Irinotecan, another topoisomerase I inhibitor, is part of the FOLFIRINOX regimen approved for the treatment of advanced pancreatic cancer. Therefore, combination of GRP78 inhibitors and topotecan could be a new treatment option for pancreatic cancer. Treatment strategies combining HDAC inhibitors with gemcitabine, radiation therapy, 5-FU or bortezomib have failed to improve survival outcomes. YUM70 in combination with vorinostat showed a promising synergistic cell killing effect in vitro. Although the underlying mechanism of this synergy was not assessed in this study, a strong similarity in molecular pathways was observed in CMap analysis (Supplementary Table S15). HDAC inhibitors (e.g. vorinostat) were reported to cause GRP78 acetylation that disrupts GRP78 function and induces ER stress (17). Moreover, previous studies showed that vorinostat also promotes cell cycle arrest, inhibits growth, and induces apoptosis in cancer (61) by ATF4 and CHOP upregulation (62). Thus, these novel combinations including YUM70 may improve pancreatic cancer treatment response.
In conclusion, we demonstrated that YUM70 treatment induces ER stress and triggers UPR by inhibiting GRP78. As a result, eIF2α is phosphorylated leading to the induction of CHOP and apoptosis in both cell culture and xenograft models. Importantly, YUM70 slowed tumor growth in a pancreatic cancer xenograft model. Although YUM70 is moderately effective as a single agent in pancreatic cancer, it can be safely combined with topotecan and vorinostat. YUM70 is an excellent tool compound to further interrogate the role of GRP78 inhibition in pancreatic cancer. Altogether, our study highlights GRP78 as a promising target to treat KRAS mutant pancreatic cancer and YUM70 as a novel anticancer agent that could be used in combination with select drugs to improve treatment efficacy and overcome drug resistance.
Supplementary Material
SIGNIFICANCE.
This study identifies a novel ER stress inducer that binds GRP78 and inhibits pancreatic cancer cell growth in vitro and in vivo, demonstrating its potential as a therapeutic agent for pancreatic cancer.
ACKNOWLEDGMENTS
We thank the In-vivo Animal Core (IVAC), Unit for Laboratory Animal Medicine, University of Michigan Medical School, for assistance in immunohistochemistry.
This study was supported by funding from NIH grant CA188252 (NN), the Department of Defense Breast Cancer Research Program W81XWH-11-10430 (NN, ASL), the University of Michigan Forbes Institute for Cancer Discovery (NN, ML), and from the L.K. Whittier Foundation (ASL).
Footnotes
DECLARATION OF INTERESTS: The authors declare no competing interests
REFERENCES
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21 [DOI] [PubMed] [Google Scholar]
- 2.Raufi AG, Manji GA, Chabot JA, Bates SE. Neoadjuvant treatment for pancreatic cancer. Semin Oncol 2019;46:19–27 [DOI] [PubMed] [Google Scholar]
- 3.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet 2016;388:73–85 [DOI] [PubMed] [Google Scholar]
- 4.Binet F, Sapieha P. ER stress and angiogenesis. Cell Metab 2015;22:560–75 [DOI] [PubMed] [Google Scholar]
- 5.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011;334:1081–6 [DOI] [PubMed] [Google Scholar]
- 6.Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov 2015;5:586–97 [DOI] [PubMed] [Google Scholar]
- 7.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 2013;32:805–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016;529:326–35 [DOI] [PubMed] [Google Scholar]
- 9.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 2005;115:2656–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002;415:92–6 [DOI] [PubMed] [Google Scholar]
- 11.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001;107:881–91 [DOI] [PubMed] [Google Scholar]
- 12.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 2000;6:1355–64 [DOI] [PubMed] [Google Scholar]
- 13.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer 2014;14:263–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem 2003;278:20915–24 [DOI] [PubMed] [Google Scholar]
- 15.Pyrko P, Schonthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res 2007;67:9809–16 [DOI] [PubMed] [Google Scholar]
- 16.Jiang CC, Chen LH, Gillespie S, Wang YF, Kiejda KA, Zhang XD, et al. Inhibition of MEK sensitizes human melanoma cells to endoplasmic reticulum stress-induced apoptosis. Cancer Res 2007;67:9750–61 [DOI] [PubMed] [Google Scholar]
- 17.Garg AD, Maes H, van Vliet AR, Agostinis P. Targeting the hallmarks of cancer with therapy-induced endoplasmic reticulum (ER) stress. Mol Cell Oncol 2015;2:e975089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen J, Ha DP, Zhu G, Rangel DF, Kobielak A, Gill PS, et al. GRP78 haploinsufficiency suppresses acinar-to-ductal metaplasia, signaling, and mutant Kras-driven pancreatic tumorigenesis in mice. Proc Natl Acad Sci U S A 2017;114:E4020–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke WR, Amundadottir L, James MA. CLPTM1L/CRR9 ectodomain interaction with GRP78 at the cell surface signals for survival and chemoresistance upon ER stress in pancreatic adenocarcinoma cells. Int J Cancer 2019;144:1367–78 [DOI] [PubMed] [Google Scholar]
- 20.Niu Z, Wang M, Zhou L, Yao L, Liao Q, Zhao Y. Elevated GRP78 expression is associated with poor prognosis in patients with pancreatic cancer. Sci Rep 2015;5:16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee E, Nichols P, Spicer D, Groshen S, Yu MC, Lee AS. GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res 2006;66:7849–53 [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Yang H, Abel EV, Ney GM, Palmbos PL, Bednar F, et al. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev 2015;29:171–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Tsan YC, et al. Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc Natl Acad Sci U S A 2013;110:2240–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramkumar K, Samanta S, Kyani A, Yang S, Tamura S, Ziemke E, et al. Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor. Nat Commun 2016;7:13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serrao E, Xu ZL, Debnath B, Christ F, Debyser Z, Long YQ, et al. Discovery of a novel 5-carbonyl-1H-imidazole-4-carboxamide class of inhibitors of the HIV-1 integrase-LEDGF/p75 interaction. Bioorg Med Chem 2013;21:5963–72 [DOI] [PubMed] [Google Scholar]
- 26.Maust JD, Frankowski-McGregor CL, Bankhead A 3rd, Simeone DM, Sebolt-Leopold JS. Cyclooxygenase-2 influences response to cotargeting of MEK and CDK4/6 in a subpopulation of pancreatic cancers. Mol Cancer Ther 2018;17:2495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartoszewski R, Gebert M, Janaszak-Jasiecka A, Cabaj A, Kroliczewski J, Bartoszewska S, et al. Genome-wide mRNA profiling identifies RCAN1 and GADD45A as regulators of the transitional switch from survival to apoptosis during ER stress. FEBS J 2020;287:2923–47 [DOI] [PubMed] [Google Scholar]
- 28.Scheffer D, Kulcsar G, Nagyeri G, Kiss-Merki M, Rekasi Z, Maloy M, et al. Active mixture of serum-circulating small molecules selectively inhibits proliferation and triggers apoptosis in cancer cells via induction of ER stress. Cell Signal 2020;65:109426 [DOI] [PubMed] [Google Scholar]
- 29.Crawford RR, Prescott ET, Sylvester CF, Higdon AN, Shan J, Kilberg MS, et al. Human CHAC1 protein degrades glutathione, and mRNA induction is regulated by the transcription factors ATF4 and ATF3 and a bipartite ATF/CRE regulatory element. J Biol Chem 2015;290:15878–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 2009;182:466–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao SJ, Gong Q, Chen XR, Ye LX, Ding Q, Zeng JS, et al. Netrin-1 rescues neuron loss by attenuating secondary apoptosis in ipsilateral thalamic nucleus following focal cerebral infarction in hypertensive rats. Neuroscience 2013;231:225–32 [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Chen XA, Hu J, Jiang JK, Li Y, Chan-Salis KY, et al. ATF4 gene network mediates cellular response to the anticancer PAD inhibitor YW3–56 in triple-negative breast cancer cells. Mol Cancer Ther 2015;14:877–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farooqi AA, Li KT, Fayyaz S, Chang YT, Ismail M, Liaw CC, et al. Anticancer drugs for the modulation of endoplasmic reticulum stress and oxidative stress. Tumour Biol 2015;36:5743–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saveljeva S, Cleary P, Mnich K, Ayo A, Pakos-Zebrucka K, Patterson JB, et al. Endoplasmic reticulum stress-mediated induction of SESTRIN 2 potentiates cell survival. Oncotarget 2016;7:12254–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A 1997;94:7245–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Su L, Zhong N, Hao X, Zhong D, Singhal S, et al. Salinomycin induces cell death with autophagy through activation of endoplasmic reticulum stress in human cancer cells. Autophagy 2013;9:1057–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Chen S, Liu H, Zhang Z, Ni Z, Chen J, et al. Tunicamycin specifically aggravates ER stress and overcomes chemoresistance in multidrug-resistant gastric cancer cells by inhibiting N-glycosylation. J Exp Clin Cancer Res 2018;37:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2000;2:326–32 [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Wey S, Zhang Y, Ye R, Lee AS. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid Redox Signal 2009;11:2307–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu S, Butkevich AN, Yamada R, Zhou Y, Debnath B, Duncan R, et al. Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc Natl Acad Sci U S A 2012;109:16348–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burslem GM, Smith BE, Lai AC, Jaime-Figueroa S, McQuaid DC, Bondeson DP, et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol 2018;25:67–77 e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassan AQ, Kirby CA, Zhou W, Schuhmann T, Kityk R, Kipp DR, et al. The novolactone natural product disrupts the allosteric regulation of Hsp70. Chem Biol 2015;22:87–97 [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Nune M, Zong Y, Zhou L, Liu Q. Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 2015;23:2191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurusinghe K, Mishra A, Mishra S. Glucose-regulated protein 78 substrate-binding domain alters its conformation upon EGCG inhibitor binding to nucleotide-binding domain: Molecular dynamics studies. Sci Rep 2018;8:5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pallmann N, Livgard M, Tesikova M, Zeynep Nenseth H, Akkus E, Sikkeland J, et al. Regulation of the unfolded protein response through ATF4 and FAM129A in prostate cancer. Oncogene 2019;38:6301–18 [DOI] [PubMed] [Google Scholar]
- 46.Park HS, Jun do Y, Han CR, Woo HJ, Kim YH. Proteasome inhibitor MG132-induced apoptosis via ER stress-mediated apoptotic pathway and its potentiation by protein tyrosine kinase p56lck in human Jurkat T cells. Biochem Pharmacol 2011;82:1110–25 [DOI] [PubMed] [Google Scholar]
- 47.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 2004;11:381–9 [DOI] [PubMed] [Google Scholar]
- 48.Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 2014;3:e02523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin S, Lamb HK, Brady C, Lefkove B, Bonner MY, Thompson P, et al. Inducing apoptosis of cancer cells using small-molecule plant compounds that bind to GRP78. Br J Cancer 2013;109:433–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen TC, Wang W, Golden EB, Thomas S, Sivakumar W, Hofman FM, et al. Green tea epigallocatechin gallate enhances therapeutic efficacy of temozolomide in orthotopic mouse glioblastoma models. Cancer Lett 2011;302:100–8 [DOI] [PubMed] [Google Scholar]
- 51.Wang CY, Guo ST, Wang JY, Liu F, Zhang YY, Yari H, et al. Inhibition of HSP90 by AUY922 preferentially kills mutant KRAS colon cancer cells by activating Bim through ER stress. Mol Cancer Ther 2016;15:448–59 [DOI] [PubMed] [Google Scholar]
- 52.Carew JS, Espitia CM, Zhao W, Kelly KR, Coffey M, Freeman JW, et al. Reolysin is a novel reovirus-based agent that induces endoplasmic reticular stress-mediated apoptosis in pancreatic cancer. Cell Death Dis 2013;4:e728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croft A, Tay KH, Boyd SC, Guo ST, Jiang CC, Lai F, et al. Oncogenic activation of MEK/ERK primes melanoma cells for adaptation to endoplasmic reticulum stress. J Invest Dermatol 2014;134:488–97 [DOI] [PubMed] [Google Scholar]
- 54.Tang Y, Zhang Z, Tang Y, Chen X, Zhou J. Identification of potential target genes in pancreatic ductal adenocarcinoma by bioinformatics analysis. Oncol Lett 2018;16:2453–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee AS, Brandhorst S, Rangel DF, Navarrete G, Cohen P, Longo VD, et al. Effects of prolonged GRP78 haploinsufficiency on organ homeostasis, behavior, cancer and chemotoxic resistance in aged mice. Sci Rep 2017;7:40919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerezo M, Lehraiki A, Millet A, Rouaud F, Plaisant M, Jaune E, et al. Compounds triggering ER stress exert anti-melanoma effects and overcome BRAF inhibitor resistance. Cancer Cell 2016;29:805–19 [DOI] [PubMed] [Google Scholar]
- 57.Bakewell SJ, Rangel DF, Ha DP, Sethuraman J, Crouse R, Hadley E, et al. Suppression of stress induction of the 78-kilodalton glucose regulated protein (GRP78) in cancer by IT-139, an anti-tumor ruthenium small molecule inhibitor. Oncotarget 2018;9:29698–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Carbonero N, Li W, Cabeza-Morales M, Martinez-Useros J, Garcia-Foncillas J. New hope for pancreatic ductal adenocarcinoma treatment targeting endoplasmic reticulum stress response: a systematic review. Int J Mol Sci 2018;19:2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parmakhtiar B, Burger RA, Kim JH, Fruehauf JP. HIF inactivation of p53 in ovarian cancer can be reversed by topotecan, restoring cisplatin and paclitaxel sensitivity. Mol Cancer Res 2019;17:1675–86 [DOI] [PubMed] [Google Scholar]
- 60.Thomas A, Pommier Y. Targeting topoisomerase I in the era of precision medicine. Clin Cancer Res 2019;25:6581–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zagni C, Floresta G, Monciino G, Rescifina A. The search for potent, small-molecule HDACIs in cancer treatment: a decade after vorinostat. Med Res Rev 2017;37:1373–428 [DOI] [PubMed] [Google Scholar]
- 62.Kikuchi S, Suzuki R, Ohguchi H, Yoshida Y, Lu D, Cottini F, et al. Class IIa HDAC inhibition enhances ER stress-mediated cell death in multiple myeloma. Leukemia 2015;29:1918–27 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.