Abstract
PI3Kα inhibitors have shown clinical activity in PIK3CA-mutated estrogen receptor-positive (ER+) breast cancer patients. Using whole genome CRISPR/Cas9 sgRNA knockout screens, we identified and validated several negative regulators of mTORC1 whose loss confers resistance to PI3Kα inhibition. Among the top candidates were TSC1, TSC2, TBC1D7, AKT1S1, STK11, MARK2, PDE7A, DEPDC5, NPRL2, NPRL3, C12orf66, SZT2 and ITFG2. Loss of these genes invariably results in sustained mTOR signaling under pharmacological inhibition of the PI3K-AKT pathway. Moreover, resistance could be prevented or overcome by mTOR inhibition, confirming the causative role of sustained mTOR activity in limiting the sensitivity to PI3Kα inhibition. Cumulatively, genomic alterations affecting these genes are identified in about 15% of PIK3CA-mutated breast tumors and appear to be mutually exclusive. This study improves our understanding of the role of mTOR signaling restoration in leading to resistance to PI3Kα inhibition and proposes therapeutic strategies to prevent or revert this resistance.
Keywords: CRISPR/Cas9 sgRNA knockout screen, drug resistance, PIK3CA, mTORC1, breast cancer
Introduction
The phosphoinositide 3-kinase (PI3K) pathway regulates many cellular processes, including cell growth, motility, survival, apoptosis and metabolism. Class IA PI3Ks consist of heterodimers of the p110 catalytic subunit and the p85 regulatory subunit. The p110 catalytic subunit isoforms are encoded by the PIK3CA, PIK3CB, and PIK3CD genes (p110α, p110β, and p110δ, respectively). Activation of PI3K catalyzes the phosphorylation of PI(4,5)P2 (PIP2) to PI(3,4,5)P3 (PIP3), which triggers downstream signaling and is tightly regulated by phosphatase and tensin homolog (PTEN) that converts PIP3 back to PIP2. Accumulation of PIP3 at the plasma membrane leads to the recruitment of the pleckstrin homology domain-containing proteins PDK1 and AKT. PDK1 phosphorylates AKT, which further activates mTORC1 (1). Mechanisms that hyperactivate this pathway include activating mutations of PIK3CA and PIK3CB, and AKT1, and loss-of-function alterations of PTEN, PIK3R1 (coding for p85), TSC1, TSC2, STK11, and other modulators of the mTOR complex (2-4).
As the most frequent genomic alterations in estrogen receptor-positive (ER+) breast cancer, inhibitors targeting PI3K p110α (PI3Kα) have shown promising clinical activity in patients with PIK3CA-mutated tumors, particularly when given in combination with endocrine therapy (5). This led to FDA approval of alpelisib in combination with the ER degrader fulvestrant in PIK3CA-mutated ER+ advanced breast cancer patients. However, clinical response to PI3K inhibitors is variable, with some patients achieving prolonged clinical benefit and others showing intrinsic resistance to therapy. While PIK3CA mutations are a confirmed biomarker of sensitivity to specific PI3Kα inhibitors (6,7), it is crucial to understand the molecular mechanisms underlying resistance to these agents and provide valid therapeutic strategies to overcome or delay such resistance.
We have previously shown that sustained activity of mTORC1 upon PI3K inhibition promotes resistance to these therapeutic agents in preclinical models and clinical samples (8). Known mechanisms to date include alternative phosphorylation of TSC2 by the kinase SGK1 (9), upregulation of Proviral Insertion site in Murine leukemia virus (PIM) kinases (10), or reactivation of the PI3K pathway through insulin growth factor receptor (IGF1R)/p110β (11,12). Other reports indicate that loss of PTEN (13,14), overexpression of FGFR1 (15) or ER (16,17) may also limit sensitivity to PI3Kα inhibitors. However, these reported mechanisms only partially explain why some breast cancer patients are or become resistant to PI3Kα inhibitors. In this work, we aimed to identify and characterize potential genetic lesions that limit the sensitivity to PI3Kα inhibitors by means of unbiased genome wide CRISPR screenings.
Materials and Methods
Cell lines, chemical reagents and plasmids
MCF7 (ATCC Cat# HTB-22, RRID: CVCL_0031) and T47D (ATCC Cat# HTB-133, RRID: CVCL_0553) cells were cultured in DMEM/F-12 (Corning) or RPMI-1640 (Corning) respectively, supplemented with 10% FBS and penicillin/streptomycin under normal oxygen conditions (5% CO2, 37 °C). 293T cells (ATCC Cat# CRL-3216, RRID: CVCL_0063) were cultured in DMEM (Corning) supplemented with 10% FBS under normal oxygen conditions (5% CO2, 37 °C). Cells within one month in culture were used for experiments. T47D alpelisib-tolerant or DMSO-tolerant cells were generated by chronic treatment with 1 μM alpelisib or DMSO for two months. All cells were tested for mycoplasma with Mycoplasma Detection Kit (Lonza).
BYL719, GDC0032, GDC0068, MK2206, AZD5363 and everolimus were purchased from Selleck Chemicals or MedChemExpress (MCE).
Cas9 expression construct (SVC9B-PS) and the whole genome sgRNA library (KOHGW-M123-P) were ordered from Cellecta. Individual sgRNAs (Table S1) were either designed using Benchling (http://www.benchling.com) or from the library and were ordered from Cellecta. Constructs expressing RagA (RRID: Addgene_73031), Rag D (RRID: Addgene_19316), RagD S77L (RRID: Addgene_19317), RagDQ121L (RRID: Addgene_19318) were ordered from Addgene. RagAT21L and RagAQ66L were generated using Q5 site-directed mutagenesis kit from NEB (E00554S) with primers listed in Table S1 following the manufacturer’s instruction and verified by DNA Sanger sequencing.
Lentivirus production
To generate the lentivirus of Cas9, the sgRNA library, individual sgRNAs or RagA and Rag D mutants, 293T cells were seeded into 15cm dishes 16 hours before transfection. For each dish, 3.5μg pMD2.G envelope vector, 7μg packaging vector pCMV-dR8.2, and 10.5μg plasmids of interest were added to 3mL jetPRIME buffer (Polyplus), and then 42μL jetPRIME transfection reagent was added. The mixture was incubated at room temperature for 10 min before being added to the cells. Medium was refreshed 6 hours post-transfection and the supernatant containing lentivirus was collected 48 hours post-transfection.
Stable cell lines generation
To obtain MCF7-Cas9 and T47D-Cas9 cells, MCF7 and T47D were transduced with Cas9 lentivirus produced as above with 8μg/mL of polybrene for 24 hours and then selected with 10μg/mL blasticidin (GIBCO) for 3 days.
To obtain MCF7 and T47D cells with the loss of target genes, MCF7-Cas9 or T47D cells were transduced with lentivirus of individual sgRNAs with 8μg/mL of polybrene for 24 hours and then selected with 2μg/mL puromycin (GIBCO) for 3 days.
RagA or RagD WT and mutant over-expressing cells were obtained by transducing parental or DEPDC5 deficient MCF7 and T47D cells with the corresponding lentivirus and selected with 2μg/mL puromycin (GIBCO) for 3 days.
Whole genome CRISPR-Cas9 knockout screen
The pooled sgRNA library comprises 150,000 individual sgRNAs targeting over 19,000 human protein-coding genes with 8 sgRNAs per gene. The sgRNA lentivirus library was generated as described above. Lentivirus titration was determined by a RT-qPCR based method according the manufacturer’s protocol (Lenti-X™ qRT-PCR Titration Kit, Cat# 631235, Clontech).
Next, lentivirus of the sgRNA library were added to 500 million MCF7- or T47D- Cas9 cells at a MOI~0.3 to reach 1000X coverage of the library. The library-transduced cells were subjected to either DMSO, alpelisib (1μM) or taselisib (60nM) treatment for 20 days. Surviving cells were pooled and genomic DNA was extracted using the Gentra Puregene kit according to the manufacturer’s protocol (Qiagen). Roughly 1000μg genomic DNA from each group was amplified by PCR to reach 600X coverage of the library and the sgRNA sequences were retrieved by sequencing the PCR products according to the manufacturer’s protocol (Cellecta, Cat# LNGS-120).
GSEA pathway enrichment analysis
Four gene lists of T47D-alpelisib_enriched, T47D-alpelisib_dropped out, MCF7-taselisib_enriched and MCF7-taselisib_dropped out were selected based on the following criteria: top 2.5% enriched or dropped-out sgRNAs (3745 sgRNAs) and at least 3 sgRNAs for each gene shown up. We conducted over-representation analysis for these four gene lists using hallmark gene set via MSigDB website and extracted the significant pathway with FDR<0.05. FDR was calculated in hypergeometric test after the correction for multiple hypothesis testing based on Benjamini and Hochberg.
Western immunoblotting
Briefly, cell lysates were prepared with RIPA lysis buffer supplemented with protease inhibitor (Roche) and phosphatase inhibitor (Thermo Scientific). Protein concentration was determined by the BCA assay (Pierce) according to the manufacturer’s protocol. Total proteins with sample buffer were separated by SDS-PAGE on 4–12% Bis-Tris gradient gels (Invitrogen) and transferred to NC membrane (Bio-Rad). SuperSignal west Pico Plus or Femto Maximum chemiluminescent detection reagents were used (ThermoFisher Scientific).
Primary antibodies used in this study include: Rabbit anti-Vinculin (Cell Signaling Technology Cat# 13901, RRID: AB_2728768), Rabbit anti-TSC1 (Cell Signaling Technology Cat# 4906, RRID: AB_220979), Rabbit anti-TSC2 (Cell Signaling Technology Cat# 4308, RRID: AB_10547134), Rabbit anti-TBC1D7 (Cell Signaling Technology Cat# 14949, RRID: AB_2749838), Rabbit anti-AKT1S1 (Cell Signaling Technology Cat# 2691, RRID: AB_2225033), Rabbit anti-DEPDC5 (Sigma-Aldrich Cat# SAB1302644, RRID: AB_2889910), Rabbit anti-NPRL2 (Cell Signaling Technology Cat# 37344, RRID: AB_2799113), Rabbit anti-NPRL3 (Sigma-Aldrich Cat# HPA011741, RRID: AB_1845577), Rabbit anti-MARK2 (Abcam Cat# ab133724, RRID: AB_2889911), Rabbit anti-PDE7A (Abcam Cat# ab154857, RRID: AB_2889912), Rabbit anti-STK11 (Abcam Cat# ab199970, RRID: AB_2889913), anti-pAKT(S473) (Cell Signaling Technology Cat# 4060, RRID: AB_2315049), anti-pS6K (T389) (Cell Signaling Technology Cat# 9205, RRID: AB_2734746), anti-pS6(S235/236) (Cell Signaling Technology Cat# 2211, RRID: AB_331679), anti-p4E-BP1(S65) (Cell Signaling Technology Cat# 9459, RRID: AB_330985).
Immunofluorescence
MCF7 WT and DEPDC5 KO cells or their derivatives expression empty vector, RagAT21L or RagAT21L/RagDQ121L were seeded on coverslips (ThermoFisher Scientific, 12CIR-1). After 24hrs, cells were treated with different conditions, including amino acid-starved, starved and followed by 5 mins amino acid stimulation, 4 hrs treatment of DMSO, 1 μM alpelisib or 20nM everolimus. Cells were then rinsed once with PBS and fixed in 4% paraformaldehyde for 10 mins at room temperature and permeabilized with 0.2% Triton X-100 in PBS for 10 mins. After two washes in PBS, cells were blocked with PBG (0.5% BSA and 0.2% gelatin from cold water fish skin in PBS) blocking solution for 1 hr, incubated with primary antibodies against mTORC1 (1:100, CST 2974) and LAMP2 (1:500, sc-18822) diluted in PBG for 1hr and incubated with goat anti-mouse Alex-647 or goat anti-rabbit Alex-488 secondary antibodies (1:500 in PBG) for 45 mins in a dark humid chamber. Cells were washed for 5 mins three times with PBG blocking solution Cells between each incubation. After staining with 0.2μg/ml DAPI for 1-2 mins, coverslips were washed in PBS twice and rinsed once in deionized water and finally mounted with Mowiol on glass slides and let dry in the dark overnight. 2 μm thick z-stack images were taken on a Zeiss LSM880 microscope with a 63x oil lens. At least 6 different fields from two independent experiments were scanned for each sample. Z-projections of the stacks were then analyzed using Fiji software, Coloc2 plugin. Approximately 50 or 100 cells per sample were analyzed.
Competitive cellular growth assay
MCF7 and T47D cells stably transduced with sgNT-GFP, sgNT-RFP or sgRNA-RFP for individual target genes were generated as described above. sgNT-GFP cells were mixed with sgNT-RFP or sgRNA-RFP cells (GFP+ and RFP+) at 1:1 ratio and RFP+/GFP+ ratios of each sample were measured as T0. sgNT-GFP cells and sgNT-RFP cells were used as control. 30,000 mixed MCF7 cells or 40,000 mixed T47D cells per well were seeded in 6-well plates. These cells were subjected to DMSO, 1μM alpelisib (BYL719) or 60nM taselisib (GDC-0032) for 6 days. RFP+/GFP+ ratios of each sample after treatment were measured by flow cytometry as T7. The change of RFP+/GFP+ ratio was scored as the impact of candidate gene KO on cell growth or drug response compared to sgNT control cells. The FlowJo Software was used for FACS data analysis.
Clonogenic-based cellular growth assay
40,000 MCF7 cells or 50,000 T47D cells per well were seeded into 12-well plates. Cells were treated with DMSO, 1μM alpelisib, 60nM taselisib, 2μM GDC0068, 2μM MK2206, 2μM AZD5363, or 20nM everolimus for 6 days. On day 7, cells were fixed with 3.7% formaldehyde and stained with 0.1% crystal violet.
CellTiter-Glo (CTG)-based cell viability assay
MCF7 cells or 4,000 T47D cells per well were seeded into 96-well plates. Cells were treated with various doses of DMSO, alpelisib, taselisib or everolimus for 4 days. Cell proliferation was determined by CellTiter-Glo assay (Promega) following the manufacturer’s protocol.
In vivo tumor xenograft
All mouse experiments were approved by RARC and IACUC at MSKCC. 0.72mg/90d-release oestrogen pellets were transplanted into 6-week-old female NSG mice 3 days prior to the tumor cell transplantation. 10 million T47D cells transduced with sgNT or sgTSC1 per mouse were orthotopically transplanted. When xenografts reach 100 mm3, mice were randomized to the alpelisib (25mg/kg p.o. QD, Monday to Friday) or untreated control groups. The tumor sizes were measured twice a week across the experiment.
TCGA and MSK datasets analysis
Datasets of TCGA breast invasive carcinoma (PanCancer Atlas) and MSK breast cancer (18) were available in the cBioPortal (http://cbioportal.org/). Genomic alterations of PIK3CA and the validated candidate genes in ER+ breast cancer patients within these two datasets were analyzed and visualized by using the cBioPortal following the tutorials (19,20). 294 PIK3CA-mutated ER+ breast cancer samples from TCGA and 443 PIK3CA-mutated ER+ breast cancer samples from MSK-IMPACT dataset were analyzed.
Mutation VCF files from Bertucci et al (21) were retrieved and annotated using the list of hotspots from Cancer Hotspots (cancerhotspots.org) (22). 129 cases harboring hotspot PIK3CA mutations were selected and mutations of the validated candidate genes were shown. Mutual exclusivity was calculated using WExT, a weighted exact test for mutual exclusivity (23).
Quantification and Statistical Analysis
All statistical analyses are shown in the appropriate method and figure legend.
Results
Genome-wide CRISPR KO screens identify regulators of PI3Kα inhibitor response
To investigate possible mediators of response to PI3Kα inhibitors, we have conducted genome-wide CRISPR/Cas9 sgRNA KO screens in PIK3CA-mutated ER+ MCF7 and T47D breast cancer cell lines, which are intrinsically sensitive to PI3Kα inhibition. The library-transduced cells were treated with either DMSO or a PI3Kα inhibitor (alpelisib or taselisib) for 20 days. The genomic DNA of each group was extracted, and the sgRNA sequences were amplified by PCR and retrieved by next-generation sequencing (NGS) (Fig. S1A). To systematically understand possible resistance mechanisms, we performed gene set enrichment analysis of our screen results using hallmark gene set and found that several pathways were significantly enriched in both cell lines (Table S2), such as MYC targets, mTORC1 signaling and PI3K-AKT-MTOR signaling.
To identify top candidate genes, the loss of which can cause resistance to PI3Kα inhibition, we used the following criteria: 1) the top 2.5% sgRNAs (3,745 out of 150,000 sgRNAs) enriched in the PI3Kα inhibitor group compared to the DMSO group (Tables S3 and S4), 2) each sgRNA had at least 150 reads in the untreated group (600 reads/sgRNA on average), and 3) at least 4 out of 8 sgRNAs targeting the same gene were enriched. The resulting lists of top candidates are shown in Tables S3 and S4.
Consistent with previous observations (12-14), PTEN is one of the top candidates mediating resistance to PI3Kα inhibition in both cell lines. Other candidates shared between MCF7 and T47D cells include TSC1, TSC2, DEPDC5 and NPRL2 (Fig. 1A-B). TSC1 and TSC2 are subunits of the TSC1-TSC2 complex, which integrates environmental signals such as stress, energy status and growth factors in mammals into TORC1 signaling. This complex is a critical negative regulator of mTORC1 by acting as a GTPase-activating protein (GAP) for the small GTPase RHEB (24,25). DEPDC5, NPRL2 and NPRL3 are components of the GATOR1 complex, which negatively regulates mTORC1 activity by acting as the GAP for RagA/B GTPases in the amino acids-sensing pathway (26). We further looked at other candidates involved in the mTORC1 pathway in either MCF7 or T47D cells. We found that STK11 (also known as LKB1), MARK2 and PDE7A in MCF7 cells, all potentially involved in the energy-sensing AMPK pathway (27-29). PRAS40, encoded by AKT1S1, an mTORC1 binding partner that mediates AKT signaling to mTOR (30,31), is another candidate in MCF7 cells. TBC1D7, a candidate found in T47D cells, is a subunit of the TSC1-TSC2 complex and maintains the association of the complex (32). NPRL3, the other component of the GATOR1 complex, and the KICSTOR complex components SZT2 and ITFG2, were also identified in T47D cells. The KICSTOR complex is required for the localization of the GATOR1 complex on the lysosome surface (33,34) (Fig. 1A-B). The other component of the KICSTOR complex C12orf66 was also included even though only 3 individual sgRNAs targeting C12orf66 were highly enriched in T47D cells (Table S2).
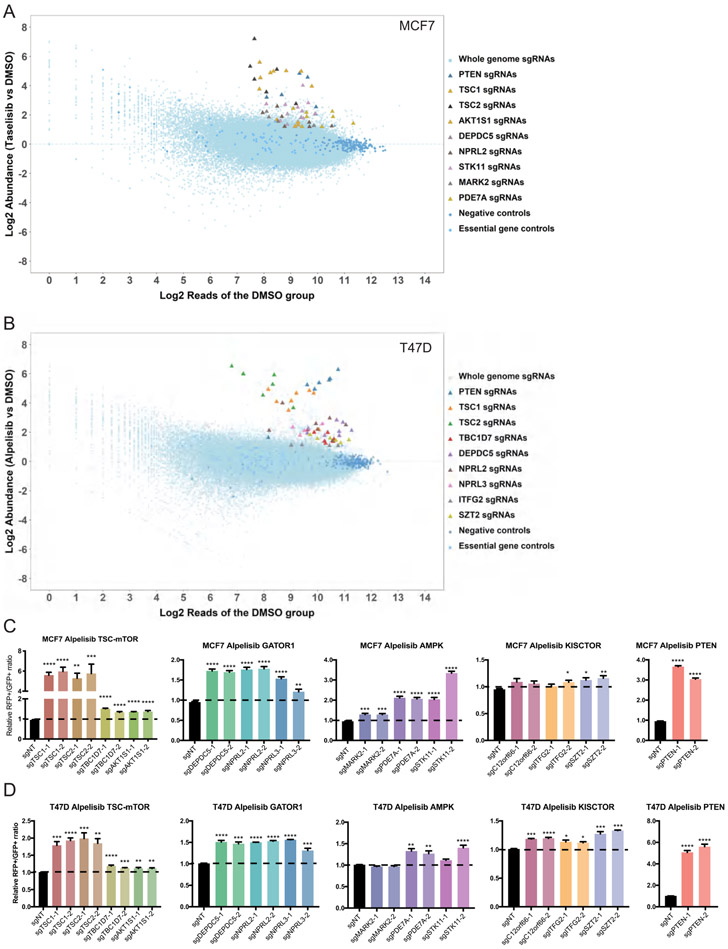
Figure 1. Genome-wide CRISPR-Cas9 KO screen identified components of the mTORC1 pathway mediate drug responses to PI3Kα inhibitor, alpelisib or taselisib, in MCF7 and T47D cells.
A. Plot of the candidate genes from CRISPR-Cas9 KO screen in MCF7 cells treated with 60nM taselisib. Top candidate genes are highlighted in the indicated colors.
B. Plot of the candidate genes from CRISPR-Cas9 KO screen in T47D cells treated with 1 μM alpelisib. Top candidate genes are highlighted in the indicated colors.
C and D. Validation results of 13 candidate genes in MCF7 (C) and T47D (D) cells by cell competition assay. The genes were presented based on their protein complexes. PTEN was used as a positive control. Cells were transduced with lentivirus of sgNT-GFP, sgNT-RFP or sgRNA-RFP targeting the candidate genes. Two sgRNAs for each gene were used. GFP and RFP labeled cells were mixed at a ratio of 1:1 and GFP-labeled cells were used as control. The mixed cells were treated with either DMSO or 1 μM alpelisib for 6 days. Ratio of RFP+/GFP+ cells in the alpelisib treated group was normalized to its DMSO treatment group. Relative RFP+/GFP+ ratios compared to control cell group were plotted. Mean and SEM are shown; n=4 experiments. P-values were calculated using Ordinary one-way ANOVA with Dunnett’s post-hoc Test. *, p<0.05; **, p<0.005; ***, p<0.0005; ****, p<0.0001.
To validate these findings, we generated MCF7 and T47D stable cell lines deficient for each of these candidates. We selected two sgRNAs with the best knockout efficiencies (Fig. S1B) and investigated whether knocking out each candidate gene would affect cell proliferation or drug response to PI3Kα inhibitors in competitive growth fitness assays (Fig. S1C). Our data shows that KO of each candidate gene did not affect per se cell proliferation in MCF7 or T47D cells, with the exceptions of TSC1 and TSC2 KO that significantly decreased cell growth in MCF7 cells (Fig. S1D-E). However, consistent with our screening data, we observed that loss of the shared candidates, the TSC1/TSC2 complex, the GATOR1 complex and the positive control PTEN, had the most dramatic effects on limiting the sensitivity to alpelisib and taselisib in both MCF7 and T47D cells (Fig. 1C-D, Fig. S2A-B). Most candidates were validated in both cell lines with significant effects in limiting the sensitivity to alpelisib or taselisib, with the exception of MARK2 in T47D cells or C12orf66 and ITFG2 in MCF7 cells (Fig. 1C-D, Fig. S2A-B). We then tested the in vivo sensitivity of TSC1-depleted MCF7-derived xenografts to alpelisib and found that loss of TSC1 was sufficient to reduce the antitumor efficacy of alpelisib (Fig. S2C). We also generated alpelisib-tolerant or DMSO-tolerant T47D cells by long-term treatment with 1μM alpelisib or DMSO for 2 months. Alpelisib-tolerant T47D cells were indeed resistant to increasing doses of alpelisib compared to DMSO-tolerant cells (Fig. S2D). More importantly, we found that DEPDC5 was dramatically decreased, and TSC2, STK11 and PDE7A were slightly decreased in alpelisib-tolerant T47D cells (Fig. S2E).
Altogether, our data identified and confirmed multiple candidate genes whose loss or reduced expression mediates resistance to PI3Kα inhibitors. These genes encode for negative regulators of mTORC1 activity, acting in different branches of the mTOR network.
Convergent mTORC1 activation dictates PI3Kα inhibitor response
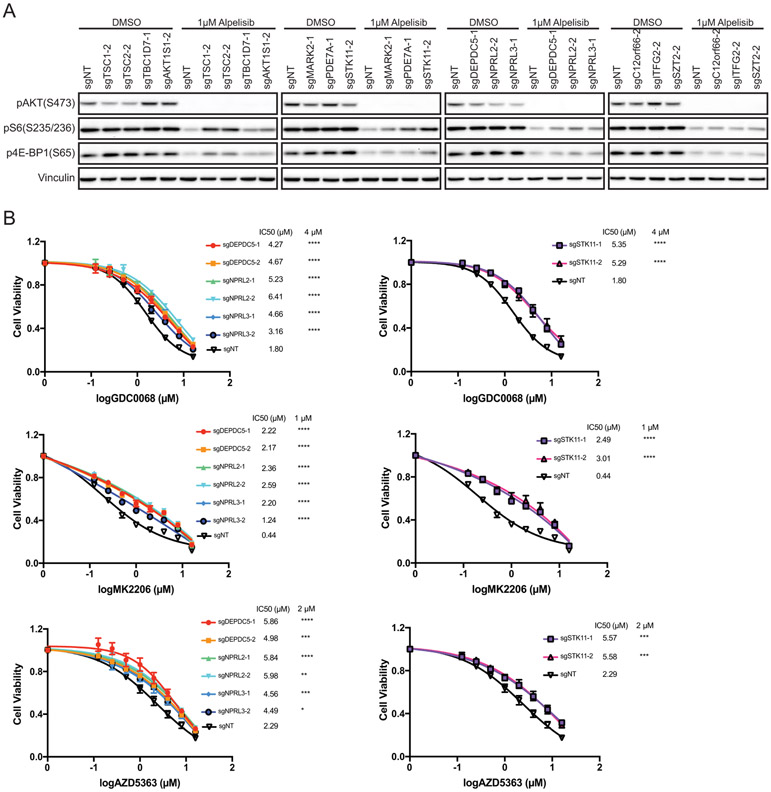
Given that these candidates are known negative regulators of mTORC1, it is conceivable that depletion of each candidate rewires mTORC1 signaling upon PI3Kα inhibition, thereby leading to the reduced antiproliferative effects of these therapeutic agents. In the absence of PI3Kα inhibition, we found that depletion of these candidates had minimal effects on the phosphorylation of the mTORC1 downstream effectors S6 ribosomal protein (S6) and 4E-BP1 compared to the control, while AKT phosphorylation (pAKT (S473)) was slightly suppressed upon loss of several candidates (Fig. 2A, Fig. S3A). These data suggest that the effects on mTORC1 caused by the loss of the candidate genes are likely compensated by a negative feedback loop that serves to restrain AKT phosphorylation (35-39). Upon alpelisib treatment, pAKT(S473) levels remained suppressed in both control and KO cells, suggesting that PI3Kα inhibition still blocks the PI3K-AKT cascade independent of the status of these genes.
Figure 2. Loss of each candidate gene maintains mTOR activity in response to PI3Kα inhibition.
A. MCF7 cells stably expressing sgNT or sgRNA targeting individual candidate genes were treated with DMSO or 1μM alpelisib for 4hrs. Cell lysates were analyzed by immunoblotting for the indicated proteins.
B. MCF7 cells stably expressing sgNT or sgRNAs targeting DEPDC5, NPRL2, NPRL3 or STK11 were treated with various doses of AKT inhibitors (GDC0068, MK2206 and AZD5363) for 4 days. Cell viability was determined by CellTiter-Glo assay and IC50 (μM) were shown. Mean and SEM are shown; n=3 experiments. P-values were calculated using two-way ANOVA with Tukey’s post-hoc tests and statistical significance for the selected doses were shown next to IC50 values.
As expected, mTORC1 downstream signaling was effectively inhibited by alpelisib treatment in cells transduced with non-targeting sgRNA (Fig. 2A, Fig. S3A). On the other hand, we observed a higher level of pS6 or p4E-BP1 in cells with individual candidate loss, especially in cells deprived of TSC1, TSC2, STK11 or the GATOR1 components, implying convergent activation of mTORC1 caused by loss of each of these candidates (Fig. 2A, Fig. S3A). These data indicate that loss of these individual genes upon treatment with PI3Kα inhibitors could result in sustained mTORC1 activity in an AKT-independent manner, which consequently limits the response to these agents. To challenge this hypothesis, we treated cells deprived of STK11 or the GATOR1 components with three different AKT inhibitors: AZD5363, GDC-0068 and MK-2206. Consistently, loss of each gene resulted in resistance to AKT inhibitors in both MCF7 and T47D cells (Fig. 2B, Fig. S3B).
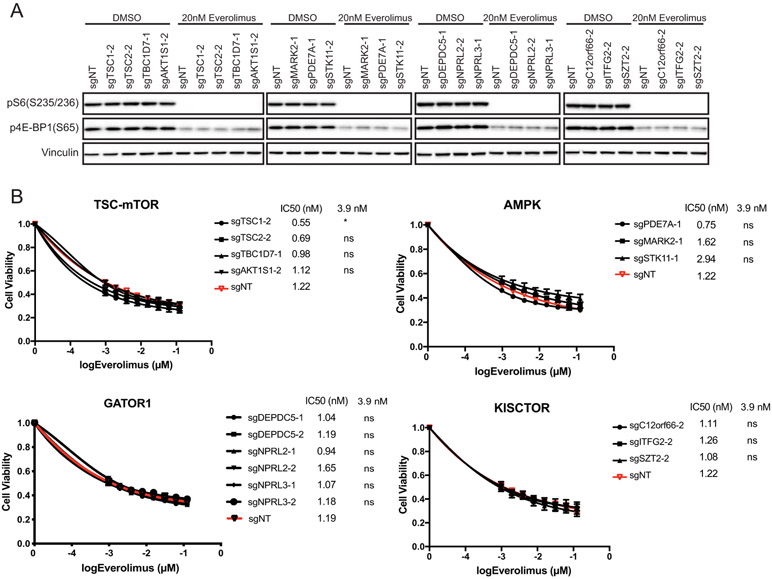
We then tested the sensitivity of each KO cell line to the mTORC1 inhibitor everolimus. As expected, everolimus effectively blocks mTORC1 signaling regardless of the status of these candidates (Fig. 3A, Fig. S4A) and elicits similar antiproliferative activity in both MCF7 and T47D cells (Fig. 3B, Fig. S4B). We found that everolimus re-sensitizes KO cells to PI3Kα inhibition by colony formation assays, underscoring the causative role of mTORC1 in limiting the effects of PI3Ra blockade (Fig. S4C).
Figure 3. mTOR inhibitor everolimus efficiently inhibits mTORC1 downstream signaling.
A. MCF7 cells stably expressing sgNT or sgRNA targeting individual candidate genes were treated with DMSO or 20nM everolimus for 4hrs. Cell lysates were analyzed by immunoblotting for the indicated proteins.
B. MCF7 cells stably expressing sgNT or sgRNAs targeting the individual candidate genes were treated with various doses of everolimus for 4 days. Cell viability was determined by CellTiter-Glo assay and IC50 (μM) were shown. Mean and SEM are shown; n=3 experiments. P-values were calculated using two-way ANOVA with Tukey’s post-hoc tests and statistical significance for the selected doses were shown next to IC50 values. ns, not significant.
In summary, our data revealed that loss of each candidate gene restores mTORC1 signaling independent of the PI3K-AKT axis upon PI3Kα inhibition, limiting the sensitivity to PI3Kα or AKT inhibitors.
The nutrient-sensing mTORC1 pathway affects PI3Kα inhibitor response
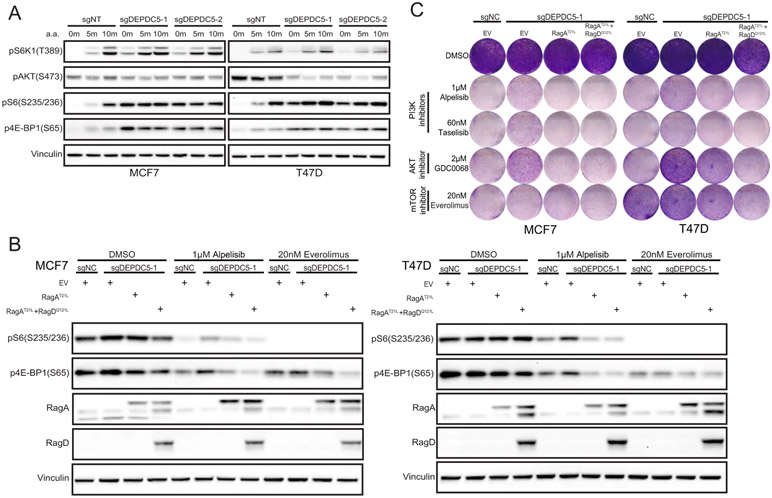
The GATOR1 (26) and KICSTOR (33,34) complexes are negative regulators of mTORC1 activity in the amino acid sensing pathway, which is a key branch within the mTOR network. Mutational inactivation or decreased expression levels of DEPDC5 or NPRL2 have been reported in glioblastoma, ovarian cancer, and gastrointestinal stromal cancer (26,40,41). Loss of GATOR1 complex, the GAP for RagA or RagB, via DEPDC5, NPRL2 or NPRL3 KO should increase levels of GTP-bound RagA or RagB and, therefore, lead to constitutive activation of mTOR (26). In line with the literature, DEPDC5 KO cells indeed displayed increased mTORC1 lysosomal localization (Fig. S5A) and mTORC1 downstream signaling (Fig. 4A), irrespective of the amino acid status, suggesting that DEPDC5 loss leads to an increase in the active form of RagA or RagB in MCF7 and T47D cells.
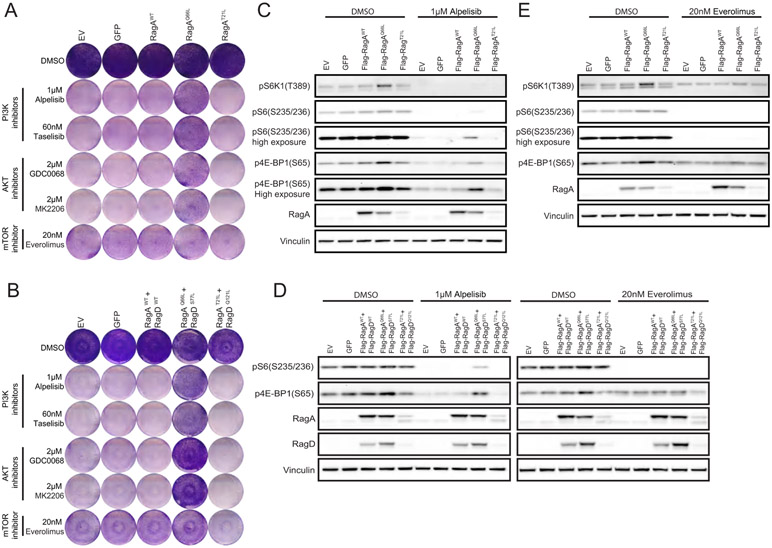
Figure 4. Introducing dominantly negative RagA or RagA/D heterodimer reverses DEPDC5 loss-induced drug resistance and mTORC1 activation.
A. WT and DEPDC5 KO MCF7 and T47D cells were starved for 6 hrs and stimulated with or without amino acids for 5 min or 10 min. Cell lysates were prepared and immunoblotted for the indicated proteins.
B. WT and DEPDC5 KO MCF7 (D) and T47D (E) cells stably expressing the indicated constructs were treated with DMSO, 1μM alpelisib or 20nM everolimus for 4 hrs. Cell lysates were prepared and immunoblotted for the indicated proteins.
C. WT and DEPDC5 KO MCF7 and T47D cells stably expressing the indicated constructs were treated with DMSO, 1μM alpelisib, 60nM taselisib, 2μM GDC0068 or 20nM everolimus for 6 days and stained with crystal violet. One representative result out of two biologically independent experiments is shown.
We next explored whether dominant negative Rag GTPases could reverse DEPDC5 loss induced drug resistance and mTORC1 restoration. We overexpressed dominant-negative RagA GTPase, RagAT21L, or dominant-negative heterodimer, RagAT21L/RagDQ121L, in DEPDC5-deficient MCF7 and T47D cells. We found that expression of RagAT21L or RagAT21L/RagDQ121L reduced mTORC1 lysosomal localization (Fig. S5B) and mTORC1 activity in DEPDC5 depleted cells (Fig. 4B), and reversed DEPDC5 loss-mediated drug resistance to PI3Kα and AKT inhibitors (Fig. 4C, Fig. S5C). Overexpression of RagAT21L alone was sufficient to block mTORC1 activation in DEPDC5-deficient cells (Fig. 4C), indicating that the deregulation of RagA or RagB GTPase induced by DEPDC5 depletion could be responsible for these phenotypes.
To further determine the impact of deregulated Rag GTPases on drug sensitivity to PI3Kα, AKT or mTOR inhibitors, we over-expressed different forms of Rag A or RagA/RagD heterodimer in MCF7 and T47D cells. Over-expression of WT RagA or RagA/D did not change drug sensitivity for any of the inhibitors tested. Conversely, over-expression of active form of RagA, RagAQ66L or RagA/D heterodimer, RagAQ66L/RagDS77L, mimicked DEPDC5 depletion-induced responses and conferred resistance to PI3Kα and AKT inhibitors but not to mTOR inhibitor (Fig. 5A-B, Fig. S6A-C). In the absence of inhibitors, changes in mTORC1 signaling were not significant upon overexpression of different forms of Rag GTPases. However, RagAQ66L and RagAQ66L/RagDS77L overexpression allows sustained mTOR signaling upon PI3Kα inhibition (Fig. 5C-D, Fig.S6D-E), while mTOR inhibition effectively blunts the signaling (Fig. 5D-E, Fig.S6E-F).
Figure 5. Over-expression of constitutively active RagA or RagA/D heterodimer mimics DEPDC5 loss-induced drug resistance and mTORC1 activation.
A and B. T47D cells expressing different forms of RagA (A) or RagA/D heterodimers (B) were treated with DMSO, 1μM alpelisib, 60nM taselisib, 2μM GDC0068, 2μM MK2206 or 20nM everolimus for 6 days and stained with crystal violet. One representative result out of two biologically independent experiments is shown.
C and D. T47D cells expressing different forms of RagA were treated with DMSO, 1μM alpelisib (C) or 20nM everolimus (D) for 4 hrs. Cell lysates were prepared and immunoblotted for the indicated proteins.
E. T47D cells expressing different forms of RagA/D heterodimers were treated with DMSO, 1μM alpelisib or 20nM everolimus for 4 hrs. Cell lysates were prepared and immunoblotted for the indicated proteins.
Genomic lesions of the candidates were identified in primary and metastatic PIK3CA-mutated ER+ breast tumors
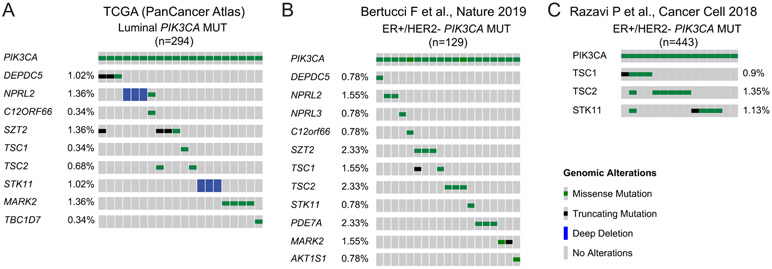
In order to study the prevalence of genomic lesions affecting the mTORC1 related genes that were found in our CRISPR screen, we first analyzed 294 PIK3CA-mutated luminal breast cancer patient samples from the Cancer Genome Atlas (TCGA) breast invasive carcinoma database (PanCancer Atlas). Truncating mutations or homozygous deletions (predicting to lead to loss of function) were observed for DEPDC5, NPRL2, SZT2 and STK11 (Fig. 6A). We then analyzed an additional cohort of 617 metastatic breast cancer samples from Bertucci et al. (21), of which 381 samples were from ER+ tumors. Of these, 129 harbored hotspot mutations in PIK3CA and, therefore, were potentially amenable to pharmacological PI3Kα inhibition. We found mutations of SZT2, TSC2, TSC1, DEPDC5, STK11, MARK2, PDE7A, NPRL3, NPRL2, AKT1S1 and C12orf66 in cumulatively 14.7% of PIK3CA mutant samples (Fig. 6B). Importantly, we observed mutual exclusivity of these genes (p = 0.0429, WexT).
Figure 6. Genomic alterations of the validated candidate genes in PIK3CA-mutated ER+ breast cancer patients.
A. Mutation heatmap was generated using cBioPortal and showing homozygous deletions and mutations of the validated candidate genes in PIK3CA-mutated luminal breast invasive carcinoma (n=294) (TCGA, PanCancer Atlas).
B. Mutation heatmap of the validated candidate genes in metastatic PIK3CA hotspot mutant samples from Bertucci et al (n=129).
C. Oncoprint were generated using cBioPortal and showing homozygous deletions and mutations of TSC1, TSC2 and STK11 in metastatic PIK3CA-mutated ER+/HER2− metastatic breast cancer patients (n=443) from the curated MSK breast cancer cohort dataset.
We then analyzed a curated cohort of 1,918 tumors from 1,756 breast cancer patients, in which 443 samples are ER+/HER2− breast cancers with PIK3CA mutations (18,42). In this cohort, genomic alterations of more than 400 cancer-associated genes were identified using the FDA-authorized Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT) platform (43). TSC1, TSC2 and STK11 are included in the list of sequenced genes. In line with the findings in the metastatic setting, we found that about 3% of PIK3CA-mutated ER+ patients carried genomic alterations of TSC1, TSC2 or STK11 (~1% for each gene, Fig. 6C)
These results suggest that genomic alterations involving the genes that potentially affect mTOR activity are present in a significant proportion of primary and metastatic PIK3CA-mutant breast tumors and may play a role in limiting the sensitivity to PI3Kα inhibition.
Discussion
Gain-of-function mutations in the PIK3CA gene are the most frequent genomic alterations in ER+ breast cancer, occurring in over 40% of cases (6,18,44). Tumors bearing these mutations are sensitive to specific PI3Kα inhibitors, particularly when given in combination with endocrine therapy (5,7,45). However, clinical responses to PI3Kα inhibitors are variable and often short-lived, suggesting that many PIK3CA-mutated ER+ breast cancer patients harbor or acquire alterations conferring resistance to PI3Kα inhibitors.
In our study, we employed an unbiased approach to identify a list of genes whose loss may affect sensitivity to PI3Kα inhibitors. Most of these genes encode components of different complexes that negatively regulate mTORC1 activity in different signaling branches of the mTOR network. Interestingly, in the absence of PI3Kα inhibitors, the presence of these genomic aberrations was insufficient to change mTORC1 downstream signaling, likely due to the activation of feedback mechanisms on AKT phosphorylation that maintained mTORC1 activity at a normal level (35-39). When the PI3K/AKT pathway was inhibited in the presence of PI3Kα inhibitors, however, loss of these negative regulators activated mTORC1 via the regulation of other parallel and/or interconnected pathways in the mTORC1 network. Our data highlights that this activation of mTORC1 is AKT-independent and suggests that loss-of-function alterations of these genes will allow a constitutively sustained activity of mTOR signaling in the presence of PI3Kα or AKT inhibitors, limiting their antitumor activity.
Genomic lesions of these identified genes were observed in different datasets of PIK3CA-mutated ER+ breast cancers. In one dataset of 129 PIK3CA-mutated ER+ breast cancer (21), 14.7% samples were found to harbor alterations involving SZT2, TSC2, TSC1, DEPDC5, STK11, MARK2, PDE7A, NPRL3, NPRL2, AKT1S1 or C12orf66, suggesting that, collectively, alterations of these genes are relatively frequent.
mTORC1 activity is sophisticatedly regulated and tailored in responding to various upstream stimuli including growth factors, energy, oxygen and amino acids. Known molecular mechanisms to date that can cause mTORC1 activation and drive resistance to PI3Kα inhibitors are mainly signaling through the tuberous sclerosis complex and the small Ras-related GTPase Rheb (9-13). Although recent studies have greatly progressed our understanding about the activation of mTORC1 by amino acids (26,33,34,40,41,46), a correlation between the amino acids-sensing pathway and cancer drug resistance has never been reported. In this work, we have dissected the role of the amino acids-sensing mTORC1 pathway in limiting the sensitivity to pharmacological PI3Kα or AKT blockade. As a critical regulator of amino acids-sensing pathway in the mTORC1 network, the GATOR1 complex regulates the GTP/GDP-bound status of RagA/B in the Rag heterodimers and determines the localization and activity of mTORC1. Consistent with a previous study (26), we found that loss of GATOR1 complex upon DEPDC5 KO resulted in high level of mTORC1 signaling irrespective of nutrient status in ER+ breast cancer cell lines, suggesting that DEPDC5 loss leads to the increase of active form of RagA/B, thereby leading to constitutive activation of mTORC1. Indeed, overexpression of dominant-negative RagA GTPase or dominant-negative RagA/D heterodimer blunts the mTORC1 activation in DEPDC5-deficient cells and reverses DEPDC5 loss-induced PI3Kα inhibitor resistance, indicating that the deregulation of RagA or RagB GTPase induced by DEPDC5 KO is responsible for these phenotypes.
A significantly higher response rate and a longer median PFS duration have been observed in patients with advanced-stage ER+, HER2-negative breast cancer treated with everolimus in combination with the aromatase inhibitor exemestane compared to placebo plus exemestane (47). However, the clinical efficacy of this treatment has proven suboptimal, at least in part due to dose-limiting toxicities (4,48,49). Thus, there is an unmet clinical need of identifying patients who are more likely to respond to mTOR inhibitors to rationalize the use of these agents. Pioneering work in this field reported that TSC1 loss-of-function mutations in bladder cancer predicted for everolimus sensitivity (3). More recent work suggested that lack of expression of PTEN and mutations in TSC1/2 and MTOR are enriched in renal cell carcinoma patients responding to everolimus (50). Thus, based on our data, we can hypothesize that breast cancer patients with tumors bearing one or more genomic lesions affecting mTOR activity may benefit from the treatment of everolimus or other rapalogs.
In summary, this study dissects potential mechanisms for sustained mTOR activity in the presence of PI3Kα inhibitors, leading to resistance to these agents. Our findings suggest that the identification of a specific subpopulation bearing loss of function mutations in negative regulators of mTORC1 may experience a limited response to PI3Kα inhibition and would benefit by concomitant mTOR blockade to prevent or delay drug resistance.
Supplementary Material
Significance.
Findings show that genetic lesions of multiple negative regulators of mTORC1 could limit the efficacy of PI3Kα inhibitors in breast cancer, which may guide patient selection strategies for future clinical trials.
Acknowledgements
We would like to thank all the Scaltriti lab members for their advice and support. This work has been supported by NIH grants P30 CA008748 and R01 CA190642-01A1, the Breast Cancer Research Foundation, the Geoffrey Beene Cancer Research Center (M. S. and S. C.). This work was also supported by grants from Stand Up to Cancer (Cancer Drug Combination Convergence Team) grant no. SU2C 2015-004, the V Foundation grant no. D2015-036 and the National Science Foundation grant no. PHY-1545853 (G.X. and M. Scaltriti). E.L. is supported by NCI grant no. K00CA212478. F.A. is supported by BCRF and foundation ARC. M.S. and S.C. are supported by a kind gift from Mrs. Barbara Smith.
Footnotes
Conflicts of Interest Statement
M.S. has received funds from Puma Biotechnology, AstraZeneca, Daiichi-Sankio, Immunomedics, Targimmune and Menarini Ricerche, is a cofounder of Medendi.org and is an employee of AstraZeneca. P.R. has received consultation fees from Novartis, AstraZeneca, Foundation Medicine, Inivata, Tempus, Neetera, Epic Sciences and institutional research funds from Grail Inc, Illumina, Novartis, and Epic Sciences. J.S.R.-F. is a consultant of Goldman Sachs and REPARE Therapeutics, a member of the Scientific Advisory Board of VolitionRx and Paige.AI, and an ad hoc member of the Scientific Advisory Board of Ventana Medical Systems, Roche, Genentech, Novartis and InviCRO. S.C. has performed consulting for Novartis, Lilly, and Paige.ai and receives institutional research support from Daiichi Sankyo, Sanofi, Novartis, and Lilly. K.J. has performed consulting for AstraZeneca, Novartis, Pfizer, Genentech, Lilly Pharmaceuticals, Taiho Oncology, AbbVie, Jounce Therapeutics, and received institutional research funding from Novartis, Genentech, Pfizer, AstraZeneca, Lilly Pharmaceuticals, Debio Pharmaceuticals, Zymeworks, Novita Pharmaceuticals, PUMA Biotechnology, Immunomedics. F.A. received research funding and served as speaker/advisor (compensated to the hospital) for Roche, AstraZeneca, Daiichi Sankyo, Pfizer, Novartis and Lilly. The other authors declare no competing interests.
References
- 1.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 2006;7:606–19 [DOI] [PubMed] [Google Scholar]
- 2.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov 2014;4:554–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 2012;338:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 2018;15:273–91 [DOI] [PubMed] [Google Scholar]
- 5.Juric D, Janku F, Rodon J, Burris HA, Mayer IA, Schuler M, et al. Alpelisib Plus Fulvestrant in PIK3CA-Altered and PIK3CA-Wild-Type Estrogen Receptor-Positive Advanced Breast Cancer: A Phase 1b Clinical Trial. JAMA Oncol 2019;5:e184475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med 2019;380:1929–40 [DOI] [PubMed] [Google Scholar]
- 7.Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science 2019;366:714–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, et al. mTORC1 inhibition is required for sensitivity to PI3K p110alpha inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med 2013;5:196ra99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ, et al. PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kalpha Inhibition. Cancer Cell 2016;30:229–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le X, Antony R, Razavi P, Treacy DJ, Luo F, Ghandi M, et al. Systematic Functional Characterization of Resistance to PI3K Inhibition in Breast Cancer. Cancer Discov 2016;6:1134–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leroy C, Ramos P, Cornille K, Bonenfant D, Fritsch C, Voshol H, et al. Activation of IGF1R/p110beta/AKT/mTOR confers resistance to alpha-specific PI3K inhibition. Breast Cancer Res 2016;18:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, et al. Feedback suppression of PI3Kalpha signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kbeta. Cancer Cell 2015;27:109–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 2015;518:240–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razavi P, Dickler MN, Shah PD, Toy W, Brown DN, Won HH, et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nature Cancer 2020;1:382–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drago JZ, Formisano L, Juric D, Niemierko A, Servetto A, Wander SA, et al. FGFR1 Amplification Mediates Endocrine Resistance but Retains TORC Sensitivity in Metastatic Hormone Receptor-Positive (HR(+)) Breast Cancer. Clin Cancer Res 2019;25:6443–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med 2015;7:283ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 2017;355:1324–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018;34:427–38 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature 2019;569:560–4 [DOI] [PubMed] [Google Scholar]
- 22.Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov 2018;8:174–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leiserson MD, Reyna MA, Raphael BJ. A weighted exact test for mutually exclusive mutations in cancer. Bioinformatics (Oxford, England) 2016;32:i736–i45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Molecular cell 2003;11:1457–66 [DOI] [PubMed] [Google Scholar]
- 25.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes & development 2003;17:1829–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013;340:1100–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee R, Wolda S, Moon E, Esselstyn J, Hertel C, Lerner A. PDE7A is expressed in human B-lymphocytes and is up-regulated by elevation of intracellular cAMP. Cell Signal 2002;14:277–84 [DOI] [PubMed] [Google Scholar]
- 28.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 2004;23:833–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004;6:91–9 [DOI] [PubMed] [Google Scholar]
- 30.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 2007;9:316–23 [DOI] [PubMed] [Google Scholar]
- 31.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Molecular cell 2007;25:903–15 [DOI] [PubMed] [Google Scholar]
- 32.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell 2012;47:535–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng M, Yin N, Li MO. SZT2 dictates GATOR control of mTORC1 signalling. Nature 2017;543:433–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfson RL, Chantranupong L, Wyant GA, Gu X, Orozco JM, Shen K, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 2017;543:438–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol 2009;29:5657–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 2004;166:213–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011;332:1317–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 2006;66:1500–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011;332:1322–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Ou Y, Yang Y, Li W, Xu Y, Xie Y, et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 2018;557:585–9 [DOI] [PubMed] [Google Scholar]
- 41.Pang Y, Xie F, Cao H, Wang C, Zhu M, Liu X, et al. Mutational inactivation of mTORC1 repressor gene DEPDC5 in human gastrointestinal stromal tumors. Proc Natl Acad Sci U S A 2019;116:22746–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD 2015;17:251–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res 2005;65:10992–1000 [DOI] [PubMed] [Google Scholar]
- 45.Mayer IA, Abramson VG, Formisano L, Balko JM, Estrada MV, Sanders ME, et al. A Phase Ib Study of Alpelisib (BYL719), a PI3Kalpha-Specific Inhibitor, with Letrozole in ER+/HER2− Metastatic Breast Cancer. Clin Cancer Res 2017;23:26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baselga J, Campone M, Piccart M, Burris HA 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 2012;366:520–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell 2017;170:605–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janku F. Phosphoinositide 3-kinase (PI3K) pathway inhibitors in solid tumors: From laboratory to patients. Cancer Treat Rev 2017;59:93–101 [DOI] [PubMed] [Google Scholar]
- 50.Roldan-Romero JM, Beuselinck B, Santos M, Rodriguez-Moreno JF, Lanillos J, Calsina B, et al. PTEN expression and mutations in TSC1, TSC2 and MTOR are associated with response to rapalogs in patients with renal cell carcinoma. Int J Cancer 2020;146:1435–44 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.