Abstract
Lung carcinogenesis is a complex and stepwise process involving accumulation of genetic mutations in signaling and oncogenic pathways via interactions with environmental factors and host susceptibility. Tobacco exposure is the leading cause of lung cancer, but its relationship to clinically relevant mutations and the composite tumor mutation burden (TMB) has not been fully elucidated. In this study, we investigated the dose-response relationship in a retrospective observational study of 931 patients treated for advanced stage non-small cell lung cancer (NSCLC) between April 2013 and February 2020 at the Dana Farber Cancer Institute and Brigham and Women’s Hospital. Doubling smoking pack-years was associated with increased KRASG12C mutations and less frequent EGFRdel19 and EGFRL858R mutations, while doubling smoking-free months was associated with more frequent EGFRL858R. In advanced lung adenocarcinoma, doubling smoking pack-years was associated with an increase in TMB, while doubling smoking-free months was associated with a decrease in TMB, after controlling for age, gender and stage. There is a significant dose-response association of smoking history with genetic alterations in cancer-related pathways and tumor mutation burden in advanced lung adenocarcinoma.
Keywords: tumor mutation burden, non-small cell lung cancer, tobacco use, EGFR, KRAS, biomarker
Introduction
Smoking is associated with specific genetic changes that give rise to histologically distinct lung cancers. The consistent effects of smoking on the lung cancer genome are well documented in a few oncogenes and oncogenic drivers. EGFR, KRAS and TP53 are the three most frequently mutated genes in Non-small cell lung cancer (NSCLC), with mutation incidence of up to 50% in different patient populations.
To date, most studies have focused on the relationship between smoking status and oncogenic drivers. However, lung carcinogenesis is a complex and stepwise process involving acquisition of multiple genetic mutations through interactions with environmental factors and host susceptibility (1). The incorporation of clinically relevant target sequences into Next Generation Sequencing (NGS) panels enables a more comprehensive characterization of the cancer genome alterations and provides more possibilities for individualized cancer‐patient care, especially in advanced-stage lung cancer (2). The genetic alterations can be analyzed individually or by pathways. Moreover, the number of tumor mutations per megabase found in clinically relevant genes, known as Tumor Mutation Burden (TMB), is emerging as a potential predictive biomarker for the response to immune checkpoint inhibitors (ICIs) (3). However, prolonged turnaround time, high expense for TMB assessment and variations across platforms and assays limit its standardization and widespread use. (4,5)
Molecular epidemiologic studies have examined the qualitative impact of smoking on genomic changes in NSCLC. These analyses were conducted based on the assumption that the association is constant within each smoking category conditional on relevant covariates (6). However, detailed smoking information has not yet been fully utilized through categorization of continuous variables (7). Difficulty in data collection has limited studies of quantitative effect of smoking history on somatic mutations. A dose-response analysis is needed to quantify the effect of smoking history as a continuous variable rather than simply designating patients into never/ever smokers. With more detailed information on smoking, comprehensive genomic change assessments and delicate data on patient outcomes, we investigated the dose-dependent association in a group of 931 advanced NSCLC patients with prospectively collected detailed smoking histories and clinical NGS genetic profiling of 275–447 genes from April 2013 and February 2020. Specifically, we investigated 1) how smoking metrics impact the likelihood of EGFR and KRAS mutations at both the gene and variant-specific level; 2) how smoking metrics impact the individual mutations in 10 cancer-related pathways; 3) and the dose-response relationship between smoking history and TMB.
Material and Methods
Clinical samples/patients
Demographic and clinical data including age at diagnosis, gender, ethnicity, histological subtypes, stage and smoking metrics were prospectively collected from patients with informed written consent to a correlative research study (DF/HCC protocol #02–180). We identified advanced-stage NSCLC patients (stage III or IV) whose tumors underwent successful targeted NGS between April 2013 and February 2020, at the DFCI and BWH (8). Smoking history were obtained from patients and recorded in the Thoracic Oncology Basic Assessment of Cancer and Clinical Outcomes (TOBACCO) (9). Smoking status included never smokers (< 100 cigarettes), former smokers (quit > 12 months before diagnosis) and current smokers (quit < 12 months before diagnosis or currently still smoking). Smoking pack-years, defined as packs/day (1 pack = 20 cigarettes) * years of smoking, was directly extracted from TOBACCO. Smoking-free months were calculated as from smoking cessation to diagnosis in ever smokers.
Mutation detection/OncoPanel
Sample collection and DNA extraction was performed as previously described (10). OncoPanel was designed for detection of Single Nucleotide Variants (SNV), small Insertions and Deletions (InDel), Copy Number Variation (CNV), and Structural Variant (SV) to guide treatment selection. There are three versions of OncoPanel including 275, 300, and 447 genes. OncoPanel was only conducted on tumor-derived samples. However, a series of systematic filtering procedure was conducted to remove the potential polymorphisms based on the allele frequency at the population level of greater than 0.1% in the Exome Sequencing Project (ESP) database (RRID: SCR_012761) and on an in-house panel of control samples. Details of the bioinformatic analysis and filtering procedures can be found in previous studies (10,11). TMB was defined as the number of somatic, nonsynonymous, SNV and small InDel mutations per megabase (Mb) of genome examined; TMB was calculated from the DFCI OncoPanel NGS platforms as previously described (8,10).
Statistical Analysis
Patients were classified based on smoking status, smoking pack-years and smoking-free months. In ever smokers, smoking pack-years and quit date were identified and follow-up time since smoking cessation was assigned. Categorical smoking pack-years were based on tertiles (never smokers, 1–19 [PYs], 20–39 [PYs] and > 40 [PYs]) and categorical smoking-free months were based on quartiles (0–4 [mo], 4–178 [mo], 178–364 [mo], > 364 [mo] (30.3 yrs) in ever smokers. Categorical and continuous variables were summarized descriptively using proportions and medians. Differences between continuous variables were tested using the Wilcoxon-Rank Sum test and Fisher’s exact test was utilized to test associations between categorical variables. Genomic landscape were discovered using R software, version 3.6.1 and R package maftools (12).
Base 2 log transformation was used for TMB, smoking pack-years and smoking-free months to meet the linearity assumption and to facilitate easy interpretation (Supplementary Figs. S1–3). Since smoking status was defined based on smoking-free months, only one of them was included in the analysis to avoid collinearity. Because smoking-free months and smoking pack-years are partly dependent, we conducted an adjusted analysis to examine the effect of these two parameters in ever smokers in our cohort.
The correlation between smoking metrics and mutations in cancer-related pathways was evaluated using logistic regression. Adjusted analysis was similarly conducted after controlling for age at diagnosis, gender, histological subtypes and stage.
Correlation between smoking history and TMB was assessed first by utilizing the generalized additive model (GAM) to allow for more flexibility (13). If a significant non-linear association existed, then piecewise regression was utilized after controlling for the same clinical covariates.
In all advanced-stage NSCLC patients:
In ever smokers:
Stratified analyses were similarly conducted based on histological subtypes and smoking status. All p-values were two-sided and confidence intervals were at the 95% level, with statistical significance defined as P ≤ 0.05. False discovery rate (FDR) correction was conducted to control for multiple comparison.
Results
1. Clinical and demographic characteristics by smoking status
A total of 931 advanced-stage NSCLC patients were included in this study (Table 1). There were 239 never smokers, 438 former smokers, and 254 current smokers. There were 764 patients with adenocarcinoma, 57 with squamous cell carcinoma, and 110 with other histologies. Patients with adenocarcinomas were more frequently represented because OncoPanel is routinely performed in these patients to identify the oncogenic drivers that can be effectively treated with targeted agents while these are rare in squamous cell carcinoma and other histologies. Former smokers made up the highest proportion of adenocarcinoma (350/764, 45.8%) and squamous cell carcinoma (28/57, 49.1%) patients. Current smokers had a larger median of smoking pack-years (40 [PYs]) compared to former smokers (24 [PYs]).
Table 1.
Major clinicopathological features of 931 NSCLC patients by smoking status
| Smoking Status | ||||
|---|---|---|---|---|
| Never Smoker (n = 239) | Former Smoker (n = 438) | Current Smoker (n = 254) | Total (N = 931) | |
| Age at diagnosis, median (SD), y | 61 (13) | 68 (10) | 60 (9) | 63 (11) |
| Sex, n (%) | ||||
| Male | 151 (63) | 252 (58) | 139 (55) | 542 (58) |
| Female | 88 (37) | 186 (42) | 115 (45) | 389 (42) |
| Ethnicity, n (%) | ||||
| White | 191 (80) | 402 (92) | 211 (83) | 804 (87) |
| Asian | 33 (14) | 9 (2) | 14 (6) | 56 (6) |
| Black | 5 (2) | 14 (3) | 19 (8) | 38 (4) |
| Hispanic | 4 (1) | 5 (1) | 4 (1) | 13 (1) |
| Unknown/Others | 6 (3) | 8 (2) | 6 (2) | 20 (2) |
| Pathology, n (%) | ||||
| Adenocarcinoma | 219 (92) | 350 (80) | 195 (77) | 764 (82) |
| Squamous Cell Carcinoma | 12 (5) | 28 (6) | 17 (6) | 57 (6) |
| Others | 8 (3) | 60 (14) | 42 (17) | 110 (12) |
| Stage, n (%) | ||||
| III | 43 (18) | 133 (30) | 101 (40) | 277 (30) |
| IV | 196 (82) | 305 (70) | 153 (60) | 654 (70) |
| Smoking pack-years, median (SD), py | 0 (0) | 24 (24) | 40 (20) | 20 (24) |
| Smoking-free months, median (SD), mo | NA | 261 (170) | 1 (3) | 161 (191) |
2. Genomic landscape of advanced NSCLC patients in relation to smoking status
Substantial differences in the affected genes, mutation spectrum and TMB were found across different smoking subgroups (Figure 1). A distinct difference was observed in the most frequently mutated genes across different smoking statuses. TP53 was highly mutated regardless of smoking status. In never smokers, EGFR (51%) was the most commonly mutated gene, and TET2 (8%), TSC2 (7%), ARID2 (7%), ERBB2 (7%) and PIK3CA (6%) mutations were observed at a higher prevalence than in former or current smokers. In contrast, KRAS mutations were predominant in current (33%) and former smokers (31%), and they had a higher prevalence of STK11 (former smokers 13%, current smokers 14%, respectively), NF1 (former smokers 12%, current smokers 15%, respectively), KEAP1 (former smokers 13%, current smokers 20%, respectively), and SMARCA4 mutations (former smokers 10%, current smokers 17%, respectively) (Figure 1A). C>T transitions were the most frequent type of SNV irrespective of smoking status. C>G transversions were the second-most frequent type of SNV in never smokers while C>A transversions were enriched in ever smokers. There was a statistically significant association between smoking and transversion events (P < 0.001), consistent with previous studies (14–16) (Figure 1B).
Figure 1. Mutation landscape in advanced NSCLC patients by smoking status.
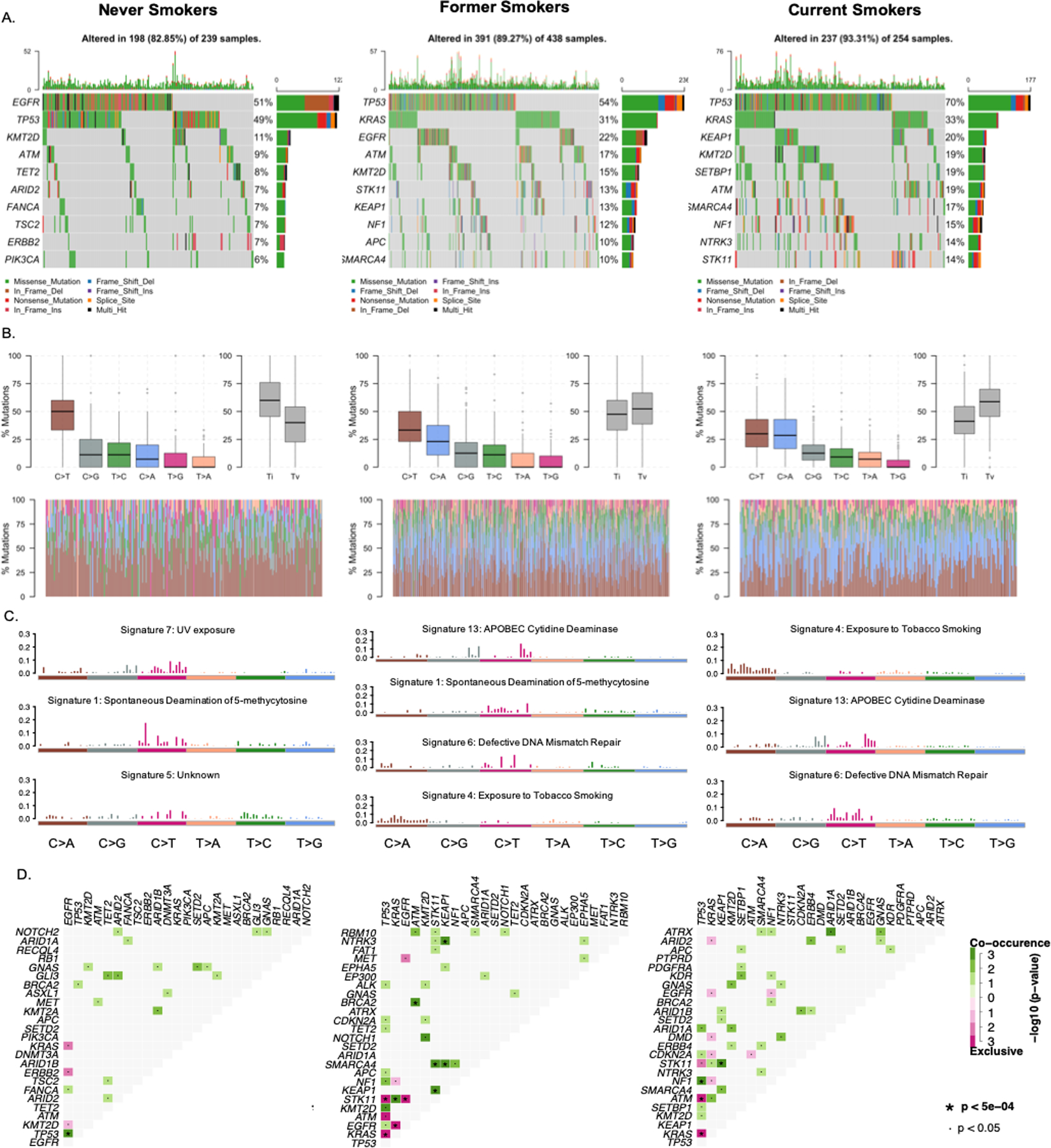
Mutation landscape in advanced NSCLC patients by smoking status. A. Oncoplot of the top 10 mutated genes in each smoking group in our cohort. Each row represents a gene and each column represents a sample. Genes are ordered by mutation frequency and are differentially colored based on different mutation types. B. Transition and transversion plot displays distribution of Single Nucleotide Variants (SNV) classified into six transition and transversion events. Stacked bar plot (bottom) shows distribution of mutation spectra for every sample. C. Mutational signatures identified in each smoking subgroup. The y-axes indicate exposure of 96 trinucleotide motifs to the overall signature. Each plot title indicates the best match against validated Catalogue of Somatic Mutations in Cancer (COSMIC) signatures and cosine similarity value along with the proposed etiology. D. Mutually exclusive and co-occurring gene pairs are displayed as a triangular matrix. Green indicates tendency toward co-occurrence, whereas pink indicates tendency toward exclusiveness.
Mutational signature
Based on the Catalogue of Somatic Mutations in Cancer (COSMIC), the mutational signature of never smokers with NSCLC in our cohort was the most similar to signature 1, spontaneous deamination of 5-methylcytosine, and signature 7, UV exposure. Signature 13, APOBEC cytidine deaminase (C>G), signature 4, exposure to tobacco (smoking) mutagens, and signature 6, defective DNA mismatch repair, were more common signatures in former and current smokers (Figure 1C).
Co-mutation/mutually exclusive patterns
In never smokers, TP53 and EGFR mutations highly co-occurred while KRAS, ERBB2 and EGFR mutations in the RAS/RTK pathway were mutually exclusive (P < 0.001). In former smokers, STK11, KRAS, KEAP1, SMARCA4 and NTRK3 mutations highly co-occurred while EGFR and TP53 were mutually exclusive with STK11 and KRAS (P < 0.001). In current smokers, in addition to the co-mutation of STK11 and KEAP1, NF1 and TP53 mutations significantly co-occurred, while ATM and KRAS mutations were mutually exclusive with TP53 mutations (P < 0.001) (Figure 1D).
3.1. Relationship between smoking metrics and EGFR and KRAS
Smoking history was inversely associated with frequency of EGFR mutation in a statistically significant dose-dependent manner, with the highest frequency observed in never smokers (50%) and in former smokers with > 364 months (30.3 years) since smoking cessation (47%). In contrast, smoking pack-years were positively associated with KRAS mutation frequency, with the highest frequency of 47% observed in smokers with > 40 pack-years (Figure 2). This dose-dependent association was also observed with EGFRL858R, EGFRdel19 and KRASG12C mutations at the variant level. EGFRL858R and EGFRdel19 had the highest mutation rates of 39.8% and 39.2% in never smokers and 31.7% and 30.8% in former smokers with > 30.3 years of smoking cessation, respectively; KRASG12C was the most common mutation (37.6%) in smokers with > 40 pack-years (Figure 2).
Figure 2. Mutation rates of EGFR and KRAS by smoking metrics.
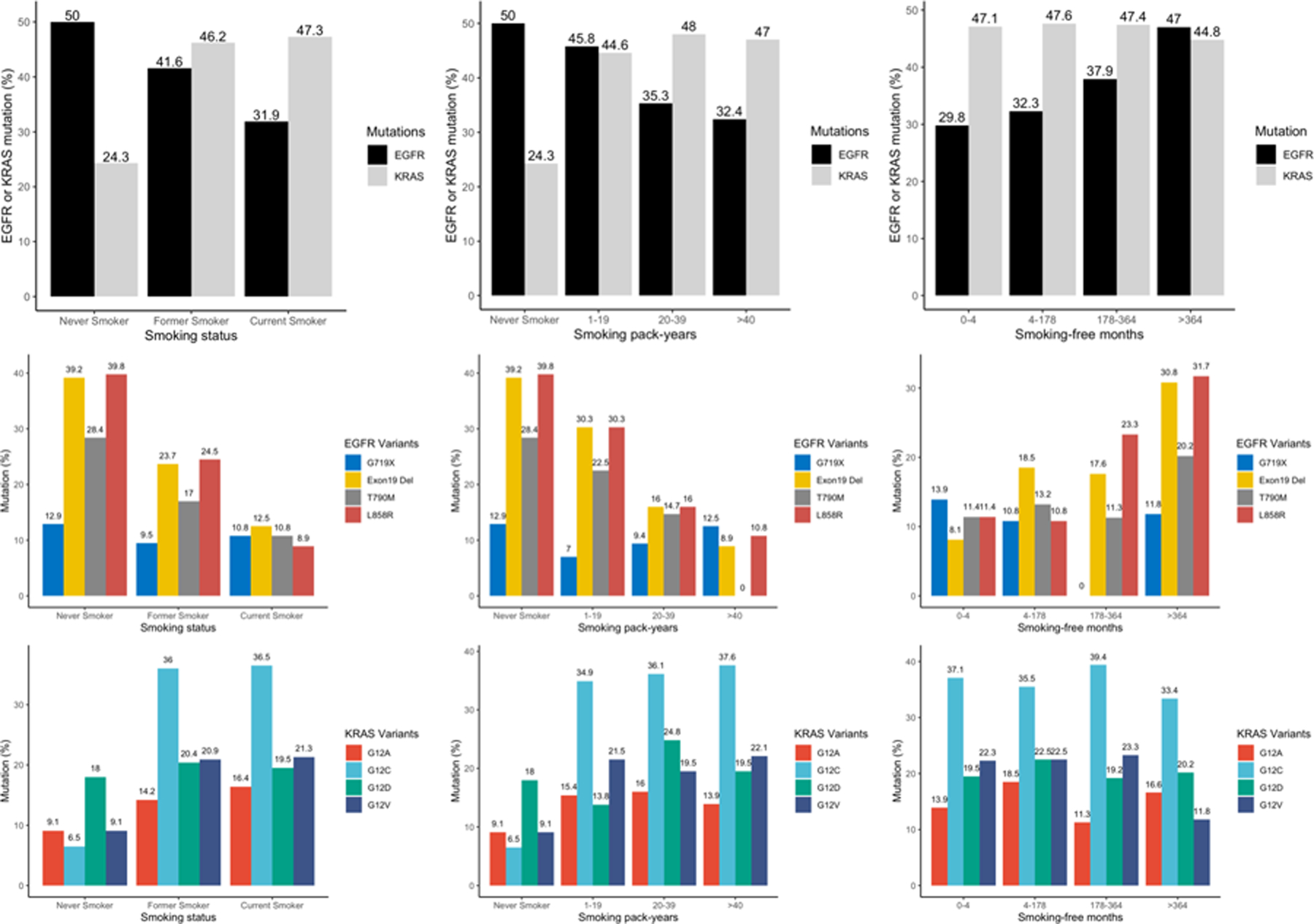
EGFR and KRAS mutation rates in different smoking subgroups based on smoking status, smoking pack-years and smoking-free months. (Upper) EGFR and KRAS mutation rates by various smoking metrics. (Middle) EGFR mutation rates by smoking metrics at the variant level. (Down) KRAS mutation rates by smoking metrics at the variant level.
In multivariable analysis, EGFRdel19, EGFRL858R mutations were most significantly enriched in never smokers, followed by former and current smokers [EGFRdel19 OR = 0.35, P < 0.001, OR = 0.09, P < 0.001, respectively; EGFRL858R OR = 0.26, P < 0.01 and OR = 0.04, P < 0.01, respectively]. Conversely, KRASG12C and KRASG12V mutations were highly enriched in former and current smokers (KRASG12C OR = 48.28, P < 0.01, OR = 54.51, P < 0.01, respectively; KRASG12V OR = 6.51, P = 0.01, OR = 6.67, P = 0.01, respectively). Doubling smoking pack-years was associated with decreased EGFRdel19 (OR = 0.47, P < 0.001) and EGFRL858R (OR = 0.62, P < 0.001) mutations in advanced NSCLC patients. In contrast, doubling smoking pack-years was associated with increased KRASG12C mutation (OR = 1.42, P < 0.001). In ever smokers, doubling smoking-free months was positively associated with EGFRL858R mutation (OR = 1.31, P = 0.03) and doubling smoking pack-years was associated with a decreased risk of EGFRdel19 mutation (OR = 0.53, P < 0.001) (Figure 3A, B). Doubling smoking pack-years was associated with increased KRASG12C mutation (OR = 1.42, P < 0.001).
Figure 3. Effect of smoking metrics on mutations and TMB.
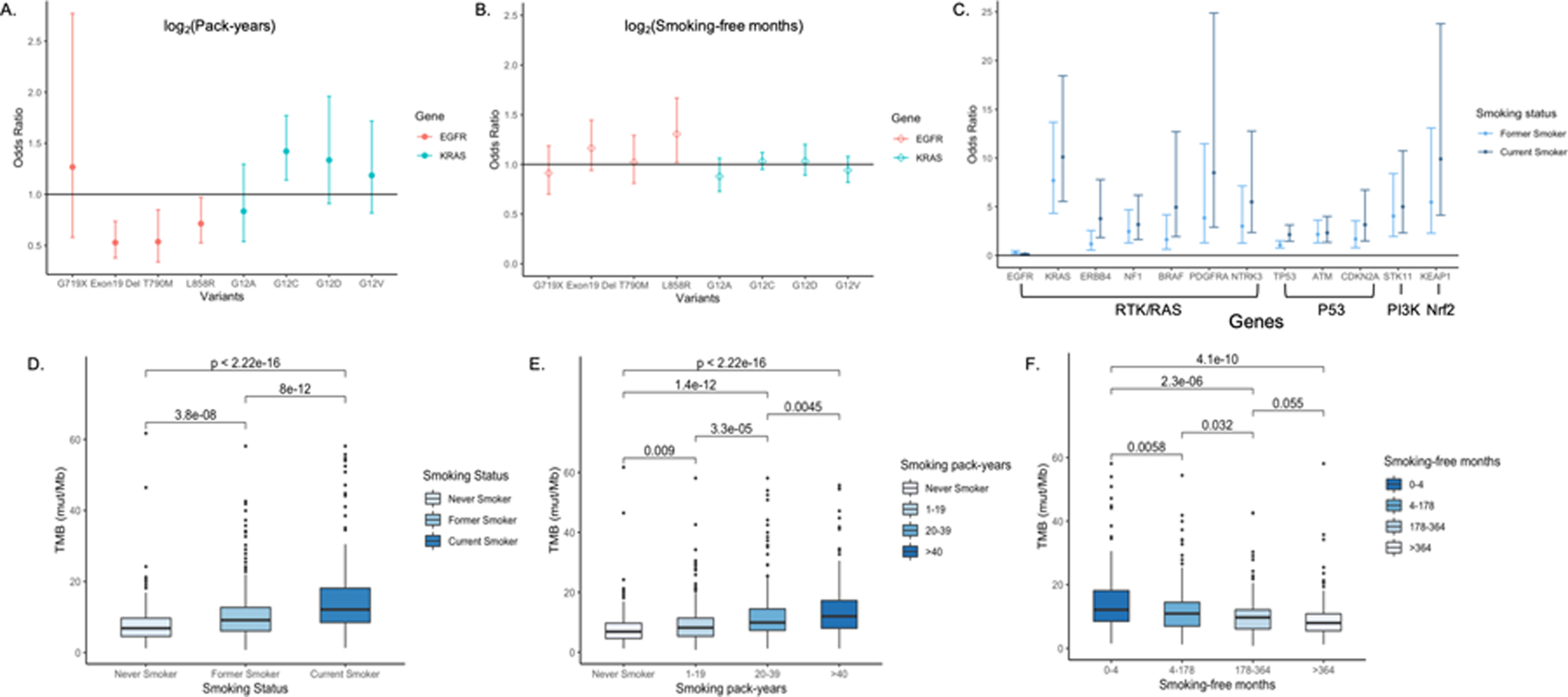
A. Odds ratios of EGFR and KRAS variant-specific mutations for smoking pack-years. B. Odds ratios of EGFR and KRAS variant-specific mutations for smoking-free months. C. Odds ratios of somatic mutations in cancer related pathways for former and current smokers obtained from multivariable logistic regression controlling for age, gender, stage and histological subtypes. D. TMB is significantly associated with smoking status, with the highest median TMB observed in current smokers (12.1 mut/Mb), followed by former and never smokers (9.1 mut/Mb and 6.8 mut/Mb, respectively). E. All patients were divided into never smokers and ever smokers and smoking pack-years in ever smokers were divided into tertiles. Smoking pack-years are significantly associated with TMB. F. Ever smokers were divided based on quartiles of smoking-free months. Smoking-free months are significantly associated with TMB. Pairwise comparisons by Wilcoxon test were conducted and FDR adjusted p-values are labeled. P ≤ 0.05 is considered statistically significant.
3.2. Relationship between smoking metrics and somatic mutations in cancer related pathways
We assessed the impact of different smoking metrics on the likelihood of somatic mutations in 10 cancer-related pathways using logistic regression controlling for related clinical variables (17). Smoking was significantly associated with mutations in the RTK/RAS, PI3K, Nrf2, and P53 pathways. KRAS (OR = 10.11, P < 0.01), ERBB4 (OR = 3.78, P < 0.01), PDGFRA (OR = 8.50, P < 0.01), NTRK3 (OR = 5.49, P < 0.01), NF1 (OR = 3.18, P < 0.01) and BRAF mutations (OR = 4.95, P < 0.01) in the RTK/RAS pathway were more likely to occur in current smokers compared to never smokers. Additionally, STK11 (OR = 5.00, P < 0.01) mutations in the PI3K pathway, KEAP1 (OR = 9.91, P < 0.01) mutations in the Nrf2 pathway, as well as CDKN2A (OR = 3.16, P < 0.01), TP53 (OR = 2.14, P < 0.01) and ATM (OR = 2.33, P < 0.01) mutations in the P53 pathway were enriched in current smokers (Figure 3C). Doubling smoking pack-years was associated with a decreased risk of EGFR (OR = 0.67, P < 0.01) and an increase in KRAS (OR = 1.46, P < 0.01) mutations while doubling smoking-free months was associated with decreased TP53 mutations (OR = 0.87, P < 0.01) (Supplementary Fig. S4).
3.3. Dose-response relationship between smoking history and TMB
Tobacco smoking was significantly associated with higher TMB and a dose-dependent association was consistently observed in different smoking metrics subgroups (Figure 3D, E, F). Smoking pack-year was positively associated with TMB by tertiles (median TMBs of 6.8 mut/Mb, 8.2 mut/Mb, 9.9 mut/Mb, and 11.9 mut/Mb in never smokers and smokers with 1–19 [PYs], 20–39 [PYs], and >40 [PYs], respectively; P < 0.001) while smoking-free months were inversely associated with TMB in a dose-dependent manner (median TMBs of 12.1 mut/Mb, 10.9 mut/Mb, 9.7 mut/Mb, and 8.4 mut/Mb in ever smokers with smoking-free months 0–4 [mo], 4–178 [mo], 178–364 [mo], and >364 [mo] (30.3 yrs), respectively; P < 0.001). The adjusted relationship between log2(pack-years) and log2(TMB) was significantly non-linear (P < 0.001 for the nonlinear contribution), while log2(smoking-free months) was negatively associated with log2(TMB) in all advanced NSCLC patients and ever smokers (Table 2, Supplementary Figs. S5 and S6).
Table 2.
Effect of smoking history in all NSCLC and in adenocarcinoma ever smokers
| All NSCLC Ever Smokers (n=692) | Adenocarcinoma Ever Smokers (n=545) | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariable Analysis | Multivariable Analysis | Univariable Analysis | Multivariable Analysis | |||||
| Parameters | Estimate (95% CI) | P | Estimate (95% CI) | P | Estimate (95% CI) | P | Estimate (95% CI) | P |
| Age | NA | NA | 1.00 (1.00–1.01) | 0.10 | NA | NA | 1.00 (1.00–1.01) | 0.02 |
| Male vs female | NA | NA | 0.95 (0.85–1.05) | 0.30 | NA | NA | 0.91 (0.81–1.03) | 0.15 |
| Squamous cell Carcinoma vs Adenocarcinoma | NA | NA | 1.2 (0.97–1.49) | 0.09 | NA | NA | NA | NA |
| Others vs adenocarcinoma | NA | NA | 1.1 (0.94–1.27) | 0.23 | NA | NA | NA | NA |
| Stage IV vs III | NA | NA | 0.93 (0.84–1.04) | 0.22 | NA | NA | 0.90 (0.79–1.02) | 0.10 |
| Doubling Smoking pack-years | 1.16 (1.10–1.23) | <0.001 | 1.14 (1.08–1.21) | <0.001 | 1.13 (1.07–1.19) | <0.001 | 1.11 (1.04–1.24) | <0.001 |
| Doubling Smoking-free months | 0.97 (0.95–0.98) | <0.001 | 0.96 (0.94–0.98) | <0.001 | 0.97 (0.95–0.99) | <0.001 | 0.95 (0.92–0.99) | <0.001 |
Only the statistically significant slopes for smoking pack-years are presented in the table.
Due to the nonlinear association between log2(pack-years) and log2(TMB), examination of the data showed that nonlinearity can be modeled using a piecewise linear model. In multivariable analysis, only the slope before the change point log2(pack-years) = 5.93 was statistically significant (effect = 1.15, P < 0.001), suggesting doubling pack-years was associated with a 1.15-times increase in TMB in all advanced NSCLC patients (Supplementary Fig. S7). To control for the potential confounding effect of histology on TMB, we also analyzed the effect of smoking history in the subset of patients with different histologies. In advanced lung adenocarcinoma, a significant linear association between log2(pack-years) and log2(TMB) was observed, suggesting doubling smoking pack-years was associated with 1.12 times increase in TMB (P < 0.001).
We restricted our analysis further to ever smokers by controlling for smoking-free months instead of smoking status in all advanced NSCLC. Similarly, only the slope before the change point of log2(pack-years) = 5.36 was statistically significant. Multivariable analysis suggested that doubling pack-years was associated with a 1.14-times increase (P < 0.001) in TMB, while doubling smoking-free months was associated with a 0.96-times decrease in TMB (P < 0.001) (Table 2). In advanced lung adenocarcinoma, doubling smoking pack-years was associated with 1.11-times increase (P < 0.001) and doubling smoking-free months was associated with 0.95 times decrease in TMB (P < 0.001) (Table 2).
Discussion
To accurately and reliably determine the association between tobacco smoking, somatic mutations and the composite TMB in advanced NSCLC cohort, we conducted this large retrospective analysis of 931 advanced NSCLC patients with OncoPanel results and prospectively collected smoking information. (i) We found distinct differences in the genomic landscapes of patients with different smoking statuses; (ii) we determined the likelihood of the two most common oncogenic drivers, EGFR and KRAS mutations, by smoking status and by smoking history in a dose-response relationship at both the gene and variant-specific level; (iii) we determined the likelihood of mutations in cancer-related pathways at the gene level by smoking status and by smoking history in a dose-response relationship; and (iv) we assessed the dose-response relationship between smoking history and TMB.
The effect of smoking status on somatic mutations in NSCLC patients has been limited to a few oncogenic driver genes in previous studies. TP53 and KRAS mutations are reported more frequently in lung cancers arising in smokers, while EGFR mutations are 6.29-fold higher in never smokers than in ever smokers among Caucasian/mixed ethnicity patients (18). Our results suggested that EGFRdel19, EGFRL858R mutations were most significantly enriched in never smokers, followed by former and current smokers while KRASG12C and KRASG12V mutations were inversely associated with smoking status. In addition to smoking status, we discovered the effect of various smoking metrics on mutations at the gene level in 10 cancer-related pathways including the RTK/RAS, PI3K, P53, and Nrf2 pathways. In previous studies, the relative risk of KRAS mutations was associated with increased tobacco consumption, with a 6-fold higher risk for smokers with more than 15 pack-years compared to never smokers (19). We determined the dose-dependent association of smoking pack-years and smoking-free months with EGFR and KRAS mutations at both the gene and variant levels. Smoking pack-years have a significant predictive value for the presence of both EGFR and KRAS mutations, and smoking-free months could predict the presence of TP53 mutation. Our multivariable analyses are consistent with the hypothesis that KRAS mutations are an early event in smokers and may lead to lung cancer as smoking pack-years increases, explaining the lack of impact of smoking-free months on the risk of lung cancer development. This is supported by the observed variant-level dose-response association in which smoking pack-years significantly increased the likelihood of KRASG12C (OR = 1.42, P < 0.01), which is the most common mutation in KRAS, while smoking-free months lacked impact (20). This finding is further supported by the observation that former and current smokers have similar proportions of KRAS mutations (Figure 1). Overall, our results support that permanent DNA damage by tobacco carcinogens acquired early on while smoking is the major source of most KRAS-mutated NSCLC. Thus, the likelihood that a patient with NSCLC develops KRAS mutations, especially KRASG12C is determined by smoking pack-years and does not decrease significantly over time upon smoking cessation. In contrast, EGFR mutations are impacted by both smoking pack-years and smoking-free months. Longer smoking pack-years is associated with a decreased risk of developing EGFRdel19 mutations, while smoking-free months increases the likelihood of harboring an EGFRL858R mutation. Both EGFRdel19 and EGFRL858R mutations have a favorable response to EGFR TKIs. However, these results should not be misinterpreted as suggesting that smoking protects against EGFR mutation in advanced NSCLC patients (20).
TMB accumulates with smoking pack-years and declines with time since smoking cessation. Previous studies focused on the differences in TMB by smoking status, with a consistent conclusion that smokers have a higher median TMB than never smokers (21–24). TMB, as a potential predictor for ICIs, was mostly defined as a categorical variable based on various thresholds (21,25–27). In NSCLC, TMB > 15 mut/Mb is more common in current/former smokers compared to never smokers (21,22,25–28). Our study, for the first time, illustrates a dose-dependent association between quantitative smoking history and TMB in an advanced NSCLC cohort. Adjusted analysis showed that smoking pack-years and smoking-free months were independent predictive factors for TMB. Although smoking pack-years has a non-linear association with TMB, this could be explained by the heterogeneity of histology and the limited sample size of patients with extreme large smoking pack-years. In stratified analysis by histology, significant linear associations were observed in adenocarcinoma, as doubling smoking pack-years was associated with a 1.11-times increase and doubling smoking-free months lead to a 0.95-times decrease in TMB (Table 2). A non-significant linear association between smoking history and TMB was observed in squamous cell carcinoma due to the limited sample size. This association needs to be confirmed in a larger cohort of patients with this smoking-related malignancy. Similar results of smoking pack-years on TMB were observed in The Cancer Genome Atlas (TCGA) data, but with smoking-free interval undiscovered (29). Smoking pack-years had a larger effect on TMB in current smokers than in former smokers, which is supported by the observed significant alleviating effect of smoking-free months (Supplementary Fig. S8–S9).
Admittedly, our study has several limitations. Our analysis was based on the TMB calculated from Oncopanel and harmonization of TMB from different panels and assays before generalizing the association is necessary. Limited by the characteristic of our study population, squamous cell carcinoma was underrepresented and the association needs to be confirmed in a larger cohort.
In conclusions, our study first clarifies the dose-dependent association between detailed smoking history and TMB in advanced lung adenocarcinoma. Our results support the public health effort on a non-smoking lifestyle and confirms the benefit of quitting smoking early. Moreover, it provides important implications that smoking history may be utilized as an easily obtainable surrogate for TMB to make prompt treatment decision and enhance the proportion of patients who may benefit from ICIs. Finally, detailed smoking history should be prospectively collected in clinical practice and the clinical utility should be further studied.
Supplementary Material
Significance.
This study clarifies the relationship between smoking history and clinically relevant mutations in non-small cell lung cancer, revealing the potential of smoking history as a surrogate for tumor mutation burden.
Acknowledgements
X.W, X.Lin, B.E.J, and D.C.C designed the experiments. X.W, X.Lin, B.E.J, and D.C.C performed the experiments. X.W, B.R, X.Li, T.N, M.S.R, M.M.A, X.Lin, B.E.J, and D.C.C acquired, analyzed or interpreted the data. X.W, X.Lin, B.E.J, and D.C.C drafted the manuscript and revised it according to suggestions by the coauthors. All authors critically reviewed the manuscript, suggested revisions as needed and approved the final version.
Funding:
This work was supported by the National Cancer Institute (5U01CA209414 to X.W, B.E.J, D.C.C, X. Lin); Sola Fund for Lung Cancer Research, LUNGSTRONG, the National Cancer Institute (P30-CA006516), Dana-Farber/Harvard Cancer Center Grant, Elaine and Gerald Schuster Fund for Lung Cancer Research to B.E.J and the National Institutes of Health (R35-CA197449 and U01HG009088 to X. Lin).
Footnotes
Disclosure: The authors declare no potential conflicts of interest
References
- 1.Gomperts BN, Spira A, Massion PP, Walser TC, Wistuba II, Minna JD, et al. Evolving concepts in lung carcinogenesis. Semin Respir Crit Care Med 2011;32:32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagahashi M, Shimada Y, Ichikawa H, Kameyama H, Takabe K, Okuda S, et al. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci 2019;110:6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu ET, Mockus SM. Tumor Origins Through Genomic Profiles. JAMA oncology 2020;6:33–4 [DOI] [PubMed] [Google Scholar]
- 4.Sholl LM, Hirsch FR, Hwang D, Botling J, Lopez-Rios F, Bubendorf L, Mino-Kenudson M, Roden AC, Beasley MB, Borczuk A and Brambilla E. The Promises and Challenges of Tumor Mutation Burden as an Immunotherapy Biomarker: A Perspective from the International Association for the Study of Lung Cancer Pathology Committee. Journal of Thoracic Oncology 2020;15(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buttner R, Longshore JW, Lopez-Rios F, Merkelbach-Bruse S, Normanno N, Rouleau E, et al. Implementing TMB measurement in clinical practice: considerations on assay requirements. ESMO Open 2019;4:e000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurston SW, Liu G, Miller DP, Christiani DC. Modeling lung cancer risk in case-control studies using a new dose metric of smoking. Cancer Epidemiol Biomarkers Prev 2005;14:2296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenland S. Dose-response and trend analysis in epidemiology: alternatives to categorical analysis. Epidemiology 1995;6:356–65 [DOI] [PubMed] [Google Scholar]
- 8.Ricciuti B, Kravets S, Dahlberg SE, Umeton R, Albayrak A, Subegdjo SJ, et al. Use of targeted next generation sequencing to characterize tumor mutational burden and efficacy of immune checkpoint inhibition in small cell lung cancer. J Immunother Cancer 2019;7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin JJ, Cardarella S, Lydon CA, Dahlberg SE, Jackman DM, Janne PA, et al. Five-Year Survival in EGFR-Mutant Metastatic Lung Adenocarcinoma Treated with EGFR-TKIs. J Thorac Oncol 2016;11:556–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med 2017;141:751–8 [DOI] [PubMed] [Google Scholar]
- 11.Sholl LM, Do K, Shivdasani P, Cerami E, Dubuc AM, Kuo FC, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016;1:e87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastie T, Tibshirani R. Generalized additive models. London ; New York: Chapman and Hall; 1990. xv, 335 p. p. [Google Scholar]
- 14.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 2010;465:473–7 [DOI] [PubMed] [Google Scholar]
- 15.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012;481:506–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321–37 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman AM, Sun KY, Ruestow P, Cowan DM, Madl AK. Lung cancer mutation profile of EGFR, ALK, and KRAS: Meta-analysis and comparison of never and ever smokers. Lung Cancer 2016;102:122–34 [DOI] [PubMed] [Google Scholar]
- 19.Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Taniere P, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res 2005;65:5076–83 [DOI] [PubMed] [Google Scholar]
- 20.Dogan S, Shen R, Ang DC, Johnson ML, D’Angelo SP, Paik PK, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vokes NI, Liu D, Ricciuti B, Jimenez-Aguilar E, Rizvi H, Dietlein F, et al. Harmonization of Tumor Mutational Burden Quantification and Association With Response to Immune Checkpoint Blockade in Non-Small-Cell Lung Cancer. JCO Precis Oncol 2019;3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354:618–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berland L, Heeke S, Humbert O, Macocco A, Long-Mira E, Lassalle S, et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis 2019;11:S71–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagahashi M, Sato S, Yuza K, Shimada Y, Ichikawa H, Watanabe S, et al. Common driver mutations and smoking history affect tumor mutation burden in lung adenocarcinoma. J Surg Res 2018;230:181–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nature medicine 2018;24:1441–8 [DOI] [PubMed] [Google Scholar]
- 26.Heeke S, Hofman P. Tumor mutational burden assessment as a predictive biomarker for immunotherapy in lung cancer patients: getting ready for prime-time or not? Translational Lung Cancer Research 2018;7:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ready N, Hellmann MD, Awad MM, Otterson GA, Gutierrez M, Gainor JF, et al. First-line nivolumab plus ipilimumab in advanced non–small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. Journal of Clinical Oncology 2019;37:992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis AA, Chae YK, Agte S, Pan A, Mohindra NA, Villaflor VM, et al. Association of tumor mutational burden with smoking and mutation status in non-small cell lung cancer (NSCLC). American Society of Clinical Oncology; 2017. [Google Scholar]
- 29.Sharpnack MF, Cho JH, Johnson TS, Otterson GA, Shields PG, Huang K, et al. Clinical and Molecular Correlates of Tumor Mutation Burden in Non-Small Cell Lung Cancer. Lung Cancer 2020;146:36–41 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


