Abstract
Background
To assess inter-reader agreement for US BI-RADS descriptors using S-Detect: a computer-guided decision-making software assisting in US morphologic analysis.
Methods
73 solid focal breast lesions (FBLs) (mean size: 15.9 mm) in 73 consecutive women (mean age: 51 years) detected at US were randomly and independently assessed according to the BI-RADS US lexicon, without and with S-Detect, by five independent reviewers. US-guided core-biopsy and 24-month follow-up were considered as standard of reference. Kappa statistics were calculated to assess inter-operator agreement, between the baseline and after S-Detect evaluation. Agreement was graded as poor (≤ 0.20), moderate (0.21–0.40), fair (0.41–0.60), good (0.61–0.80), or very good (0.81–1.00).
Results
33/73 (45.2%) FBLs were malignant and 40/73 (54.8%) FBLs were benign. A statistically significant improvement of inter-reader agreement from fair to good with the use of S-Detect was observed for shape (from 0.421 to 0.612) and orientation (from 0.417 to 0.7) (p < 0.0001) and from moderate to fair for margin (from 0.204 to 0.482) and posterior features (from 0.286 to 0.522) (p < 0.0001). At baseline analysis isoechoic (0.0485) and heterogeneous (0.1978) echo pattern, microlobulated (0.1161) angular (0.1204) and spiculated (0.1692) margins and combined pattern (0.1549) for posterior features showed the worst agreement rate (poor). After S-Detect evaluation, all variables but isoechoic pattern showed an agreement class upgrade with a statistically significant improvement of inter-reader agreement (p < 0.0001).
Conclusions
S-Detect significantly improved inter-reader agreement in the assessment of FBLs according to the BI-RADS US lexicon but evaluation of margin and echo pattern needs to be further improved, particularly isoechoic pattern.
Keywords: Breast neoplasms, Ultrasonography, BI-RADS, Problem-solving, Decision-making, Computer-assisted diagnosis
Background
High-resolution ultrasonography (US) is a valuable tool in breast imaging, and it is currently considered a useful adjunct to mammography and magnetic resonance (MR) [1–3].
Nevertheless, US is an operator-dependent technique: to overcome this issue, the American College of Radiology (ACR) has developed the Breast Imaging-Reporting and Data System (BI-RADS) US lexicon, firstly released in 2003, aiming to standardize image interpretation, reporting and teaching breast imaging [4]. Despite this effort, the variability in the assessment of Focal Breast Lesions (FBLs) with the use of BI-RADS US lexicon is still an issue, in particular for some of the specific descriptors, such as margin and echo pattern of lesion [5, 6].
Within this framework, evidence shows that computer-aided image analysis may be effective in improving the radiologist’s assessment of FBLs [7]. In particular, S-Detect is a newly developed image-analytic computer program that provides assistance in morphologic analysis of FBLs seen on US according to the BI-RADS US lexicon [8]. Some preliminary studies have showed that the use of S-Detect can lead to an increase of the number of correctly characterized breast masses, especially for less experienced radiologist [8–11]. However, in those studies, the analysis of US features was not based on the latest edition of the BI-RADS US lexicon, which was recently revised in 2013 [12].
Hence, we undertook this study to assess inter-observer variability in the sonographic characterization of FBLs using S-Detect, with a specific analysis of each descriptor feature according to the updated BI-RADS US lexicon and radiologist’s experience.
Materials and methods
Study design
The whole study has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). Institutional review board approval and informed patient consent were obtained for this prospective study. Permission was obtained from the hospital for patients’ medical records review.
Between December 2016 and March 2017, 73 solid FBLs (size range: 5–40 mm; mean 15.9 mm ± 8.84 SD) were detected in 73 consecutive women (age range: 13–98 years; mean 51 years ± 14 SD) who underwent high-resolution US, by means of a Samsung ultrasound unit provided with a 3–12-MHz linear transducer (RS80A with Prestige, Samsung Medison, Co. Ltd.). Indications for breast US included: (1) a palpable mass detected on physical examination; (2) dense breasts; (3) detected lesions from adjunct mammography examination; (4) patients with mastodynia; (5) young patients having family history or (6) in a follow-up for benign breast nodules.
Exclusion criteria included lack of adequate standard of reference (refusal to undergo biopsy or inconsistent follow-up). No FBLs were excluded due to US imaging being not appropriate for S-Detect evaluation.
Computer-guided decision-making support software
S-Detect™ is a built-in, commercially available dedicated software installed and running on the ultrasound system RS80A. The software is licensed for clinical use: by placing an operator-defined marker within one FBL, it draws the contour of the lesion, analyses it according to the BI-RADS US descriptors and provides a categorization into possibly benign or possibly malignant. The sonographer can either accept the indications as provided by the software or choose and record the most appropriate term for each descriptor as deemed necessary, including the final categorization [8, 9].
S-Detect applies a novel feature extraction technique and support vector machine classifier that classifies breast masses into benign or malignant according to the proposed feature combinations integrated according to the US BI-RADS. Features used for US feature analysis in S-Detect are as follows: shape differences, echo and texture features using spatial gray-level dependence matrices, intensity in the mass area, gradient magnitude in the mass area, orientation, depth–width ratio, distance between mass shape and best t ellipse, average gray changes or histogram changes between tissue/mass area, comparison of gray value of left, posterior, and right under the lesion, the number of lobulated areas/protuberances/depressions, lobulation index, and elliptic-normalized circumference [13].
Image selection and evaluation
An independent experienced radiologist (more than twenty years of breast US), not involved in the final image reading, chose the most representative B-mode image for each of the 73 FBLs. All the selected images in DICOM format were evaluated off-line in the same ultrasound unit used for clinical breast scan and provided with S-Detect V2 (RS80A with Prestige, Samsung Medison).
All the 73 FBLs were then randomly and independently assessed, without and with S-Detect, in four separate sessions, to avoid recall bias, by five independent reviewers with different expertise and skills in breast sonography: one Experienced Radiologist (ER, more than 20 years of experience), two General Radiologists (GR#1 and GR#2, 5–10 years of experience) and two last year Radiology Senior Residents (R#1 and R#2). Each reviewer classified the 73 FBLs into 4 categories: (1) BI-RADS 2: benign; (2) BI-RADS 3: probably benign; (3) BI-RADS 4: suspicious; and (4) BIRADS 5: highly suggestive of malignancy.
The classification was based on the BI-RADS US Atlas, using US descriptors recommended by the 5th edition of ACR BI-RADS (2013): (1) shape; (2) margin; (3) echo pattern; (4) orientation; and (5) posterior features [4].
Information about patient’s age, family or personal history of breast cancer and previous US or other radiological investigations were not available to the investigators. Reviewers were also blinded to pathologic findings.
One week before starting the image analysis process, all the readers involved in this study had to attend a separate 5-h training session and all readers had to evaluate individually and separately by means of S-Detect further 20 FBLs (encompassing both benign and malignant mass) not included in the final study.
Standard of reference (SOR)
For the purpose of this study, US-guided core-biopsy was considered as SOR for all the FBLs classified as BI-RADS 3, 4 or 5 either before or after S-detect assessment. For all lesions classified as BI-RADS 2 both before and after S-detect assessment, US findings at 6-, 12- and 24-month follow-up have been available at moment of evaluation and considered as SOR. In particular, stability or size decrease during follow-up and typically benign US findings, such as cysts, have been considered.
Statistical analysis
Statistical analysis was performed by a biostatistician involved in the study design using a computer software package (STATA/SE 14 and R; https://cran.r-project.org/).
Kappa statistics were calculated to assess inter-operator agreement, between the baseline and after S-Detect evaluation. Agreement was graded as poor (≤ 0.20), moderate (0.21–0.40), fair (0.41–0.60), good (0.61–0.80), or very good (0.81–1.00) [14].
Results
According to the SOR, 33/73 (45.2%) FBLs were malignant, and 40/73 (54.8%) FBLs were benign (Table 1).
Table 1.
Final diagnosis of 73 FBLs as by SOR
| Benign (40) | Malignant (33) |
|---|---|
| Fibroadenoma (15) | Invasive ductal Carcinoma (26) |
| Fibrosis (6) | Invasive lobular Carcinoma (2) |
| Usual ductal hyperplasia (5) | In situ ductal Carcinoma (1) |
| Adenosis (4) | Mucinous carcinoma (1) |
| Xanthogranulomatosis (3) | Medullary carcinoma (1) |
| Sclerosing Adenosis (2) | Adenoid cystic carcinoma (1) |
| Granuloma (1) | Myxoid chondrosarcoma (1) |
| Corpuscular cyst (1) | |
| Myofibroblastoma (1) | |
|
Lymphocytic mastitis (1) Periductal mastitis (1) |
Figures in parentheses are actual number of lesions
Overall, inter-reader agreement for final assessment showed a statistically significant improvement of k-score for all readers: from moderate (0.30) without the use of S-Detect to good (0.63) with the use of S-Detect (p < 0.0001) (Table2).
Table 2.
Interobserver agreement for US-BI-RADS descriptors
| All readers | P value | ER, GR1 and GR2 | P value | Senior Residents | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Without S-Detect V2 | With S-Detect V2 | Without S-Detect | With S-Detect V2 | Without S-Detect V2 | With S-Detect V2 | ||||
| Kappa (95% CI) | Kappa (95% CI) | Kappa (95%CI) |
Kappa (95%CI) |
Kappa (95%CI) |
Kappa (95%CI) |
||||
| Shape |
0.4217 (0.3465; 0.4969) |
0.6118 (0.5522; 0.6713) |
< 0.0001 | 0.6154 (0.4491;0.7816) |
0.6389 (0.4489; 0.8289) |
0.4296 |
0.3104 (0.1165;0.5044) |
0.553 (0.4431; 0.6629) |
< 0.0001 |
| Margin | 0.2041 (0.1524;0.2559) |
0.4821 (0.4360; 0.5282) |
< 0.0001 | 0.2791 (0.1302;0.4280) |
0.4078 (0.2659; 0.5502) |
< 0.0001 |
0.1494 (− 0.0028;0.3016) |
0.4477 (0.3619; 0.5339) |
< 0.0001 |
| Echo Pattern |
0.2604 (0.2006;0.3201) |
0.3907 (0.3436; 0.4377) |
< 0.0001 | 0.3313 (0.1319;0.5307) |
0.3406 (0.1825; 0.4986) |
0.7559 |
0.2912 (0.0734;0.5090) |
0.3737 (0.2865; 0.4610) |
0.0032 |
| Orientation | 0.4179 (0.3236;0.5122) | 0.7000 (0.6274;0.7725) | < 0.0001 | 0.7637 (0.6094;0.9181) |
0.8577 (0.6283; 1.0871) |
0.0044 |
0.1667 (− 0.0611; 0.3944) |
0.7262 (0.5937; 0.8586) |
< 0.0001 |
|
Posterior features |
0.2868 (0.2183;0.3553) |
0.5221 (0.4718; 0.5725) |
< 0.0001 | 0.3588 (0.0573;0.6603) |
0.4113 (0.2523; 0.5703) |
0.1909 |
0.3648 (0.1809; 0.5488) |
0.4697 (0.3776; 0.5617) |
< 0.0001 |
| Final assessment | 0.3019 (0.2332;0.3706) | 0.631 (0.5733;0.6887) | < 0.0001 | 0.2695 (0.0808;0.4582) | 0.6866 (0.5390;0.8342) | < 0.0001 | 0.2684 (0.0897;0.4470) | 0.4510 (0.3427;0.5594) | < 0.0001 |
ER expert radiologist, GR general radiologists
A statistically significant improvement of inter-reader agreement from fair to good with the use of S-Detect when compared to baseline evaluation was observed for shape (from 0.421 to 0.612) and orientation (from 0.417 to 0.7) (p < 0.0001) (Table 2) (Figs. 1, 2).
Fig. 1.
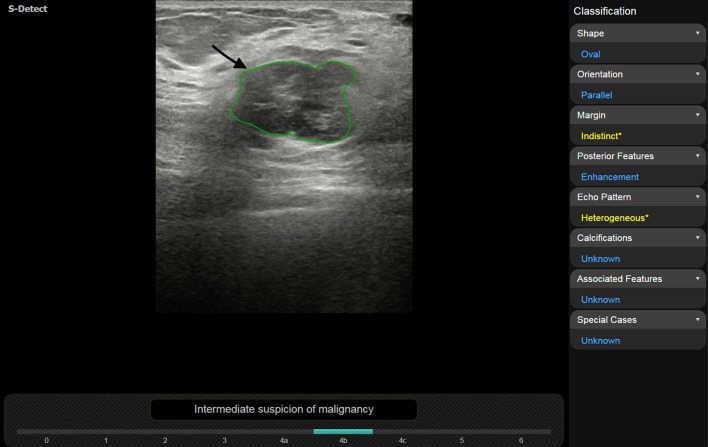
Malignant phyllodes tumor in a 63-year-old woman. Both Radiology Residents assessed shape as irregular without S-Detect, whereas with the support of S-Detect resident 1 evaluated shape as oval. Inter-reader agreement increased from fair to good
Fig. 2.
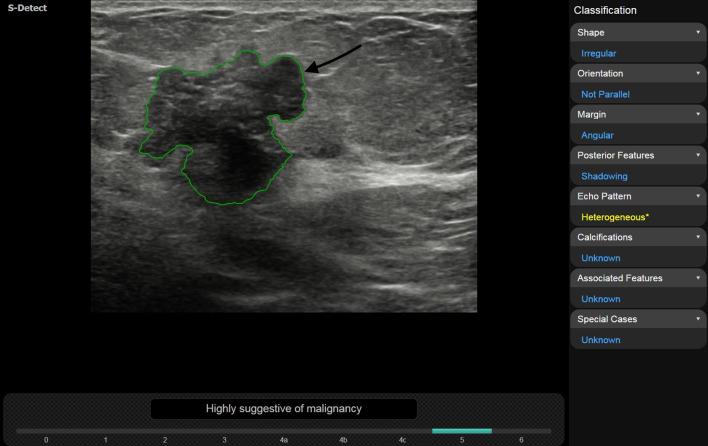
Invasive ductal carcinoma, luminal B, in a 72-year-old woman. With the support of S-Detect, interobserver agreement on lesion orientation increased from fair to good, leading from 3/5 (without S-Detect) to 4/5 readers choosing “not parallel” as descriptor
A statistically significant improvement of inter-reader agreement from moderate to fair with the use of S-Detect when compared to baseline evaluation was observed for margin (from 0.204 to 0.482) and posterior features (from 0.286 to 0.522) (p < 0.0001) (Table 2) (Figs. 3, 4).
Fig. 3.
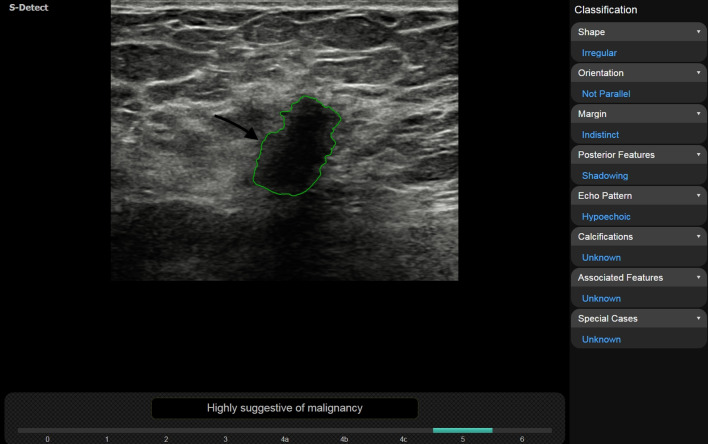
Invasive lobular carcinoma in a 57-year-old woman. One General Radiologist assessed lesion margins as “spiculated” without S-Detect and “indistinct” with S-Detect accordingly with the other 4 readers. Inter-reader agreement increased from moderate to fair with the use of S-Detect
Fig. 4.
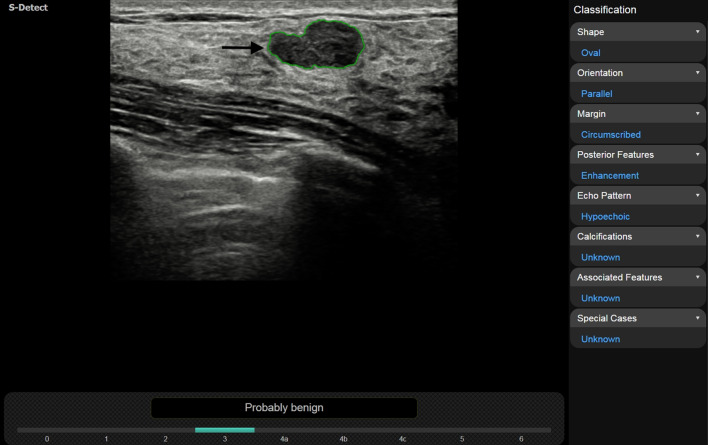
Fibroadenoma in a 32-year-old woman. Posterior feature “enhancement” was chosen by 4 out of 5 reviewers with the use of S-Detect and by 3 out of 5 reviewers without the use of S-Detect, respectively. Inter-reader agreement increased from moderate to fair
Echo pattern showed a statistically significant increase in k-score value with the use of S-Detect (from 0.260 to 0.390) (p < 0.0001) but did not change the correspondent inter-reader agreement class (moderate) (Table 2).
The improvement in agreement score for the last-year residents was statistically significant for all the evaluated descriptors (p < 0.05) (Table 2). The improvement in k-score also reached statistical significance for expert and general radiologists but only for margin and orientation (p < 0.0001 and p = 0.0044); whereas, it remained below the statistical significance for shape (p = 0.4296), echo pattern (p = 0.7559) and posterior features (p = 0.1909).
Table 3 details k-score evaluation for any possible option in the assessment of US BI-RADS descriptors: among all readers, a statistically significant improvement of inter-reader agreement was observed for all options (p < 0.0001). The variables which at baseline analysis showed the worst agreement rate (poor) were isoechoic (0.0485) and heterogeneous (0.1978) echo pattern, microlobulated (0.1161) angular (0.1204) and spiculated (0.1692) margins and combined pattern (0.1549) for posterior features (Table 3). Of these latter variables, only isoechoic pattern remained in the same agreement class (poor) even after S-Detect evaluation; whereas, all the others showed an agreement class upgrade (Table 3).
Table 3.
Interobserver agreement for the different variables of US BI RADS descriptors
| US BI-RADS descriptor | Kappa (95% CI) without S-detect | Kappa (95% CI) with S-detect | p value |
|---|---|---|---|
| Shape | |||
| Oval | 0.4581 (0.3638;0.5524) | 0.6330 (0.5604;0.7055) | < 0.0001 |
| Round | 0.2062 (0.1119;0.3005) | 0.3001 (0.2275;0.3726) | < 0.0001 |
| Irregular | 0.4674 ( 0.3731;0.5617) | 0.6478 (0.5752;0.7203) | < 0.0001 |
| Echo pattern | |||
| Hypoechoic | 0.2713 (0.1770;0.3656) | 0.4114 (0.3389;0.4840) | < 0.0001 |
| Isoechoic | 0.0485 (− 0.0458; 0.1428) | 0.1951 (0.1225; 0.2676) | < 0.0001 |
| Iperechoic | 0.7890 (0.6947;0.8833) | − 0.006 (− 0.0808;0.0643) | < 0.0001 |
| Complex echogenicity | 0.2123 (0.1180; 0.3066) | 0.3681 (− 0.5985; 1.3346) | < 0.0001 |
| Heterogeneous | 0.1978 (0.1035;0.2921) | 0.3478 (0.0339;0.6618) | < 0.0001 |
| Margin | |||
| Circumscribed | 0.3011 (0.2068;0.3954) | 0.5970 (0.5184;0.6755) | < 0.0001 |
| Indistinct | 0.2029 (0.1086;0.2972) | 0.3917 (0.2711;0.5122) | < 0.0001 |
| Microlobulated | 0.1161 (0.0218;0.2104) | 0.4830 (0.3242;0.6419) | < 0.0001 |
| Angular | 0.1204 (0.0261;0.2147) | 0.3830 (0.0644;0.8304) | < 0.0001 |
| Spiculated | 0.1692 (0.0749;0.2635) | 0.2332 (0.1607; 0.3057) | < 0.0001 |
| Orientation | |||
| Parallel | 0.4179 (0.3236;0.5122) | 0.700 (0.5606;0.8392) | < 0.0001 |
| Not parallel | 0.4179 (0.3236;0.5122) | 0.700 (0.5606;0.8392) | < 0.0001 |
| Posterior features | |||
| None | 0.2699 (0.1756;0.3642) | 0.4568 (0.3699;0.5438) | < 0.0001 |
| Enhancement | 0.3032 (0.2089;0.3975) | 0.3876 (0.2049;0.5703) | 0.0006 |
| Shadowing | 0.3269 (0.2326;0.4212) | 0.7410 (0.5490; 0.9330) | < 0.0001 |
| Combined pattern | 0.1549 (0.0606;0.2492) | 0.6719 (0.1034; 1.2405) | < 0.0001 |
Among all readers, with respect to BI-RADS classification, inter-reader agreement was statistically significant superior (p < 0.001) with the use of S-Detect when compared to baseline assessment for BI-RADS categories 3—k score (95% CI) values of 0.6148 (0.5418; 0.6869) vs 0.4128 (0.3185;0.5071); 4—0.4838 (0.3981; 0.5432) vs 0.1851 (0.0908;0.2794); and 5—0.4517 (0.3437; 0.4888) vs 0.3416 (0.2473;0.4359), respectively.
Discussion
Inter-reader variability in the assessment of FBLs with US is still an important issue in clinical practice. To overcome this limitation, the ACR conceived the US BI-RADS as standardized lexicon for image interpretation. Nevertheless, several studies have shown only fair to moderate agreement for overall final assessment either for the US BI-RADS 2003 edition, with k-score values ranging from 0.28 to 0.53 [15–19] or the latest US BI-RADS 2013 edition, with only moderate to fair agreement for overall final assessment and k-score values ranging from 0.22 to 0.51 [20–23]. Baseline analysis of our series confirmed those literature data, showing only moderate agreement for overall final assessment (k-score: 0.30) among all the readers.
Recent advancements of artificial intelligence techniques have led to the development of computer aided diagnostic tools, which are reported to be effective in improving the radiologist’s US assessment of FBLs, both in terms of detection and characterization [24, 25].
S-Detect is a newly developed image-analytic computer program, based on a deep learning algorithm and capable of auto segmentation and interpretation of US images, allowing classification of FBLs in a dichotomic form, according to US BI-RADS lexicon [26]. The use of S-Detect has been reported to increase the specificity and accuracy in the US assessment of FBLs, thus leading to a potential reduction of the number of unnecessary breast biopsies and related medical costs [10, 11, 27].
In our study, with the use of S-Detect, we observed a statistically significant increase of inter-reader agreement for overall final assessment, with correspondent level upgrade from moderate to good (k: 0.63, p < 0.001). This finding is comparable to the results of other two studies, which have also reported good agreement for final assessment with k-score values reaching 0.75 and 0.77 with the use of S-Detect, respectively, depending on the reader experience [10, 11]. Of note, other authors have also reported a statistically significant increase of overall inter-reader agreement with S-Detect, either for experienced or less experienced readers, but with lower observed level of agreement, ranging from moderate to fair and correspondent k-score values ranging from 0.320 to 0.457 [23, 24, 26–28]. Although not entirely discordant, the above-mentioned results suggest the need for more extensive evaluation of larger series of patients.
High variability rate is reported in literature for descriptor assessment (k-score: 0.19–0.66), with worse k-scores for margin, with only poor to moderate agreement (k-score: 0.19–0.31) and echo pattern, with moderate agreement (k-score: 0.26–0.30), respectively [20–23]. In our study, moderate to fair (0.20–0.42) agreement was observed for US descriptors among all readers, which is substantially comparable with the other available studies [20–23]. In particular, our baseline analysis showed only moderate agreement for both margin (0.20) and echo pattern (0.26) assessment. With the use of S-Detect, we observed a statistically significant increase of k-score for both margin (0.48, p < 0.001) and echo pattern (0.39 p < 0.001). Accordingly, margin assessment upgraded to the superior agreement category (fair), although echo pattern remained in the same category (moderate). These findings are consistent with previously data reported by Park et al., which showed a statistically significant increase from poor (k-score: 0.19) to moderate (k-score: 0.28) for margin assessment and from moderate (0.30) to fair (k-score: 0.401) with the use of S-Detect [23]. Overall, we observed a statistically significant increase of k-score values for all descriptors, including shape, orientation and posterior features, ranging from moderate to good (0.39–0.70). A one level upgrade was observed for shape and orientation (from fair to good), as well as for margin and posterior features (from moderate to fair), whereas echo pattern showed no change. This agreement improvement is relatively higher than previously reported by Park et al. with the same software, with their maximum k-score of 0.546 for orientation [23]. This finding may be at least in part explained by the training session that we held for all the readers before the reading session.
Among the possible options in the assessment of US BI-RADS descriptors, those with the worst agreement rate (poor) baseline analysis were isoechoic (0.0485) and heterogeneous (0.1978) echo pattern, microlobulated (0.1161) angular (0.1204) and spiculated (0.1692) margins and combined pattern (0.1549) for posterior features. These data confirm the high variability of margin and echo pattern assessment. Noteworthy, we observed a statistically significant improvement of inter-reader agreement with S-Detect evaluation (p < 0.0001) with concurrent class upgrade for all options, but isoechoic pattern, which remained in the same agreement class (poor).
The increase in inter-reader agreement was more evident for the less experienced readers, which achieved statistically significance (p < 0.001) for all the examined descriptors; whereas, it was less pronounced for the more experienced readers, with statistically significance (p < 0.001) observed for margin and orientation assessment. Inter-observer agreement improvement with S-Detect has been mainly reported for less experienced readers, but data also exist for more experienced ones, showing upgrade from poor to fair and from moderate to fair, respectively [11, 23].
Our study has limitations. First, it is a single-institution study and our series does not necessarily reflect a general population screened for breast cancer, with different values of prevalence of malignancy. Second, the experience of our readers is not defined by any precise quality standard.
Conclusion
In conclusion, in our experience, S-Detect significantly improved inter-reader agreement in the assessment of FBLs with the use US BI-RADS US lexicon, especially for less experienced readers. In particular, all evaluated descriptors resulted positively influenced by S-Detect assessment, but margin and echo pattern evaluation may need to be further refined, particularly isoechoic pattern.
Compliance with ethical standards
Conflict of interest
Tommaso Vincenzo Bartolotta is lecturer and scientific advisor for Samsung. Other authors declare that they have no conflict of interest.
Ethics approval
This research was supported by research equipment from SAMSUNG MEDISON Co., Ltd. Institutional Review Board approval was granted for the use of all data in this study. The procedures used in this study adhere to the tenets of the Declaration of Helsinki.
Informed consent
Informed consent was obtained from all individual participants included in the study. Permission was obtained from the hospital for patients’ medical records review.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hooley RJ, Scoutt LM, Philpotts LE. Breast ultrasonography: state of the art. Radiology. 2013;268(3):642–659. doi: 10.1148/radiol.13121606. [DOI] [PubMed] [Google Scholar]
- 2.Bartolotta TV, Ienzi R, Cirino A, et al. Characterisation of indeterminate focal breast lesions on grey-scale ultrasound: role of ultrasound elastography. Radiol Med (Torino) 2011;116(7):1027–1038. doi: 10.1007/s11547-011-0648-y. [DOI] [PubMed] [Google Scholar]
- 3.Carlino G, Rinaldi P, Giuliani M, et al. Ultrasound-guided preoperative localization of breast lesions: a good choice. J Ultrasound. 2019;22:85–94. doi: 10.1007/s40477-018-0335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendelson EB, Böhm-Vélez M, Berg WA, et al (2013) ACR BI-RADS® Ultrasound. In: ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; Reston, VA, American College of Radiology
- 5.Hong AS, Rosen EL, Soo MS, Baker JA. BI-RADS for sonography: positive and negative predictive values of sonographic features. Am J Roentgenol. 2005;184(4):1260–1265. doi: 10.2214/ajr.184.4.01841260. [DOI] [PubMed] [Google Scholar]
- 6.Raza S, Goldkamp AL, Chikarmane SA, Birdwell RL. US of breast masses categorized as BI-RADS 3, 4, and 5: pictorial review of factors influencing clinical management. Radiographics. 2010;30(5):1199–1213. doi: 10.1148/rg.305095144. [DOI] [PubMed] [Google Scholar]
- 7.Jalalian A, Mashohor SB, Mahmud HR, Saripan MI, Ramli AR, Karasfi B. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: a review. ClinImaging. 2013;37(3):420–426. doi: 10.1016/j.clinimag.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 8.Bartolotta TV, Orlando AAM, Spatafora L, Dimarco M, Gagliardo C. Taibbi A (2020) S-detect characterization of focal breast lesions according to the US BI RADS lexicon: a pictorial essay. J Ultrasound. 2020 doi: 10.1007/s40477-020-00447-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K, Song MK, Kim EK, Yoon JH. Clinical application of S-Detect to breast masses on ultrasonography: a study evaluating the diagnostic performance and agreement with a dedicated breast radiologist. Ultrasonography. 2017;36(1):3–9. doi: 10.14366/usg.16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Segni M, De Soccio V, Cantisani V, et al. Automated classification of focal breast lesions according to S-detect: validation and role as a clinical and teaching tool. J Ultrasound. 2018;21(2):105–118. doi: 10.1007/s40477-018-0297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartolotta TV, Orlando A, Cantisani V, et al. Focal breast lesion characterization according to the BI-RADS US lexicon: role of a computer-aided decision-making support. RadiolMed. 2018;123(7):498–506. doi: 10.1007/s11547-018-0874-7. [DOI] [PubMed] [Google Scholar]
- 12.Mercado CL. BI-RADS update. RadiolClin North Am. 2014;52(3):481–487. doi: 10.1016/j.rcl.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Lee JH, Seong YK, Chang CH, et al. (2013) Computer- aided lesion diagnosis in B-mode ultrasound by border irregularity and multiple sonographic features. In: Proc. SPIE 8670, Medical Imaging 2013: Computer-Aided Diagnosis, 86701; 2013 Feb 28; Lake Buena Vista, FL, USA
- 14.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 15.Lazarus E, Mainiero MB, Schepps B, Koelliker SL, Livingston LS. BI-RADS lexicon for US and mammography: interobserver variability and positive predictive value. Radiology. 2006;239(2):385–391. doi: 10.1148/radiol.2392042127. [DOI] [PubMed] [Google Scholar]
- 16.Park CS, Lee JH, Yim HW et al. (2007) Observer agreement using the ACR breast imaging reporting and data system (BI-RADS)-ultrasound, First Edition (2003). Korean J Radiol 8(5):397–402. [DOI] [PMC free article] [PubMed]
- 17.Lee HJ, Kim EK, Kim MJ, et al. Observer variability of breast imaging reporting and data system (BI-RADS) for breast ultrasound. Eur J Radiol. 2008;65(2):293–298. doi: 10.1016/j.ejrad.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Abdullah N, Mesurolle B, El-Khoury M, Kao E. Breast imaging reporting and data system lexicon for US: interobserver agreement for assessment of breast masses. Radiology. 2009;252(3):665–672. doi: 10.1148/radiol.2523080670. [DOI] [PubMed] [Google Scholar]
- 19.Park CS, Kim SH, Jung NY, Choi JJ, Kang BJ, Jung HS. Interobserver variability of ultrasound elastography and the ultrasound BI-RADS lexicon of breast lesions. Breast Cancer. 2015;22(2):153–160. doi: 10.1007/s12282-013-0465-3. [DOI] [PubMed] [Google Scholar]
- 20.Youk JH, Jung I, Yoon JH, et al (2016) Comparison of inter-observer variability and diagnostic performance of the fifth edition of BI-RADS for breast ultrasound of static versus video images. Ultrasound Med Biol. 42(9):2083–8. [DOI] [PubMed]
- 21.Lee YJ, Choi SY, Kim KS, Yang PS. Variability in observer performance between faculty members and residents using breast imaging reporting and data system (BI-RADS)-ultrasound Fifth Edition. Iran J Radiol. 2016;13(3):e28281. doi: 10.5812/iranjradiol.28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon JH, Lee HS, Kim YM, et al. Effect of training on ultrasonography (US) BI-RADS features for radiology residents: a multicentre study comparing performances after training. EurRadiol. 2019 doi: 10.1007/s00330-018-5934-9. [DOI] [PubMed] [Google Scholar]
- 23.Park HJ, Kim SM, La Yun B, et al. A computer-aided diagnosis system using artificial intelligence for the diagnosis and characterization of breast masses on ultrasound: added value for the inexperienced breast radiologist. Medicine. 2019;98(3):e14146. doi: 10.1097/MD.0000000000014146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen WC, Chang RF, Moon WK. Computer aided classification system for breast ultrasound based on breast imaging reporting and data system (BI-RADS) Ultrasound Med Biol. 2007;33(11):1688–1698. doi: 10.1016/j.ultrasmedbio.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Catalano O, Varelli C, Sbordone C, et al. A bump: what to do next? Ultrasound imaging of superficial soft-tissue palpable lesions. J Ultrasound. 2019 doi: 10.1007/s40477-019-00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C, Xiao M, Jiang Y, et al. Feasibility of computer-assisted diagnosis for breast ultrasound: the results of the diagnostic performance of S-detect from a single center in China. Cancer Manag Res. 2019;11:921–930. doi: 10.2147/CMAR.S190966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi JH, Kang B, Baek JE, Lee HS, Kim SH. Application of computer-aided diagnosis in breast ultrasound interpretation: improvements in diagnostic performance according to reader experience. Ultrasonography. 2018;37(3):217–225. doi: 10.14366/usg.17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu JY, Zhao ZZ, Zhang WY, Liang M, Ou B, Yang HY, Luo BM. Computer-aided diagnosis of solid breast lesions with ultrasound: factors associated with false-negative and false-positive results. J Ultrasound Med. 2019 doi: 10.1002/jum.15020. [DOI] [PubMed] [Google Scholar]