Abstract
Carbonic anhydrases (CAs) are a family of metalloenzymes that can catalyze the reversible interconversion of CO2/HCO3–, ubiquitously present in both prokaryotes and eukaryotes. In the present study, a CA II (designated as HdhCA II) was sequenced and characterized from the mantle tissue of the Pacific abalone. The complete sequence of HdhCA II was 1,169 bp, encoding a polypeptide of 349 amino acids with a NH2-terminal signal peptide and a CA architectural domain. The predicted protein shared 98.57% and 68.59% sequence identities with CA II of Haliotis gigantea and Haliotis tuberculata, respectively. Two putative N-linked glycosylation motifs and two cysteine residues could potentially form intramolecular disulfide bond present in HdhCA II. The phylogenetic analysis indicated that HdhCA II was placed in a gastropod clade and robustly clustered with CA II of H. gigantea and H. tuberculata. The highest level of HdhCA II mRNA expression was detected in the shell forming mantle tissue. During ontogenesis, the mRNA of HdhCA II was detected in all stages, with larval shell formation stage showing the highest expression level. The in situ hybridization results detected the HdhCA II mRNA expression in the epithelial cells of the dorsal mantle pallial, an area known to express genes involved in the formation of a nacreous layer in the shell. This is the first report of HdhCA II in the Pacific abalone, and the results of this study indicate that this gene might play a role in the shell formation of abalone.
Keywords: Haliotis discus hannai, carbonic anhydrase, qRT-PCR, ontogenesis, in situ hybridization
Introduction
Carbonic anhydrases (CAs) are zinc ion-containing metalloenzymes that can catalyze the essential hydration of CO2 through the simple chemical reaction: CO2 + H2O ⇄ HCO3– + H+ (Lindskog and Silverman, 2000). CAs play an essential role in multiple physiological processes such as pH regulation, electrolyte balance, ionic transportation, carboxylation or decarboxylation reactions, biocalcification, and tumorigenicity (Supuran, 2008, 2011; Alterio et al., 2009). CAs are important components of the CO2-concentrating mechanisms in different groups of algae. They could increase the rate of photosynthesis (Qu et al., 2018). In erythrocytes, CA is a superabundant enzyme that plays an indispensable role in CO2 transport by catalyzing the dehydration of plasma HCO3– ions (Geers and Gros, 2000; Henry and Swenson, 2000; Perry and Gilmour, 2006). The cytosolic CA in gill may contribute to provide counter ions for maintaining pH balance and ionic regulation in fish (Henry and Swenson, 2000; Marshall, 2002). In mollusk, CA seems to be contributed to shell formation via catalyzing the hydration of CO2 (Nielsen and Frieden, 1972). This enzyme has been shown to be an effective catalyst in the calcification mechanism of coral (Rahman and Oomori, 2010).
Carbonic anhydrase isozymes were isolated in the erythroid cells of mammals and have been subsequently identified in most organisms (Meldrum and Roughton, 1933; Rudenko et al., 2015). Eight evolutionarily distinct families of CAs, including α, β, γ, δ, ζ, η, θ, and ι, have been reported in unicellular and multicellular organisms (Zolfaghari et al., 2020). Their amino acid residues share no significant identities and seem to be evolved independently from distinct inherited genes (Krishnamurthy et al., 2008; Bertucci et al., 2009; Del Prete et al., 2015; Supuran and Capasso, 2015; Kikutani et al., 2016). Among these families, α-CA is widely distributed in animals and plants (Aspatwar et al., 2010). α-CA exhibits the highest catalytic activity in the hydration reaction than β- and η-CA. On the contrary, γ-, δ-, and ζ- CA isozymes possess the lowest enzymatic efficiencies (Capasso and Supuran, 2015; Supuran and Capasso, 2015). In mammals, 16 isoforms of α-CA isozymes have been explored, of which 13 are catalytically effective and 3 are non-catalytic due to the absence of one or more functionally active histidine amino acid residues (Sly and Hu, 1995; Tashian et al., 2000). The functions of each isozyme vary pursuant to their molecular sequences, kinetic attributes, sensitivities to inhibitors, tissue distributions, and subcellular localizations (Hewett-Emmett, 2000; Lehtonen et al., 2004).
Carbonic anhydrase II is a secreted and membrane-bound α-CA that can catalyze carboxylation and decarboxylation reactions. The typical structure of CA II contains three histidine (His) residues that can bind to Zn2+ ion, and a proton (H+) shuttling residue that is responsible for converting a Zn-bound water molecule to hydroxide ion. In addition, gate-keeping signature residues (namely, Glu-106 and Thr-199 in human CA) allow excellent orientation of Zn-bound hydroxide ion to increase the nucleophilic attack of a substrate (Christianson and Fierke, 1996; Lindskog and Silverman, 2000). CA II not only participates in the hydration reaction but also plays an important role in the osmoregulatory functions of fish (Grosell et al., 2007).
The Pacific abalone is a commercially important molluscan bioresources in China, Japan, and Korean Peninsula. Haliotis discus hannai is considered as a popular seafood item worldwide due to its contents of health beneficial bioactive molecules (Suleria et al., 2017). Previous studies have characterized cytosolic CA isozymes in vertebrates and invertebrates (Pongsomboon et al., 2009; Le Roy et al., 2012; Ali et al., 2015; Pan et al., 2016; Sumi et al., 2019). However, the characterization and expression analysis of CA isozymes in Pacific abalone have not yet been reported. In this study, the complete sequence of CA II isozyme was first cloned from the mantle of H. discus hannai, and its spatiotemporal expression was determined using the molecular assay.
Materials and Methods
Animals and Sample Collection
Three-year-old adult male and female Pacific abalone, H. discus hannai (total body mass: 128.2 ± 0.86 g; shell length: 10.5 ± 0.12 cm) were collected from Jindo Island, South Korea and transferred to the laboratory, College of Fisheries and Ocean Science, Chonnam National University (CNU). The tissues from the cerebral ganglion, mantle, gill, heart, shell muscle, hemocyte, testis, and ovary were collected, immediately frozen in liquid nitrogen, and kept at − 80°C for further RNA isolation. All experimental embryonic and larval samples were collected as described previously (Sharker et al., 2020a). The cryosection from the mantle tissue was prepared following the previous protocol (Sharker et al., 2020b, c, d). The experimentation was performed according to the guidelines of the Institutional Animal Care and Use Committee of CNU (approval number: CNU IACUC-YS-2020-5).
RNA Extraction and cDNA Synthesis
Total RNA was isolated from different tissues of an experimental animal using an RNeasy mini kit (Qiagen, Hilden, Germany) following the kit protocol. The quality of each RNA sample was evaluated using 1% (w/v) agarose gel electrophoresis and quantified by spectrophotometry on a NanoDrop® NP 1000 device (Thermo Fisher Scientific, Waltham, MA, United States). Subsequently, 1 μg of RNA was transformed into cDNA employing Superscript® III cDNA synthesis kit (Invitrogen, Carlsbad, CA, United States) as per the kit instruction.
Cloning and Sequencing of Full-Length cDNA of CA II
A pair of primer (forward: 5′-GTGGCAGTCTTCCTATCTAC-3′; reverse: 5′-GCTGCATCATCACCTGCCA-3′) was designed based on the nucleotide sequence of Haliotis gigantea CA isozyme (GenBank accession no. AB500104.1). Reverse transcription polymerase chain reaction (RT-PCR) amplification reactions were carried out using the following amplification program: 3 min at 95°C, followed by 35 cycles of 2 min at 94°C, 1 min at 58°C, 1 min at 72°C, with a final extension step at 72°C for 5 min. The purification was carried out using the PCR purification kit as per the kit protocol. Subsequently, the purified fragments were cloned into pTOP Blunt V2 vector (Enzynomics, Daejeon, South Korea) and transformed into DH5α-competent Escherichia coli cells (Enzynomics). Then Plasmid DNA from selected clones was isolated using plasmid miniprep kit (Qiagen, Hilden, Germany) and sequenced by a sequencing company (Macrogen, South Korea). Complete sequence of CA II was obtained from H. discus hannai by rapid amplification of cDNA ends (RACE) using a Smarter® RACE cDNA Kit (Clontech Laboratories, Inc., United States) as per the protocol provided by the manufacturer. The touchdown PCR was carried out with 25 cycles for 3′-RACE and 30 cycles for 5′-RACE using gene-specific primers (GSPs) set (antisense primer: 5′-GATT ACGCCAAGCTTCCATGGCTCCTGTACACGGTTCTTCC-3′, sense primer: 5′-GATTACGCCAAGCTTCACTTTGTCTGAG AGCGTCCTGTGGC-3′), a universal primer mix (UPM), and SeqAmp DNA Polymerase in 50 μL of reaction volume following the instruction provided by the manufacturer. The resultant PCR products were purified, ligated into linearized pRACE vector, transformed into Stellar Competent Cells, and finally sequenced as described earlier.
Sequence and Phylogenetic Analysis
The nucleotide and amino acid sequence of Pacific abalone CA II was analyzed with BLAST at the NCBI database. A web-based tool “SMART” was used for the prediction of CA domain architecture (Letunic and Bork, 2018). Expert protein analysis system was used to evaluate the physiochemical properties and subcellular localization of this gene (Gasteiger et al., 2003). Multiple sequence alignment was created using Clustal Omega package (Sievers et al., 2011; Alva et al., 2016). The Jalview Java alignment editor was employed to edit and visualize multiple sequence alignment (Waterhouse et al., 2009). Predictions of the N-linked glycosylation sites and serine/threonine phosphorylation sites were performed with NetNGlyc 1.0 server (Chuang et al., 2012) and NetPhosK 3.1 server (Blom et al., 1999), respectively. The N-terminal signal peptide and disulfide bond were predicted using SignalP 4.1 (Petersen et al., 2011) and CYSPRED (Fariselli et al., 1999), respectively. To generate a phylogram, vertebrate and molluscan CAs were curated from NCBI using BLASTP program. A phylogenetic analysis was conducted with MEGA software (version 7.0) using bootstrap analysis for 1,000 replicates (Kumar et al., 2016).
Template Identification and Three-Dimensional Homology Modeling of H. discus hannai CA II
Modeler1 was used for the analysis of high-resolution three-dimensional (3D) homology modeling of H. discus hannai CA II isozyme by optimally satisfying spatial restraints (Šali and Blundell, 1993). Human CA II 3D structure (1.07 Å) template was considered to generate the 3D model of Pacific abalone CA II. Protein Quality Predictor (Wallner and Elofsson, 2003), Verify3D (Eisenberg et al., 1997), and ERRAT tools were used for assessing the stereochemical quality of the predicted protein model (Colovos and Yeates, 1993). UCSF Chimera program was used for interactive visualization and analysis of the predicted CA II 3D structure (Pettersen et al., 2004).
Semiquantitative RT-PCR
A primer set (forward: 5′-GAACAGGGTGTGTGACACG-3′ and reverse: 5′-GCAGAACGATGTCCGAAATAG-3′) designed from the cloned sequence was applied to conduct semiquantitative RT-PCR. Ribosomal protein L-5, RPL-5 (GenBank accession no JX002679.1) (forward: 5′-TGTCCGTTTCACCAACAAGG-3′ and reverse: 5′-AGATGGAATCAAGTTTCAATT-3′), was selected as a reference gene based on its expression stability (Wan et al., 2011). The PCR amplification conditions were similar to those described earlier.
Quantitative RT-PCR Analysis
The quantitative RT-PCR (qRT-PCR) was carried out in triplicates using 2 × qPCRBIO SyGreen Mix Lo-Rox on a LightCycler® 96 System (Roche, Germany) in a 20-μL reaction mixture. Three biological replicates (N = 3) were used for each tissue and ontogenetic sample. The same gene-specific and RPL-5 primers used for semiquantitative RT-PCR analysis were used for qRT-PCR. The PCR amplification programs were subjected to a predenaturation step at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 30 s and 72°C for 1 min. Relative mRNA expression was assessed using the 2–ΔΔCT method.
Statistical Analysis
Data were statistically analyzed using one-way ANOVA followed by Tukey’s multiple comparisons using SPSS (version 16.0) to assess whether the means were significantly different. Statistically significant difference was set at p < 0.05.
In situ Hybridization
Digoxenin (DIG)-labeled RNA antisense and sense probes were synthesized from the CDS region of CA II sequence by in vitro transcription as described earlier (Sharker et al., 2020e, f). The hybridized tissue sections of the mantle were incubated with a blocking solution at RT for 1 h and then treated with an antibody at − 20°C overnight. Subsequently, the tissue sections were incubated with a labeling mix and kept in a dark place to attain color. Finally, the slides were examined under a stereomicroscope.
Results
Identification and Characterization of CA II From H. discus hannai
The complete cDNA sequence of CA II was isolated and cloned from the mantle tissue of H. discus hannai and referred to as HdhCA II (GenBank accession number MT876410). Its nucleotide sequence was 1,169 bp in length encoding a polypeptide of 349 amino acids with the calculated molecular mass and isoelectric point (pI) of 38.93 kDa and 8.58, respectively (Figure 1). The protein domain analysis revealed that HdhCA II (from 50Y to 344C) showed similarity with a potential CA isoform II. Its coding region comprised a predicted signal peptide (18 amino acids) followed by a cleavage site between Ala18 and Asp19. The cloned sequence contained two N-linked glycosylation sites and eight phosphorylation sites at positions 49S, 55S, 67T, 137S, 142S, 150T, 202S, and 319S. Two cysteine residues (Cys-40 and Cys-247) in this sequence are likely to form intramolecular disulfide bond for the enzyme biosynthesis.
FIGURE 1.
Nucleotide and amino acid sequence of HdhCA II. The start codon, stop codon (asterisks), and polyadenylation signal (AATAAA) are marked in bold. The N-terminal signal peptide is underlined. The N-linked glycosylation site is enclosed in a rectangular box. The circles indicate potential phosphorylation sites in the mature protein. Two cysteine residues (Cys-40 and Cys-247) could potentially form intramolecular disulfide bond and are shaded in gray.
The protein BLAST analysis demonstrated that the predicted CA II sequence shared the highest identities with H. gigantea and Haliotis tuberculata CA II. The alignment of fish and mammalian vertebrate CA sequences revealed that the cloned Pacific abalone CA II sequence shared 29.03%, 28.34%, and 29.67% sequence identities with human (Homo sapiens, NP_000058.1), mouse (Mus musculus, NP_033931.4), and zebra fish (NP_954685.1, Danio rerio) CA II, respectively (Table 1).
TABLE 1.
Amino acid sequence identities of HdhCA II with CA IIs of other gastropod mollusk, eutherian mammals, and piscine vertebrates.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 81.15 | 80.77 | 62.69 | 62.65 | 34.91 | 30.34 | 29.13 | 29.03 | 1. Human | |
| 93.46 | 60.38 | 61.09 | 36.64 | 30.72 | 28.99 | 28.34 | 2.Mouse | ||
| 60.77 | 60.70 | 34.91 | 29.10 | 28.16 | 27.74 | 3.Rat | |||
| 72.76 | 32.33 | 30.10 | 30.07 | 29.67 | 4. Zebra fish | ||||
| 32.13 | 29.97 | 30.51 | 29.43 | 5. Rainbow trout | |||||
| 31.29 | 29.72 | 28.75 | 6. Sea hare | ||||||
| 69.16 | 68.59 | 7. Green ormer | |||||||
| 98.57 | 8. Giant abalone | ||||||||
| 9. Pacific abalone |
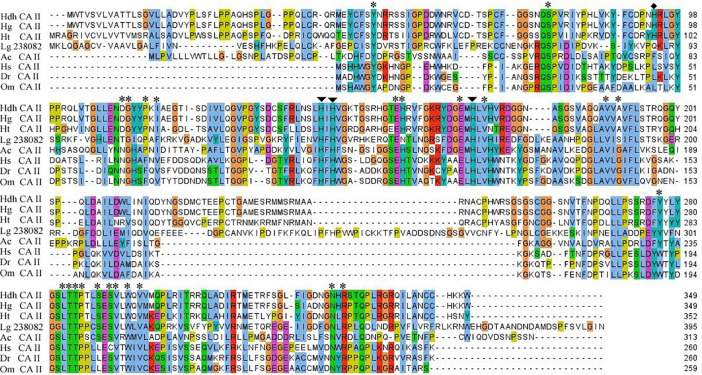
The in silico analysis indicated that this protein might be an extracellular (secreted) protein. The active site amino acid residues in CA domain of Pacific abalone and other cytoplasmic CAs of vertebrates and invertebrates are highly conserved (Figure 2). The three histidine residues predicted to form Zn2+ in the active site are also conserved in all CA isoforms. The histidine residue (94H) important for proton shuttling is also conserved in molluscan CAs. In addition, several other highly conserved amino acids are found in this cloned sequence.
FIGURE 2.
Multiple sequence alignment of HdhCA II and CA IIs of other representative invertebrate and vertebrate species. Three zinc ligand histidine residues and proton shuttling residues are indicated by arrows and diamond circle, respectively. Amino acid residue in the catalytic site involved in the hydrogen bond formation is denoted by asterisks. Hdh, H. discus hannai; Hg, H. gigantea; Ht, H. tuberculata; Lg, Lottia gigantea; Ac, Aplysia californica; Hs, Homo sapiens; Dr, Danio rerio; and Om, Oncorhynchus mykiss.
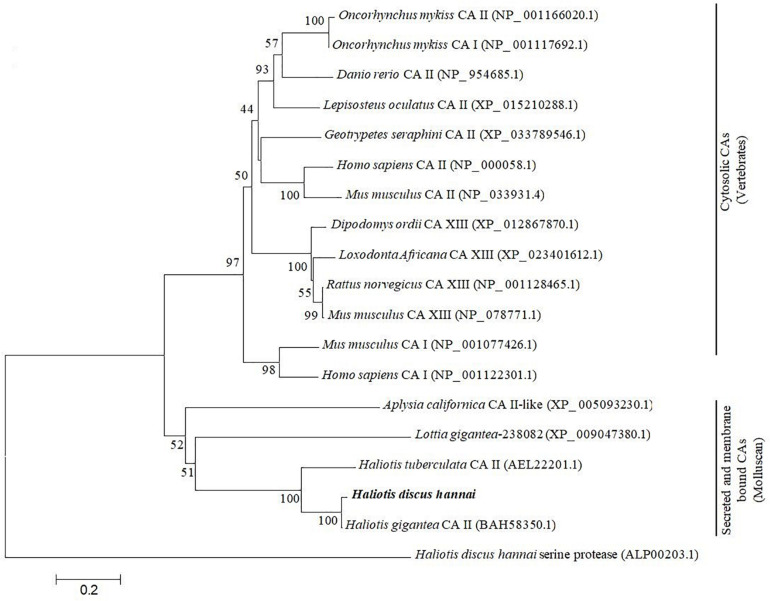
The phylogenetic analysis was performed using CAs of representative species of vertebrates, and molluscan with the neighbor joining (NJ) method to infer evolutionary connections. The phylogenetic tree showed two major clades: (1) cytosolic CAs in vertebrates and (2) secreted and membrane-bound CAs in mollusk. The CA II of H. discus hannai was placed in the molluscan clade and phylogenetically clustered with H. gigantea CA II with a high bootstrap value (Figure 3).
FIGURE 3.
Molecular phylogenetic analysis of carbonic anhydrase isoform was constructed using NJ approach with 1,000 bootstrap replications. The scale bar at the bottom represents the amino acid divergence per site. The numbers in phylogram nodes indicate percentage bootstrap values for the phylogeny. HdhCA II is highlighted in bold.
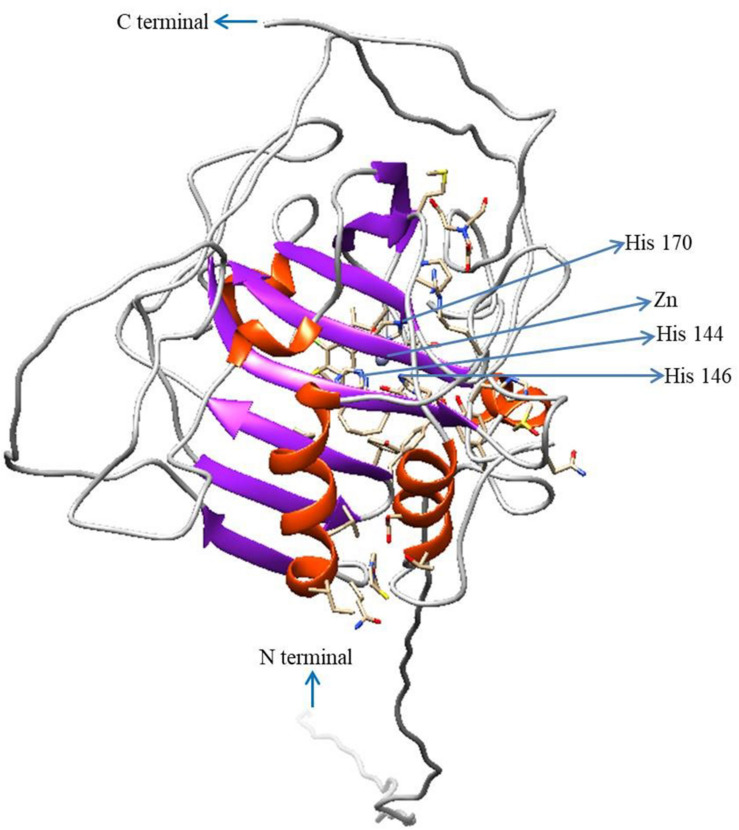
To predict the 3D model of CA II, the crystal structure of human CA II (PDB 4Q08) was selected based on the high identities of several amino acid signatures (Figure 4). The evaluation results of this predicted model were as follows: ProQ, LG score of 2.780 (value > 1.5 indicates very good model), and MaxSub sore of 0.585 (value > 0.5 indicates a very good model); Verify3D: 3D/1D profile score of 89.34%; and ERRAT quality factor of 92.94%.
FIGURE 4.
Three-dimensional structure of HdhCA II. The N- and C-termini are marked with blue arrows. Zn2+ and coordinated three histidine residues are indicated by blue arrows. The structure was generated using UCSF Chimera software.
Expression Analysis of HdhCA II mRNA
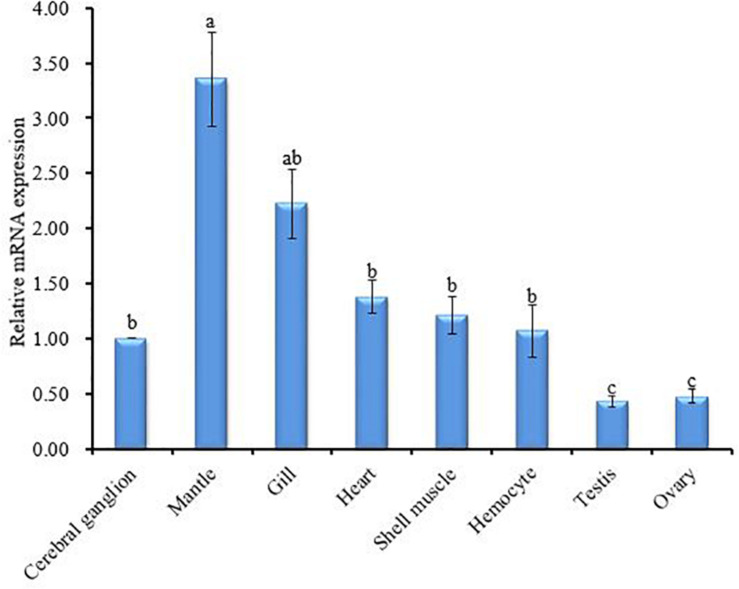
The tissue-specific expression profile of CA II was analyzed by qRT-PCR. The mantle tissue exhibited the highest level of HdhCA II mRNA expression than other tested tissues (Figure 5). The expression of HdhCA II mRNA among cerebral ganglion, heart, shell muscle, and hemocyte showed no significant differences. A significantly lower expression was found in gonadal tissues (i.e., testis and ovary). The supporting data are shown in Supplementary Figure 1.
FIGURE 5.
HdhCA II mRNA expression (means ± SD, N = 3) in different tissues of Pacific abalone is based on quantitative real-time PCR (qRT-PCR). The expression pattern of HdhCA II mRNA in all tissues is calibrated by the expression in the cerebral ganglion (1). Different characters in vertical bar indicate significantly (p < 0.05) different.
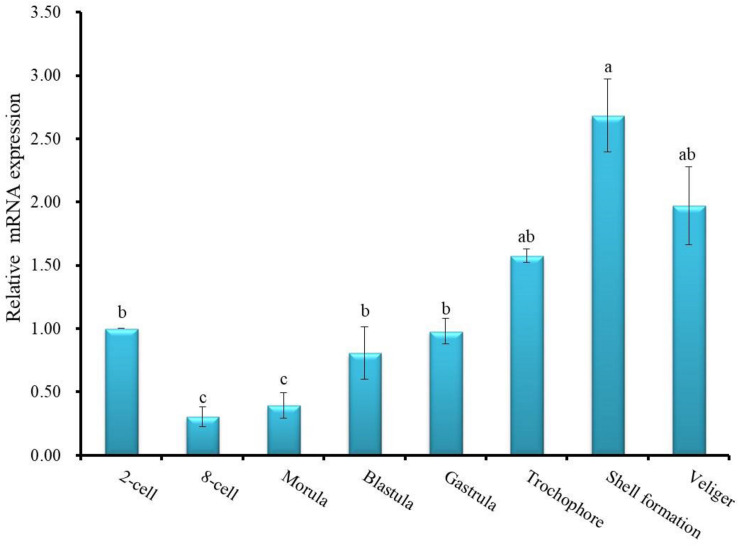
To investigate the functional role of HdhCA II during ontogenetic development of the Pacific abalone, the expression patterns of CA II mRNA transcript in different stages of development were determined using the qRT-PCR assay. The results of the analysis revealed that HdhCA II mRNA was expressed throughout the early developmental stages in a ubiquitous fashion (Figure 6). The HdhCA II mRNA levels were relatively low in multicellular stages until gastrula. The expression level was highest in the shell formation stage compared with other examined stages.
FIGURE 6.
HdhCA II mRNA expression (means ± SD, N = 3) in different ontogenetic stages of the Pacific abalone. The expression levels of HdhCA II mRNA in various stages are calibrated by its expression in the two-cell stage (1). Different letters indicate significantly (p < 0.05) different.
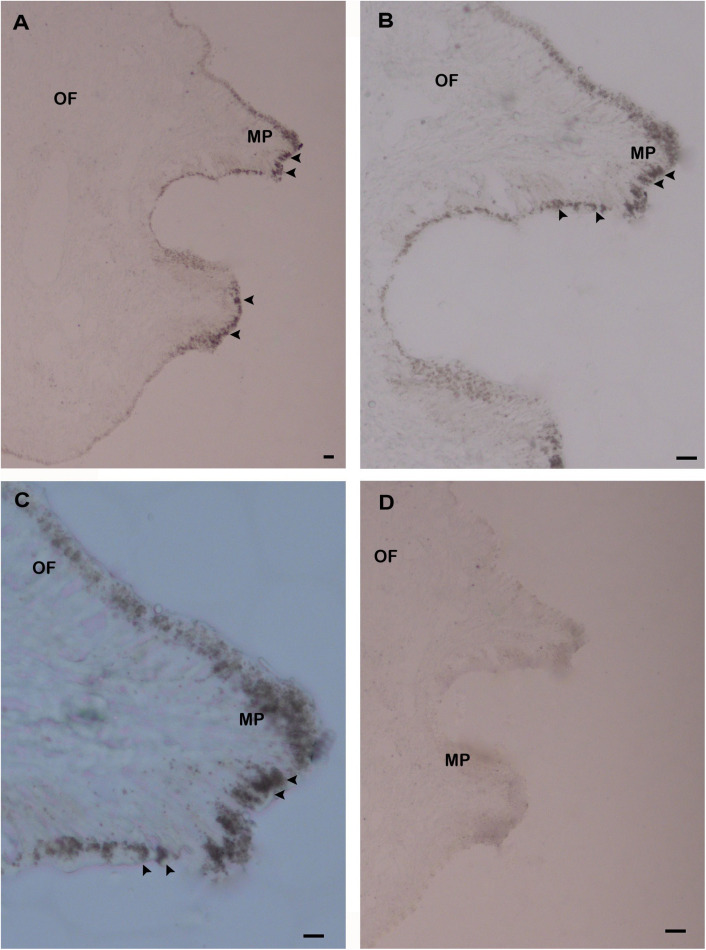
The in situ hybridization (ISH) was carried out using the mantle tissue sections to elucidate the functional role of HdhCA II mRNA in the shell formation of H. discus hannai. The HdhCA II mRNA hybridized signal was found in epithelial cells of the dorsal mantle pallial, an area known to express genes involved in the nacreous layer synthesis of the shell (Figures 7A–C). However, the negative control (sense probe) showed no hybridization signal (Figure 7D).
FIGURE 7.
The localization of HdhCA II mRNA in the mantle tissue of Pacific abalone detected by ISH. (A) The positive hybridization signals were detected in the epithelial cells of the dorsal mantle pallial (MP), (B) the medium magnification of A, (C) the higher magnification of A, and (D) the negative control section showed no hybridization signals. The positive signals are marked with black arrowheads. Scale bar, 100 μm. OF, outer fold; MP, mantle pallial.
Discussion
Carbonic anhydrases play an important role in many physiological processes by catalyzing the hydration reaction. In mollusks, CAs have been previously identified in Tridacna squamosa (Ip et al., 2017), Mytilus galloprovincialis (Perfetto et al., 2017), and H. tuberculata (Le Roy et al., 2012). To date, the identification and biomolecular characterization of CA II isoform from the Pacific abalone have not yet been reported. For the first time, the complete sequence of CA II was cloned from the mantle tissue of H. discus hannai and the molecular properties of this protein with its expression profile were determined in this study. An 18-amino-acid NH2-terminal signal sequence was found in the CA II isozyme followed by a cleavage site, suggesting that HdhCA II might be an extracellular secretory protein (Figure 1). The N-terminal signal sequence is a key characteristic of CA secretory protein (Aldred et al., 1991). A secretory CA has been cloned from the scleractinian coral, Stylophora pistillata, and this CA is localized in calicodermis, which is responsible for the precipitation of the skeleton (Moya et al., 2008). One CA isoform was isolated from the sea urchin embryo and described as an extracellular secreted protein (Karakostis et al., 2016). The cloned sequence of HdhCA II also possesses several key features including phosphorylation sites and N-linked glycosylation sites. These phosphorylation sites are crucial for several signal transduction cascades (Ali et al., 2016). Two potential N-linked glycosylation motifs were found in HdhCA II, suggesting that HdhCA II might be a glycoprotein. Two cysteine residues found in HdhCA II might form disulfide link that is crucial for stabilizing its protein structure and regulating biological functions of this protein (Kadokura et al., 2004; Inaba et al., 2006).
The amino acid sequences encoded by HdhCA II displayed high identities in the functional site of the CA domain (Figure 2). The molecular structure of HdhCA II isozyme contained important functional sites, such as zinc binding ligand, proton shuttling ligand, substrate associated pocket, and Thr-199 loop site, which are known to be involved in the enzymatic activity of this protein (Esbaugh and Tufts, 2006). The active site of CA contained a hydrophobic pocket (i.e., the catalytically productive site) that could interact with a non-polar CO2 substrate (Liang and Lipscomb, 1990) and facilitate its reaction with highly nucleophilic Zn2+-bound OH– (Alterio et al., 2012). The hydrophobic binding pocket residue and histidine residues that could bind with a catalytic zinc ion were conserved in CA II of H. discus hannai (Figure 2). This suggests that HdhCA II is a functionally active CA. It has been well established that the proton-shuttling residue (His-64) is responsible for the efficient proton transfer and the high catalytic rate of CO2 hydration in CA II of human (Supuran, 2008). A site-specific mutation (His-64 replaced by alanine) results in 20- to 30-fold decrease in the catalytic activity of CA II (Tu et al., 1989). This residue is also conserved among different species of abalone CA II.
The results of phylogenetic analysis indicated that CA II gene of H. discus hannai was evolutionarily closer to H. gigantea CA II (Figure 3). Previous studies reported that the CA II of H. tuberculata (htCA2) is placed in the molluscan CA clade and more closely linked to CA II isoform of H. gigantea (Le Roy et al., 2012).
The homology modeling of HdhCA II was performed using the 3D homology structure of human CA II with a resolution of 1.07 Å as template (Figure 4). The Zn2+ coordinated with three conserved histidine residues comprise the zinc-binding site (Christianson and Alexander, 1989). The evaluation results also supported the structural conservation of the cloned HdhCA II gene, with amino acids in favorable positions.
The expression of HdhCA II mRNA was detected in all tested tissues with mantle as the site of highest expression (Figure 5) which is in agreement with the previous report (Le Roy et al., 2012; Ip et al., 2017). The expression analysis suggests that HdhCA II could involve in mantle function such as shell formation. HdhCA II might also involve in acid–base regulation, ion transport, and modulation of ionic concentration (Miyashita et al., 2012).
The temporal expression profile of HdhCA II during ontogenesis revealed that HdhCA II mRNA was expressed throughout the early developmental stages, with shell formation stage having the highest level (Figure 6). This result of analysis suggests that HdhCA II plays an important role during larval shell formation. Previous studies have reported that the functional inhibition of CA in Paracentrotus lividus and Heliocidaris tuberculata can prevent the deposition of calcium carbonate in the larval skeleton formation (Zito et al., 2015).
The expression of HdhCA II mRNA in the mantle tissue was examined with ISH using an antisense CA II mRNA as a probe. The gene distributed in the mantle can speculate the participation of these genes in the biomineralization process during shell formation (Suzuki and Nagasawa, 2013). The expression of genes at the mantle edge and mantle pallial have participated in the synthesis of prismatic and nacreous layers, respectively (Takeuchi and Endo, 2006; Inoue et al., 2010). The expression patterns of HdhCA II transcript were detected in the epithelium layer of the mantle and mantle pallial (Figure 5). The mantle can secrete biomineralization protein in outer epithelial cells to modulate shell formation (Jablonski, 1990; Werner et al., 2013). Based on the in situ results of the present study, we speculated that HdhCA II might be involved in the shell formation by catalyzing the hydration of CO2.
Conclusion
This is the first study of molecular characterization and expression of HdhCA II mRNA in different tissues and developmental stages of the Pacific abalone. HdhCA II was highly expressed in the mantle tissue implying that it might be participated in the shell formation process. The expression patterns of HdhCA II during larval developmental stages imply that this enzyme is involved in the shell germination of abalone. The ISH results demonstrated that the signals were found in the mantle epithelial cells, indicating that this gene might be essential for the shell formation by controlling the regular deposition of CaCO3. The findings of our current research could help us to understand the functional role of CA in the shell formation of abalone or be useful for the development of aquaculture methods in this abalone species.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI GenBank, accession number: MT876410, https://www.ncbi.nlm.nih.gov/nuccore/MT876410.1.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of CNU (approval number: CNU IACUC-YS-2020-5).
Author Contributions
KK conceptualized and designed the experiments and prepared the manuscript. MS designed and conducted the experiment, analyzed the data, and wrote the manuscript. ZS conducted the ISH, analyzed the data, and prepared graphs. KS analyzed the data. SC and KC helped to plan the experiment, revised the manuscript, and gave intellectual input to improve it. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The present research was supported by the Ministry of Oceans and Fisheries, South Korea (grant no. 2018-2129). This study was part of the project entitled “Development of technology for abalone aquaculture using sperm cryopreservation (grant no. 2018-2129).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.669235/full#supplementary-material
HdhCA II mRNA expression in the cerebral ganglion, shell muscle, mantle, gill, heart, hemocyte, testis, and ovary was determined by semiquantitative reverse transcription (RT)-PCR. RPL-5 was used as reference gene.
References
- Aldred P., Fu P., Barrett G., Penschow J. D., Wright R. D., Coghlan J. P., et al. (1991). Human secreted carbonic anhydrase: cDNA cloning, nucleotide sequence, and hybridization histochemistry. Biochemistry 30 569–575. 10.1021/bi00216a035 [DOI] [PubMed] [Google Scholar]
- Ali E. S., Hua J., Wilson C. H., Tallis G. A., Zhou F. H., Rychkov G. Y., et al. (2016). The glucagon-like peptide-1 analogue exendin-4 reverses impaired intracellular Ca2 + signalling in steatotic hepatocytes. Biochim. Biophys. Acta - Mol. Cell Res. 1863 2135–2146. 10.1016/j.bbamcr.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Ali M. Y., Pavasovic A., Mather P. B., Prentis P. J. (2015). Analysis, characterisation and expression of gill-expressed carbonic anhydrase genes in the freshwater crayfish Cherax quadricarinatus. Gene 564 176–187. 10.1016/j.gene.2015.03.074 [DOI] [PubMed] [Google Scholar]
- Alterio V., Di Fiore A., D’Ambrosio K., Supuran C. T., De Simone G. (2009). “X-Ray crystallography of carbonic anhydrase inhibitors and its importance in drug design,” in Drug Design of Zinc-Enzyme Inhibitors, (Hoboken, NJ: John Wiley & Sons, Inc; ), 138–166. 10.1002/9780470508169.ch4 [DOI] [Google Scholar]
- Alterio V., Di Fiore A., D’Ambrosio K., Supuran C. T., De Simone G. (2012). Multiple binding modes of inhibitors to carbonic anhydrases: How to design specific drugs targeting 15 different isoforms? Chem. Rev. 112 4421–4468. 10.1021/cr200176r [DOI] [PubMed] [Google Scholar]
- Alva V., Nam S.-Z., Söding J., Lupas A. N. (2016). The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 44 W410–W415. 10.1093/nar/gkw348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspatwar A., Tolvanen E. E., Ortutay M. C., Parkkila S. (2010). Carbonic Anhydrase Related Protein VIII and its Role in Neurodegeneration and Cancer. Curr. Pharm. Des. 16 3264–3276. 10.2174/138161210793429823 [DOI] [PubMed] [Google Scholar]
- Bertucci A., Innocenti A., Zoccola D., Scozzafava A., Tambutté S., Supuran C. T. (2009). Carbonic anhydrase inhibitors. Inhibition studies of a coral secretory isoform by sulfonamides. Bioorganic Med. Chem. 17 5054–5058. 10.1016/j.bmc.2009.05.063 [DOI] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294 1351–1362. 10.1006/jmbi.1999.3310 [DOI] [PubMed] [Google Scholar]
- Capasso C., Supuran C. T. (2015). An overview of the alpha-, beta- and gamma-carbonic anhydrases from bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzyme Inhib. Med. Chem. 30 325–332. 10.3109/14756366.2014.910202 [DOI] [PubMed] [Google Scholar]
- Christianson D. W., Alexander R. S. (1989). Carboxylate–histidine–zinc interactions in protein structure and function. J. Am. Chem. Soc. 111 6412–6419. 10.1021/ja00198a065 [DOI] [Google Scholar]
- Christianson D. W., Fierke C. A. (1996). Carbonic Anhydrase: Evolution of the zinc binding site by nature and by design. Acc. Chem. Res. 29 331–339. 10.1021/ar9501232 [DOI] [Google Scholar]
- Chuang G. Y., Boyington J. C., Gordon Joyce M., Zhu J., Nabel G. J., Kwong P. D., et al. (2012). Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics 28 2249–2255. 10.1093/bioinformatics/bts426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C., Yeates T. O. (1993). Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 2 1511–1519. 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete S., Vullo D., De Luca V., Alothman Z., Osman S. M., Supuran C. T., et al. (2015). Biochemical characterization of recombinant β-carbonic anhydrase (PgiCAb) identified in the genome of the oral pathogenic bacterium Porphyromonas gingivalis. J. Enzyme Inhib. Med. Chem. 30 366–370. 10.3109/14756366.2014.931383 [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Lüthy R., Bowie J. U. (1997). VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 277 396–404. 10.1016/S0076-6879(97)77022-8 [DOI] [PubMed] [Google Scholar]
- Esbaugh A. J., Tufts B. L. (2006). The structure and function of carbonic anhydrase isozymes in the respiratory system of vertebrates. Respir. Physiol. Neurobiol. 154 185–198. 10.1016/j.resp.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Fariselli P., Riccobelli P., Casadio R. (1999). Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins Struct. Funct. Genet. 36 340–346. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31 3784–3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geers C., Gros G. (2000). Carbon dioxide transport and carbonic anhydrase in blood and muscle. Physiol. Rev. 80 681–715. 10.1152/physrev.2000.80.2.681 [DOI] [PubMed] [Google Scholar]
- Grosell M., Gilmour K. M., Perry S. F. (2007). Intestinal carbonic anhydrase, bicarbonate, and proton carriers play a role in the acclimation of rainbow trout to seawater. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 293 R2099–R2111. 10.1152/ajpregu.00156.2007 [DOI] [PubMed] [Google Scholar]
- Henry R. P., Swenson E. R. (2000). The distribution and physiological significance of carbonic anhydrase in vertebrate gas exchange organs. Respir. Physiol. 121 1–12. 10.1016/S0034-5687(00)00110-9 [DOI] [PubMed] [Google Scholar]
- Hewett-Emmett D. (2000). Evolution and distribution of the carbonic anhydrase gene families. EXS 20 29–76. 10.1007/978-3-0348-8446-4_3 [DOI] [PubMed] [Google Scholar]
- Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., et al. (2006). Crystal structure of the DsbB–DsbA complex reveals a mechanism of disulfide bond generation. Cell 127 789–801. 10.1016/j.cell.2006.10.034 [DOI] [PubMed] [Google Scholar]
- Inoue N., Ishibashi R., Ishikawa T., Atsumi T., Aoki H., Komaru A. (2010). Gene expression patterns and pearl formation in the Japanese pearl oyster (Pinctada fucata): A comparison of gene expression patterns between the pearl sac and mantle tissues. Aquaculture 308 S68–S74. 10.1016/j.aquaculture.2010.06.036 [DOI] [Google Scholar]
- Ip Y. K., Koh C. Z. Y., Hiong K. C., Choo C. Y. L., Boo M. V., Wong W. P., et al. (2017). Carbonic anhydrase 2-like in the giant clam, Tridacna squamosa: Characterization, localization, response to light, and possible role in the transport of inorganic carbon from the host to its symbionts. Physiol. Rep. 5 13494. 10.14814/phy2.13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski D. (1990). On Biomineralization. Heinz A. Lowenstam, Stephen Weiner. J. Geol. 98 977–977. 10.1086/629466 [DOI] [Google Scholar]
- Kadokura H., Tian H., Zander T., Bardwell J. C. A., Beckwith J. (2004). Snapshots of DsbA in ACTION: Detection of proteins in the process of oxidative folding. Science 303 534–537. 10.1126/science.1091724 [DOI] [PubMed] [Google Scholar]
- Karakostis K., Costa C., Zito F., Brümmer F., Matranga V. (2016). Characterization of an alpha type carbonic anhydrase from Paracentrotus lividus Sea Urchin embryos. Mar. Biotechnol. 18 384–395. 10.1007/s10126-016-9701-0 [DOI] [PubMed] [Google Scholar]
- Kikutani S., Nakajima K., Nagasato C., Tsuji Y., Miyatake A., Matsuda Y. (2016). Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. U. S. A. 113 9828–9833. 10.1073/pnas.1603112113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy V. M., Kaufman G. K., Urbach A. R., Gitlin I., Gudiksen K. L., Weibel D. B., et al. (2008). Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein-ligand binding. Chem. Rev. 108 946–1051. 10.1021/cr050262p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy N., Marie B., Gaume B., Guichard N., Delgado S., Zanella-Cléon I., et al. (2012). Identification of two carbonic anhydrases in the mantle of the European abalone Haliotis tuberculata (Gastropoda, Haliotidae): Phylogenetic implications. J. Exp. Zool. Part B Mol. Dev. Evol. 318 353–367. 10.1002/jez.b.22452 [DOI] [PubMed] [Google Scholar]
- Lehtonen J., Shen B., Vihinen M., Casini A., Scozzafava A., Supuran C. T., et al. (2004). Characterization of CA XIII, a novel member of the carbonic anhydrase isozyme family. J. Biol. Chem. 279 2719–2727. 10.1074/jbc.M308984200 [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46 D493–D496. 10.1093/nar/gkx922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. Y., Lipscomb W. N. (1990). Binding of substrate CO2 to the active site of human carbonic anhydrase II: A molecular dynamics study. Proc. Natl. Acad. Sci. U. S. A. 87 3675–3679. 10.1073/pnas.87.10.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindskog S., Silverman D. N. (2000). The catalytic mechanism of mammalian carbonic anhydrases. EXS 10 175–195. 10.1007/978-3-0348-8446-4_10 [DOI] [PubMed] [Google Scholar]
- Marshall W. S. (2002). Na+, Cl–, Ca2+ and Zn2+ transport by fish gills: retrospective review and prospective synthesis. J. Exp. Zool. 293 264–283. 10.1002/jez.10127 [DOI] [PubMed] [Google Scholar]
- Meldrum N. U., Roughton F. J. W. (1933). Carbonic anhydrase. Its preparation and properties. J. Physiol. 80 113–142. 10.1113/jphysiol.1933.sp003077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T., Takami A., Takagi R. (2012). Molecular cloning and characterization of the 5′-flanking regulatory region of the carbonic anhydrase nacrein gene of the pearl oyster Pinctada fucata and its expression. Biochem. Genet. 50 673–683. 10.1007/s10528-012-9510-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Tambutté S., Bertucci A., Tambutté E., Lotto S., Vullo D., et al. (2008). Carbonic anhydrase in the scleractinian coral Stylophora pistillata: Characterization, localization, and role in biomineralization. J. Biol. Chem. 283 25475–25484. 10.1074/jbc.M804726200 [DOI] [PubMed] [Google Scholar]
- Nielsen S. A., Frieden E. (1972). Carbonic anhydrase activity in molluscs. Comp. Biochem. Physiol. – Part B Biochem. 41 461–468. 10.1016/0305-0491(72)90107-1 [DOI] [PubMed] [Google Scholar]
- Pan L., Hu D., Liu M., Hu Y., Liu S. (2016). Molecular cloning and sequence analysis of two carbonic anhydrase in the swimming crab Portunus trituberculatus and its expression in response to salinity and pH stress. Gene 576 347–357. 10.1016/j.gene.2015.10.049 [DOI] [PubMed] [Google Scholar]
- Perfetto R., Del Prete S., Vullo D., Carginale V., Sansone G., Barone C. M. A., et al. (2017). Cloning, expression and purification of the α-carbonic anhydrase from the mantle of the Mediterranean mussel. Mytilus galloprovincialis. J. Enzyme Inhib. Med. Chem. 32 1029–1035. 10.1080/14756366.2017.1353502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. F., Gilmour K. M. (2006). Acid-base balance and CO2 excretion in fish: Unanswered questions and emerging models. Respir. Physiol. Neurobiol. 154 199–215. 10.1016/j.resp.2006.04.010 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., Von Heijne G., Nielsen H. (2011). SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pongsomboon S., Udomlertpreecha S., Amparyup P., Wuthisuthimethavee S., Tassanakajon A. (2009). Gene expression and activity of carbonic anhydrase in salinity stressed Penaeus monodon. Comp. Biochem. Physiol. - A Mol. Integr. Physiol. 152 225–233. 10.1016/j.cbpa.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Qu C., He Y., Zheng Z., An M., Li L., Wang X., et al. (2018). Cloning, expression analysis and enzyme activity assays of the α-carbonic anhydrase gene from Chlamydomonas sp. ICE-L. Mol. Biotechnol. 60 21–30. 10.1007/s12033-017-0040-9 [DOI] [PubMed] [Google Scholar]
- Rahman M. A., Oomori T. (2010). “The role of carbonic anhydrase enzyme in the biocalcification process of coral and its resilience to global climate change,” in OCEANS’10 IEEE Sydney, OCEANSSYD 2010, 10.1109/OCEANSSYD.2010.5603507 ∗∗. [DOI] [Google Scholar]
- Rudenko N. N., Ignatova L. K., Fedorchuk T. P., Ivanov B. N. (2015). Carbonic anhydrases in photosynthetic cells of higher plants. Biochem. 80 674–687. 10.1134/S0006297915060048 [DOI] [PubMed] [Google Scholar]
- Šali A., Blundell T. L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Sharker M. R., Hossen S., Nou I.-S., Kho K. H. (2020a). Characterization of insulin-like growth factor binding protein 7 (Igfbp7) and its potential involvement in shell formation and metamorphosis of Pacific abalone, Haliotis discus hannai. Int. J. Mol. Sci. 21 6529. 10.3390/ijms21186529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharker M. R., Kim S. C., Hossen S., Kho K. H. (2020b). Characterization of insulin-like growth factor binding protein-5 (IGFBP-5) gene and its potential roles in ontogenesis in the Pacific abalone, Haliotis discus hannai. Biology (Basel). 9 216. 10.3390/biology9080216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharker M. R., Kim S. C., Sumi K. R., Sukhan Z. P., Sohn Y. C., Lee W. K., et al. (2020c). Characterization and expression analysis of a GnRH-like peptide in the Pacific abalone, Haliotis discus hannai. Agri Gene 15 100099. 10.1016/j.aggene.2019.100099 [DOI] [Google Scholar]
- Sharker M. R., Nou I. S., Kho K. H. (2020d). Molecular characterization and spatiotemporal expression of prohormone convertase 2 in the Pacific abalone, Haliotis discus hannai. PLoS One 15:231353. 10.1371/journal.pone.0231353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharker M. R., Sukhan Z. P., Kim S. C., Lee W. K., Kho K. H. (2020e). Identification, characterization, and expression analysis of a serotonin receptor involved in the reproductive process of the Pacific abalone. Haliotis discus hannai. Mol. Biol. Rep. 47 555–567. 10.1007/s11033-019-05162-2 [DOI] [PubMed] [Google Scholar]
- Sharker M. R., Sukhan Z. P., Kim S. C., Lee W. K., Kho K. H. (2020f). Molecular identification, characterization, and expression analysis of a gonadotropin-releasing hormone receptor (GnRH-R) in Pacific abalone, Haliotis discus hannai. Molecules 25 10.3390/molecules25122733 ∗∗, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7 10.1038/msb.2011.75 ∗∗, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sly W. S., Hu P. Y. (1995). Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu. Rev. Biochem. 64 375–401. 10.1146/annurev.bi.64.070195.002111 [DOI] [PubMed] [Google Scholar]
- Suleria H. A. R., Masci P. P., Gobe G. C., Osborne S. A. (2017). Therapeutic potential of abalone and status of bioactive molecules: A comprehensive review. Crit. Rev. Food Sci. Nutr. 57 1742–1748. 10.1080/10408398.2015.1031726 [DOI] [PubMed] [Google Scholar]
- Sumi K. R., Kim S. C., Howlader J., Sharker M. R., Choi K. S., Choi S. K., et al. (2019). Molecular identification and expression analysis of carbonic anhydrase VII in Pufferfish (Takifugu rubripes). Ocean Sci. J. 54 363–374. 10.1007/s12601-019-0020-z [DOI] [Google Scholar]
- Supuran C. (2008). Carbonic Anhydrases An Overview. Curr. Pharm. Des. 14 603–614. 10.2174/138161208783877884 [DOI] [PubMed] [Google Scholar]
- Supuran C. T. (2011). Carbonic anhydrase inhibitors and activators for novel therapeutic applications. Future Med. Chem. 3 1165–1180. 10.4155/fmc.11.69 [DOI] [PubMed] [Google Scholar]
- Supuran C. T., Capasso C. (2015). The η-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin. Ther. Targets 19 551–563. 10.1517/14728222.2014.991312 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Nagasawa H. (2013). Mollusk shell structures and their formation mechanism. Can. J. Zool. 91 349–366. 10.1139/cjz-2012-0333 [DOI] [Google Scholar]
- Takeuchi T., Endo K. (2006). Biphasic and dually coordinated expression of the genes encoding major shell matrix proteins in the pearl oyster Pinctada fucata. Mar. Biotechnol. 8 52–61. 10.1007/s10126-005-5037-x [DOI] [PubMed] [Google Scholar]
- Tashian R. E., Hewett-Emmett D., Carter N., Bergenhem N. C. (2000). Carbonic anhydrase (CA)-related proteins (CA-RPs), and transmembrane proteins with CA or CA-RP domains. EXS 105–120. 10.1007/978-3-0348-8446-4_6 ∗∗∗, [DOI] [PubMed] [Google Scholar]
- Tu C., Silverman D. N., Foreman C., Jonsson B.-H., Lindskog S. (1989). Role of Histidine 64 in the Catalytic Mechanism of Human Carbonic Anhydrase II Studied with a Site-Specific Mutant. Biochemistry 28 7913–7918. 10.1021/bi00445a054 [DOI] [PubMed] [Google Scholar]
- Wallner B., Elofsson A. (2003). Can correct protein models be identified? Protein Sci. 12 1073–1086. 10.1110/ps.0236803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Whang I., Choi C. Y., Lee J. S., Lee J. (2011). Validation of housekeeping genes as internal controls for studying biomarkers of endocrine-disrupting chemicals in disk abalone by real-time PCR. Comp. Biochem. Physiol. - C Toxicol. Pharmacol. 153 259–268. 10.1016/j.cbpc.2010.11.009 [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. (2009). Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner G. D. A., Gemmell P., Grosser S., Hamer R., Shimeld S. M. (2013). Analysis of a deep transcriptome from the mantle tissue of Patella vulgata Linnaeus (Mollusca: Gastropoda: Patellidae) reveals candidate biomineralising genes. Mar. Biotechnol. 15 230–243. 10.1007/s10126-012-9481-0 [DOI] [PubMed] [Google Scholar]
- Zito F., Koop D., Byrne M., Matranga V. (2015). Carbonic anhydrase inhibition blocks skeletogenesis and echinochrome production in Paracentrotus lividus and Heliocidaris tuberculata embryos and larvae. Dev. Growth Differ. 57 507–514. 10.1111/dgd.12229 [DOI] [PubMed] [Google Scholar]
- Zolfaghari E. R., Kuuslahti M., Nosrati H., Lohi H., Parkkila S. (2020). Assessment of databases to determine the validity of β-and γ-carbonic anhydrase sequences from vertebrates. BMC genomics 21:1–8. 10.1186/s12864-020-6762-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HdhCA II mRNA expression in the cerebral ganglion, shell muscle, mantle, gill, heart, hemocyte, testis, and ovary was determined by semiquantitative reverse transcription (RT)-PCR. RPL-5 was used as reference gene.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI GenBank, accession number: MT876410, https://www.ncbi.nlm.nih.gov/nuccore/MT876410.1.