Abstract
This study aimed to determine the performance of a rapid, point-of-care testing device (HemotypeSC)™ for diagnosing sickle cell disease (SCD) relative to 2 commonly-used methods compared to DNA polymerase chain reaction (PCR) as the reference standard. The diagnostic performance of (HemotypeSC)™ in diagnosing SCD and determining various other Hb genotypes relative to high performance liquid chromatography (HPLC) and cellulose acetate Hb electrophoresis in alkaline buffer (CAE) was investigated among 156 participants aged 4 to 23 years in Ekiti, Southwest Nigeria. PCR was considered as the reference method/gold standard. The sensitivity and specificity for SS, SC, AS, AC, and AA genotypes by HemotypeSC and HPLC when compared with PCR, were each 100%. Similarly, their positive and negative predictive values were each 100%. However, sensitivity and specificity for identifying these Hb genotypes by CAE were 100, 100, 96.5, 0, 99.2%, and 99, 100, 92.9, 0, 91.7%. Also, CAE did not identify any of the 2 HbAC individuals that were correctly identified by PCR and both HemotypeSC, and HPLC, thus representing 100% HbAC misdiagnosis. In conclusion, this study shows that HemotypeSC has perfect concordance with PCR and 100% accuracy in diagnosing SCD in the population tested. Its ease of use, accuracy and other attributes make it suitable for use in sub-Saharan Africa for rapid determination of Hb genotypes.
Keywords: sickle cell disease, early diagnosis, point-of care testing, sub-Saharan Africa, Nigeria
Highlights
What do we already know about this topic?
Diagnostic delay is a key factor contributing to the gloomy outlook of sickle cell disease (SCD) in the sub-Saharan Africa
How does your research contribute to the field?
In this manuscript, we found very good agreements between a point- of -care device (HemotypeSC) performance and PCR, as well as HPLC which are considered as the gold standards for SCD diagnosis in ideal settings and HemotypeSC exhibited superior performance compared to Cellulose acetate electrophoresis (CAE) which is the most common methods in most SSA settings.
What are your research’s implications toward theory, practice, or policy?
Our findings show that HemotypeSC has a very high diagnostic accuracy with regard to SCD diagnosis and can offset diagnostic delay of SCD in the resource-poor settings of the sub-Saharan Africa if adopted for use.
Introduction
Sickle cell disease (SCD) is a monogenic disorder caused by qualitative defects in the β-globin chain as a result of a mutation [glu(E)6valA; GAG-GTG; rs334] in the HBB gene, which produces the sickle hemoglobin (HbS).1,2 SCD is associated with multiple organ damage and dysfunction because of repeated vaso-occlusion and hemolysis that are the hallmarks of the disease.1,2 As a result of these pathological processes, SCD has several manifestations, which include frequent painful episodes, bone infection and infarction, stroke, renal, cardiopulmonary, and other chronic organ disorders.1,2 Also, they are more prone to severe anemia and infections, all of which often lead to increased morbidity and early mortality, especially among patients in resource-poor parts of world.
Sub-Saharan Africa (SSA) has the highest burden of SCD, with Nigeria having the highest burden of the disease worldwide.1-4 Unfortunately, the outlook of SCD in most African countries is very gloomy and this has been linked to the lack of proper policies and inadequate care.3,4 In recognition of these limitations, the World Health Organisation (WHO) estimates that 70% of SCD deaths in Africa are preventable with simple, cost-effective interventions and recommended that “SCD should be managed at different levels of care through the use of simple, cost-effective, and affordable strategies and technologies that are accessible to all patients including those in the rural communities, where the bulk of Africans reside.”5 Early identification of SCD patients and subsequent provision of comprehensive care enhance the survival of children with SCD. There has been significant reduction in SCD-related mortality in countries where these practices are being implemented and most affected children now survive to adulthood in these developed countries.5,6
Newborn screening and point-of-care testing for SCD are 2 key approaches to ensure early diagnosis of SCD.7 In Nigeria and many other countries in SSA, early diagnostic measures are lacking.7,8 There are many clinical and laboratory methods to identify Hb genotypes and diagnose SCD. These include gel-based or capillary electrophoresis, isoelectric focusing (IEF), high-performance liquid chromatography (HPLC) and molecular diagnosis.7-9 All of these require some degree of moderate/advanced technologies, highly-skilled personnel, stable electricity, need to transport samples to central laboratories, and are usually costly. All these make standard testing for SCD inaccessible to many, thereby delaying diagnosis. These observations raise the need for inexpensive and reliable point-of-care testing methods (POCT) that are suitable for use in patients with SCD in rural settings.
Many POCT devices are now available to diagnose SCD. However, they are fraught with one deficiency or the other, which limit their use and applicability in resource-poor settings. For example, although the Sickledex10 can indicate the absence or presence of HbS, it cannot differentiate SCD from sickle cell trait. It also gives unreliable results in the newborn. Another variant of the Sickledex method that uses the differential diffusion of the various hemoglobin genotypes requires other expensive accessories to function optimally and may not be able to clearly differentiate the diffusion patterns of some Hb variants.11 Similarly, SickleSCAN12 has been associated with inconsistent visualization of HbA while the recently-developed HemeChip requires continuous electricity supply and huge financial cost to function.13
HemoTypeSC™ uses competitive lateral-flow immunoassay technique involving monoclonal antibodies to detect hemoglobins A, S, and C in a very small amount of blood (<2.0-μL).7 It is cheap and costs <2 USD per test, does not require electricity to function, reliable in the newborn period and requires no instrumentation, thus making it ideal for most low-resource settings. Prototype versions of the test have been described.7,8,14 Its accuracy for detecting all relevant hemoglobin phenotypes/genotypes have been validated in pilot studies from the USA, Europe, and Africa.7,8,14 A previous study from Nigeria8 used PCR to adjudicate the few discordant results with HPLC.
In this study, we set out to further determine the diagnostic accuracy of HemoTypeSC in relation to 3 commonly-used methods: CAE, HPLC and PCR in diagnosing SCD. We considered the latter the gold standard for SCD diagnosis and determination of other Hb genotypes.15 We hypothesized that the performance of HemoTypeSC relative to the gold standard, is not significantly different from that of the 2 other methods. Findings from this study will further establish the accuracy and usefulness or otherwise of HemoTypeSC™ in diagnosing SCD and other Hb genotypes in limited-resource settings like SSA.
Patients and Methods
Study Design, and Location and Participants
This was a blinded, diagnostic-accuracy, validation study of HemoTypeSC™ conducted among a population of young Nigerians (children and adolescents) with SCD and their healthy non-SCD counterparts, attending a tertiary hospital in Southwest Nigeria. The hospital is the largest and main center where children and other patients with SCD are referred and cared for in the state. It has many specialists providing different care needs for patients with SCD. The pediatric hematology unit as well as other specialist units run weekly clinics in addition to providing in-patients’ care. The non-SCD participants attended the pediatric outpatient clinics while the SCD participants were those being managed at the Pediatric Hematology unit of the hospital.
PCR was done at the Hematology and Hemotherapy Centre, University of Campinas, Brazil as part of the collaborative study on SCD in Africa using genomic DNA extracted from whole blood (leucocytes) using QIAamp DNA blood mini kit in Nigeria at the Institute for Advanced Medical Research and Training (IMRAT), College of Medicine, University of Ibadan, Nigeria, before being shipped to Brazil for PCR. The PCR was performed specifically to look for the presence or absence of HbS and HbC mutations. The CAE was done at the hospital while the HPLC was done at the IMRAT. Both CAE, and HPLC as well as the PCR method used to diagnose the SCD and other Hb genotypes, have been previously described.16,17
Participants and Sampling
Parents and guardians of participants in a previous study on the clinical evolution of SCD among a young Nigerian cohort17 at the study center were contacted through their telephone numbers and were asked to bring their wards for testing with the HemoTypeSC™ kits. The purpose of the study and the procedures were explained to the parents and participants in plain language. Only those parents/guardians who consented to participate had their wards consecutively recruited and tested between September and December 2019. Participants on chronic blood transfusion or those who received blood transfusion in the 100 days prior to recruitment were exempted.
There is no previous hospital-based study on the prevalence of SCD in the state. Therefore, the sample size for the study was estimated based on a study conducted in a neighouring state (Ondo State)18 and the expected test sensitivity and absolute precision.19 According to the study,18 the prevalence of HbSS was 0.72% and it has been shown previously that the accuracy of HemotypeSC relative to gold standard ranged between 97% and 100%.7,14 The sample size for this study was therefore based on HemotypeSC sensitivity of 97%, HbSS prevalence of 0.70 and absolute precision of 0.05.18,19 This translated to a minimum required sample size of 85 participants with HbSS. However, a total of 99 HbSS participants were studied. In addition, 5 HbSC, and 52 normal healthy participants (36 HbAA, 14 HbAS, & 2 HbAC) were recruited and studied.
Testing for Hb Genotypes among Participants using HemoTypeSC™
The HemoTypeSC kits were procured from the Silver Lake Research Corporation Ltd USA. Three resident doctors (raters) who were trained on how to use the HemoTypeSC, performed the tests, according to the manufacturer’s protocol. Briefly, 6 drops of water obtained from bottled drinking water were dropped into a plain tube using the pipette supplied by the manufacturer. Thereafter, finger prick was performed following aseptic procedure and approximately 1.5 µL of blood was applied to the device absorbent pad and this was put into the tube containing the 6 drops of water and swirled to elute the blood. This was followed by the insertion of the test strip, into the solution after which the result was read off and interpreted visually after 10 minutes. The interpretation guidelines provided by the kit manufacturer and available at www.hemotype.com were followed. Results for each participant were read visually and interpreted independently by at least 2 of the 3 doctors (raters) which, in most cases 93% (145 out of 156 participants) involved all the 3 performing the testing while 2 of them performed the test on all the participants. The resident doctors performed and reported the tests independently and the result was only accepted for any participant if all the interpretations were in agreement. The results were collated, anonymized and sent by the lead researcher (OSO) for analysis by a statistician who was also blinded to the participants.
Quality Control
The guidelines for evaluation of qualitative tests by the Clinical and Laboratory Standards Institute (CLSI, https://clsi.org/) were followed in testing the kit.
Research assistant training
The resident doctors who performed the tests were trained on how to use the kit through a video presentation, and reading the instruction sheet in the package supplied with the kits. In addition, lectures, and hands-on performances supervised by the lead author, a pediatric hematologist, were conducted. The study was only commenced after the trainees were able to satisfactorily perform the procedures and interpret the test results correctly.
Reproducibility of results
At regular intervals (after testing every 30 participants), and/or when a new kit is opened, randomly-selected individuals (who were not included in this study) whose genotypes had been confirmed by both HemoTypeSC™ and PCR, were selected for re-testing and validation of previous results in order to confirm the reliability of the testing kits and the procedure. The HemotypeSC results conformed with the known referenced results in all occasions.
Storage and safekeeping of HemoTypeSC™ kits
The box containing the test strips was only opened to pick up the strip and was closed immediately thereafter and any unused test strips were discarded after 30 days of opening the kit. The kits were stored at room temperature away from water and other contaminants. The average ambient temperature in the study area ranged between 22°C and 33°C during the study period. According to the manufacturer, the kit does not require refrigeration. The kit has a shelf life of >2 years in ambient temperatures of up to 40°C.14 The universal precaution for safety in handling biomedical materials and human samples were adhered to throughout the study.
Ethical Considerations
The study protocol was approved by the Research and Ethics Committee of the Institution. Informed and written consent were obtained from the parents. Also, assents were obtained, as applicable, from participants older than 7 years. Refusal of parents to allow their wards participate in the study did not affect the care given to such patient(s) as all the patients received the same standard care at the hospital regardless of their participation or not in the current study.
Data Analysis
The characteristics of the study participants were described using basic descriptive statistics. Diagnostic tests were used to test our hypothesis that the 3 screening methods (CAE, HPLC, and HemoTypeSC) were not significantly different when compared to the gold standard using the specificity, sensitivity, positive predictive value (PPV), and negative predictive value (NPV) in diagnosing individuals with different Hb phenotypes (AA, AC, AS, SC, and SS). The Receiver Operating Curves (ROCs) were computed with their corresponding Area Under Curves (AUCs) so as to compare the performances of each of the diagnostic methods. The Kappa interrater test was used to determine agreement among the 3 raters. All analyses were carried out blinded regarding the participants’ identities and their results by AFF using Stata Version 16 and P-values <.05 were considered significant. The following notations and definitions were used:
a. True Positive (TP): Number of true positive events
b. True Negative (TN): Number of true negative events
c. False Positive (FP): Number of false positive events
d. False Negative (FN): Number of false negative events
e. Sensitivity: TP/(TP+FN)
f. Specificity: TN/(TN+FP)
g. Positive Predictive Value (PPV): TP/(TP+FP)
h. Negative Predictive Value (NPV): TN/(TN+FN)
Results
A total of 158 participants were initially included in the study however, 2 participants’ results were suspected to have been mixed up due to clerical errors and were therefore excluded from the final analysis thus making the total final participants to stand at 156: 99 HbSS, 5 HbSC, and 52 normal healthy participants (36 HbAA, 14 HbAS, & 2 HbAC) among which 101 (64.7%) were males. The participants were aged 4 to 23 years with a mean of 10.9 ± 4.4 years. The HbF level in the study participants with HbSS averaged 10.2% ± 6% with a median of 9.2% and inter quartile range (IOR) of 4.9% to 14.8%. The distribution of HbF of the other group of participants is shown in Table 1.
Table 1.
Distribution of Participants’ Hemoglobin Types and their HBF Values.
| Participants | SS (n = 99) | SC (n = S) | AS (n = l4) | AC (n = 2) | AA (n = 36) |
|---|---|---|---|---|---|
| HBF% (Mean ± SD) | 10.2 ± 6.1 | 1.7 ± 1.3 | 1.4 ± 1.1 | 0.7 ± 0.2 | 0.7 ± 0.3 |
| HBF% (Median, IOR) | 9.2 (4.9-14.8) | 0.9(0.7-2.9) | 0.6 (0.5-1.5) | 0.6 (0.5-0.8) | 0.6 (0.3-1.0) |
Abbreviations: IOR, inter quartile range; HbF, fetal hemoglobin; SS-HbSS, SC-HbSC, AS-HbAS, AC-HbAC, AA-HbAA.
The HemotypeSC
The sensitivity and specificity for SS, SC, AS, AC, and AA genotypes, when compared with the gold standard, were each 100%. Similarly, their positive predictive values and negative predictive values were each 100%, indicating that the HemotypeSC perfectly agrees with the gold standard and has 100% accuracy in respect of all the Hb types among the participants. Altogether, HemotypeSC displayed 100% accuracy for participants with SCD (99 HbSS and 5 HbSC) (Table 2).
Table 2.
The Sensitivity Analysis of Cellulose Acetate Electrophoresis, HemotypeSC and HPLC Methods versus PCR in the Diagnosis of Phenotypes and Hb Variants.
| Phenotype | Method | TN | FP | FN | TP | Sensitivity | 95% CI | Specificity | 95% CI | PPV | NPV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | Electrophoresis | 119 | 1 | 3 | 33 | 0.992 | 0.948-0.999 | 0.917 | 0.764-0.978 | 0.971 | 0.976 |
| HemotypeSC | 120 | 0 | 0 | 36 | 1.000 | 0.962-1.000 | 1.000 | 0.880-1.000 | 1.000 | 1.000 | |
| HPLC | 120 | 0 | 0 | 36 | 1.000 | 0.962-1.000 | 1.000 | 0.879-1.000 | 1.000 | 1.000 | |
| AC | HemotypeSC | 154 | 0 | 0 | 2 | 1.000 | 0.970-1.000 | 1.000 | 0.197-1.000 | 1.000 | 1.000 |
| HPLC | 154 | 0 | 0 | 2 | 1.000 | 0.970-1.000 | 1.000 | 0.197-1.000 | 1.000 | 1.000 | |
| AS | Electrophoresis | 137 | 5 | 1 | 13 | 0.965 | 0.916-0.996 | 0.929 | 0.642-0.996 | 0.722 | 0.993 |
| HemotypeSC | 142 | 0 | 0 | 14 | 1.000 | 0.967-1.000 | 1.000 | 0.732-1.000 | 1.000 | 1.000 | |
| HPLC | 142 | 0 | 0 | 14 | 1.000 | 0.967-1.000 | 1.000 | 0.732-1.000 | 1.000 | 1.000 | |
| SC | Electrophoresis | 151 | 0 | 0 | 5 | 1.000 | 0.969-1.000 | 1.000 | 0.463-1.000 | 1.000 | 1.000 |
| HemotypeSC | 151 | 0 | 0 | 5 | 1.000 | 0.969-1.000 | 1.000 | 0.463-1.000 | 1.000 | 1.000 | |
| HPLC | 151 | 0 | 0 | 5 | 1.000 | 0.969-1.000 | 1.000 | 0.463-1.000 | 1.000 | 1.000 | |
| SS | Electrophoresis | 57 | 0 | 0 | 99 | 1.000 | 0.921-1.000 | 1.000 | 0.953-1.000 | 1.000 | 1.000 |
| HemotypeSC | 57 | 0 | 0 | 99 | 1.000 | 0.921-1.000 | 1.000 | 0.953-1.000 | 1.000 | 1.000 | |
| HPLC | 57 | 0 | 0 | 99 | 1.000 | 0.921-1.000 | 1.000 | 0.953-1.000 | 1.000 | 1.000 | |
| Hb variant | |||||||||||
| A | Electrophoresis | 104 | 0 | 0 | 52 | 1.000 | 0.956-1.000 | 1.000 | 0.914-1.000 | 1.000 | 1.000 |
| HemotypeSC | 104 | 0 | 0 | 52 | 1.000 | 0.956-1.000 | 1.000 | 0.914-1.000 | 1.000 | 1.000 | |
| HPLC | 104 | 0 | 1 | 51 | 1.000 | 0.956-1.000 | 0.981 | 0.884-0.999 | 1.000 | 0.990 | |
| S | Electrophoresis | 33 | 5 | 1 | 117 | 0.868 | 0.711-0.951 | 0.992 | 0.947-0.999 | 0.959 | 0.971 |
| HemotypeSC | 38 | 0 | 0 | 118 | 1.000 | 0.885-1.000 | 1.000 | 0.961-1.000 | 1.000 | 1.000 | |
| HPLC | 38 | 0 | 0 | 118 | 1.000 | 0.885-1.000 | 1.000 | 0.961-1.000 | 1.000 | 1.000 | |
| C | Electrophoresis | 148 | 0 | 3 | 5 | 1.000 | 0.968-1.000 | 0.625 | 0.259-0.897 | 1.000 | 0.980 |
| HemotypeSC | 148 | 0 | 0 | 8 | 1.000 | 0.968-1.000 | 1.000 | 0.598-1.000 | 1.000 | 1.000 | |
| HPLC | 148 | 0 | 1 | 7 | 1.000 | 0.968-1.000 | 0.875 | 0.467-0.993 | 1.000 | 0.993 | |
Abbreviations: HPLC, high performance liquid chromatography; TP, true positive; TN, true negative; FP, false positive; FN, false negative; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.
Interrater Agreement for HemotypeSC Testing
All the 3 raters performed testing for 145 participants, representing 93% of the total participants, as one of the raters (rater 2) did not perform for 11 partici-pants while the remaining 2 raters (raters 1and 3) performed for all 156 participants. There was 100% concordance among the 3 raters. (Supplemental Tables 1A and B).
The HPLC
This method correctly diagnosed all the participants (100%) with hemoglobin types AA, AC, AS, SC, and SS as identified by the gold standard, which translates to 100% sensitivity, specificity, positive predictive value and negative predictive value for each of these phenotypes (Table 2).
Cellulose Acetate Electrophoresis (CAE) in Alkaline Buffer
This method misdiagnosed 2 out of a total of 36 HbAA (5.6%) identified by PCR. They were wrongly diagnosed as HbAS samples. This represents 99.2% and 91.7% sensitivity and specificity respectively as well as positive and negative predictive values of 97.1% and 97.6%, respectively. Similarly, this method missed the diagnosis of the 2 participants with HbAC (by HPLC and PCR), thus representing 100% misdiagnosis as the 2 were returned as HbAS. Furthermore, CAE also falsely identified 1 participant out of 14 HbAS as having HbAA instead of the correct diagnosis as AS. This represents a misdiagnosis of 7.1% for the HbAS phenotype. These give sensitivity, specificity, positive predictive and negative predictive values of 96.5%, 92.9%, 72.2%, and 99.3% respectively for the HbAS. (Table 2).
The AUCs of the ROC and test of equality of the AUCs are presented in Table 3. As shown, the AUC for the phenotype AA using cellulose acetate electrophoresis, HemotypeSC, and HPLC methods were 0.954, 1.000, and 1.000, respectively. These figures showed a high level of accuracy for both the HPLC and the HemotypeSC, but some inconsistency with the CAE method and this difference almost attained statistical significance (P = .053). However, there was a significant difference (P = .0128) in the accuracy of the CAE, when compared to both HemotypeSC and HPLC methods for HbS variant (P = .0128) Table 3.
Table 3.
Comparison of Area under Curves of the ROCs for the Performance of the 3 Methods Compared with the Gold Standard in the Diagnosis of SCD Phenotypes and Hb Variants.
| Phenotypes | Method | Obs | AUC of ROC | Std. Err | [95% CI] | [95% CI] | X2 | P-value |
|---|---|---|---|---|---|---|---|---|
| AA | Electrophoresis | 156 | 0.954 | 0.024 | 0.908 | 1.000 | 3.73 | .0535 |
| HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| HPLC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| Electrophoresis | 156 | 0.500 | 0.000 | 0.500 | 0.500 | |||
| AC | HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | 0.00 | 1.0000 |
| HPLC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| AS | Electrophoresis | 156 | 0.947 | 0.037 | 0.875 | 1.000 | 2.13 | .144 |
| HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| HPLC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| SC | Electrophoresis | 156 | 1.000 | 0.000 | 1.000 | 1.000 | 0.00 | 1.000 |
| HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| HPLC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| SS | Electrophoresis | 156 | 1.000 | 0.000 | 1.000 | 1.000 | 0.00 | 1.000 |
| HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| HPLC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| Hb variants | ||||||||
| A | Electrophoresis | 156 | 1.000 | 0.000 | 1.000 | 1.000 | 1.00 | .3173 |
| HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| HPLC | 156 | 0.990 | 0.010 | 0.971 | 1.000 | |||
| S | Electrophoresis | 156 | 0.930 | 0.028 | 0.875 | 0.985 | 6.21 | .0127 |
| HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| HPLC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| C | Electrophoresis | 156 | 0.813 | 0.092 | 0.633 | 0.992 | 4.20 | .1225 |
| HemotypeSC | 156 | 1.000 | 0.000 | 1.000 | 1.000 | |||
| HPLC | 156 | 0.938 | 0.063 | 0.815 | 1.000 | |||
Abbreviations: HPLC, high performance liquid chromatography; AUC, area under curves; ROC, receptor operatives characteristic curves; CI, confidence interval.
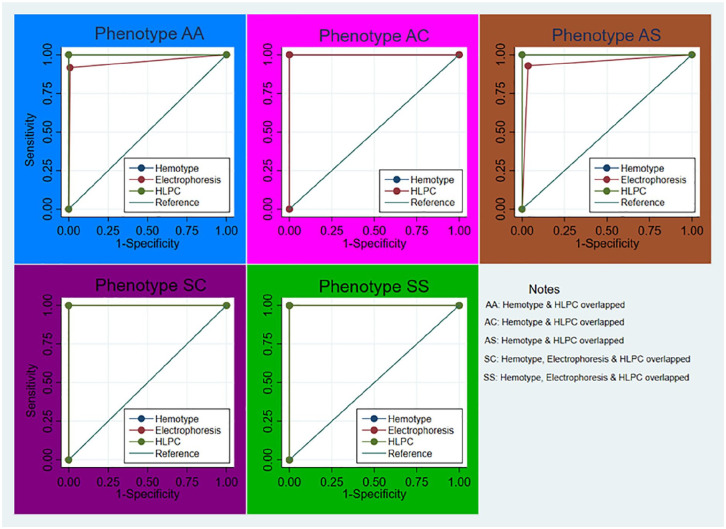
Furthermore, Figure 1, shows the ROC of HemotypeSC, CAE, and HPLC versus PCR for SCD phenotypes and other Hb variants respectively. It complemented the quantitative tests of difference of AUC of the ROCs shown in Table 3. As shown in Figure 1, there is perfect prediction while using the HemotypeSC and HLPC methods for diagnosing phenotype AC compared to the CAE while there are also agreements between the HemotypeSC and HPLC methods in the other phenotypes.
Figure 1.
ROC of HemotypeSC, electrophoresis (CAE), & HPLC versus gold standard.
Discussion
Children suffering from SCD are often identified late in SSA countries, and this is usually after hospitalization while some actually die of the disease complications without a diagnosis.20 This gloomy outlook is due to the lack of accessible, easy to use, accurate, simple, and yet, affordable diagnostic tools.5,6,20-22
The POCT kit evaluated in this study (HemotypeSC™), showed a very good agreement with the gold standard in respect of all the SCD phenotypes and other Hb genotypes found among the study participants and this is in agreement with previous reports on the evaluation of its performance.7,8,14 Hence, these observations are affirmations of its reliability and usefulness as a POCT device.
Certain attributes and qualities of the POCT make it very suitable for use in SSA. Firstly, the kit does not require rigorous procedures, laboratory reagents, or special skills. Also, it does not require external power (electricity or battery) to function, thus making it ideal for the resource-poor settings where stable electricity is not available or regular. Furthermore, the results of tests performed with the device are available in real-time and on-the-spot within ten minutes, thus averting the likelihood of missed SCD diagnosis opportunities and perhaps any possibility of loss to follow up while the patient`s result is in transit. Also, the immediacy of its results gives little or no room for sample mix up due to clerical errors as observed with the 2 cases excluded from the study. In addition, the POCT is cost-friendly, as the overall costs per test for each participant is <2 USD. These observations make the POCT device ideal for use in SSA.7,8,14,20-23
Nothing drives home the reliability of the POCT for detecting and diagnosing HbSS among patients than its 100% agreement with the gold standard in this study. This high accuracy as found in this study, has also been previously reported regarding the performance of HemotypeSC among different study populations.7,14,24 In addition, participants with various other Hb genotypes such as AC and SC were correctly diagnosed compared to CAE, thus adding to the limited data on these Hb types vis-à-vis the clinical utility of HemotypeSC in determining various Hb variants. For example, Mukherjee et al.25 in India and Steele et al.7 among participants from Ghana, France and USA had low prevalences of Hemoglobin AC and SC while Nankanja et al.24 in Uganda, reported no HbC. These observations further highlight the relevance of HemotypeSC in diagnosing SCD and other common Hb variants among diverse populations and in the SSA.
As observed in this study, even though CAE is the routine testing method in most clinical settings of the SSA, quite a number of the Hb genotypes were misdiagnosed by the CAE method. This is quite surprising given the status of the hospital (tertiary) where the tests were conducted and assumption that the migration patterns of the various Hb patterns are quite distinct so as not to be readily mixed up. These observations may reflect human error or due to lack of expertise and, or inconsistencies in the skills of the people who performed the test.16 In addition, CAE is also dependent on electricity supply which is not regular in most SSA settings.
HPLC did not only perform better than CAE, it also agreed perfectly with the reference method. This is not surprising given the expected high discriminatory nature of HPLC in identifying SCD phenotypes and Hb variants. Nevertheless, HPLC is an advanced, more technical, and elitist testing method, requiring skilled personnel, which are not available in most parts of SSA. The columns and reagents are expensive and not always available. Also, because samples are usually pooled together to run in most busy laboratories, there can be clerical errors which can lead to discordant or results mix up. HemoTypeSC does not have these limitations as its testing is usually on-the- spot, avoiding possible clerical and other human errors or mix-ups.
In a previous study,8 some discordant results between HPLC and HemotypeSC were resolved in favor of HemotypeSC with PCR adjudication thus making our study the first from Nigeria to relate the performance of HemotypeSC to PCR as the gold standard. This further affirms the reliability and applicability of HemotypeSC in determining the SCD phenotypes and other Hb genotypes in resource-limited settings.
HemoTypeSC has also been validated to identify correctly SCD phenotypes in newborns.7,8 As found in this study, HbF averaged 10.2% ± 6% with a median of 9.2% and IOR of 4.9% to 14.8% among patients with HbSS, further highlighting its potential for use in individuals with high HbF or newborn screening for SCD and subsequent enrollment of affected children into comprehensive care. This approach has been found to be crucial in reducing the mortality and morbidity of SCD and improving the quality of life of SCD patients.26-29 These observations further highlight the uniqueness, suitability and potential of HemotypeSC in helping to lessen the burden of SCD in SSA.21,23,26-29
Despite the high accuracy and other attributes of HemotypeSC in this study and other previous reports,7,8,14,24,25 its use may be impacted by certain factors. Firstly, the test result is counter-intuitive in which case, the presence of a band indicates the absence of that hemoglobin variant. This could cause confusion and lead to misinterpretation while using it in the field thus leading to some errors. However, this can be averted by properly training users to ensure competency prior to testing as was done in this study. The high agreement rates observed across the raters in this study, suggest they acquired the needed competence. This is not surprising given their professional status and level of training. Secondly, users need to strictly adhere to manufacturers specifications regarding the use of the kit and also ensure repeated and frequent quality control measures. Lastly, users of HemotypeSC should bear in mind that certain clinically-significant Hb variants like HbD, HbE, and HbO cannot be distinguished from HbA due to the fact that the kit detects HbA through sensing a single amino acid on the β-globin chain and as such, other Hb variants like those mentioned above that share the same amino acid will be identified as HbA by the device. However, these variants are scarce in SSA where the high burden of SCD abound.1,2,7,8,24 Furthermore, interpretation of results for individuals with beta-thalassemia may be confusing as sickle-β+-thalassemia is identified as HbAS which will require further testing to classify appropriately. Also, sickle-β°-thalassemia is identified as HbSS by HemotypeSC however, this misclassification is of little or no clinical significance as both phenotypes have similar disease profiles and are usually clinically indistinguishable and as such, managed similarly.
Conclusion
The POCT device (HemotypeSC) showed high accuracy and agreement when compared to PCR, the gold standard in the examined population setting. In addition, it showed superiority over CAE in its applicability, reliability and usefulness. It is affordable, easy to use and seems more applicable and practicable in SSA. Its adoption and use may result in improved outcomes for patients and lessen the burden of delay SCD diagnosis in SSA. Findings from this study suggest that, HemotypeSC could be of high clinical utility in diagnosing SCD in resource-poor settings of the SSA.
Study Limitation
This study is from a single center and with small sample size, which may affect its statistical power. Nevertheless, the sample size exceeds the minimum required for methods validation studies of this nature.19,30,31 Also, the raters performed and reported the tests independently thus minimizing the risk for bias. Furthermore, to the best of our knowledge, this study is the first from Nigeria to relate the accuracy of the HemotypeSC to PCR as the gold standard, further expanding its scope and reliability of HemotypeSC as a clinical utility tool in diagnosing SCD, especially in resource-limited setting of SSA.
Supplemental Material
Supplemental material, sj-pdf-1-gph-10.1177_2333794X211016789 for Diagnostic Accuracy of HemotypeSC as a Point-of-Care Testing Device for Sickle Cell Disease: Findings from a Southwestern State in Nigeria and Implications for Patient Care in Resource-Poor Settings of sub-Saharan Africa by Oladele S. Olatunya, Dulcinea M. Albuquerque, Adeniyi F. Fagbamigbe, Opeyemi A. Faboya, Ayotunde E. Ajibola, Oluwatoyin A. Babalola, Adewale O. Adebisi, Adeyinka G. Falusi, Adekunle Adekile and Fernando F. Costa in Global Pediatric Health
Acknowledgments
Authors also acknowledge with thanks the supports received from participants and their caregivers/parents during the study.
Footnotes
Authors’ Contribution: All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study received partial supports from grants No 2014/00984-3 from FAPESP, and grants No 2015/141693-0 from CNPq, Brazil.
Ethical Approval: The study protocol was approved by the Research and Ethics Committee of the Ekiti State University Teaching Hospital (EKSUTH) protocol number (EKSUTH/A67/2019/05/003).
ORCID iD: Oladele S. Olatunya  https://orcid.org/0000-0003-2564-3064
https://orcid.org/0000-0003-2564-3064
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Piel FB, Steinberg MH, Rees DC. Sickle cell disease. N Engl J Med. 2017;376;1561-1573. [DOI] [PubMed] [Google Scholar]
- 2. Kato GJ, Piel FB, Reid CD, et al. Sickle cell disease. Nat Rev Dis Primers. 2018;4:e18010:doi: 10.1038/nrdp/2018.10 [DOI] [PubMed] [Google Scholar]
- 3. Ware RE. Is sickle cell anemia a neglected tropical disease? PLoS Negl Trop Dis. 2013;7:e2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olatunya OS, Ogundare EO, Fadare JO, et al. The financial burden of sickle cell disease on households in Ekiti, Southwest Nigeria. Clinicoecon Outcomes Res. 2015;7:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organisation (WHO). WHO Africa Region on sickle cell disease prevention and control. 2015. Accessed December, 2019. www.afro.who.int
- 6. Makani J, Soka D, Rwezaula S, et al. Health policy for sickle cell disease in Africa: experience from Tanzania on interventions to reduce under-five mortality. Trop Med Int Health. 2015;20(2):184-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Steele C, Sinski A, Asibey J, et al. Point of care testing for sickle cell disease in low resource settings: a multi-centre evaluation of HemoTypeSC, a novel rapid test. Am J Hematol. 2019;94(1):39-45. doi: 10.1002/ajh.25305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nnodu O, Isa H, Nwegbu M, et al. HemotypeSC, a low-cost point of care testing device for sickle cell disease: promises and challenges. Blood Cells Mol Dis. 2019;78: 22-28. doi: 10.1016/j.bmcd.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ryan K, Bain BJ, Worthington D, et al. Significant haemoglobinopathies: guidelines for screening and diagnosis. Br J Haematol. 2010;149:35-49. [DOI] [PubMed] [Google Scholar]
- 10. Canning DM, Huntsman RG. An assessment of Sickledex as an alternative to the sickling test. J Clin Pathol. 1970;23:736-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X, Kanter J, Piety NZ, et al. A simple, rapid, low-cost diagnostic test for sickle cell disease. Lab Chip. 2013;13:1464-1467. [DOI] [PubMed] [Google Scholar]
- 12. McGann PT, Schaefer BA, Paniagua M, et al. Characteristics of a rapid, point-of-care lateral flow immunoassay for the diagnosis of sickle cell disease. Am J Hematol. 2016;91:205-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alapan Y, Kim C, Adhikari A, et al. Sickle cell disease biochip: a functional red blood cell adhesion assay for monitoring sickle cell disease. Transl Res. 2016;173:74-91.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quinn CT, Paniagua MC, DiNello RK, et al. A rapid, inexpensive and disposable point-of-care blood test for sickle cell disease using novel, highly specific monoclonal antibodies. Br J Haematol. 2016;175:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Martino CC, Alencar CS, Loureiro P, et al. Use of an automated pyrosequencing technique for confirmation of sickle cell disease. PLoS One. 2019;14(12):e 0216020. doi: 10.1371/journal.pone0216020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schneider RG, Hightower B, Hosty TS, et al. Abnormal hemoglobin in a Quarter million people. Blood. 1976;48(5):629-637. [PubMed] [Google Scholar]
- 17. Olatunya OS, Albuquerque DM, Akanbi GO, et al. Uridine diphosphate glucuronosyl transferase 1A (UGT1A1) promoter polymorphism in young patients with sickle cell anaemia: report of the first cohort study from Nigeria. BMC Med Genet. 2019;20:160. doi: 10.1186/s12881-019-0899-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akinboro A, Komolafe EK, Azeez MA. A retrospective study on fourteen year hemoglobin genotype variants recorded at five government hospitals in Akure, Ondo State, Southwestern Nigeria in Ondo State. Egypt J Med Hum Genet. 2016;17:377-381. [Google Scholar]
- 19. Malhotra RK, Indrayan A. A simple nomogram for sample size for estimating sensitivity and specificity of medical tests. Indian J Ophthalmol. 2010;58:519-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grosse SD, Odame I, Atrash HK, et al. Sickle cell disease in Africa: a neglected cause of early childhood mortality. Am J Prev Med. 2011;41:S398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aygun B, Odame I. A global perspective on sickle cell disease. Pediatr Blood Cancer. 2012;59:386-390. [DOI] [PubMed] [Google Scholar]
- 22. Olatunya OS, Olu-Taiwo A, Ogundare EO, et al. Evaluation of a portable haemoglobin metre performance in children with sickle disease and implications for healthcare in resource-poor settings. J Trop Paed. 2016;62:316-323. [DOI] [PubMed] [Google Scholar]
- 23. Weatherall DJ. The inherited diseases of hemoglobin are an emerging global health burden. Blood. 2010;115:4331-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nankanja R, Kadhumbula S, Tagoola A, et al. HemoTypeSC demonstrates >99% field accuracy in a sickle cell disease screening initiative in children of southeastern Uganda. Am J Hematol. 2019;94(6):E164-E166. doi: 10.1002/ajh.25458 [DOI] [PubMed] [Google Scholar]
- 25. Mukherjee MB, Colah RB, Mehta PR, et al. Multicenter evaluation of HemoTypeSC as a point-of-care sickle cell disease rapid diagnostic test for newborns and adults across India. Am J Clin Pathol. 2020;153(1):82-87. doi: 10.1093/ajcp/aqz108 [DOI] [PubMed] [Google Scholar]
- 26. Telfer P, Coen P, Chakravorty S, et al. Clinical outcomes in children with sickle cell disease living in England: a neonatal cohort in East London. Haematologica. 2007;92:905-912. [DOI] [PubMed] [Google Scholar]
- 27. Quinn CT, Rogers ZR, McCavit TL, Buchanan GR. Improved survival of children and adolescents with sickle cell disease. Blood. 2010;115:3447-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Memish ZA, Saeedi MY. Six-year outcome of the national premarital screening and genetic counseling program for sickle disease and beta-thalassemia in Saudi Arabia. Ann Saudi Med. 2011;31(3):229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Al Arrayed S, Al Hajeri A. Newborn screening services in Bahrain between 1985 and 2010. Adv Hematol. 2012:903219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou X, Yan H, Xing Y, et al. Evaluation of portable haemoglobin photometer in pregnant women in a high altitude area: a pilot study. BMC Public Health. 2009;9(1):228. doi: 10.1186/147-2458-9-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bland JM, Altman DG. Measuring agreement in method comparison study. Stat Methods Med Res. 1999;8(2):135-160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-gph-10.1177_2333794X211016789 for Diagnostic Accuracy of HemotypeSC as a Point-of-Care Testing Device for Sickle Cell Disease: Findings from a Southwestern State in Nigeria and Implications for Patient Care in Resource-Poor Settings of sub-Saharan Africa by Oladele S. Olatunya, Dulcinea M. Albuquerque, Adeniyi F. Fagbamigbe, Opeyemi A. Faboya, Ayotunde E. Ajibola, Oluwatoyin A. Babalola, Adewale O. Adebisi, Adeyinka G. Falusi, Adekunle Adekile and Fernando F. Costa in Global Pediatric Health