Abstract
This study aimed to evaluate the changes in aromatic components and other chemical properties of Tainong mango during fruit development, ripening, and storage. As the volatiles of Tainong mango and their related molecular mechanisms remain unclear, volatile profile, metabonomics, and transcriptome analyses were applied to investigate the molecular determinants of the synthesis of aroma components in mango during fruit development and storage. Total acids, total sugar, total carotenoids, enzyme activities of the mango pulp samples were also determined. Volatile components of the mango pulp samples were identified using a gas chromatography-mass spectrometric method. Ribonucleic acid (RNA) sequences of the samples were analyzed by real-time polymerase chain reaction. The results showed that 181 volatiles were isolated and identified in the fruit at seven stages. Compared to the other stages, mango collected on day 8 and day 12 had higher concentrations of 17 volatile components, especially (E,Z)-2,6-nonadienal, 53384 transcripts were also detected through RNA sequencing. The differentially expressed genes analyses included catalytic activity, transferase activity, adenosine diphosphate binding, transcription factor activity, and oxidoreductase activity. α-Pinene content and expression of the differentially expressed genes involved in terpenoid metabolism and enzyme activities in the terpenoid metabolic pathways gradually increased during the maturity of the fruit, and had maximum values at day 8 of storage. Moreover, the integrative analyses revealed potential molecular insights of mango development and aroma formation in the fruit.
Keywords: aromatic compound, de novo transcriptome assembly, gene ontology, RT-PCR, volatile profile
Introduction
Mango (Mangifera indica L.), the king of tropical fruit, is native to South Asia (Munafo et al., 2014). It is particularly rich in β-carotene. The compound is a precursor of vitamin A, which is rare in many fruits. Mango pulp is a typical source for the productions of fruit jam, canned food, pickled, sour, spicy pickles, and beverages, and more, besides being suitable for immediate consumption. It also exhibits tempting fragrance derived from the volatile components (San et al., 2017). The quality and acceptability of mango pulp and juice are mainly assessed based on the flavor (Zhang et al., 2019a). The volatile profile varies considerably among different mango cultivars.
To date, a broad scope of investigations on mango volatiles has been implemented, resulting in the isolating and identification of more than 400 compounds in different varieties of mango, most of which are esters, ketones, aldehydes, alcohols, and terpenes (Andrade et al., 2000; Pino and Mesa, 2006; Shivashankara et al., 2006). Literature has shown that a relatively high amount of α-terpinolene has been determined in Kensington Pride, Chana, Bacuri, Coquinho, Gojoba, Cametá, Cheiro, Comum, and Carlota mangoes (Andrade et al., 2000; Lalel et al., 2003). It is one of the main volatiles in these fruits. A previous study also reported that 4-methoxy-2,5-dimethyl-3(2H)-furanone (MDMF), β-damascenone, ethyl butanoate, ethyl hexanoate, (R)-linalool, (E,Z)-2,6-nonadienal, and terpinolene are the essential aromatic compounds determined in yellow Thai Keaw mangoes (Boonbumrung et al., 2001). Hence, ethyl butanoate is one of the important aromatic compounds found in different cultivars of mango grown in Brazil (Tommy Atkins, Coracao de Boi, Carlota, Rubi, Haden, and Espada). Moreover, (E)-2-decenal, hexanal, (E)-β-ocimene, (E)-2-hexenal, γ-terpinene and (Z)-3-hexenal have been detected as the main aromatic components in Thai Khieo Sawoei mangoes (Tamura et al., 2001).
Based on the odor activity values, which represents the ratio of concentration to odor threshold, MDMF, ethyl butanoate, methyl benzoate, ethyl 2-methylpropanoate, decanal, (E,Z)-2,6-nonadienal, (E)-β-ionone, and (E)-2-nonenal have been potentially identified as the most important aroma compounds in the 20 cultivars of mango (Pino and Mesa, 2006). Literature also shows that terpinolene, (E,Z)-2,6-nonadienal, (E)-β-ionone, β-damascenone, (E)-2-nonenal, ethyl butanoate, and ethyl 2-methylpropanoate are the odor-active compounds of Corazon mangoes (Pino, 2012). In addition to the small number of these aromatic compounds identified in the mango pulp samples, many other volatiles could not be quantified due to their low detection limits (Zhang et al., 2019a).
The maturity of mango had greatly affected the aroma profile of the fruit besides the geographical origin, which was related to the growing environment and the variation in cultivar (Lebrun et al., 2008; Pandit et al., 2009b; Chauhan et al., 2010; Kulkarni et al., 2012). A previous study identified limonene and p-cymene, α-terpinene, and ethyl octanoate as the dominant volatile components in Kensington Pride mango samples collected during the pre-climacteric, climacteric stage, and fully ripe stages (Lalel et al., 2003). Although many volatile components have been isolated and identified in mangoes, studies on the molecular mechanism of aroma compound biosynthesis are limited. Acyl-CoA-oxidase (MiACO), 9-lipoxygenase (Mi9LOX), hydroperoxide lyase (MiHPL), peroxygenase (MiPGX1), and epoxide hydrolase 2 (MiEH2) genes involved in lactone biosynthesis have also been isolated from mango, and the transcript profiling of these genes was analyzed during various developmental stages in fruits of three mango cultivars (Kent, Pairi, and Alphonso) with different levels of lactones (Deshpande et al., 2017b). The transcriptome analysis has also been used to explore the distinct aroma characteristics in Alphonso mango, where transcripts for the biosynthesis of furanones, sesqui-terpenes, lactones, monoterpenes, and diterpenes were identified.
Aroma is the most critical organoleptic quality of a mango. Volatile components give aroma to the fruit. These compounds are subjected to changes during fruit ripening and postharvesting. Although volatile components of fruits are widely studied, the associated molecular mechanisms remain unclear. It undergoes rapid and substantial changes during ripening and storage. Although some of the aromatic components have been isolated and identified in different cultivars of mango, little is known about the volatile profile in the “Tainong” variety of mango, which is widely grown in southern China. The ripe fruit of this variety has a special aroma because it contains certain aromatic compounds that give it a flavor. In this study, an integrative analysis of volatile profile and transcriptome was employed to identify molecular mechanism, as well as the changes in volatile components during the stages of fruit development and storage.
Materials and Methods
Plant Materials and Growth Conditions
Tainong mango trees have been planted in the Tiandong National Mango Germplasm Resources Nursery in Guangxi (Tiandong County, Baise City, Guangxi Province, China, 23°16′N, 107°26′E) since 6 years ago. The ambient temperatures of the nursery ranged between 25 and 28°C, and the altitude is 110 m. Mango samples were collected from the nursery at different stages of fruit development (40, 60, 80, and 90 days after the flowering stage began). Fruit samples harvested at 90 days after the flowering were also kept for 4, 8, and 12 days in the laboratory at a controlled temperature of 25°C and a humidity level of 95%. Seven mangoes of an average weight of 100 ± 10 g were sampled. The fruits were picked from different trees at the same positions on the day of fruit collection. Mango peels were removed, and the pulps were directly used for volatile analysis. All pulp samples were stored at –80°C before extraction. Data of triplicate analyses were obtained for each analysis.
Analysis of Total Acid Content, Total Sugar Content, and Carotenoid Content
Total acid content, total sugar content, and carotenoid content of the mango pulp samples were measured according to the methods described by Shi et al. (2013); Liao et al. (2019), and Zhang et al. (2019a), respectively. The total acid content of mango pulp samples was determined by following the Official Methods of Analysis for Vinegar in China (GB/T 5009.41-2003), while total sugars and total carotenoids of the pulp were determined using HPLC.
Determination of Enzyme Activities
Enzyme activities of 1-deoxy-D-xylose-5-phosphate reductase (DXS), 1-deoxyxylose-5-phosphate synthase (DXR), geranyl pyrophosphate synthetase (GPPS), geranylgeranyl pyrophosphate synthetase (GGPPS), pyruvate carboxylase (PC), diacylglyceryl transferase (DGAT), farnesyl diphosphate synthase (FPS), and hydroxymethyl glutarate monoacyl CoA reductase (HGMR) were measured using plant ELISA kits, where the all the ELISA kits were purchased from a local chemical supplier (Jianglai Biotechnology Co., Ltd, Shanghai, China). A microplate reader (BioTek Instruments, Inc., Winooski, VT, United States) was used to obtain the absorbance readings of each test; the analyses were performed according to the instructions provided by the manufacturers. The results were calculated based on the formula provided in the manufacturers’ instructions.
Volatile Profile Analysis
Exactly 2.5 g of homogenized mango pulp was added with 2.5 mL of saturated sodium chloride and 100 μL of internal standard solution (32.88 μg/mL 2-octanol, Sigma-Aldrich) in a 20 mL headspace bottle (ANPEL Laboratory Technologies Inc., Shanghai, China). The bottle was sealed with a crew cap fitted with a PTFE/silicone septum. After a 15-min agitation at 50°C and 250 rpm, volatile compounds were extracted with 50/30 μm DVB/CAR/PDMS as the extraction fiber, which was later exposed to the headspace for 40 min.
The gas chromatography-mass spectrometry (GC-MS) analysis was performed using a GC-mass spectrometer (7890B-5977B, Agilent Technologies) to quantify the volatile components of mango pulp samples. Separation of the volatile compounds was performed using a DB-Wax column (30 m × 0.25 mm × 0.25 μm, Agilent Technologies, Shanghai, China). The extract was injected in splitless mode and desorbed at 260°C for 5 min. Helium was used as the carrier gas with a constant flow rate of 1 mL/min. The initial oven temperature was 40°C, and then programmed at 5°C/min to 220°C, followed by an increase of 20°C/min to 250°C, and finally held at 250°C for another 2.5 min. Electron ionization (EI+) was set at 70 eV, and the data were recorded in scan mode of m/z 20–400.
Based on the MS fragmentation patterns and linear retention indices, volatile compounds were identified and quantified through comparisons with the NIST14 library. The differential volatiles in each group were screened according to the following criteria: fold change ≥ 1.5 or fold change ≤ 0.67; variable importance in project (VIP) ≥ 1.
Real-Time PCR Analysis
The real-time quantitative PCR (qRT-PCR) was performed using the fluorescent intercalating dye SYBR Green in a detection system (MJ Research, Opticon 2), and MiACT was used as a standard control (Luo et al., 2013). The two-step RT-PCR procedure was performed according to the method described by Li et al. (2005).
Ribonucleic Acid Sequencing
RNAprep pure plant plus kit (TIANGEN Biotech Co., Ltd., Beijing China) was used to purify total ribonucleic acid (RNA) in mango pulp samples. The purification steps were done in accordance with the manufacturer’s instructions. After a quality checked using NanoPhotometer® spectrophotometer and Agilent 2100 bioanalyzer, high-quality mRNA was enriched by poly-T oligo-attached magnetic beads. The library was constructed using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States). After the fragmentation of the purified mRNA by divalent cations at elevated temperature, the first-strand complementary DNA (cDNA) was generated using a random hexamer primer. The second strand of cDNA was generated using DNA polymerase I of M-MuLV reverse transcriptase (RNase H-) and RNase H (Sigma-Aldrich, Shanghai, China). After methylating the 3’ ends of DNA fragments and ligating the adaptor for hybridization, the library fragments were purified using AMPure XP beads (Beckman Coulter, Beverly, United States). Under the action of high-fidelity DNA polymerase, Universal PCR primers, and Index (X) primer, PCR was performed, and the PCR products were purified with AMPure XP system. The list of primers used is showed in Supplementary Table 1. Then, match quality of the library was assessed using the Agilent 2100 bioanalyzer system (Waldbronn, Germany). The library sequencing was performed using the Illumina HiSeq 2500TM platform (PE125, paired-end).
De novo Transcriptome Assembly, Gene Expression, and Differential Expression Analysis
After the quality check and adaptor trimming, clean reads were assembled using the Trinity software, and the transcripts were generated. The reads contained unknown and over 50% low-quality nucleotides (Qphred ≤ 20) were removed. Quality of the transcripts was evaluated using the Benchmarking Universal Single-Copy Orthologs. The coding sequences (CDs) were predicted through comparisons with the NR and Swissprot protein libraries, and using ESTScan v3.0.3 software.
The clean reads were mapped to the transcripts using RSEM v1.1.17 software (Li and Dewey, 2011). The gene expression level was quantified using FPKM (fragments per kilobase of transcript per million fragments mapped) as an indicator. FPKM was calculated as follows:
| (1) |
Differential expression analysis was done using the DEGSeq2, which was according to the criteria as follows: log2 (fold change) ≥ 1 and padj < 0.05.
Gene Ontology Enrichment Analysis
The gene ontology (GO) enrichment analysis of differentially expressed transcripts in mango pulp samples was performed using the KOBAS 2.0, GOseq, and GO database1.
Correlation Analysis of Volatiles and Transcriptome Profile
The correlation analyses of differential volatiles and differential expression transcripts are achieved using the Pearson correlation analysis (SPSS version 15.0).
Results
Volatile Profiles in Mango at Different Development and Storage Stages
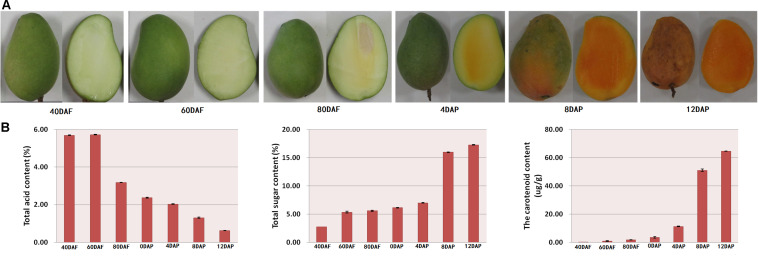
The mango morphology was observed. Total acid, total sugar, and carotenoid contents of the fruit pulp were determined at different stages of fruit development and storage. As shown in Figure 1A, the mangoes were immature and green in color at 40, 60, and 80 days after flowering (DAF). Days after postharvest storage, the peel turned yellow, especially after 4 days of storage. Moreover, the pulps of mango harvested after 40 and 60 days of flowering looked light-green in color. The inner mesocarp of mango started to turn yellow on day 80 (Figure 1A). During the postharvest storage period, especially at 8 and 12 days after picking (DAP), the mango pulp turned orange (Figure 1A). The color hues were determined using a colorimeter (data not shown).
FIGURE 1.
(A) Morphology of mango at different developmental stages and (B) total acid content, total sugar content, and total carotenoid content of mango peel. Samples labeled 40 DAF, 60 DAF, and 80 DAF are the peel samples of mango harvested at 40, 60, 80, and 90 days after the flowering, respectively; samples labeled 0 DAP, 4 DAP, 8 DAP, and 12 DAP are the peel samples of mango harvested at 90 days after flowering which kept for 0, 4, 8, and 12 days, respectively. DAF, days after flowering; DAP, days after picking.
As shown in Figure 1B, there is a gradual decline in total acid content during the fruit development. The total acid content started to reduce after 60 DAF. The total acid content of the pulp samples of 80 DAF dropped to almost half compared to the value determined for pulp samples of 60 DAF. A linear decrease in total acid content was found for the mango pulp samples during the 12 days of storage. Also, there was a gradual decline in total acid content in the developing fruits after 60 DAF (Figure 1B). The total acid content was higher than 5.6% at 40 and 60 DAF. However, the total acid content was 0.63% at 12 DAP (Figure 1B). Moreover, the content of total sugar and total carotenoid gradually increased with the fruits ripening. There was a higher increase in total acid content of the pulp samples of 8 DAP (Figure 1B).
Differential Analysis of Volatiles Among Different Development and Storage Stage
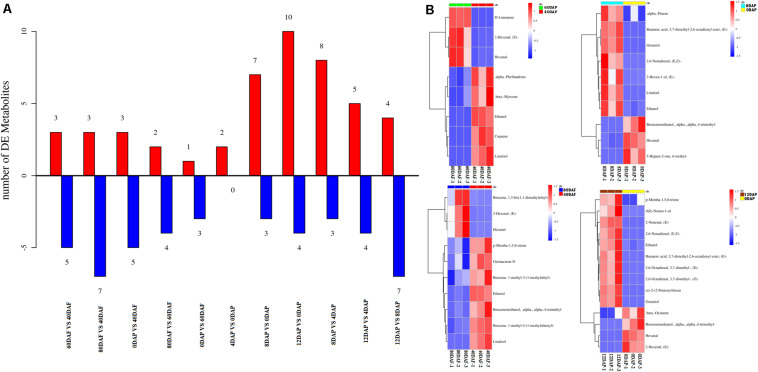
Volatile profiles of mango pulp samples were determined based on the headspace solid-phase microextraction method and GC-MS. The differential volatiles in mango pulp samples of different groups were screened according to the following criteria: fold change ≥ 1.5 or fold change ≤ 0.67; VIP≥ 1. The numbers of significantly different volatiles in each group are shown in Figure 2A. The result showed that significant differences were found for the numbers of differential volatiles between the two groups of samples (p < 0.05). However, the highest significant difference in the numbers of differential volatiles was determined between 12 DAP and 0 DAP samples (p < 0.01).
FIGURE 2.
(A) The numbers of volatiles (metabolites) in mango pulp samples at different developmental and ripening stages and (B) Hierarchical clustering analysis of the differential volatiles in the samples. Samples labeled 40 DAF, 60 DAF, and 80 DAF are the peel samples of mango harvested at 40, 60, and 80 days after the flowering, respectively; samples labeled 0 DAP, 4 DAP, 8 DAP, and 12 DAP are the peel samples of mango harvested at 90 days after flowering which kept for 0, 4, 8, and 12 days, respectively. DAF, days after flowering; DAP, days after picking.
The data determined for the 12 DAP and 0 DAP group included ten up-regulated volatiles and four down-regulated volatiles. As shown in Figure 2B, three up-regulated volatiles determined for the 60 DAF vs. 40 DAF group were D-limonene, (E)-2-hexenal, and hexanal; three up-regulated volatiles found in the 80 DAF vs. 40 DAF group were (E)-2-hexenal, hexanal, and 1,3-bis(1,1-dimethylethyl)-benzene; ten up-regulated volatiles determined in the 12 DAP and 0 DAP group were (2E)-nonenal, (2E)-(2-pentenyl)furan, geraniol, (E,Z)-2,6-nonadienal, ethanol, butanoic acid, p-mentha-1,5,8-triene, (E)-3,7-dimethyl-2,6-octadienal, (Z)-3,7-dimethyl-2,6-octadienal, and (6Z)-nonen-1-ol; seven up-regulated volatiles determined in the 8 DAP vs. 0 DAP group were (E)-butanoic acid 3,7-dimethyl-2,6-octadienyl ester, geraniol, linalool, ethanol, (E,Z)-2,6-nonadienal, α-pinene, and (E)-2-hexen-1-ol. These up-regulated volatiles might be the dominating aromatic components of Tainong mango.
As shown in Dataset 1, 181 volatiles were isolated and identified. The major volatiles detected in the ripen fruit samples are α-pinene, β-phellandrene, β-ocimene, D-limonene, β-myrcene, γ-terpinene, 2-carene, 3-carene, 4-carene, copaene hexanal, and (E)-2-hexenal (Dataset 1). In this study, the amounts of (E)-2-nonenal, (2E)-(2-pentenyl)furan, geraniol, (E,Z)-2,6-nonadienal, ethanol, butanoic acid, p-mentha-1,5,8-triene, (E)-3,7-dimethyl-2,6-octadienal, (Z)-3,7-dimethyl-2,6-octadienal, and (6Z)-nonen-1-ol in the mango pulp samples of 12 DAP were higher than that of the sample at baseline (0 DAP) (Figure 2B), whereas the amounts of (E)-3,7-dimethyl-2,6-octadienyl-butanoate, geraniol, linalool, ethanol, (E,Z)-2,6-nonadienal, α-pinene, and (E)-2-hexen-1-ol in the mango pulp samples of 8 DAP were higher than the sample at baseline.
RNA Sequencing Analysis of Mango During Different Development and Storage Stages
Molecular insight of volatile biosynthesis in mango pulp during the fruit development and storage can be obtained from RNA sequencing (RNA-Seq) analysis. Seven cDNA of the fruit samples, collected after 40, 60, 80, and 90 DAF, and at 4, 8, and 12 days of storage were constructed and large-scale sequenced. 47.29-79.61 million raw reads and 46.48-78.33 million cleaned reads were generated in each sample (Table 1). The Q20 (the percentage of bases with a Phred score greater than 20) and Q30 (the percentage of bases with a Phred score greater than 30) were higher than 95% (Table 1).
TABLE 1.
Reads numbers, Q20 values, Q30 values, and gas chromatography (GC) content of mango pulp based on the RNA sequencing (RNA-Seq) data in the libraries of A, B, C, D, E, F, and G.
| Sample | Raw reads | Clean reads | Q20 (%) | Q30 (%) | GC content (%) | Total mapped reads |
| A_1 | 54542030 | 53752304 | 98.71 | 95.68 | 45.04 | 41320416 (76.87%) |
| A_2 | 79607262 | 78327428 | 98.68 | 95.59 | 44.67 | 60458266 (77.19%) |
| A_3 | 66307584 | 65005808 | 98.69 | 95.6 | 44.67 | 50076660 (77.03%) |
| B_1 | 61825070 | 60940514 | 98.55 | 95.17 | 44.91 | 47642222 (78.18%) |
| B_2 | 61776038 | 60874120 | 98.71 | 95.66 | 44.81 | 47623800 (78.23%) |
| B_3 | 62783660 | 61896096 | 98.72 | 95.65 | 44.75 | 48219096 (77.90%) |
| C_1 | 65584430 | 64443166 | 98.74 | 95.73 | 44.65 | 50675552 (78.64%) |
| C_2 | 66911796 | 65761656 | 98.67 | 95.52 | 44.68 | 51653690 (78.55%) |
| C_3 | 70930988 | 69608456 | 98.58 | 95.29 | 44.2 | 54910004 (78.88%) |
| D_1 | 64323328 | 63325728 | 98.66 | 95.46 | 44.22 | 48845080 (77.13%) |
| D_2 | 53106480 | 52366662 | 98.73 | 95.72 | 44.51 | 40206360 (76.78%) |
| D_3 | 61134368 | 60176382 | 98.66 | 95.47 | 44.35 | 46130222 (76.66%) |
| E_1 | 64246102 | 63464652 | 98.69 | 95.53 | 44.45 | 49456492 (77.93%) |
| 294E_2 | 47298170 | 46481888 | 98.68 | 95.48 | 44.36 | 36320930 (78.14%) |
| E_3 | 58093350 | 57090472 | 98.75 | 95.69 | 44.49 | 44561666 (78.05%) |
| F_1 | 59865570 | 58791448 | 98.55 | 95.12 | 44.19 | 45797898 (77.90%) |
| F_2 | 67186956 | 66240678 | 98.75 | 95.71 | 44.1 | 51789204 (78.18%) |
| F_3 | 60845708 | 59572020 | 98.72 | 95.6 | 44.13 | 46554538 (78.15%) |
| G_1 | 73520470 | 72003528 | 98.78 | 95.73 | 43.98 | 56418766 (78.36%) |
| G_2 | 54733094 | 53300870 | 98.79 | 95.77 | 44.01 | 42180466 (79.14%) |
| G_3 | 68004602 | 67124008 | 98.65 | 95.35 | 43.96 | - |
A, 40 DAF; B, 60 DAF; C, 80 DAF; D, 0 DAP; E, 4 DAP; F, 8 DAP; G, 12 DAP. _1, _2, and _3 represent the triplicate analyses of each sample. Q20 is the percentage of bases with a Phred score greater than 20, whereas Q30 is the percentage of bases with a Phred score greater than 30.
After assembled using the Trinity software, the transcripts of each sample were acquired. The frequency and numbers of transcripts and unigene in the corresponding length are shown in Supplementary Figure 1A. The CDs were predicted by comparing them with NR protein library, Swissprot protein library, and ESTScan v3.0.3 software. The counts of CDs with different lengths were also predicted based on the corresponding method (Supplementary Figure 1B).
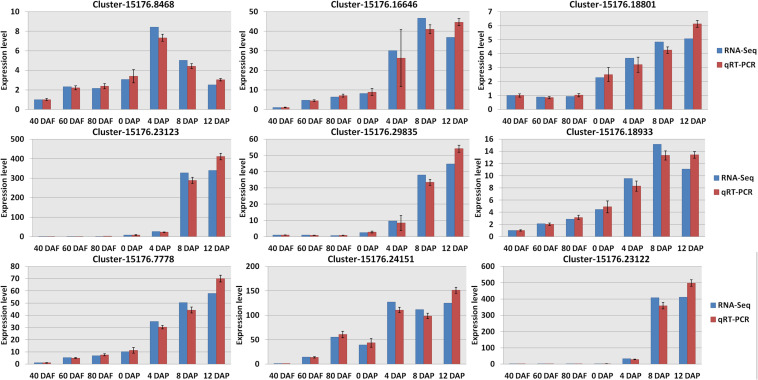
Hierarchical clustering analysis of Pearson correlation according to the level of gene expression levels revealed that a high correlation was found for the gene expression in mango pulp samples between different development stages (Supplementary Figure 1C). Besides the three replicates of mango samples collected after 40 days of flowering, the Pearson correlation coefficients among the three repetitions of the other samples collected were higher than 0.95, which showed good reliability and repeatability of RNA-Seq data. Verification of the RNA-Seq data was done based on the nine selected genes (Figure 3 and Dataset 2).
FIGURE 3.
Verification the RNA sequencing (RNA-Seq) results by real-time quantitative PCR (qRT-PCR). All data were presented as mean ± standard error of the mean of triplicate analyses. DAF, days after flowering; DAP, days after picking. Gene names (Supplementary Table 2): Cluster-15176.8468 (wrbA); Cluster-15176.23123 (LOX1_5); Cluster-15176.7778 (accA); Cluster-15176.16646 (4CL); Cluster-15176.29835 (SXD1); Cluster-15176.24151 (fabI); Cluster-15176.18801 (VTE1); Cluster-15176.18933 (TAT); and Cluster-15176.23122 (LOX1_5).
Differentially Expressed Transcripts and Gene Ontology Enrichment Analysis
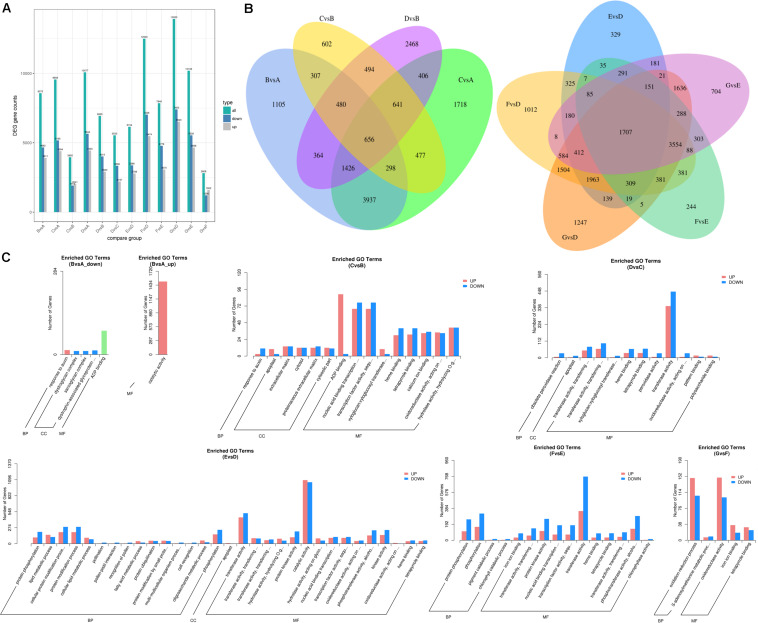
The differentially expressed transcripts in the groups of 60 DAF vs. 40 DAF, 80 DAF vs. 40 DAF, 90 DAF vs. 40 DAF, 80 DAF vs. 60 DAF, 90 DAF vs. 60 DAF, 90 DAF vs. 80 DAF, 4 DAP vs. 0 DAP, 8 DAP vs. 0 DAP, 12 DAP vs. 0 DAP, 8 DAP vs. 4 DAP, 12 DAP vs. 4 DAP, and 12 DAP vs. 8 DAP were identified based on the DEGSeq analysis for obtaining an overview of the interesting genes. The numbers of differentially expressed transcripts in each group are shown in Figure 4A and Dataset 2. The results showed that the most differentially expressed transcripts were found in the group of 12 DAP vs. 0 DAP (including 6,520 up-regulated transcripts and 7,400 down-regulated transcripts), where the data are in line with the results obtained from the determination of differential volatiles (Figure 2A). The Venn diagram also shows that 24 mutually differentially expressed transcripts existed in 60 DAF vs. 40 DAF, 80 DAF vs. 40 DAF, 80 DAF vs. 60 DAF, and 90 DAF vs. 60 DAF, whereas the number of commonly differentially expressed transcripts in 4 DAP vs. 0 DAP, 8 DAP vs. 0 DAP, 12 DAP vs. 0 DAP, 8 DAP vs. 4 DAP, and 12 DAP vs. 4 DAP (Figure 4B). These differentially expressed transcripts might be relevant to the mango development and formation of aromatic compounds.
FIGURE 4.
Differentially expressed transcripts of mango pulp samples at different developmental and ripening stages. (A) Total numbers of the differentially expressed genes (DEGs), up-regulated genes, and down-regulated genes between different samples, B vs. A, C vs. A, D vs. A, C vs. B, D vs. B, D vs. C, E vs. D, F vs. D, G vs. D, G vs. E, F vs. E, and G vs. F; (B) Venn diagram of the DEGs between different samples, B vs. A, C vs. A, C vs. B, D vs. B (left) and E vs. D, F vs. D, G vs. D, G vs. E, F vs. E (right); (C) GO classification of the up-regulated genes and down-regulated genes between different samples, B vs. A, C vs. B, D vs. C, E vs. D, F vs. E, and G vs. F; A–C are the samples collected after 40, 60, and 80 days of flowering, respectively; D–G are the samples stored for 0, 4, 8, and 12 days which collected after 90 days of flowering.
The GO analysis was performed to show the comprehensiveness of functions of differentially expressed transcripts. As shown in Figure 4C, the gene functions are described based on the cellular components, molecular functions, and biological processes. During the fruit development, the most enriched GO terms for the groups of 60 DAF vs. 40 DAF and 90 DAF vs. 80 DAF were catalytic activity and transferase activity, respectively. Adenosine diphosphate (ADP) binding and transcription factor activity were the two most enriched GO terms for 80 DAF vs. 60 DAF. During storage of mango, the most enriched GO terms found were catalytic activity (4 DAP vs. 0 DAP), transferase activity (8 DAP vs. 4 DAP), and oxidoreductase activity (12 DAP vs. 8 DAP).
Correlation Analysis of Differential Volatiles and Differentially Expressed Transcripts
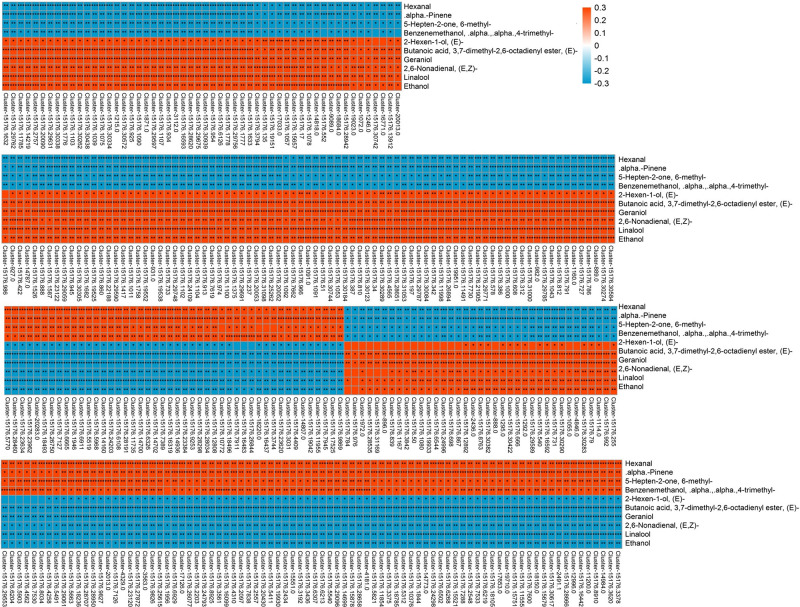
Correlation analysis was performed to explore the transcripts related to aroma biosynthesis during the development and storage of Tainong mango (Figure 5, Dataset 3). The results showed that 304 differentially expressed transcripts determined in the group of 8 DAP vs. 0 DAP were either positively or negatively correlated with 3,7-dimethyl-2,6-octadienyl ester, (E)-butanoic acid, geraniol, hexanal, 6-methyl-5-hepten-2-one, linalool, ethanol, α,α,4-trimethyl-benzenemethanol, (E,Z)-2,6-nonadienal, α-pinene, and (2E)-hexen-1-ol; 420 differentially expressed transcripts determined in the group of 4 DAP vs. 0 DAP were either positively or negatively correlated with α,α,4-trimethyl-benzenemethanol and (E,Z)-2,6-nonadienal; 601 differentially expressed transcripts determined in the group of 12 DAP vs. 0 DAP were either positively or negatively correlated with (2E)-nonenal, hexanal, (2E)-(2-pentenyl)furan, geraniol, (2E)-hexenal, (E,Z)-2,6-nonadienal, ethanol, (E)-3,7-dimethyl-2,6-octadienal, (Z)-3,7-dimethyl-2,6-octadienal, butanoic acid, α,α,4-trimethyl-benzenemethanol, (6Z)-nonen-1-ol, p-mentha-1,5,8-triene, and β-ocimene; 277 differentially expressed transcripts determined in the group of 80 DAF vs. 60 DAF were either positively or negatively correlated with α,α,4-trimethyl-benzenemethanol, 1-(4-methylphenyl)- ethanone, 1-methyl-4-(1-methylethenyl)-benzene, p-mentha-1,5,8-triene, alloaromadendrene, and 3-hexenal; 381 differentially expressed transcripts determined in the group of 60 DAF vs. 40 DAF were either positively or negatively correlated with D-limonene, ethanol, copaene, linalool, α-phellandrene, (2E)-hexenal, hexanal, and β-myrcene (Dataset 3).
FIGURE 5.
Correlation analysis of transcriptome and volatile profiles of mango pulp samples from the 8 DAP vs. 0 DAP group. The X-coordinate is the transcript name, and the Y-coordinate is the metabolite name. Each square represents the correlation and significance of the transcript and the metabolite. The correlation coefficient is presented based on different colors and shades. Red represents a positive correlation and blue represents a negative correlation. The darker the color, the higher the correlation. The names of the identified compounds and their related unigene-id are tabulated in Supplementary Table 1.
Genes and Enzymes Related to the Metabolism of Aromatic Compounds
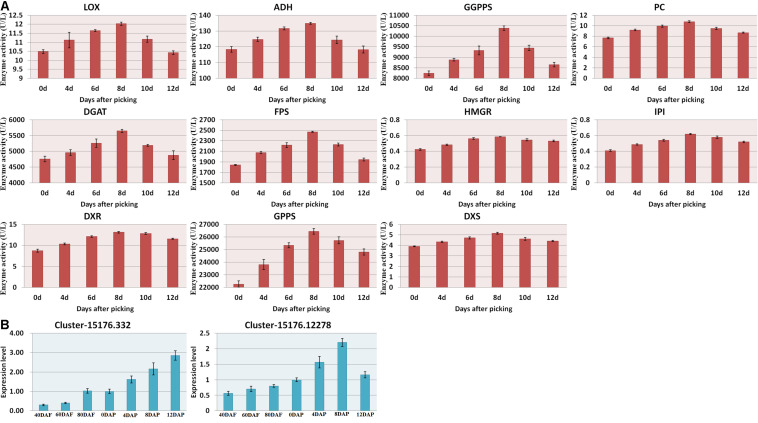
As shown in Figure 2B, the amount of α-pinene in the fruit sample stored on day 8 was significantly higher than that on the other days (p < 0.05). It indicated that α-pinene is one of the key aroma components of mango. The results showed that a total of eight genes involved in the diterpenoid metabolism pathways (Figure 6 and Supplementary Table 3). The transcriptome analysis manifested that the genes, such as Cluster-15176.332 (E5.5.1.13) and Cluster-15176.12278 (KAO), were expressed at the highest levels in the mango pulp samples of 8 DAP. The high expressions of Cluster-15176.3381 (E1.14.11.13), Cluster-15176.3380 (E1.14.11.13), Cluster-19253.0 (GA3,CYP701), Cluster-15991.0 (E1.14.11.13), Cluster-3324.0 (E1.14.11.13), and Cluster-15176.1075 (CYP82G1) were also detected in mango pulp samples during the fruit development and ripening. These genes involved in diterpenoid biosynthesis. The qRT-PCR analysis also verified the highest expressions of E5.5.1.13 (ent-copalyl diphosphate synthase) and KAO (ent-kaurenoic acid hydroxylase) in the samples of 8 DAP compared with the other stages. These enzymes have been reported to regulate the biosynthesis of gibberellin (Su et al., 2016; Szymczyk et al., 2020). Only one gene was found to be involved in monoterpenoid biosynthesis. It was K15095 (Cluster-4351.0). The gene is related to the biosynthesis of nerolidol in the fruit. However, nerolidol was not detected in the fruit samples.
FIGURE 6.
(A) Enzyme activities of the mango pulp samples and (B) gene expressions of E5.5.1.13 (Cluster-15176.332) and KAO (Cluster-15176.12278) in the samples during the fruit development and ripening. The enzyme activities of 1-deoxyd-D-xylose-5-phosphate reductase (DXS), 1-deoxyxylose-5-phosphate synthase (DXR), geranyl pyrophosphate synthetase (GPPS), geranyl geranyl pyrophosphate synthetase (GGPPS), pyruvate carboxylase (PC), diacylglyceryltransferase (DGAT), farnesyl diphosphate synthase (FPS), and hydroxymethyl glutarate monoacyl CoA reductase (FPS) in the mango pulp samples stored at different days were determined using ELISA kits. All data are presented as means ± standard errors of the means. Triplicate analyses were performed for each sample. DAF, day after flowering; DAP, days after picking.
The gene names of the respective UniGene IDs are depicted in Table 2. Ent-kaurene oxidase, gibberellin 2-β-dioxygenase, and gibberellin 2-β-dioxygenase 8 isoform X3 were highly expressed during the fruit development and ripening. Only ent-kaurenoic acid oxidase 1-like was highly expressed in the matured fruit, and it was the main enzyme expressed in the terpenoid metabolism. Moreover, 18 others genes detected were known to be involved in ubiquinone and other terpenoid-quinone biosynthesis. Gibberellin is known to be synthesized via terpenoid biosynthesis pathway (Boba et al., 2020), and kaurene oxidase catalyzes the gibberellin biosynthesis. Besides, ent-kaurene is a tetracyclic hydrocarbon precursor for gibberellins. Moreover, the enzyme activities measured using ELISA kits, including DXS, DXR, GPPS, GGPPS, PC, DGAT, FPS, and HGMR were found to be increased gradually during the storage, and generally attained maximum levels at 8 DAP (Figure 6A).
TABLE 2.
UniGene ID of the transcripts and compounds identified in the mango pulp samples.
| UniGene ID | Gene details |
| Cluster-15176.1075 | GAY40054.1| hypothetical protein |
| Cluster-15176.3381 | KDP37976.1| hypothetical protein |
| Cluster-15176.3380 | XP_015574955.1| gibberellin 2-beta-dioxygenase |
| Cluster-15991.0 | PSS05904.1| gibberellin 2-beta-dioxygenase |
| Cluster-19253.0 | XP_006431588.1| ent-kaurene oxidase |
| Cluster-15176.332 | ESR57297.1| ent-copalyl diphosphate synthase |
| Cluster-3324.0 | XP_006467978.1| gibberellin 2-beta-dioxygenase 8 isoform X3 |
| Cluster-15176.12278 | XP_021808327.1| ent-kaurenoic acid oxidase 1-like, |
| Cluster-15176.13875 | GAV73636.1| patatin domain-containing protein |
| Cluster-15176.7685 | PIA28198.1| hypothetical protein |
| Cluster-15176.7686 | EEF47638.1| diacylglycerol cholinephosphotransferase |
| Cluster-8978.0 | XP_006433476.1| phospholipase D zeta 2 isoform X2 |
| Cluster-15176.21081 | XP_022878380.1| choline..ethanolaminephosphotransferase 1-like |
| Cluster-15176.17421 | OAY57973.1| hypothetical protein |
| Cluster-15176.14147 | XP_024036163.1| phospholipase D alpha 1 |
| Cluster-15176.27952 | KDO81980.1| hypothetical protein |
| Cluster-15176.22073 | ESR39921.1| hypothetical protein |
| Cluster-15176.23120 | AQZ55551.1| 9-lipoxygenase |
| Cluster-15176.23122 | AQZ55556.1| linoleate 9S-lipoxygenase |
| Cluster-15176.23123 | ANF89411.1| linoleate 9S-lipoxygenase |
| Cluster-15176.19505 | ESR33138.1| hypothetical protein |
| Cluster-15176.23124 | AQZ55551.1| 9-lipoxygenase |
| Cluster-15176.13300 | ESR59166.1| hypothetical protein |
| Cluster-15176.23657 | GAV90434.1| adh_short domain-containing protein |
| Cluster-15176.4077 | ESR36401.1| hypothetical protein |
| Cluster-15176.7778 | GAV68539.1| acetyl-CoA carboxylase carboxyl transferase subunit alpha |
| Cluster-15176.5176 | XP_021897331.1| acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha |
| Cluster-15176.21236 | AIU99499.1| hydroxyacyl-ACP dehydrase |
| Cluster-15176.15013 | GAY44796.1| hypothetical protein |
| Cluster-15176.19615 | KDO86915.1| hypothetical protein |
| Cluster-15176.19806 | KRG93630.1| hypothetical protein |
| Cluster-15176.19807 | OMP10121.1| biotin..lipoyl |
| Cluster-15176.13810 | XP_006488591.1| malonyl CoA-acyl carrier protein transacylase |
| Cluster-15176.23708 | GAY52806.1| hypothetical protein |
| Cluster-15176.8090 | KVI00409.1| Fatty acid desaturase |
| Cluster-15176.15658 | ESR56644.1| hypothetical protein |
| Cluster-15176.20399 | OWM86900.1| hypothetical protein |
| Cluster-15176.6573 | GAY39911.1| hypothetical protein |
| Cluster-15176.17613 | XP_006492369.1| long chain acyl-CoA synthetase 1 |
| Cluster-15176.1088 | PON38544.1| fatty acid desaturase |
| Cluster-15176.8360 | XP_006482734.1| biotin carboxyl carrier protein of acetyl-CoA carboxylase 2 |
| Cluster-15176.27073 | KJB64462.1| hypothetical protein |
| Cluster-15176.27077 | XP_006434188.1| long chain acyl-CoA synthetase 9 |
| Cluster-15176.27074 | XP_008784201.1| long chain acyl-CoA synthetase 9 |
| Cluster-15176.27075 | GAY41784.1| hypothetical protein |
| Cluster-15176.11597 | AOR17397.1| palmitoyl-acyl carrier protein thioesterase |
| Cluster-15176.16995 | XP_024045530.1| 3-oxoacyl-[acyl-carrier-protein] synthase III |
| Cluster-15176.22688 | ESR59166.1| hypothetical protein |
| Cluster-15176.13123 | GAY44882.1| hypothetical protein |
| Cluster-15176.16081 | XP_022757277.1| acetyl-coenzyme A carboxylase carboxyl transferase |
| Cluster-15176.21237 | XP_006492623.1| 3-hydroxyacyl-[acyl-carrier-protein] dehydratase |
| Cluster-15176.15551 | AKA09592.1| stearoyl-ACP desaturase |
| Cluster-15176.15554 | GAY44796.1| hypothetical protein |
| Cluster-15176.17196 | GAV92051.1| acyl-ACP_TE domain-containing protein |
| Cluster-15176.7526 | XP_021284424.1| stearoyl-[acyl-carrier-protein] 9-desaturase 6 |
| Cluster-15176.24109 | ESR56805.1| hypothetical protein |
| Cluster-15176.16301 | PON38544.1| fatty acid desaturase |
| Cluster-15176.24027 | PPS18419.1| hypothetical protein |
| Cluster-15176.24028 | XP_006480952.1| acetyl-coenzyme A carboxylase carboxyl transferase |
| Cluster-15176.18609 | XP_009413949.1| long chain acyl-CoA synthetase 6 |
| Cluster-15176.18608 | PIN10227.1| long-chain acyl-CoA synthetases |
| Cluster-15176.15400 | CBI26222.3| unnamed protein product |
| Cluster-15176.24150 | XP_024443812.1| enoyl-[acyl-carrier-protein] reductase [NADH] 1 |
| Cluster-15176.24151 | XP_023914015.1| enoyl-[acyl-carrier-protein] reductase [NADH] 1 |
| Cluster-15176.24152 | AIS93131.1| enoyl-acyl carrier protein reductase [NAD+] |
| Cluster-15176.24153 | POF25828.1| enoyl-[acyl-carrier-protein] reductase [NADH] |
| Cluster-4351.0 | XP_006467945.1| (3S,6E)-nerolidol synthase 1-like |
| Cluster-15176.10743 | PHU22294.1| acyl-[acyl-carrier-protein] desaturase |
| Cluster-15176.23658 | GAY44036.1| hypothetical protein |
| Cluster-15176.14875 | OMO53044.1| thiolase |
| Cluster-19268.0 | XP_021679005.1| very-long-chain enoyl-CoA reductase-like |
| Cluster-15176.19912 | ESR59974.1| hypothetical protein |
| Cluster-13603.0 | XP_013702088.1| palmitoyl-monogalactosyldiacylglycerol delta-7 desaturase |
| Cluster-15176.9059 | XP_021736703.1| palmitoyl-monogalactosyldiacylglycerol delta-7 desaturase |
| Cluster-15176.16917 | AQZ55555.1| acyl-CoA-oxidase-1 |
| Cluster-15176.6546 | GAY56458.1| hypothetical protein |
| Cluster-15176.20306 | AIE48274.1| omega-6 fatty acid desaturase |
| Cluster-15176.12892 | AIC34706.1| omega-6 fatty acid desaturase |
| Cluster-15176.24984 | KYP77364.1| chalcone synthase 1 |
| Cluster-15176.2365 | XP_006437739.2| leucoanthocyanidin reductase |
| Cluster-15176.2366 | POE95046.1| leucoanthocyanidin reductase |
| Cluster-15176.17061 | AIY25000.1| flavanone 3’-hydroxylase |
| Cluster-15176.4728 | AIY24979.1| cinnamate 4-mono-oxygenase |
| Cluster-15176.15897 | APR63682.1| p-coumarate 3-hydroxylase 3 |
| Cluster-15176.9748 | XP_021664632.1| 4-coumarate–CoA ligase |
| Cluster-15176.14112 | OMO89673.1| oxoglutarate..iron-dependent dioxygenase |
| Cluster-15176.13889 | AIY24995.1| flavanone 3-hydroxylase 2 |
| Cluster-15176.6688 | AIY25002.1| dihydroflavonol 4-reductase |
| Cluster-15176.2805 | BAX25479.1| alkyldiketide-CoA synthase |
| Cluster-15176.19426 | XP_022862158.1| 4-coumarate–CoA ligase-like 7 |
| Cluster-15176.1022 | KDO60238.1| hypothetical protein |
| Cluster-15176.4870 | XP_021294689.1| 4-coumarate–CoA ligase 1-like |
| Cluster-19931.0 | KDO46519.1| hypothetical protein |
| Cluster-15176.18933 | TAT| tyrosine aminotransferase |
| Cluster-15176.29835 | AAP97931.1| tocopherol cyclase |
| Cluster-15176.18801 | XP_018842576.1| tocopherol cyclase |
| Cluster-15176.8468 | PSR96325.1| NAD(P)H dehydrogenase FQR1-like |
| Cluster-15176.1733 | PRQ41134.1| putative 1,4-dihydroxy-2-naphthoyl-CoA synthase |
| Cluster-15176.12108 | XP_011082392.1| probable NAD(P)H dehydrogenase (quinone) FQR1-like 3 isoform X3 |
| Cluster-15176.6725 | ESR60477.1| hypothetical protein |
| Cluster-15176.6726 | GAY65474.1| hypothetical protein |
| Cluster-15176.6727 | XP_022732769.1| probable aminotransferase |
| Cluster-15176.26169 | PSR95337.1| nicotianamine aminotransferase |
| Cluster-15176.16646 | XP_006488032.1| 4-coumarate–CoA ligase-like 7 |
| Cluster-15176.7245 | XP_006469904.1| tyrosine aminotransferase |
| Cluster-18980.0 | XP_006484288.1| protein PHYLLO |
Discussion
Volatile Components of Mango at Different Stages
Volatile components in mango account for the tempting aroma of the fruit, which is also the most important quality of mango pulp and processed mango products. In common with nutritional quality, texture, and color, and the aroma is generally shaped by the coordination of biochemical and developmental pathways (Fujisawa et al., 2013). Many volatile components have been isolated and identified in mature mango and its juice of different cultivars, and some aroma-contributing compounds have also been confirmed in the previous studies (Andrade et al., 2000; Lalel et al., 2003; Pino and Mesa, 2006; Shivashankara et al., 2006; Munafo et al., 2014; Zhang et al., 2019).
In line with a previous study, volatile profiles of Tainong mango of different stages of harvesting and storage, including three stages during the fruit development and four stages during storage, were determined. Literature shows that the transcriptomes of Alphonso mango pulp and flower collected from the seven stages of fruit development and ripening were determined (Deshpande et al., 2017a). The volatile profiling and transcriptome were integrated for the identification of genes related to the metabolism of volatiles.
A previous study reported that 4-hydroxy-2,5-dimethyl-3(2H)-furanone was an important aromatic compound detected in mango cultivars Haden, White Alfonso, Praya Sowoy, Royal Special, and Malindi (Munafo et al., 2014). However, the GC-MS data showed that the mango pulp samples had a low amount of 4-hydroxy-2,5-dimethyl-3(2H)-furanone. Ethyl octanoate, 3-carene, limonene, α-terpinene, α-terpinolene, hexanal, and p-cymene were also the key volatiles detected in the different cultivars of Australian mango (San et al., 2017). Some of these compounds, such as ethyl octanoate, terpinolene, and cymene, were not detected in Tainong mango. A total of 12 components, including 2,4-dimethylstyrene, were identified as the major aroma active compounds in Keitt mango juice (Zhang et al., 2019).
The mango stored at these two stages had a more intense aroma than the other five stages. It could be due to a gradual increase in the total sugar content and a gradual decrease in total acid content during fruit maturity (Figure 1B). The results also showed that 17 volatile components were the key aroma active compounds in Tainong mango, especially ethanol and (E,Z)-2,6-nonadienal. Moreover, three up-regulated volatiles in both groups of 60 DAF vs. 40 DAF and 80 DAF vs. 40 DAF, and about 180 volatile components identified in the mango pulp samples might be the potential aromatic compounds.
Genes and Enzymes Involved in Metabolism of Aromatic Components
The transcriptome studies have put forth important information concerning the development of mango of different cultivars, such as Zill (Wu et al., 2014), Langra (Azim et al., 2014), Kent (Dautt-Castro et al., 2015), Dashehari (Srivastava et al., 2016), and Alphonso (Deshpande et al., 2017a). Most of these studies reported the general metabolic pathways involved in the biosynthesis of metabolites in the mango. The genes encoding multiple enzymes related to gluconeogenesis from carbohydrate metabolism, glycolysis, fatty acid biosynthesis and beta-oxidation, salicylic acid biosynthesis, citrate cycle, ethylene biosynthesis, amino acids biosynthesis and degradation, β-carotene biosynthesis, α-tocopherol biosynthesis, flavonoid biosynthesis, and terpenoid backbone synthesis have also been reported in the literature.
In this study, RNA-Seq was performed to explore the molecular mechanism of aroma compounds biosynthesis in mango during the fruit development, ripening, and storage. A large number of differentially expressed transcripts involved in multiple pathways have been identified among these samples. Quantitative RT-PCR analysis also showed that the relative expression patterns of eleven genes were consistent with the RNA-Seq data. The results indicate that the transcriptome data are reliable. The number of differentially expressed transcripts in the samples consisted of over 50,000 up-regulated and down-regulated transcripts. These genes are related to the development of mango. Only three genes were highly expressed and these genes are closely related to the biogenesis of aromatic components during the fruit development. These two genes, E5.5.1.13 and KAO, have never been reported in the transcriptome analysis and metabolic profiling of mango. Another gene that was also found to be highly expressed in the mango pulp samples of 8 DAP. K15095 was the only gene found to be highly expressed during the storage of mango.
A large number of transcription factors (TFs) has been identified in the fruit samples as these factors are known to regulate the fruit development and formation of the aroma of the fruit (Bastías et al., 2011; Hong et al., 2012; Shen et al., 2016; Liu et al., 2017; Lü et al., 2018; Zhang et al., 2018). These TFs might have been participated in controlling the mango development and the biosynthesis of aromatic components of the fruit. The contents of terpenes in mango have been quantified and reported in the literature (Pandit et al., 2009a,b). The expression of genes involved in GPP, FPP, and GGPP synthesis have also been studied (Azim et al., 2014; Wu et al., 2014; Dautt-Castro et al., 2015; Srivastava et al., 2016; Deshpande et al., 2017a). As reported by Ma et al. (2006), DXS, DXR, GPPS, GGPPS, PC, DGAT, FPS, and HGMRs are the key enzymes for terpenoid-isoprenoid biosynthesis. The results obtained from ELISA assays revealed that the activities of these enzymes increased gradually after harvested and reached a maximum level on day 8. On the contrary, the PCR analysis did not show expression of the genes related to these enzymes in the mango pulp samples. In this study, the two highly expressed genes are known to be involved in the pathways of diterpenoid biosynthesis.
Conclusion
Mango is favorable due to its pleasant sensory quality and high nutritional values. Although some of the aromatic components have been identified in different cultivars of mango, little is known about the volatile profile in the Tainong mango. 181 volatiles were isolated and identified in fruits collected at seven stages. These components, especially ethanol and (E,Z)-2,6-nonadienal, were the key aroma active compounds in Tainong mango. RNA-Seq and comparative analysis showed a large number of DEGs during development and after picking. These involved in catalytic activity, transferase activity, ADP binding, transcription factor activity, and oxidoreductase activity. The content of α-pinene, expression of genes involved in terpenoid metabolism, and enzyme activities in terpenoid metabolic pathways gradually increased after picking and generally attained their maximum levels on day-8. The integrative analyses also revealed potential molecular insights into fruit development and aroma formation. This study provides important cues for future work on mango quality improvement.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: (https://www.ncbi.nlm.nih.gov/), and the data accession number is PRJNA697524.
Author Contributions
CL, MX, PY, and YT performed the experiments. CL conceived and designed the research. LL and JS analyzed data. MX and XH prepared the manuscript. CL, JS, and HEK checked and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was financially supported by the Guangxi Natural Science Foundation (2018GXNSFAA281149), the Major Program of Science and Technology in Guangxi (Guike AA17204038 and Guike AA17204042), the Key Research and Development Programs of Guangxi (Guike AB18221110 and Guike AB18294027), and Special Fund for “Bagui Scholars” of Guangxi ([2016]21).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.666805/full#supplementary-material
Overview the transcriptome analysis of mango pulp samples at different developmental and ripening stages. (A) The frequency and numbers of transcripts with different length; (B) The count of CDs with different length predicted by comparing with NR protein library and Swissprot protein library (left) or by estscan (3.0.3) software (right); (C) The hierarchical clustering analysis of Pearson correlation according to the level of gene expression levels in fruits of A, B, C, D, E, F, and G. A, 40 DAF; B, 60 DAF; C, 80 DAF; D, 0 DAP; E, 4 DAP; F, 8 DAP; G, 12 DAP. _1, _2 and _3 represent the Triplicate analyses of each sample.
References
- Andrade E. H. A., Maia J. G. S., Maria das Graças B. Z. (2000). Aroma volatile constituents of Brazilian varieties of mango fruit. J. Food Compos. Anal. 13 27–33. 10.1006/jfca.1999.0841 [DOI] [Google Scholar]
- Azim M. K., Khan I. A., Zhang Y. (2014). Characterization of mango (Mangifera indica L.) transcriptome and chloroplast genome. Plant Mol. Biol. 85 193–208. 10.1007/s11103-014-0179-8 [DOI] [PubMed] [Google Scholar]
- Bastías A., López-Climent M., Valcárcel M., Rosello S., Gómez-Cadenas A., Casaretto J. A. (2011). Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol. Plant. 141 215–226. 10.1111/j.1399-3054.2010.01435.x [DOI] [PubMed] [Google Scholar]
- Boba A., Kostyn K., Kozak B., Wojtasik W., Preisner M., Prescha A., et al. (2020). Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 251 1–14. 10.1007/s00425-020-03339-9 [DOI] [PubMed] [Google Scholar]
- Boonbumrung S., Tamura H., Mookdasanit J., Nakamoto H., Ishihara M., Yoshizawa T., et al. (2001). Characteristic aroma components of the volatile oil of yellow keaw mango fruits determined by limited odor unit method. Food Sci. Technol. Res. 7 200–206. 10.3136/fstr.7.200 [DOI] [Google Scholar]
- Chauhan O. P., Raju P. S., Bawa A. S. (2010). “Mango flavor,” in Handbook of Fruit and Vegetable Flavors, ed. Hui Y. H. (Hoboken, NJ: Wiley; ), 319–343. 10.1002/9780470622834.ch19 [DOI] [Google Scholar]
- Dautt-Castro M., Ochoa-Leyva A., Contreras-Vergara C. A., Pacheco-Sanchez M. A., Casas-Flores S., Sanchez-Flores A., et al. (2015). Mango (Mangifera indica L.) cv. Kent fruit mesocarp de novo transcriptome assembly identifies gene families important for ripening. Front. Plant Sci. 6:62. 10.3389/fpls.2015.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A. B., Anamika K., Jha V., Chidley H. G., Oak P. S., Kadoo N. Y., et al. (2017a). Transcriptional transitions in Alphonso mango (Mangifera indica L.) during fruit development and ripening explain its distinct aroma and shelf life characteristics. Sci. Rep. 7:8711. 10.1038/s41598-017-08499-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A. B., Chidley H. G., Oak P. S., Pujari K. H., Giri A. P., Gupta V. S. (2017b). Isolation and characterization of 9-lipoxygenase and epoxide hydrolase 2 genes: Insight into lactone biosynthesis in mango fruit (Mangifera indica L.). Phytochemistry 138 65–75. 10.1016/j.phytochem.2017.03.002 [DOI] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Shima Y., Ito Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25 371–386. 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G. J., Xue X. Y., Mao Y. B., Wang L. J., Chen X. Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24 2635–2648. 10.1105/tpc.112.098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni R. S., Chidley H. G., Pujari K. H., Giri A. P., Gupta V. S. (2012). Geographic variation in the flavour volatiles of Alphonso mango. Food Chem. 130 58–66. 10.1016/j.foodchem.2011.06.053 [DOI] [Google Scholar]
- Lalel H. J. D., Singh Z., Tan S. C. (2003). Aroma volatiles production during fruit ripening of ‘Kensington Pride’ mango. Postharvest Biol. Technol. 27 323–336. 10.1016/S0925-5214(02)00117-5 [DOI] [Google Scholar]
- Lebrun M., Plotto A., Goodner K., Ducamp M. N., Baldwin E. (2008). Discrimination of mango fruit maturity by volatiles using the electronic nose and gas chromatography. Postharvest Biol. Technol. 48 122–131. 10.1016/j.postharvbio.2007.09.010 [DOI] [Google Scholar]
- Li B., Dewey C. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. B., Fan X. P., Wang X. L., Cai L., Yang W. C. (2005). The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17 859–875. 10.1105/tpc.104.029629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L., Dong T., Qiu X., Rong Y., Wang Z., Zhu J. (2019). Nitrogen nutrition is a key modulator of the sugar and organic acid content in citrus fruit. PLoS One 14:e0223356. 10.1371/journal.pone.0223356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Y., Cao X. M., Liu X. H., Xin R., Wang J. J., Gao J., et al. (2017). UV-B irradiation differentially regulates terpene synthases and terpene content of peach. Plant Cell Environ. 40 2261–2275. 10.1111/pce.13029 [DOI] [PubMed] [Google Scholar]
- Lü P., Yu S., Zhu N., Chen Y. R., Zhou B., Pan Y., et al. (2018). Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants 4 784–791. 10.1038/s41477-018-0249-z [DOI] [PubMed] [Google Scholar]
- Luo C., He X. H., Chen H., Hu Y., Ou S. J. (2013). Molecular cloning and expression analysis of four actin genes (MiACT) from mango. Biol. Plant. 57 238–244. 10.1007/s10535-012-0278-9 [DOI] [Google Scholar]
- Ma L., Ding P., Yang G., He G. (2006). Advances on the plant terpenoid isoprenoid biosynthetic pathway and its key enzymes. Biotechnol. Bull. 2006, S 1 22–30. [Google Scholar]
- Munafo J. P., Jr., Didzbalis J., Schnell R. J., Schieberle P., Steinhaus M. (2014). Characterization of the major aroma-active compounds in mango (Mangifera indica L.) cultivars Haden, White Alfonso, Praya Sowoy, Royal Special, and Malindi by application of a comparative aroma extract dilution analysis. J. Agric. Food Chem. 62 4544–4551. 10.1021/jf5008743 [DOI] [PubMed] [Google Scholar]
- Pandit S. S., Chidley H. G., Kulkarni R. S., Pujari K. H., Giri A. P., Gupta V. S. (2009a). Cultivar relationships in mango based on fruit volatile profiles. Food Chem. 114 363–372. 10.1016/j.foodchem.2008.09.107 [DOI] [Google Scholar]
- Pandit S. S., Kulkarni R. S., Chidley H. G., Giri A. P., Pujari K. H., Koöllner T. G., et al. (2009b). Changes in volatile composition during fruit development and ripening of ‘Alphonso’ mango. J. Sci. Food Agr. 89 2071–2081. 10.1002/jsfa.3692 [DOI] [Google Scholar]
- Pino J. A. (2012). Odour-active compounds in mango (Mangifera indica L. cv. Corazoìn). Int. J. Food Sci. Technol. 47 1944–1950. 10.1111/j.1365-2621.2012.03054.x [DOI] [Google Scholar]
- Pino J. A., Mesa J. (2006). Contribution of volatile compounds to mango (Mangifera indica L.) aroma. Flavour Fragr. J. 21 207–213. 10.1002/ffj.1703 [DOI] [Google Scholar]
- San A. T., Joyce D. C., Hofman P. J., Macnish A. J., Webb R. I., Matovic N. J., et al. (2017). Stable isotope dilution assay (SIDA) and HS-SPME-GCMS quantification of key aroma volatiles for fruit and sap of Australian mango cultivars. Food Chem. 221 613–619. 10.1016/j.foodchem.2016.11.130 [DOI] [PubMed] [Google Scholar]
- Shen S. L., Yin X. R., Zhang B., Xie X. L., Jiang Q., Grierson D., et al. (2016). CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J. Exp. Bot. 67 4105–4115. 10.1093/jxb/erw189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zou X., Huang X., Zhao J., Li Y., Hao L., et al. (2013). Rapid detecting total acid content and classifying different types of vinegar based on near infrared spectroscopy and least-squares support vector machine. Food Chem. 138 192–199. 10.1016/j.foodchem.2012.10.060 [DOI] [PubMed] [Google Scholar]
- Shivashankara K. S., Isobe S., Horita H., Takenaka M., Shiina T. (2006). Volatile aromatic constituents of tree ripened and mature green ‘Irwin’ mango fruits during low temperature storage. J. Jpn. Soc. Hortic. Sci. 75 209–212. 10.2503/jjshs.75.209 [DOI] [Google Scholar]
- Srivastava S., Singh R. K., Pathak G., Goel R., Asif M. H., Sane A. P., et al. (2016). Comparative transcriptome analysis of unripe and mid-ripe fruit of Mangifera indica (var.“Dashehari”) unravels ripening associated genes. Sci. Rep. 6:32557. 10.1038/srep32557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P., Tong Y., Cheng Q., Hu Y., Zhang M., Yang J., et al. (2016). Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci. Rep. 6:23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyk P., Szymańska G., Lipert A., Weremczuk-Je/.zyna I., Kochan E. (2020). Computer-aided saturation mutagenesis of Arabidopsis thaliana ent-copalyl diphosphate synthase. Interdiscip. Sci. 12 32–43. 10.1007/s12539-019-00342-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura H., Boonbumrung S., Yoshizawa T., Varanyanond W. (2001). The volatile constituents in the peel and pulp of a green Thai mango, Khieo Sawoei cultivar (Mangifera indica L.). Food Sci. Technol. Res. 7 72–77. 10.3136/fstr.7.72 [DOI] [Google Scholar]
- Wu H. X., Jia H. M., Ma X. W., Wang S. B., Yao Q. S., Xu W. T., et al. (2014). Transcriptome and proteomic analysis of mango (Mangifera indica L.) fruits. J. Proteomics 105 19–30. 10.1016/j.jprot.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang Q., Li W., Zhang S., Xi W. (2019). Identification of key genes and regulators associated with carotenoid metabolism in apricot (Prunus armeniaca) fruit using weighted gene coexpression network analysis. BMC Genom. 20:876. 10.1186/s12864-019-6261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Dong P., Lao F., Liu J., Liao X., Wu J. (2019a). Characterization of the major aroma-active compounds in Keitt mango juice: comparison among fresh, pasteurization and high hydrostatic pressure processing juices. Food Chem. 289 215–222. 10.1016/j.foodchem.2019.03.064 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yin X., Xiao Y., Zhang Z., Li S., Liu X., et al. (2018). An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol. 178 189–201. 10.1104/pp.18.00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview the transcriptome analysis of mango pulp samples at different developmental and ripening stages. (A) The frequency and numbers of transcripts with different length; (B) The count of CDs with different length predicted by comparing with NR protein library and Swissprot protein library (left) or by estscan (3.0.3) software (right); (C) The hierarchical clustering analysis of Pearson correlation according to the level of gene expression levels in fruits of A, B, C, D, E, F, and G. A, 40 DAF; B, 60 DAF; C, 80 DAF; D, 0 DAP; E, 4 DAP; F, 8 DAP; G, 12 DAP. _1, _2 and _3 represent the Triplicate analyses of each sample.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: (https://www.ncbi.nlm.nih.gov/), and the data accession number is PRJNA697524.