Abstract
The COVID‐19 epidemic is one of the most influential epidemics in history. Understanding the impact of coronaviruses (CoVs) on host cells is very important for disease treatment. The SARS‐CoV‐2 envelope (E) protein is a small structural protein involved in many aspects of the viral life cycle. The E protein promotes the packaging and reproduction of the virus, and deletion of this protein weakens or even abolishes the virulence. This review aims to establish new knowledge by combining recent advances in the study of the SARS‐CoV‐2 E protein and by comparing it with the SARS‐CoV E protein. The E protein amino acid sequence, structure, self‐assembly characteristics, viroporin mechanisms and inhibitors are summarized and analyzed herein. Although the mechanisms of the SARS‐CoV‐2 and SARS‐CoV E proteins are similar in many respects, specific studies on the SARS‐CoV‐2 E protein, for both monomers and oligomers, are still lacking. A comprehensive understanding of this protein should prompt further studies on the design and characterization of effective targeted therapeutic measures.
Keywords: coronavirus, COVID‐19, E protein, inhibitors, viroporin
1. INTRODUCTION
COVID‐19 is a severe and highly contagious respiratory disease caused by a new coronavirus (CoV) called SARS‐CoV‐2. The World Health Organization (WHO) declared a global COVID‐19 pandemic in March 2020. As of February 2021, hundreds of millions of patients have been diagnosed, and millions of patients have died. The mortality rate in different countries ranges from 1 to 10%, but this disease is more contagious than SARS and MERS. 1 , 2 At present, the virus with the spike (S) protein mutation D614G shows stronger infectiousness than wild‐type virus. 3 , 4 , 5 , 6 Researchers speculate that the virus was transmitted to humans through some type of wildlife. The symptoms include fever, general malaise, dry cough, shortness of breath, and respiratory distress, 7 as well as a weakened sense of taste and smell, gastrointestinal discomfort, and certain effects on the cardiovascular and nervous systems. 8 , 9 , 10 , 11
The nucleic acid sequence of SARS‐CoV‐2, which includes a long single‐stranded (30 kb) RNA, generally ranging in size from 26.4 to 31.7 kD, was identified as that of a β‐CoV. 12 The viral genomic RNA strand attaches to the nucleocapsid protein to produce a nucleoprotein complex. 13 The SARS‐CoV‐2 genome shares approximately 82% sequence identity with SARS‐CoV, while the identity of essential enzymes and structural proteins is greater than 90%. 14 The SARS‐CoV‐2 genome consists of 10 open reading frames (ORFs). Approximately two‐thirds of the viral RNA in the ORFs encodes nonstructural proteins (nsp), and one‐third encodes structural proteins such as the S, membrane (M), envelope (E), and nucleocapsid (N) proteins. 15 The results of recent cryoelectron microscopy experiments showed that there were three size distributions of SARS‐CoV‐2 virus particles, and the diameters were mainly 64.8 ± 11.8, 85.9 ± 9.4, and 96.6 ± 11.8 nm. Viral structural proteins, including the S, E, M, and N proteins, are embedded in viral lipids. 16 , 17
The S protein has an immune recognition site and binds to the surface receptor during the process of entering the host cell; it is the most important protein for viral entry into cells. 18 , 19 The S protein binds to the host receptor through the receptor‐binding domain in the S1 subunit, and then, the S2 subunit is fused to the cell membrane. Different CoVs have different host entry mechanisms, indicating that changes in the amino acid residues of the S protein determine whether the virus enters the host cell, and this property is used to design vaccines. 20 , 21 , 22
The main function of the N protein is to package the viral RNA into the helical ribonucleocapsid and interact with the other structural proteins during virion assembly. 23 The viral genome is filled with N proteins, which form a helical nucleocapsid protected by a lipid envelope. 24 , 25 The N protein is involved in the CoV replication cycle and the host cell's response to viral infection. 26 Therefore, it is also a potential target of SARS‐CoV‐2. In addition, the transient expression of N has been shown to substantially increase the production of virus‐like particles (VLPs) in some CoVs, which indicates that the N protein is of great significance in the process of viral envelope conformation. 27 , 28
The M protein defines the shape of the viral envelope and is the most abundant structural protein. 29 It is a 222‐amino‐acid transmembrane protein with N‐ and C‐termini (exposed inside and outside the viral particle, respectively) and three transmembrane domains (TMD1–TMD3). 30 The C‐terminus of the M protein can interact with the N and E proteins, and the TMD can bind with the S protein. The TMD of the M protein is also closely related to the homotypic interaction of the M protein itself. 31 , 32 Interactions with other structural proteins are essential for membrane bending and germination. 30 , 33 In summary, stable binding of the M and N, M and E, and M and S proteins leads to the formation of SARS‐CoV‐2's inner core of VLPs, which promotes the assembly of viruses. 34
The E protein is the smallest of the major structural proteins, but it is also the most poorly understood. It is highly conserved in different viral subtypes. Studies have not fully elucidated the role of the E protein in viral invasion, replication and release. The E protein in the viral particle envelope functions by interacting with other structural proteins. The interaction of the E and M proteins maintains the shape of the viral particle and promotes release. 35 , 36 When E and M are coexpressed in host cells, the S protein relocates to the endoplasmic reticulum (ER)‐Golgi intermediate region (ERGIC) or Golgi region. 37 Mutation of the gene encoding the E protein promotes apoptosis. 38
Interestingly, previous studies have found that during the CoV replication cycle, the E protein is expressed at a high level in each infected cell, but only a small amount of this expressed E protein is inserted into the viral membrane. 39 , 40 Most of the protein is located at intracellular transport sites, namely, the ER, Golgi and ERGIC, which are involved in CoV assembly and budding. 41 Recombinant CoVs lacking the E protein show a significant reduction in viral titer and maturity or produce incompetent offspring, indicating the importance of the E protein in virus production and maturation. 42 The E protein can also bind to PALS1, which leads to enhanced destruction of the epithelial barrier, an enhanced inflammatory process and promotion of tissue remodeling, suggesting that the role of the E protein in viral infection may be important. 43
Recent studies have indicated that several SARS‐CoV‐2 proteins, including E, ORF3a, and ORF8a, can self‐assemble into oligomers and generate ion channels (ICs). 44 , 45 The SARS‐CoV E protein has higher ion permeability than the 3a and 8a channels, and blockade of the E protein channel can significantly reduce viral pathogenicity. 46 The transmembrane voltage (TMV) can promote E protein pentamer IC activity, 47 and the mutations N15A and V25F may disrupt IC activity. 48 , 49 , 50 Therefore, the E protein may play an important role in regulating the ion balance and microenvironment of host cells (Figure 1).
FIGURE 1.
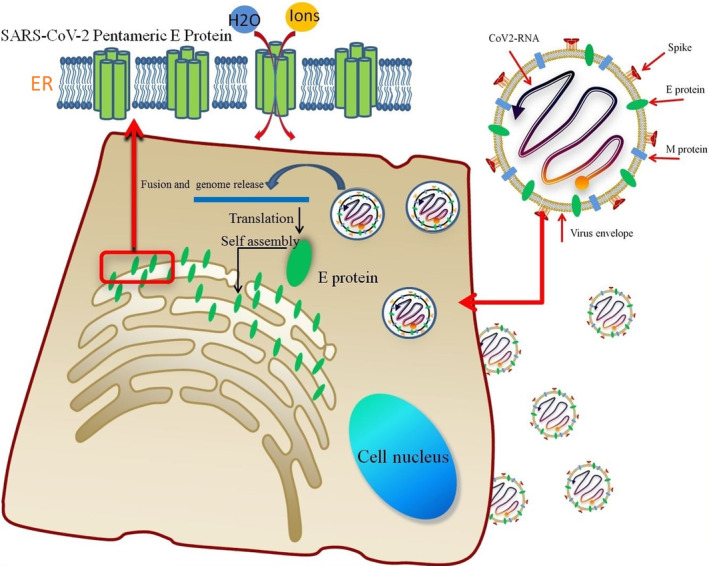
The lipid envelope encloses the virus and facilitates the entry of the SARS‐coronavirus (CoV)‐2 E protein into the host cell. The E protein is translated in the endoplasmic reticulum (ER) and accumulates in the Golgi. Then, the E protein monomer self‐assembles into an oligomer that functions as an ion channel
2. DIFFERENCES IN AMINO ACID SEQUENCE AND STRUCTURE BETWEEN THE SARS‐COV AND SARS‐COV‐2 E PROTEINS
The size of CoV E proteins ranges from 8.4 to 12 kDa. 51 NMR analysis of the SARS‐CoV‐2 E protein showed that there was a single TMD. This result was consistent with the fragment L18‐L39 found in sodium dodecyl sulfate (SDS) micelles of SARS‐CoV. 52 The conformation of the SARS‐CoV‐2 E protein TMD pentamer has been identified, while other domains have not yet been identified. SARS‐CoV and SARS‐CoV‐2 are composed of 76 and 75 amino acids, respectively. Both of them consist of three main domains: one TMD (residues 17–37), an intermediate helical domain, and N‐ and C‐terminal domains. The N‐terminus of the CoV E protein has a negatively charged hydrophilic amino terminal. In addition, the protein has an uncharged (nonpolar) hydrophobic TMD (Figure 2). Compared with the TMD, the C‐terminus shows low hydrophobicity, but it also has 20 hydrophobic residues out of a total of 37 amino acids that are positively charged. The total charge on the E protein is zero. 46 , 53 The E protein of SARS‐CoV‐2 differs in only three amino acid substitutions and one deletion: T55S, V56F, E69R, and G70‐GAP (red box in Figure 2). 54 The amino acid sequences of the TMDs of the two CoVs were identical. Two nonpolar, neutral amino acids, namely, valine and leucine, confer strong hydrophobicity to the E protein TMD. 55 The E protein may not exhibit a uniform membrane topology, and its orientation depends on the level of protein expression or oligomerization. 56
FIGURE 2.
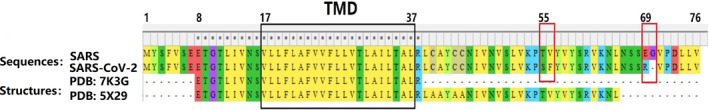
Multiple sequence alignment of SARS‐coronavirus (CoV) and SARS‐CoV‐2. Sequences 1 and 2 represent whole E proteins, and 3 and 4 represent 3D structures with identified amino acid regions (black box: transmembrane domain (TMD), red box: substituted residue or deletion)
The exact function of the N‐terminus of the E protein has not been clearly identified. This region is located inside the Golgi/ER membrane, and Asn15 in the domain forms an H‐bond, which maintains the stability of the N‐terminus. 57 , 58 Studies have shown that homotypic interactions mediated by the N‐terminus of alternating E protein molecules may interact with each other through the luminal loops between the TMD of the proteins to initiate the rearrangement of ER membranes and induce membrane curvature. Similar mechanisms have been found for the interactions of the CoV nonstructural proteins nsp3 and nsp4, which mediate membrane bending. 59 , 60 The E protein also has a predicted signal peptide cleavage site and two glycosylation sites, which are involved in the interaction between the E protein and other membrane proteins. This protein aids in the proper folding and trafficking of cellular and viral proteins. 61 , 62 Other targeting information can be found at the N‐terminus. 63
Four mutations (including one deletion mutation) in the C‐terminus of the SARS‐CoV‐2 E protein, at positions 56, 57, 69, and 70, have an additional amino acid with an alkaline R group compared to the E protein of SARS‐CoV. 64 Many functional properties of the protein can be predicted from the amino acid sequence of the C‐terminus of the E protein. According to reports, the C‐terminus of both SARS‐CoV and SARS‐CoV‐2 contains a conserved DLLV motif (the PDZ domain of the viral E protein), which can bind to PALS1 in the host cell to promote infection. 65 , 66 This region also includes the conserved “FYXY” motif, which could be associated with the high propensity for amyloid formation. 67 , 68 Another highly conserved cysteine motif, “CxxC,” can isomerize disulfide bonds. The oxidized CxxC motif can maintain the structural topology of the TMD and its adjacent cytoplasmic domain, including glycosylation sites, and participate in signal transduction and protein–protein interactions. 69 In addition, the activation of thiol in this motif could trigger other Cys residues to participate in disulfide bond formation between subunits to promote the directional conformation of the membrane. 70
Hassan, Khan, et al. also performed multiple sequence alignments of various SARS‐CoV‐2 mutations before June 18, 2020. They found that in the NCBI database, there are only 16 different E proteins among the 4,917 SARS‐CoV‐2 genomes, and most variations and mutations occur at the C‐terminus. 71 , 72 Mutation at the C‐terminus of the SARS‐CoV‐2 E protein results in the nonsynonymous R group, which may affect the interaction of the E protein with the host protein. In CoV and CoV‐2, there are two cysteine residues at the C‐terminus next to the TMD, which are essential for oligomerization and for the membrane‐directed amyloidogenic propensity. 40 , 73 , 74
3. E PROTEIN VIROPORIN PORE FORMATION AND ION PERMEATION
The formation of homo‐oligomers by the E protein depends on the TMD. Synthetic peptides corresponding to SARS‐CoV and SARS‐CoV‐2 E protein TMDs can form dimers, trimers, and pentamers, proving the importance of the TMD in homotypic interactions 75 , 76 of the mutations F26L, L39M, A36V, and L37H in the TMD were found in different countries. These mutations or deletions and mutations have been shown to attenuate the virus in the body and reduce the viral titer. 77 The substitution of charged residues for some hydrophobic residues in the TMD can significantly change the electrophoretic migration rate. 78 So far, research to identify the TMD residues that are required for the CoV E homotypic interactions has proven inconclusive. Many studies have suggested that the E protein can self‐assemble into oligomers through the interaction between the GxxxG motif and leucine–isoleucine zipper. 79 , 80 , 81 The TMD sequences of both the SARS‐CoV and SARS‐CoV‐2 E proteins contain repeated Leu–Ile sequences, which may be closely related to their homotypic interactions. Wang et al. also indicated that the glycosylation site of the E protein might affect its oligomerization ability, and glycosylated amino acids might prevent oligomerization of the E protein. 82 Asparagine 15 (N15) is mutated to alanine (N15A) and valine 25 (V25) is mutated to phenylalanine F (V25F) in the TMD. The mutations seem to abolish, or at least reduce to some extent, CoV‐E oligomerization. However, the V25F mutant showed apparent monomer formation, in contrast to the N15A mutant. V25F plays a more critical role in oligomerization, while N15A only reduces pentamerization of the E protein. 83
To control the transport of intracellular ions through lipid membranes, the optimal ion conditions of subcellular compartments can differ from those of extracellular media, and different organelles present specific ion components. These asymmetrical distributions of ions between produce electrochemical gradients, which are essential for maintaining normal cellular functions. 84 , 85 The key functions of cells are controlled by the membrane potential, Ca2+ storage in the ER/Golgi, and different pH conditions in the secretory pathway organelles, mainly due to the ion concentration gradient. The coordinated actions of multiple ICs and transporters produce and control the intracellular ionic environment. Recent studies have reported that most viral viroporins can induce inflammasome activity and the production of interleukin‐1β (IL‐1β). 86
It is well known that viruses utilize and modify host cell ion homeostasis in different ways to facilitate viral infection. One of the important ways is via the activity of viroporins. Various viruses encode viroporins, which constitute a broad family of multifunctional proteins, are widely distributed in different virus families, and are mainly found in RNA viruses. 87 , 88 Highly pathogenic human viruses, such as influenza A virus, human immunodeficiency virus 1, hepatitis C virus, and CoVs (SARS‐CoV, MERS virus, and SARS‐CoV‐2), encode at least one viroporin. Viroporins are generally composed of small hydrophobins encoded by viral RNA. These transmembrane proteins can stimulate key aspects of the viral lifecycle through various mechanisms, such as viral entry, trafficking, morphogenesis, maturation, and even virulence. When proteins oligomerize in the ER/Golgi of host cells, they form a channel‐like topology and disrupt a number of physiological properties of the cell by improving the membrane permeability of the host cell, membrane remodeling, and interactions with other normal membrane proteins. 87 , 88 , 89
Deleting the E protein of SARS‐CoV and SARS‐CoV‐2 causes the attenuation of viral virulence, suggesting that the E protein is a potential antiviral and vaccine target. Although it is important for the pathogenesis of viruses, the structure of the SARS‐CoV‐2 E protein was unclear until September 2020, when Mandala et al. analyzed the SARS‐CoV‐2 TMD (PDB: 7K3G), but the structures of the N‐ and C‐termini have remained elusive. 90 Sedimentation equilibrium and gel electrophoresis data indicate that the TMD of the E protein assembles into a homopentamer in detergents such as SDS and perfluorooctanoic acid, but the topology is controversial. NMR analysis of the SARS‐CoV‐2 E protein showed that residues 14–34 form the α‐helical core of the E protein pentamer. 75 , 90 , 91 , 92 Comparison of the spectra of the E protein at different temperatures showed that the N‐terminus is dynamic (flexible) at high temperatures, while the C‐terminus is more rigid but exhibits a temperature‐dependent conformation. 52 , 67 The self‐assembly and oligomer formation mediated by the E protein on the host membrane can also affect the integrity of the lipid bilayer, which is conducive to viral reproduction. 93 The TMD of the CoV E protein can form stable dimers, trimers, and pentamers through self‐assembly via the Leu–Ile zipper motif, 75 and the homopentameric E protein was identified as the CoV viroporin. The viroporin is involved in many functions of CoVs, including facilitating the release of viral particles from host cells. 94
Viroporins regulate the host cell microenvironment (including ion concentration and pH) through the pentameric hydrophilic pores of the cell membrane. The channels allow water and ions to pass through the cell membrane, leading to the disruption of membrane potential, collapse of ionic gradients and release of essential compounds from the cell; this condition facilitates viral dissemination and continuous virion production. 47 , 78 They affect host cell functions, including membrane vesiculation, glycoprotein tracking, and membrane permeability. 95 These characteristics suggest that the viroporin is important for regulation of the viral replication circle.
Previous works have indicated that most viroporins have specific ion selectivity, including the viroporins of human papillomavirus, rotavirus, hepatitis E virus, influenza virus, and CoVs. 96 , 97 , 98 , 99 , 100 Viroporins can be formed by the E, 3a, and 8a proteins in SARS‐CoV, and the IC activity of only the E protein causes viral pathogenicity. 101 The E protein of SARS‐CoV and avian infectious bronchitis virus (IBV) viroporin function as cation‐selective channels in a planar lipid bilayer. 102 , 103 Surya et al. showed that the SARS‐CoV E protein could form pentameric ICs in LMPG micelles. 52 The model included contraction of a >0.2 nm radius formed by the side chains of residues V25 and V28. This can be considered a gate that can expand the central pore with a diameter of less than 0.6 nm. 104 It has been found that the V25F mutation abolish channel permeability, suggesting that the channel's IC activity depends on the homopentameric conformation. 48 , 105 Pervushin et al. suggested that N15 plays an important role in IC activity, and the side chain of the TMD pore forms a pore space with a diameter of approximately 0.4 to 0.5 nm, which can accommodate a single dehydrated Na+ or K+ ion and form a cation selectivity filter for the SARS‐CoV viroporin. 106 The cation permeability of the E protein viroporin is different in high‐concentration KCl and NaCl solutions, the order of selective permeability is Na+ > K+ > Cl−, and is the permeability of Na+ is 10 times higher than that of K+. 104 Cao et al. indicated that the gating amino acids of the SARS‐CoV‐2 E protein viroporin are L10 and F19. The changes in pore radius at L10 and F19 range from 0.2 to 0.4 nm and 0.1 to 0.3 nm, respectively, under different TMVs. The results demonstrated that the SARS‐CoV‐2 E protein viroporin is a voltage‐dependent channel with a certain ion selectivity. 47 The ion‐selective permeability of the viroporin also depends on the composition of the cell membrane. The E protein is less likely to form a viroporin in membranes containing lipid components with a negative intrinsic curvature (such as DOPE), but membranes with a positive intrinsic curvature provide a more favorable environment. 105 , 107 Electrophysiological experiments in the DPhPC/DPhPS membrane further confirmed that the charge contained in lipids may affect IC activity. For example, the channel conductance in completely negatively charged lipids (DPhPS) is much lower than that in neutral lipids (DPhPC). 108
The conductivity and ion selectivity of the E protein viroporin are also influenced by different pH values. Investigation of the SARS‐CoV E protein pentamer inserted into the ERGIC showed that the carboxyl group of the Glu8 side chain in TMD was deprotonated at neutral pH but protonated at acidic pH. The proton balance of Glu8 and the charged lipids in the ERGIC membrane may adjust the ion selectivity at the entrance of the channel. 90 An intriguing study recently reported that the influenza B virus M2 proton channel is pH gated and is activated by acidic pH to mediate virus uncoating. 109 , 110 This suggests that viroporins are sensitive to changes in cellular pH and can have therapeutic value. Furthermore, the E protein viroporins of both SARS‐CoV and SARS‐CoV‐2 are capable of Ca2+ permeation. This may be due to the large Ca2+ concentration gradient between the two sides of the membrane and because a negative charge interaction occurs between Ca2+ and the protein‐lipid pore, which affects the IC activity of the channel. 111 A computational model revealed the mechanisms of Ca2+ permeation of CoV‐2 E protein pores. At a high TMV (≥0.4 V), the pore radius greatly expands, making it sufficiently large to accommodate Ca2+. However, a higher TMV may cause loss of the gating ability of the viroporin. 47 This is also consistent with other viral viroporins. 98 , 112 , 113 , 114 The leakage of Ca2+, K+, and H+ via the viroporin induces the activation of NLRP3 inflammasomes, leading to the overproduction of IL‐1β, which affects the occurrence and development of respiratory inflammation. 86 Ca2+ homeostasis plays a key role in the pathogenicity of SARS‐CoV and SARS‐CoV‐2. 115 , 116 , 117
The IC activity of the SARS‐CoV‐2 E protein causes injury to numerous vital organs. Xia et al. injected purified SARS‐CoV‐2 E protein into mice intravenously and produced lung and spleen ARDS‐like pathological damage. 118 This indicates that the cation channel formed by the overexpressed E protein can eventually rupture the host cell membrane. Polina et al. suggested that the K+ IC activity of SARS‐CoV‐2 may abnormally regulate the transmission of AP and Ca2+ in cardiomyocytes, promoting a decrease in cardiac contractility and an increase in sensitivity to arrhythmia. 119 Therefore, targeted inhibition of the SARS‐CoV‐2 E viroporin can reduce the risk of sudden cardiac death and heart damage in COVID‐19 patients with cardiovascular disease.
Although the residues affect IC activity, they also affect different aspects of the virus life cycle. The relationship between CoV IC activity and pathogenesis remains to be further explored.
4. SARS‐COV‐2 E PROTEIN VIROPORIN INHIBITORS
Specific inhibition of the CoV E protein's IC activity causes virus attenuation. 46 Mandala et al. identified the SARS‐CoV‐2 E protein pentamer structure by using NMR spectroscopy. 90 The SARS‐CoV‐2 E protein pentamer is a CoV viroporin, and some inhibitors of viroporins are considered to be equally effective in inhibiting the SARS‐CoV‐2 E protein viroporin. 120
4.1. Hexamethylene amiloride and amantadine
One potential therapeutic approach is to target the pentameric E protein channels through hexamethylene amiloride (HMA) and amantadine (AMT) and their combinations. Importantly, HMA and AMT represent a broad range of viroporin inhibitors, and their chemical formula and 3D structure are shown in Figure 3a,b. In addition to their effect on CoVs, they have good inhibitory effects on HIV, IBV, and MHV. 121 , 122 , 123 AMT and its derivatives can inhibit the IC activity of influenza virus M2 ICs and have been used in the clinical treatment of influenza A infection. 124 , 125
FIGURE 3.
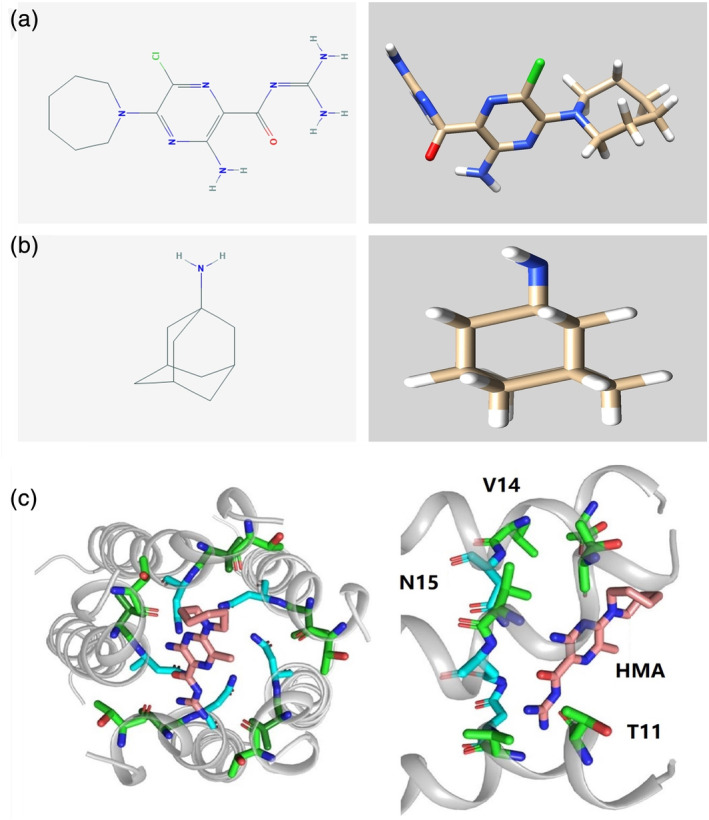
Molecular formula and 3D chemical structure of hexamethylene amiloride (HMA) and amantadine (AMT) (a and b). Docking region for HMA in the coronavirus (CoV)‐2 transmembrane domain (TMD) (c)
Specifically, HMA is exchanged between the helices of the pentameric SARS‐CoV‐2 E protein and disturbs 5 residues at the N‐terminus (namely, Thr9, Gly10, Thr11, Ile13, and Ser16, shown in Figure 3c), blocking IC conductance. It has high affinity for both acidic Glu8 and polar Asn15 to seal the N‐terminal entrance of the protein. 90 , 92 AMT can block the conductance of the SARS‐CoV wild‐type (WT) and F23A mutant proteins and exhibits significant binding only with F23A, showing nonspecific agglutination with N15A, V25F, and A32F. The conductance of E protein double mutants (N15A‐V25F and V25A‐A32F) is not affected by AMT. 48 , 77 Moreover, the combination of HMA and AMT inhibited SARS‐CoV‐2 E protein channel conductance, and HMA had a stronger affinity than AMT. Some clinical reports have indicated that HMA and AMT have some preventive and therapeutic effects on SARS‐CoV‐2. 126 , 127 , 128
4.2. Memantine and gliclazide
Memantine is a derivative of AMT (Figure 4a), which can inhibit the virus by blocking the M2 proton channel of influenza virus. It is the earliest antiviral drug used to inhibit influenza virus. 125 It is also inhibits the NMDA receptor, similar to the inhibitory effect of AMT on the virus. 129 Memantine has anti‐inflammatory effects. It reduces the release of cytokines (TNF‐α, IL‐6, and IFN) through several mechanisms, inhibits the expression of NMDA receptor 1, and has a regulatory effect on Ca2+. 130 It prevents and reverses the expression of some inflammatory factors, restores them to normal levels, and alleviates inflammation. 131
FIGURE 4.
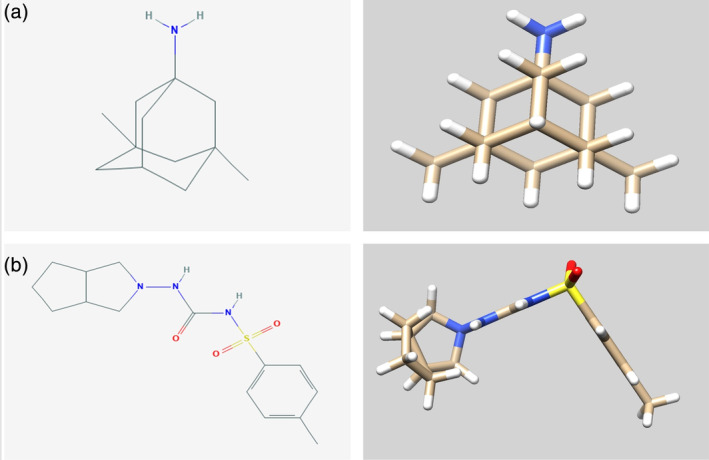
Molecular formulas and 3D chemical structures of memantine and gliclazide (a and b)
Gliclazide, the molecular formula of which is C15H21N3O3S (Figure 4b), is a sulfonylurea oral antidiabetic drug that is widely used in the treatment of Type 2 diabetes. 132 There is evidence that gliclazide and some diabetes drugs have a potential therapeutic effect on neocoronary pneumonia, but the exact mechanism remains unknown. 133 , 134
Tomar et al. screened 372 compounds in the MedChemExpress library and found that memantine and gliclazide are potential E protein channel inhibitors. Tipton et al. suggested that the inexpensive drug memantine could be useful in COVID‐19 therapy. 135 The negative/positive comparison experiment showed that the blockade of channel conductance by the compound had a significant effect on the growth of bacteria expressing the SARS‐CoV‐2 E protein. However, while the E protein is essential for bacteria, gliclazide and memantine are harmful to bacterial growth. This result suggested that the two compounds are potential channel inhibitors.
4.3. ZINC23221929 and ZINC06220062
Tomar et al. used virtual screening methods to analyze 7,000 different compounds in the ZINC database 136 and obtained 10 possible inhibitors of the E protein viroporin. Then, they identified two compounds (ZINC ids ZINC23221929 and ZINC06220062) by using saturation transfer difference (STD) NMR spectroscopy. 137 The chemical names of the two potential inhibitors are [2‐[(3S)‐2,3‐dihydro‐1,4‐benzodioxin‐3‐yl]methylamino]‐2‐oxo‐ethyl](2S)‐2‐(1,3‐dioxoisoindolin‐2‐yl)‐4‐methylsulfanyl‐butanoate (ZINC23221929) and 2‐(2‐amino‐2‐oxo‐ethoxy)‐N‐benzyl‐benzamide (ZINC06220062) (Table 1).
TABLE 1.
Potential inhibitors of the SARS‐CoV‐2 E protein
| Compound name | ZINC id | PubChem id | Chemical formula | Structure | Rings | Binding site |
|---|---|---|---|---|---|---|
| Amiloride (HMA) | ZINC4340269 | 16231 | C6H8ClN7O |
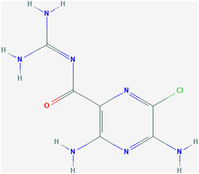
|
1 | Thr9, Glu8, Gly10, Thr11, Ile13, Asn15, Ser16 |
| Amantadine (AMT) | ZINC968256 | 2130 | C10H17N |
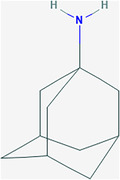
|
4 | Thr9, Gly10, Thr11, Ile13 |
| Memantine | ZINC3812933 | 4054 | C12H21N |
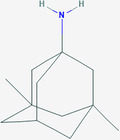
|
4 | / |
| Gliclazide | ZINC523925 | 3475 | C15H21N3O3S |
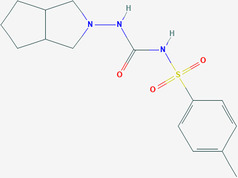
|
3 | / |
| Tretinoin | ZINC22066351 | 444795 | C20H28O2 |
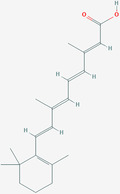
|
1 | Leu18, Leu21, Ala22, Val25, Phe26, Ala22, Val25, Phe26, Phe4 |
| Rutin | ZINC4096846 | 5280805 | C27H30O16 |
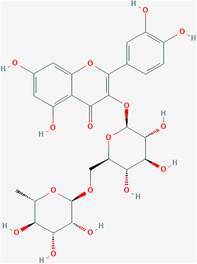
|
5 |
Leu31, Thr35, Ser55, Arg69, Pro71 |
| Doxycycline | ZINC16052277 | 54671203 | C22H24N2O8 |
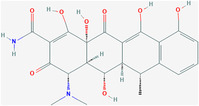
|
4 | Leu31, Thr35, Val52, Ser55 |
| Nimbolin A | / | 6443004 | C39H46O8 |
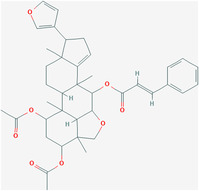
|
6 | Leu18, Leu19, Leu21, Ala22, Val25, Phe26 |
| / | ZINC23221929 | 30927842 | C24H24N2O7S |
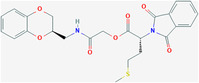
|
4 | Ile46, Leu51, P54 S60, C44 Val47, Leu31, Leu34, Cys40 |
| / | ZINC06220062 | 7917802 | C16H16N2O3 |
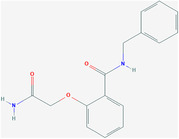
|
2 | Leu28, Leu31, Pro51, Tyr57 Val42, Cys40, Cys44, Thr35 |
The molecular formula of ZINC23221929 is C24H24N2O7S. There are two loop systems of this inhibitor that can bind with the I46, L51, and P54 residues of the E protein, found to be engaged in hydrophobic interactions and with S60, C44, and V47 via H‐bonding interactions. The 4‐methylthiobutyrate fragment and L31, L34, and C40 maintain the hydrophobic interaction of the alkyl group. The molecular formula of ZINC06220062 is C16H16N2O3, in which the 2‐oxo functional group in the 2‐amino‐2‐oxo‐ethoxy fragment is engaged in a crucial H‐bonding interaction with the S60 residue of the C‐terminal domain of the E protein. The phenyl moiety anchoring the ethoxy amine fragment and the N‐phenyl moiety are hydrophobic interactions with the V47 and C40 residues and L28, L31, and Y57 residues, respectively. STD NMR showed that ZINC06220062 has a stronger binding affinity than ZINC23221929, and this difference may be related to the structural orientation of aromatic groups. The stable binding of these two inhibitors to the C‐terminus of the E protein may inhibit its function, which also shows that the C‐terminus can be potential targeted by inhibitors, thereby preventing viral assembly and replication.
4.4. In silico prediction of SARS‐CoV‐2 viroporin inhibitors
The SARS‐CoV‐2 viroporin inhibitors were predicted by computational methods. Many studies predicted SARS‐CoV‐2 E protein viroporin inhibitors based on high‐throughput computer screening and computational modeling methods. 138 Although the inhibitors have not been experimentally verified, they can save a large amount of money and investment required by laboratory methods to identify new drugs. 139
4.4.1. Tretinoin
Tretinoin, also known as retinoic acid, is a metabolic intermediate product of vitamin A in the body that mainly affects bone growth and promotes epithelial cell proliferation, differentiation, and keratolysis. It is generally used to treat skin diseases. 140 Tretinoin is also considered an effective HBV inhibitor. In the RAR agonist family, the biological activity of retinoic acid is demonstrated through different RAR receptors. All‐trans retinoic acid can restore the HBV core‐specific response of T cells by targeting myeloid‐derived suppressor cells in isolated peripheral blood mononuclear cells in patients and mice. 141 Based on experimental evidence, tretinoin is an inhibitor of influenza virus A. 142
The composition of SARS‐CoV‐2 E protein TMD residues (from 18 to 26) is characterized by hydrophobic amino acids. Dey et al. applied a combination of all‐atom molecular dynamics simulations, including H‐bond and binding energy analysis (MM‐PBSA), to evaluate binding affinity, and tretinoin showed the strongest hydrophobic interactions in the viroporin pore. Tretinoin exhibits the ability to form extensive H‐bond interactions and shows high binding energy (−412.8 kJ/mol) with the hydrophobic side chains of the E protein pores; it has been selected as the best candidate viroporin inhibitor. 143
4.4.2. Rutin and doxycycline
Bhowmik et al. used molecular docking to screen rutin and the antibiotic doxycycline as the strongest inhibitors of the SARS‐CoV‐2 E protein viroporin among 548 antiviral compounds (Figures 5 and 6). 144 Rutin is a natural flavonoid glycoside. Studies have shown that it can inhibit many viruses. 145 , 146 , 147 Doxycycline is a tetracycline antibiotic that can be used to treat Mycoplasma and Chlamydia infections and is also an antiviral. 148 , 149 Rutin binds to the five active‐site residues of the E protein TMD via H‐bonds, while doxycycline interacts with the other four residues, which can effectively bind to the E protein to inhibit the formation of viroporin. 150
FIGURE 5.
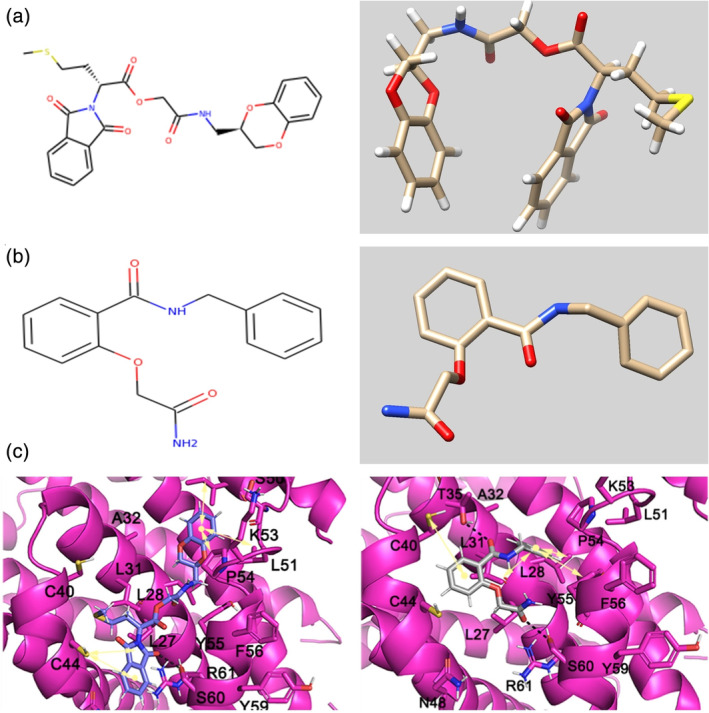
Molecular formulas and 3D chemical structures of ZINC23221929 (a) and ZINC06220062 (b). Cartoon representation of the binding mode of two ligands with the E protein (c)
FIGURE 6.
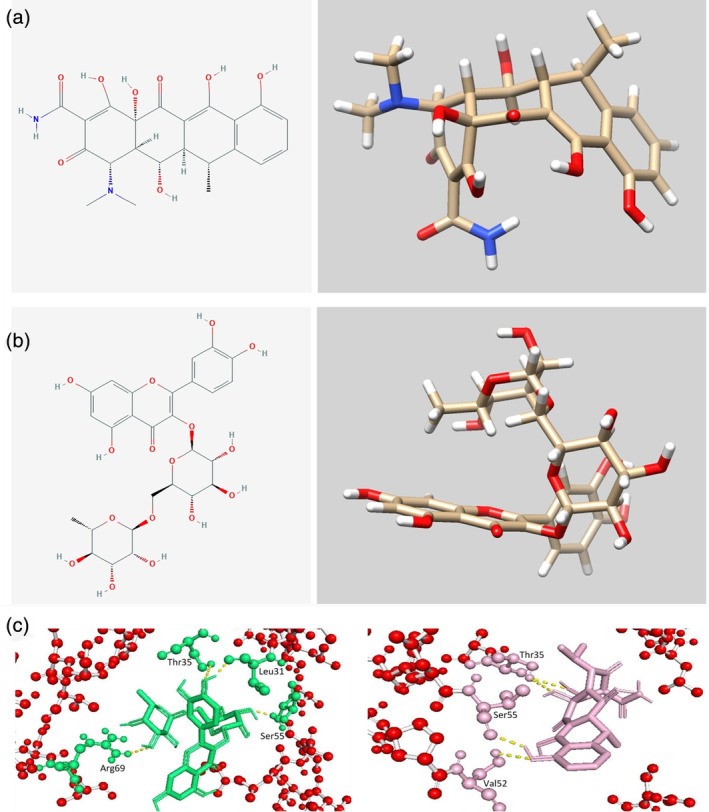
Molecular formulas and 3D chemical structures of rutin and doxycycline (a and b). Docking region for rutin and doxycycline in the coronavirus (CoV)‐2 E protein (c)
4.4.3. Nimbolin A and others
The computational model suggested that nimbolin A, nimocin, 7‐deacetyl‐7‐benzoylgedunin, 24‐methylenecycloartanol, and cycloeucalenone are potential E protein viroporin inhibitors. Among them, the binding energy of nimbolin A with viroporin is the highest. 151
An in silico method can predict SARS‐CoV‐2 E protein pentamer inhibitors quickly and at low cost. However, candidate inhibitors based on in silico approaches must be subjected to further experimental investigations.
5. CONCLUSION
Studies on the SARS‐CoV and SARS‐CoV‐2 E proteins indicate that the E protein could be involved in multiple important aspects, from the assembly and induction of membrane curvature to division or budding and release to apoptosis, inflammation and even autophagy. 152 Because the amino acid sequence and protein structure of the SARS‐CoV and SARS‐CoV‐2 E proteins are highly homologous, their multimer formation mechanisms could be similar, which is reflected in their characteristics. The E protein is involved in many aspects of the viral replication cycle through the formation of oligomers and viroporins. The ion selectivity of the viroporin in the ER/Golgi membrane strongly affects the maintenance of the ion balance of the host cell microenvironment, pH and TMV. There are limited studies on the mechanism of the E protein viroporin in virus‐infected cells according to the current understanding. Although in vitro experiments have shown that some inhibitors can effectively block the E protein viroporin of SARS‐CoV‐2 and weaken or abolish the virulence of the virus, their exact therapeutic effect remains to be further explored.
AUTHOR CONTRIBUTIONS
Yipeng Cao: Conceptualization; funding acquisition; writing‐original draft. Rui Yang: Data curation; investigation. Imshik Lee: Funding acquisition; investigation; writing‐review and editing. Wenwen Zhang: Data curation; investigation. Jiana Sun: Investigation; writing‐original draft. Wei Wang: Conceptualization; project administration. Xiangfei Meng: Investigation; project administration.
ACKNOWLEDGMENT
This study was supported by grants from the National Natural Science Foundation of China (NSFC‐31900894, NSFC‐81872472 and NSFC‐31870843).
Cao Y, Yang R, Lee I, et al. Characterization of the SARS‐CoV‐2 E Protein: Sequence, Structure, Viroporin, and Inhibitors. Protein Science. 2021;30:1114–1130. 10.1002/pro.4075
Funding information National Natural Science Foundation of China, Grant/Award Numbers: NSFC‐31900894, NSFC‐81872472: NSFC‐31870843
Contributor Information
Yipeng Cao, Email: vgsplayer1@163.com.
Wei Wang, Email: weiwang_2@126.com.
REFERENCES
- 1. Baud D, Qi X, Nielsen‐Saines K, Musso D, Pomar L, Favre G. Real estimates of mortality following COVID‐19 infection. Lancet Infect Dis. 2020;20:773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mahase E. Coronavirus: Covid‐19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ. 2020;368:m641. [DOI] [PubMed] [Google Scholar]
- 3. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: Evidence that D614G increases infectivity of the COVID‐19 virus. Cell. 2020;182:812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS‐CoV‐2 spike protein variant. Cell. 2020;183:739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grubaugh ND, Hanage WP, Rasmussen AL. Making sense of mutation: What D614G means for the COVID‐19 pandemic remains unclear. Cell. 2020;182:794–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ozono S, Zhang Y, Ode H, et al. SARS‐CoV‐2 D614G spike mutation increases entry efficiency with enhanced ACE2‐binding affinity. Nat Commun. 2021;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marini JJ, Gattinoni L. Management of COVID‐19 respiratory distress. J Am Med Assoc. 2020;323:2329–2330. [DOI] [PubMed] [Google Scholar]
- 8. Wang H‐Y, Li X‐L, Yan Z‐R, Sun X‐P, Han J, Zhang B‐W. Potential neurological symptoms of COVID‐19. Ther Adv Neurol Disord. 2020;13:1756286420917830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Russell B, Moss C, Rigg A, Hopkins C, Papa S, van Hemelrijck M. Anosmia and ageusia are emerging as symptoms in patients with COVID‐19: What does the current evidence say? Ecancermedicalscience. 2020;14:ed98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han C, Duan C, Zhang S, et al. Digestive symptoms in COVID‐19 patients with mild disease severity: Clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115:916–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng Y‐Y, Ma Y‐T, Zhang J‐Y, Xie X. COVID‐19 and the cardiovascular system. Nat Rev Cardiol. 2020;17:259–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woo PCY, Lau SKP, Lam CSF, et al. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J Virol. 2009;83:908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou G, Chen S, Chen Z. Advances in COVID‐19: The virus, the pathogenesis, and evidence‐based control and therapeutic strategies. Front Med. 2020;14:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naqvi AAT, Fatima K, Mohammad T, et al. Insights into SARS‐CoV‐2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 1866;2020:165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim D, Lee J‐Y, Yang J‐S, Kim JW, Kim VN, Chang H. The architecture of SARS‐CoV‐2 transcriptome. Cell. 2020;181:914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yao H, Song Y, Chen Y, et al. Molecular architecture of the SARS‐CoV‐2 virus. Cell. 2020;183:730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Müller MA, Raj VS, Muth D, et al. Human coronavirus EMC does not require the SARS‐coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio. 2012;3:e00515–e00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walls AC, Park Y‐J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. [DOI] [PubMed] [Google Scholar]
- 20. Bangaru S, Ozorowski G, Turner HL, et al. Structural analysis of full‐length SARS‐CoV‐2 spike protein from an advanced vaccine candidate. Science. 2020;370:1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Waqas M, Haider A, Sufyan M, Siraj S, Sehgal SA. Determine the potential epitope based peptide vaccine against novel SARS‐CoV‐2 targeting structural proteins using immunoinformatics approaches. Front Mol Biosci. 2020;7:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vangelista L, Secchi M. Prepare for the future: Dissecting the spike to seek broadly neutralizing antibodies and universal vaccine for pandemic coronaviruses. Front Mol Biosci. 2020;7:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arya R, Kumari S, Pandey B, et al. Structural insights into SARS‐CoV‐2 proteins. J Mol Biol. 2021;433:166725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlson CR, Asfaha JB, Ghent CM, et al. Phosphoregulation of phase separation by the SARS‐CoV‐2 N protein suggests a biophysical basis for its dual functions. Mol Cell. 2020;80:1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye Q, West AMV, Silletti S, Corbett KD. Architecture and self‐assembly of the SARS‐CoV‐2 nucleocapsid protein. Protein Sci. 2020;29:1890–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Savastano A, de Opakua AI, Rankovic M, Zweckstetter M. Nucleocapsid protein of SARS‐CoV‐2 phase separates into RNA‐rich polymerase‐containing condensates. Nat Commun. 2020;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu R, Shi M, Li J, Song P, Li N. Construction of SARS‐CoV‐2 virus‐like particles by mammalian expression system. Front Bioeng Biotechnol. 2020;8:862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tseng Y‐T, Wang S‐M, Huang K‐J, Wang C‐T. SARS‐CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus‐like particle production. J Biomed Sci. 2014;21:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neuman BW, Kiss G, Kunding AH, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tseng Y‐T, Chang C‐H, Wang S‐M, Huang K‐J, Wang C‐T. Identifying SARS‐CoV membrane protein amino acid residues linked to virus‐like particle assembly. PLoS One. 2013;8:e64013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tseng Y‐T, Wang S‐M, Huang K‐J, Lee AI‐R, Chiang C‐C, Wang C‐T. Self‐assembly of severe acute respiratory syndrome coronavirus membrane protein. J Biol Chem. 2010;285:12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ujike M, Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang JQ, Fang S, Yuan Q, et al. N‐linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus‐induced ER stress response, apoptosis and pathogenesis. Virology. 2019;531:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. el Omari K, Li S, Kotecha A, et al. The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins. Nat Commun. 2019;10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alsaadi EAJ, Neuman BW, Jones IM. Identification of a membrane binding peptide in the envelope protein of MHV coronavirus. Viruses. 2020;12:1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boson B, Legros V, Zhou B, et al. The SARS‐CoV‐2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus‐like particles. J Biol Chem. 2021;296:100111. 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parthasarathy K, Ng L, Lin X, et al. Structural flexibility of the pentameric SARS coronavirus envelope protein ion channel. Biophys J. 2008;95:L39–L41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Venkatagopalan P, Daskalova SM, Lopez LA, Dolezal KA, Hogue BG. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mukherjee S, Bhattacharyya D, Bhunia A. Host‐membrane interacting interface of the SARS coronavirus envelope protein: Immense functional potential of C‐terminal domain. Biophys Chem. 2020;266:106452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nieto‐Torres JL, DeDiego ML, Álvarez E, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Curtis KM, Yount B, Baric RS. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J Virol. 2002;76:1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Asghari A, Naseri M, Safari H, Saboory E, Parsamanesh N. The novel insight of SARS‐CoV‐2 molecular biology and pathogenesis and therapeutic options. DNA Cell Biol. 2020;39:1741–1753. [DOI] [PubMed] [Google Scholar]
- 44. Kern DM, Sorum B, Hoel CM, et al. Cryo‐EM structure of the SARS‐CoV‐2 3a ion channel in lipid nanodiscs. BioRxiv. 2020;6(17):15644. 10.1101/2020.06.17.156554. [DOI] [Google Scholar]
- 45. Yue Y, Nabar NR, Shi C‐S, et al. SARS‐coronavirus open reading frame‐3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nieto‐Torres JL, DeDiego ML, Verdiá‐Báguena C, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5):e1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao Y, Yang R, Wang W, et al. Computational study of the ion and water permeation and transport mechanisms of the SARS‐CoV‐2 pentameric E protein channel. Front Mol Biosci. 2020;7:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Torres J, Maheswari U, Parthasarathy K, Ng L, Liu DX, Gong X. Conductance and amantadine binding of a pore formed by a lysine‐flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16:2065–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang K, Xie S, Sun B. Viral proteins function as ion channels. Biochim Biophys Acta Biomembr. 1808;2011:510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. To J, Surya W, Torres J. Targeting the channel activity of viroporins. Adv Prot Chem Struct Biol. 2016;104:307–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu DX, Yuan Q, Liao Y. Coronavirus envelope protein: A small membrane protein with multiple functions. Cell Mol Life Sci. 2007;64:2043–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Surya W, Li Y, Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim Biophys Acta Biomembr. 1860;2018:1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruch TR, Machamer CE. The coronavirus E protein: Assembly and beyond. Viruses. 2012;4:363–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tilocca B, Soggiu A, Sanguinetti M, et al. Immunoinformatic analysis of the SARS‐CoV‐2 envelope protein as a strategy to assess cross‐protection against COVID‐19. Microbes Infect. 2020;22:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu Q, Zhang Y, Lü H, et al. The E protein is a multifunctional membrane protein of SARS‐CoV. Genom Proteom Bioinformat. 2003;1:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kuo L, Hurst KR, Masters PS. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J Virol. 2007;81:2249–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sarkar M, Saha S. Structural insight into the role of novel SARS‐CoV‐2 E protein: A potential target for vaccine development and other therapeutic strategies. PLoS One. 2020;15:e0237300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Duart G, García‐Murria MJ, Grau B, Acosta‐Cáceres JM, Martínez‐Gil L, Mingarro I. SARS‐CoV‐2 envelope protein topology in eukaryotic membranes. Open Biol. 2020;10:200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double‐membrane vesicles. MBio. 2013;4:e00524–e00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hagemeijer MC, Monastyrska I, Griffith J, et al. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vigerust DJ, Shepherd VL. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fung TS, Liu DX. Post‐translational modifications of coronavirus proteins: Roles and function. Future Virol. 2018;13:405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Maeda J, Repass JF, Maeda A, Makino S. Membrane topology of coronavirus E protein. Virology. 2001;281:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li S, Yuan L, Dai G, Chen RA, Liu DX, Fung TS. Regulation of the ER stress response by the ion channel activity of the infectious bronchitis coronavirus envelope protein modulates virion release, apoptosis, viral fitness, and pathogenesis. Front Microbiol. 2020;10:3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Teoh K‐T, Siu Y‐L, Chan W‐L, et al. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21:3838–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Toto A, Ma S, Malagrinò F, et al. Comparing the binding properties of peptides mimicking the envelope protein of SARS‐CoV and SARS‐CoV‐2 to the PDZ domain of the tight junction‐associated PALS1 protein. Protein Sci. 2020;29:2038–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ghosh A, Bhattacharyya D, Bhunia A. Structural insights of a self‐assembling 9‐residue peptide from the C‐terminal tail of the SARS corona virus E‐protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study. Biochim Biophys Acta Biomembr. 1860;2018:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ghosh A, Pithadia AS, Bhat J, et al. Self‐assembly of a nine‐residue amyloid‐forming peptide fragment of SARS corona virus E‐protein: Mechanism of self aggregation and amyloid‐inhibition of hIAPP. Biochemistry. 2015;54:2249–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chivers PT, Laboissiere MC, Raines RT. The CXXC motif: Imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 70. Brazin KN, Mallis RJ, Li C, et al. Constitutively oxidized CXXC motifs within the CD3 heterodimeric ectodomains of the T cell receptor complex enforce the conformation of juxtaposed segments. J Biol Chem. 2014;289:18880–18892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hassan SS, Choudhury PP, Roy B. Molecular phylogeny and missense mutations at envelope proteins across coronaviruses. Genomics. 2020;112:4993–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Khan MI, Khan ZA, Baig MH, et al. Comparative genome analysis of novel coronavirus (SARS‐CoV‐2) from different geographical locations and the effect of mutations on major target proteins: An in silico insight. PLoS One. 2020;15:e0238344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kupke T, Klare JP, Brügger B. Heme binding of transmembrane signaling proteins undergoing regulated intramembrane proteolysis. Commun Biol. 2020;3:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lopez LA, Riffle AJ, Pike SL, Gardner D, Hogue BG. Importance of conserved cysteine residues in the coronavirus envelope protein. J Virol. 2008;82:3000–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Torres J, Wang J, Parthasarathy K, Liu DX. The transmembrane oligomers of coronavirus protein E. Biophys J. 2005;88:1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bordag N, Keller S. α‐Helical transmembrane peptides: a “divide and conquer” approach to membrane proteins. Chem Phys Lipids. 2010;163:1–26. [DOI] [PubMed] [Google Scholar]
- 77. Alam I, Kamau AA, Kulmanov M, et al. Functional pangenome analysis shows key features of e protein are preserved in sars and sars‐cov‐2. Front Cell Infect Microbiol. 2020;10:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liao Y, Yuan Q, Torres J, Tam JP, Liu DX. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349:264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix‐helix association. J Mol Biol. 2000;296:911–919. [DOI] [PubMed] [Google Scholar]
- 80. Teese MG, Langosch D. Role of GxxxG motifs in transmembrane domain interactions. Biochemistry. 2015;54:5125–5135. [DOI] [PubMed] [Google Scholar]
- 81. Cao Y, Wu X, Yang R, Wang X, Sun H, Lee I. Self‐assembling study of sarcolipin and its mutants in multiple molecular dynamic simulations. Proteins. 2017;85:1065–1077. [DOI] [PubMed] [Google Scholar]
- 82. Wang B, Wang Y, Frabutt DA, et al. Mechanistic understanding of N‐glycosylation in Ebola virus glycoprotein maturation and function. J Biol Chem. 2017;292:5860–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Torres J, Surya W, Li Y, Liu DX. Protein‐protein interactions of viroporins in coronaviruses and paramyxoviruses: New targets for antivirals? Viruses. 2015;7:2858–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Green WN, Andersen OS. Surface charges and ion channel function. Annu Rev Physiol. 1991;53:341–359. [DOI] [PubMed] [Google Scholar]
- 85. Tillman TS, Cascio M. Effects of membrane lipids on ion channel structure and function. Cell Biochem Biophys. 2003;38:161–190. [DOI] [PubMed] [Google Scholar]
- 86. Farag NS, Breitinger U, Breitinger HG, el Azizi MA. Viroporins and inflammasomes: A key to understand virus‐induced inflammation. Int J Biochem Cell Biol. 2020;122:105738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nieva JL, Madan V, Carrasco L. Viroporins: Structure and biological functions. Nat Rev Microbiol. 2012;10:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gonzalez ME, Carrasco L. Viroporins. FEBS Lett. 2003;552:28–34. [DOI] [PubMed] [Google Scholar]
- 89. Suzuki T, Orba Y, Makino Y, et al. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc Natl Acad Sci U S A. 2013;110:18668–18673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and drug binding of the SARS‐CoV‐2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol. 2020;27:1202–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Parthasarathy K, Lu H, Surya W, Vararattanavech A, Pervushin K, Torres J. Expression and purification of coronavirus envelope proteins using a modified β‐barrel construct. Protein Expr Purif. 2012;85:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li Y, Surya W, Claudine S, Torres J. Structure of a conserved Golgi complex‐targeting signal in coronavirus envelope proteins. J Biol Chem. 2014;289:12535–12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Berterame NM, Porro D, Ami D, Branduardi P. Protein aggregation and membrane lipid modifications under lactic acid stress in wild type and OPI1 deleted Saccharomyces cerevisiae strains. Microb Cell Fact. 2016;15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nieto‐Torres JL, Verdiá‐Báguena C, Castaño‐Rodriguez C, Aguilella VM, Enjuanes L. Relevance of viroporin ion channel activity on viral replication and pathogenesis. Viruses. 2015;7:3552–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Carrasco L. Modification of membrane permeability induced by animal viruses early in infection. Virology. 1981;113:623–629. [DOI] [PubMed] [Google Scholar]
- 96. Wetherill LF, Wasson CW, Swinscoe G, et al. Alkyl‐imino sugars inhibit the pro‐oncogenic ion channel function of human papillomavirus (HPV) E5. Antiviral Res. 2018;158:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wetherill LF, Holmes KK, Verow M, et al. High‐risk human papillomavirus E5 oncoprotein displays channel‐forming activity sensitive to small‐molecule inhibitors. J Virol. 2012;86:5341–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pham T, Perry JL, Dosey TL, Delcour AH, Hyser JM. The rotavirus NSP4 viroporin domain is a calcium‐conducting ion channel. Sci Rep. 2017;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ding Q, Heller B, Capuccino JMV, et al. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc Natl Acad Sci U S A. 2017;114:1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. To J , Torres J. Viroporins in the influenza virus. Cell. 2019;8:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Castaño‐Rodriguez C, Honrubia JM, Gutiérrez‐Álvarez J, et al. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. MBio. 2018;9:e02325–e02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McClenaghan C, Hanson A, Lee S‐J, Nichols CG. Coronavirus proteins as ion channels: Current and potential research. Front Immunol. 2020;11:2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ruch TR, Machamer CE. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J Virol. 2011;85:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wilson L, Mckinlay C, Gage P, Ewart G. SARS coronavirus E protein forms cation‐selective ion channels. Virology. 2004;330:322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Verdiá‐Báguena C, Nieto‐Torres JL, Alcaraz A, et al. Coronavirus E protein forms ion channels with functionally and structurally‐involved membrane lipids. Virology. 2012;432:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pervushin K, Tan E, Parthasarathy K, et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sobko AA, Kotova EA, Antonenko YN, Zakharov SD, Cramer WA. Effect of lipids with different spontaneous curvature on the channel activity of colicin E1: Evidence in favor of a toroidal pore. FEBS Lett. 2004;576:205–210. [DOI] [PubMed] [Google Scholar]
- 108. Aguilella VM, Verdiá‐Báguena C, Alcaraz A. Lipid charge regulation of non‐specific biological ion channels. Phys Chem Chem Phys. 2014;16:3881–3893. [DOI] [PubMed] [Google Scholar]
- 109. Wang J, Pielak RM, McClintock MA, Chou JJ. Solution structure and functional analysis of the influenza B proton channel. Nat Struct Mol Biol. 2009;16:1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Mandala VS, Loftis AR, Shcherbakov AA, Pentelute BL, Hong M. Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism. Nat Struct Mol Biol. 2020;27:160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nieto‐Torres JL, Verdiá‐Báguena C, Jimenez‐Guardeño JM, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Dey D, Siddiqui SI, Mamidi P, et al. The effect of amantadine on an ion channel protein from Chikungunya virus. PLoS Negl Trop Dis. 2019;13:e0007548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Largo E, Verdiá‐Báguena C, Aguilella VM, Nieva JL, Alcaraz A. Ion channel activity of the CSFV p7 viroporin in surrogates of the ER lipid bilayer. Biochim Biophys Acta Biomembr. 1858;2016:30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Surya W, Li Y, Torres J. Pentameric viral ion channels: From structure to function. J Receptor Ligand Channel Res. 2014;8:9–18. [Google Scholar]
- 115. Freeman TL, Swartz TH. Targeting the NLRP3 inflammasome in severe COVID‐19. Front Immunol. 2020;11:1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Chen I‐Y, Moriyama M, Chang M‐F, Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol. 2019;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Siu K, Yuen K, Castano‐Rodriguez C, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3‐dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gao Z, Xia B, Shen X, et al. SARS‐CoV‐2 envelope protein causes acute respiratory distress syndrome (ARDS)‐like pathological damage and constitutes an antiviral target. bioRxiv. 2020;6(27):174953. 10.1101/2020.06.27.174953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Polina I, Sung JH, Jhun BS, Ouchi J. SARS‐Cov‐2 genes encode Ito‐like potassium channels: Linkage between viroporins and high mortality rate in COVID‐19 patients with pre‐existing cardiovascular diseases. Circulation. 2020;142:A15857. [Google Scholar]
- 120. Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen K‐Y. Coronaviruses—Drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wilson L, Gage P, Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hong M, DeGrado WF. Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Sci. 2012;21:1620–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ewart GD, Mills K, Cox GB, Gage PW. Amiloride derivatives block ion channel activity and enhancement of virus‐like particle budding caused by HIV‐1 protein Vpu. Eur Biophys J. 2002;31:26–35. [DOI] [PubMed] [Google Scholar]
- 124. Fleming DM. Managing influenza: amantadine, rimantadine and beyond. Int J Clin Pract. 2001;55:189–195. [PubMed] [Google Scholar]
- 125. Cady SD, Schmidt‐Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rejdak K, Grieb P. Adamantanes might be protective from COVID‐19 in patients with neurological diseases: Multiple sclerosis, parkinsonism and cognitive impairment. Mult Scler Relat Disord. 2020;42:102163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Aranda‐Abreu GE, Aranda‐Martínez JD, Araújo R, Hernández‐Aguilar ME, Herrera‐Covarrubias D, Rojas‐Durán F. Observational study of people infected with SARS‐Cov‐2, treated with amantadine. Pharmacol Rep. 2020;72:1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Jiménez‐Jiménez FJ, Alonso‐Navarro H, García‐Martín E, Agúndez JAG. Anti‐inflammatory effects of amantadine and memantine: Possible therapeutics for the treatment of Covid‐19? J Pers Med. 2020;10:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: Memantine and beyond. Nat Rev Drug Discov. 2006;5:160–170. [DOI] [PubMed] [Google Scholar]
- 130. Mishra SK, Hidau M, Rai S. Memantine and ibuprofen pretreatment exerts anti‐inflammatory effect against streptozotocin‐induced astroglial inflammation via modulation of NMDA receptor‐associated downstream calcium ion signaling. Inflammopharmacology. 2021;29:183–192. [DOI] [PubMed] [Google Scholar]
- 131. Cheng Q, Fang L, Feng D, et al. Memantine ameliorates pulmonary inflammation in a mice model of COPD induced by cigarette smoke combined with LPS. Biomed Pharmacother. 2019;109:2005–2013. [DOI] [PubMed] [Google Scholar]
- 132. Tessier D, Dawson K, Tetrault JP, Bravo G, Meneilly GS. Glibenclamide vs gliclazide in type 2 diabetes of the elderly. Diabet Med. 1994;11:974–980. [DOI] [PubMed] [Google Scholar]
- 133. Penlioglou T, Papachristou S, Papanas N. COVID‐19 and diabetes mellitus: May old anti‐diabetic agents become the new philosopher's stone? Diabetes Ther. 2020;11:1195–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Bułdak Ł, Machnik G, Bułdak RJ, et al. Exenatide (a GLP‐1 agonist) expresses anti‐inflammatory properties in cultured human monocytes/macrophages in a protein kinase A and B/Akt manner. Pharmacol Rep. 2016;68:329–337. [DOI] [PubMed] [Google Scholar]
- 135. Tipton PW, Wszolek ZK. What can Parkinson's disease teach us about COVID‐19? Neurol Neurochir Pol. 2020;54:204–206. [DOI] [PubMed] [Google Scholar]
- 136. Irwin JJ, Shoichet BK. ZINC—A free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Mukherjee S, Harikishore A, Bhunia A. Targeting C‐terminal helical bundle of NCOVID19 envelope (E) protein. Int J Biol Macromol. 2021;175:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ferraz WR, Gomes RA, Novaes ALS, Goulart Trossini GH. Ligand and structure‐based virtual screening applied to the SARS‐CoV‐2 main protease: An in silico repurposing study. Future Med Chem. 2020;12:1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Riva L, Yuan S, Yin X, et al. Discovery of SARS‐CoV‐2 antiviral drugs through large‐scale compound repurposing. Nature. 2020;586:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gillis JC, Goa KL. Tretinoin. Drugs. 1995;50:897–923. [DOI] [PubMed] [Google Scholar]
- 141. Fang Z, Li J, Yu X, et al. Polarization of monocytic myeloid‐derived suppressor cells by hepatitis B surface antigen is mediated via ERK/IL‐6/STAT3 signaling feedback and restrains the activation of T cells in chronic hepatitis B virus infection. J Immunol. 2015;195:4873–4883. [DOI] [PubMed] [Google Scholar]
- 142. Patel H, Kukol A. Prediction of ligands to universally conserved binding sites of the influenza a virus nuclear export protein. Virology. 2019;537:97–103. [DOI] [PubMed] [Google Scholar]
- 143. Dey D, Borkotoky S, Banerjee M. In silico identification of Tretinoin as a SARS‐CoV‐2 envelope (E) protein ion channel inhibitor. Comput Biol Med. 2020;127:104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bhowmik D, Nandi R, Jagadeesan R, Kumar N, Prakash A, Kumar D. Identification of potential inhibitors against SARS‐CoV‐2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect Genet Evol. 2020;84:104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Keivan Z, Teoh B‐T, Sam S‐S, Wong P‐F, Mustafa MR, AbuBakar S. In vitro antiviral activity of fisetin, rutin and naringenin against dengue virus type‐2. J Med Plants Res. 2011;5:5534–5539. [Google Scholar]
- 146. Savov VM, Galabov AS, Tantcheva LP, et al. Effects of rutin and quercetin on monooxygenase activities in experimental influenza virus infection. Exp Toxicol Pathol. 2006;58:59–64. [DOI] [PubMed] [Google Scholar]
- 147. Nizer WSDC, Ferraz AC, Moraes TDFS, et al. Lack of activity of rutin isolated from Tontelea micrantha leaves against Vero and BHK, fungi, bacteria and Mayaro virus and its in silico activity. J Pharm Negat Results. 2020;11:9–14. [Google Scholar]
- 148. Ng HH, Narasaraju T, Phoon MC, Sim MK, Seet JE, Chow VT. Doxycycline treatment attenuates acute lung injury in mice infected with virulent influenza H3N2 virus: Involvement of matrix metalloproteinases. Exp Mol Pathol. 2012;92:287–295. [DOI] [PubMed] [Google Scholar]
- 149. Rothan HA, Mohamed Z, Paydar M, Abd Rahman N, Yusof R. Inhibitory effect of doxycycline against dengue virus replication in vitro. Arch Virol. 2014;159:711–718. [DOI] [PubMed] [Google Scholar]
- 150. Bonzano C, Borroni D, Lancia A, Bonzano E. Doxycycline: From ocular rosacea to COVID‐19 anosmia. New Insight into the coronavirus outbreak. Front Med. 2020;7:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Borkotoky S. Banerjee M (2020) a computational prediction of SARS‐CoV‐2 structural protein inhibitors from Azadirachta indica (neem). J Biomol Struct Dyn. 2020;1–11. 10.1080/07391102.2020.1774419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Schoeman D, Fielding BC. Coronavirus envelope protein: Current knowledge. Virol J. 2019;16:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]


